Summary
Background
Rheumatoid arthritis (RA) is a common autoimmune disease with emerging environmental and microbiome risk factors. The western diet is typically deficient in magnesium (Mg), and there is some evidence suggesting that Mg may have anti-inflammatory properties. But the actual role of Mg supplementation in arthritis or in T cell subsets has not been explored.
Methods
We investigated the role of a high Mg diet in two different mouse models of RA induced with the KRN serum, and collagen-induced arthritis. We also characterized the phenotypes of splenocytes, gene expression, and an extensive intestinal microbiome analyses including fecal material transplantation (FMT).
Findings
The high Mg diet group was significantly protected with reduced arthritis severity and joint damage, and reduced expression of IL-1β, IL-6, and TNFα. The high Mg group also had increased numbers of Foxp3+ Treg cells and IL-10-producing T cells. The high Mg protective effect disappeared in IL-10 knockout mice. FMT from the high Mg diet mice recreated the phenotypes seen in the diet-treated mice, with reduced arthritis severity, increased Foxp3+ Treg, and increased IL-10-producing T cells. Intestinal microbiome analyses using 16S rDNA sequencing revealed diet-specific changes, including reduced levels of RA-associated Prevotella in the high Mg group, while increasing levels of Bacteroides and other bacteria associated with increased production of short-chain fatty acids. Metagenomic analyses implicated additional pathways including L-tryptophan biosynthesis and arginine deiminase.
Interpretation
We describe a new role for Mg in suppressing arthritis, in expanding Foxp3+ T reg cells and in the production of IL-10, and show that these effects are mediated by the intestinal microbiome. Our discoveries suggest a novel strategy for modifying the intestinal microbiome to treat RA and other autoimmune and inflammatory diseases.
Funding
None.
Keywords: Rheumatoid arthritis, High magnesium diet, Microbiome, Treg, IL-10, Inflammation, Autoimmunity
Research in context.
Evidence before this study
Little is known about environmental and dietary risk factors for autoimmune diseases such as rheumatoid arthritis (RA). The US diet is typically deficient in magnesium, and some studies have suggested that magnesium reduces cytokine expression in vitro.
Added value of this study
In the present study we show that oral magnesium supplementation reduced disease severity and joint damage, and increased T cell subsets known to suppress autoimmune responses (Tregs and Tr1), in a microbiome and IL-10-dependent manner.
Implications of all the available evidence
These new discoveries raise the possibility that magnesium supplementation has the potential to become a low risk and low cost adjuvant to the treatment of RA and other autoimmune and inflammatory diseases.
Introduction
Rheumatoid arthritis (RA) affects nearly 1% of the population and is associated with increased risk for disability and increased mortality.1 RA has both genetic and environmental contributions to disease.2 While knowledge about genes implicated in disease susceptibility has expanded significantly,3 little is known about environmental and dietary contributions to disease.4 Despite the development of novel and more effective therapies in the past twenty years, disease remission remains uncommon in RA.5 Therefore, identification and modification of at risk environmental and dietary factors has the potential to further improve disease control.
Magnesium (Mg) is the second most abundant intracellular cation and plays an important role in many intracellular biochemical functions.6 Short-term in vitro studies showed that increased concentrations of Mg reduce LPS-induced production of pro-inflammatory cytokines including TNFα, IL-6, and IL-8 and suppress NFκB activation in monocytes and endothelial cells,7,8 while reduced concentrations of Mg were reported to be pro-inflammatory and to activate NFκB.9 Levels of TNFα, IL-6, and IL-8 and NFκB activity are increased in RA and have been associated with joint inflammation and implicated in synovial hyperplasia and joint damage.1,10 Furthermore, Mg is typically deficient in the US diet with nearly 40% of the population consuming less than the required amounts.11 Therefore, we considered that the commonly Mg deficient US diet might favor pro-inflammatory pathways contributing to RA susceptibility or disease severity. We hypothesized that the administration of Mg might be beneficial in the treatment of RA or rodent models of RA.
The gut microbiota has a crucial role in driving both local and systemic immune responses.12,13 The balance between different bacterial populations in the gut can differentially affect the number of T cell subsets such as Foxp3+ Tregs and Tr1, which have central roles in controlling autoimmune and inflammatory responses. Indeed, changes in the intestinal microbiome have been associated with susceptibility to human autoimmune diseases, including RA,14, 15, 16 systemic lupus erythematosus,17 diabetes,18 multiple sclerosis,19 and others. Microbial composition in subjects with RA differs from healthy controls with a reduction of bacteria belonging to the families Bacteroides and Bifidobacterium and a marked increase of Prevotella, among others.14,20,21 To date we are not aware of any strategies that have successfully modified the gut microbiome to treat RA.
In this study, we describe for the first time that a high Mg diet reduces disease severity and joint damage in two well-established models of RA. Furthermore, we demonstrate that high Mg diet modulates arthritis via changes induced in the gut microbiome that increase the numbers of Foxp3+ Treg cells and the number of IL-10-producing T cells. We further describe that these Mg-induced changes are mediated by IL-10. Our discoveries raise the possibility that oral Mg treatment has the potential to become a new, inexpensive and benign treatment for RA, and perhaps for other inflammatory and autoimmune diseases as well.
Methods
Mice
Male C57BL/6 mice were purchased from Taconic (Rensselaer, NY). Both male and female C57BL/6J, B6.129P2-IL10tm1Cgn/J (IL-10 KO), GFPinducible- C57BL/6-Il17atm1Bcgen/J, male DBA/1J mice, and NOD/ShiLtJ were purchased from Jackson Laboratories (Farmington, CT). Given the C57BL/6 known difference in Th17-inducing intestinal levels of Segmented filamentous bacteria (SFB)22 we tested both Taconic and Jackson mice and they were similarly protected by the high Mg2800 diet in KSIA. Therefore, all the microbiome and FMT experiments used only Taconic mice. KBxN (KRN) TCR transgenic mice (gift from Dr. C. Benoist, Boston, MA) were bred and maintained at Mount Sinai. Mice were housed under specific pathogen-free conditions and all experiments conducted under a protocol approved by the Mount Sinai Institutional Animal Care and Use Committee (protocol number 2014-0283). Mouse experiments were planned according to the ARRIVE guidelines (www.ARRIVEguidelines.org).
Arthritis induction and scoring
Collagen-induced arthritis (CIA)
Eight to twelve week-old male DBA/1J mice were injected subcutaneously in the tail on day 0 with a 50 μL of 1 mg/ml of chicken type II collagen emulsion in Complete Freund’s Adjuvant (Hooke laboratories, Lawrence, MA), and received a booster containing 50 μL emulsion of 1 mg/ml of chicken type II collagen emulsion in Incomplete Freund’s Adjuvant (Hooke laboratories) on day 21.23,24 Mice were followed for 71 days and scored 3 times a week.
KRN serum-induced arthritis (KSIA)
KRN TCR transgenic mice were crossed with NOD (KRN × NOD F1) and the arthritogenic serum was collected from 60-day old arthritic mice. Serum from different batches was pooled and administered to male C57BL/6 mice at 100 μL IP on days 0 and 2. Mice typically develop arthritis on day 3, and were followed for 6–15 days and scored three times a week.25,26
Arthritis activity and severity scoring
The clinical arthritis score was determined according to a scoring scale ranging from 0 to 16 per mouse per day as previously reported where 1 = swelling and erythema in a single joint, 2 = swelling and erythema in more than one joint, 3 = swelling of the entire paw, and 4 = swelling of paw and inability to bear weigh.26,27
Histology and histological scoring
At the end of the arthritis observation period mice were euthanized, the paws fixed in 10% formaldehyde and then decalcified with a solution containing hydrochloric acid and 0.1 M EDTA (Cal-Ex; Fisher Scientific, Fairlawn, NJ). Tissues were embedded in paraffin, and slides prepared and stained with hematoxylin-eosin. The slides were evaluated in a blinded manner with a combined scoring system that included the following parameters: synovial inflammation, synovial hyperplasia, cartilage, and bone erosions (Supplemental Table S1).28, 29, 30, 31
Small intestine and colon were collected and “Swiss-rolled” for histology.32 Immunohistochemistry (IHC) was done with the Ventana Discovery Ultra (Roche, Switzerland). Mouse anti-Foxp3 monoclonal antibody (ab20034, Abcam, Cambridge, UK) was used at a 1:6000 dilution and incubated for 30 min. Discovery OmniMap anti-mouse-HRP (Roche 760-4310) was used as secondary antibody and the signal acquired using Discovery ChromoMap DAB RUO (Roche 760-2513). Tissues were then stained with Hematoxylin to visualize the nuclei.
Diet chow composition and regimens
Dietary chow
The diets were purchased from Teklad-Envigo Laboratories (Somerset, NJ). Mice received identical diets, except for the amount of magnesium. Specifically, the diets were irradiated and had the following contents (g/kg): protein (17.7), carbohydrates (64.4), fat (6.2), casein (200), DL-methionine (3.0), sucrose (415), corn starch (250), soybean oil (60), cellulose (30), vitamin mix (Teklad 40060), ethoxyquin (antioxidant) (0.01), calcium phosphate, dibasic (13.7), potassium citrate (monohydrate) (7.7), calcium carbonate (4.8), sodium chloride (2.6), potassium sulfate (1.82), ferric citrate (0.25), manganous carbonate (0.12), zinc carbonate (0.056), chromium potassium sulfate (dodecahydrate) (0.02), cupric carbonate (0.012), potassium iodate (0.0004), and sodium selenite (pentahydrate) (0.0004). The regular (normal) mouse Mg diet had Mg oxide 0.822 g/kg of chow (Mg 500 ppm) (as defined by the Subcommittee on Laboratory Animal Nutrition, Committee on Animal Nutrition, Board on Agriculture, National Research Council)33 and the high Mg diet had Mg oxide 2.3 g/kg of chow (Mg 2800 ppm).
Mg dietary regimen in KSIA
Male C57BL/6 mice were randomly assigned to be fed either normal Mg diet Mg500 or a high Mg diet Mg2800 for 14 days before the induction of KSIA, and continued on that diet until days 6–9 for tissue collection (Supplemental Fig. S1A). A longer experiment Mg2800 diet was continued until day 15 after the induction of KSIA (Fig. 1A). Fecal samples were collected at the beginning of the experiments (day-14), before induction of KSIA (day 0) and at the end of the experiments (day 6 or day 15, respectively).
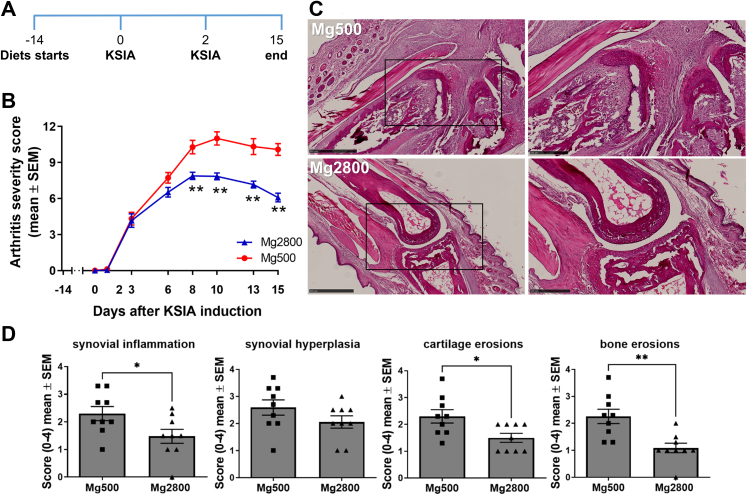
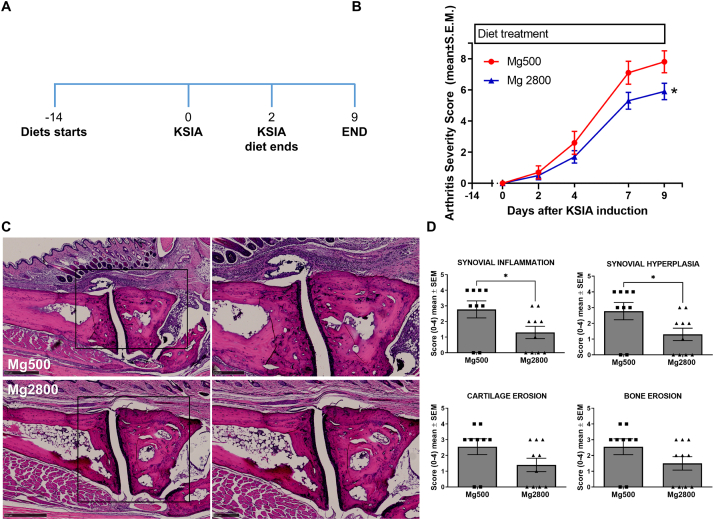
Fig. 1.
