Abstract
Efficient transcriptional terminators are essential for the performance of genetic circuitry in microbial SynBio hosts. In recent years, several libraries of characterized strong terminators have become available for model organisms such as Escherichia coli. Conversely, terminator libraries for nonmodel species remain scarce, and individual terminators are often ported over from model systems, leading to unpredictable performance in their new hosts. In this work, we mined the genomes of Pseudomonas infecting phages LUZ7 and LUZ100 for transcriptional terminators utilizing the full-length RNA sequencing technique “ONT-cappable-seq” and validated these terminators in three Gram-negative hosts using a terminator trap assay. Based on these results, we present nine terminators for E. coli, Pseudomonas putida, and Pseudomonas aeruginosa, which outperform current reference terminators. Among these, terminator LUZ7 T50 displays potent bidirectional activity. These data further support that bacteriophages, as evolutionary-adapted natural predators of the targeted bacteria, provide a valuable source of microbial SynBio parts.
Introduction
In recent years, the importance of transcriptional termination in genetic circuitry has become fully acknowledged by the scientific community. Many agree that proper transcriptional termination ensures minimal read-through of synthetic circuitry into neighboring sequences, stabilization of mRNA transcripts, and sufficient insulation of surrounding promoter sequences.1−5 Despite this awareness, the number of terminator libraries and prediction tools, especially for nonmodel hosts, remains massively underrepresented in available promoter libraries.6,7 This limited availability of characterized terminator sequences sharply contrasts the high demand for bidirectional, sequence-diverse terminators for metabolic engineering applications.8 Indeed, because of the rapid development of large-scale genome engineering tools for species all across the tree of life, it has become straightforward to metabolically engineer a host strain in a genome-wide manner, requiring an abundance of characterized, species-specific genetic parts. Furthermore, the integral transfer of terminators from one host to another without proper characterization can come with significant impairment of their function. This was also recently addressed in a revisit of terminators for the well-established SEVA vectors and underlies the need for libraries of terminators that can be used across different hosts.9
The lack of characterized terminators has not gone unnoticed, and several terminator libraries for nonmodel bacterial hosts have recently been published.3,4,10−12 While some terminator libraries contain sequence-diverse terminators mined from environmental or bacterial genomic DNA,3,4,10 others are fully synthetic with an invariable core sequence.8 The downside of these invariable sequences is the increased risk of undesired recombination events and difficulties during the cloning process.13 Therefore, identification of native transcriptional terminators from prokaryotic and viral genomes is preferred due to their inherent sequence diversity.
Recently, various state-of-the-art transcriptomics techniques have emerged that facilitate the identification of prokaryotic and viral transcriptional terminators at a genome scale. In contrast to Illumina-based, short-read RNA-seq methods,14,15 long-read transcriptomic approaches allow sequencing of full-length transcripts and hence provide a comprehensive manner to delineate the 5′ and 3′ transcript boundaries.16−18 For example, ONT-cappable-seq, a recently established nanopore-based cDNA sequencing method, enables end-to-end sequencing of primary prokaryotic transcripts, yielding a genome-wide map of key regulatory features encoded by viruses and their bacterial hosts, including transcription start sites (TSSs) and transcription termination sites (TTSs).18 By specifically capturing intact, full-length primary RNA molecules by their triphosphate 5′ end during the ONT-cappable-seq library preparation, this method is ideally suited to accurately identify RNA 5′ and 3′ ends.18,19 In addition to mapping transcription termination events, ONT-cappable-seq provides insights in their apparent termination strengths and dynamics. This approach has been established usingPseudomonas viruses LUZ7 and LUZ100.18,19
In this work, we mine bacterial virus genomes using ONT-cappable-seq to identify short, highly potent, and sequence-diverse terminators for nonmodel hosts. Bacteriophages, or phages in short, have a compact genome which, from an evolutionary perspective, is fully adapted to their host in terms of GC content and codon usage. Many phage genomes encode potent terminators to counter and regulate transcript elongation at high transcriptional levels from phage promoters.18,20 As such, phage genomes are a potential treasure trove of potent sequence-diverse termination sequences for their respective host. Here we evaluate the in vivo termination activity of these phage-derived terminator sequences and explore their potential as novel genetic parts to control gene expression in various hosts to ultimately expand the synthetic biologist’s toolbox for nonmodel bacteria.
