Abstract
Climate change is increasing the frequency of extreme heat events that aggravate its negative impact on plant development and agricultural yield. Most experiments designed to study plant adaption to heat stress apply homogeneous high temperatures to both shoot and root. However, this treatment does not mimic the conditions in natural fields, where roots grow in a dark environment with a descending temperature gradient. Excessively high temperatures severely decrease cell division in the root meristem, compromising root growth, while increasing the division of quiescent center cells, likely in an attempt to maintain the stem cell niche under such harsh conditions. Here, we engineered the TGRooZ, a device that generates a temperature gradient for in vitro or greenhouse growth assays. The root systems of plants exposed to high shoot temperatures but cultivated in the TGRooZ grow efficiently and maintain their functionality to sustain proper shoot growth and development. Furthermore, gene expression and rhizosphere or root microbiome composition are significantly less affected in TGRooZ-grown roots than in high-temperature-grown roots, correlating with higher root functionality. Our data indicate that use of the TGRooZ in heat-stress studies can improve our knowledge of plant response to high temperatures, demonstrating its applicability from laboratory studies to the field.
Key words: heat stress, root, temperature gradient, nutrition, gene expression, microbiome
Climate change and heat are limiting factors for plant growth and productivity. This study provides new insight into plant heat response using an innovative device that generates a temperature gradient in the root system, simulating natural soil conditions.
Introduction
High temperature is an adverse condition that enormously impacts plant growth and fitness (Bhattacharya, 2019). Nowadays, climate change is leading to more frequent high-temperature extremes, such as heat waves, that aggravate the negative impact of heat on plant development and agricultural yield (Gray and Brady, 2016; Miller et al., 2021). In comparison with shoot responses, root responses to high temperatures have been understudied, and the majority of studies have focused on the response to warming, which induced thermomorphogenesis responses, rather than heat stress (Nagel et al., 2009; Bellstaedt et al., 2019; Gaillochet et al., 2020; Lee et al., 2021b; Ai et al., 2022). However, there is an increasing interest in deciphering the role of roots in plant adaptation to heat stress (Huang et al., 2012; Calleja-Cabrera et al., 2020; Tiwari et al., 2022), driven primarily by general concerns about how climate change might affect crop production in agricultural systems (Fahad et al., 2017). Heat stress causes significant damage to proteins, disturbing their synthesis and folding, changing the activity of enzymes, and damaging membranes and cellular structures by massive oxidation (Hasanuzzaman et al., 2013). To cope with these heat-associated challenges and ensure plant fitness, plants have evolved several adaptive strategies that involve, among other responses, gene expression changes, metabolic adjustments, and modifications of morphological structures and organs (Zhao et al., 2020). High temperatures significantly alter gene expression and protein accumulation in soybean (Valdés-López et al., 2016), Arabidopsis (Bellstaedt et al., 2019; Gaillochet et al., 2020; Sriden and Charoensawan, 2022), and Agrostis grass species (Xu et al., 2008), among others. They also affect cell division and differentiation, reducing plant growth and development (Qi and Zhang, 2020; Liu et al., 2022). It should be noted that the majority of studies carried out to understand shoot and/or root responses to heat or warming have grown whole plants (shoots and roots) at homogeneous high temperatures and, in some cases, have heated detached shoots and roots (Heckathorn et al., 2013; Valdés-López et al., 2016; Chen and Li, 2017; Estravis-Barcala et al., 2021). However, waves of high atmospheric temperature have different effects on shoots than on roots. Because of soil geothermal properties, a decreasing temperature gradient is formed from the topsoil to deeper layers (Lynch et al., 2012), preventing overwarming of the root system and likely contributing to the maintenance of its functionality and growth (Michaletz et al., 2015).
Root system architecture (RSA), defined as the three-dimensional organization of roots in the soil, plays an essential role in water and nutrient absorption and communication with microbiota (de la Fuente Cantó et al., 2020), processes that are affected by soil temperature changes (Giri et al., 2017; Hatfield and Prueger, 2015; Onwuka, 2018; Sabri et al., 2018). RSA changes in response to heat have a genetic basis that may differ between species that have adapted to different optimum temperatures for root growth and lateral root production (Zhang et al., 2015; Gray and Brady, 2016). Depending on the species, a temperature increase has been shown to promote (Lahti et al., 2005) or inhibit (Larkindale et al., 2005; Takahashi et al., 2019) root growth.
To properly study the effect of extreme temperatures on plant growth, we have engineered a new device called the Temperature Gradient in the Root Zone (TGRooZ). The TGRooZ generates a controlled temperature gradient in the root growth zone while keeping the aerial tissues exposed to the environmental temperature, generating conditions similar to those in natural ecosystems. This device is adaptable for in vitro analyses using agar-based systems on Petri dishes or soil-containing pots for greenhouse experiments. In in vitro experiments, we demonstrated that seedlings growing under homogenous high temperatures (applied equally to the shoot and root) develop a significant shorther roots by decreasing cell division in the meristem, induce root stem cell replacement, and block lateral root emergence. Remarkably, although shoots of seedlings growing in the TGRooZ are exposed to a similar high temperature, they recover almost all parameters to values observed in seedlings cultivated at standard temperatures. We also defined the transcriptomic profiles of roots and shoots of plants grown under heat stress (homogeneous 32°C in the shoot and root), at a standard temperature (22°C in the shoot and root), and at 32°C in the TGRooZ. Gene ontology analyses indicated a role for auxin in the heat response of shoots. Genetic analyses showed that auxin is important for the maintenance of shoot growth under high-temperature conditions. We identified genes necessary for normal root growth in response to the gradient of temperature imposed with the TGRooZ. Morphological analyses of the corresponding mutants growing in the TGRooZ showed that root development was not completely recovered, suggesting that the TGRooZ is an important tool for understanding the response to heat in the whole plant. In addition, our results clearly show that a temperature gradient in the root ecosystem has beneficial properties for the plant under heat-stress conditions; it facilitates the recruitment of specific microbial communities and the differential accumulation of mineral nutrients in the plant or modulates specific gene expression, as well as increasing plant biomass production. However, a high homogeneous temperature compromises root growth and functional capacities, reducing plant adaptation to heat stress. Taken together, our data clearly indicate that by cultivating plants with their root system in a temperature gradient in the dark, we can reproduce a growth environment similar to that found in the field. Based on the results presented here, we envision that the use of TGRooZ technology will help researchers to obtain reliable and close-to-field data and improve our understanding of how plants respond to heat stress.
Results
Roots are sensitive to high temperatures
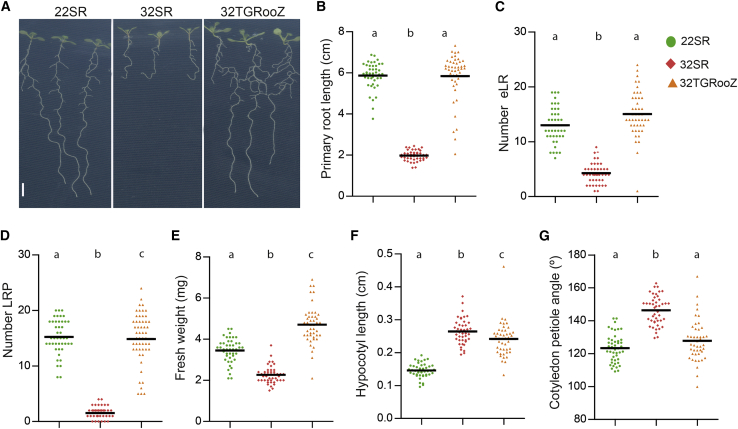
We wanted to analyze the effect of a high temperature over a long period of time on plant growth. First, we noticed that in natural soils, roots normally grow in a lower temperature than aerial plant parts, and this likely protects the root system from excessive high temperatures. In fact, a decreasing temperature gradient is formed from the topsoil to the deeper layers in a natural soil (Supplemental Figure 1A). We analyzed changes in the temperature gradient in the field during day and night periods for 4 days and found that the temperature variations were small (Supplemental Figure 1B and 1C). Taking into account these observations, we engineered a device called the TGRooZ to generate a temperature gradient in the root-growing zone for in vitro assays or greenhouse analyses. The TGRooZ consists of a metallic growth box with a cold-regulable bottom container (Supplemental Figure 1D); the temperature difference between the bottom and the top of the device generates a gradient that can be controlled by adjusting the temperature at the bottom (Supplemental Figure 1D–1F). As indicated, the day-night temperature variation at a 5- or 15-cm soil depth was small. Therefore, although the TGRooZ device generates a day-night static gradient, it can offer the advantage of studying the plant response to heat stress in a controlled environment. Seeds of Arabidopsis SKP2Bp::GUS, a marker line for lateral root primordia formation, were germinated at 22°C, using the D-Root device (Supplemental Figure 1E) to maintain the roots in darkness (Silva-Navas et al., 2015). Four days after germination, seedlings were exposed to either homogeneous 22°C (shoot and root at 22°C: 22SR seedlings), homogeneous high temperature of 32°C (shoot and root at 32°C: 32SR seedlings), or 32°C in the shoot and a temperature gradient in the root zone (32TGRooZ seedlings) (Supplemental Figure 1F) for 6 more days. The high-temperature treatment (32SR) significantly decreased root growth (Figure 1A and 1B) and lateral root (LR) number, both the number of emerged LRs (Figure 1C) and that of LR primordia (LRP) (Figure 1D). Consistent with the decrease in LR emergence in 32SR seedlings, we found that, in the root portion growing at 32°C after the transfer, the few LRP that were formed did not progress beyond stage I, whereas LRP developed normally in roots growing at 22°C (22SR) or in a temperature gradient (32TGRooZ) (Supplemental Figure 2). Strikingly, all deleterious root phenotypes associated with heat stress were reversed or improved to different levels in the 32TGRooZ seedlings (Figure 1A–1D).
Figure 1.
Effect of high temperature and a gradient in the root zone on seedling development.
(A) Representative images of 10-day old Arabidopsis seedlings grown at homogeneous 22°C (22RS), 32°C (32SR), or 32°C in the shoot with the 32TGRooZ. Scale bar, 0.5 cm.
(B) Quantification of primary root length.
(C) Quantification of the number of emerged LRs (eLR).
(D) Quantification of the number of LRP before emergence.
(E) Quantification of the fresh weight per seedling.
(F) Hypocotyl length from the root-shoot junction to the base of the shoot meristem.
(G) Cotyledonary angle. (B–G) n ≥ 25. Significance was determined by ANOVA and Tukey’s honestly significant difference (HSD) post-test. p < 0.05. Different letters indicate significant differences.
Shoot responses to prolonged heat stress depend partially on root growth
We also analyzed the effect of prolonged high temperatures on shoot development using the TGRooZ. We found that 32SR plants produced a lower shoot biomass (Figure 1E). However, the shoot biomass of 32TGRooZ seedlings was higher than that of 22SR (Figure 1E), indicating a positive effect of high temperatures on shoot growth when the root system was growing in the TGRooZ device. We also quantified the elongation of the hypocotyl and the cotyledonary petiole angle, two responses associated with increased temperature in Arabidopsis (Gray et al., 1998; Sun et al., 2012). The hypocotyl length of 32SR seedlings increased by more than 80% compared with 22SR, whereas the increase in 32TGRooZ seedlings was slightly smaller (Figure 1F). The cotyledonary petiole angle was significantly bigger in 32SR seedlings than in 22SR or 32TGRooZ seedlings (Figure 1G). These results demonstrate that the impairment of root development linked to high temperatures modulates some responses in the shoot, such as biomass production or the cotyledonary petiole angle, but not hypocotyl elongation, a response that seems to be independent of root temperature.
We also analyzed the effect of heat and soil-root temperature on plants growing in pots that contained soil. To do this, the TGRooZ was adapted to hold soil-containing pots (Supplemental Figure 1G) and generate a temperature gradient along the soil layers (Supplemental Figure 1H). First, we tested the effect of high temperature on leaf transpiration and leaf temperature. We observed that 32SR plants had reduced stomatal conductance compared with 22SR control plants, and the conductance levels were restored in 32TGRooZ plants even though their leaves were also growing at 32°C (Supplemental Figure 3A). For these experiments, the soil water content was maintained at similar levels across all treatments (Supplemental Figure 4), ensuring that the decrease in stomatal conductance was not due to water scarcity in the soil. We also found that the leaves of 32SR plants had a significantly higher temperature than those of 22SR or 32TGRooZ plants (Supplemental Figure 3B and 3C). Although the recovery was not total, leaves from 32TGRooZ plants were more efficient in downregulating their temperature than those from 32SR (Supplemental Figure 3B and 3D). These data indicate that leaf temperature is influenced by both atmospheric and root-zone temperatures.
