Abstract
Vitamin B12 (cobalamin, herein B12) is an essential cofactor involved in amino acid synthesis and carbon resupply to the TCA cycle for most prokaryotes, eukaryotic microorganisms, and animals. Despite being required by most, B12 is produced by only a minor fraction of prokaryotes and therefore leads to complex interaction between prototrophs and auxotrophs. However, it is unknown how B12 is provided by prototrophs to auxotrophs. In this study, 33 B12 prototrophic alphaproteobacterial strains were grown in co-culture with Thalassiosira pseudonana, a B12 auxotrophic diatom, to determine the bacterial ability to support the growth of the diatom by sharing B12. Among these strains, 18 were identified to share B12 with the diatom, while nine were identified to retain B12 and not support growth of the diatom. The other bacteria either shared B12 with the diatom only with the addition of substrate or inhibited the growth of the diatom. Extracellular B12 measurements of B12-provider and B12-retainer strains confirmed that the cofactor could only be detected in the environment of the tested B12-provider strains. Intracellular B12 was measured by LC-MS and showed that the concentrations of the different B12-provider as well as B12-retainer strains differed substantially. Although B12 is essential for the vast majority of microorganisms, mechanisms that export this essential cofactor are still unknown. Our results suggest that a large proportion of bacteria that can synthesise B12 de novo cannot share the cofactor with their environment.
Subject terms: Water microbiology, Microbial ecology, Symbiosis, Microbial ecology, Metabolomics
Introduction
Vitamin B12 (cobalamin, herein B12) is a water-soluble cobalt-containing compound and is required by the vast majority of prokaryotic and about half of the eukaryotic marine microorganisms that are isolated or genome sequenced [1]. B12 functions as coenzyme for the methylcobalamin-dependent methionine synthase and adenosylcobalamin-dependent methylmalonyl-CoA mutase, which are involved in amino acid synthesis and carbon resupply to the TCA cycle, respectively [2, 3]. However, de novo synthesis can only be carried out by a minor fraction of prokaryotes [1, 4, 5]. More than 30 genes are required for the complete biosynthesis of this important cofactor, which makes up about 1% of an average bacterial genome [4, 6]. This is an energetically and metabolically expensive biosynthetic process, which may explain why considerably fewer than half of all prokaryotes encode the genes for complete biosynthesis of B12 or other cobamides [1, 4, 7, 8]. To gain an evolutionary benefit in an environment where there is still a sustainable supply from distinct substrates or growth factors such as vitamins, loss of biosynthetic genes often occurs in bacteria, leading to a reduction in the size of the genome, known as genome streamlining [9, 10]. This process is believed to reduce the metabolic cost and thus provide a selective advantage, as long as sufficient quantities of the essential compound are freely available. Less than 10% of soil bacteria are capable of de novo synthesise of B12 [8]. In marine ecosystems only select heterotrophic bacteria and Thaumarchaeota can produce it [11] and the share of B12 prototrophs can be as low as one-fifth of the bacterial community, so that the vast majority of microorganisms depend on the cofactor [7]. This gap between supply and demand of B12 can result in complex microbial interactions between prokaryotes and eukaryotes [12–16]. Approximately half of known phytoplankton encode the B12-dependent methionine synthase (metH) [17], which is why they require this pivotal cofactor from the environment. Concentrations of dissolved B12 undergo strong fluctuations in the sea, are in some cases below the detection limit of a few pM, and their presence has been shown to influence marine microbial communities [11, 18–23]. In marine environments, bacteria often live in close association with phytoplankton [24, 25]. In several studies, the provision of B12 by individual heterotrophic bacteria to B12 auxotrophic phytoplankton in exchange for organic carbon was demonstrated [13, 14, 16, 26–29]. Yet, it is still debatable whether this exchange of metabolic products represents a mutualistic symbiosis between B12 prototrophs and B12 auxotrophic, microbial eukaryotes, or whether its release is unintentional [13, 30, 31]. In fact, mechanisms that lead to the provision of the cofactor and factors that favor this exchange are still largely unknown. Despite varying lower and upper ligands attached to the corrinoid ring, B12-family metabolites, including cobalamin, are always larger than 1,350 Daltons. Therefore, diffusion through the cell membrane appears to be almost impossible [32]. The relatively well studied uptake of B12 by gram-negative bacteria, requires the binding of B12 to the outer membrane protein BtuB. Then, B12 is transported into the cell by the inner membrane protein complex TonB via an electrochemical gradient of protons [33]. The mechanism of B12 export, essential for microbial interactions, still remains unexplored.
Given the available knowledge, we hypothesise that not all B12 prototrophic bacteria share the essential cofactor with other microorganisms. Our aim is to provide first indications of the requirement of an active export mechanism in order to draw conclusions on B12 driven mutualistic interactions and the global provision of B12 in marine ecosystems.
In order to achieve this goal, we co-cultured Thalassiosira pseudonana, a B12 auxotrophic diatom, with 33 B12 prototrophic bacteria of the alphaproteobacterial class to test the bacterial ability to share B12 with other microorganisms. Furthermore, we determined intra- and extracellular B12 concentrations of B12-provider and B12-retainer strains by means of liquid chromatography coupled with mass spectrometry (LC-MS) [34] and studied patterns that both B12-provider and B12-retainer strains had in common.
Methods and materials
Identification of B12 prototrophs
To study the ability of heterotrophic bacteria to share B12 with surrounding microorganisms, we selected 33 prototrophic marine representatives. As a prerequisite, all selected bacteria had to grow on synchronized artificial seawater (syn-ASW) media (Supplementary table S1) as it promotes the growth of most phototrophic eukaryotes as well as prokaryotes with equal growth facilitating conditions. Further, the genome sequence had to be complete and accessible at IMG (integrated microbial genomes; https://img.jgi.doe.gov/cgi-bin/mer/main.cgi). The ability for de novo B12 synthesis was verified based on a complete B12 pathway and growth (determined by OD) in minimal medium without the addition of B12. The genetic verification of the B12 biosynthesis pathway was assumed if at least 95% of the B12 biosynthesis pathway of a strain was annotated (Supplementary table S2). As B12 auxotrophic (recipient) organism, we selected the genome sequenced diatom, T. pseudonana (CCMP 1335).
B12 cross-feeding co-culture experiment
To establish a B12 deficient diatom culture, T. pseudonana was first cultured in F/2 media and subsequently transferred (twice) to B12-free syn-ASW media (supplemented with thiamine (vitamin B1), riboflavin (vitamin B2), nicotinic acid (vitamin B3), pantothenic acid (vitamin B5), pyridoxine hydrochloride (vitamin B6), and biotin (vitamin B7); 500 pM each). Once B12 was depleted, the final diatom pre-culture for the inoculation of the main experiment was supplemented with 10 pM B12 to ensure growth, yet growth was limited upon B12 depletion. Bacterial pre-cultures were grown in Marine Broth (MB) media at 20 °C, 100 rpm. Cultures of the late exponential growth phase were washed three times (5974 g, five minutes) with B12-free syn-ASW media prior to inoculation. All diatom pre-cultures and the experimental co-cultures were illuminated at 70 µE m-2 s−1 and incubated at 20 °C with a 12:12 h light-dark cycle (RUMED). The diatom-bacteria co-cultures were grown at three varying conditions. First, bacterial isolates were co-cultured with the diatom T. pseudonana without further additions to determine whether growth of T. pseudonana upon bacterial B12 release is enabled. Second, to eliminate the possibility that individual bacterial isolates are unable to utilize diatom derived organic carbon and thus are unable to share B12, the co-culture was supplemented with an organic carbon mixture (120 µM C), containing glucose, glutamate, and acetate (each substrate at 40 µM C). Third, B12 (1 nM) was added to the bacteria-diatom co-culture, to ensure that the growth of the diatom was not inhibited by the bacteria, thus resulting in a false consideration as B12-retainer. Alongside each experimental run, a negative control, axenic T. pseudonana grown without B12 addition and a positive control, T. pseudonana grown with the addition of 1 nM B12 was considered. All treatments were run as triplicates. To ensure that only the bacterially provided B12, but not the possible provision of methionine, enables the growth of T. pseudonana in co-culture, we cultivated T. pseudonana with only the addition of methionine and did not observe growth (Supplementary Fig. S1). For all co-culture treatments containing bacteria, the initial bacterial inoculum was calculated to be at 500,000 cells ml−1 (based on flow cytometric cell counts), initial T. pseudonana cells were estimated to be about 4,000 cells ml−1 (microscopic enumeration). Bacterial-diatom co-cultures were illuminated at 70 µE m−2 s−1 and incubated at 20 °C with a 12:12 h light-dark cycle. Growth of T. pseudonana was determined throughout the experiment by means of relative fluorescence (TD 700 fluorometer, Turner Designs, California, USA). Samples for diatom and bacterial cell count were collected after inoculation and during the early stationary growth phase of T. pseudonana. For bacterial cell counts, samples were fixed with GDA at a final concentration of 1%, incubated at 4 °C for 30 min, and stored at −20 °C until enumeration by flow cytometry [35]. Diatom samples for cell enumeration were fixed with lugol (final concentrations of 0.15% iodine and 0.29% potassium iodide) and stored at 4 °C until further analysis.
