Abstract
The herpes simplex virus type 1 (HSV-1) immediate-early protein ICP0 interacts with several cellular proteins and induces the proteasome-dependent degradation of others during infection. In this study we show that ICP0 is required for the proteasome-dependent degradation of the ND10 protein Sp100 and, as with the other target proteins, the ICP0 RING finger domain is essential. Further, comparison of the kinetics and ICP0 domain requirements for the degradation of PMI and Sp100 suggests that a common mechanism is involved. Homologues of ICP0 are encoded by other members of the alphaherpesvirus family. These proteins show strong sequence homology to ICP0 within the RING finger domain but limited similarity elsewhere. Using transfection assays, we have shown that all the ICP0 homologues that we tested have significant effects on the immunofluorescence staining character of at least one of the proteins destabilized by ICP0, and by using a recombinant virus, we found that the equine herpesvirus ICP0 homologue induced the proteasome-dependent degradation of endogenous CENP-C and modified forms of PML and Sp100. However, in contrast to ICP0, the homologue proteins had no effect on the distribution of the ubiquitin-specific protease USP7 within the cell, consistent with their lack of a USP7 binding domain. We also found that ICP0 by itself could induce the abrogation of SUMO-1 conjugation and then the proteasome-dependent degradation of unmodified exogenous PML in transfected cells, thus demonstrating that other HSV-1 proteins are not required. Surprisingly, the ICP0 homologues were unable to cause these effects. Overall, these data suggest that the members of the ICP0 family of proteins may act via a similar mechanism or pathway involving their RING finger domain but that their intrinsic activities and effects on endogenous and exogenous proteins differ in detail.
The herpes simplex virus type 1 (HSV-1) immediate-early (IE) protein ICP0 (Vmw110) is a RING finger protein encoded by IE gene 1 and is a strong and promiscuous activator of gene expression in transfection assays (reviewed in reference 18). Upon primary exposure, HSV-1 initiates a lytic infection in the epithelium and subsequently establishes a lifelong latent infection in sensory neurons (reviewed in reference 67), and ICP0 has been implicated in the regulation of both the lytic cycle and reactivation from latency. Several lines of evidence indicate that ICP0 might play a specific role in the control of the balance between the latent and lytic states, such that in its presence the latter is favored (7, 11, 34, 46, 68, 75, 76, 86). It is likely that ICP0 carries out its role in activation of transcription and reactivation from latency by interacting with cellular proteins. Consistent with this, ICP0 has been found to bind strongly and specifically to the cellular ubiquitin-specific protease USP7 (formerly called herpesvirus-associated ubiquitin-specific protease [HAUSP]) (24, 55, 56) and to interact with and stabilize cyclin D3 (43). Furthermore, ICP0 induces the proteasome-dependent degradation of a number of cellular proteins, which suggests that changes in the intranuclear environment may be involved in the function of ICP0 (25, 27, 66).
At early times of infection ICP0 localizes to specific nuclear structures called ND10 domains, PML nuclear bodies, or promyelocytic oncogenic domains (PODs) (53). These domains of unknown function are associated with the nuclear matrix and contain at least six cellular proteins, of which the most widely studied is PML (a protein implicated in promyelocytic leukemia) (4, 5, 14, 44, 77). Interestingly, USP7 is a component of a subset of ND10, and during infection the interaction of ICP0 with USP7 leads to an increased proportion of ND10 containing this USP (24). However, the consequence of the localization of ICP0 at ND10 is their disruption (21, 54), and it has recently been found that this correlates with the virus-induced and ICP0-dependent degradation of several high-molecular-weight isoforms of PML (25). Other recent studies have shown that these isoforms of PML are very likely to comprise covalent conjugates with the small ubiquitin-like protein SUMO-1 (also known as GMP1, PIC1, Sentrin, and UBL-1 [reviewed in references 39 and 70; see also references 25, 40, 62, and 74]) and that virus infection leads to the degradation of a large number of uncharacterized SUMO-1-conjugated proteins in an ICP0-dependent manner (25). Other cellular proteins targeted for degradation in an ICP0-dependent manner are the catalytic subunit of the DNA-degradation protein kinase (45, 66) and the centromeric protein CENP-C (27). Additionally Sp100, another ND10 protein, is rapidly degraded in a proteasome-dependent manner during HSV-1 infection (8). Although the identification of the viral protein(s) which causes Sp100 degradation was not determined, it is likely that ICP0 is involved, especially as it has since been reported that in transfection studies ICP0 abrogates the SUMO-1 modification of exogenous Sp100 (63).
Homologues of ICP0 exist in other members of the alphaherpesvirus family: BICP0 in bovine herpesvirus 1 (BHV-1) (84); the product of gene 63, Eg63, in equine herpesvirus 1 (EHV-1) (79); the product of gene 61, Vg61, in varicella-zoster virus (VZV) (12); and EP0 in pseudorabies virus (PRV) (9). The homologues are related to ICP0 by virtue of the location of their genes within the viral genome and the fact that they have all been shown to activate or influence gene expression (18, 23, 58, 60, 64, 84). Additionally, the VZV and EHV homologues have been shown to fully and partially complement an ICP0-deficient virus, respectively (23, 57), and the growth defect of a PRV EP0 deletion mutant is complemented in cells expressing VZV Vg61 or HSV-1 ICP0 (60). However, sequence similarity between these proteins is very limited except for the RING finger domain near their N termini. This region in ICP0 has been found to be essential for its functions in regulating gene expression, stimulating lytic infection and reactivation from quiescence, disruption of ND10 and centromeres, induced proteasome-dependent degradation of cellular proteins, and binding to and stabilization of cyclin D3 (17, 21, 22, 25, 26, 27, 34, 66, 81, 82).
Given this background, we set out to determine whether ICP0 was responsible for the HSV-1-induced degradation of Sp100 and if so, to compare the factors governing the degradation of PML and Sp100. A second objective of this study, in view of the importance of the RING finger in the functions of ICP0, was to determine whether the alphaherpesvirus ICP0 homologues also have the same effect on cellular protein stability. In addition, since during infections ICP0 has been shown to lead to an increased number of ND10 domains that contain USP7 early in infection (24), we were interested in determining whether transfected ICP0 and its homologues had any effect on USP7 during transient transfection. Although no obvious sequence homology for the USP7 binding domain of ICP0 is present in the other family members, in principle they could still affect USP7 distribution by interacting through their own different binding domains.
In this paper we show that ICP0 is indeed required for the effect on Sp100 and that the time course and ICP0 sequence requirements for the degradation of Sp100 and PML are indistinguishable. Because these proteins are related only by their modification by SUMO-1, we suggest that degradation occurs via a common pathway rather than by specific targeting of the individual susceptible proteins. Transfection experiments using epitope-tagged versions of the ICP0 homologue proteins demonstrated that all had some effect on the intracellular distribution of at least one of the proteins affected by ICP0 and that in the case of the EHV-1 homologue, this was due to induced proteasome-mediated degradation. Finally, we found that in addition to its ability to induce the abrogation of SUMO-1 conjugation of exogenous PML in transfection assays (63), ICP0 by itself is able to induce the subsequent degradation of the unmodified exogenous PML in a RING finger-dependent manner. Surprisingly, despite their effects on endogenous PML, the ICP0 homologue proteins were unable to affect either the SUMO-1 conjugation or stability of exogenous PML in our assays. These results suggest that there are many similarities between the biological activities of the ICP0 family of proteins, probably as a consequence of their conserved RING finger domains, but that their intrinsic activities differ in detail.
MATERIALS AND METHODS
Cells and viruses.
HEp-2 cells were grown in Dulbecco's modified Eagle's medium with 10% fetal calf serum and 100 U of penicillin/ml and 100 μg of streptomycin/ml. Human fetal lung (HFL) cells (Flow Laboratories) were grown in Dulbecco's modified Eagle's medium, 5% fetal calf serum, 5% newborn calf serum, 4 mM glutamine, nonessential amino acids, and antibiotics as described above. All viruses were grown and titrated in baby hamster kidney (BHK) cells propagated in Glasgow modified Eagle's medium supplemented with 10% newborn calf serum, 10% tryptose phosphate broth, and antibiotics as described above. Viruses used were the wild-type HSV-1 strain 17syn+; 17Eg63, a recombinant 17syn+ virus containing the EHV-1 g63 gene in the place of the HSV-1 IE1 gene (23); and the ICP0 mutant 17syn+ viruses dl1403 FXE, E52X, D14, and M1 (17, 26, 56, 76).
Plasmids.
pp65-ICP0 was created by cloning a human cytomegalovirus (HCMV) pp65 epitope tag (produced by annealing the following primers, 5′CATGACTGAGCGCAAGACGCCCCGCGTCACCGGCGGCAC3′ and 5′CATGGTGCCGCCGGTGACGCGGGGCGTCTTGCGCTCAGT3′) into the NcoI site at the ATG of the ICP0 coding region of plasmid p111 (15) to create plasmid pp65-ICP0. The design of the 5′ and 3′ ends of the tag was such that upon its insertion into p111, the original NcoI site at the initiation codon of ICP0 was destroyed, while a new NcoI site was introduced with a second in-frame ATG downstream of the tag. This therefore maintained all IE1 5′ sequences. Plasmid pp65-ICP0 also contains a HincII site close to the end of the ICP0 coding region. This enabled the ICP0 coding sequence of pp65-ICP0 to be removed as a NcoI-HincII fragment, leaving the 5′ sequences upstream and 3′ sequences downstream intact and allowing insertion of another open reading frame (ORF) into the IE1 transcription unit. The fragments to be inserted were selected to use either the NcoI site naturally occurring at the initiating ATG of the homologues or an NcoI site engineered into this position and a blunt-ended restriction site 3′ of the ORF. These were as follows: (i) an NcoI partial-EcoRV DNA fragment from pEHVg63 (23) containing EHV-1 gene 63 to create pp65-Eg63, (ii) an NcoI-BssHI fragment from the PRV genomic KpnI F fragment (a gift from A. Davison) containing the PRV EP0 gene to create pp65-EP0, (iii) an NcoI partial-AccI DNA fragment from VZV KpnI 23 (a gift from A. Davison) containing VZV gene 61 to create pp65-Vg61, and (iv) an NcoI-StuI PCR fragment of the N terminus of the BHV-1 BICP0 gene in plasmid pBCMV26 (a gift from M. Schwyzer) (created using oligonucleotide primers complementary to the 5′ terminus of the BHV-1 BICP0 gene containing an engineered NcoI site at the initiation codon and complementary to antisense sequences around and including the StuI site present in the BICP0 gene), and a StuI-SspI fragment from the C terminus of the BICP0 gene pBCMV26 to create pp65-BICP0. The resulting plasmids have the IE1 promoter and 5′ untranslated region, an initiating ATG followed by the tag region linked in frame to the complete homologue ORF, followed by the IE1 3′ region and regulatory signals. Further plasmids used were pPML(F), which expresses F-tagged PML [PML(F)] (43); pCIPIC1, which expresses myc-tagged SUMO-1 (25); pCIUSP7, which expresses the USP7 coding region (24) from the pCIneo vector (Promega); pCIM1, which expresses ICP0 with R623L and K621I mutations in the USP7 binding domain (26); p110FXE, which expresses an ICP0 RING finger mutant (16); p110D12, which expresses ICP0 with amino acids 594 to 632 deleted (16); p110 RING finger single-point mutations p110K144E, p110N151D, and p110Q148E; and double-point mutations p110K144E N151D, and p110K144E Q148E (22). pCImyc-Sp100 expressing myc-tagged Sp100 was made by cloning the Sp100 coding region as NcoI-XhoI and XhoI-EcoRV fragments into the NcoI-HindIII site of plasmid pmyc-ICP0, a pUC9 vector expressing myc epitope-tagged ICP0, to create plasmid pmyc-Sp100. This digest of pmyc-ICP0 removed the ICP0 ORF while leaving the myc epitope tag with an NcoI at its 3′ end. The tagged Sp100 was then removed from pmyc-Sp100 as an EcoRI-PuvII fragment and cloned into pCIneo cut with EcoRI and SmaI, creating pCImyc-Sp100.
