Abstract
Viral infection-induced cell death has long been considered as a double-edged sword in the inhibition or exacerbation of viral infections. Patients with severe Coronavirus Disease 2019 (COVID-19) are characterized by multiple organ dysfunction syndrome and cytokine storm, which may result from SARS-CoV-2-induced cell death. Previous studies have observed enhanced ROS level and signs of ferroptosis in SARS-CoV-2 infected cells or specimens of patients with COVID-19, but the exact mechanism is not clear yet. Here, we find SARS-CoV-2 ORF3a sensitizes cells to ferroptosis via Keap1-NRF2 axis. SARS-CoV-2 ORF3a promotes the degradation of NRF2 through recruiting Keap1, thereby attenuating cellular resistance to oxidative stress and facilitated cells to ferroptotic cell death. Our study uncovers that SARS-CoV-2 ORF3a functions as a positive regulator of ferroptosis, which might explain SARS-CoV-2-induced damage in multiple organs in COVID-19 patients and imply the potential of ferroptosis inhibition in COVID-19 treatment.
Keywords: SARS-CoV-2, ORF3a, Ferroptosis, Keap1, NRF2
Highlights
-
•
Pathogenic strain of SARS-CoV-2, such as Delta, triggers the sign of ferroptosis in lung tissue of K18-hACE2 mice.
-
•
SARS-CoV-2 ORF3a sensitizes cell to ferroptosis through NRF2-ARE pathway.
-
•
SARS-CoV-2 ORF3a promotes the degradation of NRF2 via increasing the binding affinity between Keap1 and NRF2.
1. Introduction
SARS-CoV-2, a novel coronavirus, was identified in early 2020 as the cause of the global epidemic Coronavirus disease 2019 (COVID-19) [[1], [2], [3]]. Since its outbreak, many studies have been performed to better understand the basic mechanisms and clinical features of this disease. COVID-19 is now considered as a multi-organ dysfunction disease with a broad spectrum of manifestations, such as hyperactivation of the immune system, which leads to a ‘‘cytokine storm’’ [4]. However, the underlying mechanisms of multiple organ failure remain unclear. One possible mechanism linking cytokine storm to organ damage is the programmed cell death, which is regarded as a common outcome of virus infection [5]. Previous studies had shown that viral infections could trigger cell death via various mechanisms, depending on the viral species [6,7]. Cell death can be a double-edged sword during pathogenic infections [8,9]. On one hand, virus-associated cell death protects the host from additional infection, while on the other hand, it contributes to the progression of many infections and pathogenesis [8,10,11]. Three major forms of programmed cell death are observed following viral infections, including apoptosis, necroptosis, and pyroptosis, which were also observed in specimens from patients with severe COVID-19 [[12], [13], [14], [15]].
Ferroptosis is a newly discovered programmed cell death, which is mainly caused by iron-mediated lipid free radical accumulation and dysregulation of antioxidation responses [16,17]. Unlike the immunologically silent apoptosis, studies showed that ferroptosis is immunogenic and ferroptotic cells release damage-associated molecular patterns (DAMPs) and alarmins, which not only leads to amplified cell death but also promotes a series of reactions associated with inflammation [[18], [19], [20]]. Ferroptosis has been shown to contribute important pathological roles in multiple system diseases involving acute kidney injury (AKI), acute lung injury (ALI) and ischaemia–reperfusion injury (IRI) [17], which are typical clinical symptoms of multiple organ involvement and failure in patients with severe COVID-19 [21]. Several evidences report the ferroptosis signatures in COVID-19 patients, implying the involvement of ferroptosis in SARS-CoV-2 infection [22]. Role of Glutathione Peroxidase 4 (GPX4) and glutathione (GSH) oxidation has been associated with SARS-CoV-2 mediated lipid ROS generation from Fenton reaction [[22], [23], [24]]. The mRNA level of GPX4 was significantly decreased upon patient-derived SARS-CoV-2 (SZ005) infection [25]. 4-Hydroxynonenal (4-HNE), a reactive breakdown product of the lipid peroxides or oxidized phosphatidylcholine, which is regarded as a biomarker of ferroptosis, was found to be positive in COVID-19 patient [26]. The abundance of lipid peroxides observed in myocardium and renal cortex of patients with COVID‐19 might be triggered by ischaemia and ischaemia–reperfusion injury in these organs as seen in multiple organ dysfunction syndromes (MODS) [26]. Recently, SARS-CoV-2 infection has been reported to trigger ferroptosis in human sinoatrial node (SAN)-like pacemaker cells and human iPSC-derived cardiac cells [22,27]. In addition, iron metabolism is dysregulated in COVID-19 patients [28]. These studies demonstrated the involvement of ferroptosis in SARS-CoV-2 infection and its potential contribution to the multiple organ damage.
Coronavirus accessory proteins are a series of proteins whose genes are interspersed among or within the genes encoding structural proteins. They are not essential to the replication of coronavirus in vitro, but participate in mediating virus–host interactions, host immune responses and pathogenicity [29]. Elucidating how SARS-CoV-2-encoded proteins, especially the accessory proteins not conserved in other coronaviruses, interact with host factors is crucial for us to understand the high infectivity and pathogenicity of this virus and also to develop potential COVID-19 treatments. In this study, we found that ORF3a, an accessory protein specific to the coronavirus SARS and SARS-CoV-2, sensitizes cells to ferroptosis via Keap1-NRF2 axis. ORF3a triggers the degradation of NRF2 by recruiting Keap1, thus enhancing the accumulation of the level of lipid reactive oxygen species (ROS) and leading to ferroptosis. According to our data, it will be valuable to develop potential COVID-19 treatments toward ferroptosis inhibition and NRF2 activation, which may mitigate the adverse effects associated with the sequela of infection during the current COVID-19 pandemic.
2. Results
2.1. SARS-CoV-2 infection increased the transcription level of PTGS2
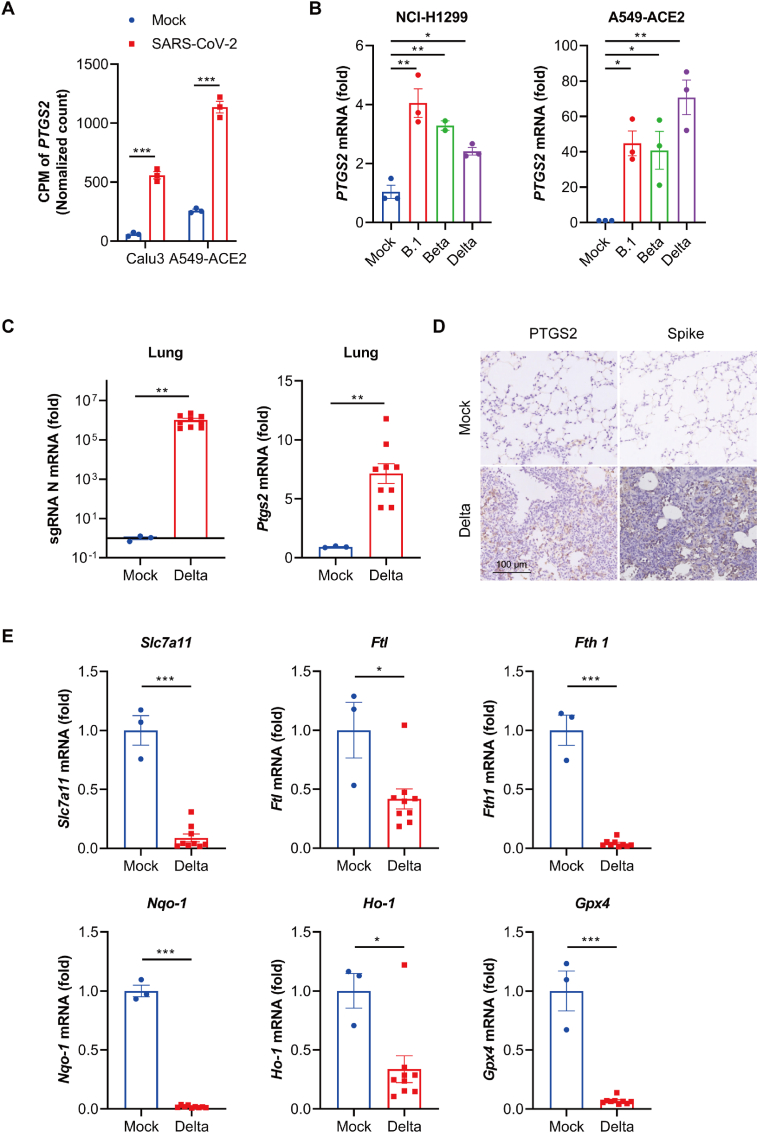
To assess whether ferroptosis is involved in SARS-CoV-2 infection, we reanalyzed an RNA-seq database of several cell lines infected with SARS-CoV-2 (GSE147507) [30]. We found that the mRNA level of prostaglandin-endoperoxide synthase 2 (PTGS2, a biomarker associated with ferroptotic cell death) [31], was significantly increased in lung cells infected with SARS-CoV-2 (Fig. 1A). To confirm the upregulation of PTGS2 by SARS-CoV-2, two human lung cell lines, NCI–H1299 and A549 with angiotensin-converting enzyme 2 (ACE2) overexpression were treated with PBS (mock-infected), or infected with three SARS-CoV-2 variants: the early strain, B.1, Beta (B.1.351) and Delta (B.1.617.2), and successful infection was confirmed by quantitative real-time polymerase chain reaction (qRT-PCR) analysis of SARS-CoV-2 subgenomic RNA (sgRNA), the indicator of coronavirus replication (Fig. S1). Meanwhile, a primary lung cell Human Bronchial Epithelial Cell (NHBE) was used to be infected with Omicron BA.2, which exhibits greater infection efficiency than other variants (B.1, Beta, Delta). As shown in Fig. S2, we didn't observe significant infection in NHBE, so we finally chose cancerous cell lines such as NCI–H1299 and A549-ACE2 to conduct the research. Result of qRT-PCR analysis verified that PTGS2 was significantly upregulated by the infection of all the variants of SARS-CoV-2 (Fig. 1B), which was consistent with data from GSE147507. Next, ferroptosis after SARS-CoV-2 infection was assessed in vivo. As SARS-CoV-2 B.1.617.2 (delta) variant had been proved to trigger severe infection, we chose this strain to evaluate ferroptosis after in vivo infection. K18-hACE2 transgenic C57BL/6 mice were mock-infected (PBS), or infected with 500 PFU of SARS-CoV-2 B.1.617.2 (delta) variant. Three days post infection, the transcriptional level and protein level of PTSG2 in the lung tissue were assessed. Result of qRT-PCR analysis of SARS-CoV-2 sgRNA in the lung showed successful infection (Fig. 1C). Compared with mock group, transcriptional level of Ptgs2 increased nearly seven folds in SARS-CoV-2 infection group (Fig. 1C). IHC results also proved the enhanced protein level of PTSG2 in lung tissue of SARS-CoV-2-infected hACE2 transgenic mice, which also showed the symptom of tissue damage (Fig. 1D). Next, we detected the transcriptional levels of relative regulators (Solute Carrier Family 7 Member 11/SLC7A11, Ferritin Light Chain/FTL, Ferritin Heavy Chain1/FTH1, NAD(P)H quinone dehydrogenase 1/NQO1, Heme Oxygenase-1/HO-1 and GPX4) of ferroptosis in lung tissue, which significantly decreased upon SARS-CoV-2 infection (Fig. 1E). These results indicate that there is a significant association between SARS-CoV-2 infection and ferroptosis.