Enumeration of bacteria and diatom
Prior to counting with the flow cytometer, bacterial cells were detached from the diatom cells using glass beads (2.3 mm) and ultrasonication (35 °C, 70%, 4 ×5 minutes, Sonorex digital 10P, Bandelin, Berlin, Germany) following by a short vortexing step (2 × 2 seconds, Vortex Genie2, Scientific Industries Inc., New York, USA) after each ultrasonic interval. This method was a further development of the detachment method described elsewhere [36]. Afterwards, bacterial cells were stained with SybrGreen I and enumerated by flow cytometry (BD Accuri C6, BD biosciences, Franklin Lakes NJ, USA) as described elsewhere [35]. Diatom samples, fixed with lugol, were loaded on a hemocytometer and enumerated by microscopy (AXIO, Lab.A1, objective lens Carl Zeiss, 40x).
Measurement of intra- and extracellular B12 concentrations
Intracellular B12 concentrations of 20 bacterial strains were measured using LC-MS. Selected strains were grown in B12-free syn-ASW media (see above) and supplemented with an organic substrate mix (30 mM C) containing glucose, acetate, and glutamate (each having 10 mM C). Cell pellets of 2 × 50 ml culture were harvested from each replicate during the late exponential or early stationary growth phase, monitored by means of optical density (OD; Tables 1 and 2, Supplementary Fig. S2 and S3). The samples were then washed twice with B12-free syn-ASW medium (3,213 g, five minutes at 4 °C) and cell pellets were stored at −20 °C until further analysis. Alongside, samples for bacterial cell counts were withdrawn, fixed with GDA (final concentration 1%), and enumerated by flow cytometry (see above).
Table 1.
Intra- and extracellular vitamin B12 concentrations obtained from B12-provider strains when grown in mono-culture and growth rates of T. pseudonana in co-culture with respective bacterial strain without and with the addition of B12 and with the addition of B12 in monoculture.
| B12 provider strains | Strain designation | Cell collection method used | Intracellular B12 molecules/ cell | Extracellular B12 molecules / cell | Growth rate of T. pseudonana grown in co-culture without addition of B12 (day) | Growth rate of T. pseudonana grown in co-culture with addition of B12 (day) | Growth rate of T. pseudonana only with addition of B12 (day) |
|---|---|---|---|---|---|---|---|
| Antarctobacter heliothermus EL-219 | DSM 11445 | Cell pellet | 753 ± 232 | N/A | 3.7 ± 1.4 | 2.6 ± 0.04 | 2.8 ± 0.02 |
| Dinoroseobacter shibae | DSM 16493 | Filtration | 7,194 ± 1,105 | N/A | 9.6 ± 0.9 | 8.8 ± 0.3 | 8.1 ± 0.1 |
| Marinovum algicola FF3 | DSM 10251 | Cell pellet | 7,171 ± 792 | 936 ± 363 | 2.8 ± 0.04 | 2.8 ± 0.04 | 2.8 ± 0.02 |
| Nautella italica R11 | DSM 26436 | Cell pellet | 1,986 ± 273 | N/A | 5.6 ± 0.2 | 2.7 ± 0.02 | 2.8 ± 0.02 |
| Phaeobacter inhibens | DSM 17395 | Cell pellet | 2,821 ± 540 | 11 ± 3 | 10.3 ± 1.1 | 8.2 ± 1.1 | 8.1 ± 0.1 |
| Phaeobacter inhibens T5 | DSM 16374 | Cell pellet | 664 ± 88 | N/A | 5.6 ± 0.5 | 2.5 ± 0.05 | 3.4 ± 0.4 |
| Ponticoccus litoralis CL-GR66 | DSM 18986 | Cell pellet | 4,622 ± 2,227 | N/A | 6.5 ± 1.2 | 3.8 ± 0.04 | 3.4 ± 0.4 |
| Aliiroseovarius crassostreae CV919-312 | DSM 16950 | Cell pellet | 26,619 ± 13,140 | N/A | 5.8 ± 1.7 | 4.7 ± 0.9 | 3.4 ± 0.4 |
| Roseovarius mucosus DFL-24 | DSM 17069 | Cell pellet | 4,028 ± 708 | N/A | 6.0 ± 0.1 | 3.8 ± 0.06 | 3.4 ± 0.4 |
| Silicibacter sp. | TM1040 | Cell pellet | 15,214 ± 3,566 | N/A | 7.7 ± 0.5 | 4.3 ± 1.0 | 3.4 ± 0.4 |
| Sulfitobacter sp. | DFL-14 | Cell pellet | 1546 ± 124 | N/A | 5.9 ± 0.8 | 5.4 ± 1.2 | 3.4 ± 0.4 |
| Sulfitobacter sp. | M22 | Filtration | 13,982 ± 6,646 | N/A | 2.9 ± 0.2 | 2.7 ± 0.1 | 2.8 ± 0.02 |
*N/A not available.
Table 2.
Intra- and extracellular vitamin B12 concentrations obtained from B12-retainer strains when grown in mono-culture and growth rates of T. pseudonana in co-culture with respective bacterial strain with the addition of B12 and with the addition of B12 in mono-culture.
| B12 retainer strains | Strain designation | Cell collection method used | Intracellular B12 molecules/ cell | Extracellular B12 molecules / cell | Growth rate of T. pseudonana grown in co-culture without addition of B12 (day) | Growth rate of T. pseudonana grown in co-culture with addition of B12 (day) | Growth rate of T. pseudonana only with addition of B12 (day) |
|---|---|---|---|---|---|---|---|
| Pseudodongicola xiamenensis Y2 | DSM 18339 | Filtration | 2,656 ± 421 | ND | N/A | 7.2 ± 1.5 | 8.1 ± 0.1 |
| Jannaschia helgolandensis Hel10 | DSM 14858 | Filtration | ND | ND | N/A | 2.3 ± 0.03 | 3.8 ± 0.9 |
| Loktanella salsilacus R-8904 | DSM 16199 | Cell pellet | 671 ± 281 | N/A | N/A | 2.8 ± 0.02 | 2.8 ± 0.02 |
| Phaeobacter gallaeciensis BS 107 | DSM 26640 | Filtration | ND | N/A | N/A | 2.8 ± 0.01 | 2.8 ± 0.02 |
| Sulfitobacter sp. | DFL-23 | Filtration | ND | N/A | N/A | 7.5 ± 0.1 | 8.1 ± 0.1 |
| Sulfitobacter sp. | M39 | Filtration | 4,599 ± 122 | N/A | N/A | 2.7 ± 0.04 | 2.8 ± 0.02 |
| Loktanella sp. | M215 | Cell pellet | ND | N/A | N/A | 2.4 ± 0.00 | 3.8 ± 0.9 |
| Sulfitobacter sp. | M220 | Cell pellet | 4,558 ± 106 | N/A | N/A | 2.8 ± 0.08 | 2.8 ± 0.02 |
*ND not detected, N/A not available.
