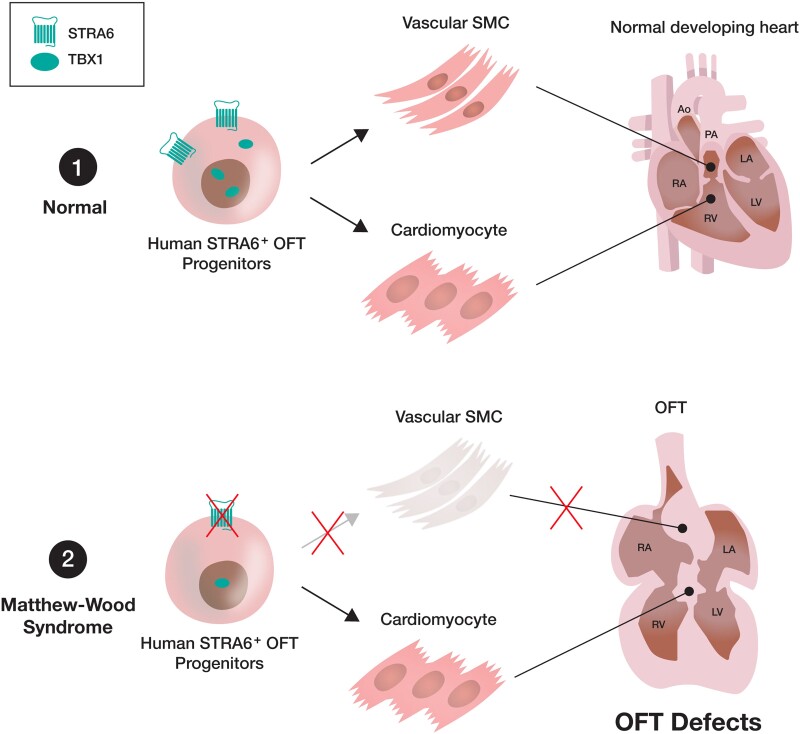
Abstract
Aims
Retinoic acid (RA) signalling is essential for heart development, and dysregulation of the RA signalling can cause several types of cardiac outflow tract (OFT) defects, the most frequent congenital heart disease (CHD) in humans. Matthew-Wood syndrome is caused by inactivating mutations of a transmembrane protein gene STRA6 that transports vitamin A (retinol) from extracellular into intracellular spaces. This syndrome shows a broad spectrum of malformations including CHD, although murine Stra6-null neonates did not exhibit overt heart defects. Thus, the detailed mechanisms by which STRA6 mutations could lead to cardiac malformations in humans remain unclear. Here, we investigated the role of STRA6 in the context of human cardiogenesis and CHD.
Methods and results
To gain molecular signatures in species-specific cardiac development, we first compared single-cell RNA sequencing (RNA-seq) datasets, uniquely obtained from human and murine embryonic hearts. We found that while STRA6 mRNA was much less frequently expressed in murine embryonic heart cells derived from the Mesp1+ lineage tracing mice (Mesp1Cre/+; Rosa26tdTomato), it was expressed predominantly in the OFT region-specific heart progenitors in human developing hearts. Next, we revealed that STRA6-knockout human embryonic stem cells (hESCs) could differentiate into cardiomyocytes similarly to wild-type hESCs, but could not differentiate properly into mesodermal nor neural crest cell-derived smooth muscle cells (SMCs) in vitro. This is supported by the population RNA-seq data showing down-regulation of the SMC-related genes in the STRA6-knockout hESC-derived cells. Further, through machinery assays, we identified the previously unrecognized interaction between RA nuclear receptors RARα/RXRα and TBX1, an OFT-specific cardiogenic transcription factor, which would likely act downstream to STRA6-mediated RA signalling in human cardiogenesis.
Conclusion
Our study highlights the critical role of human-specific STRA6 progenitors for proper induction of vascular SMCs that is essential for normal OFT formation. Thus, these results shed light on novel and human-specific CHD mechanisms, driven by STRA6 mutations.
Keywords: Congenital heart disease, Heart development, Cardiac outflow tract, Matthew-Wood syndrome, Retinoic acid, Single-cell RNA-seq, Smooth muscle cell
Graphical Abstract
Graphical Abstract.
1. Introduction
Heart formation is one of the earliest and most critical steps in embryogenesis. The morphogenetic process of mammalian heart development is complex and tightly regulated by a diverse set of heart progenitors and paracrine molecular cues.1,2 These progenitors contribute to forming the distinct heart regions, such as atria, right ventricle (RV) and left ventricle (LV), and cardiac outflow tract (OFT). Multiple paracrine signalling pathways control the heart progenitors’ specification, commitment, proliferation, and differentiation into various cardiac cell lineages in a sequential and co-ordinated fashion.3 For instance, the retinoic acid (RA) signalling regulates the patterning of the second heart field (SHF) progenitors and is thereby essential for cardiac OFT development.4,5 Either an excess or lack of RA, a biologically active metabolite of vitamin A (retinol), causes cardiac OFT defects that are the most common congenital heart disease (CHD) in humans.6 In fact, a failed septation and/or a mal-alignment of the OFT lead to persistent truncus arteriosus (PTA), double outlet right ventricle, transposition of the great arteries, overriding aorta, ventricular septal defect (VSD), tetralogy of Fallot (ToF), or other anomalies. In regard to genetic variation of the RA signalling-related genes, inactivating mutations of the STRA6 gene that encodes a transmembrane protein implementing cellular uptake of vitamin A is known to cause Matthew-Wood (also termed PDAC) syndrome, which is a rare autosomal recessive disorder and shows a broad spectrum of congenital malformations including pulmonary hypoplasia/agenesis, diaphragmatic hernia, anophthalmia/microphthalmia, and cardiac defects.7,8 Cardiovascular malformations observed in Matthew-Wood syndrome involve atrial septal defects, persistent ductus arteriosus, aortic arch abnormalities, and ToF,9,10 which are similar to those in other RA signalling disorders. Congenital heart defects are observed in around 50% of patients with this syndrome, with varying levels of severity.8 In contrast, interestingly, murine Stra6-null embryos and neonates did not exhibit overt heart defects.11–13 Although this implies a species-specific cardiac malformation program associated with dysregulation of the RA signalling, the precise mechanisms by which STRA6 mutations can cause cardiac malformations such as OFT defects in humans are still undetermined, which would further require rigorous and cross-species comparison analysis.
On the other hand, a recent sophisticated biotechnology such as single-cell RNA sequencing (RNA-seq) has been able to reconstruct complex cellular heterogeneity in vivo and identify developmental cellular trajectories and molecular signatures in various types of tissues and organs.14 This approach examining murine and human developing hearts has provided novel insights on mammalian cardiogenesis at higher resolution than before and identified previously unrecognized heart progenitors and/or molecules that would play certain roles spatiotemporally in developing hearts.15–17 Here we compared the single-cell RNA-seq datasets uniquely obtained from human and murine embryonic hearts, respectively, and focused on the STRA6 mRNA expression at the single-cell level in each. Importantly, we found that while STRA6 was much less frequently expressed in murine embryonic heart cells derived from the Mesp1+ lineage tracing mice, it was expressed predominantly in the cono-ventricular region (i.e. proximal OFT)-specific heart progenitors in the human embryonic heart. Next, we generated STRA6-knockout (STRA6-KO) human embryonic stem cell (hESC) lines via CRISPR-Cas9 technology18 and tested their capabilities to differentiate into cardiomyocytes (CMs) and vascular smooth muscle cells (SMCs) in vitro using the established protocols.19–21 Of particular interest, the STRA6-KO hESCs could differentiate into CMs similarly to wild-type (WT) hESCs but could not differentiate properly into mesodermal lineage-derived nor neural crest cell (NCC) lineage-derived SMCs in vitro. This is corroborated by the transcriptomics data showing down-regulation of a number of SMC differentiation-related and SMC marker genes in STRA6-KO cells during CM and SMC differentiation. Further, through mechanistic analyses using chromatin immunoprecipitation (ChIP) and immunoblotting assays, we newly identified the previously unrecognized gene regulatory network, involving an interaction between RA nuclear receptors RARα/RXRα and TBX1, an OFT-specific cardiogenic transcription factor (TF),22 which would likely act downstream to STRA6-mediated RA signalling in human cardiogenesis.
Collectively, these results not only give us novel insights on understanding mammalian cardiogenesis but also shed light on previously unappreciated and human-specific CHD mechanisms, driven by the STRA6 mutations.
2. Methods
2.1. Isolation of human and murine embryonic heart-derived single cells
The study was performed in accordance with the Declaration of Helsinki and the guidelines from Directive 2010/63/EU, and all the protocols including handling of human samples and animal works were approved by the Institutional Review Board at Karolinska Institutet with ethical permission numbers (Dnr 2015/1369-31/2 and N227-14). For the analysis of human developing hearts, we utilized our previously published single-cell RNA-seq dataset, involving a total of 458 individual cardiac cells derived from micro-dissected heart regions, i.e. OFT, atria, and ventricles of human embryonic/foetal hearts at 4.5–10 weeks of the gestation stages. Prior to inclusion in the study, informed written consent was obtained, as described previously.16
For the analysis of murine developing hearts, we first established the Mesp1+ mesodermal lineage tracing mice. For this, using CRISPR-Cas9 and micro-injection into zygotes followed by embryo transfer into pseudo-pregnant female mice (CD1), we generated a Mesp1-IRES2-Cre knock-in mouse line (unpublished observation) and cross-bred them with reporter mice (Rosa26tdTomato). On the Mesp1+ lineage tracing mice (Mesp1Cre/+; Rosa26tdTomato), we observed that more than 95% of the heart cells on an embryonic day 10.5 (E10.5) and at neonatal P1 were the mesodermal lineage (Mesp1+) (see Supplementary material online, Figure S1). The hearts of the Mesp1Cre/+; Rosa26tdTomato embryos were harvested at the various time points such as E9.5, E10.5, and E14.5 and micro-dissected as a whole heart tube on E9.5 or 4 divided compartments such as OFT, RV, LV, and atria on E10.5 and E14.5. Each of the divided heart regions was cut into small pieces and dissociated into single cells, as described previously.16 Separately, we also harvested the whole embryos (Mesp1Cre/+; Rosa26tdTomato) at E8.25 and dissociated them into single cells as well. After staining the dissociated cells with DAPI (Thermo Fisher Scientific), the DAPI-negative (live) and Mesp1-tdTomato+ single cardiac cells were sorted into 384-well plates containing cell lysis buffer, customized for the Smart-seq2 approach,23 using a fluorescence-activated cell sorter (FACSARIA III; BD Biosciences).
At the embryos’ transfer for generation of a Mesp1-IRES2-Cre knock-in mouse line, we used 2.5% of isoflurane to the recipient female mice one time as inhalation anaesthesia. Euthanasia was performed by overexposure to CO2 gas in a closed chamber followed by cervical dislocation.
2.2. Single-cell RNA library construction and RNA sequencing
cDNA libraries of the sorted single cardiac cells were prepared with the Smart-seq2 approach composed of the sequential steps such as denature, reverse transcription, PCR pre-amplification, PCR purification, tagmentation reaction, enrichment PCR, re-PCR purification, quality check, and library pooling.23 The pooled libraries were sequenced at 125 bp paired-end or 50 bp single-end to a mean read depth of around 700 000 total aligned reads per cell on the Hiseq 2500 or 4000 instrument (Illumina).
2.3. Single-cell RNA sequencing data analysis
Raw reads were pre-processed with the sequence-grooming tools FASTQC and Cutadapt and followed by sequence alignment with the STAR aligner and Samtools onto human (hg38) or murine (UCSC mm10) genome reference with default settings (unique mapping rate: around 75–85%).14 Mapped gene counts were carried out with HTSeq, and transcript levels were quantified for each transcript as fragments or reads per kilo base of transcript per million mapped reads (FPKM or RPKM). The average number of expressed genes (FPKM/RPKM≥1) was ∼7000–8000 per cell, and the average number of counts was ∼15–20 per gene. After filtration of low expression genes and poor-quality cells, depth-normalization of the filtered cells’ reads was conducted using edgeR or DESeq program, implemented in R/Bioconductor. To overview the entire single-cell transcriptomics to be analysed, principal component analysis (PCA), hierarchical clustering, and diffusion map dimensionality reduction were performed. To cluster the analysed single cells, dimensionality reduction methods such as two-dimensional t-distributed stochastic neighbour embedding (tSNE) and uniform manifold approximation and projection were performed using the Seurat program.24 Differential expression analysis of the defined clusters was conducted using edgeR, Limma, and Seurat programs.16
2.4. Human ESC culture and in vitro cardiomyocyte differentiation
The hESC line H9 was purchased from WiCell Research Institute and maintained on feeder-free and 0.3 mg/mL Matrigel (BD Biosciences)-coated plates in mTeSR1 medium (STEMCELL Technologies), according to manufacturers’ instructions. Cells were fed daily and passaged every 4–5 days with Accutase (STEMCELL Technologies). Media was supplemented with 5 μM ROCK inhibitor Y-27632 (Tocris) for 24 h after splitting.
