Abstract
Basal cell carcinomas (BCCs) and cutaneous squamous cell carcinomas (SCCs) are the most frequent types of cancer, and both originate from the keratinocyte transformation, giving rise to the group of tumors called keratinocyte carcinomas (KCs). The invasive behavior is different in each group of KC and may be influenced by their tumor microenvironment. The principal aim of the study is to characterize the protein profile of the tumor interstitial fluid (TIF) of KC to evaluate changes in the microenvironment that could be associated with their different invasive and metastatic capabilities. We obtained TIF from 27 skin biopsies and conducted a label-free quantitative proteomic analysis comparing seven BCCs, 16 SCCs, and four normal skins. A total of 2945 proteins were identified, 511 of them quantified in more than half of the samples of each tumoral type. The proteomic analysis revealed differentially expressed TIF proteins that could explain the different metastatic behavior in both KCs. In detail, the SCC samples disclosed an enrichment of proteins related to cytoskeleton, such as Stratafin and Ladinin-1. Previous studies found their upregulation positively correlated with tumor progression. Furthermore, the TIF of SCC samples was enriched with the cytokines S100A8/S100A9. These cytokines influence the metastatic output in other tumors through the activation of NF-kB signaling. According to this, we observed a significant increase in nuclear NF-kB subunit p65 in SCCs but not in BCCs. In addition, the TIF of both tumors was enriched with proteins involved in the immune response, highlighting the relevance of this process in the composition of the tumor environment. Thus, the comparison of the TIF composition of both KCs provides the discovery of a new set of differential biomarkers. Among them, secreted cytokines such as S100A9 may help explain the higher aggressiveness of SCCs, while Cornulin is a specific biomarker for BCCs. Finally, the proteomic landscape of TIF provides key information on tumor growth and metastasis, which can contribute to the identification of clinically applicable biomarkers that may be used in the diagnosis of KC, as well as therapeutic targets.
Keywords: basal cell carcinoma, squamous cell carcinoma, proteomics, tumor interstitial fluid, keratinocyte carcinomas, S100A9
Graphical abstract
Highlights
-
•
SCC samples disclosed an enrichment of proteins related to cytoskeleton.
-
•
Secreted cytokines such as S100A9 may help explain the higher aggressiveness of SCC.
-
•
The proteomic landscape of TIF provides key information on tumor growth & metastasis.
In Brief
In brief, the principal aim of the study is to characterize the protein profile of the TIF of KC to evaluate changes in the microenvironment that could be associated with their invasive and metastatic capabilities. In detail, the SCC samples disclosed an enrichment of proteins related to cytoskeleton, such as Stratafin and Ladinin-1, previously correlated with tumor progression. Furthermore, the TIF of SCC samples was enriched with the cytokines S100A8/S100A9 that may help explain the higher aggressiveness of SCCs.
Keratinocyte carcinomas (KCs) are the most common malignancy worldwide. Moreover, a sharp increase in their incidence has been reported in all age groups (1, 2). KCs group basal cell carcinoma (BCC) and squamous cell carcinoma (SCC), both originated from skin keratinocytes. BCC rarely metastasizes (0.03–0.55%) (3), but when grown locally, it often destroys surrounding tissues. In contrast, SCC may associate with a substantial risk of local recurrence, metastasis, and death (1.5–4%) (4). Taken together, KCs pose an enormous impact on global health problem in terms of morbidity and health expenditure. Their diagnosis is visual and confirmed by histology and immunohistochemistry (IHC) on the surgically removed tissue.
Extracellular signals from tumor cells are involved in most of the steps triggering metastasis. This prometastatic signaling is mediated by proteins secreted by cancer cells, which constitute the tumor secretome and can be obtained in vivo from tumor interstitial fluid (TIF). This fluid baths the tumor and stromal cells and characterizes the tumor microenvironment (5). The proteomic quantitative study of TIF can help to understand all biologicals process dysregulated in both skin cancers, as well as to identify new biomarkers associated with metastatic potential of SCC tumor. However, there is little data regarding TIF directly from melanoma or KC skin biopsies. Alternatively, in melanoma, the role of the tumor microenvironment has been extensively studied from plasma components (6, 7, 8, 9, 10, 11, 12), providing evidence of the secretome and exosomes affecting the course of tumorigenesis, metastasis, and responsiveness to therapy (10). In contrast, the tumor microenvironment from KC has been poorly studied. It has been described the profile of cytokines and miRNAs in the exosomes from SCC cell lines (13, 14). The plasmatic profile of LncRNAs and miRNAs from patients with cutaneous SCC has also been reported (15, 16). In a previous report, we published a protocol to isolate TIF from skin biopsies (17) that allows us to analyze the protein composition of TIF from KC.
A better understanding of the molecular alterations leading to local invasion, aggressiveness, and metastasis is needed in KC. There are still many questions concerning BCC and SCC, such as their differential ability to invade and cause metastasis, despite both originating from the same cell type. The first and most important step in the treatment of BCCs and SCCs is the radical surgical excision, when possible. However, it is difficult to control recurrences (even with unaffected surgical margins) and aggressive behaviors. For these tumors, there are no effective local biomarkers to routinely classify aggressiveness, although they have been profusely studied. Since the tumor microenvironment is a relevant factor in the ability of tumors to spread and occasionally metastasize, we thought that the description of the proteomic pattern of TIF could be a source for detecting biomarkers. The search for biomarkers among the secreted proteins present in the tumor microenvironment of BCC and SCC had not yet been addressed.
In this paper, we have analyzed the proteomic composition of the TIF from BCC and SCC tumor biopsies and discovered a new set of extracellular proteins that could be useful as differential biomarkers. In addition, these proteins may help to understand the distinct invasive and metastatic abilities of the KC.
Experimental Procedures
Sample Information
The study follows the principles of the Declaration of Helsinki. Samples came from a diagnostic tissue surplus and were collected after obtaining the written informed consent of the patient and with the approval of the Ethics Committee for Scientific Research of HUAV-UdL (CEIC-1958). The samples were managed by the Biobank of IRB Lleida, authorized by the Department of Health of Catalonia dated 29 April, 2013 and registered in the National Register of Biobanks of the Carlos III Institute of Health with B.0000682 reference number.
Preparation of Tissue Interstitial Fluid Samples
The interstitial fluid extraction by centrifugation was performed following the protocol that we previously published (17). Briefly, the biopsies were obtained after surgical resection and extensively washed with a sterile dressing to eliminate blood clots. The size of the biopsies was 4 mm. The tumor samples were from the tumor surface, in an area with no apparent necrosis or inflammation. Immediately, the tissue was collected with sterile forceps and put on a plate (6-well) with 1 ml of PBS. The forceps was used to shake the tissue in the PBS and the washing was repeated three times. The biopsy was blotted gently with tissue paper to remove excess PBS and transferred to 2 ml centrifuge tubes (Sterile ClearLine, ref 007859ACL). Immediately, the samples were centrifuged at 10,000g for 20 min at 4 °C (Eppendorf, 5415R), recovering 5 to 15 μl accumulated at the bottom of the tube. PBSi (PBS containing protease and phosphatase inhibitors) was added for a final volume of 50 μl. This last sample was stored at −80 °C until the proteomic analysis.
Proteomic Analyses
Experimental Design and Statistical Rationale
A label-free quantitative proteomic analysis was applied to characterize the proteomic composition of the TIF from 27 skin samples included in this study (16 SCCs, seven BCCs, and four Normal/Healthy skins (NS)). Features of the patients and tumors are summarized in supplemental Table S1.
The data processing and statistical analysis was assessed in R in different sequential steps (see “Data Processing and Bioinformatics Analysis”). The imputation of missing values was addressed using the kNN algorithm (‘impute’ R-package) which assumes that the missing values can be approximated by the real values that are closest to it, based on other variables (proteins). As we consider that different proteins but closely related one to each other should behave similar, we decided to use this approximation to treat the missing values.
To determine the differentially expressed proteins, we used the ‘limma’ R-package to look for a linear relationship between the response variable (disease) and predictor variables (proteins). A p-value was calculated to determine whether the linear relations were significantly different among the experimental groups (NS, SCC, and BCC). The multiple testing problem was addressed by adjusting the p-values using the Benjamini–Hochberg algorithm which introduces less false negatives than other, more restrictive methods such as the Bonferroni correction.
