Abstract
Herpes simplex virus type 1 (HSV-1) immediate-early protein ICP0 is a general activator of viral gene expression which stimulates the initiation of lytic infection and reactivation from quiescence and latency. The importance of ICP0 to the biology of HSV-1 infection has stimulated interest in its mode of action. Previous studies have reported its interactions with other viral regulatory molecules, with the translation apparatus, with cyclin D3, and with a ubiquitin-specific protease. It has been demonstrated that ICP0 is able to induce the proteasome-dependent degradation of a number of cellular proteins, including components of centromeres and small nuclear substructures known as ND10 or PML nuclear bodies. ICP0 has a RING finger zinc-binding domain which is essential for its functions. In view of several recent examples of other RING finger proteins which modulate the stability of specific target proteins by acting as components of E3 ubiquitin ligase complexes, this study has explored whether ICP0 might operate via a similar mechanism. Evidence that the foci of accumulated ICP0 in transfected and infected cells contain enhanced levels of conjugated ubiquitin is presented. This effect was dependent on the RING finger region of ICP0, and comparison of the properties of a number of ICP0 mutants revealed an excellent correlation between previously established functions of ICP0 and its ability to induce concentrations of colocalizing conjugated ubiquitin. These results strongly support the hypothesis that a major factor in the mechanism by which ICP0 influences virus infection is its ability to induce the degradation of specific cellular targets by interaction with the ubiquitin-proteasome pathway.
A definitive feature of herpes simplex virus type 1 (HSV-1) is the contrast between episodic lytic infection in the epithelia and periods of latency when the viral genome resides in a mainly quiescent state in neurons (for general reviews, see reference 22). Lytic infection involves the initial synthesis of immediate-early (IE) regulatory proteins: ICP4 functions at the level of transcription by interacting with components of the basal transcription apparatus (4), ICP27 is a multifunctional protein which acts at both the transcriptional and posttranscriptional levels (reviewed in reference 45), ICP22 enhances levels of late gene expression in selected cell types (49), and ICP0 enhances gene expression from all classes of viral genes (reviewed in references 11 and 13). The combined activities of the IE proteins lead to transcription of the early and late classes of genes throughout the whole viral genome. In contrast, abundant transcription during latency is limited to the family of latency-associated transcripts of uncertain function (reviewed in reference 55).
Attempts to understand the basis of the contrasting patterns of viral transcription during lytic and latent infection have concentrated on the properties of the IE regulatory proteins. ICP0 was originally defined as a promiscuous activator of gene expression in transfection assays, but then it was found that HSV-1 mutants deficient in functional ICP0 have a decreased probability of initiating the lytic cycle, especially in low-multiplicity infections of limited-passage human fibroblasts. Infection under these conditions appears to result in repression of viral transcription and establishment of quiescent viral genomes which are refractory to activation by transcriptional activators such as VP16 and ICP4. However, provision of exogenous ICP0 allows reactivation of the quiescent genomes and entry into a normal lytic cycle (reviewed in references 13 and 47). The results of more recent studies using viruses with lesions in several IE genes have strengthened the hypothesis that ICP0 might be involved in the control of the balance between lytic and latent infection, such that in its absence the latent state is favored (46, 48). These conclusions have been further supported by the finding that in the absence of VP16 to stimulate IE gene expression, inactivation of ICP0 leads to a profound defect in the ability to proceed to lytic infection (40).
The mechanism by which ICP0 achieves these effects has been the subject of considerable controversy and speculation. It has been shown that ICP0 results in increased transcriptional activity of the viral genome, rather than acting via posttranslational effects on viral protein expression (28), but this could be by either direct or indirect mechanisms. Direct transcriptional activators either bind to specific response elements in target promoters or interact with host transcription factors which form part of the basal transcriptional machinery or associated activator complexes. As yet there is no clear evidence for ICP0 exhibiting either of these properties; indeed, ICP0 does not bind to DNA efficiently (12). Viral proteins which modulate gene expression frequently function by interaction with cellular proteins; in the case of ICP0, interactions with other viral regulatory molecules, translation elongation factor EF-1δ, cyclin D3, and the ubiquitin-specific protease USP7 have been reported (17, 29, 30, 54, 60). It has also been established that ICP0 associates with and then disrupts specific nuclear substructures known as ND10, promyelocytic leukemia oncogenic domains (PODs), or PML nuclear bodies (reviewed in references 10 and 36). Similar events occur at centromeres, with profound consequences to the cell cycle and cell viability (20, 33).
The destruction of ND10 and centromeres during HSV-1 infection is caused by the ICP0-induced, proteasome-dependent degradation of cellular proteins that are important components of these structures (5, 18, 41, 44); these events are directly caused by ICP0 alone (15, 20, 41, 44). Initial studies found that a specific class of cellular proteins (those modified by the ubiquitin-like protein SUMO-1) were preferential targets for ICP0-induced degradation (4, 18), but not all proteins that can be modified by SUMO-1 are degraded in response to ICP0 expression, while other proteins which are not known to be so modified are also targeted by ICP0 (20, 43; P. Lomonte and R. D. Everett, submitted). Therefore, the mechanism by which ICP0 induces proteolysis of specific target proteins remains an important issue.
Mutagenesis studies from several laboratories have consistently found that a zinc-binding RING finger domain and surrounding sequences near the N terminus of ICP0 are essential for its functions in regulating gene expression, stimulating lytic infection and reactivation from quiescence, disruption of ND10 and centromeres, induced degradation of cellular proteins, and binding to and stabilization of cyclin D3 (9, 15, 16, 20, 25, 54, 56). RING finger domains have been found in a large and diverse number of proteins from many different organisms, but until recently there has been little indication that these domains could have related functions. However, a spate of reports have described a variety of RING finger proteins being involved with the control of the stability of specific target proteins by virtue of their association in ubiquitin ligase complexes (reviewed in references 3 and 23). The process of proteasome-mediated proteolysis involves a cascade of enzyme activities, which results in the formation of polyubiquitin chains on the target protein; these polyubiquitinated proteins are then recognized by the proteasome and degraded. The final step in the ubiquitination pathway often involves a complex termed an E3 or ubiquitin ligase, which consists of the substrate, an E2 ubiquitin-conjugating enzyme (Ubc), and one or more other proteins which bind the substrate, the Ubc, or both (58). Several RING finger proteins are now known to be involved in ubiquitination, either by their presence in known E3 complexes, by their interaction with one or more Ubcs, or by biochemical characterization of ubiquitin ligase activity. In all cases the RING finger was essential for the activity (3, 23).
Because of the compelling analogies between the role of ICP0 in the control of protein stability and the recent studies of other RING finger proteins with ubiquitin ligase activity, the possibility that ICP0 could have a similar activity was investigated. These studies were enabled by the use of monoclonal antibodies (MAbs) which recognize conjugated but not free ubiquitin. It was found that ICP0 induces a striking accumulation of conjugated ubiquitin at ND10, centromeres, and other locations where ICP0 accumulates and that the RING finger region of ICP0 was essential for this activity. Investigation of the ability of an extensive array of insertion and deletion mutations in the N-terminal third of ICP0 to induce colocalization of conjugated ubiquitin demonstrated a clear correlation between this activity and previously characterized functions in regulation of gene expression and stimulation of viral lytic infection. The data strongly support the hypothesis that ICP0 functions through the ubiquitin-proteasome pathway to induce the targeted destruction of specific cellular proteins.
MATERIALS AND METHODS
Viruses and cells.
HSV-1 strain 17+ and derivatives with defined lesions in ICP0 have been described previously: FXE, D22, and E32 (9); E52X and D12 (37); K144E, Q148E, and N151D (16); M1 (21); and the enhanced green fluorescent protein (EGFP)-linked ICP0 derivatives vEG-110 (wild-type ICP0), vEG-FXE, vEG-M1, vEG-D12, and vEG-E52X (33). ICP0 null mutant dl1403 (53) and the EGFP-expressing derivative vEG-dl110 (33) were also used. The characteristics of the ICP0 lesions in these viruses are listed in Table 2. Virus stocks were propagated and their titers were determined in baby hamster kidney (BHK) cells that were grown in Glasgow modified Eagle's medium containing 10% newborn calf serum (NBCS), penicillin (100 U/ml), and streptomycin (100 μg/ml). HEp-2 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and antibiotics as described above. Human fetal lung (HFL) fibroblasts and osteosarcoma U2OS cells were grown in Dulbecco's modified Eagle's medium with 5% fetal calf serum, 5% NBCS, 1% nonessential amino acids, and antibiotics as described above.
TABLE 2.
Properties of ICP0 proteins expressed by viruses used in this studya
| Virus | EGFP version | ICP0 structure | Comments |
|---|---|---|---|
| 17+ | vEG-110 | 1–775 | Wild type |
| dl1403 | vEG-dl110 | 1–105 | Null mutant |
| FXE | vEG-FXE | 1–105::150–775 | RING finger deletion |
| D22 | NAb | 1–161::189–775 | RING finger distal region deletion |
| E32 | NA | 4-aac insertion at 222 | Insertion in RING finger distal region |
| K144E | NA | aa 144 point mutation | Substitution in RING finger helix |
| Q148E | NA | aa 148 point mutation | Substitution in RING finger helix |
| N151D | NA | aa 151 point mutation | Substitution in RING finger helix |
| M1 | vEG-M1 | R623L, K624I | USP7 binding negative substitution |
| D12 | vEG-D12 | 1–593::634–775 | USP7 binding region deletion |
| E52X | vEG-E52X | 1–593 | C-terminal truncation. |
Plasmids and transfection.
