Summary
While our knowledge of gene expression in different human cell types is rapidly expanding with advances in transcriptomic profiling technologies, the next challenge is to understand gene function in each cell type. CRISPR-Cas9-based functional genomics screening offers a powerful approach to determine gene function in a high-throughput manner. With the maturation of stem cell technology, a variety of human cell types can be derived from human pluripotent stem cells (hPSCs). Recently, the integration of CRISPR screening with hPSC differentiation technologies opens up unprecedented opportunities to systematically examine gene function in different human cell types and identify mechanisms and therapeutic targets for human diseases. This review highlights recent progress in the development and applications of CRISPR-Cas9-based functional genomics screening in hPSC-derived cell types, discusses current challenges and limitations, and outlines future directions for this emerging field.
Keywords: CRISPR technology, functional genomics, human pluripotent stem cells, cell differentiation, genetic screens
Graphical abstract
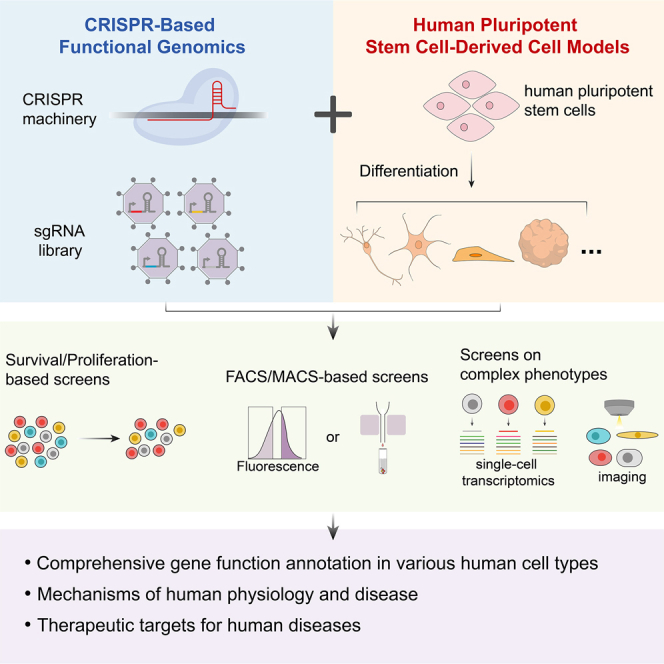
Li et al. review recent advances and existing challenges in the development and application of CRISPR-based functional genomics screening in human-pluripotent-stem-cell-derived cell models. This emerging technology holds great potential for systematically examining gene function in various human cell types and identifying mechanisms and therapeutic targets for human diseases. This field will continue to advance with the rapid development of CRISPR technology and stem cell technology.
Introduction
The human body is composed of hundreds of different cell types, each performing a unique function. In recent years, tremendous efforts have been made to characterize the gene expression signatures of various human cell types using bulk or single-cell/single-nucleus transcriptomics, leading to the generation of molecular reference maps of human cells, represented by the Human Cell Atlas (https://www.humancellatlas.org/).
Beyond gene expression, the next challenge is to systematically characterize the function of human genes in different cell types and uncover how they regulate human biology and disease. Such studies have been made possible by recent developments in human pluripotent stem cell (hPSC) technology and CRISPR-based functional genomics.
hPSC technology
hPSCs, including human embryonic stem cells (hESCs) and human induced pluripotent stem cells (hiPSCs), are characterized by their abilities of self-renewal and differentiation. hESCs were first isolated from human embryos by James Thomson in 1998.1 Later, the revolutionary discovery that somatic cells can be reprogrammed into pluripotent stem cells by introducing four Yamanaka factors — Oct4, Sox2, Klf4, and c-Myc — led to the first generation of hiPSCs from human fibroblasts in 2007.2,3
hPSCs hold the potential to generate almost all human cell types in vitro, providing opportunities to model normal development or disease states of human cell types that are otherwise inaccessible, such as various types of brain cells. Numerous efforts have been made to develop hPSC differentiation methods. To date, many human cell types can be derived from hPSCs, including endoderm cells (e.g., hepatocytes, pancreatic cells, lung, and intestinal epithelial cells), mesoderm cells (e.g., cardiomyocytes, vascular, and hematopoietic cells), and ectoderm cells (e.g., neurons and astrocytes)4,5 (Figure 1). hPSCs can also be differentiated into 3D organoids, such as brain organoids, kidney organoids, and lung organoids4,5 These organoid models can more reliably mimic the architecture, functionality, and cellular heterogeneity of the organ or tissue of interest, providing a more physiologically relevant setting than 2D models for studying complex biological processes (such as tissue development) and disease mechanisms.
Figure 1.
CRISPR-based functional genomics screening in hPSC-derived cell models
hESCs derived from the inner cell mass of human blastocysts or hiPSCs derived from human somatic cells are engineered to express the CRISPR machinery—CRISPRn for gene knockout, CRISPRi for gene repression, or CRISPRa for gene activation—by lentiviral infection or homologous recombination-based gene integration into the safe-harbor locus. Typically, the hPSCs are then transduced with a sgRNA library and differentiated into the cell type of interest. Instead of transducing at the hPSC stage, the sgRNA library can also be delivered after differentiation. According to the phenotype of interest, different screening strategies can be employed, including survival/proliferation-based and FACS/MACS-based screens for one-dimensional phenotypes and single-cell-transcriptomics-based and imaging-based screens for complex phenotypes. TSS, transcriptional start site.