Antibodies.
Anti-ICP0 monoclonal antibody (MAb) 11060, anti-ICP4 MAb 10176, and polyclonal anti-USP7 r201 have been described elsewhere (19, 20, 24). Polyclonal anti-ICP0 r190 was raised against a glutathione S-transferase fusion protein containing residues 594 to 775 of ICP0, and MAb anti-USP7 16613 recognizes an epitope in the N-terminal 193 amino acids of USP7 (66). MAb anti-pp65 was obtained from Capricorn Products, Inc., and anti-c-myc MAb 9E10 from Santa Cruz Biotechnology, Inc. Other antibodies used were polyclonal anti-BICP0 peptide serum 11 (31), anti-PML antibodies MAb 5E10 (77) and rabbit serum r8 (5), polyclonal anti-CENP-C rabbit serum r554 (69), polyclonal anti-Sp100 rabbit serum SpGH (73), and anti-F tag MAb F3 (3).
Transfections.
HEp-2 cells were transfected using either Tfx50 (Promega) or Lipofectamine PLUS (Gibco) according to the manufacturer's instructions. With Tfx50, 105 HEp-2 cells on coverslips in 24-well plates were transfected with 1 μg of DNA at a DNA-to-Tfx50 ratio of 4.5:1. The DNA-medium-Tfx mix was applied to the cells for 30 min before 1 ml of complete medium was added for an additional 2 h and then replaced with fresh complete medium. Using Lipofectamine PLUS, 0.75 × 106, 0.25 × 106, or 105 HEp-2 cells in 60- or 35-mm dishes or on coverslips in 24-well plates, respectively, were transfected with a total of 2, 1, or 0.4 μg of DNA in serum-free medium according to the manufacturer's protocol. The DNA–serum-free medium-Lipofectamine PLUS mix was applied to the cells for 3 h before an equal volume of medium containing twice the normal amount of serum and antibiotics was added. When transfections were in 60- or 35-mm dishes, the cells were trypsinized 8 h posttransfection and counted, and then 105 cells were placed on coverslips in 24-well plates for microscopy studies or 2 × 105 cells were placed in 24-well plates for Western blot analysis. Cells were either processed for microscopy or washed in phosphate-buffered saline (PBS) and then harvested into sodium dodecyl sulfate (SDS)-gel loading buffer for immunoblot analysis at 24 h posttransfection. When the effect of MG132 on transfected proteins was determined, the medium on the transfected cells was replaced at 24 h posttransfection with medium containing 5 μM MG132 in 1% dimethyl sulfoxide (DMSO) and the cells were harvested at set intervals thereafter.
Virus infections.
HFL cells at 105 cells per well in 24-well plates were infected with virus at 10 PFU per cell. After a 1-h adsorption period, medium was added and the infections were continued for the desired time period. When proteasome inhibitors were used, medium containing 1% DMSO alone or 1% DMSO with MG132 to give a final concentration of 5 μM was used. When required, the medium was removed from the infected cells, the cell monolayer was washed in PBS, and the cells were harvested directly in SDS-gel loading buffer.
Western blot (immunoblot) analysis.
Proteins in cell extracts were analyzed by separation on SDS–7.5% polyacrylamide gels prepared and run in the Bio-Rad MiniProtean II apparatus and were then electrophoretically transferred to nitrocellulose membranes, using the compatible Bio-Rad equipment according to the manufacturer's instructions. After transfer, the membranes were blocked overnight in PBS containing 0.05% Tween 20 (PBST) and 5% dried milk at 4°C and then incubated with the primary antibody in PBST–5% dried milk at room temperature for 2 h and washed in PBST and incubated for a further hour at room temperature in PBST–5% dried milk containing horseradish peroxidase-conjugated secondary antibody. After further washing in PBST, membranes were processed by the enhanced chemiluminescence method (NEN or Amersham). Antibodies were stripped from membranes following the Amersham protocol, and the membranes were reprobed as required.
Confocal microscopy.
Cells seeded onto glass coverslips were prepared for immunofluorescence and examined by confocal microscopy as previously described (27, 66), except that samples were fixed with either formaldehyde (5% [vol/vol] of the 30% stock solution in PBS containing 2% sucrose) for 10 min or 70% acetone–30% methanol (stored at −20°C) for 5 min at room temperature and then permeabilized with 0.5% Nonidet P-40 in PBS with 10% sucrose for 5 min at room temperature. Secondary antibodies used were fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit or goat anti-mouse immunoglobulin G (IgG) (Sigma) at 1/100; cy3-conjugated goat anti-mouse or goat anti-rabbit IgG at 1/1,000 and 1/5,000, respectively; or cy5-conjugated goat anti-mouse IgG at 1/500 (Amersham). Stained cells were examined using a Zeiss LSM 510 confocal microscope system, with three lasers giving excitation lines at 488 nm (FITC) and 543 nm (cy3) or 633 nm (cy5).
RESULTS
Comparison of the degradation of Sp100 and PML induced by ICP0 during HSV-1 infection.
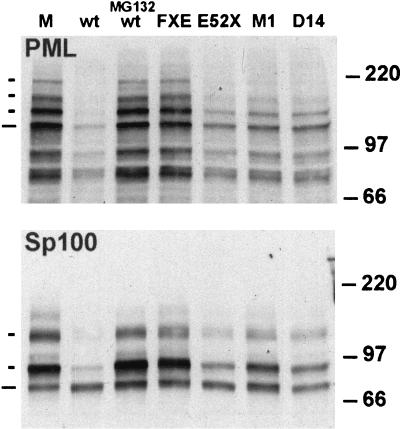
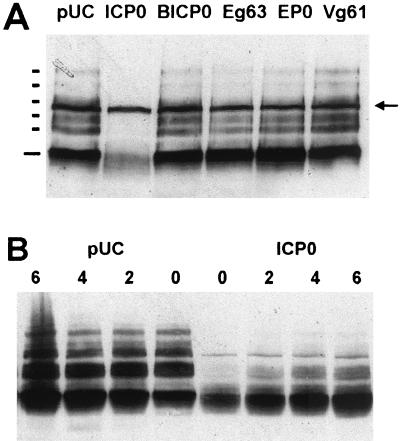
We were initially interested in determining whether, as suggested from the work of Chelbi-Ali and de Thé (8), the induced degradation of Sp100 during HSV-1 infection is dependent on the expression of ICP0. We confirmed that Sp100 was degraded during HSV-1 infection (Fig. 1, compare mock and wild-type tracks) and found that ICP0 was indeed required for this effect, since both ICP0 null mutant dl1403 and RING finger deletion mutant FXE were unable to induce the degradation (Fig. 1 and see also Fig. 7B). The most striking effect of ICP0 was on the higher-molecular-weight isoforms of Sp100 (those demonstrated to be modified by conjugation to SUMO-1 [74]), while in these experiments at this time point there was no reduction in the presumed unmodified form of Sp100.
FIG. 1.
Comparative demodification and degradation of PML and Sp100 induced by ICP0 during HSV-1 infection. HFL cells were mock infected (lane M) or infected at 10 PFU per cell with HSV-1 strain 17syn+ in the absence (lane wt) or presence (lane MG132 wt) of proteasome inhibitor MG132 (5 μM final concentration) and the ICP0 mutant viruses FXE, E52X, M1, and D14, as indicated. The cells were harvested for Western blotting 4 h postabsorption. Western blots were probed for PML using MAb 5E10 at a dilution of 1/5 (upper panel), and the filter was then stripped and reprobed for Sp100 using rabbit serum SpGH at a dilution of 1/1,000 (lower panel). To the left of the panels, the short dashes indicate the major SUMO-1-modified isoforms of PML and Sp100 and the longer ones indicate the major, presumed unmodified, forms of the proteins. The positions of the 220-, 97-, and 66-kDa molecular mass markers are indicated to the right.
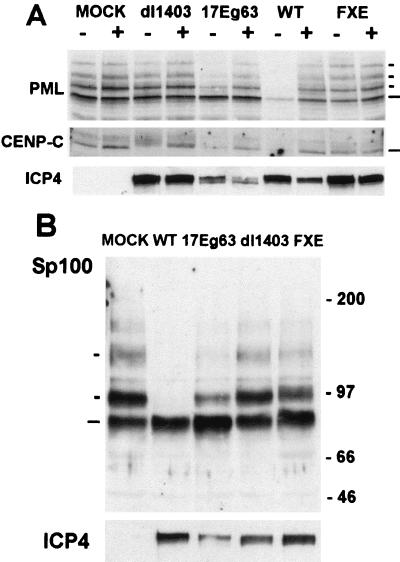
FIG. 7.
Effect of 17Eg63 on cellular proteins. (A) HFL cells were mock infected or infected with wild-type 17syn+ virus (lane WT) or ICP0 mutants and recombinant as shown at 10 PFU per cell in the presence (+) or absence (−) of 5 μM MG132. Cells were harvested into SDS-gel loading buffer at 4 h postadsorption and analyzed by Western blotting. The blot was probed with anti-PML antibody E510 at 1/5 and reprobed with anti-CENP-C antibody r554 at 1/1,000 and anti-ICP4 MAb 10176 at 1/5,000. (B) A further blot was probed with polyclonal anti-Sp100 SpGH at 1/1,000 and reprobed with MAb anti-ICP4 10176 at 1/5,000. The positions of the PML and Sp100 isoforms and CENP-C are indicated, as are molecular weight markers. Long and short dashes indicate unmodified and modified proteins, respectively.
