Abstract
Unprecedented insights into the biology and functions of bacteria have been and continue to be gained through studying bacterial secretion systems in isolation. This method, however, results in our understanding of the systems being primarily based on the idea that they operate independently, ignoring the subtleties of downstream interconnections. Gram-negative bacteria are naturally able to adapt to and navigate their frequently varied and dynamic surroundings, mostly because of the covert connections between secretion systems. Therefore, to comprehend some of the linked downstream repercussions for organisms that follow this discourse, it is vital to have mechanistic insights into how the intersecretion system functions in bacterial rivalry, virulence, and survival, among other things. To that purpose, this paper discusses a few key instances of molecular antagonistic and interdependent relationships between bacterial secretion systems and their produced functional products.
Keywords: Secretion systems, intersecretion system crosstalk , nutrient acquisition, horizontal gene transfer, bacteria-host interaction
Introduction
Systems of molecular secretion in bacteria promote pathogenicity and disease in diverse animal, human, and plant hosts. Depending on the lifestyle of the bacteria, secretion systems assume numerous key roles commonly entailing the transport of small molecules, nucleic acids, and proteins [1]. A suit of these secretory pathways dictates many aspects of bacterial biology that often maximize the success of bacteria and how pathogens impose adverse consequences on public, livestock, and plant health. For these reasons, knowledge behind the molecular mechanisms of these pathways has great implications for effective antivirulence drug discovery and subsequent management of bacterial infections.
Secretion systems are important for the biology of bacteria as transporters of proteins from the cytoplasm to the outer membrane and transporters of proteins from a donor cell to the environment or a recipient cell [1]. The latter type of transporters, which have evolved over time as nanomachines facilitating competition for resources and space, is the subject of a large number of excellent reviews that address, at large, how they facilitate bacterial competition and interaction with the environment, other bacteria, and hosts [1–3].
Secretion systems have mostly been analysed in Gram-negative bacteria (GNB) and, to some extent, Gram-positive bacteria (GPB) [4–8]. They are often designated as TXSSs, where X stands for any number from 1 to 11, including outer membrane vesicles (OMVs), sometimes called a type zero secretion system (T0SS) [2, 6, 9, 10]. A major unifying thread among pathogenic bacterial secretory pathways, not related to type, is how they primarily assemble into channels for the transport of proteins including different virulence factors. Upon release, these proteins travel into and through biological barriers to interact with host components and specific immune factors, and subsequently promote the reprogramming of several important cellular processes, ultimately leading to disease.
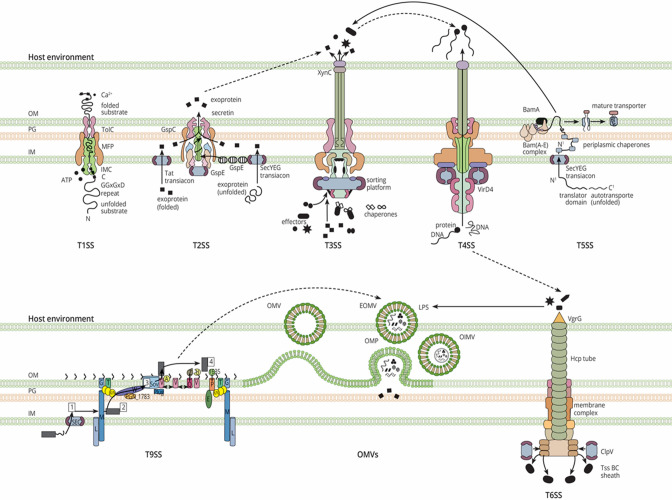
In light of recent advances in the understanding of bacterial secretion systems, the functional cooperation (i.e. several specialised secretion systems with a common goal) among them or their products is an interesting research area as it contributes to bacterial survival, rivalry, and virulence. However, the field of bacterial interaction barely comprehends this notion, given the lack of knowledge on the elaborate bacterial responses through multiple secretion pathways. As a result, the prevailing mind-set on multisecretion system-mediated bacterial response may currently be slanted among the research community. Secretion systems are well organized along a bacterial cell where they can operate either independently or interdependently of each other. Additionally, and as we later discuss, these secretion systems can also communicate remotely via the release of their functional molecules (Fig. 1).
Fig. 1.
Secretion systems (type 1–type 6 secretion system) of Gram-negative bacteria and available evidence of crosstalk with other secretion systems. The figure shows the six secretion systems of GNB (T1SS-T6SS) and the dialogs they are involved in with other secretion systems. The dotted lines indicate interaction at both substrate and system levels, while solid lines indicate interaction at either substrate or system level.
Given that other reviews (e.g. [11–19]) have already covered the fundamental functions of secretion systems, we focused heavily in our review on the interconnections between bacterial secretion systems and how they impact bacterial biology. In order to further emphasize the importance of investigating bacterial secretion system interaction, we additionally offered a few previously underestimated functions that are mostly apparent during the interaction of the secretion systems, and provided their possible downstream implications.
The general functions and biology of bacterial secretion systems
TXSSs
With the exception of the T1SS, whose function is restricted to the dispersal of substrates into the external environment, the T3SS, T4SS, and T6SS of GNB are widespread double membrane spanning one step translocators of proteins into the environment or target cells without periplasmic passage (Fig. 1). The latter are contact-dependent injectosomes of GNB [20]. Recently, it has been shown that the mechanism for T1SS in some cases is a two-step process, and through bacteriocin-like protein surface aggregates, T1SS can facilitate antibacterial competition in a contact dependent manner [21–23]. This system typically secretes diffusible bacteriocins, adhesins, and proteins required during nutrient acquisition, such as iron scavenger proteins, lipases, proteases, and pore-forming toxins important for survival and pathogenesis [24, 25]. The T3SS and T6SS secrete effectors primarily involved in host immune subversion or manipulation and bacterial competition, respectively [22, 26–30]. Both these systems are also involved in other roles such as symbiotic interactions including mutualism, commensalism, and pathogenesis [4, 31–36]. As recently hypothesized, pathogenic bacteria may also use the T3SS to indirectly target microbiota populations [37]. On the other hand, additional roles of the T6SS have come to light in recent years, including anti-fungal activities and extracellular metal uptake [38–41]. The T6SS has recently been demonstrated to affect the competition between predator and prey cells inside the biofilm [41] and to transport a DNase that kills fungus by damaging their DNA via a Mg2+-dependent mechanism [42]. Unlike other TXSSs, excluding membrane vesicles (T0SS), the T4SS appears to have a unique function of delivering DNA and protein-DNA complexes, suggesting it is a tool for horizontal gene transfer (HGT) and, importantly, for spreading resistance genes among bacterial communities [43, 44]. However, the T4SS also shares some functions with other TXSSs, including bacterial killing and delivering effectors into host cells, functions previously thought to be unique to the T6SS and T3SS, respectively [45, 46]. Recent studies have revealed insights into the structure, function, regulation, and role of the T4SS in bacterial pathogenesis and host-pathogen interactions [47, 48]. The T2SS, T5SS, T8SS, T9SS, and T11SS are the GNB two-step transporters, among which, only the T2SS and T9SS act as transenvelope machines, spanning both the inner and outer membrane of the bacterial cell envelope [1, 49, 50]. Components of the T10SS that facilitate the release of substrates from the periplasm to the extracellular space have not been described [51]. The T2SS substrate repertoire continues to expand and includes typical substrates for bacterial adaptation and nutrient acquisition such as hydrolytic enzymes and toxins [2, 52]. Most substrates of the T5SS, and similarly of the T9SS, remain attached to the outer membrane and a few are released into the extracellular environment [7].
Membrane vesicles
Bacterial membrane vesicles are arguably some of the most impressive features of microbes that have recently intrigued many scientists in the field of bacterial interactions. As vesicles exhibit some differences in transmission of molecular cargo compared to TXSSs, a brief discussion is warranted. Traditionally regarded as an inert membrane anomaly, membrane vesicles are frequently released by species in all three branches of life as subcellular lipid-bilayers (20–500 nm in diameter) [53]. According to a decades’ worth of research, membrane vesicles play a significant role in cell-cell and interorganismal communication due to their ability to internalize cellular contents (e.g. signalling molecules, toxins, proteins, metabolites, and nucleic acids) in their lumen. In this way, enclosed luminal contents maintain their potency because they are protected from extracellular degradative enzymes, making them highly concentrated and easy to deliver over long distances [54]. Upon release, membrane vesicles can either persist in the extracellular environment for long periods of time or fuse with prokaryotic or eukaryotic cells, for instance, via raft-dependent endocytosis, eliminating the need for microbes to have close contact with the host in order to rapidly transfer their cargo [55].
Bacterial membrane vesicles include outer membrane vesicles (OMVs), explosive outer membrane vesicles (EOMVs), cytoplasmic membrane vesicles (CMVs), and outer-inner membrane vesicles (OIMVs), mainly containing outer membrane (OM), cytoplasmic membrane, and outer and inner membrane (IM) components, respectively [56]. CMV, EOMV, and OIMV biogenesis happens through several proposed routes including explosive cell lysis via cryptic prophage endolysin activity [56]. OMV production entails unbalanced biosynthesis of cell wall components or the intercalation of hydrophobic molecules with the outer leaflet of the OM, leading to membrane curvature and eventual outgrowth of the OM [56–58]. For the purpose of this review, here on, all the vesicle types will be referred to as OMVs, for which formation is accompanied by the internalization of cellular contents delimited from extracellular enzymes degradation enclosed in the vesicle membrane [54].
Intersecretion system-mediated response in bacteria
In this section, we explore some of the molecular mechanisms underlying the interconnectivity of the different secretion systems, and we show how this leads to several important functions, such as collaborative assault, setting-up intimate bacteria-host and bacteria-bacteria contact, exploitative competition, and horizontal gene transfer (HGT) in plant and animal bacterial pathogens as well as non-pathogenic bacteria. In Fig. 1 and Table 1, the interaction-mediated functions have been summarized, which, for the purpose of discussions in this section, are described in terms of substrate and system level interaction.