The rapid evolution of hPSC technology has opened up a new era for disease modeling and drug discovery. For example, hiPSCs can be derived from patients and differentiated into disease-relevant cell types. These cellular models derived from patient hiPSCs have been proven to recapitulate major pathological phenotypes of the disease seen in clinical samples.4,5 For example, iPSC-derived neurons from patients with familial or sporadic Alzheimer’s disease (AD) exhibit pathological hallmarks of AD such as enlarged early endosomes, increased amyloid-β, and phosphorylated tau.6,7 iPSC-derived cardiac myocytes from patients carrying pathogenic mutations of long-QT syndrome, a cardiac electrophysiologic disorder, recapitulate the electrophysiological features of the disorder.8 For more detailed discussions of the applications of hPSC technology in disease modeling and drug discovery, see reviews by Shi et al.4 and Rowe et al.5
CRISPR-based functional genomics screening
Large-scale functional genomics screening is a powerful approach to systematically probe gene function. Its implementation in human cells has been substantially facilitated by the advent of new CRISPR-based technologies. Adapted from a bacterial innate immune system, CRISPR-Cas9 and its derivatives are robust and versatile tools for gene editing and manipulation in human cells.9 Attributed to the programmable and multiplexable nature of CRISPR-based technologies, they have been widely applied to introduce massively parallel genetic perturbations in large-scale functional genomics screens.10,11,12 Currently, three types of CRISPR-based perturbations are commonly used in such screens (as shown in Figure 1): (1) CRISPR knockout (CRISPRn), which utilizes the Cas9 nuclease to disrupt a target gene by introducing frameshift indels, (2) CRISPR interference (CRISPRi), which utilizes a catalytically dead Cas9 (dCas9) fused with a transcriptional repressor domain to silence the transcription of a target gene, and (3) CRISPR activation (CRISPRa), which utilizes dCas9 fused with transcriptional activator domains to activate the transcription of a target gene. Compared with CRISPRn, CRISPRi does not induce DNA double-strand breaks and thus is less toxic to cells that are sensitive to DNA damage, such as hPSCs.13
Different screening strategies can be employed depending on phenotypes of interest10,11,12 (Figure 1). For example, survival/proliferation-based screens can be used to identify essential genes and genes that modify the sensitivity of cells to a certain insult, such as drug treatment. Fluorescence-activated cell sorting (FACS)-based screens can be used to identify regulators of cellular phenotypes that can be reflected by fluorescent signals, such as fluorescent dyes, genetically encoded fluorescent reporters, or fluorescent antibodies. Phenotypes focusing on the levels of cell surface protein of interest can be screened using magnetic-activated cell sorting (MACS). Additionally, when combined with other high-dimensional profiling technologies, CRISPR-based screens can be used for more complex phenotypes.14 For example, CROP-seq, or other similar technologies such as Perturb-seq and CRISP-seq, combines CRISPR screening with single-cell RNA sequencing, allowing screens using transcriptomic changes as readout.11,14 The integration with high-content imaging enables CRISPR-based screens for other complex phenotypes, such as cell morphology, protein localization, and cell-cell interaction.11,14
This review focuses on the specific applications of CRISPR-based functional genomics screening in hPSC-derived models. For more general discussions on CRISPR-based genetic manipulation tools and screening methods, please refer to other recent reviews.10,11,12,14
CRISPR-based screens in hPSC-derived cell types
While previous CRISPR-based screens were performed mainly in cancer or immortalized cell lines, as well as in stem cells,11 recent efforts in combining CRISPR technology with hPSC differentiation methods have enabled such screens in various types of differentiated human cells. We summarize the CRISPR-based screens that have been carried out in hPSC-derived cell types so far, as listed in Table 1 and discussed in the sections below.
Table 1.
