Abstract
Several determinants that appear to promote the dimerization of murine retroviral genomic RNA have been identified. The interaction between these determinants has not been extensively examined. Previously, we proposed that dimerization of the Moloney murine sarcoma virus genomic RNAs relies upon the concentration-dependent interactions of a conserved palindrome that is initiated by separate G-rich stretches (H. Ly, D. P. Nierlich, J. C. Olsen, and A. H. Kaplan, J. Virol. 73:7255–7261, 1999). The cooperative action of these two elements was examined using a combination of genetic and antisense approaches. Dimerization of RNA molecules carrying both the palindrome and G-rich sequences was completely inhibited by an oligonucleotide complementary to the palindrome; molecules lacking the palindrome could not dimerize in the presence of oligomers that hybridize to two G-rich sequences. The results of spontaneous dimerization experiments also demonstrated that RNA molecules lacking either of the two stretches of guanines dimerized much more slowly than the full-length molecule which includes the dimer linkage structure (DLS). However, the addition of an oligonucleotide complementary to the remaining stretch of guanines restored the kinetics of dimerization to wild-type levels. The ability of this oligomer to rescue the kinetics of dimerization was dependent on the presence of the palindrome, suggesting that interactions within the G-rich regions produce changes in the palindrome that allow dimerization to proceed with maximum efficiency. Further, unsuccessful attempts to produce heterodimers between constructs lacking various combinations of these elements indicate that the G-rich regions and the palindrome do not interact directly. Finally, we demonstrate that both of these elements are important in maintaining efficient viral replication. Modified antisense oligonucleotides targeting the DLS were found to reduce the level of viral vector titer production. The reduction in viral titer is due to a decrease in the efficiency of viral genomic RNA encapsidation. Overall, our data support a dynamic model of retroviral RNA dimerization in which discrete dimerization elements act in a concerted fashion.
Each retroviral particle contains two plus-strand molecules of the viral genomic RNA. The two molecules are noncovalently linked near their 5′ ends in a region known as the dimer linkage structure (DLS); numerous studies have suggested that maintenance of the RNA dimer plays an important role in a number of steps in viral replication (reviewed in reference 22).
Despite the preponderance of evidence supporting a requirement that retroviral RNAs form dimers, the exact nature of the DLS has not been clearly defined. The results of electron microscopy and sedimentation studies indicate that the two RNAs are held together noncovalently near their 5′ ends (3, 9, 11, 18). Several investigators have identified a number of potential cis-acting determinants of dimerization in this region, although the precise role(s) played by each determinant in promoting dimer formation remains obscure. A phylogenetically conserved palindrome has been identified in the 5′ untranslated region and has been demonstrated to be an important determinant of dimerization in some studies (5, 7, 8, 17, 23–26). Other investigators have emphasized the role of conserved purine motifs in promoting dimerization through the formation of purine quartets (1, 2, 15, 29). Finally, Oroudjev and colleagues identified a conserved stem-loop structure upstream of the palindrome that appears to support dimerization in vitro (20).
Recently, we proposed a model for the dimerization of Moloney murine sarcoma virus (MuSV) DLS RNA in which two downstream G-rich regions act in concert with the palindrome to allow dimerization to proceed (12). This study extends those observations by examining the function of these determinants in an in vitro dimerization system and the impact these determinants have on viral replication. Using a combination of genetic and antisense approaches, we demonstrate that interactions in the G-rich regions enhance the rate of dimerization and that this effect is mediated through the palindrome. Overall, our experimental results suggest that dimerization of the MuSV DLS RNA is promoted by the coordinated action of several redundant RNA determinants and that the presence of these determinants plays an important role in viral replication.
MATERIALS AND METHODS
Preparation of RNA samples.
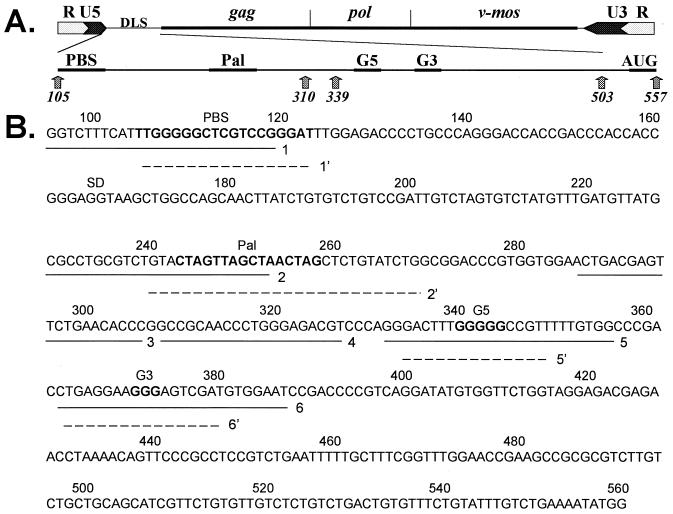
The pLNBS plasmid is a modified version of the pLNL6-based pLN plasmid (J. C. Olsen, unpublished data) containing the MuSV DLS and a neomycin resistance gene (described previously [12]). Numbering of the nucleotides is assigned according to the recently submitted MuSV sequence to GenBank (AF033813).
DNA fragments used as templates in the in vitro transcription reaction were synthesized via PCR amplification using the MuSV DLS region of pLNBS as a template. The forward primers contain a T7 promoter sequence and sequences identical to either the primer-binding site (PBS) at nucleotide (nt) 105 (5′TAATACGACTCACTATAGGGCGATGGGGGCTCGTCCGGGAT3′) or the BsiE I site at nt 310 (5′GAACATAATACGACTCACTATAGGGCGACCCGGCCGCA3′). The reverse primer begins at the putative initiation site of the gag gene (position 557) and ends at position 535 (5′CTAATTTTCAGACAAATACAGAAAC3′). PCR products were analyzed on a 1.0% agarose gel prior to purification using a QIAquick PCR purification kit (Qiagen, Valencia, Calif.). Samples were eluted from the columns with 50 μl of RNase-free water (Sigma, St. Louis, Mo.).