To generalize the temperature-dependent root growth phenotypes to other plant species, we carried out similar analyses in tomato plants using a modified TGRooZ device and increasing the temperature from 26°C (optimal for tomato in in vitro assays) to 34°C (Supplemental Figure 5A). We found that root growth and the number of LRs were significantly decreased when plants were exposed to homogeneous high temperatures of 34°C (34SR) compared with 26°C (26SR), as occurred in Arabidopsis (Supplemental Figure 5B). Likewise, when tomato plants were cultivated at 34TGRooZ, root growth and LR number were slightly increased. Compared with 26SR tomato seedlings, shoot and root biomass were decreased in 34SR seedlings, but the biomass was unchanged in 34TGRooZ (Supplemental Figure 5B). These data indicate that tomato root and shoot growth are also sensitive to excessively high temperatures, and the use of TGRooZ can generate a better environmental condition to study heat stress in crops.
High temperature affects cell division in the root apical meristem
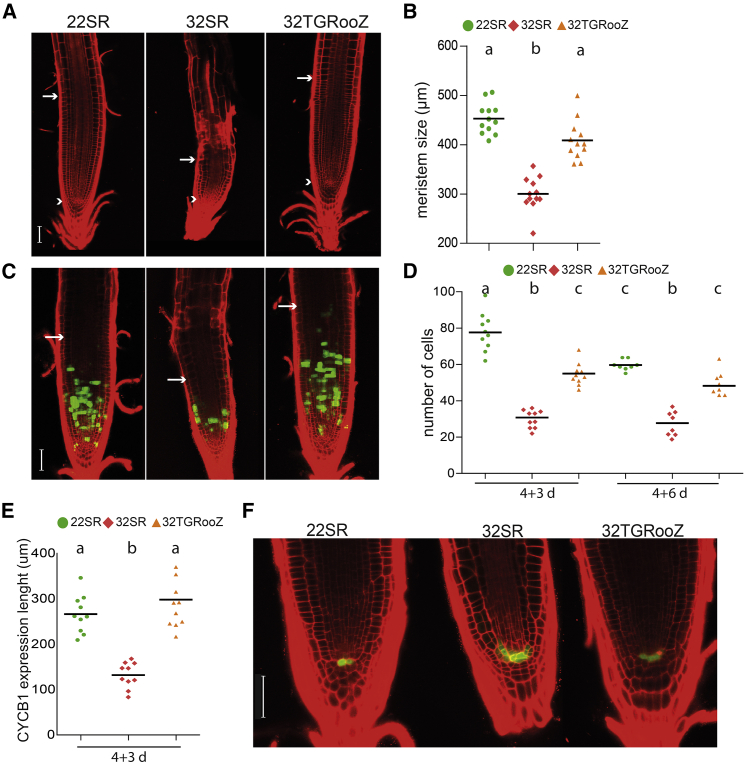
Next, we decided to investigate the causes of the root growth reduction in response to heat by analyzing the structure of the Arabidopsis root apical meristem (RAM). The RAM of 32SR seedlings showed a significantly smaller meristem size with fewer dividing cells that expressed the cell division marker CYCB1;1:CYCB1;1-GFP (Ubeda-Tomas et al., 2009) compared with 22SR plants (Figure 2A and 2B). The Arabidopsis root meristem size was larger in 32TGRooZ than 32SR seedlings, but it did not completely recover the size of 22SR. These results indicate that a temperature gradient promotes optimal root growth, but some other signals may also control the size of the meristem.
Figure 2.
High temperatures decrease cell division but activate QC proliferation.
(A) Confocal images of root meristems of 22RS, 32SR, or 32TGRooZ seedlings grown for 6 days after the transfer. White arrowheads indicate the QC, and arrows indicate the end of the meristem. Scale bar, 50 μm.
(B) Meristem size from the QC to the last dividing cell (n = 12).
(C) Confocal images of a z-stack of PI-stained RAMs from 22SR, 32SR, or 32TGRooZ CYCB1;1:CYCB1;1-GFP seedlings grown for 6 days after transplant. White arrows indicate the end of the meristem. Scale bar, 50 μm.
(D) Number of cells showing a GFP signal at 3 days (n = 10) or 6 days (n = 8) after transfer to different temperatures.
(E) CYCB1;1 expression domain in the root apical zone of seedlings grown for 6 days after transfer at different temperatures.
(F) Confocal images of the QC labelled with the WOX5p:GFP marker 6 days after transfer to 22SR, 32SR, or 32TGRooZ conditions. Scale bars, 50 μm. Significance was analyzed by ANOVA and Tukey’s honestly significant difference (HSD) post-test. p < 0.05. Different letters indicate significant differences.
Delving deeper, we explored whether the decrease in root meristem size observed at high temperature could derive from defects in the stem cell niche. The quiescent center (QC), located in the RAM, harbors a group of cells with a low division rate that replenish the stem cell niche (Dolan et al., 1993; Cruz-Ramírez et al., 2013). Interestingly, using the WOX5:GFP marker to visualize QC cells, we found more WOX5:GFP-expressing cells in 32SR seedlings compared with 22SR or 32TGRooZ seedlings (Figure 2C), indicating that QC cell division was activated in response to heat stress, likely to replace meristematic stem cells that prematurely stopped proliferation or were damaged because of the stress.
A temperature gradient in the root growth zone is required to maintain optimal levels of ERF115 in the RAM
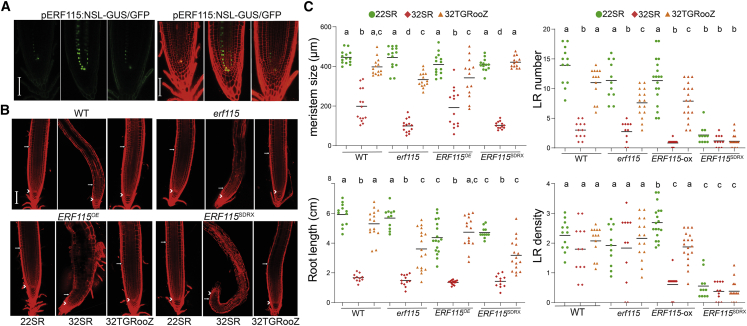
ERF115 is a transcription factor that promotes cell renewal after stem cell damage and responds to moderate changes in root temperature to control cell division and stem cell replenishment in the QC (Heyman et al., 2013). Using the D-Root system to grow roots in darkness, we found that pERF115:NLS-GUS/GFP was slightly expressed in QC cells and vascular cells of 22SR seedlings (Figure 3A). However, this expression significantly increased in response to 32°C in QC, vasculature, and pericycle cells. By contrast, the expression of ERF115 in 32TGRooZ seedlings was similar to that in 22SR (Figure 3A), indicating a local response to the temperature in the root growth zone. The loss-of-function erf115 mutant grown at 22°C showed root meristem size and root length similar to those of 22SR wild-type (WT) seedlings. However, at 32°C, the root meristem size of erf115 was significantly smaller (Figure 3B and 3C). Furthermore, the meristem size recovery of 32TGRooZ was limited for erf115 (72%) compared with WT seedlings (90%) (Figure 3C). This effect might be due to a higher sensitivity of the erf115 root meristem to the temperature variations generated along the gradient, correlating with a reduction in root growth of the mutant. As expected, WT and erf115 seedlings showed a similar trend in the number of emerged LRs in response to changes in temperature in the root zone (Figure 3C), as mutation of ERF115 does not seem to affect the number of emerged LRs (Canher et al., 2021). However, over-expression of ERF115 slightly increases the LR density, a parameter that is not fully recovered in the TGRooZ. Whether this decrease is due to a decrease in LR emergence or LR specification is not known and should be analyzed in the future. It is possible that, in response to high temperatures, over-expression of ERF115 activates a response that decreases cell division in the primordia and slows LR emergence.
Figure 3.
ERF115 is needed for root growth recovery in the TGRooZ.
(A) Confocal images of meristems of 22SR, 32SR, and 32TGRooZ pERF115:NSL-GUS/GFP. (Left) GFP-tagged expression in green. (Right) GFP overlapped with PI-stained roots. Scale bar, 50 μm.
(B) Confocal images of PI-stained root meristems of 22SR, 32SR, and 32TGRooZ wild type (WT), erf115 mutant, ERF115 over-expressing (ERF115OE), or ERF115SDRX seedlings. White arrowheads indicate the QC, and arrows indicate the end of the meristem. Scale bar, 100 μm.
(C) Quantification of root meristem size (n > 12), root length, number of LRs, and lateral root density of 22SR, 32SR, or 32TGRooZ WT, erf115 mutant, ERF115OE, or ERF115SDRX seedlings at 6 days after transfer to different temperatures (n ≥ 12). Significance was analyzed by ANOVA and Tukey’s honestly significant difference (HSD) post-test. p < 0.05. Different letters indicate significant differences.
Root temperature drives transcriptional response to heat stress in the plant
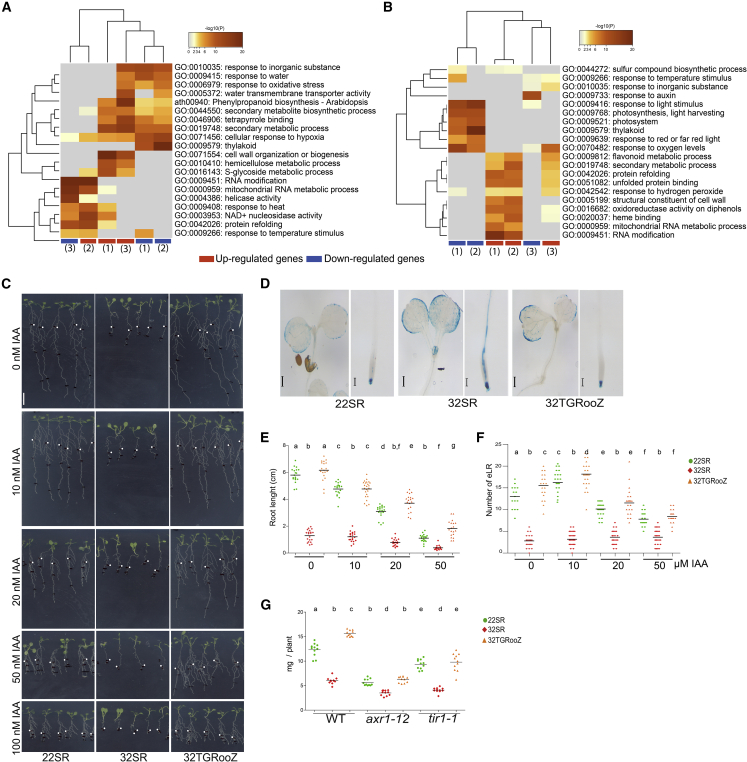
Based on our observations, we hypothesized that plant molecular responses to high temperature might be influenced by the temperature in the root zone. To demonstrate this, we carried out comparative pairwise transcriptomic analysis of shoots or roots from WT Arabidopsis seedlings grown under 22SR, 32SR, and 32TGRooZ conditions. We found that high temperatures of 32°C compared with 22°C had a strong effect on gene expression in both root and shoot, affecting more than 7000 genes (Supplemental Table 1A–1F), including many transcription factors (Supplemental Table 1G). Interestingly, 34 of them belong to the ERF family, indicating that this family probably plays an important role in the response to heat, as demonstrated above by the function of ERF115. Hierarchical clustering of the genes with the greatest variance showed that gene expression in the roots of 32SR seedlings was significantly different from that in roots of 22SR or 32TGRooZ seedlings (Supplemental Figure 6A and Supplemental Table 1A–1C). Gene ontology analyses of differentially expressed genes in the root showed an enrichment in heat stress, water deficiency, response to temperature, RNA modification, oxidative stress, and response to hypoxia, among many other terms (Figure 4A). We also noticed that gene expression in the shoot of 32SR or 32TGRooZ seedlings was significantly different from that in the shoot of 22SR seedlings (Supplemental Figure 6B and Supplemental Table 1D–1F). Gene ontology analyses of differentially expressed genes in the shoot showed an enrichment in photosynthesis, light and auxin signaling, response to temperature, and protein folding, among other categories (Figure 4B). Interestingly, although the shoots of 32SR and 32TGRooZ seedlings were grown at similar high temperatures, almost 700 genes showed a significantly different expression level (Supplemental Table 1F). These genes were enriched in auxin response, oxygen levels, protein refolding, and response to nutrient starvation and iron transport, among other terms (Supplemental Figure 6C and Supplemental Table 1F). Because auxin regulates growth, we decided to analyze the role of this hormone in response to high temperature. First, using the DR5:GUS marker, we found higher GUS activity in both shoot and root apical areas of seedlings grown at homogeneous 32°C compared with 22SR seedlings (Figure 4D). Interestingly, this higher GUS activity was decreased in 32TGRooZ seedlings to basal levels, even when the shoots were exposed to high temperatures. These data indicate that auxin signaling in response to heat stress, in both shoots and roots, is influence by the temperature in the root zone. This DR5:GUS activation correlates with the lower expression of some Aux/IAA and SAUR genes in the shoots of the 32SR seedlings (Supplemental Table 1F), which could negatively regulate the auxin response (Powers and Strader, 2020) and growth, respectively (Ren and Gray, 2015; Stortenbeker and Bemer, 2018). In addition, we found that a combination of exogenous auxin application and high temperature enhanced the root growth inhibition phenotype. However, this root growth inhibition was lower in 32TGRooZ seedlings supplemented with 20 or 50 nM of auxin than in roots from 22SR or 32SR seedlings (Figure 4E). Furthermore, we found that a low concentration of exogenous auxin (10 nM) increased LR emergence in 22SR or 32TGRooZ seedlings but not in 32SR seedlings (Figure 4F). We also analyzed the effect of high temperature on axr1-12 (Lincoln et al., 1990) and tir1-1 (Ruegger et al., 1998), two mutants with decreased auxin signaling. In contrast to WT plants, neither mutant showed an increase in shoot growth when cultivated at 32TGRooZ compared with 22SR (Figure 4G). Taken together, our data suggest that correct auxin signaling is needed for shoot growth in response to heat and that shoot responses to high temperatures are partially influenced by soil-root temperature and seem to be mediated, at least in part, by auxin signaling.
