Abstract
Environmental chemical (EC) exposures and our interactions with them has significantly increased in the recent decades. Toxicity associated biological characterization of these chemicals is challenging and inefficient, even with available high-throughput technologies. In this report, we describe a novel computational method for characterizing toxicity, associated biological perturbations and disease outcome, called the Chemo-Phenotypic Based Toxicity Measurement (CPTM). CPTM is used to quantify the EC “toxicity score” (), which serves as a holistic metric of potential toxicity and disease outcome. CPTM quantitative toxicity is the measure of chemical features, biological phenotypic effects, and toxicokinetic properties of the ECs. For proof-of-concept, we subject ECs obtained from the Environmental Protection Agency’s (EPA) database to the CPTM. We validated the CPTM toxicity predictions by correlating ‘’ scores with known toxicity effects. We also confirmed the CPTM predictions with in-vitro, and in-vivo experiments. In in-vitro and zebrafish models, we showed that, mixtures of the motor oil and food additive ‘Salpn’ with endogenous nuclear receptor ligands such as Vitamin D3, dysregulated the nuclear receptors and key transcription pathways involved in Colorectal Cancer. Further, in a human patient derived cell organoid model, we found that a mixture of the widely used pesticides ‘Tetramethrin’ and ‘Fenpropathrin’ significantly impacts the population of patient derived pancreatic cancer cells and 3D organoid models to support rapid PDAC disease progression. The CPTM method is, to our knowledge, the first comprehensive toxico-physicochemical, and phenotypic bionetwork-based platform for efficient high-throughput screening of environmental chemical toxicity, mechanisms of action, and connection to disease outcomes.
Keywords: Environmental chemicals, Mixtures, Pesticides, Food additive, Cancer
1. Introduction
Environmental Chemical (EC) exposures due to social and general living conditions, including air pollution, occupation, travel, and proximity to industrial byproducts, is ubiquitous and implicated in human pathology. More than 100,000 ECs exist and act dynamically with the human body resulting in multiple phenotypic effects. Understanding the biological interplay between these exposures and diseases is crucial for public health. To better understand the biological and clinical effects of the myriad of chemicals released into the environment, the U.S. Environmental Protection Agency (EPA) initiated the ToxCast program for high-throughput in vitro screening of chemicals across hundreds of phenotypic assays (Dix et al., 2007). Since the inception of ToxCast, thousands of ECs have been screened and characterized by the molecular and cellular pathways they affect. This mechanistic understanding helps elucidate the pathophysiology underlying exposure to these chemicals (Kleinstreuer et al., 2014; Rotroff et al., 2014; Wambaugh et al., 2013).
While this rich in vitro data can be leveraged to create in vivo toxicity models (Judson et al., 2014), full coverage of all potential ECs remains expensive and time-consuming. In addition, currently available high-throughput biochemical and cellular assays are limited in their modeling of phenotypes relevant to human health as they are devoid of mechanistic information. Computational methods help to address these issues and streamline toxicologic studies (Raunio, 2011; Modi et al., 2012). In silico tools borne out of bioinformatics and chemo-informatics have been widely employed in the drug development sector and can be valuable predictive tools in toxicology (Valerio, 2011). The Food and Drug Administration (FDA) developed the Critical Path Initiative for improving the assessment of the environmental impact of drugs, with in silico toxicology models providing a core set of tools (〈http://www.fda.gov/ScienceResearch/SpecialTopics/CriticalPathInitiative 〉; McRobb et al., 2014; Contrera, 2011; Honorio et al., 2013; Cronin and Dearden, 1995). Such in silico tools have been successful in identifying novel endocrine disruptors (McRobb et al., 2014), predicting carcinogenic potential (Contrera, 2011), and determining structure-activity relationships (Cronin and Dearden, 1995), as well as improving the modeling of toxicokinetic absorption, distribution, metabolism, and excretion (ADME) properties (Honorio et al., 2013).
Computational tools in toxicology are invaluable for the mechanistic understanding of toxicity and prediction of the impact of new chemicals released into the environment. Information derived from computational models is critical for regulatory agencies and healthcare providers for policy-making and clinical interventions. To date, most computational models are QSAR-based and derived from limited chemical sets (Schultz and Seward, 2000). Though these models may be accurate with respect to chemicals that are similar to the training set, this imposes a limitation on the applicability domain of the model (Cronin, 2002). A generalized model that can incorporate any chemical would have greater utility.
In addition, most models are built from structural, topological, and chemical properties of ECs. To date, no model exists that integrates the phenotypic and mechanistic aspects of EC toxicity. Here, we present an integrative computational platform called Chemo-Phenotypic Based Toxicity Measurement (CPTM) for the predictive toxicity and biological assessment of ECs obtained from public data sets including the EPA ToxCast chemicals. CPTM uniquely considers EC-target and other higher-order biological interactions along with EC chemical reactivity, toxico-physicochemical properties, absorption, and metabolism parameters for toxico-kinetics.
We applied the CPTM method to quantify the “toxicity potential” of ECs and validated the method in cytotoxicity assays of cultured cells and zebrafish, along with nuclear receptor and transcriptional pathway activation assays by correlating ‘Zts’ scores with toxicity effects documented in the Hazardous Substance Data Bank (HSDB). We validated the EC-molecular signatures predicted by our Tox-TMFS method (Dakshanamurthy et al., 2012; Issa et al., 2015), an integral portion of CPTM, in vitro, and against the Comparative Toxicogenomics Database (CTD) (Davis et al., 2013). Using our systems bionetwork platform GenEx-Net (Issa et al., 2016; Wathieu et al., 2017) built into CPTM, we further assessed systems-level effects of EC-protein signatures and validated them through higher-order network analysis of pathway and functional effects resulting in clinical phenotypes both in vitro, and against the CTD database. Lastly, we investigated diseases that are causally linked to perturbations of gene products/pathways such as Wnt and vitamin D receptor signaling to hypothesize EC exposure links to colorectal cancer (CRC). We showed that a food and motor oil additive ‘salpn’ in concert with vitamin D3 (colecalciferol) dysregulates vitamin D receptor (VDR) signaling, important in CRC. We also proposed a model of the salpn plus D3 mixture interaction with nuclear receptors, that provides a mechanistic explanation of the dysregulation of signaling pathways important in CRC.
Unlike current computational methods that attempt to predict lethal doses, CPTM provides a guideline for assessing an EC’s toxicity and disease outcome. In its current form, it makes no links to lethal dosages, but we intend to integrate the CPTM model with machine-learning algorithms to include lethal dosages. The CPTM method is the first platform of its kind in the toxicological sciences that is rooted in physicochemical parameters, chemical reactivity, toxico-kinetics, and biological networks. As the EC space grows exponentially with environmental pollutants, new commercial chemicals, environmental waste products and pharmaceuticals, our platform will be useful to streamline the assessment of ECs for prioritization in subsequent toxicology assays, predictions of associated diseases, mechanisms, and health monitoring.
2. Results and discussion
2.1. Chemo-phenotypic based toxicity measurement (CPTM): a novel quantitative method to determine EC toxicity, disease connection and mechanisms
The CPTM method provides a metric defining potential toxicity for a given EC that is derived from predicted EC-protein interactions, and biological network perturbation signatures along with relevant toxicology related toxico-physicochemical parameters, reactivity, and ADME properties. CPTM integrates four major modules: (1) empirical EC-protein associations predicted by our tuned proteochemometric method (herein referred to as Tox-TMFS (Dakshanamurthy et al., 2012; Issa et al., 2015)), (2) systems gene network analysis using GenEx-Net modeled after our previous work (Issa et al., 2016; Wathieu et al., 2017), (3) EC-target biological effects with toxico-kinetic, and intrinsic chemical reactivity properties, and (4) normalized perturbation scores combined into a quantifiable rank ordered “toxicity score” denoted by ‘’ (Fig. 1). Examples of values generated for all potential EC biological, physicochemical, and metabolism-related parameters are shown in Fig. S1, wherein colored headings correspond to the components ultimately used for the CPTM calculation. Table S1 depicts the CPTM-calculated , a quantitation of an EC’s potential biological effects, chemical reactivity, bioavailability, and metabolism (see Methods; Eqs. (1)–(8)). reflects the overall toxicity score for the prioritization of the top toxic ECs.
Fig. 1. Schematic of the chemo-phenotypic toxicity measurement (CPTM) for the generation of toxicity scores ().
An environmental chemical (EC) is given a toxicity score based on the combination of biological, chemical, and toxico-kinetic terms (see Section 14). Tox-TMFS is used to predict novel EC-protein target associations in addition to experimentally determined associations obtained from the Comparative Toxicogenomics Database (CTD). The chemical promiscuity score is the total number of unique known and predicted targets. ECs are then associated with higher-order biological effects through those targets, which include protein- protein interactions, pathways, and functions. The number of these elements contributes to an EC’s biological perturbation score. The toxicokinetic (TK) score is determined by the total number of potential reactions that an EC can undergo into metabolites and the predicted percent human oral absorption. The chemical reactivity index is composed of physico-toxico chemical properties associated with general mechanisms of toxicity, which include: log P, electrophilic Fukui index (f−), HOMO-LUMO energy band gap, electron affinity, ionization potential, chemical hardness, chemical potential, and electrophilicity index. The individual score terms are combined to give a comprehensive toxicity score () to rank ECs, where higher scores denote greater toxicity.
3. Validation of CPTM toxicity predictions
The Hazardous Substance Data Bank (HSDB) 〈http://www.fda.gov/ScienceResearch/SpecialTopics/CriticalPathInitiative 〉, was queri ed to obtain published toxicity data. To validate predictions derived from the CPTM method, HSDB data from the “Animal Toxicity Studies” section was curated and a quantification procedure for measuring toxicity was devised. The following subsections used: (1) subchronic exposure, (2) chronic exposure, (3) carcinogenicity, (4) developmental or reproductive toxicity, and (5) genotoxicity. Within each subsection, each unique pathological phenotype occurrence counted to give a tabulated total for that subsection. If a given pathology occurred in more than one subsection, it is considered to occur only once. The totals for each subsection were then summed to provide a final value for toxicity quantification, which was then paired with the ‘’ toxicity score for a given EC. As an example, the pesticide zoxamide has a score 1.354. Under subacute exposure, it causes irritation at the sites of application (skin and eyes), erythema and conjunctivitis (total = 3). Chronic exposure resulted in cases of polyarteritis syndrome (total = 1). Decreased extra medullary hematopoiesis was found in reproductive and development studies (total = 1). No adverse outcomes were reported for under carcinogenicity or genotoxicity. Thus, the grand total of adverse pathological outcomes for zoxamide is 5. This process was manually performed for 35 random ECs to generate toxicity scores () plotted, against their total number of documented toxicity effects.
The plant derived molecules that yielded one of the top are daidzein and genistein (Fig. S2a; Table S1). Daidzein and genistein are isoflavones typified as endocrine disruptors and found in soy-based products. These isoflavones have been shown can regulate the activity of many proteins and are thought to elicit multiple effects. The molecules with the lowest two are fenaminosulf, a fungicide, and benoxacor, a pesticide safener, implying that they are relatively safe for human use. To validate CPTM, we queried the HSDB 〈http://www.fda.gov/ScienceResearch/SpecialTopics/CriticalPathInitiative〉 for a subset of ECs to obtain relevant toxicological data (see Methods). A direct correlation between an EC’s score and the total number of observed toxic effects was found (Fig. S2b and Fig. S3). ‘’ is, therefore, a reliable quantitative metric for predicting the extent of EC toxic effects.
As the of genistein, a frequently encountered molecule is among the highest, this EC was further assessed considering all predicted genistein-target associations, enriched pathways, and functions including those involved in cancers and cardiovascular illnesses (Table S2). Pathways include peroxisome proliferator-activating receptor (PPAR) signaling, T cell receptor signaling, and pathways in cancer. Functions include negative regulation of cholesterol storage, lipid storage, and foam cell differentiation, all of which promote cardiovascular health, as well as regulation of apoptosis and cell proliferation, which are cancer-mediating functions. These predicted associations are also supported by experimental evidence (Amado et al., 2011; Messing et al., 2012; Squadrito et al., 2013; de Pascual-Teresa et al., 2010).
In addition to validating the CPTM method, we sought to deconstruct the EC chemical space to see if there are any structural correlations. The EC-chemical scaffold bipartite network reveals a diverse chemical structure space with ECs clustering around particular scaffolds (Fig. S4). This is expected, as the ToxCast dataset is a broad collection of ECs comprising many chemical classes. However, ECs that clustered by scaffolds exhibited similar ‘Zts’ scores. For example, ECs containing a pyranose scaffold (DSSTox_RIDs: 72627, 71887, 72127) have similarly low predicted Zts of 1.179, 1.039 and 1.254, respectively (Fig. S4). This is strengthened by the fact that carbohydrate-based molecules generally tend to exhibit low toxicity in humans.
4. ECOTOX validation of CPTM
Next, we validated the CPTM predicted toxicity using biological data from the ECOTOXicology knowledgebase (ECOTOX). Lethal concentration data for the adult fathead minnow (Pimephales promelas), a canonical model organism for aquatic toxicity, were curated and all chemicals were categorized by level of toxic concern in accordance with established EPA thresholds based on either molar or mass-based median lethal concentration (LC50). The scheme used is identical to that used by Judson et al. (2014), with EPA standard categories of “low”, “moderate”, and “high” concern, and an added “no concern” category. Exact LC50 cutoffs are, respectively, 100–500 mg/L, 1–100 mg/L, < 1 mg/L, and > 500 mg/L (Fig. 2). It is important to note here that “biological perturbation” refers to both the “P” (promiscuity) and “N” (cellular network perturbation) terms of the CPTM equation. For the purposes of this biological analysis, Log P is considered as the primary constituent of the “bioavailability” index, and not as part of the chemical reactivity index. Our findings indicate that each biological, intrinsic and kinetic chemical parameter follows an expected and acceptable trend with regard to differences between categories of aquatic toxicity in the fathead minnow (Fig. 2). In particular, of the 11 included parameters, six of them have non-overlapping error bars for nonadjacent levels of toxic concern and three of those six have non-overlapping error bars for adjacent levels of toxic concern. These three categories of particularly strong concordance are normalized electron affinity, normalized hardness, and normalized bioavailability (Log P). The chemical reactivity, ADME-T, and bioavailability indices all exhibit non-overlapping error bars for nonadjacent levels of toxic concern (Fig. 2). The results of our ECOTOX-based biological validation revealed that the CPTM model correctly predicts toxicity concern levels of environmental chemicals in terms of low to high concern.