Fig. 1.
SARS-CoV-2 infection increased the transcription level of PTSG2. (A) Reanalysis of data from Blanco-melo et al. (GSE147507)(30). Data from cell lines Calu3 and A549-ACE2 infected with SARS-CoV-2 was normalized against mock treated cells. Expression and p values were calculated with edgeR2 using Benjamini-Hochberg adjusted two-sided Wald test statistic. (B) NCI–H1299 cells and A549-ACE2 cells were infected with three indicated strains of SARS-CoV-2 variant (B.1, Beta and Delta) at a MOI of 0.05 for 48 h. Gene expression of PTGS2 was analyzed by qRT-PCR. Data show mean ± SEM from three independent experiments. (C) K18-hACE2 mice were intranasally inoculated with vehicle (n=3) or with SARS-CoV-2 Delta variant (500 plaque forming units (PFU) virus per mouse (n=9). Gene expression of sgRNA and Ptgs2 of lung tissues were harvested at 3 dpi and analyzed by qRT-PCR. (D) Lung tissues from three mice in each group were fixed to stain with PTGS2 and spike protein of SARS-CoV-2, the representative images were shown. (E) Total RNA in lung tissues from three mice in indicated groups were extracted and subjected to qRT-PCR analysis. Data represent mean ± SEM from three individual mouse. *P ≤ 0.05, **P ≤ 0.01, ***p ≤ 0.001.
2.2. SARS-CoV-2 ORF3a sensitizes cells to ferroptosis
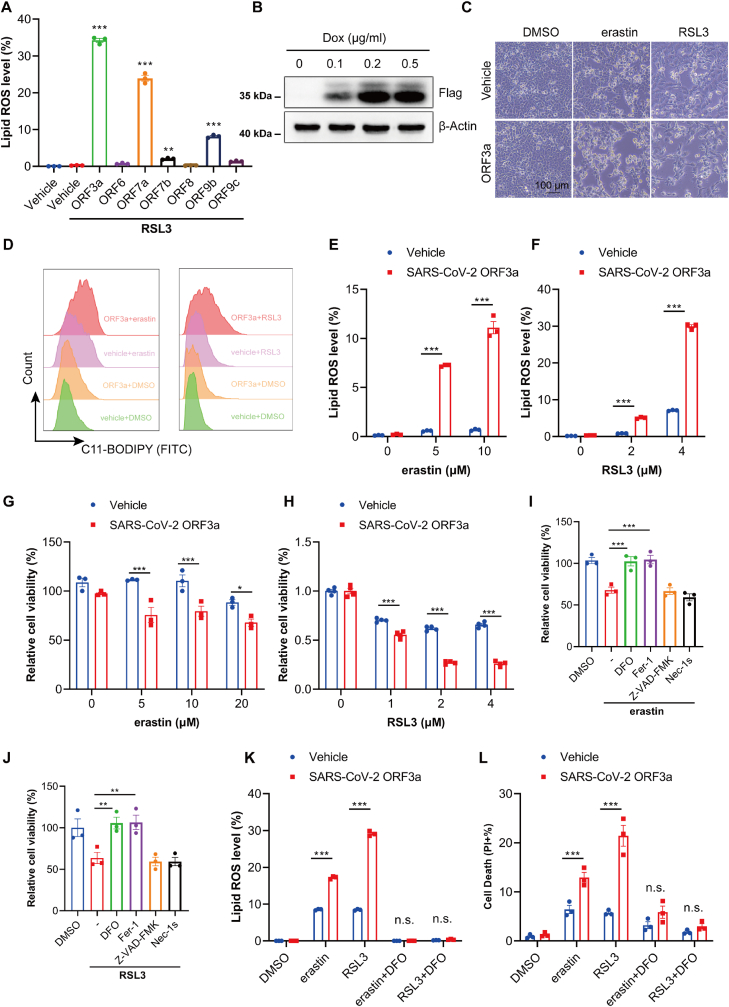
Coronavirus-encoded accessory proteins play important roles in virus–host interactions and regulate host immune responses, thereby contributing to pathogenicity of coronavirus [29,32]. Therefore, we surveyed the effect of seven accessory proteins of SARS-CoV-2 on GPX4 inhibitor RSL3 (RAS-selective lethal 3)-induced ferroptosis by detecting the accumulation of lipid ROS, a well-known ferroptosis marker [33]. Among the seven accessory proteins, SARS-CoV-2 ORF3a overexpressing cells demonstrated the highest lipid ROS level induced by RSL3 (Fig. 2A). Thus, we focused on SARS-CoV-2 ORF3a, which is simply referred to ORF3a hereafter unless the viral origin is stated. To further confirm the role of ORF3a on ferroptosis, we established a doxycycline (Dox)-regulated cell line with inducible, stable expression of flag-tagged ORF3a from NCI–H1299 cells. The stable protein expression of ORF3a in the presence of 0.1-0.5 μg/ml Dox was confirmed by Western blot (Fig. 2B). Next, we detected whether ORF3a expression renders cells susceptible to erastin and RSL3-induced ferroptosis. As shown in Fig. 2C & D, both erastin and RSL3 promoted significant cell death and enhanced lipid ROS level. Meanwhile, we also observed enhanced lipid ROS level and cell death upon RSL3 treatment in SARS ORF3a overexpressed cells, which could be rescued by deferoxamine mesylate (DFO) incubation (Figs. S3A and S3B). Furthermore, compared to control cells, erastin and RSL3-induced cell growth inhibition and lipid ROS formation were significantly increased in a dose-dependent manner in the Dox-inducible cells (Fig. 2E–H). And the effects could be completely reverted by ferroptosis inhibitors (e.g., DFO; ferrostatin-1, Fer-1), but not by inhibitors of apoptosis (e.g., Z-VAD-FMK) or necroptosis (e.g., Nec-1s) (Fig. 2I–L). These results indicate that SARS-CoV-2 ORF3a is a positive regulator of ferroptosis in cells.
Fig. 2.