Current studies using CRISPR-based screens in hPSC-derived cells
| Year | Stem cell type | Differentiated cell type | CRISPR type | Screening strategy | Phenotype | Library size | Reference |
|---|---|---|---|---|---|---|---|
| 2018 | hiPSC | hepatocyte-like cell | CRISPRn | FACS | hepatocytic differentiation | genome-wide | Li et al.15 |
| 2019 | hiPSC & hESC | neural progenitor | CRISPRn | survival | cytotoxicity of Zika virus infection | genome-wide | Li et al.16 |
| 2019 | hiPSC | glutamatergic neuron | CRISPRi | survival | neuronal survival | CRISPRi-v2 H1 library (∼2,000 genes encoding kinases, phosphatases, and drug targets) | Tian et al.17 |
| single-cell transcriptomics | transcriptome changes in response to gene knockdown | 27 hit genes | |||||
| imaging | neurite morphology | 23 hit genes | |||||
| 2020 | hESC | cerebral organoid | CRISPRn | proliferation | cerebral organoid growth | 172 microcephaly candidate genes | Esk et al.18 |
| 2020 | hESC | cardiac mesoderm and progenitor | CRISPRn | FACS | formation of cardiac mesoderm and progenitors (MSP1 and ISL1 staining) | ∼6,000 genes | Xu et al.19 |
| 2021 | hiPSC | glutamatergic neuron | CRISPRi/a | survival | neuronal survival under normal and oxidative stress conditions | genome-wide (hCRISPRi/a-v2 library) | Tian et al.20 |
| FACS | levels of ROS and lipid peroxidation | genome-wide & a focused library against 730 hit genes | |||||
| single-cell transcriptomics | transcriptome changes in response to gene knockdown | 184 hit genes for CRISPRi and 100 hit genes for CRISPRa | |||||
| 2021 | hiPSC | cardiomyocyte | CRISPRn | survival | doxorubicin-induced cardiotoxicity | genome-wide | Sapp et al.21 |
| 2022 | hiPSC | neural stem cell | CRISPRi | proliferation; FACS | cell proliferation during neuronal induction; neural differentiation (PAX6 staining) | genome-wide libraries against coding and lncRNA genes | Wu et al.22 |
| single-cell transcriptomics screens | transcriptome changes in response to gene knockdown | 240 targets (120 coding genes and 120 lncRNA genes) | |||||
| 2022 | hiPSC | human subpallial organoid | CRISPRn | FACS | interneuron differentiation | 425 neurodevelopmental disorder related genes | Meng et al.23 |
| human forebrain assembloid | interneuron migration | ||||||
| 2022 | hiPSC | cortical neuron | CRISPRn | survival | cytotoxicity of PR20 dipeptide repeats | kinome-wide (736 kinases) | Guo et al.24 |
| 2022 | hiPSC | astrocyte | CRISPRi | FACS | inflammatory reactivity of astrocytes (synaptosome phagocytosis; cell-surface VCAM1 levels) | human transcription factors and druggable genome (~4,000 targets) | Leng et al.25 |
| single-cell transcriptomics | transcriptome changes in response to gene knockdown | 30 hit genes | |||||
| 2022 | hiPSC | microglia | CRISPRi/a | survival/proliferation | microglia survival and proliferation | CRISPRi-v2 H1 library (∼2,000 genes encoding kinases, phosphatases, and drug targets) | Dräger et al.26 |
| FACS | microglia activation (CD38 staining) | ||||||
| microglia phagocytosis of pHrodo-Red-labeled synaptosomes | |||||||
| single-cell transcriptomics | transcriptome changes in response to gene knockdown | 39 hit genes | |||||
| 2022 | hiPSC | kidney organoid | CRISPRn | proliferation | kidney development and cell proliferation | genome-wide | Ungricht et al.27 |
This table summarizes current studies using CRISPR-based screens in hPSC-derived cells. CRISPRn, CRISPR knockout; CRISPRi, CRISPR interference; CRISPRa, CRISPR activation; hPSC, human pluripotent stem cell; hESC, human embryonic stem cell; hiPSC, human induced pluripotent stem cell; FACS, fluorescence-activated cell sorting.
Survival/proliferation-based screens
Cell survival or proliferation is the simplest and most scalable readout for a pooled CRISPR screen, and it is also directly relevant to many human diseases. For example, neurodegenerative diseases, such as AD, are characterized by progressive neuronal loss in patients. In order to identify genes regulating human neuronal survival, Tian et al. developed a CRISPRi/a-based screening platform in hiPSC-derived neurons.17,20 In this system, hiPSCs were engineered to stably express the CRISPRi or CRISPRa machinery as well as an inducible expression cassette for NGN2, a transcription factor that can drive neuronal differentiation when overexpressed.28 Using this platform, Tian et al. performed the first genome-wide CRISPRi and CRISPRa-based survival screens in human neurons and identified neuronal-specific essential genes.17,20 As neurons are particularly vulnerable to oxidative stress in aging and neurodegenerative diseases, Tian et al. also conducted a genome-wide CRISPRi screen in hiPSC-derived neurons under chronic oxidative stress and identified the selenoprotein GPX4, a master suppressor of ferroptosis, and the genes PSTK, SEPHS2, and SEPSECS, responsible for selenoprotein synthesis, as top essential genes for neurons to survive under oxidative stress.20
Survival/proliferation-based screens have been used to identify modifiers and potential therapeutic targets for human diseases. In the context of frontotemporal dementia (FTD) and amyotrophic lateral sclerosis (ALS), Guo et al. performed a kinome-wide CRISPRn screen in hiPSC-derived cortical neurons to identify modifiers of polyPR toxicity associated with repeat expansion in C9ORF72, which is a major genetic cause of ALS and FTD.24 From the screen, NEK6 was identified and validated as a key regulator and a potential therapeutic target.24
Survival/proliferation-based screens have also been used to study viral infection. Li et al. conducted a genome-wide survival-based CRISPRn screen in hPSC-derived neural progenitor cells to uncover host factors mediating the toxicity of Zika virus infection.16 Among the top hits were genes involved in endocytosis and interferon signaling.16
Compared to 2D cell cultures, 3D organoids can more reliably model in vivo cell environments.29 In 2020, Esk et al. conducted a CRISPRn screen in hESC-derived cerebral organoids using a focused sgRNA library targeting 173 microcephaly candidate genes to find regulators for cerebral organoid growth.18 In order to improve screening quality in organoids, they developed a new method termed CRISPR-LICHT, which uses barcoded sgRNAs, enabling lineage tracing to control for inherent variability in cell growth of heterogeneous populations in the organoids.18 From the screen, they identified that IER3IP1, a regulator of ER function and extracellular matrix protein secretion, is crucial for tissue integrity and brain-size control.18
In addition to the nervous system, survival/proliferation-based screens have also been performed in cell types from other systems. For example, Sapp et al. conducted a genome-wide CRISPRn screen in hiPSC-derived cardiomyocytes to investigate the mechanism of doxorubicin-induced cardiotoxicity.21 Two transporters—SLCO1A2 and SLCO1B3—were identified as important modulators of this process.