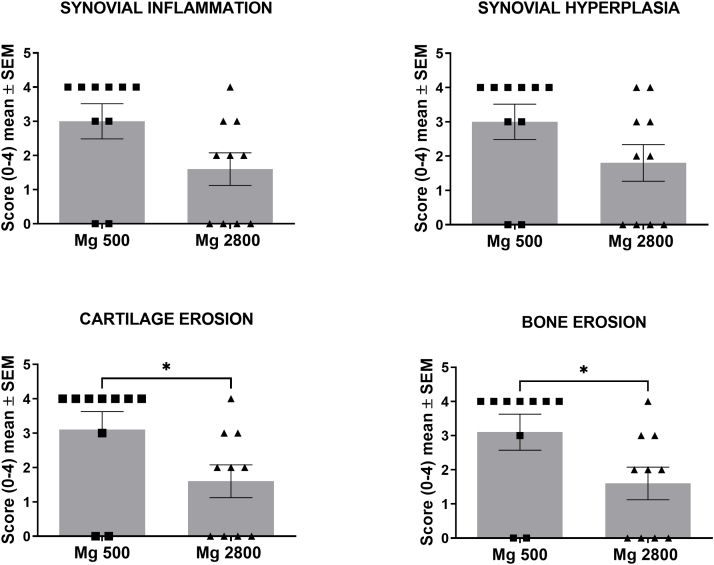
Mice on Mg2800 diet had reduced arthritis severity in KSIA. A) Timeline of the experiment: relative to the induction of KSIA: day-14 diet starts; day 0 and day 2 KRN serum injection, and day 15 end of the diet and KSIA scoring period; B) Arthritis severity scores were significantly decreased in mice receiving the Mg2800 diet compared with Mg500 (n = 25 per treatment group; ∗p = 0.0005 and ∗∗p < 0.0001). C) Representative images of joints from both Mg diet groups at day 15 (hematoxylin/eosin, H&E, staining; left panels are lower magnification, and box area are magnified on the right panels; day 12, H&E, size bars = 500 μM and 250 μM); D) Articular histology scoring for synovial inflammation, hyperplasia, and erosions of cartilage (∗p ≤ 0.03; n = 9–10 per group) and bone (∗∗p < 0.002; n = 9–10 per group).
Mg dietary regimen in CIA
Male DBA/1J mice were randomly assigned to either Mg500 or Mg2800 diets starting 14 days before induction of CIA (day-14). The diets were continued for three additional days after the initial immunization for a total duration of 17 days. On day 3 (three days after the induction of CIA) mice in the Mg2800 groups were switched to the Mg500 diet, and kept on it until the end of the experiment (Day 71). Fecal samples were collected at the beginning of the experiment (day-14), after the 17 days of the different diets (day 3) and at the end of the experiment (day 71) (Fig. 2A).
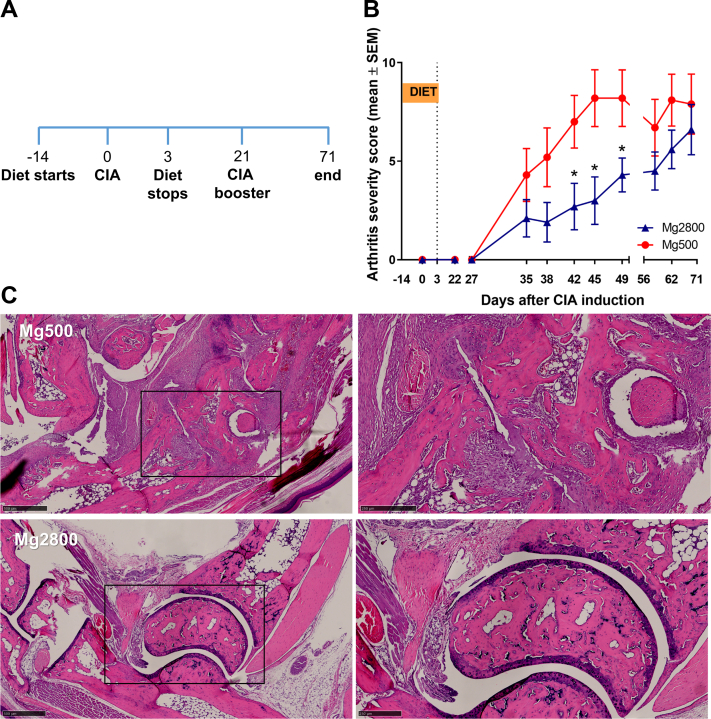
Fig. 2.
Mice on the Mg2800 diet had reduced disease severity in CIA. A) Timeline of the experiment: relative to CIA induction: day-14 diets start; day 0 CIA induction; day 3 Mg2800 diet is stopped and mice switched to regular Mg500 diet; day 21 CIA booster and day 71 end of the experiment; B) Arthritis severity scores were significantly lower in mice treated with the Mg2800 diets compared with Mg500 (n = 10 per group; ∗p ≤ 0.03); C) Representative articular histology of mice treated with Mg500 and Mg2800 diets (day 72, H&E staining; left panels are lower magnification, and box area are magnified on the right panels; day 12, H&E, size bars = 500 μM and 250 μM).
Fecal microbiota collection and transplantation
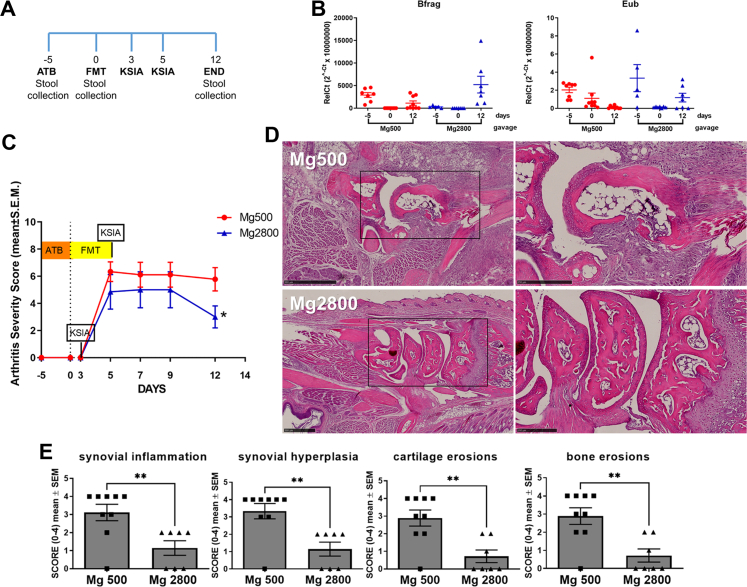
Naïve male C57BL/6 mice (donors) were placed on one of the two diets described above for 16 days. Fecal samples were collected on days 14 and 15, homogenized in water (75 gr/L) and filtered using a 40 μM mesh. Recipient male C57BL/6 received daily antibiotic treatment by oral gavage for five days consisting of a combination of Metronidazole (140 mg/kg), Neomycin (140 mg/kg), and Vancomycin (10 mg/kg). To ensure depletion of the microbiome, qPCR (fecal DNA) was done using a primer for Eubacterium (Eub) and Bacteroides fragilis (Bfrag). Donor fecal material homogenate (200 μL/mouse/day) was transplanted into recipients via daily oral gavage for 5 days. Recipient mice were kept on a Mg500 diet for the duration of the experiment. KRN arthritogenic serum was administered three and five days after stool gavage started and were scored over 12 days. Fecal samples were collected before (day-5) and after antibiotic treatment (day 0) and at end of the arthritis experiment (day 12). Spleen, synovium, and intestines were collected for histology, gene expression and flow cytometry.
Quantitative PCR (qPCR)
Spleen, intestine and ankle synovial tissues were collected after the completion of the arthritis scoring period and immediately frozen in liquid nitrogen. Tissues were homogenized, and total RNA isolated with the RNeasy Kit (Qiagen, Germantown, MD). 200 ng of total RNA from each sample was used for cDNA synthesis (ABI high-capacity reverse transcription kit, Thermo Fisher, Waltham, MA; see primers on Supplemental Table S2). Mouse Actin was used as endogenous control.
Mouse naïve CD4+ T cell isolation and differentiation
Naïve CD4+ T cells were isolated from C57BL/6 spleens by immunomagnetic negative selection using EasyStep Mouse Naïve CD4+ T cell isolation kit (StemCell Technologies, Vancouver, Canada) according to the manufacturer’s instructions. Cells were then re-suspended in RPMI to 1 × 106 cells/ml and plated in 96-well plates pre-coated with 1 μg/ml of anti-CD3 antibody (clone: 145-2C11; Biolegend, San Diego, CA). Culture media supplemented with cytokines to induce differentiation of different T cell subsets: Treg: TGFβ1 5 ng/ml (R&D Systems, Minneapolis, MN) plus mIL-2 100 U/ml (R&D); Th17: mIL-6 20 ng/ml (R&D) plus TGFβ1 5 ng/ml; Tr1: mIL-27 50 ng/ml (Biolegend); Tfh: mIL-6 100 ng/ml plus mIL-21 50 ng/ml (R&D).34 1 μg/ml of soluble anti-CD28 (clone: 37.51, Biolegend) was added to each well. These conditions were used in the in vitro experiments (below) run in triplicate at 37 °C for 5 days prior to flow cytometry analysis.
In vitro treatments
Three different treatments were used to test the ability of the CD4+ naïve cells to differentiate into T cells subsets, and to examine the ability of Tregs to suppress Tconv proliferation: a. Effect of different concentrations of Mg (MgCl2) (ThermoFisher): different concentrations of MgCl2 (low = 0.4 mM or 0.972 mg/dl; normal = 0.8 mM or 1.944 mg/dl; high = 1.6 mM or 3.889 mg/dl) during the experiments (5 days for differentiation and 72 h for Treg suppression); b. Effect of agonists and antagonists of selected Mg channels: Rocaglamide (TRPM6 agonist) 5 μM (Millipore-Sigma, Burlington, MA), Mesendogen (TRMP6 antagonist) 10 μM (Aobious, Gloucester, MA), Naltriben (TRPM7 agonist) 50 μM (Millipore-Sigma) or NS8593 (TRPM7 antagonist) 20 μM (Millipore-Sigma) were added to the wells, in triplicate for the duration indicated above; c. Effect of siRNA knockdown of selected Mg channels: 1 μM siRNA for TRPM6, TRPM7, MAGT1, or siRNA control (Dharmacon Accell, Cambridge, UK) was added to the cells for 72 h prior to starting the differentiation or suppression experiments, and knockdowns confirmed by qPCR.