Figure 4.
Temperature gradient in the root zone modifies gene expression in roots and shoots in response to heat.
Gene ontology category heatmap of genes differentially expressed in roots (A) or shoots (B) in response to temperature. (1) 32TGRooZ vs 22SR; (2) 32SR vs. SR22; and (3) 32SR vs. 32TGRooZ comparisons. Red and blue boxes correspond to induced and repressed genes, respectively.
(C) Representative pictures of DR5:GUS seedlings grown for 4 days at 22°C and then transferred to fresh medium containing 0, 10, 20, 50, or 100 nM indole acetic acid (IAA, auxin) and to 22°, 32°C, or 32TGRooZ for 6 more days. The white dots indicate the point of the RAM at the transfer. Scale bar, 1 cm.
(D) Shoot and root apical zone of DR5:GUS seedlings grown as in (C) and stained for GUS activity. Scale bar, 400 μm for shoot pictures and 100 μm for root pictures.
(E) Root length of seedlings grown as in (C).
(F) Number of LRs of seedlings grown as in (C).
(G) Fresh weight of WT, axr1-12, or tir1-1 grown for 4 days at 22°C and then transferred to 22SR, 32SR, or 32TGRooZ conditions for 6 additional days. Significance was analyzed by ANOVA and Tukey’s honestly significant difference (HSD) post-test. p < 0.05. Different letters indicate significant differences.
Chaperone function modulates root development under heat stress
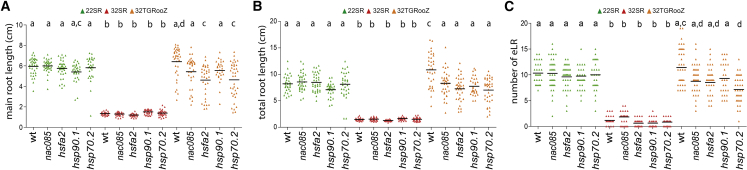
In the transcriptomic comparisons, we identified genes involved in the response to extreme temperature that were induced in both roots and shoots in all comparisons (32SR vs 22SR, 32SR vs 32TGRooZ, and 32TGRooZ vs 22SR) (Supplemental Figure 7A and 7B, Supplemental Table 1J). A gene ontology analysis of this gene set identified protein folding as a common functional category (Supplemental Figure 7C and 7D). Among the induced root and shoot common genes, we identified the HSFA2 transcription factor and the two heat shock proteins (HSPs) HSP90.1 and HSP70.2, which are involved in stress responses, thermomemory, and protein folding (Schramm et al., 2006; Wang et al., 2016; Leng et al., 2017; Friedrich et al., 2021), as well as the NAC085 transcription factor, which is involved in G2 arrest in response to heat stress (Takahashi et al., 2019). Interestingly, a gene network identified several HSP genes as targets of ERF115, including HSP90.1 (Supplemental Figure 7E), suggesting a direct connection between ERF115 and the heat responses mediated by HSPs. Analyses of the homozygous mutants for these genes at 22°C showed that their primary root length was similar to that of WT seedlings (Figure 5A). At 32°C, root growth was severely compromised in all genotypes (Figure 5A). Using the TGRooZ, we found that the root growth of nac085 and hsp90.1 seedlings recovered to levels similar to WT plants, while the root length of hsf2a and hsp70.2 remained significantly smaller (Figure 5A), indicating that they may function in the control of main root growth during heat stress. A similar trend was observed for total root length (length of primary root and LRs), but for this trait, the recovery was significantly smaller in all mutants compared with WT plants (Figure 5B), indicating an important role of chaperones in LR growth. Indeed, the LR number was significantly decreased by the effect of high temperature in all genotypes (Figure 5C), but this parameter was not fully recovered in these mutants when grown at 32TGRooZ, except for hsp90.1 (Figure 5C). By contrast, when growing in 32TGRooZ, the hsp70.2 mutant showed a significantly lower number of LRs than control seedlings (Figure 5C), suggesting an important role of this gene in LR development under heat stress.
Figure 5.
Effect of high temperature and a temperature gradient in the root zone on heat-response mutants.
(A) Root lengths of 22SR, 32SR, and 32TGRooZ WT and mutant seedlings.
(B) Total root length (main plus LR length) of 22SR, 32SR, and 32TGRooZ WT and mutant seedlings.
(C) Number of emerged LRs of 22SR, 32SR, and 32TGRooZ WT and mutant seedlings (n ≥ 24). Significance was analyzed by ANOVA and Tukey’s honestly significant difference (HSD) post-test. p < 0.05. Different letters indicate significant differences.
We also analyzed the effect of the different temperature treatments on hypocotyl length (Supplemental Figure 8A) and cotyledonary petiole angle (Supplemental Figure 8B) in these mutants. In general, we did not find any significant difference in either response between WT plants and mutants in the 22SR, 32SR, or 32TGRooZ treatments, with the exception of the hsfa2 mutant, which did not increase the cotyledonary petiole angle in response to heat (32SR) (Supplemental Figure 8B). These data suggest that HSFA2 functions in the leaf hyponastic response needed to elevate the cotyledons from the soil in response to heat.
Heat stress affects plant phosphate nutrition
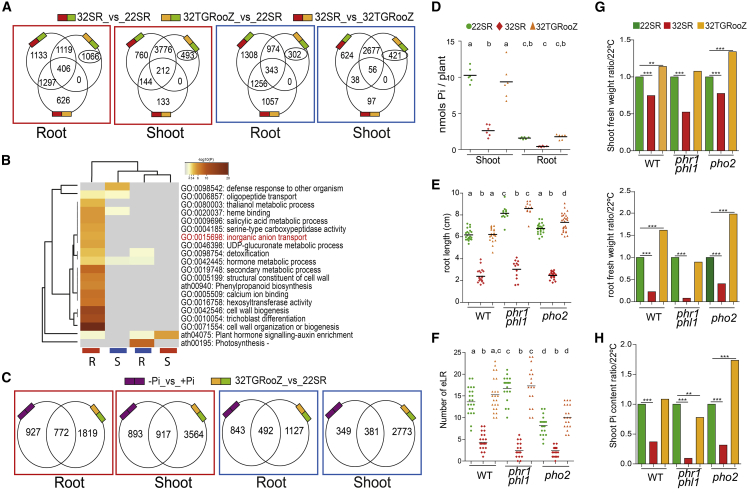
To better understand the influence of the TGRooZ on the plant transcriptome, we overlapped the differentially expressed genes between all comparisons among treatments (Figure 6A and Supplemental Table 1H). We focused on those genes that were differentially expressed in root and shoot between 22SR and 32TGRooZ experimental conditions because they were most likely to reflect the response to a heat wave in a natural field. Gene ontology analyses showed an enrichment in terms related to hormone signaling (auxin and salicylic acid), the cell wall, and anion transport, among others (Figure 6B). Analyzing the inorganic anion transport category, we found several phosphate (Pi) transporters, an observation that is in agreement with the suggested interconnection between Pi and temperature (Pacak et al., 2016; Giri et al., 2017; Singh et al., 2018). Furthermore, we found a significant overlap between genes deregulated by Pi starvation in roots (Silva-Navas et al., 2019) and shoots (del Pozo, unpublished) and genes deregulated by heat stress in both roots and shoots (Figure 6C and Supplemental Table 1I). Among them, we found several Pi starvation transporters and signaling genes that were upregulated by the effect of heat (Supplemental Figure 9A and Supplementary Table 1I). A recent study reported a curated collection of genes (the known ionome gene [KIG] list) that were experimentally demonstrated to participate in uptake, accumulation, or distribution of mineral nutrients in plants (Whitt et al., 2020). We found a significant overlap between this KIG gene set and our temperature-deregulated gene set (Supplemental Table 2). Remarkably, PHR1 and PHL1, two key regulators of the Pi response, and PHO2, a regulator of Pi transport to the xylem, were downregulated in response to heat in almost all comparisons (Supplemental Figure 9A), whereas several Pi transporters were upregulated in roots or shoots in response to heat (Supplemental Figure 9B). It is likely that the plant differentially adjusted the expression of these Pi starvation response regulators and transporters to establish a new Pi homeostasis during heat stress. Next, we measured the cellular free Pi content in plants exposed to the different temperature treatments. We found that Pi levels in 32SR seedlings were significantly lower, in both shoots and roots, than those in 22SR seedlings (Figure 6D). Interestingly, Pi levels were recovered in both shoots and roots of 32TGRooZ seedlings (Figure 6D), indicating that, during heat stress, correct Pi homeostasis is maintained if the roots are cultivated in a dark environment with a temperature gradient. We analyzed the effect of heat stress on the phr1 phl1 double mutant, which exhibits a decreased Pi starvation response (Bustos et al., 2010), and pho2, which shows an over-loading of Pi from root to shoot (Liu et al., 2012). We found that the heat response of the phr1 phl1 mutant was similar to that of WT plants in terms of root length and LR emergence (Figure 6E, 6F and Supplemental Figure 9C). However, pho2 root elongation (but not LR number) was recovered in the TGRooZ and was higher than that of WT seedlings (Figure 6E and 6F). The shoot weight of the phr1 phl1 mutant showed less recovery in TGRooZ than that of control seedlings, whereas the shoot weight of the pho2 mutant showed significantly higher recovery, consistent with its higher shoot Pi content (Figure 6G). This phenotype was more noticeable in the recovery of root biomass (Figure 6G). Interestingly, the amount of free Pi was significantly decreased in the phr1 phl1 mutant in response to heat, probably because it was unable to fully activate the Pi starvation response, whereas Pi accumulation in the pho2 mutant increased when it was grown in the TGRooZ. Taken together, these data suggest that a reduction in Pi responses or lower Pi accumulation contributes, at least in part, to the decrease in shoot growth during heat stress.
Figure 6.
High temperatures affect Pi nutrition.
(A) Venn diagrams of differentially expressed genes (DEGs) between different comparisons (up- or downregulated genes) in roots or shoots. Circled numbers indicate the number of genes specifically deregulated between 32TGRooz and 22SR but not under other conditions.