Table 1.
Interactions among secretion systems in bacteria
|
Interacting system(s)* |
Level of interaction† |
Outcome and mechanism of interaction‡ |
Species |
Ref(s) |
|---|---|---|---|---|
|
T3SS-T5SS |
Substrate level interaction (substrates: Tir and intimin) |
Cooperative. Translocated Intimin Receptor (Tir) acts as a surface receptor for intimin, a T5SS substrate, owing to T3SS-mediated translocation of Tir. |
Enteropathogenic Escherichia coli |
[59] |
|
T3SS-T4SS |
Substrate level (BadA) and system level (T4SS) interaction |
Antagonistic. BadA’s effective length creates a physical barrier between the pathogen and the host cell membrane, preventing T4SS activity. |
[65] |
|
|
T2SS-T3SS |
Substrate level (PCWDEs) and system level (T3SS) interaction |
Cooperative. T2SS releases PCWDEs that alter plant cell wall integrity, thereby permitting T3SS-dependent effector protein translocation through. |
Xanthomonas citri pv. vesicatoria |
[74] |
|
T1SS-T2SS |
Substrate level interaction (substrates PrtA and PelI2) |
Cooperative. T1SS substrate PrtA post-translationally modifies T2SS substrate PelI-2 to PelI-3 |
[73] |
|
|
T4SS-T6SS |
Substrate and system level interaction. |
Cooperative. T4SS T4P promotes contact-dependent killing by T6SS by bringing adjacent prey closer to the attacker. |
Pseudomonas aeruginosa; |
[86, 87] |
|
T0SS-T6SS |
Substrate level interaction (substrates: TseF and TeoL) |
Cooperative. T6SS recruits T0SS (outer membrane vesicles or OMVs) either through its effector Tsef ( Cupriavidus necator ) or TeoL effector and PQS present in Pseudomonas aeruginosa OMVs. |
Pseudomonas aeruginosa; Cupriavidus necator |
[80, 81] |
|
T0SS-T9SS |
Substrate and system level interaction |
Cooperative. T9SS effectors, typically extracellular cysteine proteases (gingipains), are anchored on outer membrane vesicles through anionic lipopolysaccharide (A-LPS) moiety of Porphyromonas gingivalis OM. This interaction results in a virulence coat contributing to P. gingivalis infections. |
[101] |
*Type X secretion systems, where X stands for any number from 1 to 10, with outer membrane vesicles designated as a T0SS.
†Full names of molecules and components: BadA, Bartonella adhesin A; Tir, Translocated Intimin Receptor; TseF, T6SS effector for Fe uptake; TeoL, T6SS effector for recruitment of OMVs via lipopolysaccharide (LPS); PCWDEs, plant cell wall degrading enzymes; PQS, Pseudomonas quinolone signal; T4P, T4SS pilus. ‘Substrate’ refers to any molecule released by TXSSs (e.g. effectors, adhesins, etc.).
‡The term ‘outcome’ here refers to the way systems interact, which can be either synergistic or antagonistic.
Host cell attachment and damage coupled to effector translocation
Contact with the host surface is perhaps one of the most important features of bacteria in initiating an infection, and it can trigger events in the host cell that promote a pathogen’s internalization and continuation of its life cycle [59, 60]. This is typically carried out by several T5SS classes, namely classical autotransporters (T5aSS), two-partner secretion (T5bSS), trimeric autotransporter adhesins (T5cSS), T5dSS, inverse autotransporters (T5eSS), and T5fSS [61]. Several of these T5SS classes reportedly function together with the T3SS and T4SS in establishing contact between the pathogen and the cognate adhesin receptor found on the host surface, and in some cases even driving cytoskeletal rearrangements and effector-triggered host cell invasion [62]. A classic example is the T3SS-dependent translocation of Tir (Translocated Intimin Receptor), a protein typically produced by enteropathogenic Escherichia coli (EPEC) bacteria; EPEC causes diarrhoea, the primary cause of morbidity and mortality among children in developing countries [63]. Tir plays a crucial role in the pathogenesis of EPEC infections and subsequent development of paediatric diarrhoea by promoting attachment of pathogenic E. coli cells to host intestinal cells and inducing changes in host cell behaviour. Following attachment, the T3SS of EPEC delivers a suit of effector proteins into the host cell, including Tir [59]. Tir becomes localized to the plasma membrane via the host Golgi apparatus [60], then acts as a receptor for a T5eSS substrate of the bacterial protein, intimin [59]. The established Tir–intimin linkage allows EPEC to form a tight adherence to the host cell, known as attaching and effacing (A/E) lesions, which emerge as a result of the effacing of the small intestine lining, a hallmark of EPEC infections.
The Yersinia adhesin A (YadA) from Yersinia enterocolitica and BadA ( Bartonella adhesin A) from Bartonella henselae both fall under the T5eSS class. They engage in an intersecretion system-mediated translocation of effector molecules via the T3SS and T4SS [62, 64]. In particular, system interaction between BadA (T5cSS) and a T4SS core VirB/D-like subcomplex in B. henselae reduces T4SS mediated pathogen virulence [65], meaning the interaction is antagonistic unlike in EPEC [59]. Thus, most human B. henselae isolates were observed to have lost either BadA or VirB/D4 T4SS [65]. VirB/D4 denotes a structure of the T4SS that entails VirB1-11 proteins that assemble to form a secretion machinery and a pilus (T4P), and the VirD4 protein that is liable for substrate recruitment to the T4SS for secretion through the translocation channel [66]. The VirB/D4 T4SS function is thus interrupted by BadA which, through its effective length, enforces a physical distance (space) between the pathogen’s outer membrane (OM) and host cell membrane [65]. Subsequently, this negatively impacts the VirB/D4 T4SS effector translocation efficiency and ultimately impedes pathogen virulence. Considerations were made for possible shortfalls of the study resulting in the observed antagonism. It is possible that, in isolates where BadA and VirB/D4 co-exist to ensure persistence and effective infection, the strains likely express BadA or VirB/D4 genes at different stages of host infection to, in part, minimise a speculated risk of cell wall instability due to protein overload in the OM. Also, given that most strains with either one of these virulence factors are observed, it is highly likely that they are antagonistic in these strains or the VirB/D4 is barely useful to retain in the presence of BadA. Such potential interactions have been missed in the literature as a result of isolated analysis of key virulence factors or the assumption that all secretion systems follow the same virulence factor-interaction strategies [65]. Observations relating to similar bacteria-host cell contact mechanisms that are mediated by secretion system interplay have been made for Helicobacter pylori , Pseudomonas aeruginosa , and Salmonella enterica [62, 67–72].
The T2SS and T3SS are identified as some of the key virulent determinants in bacteria and have been reported multiple times to promote host cell damage and virulence, respectively, in Gram-negative pathogens of plants and animals. It is not surprising that these two systems act in concert to achieve a bunch of objectives in bacteria. Specifically in plant infections, the T2SS of bacterial pathogens primarily export an arsenal of plant cell wall degrading enzymes (PCWDEs), such as pectate lyase, pectin lyase, polygalacturonase, cellulase, and protease [52]. In Dickeya dadantii , a broad-host-range enterobacterium belonging to plant pathogenic soft rot pathogens, there is an interplay between the T1SS and T2SS at the substrate level. During this interplay, T1SS extracellular protease PrtA post-translationally modifies the T2SS-dependent pectate lyase (PelI-2) by cleaving its N-terminal amino acids [73]. The resultant protein is a small, slightly more basic, and more efficient necrosis-inducing protein called PelI-3 [73]. PCWDEs such as PelI are delivered to the host cell surface whereby they act to reduce cell wall integrity, thereby establishing a pathogen’s nutrient supply line that is based on the cell wall components of the host plant. For this reason, it is conceivable that a portion of the cell wall-derived components will be utilized in the assembly of bacterial weapons including secretory channels of virulence factors, including the T3SS [26]. Here, we also demonstrate an interaction between this system and the T2SS in Xanthomonas campestris pv. vesicatoria, the causative agent of bacterial spot of pepper and tomato. The T2SS activity is thought to disrupt the plant cell wall by releasing hydrolytic enzymes that allow T3SS-dependent effector protein translocation [74]. The cell wall-degrading activity in X. campestris pv. vesicatoria is specifically carried out by the Xps-T2SS, one of the two T2SSs spanning the envelope of this pathogen [74]. Szczesny and co-workers [74] hypothesised that degradation of the plant cell wall helps in nutrient acquisition and that T2SS PCWDEs might facilitate the assembly of extracellular components of T3SS pili for effector injection into the host cell. The study highlighted that mutation of the Xps system reduced translocation of T3 effectors but did not markedly affect the T3SS or the synthesis of its components [74]. Traces of plant cell wall could be observed in some studies after it has been degraded. It is possible that PCWDEs weaken the plant cell wall for effective T3SS effector translocation. Further investigation is required to validated these postulated synergistic functions. Pili are often used for host penetration by bacteria since they can span the thick plant cell wall to translocate effectors into the target plant. Similar structures that are conserved in bacterial pathogens suggest that the T2SS shares composition and structural features with the T4SS pili (T4P) [75], important in bacterial adherence to host cells and other surfaces [76]. This enforces the notion of a common origin and potentiates pilus-mediated secretion, which is also involved in secretion system interaction (discussed later in this section). The interaction of the T2SS and T3SS is also tightly linked to the regulation of host defences, as well as the expression of genes and substrates of both systems, which is regulated by the HrpG/HrpX regulon [74, 77–79].