Samples Quantification and Digestion
A total of 27 TIF fluid samples were supplemented with urea to reach a final concentration of 6 mol/l and then quantified with the colorimetric RC DCTM protein assay quantification kit. Twelve micrograms of protein of each sample were digested with Lys-C and Trypsin. Prior to digestion, samples were reduced with 10 mM DL-DTT (Sigma-Aldrich, P/N D9163-25G) and alkylated with 55 mM IAA (Sigma-Aldrich, P/N C0267); then the samples were diluted with Tris 0.1 M to reach urea 2 mol/l. Lys-C (Wako Chemicals) was added at 1:25 (w/w) (enzyme-to-protein ratio), and protein digestion was carried out at 30 °C for 16 h. The samples were subsequently diluted again with Tris 0.1 M to reach urea 0.8 mol/l. Trypsin (Promega, P/N V5117) was added at 1:25 (w/w) (enzyme-to-protein ratio), and protein digestion was carried out at 30 °C for 8 h. The enzymatic reaction was stopped with formic acid (FA; Sigma-Aldrich, P/M 1.00264.0100) (10% (v/v) final concentration). The digested samples were desalted using C18 microspin column (The Nestgroup). Desalted peptides were dried in the speedvac.
Mass Spectrometry Analysis
A total of one microgram of each sample was loaded to a 300 μm × 5 mm C18 PepMap100, 5 mm, 100 Å (Thermo Fisher Scientific) at a flow rate of 15 μl/min using a Thermo Scientific Dionex Ultimate 3000 chromatographic system (Thermo Fisher Scientific). Peptides were separated using a C18 analytical column (nanoEaseTM M/Z HSS C18 T3 (75 μm × 25 cm, 100 Å, Waters)) with a 90 min run, comprising three consecutive steps with linear gradients from 3% to 35% B in 120 min, from 35% to 50% B in 5 min, from 50% to 85% B in 2 min, followed by isocratic elution at 85% B in 5 min and stabilization to initial conditions (A = 0.1% FA in water, B = 0.1% FA in CH3CN) at 250 nl/min flow rate. The column outlets were directly connected to an Advion TriVersa NanoMate (Advion) fitted on an Orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo Fisher Scientific). The mass spectrometer was operated in the data-dependent acquisition (DDA) mode. Survey MS scans were acquired in the orbitrap with the resolution (defined at 200 m/z) set to 120,000. The lock mass was user-defined at 445.12 m/z in each orbitrap scan. The top speed (most intense) ions per scan were fragmented by collision-induced dissociation. The MS/MS was detected in the Ion Trap (with Max Injection time of 35 ms). The ion count target value was 400,000 for the survey scan and 10,000 (collision-induced dissociation) for the MS/MS scan. Target ions already selected for MS/MS were dynamically excluded for 15 s. Spray voltage in the NanoMate source was set to 1.70 kV. RF Lens were tuned to 30%. The minimal signal required to trigger MS to MS/MS switch was set to 5000 and activation Q was 0.250. The spectrometer was working in positive polarity mode, and singly charge state precursors were rejected for fragmentation. Data was acquired with Xcalibur software versus 4.0.27.10 (Thermo Fisher Scientific; https://thermo.flexnetoperations.com/control/thmo/login). All the samples were injected consecutively in one single batch with quality controls (tryptic BSA digest) in between, every three samples.
Data Processing and Bioinformatic Analysis
The .RAW files were processed using the Maxquant 1.6.3.4 software (http://www.maxquant.org). The peaks lists were searched against a SwissProt Human Database downloaded in March 2018 (20,316 entries) with the help of the MaxQuant built-in search engine Andromeda. The false discovery rate was assessed by using a decoy database. Trypsin was selected as enzyme and a maximum of two missed cleavages were allowed. Carbamidomethylation in cysteines was set as a fixed modification, whereas oxidation in methionines and acetylation at the protein N-terminal were used as variable modifications. Searches were performed using 20 ppm and 4 ppm peptide tolerances for the first and main searches and MS/MS match tolerance of 0.5 Da. The quantification of the proteins was performed with the MaxLFQ algorithm integrated into the MaxQuant software. A minimum ratio count of two unique peptides was required for quantification. Finally, the protein list was filtered at a false discovery rate <1%.
The output generated by the Maxquant software (‘proteinGroups.txt’ file) was then filtered to remove the ‘reverse’, ‘potential contaminant’, and ‘only identified by site’ proteins obtaining a list of 2945 proteins. Among them, only 511 proteins showed valid quantification values in more than half of the samples of each tumoral type (BCC, SCC, and NS). This list of proteins was imputated using the KNN algorithm to obtain the final dataset used in the subsequent analyses.
The statistical analysis was achieved in R (https://cran.r-project.org/) and Rstudio (https://www.rstudio.com/) with the help of ‘limma’ (18) and ‘impute’ (https://bioconductor.org/packages/release/bioc/html/impute.html) packages. Both the p-values and Benjamini-Hochberg–adjusted p-values (for multiple testing correction) were obtained with limma by fitting the dataset to a linear model. We chose this method for p-values correction as it introduces less false negatives than other more restrictive methods such as Bonferroni correction. Only the proteins with an adjusted p-value below 0.05 were considered statistically significant. For a deeper analysis of our dataset, we performed a functional enrichment analysis using the ‘ClusterProfiler’ (19) and ‘ReactomePA’ (20) packages in R. Only the upregulated or downregulated statistically significant proteins (adjusted p-value < 0.05) from a pairwise comparison were included in these analyses. The raw data and the processed files were uploaded to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (21) with the project accession code PXD036900.
Immunohistochemistry
To estimate the minimum number of samples needed for the IHC assays, we used the GRANMO sample size calculator (version 7.12 April 2012). Previously published works on the expression of the selected proteins in various tumors were taken as reference, using the most conservative data. Accepting an alpha risk of 0.05 and a beta risk of less than 0.2, in a bilateral contrast, 22 subjects are needed in the first group and 22 in the second to detect statistically significant differences between two proportions (expected to be of 0.8 for group 1 and of 0.4 for group 2). A rate of loss to follow-up of 0% was estimated. The ARCSINUS approximation was used. In addition, to study by IHC the expression of proteins in the tissues using the histoscore, comparing their means, accepting an alpha risk of 0.05 and a beta risk of less than 0.2, in a bilateral contrast, 23 subjects are needed in the first group and 23 in the second group, to detect a difference equal to or greater than 50 histoscore units. The SD was assumed to be 60. A loss of follow-up rate of 0% was estimated.
Tissue microarray blocks were sectioned at a thickness of 3 mm and dried for 1 h at 65 °C before being dewaxed in xylene and rehydrated through a graded ethanol series and washed with PBS. Epitope retrieval was performed in the pretreatment module, PT-LINK (Dako), at 95 °C for 20 min in 50× Tris/EDTA buffer, pH 9. Before staining the sections, endogenous peroxidase was blocked. The antibodies used were 14-3-3 sigma/Stratafin (SFN) (mouse monoclonal, Abcam), Ladinin-1 (LAD1, rabbit polyclonal, Sigma-Aldrich, #HPA028732), Cornulin (CRNN, rabbit polyclonal, Proteintech#11799-1-AP), Calgranulin A (mouse monoclonal, Santa Cruz Biotechnology#SC-48352), Calgranulin B (mouse monoclonal, Santa Cruz Biotechnology, #SC-376772). After incubation, the reaction was visualized with the EnVision Flex Detection Kit (Dako), using diaminobenzidine chromogen as a substrate. Sections were counterstained with hematoxylin. Positive and negative controls were also tested.
A histological score (“histoscore” HSC) was obtained for each sample, ranging from 0 (no immunoreactivity) to 300 (maximum immunoreactivity). The intensity of the staining (1–3) and the % of positive cells (x) were both recorded by applying the following formula: histoscore = 1 × (% light staining) + 2 × (% moderate staining) + 3 × (% strong staining). The histoscore was determined by two independent investigators, and the mean was used for the statistical analysis.
The anonymized databases were assimilated and analyzed with SPSS v20.0 (IBM Corporation). Categorical variables were described using absolute and relative frequencies and continuous variables using mean and SD. Categorical variables were analyzed using the χ2-test or Fisher’s exact test when the expected observations were <5. The normal distribution of the variables was studied with the Shapiro–Wilks test. The comparison of two means was analyzed using Student t test when normally distributed data or Mann-Whitney U test when non-normally distributed data. Comparison of multiple means (>2) was studied using the ANOVA test when normal variables or Kruskal–Wallis test when non-normal variables. As variables were non-normally distributed, multiple comparisons to a reference category were performed using Scheffé test, Mann-Whitney U test, and Wilcoxon test, adjusting the p-value with the Holm method. The Spearman test was used to analyze the relationship between S100A9 and nuclear p65. In all analyses, the selected p-value for considering differences as statistically significant was p < 0.05. The “Bluesky statistics" program was used to create the box plots showing the individual data points.
