Abstract
Background:
Postoperative infections constitute more than half of all postoperative complications. Surveillance of these complications is primarily done through manual chart review, which is time consuming, expensive, and typically only covers 10% to 15% of all operations. Automated surveillance would permit the timely evaluation of and reporting of all operations.
Methods:
The goal of this study was to develop and validate parsimonious, interpretable models for conducting surveillance of postoperative infections using structured electronic health records data. This was a retrospective study using 30,639 unique operations from 5 major hospitals between 2013 and 2019. Structured electronic health records data were linked to postoperative outcomes data from the American College of Surgeons National Surgical Quality Improvement Program. Predictors from the electronic health records included diagnoses, procedures, and medications. Infectious complications included surgical site infection, urinary tract infection, sepsis, and pneumonia within 30 days of surgery. The knockoff filter, a penalized regression technique that controls type I error, was applied for variable selection. Models were validated in a chronological held-out dataset.
Results:
Seven percent of patients experienced at least one type of postoperative infection. Models selected contained between 4 and 8 variables and achieved >0.91 area under the receiver operating characteristic curve, >81% specificity, >87% sensitivity, >99% negative predictive value, and 10% to 15% positive predictive value in a held-out test dataset.
Conclusion:
Surveillance and reporting of postoperative infection rates can be implemented for all operations with high accuracy using electronic health records data and simple linear regression models.
Introduction
Surgical infectious complications are among the most common healthcare associated infections (HAI).1 Postoperative infectious complications include surgical site infection (SSI), sepsis, pneumonia, and urinary tract infection (UTI). The SSIs are the costliest HAI type, with an estimated annual cost of $1.6 to $3.3 billion, and are associated with nearly 1 million additional inpatient days annually in the US.2,3 Worldwide, significant hospital resources are devoted to surveillance of postoperative infections, which is often accomplished by manual chart review.4 Due to the time and cost required to perform these reviews, typically only a small subset of all patients is reviewed, and delays exist between data abstraction and analysis. As a result of these limitations, the data are primarily used to compare hospitals and are not fed back to surgeons and other providers.
The American College of Surgeons National Surgical Quality Improvement Program (ACS-NSQIP) provides accurate reporting of postoperative complications on a systematic sample of major operations at >800 hospitals.5 Infectious complications are the most frequent type, constituting over half of all postoperative complications.6 At each participating hospital, trained surgical nurse reviewers use the ACS-NSQIP protocol and standardized definitions to collect preoperative risk factors, operative variables, and 30-day postoperative outcomes on a systematic sample of surgical patients. These curated data are subsequently risk-adjusted centrally, and results are reported back to participating programs to facilitate tracking of outcomes over time and identifying best practices. Despite its widespread acceptance, the ACS-NSQIP has a number of important limitations: (1) it covers only a sample of operations at each participating center (10%–15% at higher volume hospitals), (2) the manual data collection burden is significant and costly, (3) the data are not reported at the individual surgeon level, and (4) there is a delay of at least 6 to 18 months between the surgical procedure and when the ACS reports back to participating programs.7
Additionally, the Centers for Disease Control’s National Healthcare Safety Network (NHSN) is the largest HAI surveillance system in the US. The goal of NHSN is to identify problems at certain facilities, track progress, and facilitate mandatory reporting of HAIs to federal agencies.8 For many large facilities, the NHSN indicators, which include SSI, pneumonia, and UTI, are only collected for specific operations, because the review process is laborious and expensive. Despite their limitations, both the ACS-NSQIP and the NHSN tracking systems are robust sources of information on HAIs.
Given the time and costs involved in postoperative infection surveillance, some institutions have considered the use of statistical models applied to the electronic health record (EHR) to identify infectious complications of surgery, specifically for SSIs,9–13 UTIs,14–24 sepsis,25 and pneumonia.12,26,27 In these studies, researchers used structured data or natural language processing (NLP) of text data to develop models for postoperative infection detection. The most commonly used data for patient outcomes were ACS-NSQIP and Veterans Administration Surgical Quality Improvement Program. In many of these previous studies, low-dimensional datasets were used because the statistical techniques applied could not handle a large number of predictors. Natural language processing has shown promise in various studies,14,20,28–30 but not all institutions have the capacity to implement NLP, limiting its reach. Knepper et al created an algorithm for flagging NHSN SSIs using EHR data.31 They showed that a single International Classification of Disease version 9 (ICD-9) code, 998.59, could detect SSIs in seven specific procedures with 64% sensitivity and 97% specificity, and their overall model, which included additional laboratory data and diagnosis codes, had 100% sensitivity and 88% specificity. Importantly, they estimated that the algorithm reduced the number of cases requiring manual review by 80%.
The primary objective of this study was to create an automated, reliable, and generalizable method to identify postoperative infections, including SSI, UTI, pneumonia, and sepsis/septic shock, using machine learning applied to EHR data. By automating the identification of infections using this approach, we believe that the following goals can be accomplished: (1) all operations can be evaluated for infectious complications, not just a limited subset, (2) the charts requiring manual review can be identified and will constitute only a small percentage of all operations, (3) complication rates can be fed back to surgeons in a timely manner, and (4) reliable results can be obtained at the level of the individual surgeon. In this article, parsimonious, interpretable models for SSI, UTI, sepsis, and pneumonia that only used structured EHR data are presented. Unique features of the approach include the use of disease phenotypes32,33 and implementation of the knockoff filter34,35 for controlled variable selection.
Methods
Study design
Retrospective observational data from surgeries that were done at 5 major hospitals within a large healthcare system, University of Colorado Health, between 2013 and 2019, were included. The hospitals included 3 academic and 2 community hospitals that participated in the ACS-NSQIP program. Approximately 80,000 total surgeries are performed at these hospitals per year. The EHR data were linked to outcomes data from the ACS-NSQIP using medical record numbers and other personal health identifiers by the University of Colorado’s Health Data Compass Data Warehouse project. All hospitals and affiliated clinics within this healthcare system used Epic (Epic Systems Corp, Verona, WI). This study was approved by the Colorado Multiple Institutional Review Board (#20–1862).
