Abstract
Isoprenoids are a very large and diverse family of metabolites required by all living organisms. All isoprenoids derive from the double-bond isomers isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP), which are produced by the methylerythritol 4-phosphate (MEP) pathway in bacteria and plant plastids. It has been reported that IPP and DMAPP feedback-regulate the activity of deoxyxylulose 5-phosphate synthase (DXS), a dimeric enzyme that catalyzes the main flux-controlling step of the MEP pathway. Here we provide experimental insights into the underlying mechanism. Isothermal titration calorimetry and dynamic light scattering approaches showed that IPP and DMAPP can allosterically bind to DXS in vitro, causing a size shift. In silico ligand binding site analysis and docking calculations identified a potential allosteric site in the contact region between the two monomers of the active DXS dimer. Modulation of IPP and DMAPP contents in vivo followed by immunoblot analyses confirmed that high IPP/DMAPP levels resulted in monomerization and eventual aggregation of the enzyme in bacterial and plant cells. Loss of the enzymatically active dimeric conformation allows a fast and reversible reduction of DXS activity in response to a sudden increase or decrease in IPP/DMAPP supply, whereas aggregation and subsequent removal of monomers that would otherwise be available for dimerization appears to be a more drastic response in the case of persistent IPP/DMAPP overabundance (e.g., by a blockage in their conversion to downstream isoprenoids). Our results represent an important step toward understanding the regulation of the MEP pathway and rational design of biotechnological endeavors aimed at increasing isoprenoid contents in microbial and plant systems.
Key words: isoprenoids, DXS, feedback regulation, allosteric, monomerization
The MEP pathway synthesizes the universal precursors of isoprenoids in plant plastids and most bacteria. This study shows that these precursors feedback-regulate their own production by direct allosteric binding to the first enzyme of the pathway, a dimeric enzyme that becomes monomeric and eventually aggregates when levels of MEP pathway products increase.
Introduction
Isoprenoids (also known as terpenoids or terpenes) are a vast family of natural compounds produced in all free-living forms of life. Despite their astonishing variety at the structural and functional levels, all isoprenoids derive from the universal building blocks isopentenyl diphosphate (IPP) and its isomer dimethylallyl diphosphate (DMAPP). In nature, two different pathways are responsible for the synthesis of IPP and DMAPP (Rodriguez-Concepcion and Boronat, 2015). The mevalonate (MVA) pathway is present mainly in eukaryotes and archaea and uses acetyl-coenzyme A as an initial substrate, whereas the methylerythritol 4-phosphate (MEP) pathway is found in most bacteria and produces IPP and DMAPP from pyruvate and glyceraldehyde 3-phosphate (GAP). In plants, both pathways coexist but in different subcellular compartments. The MVA pathway produces IPP and DMAPP in the cytosol to synthesize sterols and sesquiterpenes, whereas the MEP pathway is located in plastids and produces the precursors of plastidial isoprenoids such as monoterpenes, carotenoids, and the side chains of chlorophylls, tocopherols, phylloquinones, and plastoquinone.
The first step of the MEP pathway is catalyzed by the enzyme deoxyxylulose 5-phosphate (DXP) synthase (DXS), which uses pyruvate and GAP to generate DXP (Figure 1). DXP is converted into MEP by the next enzyme of the pathway, DXP reductoisomerase (DXR). Metabolic control analyses carried out in bacteria and plants have shown that DXS is the enzyme with the highest flux control coefficient and is consequently the primary rate-limiting step of the MEP pathway (Wright et al., 2014; Volke et al., 2019). Consistent with this central role in the control of pathway flux, DXS activity is regulated at multiple levels, including transcriptional, post-transcriptional, and post-translational (Pulido et al., 2013, 2016; Wright et al., 2014; Pokhilko et al., 2015; Rodríguez-Concepción and Boronat, 2015; Rodriguez-Concepcion et al., 2019; Volke et al., 2019; Mitra et al., 2021; Yu et al., 2021). Several reports have highlighted the importance of metabolic regulation of the MEP pathway (Banerjee and Sharkey, 2014; Pokhilko et al., 2015; Rodríguez-Concepción and Boronat, 2015). Labeling experiments suggested negative feedback regulation of plant DXS activity by the MEP pathway products IPP and DMAPP (Wolfertz et al., 2004), which was later confirmed in vitro and in vivo (Banerjee et al., 2013, 2016; Ghirardo et al., 2014; Wright et al., 2014). Interestingly, MEP pathway flux has also been observed to modulate DXS protein levels in plants (Han et al., 2013; Ghirardo et al., 2014; Pokhilko et al., 2015). The feedback regulation of DXS levels and activity has been shown to stabilize the plant pathway flux against changes in substrate supply and adjust it according to product demand under normal growth conditions (Pokhilko et al., 2015). More recently, in vitro assays have shown similar negative feedback regulation for several bacterial DXS enzymes (Kudoh et al., 2017a), but in vivo confirmation is still missing.
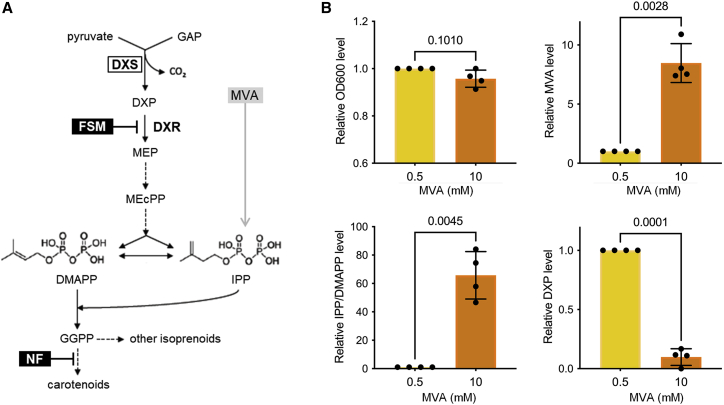
Figure 1.
Representation of the MEP pathway and levels of metabolites in MVA-supplemented cells.
(A) The MEP pathway simultaneously produces IPP and DMAPP for isoprenoid biosynthesis in bacteria and plant plastids. A synthetic operon that transforms exogenously supplied MVA into IPP and DMAPP allows survival of MEP-defective E. coli strains such as EcAB4-10, which lacks DXR activity. Dashed closed arrows represent several steps. GAP, glyceraldehyde 3-phosphate; DXP, deoxyxylulose 5-phosphate; MEP, methylerythritol 4-phosphate; MEcPP, methylerythritol cyclodiphosphate; IPP, isopentenyl diphosphate; DMAPP, dimethylallyl diphosphate; GGPP, geranylgeranyl diphosphate; MVA, mevalonic acid. Enzymes are shown in bold: DXS, DXP synthase; DXR, DXP reductoisomerase. Inhibitors are boxed in black: FSM, fosmidomycin; NF, norflurazon.
(B) Metabolite levels in DXR-defective EcAB4-10 cells supplemented with MVA. E. coli cells of the EcAB4-10 strain were grown in the presence of 0.5 or 10 mM MVA and collected during exponential phase (steady state) for metabolite extraction and quantification by LC-MS. Bar plots represent values of cell growth measured by optical density at 600 nm (OD600) and intracellular levels of MVA, DXP, and IPP/DMAPP. All values are represented relative to those in 0.5-mM samples in each replicate. Dots represent individual values. Mean and standard deviation of four replicates are shown. Numbers in the plots indicate p values of Student’s t-test analyses (paired, two tailed).
The initial crystallization of a truncated Escherichia coli DXS (Xiang et al., 2007) was recently followed by reported structures of DXS enzymes from another bacterium, Deinococcus radiodurans (Chen et al., 2019), and from the model plant Arabidopsis thaliana (Yu et al., 2021). From these studies, it was concluded that the active enzyme is a dimer and that a highly hydrophobic surface is present in the interface of the two monomers. Computational analysis of the monomer structure suggested that these hydrophobic domains increase the aggregation propensity of the protein (Pulido et al., 2016). Arabidopsis DXS was actually found to easily aggregate, leading to subsequent degradation by the plastidial Clp protease complex. When stress conditions compromise the proteolytic capacity of the plastid, however, disaggregation rather than degradation is promoted for spontaneous refolding and reactivation of DXS (Pulido et al., 2013, 2016). The high aggregation propensity of DXS was also confirmed in cyanobacteria (Kudoh et al., 2017b) and E. coli (Kudoh et al., 2017a), and involvement of the Clp protease in DXS degradation has also been described in tobacco (Nicotiana tabacum) leaves (Moreno et al., 2018), tomato (Solanum lycopersicum) fruit (D'Andrea et al., 2018), the malaria parasite Plasmodium falciparum (Florentin et al., 2017), and E. coli (Ninnis et al., 2009).
