Abstract
Uridine diphosphate-dependent glycosyltransferases (UGTs) mediate the glycosylation of plant metabolites, thereby altering their physicochemical properties and bioactivities. Plants possess numerous UGT genes, with the encoded enzymes often glycosylating multiple substrates and some exhibiting substrate inhibition kinetics, but the biological function and molecular basis of these phenomena are not fully understood. The promiscuous monolignol/phytoalexin glycosyltransferase NbUGT72AY1 exhibits substrate inhibition (Ki) at 4 μM scopoletin, whereas the highly homologous monolignol StUGT72AY2 is inhibited at 190 μM. We therefore used hydrogen/deuterium exchange mass spectrometry and structure-based mutational analyses of both proteins and introduced NbUGT72AY1 residues into StUGT72AY2 and vice versa to study promiscuity and substrate inhibition of UGTs. A single F87I and chimeric mutant of NbUGT72AY1 showed significantly reduced scopoletin substrate inhibition, whereas its monolignol glycosylation activity was almost unaffected. Reverse mutations in StUGT72AY2 resulted in increased scopoletin glycosylation, leading to enhanced promiscuity, which was accompanied by substrate inhibition. Studies of 3D structures identified open and closed UGT conformers, allowing visualization of the dynamics of conformational changes that occur during catalysis. Previously postulated substrate access tunnels likely serve as drainage channels. The results suggest a two-site model in which the second substrate molecule binds near the catalytic site and blocks product release. Mutational studies showed that minor changes in amino acid sequence can enhance the promiscuity of the enzyme and add new capabilities such as substrate inhibition without affecting existing functions. The proposed subfunctionalization mechanism of expanded promiscuity may play a role in enzyme evolution and highlights the importance of promiscuous enzymes in providing new functions.
Key words: glycosyltransferase, substrate inhibition, hydrogen/deuterium exchange mass spectrometry, protein morphing, protein conformer, scopoletin
The promiscuous monolignol/phytoalexin glucosyltransferase NbUGT72AY1 from Nicotiana benthamiana shows scopoletin substrate inhibition at low μM concentrations, whereas the highly homologous StUGT72AY2 from Solanum tuberosum is inhibited only at 50-fold higher levels. Hydrogen/deuterium exchange mass spectrometry identified amino acids involved in ligand binding, and reciprocal mutation of these candidates confirmed the importance of F87I, located in the active site, and a sequence segment distant from the catalytic center for substrate inhibition. Subfunctionalization of a monolignol glucosyltransferase in Nicotiana thus results in increased promiscuity and substrate inhibition.
Introduction
Glycosylation catalyzed by nucleoside diphosphate sugar-dependent glycosyltransferases (GTs) is an important physiological reaction that affects the solubility, stability, transport, storage, reactivity, and bioactivity of sugar acceptors such as proteins, lipids, saccharides, and small molecules (Kurze et al., 2021; Putkaradze et al., 2021). In plants and animals, uridine diphosphate (UDP) sugar-dependent GTs (UGTs) catalyze the glycosylation, e.g., the glucosylation and glucuronidation, of natural products and xenobiotics (Meech et al., 2019; Wilson and Tian, 2019). Plant UGTs involved in carbohydrate transfer to small molecules are encoded by large multigene families and show a conserved signature motif in their amino acid sequences known as the plant secondary (specialized) product glycosyltransferase (PSPG) box, which groups them among the family 1 glycosyltransferases (GT1) according to the CAZy database (www.cazy.org/GlycosylTransferases.html) (Kurze et al., 2021). Glycosyltransferases can be classified into more than 100 families, of which GT1, containing only UGTs, is the most numerous in the plant kingdom (Yonekura-Sakakibara and Hanada, 2011). Plant and animal UGTs are often promiscuous enzymes and glycosylate more than one substrate, but multiple UGT enzymes can also convert the same substrate. This fact suggests that substrate availability is a critical factor for product formation in a cellular context and that redundancy reflects the plasticity of glycosylation (Bowles et al., 2006).
Protein promiscuity plays a central role in evolution, and this is reflected in proposed models of enzyme evolution, as three out of four models start from promiscuous enzymes (Glasner et al., 2020). Two subfunctionalization models have been proposed, differing in their mechanism of functional specialization, as well as an innovation-amplification-divergence model, and all three build on multifunctional predecessors. Although these models predict that the endpoints of each evolutionary pathway are functionally specialized, examples from the literature show that endpoints can still be promiscuous for the ancestral activity (Noda-Garcia and Tawfik, 2020).
The 3D crystal structures of glycosyltransferases published to date mainly adopt one of two folds, termed GT-A and GT-B. GT-C and GT-D folds have also been identified but have few structural representatives (Zhang et al., 2014). Plant glycosyltransferases categorized as GT1 in the CAZy database (Drula et al., 2022) show a GT-B fold and catalyze the enzymatic reaction using an inverting glycosylation mechanism (Liang et al., 2015). Regardless of the individual topology, the reaction usually proceeds according to a sequential bi-bi mechanism, during which the carbohydrate donor substrate and aglycone acceptor substrate are sequentially bound, followed by sugar transfer to the latter (Luukkanen et al., 2005). The glycoside product is set free, followed by release of the nucleotide. The N- and C-terminal domains of GT-B members bind the acceptor and carbohydrate donor, respectively, and the GT-B protein undergoes a series of conformational changes during the reaction (Albesa-Jové and Guerin, 2016). Binding of the carbohydrate donor, which allows the pyrophosphate to interact by hydrogen bonds with the N- and C-terminal domains and thus stabilize the catalytically active conformation, triggers the switch from open to closed conformation (Qasba et al., 2005). The conformational closure changes the size and shape of the active site and forms the actual acceptor binding sites, which are stabilized by entropic effects, in accordance with the induced-fit mechanism (Liang et al., 2015). A histidine residue at about position 20 in the N-terminal domain of GT1 enzymes is regarded as the catalytic base for deprotonation of the hydroxyl group at the acceptor molecule to enable nucleophilic attack at the anomeric center of the sugar donor (Offen et al., 2006). The reaction corresponds to a single displacement mechanism. Protonation of the histidine is subsequently stabilized through hydrogen-bonding interactions with an aspartate residue around position 120.
Plant UGTs are involved in various pathways of specialized metabolism and therefore play a central role in growth and development. They are implicated in homeostasis of plant hormones such as abscisic acid and auxins, defense against plant pathogens by glucosylation of phytoalexins, and biosynthesis of lignin, although their exact roles in lignin formation have not yet been clarified (Yonekura-Sakakibara and Hanada, 2011; Wilson and Tian, 2019). For lignin biosynthesis, the lignin monomers 4-coumaryl, coniferyl, and sinapyl alcohols/aldehydes (monolignols) must be translocated into the cell wall for polymerization to lignin (Le Roy et al., 2016). UGT72E2 and UGT72E3, two glycosyltransferases from Arabidopsis thaliana, glucosylate monolignols at the 4-O position, suggesting that these enzymes might play a role in lignin biosynthesis (Lanot et al., 2006, 2008). Downregulation of the corresponding genes led to reduced monolignol glucoside levels in transformed Arabidopsis plants, and UGT72E1 and UGT72E2 were co-expressed with the peroxidases PRX49 and PRX72, which are major actors in lignin polymerization in the cell wall (Le Roy et al., 2016). Although the detailed mechanism underlying the relationship between lignin biosynthesis and monolignol glycosylation is not yet clear, it is generally accepted that UGTs are essential for lignification of the plant cell wall (Le Roy et al., 2016).
Inhibition of enzyme activity at high substrate and/or cofactor concentrations, also known as substrate inhibition, is a common phenomenon observed in over 20% of known enzymes, including dehydrogenases, P450 enzymes, and UGTs (Reed et al., 2010). A two-site binding model has been proposed, in which one substrate binding site is productive (catalytic site) whereas the other (inhibitory site) is suppressive (Wu, 2011; Dong and Wu, 2012). Formation of a ternary dead-end complex has also been discussed as another substrate inhibition mechanism whereby accumulation of the non-productive ternary complex slows catalysis to a significant extent (Luukkanen et al., 2005). However, these models do not account for conformational change of the proteins upon ligand binding.
In two previous studies, we investigated the promiscuous monolignol/phytoalexin glycosyltransferase NbUGT72AY1 from the tobacco plant Nicotiana benthamiana (Sun et al., 2019, 2020). In this work, we observed pronounced substrate inhibition kinetics of the enzyme with scopoletin as the acceptor substrate, whereas its most similar homolog StUGT72AY2 from the potato plant Solanum tuberosum was only slightly inhibited at high scopoletin concentrations and showed a different substrate spectrum. Examination of NbUGT72AY1 by hydrogen/deuterium exchange mass spectrometry (HDX-MS) revealed amino acids putatively involved in scopoletin and UDP binding. Mutational analyses of NbUGT72AY1 and StUGT72AY2 in combination with in silico modeling and morphing studies identified an allosteric site formed after the transition from open to closed protein conformer and highlighted amino acids important for substrate preference and inhibition. Although members of the UGT72 family preferentially glucosylate monolignols (Speeckaert et al., 2022), our results show that single amino acid substitutions can contribute to the subfunctionalization of UGT72s (Glasner et al., 2020), resulting in additional conversion of hydroxycoumarins but with substrate inhibition.
Results
NbUGT72AY1 and StUGT72AY2 share high sequence identity but exhibit distinct substrate preferences and enzyme kinetics
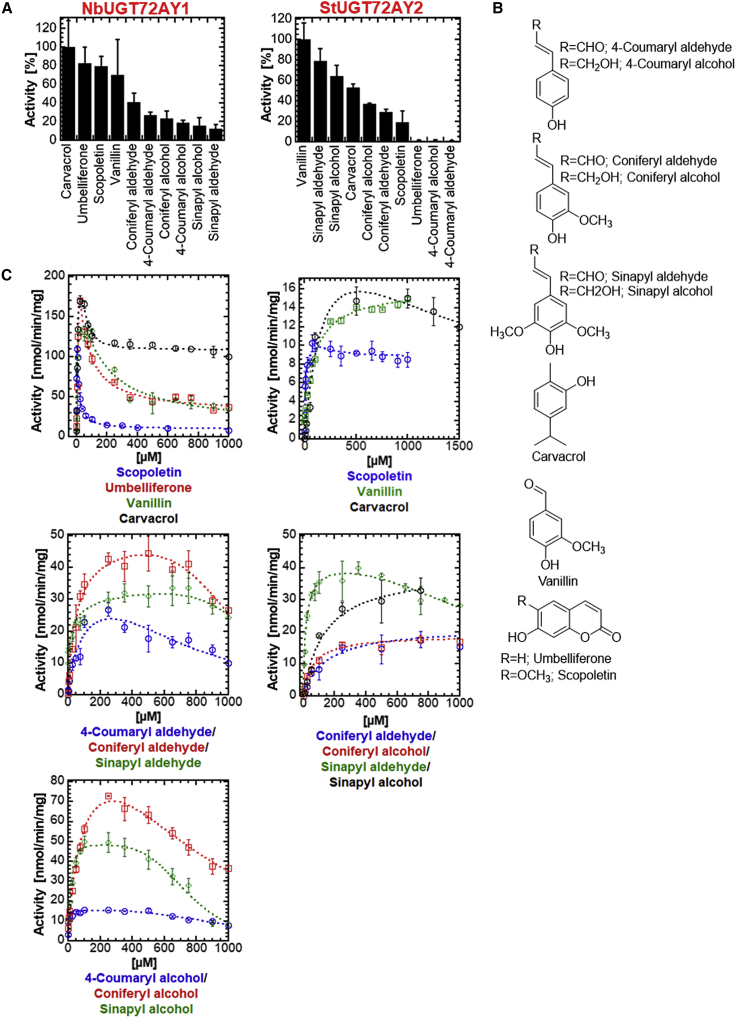
UGT72 members catalyze the glycosylation of monolignols such as 4-coumaryl alcohol, coniferyl alcohol, sinapyl alcohol, and their respective aldehydes and are therefore likely to be involved in lignin formation (Lim et al., 2005; Speeckaert et al., 2020, 2022). During comparative functional characterization studies on UGTs from N. benthamiana, NbUGT72AY1 attracted our particular attention because of its distinct, unusually strong scopoletin glycosylation activity (Sun et al., 2019). Recombinant NbUGT72AY1 produced in E. coli glycosylated six monolignols in vitro (Figures 1A and 1B); it is expressed primarily in stem tissue (Sun et al., 2019) (Supplemental Figure 1) and may thus play a role in lignification of this plant part.
Figure 1.
Substrate preferences and kinetics of NbUGT72AY1 and StUGT72AY2.
The first and second columns show the results for NbUGT72AY1 and StUGT72AY2, respectively.
(A) Substrate screening of NbUGT72AY1 and StUGT72AY2 using the UDP-Glo glycosyltransferase assay.
(B) Chemical structures of the acceptor substrates.
(C) Plots of acceptor substrate concentration versus reaction rate. Substrates are color coded. In (A and C), data represent mean ± SD of n = 3 technical replicates.
Additional biochemical assays showed that the monoterpene alcohol carvacrol, the coumarin derivatives umbelliferone and scopoletin, and the phenol vanillin are preferred substrates of NbUGT72AY1, but they strongly inhibited the enzymatic activity of NbUGT72AY1 at higher concentrations (Figures 1A and 1C). NbUGT72AY1 exhibited atypical Michaelis–Menten kinetics, and the data did not conform to the typical substrate inhibition equation, implying that the uncompetitive inhibition was incomplete. In particular, the substrate scopoletin showed unusually strong substrate inhibition kinetics for NbUGT72AY1 (Figure 1C).
The equation (Equation 4) that best fit the data of all substrates combines the two-site kinetic model for sequential ordered binding (Equation 3) (Wu, 2011) and the Hill equation (Equation 2) (LiCata and Allewell, 1997). The equation contains two Hill coefficients, n and x, where x takes into account the possibility that binding of the substrate in the inhibitory mode can also be cooperative. The model (Scheme 1) and corresponding equation also explained well the kinetics of an anthocyanidin UGT from strawberry (Fragaria vesca) (Peng et al., 2016). The high activity and strong substrate inhibition of carvacrol, umbelliferone, scopoletin, and vanillin can also be inferred from their kinetics, as the preferred substrates exhibited higher maximal reaction rates (Vmax) but lower inhibition constants (Ki) compared with the values for the monolignols (Supplemental Table 1).
Scheme 1.
