Abstract
Introduction
Despite the widespread use of electronic cigarettes, the long-term health consequences of vaping are largely unknown.
Aims and Methods
We investigated the DNA-damaging effects of vaping as compared to smoking in healthy adults, including “exclusive” vapers (never smokers), cigarette smokers only, and nonusers, matched for age, gender, and race (N = 72). Following biochemical verification of vaping or smoking status, we quantified DNA damage in oral epithelial cells of our study subjects, using a long-amplicon quantitative polymerase chain reaction assay.
Results
We detected significantly increased levels of DNA damage in both vapers and smokers as compared to nonusers (p = .005 and p = .020, respectively). While the mean levels of DNA damage did not differ significantly between vapers and smokers (p = .522), damage levels increased dose-dependently, from light users to heavy users, in both vapers and smokers as compared to nonusers. Among vapers, pod users followed by mod users, and those who used sweet-, mint or menthol-, and fruit-flavored e-liquids, respectively, showed the highest levels of DNA damage. The nicotine content of e-liquid was not a predictor of DNA damage in vapers.
Conclusions
This is the first demonstration of a dose-dependent formation of DNA damage in vapers who had never smoked cigarettes. Our data support a role for product characteristics, specifically device type and e-liquid flavor, in the induction of DNA damage in vapers. Given the popularity of pod and mod devices and the preferability of sweet-, mint or menthol-, and fruit-flavored e-liquids by both adult- and youth vapers, our findings can have significant implications for public health and tobacco products regulation.
Implications
We demonstrate a dose-dependent formation of DNA damage in oral cells from vapers who had never smoked tobacco cigarettes as well as exclusive cigarette smokers. Device type and e-liquid flavor determine the extent of DNA damage detected in vapers. Users of pod devices followed by mod users, and those who use sweet-, mint or menthol-, and fruit-flavored e-liquids, respectively, show the highest levels of DNA damage when compared to nonusers. Given the popularity of pod and mod devices and the preferability of these same flavors of e-liquid by both adult- and youth vapers, our findings can have significant implications for public health and tobacco products regulation.
Introduction
Electronic cigarette (e-cig) use, otherwise known as “vaping,” is highly popular among adolescent never smokers and adult smokers seeking a less-harmful alternative to tobacco cigarettes.1–3 E-cigs are handheld battery-powered devices, which exploit the “heating” of a liquid to produce vapor for inhalation.4,5 The liquid also called “e-liquid or e-juice,” is a mixture of propylene glycol (PG), glycerin or vegetable glycerin (VG), flavors, and varying concentrations of nicotine, although nicotine-free e-liquid is also available.6,7 Chemical analysis has shown that many of the same toxicants and carcinogens present in cigarette smoke are also found in e-cig vapor, albeit mostly at substantially lower levels.5–7 E-cig vapor also contains chemicals that are not detected in cigarette smoke.8,9 The latter compounds likely arise from the mixing of the e-liquid ingredients or vaporization of humectants (PG or VG), flavorings, or chemicals leached from the device components.10,11 To date, however, the long-term health consequences of vaping are largely unknown.12,13
Many toxic and carcinogenic chemicals present in e-cig vapor or cigarette smoke exert their biological effects through the induction of DNA damage leading to mutagenesis and genome instability.5,14 DNA damage has been implicated in a wide variety of tobacco-related diseases, including cancer in multiple organ sites.15 Quantification of DNA damage in cells and tissues of e-cig users versus cigarette smokers can help determine the genotoxic potential of vaping relative to smoking. In the present study, we have compared the DNA-damaging effects of vaping to smoking by measuring the level of DNA lesions in oral cells of e-cig users and cigarette smokers as compared to nonusers. The study population consisted of healthy adult “exclusive” vapers (never smokers), cigarette smokers only, and nonusers of any tobacco products, matched for age, gender, and race. We have used the extensively validated and highly sensitive long-amplicon quantitative polymerase chain reaction (LA-QPCR) assay16 to quantify DNA damage in oral epithelial cells collected by brushing from our study subjects. Of significance, the oral epithelium is a major target for cancer and other diseases associated with tobacco product use.17 Moreover, we have investigated the influence of use frequency and duration (ie, vaping or smoking dose) on the induction of DNA damage in e-cig users and cigarette smokers. Among e-cig users, we have also determined the impact of product characteristics, including device type and e-liquid flavor and nicotine content, on the extent of DNA damage detected. In addition, we have biochemically verified the vaping or smoking status of the study subjects by measuring plasma cotinine, a major metabolite of nicotine, exhaled carbon monoxide (CO), and carboxyhemoglobin (COHb) levels.6
Methods
Ethics Declarations
The study was approved by the Health Sciences Institutional Review Board (IRB) of the University of Southern California (Protocol No: HS-16-00175). Written informed consent was obtained from participants prior to inclusion in the study. All research was performed in accordance with the approved IRB protocol and relevant guidelines & regulations, including the Declaration of Helsinki.
Study Population
Eligible candidates for the study included healthy adults—both males and females of diverse ages, races, and ethnicities—who could read and write in English and understand and give informed consent. The catchment area for this study was the Greater Los Angeles Area. The study population consisted of 72 subjects divided equally into three groups, including group 1: Current exclusive vapers (never smokers); group 2: Current exclusive smokers; and group 3: Nonsmokers non vapers (nonusers). Detailed characteristics of the study population are listed in Table 1. Dual users of e-cigs and combustible cigarettes, poly users of e-cigs, cigarettes, or other tobacco products, and former smokers or vapers were excluded from the study. Criteria for classification of the study subjects as vapers, cigarette smokers, or nonusers, consistent with national surveys,18 were as follows: Vapers were those who reported current use of e-cigs for at least 3 times a week for a minimum of 6 months, and no use of combustible cigarettes or any other tobacco products in their lifetime. Smokers were those who reported currently smoking tobacco cigarettes at least 3 times per week for a minimum of 1 year, no or less than five vaping sessions in their lifetime, and no use of any other tobacco products (except for combustible cigarettes) in the past 6 months. Nonusers were those who reported no use of any tobacco product (e-cigs or tobacco cigarettes) more than 5 times in their life; nonusers reported smoking no or fewer than 100 cigarettes or having no or less than five vaping sessions in their lifetime (no vaping or smoking in the past 6 months). We note that participants in this study had equal opportunity to self-identify as former smokers or vapers and participate in other existing studies in our laboratory. This is important in view of the fact that all of our study participants underwent stringent screening and comprehensive personal interviews complemented with biochemical verification of their vaping or smoking status. Altogether, our enrollment strategy, inclusion and exclusion criteria, and verification analysis have ensured reliable and accurate classification of the study subjects in this report. We note that, unlike combustible cigarettes that have been on the market for many years, e-cigs are a relatively new tobacco product.5–7 Thus, we set the minimum use criteria for vapers and smokers to 6 months and 1 year, respectively, to allow enrollment of a sufficient number of subjects into this study. More detailed information about subject recruitment and enrollment, inclusion and exclusion criteria, and sample collection and processing are provided in 17,19 and Supplementary Materials.
