Abstract
Since the start of the Anthropocene, northern seasonally frozen peatlands have been warming at a rate of 0.6 °C per decade, twice that of the Earth’s average rate, thereby triggering increased nitrogen mineralization with subsequent potentially large losses of nitrous oxide (N2O) to the atmosphere. Here we provide evidence that seasonally frozen peatlands are important N2O emission sources in the Northern Hemisphere and the thawing periods are the hot moment of annual N2O emissions. The flux during the hot moment of thawing in spring was 1.20 ± 0.82 mg N2O m−2 d−1, significantly higher than that during the other periods (freezing, −0.12 ± 0.02 mg N2O m−2 d−1; frozen, 0.04 ± 0.04 mg N2O m−2 d−1; thawed, 0.09 ± 0.01 mg N2O m−2 d−1) or observed for other ecosystems at the same latitude in previous studies. The observed emission flux is even higher than those of tropical forests, the World’s largest natural terrestrial N2O source. Furthermore, based on soil incubation with 15N and 18O isotope tracing and differential inhibitors, heterotrophic bacterial and fungal denitrification was revealed as the main source of N2O in peatland profiles (0–200 cm). Metagenomic, metatranscriptomic, and qPCR assays further revealed that seasonally frozen peatlands have high N2O emission potential, but thawing significantly stimulates expression of genes encoding N2O-producing protein complexes (hydroxylamine dehydrogenase (hao) and nitric oxide reductase (nor)), resulting in high N2O emissions during spring. This hot moment converts seasonally frozen peatlands into an important N2O emission source when it is otherwise a sink. Extrapolation of our data to all northern peatland areas reveals that the hot moment emissions could amount to approximately 0.17 Tg of N2O yr−1. However, these N2O emissions are still not routinely included in Earth system models and global IPCC assessments.
Subject terms: Biogeochemistry, Soil microbiology
Introduction
Northern peatlands contain about 9–16% and 30% of the global soil-associated total organic nitrogen (N) [1, 2] and carbon (C) [3, 4], respectively. Peatland disturbances can trigger the loss of peat and the release of greenhouse gases and thus exert a significant influence on global climate change and the atmospheric C and N budget [5]. More than half of the peatlands in the Northern Hemisphere are subjected to annual freeze-thaw cycles, including perennially, seasonally, and intermittently frozen regions [6]. These regions are characterized by their soil freezing time, i.e., two years or more, more than 15 days per year, and between one and 15 days a year [6]. At least 38% of the northern peatland area is seasonally frozen [2, 6]. Seasonally frozen peatlands are characterized by unidirectional freezing and bidirectional thawing, which are vulnerable to warming and precipitation [7]. Under the influence of global warming, the active-layer depths of permafrost have continuously increased during recent decades [8, 9]. Degraded permafrost peatlands can be transformed into seasonally frozen peatlands [10], which could have a great impact on the global GHG budget [3, 5].
Nitrous oxide (N2O) emissions from soils in the Northern Hemisphere have traditionally been considered negligible as these soils were considered to be N limited [5, 11, 12], and plants competed favorably for N with microorganisms [13–15]. However, increasing evidence shows that thawing permafrost can produce and release substantial amounts of carbon dioxide, methane (CH4), and N2O [12, 16–19]. Compared with areas that experience thawing only in the active layer of the permafrost, seasonally frozen peatlands experience a more substantial freeze-thaw cycle over the whole soil profile and have a greater redox potential gradient; [3, 20] hence, they are potentially important drivers of N2O emissions. Yet, both the contribution of this effect to global change and the biogeochemical mechanism of the N cycle in seasonally frozen peatlands have not been well explored.
Seasonally frozen peatlands have warmed at a rate of 0.6 °C per decade over the past 30 years, twice the global average surface temperature [21]. This warming has increased bioavailable N by enhancing soil organic matter decomposition and N mineralization [22, 23]. Soil thawing promotes frozen N release and influences microbial N transformation rates, which could alter soil N availability and regulate ecosystem functions [24–26]. Waterlogged conditions during the thawing process can produce oxygen-limited conditions throughout the soil profile and increase the redox conditions of the soil [27], further promoting microbial N-reduction pathways, such as the heterotrophic denitrification process that generates N2O, especially at a lower pH [28–30]. Moreover, during the thawing season, there is relatively week competition for bioavailable N between plants and microorganisms [18, 31]. These processes could cause increased N2O emissions during the thawing of seasonally frozen peatlands. Based on the studies described above [18, 24–27, 31], as well as our previous microbial-N cycle studies in wetland and permafrost ecosystems [12, 26, 32–36], we put forward the hypothesis that seasonally frozen peatlands can be an important N2O emission source and that thawing periods in spring can be a hot moment of annual N2O emissions.
Microbial denitrification and nitrification are considered the most important N2O sources in soils [37–39]. There are many microbial N2O production pathways in fungi and bacteria that are involved in denitrification and nitrification, including nitrifier nitrification, nitrifier denitrification, nitrification-coupled denitrification, and heterotrophic denitrification [40–44]. These pathways have different mechanisms and contributions to N2O production in different habitats [40–44]. Previous studies showed that nitrifier denitrification is the dominate process of N2O production of soils with low oxygen availability using 15N and 18O isotope tracing techniques [40–42]. Fungal denitrification can be a significant source of N2O across diverse ecosystems, including forest, drylands, and coastal sediments [43, 44]. Although all these processes can produce N2O, little is known about their relative contribution to N2O production in seasonally frozen peatlands.
The Antu peatland, located in the Northeast Plain of China, is one of three black soil regions in the World. As it experiences a strong degree of seasonal freezing and thawing through a frozen layer depth of 100 cm, it was selected as the study site [45–47]. In this study, we first monitored the flux of N2O emissions of a seasonally frozen peatland using a closed chamber technique over a whole year, including peatland thawing, thawed, freezing, and frozen periods. Then, we conducted a series of measurements along the Gudong River (river length, 156.6 km; drainage basin area, 4303 km2) at the regional scale with a focus on the thawing season. Next, we tested our hypothesis along a soil/peat in-situ successional transect (0–200 cm). The soil profile samples were used to investigate the processes responsible for microbial N2O production during laboratory incubation using isotopic 15N and 18O tracing, inhibitor methods, metagenomic sequencing, and metatranscriptomic sequencing.
Materials and methods
Site-scale sampling
Xinhe (sampling site E (Fig. 1C), 597.0 m above sea level; N 42°56′50″, E 128°28′49″) is located in the Yanbian Korean Autonomous Prefecture in Jilin Province, in northeastern China. It is located in one of the three biggest peatland regions in the world [45–47]. Gas (to measure N2O flux) and soil samples (to determine the mechanism of N2O production) were collected from three separate profiles in March, May, July, September, and November 2019, as well as in January 2020. The sampling depth was from 0 to 200 cm, with samples taken at 20-cm intervals (0–20, 20–40, 40–60, 60–80, 80–100, 100–120, 120–140, 140–160, 160–180, and 180–200 cm) (n = 180). Within the whole peat/soil profile, the first 0–120 cm was peat, the 120–160 cm layer was mainly clay, and the 160–200 cm layer was gravel. The freezing depth was 100 cm in winter, and peat or soil below 100 cm remained frost-free. The depth of the frozen layer varied with the seasons: 0–40 cm in January, 0–100 cm in March, 20–60 cm in May, 40–60 cm in July, and 0–20 cm in November. It should be noted that the entire peat/soil profile remained unfrozen in September.
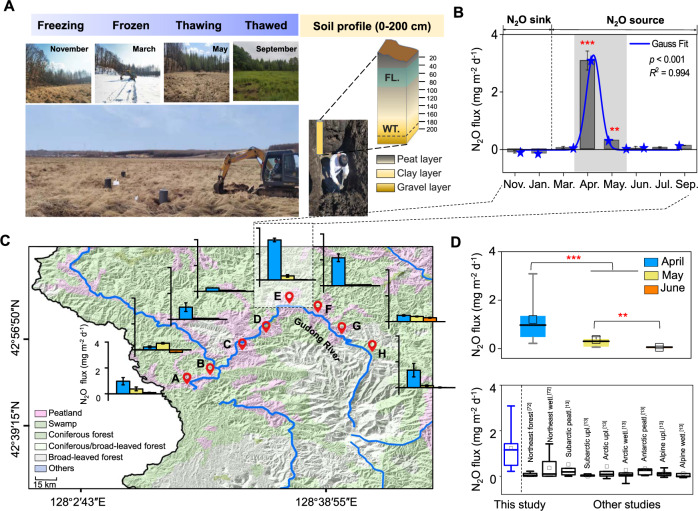
Fig. 1. Hot moment of N2O emissions in seasonally frozen peatland.
A Photos showing the sampling site in different periods and the 200 cm-soil profile of sampling site E. “FL.” and “WT.” indicate the frozen layer and water table, respectively. B Site-scale in-situ N2O fluxes in the seasonally frozen peatland. The blue curve is the Gaussian fit of the N2O fluxes. Gas samples were collected in March, May, July, September, and November 2019, as well as in January 2020. C Regional scale in-situ N2O fluxes in April, May, and June in 2020 at sites along the Gudong River. D Box plots showing the N2O fluxes of the regional-scale sites (above), and box plots showing N2O fluxes from this study compared with other ecosystems at the same latitude (below) (the horizontal line indicates the median, while the box shows the 25th and 75th percentiles). The abbreviations “wetl.,” “peatl.,” and “upl.” indicate wetland, peatland, and upland, respectively [13, 96]. Two and three asterisks indicate p < 0.01 and p < 0.001, respectively.
Three parallel quadrats (1.5 m length × 1 m width × 2 m depth) with an equal spacing of 10 m were dug each time. A new quadrat was dug next to the old quadrat each sampling time. For each soil quadrat, the soil profile was sampled at four side pits and sliced at specific intervals (20-cm intervals) along the depth from 0 to 200 cm. Soil samples collected from the same depth in all four side pits were thoroughly mixed to generate a composite sample, generating 10 samples per soil pit (n = 30) and sampling occasion. Soil samples for RNA isolation were immediately mixed with RNAlater (Thermo Fisher Scientific, Waltham, MA, USA) (6 mL of solution with 2 g of soil) according to the manufacturer’s instructions. The remaining soil samples were sealed in separate sterile plastic bags and placed on ice. Samples were transported to the laboratory immediately. Prior to experimentation, all visible plant residues were removed. All replicate samples were analyzed separately, and their assay results were averaged to represent the site conditions. One subsample was incubated to determine microbial activity, and another subsample was prepared for chemical analyses. A sub-fraction was stored at −80 °C for DNA extraction and molecular analysis.
