Abstract
A permuted whole-genome construct of a TT virus (TTV), named VT416, had 3,852 nucleotides (nt) 98.2% similar to the prototype TA278 genome. To allow the transcription of TTV from the internal promoter, pBK*VT416(1.3G), carrying 1.3 units of VT416, was constructed. The poly(A)+ RNAs expressed in COS1 cells 48 h posttransfection contained three TTV mRNA species 3.0, 1.2, and 1.0 kb in length, which were recovered in the 13 DNA clones from a λ phage cDNA library. These mRNAs in the antigenomic orientation possessed in common the 3′ terminus downstream of a poly(A) signal (A3073ATAAA) and the 5′ terminus downstream of a cap site (C98ACTTC). A common splicing to join nt 185 with nt 277 was detected in all mRNAs. The coding region of the largest open reading frame (ORF) was maintained in 3.0-kb mRNA, because this splicing was located upstream of its initiation codon (A589TG). The second splicing was detected in 1.2-kb mRNA to join nt 711 with nt 2374 and in 1.0-kb mRNA to bind nt 711 to nt 2567. They linked a proposed ORF2 to another ORF for creating new ORFs over nt 2374 to 2872 in frame 2 and nt 2567 to 3074 in frame 3. The donor and acceptor sites of all three splicings matched the consensus sequence and were conserved in most of the 16 TTVs of distinct genotypes retrieved from the database. The observed transcription profile is unique to TTV among known members in the family Circoviridae.
A novel DNA virus, named TT virus (TTV), was found in a patient with posttransfusion hepatitis of unknown etiology (14) by representational difference analysis (7). Subsequently, Okamoto et al. (18) reported that TTV is an unenveloped virus with a buoyant density of 1.31 to 1.32 g/cm3 in CsCl and a single-stranded linear DNA genome 3,739 nucleotides (nt) in length. Recently, Mushahwar et al. (12) and Miyata et al. (10) independently found an additional 113 nt to circularize and complete the 3,852-nt-long genome and proposed TTV as the first human virus similar to members of the family Circoviridae (10). TTV has an extremely wide range of sequence divergence, by which at least 16 genotypes are classified (16).
Among vertebrate viruses in Circoviridae, chicken anemia virus (CAV) in the genus Gyrovirus (21) has a negative-strand DNA (15) as is the case for TTV (12). Although the genome of CAV is 2.3 kb long and thus much shorter than the genome of TTV (3.9 kb), CAV resembles TTV in genome structure (10, 15, 17). By contrast, porcine circovirus (PCV) and beak feather disease virus in the genus Circovirus have an ambisense genome (13). A capsid protein is encoded by the antigenomic strand for CAV (8, 13). RNA transcripts of three vertebrate viruses in Circoviridae have been analyzed in detail. A single nonspliced 2.0-kb mRNA is recognized in CAV (20), although other spliced mRNAs may encode three open reading frames (ORFs) (15). Of the three transcripts of PCV, one for the Rep protein is spliced and in the genomic orientation, and the other two are in the antigenomic orientation (8). There have been no reports on mRNAs of TTV, which is proposed to encode two putative ORFs, named ORF1 and ORF2 (5, 17, 24), primarily due to the lack of an appropriate culture system to support viral replication.
To analyze the transcription profile of TTV, we designed a construct containing a linearized TTV genome which could multiply in transfected COS1 cells with help of simian virus 40 (SV40) T antigen. The obtained results suggest that at least three mRNAs of TTV would be transcribed from a common promoter and that splicing of these mRNAs may create two novel coding regions.
MATERIALS AND METHODS
Preparation of a plasmid containing the entire TTV genome.
Nucleic acids extracted from the serum (kindly supplied by M. Mayumi, Jichi Medical School, Tochigi, Japan) from which the prototype TTV (TA278) was cloned (17) were used as the template for amplifying the two overlapping parts of genomic DNA in a TTV of genotype 1 (VT416) by a modification of the PCR protocol described previously (10). Briefly, we amplified part A (nt 971 to 2877) by a double PCR using a primer pair (5′-CGC GGA TCC A946TG ACT ACA GAC AAA TTT ACT TTA A970-3′ and 5′-AAG GAA AAA AGC GGC CGC T2901TA TGG TGC TAT TTG AAA TAA GGA2878-3′) and part B (nt 2093 to 3852 linked to nt 1 to 997) by PCR with an outer primer pair (5′-GGG1274 GTA CCC CCA AAC AGA TCT TTG TGA CAT GGT GCT TCT AAC TG1315-3′ and 5′-G1281GG GTA CCA TTT ATC AGT GAA CAT TTT TGG TGC CCC TAC TCT TA1238-3′) and a nested inner primer pair (5′-CGC GGA TCC2067 ATG AAT GCC AGG CTA CTA ATA AGA A2092-3′ and 5′-AAG GAA AAA AGC GGC CGC TTA A1020GA TGC TGT CCA GTA GTT CAT AA998-3′ (flanking restriction sequences added for cloning are in boldface; the italicized A in the last primer sequence was introduced to create a stop codon for truncating the product of ORF1 when ligated to an expression vector). Each PCR product was digested with BamHI-NotI and subcloned into the corresponding site in expression vector pGEX4T3 (Amersham Pharmacia Biotech, Little Chalfont, United Kingdom). To minimize mutations during PCR, we selected for each of the two parts one clone that produced a glutathione S-transferase–ORF1 fusion protein with the predicted size (data not shown). Using the unique EcoRI site at nt 971 and PstI site at nt 2761 in parts A and B, respectively, a permuted complete genome of TTV (VT416) was subcloned into the PstI site in pTZ18U (Worthington Biochemical, Lakewood, N.J.), to create a plasmid named pTZVT416.