SARS-CoV-2 ORF3a sensitizes cells to ferroptosis. (A) MEF cells were transfected with indicated plasmids for 24 h and treated with 0.2 μM RSL3 for 8 h, lipid ROS was detected using BODIPY-C11 by flow-cytometry. Data show mean ± SEM from three independent experiments. (B) Tet-on inducible NCI–H1299 cells were induced with different doses of doxycycline (Dox) for 24 h and the protein expression of ORF3a was assessed by Western blot. (C) Tet-on inducible NCI–H1299 cells were treated with erastin (5 μM) or RSL3 (1 μM) for 24 h, and visualized by microscope (Scale Bar =100 μm). (D) Tet-on inducible NCI–H1299 cells were treated with erastin (5 μM) or RSL3 (1 μM) for 24 h, lipid ROS were determined by flow-cytometry and represent image were shown. (E&F) Tet-on inducible NCI–H1299 cells were treated with indicated doses of erastin or RSL3 for 24 h and then cell lipid ROS was detected using BODIPY-C11 by flow-cytometry. Data were analyzed and showed as mean ± SEM from three independently experiments. (G&H) Tet-on inducible NCI–H1299 cells were treated with indicated doses of erastin or RSL3 for 24 h, cell viability was determined by CCK8 assay. Data show mean ± SEM from three independent experiments. (I&J) Tet-on inducible NCI–H1299 cells were treated with erastin (5 μM) or RSL3 (1 μM) in the presence of DFO (100 μM), Fer-1 (1 μM), Z-VAD-FMK (10 μM) and Nec1-s (10 μM) and then cell viability was assessed by CCK8 assay 24 h thereafter. Data show mean ± SEM from three independent experiments. (K) Tet-on inducible NCI–H1299 cells were treated with erastin (5 μM), RSL3 (1 μM) and DFO (100 μM) for 8 h and then lipid ROS level were determined by flow-cytometry. Data show mean ± SEM from three independent experiments. (L) Tet-on inducible NCI–H1299 cells were treated with erastin (5 μM), RSL3 (1 μM) and DFO (100 μM) for 24 h and then cell death was determined with PI staining by flow-cytometry. Data show mean ± SEM from three independent experiments. *P ≤ 0.05, **P ≤ 0.01, ***p ≤ 0.001, n.s. = not significant.
2.3. ORF3a-mediated ferroptosis is regulated by NRF2-ARE pathway
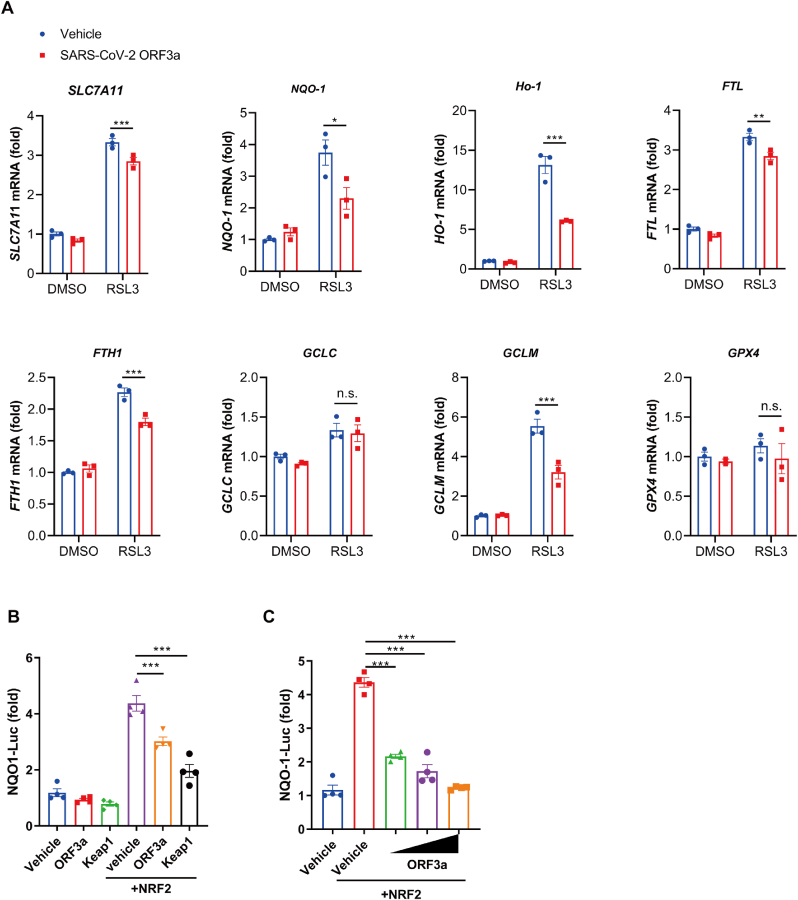
To determine the mechanism through which ORF3a induced ferroptosis, we detected the expression level of various genes involving ferroptosis. Interestingly, we found that in response to RSL3, ORF3a expression significantly decreased the mRNA levels of several antioxidant proteins (Fig. 3A & Fig. S4), especially heme oxygenase 1 (HO-1), NQO-1, FTL, FTH1 and SLC7A11, which are downstream target genes of NRF2 [34]. We questioned whether the NRF2-antioxidant response element (ARE) pathway, which is vital to cell survival response to environment, is involved in the regulation of ORF3a-mediated ferroptosis. Therefore, an ARE luciferase reporter, the NQO-1 luciferase reporter, was used to assess the ability of ORF3a to regulate NRF2-ARE pathway. Kelch-like ECH-associated protein 1 (Keap1) , a previous reported negative regulator of NRF2 was used as a positive control [35]. Expression of ORF3a significantly suppressed the luciferase expression of NQO-1, and the inhibition effect was dose-dependent (Fig. 3B & C). Thus, we speculate that ORF3a might regulate cellular ferroptotic sensitivity through NRF2-ARE pathway.
Fig. 3.
ORF3a-mediated ferroptosis is regulated by NRF2-ARE pathway. (A) Tet-on inducible NCI–H1299 cells were treated with or without doxycycline for 24h, followed by treatment with RSL3 (0.5 μM) for 12h, mRNA levels of indicated genes were quantified via qRT-PCR analysis. Quantitations are normalized to actin. Data show mean ± SEM from three independent experiments. (B&C) HEK 293T cells were transfected with indicated plasmids along with NQO-1-luc plasmid and pRL-TK for 36 h and then luciferase activity was determined by Dual Luciferase Reporter Assay. Data show mean ± SEM from three independent experiments. *P ≤ 0.05, **P ≤ 0.01, ***p ≤ 0.001, n.s. = not significant.
2.4. NRF2-driven ferroptotic inhibition genes are suppressed in SARS-CoV-2 infected cells
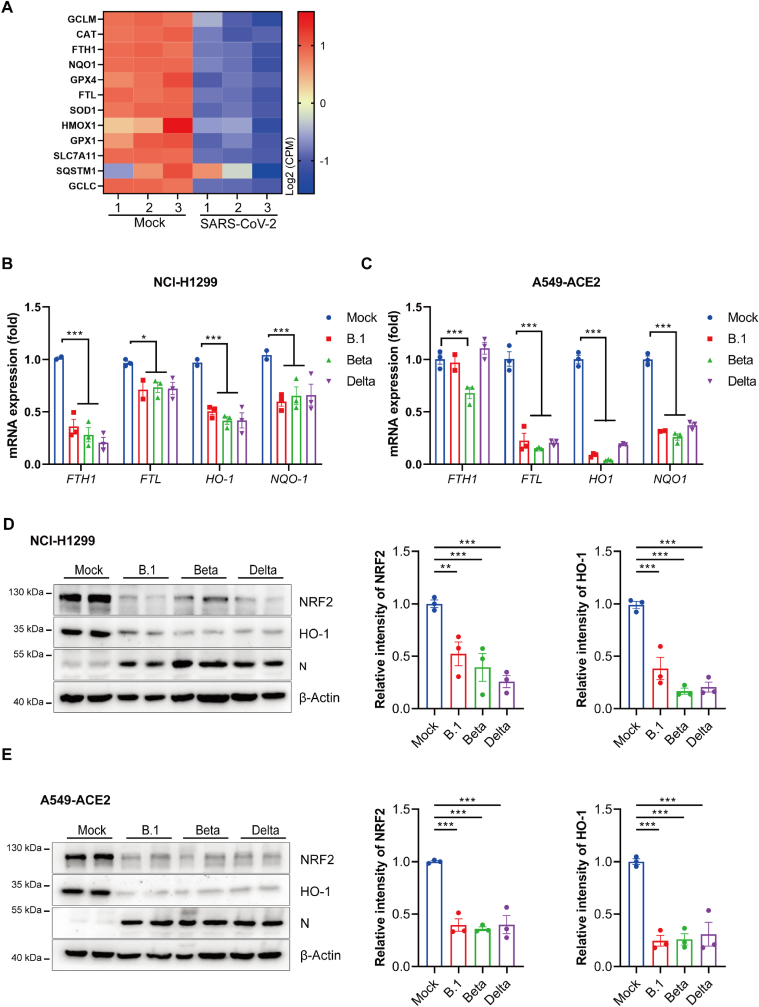
Previous study had verified that the NRF2-response is suppressed in COVID-19 patient and the expression of NRF2-dependent genes is suppressed in cells infected with SARS-CoV-2 [36]. By re-analysis of data published by Blanco-Melo et al. [30], we found that genes linked with the NRF2-dependent antioxidant response were repressed in A549-ACE2 cells during SARS-CoV-2 infection (Fig. 4A). The suppression of NRF2 pathway by SARS-CoV-2 was confirmed as the mRNA expression of NRF2 downstream target genes including FTH1, FTL, NQO-1 and HO-1 were repressed in SARS-CoV-2 infected NCI–H1299 cells and ACE2-overexpressing A549 cells (Fig. 4B & C). Correspondingly, the protein levels of NRF2 and HO-1 were also decreased upon infection of the three variants of SARS-CoV-2 both in H1299 and A549-ACE2 cells (Fig. 4D & E). Taken together, these data indicate that SARS-CoV-2 attenuates the NRF2 antioxidant pathway and SARS-CoV-2 ORF3a may play a vital role in this process, thus sensitizing cells to ferroptosis. Interestingly, we reanalyzed the data from a bioproject (https://www.ncbi.nlm.nih. gov/bioproject/PRJNA832132) and observed Omicron variant infection hardly affect the target genes of NRF2 in ferroptosis regulation in human peripheral blood mononuclear cells (PBMCs) (Fig. S5). Then we infected NCI–H1299 cells and ACE2-overexpressing A549 cells with SARS-CoV-2 variant (BA.2). In A549-ACE2, only HO-1 decreased upon infection. Although mRNA expression of FTH1, FTL and NQO-1 were repressed in NCI–H1299 but not as significantly as other variants (B.1, Beta, Delta) (Fig. S6&Fig.4B). Meanwhile, the infection slightly affected the protein level of NRF2 (Fig. S7). The weaker induction of ferroptosis sensitivity upon BA.2 infection may be one of the reasons of mild or asymptomatic infections observed in omicron variant infection.
