Summary
Background
Partial artemisinin resistance is suspected if delayed parasite clearance (ie, persistence of parasitaemia on day 3 after treatment initiation) is observed. Validated markers of artemisinin partial resistance in southeast Asia, Plasmodium falciparum kelch13 (Pfkelch13) R561H and P574L, have been reported in Rwanda but no association with parasite clearance has been observed. We aimed to establish the efficacy of artemether–lumefantrine and genetic characterisation of Pfkelch13 alleles and their association with treatment outcomes.
Methods
This open-label, single-arm, multicentre, therapeutic efficacy study was done in 2018 in three Rwandan sites: Masaka, Rukara, and Bugarama. Children aged 6–59 months with P falciparum monoinfection and fever were eligible and treated with a 3-day course of artemether–lumefantrine. Treatment response was monitored for 28 days using weekly microscopy screenings of blood samples for P falciparum. Mutations in Pfkelch13 and P falciparum multidrug resistance-1 (Pfmdr1) genes were characterised in parasites collected from enrolled participants. Analysis of flanking microsatellites surrounding Pfkelch13 was done to define the origins of the R561H mutations. The primary endpoint was PCR-corrected parasitological cure on day 28, as per WHO protocol.
Findings
228 participants were enrolled and 224 (98·2%) reached the study endpoint. PCR-corrected efficacies were 97·0% (95% CI 88–100) in Masaka, 93·8% (85–98) in Rukara, and 97·2% (91–100) in Bugarama. Pfkelch13 R561H mutations were present in 28 (13%) of 218 pre-treatment samples and P574L mutations were present in two (1%) pretreatment samples. 217 (90%) of the 240 Pfmdr1 haplotypes observed in the pretreatment samples, had either the NFD (N86Y, Y184F, D1246Y) or NYD haplotype. Eight (16%) of 51 participants in Masaka and 12 (15%) of 82 participants in Rukara were microscopically positive 3 days after treatment initiation, which was associated with pre-treatment presence of Pfkelch13 R561H in Masaka (p=0·0005). Genetic analysis of Pfkelch13 R561H mutations suggest their common ancestry and local origin in Rwanda.
Interpretation
We confirm evidence of emerging artemisinin partial resistance in Rwanda. Although artemether–lumefantrine remains efficacious, vigilance for decreasing efficacy, further characterisation of artemisinin partial resistance, and evaluation of additional antimalarials in Rwanda should be considered.
Funding
The US President’s Malaria Initiative.
Introduction
Artemisinin-based combination therapies (ACTs) are currently the most effective and widely used treatments for uncomplicated malaria caused by Plasmodium falciparum.1 ACTs combine artemisinin derivatives, short-acting drugs that clear most of the parasite biomass within 3 days of treatment initiation, with long-acting partner drugs clearing the remaining parasitaemia. In 2006, Rwanda introduced artemether–lumefantrine combination therapy as the first-line treatment for uncomplicated malaria.2
WHO recommends therapeutic efficacy studies (TES) at least every 2 years for monitoring the efficacy of ACTs and the tracking of resistance through molecular markers.3 When ACT efficacy is confirmed to be below 90% based on adequate clinical and parasitological response (ACPR) during an observation period (28 days or 42 days, depending on the partner drug), WHO recommends further confirmation and replacement with an effective antimalarial.1 A TES using artemether–lumefantrine in Rwandan children aged 1–14 years was done between 2013 and 2015 in Ruhuha and Masaka, and the PCR-adjusted ACPR on day 28 was more than 97% in both sites.4
Partial resistance to the artemisinin component of ACT is suspected if delayed parasite clearance, defined as the persistence of parasitaemia on day 3 after treatment initiation, is observed.1 First identified in Cambodia, artemisinin resistance is well documented in many southeast Asian countries5–9 and is associated with parasites that have mutations in the Pfkelch13 gene.10 Ten single nucleotide polymorphisms in the Pfkelch13 propeller domain have been validated as molecular markers of artemisinin partial resistance: F446I, N458Y, M476I, Y493H, R539T, I543T, P553L, R561H, P574L, and C580Y.1 Additionally, several other mutations in this gene, referred to as candidate markers, have been identified, but additional evidence is required to validate their association with artemisinin partial resistance.1
The R561H mutation was observed in 7·4% of P falciparum parasites collected in Masaka, Rwanda, between 2013 and 201511 and in 4·5% of parasites collected in Huye district, Rwanda, in 2019.12 Additionally, a low prevalence of the P574L mutation was reported in isolates collected in Masaka and Ruhuha (2013–15)11,12 and in Huye (2015).13 However, the presence of these mutations was not found to be associated with day 3 parasitaemia but with increased survival rate expressed in vitro by the ring stage survival assay.11 Nevertheless, these findings are concerning because an increase in the prevalence of these mutations could result in more patients having delayed parasite clearance, which could lead to an increased risk of selection and spreading of partner drug-resistant parasites and eventual ACT failure.
The Malaria and Other Parasitic Diseases Division of the Rwanda Biomedical Center did a routine TES in 2018 as per a WHO recommendation to do periodic antimalarial efficacy monitoring. We report on these TES results, including day 28 efficacy, day 3 persistence of parasitaemia, and the presence of molecular markers of artemisinin partial resistance and tolerance to lumefantrine in the Pfkelch13 and P falciparum multidrug resistance-1 (Pfmdr1) genes.