Immunoblot
The proteins from the TIF samples were resolved by 10% SDS-PAGE), transferred to PVDF membrane (Millipore), and analyzed by immunoblotting. The primary antibodies used were the same as for IHC. Appropriate peroxidase-linked secondary antibodies (GE Healthcare UK Ltd) were detected using the chemiluminescent horseradish peroxidase substrate Immobilon Western (Millipore). Chemiluminescence originated from Western blots was recorded with the aid of a charge-coupled device-based camera (Chemidoc; Bio-Rad). Band quantification was performed using the Image Lab software (Bio-Rad; https://www.bio-rad.com/es-es/product/image-lab-software?ID=KRE6P5E8Z). The results were expressed as relative values.
Results
Proteomic Analyses of the TIF From KC Biopsies
A total of 27 TIF samples have been analyzed by label-free quantitative proteomics to obtain the protein profiles of BCC and SCC. The mean of proteins detected per sample was 944, ranging from 264 to 1426 (Fig. 1A). The samples with the highest number of proteins were those of the BCC, followed by the SCC and healthy skin (Fig. 1B). Interestingly, the total intensity observed in all three tissue types was similar, suggesting that the differences in protein number can be explained by changes in its composition (supplemental Fig. S1). As we had three samples with a very low number of identified proteins, we decided to remove them from the final analysis. These samples were NS14 (264 proteins), SSC7 (452 proteins), and SSC23 (399 proteins). The principal component analysis showed how the samples cluster in the three different groups (SCC, BCC, NS) strengthening the quality of the experiment (Fig. 1C). The raw data of all the proteins obtained from the samples was uploaded to the PRIDE repository with the project accession code PXD036900. When comparing with NS, we detected 199 and 137 proteins significantly enriched (adjusted p-value < 0.05) in BCCs and SCCs, respectively (supplemental Table S2). Moreover, we detected 354 and 118 additional proteins which were quantified only in BCC and SCC, respectively (supplemental Table S3). Therefore, we gathered two lists of proteins enriched in SCC and BCC containing 553 and 255 proteins each. Of these proteins, 196 were common between BCC and SCC, however, 59 were distinctive of SCC and 357 of BCC (supplemental Fig. S2A). These data show that BCC and SCC have different profiles of secreted proteins (supplemental Fig. S2, B and C).
Figure 1.
Proteomic analysis of the tumor interstitial fluid from keratinocyte carcinomas.A, bar graphics representing the number of proteins recovered per sample. B, box diagram representing the proteins recovered per each type of sample. C, PCA graphics with the proteins quantified in at least 50% of the samples. D, heat map showing the protein profile of each sample. The expression levels of the proteins are stated by color changes. The highest expression is stated in red and the lowest is in green. PCA, principal component analysis.
The principal aim of this work was to compare the proteome TIF profiling of SCC and BCC tumors. TIF proteins are secreted principally from different tumor cells, thus, the TIF proteome in both KC could explain their pathophysiology and differences. Therefore, a direct comparison was made between the 14 SCC and the seven BCC samples. The specific protein profile comparing SCC and BCC is shown in the heat map in Figure 1D. We have obtained a list of 36 proteins significantly upregulated in SCC versus BCC (adjusted p-value < 0.05) plus three additional proteins only quantified in the SCC samples, giving a final list of 39 proteins enriched in SCC. In addition, we obtained another list of 104 proteins significantly upregulated in BCC versus SCC (adjusted p-value < 0.05) plus 32 proteins only quantified in the BCC samples, giving a second final list of 136 proteins enriched in BCC. To investigate specific biological features of both groups, we performed a Gene Ontology analysis (Fig. 2) from these lists of proteins identified.
Figure 2.
Gene ontology cellular component of KC. Bar graphics showing the GO cellular component of SCC- (A) and BCC- (B) enriched proteins. BCC, basal cell carcinoma; GO, Gene Ontology; KC, keratinocyte carcinoma; SCC, squamous cell carcinoma.
As expected, cellular component analysis (Fig. 2) showed that both SCC and BCC were enriched in extracellular proteins. However, the protein patterns observed were clearly different between these two skin tumors. We can highlight two main components of these patterns, those involved in the immune response and those related to the actin cytoskeleton function. Regarding the immune response, the TIFs of BCC and SCC samples were enriched with proteins localized in the lumen of secretory and ficolin granules. This is consistent with the Reactome analysis where neutrophil degranulation is highlighted also in both SCC and BCC (supplemental Fig. S3). Neutrophil degranulation is a relevant step in cancer development where the secretion of different kinds of granules (vesicles) modifies the tumor microenvironment. Nevertheless, in contrast to the BCCs, the SCCs samples were enriched in granule proteins associated with tumor-induced immunosuppression and metastatic response such as CD44 (22) (supplemental Table S4). Consistently, previous results show that immune response differs in SCC versus BCC (23).
Gene Ontology analysis also revealed other meaningful differences between SCC and BCC TIFs. Samples of TIFs from SCC were enriched in actin-cytoskeleton proteins (Fig. 2) which are considered potential biomarkers of cancer aggressiveness (16). Changes in actin-cytoskeleton dynamics and the formation of actin-rich protrusions are required by tumor cells to disseminate and colonize other organs (24). Differing from SCCs, the BCCs’ TIF samples exhibited significant enrichment in focal adhesion and cell-substrate junction proteins (Fig. 2). These proteins are functionally related to the connection between cells and the extracellular matrix. Therefore, the variations observed in the protein patterns may also reflect different behaviors in the physiopathology of these tumors.
To corroborate the results from the proteomic approach, we tested by immunoblot the presence of several candidates in the TIF samples (Fig. 3). We focused the following part of our study on the candidates enriched in SCC since these could help explain the dissemination capacity of this tumor compared with BCC. Based on the findings from the proteomic approach, we selected among the enriched candidates those related to cytoskeleton cell signaling and immune response, such as LAD1, SFN, S100A8, and S100A9 (Calgranulins A and B, respectively). We also tested CRNN as a specific biomarker in BCC. The immunoblot data also confirmed that LAD1, SFN, S100A8, and S100A9 were enriched in SCC TIFs and CRNN in BCC TIFs (Fig. 3).
Figure 3.
The enrichment of the TIF samples from SCC, BCC, and NS was analyzed by immunoblot. Pictures of the gels stained with Coomassie Blue are shown as a loading control. BCC, basal cell carcinoma; SCC, squamous cell carcinoma; TIF, tumor interstitial fluid; NS, normal skin.
Characterization of CRNN, LAD1, and SFN as Specific Biomarkers of KC
We were interested in studying whether the expression and secretion of specific proteins such as CRNN, LAD1, and SFN may act as differential tumoral tissue biomarkers among KC. Although TIF proteins are secreted principally from tumor cells, other local cell compartments may secrete proteins to the TIF. Therefore, the origin of the candidates may be the tumor cells, but also the stromal cells or the immune cells. Hence, we studied the expression and localization of the candidate proteins by IHC in primary tumor samples (30 SCCs, 30 BCCs, 57 adjacent NSs). The features of the patients and tumors are summarized in supplemental Tables S5 and S6.
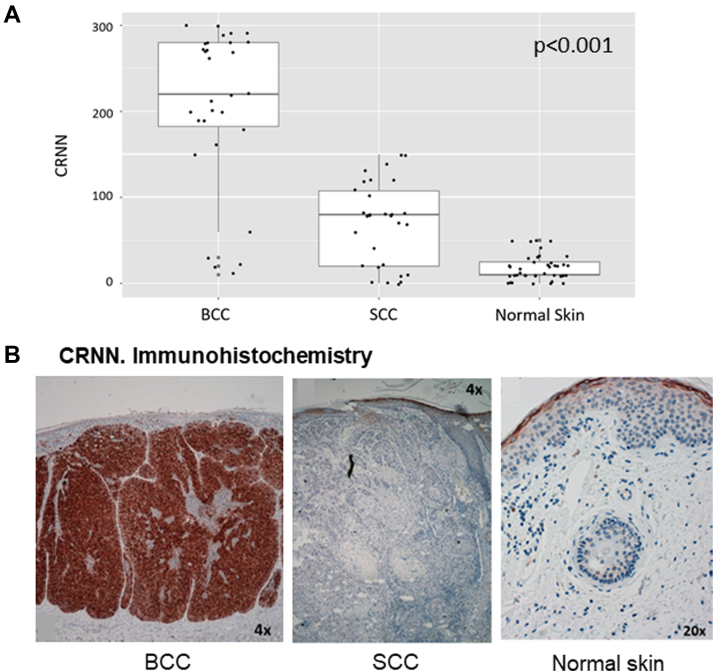
We found the CRNN intensely expressed in the neoplastic cells of BCC, with a mean histoscore of 206.3 (Fig. 4). CRNN was also expressed in the granular layer of the healthy epidermis and mildly expressed in the neoplastic cells of SCC (mean histoscore of 70.0). However, CRNN was not detected in the SCC TIF (Fig. 3). Then, the data suggest that the secretion of CRNN is characteristic of tumor cells from BCC.