Transfection and RNA extraction.
COS1 cells constitutionally expressing SV40 T antigen were cultured in Dulbecco's minimum essential medium supplemented with 10% (vol/vol) fetal bovine serum (Filtron Pty., Brooklyn, Victoria, Australia). Plasmid DNA in an amount of 24 μg was transfected into 2 × 106 COS1 cells by the calcium phosphate method (3). Total cellular RNAs in COS1 cells 48 h posttransfection were extracted with guanidinium thiocyanate and phenol chloroform (4), and poly(A)+ mRNAs were selected by Oligotex-dT30 (TaKaRa Biomedicals, Kyoto, Japan).
TTV-specific probes.
Three probes were transcribed in vitro: 148/Bam (full genome length), 627/Eco (nt 1522 to 972) in the antisense orientation, and 148/Not (full genome length) in the sense orientation. They were labeled with digoxigenin by a DIG RNA labeling kit (Roche Diagnostics, Mannheim, Germany) and used in Northern blotting. An additional 1.8-kb DNA probe, EP1.8 (nt 972 to 2759), was labeled with [α-32P]dCTP (Amersham Pharmacia Biotech) by a random priming DNA labeling kit (version 2; TaKaRa) and used for plaque hybridization.
Northern blotting.
Poly(A)+ mRNAs (1 μg) were denatured with 50% (vol/vol) formamide and electrophoresed on a 1% SeaKem ME agarose (FMC BioProducts, Rockland, Maine). They were transferred onto a nylon membrane (Hybond N+; Amersham Pharmacia Biotech) and fixed by UV irradiation (22). The filter was prehybridized at 68°C for 1 h in 5× SSPE (1× SSPE contained 180 mM NaCl, 10 mM sodium phosphate [pH 7.7], and 1 mM EDTA)–50% formamide–5× Denhardt's solution (1)–0.5% (wt/vol) sodium dodecyl sulfate (SDS)–20 μg of denatured sermon sperm DNA per ml, hybridized at 68°C for 16 h in the prehybridization solution, washed once in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% SDS at 25°C for 15 min, and then washed once in 0.2× SSC–0.1% SDS at 68°C for 30 min. Thereafter, the filter was incubated with antidigoxigenin antibody labeled with alkaline phosphatase, and mRNAs on it were visualized with a DIG luminescent detection kit (Roche).
cDNA cloning.
On poly(A)+ mRNAs from COS1 cells transfected with TTV DNA clones, cDNAs were reverse transcribed by Moloney murine leukemia virus reverse transcriptase (Stratagene, La Jolla, Calif.) using a primer, 5′-(GAGA)5 ACT AGT CTC GAG T17-3′, carrying a XhoI restriction site (in boldface) and a poly(dT) stretch. The antisense strand was synthesized with a mixture of dATP, dGTP, dTTP, and 5′-methyl-dCTP that protected the products from digestion with restriction endonucleases in a later step. The sense strand was synthesized with DNA polymerase I and RNase H. Obtained cDNAs were ligated to an EcoRI adapter and digested with EcoRI and XhoI. cDNA fragments greater than 500 nt in size were fractionated by a Sephacryl S-500 column, ligated into the EcoRI and XhoI sites in a λ phage vector (Zap Express; Stratagene), and packaged with Gigapack II Gold packaging extracts (Stratagene). Escherichia coli XL1-Blue MRF (Stratagene) was infected with phages, and the plaques obtained were lifted onto a nitrocellulose membrane (Protran BA85; Schleicher & Schuell, Dassel, Germany) and hybridized with the TTV EP1.8 probe. The candidate phages were clone purified three times. Recovered cDNA fragments were subcloned into pBK-CMV by in vivo excision through superinfection with the ExAssist helper phage (Stratagene).
Nucleotide sequencing.
DNA was sequenced by the dideoxy-sequencing method (23) using a Cy5 AutoCycle sequencing kit (Amersham Pharmacia Biotech). Some regions were subcloned into M13mp18, as required, and sequenced by the single-stranded M13 DNA method with use of a SequiTherm EXCEL II Long-Read DNA sequencing kit-ALF (Epicentre, Madison, Wis.).
Computer analyses.
Database (DDBJ) searches and sequence analyses were performed by using FASTA (19) and Genetyx (version 9.0). The alignment of DNAs was carried out with Sequencher (version 3.0).
The sequences of the following 16 TTV genomes were retrieved from the database: nine TTV isolates of genotype 1 (CHN1 [accession no. AF079173], BDH1 [AF116842], JA9 [AF122915], TRM1 [AB026345], TK16 [AB026346], TP1-3 [AB026347], GH1 [AF122913], JA20 [AF122914], and CHN2 [AF129887]), three of genotype 2 (JA1 [AF122916], US35 [AF122920], and US32 [AF122921]), two of genotype 3 (JA2B [AF122918] and JA10 [AF122919]), one of genotype 11 (TUS01 [AB017613]), and one of genotype 13 (SANBAN [AB025946]).
Nucleotide sequence accession numbers.