Transcription mixtures (20 μl) contained 1 μg of DNA template, 0.5 mM (each) unlabeled ATP, GTP, UTP, 50 μM unlabeled CTP, and 2.5 μM [α-32P]CTP (Amersham, Arlington Heights, Ill.). The reaction was performed with a T7 MAXIscript kit as suggested by the manufacturer (Ambion, Austin, Tex.). RNA transcripts were separated by electrophoresis on 5% polyacrylamide (PA)–8 M urea gels at 250 V for 2 h in 1× TBE buffer (90 mM Tris-borate, 2 mM EDTA). RNA samples were eluted from gels with elution buffer (Ambion). The purified RNAs were then ethanol precipitated and resuspended in 100 μl of water (Sigma) and stored at −70°C. The products were quantified with a Beckman LS6500 scintillation counter. Full-length RNA transcript refers to in vitro-transcribed MuSV RNA that contains an unaltered DLS signal.
RNA dimerization assay.
For a standard dimerization assay, 8-μl portions of ∼1.5 nM α-32P-labeled RNA were heated at 95°C for 3 min, chilled on ice for 3 min, and 2 μl of D5× buffer (500 mM NaCl, 250 mM Tris-HCl [pH 7]) were added. The samples were then incubated at 60°C for 1 h. The samples were chilled and separated by electrophoresis on 5% nondenaturing polyacrylamide gels in the presence of 1× TBE buffer. Gels were dried and exposed directly on a phosphorimager screen for 18 to 20 h. The amount of dimer formation was quantitated using a model 445SI PhosphorImager system and ImageQuaNT image analysis software. The fraction of RNA in dimeric form was estimated by taking the ratio of dimeric RNA to the sum of both the monomeric and dimeric species. Dimer inhibition assays were carried as outlined above except that antisense oligonucleotides (60 μM) (Table 1) were added before heat denaturation. In some instances, the oligonucleotides were 5′ end labeled with [γ-32P]ATP using bacteriophage T4 polynucleotide kinase (Gibco-BRL, Grand Island, N.Y.). The labeled oligomers were purified through a G-25 column (Amersham). The kinetic comparisons of f105-557, f105-557ΔPal, and f105-557ΔG5 RNAs were done in a manner similar to the standard dimerization reaction, except that the incubation times varied as indicated in the figures and that f105-557ΔPal was incubated at 55°C rather than at 60°C (see Fig. 4A). To assay heterodimer formation, various RNA fragments, each at 1.5 nM concentration, were combined and used in the standard dimerization assay (see Fig. 5).
TABLE 1.
Sense and antisense oligonucleotides
| Oligonucleotidea | Sequence (5′-3′)b | Tm (°C)c |
|---|---|---|
| Unmodified | ||
| antiPBS (106–123) | ATCCCGGACGAGCCCCCA | 74 |
| antiPal-30 (240–269) | CAGATACAGAGCTAGTTAGCTAACTAGTAC | 75 |
| antiPal-21 (240–260) | AGCTAGTTAGCTAACTAGTAC | 65 |
| antiG5 (334–349) | AACGGCCCCCAAAGTC | 65 |
| antiG3 (364–380) | ATCGACTCCCTTCCTCA | 64 |
| Morpholino | ||
| antiPBS (95–119) | CGGACGAGCCCCCAAATGAAAGACC | 79 |
| antiPal (229–253) | TAGCTAACTAGTACAGACGCAGGCG | 76 |
| antiG5 (332–356) | CCACAAAAACGGCCCCCAAAGTCCC | 79 |
| antiG3 (363–387) | ATTCCACATCGACTCCCTTCCTCAG | 76 |
| antiCon1 (285–306) | GGGTGTTCAGAACTCGTCAGT | 71 |
| antiCon2 (308–327) | ACGTCTCCCAGGGTTGCGGC | 76 |
| revNeo (1651–1675) | AAGCGTACTAACTTGTTCTACCTAA | 69 |
| antiNeo (1651–1675) | AATCCATCTTGTTCAATCATGCGAA | 69 |
| Standard | CCTCTTACCTCAGTTACAATTTATA |
Numbering in parentheses is with respect to the MuSV viral mRNA (GenBank accession no. AF33813). For revNeo and antiNeo, the positions of the nucleotides are with respect to those of the retroviral vector pLNL6 (GenBank accession no. M63653). The sequence of revNeo is identical to that of the antiNeo oligomer, but the orientation of revNeo is opposite that of antiNeo. antiNeo is an antisense oligonucleotide that hybridizes to the initiation site of the neomycin phosphotransferase gene. Standard, control oligonucleotide of randomized sequence, supplied by GeneTools, LCC.
Sequences are antisense in polarity with respect to the viral mRNA except where indicated (i.e., revNeo).
Melting temperature of oligonucleotides is calculated by the %GC method.
FIG. 4.
Dimerization of f105-557ΔG5 is delayed compared to that of the full-length f105-557, but it can be kinetically enhanced by the antiG3 (αG3) oligonucleotide. (A) The standard dimer inhibition assay was conducted as outlined in Materials and Methods, except that the individual reaction was terminated by chilling on ice at the end of the appropriate incubation times (0, 7.5, 15, 30, and 60 min) until separation on a 5% PA gel. (B) Kinetics of dimerization. The mean values (as percentages) of the dimeric species of the RNA fragments in the presence or absence of the antiG3 oligomer (αG3) were plotted and compared. The standard deviation at each time point is less than 8.0% for at least a duplicate set of reaction mixtures. WT, wild type (f105-557); ΔPal, f105-557ΔPal; ΔG5, f105-557ΔG5.
FIG. 5.
Heterodimer formation shows no direct interaction between the kissing loop and the G-rich sequences. The standard dimerization reaction was performed as outlined (Materials and Methods) except that, in some instances, a combination of different RNAs (1.5 nM [each] RNA) was codimerized in a single reaction mixture (lanes 6, 10, and 12). Also, reaction mixtures containing f105-339 RNA are supplemented with 0.5 μg of identical but unlabeled product. The different RNAs are indicated by numbers as follows: 1, f105-339; 2, f310-557; 3, f105-557. The positions of monomeric RNA (M), dimeric RNA (D), and heterodimers (HD) are shown to the left and right of the gel.