(B) Gene ontology (GO) heatmap of specific genes (circled in A) in roots (R) or shoots (S) that were up- or downregulated (red and blue rectangles, respectively).
(C) Venn diagrams of DEGs deregulated by Pi starvation and by the effect of temperature between 32TGRooZ and 22SR in both roots and shoots. Red and blue boxes correspond to up- and downregulated genes, respectively.
(D) Free Pi level in roots or shoot of SR22, SR32, or 32TGRooZ Arabidopsis seedlings grown in a medium containing Pi for4 days and then transferred to different temperatures for 6 additional days. Values correspond to nmoles Pi per plant.
(E and F) Root length (E) and emerged LRs (eLR) (F) in WT, phr1 phl1, or pho2 mutants grown for 4 days at 22°C and then transferred to 22SR, 32SR, or 32TGRooZ conditions for 6 additional days. Significance was analyzed by ANOVA and Tukey’s honestly significant difference (HSD) post-test. Different letters indicate significant differences.
(G) Shoot or root fresh weight of WT, phr1 phl1, or pho2 mutants grown for 4 days at 22°C and then transferred to 22SR, 32SR, or 32TGRooZ conditions for 6 additional days. Values are expressed relative to those at 22°C.
(H) Shoot Pi level (nmoles/plant) in WT, phr1 phl1, or pho2 mutants grown as in (G). Values are expressed relative to those at 22°C. Asterisks indicate significant differences with respect to 22°C by t-test. ∗∗p < 0.01; ∗∗∗p < 0.001.
High temperature in the root system affects bacterial community assembly and root functionality
To further understand the relationship between high temperature and root function, we compared the bacterial community composition and ionome of tomato plants whose root growth was limited by excessive high temperature. Tomato seedlings were grown in natural soil and exposed to three different temperature regimes: 22SR, 36SR, and 36TGRooZ (a root temperature gradient that partially mimicked real soil conditions). We determined their shoot mineral nutrient accumulation (ionome) and found that, in general, mineral nutrient accumulation patterns were similar in 22SR and 36TGRooZ leaves but differed in SR36 leaves (Supplemental Figure 10A). Consistent with the transcriptional analysis, in which the expression of several ion transporters was deregulated, different temperature regimes altered the levels of various mineral nutrients in leaves, including P, Cu, B, Mo, As, Rb, and K (Supplemental Figure 10B). These results suggest that changes in tomato mineral nutrition are influenced by soil temperature and likely by root functionality.
We compared bacterial community profiles in the rhizosphere and root using 16S rRNA amplicon sequencing. Across all temperatures used, soil and rhizosphere samples supported higher bacterial alpha diversity and richness indexes compared with roots (Supplemental Figure 11A and 11B), a difference that has been systematically observed in natural environments where the species richness is lower in the plant than in the soil (Castrillo et al., 2017; Finkel et al., 2019).
As in previous experiments (Castrillo et al., 2017; Finkel et al., 2019), the original soil (inoculum), rhizosphere, and roots assembled diverse bacterial communities; the differences among these fractions explained most of the variance in community composition (Supplemental Figure 12A). Compared with the rhizosphere and soil fractions, the tomato root samples were mainly enriched in Proteobacteria and Bacteroidetes and depleted in Acidobacteria, Gemmatimonadetes, Firmicutes, Chloroflexi, and Verrucomicrobia (Supplemental Figure 12B and 12C). Consistent with the ionomic data, we noticed that the bacterial profile in roots of high-temperature-treated plants (36SR) differed from that of 22SR and 36TGRooZ plants (Supplemental Figure 12B and 12C). Furthermore, 36SR tomato roots showed a clear increase in Actinobacteria, a phylum that is normally decreased in healthy roots (Castrillo et al., 2017; Finkel et al., 2019) (Supplemental Figure 12B and 12C).
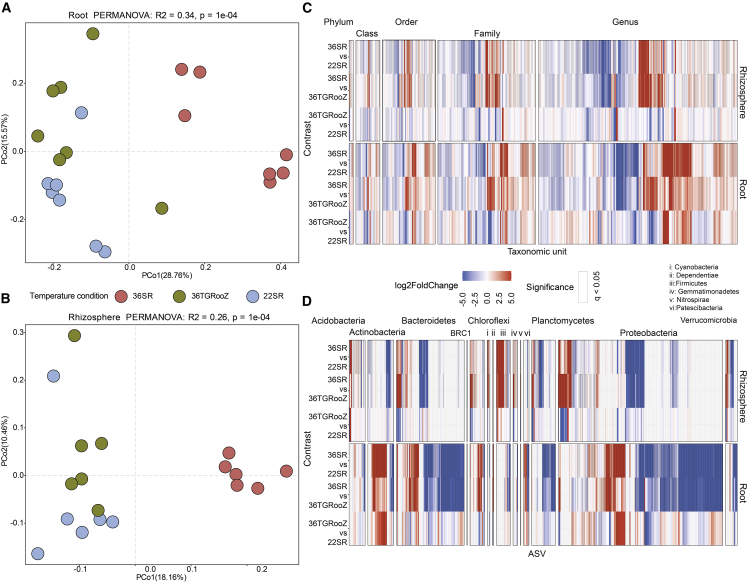
We further investigated the effect of high temperature on root bacterial community composition. Canonical analysis of principal coordinates showed significant differences in root bacterial community composition across the temperature treatments (Figure 7A). These differences were robust in both fractions, the rhizosphere and the root (Figure 7A and 7B). We noticed that the rhizosphere and root microbiome compositions were similar in tomato 22SR and 36TGRooZ plants and differed significantly from those of 36SR plants (Figure 7). This observation was consistent at different phylogenetic levels (Figure 7C and 7D), indicating that high temperature in the soil, but not in the shoot (comparison between 36SR and 36TGRooZ), influences the capacity of the root to assemble an intact microbiome. The high-temperature effect of 36°C on root microbiome composition and plant mineral nutrient accumulation, in contrast to 22°C or TGRooZ in heat-stressed plants, emphasizes the protective role of the soil in buffering high temperature fluctuations to preserve root function. Taken together, these results suggest that changes in the root function of tomato are likely to be a direct consequence of high temperature on the soil, the alterations in the microbiota, or the interaction of both.
Figure 7.
High soil temperatures select different rhizosphere and root microbiome compositions.
Rhizosphere and root microbiome compositions of tomato plants grown at 22°C (22SR) and 36TGRooZ are similar, and both are significantly different from the microbiomes found in plants grown at 36°C (36SR).
(A) Principal coordinate analysis (PCoA) based on Bray-Curtis dissimilarities between bacterial communities in roots or (B) rhizospheres of tomato plants grown at 22°C (22SR), 36°C (36SR), or 36TGRooZ for 4 weeks.
(C) Heatmaps showing root and rhizosphere enrichment patterns of different taxonomic units (phylum, class, order, family, genus) or (D) amplicon sequence variants (ASVs) across all versus all contrasts between temperature conditions used in (A). The cells of the heatmaps are colored based on the log2 fold change estimated from a generalized linear model contrasting the abundance of each taxonomic unit at each temperature with respect to another in each contrast. Squares outlined in black represent taxonomic units at each taxonomic level that were significantly enriched (red) and depleted (blue) in each comparison (q < 0.05).
Discussion
In this work, we used a novel approach that reflects natural soil properties in many aspects to demonstrate that a temperature gradient in the root ecosystem is essential for studying and understanding the whole-plant response to heat stress. In general, in vitro and greenhouse studies have not considered the temperature in the root zone when analyzing the effect of heat stress (Heckathorn et al., 2013; Valdés-López et al., 2016; Chen and Li, 2017; Estravis-Barcala et al., 2021; Pei et al., 2021). In response to warming temperatures, which are normally optimal and non-detrimental for plant growth, one of the thermomorphogenetic responses is the elongation of roots. By contrast, when entire plants are cultivated at high temperatures (above optimal), root growth is arrested and LR formation is blocked, decreasing root functionality and negatively affecting shoot biomass. Here, we present robust data showing that a high temperature in the root growing zonehas a strong impact on plant development, physiological and molecular responses, and microbiome assembly. The TGRooZ provides a novel and useful approach for studying heat stress in the shoot and roots. This approach can help to identify new solutions and facilitate plant breeding programs to obtain more heat-tolerant crops or to test new microbes to enhance plant tolerance to heat under closer-to-field conditions. Thus, the knowledge generated using the TGRooZ can be useful for coping with the negative effects of global warming and heat waves on crops.
Soil temperature in the root growing zone is important for plant growth and development
In natural ecosystems, soil temperature is affected by several factors, such as environmental temperature, water content, soil compaction, and organic material, among others (Elias et al., 2004). Soil temperature affects many soil physical-chemical properties and living processes that have an impact on plant growth and environmental adaptation (onwuka, 2018; Smith, 2000). A recent work found that soil surface temperature increases, proportionally, more rapidly than air temperature (Zhang et al., 2016). This work predicted that soil respiration will increase by up to 28%, reinforcing the idea that climate warming affects the soil ecosystem and perturbs plant growth. To analyze the effect of heat on soil temperature, in July 2019 in Spain (with an average atmospheric temperature of 34°C but with heat wave peaks over 40°C), we measured the temperature of natural soil at different depths from the top surface down to 14 cm in order to estimate the temperature gradient that formed naturally in the soil. However, we should take into account that this gradient will depend on physical soil characteristics such as humidity or compaction, among others. We observed that a decreasing temperature gradient was formed in the root growth zone (Supplemental Figure 1A). These data were used to engineer the TGRooZ device that generates a temperature gradient in the plant growth substrate (agar-containing plates or soil) to simulate natural conditions during a heat wave in an enclosed ecosystem (plant growth chamber or greenhouse), where we can control the environmental temperature. In addition, we measured the day-night fluctuation over 4 days in a natural soil in Spain in June 2022. We found that the soil has a stronger temperature buffering capacity at a 15-cm depth than at a 5-cm depth, whereas the temperature fluctuation is significantly greater at the top surface (Supplemental Figure 1B and 1C). This is in agreement with soil thermal diffusivity analyses showing that superficial soil (5 cm) experiences greater temperature variation, whereas at 15 cm or below, temperature changes are buffered across the different seasons (Brunetti et al., 2021). Using the TGRooZ, we provide strong evidence that the soil-root temperature is essential for the plant to activate a full response to heat, conditions that cause severe cellular damage and organ failure (Hasanuzzaman et al., 2013; Qi and Zhang, 2020; Liu et al., 2022). However, the severity of the damage depends on the species, the genotype, and the context of the stress, as these responses are different if the plant encounters gradual temperature variation or if it suddenly faces a high-temperature wave (Larkindale and Vierling, 2008; Gomaa et al., 2011; Zhang et al., 2015). During their lifetime, roots face many hostile environmental conditions that affect growth and development. In response to high temperatures, plants decrease root growth and root meristem size in vitro (Yang et al., 2017; Liu et al., 2022). It was reported that heat stress shortens the cell elongation zone, decreasing cell flux from the meristem to the elongation area and stopping root growth (Baskin et al., 1992), likely by decreasing cell division in the root meristem. We found similar phenotypes in plants exposed to a shoot-root homogeneous temperature of 32°C. However, if seedlings were grown in a TGRooZ, with conditions more similar to the field, root growth was not affected, and meristem size was recovered to the level found at optimal temperatures. Hypocotyl elongation, a trait associated with warming (Gray et al., 1998; Bellstaedt et al., 2019; Gaillochet et al., 2020; Lee et al., 2021b), was the unique parameter that was slightly recovered in TGRooZ plants. This suggests that this response mainly depends on the atmospheric temperature surrounding the shoot, although the root temperature can partially influence it.