Regional-scale sampling
We first selected site-scale sampling site (E) for the annual N2O flux measurements throughout a whole year in order to determine the hot moment of N2O emissions for the study site (n = 18), namely, the spring thawing period. Then, regional-scale N2O flux measurements were conducted in eight peatlands along the Gudong River with a focus on April, May, and June in 2020 (n = 72). The aim of this regional-scale study was to confirm the hot moment identified in the site-scale study; thus, only gas samples were collected in these regional-scale sampling sites.
The Gudong River, the largest tributary of the Di’er Songhua Jiang River, originates from the East Valley of Laoling, which is also located in the Northeast Plain of China, one of three black soil regions worldwide [45–47]. The river is 156.6 km long, and its drainage basin area is 4303 km2. All peatland sites were located in the Changbai Mountain region, where the total area of peatland was estimated at 463.31 km2, and the estimated soil organic C storage was 47.64 Tg [48]. The plants of these peatlands are similar, generally dominated by hummock-forming Carex species [49]. The height of the hummocks is generally 30–40 cm, and the vegetation coverage is 60–70% [50]. Eight peatland sampling points were selected along the Gudong River; Liangjiang (A, 640.3 m above sea level; N 42°39′15″, E 128°2′43″), Lixin (B, 625.6 m above sea level; N 42°42′13″, E 128°7′40″), Dongqing (C, 636.3 m above sea level; N 42°47′26″, E 128°13′52″), Wanbao (D, 652.2 m above sea level; N 42°50′32″, E 128°18′55″), Xinhe (E), Daba (F, 710.4 m above sea level; N 42°55′27″, E 128°33′25″), Jifang Zi (G, 612.1 m above sea level; N 42°52′21″, E 128°38′55″), and Gudong (H, 656.2 m above sea level; N 42°52′57″, E 128°36′55″) (Fig. 1C).
N2O flux measurements
The in-situ N2O fluxes were determined by the closed-chamber technique (volume of 4.8 × 104 mL, area of 0.08 m2) [32]. At each sampling point and each time, three parallel static chambers were established. Approximately 300-mL gas samples were extracted from the top of the chamber at 0, 5, 10, 15, 20, and 30 min. The N2O concentrations were measured upon arrival at the laboratory using gas chromatography (7890A; Agilent Technologies, Basel, Switzerland) with micro-electron capture detection (μECD) [32]. The carrier gas was N2 with a flow rate of 25 mL min−1. The temperatures of the oven, injection port, and μECD were 70 °C, room temperature, and 330 °C, respectively. The relative error for N2O measurements by this instrument was <1.5%. N2O flux was calculated based on the linear variation of its concentration in the chamber headspace as a function of time, base area, chamber volume, and molar volume of N2O at the chamber headspace air temperature. For all data collected, the coefficient of determination (R2) of the linear regression of concentration change over time was greater than 0.85.
15N and 18O tracer assay to determine N2O production source
In this study, 15N and 18O tracing was adopted to distinguish the sources of microbial N2O in the peatland of sampling site E [40–42]. The surface peat of each sampling month as well as three soil layers (40–60, 100–120, and 160–180 cm) in May (n = 27) were chosen. Fresh and pre-processed peat/soil were pre-incubated for approximately 2 d to increase the homogeneity of the samples and to avoid respiration pulses from the soil samples. Five-gram pre-incubated samples were evenly distributed on the bottom of 60-mL glass serum vials (Ochs Laborbedarf, Bovenden, Germany), which were then sealed with plugs (Ochs Laborbedarf) and aluminum crimp caps (Agilent, Santa Clara, CA, USA). Four treatments enriched with 18O and 15N were applied:[40–42] (i) H218O (18O at 97.2%) + NH4+ + NO3−; (ii) N18O3− (18O at 96.3%) + NH4+ + NO3−; (iii) 15NO3− (15N at 99.19%) + NH4+ + NO3−; and (iv) 15NH4+ (15N at 99.16%) + NH4+ + NO3−. All treatments were conducted in triplicate. For all treatments, final enrichments of 18O-H2O/NO3− and 15N-NH4+/NO3− were added to 1.0 atom% 18O and 30 atom% 15N excesses, respectively. The final N concentrations of both NH4+ and NO3− were maintained at 50 mg/kg NH4+-N/NO3−-N [40–42]. Subsequently, the gas in the vials was evacuated and then balanced using high-purity Ar (99.99%; Beijing Huayuan Gas, Beijing, China) at standard atmospheric pressure. Treatments were incubated at 60 rpm at different in-situ air temperatures (January, 0–4 °C; March, 4 °C; May, 10 °C; July, 15 °C; September, 20 °C; November, 5 °C). The incubation temperatures of all the following experiments were the same as the temperatures used in this 15N-18O tracer experiment.
After one week, the incubation was terminated using 7 M ZnCl2 solution (500 μL each), and then, the gas samples were transferred to 12-mL vacuum exetainers (Labco, Lampeter, UK). N2O gas was quantified using gas chromatography. The 15N and 18O signatures of N2O in the gas samples were measured using an isotope-ratio mass spectrometer (IRMS and Precon, Delta V Advantage, Thermo Fisher Scientific, Bremen, Germany; with precisions of <0.04 ‰ δ15N and <0.07‰ δ18O, respectively). The 15N enrichments of mineral N in the soil samples from different treatments were determined using the chemical transformation method [40–42].
The relative contributions of microbial N2O production pathways (nitrifier nitrification, NN; nitrifier denitrification, ND; nitrification-coupled denitrification, NCD; heterotrophic denitrification, HD) (Supplementary Fig. 1) were calculated according to the method of Kool et al. [40–42].
15N-tracer assay for N-conversion rate measurement
Potential rates of anammox, denitrification, and dissimilatory nitrate reduction to ammonium (DNRA) were measured with the 15N-tracer technique, as described by our previous publications [32, 51]. The surface peat of each sampling month (n = 18) was used to perform this experiment. The yield rates of the related products were determined as the potential rates of the three processes, briefly, treatments (1) 15NH4+ [15N at 99.16%]; (2) 15NH4+ + 14NO3–; (3) 15NO3– [15N at 99.19%] + 14NH4+. In the case of the soil amended solely with 15NH4+ (treatment 1), no significant accumulation of 15N2-labeled gas (29N2 and/or 30N2) was observed in any sample, indicating that all ambient 14NOx− had been consumed. When both 15NH4+ and 14NO3− were added (treatment 2), 29N2 accumulated in every sample without the accumulation of 30N2. This pattern was reproducible. The results showed that anammox was detectable in the soil. Samples amended solely with 15NO3− (treatment 3) showed significant anammox and denitrification rates. The rates and potential contributions to N2 formation from either anammox or denitrification were calculated from the produced 29N2 and 30N2 in treatment 3. Two hundred microliters of ZnCl2 solution (7 M) was added to terminate the incubation at each time point (0, 6, 12, 24, and 36 h). For DNRA, 100 μL of diluted 15NO2– (15N at 98.17%) isotopic stock solution was added to the samples. At defined intervals (0, 6, 12, 24, and 36 h), the reaction was stopped, and hypobromite was added followed by incubation for more than 12 h to convert produced 15NH4+ into 30N2 completely.
Acetylene inhibition experiment
An acetylene inhibition experiment was used to preliminarily judge the N2O source (biotic nitrification and denitrification or abiotic processes). The surface peat of each sampling month (n = 18) was used to perform this experiment. Three treatments were performed. One treatment without acetylene (C2H2) was used to measure the potential rate of total N2O production. The second treatment with 0.01% C2H2 (v/v) was used to measure the potential rate of the heterotrophic denitrification pathway and abiotic processes [52]. The last treatment used sterilized samples to determine the potential rate of abiotic processes. All treatments were conducted in triplicate. Gas samples were withdrawn from the headspace of the vials at different intervals (0, 3, 6, 12, 24, and 36 h) and transferred to evacuated exetainers (Labco) using a syringe with a three-port valve. The N2O concentrations of the gas samples were also measured using gas chromatography. The potential rate was calculated from the linear change of N2O concentrations over the sampling time, and a linear regression with a coefficient (R2) above 0.9 was selected.
Substrate-induced respiration inhibition
The substrate-induced respiration-inhibition (SIRIN) method was used to estimate the contribution from fungi and bacteria [44]. The inhibitor additivity ratio (IAR) was used to evaluate whether antibiotics exerted non-target effects [44]. It was estimated using Eq. (1):
| 1 |
Here, , , , and .. represent CO2 fluxes in antibiotic-free control soil, cycloheximide (CYH)-amended soil, streptomycin (STP)-amended soil, and soil amended with both antibiotics, respectively. A preliminary experiment was designed to determine the optimal concentrations of antibiotics at 0, 4, 6, 8, 12, and 24 h following substrate amendments. Four treatments were performed: (A) antibiotic-free control, soil with no antibiotic addition; (B) CYH, soil with the addition of CYH at 3.0, 6.0, 10.0, and 15.0 mg g−1 soil; (C) STP, soil with the addition of STP at 3.0, 6.0, and 10.0 mg g−1 soil; and (D) both antibiotics, soil with the addition of both CYH (3.0, 6.0, 10.0, and 15.0 mg g−1 soil) and STP (3.0, 6.0, and 10.0 mg g−1 soil). Each sample received an addition of glucose (5.0 mg g−1 soil) as a C source. N2O and CO2 concentrations were determined by gas chromatography. Finally, we found that peatland bacterial and fungal activities were largely terminated when STP and CYH concentrations were 10 mg g−1 and 3 mg g−1, respectively.