Results and Discussion
ONT-cappable-seq Allows Straightforward Terminator Identification
The ONT-cappable-seq method enables full-length sequencing of primary prokaryotic transcripts and can be readily leveraged to mine regulatory features from densely coded phage genomes, including transcriptional terminators (Figure 1).18 Indeed, the transcriptional landscapes ofPseudomonasphages LUZ7 and LUZ100 obtained by ONT-cappable-seq were leveraged to identify 61 and 22 transcription termination sites, respectively.18,19 These phage-encoded terminator sequences are short and highly diverse and display varying levels of transcription read-through, making them interesting candidate terminator parts for SynBio applications. About 20% of the terminator sequences from LUZ7 and LUZ100 are predicted to be intrinsic, factor-independent terminators, which are hallmarked by a canonical GC-rich hairpin and an uridine-rich terminal segment to destabilize the RNA polymerase (RNAP) elongation complex.21 In contrast, factor-dependent terminators rely on ATP-dependent translocases to promote transcript release from the RNAP.21 Based on the ONT-cappable-seq transcriptomic data of LUZ7 and LUZ100, we selected 11 putative intrinsic and eight apparent nonintrinsic phage-encoded terminators that show a wide range of termination efficiencies (TEs) (Table 1).18,19 The phage terminator library was subsequently evaluated in vivo to identify potent terminator parts for robust synthetic circuit design beyondEscherichia coli.
Figure 1.
ONT-cappable-seq delineates transcriptional boundaries. IGV data track of ONT-cappable-seq data of Pseudomonas phage LUZ7 5 min postinfection is shown.18 Only the leftmost region of the LUZ7 genome is shown. ONT-cappable-seq can accurately define transcriptional start sites (green arrows) and transcription terminators (red ‘T’) across the genome.
Table 1. Overview of Phage Terminator Librarya.
| phage | ID | terminator sequence | genomic location TTS | free energy(kcal/mol) | stem-loop free energy (kcal/mol) | average ONT-cappable-seq TE (%) |
|---|---|---|---|---|---|---|
| LUZ7 | LUZ7 T1 | ugccaaguccuaacucucuauccauccucugccucacuccuucauuggaguggggc | 465(+) | –21.50 | – | 99 |
| uuaccuuuucaagcacacauaagaucaauagcacacaaagagca | ||||||
| LUZ7 | LUZ7 T7 | agagcgcgcgcucgaccuguaacaccaugcccaaccuccucuuggggguugggcgu | 4098(+) | –40.50 | –14.5 | 95 |
| guucuuugugcuauccgucaguccguaaggaguuccaacaugaa | ||||||
| LUZ7 | LUZ7 T25 | cacgagaucacugucacgcaaggucucauugaggucaggcgugauggugaucucgu | 21202(+) | –51.40 | – | 38 |
| acaugaauguggcggccugugggcugccugcgacuuugcacgag | ||||||
| LUZ7 | LUZ7 T27 | ucguucaugaggaaucccgcaugacgcauaccuggguaaugggccaagguccagua | 22088(+) | –28.60 | – | 70 |