Plasmids expressing wild-type or mutant forms of ICP0 were either from the pCI series, which uses the human cytomegalovirus promoter-enhancer (21), or the p110 series, which uses the HSV-1 IE-1 transcriptional control signals. The plasmids were originally described as follows: p110K144E, p110Q148E, p110N151D, p110DR36c, and p110DR40 (16, 42); p110F1, p110R2, p110E8, p110E13, p110R3, and p110E32-1 (7); and p110D13/32 (8). Plasmid pCIQEUb52 was constructed by taking the coding region of the ubiquitin precursor protein Ub52 from plasmid pGEX-Ub52 (17), fusing it to the MRGS-6×His epitope tag of plasmid pQE30 (Qiagen) to produce an N-terminally tagged ubiquitin precursor gene, and then inserting this fusion cDNA into vector plasmid pCIneo (Promega). Plasmid pCW7 (kindly provided by C. Ward and R. Kopito) expresses a myc-tagged monoubiquitin protein which can be utilized in the ubiquitination pathway without cleavage from a precursor. Plasmids were transfected into HEp-2 cells with Lipofectamine PLUS reagent (GibcoBRL), using 0.4 μg of total DNA and 1 μl of reagent per 105 cells. Transfected cells were fixed and stained for immunofluorescence about 24 h later.
Antibodies.
MAbs FK1 and FK2, which recognize conjugated but not free ubiquitin (24), were obtained from International Bioscience, Inc. Anti-ICP0 MAb 11060 (14), anti-PML rabbit serum r8 (2), anti-ICP0 polyclonal rabbit serum r190 (43), and anti-Sp100 polyclonal rabbit serum SpGH (52) have been described previously. MAb MRGS, which recognizes the MRGS-6×His tag, was obtained from Qiagen, and MAb 9E10, which recognizes the myc tag, was obtained from Santa Cruz Biotechnology, Inc. Secondary antibodies were used at the indicated dilutions: fluorescein isothiocyanate-conjugated sheep anti-rabbit immunoglobulin G (IgG) (1/100) and goat anti-mouse IgM (1/100) (Sigma), Cy3-conjugated goat anti-mouse IgG (1/500) and goat anti-rabbit IgG (1/5,000), and Cy5-conjugated goat anti-rabbit IgG (1/500) (all from Amersham).
Immunofluorescence.
HEp-2 or HFL cells were seeded onto coverslips in Linbro wells at a density of 105 cells per well 1 day prior to infection or transfection. After appropriate infection or transfection, cells were fixed with formaldehyde (5% [vol/vol] in phosphate-buffered saline [PBS] containing 2% sucrose) and then permeabilized with 0.5% NP-40 in PBS with 10% sucrose. The primary antibodies were diluted in PBS containing 1% NBCS. Antibodies were used at the following dilutions: 11060, 1/1,000; r8, 1/1,000; r190, 1/200; FK1, 1/1,000; FK2, 1/10,000; SpGH, 1/1,000; MRGS, 1/1,000; and 9E10, 1/1,000. After incubation at room temperature for 1 h, the coverslips were washed at least six times and then treated with secondary antibodies. After a further 60-min incubation, the coverslips were again washed at least six times and mounted using Citifluor AF1. Experimental combinations were checked for artifactual results by using different combinations of primary and secondary antibodies, by not adding one of the secondary antibodies in dual-labeling experiments, by repeating experiments using different combinations of fluorophores, and frequently by using EGFP-linked ICP0 proteins as an alternative approach, so that staining with only one antibody was necessary.
Confocal microscopy.
Samples were examined using a Zeiss LSM 510 confocal microscope with three lasers giving excitation lines at 633, 543, and 488 nm. The data from the channels were collected sequentially using the appropriate band-pass filters built into the instrument. Data were collected with fourfold averaging at a resolution of 1,024 × 1,024 pixels, using optical slices of between 0.5 and 1 μm. The microscope was a Zeiss Axioplan utilizing a 63× objective oil immersion lens, NA 1.4. Data sets were processed using the LSM 510 software and then exported for preparation for printing using Photoshop. The scanning conditions used ensured that signal overlap between channels was essentially eliminated.
The data presented are representative of a large number of images from independent experiments. Because of space constraints, it has not been possible to show all the control data or examples of images with all the mutants analyzed and other experimental variations. Examples of “data not shown” are of a quality similar to that of those presented. Greyscale images of the separate channels have been presented, because at the level of resolution required it is often difficult to reproduce the necessary detail and color balance due to the processes used for CMYK color printing. The original color images of the double- and triple-labeled samples may be viewed at http://www.vir.gla.ac.uk/staff/everettrd/JVI832.shtml.
RESULTS
ICP0 induces colocalization of conjugated ubiquitin.
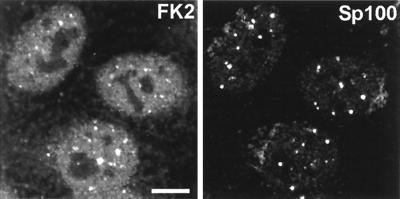
The approach that we have adopted in this study is to take advantage of MAb FK2, which specifically recognizes ubiquitin conjugated to other proteins but not free-monomer ubiquitin (24). This antibody has been extensively characterized by Western blotting and enzyme-linked immunosorbent assay (24) and has also been used to detect intracellular conjugated ubiquitin by immunofluorescence (1). In uninfected cells, free ubiquitin is limiting but rapidly turned over via conjugation to proteins targeted for proteasome-mediated degradation. The polyubiquitin chains released after degradation of the substrate are cleaved by the action of ubiquitin isopeptidases, thereby regenerating monomeric ubiquitin. Most of the conjugated ubiquitin detected by MAb FK2 is generally spread throughout HEp-2 cells, but there are local accumulations which vary in intensity from cell to cell (Fig. 1). In agreement with the results of Anton et al. (1), double staining with rabbit sera recognizing ND10 protein Sp100 showed that in most cells there are small amounts of conjugated ubiquitin in association with a proportion of ND10 domains (Fig. 1).
FIG. 1.
Distribution of conjugated ubiquitin in uninfected HEp-2 cells and its presence in some ND10. HEp-2 cells were simultaneously stained for conjugated ubiquitin with MAb FK2 (left panel) and for the ND10 component Sp100 with rabbit serum SpGH (right panel). A proportion of the major ND10 foci contain local accumulations of conjugated ubiquitin. Bar, 10 μm. The magnifications of most panels in other figures are similar, but in all cases the average width of a HEp-2 cell nucleus is 10 to 15 μm. The original colored images of this and all other multichannel figure panels may be viewed at http://www.vir.gla.ac.uk/staff/everettrd/JVI832.shtml.
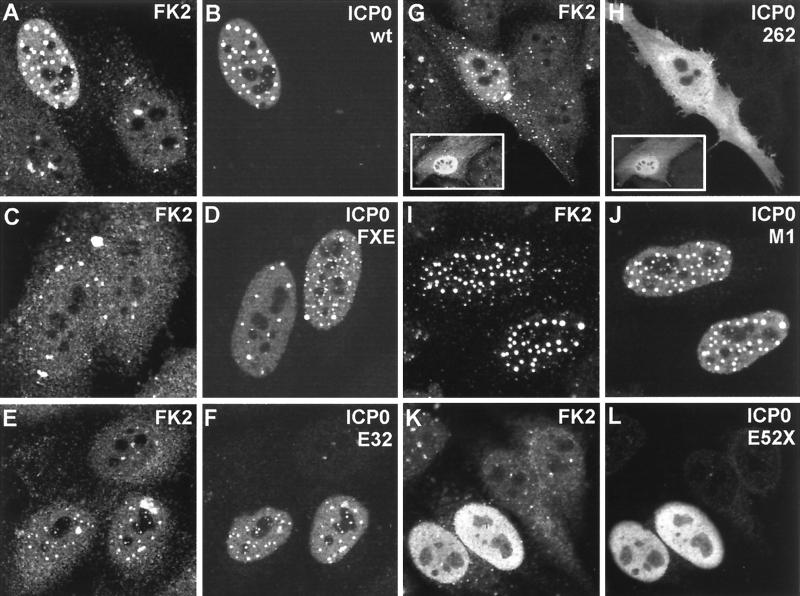
Transfection of HEp-2 cells with plasmid pCI110, which expresses wild-type ICP0, caused the dramatic appearance of conjugated ubiquitin staining which colocalized with all punctate ICP0 foci (Fig. 2A and B). Since previous data have shown that ICP0 induces the proteasome-dependent degradation of several cellular proteins (18, 20, 43) and given that there have been several recent reports of RING finger proteins being components of E3 ubiquitin ligase complexes (3, 23), a likely explanation of this result is that ICP0 is inducing the conjugation of ubiquitin to substrate proteins.
FIG. 2.
Expression of ICP0 in transfected cells leads to accumulation of colocalizing conjugated ubiquitin. HEp-2 cells were transfected with plasmids expressing wild-type and mutant forms of ICP0 and then stained to detect ICP0 with rabbit serum r191 (r95 for plasmids 110262 and E52X) and conjugated ubiquitin with MAb FK2. Each pair of panels shows the FK2 (left) and r191 (right) staining of the same field of view. (A and B) Wild-type ICP0. Note the difference in FK2 staining between the transfected (upper left) and untransfected (lower two) cells. Superimposition of the two images in the original data showed precise colocalization of the FK2 and ICP0 foci. (C and D) ICP0 RING finger deletion mutant FXE. (E and F) Exon 2 insertion mutant E32. The transfected cells are clearly distinguishable from the untransfected cells in the FK2 staining, but the result is not as striking as with the wild-type protein. (G and H) Exon 1 and exon 2 truncation mutant 110262 (ICP0R). The transfected cells have clearly altered FK2 staining patterns that in some cells had a punctate character (main panel) but was sometimes diffuse (inset). The 110262 staining pattern was generally diffuse throughout the whole cell. (I and J) USP7 binding-deficient point mutant M1. (K and L) C-terminal truncation mutant E52X. Although this mutant has a generally diffuse nuclear staining pattern, the transfected cells were easily distinguishable by examining the FK2 staining.