Fig. 4.
NRF2-driven genes involving ferroptosis is suppressed in SARS-CoV-2 infected cells. (A) Indicated gene expression levels were reanalyzed from a data set conducted by Blanco-melo et al (GSE147507) (30). Heatmap shows NRF2-driven genes in SARS-CoV-2 infected versus mock treated A549-ACE2 cells. The color intensity indicates the row-scaled-normalized log2 (CPM) expression values, the columns display the data for each of the 3 replicates. (B&C) NCI–H1299 cells (B) and A549-ACE2 cells (C) were infected with three indicated strains of SARS-CoV-2 variant (B.1, Beta and Delta) at a MOI of 0.05 for 48 h mRNA levels of indicated genes were analyzed by qRT-PCR. Data show mean ± SEM from three independent experiments. (D&E) NCI–H1299 cells (D) and A549-ACE2 cells (E) were infected with three indicated strains of SARS-CoV-2 variant (B.1, Beta and Delta) at a MOI of 0.05 for 48 h. Protein levels were analyzed by immunoblotting. Quantitation was normalized to actin. The representative immunoblotting images were shown as left panel, the intensity of NRF2 and HO-1 were quantified by ImageJ software and mean ± SEM from three independent replicates were shown as right panel. *P ≤ 0.05, **P ≤ 0.01, ***p ≤ 0.001. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
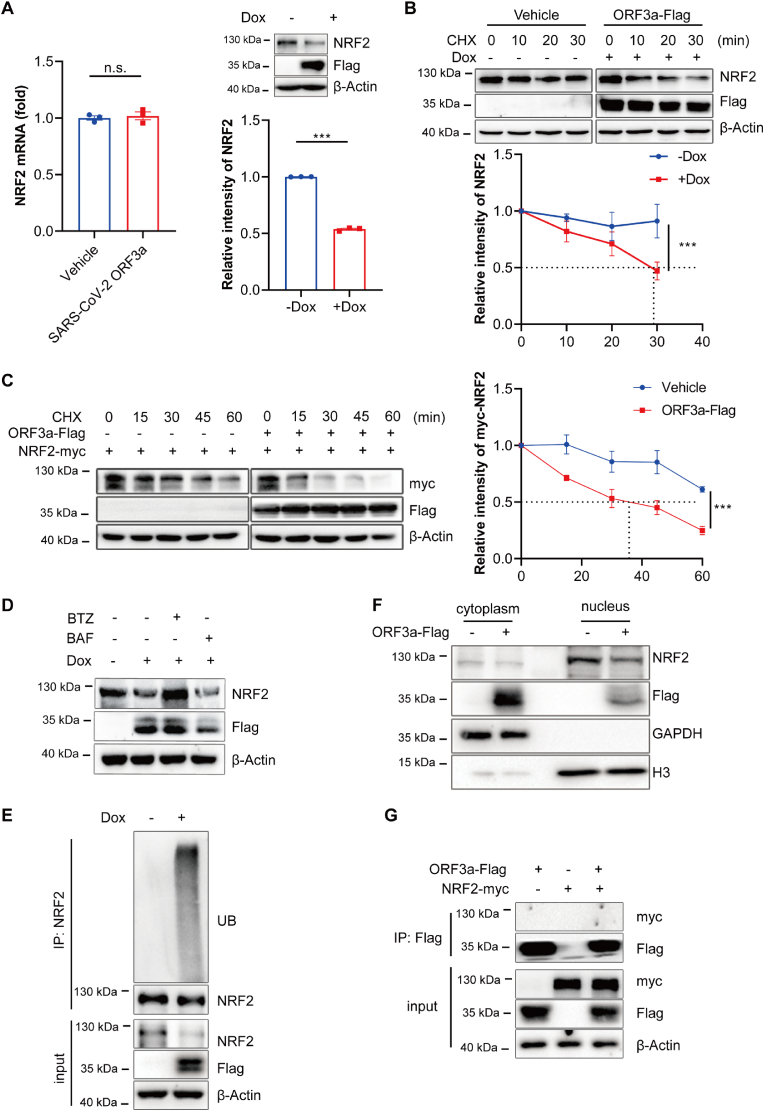
2.5. SARS-CoV-2 ORF3a promotes the degradation of NRF2
As intracellular protein levels are determined by the balance between protein synthesis and degradation, we examined these parameters of NRF2 in cells with or without ORF3a overexpression. Results revealed that ORF3a overexpression had no effect on the mRNA level of NRF2 but decreased the protein level of NRF2 (Fig. 5A), indicating that ORF3a influences the stability of NRF2. To test this possibility, we treated cells with cycloheximide (CHX, the protein synthesis inhibitor) and analyzed the degradation rate of NRF2 by chase assay. As shown in Fig. 5B, ORF3a overexpression promoted the degradation of endogenous NRF2. Meanwhile, HEK 293T cells were transiently transfected with Myc-tagged NRF2 and Flag-tagged ORF3a, and before harvest, cells were incubated with CHX and analyzed the degradation rate of NRF2. As expected, the degradation of NRF2 was accelerated upon ORF3a overexpression (Fig. 5C). To further elucidate whether ORF3a promotes NRF2 degradation via the ubiquitin-proteasome pathway, we treated Dox-inducible NCI–H1299 cells with the proteasome inhibitor MG132 and autophagy inhibitor Bafilomycin A1 (BAF). The result showed that only MG132 treatment rescued the protein level of NRF2 in ORF3a-expressing cells (Fig. 5D). Moreover, ubiquitylation level of NRF2 was dramatically increased upon ORF3a overexpression (Fig. 5E), suggesting that ORF3a destabilized NRF2 by promoting its degradation through ubiquitin-proteasome pathway.
Fig. 5.
SARS-CoV-2 ORF3a promotes the degradation of NRF2. (A) Tet-on inducible NCI–H1299 cells were treated with or without doxycycline for 24 h, mRNA expression levels and protein levels of NRF2 were assayed. The relative intensities of NRF2 were quantified by ImageJ software. Data show mean ± SEM from three independent experiments. (B) Tet-on inducible NCI–H1299 cells were treated with doxycycline followed by treatment with cycloheximide (CHX) for the indicated time courses. Lysates were analyzed by immunoblotting. The relative intensities of NRF2 were quantified by ImageJ software and and mean ± SEM from three independent replicates were shown. (C) HEK 293T cells were transfected with NRF2-myc alone or with ORF3a-Flag for 24h followed by CHX treatment for the indicated time courses. Lysates were analyzed by immunoblotting. The relative intensities of NRF2-myc were quantified by ImageJ software and mean ± SEM from three independent replicates were shown. (D) Tet-on inducible NCI–H1299 cells were treated with or without doxycycline for 24 h, followed by treated with 1 μM BTZ or 200 nM BAF for 8 h and lysates were analyzed by immunoblotting. (E) Indicated NCI–H1299 cells were treated with 1uM BTZ for 8h and subjected to an in vivo ubiquitination assay. Cell lysates were subjected to immunoprecipitation with anti-NRF2 antibody and analyzed with indicated antibodies. (F) Subcellular fractionation analysis was performed, followed by the immunoblotting analysis of cytoplasmic and nuclear fractions. (G) HEK 293T cells were transfected with ORF3a-Flag and NRF2-Myc for 36 h. Cell lysates were immunoprecipitated with anti-Flag beads and the precipitated proteins were detected with indicated antibodies. ***p ≤ 0.001, n.s. = not significant.