Fresh, pre-processed, and homogenized peat/soil (0–20, 30–40, 40–60, 100–120, and 160–180 cm of each sampling month, n = 90) were distributed on the bottom of 60-mL glass serum vials, which were then sealed with plugs and aluminum crimp caps. Six treatments in triplicate were applied to random vials to assess the potential rate and contribution to soil N2O production by bacterial denitrification, fungal denitrification, nitrification, and abiotic processes: (A) antibiotic-free control, soil with no antibiotic addition; (B) CYH, soil with the addition of CYH at 3 mg g−1; (C) STP, soil with the addition of STP at 10 mg g−1; (D) both antibiotics, soil with the addition of both CYH and STP; (E) C2H2 at 0.01% (v/v; ammonia oxidation inhibitor), and (F) ZnCl2 (600 μL, 7 M; biotic process inhibitor). The headspace gas was sampled with a syringe equipped with a Luer lock valve (25.0 mL; Agilent) and injected into a 12.0-mL vacuumed glass serum vial after 0, 4, 6, 8, 12, and 24 h. The gas samples were also measured using gas chromatography. The potential rate and contribution to soil N2O production by bacterial and fungal denitrification were estimated by Eqs. (2) and (3):
| 2 |
| 3 |
Here, is the soil N2O production rate in the antibiotic-free control, , , and represent the soil N2O production rate in soils treated with cycloheximide (3 mg g−1), streptomycin (10 mg g−1), and both streptomycin (10 mg g−1) and cycloheximide (3 mg g−1), respectively. The potential rate and contribution to soil N2O production by nitrification and abiotic process were assessed based on treatments (A), (E), and (F) according to previous studies [32, 44].
Extraction of soil total DNA and RNA
DNA and RNA extraction was conducted immediately after transport back to the laboratory. Total DNA and RNA were extracted from 2.0 g of soil using the DNeasy PowerSoil DNA Isolation Kit (QIAGEN, Hilden, Germany) and the RNeasy PowerMicrobiome RNA Isolation Kit (QIAGEN), respectively, according to the manufacturer’s instructions [53]. All extracted DNA and RNA samples were stored at −80 °C.
Molecular analysis
Quantitative PCR (qPCR) targeting the ammonia monooxygenase sub-unit A gene (archaeal amoA and bacterial amoA) [54] and the nitrite reductase gene (nirK and nirS) [52] were performed on a sequence detection system (ABI 7500; Applied Biosystems, Foster City, CA, USA) with SYBR-Green fluorescent dye (TaKaRa, Dalian, China) (Supplementary Table 1). Standard curves were obtained using 10-fold dilutions of standard plasmids containing archaeal-amoA, bacterial-amoA, nirK, and nirS genes, respectively. Negative controls without DNA template were included in each amplification reaction. The concentration of the primer was 10 μM. The amplification mixture (25 μL) contained the following: 10 μL of SYBR Premix Ex Taq (TaKaRa Biotechnology, Dalian, China), 0.4 μL of ROX reference dye, 0.5 μL of each primer (10 μM), 0.4 μL of bovine serum albumin (BSA), 2 μL of DNA, and 6.2 μL of ddH2O. Reactions with amplification efficiencies ranging from 90% to 110% and correlation coefficients (R2) above 0.99 were selected for further analysis.
Metatranscriptomic library sequencing and analysis
Ribosomal RNA (rRNA) sequences in the total RNA extract were removed using the Ribo-zero rRNA Removal Kit (Epicentre, Madison, WI, USA). The RNA fragments were reverse-transcribed to create cDNA. Subsequently, the cDNA library was constructed with an average insert size of 300 bp (±50 bp). Finally, 150-bp paired-end sequencing was performed on the Illumina HiSeq 4000 (Illumina, San Diego, CA, USA) at LC-BIO TECHNOLOGIES (Hangzhou, China) (Supplementary Table 2). RNA concentrations for samples from January, obtained from soil depths of 40–60 cm, 60–80 cm, and 80–100 cm, as well as all samples below 1 m, were below the detection limit.
Clean data were generated from raw metatranscriptomic reads after filtering low-quality nucleotides and reads with any ambiguous base calls using Cutadapt (v1.9) [55] and Fqtrim (v0.94). Subsequently, clean data were quality-checked using FastQC [56]. The clean data from each sample were de novo assembled and integrated using Trinity [57]. A non-redundant UniGene catalog was constructed after removing redundant sequences using CD-HIT [58].
Metagenomic library sequencing and analysis
Approximately 1.5 μg of extracted DNA (per sample) was used for metagenomic library preparation and subsequent sequencing on the Illumina NovaSeq 6000 (150-bp paired-end) at LC-BIO TECHNOLOGIES (Hangzhou, China) (Supplementary Table 3). Clean data were generated from raw metagenomic reads after filtering out low-quality nucleotides and reads with any ambiguous base calls using Cutadapt (v1.9) [55] and Fqtrim (v0.94). Valid data from each sample were assembled using IDBA-UD (v1.1.1) [59]. The gene prediction of the assembled contigs was performed by CDS MetaGeneMark (v3.26); [60] then, predicted genes were clustered, and redundant predicted genes were removed using CD-HIT [58] (at 95% identity and 90% coverage thresholds). The non-redundant gene catalog (UniGenes) was obtained after removing the redundant genes. The relative abundance (transcripts per million, TPM) of UniGenes was calculated by comparing the reads of each sample with the CDS library using Bowtie2 (v2.2.0) [61]. Gene taxonomic and functional annotations were obtained using BLASTp [56] against the taxonomic NCBI-NR (v2016-07-12) database and the functional databases eggnog (v2016-06), KEGG (v2016-05), and NCyc (v2019-03) [62] at an e-value <10−5 threshold.
Genome binning, taxonomic classification, and functional annotation
The clean data obtained from metagenomic sequencing were used to retrieve metagenome-assembled genomes (MAGs) using MetaWRAP [63]. After quality control, clean data were assembled into contigs using MEGAHIT [64]. The assembled contigs were clustered to recover MAGs using MetaBAT2 [65], MaxBin2 [66], and CONCOCT [67]. The resulting three bins were consolidated and filtered with the Bin_refinement module (completeness >50%, contamination <10%) (Supplementary Table 4). CheckM [68] was used to evaluate the completeness and contamination of the MAGs. The taxa corresponding to each of the genome bins were annotated using NCBI_nt, NCBI_tax, and GTDB-Tk (v2.1.15) [69], and their corresponding abundances were estimated using Salmon [70]. Protein-coding genes of each MAG were assigned using the eggNOG, KEGG, and NCyc [62] databases for functional annotation with PROKKA.
Analytical procedures for environmental variables
The soil variables, namely moisture, pH, total organic carbon (TOC), NH4+, NO3−, total organic matter (TOM), total carbon (TC), total nitrogen (TN), and total sulfur (TS), were investigated as described previously [32, 51, 54] (Supplementary Fig. 2). The dissolved oxygen (DO) concentration was measured using a Pocket Oxygen Meter (PyroScience GmbH, Aachen, Germany) equipped with an optical oxygen sensor accordingly.
Statistical analysis
Spearman correlation analysis and one-way ANOVA were conducted using SPSS 10.0 (SPSS Inc., Chicago, IL, USA). Redundancy analysis (RDA) and non-metric multidimensional scaling analysis (nMDS) were performed with CANOCO 5.0. Network analysis controlling for the false discovery rate (FDR) was conducted using the psych package in R and visualized in Gephi [71]. Linear regression was conducted using Origin 9.0 (OriginLab, Northampton, MA, USA). A p value of less than 0.05 indicated statistical significance. All bar charts, scatter plots, and pie charts were generated using Origin.
Results
Hot moment of N2O flux in peatland
Based on the site-scale investigation of sampling site E (Fig. 1A), the peak N2O emissions occurred in April (3.08 ± 0.01 mg N2O m−2 d−1) and May (0.30 ± 0.00 mg N2O m−2 d−1) and were significantly higher than N2O emissions during the other months (−0.002 ± 0.04 mg m−2 d−1) (one-way ANOVA, p < 0.001) (Fig. 1B). Moreover, in January and November, the N2O fluxes were negative, indicating that the frozen peatland during this period was a sink for N2O. The cumulative amount of N2O emissions during the thawing period accounted for 103% of the annual net emissions. These results indicate that the seasonally frozen peatland had a high rate of N2O emission during a hot moment in spring.
The N2O emissions of the seasonally frozen peatland were further investigated based on regional investigation, including eight peatland sampling sites separated by an interval of 15 km (Fig. 1C). N2O flux measurements in April, May, and June showed substantial temporal and spatial heterogeneity of N2O emissions (one-way ANOVA, p < 0.05), with an average flux of 0.55 ± 0.29 mg N2O m−2 d−1. However, the N2O emission fluxes of all sampling sites peaked in April (1.20 ± 0.88 mg N2O m−2 d−1) and May (0.38 ± 0.15 mg N2O m−2 d−1), and were significantly higher than that in June (0.05 ± 0.05 mg N2O m−2 d−1) (Fig. 1D, one-way ANOVA, p < 0.05) (Supplementary Fig. 3). This is in accordance with the site-scale investigation. These results indicate that the thawing period is the hot moment of N2O emissions in the seasonally frozen peatland. The occurrence of this N2O emission hot moment may convert the seasonally frozen peatland into an important N2O emission source (Fig. 1D). If the contribution of the hot moment of these peatlands is not considered, the N2O emission flux from northern peatlands will be underestimated.
Microbial source of N2O production
Sources of N2O production were further investigated in soil cores (0–200 cm cores from site E split at 20-cm intervals) using inhibitor methods (Fig. 2). We found that microbial processes contributed 99.1 ± 0.2% to the total N2O production, while the remainder was attributed to abiotic processes (Fig. 2A). Among the microbial N2O production rates, bacterial denitrification was the greatest source (0.27 ± 0.07 ug N2O g−1 d−1, one-way ANOVA p < 0.01) and accounted for 52.9% of the peatland N2O production (Fig. 2A), followed by fungal denitrification (0.15 ± 0.05 ug N2O g−1 d−1, 30.2%) and microbial nitrification (0.07 ± 0.03 ug N2O g−1 d−1, 15.8%). During the interval from the frozen period to the thawed period of the peatland, the contribution to N2O production from fungal denitrification and nitrification increased, while that from bacterial denitrification decreased (Fig. 2A). For each sampling time, bacterial and fungal denitrification together contributed 83.1 ± 8.4% to N2O production. Moreover, their production was highly associated with the N2O emission flux (R2 = 0.63 and R2 = 0.80, for bacterial and fungal denitrification, respectively (Supplementary Fig. 4)). Hence, denitrification was the main source of N2O production in the seasonally frozen peatland.