The RING finger of ICP0 is essential for induction of colocalizing conjugated ubiquitin.
Transfection of HEp-2 cells with plasmids expressing mutant forms of ICP0 demonstrated that the RING finger and sequences downstream of this motif in the second exon were essential for the induction of an increased amount of colocalizing conjugated ubiquitin. Because RING finger mutants of ICP0 do not disrupt ND10 domains (15) and some conjugated ubiquitin is normally present in these domains (Fig. 1), cells expressing the FXE mutant protein exhibited a degree of colocalization of the two signals. However, comparison with cells expressing the wild-type protein clearly showed that the amount of conjugated ubiquitin colocalizing with the FXE mutant protein was trivial compared to that in pCI110-transfected HEp-2 cells (Fig. 2C and D).
Analysis in plasmid transfection assays of a number of previously characterized insertion, deletion, and substitution mutants in the second exon of ICP0 showed that the severity of loss of function in other assays gave an excellent correlation with the ability of these mutants to induce accumulations of conjugated ubiquitin (Table 1). Notably, the mutations in the RING finger and distal region which had been found previously to affect ICP0 function all essentially eliminated the prominent colocalizing conjugated ubiquitin signal seen with the wild-type protein. In contrast, mutant W146A retains wild-type levels of activity in all assays, while mutant Q148E is positive but less active than the wild type in all assays. Mutant E32-1 was one of the least deleterious mutants described in an early cohort of ICP0-defective viruses (9), and this mutant protein was clearly active, but to a lesser degree than the wild type, in these latest experiments (Fig. 2E and F; Table 1). Coupled with the observation that a truncated form of ICP0 containing the 241 residues derived from the first and second exons (plus 21 residues from the second intron) induced accumulations of conjugated ubiquitin in transfected cells (Fig. 2G and H), this result places the C-terminal boundary of the active domain around the last 20 residues encoded by exon 2. We conclude that expression of ICP0 over several hours in transfected cells leads to significant accumulations of colocalizing conjugated ubiquitin and that this activity correlates with the function of the RING finger region in other assays. Because a range of different insertion, deletion, and substitution mutants fail to induce the colocalizing conjugated ubiquitin signal, these results discount the possibility that MAb FK2 cross-reacts with an epitope in the ICP0 RING finger, unless the cross-reacting epitope is conformational and disrupted by all of the defective mutants analyzed.
TABLE 1.
Plasmids used in this study
| Name | Mutationf | Activationa | Repressionb | Increased FK2 detection of conjugated ubiquitinc |
|---|---|---|---|---|
| pCI110 | Wild type | +++ | NAd | +++ |
| pCIFXE | del 106–149 | − | − | − |
| p110DR40 | del 129–130 | − | NDe | − |
| p110K144E | Substitution | − | ND | − |
| p110W146A | Substitution | +++ | ND | +++ |
| p110Q148E | Substitution | ++ | ND | + |
| p110N151D | Substitution | + | ND | − |
| p110F1 | ins 150 | − | − | − |
| p110D22 | del 162–188 | − | − | − |
| p110R2 | ins 162 | − | − | − |
| p110E8 | ins 188 | − | − | − |
| p110E13 | ins 197 | − | − | − |
| p110R3 | ins 212 | − | − | − |
| p110E32-1 | ins 222 | − | − | + |
| p110D13/32 | del 197–222 | ND | ND | − |
| p110262 | 1–241 | ND | + | ++ |
| pCIM1 | R623L, K624I | − | NA | +++ |
| p110D12 | del 594–633 | + | NA | +++ |
| p110E52X | 1–593 | ++ | NA | +++ |
Data taken from references 8, 16, 21, and 42. The effects of the mutations on the activation of a target expression cassette by ICP0 alone were estimated, using slightly different conditions in the three series of experiments; −, less than 10% of wild-type activity; +, 10 to 40%; ++, 40 to 70%; +++, >70%.
Truncated forms of ICP0 containing the RING finger region repress expression from reporter constructs (50). p110262 is a plasmid equivalent to the repression construct used in those studies; + and − indicate the effects of these same mutations in this region in their assay.
Ability of plasmid construct to induce increased conjugated ubiquitin staining detected by MAb FK2 in transfected cells.
NA, not applicable.
ND, not done.
ins, insertion; del, deletion.
Variants of ICP0 with lesions in the C-terminal region are also relevant to this study because a motif in the region 594 to 633 has been shown to bind very strongly to the cellular ubiquitin-specific protease USP7 (17). Mutant D12 has a deletion of this region, mutant M1 has a double substitution in residues essential for binding to USP7, and mutant E52X has a deletion of the whole C-terminal end of Vmw110, which is required for efficient localization at ND10. All these mutants strongly induced the conjugated ubiquitin signal (Table 1), demonstrating that the ability to bind USP7 is not required for this activity. Mutant M1 gave strong colocalization of conjugated ubiquitin in nuclear foci (Fig. 2I and J), while the diffuse nuclear localization of mutant E52X correlated with a significantly increased nuclear conjugated ubiquitin signal (Fig. 2K and L). The issue of why ICP0 apparently associates with opposing functions in its RING finger and C-terminal regions (the former induces colocalizing conjugated ubiquitin while the latter binds to a ubiquitin-specific protease) is considered below (see Discussion).
Concentration of colocalizing epitope-tagged ubiquitin induced by ICP0.
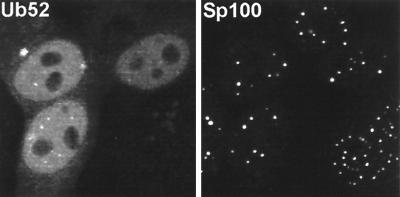
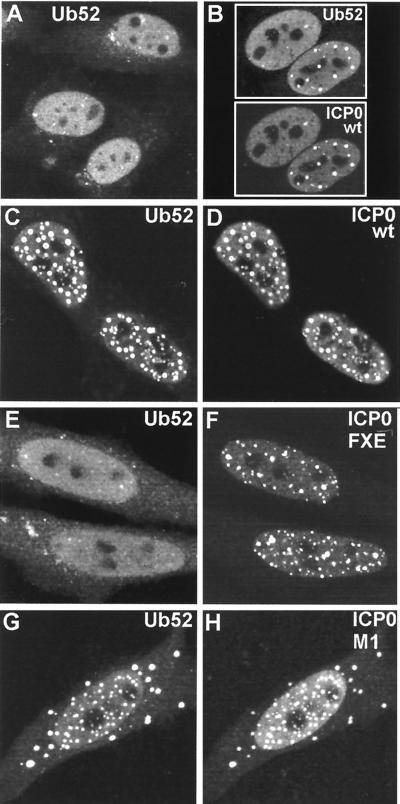
Although the reactivity of MAb FK2 has been established previously, interpretation of the above results depends on the authenticity of its recognition of conjugated ubiquitin in cells fixed for immunofluorescence. As an alternative approach, we transfected HEp-2 cells with plasmid pCIQEUb52, which expresses the Ub52 ubiquitin precursor protein linked to an N-terminal MRGS epitope tag (17). Providing that expression levels were limited by using small amounts of plasmid DNA, transfected cells exhibited a generally diffuse distribution of tagged ubiquitin throughout the cell but particularly in the nucleus (Fig. 3). Interestingly, some cells exhibited brighter foci embedded in the nuclear signal, and these colocalized with ND10 (Fig. 3). Cotransfection of pCIQEUb52 with pCI110 caused a dramatic rearrangement of the tagged ubiquitin signal, which now strongly colocalized with punctate ICP0 foci when present (Fig. 4B to D). This result did not occur with the RING finger mutant FXE (Fig. 4E and F); the faint colocalization of the FXE mutant with the tagged ubiquitin shown in the upper cell of Fig. 4E can be attributed to the presence of the mutant ICP0 at undisrupted ND10 and the presence of tagged ubiquitin in some ND10 in singly transfected cells, as described above. Cells cotransfected with mutant M1 frequently had both cytoplasmic and nuclear foci, and these colocalized with the tagged ubiquitin (Fig. 4G and H). The precise parallels between the results for Fig. 2 and 4 obtained by using both endogenous and exogenous ubiquitin and different antibodies strongly support the interpretation that ICP0 induces accumulations of colocalizing conjugated ubiquitin. Furthermore, similar results were obtained in cotransfection experiments using plasmid pCW7, which expresses myc-tagged monomeric ubiquitin, which can be conjugated to substrate proteins directly, without prior cleavage from a precursor (data not shown).
FIG. 3.
Distribution of an epitope-tagged ubiquitin precursor in transfected HEp-2 cells. Cells transfected with plasmid pCIQEUb52 were stained simultaneously with MAb MRGS to detect the tagged ubiquitin (left panel) and with SpGH to detect Sp100 (right panel). MAb MRGS gives no significant signal in the untransfected cells (upper and lower cells on the right of the left panel) and a diffuse nuclear distribution with some local accumulations which often coincide with ND10.
FIG. 4.
Cotransfection with plasmids expressing ICP0 and a ubiquitin precursor leads to colocalization of accumulations of the two proteins. HEp-2 cells were cotransfected with pCIQEUb52 (expressing the tagged ubiquitin precursor Ub52) and either vector alone (A) or plasmids expressing wild-type and mutant forms of ICP0 (B to H, as marked). (A) Tagged ubiquitin in singly transfected cells has a diffuse nuclear distribution with a number of small foci, some of which colocalize with ND10 (not shown). (B) Cells expressing ICP0 have a variable degree of punctate accumulation of ICP0; in cotransfected cells the ubiquitin precursor colocalizes with the punctate ICP0 foci. wt, wild type. (C to H) Each pair of images shows the tagged ubiquitin staining (left) and the ICP0 staining (right) of the same field of view. (C and D) Wild-type (wt) ICP0; (E and F) RING finger mutant FXE; (G and H) USP7 binding mutant M1.