Table 1.
Characteristics of the Study Population
| Vapers (N = 24) |
Smokers (N = 24) |
Nonusers (N = 24) |
||
|---|---|---|---|---|
| Age* | 24.3 ± 0.8 | 26.0 ± 0.7 | 25.3 ± 0.6 | |
| Gender† | Male | 20 (83.3%) | 20 (83.3%) | 20 (83.3%) |
| Female | 4 (16.7%) | 4 (16.7%) | 4 (16.7%) | |
| Race† | White | 7 (29.2%) | 7 (29.2%) | 7 (29.2%) |
| Hispanic | 5 (20.8%) | 5 (20.8%) | 5 (20.8%) | |
| African American | 2 (8.3%) | 2 (8.3%) | 2 (8.3%) | |
| Asian | 8 (33.3%) | 8 (33.3%) | 8 (33.3%) | |
| Other‡ | 2 (8.3%) | 2 (8.3%) | 2 (8.3%) | |
| Marital status† | Single and never married | 20 (83.3%) | 19 (79.2%) | 21 (87.5%) |
| Married | 2 (8.3%) | 2 (8.3%) | 1 (4.2%) | |
| Currently living with someone | 2 (8.3%) | 1 (4.2%) | 2 (8.3%) | |
| Widowed | 0 (0%) | 0 (0%) | 0 (0%) | |
| Separated | 0 (0%) | 0 (0%) | 0 (0%) | |
| Divorced | 0 (0%) | 2 (8.3%) | 0 (0%) | |
| Education† | Less than high school | 0 (0%) | 0 (0%) | 0 (0%) |
| High school diploma or GED | 5 (20.8%) | 3 (12.5%) | 0 (0%) | |
| Some college completed or currently enrolled in college | 13 (54.2%) | 6 (25.0%) | 3 (12.5%) | |
| College degree or higher | 6 (25.0%) | 15 (62.5%) | 21 (87.5%) | |
| Employment status† | Full time | 13 (54.2%) | 15 (62.5%) | 14 (58.3%) |
| Part time | 9 (37.5%) | 7 (29.2%) | 7 (29.2%) | |
| Retired or disability | 1 (4.2%) | 0 (0%) | 0 (0%) | |
| Unemployed | 1 (4.2%) | 2 (8.3%) | 3 (12.5%) | |
| Pretax annual income† | <$15 000 | 1 (4.2%) | 7 (29.2%) | 7 (29.2%) |
| ≥$15 000–<$30 000 | 4 (16.7%) | 4 (16.7%) | 6 (25.0%) | |
| ≥$30 000–<$45 000 | 5 (20.8%) | 6 (25.0%) | 1 (4.2%) | |
| ≥$45 000–<$60 000 | 4 (16.7%) | 1 (4.2%) | 4 (16.7%) | |
| ≥$60 000–<$75 000 | 1 (4.2%) | 5 (20.8%) | 1 (4.2%) | |
| ≥$75 000–<$90 000 | 1 (4.2%) | 0 (0%) | 1 (4.2%) | |
| ≥$90 000–<$105 000 | 1 (4.2%) | 0 (0%) | 2 (8.3%) | |
| ≥$105 000–<$120 000 | 2 (8.3%) | 0 (0%) | 0 (0%) | |
| ≥$120 000 | 5 (20.8%) | 1 (4.2%) | 2 (8.3%) | |
| BMI*,§ | 26.6 ± 1.2 | 26.9 ± 1.1 | 23.9 ± 1.0 | |
| Years smoked* | NA | 7.3 ± 1.1 | NA | |
| Pack year*,¶ | NA | 3.1 ± 0.6 | NA | |
| Years vaped* | 2.9 ± 0.4 | NA | NA | |
| Cumulative e-liquid (ml)*,# | 5,780.1 ± 2,017.5 | NA | NA | |
| E-cig device type† | Cigalike | 1 (4.2%) | NA | NA |
| Mod | 11 (45.8%) | NA | NA | |
| Pod | 8 (33.3%) | NA | NA | |
| Multiple | 4 (16.7%) | NA | NA | |
| E-liquid flavor† | Fruit | 7 (29.2%) | NA | NA |
| Sweet | 3 (12.5%) | NA | NA | |
| Mint or Menthol | 5 (20.8%) | NA | NA | |
| Tobacco | 1 (4.2%) | NA | NA | |
| Multiple | 8 (33.3%) | NA | NA | |
| Plasma cotinine (ng/ml)* | 84.9 ± 13.1** | 76.7 ± 8.6** | 2.6 ± 0.1 | |
| Breath CO (ppm)* | 2.0 ± 0.3 | 12.0 ± 1.6†† | 1.9 ± 0.3 | |
| %COHb* | 0.9 ± 0.07 | 2.5 ± 0.2‡‡ | 0.9 ± 0.05 | |
| Vitamin or multi-vitamin use‖ | 7 (29.2%) | 5 (20.8%) | 6 (25.0%) | |
*Results are expressed as Mean ± SE.
†Numbers and percentages (inside brackets) are indicated.
‡Other = Multiracial or Native American.
§BMI = Body mass index [Weight (kg) ÷ Height2(m)].
¶Pack year is calculated by multiplying the number of packs of cigarettes a person smoked per day by the number of years he or she smoked.
#Cumulative e-liquid is calculated as the total volume of e-liquid (in milliliters) vaped by a person during his or her lifetime.
‖Defined as those who used vitamin or multi-vitamin regularly (≥3 times per week in the past year).
Plasma cotinine levels were measured using a solid-phase competitive ELISA (Abnova Corp.) and exhaled breath CO levels and %COHb were quantitated by a Bedfont Micro Smokerlyzer Breath CO monitor (Bedfont Scientific Ltd.) (see, Supplementary Materials).
**Statistically significant as compared to nonusers, p < .0001.
††Statistically significant as compared to nonusers, p = .0005.
‡‡Statistically significant as compared to nonusers, p = .0002.
GED = General Education Development or General Education Diploma, The GED or High School Equivalency Certificate shows that one has a level of knowledge equivalent to a high school graduate, CO = carbon monoxide, ppm = parts per million, COHb = carboxyhemoglobin, NA = Not applicable.
Quantification of DNA Damage by LA-QPCR
LA-QPCR quantification of DNA damage was performed as described in Ref.16 with few modifications. The LA-QPCR analysis interrogated a 12.2 kb region of the DNA polymerase beta (POLB) gene.16,20,21 For validation purposes, we also interrogated an additional gene target, hypoxanthine phosphoribosyltransferase 1 (HPRT). We used the same protocol to amplify a 10.4 kb fragment encompassing exons 2–5 of the HPRT gene.16,20,21 Detailed information about the LA-QPCR assay, a description of the protocol, and quantification of the results are provided in Supplementary Materials.