To also analytically analyse whether B12 prototrophic bacteria share B12, we sampled the exometabolome of four representative bacterial strains. We chose two isolates (Marinovum algicola FF3 DSM 10251, Phaeobacter inhibens DSM 17395) that promoted the growth of T. pseudonana in co-culture, while the other two isolates (Pseudodongicola xiamenensis DSM 18339, Jannaschia helgolandensis DSM 14858) did not. The isolates were grown as described above and growth was monitored by OD. The exudate was collected by filtering the culture with 0.2 µm filter (Sartorius, Minisart syringe filter) during late exponential or early stationary growth phase. Samples were stored at -20 °C until further analysis.
Bacterial cell pellets for intracellular B12 analysis were extracted with bead beating, as described elsewhere [37]. To assess the recoveries, the work-up procedure was performed with known amounts of reference standards of the respective vitamins, and the amounts after the work-up were compared to the theoretical amounts without losses for each analyte. Recoveries of the different B12 forms (cyano-, adenosyl-, methyl- and hydroxycobalamin) were 97-99%. Extracellular B12 was concentrated on a solid phase extraction column (HLB, 1 g, Macherey-Nagel) at pH 6 and eluted with methanol [38]. The solvent extracts were dried down under nitrogen stream and redissolved in 200 µl of water. Concentrations of individual intra- and extracellular B12 forms were analyzed by LC-MS as described elsewhere [34] and summarized as total B12. For HPLC separation with an Ultimate 3000 (ThermoFisher Scientific) on a Kinetex Evo C18 column (100 × 2.1 mm, 2.6 µm pore size, Phenomenex, Torrance, CA, USA) 10 mM ammonium formate (pH 6.0) (A) and acetonitrile (B) were used with the following solvent gradient: 0–13 min 100–75% A; 13–15 min 75–0% A; 15–19 min 0% A; 19–21 min 0–100% A; 21–26 min 100% A. Parameters for selected reaction monitoring mode on a TSQ Quantum Ultra triple quadrupole mass spectrometer (ThermoFisher Scientific) can be found in supplementary table S3.
Determination of the B12 requirement of T. pseudonana
In order to get a better insight into the B12 requirement of T. pseudonana, we grew the axenic diatom in cultures of triplicates in syn-ASW-medium with the addition of different B12 concentrations (5, 10, 25, 50, and 100 pM) and without any addition. All cultures were illuminated at 70 µE m−2 s−1 and incubated at 20 °C with a 12:12 h light-dark cycle. The growth was determined every two to three days by means of relative fluorescence and the determination of the cell numbers (see above).
Results
Growth of T. pseudonana at varying B12 concentrations
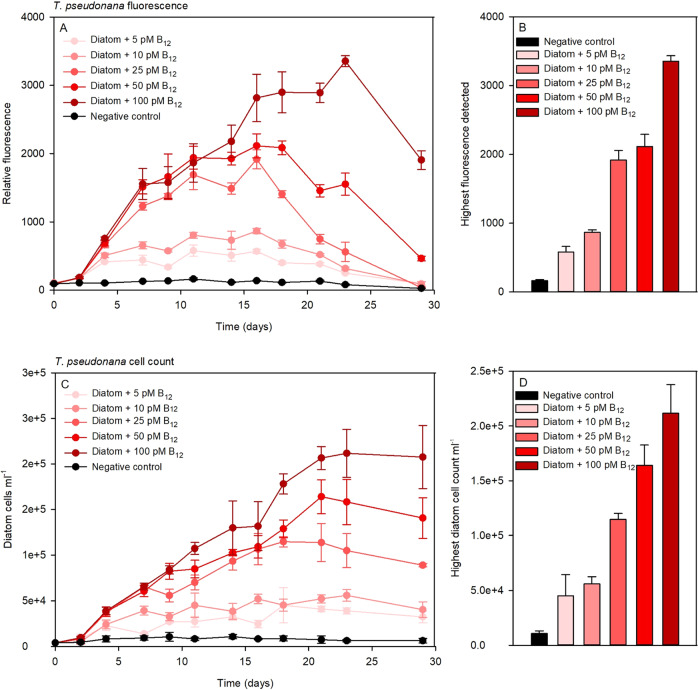
We observed similar growth yield patterns by adding different B12 concentrations both by relative fluorescence (Fig. 1A) and by cell number determination (Fig. 1C). However, especially in the later growth phase (days 18 - 29), a strong decrease in relative fluorescence occurred, whereas at the same time points microscopically counted cell numbers still increased strongly (Fig. 1A, C). Highest relative fluorescence and also T. pseudonana cell density was achieved with the addition of 100 pM B12 (Fig. 1B, D). All other measured values rank according to the added concentration. Even the addition of fairly low B12 concentrations (five pM) resulted in significant growth compared to the negative control, which was detected by means of relative fluorescence as well as cell enumeration.
Fig. 1. T. pseudonana vitamin B12 bioassay.
Shown is the growth of T. pseudonana at varying B12 concentrations, monitored by relative fluorescence (A, B) and cell count (C, D). In A the Y-axis represents the relative fluorescence and in C the Y-axis represents diatom cell count/ml. Depicted are maximum growth determined by relative fluorescence (B) and cell count (D) of the diatom at respective B12 concentrations.
Growth of T. pseudonana in co-culture
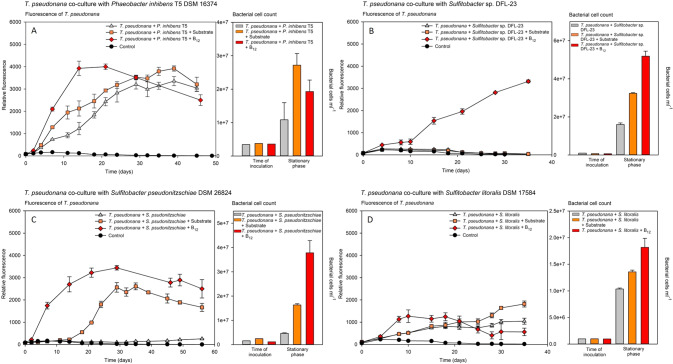
To analyze the B12 sharing between B12 prototrophic bacteria and the auxotrophic diatom T. pseudonana, both microorganisms were co-cultured. Among 33 B12 prototrophic bacterial strains, 18 promoted the growth of the diatom. Growth of T. pseudonana in co-culture with B12-providing bacteria mostly achieved the same growth yield as the positive control, where the alga was grown with addition of 1 nM B12, however with a slightly delayed growth (Fig. 2A, Fig. 3 and Supplementary Fig. S4). In the following, this group of bacteria is referred to as B12-provider (Table 1). Co-cultivation with nine other B12 prototrophic bacteria did not result in distinct growth of the diatom, although the bacterial cell counts increased significantly over the course of the co-culture. The addition of substrate to exclude the possibility that the respective bacteria cannot utilise the diatom derived dissolved organic carbon did not lead to growth of the diatom either (Fig. 2B, Fig. 4, and Supplementary Fig. S5). However, the additional supply of B12 to the co-culture led to growth of T. pseudonana, which eliminates possible growth inhibition of the diatom induced by the tested bacteria. The group of these B12 prototrophic bacteria is hereafter referred to as B12-retainer (Table 2). Apart from the B12-provider and B12-retainer strains, co-cultivation with six additional B12 prototrophic bacterial isolates did not show distinct results, that would clearly favour one of the two groups. Five bacterial isolates were particularly growth-promoting when additional substrate was added to the co-culture. This observation suggests that these bacterial isolates cannot utilise the diatom derived DOM. In fact, most of these isolates (four out of five) were isolated from a source other than diatoms (Supplementary table S4). It is quite possible that these bacteria also share the synthesised B12 with their environment (Fig. 2C and Supplementary Fig. S6 and S7). Nevertheless, we have not considered this group for follow-up analysis. In co-cultivation with one bacterial strain, S. litoralis, the growth of T. pseudonana was inhibited under all three culture conditions. Growth yield of T. pseudonana remained at only half the level seen when T. pseudonana was grown in monoculture with the addition of B12 (Fig. 2D). Again, it can be assumed that S. litoralis shares B12 with the diatom, but again, we did not take this strain into account for further investigations.