The striking colocalization depicted here was achieved by using relatively small amounts of the plasmid expressing the tagged ubiquitin. If larger amounts of this plasmid were used, the highly expressed additional free ubiquitin was spread throughout the cell and a smaller proportion of the tagged ubiquitin colocalized with ICP0 (data not shown). This result illustrates that the activity of ICP0 in sequestering ubiquitin in its vicinity can be saturated or is dependent on the relative amounts of ICP0 and free ubiquitin available.
ICP0 induces the accumulation of polyubiquitin chains at ND10 and centromeres during the early stages of virus infection.
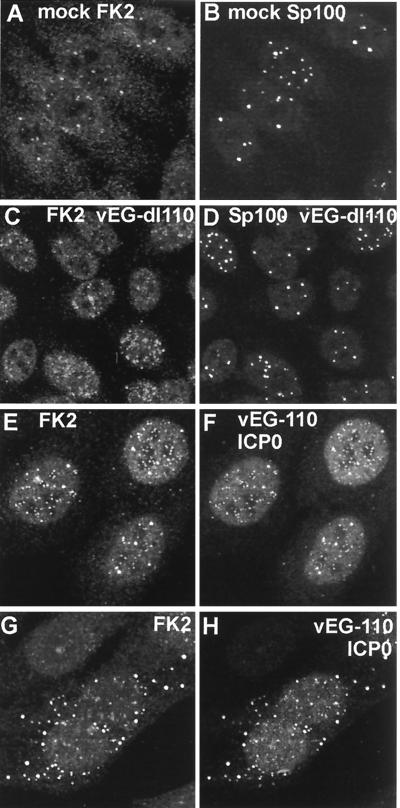
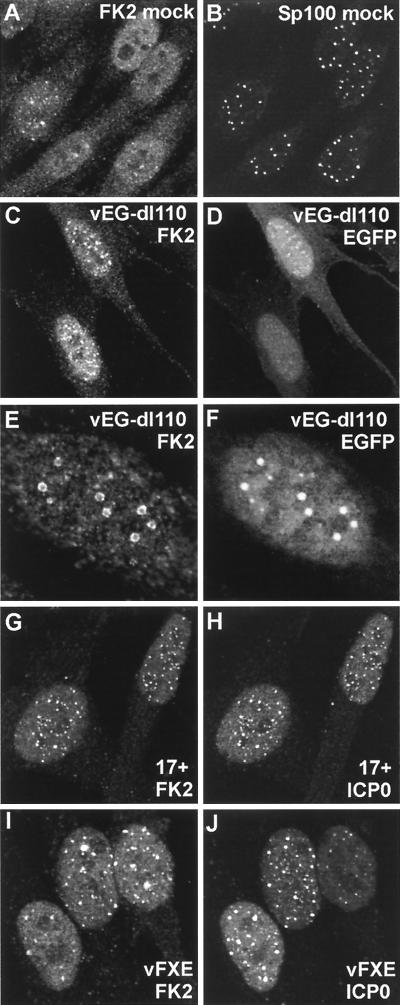
We next considered the relevance of the results described above to ICP0 function during the early stages of HSV-1 infection. Since virus infection induces a stress response and expression of ICP4 has been shown to induce transcription of a ubiquitin precursor gene (32), it was first necessary to assess the effect of HSV-1 infection on the distribution of conjugated ubiquitin by using an ICP0-deficient virus. Mutant vEG-dl110 expresses EGFP from the IE-1 promoter in place of ICP0 (33) and is therefore very convenient for these experiments, since it is ICP0 negative and expresses EGFP at IE times. As noted above, uninfected cells showed a variable number of nuclear foci of conjugated ubiquitin, some of which colocalized with the ND10 protein Sp100 (Fig. 5A and B). Infection with virus vEG-dl110 changed the distribution and apparent amount of conjugated ubiquitin as early as 2 h postabsorption, with some (but not all) cells having increased numbers of blotchy foci. However, in general these did not coincide with ND10 (Fig. 5C and D).
FIG. 5.
Effect of HSV-1 infection on the distribution of conjugated ubiquitin in HEp-2 cells at 2 h postinfection. Each pair of images shows the same field of view with the conjugated ubiquitin (FK2) staining on the left (A, C, E, G) and either ND10 protein Sp100 (B and D) or ICP0 (F and H) staining on the right. (A and B) The distribution of conjugated ubiquitin in uninfected cells. A few foci are embedded in a generally diffuse distribution throughout the cell. Superimposition of the two images showed that some of the conjugated ubiquitin foci colocalize with ND10 (not shown). (C and D) Infection with ICP0 null mutant vEG-dl110 results in increased numbers of conjugated ubiquitin foci in the nucleus. The original triple-labeled image with the EGFP signal to identify infected cells is available on http://www.vir.gla.ac.uk/staff/everettrd/JVI832.shtml. (E and F) Infection with virus vEG-110 expressing wild-type ICP0 fused to EGFP results in induction of conjugated ubiquitin foci that colocalize with ICP0. Infected cells were easily detected on the basis of the FK2 staining alone. (G and H) When ICP0 accumulates in foci in the cytoplasm, it induces the formation of colocalizing conjugated ubiquitin here also.
The changes in polyubiquitin distribution brought about by virus infection itself complicate the interpretation of additional changes caused by ICP0 activity. However, infection of HEp-2 cells with virus vEG-110, which expresses wild-type ICP0 fused to EGFP (33), caused changes in polyubiquitin distribution which were clearly distinguishable from distribution in both uninfected and vEG-dl110-infected cells. The foci of ICP0 were very frequently associated with an increased conjugated ubiquitin signal (Fig. 5E and F), and at early times of infection before ND10 domains were disrupted, there was excellent triple colocalization of ICP0, Sp100, and conjugated ubiquitin at ND10 (Fig. 6A to C). Later in infection ICP0 forms cytoplasmic foci in some cells; these foci also contained significant amounts of colocalizing conjugated ubiquitin (Fig. 5G and H). This effect of ICP0 inducing association of conjugated ubiquitin was particularly striking at centromeres in infected mitotic cells (Fig. 7A and B), and other triple-staining experiments showed that ICP0 induced the appearance of conjugated ubiquitin at interphase centromeres as well (Fig. 6D to F). The accumulation of conjugated ubiquitin at centromeres of infected cells could also be detected by MAb FK1, another antibody specific for conjugated ubiquitin (Fig. 7C and D). Neither FK1 nor FK2 detected foci of conjugated ubiquitin at centromeres of uninfected cells (Fig. 7E and F). Furthermore, in a rare example of a HEp-2 cell transfected with plasmid pCIQEUb52 and attempting mitosis after infection with HSV-1 strain 17+, the tagged ubiquitin was present at centromeres, just like the conjugated ubiquitin detected by FK1 and FK2 (Fig. 7G and H). These results correlate very well with the proteasome-dependent degradation of centromeric protein CENP-C induced by ICP0 (20). Interestingly, polyubiquitin was detectable in centromeres in infected mitotic cells at later times of infection when CENP-C would have been degraded, which suggests either that there may be other centromere components which are targeted for degradation by ICP0 or that the conjugated ubiquitin induced by ICP0 is only slowly turned over. Thus, cells infected with viruses expressing active ICP0 were clearly distinguishable from uninfected cells and vEG-dl110-infected cells by the increased number and intensity of conjugated ubiquitin foci in the nucleus and sometimes in the cytoplasm (which closely colocalized with ICP0) and by the clear induction of conjugated ubiquitin in mitotic centromeres.
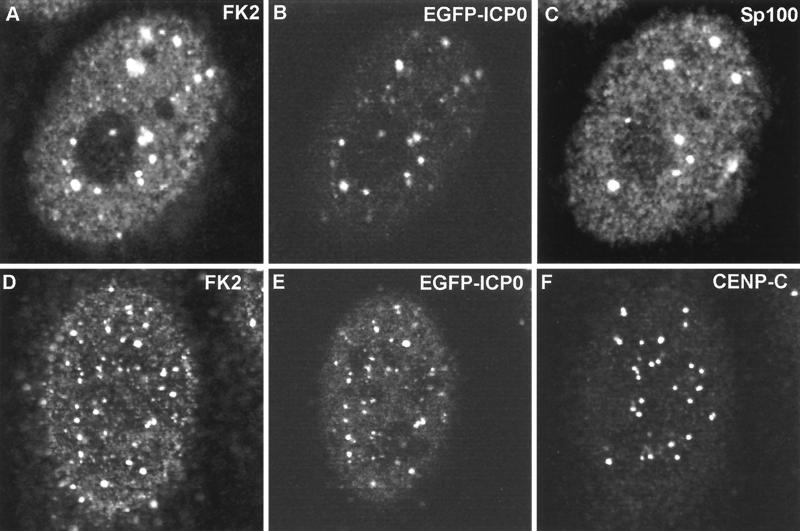
FIG. 6.
Triple colocalization of ICP0 and conjugated ubiquitin at ND10 and interphase centromeres in the early stages of infection. HEp-2 cells were infected with vEG-110 and stained for FK2 and either Sp100 or CENP-C as indicated. ICP0 was detected by the linked EGFP signal. All the major Sp100 foci (C) contain ICP0 (B), and all the ICP0 foci (B) react with MAb FK2 (A). In a certain proportion of cells, ICP0 also colocalizes with interphase centromeres, as shown here (D through F). The major ICP0 foci (E) are likely to be at ND10, but many of the minor foci are at centromeres (F). Of the 27 centromere foci visible in this confocal optical slice (F), at least 11 both contain ICP0 (E) and react with MAb FK2 (D). Because of the difficulty of reproducing the correct color balance of triple-colored images in printed journals, the single-channel data are shown here in greyscale. The original triple-colored images may be viewed at http://www.vir.gla.ac.uk/staff/everettrd/JVI832.shtml.