Proliferation-based screens have also been done in 3D organoid models to study tissue development. For example, Ungricht et al. performed a genome-wide CRISPRn screen in hiPSC-derived kidney organoids to uncover proliferation regulators of different lineages during kidney development.27 In this study, longitudinal sampling and FACS sorting were used to determine sgRNA enrichment in different types of kidney cells at different differentiation stages of kidney organoids.27 A large dataset of genes regulating kidney development, including those associated with human kidney diseases, was generated from the screen.27
FACS/MACS-based screens
The application of FACS and MACS greatly expands the range of phenotypes that can be screened by CRISPR-based functional genomics. FACS- or MACS-based screens can be used for any phenotypes that can be monitored by fluorescent signals via flow cytometry or distinguished by the expression of specific cell surface proteins via antibody-conjugated magnetic beads, respectively.11
FACS/MACS-based screens have been used to identify genes driving cell fate determination. Li et al. performed a genome-wide CRISPRn screen to identify regulators of hepatic differentiation.15 This study established a reporter hiPSC line by tagging the endogenous hepatocyte marker ALB with a fluorescent protein, Venus. After inducing the differentiation of the reporter cells toward hepatocyte-like cells (HLCs), a FACS-based screen on Venus intensity was performed. From the screen, they identified and validated HDAC3 as a master regulator of hepatic differentiation.15 In another study, Xu et al. performed FACS-based CRISPRn screens focusing on the generation of cardiac mesoderm and progenitors, using immunofluorescence staining against cardiac mesoderm marker MESP1 and cardiac progenitor marker ISL1 as readouts.19 The screens identified ZIC2 as a necessary factor for cardiac progenitor formation.19
In addition to protein-coding genes, non-coding RNAs, especially lncRNAs, play important roles in various cellular processes. Previously, a CRISPRi-based screening platform has been established to investigate lncRNA function in hiPSCs and cancer cell lines.30 In a recent study by Wu et al., dual genome-wide screens against all coding and lncRNA genes in the human genome were performed to identify regulators of neural induction, using immunostaining against the canonical neural stem cell marker PAX6 as a readout.22 The screens identified several coding and lncRNA hits that modulate neural differentiation.22
3D organoids provide a more physiological environment for cell differentiation.29 Multiple organoids representing different tissue types or regions can be further assembled into assembloids.29 In a recent study, Meng et al. developed a CRISPR-based screening strategy in human brain assembloids.23 Using an interneuron reporter, Dlxi1/2beGFP, they conducted FACS-based CRISPRn screens in hiPSC-derived human subpallial organoids (hSO) and human forebrain assembloids (hFA), which were generated by the integration of hSO with human cortical organoids (hCO), for regulators of interneuron generation and migration, respectively. From a sgRNA library targeting 425 genes associated with autism spectrum disorder and other neurodevelopmental disorders, they identified genes affecting human interneuron differentiation, including the RNA-binding protein CSDE1 and the activator of TGFβ signaling SMAD4, and genes affecting interneuron migration, including cytoskeleton genes and the endoplasmic reticulum (ER)-related gene LNPK.23 They also demonstrated an interesting role of ER displacement in interneuron migration.23
FACS/MACS-based screens are also potent at identifying genes regulating cellular homeostasis and response to stimuli. Using a fluorescent dye for reactive oxygen species (ROS), Tian et al. performed a genome-wide FACS-based CRISPRi screen in hiPSC-derived neurons focusing on neuronal redox homeostasis.20 The lysosomal gene PSAP, which is associated with PD, was identified as a top hit for ROS level regulation in human neurons. Follow-up mechanistic investigations uncovered that loss of PSAP impairs sphingolipid degradation in the lysosome, leading to the formation of lipofuscins, which trap iron, generating ROS and triggering neuronal ferroptosis.20
Glial cells, including microglia and astrocytes, play critical roles in supporting neurons and maintaining homeostasis in brain development and brain function. They are sensitive to environmental stimuli and can change their cellular status in response. Activation of glial cells is considered a major driver for many neurological diseases, such as AD. To understand the mechanism underlying microglia activation, Dräger et al. developed a novel microglia differentiation method based on the overexpression of 6 transcription factors in hiPSCs and established a CRISPRi/a screening platform in these cells.26 FACS-based screens were conducted to identify regulators for microglia activation and phagocytosis using CD38 immunofluorescent staining and uptake of pHrodo-Red-labeled synaptosomes as readouts, respectively.26 Similarly, Leng et al. established a CRISPRi screening platform in hiPSC-derived astrocytes and used it to investigate astrocyte reactivity induced by inflammatory cytokines. FACS-based screens were performed focusing on phagocytosis as measured by the uptake of pHrodo-labeled synaptosomes and cell-surface levels of VCAM1 as measured by immunofluorescent staining.25
Screens on complex phenotypes
Coupling CRISPR-based screening with high-dimensional profiling technologies has enabled functional genomics screens on more complex phenotypes in hPSC-derived cell types. Two major technologies for complex phenotype screens are single-cell transcriptomics-based screening and imaging-based screening, which are developed for transcriptomic and optical phenotypes, respectively.