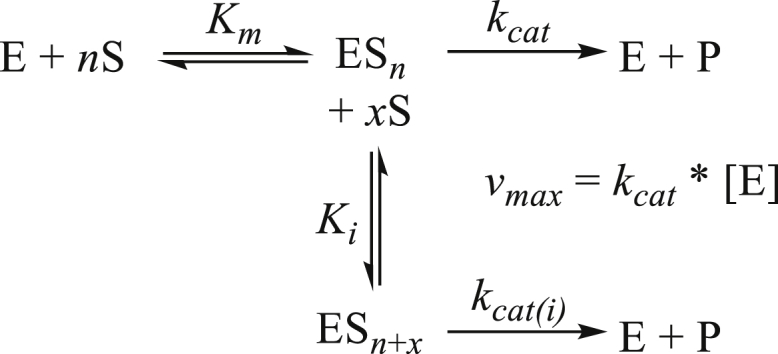
General reaction scheme for partial uncompetitive substrate inhibition
The inhibition constant Ki is the concentration of inhibitor required to decrease the maximal rate of the reaction to half of the uninhibited value. The glucosylation of monolignols is inhibited at much higher substrate concentrations (Ki > 500 μM), and the inhibition appears to be complete (Vi = 0 nmol/min/mg).
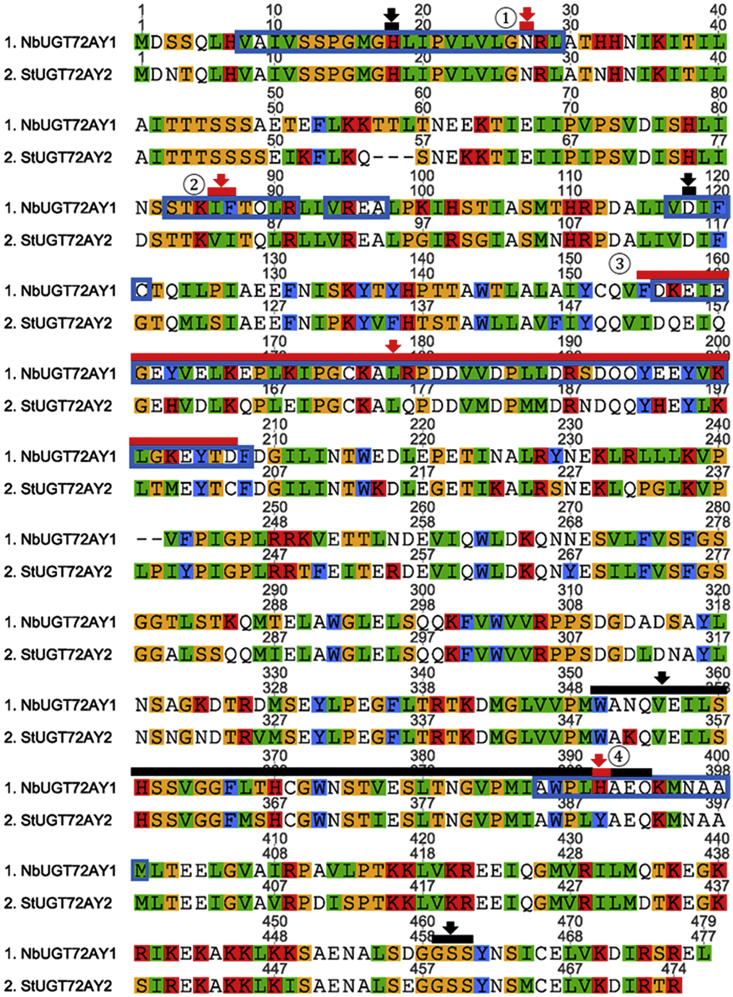
To analyze the remarkable properties of NbUGT72AY1 in more detail by comparative analysis, we searched for similar enzymes using BLAST (blast.ncbi.nlm.nih.gov) and identified StUGT72AY2 from potato (S. tuberosum) (Figure 2), whose gene was also strongly expressed in stem tissue regardless of whether TPM or FPKM values were used (www.ebi.ac.uk/gxa/experiments/E-MTAB-552/; PGSC0003DMG401004500) (Supplemental Figure 1). NbUGT72AY1 and StUGT72AY2 consist of 477 and 474 amino acids, 383 of which are shared (80.3% identity), and have isoelectric points of 6.00 and 5.67, respectively. Both contain the catalytically active H18 as well as D118 that activates H18, the PSPG box (W352–Q395; consensus sequence), and the GSS motif, a feature of mono-glucosyltransferases (Kurze et al., 2021). However, in vitro enzyme activity assays revealed distinct substrate preferences and kinetics for StUGT72AY2 compared with NbUGT72AY1 (Figure 1A). StUGT72AY2 showed high catalytic activity toward 2-methoxy-phenols (vanillin, sinapyl aldehyde, sinapyl alcohol, coniferyl alcohol, coniferyl aldehyde, and scopoletin), as well as carvacrol (in which the methoxy group is replaced by a methyl group), which was also converted. By contrast, NbUGT72AY1 favored short-chain phenols (carvacrol and vanillin) and cyclic hydroxyl phenylpropanoids (umbelliferone and scopoletin), whereas open-chain hydroxyl phenylpropanoids were slowly transformed. StUGT72AY2 showed weak substrate inhibition and even Michaelis–Menten curves were obtained for vanillin, coniferyl alcohol, coniferyl aldehyde, and sinapyl alcohol (Figure 1C; Supplemental Table 1). Both enzymes produced exclusively 4-O-glucosides of monolignols, as confirmed by LC-MS analysis (Supplemental Figure 2).
Figure 2.
Amino acid sequence alignment of NbUGT72AY1 and StUGT72AY2 from Nicotiana benthamiana and Solanum tuberosum, respectively, and HDX results.
The catalytically active H18, activating D118, plant secondary product glycosyltransferase (PSPG) consensus sequence position 352–395, and G461/S462/S463 motif are highlighted by black bars and arrows; mutations ① N27D, ② IF/VI double mutant, ③ chimeric mutant, and ④ H/Y mutant are highlighted by red bars and arrows. Amino acids of NbUGT72AY1 that showed reduced hydrogen/deuterium exchange in HDX experiments in the presence of scopoletin are framed with blue boxes (Supplemental Figure S9A).
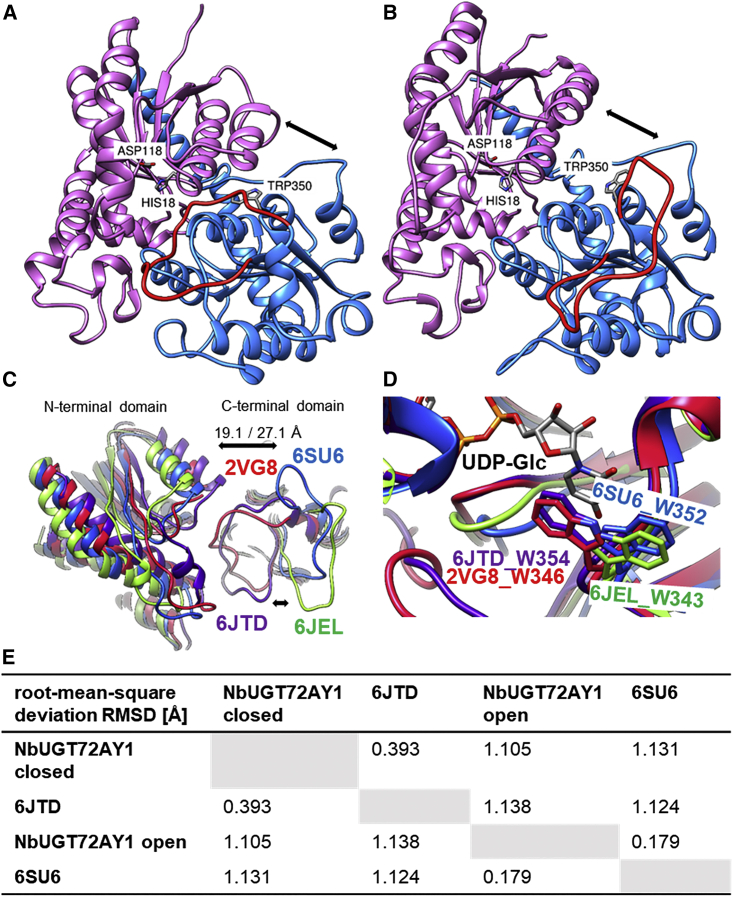
Homology modeling of NbUGT72AY1 identified closed and open conformers
To understand the structural requirements and conformational changes that accompany substrate inhibition, we generated a homology model of NbUGT72AY1. The amino acid sequence of NbUGT72AY1 was submitted to the IntFOLD web server (https://www.reading.ac.uk/bioinf/IntFOLD/). Among the top five 3D models, model IntFOLD5_MUSTER_multi8_TS1.bfact (template 6JTD_A; coverage of target: 0.9937107; RMSD: 0.72; TM-score: 0.98727) was chosen on the basis of its high Global Model Quality Score (GMQS) of 0.59 and confidence p-value of 1.818 × 10−9.
Scores greater than 0.4 are characteristic of more complete and confident models; thus, the predicted model should closely reflect the native protein 3D structure (McGuffin et al., 2019). With the exception of the first five amino acids at the N terminus and amino acids 310 to 315, no residue exceeded the disorder/order probability score of 0.5 (Supplemental Figure 3), and conserved regions of UGTs (catalytic H18, activating D118, and the PSPG box from W350–Q393) were located in highly ordered regions of the protein. Accordingly, these residues and regions also show low per-residue errors (Supplemental Figure 4). NbUGT72AY1 is a typical GT1 family member, as it adopts the GT-B fold, and it should follow an inverting mechanism (Figure 3A).
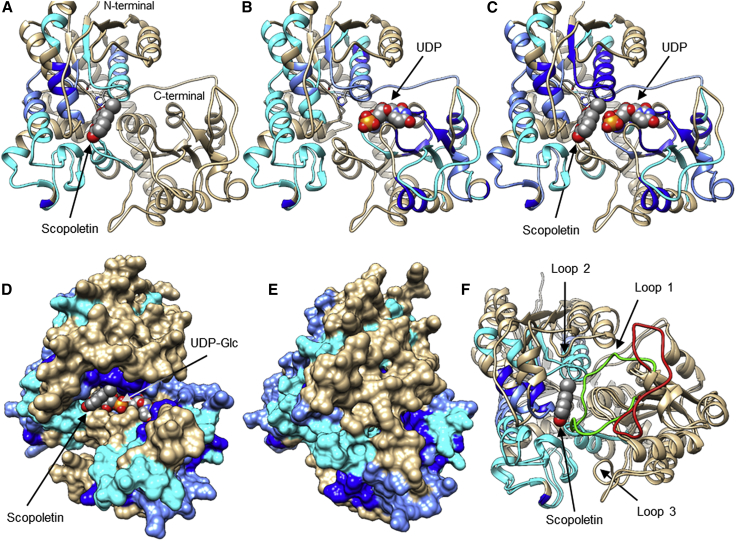
Figure 3.
3D UGT structures showing closed and open conformers.
(A) Prediction of the 3D structure of NbUGT72AY1 (closed conformer) was performed by the IntFOLD Integrated Protein Structure and Function Prediction Server (https://www.reading.ac.uk/bioinf/IntFOLD/) with default values based on 6JTD (www.rcsb.org). The result was visualized with UCSF Chimera (https://www.cgl.ucsf.edu/chimera). N- and C-terminal domains are shown in purple and blue, respectively. Important amino acids are marked, and the flexible loop covering the catalytic site is highlighted in red.
(B) Prediction of the 3D structure of NbUGT72AY1 (open conformer) was performed by the SWISS-MODEL Server (https://swissmodel.expasy.org) with default values based on 6SU6 (www.rcsb.org).
(C) Close-up of the superimposition of two putative open UGT conformers (6SU6 and 6JEL) and two closed UGT conformers (6JTD and 2VG8). Distances were measured between the N- and C-terminal domains of 6JTD and 6SU6 (S62.A CB to E263.A CB in 6JTD: 19.1 Å, in comparison to S62.A CB to E262.A CB in 6SU6: 27.1 Å).
(D) Tryptophan/uridine π-stacking interaction of the first amino acid of the PSPG box and UDP-Glc. In the crystal structures of the putative open protein conformers (6SU6 and 6JEL), W is rotated by 180° in comparison to the 3D structures of the closed conformers (6JTD and 2VG8).
(E) Calculation of mutual root-mean-square deviation (RMSD) values using UCSF Chimera.
The IntFOLD5-TS method works via iterative multi-template-based modeling (Buenavista et al., 2012) and uses target–template alignments. The templates (www.rcsb.org) 6JTD (He et al., 2019) and 2VG8 (identical in sequence to 2VCE and 2VCH) (Brazier-Hicks et al., 2007) were preferred for generation of 3D structures because of their high amino acid sequence identities of 47.0% and 41.4%, respectively, with NbUGT72AY1. A search for suitable templates at the SWISS-MODEL server (https://swissmodel.expasy.org) and a detailed analysis revealed two additional protein sequences highly similar to NbUGT72AY1 whose 3D structures have been elucidated (Supplemental Figure 5).
The sequences of 6SU6 (identical to 6SU7 and 5NLM) (Hsu et al., 2018; Teze et al., 2021) and 6JEL (identical to 6JEN and 6JEM) (Maharjan et al., 2020) share 41.2% and 39.4% identical amino acids, respectively, with NbUGT72AY1. A closer inspection of the UGT protein structures suggested that 6JTD and 2VG8 show the enzyme in the closed form, whereas 6SU6 and 6JEL display the 3D model of the open conformer (Figure 3C). This assumption is supported by the greater distance between the N- and C-terminal domains of 2VG8/6JTD and 6SU6/6JEL (S62.A CB to E263.A CB in 6JTD: 19.1 Å, compared with S62.A CB to E262.A CB in 6SU6: 27.1 Å) and the opposite orientation of a loop in 2VG8/6JTD and 6SU6/6JEL, which is located between the domains. Furthermore, the side chain of the first amino acid of the PSPG box, Trp, orients differently to the donor substrate (Figure 3D). In the crystal structures of the putative open protein conformers (6SU6 and 6JEL), Trp is rotated about 180° compared with the 3D structures of the closed conformers (6JTD and 2VG8). This can explain why all soaking experiments with (closed) 6JTD were unsuccessful (He et al., 2019), whereas (open) 6SU6 (with UDP-Glc) and 6SU7 (with 3,4-dichloroaniline) complex structures were readily obtained after soaking of pre-formed Polygonum tinctorium UGT1 crystals with the ligands (Teze et al., 2021). Consequently, we also generated the 3D structure of the open conformer of NbUGT72AY1 based on 6SU6 using the SWISS-MODEL server (https://swissmodel.expasy.org), as the IntFOLD server does not accept predefined templates (Figure 3B). The GMQE and Qualitative Model Energy Analysis (QMEA) values were 0.71 and 0.71 ± 0.05 (Supplemental Figures 6 and 7), respectively. Among other differences, the open and closed NbUGT72AY1 conformers differ in the orientation of W350 and the flexible loop (amino acids 306 to 325, shown in red) (Figures 3A and 3B). The root-mean-square deviations (RMSDs) of atomic positions of closed and open NbUGT72AY1 compared with 6JTD and 6SU6 were 0.393 and 0.179 Å, respectively (Figure 3E).