Cardiac-directed differentiation was performed using a well-established hESC-CM differentiation protocol based on Wnt signalling modulation.19,25 Briefly, hESCs were dissociated into single cells with Accutase and seeded onto Matrigel-coated 12-well plates at 750 000–1.2 million cells per well in mTeSR1 supplemented with 5 μM Y-27632 for 24 h (day −2). At Day 0, cells were treated with 12 μM GSK-3β inhibitor CHIR99021 (Sigma) in RPMI medium supplemented with B27 minus insulin (RPMI/B27-ins; Thermo Fisher Scientific) for 24 h. At Day 1, the medium was replaced with fresh RPMI/B27-ins. At Day 3, half of the medium in each well was changed to RPMI/B27-ins supplemented with 5 μM Wnt inhibitor IWP-2 (Tocris), all of which was replaced with fresh RPMI/B27-ins at Day 5. At Day 7, the medium was switched to fresh RPMI medium with B27 supplement (RPMI/B27). Thereafter, the medium was replaced with fresh RPMI/B27 every other day. Beating CMs start to appear in the culture typically at Day 8 or 9. Robust and broad spontaneous contractions are observed from Day 10 to 12 onward. In a separate experiment, cells were simultaneously treated with 0.5 μM RA (Sigma) during Days 3–7, in order to examine the effects of the enhanced RA signalling pathway in CM differentiation.
2.5. In vitro smooth muscle cell differentiation
In vitro mesodermal lineage-derived vascular SMC differentiation from hESCs was performed using a well-established protocol with minor modification.20 Briefly, dissociated hESCs were seeded onto Matrigel-coated 6-well plates at 400 000 cells per well in mTeSR1 supplemented with 5 μM Y-27632 for 24 h (Day 0). At Day 1, the medium was replaced with N2/B27 medium [1:1 of DMEM/F12 and Neurobasal medium (Gibco) with B27 and N2 supplements (Thermo Fisher Scientific) and 0.1% β-mercaptoethanol] supplemented with 8 μM CHIR99021 and 25 ng/mL human BMP4 (R&D), which was maintained for 3 days. At Days 4 and 5, the medium was replaced with fresh N2/B27 medium supplemented with 10 ng/mL PDGF-BB (Peprotech) and 2 ng/mL Activin-A (Peprotech). Typically, mesodermal vascular SMCs (PDGFRB+SM22+) show up from Day 6 onward.
In vitro NCC lineage-derived vascular SMC differentiation from hESCs was performed using another established protocol.21 Briefly, dissociated hESCs were seeded onto Matrigel-coated 6-well plates at 200 000 cells per well in mTeSR1 supplemented with 5 μM Y-27632 for 24 h (day −1). On Day 0, the medium was switched to NCC differentiation basic medium [DMEM/F12 supplemented with N2 supplement (Thermo Fisher Scientific), 0.1% BSA (Sigma) and 1% pen/strep] with 10 μM of a Smad2/3-specific inhibitor SB4315421 (Stemgent) and 1 μM of an Alk2/3 inhibitor LDN193189 (Stemgent). At Day 1, 3 μM CHIR99021 was added to the above medium. The medium was kept for 6 days without exchange. At Day 6, NCCs were showing up in culture and separated into single cells with Accutase. The dissociated cells were then transferred to Matrigel-coated 6-well plates at 1 million cells per well in NCC differentiation basic medium with 5 μM Y-27632. Twenty-four hours later, the medium was replaced with NCC-SMC differentiation basic medium [DMEM/F12, 20% knockout serum replacement (Thermo Fisher Scientific), 1% pen/strep] supplemented with 2 ng/mL TGF-β1 (Peprotech). The medium was changed to the fresh NCC-SMC medium with TGF-β1 every other day for 8 days. In a separate experiment, cells were simultaneously treated with 0.5 μM RA (Sigma) during Days 1–6 in mesodermal SMC differentiation or during Days 6–10 in NCC-derived SMC differentiation, in order to examine the effects of the enhanced RA signalling pathway in SMC differentiation.
2.6. Flow cytometry analysis
Flow cytometry analysis was performed for cells at Days 6 and 12 in CM differentiation, at Day 6 in mesodermal SMC differentiation, and at Day 14 in NCC-derived SMC differentiation. Cells were dissociated into single cells with Accutase for 5–10 min, washed in phosphate buffered saline, and blocked for 30 min in fluorescence-activated cell sorting (FACS) buffer (1% bovine serum albumin and 10% horse serum in PBS) at 4°C. For cells in SMC differentiation, staining for a cell surface antigen was first performed for 30 min at 4°C with a primary antibody, anti-platelet derived growth factor receptor-β (PDGFRB, an SMC marker; BD Biosciences). Cells were then fixed with 4% paraformaldehyde, permeabilized, blocked, and stained for intracellular antigens for 30 min at room temperature with the following primary antibodies: (CM differentiation) anti-Ki67-FITC (a proliferative marker; BD Biosciences), anti-ISL1-PE (a cardiac progenitor marker; BD Biosciences), and anti-TNNT2-APC (a CM marker; Miltenyi Biotec); and (SMC differentiation) anti-SM22 (an SMC marker; Abcam) followed by staining with an Alexa-Fluor 647-conjugated secondary antibody (BD Biosciences) for 15 min at 4°C. Flow cytometry analysis was then conducted with a flow cytometer (FACSARIA III) and FACS Diva software (Beckton Dickinson). The detailed gating strategies on flow cytometry analyses were previously described elsewhere.25 Flow cytometry data were analysed with FACS Diva and FlowJo software (Tree Star). All the primary antibodies used in flow cytometry analyses are listed in Supplementary material online, Table S1.
2.7. Clonal assay
Human ESC-derived STRA6+ single cells were sorted with FACS after cell staining with an anti-STRA6 primary antibody (Novus) followed by an Alexa-Fluor 647-conjugated secondary antibody on Day 3 in CM differentiation. The sorted cells were seeded onto fibronectin-coated 96-well plates at 1 cell per well in DMEM/F12 medium supplemented with 5 μM Y-27632 and 10% KnockOut Serum Replacement (KO-SR; Thermo Fisher Scientific). Growing clones from single cells were picked after 7 days in culture and trypsinized. Single clone-derived cells were plated into 3 wells of a 96-well plate for differentiation experiments and further cultured under the 3 different culture conditions, customized for the CM (RPMI/B27 with 2% KO-SR), SMC (SmGM-2, Lonza), or endothelial cell (EC) (EGM2, Lonza) differentiation, respectively, for additional 14 days.16,26 Medium was replaced thereafter every other day. After 14 days, cells were fixed and stained using primary antibodies of the specific markers for each cell type (see Supplementary material online, Table S1).
2.8. Immunostaining
Human embryonic hearts were snap-frozen and cryosectioned at 10 μm thickness. The heart sections or the hESC-derived cells were fixed with 4% paraformaldehyde, permeabilized in PBS with 0.1% saponin, and blocked in PBS with 1% bovine serum albumin and 10% horse serum. Samples were then stained with primary antibodies at 4°C overnight, followed by three washes with PBS and incubation with Alexa-Fluor 488-, 594-, and/or Alexa-Fluor 647-conjugated secondary antibodies (Molecular Probes) specific to the appropriate species for 60 min at room temperature. After three washes with PBS, nuclei were counterstained with DAPI (Sigma) or the slides were mounted in Vectashield Mounting Medium with DAPI (Vector Laboratories). All images were obtained using a Zeiss 710 confocal microscope and its imaging system. All the primary antibodies used at immunostaining are listed in Supplementary material online, Table S1.
2.9. Generation of STRA6-KO and TBX1 promoter-mutant hESCs by CRISPR-Cas9
The following sequence was selected as a single guide-RNA (sgRNA) which targets the second exon of the STRA6 gene locus and has minimal off-target activity, using the CHOPCHOP software (http://chopchop.cbu.uib.no/): 5′-AGGGAACCAGACCTCCCCCG(GGG)-3′. The sgRNA was cloned into a bicistronic expression vector pX459 expressing Streptococcus pyogenes Cas9, following a previously published protocol.14 Human ESCs (H9) were transiently co-transfected with pX459-sgRNA and a plasmid encoding a puromycin resistance gene using Human Stem Cell Nucleofector Kit (Lonza), according to the manufacturer’s instructions. After drug selection with 0.5 μg/mL puromycin for 2 days, single clones were obtained by re-plating transfected cell pools at low density (5000 cells per dish) on Matrigel-coated 10 cm dishes. Cells were allowed to grow for 6–10 days, until single colonies were big enough to pick and transferred to a 96-well plate. Monoclonal cell lines were then expanded, and genomic DNAs of each clone were isolated using the GeneJET Genomic DNA Purification Kit (Thermo Fisher Scientific). The CRISPR/Cas9-mediated gene edition on the STRA6 gene locus in each clone was confirmed by Sanger sequencing (Eurofins) and aligned to a WT sequence of the target region with SnapGene (GSL Biotech LLC) (see Supplementary material online, Figure S2). To confirm the absence of STRA6 protein expression of STRA6-KO hESC-derived cells on Day 6 in CM differentiation, flow cytometry and western blotting analyses were performed with an anti-STRA6 antibody (Novus).
Separately, to delete a specific region of 1953–1769 bp upstream from the transcription start site (TSS) of the human TBX1 gene, two sgRNAs were selected as follows: 5′-CCAACACCAAGGAGAACACG(TGG)-3′ and 5′-ATCGCAGGCAGTGTTTGCGG(TGG)-3′. As noted above, the two pX459-sgRNAs were then co-transfected into hESCs. Monoclonal cell lines were expanded in the same fashion and tested for the CRISPR/Cas9-mediated deletion of 184 bp fragment by PCR and Sanger sequencing.
2.10. Western blotting
Total cellular protein was extracted from cultured cells using radioimmunoprecipitation assay (RIPA) lysis buffer (Sigma) with a protease and phosphatase inhibitor cocktail (Thermo Scientific). Protein concentration was determined with a BCA protein assay kit (Thermo Scientific); 20 μg of protein lysates were loaded on 4–12% Bis-Tris gel (Thermo Scientific), separated by electrophoresis, and transferred onto a 0.2 μm nitrocellulose membrane with the Trans-Blot Turbo system (Biorad). The membranes were subsequently blocked in 5% skim milk in TBS-T (TBS with 0.1% Tween20) for 30 min at room temperature and incubated with primary antibodies in TBS-T containing 5% bovine serum albumin overnight at 4°C. The used primary antibodies were as follows: anti-β-actin-HRP (Cell Signaling Technology, 5125S), anti-GAPDH-HRP (Cell Signaling Technology, 8884S); anti-RARα (Cell Signaling Technology, 62294S); anti-RXRα (Cell Signaling Technology, 3085S); anti-STRA6 (Novus, NBP1-83719); and anti-TBX1 (Abcam, ab18530) (see Supplementary material online, Table S1). After washing with TBS-T, membranes were incubated with anti-mouse IgG or anti-rabbit IgG secondary antibodies conjugated with HRP (1:2000) in TBS-T containing 5% bovine serum albumin for 1 h at room temperature. After washing, membranes were incubated with SuperSignal West Femto Chemiluminescent Substrate (Thermo Scientific) for 1–5 min and imaged on a Chemidoc (Biorad). Image analysis and quantification was performed on Image Lab software version 6.1 (Biorad). The signal intensity was normalized to the expression of a loading control protein (β-actin or GAPDH) and translated to relative values.
2.11. In vitro population RNA sequencing and data analysis
cDNA libraries of the harvested population RNA samples, which were derived from both WT and STRA6-KO hESC-derived cells at Days 3, 6, and 12 in CM differentiation, were generated using Illumina TrueSeq mRNA (poly-A selection) kits. Each library was sequenced at 150 bp paired-end on an Illumina NovaSeq 6000 S4 instrument to a depth of 2–4 × 107 reads per sample. The quality of the fastq-format sequenced data was assessed using FASTQC, and raw reads were further trimmed and aligned onto human genome reference (hg38) using Cutadapt and STAR. Transcript levels were quantified as FPKM. Further normalization and differential expression analysis were conducted using edgeR and Limma programs on R/Bioconductor.16 The gene set enrichment analysis (GSEA) was performed on the GSEA software v4.1.0 (Broad institute).