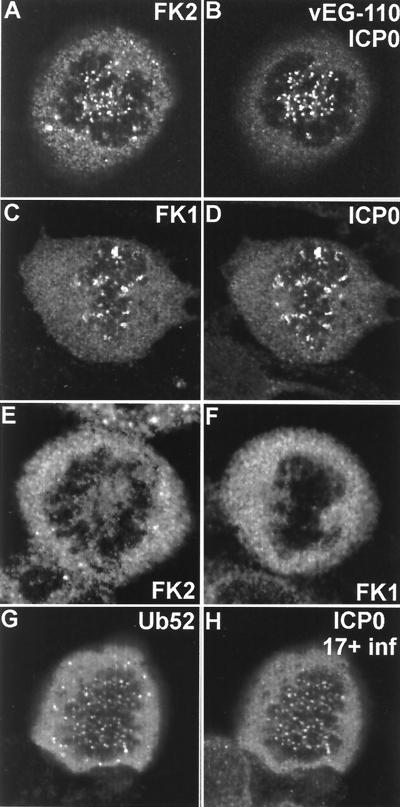
FIG. 7.
ICP0 induces the accumulation of conjugated ubiquitin at mitotic centromeres. (A and B) A mitotic cell infected with vEG110 and stained with MAb FK2. (C and D) A mitotic cell infected with HSV-1 strain 17+ costained with MAb FK1 and r191 to detect ICP0. (E and F) Corresponding uninfected controls. (G and H) HEp-2 cells singly transfected with pCIQEUb52 and later infected with HSV-1 strain 17+. This view shows a rare transfected cell that was infected in the late G2 phase of the cell cycle and has arrested in mitosis with the ICP0-induced localization of tagged ubiquitin at centromeres.
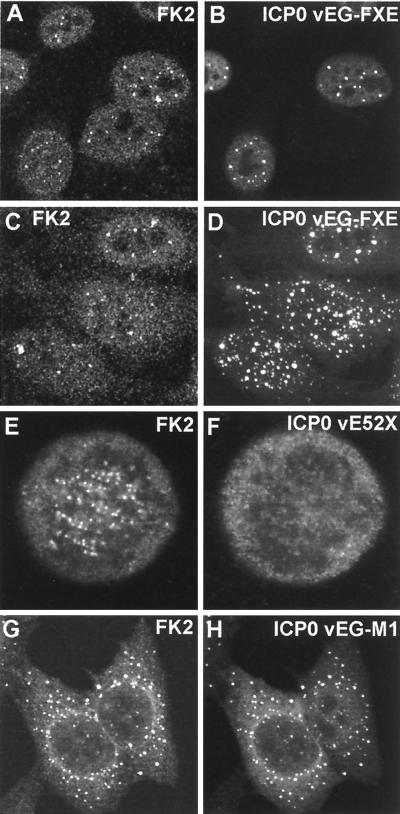
A number of viruses with lesions throughout ICP0 were available for comparison of the results of the transfection experiments detailed above with the situation during virus infection of HEp-2 cells; in all cases the results were analogous. Like vEG-dl110, virus vEG-FXE expressing a GFP-tagged ICP0 RING finger deletion caused general changes in the overall distribution of polyubiquitin in some cells as infection progressed. However, the conjugated ubiquitin was relatively weakly associated with the mutant ICP0 protein (Fig. 8A and B) at sites that correspond to undisrupted ND10 (data not shown). When the mutant ICP0 was present in the cytoplasm at later times of infection, it was not as well associated with conjugated ubiquitin as in the case of normal ICP0 (Fig. 8C and D) and never induced the appearance of conjugated ubiquitin at centromeres (data not shown). Similar results were obtained with viruses with deletion, substitution, and insertion mutations in the RING finger region which inactivated ICP0 functions in other assays (Tables 2 and 3). Viruses expressing ICP0 mutants not linked to EGFP gave results essentially identical to those obtained with the EGFP-linked variants (data not shown).
FIG. 8.
Effect of expression of mutant ICP0 proteins on the distribution of conjugated ubiquitin during HSV-1 infection of HEp-2 cells. The paired images show conjugated ubiquitin (left) and ICP0 fused to EGFP (right) except for the E52X results, which were obtained by staining for ICP0 with rabbit serum r95. (A and B) At early times of infection (2 h), RING finger mutant protein FXE accumulates at ND10 and there is variable colocalization with conjugated ubiquitin. (C and D) At later times of infection (4 h), foci of FXE protein in both nucleus and cytoplasm are less commonly associated with conjugated ubiquitin (compare with wild-type ICP0, Fig. 3I and J). (E and F) Although mutant ICP0 protein E52X inefficiently localizes at centromeres in mitotic cells, it clearly induces the accumulation of conjugated ubiquitin at these structures. (G and H) EGFP-linked ICP0 mutant M1 has a marked cytoplasmic punctate and perinuclear distribution that is mirrored by the induced colocalizing conjugated ubiquitin.
TABLE 3.
Plaque-forming ability of ICP0 mutant viruses and induction of conjugated ubiquitin at centromeres during infectiona
| Virus | Relative plaque-forming ability on:
|
Detection of conjugated ubiquitin at centromeres by FK2 | ||
|---|---|---|---|---|
| U2OS | BHK | HFL | ||
| 17+ | 1 | 0.60, 0.68 | 0.70, 0.73 | + |
| dl1403 | 1 | 0.08, 0.04 | 0.000, 0.0004* | − |
| FXE | 1 | 0.08, 0.04 | 0.006, 0.008* | − |
| D22 | 1 | 0.05, 0.03 | 0.01, 0.003* | − |
| E32 | 1 | 0.19, 0.31 | 0.20, 0.14 | + |
| K144E | 1 | 0.05, 0.10 | 0.03, 0.08 | − |
| Q148E | 1 | 0.24, 1.08 | 1.00, 1.50 | + |
| N151D | 1 | 0.10, 0.14 | 0.38, 0.68 | − |
| M1 | 1 | 0.05, 0.04 | 0.31, 0.44 | + |
| D12 | 1 | 0.08, 0.24 | 0.25, 0.74 | + |
| E52X | 1 | 0.07, 0.20 | 0.10, 0.47 | + |
Titers of virus stocks were initially determined on U2OS cells, in which HSV-1 requires little ICP0 to stimulate virus growth. Dilutions of these virus stocks were then titrated simultaneously on all three cell lines, and the ratio of the titers in BHK and HFL cells to those in U2OS cells was calculated. The values given represent the results from two totally independent determinations. Asterisks indicate that the data for these viruses in HFL cells were nonlinear (as previously reported for ICP0-deficient viruses). The data in the rightmost column indicate the ability (+) or inability (−) of the ICP0 proteins to induce the formation of conjugated ubiquitin at centromeres in infected cells.
It was found previously that the ICP0 C-terminal truncation mutant protein E52X localized at centromeres in much reduced amounts but nonetheless in sufficient quantities to induce the degradation of CENP-C (20). Consistent with this result, we found that virus E52X induced conjugated ubiquitin deposits in mitotic centromeres, even though only trace amounts of the E52X protein were present in these structures (Fig. 8E and F). The USP7 binding mutant M1 and its EGFP derivative vEG-M1 presented an additional interesting phenotype. As previously reported (21), the ICP0 protein expressed by virus M1 frequently forms foci in the cytoplasm, and in all cases these were strongly marked by colocalizing conjugated ubiquitin (data not shown). The effect was particularly marked with virus vEG-M1, since the fusion protein is retained in the cytoplasm to an even greater extent (Fig. 8G and H). However, M1 and vEG-M1 both very strongly induced the conjugated ubiquitin signal at centromeres in mitotic cells, and consistent results were obtained with the USP7 binding-defective mutant D12 (data not shown).
The consistent pattern of the results in transfected cells, cotransfected cells, and infected HEp-2 cells strongly suggests that the previously characterized functional defects caused by mutations in the ICP0 RING finger and distal exon 2 sequences correlate very well with the induction of colocalizing conjugated ubiquitin. These results are also highly consistent with the ability of ICP0 to induce the proteasome-dependent degradation of a number of cellular proteins present at ND10 and centromeres.
The distribution of conjugated ubiquitin during infection of HFL cells.
The requirement for ICP0 to stimulate virus infection varies with cell type and is particularly marked in HFL cells (53). Since the implication of the work described above is that ICP0 function correlates with the formation of colocalizing conjugated ubiquitin, it was appropriate to investigate this assay in other cell types. In uninfected HFL cells, the MAb FK2 signal gave a diffuse staining pattern embedded with micropunctate foci, some of which colocalized with ND10 (Fig. 9A and B and data not shown). Infection of HFL cells with virus vEG-dl110 resulted in a rapid increase in the focal accumulations of the MAb FK2 signal, and frequently these colocalized or were associated with ND10 (Fig. 9C and D). At later times of infection, ringlike structures of conjugated ubiquitin appeared in many cells, which in the case of virus vEG-dl110 frequently surrounded foci of the EGFP signal (Fig. 9E and F). Similar ring structures of conjugated ubiquitin could be detected in cells infected with all the mutant viruses (but never the wild type). These structures were not associated with ICP4, ICP8, or replication compartments, but they did occasionally associate with mutant ICP0 proteins (data not shown)—it is likely that they correspond to the previously observed “hollow spheres” formed in the nuclei of some ICP0 mutant-infected cells, which were characterized by the absence of interior ICP0 and DNA staining (35).
FIG. 9.