Fig. 2. Growth of T. pseudonana in co-culture with B12 prototrophic bacteria.
Representative co-cultures of T. pseudonana with B12 prototrophic bacteria that provide B12 (A), retain B12 (B), provide only when substrate is available (C) and likely provide B12 while inhibiting growth (D). (left panels, growth curves) Growth of T. pseudonana monitored by relative fluorescence unit (RFU) over time with additions of substrate mix (orange square), B12 (red diamond), or without addition of either (grey triangle). (Right panels; bar plots) Bacterial cell counts in co-cultures at the time of inoculation and early stationary growth phase of T. pseudonana. A P. inhibens T5 supports the growth of T. pseudonana by providing B12 (Further examples in Fig. S4); B Sulfitobacter sp. DFL-23 retains B12 and does not support growth of the diatom (Further examples in Fig. S5); C S. pseudonitzschiae provides B12 only with additions of a substrate mix (Further examples in Fig. S6); and D S. litoralis provides B12 and inhibits growth of T. pseudonana.
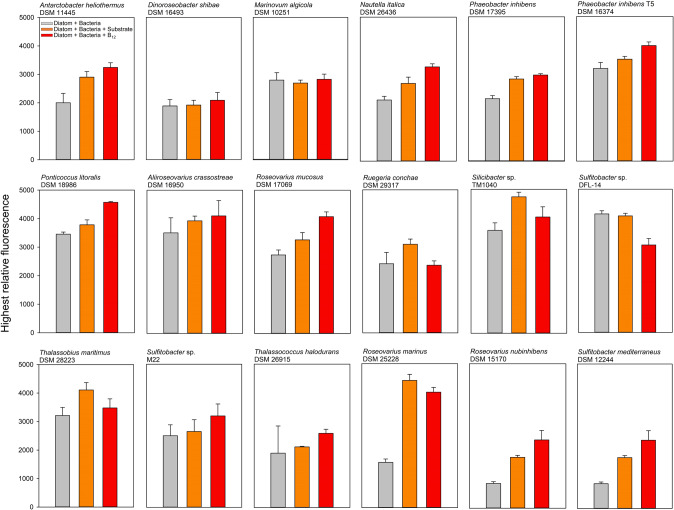
Fig. 3. Maximum growth of T. pseudonana in co-culture with B12-provider.
Bars represent the maximum relative fluorescence of T. pseudonana during growth in co-culture with 18 different B12-providers under different growth conditions (corresponding growth curves can be seen in the appendix). Grey bars represent maximum relative fluorescence of T. pseudonana in co-cultures without further additions, orange co-cultures with an additional substrate mix and red the co-cultures with B12 additions.
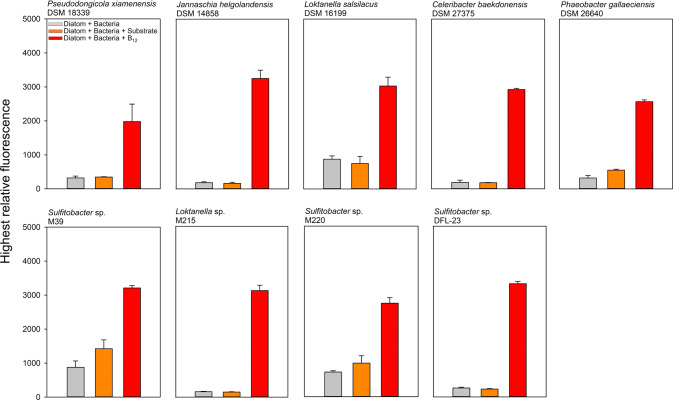
Fig. 4. Maximum growth of T. pseudonana in co-culture with B12-retainer.
Bars represent the maximum relative fluorescence of T. pseudonana during growth in co-culture with nine different B12-retainers under different growth conditions (corresponding growth curves can be seen in the appendix). Grey bars represent maximum relative fluorescence of T. pseudonana in co-cultures without further additions, orange co-cultures with an additional substrate mix and red the co-cultures with B12 additions.
Growth characteristics of bacteria and T. pseudonana in co-culture
In most B12-provider-diatom co-cultures without supplementations of substrate or B12, T. pseudonana achieved the same growth yield as with the additional supply of B12, however, mostly with a slight delay in growth (Fig. 2A and Supplementary Fig. S4). Growth rates in most co-cultures were fastest with the addition of B12, followed by co-cultures with substrate additions and mostly slowest in co-cultures without any addition. The only exception was Sulfitobacter sp. DFL-14, in which the growth rate of T. pseudonana was fastest in co-culture without the addition of B12 compared to the one with B12 (Supplementary Fig. S4). In all cultures with B12-provider strains, bacterial cell counts in the late exponential or early stationary growth phase were distinctly above those of the time point of inoculation (Fig. 2A and Supplementary Fig. S4). Bacterial cell counts of all these co-cultures for the treatments with substrate and B12 addition were mostly in the same order of magnitude, whereas co-cultures without further additions were mostly slightly below these values. The only exception was observed for the co-cultures with A. crassostreae DSM 16950, in which the highest bacterial cell counts were detected in the co-culture without further additions (Supplementary Fig. S4).
In the B12-retainer-diatom co-cultures, we observed considerable differences in growth rates and yield in co-cultures with B12 addition. The significantly increased growth yield of T. pseudonana when co-cultured with C. baekdonensis DSM 27375 and B12 additive was very noticeable (Supplementary Fig. S5). The relative fluorescence of this co-culture was almost twice as high as compared to others. In most of the B12-retainer-diatom co-cultures, the detected relative fluorescence of T. pseudonana without any and with substrate addition was comparable to negative control values of T. pseudonana, when cultivated axenically without B12 addition. Only a slight growth of T. pseudonana in co-culture with the B12-retainers L. salsilacus, Sulfitobacter sp. M39, and Sulfitobacter sp. M220 was observed (Supplementary Fig. S5). Due to the low growth, which only became apparent in the later course of the growth curve, we nevertheless classified these strains as B12-retainers. For all B12-retainer-diatom co-cultures, the bacterial cell counts sampled at the stationary phase (B12 addition) or at the end of the experiment (no addition and substrate addition) were significantly higher than the measurements at the time of inoculation (Fig. 2B and Supplementary Fig. S5). Only in B12-retainer-diatom co-cultures (without any addition), J. helgolandensis DSM 14858, Loktanella sp. M215, and P. gallaciensis, a slight to no increases in cell numbers was detected, yet an increase in cell numbers with substrate addition was detected in all co-cultures (Supplementary Fig. S5). The bacterial strains studied, divided into the groups of B12-providers and B12-retainer, are listed in supplementary table S4 with their known habitats or isolation sites. Here it can be seen that especially bacteria of the B12-provider group were isolated from or are mostly living in association with eukaryotic microorganisms.
Intra- and extracellular B12 concentration
Twenty of the bacteria that we identified as either B12-provider or B12-retainer were grown again in monoculture with the addition of substrate, to determine the intracellular concentration of B12. The growth yield of some B12-retainer strains was significantly lower, which is why their biomass sampling yield was significantly lower as well. Detected B12 concentrations were normalised against cell numbers to better distinguish between cultures with different growth rates and yields. In some cases, the intracellular B12 values differed immensely, with 40-fold deviations within the B12-provider strains. When comparing intracellular B12 values of individual B12-provider strains to their ability to impact the growth rate of T. pseudonana in co-culture through their release of B12, we cannot discern a direct correlation (Table 1). In B12-retainer strains, we were unable to detect B12 in four out of eight bacterial cultures (Table 2). Detected B12 values varied between 671 to 4,599 B12 molecules per cell. The four detected intracellular B12 values of B12-retainer strains were comparable to the average values measured for the B12-providers (Tables 1 and 2).