A previously unrecognized iron acquisition and horizontal gene transfer mechanism
Direct physical contact of one bacterial cell with cells in the vicinity is an essential component of bacterial survival within a community. The T6SS is a bacteriophage-like machinery that is usually deployed during bacteria-bacteria contact where it delivers toxic effectors directly into neighbouring cells of competitor bacteria. All things considered, the T6SS is mostly regarded as a contact-dependent system, however, as we will show later, it can also engage in contactless exercises through interaction of its substrates with OMVs. Currently, there are no general mechanisms defining exactly how bacteria release, recognize, and recruit OMVs in an intra and inter-specific manner. However, two recent studies have revealed some key molecular mechanisms that might be involved in some of these processes in P. aeruginosa and Cupriavidus necator , entailing T6SS-mediated recruitment of OMVs during species communication [80, 81]. There are some aspects of this recruitment mechanism that are shared by both bacterial pathogens, and may also be found in other species. To begin, the T6SS effectors are employed as an OMV recruitment tool, and when OMVs are brought into play, a non-contact apparatus in the form of T6SS-OMV emerges [80, 81]. This makes reasonable sense as research shows that bacterial species can respond to environmental stimuli thanks to the release of OMVs [2]. Iron is an important metal for bacterial survival and virulence. P. aeruginosa H3-T6SS promoters in PAΔ3Fe (an iron acquisition mutant strain defective in the pyoverdin biosynthetic pathway (ΔpvdA), the pyochelin synthetase (ΔpchE) and the ferrous iron transport (ΔfeoB) are induced in iron-deficiency conditions [81]. Under iron depleted environments, P. aeruginosa is able to scavenge iron from the extracellular 2-heptyl-3-hydroxy-4-quinolone (PQS), mostly found in OMVs. First, TseF, a H3-T6SS secreted effector, directly interacts with the iron acquisition receptor, FptA, then PQS, whose affinity with TseF increased in the presence of iron. Ultimately, TseF indirectly facilitates iron acquisition by delivering the OMV associated Fe3+-PQS complex iron to P. aeruginosa cells in a PQS dependent manner. More so, TseF bridging of the interaction between P. aeruginosa OMV-bound iron binding molecule PQS and P. aeruginosa cells by directly binding FptA [Fe(III)-pyochelin receptor] and porin OprF, consequently directs iron to the cells for uptake via an unknown mechanism. As expected, the growth of TseF and H3-T6SS mutants in iron depleted or Fe3+-PQS supplemented media, even in the presence of functional iron receptors, is severely affected in the absence of the recruiting effector, TseF [81].
The delivery of effectors or ions into target cells, even at a distance, from the site of colonization presents a unique population feeding advantage over traditional secretion systems that are often tightly fixed to the membrane and peptidoglycan layer of the cells. As noted previously, the T6SS primarily depends on contact to induce an effect on the recipient cells. Therefore, by recruiting OMVs through the incorporation of effectors into OMVs, the T6SS may perform its functions without proximity restrictions. Second, effectors that associate with OMVs for function or transport might also direct OMVs to the bacterial cell surface where they interact with specific OM receptors involved in iron uptake [80, 81]. Third, under iron-diluted conditions, activation of surface-associated receptors facilitates the delivery of iron to the cytosol, and once replete, dissolved iron can promote the competitive ability of P. aeruginosa and C. necator in their respective environments [80, 81]. When all of these factors are considered, a novel function of the T6SS, seen to promote the efficient utilization of iron when its sources are running low seems to be an intricate one.
We further consider the unique feature at play during the interaction of the T6SS with OMVs that is required for the bacteria to thrive in an iron-diluted environment. First, in P. aeruginosa , some aspect of iron uptake is facilitated by a newly described effector, called PA2374 or TseF, leading to the delivery of iron into P. aeruginosa cells [81]. In C. necator , a ligand (lipopolysaccharide)-receptor (CubA and CstR) interaction mediated inter-species OMV recruitment mechanism is observed. The LPS-binding effector TeoL [T6SS effector for recruitment of OMVs via lipopolysaccharide (LPS)] especially recognizes and binds the LPS on OMVs from different species (e.g. C. necator and distantly related P. aeruginosa PAO1 and Yersinipseudotuberculosis YPIII) containing the iron-chelating molecule PQS. Next, CubA (cupriabactin siderophore receptor) and CstR (catecholate siderophore receptor), both of which are part of the bacterial OM and essential for the ability of bacteria to grow and survive in iron-poor environments, are activated, eventually leading to ligand-receptor interaction-based OMV recruitment [80]. The O-antigen component of OMVs, which is a carbohydrate structural region of the LPS, and the compositional differences in LPS between bacterial cells and OMVs (e.g. partial loss of the LPS in OMVs), contribute to TeoL’s preferential binding to bacterial OMVs rather than to bacterial cells [80]. In addition to iron uptake, the OMVs recruited by TeoL were observed to be important for exploitative competition, resistance to oxidative stress, and HGT in recipient cells—key in their survival and persistence. Given the fact that PQS and LPS are involved in OMV production during a process entailing PQS-mediated anionic repulsions between the LPS molecules [82], it would be interesting to examine the possibility of effector-induced vesiculation in bacteria as a way to increase nutrient sources that are depleted. This brings up the issue of whether OMVs could be utilized as a wellspring of nutrients or factors significant for nutrient acquisition, and whether TXSS effector-mediated recruitment of OMVs is a general survival mechanism used by bacteria. Nonetheless, the discoveries of these studies certainly illuminate how we might interpret the role of bacteria in recognizing and recruiting OMVs for various survival and host invasion strategies.
T6SS-mediated recruitment of OMVs also enables bacteria to participate in HGT. OMVs can drive HGT and bacterial resistance to stress [83–85], but to our knowledge, they have seldom been associated with other TXSSs in accomplishing HGT. Recently, it was established that the T6SS can promote HGT by enabling acquisition of DNA from OMVs purified from bacterial cultures containing plasmid DNA [80]. As previously discussed, pilus-mediated interaction in terms of intersecretion system crosstalk plays a key role in bacterial contact with either other bacteria or the host. A key example of this is a pilus-mediated interaction in terms of HGT that involves a crosstalk between the T4SS and T6SS (Fig. 1) [86]. This is consistent with HGT encompassing T4SS-mediated cell-cell contact through conjugative DNA transfer in bacteria [76]. The T4SS-mediated HGT was found to activate T6SS-mediated killing of adjacent donor cells carrying parasitic foreign DNA, and is considered an ‘innate immune system’ that recognizes transfer-associated patterns instead of molecular patterns of infectious elements [86]. When contact-dependent killing via intersecretion system crosstalk is involved, this suggests that HGT is undeniably more complex than previously thought.
Intersecretion system-mediated microbe-microbe contact
The interaction between the T6SS and T4SS has recently been observed to play a role during contact between non-pathogenic Neisseria cinerea and other human pathogens [87]. As mentioned previously, T4Ps form part of the VirB/D4 T4SS design, and are found on bacteria’s surfaces, aiding in bringing bacteria into close physical association with host cells and other bacteria [88]. In competition assays involving human commensal and pathogenic strains of Neisseria , N. cinerea was instrumental in killing pathogens, N. meningitidis and N. gonorrhoeae , in a T6SS-dependent manner [87]. Prey strains of Neisseria lacking a T4P, in particular, were able to escape the T6SS-mediated killing by segregating to the edge of the colony seeded on agar medium. However, prey strains that expressed a pilus were outcompeted by the killer strain due to cellular interaction between themselves and N. cinerea , which was mediated by a T4P. This implies that T4P promotes the activity of contact-dependent TXSSs by bringing prey closer to the attacker. It is not uncommon for bacteria to influence the outcome of an infection by directly killing other bacteria through the antibacterial action of the T6SS. This has been associated with changes in microbial communities and a range of ecological consequences [89]. Therefore, intersecretion system as a function of pilus-mediated interaction may greatly contribute to microbial community structures and composition, which is reminiscent with the role of the T6SS [89].
Shared TXSS substrates highlight a conceivable interplay
Although there are a number of studies which do not directly dissect bacterial secretion system dialogue, they give significant insights to their possible interaction and set the stage for future research. For example, the P. aeruginosa T3SS and T6SS may work together to regulate transcription factors that activate unique transcriptome changes during early airway epithelial cell infection [90]. With respect to the T3SS and T4SS, we speculate functional interplay could be a possible outcome since the effectors of these secretion systems are often observed to be remarkably alike in structure and function [91]. This means that the functional secretion system might act in place of the mutant system and secrete its substrates. Alternatively, the functional secretion system substrates might substitute the mutated systems’ substrates. In addition, several studies on bacterial secretion suggest that many substrates are shared between OMVs and TXSSs. For instance, OMVs are known to export many proteins which play a role in bacterial virulence and communication, and many of these are shared with TXSSs [54]. In plant-associated bacteria, a large number of studies reported diverse proteins in OMVs that are biologically important [92]. An overlap in substrates between the T2SS and OMVs was observed, whereby X. campestris pv. vesicatoria strains lacking a functional T2SS independently secreted several substrates of the T2SS system, including extracellular protein cargoes such as lipases, proteases, and cell wall–modifying enzymes via OMVs [93]. Likewise, OMVs isolated from phytopathogens overlap in extracellular protein cargo with the T2SS and T3SS [94, 95]. Both T2SS secreted hydrolytic enzymes and the T3SS effectors whose translocation into host cells is potentially facilitated by them were found in OMVs of P . syringae pv. tomato T1 [94]. In addition, OMV-mediated transport of biologically active T2SS dependent PCWDEs has been reported in Pectobacterium spp. P . brasiliense , P. odoriferum, P. versatile and P. zantedeschiae and X. fastidiosa vesicles [96–98]. Taken together, these analyses suggest that OMVs could serve as an alternative secretory pathway for other TXSSs in bacteria or act in coordination with them.
Implications for the inter-secretion system dialogue
Interaction between secretion systems could be a common occurrence in prokaryotic organisms. How it impacts bacterial function and interactions is not yet clear, but it perhaps represents an ingenious mechanism that ensures bacteria use adequate tools at the right time to enhance their fitness potential. In this section we consider some of these specific aspects and their potential consequences.