Electronic Health Records Data
The EHR data included demographic characteristics, details about the operation, surgeon information, ICD-9 and ICD-10 codes, procedure names, laboratory data, and medications. Diagnosis codes and laboratory data were included if they occurred up to 30 days after the primary operation. Medications were included only if they were prescribed between 2 and 30 days after the operation to avoid inclusion of perioperative prophylactic antibiotics prescribed at the time of the operation. Current procedural terminology and Logical Observation Identifiers Names and Codes were often missing or mis-coded, so procedure and laboratory names were used instead. Due to the longitudinal nature of the data and the multiple sites, these names appeared in a variety of forms (eg, “AEROBIC CULTURE [ie, TISSUE, ABSCESS, WOUND, SINUS, ETC]” and “ANAEROBIC CULTURE, [ie, TISSUE, ABSCESS, WOUND, SINUS, ETC],” and “CULTURE ANAEROBIC”). Therefore, to avoid having to create logical groupings of these names for the entire set, a 2-stage process was implemented, where individual names were included as predictors on the first pass, and for those selected during the model selection process (described in the statistical methods section), similar names were classified into 1 predictor (eg, “blood culture”) and the models were refit. If the beginning of the laboratory result started with “no [space]” or “negative [space],” the result was coded as negative. For the medication data, we first subset the pharmaceutical classes into only those in the therapeutic class of antibiotics and created a binary indicator of at least one antibiotic prescribed versus none. Antifungals were also included, and for the sepsis models, the therapeutic class of “cardiac” was included to capture vasoactive and inotropic agents. We used phecode mappings to reduce the number of diagnosis codes as predictors and to combine similar diagnoses into diagnosis phenotypes.32,33 This also permitted us to include data that spanned the change from ICD-9 to ICD-10 codes, which occurred in October 2015. The phecode mapping versions used were 1.2 (ICD-9), 1.2 b1 (ICD-10), and 1.2 b1 CM (ICD-10 CM). For hierarchical phecodes, we rounded them to whole numbers and labeled their extension as “X.” These were phecodes that clinically could be combined into a single predictor representing a specific diagnosis related to one of the infections. We assumed that if a patient did not have a specific code, then they did not experience that exposure (ie, there were no missing covariates).
ACS-NSQIP Outcomes Data
Postoperative infection outcomes collected by the ACS-NSQIP included the following complications occurring within 30 days of the index operation: superficial SSI, deep incisional SSI, organ space SSI, wound disruption, UTI, sepsis, septic shock, and pneumonia. We grouped the different SSI complications plus wound disruption into one SSI variable, and sepsis and septic shock into one variable, because the process for diagnosing each infection within those groups is similar.
Statistical analysis
Data were divided into training and test datasets for model development and validation using a temporal split of 70%/30%, respectively. Supervised learning was used to develop and validate models for identifying infectious complications in patients who underwent surgery using EHR covariate data and ACS-NSQIP outcomes data.
To identify a parsimonious and generalizable model, we used the “knockoffs” framework described by Barber and Candes34 and Candes et al,35 which is a variable selection technique that controls type I error using false discovery rate (FDR) correction. Previously, Colborn et al developed and validated models for SSI,10 UTI,16 and overall morbidity36 using data only from one hospital, and they used penalized regression techniques to carry out variable selection. Although penalized regression techniques are suited to handle problems with a large number of predictors, it is difficult to choose a value of λ (ie, the penalty term) such that type I error is controlled. Knockoffs solve the variable selection problem by providing a negative control group for the predictors. These “knockoff” variables mimic the original independent variables, except they are known to be null (ie, not associated with the outcome). When the effect estimates of the true variables are sufficiently larger than their negative controls, they are selected by the knockoff filter. The threshold for this difference is determined by the FDR rate, which is set by the user.
We created 63 training scenarios. For each scenario, we varied the cutoff of frequency of each variable between 5 and 50 by increments of 5. This preprocessing step allowed us to remove exposures that were only seen in a small number of patients, which sped up model fitting. We also treated the FDR rate as a tuning parameter, testing values between 0.1 and 0.25 by increments of 0.025, as the choice of FDR rate is not obvious before model fitting. Next, lasso models were used to estimate regression coefficients for the knockoff filter. Models that exhibited parsimony, that had high performance in the training set and made sense clinically, were selected for each infection outcome. As mentioned in the methods section, for any procedures or laboratory tests that were selected, we went back to the original data and searched for similar names so that we could regroup these into one variable. Finally, multiple logistic regression was used to estimate unbiased coefficients and 95% CIs of the selected variables (lasso coefficients are biased). The logistic regression models were applied to the training and test datasets to report final model performance. Performance statistics of these models were computed in the test dataset using the Youden’s J37 threshold estimated in the training dataset. Confidence intervals of the performance statistics were estimated using bootstrap methods. The model estimated prevalence for each outcome was calculated as the sum of the predicted probabilities across all patients. Calibration plots were created by plotting predicted values for each outcome divided into 5 groups determined by equally spaced cut-points between 0% and 100%.
All analyses were performed in R Version 3.6.1 (R Project for Statistical Computing, Vienna, Austria). We used the knockoff38 and glmnet39 packages for model fitting, and the pROC40 package for estimating model performance statistics. Example R code for implementing the models is provided in a GitHub repository.41 The methods for developing and validating the models in this study follow the “Type 2b: Nonrandom split-sample development and validation” guidelines described in the Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis.42 A completed copy of the Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis checklist for this study can be found in Supplementary Table S1.
Results
There were 31,579 operations in the ACS-NSQIP data from these institutions occurring between June 30, 2013 and October 16, 2019. Of these operations, 940 (3%) were not identified in the EHR data. Therefore, 30,639 operations were included in the analyses presented herein (STrengthening the Reporting of OBservational studies in Epidemiology diagram provided in Figure 1). The training dataset consisted of operations that occurred between June 30, 2013 and March 1, 2017 (N = 21,450 [70%]), and the test dataset included operations between March 2, 2017 and October 16, 2019 (N = 9,189 [30%]). There were >3,000 candidate predictors in each analytic dataset.
Figure 1.