Figure 4.
Cornulin expression in KCs and normal skin.A, box diagram showing CRNN expression in the neoplastic cells of BCC, SCC, and healthy skin according to the histoscore values [0–300]. Differences in the expression were statistically significant (p < 0.001) for the three types of samples. B, representative images of immunohistochemistry. CRNN expression in BCC, SCC, and normal skin. BCC, basal cell carcinoma; CNN, Cornulin; KC, keratinocyte carcinoma; SCC, squamous cell carcinoma.
LAD1 was highly expressed only in neoplastic cells from SCC with a mean histoscore of 242.3 (Fig. 5, A–C). BCCs express little or no LAD1 in the neoplastic or stromal cells, with a mean histoscore of 39.2 (Fig. 5, A and B). In healthy skin, we found LAD1 intensely expressed in all epidermic layers, with a mean histoscore of 205.4 (Fig. 5D). Interestingly, the expression of LAD1 was mainly in the membrane of SCC cells, but in healthy skin appeared diffused in the cytoplasm (Fig. 5, C and D).
Figure 5.
Ladinin-1 expression in KCs and normal skin.A, box diagram showing LAD1 expression in the neoplastic cells of BCC, SCC, and healthy skin according to the histoscore values. Differences in the expression were statistically significant (p < 0.01) for the three types of samples. B–D, representative images of immunohistochemistry. LAD1 expression in BCC (B), SCC (C), and normal skin (D). BCC, basal cell carcinoma; KC, keratinocyte carcinoma; SCC, squamous cell carcinoma.
Regarding SFN, this protein was intensely expressed in the neoplastic cells of SCC, with a mean histoscore of 276.0 (supplemental Fig. S4). We only found SFN at the stroma in 19% of the SCC samples. We also found an intense expression of SFN in healthy skin, with a mean histoscore of 259.5. However, BCCs express low SFN in the neoplastic or stromal cells, with a mean histoscore of 45.3 (supplemental Fig. S4). In general, SFN was located in the membrane and cytoplasm. It should be noted that, unlike healthy skin and BCC, we found SFN also expressed in the nucleus in 66.7% (20/30) of the SCC samples, regardless of the histological aggression of the tumor.
The Presence of S100A9-Dependent Signaling Differentiates SCC From BCC
S100A8 and S100A9 are cytokines we found enriched in the SCC TIF samples (supplemental Table S2 and supplemental Fig. S3). These cytokines are usually coexpressed and can form homodimers and heterodimers (25), and both proteins regulate the behavior of cancer cells by inducing premetastatic cascades associated with cancer spread (26). S100A9 has been described to have a relevant role in cell signaling and immune response in the skin and melanomas (27). To better characterize the cytokine S100A9 as an SCC biomarker, we analyzed its expression and localization by IHC. We observed that S100A9 was present in the neoplastic cells of SCC (mean histoscore of 105.9), but no expression of S100A9 was detected in the neoplastic cells of BCC (Fig. 6A). For this protein, we also found differences in the stromal expression. The SCC neoplastic cells were surrounded by an intense inflammatory infiltrate, with a mean of 19.3% of intensely stained cells in this infiltrate (Fig. 6B). In contrast, the BCC neoplastic cells were surrounded by a low-mild inflammatory infiltrate, with a mean of 11.19% of intensely stained cells in this infiltrate. Moreover, we found a positive correlation between the tumoral expression of S100A9 and the quantity of inflammatory infiltrate in the KCs (Fig. 6C). Then, both the expression of S1009A by the neoplastic cells and a high number of infiltrating cells in the stroma could account for the enriched presence of this cytokine in the SCC TIF.
Figure 6.
S100A9 expression in KCs and normal skin.A, box diagram and IHC comparing S100A9 expression in the neoplastic cells of BCC, SCC, and NS (p < 0.0000001). B, box diagram and IHC showing S100A9 expression in the inflammatory infiltrate surrounding BCC and SCC (p = 0.0002). C, graphics showing the positive correlation between the tumoral expression of S110A9 and the surrounding inflammatory infiltrate. D, IHC showing S100A9 expression in NS. BCC, basal cell carcinoma; IHC, immunohistochemistry; KC, keratinocyte carcinoma; NS, normal skin; SCC, squamous cell carcinoma.
It is well known that S100A9 promotes activation of the NF-kB pathway (nuclear p65), altering immune response and enhancing tumor dissemination (28). To study S100A9 activity, we determined nuclear p65 in the primary KC samples. We found a mean of 9.17% of nuclear-p65 cells in SCC and only 2.89% in BCC (Fig. 7, A–C). These results represent a significant increase in the number of cells with nuclear p65 in SCC versus BCC (p = 0.000002). Consistent with the S100A9 data, there was a positive correlation between S100A9 expression and nuclear p65 in KCs (Fig. 7B). Overall, our results show that the presence of S100A9 in the extracellular environment is one crucial distinction between SCC and BCC that could explain the differences in the amount of inflammatory infiltrate and tumor dissemination.
Figure 7.
NF-kB-p65 localization in KCs and NS.A, box diagram comparing nuclear p65 expression (%) in the neoplastic cells of BCC and SCC and also in NS (p = 0.00002). B, graphics showing the positive correlation between the tumoral expression of S110A9 and their nuclear p65. C, IHC showing nuclear p65 expression in the neoplastic cells of BCC and SCC and in NS. BCC, basal cell carcinoma; IHC, immunohistochemistry; KC, keratinocyte carcinoma; NS, normal skin; SCC, squamous cell carcinoma.
Discussion
Significant efforts have been focused on the characterization of the TIF proteomes from KC samples. The proteomic analysis revealed that the samples with the highest protein levels were those of the BCC, followed by the SCC and finally, those of the healthy skin (Fig. 1, A and B). That fact agrees with the already published proteomic studies carried out in other tumors, where the authors also found more proteins in the tumor samples than in the adjacent healthy samples (29). These data support, as the previous authors hypothesized, that this difference in the number of detected proteins shows the greater variety of cell phenotypes among tumor cells and the greater complexity of their cellular activities.
From the proteomic analysis of TIF from BCC and SCC biopsies, we have obtained a list of differential biomarkers. The TIF of both types of tumors was enriched with proteins involved in the immune response, suggesting the relevance of this process in the tumor microenvironment. According to this, immune response proteins were also found enriched in plasma and exosomes of melanoma samples (9, 11). Notably, the list of proteins involved in the immune response was different among SCC and BCC samples. For instance, the TIF from SCC but not from BCC showed S100A8 and S100A9 cytokines, also present in the melanoma microenvironment (11, 12, 30) (see below). Another difference was that SCC samples were enriched in cytoskeleton proteins, while BCC samples were not. Most of these cytoskeleton proteins have been previously described as present in the exosomes of diverse tumors (ExoCarta) (31) and have been related to poor prognoses such as LAD1, SFN, and JUP (32, 33, 34, 35). Then, we propound that the differences between SCC and BCC-TIF composition could explain SCC aggressiveness and metastatic output.
In our proteomic analysis, we found secreted LAD1 only in the SCC-TIF samples. Nonetheless, there are no publications regarding LAD1 secretion or expression of healthy skin or cutaneous tumors. Indeed, LAD1 has been detected in exosomes from ovarian cancer (36), but the function of secreted LAD1 is still unknown. LAD1 is a substrate of the ERK kinase downstream of the EGFR pathway involved in cell motility and cell cycle progression (37). The other way around, LAD1 regulates the ERK–MEK pathway through the SFN adapter protein, which we also have found in our TIF analysis. SFN acts as a soluble cytoskeleton cofactor and binds to LAD1, and their overexpression is associated with increased MEK–ERK pathway activity. These data could explain why in our work, the expressions of LAD1 and SFN were low in BCC samples but high in SCC samples, as the ERK pathway has been reported activated in SCC (38), but not in BCC (39). Moreover, it is described that SCC has epithelial-mesenchymal transition (EMT), which confers aggressive capacities to the tumor. In this way, Klobuclar et al. (32) detected overexpression of LAD1 in laryngeal cancer metastatic tissues but not in nonmetastatic tumors, correlating it to EMT.