Membrane vesicles shared as ‘public goods’
Although OMVs were originally thought to be merely membrane artefacts with no clear cellular importance, in recent decades, an enormous number of investigations gave an account of their functions including in nutrient acquisition and exploitative competition. For example, OMVs from Mycobacterium tuberculosis can carry high amounts of an iron chelating molecule, myobactin, which forms iron-scavenging OMVs [99]. Once released into the environment, these myobactin-OMVs can be shared bona publica as they will be relatively easy to access by neighbour bacteria as a community resource, thereby contributing to the social life of that community [100]. In essence, OMVs can serve an ecologically significant role for the successful coexistence of different bacterial species in the same habitat or within biofilm–structured communities of microbial aggregates enclosed in a self-produced polymeric matrix and attached to biological and non-biological surfaces. Here, we consider these ideas in the context of OMV interaction, which so far have been reported with the T6SS and T9SS. As previously noted, P. aeruginosa secretes OMVs carrying on their surface the PQS molecule, which, like myobactin, also strongly binds iron. These PQS-containing OMVs can be recruited by the T6SS effector TseF [81], and possibly by other members in the bacterial community that express the T6SS effector. Although further research is needed to determine the entire range of bacteria that secrete this effector, it is possible that other bacteria may also produce the effector. Additional proof of OMVs as potential public goods relates to features of the bacterial OM. The LPS, which is one of the major constituents of bacterial cells and OMVs, is important for the cell envelope of GNB, serving principally as a structural component of OM and released OMVs. Likewise, there is solid support of OMV-mediated conveyance of the LPS among bacterial species which can also trigger important host cellular processes [101–103]. This solidly addresses the ability of bacterial cells to disperse significant surface-confined particles even to locales where OMV donor cells themselves cannot reach. Along these lines, the T6SS-mediated recruitment of OMVs may hypothetically be an element of bacteria traded among neighbours and as a public good. Considering that the T6SSs are present in a wide spectrum of Gram-negatives (>25 % of Proteobacteria) [104], the T6SS-mediated recruitment of OMVs could therefore be an alternative mechanism of utilizing scarce iron from its dilute sources exploited by many bacteria. In addition, iron acquisition in this manner also brings about other benefits, as we have seen that the capacity of the T6SS effectors to recruit OMVs also prompts other important functions, including HGT and stress tolerance, that are critical for bacterial rivalry and survival while sharing the same habitat [80, 81, 105]. In a similar scenario, the T9SS, a translocon in a few Bacteroidetes spp. including Porphyromonas gingivalis , interacts with OMVs via a battery of virulence effectors (Fig. 1) [101]. These include extracellular cysteine proteases, representing hallmark virulence factors of this pathogen, commonly called gingipains (RgpA, RgpB, and Kgp) [101]. These virulence factors carry a conserved C-terminal domain (CTD) that is used as an extracellular OM translocation signal by the T9SS [106]. Following translocation, the CTD is cleaved off by a bifunctional C-terminal signal peptidase and sortase enzyme, PorU. The sortase is also released via the T9SS but, unlike gingipains, it retains the CTD signal [106–108], after which its replacement, as well as that of gingapins, with a unique anionic lipopolysaccharide (A-LPS) moiety of P. gingivalis OM effectively anchors the released proteins to the OM, forming a virulence coat on the cells and OMVs [101, 108, 109]. Because the LPS can be exchanged between bacterial strains, the OMVs coated with virulence factors (virulence factor-coated) may be released into the extracellular space, where they will most likely be shared as public goods [101, 103]. In this way, the virulence coat can be spread by released OMVs, and in some cases even to other non-pathogenic P. gingivalis strains within the pathogen population. Noteworthy, bacteria can trade certain traits through the use of OMVs [103]. Such a vesicle-mediated exchange of cargo or traits is not confined to bacteria. In the yeast genus Cryptococcus, for instance, extracellular vesicles released by virulent strains can trigger a rapid intracellular proliferation of non-virulent yeast strains residing within the macrophages [110]. This results in pathogenic ‘division of labour’, which occurs remotely where vesicles diffuse over large distances, in the process possibly transmitting virulence factors to these strains. Alternatively, it is possible that the Cryptococcus vesicles can modify the host environment to allow less pathogenic strains to thrive, which would also facilitate the pathogenic ‘division of labour’. The same ‘division of labour’ phenomenon may apply in P. gingivalis strains when the T9SS virulence coat is distributed from a few pathogenic members to otherwise latent and non-virulent strains.
Conjugation is a widespread channel of HGT, a consequence of which is rapid evolution and adaptation of bacterial strains through the spread of antimicrobial resistance genes [111]. Plant-derived bioactive compounds identified to inhibit T4SS-mediated conjugal transfer of plasmids without perturbing GNB growth were reported not long ago [112]. However, in addition to conjugation and transformation by the T4SS, OMVs are also disseminators of antibiotic resistance genes in GNB, and cultures supplemented with purified vesicles are protected from antimicrobial compounds by several strategies such as drug binding [113–116]. In Vibrio cholerae , a protease essential in bacterial resistance to host antimicrobial peptides and conventionally secreted via the T2SS, is associated with OMVs [117]. Being evolutionarily conserved, constitutive, and primarily produced in response to stress, dissemination of resistance via OMVs may soon become notably problematic as resistance genes continue to be availed to complex microbial communities [118]. We are likely to observe roles beyond OMV-T2SS, -T3SS, -T4SS, -T6SS, and -T9SS associations as vesicles are handy and convenient sources (i.e. public goods) of protected and concentrated bioactive compounds for mixed communities [3, 119].
Differential secretion of substrates
The focus of profiling secretion systems includes proteomic research. To demonstrate secretomes that are representative of the function of the missing system, secretion system knockout mutants are often constructed. In proteomic investigations, it has been revealed that proteins are downregulated in TXSS mutant strains compared to wild-type strains in the hunt for the TXSSs' major substrates. It is possible that the differentially secreted proteins are an indication of secondary protein secretion or regulation by the disrupted system. The key to bacterial fitness and survival in the environment is synchrony. Bacteria intentionally stimulate protein secretion for this reason so they can react quickly to environmental changes [120]. In this way, regulatory networks, and secretion systems along with their crosstalk play a critical part in the communication, which is in turn critical for fitness in all forms of life [121]. For instance, a recent study identified new D. dadantii T2SS substrates displaying a band of low intensity and thickness for one protein, VirK, in the supernatant of the T2SS inactive strain as compared to the supernatant of the T2SS active strain or the complement [122]. VirK could have leaked, or in our judgement was a subject of secondary secretion by OMVs or another system thus the cause of its appearance in T2SS mutant secretomes. Similarly VirK and a cellulase were previously only discovered in the minimum media with extract of the T3SS active strain secretome of the phytopathogen X. citri subsp . citri [123]. The study attributed the outcome to the dysfunctional T3SS system in the mutant strain or that the T3SS apparatus had pleiotropic effects on the expression and secretion of some proteins. However, since another study demonstrated in Ralstonia solanacearum that HrpG of the T3SS regulated VirK, it was concluded that the T3SS controls T2SS secretion [123]. A study in Salmonella identified the crosstalk between the T6SS encoding SPI-19 and the T3SS in avian infection, which is another recent discovery [124]. The research revealed that T3SS regulators made up part of the T6SS island. Genes that make up the T6SS core components were not a part of the observed regulation [124]. All things considered, regulatory or polar effects of some mutations may be at play. Nonetheless, it is crucial to keep in mind that secretion systems could be involved in co-secretion and species dependent secretion of substrates in addition to substrate and systems level interaction. The most recent computational resource for identifying novel substrates is called BastionX. It has predictors for the secretion systems T1SS-T4SS and T6SS and calculates probabilities of secreting a given protein by each system. Some proteins seem to fit profiles with the best possible score using this method as substrates of at least one secretion system [125].
Bacteria must have mechanisms in place to carefully control secretion in order to produce smooth and effective interaction mechanisms, and we expect to be able to properly respond in case a system fails. In this review, the effects of secretion systems crosstalk have been emphasized to show potential consequences of both harmonious and antagonistic action. It is obvious that specific TXSSs' activities have an impact on how well other secretion systems function. One potential effect of a dialogue is compensatory behaviour among secretory systems to attain collective goals in light of the impacts. It is a frequent misconception that a secretion system only carries one protein [1]. Bacteria likely modify these underutilized systems to transport additional substrates with a similar structure to their principal substrates. Therefore, it is essential to develop methods to study the full-circle circuitry of protein secretion. Outputs will reveal data required to understand the complexities behind secretion system overlap and implications in crucial areas like the use of secretion system inhibitory strategies for disease control.