Empowered by high-throughput single-cell transcriptomics, single-cell transcriptomics-based screening (as is represented by CROP-seq) can reveal gene expression changes in cells in response to a large number of genetic perturbations in parallel at single-cell resolution. For example, Tian et al. performed CROP-seq as secondary screens to characterize transcriptome changes in hiPSCs and hiPSC-derived neurons in response to the knockdown of 27 hit genes identified from the primary survival/proliferation screens.17 They uncovered striking neuron-specific responses to several gene knockdowns, such as that of MAT2A. Dräger et al. performed CROP-seq in hiPSC-derived microglia for 39 hit genes selected from their primary screens on microglia survival and activation. They uncovered genes whose knockdown could change microglia cell states.17 Leng et al. used CROP-seq to characterize the effect of knocking down 30 selected genes on the inflammatory reactivity of hiPSC-derived astrocytes.25
Methods for imaging-based screening in pooled or arrayed formats have been developed for rich phenotypes such as cellular structure morphology, organelle dynamics, and protein/RNA localization.11 Tian et al. performed a longitudinal arrayed imaging screen in hiPSC-derived neurons on neuronal morphology and uncovered genes whose knockdown could affect neurite length and branching.17
Current challenges and future directions
While the technology for utilizing CRISPR-based functional genomics screening in hPSC-derived models shows great promise, it is still in an early stage and requires further development. In the following sections, we highlight some of the current challenges, point out several considerations when conducting such screens, and outline several directions for future research.
hPSC differentiation
Although hPSCs have the potential to be differentiated into almost all cell types in three germ layers, only a few of them (mostly brain cell types) have been used in CRISPR-based screens so far, as summarized above. This is in part because of the limitation of the current hPSC differentiation methods, which only allow for the robust generation of a limited number of cell types, such as neurons and cardiomyocytes. A successful CRISPR-based screen relies on a homogeneous starting cell population and sufficient sgRNA coverage (typically 100–1,000 cells per sgRNA) for ensuring screening quality and reproducibility. However, the existing differentiation methods for some cell types suffer from low efficiency, high variability between batches, high heterogeneity within differentiated cells, and high reagent and labor costs, thus limiting the applications of these cell types in large-scale CRISPR screens.
Another challenge in using hPSC-derived cell models is the maturity of differentiated cells. The majority of current hPSC differentiation protocols produce immature/fetal-like cells. This property is suitable for studying early tissue development and cell-fate specification, as exemplified by some of the above-mentioned studies.15,18,19,22,23,27 However, it limits the applications of hPSC-derived models in screens for phenotypes that require mature or aged cells, such as phenotypes involved in degenerative diseases.
To address these challenges, new hPSC differentiation methods that can robustly generate new cell types and matured or aged cells in a cost-efficient way need to be developed. One attractive approach would be to identify single or combinations of transcription factors (TFs) that can drive hPSC differentiation into specific cell types or accelerate the maturation of hPSC-derived cells. Compared with chemical-based differentiation methods, TF-based differentiation methods have advantages, including shorter differentiation periods and higher homogeneity,31 and thus may be preferred for large-scale genetic screens.
CRISPR-based technologies and functional genomics screening methods
A technical consideration when conducting CRISPR screens in hPSC-derived cells is the timing of sgRNA library delivery—whether at the hPSC stage or after differentiation. Since hPSCs are typically more efficient for transduction by lentivirus and easier to expand to sufficient quantities than differentiated cells, it is more common to deliver an sgRNA library at the hPSC stage. However, one potential caveat of doing so is that some genetic perturbations in hPSCs may interfere with differentiation, thus confounding the screening results. To overcome this challenge, inducible CRISPR systems can be used to allow gene perturbation to be initiated only at the desired stage, as has been applied in many above-mentioned studies.17,18,20,26,27,30 Further optimization of the inducible CRISPR tools to reduce their leakiness and improve their efficiencies is an important direction for future research.