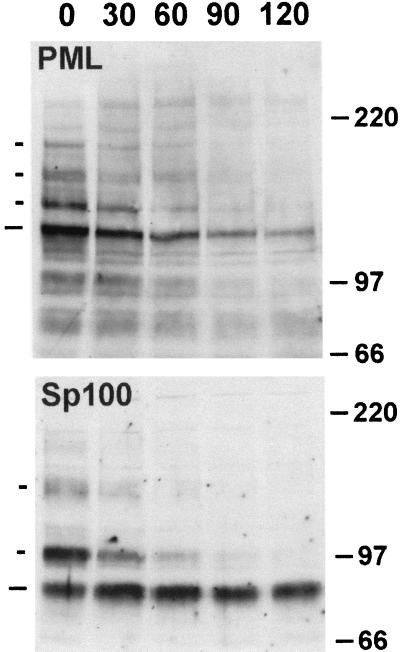
To analyze this induced degradation of Sp100 in more detail, we compared the effects of various mutations in ICP0 on the degradation of PML and Sp100 in parallel and also monitored the time course of these effects. HFL cells were infected with strain 17syn+ and a panel of ICP0 mutant viruses, and then total cell proteins were analyzed by Western blotting. Probing for PML confirmed previous results (25), demonstrating that all isoforms of PML are sensitive to ICP0 during infection of this cell type (Fig. 1 and see also Fig. 7A, compare mock, wild-type, and null mutant tracks); the loss of protein was inhibited by the proteasome inhibitor MG132 and required the RING finger of ICP0. Mutations in the C-terminal region of ICP0 E52X (deletion of residues 594 to 775), M1 (a double-substitution mutant inactivating the USP7 binding motif), and D14 (a deletion affecting ICP0 multimerization) all induced degradation of the PML isoforms but at a lesser efficiency. Reprobing of the blot for Sp100 gave an identical pattern of results, which suggests that PML and Sp100 are degraded by the same ICP0-dependent mechanism. However, as with the wild-type virus, the modified forms of Sp100 were more sensitive to degradation than the presumed unmodified form. The implications from these results were supported by examination of the time course of degradation of PML and Sp100 during wild-type virus infection (Fig. 2). Careful examination of the Sp100 data from these and other repeat experiments suggested that the presumed unmodified form of Sp100 was in fact slightly increased in intensity at the early time points shown, rather than being degraded; this suggests that the SUMO-1-modified forms of Sp100 are particularly sensitive to the effects of ICP0 and that deconjugation of SUMO-1 without degradation can occur.
FIG. 2.
Time course of demodification and degradation of PML and Sp100 induced by HSV-1 infection. HFL cells were infected with HSV-1 strain 17syn+ at 10 PFU per cell and then harvested 30, 60, 90, and 120 min postabsorption, as indicated. The lane marked 0 contains proteins from a mock-infected sample. Total cell proteins were analyzed for PML and Sp100, and the results were annotated as described for Fig. 1.
ICP0 homologue protein sequence comparisons.
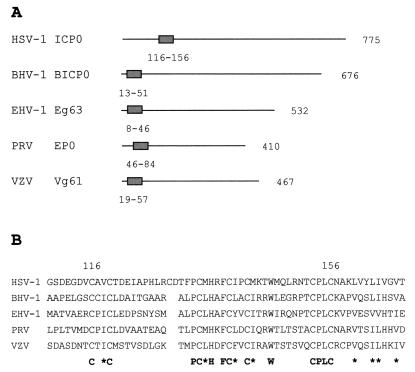
Sequence comparisons of ICP0 and its alphaherpesvirus homologues have previously revealed the presence of homologous RING finger domains but no other obvious similarities. Recently other regions of importance have been more precisely located within the sequence of ICP0, namely the nuclear localization signal, the USP7 binding domain (594 to 633) (55), and the multimerization C-terminal sequences (633 to 711) (10, 56). Thus, to determine more specifically the extent of sequence similarity between ICP0 and its homologues BICP0, Eg63, Vg61, and EP0, the amino acid sequences were searched for homology to these regions of ICP0. RING finger domains were identified in all the homologues (Fig. 3), as were nuclear localization signals, but no other regions of similarity were found, and in particular, none of the ICP0-related proteins contained a sequence similar to the now well-defined USP7 binding region. This analysis confirms that outside the RING finger domains, the related proteins expressed by the other alphaherpesviruses are not at all similar to ICP0.
FIG. 3.
Comparisons of ICP0 and the alphaherpesvirus homologues. (A) Diagram to show the position of the RING finger domain in ICP0 and the homologues. (B) Alignment of the RING finger domains. The first and last cysteine residues of the ICP0 RING finger are numbered, and the conserved residues and positions of similar hydrophobic residues (∗) are shown underneath.
Construction and characterization of epitope-tagged ICP0 proteins.
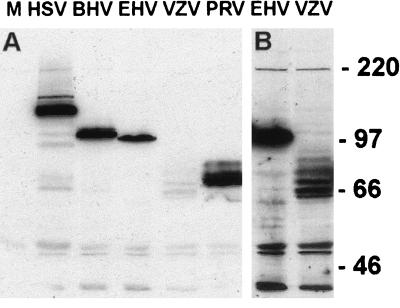
As the RING finger domain of ICP0 is required for the ICP0-induced degradation of several nuclear proteins, it is possible that the RING finger regions of the homologues also serve this function. To determine the effect of the individual viral proteins in the absence of a viral infection, plasmids encoding HCMV pp65 epitope-tagged versions of the proteins were created. Briefly, a pp65 epitope tag was inserted into the plasmid p111 (15) upstream and fused in frame to the ICP0 coding region, creating plasmid pp65-ICP0 (see Materials and Methods). The ICP0 coding region was then replaced with DNA encoding the other ICP0 family members, creating plasmids which express these proteins with an N-terminal pp65 tag. Western blot analysis of transfected HEp-2 cells indicated that tagged proteins of the expected size were produced from the plasmids (Fig. 4A), although the level of expression of full-length pp65-Vg61 was low and a number of apparent degradation products were produced. Figure 4B shows a longer exposure of the pp65-Vg61 track with the pp65-Eg63 track for comparison.
FIG. 4.
Expression of tagged ICP0 homologues. (A) HEp-2 cells were transfected with pUC9 (lane M) or plasmids expressing pp65-ICP0 or the pp65 homologues, as indicated. Samples were harvested into SDS-gel loading buffer at 24 h posttransfection and analyzed by Western blotting using MAb anti-pp65 at a dilution of 1/750. The positions of the molecular weight markers are indicated. (B) Longer exposure of the EHV and VZV tracts of the blot.
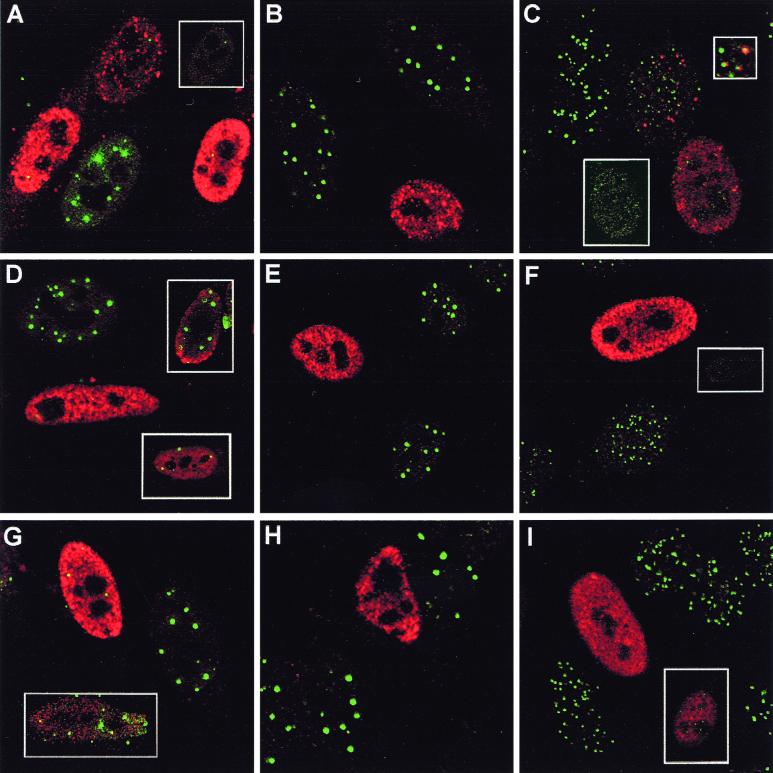
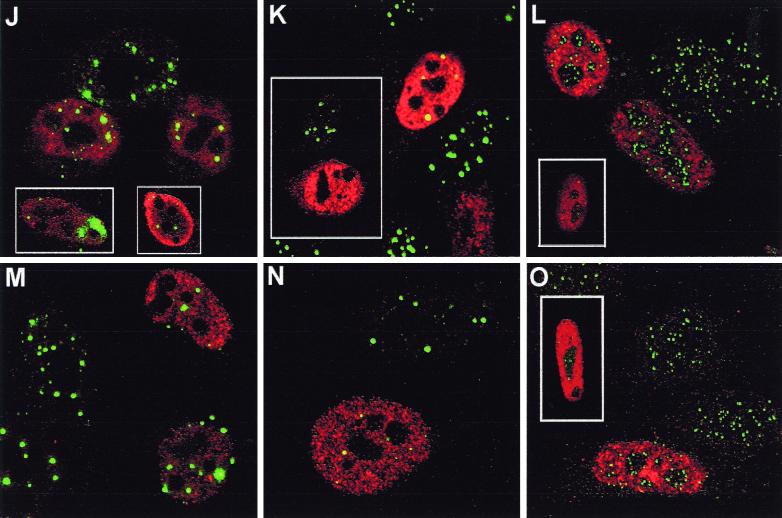
The location of the tagged proteins within transfected HEp-2 cells was also determined by confocal microscopy (Fig. 5). The distribution of tagged ICP0 was indistinguishable from that of untagged ICP0, giving a number of punctate foci within a diffuse nuclear background, although the level of expression varied between cells and in those with very high levels of expression, ICP0 could be found in large globs in the cytoplasm and nucleus. These latter cells were not analyzed in later microscopic studies. BICP0, Eg63, Vg61, and EP0 were generally expressed diffusely throughout the nucleus but with some local accumulations or dots, and again the level of expression varied between cells. Staining was identical with both formaldehyde or acetone-methanol fixation conditions, except that nuclear dots, especially in some cells transfected with BICP0, Eg63, and Vg61, were easier to distinguish after acetone-methanol fixation as the nuclear diffuse staining was reduced (data not shown).