Prediction and screening of secretion system substrates
Secreted proteins and their functions delineate what a secretion system is used for [7]. A portion of secretomes typically studied in vitro includes effectors that facilitate interactions between producing bacteria and their hosts. Bioinformatics, proteomics, and biochemical approaches are used to study secretomes. In addition, databases such as Bacterial Secreted Effector Protein DataBase (SecretEPDB) harbour pre-calculated knowledge bases of reference effectors [91]. Computational pipelines use the available secretion systems substrate data (sequence features, e.g. secretion signals and conserved motifs) to predict candidate effectors from available microbiota genomes and proteomes. To date, over 40 bioinformatics tools are available to predict GNB T1SS-T9SS substrates [126]. Bioinformatic tools often rely on two strategies: sequence similarity to known effectors and identification and analysis of unique gene islands of a pathogen. However, there is a limited number of experimentally validated secretion systems cargo, and the poor specificity of prediction tools poses significant challenges to the similarity and unique island predictions' efficiency [126, 127]. Poor specificity is partly due to the construction of some algorithms and tools for specific bacteria or substrates [126]. Another limitation of in silico screening of effectors is that most predictors are designed to screen proteomes for substrates of a single secretion system, further limited by a bias towards specific effector groups. Hui and co-workers [126] presented 45 representative single secretion system substrate predictors. This group is populated with machine learning algorithms based T3SS and T4SS predictors. Recent advances seek to delimit computational prediction limitations by constructing tools that predict effectors for several secretion systems. One recent semi-knowledge based multiple secretion systems substrate predictor is PREFFECTOR [128]. The most recent computational resource for screening for novel TXSS secreted substrates is BastionX [125]. It has substrate predictors for the secretion systems T1SS-T4SS and T6SS and calculates probabilities of each system secreting a given protein. The upside of this tool is that it predicts both homologs of experimentally validated effectors and novel distant candidates [125]. Computational pipelines have been utilized to some degree to predict host-pathogen-effector triggered susceptibility agents and pathogen associated molecular patterns or effector triggered immunity protein interactions [129]. Developing predictors with broad prediction capabilities will go a long way in screening GNB proteomes for candidate effectors. Bioinformatic approaches used alongside approaches such as proteomics and phage display technologies take the screening process for candidate effectors a step further. Mass spectrometry coupled with bioinformatics is used to identify and annotate proteins in secreted protein sample preparations, thus further filtering the in silico predicted candidates and identifying secretomes unbiasedly. Phage display affinity screening identifies proteins implicated in interactions of microbes with their environments or hosts through virion facilitated physical interactions with target proteins [130]. In our view, these platforms offer a means to identify, functionally annotate proteins, and establish protein-protein interaction networks between self (in this case, secreted effectors of multiple systems) and environmental or host proteins. The networks might give insight into probable functions and functional links of proteins secreted by different systems that would guide downstream undertaking applications, i.e. disease management. To study substrate cooperation or co-secretion by OMVs, T1SS-T4SS, and T6SS, reporter-based assays can be used. Guided by the secretion systems under inspection, substrates would be cloned as fusions with detectable tags that do not impact secretion (i.e. neutral tags) and expressed by transformed strains for selective identification using affinity chromatography, fluorescence microscopy, enzymatic assays, or Western blot from the wild-type and single or multiple secretion system inactive strain secretomes. Tags associated with an enzymatic activity provide quantitative data on secretion. Densitometry can be used to quantify relative band intensities [131]. The final step would be to seek experimental evidence to support the proposed biological roles of the secreted proteins as effectors.
Conclusion
We have emphasized that secretion systems can be interconnected and, in some cases, interdependent through physical contact or by delivering molecular payloads – like effectors and other key proteins into the environment or target cells. By retaining such an elaborate capacity to enable interactive system communication in their cells partly helps us explain why bacteria can thrive in their natural habitats through a number of functions; for one, bacteria, among many roles, could derive an agile response to cope with a dynamically changing environment. In other words, this phenomenon could be behind some of the complex phenotypes commonly encountered when multigene pathways are involved, such as bacterial resilience to stress and antimicrobial agents. Be that as it may, this review in no way implies that any of the discussed interactions or their effects serve as a universal mechanism for bacteria to interact with one another or with their hosts. The review only seeks to highlight the magnitude of any beneficial or harmful effects resulting from interactions amongst secretion systems and the potential consequences as these interactions evolve. The absence of empirical data for a crosslink between these secretion systems represents a significant barrier to analyse their effects in bacteria. Therefore, it is envisioned that taking advantage of the complexity of these linked operations holds out the prospect of new horizons to be explored, such as new treatment strategies for complex bacterial diseases in animals or new plant improvement prospects.
New direction
Considerable efforts are necessary to unpack the molecular mechanisms underlining the convergent function of secretion systems, which may open up new possibilities. As this area is one of a few enigmas in bacterial interactions, research behind the system interaction is likely to gain traction, and a broader consequences of cooperative or antagonistic behaviour that is primarily directed at maintaining adaptability in bacterial responses in the natural environment would emerge. Such details could present an attractive model for developing broad spectrum or multisecretion system antivirulence compounds to alleviate the negative consequences of bacteria, or simply to study the well-coordinated phenotypic developments in prokaryotic species.
Funding information
This work received no specific grant from any funding agency.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: A/E, attaching and effacing; A-LPS, anionic lipopolysaccharide; BadA, Bartonella adhesin A; B. henselae, Bartonella henselae; CMVs, cytoplasmic membrane vesicles; C. necator, Cupriavidus necator; CTD, C-terminal domain; EOMVs, explosive outer membrane vesicles; EPEC, enteropathogenic Escherichia Coli; GNB, gram-negative bacteria; GPB, gram-positive bacteria; HGT, horizontal gene transfer; IM, inner membrane; LPS, lipopolysaccharide; N. cinerea, Neisseria cinerea; N. gonorrhoeae, Neisseria gonorrhoeae; N. meningitidis, Neisseria meningitidis; OIMVs, outer inner membrane vesicles; OM, outer membrane; OMVs, outer membrane vesicles; P. aeruginosa, Pseudomonas aeruginosa; P. brasiliense, Pectobacterium brasiliense; PCWDEs, plant cell wall degrading enzymes; P. gingivalis, Porphyromonas gingivalis; P. odoriferum, Pectobacterium odoriferum; PQS, pseudomonas quinolone signal; P. syringae, Pseudomonas syringae; P. versatile, Pectobacterium versatile; P. zantedeschiae, Pectobacterium zantedeschiae; SecretEPDB, bacterial secreted effector protein database; TeoL, type 6 effector for recruitment of OMVs via lipopolysaccharide; Tir, translocated intimin receptor; T4P, type 4 secretion system pilus; TseF, type 6 secretion system effector for Fe uptake; TXSS, type X secretion system; X. campestris pv. vesicatoria, Xanthomonas campestris pv. vesicatoria; X. citri subsp. citri, Xanthomonas citri subspecies citri; X. fastidiosa, Xylella fastidiosa; YadA, Yersinia adhesin A; Y. pseudotuberculosis, Yersinia pseudotuberculosis.
References
- 1.Green ER, Mecsas J. Bacterial secretion systems: an overview. Virulence Mechan Bacterial Pathogens. 2016:213–239. doi: 10.1128/microbiolspec.VMBF-0012-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Costa TRD, Felisberto-Rodrigues C, Meir A, Prevost MS, Redzej A, et al. Secretion systems in Gram-negative bacteria: structural and mechanistic insights. Nat Rev Microbiol. 2015;13:343–359. doi: 10.1038/nrmicro3456. [DOI] [PubMed] [Google Scholar]
- 3.Lynch JB, Alegado RA. Spheres of hope, packets of doom: the good and bad of outer membrane vesicles in interspecies and ecological dynamics. J Bacteriol. 2017;199:e00012–00017. doi: 10.1128/JB.00012-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernal P, Llamas MA, Filloux A. Type VI secretion systems in plant-associated bacteria. Environ Microbiol. 2018;20:1–15. doi: 10.1111/1462-2920.13956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowman L, Palmer T. The type VII secretion system of Staphylococcus . Annu Rev Microbiol. 2021;75:471–494. doi: 10.1146/annurev-micro-012721-123600. [DOI] [PubMed] [Google Scholar]
- 6.Bunduc CM, Fahrenkamp D, Wald J, Ummels R, Bitter W, et al. Structure and dynamics of a mycobacterial type VII secretion system. Nature. 2021;593:445–448. doi: 10.1038/s41586-021-03517-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Filloux A. Bacterial protein secretion systems: game of types. Microbiology. 2022;168:001193. doi: 10.1099/mic.0.001193. [DOI] [PubMed] [Google Scholar]
- 8.Rivera-Calzada A, Famelis N, Llorca O, Geibel S. Type VII secretion systems: structure, functions and transport models. Nat Rev Microbiol. 2021;19:567–584. doi: 10.1038/s41579-021-00560-5. [DOI] [PubMed] [Google Scholar]
