Abstract
Background:
The pathogenic effect of colorectal tumor molecular features may be influenced by several factors, including those related to microbiota, inflammation, metabolism, and epigenetics, which may change along colorectal segments. We hypothesized that the prognostic association of colon cancer location might differ by tumor molecular characteristics.
Methods:
Utilizing a consortium dataset of 13,101 colorectal cancer cases, including 2,994 early-onset cases, we conducted survival analyses of detailed tumor location stratified by statuses of microsatellite instability (MSI), CpG island methylator phenotype (CIMP), and KRAS and BRAF oncogenic mutation.
Results:
There was a statistically significant trend for better colon cancer-specific survival in relation to tumor location from the cecum to sigmoid colon (Ptrend = 0.002), excluding the rectum. The prognostic association of colon location differed by MSI status (Pinteraction = 0.001). Non-MSI-high tumors exhibited the cecum-to-sigmoid trend for better colon cancer-specific survival [Ptrend <0.001; multivariable hazard ratio (HR) for the sigmoid colon (vs. cecum), 0.80; 95% confidence interval (CI), 0.70–0.92], whereas MSI-high tumors demonstrated a suggestive cecum-to-sigmoid trend for worse survival (Ptrend = 0.020; the corresponding HR, 2.13; 95% CI, 1.15–3.92). The prognostic association of colon tumor location also differed by CIMP status (Pinteraction = 0.003) but not significantly by age, stage, and other features. Furthermore, MSI-high status was a favorable prognostic indicator in strata of stage.
Conclusions:
Both detailed colonic location and tumor molecular features need to be accounted for colon cancer prognostication to advance precision medicine. Our study indicates the important role of large-scale studies to robustly examine detailed colonic subsites in molecular oncology research.
Keywords: biogeography, epigenetics, mismatch repair, molecular pathological epidemiology, young-onset cancer
BACKGROUND
Colorectal cancer consists of a heterogeneous group of tumors with different molecular features by anatomical location [1, 2]. The colon is largely divided into two segments, namely the proximal and distal anatomic segments, using the splenic flexure as a cutpoint. However, it is conceivable that biological characteristics of colorectal cancer may be influenced by a variety of factors, including those related to microbiota, inflammation, metabolism, and epigenetics, which may vary along detailed colorectal location [3–6]. Accumulating evidence indicates that the proportions of colorectal carcinomas positive for high-level microsatellite instability (MSI), CpG island methylator phenotype (CIMP), and BRAF mutation gradually increase along more detailed location from the rectum to ascending colon [1, 7–9]. Both MSI status and BRAF mutation are established prognostic and predictive biomarkers in colorectal cancer [10–12]. These facts underscore the importance of examining detailed location and molecular biomarkers of colorectal cancer.
Primary tumor location has recently attracted considerable attention as a potential prognostic feature in colon cancer. A meta-analysis has shown that distal colon cancer patients in average survive longer than proximal colon cancer patients [13]; however, a vast majority of published studies used analyses based on the dichotomy design (proximal/right-sided vs. distal/left-sided colon) but not on detailed colonic subsites. Furthermore, whether the prognostic association of detailed colon cancer location differs by clinical and molecular characteristics remains uncertain with limited literature data [14–18]. In our current study, we tested a hypothesis that the prognostic association of detailed tumor location might differ by key tumor molecular characteristics.
The incidence of early-onset cancers diagnosed before age 50 in many body sites including the colorectum has been increasing globally for unknown reasons [19–21]. Early-onset colorectal cancer tends to occur more frequently in the distal colon and rectum compared to later-onset colorectal cancer [22, 23]. Considering this intriguing association between colorectal cancer location and age of diagnosis, we examined whether the prognostic association of tumor location might differ by age of diagnosis.
To test our hypotheses, we leveraged a large pooled-consortium dataset of colorectal cancer cases from the Cancer Genome Atlas (TCGA), and 10 studies participating in the Genetics and Epidemiology of Colorectal Cancer Consortium (GECCO).
METHODS
Study Sample
We used a pooled database of 13,101 colorectal cancer cases, including 2,994 early-onset cases diagnosed before age 50 years, which had available data on patient survival, tumor location, and tumor molecular characteristics. The database consisted of TCGA and the following 10 GECCO studies: the Colon Cancer Family Registry (CCFR) [24], Cancer Prevention Study II (CPS-II) [25], the German Darmkrebs: Chancen der Verhütung durch Screening Study (DACHS) [26], the Diet, Activity and Lifestyle Study (DALS) [27], the Early Detection Research Network (EDRN) [28], the European Prospective Investigation into Cancer - Sweden (EPIC_Sweden) [29], the Health Professionals Follow-up Study (HPFS) [30], the Melbourne Collaborative Cohort Study (MCCS) [31], the Nurses’ Health Study (NHS) [32], and the Northern Sweden Health and Disease Study (NSHDS) [33]. Each study was approved by their relevant research ethics committee or institutional review board, and all study participants provided informed consent. All colorectal adenocarcinoma cases were identified and confirmed by review of medical records, pathological reports, and/or death certificates. Participant demographics were obtained by review of medical records. Protocols for assessing colorectal cancer-specific and overall mortality in each study have been described previously [12]. Most studies ascertained mortality status through state or national death registries, or state cancer registries, with cause of death verified by death certificates and/or medical records. NSHDS and TCGA lacked data on colorectal cancer-specific mortality. Details of the study designs and the study populations were described in previous papers [12, 34]. Participant demographics and colorectal cancer molecular characteristics according to study are shown in Supplementary Table 1.
Assessment of Tumor Location and Tumor Molecular Characteristics
Primary tumor location data (derived from medical records in each study) were documented based on the International Classification of Disease (ICD). In this analysis, cases diagnosed with synchronous carcinomas at multiple sites (N=45) were excluded. To harmonize the location data across studies, we included hepatic flexure in the transverse colon, splenic flexure in the descending colon, and rectosigmoid junction in the rectum. The location variable had 6 anatomical subsites, namely cecum, ascending colon, transverse colon, descending colon, sigmoid colon, and rectum.
Testing for tumor molecular markers was performed by each study and according to individual study protocols [12, 35]. Detailed analysis methods for MSI, CIMP, BRAF mutation, and KRAS mutation are described in Supplementary Methods and Supplementary Tables 2 to 5. We compared the results of MSI status, BRAF mutation, and KRAS mutation for participants that had both existing tumor marker data and targeted sequencing data [36]. The tumor molecular marker status by both approaches were highly concordant with more than 90% agreement for MSI status, BRAF mutation, and KRAS mutation [36].
Statistical Analyses
All statistical analyses were conducted using STATA software (version 15.1, Stata Corp., College Station, TX). All P values were two-sided and the two-sided α level of 0.005 was used as recommended by the expert statisticians [37].
We used multivariable Cox proportional hazards models to estimate hazards ratio (HR) for colorectal cancer-specific mortality (a primary outcome endpoint) and overall mortality (a secondary endpoint) according to detailed subsites. The time axis was defined as days from diagnosis, with left truncation for participating studies that enrolled cases after colorectal cancer diagnosis. Proportional hazards assumptions were assessed by Schoenfeld residuals and found to be justified. We conducted analyses in two ways. The first model (minimally adjusted model) was adjusted for a limited number of variables, including sex (female vs. male), age at diagnosis (continuous), disease stage (I vs. II-III vs. IV), and study (as a stratification variable). The second model (multivariable “fully-adjusted” model) was adjusted for the aforementioned variables, family history of colorectal cancer in any first degree relative (yes vs. no), year of diagnosis (before 1995 vs. 1995–2000 vs. after 2000), MSI status (MSI-high vs. non-MSI-high), CIMP status (high vs. low/negative), BRAF mutation (mutant vs. wild-type), and KRAS mutation (mutant vs. wild-type). We conducted tests of heterogeneity using the Q statistic and observed no statistically significant heterogeneity between studies in the prognostic association of detailed primary tumor location (Pheterogeneity > 0.15). We therefore pooled data from the studies and adjusted for each study as a stratification variable. Missing values for covariates were treated as separate indicator variables in the models. To assess differences in categorical variables across tumor subsites, the chi-square test was performed. To assess differences in continuous variables across tumor subsites, an analysis of variance assuming equal variances was performed.
Our primary hypothesis testing was to assess whether the prognostic association of an ordinal colon location variable (cecum, 1; ascending, 2; transverse, 3; descending, 4; and sigmoid, 5) might differ by clinical and key tumor molecular characteristics. We used the Wald test on an interaction term between the ordinal colon location variable and each tumor marker in the multivariable model excluding rectal cancer cases. We further examined the statistical interaction between the ordinal subsite variable and each of these variables [age (continuous), sex (female vs. male), and disease stage (I to III vs. IV)]. Considering different patient management practice for rectal cancer, we did assess the statistical trend from the cecum to sigmoid colon (excluding rectum). Nonetheless, the analysis model included rectal cancer patients with the rectal location variable as a separate indicator variable. In this fashion, we could include exactly the same population (i.e., all available colorectal cancer patients) as that used in our secondary analyses to assess HR for each of the colorectal subsites including the rectum. In our secondary analyses, we assessed an HR for each site (including the rectum) compared to the cecum.
We also conducted a meta-analysis with random-effects models as a sensitivity analysis. We assessed the prognostic association of colon cancer in each study separately. Then, each study-specific HR was pooled using the random-effect meta-analysis method.
RESULTS
To examine the prognostic role of colorectal tumor location, we analyzed the pooled dataset of 13,101 colorectal cancer cases. Table 1 summarizes clinical and molecular characteristics (the statuses of MSI, CIMP, KRAS mutation, and BRAF mutation) according to primary tumor location. Higher fractions of early-onset colorectal cancers were located in the rectum (38%) compared to later-onset colorectal cancers (28%). Proportions of stage IV cases were similar across primary tumor location. Molecular characteristics by disease stage and age at diagnosis are presented in Supplementary Table 6. Compared to later-onset cases, early-onset cases showed higher prevalence of MSI-high status and lower prevalence of CIMP-high status and BRAF mutations.
Table 1.