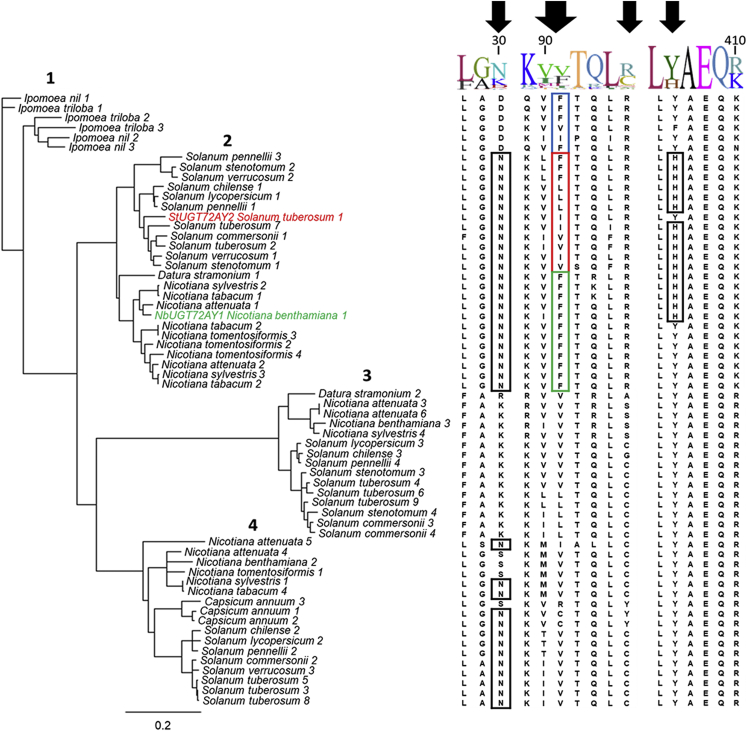
Figure 7.
Phylogenetic analysis of the closest homologs of NbUGT72AY1.
The tree shows the plant species from which NbUGT72A1-like protein sequences were extracted by BLAST (blast.ncbi.nlm.nih.gov/Blast.cgi). Regions of the amino acid sequence alignment are shown, with the positions of the mutated amino acids indicated by black arrows. Accession numbers are included in the supplementary information (Supplemental Figure 20).
HDX-MS analysis of NbUGT72AY1 revealed conformational changes upon substrate binding
It has been suggested that two substrate binding sites may contribute to substrate inhibition in the glycosylation reaction (Wu, 2011; Dong and Wu, 2012). To probe for substrate binding to NbUGT72AY1, we performed HDX-MS in solution. HDX-MS relies on exchange of the amide hydrogens of the protein backbone for hydrogens from the solvent, which can be quantified by MS using a solvent containing D2O after proteolytic digestion of the protein owing to peptide mass shifts. The rate of H/D isotope substitution depends on the folded state and the dynamics of the protein (the stability of hydrogen bridge networks), and it may thus provide insights into protein structure or conformational changes upon ligand binding (Konermann et al., 2011; Masson et al., 2019).
NbUGT72AY1 was subjected to HDX-MS in the absence and presence of scopoletin or UDP ligands. After deuterium labeling and pepsin digestion, 150 peptides covering approximately 92% of the GST-tagged NbUGT72AY1 protein were obtained (Supplemental Figure 8A; Supplemental Table 2_HDX). The pattern of deuterium incorporation by NbUGT72AY1 agreed well with the predicted secondary structure, as rapid and high deuteration was apparent in protein parts without assigned secondary structures, e.g., the linker between the GST tag and NbUGT72AY1 and NbUGT72AY1 residues 244 to 255 and 305 to 328 (Supplemental Figure 8B).
Upon incubation with scopoletin, reduced HDX was observed in several areas of the N-terminal domain of NbUGT72AY1 (Supplemental Figure 9A), i.e., β-strand 1 and helix 1 (residues 8–29), the N-terminal portion of helix 4 (residues 83–91 and 94–98), the loop between β4 and α5 (residues 117–121), and parts of helices 6–10 and their interconnecting loops (residues 156–208). Most of these alterations should be caused by scopoletin binding to the enzyme active site, which is reflected in HDX changes surrounding the active site residues His18 and Asp118 (Figure 2). Correspondingly, no scopoletin-dependent perturbations in HDX were observed in peptides located in the GST tag or C-terminal domain of NbUGT72AY1. Changes in the HDX profile with respect to UDP were apparent for both N-terminal and C-terminal domains of NbUGT72AY1 (Supplemental Figure 9B) and similarly encompassed residues critical for coordination of this substrate (e.g., α-helix 17 with residues 371–376, and W350 located in the PSPG box, Figure 2). When both ligands (scopoletin/UDP) were applied at the same time, the effects were additive (Supplemental Figure 10). Collectively, these data suggest major conformational changes in NbUGT72AY1 caused by substrate binding, with scopoletin binding leading almost exclusively to transitions in the N-terminal part and UDP binding affecting both domains.
Scopoletin docking experiments on the predicted 3D model of NbUGT72AY1 to identify binding sites
Scopoletin docking was performed on the predicted closed 3D model of NbUGT72AY1 using the AutoDock Vina tool implemented in UCSF Chimera and the protein 3D structure 2VCE (www.rcsb.org) with the acceptor and donor ligands trichlorophenol and UDP-2-deoxy-2-fluoro-glucose, respectively. For the correct placement of UDP-Glc, 6SU6 (www.rcsb.org) was used as a template. The ligand scopoletin was installed in a plane with trichlorophenol in the active site (Supplemental Figure 11A). Comparison of the arrangement of the phenolic acceptor substrates in the active sites of the crystallized UGTs showed that they were all perfectly coplanar, with the hydroxyl group pointing toward the catalytically active His (Supplemental Figure 11B). The bond lengths (3.1 and 3.3 Å) and angle (102°) of the arrangement H18/scopoletin-OH/C1-UDP-Glc correspond well to the lengths (2.3 and 3.8 Å) and angle (111°) of the catalytic conformation in the crystal structure of 2VCE. A closer look at the active site shows that it is lined mainly with nonpolar amino acids (P14, G15, I86, F87, L90, I119, F120, P186, and A391), which presumably interact in a nonpolar (alkyl and π-π) and polar (van der Waals and carbon hydrogen) manner with the coumarin derivative (Supplemental Figure 11C). Scopoletin further forms a hydrogen bond with H18 that is important for catalysis, and D185, Q194, and H390 interact with the acceptor molecule via van der Waals forces and π-π stacking. The reaction between the acceptor and donor substrate takes place in a tunnel with openings at both ends (Supplemental Figure 11D).
The amino acids of NbUGT72AY1 that showed reduced HDX after the addition of scopoletin, UDP, and combinations of both were projected onto the predicted 3D structure of the open conformer of UGT72AY1 (Figure 4). The colors reflect the maximum amount of HDX regardless of the time course (Supplemental Figures 9 and 10). Most of the residues affected in their HDX by scopoletin (Figure 4A) are localized in the N-terminal domain and are part of α-helices (8–29, 83–91, 94–98, 117–121, and 386–399) that extend into the catalytic center. Interactions of amino acids with scopoletin in the active site stabilizes the hydrogen bonding networks, in particular the intra-helical hydrogen bonds, which results in significant decreases in deuterium incorporation (Skinner et al., 2012). However, residues 156–208 are rather remote from the active site and separated from it by α-helix 4 (Supplemental Figure 8A; Figure 4A). The effect of UDP on HDX in NbUGT72AY1 was more pronounced because the H/D exchange in amino acids of the N- and C-terminal domains was reduced (Figure 4B). In addition to amino acids whose HDX was also affected by scopoletin (N-terminal domain), numerous amino acids were affected at positions 243–377, a region that overlaps with the PSPG box, which is already known to interact with the donor substrate. Thus, UDP already stabilizes the hydrogen bonding networks in both domains. After co-addition of scopoletin and UDP, the effects were additive (Figure 4C).
Figure 4.
HDX results mapped on NbUGT72AY1.
Differential HDX results obtained after the addition of scopoletin, UDP, and a combination of both to the protein were color-mapped onto the predicted 3D structure of the open conformer of NbUGT72AY1.
(A) After addition of scopoletin (in sphere display), (B) after addition of UDP, and (C) after addition of scopoletin and UDP.
(D) Scopoletin and UDP-Glc (both in sphere display) are shown in the open conformer of the predicted structure of NbUGT72AY1 (HDX results of UDP/scopoletin are color-mapped).
(E) Scopoletin and UDP-Glc are buried in the closed conformer of NbUGT72AY1.
(F) Flexible loop (1) is highlighted in the NbUGT72AY1 model (green, closed conformation; red, open conformation; HDX results of scopoletin are color-mapped). Two additional closing loops are displayed.
Generation of NbUGT72AY1 and StUGT72AY2 mutants and their analysis highlight residues involved in substrate inhibition and promiscuity
To identify amino acids involved in scopoletin substrate inhibition and preference in NbUGT72AY1, the protein sequences of NbUGT72AY1 and StUGT72AY2 were compared in regions identified by HDX-MS as putatively important for ligand binding, and reverse mutations were generated when the sequences differed (Figure 2).
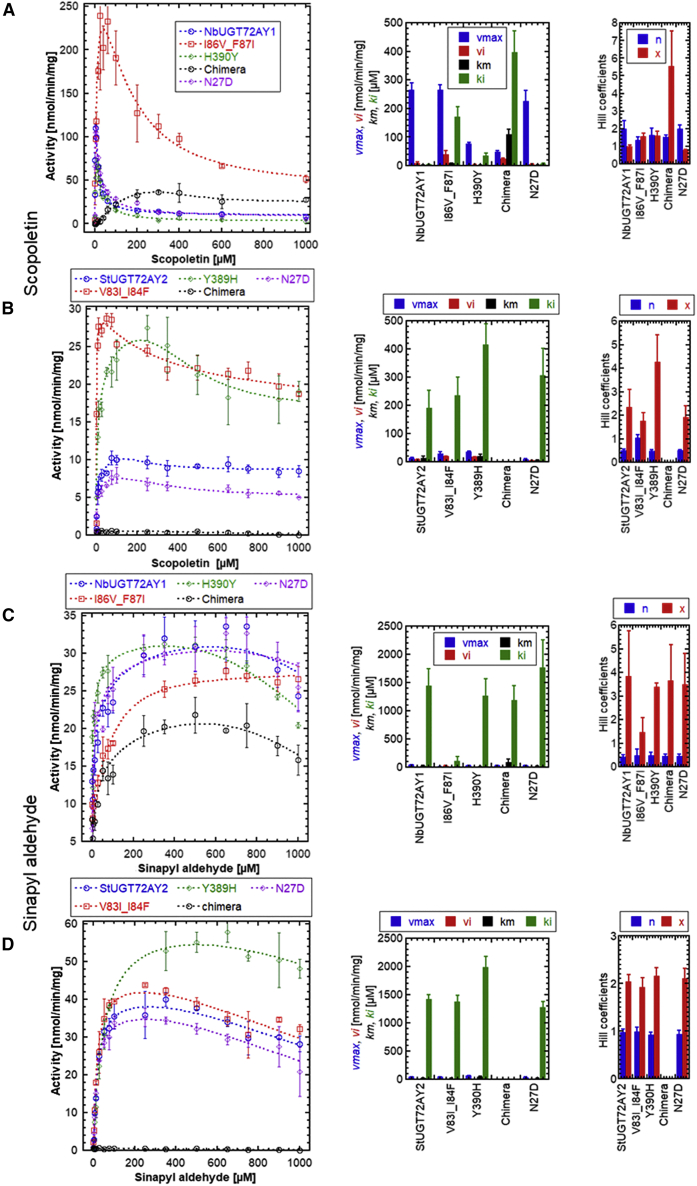
Deuterium incorporation of amino acids 87–97 in NbUGT72AY1 was strongly affected, but both sequences were identical in this region, except for position 87. Thus, the double mutants I86V_F87I of NbUGT72AY1 and V83I_I84F of StUGT72AY2 were generated, as these residues are within 5 Å of the catalytic center (② in Figure 2; Supplemental Figure 11C) and differ in both enzymes. In addition, a region from amino acids 156–208 of NbUGT72AY1 showed reduced HDX, and several residues were different in the corresponding region of StUGT72AY2. Therefore, this entire sequence part was exchanged, and chimeric mutant proteins were generated (③ in Figure 2). Scopoletin binding also affected deuterium incorporation of amino acids 386–399 located at the C-terminal end of the PSPG box. In this sequence segment, only the amino acid H390Y was different, and H390 was predicted to interact with scopoletin via π-stacking (Supplemental Figure 11C); therefore, a single mutant was created (④ in Figure 2). Because preliminary scopoletin docking experiments had revealed the N-terminal α-helix extending from amino acids 15–29 as a possible second interaction site, the polarity and charge of this site was altered by generating the single mutant N27D for both proteins (① in Figure 2). The kinetics of the four mutants of NbUGT72AY1 and StUGT72AY2 were analyzed with the substrates scopoletin and sinapyl aldehyde using the UDP-Glo assay (Figure 5).
Figure 5.
Kinetic parameters of NbUGT72AY1, StUGT72AY2, and mutant enzymes using scopoletin and sinapyl aldehyde as substrates.
(A) NbUGT72AY1 and its mutants were used to glucosylate scopoletin.
(B) StUGT72AY2 and its mutants were used to glucosylate scopoletin.
(C) NbUGT72AY1 and its mutants were used to glucosylate sinapyl aldehyde.