Methods
Study design and participants
This open-label, single-arm, multicentre, therapeutic efficacy study (TES) was done in three health centres: Rukara (Kayonza district, Eastern Province), Masaka (Kicukiro district, Kigali Province), and Bugarama (Rusizi district, Western Province) (figure 1). Rukara and Bugarama have rural populations, and Masaka, located near the capital city Kigali, is an urban and periurban location with low transmission of malaria. These three sites were selected because they have varying transmission intensities and established study centres that have been used in previous TESs. In all three sites, P falciparum accounts for most malaria cases, with transmission seasons from May to July and November to December. Based on an assumption of 95% efficacy of artemether–lumefantrine, 95% CI, a precision of 10%, and 20% loss to follow-up, we estimated that 88 children needed to be enrolled per site.
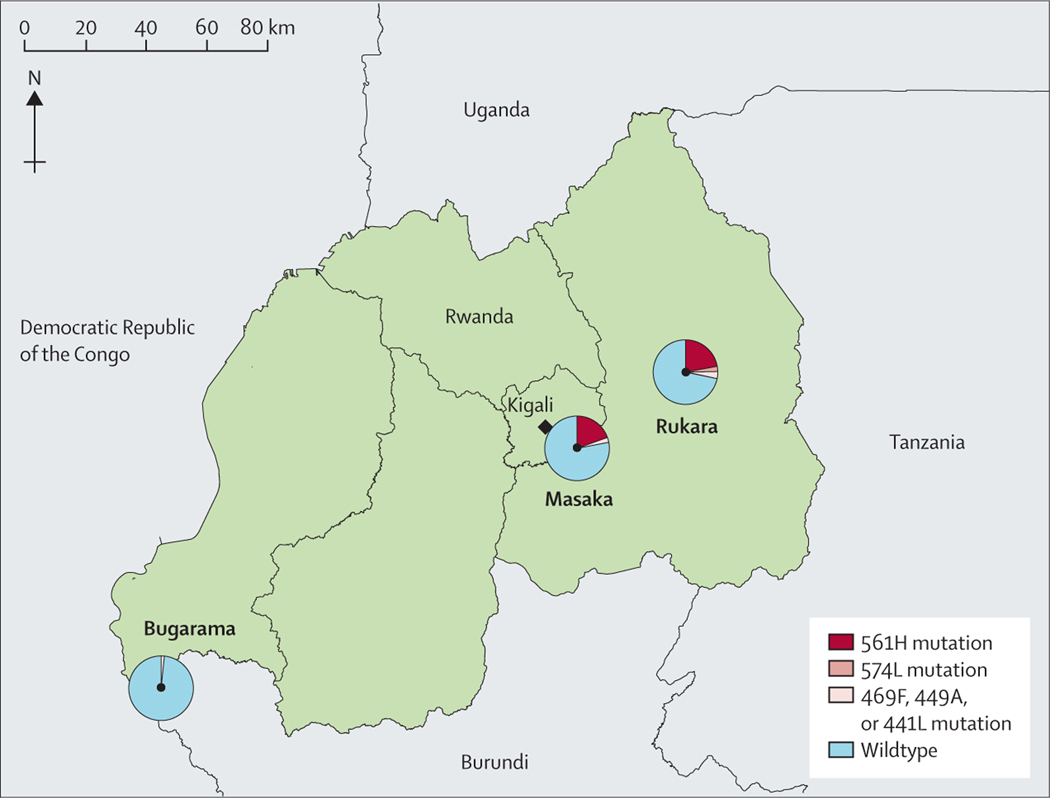
Figure 1: Study sites and prevalence of Pfkelch13 mutations in pre-treatment samples by study site.
This study was done in three Rwandan health centres: Rukara (Kayonza Distric), Masaka (Kicukiro District) and Bugarama (Rusizi District). The prevalence of the Pfkelch13 R561H mutation was found in ten (20%) of 51 samples in Masaka, and eight (22%) of 82 in Rukara; that of the P574L marker was two (1%) of 82 samples in Rukara. Prevalence of the candidate artemisinin partial resistance markers was one (2%) of 51 samples in Masaka, three (4%) of 82 samples in Rukara, and one (1%) of 85 samples in Bugarama.
Participants were eligible for enrolment if they were aged 6–59 months, had a fever at presentation (axillary temperature ≥37·5°C), history of fever in the past 24 h, or both, and parasitaemia of 1000–100 000 parasites per μL by microscopy. Other inclusion and exclusion criteria were assessed according to the standard WHO TES protocol.3 Ineligible children were treated in accordance with national malaria treatment guidelines.2
This study was approved by the Rwanda National Ethics Committee (reference 195/RNEC/2017), Rwanda National Health Research Committee, and the Johns Hopkins Bloomberg School of Public Health Institutional Review Board. The US Centers for Disease Control and Prevention determined the laboratory work to be non-research (programme evaluation). Written informed consent was obtained from the parents or guardians of eligible children in the local language (Kinyarwanda).
Procedures
Microscopy screening
At each study site, potential participants were screened for malaria parasites using microscopy. Both thin and thick blood slides were prepared and stained with 5% Giemsa for 10–15 min and examined by two trained expert microscopists to detect and estimate density of malaria parasites. A blood slide was declared negative when examination of 100 high-power fields did not suggest the presence of P falciparum parasites.