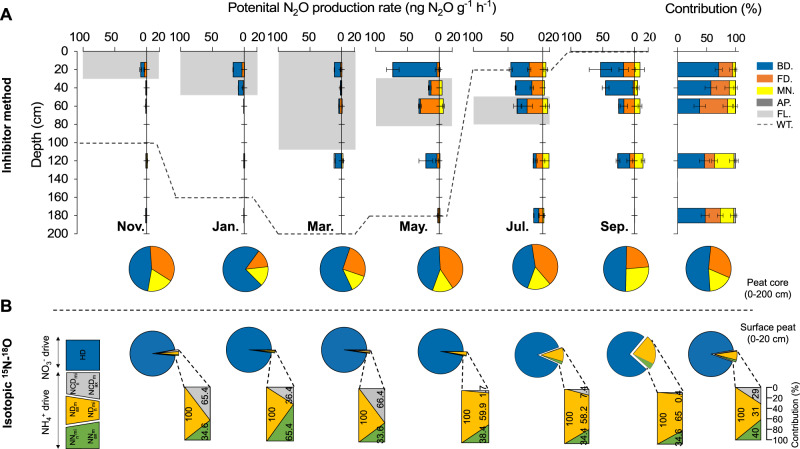
Fig. 2. Rates and contributions of bacterial, fungal denitrification and nitrification to N2O production in the seasonally frozen peatland.
A Potential rates of bacterial denitrification, fungal denitrification, and nitrification pathways to the total N2O production over a whole year in the seasonally frozen peatland, as determined using inhibitor methods. For each sampling month, the pie chart represents the average proportions of the three processes for the 2-m soil core. “BD.,” “FD.,” “MN.,” “AP.,” “FL.,” and “WT.” indicate bacterial denitrification, fungal denitrification, microbial nitrification, abiotic processes, frozen layer, and water table, respectively. The stacked area plot shows the contributions of bacterial denitrification, fungal denitrification, nitrification, and abiotic processes to N2O production. B Contributions of nitrifier nitrification (NN), nitrifier denitrification (ND), nitrification-coupled denitrification (NCD), and heterotrophic denitrification (HD) pathways in the surface peat to N2O production, as determined using isotopic 15N−18O techniques.
There were significant differences in N2O production in the soil cores through the 200-cm profiles. The total N2O production rates in different months decreased with soil depth (Fig. 2A). Surface peat (0–20 cm) was the main source of N2O, contributing 49.1% to N2O production (Fig. 2A, one-way ANOVA, p < 0.05). The contribution from deep soil (100–120 and 160–180 cm) was lower, but not negligible (17.1%). In the surface and sub-surface peat (20–40 cm), the production of N2O was mainly attributed to bacterial denitrification. As the depth increased (40–60 cm peat), N2O was mainly produced via fungal denitrification, accounting for 48.3 ± 8.2% of the N2O (Fig. 2A). In all the layers, the contribution from nitrification (17.1 ± 5.0%) was lower than that from denitrification. TOC and TOM were the key influencing factors for N2O production at all soil depths (Supplementary Fig. 5). Overall, the above results demonstrated that the hot moment of N2O emissions of the seasonally frozen peatland was mainly driven by denitrification.
As surface peat is the main N2O source of peatlands, peat samples were chosen to furtherly verify the above results using 15N and 18O tracing technology. In this analysis, denitrification was still the dominant process contributing to N2O production (88.6 ± 4.7%). Furthermore, this process was significantly higher than the contribution of nitrification, which includes NN, ND, and NCD (one-way ANOVA, p < 0.001, Fig. 2B). Although N2O production from ammonia oxidation pathways accounted for a small part, the distributions of NN, ND, and NCD differed among the months (Fig. 2B). ND and NCD pathways were more active in July and September, accounting for over 95% of the ammonia oxidation-derived N2O (96.3% and 99.8% for July and September, respectively) (Fig. 2B). In the soil profile, the N2O production from ammonia oxidation was still significantly lower than that of denitrification (Supplementary Fig. 6). The 15N and 18O isotope tracing further confirmed that N2O production in the seasonally frozen peatland was mainly produced by heterotrophic NO3- reduction.
The inhibitor assay and isotopic 15N-18O tracing results demonstrated that NO3− reduction was the main driver of the hot moment of N2O emissions of the seasonally frozen peatland. The NO3− reduction process includes heterotrophic denitrification, DNRA, and anammox, and their individual contributions were further quantified using 15N tracing. The contribution of heterotrophic denitrification to NO3− reduction (31.6 ± 2.0 nmol N g−1 h−1) was significantly higher than that of both anammox (1.14 ± 0.2 nmol N g−1 h−1) and DNRA (1.52 ± 0.4 nmol N g−1 h−1), demonstrating the dominant role of denitrification in the NO3- reduction process. However, in contrast with the N2O emission flux, the denitrification rate had no obvious hot moment (Supplementary Fig. 7). The denitrification rate increased with temperature and peaked during the fully thawed period. This result conflicts with the N2O emission hot moment observed during the thawing period. Therefore, it was necessary to further explore the microbial mechanism of N2O production.
Thawing stimulated the expression of genes associated with N2O production
Metatranscriptomic and metagenomic sequencing were used to analyze functional genes associated with N2O production (norB, p450nor, and hao) and reduction (nosZ) in bacterial denitrification, fungal denitrification, and nitrification (Fig. 3). In the soil profile, both the DNA- and RNA-based relative abundances of norB showed a slight decrease, while that of hao increased with soil depth. For p450nor, there was no significant difference in abundance (Supplementary Fig. 8). At different sampling depths, both the DNA- and RNA-based relative abundances of nor in the bacterial and fungal denitrification pathway were significantly higher than that of hao in the nitrification pathway, which was further confirmed by DNA-based qPCR results (one-way ANOVA, p < 0.05, Fig. 3A, Supplementary Fig. 8, Supplementary Fig. 9). These multi-omics results indicate that denitrification was the dominant N2O production route in the seasonally frozen peatland and are consistent with the inhibitor and isotopic tracing results.
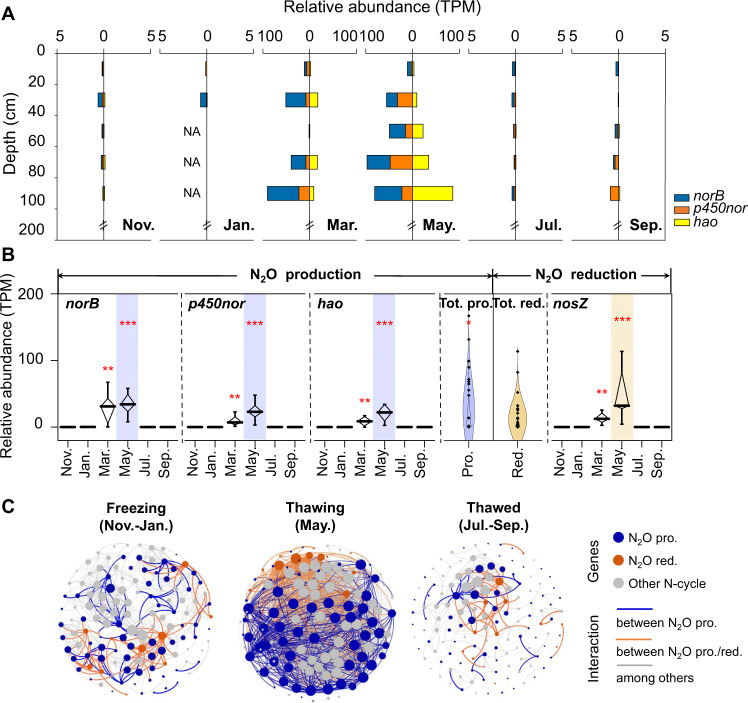
Fig. 3. N2O production- and reduction-related functional gene transcripts identified in seasonally frozen peatland.
A Histogram showing the transcripts of N2O production genes in the seasonally frozen peatland across a whole year as derived from metatranscriptomic data. “NA” indicates RNA concentrations that were below the detection limit for metatranscriptomic sequencing. TPM means transcripts per million. B The box plots show the relative abundance of N2O production- and reduction-related functional gene based on RNA data (the horizontal line indicates the median, while the box shows the 25th and 75th percentiles). “Tot. pro.” and “Tot. red.” indicate total production and total reduction, respectively. One, two, and three asterisks indicate p < 0.05, p < 0.01, and p < 0.001, respectively. The months of peak abundance were highlighted for N2O production (light purple) and reduction (light yellow) genes. C Co-occurrence network analysis showing the interaction among active N2O production- and reduction-related microorganisms during different periods of the seasonally frozen peatland. Node colors indicate the types of genes. Edges are colored according to the interactions among different genes. The abbreviations “pro.” and “red.” indicate production and reduction, respectively.
Metatranscriptomic and metagenomic analysis showed that the relative abundances of genes encoding N2O production protein complexes (i.e., nor and hao genes) were significantly higher than those of genes encoding N2O reduction proteins (i.e., nosZ). However, the abundances of these genes during the thawing period were 300–500 times higher than those during the thawed and freezing periods as RNA transcripts, but there was no significant difference across all the periods at the DNA level (Fig. 3B, Supplementary Fig. 8, one-way ANOVA, p > 0.05). The large difference between N2O production and reduction (16–32 times) implies that the seasonally frozen peatland had high N2O emission potential during all the periods, while only the soil properties and physical state during the thawing period stimulated the observed N2O burst.
The high expression of genes associated with N2O production in March indicates that a large amount of N2O may also be produced in March. Because the peatland is completely frozen at this time (frozen-depth, 100 cm), the N2O produced during the frozen period may be trapped by ice in the soil. This N2O can then be released with soil thawing [19, 27] and thus may also contribute to the hot moment of N2O emissions in spring (April and May). However, such a physical release of N2O is unlikely to be the direct reason for the occurrence of the observed hot moment, because the potential N2O production rate was low during the frozen period (Fig. 2A). Moreover, both the N2O production rate and gene expression during the thawing period were significantly higher than those during the frozen period (Figs. 2A and 3A). Hence, microbial activity during thawing periods is expected to be the main cause of the hot moment of N2O emissions.