The nucleotide sequence data in this report have been deposited in the DDBJ/EMBL/GenBank nucleotide sequence databases under accession no. AB041007 for the whole TTV genome (VT416) and AB041821, AB041822, and AB041823 for cDNA clones 302 (representative of 3.0-kb mRNA), 1031 (1.2-kb mRNA), and 19 (1.0-kb mRNA), respectively.
RESULTS
Construction of plasmids.
pTZVT416 (Fig. 1A) contained the permuted whole genome of a TTV DNA (VT416) with 3,852 nt and 98.2% sequence similarity to TA278 (accession no. AB017610). The major ORFs and motifs in TA278, including a 113-base-long GC-rich region, five ATF (CREB) binding sites, and one TATA box (10), were conserved in VT416. However, VT416 had only one poly(A) signal at A3073ATAAA, and A1722ATAAA in TA278 was replaced with A1722AAACA.
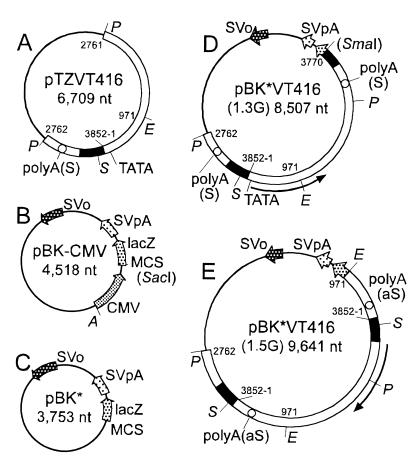
FIG. 1.
Schematic diagrams of plasmid constructs. (A) pTZVT416 containing a permuted full genome of VT416 (nt 2762 to 3852 and nt 1 to 2761), carrying a putative promoter (black area), TATA box, and the poly(A) signal (open circle) in the sense orientation [polyA(S)]; (B) pBK-CMV containing the SV40 replication origin (SVo), SV40 poly(A) signal (SVpA), lacZ, multiple cloning sites (MCS), and CMV promoter (CMV); (C) pBK* lacking the CMV promoter from pBK-CMV; (D) pBK*VT416(1.3G) containing 1.3 genome lengths of VT416 (nt 2762 to 3852 and nt 1 to 3770) (terminal SmaI site in parentheses because of its interruption in the construct); (E) pBK*VT416(1.5G) containing 1.5 genome lengths of VT416 (nt 2762 to 3852 nt 1 to 3852 and nt 1 to 971) with a putative promoter region and a poly(A) site in the antisense orientation [polyA(aS)]. The predicted orientations of mRNAs are shown by curved arrows in the peripheries of panels D and E. Restriction sites are shown for EcoRI (E), PstI (P), SmaI (S), and AseI (A).
Attempts were made to amplify TTV DNA by transfection of COS1 cells expressing SV40 T antigen with pBK-CMV carrying the SV40 replication origin (Fig. 1B). To preclude transcription driven by the cytomegalovirus (CMV) promoter in pBK-CMV, we prepared pBK*, in which the CMV promoter was removed from pBK-CMV by digestion with AseI and SacI and self-ligation (Fig. 1C). A genome-length TTV DNA was excised by digestion of pTZVT416 with PstI, and a downstream 0.3-genome-length fragment was obtained by digestion with PstI and SmaI. To allow the transcription of TTV RNA, these two fragments were introduced into pBK* in tandem, to generate pBK*VT416(1.3G), which contained a 1.3-genome-length TTV DNA (nt 2762 to 3852 and nt 1 to 3770) starting from the putative promoter region (Fig. 1D). To evaluate possible transcripts in the genomic orientation, we also constructed pBK*VT416(1.5G), which carried a 1.5-genome-length TTV DNA (nt 2762 to 3852, nt 1 to 3852, and nt 1 to 971) starting from the putative promoter for mRNAs in the antisense orientation (Fig. 1E).
Expression of TTV mRNAs in COS1 cells.
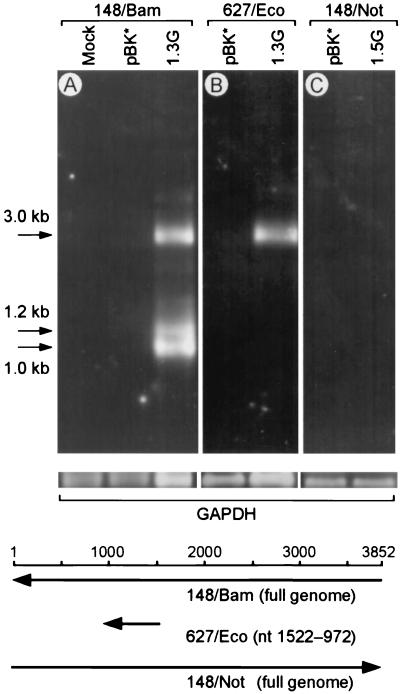
The poly(A)+ RNAs obtained from COS1 cells 48 h posttransfection were tested by Northern blotting. As shown in Fig. 2A (upper panel, lane 1.3G), the cells transfected with pBK*VT416(1.3G) revealed three distinct RNA bands at 3.0, 1.2, and 1.0 kb which were hybridized with the full-genome-length probe in the genomic orientation (148/Bam). No visible bands were observed for mock- or pBK*-transfected cells (lanes Mock and pBK*). The quality and amount of poly(A)+ RNA applied in each lane were confirmed in a parallel run probed for a constitutional mRNA expressed in COS1 cells (glyceraldehyde-3-phosphate dehydrogenase) (Fig. 2, middle panel). In contrast, only the 3.0-kb band was visible by hybridization with the 627/Eco probe representing a partial genome (nt 1522 to 972) (Fig. 2B, lane 1.3G), suggesting a splicing in both 1.2- and 1.0-kb mRNAs over the corresponding region. The cells transfected with pBK*VT416(1.5G), for expression of mRNAs in the genomic orientation, did not produce any positive signals by hybridization with the full-genome-length probe in the antigenomic orientation (148/Not) (Fig. 2C, lane 1.5G).