Participant Demographics and Colorectal Cancer Molecular Characteristics by Primary Tumor Location
| Characteristicsa | Total No. | Cecum | Ascending colon | Transverse colon | Descending colon | Sigmoid colon | Rectum | P valueb |
|---|---|---|---|---|---|---|---|---|
| All cases | 13,101 | 1,968 | 1,871 | 1,301 | 968 | 3,057 | 3,936 | |
| Sex | <0.001 | |||||||
| Female | 6,339 (48%) | 1,036 (53%) | 1,044 (56%) | 663 (51%) | 481 (50%) | 1,460 (48%) | 1,655 (42%) | |
| Male | 6,762 (52%) | 932 (47%) | 827 (44%) | 638 (49%) | 487 (50%) | 1,597 (52%) | 2,281 (58%) | |
| Mean age ± SD | 61.3 ± 13.3 | 63.2 ± 13.0 | 64.3 ± 13.4 | 62.7 ± 13.8 | 60.1 ± 13.6 | 61.1 ± 12.5 | 58.8 ± 13.2 | <0.001 |
| <50 (Early-onset) | 2,994 (23%) | 360 (18%) | 315 (17%) | 261 (20%) | 256 (26%) | 662 (22%) | 1,140 (29%) | |
| 50–69 | 6,071 (46%) | 879 (45%) | 793 (42%) | 561 (43%) | 437 (45%) | 1,512 (49%) | 1,889 (48%) | |
| ≥70 | 4,036 (31%) | 729 (37%) | 763 (41%) | 479 (37%) | 275 (28%) | 883 (29%) | 907 (23%) | |
| Year of diagnosis | <0.001 | |||||||
| Before 1995 | 1,963 (15%) | 345 (18%) | 288 (16%) | 231 (18%) | 173 (18%) | 571 (19%) | 355 (9%) | |
| 1995–2000 | 5,522 (43%) | 915 (48%) | 724 (39%) | 568 (44%) | 398 (42%) | 1,284 (43%) | 1,633 (43%) | |
| After 2000 | 5,345 (42%) | 666 (35%) | 824 (45%) | 486 (38%) | 376 (40%) | 1,145 (38%) | 1,848 (48%) | |
| Family history of colorectal cancer | <0.001 | |||||||
| Absent | 9,531 (75%) | 1,404 (74%) | 1,289 (72%) | 898 (71%) | 676 (72%) | 2,303 (78%) | 2,961 (78%) | |
| Present | 3,125 (25%) | 495 (26%) | 510 (28%) | 360 (29%) | 261 (28%) | 659 (22%) | 840 (22%) | |
| AJCC disease stage | <0.001 | |||||||
| I | 3,062 (25%) | 425 (23%) | 415 (23%) | 248 (20%) | 171 (19%) | 775 (28%) | 1,028 (29%) | |
| II and III | 7,677 (63%) | 1,206 (65%) | 1,187 (67%) | 836 (69%) | 630 (68%) | 1,686 (60%) | 2,132 (60%) | |
| IV | 1,378 (11%) | 232 (12%) | 166 (8.8%) | 130 (11%) | 122 (13%) | 333 (12%) | 395 (11%) | |
| MSI status | <0.001 | |||||||
| Non-MSI-high | 10,141 (83%) | 1,311 (71%) | 1,093 (62%) | 786 (67%) | 764 (84%) | 2,710 (94%) | 3,477 (95%) | |
| MSI-high | 2,081 (17%) | 528 (29%) | 669 (38%) | 393 (33%) | 148 (16%) | 170 (5.9%) | 173 (4.7%) | |
| CIMP status | <0.001 | |||||||
| Low/negative | 7,701 (84%) | 1057 (72%) | 838 (62%) | 664 (71%) | 589 (90%) | 2,024 (94%) | 2,529 (95%) | |
| High | 1,509 (16%) | 401 (28%) | 520 (38%) | 275 (29%) | 64 (10%) | 124 (5.8%) | 125 (4.7%) | |
| BRAF | <0.001 | |||||||
| Wild-type | 10,252 (89%) | 1,425 (82%) | 1,236 (74%) | 887 (79%) | 773 (92%) | 2,576 (95%) | 3,355 (96%) | |
| Mutant | 1,315 (11%) | 314 (18%) | 440 (26%) | 234 (21%) | 68 (8.1%) | 137 (5.1%) | 122 (3.5%) | |
| KRAS | <0.001 | |||||||
| Wild-type | 6,494 (67%) | 828 (55%) | 936 (67%) | 674 (71%) | 480 (67%) | 1,641 (70%) | 1,935 (68%) | |
| Mutant | 3,269 (33%) | 681 (45%) | 467 (33%) | 274 (29%) | 241 (33%) | 701 (30%) | 905 (32%) |
Percentage indicates the proportion of patients with a specific patient molecular characteristic among all patients or in strata of tumor location (cecum, ascending colon, transverse colon, descending colon, sigmoid colon, and rectum).
To compare categorical data between subgroups classified by the tumor location, the chi-square test was performed. To compare continuous variables, an analysis of variance was performed.
Abbreviations: AJCC, American Joint Committee on Cancer; CIMP, CpG island methylator phenotype; MSI, microsatellite instability; SD, standard deviation.
There was a statistically significant trend for better colon cancer-specific survival in relation to tumor location from the cecum to sigmoid colon (Ptrend = 0.002; Table 2 and Figure 1). In contrast, compared to the cecum, rectal location was not significantly associated with colorectal cancer-specific survival [multivariable hazard ratio (HR), 1.10; 95% confidence interval (CI), 0.97–1.25; P = 0.12].
Table 2.
Colorectal Cancer-Specific and Overall Mortality in Relation to Primary Tumor Location by Tumor Molecular Features
| Cecum | Ascending colon | Transverse colon | Descending colon | Sigmoid colon | Ptrendc | Pinteractiond | Rectum | |
|---|---|---|---|---|---|---|---|---|
| CRC-specific mortality | ||||||||
| All cases | ||||||||
| No. of cases | 1,846 | 1,732 | 1,241 | 925 | 2,866 | 3,685 | ||
| No. of events | 418 | 335 | 246 | 194 | 592 | 884 | ||
| Minimally-adjusted HR (95% CI)a | 1 (reference) | 0.91 (0.79–1.06) | 0.88 (0.75–1.03) | 0.90 (0.76–1.07) | 0.93 (0.82–1.05) | 0.12 | 1.21 (1.07–1.37) | |
| Multivariable-adjusted HR (95% CI)b | 1 (reference) | 0.99 (0.86–1.15) | 0.90 (0.77–1.06) | 0.85 (0.71–1.01) | 0.84 (0.74–0.96) | 0.002 | 1.10 (0.97–1.25) | |
| MSI status | 0.001 | |||||||
| Non-MSI-high | ||||||||
| No. of cases | 1,224 | 1,000 | 752 | 726 | 2,532 | 3,240 | ||
| No. of events | 363 | 258 | 201 | 167 | 549 | 813 | ||
| Minimally-adjusted HR (95% CI)a | 1 (reference) | 0.94 (0.80–1.11) | 0.88 (0.74–1.05) | 0.74 (0.62–0.90) | 0.75 (0.66–0.86) | <0.001 | 0.99 (0.87–1.13) | |
| Multivariable-adjusted HR (95% CI)b | 1 (reference) | 0.96 (0.81–1.13) | 0.90 (0.75–1.07) | 0.78 (0.65–0.94) | 0.80 (0.70–0.92) | <0.001 | 1.07 (0.94–1.22) | |
| MSI-high | ||||||||
| No. of cases | 499 | 626 | 374 | 147 | 163 | 169 | ||
| No. of events | 33 | 55 | 27 | 14 | 17 | 21 | ||
| Minimally-adjusted HR (95% CI)a | 1 (reference) | 1.43 (0.91–2.26) | 1.13 (0.66–1.93) | 1.79 (0.94–3.38) | 2.06 (1.13–3.77) | 0.028 | 2.26 (1.24–4.09) | |
| Multivariable-adjusted HR (95% CI)b | 1 (reference) | 1.53 (0.97–2.42) | 1.20 (0.70–2.06) | 1.92 (1.01–3.66) | 2.13 (1.15–3.92) | 0.020 | 2.33 (1.27–4.30) | |
| CIMP status | 0.003 | |||||||
| CIMP-low/negative | ||||||||
| No. of cases | 993 | 759 | 633 | 556 | 1,853 | 2,321 | ||
| No. of events | 256 | 167 | 155 | 109 | 395 | 592 | ||
| Minimally-adjusted HR (95% CI)a | 1 (reference) | 0.82 (0.67–0.99) | 0.88 (0.72–1.08) | 0.69 (0.55–0.87) | 0.77 (0.66–0.90) | 0.002 | 1.10 (0.95–1.28) | |
| Multivariable-adjusted HR (95% CI)b | 1 (reference) | 0.85 (0.70–1.03) | 0.89 (0.73–1.09) | 0.68 (0.54–0.86) | 0.75 (0.64–0.88) | <0.001 | 1.07 (0.92–1.25) | |
| CIMP-high | ||||||||
| No. of cases | 355 | 470 | 253 | 59 | 115 | 115 | ||
| No. of events | 63 | 91 | 49 | 20 | 30 | 31 | ||
| Minimally-adjusted HR (95% CI)a | 1 (reference) | 1.50 (1.07–2.11) | 1.48 (1.01–2.18) | 1.91 (1.12–3.25) | 1.53 (0.97–2.41) | 0.024 | 2.16 (1.36–3.41) | |
| Multivariable-adjusted HR (95% CI)b | 1 (reference) | 1.54 (1.10–2.17) | 1.68 (1.13–2.48) | 1.93 (1.12–3.35) | 1.27 (0.80–2.02) | 0.10 | 1.70 (0.80–2.02) | |
| BRAF mutation status | 0.021 | |||||||
| BRAF wild-type | ||||||||
| No. of cases | 1,338 | 1,140 | 852 | 738 | 2,406 | 3,128 | ||
| No. of events | 336 | 230 | 180 | 154 | 514 | 750 | ||