Cell culture, transfections, and infections.
GP + E86 murine ecotropic packaging cells (12), PA-317 murine amphotropic packaging cells (ATCC CRL 9078), and NIH 3T3 murine fibroblast cells (ATCC CRL 1658) were maintained in Dulbecco's modified Eagle's medium containing a high level of glucose (4.5 g/liter) and 10% newborn calf serum, 10% fetal calf serum, and 10% calf serum, respectively. The polyclonal cell line expressing the wild-type MuSV DLS sequence was made by first transfecting 5.0 × 105 PA-317 cells with 20 μg of pLNBS DNA by calcium phosphate precipitation (Calcium Phosphate Transfection System; Gibco-BRL).
Virus particles were collected from the transfection supernatant 48 h posttransfection. Various concentrations of the viral supernatants (102- to 105-fold dilutions) were used to infect 2.5 × 105 GP + E86 cells in the presence of 8 μg of Polybrene per ml as described previously (19). G418-resistant colony formation was measured in medium supplemented with 500 μg of Geneticin (Gibco-BRL) per ml (19). Approximately 200 G418-resistant colonies were trypsinized, pooled, and stored in freezing solution Dulbecco's modified Eagle's medium containing a high level of glucose [4.5 g/liter], 10% fetal calf serum, 10% dimethyl sulfoxide).
Prior to treatment with morpholino oligonucleotides (GeneTools, LLC, Corvallis, Oreg.), 5 × 105 polyclonal cells expressing the wild-type MuSV DLS sequence (passage 39) were seeded into each well of a six-well culture plate in 1 ml of growth medium lacking Geneticin. Morpholino oligonucleotide is a modified nucleotide analog in which the deoxyribose moiety of DNA is replaced by a six-membered morpholine ring and the charged phosphodiester nucleoside linkage is substituted by an uncharged phosphoramidate bond (28). The oligonucleotides were introduced into the polyclonal cells by the method of Summerton et al. (27). Briefly, 24 h postseeding, the cell medium was replaced with 1 ml of fresh medium containing 10 μM morpholino oligonucleotide. Cells were gently scraped off the surface of the well with a sterile cell scraper (Costar, Cambridge, Mass.) and transferred to a new well. The medium, containing released virus particles, was collected 24 h later and filtered through a 0.2-μm-pore-size filter (Schleicher & Schuell, Keene, N.H.). Viral titers were determined by measuring G418-resistant colony formation on NIH 3T3 cells as described previously (12, 19).
RT-PCR analyses.
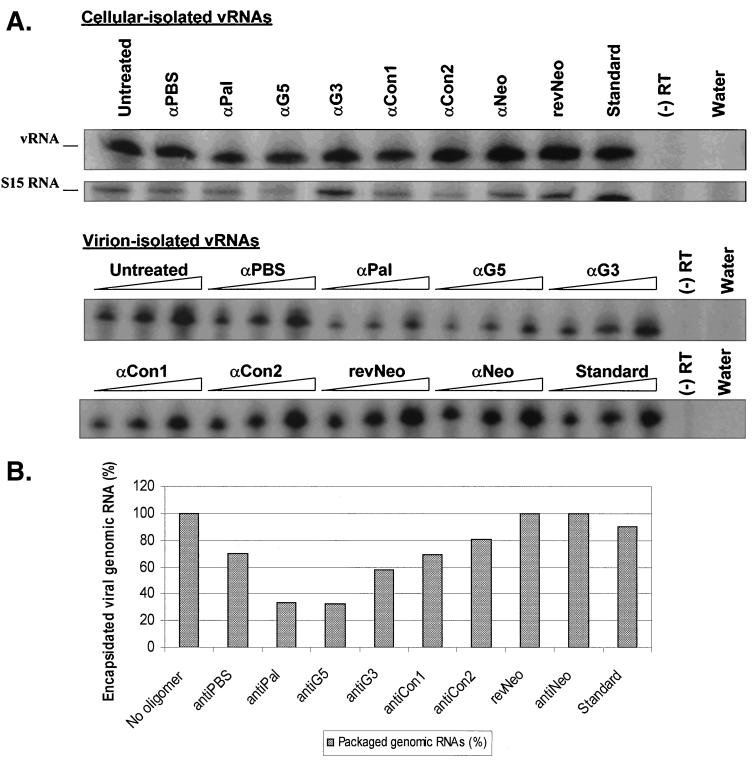
Viral genomic RNAs from oligomer-treated and untreated samples were isolated from cells and viral supernatants using the Trizol reagent (Gibco-BRL) and the Qiagen viral RNA miniprep kit (Qiagen), respectively. Prior to RNA extraction, the virus supernatants were concentrated via centrifugation at 25,000 rpm (Tomy TX-160) for 30 min. The sample was treated with 10 U of RQ1 DNase in 1× DNase buffer for 1 h at 37°C (Promega, Madison, Wis.). The RNAs were extracted with phenol-chloroform and precipitated with ammonium acetate as previously described (12). Similar DNase treatment (minus the phenol-chloroform extraction step) was applied to the virus supernatants prior to RNA extraction. Cell-isolated RNA (1.0 μg) (optical density at 260 nm) and various volumes of virion-isolated RNA (5, 10, and 20 μl) were each used as the template in the single-step reverse transcriptase PCR (RT-PCR) as suggested by the manufacturer (18 cycles at 55°C annealing temperature) (Ambion). The oligonucleotides used in the RT-PCR are RTPCRup primer (5′GTTATGCGCCTGCGTCTGTACT3′) and RTPCRdn primer (5′CATATCCTGACGGGGTCGGA3′), which correspond to viral nt 223 to 244 and 387 to 406, respectively. RTPCRup primer (50 μM) was 5′ end labeled with [γ-32P]ATP using bacteriophage T4 polynucleotide kinase (Gibco-BRL). A mixture of control primers (50 μM) (Ambion), which correspond to the constitutively expressed cellular ribosomal protein S15 subunit, was also 5′ end labeled. The labeled products were purified through a G-25 column (Amersham). Each primer (5 μM) was used in the one-step RT-PCR as suggested by the manufacturer (Ambion). A quarter of the PCR product was denatured (95°C for 5 min) and separated on a 5% PA–8 M urea gel at 150 V. PhosphorImager and related softwares (Molecular Dynamics) were used to quantify the PCR products on the gels (see Fig. 6A).