(D) StUGT72AY2 and its mutants were used to glucosylate sinapyl aldehyde. Experimental data were obtained by UDP-Glo glycosyltransferase assay and fitted to the partial uncompetitive inhibition model (Equation 4). The amino acid sequence information of the mutants is shown in Figure 2. Data represent mean ± SD of n = 3 technical replicates.
Comparison of the Vmax, Vi, Km, and Ki values for scopoletin of the N27D mutant and the wild-type NbUGT72AY1 showed that the data were virtually identical. Although the I86 and F87 residues in NbUGT72AY1 are located near the substrate binding site, Vmax was unchanged for I86V_F87I. However, Vi and Ki were significantly increased, resulting in a substantial decrease in substrate inhibition, similar to the chimeric mutant, in which Km and Vmax were also significantly increased and reduced, respectively, leading to an overall lower enzymatic activity. The elevated Ki value of the I86V_F87I mutant indicates that the two mutant amino acids are also located near the allosteric site. The catalytic activity of the H390Y mutant was reduced owing to a significantly lower Vmax value, but substrate inhibition (Ki) was only slightly affected.
Interestingly, the cooperativity in the inhibitory mode (Hill coefficient x) was significantly increased for the chimeric mutant (Figure 5A). By contrast, the chimeric mutant of StUGT72AY2 showed no enzymatic activity, and the kinetic data of the N27D mutant of StUGT72AY2 closely resembled those of the wild-type enzyme.
For the double mutant V83I_I84F, increased Vmax and Vi and a lower Km value resulted in higher catalytic activity, whereas for the Y389H mutant, Vmax, Vi, Km, and Ki values increased along with the Hill coefficient x, leading to higher enzymatic activity.
LC-MS was used as a second independent measurement method because it directly quantifies the glucoside product of enzymatic catalysis. Although in the case of NbUGT72AY1 and its mutants, the kinetic data obtained by the UDP-Glo assay were confirmed, for StUGT72AY2 and its mutants, LC-MS revealed lower enzymatic activities for all enzymes in the concentration range of less than 200 μM scopoletin (Supplemental Figure 12). We therefore hypothesize that the UDP-glucose hydrolase side activity of StUGT72AY2 and its mutants is responsible for the higher overall activity in the UDP-Glo assay at low scopoletin concentrations. However, the general conclusion from the experiments was not compromised.
When sinapyl aldehyde was used as a substrate for the various mutant enzymes, the kinetic data differed less than with scopoletin. For I86V_F87I, substrate inhibition was completely abolished because Vi was greater than Vmax, and Ki and the second Hill coefficient were significantly reduced. The enzymatic activity of the chimeric mutant was decreased owing to a higher Km value. As with scopoletin, the chimeric mutant of StUGT72AY2 was inactive with sinapyl aldehyde, whereas the other mutants, except for Y389H, showed similar kinetics to their wild-type enzyme. Y389H exhibited an increased Ki value, resulting in higher enzymatic activity. Overall, these results highlighted the role of I86–F87, H390, and the sequence segment F155–D207 in substrate inhibition of scopoletin in NbUGT72AY1.
Morphing of open and closed NbUGT72AY1 conformers reveals conformational changes important for catalysis and substrate inhibition
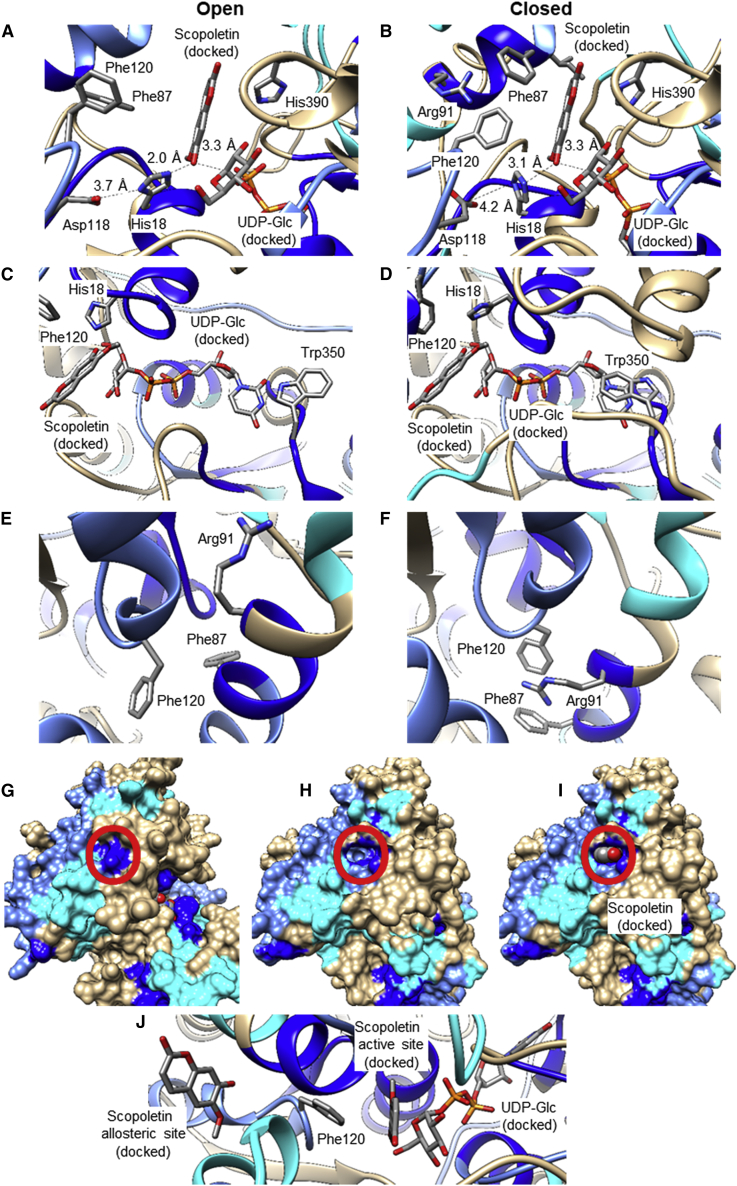
Major changes in secondary, tertiary, and quaternary structure are essential for the functions of UGTs (Qasba et al., 2005, 2008). Because we obtained the 3D structures of the open and closed conformers of NbUGT72AY1 by homology modeling based on X-ray structures of proteins with sequence identities of >40%, we performed morphing of both extreme forms. Morphing involves calculation of a series of intermediate, interpolated structures between the original input structures. The series of structures can be rendered as a movie (Weiss and Levitt, 2009). The HDX results for scopoletin/UDP were color-mapped onto the 3D morphing structures to visualize structural and dynamic aspects of NbUGT72AY1 motions. The moving model also includes the ligands scopoletin and UDP-Glc in the active site to identify the second scopoletin binding site (Supplemental Video 1; Figures 4D and 4E). The animation shows that both the C- and N-terminal domains move toward each other, thus closing the cleft, which represents the active site between the two domains. In the animation, all amino acid residues are in motion, not only the amino acids, which are marked according to their HDX results. However, two regions (amino acids 87–91 and 346–353, labeled in dark blue, indicating strongly reduced deuterium incorporation, in the left and right corners of the active site, respectively) show strong conformational changes and are covered in the closed conformer. Three loops are primarily responsible for closure of the catalytic center (Supplemental Video 2; Figure 4F). A loop of amino acids 305–325 (right) approaches an opposite loop (amino acids 71–82) in the N-terminal domain (left), while the third loop (amino acids 410–420) covers the active site from below (Supplemental Figure 13). Overall, the N-terminal domain (left) undergoes a much stronger conformational change than the C-terminal part (right). A closer look at the catalytic center reveals a minimal distance between the proton acceptor Nε2-His and OH-scopoletin of 2.0 Å in the open enzyme conformer, whereas the distance to the accessory D118 (Nδ1-His to COOH-Asp) is 3.7 Å (Supplemental Video 3; Figures 6A and 6B). During closure, D118 approaches H18 to within 3.1 Å, and H18 simultaneously rotates and thus moves away from the OH of scopoletin (to within 3.1 Å), consistent with the presumed deprotonation of the hydroxyl group. D118 accepts a proton on its closest approach to H18 and then distances itself again, supporting the proton transfer to H18. After the acceptor substrate anion attacks the donor substrate UDP-Glc (not shown), the amino acids involved in catalysis return to their positions in the open UGT conformer.
Figure 6.
Visualization of conformational differences in the open and closed conformer of NbUGT72AY1.
(A) Distance of Nε2-His to OH-scopoletin, Nδ1-His to COOH-Asp, and OH-scopoletin to C1-UDP-Glc in the ternary complex (NbUGT72AY1•scopoletin•UDP-Glc); HDX results with scopoletin/UDP are color-mapped, and the open conformer is shown.
(B) Same as in (A), but the closed conformer is shown.
(C) UDP-Glc binding pocket, showing the rotation of Trp350; HDX results with scopoletin/UDP are color-mapped, and the open conformer is shown.
(D) Same as in (C), but the closed conformer is shown.
(E) Putative allosteric binding site formed after rotation of Arg91; HDX results with scopoletin/HDX are color-mapped, and the open conformer is shown.
(F) Same as in (E), but the closed conformer is shown.
(G) Surface presentation of the open conformation of NbUGT72AY1; HDX results with scopoletin/UDP are color-mapped. Red circle shows the putative allosteric site, which is closed in the open conformer.
(H) Surface presentation of the closed conformation of NbUGT72AY1. The new binding site is marked by a red circle.
(I) Same as in (H), with scopoletin bound in the putative allosteric binding site.
(J) Phe120 is located between the catalytic and allosteric sites.
Scopoletin and UDP-glucose occupy the active site. NbUGT72AY1 is color-coded according to the HDX-MS results for scopoletin/UDP.
Scopoletin and UDP-glucose occupy the active site. NbUGT72AY1 is color-coded according to the HDX-MS results for scopoletin/UDP.
Scopoletin and UDP-glucose occupy the active site. NbUGT72AY1 is color-coded according to the HDX-MS results for scopoletin/UDP.
From the animation, it can be concluded that catalysis already starts when the catalytic center closes. In addition, F87 and I86 mutated in the double mutant are involved in formation of the scopoletin binding pocket, which is completed only during active site closure. They increase the hydrophobicity of the catalytic center, facilitating acceptor substrate incorporation and catalysis. A feature that distinguishes closed UGT conformers from open ones is the orientation of the first amino acid Trp of the PSPG box to the uridine part of the donor substrate (Figure 3; Supplemental Video 4; Figure 6C and 6D). The animation reveals the approximate 180° rotation of W350 during active site closure, resulting in formation of a π-π stacking interaction of the phenyl and uracil rings (Harrus et al., 2018). In addition to the motions of H18, I86, and F87, the animation also shows the rotation of H390, an amino acid that was mutated because HDX results and comparative biochemical assays suggested that it might be involved in catalysis and substrate inhibition. H390 is located near the donor and acceptor substrate, I86, and the loop (amino acids 306–325) that covers the active site.
Scopoletin and UDP-glucose occupy the active site. NbUGT72AY1 is color-coded according to the HDX-MS results for scopoletin/UDP.
In search of the allosteric binding site, we took a closer look at the vicinity of F87 and I86 because the substrate inhibition of NbUGT72AY1 was significantly reduced in the double mutant (Supplemental Video 5; Figures 6E and 6F). R91, located at the same α-helix as F87, undergoes a dramatic conformational change during transition from the open to the closed conformer and eventually forms a cation-π interaction with F87 and F120 (Flocco and Mowbray, 1994; Gallivan and Dougherty, 1999; Steiner and Koellner, 2001). The dislocation opens a new binding site in the closed NbUGT72AY1 conformer, allowing attachment of a second scopoletin molecule (Supplemental Video 6; Figure 6G–6I). Formation of the allosteric binding site upon binding of scopoletin in the catalytic center is consistent with the uncompetitive substrate inhibition model (Scheme 1).
NbUGT72AY1 is color-coded according to the HDX-MS results for scopoletin/UDP.
Scopoletin and UDP-glucose occupy the active site. NbUGT72AY1 is color-coded according to the HDX-MS results for scopoletin/UDP.
To confirm the detection of the allosteric site experimentally, we generated three R91 mutants, R91A, R91F, and R91M. Because A91 cannot block the allosteric site and F91, similar to R91, can form π-interactions with F87, we expected that only M91 would reduce substrate inhibition in comparison with the wild-type enzyme. The results showed that the mutants had similar Vmax and Km values, and therefore the catalytic efficiency of the mutants was not affected, whereas the Vi and in particular the Ki data differed (Supplemental Figures 14A and 14B). Overall, R91M exhibited reduced substrate inhibition, as Vi (13.1 ± 6.1 nmol/min/mg) and Ki (27.3 ± 4.2 μM) exceeded those of the wild-type enzyme (R91; 4.4 ± 2.3 nmol/min/mg and 10.7 ± 2.2 μM, respectively). R91A and R91F showed significantly stronger inhibition by scopoletin than the wild-type enzyme, reflected in lower Ki values (3.3 ± 0.7 μM and 0.9 ± 0.3 μM, respectively). After molecular docking of scopoletin in the putative allosteric site, the binding energies ΔG were used to calculate equilibrium constants KD (Supplemental Figures 14C–14G). The KD values determined in silico were considerably higher than the experimentally determined Ki values, but the ranking of the values was identical (R91F < R91A < R91 < R91M). Finally, we generated the single I86V and F87I mutants of NbUGT72AY1 and determined their kinetic parameters (Supplemental Figures 15A and 15B). Whereas I86V showed similar data to the wild-type enzyme, F87I exhibited lower substrate inhibition, similar to the double mutant I86V_F87I. Thus, only F87 is involved in formation of the second scopoletin binding site.
Subdivision of the chimeric mutants narrowed down the amino acids responsible for substrate inhibition
Because both chimeric mutants each contain 53 amino acids of the homologous UGT, we generated additional mutants to identify amino acid sequences that cause substrate inhibition in NbUGT72AY1. The newly produced mutants each carried only one half of the 53-amino-acid chimera sequence (Supplemental Figure 16C). Enzyme assays revealed that NbUGT72AY1-chimera A, carrying I152–N188 of StUGT72AY2, exhibited significantly reduced substrate inhibition kinetics, whereas the reverse mutant StUGT72AY2-chimera A showed atypical Michaelis–Menten kinetics in contrast to StUGT72AY2 and StUGT72AY2-chimera B. The reduction in substrate inhibition resulted in significantly increased Vi and Ki values in NbUGT72AY1-chimera A. The kinetics of StUGT72AY2 and its chimera B mutant can best be described by the Michaelis–Menten equation (Equation 1) but also by Equation 4 if Vmax = Vi, Km = Ki, and n = x = 1 are assumed. None of the amino acids that differ in the chimera A sequence in NbUGT72AY1 and StUGT72AY2 are components of the catalytic or postulated allosteric site (Supplemental Figure 16D). Because the different mutants examined in the present study could not completely reverse the substrate inhibition kinetics of the enzymes from tobacco and potato in each other, more than one amino acid exchange must be responsible.