The gene expression response to heat is strongly conditioned by root temperature (Figure 4). Here, we show that the gene expression of shoots exposed to high temperature is modified when the root system is cultivated in a temperature gradient. It should be noted that many of these differentially expressed genes are related to auxin signaling and are downregulated in plants subjected to homogeneous 32°C compared with those growing in the TGRooZ. A large portion of these genes correspond to members of the SAUR family that are involved in growth control mediated by auxin (Stortenbeker and Bemer, 2018). Auxin plays an essential role in shoot growth and biomass production (Tivendale and Millar, 2022) and heat stress-induced thermomorphogenetic responses, including hypocotyl elongation and leaf hyponasty (Gray et al., 1998; Küpers et al., 2020). Thus, a decrease in auxin signaling might explain, at least in part, the observed decrease in shoot biomass. In this sense, axr1-12 and tir1-1, two mutants with impaired auxin responses, do not fully recover the shoot growth, suggesting an important role of auxin in this process during heat stress. It should be noted that auxin responses require the activity of HSP90 because this chaperone stabilizes TIR1, one of the auxin co-receptors, at high temperatures (Wang et al., 2016), thereby connecting heat stress and auxin signaling. We found that several chaperones, including HSP90.1, were induced by heat. It is also possible that some of these chaperones are regulated by ERF115, connecting DNA damage, activation of QC cell division, and auxin signaling. However, further experiments need to be done to clarify this possibility.
Based on our data, we strongly believe that use of the TGRooZ can improve our understanding of how plants respond, as entire organisms (shoots and roots), to heat stress and also improve the translation of laboratory data to field trials.
Heat stress affects stem cell activity
Plant root growth and development rely on a coordinated balance between cell division and differentiation. In the RAM, the stem cell niche supplies new cells to the root pool by continuous cell divisions to sustain growth. This stem cell niche is organized and maintained by a small group of cells called the QC that rarely divide (Dolan et al., 1993; van den Berg et al., 1997). The QC cells can replenish the stem cell niche when its cells are damaged or prematurely differentiated (Cruz-Ramírez et al., 2013). It was shown that ERF115 functions in slowing QC cell division and ensures the longevity of the stem cell niche (Heyman et al., 2013), whereas cytokinin activates the cell division of QC cells in Arabidopsis roots (Zhang et al., 2013). In dark-grown roots exposed to excessive high temperature, ERF115 expression is induced in QC, pericycle, and vascular cells, probably to prevent their collapse. Interestingly, if ERF115 is ectopically over-expressed, the resulting plants do not fully recover root growth in the temperature gradient. A possible explanation for this effect could be that ERF115OE roots are temporarily exposed to high temperature in the upper part of the gradient, activating a stress-response system that, combined with an over-accumulation of ERF115, delays root growth. In addition, acute high temperature has strong effects on the root meristem of ERF115 over-expressing plants, likely because of a constitutive over-response of ERF115 signaling. It should be noted that, as a general trend, stronger propidium iodide staining was observed in erf115 mutant than in 32SR WT seedlings, suggesting a higher rate of cell death in the meristematic area. The response to cell death and DNA damage is controlled by the SOG1 pathway (Bourbousse et al., 2018), which seems to control the transcription of ERF115, as well as ANAC085 and HSP70.2, among others. These data suggest that high temperatures in the root meristem lead to cell death and/or DNA damage, inducing a rescue system that involves ERF115, ANAC085, and HSP. In this work, we show that several ERF transcription factors with functions in cytokinin signaling (Rashotte et al., 2006) are activated, but only by the effect of high homogeneous temperatures (Supplementary Table 1G), suggesting that they might regulate QC cell division via ERF-cytokinin signaling (Zhang et al., 2013). ERF members related to ethylene signaling such as ERF109 or ERF114 (Kong et al., 2018) are repressed and upregulated, respectively, indicating a complex balance of ERF family members in response to heat. The maintenance of the stem cell subpopulation to replace damaged stem cells might represent a general mechanism for ensuring a functional stem cell niche under stress conditions, including extremely high temperatures (Ubogoeva et al., 2021). Stress signals activate QC cell proliferation with the aim of restoring root growth in the case of major damage. In rice, the ERF115 ortholog is involved in heat and drought stress tolerance (Park et al., 2021). In recent years, an emerging role for plant ERF transcription factors in stress responses is becoming evident. In our transcriptomic data, we identified a greater number of ERF factors (Supplemental Table 1) differentially expressed in response to heat stress than previously indicated (Heyman et al., 2018). Our data suggest that ERF115 is needed to sustain root growth in response to high temperatures and also to maintain meristem activity when the shoot is subjected to high temperatures, as erf115 seedlings do not fully recover root meristem size to control levels when grown in the TGRooZ. Similarly, root growth does not fully recover in the hsfa2 mutant grown in a root temperature gradient. Interestingly, a gene regulatory network for cellular reprogramming in plant regeneration shows that ERF115 and HSFA2 are co-regulated by similar transcription factors of the LOB/AS2 family (Ikeuchi et al., 2018), suggesting an interconnection between these two genes. HSFA2 regulates the expression of HSPs and is required for the acquisition and extension of plant thermotolerance (Charng et al., 2006). Analyses of mutants of individual members of the HSP 70 (HSP70.2) family did not reveal morphological phenotypes (Leng et al., 2017). Under these conditions, heat stress decreases root growth and the number of LRs formed in 32TGRooZ hsp70.2 mutant seedlings, indicating that this gene, similar to HSFA2, might connect shoot heat stress responses with root development. Using the TGRooZ, we have been able to identify new genes that function in root responses to heat stress. This has been possible because the root responses are not blocked by the excessively high temperature that is normally used in heat stress experiments, supporting the idea that plants under heat stress should be cultivated with the root system in a temperature gradient like that observed in the field.
Heat stress affects plant Pi nutrition
Different transcriptomic analyses have pointed out the interconnection between high temperature and Pi starvation responses (Pacak et al., 2016; Giri et al., 2017). Here, we show that high temperatures affect several genes related to Pi starvation and decrease the level of Pi in roots and shoots. The KIG collection of genes involved in plant ionome composition was significantly regulated by temperature in shoots and roots. Interestingly, two of the main Pi starvation regulators were downregulated in accordance with our data, showing an increase in Pi nutrition pathways and accumulation (Whitt et al., 2020). Remarkably, a significant number of these KIG genes were also regulated by temperature in the root and shoot. Mutation of PHO2 increases the Pi level in the shoot by increasing the activity of the PHO1 transporter (Bari et al., 2006; Pacak et al., 2016). Mutations of PHR1 and its homolog PHL1, key transcription factors that regulate the Pi starvation response, decrease the level of Pi in the shoot (Rouached et al., 2011). In response to high temperatures, PHR1 and PHO2 were downregulated, but levels of PHO1:3 increased. However, contrary to our expectations, these 32SR seedlings accumulated a lower Pi level in their shoots. It is possible that high temperature alters the Pi balance and that seedlings therefore respond by regulating a set of Pi-related genes to equilibrate Pi homeostasis during heat stress. As high temperatures decrease Pi nutrition, it is possible that this decrease could be one of the major limiting factors affecting plant growth. This idea is supported by the fact that pho2 accumulates a greater Pi content in response to heat, but only when plants are grown under TGRooZ conditions. Interestingly, this greater accumulation correlates with an increase in pho2 biomass production. Our results indicate a close correlation between heat stress and Pi nutrition; therefore, the TGRooZ will be useful for analyzing plant nutritional status during climate change.
Microbiome assembly and plant growth are influenced by soil temperature
Nutrient availability and plant productivity are determined to a large extent by soil properties and the soil microbial community inhabiting the root. The soil microbiota can be affected by diverse soil factors such as nutrient content, pH, and soil texture (Chaparro et al., 2012). The effects of climate change factors such as elevated CO2, drought, and higher temperature on beneficial plant–microbe interactions are increasingly being explored. For example, soil temperature affects the recruitment of microbiota by plant roots (Compant et al., 2010; Rousk et al., 2012), and increased soil temperature may help roots to select microbial species that possess heat tolerance mechanisms and high growth rates rather than temperature-sensitive and slow-growing microbes (van der Voort et al., 2016). This community disturbance might lead to changes in plant growth, pathogen protection, or abiotic stress responses (Hariprasad et al., 2021; Vogel et al., 2021).
A hypothetical model based on the responses of microbial communities to temperature sensitivity suggests that the growth of bacterial and fungal soil communities tends to increase under moderately high temperatures; however, microbial diversity decreases significantly when the soil is exposed to a continuous high temperature (Nottingham et al., 2019). This decrease in diversity was also observed when soil was incubated at 35°C for a long period of time (Lin et al., 2017); during a wildfire, which increases the soi temperature to abnormally high levels, leading to a decrease in microbial activity and major changes in microbial communities (Ferrenberg et al., 2013; Jolly et al., 2015); and during solarization (soil heating) to sterilize soil, a procedure widely used in agriculture to eliminate or decrease the soil microbiota (Katan and Gamliel, 2014). Here, our data clearly show that plants exposed to shoot/root homogenous high temperature accommodate a different rhizosphere- and root-associated microbiome than plants grown at a lower temperature or with a high shoot temperature of 36°C and roots in a temperature gradient, conditions that replicate soil changes in natural ecosystems. We found that homogeneous high temperature changes the beta diversity of both the rhizosphere and the root-associated microbiome, as well as mineral nutrient accumulation in leaves.
In general, the microbiome characteristics of tomato plants under optimal temperature described in this manuscript replicated general observations found in tomato plants in other published papers (Lee et al., 2021a). It is well established that plants under stress can change the root microbiome composition, enhancing the reproduction of beneficial microbes that represent an advantage for plant fitness. As high temperature can modulate the plant immune system (Huot et al., 2017), we speculate that part of the tomato plant’s response to high temperature might overlap with the activation of plant defense, with the plant sensing an increase in temperature partially as a pathogen attack. In line with this hypothesis, the abundance of Actinobacteria was higher in the rhizosphere of tomato plants that were tolerant to bacterial wilt disease than in that of susceptible plants, whereas the abundance of Proteobacteria was lower (Huot et al., 2017). Therefore, changes in Actinobacteria and Proteobacteria observed in 36SR tomato plants might be associated with the plant stress response and specifically with changes in the activation of plant immune system components. Thus, changes in the ratio of Actinobacteria to Proteobacteria indicate that high temperatures alter the capacity of the root to recruit a typical microbiota. This observation supports the idea that the root might function as a sensing hub for abiotic and biotic stresses and that high soil temperature, but not the gradient of temperature in the root, disturbs the equilibrium and affects root functionality. Nevertheless, this hypothesis must be tested in future analyses.
Taken together, our results demonstrate that over-warming the soil compromises root function, which in turn may decrease plant capacity to cope with environmental changes. We envision that the system provided here, TGRooZ, will help with the design of new experiments aimed at mitigating the coming heat waves caused by climate change. In addition, our approach will be helpful for studying root growth and adaptation of soil-grown plants in response to heat, a trait that has been understudied in the last decades and that will definitely result in improved harvests and food security.
Methods
Plant material
In this work, we used Arabidopsis ecotype Columbia and the tomato variety ‘Moneymaker.’ We used SKP2Bp:GUS (Manzano et al., 2012) as a marker for LRP, as well as the reporter line WOX5:GFP (Sarkar et al., 2007) and the marker line CYCB1;1:CYCB1;1-GFP (Ubeda-Tomas et al., 2009). The Arabidopsis mutants were obtained from the NASC stock center with the following code numbers: SALK_208662 (NAC085 AT5G14490); SALK_008978 (HSFA2 AT2G26150); SALK_085076 (HSP70.2 AT5G02490); and SALK_075596 (HSP90.1 AT5G52640).
The erf115, ERF115OE, and ERF115SDRX lines were described in (Heyman et al., 2013; Canher et al., 2021). The pho2 (Aung et al., 2006), phr1 phl1 (Bustos et al., 2010), axr1-12 (Lincoln et al., 1990), and tir1-1 (Ruegger et al., 1998) mutants were used to analyze the effect of auxin signaling or Pi starvation responses on high temperature responses.