| cccccacauuucaagcaggugaccauggaagaguacgaccucuu | ||||||
| LUZ7 | LUZ7 T31 | agagauuccaaaucccugacuccaucuaggaaaccagaaccccucuucggaggggu | 26996(+) | –32.10 | –11.8 | 91 |
| uuuuuauugucugagguuccaccaugagcaagucauacacaguu | ||||||
| LUZ7 | LUZ7 T32 | uccgugggguccgcuccuuccccauagguuucauccaaccucaacccccuacaggc | 27816(−) | –39.20 | –14.4 | 94 |
| augucgcuaccuguaggggguuuuuuuauaccuggucgaaggga | ||||||
| LUZ7 | LUZ7 T47 | gucuccuagccucugaguucacccacccuuucuagauagccagcuuuaugcuggcu | 42911(+) | –38.50 | –11.7 | 58 |
| uucuuaugucuaugucucguagacauaagggagugcucuggccc | ||||||
| LUZ7 | LUZ7 T50 | uucugggagauccuggcaaacgcauucaaaaagaaaggggagcauuagcuccccuu | 43803(+) | –26.80 | –14.7 | 90 |
| cuucuuuacagguucaucaaaccuuucaccaucggcaguucacu | ||||||
| LUZ7 | LUZ7 T56 | caagaccguagcccgccuguaagguaggcuggaggggagggaaaccuccccuuuuu | 59943(−) | –34.70 | –15.6 | 65 |
| uaacuccgcccagaggaaucaaaccaugucucucgacaacgccc | ||||||
| LUZ7 | LUZ7 T61 | gacggggaggcugggcguaagggugaaggggaaccaggacgguuccccuuuuucuu | 65450(−) | –43.40 | –15.5 | 95 |
| uuaggcuuugccgguacuaccgaacccaccuucaccucgacugg | ||||||
| LUZ100 | LUZ100 T1 | ccguagguaacacaaacggcaaggauugccgguaauccccaaggaugguggaugcc | 732(+) | –41.00 | – | 94 |
| ucaaggcacauugacugggucauucccaggaucagugcgccuug | ||||||
| LUZ100 | LUZ100 T4 | gucuugcauccgacuauccacuggcacauugaauaacccggcaucguccggguuuu | 3985(+) | –20.20 | –8.5 | 61 |
| cuuuucccaucaacuauccacugucgcaaccuuugcagaccgcu | ||||||
| LUZ100 | LUZ100 T6 | ccaauuaccacacagguauggaagaccgcccccgguaugcccagcgcugccggggg | 8683(+) | –33.10 | –14.4 | 39 |
| uuucuguuucaagacccacgacaacugugaggagaaccccauga | ||||||
| LUZ100 | LUZ100 T14 | acgcucaaaucguggggccgcuaggcagcuugcuccgucugccgaaucacggagcu | 20828(+) | –30.50 | – | 53 |
| uucacuggcauguagcucagaugguagagcacccggcuguuaac | ||||||
| LUZ100 | LUZ100 T15 | ggugcagcuacggaaacugcacaagcccuagcacgucgggagacguuacugggcaa | 21029(+) | –31.30 | – | 74 |
| ccccuaccucgaacgacgagugccucaagucgcacuggcagcaa | ||||||
| LUZ100 | LUZ100 T16 | cuguugcguaagcaacuauccaccaucgcauaggagggccagaaauggcucuccua | 22194(+) | –32.20 | –9.5 | 82 |
| uuuuuuucuuauggggguuccaugauuucuccaaccacugagcu | ||||||
| LUZ100 | LUZ100 T19 | uucugggaucaguaacaaccauucguucaacgcacaggggcggccuucgggucgccc | 31652(+) | –31.60 | –17 | 38 |
| uuuuucuuugucuggaggaaaagcauggcucuugcacgaguca | ||||||
| LUZ100 | LUZ100 T20 | ggaugguuauguagaacauggaugugacugaggggagcaggaggcuccucucgguc | 36750(+) | –33.80 | – | 82 |
| ucuuugcaagaucgcugcaagaucgacgacguucaauuaccaca | ||||||
| LUZ100 | LUZ100 T22 | ucugcgaaucgaggccggugccgggggcugccggugcuggucggggggcuggcgac | 37192(+) | –41.60 | – | 86 |
| acaagccacgacacaagaugcccgccaagccgcgccguugcugg |