Fig. 2.
Graphical representation of CPTM-computed EC biological and chemical parameters for each level of toxic concern category derived from ECOTOX fathead minnow (Pimephales promelas) LC50 animal aquatic toxicity data. Also, graphically depicted are the CPTM parameters, as they vary depending on the level of toxic concern of our assessed ECs. Brackets demonstrate the parameters included in the calculation of chemical reactivity and ADME-T toxic indices (see Section 14 for description). The other two indices, those of biological perturbation and bioavailability, are represented as single parameters. Left columns are bar graphs displaying mean values with error bars representing one standard error from the mean. Right columns are box plots reflecting the distribution of values in each category.
In the case of normalized biological perturbation, we hypothesized that toxic chemicals would have a higher overall perturbation score than non-toxic chemicals (). Consistent with this, we found a statistically significant difference in perturbation score between the two groups (Fig. 2). This finding validates the inclusion of protein binding promiscuity as predicted by CPTM as a useful parameter in assessing general toxicity. It also suggests that considering such promiscuity as a whole and promiscuity with respect to specific protein families and cellular functions, such as in the activity of endocrine disruptors, would be a worthwhile endeavor. The overall CPTM score (Zts), because it incorporates every calculated index equally, reflects in some part the unpredictable distribution of values in each of the pro-toxic (“low”, “moderate”, and “high”) concern levels, but nevertheless has a consistently increasing trend.
5. EC-protein network and disease signatures
Next, we analyzed the association of high-scoring ECs with diseases and their mechanisms of action. We started by analyzing predicted EC-proteins and their bionetwork interaction signatures. The EC-protein signature space predicted by CPTM is large and highly interconnected (Fig. S5a, Table S2). Many ECs were predicted to interact with more than one protein, likely explaining the often-multi-phenotypic manifestations of toxicities. Apparent in our predicted interactions that the presence of some false positives and false negatives, which are expected, however that does not diminish the validity or the performance of our method (Fig. S6). Additionally, using a subset of nuclear receptors which have homologous domains does not represent a broad class of proteins and therefore cannot accurately reflect the performance of our binary classification predictive platform. We addressed this limitation by utilizing PubChem database-derived biological assays (protein binding activity assays) to verify both positive (active binding) and negative (inactive non-binding) interactions that were predicted using CPTM. Only proteins for which 10 positive binders exist that were also assessed computationally were included, and EC binding activity classified only as either active or inactive were kept, resulting in 14 proteins (Fig. S6). Receiving Operating Characteristic (ROC) curves were developed to compare the false positive rate and true positive rate and whether the area under the curve (AUC) is significantly greater than it would occur by chance (at the line y = x). The average AUC for assessed proteins was 0.65 (Fig. S6). A two-tiered validation of EC-target and EC-function signatures was performed using the CTD (Davis et al., 2013). 898 (~ 14%) EC-protein interactions and 14,461 EC-function signatures were validated (Table S3). While the absolute quantity of validations is less than the total prediction space, this is expected as many of these EC associations have yet to be tested experimentally. This global approach to validating our EC-protein binding interactions validates current attempts to predict such interactions with the integrated proteochemometric methodology in the CPTM.
As the protein dataset is large and diverse, we first sought to delineate which protein classes were represented through EC-protein class bipartite network (Fig. S3b). Protein classes that are most peripheral include flavoproteins, metalloproteases, ligases, and gyrases. These proteins share the least number of ECs with the other protein classes, which is expected as these proteins exhibit highly specific functions requiring more specific binding site properties. Protein classes found more central in the network include kinases, transferases, transcription factors (e.g. estrogen receptor), and hydrolases, among others. Targets within these classes share many ECs, reflecting potentially similar binding site properties. Conversely, the central clustering of ECs reflects their promiscuity across protein classes and potential to broadly perturb physiology through many pathways and functions.
Next, we established molecular-level EC effects and related them to clinical phenotypes. ECs were linked to cell signaling pathways and molecular functions through their predicted and known proteins as well as the interacting proteins (protein-protein interactions) of those protein targets (Fig. S7 and Table S4). Networks reveal that EC promiscuity is due to their binding to various proteins/protein classes (Fig. S7). The “one EC-many protein targets” outcome results in simultaneous perturbation of pathways and functions leading to multi-phenotypic diseases. The predicted EC-target signatures result in a clustering of pathways involved in cell communication, transport and catabolism, cell growth and death, signal transduction, cancers, as well as endocrine, immune and nervous system functions (Fig. S7a). Pathways found peripherally, such as xenobiotic degradation and metabolism, are those that have the least number of associated ECs. Such pathways usually consist of a few proteins with highly specialized catalytic sites and functions. The EC-protein-function space, on the other hand, reveals the redundancy of molecular functions across protein targets as evident by the spherical structure of the network (Fig. S7b).
As proteins seldom act in isolation but rather through interacting with one another, protein-protein interactions (PPIs) were integrated into the EC network analysis (Fig. S7c). Since PPIs greatly expand the analytical space, for proof-of-concept we highlight here only cancer-associated PPIs (Fig. S7c) as they are shared among other diseases and may highlight interesting EC-disease associations (Fig. S7d). Inclusion of PPIs also revealed connections underlying unexpected EC-disease associations (Fig. S7c and d; Table S4). For example, bisphenol derivatives were linked to peripheral arterial disease, particularly vascular intimal thickness. CPTM predicted that these ECs bind to estrogen receptor alpha (ESR1), which is confirmed by an available crystal structure (Delfosse et al., 2012). ESR1 is known to interact with , a pro-inflammatory protein involved in multiple pathogenic mechanisms (Bonofiglio et al., 2005). Thus, through the interaction, CPTM infers an indirect link between bisphenols and as well as cardiovascular illness. The association between bisphenols and activity has been confirmed (Kwintkiewicz et al., 2010) and circulating bisphenols have been correlated with carotid atherosclerosis and type 2 diabetes (Lind and Lind, 2011; Shankar and Teppala, 2011). Furthermore, ECs are linked to diseases if their protein targets and PPIs have annotations in the Online Mendelian Inheritance in Man (OMIM) database (Goh et al., 2007). Table S4 contains predicted EC-protein target-biological effect associations that may explain EC mechanisms of action for known toxicity phenotypes as well as suggest new phenotypes.
6. Investigation of EC-disease associations
CPTM predicted EC-target network signatures were related to diseases to obtain a cursory perspective of the potential EC-disease signature space (Fig. S7d; Table S4). Clusters are evident among endocrine, hematological and immunological disease categories, implying shared pathophysiologic mechanisms among those categories but are collectively unique relative to the other disease categories. Neoplastic and neurological illnesses exhibit the least amount of clustering, indicating diverse yet overlapping pathophysiology. Retrospective clinical validations for some EC-disease associations pertinent to public health are described below for CRC, and other disease validations.
6.1. Autism spectrum disorder (ASD)
ASD is a complex neurodevelopmental disorder diagnosed early in life that has become a public health concern. We first identified putative ASD-associated pathways that may be perturbed by EC exposures. As the KEGG database does not currently identify ASD-relevant pathways, we obtained pathways from Skafidas et al. (2014). An EC-pathway bipartite network and waterfall plot showing ECs predicted to affect the greatest number of those pathways (Fig. S8). These ECs (apigenin, bifenazate, monobenzyl phthalate, daidzein, boscalid and aspartame) are assumed to more strongly associate with ASD than those predicted to disrupt fewer pathways. Daidzein was predicted to be a potential contributor to ASD. Epilepsy is strongly associated with ASD (Spence and Schneider, 2009) and daidzein is a soy estrogen that increases seizure propensity (Westmark, 2014). In addition, daidzein is a component of soy infant formula, which is implicated in autistic behaviors and seizures in children with ASD (Westmark et al., 2013). The ubiquity of soy infant formula in western post-natal diets merits a deeper investigation of the potential link between daidzein and ASD. Monobenzyl phthalate, a plasticizer found in many products such as children’s toys, is implicated in childhood social impairment as an endocrine disruptor (Ejaredar et al., 2015). Furthermore, maternal pesticide exposure during gestation is implicated in ASD in children (Roberts et al., 2007). Although no reports link bifenazate and boscalid pesticides to ASD, it may be important to monitor for this association. In summary, CPTM has linked a diverse EC space to the disruption of putative ASD-associated pathways with epidemiological findings substantiating these predictions.
6.2. Headache/migraine
Many ECs cause headaches or migraines in an acute setting. Such pathology is important because it is typically one of the first clinical signs indicating exposure. CPTM was used to predict ECs that may be associated with headaches/migraines (Fig. S9; Table S5). The Hazardous Substance Data Bank (HSDB) was used to validate ~ 49% (33 out of 67) of the EC-headache/migraine predictions. These results not only validate CPTM as a platform for determining EC-protein-disease interactions but also suggest other EC exposures that toxicologists and health care professionals should consider when patients present acutely with headaches or migraines.
6.3. Metabolic diseases
Metabolic diseases, such as obesity, metabolic syndrome and type 2 diabetes, have complex etiologies arising from genetics and gene-environment interactions. Endocrine dysregulation is an important contributor to these diseases (Swedenborg et al., 2009), and many ECs have the potential to interact with endocrine signaling regulators (e.g., hormone and nuclear receptors). CPTM predicted EC associations for many such regulators (Table S6). One example is octylparaben interacting with PPARγ, an association that has been experimentally determined and shown to affect adipocyte differentiation (Hu et al., 2013). Association of phthalates with both androgen receptor (AR) and PPARγ were also predicted, which may explain the obesogenic link of phthalates and their major metabolites (Kim and Park, 2014).
The ability of CPTM to recapitulate known EC interactions allows for its use in classifying new ECs as metabolic disruptors as well as providing mechanistic insight into observed effects. For example, the pesticide bifenazate causes increased body weight and hyperglycemia in animal models (Thayer et al., 2012). CPTM predicted that bifenazate would interact with multiple metabolism-associated receptors (Table S6). Furthermore, reports have linked bisphenol A (BPA) with metabolic disorders (Ben-Jonathan et al., 2009). CPTM makes a similar association for the analog bisphenol AF due to its predicted interaction with the progesterone receptor even though it contains trifluoromethyl groups. The associations mentioned here are just a few among many notable potential interactions.
6.4. Colorectal cancer (CRC)
CRC is a solid malignancy generally diagnosed in late adulthood although in recent years there has been a remarkable increase in its incidence in younger individuals (Goh et al., 2007). EC exposures over many years and/or exposure at younger ages may contribute to a subset of sporadic CRCs. A CRC EC-biological effect network was generated through CPTM predicted CRC-related protein signatures (Fig. S10a; Table S7). ECs were associated with CRC if their protein targets were annotated with CRC in the OMIM database (Goh et al., 2007). Centrally clustered ECs include butyl benzyl phthalate, oxazepam, bifenazate, sulfaquinoxaline, and genistein. The uses of these compounds vary, and they are found in distinct environment spaces (e.g., plasticizers, pesticides, etc.). However, their chemical structures are similar in that they contain two substituted benzyl groups that are consistently spaced apart (Fig. S10b). This structure is similar to polychlorinated biphenyls, which are pesticides that correlate with CRC. As molecules with structural similarities tend to exhibit similar biological properties, these molecules may be associated with CRC through similar mechanisms. Alternatively, molecules such as oleic acid and progesterone are less clustered and found peripherally in the network. These molecules are of distinct chemical structure classes (fatty acid and steroidal hormone, respectively; Fig. S10c) that tend to bind to more specific protein targets.
Of the predicted CRC-associated ECs, literature validation revealed some to be harmful. A CRC-promoting association was found for the laxative phenolphthalein (Coogan et al., 2000). However, there is no consensus yet on this association as epidemiological data is limited (Anon, 2022a). However, the validated space is small as many of the predicted ECs have not been studied for CRC. Moreover, CRC is usually a late-onset adult cancer with an important time-course consideration for human epidemiological evaluation of ECs. Overall, the CPTM platform provides biologically plausible hypotheses for ECs to focus toxicology studies for CRC as well as investigate environmental molecules such as phytoestrogens to prevent or treat CRC.
7. Salpn induced disruption of key pathways involved in CRC
The CPTM model predicted that the motor oil additive salpn, is toxic. Salpn is a salen-type ligand used as fuel additive in motor and jet fuels; meta deactivating agent; as a lubricant; as a chelating agent; as an antioxidant in food contact rubber articles for repeated use and as an indirect additive used in food contact substances (〈https://gcomply.decernis.com/reference/document/indirects_2011nov.pdf 〉, 〈https://www.cfsanappsexternal.fda.gov/scripts/fdcc/index.cfm?set=IndirectAdditives&id=DISALICYLIDENEPROPYLENEDIAMINE〉, 〈https://pubchem.ncbi.nlm.nih.gov/compound/N_N_-Disalicylidene-1_2-diamin opropane#section=Use-and-Manufacturing〉). Salpn exposure has been linked to autoimmune diseases such as rheumatoid arthritis (Sverdrup et al., 2005), pro-inflammatory skin conditions (Wolf et al., 1996), skin cancers (Mehlman, 1992), and other diseases. Occupations such as automobile mechanics, deployed military personnel, aerospace workers, and consumers who frequently use food packed with plastic or rubber articles are exposure increased levels of salpn. Exposure usually occurs via direct contact, orally or through inhalation. The increase in the prevalence of certain pathologies-lung cancer and malignant melanoma in aerospace workers (Zhao et al., 2005) and rectal cancer in automobile manufacturing workers (Malloy et al., 2007) may be attributed to multiple effects from salpn - induced dysregulation of cancer regulating signaling.