Discussion
UGTs represent a superfamily of enzymes found in all kingdoms of life. They comprise several groups and subgroups owing to the large number of acceptor substrates (Bowles et al., 2006). N. benthamiana transcribes about 290 UGTs (https://sefapps02.qut.edu.au/atlas/benthgenereturn6.php), some of which were recently characterized (Sun et al., 2019, 2020). Among them, one translated enzyme (NbUGT72AY1) stood out because it exhibited strong substrate inhibition with several substrates (Figure 1).
UGT72 enzymes are probably involved in lignin biosynthesis
NbUGT72AY1 is a homolog of AT5G66690.1 from Arabidopsis (AtUGT72E2), an enzyme shown to be involved in lignin metabolism (Baldacci-Cresp et al., 2020). AtUGT72E2 glucosylates sinapyl and coniferyl aldehydes as well as their corresponding alcohols. A knockdown mutant line (72E2KD) produced by RNAi silencing showed a two-fold reduction in coniferyl alcohol 4-O-glucoside and sinapyl alcohol 4-O-glucoside compared with the wild type (Lanot et al., 2006, 2008). Similarly, NbUGT72AY1 and its ortholog from potato, StUGT72AY2, transfer glucose to monolignols, preferring coniferyl and sinapyl derivatives (Figure 1A), and their products were identified as 4-O-glucosides (Supplemental Figure 2). Because of their high sequence identity, the two enzymes have been assigned consecutive numbers by the UGT Nomenclature Committee (https://prime.vetmed.wsu.edu/resources/udp-glucuronsyltransferase-homepage). They are also likely to be involved in lignin biosynthesis because their sequences are highly similar to those of monolignol UGTs (Speeckaert et al., 2020, 2022) and they efficiently glucosylate phenylpropanoids in vitro (Figure 1). They are constitutively expressed in the stem (Supplemental Figure 1), and the potato enzyme interacts with nine peroxidases that are thought to be required for lignin formation (https://string-db.org/network/4113.PGSC0003DMT400011466). However, UGT72 members are promiscuous and also glycosylate flavonoids and other phenolics (Speeckaert et al., 2022).
Identification of open and closed protein conformers enabled the observation of enzyme dynamics by morphing
Because of the high substrate similarity of NbUGT72AY1 and StUGT72AY2 but their drastically different enzyme kinetics toward scopoletin, HDX analyses in combination with mutation studies appeared to be a promising approach for elucidating the molecular mechanism of substrate inhibition. GT1 family members catalyze the reaction by a compulsory ordered bi-bi mechanism in which UDP-sugar is the first binding substrate (Luukkanen et al., 2005; Hashimoto et al., 2010; Liang et al., 2015). However, similar to (Albesa-Jové et al., 2017), our HDX results demonstrated that acceptor substrate binding is possible in the absence of UDP/UDP-Glc (Figure 4A; Supplemental Figure 9A). NbUGT72AY1 was found to have more than 40% sequence identity with four crystallized UGTs whose 3D structures have already been elucidated (Supplemental Figure 5). Interestingly, two structures (6SU6 and 6JEL) represented UGTs with an open catalytic site, whereas the other two structures (6JTD and 2VG8) showed closed conformers. These templates provided a unique opportunity to model both conformer states of NbUGT72AY1 and visualize the dynamics of enzyme catalysis by morphing (Supplemental Videos 1 and 2; Figure 4).
Conformational changes and enzyme catalysis
Conformational changes of UGTs during catalysis have been reported for GT-A enzymes, including the human beta-1,4-galactosyltransferase (B4GalT1) (Harrus et al., 2018) and the glucosyl-3-phosphoglycerate synthase (GpgS) from Mycobacterium tuberculosis (Albesa-Jové et al., 2017). These proteins contain short dynamic loops inserted between two Rossmann fold domains, which undergo an open-to-closed motion essential for acceptor and donor substrate recognition and assembly of the reaction center. A limited number of GT-B-folded UGT structures show that these enzymes often exhibit global domain motion upon substrate binding and differ in the type and extent of movement (Chang et al., 2011). It was hypothesized that this open-to-closed conformational transition brings acceptor and donor substrate into close proximity and is accompanied by multiple loop displacements (Bolam et al., 2007). We observed that the distance between the catalytically active His and C1 of the acceptor substrate scopoletin is smaller in the open enzyme conformer (2.0 Å) than in the closed form (3.1 Å; Supplemental Video 3; Figures 6A and 6B). This indicates that deprotonation and therefore enzyme catalysis already starts with the motion of the loop, which is suggested by distance measurements between Nε2-His and C1 of UDP-Glc in the structures 6SU6 (open), 2ACW (closed), and 6LFZ (closed) of 4.6, 5.5, and 5.6 Å, respectively.
Particularly striking in the transition from open to closed form is the movement of a loop (amino acids 305–325; Supplemental Figure 13) toward the active center (Supplemental Videos 1 and 2, Figure 4F). Consequently, water is likely to be displaced from the active site, leading to the conclusion that the two small openings that can be identified in the closed enzyme conformer (Supplemental Figure 11D) are not substrate entry ports but probably drainage channels (Breton et al., 2006). Amino acids located at the two apertures could affect the efflux of water from the active site and thus control the inherent UDP-Glc hydrolase activity of UGTs (Sheikh et al., 2017). The closing loop, albeit of varying length, is present in various already crystallized GT-B-fold UGTs from plants (Supplemental Figure 5) and should behave similarly in them, as shown for flexible structures in GT-A-fold enzymes (Chang et al., 2011). In structures that do not contain this loop, alternative loops can be found at other positions, which presumably have a similar function.
When comparing the open and closed NbUGT72AY1 structural models, the position of the first amino acid of the PSPG box W350, which participates in fixation of the donor substrate by π-π interaction with the uracil group, is apparent (Supplemental Video 4; Figures 6C and 6D). This arrangement, together with the position of the loop (amino acids 305–325), can be used to distinguish open and closed GT-B folded UGTs based on their crystal structure. The Trp rotation could guide the donor substrate into the active site, but rotation could also be triggered only by the incorporation of the donor substrate into the active site, and/or the back rotation of W350 could facilitate the export of the reaction product UDP (Harrus et al., 2018).
An amino acid that affects mainly catalytic activity, but to a lesser extent substrate inhibition, of NbUGT72AY1 is H390. It moves during the transition from the open to the closed form (Supplemental Video 4) and interacts with the phosphate group adjacent to glucose in UDP-Glc, F87 in the active site, and the flexible loop (amino acids 303–325). Mutation to Tyr, as in the enzyme from potato, decreases catalytic activity by decreasing Vmax (75.5 versus 264.3 nmol/min/mg) and increasing Ki (36.2 versus 4.5 μM), explainable by reduced stabilization of the donor substrate and altered enzyme dynamics due to interaction with amino acids of the flexible loop (S315 and A316).
Identification of the second binding site
It has been proposed that two substrate binding sites and the formation of a dead-end ternary complex might contribute to substrate inhibition in the glycosylation reaction (Wu, 2011; Dong and Wu, 2012). Our results indicate two dependent substrate binding sites: after binding of scopoletin in the active site, conformational changes in which R91 plays an important role result in formation of a second allosteric binding site (Figures 6E and 6F). The conformational change is promoted by the formation of two cation-π interactions, structures commonly found in 3D protein structures (Supplemental Videos 5 and 6) (Flocco and Mowbray, 1994; Gallivan and Dougherty, 1999; Steiner and Koellner, 2001). This mechanism is consistent with the partial uncompetitive inhibition model used for calculating the kinetic parameters (Scheme 1) (Peng et al., 2016). The importance of R91 as a key amino acid for substrate inhibition has been clearly demonstrated by mutational studies (Supplemental Figure 14). Therefore, our study provides new insights into the dynamics of the UGT catalytic mechanism and the conformational changes that lead to substrate inhibition. The arrangement of the three V-shaped helices enclosing the allosteric center in NbUGT72AY1 is evident in 3D structures of GT-B-folded UGTs (Supplemental Figure 17). Because substrate inhibition is observed in numerous plant (Peng et al., 2016) and human UGTs (Wu, 2011), it can be assumed that this conserved spatial structure is also used as an allosteric center in other UGTs. The allosteric binding site is formed by R91 spatially approaching and forming a π-stacking interaction with F87 during the transition from the open to the closed conformation. Biochemical analysis of the single mutants I86V and F87I confirmed this hypothesis (Supplemental Figure 15). In StUGT72AY1, F87 is replaced by I84, which is not able to make this kind of interaction with R91 (Figure 2; Supplemental Figure 18).
Our studies show that the allosteric site is presumably located in close proximity to the catalytic center so that, for example, Phe120 is within 5 Å of both binding sites (Figure 6J). This means that mutations to reduce substrate inhibition will inevitably have an effect on catalytic activity, as has been shown for UGTs and other enzymes (Barnett et al., 2004; Miller et al., 2008; Chen et al., 2014). In our study, based on structural information and structure-activity experiments, we identified a position that exclusively affects substrate inhibition.
Chimeric mutants and sinapyl aldehyde glucosylation
All four parameters, Vmax, Vi, Km, and Ki, were severely altered in the chimeric mutant of NbUGT72AY1, providing the explanation for its decreased substrate inhibition and reduced catalytic efficiency (Figure 5A). Because the amino acids exchanged in the chimeric mutant are not located directly in the catalytic and allosteric centers, we assume that the dynamics of protein conformational change during catalysis are altered in this mutant, e.g., by changing the quaternary structure. The high Hill coefficient for the inhibited reaction, which is only observed in the chimeric mutant of NbUGT72AY1, hints at changes in quaternary structure (Figure 5A). In this regard, there are multiple examples in which UGTs adopt a quaternary structure, e.g., by homodimerization (Liu et al., 2016; Wang et al., 2020), and in which substrate specificities, reaction rates, and types of reaction products are influenced by oligomerization (Fujiwara et al., 2016; Smith et al., 2020). The number and/or oligomerization positions of protomers may have been altered by the mutation. The chimeric mutant of StUGT72AY2, on the other hand, was enzymatically inactive, potentially because of similar alterations in oligomerization (Figure 5B).
Additional mutants that split the chimera mutant into two parts were generated and analyzed. NbUGT72AY1-chimera A showed significantly reduced substrate inhibition, whereas the reciprocal StUGT72AY2-chimera A was inhibited by scopoletin (Supplemental Figure 16). This result indicates that amino acids F155–S191 contribute to substrate inhibition in NbUGT72AY1 in addition to F87.
Overall, scopoletin substrate inhibition was not completely abolished by the generated mutants. The strongest effects on substrate inhibition were observed with the double and especially the chimera/chimera A mutants of the tobacco enzyme, as evidenced by their increased Vi values. The postulated allosteric site of NbUGT72AY1 differs from the corresponding site in StUGT72AY2 by 87Phe and amino acids in the C-terminal sequence of the chimera (Glu196, Val199, Gly202, Lys203, and Asp207) (Figure 2). However, the side chains of the latter amino acids are located outside the putative allosteric site, and the reciprocal chimera potato mutant was inactive, whereas the double mutant exhibited apparently enhanced substrate inhibition due to increased Vmax and lower Km values. These results illustrate that no single amino acid alone is responsible for substrate inhibition, which instead arises from the interplay of different amino acid residues that influence the dynamics of protein movement and interaction with additional substrate molecules. The different mutations could contribute additively or synergistically to substrate inhibition.
The mutants of the two UGTs did not exhibit strong changes in kinetic data for the monolignol sinapyl aldehyde, which showed little substrate inhibition of the two wild-type enzymes (Figures 5C and 5D). However, results obtained with the I86V_F87I mutant of NbUGT72AY1 and the chimeric mutant of StUGT72AY2 using sinapyl aldehyde as a substrate confirmed the importance of the mutant amino acids for catalytic reaction and allosteric control.
Subfunctionalization of NbUGT72AY1
Evolutionary innovations arise from gene duplications whereby the copies undergo different fates during evolution. First, one copy can retain the original function so that the other copy is freed from purifying selection and can be altered by mutations that create a new function for the gene (neofunctionalization). Second, if the original gene had multiple activities, duplication can result in the different functions being shared, and sometimes optimized, among the different copies (subfunctionalization). Third, the two copies may retain the same function, resulting in redundancy and/or increased activity of the gene (gene dosage) (Voordeckers and Verstrepen, 2015).
NbUGT72AY1 likely arose from a promiscuous monolignol UGT and probably optimized some of its additional activities (subfunctionalization) (Glasner et al., 2020), as this enzyme can also effectively glucosylate coumarin derivatives, although it exhibits strong substrate inhibition for these substrates (Figure 1). Both the double mutant V83I_I84F and the single mutant Y389H of StUGT72AY2 (monolignol UGT) had significantly increased scopoletin glycosylation activity, but both modifications also led to substrate inhibition (Figure 5B). By contrast, monolignol UGT activity of StUGT72AY2 was almost unaffected (Figure 5D). The NbUGT72AY1/StUGT72AY2 example shows that minor changes in amino acid sequences can increase the catalytic activity of a substrate without compromising existing functions. Subfunctionalization in the case of NbUGT72AY1 results in a multifunctional enzyme with altered substrate preference, activity, and kinetics from a promiscuous UGT that can be shared by lignin and scopoletin metabolism. This result supports the hypothesis that the enzymes of natural product biosynthesis are trapped in the generalist state (Noda-Garcia and Tawfik, 2020).