Considering that (1) reduced IPP and DMAPP levels result in higher DXS activity and protein levels, (2) DXS activity requires dimerization, (3) monomers expose aggregation-prone hydrophobic domains, and (4) aggregation normally leads to DXS degradation, we hypothesized that high IPP or DMAPP levels might displace the equilibrium toward the monomeric conformation of the enzyme. If sustained, high IPP and DMAPP would then lead to DXS monomer aggregation and eventual degradation. Here we tested this hypothesis using bacterial (E. coli) and plant (tomato) DXS enzymes.
Results
IPP/DMAPP inhibit EcDXS activity in vivo
Feedback regulation of DXS by MEP pathway products (IPP/DMAPP) has been described in vitro and in vivo in plants (Wolfertz et al., 2004; Banerjee et al., 2013; Wright et al., 2014) but only in vitro in bacteria (Kudoh et al., 2017a). To validate the effect of IPP and DMAPP on the activity of E. coli DXS (EcDXS) in vivo, we used engineered strains harboring a synthetic MVA operon that allows the production of IPP and DMAPP when MVA is supplied to the growth medium (Sauret-Güeto et al., 2006; Figure 1A). To accurately detect changes in DXS activity, we used the EcAB4-10 strain, which lacks DXR; hence, it is unable to convert DXP into MEP and downstream IPP and DMAPP (Sauret-Güeto et al., 2006; Sangari et al., 2010). Survival of EcAB4-10 cells was enabled by supplementing the growth medium with MVA, which is converted into IPP and DMAPP by the synthetic MVA operon (Figure 1A). EcAB4-10 cells were grown at 37°C in Luria broth (LB) medium in the presence of 0.5 or 10 mM MVA and collected during the exponential phase to ensure steady-state conditions. Intracellular metabolites were then extracted and measured by liquid chromatography-mass spectrometry (LC-MS) (Figure 1B). IPP and DMAPP were quantified together because of the technical difficulty of separating these double-bond isomers. As expected, increasing amounts of intracellular MVA were measured in MVA-supplemented cultures, demonstrating uptake of the molecule from the medium (Figure 1B). Also as expected, IPP/DMAPP levels were markedly increased in cells growing with 10 mM MVA compared with those growing with 0.5 mM MVA (Figure 1B). By contrast, DXP levels were much lower with 10 mM MVA, which is consistent with the conclusion that enhanced IPP/DMAPP levels cause a reduction in EcDXS activity in living cells (Figure 1B). It is important to note that EcAB4-10 cells growing with 10 mM MVA did not show any growth impairment compared with those growing with 0.5 mM MVA (Figure 1B), suggesting that the amounts of MVA-derived IPP/DMAPP were not reaching the very high levels previously reported to trigger toxicity (George et al., 2018; Bongers et al., 2020). Taken together, these results confirm that the negative feedback mechanism reported to control the activity of DXS also operates in vivo to decrease the activity of the bacterial EcDXS enzyme when IPP/DMAPP levels are increased.
IPP/DMAPP directly interact with DXS
Based on kinetic analyses and structural modeling, it was suggested that IPP and DMAPP might inhibit the activity of DXS by competing with its cofactor, thiamine diphosphate (TPP), for its binding site (Banerjee et al., 2013, 2016). To experimentally test this model, the interaction of DXS with TPP, IPP, and DMAPP was assessed by isothermal titration calorimetry (ITC). Specifically, we tested purified 6×His-tagged versions of the full-length EcDXS protein or a truncated version of the tomato SlDXS1 isoform lacking the N-terminal plastid-targeting peptide. However, the purified EcDXS protein was found to rapidly aggregate in solution, and we therefore decided to use only the plant SlDXS1 enzyme for the ITC experiments, together with TPP, IPP, or/and DMAPP as ligands. Based on the thermograms obtained and the corresponding binding isotherms using 20 μM SlDXS1 in the sample cell solution and 200 μM of the ligand in the syringe solution, we confirmed the interaction with TPP, as expected, with a dissociation constant (KD) of 4.2 μM (Table 1; Supplemental Figure 1). IPP and DMAPP were also found to interact with the plant enzyme, showing similar KD values (2.2 μM for IPP and 1.7 μM for DMAPP). Similar KD values were also found when the SlDXS1 protein was preincubated with 100 μM TPP in the sample cell (1.2 μM for IPP and 4.3 μM for DMAPP), suggesting the absence of competition for the TPP binding (i.e., active) site and thus the existence of an unrelated binding site for IPP/DMAPP (Table 1; Supplemental Figure 1).
Table 1.
ITC-calculated KA and KD values.
| KA (M−1) | KD (μM) | ||
|---|---|---|---|
| SlDXS1 | TPP | 2.3·105 ± 0.4·105 | 4.2 ± 0.7 |
| IPP | 4.5·105 ± 0.7·105 | 2.2 ± 0.3 | |
| DMAPP | 5.8·105 ± 0.8·105 | 1.7 ± 0.2 | |
| SlDXS1+TPP | IPP | 8.1·105 ± 0.9·105 | 1.2 ± 0.1 |
| DMAPP | 2.3·105 ± 0.5·105 | 4.3 ± 0.9 |
Mean and standard deviation values are shown.
To identify DXS regions where IPP and DMAPP might bind, we performed in silico docking calculations (Figure 2). First, we used the crystal structure information available for EcDXS to carry out an exhaustive protein surface characterization aimed at detecting all cavities within the DXS dimeric structure that could act as ligand binding sites. Two potential sites were found (Figure 2A). One of them corresponds to the active site region (ASR), whereas a second site was located in the contact area between the two EcDXS monomers (named the monomer interaction region [MIR]). Next, rigid (receptor) docking calculations for TPP generated a top docking pose in the ASR that perfectly matches the available conformation with the X-ray EcDXS structure (Xiang et al., 2007), recovering all key interactions and therefore validating our methodology (Supplemental Figure 2). IPP and DMAPP showed a similar top docking pose in the ASR, with a calculated interaction energy of −6.57 kcal/mol and −7.01 kcal/mol, respectively (Supplemental Figure 2). The ligand–protein interaction energy, however, was lower than that for TPP (−13.49 kcal/mol). In the case of the MIR, rigid docking calculations predicted a similar binding mode for TPP, IPP, and DMAPP, but the interaction energy for TPP (−5.83 kcal/mol) indicated a clear preference of the cofactor for the active site (Supplemental Figure 2). Several residues potentially involved in IPP/DMAPP binding to the MIR (such as H122, Q397, R398, and F435) are conserved between EcDXS and plant DXS enzymes, including SlDXS1. Consistent with this observation, rigid docking calculation of DMAPP with the Alpha-Fold2 model obtained for SlDXS1 (which showed high structural similarity to EcDXS) indicated a similar binding mode to the MIR of SlDXS1 compared with EcDXS (Figure 2B).
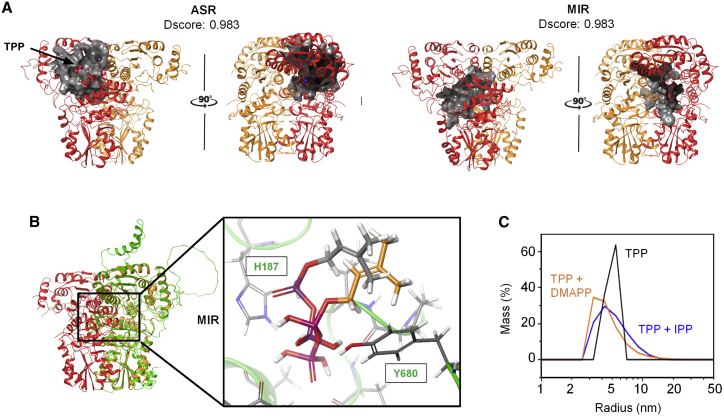
Figure 2.
Docking and DLS analysis of DXS–ligand binding.
(A) Docking simulation of the ligand binding site for EcDXS. The two predicted binding sites for ligands with high Sitemap scores (Dscores) are shown in gray: the active site region (ASR) and monomer interaction region (MIR).
(B) Structural superposition of EcDXS and SlDXS1. EcDXS (PDB: 2O1S) is marked with orange-red structures, and SlDXS1 (Alpha-Fold2 model AF-Q9XH50-F1) is represented in green. Magnification shows the MIR binding mode prediction from docking calculations of DMAPP for EcDXS (gray carbon atoms) and SlDXS1 (orange carbon atoms). SlDXS1 residues interacting with DMAPP are indicated.
(C) DLS analysis of purified SlDXS1 with the indicated ligands. Recombinant SlDXS1 (20 μM) was pre-mixed with 50 μM TPP and analyzed in the absence (black line) or presence of 50 μM IPP (blue line) or 50 μM DMAPP (orange line). For each measurement, 5 acquisitions of 5 s were taken.