2.12. RNA extraction and quantitative PCR
Total RNA was isolated using Direct-zol RNA Miniprep kit (Zymo Research) from cells at Days 3, 6, and 12 in CM differentiation, at Days 4 and 6 in mesodermal SMC differentiation, and at Days 6 and 14 in NCC-derived SMC differentiation. cDNA was synthesized from 0.5 μg of isolated total RNA with Maxima H Minus reverse transcriptase (Thermo Scientific) and used as a template for PCR amplification using primers specific to each of genes listed in Supplementary material online, Table S2. Quantitative PCR was performed using Powerup SYBR green master mix (Applied Biosystems) for 40 cycles on a 7500 Fast Real-Time PCR System (Applied Biosystems) under standard manufacturer’s conditions. Threshold cycles of each gene were normalized to the housekeeping gene GAPDH and translated to relative values.
2.13. Chromatin immunoprecipitation
For ChIP experiments, the nuclei from WT and STRA6-KO hESC-derived cells on Days 3 and 6 in CM differentiation were prepared, and the following chromatin digestion was performed using SimpleChIP Plus Enzymatic Chromatin IP Kit (Cell Signaling Technology), according to the manufacturer’s instructions. The lysates were immunoprecipitated with normal rabbit IgG, anti-RARα antibody (Cell Signaling Technology, 62294S), or anti-RARα antibody together with anti-RXRα antibody (Cell Signaling Technology, 3085S) at 4°C overnight. Immune complexes were incubated with Protein G Magnetic Beads for 2 h at 4°C with rotation. After washing and eluting chromatin from the magnetic beads, protein-DNA cross-linking was reversed in 5 M NaCl with 40 mg proteinase K by overnight incubation at 65°C. After a purification process, the precipitated DNA was amplified for fragments of the retinoic acid-response element (RARE) sites on the TBX1 promoter by quantitative PCR for 40 cycles on a 7500 Fast Real-Time PCR System (Applied Biosystems) under standard manufacturer’s conditions. Data are presented as fold enrichment compared with the IgG negative control. The following PCR primers were used: (Forward) 5′-GAGTAAAGGCCCAACACCAA-3′ and (Reverse) 5′-AAGGAGGCCGTTCCTGTTAC-3′; or (Forward) 5′-AACTAATCTCTCCAGGCCCC-3′ and (Reverse) 5′-GACCCTGCTCATATCTCCCC-3′, which were for detection of the putative RARE regions of 1884–1873 and 4496–4472 bp upstream from the TSS of the human TBX1 gene, respectively.
2.14. Karyotyping
WT and STRA6-KO hESC-derived cells were harvested and processed for standard G-Band karyotype analysis following the manufacturer’s instructions (Cell Guidance Systems).
2.15. Statistical analysis
Data are presented as mean ± standard deviation. Differences between groups were examined with Student’s t-test or one-way ANOVA followed by Tukey–Kramer post hoc test. Statistical significance is defined as P < 0.05. All bioinformatics analyses were performed using R/Bioconductor, as described above.
3. Results
3.1. Human embryonic heart single-cell RNA-seq analysis identified STRA6 as a novel OFT progenitor-associated gene
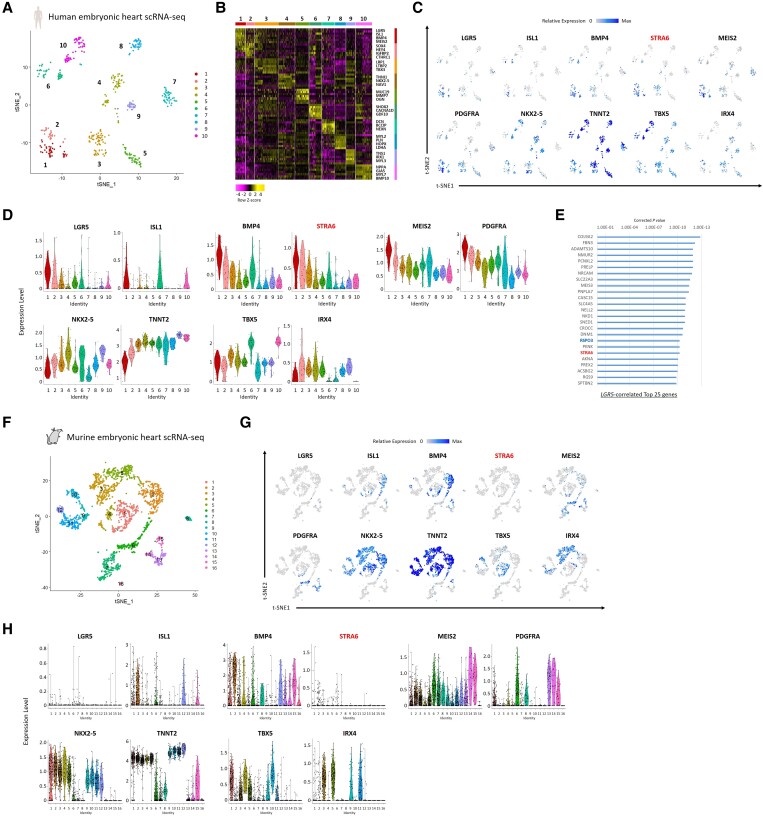
To elucidate the potential role of the STRA6 gene at the single-cell level in human cardiogenesis, we utilized a single-cell RNA-seq dataset, previously obtained from human embryonic heart samples (4.5–10 weeks of foetal ages).16,25 A total of the 458 individual cardiac cells derived from micro-dissected heart regions, i.e. OFT, atria, and ventricles, were segregated into 10 clusters including a cono-ventricular region-specific heart progenitor (CVP; Cluster #1) that appeared predominantly in OFT at the earlier stage (4.5–5.5 weeks of foetal age), by a dimensionality reduction method such as tSNE based on the differentially expressed genes’ profiles (Figure 1A and B). We then focused on the CVP-specific genes and found that interestingly STRA6 was differentially enriched in the CVP cluster, similarly to other early cardiogenic and SHF/OFT progenitor markers, such as ISL1,26BMP4,27MEIS2,28PDGFRA,29 and LGR516 (Figure 1C and D). To assess the significance of the co-expression of genes in all cells, we performed Guilt-by-Association and correlation analysis14 and identified that STRA6, as well as RSPO3 (a ligand for a Wnt signalling receptor LGR5), was highly co-expressed in the LGR5+ cells that emerged specifically in the early-staged OFT,16 suggesting that STRA6 would play a role in OFT development on human cardiogenesis (Figure 1E). To validate the findings of single-cell RNA-seq data obtained from human embryonic hearts, we also analysed the previously obtained single-cell RNA-seq dataset16 of in vitro hESC-derived cells during CM differentiation (Days 3–15) based on a well-established CM differentiation protocol with Wnt signalling modulation.19,25 The 366 high-quality individual cells were segregated into 6 clusters (see Supplementary material online, Figure S3A), in which we identified that Cluster #2 was occupied by cells at Day 3 and expressed Mesp1 entirely, thereby termed as ‘Mesp1+ cardiac precursors’ (see Supplementary material online, Figure S3B–D). Of interest, the majority of cells (≈60%) in Cluster #2 also expressed STRA6 (see Supplementary material online, Figure S3C and D), which supports the notion that STRA6+ progenitors would play a certain role in human cardiogenesis.
Figure 1.
Single-cell RNA-seq analyses of human and murine embryonic hearts. (A) The tSNE analysis using the single-cell RNA-seq dataset, which was obtained from human embryonic hearts (4.5–10 weeks of foetal ages),16 segregated 458 individual cardiac cells into 10 molecularly distinct clusters, including a cono-ventricular region-specific heart progenitor (CVP; Cluster #1). (B) A heatmap image showing the representative differential expression genes in each of the 10 clusters in (A). (C) Feature plots of the SHF/OFT marker (CVP-enriched) genes as well as the pan-cardiac/FHF/developing CM marker genes with STRA6 on the tSNE plots in (A). (D) Violin plots of the same genes as in (C), in the segregated 10 clusters of the human embryonic heart-derived single cells. (E) The top 25 genes correlated with the expression of the CVP-specific gene LGR5 in single-cell RNA-seq data of human embryonic hearts. Corrected P-value for each gene was calculated by Guilt-by-Association and correlation analysis.14 (F) The tSNE analysis segregated a total of 2079 single cardiac cells, obtained from embryonic hearts (E9.5, E10.5, and E14.5) of the Mesp1+ lineage tracing mice (Mesp1Cre/+; Rosa26tdTomato), into 16 clusters including FHF progenitors (Cluster #1) and SHF progenitors (Cluster #2). (G) Feature plots of the SHF/pan-cardiac/FHF/developing CM marker genes with STRA6 on the tSNE plots in (F). (H) Violin plots of the same genes as in (G), in the segregated 16 clusters of the murine embryonic heart-derived single cells.
To further gain insights in regard to the STRA6 gene’s role in mammalian cardiogenesis, we next analysed single-cell RNA-seq data that were uniquely obtained from embryonic hearts of the Mesp1+ lineage tracing mice (Mesp1Cre/+; Rosa26tdTomato) (see Supplementary material online, Figure S1), harvested on E9.5, E10.5, and E14.5. A total of 2079 single cardiac cells that passed the quality control tests were segregated into 16 clusters, including first heart field (FHF; Cluster #1) and SHF (Cluster #2) progenitors, by tSNE using the Seurat program24 (Figure 1F). A pan-cardiac marker NKX2-5 was broadly expressed in early and late CM clusters (Clusters #1–5 and #9–12, respectively) including the FHF and SHF, while an FHF marker TBX530 and a developing ventricular CM marker IRX431 were enriched predominantly in the FHF (Cluster #1) and the early/late ventricular CMs (Clusters #3, 5, 9, and 11), respectively (Figure 1G and H). As expected, the SHF/OFT progenitor markers ISL1 and BMP4 were enriched in the SHF (Cluster #2). Importantly, we found that as contrasted to the expression patterns in human embryonic hearts, STRA6 was much less frequently expressed in murine embryonic heart cells, irrespective of the heart regions and stages (Figure 1G and H). We then analysed single-cell RNA-seq data of the Mesp1+ lineage cells obtained from murine whole embryos (Mesp1Cre/+; Rosa26tdTomato) at the earlier stage (E8.25). A total of 768 single cells were segregated into 11 clusters, including heart cells (Cluster #4) and other organ cells (see Supplementary material online, Figure S4A and B). We observed that although STRA6 was often expressed in cells of somite/neural tube (Cluster #3), it was less frequently expressed in the heart cells at this early stage again. STRA6 was only expressed in cells that were <5% of the FHF (NKX2-5+) or the SHF (ISL1+) cells, respectively (see Supplementary material online, Figure S4C and D). These results suggest that the role of STRA6 in murine embryonic heart development would not be as essential as that in human embryonic heart development.