As an important nuclear transcription factor, NRF2 is activated in the cytoplasm and subsequently translocated into the nucleus when cells are under oxidative and/or electrophilic stimuli [35]. As shown in Fig. 5F, ORF3a overexpressing cells had lower levels of nuclear NRF2. Next, we checked whether ORF3a directly interacted with NRF2 and increased its degradation. However, immunoprecipitation assay showed that ORF3a had no interaction with NRF2 (Fig. 5G). Taken together, these data suggest that ORF3a destabilizes NRF2 by indirectly promoting its ubiquitin-proteasome degradation.
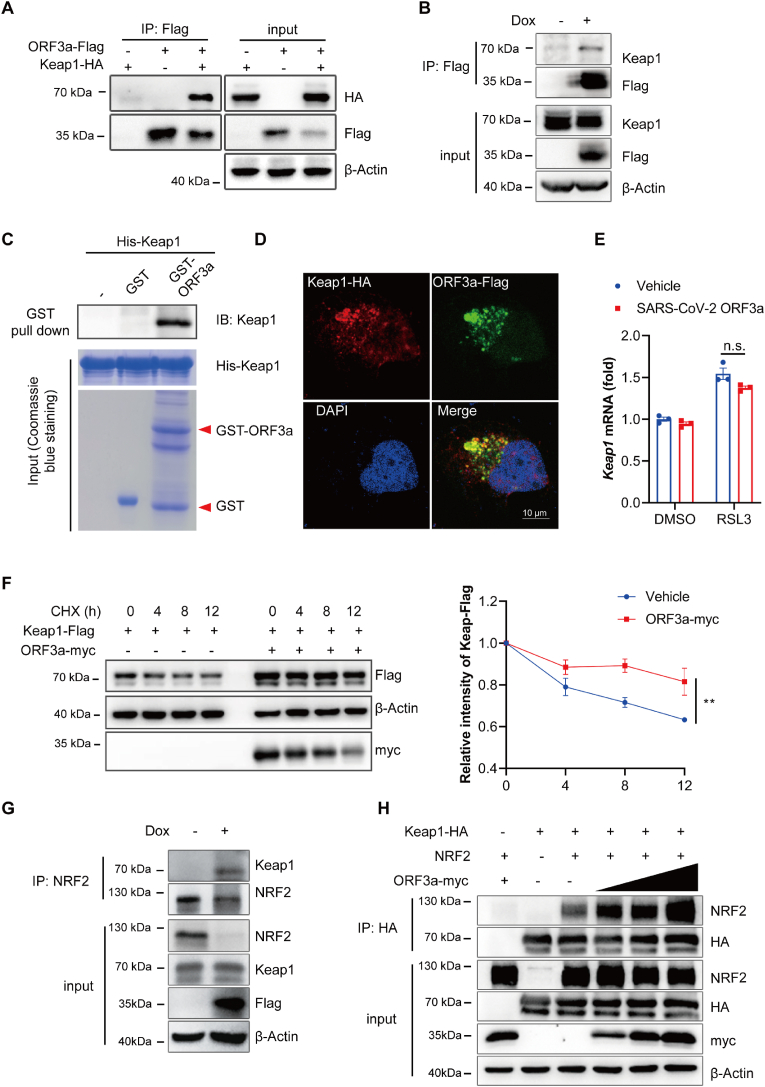
2.6. SARS-CoV-2 ORF3a enhances the binding affinity of Keap1 with NRF2
Kelch-like ECH-associated protein 1 (Keap1) is well known to act as a substrate adaptor to bring NRF2 into the Cul3-dependent E3 ubiquitin ligase complex, resulting in the rapid proteasome-mediated degradation of NRF2 [37]. Besides that, Keap1-NRF2 axis has been proved to protect against ferroptosis [38]. Thus, we explored whether Keap1 was involved in ORF3a-induced NRF2 degradation. Immunoprecipitation assay showed that ORF3a interacted with Keap1 (Fig. 6A). We also performed an immunoprecipitation assay in Dox inducible NCI–H1299 cells and found that ORF3a bound to endogenous Keap1 (Fig. 6B). In vitro GST pulldown assay verified that ORF3a directly bind to Keap1 (Fig. 6C). We also observed colocalization between Keap1 and ORF3a by immunofluorescence assay (Fig. 6D). In addition, the binding affinity between ORF3a and Keap1 was enhanced in cells treated with RSL3 (Fig. S8), further confirming that RSL3-induced NRF2 degradation was mediated by Keap1.
Fig. 6.
SARS-CoV-2 ORF3a enhances the binding affinity of Keap1 with NRF2. (A) HEK 293T cells were transfected with ORF3a-Flag and Keap1-HA for 36 h. Whole-cell lysates were immunoprecipitated with anti-Flag beads and the precipitated proteins were detected with indicated antibodies. (B) Tet-on inducible NCI–H1299 cells were treated with or without doxycycline for 36 h, cell lysates were immunoprecipitated with anti-Flag beads and detected with immunoblotting. (C) Purified fusion proteins GST-ORF3a and His-Keap1 were used for the in vitro GST pull-down assay, GST protein was used as negative control. His-Keap1 was incubated with GST or GST-ORF3a at 4 °C overnight, and then the protein was detected by immunoblotting. (D) HeLa cells were transfected with Keap1-HA and ORF3a-Flag for 24 h, then cells were fixed, stained and visualized by confocal microscopy (scale bar = 10 μm). (E) Tet-on inducible NCI–H1299 cells were treated with or without doxycycline for 24 h, followed by treatment with RSL3 (0.5 μM) for 12 h, mRNA levels of Keap1 were quantified via qRT-PCR analysis and data show mean ± SEM from three independent experiments. (F) HEK 293T cells were transfected with Keap1-Flag alone or with ORF3a-myc for 24 h followed by CHX treatment for the indicated time course. Lysates were analyzed by immunoblotting. The relative intensities of Keap1-Flag were quantified by ImageJ software and mean ± SEM from three independent replicates were shown. (G) Tet-on inducible NCI–H1299 cells were treated with or without doxycycline for 24 h, followed by treatment with BTZ (1 μM) for 8 h. Then cells were harvested and the cell lysates were immunoprecipitated with anti-NRF2 antibody and analyzed with indicated antibodies. (H) HEK 293T cells were transfected with Keap1-HA, NRF2-myc and increased dose of ORF3a-Flag for 24 h, followed by treatment with BTZ (1 μM) for 8 h. Then cells were harvested and the cell lysates were immunoprecipitated with HA beads and immunoblotted with indicated antibodies. **P ≤ 0.01, n.s. = not significant.
Then, we explored the effect of ORF3a on the expression of Keap1. Results showed that ORF3a had no effect on the mRNA level of Keap1 (Fig. 6E), but it stabilized the protein level of Keap1 and attenuated its degradation (Fig. 6F). Meanwhile, we infected NCI–H1299 cells and ACE2-overexpressing A549 cells with SARS-CoV-2 variants (B.1, Beta, Delta, BA.2). In A549-ACE2 cells, protein level of keap1 significantly increased upon SARS-CoV-2(B.1, Beta, Delta) infection, except for Omicron BA.2 strain (Fig. S9). This phenomenon was also observed in NCI–H1299 cells, although not obvious as in A549-ACE2 cell. These results were consistent with our previous data, which may explain that endogenous NRF2 degraded upon SARS-CoV-2 variants (B.1, Beta, Delta) infection (Fig. 4D and E) but not in cells infected with SARS-CoV-2 (BA.2) (Fig. S7). Next, we observed much stronger endogenous interaction between NRF2 and Keap1 in the presence of ORF3a in NCI–H1299 cells (Fig. 6G). Moreover, results of co-immunoprecipitation showed that NRF2-Keap1 interaction was enhanced in a dose-dependent manner of ORF3a (Fig. 6H). These results imply that ORF3a directly binds to Keap1 and enhances the stability of Keap1, thus promoting Keap1-NRF2 interaction and facilitating the degradation of NRF2.