Flow cytometry analysis
Spleens were harvested and individually analyzed. Single cell suspensions (3 × 106) were stained with fluorescent-labelled monoclonal antibodies for cell surface antigens and incubated for 10 min at 25 °C. For intracellular staining cells were fixed with Cytofix/Cytoperm™ (BD Bioscience, San Jose, CA) for 20 min at 4 °C, followed by permeabilization in 1X Perm/Wash™ solution (BD Bioscience) for 15 min. Fixed cells were stained with fluorochrome-conjugated antibodies for 1 h at 4 °C in the dark. CD4+CD25+Foxp3+ regulatory T cells (Tregs) were stained using anti-CD4-Vioblue (clone: VIT4), anti-CD25-APC (clone: 4E3), and anti-Foxp3-PE (clone: 3G3) (all from Miltenyi, Bergisch Gladbach, Germany). Th17 cells were analyzed using anti-CD4-Pacific blue (clone: RM4-5), and anti-IL-17-PerCP/Cy5.5 (clone: TC11-18H10.1). Tr1 cells were identified by anti-CD4-Pacific blue (clone: RM4-5), anti-CD45RA-PE (clone: 14.8), anti-CD49B-PeCy7 (clone: DX-5), anti-LAG3-APC (clone: C9B7W), and anti-IL-10-PerCP/Cy5.5 (clone: JES5-16E3). Lastly, Tfh cells were stained with anti-CD4-Pacific blue (clone: RM4-5), anti-CXCR5-biotin-APC (clone 2G8), anti-PD1-PeCy7 (clone: RMP1-30), anti-BCL6-PE (Clone: IG191E/A8), and anti-IL-10-PerCP/Cy5.5 (clone: JES5-16E3) (antibodies from Biolegend or BD Bioscience). At least 50,000 cells were acquired per sample. All samples were acquired on a BD LSRII (BD Biosciences) or an Attune (Thermo Fisher) flow cytometer, and analyzed with De Novo FCS Express (Pasadena, CA).
Fecal DNA extraction, 16S rDNA amplification, multiplex sequencing, and metagenomics analyses
DNA extraction buffer (4.3 mM Tris, pH 8; 0.4 mM EDTA, 43 mM NaCl, 3% v/w SDS, 42% v/v phenol/chloroform/IAA and 21% v/v QIAquick PM buffer) containing 400 μL 0.1 mm Zirconia/Silica Beads was added to fecal samples and the samples were bead-beat on a BioSpec Mini-Beadbeater-96 for 5 min as previously described.35 After centrifugation (5 min, 4000 rpm), the ∼400 μL of aqueous layer was mixed with 650 μL QIAquick PM buffer, and DNA was purified using the QIAquick 96 PCR Purification Kit (Qiagen). DNA concentration was determined with the Quant-iT dsDNA Assay Kit, broad range (Life Technologies) and normalized to 2 ng/μL on a Beckman liquid handling robot. Amplicon preparation and sequencing was performed as previously described.36 Briefly, bacterial 16S rDNA PCR including no template controls were setup in a separate PCR workstation using dual-indexed primers. PCR reactions contained 1 μM for each primer, 4 ng DNA, and Phusion Flash High-Fidelity PCR Master Mix (Thermo Fisher Scientific). Reactions were run at 98 °C for 30 s, proceeding to 50 cycles at 98 °C for 10 s, 45 °C for 30 s, and 72 °C for 30 s and a final extension of 2 min at 72 °C. Amplicons were evaluated by gel electrophoresis. The sequencing library was prepared by combining equivalent volume amounts of each amplicon, size-selected and concentrated using AMPure XP beads (0.8X, Beckman). Library concentration was quantified by Qubit and qPCR, mixed with 15% PhiX, diluted to 4 pM and subjected to paired-end sequencing (Reagent Kit V2, 2 × 150bp) on an Illumina MiSeq sequencer.
Statistics
We calculated that 7–15 mice per treatment group would be able to detect a 30% reduction in arthritis scores with an 80%–93% power, and a p ≤ 0.05. Means were compared with the t-test or paired t-test and medians with the rank sum test whenever indicated using GraphPad Prism 6 (San Diego, CA). The arthritis scores of each treatment group at each time-point during the arthritis experiments were compared with the scores of the other group at the same time point in the experiment using the t-test or the rank sum test. QPCR analyses used Ct (threshold cycle) obtained with the StepOne software v2.3 (Thermo Fisher; primers and probes on Supplemental Table S2). Ct was adjusted for Actin in each sample (ΔCt). RelCt (2−ΔCt × 10,000) values were compared by t-test, and fold differences in gene expression determined by the 2−ΔΔCt.37
For the microbiome analyses, sequencing fastq files (average counts for each sample 42,360 ± 10,250) were analyzed using QIIME2 version 2019.1038 and the DADA239 denoised-paired plugin with a truncation length of 150bp and 145bp for the forward and reverse read, respectively. Amplicon Sequence Variants (ASV) were classified using the Scikit-Learn plug-in40 using the Naive Bayes classifiers trained on the silva-132-99-515-806-nb-classifier. Resulting ASV tables were filtered using a minimum depth of 5000, a minimal ASV frequency of 10 and minimal sample frequency of 2 and core metrics were calculated using the Qiime2 core metrics plugin. Statistical analysis was performed using LEfSe41 v1.0.8 post1 or Kruskal–Wallis and Wilcoxon as implemented in R v3.6.1. All plots were generated using R/ggplot242 or R/pheatmap. Metagenomics functions were predicted using PICRUSt2.43 We considered statistically significant the comparisons that had a false discovery rate (FDR)-corrected p value < 0.05 for on ANOVA and a Tukey’s post-hoc test p < 0.05.
Results
High Mg diet (Mg2800) significantly reduces joint inflammation and arthritis severity scores in the KSIA and CIA models
High Mg diet is protective in KSIA
As early as eight and nine days after the induction of KSIA, mice treated with Mg2800 (high Mg) diet had significantly lower arthritis severity scores compared with Mg500 (normal) diet33,44 (Fig. 1A and B, and Supplemental Fig. S1A–D). The protective effect persisted through the end of the experiment at day 15 with a significant reduction in arthritis severity scores ranging from 20% to 40% (Fig. 1A and B). On histology the Mg2800 group had a 54% lower synovial inflammation score, and 53% lower synovial hyperplasia on day nine (Supplemental Fig. S1D), and reduced inflammation and erosive changes by day 15 (Fig. 1C and D). Oral Mg is absorbed in the intestines, and non-absorbed Mg is eliminated with the feces. Absorbed and excessive Mg is excreted by the kidneys. Mice on the Mg500 and Mg2800 groups had similar feeding patterns and had no significant changes in weight (Supplemental Fig. S2A), and no difference in serum levels of Mg (Supplemental Fig. S2B), as expected for mice with normal renal function.
High Mg diet is protective in CIA and reduces erosive joint damage
To examine the duration of the protective effect of the high Mg diet (Mg2800), and its effect in erosive joint changes, we used CIA, which is a chronic and more erosive model of RA. Mice were placed on a 17-day Mg2800 diet starting 14 days before the induction of CIA, and then switched to Mg500 (same control diet group) and kept on it until the end of the experiment on day 71 (Fig. 2A). The short duration Mg2800 group was protected and reached a 61% reduction in arthritis severity scores compared with control (p = 0.03 at day 42; Fig. 2B). The Mg2800 protective effect persisted until day 49 (46 days after the discontinuation of the Mg2800 diet) reaching statistical significance from day 42 through day 49 (Fig. 2B). The joint protective effects persisted and included preservation of the normal joint architecture in the Mg2800 group with reduced cartilage erosions (48%, p = 0.05) and bone erosions (48%, p = 0.05) compared with controls (Fig. 2C and Supplemental Fig. S3).
High Mg diet reduced the expression of pro-inflammatory and chemotaxis genes, while increasing levels of IL-10
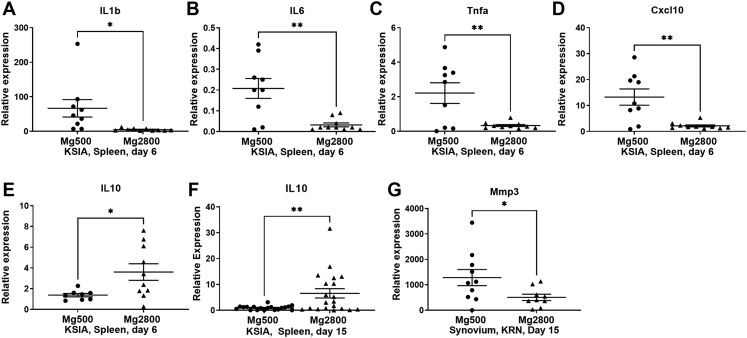
Splenocytes obtained before disease peak (day 6) of KSIA mice and analyzed by qPCR revealed a significant reduction in levels of pro-inflammatory cytokines IL-1β, IL-6, TNFα, and CXCL10 (Fig. 3A–D). Levels of IL-10 were increased in the Mg2800 diet group compared with controls, and were even higher at later stages of disease (by day 15) (Fig. 3E and F).
Fig. 3.
Mg2800 diet significantly affects levels of cytokines and other inflammatory mediators (qPCR). Levels of A) IL1β, B) IL6, C) Tnfα, and D) Cxcl10 were significantly decreased in the Mg2800 group. E) IL10 was increased in the Mg2800 diet at an early time point (day 6) and F) at a later stage (day 15) of KSIA. G) MMP3 was decreased in the synovium of Mg2800 group (data shown as mean ± SEM; (∗p ≤ 0.04; ∗∗p ≤ 0.005)).
MMP3 expression was decreased in the synovial tissues of Mg2800 mice in the KSIA model (Fig. 3G).
Dietary Mg content affect the numbers of T cell subsets
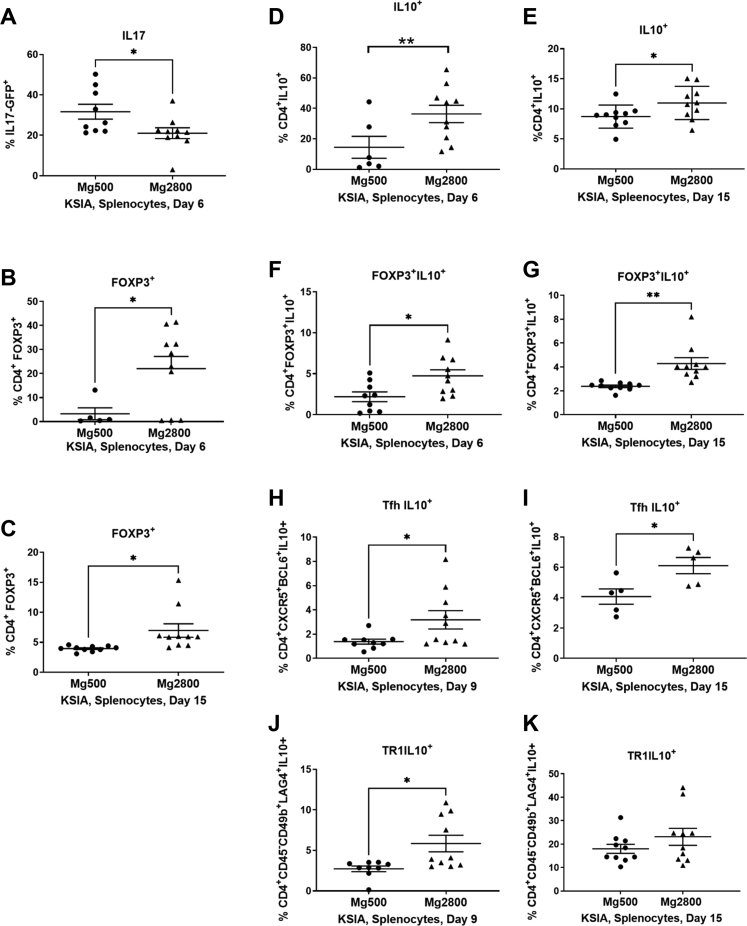
KSIA mice on the Mg2800 diet had a lower number of IL17GFP+ cells compared with control Mg500 at on day 6 (33.49%; p = 0.03; Fig. 4A). At day 6 there was also a nearly six-fold increase in the numbers of CD4+Foxp3+ Treg cells compared with the Mg500 diet (average 22.05 versus 3.29; p = 0.03; Fig. 4B). At day 15 numbers of Foxp3+ Treg cells remained two-fold higher than in the Mg500 group (Fig. 4C; see Supplemental Fig. S4 for gating strategies).