3.2. Human STRA6+ heart progenitors differentiate into CMs and vascular SMCs in vitro and in vivo
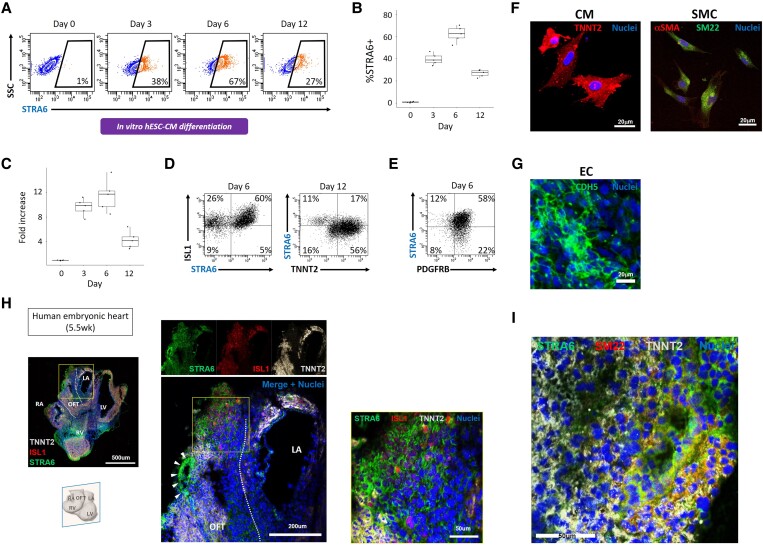
We subsequently investigated a sequential expression pattern of STRA6 protein in the in vitro hESC cardiogenesis. We observed that STRA6+ cells started to emerge at Days 2 and 3 in hESC-CM differentiation and peaked at Day 6 when those cells occupied ∼60% among the total cells in culture (Figure 2A and B). These results in flow cytometry analysis were corroborated by a sequential expression pattern of STRA6 mRNA in cells during CM differentiation, measured with quantitative PCR (Figure 2C). This expression pattern of STRA6 is quite similar to that of the SHF/OFT progenitor marker ISL1, as shown below and in the previous study.16 In fact, the majority of STRA6+ cells were co-expressing ISL1 at Day 6 in CM differentiation (Figure 2D). On Day 12 when STRA6 expression was already peaked out but a CM marker TNNT2 expression reached its peak, more than half of STRA6+ cells were also co-expressing TNNT2 (Figure 2D). On the other hand, during mesodermal SMC differentiation, the majority of STRA6+ cells were also co-expressing an SMC marker PDGFRB at Day 6 (Figure 2E; also see below). To explore the multipotency for STRA6+ cells to differentiate into the three major cardiac cell lineages such as CMs, SMCs, and ECs, we performed a clonal assay using live STRA6+ cells that were harvested with FACS at Day 3 in hESC-CM differentiation and cultured under the three different culture conditions, customized for CM, SMC, and EC differentiation.16,26 We found that the STRA6+ single cell-derived clones could differentiate easily into CMs and SMCs (Figure 2F), but could not differentiate into ECs. In contrast, STRA6-negative cell-derived clones could differentiate into ECs (Figure 2G), as well as CMs and SMCs. This suggests that STRA6+ cells would have dual potency to differentiate into the CM and SMC lineages but not an EC lineage in in vitro cardiogenesis. To corroborate the in vitro findings, we then employed immunostaining of human embryonic sectioned hearts (Figure 2H). We observed that STRA6+ cells appeared predominantly in the OFT region of the early-staged heart (5.5 weeks of foetal age), often co-localizing with an SHF/OFT progenitor marker ISL1 and/or a CM marker TNNT2 (Figure 2H, middle and right). Of particular interest, some of ISL1−TNNT2−STRA6+ cells in OFT were shown to construct a vascular structure in this region (Figure 2H, arrowheads in middle). In fact, these ISL1−TNNT2−STRA6+ cells co-expressed an SMC marker SM22 and were thereby considered as vascular SMCs (Figure 2I). This may reflect the myocardial-to-arterial phenotypic change associated with the OFT development, i.e. progression of OFT septation and remodelling to form early aorta and pulmonary artery, as indicated in the previous reports.32,33
Figure 2.
Human STRA6+ heart progenitors in in vitro and in vivo cardiogenesis. (A) Flow cytometry analysis showing the time course of STRA6 protein expression levels in in vitro hESC-derived cells during CM differentiation. SSC, side scatter. (B) Statistical analysis of the %STRA6+ population on FACS in (A). (C) Quantitative PCR results of STRA6 mRNA expression in hESC-derived cells during CM differentiation. (D) Flow cytometry analysis for detection of ISL1+ and/or STRA6+ cells at Day 6 (left) and of TNNT2+ and/or STRA6+ cells at Day 12 (right) in CM differentiation. (E) Flow cytometry analysis for detection of PDGFRB+ and/or STRA6+ cells at Day 6 in mesodermal SMC differentiation. (F) Differentiated CMs (left) and SMCs (right) derived from the STRA6+ progenitor clones that were initially harvested on Day 3 in hESC-CM differentiation in the clonal assay. (G) Differentiated ECs derived from the STRA6− cell-derived clone initially harvested on Day 3 in the clonal assay. (H) Immunohistochemistry of the sectioned human embryonic heart at 5.5 weeks of foetal age. The middle images are the enlarged ones of a yellow square in the left image. The right image is the enlarged one of a yellow square in the middle image. Coronal view. The white dotted lines (left and middle) indicate the OFT structure. Arrowheads (middle) point to the ISL1−TNNT2−STRA6+ vascular structure, indicating dividing OFT. In contrast, a number of STRA6+ cells co-expressing ISL1 and/or TNNT2 are also seen in the OFT region (right). LA, left atria; LV, left ventricle; OFT, outflow tract; RA, right atria; RV, right ventricle. (I) The ISL1−TNNT2−STRA6+ cells in the vascular structure in OFT in (H) co-expressed an SMC marker SM22.
3.3. STRA6 deletion in hESCs attenuates induction of vascular SMCs, but not of CMs, in the in vitro differentiation
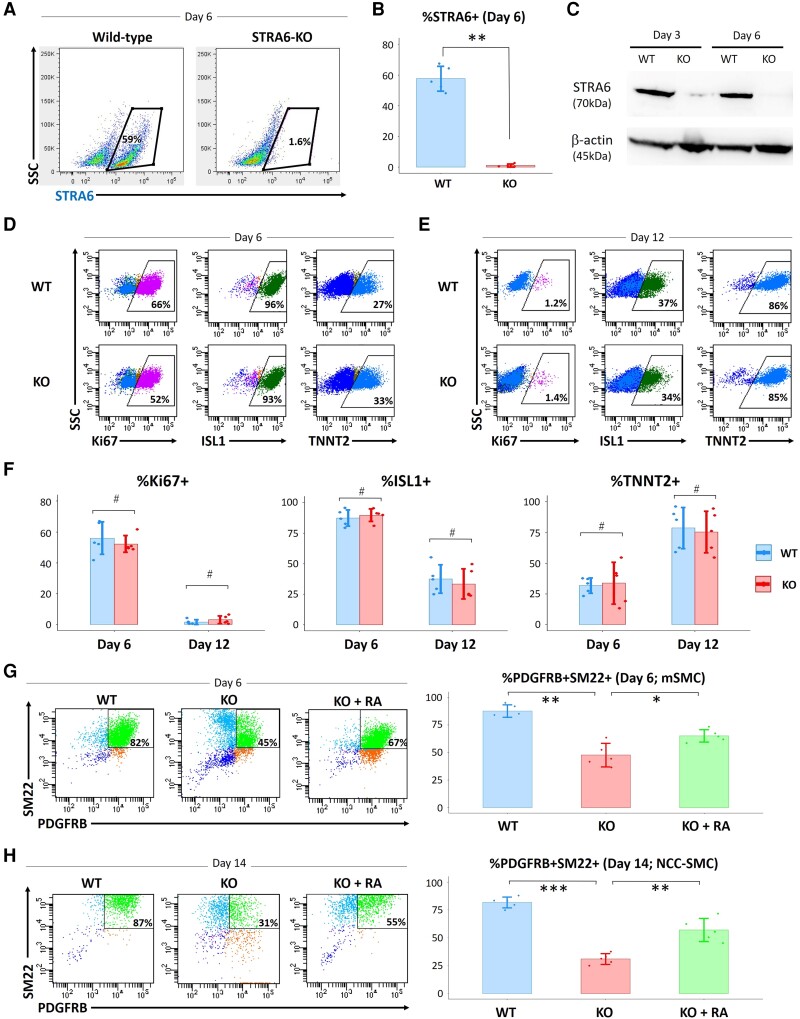
Next, to test the essential role of STRA6 in human cardiogenesis and vasculogenesis, we generated STRA6-knockout (STRA6-KO) hESC lines for the loss-of-function experiments. Through CRISPR-Cas9 technology,18 we established the STRA6-KO hESC clone, which had frameshift mutations in the second exon of the STRA6 gene in both alleles (see Supplementary material online, Figure S2). Little or no expression of STRA6 protein in the STRA6-KO hESC-derived cells on Day 6 in CM differentiation was confirmed by flow cytometry (Figure 3A and B) and western blotting analyses (Figure 3C). We also validated a normal karyotype for both the WT and STRA6-KO hESC lines used for further differentiation experiments (see Supplementary material online, Figure S5A). In addition, no mutations on putative off-target sites that had the highest similarity with three base pair mismatches to the target sequence of the sgRNA were observed in the STRA6-KO hESCs (see Supplementary material online, Figure S5B). We then employed the STRA6-KO hESC clone into the same CM differentiation protocol,19,25 to detect any cardiogenic phenotype in regard to STRA6 deletion. For this, we measured the ratios of cells that were positive for a cell proliferation marker Ki67, an SHF/heart progenitor marker ISL1, and a differentiated CM marker TNNT2 on Days 6 and 12 in CM differentiation by flow cytometry analysis (Figure 3D and E). In this protocol, while ISL1 expression peaks on Day 6, occupying around 90% of the total cells in culture, TNNT2 expression reached around 80% of the total cells on Day 12. Contrary to our expectation, %ISL1+ and %TNNT2+ on Days 6 and 12 did not change between WT and STRA6-KO hESC-derived cells, although STRA6-KO cells showed a bit lower %Ki67+ than WT cells on Day 6 (but not statistically significant) (Figure 3D–F). Likewise, the numbers of generated beating CMs (TNNT2+) derived from WT or STRA6-KO hESCs were comparable (data not shown).
Figure 3.
Impacts of STRA6 deletion in the in vitro hESC differentiation into CMs and SMCs. (A and B) Flow cytometry analysis showing the ratios of wild-type (WT) and STRA6-KO hESC-derived STRA6+ cells at Day 6 in CM differentiation. SSC, side scatter. **P < 0.01. (C) Western blotting analysis for expression of STRA6 protein with β-actin protein (a loading control) in WT and STRA6-KO cells at Days 3 and 6 in hESC-CM differentiation. (D and E) Representative images on flow cytometry analysis showing the ratios of a cell proliferation marker Ki67+ (left), an SHF/heart progenitor marker ISL1+ (middle), and a differentiated CM marker TNNT2+ (right) in WT (top) and STRA6-KO (bottom) cells at Days 6 (D) and 12 (E) in hESC-CM differentiation. (F) Statistical data of the ratios of %Ki67+ (left), %ISL1+ (middle), and %TNNT2+ (right) in (D) and (E). #P = not significant. (G) Flow cytometry analysis and statistical data showing the ratios of vascular SMCs (PDGFRB+SM22+) at Day 6 in mesodermal SMC (mSMC) differentiation of WT and STRA6-KO hESCs with or without treatment with retinoic acid (RA, 0.5 μM) during Days 1–6. *P < 0.05 and **P < 0.01. (H) Flow cytometry analysis and statistical data showing the ratios of vascular SMCs (PDGFRB+SM22+) at Day 14 in NCC-derived SMC differentiation of WT and STRA6-KO hESCs with or without treatment with RA (0.5 μM) during Days 6–10. **P < 0.01 and ***P < 0.001. Differences between groups were examined with one-way ANOVA followed by Tukey–Kramer post hoc test.
On the other hand, we also tested WT and STRA6-KO hESCs for evaluating their capabilities to differentiate into vascular SMCs, using the previously established protocols for SMC differentiation.20,21 In line with the previous reports, the mesodermal lineage-derived or the NCC linage-derived vascular SMCs (PDGFRB+SM22+) from WT hESCs are obtained at Day 6 in mesodermal SMC or at Day 14 in NCC-SMC differentiation, respectively, occupying ≥80% of the total cultured cells in both differentiation (Figure 3G and H). Of particular interest, STRA6-KO hESC-derived cells exhibited lower efficacies for induction of both the mesodermal and NCC lineage-derived SMCs [mesodermal lineage-SMCs (Day 6): WT, 87.7 ± 5.7% vs. KO_clone, 47.7 ± 10.6%, P < 0.01 (Figure 3G); NCC lineage-SMCs (Day 14): WT 81.8 ± 4.8% vs. KO_clone, 31.1 ± 4.9%, P < 0.001 (Figure 3H)]. Interestingly, treatment with RA (0.5 μM) in STRA6-KO cells during Days 1–6 in mesodermal SMC and Days 6–10 in NCC-SMC differentiation partially rescued for induction of both SMCs, respectively (Figure 3G and H). These results suggest that STRA6 would be essential for induction of both the mesodermal and NCC-derived vascular SMCs from hESCs in vitro.