Growing conditions
Arabidopsis seedlings were germinated in half-strength Murashige and Skoog (MS) medium (MS1/2) with vitamins plus 1% sucrose, 1% Difco Agar, and 0.05% MES (pH 5.8) using the D-Root device to prevent root illumination (Silva-Navas et al., 2015) or in the TGRooZ device (Supplemental Figure 1) to generate a temperature gradient. The top of the TGRooZ device was closed with an iron cover that has rectangular holes to fit a 12 × 12-cm square in vitro plate or holes for a 30 × 40-cm zip bag containing a germination paper. The plant growth chamber was set to 22°C (standard) or 32°C (heat stress) for Arabidopsis and 26°C and 36°C for tomato using the germination paper system (Supplemental Figure 3). To generate the temperature gradient, the TGRooZ refrigerated liquid was cooled to 13°C for Arabidopsis or 10.5°C for tomato. Seedlings were germinated in MS1/2 for 4 days at 22°C and then transferred to 22°C (22SR seedlings), 32°C (32SR seedlings), or 32°C with a temperature gradient (32TGRooZ) for 6 more days. To analyze heat stress in tomato grown in soil, pots containing tomato plants were placed in the adapted TGRooZ (Supplemental Figure 1), and the growth chambers were set to 22°C or 36°C. For the 36TGRooZ condition, to generate the gradient the refrigerant liquid in the base of the modified TGRooZ (Supplemental Figure 1D) was cooled at 10°C. Tomato seedlings were germinated in vermiculite at 22°C, and tomato seedlings of similar size were transplanted to soil. They were grown at 22°C for 2 more days and then moved to 22°C (22SR seedlings), 36ºC (36SR seedlings), or 36°C with a temperature gradient (36TGRooZ seedlings) for 3 more weeks at a relative humidity of 40%. Plants were grown under a 16-h light/8-h dark photoperiod with a light fluency rate of 130 μM m–1 sec–1.
Root morphological analyses
The 22SR, 32SR, and 32TRGooZ seedlings were grown as described above. They were scanned at a high resolution with an Epson 600V scanner, and root and hypocotyl length and cotyledon epinasty angle were quantified using Fiji software. To quantify LRP, SKP2Bp:GUS seedlings were stained for GUS activity as described by Silva-Navas (Silva-Navas et al., 2015), and GUS-stained LRP were quantified using a Leica Z9 stereomicroscope.
Mitotic cell quantification and root meristem size
The 22SR, 32SR, and 32TRGooZ Arabidopsis seedlings were grown as described above using in vitro plates. Root meristem size was quantified from confocal images taken with a Leica Z8 microscope of the RAM stained with propidium iodide as described by González-García et al. (2011). PI and GFP were detected with a band-pass 570–670-nm filter and 500–545-nm filter, respectively, and observed under a confocal microscope (Leica TCS-SP8). Size was calculated based on the number of meristematic cortical cells and/or the distance from the QC to the last meristematic cell. The end of the meristem zone was taken as the point where a meristematic cortical cell doubled in size from the previous one. Mitotic cell numbers were quantified using the CYCB1;1:CYCB1;1-GFP marker line (Ubeda-Tomas et al., 2009). The number of fluorescent cells was quantified through all stacks.
Root length, number of LRs, hypocotyl length, leaf epinasty angle, and root meristem size in Arabidopsis were assessed in at least three independent experiments. Roots were scanned and root length measured with ImageJ software (http://rsb.info.nih.gov/ij/). For comparisons, significance was analyzed by ANOVA and Tukey’s honestly significant difference post-test using a p value <0.05 in all cases.
Transcriptomic analyses
To carry out the transcriptomic analyses, total RNA was extracted from roots and shoots of Arabidopsis seedlings that were germinated for 4 days at 22°C and then transferred for 7 more days to 22°C (22SR), 32°C (32SR), or 32°C with a temperature gradient in the root zone (32TGRooZ) (32°C at the top of the plate and 18°C at the bottom). In the case of 22°C, we had only two replicates because the third degraded during processing. Library construction and RNA sequencing were performed by Beijing Genomics Institute (BGI-Shenzhen), and 30 M reads were obtained (100 PE; average quality 96%). Approximately 20 μg total RNA was subjected to poly-A+RNA isolation by oligo-dT chromatography, followed by RNA fragmentation. Fragmented RNA was converted to double-stranded cDNA using random hexamer primers followed by end repair, 3′ end adenylation, and adapter ligation. cDNA fragments were selected by agarose gel extraction and enriched by PCR amplification. The library was loaded onto an Illumina HiSeq 2000 instrument for paired-end sequencing (average raw read length, 100 bp).
Before assembly, FastQC (Andrews, 2010) (v0.11.9) was used to obtain information about the quality of the sequencing data. This information was used for the initial filtering of sequences by Trimmomatic (v0.36) (Bolger et al., 2014). For each sample, RNA sequencing (RNA-seq) raw reads (paired-end, 100 bp) were trimmed to remove potential Illumina adaptor contamination, followed by read trimming and clipping of low-quality bases. The remaining reads were aligned to the A. thaliana TAIR10 reference genome using the Araport11 annotation (Cheng et al., 2017) with the STAR aligner (v2.5.3a) (Dobin et al., 2013) and the following command-line parameters: --outFilterMultimapNmax 20 --alignSJoverhangMin 8 --alignSJDBoverhangMin 8 --outFilterMismatchNmax 8 --alignIntronMin 35 --alignMatesGapMax 100 000 --alignIntronMax 20 000. Based on the RNA-seq mapped reads and the Araport11 annotation, HTSeq (v1.99.2) (Anders and Huber, 2010) with the intersection ‘union’ option was used to generate the read counts per gene. Normalization and statistical analyses of differential gene expression were performed with the DESeq2 Bioconductor package in R (Anders and Huber, 2010; Love et al., 2014). A multiple-testing-corrected p value (q value) (Benjamini and Hochberg, 1995) of 0.05 was used. Differentially expressed genes were defined as those genes with a corrected p value of <0.05 and a log2(fold-change) >0.8 or <–0.8.
Gene ontology and statistical analyses
Gene ontology analyses were performed using the Metascape tool (https://metascape.org/gp/index.html#/main/step1) with the following parameters: minimum overlapping of 3, p value cutoff of 0.05, and minimum enrichment of 1.5. The overall data were statistically analyzed using GraphPad5 software. Comparisons between two groups were made with Student’s t-test, and multigroup comparisons were made using one-way ANOVA followed by Tukey’s test. A p value of less than 0.05 was considered statistically significant, and significant differences are indicated by different letters.
Free Pi quantification
Arabidopsis plants were germinated in MS1/2 medium for 4 days at 22°C and then transferred to 22°C (22SR), 32°C (32SR), or 32°C with a temperature gradient in the root zone (32TGRooZ) for 6 moredays. Roots and shoots were collected separately, and inorganic Pi was quantified as described previously (Ames, 1966). Free Pi was expressedper plant or per mg. For every plate analyzed, the total amount of Pi (nmoles) was divided by the number of plants in the plate or divided by the total fresh weight (n = 6 plates).
For the phr1 phl1 and pho2 mutants, free Pi (nmoles of Pi) and fresh weight (mg) wereexpressed per plant. Afterwards, shoot and root fresh weights or Pi levels of seedlings grown at different temperatures were standardized with respect to those of plants grown at 22°C.
Funding
The authors thank Malcolm Bennet for CYCB1;1:CYCB1;1-GFP, Lieven De Veylder for the erf115 mutant (SALK_021981), ERF115SDRX, ERF115OE, and pERF115:NLS-GUS/GFP (Heyman et al., 2013), and Javier Paz Ares for the phr1 phl1 double mutant and pho2. We also thank the CBGP’s Plant Facility Service and Bioinformatic Unit for help with plant growth, treatments, and bioinformatics analyses. This research was supported by grants from the Spanish Government BIO2017-82209-R and PID2020-113479RB-I00 granted by MCIN/AEI/10.13039/501100011033/ to J.C.P and by the “Severo Ochoa Program for Centres of Excellence in R&D” from the Agencia Estatal de Investigación of Spain (grant SEV-2016-0672; 2017–2021) to the C.B.G.P. M.P.G.G. is supported by a postdoctoral contract associated with the “Severo Ochoa Program” and a UPM talent attraction contract. C.M.C. and M.S.-B. are supported by a predoctoral fellowship (BES-2017-082152 and PRE2019-088076 respectively) associated with the Severo Ochoa Program. V.B.G. is supported by the Ministry of Universities (predoctoral fellowship FPU20/07 453). G.C. was supported by the Biotechnology and Biological Sciences Research Council and the National Science Foundation (BBSRC-NSF), grant no. BB/V011294/1, and the Leverhulme Trust, grant no. RPG-2019-337.
Author contributions
M.P.G.G., C.M.C., A.L., B.S., S.N.N., V.B., and M.S. performed the experiments, and M.P.G.G., C.M.C., G.C., and J.C.P. planned and designed the research. I.S.-G., G.C., E.C., and J.C.P. analyzed the data. M.P.G.G., E.C., I.S.-G., G.C., and J.C.P. wrote and edited the manuscript.
Acknowledgments
No conflict of interest declared.
Published: December 30, 2022
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Supplemental information is available at Plant Communications Online.
Supplemental information
Data availability
RNA-Seq data are deposited in the GEO Data Bank (GSE214280).
References
- Ai H., Bellstaedt J., Bartusch K.S., Eschen-Lippold L., Babben S., Balcke G.U., Tissier A., Hause B., Andersen T.G., Delker C., et al. Auxin-dependent acceleration of cell division rates regulates root growth at elevated temperature. bioRxiv. 2022 doi: 10.1101/2022.06.22.497127. Preprint at. [DOI] [Google Scholar]
- Ames B.N. In Methods in Enzymology. Academic Press; 1966. Assay of inorganic phosphate, total phosphate and phosphatases; pp. 115–118. [DOI] [Google Scholar]
- Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. http://www.bioinformatics.babraham.ac.uk/projects/fastqc
- Aung K., Lin S.I., Wu C.C., Huang Y.T., Su C.L., Chiou T.J. pho2, a phosphate overaccumulator, is caused by a nonsense mutation in a microRNA399 target gene. Plant Physiol. 2006;141:1000–1011. doi: 10.1104/pp.106.078063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari R., Datt Pant B., Stitt M., Scheible W.R. PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol. 2006;141:988–999. doi: 10.1104/pp.106.079707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin T., Betzner A., Hoggart R., Cork A., Williamson R. Root morphology mutants in <I>Arabidopsis thaliana</I>. Functional Plant Biol. 1992;19:427–437. doi: 10.1071/PP9920427. [DOI] [Google Scholar]
- Bellstaedt J., Trenner J., Lippmann R., Poeschl Y., Zhang X., Friml J., Quint M., Delker C. A mobile auxin signal connects temperature sensing in cotyledons with growth responses in hypocotyls. Plant Physiol. 2019;180:757–766. doi: 10.1104/pp.18.01377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Stat. Soc. B. 1995;57:289–300. [Google Scholar]
- Bhattacharya A. In: In Effect of High Temperature on Crop Productivity and Metabolism of Macro Molecules. Bhattacharya A., editor. Academic Press; 2019. Chapter 1 - effect of high-temperature stress on crop productivity; pp. 1–114. [DOI] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourbousse C., Vegesna N., Law J.A. SOG1 activator and MYB3R repressors regulate a complex DNA damage network in <i>Arabidopsis</i>. Proc. Natl. Acad. Sci. USA. 2018;115:E12453–E12462. doi: 10.1073/pnas.1810582115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunetti C., Lamb J., Wielandt S., Uhlemann S., Shirley I., McClure P., Dafflon B. Estimation of depth-resolved profiles of soil thermal diffusivity from temperature time series and uncertainty quantification. Earth Surf. Dynam. Discuss. 2021;2021:1–25. doi: 10.5194/esurf-2021-68. [DOI] [Google Scholar]
