Abstract
This review discusses the current understanding of biomarkers of immune quiescence based on reviews of published literature in kidney transplant operational tolerance and mechanistic studies based on a better characterization of the stable, well-functioning renal allograft.
Keywords: Immune tolerance, Tolerant footprint, Molecular biomarkers, Regulatory dendritic cells, Regulatory T cells
1. Introduction
The concept of transplant tolerance encompasses the presence of a well-functioning graft, lacking histological signs of rejection, in the absence of any immunosuppressive (IS) drugs, in an immunocompetent host [1,2]. Most reports use a cut-off point of 1 year after IS withdrawal to see if stable (or metastable) tolerance has been achieved [1–3]. Spontaneous operational tolerance has incidentally been found in patients, who are either non-adherent or are under physician-directed IS minimization at the time of clinically evident over IS, such as in the context of malignancy and severe infections [3,4]. On the contrary, induction of deliberate tolerance has occasionally been observed in humans; for example, with induced mixed chimerism seen after adoptive transfer of tolerogenic regulatory cells [4–6]. Selecting which patient will achieve this state and when drugs should or can be withdrawn safely for deliberate tolerance induction, remains difficult, as no single tolerance specific biomarker has been validated sufficiently for clinical use [4]. Benefits from IS withdrawal are very attractive, such as less IS-related complications, lower drug costs, and resulting in a better quality of life [7]. Therefore, considerable interest has been garnered in the community for detection of marker “states” for kidney transplant tolerance, so as to identify the patient and the timing for IS withdrawal, rather than the current ad hoc, trial and error approach [8].
Stable transplant tolerance requires both a state of donor-specific hyporesponsiveness and active immune regulation [9], inclusive of suppression or apoptosis of donor-reactive inflammatory cells and expansion in the number/activation state of regulatory cells. Harnessing the pathophysiology and clinical definitions of transplant tolerance to develop diagnostic biomarkers of metastable tolerogenic states, as surrogate biomarkers of immune quiescence, has been one approach to better assess and detect a state of ongoing/active immune acceptance, that would be amenable to IS manipulation and minimization, without rebound graft rejection. The process for development of these diagnostic markers faces challenges of patient selection, clinical phenotyping, sample numbers, false discovery rates during unbiased approaches, and difficulty in obtaining replicate or equivalent validation and cross-validation cohorts (Fig. 1). Additionally, assays and clinical development processes cannot translate into clinical benefit without continued support from funding agencies and clinical collaborations. Finally, during the clinical development phase, multi-step trials are needed to be approved by regulatory agencies before applying these discoveries back to the clinic, where they can be used to change practice guidelines, and support acquisition of reimbursement, and development of new or revised ICD-9 codes (Fig. 1).
Fig. 1.
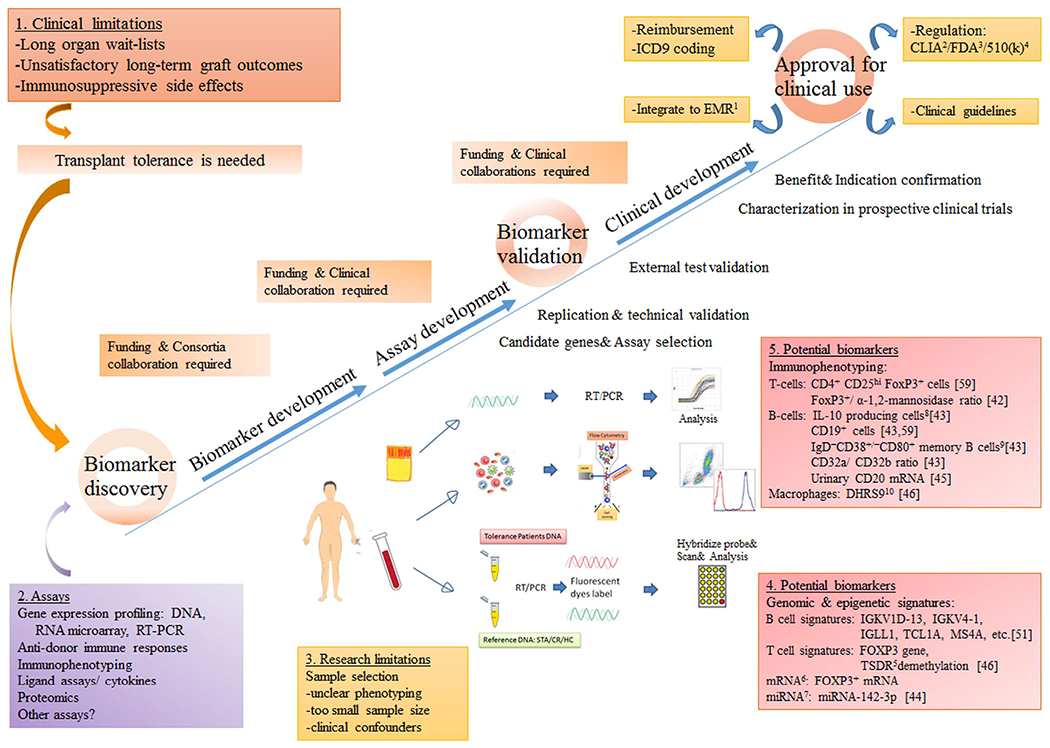
A summary of different components of successful biomarker discovery and validation for transplant tolerance. Abbreviation: 1EMR: electronic medical record, 2CLIA: The Clinical Laboratory Improvement Amendments, 3FDA: the Food and Drug Administration, 4510(k): section of the Food, Drug and Cosmetic Act requires device manufacturer who must register, to notify FDA of their intent to market a medical device at least 90 days in advance., 5TSDR: Regulatory T cells-specific demethylated region (TSDR), 6mRNA: messenger ribonucleic acid, 7miRNA: micro-ribonucleic acid, 8IL-10: Interleukin-10, 9IgD: immunoglobulin G, 10DHRS9: dehydrogenase/reductase 9. References numbers: provided in the brackets.