Effect of expression of mutant ICP0 proteins on the distribution of conjugated ubiquitin during HSV-1 infection of HFL cells. The panel pairs show on the left the conjugated ubiquitin staining and on the right either Sp100 staining (B), EGFP (D and F), or EGFP-linked ICP0 (H and J). (A and B) The relationship between the distributions of conjugated ubiquitin and ND10 is similar to that in HEp-2 cells. (C and D) Infection of HFL cells by ICP0 null mutant vEG-dl110 for 2 h has a more marked effect on conjugated ubiquitin than in HEp-2 cells, with more prominent induction of nuclear foci, which are commonly associated with ND10 (not shown). EGFP itself forms foci in some cells, which can be associated with the conjugated ubiquitin foci. (E and F) At later times of infection, conjugated ubiquitin forms ringlike structures in some cells, and these frequently contain internal EGFP in vEG-dl110 infections. The faint clouds of EGFP in panel F correspond to replication compartments, which are generally not associated with the conjugated ubiquitin ring structures (which can occur in all mutant ICP0 virus infections but never in wild-type infections). (G to J) At 3 h postadsorption, EGFP-linked wild-type ICP0 is strongly associated with conjugated ubiquitin foci, but the RING finger deletion mutant FXE shows a variable degree of colocalization.
Infection of HFL cells with wild-type virus strain 17+ resulted in extensive colocalization of induced conjugated ubiquitin with ICP0 foci (Fig. 9G and H, 3 h postinfection), but the situation with the RING finger mutant FXE was more complex. At the early stages of infection there appeared to be little difference between this mutant and the wild type, as foci of conjugated ubiquitin appeared at ND10 in an even more marked manner than that observed with the deletion mutant vEG-dl110 (not shown). However, by 3 h postinfection, although many foci of the mutant ICP0 colocalized with conjugated ubiquitin, there were many foci of both proteins that were separate from each other (Fig. 9I and J). As in HEp-2 cells, wild-type and EGFP-linked ICP0 induced the appearance of conjugated ubiquitin at centromeres in HFL cells, but the RING finger mutants never did (data not shown). Therefore the changes in conjugated ubiquitin distribution in HFL cells caused by virus infection itself, and the expression of the RING finger deletion mutant ICP0, complicate the interpretation of these results. It appears that virus infection induces substantial alterations in at least the location of ubiquitin metabolism in HFL cells and that ICP0 induces additional alterations which, except those at centromeres, are less clear-cut than in HEp2 cells.
Comparison of the effects of mutations in ICP0 on viral infectivity and the induction of colocalizing conjugated ubiquitin.
Although all the ICP0 mutant viruses used in the above experiments have been described before, the growth efficiencies of many of them have been compared only by the relatively crude assay of growth curves in BHK cells. Because some of the mutants, such as E32, gave intermediate results in the conjugated ubiquitin assays, a more rigorous comparison of all of the viruses was conducted. Initially, the viral stocks were titrated on U2OS cells to determine their titers in the absence of a requirement for ICP0 (61). Subsequently, virus titers on U2OS, BHK, and HFL cells were determined for all stocks in parallel. Whereas wild-type virus had titers within about a factor of 2 of each other in all cell types, the null mutant dl1403 produced 10- to 20-fold fewer plaques on BHK cells and was reduced a further 100-fold in HFL cells (Table 3). Titer reductions in the 10- to 20-fold range in BHK cells were observed for all the other viruses with lesions in the ICP0 RING finger region, except E32 and Q148E; these were the only viruses of this group which induced the formation of conjugated ubiquitin at centromeres (Table 3). Similarly, of the mutants in the RING finger region, E32 and Q148E were the most efficient at plaque formation in HFL cells. Therefore, with the possible exception of N151D (discussed below), there is an excellent correlation between the relative defects caused by lesions in the ICP0 RING finger region in the virus infection and induction of conjugated ubiquitin assays.
DISCUSSION
This study used a combination of indirect approaches to demonstrate that ICP0 stimulates the production of colocalizing conjugated ubiquitin, particularly at cellular substructures containing proteins which are known to be degraded by the ubiquitin-proteasome pathway after ICP0 expression. Coupled with previous results, these data greatly strengthen the hypothesis that a principal biochemical activity of ICP0 is to stimulate the ubiquitination and degradation of a number of cellular target proteins. The experiments using MAb FK2, which recognizes mono- and polyconjugated ubiquitin, are completely consistent with an independent approach using transfected tagged ubiquitin precursors. Furthermore, supporting data were obtained using MAb FK1, which recognizes only polyconjugated ubiquitinated proteins (24). Although MAb FK1 is a less effective reagent in immunofluorescence experiments than MAb FK2, such that increased MAb FK1 staining colocalizing with ICP0 was weaker than that with MAb FK2, analogous results were obtained in infected mitotic cells. Thus, the essential features of these results have been repeated with two independent MAbs recognizing endogenous conjugated ubiquitin and two different transfected ubiquitin precursors, each recognized by different tag MAbs.
Several RING finger proteins have been found to function by inducing the degradation of specific targets by forming an essential component of an E3 ubiquitin ligase complex. Examples include members of both the so-called RING-H2 family (which have a histidine residue in place of the more normal cysteine at the fourth zinc coordinating residue of the motif) and also the original canonical RING finger family, of which ICP0 is a member. Furthermore, interaction screens with several RING finger proteins have defined E2 Ubcs as partners, and vice versa (3, 23). These examples provide ample precedent for the proposed biochemical function of ICP0 put forward here. At first sight the suggestion that ICP0 is a component of an E3 ubiquitin ligase complex seems incongruous with its interaction with USP7, an enzyme which has an opposing activity. An analogous situation could occur with BRCA1, a RING finger protein which interacts with a ubiquitin hydrolase (27). Free ubiquitin is limiting in the cell, particularly in the nucleus (38), so it is possible that the role of USP7 is to release free ubiquitin so that it can be used for the conjugation reactions occurring elsewhere in the ICP0 complex. It is easy to envisage a structural arrangement by which USP7 activity could increase local concentrations of substrate ubiquitin without competing with an associated conjugation activity. If so, the role of USP7 binding to ICP0 could be to regulate ICP0 activity; this suggestion is consistent with previous data (21) and preliminary work on more detailed aspects of ICP0-induced degradation of specific substrates (44).
One of the notable aspects of the results is the compelling correlation between the effects of mutations in the RING finger and neighboring region on the ability of ICP0 to induce the formation of increased colocalizing conjugated ubiquitin and the phenotypes of the mutants in other functional assays. Some of the available ICP0 mutant viruses had been characterized only by the relatively crude method of growth curve comparisons in BHK cells. To enable better comparisons, particularly of the partially affected mutants, the plaque-forming efficiencies of all the mutants in both BHK and HFL cells have been reexamined in comparison with “complementing” U2OS cells, in which ICP0 mutant viruses do not exhibit a multiplicity-dependent growth defect (61). These data clarify the deficiencies of the partially affected mutants, such as Q148E and E32; such mutants gave positive results in the experiments described in this work but were clearly not as active as the wild type. The one mutant virus which is apparently inconsistent with the others is N151D. This virus grew relatively well in HFL (but not BHK) cells, although the N151D mutant ICP0 protein was clearly defective in the assays described here. It is possible that this mutant protein remains able to assemble a complex with the ubiquitination machinery and a critical target protein in HFL cells, but the change in the RING finger renders the complex inactive. In this case, the target protein would be inactivated through sequestration rather than degradation. Whatever the explanation for the properties of this mutant, overall these findings support the conclusion that the effects on conjugated ubiquitin observed here are a major factor in ICP0 function during infection.
These results also shed considerable light on a series of papers concerning the transrepression property of a truncated form of ICP0 containing sequences only from exons 1 and 2. This truncated protein, which is expressed in small quantities late in infection (14), is a potent transrepressor when expressed in high amounts in transfection assays (51, 56). The panel of mutations used in this study had been analyzed in the transrepression assay, and again the correlation between the two sets of results is excellent (Table 1). Furthermore, it had been demonstrated that the truncated form of ICP0 was modified by covalent addition of a single ubiquitin to a lysine residue derived from sequences normally excised in the second intron (57). The appearance of this ubiquitinated protein depended on the integrity of the RING finger motif, and again the correlation between the formation of monoubiquitinated mutant proteins and the activity described here is excellent (Table 1). It is likely that the truncated form of ICP0 is catalyzing its own ubiquitination (as well as, probably, that of other proteins) and that this deregulated activity causes interference with the normal activities of ICP0 and thus the transrepression.
It has been reported that wild-type ICP0 reduces the rate at which infection induces the degradation of cyclin D3 during infection via an interaction requiring sequences in the second exon downstream of the RING finger; mutation of aspartic acid residue 198 inhibited these activities and decreased ICP0 function in vivo (30, 50, 54). The data presented here add an extra consideration to the interpretation of these results. The E13 insertion at residue 197 inactivates ICP0 in the colocalizing conjugated ubiquitin assay (Table 1), so the substitution at residue 198 may have a similar effect. If so, the interpretation that the interaction with cyclin D3 is an important factor in ICP0 activity (30, 54) must be treated with caution, since this region of the protein is clearly very important in the assays described here and also in induced disruption of ND10 and centromeres (data not shown). Furthermore, it is possible that by introducing a novel and apparently powerful ubiquitination activity, the availability of substrate ubiquitin for other cellular processes may be altered. Thus, in addition to destabilizing its target proteins, by sequestering components of the ubiquitin pathway, ICP0 may decrease the rate at which normally very unstable proteins (such as cyclin D3) are turned over. This indirect effect could explain the partial stabilization of cyclin D3 during infection and perhaps may also contribute to the G1/S block caused by ICP0 (26, 33), because the turnover of other G1 cyclins may also be affected. This possibility awaits further experimentation.