FIG. 2.
Expression of TTV mRNA in COS1 cells. In the upper panel, poly(A)+ RNAs of COS1 cells harvested 48 h posttransfection were hybridized with the genomic 148/Bam probe (A), the genomic 627/Eco probe (nt 1522 to 972) (B), or the antigenomic 148/Not probe (C). Lanes: Mock, mock transfection; pBK*, transfection with pBK*; 1.3G, transfection with pBK*VT416(1.3G); 1.5G, transfection with pBK*VT416(1.5G). The positions of 3.0-, 1.2-, and 1.0-kb mRNAs are indicated by arrows on the left. In the middle panel, parallel runs were hybridized with the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probe as a loading control. Positions and orientations of the probes are shown below the gels.
cDNA clones of TTV mRNAs.
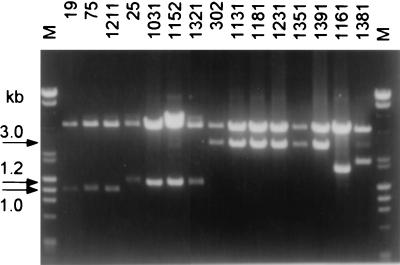
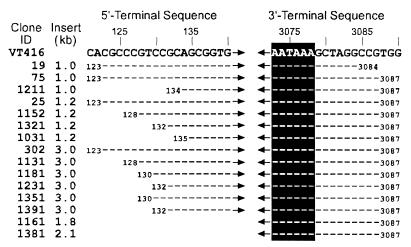
A λ phage cDNA library was obtained on a preparation of poly(A)+ RNAs from COS1 cells transfected with pBK*VT416(1.3G). From 3 × 105 plaques screened by the TTV DNA probe (EP1.8 [nt 972 to 2761]), 15 clones which carried TTV sequences were recovered. The sizes of TTV inserts were 1.0 kb in three clones (clones 19, 75, and 1211), 1.2 kb in four clones (clones 25, 1031, 1152, and 1321), 3.0 kb in six clones (clones 302, 1131, 1181, 1231, 1351, and 1391), and 1.8 (clone 1161) and 2.1 (clone 1381) kb in one clone each (Fig. 3). Both 3′ and 5′ termini of each insert were sequenced. The 3′ terminus of every insert had poly(A) starting at 7 or 10 nt downstream of the poly(A) signal (A3073ATAAA), suggesting that all mRNAs used this poly(A) signal (Fig. 4). In each of the 13 clones of 1.0, 1.2, or 3.0 kb, the 5′-terminus was at a position from 123 to 135 nt downstream of the TATA box (A85TATAA). Hence, they would use a common promoter and C98ACTTC that was the only cap site between the promoter and the 5′ terminus in each of these clones.
FIG. 3.
Sizes of mRNA inserts in the 15 TTV cDNA clones in pBK-CMV. TTV mRNA inserts were excised by digestion with XhoI and SalI. Lanes are shown for 15 clones (numbered) and a DNA size marker (M) consisting of a mixture of λ phage DNA digested with HindIII and φX174 DNA with HaeIII.
FIG. 4.
The 5′- and 3′-terminal sequences of the 15 TTV cDNA clones. The sequence of VT416 (identical to that of TA278 [accession no. AB016710]) is shown at the top as the reference. Locations of the TATA box and cap site in VT416 are A85TATAA and C98ACTTC, respectively. White-on-black letters indicate the poly(A) signal at nt 3073; hyphens indicate nucleotides identical to those of VT416.
The 5′ termini of two exceptional clones 1.8 and 2.1 kb in size (clones 1161 and 1381) were positioned at nt 1309 and 1023, respectively (data not shown). Because their sizes were consistent with the length of reverse transcripts starting from the poly(A) site and their restriction patterns were compatible with those predicted (data not shown), these two clones represented incompletely reverse-transcribed 3.0-kb mRNAs and hence were not pursued further.
The 3.0-kb mRNA.
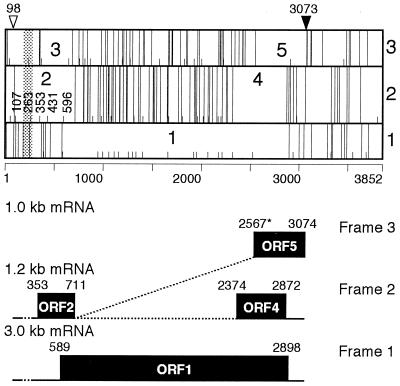
Clone 302 representing 3.0-kb mRNA covered the entire ORF1 (nt 589 to 2898) in the TTV genome (10, 12, 17). It had a single splicing to join nt 185 with nt 277. The splicing did not affect ORF1, however, because of its location upstream of the first A589TG and the presence of multiple stop codons between the splicing and A589TG in frame 1 (Fig. 5).