2. How do we define immune quiescence?
An unanswered, yet important, a question is to re-evaluate our understanding of immune quiescence and its actual definition. A lack of coherence for this definition among clinical and research groups results in misleading results from different studies. The definition of immune quiescence, in the context of the kidney allograft, faces challenges from insensitive clinical diagnosis (with the redundancy of the serum creatinine for detecting early injury), the variability of tissue sampling by biopsy, the invasiveness of the biopsy, and the high inter-intraobserver variability in pathological diagnoses [10–12]. Our group and others have shown that normal “clinical” graft function cannot be quarantined from subclinical tissue injury and normal histology cannot entirely preclude patchy inflammatory molecular changes in the same kidney [13–16]. Thus, a clinical diagnosis of non-rejection is not necessarily a lack of inflammation; and stable graft function is not necessarily immune quiescence.
As the majority of genomic studies in kidney transplant tolerance have used a clinical diagnosis for stable graft function [17–23], it is likely that incorrect input phenotype diagnoses in those studies may be another reason why inconsistent gene signature patterns were found in different microarray analysis [17–23]. Before moving forward, the first hurdle to overcome is the lack of standardized molecular testing in order to discriminate stable graft function, or a control group, from a rejection group and other injuries. We would suggest that the absence of any of the validated biomarkers for graft injury and rejection from blood, such as donor-derived cell-free DNA, and the monocyte-specific 17 gene-set called the kidney solid organ response test, or kSORT, will support selection of stable transplant patients and more precise phenotyping of patients to be included in tolerance studies for finding the most sensitive and specific biomarkers for immune quiescence.
3. The kidney: resistant to tolerance induction
The kidney is vulnerable to immune injury from many events as seen in immune-mediated glomerular diseases, which are common causes of end-stage failure [24]. Even under IS therapy after transplantation, the kidney graft carries a high risk of immune injury which gates graft life expectancy. When compared with the liver graft, the most tolerogenic transplanted organ, with 20–42.6% being tolerant after deliberate IS withdrawal [25–30], the rates of operational tolerance observed in kidney transplantation are closer to 7% [8,31]. Studies also indicate that the kidney graft is more likely to be resistant to tolerance induction [32]. Some kidney transplant trials have found that T cell depletion results in the subsequent repopulation of activated memory T cells which are resistant to suppression by regulatory T cells [32,33].
4. Understanding pathways in human studies of induced, deliberate transplant tolerance: clues for immune monitoring for graft accommodation
Successful tolerance induction in animal models have been reported through several combinatorial mechanisms including hematopoietic mixed chimerism [34,35], regulatory cell transfer [36–38], depleting antibodies [34], and costimulatory blockades; with some reported success in selected human trials [39–41]. However, stable mixed chimerism has been more difficult to achieve in the human setting [32], and usually requires some modification of the recipient immune environment either by myeloablative [34] or non-myeloablative IS protocols [42,43] (Fig. 2). Though CD34+ monocyte stem cells may be important in some types of tolerance induction strategies [34,44], the nature of engraftment can still be unpredictable. Therefore, using samples from patients who are undergoing various tolerance induction protocols, with varying degrees of success, can be problematic, as a potential biomarker for stable tolerance will be difficult to define due to the underlying heterogeneity of induction, approaches, and likely mechanisms that underlie the tolerogenic process. Additionally, as there are very few patients that actually achieve “success” in each designated protocol, there are insufficient sample numbers to develop regimen specific biomarkers.
Fig. 2.
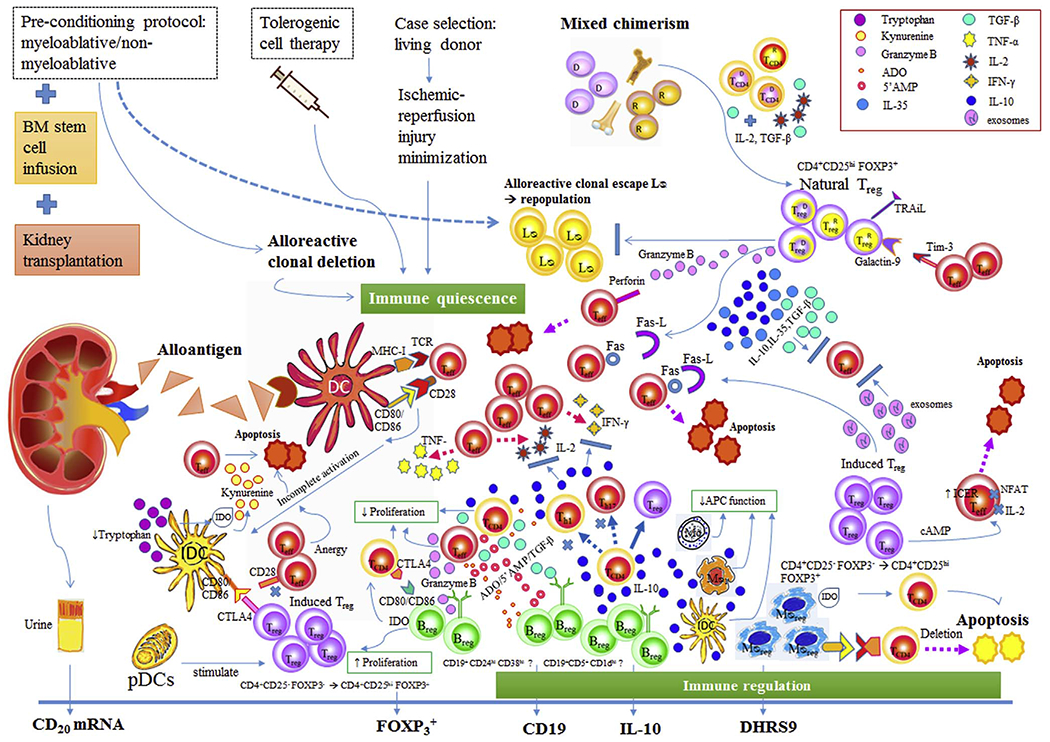
A summary of potential pathways to suppress donor-specific responsiveness by apoptosis of donor-reactive inflammatory cells and expansion of regulatory cells. 2A: In tolerance milieu, incomplete activated T cells go to anergy state or apoptosis while immature dendritic cells fail to develop maturity [45,48]. The immature dendritic cells can be activated to regulatory phenotype by IL-10 which are released from the apoptotic T cells [51,52]. Tregs modulate dendritic cell activation via CTLA-4 which binds CD80 (and CD86) with high affinity and resulting in the secretion of the enzyme IDO which catalyzes the breakdown of the amino acid tryptophan into N-formyl-kynurenine [52]. Tryptophan depletion leads to profound T lymphocyte apoptosis [52]. Another dendritic cells mechanism is through pDCs which indirectly operate Tregs differentiation [55]. 