To explore whether potential binding of IPP and DMAPP to the MIR had any effect on SlDXS1 dimerization, we initially tried size exclusion chromatography. However, the propensity of the protein to aggregate made it very challenging to keep it soluble while being carried through the stationary phase. As an alternative, we used dynamic light scattering (DLS), which is much faster and more reliable for this type of protein. Purified SlDXS1 was mixed with 50 μM TPP in the absence or presence of 50 μM IPP or DMAPP and then analyzed by DLS. In the absence of IPP/DMAPP, the estimated radius of the protein (5.8 nm) correlates with the dimeric form of the enzyme of about 150 kDa. By contrast, in the presence of either IPP or DMAPP, the peak shifted to a smaller radius, and the range of particle sizes increased (Figure 2C). From these data, we speculated that IPP and DMAPP binding to the MIR might somehow interfere with the dimerization of SlDXS1, displacing the monomer–dimer equilibrium to the monomeric forms that might eventually aggregate, forming the high-molecular-weight particles detected by DLS (Figure 2C).
IPP/DMAPP promote monomerization of bacterial and plant DXS enzymes
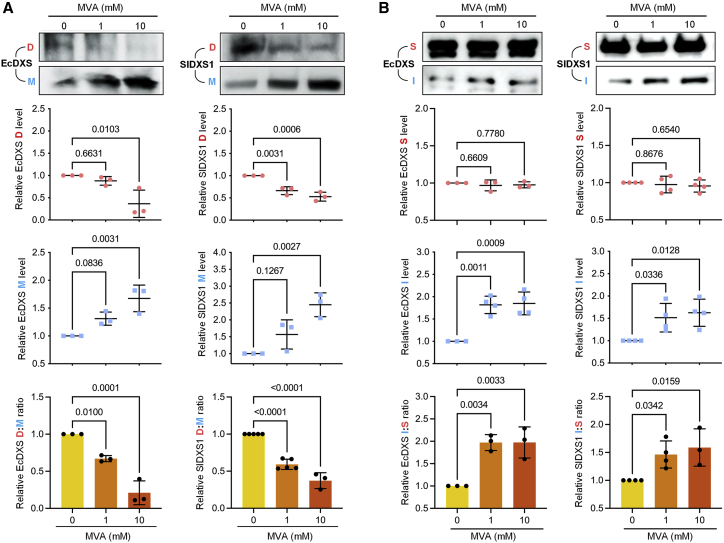
To validate our interpretation of the DLS experiments, we analyzed whether the oligomeric state of EcDXS and SlDXS1 changed in the presence of high levels of IPP/DMAPP in vivo. We initially expressed constructs encoding 6×His-tagged versions of the full-length EcDXS protein or the truncated SlDXS1 protein lacking the N-terminal plastid-targeting peptide in the E. coli strain EcAM5-1, which harbors the MVA operon in a BL21(DE3) background (Rodriguez-Villalon et al., 2008). Transformed cells were grown at 37°C until the exponential phase, and then arabinose and IPTG were added to induce expression of the MVA operon and the recombinant DXS enzyme, respectively. Cultures were also supplemented with 0, 1, or 10 mM MVA and then incubated for another 3 h at 26°C. The quaternary structure of recombinant DXS proteins in the cultures was analyzed by SDS-PAGE followed by immunoblot analysis with an anti-6×His antibody after incubating total protein extracts with the crosslinker dimethyl suberimidate. In the presence of increasing concentrations of MVA (i.e., as IPP/DMAPP levels increased), the proportion of dimers decreased, whereas monomeric forms of EcDXS and SlDXS1 increased (Figure 3A). The structural change was even clearer when representing the dimer-to-monomer ratio (Figure 3A). These results demonstrate that high IPP and DMAPP levels decrease the proportion of active DXS dimers and increase the amounts of monomeric forms of bacterial and plant enzymes when expressed in E. coli cells.
Figure 3.
Increasing IPP/DMAPP levels promote monomerization and aggregation of bacteria and plant DXS enzymes in E. coli.
EcAM5-1 cells were transformed with constructs to express 6×His-tagged versions of bacterial (EcDXS) and plant (SlDXS1) enzymes, and positive transformants were grown in medium supplemented with the indicated amounts of MVA. Samples were then taken to analyze (A) the accumulation of dimers (D) and monomers (M) of the enzymes or (B) soluble (S) and insoluble/aggregated (I) proteins. Top panels show representative images of immunoblot analyses with anti-6×His antibodies. Quantification of abundance from immunoblot analyses of three or more independent experiments is presented in the plots. Individual data points as well as mean and standard deviation are presented. Numbers in the plots indicate p values (one-way ANOVA with Dunnett’s multiple comparison test).
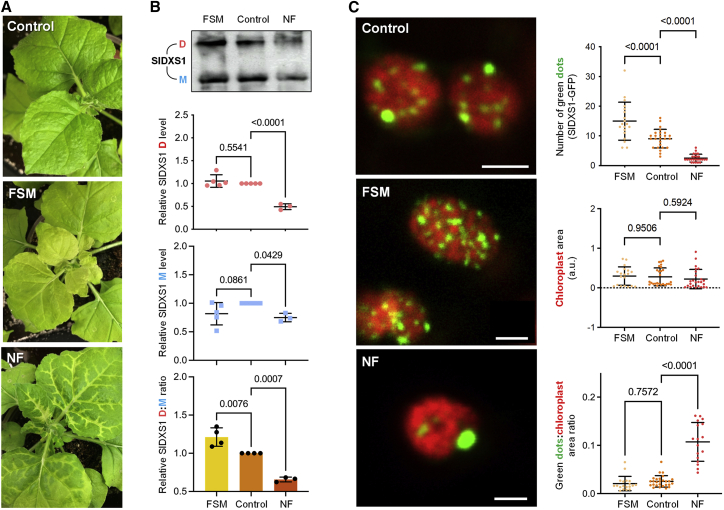
To validate the results obtained with the tomato SlDXS1 protein using a plant system, we agroinfiltrated constructs encoding the full-length protein into tobacco (Nicotiana benthamiana) leaves. The amounts of IPP/DMAPP in leaf cells were modulated using the MEP pathway inhibitor fosmidomycin (FSM) and the carotenoid biosynthesis inhibitor norflurazon (NF) (Figure 1A). Although both inhibitors cause a bleaching phenotype because of photooxidative damage associated with the loss of carotenoids (Figure 4A), FSM causes a drop in IPP/DMAPP levels, whereas NF is expected to increase the amount of these isoprenoid precursors by preventing their consumption by the carotenoid pathway (Figure 1A). Leaf bleaching associated with the activity of these inhibitors, which were added to the agroinfiltration mixture, was clearly observed at 5 dpi (days post inoculation) (Figure 4A). This time point was selected to take samples for LC-MS analysis of MEP pathway intermediates and for protein extraction and immunoblot analysis using an anti-DXS serum (Pulido et al., 2013). Our LC-MS method was not sensitive enough to detect IPP/DMAPP in tobacco leaves agroinfiltrated with the SlDXS1 construct, but we could successfully quantify the levels of DXP, MEP, and the downstream intermediate methylerythritol-cyclodiphosphate (ME-cPP). FSM treatment caused an accumulation of DXP and a drop in MEP and MEcPP levels compared with untreated controls (Table 2; Supplemental Figure 3). By contrast, NF treatment reduced the levels of DXP, MEP, and MEcPP (Table 2; Supplemental Figure 3), as expected, considering that increased IPP/DMAPP levels should result in lower DXS activity. Consistent with our interpretation, the dimer-to-monomer ratio of SlDXS1 decreased in plant leaves treated with NF (Figure 4B). Also in agreement with the results obtained previously in E. coli, reduced IPP/DMAPP levels in FSM-treated samples led to an increased dimer-to-monomer ratio compared with mock-treated controls (Figure 4B). Although bleaching and increased oxidative stress derived from FSM and NF treatments may affect DXS activity by mechanisms other than changes in IPP/DMAPP contents (Mitra et al., 2021), the negative correlation between the dimer-to-monomer ratio and IPP/DMAPP levels strongly supports our conclusion that IPP and DMAPP are able to modulate DXS activity in vivo by interfering with formation of the active dimeric form of the enzyme in bacterial and plant systems.
Figure 4.
Changing IPP/DMAPP levels modulate monomerization and aggregation of SlDXS1 in N. benthamiana.
Leaves from N. benthamiana plants were infiltrated with constructs to produce SlDXS1 together with either water (control) or inhibitors, and samples were collected 5 days later.
(A) Representative images of the leaves.
(B) Immunoblot analyses of protein extracts with anti-DXS antibodies. The position of dimers (D) and monomers (M) is indicated in the top panel, which shows the result of a representative experiment. Quantification of D and M abundance from immunoblot analyses of three or more independent experiments is presented in the plots.
(C) Representative confocal microscopy images of SIDXS1-GFP (green) and chlorophyll (red) fluorescence in chloroplasts (scale bar, 2 μm) and quantitative values from seven or more pictures corresponding to different leaves. Individual data points as well as mean and standard deviation are presented in the plots. Numbers in the plots indicate p values (one-way ANOVA with Dunnett’s multiple comparison test).
Table 2.