FIG. 5.
Effect of tagged ICP0 and the tagged homologues on cellular proteins seen by confocal microscopy. HEp-2 cells were transfected with pp65-ICP0 (A to C), pp65-BICP0 (D to F), pp65-Eg63 (G to I), pp65-EP0 (J to L), or pp65-Vg61 (M to O). At 24 h posttransfection, cells were processed for confocal microscopy and costained with MAb anti-pp65 at a dilution of 1/1,000 and either polyclonal anti-PML r8 at a dilution of 1/1,000 (A, D, G, J, and M), polyclonal anti-Sp100 SpGh at a dilution of 1/1,000 (B, E, H, K, and N), or polyclonal anti-CENP-C r554 at a dilution of 1/500 (C, F, I, L, and O). Secondary antibodies used were FITC-conjugated goat anti-rabbit IgG (Sigma) at 1/100 and Cy3-conjugated goat anti-mouse (Amersham) at 1/1,000.
The ICP0 homologues affect cellular proteins.
The centromere protein CENP-C and the ND10 proteins PML and Sp100 are degraded in an ICP0-, RING finger-, and proteasome-dependent manner during HSV-1 infection. To determine the effect of the homologues on these proteins, HEp-2 cells were transfected with the tagged plasmids and analyzed by confocal microscopy.
Epitope-tagged ICP0 affected cellular proteins in a manner similar to that shown in published results for untagged protein; however, in all experiments there was a dose-dependent effect such that intermediate results were seen in cells expressing low amounts of the protein (Fig. 5A to C). Minor foci of undispersed PML remained in some transfected cells (Fig. 5A, inset), while CENP-C was generally completely dispersed in transfected cells (Fig. 5C, lower inset), although in cells expressing lesser amounts of tagged ICP0, association of ICP0 with remaining centromeres could occasionally be seen (Fig. 5C, middle cell main panel; upper inset shows a region of this cell magnified). Sp100 was also completely dispersed and its staining disappeared (Fig. 5B), strongly supporting the observations on the effect of ICP0 on Sp100 degradation during HSV-1 infection (Fig. 1 and 2) (see also Fig. 7B) (8).
Of the homologues, BICP0 had an effect similar to that of ICP0 (Fig. 5D to F), except that in some cells PML was redistributed into strange, globular, sometimes arc-like structures, some of which were coincident with BICP0 (Fig. 5D, upper inset; also data not shown). Punctate Sp100 and CENP-C staining was always lost from transfected cells, even when low amounts of BICP0 were expressed (Fig. 5E and F; the inset shows the CENP-C staining alone of the transfected cell in the main panel). In turn, Eg63 had a similar effect to BICP0, with PML remaining in abnormal foci in some transfected cells (Fig. 5G, inset) but extensively dispersed in others (Fig. 5G, main panel). Remaining PML, however, was never associated with Eg63 nuclear foci. Sp100 staining was always lost in pp65-Eg63-expressing cells (Fig. 5H), but punctate CENP-C staining was retained at lower intensity in some (Fig. 5I, inset). In contrast, EP0 associated with PML foci in some cells and redistributed PML in others but in general had a less dramatic effect than ICP0, BICP0, and Eg63 (Fig. 5J). The insets show other examples of EP0-expressing cells. Sp100 was either completely (Fig. 5K, inset) or partially (Fig. 5K, main panel) dispersed, while CENP-C was either unaffected or displayed reduced intensity (Fig. 5L, inset). Finally, Vg61 was the least active of the proteins in these assays. Many transfected cells had no obvious abnormality in PML staining (Fig. 5M), although Vg61 associated with some PML foci, but Sp100 was affected to a greater degree, although most cells retained some Sp100 foci (Fig. 5N). Any effect on CENP-C was limited to apparent reduced intensity in some transfected cells (Fig. 5O, inset).
All the proteins exhibited dose-dependent effects, and while EP0 and Vg61 were the least active in these assays, they were also the least efficiently expressed. However, the results show that all the homologues have significant effects on the immunofluorescence staining character of at least one of the proteins destabilized by ICP0.
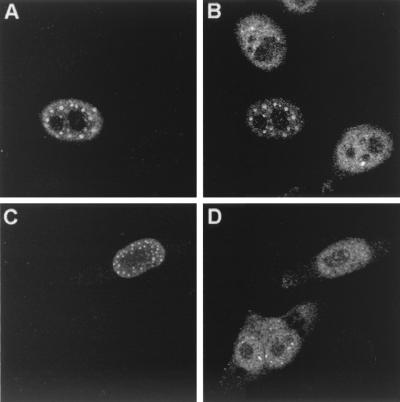
Only ICP0 affects the distribution of USP7.
The effect of ICP0 on USP7 has been assessed during viral infections (24) but not during transient transfections. To investigate this, HEp-2 cells were transfected with tagged ICP0, untagged ICP0, the ICP0 RING finger mutant FXE (which binds USP7 and accumulates at ND10 domains but fails to cause their redistribution [21]), or the ICP0 point mutant M1 (which fails to interact with USP7 in binding assays [26]). Tagged and untagged ICP0 had a marked and similar effect on the distribution of USP7 in the cell (Fig. 6B, tagged ICP0). The USP7 staining pattern changed from being nuclear diffuse with a few dots to punctate nuclear, and the majority of dots coincided with ICP0 dots (Fig. 6, compare the central transfected cell with the untransfected cells). This reorganization of USP7 was dramatically seen with FXE (data not shown), while in contrast the USP7 ICP0 binding mutant M1 failed to reorganize USP7, indicating that the binding of ICP0 to USP7 is essential for this effect to occur (Fig. 6D). Similar transfections using the tagged homologues showed that none had any effect on the distribution of USP7 (data not shown), a result that is consistent with the lack of any obvious USP7 binding motif in these proteins. This verifies that direct binding to USP7 is required for its redistribution to occur (24), and although the effect of the homologues on USP7 was not investigated in the context of a viral infection, it is likely that the corresponding alphaherpesviruses fail to affect USP7, as in HSV-1 infections only ICP0 was found to bind USP7. Thus, HSV-1 is unique in this respect amongst the alphaherpesviruses studied, and its effect on USP7 presumably reflects a difference in its life cycle.
FIG. 6.
Effect of ICP0 on USP7. HEp-2 cells were transfected with pp65-ICP0 (A and B) or pCIM1 (C and D), processed for confocal microscopy after 24 h, and costained with MAb anti-ICP0 11060 at 1/5,000 (A and C) and polyclonal anti-USP7 r201 at 1/200 (B and D). Secondary antibodies used were FITC-conjugated goat anti-rabbit IgG (Sigma) at 1/100 and Cy3-conjugated goat anti-mouse (Amersham) at 1/1,000 or Cy5-conjugated goat anti-mouse (Amersham) at 1/500.
EHV gene 63 proteasome-dependent degradation of cellular proteins during infection.
The loss of staining of the nuclear proteins described above could be due to an alteration in the stability of the proteins and their modified forms caused by the ICP0 homologues. However, the transfection approach is limited to visualizing only the effect on the immunofluorescent staining pattern and intensity of the endogenous cellular proteins, since insufficient cells are transfected to allow analysis by Western blotting. In a previous study, we had constructed a variant of HSV-1 strain 17syn+ which expresses Eg63 in place of ICP0 (17Eg63) (23); therefore in this case, the fate of the cellular proteins during infection could be studied by Western blotting. HFL cells were mock infected or infected with wild-type 17syn+, 17Eg63, FXE, or dl1403 (ICP0 deletion mutant) viruses in the presence or absence of the proteasome inhibitor MG132 and analyzed by Western blotting (Fig. 7).
The results showed that, similar to the wild-type 17syn+ virus, virus 17Eg63 induced the degradation of CENP-C and the PML and Sp100 isoforms during infection (albeit less efficiently than strain 17syn+) and that this was dependent on proteasome activity (Fig. 7A and B and data not shown for inhibition for Sp100 degradation by MG132). Since the ICP0 null mutant dl1403 does not induce degradation (although in this experiment dl1403 slightly altered the mobility of CENP-C-related bands), this effect must be due to the expression of Eg63 by this virus. Reprobing of the blot for ICP4 indicated that the cells had been successfully infected and that the infections had not been inhibited by MG132 at the high multiplicities used. However, expression of ICP4 by 17Eg63 was consistently lower than that of the other viruses despite the identical amount of titered input virus used; this may reflect the failure of Eg63 to completely complement for the absence of ICP0 (22).
Effect of ICP0 and its homologues on the SUMO-1 conjugations of exogenous PML.
As previously mentioned, ICP0 has been shown to abrogate the SUMO-1 modification of Sp100 and PML during transfection (63). We were interested in determining whether the homologues also have this effect, since if so, this would indicate that they function via a common pathway or mechanism. To this end, HEp-2 cells were transfected with plasmid pPML(F), which expresses an F epitope-tagged version of PML [PML(F)] (42), together with plasmids expressing either pp65-ICP0, the pp65-homologues, or pUC9 control. Transfections were checked by immunofluorescence to determine that the exogenously expressed PML was correctly targeted to the nucleus and the ND10 domains, indicating that it had been SUMO-1 conjugated (62) and that the homologues were sufficiently expressed. Controls of transfected cells costained for either ICP0 or BICP0 and exogenous PML showed that cotransfection was successful (costaining was not possible for the other homologues due to the lack of a suitable antibody). Cellular extracts were analyzed by immunoblotting with antibodies to detect PML(F) and the tagged ICP0 and its homologues.
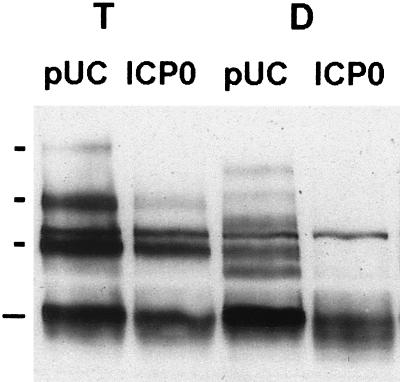
When a pUC plasmid was introduced with pPML(F), five exogenous modified PML bands were observed (Fig. 8A, indicated by short bars). These have previously been interpreted as SUMO-1 or SUMO-1-like modified PML (40, 62, 74). Cotransfection of a plasmid expressing tagged ICP0 with pPML(F) caused, as expected, a significant decrease in the intensity of modified PML(F) bands. Muller and Dejean reported that the presumedly unmodified form of PML was not affected by cotransfected ICP0 (63). However, we found that if PML(F) expression levels were decreased to low levels by reducing the amount of plasmid pPML(F), a clear reduction in the amount of unmodified PML(F) was observed in the ICP0 cotransfection (indicated by long bars in Fig. 8A). Furthermore, there appeared to be microheterogeneity and smearing of the unmodified PML(F) material, which implies that ICP0 may be affecting other aspects of PML, such as phosphorylation. These important results suggest that ICP0 alone is responsible for inducing the abrogation of SUMO-1 conjugation of PML and the subsequent degradation of unmodified PML.