As substrates for microbial growth, TOC (p = 0.004, F = 11) and TC (p = 0.004, F = 8) were the dominant factors influencing the abundance of N2O production- and reduction-related functional genes according to RDA (Supplementary Fig. 10). TOC and TC explained 30.6% and 24.2% of the variation in gene abundances, respectively, as supported by correlation analysis (Supplementary Table 5). Repeated freezing and thawing of the peatland can lead to the breakdown of soil aggregates and render decomposable organic C more easily accessible to microorganisms, thereby activating N2O producers [27, 72, 73]. The acidic peatland environment (pH 3.3–4.5) could inhibit N2O reductase and thus cause N2O emissions [28, 30]. These results show that the freeze-thaw cycle during the thawing period stimulated microbial activities related to N2O production and reduction and further promoted peatland N2O emissions.
The microbial community structure was then further analyzed. Metatranscriptomic analysis revealed that the samples from the thawing period not only showed a clear separation from those of other periods, but also had significantly higher microbial diversity based on RNA, while there was no significant difference between periods in terms of microbial community and diversity based on DNA (Supplementary Figs. 11 and 12), which indicates that thawing altered the active microbial community structure. Co-occurrence networks from the different seasonal periods revealed that thawing increased the complexity of the N2O-related microbial networks (Fig. 3C, Supplementary Tables 6 and 7), thereby reflecting the strengthened interactions between N2O-related communities. These results indicated that thawing promoted synergy among active microorganisms.
Metabolic potential related to N2O production
The potential denitrification metabolic pathway related to the N2O hot moment was further revealed by metagenome analysis (Fig. 4). The assembly and binning processes generated 77 metagenome-assembled genomes (MAGs), of which 41 genomes encode genes involved in N2O production and reduction (Supplementary Fig. 13). A total of 22 genomes possessed denitrifying nirK or nirS genes (encoding proteins involved in NO2− reduction to NO), while no MAGs contained genes involved in ammonia oxidation (amoA/B/C) (Fig. 4, Supplementary Fig. 13), further indicating a dominant role of denitrification in N2O production. No MAGs encoded capabilities for all denitrification genes, i.e., genes that encode proteins involved in NO3− reduction to NO2−, NO2− reduction to NO, NO reduction to N2O, and finally N2O reduction to N2 (Fig. 4). This suggested that denitrification was a sequential process catalyzed by different enzymes and microorganisms as was observed in other studies [74, 75], and during the thawing period, the interaction among N2O producing microorganisms was significantly stronger than that between N2O producing and reducing microorganisms (Fig. 3C, Supplementary Table 6). Additionally, the abundance and expression of N2O production genes were also significantly higher than those of N2O reduction genes (Fig. 3B, Supplementary Fig. 8). These dual effects resulted in the occurrence of an N2O emission hot moment during the thawing period.
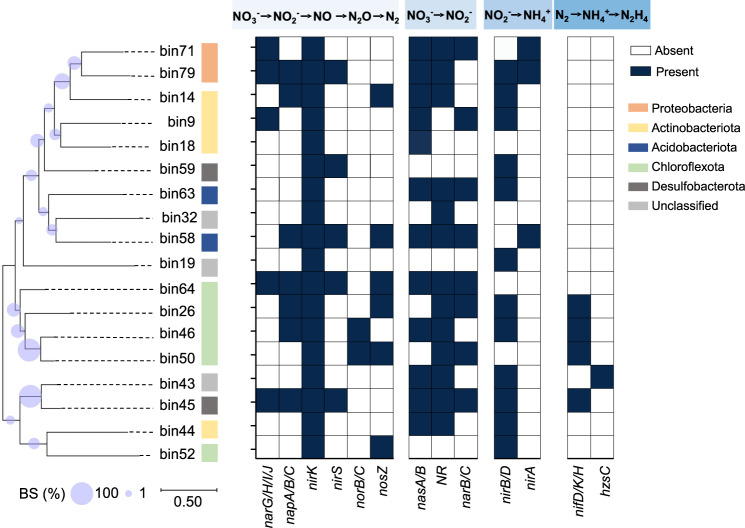
Fig. 4. Metabolic reconstruction and features of N2O production- and reduction-related microorganisms identified in seasonally frozen peatland.
Metabolic features across selected metagenome assembled genome (MAG) phylogenetic clusters. MAGs with the nirK gene were selected for phylogeny construction. Having any one or more functional genes in a MAG that performs the same process is indicated by “Present.” Bootstrap support values for 1000 replicates are indicated at each node.
The N-cycle metabolic pathway potentially related to the N2O hot moment was further examined. Among the MAGs, 76.9% contained genes (narG/H/I, napA/B, narB, nasA/B, and NR) involved in NO3− reduction to NO2− (Supplementary Fig. 13), indicating that under limited oxygen conditions, NO3− reduction may be an appropriate alternative pathway for energy conservation, which supports our previous results [51]. Additionally, 62.3% of the MAGs contained genes that encode proteins with capabilities for NO2− reduction to NH4+ (nirB/D, nirA, and nrfA/H) through assimilatory/dissimilatory nitrate reduction (Supplementary Fig. 13). The gene abundances for NO2− reduction to NH4+ (99 ± 6 TPM) were significantly higher than that for NH4+ oxidation (3 ± 1 TPM). In addition, the gene abundances for organic nitrogen mineralization (138 ± 55 TPM) were significantly higher than that for nitrogen assimilation (78 ± 26 TPM). This resulted in the accumulation of NH4+ in the peatland (median 15.0 mg kg−1, IQR (3.6, 21.5) mg kg−1), in accordance with our hypothesis of an increased microbial mineralization rate.
However, the driver of the N2O emission hot moment was heterotrophic denitrification rather than ammonia oxidation, despite the high NH4+ content of the peatland (Supplementary Fig. 2). While acidophilic nitrification occurs in soils [76], oxygen-limitation (DO 0.05 ± 0.04 mg L−1) in the peatland environment is unlikely to be conducive to ammonia oxidation [77]. Denitrifying community members in the peatland may exhibit acid tolerance under a low pH value. For example, denitrifying genome bin 79, contains genes encoding urease, which can move cytoplasmic NH3 through to the periplasm and buffer the periplasm through the reaction converting H+ and NH3 to NH4+, resulting in a relatively neutral periplasmic pH (Supplementary Fig. 14). All these results indicated that heterotrophic denitrification driven by the thaw-freeze cycle was the predominant factor causing the hot moment of N2O emissions occurring in the seasonally frozen peatland.
Discussion
Northern peatlands are sensitive to climate change, while their vulnerability to climate warming is uncertain [2–4]. Our results reported that seasonally frozen peatlands can be important N2O emission sources in the Northern Hemisphere and that thawing periods are a hot moment of N2O emissions driven by freezing-thawing cycles. These emission rates matched the highest N2O emissions recorded in natural terrestrial ecosystems [78], corresponding to about 6% of the global warming potential of natural northern peatlands CH4 emissions [79]. The hot moment of N2O emissions that occurred in the seasonally frozen peatlands (0.79 ± 0.29 mg N2O m−2 d−1) was higher than those from drained peatland forests (spring, 0.58 mg N2O m−2 d−1) [13], permafrost (growing season, 0.047 mg N2O m−2 d−1) [13], and non-managed northern peatland (0.22–0.65 mg N2O m−2 d−1) [80]. It was comparable to N2O emissions from managed northern peatlands influenced by freeze-thaw cycles (0.13–10.2 mg N2O m−2 d−1) [81], with equivalent N2O production rates. Neglecting these hot moment emissions may lead to underestimation of N2O emissions representing about 1.1% of N2O emissions from natural sources [82].
The hot moment release characteristics of N2O emissions from seasonally frozen peatlands should be mainly determined by the freeze-thaw state of the soil. During the freezing (fall), frozen (winter), and thawed (summer) periods of the seasonally frozen peatland, N2O emissions were relatively low, and during these periods, the study site even acted as a sink rather than an emission source. However, the flux during the hot moment of thawing (spring) was significantly higher than that during the other periods. The ice-covered peat surface in the freezing and frozen periods prevents N2O from escaping until peat thaw commences [27]. These N2O emissions can be physically released with peat thawing, but are unlikely to be the direct reason for the occurrence of a hot moment, because the potential N2O production rate during winter was relatively low. In contrast with previous studies, in which temperature was determined to be the most critical factor affecting N2O emission flux [13, 17], in our study, the N2O fluxes in both high (summer, 0.04 ± 0.01 mg N2O m−2 d−1) and low temperature seasons (winter, -0.15 ± 0.03 mg N2O m−2 d−1) were significantly lower than that during the thawing seasons (spring, 1.69 ± 0.98 mg N2O m−2 d−1), indicating the key role of thawing in N2O emission. The freezing-thawing cycles in the thawing periods increased both the quantities and availabilities of soil carbon and nitrogen compounds [20, 24], which supply substrates necessary for microbial N2O production, especially via bacterial and fungal denitrification, underlying the hot moment of N2O emission.
Heterotrophic denitrification driven by the thaw-freeze cycle was the predominant cause of the hot moment of N2O emissions from seasonally frozen peatlands. Consistent with this study, denitrification can generate large amounts of N2O in northern peat soils [11, 38], especially within deeper peat layers [83]. However, nitrification can be a key driver of high N2O emissions from arctic peat soils [84, 85], as it regulates the supply of NO2− and NO3− for N2O production through denitrification, especially in bare peat during dry years [83]. In this study, corresponding with the N2O production rate, the abundances of nitrification-related genes were significantly lower than those of denitrification-related genes, indicating that nitrification may not provide enough NO3− for denitrification. The dominant vegetation type is often used to judge the nutritional status of peatlands [86, 87]. In this study, Carex is the dominant plant in the peatlands, which indicates a mesotrophic peat environment. The nutrient supply here, including NO3−, comes from rainfall, surface water, and groundwater [86, 87], not mainly from nitrification.