Treatment and clinical monitoring during follow-up
Enrolled participants were admitted as inpatients and treated with a weight adjusted, six dose, 3-day course of artemether–lumefantrine (Coartem, Novartis). Each dose was given with water and fatty foods such as ibitoki (a local delicacy prepared from peanuts and bananas) to optimise bioavailability of the drug. Participants were observed for 30 min following administration of each dose to ensure they did not vomit. If vomiting occurred, a repeat dose was given after vomiting stopped. Parasitaemia was determined on the day of enrolment (day 0) and on days 2 and 3. Patients were discharged on day 3 after enrolment if they had a slide result that was negative for P falciparum malaria. Participants who were slide malaria positive on day 3 were kept in hospital until two consecutive malaria negative slides, prepared daily, were obtained. Treatment response was monitored for 28 days with scheduled visits on days 7, 14, 21, 28, and at any other time when patients were unwell (unscheduled visit). During every visit, clinical (evaluation of vital signs) and parasitological (malaria diagnosis by microscopy) assessments were done. Blood samples, for microscopy and preparation of dried blood spots on Whatman grade 3 filter papers (GE Healthcare Life Sciences, PA, USA), were collected from a finger prick. Patients with recurrent infections on day 7 and afterward were treated with quinine (tablets or injection) or artesunate injection (in patients with severe malaria), as per national treatment guidelines.2
DNA extraction and molecular analysis
Molecular analysis was done on all samples collected upon enrolment (pre-treatment samples) and during follow-up in the case of recurrent infections (post-treatment samples). Parasite genomic DNA was extracted from dried blood spots using a QIAamp DNA Mini Kit (Qiagen; Hilden, Germany). Molecular markers of antimalarial drug resistance and microsatellite markers were analysed by Rwandan laboratory technicians with support from US Centers for Disease Control and Prevention laboratory staff (Atlanta, GA, USA).14 The Pfkelch13 propeller domain (codon positions 440–600) and Pfmdr1 (codon positions 86, 184, and 1246) were analysed for single nucleotide polymorphisms.15,16 Sanger sequences were analysed using Geneious (version R11) software package (Biomatters; San Francisco, CA, USA) using the 3D7 Pfkelch13 (PF3D7_1343700) and Pfmdr1 sequences (PF3D7_0523000) as references. Raw sequence reads were cleaned using default settings and reads with high-quality scores (>30%) were further analysed.
Differentiation between recrudescence and reinfection
PCR correction, differentiating recrudescence from reinfection in recurrent infection samples, was done by genotyping seven neutral microsatellites (TA1 on chromosome 6, Poly-α on chromosome 4, PfPK2 on chromosome 12, 2490 on chromosome 10, C2M34–313 on chromosome 2, C2M69–383 on chromosome 3, and TA109 on chromosome 6) in the paired pre-treatment and post-treatment samples. The sizes of the amplification products were determined by capillary electrophoresis on an Applied Biosystems 3130xl Genetic Analyzer (Applied Biosystems; Foster City, CA, USA). Microsatellite data were used to assign each recurrent infection a posterior probability of recrudescence using a Bayesian algorithm.17 This analysis allowed for classification of recrudescent infections (if the posterior probability of recrudescence based on the algorithm was ≥50%) versus reinfections (<50% posterior probability) for tabulation. The posterior probabilities were used in the calculation of PCR-corrected per protocol and Kaplan-Meier estimates.
Pfkelch13 flanking microsatellite genotyping and genetic diversity
Seven microsatellite loci flanking the Pfkelch13 gene on chromosome 13 (PF3D7_1343700; downstream of 3·4 kb, 8·6 kb, and 72·0 kb; upstream of –0·15 kb, –3·7 kb, –6·3 kb, and –31·9 kb) were used to characterise regions flanking Pfkelch13 to assess for origin, genetic diversity, and patterns consistent with selection.18 All isolates from Rwanda and four R561H mutant samples from Thailand were subjected to the Pfkelch13 flanking sequence microsatellite analysis.18 The heterozygosity on the flanking Pfkelch13 microsatellites within each group (wildtype and mutant, in each population) was estimated using the Nei’s index of genetic diversity ().19
was obtained by taking the sum of identifiable alleles, and Pi was the relative frequency of the i-th allele () in all genotyped samples for each locus. gave the average probability that a pair of alleles randomly selected from the population was different. Samples with mixed infections or missing data at any loci were excluded from the construction of the heterozygosity figure.
Outcomes
The primary endpoint was PCR-corrected parasitological cure on day 28 as per WHO protocol.3 Secondary endpoints included parasitaemia on day 3 following treatment, which was assessed by microscopy and the prevalence of molecular markers of antimalarial drug resistance in Pfmdr1 and Pfkelch13 genes. Treatment outcomes were classified as early treatment failure (ie, development of signs of severe malaria, no rapid resolution of clinical symptoms, and slow clearance of slide parasitaemia), recurrent infections (ie, having detectable parasitaemia between day 4 and 28 during the follow-up) that included recrudescent infections and reinfections, or ACPR.3
Data management and statistical analysis
Data were entered into paper-based clinical record forms by trained staff at study sites, after which electronic double data entry was done at the national reference laboratory. Uncorrected and PCR-corrected per protocol3 (proportional) and Kaplan-Meier (cumulative survival) estimates were calculated per site. Reinfections were calculated as the total number of recurrent infections minus the sum of the posterior probabilities of recrudescence among the recurrent infections and removed from the PCR-corrected per-protocol estimates. To calculate the Kaplan-Meier estimates and corresponding 95% CI, we did posterior sampling using the posterior probabilities of recrudescence. Prevalence of Pfkelch13 mutations, Pfmdr1 mutations, and Pfmdr1 haplotypes were described by treatment outcome and site. Comparison analyses were done using Fisher’s exact test and adjusted comparisons using logistic regression. All possible haplotypes from mixed infections (both wildtype and mutants) were included in construction of the Pfmdr1 haplotype. Differences in the heterozygosity between the wildtype and R561H mutant haplotypes were measured using Wright’s F statistics ranging from 0 to 1, by which 0 represents no differentiation.20
Role of the funding source
The sponsor of the study had no role in the recruitment of subjects and implementation of the TES but provided technical assistance in data analysis, data interpretation, and the writing of the report. At least five authors, including the corresponding authors had full access to all data in the study and had final responsibility for the decision to submit for publication.