The CPTM model predicted the binding of salpn to a number of proteins including nuclear receptors such as the Vitamin D receptor (VDR) (Table S8). Dysregulation caused by agonism or antagonism of nuclear receptors by chemical agents can lead to acute and chronic toxic effects such as those associated with CRC and other cancers. Some of the nuclear receptors are related in some way to the VDR and Wnt mediated pathways, and others affect cell death, cell proliferation, hormone signaling, or metabolic response to toxic substances (Kwintkiewicz et al., 2010). Binding of the active form of vitamin D3 (calcitriol) activates VDR and typically has pleiotropic beneficial health effects (Byers et al., 2012; Lai and Fang, 2013). However excess activation results in increased expression of catabolic enzymes such as CYP24A1. Consequently, exposure to certain environmental chemicals may pathologically dysregulate VDR signaling, which is highly complex and implicated in many diseases processes (Froicu and Cantorna, 2007; Laragione et al., 2012; Tremezaygues and Reichrath, 2011). Perturbation of the VDR network or decreased VDR protein activation through inappropriate antagonism or excessive agonism is pathological. CPTM links the toxic effects of some ECs through their potential to disrupt VDR signaling. CPTM predicted that salpn, and a structurally similar natural compound nordihydroguaiaretic acid (NDGA, with high ), can bind VDR (Teske et al., 2014). Moreover, medicinal use of NDGA is controversial as NDGA exhibits pro-oxidant activity resulting in hepato- and renal toxicity (Lambert et al., 2004; Sahu et al., 2006).
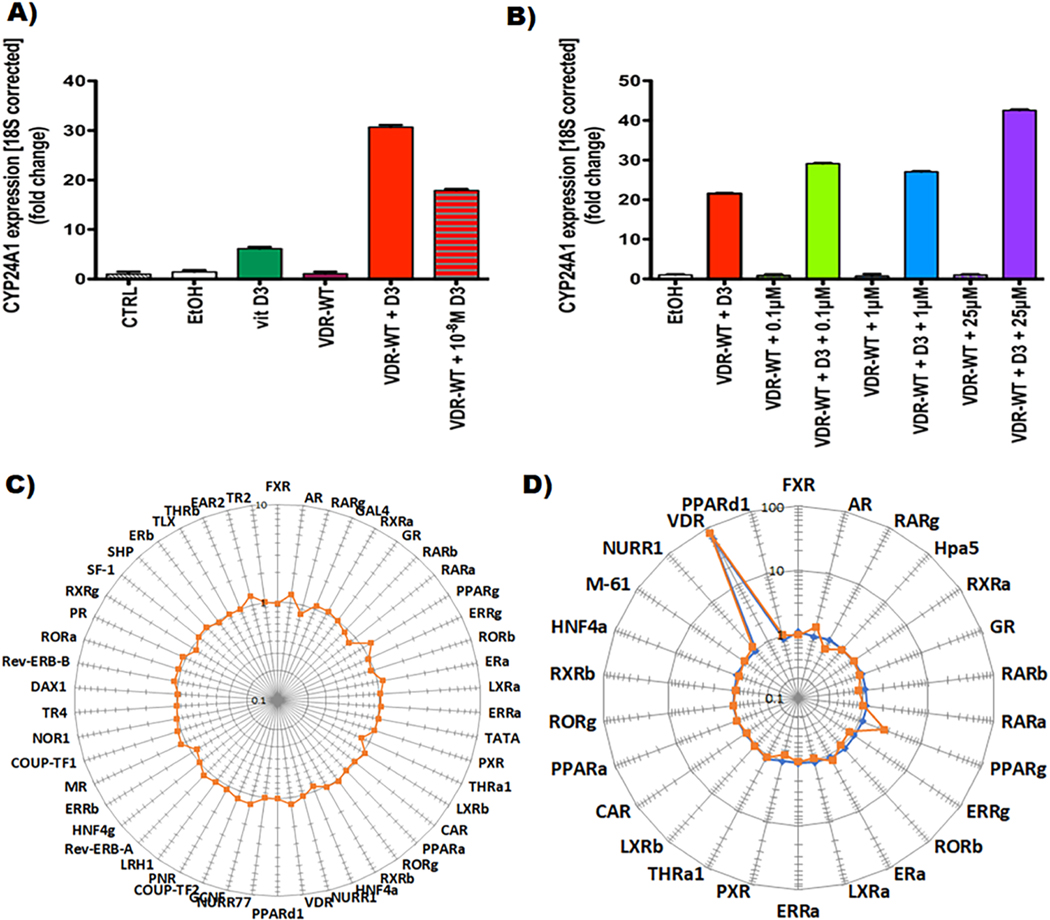
We hypothesize that at high concentrations, salpn binds to the secondary binding site of the nuclear receptors (NRs), stabilizes helix 12 (H12), and enhances the activation of VDR and cooperate with D3, by recruiting coactivators. Alternate binding sites for salpn exist in other NRs and, in combination with their native or synthetic ligands, could potentially contribute to regulation of these transcription factors and their pathways. We tested the ability of salpn to bind VDR and activate a responsive reporter (CYP24A1) in human embryonic kidney 293 cells (HEK293) and human liver carcinoma cells (HepG2 cells). Salpn weakly activates VDR, alone at 25 μM in HEK293 cells (Fig. 3a, b) and up to 1 μM in HepG2 cells (Fig. 3c). However, at 1 μM salpn super activates VDR in combination with the endogenous vitamin D3 ligand at 500 nM in HepG2 cells (Fig. 3d), and at 25 μM of salpn with vitamin D3 at 10−7 M in HEK293 cells (Fig. 3b). This super activation results in increased expression of the catabolic enzyme CYP24A1. Salpn induces CYP24A1 expression in a dose-dependent manner in the presence of vitamin D3, presenting an additive effect over vitamin D3 alone. It is likely that salpn binds at the allosteric second site at low affinity, and in the presence of endogenous D3, it super activates the VDR. Our molecular modeling studies show that the salpn docks at the VDR allosteric lithocholic acid (LCA) binding site (Fig. 4a; VDR-LCA binding complex PDB: 4Q0A). This alternate allosteric site binding of salpn, can provide additional stabilization of the VDR AF-2 surface and significantly enhance coregulator recruitment.
Fig. 3.
(A) and (B) Treatment with vitamin D3 in combination with salpn super-activates VDR signaling in HEK293T cells. CYP24A1 is induced upon activation of VDR signaling, and its expression was assessed in cells with native VDR or cells transfected with wild type VDR (VDR-WT). Upon transfection, HEK293T cells were treated for 24 h with vitamin D3 with or without salpn at various concentrations. Vitamin D3 at 10− 7 M was used unless stated otherwise. 40x diluted 100% ethanol (EtOH) in cell growth media was used as vehicle control. The bars represent the mean value of triplicate treatments, and error bars represent standard error of the mean. (A) Treatment with 10− 7 M of vitamin D3 induces CYP24A1 expression even further when cells are transfected with VDR-WT. Additionally, 10− 8 M of vitamin D3 increases CYP24A1 expression, showing a dose-dependent effect. Control (CTRL) and vehicle (EtOH) treated or VDR-WT transfected cells did not increase CYP24A1 expression in the absence of vitamin D3. (B) Salpn induces CYP24A1 expression in a dose-dependent manner when vitamin D3 is present, showing an additive effect over vitamin D3 effect by itself. (C) Effect of salpn alone (orange) at 10 μM, 16 h, (D) vitamin D3 alone (blue), and (D) salpn + Vitamin D3 mixture (orange) on nuclear receptor activity in HepG2 cells. HepG2 cells were transiently transfected with optimized trans-FACTORIALTM library. Twenty-four hours after transfection cells were washed and supplied with fresh low serum (1% FBS, charcoal stripped) culture medium and treated with inducer for 16 h. Profile of the trans- FACTORIALTM activities was determined as fold of induction values versus vehicle-treated (DMSO) control cells. Graph shows fold-induction data plotted in logarithmic scale.
Fig. 4.
(A) Vitamin D3 receptor, VDR (white ribbon) with D3 (magenta) docked at its orthosteric site and salpn (green) docked at the LCA (yellow) allosteric binding site. (B) Downstream effects of VDR activation by its cognate ligand calcitriol (activated Vitamin D3). Arrows denote directionality of biological response. (C) Entire Cellular Network Model for salpn with highlighted VDR subnetwork. The green node is salpn, blue nodes are direct protein interactions, light pink nodes are PPIs, orange nodes are pathways, and purple nodes are molecular functions. Dark pink node is the direct interacting protein VDR, with protruding edges in red to signify direct perturbation network of VDR. This network excludes disease associations from the OMIM database, instead describing all possible disease-associated biological activity perturbed from an EC-centric viewpoint. (D) Effect of salpn (10 μM, 16 h) on transcription factor activity in HepG2 cells. HepG2 cells were transiently transfected with optimized cis-FACTORIALTM library. Twenty-four hours after transfection cells were washed and supplied with fresh low serum (1% FBS, charcoal stripped) culture medium and treated with inducer for 16 h. Profile of the cis-FACTORIALTM activities was determined as fold of induction values versus vehicle-treated (DMSO) control cells. Graph shows fold-induction data plotted in logarithmic scale. Insert table is a list of transcriptional pathways tested for their activity.
Additionally, several studies have been reported the existence of allosteric ligand binding sites for VDR (Mizwicki et al., 2004, 2007, 2005; Mizwicki and Norman, 2009; Menegaz et al., 2011). Anna et al. showed that the recognition of two molecules of lithocholic acid (LCA), a promoter of colon carcinogenesis in mammals is crucial for VDR agonism (Belorusova et al., 2014). They showed that one LCA molecule binds at the canonical VDR ligand binding domain (LBD) site and a second LCA molecule at the allosteric surface site. Despite the low affinity of the LCA at the allosteric site, the binding of the second LCA molecule promotes stabilization of the VDR active conformation (Belorusova et al., 2014). This finding is consistent with our data that salpn binds to VDR with low affinity and by itself does not activate VDR well (Fig. 3, and Fig. 4a). However, in the presence of the endogenous ligand D3 it results in excessive agoniztic activity. The super-agoniztic effect of salpn plus vitamin D3, influences the beneficial effects of vitamin D3. Perturbation of the VDR network through excessive agonism is pathological. Consequently, exposure to salpn may pathologically dysregulate VDR signaling (Fig. 4b). In real-life, as sunlight provides abundant vitamin D3-synthetic activity and D3 supplementation is also common, mixtures of vitamin D3 plus salpn, may super activate VDR, thereby increasing its catabolism by increasing CYP24 and causing toxicity.
The CPTM method predicted that salpn effects may also contribute to the dysregulation of other important signaling pathways related to CRC development such as those mediated downstream of VDR response elements (VDRE), , HIF1, and (Table S8, Fig. 4b, c). We show that salpn exhibits similar super-activation of 46 other nuclear receptors in combination with their endogenous ligands. In the nuclear receptor transcriptional pathway assay, as predicted by CPTM, at 1 μM in HepG2 cells, salpn dysregulates several pathways including VDRE, , , NFkB and HIF1 (Table S8, Fig. 4d). NRs cross-talk directly (Fig. 5a) or indirectly (Fig. 5b), and interaction with other protein family targets could also act as CRC driver pathways (Table S8, Fig. 5a, b). Nonetheless, for salpn alone, we noticed a significant transcriptional pathway upregulation of VDRE, RORE, MRE, HIF1a, Oct-MLP, MRE, TAL and down regulation of ERE, , (Table S8, Fig. 4d). Notably, compared to salpn alone, the combination of 1 μM of salpn and 500 nM of D3 in HepG2 cells, enhanced the dysregulation of several pathways including VDRE, ERE, Ahr, , , NFkB and, RORE, MRE, HIF1a, Oct-MLP, MRE, TAL (Fig. 5c). In addition to the effect of the salpn plus D3 combination on VDR super activation, salpn may also interact with other NRs and enhance cross-talk directly (Fig. 5a) or indirectly (Fig. 5b) to drive multiple adverse outcome pathways in CRC (Fig. 5a–c).
Fig. 5.
(A) Protein-protein STRING Interaction map of nuclear receptors cross-talk with each other including VDR directly or indirectly (B). (C) Effect of salpn (10 μM, 16 h) in combination with D3 (orange) or D3 alone (blue) on transcription factor activity in HepG2 cells. HepG2 cells were transiently transfected with the optimized cis-FACTORIALTM library. Twenty-four hours after transfection cells were washed and supplied with fresh low serum (1% FBS, charcoal stripped) culture medium and treated with inducer for 16 h. Profile of the cis-FACTORIALTM activities was determined as fold of induction values versus vehicle-treated (DMSO) control cells. The graph shows fold-induction data plotted on a logarithmic scale. Insert table is a list of transcriptional pathways tested for their activity. (G) Wnt signaling modulation of salpn in zebrafish. (E) Wnt reporter activity was visualized in zebrafish using whole-mount in situ hybridization (WISH) for GFP expression.
To further confirm the CPTM model Wnt signaling marker predictions, and transcriptional pathway assay results, we tested salpn in a zebrafish transgenic Wnt reporter assay. In transgenic Wnt reporter zebrafish embryos,Tg(TOP:GFP)w25, Wnt signaling is markedly decreased by 5 μM salpn. Interestingly, at 8 μM, localized Wnt signaling appears to be increased, however, this may reflect the rather severe developmental delay in these embryos (Fig. 5d). Taken together, salpn in concert with D3, likely contributes to dysregulation of key CRC signaling pathways. These results also suggest that an extended time-course (chronic) exposure study with higher concentration is needed to confirm the toxicity of salpn in the dysregulation of Wnt and VDR signaling and its association with CRC prevalence.