- 9.Chang JH, Desveaux D, Creason AL. The ABCs and 123s of bacterial secretion systems in plant pathogenesis. Annu Rev Phytopathol. 2014;52:317–345. doi: 10.1146/annurev-phyto-011014-015624. [DOI] [PubMed] [Google Scholar]
- 10.Guerrero-Mandujano A, Hernández-Cortez C, Ibarra JA, Castro-Escarpulli G. The outer membrane vesicles: secretion system type zero. Traffic. 2017;18:425–432. doi: 10.1111/tra.12488. [DOI] [PubMed] [Google Scholar]
- 11.Bhoite S, van Gerven N, Chapman MR, Remaut H. Curli biogenesis: bacterial amyloid assembly by the type VIII secretion pathway. EcoSal Plus. 2019;8 doi: 10.1128/ecosalplus.ESP-0037-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christie PJ, Whitaker N, González-Rivera C. Mechanism and structure of the bacterial type IV secretion systems. Biochim Biophys Acta. 2014;1843:1578–1591. doi: 10.1016/j.bbamcr.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cianfanelli FR, Monlezun L, Coulthurst SJ. Aim, load, fire: the type VI secretion system, a bacterial Nanoweapon. Trends Microbiol. 2016;24:51–62. doi: 10.1016/j.tim.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 14.Deng W, Marshall NC, Rowland JL, McCoy JM, Worrall LJ, et al. Assembly, structure, function and regulation of type III secretion systems. Nat Rev Microbiol. 2017;15:323–337. doi: 10.1038/nrmicro.2017.20. [DOI] [PubMed] [Google Scholar]
- 15.Grohmann E, Christie PJ, Waksman G, Backert S. Type IV secretion in Gram-negative and Gram-positive bacteria. Mol Microbiol. 2018;107:455–471. doi: 10.1111/mmi.13896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nivaskumar M, Francetic O. Type II secretion system: a magic beanstalk or a protein escalator. Biochim Biophys Acta. 2014;1843:1568–1577. doi: 10.1016/j.bbamcr.2013.12.020. [DOI] [PubMed] [Google Scholar]
- 17.Palmer T, Finney AJ, Saha CK, Atkinson GC, Sargent F. A holin/peptidoglycan hydrolase-dependent protein secretion system. Mol Microbiol. 2021;115:345–355. doi: 10.1111/mmi.14599. [DOI] [PubMed] [Google Scholar]
- 18.Thomas S, Holland IB, Schmitt L. The Type 1 secretion pathway - the hemolysin system and beyond. Biochim Biophys Acta. 2014;1843:1629–1641. doi: 10.1016/j.bbamcr.2013.09.017. [DOI] [PubMed] [Google Scholar]
- 19.van Ulsen P, Zinner KM, Jong WSP, Luirink J. On display: autotransporter secretion and application. FEMS Microbiol Lett. 2018;365:fny165. doi: 10.1093/femsle/fny165. [DOI] [PubMed] [Google Scholar]
- 20.Galán JE, Waksman G. Protein-injection machines in bacteria. Cell. 2018;172:1306–1318. doi: 10.1016/j.cell.2018.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.García-Bayona L, Guo MS, Laub MT. Contact-dependent killing by Caulobacter crescentus via cell surface-associated, glycine zipper proteins. Elife. 2017;6:e24869. doi: 10.7554/eLife.24869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith WPJ, Vettiger A, Winter J, Ryser T, Comstock LE, et al. The evolution of the type VI secretion system as a disintegration weapon. PLoS Biol. 2020;18:e3000720. doi: 10.1371/journal.pbio.3000720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spitz O, Erenburg IN, Beer T, Kanonenberg K, Holland IB, et al. Type I Secretion Systems-One Mechanism for All? Microbiol Spectr. 2019;7:7. doi: 10.1128/microbiolspec.PSIB-0003-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanonenberg K, Schwarz CKW, Schmitt L. Type I secretion systems - a story of appendices. Res Microbiol. 2013;164:596–604. doi: 10.1016/j.resmic.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 25.Kanonenberg K, Spitz O, Erenburg IN, Beer T, Schmitt L. Type I secretion system-it takes three and a substrate. FEMS Microbiol Lett. 2018;365:fny094. doi: 10.1093/femsle/fny094. [DOI] [PubMed] [Google Scholar]
- 26.Büttner D, He SY. Type III protein secretion in plant pathogenic bacteria. Plant Physiol. 2009;150:1656–1664. doi: 10.1104/pp.109.139089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Durand E, Cambillau C, Cascales E, Journet L. VgrG, Tae, Tle, and beyond: the versatile arsenal of type VI secretion effectors. Trends Microbiol. 2014;22:498–507. doi: 10.1016/j.tim.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 28.Galán JE, Lara-Tejero M, Marlovits TC, Wagner S. Bacterial type III secretion systems: specialized nanomachines for protein delivery into target cells. Annu Rev Microbiol. 2014;68:415–438. doi: 10.1146/annurev-micro-092412-155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Russell AB, Hood RD, Bui NK, LeRoux M, Vollmer W, et al. Type VI secretion delivers bacteriolytic effectors to target cells. Nature. 2011;475:343–347. doi: 10.1038/nature10244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wagner S, Grin I, Malmsheimer S, Singh N, Torres-Vargas CE, et al. Bacterial type III secretion systems: a complex device for the delivery of bacterial effector proteins into eukaryotic host cells. FEMS Microbiol Lett. 2018;365:fny201. doi: 10.1093/femsle/fny201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Borrero de Acuña JM, Bernal P. Plant holobiont interactions mediated by the type VI secretion system and the membrane vesicles: promising tools for a greener agriculture. Environ Microbiol. 2021;23:1830–1836. doi: 10.1111/1462-2920.15457. [DOI] [PubMed] [Google Scholar]
- 32.Egan F, Barret M, O’Gara F. The SPI-1-like type III secretion system: more roles than you think. Front Plant Sci. 2014;5:34. doi: 10.3389/fpls.2014.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hachani A, Wood TE, Filloux A. Type VI secretion and anti-host effectors. Curr Opin Microbiol. 2016;29:81–93. doi: 10.1016/j.mib.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 34.Hu Y, Huang H, Cheng X, Shu X, White AP, et al. A global survey of bacterial type III secretion systems and their effectors. Environ Microbiol. 2017;19:3879–3895. doi: 10.1111/1462-2920.13755. [DOI] [PubMed] [Google Scholar]
- 35.Lucke M, Correa MG, Levy A. The role of secretion systems, effectors, and secondary metabolites of beneficial rhizobacteria in interactions with plants and microbes. Front Plant Sci. 2020;11 doi: 10.3389/fpls.2020.589416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Speare L, Cecere AG, Guckes KR, Smith S, Wollenberg MS, et al. Bacterial symbionts use a type VI secretion system to eliminate competitors in their natural host. Proc Natl Acad Sci U S A. 2018;115:E8528–E8537. doi: 10.1073/pnas.1808302115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu X, Cai J, Li X, Yu F, Wu D. Can bacterial type III effectors mediate pathogen-plant-microbiota ternary interactions? Plant Cell Environ. 2022;45:5–11. doi: 10.1111/pce.14185. [DOI] [PubMed] [Google Scholar]
- 38.Coulthurst S. The type VI secretion system: a versatile bacterial weapon. Microbiology. 2019;165:503–515. doi: 10.1099/mic.0.000789. [DOI] [PubMed] [Google Scholar]
- 39.Lien Y-W, Lai E-M. Type VI secretion effectors: methodologies and biology. Front Cell Infect Microbiol. 2017;7:254. doi: 10.3389/fcimb.2017.00254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Russell AB, Peterson SB, Mougous JD. Type VI secretion system effectors: poisons with a purpose. Nat Rev Microbiol. 2014;12:137–148. doi: 10.1038/nrmicro3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Teschler JK, Jiménez-Siebert E, Jeckel H, Singh PK, Park JH, et al. VxrB influences antagonism within biofilms by controlling competition through extracellular matrix production and type 6 secretion. mBio. 2022;13:e0188522. doi: 10.1128/mbio.01885-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luo J, Chu X, Jie J, Sun Y, Guan Q, et al. Acinetobacter baumannii kills fungi via a type VI DNase effector. mBio. 2023;14:e03420–03422. doi: 10.1128/mbio.03420-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huddleston JR. Horizontal gene transfer in the human gastrointestinal tract: potential spread of antibiotic resistance genes. Infect Drug Resist. 2014;7:167–176. doi: 10.2147/IDR.S48820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Juhas M, Crook DW, Hood DW. Type IV secretion systems: tools of bacterial horizontal gene transfer and virulence. Cell Microbiol. 2008;10:2377–2386. doi: 10.1111/j.1462-5822.2008.01187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bayer-Santos E, Cenens W, Matsuyama BY, Oka GU, Di Sessa G, et al. The opportunistic pathogen Stenotrophomonas maltophilia utilizes a type IV secretion system for interbacterial killing. PLoS Pathog. 2019;15:e1007651. doi: 10.1371/journal.ppat.1007651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Souza JAM, Baltazar L de M, Carregal VM, Gouveia-Eufrasio L, de Oliveira AG, et al. Corrigendum: characterization of Aspergillus fumigatus extracellular vesicles and their effects on macrophages and neutrophils functions. Front Microbiol. 2019;10:2334. doi: 10.3389/fmicb.2019.02334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Purtschert-Montenegro G, Cárcamo-Oyarce G, Pinto-Carbó M, Agnoli K, Bailly A, et al. Pseudomonas putida mediates bacterial killing, biofilm invasion and biocontrol with a type IVB secretion system. Nat Microbiol. 2022;7:1547–1557. doi: 10.1038/s41564-022-01209-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sheedlo MJ, Ohi MD, Lacy DB, Cover TL. Molecular architecture of bacterial type IV secretion systems. PLoS Pathog. 2022;18:e1010720. doi: 10.1371/journal.ppat.1010720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sato K, Naito M, Yukitake H, Hirakawa H, Shoji M, et al. A protein secretion system linked to bacteroidete gliding motility and pathogenesis. Proc Natl Acad Sci U S A. 2010;107:276–281. doi: 10.1073/pnas.0912010107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Veith PD, Glew MD, Gorasia DG, Reynolds EC. Type IX secretion: the generation of bacterial cell surface coatings involved in virulence, gliding motility and the degradation of complex biopolymers. Mol Microbiol. 2017;106:35–53. doi: 10.1111/mmi.13752. [DOI] [PubMed] [Google Scholar]