Results
Of 986 children screened at the three sites, 228 children with symptomatic uncomplicated P falciparum malaria were enrolled: 88 (39%) in Rukara, 52 (22%) in Masaka, and 88 (39%) in Bugarama. Lower than expected number of participants were enrolled in Masaka, probably because Masaka is located in a low transmission peri-urban site. Baseline characteristics and treatment outcomes of participants are shown in table 1. Four (2%) of the 228 participants withdrew, three between days 4 and 6, and one on day 28. Only the participants who reached the study endpoints (224 [98%] of 228 participants) were included in the per-protocol analysis (table 1). No early treatment failures were observed, and no medication-related serious adverse events occurred. 37 recurrent infections were observed and 187 (84%) of 224 patients had ACPR. Of the recurrent infections, eight (22%) of 37 were classified as recrudescent infections and 29 (78%) of 37 as reinfections (table 1). The per-protocol PCR-corrected drug efficacies were 93·8% (95% CI 85–98) in Rukara, 97·0% (95% CI 88–100) in Masaka, and 97·2% (95% CI 91–100) in Bugarama. The uncorrected and PCR-corrected efficacies per study site are summarised in table 2 and appendix 2 p 1).
Table 1:
Baseline characteristics of participants and classification of treatment outcomes
| Rukara | Masaka | Bugarama | Total | |
|---|---|---|---|---|
|
| ||||
| Baseline characteristics | ||||
| Enrolled, n | 88 | 52 | 88 | 228 |
| Age, years | 2·9 (1·1) | 3·1 (1·1) | 3·2 (1·0) | 3·1 (1·1) |
| Gender | ||||
| Male (%) | 47 (53%) | 23 (44%) | 41 (47%) | 111 (49%) |
| Female (%) | 41 (47%) | 29 (56%) | 47 (53%) | 117 (51%) |
| Weight, kg | 13 (2·6) | 13·8 (2·5) | 13·3 (2·5) | 13·3 (2·5) |
| Height, cm | 86·5 (9·7) | 92·6 (11·3) | 93·9 (11·1) | 90·7 (11·1) |
| Parasite density, parasites per μl at enrollment, median (range) | 44 560 (1120–99 920) | 41 440 (1160–99 200) | 39 100 (1080–98 480) | 42 760 (1080–99 920) |
| Treatment outcomes | ||||
| Early treatment failure | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) |
| Recurrent infections | 21 (24%) | 2 (4%) | 14 (16%) | 37 (16%) |
| Recrudescence | 4 (19%) | 2 (100%) | 2 (14%) | 8 (22%) |
| Days 8–14 | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) |
| Days 15–21 | 4 (100%) | 1 (50%) | 2 (100%) | 7 (88%) |
| Days 22–28 | 0 (0%) | 1 (50%) | 0 (0%) | 1 (13%) |
| Reinfection | 17 (81%) | 0 (0%) | 12 (86%) | 29 (78%) |
| Days 4–7 | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) |
| Days 8–14 | 1 (6%) | 0 (0%) | 1 (8%) | 2 (7%) |
| Days 15–21 | 10 (59%) | 0 (0%) | 5 (42%) | 15 (52%) |
| Days 21–28 | 6 (35%) | 0 (0%) | 6 (50%) | 12 (41%) |
| ACPR | 65 (74%) | 48 (92%) | 74 (84%) | 187 (82%) |
| Excluded* or lost to follow-up | 2 (2%) | 2 (4%) | 0 (0%) | 4 (2%) |
Baseline characteristics data are mean (SD) unless otherwise stated. ACPR=adequate clinical and parasitological response.
Three patients excluded between days 4–6 because they received additional antimalarial treatment.
Table 2:
Uncorrected and PCR-corrected Kaplan-Meier and per-protocol estimates of artemether–lumefantrine efficacy
| Kaplan-Meier |
Per protocol |
|||
|---|---|---|---|---|
| Uncorrected (95% CI) | PCR-corrected (95% CI) | Uncorrected (95% CI) | PCR-corrected (95% CI) | |
|
| ||||
| Rukara (n=88) | 75·6% (68–85) | 94·5% (90–100) | 75·6% (65–84) | 93·8% (85–98) |
| Masaka (n=52) | 96·0% (91–100) | 97·0% (93–100) | 96·0% (86–100) | 97·0 % (88–100) |
| Bugarama (n=88) | 84·1% (77–92) | 97·6% (91–100) | 84·1% (75–91) | 97·2% (91–100) |
Data from follow-up at day 28.
The Pfkelch13 gene was successfully sequenced from 254 (96%) of 265 samples (218 pre-treatment and 36 post-treatment: 28 from reinfected patients and eight patients with recrudescent infections). 38 (15%) of 254 samples had the validated artemisinin partial resistance marker, of which 36 had a R561H mutation and two had a P574L mutation. Eight of the 36 isolates with the R561H marker were mixed infections (with wildtype and mutant strains). The R561H mutation was found in 28 (12·8%) of 218 pre-treatment samples, four (14·3%) of 28 reinfection samples, and four (50·0%) of eight recrudescent samples. Three candidate artemisinin partial resistance markers were found in six isolates: C469F in three isolates (two pre-treatment and one recrudescence), P441L in one pre-treatment isolate, and G449A in two pre-treatment isolates. Additional Pfkelch13 mutations, not known to be associated with artemisinin resistance, were observed in four isolates in Rukara: V555A (three isolates) and R575K (one isolate). The prevalence of the Pfkelch13 mutants associated with artemisinin partial resistance was determined per site in pre-treatment isolates (figure 1).