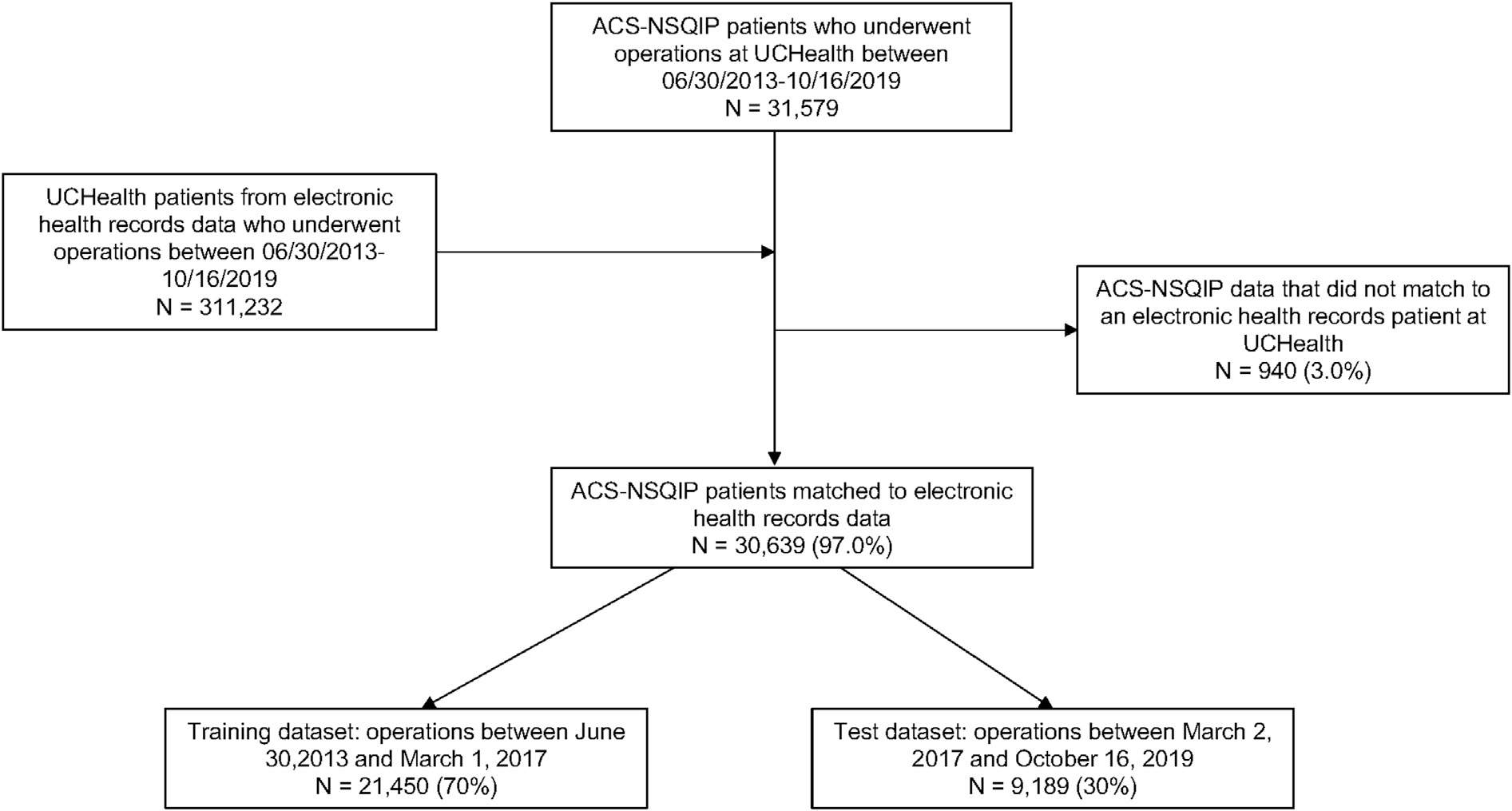
STrengthening the Reporting of OBservational studies in Epidemiology diagram. ACS-NSQIP, American College of Surgeons National Surgical Quality Improvement Program; UCHealth, University of Colorado Health.
Table I summarizes the patient characteristics and outcomes in this population, overall and by hospital. Because the outcomes are not risk-adjusted, we have chosen to keep the names of the hospitals anonymous. Five hospitals were included but 2 are not distinguishable from one another in the data, so the data are summarized into 4 hospitals. Overall, the average age was 55.3 ± 16.6, and 56% of the patients were female. All of the hospitals served mostly White patients (76%–93%). Black patients were seen more at 2 of the hospitals (5% and 7%) but were not seen commonly at the other 2 hospitals (1% each). Hispanic patients made up between 7% to 12% of patients across the hospitals. Most of the operations were from orthopedic (33%), general (28%), gynecology (11%), urology (9%), and neurosurgery (8%). The overall infection rate was 7% and varied from 4% to 9% at individual hospitals.
Table I.