Fig. 4.
Early and sustained effect of the Mg2800 diet on T cell subsets. The Mg2800 diet group with KSIA had the following changes in splenocyte T cell subpopulations: A) reduced percentage of GFP+IL17+ cells; B) increased percentage of CD4+Foxp3+ T cells on day 6; C) CD4+Foxp3+ cells remained increased at day 15; D) Increased percentage of CD4+IL10+ at day 6 and E) at day 15; F) CD4+Foxp3+IL10+ cells were increased at day 6; G) and remained increased at day 15; H) Tfh IL10+ cells were increased at day 9 and I) day 15; J) Increase in Tr1 IL10+ cells on day 9 (results shown as mean ± SEM; ∗p ≤ 0.03; ∗∗p = 0.001); K) Numbers of Tr1 IL10+ remained increased at day 15, but that difference did not reach statistical significance.
The Mg2800 group also had a nearly two-fold increase in numbers of CD4+IL10+ cells at day 6 (p = 0.03; Fig. 4D), with numbers persisting elevated by day 15 (p < 0.05; Fig. 4E). A subset of the CD4+IL10+ expressing Foxp3 was also expanded in the Mg2800 diet group at both days 6 and 15 (p = 0.02 and p = 0.001, respectively; Fig. 4F and G).
We identified a subset of the Tfh cells expressing IL10 that was expanded in the Mg2800 diet at both the early stage day 9 and later stage day 15 in KSIA (Fig. 4H and I). These IL10+ cells have been recently described as Tfh with regulatory properties (Tfr).45,46 Numbers of Tfh (CD4+CXCR5+PD1+BCL6+) cells were not affected by the Mg2800 diet (data not shown).
IL10+Tr1 cells were also increased in splenocytes from the Mg2800 group at day 9 (Fig. 4J). By day 15, when KSIA is starting its resolution phase, numbers of IL10+Tr1 were increased in both diet groups. While mean percentage of IL10+Tr1 cells was still higher in the Mg2800 group, the difference was no longer statistically significant (Fig. 4K). This observation raised the possibility that IL10+Tr1 cell may have an important role in the resolution of KSIA and that the Mg2800 diet induced an earlier increase in their numbers. CD4-IL10+ cells were not significantly different between the groups (data not shown).
We also examined the effect of the Mg diets on percentages of different splenocyte cell populations in naïve C57BL/6 mice. Percentages of T cells sub-populations (Supplemental Fig. S5A–F), CD19+ B cells (Supplemental Fig. S5G and H), and IL10-producing cells (Supplemental Fig. S5I) were not significantly different between the two diet groups, suggesting a more pronounced effect of the Mg2800 diet in the context of inflammation. However, there was a trend towards increasing numbers of IL10+Tr1 cells in the Mg2800 group (Supplemental Fig. S5F; not significant).
In vitro changes in the Mg concentration, or interference with selected Mg channels did not affect CD4+ naïve T cell differentiation into Foxp3+ Treg or other T cell subsets
We next examined the role of changes on extracellular Mg concentrations in vitro differentiation of CD4+ naïve T cells into different cell subsets. Increased concentrations of Mg had no significant effect on the in vitro differentiation of naïve T cells into Treg, Th17, Tr1, or Tfh cells (data not shown). We also examined the role of specific Mg channels on the differentiation of naïve T cells into T cell subsets using siRNA gene silencing (TRPM6, TRPM7, or MAGT1), or treatment with agonists or antagonists of TRPM6 and TRPM7. None of these treatments induced significant changes in naïve T cell differentiation into T cells subset (data not shown).
Fecal microbiota transplant (FMT) from donor mice receiving Mg2800 diet transfers arthritis protection and increases numbers of Foxp3+ Treg cells in recipient mice
To determine whether the high Mg diet-induced changes in arthritis severity and on T cell subsets was dependent on the intestinal microbiota we conducted a FMT experiment (Fig. 5A). Fecal material was collected from healthy mice that had received either Mg500 or Mg2800 diets for 14 days. Recipient mice were treated with antibiotics given by daily oral gavage for five days. Antibiotic treatment significantly depleted levels of bacteria, followed by bacteria repopulation after FMT (Fig. 5B).
Fig. 5.
Recipients of fecal material transplantation (FMT) from the Mg2800 diet group were protected in KSIA. A) Timeline, with day 0 being the first day of the FMT (ATB = antibiotic); B) Fecal DNA qPCR for Bacteroidis fragilis (Bfrag) and Eubacteria (Eub) confirming bacteria depletion following antibiotics (D0); C) Arthritis severity scores are significantly decreased in recipients of FMT from Mg2800 diet group (n = 7), compared with the Mg500 group (n = 9); D) Representative histology images showing that recipients of FMT from Mg2800-treated mice preserved near normal structures while recipients from Mg500 transplantation had erosive changes with pronounced synovial hyperplasia, cellular infiltrate and angiogenesis (left panels are lower magnification, and box area are magnified on the right panels; day 12, H&E, size bars = 500 μM and 250 μM); E) Histology scoring revealed significantly reduced synovial inflammation, reduced synovial hyperplasia, and reduced cartilage and bone erosions in recipients (Mg2800: n = 7, Mg500: n = 9; ∗∗p < 0.007).
Recipients of FMT from mice treated with Mg2800 diet had a nearly 50% reduction in arthritis severity by day 12 of KSIA (p = 0.04; Fig. 5C), which was similar to the effect observed in the diet experiment (Fig. 1B). The Mg2800 FMT recipients also had reduced synovial inflammation, reduced synovial hyperplasia, and reduced cartilage and bone erosions (53% or greater reduction; p < 0.007) on histology (Fig. 5D and E).
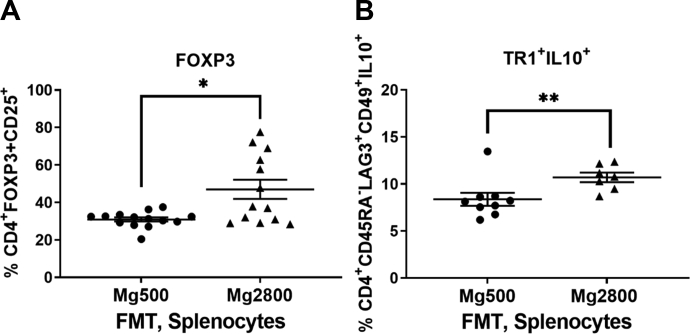
Recipients of FMT from Mg2800 had increased percentage of splenic Foxp3+ Treg cells (Fig. 6A, p = 0.002; for gating strategies see Supplemental Fig. S4), and increased percentage of Tr1 cells (p = 0.02; Fig. 6B), matching the changes seen in KSIA mice that had received the Mg2800 diet (Fig. 4B and J, respectively).
Fig. 6.
Recipients of FMT from the Mg2800 diet group have increased numbers of Foxp3+ Treg and IL10+ Tr1 cells. A) Increased numbers of Foxp3+ Treg cells in recipients of FMT from Mg2800 groups compared with Mg500 (∗p = 0.002); B) Increased numbers of IL10+ Tr1 cells in the FMT recipients of the Mg2800 group (∗∗p = 0.02 (results shown as mean ± SEM)).
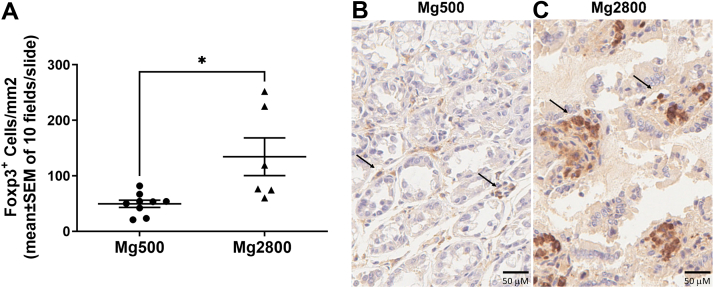
We next examined the intestines and detected increased numbers of Foxp3+ cells in the colonic villi of recipients from Mg2800 FMT (p = 0.01, Mg2800 versus Mg500; Fig. 7), but not in the small intestine (data not shown).
Fig. 7.
Increased numbers of Foxp3+ cells in the colon of recipients of FMT from Mg2800 group in KSIA. A) Histologic analyses and counting of numbers of Foxp3+ cells per mm2 on H&E-stained slides counter stained with Foxp3 immunoperoxidase (scored 10 fields per slide, shown as Mean ± SEM; ∗p = 0.01); Representative slides of the colon of recipients of FMT from the B) Mg500 and C) Mg2800 groups; arrows indicate Foxp3+ cells (size bars = 50 μM).
High Mg diet alters intestinal microbiota associated with Th17 and Treg cell differentiation
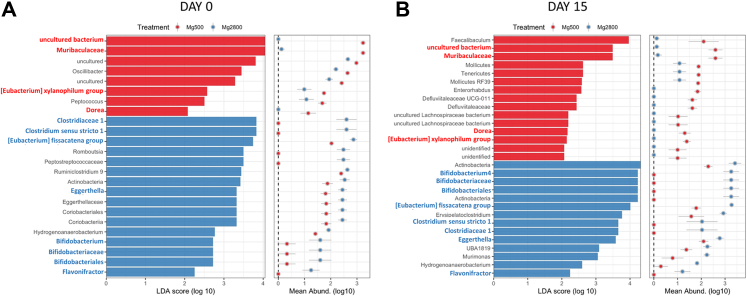
Based on the similarities of changes observed in the Mg diet and the respective FMT recipient in arthritis and T cell subsets we hypothesized that the high Mg diet was affecting the intestinal microbiota. Fecal bacterial DNA was examined with 16S rDNA amplification and sequencing. Alpha diversity profiles were similar in both groups before the initiation of the Mg diets (day 14), while on the diet (day zero) and at the end of the KSIA arthritis experiment (day 15) (Supplemental Fig. S6A).
The LDA (Linear Discriminant Analysis) effect size (LEfSe) analysis was used to identify differences in bacteria representation between the two diet groups using relative abundances. On the day of arthritis induction (day 0), 24 clades were significantly different between the two diets, where 8 clades were higher in the Mg500 group and 16 clades were more abundant in the Mg2800 group (Fig. 8A). On day 15, twenty-nine clades were significantly different where 15 clades had increased representation in the Mg500 group and 14 were more abundant in the Mg2800 group (Fig. 8B). There were specific groups of bacteria that remained differentially represented both at day 0 and day 15 in each diet group, with four in the Mg500, and eight in the Mg2800 group (Fig. 8A and B, marked in bold red and blue, respectively).
Fig. 8.