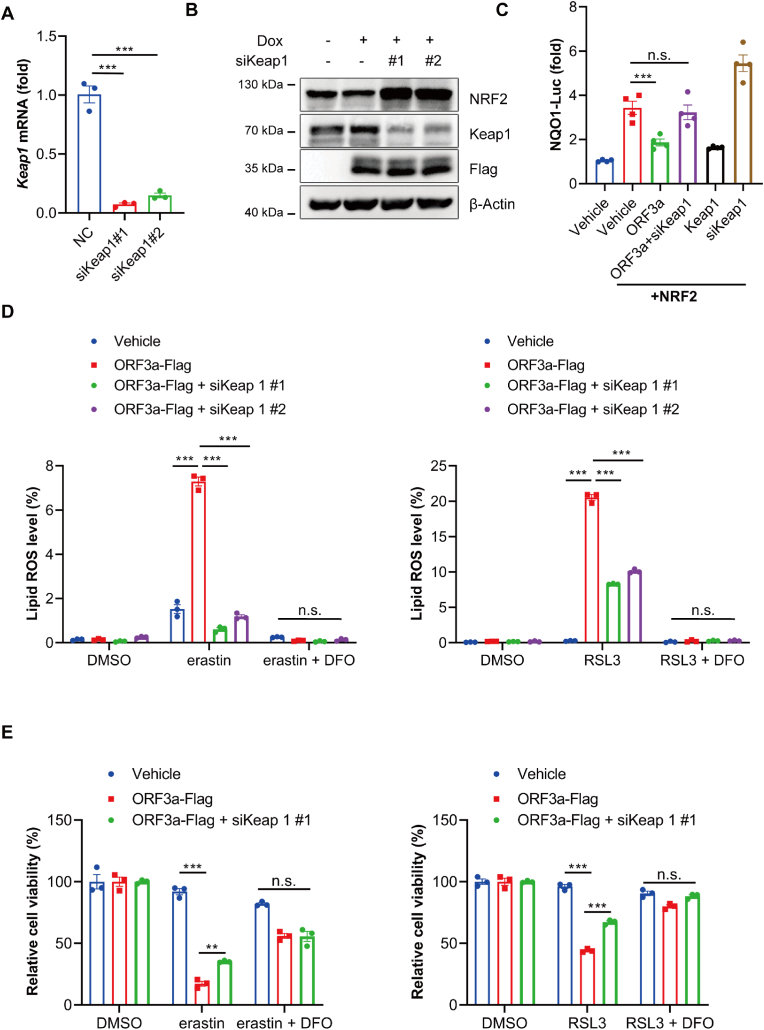
2.7. SARS-CoV-2 ORF3a sensitizes cellular ferroptosis through Keap1-NRF2 axis
To further explore the role of Keap1-NRF2 pathway in ORF3a-induced ferroptosis, we knocked down Keap1 by using siRNAs in ORF3a-overexpressing NCI–H1299 cells (Fig. 7A&7B). Next, we employed NQO1-Luc reporter assay to detect the activity of NRF2 on downstream target genes. The result showed that the overexpression of ORF3a downregulated the NQO1 luciferase activity by about 50%, which could be abolished when Keap1 was suppressed (Fig. 7C), indicated that downregulation of NRF2 mediated by ORF3a overexpression is rescued by Keap1 knockdown. To examine the effects of Keap1 on ORF3a-induced ferroptosis, we downregulated Keap1 in ORF3a-expressing cells and observed the cell death and lipid ROS induced by ferroptosis inducers. ORF3a significantly increased the lipid ROS and cell death induced by both erastin and RSL3 treatment, whereas Keap1 knockdown partially rescued the lipid ROS in ORF3a overexpressed cells and protected cells from ferroptotic cell death. (Fig. 7D & E). These results indicate that the ability of ORF3a to promote ferroptosis depends on the NRF2-Keap1 axis.
Fig. 7.
SARS-CoV-2 ORF3a sensitizes cellular ferroptosis through Keap1-NRF2 axis. (A) NCI–H1299 cells were transfected with siKeap1 for 48 h, mRNA level was assayed by qRT-PCR. (B) Tet-on inducible NCI–H1299 cells were transfected with siRNA negative control or siKeap1 for 24 h, then treated with doxycycline for 24 h. Protein level was assayed by immunoblotting. (C) HEK 293T cells were transfected with siRNA negative control or siKeap1 and indicated plasmids for 48 h, and then luciferase activity was determined by Dual Luciferase Reporter Assay. Data show mean ± SEM from three independent experiments. (D) Tet-on inducible NCI–H1299 cells were transfected with siRNA negative control or siKeap1 for 24 h, followed by incubation with doxycycline for 24 h, then cells were treated with erastin, RSL3 and DFO for 8 h, and lipid ROS were determined by flow-cytometry. Data show mean ± SEM from three independent experiments. (E) Tet-on inducible NCI–H1299 cells were transfected with siRNA negative control or siKeap1 for 24 h, followed by treatment with doxycycline for 24 h, then cells were incubated with erastin, RSL3 and DFO for 12 h, and cell viability was determined by CCK8 assay. Data show mean ± SEM from three independent experiments. **P ≤ 0.01, ***P ≤ 0.001, n.s. = not significant.
3. Discussion
Infection with SARS-CoV-2 has resulted in a pandemic that has widespread effects all over the world. The clinical symptoms of severe COVID-19 includes a broad spectrum of clinical disease, and the typical clinical presentations of severe patients contain acute respiratory distress syndrome, cytokine release syndrome (CRS), multiorgan failure, and death [39,40]. Cytokine storm induced by hyperactivation of the immune system and multiple organ failure have been suggested as contributory factors in COVID-19 disease severity [41]. Programmed cell death are demonstrated as a major trigger of inflammatory process, linking cytokine storm to organ damage. Several types of programmed cell death, such as apoptosis, necroptosis and pyroptosis have been observed in specimens from patients with COVID-19 [15,42,43]. Here, we provide evidences that a novel type of programmed cell death, ferroptosis, is also involved in SARS-CoV-2 infection. We demonstrated that ORF3a, an accessory protein encoded by SARS-CoV-2, contributes to the induction of ferroptosis, which at least in part, explains the tissue damage and multiple organ failure caused by SARS-CoV-2.
The ORF3a protein is unique to SARS-CoV and SARS-CoV-2, which has multiple functions in critical aspects of virus pathogenicity. The different functional domains of SARS-CoV-2 ORF3a have been demonstrated to associate with virulence, infectivity, and virus release [44]. In animal models of SARS-CoV-2 infection, genomic deletion of ORF3a showed a general decrease of the cytokine storm, the reduction of virus infection and tissue damage in lungs, indicating a major role of ORF3a in viral pathogenesis [45]. Previous studies implicated that ORF3a is an inducer of cell death including apoptosis, necrosis and pyropotsis [[46], [47], [48]]. In this study, we identified that ORF3a of SARS-CoV-2 could mediate the down-regulation of NRF2 and its downstream antioxidant response, therefore sensitizing cells to ferroptosis. In the context, it is speculated that ORF3a induces various cell death through different strategies, which leads to tissue damage that affects the severity of COVID-19.
Iron overload induced lipid free radical accumulation and dysregulation of antioxidative responses are considered as two major reasons for ferroptosis [17]. Several studies reported the dysregulated iron homeostasis in COVID-19 patients. Clinical studies showed that serum ferritin levels in severe COVID-19 patients were significantly higher than the mild cases [49,50], which is likely caused by releasing the intracellular ferritin due to cell death and tissue damage. Virus replication requires iron to serve as a raw material [51]. Transferrin receptor is responsible for transporting iron into the cell. A study showed that the expression of transferrin is higher in males and age-related, which is correlation with the severity and mortality of male and elderly COVID-19 patients [52]. Therefore, overwhelming iron uptake upon virus infection would result in the accumulation of intracellular labile iron, triggering Fenton reaction and generating excessive lipid ROS, which cannot be eliminated by the cellular antioxidation system. Viral infections are associated with a decrease in antioxidant defenses. A previous study found that oxidative burst recruits neutrophils to sites of infection where they kill the pathogens, which may contribute to organ damage and mortality in COVID-19 [53]. Consequently, ferroptosis may occur due to the dysregulation of iron homeostasis in COVID-19 patients.
Differing from apoptosis, ferroptosis is a form of immunogenic cell death that can trigger inflammatory responses through releasing DAMPs [54]. Inhibiting ferroptosis prevents certain diseases by anti-inflammatory mechanisms [55]. As ferroptosis inhibitors such as deferoxamine and deferiprone are usually iron chelators, which are generally safe and possess antiviral activities, further efforts should be devoted to assess whether anti-ferroptosis treatment can be considered as the therapeutic strategies to treat COVID-19. Indeed, some strategies inhibiting ferroptosis showed potentiality in COVID-19 treatment. Two ferroptosis inhibitors, rosiglitazone and rosiglitazone, could reduce viral yields of coronavirus, including SARS-CoV-2 [56].
In general, NRF2 and its responsible genes protect against stress-induced cell death and regulate the inflammatory response and tissue damage during infection. Under basal conditions, NRF2 in the cytosol interacts with its negative regulator protein Keap1 that targets NRF2 for ubiquitination and proteasomal degradation [57]. Redox products inactivate Keap1 by modifying specific sensor cysteine residues, promoting its dissociation with NRF2, and then NRF2 translocates into the nuclear to activate antioxidant defenses [58]. The NRF2 pathway has been found to be repressed in lung biopsies of COVID-19 patients, and pharmacological inducer of NRF2 inhibits the replication of SARS-CoV-2 and the inflammatory response [36]. Consistent with previous studies, our findings supported the suppression of the NRF2 pathway during infection with SARS-CoV-2, highlighting the potential of activation of NRF2 to be an attractive antiviral approach against COVID-19 while limiting overflowing inflammatory host responses. Indeed, experimental evidence is beginning to emerge and NRF2 activators such as sulforaphane and bardoxolone methyl are under clinical trials. For example, NRF2 activator SFN (phytochemical sulforaphane) has been reported to exhibit in vitro and in vivo antiviral activity against SARS-CoV-2 [59]. Some natural chemical compounds targeting the Keap1-NRF2 axis may have the potential for anti-SARS-CoV-2 therapy, such as andrographolide [60]. Bardoxolone and bardoxolone methyl, two NRF2 activators in clinical trials, showed their SARS-CoV-2 inhibition activity [61]. Recently, NRF2 agonists 4-octyl-itaconate (4-OI) and dimethyl fumarate (DMF) have also been proved to inhibit the replication of SARS-CoV-2 and suppress the inflammatory responses to SARS-CoV-2 in different cell types [36].