Extracellular B12 was measured additionally in two selected bacterial strains from the groups of B12-provider and B12-retainer, each of which exhibited a comparably high growth yield. B12 was detected in both B12-provider cultures (M. algicola and P. inhibens), while no B12 was measured in both B12-retainer cultures (P. xiamenensis and J. helgolandensis, Tables 1 and 2). Extracellular B12 concentrations of the two B12-provider strains were approximately 8 and 256 times lower than the corresponding intracellularly detected values (Table 1). However, when evaluating these values and drawing conclusions for the observations from the co-cultures, it must be considered that the values were obtained from monocultures with a significantly shortened growth phase. B12 production by prototrophic bacteria can vary in co-culture with the diatom, as it is known that algal metabolism upregulates bacterial production of B12 [13].
Discussion
Vitamin B12 biosynthesis potential of different bacteria
B vitamins play a key role in complex marine microbial interactions as they are obligatory cofactors in various essential metabolic reactions in all living organism [13, 14, 39–41]. An exciting fact about B12 is that genes for synthesis of this complex cofactor have never made the transition to the eukaryotic kingdom, although it is required by both prokaryotes and eukaryotes. De novo synthesis is restricted to a minor fraction of bacteria and archaea, thus, suggesting that the ability to synthesise B12 is disproportionate to its demand in nature [1, 4]. This phenomenon can be observed in various habitats, for example in the soil microbiome, where the proportion of B12 producers is less than one tenth [8]. Similar findings have been shown for the microbiome on human skin, where only 1% of the core species are predicted to produce B12 de novo, while 39 % of the species are predicted to use B12 for metabolism [42]. In order to adequately answer this fundamental question regarding the balance between B12 availability and consumption, we should aim to better understand the synthesis potential of individual prototrophic prokaryotes.
Here we present intra- and extracellular B12 concentrations of various B12 prototrophic, alphaproteobacterial strains. The concentration of intracellular B12 differs widely between the various heterotrophic bacteria examined. Converted, B12 molecules detected per cell ranged between 664 to 26,619 in the analysed bacterial cultures, including B12-provider and B12-retainer. Such strong variation in intracellular B12 concentrations have already been shown for a number of other prokaryotes, including Archaea, heterotrophic bacteria, and cyanobacteria [11, 34]. Also, in these studies, the detected intracellular B12 values differed up to three orders of magnitude and showed values similar to the ones we detected. Whether factors such as cell size, which we did not consider in our analysis, or the exact growth phase in which we took the samples had an influence on the strong variation cannot be clarified here. It is quite conceivable that different B12 requirements of the individual cells or different regulatory mechanisms of B12 synthesis played a decisive role for the intracellular B12 concentrations. Nevertheless, we can conclude that not only the genetic B12 biosynthetic potential within a microbial community is decisive, but rather which prokaryote is actually present is crucial for the availability of B12.
The extracellular concentrations of B12 detected in M. algicola and P. inhibens were about 8 and 256 times lower than respective intracellular levels. For example, M. algicola secreted about 936 B12 molecules per cell, which was roughly 85 times more as detected for P. inhibens. On the basis of the detected B12 demand of T. pseudonana determined by the bioassay, we can calculate that the eukaryote requires roughly 135,000 B12 molecules per cell, if we base the limitation of cell number solely on B12 availability. Thus, it would take about 144 living M. algicola cells that release B12 to cover the requirements for the growth of one T. pseudonana cell. In fact, the bacterial cell numbers in the stationary phase of the B12-provider-diatom co-cultures were at least 110 times higher than the cell numbers of T. pseudonana. These calculations are all based on ideal laboratory conditions, with sufficient supply of inorganic nutrients and organic substrates and may differ in natural environments where viral infections or sloppy feeding can lead to cell disruption and subsequent release of intracellular B12 [43, 44]. Also, B12 requirement of T. pseudonana cells can vary under different growth conditions. For example, it has been shown that growth of T. pseudonana even with 1 pM of B12 can result in a significant change in the metabolite pool of the diatom, which in turn may have implications for the interaction with bacteria [45]. Nevertheless, our data give a first approximate insight into the interplay between B12-producers and -consumers in the world of microorganisms.
Bacterial effects on the growth of T. pseudonana
Growth characteristics of T. pseudonana in co-culture show not only the obligatory provision of B12 by bacteria but also other bacterial factors that influence growth. For example, we observed that Sulfitobacter litoralis, a representative of the Roseobacter group, showed inhibitory behaviour towards the diatom. Other studies have shown that Roseobacter group isolates can produce inhibitory substances, roseobacticides, which can suppress the growth of eukaryotic phototrophs [46]. The provision of B12 leads to a promotion in growth and, at the same time, growth of the diatom is inhibited. One reason for the different growth characteristics of the diatoms observed in co-culture with different bacteria could be the adaptation to different habitats where the bacterial isolates naturally occur.
In contrast to these observations, Celeribacter baekdonensis DSM 27375 significantly stimulated the growth of T. pseudonana. Even though C. baekdonensis did not provide B12 despite being synthesized, its presence in co-culture with B12 addition significantly increased the growth rate and growth yield of T. pseudonana compared to the positive control of the corresponding experimental run. In previous bacterial-diatom co-culture experiments, it has been shown that the excretion of cyclic peptides, diketopiperazines, by a bacterium, significantly increased diatom cell numbers [47]. Another plausible scenario is the synthesis and excretion of indoleacetic acid (IAA) by C. baekdonensis, which is a growth-promoting hormone for diatoms [48]. A similar effect is also conceivable for C. baekdonensis and would be exciting to explore in greater depth.
A finding that appears to be overlooked in the context of our actual question is the fact that the expected bacterial cell death does not necessarily lead to the release of B12, which would promote the growth of T. pseudonana, and thus promote the interaction. Even after up to six weeks in co-culture, we cannot observe significant growth of T. pseudonana despite the presence of a bacterial B12 prototroph. This fact highlights the importance of cell lysis mechanisms in nature, for example caused by viral infections or sloppy feeding. Already today, these two natural processes are considered to play a significant role in the turnover of dissolved organic matter [44, 49–51] and are likely to also have a decisive influence on the release of B-vitamins in marine ecosystems [23]. Additionally, T. pseudonana is known to secret a B12 binding protein under B12 deficient conditions that has an affinity constant of 2 × 1011 M−1. This protein might help them to acquire B12 from the surroundings, when it is released through bacterial cell lysis mechanism [52]. Other phytoplankton might also have a similar strategy to scavenge B12 from the environment. When intracellular B12 is considered as a reservoir for other B12 auxotrophic microorganisms, then, for example, already 19 M. algicola cells would be sufficient to enable the growth of one T. pseudonana cell.
The vital cofactor B12 is not shared by all prototrophic bacteria
About half of the marine phytoplankton species are B12 auxotrophs and rely on prototrophic prokaryotes to obtain this essential vitamin [1, 53]. Several co-culture experiments have confirmed that individual marine bacterial isolates, mainly Alphaproteobacteria, enable phytoplankton species to overcome their auxotrophy by providing the essential cofactor [13–16, 27, 28]. In our study we hypothesised that not all B12 prototrophs share B12 with other microorganisms and to prove that we performed individual co-culture experiments between T. pseudonana and 33 B12 prototrophic bacteria. B12 prototrophy of the bacterial isolates was confirmed by their genetic ability to synthesize B12 (Supplementary table S2) and their ability to grow in B12-free medium. The results of our study support this hypothesis, as we were able to identify one group of bacteria that enables growth of T. pseudonana by the supply of the essential cofactor, B12-providers. On the other hand, we also identified a second group of B12 prototrophic bacteria that did not support the growth of the diatom, the B12-retainers. Moreover, while categorizing them into B12-providers and B12-retainers, we observed that there are species within one genus, such as P. inhibens and P. galleciensis, in which one is a B12-provider and the other is a B12-retainer, respectively, although both of them possess the necessary genes for B12 biosynthesis. Yet, the question remains why some bacteria share the cofactor, and others, despite an obligatory interaction enforced in co-culture, do not. In the following, we describe and discuss three scenarios that we consider plausible, whereby not only one scenario has to be correct, but rather all three can take place in the B12-retainer strains that we have identified.