2B: Tregs suppressor effects include (1) anti-inflammatory cytokines secretion (IL-10, IL-35, TGF-β) to inhibit T cell activation, (2) exosome carried-miRNA production to silence T cells gene specific to cytokine production, (3) contact-dependent suppression with CD80/CD86 on APC by CTLA4 on Tregs to signal T cell inhibition, and (4) induction of apoptosis by multiple pathways such as via granzyme A/B and perforin, ICER, TRAiL, the Fas/Fas-ligand pathway, the galectin-9/TIM3 pathway [58]. 2C: Bregs mechanisms in tolerance milieu via IL-10 are suppression the differentiation of naive T cells into Th1 and Th17, promotion CD4+ CD25−T cells conversion into Tregs, downregulation of antigen presentation by macrophages, dendritic cells and monocytes, suppression of production of proinflammatory cytokines by CD4+ T cells, monocytes, and macrophages. In addition, IL-10 independent mechanisms are inhibiting CD4+ T cell functions via CD80/CD86-CD28/CTLA-4, induce T cell apoptosis via Fas/Fas-L cascade [61], suppress T cells by granzyme B, IDO, ADO, 5′ AMP and TGF-β. 2D: Mregs have shown potent suppressive effects on T cells proliferation via IFN-γ induced IDO activity and contact-dependent deletion of activated T cells. Abbreviation: Treg: Regulatory T cells, Breg: Regulatory B cells, MØreg: Regulatory macrophages, Teff: Effector T cells, TCD4 CD4+ T cells, IDC: immature dendritic cells, pDCs: Plasmacytoid dendritic cells, LØ: lymphocytes, Th1: Helper 1T cells, Th17: Helper 17T cells, CTLA-4: Cytotoxic T lymphocyte antigen-4, TNF-α: Tumor necrotic factor-α, IL-2: Interleukin-2, IFN-γ: Interferon-γ, D: Donor, R: Recipient, TCR: T cell receptor, MHC-I: Major histocompatibility complex class I, FOXP3: Forkhead box protein 3, IDO: Indoleamine 2,3-dioxygenase, ADO: Adenosine, 5′AMP: 5′ Adenosine monophosphate, TGF-β: Transforming growth factor-beta, IL-10: Interleukin-10, Fas, Fas-L: Fas cell surface death receptor, Fas cell surface death receptor-ligand, Mo: Monocyte, MØ1: Classical activated macrophages, APC: Antigen presenting cells, TRAiL: TNF-related apoptosis-inducing ligand, Tim-3: T cell immunoglobulin and mucin domain-3, NFAT: Nuclear factor of activated T-cells, cAMP: Cyclic adenosine monophosphate, ICER: Inducible cyclic adenosine monophosphate (cAMP) early repressor, mRNA: messenger ribonucleic acid, DHRS9: dehydrogenase/reductase 9.
Development of transplant tolerance can be categorized into three different phases: as an induction phase, as a metastable phase, and lastly, a stable phase [45]. In the induction phase, donor-reactive clones are depleted from the recipient reticuloendothelial (RE) system providing a space for donor-derived stem cells [45,46]. Antigen recognition by the adaptive immune system is declined by intensified conditioning protocols [46]. Antigen presentation to naïve T cells by immature dendritic cells ends up with incomplete T cells activation on the grounds of insufficient pro-inflammatory cytokines such as tumor necrosis factor (TNF), interleukin-6 (IL-6), interleukin-1β (IL-1β) or prostaglandin E2 (PGE2) [47]. By bidirectional communication, activated T cells provide an activation signal for dendritic cell maturation and survival via CD-40/CD-40 ligand and Tumor necrosis factor (TNF)-related activation-induced cytokine (TRANCE)/TRANCE receptor interactions [48–50]. In the tolerogenic milieu (Fig. 2, legend 2A), incompletely activated T cells go to an anergic state or undergo apoptosis while immature dendritic cells fail to develop [45,48]. Immature dendritic cells can be activated by anti-inflammatory cytokines like interleukin-10 (IL-10) which are released from the apoptotic T cells, adapting them to a regulatory phenotype [51,52]. The immunoregulatory dendritic cells mediate graft acceptance by decreased cytokine production, a loss of capacity to stimulate CD4+ and CD8+ T cells in response to donor alloantigen [53,54]. Plasmacytoid dendritic cells (pDCs) also indirectly drive regulatory T cells (Tregs) differentiation [55]. A small number of leukocytes that are contained within kidney graft migrate into the recipient and establish microchimerism concurrent with an immunologic ignorance in the metastable phase [45]. The factors that lead to the outcome of this chimerism whether they go to rejection or tolerance have as yet to be fully determined [56]. Immune quiescence from the previous phase cannot ensure stable tolerance because some peripheral donor-reactive lymphocytes can escape from the induction process and the mature immune system can repopulate new alloreactive cells [45]. To obtain life-long tolerance, active immunoregulation is required to overcome newly developed donor-reactive clones [45,46]. The regulatory phenotypes of T cells, B cells, macrophages, and dendritic cells identified from human and animal models are further discussed below for their potential roles in the development of tolerance. Monitoring of different cell subsets, though feasible, has been difficult to apply for clinically relevant monitoring, given the different mechanisms involved in different tolerance induction protocols and the small patient numbers enrolled in each of them. Nevertheless, we discuss some of the important cell subsets involved in tolerance mechanisms based on published studies.