Linear discriminant analysis (LDA) effect size (LEfSe) analysis of gut microbiota changes following the Mg500 or Mg2800 diets. Specific phylotypes of gut bacteria in response to Mg diet using LEfSe. The histogram shows the significant LDA scores computed for features at the clades levels. The colors represent the group where the taxa was more abundant compared to the other group (red = Mg500, blue = Mg2800). A) day 0 and B) day 15. Bold font marks bacteria persistently more abundant in the Mg500 group (red), and in the Mg2800 group (blue).
We analyzed the bacterial composition and the individual bacterial counts/levels according to diet. At day 0, after the mice had received the diets for 14 days, but before the induction of KSIA, twenty six bacteria counts were different between the two diets. Eight were increased in Mg2800 (Supplemental Fig. S7A–H) and eighteen were increased in Mg500 (Supplemental Fig. S7I–Z), all belonging to the Firmicutes-Clostridia.
At day 15, after the mice had established arthritis, fourteen bacteria families were significantly different, six increased in Mg2800 (Supplemental Fig. S8A–F) all belonging to the Firmicutes-Clostridia, and eight increased in Mg500 (Supplemental Fig. S8G–O), and most from Firmicutes-Clostridia (Supplemental Fig. S8G–L), in addition to one Bacteriodetes-Bacteroidia (Supplemental Fig. S8M), and one Actinobacteria-Coriobacterilia (Supplemental Fig. S8N).
Eleven bacteria families remained significantly differentially represented at both days 0 and 15, and therefore were considered to be of potential greater relevance to the long-term effect of the diets in arthritis severity. These bacteria included different Clostridia (Supplemental Fig. S9A–D for Mg2800, and Supplemental Fig. S9F–J for Mg500), Actinobacteria (Supplemental Fig. S9E), and Bacteroidia (Supplemental Fig. S9K).
FMT modulate gut microbiota composition in recipient mice
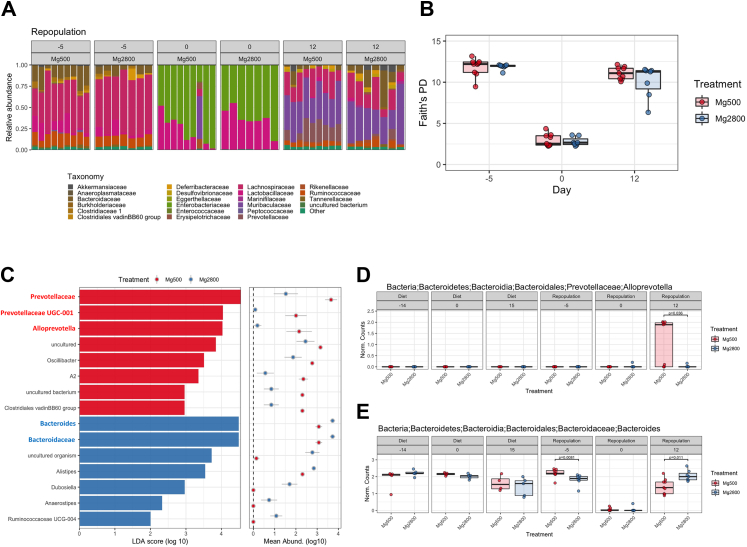
We next examined the bacterial composition of the recipient mice following FMT. Relative abundance analysis showed that before antibiotics or diets the microbiota composition (Fig. 9A) and alpha diversity (Fig. 9B; day 5) were similar and that antibiotic treatment depleted most of the fecal microbiota in both groups (Fig. 9A and B; day 0). FMT from the two Mg diet groups reconstituted the intestinal microbiome diversity (Fig. 9A and B; day 12). 15 clades were significantly altered in the two groups at day 12 (after FMT), with 8 clades with increased abundance in the Mg500 and 7 in the Mg2800 (Fig. 9C).
Fig. 9.
Variations in Relative abundance and alpha diversity of gut microbiome in recipients of the FMT from animals fed Mg500 or Mg2800 diets. A) Mice originally displaying similar microbiome relative abundance (day 5). The relative abundance was significantly diminished after antibiotic treatment (day 0). B) Alpha diversity (Faith’s PD) is reduced after antibiotics (day 0) but recovers following FMT repopulation (day 12). C) Linear discriminant analysis (LDA) effect size (LEfSe) analysis of fecal microbiota changes following FMT of Mg500 (red bars) or Mg2800 (blue bars) derived stool revealed increased representation of specific phylotypes of gut bacteria for each group. The histogram shows the LDA scores computed for features at the clade level. The colors represent which group the taxa was more abundant compared to the other group (p values range from 0.0000832 to 0.0492). Shown in bold red are Prevotellaceae, which were increased in the recipients of FMT from the Mg500 group (C and D) and Bacterioides, which were increased in recipients of FMT from the Mg2800 group (C and E).
The Prevotellaceae family (Alloprevotella genera) was the most significantly overrepresented family in the recipients of FMT from regular Mg500, while their levels were reduced in recipients of FMT from the Mg2800 group (Fig. 9A, day 12; Fig. 9C and D). The recipients of Mg2800 FMT had increased levels of Bacterioides, Bacteroidaceae (Fig. 9C and E) among others (Fig. 9A and C). No significant difference was detected in levels of SFB (Candidatus Arthromitus) following FMT from Mg2800 (Supplemental Fig. S6B).
Metagenomic function analyses reveals increased L-tryptophan biosynthesis and others processes in the high Mg group
Metagenomic analyses identified 313 pathways, enzymes or processes differentially represented or regulated between the microbiome of Mg2800 and Mg500 diets (Supplemental Table S3). Of these, 271 had significantly increased representation in the Mg2800 FMT recipients, including a) superpathway of L-tryptophan biosynthesis, b) tryptophanase, c) L-methionine biosynthesis, d) transcriptional regulator, propionate catabolism operon regulatory protein, e) mannan degradation, f) mannuronate biosynthesis, g) propionate catabolism, h) arginine deiminase, and others (Table 1; Supplemental Table S4). 42 had decreased abundance in recipients of Mg2800 FMT and included a) super pathway of arginine and polyamine biosynthesis, b) superpathway of heme biosynthesis from glycine, and c) superpathway of L-lysine, L-threonine, and L-methionine biosynthesis I (Table 1; Supplemental Table S3).
Table 1.
Selected list of abundant metagenomic pathways identified in the FMT recipients (from a list of 313).a
| Pathway description | ID number | p-value | Mg2800 abundance | Mg500 abundance | fold difference |
|---|---|---|---|---|---|
| Increased abundance in recipients from Mg2800 FMT | |||||
| coxM, cutM; aerobic carbon-monoxide dehydrogenase medium subunit [EC:1.2.5.3] | K03519 | 0.0021 | 1140.86 | 16.89 | 67.6 |
| rspA, manD; mannonate dehydratase [EC:4.2.1.8] | K08323 | 0.0022 | 2196.71 | 43.33 | 50.7 |
| rspB; L-gulonate 5-dehydrogenase [EC:1.1.1.380] | K08322 | 0.0010 | 1663.00 | 42.33 | 39.3 |
| phnK; putative phosphonate transport system ATP-binding protein | K05781 | 0.0010 | 21.50 | 0.61 | 35.2 |
| phnM; alpha-D-ribose 1-methylphosphonate 5-triphosphate diphosphatase [EC:3.6.1.63] | K06162 | 0.0010 | 21.50 | 0.61 | 35.2 |
| D-arabinitol dehydrogenase (NADP (+)) | EC:1.1.1.287 | 0.0010 | 41.86 | 1.22 | 34.2 |
| 4-hydroxyphenylacetate decarboxylase | EC:4.1.1.83 | 0.0010 | 125.57 | 3.67 | 34.2 |
| asrB; anaerobic sulfite reductase subunit B | K16951 | 0.0013 | 1273.07 | 39.03 | 32.6 |
| Methylmalonate-semialdehyde dehydrogenase (CoA acylating) | EC:1.2.1.27 | 0.0008 | 588.57 | 24.33 | 24.2 |
| mmsA, iolA, ALDH6A1; malonate-semialdehyde dehydrogenase (acetylating)/methylmalonate-semialdehyde dehydrogenase [EC:1.2.1.18 1.2.1.27] | K00140 | 0.0008 | 588.57 | 24.33 | 24.2 |
| asrC; anaerobic sulfite reductase subunit C | K00385 | 0.0011 | 744.50 | 32.69 | 22.8 |
| ulaG; L-ascorbate 6-phosphate lactonase [EC:3.1.1.-] | K03476 | 0.0008 | 1771.64 | 81.10 | 21.8 |
| dcyD; D-cysteine desulfhydrase [EC:4.4.1.15] | K05396 | 0.0007 | 710.57 | 38.11 | 18.6 |
| 2-deoxy-D-gluconate 3-dehydrogenase | EC:1.1.1.125 | 0.0008 | 1464.79 | 133.94 | 10.9 |
| kduD; 2-dehydro-3-deoxy-D-gluconate 5-dehydrogenase [EC:1.1.1.127] | K00065 | 0.0008 | 1464.79 | 133.94 | 10.9 |
| TC.CITMHS; citrate-Mg2+:H+ or citrate-Ca2+:H+ symporter, CitMHS family | K03300 | 0.0007 | 143.00 | 14.22 | 10.1 |
| prpR; transcriptional regulator, propionate catabolism operon regulatory protein | K02688 | 0.0010 | 85.14 | 23.33 | 3.6 |
| Superpathway of L-tryptophan biosynthesis | PWY-6629 | 0.0023 | 588.03 | 181.67 | 3.2 |
| UDP-2,3-diacetamido-2,3-dideoxy-α-D-mannuronate biosynthesis | PWY-7090 | 0.0004 | 184.03 | 67.61 | 2.7 |
| mannan degradation | PWY-7456 | 0.0001 | 13612.67 | 5866.52 | 2.3 |
| tnaA; tryptophanase [EC:4.1.99.1] | K01667 | 0.0004 | 2503.02 | 1191.11 | 2.1 |
| Acetaldehyde dehydrogenase (acetylating) | EC:1.2.1.10 | 0.0013 | 7466.29 | 7069.63 | 1.1 |
| L-methionine biosynthesis I | HOMOSER-METSYN-PWY | 0.0018 | 1377.44 | 1304.59 | 1.1 |
| superpathway of S-adenosyl-L-methionine biosynthesis | MET-SAM-PWY | 0.0016 | 2072.66 | 1985.72 | 1.0 |
| arcA; arginine deiminase [EC:3.5.3.6] | K01478 | 0.0025 | 1221.59 | 1182.67 | 1.0 |
| superpathway of L-methionine biosynthesis (transsulfuration) | PWY-5347 | 0.0015 | 2973.71 | 2886.48 | 1.0 |
| Decreased abundance in recipients from Mg2800 FMT | |||||
| SCD, desC; stearoyl-CoA desaturase (Delta-9 desaturase) [EC:1.14.19.1] | K00507 | 0.0011 | 13.71 | 81.67 | −6.0 |
| FAH, fahA; fumarylacetoacetase [EC:3.7.1.2] | K01555 | 0.0010 | 13.71 | 81.11 | −5.9 |
| FAB2, SSI2, desA1; acyl-[acyl-carrier-protein] desaturase [EC:1.14.19.2 1.14.19.11 1.14.19.26] | K03921 | 0.0010 | 13.71 | 81.11 | −5.9 |
| K09992; uncharacterized protein | K09992 | 0.0010 | 13.71 | 81.11 | −5.9 |
| lcyB, crtL1, crtY; lycopene beta-cyclase [EC:5.5.1.19] | K06443 | 0.0010 | 13.71 | 81.11 | −5.9 |
| Aldehyde dehydrogenase (NADP (+)) | EC:1.2.1.4 | 0.0011 | 14.29 | 81.11 | −5.7 |
| chlI, bchI; magnesium chelatase subunit I [EC:6.6.1.1] | K03405 | 0.0011 | 14.29 | 81.11 | −5.7 |
| aldH; NADP-dependent aldehyde dehydrogenase [EC:1.2.1.4] | K14519 | 0.0011 | 14.29 | 81.11 | −5.7 |
| Magnesium chelatase | EC:6.6.1.1 | 0.0011 | 14.29 | 81.11 | −5.7 |
| L-tyrosine degradation I | TYRFUMCAT-PWY | 0.0008 | 205.67 | 334.25 | −1.6 |
| coaW; type II pantothenate kinase [EC:2.7.1.33] | K09680 | 0.0009 | 7363.63 | 9041.14 | −1.2 |
| 4-hydroxy-2-oxoheptanedioate aldolase | EC:4.1.2.52 | 0.0021 | 841.86 | 1023.44 | −1.2 |
| Superpathway of heme biosynthesis from glycine | PWY-5920 | 0.0002 | 2303.94 | 2707.64 | −1.2 |
| coxC, ctaE; cytochrome c oxidase subunit III [EC:1.9.3.1] | K02276 | 0.0002 | 1290.71 | 1508.11 | −1.2 |
| Superpathway of L-lysine, L-threonine and L-methionine biosynthesis I | P4-PWY | 0.0018 | 4131.51 | 4676.90 | −1.1 |
| Superpathway of arginine and polyamine biosynthesis | ARG + POLYAMINE-SYN | 0.0021 | 2165.21 | 2257.81 | −1.0 |
Day 12 post FMT; ordered according to fold-difference; p-values based on ANOVA; Bold entries represent pathways and processes mentioned in the manuscript.