Cotinine Measurement by ELISA
Plasma cotinine was measured by a solid-phase competitive enzyme-linked immunosorbent assay (ELISA) (Abnova Corp) (Supplementary Materials).
CO and COHb Quantification by Breath Monitor
Exhaled CO levels and %COHb were measured using the Bedfont Micro Smokerlyzer according to the manufacturer’s instructions (Bedfont Scientific Ltd) (Supplementary Materials).
Statistical Analysis
The distribution of data were evaluated by the Shapiro–Wilk test. Results are expressed as mean ± SE. Comparisons of all variables between the two groups were performed by the Student’s t test. Specifically, DNA damage levels between two independent groups, namely vapers and nonusers, smokers and nonusers, or vapers and smokers, were compared by the Student’s t test. To compare variables in three or more groups, we used a one-way Analysis of Variance (ANOVA) followed by a post hoc Tukey HSD test. The multi-group comparison by ANOVA was used to compare damage levels among heavy vapers, light vapers, and nonusers, as well as heavy smokers, light smokers, and nonusers. Similarly, we used this multiple-group comparison to assess DNA damage levels in vapers who used different devices or e-liquids and nonusers. Relationships between different variables were examined by Pearson correlation coefficient analysis. Other statistical tests used are specified in the text. All statistical tests were two-sided. p values <.05 were considered statistically significant. All statistical analyses were performed using the R environment for statistical computing, available at RStudio (https://rstudio.com/), which is a free and open-source software.
Results
DNA Damage Quantification
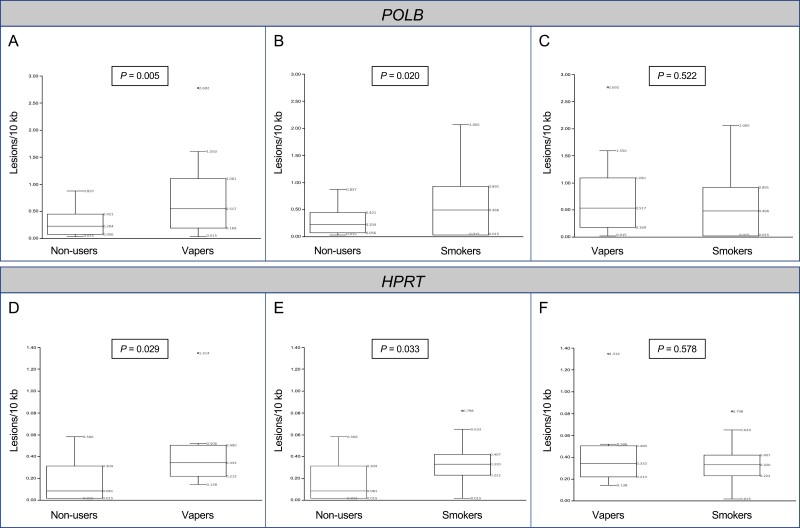
As shown in Figure 1, both vapers and smokers had significantly higher levels of DNA damage in their oral cells as compared to nonusers. There were 2.6- and 2.2-fold increases, respectively, in mean levels of DNA damage in the POLB gene in the oral cells of vapers and smokers as compared to nonusers (p = .005 and p = .020, respectively). The levels of DNA damage in the POLB gene in vapers’ oral cells were not statistically significantly different from those in smokers (p = .522) (Figure 1A–C). To validate these results, we have used a subset of samples from which extra DNA was available (N = 36 total, 12 per group) for LA-QPCR analysis of an independent gene target (HPRT). Similar to LA-QPCR results in the POLB gene, the levels of DNA damage in the HPRT gene, as quantified by LA-QPCR, were significantly higher in both vapers and smokers as compared to nonusers (p = .029 and p = .033, respectively). Furthermore, DNA damage levels in the HPRT gene in vapers were not significantly different from those in smokers (p = .578) (Figure 1D–F). In addition, there was a significant correlation between DNA damage levels in the POLB and HPRT genes in the tested samples (r = 0.647, p < .0001). Given the confirmatory results of LA-QPCR in the HPRT gene, we have performed an in-depth analysis of DNA damage data in the larger samples that were assayed for the POLB gene (N = 72). This was to allow a more meaningful comparison of variables among subgroups of vapers and smokers relative to nonusers. Henceforth, the following sections exclusively present the results and discussion of DNA damage data obtained by LA-QPCR in the POLB gene.
Figure 1.
Comparison of DNA damage levels between vapers and nonusers, smokers and nonusers, and vapers and smokers. DNA damage levels were determined in genomic DNA of oral epithelial cells from healthy adult “exclusive” vapers (never smokers), cigarette smokers only, and nonusers by LA-QPCR, as described in the text. Panels (A–C) show the LA-QPCR results in the POLB gene whereas Panels (D–F) displays the respective results in the HPRT gene. Distribution of data within each group is shown by box and whisker plots whereby “lower” and “upper” edges of the boxes represent the first and third quartiles, respectively, and horizontal lines within the boxes indicate the second quartile. The “lower” and “upper” vertical lines extending from the boxes, also known as the “whiskers,” represent the lowest and highest data points, respectively, in the set (minimum and maximum values, respectively, excluding values outside the whiskers’ range). The five measures of box and whisker plots are all labeled within the graphs. All samples were assayed independently up to 2 times and results were averaged for each sample. DNA damage levels were compared between each two independent groups, as described in the text; p values are indicated for all comparisons.
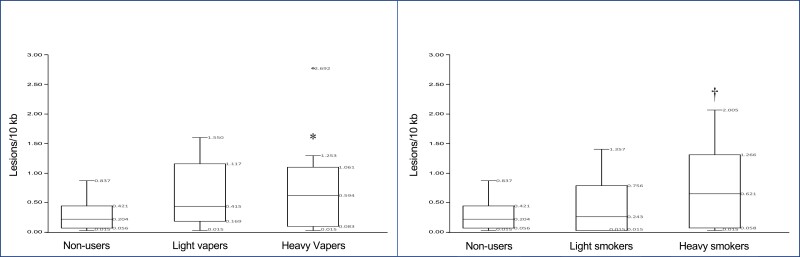
To examine the dose dependency of the induced DNA damage in vapers and smokers, we have further analyzed the DNA damage data (in the POLB gene) using indicators of intensity and duration of e-cig- and cigarette use, expressed as cumulative e-liquid (cum e-liq) and pack year, respectively. Whereas cumulative e-liquid consumption was calculated as the total volume of e-liquid (in milliliters) vaped by a person during his or her lifetime, pack year was computed by multiplying the number of packs of cigarettes a person smoked per day by the number of years he or she smoked.22 Using the second quartile as a cutoff point, we have divided both vapers and smokers into two groups, including “light” and “heavy” users. As illustrated in Figure 2A, levels of DNA damage increased dose-dependently, from light vapers to heavy vapers, as compared to nonusers (F = 4.571, p = .0156 | Tukey’s HSD p = .0195). A similar trend was found in smokers wherein DNA damage levels were increased from light smokers to heavy smokers when compared to nonusers (F = 4.368, p = .0185 | Tukey’s HSD p = .0135) (Figure 2B).