| Minimally-adjusted HR (95% CI)a | 1 (reference) | 0.89 (0.75–1.05) | 0.82 (0.68–0.98) | 0.78 (0.65–0.95) | 0.85 (0.74–0.98) | 0.030 | 1.14 (1.00–1.30) | |
| Multivariable-adjusted HR (95% CI)b | 1 (reference) | 0.96 (0.81–1.13) | 0.86 (0.71–1.03) | 0.77 (0.63–0.93) | 0.81 (0.70–0.93) | 0.001 | 1.08 (0.95–1.24) | |
| BRAF mutant | ||||||||
| No. of cases | 283 | 403 | 215 | 63 | 127 | 116 | ||
| No. of events | 49 | 85 | 50 | 16 | 37 | 32 | ||
| Minimally-adjusted HR (95% CI)a | 1 (reference) | 1.16 (0.80–1.68) | 1.30 (0.87–1.95) | 1.33 (0.74–2.41) | 1.62 (1.03–2.54) | 0.034 | 1.68 (1.04–2.70) | |
| Multivariable-adjusted HR (95% CI)b | 1 (reference) | 1.14 (0.79–1.66) | 1.30 (0.86–1.96) | 1.12 (0.61–2.05) | 1.12 (0.70–1.80) | 0.58 | 1.25 (0.76–2.05) | |
| KRAS mutation status | 0.86 | |||||||
| KRAS wild-type | ||||||||
| No. of cases | 769 | 858 | 640 | 451 | 1,520 | 1,780 | ||
| No. of events | 170 | 164 | 137 | 99 | 337 | 438 | ||
| Minimally-adjusted HR (95% CI)a | 1 (reference) | 0.89 (0.71–1.10) | 0.93 (0.74–1.16) | 0.95 (0.74–1.21) | 0.90 (0.75–1.09) | 0.49 | 1.16 (0.96–1.39) | |
| Multivariable-adjusted HR (95% CI)b | 1 (reference) | 0.91 (0.73–1.13) | 0.91 (0.72–1.14) | 0.87 (0.68–1.13) | 0.79 (0.65–0.96) | 0.015 | 1.03 (0.85–1.24) | |
| KRAS mutant | ||||||||
| No. of cases | 625 | 413 | 254 | 228 | 639 | 822 | ||
| No. of events | 187 | 108 | 70 | 64 | 145 | 238 | ||
| Minimally-adjusted HR (95% CI)a | 1 (reference) | 1.04 (0.82–1.32) | 0.88 (0.67–1.17) | 0.81 (0.61–1.08) | 0.83 (0.67–1.04) | 0.039 | 1.25 (1.02–1.52) | |
| Multivariable-adjusted HR (95% CI)b | 1 (reference) | 1.06 (0.83–1.35) | 0.94 (0.71–1.24) | 0.81 (0.61–1.08) | 0.81 (0.65–1.01) | 0.019 | 1.20 (0.98–1.47) | |
| Overall mortality | ||||||||
| All cases | ||||||||
| No. of cases | 1,968 | 1,871 | 1,301 | 968 | 3,057 | 3,936 | ||
| No. of events | 872 | 796 | 536 | 394 | 1,253 | 1,680 | ||
| Minimally-adjusted HR (95% CI)a | 1 (reference) | 0.98 (0.89–1.08) | 0.96 (0.86–1.07) | 0.94 (0.83–1.06) | 0.94 (0.86–1.03) | 0.056 | 1.15 (1.05–1.25) | |
| Multivariable-adjusted HR (95% CI)b | 1 (reference) | 1.02 (0.92–1.12) | 0.97 (0.87–1.08) | 0.93 (0.83–1.05) | 0.91 (0.83–0.99) | 0.004 | 1.10 (1.01–1.20) | |
| MSI status | <0.001 | |||||||
| Non-MSI-high | ||||||||
| No. of cases | 1,311 | 1,093 | 786 | 764 | 2,710 | 3,477 | ||
| No. of events | 652 | 489 | 370 | 325 | 1,130 | 1,511 | ||
| Minimally-adjusted HR (95% CI)a | 1 (reference) | 0.92 (0.81–1.03) | 0.90 (0.79–1.03) | 0.84 (0.73–0.96) | 0.81 (0.74–0.90) | <0.001 | 1.00 (0.91–1.10) | |
| Multivariable-adjusted HR (95% CI)b | 1 (reference) | 0.93 (0.83–1.05) | 0.92 (0.81–1.05) | 0.86 (0.76–0.99) | 0.84 (0.76–0.93) | <0.001 | 1.04 (0.94–1.14) | |
| MSI-high | ||||||||
| No. of cases | 528 | 669 | 393 | 148 | 170 | 173 | ||
| No. of events | 171 | 260 | 123 | 47 | 64 | 61 | ||
| Minimally-adjusted HR (95% CI)a | 1 (reference) | 1.27 (1.04–1.56) | 1.20 (0.95–1.53) | 1.08 (0.77–1.50) | 1.58 (1.18–2.13) | 0.020 | 1.37 (1.00–1.89) | |
| Multivariable-adjusted HR (95% CI)b | 1 (reference) | 1.32 (1.08–1.62) | 1.21 (0.95–1.54) | 1.14 (0.81–1.58) | 1.70 (1.26–2.29) | 0.006 | 1.48 (1.07–2.06) | |
| CIMP status | 0.019 | |||||||
| CIMP-low/negative | ||||||||
| No. of cases | 1,057 | 838 | 664 | 589 | 2,024 | 2,529 | ||
| No. of events | 491 | 350 | 296 | 244 | 849 | 1,140 | ||
| Minimally-adjusted HR (95% CI)a | 1 (reference) | 0.83 (0.73–0.96) | 0.90 (0.78–1.04) | 0.81 (0.70–0.95) | 0.81 (0.72–0.90) | 0.001 | 1.03 (0.93–1.15) | |
| Multivariable-adjusted HR (95% CI)b | 1 (reference) | 0.86 (0.75–0.99) | 0.92 (0.79–1.06) | 0.81 (0.69–0.94) | 0.79 (0.71–0.89) | <0.001 | 1.01 (0.90–1.13) | |
| CIMP-high | ||||||||
| No. of cases | 401 | 520 | 275 | 64 | 124 | 125 | ||
| No. of events | 183 | 268 | 123 | 38 | 58 | 54 | ||
| Minimally-adjusted HR (95% CI)a | 1 (reference) | 1.42 (1.14–1.74) | 1.29 (1.02–1.63) | 1.33 (0.93–1.91) | 1.18 (0.87–1.59) | 0.17 | 1.65 (1.19–2.27) | |
| Multivariable-adjusted HR (95% CI)b | 1 (reference) | 1.43 (1.17–1.75) | 1.27 (1.00–1.62) | 1.34 (0.93–1.94) | 1.11 (0.81–1.51) | 0.33 | 1.51 (1.09–2.10) | |
| BRAF mutation status | 0.024 | |||||||
| BRAF wild-type | ||||||||
| No. of cases | 1,425 | 1,236 | 887 | 773 | 2,576 | 3,355 | ||
| No. of events | 647 | 510 | 373 | 315 | 1,054 | 1,443 | ||
| Minimally-adjusted HR (95% CI)a | 1 (reference) | 0.96 (0.86–1.08) | 0.93 (0.81–1.05) | 0.90 (0.78–1.03) | 0.90 (0.81–0.99) | 0.020 | 1.12 (1.02–1.23) | |
| Multivariable-adjusted HR (95% CI)b | 1 (reference) | 1.00 (0.89–1.13) | 0.95 (0.84–1.08) | 0.90 (0.78–1.03) | 0.88 (0.79–0.97) | 0.002 | 1.09 (0.99–1.21) | |
| BRAF mutant | ||||||||
| No. of cases | 314 | 440 | 234 | 68 | 137 | 122 | ||
| No. of events | 145 | 220 | 115 | 36 | 74 | 58 | ||
| Minimally-adjusted HR (95% CI)a | 1 (reference) | 1.16 (0.93–1.45) | 1.19 (0.93–1.53) | 1.19 (0.82–1.74) | 1.28 (0.96–1.72) | 0.092 | 1.36 (0.99–1.87) | |
| Multivariable-adjusted HR (95% CI)b | 1 (reference) | 1.15 (0.92–1.44) | 1.15 (0.89–1.48) | 1.11 (0.76–1.63) | 1.14 (0.84–1.55) | 0.37 | 1.29 (0.91–1.82) | |
| KRAS mutation status | 0.99 | |||||||
| KRAS wild-type | ||||||||
| No. of cases | 828 | 936 | 674 | 480 | 1,641 | 1,935 | ||
| No. of events | 363 | 437 | 310 | 208 | 684 | 847 | ||
| Minimally-adjusted HR (95% CI)a | 1 (reference) | 1.09 (0.95–1.26) | 1.08 (0.93–1.26) | 1.03 (0.86–1.22) | 0.91 (0.80–1.04) | 0.026 | 1.12 (0.99–1.27) | |
| Multivariable-adjusted HR (95% CI)b | 1 (reference) | 1.08 (0.94–1.25) | 1.06 (0.91–1.23) | 1.01 (0.85–1.21) | 0.88 (0.77–1.01) | 0.013 | 1.07 (0.94–1.23) | |
| KRAS mutant | ||||||||
| No. of cases | 681 | 467 | 274 | 241 | 701 | 905 | ||
| No. of events | 344 | 219 | 128 | 118 | 307 | 422 | ||
| Minimally-adjusted HR (95% CI)a | 1 (reference) | 1.05 (0.88–1.24) | 0.89 (0.73–1.10) | 0.90 (0.73–1.11) | 0.85 (0.73–1.00) | 0.018 | 1.11 (0.95–1.28) | |
| Multivariable-adjusted HR (95% CI)b | 1 (reference) | 1.06 (0.89–1.26) | 0.92 (0.75–1.14) | 0.92 (0.74–1.13) | 0.84 (0.72–0.99) | 0.012 | 1.10 (0.95–1.28) | |
Adjusted for sex, age, disease stage, and study (as strata).
Adjusted for sex, age, disease stage, study (as strata), family history of colorectal cancer, year of diagnosis, MSI status, CIMP status, BRAF mutation, and KRAS mutation except for stratification factors.
Trend tests from cecum to sigmoid colon were performed the ordinal subsite variable (cecum: 1, ascending: 2, transverse: 3, descending: 4, and sigmoid: 5).
Pinteraction was calculated using the Wald test for the cross-product of the ordinal subsite variable and molecular marker variables (binary) in the multivariable Cox regression model. Abbreviations: CI, confidence interval; CIMP, CpG island methylator phenotype; CRC, colorectal cancer; HR, hazard ratio; MSI, microsatellite instability.
Figure 1.