Patient sample stratified by hospital and overall
| Hospital 1 | Hospital 2 | Hospital 3 | Hospital 4 | Overall | |
|---|---|---|---|---|---|
|
|
|
|
|
|
|
| (N = 6,440) | (N = 6,264) | (N = 6,469) | (N = 11,466) | (N = 30,639) | |
|
| |||||
| Age* | 57.0(16.5) | 55.1 (16.6) | 57.1 (16.9) | 53.4 (16.3) | 55.3 (16.6) |
| Sex | |||||
| Female | 3,199 (49.7%) | 3,793 (60.6%) | 3,807 (58.8%) | 6,390 (55.7%) | 17,189 (56.1%) |
| Male | 3,241 (50.3%) | 2,471 (39.4%) | 2,662 (41.2%) | 5,076 (44.3%) | 13,450 (43.9%) |
| Race | |||||
| American Indian and Alaska Native | 17 (0.3%) | 25 (0.4%) | 28 (0.4%) | 41 (0.4%) | 111 (0.4%) |
| Asian | 24 (0.4%) | 92 (1.5%) | 38 (0.6%) | 230 (2.0%) | 384 (1.3%) |
| Black or African American | 33 (0.5%) | 331 (5.3%) | 57 (0.9%) | 843 (7.4%) | 1,264 (4.1%) |
| Multiple race | 82 (1.3%) | 93 (1.5%) | 78 (1.2%) | 343 (3.0%) | 596 (1.9%) |
| Native Hawaiian and Other Pacific Islander | 8 (0.1%) | 18 (0.3%) | 8 (0.1%) | 19 (0.2%) | 53 (0.2%) |
| Null/unknown | 114(1.8%) | 158 (2.5%) | 83 (1.3%) | 282 (2.5%) | 637 (2.1%) |
| Other | 223 (3.5%) | 504 (8.0%) | 136 (2.1%) | 998 (8.7%) | 1,861 (6.1%) |
| White or Caucasian | 5,939 (92.2%) | 5,043 (80.5%) | 6,041 (93.4%) | 8,710 (76.0%) | 25,733 (84.0%) |
| Ethnicity Hispanic |
704 (10.9%) | 625 (10.0%) | 475 (7.3%) | 1,377 (12.0%) | 3,181 (10.4%) |
| Non-Hispanic | 5,628 (87.4%) | 5,398 (86.2%) | 5,904 (91.3%) | 9,807 (85.5%) | 26,737 (87.3%) |
| Patient refused | 41 (0.6%) | 49 (0.8%) | 41 (0.6%) | 53 (0.5%) | 184 (0.6%) |
| Unknown | 67 (1.0%) | 192 (3.1%) | 49 (0.8%) | 229 (2.0%) | 537 (1.8%) |
| Surgeon specialty General surgery | 2,055 (31.9%) | 1,630 (26.0%) | 1,859 (28.7%) | 2,932 (25.6%) | 8,476 (27.7%) |
| Gynecology | 408 (6.3%) | 1,113 (17.8%) | 722 (11.2%) | 1,078 (9.4%) | 3,321 (10.8%) |
| Neurosurgery | 549 (8.5%) | 409 (6.5%) | 313 (4.8%) | 1,036 (9.0%) | 2,307 (7.5%) |
| Orthopedics | 1,958 (30.4%) | 2,398 (38.3%) | 2,814 (43.5%) | 2,978 (26.0%) | 10,148 (33.1%) |
| Otolaryngology, Head and Neck | 94 (1.5%) | 194(3.1%) | 321 (5.0%) | 993 (8.7%) | 1,602 (5.2%) |
| Plastics | 79(1.2%) | 103 (1.6%) | 134 (2.1%) | 445 (3.9%) | 761 (2.5%) |
| Thoracic | 274 (4.3%) | 1 (0.0%) | 0(0%) | 540 (4.7%) | 815 (2.7%) |
| Urology | 943 (14.6%) | 232 (3.7%) | 306 (4.7%) | 1,128 (9.8%) | 2,609 (8.5%) |
| Vascular | 80(1.2%) | 184 (2.9%) | 0 (0%) | 336 (2.9%) | 600 (2.0%) |
| Infectious complications Overall infections | 371 (5.8%) | 419 (6.7%) | 253 (3.9%) | 1,075 (9.4%) | 2,118 (6.9%) |
| Surgical site infections | 170 (2.6%) | 185 (3.0%) | 135 (2.1%) | 579 (5.0%) | 1,069 (3.5%) |
| Urinary tract infections | 83 (1.3%) | 78 (1.2%) | 61 (0.9%) | 244 (2.1%) | 466 (1.5%) |
| Sepsis or septic shock | 126 (2.0%) | 208 (3.3%) | 82 (1.3%) | 352 (3.1%) | 768 (2.5%) |
| Pneumonia | 82 (1.3%) | 43 (0.7%) | 23 (0.4%) | 137 (1.2%) | 285 (0.9%) |
Mean (SD); otherwise, N (%).
Coefficients, 95% CIs, and intercepts for each of the models are provided in Table II. A total of 4 variables were chosen for the SSI model, 7 for the UTI model, 7 for sepsis, and 8 for pneumonia. All of the variables chosen by the knockoff filter made clinical sense.
Table II.
Logistic regression model estimates
| Model | Estimates | ||||
|---|---|---|---|---|---|
|
|
|||||
| Beta | OR | LCL | UCL | P value | |
|
| |||||
| Surgical site infections | |||||
| (Intercept) | −5.1187 | ||||
| Phecode 080: “Postoperative infection” | 2.7245 | 15.25 | 11.82 | 19.67 | < .001 |
| Phecode 1011: “Complications of surgical and medical procedures” | 0.886 | 2.43 | 1.85 | 3.18 | < .001 |
| At least 1 antibiotic prescribed between 2–30 d after surgery | 2.0891 | 8.08 | 6.51 | 10.02 | < .001 |
| Laboratory procedure: Blood culture | 2.041 | 7.7 | 6.38 | 9.29 | < .001 |
| Urinary tract infections | |||||
| (Intercept) | −6.1696 | ||||
| Phecode 590: “Pyelonephritis” | 2.3597 | 10.59 | 4.44 | 25.23 | < .001 |
| Phecode 591: “Urinary tract infection” | 1.9223 | 6.84 | 5.16 | 9.05 | < .001 |
| Phecode 592.X: “Cystitis,” “Urethritis,” “Urethral stricture due to infection” | 1.7764 | 5.91 | 3.6 | 9.7 | < .001 |
| Phecode 599.X: Various symptoms involving the urinary system | 1.1322 | 3.1 | 2.36 | 4.09 | < .001 |
| At least 1 antibiotic prescribed between 2—30 d after surgery | 1.2781 | 3.59 | 2.58 | 5 | < .001 |
| Laboratory procedure: Urine culture | 1.6599 | 5.26 | 3.78 | 7.32 | < .001 |
| Laboratory procedure: Clostridioides difficile PCR | 0.4246 | 1.53 | 0.92 | 2.55 | 0.1 |
| Sepsis | |||||
| (Intercept) | −7.2263 | < .001 | |||
| Phecode 540.X: “Acute appendicitis,” “Appendicitis,” “Appendiceal conditions” | 2.0447 | 7.73 | 5.53 | 10.79 | < .001 |
| Phecode 994.X: “Sepsis,” “SIRS” | 2.4980 | 12.16 | 9.23 | 16.02 | < .001 |
| At least 1 antibiotic prescribed between 2—30 d after surgery | 1.6909 | 5.42 | 4.14 | 7.11 | < .001 |
| Laboratory procedure: CBC auto diff | 1.3637 | 3.91 | 2.07 | 7.38 | < .001 |
| Laboratory procedure: Blood culture | 1.9005 | 6.69 | 5.36 | 8.34 | < .001 |
| Laboratory procedure: Magnesium serum | 1.2173 | 3.38 | 2.65 | 4.30 | < .001 |
| Laboratory procedure: Peripheral blood smear | 1.4547 | 4.28 | 3.00 | 6.12 | < .001 |
| Pneumonia | |||||
| (Intercept) | −7.3366 | ||||
| Phecode 480.X: Bacterial, viral, and fungal pneumonias | 2.5952 | 13.4 | 9.17 | 19.57 | < .001 |
| Phecode 501: “Pneumonitis due to inhalation of food or vomitus” | 1.7072 | 5.51 | 3.03 | 10.03 | < .001 |
| Phecode 1013: “Asphyxia and hypoxemia” | 0.9214 | 2.51 | 1.67 | 3.78 | < .001 |
| At least 1 antibiotic prescribed between 2—30 d after surgery | 1.812 | 6.12 | 3.59 | 10.44 | < .001 |
| Laboratory procedure: Magnesium serum | 0.7905 | 2.2 | 1.37 | 3.56 | .001 |
| Laboratory procedure: Vancomycin trough | 0.9937 | 2.7 | 1.84 | 3.96 | < .001 |
| Laboratory procedure: Respiratory culture | 1.4024 | 4.06 | 2.52 | 6.55 | < .001 |
| Laboratory procedure: Blood gasses | 1.511 | 4.53 | 3.02 | 6.79 | < .001 |
CBC, complete blood count; LCL, lower confidence limit; OR, odds ratio; PCR, polymerase chain reaction; SIRS, systemic inflammatory response syndrome; UCL, upper confidence limit.