Tregs are naturally occurring or can be induced [57] (Fig. 2, legend 2B). The suppressive effects of Tregs include mechanisms involved in secretion of anti-inflammatory cytokines (IL-10, IL-35), tumor growth factor-β (TGF-β) mediated inhibition T cell activation, exosome carried-microRNA (miRNA) production to silence T cell genes specific to cytokine production, and contact-dependent suppression with CD80/CD86 on antigen-presenting cell (APC) by Cytotoxic T-Lymphocyte Associated Protein 4 (CTLA4) on Tregs to signal T cell inhibition. In addition Tregs are also involved in induction of apoptosis by multiple pathways via granzyme A/B and perforin, inducible cyclic adenosine monophosphate (cAMP) early repressor (ICER), TNF-related apoptosis-inducing ligand (TRAIL), the Fas/Fas-ligand pathway, the galectin-9/T cell immunoglobulin, and the mucin domain-3 (TIM3) pathway [58].
Our understanding of the role of regulatory B cells (Bregs) in human transplant tolerance is evolving [59]. These Bregs are IL-10 producing B cells and can be CD19+CD24hiCD38hi transitional B cells or CD19+CD5+CD1dhi B cells [60] (Fig. 2, legend 2C). Bregs provide IL10 dependent suppression of differentiation of naïve T cells into T helper 1(Th1) and Th17 cells, promote conversion of CD4+CD25− T cells into Tregs, downregulate antigen presentation by macrophages, dendritic cells, and monocytes, and suppress production of proinflammatory cytokines such as IL-1, IFN-γ, and TNF-α by CD4+ T cells, monocytes, and macrophages [60]. On the other hand, IL-10 independent mechanisms can also inhibit CD4+ T cell functions via CD40-CD40 ligand and CD80/CD86-CD28/CTLA-4, inducing T cell apoptosis via the Fas/Fas-L cascade [61], and suppression T cells by granzyme B, Indoleamine 2,3-dioxygenase (IDO), adenosine (ADO), 5′ adenosine monophosphate (5′ AMP) and TGF-β [60].
Another immunoregulatory cell in kidney transplantation is the macrophage. Regulatory macrophages (Mregs) have shown promising results in pilot human clinical trials, namely the transplant acceptance-inducing cell II trial (TAIC-II) [62]. In mice, Mregs have shown deletion of cocultured allogeneic T cells via phagocytosis and inhibit T cell activity in vitro via inducible nitric oxide synthase (iNOS) [63]. In human, Mregs have shown potent suppressive effects on T cells proliferation via interferon (IFN)-gamma induced indoleamine 2,3-dioxygenase (IDO) activity and contact-dependent deletion of activated T cells [38] (Fig. 2, legend 2D).
Kidney transplantation concurrent with hematopoietic stem cell transplantation or tolerogenic cell therapy permits an opportunity for us to explore the tolerance atmosphere and define possible intracellular signaling and cell surface biomarkers (Fig. 1), for instance: forkhead box protein 3 (FoxP3+), CD4+CD25hiFOXP3+ cells, FOXP3/α-1,2-mannosidase ratio [21], Perforin-Granzyme A/B, IL-10 producing B cells, CD19+ cells, IgD−CD38+/−CD80+ memory B cells [64], CD32a/CD32b ratio [64], microRNA (miRNA)-142-3p [65], CD20 messenger RNA (mRNA) [20], and dehydrogenase/reductase 9 (DHRS9) [66].
5. Biomarkers for immune quiescence in spontaneous, operational transplant tolerance
Transcriptional studies have been applied for an unbiased, hypothesis-generating approach to identify novel signature gene transcripts in peripheral blood as putative new biomarkers to detect operationally tolerant patients, and by extension, apply these to monitor for graft immune quiescence. Single center and collaborative research groups in the US and EU (Immune Tolerance Network, ITN) and Europe (Indices of Tolerance, IOT) [67]) have worked together to focus on developing transplant tolerance gene footprints from peripheral blood samples. Conceptually, the process is to find the targeted gene which shows differential expression in the tolerance group compared to different control groups- the target genes differ based on which control group is selected- either stable graft function, acute rejection, chronic rejection, or healthy non-transplant volunteers. The most common process to identify such genes are microarray gene profiling, quantitative polymerase chain reaction (qPCR) validation, and immunophenotyping [17–23] (Fig. 1). As the gene lists differ, based on the control group selected, the challenge remains as to how to choose the best controls, and how to normalize the analysis across multiple clinical confounders, particularly the confounding effect of immunosuppression, so as to select the best gene-set that will be most clinically informative for immune quiescence monitoring for transplant patients on standard maintenance IS therapy.