FIG. 6.
Reduction of viral genomic RNA encapsidation due to antisense DNA oligomers. (A) RT-PCR amplification of viral genomic RNAs. Polyclonal cells expressing the wild-type DLS sequence were treated either with one of a series of antisense DNA oligomers or with a medium control. RNAs were isolated from cells or virions. Various volumes of virion-isolated RNAs (5, 10, and 20 μl [indicated by the height of the triangle over each set of three lanes]) and 1.0 μg of cell-isolated RNAs were RT-PCR amplified. vRNA and S15 are PCR products amplified from viral RNA and control ribosomal protein S15 message, respectively. α, anti; (−) RT, one-step RT-PCR mixture lacking MuLV RT; Water, one-step RT-PCR mixture lacking the RNA substrate. (B) Relative reduction of genomic RNA packaging. Percentage reductions in genomic RNA encapsidation of the oligomer-treated samples relative to that of the untreated mixture were compared.
RNase H cleavage assay.
Mixtures of DLS RNA transcripts (1.5 nM) and either modified (morpholino) or unmodified DNA oligomers (60 μM) were allowed to anneal by heating the samples to 95°C for 3 min and cooling on ice. After adding the First-Strand MuLV RT buffer (1×) (Gibco-BRL), the samples were allowed to dimerize at 60°C for 1 h. At the end of the incubation time, samples were again chilled on ice for 3 min prior to the addition of 10 U of MuLV RT enzyme (Gibco-BRL). The mixtures were then incubated at 37°C for an additional hour before being denatured and separated on 5% PA–8 M urea gels (see Fig. 7).
FIG. 7.
Effects of modified (Morph) and unmodified (Phos) antisense DNA oligomers (oligo) on the stability of the target RNA. Viral RNAs are cleaved by the MuLV RT enzyme in the presence of unmodified DNA oligomers (lanes 3, 5, and 7) into the expected RNA products (antiPal [α Pal], 300- and 130-nt fragments; antiG5 [α G5], 210- and 220-nt fragments; antiCon1 [α Con1], 200- and 240-nt fragments). Viral RNAs are not altered in the presence of modified oligonucleotides (lanes 4, 6, and 8). The presence (+) or absence (−) of the RT enzyme is indicated over the gel.
RESULTS
Antisense DNA oligonucleotides binding to the palindrome or G-rich regions alter in vitro dimerization of MuSV DLS RNA.
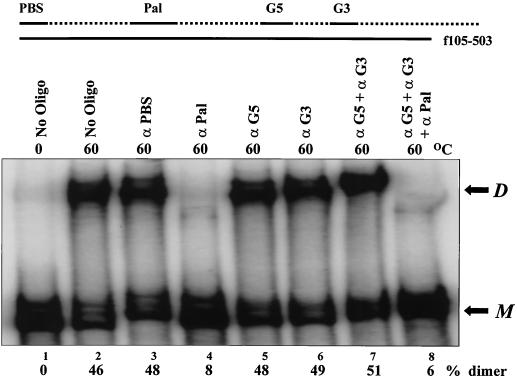
We began by examining the effects of antisense DNA oligonucleotides on spontaneous dimer formation of a region of the MuSV genome containing the DLS (Fig. 1). Initially, antisense DNA oligonucleotides binding to various regions of the DLS were tested to determine whether they would interfere with the dimerization of the full-length DLS RNA (f105-503), which contains both the palindrome and G-rich sequences (Fig. 2). Although oligonucleotides binding to the palindrome (antiPal) effectively inhibited dimer formation of the full-length DLS RNAs, other oligonucleotides targeting either the PBS or the G-rich sequences had little effect (Fig. 2, compare lane 4 to lanes 3, 5, and 6).
FIG. 1.
(A) Representation of the MuSV genome. Numbering is with respect to the viral mRNA, which begins at the transcriptional start site and ends at the poly(A) site (GenBank accession no. AF033813). Viral long terminal repeats, the gag, pol, and v-mos genes, and the sequences of interest (i.e., PBS, DLS, the Pal palindrome, G5 and G3 G-rich motifs, and the gag initiation site AUG) are shown. (B) The primary DLS sequence and antisense oligonucleotides are shown. Solid underlines indicate the positions of the modified (morpholino) oligonucleotides relative to the primary sequence. Broken underlines show the positions of the unmodified oligonucleotides. Oligonucleotides: 1 and 1', modified and unmodified antiPBS, respectively; 2 and 2', modified and unmodified antiPal, respectively; 3, modified antiCon1; 4, modified antiCon2; 5 and 5', modified and unmodified antiG5; and 6 and 6', modified and unmodified antiG3, respectively. See Table 1 for the sequences of the antisense oligonucleotides. The positions of the PBS, Pal, G5, and G3 sequences are shown in boldface and are noted above the sequences.
FIG. 2.
Dimerization of the full-length RNA (f105-503) is inhibited by the antiPal oligonucleotide. α-32P-labeled RNA (1.5 nM) was incubated with various unlabeled oligonucleotides (60 μM) at 60°C for 1 h before separation on a 5% nondenaturing PA gel as described in Materials and Methods. Whenever a combination of oligonucleotides was used (lanes 7 and 8), the final concentration of each oligonucleotide was 30 μM. Pal palindromic sequence and G5 and G3, two consecutive guanines, are shown at the top. The positions of monomeric RNA (M) and dimeric RNA (D) are shown to the right of the gel. The oligonucleotides are given over the lanes (No Oligo, no oligonucleotides; α, anti).