Performance of the models applied to both the training and the test datasets are summarized in Table III. Estimated prevalence in the test dataset was 3.3% for SSI, 1.7% for UTI, 2.5% for sepsis, and 0.9% for pneumonia, compared to 3.1% for SSI, 1.6% for UTI, 2.0% for sepsis, and 0.7% for pneumonia reported by the NSQIP nurses, which showed strong agreement. When applied to the test dataset, the models achieved between 0.91 to 0.96 AUC, 82% to 95% specificity, 87% to 93% sensitivity, 82% to 95% accuracy, 99% negative predictive value, and 10% to 15% positive predictive value (PPV). All the models performed similarly in the test dataset as they did in the training dataset.
Table III.
Performance statistics for each model in both the training and test datasets
| Statistic | SSI | UTI | Sepsis | Pneumonia | ||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|
||||
| Data set | Training | Test | Training | Test | Training | Test | Training | Test |
|
| ||||||||
| NSQIP prevalence | 784 (3.7%) | 285 (3.1%) | 322 (1.5%) | 144 (1.6%) | 584 (2.7%) | 184 (2.0%) | 222 (1.0%) | 63 (0.7%) |
| Model prevalence | 3.7% | 3.3% | 1.5% | 1.7% | 2.7% | 2.5% | 1.0% | 0.9% |
| FDR level | 0.175 | NA | 0.1 | NA | 0.125 | NA | 0.125 | NA |
| No. of predictors selected | 4 | NA | 7 | NA | 7 | NA | 8 | NA |
| Threshold for classification* | 2.9 | 2.9 | 1.1 | 1.1 | 4.5 | 4.5 | 1.6 | 1.6 |
| AUC | 0.91 | 0.91 | 0.91 | 0.93 | 0.95 | 0.95 | 0.96 | 0.96 |
| Specificity* | 81 (80, 81) | 82 (81,82) | 90 (90,91) | 90 (89,91) | 89 (89, 90) | 89 (88, 90) | 95 (94, 95) | 95 (94, 95) |
| Sensitivity* | 89 (87, 91) | 89 (85, 93) | 82 (78, 86) | 90 (85, 94) | 89 (87, 92) | 93 (90, 97) | 88, (84, 92) | 87 (79, 95) |
| Accuracy* | 81 (81, 82) | 82 (81,83) | 90 (89,91) | 90 (89,91) | 89 (89, 90) | 89 (88, 90) | 94 (94, 95) | 95 (94, 95) |
| NPV* | 99 (99, 99) | 99 (99, 99) | 99 (99, 99) | 99 (99, 99) | 99 (99, 99) | 99 (99, 99) | 99 (99, 99) | 99 (99, 99) |
| PPV* | 15 (15, 15) | 13 (13, 14) | 11 (10, 11) | 12 (11, 13) | 19(18, 20) | 15 (14, 16) | 14(13, 15) | 10(9, 11) |
AUC, area under the curve; FDR, false discovery rate; NA, not applicable; NPV, negative predictive value; NSQIP, National Surgical Quality Improvement Program; PPV, positive predictive value; SSI, surgical site infection; UTI, urinary tract infection.
Values multiplied by 100.
Calibration plots are provided in Figures 2A to 2D; the dashed lines indicate the training data, and the solid lines indicate the test data. Calibration was very good in the training and test data for each outcome, but there was a slight tendency for models to overestimate risk. This could be because prevalence of infectious complications has decreased over time (Table III).
Figure 2.