N2O can be produced at different depths in the soil profile. In this study, surface peat was the main source of N2O attributed to denitrification. The relatively high oxygen concentration in the surface peat may impair the reduction of N2O [88], leading to N2O accumulation and emission. The subsurface peat is an anoxic environment with a high concentration of organic matter, which may promote complete denitrification [89]. The final product is mainly N2, not N2O. Low nutrient contents in deeper soil layers result in a decrease in denitrifying microbial abundance and a limitation of N2O production by denitrification, which is consistent with a previous study [90].
By using combined metagenomic and metatranscriptomic sequence data designed to capture the genes responsible for key functions of the N cycle, we observed significant increases in the relative abundance of N2O production genes related to bacterial denitrification, fungal denitrification, and nitrification during the thawing period, which was in line with previous findings [11, 91, 92]. The RNA transcripts of N2O production genes in the thawed period were significantly lower than those in the thawing period, but not the potential rate of N2O production and denitrification. On the one hand, this could be the result of intense competition for available N sources between plants and microorganisms [11, 17]. DNA-based qPCR and metagenomic data indicated high N2O production potential across all periods. However, plants meet their N requirements by absorbing small compounds such as oligopeptides, amino acids, and NO3−. Insufficient NO3− and TOC may lead to low activities of denitrification in the in-situ environment, which is manifested as low RNA transcript abundance and N2O emission flux. On the other hand, laboratory microcosm incubation methods may disrupt mass transfer and increase substrate utilization, which just characterizes the potential rate of N2O production and denitrification [93]. This highlights that short-term laboratory experiments used to examine the microbial limitations of the N cycles of seasonally frozen peatlands [11, 94] do not well represent the real changes in the microbial community and their functioning over time. These results further indicated that thawing provided ideal conditions for the development of active N2O-producing microbial communities, resulting in high N2O release in seasonally frozen peatlands.
Conclusion
With the intensification of global warming [21], the rate of carbon degradation and nitrogen mineralization in peatlands will be greatly increased [22, 23]. It can be predicted from our study that the warming-effect caused by hot moment N2O emissions in seasonally frozen peatlands may make peatlands a net source of warming; however, both anthropogenic and natural disturbances may accelerate this occurrence [95]. Thus, it is important to protect peatlands for the sake of ecological security and climate change at a global scale. Overall, patterns and drivers of seasonally frozen peatland N2O emission and production rates observed in this study offer new insights into potential nitrogen release upon peatland thawing and provide important information for use in Earth system models to better predict northern peatland biogeochemical cycles under a warming climate.
Supplementary information
Acknowledgements
The authors would like to thank Mr. Dachun Zhu, Mr. Bangrui Lan, Mr. Longbin Yu, Mr. Shiguang Liu, Mr. Yuantao Wang, Mr. Libo Sun, and Miss. Gawhar Armanbek for sampling peatland soil cores in Antu. This research was financially supported by the National Natural Science Foundation of China (Nos. 91851204 and 92251304), Local Innovative and Research Teams Project of Guangdong Pearl River Talents Program (2017BT01Z176), National Key R&D Program (2016YFA0602303), and special fund from the State Key Joint Laboratory of Environment Simulation and Pollution Control (Research Center for Eco-environmental Sciences, Chinese Academy of Sciences) (18Z02ESPCR). The author GZ gratefully acknowledges the Program of the Youth Innovation Promotion Association of Chinese Academy of Sciences.
Author contributions
GZ designed the project. XW, GZ, and SW contributed to sample analysis. XW and GZ wrote the manuscript with contributions from SW, YY, HT, MSMJ, and CS. All authors discussed and interpreted the results and contributed to the manuscript. All authors discussed and commented on the manuscript. Correspondence and requests for materials should be addressed to GZ. (gbzhu@rcees.ac.cn).
Data availability
The metagenomic data and the metatranscriptomic data were deposited in the NCBI Sequence Read Archive under accession numbers SAMN26424243–SAMN26424272 and SRR14251110–SRR14251136, respectively.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-023-01389-x.
References
- 1.Loisel JL, Yu ZC, Beilman DW, Camill P, Alm J, Amesbury MJ, et al. A database and synthesis of northern peatland soil properties and Holocene carbon and nitrogen accumulation. Holocene. 2014;24:1028–42. doi: 10.1177/0959683614538073. [DOI] [Google Scholar]
- 2.Hugelius G, Loisel J, Chadburn S, Jackson RB, Jones M, MacDonald G, et al. Large stocks of peatland carbon and nitrogen are vulnerable to permafrost thaw. Proc Natl Acad Sci USA. 2020;117:20438–46. doi: 10.1073/pnas.1916387117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frolking S, Talbot J, Jones MC, Treat CC, Kauffman JB, Tuittila E-S, et al. Peatlands in the Earth’s 21st century climate system. Environ Rev. 2011;19:371–96. doi: 10.1139/a11-014. [DOI] [Google Scholar]
- 4.Yu Z, Loisel JL, Brosseau DP, Beilman DW, Hunt SJ. Global peatland dynamics since the Last Glacial Maximum. Geophys Res Lett. 2010;37:13402. doi: 10.1029/2010GL043584. [DOI] [Google Scholar]
- 5.Leifeld J, Menichetti L. The underappreciated potential of peatlands in global climate change mitigation strategies. Nat Commun. 2018;9:1–7. doi: 10.1038/s41467-018-03406-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang T, Barry RG, Knowles K, Ling F, Armstong RL. Distribution of seasonally and perennially frozen ground in the Northern Hemisphere. In: ICOP 2003 Permafrost: proceedings of the 8th International Conference on Permafrost. Netherlands: A.A. Balkma Publishers; 2003. 2, p. 1289–94.
- 7.Dai LC, Guo XW, Zhang FW, Du Y, Ke X, Li YK, et al. Seasonal dynamics and controls of deep soil water infiltration in the seasonally frozen region of the Qinghai-Tibet Plateau. J Hydrol. 2019;571:740–8. doi: 10.1016/j.jhydrol.2019.02.021. [DOI] [Google Scholar]
- 8.Åkerman HJ, Johansson M. Thawing permafrost and thicker active layers in sub-arctic Sweden. Permafr Periglac Process. 2008;19:279–92. doi: 10.1002/ppp.626. [DOI] [Google Scholar]
- 9.Quinton WL, Baltzer J. The active-layer hydrology of a peat plateau with thawing permafrost (Scotty Creek, Canada). Hydrol J. 2013;21:201–20. 10.1007/s10040-012-0935-2.
- 10.Swindles GT, Morris PJ, Mullan D, Watson EJ, Turner TE, Roland TP, et al. The long-term fate of permafrost peatlands under rapid climate warming. Sci Rep. 2015;5:17951. doi: 10.1038/srep17951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marushchak ME, Kerttula J, Diáková K, Faguet A, Gil J, Grosse G, et al. Thawing Yedoma permafrost is a neglected nitrous oxide source. Nat Commun. 2021;12:1–10. doi: 10.1038/s41467-021-27386-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kou D, Yang GB, Li F, Feng XH, Zhang DY, Zhang QW, et al. Progressive nitrogen limitation across the Tibetan alpine permafrost region. Nat Commun. 2020;11:3331. doi: 10.1038/s41467-020-17169-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Voigt C, Marushchak ME, Abbott BW, Biasi C, Elberling B, Siciliano SD, et al. Nitrous oxide emissions from permafrost-affected soils. Nat Rev Earth Environ. 2020;1:420–34. doi: 10.1038/s43017-020-0063-9. [DOI] [Google Scholar]
- 14.Larmola T, Leppänen SM, Tuittila E-S, Aarva M, Merilä P, Fritze H, et al. Methanotrophy induces nitrogen fixation during peatland development. Proc Natl Acad Sci USA. 2014;111:734–9. doi: 10.1073/pnas.131428411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kox MAR, van den Elzen E, Lamers LPM, Jetten MSM. Microbial nitrogen fixation and methane oxidation are strongly enhanced by light in Sphagnum mosses. AMB Expr. 2020;10:61. doi: 10.1186/s13568-020-00994-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Jong AEE, in’t Zandt MH, Meisel OH, Jetten MSM, Dean JF, Rasigraf O, et al. Increases in temperature and nutrient availability positively affect methane-cycling microorganisms in Arctic thermokarst lake sediments. Environ Microbiol. 2018;20:4314–27. doi: 10.1111/1462-2920.14345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Voigt C, Marushchak ME, Lamprecht RE, Jackowicz-Korczyński M, Lindgren A, Mastepanov M, et al. Increased nitrous oxide emissions from arctic peatlands after permafrost thaw. Proc Natl Acad Sci USA. 2017;114:6238–43. doi: 10.1073/pnas.1702902114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Repo ME, Susiluoto S, Lind SE, Jokinen S, Elsakov V, Biasi C, et al. Large N2O emissions from cryoturbated peat soil in tundra. Nat Geosci. 2009;2:189–92. doi: 10.1038/NGEO434. [DOI] [Google Scholar]
- 19.Elberling B, Christiansen HH, Hansen BU. High nitrous oxide production from thawing permafrost. Nat Geosci. 2010;3:332–5. doi: 10.1038/NGEO803. [DOI] [Google Scholar]
- 20.Wang JY, Song CC, Hou AX, Miao YQ, Yang GS, Zhang J. Effects of freezing-thawing cycle on peatland active organic carbon fractions and enzyme activities in the Da Xing’anling Mountains, Northeast China. Environ Earth Sci. 2014;72:1853–60. doi: 10.1007/s12665-014-3094-z. [DOI] [Google Scholar]
- 21.Masson-Delmotte V, Zhai P, Pirani A, Connors SL, Pean C, Berger S, et al., editors. and Intergovernmental Panel on Climate Change (IPCC). IPCC, 2021: Climate Change 2021: The Physical Science Basis. Contribution of Working Group I to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change. Open Access. UK, Cambridge: Cambridge University Press; 2021. pp. 2391. 10.1017/9781009157896.