Mg2800 diet protective effect disappears in the IL-10-knockout mice
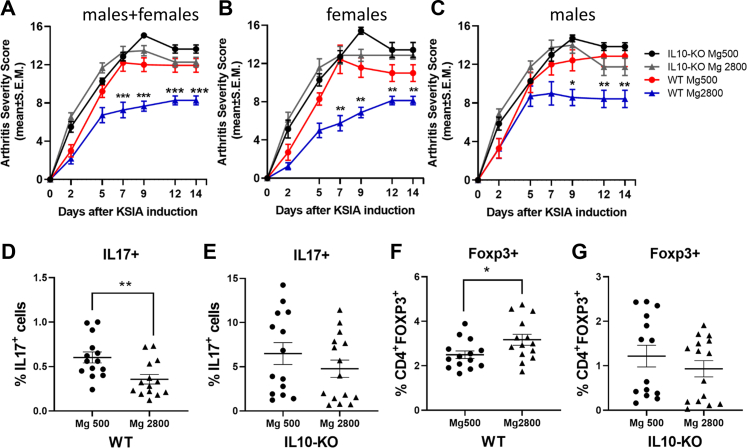
Given the increased frequency of different types of IL-10-producing T cells, including CD4+Foxp3+ both in mice receiving the Mg2800 diet as well as in the recipients of FMT from these mice, we considered that the Mg2800 diet might mediate its arthritis suppressive activity via the induction of this cytokine. IL-10 knockout mice were placed on the Mg diets for 14 days, followed by the induction of KSIA. IL-10-expressing male and female wild-type mice receiving the Mg 2800 diet were consistently protected in the KSIA model. However, the IL-10 KO mice were not protected by the Mg2800 diet, and these mice developed arthritis as severe as the mice receiving the Mg500 diet (Fig. 10A–C). While the wild-type IL10+ mice receiving the Mg2800 diet had decreased numbers of IL17+ cells and increased numbers of CD4+Foxp3+ cells, that was not observed in the IL-10 KO mice (Fig. 10D–G).
Fig. 10.
Male and female IL-10 KO mice are not protected by the Mg2800 diet. A–C) The Mg2800 diet was protective in wild-type (WT) C57BL/6 mice, but was not protective in the IL10 KO mice. The IL10 KO mice on the Mg2800 diet developed arthritis as severe as those of the WT and IL10 KO mice receiving the Mg500 diet (7 females and 8 males per group); D) The Mg2800 diet group was associated with reduced numbers of IL17+ cells both in WT and E) but not in the IL10 KO mice; F) CD4+Foxp3+ cells were increased in the Mg2800 diet in WT mice; G) but not in the IL10 KO (∗p ≤ 0.015, ∗∗p ≤ 0.009; ∗∗∗p ≤ 0.0001 shown as mean ± SEM).
Discussion
In the present study, we describe for the first time that increased dietary intake of Mg (Mg2800), even in the absence of Mg deficiency, reduces disease severity, joint inflammation, and joint damage in two different mouse models of RA. Our studies show that both a continuous and an intermittent high Mg diet reduce disease severity. The high Mg diet-induced reduction in arthritis severity was associated with reduced levels of key inflammatory mediators in RA pathogenesis and joint damage, such as IL-1β, IL-6, TNFα, CXCL10, and MMP3, and with increased levels of the anti-inflammatory cytokine IL-10.
The high Mg diet was also associated with increased numbers of CD4+IL10+ cells, CD4+Foxp3+ Tregs, CD4+Foxp3+IL10+ Tregs, and IL10+Tfh (Tfr), and CD4+Foxp3+ Treg cells have been implicated in the regulation of disease severity in CIA,47 in antibody-induced arthritis,48 and other autoimmune diseases.49, 50, 51, 52 Additionally, numbers of CD4+Foxp3+ Treg cells strongly correlate with disease remission in RA.53 The high Mg diet also reduced numbers of pathogenic Th17 cells implicated in CIA,54,55 and in the regulation of disease severity in KSIA.56
Here we also discovered a more specific mechanistic explanation and show that the high Mg diet effect on Foxp3+ Treg cell numbers and in arthritis severity is mediated by and dependent on IL-10. IL-10 inhibits several pro-inflammatory cytokines and has been previously shown to suppress CIA57,58 and other autoimmune diseases.49,50 While the KSIA model is predominantly mediated by the innate immunity, IL-10 is known to also regulate complement59 and neutrophil activation,60 two central components in the pathogenesis of KSIA.61,62
The lack of a direct effect of different Mg concentrations on naïve T cell in vitro differentiation, combined with similar serum levels of Mg in both treatment groups, and with the oral administration of Mg led to the consideration that the diet effect on arthritis might be mediated by changes in the intestinal microbiome. And indeed, FMT from the high Mg diet mice into antibiotic-treated recipients induced similar effects as the diet itself in arthritis protection, expansion of Foxp3+ and Foxp3+IL10+ Tregs and CD4+IL10+ (Tr1) cells. A comprehensive microbiome analyses revealed significant changes associated with the diets and with the FMT, demonstrating that the high Mg groups had an increased levels of bacteria known to produce elevated levels of intestinal short-chain fatty acid (SCFA), tryptophan metabolites, and other bacterial products implicated in the regulation of immune cells.
Prevotellaceae, particularly Prevotella copri, have been consistently associated with RA in different populations.14,20,63 Similar to RA, the non-protected recipients of FMT from the normal Mg500 diet had increased levels of Prevotellaceae, while the levels in the protected recipients of FMT from the high Mg2800 diet were significantly decreased. While the precise pathways regulated by Prevotella in RA and its role in susceptibility and/or severity remain incompletely understood, Prevotellaceae administered to mice can increase the number of colonic Th17 cells and serum levels of IL-6,64 and also increase arthritis severity in rodent models,65 similarly to our observations. Prevotella has also been associated with reduced levels of SCFA.66
SCFA are known to induce the differentiation and expansion of Foxp3+ Tregs,67, 68, 69 and recipients of FMT from high Mg2800 had increased levels of bacteria associated with increased levels of SCFA such as Alistipes,70 Anaerostipes,71 Bacterioides,71 and Ruminococcaceae,71 and correlated with disease protection and increased number of Foxp3+ Tregs. Bacteroides species are reduced in RA14,20,63 and were also reduced in recipients of the Mg500 FMT with severe disease, while numbers were increased in the high Mg2800 diet FMT recipients. In addition to SCFA, Bacteroides (particularly B. fragilis) produces polysaccharide A (PSA) which also promotes Treg cells expansion72 and enhances Treg secretion of IL-10,73 which were two phenomena observed in this study.
Additional intestinal bacteria can produce increased levels of SCFA and regulate IL-10-producing T cells74, 75, 76 and several of them were detected in increased levels in the Mg2800 diet group, including Bifidobacterium,74 Clostidieaceae, Eubacterium, and Actinobacteria.66
The differences in fecal bacteria composition between mice receiving diets and the recipients of FMT have been reported by other groups and are considered to be related to the duration of the dietary treatments, and with the time for microbiome reconstitution.77 FMT was preceded by antibiotic treatment to significantly deplete all intestinal bacteria followed by a short five-day period for the reconstitution of the intestinal flora, perhaps reducing the diversity and selecting for those bacteria more likely to thrive and repopulate in a short period of time. Other studies have also found that using or not antibiotic treatment prior to FMT did not increase the overall similarity of the recipient’s microbiota and the donor’s.77 The same concept applies to our studies. Yet, the differences in bacterial engraftment following different dietary Mg content is of great relevance and of potential applicability for therapeutics.
Nevertheless, despite the differences in microbiome representation we still observed similar effects in clinical and immunologic phenotypes in the diet groups and in the respective recipients of FMT. Both the Mg2800 diet and the FMT recipients shared expansion of bacteria known to produce increased levels of SCFA, providing a unifying explanation for the increased Tregs and reduced arthritis severity.
The metagenomics analyses identified several pathways differentially represented and associated with the protective effects of the high Mg diet and respective FMT. “Superpathway of L-tryptophan biosynthesis” is a pathway of interest as tryptophan and its metabolites can activate the aryl hydrocarbon receptor to suppress immune responses,78 and are capable or increasing the differentiation of Treg cells.79,80 The tryptophan metabolite 3-hydroxyanthranillic acid (3-HAA) was reported to enhance the percentage of Tregs, inhibit Th17 cells, and ameliorate experimental autoimmune encephalomyelitis (EAE).81 A tryptophan deficient diet was protective in EAE, and was associated with reduced fecal levels of Prevotellaceae and increased levels of Clostridium,82 similar to changes detected in the present study. “Superpathway of arginine and polyamine biosynthesis”, present in increased abundance in the Mg2800 FMT recipients, is also of interest as increased levels of arginine can regulate the balance between T effector cells and Tregs activity.83,84 “Mannan degradation” involves mannan oligosaccharides, which broadly activate the immune system including complement and neutrophils, two central components in arthritis pathogenesis.85 Bacterial-derived “Arginine deiminase” has anti-inflammatory properties, reducing LPS-induced responses86 and colitis in mice.87 “Propionate catabolism” is another important metagenome pathway that was abundant in the Mg2800 group, and implicated in SCFA.71,88 Other pathways were differentially represented in the Mg diets and may lead to new discoveries and links between the microbiome, immune responses, and arthritis.
We acknowledge that direct measurement of metabolites in the fecal materials was not done, and this is something that will be done in future studies in rodents or in a future trial of Mg in RA patients to confirm and validate the metagenomic analyses. Additionally, while we hypothesize that changes in intra-luminal concentration of Mg in the intestines may cause the changes observed in the microbiome, future studies will be required to confirm.