FIG. 5.
Schematic diagram of the VT416 genome and its mRNAs. Three reading frames of the VT416 genome are shown in the upper panel. The open triangle indicates the position of the cap site, and the closed triangle indicates that of the poly(A) signal. Short and long vertical lines indicate ATGs and stop codons, respectively. Predicted ORFs are indicated by numbers. The shaded area represents the first splicing common to all mRNAs in three different sizes. The lower panel indicates used frames and configurations of 3.0-, 1.2-, and 1.0-kb mRNAs. Solid lines indicate exons, dotted lines represent introns, and boxes stand for coding regions. Because of alternative splicing for 1.0-kb mRNA, the 5′-terminal nt 2567 is labeled with an asterisk.
The 1.2-kb mRNA.
Clone 1031 representing 1.2-kb mRNA had two splicings to join nt 185 with nt 277 and nt 711 with nt 2374 (Fig. 5). The first splicing was identical to that in 3.0-kb mRNA; the second, at nt 711, was located 2 nt upstream of the termination codon (T713AA) of ORF2 that is predicted in frame 2 (10, 17). Due to this splicing, however, the predicted coding region of ORF2 was linked to a new ORF (ORF4 [nt 2374 to 2872]) in the same frame. The other two clones carrying 1.2-kb mRNA (clones 1152 and 1321) had the same two splicings. Clone 25 carrying 1.2-kb mRNA had an additional splicing to join nt 2875 with nt 2894.
The 1.0-kb mRNA.
The complete sequence of clone 75, one of the three 1.0-kb mRNA clones, had two splicings to join nt 185 with nt 277 and nt 711 with nt 2567 (Fig. 5). As in 1.2-kb mRNA, the first splicing was identical to that in 3.0-kb mRNA. The second splicing donor was located at the same position as that in 1.2-kb mRNA, but the coding region abutted another ORF (ORF5 [nt 2567 to 3074]) in frame 3 and terminated in T3075AA that overlapped with the poly(A) signal starting at nt 3073 (Fig. 5). The second splicing in the other two 1.0-kb mRNAs (clones 1211 and 19) spanned nt 711 to 2564 and was 3 nt shorter than those in clone 75.
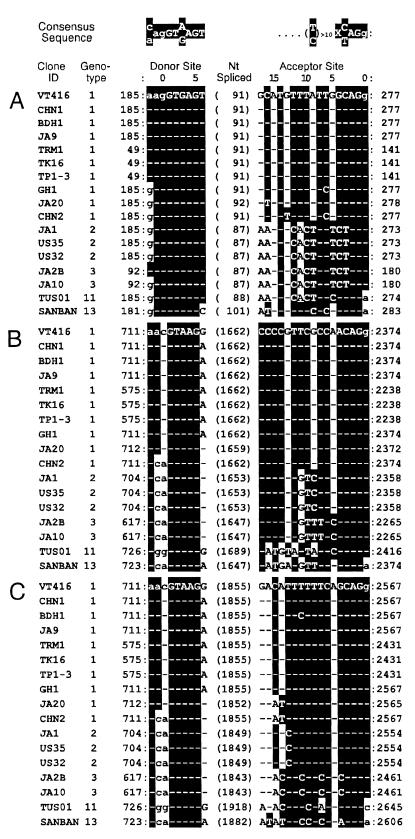
Consensus sequence of splicing.
We searched the database for the three splicings in mRNAs from 16 TTVs of five distinct genotypes, using the GT-AG splicing rule (2) and the intron consensus rule (11) (indicated at the top of Fig. 6).
FIG. 6.
Sequences of donor and acceptor sites in 16 TTVs of various genotypes retrieved from the database. The consensus sequence of the splice junction (2, 11) is shown at the top. (A) The first splicing shared by 3.0-, 1.2-, and 1.0-kb mRNAs of VT416; (B) the second splicing in 1.2-kb mRNA; (C) the second splicing in 1.0-kb mRNA. Splicings in TTVs retrieved from the database were predicted on the basis of consensus sequence. Nucleotide numbers of the donor and acceptor sites are counted from the last and first nucleotides in the exon, respectively, which are assigned position 0. The figure in parentheses indicates the number of nucleotides spliced out. White-on-black letters indicate nucleotides matching the consensus sequence shown at the top; hyphens indicate nucleotides identical to those in VT416. The sequence of TA278 (accession no. AB106710) is identical to that shown for VT416. Nucleotides for exons are in lowercase; those for introns are in uppercase.
The sequences of the first splicing common to three TTV mRNAs are shown in Fig. 6A. The donor site of the first splicing at nt 185, or the position corresponding to that in VT416, was conserved in all 16 TTVs examined, as was the splicing acceptor site at nt 277 in VT416. In eight of the nine TTVs of genotype 1, 91 nt were spliced out; the exception was JA20, in which 92 nt were spliced. At the acceptor site, all five TTVs of genotype 2 or 3 had the same sequence but were different in 9 nt from that in VT416. The acceptor site in TUS01 of genotype 11 had substitutions of 2 nt in the sequences of genotypes 2 and 3. The acceptor site in SANBAN of genotype 13 had substitutions of 4 nt in that of VT416, and the splicing was 10 nt longer than that of VT416.