- 22.Koven CD, Lawrence DM, Riley WJ. Permafrost carbon climate feedback is sensitive to deep soil carbon decomposability but not deep soil nitrogen dynamics. Proc Natl Acad Sci USA. 2015;112:3752–7. doi: 10.1073/pnas.1415123112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen LY, Liu L, Mao C, Qin SQ, Wang J, Liu FT, et al. Nitrogen availability regulates topsoil carbon dynamics after permafrost thaw by altering microbial metabolic efficiency. Nat Commun. 2018;9:1–11. doi: 10.1038/s41467-018-06232-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salmon VG, Soucy P, Mauritz M, Celis G, Natali SM, Mack MC, et al. Nitrogen availability increases in a tundra ecosystem during five years of experimental permafrost thaw. Glob Change Biol. 2016;22:1927–41. doi: 10.1111/gcb.13204. [DOI] [PubMed] [Google Scholar]
- 25.Finger RA, Turetsky MR, Kielland K, Ruess RW, Mack MC, Euskirchen ES. Effects of permafrost thaw on nitrogen availability and plant-soil interactions in a boreal Alaskan lowland. J Ecol. 2016;104:1542–54. doi: 10.1111/1365-2745.12639. [DOI] [Google Scholar]
- 26.Mao C, Kou D, Qin S, Yang YH. Permafrost nitrogen status and its determinants on the Tibetan Plateau. Glob Change Biol. 2020;26:5290–302. doi: 10.1111/gcb.15205. [DOI] [PubMed] [Google Scholar]
- 27.Congreves KA, Wagner-Riddle C, Si BC, Clough TJ. Nitrous oxide emissions and biogeochemical responses to soil freezing-thawing and drying-wetting. Soil Biol Biochem. 2018;117:5–15. doi: 10.1016/j.soilbio.2017.10.040. [DOI] [Google Scholar]
- 28.Frostegård Å, Vick SHW, Lim NYN, Bakken LR, Shapleigh JP. Linking meta-omics to the kinetics of denitrification intermediates reveals pH-dependent causes of N2O emissions and nitrite accumulation in soil. ISME J. 2022;16:26–37. doi: 10.1038/s41396-021-01045-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van den Heuvel RN, van der Biezen E, Jetten MS, Hefting MM. Denitrification at pH 4 by a soil-derived Rhodanobacter-dominated community. Environ Microbiol. 2010;12:3264–71. doi: 10.1111/j.1462-2920.2010.02301.x. [DOI] [PubMed] [Google Scholar]
- 30.Hénault C, Bourennane H, Ayzac A, Ratié C, Saby NPA, Cohan J, et al. Management of soil pH promotes nitrous oxide reduction and thus mitigates soil emissions of this greenhouse gas. Sci Rep. 2019;9:20182. doi: 10.1038/s41598-019-56694-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jonasson S, Michelsen A, Schmidt IK. Coupling of nutrient cycling and carbon dynamics in the Arctic, integration of soil microbial and plant processes. Appl Soil Ecol. 1999;11:135–46. doi: 10.1016/S0929-1393(98)00145-0. [DOI] [Google Scholar]
- 32.Zhu GB, Wang SY, Wang WD, Wang Y, Zhou LL, Jiang B, et al. Hotspots of anaerobic ammonium oxidation at land-freshwater interfaces. Nat Geosci. 2013;6:103–7. doi: 10.1038/NGEO1683. [DOI] [Google Scholar]
- 33.Chen LY, Liang JY, Qin SQ, Liu L, Fang K, Xu YP, et al. Determinants of carbon release from the active layer and permafrost deposits on the Tibetan Plateau. Nat Commun. 2016;7:13046. doi: 10.1038/ncomms13046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ding JZ, Chen LY, Ji CJ, Hugelius G, Li YN, Li L, et al. Decadal soil carbon accumulation across Tibetan permafrost regions. Nat Geosci. 2017;10:420–4. doi: 10.1038/NGEO2945. [DOI] [Google Scholar]
- 35.Kartal B, Maalcke WJ, de Almeida NM, Cirpus I, Gloerich J, Geerts W, et al. Molecular mechanism of anaerobic ammonium oxidation. Nature. 2011;479:127–30. doi: 10.1038/nature10453. [DOI] [PubMed] [Google Scholar]
- 36.Song CC, Xu XF, Tian HQ, Wang YY. Ecosystem–atmosphere exchange of CH4 and N2O and ecosystem respiration in wetlands in the Sanjiang Plain, Northeastern China. Glob Change Biol. 2009;15:692–705. doi: 10.1111/j.1365-2486.2008.01821.x. [DOI] [Google Scholar]
- 37.Butterbach-Bahl K, Baggs EM, Dannenmann M, Kiese R, Zechmeister-Boltenstern S. Nitrous oxide emissions from soils: how well do we understand the processes and their controls? Philos Trans R Soc B Biol Sci. 2013;368:20130122. doi: 10.1098/rstb.2013.0122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Palmer K, Biasi C, Horn MA. Contrasting denitrifier communities relate to contrasting N2O emission patterns from acidic peat soils in arctic tundra. ISME J. 2012;6:1058–77. doi: 10.1038/ismej.2011.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siciliano SD, Ma WK, Ferguson S, Farrell RE. Nitrifier dominance of Arctic soil nitrous oxide emissions arises due to fungal competition with denitrifiers for nitrate. Soil Biol Biochem. 2009;41:1104–10. doi: 10.1016/j.soilbio.2009.02.024. [DOI] [Google Scholar]
- 40.Kool DM, Wrage N, Zechmeister-Boltenstern S, Pfeffer M, Brus D, Oenema O, et al. Nitrifier denitrification can be a source of N2O from soil: a revised approach to the dual-isotope labelling method. Eur J Soil Sci. 2010;61:759–72. doi: 10.1111/j.1365-2389.2010.01270.x. [DOI] [Google Scholar]
- 41.Kool DM, Dolfing J, Wrage N, Groenigen JWV. Nitrifier denitrification as a distinct and significant source of nitrous oxide from soil. Soil Biol Biochem. 2011;43:174–8. doi: 10.1016/j.soilbio.2010.09.030. [DOI] [Google Scholar]
- 42.Zhu X, Burger M, Doane TA, Horwath WR. Ammonia oxidation pathways and nitrifier denitrification are significant sources of N2O and NO under low oxygen availability. Proc Natl Acad Sci USA. 2013;110:6328–33. doi: 10.1073/pnas.1219993110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen HH, Mothapo NV, Shi W. The significant contribution of fungi to soil N2O production across diverse ecosystems. Appl Soil Ecol. 2014;73:70–77. doi: 10.1016/j.apsoil.2013.08.011. [DOI] [Google Scholar]
- 44.Aldossari N, Ishii A. Fungal denitrification revisited-Recent advancements and future opportunities. Soil Biol Biochem. 2021;157:108250. doi: 10.1016/j.soilbio.2021.108250. [DOI] [Google Scholar]
- 45.Xu XZ, Xu Y, Chen SC, Xu SG, Zhang HW. Soil loss and conservation in the black soil region of Northeast China: a retrospective study. Environ Sci Policy. 2010;13:793–800. doi: 10.1016/j.envsci.2010.07.004. [DOI] [Google Scholar]
- 46.Li H, Pei JB, Wang JK, Li SY, Gao GW. Organic carbon density and storage of the major black soil regions in Northeast China. J Soil Sci Plant Nut. 2013;13:883–93. doi: 10.4067/S0718-95162013005000070. [DOI] [Google Scholar]
- 47.Wang BW, Zhao XL, Wang X, Zhang ZX, Yi L, Hu SG. Spatial and temporal variability of soil erosion in the black soil region of Northeast China from 2000 to 2015. Environ Monit Assess. 2020;192:1–14. doi: 10.1007/s10661-020-08298-y. [DOI] [PubMed] [Google Scholar]
- 48.Ma X, Yin C, Wen B, Wang M, Wang G, Wang D, et al. The carbon reserves and emissions of peatlands in China, estimation of organic carbon reserves of peatlands in China. Beijing: Chinese Forestry Press; 2013.