Concentrations of MEP pathway intermediates in leaves infiltrated with SlDXS1 and the indicated inhibitors.
| Metabolite concentration (pg/mg dry weight) | ||||
|---|---|---|---|---|
| m/z | 213.016963 | 215.032613 | 276.988379 | |
| RT | 7.42 | 7.95 | 6.79 | |
| DXP | MEP | MEcPP | ||
| FSM | 641.99 | 7.67 | 31.03 | |
| Control | 7.69 | 351.35 | 671.68 | |
| NF | 5.73 | 175.33 | 405.61 | |
Mass-to-charge (m/z) ratios and retention times (RT) used for identification of individual metabolites are shown.
IPP/DMAPP promote DXS aggregation in bacteria and plant cells
DXS monomers expose highly hydrophobic domains that make them prone to aggregation (Pulido et al., 2016). To test whether IPP/DMAPP-induced monomerization of EcDXS and SlDXS1 proteins favored their aggregation, we expressed 6×His-tagged versions of both proteins in EcAM5-1 cells, induced IPP/DMAPP accumulation in vivo by supplementing the growth medium with 1 or 10 mM MVA, and then isolated proteins from soluble and insoluble fractions for immunoblot analysis with anti-6×His serum (Figure 3B). Most of the recombinant EcDXS and SlDXS1 proteins were found in the soluble fractions, with no major differences in the presence or absence of MVA (Figure 3B). In the insoluble fraction, however, the amounts of EcDXS and SlDXS1 increased in MVA-supplemented cells (i.e., when IPP/DMAPP levels increased), resulting in a higher ratio of insoluble (aggregated) to soluble (disaggregated) protein (Figure 3B).
The aggregation status of the SlDXS1 protein was confirmed in plant cells by observing the accumulation of a GFP-tagged fusion, SlDXS1-GFP. Arabidopsis DXS enzymes fused to GFP are localized in the plastid stroma, but they can also be observed forming a spotted distribution corresponding to aggregates when overexpressed (Pulido et al., 2013, 2016; Perello et al., 2016). As expected, transient overexpression of SlDXS1-GFP in N. benthamiana leaves also led to formation of fluorescent speckles identified by confocal laser scanning microscopy (Figure 4C). Quantification of the number of fluorescent spots (i.e., SlDXS1-GFP aggregates) per chloroplast in leaves treated with inhibitors that alter IPP/DMAPP levels showed higher amounts with FSM and lower amounts with NF compared with untreated controls (Figure 4C). The size of the spots, however, was larger in NF samples, in agreement with the conclusion that SlDXS1 aggregation increases when IPP/DMAPP levels increase.
Discussion
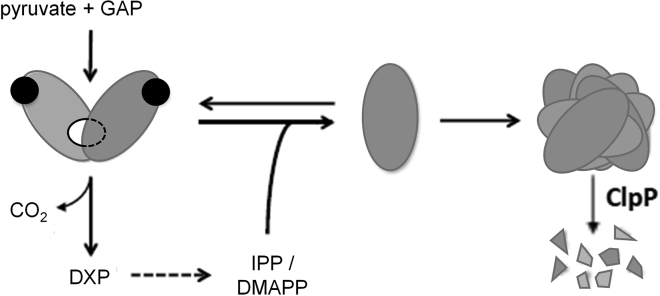
Regulation of the MEP pathway to produce the building blocks required for biosynthesis of isoprenoids (IPP and DMAPP) has been revealed to be rather complex, encompassing transcriptional, post-transcriptional, post-translational, and metabolic levels (Hemmerlin 2013; Vranová et al., 2013; Banerjee and Sharkey, 2014; Rodriguez-Concepcion and Boronat, 2015). This is especially relevant for the first enzyme of the pathway (DXS), which catalyzes the TPP-dependent conversion of pyruvate and GAP into DXP in the main rate-limiting step of the pathway (Wright et al., 2014; Volke et al., 2019). Here we propose that a mechanism by which accumulation of intracellular IPP and DMAPP inhibits the activity of DXS is conserved in bacteria and plant plastids. This mechanism involves (1) direct binding of IPP/DMAPP to DXS in a location different from the TPP-binding active site, followed by (2) enhanced monomerization of the enzyme, and eventually (3) aggregation of the inactive monomers (Figure 5). The proposed mechanism enables a short-term response to a sudden increase or decrease in IPP/DMAPP supply (by rapidly shifting the dimer-monomer equilibrium accordingly) but also a long-term response if IPP/DMAPP abundance persists (as monomers aggregate and become unavailable to form active DXS dimers).
Figure 5.
Model of the molecular mechanism involved in allosteric feedback inhibition of DXS.
The first step of the MEP pathway is production of DXP from pyruvate and GAP, a reaction catalyzed by DXS. The active DXS enzyme is a dimer and requires binding of the cofactor TPP (represented as black circles) to the active site present in each of the subunits. Allosteric changes induced by binding of IPP/DMAPP (likely to a site in the contact area between the two monomers, represented as a white circle) shifts the dimer:monomer equilibrium towards the monomeric (inactive) forms, causing immediate downregulation of enzyme activity. If monomers are not used to make new dimers (e.g., because of the presence of high IPP/DMAPP levels), exposed hydrophobic domains can lead to monomer aggregation and eventual degradation by the Clp protease complex (ClpP), resulting in sustained removal of DXS activity.
DXS, in its dimeric active form, has been described as a three-domain polypeptide, with domains I and II from the same chain involved in formation of the active site and binding of TPP. Domain III is involved in formation of the dimer interface, which includes a highly hydrophobic surface that remains unexposed to the solvent in the dimer (Xiang et al., 2007; Chen et al., 2019; Yu et al., 2021). The reaction mechanism involves formation of a covalent intermediate between enzyme-bound TPP and pyruvate, followed by GAP-stimulated decarboxylation of the bound pyruvate and incorporation of GAP into the remaining fragment to generate DXP (Xiang et al., 2007; Patel et al., 2012; Chen et al., 2019). Using a recombinant DXS protein from the tree Populus trichocarpa, it was proposed that IPP and DMAPP might compete with TPP for its pocket in the active site formed in each of the two subunits of the homodimer, with inhibitor constant values of ∼65 μM for IPP and 81 μM for DMAPP (Banerjee et al., 2013). This competition is striking, considering that TPP is generally thought to be tightly bound as an integral part of DXS but also that it functions as a cofactor of many enzymes unrelated to isoprenoid synthesis for whom regulation by IPP/DMAPP might not be physiologically meaningful. Experimental data from enzyme activity assays could not be fit to standard competitive inhibition kinetics, and instead, a negative cooperative model was proposed, in which binding of IPP/DMAPP to one of the DXS monomers would somehow hamper the binding of a second molecule to the other subunit of the homodimer (Banerjee et al., 2013). Furthermore, mutation of residues at the active site of P. trichocarpa DXS that were predicted to be critical for binding the IPP/DMAPP carbon chain was found to have opposite effects on IPP-mediated inhibition of enzyme activity (Banerjee et al., 2016). Our ITC experiments using SlDXS1 in the presence of an excess of TPP, IPP, and DMAPP confirmed that these three small diphosphate metabolites could indeed interact with the enzyme but showed KD values for IPP and DMAPP that were very similar in the absence or presence of TPP (Table 1; Supplemental Figure 1). These results argue against direct competition of IPP/DMAPP for the TPP binding site in DXS and instead suggest the existence of an alternative binding site for the MEP pathway products. The inconsistencies between these two models of DXS inhibition by IPP/DMAPP might derive from the underlying methodology; i.e., enzymatic versus calorimetric assays. ITC is a powerful tool for studying protein/ligand binding interactions that involves directly measuring the heat that is released or absorbed in real time when one solution is titrated into another (Wang et al., 2020). ITC is capable of providing information on enzymatic reactions that is difficult to obtain using traditional (indirect) biochemical assays. In particular, use of DXS substrates (pyruvate) together with additional metabolites (dihydroxyacetone phosphate) and enzymes (triose-phosphate isomerase from rabbit muscle) in the reaction mixtures used previously to determine the inhibition dynamics of IPP/DMAPP (Banerjee et al., 2013) might have resulted in interference that influenced the final interpretation of the data. Although our docking analyses identified a binding site for IPP and DMAPP in the active site, they also unveiled a second potential binding domain for these MEP pathway products that might function as the allosteric site (Figure 2).