- 51.Hamilton JJ, Marlow VL, Owen RA, Costa M de AA, Guo M, et al. A holin and an endopeptidase are essential for chitinolytic protein secretion in Serratia marcescens . J Cell Biol. 2014;207:615–626. doi: 10.1083/jcb.201404127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cianciotto NP, White RC. Expanding role of type II secretion in bacterial pathogenesis and beyond. Infect Immun. 2017;85:e00014–00017. doi: 10.1128/IAI.00014-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gill S, Catchpole R, Forterre P. Extracellular membrane vesicles in the three domains of life and beyond. FEMS Microbiol Rev. 2019;43:273–303. doi: 10.1093/femsre/fuy042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bonnington KE, Kuehn MJ. Protein selection and export via outer membrane vesicles. Biochim Biophys Acta. 2014;1843:1612–1619. doi: 10.1016/j.bbamcr.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bomberger JM, Maceachran DP, Coutermarsh BA, Ye S, O’Toole GA, et al. Long-distance delivery of bacterial virulence factors by Pseudomonas aeruginosa outer membrane vesicles. PLoS Pathog. 2009;5:e1000382. doi: 10.1371/journal.ppat.1000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Toyofuku M, Nomura N, Eberl L. Types and origins of bacterial membrane vesicles. Nat Rev Microbiol. 2019;17:13–24. doi: 10.1038/s41579-018-0112-2. [DOI] [PubMed] [Google Scholar]
- 57.McMillan HM, Kuehn MJ. The extracellular vesicle generation paradox: a bacterial point of view. EMBO J. 2021;40:e108174. doi: 10.15252/embj.2021108174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schertzer JW, Whiteley M. A bilayer-couple model of bacterial outer membrane vesicle biogenesis. mBio. 2012;3:e00297–00211. doi: 10.1128/mBio.00297-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kenny B, DeVinney R, Stein M, Reinscheid DJ, Frey EA, et al. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell. 1997;91:511–520. doi: 10.1016/s0092-8674(00)80437-7. [DOI] [PubMed] [Google Scholar]
- 60.Mao C, Gu J, Wang HG, Fang Y, Yang P, et al. Translocation of enterohemorrhagic Escherichia coli effector Tir to the plasma membrane via host Golgi apparatus. Mol Med Rep. 2017;16:1544–1550. doi: 10.3892/mmr.2017.6763. [DOI] [PubMed] [Google Scholar]
- 61.Meuskens I, Saragliadis A, Leo JC, Linke D. Type V secretion systems: an overview of passenger domain functions. Front Microbiol. 2019;10:1163. doi: 10.3389/fmicb.2019.01163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mix A-K, Goob G, Sontowski E, Hauck CR. Microscale communication between bacterial pathogens and the host epithelium. Genes Immun. 2021;22:247–254. doi: 10.1038/s41435-021-00149-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet. 2013;382:209–222. doi: 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- 64.Keller B, Mühlenkamp M, Deuschle E, Siegfried A, Mössner S, et al. Yersinia enterocolitica exploits different pathways to accomplish adhesion and toxin injection into host cells. Cell Microbiol. 2015;17:1179–1204. doi: 10.1111/cmi.12429. [DOI] [PubMed] [Google Scholar]
- 65.Lu Y-Y, Franz B, Truttmann MC, Riess T, Gay-Fraret J, et al. Bartonella henselae trimeric autotransporter adhesin BadA expression interferes with effector translocation by the VirB/D4 type IV secretion system. Cell Microbiol. 2013;15:759–778. doi: 10.1111/cmi.12070. [DOI] [PubMed] [Google Scholar]
- 66.Redzej A, Ukleja M, Connery S, Trokter M, Felisberto-Rodrigues C, et al. Structure of a VirD4 coupling protein bound to a VirB type IV secretion machinery. EMBO J. 2017;36:3080–3095. doi: 10.15252/embj.201796629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Backert S, Selbach M. Role of type IV secretion in Helicobacter pylori pathogenesis. Cell Microbiol. 2008;10:1573–1581. doi: 10.1111/j.1462-5822.2008.01156.x. [DOI] [PubMed] [Google Scholar]
- 68.Backert S, Clyne M, Tegtmeyer N. Molecular mechanisms of gastric epithelial cell adhesion and injection of CagA by Helicobacter pylori . Cell Commun Signal. 2011;9:28. doi: 10.1186/1478-811X-9-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gerlach RG, Cláudio N, Rohde M, Jäckel D, Wagner C, et al. Cooperation of Salmonella pathogenicity islands 1 and 4 is required to breach epithelial barriers. Cell Microbiol. 2008;10:2364–2376. doi: 10.1111/j.1462-5822.2008.01218.x. [DOI] [PubMed] [Google Scholar]
- 70.Ishijima N, Suzuki M, Ashida H, Ichikawa Y, Kanegae Y, et al. BabA-mediated adherence is a potentiator of the Helicobacter pylori type IV secretion system activity. J Biol Chem. 2011;286:25256–25264. doi: 10.1074/jbc.M111.233601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Miyake M, Hanajima M, Matsuzawa T, Kobayashi C, Minami M, et al. Binding of intimin with Tir on the bacterial surface is prerequisite for the barrier disruption induced by enteropathogenic Escherichia coli . Biochem Biophys Res Commun. 2005;337:922–927. doi: 10.1016/j.bbrc.2005.09.130. [DOI] [PubMed] [Google Scholar]
- 72.Sundin C, Wolfgang MC, Lory S, Forsberg A, Frithz-Lindsten E. Type IV pili are not specifically required for contact dependent translocation of exoenzymes by Pseudomonas aeruginosa . Microb Pathog. 2002;33:265–277. doi: 10.1006/mpat.2002.0534. [DOI] [PubMed] [Google Scholar]
- 73.Shevchik VE, Boccara M, Vedel R, Hugouvieux-Cotte-Pattat N. Processing of the pectate lyase PelI by extracellular proteases of Erwinia chrysanthemi 3937. Mol Microbiol. 1998;29:1459–1469. doi: 10.1046/j.1365-2958.1998.01028.x. [DOI] [PubMed] [Google Scholar]
- 74.Szczesny R, Jordan M, Schramm C, Schulz S, Cogez V, et al. Functional characterization of the Xcs and Xps type II secretion systems from the plant pathogenic bacterium Xanthomonas campestris pv vesicatoria. New Phytol. 2010;187:983–1002. doi: 10.1111/j.1469-8137.2010.03312.x. [DOI] [PubMed] [Google Scholar]
- 75.Korotkov KV, Sandkvist M, Hol WGJ. The type II secretion system: biogenesis, molecular architecture and mechanism. Nat Rev Microbiol. 2012;10:336–351. doi: 10.1038/nrmicro2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hospenthal MK, Costa TRD, Waksman G. A comprehensive guide to pilus biogenesis in Gram-negative bacteria. Nat Rev Microbiol. 2017;15:365–379. doi: 10.1038/nrmicro.2017.40. [DOI] [PubMed] [Google Scholar]
- 77.Alvarez-Martinez CE, Sgro GG, Araujo GG, Paiva MRN, Matsuyama BY, et al. Secrete or perish: the role of secretion systems in Xanthomonas biology. Comput Struct Biotechnol J. 2021;19:279–302. doi: 10.1016/j.csbj.2020.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Guo Y, Figueiredo F, Jones J, Wang N. HrpG and HrpX play global roles in coordinating different virulence traits of Xanthomonas axonopodis pv. citri. Mol Plant Microbe Interact. 2011;24:649–661. doi: 10.1094/MPMI-09-10-0209. [DOI] [PubMed] [Google Scholar]
- 79.Jha G, Rajeshwari R, Sonti RV. Functional interplay between two Xanthomonas oryzae pv. oryzae secretion systems in modulating virulence on rice. MPMI. 2007;20:31–40. doi: 10.1094/MPMI-20-0031. [DOI] [PubMed] [Google Scholar]
- 80.Li C, Zhu L, Wang D, Wei Z, Hao X, et al. T6SS secretes an LPS-binding effector to recruit OMVs for exploitative competition and horizontal gene transfer. ISME J. 2021;16:500–510. doi: 10.1038/s41396-021-01093-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lin J, Zhang W, Cheng J, Yang X, Zhu K, et al. A Pseudomonas T6SS effector recruits PQS-containing outer membrane vesicles for iron acquisition. Nat Commun. 2017;8:14888. doi: 10.1038/ncomms14888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mashburn-Warren L, Howe J, Garidel P, Richter W, Steiniger F, et al. Interaction of quorum signals with outer membrane lipids: insights into prokaryotic membrane vesicle formation. Mol Microbiol. 2008;69:491–502. doi: 10.1111/j.1365-2958.2008.06302.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fulsundar S, Harms K, Flaten GE, Johnsen PJ, Chopade BA, et al. Gene transfer potential of outer membrane vesicles of Acinetobacter baylyi and effects of stress on vesiculation. Appl Environ Microbiol. 2014;80:3469–3483. doi: 10.1128/AEM.04248-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Klieve AV, Yokoyama MT, Forster RJ, Ouwerkerk D, Bain PA, et al. Naturally occurring DNA transfer system associated with membrane vesicles in cellulolytic Ruminococcus spp. of ruminal origin. Appl Environ Microbiol. 2005;71:4248–4253. doi: 10.1128/AEM.71.8.4248-4253.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mozaheb N, Mingeot-Leclercq M-P. Membrane vesicle production as a bacterial defense against stress. Front Microbiol. 2020;11:600221. doi: 10.3389/fmicb.2020.600221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ho BT, Basler M, Mekalanos JJ. Type 6 secretion system-mediated immunity to type 4 secretion system-mediated gene transfer. Science. 2013;342:250–253. doi: 10.1126/science.1243745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Custodio R, Ford RM, Ellison CJ, Liu G, Mickute G, et al. Type VI secretion system killing by commensal Neisseria is influenced by expression of type four pili. Elife. 2021;10:e63755. doi: 10.7554/eLife.63755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Craig L, Forest KT, Maier B. Type IV pili: dynamics, biophysics and functional consequences. Nat Rev Microbiol. 2019;17:429–440. doi: 10.1038/s41579-019-0195-4. [DOI] [PubMed] [Google Scholar]
- 89.Gallegos-Monterrosa R, Coulthurst SJ. The ecological impact of a bacterial weapon: microbial interactions and the Type VI secretion system. FEMS Microbiol Rev. 2021;45:fuab033. doi: 10.1093/femsre/fuab033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sen-Kilic E, Huckaby AB, Damron FH, Barbier M. P. aeruginosa type III and type VI secretion systems modulate early response gene expression in type II pneumocytes in vitro. BMC Genomics. 2022;23:345. doi: 10.1186/s12864-022-08554-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.An Y, Wang J, Li C, Revote J, Zhang Y, et al. SecretEPDB: a comprehensive web-based resource for secreted effector proteins of the bacterial types III, IV and VI secretion systems. Sci Rep. 2017;7:1–10. doi: 10.1038/srep41031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Janda M, Robatzek S. Extracellular vesicles from phytobacteria: properties, functions and uses. BiotechnolAdv. 2022;58:107934. doi: 10.1016/j.biotechadv.2022.107934. [DOI] [PubMed] [Google Scholar]