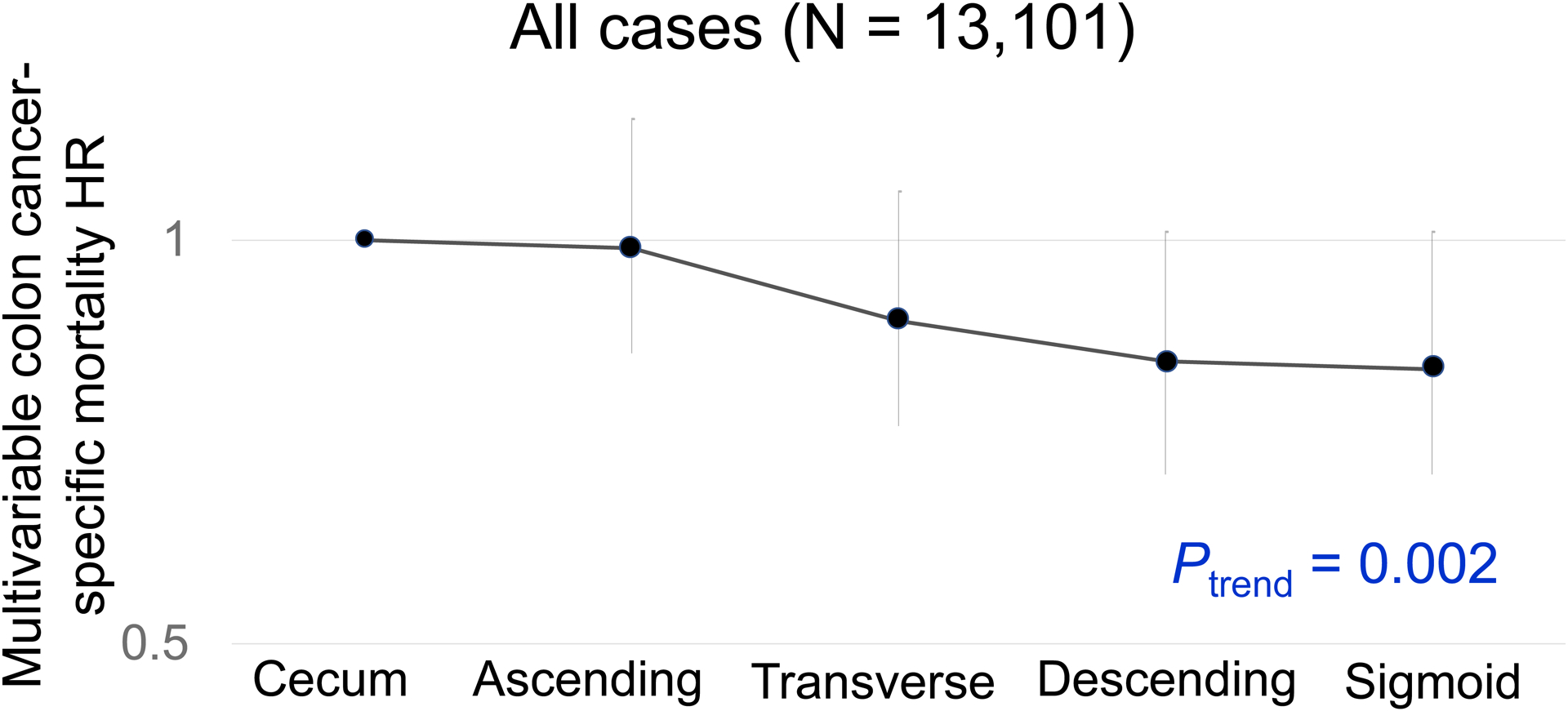
Colon Cancer-Specific Mortality in Relation to Primary Tumor Location.
Abbreviations: HR, hazard ratio.
In our primary hypothesis testing, there was a statistically significant interaction between colonic location and MSI status for colon cancer-specific survival (Pinteraction = 0.001; Table 2). Non-MSI-high tumors exhibited the cecum-to-sigmoid trend for better colon cancer-specific survival [Ptrend <0.001; multivariable HR for the sigmoid colon (vs. cecum), 0.80; 95% confidence interval (CI), 0.70–0.92; Figure 2], whereas MSI-high tumors demonstrated a suggestive opposite cecum-to-sigmoid trend for worse survival (Ptrend = 0.020; the corresponding HR, 2.13; 95% CI, 1.15–3.92; Figure 3). Similar results were observed in analyses using overall survival as a secondary endpoint. A statistically significant interaction between colonic location and CIMP status was also observed for colon-cancer specific mortality (Pinteraction = 0.003). CIMP-low/negative tumors exhibited the cecum-to-sigmoid trend for better colon cancer-specific survival [Ptrend <0.001; multivariable HR for the sigmoid colon (vs. cecum), 0.75; 95% CI, 0.64–0.88], whereas CIMP-high tumors did not demonstrate such a trend (Ptrend = 0.10). No significant interaction was observed between tumor location and BRAF or KRAS mutation status at the stringent α level of 0.005 (Pinteraction > 0.020).
Figure 2.
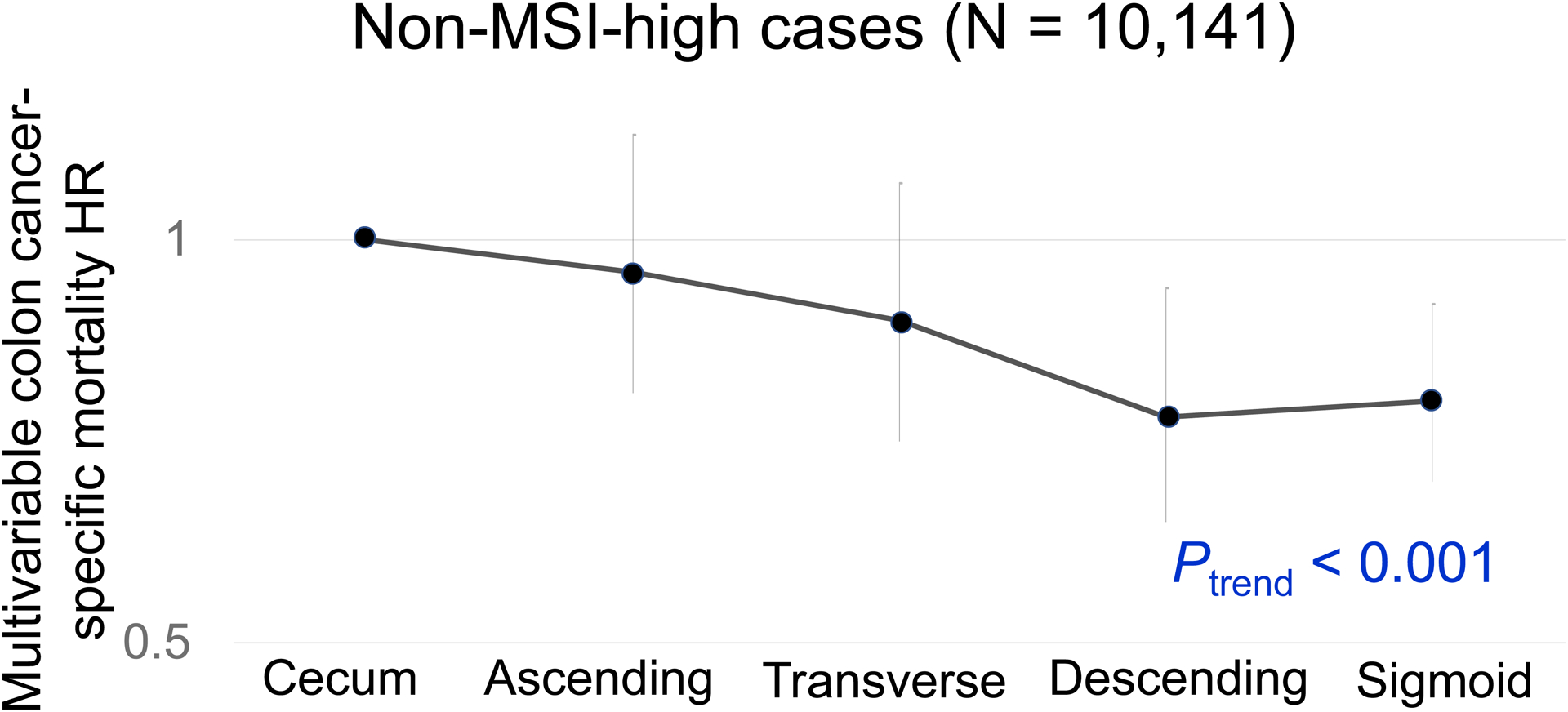
Colon Cancer-Specific Mortality in Relation to Primary Tumor Location in non-MSI-high tumors.
Abbreviations: HR, hazard ratio; MSI, microsatellite instability.
Figure 3.
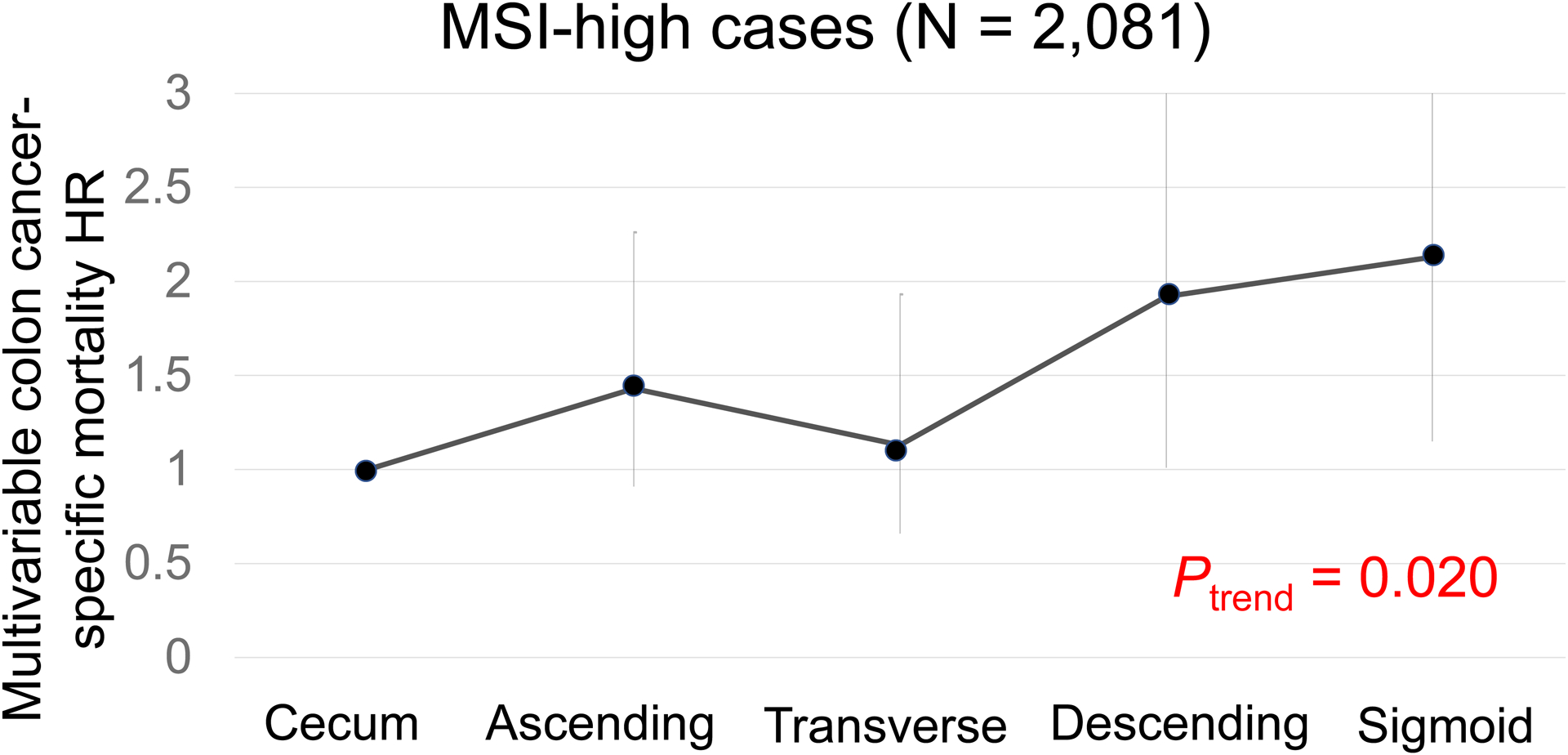
Colon Cancer-Specific Mortality in Relation to Primary Tumor Location in non-MSI-high tumors.