FIG. 8.
Effect of ICP0 and its homologues on the SUMO-1 conjugation of PML. Using Lipofectamine PLUS, HEp-2 cells were (A) cotransfected with pPML(F) and either pUC9 (lane pUC), pp65-ICP0 (lane ICP0), or the pp65-homologue plasmids (remaining lanes) shown, and harvested into SDS-gel loading buffer at 24 h posttransfection and analyzed by Western blotting using MAb anti-F at a dilution of 1/5,000 or (B) cotransfected with pPML(F) and either pUC9 or pp65-ICP0 and treated with 5 μM MG132 at 24 h posttransfection and harvested over the hourly time course as indicated. The positions of unmodified and SUMO-1-conjugated PML(F) bands are indicated by the long and short bars, respectively, in panel A, and the arrow (A) indicates a background antibody-detected band present in untransfected cells.
As a further control to show that the effect of ICP0 on PML(F) was not due to inhibition of transcription or translation from plasmid pPML(F), cotransfected cells were prepared as described above and then treated with MG132 to inhibit proteasome activity, at a time when ICP0 had abrogated the SUMO-1 conjugation and led to degradation of PML(F). Analysis of PML(F) expression at various times after MG132 treatment showed a time-dependent increase in both unmodified and modified forms of PML(F) (Fig. 8B). This suggests that there is continued transcription and translation from plasmid pPML(F) but that in the presence of ICP0 the newly synthesized PML(F) protein is subject to degradation.
Analysis of the effects of the ICP0 homologue proteins in this assay revealed that none were able to affect either the expression of modified or unmodified forms of PML(F) (Fig. 8A). Except in the case of Vg61, this result could not be explained by poor expression of the homologues, as all were expressed at high levels (data not shown), nor was it due to saturation by overexpression of PML(F), as no effect was seen even when PML(F) expression was reduced to the lowest possible levels. Costaining by immunofluorescence showed that virtually all cells which were transfected with pPML(F) were also expressing ICP0 or BICP0 in the relevant experiments, and although it was not possible to detect coexpression of EP0 and Eg63 in this system because of a lack of suitable antibodies, the proportions of transfected cells expressing all these proteins in transfections stained singly were similar.
Further, HEp-2 cells were triple transfected with plasmids pPML(F); pCIPIC1, a plasmid expressing a myc-tagged version of SUMO-1 (25); and pp65-ICP0 to determine whether overexpression of SUMO-1 altered the effect of ICP0 on PML(F). In these triple transfections it is clear that the size of PML(F) is altered by the exogenous SUMO-1 (Fig. 9, bands in track T pUC indicated by short bars) and that ICP0 affects this SUMO-1-conjugation of PML(F) (Fig. 9, compare track T pUC with track T ICP0), although not to such a dramatic effect as for the double transfection without the SUMO-1 plasmid (Fig. 9, tracks D). This is probably due to the increased SUMO-1 in the cell affecting the equilibrium of the reaction. It was also interesting to note that the size of the SUMO-1-conjugated PML(F) bands is altered in the presence of exogenous SUMO-1. The reason for this effect is unclear, although it could reflect increased multiply modified forms of PML created by unexplained posttranslational modification or it could be that the exogenous SUMO-1 imparts a different mobility shift on PML due to the presence of the tag sequence. Once again the homologues failed to have any effect on the SUMO-1 conjugation of this modified PML(F) (data not shown).
FIG. 9.
Effect of ICP0 on the SUMO-1 conjugation of PML in the presence of exogenous SUMO-1. Using Lipofectamine PLUS, HEp-2 cells were cotransfected with pPML(F) and either pUC9 or pp65-ICP0 (lanes D) or triple transfected with pPML(F), pCIPIC1, and either pUC9 or pp65-ICP0 (lanes T). Samples were harvested into SDS-gel loading buffer at 24 h posttransfection and analyzed by Western blotting using MAb anti-F at a dilution of 1/5,000. The positions of unmodified PML(F) and PML(F) modified by exogenous SUMO-1 in the triple transfections are indicated by long and short bars, respectively.
Importance of the RING finger of ICP0 on modification and stability of PML.
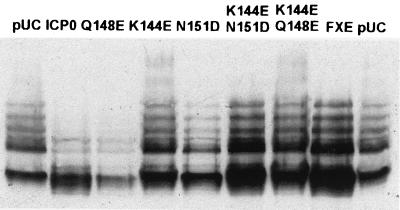
To investigate further the degradation of modified and unmodified forms of PML caused by ICP0, HEp-2 cells were transfected with pPML(F) and either pCIneo, p111 (untagged ICP0), p110FXE (RING finger mutant), p110K144E, p110N151D, p110Q148E, (single-point mutations in the RING finger), p110K144E N151D, or p110K144E Q148E (double-point mutations in the RING finger). Untagged ICP0 abrogated PML(F) conjugation and induced its degradation, as did mutant ICP0 RING finger mutant p110Q148E (Fig. 10). In contrast, deletion of the RING finger domain and mutation of RING finger amino acid residues K144E and N151D, either singly or in combination with other RING finger point mutations, eliminated the effect of ICP0 on PML SUMO-1 conjugation. This indicates that the RING finger and specific critical residues within it are important in abrogating the SUMO-1 conjugation of PML and causing its subsequent destabilization.
FIG. 10.
The RING finger of ICP0 is required for abrogation of SUMO-1 conjugation of PML. HEp-2 cells were cotransfected with plasmids as shown and pPML(F), using Lipofectamine PLUS. Samples were harvested into SDS-gel loading buffer at 24 h posttransfection and analyzed by Western blotting using MAb anti-F at a dilution of 1/5,000. p110N151D caused a significant level of transfected-cell mortality in this experiment, which explains why the level of PML(F) is reduced in this lane (lane N151D); however, staining for ICP0 indicated that the remaining cotransfected cells expressed ICP0.
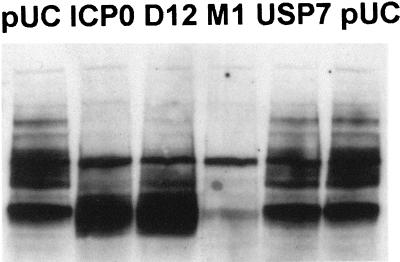
To determine the effect of USP7 binding to ICP0 on PML(F) stability, we used plasmids pCIM1 and p110D12, which contain substitutions and deletions in the USP7 binding regions, respectively. ICP0 mutant protein D12 gave results similar to those of the wild-type ICP0 in this assay, but the point mutant M1 was surprisingly exceptionally active, routinely almost completely eliminating PML(F) protein accumulation (Fig. 11). As with wild-type ICP0, addition of MG132 allowed the reaccumulation of PML(F) in pCIM1-cotransfected cells (data not shown), which confirms that the effect is on protein stability rather than on transcription and translation. Although mutant D12 gave similar results to the wild type, the behavior of mutant M1 suggests that binding to USP7 might in some way regulate ICP0 activity. Cotransfection of USP7 itself had no effect on the PML(F).
FIG. 11.
Effect of ICP0 USP7 binding mutants on the SUMO-1 conjugation of PML. HEp-2 cells were cotransfected with pPML(F) and the plasmids as shown, using Lipofectamine PLUS. Samples harvested 24 h posttransfection were analyzed by Western blotting using MAb anti-F at a dilution of 1/5,000.
DISCUSSION
This paper demonstrates that the homologues of HSV-1 protein ICP0 expressed by the related alphaherpesviruses BHV-1, EHV-1, PRV, and VZV all, like ICP0, cause changes to ND10 structures in transfected cells and, with the possible exceptions of PRV EP0 and VZV Vg61, also disrupt centromeres. Further, of the cellular proteins studied, all the ICP0 proteins affected the ND10 protein Sp100 more readily than they affected PML and even more so than they affected CENP-C. The severity of these effects, however, varied between the different ICP0 homologous proteins and was dose dependent; the apparently lesser effects of VZV Vg61 may be due to the poor expression of this protein in our systems. Previous observations that these ICP0 family members regulate gene expression and can in some cases at least partially complement one another may be related to our current findings on their similar effects on cellular nuclear structures.
To investigate the basis of these effects, we have expanded the studies on cellular protein stability and have shown that ICP0 induces the proteasome-dependent degradation of Sp100, particularly its SUMO-1-modified isoforms during infection and that this mirrors in both time course and sequence requirements the induced degradation of PML. Recombinant virus 17Eg63 demonstrated that EHV-1 protein Eg63 has an effect similar to that of ICP0 on cellular proteins PML, Sp100, and CENP-C during virus infection, which suggests that the effect of transfected Eg63 on ND10 and centromere structures may also be due to induced proteasome-dependent degradation of the endogenous cellular proteins. The RING finger domain of ICP0 is essential to induce the proteasome-dependent degradation of the cellular proteins (25, 27, 66). It is possible that this region interacts with other cellular proteins leading to the activation of the degradation pathway (discussed below). The fact that the homologues also affect, to differing extents, these cellular proteins and that the only region of homology between them is the RING finger domain indicates that this region is of major importance in the induced disruption. This suggests that a similar or related pathway may be involved in all cases.
The homologues are thus related to ICP0 in function but are not identical. This is hardly surprising, due to their limited homology outside the RING finger domain, as demonstrated by the fact that only ICP0 has a USP7 binding domain and through this domain alters the distribution of USP7 in transfected cells. The RING finger could act as an instability-inducing domain targeting a protein instability pathway, with differences in its sequence accounting for the different intrinsic activity of the homologues, while other regions of the proteins could be important for the specificity of proteins targeted for degradation. Indeed, there is evidence from domain swap chimera experiments that the RING finger domain might have to be in the context of a larger portion of the parent protein in order to be active (59). So although targeting of a protein instability pathway or function is likely to be a common function of ICP0 family members, there is variation in subsequent substrate specificity. The difference in the specificity of the homologues probably has a biological basis and could reflect the differences in the hosts and cell types infected by the viruses and their pathological properties.