8. Structural basis of salpn mediated super-activation of VDR
To explore the binding modes of the salpn, docking studies were performed with the VDR LBD crystal structure using Glide software (Schrodinger Inc). Several binding sites were located and their binding modes are shown (Fig. S11a) with salpn and D3 occupying a similar overall orientation and binding location within the LBD (Fig. S11b; (Hughes et al., 2016, 2014; Mosure et al., 2018)). The existence of allosteric ligand binding sites is reported for other nuclear receptors (Moore et al., 2010) including VDR (Mizwicki et al., 2004, 2007, 2005; Mizwicki and Norman, 2009; Menegaz et al., 2011), ER (Wang et al., 2006; Kojetin et al.; Jensen and Khan, 2004), AR (Buzon et al., 2012; Grosdidier et al., 2012; Estébanez-Perpiñá et al., 2007), and TR (Estébanez-Perpiñá et al., 2007; Arnold et al., 2005; Sadana et al., 2011). We hypothesize that the super-activation characteristics of salpn are typical of agonist or partial agonist mixture effects by binding to the VDR LBD or to the non-canonical binding site distal to the VDR LBD.
We propose several hypothetical models for salpn mediated super activation of the VDR. In the first model, salpn binds to VDR, either near the conventional orthosteric binding site or at the orthosteric binding site, and in the presence of vitamin D3, cooperatively activates the VDR (Fig. S11a, b). Given that the VDR ligand binding pocket (LBP) volume is large (around 900A3), it is likely that it could accommodate a second molecule. This model is supported by the highly dynamic nature of the LBD helices, and also by previous studies that showed that the LBPs are flexible enough to adjust their size to adopt different-sized compounds ((Meijer et al., 2019; Delfosse et al., 2015; Färnegårdh et al., 2003 ), PDB: 3K8S, and 4E4K). Delfosse et al showed that the structures of the PXR agonizts 17α-ethinylestradiol (EE2), and the trans-nonachlor (TNC), that TNC occupies a portion of the PXR LBP that is only accessible when EE2 is bound (Delfosse et al., 2015). In the second model (Fig. 4a, Fig. S11a, b), salpn binds to the allosteric VDR AF3 surface site and interacts with Helix-12 and potentiates vitamin D3 agonist effect. In the third model, at high concentrations (25 μM) salpn binds at the VDR orthosteric site and competes with the vitamin D3, thus vitamin D3 tends to bind at the allosteric surface site (Fig. S11c). In second and third model, it is likely that salpn or vitamin D3 occupies the orthosteric binding site and sequentially the vitamin D3 or salpn binds to the low-affinity site (Fig. 4a, Fig. S11a–c). In support of the second and third models, Belorusova et al. (2014) showed that two molecules of lithocholic acid (LCA, a natural bile acid) bind to VDR, one located in the orthosteric site recognized by vitamin D3, and a second site located on the allosteric VDR surface site (the same site we located) between loops 1–3, Helix 2, and Helix 3 (Fig. 4a, Fig. S11a–c). The binding of LCA to the second binding site by direct interaction with Helix 12 stabilizes the active agonist conformation of the complex (Belorusova et al., 2014). Our VDR-salpn and vitamin D3 structural model showed a similar binding mode at exactly the same VDR surface site with Vitamin D3 shares high structural similarity with LCA (Fig. 4a, Fig. S11a–c).
9. Salpn activates other nuclear receptors
As predicted by our CPTM model, we showed in vitro in the nuclear receptor (NR) assay, that salpn alone activates other NRs such as , AR, , and (Fig. 4d). This direct agonist activity of salpn serves as an early indicator of the adverse health effects. Further, upon analysis of the nuclear receptor ligands including AR, and , we found that ligands similar in structure to salpn bound at the allosteric surface site of AR (PDB: 3ZQT), (PDB: 4BCR) and (PDB: 4LNW) with low affinity and that the endogenous ligand bound at the NR LBP in a two-mode mechanism involving both ligands. Previous studies showed that several ligands bind to the allosteric pocket on the NRs (Meijer et al., 2019; Bona et al., 1979; Tanenbaum et al., 1998). We also located salpn in other NR allosteric sites using docking studies. For example, we found that salpn docked to a novel site on the Androgen Receptor (AR) called binding function 3 (BF3) that is involved into AR transcriptional activity (PDB: 3ZQT). Salpn is superimposed well with a similar AR allosteric BF3 site binding ligand called ‘30Z’ with testosterone binding at the AF3 orthosteric site (Fig. S11d). Further, Capelli et al. showed that can host two molecules of (S)-16 when present at a high concentration (Capelli et al., 2016; Laghezza et al., 2018). Consistently, salpn + D3 treatment showed activation of AR and in the NR activation and transcription pathway activation assays (Figs. 4d, 5c). These results led us to speculate that interaction of two distinct ligands salpn and D3, and/or two salpn ligands at high concentrations in the orthosteric and allosteric binding site could super activate the VDR and other NRs. This super activation would lead to potential recruitment of the co-activators which dysregulate the VDRE, , TCF, and other signaling pathways important in CRC. Taken together, we hypothesize that salpn has the potential to bind at the alternate site with weak affinity but in the presence of orthosteric or allosteric site endogenous or synthetic ligands, could super-activate the nuclear receptors. To confirm this hypothesis our next step is to perform the salpn plus NR endogenous or synthetic ligands combinatory studies.
10. Salpn is toxic to cultured skin cells in vitro, and zebrafish embryos in vivo
To assess the ability of the CPTM to predict overall toxicity, we measured cell viability using XTT metabolic assays and apoptotic activity using Annexin-PI staining and caspase 3 activity assays. Humans are readily exposed to motor oil, either topically, orally, or by respiration because of its widespread use in industry and for consumer-owned machinery. Thus, we assessed salpn cytotoxic effects in normal human skin cells, such as HaCaT human keratinocytes. HaCaT cells were chosen as they are spontaneously immortalized skin keratinocytes derived from adult human skin, and have been extensively used in diverse cell culture experiments.
In vitro cytotoxicity studies revealed that salpn is cytotoxic to HaCaT cells in a dose-dependent manner (Fig. 6a, b). Dose-response curves show a significant decrease in cell viability in HaCaT cells exposed to salpn, with an IC50 value of 84 μM (Fig. 6b). Decreased cell viability is due to increased apoptotic cell death (Fig. 6d). Annexin PI staining and flow cytometry data show a significant increase in early apoptosis after exposure to salpn, as well as a significant increase in total apoptotic death (Fig. 6d). We also showed that the activity of the apoptotic cell cysteine proteases recognizing aspartate motifs (esp. DEVD), caspase 3, was proteolytically induced dose-dependently with increasing salpn concentrations, confirming that the decrease in cell viability is largely due to apoptotic cell death (Fig. 6e). Importantly, the result of caspase-3 activity assays directly correlated with the Annexin/PI staining assay data, which is dependent on exposure of phosphatidylserine, a signal for phagocytosis, on the outer leaflet of the cell membranes of apoptotic cells. These results suggest that salpn is a potential environmental toxicant to humans, even at low concentrations, through the induction of apoptosis, in normal adult skin keratinocytes.
Fig. 6. XTT assay of HaCaT Cells treated with increasing concentrations of salpn for 72 h (A & B).
(A) Representative images of HaCaT human skin keratinocytes incubated with indicated concentrations of salpn for 72 h in 96-well plates. (B) Dose-response for HaCaT skin keratinocytes cells after treatment with salpn showing a marked decrease in cell viability after exposure to salpn. The (C) for salpn is 84 μM. Annexin/PI Apoptotic Assay of HaCaT cells treated by salpn for 72 h (C & D). (C) Representative images of HaCaT human skin keratinocytes in 6-well plates showing effects of exposure to increasing concentrations of salpn for 72 h. (D) Annexin-PI staining apoptosis data after treatment with salpn showing % of live cells, early, late and total apoptosis, respectively. Caspase-3 Activity Assay of HaCaT Cells treated with increasing concentrations of salpn for 72 h (E). Induction of caspase-3 activity in Jurkat positive control cells exposed to etoposide (upper panel) and cells treated with different concentrations of salpn (lower panel). Zebrafish morphological assay (F). Dose-dependent response toxicity of salpn measured as the lowest dose leading to significant developmental abnormalities at 2 days after post fertilization (2 dpf). TUNEL TMR apoptosis assay (G) with salpn treated embryos, dark dim background staining has either no staining (Negative control) or very few apoptotic cells (DMSO control). Arrows on the picture show the positive cells. Apoptotic cells in 2 dpf embryos were labeled using the in-situ Cell Death Detection Kit-TMR.
We then measured dose dependent response toxicity of salpn using a zebrafish embryo morphological assay (Fig. 6f). Toxicity is measured as the lowest dose leading to significant developmental abnormalities at 2 days post fertilization (2 dpf). Salpn is toxic at 5 μM with delayed development displaying morphological characteristics of a 36 h post-fertilization (hpf) stage embryo, slight ventral curve of the body axis, and an emaciated phenotype. Embryos were dead at 10 μM (Fig. 6f). The terminal deoxynucleotidyl transferase TMR-dUTP nick end labeling assay (TUNEL TMR), detecting apoptotic DNA breaks, revealed an increase in apoptosis in the head of salpn-exposed embryos, most likely localized to olfactory pits, as well as increased apoptosis in the tail (Fig. 6g). Taken together, with the effects of salpn on NR and wnt pathways, these data strongly indicate that salpn is toxic in cultured human cells and in vivo systems with the potential for increased CRC incidence in adults.
11. Tetramethrin and fenpropathrin is toxic to Zebrafish embryo in vivo
The CPTM model predicted that Tetramethrin (T) and Fenpropathrin (F), two widely used pesticides, are toxic. We measured the dose-dependent response toxicity of Tetramethrin and Fenpropathrin using a zebrafish embryo morphological assay (Fig. S12). We used 0.1% DMSO and fresh water as vehicle control (Fig. S12). Toxicity was defined as the lowest dose leading to significant developmental abnormalities at 2 dpf in a dose-response zebrafish live assay. Fenpropathrin was determined to be toxic at 5 μM, with a slight developmental delay and variable dorsal arching. Prominent dorsal arching and a distinctive twitching phenotype were seen at 10 μM and 15 μM. The twitching phenotype suggests specific neuronal toxicity (Fig. S12b). Tetramethrin was determined to be toxic at 5 μM, with a slight developmental delay and increased cardiac edema. At 10 μM, development is further delayed, with a pronounced dorsal arch and cardiac edema. Exposure to 15 μM, resulted in ventral curvature and variable lethality at 2 dpf (Fig. S12c). Exposure to a mixture of 5 μM Tetramethrin and 5 μM Fenpropathrin resulted in 100% lethality, indicating a potentiating interaction of these two compounds in zebrafish embryos.
11.1. Tetramethrin and fenpropathrin mixture promotes pancreatic ductal adenocarcinoma (PDAC) progression
CPTM model predicted that several of the cancer pathways including pancreatic cancer are dysregulated by T and F. Therefore, we decided to use our patient derived pancreatic ductal adenocarcinoma (PDAC) cell model to assess the tumor-promoting effect of the T, F, and T plus F mixture. PDAC is a highly aggressive disease, with less than a 10% overall five-year survival rate. Using the patient derived PDAC cells that we developed (Parasido et al., 2019), we compared the effects of exposure to 3 μM T or 1 μM F, alone or in a combination of the two with F at .25 μM and T .75 uM, in both the treatment-naive Parental line 1 (Patient 1-CR) and the isogenic n-PTX resistant (P1-nPTX-R7) cells in 2-dimensional cultures. T or F, alone or in combination only modestly increased cell numbers in the P1 parental (Fig. 7a) cells, more substantial increases in cell number were seen in the P1-n-PTX-R7 cells when exposed to both pesticides in combination (Fig. 7b) despite the significantly reduced concentration of the chemicals. Interestingly, while modest increases in cell number were seen with Fenpropathrin in the P1-n-PTX-R7, exposure both pesticides in combination resulted in increased cell numbers and colony formation despite the significantly reduced concentration of the chemicals, when used in combination versus either chemical alone or control (Fig. 7a,b).
Fig. 7.
(A) Patient-1-derived (Patient-1-CR) cells and (B) n-PTX-resistant (P1-n-PTX-R7) cells were continuously exposed to fenpropathrin (F) and tetramethrin (T). To evaluate the effect of F and T on growth rate, 10,000 cells were seeded for each condition. Cells were counted at 4, 8, 11, and 18 days. The chemicals are treated at the following concentrations with F (1 μM); T (3 μM) and the mixture F (0.25 μM) + T (0.75 μM). (C) and (D) Patient-1-CR cells, continuously exposed to F and T were used to evaluate the spheroid formation. 4000 cells per condition were seeded in a low adherent 96 well plate. F and T were added alone or in combination to 3D media. Spheroids volume was measured at day 4 using ImageJ and ellipsoid formula. (D) Representative images of the spheroids shown with control, treated F, T, and F + T. The spheroids were originated seeding 1000 cells in a U shape low adherent plate, using 3D-conditioned media, as per protocol, with a volume of 50 μL. Every 2 days, the medium was replaced with fresh medium containing fresh T, F or T + F. Final measurements were taken after 13 days. (E) Patient-1-CR cells and P1-n-PTX-R7 cells were continuously exposed to F and T for over a month. To evaluate the effect of F and T exposure on n-PTX resistance, cells were exposed to 1 μM of n-PTX. After 72 h, trypan blue exclusion assay was used to evaluate the % of dead cells.
Next, spheroids were generated from the two-dimensional cultures. Tetramethrin and Fenpropathrin were added alone or in combination as described above to 3D media. Spheroid volumes were measured at day 4 using ImageJ, and the ellipsoid formula was used to define spheroid volume. The combination treatment significantly enhanced the volume of the spheroids (*p = 0.039; Fig. 7c–d). These data clearly show that these two frequently used pesticides can act in an additive or synergistic manner to enhance PDAC cell proliferation and/or survival. We have previously established that the nab-paclitaxel resistant P1-n-PTX-R7 cells exhibit significant and persistent upregulation of C-MYC, which is a known cancer stem cell marker and is associated with profound alterations in cellular metabolism and cellular function (Parasido et al., 2019). The differential responses of the drug sensitive Patient 1 cells in 2D vs 3D cultures may be the result of the more complex interactions of the PDAC cells associated with stem cell marker genes such as LDH1A1, ALDH1B1, and LRIG1 (Nelson et al., 2020). Finally, the P1-Parental and P1-n-PTX-R7 cells were exposed to T, F or T + F for one month at the concentrations 3 μM of T, 1 μM of F, alone and in a combination of the two with F at .25 μM and T .75 μM. These cells were then treated with 1uM nab-Paclitaxel and cell viability was assessed. Long-term exposure significantly reduced drug sensitivity in both the P1-Parental and P1-n-PTX-R7 cells (Fig. 7d). Collectively, our data suggest that exposure to Tetramethrin and Fenpropathrin may significantly impact the populations of pancreatic cancer cells that support rapid PDAC progression, further complicating treatment and resulting in increased mortality in an already difficult to treat disease. Nevertheless, further explorative studies are needed for the mechanistic understanding of T + F PDAC tumor promotion potential.