Reports from ours and other groups have identified signature gene assays from tolerant kidney recipients [17–23] and have highlighted roles for immature B cells and myeloid and plasmacytoid dendritic cells in operational tolerance, but given the small numbers of patients with either induced or operational tolerance, national and international collaborations across patient and physician groups are imperative to advance the field. Publicly accessible online gene expression datasets of tolerance, such as Gene Expression Omnibus (GEO) datasets or Array Express, are also an indirect form of collaborations which provide more sources for validation sets (https://www.ebi.ac.uk/arrayexpress/search.html?query=kidney+transplantation+tolerance). Nonetheless, the nature of multi-center studies brings about a variety of clinical characteristics of samples and laboratory incongruity. Different clinical and IS variables are important confounding factors that can limit the identification of successful and reproducible biomarkers [68]. Recently, the meta-analysis of multiple microarray studies in kidney transplant tolerance has shown, when the controls group consists of stable transplant patients on maintenance IS, B cell-related genes are a center-point of tolerance gene signatures [22]. From five studies, there were 14 common genes recognized (0.08%). Neither those 14 genes nor the unique gene lists from the different five studies were able to discriminate tolerance from stable graft function in the pooled dataset. The author selected the top 20 genes from statistical analysis for validation and significant discrimination between the tolerance phenotype, and stable graft phenotype was found, as expected. In a recent publication, B cell signatures of tolerance associated genes, IGKV1D-13 and IGKV4-1, were persistently found in a multi-center prospective cohort of kidney transplant recipients on IS maintenance therapy over a two-year period [69]. Further validation studies will be needed to refine and confirm these findings. It is currently unknown how if these gene sets can actually be used to minimize or withdraw IS in patients showing high scores for operational tolerance- to date, this has not been tested in kidney transplant patients and would need the conduct of a carefully designed, regulated, and adaptive designed clinical trial.
Variations in experimental platform probe design and non-uniform computational analysis among tolerance microarray studies may be another reason for inconclusive results of multiple tolerance studies [17–23]. The differences in fold change and p-value cutoffs also affect microarray interpretation [70]. Furthermore, how one narrows the group of genes is critical for candidate gene selection in the assay development process. Key genes with low expression level may be removed by analytic processes that use simple fold changes.
6. Conclusions
Advanced genomics and transplantomics have gained insights in the tolerance niche, but further work needs to be done to develop a clinically validated and reliable immune quiescence biomarker that can also identify the acquisition of operational tolerance, while a patient is on stable maintenance IS with a well-functioning graft. Though different research groups have tried to identify signature gene assays from tolerant kidney recipients, results have found only a few overlaps among these gene patterns. We have discussed in our review that some of these results can be explained by many factors such as limited eligible tolerant cases, various criteria for phenotype selection, clinical confounders, and inconsistent analytic methodologies that contribute to variations in gene discovery. In addition to focusing on biomarkers of immune quiescence, also including the typing of samples for biomarkers of graft injury and rejection, and noting the absence of the latter, would better classify a stable, immune quiescent allograft. A combination of biomarkers of rejection and quiescence would provide the best biomarker combination to support IS customization and minimization trials.
This review did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Acknowledgements
The author would like to thank Assistant Professor Tara K. Sigdel for English editing.
Abbreviations:
- 5′AMP
5′ adenosine monophosphate
- ADO
adenosine
- DSAs
anti-donor specific antibodies
- APC
antigen-presenting cell
- CXCL-9
chemokine (C-X-C motif) ligand 9
- cAMP
cyclic adenosine monophosphate
- CTLA4
cytotoxic T-lymphocyte associated protein 4
- ELISAs
enzyme-linked immunosorbent assays
- Fas/Fas-L
fas cell surface death receptor, fas cell surface death receptor-ligand
- FoxP3+
forkhead box protein 3
- GEO
gene expression omnibus
- IS
immunosuppressive
- IOT
indices of tolerance
- IDO
indoleamine 2,3-dioxygenase
- ICER
inducible cyclic adenosine monophosphate (cAMP) early repressor
- iNOS
inducible nitric oxide synthase
- IFN
interferon
- IL-1β
interleukin -1β
- IL-10
interleukin-10
- IL-35
interleukin-35
- IL-6
interleukin-6
- kSORT
kidney solid organ response test
- mRNA
messenger RNA
- miRNA
microRNA
- pDCs
plasmacytoid dendritic cells
- PGE2
prostaglandin E2
- qPCR
quantitative polymerase chain reaction
- Bregs
regulatory B cells
- Mregs
regulatory macrophages
- Tregs
regulatory T cells
- RE
reticuloendothelial
- TIM3
T cell immunoglobulin and mucin domain-3
- Th
Helper T cells
- CLIA
the clinical laboratory improvement amendments
- FDA
the food and drug administration
- ITN
the immune tolerance network
- TAIC-II
the transplant acceptance-inducing cell II trial
- TRAIL
TNF-related apoptosis-inducing ligand
- TGF-β
tumor growth factor-β
- TNF
tumor necrosis factor
- TRANCE
tumor necrosis factor (TNF)-related activation-induced cytokine
Footnotes
Conflicts of interest
None.