A total of 243 (95%) of 255 samples were successfully sequenced in the Pfmdr1 gene: 208 pre-treatment and 35 post-treatment (appendix 2 p 4). The N86Y, Y184F, D1246Y (NYD) haplotype was observed in 114 (48%) of 240 of pre-treatment and 20 (50%) of 40 post-treatment and the N86, 184F, D1246 (NFD) haplotype was observed in 103 (43%) of 240 of pre-treatment and 16 (40%) of 40 of post-treatment total observed haplotypes (appendix 2 p 4). No difference was seen in the prevalence of the NFD and NYD haplotype in participants with recrudescent infection (eight [100%] of eight) compared with those with ACPR, (177 [91%] of 194; Fisher’s exact test, p=1·00).
A total of 20 participants (eight of [16%] 51 participants in Masaka and 12 [15%] of 82 participants in Rukara) had detectable parasitaemia by microscopy on day 3 post-treatment, a WHO criterion for artemisinin partial resistance.1 11 (55%) of these 20 participants also carried the R561H mutation (table 3). Presence of day 3 parasitaemia was compared between pre-treatment samples with (n=28) and without (n=190) the R561H mutation. Day 3 parasitaemia was observed in 11 (39%) of isolates with the R561H mutation and in nine (5%) of 190 isolates without the mutation (Fisher’s exact test, p<0.0001). When adjusting for initial parasitaemia, the association between day 3 parasitaemia and presence of the R561H mutation remained (adjusted OR 14·2, 95% CI 5·1–41·5, p<0·0001). When stratified by site, this association was only observed in Masaka (p=0.0005) and not in Rukara (p=0·063). Two pre-treatment isolates had the P574L mutation, one from Masaka and one from Rukara, neither of which had day 3 parasitaemia and both were classified as ACPR. Three of the five pre-treatment isolates with the candidate mutations, (one with C469F and two with G449A), all from Rukara, also had day 3 parasitaemia; however, additional association analyses were not done because of the small sample size.
Table 3:
Prevalence of day 3 parasitaemia and Pfkelch13 R561H mutation in pre-treatment isolates in the three study sites
| Day 3 parasitaemia positive |
Day 3 parasitaemia negative |
|||
|---|---|---|---|---|
| 561H mutation | R561 wildtype | 561H mutation | R561 wildtype | |
|
| ||||
| Rukara (n=82) | 5 (6·1%) | 7 (8·5%) | 13 (15·9%) | 57 (69·5%) |
| Masaka (n=51) | 6 (11·8%) | 2 (3·9%) | 4 (7·8%) | 39 (76·5%) |
| Bugarama (n=85) | 0 (0·0%) | 0 (0·0%) | 0 (0·0%) | 85 (100%) |
Data are n (%). Pfkelch13=Plasmodium falciparum kelch 13.
To determine whether Pfkelch13 R561H mutation was associated with the 28-day treatment outcome, proportions of pre-treatment isolates with this mutation were compared in participants with parasite clearance with those who were classified as having recrudescent infection. Samples from four (50%) of eight participants with a recrudescent infection had a mutation compared with samples from 22 (12·4%) of 178 participants with parasite clearance (Fisher’s exact test, p=0·014).
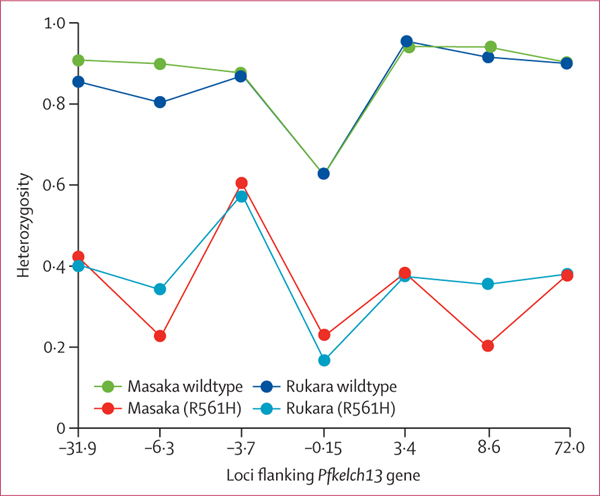
Analysis of flanking microsatellites surrounding Pfkelch13 was investigated in 82 isolates (33 R561H mutants and 49 R561H wildtypes). Recrudescent infections and isolates with multiple infections were excluded in the haplotype analysis. The sizes of the microsatellites were considered unique if they differed by two base pairs except for the −6·3 kb locus, for which a difference in three base pairs was used. Similar haplotypes, differing only in a few loci, were observed in the R561H mutation from Rwanda, which were different from haplotypes observed in the R561H mutation from Thailand (table 4, appendix 2 p 5). Genetic diversity in the R561H mutant, as estimated by the heterozygosity on the flanking Pfkelch13 microsatellites, was lower than that observed in wildtype parasites (figure 2). Wildtype genotypes were highly differentiated from their sympatric R561H mutation in both populations (average F 0·847 between the R561H wildtype and R561H mutation in Masaka, p=0·0021; F 0·841 in Rukara, p=0·0006).