A fundamental question that remains to be investigated is the identity and full catalogue of the proteins targeted for ubiquitination by ICP0. It is already known that components of ND10 and centromeres are targeted for ubiquitin-proteasome pathway degradation by ICP0, and these are likely candidate targets for a proportion of the accumulated conjugated ubiquitin observed here. The apparent absence of such modified forms of PML and CENP-C in previous experiments could be explained by the low amounts of the proteins themselves in the cell, the short-lived nature of ubiquitinated proteins, and the fact that ubiquitinated derivatives of many large proteins tend to form smears on a gel rather than a discrete ladder of ubiquitinated products (39). However, ICP0 seems to induce colocalizing conjugated ubiquitin at other locations in the cell, including the cytoplasm. There are a number of possible explanations: the proposed E3 ubiquitin ligase activity could be relatively nonspecific, and many proteins could be substrates providing that they are in the vicinity of accumulations of ICP0; the observed conjugated ubiquitin could comprise aggregates of unanchored polyubiquitin released after degradation of the substrates (this might explain why foci of ICP0, particularly in transfected cells, correspond to phase-dense bodies observable by light microscopy); and, finally, ICP0 itself might be conjugated to ubiquitin. There is no Western blot evidence in favor of the last possibility, and the monoubiquitinated lysine in the truncated form of ICP0 is not present in the full-length protein (57). Attempts to detect specific changes in ubiquitinated proteins in whole-cell extracts of infected cells were not informative; certainly, no major new ubiquitinated species were detected (data not shown). Because the amount of free ubiquitin in the cell (particularly in the nucleus [38]) is limiting and ubiquitinated proteins are rapidly turned over, it is perhaps not surprising that we did not observe any global changes in ubiquitinated proteins via Western blotting; it is likely that the microscopy reveals changes in intracellular location and concentration of conjugated ubiquitin rather than increases in the total amounts of conjugated proteins in general. Therefore identification of the nature of the conjugated ubiquitin deposits observed here awaits more detailed (and difficult) experimentation.
These results lend further weight to the hypothesis that ICP0 stimulates viral infection by inducing the proteasome-dependent degradation of cellular proteins. It has been suggested that one or more of the target proteins could be involved in a repression mechanism that would otherwise repress viral transcription (11, 19, 20). While this hypothesis can be verified only by further characterization of the substrate targets of ICP0 and their roles in the cell, in principle this suggestion is analogous to the regulation of NF-κB-responsive genes, which can be activated only after the SCF E3 ubiquitin ligase complex has degraded the I-κB inhibitor (34). The proposed activity of ICP0 also has some parallels with events occurring during at least two other viral infections: first, poliovirus 2A and 3C proteases cleave not only the viral precursor polyprotein but also a number of cellular proteins whose loss could be beneficial to viral infection (31, 59); and second, paramyxovirus simian virus 5 inhibits the interferon response by targeting STAT1 for proteasome-mediated degradation (6).
The next steps in the characterization of ICP0 function and its role in HSV-1 infection must be to establish in vitro assays to confirm and explore its role in inducing conjugated ubiquitin, to define the full catalogue of substrate proteins, and to understand why degradation of one or more of these proteins can result in increased viral gene expression. These studies are in progress.
ACKNOWLEDGMENTS
This work was supported by the Medical Research Council.
Jon Yewdell (National Institutes of Health) initiated discussions which led to the start of this study, Patrick Lomonte provided viruses expressing EGFP-linked ICP0 proteins, Thomas Sternsdorf provided antiserum SpGH, and Paul Freemont donated antiserum r8. The technical assistance of Anne Orr and constructive comments on the text from Duncan McGeoch, Patrick Lomonte, and Jane Parkinson were greatly appreciated.
REFERENCES
- 1.Anton L C, Schubert U, Bacik I, Princiotta M F, Wearsch P A, Gibbs J, Day P M, Realini C, Rechsteiner M C, Bennink J R, Yewdell J W. Intracellular localization of proteasomal degradation of a viral antigen. J Cell Biol. 1999;146:113–124. doi: 10.1083/jcb.146.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boddy M N, Howe K, Etkin L D, Solomon E, Freemont P S. PIC1, a novel ubiquitin-like protein which interacts with the PML component of a multiprotein complex that is disrupted in acute promyelocytic leukaemia. Oncogene. 1996;13:971–982. [PubMed] [Google Scholar]
- 3.Borden K L B. RING domains: master builders of molecular scaffolds? J Mol Biol. 2000;295:1103–1112. doi: 10.1006/jmbi.1999.3429. [DOI] [PubMed] [Google Scholar]
- 4.Carrozza M J, DeLuca N A. Interaction of the viral activator protein ICP4 with TFIID through TAF250. Mol Cell Biol. 1996;16:3085–3093. doi: 10.1128/mcb.16.6.3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chelbi-Alix M K, de The H. Herpes virus induced proteasome-dependent degradation of the nuclear bodies-associated PML and Sp100 proteins. Oncogene. 1999;18:935–941. doi: 10.1038/sj.onc.1202366. [DOI] [PubMed] [Google Scholar]
- 6.Didcock L, Young D F, Goodbourn S, Randall R E. The V protein of simian virus 5 inhibits interferon signalling by targeting STAT1 for proteasome-mediated degradation. J Virol. 1999;73:9928–9933. doi: 10.1128/jvi.73.12.9928-9933.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Everett R D. A detailed mutational analysis of Vmw110, a trans-acting transcriptional activator encoded by herpes simplex virus type 1. EMBO J. 1987;6:2069–2076. doi: 10.1002/j.1460-2075.1987.tb02472.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Everett R D. Analysis of the functional domains of herpes simplex virus type 1 immediate-early polypeptide Vmw110. J Mol Biol. 1988;202:87–96. doi: 10.1016/0022-2836(88)90521-9. [DOI] [PubMed] [Google Scholar]
- 9.Everett R D. Construction and characterisation of herpes simplex virus type 1 mutants with defined lesions in immediate-early gene 1. J Gen Virol. 1989;70:1185–1202. doi: 10.1099/0022-1317-70-5-1185. [DOI] [PubMed] [Google Scholar]
- 10.Everett R D. A surprising role for the proteasome in the regulation of herpesvirus infection. Trends Biochem Sci. 1999;24:293–295. doi: 10.1016/s0968-0004(99)01433-4. [DOI] [PubMed] [Google Scholar]
- 11.Everett R D. ICP0, a regulator of herpes simplex virus during lytic and latent infection. Bioessays. 2000;22:761–770. doi: 10.1002/1521-1878(200008)22:8<761::AID-BIES10>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 12.Everett R D, Orr A, Elliott M. High level expression and purification of herpes simplex virus type 1 immediate early polypeptide Vmw110. Nucleic Acids Res. 1991;19:6155–6161. doi: 10.1093/nar/19.22.6155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Everett R D, Preston C M, Stow N D. Functional and genetic analysis of the role of Vmw110 in herpes simplex virus replication. In: Wagner E K, editor. The control of herpes simplex virus gene expression. Boca Raton, Fla: CRC Press, Inc.; 1991. pp. 50–76. [Google Scholar]
- 14.Everett R D, Cross A, Orr A. A truncated form of herpes simplex virus type 1 immediate-early protein Vmw110 is expressed in a cell type dependent manner. Virology. 1993;197:751–756. doi: 10.1006/viro.1993.1651. [DOI] [PubMed] [Google Scholar]
- 15.Everett R D, Maul G G. HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J. 1994;13:5062–5069. doi: 10.1002/j.1460-2075.1994.tb06835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Everett R D, Barlow P N, O'Hare P, O'Rourke D, Orr A. Point mutations in the HSV-1 Vmw110 RING finger helix affect activation of gene expression, viral growth, and interaction with PML-containing nuclear structures. J Virol. 1995;69:7339–7344. doi: 10.1128/jvi.69.11.7339-7344.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Everett R D, Meredith M R, Orr A, Cross A, Kathoria M, Parkinson J. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO J. 1997;16:1519–1530. doi: 10.1093/emboj/16.7.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Everett R D, Freemont P, Saitoh H, Dasso M, Orr A, Kathoria M, Parkinson J. The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J Virol. 1998;72:6581–6591. doi: 10.1128/jvi.72.8.6581-6591.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Everett R D, Orr A, Preston C M. A viral activator of gene expression functions via the ubiquitin-proteasome pathway. EMBO J. 1998;17:7161–7169. doi: 10.1093/emboj/17.24.7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Everett R D, Earnshaw W C, Findlay J, Lomonte P. Specific destruction of kinetochore protein CENP-C and disruption of cell division by herpes simplex virus immediate-early protein Vmw110. EMBO J. 1999;18:1526–1538. doi: 10.1093/emboj/18.6.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Everett R D, Meredith M, Orr A. The ability of herpes simplex virus type 1 immediate-early protein Vmw110 to bind to a ubiquitin-specific protease contributes to its roles in the activation of gene expression and stimulation of virus infection. J Virol. 1999;73:417–426. doi: 10.1128/jvi.73.1.417-426.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd edition. Vol. 2. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. [Google Scholar]
- 23.Freemont P S. Ubiquitination: RING for destruction? Curr Biol. 2000;10:R84–R87. doi: 10.1016/s0960-9822(00)00287-6. [DOI] [PubMed] [Google Scholar]