3.4. Population RNA-seq analysis demonstrates that STRA6 deletion leads to the down-regulation of vascular smooth muscle cell-related genes’ expression in cardiac differentiating cells
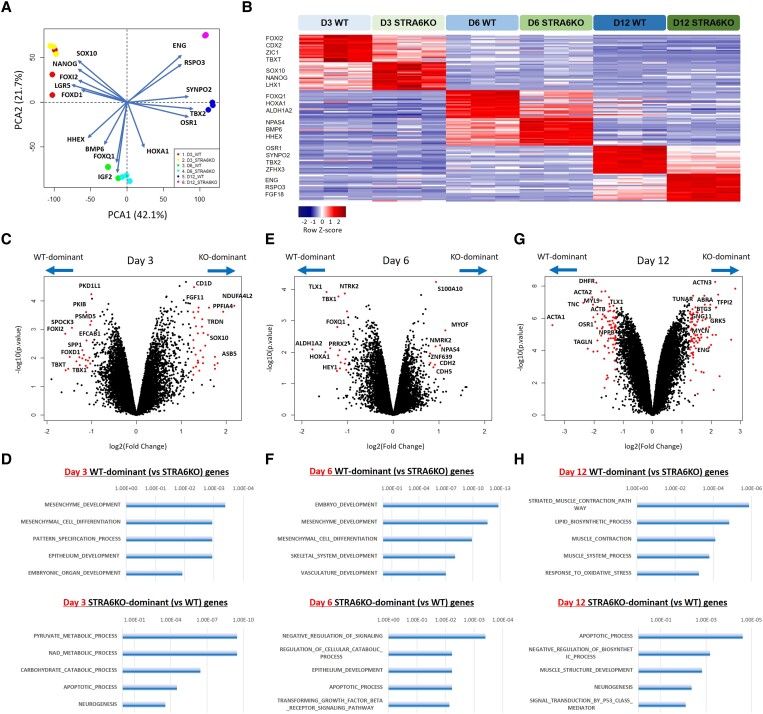
To clarify molecular signatures in human STRA6-mediated cardiac development, we analysed population RNA-seq data obtained from both WT and STRA6-KO hESC-derived cells on Days 3, 6, and 12 in CM differentiation. PCA and differential gene expression analysis clearly segregated the two cell lines in a stage-dependent manner (Figure 4A and B). We then compared directly the transcriptomes of the two cell lines on the same differentiation day with the limma package34 in R/Bioconductor and the GSEA on the GSEA software (Broad Institute; http://www.gsea-msigdb.org/gsea/) (Figure 4C–H). On Day 3, the genes up-regulated in WT cells compared with STRA6-KO cells were enriched for gene ontology (GO) terms such as ‘mesenchyme development’, ‘mesenchymal cell differentiation’, ‘pattern specification process’, and ‘embryonic organ development’, which contained genes such as FOXD1, TBXT, SEMA3D, GBX2, TBX1, ZIC1, etc. In contrast, the genes up-regulated in STRA6-KO cells compared with WT cells on Day 3 were enriched for GO terms such as ‘pyruvate metabolic process’, ‘apoptotic process’, and ‘neurogenesis’, which contained genes such as ENO1, ARNT, LHX1, MYC, NGFR, etc. (Figure 4C and D and Supplementary material online, Table S3). These findings suggest that STRA6 would be critical for mesenchyme formation and remodelling at the early stage of embryogenesis, and that STRA6 deletion might shift the hESC-derived differentiating cell trajectories towards the ectoderm lineages (e.g. neurons) from the originally destined mesoderm lineages, at least in part. This tendency can also be observed in the comparison of the two transcriptome datasets on Day 6 (Figure 4E and F). Further, on Day 6, RA signalling targets such as homeobox-containing Hox genes HOXA1, HOXB2, and HOXB3,35 as well as ALDH1A2, a key enzyme for the synthesis of endogenous RA36 reached peak expression and were significantly down-regulated in STRA6-KO cells compared with WT cells, respectively, likely reflecting the impacts of deletion of the upstream gene STRA6 (Figure 4E and F and Supplementary material online, Table S3). Importantly, on Day 12 when marker genes of differentiated cardiac cells such as CMs, SMCs, and ECs reached peak expression, the genes up-regulated in WT cells compared with STRA6-KO cells were enriched for GO terms such as ‘striated muscle contraction pathway’, ‘muscle contraction’, and ‘muscle system process’, which contained genes such as CALM1, ACTA1, MYL3, ACTA2, CNN1, MYL9, etc. (Figure 4G and H and Supplementary material online, Table S3).
Figure 4.
Population RNA-seq analysis showcases clearly differential molecular signatures between WT and STRA6-KO cells during CM differentiation. (A) The principal component analysis and the biplot using the 18 population RNA-seq data of WT and STRA6-KO cells harvested at Days 3, 6, and 12 in hESC-CM differentiation (3 biological replicates). (B) Differential gene expression analysis of the six cell groups, i.e. WT and STRA6-KO hESC-derived cells at Days 3, 6, and 12 in CM differentiation. A heatmap showing the representative differential expression genes in each of the six groups. (C, E, and G) Volcano plots visualizing differential gene expression analysis with the limma package34 between WT and STRA6-KO hESC-derived cells at Days 3 (C), 6 (E), and 12 (G) in CM differentiation, respectively. For each gene, the average difference [log2(Fold change)] between the cell groups on the same day was plotted against the power to discriminate between groups [−log10(P-value)]. Top-scoring genes for both metrics are indicated as red dots, and representative differential expression genes’ names are labelled. (D, F, and H) The gene set enrichment analysis (GSEA) was performed using the top 250 WT or STRA6-KO cell-enriched genes with the GSEA software (Broad Institute; http://www.gsea-msigdb.org/gsea/). Bar graphs showing the representative gene ontology (GO) terms specific to WT (top) or STRA6-KO cells (bottom) at Days 3 (D), 6 (F), and 12 (H), respectively.
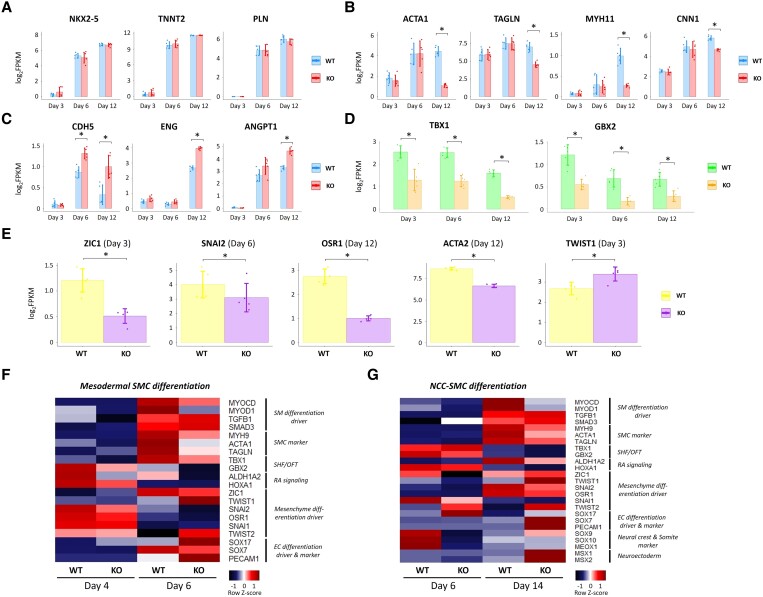
We then compared population RNA-seq data between WT cells on Day 12 in CM differentiation (around 80% TNNT2+) and the previously obtained ISL1−TNNT2+ cells (i.e. differentiated CMs) purified by FACS on Day 10 in CM differentiation,16 and found that expression levels of many of representative CM markers were quite comparable between the two populations (see Supplementary material online, Figure S6), indicating the WT cells on Day 12 in the CM differentiation protocol would nearly represent differentiated CMs, although containing minor other cell populations. Between the WT and STRA6-KO cells, of interest, while there were no differences in expression levels of the CM marker genes such as NKX2-5, TNNT2, MYH6, and PLN, the SMC marker genes such as ACTA1, ACTA2, TAGLN, MYH11, and CNN1 were significantly down-regulated in STRA6-KO cells compared with WT cells on Day 12 (Figure 5A and B). About other CM-related genes, however, several cardiac maturation marker genes (e.g. CACNA1C, MYL2, NPPB, IRX5, etc.) and several atrial CM marker genes (e.g. KCNA5, ALDH1A2, DHRS9, PITX2, etc.) were down-regulated in STRA6-KO cells than WT cells on Day 6 or 12 in CM differentiation (see Supplementary material online, Figure S7A and B), indicating partial impairment in both CM maturation and atrial CM induction in the STRA6-KO line. As contrasted to the down-regulation in the SMC marker genes, the EC marker genes such as CDH5, ENG, HHEX, and ANGPT1 were up-regulated in STRA6-KO cells on Day 12 (Figure 5C). These results suggest that STRA6 would be essential for induction of vascular SMCs from hESCs in vitro and that STRA6 deletion might shift the hESC-derived differentiating cell trajectories towards the EC lineage from the originally destined SMC lineage at the late stage in part.
Figure 5.
STRA6 deletion down-regulates expression of vascular SMC-related genes and mesenchyme differentiation drivers in cells during CM and SMC differentiation. (A–C) Comparisons of the CM (A), SMC (B), and EC (C) marker genes’ expression between WT and STRA6-KO cells at Days 3, 6, and 12 in hESC-CM differentiation. *P < 0.05. (D) Comparisons of the TBX1 and GBX2 genes’ expression between WT and STRA6-KO cells at Days 3, 6, and 12 in hESC-CM differentiation. *P < 0.05. (E) Comparisons of gene expression of ZIC1 (Day 3), SNAI2 (Day 6), OSR1 (Day 12), ACTA2 (Day 12), and TWIST1 (Day 3) between WT and STRA6-KO cells in CM differentiation. *P < 0.05. Differences between groups were examined with Student’s t-test. (F and G) Quantitative PCR-based gene expression heatmap of the representative SMC differentiation-related genes between WT and STRA6-KO cells at Days 4 and 6 in mesodermal SMC differentiation (left) and at Days 6 and 14 in NCC-SMC differentiation (right).
Next, we observed that through Days 3–12, an SHF/OFT-specific TF TBX1 and its downstream target TF GBX237 were significantly down-regulated in STRA6-KO cells compared with WT cells (Figure 5D). Among other SHF marker genes, LGR5 (Day 3), PDGFRA (Day 6), and MEIS2 (Day 6) were also down-regulated in STRA6-KO cells, whereas there were no significant differences in ISL1 and BMP4 expression between the STRA6-KO and WT cells (see Supplementary material online, Figure S7C). We did not see any differences in expression levels of a critical myogenic TF MYOD1 or MYOCD38 between the two cell lines (data not shown). Instead, of interest, a cardiac NCC migration TF ZIC139 on Day 3, an epithelial-to-mesenchymal transformation (EMT)-promoting TF SNAI240 on Day 6, and a mesenchymal differentiation-promoting TF OSR141 on Day 12, as well as an SMC marker ACTA2 on Day 12, were significantly down-regulated in STRA6-KO cells, while a myogenic differentiation-inhibiting TF TWIST142 were up-regulated in STRA6-KO cells on Day 3 (Figure 5E).
Finally, to further gain insights in regard to the impacts of STRA6 deletion in cardiovascular differentiation, we analysed expression levels of the SMC differentiation-related genes in WT and STRA6-KO cells at Days 4 and 6 in mesodermal SMC differentiation and at Days 6 and 14 in NCC-derived SMC differentiation with quantitative PCR analysis. Interestingly, SMC differentiation drivers MYOD1 and MYOCD, as well as other SMC markers, were down-regulated in STRA6-KO cells during both mesodermal SMC (Day 6) and NCC-derived SMC differentiation (Day 14) (Figure 5F and G). As seen in the RNA-seq results during CM differentiation, TBX1/GBX2 and several mesenchyme differentiation drivers (e.g. ZIC1, SNAI2, OSR1, and SNAI1) were down-regulated in STRA6-KO cells, while myogenic differentiation-inhibiting TFs (e.g. TWIST1 and TWIST2) were up-regulated in STRA6-KO cells in both mesodermal and NCC-derived SMC differentiation. In contrast with the SMC differentiation drivers and markers, the EC differentiation drivers and markers (e.g. SOX17, SOX7, and PECAM1) were up-regulated in STRA6-KO cells in both mesodermal and NCC-derived SMC differentiation, which further corroborates the RNA-seq results obtained during CM differentiation. In addition, during NCC-SMC differentiation, NCC and somite markers (e.g. SOX9, SOX10,43 and MEOX144) were down-regulated in STRA6-KO cells on Day 6, while neuroectoderm markers (e.g. MSX1 and MSX245) were up-regulated in STRA6-KO cells on Day 14, suggesting partial impairment in proper induction of NCCs in the early stage and a cell-fate shift from the SMC to the neuroectoderm lineages in the late stage due to STRA6 deletion, which may explain lesser induction of SMCs in the NCC-derived SMC differentiation protocol (Figure 3H).