- 49.Wang M, Wang SZ, Cao YW, Jiang M, Wang GD, Dong YM. The effects of hummock-hollow microtopography on soil organic carbon stocks and soil labile organic carbon fractions in a sedge peatland in Changbai Mountain, China. Catena. 2021;201:105204. doi: 10.1016/j.catena.2021.105204. [DOI] [Google Scholar]
- 50.Yang W, Zhao H, Chen XL, Yin SL, Cheng XL, An SQ. Consequences of short-term C4 plant Spartina alterniflora invasions for soil organic carbon dynamics in a coastal wetland of Eastern China. Ecol Eng. 2013;61:50–57. doi: 10.1016/j.ecoleng.2013.09.056. [DOI] [Google Scholar]
- 51.Zhu GB, Wang SY, Wang C, Zhou LG, Zhao SY, Li YX, et al. Resuscitation of anammox bacteria after >10,000 years of dormancy. ISME J. 2019;13:1098–109. doi: 10.1038/s41396-018-0316-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao SY, Wang XM, Pan HW, Wang SY, Zhu GB. High N2O reduction potential by denitrification in the nearshore site of a riparian zone. Sci Total Environ. 2022;813:152458. doi: 10.1016/j.scitotenv.2021.152458. [DOI] [PubMed] [Google Scholar]
- 53.Reck M, Tomasch J, Deng ZL, Jarek M, Husemann P, Wagner-Döbler I, et al. Stool metatranscriptomics: a technical guideline for mRNA stabilisation and isolation. BMC Genomics. 2015;16:494. doi: 10.1186/s12864-015-1694-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang XM, Wang SY, Jiang YY, Zhou JM, Han C, Zhu GB. Comammox bacterial abundance, activity, and contribution in agricultural rhizosphere soils. Sci Total Environ. 2020;727:138563. doi: 10.1016/j.scitotenv.2020.138563. [DOI] [PubMed] [Google Scholar]
- 55.He BS, Zhu RR, Yang HD, Lu QQ, Wang WW, Song L, et al. Assessing the impact of data preprocessing on analyzing next generation sequencing data. Front Bioeng Biotechnol. 2020;8:817. doi: 10.3389/fbioe.2020.00817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun X, Kop LFM, Lau MCY, Frank J, Jayakumar A, Lücker S, et al. Uncultured Nitrospina-like species are major nitrite oxidizing bacteria in oxygen minimum zones. ISME J. 2019;13:2391–402. doi: 10.1038/s41396-019-0443-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013;8:1494–512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fu LM, Niu BF, Zhu ZW, Wu ST, Li WZ. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–2. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peng, Y, Leung HCM, Yiu SM, Chin FYL. IDBA - A practical iterative de bruijn graph De Novo assembler. In: Annual International Conference on Research in Computational Molecular Biology Lisbon. 6044, 426–40 (2010). 10.1007/978-3-642-12683-3_28
- 60.Zhu W, Lomsadze A, Borodovsky M. Ab initio gene identification in metagenomic sequences. Nucleic Acids Res. 2010;38:132–132. doi: 10.1093/nar/gkq275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–60. doi: 10.1038/NMETH.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tu QC, Lin L, Cheng L, Deng Y, He ZL. NCycDB: a curated integrative database for fast and accurate metagenomic profiling of nitrogen cycling genes. Bioinformatics. 2019;35:1040–8. doi: 10.1093/bioinformatics/bty741. [DOI] [PubMed] [Google Scholar]
- 63.Uritskiy GV, DiRuggiero J, Taylor J. MetaWRAP-A flexible pipeline for genome-resolved metagenomic data analysis. Microbiome. 2018;6:1–13. doi: 10.1186/s40168-018-0541-1b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li D, Liu C, Luo R, Sadakane K, Lam T. MEGAHIT: An Ultra-fast single-node solution for large and complex metagenomics assembly via succinct de bruijn graph. Bioinformatics. 2015;31:1674–6. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 65.Kang DD, Li F, Kirton E, Thomas A, Egan R, An H, et al. MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. Peer J. 2019;7:7359. doi: 10.7717/peerj.7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu Y, Simmons BA, Singer SW. MaxBin 2.0: An Automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics. 2016;32:605–7. doi: 10.1093/bioinformatics/btv638. [DOI] [PubMed] [Google Scholar]
- 67.Alneberg J, Bjarnason BS, de Bruijin I, Schirmer M, Quick J, Ijaz UZ, et al. Binning metagenomic contigs by coverage and composition. Nat Methods. 2014;11:1144–6. doi: 10.1038/nmeth.3103. [DOI] [PubMed] [Google Scholar]
- 68.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–55. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Parks DH, Chuvochina M, Waite DW, Rinke C, Skarshewski A, Chaumeil PA, et al. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol. 2018;36:996–1004. doi: 10.1038/nbt.4229. [DOI] [PubMed] [Google Scholar]
- 70.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14:417–9. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bastian M. Gephi: an open source software for exploring and manipulating networks. In: Third international AAAI conference on weblogs and social media. 2009, vol 8, p. 361–2. 10.13140/2.1.1341.1520.
- 72.Hu H, Chen D, He J. Microbial regulation of terrestrial nitrous oxide formation: understanding the biological pathways for prediction of emission rates. FEMS Microbiol Rev. 2015;39:729–49. doi: 10.1093/femsre/fuv021. [DOI] [PubMed] [Google Scholar]
- 73.Schimel J, Balser TC, Wallenstein M. Microbial stress-response physiology and its implications for ecosystem function. Ecology. 2007;88:1386–94. doi: 10.1890/06-0219. [DOI] [PubMed] [Google Scholar]
- 74.Speth D, in’t Zandt MH, Guerrero-Cruz S, Dutilh BE, Jetten MSM. Genome-based microbial ecology of anammox granules in a full-scale wastewater treatment system. Nat Commun. 2016;7:11172. doi: 10.1038/ncomms11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pessi, IS, Viitamäki S, Rasimus EE, Delmont TO, Luoto M, Hultman J. Truncated denitrifiers dominate the denitrification pathway in tundra soil metagenomes. bioRxiv. 2020;419267. 10.1101/2020.12.21.419267.
- 76.Lehtovirta-Morley LE, Stoecker K, Vilcinskas A, Prosser JI, Nicol GW. Cultivation of an obligate acidophilic ammonia oxidizer from a nitrifying acid soil. Proc Natl Acad Sci USA. 2011;108:15892–7. doi: 10.1073/pnas.110719610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Peng YZ, Zhu GB. Biological nitrogen removal with nitrification and denitrification via nitrite pathway. Appl Microbiol Biotechnol. 2006;73:15–26. doi: 10.1007/s00253-006-0534-z. [DOI] [PubMed] [Google Scholar]
- 78.Werner C, Butterbach-Bahl K, Hass E, Hickler T, Kiese R. A global inventory of N2O emissions from tropical rainforest soils using a detailed biogeochemical model. Global Biogeochem Cy. 2007;21:GB3010. doi: 10.1029/2006GB002909. [DOI] [Google Scholar]
- 79.Harriss RC, Gorham E, Sebacher DI, Bartlett KB, Flebbe PA. Methane flux from northern peatlands. Nature. 1985;315:652–4. doi: 10.1038/315652a0. [DOI] [Google Scholar]
- 80.Martikainen P, Nykänen H, Crill P, Silvola J. Effect of a lowered water table on nitrous oxide fluxes from northern peatlands. Nature. 1993;366:51–53. doi: 10.1038/366051a0. [DOI] [Google Scholar]
- 81.Liimatainen M, Voigt C, Martikainen PJ, Hytönen J, Regina K, Óskarsson H, et al. Factors controlling nitrous oxide emissions from managed northern peat soils with low carbon to nitrogen ratio. Soil Biol Biochem. 2018;122:186–95. doi: 10.1016/j.soilbio.2018.04.006. [DOI] [Google Scholar]
- 82.Tian HQ, Xu RT, Canadell JG, Thompson RL, Winiwarter W, Suntharalingam P, et al. A comprehensive quantification of global nitrous oxide sources and sinks. Nature. 2020;586:248–56. doi: 10.1038/s41586-020-2780-0. [DOI] [PubMed] [Google Scholar]
- 83.Gil J, Pérez T, Boering K, Martikainen PJ, Biasi C. Mechanisms responsible for high N2O emissions from subarctic permafrost peatlands studied via stable isotope techniques. Global Biogeochem Cy. 2017;31:172–89. doi: 10.1002/2015GB005370. [DOI] [Google Scholar]
- 84.Siljanen HMP, Alves RJE, Ronkainen JG, Lamprecht RE, Bhattarai HR, Bagnoud A, et al. Archaeal nitrification is a key driver of high nitrous oxide emissions from arctic peatlands. Soil Biol Biochem. 2019;137:107539. doi: 10.1016/j.soilbio.2019.107539. [DOI] [Google Scholar]
- 85.Kozlowski J, Stieglmeier M, Schleper C, Klotz MG, Stein LY. Pathways and key intermediates required for obligate aerobic ammonia-dependent chemolithotrophy in bacteria and Thaumarchaeota. ISME J. 2016;10:1836–45. doi: 10.1038/ismej.2016.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Clymo RS, Hayward PM. The ecology of sphagnum. In: Bryophyte Ecology. Springer Netherlands; 1982, p. 229–89. 10.1007/978-94-009-5891-3_8.
- 87.Malmer N, Svensson BM, Wallén B. Interactions between sphagnum mosses and field layer vascular plants in the development of peat-forming systems. Folia Geobot Phytotaxonomica. 1994;29:483–96. doi: 10.1007/BF02883146. [DOI] [Google Scholar]
- 88.Morley N, Baggs EM, Dörsch P, Bakken L. Production of NO, N2O and N2 by extracted soil bacteria, regulation by NO2- and O2 concentrations. FEMS Microbiol Ecol. 2008;65:102–12. doi: 10.1111/j.1574-6941.2008.00495.x. [DOI] [PubMed] [Google Scholar]
- 89.Senbayram M, Chen R, Budai A, Bakken L, Dittert K. N2O emission and the N2O/(N2O+N2) product ratio of denitrification as controlled by available carbon substrates and nitrate concentrations. Agr Ecosyst Environ. 2012;147:4–12. doi: 10.1016/j.agee.2011.06.022. [DOI] [Google Scholar]
- 90.Chen SM, Wang FH, Zhang YM, Qin SP, Wei SC, Wang SQ, et al. Organic carbon availability limiting microbial denitrification in the deep vadose zone. Environ Microbiol. 2018;20:980–92. doi: 10.1111/1462-2920.14027. [DOI] [PubMed] [Google Scholar]
- 91.Yang GB, Peng YF, Marushchak ME, Chen YL, Wang GQ, Li F, et al. Magnitude and pathways of increased nitrous oxide emissions from uplands following permafrost thaw. Environ Sci Technol. 2018;52:9162–9. doi: 10.1021/acs.est.8b02271. [DOI] [PubMed] [Google Scholar]
- 92.Mackelprang R, Waldrop MP, DeAngelis KM, David MM, Chavarria, K L, et al. Metagenomic analysis of permafrost microbial community response to thaw. Nature. 2011;480:368–71. doi: 10.1038/nature10576. [DOI] [PubMed] [Google Scholar]
- 93.Hazard C, Prosser JI, Nicol GW. Use and abuse of potential rates in soil microbiology. Soil Biol Biochem. 2021;157:108242. doi: 10.1016/j.soilbio.2021.108242. [DOI] [Google Scholar]
- 94.Monteux S, Keuper F, Fontaine S, Gavazov K, Hallin S, Juhanson J, et al. Carbon and nitrogen cycling in Yedoma permafrost controlled by microbial functional limitations. Nat Geosci. 2020;13:794–8. doi: 10.1038/s41561-020-00662-4. [DOI] [Google Scholar]
- 95.Turetsky M, Wieder K, Halsey L, Vitt D. Current disturbance and the diminishing peatland carbon sink. Geophys Res Lett. 2002;29:11. doi: 10.1029/2001GL014000. [DOI] [Google Scholar]
- 96.Cui Q, Song C, Wang X, Shi F, Yu X, Tan W. Effects of warming on N2O fluxes in a boreal peatland of Permafrost region, Northeast China. Sci Total Environ. 2018;616-617:427–34. doi: 10.1016/j.scitotenv.2017.10.246. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The metagenomic data and the metatranscriptomic data were deposited in the NCBI Sequence Read Archive under accession numbers SAMN26424243–SAMN26424272 and SRR14251110–SRR14251136, respectively.