12. Conclusion
In this study, we assessed the ability of CPTM to predict the toxicity of a subset of ECs. CPTM integrates systems bionetworks, with the chemical reactivity and toxicokinetic properties of ECs to reliably predict toxicity through a quantitative measurement called the “toxicity score” (). CPTM predicted other clinically relevant EC toxicity and disease associations including for the phytoestrogen daidzein and plasticizer monobenzyl phthalate with ASD as well as the obesogenic potential of parabens (Kolatorova et al., 2018). CPTM predicted Salpn toxicity was confirmed in human skin cells and in zebrafish embryo model. CPTM predicted, and biological studies showed that the salpn would interact with the nuclear receptors either at their orthosteric or allosteric site. Consistently, we showed that the salpn super activates the VDR in combination with the endogenous vitamin D3. We provided the structural basis of this combinatory super activation of the VDR. The super-agoniztic effect of salpn + D3 mixture may negatively influence the beneficial health effect of D3. In the in vitro and in vivo studies, we found that dysregulation of key colorectal cancer CRC related signaling pathways, a potential link with the initiation of CRC. This is an important finding as there is no established link between salpn exposure and CRC prevalence. In real-life, as sunlight provides abundant vitamin D3-synthetic activity and D3 supplementation is also common, thus mixtures of vitamin D3 plus salpn, may super activate VDR, thereby increasing its catabolism by increasing CYP24A1 and causing toxicity. An extended time-course (chronic) low-dose salpn co-exposure study is needed to elucidate its harmful effects and association with CRC prevalence.
Additionally, our predictive modeling studies followed by in vitro assays showed that salpn likely activates other nuclear receptors with a similar allosteric mode of action. Allosteric pocket binding ligands are ignored and biased because of the high affinity of the orthosteric ligands. However, this can be of importance from a primarily toxicological perspective, and our future studies will focus on this. Our experiments are carried out with low micromolar concentrations of salpn. We expect that higher concentrations of salpn would lead to more dysregulation of pathways that promote the CRC at a faster rate. On the other hand, daily or frequent exposure to salpn such as occurs in motor or jet fuel oil workers may contribute to their increased risk towards cancer development. Further, our in-silico and in vitro human patient derived cell model data suggest that exposure to the widely used pesticides Tetramethrin and Fenpropathrin may significantly impact the populations of pancreatic cancer cells that support rapid PDAC progression, further complicating treatment and resulting in increased mortality in an already difficult to treat disease. We anticipate that the CPTM platform will evolve into a robust computational platform that aids in the rapid prioritization of ECs for more extensive toxicological studies, thus reducing costs while increasing efficiency of testing as well as identifying putative highly toxic ECs to alert clinicians and health policy officials. This further benefits future industrial endeavors as prior knowledge of potential toxicity would guide the safety-driven production of new chemicals.
12. Limitations of the study
A major assumption in CPTM is that ECs with greater protein target and biological effect promiscuity, as well as toxicity-related physicochemical features, would have a greater number of overall toxic effects. This was found to be true in our analysis. Unlike other models, the CPTM toxicity score is not meant in its current form to correlate with LD50 values or other endpoint-specific measures, which QSAR models typically strive to do. We aimed for a preliminary assessment in which the toxicity scores correlated with the number of toxic effects noted in the HSDB as a measure of general toxicity.
While CPTM in its current form is a valid method for general toxicity, makes no links to lethal dosages or of dose/potency-relationships with toxicity. Future improvements are necessary for biological adverse outcome pathways and full toxicokinetic considerations. EC-target-pathway effects are not all equal, with some resulting in different toxic effects while others remain benign. It is necessary to account for such Adverse Outcomes Pathways (AOPs) (Kleensang et al., 2014). While a central database for AOP study has been created, the full scope of such pathways has yet to be realized. Furthermore, as knowledge of AOPs increases, it will be integrated in concert with patient gene expression and survival data to fine-tune weighting of the biological component of CPTM to more accurately reflect biological effects. Furthermore, dose-dependency of EC exposures is a critical determinant of target binding and biological effects. Incorporation of binding affinities into CPTM will be helpful to differentiate toxic effects between low- and high-exposure ECs and model differential downstream biological effects. More detailed consideration of toxicokinetic effects will further increase the accuracy of CPTM. This is especially important in considering all potential routes of exposure as well as the reactivity of metabolites generated. Future iterations of CPTM will integrate all these factors including the machine learning algorithms for improved predictions of lethal doses and doses for other endpoints. While CPTM can recapitulate EC toxicity and disease associations and help explain the mechanisms behind them, more detailed disease association mechanisms to be tested in future prospective in vitro and in vivo experiments.
14. Materials and methods
14.1. Chemo-phenotypic based toxicity measurement (CPTM) method
14.1.1. Reflecting toxicity quantitatively using a “toxicity score” ()
The CPTM method (Fig. 1) is a comprehensive integration of biological interactions, reactivity, physicochemical, and toxico-kinetic (TK) properties to quantify an EC’s toxicity risk through a “toxicity score” () (Fig. S1). ECs with higher values are predicted to have higher overall human toxicity relative to ECs with lower values. An EC’s toxicity score is calculated using the following Eq. (1):
| (1) |
where the “P” term represents the normalized chemical promiscuity score of a given EC , along with its designated weight (). The promiscuity score is the sum of two factors: (1) represents predicted chemical-protein interactions using Tox-TMFS (Dakshanamurthy et al., 2012; Issa et al., 2015), and (2) represents annotated chemical-protein interactions, which are obtained from the Comparative Toxicogenomics Database ( CTD). The “N” term represents the normalized biological network perturbation score of an EC, which is derived from the total number of higher-order EC-biological effect associations (Fig. 1) at the level of protein-protein interactions (), pathways (), and functions (), along with its designated weight (). The “R” term represents the normalized score of the EC’s chemical reactive index score containing log P, HOMO-LUMO band gap energy (), ionization potential (), electron affinity (), chemical hardness (), electronic chemical potential (), electrophilic (f−) Fukui function, and electrophilicity (), along with its designated weight (). This index is composed of physicochemical properties related to reactivity associated with general mechanisms of toxicity according to the toxicology literature (Tsakovska et al., 2008). The “TK” term represents the normalized toxicokinetic score containing the number of potential metabolic reactions () that an EC can undergo as well as its percent human oral absorption (). A simplified version of the toxicity score () is given below.
14.2. Toxicity score equation
The Toxicity Score () is a quantitation of the potential toxicity for a given EC molecule (Fig. S1). It consists of three terms: (1) biological network perturbation score, (2) chemical reactivity index score, and (3) TK score. Eq. (2) represents the mathematical expression of the ‘’ for a given EC molecule :
| (2) |
14.2.1. Biological perturbation score ()
The biological perturbation score () is a metric of physiologic disturbance upon exposure to EC. BioPSi is a combination of chemical promiscuity score (“P” term) and biological network perturbation score (‘N’ term) as given in Eq. (1). To quantify this perturbation, biological network analysis was used to contextualize the physiology in terms of protein targets and their related biochemical functions, signaling pathways, and clinical diseases. Further refinement is achieved by including “unique class” terms that correct for the overrepresentation of the aforementioned disease classes. For example, if an EC is hypothetically predicted to bind 20 proteins, 15 of which are involved in breast cancer, three in ovarian cancer, and the remaining in Alzheimer’s disease, then the “unique disease classes” correction term would be weighted as “2” to represent the oncologic and neurologic disease classes. The same is done for the other two higher-order terms of function and pathway. On the protein level, the correction term is determined by the number of predicted protein families hit by that molecule. The summation of all these terms and subsequent normalization provides the final value for ‘BioPS’ for incorporation into the toxicity score (). The greater the BioPS, the greater the predicted physiological perturbation for a given EC. Eq. (3) represents the BioPS metric for a given EC :
| (3) |
where and represent the total number of proteins and unique protein families, respectively, and represent the total number of diseases and unique disease classes, respectively, and represent the total number of pathways and unique pathway classes, and and represents the total number of functions and unique function classes.
14.2.2. Chemical reactivity index score ()
The chemical reactivity index score (ChemRIS) is a purely chemical-based metric. The addition of ChemRIS serves to enhance the toxicity score () by accounting for the potential damage caused by the intrinsic reactivity of a molecule irrespective of its protein targets and biological associations. Chemicals with high reactivity are generally less stable, thus contributing to the formation of deleterious substances, such as reactive oxygen species, that interact with cellular and organ components.
In the context of the Toxicity score (), we define chemical reactivity index by the following parameters: log P, HOMO-LUMO band gap energy, ionization potential (I), electron affinity (A), chemical hardness (), electronic chemical potential (), electrophilic (f−) Fukui function, and electrophilicity (). Electron affinity (A) and ionization potentials (I) were used to calculate the electrophilicity index (), chemical hardness (), and chemical potential (). We and others, previously described the use of the electrophilic (f−) condensed Fukui function, and chemical hardness (Fukui, 1982; Parr and Yang, 1984; Pearson, 1997; Subramanian et al., 1998; Sivanesan et al., 1999; Parr and Pearson, 1983) is defined as:
| (4) |
Chemical hardness () is a molecule’s propensity for resisting the change of its electron cloud as described by us and others (Pearson, 1997; Subramanian et al., 1998). Assuming the validity of Koopman’s theorem for closed-shell molecules, chemical hardness (Parr and Pearson, 1983) is defined as:
| (5) |
where I and A are the ionization potential and electron affinity of an EC molecule, respectively.
The chemical potential () is the potential energy of a molecule that can be absorbed or released when it undergoes either a physical change or a chemical reaction (Parr et al., 1978). Again, assuming the validity of Koopman’s theorem for closed-shell molecules, chemical potential () is defined as:
| (6) |
where I and A are the ionization potential and electron affinity of an EC molecule, respectively.
The electrophilicity index () is a quantification of a molecule’s likelihood of accepting electrons from a nucleophile for bond formation. Electrophilicity index () (Parr et al., 1999) is defined as:
| (7) |
where is the chemical potential and is the chemical hardness from Eqs. (5) and (6) above.
Eq. (8) represents the ChemRISi metric for a given EC :
| (8) |
The ionization potential (I) and electron affinity (A) for our set of EC molecules were calculated using QikProp (Small-Molecule Drug Discovery Suite 2013–3: QikProp, version 38, 2013). Jaguar (Schrödinger Release 2013–3: Jaguar, version 80, 2013) was used to calculate electrophilic Fukui indices and HOMO (Highest Occupied Molecular Orbital), and LUMO (Lowest Unoccupied Molecular Orbital) band gap energies.
14.2.3. Toxico-kinetic score ()
The “TK” score consists of the number of potential metabolic reactions (MR) that an EC can undergo as well as its percent human oral absorption (OAh). We assume that an EC’s bioavailability is directly correlated with its toxicity, and that ECs likely undergo metabolism to more reactive intermediates that give rise to toxicity. By way of this toxico-kinetic term, we seek to address the fact that many environmental ECs will arrive at the colonic epithelium or other disease-relevant tissue in an altered state, i.e. as a metabolite. The metabolic reaction term, as detailed in Fig. S1, is prospective in predicting the total number of reactions based on chemical structure and circumvents the issues arising from animal model genetic backgrounds (e.g., differential cytochrome P450 expression levels). While this may lead to some overestimated risks, we believe it is prudent to have this overestimation for a few ECs than to underestimate the risk of the majority of ECs. Eq. (9) represents the metric for a given EC :
| (9) |
Both properties were calculated using QikProp (Small-Molecule Drug Discovery Suite 2013–3: QikProp, version 38, 2013).
14.2.4. EC-protein target and biological effect predictions
The detailed methodological procedure of EC and Protein Target Datasets, Predicting EC-Protein Target Signatures and Relating EC-Target Signatures to Biological Effect Endpoints are described in the Supplementary information.
14.3. Human nuclear receptors and transcription factor activity assay
We used Attagene Inc. proprietary FACTORIAL TF assay (a.k.a. cis- FACTORIAL™) that enables profiling compound-induced changes of activities of multiple transcription factors (TFs) within cells and the FACTORIAL NR assay (a.k.a. trans-FACTORIAL™) that enables profiling agonist/antagonist activities of a compound across all human nuclear receptors (NRs). This is a reporter detection platform; the FACTORIAL™ that allows for a simultaneous assessment of multiple reporter constructs in a single well of cells and ensures essentially identical detection efficacies for the multiple endpoints. The detailed description of the FACTORIAL TF (a.k.a. cis-FACTORIAL™) and the FACTORIAL NR assay (a.k.a. trans-FACTORIAL™) assays are reported previously (Medvedev et al., 2018; Romanov et al., 2008). The effect of salpn (10 μM, 16 h) on NR Activity in HepG2 cells was determined. HepG2 cells were transiently transfected with an optimized trans-FACTORIAL™ library. Twenty-four hours after transfection cells were washed and supplied with fresh low serum (1% FBS, charcoal stripped) culture medium and treated with inducer for 16 h. Profile of the trans-FACTORIAL™ activities was determined as fold of induction values versus vehicle-treated (DMSO) control cells. Similarly, the effect of salpn (10 μM, 16 h) on TF Activity in HepG2 cells was determined. HepG2 cells were transiently transfected with the optimized cis-FACTORIALTM library. Twenty-four hours after transfection cells were washed and supplied with fresh low serum (1% FBS, charcoal stripped) culture medium and treated with an inducer for 16 h. Profile of the cis-FACTORIALTM activities was determined as fold of induction values versus vehicle-treated (DMSO) control cells.