First, biosynthesis of metabolites, such as the energetically costly B12 cofactor, are subject to intracellular regulation. Transcriptional regulation of the B12 biosynthesis pathway determines whether, and in what quantity B12 is synthesised in the cell. For example, sigma factors can alter the specificity of an RNA polymerase for a particular promoter, so that gene expression is enhanced or reduced [54]. In the case of the bacterial isolate Propionibacterium strain UF1, the riboswitch cbiMCbl was identified to regulate the gene expression of the cobA operon and thus controls B12 biosynthesis [55]. It is also known that sufficient availability of B12 can repress B12 biosynthesis gene expression in bacteria [56, 57]. In gram-negative proteobacteria as well as in cyanobacteria, for example, cobalamin (pseudocobalamin, in case of some bacteria) biosynthesis and B12 transport genes are regulated by inhibition of translation initiation, whereas in some gram-positive bacteria gene regulation proceeds by transcriptional antitermination [58]. The mechanisms described above are likely to also occur in the bacterial isolates that we tested. The large difference between the detected intracellular B12 concentrations could therefore be due to differences in gene regulation of the different bacteria and may also have had an influence on the release of B12 in the co-culture with T. pseudonana.
Second, cobalamin, which we referred to here as B12 for simplicity, belongs to a group of B12-like metabolites, called cobamides. Each cobamide differs in the lower ligand attached. For example, the common cobamide, cobalamin, which is bioavailable to most microorganisms, carries 5,6-dimethylbenzimidazol (DMB) as its lower ligand, whereas pseudocobalamin synthesised by cyanobacteria in high concentrations in the ocean and being less or not bioavailable to most microorganisms, has adenine attached as its lower ligand [11, 41, 59, 60]. In general, the lower ligands of cobamides can be divided into benzimidazoles, purines, and phenols, and more than a dozen cobamides and cobamide-analogs have already been discovered [61]. However, research into the synthesis and actual diversity of cobamides, especially in marine bacteria and archaea, is still in its infancy. In our study, we were unable to detect intracellular B12 in four out of eight bacterial B12-retainer strains, although the cell counts at the time of sampling should have been sufficient for its detection. However, as is generally the case, our LC-MS analysis only targets cobalamin (B12) with its different upper ligands (adenosyl-, cyano-, methyl-, and hydroxy-cobalamin). Therefore, we cannot exclude the possibility that the here studied bacteria synthesise different cobamides, which are possibly not or less bioavailable to T. pseudonana, and not covered by our analytical measurement method. This speculation was supported by the fact that one of these four B12- retainer strains, Sulfitobacter sp. DFL-23, does not possess the DMB synthesis gene bluB and there was no intracellular B12 detected in this strain (Supplementary table S2 and Table 2). Again, it is difficult to explain this phenomenon solely depending on the presence of annotated DMB synthesis gene, as for Loktanella salsilacus DSM 16199 no bluB gene was annotated, still we detected intracellular B12 in this strain using our detection method (Supplementary table S2 and Table 2).
Third, the bacteria we have identified as B12-retainer simply may not have actively released the synthesised B12 into their environment. Regardless of the importance of B12 for the vast majority of living organisms on our planet, its excretion mechanisms are to our knowledge still largely unknown. Its size of more than 1,350 Dalton does not allow sufficient diffusion through the cell membrane, which would enable microbial interactions [32]. Thus, it is likely that an unknown mechanism is required for its release. This assumption is further supported by the fact that we were able to detect intracellular B12 in four of the eight B12-retainer strains and at concentrations comparable to those detected in the B12-provider strains. In addition, we could detect intracellular B12 in P. xiamenensis, but none in its exometabolome. On the other hand, presence of extracellular B12 was detected in the exometabolome of both the provider strains examined, M. algicola and P. inhibens. Our findings show that not all bacteria share the pivotal cofactor with their environment, which has an impact on our current understanding of the marine B12 cycle and presumably in other ecosystems as well. The active exchange of B12 and thus microbial interaction plays a much smaller role than previously assumed for a relatively large number of bacteria. Consequently, for some of the B12 prototrophic bacteria within a community, it is likely that the cofactor is only released upon cell lysis.
B12 production in the marine ecosystem and ecological implications
Looking at the original source of B12 in nature, namely prototrophic bacteria and archaea, the bacteria studied here show pronounced differences between the biosynthetic potentials of the cofactors and the ability to share them with their environment. Thus, the natural source of vitamin B12 within a given ecosystem does not primarily depend on the ratio of prototrophic bacteria, but even more crucially on how much of the cofactor is synthesised by the prototrophic prokaryotes within an ecosystem and is actively released. The fact that some bacteria do not voluntarily share B12 with ambient microorganisms, significantly increases the importance of processes, such as sloppy feeding by zooplankton or virus infections [44, 49–51], for the release of vitamins in the marine and likely also other ecosystems.
Our results also contribute to the controversially discussed question of whether B12 prototrophic bacteria live in symbiosis with phototrophic microorganisms [13, 30]. Despite numerous co-cultivation experiments demonstrating the obligatory provision of B12 by individual bacteria to phototrophic microorganisms, the decisive question of the mechanism of provision has so far been overlooked [13–16, 27, 28]. In our view, however, this question is crucial when assessing whether a symbiotic interaction is taking place. Our results support the hypothesis that a bacterial mechanism for the active release is likely to exist, as our experiments distinguish between B12-provider and B12-retainer within prototrophic bacteria. Looking at the ecological niches and the isolation sites of the two respective groups, differences can be identified. Most B12-provider strains were isolated from or discovered in association with eukaryotic microorganisms, whereas most B12-retainer strains were isolated as free-living in the ocean (Supplementary table S4). Moreover, six of the tested bacterial strains were isolated from dinoflagellates and five of them were B12-provider. Since we used a diatom as a B12 auxotrophic organism in our study, it would also be interesting to know if these B12-provider strains also provide B12 to other phytoplankton, such as dinoflagellates. Also, in this study we only studied bacteria from the alphaproteobacteria class, since a large share of them are known to be B12 prototrophs and abundant in the marine ecosystem. For future studies, it would be interesting to see if a similar pattern of B12 provisioning can be observed in bacteria from other classes. Our results indicate that the B12 prototrophy of a bacterium does not necessarily indicate a mutualistic interaction with other auxotrophic microorganisms. However, the bacterial group of B12-provider in particular seems to favour living in close proximity to other microorganisms, which is why the exchange of B12 for e.g. organic compounds can establish itself as a distinct symbiotic interaction between individual microorganisms.
Supplementary information
Acknowledgements
This work was supported by Deutsche Forschungsgemeinschaft within the Transregional Collaborative Research Center Roseobacter (TRR51).
Author contributions
SS performed the experimental laboratory work, data analysis, data interpretation, and manuscript drafting. SB and HW performed the cobalamin LC-MS analyses and revised the manuscript, MS revised the manuscript. GW designed the experiments, advised data evaluation, wrote parts of and finalised the manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Data availability
The datasets generated during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-023-01391-3.