Table 4:
Pfkelch13-linked microsatellite haplotypes from Rwanda and Thailand
| −31·9 kb | −6·3 kb | −3·7 kb | −0·15 kb | 3·4 kb | 8·6 kb | 72·0 kb | |
|---|---|---|---|---|---|---|---|
|
| |||||||
| Rwanda | |||||||
| Haplotype 1 (n=8) | 209 | 277 | 152 | 194 | 128 | 262 | 236 |
| Haplotype 2 (n=8) | 209 | 277 | 150 | 194 | 128 | 262 | 236 |
| Haplotype 3 (n=1) | 197 | 277 | 150 | 194 | 128 | 262 | 236 |
| Haplotype 4 (n=1) | 209 | 271 | 152 | 194 | 128 | 262 | 236 |
| Haplotype 5 (n=1) | 209 | 277 | 150 | 194 | 128 | 262 | 232 |
| Haplotype 6 (n=1) | 209 | 277 | 152 | 194 | 132 | 262 | 236 |
| Haplotype 7 (n=1) | 219 | 277 | 152 | 194 | 128 | 262 | 236 |
| Haplotype 8 (n=1) | 209 | 277 | 150 | 194 | 120 | 278 | 232 |
| Haplotype 9 (n=1) | 209 | 277 | 150 | 194 | 128 | 262 | 252 |
| Haplotype 10 (n=1) | 209 | 277 | 152 | 194 | 128 | 262 | 232 |
| Thailand | |||||||
| Haplotype 1 (n=2) | 203 | 283 | 148 | 194 | 130 | 288 | 230 |
| Haplotype 2 (n=1) | 203 | 283 | 148 | 194 | 130 | 288 | 244 |
| Haplotype 3 (n=1) | 203 | 283 | 148 | 194 | 130 | 288 | 250 |
Data represent the sizes, in base pairs, of the flanking microsatellites surrounding the Pfkelch13. This analysis showed ten similar haplotypes in the isolates from Rwanda. By contrast, different haplotypes were observed in the R561H isolates from Thailand.
Figure 2: Reduced heterozygosity on the sampled loci around the pfk13 gene in R561H isolates.
A total of 33 R561H mutations(nine from Masaka and 24 from Rukara) and 49 wildtype isolates (24 from Masaka and 25 Rukara) are shown (samples with mixed infections or missing data at any loci were excluded). We observed distinct haplotypes with the R561H mutation(compared with the wildtype (blue and green lines), with a reduction in the heterozygosity in those with the R561H mutation. The heterozygosity of the wildtype genotypes was significantly different and higher than the one observed in the sympatric R561H mutant haplotypes in the two populations.
Discussion
This study confirms that artemether–lumefantrine remains highly efficacious in all three study sites, with PCR-corrected efficacy of 94–97%. However, the presence of two validated markers of artemisinin partial resistance, R561H and P574L, and delayed parasite clearance (parasitaemia at day 3) in more than 10% of the study participants in Masaka and Rukara are of some concern.1 Although factors such as initial parasitaemia on admission are known to affect parasite clearance rate,21 treatment with artemisinin compounds results in a rapid clearance of parasitaemia by day 3 of treatment initiation, and delayed parasite clearance is suggestive of artemisinin partial resistance.1,5 The prevalence of the Pfkelch13 R561H mutation was significantly higher in patients with recrudescent infections than in those who cleared their infection, and the association between day 3 parasitaemia and presence of the R561H mutation was significant in Masaka. However, besides artemisinin partial resistance, other factors affecting treatment efficacy include efficacy of the partner drug, host immunity, and drug absorption. These factors could explain why R561H mutations were also observed in patients who cleared their infections and the fact that the association between day 3 parasitaemia and having the R561H mutation was significant in Masaka but not in Rukara.
Results from our study confirmed the previous finding of Pfkelch13 R561H mutations in isolates collected in Masaka, Rwanda, between 2013 and 2015.11 The prevalence of Pfkelch13 R561H mutation in our study was found to be higher than previously reported in Rwanda,11,12 Uganda,22 and Tanzania,23 suggesting that this mutation is strongly selected for in Rwanda. It remains to be determined if other detected mutations, such as Pfkekch13 P574L mutations, will be selected for and increase over time. P574L mutations have also been shown to confer artemisinin resistance using in-vitro susceptibility assays, albeit to a lesser degree by comparison with the R561H mutation.11
Further investigation into the origin of Pfkelch13 R561H mutations using Pfkelch13 flanking microsatellite analysis revealed that they shared similar haplotypes distinct from Pfkelch13 R561H haplotypes observed in Thailand. Similar results were obtained, using a whole-genome sequencing (WGS) analysis for the R561H isolates collected in 2012–15 from Rwanda.11 Together, these results suggest an independent origin of the Pfkelch13 R561H mutation in Rwanda.11 Microsatellite analysis has been used for similar analyses in previous evaluations,24 including landmark studies on the evolution of chloroquine25 and sulfadoxine resistance.26 Although microsatellites are less powerful than WGS, they provide a good alternative, particularly when appropriate samples for WGS are not available, as was the case in this study.
As previously reported in Africa,12,27 we identified the candidate markers Pfkelch13 C469F, P441L, and G449A and two additional mutations, V555A and R575K, whose role in artemisinin partial resistance is unknown. Studies have also detected mutations beyond codon 600 such as A675V,22 which our study did not examine. To date, the reported prevalence of the aforementioned mutations in Africa is low, however, a study showed that the prevalence of C469Y and A675V mutations has increased at multiple sites in northern Uganda (up to 23% for C469Y and 40% for A675V).22 Therefore, monitoring of these mutations in Africa is important even if the efficacy of most of the commonly used ACTs is high. It is possible that other Pfkelch13 mutations observed in African isolates but not validated so far, or mutations in other unknown genes, will affect artemisinin resistance in Africa, but this hypothesis will have to be determined by additional studies on the continent.
Most samples in this study had either the NFD or NYD haplotype of the Pfmdr1 gene, associated in some studies with decreased sensitivity to lumefantrine.28 The prevalence of these two Pfmdr1 haplotypes in this study was similar in a 2011 study, in which 59% of the P falciparum isolates collected in a southern province of Rwanda had those haplotypes.29 In our study, the NFD and NYD haplotypes were highly present in pre-treatment isolates and did not appear to be selected in post-treatment isolates; despite this high prevalence, their presence in pre-treatment isolates was not associated with recrudescent infections.