In support of our general hypothesis that ICP0 functions via the ubiquitin-proteasome pathway is the increasing evidence that many RING finger proteins participate in E3 ubiquitin ligase complexes in the ubiquitin-dependent protein degradation pathway (reviewed in references 6 and 32) (35, 36, 38, 41, 50, 52, 61, 65, 71, 85). For example, the c-Cbl RING finger protein binds the target substrate and recruits (and possibly activates) a ubiquitin-conjugating E2 enzyme (38, 85). Other RING finger proteins, such as Rbx-1 and APC11, are components of multisubunit E3 ubiquitin ligases and are involved in E2 binding only, with other subunits binding the substrates (41, 65, 71, 72, 80). In all cases the RING finger domain is responsible for E2 interaction. It is likely that ICP0 and its homologues function in a similar manner, with their RING finger domains interacting with an E2 ubiquitin-conjugating enzyme and other regions interacting with substrate or other components of an E3 complex. Consistent with these suggestions, we have found that in both transfected and infected cells, foci of accumulated ICP0 contain enhanced levels of conjugated ubiquitin (29). This occurs in a RING finger-dependent manner, consistent with ICP0 functioning as or stimulating E3 ligase activity. Although the RING finger domains of the homologues do not appear to function exactly as does that of ICP0, it would be interesting to determine whether the homologues have the same effect on conjugated ubiquitin in transfected cells. Preliminary evidence with BICP0 suggests that this may be so (data not shown), but more reagents will be required for definitive studies, because the available antibodies do not permit this approach with the other homologues.
Further evidence of the ability of ICP0 to induce proteasome-dependent degradation of selected target proteins comes from the cotransfection studies reported here. Our experiments in part confirmed the previous results of Muller and Dejean (63), but by exploring the assay conditions we have demonstrated that ICP0 not only causes the abrogation of SUMO-1 conjugation of PML but also induces the proteasome-dependent degradation of unmodified PML protein. This is the first experiment to demonstrate directly that ICP0 acts in this way in the absence of other viral protein expression. The more detailed investigation of the role of the ICP0 RING finger in SUMO-1 abrogation and subsequent degradation of exogenous PML indicated that, of the amino acid substitutions in the ICP0 RING finger studied, only Q148E retains wild-type ICP0 activity. The behavior of the RING finger substitutions studied in this assay is consistent with their activity in other assays of ICP0 function (22, 29), indicating that the RING finger and, more specifically, certain residues within it are very important for a wide range of properties of ICP0.
In contrast, however, none of the ICP0 homologues had any effect on exogenous PML in the cotransfection assay. Given the effect of these proteins on endogenous PML in transfected cells (Fig. 5) and the effect of virus 17Eg63 on PML in infected cells (Fig. 7A), these results are surprising. Also surprising in view of the data from infected cells was our finding that in similar cotransfection experiments, wild-type ICP0 had little effect on exogenous Sp100, apart from a slight and variable alteration to the ratio between the modified and unmodified forms (data not shown). Muller and Dejean (63) reported that ICP0 reduced SUMO-1 conjugation of cotransfected Sp100, although their published data suggest that the effect was less dramatic than that seen with PML(F). However, ICP0 has the potential to affect exogenous Sp100 in cotransfection assays, since mutant M1, like its effect on PML(F), caused dramatic instability of all forms of exogenous Sp100 which could be reversed by the addition of MG132 (data not shown). Again, none of the ICP0 homologues affected exogenous Sp100 in cotransfection assays despite their effects on endogenous Sp100 distribution.
These results suggest that some aspects of the activities of ICP0 and its homologues in infected and transfected cells are as yet poorly understood. All the members of the ICP0 family of viral proteins clearly affect ND10, yet only ICP0 affected exogenous PML in cotransfection assays and exogenous Sp100 was affected only by a mutant form of ICP0. We can suggest a number of mutually inexclusive explanations for these observations.
Nonequivalence of endogenous and exogenous proteins.
Both PML and Sp100 are highly complex proteins, being multiply posttranslationally modified and derived from large families of alternatively spliced transcripts. Exogenously expressed protein represents only one of the alternatively spliced forms, may not be properly modified in all respects, and may be expressed at unnaturally high levels in the transfected cells. These factors will influence not only the quality of the exogenous protein but also its localization and assembly into the correct macromolecular complexes. If this is the case, it is not difficult to envisage differing responses of endogenous and exogenous proteins to ICP0 and to suppose that these responses would be affected by individual variations among the ICP0 family members.
Dominant effects of initial disruption of endogenous ND10.
We observed that in all ICP0- and some BICP0-cotransfected cells, exogenous Sp100 was not present in ND10 but was diffuse throughout the nucleus, which is not surprising, given the effects of ICP0 and BICP0 on endogenous PML and ND10 domains. Recent evidence suggests that PML may be essential for ND10 domain integrity (37) and that in its absence or deconjugation from SUMO-1, the ND10 domains disintegrate. The diffuse localization of exogenous Sp100 in cells also expressing ICP0 is likely to be due to loss of endogenous PML and ND10 domains; the failure of ICP0 and its homologues to degrade the exogenous Sp100 could be because it is no longer targeted to the appropriate macromolecular complexes.
The ICP0 homologues could affect an ND10 component other than PML.
While it has been shown that PML is essential for ND10 domain integrity (37), it is also possible that some other component as yet unidentified is similarly essential. If the ICP0 homologues preferentially targeted this component, it would explain why they disrupt ND10 without apparently affecting PML and Sp100 in the cotransfection assay.
Differential effects on SUMO-1-specific proteases.
Recently a number of cellular SUMO-1-specific proteases have been identified. These enzymes remove SUMO-1 from conjugated proteins (including PML) but do not themselves cause the degradation of these proteins (33, 48, 49, 78). ICP0 could possibly interact with or cause a SUMO-1 protease to deconjugate exogenous PML. Subsequently, ICP0 would induce the degradation of the unconjugated protein via the proteasome pathway. There is precedence for this model: SUMO-1-conjugated IκBα is resistant to degradation by the proteasome, and before it can be ubiquitinated and degraded, it must first be deconjugated (13). It is possible that the ICP0 homologues fail to target exogenous PML, because for some reason they are unable to influence SUMO-1 protease activity on the exogenous protein.
The concept that different regulatory proteins could interact with and disrupt ND10 by different mechanisms is not without precedent. For example, although the HCMV IE1 protein has been reported to abrogate the SUMO-1 modification of PML in transfection assays (63), its role in the disassembly of ND10 structures during HCMV infection appears to involve direct binding to PML and its sequestration elsewhere, rather than altered modification (1, 2). Furthermore, E4orf3 expression during adenovirus infection disrupts ND10, eventually giving rise to SUMO-1 demodification and the appearance of novel modified forms of PML (47), although the E4orf3 protein itself in transfection assays has no apparent effect on PML modification (63). The differences between the transfection and infection approaches in these other systems emphasize that transfected cells do not necessarily recapitulate the situation of endogenous proteins, in ways that are as yet not understood.
The fact that different viral regulatory proteins target the same nuclear structures via different mechanisms strongly implies that the ND10 structures have an important general role in the biology of virus-cell interactions. We have discussed in detail elsewhere the working hypothesis that ICP0 inactivates a cellular repression mechanism by inducing the degradation of selected cellular proteins (27, 28, 30) and we note that this hypothesis is analogous to what may be a generally common cellular regulatory strategy (6, 32, 51). The next challenges are to understand the consequences of the degradation of cellular proteins brought about by ICP0 and its homologues and to determine the precise molecular mechanisms by which the degradation is achieved.
ACKNOWLEDGMENTS
We thank Martin Schwyzer for permission to use plasmid pBCM26 (supplied by Len Bello) to obtain BHV sequences and for the BICP0 rabbit serum, Pierre Chambon for MAb anti-F tag and pPML(F), Paul Freemont for antiserum r8, and Thomas Sterndorf for antiserum SpGH. Anne Orr provided valuable technical assistance, and Duncan McGeoch provided constructive criticism.
This research was supported by the Medical Research Council.