The donor site of the second splicing at nt 711 in VT416 also was well conserved in all TTVs examined (Fig. 6B). Sequences of the acceptor sites in all TTVs of genotype 1, 2, and 3 were genotype specific and identical among the TTVs of the same genotype. Of the nine TTVs of genotype 1, seven had a G-to-A conversion at the same position and CHN2 had changes of 2 nt. Sequences of the donor sites in TTVs of genotypes 2 and 3 were identical and the same as that in CHN2 of genotype 1. Also, the acceptor site of splicing at nt 2374 in each was conserved and satisfied the rule (2, 11). Although the predicted acceptor sites in TUS01 and SANBAN had several nucleotide substitutions, their configuration matched the rule as well.
Likewise, the acceptor site at nt 2567 in ORF5 of VT416 was conserved in the examined TTVs and matched the rule (Fig. 6C). A similar matching of the acceptor site is seen at nt 2564 in Fig. 6C, represented by clones 1211 and 19, but the site ended with CAGc instead of CAGg in most examined TTVs spanning positions 6 to 3 in the acceptor site. The acceptor sites in JA20 and CHN2 of genotype 1 differed by 2 nt from those of the other eight TTVs of genotype 1. The sequences of this site in all three TTVs of genotype 2, as well as both of genotype 3, were identical and different by only 1 nt from that in VT416. Both TTVs of genotype 3 had substitutions of 5 nt. TUS01 of genotype 11 and SANBAN of genotype 13 also possessed a motif matching the rule.
DISCUSSION
Three classes of mRNAs driven from the internal promoter in the TTV genome were recovered from COS1 cells transiently transfected with the plasmid carrying 1.3 genome lengths of a TTV of genotype 1 (VT416). Their sizes were 3.0, 1.2, and 1.0 kb, and the 3′ termini of all were 7 or 10 nt downstream of the only poly(A) signal in VT416 (T3073ATAAA). Identification of the exact 5′ terminus of each mRNA was not feasible with the experimental protocol used in this present study. However, the 5′ termini of all 13 cDNA clones reverse transcribed from TTV mRNAs were located at a position from nt 123 to 135 immediately downstream of the putative promoter, TATA box (A85TATAA), and cap site (C98ACTCC). The results suggest strongly that the three TTV mRNAs would use the same internal promoter, the TATA box and cap site in VT416.
mRNAs in the genomic orientation were not detected in COS1 cells transfected with pBKVT*416(1.5G) carrying a TTV genome in the antisense orientation. This had to be evaluated in view of the report of one genomic and two antigenomic mRNAs in PCV (8), along with several transcription promoter motifs identified in a region (nt 521 to 13) in the genomic orientation of VT416. Although VT416 had a poly(A) signal (A271ATAAA) in the antisense orientation, the signal was not conserved in most 16 TTVs from the database. Furthermore, no TATA boxes were found downstream of this putative promoter sequence in any TTVs examined. The results suggest that TTV would not transcribe mRNAs in the genomic orientation. In this respect, TTV is distinct from PCV, which has an ambisense genome (8, 9, 13). TTV is also different from CAV in that it transcribes three species of mRNAs (20).
The first splicing, joining nt 185 with nt 277, was shared by the three mRNAs. The consensus sequence for the splicing is preserved in all 16 examined TTVs of genotype 1, 2, 3, 11, or 13, although there were minor differences in the length of splicing. Based on the obtained results, the first splicing would be ubiquitous in TTVs of all genotypes.
Only 3.0-kb mRNA possessed ORF1 (nt 589 to 2898), which has been assigned to a putative capsid protein of TTV; it had the first splicing alone. By contrast, 1.2- and 1.0-kb mRNAs had a second splicing, in addition to the first splicing in 3.0-kb mRNA, which joined nt 711 with nt 2374 and nt 711 with nt 2567. The stop codon (T713AA) predicted on ORF2 in frame 2 (nt 95 to 712) (14, 17, 24) cannot be used due to the second splicings in 1.2- and 1.0-kb mRNAs (Fig. 5, upper panel). The coding region of the 1.2-kb mRNA adjoined ORF4 (nt 2374 to 2872) in the frame 2, and that of the 1.0-kb mRNA adjoined ORF5 (nt 2553 to 3074) in frame 3 (Fig. 5, lower panel). As a result, two novel coding regions which may encode distinct TTV proteins were generated. The results of this study could not determine, however, which of the two second acceptor sites at nt 2567 and 2564 is actually operating.
The 16 TTVs of various genotypes retrieved from the database had similar ORFs in frames 2 and 3 corresponding to ORF4 and ORF5, respectively, in VT416. Furthermore, the donor and acceptor sites for the second splicings in 1.2- and 1.0-kb mRNAs of VT416 were conserved in all 16 TTVs examined. The obtained data suggest that these second splicings also would be common to TTVs of all genotypes.