3.5. TBX1 is a novel target of STRA6-mediated RA signalling
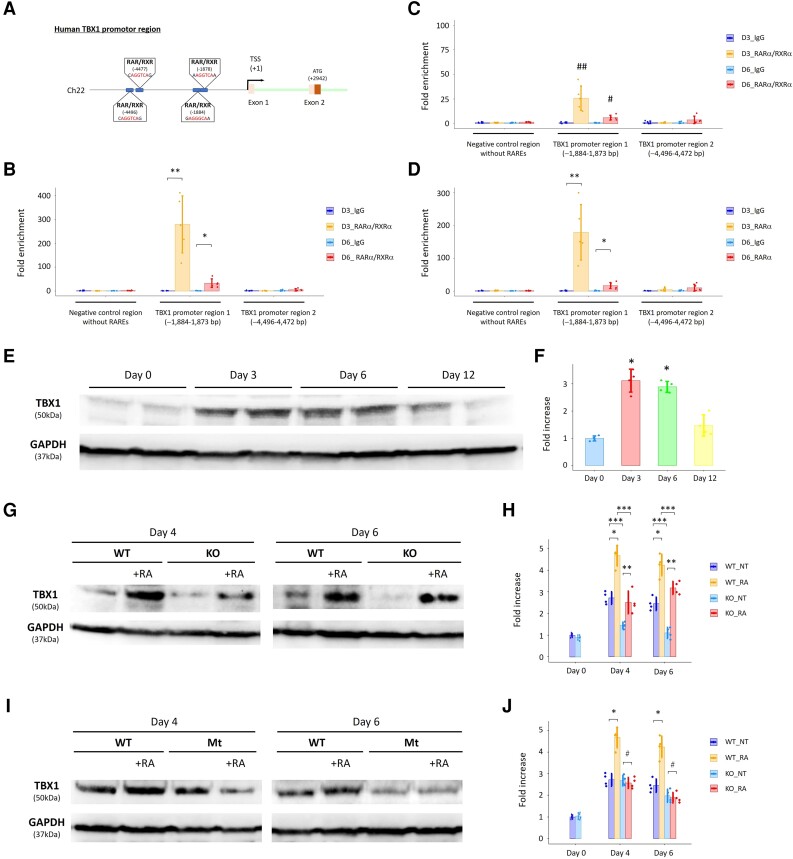
To further elucidate molecular machinery in human STRA6-mediated cardiac development, we explored a previously unrecognized transcription network behind STRA6 and RA signalling pathway. We focused on an SHF/OFT-specific cardiogenic TF TBX1,22 as TBX1 was significantly up-regulated in WT cells compared with STRA6-KO cells throughout culture periods (notably, on Days 3 and 6 in CM differentiation) (Figures 4C, E, and 5D), implying that TBX1 may function downstream of STRA6 in the in vitro cardiac differentiation. Intracellular RA synthesized from retinaldehyde by a key enzyme Aldh1a2 binds to RA nuclear receptors RAR/RXR, which bind to the RARE composed of tandem 5′-AGGTCA-3′ sites with a coactivator complex, and promotes transcription of the RA signalling target genes.36 Through the TF motif analysis with the Jaspar database (http://jaspar.genereg.net/) and the MatInspector software (Genomatix, http://www.genomatix.de/), we found the novel RARE sites, which are 4496–4472 bp and 1884–1873 bp upstream of the TSS of the human TBX1 gene on the TBX1 promoter region (Figure 6A). Then, ChIP assays with antibodies specific to RARα and RXRα were performed using extracts derived from WT hESC-derived cells on Days 3 and 6 in CM differentiation. We first confirmed successful and specific protein immunoprecipitation by anti-RARα and anti-RXRα antibodies by western blotting analysis (see Supplementary material online, Figure S8A). We subsequently revealed that recruitment of RARα/RXRα complexes onto one of the novel RARE sites on the TBX1 promoter (−1884–1873 bp) was augmented on Days 3 and 6 (notably on Day 3) (Figure 6B). The ChIP assays were then performed using extracts derived from STRA6-KO hESC-derived cells, which expressed RARα and RXRα proteins equally to WT hESC-derived cells during CM differentiation (see Supplementary material online, Figure S8B). We found that RARα/RXRα complexes were recruited to the same RARE site (−1884–1873 bp) on Days 3 and 6 but to a much lesser degree than in WT cells, suggesting less amount and function of intranuclear RA due to the STRA6 deletion (Figure 6C). To validate specific binding of RARα protein to the identified RARE site, we also performed the ChIP assays using only the anti-RARα antibody and extracts from WT cells, and similarly, showed recruitment of RARα onto the RARE site on the TBX1 promoter (−1884–1873 bp) (Figure 6D). Consistent with the findings in the ChIP assays, the Guild-by-Association and correlation analysis using the human embryonic heart single-cell RNA-seq data (Figure 1A) revealed that TBX1 expression was highly positively co-related to STRA6 expression (corrected P-value: 1.62E−14), as shown in the feature plots on the tSNE plots for TBX1 as well as other RA signalling genes (see Supplementary material online, Figure S9). These indicate that the STRA6-mediated RA signalling pathway might enhance TBX1 expression in the early stage of cardiogenesis. Of interest, this RARE site (−1884–1873 bp) can be seen on the human genome sequence, but not on the murine genome sequence, indicating evolutionary divergences among these species.
Figure 6.
ChIP assays highlight the human-specific binding site of RARα/RXRα complexes on the TBX1 promoter region. (A) Schema showing novel (human-specific) and putative RAR/RXR binding sites (i.e. RARE) on the human TBX1 promoter region. (B) The ChIP assays demonstrated that recruitment of RARα/RXRα complexes onto one of the novel RARE sites of the human TBX1 promoter (−1884–1873 bp) was augmented at Days 3 (D3) and 6 (D6) in CM differentiation of WT hESCs. *P < 0.01 and **P < 0.0001 vs. IgG (negative control). (C) The ChIP assays using STRA6-KO cells demonstrated that RARα/RXRα complexes were also recruited onto the identified RARE site of the human TBX1 promoter (−1884–1873 bp) at Days 3 and 6 in CM differentiation but to a much lesser degree compared with that in WT cells in (B). #P < 0.01 vs. IgG and vs. D6_RARα/RXRα in WT (B). ##P < 0.001 vs. IgG and vs. D3_RARα/RXRα in WT (B). (D) The ChIP assays using only the anti-RARα antibody demonstrated that recruitment of RARα onto the identified RARE site of the human TBX1 promoter (−1884–1873 bp) was augmented at Days 3 and 6 in CM differentiation of WT hESCs again. *P < 0.01 and **P < 0.0001 vs. IgG. (E) Western blotting analysis for expression of TBX1 protein with GAPDH protein (a loading control) in WT cells at Days 0, 3, 6, and 12 in hESC-CM differentiation. (F) Quantitative results in (E). *P < 0.01 vs. Day 0. (G) Comparison of expression of TBX1 protein between WT and STRA6-KO cells at Days 4 and 6 in hESC-CM differentiation with or without treatment with retinoic acid (RA, 0.5 μM) during Days 3–7. GAPDH was used as a loading control. (H) Quantitative results in (G). NT, normal treatment (without adding RA). *P < 0.01 between the NT-administered and RA-co-administered WT cells at Days 4 and 6, respectively. **P < 0.05 between the NT-administered and RA-co-administered STRA6-KO cells at Days 4 and 6, respectively. ***P < 0.01 between WT and STRA6-KO cells under the same treatment conditions at Days 4 and 6, respectively. (I) Comparison of expression of TBX1 protein between WT and TBX1 promoter-mutant (Mt) cells at Days 4 and 6 in hESC-CM differentiation with or without treatment with RA (0.5 μM) during Days 3–7. GAPDH was used as a loading control. (J) Quantitative results in (I). *P < 0.01 between the NT-administered and RA-co-administered WT cells at Days 4 and 6, respectively. #P = not significant between the NT-administered and RA-co-administered TBX1 promoter-mutant cells at Days 4 and 6, respectively. Differences between groups were examined with one-way ANOVA followed by Tukey–Kramer post hoc test.
To further clarify expression levels of TBX1 in CM differentiation, we performed western blotting analysis of TBX1 protein using WT and STRA6-KO hESC-derived cells. TBX1 protein was strongly expressed in WT cells on Days 3 and 6 to a higher degree than on Day 0 (Figure 6E and F). Consistent with the RNA-seq results, STRA6-KO cells showed lower expression of TBX1 protein than WT cells (Figure 6G and H). Interestingly, when we treated cells with RA (0.5 μM; Days 3–7), TBX1 expression was significantly increased at Days 4 and 6 in both WT and STRA6-KO cells (Figure 6G and H). These findings support the notion that TBX1 would act downstream to STRA6-mediated RA signalling in human cardiogenesis. Next, when the identified RARE site (−1884–1873 bp) was deleted by two sgRNAs and CRISPR-Cas9 technology (Section 2) (see Supplementary material online, Figure S10), TBX1 expression was not anymore enhanced with RA treatment (0.5 μM; Days 3–7) (Figure 6I and J), suggesting that RARα/RXRα complexes and the identified RARE site would function to enhance TBX1 expression in an additive or synergistic manner during cardiac differentiation.
Collectively, we newly identified the previously unrecognized gene regulatory network, involving an interaction between RARα/RXRα complexes and TBX1, which would play an indispensable role in human STRA6-mediated cardiogenesis and vasculogenesis.
4. Discussion
Human CHDs affect 1/100 live births and cause more deaths in the first year of life than any other birth defects, and cardiac OFT defects are the most common CHD with a prevalence of 30%.6 A diverse set of heart progenitors such as SHF progenitors and cardiac NCCs and their paracrine molecular cues drive cardiac OFT formation including septation and remodelling.1,2 The RA signalling plays a critical role in OFT development, and dysregulation of this signalling pathway is responsible for various types of OFT defects.3,5 Inactivating mutations in STRA6, a transmembrane receptor regulating vitamin A and RA metabolism can cause Matthew-Wood (also termed PDAC) syndrome, characterized by micro-/anophthalmia, pulmonary hypoplasia, diaphragmatic hernia, and cardiac malformation. The patients with Matthew-Wood syndrome, however, show heterogeneous phenotypes, ranging from isolated micro-/anophthalmia [100% of STRA6 mutation-confirmed/inferred cases (40/40) showed ocular malformations] to complex presentations involving other malformations in cardiac (53.5%), pulmonary (44.2%), diaphragmatic (25.6%), and renal (20.9%) systems.7,8 Although the Human Gene Mutation Database listed more than 27 unique STRA6 mutations, represented by either missense or frameshift changes,46 the previous analysis has failed to demonstrate a clear correlation between genotype and phenotype,8 indicating the variable spectrum of malformations and penetrance in this syndrome, likely affected by not only genetic but also environmental perturbations. The cardiac defects in Matthew-Wood syndrome involve OFT and pharyngeal arch arteries (PAAs) malformations, with varying levels of their severity.8–10 In contrast, Stra6 knockout mice did not have overt cardiac malformations themselves, although the ocular phenotypes were recapitulated in the murine models.11–13 Consequently, the detailed mechanisms of how the STRA6 mutations could lead to variable and species-specific phenotypes and cause cardiac malformations such as OFT defects in humans still remained unclear.
Through the cross-species comparison analysis of single-cell RNA-seq datasets derived from human and murine embryonic hearts uniquely obtained, here we show a clear difference in STRA6+ cells’ appearance and localization between human and murine embryonic hearts (Figure 1), likely explaining the observed phenotypic differences between Stra6 knockout mice and human foetus/children with Matthew-Wood syndrome. We found that STRA6+ progenitors appeared specifically in the early-staged OFT region of human embryonic hearts (Figures 1 and 2), suggesting a potential role of STRA6 for OFT development in humans. Then our interests were to evaluate whether STRA6 mutation could potentially affect cardiomyogenesis and/or vasculogenesis (i.e. induction of CMs and/or vascular SMCs) using the established protocols for in vitro CM and SMC differentiation.19–21 Our data revealed that while STRA6 deletion did not alter the efficacy of in vitro hESC-CM differentiation, albeit being accompanied with partial impairment in both CM maturation and atrial CM induction, it attenuated significantly the efficacies of both in vitro mesodermal and NCC lineage-derived vascular SMC differentiation (notably, the induction of NCC-SMCs was affected) (Figure 3). These findings are supported by the transcriptomics data from population RNA-seq and quantitative PCR analyses of STRA6-KO hESC-derived cardiac differentiating cells, which revealed that STRA6 deletion impaired both mesenchyme formation and mesenchymal cell differentiation at the early stage and muscle system process and contraction at the late stage, accompanied with down-regulation of expression of the SMC differentiation-related genes (Figures 4 and 5).