Allosteric regulation of protein function occurs when binding of a molecule that either activates or inhibits the activity of the protein takes place away from the active site. This differs from competitive inhibition, in which the inhibitor binds to the active site and prevents the protein’s natural substrate from gaining access. In contrast to the previously proposed model of negative cooperative inhibition of TPP binding by IPP/DMAPP (Banerjee et al., 2013, 2016), our data support the conclusion that IPP/DMAPP-mediated inhibition of DXS is truly allosteric because the modulation of enzyme activity involves binding outside of the active site. Many proteins are allosterically regulated by a variety of mechanisms (Laskowski et al., 2009). Our model of IPP/DMAPP inhibition of DXS activity is a typical example of allosteric regulation, in which the downstream products of a biosynthetic pathway downregulate the activity of the enzyme that catalyzes the first committed step through feedback inhibition, hence ensuring that pathway flux is adjusted to end-product usage (Gerhart 2014; Ruszkowski 2018; Wang et al., 2020). In most cases, binding of the ligand inhibits enzyme activity by promoting a conformational change (Laskowski et al., 2009). Because prediction of the energetics and mechanisms of protein conformational changes by ITC remains very challenging, we used DLS to initially address this possibility. Indeed, our results showed that IPP/DMAPP binding disrupts the dimeric structure of the active SlDXS1 enzyme, causing a peak shift toward smaller (monomeric) forms (Figure 2). The presence of a likely IPP/DMAPP binding site in the region where DXS monomers interact (Figure 2; Supplemental Figure 2) is in agreement with a model in which binding of these ligands to this site (MIR) interferes with dimerization. Further analysis of EcDXS and SlDXS1 dimeric and monomeric forms by immunoblotting experiments confirmed a decreased dimer-to-monomer ratio when levels of IPP and DMAPP increased in living bacteria (Figure 3A) and plant cells (Figure 4B). Changes in the dimer-to-monomer ratio triggered by allosteric regulation have been observed in different proteins, including receptors (Petersen et al., 2017), membrane translocators (Jaipuria et al., 2017), and enzymes (Seetoh and Abell, 2016). In the case of DXS, however, IPP/DMAPP-mediated monomerization was found to be linked to increased aggregation in bacteria (Figure 3B) and plastids (Figure 4C). DXS exists in a bimodal distribution of open and closed conformations. In the catalytic reaction mechanism described for DXS, the dimeric enzyme adopts a closed conformation when pyruvate binds the TPP-containing active site and then changes to an open state upon GAP binding (Zhou et al., 2017; Chen et al., 2019; DeColli et al., 2019). In the open conformation, solvent-accessible areas are increased in different segments near the active site, exposing hydrophobic domains in the interface of the two monomers that are normally buried in the quaternary structure of the closed dimer. This might partially explain the previously observed propensity of this protein to aggregate in plants (Pulido et al., 2013, 2016) and microbes (Kudoh et al., 2017a, 2017b). Most interestingly, DXS monomers fully expose the aggregation-prone domains (Pulido et al., 2016). We therefore conclude that high IPP/DMAPP levels shift the dimer–monomer equilibrium of DXS to monomeric forms for rapid downregulation of enzyme activity. Monomers would expectedly be ready for immediate dimerization and hence enzyme reactivation upon return of IPP/DMAPP to steady-state levels. However, if IPP/DMAPP levels remain high (e.g., because of a blockage in their consumption by downstream isoprenoid biosynthetic pathways), then monomers would remain available for aggregation to drastically reduce DXS activity.
Our model, summarized in Figure 5, not only provides a mechanistic explanation of how MEP-derived IPP and DMAPP supplies can be adapted to changes in their demand but also explains the changes in DXS protein levels observed after long-term interference in the MEP pathway flux (Han et al., 2013; Ghirardo et al., 2014; Pokhilko et al., 2015). In particular, genetic or pharmacological reduction of IPP/DMAPP levels causes upregulation of DXS protein levels without concomitant changes in gene expression (Han et al., 2013; Ghirardo et al., 2014; Pokhilko et al., 2015), whereas mutants that are unable to produce carotenoids and are thus expected to accumulate higher IPP/DMAPP levels (similar to NF treatments) showed an opposite phenotype of reduced DXS protein levels (Pokhilko et al., 2015). Under non-stressed conditions, DXS aggregates are degraded by the Clp protease complex in different organisms, including plants but also microorganisms (Ninnis et al., 2009; Pulido et al., 2013; Pulido et al., 2016; Florentin et al., 2017; D'Andrea et al., 2018; Moreno et al., 2018). We therefore propose that a prolonged shortage of IPP/DMAPP might lead to mostly dimeric (i.e., soluble and enzymatically active) DXS, whereas sustained IPP/DMAPP abundance would cause enhanced monomerization and aggregation of the protein, eventually resulting in efficient removal by the Clp protease (Figure 5).
In summary, our data support an evolutionarily conserved and mechanistically simple system in which the MEP pathway products IPP and DMAPP (the universal precursors of all isoprenoids) allosterically control the activity of DXS (the main rate-limiting enzyme of the pathway) by modulating the equilibrium between dimeric (active) and monomeric/aggregated (inactive) forms of the enzyme. Peak demand for MEP pathway products (e.g., to produce monoterpenes in response to a pathogen attack or carotenoids upon high light stress) would decrease the amount of IPP/DMAPP, promoting DXS dimerization and hence maximizing activity to increase pathway flux. By contrast, build-up of IPP/DMAPP caused by reduced consumption of these metabolites would promote DXS monomerization, which could eventually result in aggregation and subsequent proteolytic removal of the protein (Figure 5). Although these results do not exclude other mechanisms for regulation of DXS activity through binding of other metabolites, they improve our surprisingly scarce knowledge about how the MEP pathway is regulated (Hemmerlin 2013; Vranová et al., 2013; Banerjee and Sharkey, 2014; Pokhilko et al., 2015; Mitra et al., 2021). The MEP pathway, probably the main metabolic pathway elucidated in this century (Rodríguez-Concepción and Boronat 2002), provides the precursors for a large diversity of isoprenoids with high added value from industrial and nutritional points of view. Understanding its regulation, in particular that of its main enzyme, DXS, is therefore necessary for rational design of biotechnological endeavors aimed at increasing isoprenoid contents in microbial and plant systems.
Methods
Bacterial strains, plant material, and growth conditions
E. coli and Agrobacterium tumefaciens strains were grown in LB (10 g/l Bacto Tryptone, 5 g/l yeast extract, and 10 g/l NaCl, plus 15 g/l Bacto Agar for plates) supplemented with antibiotics to a final concentration of 100 μg/ml ampicillin, 50 μg/ml kanamycin, 34 μg/ml chloramphenicol, 100 μg/ml rifampicin, 30 μg/ml gentamicin, or/and 100 μg/ml spectinomycin when required. Bacterial growth in liquid medium was monitored by measuring optical density at 600 nm (OD600). The strains used in this study are described in Supplemental Table 1. N. benthamiana plants were grown under standard greenhouse conditions.
Gene constructs
To obtain 6×His-tagged proteins, we cloned DXS-encoding bacterial and plant sequences into the pET23 vector (Novagen). The sequence of the E. coli gene encoding EcDXS was PCR amplified from wild-type strain MG1655 genomic DNA with primers EcDXS-NdeI-F and EcDXS-XhoI-R (Supplemental Table 2), and the fragment obtained was digested with NdeI and XhoI for ligation to a pET23 backbone (previously digested with the same enzymes) to generate the construct pET23-EcDXS. The coding sequence of SlDXS1 was amplified from an S. lycopersicum ripe fruit cDNA library using primers NheI-myc-SlDXS1-F and SlDXS1-XhoI-R (Supplemental Table 2). The DNA fragment of the expected size was digested with NheI and XhoI and ligated to a pET23 vector digested previously with the same enzymes to yield the construct pET23-SlDXS1 for expression in E. coli. Constructs pGWB420-SlDXS1 and pGWB405-SlDXS1-GFP for expression in plants were obtained following a two-step (BP/LR) Gateway reaction, yielding proteins fused to C-terminal myc or GFP tags, respectively. Primers and constructs are listed in Supplemental Table 2. All constructs were sequenced to confirm the identity of the genes and the absence of undesired mutations.
Production and purification of recombinant proteins
Following transformation of E. coli BL21(DE3) pLysS cells with the required construct, production of recombinant DXS proteins was induced by adding 50 μM IPTG to cultures with an OD600 ≈ 0.6. After growth for 14 h at 28°C, bacterial cells were recovered by centrifugation, and the recombinant protein was purified. The cell pellet was resuspended in buffer A (50 mM HEPES, 150 mM NaCl, pH 7.5) supplemented with 1 mg/ml lysozyme, 0.5 mM EDTA, and one tablet of cOmplete protease inhibitor cocktail (Roche) for every 10 ml of buffer. The resuspended pellet was incubated on ice for 20 min with light shaking, and after brief sonication (6 pulses of 30 s at 10% with a 45-s pause between pulses), the cell lysate was centrifuged at 27 000 g for 20 min to separate broken cells. A 1:7 volume of protamine sulphate (1% in water) was added to the supernatant, and the mixture was centrifuged at 39 000 g for 1 h. The cleaned and filtered supernatant was loaded into a Poly-Prep chromatography column (Bio-Rad) containing 1.5 ml of Ni-NTA agarose (Qiagen) previously cleaned and equilibrated with buffer A. After adding 10 ml of buffer A and 5 ml of washing buffer (buffer A containing 10 mM imidazole), the recombinant protein was eluted with 1.5-ml aliquots of elution buffer (buffer A supplemented with 150 mM imidazole). Fractions containing the purified protein were pooled, and imidazole was removed using a PD10 desalting column (Cytiva) equilibrated with buffer A. The protein was collected after centrifugation at 1000 g for 2 min and stored as single-use aliquots at −80°C.