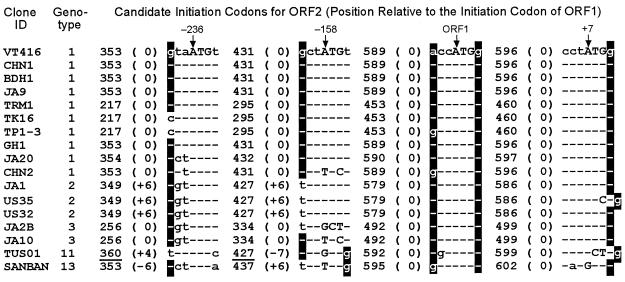
Since the initiation codon proposed for ORF2 (14, 17, 24) is no longer valid, we searched for it in TTVs of various genotypes (Fig. 5). The first A107TG in frame 2 after the cap site in VT416 should be out of frame, because the first splicing spanned 91 nt. No ATGs are found between the cap site and the first splicing donor site in frame 3. Moreover, the A263TG in frame 2 that was proposed for the initiation codon (14, 17, 24) is lost by the first splicing. Hence, three ATGs at nt 353, 431, and 596, located between the first splicing acceptor site and the end of ORF2, would be candidates (Fig. 5). Their positions in reference to the A589TG of ORF1 were at −236, −158 and +7, respectively, in all TTVs examined (Fig. 7). They were evaluated on the basis of Kozak's rule, which prefers A−3 or G−3 and G+4 with reference to ATG (6). Although A−3 and G+4 in A589TG of ORF1 satisfy the rule (Fig. 7), those in the three candidate ATGs for ORF2 do so only partially. The first A−236TG is conserved in 16 of the 17 TTVs (VT416 included); the exception is TUS01, although its position was shifted in the three TTVs of genotype 2 (JA1, US35, and US32) and the one of genotype 13 (SANBAN). The second A−158TG is conserved in 12 of the 17 TTVs used for comparison, the five exceptions being CHN2 of genotype 1, JA2B and JA10 of genotype 3, TUS01 of genotype 11, and SANBAN of genotype 13. The third A+7TG is conserved in 14 of the 17 TTVs, the exceptions being US35 (genotype 2), TUS01 (genotype 11), and SANBAN (genotype 13). However, ATGs of US35 and TUS01 may be reconstituted if C588 and C600, respectively, were deleted. In that event, 16 of the 17 TTVs of various genotypes would have A+7TG at the expected position. These speculations, however, do not specify which of the first and third ATGs serves as the authentic initiation codon for ORF2 in 1.2- and 1.0-kb mRNAs.
FIG. 7.
Sequences of seven nucleotides around the three candidate ATGs for ORF2. A353TG at nt −236 with respect to the initiation codon of ORF1 (A589TG), A431TG at nt −158, and A596TG at nt +7 are shown. Uppercase letters represent nucleotides constituting ATG, and lowercase letters represent those in flanking regions. Hyphens indicate nucleotides identical to those in VT416. White-on-black letters indicate nucleotides matching Kozak's rule. Nonsense ATGs due to frameshifts are underlined.
Because of the second splicing, 1.2- and 1.0-kb mRNAs would code for two proteins corresponding to the regions ORF2-ORF4 and ORF2-ORF5. The presence of ORF4 and ORF5 in all TTVs in the database lends support to this view. Should the ATG at nt 353 be used, the joint polypeptides predicted in VT416 would have 286 and 290 amino acids (aa) coded for by 1.2- and 1.0-kb mRNAs, respectively. The predicted protein encoded by the 1.2-kb mRNA has a serine-rich region (aa 213 to 264). Proteins with amino acid sequences similar to the two joint proteins of TTV were searched for by FASTA. Those resembling the ORF2–ORF4 protein carried transcription factors and mammalian RNA-binding motifs with serine-rich residues. Likewise, those similar to the ORF2-ORF5 protein possessed transcription factors. Hence, both novel TTV proteins, encoded by ORF2-ORF4 and ORF2-ORF5, respectively, may be speculated to serve as regulators in the transcription of TTV DNA. The expression of these two novel proteins encoded by joint ORFs, as well as the product of putative ORF1, would have to be demonstrated by immunoassays with antibodies specific to each of them.
There is the possibility that both 1.2- and 1.0-kb mRNAs use the same A589TG in frame 1 bearing ORF1. Should this be the case, 1.2-kb mRNA would code for a 216-aa protein having the same head and tail as the product of ORF1 but lacking the middle 554 aa out of the encoded 770 aa. Likewise, 1.0-kb mRNA would code for a 144-aa protein joining ORF1 with ORF2. However, a translation start in frame 2 would be preferred, since those from frame 1 would not support the maintenance of ORF5 shared by most TTVs from the database, despite continuous evolutionary pressure in the past.
In conclusion, three distinct species of mRNA were transcribed by TTV in COS1 cells. Their profiles are distinct from those of the members of Circoviridae, such as a single nonspliced transcript for CAV (20) and two antigenomic and one genomic mRNA for PCV (8, 13). The two putative proteins encoded by novel joint ORFs, borne by 1.2- and 1.0-kb mRNAs, warrant analysis for roles in the replication of TTV.
ACKNOWLEDGMENTS
We thank D. Nameki, R. Okamoto, and A. Yamada, undergraduate students of the faculty, for technical assistance.