- 24.Fujimuro M, Sawada H, Yokosawa H. Production and characterisation of monoclonal antibodies specific to multi-ubiquitin chains of polyubiquitinated proteins. FEBS Lett. 1994;349:173–180. doi: 10.1016/0014-5793(94)00647-4. [DOI] [PubMed] [Google Scholar]
- 25.Harris R A, Everett R D, Zhu X, Silverstein S, Preston C M. Herpes simplex virus type 1 immediate-early protein Vmw110 reactivates latent herpes simplex virus type 2 in an in vitro latency system. J Virol. 1989;63:3513–3515. doi: 10.1128/jvi.63.8.3513-3515.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hobbs W E, DeLuca N A. Perturbation of cell cycle progression and cellular gene expression as a function of herpes simplex virus ICP0. J Virol. 1999;73:8245–8255. doi: 10.1128/jvi.73.10.8245-8255.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jensen D E, Proctor M, Marquis S T, Gardner H P, Ha S I, Chodosh L A, Ishov A M, Tommerup N, Vissing H, Sekido Y, Minna J, Borodovsky A, Schultz D C, Wilkinson K D, Maul G G, Barlev N, Berger S L, Prendergast G C, Rauscher F J. BAP1: a novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene. 1998;16:1097–1112. doi: 10.1038/sj.onc.1201861. [DOI] [PubMed] [Google Scholar]
- 28.Jordan R, Schaffer P A. Activation of gene expression by herpes simplex virus type 1 ICP0 occurs at the level of mRNA synthesis. J Virol. 1997;71:6850–6862. doi: 10.1128/jvi.71.9.6850-6862.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kawaguchi Y, Bruni R, Roizman B. Interaction of herpes simplex virus 1 α regulatory protein ICP0 with elongation factor 1δ: ICP0 affects translational machinery. J Virol. 1997;71:1019–1024. doi: 10.1128/jvi.71.2.1019-1024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kawaguchi Y, van Sant C, Roizman B. Herpes simplex virus regulatory protein ICP0 interacts with and stabilizes the cell cycle regulator cyclin D3. J Virol. 1997;71:7328–7336. doi: 10.1128/jvi.71.10.7328-7336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krausslich H G, Nicklin M J, Toyoda H, Etchison D, Wimmer E. Poliovirus proteinase 2A induces cleavage of eucaryotic initiation factor 4F polypeptide p220. J Virol. 1987;61:2711–2718. doi: 10.1128/jvi.61.9.2711-2718.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Latchman D S, Estridge J K, Kemp L M. Transcriptional induction of the ubiquitin gene during herpes simplex virus infection is dependent upon the viral immediate-early protein ICP4. Nucleic Acids Res. 1987;15:7283–7293. doi: 10.1093/nar/15.18.7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lomonte P, Everett R D. Herpes simplex virus regulatory protein Vmw110 inhibits cell cycle progression. J Virol. 1999;73:9456–9467. doi: 10.1128/jvi.73.11.9456-9467.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maniatis T. A ubiquitin ligase complex essential for the FK-κB, Wnt/Wingless and Hedgehog signalling pathways. Genes Dev. 1999;13:505–510. doi: 10.1101/gad.13.5.505. [DOI] [PubMed] [Google Scholar]
- 35.Maul G G, Everett R D. The nuclear location of PML, a cellular member of the C3HC4 zinc binding domain protein family, is rearranged during herpes simplex virus infection by the C3HC4 viral protein ICP0. J Gen Virol. 1994;75:1223–1233. doi: 10.1099/0022-1317-75-6-1223. [DOI] [PubMed] [Google Scholar]
- 36.Maul G G. DNA virus transcription and replication at a specific nuclear domain, ND10. Bioessays. 1998;20:660–667. doi: 10.1002/(SICI)1521-1878(199808)20:8<660::AID-BIES9>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 37.Meredith M R, Orr A, Elliott M, Everett R D. Separation of the sequence requirements for HSV-1 Vmw110 multimerisation and interaction with a 135-kDa cellular protein. Virology. 1995;209:174–187. doi: 10.1006/viro.1995.1241. [DOI] [PubMed] [Google Scholar]
- 38.Mimnaugh E G, Chen H Y, Davie J R, Celis J E, Neckers L. Rapid deubiquitination of nucleosomal histones in human tumor cells caused by proteasome inhibitors and stress response inducers: effects on replication, transcription, translation and the cellular stress response. Biochemistry. 1997;36:14418–14429. doi: 10.1021/bi970998j. [DOI] [PubMed] [Google Scholar]
- 39.Mimnaugh E G, Bonvini P, Neckers L. The measurement of ubiquitin and ubiquitinated proteins. Electrophoresis. 1999;20:418–428. doi: 10.1002/(SICI)1522-2683(19990201)20:2<418::AID-ELPS418>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 40.Mossman K L, Smiley J R. Truncation of the C-terminal acidic transcriptional activation domain of herpes simplex virus VP16 renders expression of the immediate-early genes almost entirely dependent on ICP0. J Virol. 1999;73:9726–9733. doi: 10.1128/jvi.73.12.9726-9733.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muller S, Dejean A. Viral immediate-early proteins abrogate the modification by SUMO-1 of PML and Sp100 proteins correlating with nuclear body disruption. J Virol. 1999;73:5137–5143. doi: 10.1128/jvi.73.6.5137-5143.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O'Rourke D, Elliott G, Papworth M, Everett R, O'Hare P. Examination of determinants for intranuclear localisation and transactivation within the RING finger of herpes simplex virus type 1 IE110k protein. J Gen Virol. 1998;79:537–548. doi: 10.1099/0022-1317-79-3-537. [DOI] [PubMed] [Google Scholar]
- 43.Parkinson J, Lees-Miller S P, Everett R D. Herpes simplex virus type 1 immediate-early protein Vmw110 induces the proteasome-dependent degradation of DNA-dependent protein kinase. J Virol. 1999;73:650–657. doi: 10.1128/jvi.73.1.650-657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parkinson J, Everett R D. Alphaherpesvirus proteins related to herpes simplex virus type 1 ICP0 affect cellular structures and proteins. J Virol. 2000;74:10006–10017. doi: 10.1128/jvi.74.21.10006-10017.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Phelan A, Clements J B. Post-transcriptional regulation in herpes simplex virus. Semin Virol. 1998;8:309–318. [Google Scholar]
- 46.Preston C M, Nicholl M J. Repression of gene expression upon infection of cells with herpes simplex virus type 1 mutants impaired for immediate-early protein synthesis. J Virol. 1997;71:7807–7813. doi: 10.1128/jvi.71.10.7807-7813.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Preston C M. Repression of viral transcription during herpes simplex virus latency. J Gen Virol. 2000;81:1–19. doi: 10.1099/0022-1317-81-1-1. [DOI] [PubMed] [Google Scholar]
- 48.Samaniego L A, Neiderhiser L, DeLuca N A. Persistence and expression of the herpes simplex virus genome in the absence of immediate-early proteins. J Virol. 1998;72:3307–3320. doi: 10.1128/jvi.72.4.3307-3320.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sears A E, Halliburton I W, Meignier B, Silver S, Roizman B. Herpes simplex virus 1 mutant deleted in the α22 gene: growth and gene expression in permissive and restrictive cells and establishment of latency in mice. J Virol. 1985;55:338–346. doi: 10.1128/jvi.55.2.338-346.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Song B, Liu J, Yeh K C, Knipe D M. Herpes simplex virus infection blocks events in G1 phase of the cell cycle. Virology. 2000;267:326–334. doi: 10.1006/viro.1999.0146. [DOI] [PubMed] [Google Scholar]
- 51.Spatz S J, Norby E C, Weber P C. Mutational analysis of ICP0R, a transrepressor protein created by alternative splicing of the ICP0 gene of herpes simplex virus type 1. J Virol. 1996;70:7360–7370. doi: 10.1128/jvi.70.11.7360-7370.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sternsdorf T, Guldner H H, Szostecki C, Grötzinger T, Will H. Two nuclear-dot associated proteins, PML and Sp100, are often co-autoimmunogenic in patients with primary biliary cirrhosis. Scand J Immunol. 1995;42:257–268. doi: 10.1111/j.1365-3083.1995.tb03652.x. [DOI] [PubMed] [Google Scholar]
- 53.Stow N D, Stow E C. Isolation and characterisation of a herpes simplex virus type 1 mutant containing a deletion within the gene encoding the immediate-early polypeptide Vmw110. J Gen Virol. 1986;67:2571–2585. doi: 10.1099/0022-1317-67-12-2571. [DOI] [PubMed] [Google Scholar]
- 54.Van Sant C, Kawaguchi Y, Roizman B. A single amino acid substitution in the cyclin D binding domain of the infected cell protein no. 0 abrogates the neuroinvasiveness of herpes simplex virus without affecting its ability to replicate. Proc Natl Acad Sci USA. 1999;96:8184–8189. doi: 10.1073/pnas.96.14.8184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wagner E K, Bloom D C. Experimental investigation of herpes simplex virus latency. Clin Microbiol Rev. 1997;10:419–443. doi: 10.1128/cmr.10.3.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weber P C, Wigdahl B. Identification of dominant-negative mutants of the herpes simplex virus type 1 immediate-early protein ICP0. J Virol. 1992;66:2261–2267. doi: 10.1128/jvi.66.4.2261-2267.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weber P C, Spatz S J, Norby E C. Stable ubiquitination of the ICP0R protein of herpes simplex virus type 1 during productive infection. Virology. 1999;253:288–298. doi: 10.1006/viro.1998.9502. [DOI] [PubMed] [Google Scholar]
- 58.Wilkinson K D. Roles of ubiquitination in proteolysis and cellular regulation. Annu Rev Nutr. 1995;15:161–189. doi: 10.1146/annurev.nu.15.070195.001113. [DOI] [PubMed] [Google Scholar]
- 59.Yalamanchili P, Datta U, Dasgupta A. Inhibition of host cell transcription by poliovirus: cleavage of transcription factor CREB by poliovirus-encoded protease 3Cpro. J Virol. 1997;71:1220–1226. doi: 10.1128/jvi.71.2.1220-1226.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yao F, Schaffer P A. Physical interaction between the herpes simplex virus type 1 immediate-early regulatory proteins ICP0 and ICP4. J Virol. 1994;68:8158–8168. doi: 10.1128/jvi.68.12.8158-8168.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yao F, Schaffer P A. An activity specified by the osteosarcoma line U2OS can substitute functionally for ICP0, a major regulatory protein of herpes simplex virus type 1. J Virol. 1995;69:6249–6258. doi: 10.1128/jvi.69.10.6249-6258.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]