In vivo inhibition assays
E. coli strain EcAB4-10 was grown on LB plates supplemented with kanamycin, chloramphenicol, and MVA. Five colonies were grown in liquid medium at 37°C and 200 rpm overnight to inoculate a 15-ml fresh culture containing 0.1% arabinose and either 0.5 or 10 mM MVA at an initial OD600 of 0.1. Cells were grown at 37°C and 200 rpm until OD was ≈1 to ensure a steady state for proper analysis, collected by centrifugation, and stored at −80°C. For metabolite analysis, samples were thawed on ice, immediately quenched with 400 μl methanol, and lysed, mixing thoroughly. Then, 400 μl of water was added and mixed in before centrifugation at 13 000 g for 5 min at 4°C. About 750 μl of the supernatant was collected, and macromolecules were removed using a MilliporeSigma 3-kDa Amicon Ultra centrifuge filter tube at 4°C and 13 000 g for 1 h. Then, 700 μl of the filtrate was mixed with 1 ml of water and flash frozen in liquid nitrogen. Frozen samples were lyophilized and stored at −80°C until reconstitution with 100 μl of methanol:water (1:1 [v/v]) for LC-MS analysis as described by Amer et al. (2022). For plant metabolite extraction, the lyophilized leaf tissue (50–80 mg) was resuspended in 1 ml of cold methanol (kept on ice) and vortexed for 5 min with a TOMY Micro Tube Mixer MT-400 in a 4°C cold room. The methanol extract was centrifuged at 13 000 g and 4°C for 3 min. Then, a 700-μl volume of supernatant was transferred to a Falcon tube. These steps were repeated twice, with 1 ml of the supernatant (instead of 700 μl) transferred to the same Falcon tube each time, resulting in a total volume of 2.7 ml. The pelleted plant tissue was resuspended in 1 ml of cold water (kept on ice) and vortexed for 5 min as described above. The water extract was centrifuged at 13 000 g and 4°C for 3 min, and then 1 ml of supernatant was transferred to a separate Falcon tube. The water extraction steps were repeated twice, with 950 μl of the supernatant (instead of 1 ml) transferred to the same Falcon tube each time, resulting in a total volume of 2.9 ml. Then, 1 ml of the methanol extraction supernatant was mixed with 1 ml of the water extraction supernatant and transferred to a MilliporeSigma Amicon Ultra centrifuge filter tube (i.e., 2 ml of metabolite extract in 1:1 methanol:water). After centrifugation at 4000 g in a swing bucket centrifuge (Allegra X-15 centrifuge, Beckman Coulter) for 1 h at 4°C, the filtrate was flash frozen with liquid nitrogen, lyophilized, and reconstituted in 100 μl of 1:1 methanol:water for LC-MS analysis as described previously (Amer et al., 2022).
ITC
The interaction between the DXS proteins and TPP, IPP, or DMAPP was assessed using a high-precision Auto-iTC200 calorimeter (MicroCal, Malvern Panalytical). Protein solutions in the calorimetric cell at 20 μM were titrated with the different ligands (TPP, IPP, or DMAP) at 200 μM in the injecting syringe using buffer A (50 mM HEPES, 150 mM NaCl, pH 7.5) as the common buffer solvent. A sequence of 19 injections of 2 μl each was programmed, with a stirring speed of 750 rpm, a 150-s spacing, and an applied reference power of 10 μcal/s. The association constant (KA) was estimated by non-linear least-squares regression analysis of the data with a model that considered a single ligand binding site (1:1 protein:TPP/IPP/DMAPP) using a user-defined fitting routine implemented in Origin 7.0 (OriginLab, Northampton, MA). KD was calculated as the inverse of KA.
DLS
DLS measurements were performed in a DynaPro Plate Reader III (Wyatt Technology) using a 384-multiwell plate (Aurora Microplates). The hydrodynamic radius of SlDXS1 was measured before and after addition of TPP, DMAPP, and IPP to estimate the size distribution of the protein. A second measurement was performed after 10 min of incubation to assess the evolution of the complex. For each measurement, 5 acquisitions of 5 s were taken, and the apparent hydrodynamic radius was estimated from the experimental diffusion coefficient, obtained by the cumulant fit of the translational autocorrelation function, assuming an equivalent Rayleigh sphere model. Experiments were performed with a fixed protein concentration of 20 μM, and the ligands IPP, DMAPP, and TPP at 50 μM in the same buffer were used for ITC experiments.
In vivo DXS oligomeric state analysis in bacteria
E. coli EcAM5-1 cells were transformed with construct pET23-EcDXS or pET23-SlDXS1, and single colonies were grown overnight in LB medium supplemented with ampicillin and kanamycin at 37°C. Fresh medium (150 ml) was inoculated at 1.5% and grown at 37°C and 200 rpm until OD600 ≈ 0.6. Following addition of 0.1% arabinose and 0.05 mM IPTG, the culture was split in three, and 0 mM, 1 mM, or 10 mM MVA was added to each of the three subcultures. All subcultures were then grown at 26°C and 200 rpm for 3 h, and cells were collected by centrifugation. Samples were ground in liquid nitrogen and extracted with 0.2 M triethanolamine (pH 8). The mixture was centrifuged at 13 000 g for 10 min at 4°C, and the supernatant was treated for 40–50 min at room temperature with a 10-fold molar excess of dimethyl suberimidate. The crosslinking reaction was stopped by addition of 20 mM Tris. To separate soluble and insoluble (aggregate) protein fractions, samples were ground in liquid nitrogen, extracted with 0.2 M triethanolamine (pH 8), and centrifuged at 13 000 g for 10 min at 4°C, and the supernatant was collected as the soluble protein fraction. The cell pellet was washed with 0.2 M triethanolamine and resuspended in insoluble protein extraction buffer (8 M urea, 10 mM Tris, and 100 mM NaH2PO4) for 20 min at room temperature. After centrifugation at 13 000 g for 10 min, the supernatant was collected as the insoluble protein fraction. The concentrations of soluble and insoluble protein fractions were determined using the Bio-Rad protein assay. Samples were resolved by 7% SDS-PAGE, transferred to a polyvinylidene fluoride membrane (Amersham), and used for immunoblot analysis with 1:2500 anti-6×His antibody (Proteintech) for 10 min at room temperature. Detection of immunoreactive bands was performed using SuperSignal West Pico PLUS (Thermo Scientific). Chemiluminescent signals were visualized using an ImageQuant 800 biomolecular imager (Amersham) and quantified using ImageJ software. Statistical significance of quantified differences was analyzed with GraphPad.
In vivo DXS oligomeric state analysis in planta
A. tumefaciens GV3101 cells were transformed with the construct pGWB420-SlDXS1 or pGWB405-SlDXS1-GFP, and a single transformed colony was grown overnight in 3 ml LB medium supplemented with rifampicin, gentamycin, and spectinomycin at 28°C. Fresh medium (25 ml) was inoculated with the preculture and grown overnight at 28°C. The culture was centrifuged to collect cells, and the pellet was resuspended in agroinfiltration buffer (10 mM MgCl2, 10 mM MES, 150 μM acetosyringone, pH 5.6) to achieve OD600 = 0.8. The mixture was incubated at 28°C and 200 rpm for 1.5 h and then mixed with a similar culture harboring vector pGWB702-HCProWMV to prevent silencing (Andersen et al., 2021). The 9:1 (DXS:HcProWMV) mix was split in three, and the individual fractions were supplemented with FSM (200 μM), NF (40 μM), or water before infiltration of leaves from 4-week-old N. benthamiana plants as described previously (Andersen et al., 2021). Leaf samples agroinfiltrated with the pGWB420-SlDXS1 construct were collected at 5 dpi, snap-frozen in liquid nitrogen, and stored at −80°C. To extract proteins, around 300 mg of frozen tissue was ground in liquid nitrogen and mixed with 450 μl TKMES buffer (Pulido et al., 2013) supplemented with 20 μl/ml protease inhibitor cocktail and 20 μl/ml 10% Triton-100 (v/v). The mixture was centrifuged at 2800 g for 10 min at 4°C, and the supernatant was further cleared with two additional centrifugation steps. The protein concentration of the extract was measured with the Bio-Rad protein assay. Samples were mixed with non-denaturing loading buffer, separated by 7% SDS-PAGE, and then transferred to a polyvinylidene fluoride membrane. The membrane was incubated overnight at 4°C with 1:500 anti-DXS antibody (Pulido et al., 2013). Chemiluminescent signals and statistical analysis were performed as described above. Subcellular localization of GFP-tagged SlDXS1 was observed by direct examination of agroinfiltrated leaf tissue at 3 dpi using a confocal microscope (Axio Observer 780, Carl Zeiss) with a BP515-525 filter after excitation at 488 nm. The areas of the whole chloroplast and the SlDXS1-GFP green fluorescent dots were quantified from pictures using ImageJ.