References
- [1].Ashton-Chess J, Giral M, Brouard S, Soulillou JP, Spontaneous operational tolerance after immunosuppressive drug withdrawal in clinical renal allotransplantation, Transplantation 84 (2007) 1215. [DOI] [PubMed] [Google Scholar]
- [2].Orlando G, Hematti P, Stratta RJ, Burke GW 3rd, Di Cocco P, Pisani F, et al. , Clinical operational tolerance after renal transplantation: current status and future challenges, Ann. Surg 252 (2010) 915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Massart A, Pallier A, Pascual J, Viklicky O, Budde K, Spasovski G, et al. , The DESCARTES-Nantes survey of kidney transplant recipients displaying clinical operational tolerance identifies 35 new tolerant patients and 34 almost tolerant patients, Nephrol. Dial. Transplant 31 (2016) 1002. [DOI] [PubMed] [Google Scholar]
- [4].Brouard S, Pallier A, Renaudin K, Foucher Y, Danger R, Devys A, et al. , The natural history of clinical operational tolerance after kidney transplantation through twenty-seven cases, Am. J. Transplant 12 (2012) 3296. [DOI] [PubMed] [Google Scholar]
- [5].Lerut J, Sanchez-Fueyo A, An appraisal of tolerance in liver transplantation, Am. J. Transplant 6 (2006) 1774. [DOI] [PubMed] [Google Scholar]
- [6].Bishop GA, Bertolino PD, Bowen DG, McCaughan GW, Tolerance in liver transplantation, Best Practice Res. Clin. Gastroenterol 26 (2012) 73. [DOI] [PubMed] [Google Scholar]
- [7].Madariaga MLL, Spencer PJ, Shanmugarajah K, Crisalli KA, Chang DC, Markmann JF, et al. , Effect of tolerance versus chronic immunosuppression protocols on the quality of life of kidney transplant recipients, JCI Insight 1 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Sani KB, Sawitzki B, Immune monitoring as prerequisite for transplantation tolerance trials, Clin. Exp. Immunol 189 (2017) 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Yolcu ES, Shirwan H, Askenasy N, Mechanisms of tolerance induction by hematopoietic chimerism: the immune perspective, Stem Cells Transl. Med 6 (2017) 700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Szederkenyi E, Ivanyi B, Morvay Z, Szenohradszki P, Borda B, Marofka F, et al. , Treatment of subclinical injuries detected by protocol biopsy improves the long-term kidney allograft function: a single center prospective randomized clinical trial, Transplant Proc. 43 (2011) 1239. [DOI] [PubMed] [Google Scholar]
- [11].Furness PN, Taub N, International variation in the interpretation of renal transplant biopsies: report of the CERTPAP Project11A complete listing of the participants who all contributed equally to the work of the CERTPAP Project includes Furness Peter N. (Leicester, UK: ); Taub Nicholas (Leicester, United Kingdom: ); Assmann Karel J.M. (Nijmegen, The Netherlands: ); Banfi Giovanni (Milan, Italy: ); Botelis John N. (Athens, Greece: ); Carrera Marta (Barcelona, Spain: ); Cosyns Jean-Pierre (Brussels, Belgium: ); Dorman Anthony M. (Dublin, Ireland: ); Droz Dominique (Paris, France: ); Hill Claire M. (Belfast, N. Ireland: ); Iványi Bela (Kossuth, Hungary: ); Kapper Silke (Mannheim, Germany: ); Larsson Erik N. (Uppsala, Sweden: ); Laurinavicius Aryvdas (Vilius, Lithuania: ); Marcussen Niels (Aachus, Denmark: ); Martins Anna Paula (Lisbon, Portugal: ); Mihatsch Michael J. (Basel, Switzerland: ); Nakopoulou Lydia (Athens, Greece: ); Nickeleit Volker (Basel, Switzerland: ); Nöel L-H (Paris, France: ); Paavonen Timo (Helsinki, Finland: ); Perkowska Agnieszua K. (Warsaw, Poland: ); Regele Heinz (Vienna, Austria: ); Rosenthal Rafail (Riga, Latvia: ); Rossmann Pavel (Prague, Czech Republic: ); Rowinski Wotgech A. (Warsaw, Poland: ); Seron Daniel (Barcelona, Spain: ); Sund Stale (Oslo, Norway: ); Taskinen Eero I. (Helsinki, Finland: ); Tihomirova Tatjana (Riga, Latvia: ); and Waldherr Rudiger (Mannheim, Germany: ), Kidney Int. 60 (2001) 1998. [DOI] [PubMed] [Google Scholar]
- [12].Azancot MA, Moreso F, Salcedo M, Cantarell C, Perello M, Torres IB, et al. , The reproducibility and predictive value on outcome of renal biopsies from expanded criteria donors, Kidney Int. 85 (2014) 1161. [DOI] [PubMed] [Google Scholar]
- [13].Roedder S, Li L, Alonso MN, Hsieh SC, Vu MT, Dai H, et al. , A three-gene assay for monitoring immune quiescence in kidney transplantation, J. Am. Soc. Nephrol 26 (2015) 2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Naesens M, Li L, Ying LH, Sansanwal P, Sigdel TK, Hsieh SC, et al. , Expression of complement components differs between kidney allografts from living and deceased donors, J. Am. Soc. Nephrol 20 (2009) 1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Sigdel TK, Bestard O, Tran TQ, Hsieh SC, Roedder S, Damm I, et al. , A computational gene expression score for predicting immune injury in renal allografts, PLoS One 10 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Miyagi M, Ishikawa Y, Mizuiri S, Aikawa A, Ohara T, Hasegawa A, Significance of subclinical rejection in early renal allograft biopsies for chronic allograft dysfunction, Clin. Transplant 19 (2005) 456. [DOI] [PubMed] [Google Scholar]
- [17].Braud C, Baeten D, Giral M, Pallier A, Ashton-Chess J, Braudeau C, et al. , Immunosuppressive drug-free operational immune tolerance in human kidney transplant recipients: Part I. Blood gene expression statistical analysis, J. Cell Biochem 103 (2008) 1681. [DOI] [PubMed] [Google Scholar]
- [18].Brouard S, Mansfield E, Braud C, Li L, Giral M, Hsieh SC, et al. , Identification of a peripheral blood transcriptional biomarker panel associated with operational renal allograft tolerance, PNAS 104 (2007) 15448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lozano JJ, Pallier A, Martinez-Llordella M, Danger R, Lopez M, Giral M, et al. , Comparison of transcriptional and blood cell-phenotypic markers between operationally tolerant liver and kidney recipients, Am. J. Transplant 11 (2011) 1916. [DOI] [PubMed] [Google Scholar]