Figure 2.
Dose-dependent formation of DNA damage in vapers and smokers as compared to nonusers. To examine the dose dependency of DNA damage, both vapers and smokers were divided into two groups, including “light” and “heavy” users based on cumulative e-liquid consumption and pack year, respectively, as described in the text. Distribution of data within each group is shown by box and whisker plots, with the five indicating measures of each plot being labeled within the graphs (see, description in legend for Figure 1). *Statistically significant as compared to nonusers: Analysis of variance (ANOVA): F = 4.571, p = .0156 | Tukey’s HSD p = .0195. †Statistically significant as compared to nonusers: ANOVA: F = 4.368, p = .0185 | Tukey’s HSD p = .0135.
To investigate the influence of device type on the extent of DNA damage detected in e-cig users, we have divided the vapers into three groups, including users of third-generation devices (“mod”), users of “fourth” generation devices (“pod” = JUUL and JUUL alike), and users of “multiple” generation devices. We note that except for one individual who was an exclusive user of “Cigalike” devices, all vapers in our study were users of third or fourth-generation devices or users of multiple-generation devices (Table 1). As shown in Table 2, users of pod-based devices had the highest levels of DNA damage in their oral cells as compared to nonusers, followed by users of mod-based devices and multiple device users. There was a 3.3-fold increase in the mean level of DNA damage in the oral cells of pod users as compared to nonusers (F = 3.886, p = .0152 | Tukey’s HSD p = .0216). Mod users and multiple device users showed 2.6-fold and 1.6-fold increases, respectively, in mean levels of DNA damage in their oral cells as compared to nonusers. One may argue that the extent and duration of use of different generation devices might modulate the levels of DNA damage in pod users versus mod users versus multiple device users. To account for these factors, we have adjusted the DNA damage data in vapers based on (1) “cumulative e-liquid consumed,” and (2) “years vaped” per device(s). In both cases, the adjusted data showed a similar pattern of highest mean levels of DNA damage in pod users, followed by mod users and multiple device users (Table 2).
Table 2.
The Influence of E-cig Device Type and E-liquid Flavors on the Extent of DNA Damage in Vapers Versus Nonusers
| Lesions/10 kb | Lesions/10 kb/Years vaped* | Lesions/10 kb/Cum e-liq* | |||
|---|---|---|---|---|---|
| Vapers | Device | Mod | 0.684 ± 0.239 | 0.195 ± 0.059 | 1.80E-4 ± 6.22E-5 |
| Pod | 0.864 ± 0.175† | 1.077 ± 0.336 | 3.13E-2 ± 1.70E2 | ||
| Multiple | 0.423 ± 0.280 | 0.089 ± 0.044 | 1.20E-4 ± 5.10E-5 | ||
| Flavor | Fruit | 0.402 ± 0.176 | 0.295 ± 0.187 | 0.001 ± 0.001 | |
| Sweet | 1.262 ± 0.148‡ | 1.513 ± 0.836 | 0.065 ± 0.041 | ||
| Mint or Menthol | 0.692 ± 0.222 | 0.598 ± 0.224 | 0.009 ± 0.005 | ||
| Tobacco | 0.670¶ | 0.164 | 4.6E-4 | ||
| Multiple | 0.699 ± 0.320 | 0.193 ± 0.068 | 2.0E-4 ± 9.0E-5 | ||
| Nonusers | 0.262 ± 0.049 | NA | NA | ||
NA = Not applicable, ANOVA = Analysis of variance.
Summary results of LA-QPCR in the POLB gene in oral cells of vapers as compared to nonusers. Vapers were divided into three groups, including users of third-generation devices (“mod”), users of fourth-generation devices (“pod” = JUUL and JUUL alike), and users of “multiple” generation devices. E-liquid flavors consumed by vapers were divided into five categories, including (1) fruit, (2) sweet (ie, candy or desserts or other sweets), (3) mint or menthol, (4) tobacco, and (5) multiple.
*To account for the extent and duration of use of different generation devices or different e-liquid flavors, data were adjusted for “years vaped” and “cumulative e-liquid consumed.” Cumulative e-liquid (Cum e-liq) is calculated as the total volume of e-liquid (in milliliters) vaped by a person during his or her lifetime.
†Statistically significant as compared to nonusers; ANOVA: F = 3.886, p = .0152 | Tukey’s HSD p = .0216. We have also analyzed the data using the nonparametric test of Kruskal Wallis followed by post hoc Dunn’s test, which is better equipped for smaller samples with data variability.23 Analysis of the data by this nonparametric test yielded statistically significant results similar to those obtained by its parametric counterpart (ANOVA) as follows: p = .036 | Dunn’s p = .0038.
‡Statistically significant as compared to nonusers; ANOVA: F = 3.238, p = .0146 | Tukey’s HSD p < .05. We note that exclusion of ¶tobacco group from the analysis did not change the statistically significant result: ANOVA: F = 4.002, p = .0077 | Tukey’s HSD p = .0112. Furthermore, the nonparametric test of Kruskal Wallis followed by post hoc Dunn’s test yielded similar statistically significant results: Tobacco group included: p = .043 | Dunn’s p = .0041 and Tobacco group excluded: p = .033 | Dunn’s p = .0048.
Results are expressed as mean ± SE from duplicate samples assayed independently up to 2 times.
Furthermore, we investigated the impact of the chemical composition of e-liquid on the induction of DNA damage in e-cig users. To determine the role of e-liquid flavors, we have categorized the e-liquid flavors consumed by our study subjects and divided the vapers into five groups, including those who used e-liquid with (1) fruit flavors, (2) candy or desserts or other sweet flavors (hereinafter referred to as “sweet” flavors), (3) mint or menthol flavor, (4) tobacco flavor, and (5) multiple flavors. As shown in Table 2, vapers consuming sweet-flavored e-liquids, had the highest levels of DNA damage in their oral cells as compared to nonusers (F = 3.238, p = .0146 | Tukey’s HSD p < .05), followed by vapers of multiple e-liquid flavors, mint or menthol flavor and tobacco flavor, and fruit-flavored e-liquids. Adjusting the data for (1) “cumulative e-liquid consumed,” and (2) “years vaped” per flavor(s), vapers of sweet-flavored e-liquids still showed the highest mean level of DNA damage, followed by vapers of mint or menthol- and fruit-flavored e-liquids (Table 2). To substantiate these results, we have further scrutinized the data, performed additional tests, and conducted statistical analyses with or without certain subgroups. More specifically, we have demonstrated that the exclusion of small subgroups, such as “tobacco” flavor e-liquid users, from the analysis, did not change the statistically significant results obtained by comparing all subgroups to nonusers (F = 4.002, p = .0077 | Tukey’s HSD p = .0112). We have also analyzed the data using the nonparametric test of Kruskal Wallis followed by post hoc Dunn’s test, which is better equipped for smaller samples with data variability.23 Analysis of the data by this nonparametric test yielded statistically significant results similar to those obtained by its parametric counterpart (see, above) (Device type: p = .036 | Dunn’s p = .0038; Flavor type [tobacco group included]: p = .043 | Dunn’s p = .0041; Flavor type (tobacco group excluded): p = .033 | Dunn’s p = .0048) (Table 2).