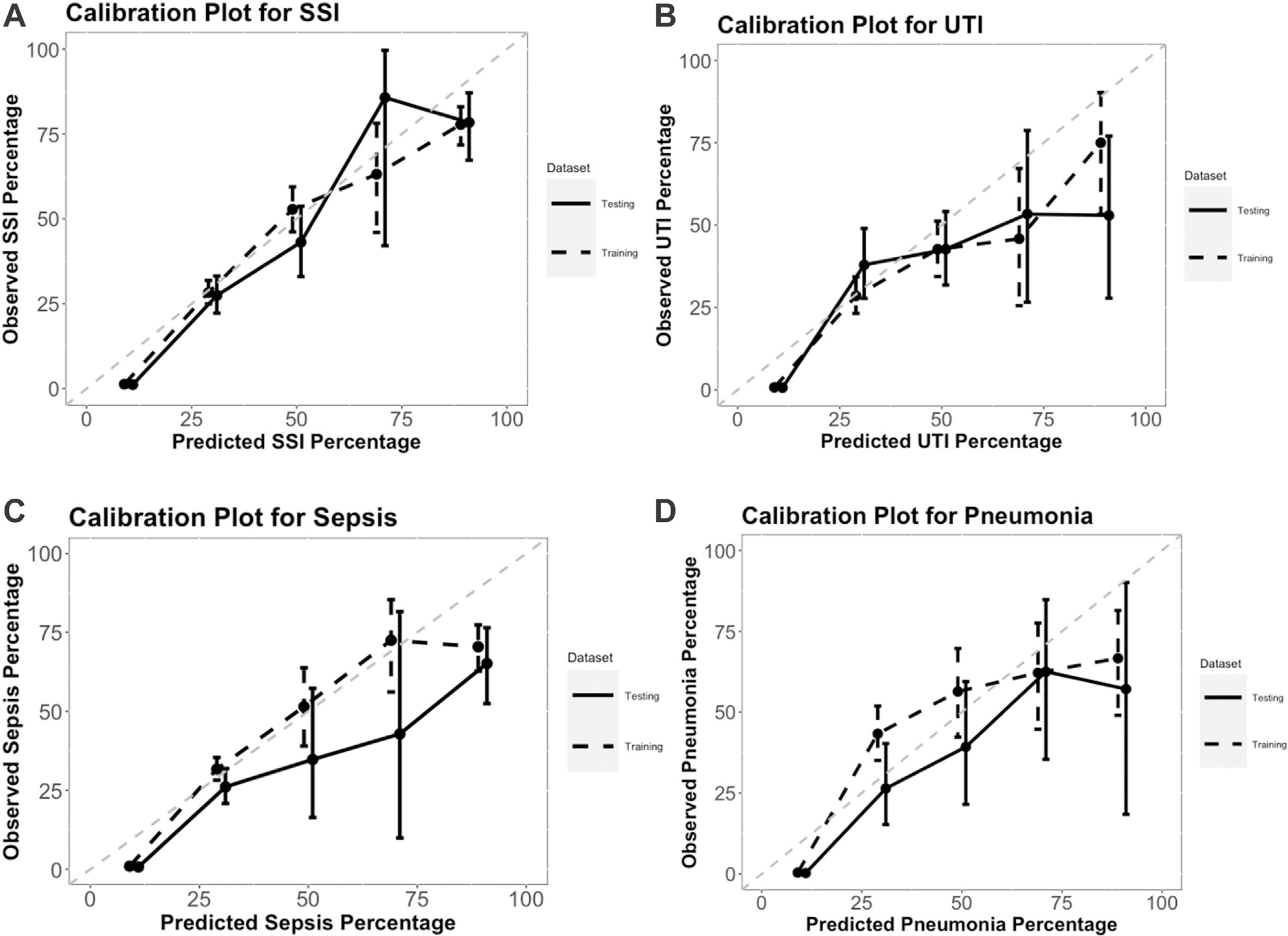
Calibration plots for each model applied to the training and test datasets: (A) surgical site infection, (B) urinary tract infection, (C) sepsis, (D) pneumonia. The dots signify the mean predicted values by the observed complication rates; whiskers represent the 95% CIs of the observed rates based on the binomial distribution; and perfect estimation is indicated in these plots by the gray 45° line. SSI, surgical site infection; UTI, urinary tract infection.
How to use the models
The models can be implemented by using the regression coefficients in Table II and information taken directly from a patient’s EHR. For example, a patient who had a diagnosis code that mapped to phecode 080, a blood culture within 30 days after their index operation, and an antibiotic prescription between 2 and 30 days after the index operation, would have the predicted log odds of an SSI: −5.1187 + 2.7245 × 1 +0.886 × 0 + 2.0891 × 1 + 2.041 × 1 =1.7359. To convert the log odds to a probability, take exp (1.7359) / 1 + exp (1.7359) = 0.85. Comparing this to a patient who did not have any of the 4 SSI variables in their chart, −5.1187 + 2.7245 × 0 + 0.886 × 0 + 2.0891 × 0 + 2.041 × 0 =−5.1187, and exp (−5.1187) / 1 + exp (−5.1187) = 0.006.
Discussion
This study suggests that simple, interpretable models can be applied with high accuracy to EHR data to perform surveillance of postoperative infections in patients who underwent operations at a large healthcare system. The final models presented are easy to implement in virtually any software and can be applied to all operations. This work is unique in that it covered 4 major postoperative infectious complications, it used phecode mappings which improved generalizability and decreased the chance of missing an important ICD-9 or ICD-10 code, and it used robust statistical methods to control type I error when selecting the most important variables to include in the models. Because prevalence of these infectious complications is relatively rare, PPV was low. Therefore, these models are most appropriate for estimating rates of complications rather than identifying specific patients who experienced an infection. To estimate rates, it is best to sum the predicted probabilities rather than introduce error by classifying patients.
Although the granularity of diagnosis codes is lost when using phecodes, those that were selected for the models contained logical mappings from a clinical perspective upon inspection. For example, ICD-9 code 519.01 “Infection of tracheostomy” and ICD-10CM code J95.02 “Infection of tracheostomy stoma” both map to phecode 080 “Postoperative infection,” as well as ICD-10CM T81.4 “Infection following a procedure.” This is the desired outcome of using phecodes because the infection is picked up in all cases. Although each postoperative infection is slightly different, they all likely indicate that a postoperative infection occurred. If the codes had been left as individual predictors, less common codes would not have been selected for inclusion in the model, and some infections would have been missed.
Previously published studies reported models for SSI detection with sensitivities between 0.65 and 0.79 and specificities between 0.92 and 1.00.9,11 A recent article showed that artificial intelligence detected UTIs in 59 patients using EHR data with 98% sensitivity and 100% specificity using artificial neural networks, but given this very small sample size, the use of a black box algorithm, and the lack of external validation, generalizability of these findings is questionable.43 The SSI model we presented is simple and is similar to that of Knepper et al, which suggests that it could also be used to augment NHSN SSI surveillance.31
A future goal is to also produce a preoperative risk model that can be used to risk-adjust the infectious complication rates by creating observed to expected event ratios at administrative levels of interest. Moreover, there is a large amount of narrative text data that might enhance the models. For example, FitzHenry et al found sensitivities and specificities of 80% and 93% for sepsis, 80% and 90% for pneumonia, 95% and 80% for UTI, and 80% and 93% for wound infections, respectively, using NLP alone on EHR data from the Veteran’s Health Administration.28 Currently, we are working with experts in NLP to use narrative text data. However, the current models presented performed well and do not require NLP or sophisticated machine learning techniques to implement them. This improves generalizability across institutions.