- Bustos R., Castrillo G., Linhares F., Puga M.I., Rubio V., Pérez-Pérez J., Solano R., Leyva A., Paz-Ares J. A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet. 2010;6:e1001102. doi: 10.1371/journal.pgen.1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calleja-Cabrera J., Boter M., Oñate-Sánchez L., Pernas M. Root growth adaptation to climate change in crops. Front. Plant Sci. 2020;11:544. doi: 10.3389/fpls.2020.00544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canher B., Lanssens F., Zhang A., Bisht A., Mazumdar S., Heyman J., Augstein F., Wolf S., Carlsbecker A., Melnyk C.W., et al. The regeneration factors ERF114 and ERF115 act as transducers of mechanical cues to developmental pathways. bioRxiv. 2021 doi: 10.1101/2021.11.29.470368. Preprint at. [DOI] [Google Scholar]
- Castrillo G., Teixeira P.J.P.L., Paredes S.H., Law T.F., de Lorenzo L., Feltcher M.E., Finkel O.M., Breakfield N.W., Mieczkowski P., Jones C.D., et al. Root microbiota drive direct integration of phosphate stress and immunity. Nature. 2017;543:513–518. doi: 10.1038/nature21417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaparro J.M., Sheflin A.M., Manter D.K., Vivanco J.M. Manipulating the soil microbiome to increase soil health and plant fertility. Biol. Fertil. Soils. 2012;48:489–499. doi: 10.1007/s00374-012-0691-4. [DOI] [Google Scholar]
- Charng Y.-y., Liu H.-c., Liu N.-y., Chi W.-t., Wang C.-n., Chang S.-h., Wang T.-t. A heat-inducible transcription factor, HsfA2, is required for extension of acquired thermotolerance in Arabidopsis. Plant Physiol. 2006;143:251–262. doi: 10.1104/pp.106.091322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Li H. Heat stress regulates the expression of genes at transcriptional and post-transcriptional levels, revealed by RNA-seq in brachypodium distachyon. Front. Plant Sci. 2017;7 doi: 10.3389/fpls.2016.02067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C.Y., Krishnakumar V., Chan A.P., Thibaud-Nissen F., Schobel S., Town C.D. Araport11: a complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 2017;89:789–804. doi: 10.1111/tpj.13415. [DOI] [PubMed] [Google Scholar]
- Compant S., van der Heijden M.G.A., Sessitsch A. Climate change effects on beneficial plant-microorganism interactions. FEMS Microbiol. Ecol. 2010;73:197–214. doi: 10.1111/j.1574-6941.2010.00900.x. [DOI] [PubMed] [Google Scholar]
- Cruz-Ramírez A., Díaz-Triviño S., Wachsman G., Du Y., Arteága-Vázquez M., Zhang H., Benjamins R., Blilou I., Neef A.B., Chandler V., et al. A SCARECROW-RETINOBLASTOMA protein network controls protective quiescence in the Arabidopsis root stem cell organizer. PLoS Biol. 2013;11:e1001724. doi: 10.1371/journal.pbio.1001724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Fuente Cantó C., Simonin M., King E., Moulin L., Bennett M.J., Castrillo G., Laplaze L. An extended root phenotype: the rhizosphere, its formation and impacts on plant fitness. Plant J. 2020;103:951–964. doi: 10.1111/tpj.14781. [DOI] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan L., Janmaat K., Willemsen V., Linstead P., Poethig S., Roberts K., Scheres B. Cellular organisation of the Arabidopsis thaliana root. Development. 1993;119:71–84. doi: 10.1242/dev.119.1.71. [DOI] [PubMed] [Google Scholar]
- Elias E.A., Cichota R., Torriani H.H., de Jong van Lier Q. Analytical soil–temperature model. Soil Sci. Soc. Am. J. 2004;68:784–788. doi: 10.2136/sssaj2004.7840. [DOI] [Google Scholar]
- Estravis-Barcala M., Heer K., Marchelli P., Ziegenhagen B., Arana M.V., Bellora N. Deciphering the transcriptomic regulation of heat stress responses in Nothofagus pumilio. PLoS One. 2021;16:e0246615. doi: 10.1371/journal.pone.0246615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahad S., Bajwa A.A., Nazir U., Anjum S.A., Farooq A., Zohaib A., Sadia S., Nasim W., Adkins S., Saud S., et al. Crop production under drought and heat stress: plant responses and management options. Front. Plant Sci. 2017;8:1147. doi: 10.3389/fpls.2017.01147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrenberg S., O'Neill S.P., Knelman J.E., Todd B., Duggan S., Bradley D., Robinson T., Schmidt S.K., Townsend A.R., Williams M.W., et al. Changes in assembly processes in soil bacterial communities following a wildfire disturbance. ISME J. 2013;7:1102–1111. doi: 10.1038/ismej.2013.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel O.M., Salas-González I., Castrillo G., Spaepen S., Law T.F., Teixeira P.J.P.L., Jones C.D., Dangl J.L. The effects of soil phosphorus content on plant microbiota are driven by the plant phosphate starvation response. PLoS Biol. 2019;17:e3000534. doi: 10.1371/journal.pbio.3000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich T., Oberkofler V., Trindade I., Altmann S., Brzezinka K., Lämke J., Gorka M., Kappel C., Sokolowska E., Skirycz A., et al. Heteromeric HSFA2/HSFA3 complexes drive transcriptional memory after heat stress in Arabidopsis. Nat. Commun. 2021;12:3426. doi: 10.1038/s41467-021-23786-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillochet C., Burko Y., Platre M.P., Zhang L., Simura J., Willige B.C., Kumar S.V., Ljung K., Chory J., Busch W. HY5 and phytochrome activity modulate shoot-to-root coordination during thermomorphogenesis in Arabidopsis. Development. 2020;147:dev192625. doi: 10.1242/dev.192625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giri A., Heckathorn S., Mishra S., Krause C. Heat stress decreases levels of nutrient-uptake and -assimilation proteins in tomato roots. Plants. 2017;6:6. doi: 10.3390/plants6010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomaa N.H., Montesinos-Navarro A., Alonso-Blanco C., Picó F.X. Temporal variation in genetic diversity and effective population size of Mediterranean and subalpine Arabidopsis thaliana populations. Mol. Ecol. 2011;20:3540–3554. doi: 10.1111/j.1365-294X.2011.05193.x. [DOI] [PubMed] [Google Scholar]
- González-García M.P., Vilarrasa-Blasi J., Zhiponova M., Divol F., Mora-García S., Russinova E., Caño-Delgado A.I. Brassinosteroids control meristem size by promoting cell cycle progression in Arabidopsis roots. Development. 2011;138:849–859. doi: 10.1242/dev.057331. [DOI] [PubMed] [Google Scholar]
- Gray S.B., Brady S.M. Plant developmental responses to climate change. Dev. Biol. 2016;419:64–77. doi: 10.1016/j.ydbio.2016.07.023. [DOI] [PubMed] [Google Scholar]
- Gray W.M., Ostin A., Sandberg G., Romano C.P., Estelle M. High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc. Natl. Acad. Sci. USA. 1998;95:7197–7202. doi: 10.1073/pnas.95.12.7197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariprasad P., Gowtham H.G., Gourav C. In: In Biocontrol Agents and Secondary Metabolites. Jogaiah S., editor. Woodhead Publishing; 2021. Beneficial plant-associated bacteria modulate host hormonal system enhancing plant resistance toward abiotic stress; pp. 113–151. [DOI] [Google Scholar]
- Hasanuzzaman M., Nahar K., Alam M.M., Roychowdhury R., Fujita M. Physiological, biochemical, and molecular mechanisms of heat stress tolerance in plants. Int. J. Mol. Sci. 2013;14:9643–9684. doi: 10.3390/ijms14059643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfield J.L., Prueger J.H. Temperature extremes: effect on plant growth and development. Weather Clim. Extrem. 2015;10:4–10. doi: 10.1016/j.wace.2015.08.001. [DOI] [Google Scholar]
- Heckathorn S.A., Giri A., Mishra S., Bista D. In Climate Change and Plant Abiotic Stress Tolerance. 2013. Heat stress and roots; pp. 109–136. [DOI] [Google Scholar]
- Heyman J., Canher B., Bisht A., Christiaens F., De Veylder L. Emerging role of the plant ERF transcription factors in coordinating wound defense responses and repair. J. Cell Sci. 2018;131 doi: 10.1242/jcs.208215. [DOI] [PubMed] [Google Scholar]
- Heyman J., Cools T., Vandenbussche F., Heyndrickx K.S., Van Leene J., Vercauteren I., Vanderauwera S., Vandepoele K., De Jaeger G., Van Der Straeten D., et al. ERF115 controls root quiescent center cell division and stem cell replenishment. Science. 2013;342:860–863. doi: 10.1126/science.1240667. [DOI] [PubMed] [Google Scholar]
- Huang B., Rachmilevitch S., Xu J. Root carbon and protein metabolism associated with heat tolerance. J. Exp. Bot. 2012;63:3455–3465. doi: 10.1093/jxb/ers003. [DOI] [PubMed] [Google Scholar]
- Huot B., Castroverde C.D.M., Velásquez A.C., Hubbard E., Pulman J.A., Yao J., Childs K.L., Tsuda K., Montgomery B.L., He S.Y. Dual impact of elevated temperature on plant defence and bacterial virulence in Arabidopsis. Nat. Commun. 2017;8:1808. doi: 10.1038/s41467-017-01674-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi M., Shibata M., Rymen B., Iwase A., Bågman A.M., Watt L., Coleman D., Favero D.S., Takahashi T., Ahnert S.E., et al. A gene regulatory network for cellular reprogramming in plant regeneration. Plant Cell Physiol. 2018;59:765–777. doi: 10.1093/pcp/pcy013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly W.M., Cochrane M.A., Freeborn P.H., Holden Z.A., Brown T.J., Williamson G.J., Bowman D.M.J.S. Climate-induced variations in global wildfire danger from 1979 to 2013. Nat. Commun. 2015;6:7537. doi: 10.1038/ncomms8537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katan J., Gamliel A. In: In Encyclopedia of Agriculture and Food Systems. Van Alfen N.K., editor. Academic Press; Oxford: 2014. Plant health management: soil solarization; pp. 460–471. [DOI] [Google Scholar]
- Kong X., Tian H., Yu Q., Zhang F., Wang R., Gao S., Xu W., Liu J., Shani E., Fu C., et al. PHB3 maintains root stem cell niche identity through ROS-responsive AP2/ERF transcription factors in Arabidopsis. Cell Rep. 2018;22:1350–1363. doi: 10.1016/j.celrep.2017.12.105. [DOI] [PubMed] [Google Scholar]
- Küpers J.J., Oskam L., Pierik R. Photoreceptors regulate plant developmental plasticity through auxin. Plants. 2020;9:940. doi: 10.3390/plants9080940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahti M., Aphalo P.J., Finér L., Ryyppö A., Lehto T., Mannerkoski H. Effects of soil temperature on shoot and root growth and nutrient uptake of 5-year-old Norway spruce seedlings. Tree Physiol. 2005;25:115–122. doi: 10.1093/treephys/25.1.115. [DOI] [PubMed] [Google Scholar]
- Larkindale J., Vierling E. Core genome responses involved in acclimation to high temperature. Plant Physiol. 2008;146:748–761. doi: 10.1104/pp.107.112060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkindale J., Hall J.D., Knight M.R., Vierling E. Heat stress phenotypes of Arabidopsis mutants implicate multiple signaling pathways in the acquisition of thermotolerance. Plant Physiol. 2005;138:882–897. doi: 10.1104/pp.105.062257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Wang W., Huq E. Spatial regulation of thermomorphogenesis by HY5 and PIF4 in Arabidopsis. Nat. Commun. 2021;12:3656. doi: 10.1038/s41467-021-24018-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.-M., Kong H.G., Song G.C., Ryu C.-M. Disruption of Firmicutes and Actinobacteria abundance in tomato rhizosphere causes the incidence of bacterial wilt disease. ISME J. 2021;15:330–347. doi: 10.1038/s41396-020-00785-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng L., Liang Q., Jiang J., Zhang C., Hao Y., Wang X., Su W. A subclass of HSP70s regulate development and abiotic stress responses in Arabidopsis thaliana. J. Plant Res. 2017;130:349–363. doi: 10.1007/s10265-016-0900-6. [DOI] [PubMed] [Google Scholar]
- Lin Y.T., Jia Z., Wang D., Chiu C.Y. Effects of temperature on the composition and diversity of bacterial communities in bamboo soils at different elevations. Biogeosciences. 2017;14:4879–4889. doi: 10.5194/bg-14-4879-2017. [DOI] [Google Scholar]