REFERENCES
- 1.Ahn J-H, Hayward G S. The major immediate-early proteins IE1 and IE2 of human cytomegalovirus colocalize with and disrupt PML-associated nuclear bodies at very early times in infected permissive cells. J Virol. 1997;71:4599–4613. doi: 10.1128/jvi.71.6.4599-4613.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn J-H, Brignole III E J, Hayward G S. Disruption of PML subnuclear domains by the acidic IE1 protein of human cytomegalovirus is mediated through interaction with PML and may modulate a RING finger-dependent cryptic transactivator function of PML. Mol Cell Biol. 1998;18:4899–4913. doi: 10.1128/mcb.18.8.4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ali S, Lutz Y, Bellocq J P, Chenard-Neu M P, Rouyer N, Metzger D. Production and characterization of monoclonal antibodies recognising defined regions of the human oestrogen receptor. Hybridoma. 1993;12:391–405. doi: 10.1089/hyb.1993.12.391. [DOI] [PubMed] [Google Scholar]
- 4.Ascoli C A, Maul G G. Identification of a novel nuclear domain. J Cell Biol. 1991;112:785–795. doi: 10.1083/jcb.112.5.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boddy M N, Howe K, Etkin L D, Solomon E, Freemont P S. PIC1, a novel ubiquitin-like protein which interacts with the PML component of a multiprotein complex that is disrupted in acute promyelocytic leukaemia. Oncogene. 1996;13:971–982. [PubMed] [Google Scholar]
- 6.Borden K L B. RING domains: master builders of molecular scaffolds? J Mol Biol. 2000;295:1103–1112. doi: 10.1006/jmbi.1999.3429. [DOI] [PubMed] [Google Scholar]
- 7.Cai W, Astor T D, Liptak L M, Cho C, Coen D, Schaffer P A. The herpes simplex virus type 1 regulatory protein ICP0 enhances replication during acute infection and reactivation from latency. J Virol. 1993;67:7501–7512. doi: 10.1128/jvi.67.12.7501-7512.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chelbi-Alix M K, de Thé H. Herpes virus induced proteasome-dependent degradation of the nuclear bodies-associated PML and Sp100 proteins. Oncogene. 1999;18:935–941. doi: 10.1038/sj.onc.1202366. [DOI] [PubMed] [Google Scholar]
- 9.Cheung A K. DNA nucleotide sequence analysis of the immediate-early gene of pseudorabies virus. Nucleic Acids Res. 1989;17:4637–4646. doi: 10.1093/nar/17.12.4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ciufo D M, Mullen M, Hayward G S. Identification of a dimerization domain in the C-terminal segment of the IE110 transactivator protein from herpes simplex virus. J Virol. 1994;68:3267–3282. doi: 10.1128/jvi.68.5.3267-3282.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clements G B, Stow N D. A herpes simplex virus type 1 mutant containing a deletion within immediate-early gene 1 is latency competent in mice. J Gen Virol. 1989;70:2501–2506. doi: 10.1099/0022-1317-70-9-2501. [DOI] [PubMed] [Google Scholar]
- 12.Davison A J, Scott J E. The complete DNA sequence of varicella-zoster virus. J Gen Virol. 1986;67:1759–1816. doi: 10.1099/0022-1317-67-9-1759. [DOI] [PubMed] [Google Scholar]
- 13.Desterro J M P, Rodriguez M S, Hay R T. SUMO-1 modification of IkBα inhibits NF-κB activation. Mol Cell. 1998;2:233–239. doi: 10.1016/s1097-2765(00)80133-1. [DOI] [PubMed] [Google Scholar]
- 14.Dyck J A, Maul G G, Miller W H, Jr, Chen J D, Kakizuka A, Evans R M. A novel macromolecular structure is a target of the promyelocytic-retinoic acid receptor oncoprotein. Cell. 1994;76:333–343. doi: 10.1016/0092-8674(94)90340-9. [DOI] [PubMed] [Google Scholar]
- 15.Everett R D. A detailed mutational analysis of Vmw110, a trans-acting transcriptional activator encoded by herpes simplex virus type 1. EMBO J. 1987;6:2069–2076. doi: 10.1002/j.1460-2075.1987.tb02472.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Everett R D. Analysis of the functional domains of herpes simplex virus type 1 immediate-early polypeptide Vmw110. J Mol Biol. 1988;202:87–96. doi: 10.1016/0022-2836(88)90521-9. [DOI] [PubMed] [Google Scholar]
- 17.Everett R D. Construction and characterisation of herpes simplex virus type 1 mutants with defined lesions in immediate-early gene 1. J Gen Virol. 1989;70:1185–1202. doi: 10.1099/0022-1317-70-5-1185. [DOI] [PubMed] [Google Scholar]
- 18.Everett R D, Preston C M, Stow N D. Functional and genetic analysis of the role of Vmw110 in herpes simplex virus replication. In: Wagner E K, editor. The control of herpes simplex virus gene expression. Boca Raton, Fla: CRC Press, Inc.; 1991. pp. 50–76. [Google Scholar]
- 19.Everett R D, Cross A, Tyler J K, Orr A. An epitope within the DNA binding domain of the herpes simplex virus immediate-early protein Vmw175 is conserved in the varicella-zoster virus gene 62 protein. J Gen Virol. 1993;74:1955–1958. doi: 10.1099/0022-1317-74-9-1955. [DOI] [PubMed] [Google Scholar]
- 20.Everett R D, Cross A, Orr A. A truncated form of herpes simplex virus type 1 immediate-early protein Vmw110 is expressed in a cell-type dependent manner. Virology. 1993;197:751–756. doi: 10.1006/viro.1993.1651. [DOI] [PubMed] [Google Scholar]
- 21.Everett R D, Maul G G. HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J. 1994;13:5062–5069. doi: 10.1002/j.1460-2075.1994.tb06835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Everett R D, Barlow P N, O'Hare P, O'Rourke D, Orr A. Point mutations in the HSV-1 Vmw110 RING finger helix affect activation of gene expression, viral growth, and interaction with PML-containing nuclear structures. J Virol. 1995;69:7339–7344. doi: 10.1128/jvi.69.11.7339-7344.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Everett R D, Orr A, Elliott M. The equine herpesvirus 1 gene 63 RING finger protein partially complements Vmw110, its herpes simplex virus type 1 counterpart. J Gen Virol. 1995;76:2369–2374. doi: 10.1099/0022-1317-76-9-2369. [DOI] [PubMed] [Google Scholar]
- 24.Everett R D, Meredith M R, Orr A, Cross A, Kathoria M, Parkinson J. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO J. 1997;16:1519–1530. doi: 10.1093/emboj/16.7.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Everett R D, Freemont P, Saitoh H, Dasso M, Orr A, Kathoria M, Parkinson J. The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J Virol. 1998;72:6581–6591. doi: 10.1128/jvi.72.8.6581-6591.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Everett R D, Meredith M, Orr A. The ability of herpes simplex virus type 1 immediate-early protein Vmw110 to bind to a ubiquitin-specific protease contributes to its roles in the activation of gene expression and stimulation of virus replication. J Virol. 1999;73:417–426. doi: 10.1128/jvi.73.1.417-426.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Everett R D, Earnshaw C E, Findley J, Lomonte P. Specific destruction of kinetochore protein CENP-C and disruption of cell division by herpes simplex virus immediate-early protein Vmw110. EMBO J. 1999;18:1526–1538. doi: 10.1093/emboj/18.6.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Everett R D. A surprising role for the proteasome in the regulation of herpesvirus infection. Trends Biochem Sci. 1999;24:293–295. doi: 10.1016/s0968-0004(99)01433-4. [DOI] [PubMed] [Google Scholar]
- 29.Everett R D. ICP0 induces the accumulation of conjugated ubiquitin. J Virol. 2000;74:9994–10005. doi: 10.1128/jvi.74.21.9994-10005.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Everett R D. ICP0, a regulator of herpes simplex virus during lytic and latent infection. Bioessays. 2000;22:761–770. doi: 10.1002/1521-1878(200008)22:8<761::AID-BIES10>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 31.Fraefel C, Zeng J, Choffat Y, Engels M, Schwyzer M, Ackermann M. Identification and zinc dependence of the bovine herpesvirus 1 transactivator protein BICP0. J Virol. 1994;68:3154–3162. doi: 10.1128/jvi.68.5.3154-3162.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Freemont P S. Ubiquitination: RING for destruction? Curr Biol. 2000;10:R84–R87. doi: 10.1016/s0960-9822(00)00287-6. [DOI] [PubMed] [Google Scholar]
- 33.Gong L, Stefanos M, Maul G G, Yeh E T H. Differential regulation of sentirinized proteins by a novel sentin-specific protease. J Biol Chem. 2000;275:3355–3359. doi: 10.1074/jbc.275.5.3355. [DOI] [PubMed] [Google Scholar]
- 34.Harris R A, Everett R D, Zhu X, Silverstein S, Preston C M. Herpes simplex virus type 1 immediate early protein Vmw110 reactivates latent herpes simplex virus type 2 in an in vitro latency system. J Virol. 1989;63:3513–3515. doi: 10.1128/jvi.63.8.3513-3515.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Honda R, Hirofumi T, Hideyo Y. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 36.Hu G, Fearon E R. Siah-1 N-terminal RING domain is required for proteolysis function, and C-terminal sequences regulate oligomerization and binding to target proteins. Mol Cell Biol. 1999;19:724–732. doi: 10.1128/mcb.19.1.724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ishov A M, Sotnikov A G, Negorev D, Vladimirova O V, Neff N, Kamitani T, Yeh E T H, Strauss III J F, Maud G G. PML is critical for ND10 formation and recruits PML-interacting protein Daxx to this nuclear structure when modified by SUMO-1. J Cell Biol. 1999;147:221–233. doi: 10.1083/jcb.147.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Joazeiro C A P, Wing S S, Huang H, Leverson J D, Hunter T, Liu Y. The tyrosine kinase negative regulator c-Cbl as a RING-type E2-dependent ubiquitin-protein ligase. Science. 1999;286:309–312. doi: 10.1126/science.286.5438.309. [DOI] [PubMed] [Google Scholar]
- 39.Johnson P R, Hochstrasser M. SUMO-1: Ubiquitin gains weight. Trends Cell Biol. 1997;7:408–413. doi: 10.1016/S0962-8924(97)01132-X. [DOI] [PubMed] [Google Scholar]
- 40.Kamitani T, Nguyen H P, Kito K, Fukuda-Kamitani T, Yeh E T H. Covalent modification of PML by the Sentrin family of ubiquitin-like proteins. J Biol Chem. 1998;273:3117–3120. doi: 10.1074/jbc.273.6.3117. [DOI] [PubMed] [Google Scholar]
- 41.Kamura T, Koepp D M, Conrad M N, Skowyra D, Moreland R J, Iliopoulos O, Lane W S, Kaelin W G, Jr, Elledge S J, Conaway R C, Harper J W, Conaway J W. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science. 1999;284:657–661. doi: 10.1126/science.284.5414.657. [DOI] [PubMed] [Google Scholar]