Abbreviations: CRC, colorectal cancer; HR, hazard ratio; MSI, microsatellite instability.
We conducted stratified analyses by age at diagnosis (<50, 50–69, ≥70 years), sex (female vs. male) or disease stage (I to III vs. IV) (Table 3 and Supplementary Tables 7–8). The prognostic association of tumor location did not significantly differ by age of diagnosis, sex, or stage (Pinteraction > 0.17).
Table 3.
Colorectal Cancer-Specific and Overall Mortality in Relation to Primary Tumor Location According to Age at Diagnosis
| Cecum | Ascending colon | Transverse colon | Descending colon | Sigmoid colon | Ptrendc | Pinteractiond | Rectum | |
|---|---|---|---|---|---|---|---|---|
| CRC-specific mortality | 0.67 | |||||||
| Age <50 (early-onset) | ||||||||
| No. of cases | 350 | 307 | 254 | 250 | 649 | 1,106 | ||
| No. of events | 75 | 54 | 38 | 46 | 140 | 266 | ||
| Minimally-adjusted HR (95% CI)a | 1 (reference) | 0.98 (0.69–1.40) | 0.66 (0.44–0.97) | 0.88 (0.61–1.27) | 1.01 (0.76–1.35) | 0.82 | 1.24 (0.96–1.62) | |
| Multivariable-adjusted HR (95% CI)b | 1 (reference) | 1.06 (0.75–1.51) | 0.68 (0.46–1.01) | 0.82 (0.57–1.19) | 0.86 (0.64–1.14) | 0.39 | 1.09 (0.83–1.42) | |
| Age 50–69 | ||||||||
| No. of cases | 814 | 736 | 532 | 411 | 1,400 | 1,740 | ||
| No. of events | 193 | 142 | 127 | 87 | 277 | 405 | ||
| Minimally-adjusted HR (95% CI)a | 1 (reference) | 0.83 (0.66–1.03) | 0.98 (0.78–1.23) | 0.80 (0.62–1.03) | 0.84 (0.70–1.01) | 0.11 | 1.04 (0.87–1.24) | |
| Multivariable-adjusted HR (95% CI)b | 1 (reference) | 0.87 (0.70–1.09) | 1.00 (0.80–1.25) | 0.73 (0.56–0.94) | 0.74 (0.62–0.90) | 0.002 | 0.93 (0.78–1.12) | |
| Age ≥70 | ||||||||
| No. of cases | 682 | 689 | 455 | 264 | 817 | 839 | ||
| No. of events | 150 | 139 | 81 | 61 | 175 | 213 | ||
| Minimally-adjusted HR (95% CI)a | 1 (reference) | 0.99 (0.78–1.26) | 0.83 (0.63–1.10) | 1.04 (0.77–1.40) | 0.97 (0.78–1.21) | 0.86 | 1.46 (1.18–1.82) | |
| Multivariable-adjusted HR (95% CI)b | 1 (reference) | 1.05 (0.83–1.34) | 0.82 (0.62–1.08) | 1.04 (0.77–1.41) | 0.89 (0.71–1.11) | 0.28 | 1.32 (1.06–1.65) | |
| Overall mortality | 0.73 | |||||||
| Age <50 (early-onset) | ||||||||
| No. of cases | 360 | 315 | 261 | 256 | 662 | 1,140 | ||
| No. of events | 110 | 82 | 58 | 68 | 194 | 375 | ||
| Minimally-adjusted HR (95% CI)a | 1 (reference) | 0.96 (0.72–1.28) | 0.64 (0.46–0.89) | 0.86 (0.63–1.16) | 0.97 (0.77–1.23) | 0.91 | 1.17 (0.94–1.46) | |
| Multivariable-adjusted HR (95% CI)b | 1 (reference) | 1.02 (0.77–1.36) | 0.68 (0.49–0.94) | 0.82 (0.60–1.12) | 0.86 (0.68–1.10) | 0.27 | 1.06 (0.85–1.32) | |
| Age 50–69 | ||||||||
| No. of cases | 879 | 793 | 561 | 437 | 1,512 | 1,889 | ||
| No. of events | 373 | 302 | 262 | 180 | 597 | 810 | ||
| Minimally-adjusted HR (95% CI)a | 1 (reference) | 0.88 (0.75–1.03) | 1.11 (0.95–1.30) | 0.88 (0.74–1.05) | 0.90 (0.79–1.03) | 0.18 | 1.07 (0.95–1.22) | |
| Multivariable-adjusted HR (95% CI)b | 1 (reference) | 0.90 (0.77–1.06) | 1.13 (0.97–1.33) | 0.85 (0.71–1.02) | 0.86 (0.75–0.98) | 0.039 | 1.03 (0.90–1.17) | |
| Age ≥70 | ||||||||
| No. of cases | 729 | 763 | 479 | 275 | 883 | 907 | ||
| No. of events | 389 | 412 | 216 | 146 | 462 | 495 | ||
| Minimally-adjusted HR (95% CI)a | 1 (reference) | 1.06 (0.92–1.22) | 0.92 (0.77–1.08) | 1.02 (0.85–1.24) | 0.97 (0.85–1.11) | 0.46 | 1.19 (1.04–1.37) | |
| Multivariable-adjusted HR (95% CI)b | 1 (reference) | 1.11 (0.96–1.28) | 0.91 (0.77–1.08) | 1.05 (0.87–1.28) | 0.97 (0.84–1.11) | 0.25 | 1.18 (1.03–1.36) | |
Adjusted for sex, disease stage, and study (as strata).
Adjusted for sex, disease stage, and study (as strata), family history of colorectal cancer, year of diagnosis, MSI status, CIMP status, BRAF mutation, and KRAS mutation.
Trend tests from cecum to sigmoid colon were performed using the ordinal subsite variable (cecum: 1, ascending: 2, transverse: 3, descending: 4, and sigmoid: 5).
Pinteraction was calculated using the Wald test for the cross-product of the ordinal subsite variable and age (continuous) in the multivariable Cox regression model.
Abbreviations: CI, confidence interval; CIMP, CpG island methylator phenotype; CRC, colorectal cancer; HR, hazard ratio; MSI, microsatellite instability.
In a sensitivity analysis, we conducted a random-effect meta-analysis that summarized multivariable HR of each individual study (Supplementary Figures 1–2, Supplementary Table 9). In general, the meta-analysis results were similar to those in the pooled analysis.
To provide additional information on the prognostic roles of the tumor markers, we conducted survival analyses of the statuses of MSI, CIMP, KRAS mutation, and BRAF mutation in overall cases as well as strata of disease stage (Table 4). MSI was consistently associated with better survival in all stages (multivariable-adjusted CRC-specific mortality HR for MSI-high vs. non-MSI-high, 0.35; 95% CI, 0.29–0.42, for overall cases).
Table 4.
Colorectal Cancer-Specific and Overall Mortality in Relation to Tumor Molecular Features.
| CRC-specific mortality | Overall mortality | |||||
|---|---|---|---|---|---|---|
| No. of cases | No. of events | Multivariable HRa (95% CI) | No. of cases | No. of events | Multivariable HRa (95% CI) | |
| Overall | ||||||
| MSI status | ||||||
| Non-MSI-high | 9474 | 2351 | 1 (reference) | 10141 | 4477 | 1 (reference) |
| MSI-high | 1978 | 167 | 0.35 (0.29–0.42) | 2081 | 726 | 0.72 (0.66–0.80) |
| CIMP status | ||||||
| CIMP-low/negative | 7115 | 1674 | 1 (reference) | 7701 | 3370 | 1 (reference) |
| CIMP-high | 1367 | 284 | 1.10 (0.94–1.29) | 1509 | 724 | 1.14 (1.03–1.27) |
| BRAF mutation status | ||||||
| BRAF wild-type | 9602 | 2164 | 1 (reference) | 10252 | 4324 | 1 (reference) |
| BRAF mutant | 1207 | 269 | 1.37 (1.18–1.61) | 1315 | 648 | 1.14 (1.03–1.26) |
| KRAS mutation status | ||||||
| KRAS wild-type | 6018 | 1345 | 1 (reference) | 6494 | 2849 | 1 (reference) |
| KRAS mutant | 2918 | 812 | 1.11 (1.01–1.22) | 3269 | 1538 | 1.02 (0.96–1.09) |
| Stage I | ||||||
| MSI status | ||||||
| Non-MSI-high | 2239 | 148 | 1 (reference) | 2361 | 763 | 1 (reference) |
| MSI-high | 453 | 11 | 0.53 (0.27–1.04) | 473 | 136 | 1.04 (0.83–1.30) |
| CIMP status | ||||||
| CIMP-low/negative | 1693 | 104 | 1 (reference) | 1792 | 585 | 1 (reference) |
| CIMP-high | 281 | 11 | 1.31 (0.66–2.59) | 310 | 104 | 1.16 (0.88–1.54) |
| BRAF mutation status | ||||||
| BRAF wild-type | 2284 | 145 | 1 (reference) | 2400 | 783 | 1 (reference) |
| BRAF mutant | 225 | 5 | 1.40 (1.13–1.73) | 247 | 81 | 0.71 (0.54–0.94) |
| KRAS mutation status | ||||||
| KRAS wild-type | 1496 | 85 | 1 (reference) | 1582 | 535 | 1 (reference) |
| KRAS mutant | 659 | 80 | 1.24 (0.86–1.78) | 711 | 255 | 1.01 (0.87–1.18) |
| Stage II and III | ||||||
| MSI status | ||||||
| Non-MSI-high | 5428 | 1205 | 1 (reference) | 5826 | 2377 | 1 (reference) |
| MSI-high | 1287 | 108 | 0.33 (0.26–0.41) | 1362 | 472 | 0.70 (0.62–0.79) |
| CIMP status | ||||||
| CIMP-low/negative | 4189 | 891 | 1 (reference) | 4551 | 1833 | 1 (reference) |
| CIMP-high | 888 | 149 | 1.12 (0.89–1.39) | 984 | 451 | 1.29 (1.11–1.49) |
| BRAF mutation status | ||||||
| BRAF wild-type | 5628 | 1130 | 1 (reference) | 6030 | 2334 | 1 (reference) |
| BRAF mutant | 1014 | 776 | 1.51 (1.16–1.96) | 850 | 411 | 1.10 (0.95–1.26) |
| KRAS mutation status | ||||||
| KRAS wild-type | 3525 | 691 | 1 (reference) | 3833 | 1522 | 1 (reference) |
| KRAS mutant | 1762 | 438 | 1.26 (1.11–1.43) | 1929 | 837 | 1.10 (1.01–1.21) |
| Stage IV | ||||||
| MSI status | ||||||
| Non-MSI-high | 1106 | 866 | 1 (reference) | 1218 | 1018 | 1 (reference) |
| MSI-high | 64 | 31 | 0.44 (0.30–0.64) | 71 | 48 | 0.54 (0.39–0.74) |
| CIMP status | ||||||
| CIMP-low/negative | 741 | 581 | 1 (reference) | 984 | 451 | 1 (reference) |
| CIMP-high | 130 | 109 | 1.06 (0.83–1.37) | 145 | 129 | 1.02 (0.81–1.29) |
| BRAF mutation status | ||||||
| BRAF wild-type | 1014 | 779 | 1 (reference) | 1116 | 916 | 1 (reference) |
| BRAF mutant | 110 | 90 | 1.66 (0.96–2.87) | 123 | 110 | 1.55 (1.22–1.97) |
| KRAS mutation status | ||||||
| KRAS wild-type | 606 | 482 | 1 (reference) | 673 | 567 | 1 (reference) |
| KRAS mutant | 381 | 298 | 1.01 (0.86–1.19) | 432 | 352 | 1.00 (0.87–1.16) |
Adjusted for sex, age, primary tumor location, disease stage, study (as strata), family history of colorectal cancer, year of diagnosis, MSI status, CIMP status, BRAF mutation, and KRAS mutation except for the predictor.