14.4. VDR activation assay
14.4.1. Cell culture
Human embryonic kidney cells, HEK293T cells (available through ATCC, Cat#: CRL-3216) were grown in Dulbecco’s modified Eagle’s medium, high glucose, pyruvate (DMEM, Gibco, Cat#: 11995065) with 10% fetal bovine serum (FBS), and without antibiotics. Cells were grown under standard cell culture conditions in a humidified incubator at 37 °C, with 5% CO2, and tested negative for mycoplasma.
14.4.2. Cell transfection
On day 1, 1 × 105 HEK293T cells were seeded in 1 ml of media per well in three 12-well plates. Each condition was run in triplicate. The following day, cells were transfected with 100 ng/μl of wild type vitamin D receptor (VDR-WT) plasmid and FuGENE 6 Transfection Reagent (Promega, Cat#: E2691).
14.4.3. Cell treatment
On day 3 cells were treated with vitamin D3 ± salpn. The stock dilution of vitamin D3 at 10−3 M was prepared in ethanol, and it was further diluted to the final concentration of 10−7 M in cell growth media. The stock dilution of salpn at 10−3 M was prepared in ethanol and diluted to final concentrations of 0.1 μM, 1 μM or 25 μM. As a vehicle control, 40 × dilution of 100% ethanol in cell growth media was used.
14.4.4. RNA extraction, cDNA synthesis and quantitative PCR
RNA was extracted using Direct-zol RNA Miniprep Kit (Zymo Research Cat#: R2052). RNA concentration and purity was measured by NanoDrop (Thermo Scientific). cDNA was synthesized from 1 μg of RNA using TaqMan Reverse Transcription Reagents (Invitrogen, Cat#: N8080234). All targets were amplified for 40 cycles using gene specific TaqMan primer/probe sets (Applied Biosystems, CYP24A1: Hs00167999_m1, 18S: Hs99999901_s1) and TaqMan Fast Universal PCR Master Mix [2x] (Applied Biosystems, Cat#: 4352042) on a StepOnePlus Real-Time PCR System (Applied Biosystems). Relative gene expression quantification was performed using the ΔΔCt method, and expression levels were normalized to 18S rRNA.
14.5. Cell culture and maintenance of HaCaT cells
HaCaT cells, immortalized keratinocytes derived from adult human skin, were maintained in Dulbecco’s Modified Eagle Medium (DMEM, Mediatech) supplemented with 10% Fetal Bovine Serum (FBS, Mediatech) and 1% Penicillin/Streptomycin (Mediatech). Cells were plated in 100 mm dishes and cultured in a humid incubator at 37 °C and 5% CO2. Cells were passaged 1:4 (two population doublings) when they reach 70–80% confluency. Cells were cryopreserved in DMEM freezing media (DMEM media with 20% FBS and 10% DMSO) in liquid N2 tanks.
14.6. Preparation of chemicals
A stock solution (10 mM) of salpn (MW: 282.35 g/mol, Sigma-Aldrich) was prepared using DMSO. The working solution of salpn (1 mM) was prepared using DMEM media supplemented with 10% FBS and 1% Pen/Strep. Salpn was diluted and dissolved in the same final volume of DMSO, with a final concentration of 0.2% DMSO in the culture medium.
14.7. Cell viability assay (XTT assay)
Cells were plated at 5000 cells in 100 μl medium per well in 96-well plates and treated with 100 μl of different concentrations of salpn (0 μM, 0.1 μM, 1 μM, 10 μM, 100 μM and 1000 μM). XTT cytotoxicity colorimetric assays (Biotium) were performed after 72 h. XTT reagent was added, and absorbance was measured every hour for 3 h using a Perkin Elmer Victor3 plate reader set at 450 nm to obtain a slope of A450/min. Each plate included wells with salpn-treated or untreated cells in triplicate, along with six replicates each of increasing numbers of cells in medium to generate a standard curve of A450/min vs. cell number, which was then used to generate drug dose-response survival curves. Data shown represent the mean ± SD of each set of triplicate salpn-treated or untreated cells from a representative experiment.
14.8. Annexin/PI staining assay
HaCaT cells were plated at 300,000 cells per well in a 6-well plate for 24 h, and incubated with increasing concentrations of either salpn or DMSO alone for 72 h. Both live and dead cells were collected by centrifugation at 2000 rpm, 4 °C for 5 min. The cell pellets were then resuspended in PBS, washed twice, and resuspended in 100 μl working solution (10 μl of 10 × Binding Buffer, 7 μl AnnexinV-FITC dye, 5 μl PI dye (BioLegend), and 78 μl dH2O). Samples were stained with the Annexin V working solution to distinguish cells that were alive (AnnexinV-; PI-), undergoing early primary apoptosis (AnnexinV+; PI-), and those undergoing late apoptosis/secondary necrosis (AnnexinV+; PI+). The percentage of cells in early or late apoptosis was measured by fluorescence performed on a Cellometer Vision cytofluorometer (Nexcelom). Data shown in each figure represent the mean ± SD of each set of triplicate salpn-treated or untreated cells from a representative experiment.
14.9. Caspase-3 activity assay
Cells per plated at 500,000 cells per well in 6-well plates, and exposed to different concentrations of salpn. After 72 h, both live and detached dead cells were collected, resuspended in lysis buffer, and lysed using a –freeze-thaw method. Lysates were centrifuged at 5000 rpm, 4 °C for 5 min and supernatants containing cytosolic extracts were collected. Protein concentrations of supernatants were measured using a DC/Bradford assay (BioRad) to ensure equal protein is added to each well of a 96 well plate. Caspase-3 assays were performed to measure free 7-amino-4-methylcoumarin (AMC) using 100 μl reactions containing caspase-3 fluorogenic substrate Ac-DEVD-AMC (Invitrogen). Free AMC fluorescence was measured at excitation/emission of 342 nm/ 441 nm with a Wallac Victor3 fluorometer (Perkin-Elmer) in a kinetic assay measured at 3 min intervals for 30 min. Linear regions of slopes were used to determine and compare caspase-3 activities. Data shown in the figure represent the mean ± SD of each set of triplicate salpn-treated or untreated cells from a representative experiment.
13. Zebrafish toxicity assay
We used a morphological phenotypic criterion test to determine the EC50, Lowest Observed Effective Concentration (LOEC), and No Observed Effective Concentration (NOEC) for salpn. Toxicity, as defined here, was measured as the dose resulting in significant developmental abnormalities, degree of deformity, morphological changes, and survival of the fish embryos with 0.1% DMSO and Fish Water (0.3 g/L sea salt) as controls.
13.1. Zebrafish Wnt/β-catenin signaling assay
For the Wnt signaling assays, Tg(TOP:GFP) Wnt reporter fish were crossed with EK wild-type fish (Dorsky et al., 2002). Embryos were fixed at 2 dpf in 4% paraformaldehyde, overnight at 4 °C. Following fixation, the embryos were washed 2 × in PTw (PBS plus 0.1% Tween 20), and then 2 × in 100% methanol. Embryos were then stored at − 20 °C in methanol. Wnt reporter activity was visualized using whole-mount in situ hybridization (WISH) for GFP expression.
13.2. Zebrafish WISH assay
Whole-mount In Situ Hybridization (WISH) was performed as we previously described, with slight modifications (Eaton et al., 2007). In brief, embryos were incubated in 2% hydrogen peroxide in 100% methanol for 20 min at room temperature (RT), rehydrated by stepped 5 min washes in 75%, 50%, and 25% MeOH:PTw, and twice in PTw (1 × PBS, 0.1% Tween-20). Larvae was bleached to remove pigment in 0.5 ml freshly prepared 3% hydrogen peroxide, 1% KOH, and then washed twice in PTw. To improve probe penetration, the 84 hpf larvae are partially digested in 50 ug/ml Proteinase K for 17 min at RT. The 120 hpf larvae was treated with collagenase followed by Proteinase K digestion for 20 min at 37 °C. The digestion was stopped by fixation in 4% paraformaldehyde for 20 min at RT. After five washes in PTw to remove the paraformaldehyde, larvae will be incubated in hybridization solution (50% formamide, 5 × SSC, 50 ug/ml heparin, 5 mg/ml torula RNA, 0.1% Tween-20) for 2 h at 65 °C. Hybridization solution was replaced with 1 ng/ml antisense DIG-labeled riboprobe in hybridization solution and incubated at 65 °C over-night. Unbound probe was removed by 5 min washes at 65 °C in: 50% formamide, 2 × SSC, 0.1% Tween 20; 25% formamide, 2 × SSC, 0.1% Tween 20; and 2 × SSC, 0.1% Tween 20, followed by two 10 min high stringency washes in 0.2 × SSC, 0.1% Tween 20. Larvae were then incubated in blocking buffer (10% Roche blocking buffer, 5% sheep serum in 1 × PTw) for 30 min at RT, followed by incubation in 1:5000 dilution of anti-DIG-AP antibodies in 5% sheep serum in 1 × PTw for 2 h at RT. Unbound antibody was removed by six 10 min washes in PTw. Buffer will then be exchanged by two 5 min washes in NTMT (100 mM NaCl, 50 mM MgCl2, 100 mM Tris pH 9.0, 0.1% Tween-20 in dH20). Larvae were then stained in a 1:5 BM purple:NTMT solution at 37 °C for 4–10 h. Following signal development, larvae were washed twice in PTw, post-fixed for 20 min in 4% paraformaldehyde, washed twice again in PTw, and then cleared in 30%, 50%, 70%, 100% glycerol in PBS. Stained larvae were stored in 100% glycerol at 4 °C until imaging.
13.3. Zebrafish apoptosis assay – TUNEL TMR assays
TUNEL was performed using the in-situ Cell Death Detection Kit- TMR (Roche #12156792910, Sigma-Aldrich) using the following procedure. Embryos were incubated in ice-cold acetone for 20 min at − 20 °C, followed by rehydration through stepped 5 min washes in 75%, 50%, and 25% MeOH:PTw, and then twice in PTw (1 × PBS, 0.1% Tween-20). To improve enzyme penetration, the embryos were partially digested in 50 ug/ml Proteinase K for 17 min at RT. The digestion was stopped by fixation in 4% paraformaldehyde for 20 min at RT, followed by two washes in PTw. The embryos were then washed 3 times in PBS + 1% Triton-X100 for 20 min. After two quick PBS washes, the embryos were incubated for 1 h in a 1:9 Enzyme:Label solution (In Situ Cell Death Detection Kit-TMR). The reaction was stopped by two washes in PTw. Embryos were mounted in 0.8% low melt agarose for imaging. Apoptotic cells in 2 dpf embryos were labeled using the in-situ Cell Death Detection Kit-TMR. Control embryos only show occasional apoptotic cells scattered about the body and tail. Only at the end of the tail are there consistently apoptotic cells.
13.4. PDAC cell cultures
Primary PDAC cultures were established at Georgetown using the primary cell Conditional Reprogramming (CR) methodology previously described (Liu et al., 2012, 2017; Pollock et al., 2014; Ringer et al., 2014; Parasido et al., 2019; Yuan et al., 2012) For nab-pacitaxel sensitivity studies, the cells were carried in conditioned media as described (Pollock et al., 2014; Ringer et al., 2014; Parasido et al., 2019; Palechor-Ceron et al., 2013). Briefly, irradiated J2 feeder cells were plated in 175 cm2 tissue culture flasks (BD Biosciences, Franklin Lakes, NJ) in 30 ml of F media. The media was collected after three days in culture and centrifuged at 1000 g for 5 min at 4 °C to remove cellular debris, followed by filtration using a 0.22 μm Millex-GP filter unit (Millipore, Billerica, MA). For cell culture, three volumes of conditioned F media were mixed with one volume of fresh F medium, and the final working conditioned media was supplemented with 5 μM Y-27632.
13.5. Generation of isogenic nab-paclitaxel-resistant cells
The P1-Parental cells were seeded in 96 well plates in CM. 24 h after seeding, regular CM was replaced with CM containing the concentration of the chemical. CM containing media was replaced every two days. The chemical concentration was increased once a week from , , , . At , only few cells were left in the wells and regular CM without chemical was added in order to allow the resistant cells to proliferate. Once the resistant cells reached confluence, they are passed, banked, and further expanded for analysis. A chemical sensitivity assessment was performed as above to confirm the acquired resistance as previously described (Parasido et al., 2019).
Exome sequence libraries were created using the Illumina TruSeq Exome Library and subjected to 2 × 150 paired end sequencing on an Illumina NextSeq 500. The samples were aligned to the human genome version GRCh38 using the Burrows-Wheeler Alignment (BWA) tool, BWA-MEM and Single nucleotide variants (SNVs) and insertion-deletions (indels) were called using the Genome Analyses Tool Kit v3.5 as described (Parasido et al., 2019). The genetic fidelity of the n-PTX-R cells vs the parental cells was evaluated using the WES data. Jaccard Index (JI) analyses were used to define the similarity/dissimilarity in the data sets as described (Parasido et al., 2019). All JI values of the CRs remained above 0.94, indicating sustained genetic stability despite extended time in culture and selection in nab-Paclitaxel. For STR analyses of human PDAC tissue, the parental PDAC, and the chemical resistant CR cells, DNA was extracted using a Qiagen DNA extraction kit (cat # 69506, Qiagen, Germantown, MD). STR analyses were provided by Genetica DNA Laboratories (Cincinnati, OH). 15 independent genetic sites and amelogenin were to authenticate and verify the CR cells. Final match profiles were evaluated using the Percentage Match Algorithm (ANSI/ATCC ASN-0002–2011) and established the fidelity of the CR cells to the tissue of origin (Parasido et al., 2019).
Supplementary Material
Acknowledgements
The authors wish to acknowledge for the support of Department of Defense Grant CA140882; NIH R01CA129813; NIH CCSG Grant NIH-P30 CA051008; Lombardi Comprehensive Cancer Center, Georgetown University Medical Center; CCSG Grant NIH-P30 CA051008; and the GUMC Computational Chemistry Shared Resources (CCSR).