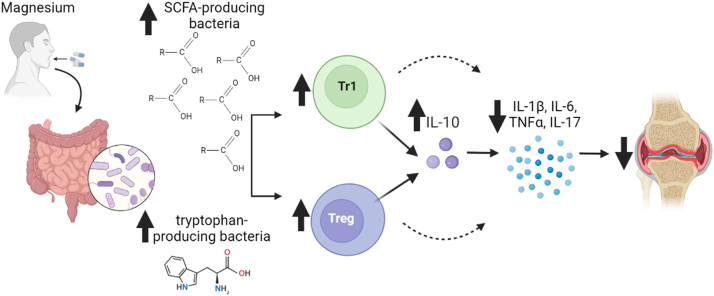
In conclusion, we describe for the first time that increasing Mg intake can have significant effects in increasing the numbers of Foxp3 Tregs and increasing levels of IL-10, reducing the expression of pathogenic cytokines to suppress inflammation and reduce arthritis severity and joint damage (Fig. 11). We also showed that these protective findings were dependent on IL-10, and mediated by the high Mg diet-induced modifications in the intestinal microbiome, and included changes that favored the presence of SCFA-producing bacteria that are typically reduced in RA. The high Mg diet also reduced levels of Prevotella, which is associated with RA. To our knowledge this is the first dietary strategy capable of inducing these beneficial changes in the intestinal microbiome to improve arthritis, and has the potential to be useful in the treatment of different autoimmune and inflammatory diseases.
Fig. 11.
Summary of the effect of the high Mg diet in arthritis. SCFA = short chain fatty acids; arrow up = increase; arrow down = decrease; joint image = inflamed and hyperplastic arthritic synovial tissue.
Contributors
Conceptualization: TL, PSG
Methodology: TL, CH, NA, GB
Investigation: TL, CH, PSG, GB, CB
Funding acquisition: PSG
Project administration: PSG
Supervision: TL, PSG
Writing—original draft: TL, PSG
Writing—review & editing: TL, CH, NA, CB, GB, PSG
Verification of the underlying data: TL and PSG
Data and materials availability and sharing
All data are available in the main text and in the supplementary materials.
Microbiome sequencing data are available upon request.
Declaration of interests
Authors declare that they have no competing interests.
Acknowledgements
Funding: National Institutes of Health (NIAMS) R01 AR 073165 (PSG).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104603.
Appendix B. Supplementary data
Supplemental Fig. S1.
Supplemental Fig. S3.
Supplemental Fig. S4.
References
- 1.Smolen J.S., Aletaha D., McInnes I.B. Rheumatoid arthritis. Lancet. 2016;388(10055):2023–2038. doi: 10.1016/S0140-6736(16)30173-8. [DOI] [PubMed] [Google Scholar]
- 2.Alpizar-Rodriguez D., Finckh A., Gilbert B. The role of nutritional factors and intestinal microbiota in rheumatoid arthritis development. Nutrients. 2020;13(1):96. doi: 10.3390/nu13010096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim K., Bang S.Y., Lee H.S., Bae S.C. Update on the genetic architecture of rheumatoid arthritis. Nat Rev Rheumatol. 2017;13(1):13–24. doi: 10.1038/nrrheum.2016.176. [DOI] [PubMed] [Google Scholar]
- 4.Jiang X., Alfredsson L. Modifiable environmental exposure and risk of rheumatoid arthritis-current evidence from genetic studies. Arthritis Res Ther. 2020;22(1):154. doi: 10.1186/s13075-020-02253-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aletaha D., Smolen J.S. Diagnosis and management of rheumatoid arthritis: a review. JAMA. 2018;320(13):1360–1372. doi: 10.1001/jama.2018.13103. [DOI] [PubMed] [Google Scholar]
- 6.Ahmed F., Mohammed A. Magnesium: the forgotten electrolyte-A review on hypomagnesemia. Med Sci. 2019;7(4):56. doi: 10.3390/medsci7040056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sugimoto J., Romani A.M., Valentin-Torres A.M., et al. Magnesium decreases inflammatory cytokine production: a novel innate immunomodulatory mechanism. J Immunol. 2012;188(12):6338–6346. doi: 10.4049/jimmunol.1101765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rochelson B., Dowling O., Schwartz N., Metz C.N. Magnesium sulfate suppresses inflammatory responses by human umbilical vein endothelial cells (HuVECs) through the NFkappaB pathway. J Reprod Immunol. 2007;73(2):101–107. doi: 10.1016/j.jri.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 9.Ferre S., Baldoli E., Leidi M., Maier J.A.M. Magnesium deficiency promotes a pro-atherogenic phenotype in cultured human endothelial cells via activation of NFkB. Biochim Biophys Acta. 2010;1802(11):952–958. doi: 10.1016/j.bbadis.2010.06.016. [DOI] [PubMed] [Google Scholar]
- 10.Miagkov A.V., Kovalenko D.V., Brown C.E., et al. NF-kappaB activation provides the potential link between inflammation and hyperplasia in the arthritic joint. Proc Natl Acad Sci U S A. 1998;95(23):13859–13864. doi: 10.1073/pnas.95.23.13859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosanoff A., Weaver C.M., Rude R.K. Suboptimal magnesium status in the United States: are the health consequences underestimated? Nutr Rev. 2012;70(3):153–164. doi: 10.1111/j.1753-4887.2011.00465.x. [DOI] [PubMed] [Google Scholar]
- 12.Li B., Selmi C., Tang R., Gershwin M.E., Ma X. The microbiome and autoimmunity: a paradigm from the gut-liver axis. Cell Mol Immunol. 2018;15(6):595–609. doi: 10.1038/cmi.2018.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown E.M., Kenny D.J., Xavier R.J. Gut microbiota regulation of T cells during inflammation and autoimmunity. Annu Rev Immunol. 2019;37:599–624. doi: 10.1146/annurev-immunol-042718-041841. [DOI] [PubMed] [Google Scholar]
- 14.Kishikawa T., Maeda Y., Nii T., et al. Metagenome-wide association study of gut microbiome revealed novel aetiology of rheumatoid arthritis in the Japanese population. Ann Rheum Dis. 2020;79(1):103–111. doi: 10.1136/annrheumdis-2019-215743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scher J.U., Nayak R.R., Ubeda C., Turnbaugh P.J., Abramson S.B. Pharmacomicrobiomics in inflammatory arthritis: gut microbiome as modulator of therapeutic response. Nat Rev Rheumatol. 2020;16(5):282–292. doi: 10.1038/s41584-020-0395-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang X., Zhang D., Jia H., et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat Med. 2015;21(8):895–905. doi: 10.1038/nm.3914. [DOI] [PubMed] [Google Scholar]
- 17.Azzouz D., Omarbekova A., Heguy A., et al. Lupus nephritis is linked to disease-activity associated expansions and immunity to a gut commensal. Ann Rheum Dis. 2019;78(7):947–956. doi: 10.1136/annrheumdis-2018-214856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vatanen T., Franzosa E.A., Schwager R., et al. The human gut microbiome in early-onset type 1 diabetes from the TEDDY study. Nature. 2018;562(7728):589–594. doi: 10.1038/s41586-018-0620-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jangi S., Gandhi R., Cox L.M., et al. Alterations of the human gut microbiome in multiple sclerosis. Nat Commun. 2016;7 doi: 10.1038/ncomms12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scher J.U., Sczesnak A., Longman R.S., et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife. 2013;2 doi: 10.7554/eLife.01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pianta A., Arvikar S., Strle K., et al. Evidence of the immune relevance of Prevotella copri, a gut microbe, in patients with rheumatoid arthritis. Arthritis Rheumatol. 2017;69(5):964–975. doi: 10.1002/art.40003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ivanov I.I., Atarashi K., Manel N., et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139(3):485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stuart J.M., Townes A.S., Kang A.H. Collagen autoimmune arthritis. Annu Rev Immunol. 1984;2:199–218. doi: 10.1146/annurev.iy.02.040184.001215. [DOI] [PubMed] [Google Scholar]
- 24.Holmdahl R., Jansson L., Gullberg D., Rubin K., Forsberg P.O., Klareskog L. Incidence of arthritis and autoreactivity of anti-collagen antibodies after immunization of Dba/1 mice with heterologous and autologous collagen-Ii. Clin Exp Immunol. 1985;62(3):639–646. [PMC free article] [PubMed] [Google Scholar]
- 25.Ji H., Gauguier D., Ohmura K., et al. Genetic influences on the end-stage effector phase of arthritis. J Exp Med. 2001;194(3):321–330. doi: 10.1084/jem.194.3.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laragione T., Cheng K.F., Tanner M.R., et al. The cation channel Trpv2 is a new suppressor of arthritis severity, joint damage, and synovial fibroblast invasion. Clin Immunol. 2015;158(2):183–192. doi: 10.1016/j.clim.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laragione T., Brenner M., Lahiri A., Gao E., Harris C., Gulko P.S. Huntingtin-interacting protein 1 (HIP1) regulates arthritis severity and synovial fibroblast invasiveness by altering PDGFR and Rac1 signalling. Ann Rheum Dis. 2018;77(11):1627–1635. doi: 10.1136/annrheumdis-2018-213498. [DOI] [PubMed] [Google Scholar]
- 28.Bendele A., McAbee T., Sennello G., Frazier J., Chlipala E., McCabe D. Efficacy of sustained blood levels of interleukin-1 receptor antagonist in animal models of arthritis: comparison of efficacy in animal models with human clinical data. Arthritis Rheum. 1999;42(3):498–506. doi: 10.1002/1529-0131(199904)42:3<498::AID-ANR15>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 29.Larsson E., Harris H.E., Larsson A., Mansson B., Saxne T., Klareskog L. Corticosteroid treatment of experimental arthritis retards cartilage destruction as determined by histology and serum COMP. Rheumatology. 2004;43(4):428–434. doi: 10.1093/rheumatology/keh073. [DOI] [PubMed] [Google Scholar]
- 30.Mould A.W., Tonks I.D., Cahill M.M., et al. Vegfb gene knockout mice display reduced pathology and synovial angiogenesis in both antigen-induced and collagen-induced models of arthritis. Arthritis Rheum. 2003;48(9):2660–2669. doi: 10.1002/art.11232. [DOI] [PubMed] [Google Scholar]
- 31.Douni E., Sfikakis P.P., Haralambous S., Fernandes P., Kollias G. Attenuation of inflammatory polyarthritis in TNF transgenic mice by diacerein: comparative analysis with dexamethasone, methotrexate and anti-TNF protocols. Arthritis Res Ther. 2004;6(1):R65–R72. doi: 10.1186/ar1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bialkowska A.B., Ghaleb A.M., Nandan M.O., Yang V.W. Improved Swiss-rolling technique for intestinal tissue preparation for immunohistochemical and immunofluorescent analyses. J Vis Exp. 2016;113 doi: 10.3791/54161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Subcommittee on Laboratory Animal Nutrition CoAN, Board on Agriculture . In: Nutrient requirements of laboratory animals. 4th ed. Council N.R., editor. National Academy Press; Washington, DC: 1995. Nutrient requirements of the mouse. [Google Scholar]
- 34.Flaherty S., Reynolds J.M. Mouse naive CD4(+) T cell isolation and in vitro differentiation into T cell subsets. J Vis Exp. 2015;98 doi: 10.3791/52739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Faith J.J., Guruge J.L., Charbonneau M., et al. The long-term stability of the human gut microbiota. Science. 2013;341(6141) doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kozich J.J., Westcott S.L., Baxter N.T., Highlander S.K., Schloss P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 2013;79(17):5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 38.Bolyen E., Rideout J.R., Dillon M.R., et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37(8):852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J., Holmes S.P. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13(7):581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pedregosa F., Varoquaux G., Gramfort A., et al. Scikit-learn: machine learning in Python. J Mach Learn Res. 2011;12:2825–2830. [Google Scholar]
- 41.Segata N., Izard J., Waldron L., et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12(6):R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wickham H. Use R; 2009. ggplot2: elegant graphics for data analysis; pp. 1–212. [Google Scholar]