Abbreviations: CI, confidence interval; CIMP, CpG island methylator phenotype; CRC, colorectal cancer; HR, hazard ratio; MSI, microsatellite instability.
DISCUSSION
Colorectal carcinoma represents a group of heterogenous neoplastic diseases that arise from colorectal epithelia, interacting with the local microenvironment that includes the microbiome [38, 39]. As the luminal contents of the colorectum move from the cecum to rectum, its microbial composition changes. Hence, investigations into heterogeneity of colorectal carcinomas according to detailed tumor location are of particular importance. In this study, non-MSI-high tumors exhibited the cecum-to-sigmoid trend for better colon cancer-specific survival, whereas MSI-high tumors showed a suggestive opposite cecum-to-sigmoid trend for worse survival, indicating the biological and clinical significance of both MSI status and detailed information on colorectal tumor location.
This study indicates that the prognostic role of tumor location differs by molecular features, in particular the MSI and CIMP statuses, which correlate with each other [40]. Notably, a significant trend for better survival from the cecum to sigmoid colon was observed in non-MSI-high cases and CIMP-low/negative cases. Most CIMP-high colorectal carcinomas exhibit hypermethylation of MLH1 promoter CpG island, which is a major cause of the MSI-high phenotype [40]. A prior analysis using the CCFR, one of the participating cohorts in the current study, showed a better prognostic association of distal (vs. proximal) location in non-MSI-high colorectal cancer, while statistical power was limited in MSI-high tumors [16]. Another previous study showed that, compared to distal tumor localization, proximal localization was associated with favorable survival in stage III RAS-mutated colon cancer, but with unfavorable survival in stage III cancer with wild-type RAS and BRAF [41]. That study [41] used the colon dichotomy design. As tumor status (MSI status, BRAF and KRAS mutations, etc.) has become crucial for patient management [12, 41], our new knowledge on the prognostic role of detailed tumor location in strata of key molecular features can likely inform future personalized oncology practice.
The colorectal continuum model [5] is a well-recognized paradigm [42–45]. Recently, the colorectal continuum model has shown its relevance to molecular pathology of early-onset colorectal cancer [46]. However, the literature data on the prognostic relevance of the colorectal continuum model remain scarce. A meta-analysis has shown better prognosis associated with distal (left-sided) colon cancer compared to proximal (right-sided) colon cancers [13]. To our knowledge, only few studies have evaluated the prognostic association of more detailed colorectal tumor location with reasonable statistical power [14–18]. One study [15] showed a cecum-to-sigmoid trend for better survival, but it included only 895 stage III colon cancer cases with no tumor molecular data. Other studies did not show an apparent cecum-to-sigmoid trend [14, 16–18]. In the present study, we showed that the prognostic association of tumor location differed by MSI (and CIMP) status. Previous inconsistent results could be due to heterogenous prognostic impact of colorectal tumor location by molecular characteristics (especially MSI status); therefore, both tumor molecular features and detailed subsite data should be integrated in clinical oncology research.
In addition to the statuses of MSI, CIMP, BRAF, and KRAS mutation, several factors might influence the prognostic association of tumor location. Evidence suggests that copy number alterations of various genes play an important role in the development and progression of colorectal cancer [47]. Copy number alterations in early-onset colorectal cancer are particularly interesting future research topics. Anti-tumor immune response and the tumor microbiota have been associated with colorectal cancer survival [48–53]. Characterization of additional somatic mutations (e.g., HRAS and NRAS mutations), copy number alterations, gene amplification/expression [e.g., ERBB2 (HER2) expression], anti-tumor immune response, microbiota, and other biomarkers will allow more refined and detailed classification of colorectal cancer subtypes in future studies, which will further elucidate the prognostic association of tumor location.
Early-onset cancers that occur in over 10 different organ systems of adults before age 50 years have shown increased incidence in many parts of the world [54]. Those include early-onset cancers in not only the colorectum but also other gastrointestinal organs such as the esophagus, stomach, liver, gallbladder, extrahepatic bile duct, and pancreas [54]. Among them, early-onset colorectal cancer has become an intensive research topic. Considering the predilection of early-onset colorectal cancer to distal location, we investigated the prognostic significance of detailed tumor location by age at diagnosis. Notably, the prognostic association of tumor location did not significantly differ by age of diagnosis. Evidence indicates that there are differences in tumor characteristics (including MSI status, consensus molecular subtypes, key driver gene mutations, epigenetic features such as LINE-1 hypomethylation, and immune cell infiltrates) between age groups [19, 55–57], which might affect prognosis of early-onset colorectal cancer. Further research is warranted to clarify prognostic factors in early-onset colorectal cancer.
Our finding could be partly due to differential presence of intratumor bacteria along colonic subsites. Compelling evidence indicates that the microbiome can contribute to colorectal tumor development and progression [43, 58–60]. The composition of the intratumoral bacteria varies according to tumor locations in the colorectum [61–63]. A study has shown that the proportion of Fusobacterium nucleatum-high colorectal cancers gradually decreases from the cecum to rectum [64]. Evidence also indicates that intratumoral F. nucleatum is associated with MSI status and worse colorectal cancer survival [51, 65]. Further research is needed to clarify possibly interactive roles of tumor location and the microbiota in colorectal cancer prognosis.
We acknowledge limitations of the current study. First, cancer treatment information was largely unavailable. However, besides rectal cancer, it is unlikely that medical treatment strategies substantially differed by tumor location in each stage stratum during the study periods [66]. Second, it is possible that distal tumors might be detected in average earlier than proximal tumors due to higher prevalence of detectable bleeding and bowel obstruction in distal colorectal cancer patients. However, proportions of stage IV cases were similar across each subsite in this study and we adjusted for stage in our survival analyses. Third, testing for tumor molecular markers were performed using different protocols across the participating studies. However, we showed good concordance in certain molecular markers (such as MSI, KRAS, and BRAF) between the previous assays and newly-designed targeted sequencing assay [36]. Fourth, there existed measurement errors in clinical and survival data, which may be heterogeneous between cohorts. Additionally, we pooled different studies with different sampling frames (e.g., population-based or clinic-based), which might have resulted in the slightly higher proportions of MSI-high tumors and KRAS wild-type tumors than those reported in previous studies [11]. Nonetheless, we observed comparable prognostic associations of detailed tumor location across studies (see Supplementary Figures 1–2). Fifth, most study participants were non-Hispanic Whites. Therefore, our findings need to be validated in other populations. Lastly, findings from our study population may not be the same as those from a contemporary cohort, as most of the cases were diagnosed before 2010. Considering prolonged survival of patients with high-stage MSI-high tumors after introduction of immune checkpoint inhibitors, our findings should be replicated in more recent datasets, especially for patients with MSI-high tumors.
This study has notable strengths. First, the analysis of individual-level data with the large sample size allowed us to evaluate the prognostic significance of colorectal subsite information in strata of tumor molecular features with adequate statistical power. Second, the large sample size using the well-annotated cohorts enabled us to analyze strata of important patient subgroups, including early-onset colorectal cancer, incidence of which has been increasing worldwide for recent decades. Third, the study participants were drawn from multiple studies in several countries, which increases the generalizability of our findings.
In conclusion, the prognostic association of primary tumor location differed by MSI (and CIMP) status. There exists the cecum-to-sigmoid trend for better colon cancer-specific survival in non-MSI-high colon cancer, whereas there may be an opposite cecum-to-sigmoid trend for worse colon cancer-specific survival in MSI-high colon cancer. Both detailed colonic location and tumor molecular features need to be accounted for colon cancer prognostication to advance precision medicine. Furthermore, our study indicates the important role of large-scale studies to robustly examine detailed colonic subsites in molecular oncology research.
Supplementary Material
Acknowledgements
CCFR: The Colon CFR graciously thanks the generous contributions of their study participants, dedication of study staff, and the financial support from the U.S. National Cancer Institute, without which this important registry would not exist.
CPS-II: The authors thank the CPS-II participants and Study Management Group for their invaluable contributions to this research. The authors would also like to acknowledge the contribution to this study from central cancer registries supported through the Centers for Disease Control and Prevention National Program of Cancer Registries, and cancer registries supported by the National Cancer Institute Surveillance Epidemiology and End Results program.
DACHS: We thank all participants and cooperating clinicians, and Ute Handte-Daub, Utz Benscheid, Muhabbet Celik and Ursula Eilber for excellent technical assistance.
EDRN: We acknowledge all the following contributors to the development of the resource: University of Pittsburgh School of Medicine, Department of Gastroenterology, Hepatology and Nutrition: Lynda Dzubinski; University of Pittsburgh School of Medicine, Department of Pathology: Michelle Bisceglia; and University of Pittsburgh School of Medicine, Department of Biomedical Informatics.
EPIC: Where authors are identified as personnel of the International Agency for Research on Cancer/World Health Organization, the authors alone are responsible for the views expressed in this article and they do not necessarily represent the decisions, policy or views of the International Agency for Research on Cancer/World Health Organization.
Harvard cohorts (HPFS, NHS): The study protocol was approved by the institutional review boards of the Brigham and Women’s Hospital and Harvard T.H. Chan School of Public Health, and those of participating registries as required. We would like to thank the participants and staff of the HPFS and NHS for their valuable contributions as well as the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY. The authors assume full responsibility for analyses and interpretation of these data.
NSHDS: NSHDS investigators thank the Biobank Research Unit at Umeå University, the Västerbotten Intervention Programme, the Northern Sweden MONICA study and Region Västerbotten for providing data and samples and acknowledge the contribution from Biobank Sweden, supported by the Swedish Research Council (VR 2017-00650).