By IHC, we observed that the expression of LAD1 was diffused in the cytoplasm of healthy skin keratinocytes, but it was intensely present along the cell periphery in the SCC cells. Changes in the cellular location of LAD1 have also been reported by other authors. Abé et al. (29) found LAD1 in both oral in situ carcinoma samples and adjacent healthy samples, but with different intracellular localizations. In in situ carcinoma samples, it locates along the cell periphery, while in healthy samples, it locates diffusely in the cytoplasm, comparable to our results. This location change could be related to LAD1 involvement in cellular motility, as LAD1 participates in the remodeling of actin fibers (33).
SFN is a scaffold protein present in the exosomes of urine, thymus, and different tumors (ExoCarta) (31). Significantly, this protein is secreted by normal keratinocytes and A431 cells derived from a skin SCC (40, 41), and it has been proposed that exosomes containing SFN have MMP-1 stimulatory activity in fibroblasts (41). This fact may be relevant for the dissemination and aggressiveness of the tumor and is consistent with the SFN enrichment in the TIF of SCC samples. In contrast with this possibility, SFN takes part in the terminal differentiation of keratinocytes. Under normal conditions, SFN sequestrates the YAP1 transcriptional regulator in the cytoplasm, promoting keratinocyte differentiation. SFN mutations in mice cause a phenotype characterized by a poorly differentiated hyperproliferative epidermis (42). In our work, SFN was present in the cytoplasm but not in the nucleus of healthy skin keratinocytes. However, we found SFN intensely present in the cytoplasm but also in the nucleus of SCC cells. As commented above, the influence of SFN on keratinocyte differentiation depends on its cytoplasmic location. Therefore, although SFN was present in both healthy skin and SCC, its nuclear localization could play a crucial role in its carcinogenesis, hindering the accurate differentiation of keratinocytes in the tumor.
Our results also showed that the cytokines S100A8/S100A9 were substantially enriched in the TIFs from SCC tumors. In line with this result, it was described the expression of these cytokines in SCC biopsies (43, 44). Also, it has been published the relevance of these cytokines in tumor growth by using a mouse xenograft model (44). In addition, S100A9 overexpression correlates with invasion and metastasis in various cancers, among which malignant melanoma and secreted S100A9 directly enhances tumor cell malignancy by activating NF-kB p65 (nuclear p65) through TLR4-mediated signaling (28). Consistent with this data, we have observed expression of S100A9 in neoplastic cells of SCC but not on BCC cells, a tumor that rarely metastasizes. We have shown that the presence of S100A9 correlates with the presence of nuclear NF-kB p65 (NF-kB activation) in KC. This result is also consistent with previously published data (45). In addition, S100A8 and S100A9 are secreted cytokines involved in the nesting of disseminated tumor cells during metastasis (26) and in promoting inflammation and immune evasion of cancer cells (46). The cytokines S100A8/S100A9 are secreted from cultured keratinocytes (47), suggesting that they could be secreted to the microenvironment by SCC neoplastic cells. However, it is relevant to consider that S100A8/S100A9 are constitutively expressed in immune cells such as neutrophils and increase the inflammation process (48). Accordingly, we have found a significantly more intense infiltrate around SCC tumors than BCC, with a higher expression of S100A9 in this infiltrate, and a positive correlation between the tumoral expression of S100A9 and the amount of inflammatory infiltrate. Then, we consider also relevant for the aggressive SCC behavior the presence of S100A9-positive cells in the tumor infiltrate. Supporting this idea, in NK/T-cell lymphomas, the overexpression of S100A9 in tumor stroma contributes to immune evasion and predicts a poorer response rate (46). Interestingly, we have also observed a positive correlation between the tumor expression of S100A9 and the quantity of surrounding inflammatory infiltrate in KCs. Therefore, it could be possible that the secretion of S100A9 by SCC neoplastic cells would promote the recruitment of a larger inflammatory infiltrate surrounding SCC in comparison with BCC.
Besides S100A8/S100A9, the SCC TIF samples showed other secreted proteins associated with immune evasion and inflammation, such as CD44, LSP1, and CD163. CD44 and LSP1 have been involved in immune suppression by regulating PD-L1/PD-1 function (22, 49). CD163 is a specific marker of M2-type macrophages. The infiltration of these macrophages in the tumor surroundings promotes an immunosuppressive environment favoring metastatic lymph nodes in colorectal cancer (50). Overall, these data reinforce the idea that the tumor microenvironment in SCC, unlike in BCC, may contain particular immune infiltrate characteristics that favor immune evasion and tumor aggressiveness.
On the other hand, we found enriched in SCC the fatty acid–binding protein 5 (FABP5) which is part of a lipid-binding protein family involved in transporting long-chain fatty acids related to metastasis in breast cancer cells (51). Among the top SCC-enriched proteins, we also have ANHAK2, SAMD9, and the nucleotide metabolism enzymes TYMP and PNP. All of them are also related to malignancy characteristics or metastasis (52, 53, 54, 55). Moreover, ANHAK2 is a giant protein involved in cell signaling that regulates EMT response in lung adenocarcinoma cells (56).
Regarding the top proteins enriched in the BCC TIF, many of them have dual characteristics on tumor cell regulation. For instance, the enzyme DDAH1 metabolizes dimethyl-Arginine, which is an inhibitor of nitric oxide synthase (57). This enzyme is overexpressed in prostate cancer, inducing tumor growth and angiogenesis by regulating the oxide nitric concentration (58). However, DDAH1 functions as a tumor suppressor and as a good prognostic marker in gastric cancer (59). Another BCC-enriched protein is the Neddylation Activating Enzyme 1, which is a regulator of protein stability and activity (60). This protein is a negative regulator of cell migration, as the inhibition of Neddylation Activating Enzyme 1 reduces migration in prostate and glioblastoma cells (60). Like these proteins, CRNN, RNA-binding protein 3, Target of Nesh-SH3, and LHPP are correlated with favorable clinic-pathological features in different cancers (61, 62, 63, 64, 65). More in detail, we found CRNN highly expressed in BCC and low-moderately in SCC. However, CRNN was described as under-expressed in several tumors and considered a tumor suppressor protein (66, 67). The difference between these reported tumors and our cutaneous tumors, in terms of CRNN expression, could be explained because of specific tissue functions. For instance, Li et al. (68) published that CRNN promotes proliferation in psoriatic keratinocytes through the activation of the PI3K/Akt pathway. These authors hypothesized that the contradiction of CRNN expression in psoriasis versus other carcinomas could be explained by the benignity-malignancy context. Considering that CRNN is present in our cutaneous tumors, we propose that the expression of CRNN would be related to the location-specific function (cutaneous or not) rather than to the condition of the pathology studied (benign or malignant). In our samples, CRNN was quantified only in BCC.
Among the BCC-enriched proteins, there is also the Myosin Light Chain 9 (MYL9), which is a regulatory protein of the Myosin II complex (Fig. 2). This protein regulates muscle contraction and actin cytoskeleton dynamics, and the expression of MYL9 has been associated with tumor invasion and metastasis (69). However, the overexpression of MYL9 in different cancers is not correlated with minor patient survival (69).
Overall, the enrichment data (supplemental Tables S2 and S3) showed that SCC samples contained many secreted proteins associated with tumor invasion and aggressiveness, but BCC proteins mainly relied on a good prognosis. This result is consistent with the major aggressiveness of SCC tumors, reinforcing the quality of our proteomic data.