Besides the Keap1-NRF2 axis, there may be other possible mechanisms of ferroptosis in relation to SARS-CoV-2 infection. In this paper, we also observed other accessory proteins, ORF7a and ORF9b, enhanced cellular ferroptosis sensitivity in MEF cells (Fig.2A). Some researchers had demonstrated that ORF7a hires anti-apoptosis protein and aggregates on the ER, resulting in ER stress and apoptosis initiation, which may trigger ferroptosis [62,63]. A study showed that SARS ORF9b protein was associated with nucleocytoplasmic export linked apoptosis [64]. SARS-CoV-2 ORF9b had been proved to bind the outer mitochondrial membrane and causes mitochondrial elongation, manipulating host cell mitochondria and its function [65]. SARS-CoV-2-encoded proteins nsp4 and ORF9b had been proved synergistically induce mtDNA release [66]. The release of mtDNA into the cytosol or the extracellular space is intricately associated with the activation of different PRRs, which may lead to cell death [67,68]. Based on these results, it worthy to further study the role of ORF7a and ORF9b in ferroptosis regulation.
In summary, we revealed that ferroptosis is associated with SARS-CoV-2 infection. ORF3a of SARS-CoV-2 sensitizes cells to ferroptosis via Keap1-NRF2 axis. Mechanistically, ORF3a acts as a molecular linker, directly binds Keap1 and recruits Keap1 to NRF2, leading to the proteasome degradation of NRF2. The degradation of NRF2 dampens the cellular antioxidant ability and sensitized cells to ferroptosis. Our study provides new evidence to promote the development of antiviral drugs specifically targeting the impairment of antioxidant pathway by ORF3a, thereby enhancing the therapeutic value and mitigating the adverse effects against SARS-CoV-2 infection.
Funding
This work was supported by the National Natural Science Foundation of China, China (81620108020 to D.G., 32041002 to D.G., 81702724 to H.P. 32200109 to S.Y.), Shenzhen Science and Technology Program, China (JSGG20200225150431472 to D.G., KQTD20180411143323605 D.G., JCYJ20190807161009621 to H.P., JSGG20220606141003007 to H.P.)
Author contributions
LL, JD, SY, BZ, and HP performed the experiments. JS, XL and LG analyzed the data. HP, DG and LL designed the experiments and wrote the manuscript. CK and XP provided the omicron strain BA.2. LC, SH, JH and CL reviewed the manuscript. All authors read and approved the final paper.
Declaration of competing interest
As the authors of this article, we accept the explanations and conditions listed below:
-
1.
We confirm that the manuscript has been read and approved by all named authors and that there are no other persons who satisfied the criteria for authorship but are not listed. We further confirm that the order of the authors listed in the manuscript has been approved by all of us.
-
2.
We confirm that the manuscript has been read and approved by all named authors and that there are no other persons who satisfied the criteria for authorship but are not listed. We further confirm that the order of the authors listed in the manuscript has been approved by all of us.
-
3.
Scientific, ethical and legal responsibility of the article belongs to us as the authors. We are responsible for the ideas, comments and accuracy of references in the article.
-
4.
For quoted parts (text, figures, tables, pictures), legal permission has been taken from the copyright holder (s) and we agree that all financial and legal responsibility belongs to us.
-
5.
The above mentioned research has been carried out as a part of our work and is open to use by the community.
-
6.
We wish to confirm that there are no known conflicts of interest associated with this publication and there has been no significant financial support for this work that could have influenced its outcome.
-
7.
We understand that the Corresponding Author is the sole contact for the Editorial process. He/she is responsible for communicating with the other authors about progress, submissions of revisions and final approval of proofs.
Acknowledgments
We thank Prof. Pei-hui Yang (School of Basic Medical Sciences, Cheeloo College of Medicine, Shandong University, Jinan) for kindly offering pCAGGS-3′Flag-ORF3a, ORF6, ORF7a, ORF7b, ORF8, ORF9b and ORF9c constructs. We thank Prof. Hui Zhang for providing SARS-CoV-2 strain B.1 and the Center for Disease Control and Prevention of Guangdong Province for providing the SARS-CoV-2 variants. We thank the Biosafety Level 3 (BSL-3) facilities of Sun Yat-sen University (Guangzhou), Guangzhou Customs District Technology Center (Guangzhou).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2023.102752.
Contributor Information
Deyin Guo, Email: guodeyin@mail.sysu.edu.cn.
Hong Peng, Email: pengh37@mail.sysu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Wu F., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou P., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu N., et al. A novel coronavirus from patients with pneumonia in China. N. Engl. J. Med. 2019;382:727–733. doi: 10.1056/NEJMoa2001017. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nalbandian A., et al. Post-acute COVID-19 syndrome. Nat. Med. 2021;27:601–615. doi: 10.1038/s41591-021-01283-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Danthi P. Viruses and the diversity of cell death. Annu Rev Virol. 2016;3:533–553. doi: 10.1146/annurev-virology-110615-042435. [DOI] [PubMed] [Google Scholar]
- 6.Castillo J.A., Urcuqui-Inchima S. Mechanisms of monocyte cell death triggered by dengue virus infection. Apoptosis. 2018;23:576–586. doi: 10.1007/s10495-018-1488-1. [DOI] [PubMed] [Google Scholar]
- 7.Kabiljo J., Laengle J., Bergmann M. From threat to cure: understanding of virus-induced cell death leads to highly immunogenic oncolytic influenza viruses. Cell Death Dis. 2020;6:48. doi: 10.1038/s41420-020-0284-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jorgensen I., Rayamajhi M., Miao E.A. Programmed cell death as a defence against infection. Nat. Rev. Immunol. 2017;17:151–164. doi: 10.1038/nri.2016.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ashida H., et al. Cell death and infection: a double-edged sword for host and pathogen survival. J. Cell Biol. 2011;195:931–942. doi: 10.1083/jcb.201108081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naidoo J., et al. Pneumonitis in patients treated with anti-programmed death-1/programmed death ligand 1 therapy. J. Clin. Oncol. 2017;35:709–717. doi: 10.1200/JCO.2016.68.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Legrand A.J., Konstantinou M., Goode E.F., Meier P. The diversification of cell death and immunity: memento mori. Mol. Cell. 2019;76:232–242. doi: 10.1016/j.molcel.2019.09.006. [DOI] [PubMed] [Google Scholar]
- 12.Jérôme Hadjadj N.Y. Laura Barnabei, Aurélien Corneau, etal, Impaired type I interferon activity and inflammatoryresponses in severe COVID-19 patient. Science (New York, N.Y.) 2020;369:7. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fox S.E., et al. Unexpected features of cardiac pathology in COVID-19 infection. Circulation. 2020;142:1123–1125. doi: 10.1161/CIRCULATIONAHA.120.049465. [DOI] [PubMed] [Google Scholar]
- 14.Li S., et al. Clinical and pathological investigation of patients with severe COVID-19. JCI Insight. 2020;5 doi: 10.1172/jci.insight.138070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andre S., et al. Cell Death Differentiation; 2022. T Cell Apoptosis Characterizes Severe Covid-19 Disease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dixon S.J., et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li J., et al. Ferroptosis: past, present and future. Cell Death Dis. 2020;11:88. doi: 10.1038/s41419-020-2298-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin-Sanchez D., et al. Ferroptosis, but not necroptosis, is important in nephrotoxic folic acid–induced AKI. J. Am. Soc. Nephrol. 2017;28:218–229. doi: 10.1681/ASN.2015121376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim E.H., Wong S.-W., Martinez J. Programmed Necrosis and Disease:We interrupt your regular programming to bring you necroinflammation. Cell Death Differ. 2019;26:25–40. doi: 10.1038/s41418-018-0179-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Proneth B., Conrad M. Ferroptosis and necroinflammation, a yet poorly explored link. Cell Death Differ. 2019;26:14–24. doi: 10.1038/s41418-018-0173-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lopes-Pacheco M., et al. Pathogenesis of multiple organ injury in COVID-19 and potential therapeutic strategies. Front. Physiol. 2021;12 doi: 10.3389/fphys.2021.593223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han Y., et al. SARS-CoV-2 infection induces ferroptosis of sinoatrial node pacemaker cells. Circ. Res. 2022;130:963–977. doi: 10.1161/CIRCRESAHA.121.320518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Y., Xu Y., Zhang K., Shen L., Deng M. Ferroptosis in COVID-19-related liver injury: a potential mechanism and therapeutic target. Front. Cell. Infect. Microbiol. 2022;12 doi: 10.3389/fcimb.2022.922511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fratta Pasini A.M., Stranieri C., Girelli D., Busti F., Cominacini L. Is ferroptosis a Key component of the process leading to multiorgan damage in COVID-19? Antioxidants. 