REFERENCES
- 1.Alwine J C, Kemp D J, Parker B A, Reiser J, Renart J, Stark G R, Wahl G M. Detection of specific RNAs or specific fragments of DNA by fractionation in gels and transfer to diazobenzyloxymethyl paper. Methods Enzymol. 1979;68:220–242. doi: 10.1016/0076-6879(79)68017-5. [DOI] [PubMed] [Google Scholar]
- 2.Breathnach R, Benoist C, O'Hare K, Gannon F, Chambon P. Ovalbumin gene: evidence for a leader sequence in mRNA and DNA sequences at the exon-intron boundaries. Proc Natl Acad Sci USA. 1978;75:4853–4857. doi: 10.1073/pnas.75.10.4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 5.Hijikata M, Takahashi K, Mishiro S. Complete circular DNA genome of a TT virus variant (isolate name SANBAN) and 44 partial ORF2 sequences implicating a great degree of diversity beyond genotypes. Virology. 1999;260:17–22. doi: 10.1006/viro.1999.9797. [DOI] [PubMed] [Google Scholar]
- 6.Kozak M. Point mutations close to the AUG initiator codon affect the efficiency of translation of rat preproinsulin in vivo. Nature. 1984;308:241–246. doi: 10.1038/308241a0. [DOI] [PubMed] [Google Scholar]
- 7.Lisitsyn N, Lisitsyn N, Wigler M. Cloning the differences between two complex genomes. Science. 1993;259:946–951. doi: 10.1126/science.8438152. [DOI] [PubMed] [Google Scholar]
- 8.Mankertz J, Buhk H J, Blaess G, Mankertz A. Transcription analysis of porcine circovirus (PCV) Virus Genes. 1998;16:267–276. doi: 10.1023/a:1008022521329. [DOI] [PubMed] [Google Scholar]
- 9.Meehan B M, Creelan J L, McNulty M S, Todd D. Sequence of porcine circovirus DNA: affinities with plant circoviruses. J Gen Virol. 1997;78:221–227. doi: 10.1099/0022-1317-78-1-221. [DOI] [PubMed] [Google Scholar]
- 10.Miyata H, Tsunoda H, Kazi A, Yamada A, Khan M A, Murakami J, Kamahora T, Shiraki K, Hino S. Identification of a novel GC-rich 113-nucleotide region to complete the circular, single-stranded DNA genome of TT virus, the first human circovirus. J Virol. 1999;73:3582–3586. doi: 10.1128/jvi.73.5.3582-3586.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mount S M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982;10:459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mushahwar I K, Erker J C, Muerhoff A S, Leary T P, Simons J N, Birkenmeyer L G, Chalmers M L, Pilot Matias T J, Dexai S M. Molecular and biophysical characterization of TT virus: evidence for a new virus family infecting humans. Proc Natl Acad Sci USA. 1999;96:3177–3182. doi: 10.1073/pnas.96.6.3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Niagro F D, Forsthoefel A N, Lawther R P, Kamalanathan L, Ritchie B W, Latimer K S, Lukert P D. Beak and feather disease virus and porcine circovirus genomes: intermediates between the geminiviruses and plant circoviruses. Arch Virol. 1998;143:1723–1744. doi: 10.1007/s007050050412. [DOI] [PubMed] [Google Scholar]
- 14.Nishizawa T, Okamoto H, Konishi K, Yoshizawa H, Miyakawa Y, Mayumi M. A novel DNA virus (TTV) associated with elevated transaminase levels in posttransfusion hepatitis of unknown etiology. Biochem Biophys Res Commun. 1997;241:92–97. doi: 10.1006/bbrc.1997.7765. [DOI] [PubMed] [Google Scholar]
- 15.Noteborn M H, de Boer G F, van Roozelaar D J, Karreman C, Kranenburg O, Vos J G, Jeurissen S H, Hoeben R C, Zantema A, Koch G, van Ormondt H, van der Eb A J. Characterization of cloned chicken anemia virus DNA that contains all elements for the infectious replication cycle. J Virol. 1991;65:3131–3139. doi: 10.1128/jvi.65.6.3131-3139.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okamoto H, Fukuda M, Tawara A, Nishizawa T, Itoh Y, Hayasaka I, Tsuda F, Tanaka T, Miyakawa Y, Mayumi M. Species-specific TT viruses and cross-species infection in nonhuman primates. J Virol. 2000;74:1132–1139. doi: 10.1128/jvi.74.3.1132-1139.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okamoto H, Nishizawa T, Kato N, Ukita M, Ikeda H, Iizuka H, Miyakawa Y, Mayumi M. Molecular cloning and characterization of a novel DNA virus (TTV) associated with posttransfusion hepatitis of unknown etiology. Hepatol Res. 1998;10:1–16. [Google Scholar]
- 18.Okamoto H, Nishizawa T, Ukita M, Takahashi M, Fukuda M, Iizuka H, Miyakawa Y, Mayumi M. The entire nucleotide sequence of a TT virus isolate from the United States (TUS01): comparison with reported isolates and phylogenetic analysis. Virology. 1999;259:437–448. doi: 10.1006/viro.1999.9769. [DOI] [PubMed] [Google Scholar]
- 19.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Phenix K V, Meehan B M, Todd D, McNulty M S. Transcriptional analysis and genome expression of chicken anaemia virus. J Gen Virol. 1994;75:905–909. doi: 10.1099/0022-1317-75-4-905. [DOI] [PubMed] [Google Scholar]
- 21.Pringle C R. Virus taxonomy at the XIth International Congress of Virology, Sydney, Australia, 1999. Arch Virol. 1999;144:2065–2070. doi: 10.1007/s007050050728. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 23.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takahashi K, Ohta Y, Mishiro S. Partial ∼2.4kb sequences of TT virus (TTV) genome from eight Japanese isolates: diagnostic and phylogenetic implications. Hepatol Res. 1998;12:111–120. [Google Scholar]