To sum up, the TIF is the proteomic signature of BCC- and SCC-associated changes on an aggregate, functional level by determining differentially metabolic regulated processes. Most of the processes differentially regulated showed a high concordance with the pathophysiology of both KC. The data revealed induction of the immune system, upregulation of cell cycle–associated processes, metastasis process and glycan degradation pathways, downregulation of cofactor-associated metabolic processes, and a pronounced induction of several signaling pathways. Hence, the results of our first proteomic approach help to shed light on the pathophysiology of the KC and their differences. In the IHC study, the SCC samples revealed an enrichment of proteins related to cytoskeleton, such as SFN and LAD-1. These proteins may help explain the invasive behavior of SCC. LAD1 is involved in cell motility and cell cycle progression. By IHC, we observed LAD1 diffuse in the cytoplasm of healthy skin keratinocytes and along the cell periphery in the SCC cells. This location change could be related to LAD1 involvement in cellular motility and the remodeling of actin fibers. Furthermore, SFN takes part in the terminal differentiation of keratinocytes depending on its cytoplasmatic location. In our work, SFN was present in the cytoplasm but not in the nucleus of healthy skin keratinocytes. However, we found SFN in the nucleus of SCC cells. Therefore, although SFN was present in both healthy skin and SCC, its nuclear localization could play a crucial role in its carcinogenesis, hindering the accurate differentiation of keratinocytes in the tumor. Finally, we propose that the expression and secretion of S100A9 would be a landmark for metastasis in cutaneous tumors favoring metastatic signals from the neoplastic and infiltrate cells. The absence of S100A9 signaling may help to explain why BCC tumors rarely metastasize.
Data Availability
The raw data and the processed files were uploaded to the PRIDE repository with the project accession code PXD036900.
Supplemental data
Conflict of interest
The authors declare no conflict of interest to disclosure.
Acknowledgments
This work was funded by the Spanish Ministry of Innovation and Science MICINN (PID2019-104859GB-I00) and by Generalitat de Catalunya (2017-SGR-569). This work was supported by IRBLleida Biobank (B.0000682) and PLATAFORMA BIOBANCOS PT20/0021. “The proteomics analyses were performed in the IJC Proteomics Unit. The IJC Proteomics Unit is part of the Spanish Platform of Molecular and Bioinformatics Resources (ProteoRed), Instituto de Salud Carlos III (PT13/0001).” For detailed “Experimental procedures” for publication, please, contact the Proteomics Unit. Special thanks to Eddie Chalecki lluMme.
Funding and additional information
M. R.-S. is the recipient of a TALENT grant from Lleida Institute for Biomedical Research-Dr Pifarré Foundation supported by Diputació de Lleida. M. G.-V. (FPU17/00229) was supported by a predoctoral fellowship from Ministerio de Educación, Cultura y Deportes.
Author contributions
C. M.-N., S. G., M. R.-S., M. G.-V., N. P., J. M. C., E. G., and R. S. A.-O. conceptualization; C. M.-N., S. G., M. R.-S., M. G.-V., N. P., J. M. C., E. G., and R. S. A.-O. methodology; C. M.-N., S. G., M. R.-S., M. G.-V., N. P., J. M. C., E. G., and R. S. A.-O. investigation; C. M.-N., S. G., M. R.-S., M. G.-V., N. P., J. M. C., E. G., and R. S. A.-O. formal analysis; C. M.-N., S. G., M. R.-S., M. G.-V., N. P., J. M. C., E. G., and R. S. A.-O. data curation; C. M.-N. writing–original draft; C. M.-N., J. J. B.-S., and C. d. l. T. G. writing–review and editing.
Contributor Information
Clara Matas-Nadal, Email: clamatas@hotmail.com.
Joan J. Bech-Serra, Email: jbech@carrerasresearch.org.
Supplemental Data
Supplemental Figure S1.
Box diagram showing the total protein intensity observed in all three tissue types.
Supplemental Figure S2.
Comparisons of the protein profiles of KCs and normal skin.A, Venn diagram showing the proteins significantly enriched or only present in SCC or BCC, compared to healthy skin. A total of 553 and 255 proteins were found for BCC and SCC, respectively. Of these proteins, 196 were common between BCC and SCC, however, 59 were distinctive of SCC and 357 of BCC. B and C, heat maps showing the protein profile of each sample, SCC versus NS (B) and BCC versus NS (C). The expression levels of the proteins are stated by color changes. The highest expression is stated in red and the lowest in green.
Supplemental Figure S3.
Bar graphic showing the reactome analysis of SCC (A) and BCC (B) -enriched proteins.
Supplemental Figure S4.
SFN expression in KCs and normal skin.A, box diagram showing SFN expression in the neoplastic cells of SCC, BCC, and healthy skin according to the histoscore values. Differences in the expression were statistically significant (p < 0.001) for the three types of samples. B, immunohistochemistry. SFN expression in normal skin, BCC, and SCC.
References
- 1.Matas-Nadal C., Sagristà M., Gómez-Arbonés X., Sobrino Bermejo C., Fernández-Armenteros J.M., Àngel Baldó J., et al. Risk factors for early-onset basal cell carcinomas and the trend towards their female predominance. J. Dtsch. Dermatol. Ges. 2021;19:364–371. doi: 10.1111/ddg.14390. [DOI] [PubMed] [Google Scholar]
- 2.Robsahm T.E., Helsing P., Veierød M.B. Cutaneous squamous cell carcinoma in Norway 1963-2011: incresasing incidence and stable mortality. Cancer Med. 2015;4:472–480. doi: 10.1002/cam4.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ionescu D.N., Arida M., Jukic D.M. Metastatic basal cell carcinoma: four case reports, review of literature, and immunohistochemical evaluation. Arch. Pathol. Lab. Med. 2006;130:45–51. doi: 10.5858/2006-130-45-MBCCFC. [DOI] [PubMed] [Google Scholar]
- 4.Venables Z.C., Autier P., Nijsten T., Wong K.F., Langan S.M., Rous B., et al. Nationwide incidence of metastatic cutaneous squamous cell carcinoma in England. JAMA Dermatol. 2019;155:298–306. doi: 10.1001/jamadermatol.2018.4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burian M., Velic A., Matic K., Günther S., Kraft B., Gonser L., et al. Quantitative proteomics of the human skin secretome reveal a reduction in immune defense mediators in ectodermal dysplasia patients. J. Invest. Dermatol. 2015;135:759–767. doi: 10.1038/jid.2014.462. [DOI] [PubMed] [Google Scholar]
- 6.Bardi G.T., Smith M.A., Hood J.L. Melanoma exosomes promote mixed M1 and M2 macrophage polarization. Cytokine. 2018;105:63–72. doi: 10.1016/j.cyto.2018.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peinado H., Alečković M., Lavotshkin S., Matei I., Costa-Silva B., Moreno-Bueno G., et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 2012;18:883–891. doi: 10.1038/nm.2753. Erratum in: Nat Med. 2016; 6;22(12):1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chavanet A., Hill K.R., Jiménez-Andrade Y., Choo M.K., White K., Park J.M. Intracellular signaling modules linking DNA damage to secretome changes in senescent melanoma cells. Melanoma Res. 2020;30:336–347. doi: 10.1097/CMR.0000000000000671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maus R.L.G., Jakub J.W., Nevala W.K., Christensen T.A., Noble-Orcutt K., Sachs Z., et al. Human melanoma-derived extracellular vesicles regulate dendritic cell maturation. Front. Immunol. 2017;29:358. doi: 10.3389/fimmu.2017.00358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Almeida N., Rodriguez J., Pla Parada I., Perez-Riverol Y., Woldmar N., Kim Y., et al. Mapping the melanoma plasma proteome (MPP) using single-shot proteomics interfaced with the WiMT database. Cancers (Basel) 2021;10:6224. doi: 10.3390/cancers13246224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pietrowska M., Zebrowska A., Gawin M., Marczak L., Sharma P., Mondal S., et al. Proteomic profile of melanoma cell-derived small extracellular vesicles in patients’ plasma: a potential correlate of melanoma progression. J. Extracell. Vesicles. 