Efficacy of artemether–lumefantrine remains high in Rwanda despite the presence of Pfkelch13 mutations and delayed parasite clearance. In general, ACTs remain efficacious even if artemisinin partial resistant mutations are present as long as the partner drug is still effective. Evidence from the Mekong region has shown that once artemisinin resistance becomes prevalent, resistance to the partner drug often follows, resulting in ACT treatment failure.30 Additional studies, including parasite clearance assays within the first 3 days of treatment, in-vitro assays to determine mutant parasites’ phenotypes, and WGS to determine relatedness of mutant parasites, will further confirm our findings in Rwanda. Heightened vigilance for artemether–lumefantrine efficacy and the evaluations of the efficacy of other ACTs in Rwanda should be considered.
Supplementary Material
Research in context.
Evidence before this study
We searched PubMed, without language restrictions, for articles published after 2005 using the terms “artemisinin”, “ACTs”, “resistance”, “kelch 13” in combination with either “Africa” or “Rwanda”. Artemisinin-based combination therapies (ACTs), introduced in the early 2000s, are the primary drugs used to treat uncomplicated malaria caused by Plasmodium falciparum. Evidence for artemisinin partial resistance was first reported in 2008 in Cambodia and subsequently confirmed in other parts of southeast Asia. Artemisinin partial resistance is characterised by delayed parasite clearance as shown by microscopically detectable parasitaemia on day 3 after treatment initiation. Despite artemisinin partial resistance, the efficacy of ACTs remains high until resistance to the partner drugs occur. Accumulation of mutations in the P falciparum kelch 13 (Pfkelch13) gene have been associated with artemisinin partial resistance. Rwanda introduced artemether–lumefantrine as the first-line treatment of malaria in 2006. Past therapeutic efficacy studies (TES) in Rwanda found that artemether–lumefantrine had more than 90% efficacy. However, a Pfkelch13 mutation in codon 561 (R to H transition) known to be associated with artemisinin partial resistance, was observed at low prevalence in studies from Rwanda, including a TES we did between 2013 and 2015. Additionally, reports in neighbouring countries, Uganda and Tanzania, have reported a low prevalence of this mutation. In the 2013–15 study, association of the mutation with day 3 parasitaemia was not found but the mutation was associated with increased survival rate expressed in vitro by the ring stage survival assay. Genomic analysis of the R561H isolates showed that the mutation had evolved independently in Rwanda.
Added value of this study
This 2018 TES showed that the overall efficacy of artemether-lumefantrine remained at more than 90%, but there was evidence of delayed parasite clearance, suggesting emergence of artemisinin partial resistance in Rwanda. Day 3 parasitaemia was observed in participants in two of the three sites and was associated with pre-treatment presence of the Pfkelch13 R561H mutation in one site. Additionally, the prevalence of the Pfkelch13 R561H mutation increased compared to previous reports. This is the first documented evidence of artemisinin partial resistance in Africa. Molecular analysis of Pfkelch13 R561H mutant samples observed in our study support the previous observation of an independent evolution in Rwanda. There was no evidence of partner drug (lumefantrine) resistance, which is consistent with the more than 90% efficacy of artemether–lumefantrine observed.
Implications of all the available evidence
Emergence of artemisinin partial resistance in Africa is a warning signal that the efficacy of ACTs could become compromised should resistance to the partner drug emerge. These results highlight the importance of additional TESs to confirm the current findings and collect additional evidence for the presence of delayed parasite clearance by more frequent monitoring (every 8 h) of parasitaemia within the first 3 days following treatment. Additional molecular surveillance in different parts of the country and in bordering countries will help monitor the extent of spread of the mutant parasites to inform public health actions to mitigate the spread of this mutation. Periodic high-quality TESs are required to detect changes in the sensitivity of parasites to artemether–lumefantrine, which could affect national malaria treatment policy.
Acknowledgments
We thank the Malaria and Other Parasitic Diseases Division of the Rwanda Biomedical Center, the Maternal and Child Survival Program at Jhpiego, all the study team members, and the study participants. We thank the Division of Vector-borne Diseases, Thailand, for sharing the Plasmodium falciparum kelch 13 mutant isolates. This study was funded by the US President’s Malaria Initiative (PMI). Molecular marker testing was funded as part of the PMI Antimalarial Resistance Monitoring in Africa network. The opinions expressed herein are those of the authors and do not necessarily reflect the views of the US Centers for Disease Control and Prevention or PMI.
Footnotes
Declaration of interests
We declare no competing interests.
Data sharing
All data will be made available upon reasonable request after publication.
For the WHO therapeutic efficacy study protocol see https://www.who.int/malaria/areas/drug_resistance/efficacy-monitoring-tools/en/
See Online for appendix 2
References
- 1.WHO. Report on antimalarial drug efficacy, resistance and response: 10 years of surveillance (2010–2019). Nov 19, 2020. https://www.who.int/publications/i/item/9789240012813 (accessed March 31, 2021).
- 2.Ministry of Health Rwanda. National guidelines for the treatment of malaria in Rwanda. June 2018. http://elearning.moh.gov.rw/pluginfile.php/3000/course/overviewfiles/National%20Guidelines%20for%20the%20Treatment%20of%20Malaria%20in%20Rwanda.pdf?forcedownload=1 (accessed Jan 29, 2021).
- 3.WHO. Methods for surveillance of antimalarial drug efficacy. November, 2009. https://www.who.int/malaria/publications/atoz/9789241597531/en/ (accessed Jan 29, 2021).