While existing screens in hPSC-derived cells utilize CRISPRn/i/a to generate genetic perturbations, the introduction of other CRISPR-based tools would unlock new screening strategies. For example, base editing and prime editing, which introduce single-nucleotide variants (SNVs) into the genome, could be used for large-scale screens in hPSC-derived cells to directly evaluate the effect of disease-associated genetic variants in relevant cell types.32,33,34
In addition, many exciting new screening methods have recently been developed for screens on complex phenotypes in cell lines, which can be readily applied to hPSC-derived models. For example, pooled optical screening, which applies imaging-based screening in a pooled format, offers unprecedented opportunities to assess the effect of gene perturbation on rich imaging phenotypes in high spatial resolution and at large scale.14
Currently, single-cell-RNA-sequencing-based screens are mainly used as secondary screens for relatively small sets of genes selected from primary screens. As the expenses for library preparation and sequencing continue to decline rapidly, it will become feasible to conduct larger-scale, even genome-wide, single-cell-RNA-sequencing-based screens in hPSC-derived models. Moreover, fueled by the development of single-cell multi-omics technologies, CRISPR screens could be coupled with multiplexed single-cell readouts, including transcriptome, epigenome, and proteome, providing comprehensive molecular characterizations of gene perturbations.
CRISPR-based screens in hiPSC-derived disease models
While existing CRISPR-based screens in hiPSC-derived cells mainly used hiPSCs from healthy donors, an attractive application of this system is to perform parallel modifier screens on disease-relevant phenotypes in cell models derived from both healthy donors and patients, including those with disease-associated genetic variants. Comparison of hits from these screens could reveal disease mechanisms and potential therapeutic targets. However, one challenge for such screens is the intrinsic variability between different donors, which may significantly reduce the accuracy of the screens. One way to overcome this challenge is to use isogenic controls, which can be generated either by correcting the pathogenic mutations in patient-derived iPSC lines or by introducing disease mutations into a “wild-type” iPSC line using CRISPR-based gene editing. One example of the latter approach is the hiPSC Neurodegenerative Disease Initiative (iNDI) project, which aims to generate and characterize isogenic iPSC lines for more than 100 genetic variants associated with Alzheimer’s disease and related dementias.35
Toward building comprehensive gene function maps for human cells
It is expected that, with the advances in both hPSC differentiation methods and CRISPR-based screening technology, a growing number of large-scale functional genomics screens will be carried out in a variety of human cell types on a range of cellular phenotypes, ultimately moving toward building comprehensive gene function maps for all human cells.
Data portals for sharing and visualizing these screening data will be of great value to the scientific community. As an example, the CRISPRbrain Data Commons20 (http://crisprbrain.org/) has been established as an open-access platform for depositing, visualizing, and exploring functional genomics screens in differentiated human cell types. These data portals will complement others, such as the Human Cell Atlas, which focuses on gene expression signatures in different human cell types, and the Cancer Dependency Map (https://depmap.org/portal/), which focuses on genetic screens in cancer cell lines.
To enhance reproducibility and facilitate data integration of screens performed in hPSC-derived models, reference hPSC lines and standardized differentiation protocols could be established and shared in the scientific community via stem cell biobanks and open-source platforms. For example, the Allen Cell Collection (https://www.allencell.org/cell-catalog.html) offers various fluorescently tagged hiPSC lines as well as CRISPRi hiPSC lines that are generated from the same parental WTC-11 line, while the iNDI project uses the KOLF2.1J line as the parental cell line for generating isogenic hiPSC lines carrying different disease-related variants, which are available through the Jackson Laboratory (https://www.jax.org/jax-mice-and-services/ipsc).35 In addition, given the potential high variability and heterogeneity in the outcomes of hPSC differentiation, it is recommended to include technical and biological replicates (e.g. different batches of differentiation or different genetic backgrounds of hPSC lines) in the screens to ensure data rigor and reproducibility.
Conclusions
The integration of CRISPR-based functional genomics screening with hPSC differentiation technology opens up new opportunities to unravel gene function in diverse human cell types and identify mechanisms and therapeutic targets for human diseases in more physiologically relevant contexts. Given the rapid development of CRISPR technology and stem cell technology, this field will continue to evolve and advance our understanding of human physiology and disease.
Acknowledgments
We thank Xiaoyan Guo for comments on this manuscript and members of the Tian lab for discussions. Research in the Tian lab is supported by the National Natural Science Foundation of China (No. 32100766 and No. 82171416), Guangdong Basic and Applied Basic Research Foundation (No. 2023B1515020075), the Science, Technology and Innovation Commission of Shenzhen Municipality (No. RCBS20210609103800006, No. JCYJ20220530112602006 and No. RCYX20221008092845052), the Lingang Laboratory Grant (No. LG-QS-202203-11) and Provincial College Students’ Innovation and Entrepreneurship Training Program (No. S202214325035).
Author contributions
K.L. performed the literature review and made Table 1 under the guidance of J.Z. and R.T. R.T. made Figure 1 with input from M.O. All authors contributed to the paper writing and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Jiangshan Zhan, Email: zhanjs@bjmu.edu.cn.
Ruilin Tian, Email: tianrl@sustech.edu.cn.