References
- 1.Sañudo-Wilhelmy SA, Gómez-Consarnau L, Suffridge C, Webb EA. The role of B vitamins in marine biogeochemistry. Ann Rev Mar Sci. 2014;6:339–67. doi: 10.1146/annurev-marine-120710-100912. [DOI] [PubMed] [Google Scholar]
- 2.Dowling DP, Croft AK, Drennan CL. Radical use of Rossmann and TIM Barrel architectures for controlling coenzyme B12 chemistry. Annu Rev Biophys. 2012;41:403–27. doi: 10.1146/annurev-biophys-050511-102225. [DOI] [PubMed] [Google Scholar]
- 3.Matthews RG, Smith AE, Zhou ZS, Taurog RE, Bandarian V, Evans JC, et al. Cobalamin-dependent and cobalamin-independent methionine synthases: are there two solutions to the same chemical problem? Helv Chim Acta. 2003;86:3939–54. doi: 10.1002/hlca.200390329. [DOI] [Google Scholar]
- 4.Warren MJ, Raux E, Schubert HL, Escalante-Semerena JC. The biosynthesis of adenosylcobalamin (vitamin B12) Nat Prod Rep. 2002;19:390–412. doi: 10.1039/b108967f. [DOI] [PubMed] [Google Scholar]
- 5.Doxey AC, Kurtz DA, Lynch MD, Sauder LA, Neufeld JD. Aquatic metagenomes implicate Thaumarchaeota in global cobalamin production. ISME J. 2015;9:461–71. doi: 10.1038/ismej.2014.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roth JR, Lawrence JG, Rubenfield M, Kieffer-Higgins S, Church GM. Characterization of the cobalamin (vitamin B12) biosynthetic genes of Salmonella typhimurium. J Bacteriol. 1993;175:3303–16. doi: 10.1128/jb.175.11.3303-3316.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shelton AN, Seth EC, Mok KC, Han AW, Jackson SN, Haft DR, et al. Uneven distribution of cobamide biosynthesis and dependence in bacteria predicted by comparative genomics. ISME J. 2019;13:789–804. doi: 10.1038/s41396-018-0304-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu X, Heal KR, Ingalls AE, Doxey AC, Neufeld JD. Metagenomic and chemical characterization of soil cobalamin production. ISME J. 2020;14:53–66. doi: 10.1038/s41396-019-0502-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giovannoni SJ, Cameron Thrash J, Temperton B. Implications of streamlining theory for microbial ecology. ISME J. 2014;8:1553–65. doi: 10.1038/ismej.2014.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D’Souza G, Waschina S, Pande S, Bohl K, Kaleta C, Kost C. Less is more: selective advantages can explain the prevalent loss of biosynthetic genes in bacteria. Evolution. 2014;68:2559–70. doi: 10.1111/evo.12468. [DOI] [PubMed] [Google Scholar]
- 11.Heal KR, Qin W, Ribalet F, Bertagnolli AD, Coyote-Maestas W, Hmelo LR, et al. Two distinct pools of B12 analogs reveal community interdependencies in the ocean. Proc Natl Acad Sci. 2017;114:364–9. doi: 10.1073/pnas.1608462114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Droop MR. A pelagic marine diatom requiring cobalamin. J Mar Biol Assoc U K. 1955;34:229–31. doi: 10.1017/S0025315400027600. [DOI] [Google Scholar]
- 13.Croft MT, Lawrence AD, Raux-Deery E, Warren MJ, Smith AG. Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature. 2005;438:90–3. doi: 10.1038/nature04056. [DOI] [PubMed] [Google Scholar]
- 14.Haines KC, Guillard RRL. Growth of vitamin B12-requiring marine diatoms in mixed laboratory cultures with vitamin B12-producing marine bacteria. J Phycol. 1974;10:245–52. [Google Scholar]
- 15.Cruz-López R, Maske H. The vitamin B1 and B12 required by the marine dinoflagellate Lingulodinium polyedrum can be provided by its associated bacterial community in culture. Front Microbiol. 2016;7:560. doi: 10.3389/fmicb.2016.00560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cooper MB, Kazamia E, Helliwell KE, Kudahl UJ, Sayer A, Wheeler GL, et al. Cross-exchange of B-vitamins underpins a mutualistic interaction between Ostreococcus tauri and Dinoroseobacter shibae. ISME J. 2019;13:334–45. doi: 10.1038/s41396-018-0274-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kazamia E, Czesnick H, Nguyen TTV, Croft MT, Sherwood E, Sasso S, et al. Mutualistic interactions between vitamin B12-dependent algae and heterotrophic bacteria exhibit regulation. Environ Microbiol. 2012;14:1466–76. doi: 10.1111/j.1462-2920.2012.02733.x. [DOI] [PubMed] [Google Scholar]
- 18.Sañudo‐Wilhelmy SA, Gobler CJ, Okbamichael M, Taylor GT. Regulation of phytoplankton dynamics by vitamin B12. Geophys Res Lett. 2006;33:L04604. doi: 10.1029/2005GL025046. [DOI] [Google Scholar]
- 19.Suffridge CP, Gómez‐Consarnau L, Monteverde DR, Cutter L, Arístegui J, Alvarez‐Salgado XA, et al. B vitamins and their congeners as potential drivers of microbial community composition in an oligotrophic marine ecosystem. J Geophys Res Biogeosci. 2018;123:2890–907. doi: 10.1029/2018JG004554. [DOI] [Google Scholar]
- 20.Suffridge C, Cutter L, Sañudo-Wilhelmy SA. A new analytical method for direct measurement of particulate and dissolved B-vitamins and their congeners in seawater. Front Mar Sci. 2017;4:11. doi: 10.3389/fmars.2017.00011. [DOI] [Google Scholar]
- 21.Menzel DW, Spaeth JP. Occurrence of vitamin B12 in the Sargasso Sea. Limnol Oceanogr. 1962;7:151–4. doi: 10.4319/lo.1962.7.2.0151. [DOI] [Google Scholar]
- 22.Bertrand EM, Saito MA, Rose JM, Riesselman CR, Lohan MC, Noble AE, et al. Vitamin B12 and iron colimitation of phytoplankton growth in the Ross Sea. Limnol Oceanogr. 2007;52:1079–93. doi: 10.4319/lo.2007.52.3.1079. [DOI] [Google Scholar]
- 23.Wienhausen G, Dlugosch L, Jarling R, Wilkes H, Giebel H-A, Simon M. Availability of vitamin B12 and its lower ligand intermediate α-ribazole impact prokaryotic and protist communities in oceanic systems. ISME J. 2022;16:2002–14. doi: 10.1038/s41396-022-01250-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seymour JR, Amin SA, Raina J-B, Stocker R. Zooming in on the phycosphere: the ecological interface for phytoplankton–bacteria relationships. Nat Microbiol. 2017;2:1–12. doi: 10.1038/nmicrobiol.2017.65. [DOI] [PubMed] [Google Scholar]
- 25.Cirri E, Pohnert G. Algae−bacteria interactions that balance the planktonic microbiome. N. Phytol. 2019;223:100–6. doi: 10.1111/nph.15765. [DOI] [PubMed] [Google Scholar]
- 26.Cruz-López R, Maske H, Yarimizu K, Holland NA. The B-vitamin mutualism between the dinoflagellate Lingulodinium polyedrum and the bacterium Dinoroseobacter shibae. Front Mar Sci. 2018;5:274. doi: 10.3389/fmars.2018.00274. [DOI] [Google Scholar]
- 27.Wang H, Tomasch J, Jarek M, Wagner-Döbler I. A dual-species co-cultivation system to study the interactions between Roseobacters and dinoflagellates. Front Microbiol. 2014;5:311. doi: 10.3389/fmicb.2014.00311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grant MA, Kazamia E, Cicuta P, Smith AG. Direct exchange of vitamin B12 is demonstrated by modelling the growth dynamics of algal–bacterial cocultures. ISME J. 2014;8:1418–27. doi: 10.1038/ismej.2014.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Durham BP, Sharma S, Luo H, Smith CB, Amin SA, Bender SJ, et al. Cryptic carbon and sulfur cycling between surface ocean plankton. Proc Natl Acad Sci. 2015;112:453–7. doi: 10.1073/pnas.1413137112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Droop MR. Vitamins, phytoplankton, and bacteria: symbiosis or scavenging? J Plankton Res. 2007;29:107–13. doi: 10.1093/plankt/fbm009. [DOI] [Google Scholar]
- 31.Wienhausen G, Bittner MJ, Paerl RW Key knowledge gaps to fill at the cell-to-ecosystem level in marine B-vitamin cycling. Front Mar Sci. 2022; 10.3389/fmars.2022.876726.