- Lincoln C., Britton J.H., Estelle M. Growth and development of the axr1 mutants of Arabidopsis. Plant Cell. 1990;2:1071–1080. doi: 10.1105/tpc.2.11.10712/11/1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Liu Y., Wang S., Cui Y., Yan D. Heat stress reduces root meristem size via induction of plasmodesmal callose accumulation inhibiting phloem unloading in Arabidopsis. Int. J. Mol. Sci. 2022;23 doi: 10.3390/ijms23042063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T.Y., Huang T.K., Tseng C.Y., Lai Y.S., Lin S.I., Lin W.Y., Chen J.W., Chiou T.J. PHO2-dependent degradation of PHO1 modulates phosphate homeostasis in Arabidopsis. Plant Cell. 2012;24:2168–2183. doi: 10.1105/tpc.112.096636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch J., Marschner P., Rengel Z. In: In Marschner's Mineral Nutrition of Higher Plants. Third Edition. Marschner P., editor. Academic Press; San Diego: 2012. Chapter 13 - effect of internal and external factors on root growth and development; pp. 331–346. [DOI] [Google Scholar]
- Manzano C., Ramirez-Parra E., Casimiro I., Otero S., Desvoyes B., De Rybel B., Beeckman T., Casero P., Gutierrez C., C Del Pozo J. Auxin and epigenetic regulation of SKP2B, an F-box that represses lateral root formation. Plant Physiol. 2012;160:749–762. doi: 10.1104/pp.112.198341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaletz S.T., Weiser M.D., Zhou J., Kaspari M., Helliker B.R., Enquist B.J. Plant thermoregulation: energetics, trait–environment interactions, and carbon economics. Trends Ecol. Evol. 2015;30:714–724. doi: 10.1016/j.tree.2015.09.006. [DOI] [PubMed] [Google Scholar]
- Miller S., Chua K., Coggins J., Mohtadi H. Heat waves, climate change, and economic output. J. Eur. Econ. Assoc. 2021;19:2658–2694. doi: 10.1093/jeea/jvab009. [DOI] [Google Scholar]
- Nagel K.A., Kastenholz B., Jahnke S., van Dusschoten D., Aach T., Mühlich M., Truhn D., Scharr H., Terjung S., Walter A., et al. Temperature responses of roots: impact on growth, root system architecture and implications for phenotyping. Funct. Plant Biol. 2009;36:947–959. doi: 10.1071/fp09184. [DOI] [PubMed] [Google Scholar]
- Nottingham A.T., Bååth E., Reischke S., Salinas N., Meir P. Adaptation of soil microbial growth to temperature: using a tropical elevation gradient to predict future changes. Glob. Chang. Biol. 2019;25:827–838. doi: 10.1111/gcb.14502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- onwuka B. Effects of soil temperature on some soil properties and plant growth. Advances in Plants & Agriculture Research. 2018;8 doi: 10.15406/apar.2018.08.00288. [DOI] [Google Scholar]
- Pacak A., Barciszewska-Pacak M., Swida-Barteczka A., Kruszka K., Sega P., Milanowska K., Jakobsen I., Jarmolowski A., Szweykowska-Kulinska Z. Heat stress affects pi-related genes expression and inorganic phosphate deposition/accumulation in barley. Front. Plant Sci. 2016;7:926. doi: 10.3389/fpls.2016.00926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.-I., Kwon H.J., Cho M.H., Song J.S., Kim B.-G., Baek J., Kim S.L., Ji H., Kwon T.-R., Kim K.-H., et al. The OsERF115/AP2EREBP110 transcription factor is involved in the multiple stress tolerance to heat and drought in rice plants. Int. J. Mol. Sci. 2021;22:7181. doi: 10.3390/ijms22137181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei X., Zhang Y., Zhu L., Zhao D., Lu Y., Zheng J. Physiological and transcriptomic analyses characterized high temperature stress response mechanisms in Sorbus pohuashanensis. Sci. Rep. 2021;11:10117. doi: 10.1038/s41598-021-89418-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers S.K., Strader L.C. Regulation of auxin transcriptional responses. Dev. Dyn. 2020;249:483–495. doi: 10.1002/dvdy.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi F., Zhang F. Cell cycle regulation in the plant response to stress. Front. Plant Sci. 2020;10:1765. doi: 10.3389/fpls.2019.01765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashotte A.M., Mason M.G., Hutchison C.E., Ferreira F.J., Schaller G.E., Kieber J.J. A subset of Arabidopsis AP2 transcription factors mediates cytokinin responses in concert with a two-component pathway. Proc. Natl. Acad. Sci. USA. 2006;103:11081–11085. doi: 10.1073/pnas.0602038103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H., Gray W.M. SAUR proteins as effectors of hormonal and environmental signals in plant growth. Mol. Plant. 2015;8:1153–1164. doi: 10.1016/j.molp.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouached H., Secco D., Arpat B., Poirier Y. The transcription factor PHR1 plays a key role in the regulation of sulfate shoot-to-root flux upon phosphate starvation in Arabidopsis. BMC Plant Biol. 2011;11:19. doi: 10.1186/1471-2229-11-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousk J., Frey S.D., Bååth E. Temperature adaptation of bacterial communities in experimentally warmed forest soils. Glob. Chang. Biol. 2012;18:3252–3258. doi: 10.1111/j.1365-2486.2012.02764.x. [DOI] [PubMed] [Google Scholar]
- Ruegger M., Dewey E., Gray W.M., Hobbie L., Turner J., Estelle M. The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast grr1p. Genes Dev. 1998;12:198–207. doi: 10.1101/gad.12.2.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabri N.S.A., Zakaria Z., Mohamad S.E., Jaafar A.B., Hara H. Importance of soil temperature for the growth of temperate crops under a tropical climate and functional role of soil microbial diversity. Microbes Environ. 2018;33:144–150. doi: 10.1264/jsme2.ME17181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar A.K., Luijten M., Miyashima S., Lenhard M., Hashimoto T., Nakajima K., Scheres B., Heidstra R., Laux T. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature. 2007;446:811–814. doi: 10.1038/nature05703. [DOI] [PubMed] [Google Scholar]
- Schramm F., Ganguli A., Kiehlmann E., Englich G., Walch D., von Koskull-Döring P. The heat stress transcription factor HsfA2 serves as a regulatory amplifier of a subset of genes in the heat stress response in Arabidopsis. Plant Mol. Biol. 2006;60:759–772. doi: 10.1007/s11103-005-5750-x. [DOI] [PubMed] [Google Scholar]
- Silva-Navas J., Conesa C.M., Saez A., Navarro-Neila S., Garcia-Mina J.M., Zamarreño A.M., Baigorri R., Swarup R., Del Pozo J.C. Role of cis-zeatin in root responses to phosphate starvation. New Phytol. 2019;224:242–257. doi: 10.1111/nph.16020. [DOI] [PubMed] [Google Scholar]
- Silva-Navas J., Moreno-Risueno M.A., Manzano C., Pallero-Baena M., Navarro-Neila S., Téllez-Robledo B., Garcia-Mina J.M., Baigorri R., Gallego F.J., del Pozo J.C. D-Root: a system to cultivate plants with the root in darkness or under different light conditions. Plant J. 2015;84:244–255. doi: 10.1111/tpj.12998. [DOI] [PubMed] [Google Scholar]
- Singh S.K., Reddy V.R., Fleisher D.H., Timlin D.J. Phosphorus nutrition affects temperature response of soybean growth and canopy photosynthesis. Front. Plant Sci. 2018;9 doi: 10.3389/fpls.2018.01116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K.A. 2nd ed. CRC Press; 2000. Soil and Environmental Analysis: Physical Methods, Revised, and Expanded. [DOI] [Google Scholar]
- Sriden N., Charoensawan V. Large-scale comparative transcriptomic analysis of temperature-responsive genes in Arabidopsis thaliana. Plant Mol. Biol. 2022;110:425–443. doi: 10.1007/s11103-021-01223-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stortenbeker N., Bemer M. The SAUR gene family: the plant’s toolbox for adaptation of growth and development. J. Exp. Bot. 2018;70:17–27. doi: 10.1093/jxb/ery332. [DOI] [PubMed] [Google Scholar]
- Sun J., Qi L., Li Y., Chu J., Li C. PIF4–Mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating Arabidopsis hypocotyl growth. PLoS Genet. 2012;8:e1002594. doi: 10.1371/journal.pgen.1002594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N., Ogita N., Takahashi T., Taniguchi S., Tanaka M., Seki M., Umeda M. A regulatory module controlling stress-induced cell cycle arrest in Arabidopsis. Elife. 2019;8:e43944. doi: 10.7554/eLife.43944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tivendale N.D., Millar A.H. How is auxin linked with cellular energy pathways to promote growth? New Phytol. 2022;233:2397–2404. doi: 10.1111/nph.17946. [DOI] [PubMed] [Google Scholar]
- Tiwari M., Kumar R., Min D., Jagadish S.V.K. Genetic and molecular mechanisms underlying root architecture and function under heat stress—a hidden story. Plant Cell Environ. 2022;45:771–788. doi: 10.1111/pce.14266. [DOI] [PubMed] [Google Scholar]
- Ubeda-Tomás S., Federici F., Casimiro I., Beemster G.T.S., Bhalerao R., Swarup R., Doerner P., Haseloff J., Bennett M.J. Gibberellin signaling in the endodermis controls Arabidopsis root meristem size. Curr. Biol. 2009;19:1194–1199. doi: 10.1016/j.cub.2009.06.023. [DOI] [PubMed] [Google Scholar]
- Ubogoeva E.V., Zemlyanskaya E.V., Xu J., Mironova V. Mechanisms of stress response in the root stem cell niche. J. Exp. Bot. 2021;72:6746–6754. doi: 10.1093/jxb/erab274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdés-López O., Batek J., Gomez-Hernandez N., Nguyen C.T., Isidra-Arellano M.C., Zhang N., Joshi T., Xu D., Hixson K.K., Weitz K.K., et al. Soybean roots grown under heat stress show global changes in their transcriptional and proteomic profiles. Front. Plant Sci. 2016;7 doi: 10.3389/fpls.2016.00517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg C., Willemsen V., Hendriks G., Weisbeek P., Scheres B. Short-range control of cell differentiation in the Arabidopsis root meristem. Nature. 1997;390:287–289. doi: 10.1038/36856. [DOI] [PubMed] [Google Scholar]
- van der Voort M., Kempenaar M., van Driel M., Raaijmakers J.M., Mendes R. Impact of soil heat on reassembly of bacterial communities in the rhizosphere microbiome and plant disease suppression. Ecol. Lett. 2016;19:375–382. doi: 10.1111/ele.12567. [DOI] [PubMed] [Google Scholar]
- Vogel C.M., Potthoff D.B., Schäfer M., Barandun N., Vorholt J.A. Protective role of the Arabidopsis leaf microbiota against a bacterial pathogen. Nat. Microbiol. 2021;6:1537–1548. doi: 10.1038/s41564-021-00997-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Zhang Y., Kieffer M., Yu H., Kepinski S., Estelle M. HSP90 regulates temperature-dependent seedling growth in Arabidopsis by stabilizing the auxin co-receptor F-box protein TIR1. Nat. Commun. 2016;7:10269. doi: 10.1038/ncomms10269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitt L., Ricachenevsky F.K., Ziegler G.Z., Clemens S., Walker E., Maathuis F.J.M., Kear P., Baxter I. A curated list of genes that affect the plant ionome. Plant Direct. 2020;4:e00272. doi: 10.1002/pld3.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Belanger F., Huang B. Differential gene expression in shoots and roots under heat stress for a geothermal and non-thermal Agrostis grass species contrasting in heat tolerance. Environ. Exp. Bot. 2008;63:240–247. doi: 10.1016/j.envexpbot.2007.11.011. [DOI] [Google Scholar]
- Yang X., Dong G., Palaniappan K., Mi G., Baskin T.I. Temperature-compensated cell production rate and elongation zone length in the root of Arabidopsis thaliana. Plant Cell Environ. 2017;40:264–276. doi: 10.1111/pce.12855. [DOI] [PubMed] [Google Scholar]
- Zhang H., Wang E., Zhou D., Luo Z., Zhang Z. Rising soil temperature in China and its potential ecological impact. Sci. Rep. 2016;6:35530. doi: 10.1038/srep35530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N., Belsterling B., Raszewski J., Tonsor S.J. Natural populations of Arabidopsis thaliana differ in seedling responses to high-temperature stress. AoB PLANTS. 2015;7 doi: 10.1093/aobpla/plv101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Swarup R., Bennett M., Schaller G.E., Kieber J.J. Cytokinin induces cell division in the quiescent center of the Arabidopsis root apical meristem. Curr. Biol. 2013;23:1979–1989. doi: 10.1016/j.cub.2013.08.008. [DOI] [PubMed] [Google Scholar]
- Zhao J., Lu Z., Wang L., Jin B. Plant responses to heat stress: physiology, transcription, noncoding RNAs, and epigenetics. Int. J. Mol. Sci. 2021;22:117. doi: 10.3390/ijms22010117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-Seq data are deposited in the GEO Data Bank (GSE214280).