There are some limitations to our methods. First, we assumed that if a code was not observed in a patient’s chart, then the event represented by that code did not occur. This may not be valid for various reasons, including the possibility that postdischarge complication care was sought outside of the University of Colorado Health network of hospitals and clinics. Second, the probability that a patient experienced a postoperative infection could still increase if the surgeon prescribed perioperative antibiotics past 24 to 48 hours for prophylaxis, against the Surgical Care Improvement Project recommendations. Third, only 2 discriminators for negative test results were used but there may have been more indicators of negative laboratory results. Fourth, if a urine or blood culture name did not contain “culture” it may have been missed. Moreover, although we would have preferred to use the Logical Observation Identifiers Names and Codes for laboratory procedures rather than panel names, because these were not completed consistently in the EHR data, they could not be used. However, because there are no phenotype mappings for procedures or laboratory codes, like the phecodes, the individual codes become cumbersome. The approach implemented in this study attempts to create that mapping using the procedure and lab names, but it is likely incomplete and may need to be modified at an external institution that uses different names. Fifth, because only one EHR system was used across all sites, it is unclear whether the results are generalizable beyond this EHR system. However, we did observe variability in the individual EHRs across the hospitals and clinics within this system, and the methodological approach implemented here captures that variability by mapping specific procedures, labs, and diagnoses to broader groups. Finally, the models should eventually be validated in an external dataset from a different healthcare system, much like Zhu et al did for their SSI detection algorithm,44 and because they were developed on retrospective data and applied to prospective data, they will need to be recalibrated over time due to model drift.45,46
In conclusion, artificial intelligence can be used for the surveillance of postoperative infections with high accuracy, specificity, and sensitivity using EHR data and simple linear regression coefficients. Although the model selection technique that was applied was complex, the final chosen models are simple to implement. Unlike HAI surveillance that relies on manual chart review, these models can be applied to all operations and results can be disseminated in a timely manner. Due to the low PPV, these models are most appropriate for tracking rates of complications over time, but they may require additional manual review to identify exact patients who experienced an infection.
Supplementary Material
Funding/Support
This project was supported by grant number R01HS027417 from the Agency for Healthcare Research and Quality. The content is solely the responsibility of the authors and does not necessarily represent the official views of the Agency for Healthcare Research and Quality.
Footnotes
Conflict of interest/Disclosure
The American College of Surgeons National Surgical Quality Improvement Program and participating hospitals are the source of the outcomes data; the American College of Surgeons has not verified, and are not responsible for, the statistical validity of the data analysis or the conclusions derived by the authors. The authors have no relevant financial disclosures.
References
- 1.Al-Tawfiq JA, Tambyah PA. Healthcare associated infections (HAI) perspectives. J Infect Public Health. 2014;7:339–344. [DOI] [PubMed] [Google Scholar]
- 2.de Lissovoy G, Fraeman K, Hutchins V, Murphy D, Song D, Vaughn BB. Surgical site infection: incidence and impact on hospital utilization and treatment costs. Am J Infect Control. 2009;37:387–397. [DOI] [PubMed] [Google Scholar]
- 3.Zimlichman E, Henderson D, Tamir O, et al. Health care-associated infections: a meta-analysis of costs and financial impact on the US health care system. JAMA Intern Med. 2013;173:2039–2046. [DOI] [PubMed] [Google Scholar]
- 4.Stone PW, Dick A, Pogorzelska M, Horan TC, Furuya EY, Larson E. Staffing and structure of infection prevention and control programs. Am J Infect Control. 2009;37:351–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.ACS NSQIP. American College of Surgeons; 2019. https://www.facs.org/quality-programs/acs-nsqip/about. Accessed February 24, 2019.
- 6.Aasen DM, Bronsert MR, Rozeboom PD, et al. Relationships between predischarge and postdischarge infectious complications, length of stay, and unplanned readmissions in the. ACS NSQIP database. Surgery. 2021;169:325–332. [DOI] [PubMed] [Google Scholar]
- 7.Hammermeister KE, Henderson WG, Bronsert MR, Juarez-Colunga E, Meguid RA. Bringing quantitative risk assessment closer to the patient and surgeon: a novel approach to improve outcomes. Ann Surg. 2016;263:1039–1041. [DOI] [PubMed] [Google Scholar]
- 8.Centers for Disease Control and Prevention. National Healthcare Safety Network; 2021. https://www.cdc.gov/nhsn/about-nhsn/index.html. Accessed May 24, 2021.
- 9.Branch-Elliman W, Strymish J, Itani KM, Gupta K. Using clinical variables to guide surgical site infection detection: a novel surveillance strategy. Am J Infect Control. 2014;42:1291–1295. [DOI] [PubMed] [Google Scholar]
- 10.Colborn KL, Bronsert M, Amioka E, Hammermeister K, Henderson WG, Meguid R. Identification of surgical site infections using electronic health record data. Am J Infect Control. 2018;46:1230–1235. [DOI] [PubMed] [Google Scholar]
- 11.Goto M, Ohl ME, Schweizer ML, Perencevich EN. Accuracy of administrative code data for the surveillance of healthcare-associated infections: a systematic review and meta-analysis. Clin Infect Dis. 2014;58:688–696. [DOI] [PubMed] [Google Scholar]
- 12.Hu Z, Simon GJ, Arsoniadis EG, Wang Y, Kwaan MR, Melton GB. Automated detection of postoperative surgical site infections using supervised methods with electronic health record data. Stud Health Technol Inform. 2015;216:706–710. [PMC free article] [PubMed] [Google Scholar]
- 13.Ju MH, Ko CY, Hall BL, Bosk CL, Bilimoria KY, Wick EC. A comparison of 2 surgical site infection monitoring systems. JAMA Surg. 2015;150:51–57. [DOI] [PubMed] [Google Scholar]
- 14.Branch-Elliman W, Strymish J, Kudesia V, Rosen AK, Gupta K. Natural language processing for real-time catheter-associated urinary tract infection surveillance: results of a pilot implementation trial. Infect Control Hosp Epidemiol. 2015;36:1004–1010. [DOI] [PubMed] [Google Scholar]