Footnotes
CRediT authorship contribution statement
Naiem T Issa: Conceptualization, Methodology, Investigation, Formal analysis, Validation, Visualization, Writing – original draft. Henri Wathieu: Conceptualization, Methodology, Data curation, Formal analysis, Validation, Visualization, Writing – original draft, Writing – review & editing. Eric Glasgow: Conceptualization, Investigation, Formal analysis, Validation, Writing – original draft, Writing – review & editing. Ivana Peran: Investigation, Validation, Writing – original draft, Writing – review & editing. Erika Parasido: Investigation, Validation. Tianqi Li: Investigation, Validation. Cynthia M. Simbulan-Rosenthal: Writing – review & editing, Resources. Dean Rosenthal: Writing – review & editing, Supervision. Alexander V Medvedev: Investigation, Validation, Formal analysis. Sergei S Makarov: Writing – review & editing. Christopher Albanese: Writing – review & editing, Supervision, Resources. Stephen W Byers: Writing – review & editing, Supervision, Resources. Sivanesan Dakshanamurthy: Conceptualization, Methodology, Writing – original draft, Supervision, Investigation, Validation, Visualization, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supporting information
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.ecoenv.2022.113330.
References
- Amado NG, Fonseca BF, Cerqueira DM, Neto VM, Abreu JG, 2011. Flavonoids: potential Wnt/beta-catenin signaling modulators in cancer. Life Sci. 89 (15–16), 545–554. [DOI] [PubMed] [Google Scholar]
- Arnold LA, et al. , 2005. Discovery of small molecule inhibitors of the interaction of the thyroid hormone receptor with transcriptional coregulators. J. Biol. Chem 280, 43048–43055. [DOI] [PubMed] [Google Scholar]
- Belorusova AY, Eberhardt J, Potier N, Stote RH, Dejaegere A, Rochel N, 2014. 12. Structural insights into the molecular mechanism of vitamin D receptor activation by lithocholic acid involving a new mode of ligand recognition. J. Med. Chem 57 (11), 4710–4719. 10.1021/jm5002524. Epub 2014 May 21. [DOI] [PubMed] [Google Scholar]
- Ben-Jonathan N, Hugo ER, Brandebourg TD, 2009. Effects of bisphenol A on adipokine release from human adipose tissue: implications for the metabolic syndrome. Mol. Cell. Endocrinol 304 (1–2), 49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bona C, Hooghe R, Cazenave PA, Leguérn C, Paul WE, 1979. Cellular basis of regulation of expression of idiotype. II. Immunity to anti-MOPC-460 idiotype antibodies increases the level of anti-trinitrophenyl antibodies bearing 460 idiotypes. J. Exp. Med 149, 815–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonofiglio D, Gabriele S, Aquila S, Catalano S, Gentile M, Middea E, et al. , 2005. Estrogen receptor alpha binds to peroxisome proliferator-activated receptor response element and negatively interferes with peroxisome proliferator-activated receptor gamma signaling in breast cancer cells. Clin. Cancer Res 11 (17), 6139–6147. [DOI] [PubMed] [Google Scholar]
- Buzon V, Carbo LR, Estruch SB, Fletterick RJ, Estebanez-Perpina E, 2012. A conserved surface on the ligand binding domain of nuclear receptors for allosteric control. Mol. Cell. Endocrinol 348, 394–402. [DOI] [PubMed] [Google Scholar]
- Byers SW, Rowlands T, Beildeck M, Bong YS, 2012. Mechanism of action of vitamin D and the vitamin D receptor in colorectal cancer prevention and treatment. Rev. Endocr. Metab. Disord 13 (1), 31–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capelli D, Cerchia C, Montanari R, Loiodice F, Tortorella P, Laghezza A, Cervoni L, Pochetti G, Lavecchia A, 2016. Structural basis for PPAR partial or full activation revealed by a novel ligand binding mode. Sci. Rep 6, 34792, 10.1038/srep34792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contrera JF, 2011. Improved in silico prediction of carcinogenic potency (TD50) and the risk specific dose (RSD) adjusted Threshold of Toxicological Concern (TTC) for genotoxic chemicals and pharmaceutical impurities. Regul. Toxicol. Pharmacol 59 (1), 133–141. [DOI] [PubMed] [Google Scholar]
- Coogan PF, Rosenberg L, Palmer JR, Strom BL, Zauber AG, Stolley PD, et al. , 2000. Phenolphthalein laxatives and risk of cancer. J. Natl. Cancer Inst 92 (23), 1943–1944. [DOI] [PubMed] [Google Scholar]
- Cronin MT, 2002. The current status and future applicability of quantitative structure-activity relationships (QSARs) in predicting toxicity. Alter. Lab Anim 30 (Suppl. 2), S81–S84. [DOI] [PubMed] [Google Scholar]
- Cronin MTD, Dearden JC, 1995. QSAR in toxicology. 2. Prediction of acute mammalian toxicity and interspecies correlations. Quant. Struct.-Act. Relatsh 14 (2), 117–120. [Google Scholar]
- Dakshanamurthy S, Issa NT, Assefnia S, Seshasayee A, Peters OJ, Madhavan S, et al. , 2012. Predicting new indications for approved drugs using a proteochemometric method. J. Med. Chem 55 (15), 6832–6848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AP, Murphy CG, Johnson R, Lay JM, Lennon-Hopkins K, Saraceni- Richards C, et al. , 2013. The comparative toxicogenomics database: update 2013. Nucleic Acids Res. 41 (Database issue), D1104–D1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delfosse V, et al. , 2015. Synergistic activation of human pregnane X receptor by binary cocktails of pharmaceutical and environmental compounds. Nat. Commun 6, 8089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delfosse V, Grimaldi M, Pons JL, Boulahtouf A, le Maire A, Cavailles V, et al. , 2012. Structural and mechanistic insights into bisphenols action provide guidelines for risk assessment and discovery of bisphenol A substitutes. Proc. Natl. Acad. Sci. USA 109 (37), 14930–14935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dix DJ, Houck KA, Martin MT, Richard AM, Setzer RW, Kavlock RJ, 2007. The ToxCast program for prioritizing toxicity testing of environmental chemicals. Toxicol. Sci 95 (1), 5–12. [DOI] [PubMed] [Google Scholar]
- Dorsky RI, Sheldahl LC, Moon RT, 2002. A transgenic Lef1/beta-catenin-dependent reporter is expressed in spatially restricted domains throughout zebrafish development. Dev. Biol 241 (2), 229–237. [DOI] [PubMed] [Google Scholar]
- Eaton JL, Glasgow E, 2007. Zebrafish orthopedia (otp) is required for isotocin cell development. Dev. Genes Evol 217 (2), 149–158 doi: 10.1007/s00427-006-0123-2. Epub 2006 Dec 19. [DOI] [PubMed] [Google Scholar]
- Ejaredar M, Nyanza EC, Ten Eycke K, Dewey D, 2015. Phthalate exposure and childrens neurodevelopment: a systematic review. Environ. Res 142, 51–60. [DOI] [PubMed] [Google Scholar]
- Estébanez-Perpiñá E, et al. , 2007. A surface on the androgen receptor that allosterically regulates coactivator binding. Proc. Natl. Acad. Sci. USA 104, 16074–16079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estébanez-Perpiñá E, et al. , 2007. Structural insight into the mode of action of a direct inhibitor of coregulator binding to the thyroid hormone receptor. Mol. Endocrinol 21, 2919–2928. [DOI] [PubMed] [Google Scholar]
- Färnegårdh M, et al. , 2003. The three-dimensional structure of the liver X receptor beta reveals a flexible ligand-binding pocket that can accommodate fundamentally different ligands. J. Biol. Chem 278, 38821–38828. [DOI] [PubMed] [Google Scholar]
- Froicu M, Cantorna MT, 2007. Vitamin D and the vitamin D receptor are critical for control of the innate immune response to colonic injury. BMC Immunol. 8, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui K, 1982. Role of frontier orbitals in chemical reactions. Science 218 (4574), 747–754. [DOI] [PubMed] [Google Scholar]
- Goh KI, Cusick ME, Valle D, Childs B, Vidal M, Barabasi AL, 2007. The human disease network. Proc. Natl. Acad. Sci. USA 104 (21), 8685–8690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosdidier S, et al. , 2012. Allosteric conversation in the androgen receptor ligand- binding domain surfaces. Mol. Endocrinol 26, 1078–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honorio KM, Moda TL, Andricopulo AD, 2013. Toxcokinetic properties and in silico ADME modeling in drug discovery. Med Chem. 9 (2), 163–176. [DOI] [PubMed] [Google Scholar]
- Hu P, Chen X, Whitener RJ, Boder ET, Jones JO, Porollo A, et al. , 2013. Effects of parabens on adipocyte differentiation. Toxicol. Sci 131 (1), 56–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes TS, Giri PK, de Vera IMS, Marciano DP, Kuruvilla DS, Shin Y, Blayo A, Kamenecka TM, Burris TP, Griffin PR, Kojetin DJ, 2014. An alternate binding site for synthetic PPARγ ligands. Nat. Commun 5, 3571. 10.1038/ncomms4571 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes TS, Shang J, Brust R, de Vera IMS, Fuhrmann J, Ruiz C, Cameron MD, Kamenecka TM, Kojetin DJ, 2016. Probing the complex binding modes of the PPARγ partial agonist 2-chloro-N-(3-chloro-4-((5-chlorobenzo[d]thiazol-2-yl)thio) phenyl)-4-(trifluoromethyl)benzenesulfonamide (T2384) to orthosteric and allosteric sites with NMR spectroscopy. J. Med. Chem 59 (22), 10335–10341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issa NT, Peters OJ, Byers SW, Dakshanamurthy S, 2015. RepurposeVS: a drug repurposing-focused computational method for accurate drug-target signature predictions. Comb. Chem. High. Throughput Screen 18 (8), 784–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issa NT, Kruger J, Wathieu H, Raja R, Byers SW, Dakshanamurthy S, 2016. DrugGenEx-Net: a novel computational platform for systems pharmacology and gene expression-based drug repurposing. BMC Bioinform. 17 (1), 202. 10.1186/s12859-016-1065-y . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen EV, Khan SA, 2004. A two-site model for antiestrogen action. Mech. Ageing Dev 125, 679–682. [DOI] [PubMed] [Google Scholar]
- Judson R, Houck K, Martin M, Knudsen T, Thomas RS, Sipes N, et al. , 2014. In-vitro and modelling approaches to risk assessment from the U.S. Environmental Protection Agency ToxCast programme. Basic Clin. Pharmacol. Toxicol 115 (1), 69–76. [DOI] [PubMed] [Google Scholar]
- Kim SH, Park MJ, 2014. Phthalate exposure and childhood obesity. Ann. Pediatr. Endocrinol. Metab 19 (2), 69–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleensang A, Maertens A, Rosenberg M, Fitzpatrick S, Lamb J, Auerbach S, et al. , 2014. Pathways of toxicity. ALTEX 31 (1), 53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstreuer NC, Yang J, Berg EL, Knudsen TB, Richard AM, Martin MT, et al. , 2014. Phenotypic screening of the ToxCast chemical library to classify toxic and therapeutic mechanisms. Nat. Biotechnol 32 (6), 583–591. [DOI] [PubMed] [Google Scholar]
- Kojetin DJ, Burris TP, Jensen EV, Khan SA Implications of the binding of tamoxifen to the coactivator recognition site. [DOI] [PubMed]
- Kolatorova L, Sramkova M, Vitku J, Vcelak J, Lischkova O, Starka L, et al. , 2018. Parabens and their relation to obesity. Physiol. Res 67 (Suppl. 3), S465–S472. [DOI] [PubMed] [Google Scholar]
- Kwintkiewicz J, Nishi Y, Yanase T, Giudice LC, 2010. Peroxisome proliferator-activated receptor-gamma mediates bisphenol A inhibition of FSH-stimulated IGF-1, aromatase, and estradiol in human granulosa cells. Environ. Health Perspect 118 (3), 400–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laghezza A, Piemontese L, Cerchia C, Montanari R, Capelli D, Giudici M, Crestani M, Tortorella P, Peiretti F, Pochetti G, Lavecchia A, Loiodice F, 2018. Identification of the first PPARα/γ dual agonist able to bind to canonical and alternative sites of PPARγ and to inhibit its Cdk5-mediated phosphorylation. J. Med. Chem 61 (18), 8282–8298. 10.1021/acs.jmedchem.8b00835. Epub 2018 Sep 18. [DOI] [PubMed] [Google Scholar]
- Lai YH, Fang TC, 2013. The pleiotropic effect of vitamin d. ISRN Nephrol. 2013, 898125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert J, Dorr R, Timmermann B, 2004. Nordihydroguaiaretic acid: a review of its numerous and varied biological activities. Pharmaceut. Biol 42 (2), 149–158. [Google Scholar]
- Laragione T, Shah A, Gulko PS, 2012. The vitamin D receptor regulates rheumatoid arthritis synovial fibroblast invasion and morphology. Mol. Med 18, 194–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind PM, Lind L, 2011. Circulating levels of bisphenol A and phthalates are related to carotid atherosclerosis in the elderly. Atherosclerosis 218 (1), 207–213. [DOI] [PubMed] [Google Scholar]
- Liu X, Ory V, Chapman S, Yuan H, Albanese C, Kallakury B, et al. , 2012. ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. Am. J. Pathol 180, 590–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Krawczyk E, Suprynowicz FA, Palechor-Ceron N, Yuan H, Dakic A, et al. , 2017. Conditional reprogramming and long-term expansion of normal and tumor cells from human biospecimens. Nat. Protoc 12, 439–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malloy EJ, Miller KL, Eisen EA, 2007. Rectal cancer and exposure to metalworking fluids in the automobile manufacturing industry. Occup. Environ. Med 64 (4), 244–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McRobb FM, Sahagun V, Kufareva I, Abagyan R, 2014. In silico analysis of the conservation of human toxicity and endocrine disruption targets in aquatic species. Environ. Sci. Technol 48 (3), 1964–1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medvedev A, Moeser M, Medvedeva L, Martsen E, Granick A, Raines L, Zeng M, Makarov S Jr, Houck KA, Makarov SS, 2018. Evaluating biological activity of compounds by transcription factor activity profiling. Sci. Adv 4 (9), eaar4666 doi: 10.1126/sciadv.aar4666. eCollection 2018 Sep. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlman MA, 1992. Dangerous and cancer-causing properties of products and chemicals in the oil refining and petrochemical industry. VIII. Health effects of motor fuels: carcinogenicity of gasoline–scientific update. Environ. Res 59 (1), 238–249. [DOI] [PubMed] [Google Scholar]