The RA signalling is known to mediate the patterning of SHF progenitors and cardiac NCCs, both of which are essential for the normal development of the OFT and PAAs.47,48 Increased or decreased RA signalling in mice leads to conotruncal malformations, such as OFT septation defects, and PAA anomalies, such as the interrupted aortic arch (IAA).49 In fact, a major embryonic RA-producing enzyme Aldh1a2-knockout mice cause abnormal patterning of the SHF progenitors and cardiac NCCs and thereby exhibit the OFT and PAA defects, leading to perinatal deaths.4,48,49 Cardiac OFT is a transient conduit during embryogenesis and is encased by a myocardial wall at the earlier stage of development.50 The OFT cushion cells are then populated by SHF-derived and NCC-derived mesenchymal cells that migrate from the pharyngeal mesoderm and dorsal neural tube at around E9.5–E10.5 in mice and subsequently differentiate into vascular SMCs, which form the septum dividing the aorta and the pulmonary artery.51,52 As this septation and remodelling in OFT is progressed, the myocardial wall of OFT is morphologically changed and starts to show arterial phenotype, which is represented by the expanded SMC layer in the wall.51,52 Thus, these mesenchymal-to-SMC transitions and myocardial-to-arterial phenotypic change,32 as well as endocardial EMT51 play critical roles in OFT septation and remodelling. To dissect the molecular pathways disrupted following STRA6 ablation, we performed the transcriptomics analyses (population RNA-seq and RT–qPCR) using STRA6-KO and WT hESC-derived cells in CM and SMC differentiation, and found significant down-regulation of expression of an important SHF/OFT cardiogenic TF TBX1 and its downstream target gene GBX237 in STRA6-KO cells (Figure 5D, F, and G). TBX1 is considered as the most causative candidate gene in human 22q11.2 deletion syndrome (DiGeorge syndrome), as Tbx1-KO mice show embryonic or perinatal lethality due to similar phenotypes to those of human 22q11.2, involving OFT and PAA defects such as ToF, PTA, and IAA.22 Murine Gbx2-KO embryos have also been shown to develop abnormal PAA derivatives, having malformations such as IAA type-B and VSD.53 Of interest, dual haploinsufficiency of Tbx1 and Gbx2 in mice caused cardiac NCC migration defects, leading to PAA abnormalities,54 supporting an indispensable role of the Tbx1–Gbx2 signal network in the regulation of cardiac NCCs and PAA/OFT development. Although we identified the recruitment of RA nuclear receptors RARα/RXRα complexes onto a novel and human-specific RARE site on the TBX1 promoter region, likely regulating TBX1 expression positively in part (Figure 6), previous reports have rather indicated a mutual inhibition of RA signalling and Tbx1 expression on murine embryogenesis.55 For instance, Tbx1 expression in the OFT and PAAs was increased in Aldh1a2+/− embryos or in pharyngeal arch tissues treated with a pharmacological RA inhibitor, while Tbx1 expression was repressed by RA treatments.56 Reversely, in Tbx1 mutant embryos, Aldh1a2 expression in the anterior SHF was increased and expanded cranially.57 The possible reason(s) explaining this discrepancy over the positive or negative correlation between RA signalling and Tbx1 may include the species differences and evolutionary divergences between mice and humans; and a putative differential role of STRA6 compared with other RA signalling modifiers (e.g. Aldh1a2) in the RA signalling pathway during embryogenesis. Nevertheless, in addition to TBX1 and GBX2, we further found significant down-regulation of expression of a cardiac NCC migration TF ZIC1,39 an EMT-promoting TF SNAI2,40 and a proper mesenchymal differentiation-promoting TF OSR1,41 and reversely, significant up-regulation of expression of a myogenic differentiation-inhibiting TF TWIST142 in STRA6-KO cells compared with WT cells (Figure 5E–G). Of interest, during NCC-SMC differentiation, not only SMC differentiation drivers (e.g. MYOD1 and MYOCD) and SMC markers (e.g. ACTA1, TAGLN, and MYH9) but also NCC and somite markers (e.g. SOX9, SOX10,43 and MEOX144) were down-regulated in STRA6-KO cells compared with WT cells (Figure 5G), suggesting partial impairment in proper induction of NCCs and somite formation, which may explain lesser induction of SMCs during NCC-SMC differentiation (Figure 3H). Consistent with this, a recent study revealed that a somite-expressing TF MEOX1 is a positive regulator of SMC differentiation and critical for proper vasculogenesis and angiogenesis during mammalian embryogenesis.58 Collectively, dynamic transcriptomics changes in one or some combination of these transcriptional mediators due to STRA6 mutations may cause and define the variable cardiovascular phenotypes, observed in patients with Matthew-Wood syndrome.
One limitation in the current study is that the generated STRA6-KO hESCs did not have the completely same mutations seen in patients with Matthew-Wood syndrome and thereby, might not precisely mimic the phenotypes of this syndrome, although the previous genetics studies have failed to identify pathogenic mutations nor to demonstrate a clear correlation between genotype and phenotype in Matthew-Wood syndrome.8,10 Future work is warranted to elucidate more comprehensively the human-specific cellular, molecular, and epigenetic mechanisms, by which STAR6 mutations could perturb developing hearts and cause cardiac OFT/PAA defects.
In conclusion, our study highlights the critical role of human-specific STRA6 progenitors for proper induction of vascular SMCs that is essential for normal OFT formation. These results not only give us novel insights on understanding mammalian cardiogenesis but also shed light on previously unappreciated and human-specific CHD programs, driven by STRA6 mutations. Thus, our study paves the way for further studies of deciphering the origins and the disease mechanisms of a rare genetic disorder Matthew-Wood syndrome, which would help us develop diagnosis, prevention, and novel treatment for the disease in the future.
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Authors’ contributions
C.Z. performed murine works, in vitro experiments, bench works, collection and assembly of data, and data analysis and interpretation. T.H. performed in vitro experiments, bench works, and collection and assembly of data. E.R. and J.S. assisted in vitro experiments and bench works. P.K., A.A., and I.A. performed and assisted murine embryo works. M.S. conceived and designed the study, provided financial support, and performed in vitro experiments, bench works, collection and assembly of data, data analysis and interpretation, and manuscript writing.
Supplementary Material
Acknowledgements
We would like to thank the National Genomics Infrastructure (NGI) Sweden and the Eukaryotic Single Cell Genomics Facility (ESCG) at Science for Life Laboratory for support of RNA sequencing and generating raw sequencing data. We would like to thank Federica Santoro and Kenneth R. Chien for their help with the experiments using human samples. pSpCas9(BB)-2A-GFP (PX458) (Addgene plasmid # 48138) and pSpCas9(BB)-2A-Puro (PX459) V2.0 (Addgene plasmid # 62988) were gifts from Feng Zhang.
Contributor Information
Chikai Zhou, Department of Cell and Molecular Biology, Karolinska Institutet, 171 77 Stockholm, Sweden.
Timm Häneke, Department of Cell and Molecular Biology, Karolinska Institutet, 171 77 Stockholm, Sweden.
Eduarde Rohner, Department of Cell and Molecular Biology, Karolinska Institutet, 171 77 Stockholm, Sweden.
Jesper Sohlmér, Department of Cell and Molecular Biology, Karolinska Institutet, 171 77 Stockholm, Sweden.
Polina Kameneva, Department of Neuroimmunology, Center for Brain Research, Medical University of Vienna, Vienna, Austria.
Artem Artemov, Department of Neuroimmunology, Center for Brain Research, Medical University of Vienna, Vienna, Austria.
Igor Adameyko, Department of Neuroimmunology, Center for Brain Research, Medical University of Vienna, Vienna, Austria; Department of Physiology and Pharmacology, Karolinska Institutet, Stockholm, Sweden.
Makoto Sahara, Department of Cell and Molecular Biology, Karolinska Institutet, 171 77 Stockholm, Sweden; Department of Surgery, Yale University School of Medicine, New Haven, CT 06510, USA.
Funding
This work was supported by research grants to M.S. from the Swedish Research Council (Dnr: 2019-01359); the Swedish Heart and Lung Foundation (Dnr: 20150421 and 20190380); and Karolinska Institutet (Strategic Research Area Stem Cells and Regenerative Medicine 2020); and to I.A. from the European Research Council (ERC Synergy grant ‘KILL-OR-DIFFERENTIATE’, Dnr: 856529).
Data availability
Human and murine RNA-seq data reported in this paper have been deposited in the ArrayExpress database at EMBL-EBI (http://www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-7537 and in the Sequence Read Archive (SRA, www.ncbi.nlm.nih.gov/sra/) under accession number PRJNA510181 and PRJNA907958.
Translational perspective.
Dysregulation of the RA signalling can cause cardiac outflow tract (OFT) defects; however, the detailed mechanisms by which STRA6 mutations lead to cardiac malformations have remained unclear. Our study highlights the critical role of human-specific STRA6 progenitors for proper induction of vascular smooth muscle cells that is essential for normal OFT formation. These results shed light on novel and human-specific congenital heart disease programs, driven by STRA6 mutations. Thus, our study paves the way for further studies of deciphering the origins and the disease mechanisms of a rare genetic disorder Matthew-Wood syndrome, which would help us develop diagnosis, prevention, and novel treatment for the disease.