SCCFR: The authors would like to thank the study participants and staff of the Seattle Colon Cancer Family Registry and the Hormones and Colon Cancer study (CORE Studies).
TCGA: The results shown here are in part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga
Funding
Genetics and Epidemiology of Colorectal Cancer Consortium (GECCO): National Cancer Institute, National Institutes of Health, U.S. Department of Health and Human Services (U01 CA137088 and R01 CA248857). This research was funded in part through the NIH/NCI Cancer Center Support Grant P30 CA015704. Scientific Computing Infrastructure at Fred Hutch funded by ORIP grant S10OD028685.
The Colon Cancer Family Registry (CCFR): The Colon Cancer Family Registry (CCFR, www.coloncfr.org) is supported in part by funding from the National Cancer Institute (NCI), National Institutes of Health (NIH) (award U01 CA167551). The Seattle Colon Cancer Family Registry was also supported in part by the National Cancer Institute (NCI) of the National Institutes of Health (NIH) under R01 CA076366 (to P.A.N.). The content of this manuscript does not necessarily reflect the views or policies of the NCI, NIH or any of the collaborating centers in the Colon Cancer Family Registry (CCFR), nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government, any cancer registry, or the CCFR.
CPS-II: The American Cancer Society funds the creation, maintenance, and updating of the Cancer Prevention Study-II (CPS-II) cohort. This study was conducted with Institutional Review Board approval.
DACHS: This work was supported by the German Research Council (BR 1704/6-1, BR 1704/6-3, BR 1704/6-4, CH 117/1-1, HO 5117/2-1, HE 5998/2-1, KL 2354/3-1, RO 2270/8-1 and BR 1704/17-1), the Interdisciplinary Research Program of the National Center for Tumor Diseases (NCT), Germany, and the German Federal Ministry of Education and Research (01KH0404, 01ER0814, 01ER0815, 01ER1505A and 01ER1505B).
DALS: National Institutes of Health (R01 CA48998 to M. L. Slattery).
EDRN: This work is funded and supported by the NCI, EDRN Grant (U01 CA 84968-06).
EPIC: The coordination of EPIC is financially supported by the European Commission (DGSANCO) and the International Agency for Research on Cancer. The national cohorts are supported by Danish Cancer Society (Denmark); Ligue Contre le Cancer, Institut Gustave Roussy, Mutuelle Générale de l’Education Nationale, Institut National de la Santé et de la Recherche Médicale (INSERM) (France); German Cancer Aid, German Cancer Research Center (DKFZ), Federal Ministry of Education and Research (BMBF), Deutsche Krebshilfe, Deutsches Krebsforschungszentrum and Federal Ministry of Education and Research (Germany); the Hellenic Health Foundation (Greece); Associazione Italiana per la Ricerca sul Cancro-AIRCItaly and National Research Council (Italy); Dutch Ministry of Public Health, Welfare and Sports (VWS), Netherlands Cancer Registry (NKR), LK Research Funds, Dutch Prevention Funds, Dutch ZON (Zorg Onderzoek Nederland), World Cancer Research Fund (WCRF), Statistics Netherlands (The Netherlands); ERC-2009-AdG 232997 and Nordforsk, Nordic Centre of Excellence programme on Food, Nutrition and Health (Norway); Health Research Fund (FIS), PI13/00061 to Granada, PI13/01162 to EPIC-Murcia, Regional Governments of Andalucía, Asturias, Basque Country, Murcia and Navarra, ISCIII RETIC (RD06/0020) (Spain); Swedish Cancer Society, Swedish Research Council and County Councils of Skåne and Västerbotten (Sweden); Cancer Research UK (14136 to EPIC-Norfolk; C570/A16491 and C8221/A19170 to EPIC-Oxford), Medical Research Council (1000143 to EPIC-Norfolk, MR/M012190/1 to EPICOxford) (United Kingdom).
Harvard cohorts (HPFS, NHS): HPFS is supported by the National Institutes of Health (P01 CA055075, UM1 CA167552, U01 CA167552, R01 CA137178, R01 CA151993, R00 CA215314, and R35 CA197735) and NHS by the National Institutes of Health (R01 CA137178, P01 CA087969, UM1 CA186107, R01 CA151993, and R35 CA197735). Additionally, laboratories of M.G. and S.O. were supported by the Cancer Research UK Grand Challenge Award (UK C10674/A27140).
MCCS: Melbourne Collaborative Cohort Study (MCCS) cohort recruitment was funded by VicHealth and Cancer Council Victoria. The MCCS was further augmented by Australian National Health and Medical Research Council grants 209057, 396414 and 1074383 and by infrastructure provided by Cancer Council Victoria. Cases and their vital status were ascertained through the Victorian Cancer Registry and the Australian Institute of Health and Welfare, including the National Death Index and the Australian Cancer Database. New South Wales (NSW) cancer registry data were obtained via the ACD with the assistance of the NSW Ministry of Health.
NSHDS: Swedish Cancer Society; Cancer Research Foundation in Northern Sweden; Swedish Research Council; J C Kempe Memorial Fund; Faculty of Medicine, Umeå University, Umeå, Sweden; and Cutting-Edge Research Grant from the County Council of Västerbotten, Sweden.
OFCCR: The Ontario Familial Colorectal Cancer Registry was supported in part by the National Cancer Institute (NCI) of the National Institutes of Health (NIH) under award U01 CA167551 and award U01/U24 CA074783 (to S.G.). Additional funding for the OFCCR and ARCTIC testing and genetic analysis was provided by Canadian Cancer Society CaRE (Cancer Risk Evaluation) program grant and Ontario Research Fund award GL201-043 (to B.W.Z.), through the Canadian Institutes of Health Research award 112746 (to T.J.H.), and through generous support from the Ontario Ministry of Research and Innovation. OSUMC: OCCPI funding was provided by Pelotonia and HNPCC funding was provided by the NCI (CA16058 and CA67941).
SCCFR: The Seattle Colon Cancer Family Registry was supported in part by the National Cancer Institute (NCI) of the National Institutes of Health (NIH) under awards U01 CA167551, U01 CA074794 (to J.D.P.), and awards U24 CA074794 and R01 CA076366 (to P.A.N.).
Other: Dr. Berndt is supported by the Intramural Research Program, NCI, National Institutes of Health.
Conflict of Interest:
A.T.C. previously served as a consultant for Bayer Healthcare and Pfizer Inc. A.T.C. is a Stuart and Suzanne Steele MGH Research Scholar. M.G. was on an advisory board for AstraZeneca and has received research funding from Bristol-Myers Squibb, Merck and Servier. R.N. is currently employed by Pfizer Inc. The other authors declare no potential conflicts of interest. The content is solely the responsibility of the authors and does not necessarily represent the official views of NIH. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations:
- AJCC
American Joint Committee on Cancer
- CCFR
Colon Cancer Family Registry
- CI
confidence interval
- CIMP
CpG island methylator phenotype
- CPS-II
Cancer Prevention Study II
- DACHS
Darmkrebs: Chancen der Verhütung durch Screening Study
- DALS
Diet, Activity and Lifestyle Study
- EDRN
Early Detection Research Network
- EPIC_Sweden
European Prospective Investigation into Cancer_Sweden
- GECCO
Genetics and Epidemiology of Colorectal Cancer Consortium
- HPFS
Health Professionals Follow-up Study
- HR
hazard ratio
- ICD
International Classification of Disease
- MCCS
Melbourne Collaborative Cohort Study
- MSI
microsatellite instability
- NHS
Nurses’ Health Study
- NSHDS
Northern Sweden Health and Disease Study
- PCR
polymerase chain reaction
- SD
standard deviation
- TCGA
The Cancer Genome Atlas
Footnotes
Use of Standardized Official Symbols: We use HUGO (Human Genome Organisation)-approved official symbols (or root symbols) for genes and gene products, including BRAF, ERBB2, KRAS, HRAS, MLH1, MSH2, MSH6, NRAS, and PMS2; all of which are described at www.genenames.org. Gene symbols are italicized whereas symbols for gene products are not italicized.