One hundred NbUGT72AY1-like protein sequences were extracted from Solanaceae databases using BLAST. After manual editing based on sequence lengths, gaps, and insertions, 62 sequences were aligned, and a phylogenetic tree was generated with Ipomoea sequences as outgroups (Supplemental Figures 19 and 20; Figure 7). The sequences grouped into four classes: the Ipomoea outgroup and three other classes that sub-clustered into Nicotiana, Solanum, Datura, and Capsicum sub-groups. NbUGT72AY1 (class 2) and the most similar UGTs from other Nicotiana species, including a sequence from Datura stramonium (class 2), have Phe exclusively at position 87 (position 91 in the consensus sequence), the variant that has the highest scopoletin UGT activity. The related UGTs from Solanum (class 2) and Ipomoea species (class 1) have Phe, Val, Ile, and Leu at this position, whereas proteins from classes 3 and 4, including other Nicotiana species, do not show Phe at position 87 (91 in the consensus sequence). Phe87 thus appears to have been fixed in NbUGT72AY1 and its closest relatives by selection pressure during evolution and arose from a precursor that probably contained a different amino acid at this position, as indicated by the variability of the amino acid in related sequences. We assume that after duplication of a promiscuous precursor gene, one gene copy could mutate owing to lack of selection pressure, resulting in different enzyme variants with altered substrate preference, activity, and kinetics (substrate inhibition). Because of the need to effectively glucosylate scopoletin, the Phe variant could then have been positively selected in Nicotiana species. The large number of NbUGT72AY1-like sequences in Nicotiana species corroborates this hypothesis (Figure 7). Similarly, the His390 mutation may have been co-fixed (position 406 in the consensus sequence) with Phe87 (Figure 7). Moreover, the promiscuity of secondary metabolism UGTs argues against the mechanism of neofunctionalization. It is therefore very likely that the precursor exhibited substrate tolerance, whereas the diversity of similar but not functionally identical UGTs contradicts the gene dosage mechanism. Co-expression analysis of the NbUGT72AY1 ortholog in an N. attenuata transcriptome database revealed a feruloyl ortho-hydroxylase-1-like gene as the most similarly expressed (Supplemental Figure 21). The encoded enzyme catalyzes the first reaction of the scopoletin biosynthetic pathway (Kai et al., 2008) and confirms the involvement of NbUGT72AY1 in the formation of scopoletin glucoside.
Tobacco plants produce the antifungal phytoalexin scopoletin in response to pathogen attack (Sun et al., 2014), and glycosylation appears to confer a selective advantage on N. benthamiana, as it probably protects the plant from the phytotoxin (Graña et al., 2017). Overexpression of a different scopoletin UGT in N. tabacum (TOGT; UGT73 homolog) resulted in early lesion formation during the hypersensitive response to tobacco mosaic virus but had no effect on virus resistance (Gachon et al., 2004). In a similar study, transgenic tobacco plants overexpressing TOGT showed similar formation of necrotic leaf lesions after inoculation with potato virus Y but significantly decreased levels of virus coat protein compared with control plants (Matros and Mock, 2004). These controversial results may have been obtained because tobacco plants express multiple UGTs that glucosylate hydroxycoumarins (Sun et al., 2019). Control mechanisms of the redundant UGT activities could have had different effects in the respective experiments.
Possible biological function of substrate inhibition
Enzymes of many classes are inhibited by their own substrates (Wu, 2011). Substrate inhibition has often been regarded as a biochemical curiosity and an experimental annoyance; this has changed, and substrate inhibition is now believed to have an important biological function (Reed et al., 2010). Scopoletin exhibits strong antifungal activity against Alternaria alternata, a necrotrophic fungus that causes severe yield losses in Nicotiana species in vitro and in vivo (Sun et al., 2014). The phenolic compound accumulates in large quantities during the hypersensitive response to plant pathogens, together with its glucoside (Gachon et al., 2004), but it is phytotoxic (Graña et al., 2017). Therefore, it is conceivable that at low scopoletin concentrations, neighboring plant cells protect themselves against the phytoalexin by glucosylation, whereas at higher pathogen infestations and scopoletin concentrations, it is more advantageous for the plants that the cells die and thus form a barrier against the invader. The role of the tobacco glucosyltransferase TOGT that catalyzes the glucosylation of scopoletin has been studied by both loss- and gain-of-function approaches (Chong et al., 2002; Gachon et al., 2004; Matros and Mock, 2004). Although the results were not consistent, it appears that TOGT activity represents an important step in the containment of viral pathogens. Similarly, leaf necrosis was recently shown to result from downregulation of the poplar glycosyltransferase UGT72A2 (Behr et al., 2022).
Although scopoletin levels ranging from 0.07 ng/g fresh weight (FW) to 139 μg/g FW have been quantified in several plant species (Gnonlonfin et al., 2012), healthy, uninfected Nicotiana leaves contain only 7.3 ng/g FW of hydroxycoumarin (Großkinsky et al., 2013). However, in tobacco plants, tissues exhibiting localized acquired resistance (LAR) induced by tobacco mosaic virus or an elicitor accumulate 20–40 μg/g FW scopoletin (Costet et al., 2002; Ménard et al., 2004). From this, conservative estimation yields a concentration of 30 nM scopoletin in uninfected tobacco leaves and 100–200 μM in LAR leaves, suggesting that substrate inhibition of NbUGT72AY1 is relevant in the natural context.
Although NbUGT72AY1 and StUGT72AY2 glucosylate monolignols with similar effectiveness, they show very different activities and kinetics when converting scopoletin and other low molecular weight phenols (Figure 1). NbUGT72AY1 shows significantly higher turnover rates at very low hydroxycoumarin concentrations but behaves similarly to StUGT72AY2 at high scopoletin concentrations. The monolignol StUGT72AY2 exhibits little side activity for scopoletin, whereas NbUGT72AY1 has been additionally adapted to hydroxycoumarin by mutations. The effective detoxification of low levels of scopoletin appears to confer a selective advantage to tobacco plants in this regard because scopoletin not only possesses antifungal, antiviral, and cytotoxic properties but also behaves in an auxin-like manner (Graña et al., 2017). Finally, with the evolutionary acquisition of the allosteric property, the ability to regulate enzyme activity is also acquired (LiCata and Allewell, 1997; Cornish-Bowden, 2014).
Materials and methods
Cloning of UGT72AY1 and UGT72AY2 and production of the mutant proteins
Cloning of NbUGT72AY1 from Nicotiana benthamiana (accession MT945401) and production of the NbUGT72AY1 protein was performed according to Sun et al. (2019). Genewiz, Leipzig, Germany (www.genewiz.com) synthesized StUGT72AY2 (PGSC0003DMG401004500). The gene was ligated via BglII at the 5′ end and the XhoI site at the 3′ end into the pGEX-4T-1 vector. NbUGT72AY1-I86V_F87I, -H390Y, -N27D, -R91A, -R91M, -R91F, -I86V, and -F87I and StUGT72AY2-V83I_I84F, -N27D, and -Y389H were generated by site-directed mutagenesis following the QuickChange protocol (Agilent Technology, Santa Clara, CA). The temperature program was 3 min at 94°C, one cycle; 30 s at 94°C, 30 s at 65°C, 8 min at 72°C, 30 cycles; 10 min at 72°C, one cycle; and a final temperature of 4°C, using appropriate primers (Supplemental Table 3). After Dpn I digestion of templates, the PCR products were transformed into E. coli NEB 10 beta, followed by colony PCR and sequence confirmation. Cloning of NbUGT72AY1-chimera (Tobacco-Potato-Tobacco) and StUGT72AY2-chimera (Potato-Tobacco-Potato) involved six steps. First, the target amino acids were divided into three parts using the following PCR program and primers shown in Supplemental Table 1: 3 min at 94°C, one cycle; 30 s at 94°C, 40 s at 60°C, 1.5 min at 72°C, 40 cycles; 10 min at 72°C, one cycle; and a final temperature of 4°C. Then, the first part and the final part were separately joined to the middle part using the same program to obtain two longer fragments. Finally, the two longer fragments were used as templates to generate the target fragments. After gel extraction and Vector PGEX-4T1 DNA digestion, ligation was performed according to (Sun et al., 2020).
Protein production
Protein expression was performed using E. coli BL21(DE3) pLysS cells transformed with pGEX-4T-1 UGT72AY1, pGEX-4T-1 UGT72AY2, or their corresponding mutants. After pre-culturing overnight at 37°C and 150 rpm in Luria-Bertani medium containing 100 μg/mL ampicillin and 34 μg/mL chloramphenicol, 10 mL of the pre-culture was added to 1 L of the main culture containing the corresponding antibiotics and incubated at 37°C and 120 rpm until OD600 reached 1 in a chicane flask. For UGT72AY2 and its mutants, the cells were cultured in a 5-L fermenter at 600 rpm and pH 7.0. Gene expression was induced with 1 mM isopropyl-β-D-thiogalactopyranoside, and cultures were incubated overnight at 18°C and 150 rpm. Cells were harvested via centrifugation and stored at −80°C. Recombinant fusion proteins with an N-terminal GST tag were purified with Novagen GST Bind Resin following the manufacturer’s instructions. After resuspension, the cells were disrupted by sonication. After centrifugation, the crude protein extract was incubated overnight at 4°C with the resin to bind the GST fusion protein and eluted with GST elution buffer containing reduced glutathione. The quality of the purified proteins was verified by SDS–PAGE (Supplemental Figure 22), and the protein concentration was determined with Roti-Nanoquant (Carl Roth, Karlsruhe, Germany) in 96-well microtiter plates according to the manufacturer’s instructions. Absorption was measured at 450 nm and 590 nm using a CLARIOstar plate reader (BMG Labtech, Germany).
UDP-Glo glycosyltransferase assay
The enzymatic reaction was performed according to (Sun et al., 2019) with minor modifications and quantified by the UDP-Glo Glycosyltransferase Assay (Promega, Mannheim, Germany). Assays with NbUGT72AY1 and its mutants were performed at 40°C for 10 min in 50 mM Tris-HCl (pH 7.5) containing 100 μM UDP-glucose, substrate (dissolved in DMSO), and 0.5 μg purified protein, made up to 100 μL with water. The assays with StUGT72AY2 and its mutants were performed at 30°C for 20 min in 50 mM Tris-HCl (pH 7.0) containing 100 μM UDP-glucose, substrate (dissolved in DMSO), and 2 μg purified protein, made up to 100 μL with water. Controls did not contain the enzyme. The reaction was stopped by addition of 12.5 μL 0.6 M HCl and further neutralization with 1 M Trizma base. Five microliters of the UGT reaction was pipetted into a 384-well plate. The luminescence reaction was started by adding 5 μL UDP-Glo detection reagent and incubated for 30 min in the dark. The luminescence signal was detected with a CLARIOstar plate reader (McGraphery and Schwab, 2020). The calculation of kinetic data was performed with KaleidaGraph (https://www.synergy.com/; v4.5.4). The pH optima of NbUGT72AY1 and StUGT72AY2 were determined using different protein amounts from 0.5 μg to 4 μg and different buffers for various pH ranges: 50 mM sodium phosphate (pH 6–8) and 50 mM Tris-HCl (pH 6–8.5) in 0.5-unit intervals. The optimal temperature was evaluated from 15 to 45°C. The optimal time was measured from 5 min to 30 min.
Enzyme kinetics analysis
Normal kinetics data were fitted to the Michaelis–Menten equation (Equation 1).
| (Equation 1) |
In this equation, [S] is the concentration of the varied substrate, Vmax represents the maximal reaction rate, and Km is the substrate concentration at which the reaction rate is half of Vmax. An enzyme is under allosteric control if the binding of a molecule at one binding site alters the affinity of the enzyme for its substrate and thus regulates the enzyme activity. In this case, the Hill equation applies (Equation 2).
| (Equation 2) |
Here, n is the Hill coefficient indicating the degree of cooperativity. Positive cooperativity (n > 1) occurs when an enzyme has more than one site to which a substrate can bind, and the binding of one molecule increases the rate of binding of other substrates. No or negative cooperativity is observed if n = 1 or n < 1, respectively. A partial uncompetitive inhibition model (Equation 3) was used to analyze atypical Michaelis–Menten substrate inhibition data (Wu, 2011).
| (Equation 3) |
Here, a two-site model is assumed to explain the substrate inhibition phenomenon (Scheme 1; n = x = 1). The parameter Vi is the reaction velocity in the presence of inhibition, Ki is the inhibition constant, which is the inhibitor concentration required to decrease the maximal rate of the reaction to half of the uninhibited value. The equation presumes the sequential binding of substrate molecules, i.e., the inhibitory site cannot be occupied until the reaction site is filled. Combining the cooperativity-describing Hill equation and the partial uncompetitive inhibition model yields Equation 4, which best described the measured data (LiCata and Allewell, 1997; Kapelyukh et al., 2008; Peng et al., 2016).
| (Equation 4) |
The superscript n is a Hill coefficient, and x is another Hill coefficient that allows for the possibility that binding of substrate in the inhibitory mode may also be cooperative (Peng et al., 2016). To obtain convergence for Equation 4, the value of x was fixed to an integral number that was determined empirically to give the best fit (lowest variance). The general reaction scheme for Equation 4 looks as follows (LiCata and Allewell, 1997).
The maximal reaction rates Vmax and Vi correspond to the catalytic constants kcat and kcat(i), respectively. The kinetic parameters were determined under optimum conditions and were calculated with KaleidaGraph v4.5.4. The data were derived from at least three repeats. Statistical analysis was performed using Student’s t test (p < 0.05).
LC-MS analysis
For the initial substrate screening, the UGT reaction was performed in a final volume of 100 μL 100 mM Tris-HCl buffer (pH 7.5) containing 5 μg of purified recombinant protein, 1 mM UDP-Glc, and 600 μM substrate dissolved in DMSO. The reaction was incubated at 30°C with constant shaking at 400 rpm overnight. The enzymatic reaction was stopped by heat inactivation for 10 min at 95°C. After centrifugation, the supernatant was analyzed via LC-MS analysis performed according to (Huang et al., 2018). Products were identified using authentic reference materials (Supplemental Table 4 and Supplemental Figure 2).