Docking simulations
The crystal structure of EcDXS in complex with TPP (PDB: 2O1S) was obtained from the RCSB Protein Data Bank (https://www.rcsb.org/). The protein structure was prepared using the Protein Preparation Wizard from the Schrödinger package. Identification of potential ligand-binding sites was performed using the SiteMap tool from Schrödinger (Halgren, 2007). All potential sites were scored according to their size, functionality, and extent of solvent exposure, and those with higher site scores were visualized and carefully analyzed. Values greater than 0.80 usually define a hit, and sites with values close to 1.0 are considered to be particularly promising. Specific ligands to be tested (TPP, IPP, and DMAPP) were prepared using the Ligand Preparation Wizard LigPrep tool from Schrödinger. More specifically, they were first converted to a 3D structure, geometrically optimized by OPLS3e force field-based minimization, and accurately protonated using Epik (Shelley et al., 2007) at pH 7 ± 2. Docking simulations were carried out against different DXS ligand-binding sites using the Glide XP module from Schrödinger with a flexible ligand sampling and the extra-precision settings.
Funding
This work was funded by grants from the Spanish MCIN/AEI/10.13039/501100011033 and European ERDF/FEDER, NextGeneration EU/PRTR and PRIMA programs (PID2020-115810GB-I00 and UToPIQ-PCI2021-121941 to M.R.-C. and BFU2016-78232-P to A.V.-C.). M.R.-C. is also supported by CSIC (202040E299) and Generalitat Valenciana (PROMETEU/2021/056). R.K. and E.E.K.B. conducted the metabolite analysis at the Joint BioEnergy Institute (http://www.jbei.org), supported by the US Department of Energy, Office of Science, Office of Biological and Environmental Research under contract DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory and the US Department of Energy. J.P.-G. was supported by a Marie Curie International Outgoing Fellowship within the EC-FP7 Program (project 627639). X.D. was supported by the China Scholarship Council and D.O.-A. by an MCIN/AEI/fellowship (BES-2017-080739).
Author contributions
M.R.-C. and J.P.-G. conceptualized the whole study. X.D., M.R.-C., and J.P.-G. designed the experiments. X.D., D.O.-A., R.K., J.I.-F., L.D., E.E.K.B., and J.P.-G. performed the experiments. X.D., D.O.-A., J.I.-F., L.D., E.E.K.B., A.V.-C., M.R.-C., and J.P.-G. analyzed, discussed, and curated the data. M.R.-C. and J.P.-G. wrote the paper. All authors read and approved the final manuscript.
Acknowledgments
We thank Mª Rosa Rodriguez-Goberna (CRAG) and the Metabolics Platform at IBMCP for technical support. No conflict of interest declared.
Published: December 26, 2022
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Supplemental information is available at Plant Communications Online.
Contributor Information
Manuel Rodríguez-Concepción, Email: manuelrc@ibmcp.upv.es.
Jordi Perez-Gil, Email: jordi.perez@cragenomica.es.
Supplemental information
References
- Amer B., Kakumanu R., Baidoo E.E.K. HILIC-MS analysis of central carbon metabolites in Gram negative bacteria. 2022. [DOI]
- Andersen T.B., Llorente B., Morelli L., Torres-Montilla S., Bordanaba-Florit G., Espinosa F.A., Rodriguez-Goberna M.R., Campos N., Olmedilla-Alonso B., Llansola-Portoles M.J., et al. An engineered extraplastidial pathway for carotenoid biofortification of leaves. Plant Biotechnol. J. 2021;19:1008–1021. doi: 10.1111/pbi.13526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A., Sharkey T.D. Methylerythritol 4-phosphate (MEP) pathway metabolic regulation. Nat. Prod. Rep. 2014;31:1043–1055. doi: 10.1039/c3np70124g. [DOI] [PubMed] [Google Scholar]
- Banerjee A., Wu Y., Banerjee R., Li Y., Yan H., Sharkey T.D. Feedback inhibition of deoxy-D-xylulose-5-phosphate synthase regulates the methylerythritol 4-phosphate pathway. J. Biol. Chem. 2013;288:16926–16936. doi: 10.1074/jbc.M113.464636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A., Preiser A.L., Sharkey T.D. Engineering of recombinant poplar deoxy-D-xylulose-5-phosphate synthase (PtDXS) by site-directed mutagenesis improves its activity. PLoS One. 2016;11:e0161534. doi: 10.1371/journal.pone.0161534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongers M., Perez-Gil J., Hodson M.P., Schrübbers L., Wulff T., Sommer M.O., Nielsen L.K., Vickers C.E. Adaptation of hydroxymethylbutenyl diphosphate reductase enables volatile isoprenoid production. Elife. 2020;9:e48685. doi: 10.7554/eLife.48685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P.Y.T., DeColli A.A., Freel Meyers C.L., Drennan C.L. X-ray crystallography-based structural elucidation of enzyme-bound intermediates along the 1-deoxy-d-xylulose 5-phosphate synthase reaction coordinate. J. Biol. Chem. 2019;294:12405–12414. doi: 10.1074/jbc.RA119.009321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Andrea L., Simon-Moya M., Llorente B., Llamas E., Marro M., Loza-Alvarez P., Li L., Rodriguez-Concepcion M. Interference with Clp protease impairs carotenoid accumulation during tomato fruit ripening. J. Exp. Bot. 2018;69:1557–1568. doi: 10.1093/jxb/erx491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeColli A.A., Zhang X., Heflin K.L., Jordan F., Freel Meyers C.L. Active site histidines link conformational dynamics with catalysis on anti-infective target 1-Deoxy-d-xylulose 5-phosphate synthase. Biochemistry. 2019;58:4970–4982. doi: 10.1021/acs.biochem.9b00878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florentin A., Cobb D.W., Fishburn J.D., Cipriano M.J., Kim P.S., Fierro M.A., Striepen B., Muralidharan V. PfClpC is an essential Clp chaperone required for plastid integrity and Clp protease stability in Plasmodium falciparum. Cell Rep. 2017;21:1746–1756. doi: 10.1016/j.celrep.2017.10.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George K.W., Thompson M.G., Kim J., Baidoo E.E.K., Wang G., Benites V.T., Petzold C.J., Chan L.J.G., Yilmaz S., Turhanen P., et al. Integrated analysis of isopentenyl pyrophosphate (IPP) toxicity in isoprenoid-producing Escherichia coli. Metab. Eng. 2018;47:60–72. doi: 10.1016/j.ymben.2018.03.004. [DOI] [PubMed] [Google Scholar]
- Gerhart J. From feedback inhibition to allostery: the enduring example of aspartate transcarbamoylase. FEBS J. 2014;281:612–620. doi: 10.1111/febs.12483. [DOI] [PubMed] [Google Scholar]
- Ghirardo A., Wright L.P., Bi Z., Rosenkranz M., Pulido P., Rodríguez-Concepción M., Niinemets Ü., Brüggemann N., Gershenzon J., Schnitzler J.P. Metabolic flux analysis of plastidic isoprenoid biosynthesis in poplar leaves emitting and nonemitting isoprene. Plant Physiol. 2014;165:37–51. doi: 10.1104/pp.114.236018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halgren T. New method for fast and accurate binding-site identification and analysis. Chem. Biol. Drug Des. 2007;69:146–148. doi: 10.1111/j.1747-0285.2007.00483.x. [DOI] [PubMed] [Google Scholar]
- Han M., Heppel S.C., Su T., Bogs J., Zu Y., An Z., Rausch T. Enzyme inhibitor studies reveal complex control of methyl-D-erythritol 4-phosphate (MEP) pathway enzyme expression in catharanthus roseus. PLoS One. 2013;8:e62467. doi: 10.1371/journal.pone.0062467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmerlin A. Post-translational events and modifications regulating plant enzymes involved in isoprenoid precursor biosynthesis. Plant Sci. 2013;203-204:41–54. doi: 10.1016/j.plantsci.2012.12.008. [DOI] [PubMed] [Google Scholar]
- Jaipuria G., Leonov A., Giller K., Vasa S.K., Jaremko Ł., Jaremko M., Linser R., Becker S., Zweckstetter M. Cholesterol-mediated allosteric regulation of the mitochondrial translocator protein structure. Nat. Commun. 2017;8:14893. doi: 10.1038/ncomms14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudoh K., Kubota G., Fujii R., Kawano Y., Ihara M. Exploration of the 1-deoxy-d-xylulose 5-phosphate synthases suitable for the creation of a robust isoprenoid biosynthesis system. J. Biosci. Bioeng. 2017;123:300–307. doi: 10.1016/j.jbiosc.2016.10.005. [DOI] [PubMed] [Google Scholar]