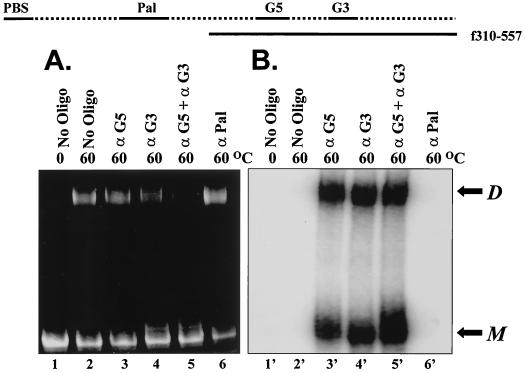
We extended these observations by examining the dimerization of a DLS fragment lacking the palindrome (f310-557) (Fig. 3). In contrast to the full-length fragment (f105-503), spontaneous dimerization of this fragment (f310-557) was inhibited by the addition of a combination of oligonucleotides directed against both G-rich regions (antiG3 and antiG5). Formation of dimers did not appear to be affected by the addition of either antiG3 or antiG5 oligonucleotides alone. However, the addition of antiG3 and antiG5 oligomers also effectively inhibited dimerization of the full-length DLS fragment containing a 10-nt deletion within the palindrome (data not shown). Of note, experiments using radiolabeled oligonucleotides with unlabeled DLS RNA indicate that the antiG3 and antiG5 oligonucleotides bind to both dimeric and monomeric forms of the genomic RNA (Fig. 3, lanes 3' to 5'). The inhibitory effect of the oligomers was not as pronounced at high concentrations of RNA, however (compare Fig. 2 and 3).
FIG. 3.
Dimerization of an RNA molecule lacking the Pal sequence (f310-557) is inhibited by a combination of the antiG-rich oligonucleotides. Unlabeled f310-557 (1 μg) was dimerized in the presence of 60 μM 5′-end-labeled DNA oligonucleotide for 1 h at 60°C prior to separation on a 5% PA gel. The RNA was stained with 100 μg of ethidium bromide per ml in 1× TBE buffer for 15 min (A). After being photographed, the gel was then dried and exposed directly onto a phosphorimager screen for 18 to 20 h (B). The positions of monomeric RNA (M) and dimeric RNA (D) are shown to the right of the gel. The oligonucleotides are shown over the gel (No Oligo, no oligonucleotides; α, anti).
RNA molecule lacking one of the G-rich sequences (f105-557ΔG5) dimerizes more slowly than wild type but can be kinetically enhanced by an antiG3 oligomer.
Our previous study indicated that at low concentration, the dimerization of MuSV DLS RNA is initiated by contact between the G-rich sequences (12). The effects of these determinants on the rate of dimerization were examined by assaying the conversion of monomers to dimers over time.
Dimerization of a full-length DLS fragment lacking either the palindrome (f105-557ΔPal) (Fig. 4A, lanes 6 to 10) or one of the G-rich stretches (f105-557ΔG5) (lanes 11 to 15) was slower than dimerization of the wild-type fragment (lanes 1 to 5). In contrast to the wild-type fragment, for which dimers were apparent by 7.5 min (lane 2), dimerization of a fragment missing the stretch of five guanines was not appreciable until 30 min had elapsed (lane 14). However, when the incubation time was extended to more than 6 h, dimerization of f105-557ΔG5 reached the same level as that of the full-length f105-557 fragment (Fig. 4B).
Remarkably, the addition of the antiG3 oligonucleotide to the construct lacking the G5 sequence (f105-557ΔG5) restored the rate of dimerization (Fig. 4A, lanes 16 to 20). Dimerization of this RNA in the presence of the antiG3 oligomer followed the same kinetics as the full-length fragment (Fig. 4B). The ability of this oligonucleotide to rescue dimerization was dependent on the presence of the palindrome; the addition of the antiG3 oligonucleotide had no effect on the rate of dimerization for either the RNA in which the palindrome had been deleted or for the wild-type DLS molecule (Fig. 4A, lanes 21 to 30).
Heterodimer formation data suggest that there is no direct physical interaction between the palindrome and G-rich sequences.
RNAs containing either one or both of the dimerization determinants were used to evaluate whether there is a direct physical interaction between the palindrome and G-rich regions. We tested the ability of various truncated and full-length DLS RNAs to form heterodimers. As expected, the truncated RNA fragments could form homodimers efficiently under the proper conditions (Fig. 5, lanes 1 to 4, 7, and 8). Further, the truncated fragments could form heterodimers with the full-length DLS RNAs (lanes 9 to 12). However, no heterodimer formation was noted between two RNA fragments which contained only the palindrome (f105-339) or the G-rich motifs (f310-557) (lane 6). These results suggest that the elements form discrete dimerization units and that there is no direct interaction between them.
Viral vector production is impaired by antisense oligonucleotide targeting the DLS sequence.
We also examined the roles of these dimerization domains by attempting to inhibit viral replication in a MuSV-based vector system using antisense oligonucleotides that bind to these domains. These experiments were performed by assessing the effects of morpholino antisense oligonucleotides on viral titer (Table 2). Large effects on titer were obtained by using the oligonucleotides that bound to either the palindrome or the G5 region (Table 2). These effects were comparable to those seen with an antisense oligonucleotide complementary to the PBS and greater than the decline in titer obtained with an antiNeo oligomer, which hybridizes to the initiation site of the neomycin phosphotransferase gene. Little or no effect on titer was seen with several control oligonucleotides that were complementary to sequences between the palindrome and G5 (antiGcon1 and antiGcon2) (Fig. 1). We tested these oligomers because other groups have suggested that these regions may play a role in dimerization in vitro (16, 30).
TABLE 2.