References
- 1.Thomson J.A., Itskovitz-Eldor J., Shapiro S.S., Waknitz M.A., Swiergiel J.J., Marshall V.S., Jones J.M. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 2.Yu J., Vodyanik M.A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J.L., Tian S., Nie J., Jonsdottir G.A., Ruotti V., Stewart R., et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 3.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 4.Shi Y., Inoue H., Wu J.C., Yamanaka S. Induced pluripotent stem cell technology: a decade of progress. Nat. Rev. Drug Discov. 2017;16:115–130. doi: 10.1038/nrd.2016.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rowe R.G., Daley G.Q. Induced pluripotent stem cells in disease modelling and drug discovery. Nat. Rev. Genet. 2019;20:377–388. doi: 10.1038/s41576-019-0100-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Israel M.A., Yuan S.H., Bardy C., Reyna S.M., Mu Y., Herrera C., Hefferan M.P., Van Gorp S., Nazor K.L., Boscolo F.S., et al. Probing sporadic and familial Alzheimer's disease using induced pluripotent stem cells. Nature. 2012;482:216–220. doi: 10.1038/nature10821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lagomarsino V.N., Pearse R.V., 2nd, Liu L., Hsieh Y.C., Fernandez M.A., Vinton E.A., Paull D., Felsky D., Tasaki S., Gaiteri C., et al. Stem cell-derived neurons reflect features of protein networks, neuropathology, and cognitive outcome of their aged human donors. Neuron. 2021;109:3402–3420.e9. doi: 10.1016/j.neuron.2021.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Itzhaki I., Maizels L., Huber I., Zwi-Dantsis L., Caspi O., Winterstern A., Feldman O., Gepstein A., Arbel G., Hammerman H., et al. Modelling the long QT syndrome with induced pluripotent stem cells. Nature. 2011;471:225–229. doi: 10.1038/nature09747. [DOI] [PubMed] [Google Scholar]
- 9.Wang J.Y., Doudna J.A. CRISPR technology: a decade of genome editing is only the beginning. Science. 2023;379 doi: 10.1126/science.add8643. [DOI] [PubMed] [Google Scholar]
- 10.Kampmann M. CRISPR-based functional genomics for neurological disease. Nat. Rev. Neurol. 2020;16:465–480. doi: 10.1038/s41582-020-0373-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Przybyla L., Gilbert L.A. A new era in functional genomics screens. Nat. Rev. Genet. 2022;23:89–103. doi: 10.1038/s41576-021-00409-w. [DOI] [PubMed] [Google Scholar]
- 12.Leng K., Kampmann M. Towards elucidating disease-relevant states of neurons and glia by CRISPR-based functional genomics. Genome Med. 2022;14:130. doi: 10.1186/s13073-022-01134-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ihry R.J., Worringer K.A., Salick M.R., Frias E., Ho D., Theriault K., Kommineni S., Chen J., Sondey M., Ye C., et al. p53 inhibits CRISPR-Cas9 engineering in human pluripotent stem cells. Nat. Med. 2018;24:939–946. doi: 10.1038/s41591-018-0050-6. [DOI] [PubMed] [Google Scholar]
- 14.Bock C., Datlinger P., Chardon F., Coelho M.A., Dong M.B., Lawson K.A., Lu T., Maroc L., Norman T.M., Song B., et al. High-content CRISPR screening. Nat. Rev. Methods Primers. 2022;2:8. doi: 10.1038/s43586-022-00098-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li S., Li M., Liu X., Yang Y., Wei Y., Chen Y., Qiu Y., Zhou T., Feng Z., Ma D., et al. Genetic and chemical screenings identify HDAC3 as a key regulator in hepatic differentiation of human pluripotent stem cells. Stem Cell Rep. 2018;11:22–31. doi: 10.1016/j.stemcr.2018.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Y., Muffat J., Omer Javed A., Keys H.R., Lungjangwa T., Bosch I., Khan M., Virgilio M.C., Gehrke L., Sabatini D.M., Jaenisch R. Genome-wide CRISPR screen for Zika virus resistance in human neural cells. Proc. Natl. Acad. Sci. USA. 2019;116:9527–9532. doi: 10.1073/pnas.1900867116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tian R., Gachechiladze M.A., Ludwig C.H., Laurie M.T., Hong J.Y., Nathaniel D., Prabhu A.V., Fernandopulle M.S., Patel R., Abshari M., et al. CRISPR interference-based platform for multimodal genetic screens in human iPSC-derived neurons. Neuron. 2019;104:239–255.e12. doi: 10.1016/j.neuron.2019.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Esk C., Lindenhofer D., Haendeler S., Wester R.A., Pflug F., Schroeder B., Bagley J.A., Elling U., Zuber J., von Haeseler A., Knoblich J.A. A human tissue screen identifies a regulator of ER secretion as a brain-size determinant. Science. 2020;370:935–941. doi: 10.1126/science.abb5390. [DOI] [PubMed] [Google Scholar]