- 32.Nikaido H, Rosenberg EY. Effect on solute size on diffusion rates through the transmembrane pores of the outer membrane of Escherichia coli. J Gen Physiol. 1981;77:121–35. doi: 10.1085/jgp.77.2.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shultis DD, Purdy MD, Banchs CN, Wiener MC. Outer membrane active transport: structure of the BtuB:TonB complex. Science. 2006;312:1396–9. doi: 10.1126/science.1127694. [DOI] [PubMed] [Google Scholar]
- 34.Bruns S, Scholz-Böttcher B, Wienhausen G, Wilkes H Simultaneous quantification of all B vitamins and selected biosynthetic precursors in seawater and bacteria by means of different mass spectrometric approaches: Anal. Bioanal Chem. 2022; 10.1007/s00216-022-04317-8. [DOI] [PMC free article] [PubMed]
- 35.Giebel HA, Wolterink M, Brinkhoff T, Simon M Complementary energy acquisition via aerobic anoxygenic photosynthesis and carbon monoxide oxidation by Planktomarina temperata of the Roseobacter group. FEMS Microbiol Ecol. 2019; 10.1093/femsec/fiz050. [DOI] [PubMed]
- 36.Lunau M, Lemke A, Walther K, Martens-Habbena W, Simon M. An improved method for counting bacteria from sediments and turbid environments by epifluorescence microscopy. Environ Microbiol. 2005;7:961–8. doi: 10.1111/j.1462-2920.2005.00767.x. [DOI] [PubMed] [Google Scholar]
- 37.Cakić N, Kopke B, Rabus R, Wilkes H. Suspect screening and targeted analysis of acyl coenzyme A thioesters in bacterial cultures using a high-resolution tribrid mass spectrometer. Anal Bioanal Chem. 2021;413:3599–610. doi: 10.1007/s00216-021-03318-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bruns S, Wienhausen G, Scholz-Böttcher B, Wilkes H Method development and quantification of all B vitamins and selected biosynthetic precursors in winter and spring samples from the North Sea and de-novo synthesized by Vibrio campbellii. Mar Chem. In review.
- 39.Paerl RW, Bertrand EM, Allen AE, Palenik B, Azam F. Vitamin B1 ecophysiology of marine picoeukaryotic algae: Strain-specific differences and a new role for bacteria in vitamin cycling. Limnol Oceanogr. 2015;60:215–28. doi: 10.1002/lno.10009. [DOI] [Google Scholar]
- 40.Carini P, Campbell EO, Morré J, Sañudo-Wilhelmy SA, Cameron Thrash J, Bennett SE, et al. Discovery of a SAR11 growth requirement for thiamin’s pyrimidine precursor and its distribution in the Sargasso Sea. ISME J. 2014;8:1727–38. doi: 10.1038/ismej.2014.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Helliwell KE, Lawrence AD, Holzer A, Kudahl UJ, Sasso S, Kräutler B, et al. Cyanobacteria and eukaryotic algae use different chemical variants of vitamin B12. Curr Biol. 2016;26:999–1008. doi: 10.1016/j.cub.2016.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Swaney MH, Sandstrom S, Kalan LR. Cobamide sharing is predicted in the human skin microbiome. mSystems. 2022; 10.1128/msystems.00677-22. [DOI] [PMC free article] [PubMed]
- 43.Bonnet S, Webb EA, Panzeca C, Karl DM, Capone DG, Wilhelmy SAS. Vitamin B12 excretion by cultures of the marine cyanobacteria Crocosphaera and Synechococcus. Limnol Oceanogr. 2010;55:1959–64. doi: 10.4319/lo.2010.55.5.1959. [DOI] [Google Scholar]
- 44.Møller EF, Nielsen TG. Production of bacterial substrate by marine copepods: effect of phytoplankton biomass and cell Size. J Plankton Res. 2001;23:527–36. doi: 10.1093/plankt/23.5.527. [DOI] [Google Scholar]
- 45.Heal KR, Kellogg NA, Carlson LT, Lionheart RM, Ingalls AE. Metabolic consequences of cobalamin scarcity in the diatom Thalassiosira pseudonana as revealed through metabolomics. Protist. 2019;170:328–48. doi: 10.1016/j.protis.2019.05.004. [DOI] [PubMed] [Google Scholar]
- 46.Seyedsayamdost MR, Case RJ, Kolter R, Clardy J. The Jekyll-and-Hyde chemistry of Phaeobacter gallaeciensis. Nat Chem. 2011;3:331–5. doi: 10.1038/nchem.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sittmann J, Bae M, Mevers E, Li M, Quinn A, Sriram G, et al. Bacterial diketopiperazines stimulate diatom growth and lipid accumulation. Plant Physiol. 2021;186:1159–70. doi: 10.1093/plphys/kiab080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Amin SA, Hmelo LR, van Tol HM, Durham BP, Carlson LT, Heal KR, et al. Interaction and signalling between a cosmopolitan phytoplankton and associated bacteria. Nature. 2015;522:98–101. doi: 10.1038/nature14488. [DOI] [PubMed] [Google Scholar]
- 49.Suttle CA, Chan AM, Cottrell MT. Infection of phytoplankton by viruses and reduction of primary productivity. Nature. 1990;347:467–9. doi: 10.1038/347467a0. [DOI] [Google Scholar]
- 50.Proctor LM, Fuhrman JA. Viral mortality of marine bacteria and cyanobacteria. Nature. 1990;343:60–2. doi: 10.1038/343060a0. [DOI] [Google Scholar]
- 51.Strom SL, Benner R, Ziegler S, Dagg MJ. Planktonic grazers are a potentially important source of marine dissolved organic carbon. Limnol Oceanogr. 1997;42:1364–74. doi: 10.4319/lo.1997.42.6.1364. [DOI] [Google Scholar]
- 52.Sahni MK, Spanos S, Wahrman MZ, Sharma GM. Marine corrinoid-binding proteins for the direct determination of vitamin B12 by radioassay. Anal Biochem. 2001;289:68–76. doi: 10.1006/abio.2000.4923. [DOI] [PubMed] [Google Scholar]
- 53.Tang YZ, Koch F, Gobler CJ. Most harmful algal bloom species are vitamin B1 and B12 auxotrophs. Proc Natl Acad Sci. 2010;107:20756–61. doi: 10.1073/pnas.1009566107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rodionov DA. Comparative genomic reconstruction of transcriptional regulatory networks in bacteria. Chem Rev. 2007;107:3467–97. doi: 10.1021/cr068309+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li J, Ge Y, Zadeh M, Curtiss R, Mohamadzadeh M. Regulating vitamin B12 biosynthesis via the cbiMCbl riboswitch in Propionibacterium strain UF1. Proc Natl Acad Sci. 2020;117:602–9. doi: 10.1073/pnas.1916576116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lundrigan MD, Köster W, Kadner RJ. Transcribed sequences of the Escherichia coli btuB gene control its expression and regulation by vitamin B12. Proc Natl Acad Sci. 1991;88:1479–83. doi: 10.1073/pnas.88.4.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Richter-Dahlfors AA, Andersson DI. Cobalamin (vitamin B12) repression of the Cob operon in Salmonella typhimurium requires sequences within the leader and the first translated open reading frame. Mol Microbiol. 1992;6:743–9. doi: 10.1111/j.1365-2958.1992.tb01524.x. [DOI] [PubMed] [Google Scholar]
- 58.Vitreschak AG, Rodionov DA, Mironov AA, Gelfand MS. Regulation of the vitamin B12 metabolism and transport in bacteria by a conserved RNA structural element. RNA. 2003;9:1084–97. doi: 10.1261/rna.5710303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tanioka Y, Yabuta Y, Yamaji R, Shigeoka S, Nakano Y, Watanabe F, et al. Occurrence of pseudovitamin B12 and its possible function as the cofactor of cobalamin-dependent methionine synthase in a cyanobacterium Synechocystis sp. PCC6803. J Nutr Sci Vitaminol. 2009;55:518–21. doi: 10.3177/jnsv.55.518. [DOI] [PubMed] [Google Scholar]
- 60.Stupperich E, Kräutler B. Pseudo vitamin B12 or 5-hydroxybenzimidazolyl-cobamide are the corrinoids found in methanogenic bacteria. Arch Microbiol. 1988;149:268–71. doi: 10.1007/BF00422016. [DOI] [Google Scholar]
- 61.Sokolovskaya OM, Shelton AN, Taga ME. Sharing vitamins: cobamides unveil microbial interactions. Science. 2020;369:6499. doi: 10.1126/science.aba0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during the current study are available from the corresponding author on reasonable request.