- 4.Uwimana A, Penkunas MJ, Nisingizwe MP, et al. Efficacy of artemether–lumefantrine versus dihydroartemisinin–piperaquine for the treatment of uncomplicated malaria among children in Rwanda: an open-label, randomised controlled trial. Trans R Soc Trop Med Hyg 2019; 113: 312–19. [DOI] [PubMed] [Google Scholar]
- 5.Dondorp AM, Nosten F, Yi P, et al. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med 2009; 361: 455–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang F, Tang L, Yang H, et al. Molecular epidemiology of drug resistance markers of Plasmodium falciparum in Yunnan Province, China. Malar J 2012; 11: 243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kyaw MP, Nyunt MH, Chit K, et al. Reduced susceptibility of Plasmodium falciparum to artesunate in southern Myanmar. PLoS One 2013; 8: e57689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Noedl H, Se Y, Schaecher K, Smith BL, Socheat D, Fukuda MM. Evidence of artemisinin-resistant malaria in western Cambodia. N Engl J Med 2008; 359: 2619–20. [DOI] [PubMed] [Google Scholar]
- 9.Phyo AP, Nkhoma S, Stepniewska K, et al. Emergence of artemisinin-resistant malaria on the western border of Thailand: a longitudinal study. Lancet 2012; 379: 1960–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ariey F, Witkowski B, Amaratunga C, et al. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature 2014; 505: 50–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uwimana A, Legrand E, Stokes BH, et al. Emergence and clonal expansion of in vitro artemisinin-resistant Plasmodium falciparum kelch13 R561H mutant parasites in Rwanda. Nat Med 2020; 26: 1602–08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bergmann C, van Loon W, Habarugira F, et al. Increase in Kelch 13 polymorphisms in Plasmodium falciparum, southern Rwanda. Emerg Infect Dis 2021; 27: 294–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tacoli C, Gai PP, Bayingana C, et al. Artemisinin resistance-associated K13 polymorphisms of Plasmodium falciparum in southern Rwanda, 2010–2015. Am J Trop Med Hyg 2016; 95: 1090–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Halsey ES, Venkatesan M, Plucinski MM, et al. Capacity Development through the US President’s Malaria Initiative-Supported Antimalarial Resistance Monitoring in Africa Network. Emerg Infect Dis 2017; 23 (suppl 1): S53–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vinayak S, Alam MT, Sem R, et al. Multiple genetic backgrounds of the amplified Plasmodium falciparum multidrug resistance (pfmdr1) gene and selective sweep of 184F mutation in Cambodia. J Infect Dis 2010; 201: 1551–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Talundzic E, Chenet SM, Goldman IF, et al. Genetic analysis and species specific amplification of the artemisinin resistance-associated Kelch propeller domain in P falciparum and P vivax. PLoS One 2015; 10: e0136099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Plucinski MM, Morton L, Bushman M, Dimbu PR, Udhayakumar V. Robust algorithm for systematic classification of malaria late treatment failures as recrudescence or reinfection using microsatellite genotyping. Antimicrob Agents Chemother 2015; 59: 6096–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Talundzic E, Okoth SA, Congpuong K, et al. Selection and spread of artemisinin-resistant alleles in Thailand prior to the global artemisinin resistance containment campaign. PLoS Pathog 2015; 11: e1004789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nei M.Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci USA 1973; 70: 3321–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCollum AM, Basco LK, Tahar R, Udhayakumar V, Escalante AA. Hitchhiking and selective sweeps of Plasmodium falciparum sulfadoxine and pyrimethamine resistance alleles in a population from central Africa. Antimicrob Agents Chemother 2008; 52: 4089–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stepniewska K, Ashley E, Lee SJ, et al. In vivo parasitological measures of artemisinin susceptibility. J Infect Dis 2010; 201: 570–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Asua V, Conrad MD, Aydemir O, et al. Changing prevalence of potential mediators of aminoquinoline, antifolate, and artemisinin resistance across Uganda. J Infect Dis 2020; jiaa687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bwire GM, Ngasala B, Mikomangwa WP, Kilonzi M, Kamuhabwa AAR. Detection of mutations associated with artemisinin resistance at k13-propeller gene and a near complete return of chloroquine susceptible falciparum malaria in southeast of Tanzania. Sci Rep 2020; 10: 3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chenet SM, Akinyi Okoth S, Huber CS, et al. Independent emergence of the Plasmodium falciparum Kelch propeller domain mutant allele C580Y in Guyana. J Infect Dis 2016; 213: 1472–75. [DOI] [PubMed] [Google Scholar]
- 25.Wootton JC, Feng X, Ferdig MT, et al. Genetic diversity and chloroquine selective sweeps in Plasmodium falciparum. Nature 2002; 418: 320–23. [DOI] [PubMed] [Google Scholar]
- 26.Vinayak S, Alam MT, Mixson-Hayden T, et al. Origin and evolution of sulfadoxine resistant Plasmodium falciparum. PLoS Pathog 2010; 6: e1000830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ocan M, Akena D, Nsobya S, et al. bK13-propeller gene polymorphisms in Plasmodium falciparum parasite population: a systematic review protocol of burden and associated factors. Syst Rev 2018; 7: 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Venkatesan M, Gadalla NB, Stepniewska K, et al. Polymorphisms in Plasmodium falciparum chloroquine resistance transporter and multidrug resistance 1 genes: parasite risk factors that affect treatment outcomes for P falciparum malaria after artemether-lumefantrine and artesunate-amodiaquine. Am J Trop Med Hyg 2014; 91: 833–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeile I, Gahutu JB, Shyirambere C, et al. Molecular markers of Plasmodium falciparum drug resistance in southern highland Rwanda. Acta Trop 2012; 121: 50–54. [DOI] [PubMed] [Google Scholar]
- 30.Woodrow CJ, White NJ. The clinical impact of artemisinin resistance in southeast Asia and the potential for future spread. FEMS Microbiol Rev 2017; 41: 34–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.