- [20].Newell KA, Asare A, Kirk AD, Gisler TD, Bourcier K, Suthanthiran M, et al. , Identification of a B cell signature associated with renal transplant tolerance in humans, J. Clin. Invest 120 (2010) 1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Sagoo P, Perucha E, Sawitzki B, Tomiuk S, Stephens DA, Miqueu P, et al. , Development of a cross-platform biomarker signature to detect renal transplant tolerance in humans, J. Clin. Invest 120 (2010) 1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Baron D, Ramstein G, Chesneau M, Echasseriau Y, Pallier A, Paul C, et al. , A common gene signature across multiple studies relate biomarkers and functional regulation in tolerance to renal allograft, Kidney Int. 87 (2015) 984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Choi JW, Kim YH, Oh JW, Comparative analyses of signature genes in acute rejection and operational tolerance, Immune Netw. 17 (2017) 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Imig JD, Ryan MJ, Immune and inflammatory role in renal disease, Compr. Physiol 3 (2013) 957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Takatsuki M, Uemoto S, Inomata Y, Egawa H, Kiuchi T, Fujita S, et al. , Weaning of immunosuppression in living donor liver transplant recipients, Transplantation 72 (2001) 449. [DOI] [PubMed] [Google Scholar]
- [26].Levitsky J, Operational tolerance: past lessons and future prospects, Liver Transpl. 17 (2011) 222. [DOI] [PubMed] [Google Scholar]
- [27].Tryphonopoulos P, Tzakis AG, Weppler D, Garcia-Morales R, Kato T, Madariaga JR, et al. , The role of donor bone marrow infusions in withdrawal of immunosuppression in adult liver allotransplantation, Am. J. Transplant 5 (2005) 608. [DOI] [PubMed] [Google Scholar]
- [28].Donckier V, Troisi R, Le Moine A, Toungouz M, Ricciardi S, Colle I, et al. , Early immunosuppression withdrawal after living donor liver transplantation and donor stem cell infusion, Liver Transpl. 12 (2006) 1523. [DOI] [PubMed] [Google Scholar]
- [29].Girlanda R, Rela M, Williams R, O’Grady JG, Heaton ND, Long-term outcome of immunosuppression withdrawal after liver transplantation, Transplant Proc. 37 (2005) 1708. [DOI] [PubMed] [Google Scholar]
- [30].Oike F, Yokoi A, Nishimura E, Ogura Y, Fujimoto Y, Kasahara M, et al. , Complete withdrawal of immunosuppression in living donor liver transplantation, Transplant Proc. 34 (2002) 1521. [DOI] [PubMed] [Google Scholar]
- [31].Dugast E, Chesneau M, Soulillou JP, Brouard S, Biomarkers and possible mechanisms of operational tolerance in kidney transplant patients, Immunol. Rev 258 (2014) 208. [DOI] [PubMed] [Google Scholar]
- [32].Goldman M, Wood K, Translating transplantation tolerance in the clinic: where are we, where do we go? Clin. Exp. Immunol 156 (2009) 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Benghiat FS, Craciun L, De Wilde V, Dernies T, Kubjak C, Lhomme F, et al. , IL-17 production elicited by allo-major histocompatibility complex class II recognition depends on CD25posCD4pos T cells, Transplantation 85 (2008) 943. [DOI] [PubMed] [Google Scholar]
- [34].Scandling JD, Busque S, Dejbakhsh-Jones S, Benike C, Millan MT, Shizuru JA, et al. , Brief report: tolerance and chimerism after renal and hematopoietic-cell transplantation, N. Engl. J. Med 358 (2008) 362. [DOI] [PubMed] [Google Scholar]
- [35].Kawai T, Cosimi AB, Spitzer TR, Tolkoff-Rubin N, Suthanthiran M, Saidman SL, et al. , Brief report: HLA-mismatched renal transplantation without maintenance immunosuppression, N. Engl. J. Med 358 (2008) 353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Nagahama K, Nishimura E, Sakaguchi S, Induction of tolerance by adoptive transfer of Treg cells, Methods Mol. Biol 380 (2007) 431. [DOI] [PubMed] [Google Scholar]
- [37].Todo S, Yamashita K, Goto R, Zaitsu M, Nagatsu A, Oura T, et al. , A pilot study of operational tolerance with a regulatory T-cell-based cell therapy in living donor liver transplantation, Hepatology 64 (2016) 632. [DOI] [PubMed] [Google Scholar]
- [38].Hutchinson JA, Riquelme P, Sawitzki B, Tomiuk S, Miqueu P, Zuhayra M, et al. , Cutting edge: immunological consequences and trafficking of human regulatory macrophages administered to renal transplant recipients, J. Immunol 187 (2011) 2072. [DOI] [PubMed] [Google Scholar]
- [39].Kawai T, Cosimi AB, Induction of tolerance in clinical kidney transplantation, Clin. Transplant 24 (2010) 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Wekerle T, Sayegh MH, Hill J, Zhao Y, Chandraker A, Swenson KG, et al. , Extrathymic T cell deletion and allogeneic stem cell engraftment induced with costimulatory blockade is followed by central T cell tolerance, J. Exp. Med 187 (1998) 2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Page E, Kwun J, Oh B, Knechtle S, Lymphodepletional strategies in transplantation, Cold Spring Harbor Perspect. Med 3 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Fudaba Y, Spitzer TR, Shaffer J, Kawai T, Fehr T, Delmonico F, et al. , Myeloma responses and tolerance following combined kidney and nonmyeloablative marrow transplantation: in vivo and in vitro analyses, Am. J. Transplant 6 (2006) 2121. [DOI] [PubMed] [Google Scholar]
- [43].Trivedi HL, Vanikar AV, Modi PR, Shah VR, Vakil JM, Trivedi VB, et al. , Allogeneic hematopoietic stem-cell transplantation, mixed chimerism, and tolerance in living related donor renal allograft recipients, Transpl. Proc 37 (2005) 737. [DOI] [PubMed] [Google Scholar]