Effects of antisense DNA oligonucleotide targeting the viral DLS region on viral replication
| Treatment | Titera | Reduction in titer (fold)b | Relative fold reductionb in:
|
Reduction in packaged RNAsc | |
|---|---|---|---|---|---|
| Genomic RNAs | Virion RNAs | ||||
| No oligonucleotide | 3.70 (0.35) | NAd | 1.00 | 1.00 | NA |
| antiPBS | 0.51 (0.05) | 6.00 | 0.95 (0.07) | 0.66 (0.08) | 1.44 |
| antiPal | 0.61 (0.01) | 5.00 | 0.83 (0.25) | 0.27 (0.02) | 3.10 |
| antiG5 | 0.50 (0.09) | 7.70 | 0.81 (0.13) | 0.26 (0.04) | 3.12 |
| antiG3 | 1.30 (0.07) | 2.60 | 1.00 (0.01) | 0.58 (0.01) | 1.72 |
| antiCon1 | 2.80 (0.63) | 1.00 | 0.80 (0.22) | 0.55 (0.05) | 1.46 |
| antiCon2 | 3.20 (0.64) | 1.20 | 0.95 (0.07) | 0.77 (0.05) | 1.23 |
| antiNeo | 0.75 (0.14) | 3.00 | 0.88 (0.18) | 0.91 (0.16) | 0.91 |
| revNeo | 2.90 (0.14) | 1.20 | 0.93 (0.11) | 0.97 (0.04) | 1.02 |
| Standard | 2.30 (0.78) | 2.00 | 0.83 (0.04) | 0.75 (0.11) | 1.11 |
Titers (105 CFU per milliliter) of the MuSV vector from a representative experiment. The numbers in parentheses represent standard deviations.
Mean values (fold reduction) normalized to the values for the untreated samples. Data are taken from at least two independent assays. The numbers in parentheses represent standard deviations.
Relative fold reduction of packaged RNAs = relative fold reduction of genomic RNAs/relative fold reduction of virion RNAs.
NA, not applicable.
Viral titer reduction can be attributed, in part, to the reduced efficiency of viral genomic RNA encapsidation.
To help define the point in the viral life cycle that was influenced by the antisense oligonucleotide treatment, we decided to determine the level of encapsidated genomic RNA (Fig. 6A). Both the antiPal and antiG5 oligomers reduced the viral genomic RNA packaging by approximately 70% relative to that of the untreated sample; treatment with the other oligomers had little or no effect on the level of RNA encapsidation (Fig. 6B). The moderate reductions in RNA packaging observed for the oligomer-treated samples are consistent with the results obtained from deletion analyses of these similar regions in the related MuLV virus (16). As expected, no marked reduction in viral RNA packaging was observed for the antiPBS and antiNeo samples, although the titers produced from cells treated with these oligomers were reduced by three- to sixfold (Table 2).
Hybridization of the antisense DNA oligomers to the target RNA does not alter the integrity of the genomic RNA inside the virions.
We were concerned that the formation of RNA-DNA hybrids produced by morpholino oligomer treatment may render the viral genome more susceptible to RNase H activity of the viral RT inside the virions. To examine this possibility, mixtures of viral DLS RNA and either modified (morpholino) or unmodified DNA oligomers were allowed to anneal. The mixtures were then treated with the MuLV RT enzyme and separated on 5% PA–8 M urea gels (Fig. 7). While RT could effectively cleave the target RNAs in the presence of the unmodified oligomers (lanes 3, 5, and 7), it could not modify the RNA samples that contained the modified oligomers (lanes 4, 6, and 8). These observations suggest that the morpholino oligomers used in our viral culture system did not render the viral genomic RNA susceptible to RT-associated RNase H cleavage.
DISCUSSION
We previously demonstrated that spontaneous dimerization of the MuSV DLS RNA is dependent upon the cooperative interactions between the sequences that comprise the palindrome and the G-rich motifs (12). Here we extend these observations by exploring the interactions between these two domains. Our results support a role for both of these determinants in promoting dimerization and suggest that the two domains function cooperatively to allow dimerization to proceed with maximum efficiency.
Our data demonstrate that an antisense oligonucleotide directed against the palindrome is sufficient to block spontaneous dimerization of the full-length DLS fragment. Although oligomers complementary to the G-rich regions were unable to block dimerization of the full-length DLS molecule, they did inhibit dimerization of RNAs lacking the palindrome. This observation is consistent with a model in which the interaction of the G-rich regions is in a pathway preceding the final state of dimerization. Overall, the influence of the antisense oligomers on in vitro dimerization is in keeping with a process in which the two domains contribute to the formation of the dimer.
It is interesting to note that when the antiG3 or antiG5 oligomers are added singly, they bind to both the dimeric and monomeric forms of the RNA and do not appear to inhibit dimerization of RNAs lacking the palindrome. Further, when the radiolabeled antiG-rich oligomers are added together, they produce a more intense monomer band (Fig. 3, lane 5'), suggesting that either G-rich region can function in dimerization. Overall, the results of these experiments combined with those reported previously indicate that dimeric RNAs lacking the Pal sequence are held together by the G-rich sequences and that simultaneous hybridization of these elements by both antiG-rich oligomers completely blocks dimer formation.
We noted that dimerization for an RNA molecule lacking one of the G-rich regions is delayed. This delay is completely rescued by the addition of a DNA oligomer that hybridizes to the other stretch of guanines. Our results also demonstrate that the ability of this antiG3 oligomer to promote dimerization is dependent on the presence of the palindrome. Therefore, it appears that an interaction in the region of the guanines is required in order for dimerization to occur at the same rate as the full-length DLS RNA and that this interaction is mediated through the presence of the palindrome. This observation supports the view of RNA dimerization as a dynamic process.
Our data are in accord with a dynamic model of the dimerization of MuLV genomic RNA proposed by several investigators (5, 8). De Tapia and colleagues have suggested that the association of the palindromes is a slow process that is kinetically enhanced by sequences downstream (5). In their model, the initial interactions involve stem-loop structures downstream from the kissing loop that allow rapid structural rearrangement of the monomeric RNA. They propose that these initial interactions increase the fraction of RNA conformers that are available to dimerize via the palindrome (5). Accordingly, the secondary structure of the MuLV DLS is predicted to undergo dimerization-induced conformational changes, as indicated by changes in reactivity to chemical probing agents in solution (30). More specifically, besides the changes observed for the kissing-loop structure, sequences downstream from it are predicted to undergo almost complete structural rearrangement in the RNA dimer (29). It is interesting to note that the five guanines deleted in the MuSV f105-557ΔG5 fragment are analogous to a MuLV sequence predicted to be part of a stem-loop structure that was proposed to participate in the initial intermolecular interactions between RNA molecules (8).