Moreover, we examined how the nicotine content of e-liquid may influence the induction of DNA damage in e-cig users. Specifically, we sought a correlation between the cumulative amounts of nicotine in e-liquids consumed by vapers and the levels of DNA damage in their oral cells. The cumulative nicotine consumption by e-cig users (in milligrams) did not correlate to the levels of DNA damage in their oral cells (r = 0.3189, p = .1288). Similarly, no correlation was found between the indicator of “recent” nicotine intake, calculated as past-week nicotine consumption (mg), and DNA damage levels in vapers’ oral cells (r = −0.0457, p = .834612).
Biochemical Verification of Vaping/Smoking
As shown in Table 1, while both vapers and smokers had significantly higher levels of plasma cotinine than nonusers (p < .0001), only smokers did show significantly increased levels of breath CO and %COHb in comparison to nonusers (p = .0005 and p = .0002, respectively); vapers had similar levels of CO and %COHb to nonusers (Table 1). Plasma cotinine levels in vapers and smokers were not significantly different from one another (p = .607). Of note, plasma cotinine, a primary metabolite of nicotine, is a validated marker of tobacco product use (both for smoking combustible cigarettes and vaping nicotine-containing e-cigs).19 However, exhaled breath CO is an objective biomarker of recent exposure to combustible products, such as tobacco cigarettes (but not vaping).6 In addition, %COHb indicates the proportion of red blood cells carrying CO instead of oxygen.6 Although the cutoff point of exhaled CO to distinguish cigarette smokers from nonsmokers varies across different studies,24 we considered the cutoff point of 7.0 ppm recommended by the manufacturer of Smokerlyzer (Bedfont Scientific Ltd), which was used in this study. Likewise, there is no unanimous consensus regarding the cutoff point of cotinine for distinguishing cigarette smokers from nonsmokers.25 Nonetheless, we confirmed the self-reported exposure status of the study subjects by demonstrating that nonusers had breath CO levels of 1.9 ± 0.3 ppm (all below the 3.0 ppm) and plasma cotinine of 2.6 ± 0.1 ng/ml, whereas the respective values for smokers and vapers were: (CO = 12.0 ± 1.6 ppm and cotinine = 76.7 ± 8.6 ng/ml [for smokers]) and (CO = 2.0 ± 0.3 ppm and cotinine = 84.9 ± 13.1 ng/ml [for vapers]) (Table 1).
Discussion
Most adult vapers have a prior history of smoking combustible cigarettes.3,5 Thus, many adults who are “current” users of e-cigs, are likely to be “former" or "ex” smokers.1–3 The existing literature on the “potential” health risks of vaping is often criticized by the fact that the study subjects in many reports consist of adult vapers whose “past” smoking history is either unspecified or ambiguously defined.3,13 This has complicated the interpretation of results as it is unclear whether the observed effects in e-cig users are solely caused by vaping or because of the persistent effects of “past” smoking.1,3,5 These uncertainties have fueled a highly contentious debate on the public health impact of vaping.3,26 The design of the present study has allowed us to tease out the biological effects of “exclusive” vaping in a thoroughly characterized population of adults.
LA-QPCR quantification of DNA damage in oral epithelial cells, a target cell type for cancer and other diseases associated with tobacco product use,17 showed significantly increased levels of polymerase-blocking lesions in both vapers and smokers as compared to nonusers. The mean levels of DNA damage did not differ significantly between vapers and smokers. Importantly, DNA damage levels in both vapers and smokers increased dose-dependently, from light users to heavy users, when compared to nonusers. These in vivo findings are novel and significant as they demonstrate, for the first time, a dose-dependent formation of DNA damage in target cells from vapers who had never smoked tobacco cigarettes. In this study, we used cumulative e-liquid consumption (Cum e-liq) and pack year17,19 as indicators of vaping- and smoking dose, respectively. We17,19,22 and others27 have previously shown the utility of Cum e-liq and PY for estimating chronic e-cig use and cigarette smoking, respectively. Soule et al.28 have argued that the amount of e-liquid consumed may be a useful indicator of the quantity of aerosol inhaled by e-cig users, but not necessarily a precise measure of exposure to nicotine and other toxicants in the aerosol. This is consistent with our choice of Cum e-liq for estimating cumulative exposure to the complex mixture of e-cig aerosol (as a whole) but not to its individual chemical constituents. We did not use puff topography to assess exposure in the present study because e-cig users are known to puff often from the same e-cig in multiple sessions, with sessions not being consistent in total puff duration or the number of puffs.28 This is in sharp contrast with cigarette smokers who typically smoke a cigarette from start to finish in a single session. It is important to note that consensus on e-cig use intensity measures that can be used for survey research has yet to be established due to great heterogeneity in e-cig device and e-liquid characteristics and user behavior, which lead to different levels of exposure to toxicants and carcinogens by e-cig users.28
We note that the comparable levels of DNA damage detected in vapers and smokers deserve further investigation. Given the similarities and differences in the chemical composition of e-cig vapor and cigarette smoke,5–7,29 it is important to uncover the identity of DNA lesions formed in vapers and smokers. Future studies should exploit the high specificity and sensitivity of mass spectrometry based assays15 to characterize the type of induced DNA damage in vapers and smokers. Identifying the chemical structure of DNA lesions formed in vapers versus smokers will have significant implications for tobacco products regulation.
Ganapathy et al.30 have shown that in vitro treatment of human oral squamous cell carcinoma cells (UM-SCC-1) with e-cig aerosol condensates resulted in the formation of DNA damage in the TP53 gene, as detected by a similar PCR-based assay. The levels of 8-hydroxy-2’-deoxyguanosine, an indicator of promutagenic oxidative DNA damage,14 were also increased significantly in treated UM-SCC-1 cells as compared to controls, as quantified by ELISA. Sundar et al.31 have shown that in vitro exposure of human gingival epithelium progenitors pooled cells and periodontal ligament fibroblasts to e-cig aerosols at air-liquid interface caused significant DNA damage, as reflected by increased phosphorylated γH2A.X Ser139 (a DNA damage marker) and/or elevated comet tail lengths in the treated cells as compared to air-exposed controls. The findings were recapitulated in a normal human 3D in vitro model of EpiGingival tissues that were similarly exposed to e-cig aerosols and afterward analyzed by immunohistochemical staining for γH2AX. Recently, Cheng et al.32 have reported significantly increased levels of the major DNA adduct of acrolein, a carcinogenic aldehyde found substantially in cigarette smoke and to a lesser extent, in e-cig vapor,6,7 in buccal cells of vapers as compared to nonusers, using mass spectrometry based analysis. Vapers in Cheng et al.’s study (N = 20) were defined as those who used e-cigs for a minimum of 3 months and, at least, 4 days per week; albeit no information was provided on the subjects’ history of smoking or other tobacco products use.32 It is important to put into context the diverse types of assay used in the above studies, which were conducted in different settings (in vitro or in vivo). Some of the applied assays in the cited studies do not measure the exact same endpoint as that quantifiable by LA-QPCR. While the detection of DNA damage in vapers and smokers in the present study is consistent with the findings reported by others in other settings, the type of assays used in some of those reports enables the quantification of specific markers of DNA damage. The specific markers detected by those assays do not necessarily reflect the formation of the same type of lesions detectable by LA-QPCR.