REFERENCES
- 1.Loree JM, Pereira AAL, Lam M, et al. Classifying Colorectal Cancer by Tumor Location Rather than Sidedness Highlights a Continuum in Mutation Profiles and Consensus Molecular Subtypes. Clinical cancer research : an official journal of the American Association for Cancer Research. 2018;24(5):1062–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khalyfa AA, Punatar S, Aslam R, et al. Exploring the Inflammatory Pathogenesis of Colorectal Cancer. Diseases. 2021;9(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giannakis M, Mu XJ, Shukla SA, et al. Genomic correlates of immune-cell infiltrates in colorectal carcinoma. Cell Rep. 2016;15:857–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Phipps AI, Chan AT, Ogino S. Anatomic subsite of primary colorectal cancer and subsequent risk and distribution of second cancers. Cancer. 2013;119(17):3140–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamauchi M, Lochhead P, Morikawa T, et al. Colorectal cancer: a tale of two sides or a continuum? Gut. 2012;61(6):794–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin Y, Kong DX, Zhang YN. Does the Microbiota Composition Influence the Efficacy of Colorectal Cancer Immunotherapy? Front Oncol. 2022;12:852194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosty C, Young JP, Walsh MD, et al. PIK3CA activating mutation in colorectal carcinoma: associations with molecular features and survival. PLoS One. 2013;8(6):e65479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamauchi M, Morikawa T, Kuchiba A, et al. Assessment of colorectal cancer molecular features along bowel subsites challenges the conception of distinct dichotomy of proximal versus distal colorectum. Gut. 2012;61(6):847–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Phipps AI, Buchanan DD, Makar KW, et al. BRAF mutation status and survival after colorectal cancer diagnosis according to patient and tumor characteristics. Cancer Epidemiol Biomarkers Prev. 2012;21(10):1792–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singh MP, Rai S, Pandey A, et al. Molecular subtypes of colorectal cancer: An emerging therapeutic opportunity for personalized medicine. Genes Dis. 2021;8(2):133–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bai J, Chen H, Bai X. Relationship between microsatellite status and immune microenvironment of colorectal cancer and its application to diagnosis and treatment. J Clin Lab Anal. 2021;35(6):e23810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Phipps AI, Alwers E, Harrison T, et al. Association Between Molecular Subtypes of Colorectal Tumors and Patient Survival, Based on Pooled Analysis of 7 International Studies. Gastroenterology 2020;158(8):2158–2168 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petrelli F, Tomasello G, Borgonovo K, et al. Prognostic Survival Associated With Left-Sided vs Right-Sided Colon Cancer: A Systematic Review and Meta-analysis. JAMA Oncol. 2017;3(2):211–219. [DOI] [PubMed] [Google Scholar]
- 14.Sinicrope FA, Mahoney MR, Yoon HH, et al. Analysis of Molecular Markers by Anatomic Tumor Site in Stage III Colon Carcinomas from Adjuvant Chemotherapy Trial NCCTG N0147 (Alliance). Clin Cancer Res. 2015;21(23):5294–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y, Ma J, Zhang S, et al. A prognostic analysis of 895 cases of stage III colon cancer in different colon subsites. Int J Colorectal Dis. 2015;30(9):1173–83. [DOI] [PubMed] [Google Scholar]
- 16.Phipps AI, Lindor NM, Jenkins MA, et al. Colon and rectal cancer survival by tumor location and microsatellite instability: the Colon Cancer Family Registry. Dis Colon Rectum. 2013;56(8):937–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jess P, Hansen IO, Gamborg M, et al. A nationwide Danish cohort study challenging the categorisation into right-sided and left-sided colon cancer. BMJ Open. 2013;3(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wray CM, Ziogas A, Hinojosa MW, et al. Tumor subsite location within the colon is prognostic for survival after colon cancer diagnosis. Dis Colon Rectum. 2009;52(8):1359–66. [DOI] [PubMed] [Google Scholar]
- 19.Akimoto N, Ugai T, Zhong R, et al. Rising incidence of early-onset colorectal cancer - a call to action. Nat Rev Clin Oncol. 2021;18(4):230–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson CA, Begi T, Parada H, Jr. Alarming recent rises in early-onset colorectal cancer. Cancer. 2022;128(2):230–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Otegbeye EE, Colditz GA, Cao Y. Prevention of Early-Onset Colorectal Cancer: Not One Size Fits All. JNCI Cancer Spectr. 2021;5(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siegel RL, Miller KD, Goding Sauer A, et al. Colorectal cancer statistics, 2020. CA Cancer J Clin. 2020;70(3):145–164. [DOI] [PubMed] [Google Scholar]
- 23.Archambault AN, Su YR, Jeon J, et al. Cumulative Burden of Colorectal Cancer-Associated Genetic Variants Is More Strongly Associated With Early-Onset vs Late-Onset Cancer. Gastroenterology. 2020;158(5):1274–1286 e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Newcomb PA, Baron J, Cotterchio M, et al. Colon Cancer Family Registry: an international resource for studies of the genetic epidemiology of colon cancer. Cancer Epidemiol Biomarkers Prev. 2007;16(11):2331–43. [DOI] [PubMed] [Google Scholar]
- 25.Campbell PT, Deka A, Briggs P, et al. Establishment of the cancer prevention study II nutrition cohort colorectal tissue repository. Cancer Epidemiol Biomarkers Prev. 2014;23(12):2694–702. [DOI] [PubMed] [Google Scholar]
- 26.Brenner H, Chang-Claude J, Seiler CM, et al. Protection from colorectal cancer after colonoscopy: a population-based, case-control study. Ann Intern Med. 2011;154(1):22–30. [DOI] [PubMed] [Google Scholar]
- 27.Slattery ML, Friedman GD, Potter JD, et al. A description of age, sex, and site distributions of colon carcinoma in three geographic areas. Cancer. 1996;78(8):1666–70. [DOI] [PubMed] [Google Scholar]
- 28.Crichton DJ, Mattmann CA, Thornquist M, et al. Bioinformatics: biomarkers of early detection. Cancer Biomark. 2010;9(1–6):511–30. [DOI] [PubMed] [Google Scholar]
- 29.Riboli E, Kaaks R. The EPIC Project: rationale and study design. European Prospective Investigation into Cancer and Nutrition. Int J Epidemiol. 1997;26 Suppl 1:S6–14. [DOI] [PubMed] [Google Scholar]
- 30.Nishihara R, Wu K, Lochhead P, et al. Long-term colorectal-cancer incidence and mortality after lower endoscopy. N Engl J Med. 2013;369(12):1095–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Giles GG, English DR. The Melbourne Collaborative Cohort Study. IARC Sci Publ. 2002;156:69–70. [PubMed] [Google Scholar]
- 32.Liao X, Lochhead P, Nishihara R, et al. Aspirin use, tumor PIK3CA mutation, and colorectal-cancer survival. N Engl J Med. 2012;367(17):1596–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Guelpen B, Hultdin J, Johansson I, et al. Low folate levels may protect against colorectal cancer. Gut. 2006;55(10):1461–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peters U, Jiao S, Schumacher FR, et al. Identification of Genetic Susceptibility Loci for Colorectal Tumors in a Genome-Wide Meta-analysis. Gastroenterology. 2013;144(4):799–807.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hidaka A, Harrison TA, Cao Y, et al. Intake of dietary fruit, vegetables, and fiber and risk of colorectal cancer according to molecular subtypes: A pooled analysis of 9 studies. Cancer Res. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zaidi SH, Harrison TA, Phipps AI, et al. Landscape of somatic single nucleotide variants and indels in colorectal cancer and impact on survival. Nat Commun. 2020;11(1):3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Benjamin DJ, Berger JO, Johannesson M, et al. Redefine statistical significance. Nat Hum Behav. 2018;2(1):6–10. [DOI] [PubMed] [Google Scholar]
- 38.Harada S, Morlote D. Molecular Pathology of Colorectal Cancer. Adv Anat Pathol. 2020;27(1):20–26. [DOI] [PubMed] [Google Scholar]
- 39.Ogino S, Nowak JA, Hamada T, et al. Insights into Pathogenic Interactions Among Environment, Host, and Tumor at the Crossroads of Molecular Pathology and Epidemiology. Annu Rev Pathol. 2019;14:83–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nosho K, Irahara N, Shima K, et al. Comprehensive biostatistical analysis of CpG island methylator phenotype in colorectal cancer using a large population-based sample. PLoS One. 2008;3(11):e3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taieb J, Kourie HR, Emile JF, et al. Association of Prognostic Value of Primary Tumor Location in Stage III Colon Cancer With RAS and BRAF Mutational Status. JAMA Oncol. 2018;4(7):e173695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Testa U, Pelosi E, Castelli G. Colorectal cancer: genetic abnormalities, tumor progression, tumor heterogeneity, clonal evolution and tumor-initiating cells. Med Sci (Basel). 2018;6(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li S, Peppelenbosch MP, Smits R. Bacterial biofilms as a potential contributor to mucinous colorectal cancer formation. Biochim Biophys Acta Rev Cancer. 2019;1872(1):74–79. [DOI] [PubMed] [Google Scholar]
- 44.Caiazza F, Ryan EJ, Doherty G, et al. Estrogen receptors and their implications in colorectal carcinogenesis. Front Oncol. 2015;5:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ito M, Mitsuhashi K, Igarashi H, et al. MicroRNA-31 expression in relation to BRAF mutation, CpG island methylation and colorectal continuum in serrated lesions. Int J Cancer. 2014;135(11):2507–15. [DOI] [PubMed] [Google Scholar]
- 46.Ugai T, Haruki K, Harrison T, et al. Molecular Characteristics of Early-onset Colorectal Cancer According to Detailed Anatomical Locations: Comparison to Later-onset Cases. Am J Gastroenterol. 2022;in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tan ES, Knepper TC, Wang X, et al. Copy Number Alterations as Novel Biomarkers and Therapeutic Targets in Colorectal Cancer. Cancers (Basel). 2022;14(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wirta EV, Seppala T, Friman M, et al. Immunoscore in mismatch repair-proficient and -deficient colon cancer. J Pathol Clin Res. 2017;3(3):203–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pagès F, Mlecnik B, Marliot F, et al. International validation of the consensus Immunoscore for the prognostic classification of colon cancer. Lancet. 2018;391(10135):2128–2139. [DOI] [PubMed] [Google Scholar]
- 50.Marisa L, Svrcek M, Collura A, et al. The Balance Between Cytotoxic T-cell Lymphocytes and Immune Checkpoint Expression in the Prognosis of Colon Tumors. J Natl Cancer Inst. 2018;110(1):djx136. [DOI] [PubMed] [Google Scholar]
- 51.Mima K, Nishihara R, Qian ZR, et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut. 2016;65(12):1973–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kamal Y, Schmit SL, Frost HR, et al. The tumor microenvironment of colorectal cancer metastases: opportunities in cancer immunotherapy. Immunotherapy. 2020;12(14):1083–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hu LF, Lan HR, Huang D, et al. Personalized Immunotherapy in Colorectal Cancers: Where Do We Stand? Front Oncol. 2021;11:769305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ugai T, Sasamoto N, Lee HY, et al. Is early-onset cancer an emerging global epidemic? Current evidence and future implications. Nat Rev Clin Oncol. 2022;19(10):656–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Willauer AN, Liu Y, Pereira AAL, et al. Clinical and molecular characterization of early-onset colorectal cancer. Cancer. 2019;125(12):2002–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lieu CH, Golemis EA, Serebriiskii IG, et al. Comprehensive Genomic Landscapes in Early and Later Onset Colorectal Cancer. Clin Cancer Res. 2019;25(19):5852–5858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ugai T, Vayrynen JP, Lau MC, et al. Immune cell profiles in the tumor microenvironment of early-onset, intermediate-onset, and later-onset colorectal cancer. Cancer Immunol Immunother. 2022;71(4):933–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yachida S, Mizutani S, Shiroma H, et al. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat Med. 2019;25(6):968–976. [DOI] [PubMed] [Google Scholar]
- 59.Yu I, Wu R, Tokumaru Y, et al. The Role of the Microbiome on the Pathogenesis and Treatment of Colorectal Cancer. Cancers (Basel). 2022;14(22). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Inamura K, Hamada T, Bullman S, et al. Cancer as microenvironmental, systemic and environmental diseases: opportunity for transdisciplinary microbiomics science. Gut. 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Flemer B, Lynch DB, Brown JM, et al. Tumour-associated and non-tumour-associated microbiota in colorectal cancer. Gut. 2017;66(4):633–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Borozan I, Zaidi SH, Harrison TA, et al. Molecular and Pathology Features of Colorectal Tumors and Patient Outcomes Are Associated with Fusobacterium nucleatum and Its Subspecies animalis. Cancer Epidemiol Biomarkers Prev. 2022;31(1):210–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Luo K, Zhang Y, Xv C, et al. Fusobacterium nucleatum, the communication with colorectal cancer. Biomed Pharmacother. 2019;116:108988. [DOI] [PubMed] [Google Scholar]
- 64.Mima K, Cao Y, Chan AT, et al. Fusobacterium nucleatum in Colorectal Carcinoma Tissue According to Tumor Location. Clin Transl Gastroenterol. 2016;7(11):e200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fong W, Li Q, Yu J. Gut microbiota modulation: a novel strategy for prevention and treatment of colorectal cancer. Oncogene. 2020;39(26):4925–4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dekker E, Tanis PJ, Vleugels JLA, et al. Colorectal cancer. Lancet. 2019;394(10207):1467–1480. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