Our data and the data of others suggest that rearrangements in RNA determinants downstream from the palindrome accompany dimerization. However, the precise nature of these interactions had not been defined and the possibility that these determinants may interact directly with each other had not been examined. We addressed this question by attempting to produce heterodimers between RNA molecules containing one or both of these elements (Fig. 5). The results of these experiments indicate that an RNA monomer containing only one of these elements can pair only with a monomer containing the same element. Therefore, although both the palindrome and the downstream elements participate in the final dimer, they do not appear to interact directly with each other. Overall, these results are consistent with the data obtained from the successful heterodimer formation between full-length DLS RNAs from the various retroviruses (15) and suggest that these elements form discrete dimerization domains.
The conclusion that both of these determinants play an important role in dimerization and/or other aspects of viral replication is further supported by our viral infectivity data. We demonstrate that an oligomer complementary to either of the stretches of guanines also has a significant effect on viral replication (Table 2). The reductions in viral vector titer can, in part, be explained by the attenuation of the viral genomic RNA encapsidation efficiency (Fig. 6). Nevertheless, the defect in RNA packaging alone could not account for the full reduction effect observed with the viral vector titer production. These results suggest that aspects of viral replication other than RNA encapsidation may be affected by changes that disrupt dimerization.
Overall, our data support a multistage model of RNA dimerization in which initial interactions between two RNA molecules involve the regions encompassing the stretches of guanines. These initial interactions induce subsequent changes in the palindrome that allow dimerization to proceed. It has been suggested that dimerization of the retroviral genomic RNA plays an important role in several aspects of viral replication including RNA encapsidation and reverse transcription (22). It has also been noted that retroviral assembly is very efficient at selectively packaging dimers of RNA of viral, but not cellular, origin (11). A mechanism for RNA dimerization that relies on multiple determinants that act sequentially would meet the challenge of maintaining specificity while providing the redundancy that might be expected for a critical biologic function. We are currently extending our evaluation of the biologic and structural roles played by these elements.
ACKNOWLEDGMENTS
This work was supported by NIH grant K11 AI01107 to A.H.K. and a seed grant from the UCLA AIDS Institute to D.P.N. H.L. was supported in part by the UCLA Presidential Research Fellowship and the UNC Graduate Research Fellowship (Ki Ha Chang Memorial Fellowship).
We thank Natalie Thornburg, John Sechelski, Nan N. Lee, Amy T. A. Tran, Timothy Moran, Liza Kim, Sami Yusuf, Peter Bui, James Matsunaga, and Steve Pettit for their help. We also thank Kevin M. Weeks for critical reading of the manuscript.
REFERENCES
- 1.Awang G, Sen D. Mode of dimerization of HIV-1 genomic RNA. Biochemistry. 1993;32:11453–11457. doi: 10.1021/bi00093a024. [DOI] [PubMed] [Google Scholar]
- 2.Baudin F, Marquet R, Isel C, Darlix J-L, Ehresmann B, Ehresmann C. Functional sites in the 5′ region of human immunodeficiency virus type 1 RNA from defined structural domains. J Mol Biol. 1993;229:382–397. doi: 10.1006/jmbi.1993.1041. [DOI] [PubMed] [Google Scholar]
- 3.Bender W, Chien Y-H, Chattopadhyay S, Vogt P K, Gardner M B, Davidson N. High-molecular-weight RNAs of AKR, NZB, and wild mouse viruses and avian reticuloendotheliosis virus all have similar dimer structures. J Virol. 1978;25:888–896. doi: 10.1128/jvi.25.3.888-896.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clever J L, Parslow T G. Mutant human immunodeficiency virus type 1 genomes with defects in RNA dimerization or encapsidation. J Virol. 1997;71:3407–3414. doi: 10.1128/jvi.71.5.3407-3414.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Tapia M, Metzler V, Mougel M, Ehresmann B, Ehresmann C. Dimerization of MoMuLV genomic RNA: redefinition of the role of the palindromic stem-loop H1 (278-303) and new roles for stem-loops H2 (310-352) and H3 (355-374) Biochemistry. 1998;37:6077–6085. doi: 10.1021/bi9800303. [DOI] [PubMed] [Google Scholar]
- 6.Fu W, Rein A. Maturation of dimeric viral RNA of Moloney murine leukemia virus. J Virol. 1993;67:5443–5449. doi: 10.1128/jvi.67.9.5443-5449.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Girard P M, Bonnet-Mathoniere B, Muriaux D, Paoletti J. A short autocomplementary sequence in the 5′ leader region is responsible for dimerization of MoMuLV genomic RNA. Biochemistry. 1995;34:9785–9794. doi: 10.1021/bi00030a016. [DOI] [PubMed] [Google Scholar]
- 8.Girard P M, de Rocquigny H, Roques B P, Paoletti J. A model of psi dimerization: destabilization of the C278-G303 stem-loop by the nucleocapsid protein (NCp10) of MoMuLV. Biochemistry. 1996;35:8705–8714. doi: 10.1021/bi952454s. [DOI] [PubMed] [Google Scholar]
- 9.Kung H J, Hu S, Bender W, Bailey J M, Davidson N, Nicolson M O, McAllister R M. RD-114, baboon, and woolly monkey viral RNAs compared in size and structure. Cell. 1976;7:609–620. doi: 10.1016/0092-8674(76)90211-7. [DOI] [PubMed] [Google Scholar]