Our findings on the role of product characteristics in the induction of DNA damage in vapers are novel and can have significant regulatory implications for electronic nicotine delivery systems. We observed that both e-cig device type and e-liquid flavor are determinants of DNA damage in oral epithelial cells in vapers. Users of pod devices followed by mod users had the highest levels of DNA damage in their oral cells as compared to nonusers. Since entering the US market around 2006–2007, e-cig devices have evolved continually and rapidly, from the first-generation “Cigalike,” to the second-generation vape pens, third-generation box mods, and the current fourth-generation pod-based devices.5,7 A combination of sleek and high-tech design, innovative salt-based nicotine delivery technology, large assortment of e-liquid flavors, and social media-oriented marketing has made pod-based e-cigs, such as JUUL and JUUL alike devices, widely popular among both novice and experienced vapers.5,7,33 The small size, lightweight, and easy-to-conceal nature of these devices together with teen-appealing e-liquid flavors have also made them a choice of preference for adolescents experimenting with tobacco products.33 The latter is believed to have contributed to the ongoing epidemic of youth vaping in the United States.34
Furthermore, we observed that users of sweet-flavored e-liquids, followed by users of mint or menthol- and fruit-flavored e-liquids had the highest levels of DNA damage in their oral cells when compared to nonusers. This finding can have significant implications for public health because these e-liquid flavors, which exhibit the highest DNA-damaging potencies, are not only popular among adult vapers but also, they are the preferred flavors for youth vapers.35–37 Common flavoring chemicals, such as ethyl maltol imparting sweet and caramel-like aromas and flavors, lactones imparting fruity and creamy flavors, piperonal, an aromatic aldehyde imparting cherry and vanilla-like flavors, and benzaldehyde, a natural fruit flavoring, are known to decompose to radicals and redox active species during vaporization.38 It has also been shown that vapor generated from JUUL pod with Cool Mint significantly increased acellular reactive oxygen species levels when compared to control filtered air.39 Moreover, in vitro experiments have confirmed that vapor produced from Classic Menthol JUUL pod caused the greatest increase in mitochondrial superoxide production in lung epithelial cells in comparison to vapors from pods with other flavors or filtered air.39 Altogether, the higher oxidative properties of sweet-, mint or menthol-, or fruit-flavored e-liquids as compared to non-flavored e-liquids may translate to a greater genotoxic potential for the former products.40 This is reinforced by our observation that vapers of e-liquids with such flavors exhibited the highest levels of DNA damage in their oral cells.
Moreover, we found that the nicotine content of e-liquid was not a predictor of DNA damage in vapers’ oral cells. This finding is in agreement with previous reports showing that in vitro e-cig aerosols or condensates induce genotoxic effects independently of nicotine concentrations in the e-liquid.30,41,42 Collectively, our data suggest that flavoring components alone or in combination with other e-liquid constituents (eg, humectant or additives) can give rise to DNA reactive species, which may, in turn, cause genotoxic effects in cells and tissues of e-cig users. Potential chemicals involved in this process may include those formed during the vaporization of e-liquid flavorings and humectants, particularly reaction products of flavoring aldehydes or formaldehyde and PG or VG, such as acetals and hemiacetals, and reactive oxygen species, among others.10,11,22 Future studies should uncover the chemical structures of the herein-detected DNA lesions in vapers. These follow-up investigations should help inform the regulation of e-liquid ingredients that are responsible for the genotoxic effects of vaping observed in this study as well as in previous studies by others.30–32,39,41,42
Strengths and Limitations
By design, the present study accounted for relevant biological variables, specifically age, gender, and race. In addition, dietary intake data obtained from the study subjects confirmed that there was no significant difference in alcohol use or consumption of grilled or roasted or broiled foods among vapers, smokers, and nonusers. Furthermore, vapers in our study were mostly young adults and likely representative of the population from which they were drawn. Young adults are known to favor pod and mod devices and prefer e-liquids with sweet-, mint or menthol-, and fruit flavors rather than tobacco-flavored e-liquids.35–37 We demonstrated the impact of device type and e-liquid flavor on the induction of DNA damage in vapers. The small sample size of certain subgroups in our study can be considered a limitation. Nevertheless, we have substantiated our findings by further scrutinizing the data, performing additional tests, and conducting statistical analyses with or without the small subgroups. We acknowledge that future studies with larger sample sizes are needed to further validate our results and allow for a more detailed characterization of the ingredients of e-liquid as well as device features that may contribute to the biological effects of vaping. These follow-up investigations are highly important given the diversity of e-cig devices and variation in e-liquid products.5,7,37
The LA-QPCR assay used in this study quantifies broad spectra of polymerase stalling or stopping DNA lesions (eg, oxidative, alkylative, and bulky lesions, and single-strand breaks) within the amplification region of the primer set designed for a specific gene.16 The assay does not provide information on the chemical structure of the detected lesions or the distribution of DNA damage across the genome.16 However, we and others have shown that DNA damage in reporter genes faithfully captures many aspects of lesion formation and repair in cancer-related genes, such as tumor suppressor genes or oncogenes.16,30,43,44 In this study, the choice of POLB as a gene target for LA-QPCR analysis was based on previous studies by others16,20,21 who have confirmed that POLB can serve as a representative gene target for DNA damage detection. In addition, the results of LA-QPCR in the POLB and other gene targets, such as HPRT, have been shown to be highly consistent and correlated,21 as reconfirmed in our study.
Of importance for this study, we stress the challenges of research in healthy volunteers with matching characteristics (ie, age, gender, and race) and strictly defined exposure, whose source materials (eg, tissues, cells, DNA and RNA) are often limited for molecular analysis. The limited source materials in these studies inevitably lead to the prioritization of endpoints or selective analysis of target gene(s)). With the same token, prioritization will be required to detect the selected endpoint(s) in specific tissues, cells, or cellular compartments (eg, nucleolus vs. mitochondria). For example, while the significance and importance of damage to the nuclear genome in disease pathophysiology are well-established, the role of mitochondrial DNA damage in disease development is beginning to be fully appreciated45,46 (see, Supplementary Materials for distinctions between nuclear and mitochondrial genomes). Given the limited source materials for molecular analysis in population-based studies,47 nuclear DNA has been extensively used as a preferred choice for direct measurement of DNA damage.15 The focus of the present study was on nuclear DNA damage because the source materials for this study had to be shared with our ongoing genomic sequencing project, which aims to detect mutations in the nuclear genome. While there is a growing recognition of the importance of mitochondria in health versus disease state,45,46,48 measuring mitochondrial DNA damage was beyond the scope of the present study and outside its prioritization scheme.