HDX-MS
Ligands were used as 50-mM concentrated stock solutions dissolved in DMSO (scopoletin) or double-distilled water (UDP). Prior to HDX-MS, 194 μL of purified NbUGT72AY1 was mixed with 2 μL of ligand stock solution or solvent to reach final concentrations of 35 μM (NbUGT72AY1) and 500 μM (ligands), then incubated for 5 min at ambient temperature. Preparation of exchange reactions for HDX-MS was aided by a two-arm robotic autosampler (LEAP Technologies). The protein solution (7.5 μL) was mixed with 67.5 μL of D2O-containing buffer (20 mM Tris-Cl [pH 7.5], 2% [v/v] DMSO), which also contained 500 μM of ligands to prevent their dilution during HDX, and incubated for 10/30/95/1000/10 000 s at 25°C. The completed HDX reactions were added to 55 μL quench solution (400 mM KH2PO4/H3PO4, 2 M guanidine-HCl, pH 2.2) kept at 1°C, and 95 μL of the resulting mixture was injected into an ACQUITY UPLC M-class system with HDX technology (Waters) (Wales et al., 2008). NbUGT72AY1 was digested online with a column (2 mm × 2 cm) packed with immobilized porcine pepsin and separated by reversed-phase HPLC followed by mass spectrometric analysis as described previously (Osorio-Valeriano et al., 2019; Skotnicka et al., 2020). Peptides were identified from the undeuterated samples with ProteinLynx Global Server (Waters) software as described previously (Osorio-Valeriano et al., 2019). For quantification of deuterium incorporation with DynamX 3.0 (Waters), peptides had to fulfill the following criteria: minimum intensity of 5000 counts; maximum length of 30 amino acids; minimum number of products of three; maximum mass error of 25 ppm; and retention time tolerance of 0.5 min. After automated data processing with DynamX 3.0, the mass spectra were manually inspected and, if necessary, peptides were omitted, e.g., in the case of low signal-to-noise ratio or presence of overlapping peptides. Raw HDX-MS data are supplied in the supplemental dataset (Masson et al., 2019).
In silico homology modeling and ligand docking
Three-dimensional structure homology models of NbUGT71AY1 (GenBank accession MT945401; Niben101Scf06112g01008.1; https://solgenomics.net/tools/blast) and StUGT72AY2 (GenBank accession XP_015164078.1, UniProt: M0ZZL3) were produced using the IntFOLD (McGuffin et al., 2015) and Swiss-Model (Waterhouse et al., 2018) servers. Four proteins with known crystal structures were found that show sequence identities between 39.4% and 47.0% with NbUGT72AY1, and StUGT72AY2 shares 80.3% identical amino acids with NbUGT72AY1. Two each were crystalized in their open (PDB: 6SU6/6SU7/5NLM and PDP: 6JEL/6JEM/6JEN) and closed (PDB: 6JTD and PDB: 2VG8/2VCE/2VCH) conformation. Therefore, PDB: 6JTD and PDB: 6SU6 were used as templates to guide the modeling of both NbUGT72AY1 and StUGT72AY2, each in the closed and open form. Models with the highest confidence scores were uploaded into UCSF Chimera 1.15 (https://www.cgl.ucsf.edu/chimera) for visualization and comparative analysis (Pettersen et al., 2004; Goddard et al., 2018). Ligand docking was performed with the AutoDock Vina tool implemented in UCSF Chimera 1.15 (Trott and Olson, 2010). Binding energies (ΔG) calculated by UCSF Chimera 1.15 were used to calculate equilibrium constants KD by Kd = e−ΔG/R/T and KD = Kd/c with R = 1.986 cal/mol/K, T = 298.15 K, and the standard reference concentration c = 1 mol/L. Videos (mp4) were recorded with UCSF Chimera 1.15.
Funding
DFG SCHW 634/34-1.
Author contributions
J.L., G.S., E.K., T.H., and W.G.S. designed experiments and interpreted data; J.L., G.S., E.K., T.H., T.D.H., C.S., and W.G.S. wrote the manuscript; J.L., G.S., and E.K. performed biochemical analyses and analyzed the proteins; J.L. and E.K. generated mutant proteins; J.L., G.S., E.K., and T.H. performed LC-MS analyses. W.S. performed HDX-MS analyses; W.G.S. performed in silico analyses; C.S., T.D.H., and W.S. contributed to materials, instruments, data analysis, and the manuscript. All authors approved the final manuscript.
Acknowledgments
We thank DFG for financial support. No conflict of interest declared.
Published: December 24, 2022
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Supplemental information is available at Plant Communications Online.
Data availability
All relevant data generated in this study are provided in the main text and the supplemental information.
Supplemental information
References
- Albesa-Jové D., Guerin M.E. The conformational plasticity of glycosyltransferases. Curr. Opin. Struct. Biol. 2016;40:23–32. doi: 10.1016/j.sbi.2016.07.007. [DOI] [PubMed] [Google Scholar]
- Albesa-Jové D., Romero-García J., Sancho-Vaello E., Contreras F.-X., Rodrigo-Unzueta A., Comino N., Carreras-González A., Arrasate P., Urresti S., Biarnés X., et al. Structural snapshots and loop dynamics along the catalytic cycle of glycosyltransferase GpgS. Structure. 2017;25:1034–1044.e3. doi: 10.1016/j.str.2017.05.009. [DOI] [PubMed] [Google Scholar]
- Baldacci-Cresp F., Le Roy J., Huss B., Lion C., Créach A., Spriet C., Duponchel L., Biot C., Baucher M., Hawkins S., et al. UDP-glycosyltransferase 72E3 plays a role in lignification of secondary cell walls in Arabidopsis. Int. J. Mol. Sci. 2020;21:6094. doi: 10.3390/ijms21176094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett A.C., Tsvetanov S., Gamage N., Martin J.L., Duggleby R.G., McManus M.E. Active site mutations and substrate inhibition in human sulfotransferase 1A1 and 1A3. J. Biol. Chem. 2004;279:18799–18805. doi: 10.1074/jbc.M312253200. [DOI] [PubMed] [Google Scholar]
- Behr M., Speeckaert N., Kurze E., Morel O., Prévost M., Mol A., Mahamadou Adamou N., Baragé M., Renaut J., Schwab W., et al. Leaf necrosis resulting from downregulation of poplar glycosyltransferase UGT72A2. Tree Physiol. 2022;42:1084–1099. doi: 10.1093/treephys/tpab161. [DOI] [PubMed] [Google Scholar]
- Bolam D.N., Roberts S., Proctor M.R., Turkenburg J.P., Dodson E.J., Martinez-Fleites C., Yang M., Davis B.G., Davies G.J., Gilbert H.J. The crystal structure of two macrolide glycosyltransferases provides a blueprint for host cell antibiotic immunity. Proc. Natl. Acad. Sci. USA. 2007;104:5336–5341. doi: 10.1073/pnas.0607897104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowles D., Lim E.-K., Poppenberger B., Vaistij F.E. Glycosyltransferases of lipophilic small molecules. Annu. Rev. Plant Biol. 2006;57:567–597. doi: 10.1146/annurev.arplant.57.032905.105429. [DOI] [PubMed] [Google Scholar]
- Brazier-Hicks M., Offen W.A., Gershater M.C., Revett T.J., Lim E.-K., Bowles D.J., Davies G.J., Edwards R. Characterization and engineering of the bifunctional N- and O-glucosyltransferase involved in xenobiotic metabolism in plants. Proc. Natl. Acad. Sci. USA. 2007;104:20238–20243. doi: 10.1073/pnas.0706421104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton C., Snajdrová L., Jeanneau C., Koca J., Imberty A. Structures and mechanisms of glycosyltransferases. Glycobiology. 2006;16:29R–37R. doi: 10.1093/glycob/cwj016. [DOI] [PubMed] [Google Scholar]
- Buenavista M.T., Roche D.B., McGuffin L.J. Improvement of 3D protein models using multiple templates guided by single-template model quality assessment. Bioinformatics. 2012;28:1851–1857. doi: 10.1093/bioinformatics/bts292. [DOI] [PubMed] [Google Scholar]
- Chang A., Singh S., Phillips G.N., Thorson J.S. Glycosyltransferase structural biology and its role in the design of catalysts for glycosylation. Curr. Opin. Biotechnol. 2011;22:800–808. doi: 10.1016/j.copbio.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Joo J.C., Brown G., Stolnikova E., Halavaty A.S., Savchenko A., Anderson W.F., Yakunin A.F. Structure-based mutational studies of substrate inhibition of betaine aldehyde dehydrogenase BetB from Staphylococcus aureus. Appl. Environ. Microbiol. 2014;80:3992–4002. doi: 10.1128/AEM.00215-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong J., Baltz R., Schmitt C., Beffa R., Fritig B., Saindrenan P. Downregulation of a pathogen-responsive tobacco UDP-Glc:phenylpropanoid glucosyltransferase reduces scopoletin glucoside accumulation, enhances oxidative stress, and weakens virus resistance. Plant Cell. 2002;14:1093–1107. doi: 10.1105/tpc.010436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish-Bowden A. Understanding allosteric and cooperative interactions in enzymes. FEBS J. 2014;281:621–632. doi: 10.1111/febs.12469. [DOI] [PubMed] [Google Scholar]
- Costet L., Fritig B., Kauffmann S. Scopoletin expression in elicitor-treated and tobacco mosaic virus-infected tobacco plants. Physiol. Plant. 2002;115:228–235. doi: 10.1034/j.1399-3054.2002.1150208.x. [DOI] [PubMed] [Google Scholar]
- Dong D., Wu B. What are the real causes of substrate inhibition in the glucuronidation reaction? Single Cell Biol. 2012;1:e105. [Google Scholar]
- Drula E., Garron M.-L., Dogan S., Lombard V., Henrissat B., Terrapon N. The carbohydrate-active enzyme database: functions and literature. Nucleic Acids Res. 2022;50:D571–D577. doi: 10.1093/nar/gkab1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flocco M.M., Mowbray S.L. Planar stacking interactions of arginine and aromatic side-chains in proteins. J. Mol. Biol. 1994;235:709–717. doi: 10.1006/jmbi.1994.1022. [DOI] [PubMed] [Google Scholar]
- Fujiwara R., Yokoi T., Nakajima M. Structure and protein-protein interactions of human UDP-glucuronosyltransferases. Front. Pharmacol. 2016;7:388. doi: 10.3389/fphar.2016.00388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gachon C., Baltz R., Saindrenan P. Over-expression of a scopoletin glucosyltransferase in Nicotiana tabacum leads to precocious lesion formation during the hypersensitive response to tobacco mosaic virus but does not affect virus resistance. Plant Mol. Biol. 2004;54:137–146. doi: 10.1023/B:PLAN.0000028775.58537.fe. [DOI] [PubMed] [Google Scholar]
- Gallivan J.P., Dougherty D.A. Cation-pi interactions in structural biology. Proc. Natl. Acad. Sci. USA. 1999;96:9459–9464. doi: 10.1073/pnas.96.17.9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasner M.E., Truong D.P., Morse B.C. How enzyme promiscuity and horizontal gene transfer contribute to metabolic innovation. FEBS J. 2020;287:1323–1342. doi: 10.1111/febs.15185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnonlonfin G.J.B., Sanni A., Brimer L. Review scopoletin – a coumarin phytoalexin with medicinal properties. Crit. Rev. Plant Sci. 2012;31:47–56. [Google Scholar]
- Goddard T.D., Huang C.C., Meng E.C., Pettersen E.F., Couch G.S., Morris J.H., Ferrin T.E. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 2018;27:14–25. doi: 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graña E., Costas-Gil A., Longueira S., Celeiro M., Teijeira M., Reigosa M.J., Sánchez-Moreiras A.M. Auxin-like effects of the natural coumarin scopoletin on Arabidopsis cell structure and morphology. J. Plant Physiol. 2017;218:45–55. doi: 10.1016/j.jplph.2017.07.007. [DOI] [PubMed] [Google Scholar]
- Großkinsky D.K., Edelsbrunner K., Pfeifhofer H., van der Graaff E., Roitsch T. Cis- and trans-zeatin differentially modulate plant immunity. Plant Signal. Behav. 2013;8:e24798. doi: 10.4161/psb.24798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrus D., Khoder-Agha F., Peltoniemi M., Hassinen A., Ruddock L., Kellokumpu S., Glumoff T. The dimeric structure of wild-type human glycosyltransferase B4GalT1. PLoS One. 2018;13:e0205571. doi: 10.1371/journal.pone.0205571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto K., Madej T., Bryant S.H., Panchenko A.R. Functional states of homooligomers: insights from the evolution of glycosyltransferases. J. Mol. Biol. 2010;399:196–206. doi: 10.1016/j.jmb.2010.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J.-B., Zhao P., Hu Z.-M., Liu S., Kuang Y., Zhang M., Li B., Yun C.-H., Qiao X., Ye M. Molecular and structural characterization of a promiscuous C-glycosyltransferase from Trollius chinensis. Angew. Chem. Int. Ed. Engl. 2019;58:11513–11520. doi: 10.1002/anie.201905505. [DOI] [PubMed] [Google Scholar]
- Hsu T.M., Welner D.H., Russ Z.N., Cervantes B., Prathuri R.L., Adams P.D., Dueber J.E. Employing a biochemical protecting group for a sustainable indigo dyeing strategy. Nat. Chem. Biol. 2018;14:256–261. doi: 10.1038/nchembio.2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F.-C., Giri A., Daniilidis M., Sun G., Härtl K., Hoffmann T., Schwab W. Structural and functional analysis of UGT92G6 suggests an evolutionary link between mono- and disaccharide glycoside-forming transferases. Plant Cell Physiol. 2018;59:857–870. doi: 10.1093/pcp/pcy028. [DOI] [PubMed] [Google Scholar]
- Kai K., Mizutani M., Kawamura N., Yamamoto R., Tamai M., Yamaguchi H., Sakata K., Shimizu B.i. Scopoletin is biosynthesized via ortho-hydroxylation of feruloyl CoA by a 2-oxoglutarate-dependent dioxygenase in Arabidopsis thaliana. Plant J. 2008;55:989–999. doi: 10.1111/j.1365-313X.2008.03568.x. [DOI] [PubMed] [Google Scholar]
- Kapelyukh Y., Paine M.J.I., Maréchal J.D., Sutcliffe M.J., Wolf C.R., Roberts G.C.K. Multiple substrate binding by cytochrome P450 3A4: estimation of the number of bound substrate molecules. Drug Metab. Dispos. 2008;36:2136–2144. doi: 10.1124/dmd.108.021733. [DOI] [PubMed] [Google Scholar]
- Konermann L., Pan J., Liu Y.-H. Hydrogen exchange mass spectrometry for studying protein structure and dynamics. Chem. Soc. Rev. 2011;40:1224–1234. doi: 10.1039/c0cs00113a. [DOI] [PubMed] [Google Scholar]