- 10.Laughrea M, Jette L. A 19-nucleotide sequence upstream of the 5′ major splice donor is part of the dimerization domain of human immunodeficiency virus 1 genomic RNA. Biochemistry. 1994;33:13464–13474. doi: 10.1021/bi00249a035. [DOI] [PubMed] [Google Scholar]
- 11.Linial M L, Miller A D. Retroviral RNA packaging: sequence requirements and implications. Curr Top Microbiol Immunol. 1990;157:125–152. doi: 10.1007/978-3-642-75218-6_5. [DOI] [PubMed] [Google Scholar]
- 12.Ly H, Nierlich D P, Olsen J C, Kaplan A H. Moloney murine sarcoma virus genomic RNAs dimerize via a two-step process: a concentration-dependent kissing-loop interaction is driven by initial contact between consecutive guanines. J Virol. 1999;73:7255–7261. doi: 10.1128/jvi.73.9.7255-7261.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maisel J, Bender W, Hu S, Duesberg P H, Davidson N. Structure of 50 to 70S RNA from Moloney sarcoma viruses. J Virol. 1978;25:384–394. doi: 10.1128/jvi.25.1.384-394.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Markowitz D, Goff S, Bank A. A safe packaging cell line for gene transfer: separating viral genes on two different plasmids. J Virol. 1988;62:1120–1124. doi: 10.1128/jvi.62.4.1120-1124.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marquet R, Baudin F, Gabus C, Darlix J-L, Mougel M, Ehresmann C, Ehresmann B. Dimerization of human immunodeficiency virus (type 1) RNA: stimulation by cations and possible mechanism. Nucleic Acids Res. 1991;91:2349–2357. doi: 10.1093/nar/19.9.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moguel M, Zhang Y, Barklis E. cis-Active structural motifs involved in specific encapsidation of Moloney murine leukemia virus RNA. J Virol. 1996;70:5043–5050. doi: 10.1128/jvi.70.8.5043-5050.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muriaux D, Girard P-M, Bonnet-Mathoniere B, Paoletti J. Dimerization of HIV-1Lai RNA at low ionic strength: an autocomplementary sequence in the 5′ leader region is evidenced by an antisense oligonucleotide. J Biol Chem. 1995;270:8209–8216. doi: 10.1074/jbc.270.14.8209. [DOI] [PubMed] [Google Scholar]
- 18.Murti K G, Bondurant M, Tereba A. Secondary structural features in the 70S RNAs of Moloney murine leukemia and Rous sarcoma viruses as observed by electron microscopy. J Virol. 1981;37:411–419. doi: 10.1128/jvi.37.1.411-419.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olsen J C, Sechelski J. Use of sodium butyrate to enhance production of retroviral vectors expressing CFTR cDNA. Hum Gene Ther. 1995;6:1195–1202. doi: 10.1089/hum.1995.6.9-1195. [DOI] [PubMed] [Google Scholar]
- 20.Oroudjev E M, Kang P C E, Kohlstaedt L A. An additional dimer linkage structure in Moloney murine leukemia virus RNA. J Mol Biol. 1999;291:603–613. doi: 10.1006/jmbi.1999.2984. [DOI] [PubMed] [Google Scholar]
- 21.Paillart J C, Marquet R, Skripkin E, Ehresmann B, Ehresmann C. Mutational analysis of the bipartite dimer linkage structure of human immunodeficiency virus type 1 genomic RNA. J Biol Chem. 1994;269:27486–27493. [PubMed] [Google Scholar]
- 22.Paillart J C, Marquet R, Skripkin E, Ehresmann C, Ehresmann B. Dimerization of retroviral genomic RNAs: structural and functional implications. Biochimie. 1996;78:639–653. doi: 10.1016/s0300-9084(96)80010-1. [DOI] [PubMed] [Google Scholar]
- 23.Paillart J C, Skripkin E, Ehresmann B, Ehresmann C, Marquet R. A loop-loop “kissing” complex is the essential part of the dimer linkage of genomic HIV-1 RNA. Proc Natl Acad Sci USA. 1996;93:5572–5577. doi: 10.1073/pnas.93.11.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paoletti J, Mougel M, Tounekti N, Girard P M, Ehresmann C, Ehresmann B. Spontaneous dimerization of retroviral MoMuLV RNA. Biochimie. 1993;75:681–686. doi: 10.1016/0300-9084(93)90099-e. [DOI] [PubMed] [Google Scholar]
- 25.Prats A-C, Roy C, Wan P, Erard M, Housset V, Gabus C, Paoletti C, Darlix J-L. cis Elements and trans-acting factors involved in dimer formation of murine leukemia virus RNA. J Virol. 1990;64:774–783. doi: 10.1128/jvi.64.2.774-783.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skripkin E, Paillart J C, Marquet R, Ehresmann B, Ehresmann C. Identification of the primary site of the human immunodeficiency virus type 1 RNA dimerization in vitro. Proc Natl Acad Sci USA. 1994;91:4945–4949. doi: 10.1073/pnas.91.11.4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Summerton J, Stein D, Huang S B, Matthews P, Weller D, Partridge M. Morpholino and phosphorothioate antisense oligomers compared in cell-free and in-cell systems. Antisense Nucleic Acid Drug Dev. 1997;7:63–70. doi: 10.1089/oli.1.1997.7.63. [DOI] [PubMed] [Google Scholar]
- 28.Summerton J, Weller D. Morpholino antisense oligomers: designs, preparation, and properties. Antisense Nucleic Acid Drug Dev. 1997;7:187–195. doi: 10.1089/oli.1.1997.7.187. [DOI] [PubMed] [Google Scholar]
- 29.Sundquist W I, Heaphy S. Evidence for interstrand quadruplex formation in the dimerization of human immunodeficiency virus type 1 genomic RNA. Proc Natl Acad Sci USA. 1993;90:3393–3397. doi: 10.1073/pnas.90.8.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tounekti N, Mougel M, Roy C, Marquet R, Darlix J-L, Paoletti J, Ehresmann B, Ehresmann C. Effect of dimerization on the conformation of the encapsidation Psi domain of Moloney murine leukemia virus RNA. J Mol Biol. 1992;223:205–220. doi: 10.1016/0022-2836(92)90726-z. [DOI] [PubMed] [Google Scholar]