In the present study, we did not collect quantitative data on the physical activity levels of the study subjects. Limited but emerging data suggest modulatory effects of exercise on DNA damage associated with lifestyle factors, such as smoking.49 Urinary levels of 8-hydroxydeoxyguanosine (8-OH-dG) in male smokers were inversely related to physical activity of moderate or rigorous intensity. A similar trend was found in female smokers between urinary 8-OH-dG levels and total physical activity.50 We note that investigating the effects of exercise on DNA damage in vapers versus smokers is beyond the scope of this study. Finally, while we underscore the importance of follow-up studies in large populations, our power calculations showed that the present study was powered at 83% and 63%, respectively, to detect statistically significant differences in DNA damage levels between vapers and nonusers, and smokers and nonusers (at α = 0.05).
Conclusions
We have demonstrated a dose-dependent formation of DNA damage in oral cells of vapers who had never smoked tobacco cigarettes as well as exclusive cigarette smokers. We have also shown that e-cig device type and e-liquid flavor determine the extent of DNA damage detected in vapers. In terms of device type, pods followed by mods, and in terms of flavor type, sweet, followed by mint or menthol- and fruit-flavored e-liquids exhibit the greatest DNA-damaging potencies in vapers. Given the popularity of pod and mod devices and the preferability of these same flavors of e-liquid by both adult- and youth vapers,33,35,36 our findings can have significant implications for public health and tobacco products regulation.
Supplementary Material
A Contributorship Form detailing each author’s specific involvement with this content, as well as any supplementary data, are available online at https://academic.oup.com/ntr.
Acknowledgments
We would like to thank Andrew Caliri and Amanda Caceres for technical support and help with laboratory assays. Special thanks to Dr. Alan Hiti and the USC-Norris Outpatient Clinical Laboratory staff for phlebotomy work. We are grateful to Drs. Bo Hang and Altaf Sarker of Lawrence Berkeley National Laboratory for sharing their expertise in LA-QPCR.
Contributor Information
Stella Tommasi, Department of Population and Public Health Sciences, USC Keck School of Medicine, University of Southern California, M/C 9603, Los Angeles, CA 90033, USA.
Hannah Blumenfeld, Department of Population and Public Health Sciences, USC Keck School of Medicine, University of Southern California, M/C 9603, Los Angeles, CA 90033, USA.
Ahmad Besaratinia, Department of Population and Public Health Sciences, USC Keck School of Medicine, University of Southern California, M/C 9603, Los Angeles, CA 90033, USA.
Funding
The work of the authors is supported by grants from the National Cancer Institute and National Institute of Dental and Craniofacial Research of the National Institutes of Health (1R21CA268197 and 1R01DE026043, resp.), and the University of California Tobacco-Related Disease Research Program (T31IR1839, and T32IR5144). The sponsors of the study had no role in study design, data collection, data analysis, data interpretation, writing of the report, or in the decision to submit for publication.
Declaration of Interests
All the authors declare no conflict of interest.
Data Availability
The data underlying this article are available in the article and in its online supplementary material.
References
- 1. Hartmann-Boyce J, McRobbie H, Lindson N, et al. Electronic cigarettes for smoking cessation. Cochrane Database Syst Rev. 2020;10:CD010216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang RJ, Bhadriraju S, Glantz SA.. E-cigarette use and adult cigarette smoking cessation: a meta-analysis. Am J Public Health. 2020;111(2):230–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Balfour DJK, Benowitz NL, Colby SM, et al. Balancing consideration of the risks and benefits of e-cigarettes. Am J Public Health. Sep 2021;111(9):1661–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Besaratinia A, Tommasi S.. Vaping epidemic: challenges and opportunities. Cancer Causes Control. 2020;31(7):663–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gordon T, Karey E, Rebuli ME, et al. E-cigarette toxicology. Annu Rev Pharmacol Toxicol. 2022;62:301–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. McDonough SR, Rahman I, Sundar IK.. Recent updates on biomarkers of exposure and systemic toxicity in e-cigarette users and EVALI. Am J Physiol Lung Cell Mol Physiol. 2021;320(5):L661–l679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Park JA, Crotty Alexander LE, Christiani DC.. Vaping and Lung Inflammation and Injury. Annu Rev Physiol. 2022;84:611–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wei B, O’Connor RJ, Goniewicz ML, Hyland A.. Emerging chemicals of health concern in electronic nicotine delivery systems. Chem Res Toxicol. 2020;33(10):2637–2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tehrani MW, Newmeyer MN, Rule AM, Prasse C.. Characterizing the chemical landscape in commercial e-cigarette liquids and aerosols by liquid chromatography-high-resolution mass spectrometry. Chem Res Toxicol. 2021;34(10):2216–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Khlystov A, Samburova V.. Flavoring compounds dominate toxic aldehyde production during e-cigarette vaping. Environ Sci Technol. 2016;50(23):13080–13085. [DOI] [PubMed] [Google Scholar]
- 11. Erythropel HC, Jabba SV, DeWinter TM, et al. Formation of flavorant-propylene glycol adducts with novel toxicological properties in chemically unstable e-cigarette liquids. Nicotine Tob Res. 2019;21(9):1248–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gotts JE, Jordt SE, McConnell R, Tarran R.. What are the respiratory effects of e-cigarettes? BMJ. 2019;366:l5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wills TA, Soneji SS, Choi K, Jaspers I, Tam EK.. E-cigarette use and respiratory disorders: an integrative review of converging evidence from epidemiological and laboratory studies. Eur Respir J. 2021;57(1):1901815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Caliri AW, Tommasi S, Besaratinia A.. Relationships among smoking, oxidative stress, inflammation, macromolecular damage, and cancer. Mutat Res. 2021;787:108365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Phillips DH, Venitt S.. DNA and protein adducts in human tissues resulting from exposure to tobacco smoke. Int J Cancer. 2012;131(12):2733–2753. [DOI] [PubMed] [Google Scholar]
- 16. Furda A, Santos JH, Meyer JN, Van Houten B.. Quantitative PCR-based measurement of nuclear and mitochondrial DNA damage and repair in mammalian cells. Methods Mol Biol. 2014;1105:419–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tommasi S, Caliri AW, Caceres A, et al. Deregulation of biologically significant genes and associated molecular pathways in the oral epithelium of electronic cigarette users. Int J Mol Sci . 2019;20(3):738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Centers for Disease Control and Prevention. Disability and Health Data System (DHDS). http://dhds.cdc.gov/guides/healthtopics/indicator?I=smokingstatus.Accessed October 20, 2022.