- [44].Leventhal JR, Mathew JM, Salomon DR, Kurian SM, Friedewald JJ, Gallon L, et al. , Nonchimeric HLA-identical renal transplant tolerance: regulatory immunophenotypic/genomic biomarkers, Am. J. Transplant 16 (2016) 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Li XC, Strom TB, Turka LA, Wells AD, T cell death and transplantation tolerance, Immunity 14 (2001) 407. [DOI] [PubMed] [Google Scholar]
- [46].Domenig C, Sanchez-Fueyo A, Kurtz J, Alexopoulos SP, Mariat C, Sykes M, et al. , Roles of deletion and regulation in creating mixed chimerism and allograft tolerance using a nonlymphoablative irradiation-free protocol, J. Immunol 175 (2005) 51. [DOI] [PubMed] [Google Scholar]
- [47].Raker VK, Domogalla MP, Steinbrink K, Tolerogenic dendritic cells for regulatory T cell induction in man, Front. Immunol 6 (2015) 569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Banchereau J, Briere F, Caux C, Davoust J, Lebecque S, Liu YJ, et al. , Immunobiology of dendritic cells, Annu. Rev. Immunol 18 (2000) 767. [DOI] [PubMed] [Google Scholar]
- [49].Cremer I, Dieu-Nosjean MC, Marechal S, Dezutter-Dambuyant C, Goddard S, Adams D, et al. , Long-lived immature dendritic cells mediated by TRANCE-RANK interaction, Blood 100 (2002) 3646. [DOI] [PubMed] [Google Scholar]
- [50].Josien R, Wong BR, Li HL, Steinman RM, Choi YW, TRANCE, a TNF family member, is differentially expressed on T cell subsets and induces cytokine production in dendritic cells, J. Immunol 162 (1999) 2562. [PubMed] [Google Scholar]
- [51].Stuart LM, Lucas M, Simpson C, Lamb J, Savill J, Lacy-Hulbert A, Inhibitory effects of apoptotic cell ingestion upon endotoxin-driven myeloid dendritic cell maturation, J. Immunol 168 (2002) 1627. [DOI] [PubMed] [Google Scholar]
- [52].Wallet MA, Sen P, Tisch R, Immunoregulation of dendritic cells, Clin. Med. Res 3 (2005) 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Steinbrink K, Wolfl M, Jonuleit H, Knop J, Enk AH, Induction of tolerance by IL-10-treated dendritic cells, J. Immunol 159 (1997) 4772. [PubMed] [Google Scholar]
- [54].Haase C, Jorgensen TN, Michelsen BK, Both exogenous and endogenous interleukin-10 affects the maturation of bone-marrow-derived dendritic cells in vitro and strongly influences T-cell priming in vivo, Immunology 107 (2002) 489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Mellor AL, Munn DH, IDO expression by dendritic cells: tolerance and tryptophan catabolism, Nat. Rev. Immunol 4 (2004) 762. [DOI] [PubMed] [Google Scholar]
- [56].Anderson CC, Matzinger P, Immunity or tolerance: opposite outcomes of microchimerism from skin grafts, Nat. Med 7 (2001) 80. [DOI] [PubMed] [Google Scholar]
- [57].Safinia N, Scotta C, Vaikunthanathan T, Lechler RI, Lombardi G, Regulatory T cells: serious contenders in the promise for immunological tolerance in transplantation, Front. Immunol 6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Vaikunthanathan T, Safinia N, Boardman D, Lechler RI, Lombardi G, Regulatory T cells: tolerance induction in solid organ transplantation, Clin. Exp. Immunol 189 (2017) 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Chesneau M, Michel L, Degauque N, Brouard S, Regulatory B cells and tolerance in transplantation: from animal models to human, Front. Immunol 4 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Carreras-Planella L, Borràs FE, Franquesa M, Tolerance in kidney transplantation: what is on the B side? Mediators Inflamm. 2016 (2016) 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Lee KM, Stott RT, Zhao G, SooHoo J, Xiong W, Lian MM, et al. , TGF-beta-producing regulatory B cells induce regulatory T cells and promote transplantation tolerance, Eur. J. Immunol 44 (2014) 1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Hutchinson JA, Riquelme P, Brem-Exner BG, Schulze M, Matthai M, Renders L, et al. , Transplant acceptance-inducing cells as an immune-conditioning therapy in renal transplantation, Transpl. Int 21 (2008) 728. [DOI] [PubMed] [Google Scholar]
- [63].Scalea JR, Tomita Y, Lindholm CR, Burlingham W, Transplantation tolerance induction: cell therapies and their mechanisms, Front. Immunol 7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Pallier A, Hillion S, Danger R, Giral M, Racape M, Degauque N, et al. , Patients with drug-free long-term graft function display increased numbers of peripheral B cells with a memory and inhibitory phenotype, Kidney Int. 78 (2010) 503. [DOI] [PubMed] [Google Scholar]
- [65].Danger R, Pallier A, Giral M, Martinez-Llordella M, Lozano JJ, Degauque N, et al. , Upregulation of miR-142-3p in peripheral blood mononuclear cells of operationally tolerant patients with a renal transplant, J. Am. Soc. Nephrol 23 (2012) 597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Riquelme P, Amodio G, Macedo C, Moreau A, Obermajer N, Ahrens N, et al. , Dhrs9 is a specific and stable marker of human regulatory macrophages, Transpl. Int 30 (2017) 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Page EK, Dar WA, Knechtle SJ, Tolerogenic therapies in transplantation, Front. Immunol 3 (2012) 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Perkins D, Verma M, Park KJ, Advances of genomic science and systems biology in renal transplantation: a review, Semin. Immunopathol 33 (2011) 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Asare A, Kanaparthi S, Lim N, Phippard D, Vincenti F, Friedewald J, et al. , B cell receptor genes associated with tolerance identify a cohort of immunosuppressed patients with improved renal allograft graft function, Am. J. Transplant 17 (2017) 2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Dalman MR, Deeter A, Nimishakavi G, Duan Z-H, Fold change and p-value cutoffs significantly alter microarray interpretations, BMC Bioinf. 13 (2012) S11. [DOI] [PMC free article] [PubMed] [Google Scholar]