- 42.Kastner P A, Perez A, Lutz Y, Rochette-Egly C, Gaub M P, Durand B, Lanotte M, Berger R, Chambon P. Structure, localization and transcriptional properties of two classes of retinoic acid receptor α fusion proteins in acute promyelocytic leukemia (APL): structural similarities with a new family of oncoproteins. EMBO J. 1992;11:629–642. doi: 10.1002/j.1460-2075.1992.tb05095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kawaguchi Y, Van Sant C, Roizman B. Herpes simplex virus regulatory protein ICP0 interacts with and stabilizes the cell cycle regulator cyclin D3. J Virol. 1997;71:7328–7336. doi: 10.1128/jvi.71.10.7328-7336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Korioth F, Gieffers C, Maul G G, Frey J. Molecular characterisation of NDP52, a novel protein of nuclear domain 10 which is redistributed upon infection and interferon treatment. J Cell Biol. 1995;130:1–14. doi: 10.1083/jcb.130.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lees-Miller S P, Long M C, Kilvert M A, Lam V, Rice S A, Spencer C A. Attenuation of DNA-dependent protein kinase activity and its catalytic subunit by the herpes simplex virus type 1 transactivator ICP0. J Virol. 1996;70:7471–7477. doi: 10.1128/jvi.70.11.7471-7477.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leib D A, Coen D M, Bogard C L, Hicks K A, Yager D R, Knipe D M, Tyler K L, Schaffer P A. Immediate early regulatory gene mutants define different stages in the establishment and reactivation of herpes simplex virus latency. J Virol. 1989;63:759–768. doi: 10.1128/jvi.63.2.759-768.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leppard K N, Everett R D. The adenovirus type 5 E1b 55K and E4 Orf3 proteins associate in infected cells and affect ND10 domains. J Gen Virol. 1999;80:997–1008. doi: 10.1099/0022-1317-80-4-997. [DOI] [PubMed] [Google Scholar]
- 48.Li S-J, Hochstrasser M. A new protease required for cell-cycle progression in yeast. Nature. 1999;398:246–251. doi: 10.1038/18457. [DOI] [PubMed] [Google Scholar]
- 49.Li S-J, Hochstrasser M. The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Mol Cell Biol. 2000;20:2367–2377. doi: 10.1128/mcb.20.7.2367-2377.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lorick K L, Jensen J P, Fang S, Ong A M, Hatakeyama S, Weissman A M. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci USA. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maniatis T. A ubiquitin ligase complex essential for NK-κB, Wnt/Wingless and Hedgehog signalling pathways. Genes Dev. 1999;13:505–510. doi: 10.1101/gad.13.5.505. [DOI] [PubMed] [Google Scholar]
- 52.Martinez-Noel G, Niedenthal R, Tamura T, Harbers K. A family of structurally related RING finger proteins interacts specifically with the ubiquitin-conjugating enzyme UbcM4. FEBS Lett. 1999;454:257–261. doi: 10.1016/s0014-5793(99)00823-6. [DOI] [PubMed] [Google Scholar]
- 53.Maul G G, Guldner H H, Spivack J G. Modification of discrete nuclear domains induced by herpes simplex virus type 1 immediate-early gene 1 product ICP0. J Gen Virol. 1993;74:2679–2690. doi: 10.1099/0022-1317-74-12-2679. [DOI] [PubMed] [Google Scholar]
- 54.Maul G G, Everett R D. The nuclear location of PML, a cellular member of the C3HC4 zinc binding domain protein family, is rearranged during herpes simplex virus infection by the C3HC4 viral protein ICP0. J Gen Virol. 1994;75:1223–1233. doi: 10.1099/0022-1317-75-6-1223. [DOI] [PubMed] [Google Scholar]
- 55.Meredith M R, Orr A, Everett R D. Herpes simplex virus type 1 immediate-early protein Vmw110 binds strongly and specifically to a 135-kDa cellular protein. Virology. 1994;200:457–469. doi: 10.1006/viro.1994.1209. [DOI] [PubMed] [Google Scholar]
- 56.Meredith M R, Orr A, Elliott M, Everett R D. Separation of the sequence requirements for HSV-1 Vmw110 multimerisation and interaction with a 135-kDa cellular protein. Virology. 1995;209:174–187. doi: 10.1006/viro.1995.1241. [DOI] [PubMed] [Google Scholar]
- 57.Moriuchi H, Moriuchi M, Smith H A, Straus S E, Cohen J I. Varicella-zoster virus open reading frame 61 protein is functionally homologous to herpes simplex virus type 1 ICP0. J Virol. 1992;66:7303–7308. doi: 10.1128/jvi.66.12.7303-7308.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moriuchi H, Moriuchi M, Straus S E, Cohen J I. Varicella-zoster virus (VZV) open reading frame 61 protein transactivates VZV gene promoters and enhances the infectivity of VZV DNA. J Virol. 1993;67:4290–4295. doi: 10.1128/jvi.67.7.4290-4295.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moriuchi H, Moriuchi M, Cohen J I. The RING finger domain of the varicella-zoster virus open reading frame 61 protein is required for its transregulatory functions. Virology. 1994;205:238–246. doi: 10.1006/viro.1994.1639. [DOI] [PubMed] [Google Scholar]
- 60.Moriuchi H, Moriuchi M, Dean H, Cheung A K, Cohen J I. Pseudorabies virus EP0 is functionally homologous to varicella-zoster virus ORF61 protein and herpes simplex virus type 1 ICP0. Virology. 1995;209:281–283. doi: 10.1006/viro.1995.1256. [DOI] [PubMed] [Google Scholar]
- 61.Moynihan T P, Ardley H C, Nuber U, Rose S A, Jones P F, Markham A F, Scheffner M, Robinson P A. The ubiquitin-conjugating enzymes UbcH7 and UbcH8 interact with RING finger/IBR motif-containing domains of HHAR1 and H7-AP1. J Biol Chem. 1999;274:30963–30968. doi: 10.1074/jbc.274.43.30963. [DOI] [PubMed] [Google Scholar]
- 62.Muller S, Matunis M J, DeJean A. Conjugation of the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. EMBO J. 1998;17:61–70. doi: 10.1093/emboj/17.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Muller S, Dejean A. Viral immediate-early proteins abrogate the modification by SUMO-1 of PML and Sp100 proteins correlating with nuclear body disruption. J Virol. 1999;73:5137–5143. doi: 10.1128/jvi.73.6.5137-5143.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nagpal S, Ostrove J M. Characterization of a potent varicella-zoster virus-encoded trans-repressor. J Virol. 1991;65:5289–5296. doi: 10.1128/jvi.65.10.5289-5296.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ohta T, Michel J J, Schottelius A J, Xiong Y. ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol Cell. 1999;3:535–541. doi: 10.1016/s1097-2765(00)80482-7. [DOI] [PubMed] [Google Scholar]
- 66.Parkinson J, Lees-Miller S P, Everett R D. Herpes simplex virus type 1 immediate-early protein Vmw110 induces the proteasome-dependent degradation of the catalytic subunit of DNA-dependent protein kinase. J Virol. 1999;73:650–657. doi: 10.1128/jvi.73.1.650-657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Roizman B, Sears A E. Herpes simplex viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2231–2296. [Google Scholar]
- 68.Sacks W R, Schaffer P A. Deletion mutants in the gene encoding the herpes simplex virus type 1 immediate-early protein ICP0 exhibit impaired growth in cell culture. J Virol. 1987;61:829–839. doi: 10.1128/jvi.61.3.829-839.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Saitoh H, Tomkiel J, Cooke C A, Ratrie H, Maurer M, Rothfield N F, Earnshaw W C. CENP-C, an autoantigen in scleroderma, is a component of the human inner kinetochore plate. Cell. 1992;70:115–125. doi: 10.1016/0092-8674(92)90538-n. [DOI] [PubMed] [Google Scholar]
- 70.Saitoh H, Pu R T, Dasso M. SUMO-1: wrestling with a new ubiquitin-related modifier. Trends Biochem Sci. 1997;22:374–376. doi: 10.1016/s0968-0004(97)01102-x. [DOI] [PubMed] [Google Scholar]
- 71.Skowyra D, Koepp D M, Kamura T, Conrad M N, Conaway R C, Conaway J W, Elledge S J, Harper J W. Reconstitution of G1 Cyclin ubiquitination with complexes containing SCFgrr1 and Rbx1. Science. 1999;284:662–665. doi: 10.1126/science.284.5414.662. [DOI] [PubMed] [Google Scholar]
- 72.Soel J H, Feldman R M R, Zachariae W, Shevchenko A, Corell C C, Lyapina S, Chi Y, Galova M, Claypool J, Sandmeyer S, Nasmyth K, Shevchenko A, Deshaies R J. Cdc53/cullin and the essential Hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes Dev. 1999;13:1614–1626. doi: 10.1101/gad.13.12.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sternsdorf T, Guldner H H, Szostecki C, Grötzinger T, Will H. Two nuclear-dot associated proteins, PML and Sp100, are often co-autoimmunogenic in patients with primary biliary cirrhosis. Scand J Immunol. 1995;42:257–268. doi: 10.1111/j.1365-3083.1995.tb03652.x. [DOI] [PubMed] [Google Scholar]
- 74.Sternsdorf T, Jensen K, Will H. Evidence for covalent modification of the nuclear dot-associated proteins PML and Sp100 by PIC1/SUMO-1. J Cell Biol. 1997;139:1621–1634. doi: 10.1083/jcb.139.7.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stow E C, Stow N D. Complementation of a herpes simplex virus type 1 deletion mutant by human cytomegalovirus. J Gen Virol. 1989;70:695–704. doi: 10.1099/0022-1317-70-3-695. [DOI] [PubMed] [Google Scholar]
- 76.Stow N D, Stow E C. Isolation and characterization of a herpes simplex virus type 1 mutant containing a deletion within the gene encoding the immediate-early polypeptide Vmw110. J Gen Virol. 1986;67:2571–2585. doi: 10.1099/0022-1317-67-12-2571. [DOI] [PubMed] [Google Scholar]
- 77.Stuurman N, DeGraaf A, Josso A, Humbel B, DeYong L, van Driel R. A monoclonal antibody recognising nuclear matrix associated nuclear bodies. J Cell Sci. 1992;101:773–784. doi: 10.1242/jcs.101.4.773. [DOI] [PubMed] [Google Scholar]
- 78.Suzuki T, Ichiyama A, Saitoh H, Kawakami T, Omata M, Chung C H, Kimura M, Shimbara N, Tanaka K. A new 30-kDa ubiquitin-related SUMO-1 hydrolase from bovine brain. J Biol Chem. 1999;274:31131–31134. doi: 10.1074/jbc.274.44.31131. [DOI] [PubMed] [Google Scholar]
- 79.Telford E A R, Watson M S, McBride K, Davison A J. The DNA sequence of equine herpesvirus 1. Virology. 1992;189:304–316. doi: 10.1016/0042-6822(92)90706-u. [DOI] [PubMed] [Google Scholar]
- 80.Tyers M, Willems A R. One RING to rule a superfamily of E3 ubiquitin ligases. Science. 1999;284:601–604. doi: 10.1126/science.284.5414.601. [DOI] [PubMed] [Google Scholar]
- 81.Van Sant C, Kawaguchi Y, Roizman B. A single amino acid substitution in the cyclin D binding domain of the infected cell protein no. 0 abrogates the neuroinvasiveness of herpes simplex virus without affecting its ability to replicate. Proc Natl Acad Sci USA. 1999;96:8184–8189. doi: 10.1073/pnas.96.14.8184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Weber P C, Wigdahl B. Identification of dominant-negative mutants of the herpes simplex virus type 1 immediate-early protein ICP0. J Virol. 1992;66:2261–2267. doi: 10.1128/jvi.66.4.2261-2267.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wilkinson K D. Roles of ubiquitination in proteolysis and cellular regulation. Annu Rev Nutr. 1995;15:161–189. doi: 10.1146/annurev.nu.15.070195.001113. [DOI] [PubMed] [Google Scholar]
- 84.Wirth U V, Fraefel C, Vogt B, Vlcek C, Paces V, Schwyzer M. Immediate-early RNA 2.9 and early RNA 2.6 of bovine herpesvirus 1 are 3′ coterminal and encode a putative zinc finger transactivator protein. J Virol. 1992;66:2763–2772. doi: 10.1128/jvi.66.5.2763-2772.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yokouchi M, Kondo T, Houghton A, Bartkiewicz M, Horne W C, Zhang H, Yoshimura A, Baron R. Ligand-induced ubiquitination of the epidermal growth factor receptor involves the interaction of the c-Cbl RING finger and UbcH7. J Biol Chem. 1999;274:31707–31712. doi: 10.1074/jbc.274.44.31707. [DOI] [PubMed] [Google Scholar]
- 86.Zhu X, Chen J, Young C S H, Silverstein S. Reactivation of latent herpes simplex virus by adenovirus recombinants encoding mutant IE-0 gene products. J Virol. 1990;64:4489–4498. doi: 10.1128/jvi.64.9.4489-4498.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]