- 19. Caliri AW, Caceres A, Tommasi S, Besaratinia A.. Hypomethylation of LINE-1 repeat elements and global loss of DNA hydroxymethylation in vapers and smokers. Epigenetics. 2020;15(8):816–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Santos JH, Meyer JN, Mandavilli BS, Van Houten B.. Quantitative PCR-based measurement of nuclear and mitochondrial DNA damage and repair in mammalian cells. Methods Mol Biol. 2006;314:183–199. [DOI] [PubMed] [Google Scholar]
- 21. Hang B, Sarker AH, Havel C, et al. Thirdhand smoke causes DNA damage in human cells. Mutagenesis. 2013;28(4):381–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tommasi S, Pabustan N, Li M, Chen Y, Siegmund KD, Besaratinia Aet al. A novel role for vaping in mitochondrial gene dysregulation and inflammation fundamental to disease development. Sci Rep. 23 2021;11(1):22773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mangiafico SS. Summary and Analysis of Extension Program Evaluation in R, version 1.19.10. New Brunswick, NJ, USA: Rutgers Cooperative Extension; 2016. [Google Scholar]
- 24. Karelitz JL, McClure EA, Wolford-Clevenger C, Pacek LR, Cropsey KL.. Cessation classification likelihood increases with higher expired-air carbon monoxide cutoffs: a meta-analysis. Drug Alcohol Depend. 2021;221:108570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Benowitz NL, Bernert JT, Caraballo RS, Holiday DB, Wang J.. Optimal serum cotinine levels for distinguishing cigarette smokers and nonsmokers within different racial/ethnic groups in the United States between 1999 and 2004. Am J Epidemiol. 2009;169(2):236–248. [DOI] [PubMed] [Google Scholar]
- 26. Tommasi S, Bates SE, Behar RZ, Talbot P, Besaratinia A.. Limited mutagenicity of electronic cigarettes in mouse or human cells in vitro. Lung Cancer. 2017;112:41–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tindle HA, Stevenson Duncan M, Greevy RA, et al. Lifetime smoking history and risk of lung cancer: results from the framingham heart study. J Natl Cancer Inst. 2018;110(11):1201–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Soule E, Bansal-Travers M, Grana R, et al. Electronic cigarette use intensity measurement challenges and regulatory implications. Tob Control. 2021;32(1):124–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Besaratinia A, Tommasi S.. An opportune and unique research to evaluate the public health impact of electronic cigarettes. Cancer Causes Control. 2017;28(10):1167–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ganapathy V, Manyanga J, Brame L, et al. Electronic cigarette aerosols suppress cellular antioxidant defenses and induce significant oxidative DNA damage. PLoS One. 2017;12(5):e0177780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sundar IK, Javed F, Romanos GE, Rahman I.. E-cigarettes and flavorings induce inflammatory and pro-senescence responses in oral epithelial cells and periodontal fibroblasts. Oncotarget. 2016;7(47):77196–77204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cheng G, Guo J, Carmella SG, et al. Increased acrolein-DNA adducts in buccal brushings of e-cigarette users. Carcinogenesis. 2022;43(5):437–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Besaratinia A, Tommasi S.. The consequential impact of JUUL on youth vaping and the landscape of tobacco products: the state of play in the COVID-19 era. Prev Med Rep. 2021;22:101374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Besaratinia A. COVID-19: a pandemic converged with global tobacco epidemic and widespread vaping-state of the evidence. Carcinogenesis. 2021;42(8):1009–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Landry RL, Groom AL, Vu TT, et al. The role of flavors in vaping initiation and satisfaction among U.S. adults. Addict Behav. 2019;99:106077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Krüsemann EJZ, Boesveldt S, de Graaf K, Talhout R.. An e-liquid flavor wheel: a shared vocabulary based on systematically reviewing e-liquid flavor classifications in literature. Nicotine Tob Res. 2019;21(10):1310–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Besaratinia A. From tobacco cigarettes to electronic cigarettes: the two sides of a nicotine coin. Front Oral Health. 2021;2:790634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bitzer ZT, Goel R, Reilly SM, et al. Effect of flavoring chemicals on free radical formation in electronic cigarette aerosols. Free Radic Biol Med. 2018;120:72–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Muthumalage T, Lamb T, Friedman MR, Rahman I.. E-cigarette flavored pods induce inflammation, epithelial barrier dysfunction, and DNA damage in lung epithelial cells and monocytes. Sci Rep. 2019;9(1):19035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Son Y, Mishin V, Laskin JD, et al. Hydroxyl Radicals in e-cigarette vapor and e-vapor oxidative potentials under different vaping patterns. Chem Res Toxicol. 2019;32(6):1087–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yu V, Rahimy M, Korrapati A, et al. Electronic cigarettes induce DNA strand breaks and cell death independently of nicotine in cell lines. Oral oncology. 2016;52:58–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sun YW, Chen KM, Atkins H, et al. Effects of e-cigarette aerosols with varying levels of nicotine on biomarkers of oxidative stress and inflammation in mice. Chem Res Toxicol. 2021;34(4):1161–1168. [DOI] [PubMed] [Google Scholar]
- 43. Besaratinia A, Pfeifer GP.. Enhancement of the mutagenicity of benzo(a)pyrene diol epoxide by a nonmutagenic dose of ultraviolet A radiation. Cancer Res. 2003;63(24):8708–8716. [PubMed] [Google Scholar]
- 44. Besaratinia A, Li H, Yoon JI, Zheng A, Gao H, Tommasi Set al. A high-throughput next-generation sequencing-based method for detecting the mutational fingerprint of carcinogens. Nucleic Acids Res. 2012;40(15):e116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ng YS, Bindoff LA, Gorman GS, et al. Mitochondrial disease in adults: recent advances and future promise. Lancet Neurol. 2021;20(7):573–584. [DOI] [PubMed] [Google Scholar]
- 46. Silva-Pinheiro P, Minczuk M.. The potential of mitochondrial genome engineering. Nat Rev Genet. 2022;23(4):199–214. [DOI] [PubMed] [Google Scholar]
- 47. Besaratinia A, Tommasi S.. Epigenetics of human melanoma: promises and challenges. J Mol Cell Biol. 2014;6(5):356–367. [DOI] [PubMed] [Google Scholar]
- 48. Taylor RW, Turnbull DM.. Mitochondrial DNA mutations in human disease. Nat Rev Genet. 2005;6(5):389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Williamson J, Davison G.. Targeted antioxidants in exercise-induced mitochondrial oxidative stress: emphasis on DNA damage. Antioxidants (Basel). 2020;9(11):1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hara M, Nishida Y, Shimanoe C, et al. Intensity-specific effect of physical activity on urinary levels of 8-hydroxydeoxyguanosine in middle-aged Japanese. Cancer Sci. 2016;107(11):1653–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article and in its online supplementary material.