References
- 1. Vincent SD, Buckingham ME. How to make a heart: the origin and regulation of cardiac progenitor cells. Curr Top Dev Biol 2010;90:1–41. [DOI] [PubMed] [Google Scholar]
- 2. Sahara M, Santoro F, Chien KR. Programming and reprogramming a human heart cell. EMBO J 2015;34:710–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Stefanovic S, Etchevers HC, Zaffran S. Outflow tract formation-embryonic origins of conotruncal congenital heart disease. J Cardiovasc Dev Dis 2021;8:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sirbu OI, Zhao X, Duester G. Retinoic acid controls heart anteroposterior patterning by down-regulating Isl1 through the Fgf8 pathway. Dev Dyn 2008;237:1627–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sakabe M, Kokubo H, Nakajima Y, Saga Y. Ectopic retinoic acid signaling affects outflow tract cushion development through suppression of the myocardial Tbx2-Tgfβ2 pathway. Development 2012;139:385–395. [DOI] [PubMed] [Google Scholar]
- 6. Reller MD, Strickland MJ, Riehle-Colarusso T, Mahle WT, Correa A. Prevalence of congenital heart defects in metropolitan Atlanta, 1998–2005. J Pediatr 2008;153:807–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pasutto F, Sticht H, Hammersen G, Gillessen-Kaesbach G, Fitzpatrick DR, Nurnberg G, Brasch F, Schirmer-Zimmermann H, Tolmie JL, Chitayat D, Houge G, Fernández-Martínez L, Keating S, Mortier G, Hennekam RC, von der Wense A, Slavotinek A, Meinecke P, Bitoun P, Becker C, Nürnberg P, Reis A, Rauch A. Mutations in STRA6 cause a broad spectrum of malformations including anophthalmia, congenital heart defects, diaphragmatic hernia, alveolar capillary dysplasia, lung hypoplasia, and mental retardation. Am J Hum Genet 2007;80:550–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Marcadier JL, Mears AJ, Woods EA, Fisher J, Airheart C, Qin W, Beaulieu CL, Dyment DA, Innes AM, Curry CJ, Care4Rare Canada Consortium . A novel mutation in two Hmong families broadens the range of STRA6-related malformations to include contractures and camptodactyly. Am J Med Genet 2016;170A:11–18. [DOI] [PubMed] [Google Scholar]
- 9. Chassaing N, Golzio C, Odent S, Lequeux L, Vigouroux A, Martinovic-Bouriel J, Tiziano FD, Masini L, Piro F, Maragliano G, Delezoide AL, Attié-Bitach T, Manouvrier-Hanu S, Etchevers HC, Calvas P. Phenotypic spectrum of STRA6 mutations: from Matthew-Wood syndrome to non-lethal anophthalmia. Hum Mutat 2009;30:E673–E681. [DOI] [PubMed] [Google Scholar]
- 10. Chassaing N, Ragge N, Kariminejad A, Buffet A, Ghaderi-Sohi S, Martinovic J, Calvas P. Mutation analysis of the STRA6 gene in isolated and non-isolated anophthalmia/microphthalmia. Clin Genet 2013;83:244–250. [DOI] [PubMed] [Google Scholar]
- 11. Perl E, Waxman JS. Reiterative mechanisms of retinoic acid signaling during vertebrate heart development. J Dev Biol 2019;7:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Berry DC, Jacobs H, Marwarha G, Gely-Pernot A, O’Byrne SM, DeSantis D, Klopfenstein M, Feret B, Dennefeld C, Blaner WS, Croniger CM, Mark M, Noy N, Ghyselinck NB. The STRA6 receptor is essential for retinol-binding protein-induced insulin resistance but not for maintaining vitamin A homeostasis in tissues other than the eye. J Biol Chem 2013;288:24528–24539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Amengual J, Zhang N, Kemerer M, Maeda T, Palczewski K, Von Lintig J. STRA6 is critical for cellular vitamin A uptake and homeostasis. Hum Mol Genet 2014;23:5402–5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Treutlein B, Brownfield DG, Wu AR, Neff NF, Mantalas GL, Espinoza FH, Desai TJ, Krasnow MA, Quake SR. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature 2014;509:371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. DeLaughter DM, Bick AG, Wakimoto H, McKean D, Gorham JM, Kathiriya IS, Hinson JT, Homsy J, Gray J, Pu W, Bruneau BG, Seidman JG, Seidman CE. Single-cell resolution of temporal gene expression during heart development. Dev Cell 2016;39:480–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sahara M, Santoro F, Sohlmér J, Zhou C, Witman N, Leung CY, Mononen M, Bylund K, Gruber P, Chien KR. Population and single-cell analysis of human cardiogenesis reveals unique LGR5 ventricular progenitors in embryonic outflow tract. Dev Cell 2019;48:475–490. [DOI] [PubMed] [Google Scholar]
- 17. de Soysa TY, Ranade SS, Okawa S, Ravichandran S, Huang Y, Salunga HT, Schricker A, Del Sol A, Gifford CA, Srivastava D. Single-cell analysis of cardiogenesis reveals basis for organ-level developmental defects. Nature 2019;572:120–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science 2013;339:819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Burridge PW, Matsa E, Shukla P, Lin ZC, Churko JM, Ebert AD, Lan F, Diecke S, Huber B, Mordwinkin NM, Plews JR, Abilez OJ, Cui B, Gold JD, Wu JC. Chemically defined generation of human cardiomyocytes. Nat Methods 2014;11:855–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Patsch C, Challet-Meylan L, Thoma EC, Urich E, Heckel T, O’Sullivan JF, Grainger SJ, Kapp FG, Sun L, Christensen K, Xia Y, Florido MH, He W, Pan W, Prummer M, Warren CR, Jakob-Roetne R, Certa U, Jagasia R, Freskgård PO, Adatto I, Kling D, Huang P, Zon LI, Chaikof EL, Gerszten RE, Graf M, Iacone R, Cowan CA. Generation of vascular endothelial and smooth muscle cells from human pluripotent stem cells. Nat Cell Biol 2015;17:994–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gong J, Zhou D, Jiang L, Qiu P, Milewicz DM, Chen YE, Yang B. In vitro lineage-specific differentiation of vascular smooth muscle cells in response to SMAD3 deficiency: implications for SMAD3-related thoracic aortic aneurysm. Arterioscler Thromb Vasc Biol 2020;40:1651–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jerome LA, Papaioannou VE. DiGeorge syndrome phenotype in mice mutant for the T-box gene, Tbx1. Nat Genet 2001;27:286–291. [DOI] [PubMed] [Google Scholar]
- 23. Picelli S, Björklund ÅK, Faridani OR, Sagasser S, Winberg G, Sandberg R. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods 2013;10:1096–1098. [DOI] [PubMed] [Google Scholar]
- 24. Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, Trombetta JJ, Weitz DA, Sanes JR, Shalek AK, Regev A, McCarroll SA. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 2015;161:1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Santoro F, Chien KR, Sahara M. Isolation of human ESC-derived cardiac derivatives and embryonic heart cells for population and single-cell RNA-seq analysis. STAR Protoc 2021;2:100339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Moretti A, Caron L, Nakano A, Lam JT, Bernshausen A, Chen Y, Qyang Y, Bu L, Sasaki M, Martin-Puig S, Sun Y, Evans SM, Laugwitz KL, Chien KR. Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell 2006;127:1151–1165. [DOI] [PubMed] [Google Scholar]
- 27. McCulley DJ, Kang JO, Martin JF, Black BL. BMP4 is required in the anterior heart field and its derivatives for endocardial cushion remodeling, outflow tract septation, and semilunar valve development. Dev Dyn 2008;237:3200–3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stankunas K, Shang C, Twu KY, Kao SC, Jenkins NA, Copeland NG, Sanyal M, Selleri L, Cleary ML, Chang CP. Pbx/Meis deficiencies demonstrate multigenetic origins of congenital heart disease. Circ Res 2008;103:702–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chong JJ, Reinecke H, Iwata M, Torok-Storb B, Stempien-Otero A, Murry CE. Progenitor cells identified by PDGFR-alpha expression in the developing and diseased human heart. Stem Cells Dev 2013;22:1932–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Herrmann F, Bundschu K, Kühl SJ, Kühl M. Tbx5 overexpression favors a first heart field lineage in murine embryonic stem cells and in Xenopus laevis embryos. Dev Dyn 2011;240:2634–2645. [DOI] [PubMed] [Google Scholar]
- 31. Nelson DO, Jin DX, Downs KM, Kamp TJ, Lyons GE. Irx4 identifies a chamber-specific cell population that contributes to ventricular myocardium development. Dev Dyn 2014;243:381–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Anderson RH, Webb S, Brown NA, Lamers W, Moorman A. Development of the heart: (3) formation of the ventricular outflow tracts, arterial valves, and intrapericardial arterial trunks. Heart 2003;89:1110–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu X, Chen W, Li W, Li Y, Priest JR, Zhou B, Wang J, Zhou Z. Single-cell RNA-Seq of the developing cardiac outflow tract reveals convergent development of the vascular smooth muscle cells. Cell Rep 2019;28:1346–1361. [DOI] [PubMed] [Google Scholar]
- 34. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Deschamps J, van Nes J. Developmental regulation of the Hox genes during axial morphogenesis in the mouse. Development 2005;132:2931–2942. [DOI] [PubMed] [Google Scholar]
- 36. Duester G. Retinoic acid synthesis and signaling during early organogenesis. Cell 2008;134:921–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Scambler PJ. 22q11 Deletion syndrome: a role for TBX1 in pharyngeal and cardiovascular development. Pediatr Cardiol 2010;31:378–390. [DOI] [PubMed] [Google Scholar]
- 38. Wang L, Qiu P, Jiao J, Hirai H, Xiong W, Zhang J, Zhu T, Ma PX, Chen YE, Yang B. Yes-associated protein inhibits transcription of myocardin and attenuates differentiation of vascular smooth muscle cell from cardiovascular progenitor cell lineage. Stem Cells 2017;35:351–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Roux M, Laforest B, Eudes N, Bertrand N, Stefanovic S, Zaffran S. Hoxa1 and Hoxb1 are required for pharyngeal arch artery development. Mech Dev 2017;143:1–8. [DOI] [PubMed] [Google Scholar]
- 40. Nieto MA, Sargent MG, Wilkinson DG, Cooke J. Control of cell behavior during vertebrate development by Slug, a zinc finger gene. Science 1994;264:835–839. [DOI] [PubMed] [Google Scholar]
- 41. Han L, Xu J, Grigg E, Slack M, Chaturvedi P, Jiang R, Zorn AM. Osr1 functions downstream of Hedgehog pathway to regulate foregut development. Dev Biol 2017;427:72–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Koutalianos D, Koutsoulidou A, Mastroyiannopoulos NP, Furling D, Phylactou LA. MyoD transcription factor induces myogenesis by inhibiting Twist-1 through miR-206. J Cell Sci 2015;128:3631–3645. [DOI] [PubMed] [Google Scholar]
- 43. Soldatov R, Kaucka M, Kastriti ME, Petersen J, Chontorotzea T, Englmaier L, Akkuratova N, Yang Y, Häring M, Dyachuk V, Bock C, Farlik M, Piacentino ML, Boismoreau F, Hilscher MM, Yokota C, Qian X, Nilsson M, Bronner ME, Croci L, Hsiao WY, Guertin DA, Brunet JF, Consalez GG, Ernfors P, Fried K, Kharchenko PV, Adameyko I. Spatiotemporal structure of cell fate decisions in murine neural crest. Science 2019;364:eaas9536. [DOI] [PubMed] [Google Scholar]
- 44. Skuntz S, Mankoo B, Nguyen MT, Hustert E, Nakayama A, Tournier-Lasserve E, Wright CV, Pachnis V, Bharti K, Arnheiter H. Lack of the mesodermal homeodomain protein MEOX1 disrupts sclerotome polarity and leads to a remodeling of the cranio-cervical joints of the axial skeleton. Dev Biol 2009;332:383–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ramos C, Robert B. msh/Msx gene family in neural development. Trends Genet 2005;21:624–632. [DOI] [PubMed] [Google Scholar]
- 46. Stenson PD, Mort M, Ball EV, Shaw K, Phillips A, Cooper DN. The human gene mutation database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum Genet 2014;133:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hochgreb T, Linhares VL, Menezes DC, Sampaio AC, Yan CY, Cardoso WV, Rosenthal N, Xavier-Neto J. A caudorostral wave of RALDH2 conveys anteroposterior information to the cardiac field. Development 2003;130:5363–5374. [DOI] [PubMed] [Google Scholar]
- 48. Robrini NE, Etchevers HC, Ryckebüsch L, Faure E, Eudes N, Niederreither K, Zaffran S, Bertrand N. Cardiac outflow morphogenesis depends on effects of retinoic acid signaling on multiple cell lineages. Dev Dyn 2016;245:388–401. [DOI] [PubMed] [Google Scholar]
- 49. Vermot J, Niederreither K, Garnier JM, Chambon P, Dollé P. Decreased embryonic retinoic acid synthesis results in a DiGeorge syndrome phenotype in newborn mice. Proc Natl Acad Sci USA 2003;100:1763–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Webb S, Qayyum SR, Anderson RH, Lamers WH, Richardson MK. Septation and separation within the outflow tract of the developing heart. J Anat 2003;202:327–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dyer LA, Kirby ML. The role of secondary heart field in cardiac development. Dev Biol 2009;336:137–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mahmoud M, Evans I, Wisniewski L, Tam Y, Walsh C, Walker-Samuel S, Frankel P, Scambler P, Zachary I. Bcar1/p130Cas is essential for ventricular development and neural crest cell remodelling of the cardiac outflow tract. Cardiovasc Res 2022;118:1993–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Byrd NA, Meyers EN. Loss of Gbx2 results in neural crest cell patterning and pharyngeal arch artery defects in the mouse embryo. Dev Biol 2005;284:233–245. [DOI] [PubMed] [Google Scholar]
- 54. Calmont A, Ivins S, Van Bueren KL, Papangeli I, Kyriakopoulou V, Andrews WD, Martin JF, Moon AM, Illingworth EA, Basson MA, Scambler PJ. Tbx1 controls cardiac neural crest cell migration during arch artery development by regulating Gbx2 expression in the pharyngeal ectoderm. Development 2009;136:3173–3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yutzey KE. DiGeorge syndrome, Tbx1, and retinoic acid signaling come full circle. Circ Res 2010;106:630–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ryckebüsch L, Bertrand N, Mesbah K, Bajolle F, Niederreither K, Kelly RG, Zaffran S. Decreased levels of embryonic retinoic acid synthesis accelerate recovery from arterial growth delay in a mouse model of DiGeorge syndrome. Circ Res 2010;106:686–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Roberts C, Ivins S, Cook AC, Baldini A, Scambler PJ. Cyp26 genes a1, b1 and c1 are down-regulated in Tbx1 null mice and inhibition of Cyp26 enzyme function produces a phenocopy of DiGeorge syndrome in the chick. Hum Mol Genet 2006;15:3394–3410. [DOI] [PubMed] [Google Scholar]
- 58. Dong K, Guo X, Chen W, Hsu AC, Shao Q, Chen JF, Chen SY. Mesenchyme homeobox 1 mediates transforming growth factor-β (TGF-β)-induced smooth muscle cell differentiation from mouse mesenchymal progenitors. J Biol Chem 2018;293:8712–8719. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Human and murine RNA-seq data reported in this paper have been deposited in the ArrayExpress database at EMBL-EBI (http://www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-7537 and in the Sequence Read Archive (SRA, www.ncbi.nlm.nih.gov/sra/) under accession number PRJNA510181 and PRJNA907958.