- 19.Xu J., Zhou C., Foo K.S., Yang R., Xiao Y., Bylund K., Sahara M., Chien K.R. Genome-wide CRISPR screen identifies ZIC2 as an essential gene that controls the cell fate of early mesodermal precursors to human heart progenitors. Stem Cell. 2020;38:741–755. doi: 10.1002/stem.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tian R., Abarientos A., Hong J., Hashemi S.H., Yan R., Dräger N., Leng K., Nalls M.A., Singleton A.B., Xu K., et al. Genome-wide CRISPRi/a screens in human neurons link lysosomal failure to ferroptosis. Nat. Neurosci. 2021;24:1020–1034. doi: 10.1038/s41593-021-00862-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sapp V., Aguirre A., Mainkar G., Ding J., Adler E., Liao R., Sharma S., Jain M. Genome-wide CRISPR/Cas9 screening in human iPS derived cardiomyocytes uncovers novel mediators of doxorubicin cardiotoxicity. Sci. Rep. 2021;11 doi: 10.1038/s41598-021-92988-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu D., Poddar A., Ninou E., Hwang E., Cole M.A., Liu S.J., Horlbeck M.A., Chen J., Replogle J.M., Carosso G.A., et al. Dual genome-wide coding and lncRNA screens in neural induction of induced pluripotent stem cells. Cell Genom. 2022;2 doi: 10.1016/j.xgen.2022.100177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meng X., Yao D., Chen X., Kelley K.W., Reis N., Thete M.V., Kulkarni S., Bassik M.C., Pașca S.P. CRISPR screens in 3D assembloids reveal disease genes associated with human interneuron development. bioRxiv. 2022 doi: 10.1101/2022.09.06.506845. Preprint at. [DOI] [Google Scholar]
- 24.Guo W., Wang H., Kumar Tharkeshwar A., Couthouis J., Braems E., Masrori P., Van Schoor E., Fan Y., Ahuja K., Moisse M., et al. CRISPR/Cas9 screen in human iPSC-derived cortical neurons identifies NEK6 as a novel disease modifier of C9orf72 poly(PR) toxicity. Alzheimers Dement. 2022 doi: 10.1002/alz.12760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leng K., Rose I.V.L., Kim H., Xia W., Romero-Fernandez W., Rooney B., Koontz M., Li E., Ao Y., Wang S., et al. CRISPRi screens in human iPSC-derived astrocytes elucidate regulators of distinct inflammatory reactive states. Nat. Neurosci. 2022;25:1528–1542. doi: 10.1038/s41593-022-01180-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dräger N.M., Sattler S.M., Huang C.T.L., Teter O.M., Leng K., Hashemi S.H., Hong J., Aviles G., Clelland C.D., Zhan L., et al. A CRISPRi/a platform in human iPSC-derived microglia uncovers regulators of disease states. Nat. Neurosci. 2022;25:1149–1162. doi: 10.1038/s41593-022-01131-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ungricht R., Guibbal L., Lasbennes M.C., Orsini V., Beibel M., Waldt A., Cuttat R., Carbone W., Basler A., Roma G., et al. Genome-wide screening in human kidney organoids identifies developmental and disease-related aspects of nephrogenesis. Cell Stem Cell. 2022;29:160–175.e7. doi: 10.1016/j.stem.2021.11.001. [DOI] [PubMed] [Google Scholar]
- 28.Wang C., Ward M.E., Chen R., Liu K., Tracy T.E., Chen X., Xie M., Sohn P.D., Ludwig C., Meyer-Franke A., et al. Scalable production of iPSC-derived human neurons to identify tau-lowering compounds by high-content screening. Stem Cell Rep. 2017;9:1221–1233. doi: 10.1016/j.stemcr.2017.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marton R.M., Pașca S.P. Organoid and assembloid technologies for investigating cellular crosstalk in human brain development and disease. Trends Cell Biol. 2020;30:133–143. doi: 10.1016/j.tcb.2019.11.004. [DOI] [PubMed] [Google Scholar]
- 30.Liu S.J., Horlbeck M.A., Cho S.W., Birk H.S., Malatesta M., He D., Attenello F.J., Villalta J.E., Cho M.Y., Chen Y., et al. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science. 2017;355 doi: 10.1126/science.aah7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fernandopulle M.S., Prestil R., Grunseich C., Wang C., Gan L., Ward M.E. Transcription factor-mediated differentiation of human iPSCs into neurons. Curr. Protoc. Cell Biol. 2018;79:e51. doi: 10.1002/cpcb.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Erwood S., Bily T.M.I., Lequyer J., Yan J., Gulati N., Brewer R.A., Zhou L., Pelletier L., Ivakine E.A., Cohn R.D. Saturation variant interpretation using CRISPR prime editing. Nat. Biotechnol. 2022;40:885–895. doi: 10.1038/s41587-021-01201-1. [DOI] [PubMed] [Google Scholar]
- 33.Hanna R.E., Hegde M., Fagre C.R., DeWeirdt P.C., Sangree A.K., Szegletes Z., Griffith A., Feeley M.N., Sanson K.R., Baidi Y., et al. Massively parallel assessment of human variants with base editor screens. Cell. 2021;184:1064–1080.e20. doi: 10.1016/j.cell.2021.01.012. [DOI] [PubMed] [Google Scholar]
- 34.Cuella-Martin R., Hayward S.B., Fan X., Chen X., Huang J.W., Taglialatela A., Leuzzi G., Zhao J., Rabadan R., Lu C., et al. Functional interrogation of DNA damage response variants with base editing screens. Cell. 2021;184:1081–1097.e19. doi: 10.1016/j.cell.2021.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramos D.M., Skarnes W.C., Singleton A.B., Cookson M.R., Ward M.E. Tackling neurodegenerative diseases with genomic engineering: a new stem cell initiative from the NIH. Neuron. 2021;109:1080–1083. doi: 10.1016/j.neuron.2021.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]