2021;10 doi: 10.1002/jev2.12063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maus R.L.G., Jakub J.W., Hieken T.J., Nevala W.K., Christensen T.A., Sutor S.L., et al. Identification of novel, immune-mediating extracellular vesicles in human lymphatic effluent draining primary cutaneous melanoma. Oncoimmunology. 2019;8 doi: 10.1080/2162402X.2019.1667742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flemming J.P., Hill B.L., Anderson-Pullinger L., Harshyne L.A., Mahoney M.G. Cytokine profiling in low- and high-density small extracellular vesicles from epidermoid carcinoma cells. JID Innov. 2021;1:100053. doi: 10.1016/j.xjidi.2021.100053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flemming J.P., Hill B.L., Haque M.W., Raad J., Bonder C.S., Harshyne L.A., et al. miRNA- and cytokine-associated extracellular vesicles mediate squamous cell carcinomas. J. Extracell. Vesicles. 2020;9:1790159. doi: 10.1080/20013078.2020.1790159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang D., Zhou X., Yin J., Zhou Y. Lnc-PICSAR contributes to cisplatin resistance by miR-485-5p/REV3L axis in cutaneous squamous cell carcinoma. Open Life Sci. 2020;15:488–500. doi: 10.1515/biol-2020-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Z., Guo H., Yang W., Li J. Exosomal circular RNA RNA-seq profiling and the carcinogenic role of exosomal circ-CYP24A1 in cutaneous squamous cell carcinoma. Front. Med. (Lausanne) 2021;8 doi: 10.3389/fmed.2021.675842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matas-Nadal C., Bech-Serra J.J., Guasch-Vallés M., Fernández-Armenteros J.M., Barceló C., Casanova J.M., et al. Evaluation of tumor interstitial fluid-extraction methods for proteome analysis: comparison of biopsy elution versus centrifugation. J. Proteome Res. 2020;19:2598–2605. doi: 10.1021/acs.jproteome.9b00770. [DOI] [PubMed] [Google Scholar]
- 18.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu T., Hu E., Xu S., Chen M., Guo P., Dai Z., et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation (Camb) 2021;2:100141. doi: 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu G., He Q.-Y. ReactomePA: an R/bioconductor package for reactome pathway analysis and visualization. Mol. Biosyst. 2016;12:477–479. doi: 10.1039/c5mb00663e. [DOI] [PubMed] [Google Scholar]
- 21.Perez-Riverol Y., Bai J., Bandla C., García-Seisdedos D., Hewapathirana S., Kamatchinathan S., et al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022;50:D543–D552. doi: 10.1093/nar/gkab1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kong T., Ahn R., Yang K., Zhu X., Fu Z., Morin G., et al. CD44 promotes PD-L1 expression and its tumor-intrinsic function in breast and lung cancers. Cancer Res. 2020;80:444–457. doi: 10.1158/0008-5472.CAN-19-1108. [DOI] [PubMed] [Google Scholar]
- 23.Mushtaq S. The immunogenetics of non-melanoma skin cancer. Adv. Exp. Med. Biol. 2022;1367:397–409. doi: 10.1007/978-3-030-92616-8_16. [DOI] [PubMed] [Google Scholar]
- 24.Mondal C., Di Martino J.S., Bravo-Cordero J.J. Actin dynamics during tumor cell dissemination. Int. Rev. Cell Mol. Biol. 2021;360:65–98. doi: 10.1016/bs.ircmb.2020.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Teigelkamp S., Bhardwaj R.S., Roth J., Meinardus-Hager G., Karas M., Sorg C., et al. Calcium-dependent complex assembly of the myeloic differentiation proteins MRP-8 and MRP-14. J. Biol. Chem. 1991;266:13462–13467. [PubMed] [Google Scholar]
- 26.Nedjadi T., Evans A., Sheikh A., Barerra L., Al-Ghamdi S., Oldfield L., et al. S100A8 and S100A9 proteins form part of a paracrine feedback loop between pancreatic cancer cells and monocytes. BMC Cancer. 2018;18:1255. doi: 10.1186/s12885-018-5161-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rad Pour S., Pico de Coaña Y., Demorentin X.M., Melief J., Thimma M., Wolodarski M., et al. Predicting anti-PD-1 responders in malignant melanoma from the frequency of S100A9+ monocytes in the blood. J. Immunother. Cancer. 2021;9 doi: 10.1136/jitc-2020-002171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang M., Wu R., Chen L., Peng Q., Li S., Zhang Y., et al. S100A9 regulates MDSCs-mediated immune suppression via the RAGE and TLR4 signaling pathways in colorectal carcinoma. Front. Immunol. 2019;10:2243. doi: 10.3389/fimmu.2019.02243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abé T., Maruyama S., Yamazaki M., Xu B., Babkair H., Sumita Y., et al. Proteomic and histopathological characterization of the interface between oral squamous cell carcinoma invasion fronts and noncancerous epithelia. Exp. Mol. Pathol. 2017;102:327–336. doi: 10.1016/j.yexmp.2017.02.018. [DOI] [PubMed] [Google Scholar]
- 30.Alegre E., Zubiri L., Perez-Gracia J.L., González-Cao M., Soria L., Martín-Algarra S., et al. Circulating melanoma exosomes as diagnostic and prognosis biomarkers. Clin. Chim. Acta. 2016;454:28–32. doi: 10.1016/j.cca.2015.12.031. [DOI] [PubMed] [Google Scholar]
- 31.Mathivanan S., Simpson R.J. ExoCarta: a compendium of exosomal proteins and RNA. Proteomics. 2009;9:4997–5000. doi: 10.1002/pmic.200900351. [DOI] [PubMed] [Google Scholar]
- 32.Klobučar M., Sedić M., Gehrig P., Grossmann J., Bilić M., Kovač-Bilić L., et al. Basement membrane protein ladinin-1 and the MIF-CD44-β1 integrin signaling axis are implicated in laryngeal cancer metastasis. Biochim. Biophys. Acta. 2016;1862:1938–1954. doi: 10.1016/j.bbadis.2016.07.014. [DOI] [PubMed] [Google Scholar]
- 33.Roth L., Srivastava S., Lindzen M., Sas-Chen A., Sheffer M., Lauriola M., et al. SILAC identifies LAD1 as a filamin-binding regulator of actin dynamics in response to EGF and a marker of aggressive breast tumors. Sci. Signal. 2018;11:eaan0949. doi: 10.1126/scisignal.aan0949. [DOI] [PubMed] [Google Scholar]
- 34.Zhou Wh, Tang F., Xu J., Wu X., Feng Z.Y., Li H.G., et al. Aberrant upregulation of 14-3-3ơ expression serves as an inferior prognostic biomarker for gastric cancer. BMC Cancer. 2011;11:397. doi: 10.1186/1471-2407-11-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spethmann T., Böckelmann L.C., Labitzky V., Ahlers A.K., Schröder-Schwarz J., Bonk S., et al. Opposing prognostic relevance of junction plakoglobin in distinct prostate cancer patient subsets. Mol. Oncol. 2021;15:1956–1969. doi: 10.1002/1878-0261.12922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liang B., Peng P., Chen S., Li L., Zhang M., Cao D., et al. Characterization and proteomic analysis of ovarian cancer-derived exosomes. J. Proteomics. 2013;80:171–182. doi: 10.1016/j.jprot.2012.12.029. [DOI] [PubMed] [Google Scholar]
- 37.Tarcic G., Avraham R., Pines G., Amit I., Shay T., Lu Y., et al. EGR1 and the ERK-ERF axis drive mammary cell migration in response to EGF. FASEB J. 2011;26:1582–1592. doi: 10.1096/fj.11-194654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu S.C., Yu H.S., Yen F.L., Chen G.S., Lan C.C. CXCR7 expression correlates with tumor depth in cutaneous squamous cell carcinoma skin lesions and promotes tumor cell survival through ERK activation. Exp. Dermatol. 2014;23:902–908. doi: 10.1111/exd.12557. [DOI] [PubMed] [Google Scholar]
- 39.Aguayo R.S., Rafel M., Santacana M., Fusté N.P., Garí E. Erk1/2 activation in stromal fibroblasts from sporadic basal cell carcinomas. Dermatol. Surg. 2015;41:677–684. doi: 10.1097/DSS.0000000000000367. [DOI] [PubMed] [Google Scholar]
- 40.Park J.E., Tan H.S., Datta A., Lai R.C., Zhang H., Meng W., et al. Hypoxic tumor cell modulates its microenvironment to enhance angiogenic and metastatic potential by secretion of proteins and exosomes. Mol. Cell. Proteomics. 2010;9:1085–1099. doi: 10.1074/mcp.M900381-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chavez-Muñoz C., Morse J., Kilani R., Ghahary A. Primary human keratinocytes externalize stratifin protein via exosomes. J. Cell. Biochem. 2008;104:2165–2173. doi: 10.1002/jcb.21774. [DOI] [PubMed] [Google Scholar]
- 42.Herron B.J., Liddell R.A., Parker A., Grant S., Kinne J., Fisher J.K., et al. A mutation in stratifin is responsible for the repeated epilation (Er) phenotype in mice. Nat. Genet. 2005;37:1210–1212. doi: 10.1038/ng1652. [DOI] [PubMed] [Google Scholar]