- Meijer Femke A., Leijten-van de Gevel Iris A., de Vries Rens M.J.M., Luc Brunsveld, 2019. Allosteric small molecule modulators of nuclear receptors. Mol. Cell. Endocrinol 485, 20–34. [DOI] [PubMed] [Google Scholar]
- Menegaz D, et al. , 2011. Vitamin D receptor (VDR) regulation of voltage-gated chloride channels by ligands preferring a VDR-alternative pocket (VDR-AP). Mol. Endocrinol 25, 1289–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing E, Gee JR, Saltzstein DR, Kim K, diSant’Agnese A, Kolesar J, et al. , 2012. A phase 2 cancer chemoprevention biomarker trial of isoflavone G-2535 (genistein) in presurgical bladder cancer patients. Cancer Prev. Res 5 (4), 621–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizwicki MT, et al. , 2004. Identification of an alternative ligand-binding pocket in the nuclear vitamin D receptor and its functional importance in 1alpha,25 (OH)2-vitamin D3 signaling. Proc. Natl. Acad. Sci. USA 101, 12876–12881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizwicki MT, Norman AW, 2009. The vitamin D sterol-vitamin D receptor ensemble model offers unique insights into both genomic and rapid-response signaling. Sci. Signal 2, re4–57. [DOI] [PubMed] [Google Scholar]
- Mizwicki MT, Bula CM, Bishop JE, Norman AW, 2005. A perspective on how the Vitamin D sterol/Vitamin D receptor (VDR) conformational ensemble model can potentially be used to understand the structure-function results of A-ring modified Vitamin D sterols. J. Steroid Biochem. Mol. Biol 97, 69–82. [DOI] [PubMed] [Google Scholar]
- Mizwicki MT, Bula CM, Bishop JE, Norman AW, 2007. New insights into Vitamin D sterol-VDR proteolysis, allostery, structure-function from the perspective of a conformational ensemble model. J. Steroid Biochem. Mol. Biol 103, 243–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modi S, Hughes M, Garrow A, White A, 2012. The value of in silico chemistry in the safety assessment of chemicals in the consumer goods and pharmaceutical industries. Drug Discov. Today 17 (3–4), 135–142. [DOI] [PubMed] [Google Scholar]
- Moore TW, Mayne CG, Katzenellenbogen JA, 2010. Minireview: Not picking pockets: nuclear receptor alternate-site modulators (NRAMs). Mol. Endocrinol 24, 683–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosure SA, Bass J, Munoz-Tello P, Lin H, Hughes TS, Tang M, Ge Q, Kamekencka TM, Kojetin DJ, 2018. Cooperative cobinding of synthetic and natural ligands to the nuclear receptor PPARγ. Elife 7. 10.7554/eLife.43320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson SR, Zhang C, Roche S, et al. , 2020. Modelling of pancreatic cancer biology: transcriptomic signature for 3D PDX-derived organoids and primary cell line organoid development. Sci. Rep 10, 2778. 10.1038/s41598-020-59368-7 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- NTP 12th Report on Carcinogens, 2011. Rep Carcinog. vol. 12, iii-499. [PubMed]
- Palechor-Ceron N, Suprynowicz FA, Upadhyay G, Dakic A, Minas T, Simic V, et al. , 2013. Radiation induces diffusible feeder cell factor(s) that cooperate with ROCK inhibitor to conditionally reprogram and immortalize epithelial cells. Am. J. Pathol 183, 1862–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parasido E, Avetian GS, Naeem A, Graham G, Pishvaian M, Glasgow E, et al. , 2019. The sustained induction of c-MYC drives nab-paclitaxel resistance in primary pancreatic ductal carcinoma cells. Mol. Cancer Res 17, 1815–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parasido E, Avetian GS, Naeem A, Graham G, Pishvaian M, Glasgow E, Mudambi S, Lee Y, Ihemelandu C, Choudhry M, Peran I, Banerjee PP, Avantaggiati ML, Bryant K, Baldelli E, Pierobon M, Liotta L, Petricoin E, Fricke ST, Sebastian A, Cozzitorto J, Loots GG, Kumar D, Byers S, Londin E, DiFeo A, Narla G, Winter J, Brody JR, Rodriguez O, Albanese C, 2019. The sustained induction of c-MYC drives nab-paclitaxel resistance in primary pancreatic ductal carcinoma cells. Mol. Cancer Res 17 (9), 1815–1827. 10.1158/1541-7786.MCR-19-0191 . Epub 2019 Jun 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parr RG, Pearson RG, 1983. Absolute hardness: companion parameter to absolute electronegativity. J. Am. Chem. Soc 105 (26), 7512–7516. [Google Scholar]
- Parr RG, Pearson RG, 1983. Absolute hardness: companion parameter to absolute electronegativity. J. Am. Chem. Soc 105 (26), 7512–7516. [Google Scholar]
- Parr RG, Yang W, 1984. Density functional approach to the frontier-electron theory of chemical reactivity. J. Am. Chem. Soc 106 (14), 4049–4050. [Google Scholar]
- Parr RG, Donnelly RA, Levy M, Palke WE, 1978. Electronegativity: the density functional viewpoint. J. Chem. Phys 68 (8), 3801–3807. [Google Scholar]
- Parr RG, Szentpály Lv, Liu S, 1999. Electrophilicity index. J. Am. Chem. Soc 121 (9), 1922–1924. [Google Scholar]
- de Pascual-Teresa S, Moreno DA, Garcia-Viguera C, 2010. Flavanols and anthocyanins in cardiovascular health: a review of current evidence. Int. J. Mol. Sci 11 (4), 1679–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson R, 1997. Chemical Hardness. Wiley-VCH. [Google Scholar]
- Pollock CB, McDonough S, Wang VS, Lee H, Ringer L, Li X, et al. , 2014. Strigolactone analogues induce apoptosis through activation of p38 and the stress response pathway in cancer cell lines and in conditionally reprogramed primary prostate cancer cells. Oncotarget 5, 1683–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raunio H, 2011. In silico toxicology - non-testing methods. Front. Pharmacol 2, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringer L, Sirajuddin P, Tricoli L, Waye S, Parasido E, Lee RJ, et al. , 2014. The induction of the p53 tumor suppressor protein bridges the apoptotic and autophagic signaling pathways to regulate cell death in prostate cancer cells. Oncotarget 5, 10678–10691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts EM, English PB, Grether JK, Windham GC, Somberg L, Wolff C, 2007. Maternal residence near agricultural pesticide applications and autism spectrum disorders among children in the California Central Valley. Environ. Health Perspect 115 (10), 1482–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanov S, Medvedev A, Gambarian M, Poltoratskaya N, Moeser M, Medvedeva L, Gambarian M, Diatchenko L, Makarov S, 2008. Homogeneous reporter system enables quantitative functional assessment of multiple transcription factors. Nat. Methods 5 (3), 253–260 doi: 10.1038/nmeth.1186. Epub 2008 Feb 24. [DOI] [PubMed] [Google Scholar]
- Rotroff DM, Martin MT, Dix DJ, Filer DL, Houck KA, Knudsen TB, et al. , 2014. Predictive endocrine testing in the 21st century using in-vitro assays of estrogen receptor signaling responses. Environ. Sci. Technol 48 (15), 8706–8716. [DOI] [PubMed] [Google Scholar]
- Sadana P, et al. , 2011. Similarities and differences between two modes of antagonism of the thyroid hormone receptor. ACS Chem. Biol 6, 1096–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu SC, Ruggles DI, O’Donnell MW, 2006. Prooxidant activity and toxicity of nordihydroguaiaretic acid in clone-9 rat hepatocyte cultures. Food Chem. Toxicol 44 (10), 1751–1757. [DOI] [PubMed] [Google Scholar]
- Schrödinger Release 2013–3: Jaguar, version 80, 2013. Schrödinger, LLC, New York, NY. ¨ [Google Scholar]
- Schultz TW, Seward JR, 2000. Health-effects related structure-toxicity relationships: a paradigm for the first decade of the new millennium. Sci. Total Environ 249 (1–3), 73–84. [DOI] [PubMed] [Google Scholar]
- Shankar A, Teppala S, 2011. Relationship between urinary bisphenol A levels and diabetes mellitus. J. Clin. Endocrinol. Metab 96 (12), 3822–3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivanesan D, Amutha R, Subramanian V, Nair BU, Ramasami T, 1999. Assessment of importance of solvent in the calculation of condensed Fukui function: a self-consistent reaction field calculation study. Chem. Phys. Lett 308, 223–228. [Google Scholar]
- Skafidas E, Testa R, Zantomio D, Chana G, Everall IP, Pantelis C, 2014. Predicting the diagnosis of autism spectrum disorder using gene pathway analysis. Mol. Psychiatry 19 (4), 504–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small-Molecule Drug Discovery Suite 2013–3: QikProp, version 38, 2013. Schrodinger, ¨ LLC, New York, NY. [Google Scholar]
- Spence SJ, Schneider MT, 2009. The role of epilepsy and epileptiform EEGs in autism spectrum disorders. Pediatr. Res 65 (6), 599–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squadrito F, Marini H, Bitto A, Altavilla D, Polito F, Adamo EB, et al. , 2013. Genistein in the metabolic syndrome: results of a randomized clinical trial. J. Clin. Endocrinol. Metab 98 (8), 3366–3374. [DOI] [PubMed] [Google Scholar]
- Subramanian V, Sivanesan D, Amutha R, Padmanabhan J, Ramasami T, 1998. Hardness profile of interaction of H–F with He, Ne and Ar: a density functional and MP2 calculation. Chem. Phys. Lett 294 (4), 285–291. [Google Scholar]
- Sverdrup B, Källberg H, Bengtsson C, Lundberg I, Padyukov L, Alfredsson L, et al. , 2005. Association between occupational exposure to mineral oil and rheumatoid arthritis: results from the Swedish EIRA case–control study. Arthritis Res. Ther 7 (6), R1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swedenborg E, Ruegg J, Makela S, Pongratz I, 2009. Endocrine disruptive chemicals: mechanisms of action and involvement in metabolic disorders. J. Mol. Endocrinol 43 (1), 1–10. [DOI] [PubMed] [Google Scholar]
- Tanenbaum DM, et al. , 1998. Crystallographic comparison of the estrogen and progesterone receptor’s ligand binding domains. Proc. Natl. Acad. Sci. USA 95, 5998–6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teske K, Nandhikonda P, Bogart JW, Feleke B, Sidhu P, Yuan N, et al. , 2014. Identification of VDR antagonists among nuclear receptor ligands using virtual screening. Nucl. Recept. Res 1, 101076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thayer KA, Heindel JJ, Bucher JR, Gallo MA, 2012. Role of environmental chemicals in diabetes and obesity: a national toxicology program workshop review. Environ. Health Perspect 120 (6), 779–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremezaygues L, Reichrath J, 2011. Vitamin D analogs in the treatment of psoriasis: where are we standing and where will we be going? Dermatoendocrinology 3 (3), 180–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsakovska I, Lessigiarska I, Netzeva T, Worth AP, 2008. A mini review of mammalian toxicity (Q)SAR models. QSAR Comb. Sci 27 (1), 41–48. [Google Scholar]
- Valerio LG Jr, 2011. In silico toxicology models and databases as FDA critical path initiative toolkits. Hum. Genom 5 (3), 200–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wambaugh JF, Setzer RW, Reif DM, Gangwal S, Mitchell-Blackwood J, Arnot JA, et al. , 2013. High-throughput models for exposure-based chemical prioritization in the ExpoCast project. Environ. Sci. Technol 47 (15), 8479–8488. [DOI] [PubMed] [Google Scholar]
- Wang Y, et al. , 2006. A second binding site for hydroxytamoxifen within the coactivator-binding groove of estrogen receptor β. Proc. Natl. Acad. Sci. USA 103, 9908–9911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wathieu H, Issa NT, Fernandez AI, Mohandoss M, Tiek DM, Franke JL, Byers SW, Riggins RB, Dakshanamurthy S, 2017. Differential prioritization of therapies to subtypes of triple negative breast cancer using a systems medicine method. Oncotarget 8 (54), 92926–92942. 10.18632/oncotarget.21669 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westmark CJ, 2014. Soy infant formula and seizures in children with autism: a retrospective study. PLoS One 9 (3), e80488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westmark CJ, Westmark PR, Malter JS, 2013. Soy-based diet exacerbates seizures in mouse models of neurological disease. J. Alzheimers Dis 33 (3), 797–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf R, Movshowitz M, Brenner S, 1996. Supplemental tests in the evaluation of occupational hand dermatitis in soldiers. Int. J. Dermatol 35 (3), 173–176. [DOI] [PubMed] [Google Scholar]
- Yuan H, Myers S, Wang J, Zhou D, Woo JA, Kallakury B, et al. , 2012. Use of reprogrammed cells to identify therapy for respiratory papillomatosis. N. Engl. J. Med 367, 1220–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Krishnadasan A, Kennedy N, Morgenstern H, Ritz B, 2005. Estimated effects of solvents and mineral oils on cancer incidence and mortality in a cohort of aerospace workers. Am. J. Ind. Med 48 (4), 249–258. [DOI] [PubMed] [Google Scholar]
Further reading
- Schrödinger Release 2013–3: LigPrep, version 28, 2013. Schrödinger, LLC, New York, NY. ¨ [Google Scholar]
- Schrödinger Release 2013–3: Protein Preparation Wizard, 2013; Epik, Schrödinger, LLC, New York, NY, 2013; Impact, Schrödinger, LLC, NewYork, NY, 2013; Prime, Schrödinger, LLC, New York, NY. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.