- 15.Choudhuri JA, Pergamit RF, Chan JD, et al. An electronic catheter-associated urinary tract infection surveillance tool. Infect Control Hosp Epidemiol. 2011;32:757–762. [DOI] [PubMed] [Google Scholar]
- 16.Colborn KL, Bronsert M, Hammermeister K, Henderson WG, Singh AB, Meguid RA. Identification of urinary tract infections using electronic health record data. Am J Infect Control. 2018;47:371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gundlapalli AV, Divita G, Redd A, et al. Detecting the presence of an indwelling urinary catheter and urinary symptoms in hospitalized patients using natural language processing. J Biomed Inform. 2017;71s:S39–S45. [DOI] [PubMed] [Google Scholar]
- 18.Hsu HE, Shenoy ES, Kelbaugh D, et al. An electronic surveillance tool for catheter-associated urinary tract infection in intensive care units. Am J Infect Control. 2015;43:592–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Landers T, Apte M, Hyman S, Furuya Y, Glied S, Larson E. A comparison of methods to detect urinary tract infections using electronic data. Jt Comm J Qual Patient Saf. 2010;36:411–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanger PC, Granich M, Olsen-Scribner R, et al. Electronic surveillance for catheter-associated urinary tract infection using natural language processing. AMIA Annu Symp Proc. 2018;2017:1507–1516. [PMC free article] [PubMed] [Google Scholar]
- 21.Shepard J, Hadhazy E, Frederick J, et al. Using electronic medical records to increase the efficiency of catheter-associated urinary tract infection surveillance for National Health and Safety Network reporting. Am J Infect Control. 2014;42:e33e–e36. [DOI] [PubMed] [Google Scholar]
- 22.Sopirala MM, Syed A, Jandarov R, Lewis M. Impact of a change in surveillance definition on performance assessment of a catheter-associated urinary tract infection prevention program at a tertiary care medical center. Am J Infect Control. 2018;46:743–746. [DOI] [PubMed] [Google Scholar]
- 23.Wald HL, Bandle B, Richard A, Min S. Accuracy of electronic surveillance of catheter-associated urinary tract infection at an academic medical center. Infect Control Hosp Epidemiol. 2014;35:685–691. [DOI] [PubMed] [Google Scholar]
- 24.Zhan C, Elixhauser A, Richards CL Jr, et al. Identification of hospital-acquired catheter-associated urinary tract infections from Medicare claims: sensitivity and positive predictive value. Med Care. 2009;47:364–369. [DOI] [PubMed] [Google Scholar]
- 25.Thottakkara P, Ozrazgat-Baslanti T, Hupf BB, et al. Application of machine learning techniques to high-dimensional clinical data to forecast postoperative complications. PLoS One. 2016;11, e0155705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu Z, Melton GB, Moeller ND, et al. Accelerating chart review using automated methods on electronic health record data for postoperative complications. AMIA Annu Symp Proc. 2017;2016:1822–1831. [PMC free article] [PubMed] [Google Scholar]
- 27.Leth RA, Moller JK. Surveillance of hospital-acquired infections based on electronic hospital registries. J Hosp Infect. 2006;62:71–79. [DOI] [PubMed] [Google Scholar]
- 28.FitzHenry F, Murff HJ, Matheny ME, et al. Exploring the frontier of electronic health record surveillance: the case of postoperative complications. Med Care. 2013;51:509–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murff HJ, FitzHenry F, Matheny ME, et al. Automated identification of postoperative complications within an electronic medical record using natural language processing. JAMA. 2011;306:848–855. [DOI] [PubMed] [Google Scholar]
- 30.Selby LV, Narain WR, Russo A, Strong VE, Stetson P. Autonomous detection, grading, and reporting of postoperative complications using natural language processing. Surgery. 2018;164:1300–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knepper BC, Young H, Jenkins TC, Price CS. Time-saving impact of an algorithm to identify potential surgical site infections. Infect Control Hosp Epidemiol. 2013;34:1094–1098. [DOI] [PubMed] [Google Scholar]
- 32.Wei WQ, Bastarache LA, Carroll RJ, et al. Evaluating phecodes, clinical classification software, and ICD-9-CM codes for phenome-wide association studies in the electronic health record. PLoS One. 2017;12:e0175508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu P, Gifford A, Meng X, et al. Mapping ICD-10 and ICD-10-CM Codes to Phecodes: workflow development and initial evaluation. JMIR Med Inform. 2019;7:e14325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barber RF, Candes EJ. Controlling the false discovery rate via knockoffs. Ann Stat. 2015;43:2055–2085. [Google Scholar]
- 35.Candes E, Fan Y, Janson L, Lv J. Panning for gold: ‘model-X’ knockoffs for high dimensional controlled variable selection. J R Stat Soc Series B Stat Methodol. 2018;80:551–577. [Google Scholar]
- 36.Bronsert M, Singh AB, Henderson WG, Hammermeister K, Meguid RA, Colborn KL. Identification of postoperative complications using electronic health record data and machine learning. Am J Surg. 2020;220:114–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Youden WJ. Index for rating diagnostic tests. Cancer. 1950;3:32–35. [DOI] [PubMed] [Google Scholar]
- 38.Patterson E, Sesia M. knockoff: The Knockoff Filter for Controlled Variable Selection. R package version 0.3.6. 2022. https://CRAN.R-project.org/package=knockoff. Accessed January 1, 2022. [Google Scholar]
- 39.Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- 40.Robin X, Turck N, Hainard A, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Colborn KL, Zhuang Y. Automated surveillance of postoperative infections (ASPIN). https://github.com/katiecolborn/ASPIN. Accessed January 10, 2022. [Google Scholar]
- 42.Moons KG, Altman DG, Reitsma JB, et al. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): explanation and elaboration. Ann Inter Med. 2015;162:W1–W73. [DOI] [PubMed] [Google Scholar]
- 43.Ozkan IA, Koklu M, Sert IU. Diagnosis of urinary tract infection based on artificial intelligence methods. Comput Methods Programs Biomed. 2018;166:51–59. [DOI] [PubMed] [Google Scholar]
- 44.Zhu Y, Simon GJ, Wick EC, et al. Applying machine learning across sites: external validation of a surgical site infection detection algorithm. J Am Coll Surg. 2021;232:963–971.e961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Davis SE, Greevy RA, Lasko TA, Walsh CG, Matheny ME. Comparison of prediction model performance updating protocols: using a data-driven testing procedure to guide updating. AMIA Annu Symp Proc. 2019;2019:1002–1010. [PMC free article] [PubMed] [Google Scholar]
- 46.Davis SE, Lasko TA, Chen G, Matheny ME. Calibration drift among regression and machine learning models for hospital mortality. AMIA Annu Symp Proc. 2017;2017:625–634. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