- 93.Solé M, Scheibner F, Hoffmeister A-K, Hartmann N, Hause G, et al. Xanthomonas campestris pv. vesicatoria secretes proteases and xylanases via the Xps type II secretion system and outer membrane vesicles. J Bacteriol. 2015;197:2879–2893. doi: 10.1128/JB.00322-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chowdhury C, Jagannadham MV. Virulence factors are released in association with outer membrane vesicles of Pseudomonas syringae pv. tomato T1 during normal growth. Biochim Biophys Acta. 2013;1834:231–239. doi: 10.1016/j.bbapap.2012.09.015. [DOI] [PubMed] [Google Scholar]
- 95.Nascimento R, Gouran H, Chakraborty S, Gillespie HW, Almeida-Souza HO, et al. The type II secreted lipase/esterase LesA is a key virulence factor required for Xylella fastidiosa pathogenesis in grapevines. Sci Rep. 2016;6:1–17. doi: 10.1038/srep18598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Feitosa-Junior OR, Stefanello E, Zaini PA, Nascimento R, Pierry PM, et al. Proteomic and metabolomic analyses of Xylella fastidiosa OMV-enriched fractions reveal association with virulence factors and signaling molecules of the DSF family. Phytopathology. 2019;109:1344–1353. doi: 10.1094/PHYTO-03-19-0083-R. [DOI] [PubMed] [Google Scholar]
- 97.Jonca J, Waleron M, Czaplewska P, Bogucka A, Steć A, et al. Membrane vesicles of Pectobacterium as an effective protein secretion system. Int J Mol Sci. 2021;22:12574. doi: 10.3390/ijms222212574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Maphosa S, Moleleki LN. Isolation and characterization of outer membrane vesicles of Pectobacterium brasiliense 1692. Microorganisms. 2021;9:1918. doi: 10.3390/microorganisms9091918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Prados-Rosales R, Weinrick BC, Piqué DG, Jacobs WR, Jr, Casadevall A, et al. Role for Mycobacterium tuberculosis membrane vesicles in iron acquisition. J Bacteriol. 2014;196:1250–1256. doi: 10.1128/JB.01090-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.West SA, Diggle SP, Buckling A, Gardner A, Griffin AS. The social lives of microbes. Annu Rev Ecol Evol Syst. 2007;38:53–77. doi: 10.1146/annurev.ecolsys.38.091206.095740. [DOI] [Google Scholar]
- 101.Glew MD, Gorasia DG, McMillan PJ, Butler CA, Veith PD, et al. Complementation in trans of Porphyromonas gingivalis lipopolysaccharide biosynthetic mutants demonstrates lipopolysaccharide exchange. J Bacteriol. 2021;203:e00631–00620. doi: 10.1128/JB.00631-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Vanaja SK, Russo AJ, Behl B, Banerjee I, Yankova M, et al. Bacterial outer membrane vesicles mediate cytosolic localization of LPS and caspase-11 activation. Cell. 2016;165:1106–1119. doi: 10.1016/j.cell.2016.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Vassallo C, Pathak DT, Cao P, Zuckerman DM, Hoiczyk E, et al. Cell rejuvenation and social behaviors promoted by LPS exchange in myxobacteria. Proc Natl Acad Sci U S A. 2015;112:E2939–46. doi: 10.1073/pnas.1503553112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bingle LE, Bailey CM, Pallen MJ. Type VI secretion: a beginner’s guide. Curr Opin Microbiol. 2008;11:3–8. doi: 10.1016/j.mib.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 105.Schwechheimer C, Kuehn MJ. Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions. Nat Rev Microbiol. 2015;13:605–619. doi: 10.1038/nrmicro3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Glew MD, Veith PD, Peng B, Chen Y-Y, Gorasia DG, et al. PG0026 is the C-terminal signal peptidase of a novel secretion system of Porphyromonas gingivalis . J Biol Chem. 2012;287:24605–24617. doi: 10.1074/jbc.M112.369223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gorasia DG, Veith PD, Chen D, Seers CA, Mitchell HA, et al. Porphyromonas gingivalis type IX secretion substrates are cleaved and modified by a sortase-like mechanism. PLoS Pathog. 2015;11:e1005152. doi: 10.1371/journal.ppat.1005152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Veith PD, Shoji M, O’Hair RAJ, Leeming MG, Nie S, et al. Type IX secretion system cargo proteins are glycosylated at the C terminus with a novel linking sugar of the Wbp/Vim pathway. mBio. 2020;11:e01497–01420. doi: 10.1128/mBio.01497-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Madej M, Nowakowska Z, Ksiazek M, Lasica AM, Mizgalska D, et al. PorZ, an essential component of the type IX secretion system of Porphyromonas gingivalis, delivers anionic lipopolysaccharide to the PorU sortase for transpeptidase processing of T9SS cargo proteins. mBio. 2021;12:e02262–02220. doi: 10.1128/mBio.02262-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bielska E, Sisquella MA, Aldeieg M, Birch C, O’Donoghue EJ, et al. Pathogen-derived extracellular vesicles mediate virulence in the fatal human pathogen Cryptococcus gattii . Nat Commun. 2018;9:1556. doi: 10.1038/s41467-018-03991-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Virolle C, Goldlust K, Djermoun S, Bigot S, Lesterlin C. Plasmid transfer by conjugation in Gram-negative bacteria: from the cellular to the community level. Genes. 2020;11:1239. doi: 10.3390/genes11111239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Oyedemi BOM, Shinde V, Shinde K, Kakalou D, Stapleton PD, et al. Novel R-plasmid conjugal transfer inhibitory and antibacterial activities of phenolic compounds from Mallotus philippensis (Lam.) Mull. Arg. J Glob Antimicrob Resist. 2016;5:15–21. doi: 10.1016/j.jgar.2016.01.011. [DOI] [PubMed] [Google Scholar]
- 113.Chatterjee S, Mondal A, Mitra S, Basu S. Acinetobacter baumannii transfers the blaNDM-1 gene via outer membrane vesicles. J Antimicrob Chemother. 2017;72:2201–2207. doi: 10.1093/jac/dkx131. [DOI] [PubMed] [Google Scholar]
- 114.Kulkarni HM, Nagaraj R, Jagannadham MV. Protective role of E. coli outer membrane vesicles against antibiotics. Microbiol Res. 2015;181:1–7. doi: 10.1016/j.micres.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 115.Manning AJ, Kuehn MJ. Contribution of bacterial outer membrane vesicles to innate bacterial defense. BMC Microbiol. 2011;11:1–15. doi: 10.1186/1471-2180-11-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rumbo C, Fernández-Moreira E, Merino M, Poza M, Mendez JA, et al. Horizontal transfer of the OXA-24 carbapenemase gene via outer membrane vesicles: a new mechanism of dissemination of carbapenem resistance genes in Acinetobacter baumannii . Antimicrob Agents Chemother. 2011;55:3084–3090. doi: 10.1128/AAC.00929-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rompikuntal PK, Vdovikova S, Duperthuy M, Johnson TL, Åhlund M, et al. Outer membrane vesicle-mediated export of processed PrtV protease from Vibrio cholerae . PLoS One. 2015;10:e0134098. doi: 10.1371/journal.pone.0134098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Li J, Azam F, Zhang S. Outer membrane vesicles containing signalling molecules and active hydrolytic enzymes released by a coral pathogen Vibrio shilonii AK1. Environ Microbiol. 2016;18:3850–3866. doi: 10.1111/1462-2920.13344. [DOI] [PubMed] [Google Scholar]
- 119.Toyofuku M, Morinaga K, Hashimoto Y, Uhl J, Shimamura H, et al. Membrane vesicle-mediated bacterial communication. ISME J. 2017;11:1504–1509. doi: 10.1038/ismej.2017.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Leung KY, Siame BA, Snowball H, Mok Y-K. Type VI secretion regulation: crosstalk and intracellular communication. Curr Opin Microbiol. 2011;14:9–15. doi: 10.1016/j.mib.2010.09.017. [DOI] [PubMed] [Google Scholar]
- 121.Yeo CC, Espinosa M, Venkova T. Editorial: prokaryotic communications: from macromolecular interdomain to intercellular talks (recognition) and beyond. Front Mol Biosci. 2021;8:670572. doi: 10.3389/fmolb.2021.670572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Condemine G, Le Derout B. Identification of new Dickeya dadantii virulence factors secreted by the type 2 secretion system. PLoS One. 2022;17:e0265075. doi: 10.1371/journal.pone.0265075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ferreira RM, Moreira LM, Ferro JA, Soares MRR, Laia ML, et al. Unravelling potential virulence factor candidates in Xanthomonas citri. subsp. citri by secretome analysis. PeerJ. 2016;4:e1734. doi: 10.7717/peerj.1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhang Y, Xian H, Jiang X, Yuan Y, Ji R, et al. Identification of two Sel1-like proteins in SPI-19 of Salmonella enterica serovar Pullorum that can mediate bacterial infection through T3SS. Microbiol Res. 2022;262:127085. doi: 10.1016/j.micres.2022.127085. [DOI] [PubMed] [Google Scholar]
- 125.Wang J, Li J, Hou Y, Dai W, Xie R, et al. BastionHub: a universal platform for integrating and analyzing substrates secreted by Gram-negative bacteria. Nucleic Acids Res. 2021;49:D651–D659. doi: 10.1093/nar/gkaa899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hui X, Chen Z, Zhang J, Lu M, Cai X, et al. Computational prediction of secreted proteins in gram-negative bacteria. CSBJ. 2021;19:1806–1828. doi: 10.1016/j.csbj.2021.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ma B, Charkowski AO, Glasner JD, Perna NT. Identification of host-microbe interaction factors in the genomes of soft rot-associated pathogens Dickeya dadantii 3937 and Pectobacterium carotovorum WPP14 with supervised machine learning. BMC Genomics. 2014;15:508. doi: 10.1186/1471-2164-15-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Dhroso A, Eidson S, Korkin D. Genome-wide prediction of bacterial effector candidates across six secretion system types using a feature-based statistical framework. Sci Rep. 2018;8:17209. doi: 10.1038/s41598-018-33874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Mishra B, Kumar N, Mukhtar MS. Systems biology and machine learning in plant-pathogen interactions. Mol Plant Microbe Interact. 2019;32:45–55. doi: 10.1094/MPMI-08-18-0221-FI. [DOI] [PubMed] [Google Scholar]
- 130.Gagic D, Ciric M, Wen WX, Ng F, Rakonjac J. Exploring the secretomes of microbes and microbial communities using filamentous phage display. Front Microbiol. 2016;7:429. doi: 10.3389/fmicb.2016.00429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Maffei B, Francetic O, Subtil A. Tracking proteins secreted by bacteria: what’s in the toolbox? Front Cell Infect Microbiol. 2017;7:221. doi: 10.3389/fcimb.2017.00221. [DOI] [PMC free article] [PubMed] [Google Scholar]