- 43.Douglas G.M., Maffei V.J., Zaneveld J.R., et al. PICRUSt2 for prediction of metagenome functions. Nat Biotechnol. 2020;38(6):685–688. doi: 10.1038/s41587-020-0548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Franceschi L., Brugnara C., Beuzard Y. Dietary magnesium supplementation ameliorates anemia in a mouse model of beta-thalassemia. Blood. 1997;90(3):1283–1290. [PubMed] [Google Scholar]
- 45.Lopez-Ocasio M., Buszko M., Blain M., Wang K., Shevach E.M. T follicular regulatory cell suppression of T follicular helper cell function is context-dependent in vitro. Front Immunol. 2020;11:637. doi: 10.3389/fimmu.2020.00637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Canete P.F., Sweet R.A., Gonzalez-Figueroa P., et al. Regulatory roles of IL-10-producing human follicular T cells. J Exp Med. 2019;216(8):1843–1856. doi: 10.1084/jem.20190493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morgan M.E., Flierman R., van Duivenvoorde L.M., et al. Effective treatment of collagen-induced arthritis by adoptive transfer of CD25+ regulatory T cells. Arthritis Rheum. 2005;52(7):2212–2221. doi: 10.1002/art.21195. [DOI] [PubMed] [Google Scholar]
- 48.Kim J.Y., Lim K., Kim K.H., Kim J.H., Choi J.S., Shim S.C. N-3 polyunsaturated fatty acids restore Th17 and Treg balance in collagen antibody-induced arthritis. PLoS One. 2018;13(3) doi: 10.1371/journal.pone.0194331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu H., Gagliani N., Ishigame H., et al. Intestinal type 1 regulatory T cells migrate to periphery to suppress diabetogenic T cells and prevent diabetes development. Proc Natl Acad Sci U S A. 2017;114(39):10443–10448. doi: 10.1073/pnas.1705599114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pot C., Apetoh L., Kuchroo V.K. Type 1 regulatory T cells (Tr1) in autoimmunity. Semin Immunol. 2011;23(3):202–208. doi: 10.1016/j.smim.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yan J.J., Lee J.G., Jang J.Y., Koo T.Y., Ahn C., Yang J. IL-2/anti-IL-2 complexes ameliorate lupus nephritis by expansion of CD4+CD25+Foxp3+ regulatory T cells. Kidney Int. 2017;91(3):603–615. doi: 10.1016/j.kint.2016.09.022. [DOI] [PubMed] [Google Scholar]
- 52.Duscha A., Gisevius B., Hirschberg S., et al. Propionic acid shapes the multiple sclerosis disease course by an immunomodulatory mechanism. Cell. 2020;180(6):1067–1080.e16. doi: 10.1016/j.cell.2020.02.035. [DOI] [PubMed] [Google Scholar]
- 53.Kanjana K., Chevaisrakul P., Matangkasombut P., Paisooksantivatana K., Lumjiaktase P. Inhibitory activity of FOXP3+ regulatory T cells reveals high specificity for displaying immune tolerance in remission state rheumatoid arthritis. Sci Rep. 2020;10(1) doi: 10.1038/s41598-020-76168-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nakae S., Nambu A., Sudo K., Iwakura Y. Suppression of immune induction of collagen-induced arthritis in IL-17-deficient mice. J Immunol. 2003;171(11):6173–6177. doi: 10.4049/jimmunol.171.11.6173. [DOI] [PubMed] [Google Scholar]
- 55.Nakae S., Saijo S., Horai R., Sudo K., Mori S., Iwakura Y. IL-17 production from activated T cells is required for the spontaneous development of destructive arthritis in mice deficient in IL-1 receptor antagonist. Proc Natl Acad Sci U S A. 2003;100(10):5986–5990. doi: 10.1073/pnas.1035999100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jacobs J.P., Wu H.J., Benoist C., Mathis D. IL-17-producing T cells can augment autoantibody-induced arthritis. Proc Natl Acad Sci U S A. 2009;106(51):21789–21794. doi: 10.1073/pnas.0912152106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walmsley M., Katsikis P.D., Abney E., et al. Interleukin-10 inhibition of the progression of established collagen- induced arthritis. Arthritis Rheum. 1996;39(3):495–503. doi: 10.1002/art.1780390318. [DOI] [PubMed] [Google Scholar]
- 58.Tanaka Y., Otsuka T., Hotokebuchi T., et al. Effect of IL-10 on collagen-induced arthritis in mice. Inflamm Res. 1996;45(6):283–288. doi: 10.1007/BF02280992. [DOI] [PubMed] [Google Scholar]
- 59.Ismail H.F., Zhang J., Lynch R.G., Wang Y., Berg D.J. Role for complement in development of Helicobacter-induced gastritis in interleukin-10-deficient mice. Infect Immun. 2003;71(12):7140–7148. doi: 10.1128/IAI.71.12.7140-7148.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Okeke E.B., Uzonna J.E. The pivotal role of regulatory T cells in the regulation of innate immune cells. Front Immunol. 2019;10:680. doi: 10.3389/fimmu.2019.00680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ji H., Ohmura K., Mahmood U., et al. Arthritis critically dependent on innate immune system players. Immunity. 2002;16(2):157–168. doi: 10.1016/s1074-7613(02)00275-3. [DOI] [PubMed] [Google Scholar]
- 62.Wipke B.T., Allen P.M. Essential role of neutrophils in the initiation and progression of a murine model of rheumatoid arthritis. J Immunol. 2001;167(3):1601–1608. doi: 10.4049/jimmunol.167.3.1601. [DOI] [PubMed] [Google Scholar]
- 63.Alpizar-Rodriguez D., Lesker T.R., Gronow A., et al. Prevotella copri in individuals at risk for rheumatoid arthritis. Ann Rheum Dis. 2019;78(5):590–593. doi: 10.1136/annrheumdis-2018-214514. [DOI] [PubMed] [Google Scholar]
- 64.Huang Y., Tang J., Cai Z., et al. Prevotella induces the production of Th17 cells in the colon of mice. J Immunol Res. 2020;2020 doi: 10.1155/2020/9607328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Maeda Y., Kurakawa T., Umemoto E., et al. Dysbiosis contributes to arthritis development via activation of autoreactive T cells in the intestine. Arthritis Rheumatol. 2016;68(11):2646–2661. doi: 10.1002/art.39783. [DOI] [PubMed] [Google Scholar]
- 66.Lucas S., Omata Y., Hofmann J., et al. Short-chain fatty acids regulate systemic bone mass and protect from pathological bone loss. Nat Commun. 2018;9(1):55. doi: 10.1038/s41467-017-02490-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Arpaia N., Campbell C., Fan X., et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504(7480):451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Furusawa Y., Obata Y., Fukuda S., et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504(7480):446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 69.Smith P.M., Howitt M.R., Panikov N., et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341(6145):569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Parker B.J., Wearsch P.A., Veloo A.C.M., Rodriguez-Palacios A. The genus Alistipes: gut bacteria with emerging implications to inflammation, cancer, and mental health. Front Immunol. 2020;11:906. doi: 10.3389/fimmu.2020.00906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Parada Venegas D., De la Fuente M.K., Landskron G., et al. Short chain fatty acids (SCFAs)-Mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front Immunol. 2019;10:277. doi: 10.3389/fimmu.2019.00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Telesford K.M., Yan W., Ochoa-Reparaz J., et al. A commensal symbiotic factor derived from Bacteroides fragilis promotes human CD39(+)Foxp3(+) T cells and Treg function. Gut Microb. 2015;6(4):234–242. doi: 10.1080/19490976.2015.1056973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Round J.L., Mazmanian S.K. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci U S A. 2010;107(27):12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jeon S.G., Kayama H., Ueda Y., et al. Probiotic Bifidobacterium breve induces IL-10-producing Tr1 cells in the colon. PLoS Pathog. 2012;8(5) doi: 10.1371/journal.ppat.1002714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sun M., Wu W., Chen L., et al. Microbiota-derived short-chain fatty acids promote Th1 cell IL-10 production to maintain intestinal homeostasis. Nat Commun. 2018;9(1):3555. doi: 10.1038/s41467-018-05901-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Luu M., Pautz S., Kohl V., et al. The short-chain fatty acid pentanoate suppresses autoimmunity by modulating the metabolic-epigenetic crosstalk in lymphocytes. Nat Commun. 2019;10(1):760. doi: 10.1038/s41467-019-08711-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Freitag T.L., Hartikainen A., Jouhten H., et al. Minor effect of antibiotic pre-treatment on the engraftment of donor microbiota in fecal transplantation in mice. Front Microbiol. 2019;10:2685. doi: 10.3389/fmicb.2019.02685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zelante T., Iannitti R.G., Cunha C., et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39(2):372–385. doi: 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 79.Curti A., Pandolfi S., Valzasina B., et al. Modulation of tryptophan catabolism by human leukemic cells results in the conversion of CD25- into CD25+ T regulatory cells. Blood. 2007;109(7):2871–2877. doi: 10.1182/blood-2006-07-036863. [DOI] [PubMed] [Google Scholar]
- 80.Fallarino F., Grohmann U., You S., et al. The combined effects of tryptophan starvation and tryptophan catabolites down-regulate T cell receptor zeta-chain and induce a regulatory phenotype in naive T cells. J Immunol. 2006;176(11):6752–6761. doi: 10.4049/jimmunol.176.11.6752. [DOI] [PubMed] [Google Scholar]
- 81.Yan Y., Zhang G.X., Gran B., et al. Ido upregulates regulatory T cells via tryptophan catabolite and suppresses encephalitogenic T cell responses in experimental autoimmune encephalomyelitis. J Immunol. 2010;185(10):5953–5961. doi: 10.4049/jimmunol.1001628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sonner J.K., Keil M., Falk-Paulsen M., et al. Dietary tryptophan links encephalogenicity of autoreactive T cells with gut microbial ecology. Nat Commun. 2019;10(1):4877. doi: 10.1038/s41467-019-12776-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lowe M.M., Boothby I., Clancy S., et al. Regulatory T cells use arginase 2 to enhance their metabolic fitness in tissues. JCI Insight. 2019;4(24) doi: 10.1172/jci.insight.129756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cobbold S.P., Adams E., Farquhar C.A., et al. Infectious tolerance via the consumption of essential amino acids and mTOR signaling. Proc Natl Acad Sci U S A. 2009;106(29):12055–12060. doi: 10.1073/pnas.0903919106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hagert C., Siitonen R., Li X.G., Liljenback H., Roivainen A., Holmdahl R. Rapid spread of mannan to the immune system, skin and joints within 6 hours after local exposure. Clin Exp Immunol. 2019;196(3):383–391. doi: 10.1111/cei.13268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kim J.E., Hur H.J., Lee K.W., Lee H.J. Anti-inflammatory effects of recombinant arginine deiminase originating from Lactococcus lactis ssp. lactis ATCC 7962. J Microbiol Biotechnol. 2007;17(9):1491–1497. [PubMed] [Google Scholar]
- 87.Oz H.S., Zhong J., de Villiers W.J. Pegylated arginine deiminase downregulates colitis in murine models. Mediators Inflamm. 2012;2012 doi: 10.1155/2012/813892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shimizu J., Kubota T., Takada E., et al. Propionate-producing bacteria in the intestine may associate with skewed responses of IL10-producing regulatory T cells in patients with relapsing polychondritis. PLoS One. 2018;13(9) doi: 10.1371/journal.pone.0203657. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.