2021;10 doi: 10.3390/antiox10111677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y., et al. SARS-CoV-2 suppresses mRNA expression of selenoproteins associated with ferroptosis, endoplasmic reticulum stress and DNA synthesis. Food Chem. Toxicol. 2021;153 doi: 10.1016/j.fct.2021.112286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacobs W., et al. ESC Heart Fail; 2020. Fatal Lymphocytic Cardiac Damage in Coronavirus Disease 2019 (COVID-19): Autopsy Reveals a Ferroptosis Signature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.K S., Perez-Bermejo Juan A., Rockwood Sarah J., et al. SARS-CoV-2 infection of human iPSC-derived cardiac cells reflects cytopathic features in hearts of patients with COVID-19. Sci. Transl. Med. 2021;13 doi: 10.1126/scitranslmed.abf7872. eabf7872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bellmann-Weiler R., et al. Prevalence and predictive value of anemia and dysregulated iron homeostasis in patients with COVID-19 infection. J. Clin. Med. 2020;9 doi: 10.3390/jcm9082429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fang P., Fang L., Zhang H., Xia S., Xiao S. Functions of coronavirus accessory proteins: overview of the state of the art. Viruses. 2021;13 doi: 10.3390/v13061139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blanco-Melo D., et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045.e1039. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang W.S., et al. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156:317–331. doi: 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Redondo N., Zaldívar-López S., Garrido J.J., Montoya M. SARS-CoV-2 accessory proteins in viral pathogenesis: knowns and unknowns. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.708264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang W.S., Stockwell B.R. Ferroptosis: death by lipid peroxidation. Trends Cell Biol. 2016;26:165–176. doi: 10.1016/j.tcb.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song X., Long D. Nrf2 and ferroptosis: a new research direction for neurodegenerative diseases. Front. Neurosci. 2020;14:267. doi: 10.3389/fnins.2020.00267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kensler T.W., Wakabayashi N., Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu. Rev. Pharmacol. Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 36.Olagnier D., et al. SARS-CoV2-mediated suppression of NRF2-signaling reveals potent antiviral and anti-inflammatory activity of 4-octyl-itaconate and dimethyl fumarate. Nat. Commun. 2020;11:4938. doi: 10.1038/s41467-020-18764-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hayes J.D., Dinkova-Kostova A.T. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem. Sci. 2014;39:199–218. doi: 10.1016/j.tibs.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 38.Sun X., et al. Activation of the p62-Keap1-NRF2 pathway protects against ferroptosis in hepatocellular carcinoma cells. Hepatology (Baltimore, Md. 2016;63:173–184. doi: 10.1002/hep.28251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mehta P., et al. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jose R.J., Manuel A. COVID-19 cytokine storm: the interplay between inflammation and coagulation. Lancet Respir. Med. 2020;8:e46–e47. doi: 10.1016/S2213-2600(20)30216-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fajgenbaum D.C., June C.H., Storm Cytokine. N. Engl. J. Med. 2020;383:2255–2273. doi: 10.1056/NEJMra2026131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tang D., Comish P., Kang R. The hallmarks of COVID-19 disease. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1008536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koupenova M., et al. SARS-CoV-2 initiates programmed cell death in platelets. Circ. Res. 2021;129:631–646. doi: 10.1161/CIRCRESAHA.121.319117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Issa E., Merhi G., Panossian B., Salloum T., Tokajian S. SARS-CoV-2 and ORF3a: nonsynonymous mutations, functional domains, and viral pathogenesis. mSystems. 2020;5 doi: 10.1128/mSystems.00266-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Silvas J.A., et al. Contribution of SARS-CoV-2 accessory proteins to viral pathogenicity in K18 human ACE2 transgenic mice. J. Virol. 2021;95 doi: 10.1128/JVI.00402-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yue Y., et al. SARS-Coronavirus Open Reading Frame-3a drives multimodal necrotic cell death. Cell Death Dis. 2018;9:904. doi: 10.1038/s41419-018-0917-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ren Y., et al. The ORF3a protein of SARS-CoV-2 induces apoptosis in cells. Cell. Mol. Immunol. 2020;17:881–883. doi: 10.1038/s41423-020-0485-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Olagnier D., et al. SARS-CoV2-mediated suppression of NRF2-signaling reveals potent antiviral and anti-inflammatory activity of 4-octyl-itaconate and dimethyl fumarate. Nat. Commun. 2020;11:4938. doi: 10.1038/s41467-020-18764-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ruan Q., Yang K., Wang W., Jiang L., Song J. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 2020;46:846–848. doi: 10.1007/s00134-020-05991-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu C., et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in wuhan, China. JAMA Intern. Med. 2020;180:934–943. doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Drakesmith H., Prentice A. Viral infection and iron metabolism. Nat. Rev. Microbiol. 2008;6:541–552. doi: 10.1038/nrmicro1930. [DOI] [PubMed] [Google Scholar]
- 52.McLaughlin K.-M., et al. COVID-19-Related coagulopathy-is transferrin a missing link? Diagnostics. 2020;10:539. doi: 10.3390/diagnostics10080539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barnes B.J., et al. Targeting potential drivers of COVID-19: neutrophil extracellular traps. J. Exp. Med. 2020;217 doi: 10.1084/jem.20200652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen D., et al. ORF3a of SARS-CoV-2 promotes lysosomal exocytosis-mediated viral egress. Dev. Cell. 2021;56:3250–3263 e3255. doi: 10.1016/j.devcel.2021.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sun Y., et al. The emerging role of ferroptosis in inflammation. Biomed. Pharmacother. 2020;127 doi: 10.1016/j.biopha.2020.110108. [DOI] [PubMed] [Google Scholar]
- 56.C H.-J., Kunga Yu-An, Lib Mei-Ling, Gonga Yu-Nong, et al. Acyl-coenzyme A synthetase long-chain family member 4 is involved in viral replication organelle formation and facilitates virus replication via ferroptosis. mBio. 2022;13 doi: 10.1128/mbio.02717-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cuadrado A., et al. Therapeutic targeting of the NRF2 and KEAP1 partnership in chronic diseases. Nat. Rev. Drug Discov. 2019;18:295–317. doi: 10.1038/s41573-018-0008-x. [DOI] [PubMed] [Google Scholar]
- 58.Ma Q. Role of nrf2 in oxidative stress and toxicity. Annu. Rev. Pharmacol. Toxicol. 2013;53:401–426. doi: 10.1146/annurev-pharmtox-011112-140320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ordonez A.A., et al. Sulforaphane exhibits antiviral activity against pandemic SARS-CoV-2 and seasonal HCoV-OC43 coronaviruses in vitro and in mice. Communications Biology. 2022;5:242. doi: 10.1038/s42003-022-03189-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schulte B., et al. Andrographolide derivatives target the KEAP1/NRF2 Axis and possess potent anti-SARS-CoV-2 activity. ChemMedChem. 2022;17 doi: 10.1002/cmdc.202100732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun Q., et al. Bardoxolone and bardoxolone methyl, two Nrf2 activators in clinical trials, inhibit SARS-CoV-2 replication and its 3C-like protease. Signal Transduct. Targeted Ther. 2021;6:212. doi: 10.1038/s41392-021-00628-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tan Y.X., et al. Induction of apoptosis by the severe acute respiratory syndrome coronavirus 7a protein is dependent on its interaction with the Bcl-XL protein. J. Virol. 2007;81:6346–6355. doi: 10.1128/JVI.00090-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhixin Liu e. Ubiquitination of SARS-CoV-2 ORF7a prevents cell death induced by recruiting BclXL to activate ER stress. American society for microbiology. 2022;10 doi: 10.1128/spectrum.01509-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sharma K., et al. SARS-CoV 9b protein diffuses into nucleus, undergoes active Crm1 mediated nucleocytoplasmic export and triggers apoptosis when retained in the nucleus. PLoS One. 2011;6 doi: 10.1371/journal.pone.0019436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Srinivasan K., Pandey A.K., Livingston A., Venkatesh S. Roles of host mitochondria in the development of COVID-19 pathology: could mitochondria be a potential therapeutic target? Mol Biomed. 2021;2:38. doi: 10.1186/s43556-021-00060-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Faizan M.I., et al. NSP4 and ORF9b of SARS-CoV-2 induce pro-inflammatory mitochondrial DNA release in inner membrane-derived vesicles. Cells. 2022;11 doi: 10.3390/cells11192969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.West A.P., Shadel G.S. Mitochondrial DNA in innate immune responses and inflammatory pathology. Nat. Rev. Immunol. 2017;17:363–375. doi: 10.1038/nri.2017.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Riley J.S., Tait S.W. Mitochondrial DNA in inflammation and immunity. EMBO Rep. 2020;21 doi: 10.15252/embr.201949799. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.