- Kurze E., Wüst M., Liao J., McGraphery K., Hoffmann T., Song C., Schwab W. Structure-function relationship of terpenoid glycosyltransferases from plants. Nat. Prod. Rep. 2021;39:389–409. doi: 10.1039/d1np00038a. [DOI] [PubMed] [Google Scholar]
- Lanot A., Hodge D., Jackson R.G., George G.L., Elias L., Lim E.-K., Vaistij F.E., Bowles D.J. The glucosyltransferase UGT72E2 is responsible for monolignol 4-O-glucoside production in Arabidopsis thaliana. Plant J. 2006;48:286–295. doi: 10.1111/j.1365-313X.2006.02872.x. [DOI] [PubMed] [Google Scholar]
- Lanot A., Hodge D., Lim E.-K., Vaistij F.E., Bowles D.J. Redirection of flux through the phenylpropanoid pathway by increased glucosylation of soluble intermediates. Planta. 2008;228:609–616. doi: 10.1007/s00425-008-0763-8. [DOI] [PubMed] [Google Scholar]
- Le Roy J., Huss B., Creach A., Hawkins S., Neutelings G. Glycosylation is a major regulator of phenylpropanoid availability and biological activity in plants. Front. Plant Sci. 2016;7:735. doi: 10.3389/fpls.2016.00735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang D.-M., Liu J.-H., Wu H., Wang B.-B., Zhu H.-J., Qiao J.-J. Glycosyltransferases: mechanisms and applications in natural product development. Chem. Soc. Rev. 2015;44:8350–8374. doi: 10.1039/c5cs00600g. [DOI] [PubMed] [Google Scholar]
- LiCata V.J., Allewell N.M. Is substrate inhibition a consequence of allostery in aspartate transcarbamylase? Biophys. Chem. 1997;64:225–234. doi: 10.1016/s0301-4622(96)02204-1. [DOI] [PubMed] [Google Scholar]
- Lim E.-K., Jackson R.G., Bowles D.J. Identification and characterisation of Arabidopsis glycosyltransferases capable of glucosylating coniferyl aldehyde and sinapyl aldehyde. FEBS Lett. 2005;579:2802–2806. doi: 10.1016/j.febslet.2005.04.016. [DOI] [PubMed] [Google Scholar]
- Liu Y.-Q., Yuan L.-M., Gao Z.-Z., Xiao Y.-S., Sun H.-Y., Yu L.-S., Zeng S. Dimerization of human uridine diphosphate glucuronosyltransferase allozymes 1A1 and 1A9 alters their quercetin glucuronidation activities. Sci. Rep. 2016;6:23763. doi: 10.1038/srep23763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luukkanen L., Taskinen J., Kurkela M., Kostiainen R., Hirvonen J., Finel M. Kinetic characterization of the 1A subfamily of recombinant human UDP-glucuronosyltransferases. Drug Metab. Dispos. 2005;33:1017–1026. doi: 10.1124/dmd.105.004093. [DOI] [PubMed] [Google Scholar]
- Maharjan R., Fukuda Y., Shimomura N., Nakayama T., Okimoto Y., Kawakami K., Nakayama T., Hamada H., Inoue T., Ozaki S.-I. An ambidextrous polyphenol glycosyltransferase PaGT2 from Phytolacca americana. Biochemistry. 2020;59:2551–2561. doi: 10.1021/acs.biochem.0c00224. [DOI] [PubMed] [Google Scholar]
- Masson G.R., Burke J.E., Ahn N.G., Anand G.S., Borchers C., Brier S., Bou-Assaf G.M., Engen J.R., Englander S.W., Faber J., et al. Recommendations for performing, interpreting and reporting hydrogen deuterium exchange mass spectrometry (HDX-MS) experiments. Nat. Methods. 2019;16:595–602. doi: 10.1038/s41592-019-0459-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matros A., Mock H.-P. Ectopic expression of a UDP-glucose:phenylpropanoid glucosyltransferase leads to increased resistance of transgenic tobacco plants against infection with Potato Virus Y. Plant Cell Physiol. 2004;45:1185–1193. doi: 10.1093/pcp/pch140. [DOI] [PubMed] [Google Scholar]
- McGraphery K., Schwab W. Comparative analysis of high-throughput assays of family-1 plant glycosyltransferases. Int. J. Mol. Sci. 2020;21:2208. doi: 10.3390/ijms21062208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuffin L.J., Adiyaman R., Maghrabi A.H.A., Shuid A.N., Brackenridge D.A., Nealon J.O., Philomina L.S. IntFOLD: an integrated web resource for high performance protein structure and function prediction. Nucleic Acids Res. 2019;47:W408–W413. doi: 10.1093/nar/gkz322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuffin L.J., Atkins J.D., Salehe B.R., Shuid A.N., Roche D.B. IntFOLD: an integrated server for modelling protein structures and functions from amino acid sequences. Nucleic Acids Res. 2015;43:W169–W173. doi: 10.1093/nar/gkv236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meech R., Hu D.G., McKinnon R.A., Mubarokah S.N., Haines A.Z., Nair P.C., Rowland A., Mackenzie P.I. The UDP-Glycosyltransferase (UGT) superfamily: new members, new functions, and novel paradigms. Physiol. Rev. 2019;99:1153–1222. doi: 10.1152/physrev.00058.2017. [DOI] [PubMed] [Google Scholar]
- Ménard R., Alban S., de Ruffray P., Jamois F., Franz G., Fritig B., Yvin J.-C., Kauffmann S. Beta-1, 3 glucan sulfate, but not beta-1, 3 glucan, induces the salicylic acid signaling pathway in tobacco and Arabidopsis. Plant Cell. 2004;16:3020–3032. doi: 10.1105/tpc.104.024968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G.P., Lichti C.F., Zielinska A.K., Mazur A., Bratton S.M., Gallus-Zawada A., Finel M., Moran J.H., Radominska-Pandya A. Identification of hydroxywarfarin binding site in human UDP glucuronosyltransferase 1a10: phenylalanine90 is crucial for the glucuronidation of 6- and 7-hydroxywarfarin but not 8-hydroxywarfarin. Drug Metab. Dispos. 2008;36:2211–2218. doi: 10.1124/dmd.108.022863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda-Garcia L., Tawfik D.S. Enzyme evolution in natural products biosynthesis: target- or diversity-oriented? Curr. Opin. Chem. Biol. 2020;59:147–154. doi: 10.1016/j.cbpa.2020.05.011. [DOI] [PubMed] [Google Scholar]
- Offen W., Martinez-Fleites C., Yang M., Kiat-Lim E., Davis B.G., Tarling C.A., Ford C.M., Bowles D.J., Davies G.J. Structure of a flavonoid glucosyltransferase reveals the basis for plant natural product modification. EMBO J. 2006;25:1396–1405. doi: 10.1038/sj.emboj.7600970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio-Valeriano M., Altegoer F., Steinchen W., Urban S., Liu Y., Bange G., Thanbichler M. ParB-type DNA segregation proteins are CTP-dependent molecular switches. Cell. 2019;179:1512–1524.e15. doi: 10.1016/j.cell.2019.11.015. [DOI] [PubMed] [Google Scholar]
- Peng H., Yang T., Whitaker B.D., Shangguan L., Fang J. Calcium/calmodulin alleviates substrate inhibition in a strawberry UDP-glucosyltransferase involved in fruit anthocyanin biosynthesis. BMC Plant Biol. 2016;16:197. doi: 10.1186/s12870-016-0888-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Putkaradze N., Teze D., Fredslund F., Welner D.H. Natural product C-glycosyltransferases - a scarcely characterised enzymatic activity with biotechnological potential. Nat. Prod. Rep. 2021;38:432–443. doi: 10.1039/d0np00040j. [DOI] [PubMed] [Google Scholar]
- Qasba P.K., Ramakrishnan B., Boeggeman E. Substrate-induced conformational changes in glycosyltransferases. Trends Biochem. Sci. 2005;30:53–62. doi: 10.1016/j.tibs.2004.11.005. [DOI] [PubMed] [Google Scholar]
- Qasba P.K., Ramakrishnan B., Boeggeman E. Structure and function of β-1, 4-galactosyltransferase. Curr. Drug Targets. 2008;9:292–309. doi: 10.2174/138945008783954943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed M.C., Lieb A., Nijhout H.F. The biological significance of substrate inhibition: a mechanism with diverse functions. Bioessays. 2010;32:422–429. doi: 10.1002/bies.200900167. [DOI] [PubMed] [Google Scholar]
- Sheikh M.O., Halmo S.M., Patel S., Middleton D., Takeuchi H., Schafer C.M., West C.M., Haltiwanger R.S., Avci F.Y., Moremen K.W., et al. Rapid screening of sugar-nucleotide donor specificities of putative glycosyltransferases. Glycobiology. 2017;27:206–212. doi: 10.1093/glycob/cww114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner J.J., Lim W.K., Bédard S., Black B.E., Englander S.W. Protein hydrogen exchange: testing current models. Protein Sci. 2012;21:987–995. doi: 10.1002/pro.2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skotnicka D., Steinchen W., Szadkowski D., Cadby I.T., Lovering A.L., Bange G., Søgaard-Andersen L. CdbA is a DNA-binding protein and c-di-GMP receptor important for nucleoid organization and segregation in Myxococcus xanthus. Nat. Commun. 2020;11:1791. doi: 10.1038/s41467-020-15628-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A.D., Page B.D.G., Collier A.C., Coughtrie M.W.H. Homology modeling of human uridine-5'-diphosphate-glucuronosyltransferase 1A6 reveals insights into factors influencing substrate and cosubstrate binding. ACS Omega. 2020;5:6872–6887. doi: 10.1021/acsomega.0c00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speeckaert N., Adamou N.M., Hassane H.A., Baldacci-Cresp F., Mol A., Goeminne G., Boerjan W., Duez P., Hawkins S., Neutelings G., et al. Characterization of the UDP-glycosyltransferase UGT72 family in poplar and identification of genes involved in the glycosylation of monolignols. Int. J. Mol. Sci. 2020;21:5018. doi: 10.3390/ijms21145018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speeckaert N., El Jaziri M., Baucher M., Behr M. UGT72, a major glycosyltransferase family for flavonoid and monolignol homeostasis in plants. Biology. 2022;11:441. doi: 10.3390/biology11030441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner T., Koellner G. Hydrogen bonds with pi-acceptors in proteins: frequencies and role in stabilizing local 3D structures. J. Mol. Biol. 2001;305:535–557. doi: 10.1006/jmbi.2000.4301. [DOI] [PubMed] [Google Scholar]
- Sun G., Putkaradze N., Bohnacker S., Jonczyk R., Fida T., Hoffmann T., Bernhardt R., Härtl K., Schwab W. Six uridine-diphosphate glycosyltransferases catalyze the glycosylation of bioactive C13-apocarotenols. Plant Physiol. 2020;184:1744–1761. doi: 10.1104/pp.20.00953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G., Strebl M., Merz M., Blamberg R., Huang F.-C., McGraphery K., Hoffmann T., Schwab W. Glucosylation of the phytoalexin N-feruloyl tyramine modulates the levels of pathogen-responsive metabolites in Nicotiana benthamiana. Plant J. 2019;100:20–37. doi: 10.1111/tpj.14420. [DOI] [PubMed] [Google Scholar]
- Sun H., Wang L., Zhang B., Ma J., Hettenhausen C., Cao G., Sun G., Wu J., Wu J. Scopoletin is a phytoalexin against Alternaria alternata in wild tobacco dependent on jasmonate signalling. J. Exp. Bot. 2014;65:4305–4315. doi: 10.1093/jxb/eru203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teze D., Coines J., Fredslund F., Dubey K.D., Bidart G.N., Adams P.D., Dueber J.E., Svensson B., Rovira C., Welner D.H. O -/N -/S -specificity in glycosyltransferase catalysis: from mechanistic understanding to engineering. ACS Catal. 2021;11:1810–1815. [Google Scholar]
- Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voordeckers K., Verstrepen K.J. Experimental evolution of the model eukaryote Saccharomyces cerevisiae yields insight into the molecular mechanisms underlying adaptation. Curr. Opin. Microbiol. 2015;28:1–9. doi: 10.1016/j.mib.2015.06.018. [DOI] [PubMed] [Google Scholar]
- Wales T.E., Fadgen K.E., Gerhardt G.C., Engen J.R. High-speed and high-resolution UPLC separation at zero degrees Celsius. Anal. Chem. 2008;80:6815–6820. doi: 10.1021/ac8008862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.-L., Gao H.-M., Wang S., Zhang M., Chen K., Zhang Y.-Q., Wang H.-D., Han B.-Y., Xu L.-L., Song T.-Q., et al. Dissection of the general two-step di-C-glycosylation pathway for the biosynthesis of (iso)schaftosides in higher plants. Proc. Natl. Acad. Sci. USA. 2020;117:30816–30823. doi: 10.1073/pnas.2012745117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., de Beer T.A.P., Rempfer C., Bordoli L., et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss D.R., Levitt M. Can morphing methods predict intermediate structures? J. Mol. Biol. 2009;385:665–674. doi: 10.1016/j.jmb.2008.10.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A.E., Tian L. Phylogenomic analysis of UDP-dependent glycosyltransferases provides insights into the evolutionary landscape of glycosylation in plant metabolism. Plant J. 2019;100:1273–1288. doi: 10.1111/tpj.14514. [DOI] [PubMed] [Google Scholar]
- Wu B. Substrate inhibition kinetics in drug metabolism reactions. Drug Metab. Rev. 2011;43:440–456. doi: 10.3109/03602532.2011.615320. [DOI] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K., Hanada K. An evolutionary view of functional diversity in family 1 glycosyltransferases. Plant J. 2011;66:182–193. doi: 10.1111/j.1365-313X.2011.04493.x. [DOI] [PubMed] [Google Scholar]
- Zhang H., Zhu F., Yang T., Ding L., Zhou M., Li J., Haslam S.M., Dell A., Erlandsen H., Wu H. The highly conserved domain of unknown function 1792 has a distinct glycosyltransferase fold. Nat. Commun. 2014;5:4339. doi: 10.1038/ncomms5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Scopoletin and UDP-glucose occupy the active site. NbUGT72AY1 is color-coded according to the HDX-MS results for scopoletin/UDP.
Scopoletin and UDP-glucose occupy the active site. NbUGT72AY1 is color-coded according to the HDX-MS results for scopoletin/UDP.
Scopoletin and UDP-glucose occupy the active site. NbUGT72AY1 is color-coded according to the HDX-MS results for scopoletin/UDP.
Scopoletin and UDP-glucose occupy the active site. NbUGT72AY1 is color-coded according to the HDX-MS results for scopoletin/UDP.
NbUGT72AY1 is color-coded according to the HDX-MS results for scopoletin/UDP.
Scopoletin and UDP-glucose occupy the active site. NbUGT72AY1 is color-coded according to the HDX-MS results for scopoletin/UDP.
Data Availability Statement
All relevant data generated in this study are provided in the main text and the supplemental information.