- Kudoh K., Hotta S., Sekine M., Fujii R., Uchida A., Kubota G., Kawano Y., Ihara M. Overexpression of endogenous 1-deoxy-d-xylulose 5-phosphate synthase (DXS) in cyanobacterium Synechocystis sp. PCC6803 accelerates protein aggregation. J. Biosci. Bioeng. 2017;123:590–596. doi: 10.1016/j.jbiosc.2017.01.001. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., Gerick F., Thornton J.M. The structural basis of allosteric regulation in proteins. FEBS Lett. 2009;583:1692–1698. doi: 10.1016/j.febslet.2009.03.019. [DOI] [PubMed] [Google Scholar]
- Mitra S., Estrada-Tejedor R., Volke D.C., Phillips M.A., Gershenzon J., Wright L.P. Negative regulation of plastidial isoprenoid pathway by herbivore-induced β-cyclocitral in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2008747118. e2008747118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno J.C., Martínez-Jaime S., Schwartzmann J., Karcher D., Tillich M., Graf A., Bock R. Temporal proteomics of inducible RNAi lines of Clp protease subunits identifies putative protease substrates. Plant Physiol. 2018;176:1485–1508. doi: 10.1104/pp.17.01635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninnis R.L., Spall S.K., Talbo G.H., Truscott K.N., Dougan D.A. Modification of PATase by L/F-transferase generates a ClpS-dependent N-end rule substrate in Escherichia coli. EMBO J. 2009;28:1732–1744. doi: 10.1038/emboj.2009.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel H., Nemeria N.S., Brammer L.A., Freel Meyers C.L., Jordan F. Observation of thiamin-bound intermediates and microscopic rate constants for their interconversion on 1-deoxy-D-xylulose 5-phosphate synthase: 600-fold rate acceleration of pyruvate decarboxylation by D-glyceraldehyde-3-phosphate. J. Am. Chem. Soc. 2012;134:18374–18379. doi: 10.1021/ja307315u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perello C., Llamas E., Burlat V., Ortiz-Alcaide M., Phillips M.A., Pulido P., Rodriguez-Concepcion M. Differential subplastidial localization and turnover of enzymes involved in isoprenoid biosynthesis in chloroplasts. PLoS One. 2016;11:e0150539. doi: 10.1371/journal.pone.0150539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen J., Wright S.C., Rodríguez D., Matricon P., Lahav N., Vromen A., Friedler A., Strömqvist J., Wennmalm S., Carlsson J., et al. Agonist-induced dimer dissociation as a macromolecular step in G protein-coupled receptor signaling. Nat. Commun. 2017;8:226. doi: 10.1038/s41467-017-00253-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokhilko A., Bou-Torrent J., Pulido P., Rodríguez-Concepción M., Ebenhöh O. Mathematical modelling of the diurnal regulation of the MEP pathway in Arabidopsis. New Phytol. 2015;206:1075–1085. doi: 10.1111/nph.13258. [DOI] [PubMed] [Google Scholar]
- Pulido P., Toledo-Ortiz G., Phillips M.A., Wright L.P., Rodríguez-Concepción M. Arabidopsis J-protein J20 delivers the first enzyme of the plastidial isoprenoid pathway to protein quality control. Plant Cell. 2013;25:4183–4194. doi: 10.1105/tpc.113.113001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulido P., Llamas E., Llorente B., Ventura S., Wright L.P., Rodríguez-Concepción M. Specific Hsp100 chaperones determine the fate of the first enzyme of the plastidial isoprenoid pathway for either refolding or degradation by the stromal Clp protease in Arabidopsis. PLoS Genet. 2016;12:e1005824. doi: 10.1371/journal.pgen.1005824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Concepción M., Boronat A. Elucidation of the methylerythritol phosphate pathway for isoprenoid biosynthesis in bacteria and plastids. A metabolic milestone achieved through genomics. Plant Physiol. 2002;130:1079–1089. doi: 10.1104/pp.007138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Concepción M., Boronat A. Breaking new ground in the regulation of the early steps of plant isoprenoid biosynthesis. Curr. Opin. Plant Biol. 2015;25:17–22. doi: 10.1016/j.pbi.2015.04.001. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Concepcion M., D'Andrea L., Pulido P. Control of plastidial metabolism by the Clp protease complex. J. Exp. Bot. 2019;70:2049–2058. doi: 10.1093/jxb/ery441. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Villalón A., Pérez-Gil J., Rodríguez-Concepción M. Carotenoid accumulation in bacteria with enhanced supply of isoprenoid precursors by upregulation of exogenous or endogenous pathways. J. Biotechnol. 2008;135:78–84. doi: 10.1016/j.jbiotec.2008.02.023. [DOI] [PubMed] [Google Scholar]
- Ruszkowski M. Guarding the gateway to histidine biosynthesis in plants: medicago truncatula ATP-phosphoribosyltransferase in relaxed and tense states. Biochem. J. 2018;475:2681–2697. doi: 10.1042/BCJ20180289. [DOI] [PubMed] [Google Scholar]
- Sangari F.J., Pérez-Gil J., Carretero-Paulet L., García-Lobo J.M., Rodríguez-Concepción M. A new family of enzymes catalyzing the first committed step of the methylerythritol 4-phosphate (MEP) pathway for isoprenoid biosynthesis in bacteria. Proc. Natl. Acad. Sci. USA. 2010;107:14081–14086. doi: 10.1073/pnas.1001962107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauret-Güeto S., Urós E.M., Ibáñez E., Boronat A., Rodríguez-Concepción M. A mutant pyruvate dehydrogenase E1 subunit allows survival of Escherichia coli strains defective in 1-deoxy-D-xylulose 5-phosphate synthase. FEBS Lett. 2006;580:736–740. doi: 10.1016/j.febslet.2005.12.092. [DOI] [PubMed] [Google Scholar]
- Seetoh W.G., Abell C. Disrupting the constitutive, homodimeric protein-protein interface in CK2β using a biophysical fragment-based approach. J. Am. Chem. Soc. 2016;138:14303–14311. doi: 10.1021/jacs.6b07440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelley J.C., Cholleti A., Frye L.L., Greenwood J.R., Timlin M.R., Uchimaya M. Epik: a software program for pK( a ) prediction and protonation state generation for drug-like molecules. J. Comput. Aided Mol. Des. 2007;21:681–691. doi: 10.1007/s10822-007-9133-z. [DOI] [PubMed] [Google Scholar]
- Volke D.C., Rohwer J., Fischer R., Jennewein S. Investigation of the methylerythritol 4-phosphate pathway for microbial terpenoid production through metabolic control analysis. Microb. Cell Fact. 2019;18:192. doi: 10.1186/s12934-019-1235-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vranová E., Coman D., Gruissem W. Network analysis of the MVA and MEP pathways for isoprenoid synthesis. Annu. Rev. Plant Biol. 2013;64:665–700. doi: 10.1146/annurev-arplant-050312-120116. [DOI] [PubMed] [Google Scholar]
- Wang Y., Wang G., Moitessier N., Mittermaier A.K. Enzyme kinetics by isothermal titration calorimetry: allostery, inhibition, and dynamics. Front. Mol. Biosci. 2020;7:583826. doi: 10.3389/fmolb.2020.583826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfertz M., Sharkey T.D., Boland W., Kühnemann F. Rapid regulation of the methylerythritol 4-phosphate pathway during isoprene synthesis. Plant Physiol. 2004;135:1939–1945. doi: 10.1104/pp.104.043737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright L.P., Rohwer J.M., Ghirardo A., Hammerbacher A., Ortiz-Alcaide M., Raguschke B., Schnitzler J.P., Gershenzon J., Phillips M.A. Deoxyxylulose 5-phosphate synthase controls flux through the methylerythritol 4-phosphate pathway in Arabidopsis. Plant Physiol. 2014;165:1488–1504. doi: 10.1104/pp.114.245191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang S., Usunow G., Lange G., Busch M., Tong L. Crystal structure of 1-deoxy-D-xylulose 5-phosphate synthase, a crucial enzyme for isoprenoids biosynthesis. J. Biol. Chem. 2007;282:2676–2682. doi: 10.1074/jbc.M610235200. [DOI] [PubMed] [Google Scholar]
- Yu C., Leung S.K.P., Zhang W., Lai L.T.F., Chan Y.K., Wong M.C., Benlekbir S., Cui Y., Jiang L., Lau W.C.Y. Structural basis of substrate recognition and thermal protection by a small heat shock protein. Nat. Commun. 2021;12:3007. doi: 10.1038/s41467-021-23338-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Yang L., Decolli A., Freel Meyers C., Nemeria N.S., Jordan F. Conformational dynamics of 1-deoxy-d-xylulose 5-phosphate synthase on ligand binding revealed by H/D exchange MS. Proc. Natl. Acad. Sci. USA. 2017;114:9355–9360. doi: 10.1073/pnas.1619981114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.