- 43.Martinsson H., Yhr M., Enerbäck C. Expression patterns of S100A7 (psoriasin) and S100A9 (calgranulin-B) in keratinocyte differentiation. Exp. Dermatol. 2005;14:161–168. doi: 10.1111/j.0906-6705.2005.00239.x. [DOI] [PubMed] [Google Scholar]
- 44.Choi D.K., Li Z.J., Chang I.K., Yeo M.K., Kim J.M., Sohn K.C., et al. Clinicopathological roles of S100A8 and S100A9 in cutaneous squamous cell carcinoma in vivo and in vitro. Arch. Dermatol. Res. 2014;306:489–496. doi: 10.1007/s00403-014-1453-y. [DOI] [PubMed] [Google Scholar]
- 45.Weng H., Deng Y., Xie Y., Liu H., Gong F. Expression and significance of HMGB1, TLR4 and NF-κB p65 in human epidermal tumors. BMC Cancer. 2013;13:311. doi: 10.1186/1471-2407-13-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou Z., Chen X., Li Z., Wang X., Zhang M. Overexpression of S100A9 in tumor stroma contribute to immune evasion of NK/T cell lymphoma and predict poor response rate. Sci. Rep. 2021;11 doi: 10.1038/s41598-021-90794-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thorey I.S., Roth J., Regenbogen J., Barerra L., Al-Ghamdi S., Oldfield L., et al. The Ca2+-binding proteins S100A8 and S100A9 are encoded by novel injury-regulated genes. J. Biol. Chem. 2001;276:35818–35825. doi: 10.1074/jbc.M104871200. [DOI] [PubMed] [Google Scholar]
- 48.Wang S., Song R., Wang Z., Jing Z., Wang S., Ma J. S100A8/A9 in inflammation. Front. Immunol. 2018;9:1298. doi: 10.3389/fimmu.2018.01298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cao J.Y., Guo Q., Guan G.F., Zhu C., Zou C.Y., Zhang L.Y., et al. Elevated lymphocyte specific protein 1 expression is involved in the regulation of leukocyte migration and immunosuppressive microenvironment in glioblastoma. Aging (Albany NY) 2020;12:1656–1684. doi: 10.18632/aging.102706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen F.L., Wang Y.Y., Liu W., Xing B.C. Neoadjuvant chemotherapy improves overall survival in resectable colorectal liver metastases patients with high clinical risk scores-- a retrospective, propensity score matching analysis. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.973418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Powell C.A., Nasser M.W., Zhao H., Wochna J.C., Zhang X., Shapiro C., et al. Fatty acid binding protein 5 promotes metastatic potential of triple negative breast cancer cells through enhancing epidermal growth factor receptor stability. Oncotarget. 2015;6:6373–6385. doi: 10.18632/oncotarget.3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lu D., Wang J., Shi X., Yue B., Hao J. AHNAK2 is a potential prognostic biomarker in patients with PDAC. Oncotarget. 2017;8:31775–31784. doi: 10.18632/oncotarget.15990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ma W., Jin H., Liu W., Li X., Zhou X., Guo X., et al. Homeobox B8 targets sterile alpha motif domain-containing protein 9 and drives glioma progression. Neurosci. Bull. 2020;36:359–371. doi: 10.1007/s12264-019-00436-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Q., Zhang Y., Hu X., Qin Y., Zhong W., Meng J., et al. Thymidine phosphorylase promotes metastasis and serves as a marker of poor prognosis in hepatocellular carcinoma. Lab. Invest. 2017;97:903–912. doi: 10.1038/labinvest.2017.51. [DOI] [PubMed] [Google Scholar]
- 55.Kojima S., Chiyomaru T., Kawakami K., Yoshino H., Enokida H., Nohata N., et al. Tumour suppressors miR-1 and miR-133a target the oncogenic function of purine nucleoside phosphorylase (PNP) in prostate cancer. Br. J. Cancer. 2012;106:405–413. doi: 10.1038/bjc.2011.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu G., Guo Z., Zhang Q., Liu Z., Zhu D. AHNAK2 promotes migration, invasion, and epithelial-mesenchymal transition in lung adenocarcinoma cells via the TGF-β/Smad3 pathway. Onco Targets Ther. 2020;13:12893–12903. doi: 10.2147/OTT.S281517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kostourou V., Robinson S., Cartwright J., Whitley G.S. Dimethylarginine dimethylaminohydrolase I enhances tumour growth and angiogenesis. Br. J. Cancer. 2002;87:673–680. doi: 10.1038/sj.bjc.6600518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reddy K.R.K., Dasari C., Duscharla D., Supriya B., Ram N.S., Surekha M.V., et al. Dimethylarginine dimethylaminohydrolase-1 (DDAH1) is frequently upregulated in prostate cancer, and its overexpression conveys tumor growth and angiogenesis by metabolizing asymmetric dimethylarginine (ADMA) Angiogenesis. 2018;21:79–94. doi: 10.1007/s10456-017-9587-0. [DOI] [PubMed] [Google Scholar]
- 59.Ye J., Xu J., Li Y., Huang Q., Huang J., Wang J., et al. DDAH1 mediates gastric cancer cell invasion and metastasis via Wnt/β-catenin signaling pathway. Mol. Oncol. 2017;11:1208–1224. doi: 10.1002/1878-0261.12089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Park S.Y., Park J.W., Lee G.W., Li L., Chun Y.S. Inhibition of neddylation facilitates cell migration through enhanced phosphorylation of caveolin-1 in PC3 and U373MG cells. BMC Cancer. 2018;18:30. doi: 10.1186/s12885-017-3942-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Al-Astal H.I., Massad M., AlMatar M., Ekal H. Cellular functions of RNA-binding motif protein 3 (RBM3): clues in hypothermia, cancer biology and apoptosis. Protein Pept. Lett. 2016;23:828–835. doi: 10.2174/0929866523666160628090340. [DOI] [PubMed] [Google Scholar]
- 62.Zhou R.B., Lu X.L., Zhang C.Y., Yin D.C. RNA binding motif protein 3: a potential biomarker in cancer and therapeutic target in neuroprotection. Oncotarget. 2017;8:22235–22250. doi: 10.18632/oncotarget.14755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Saleem S., Aleem I., Atiq A., Ariq S., Babar A., Bakar M.A., et al. Expression of cornulin in tongue squamous cell carcinoma. Ecancermedicalscience. 2021;15:1197. doi: 10.3332/ecancer.2021.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Govindaraj P.K., Kallarakkal T.G., Mohd Zain R., Tilakaratne W.M., Lew H.L. Expression of Ki-67, Cornulin and ISG15 in non-involved mucosal surgical margins as predictive markers for relapse in oral squamous cell carcinoma (OSCC) PLoS One. 2021;16 doi: 10.1371/journal.pone.0261575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hou B., Li W., Xia P., Zhao F., Liu Z., Zeng Q., et al. LHPP suppresses colorectal cancer cell migration and invasion in vitro and in vivo by inhibiting Smad3 phosphorylation in the TGF-β pathway. Cell Death Discov. 2021;7:273. doi: 10.1038/s41420-021-00657-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xiao H., Langerman A., Zhang Y., Khalid O., Hu S., Cao C.X., et al. Quantitative proteomic analysis of microdissected oral epithelium for càncer biomarker discovery. Oral Oncol. 2015;51:1011–1019. doi: 10.1016/j.oraloncology.2015.08.008. [DOI] [PubMed] [Google Scholar]
- 67.Pawar H., Maharudraiah J., Kashyap M.K., Sharma J., Srikanth S.M., Choudhary R., et al. Downregulation of cornulin in esophageal squamous cell carcinoma. Acta Histochem. 2012;115:89–99. doi: 10.1016/j.acthis.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 68.Li C., Xiao L., Jia J., Li F., Wang X., Duan Q., et al. Cornulin is induced in psoriasis lesions and promotes keratinocyte proliferation via phosphoinositide 3-kinase/akt pathways. J. Invest. Dermatol. 2019;139:71–80. doi: 10.1016/j.jid.2018.06.184. [DOI] [PubMed] [Google Scholar]
- 69.Lv M., Luo L., Chen X. The landscape of prognostic and immunological role of myosin light chain 9 (MYL9) in human tumors. Immun. Inflamm. Dis. 2022;10:241–254. doi: 10.1002/iid3.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Amici J.M., Dousset L., Battistella M., Vergier B., Bailly J.Y., Cogrel O., et al. Clinical factors predictive for histological aggressiveness of basal cell carcinoma: a prospective study of 2274 cases. Ann. Dermatol. Venereol. 2021;148:23–27. doi: 10.1016/j.annder.2019.10.028. [DOI] [PubMed] [Google Scholar]
- 71.Waldman A., Schmults C. Cutaneous squamous cell carcinoma. Hematol. Oncol. Clin. North Am. 2019;33:1–12. doi: 10.1016/j.hoc.2018.08.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data and the processed files were uploaded to the PRIDE repository with the project accession code PXD036900.