An overview of the selected transcription termination sites and their associated terminator sequences of phage LUZ7 and LUZ100 is presented. Intrinsic terminators predicted by ARNold are indicated in bold, and predicted hairpins are underlined. Free energies of the entire terminator region and the predicted stem loop structure are computed using RNAfold and ARNold, respectively.22,23 TTS, transcription termination site; TE, termination efficiency as calculated by the ONT-cappable-seq pipeline.18
A Library of Potent Phage Terminators for Gram-Negative Hosts
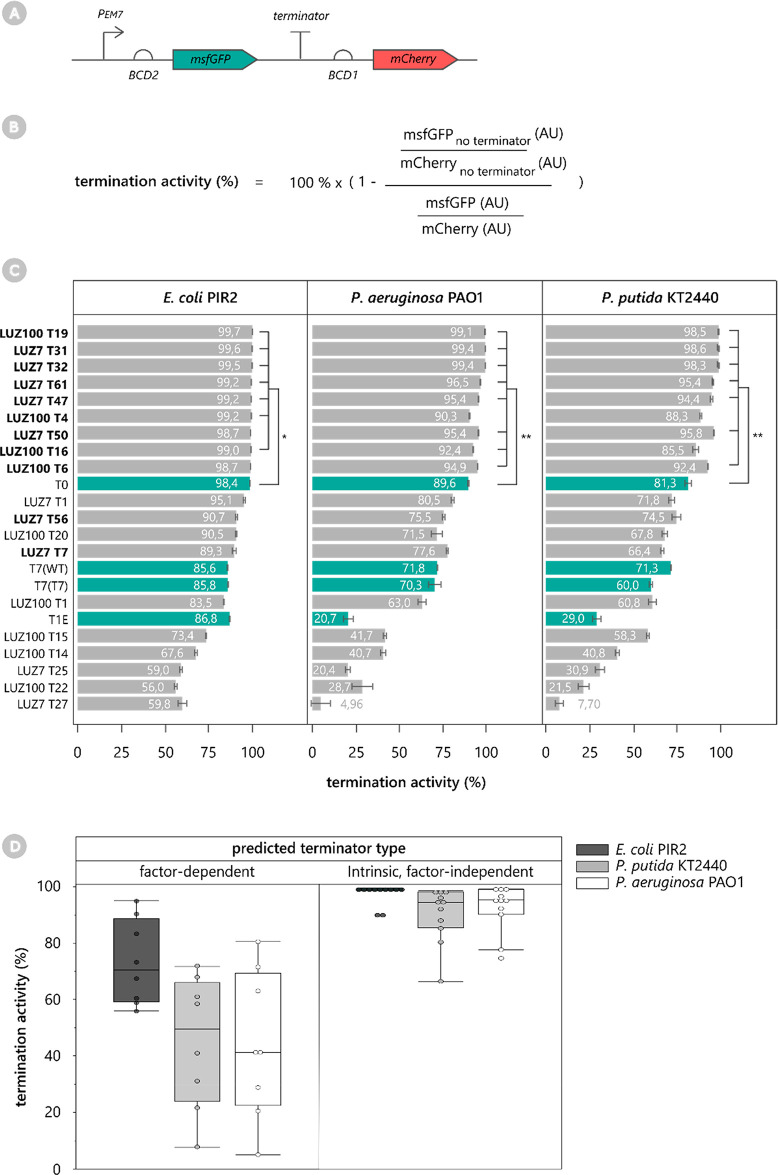
The termination activity of the predicted phage terminators was validated in vivo using a “terminator trap” design in three Gram-negative hosts: E. coli PIR2, Pseudomonas aeruginosa PAO1, and Pseudomonas putida KT2440.8,20,24 In addition to the universal bacterial model organism E. coli, the other hosts represent the native host of LUZ7 and an industrially relevant nonmodel organism, respectively. Within the terminator trap, the terminator is placed between two fluorescent reporters to quantitatively determine its termination activity (Figure 2A). More specifically, the terminator is located downstream of an msfgfp reporter gene, which is transcribed and translated by the strong constitutive promoter PEM7 and potent ribosomal binding site BCD2.25 Downstream of the terminator, a second ribosomal binding site BCD1 and mCherry reporter allow indirect quantification of read-through over the tested terminator. The termination activity (%) is then obtained by calculating the ratio of msfGFP and mCherry levels, normalizing for a control construct without terminator, and converting this value to % (Figure 2). The control construct (Pem7-BCD2 msfgfp-BCD1-mcherry) therefore represents the baseline msfGFP level and the maximal mCherry level when no terminator is inserted between msfgfp and BCD1. Apart from the phage terminators, four well-known and validated terminators were included in the assay as controls: (1) lambda terminator T0 (T0), a standard part in established SEVA vectors;26 (2) the wild-type T7 phage terminator (T7 (WT)), allowing strong transcriptional termination in pET vectors;27 (3) optimized T7 phage terminator variant 7 (T7 (T7));20 and (4) potent terminator T1 from the broad-host-range terminator library of environmental DNA samples (T1E).10
Figure 2.
Library of phage terminators for Gram-negative hosts. (A) The efficiency of ONT-cappable-seq-identified phage terminators was analyzed with a terminator trap. The terminator is flanked by an upstream msfGFP reporter (green) and downstream mCherry reporter (red). Quantitative measurements of mCherry levels correspond to the level of read-through from the terminator, while msfGFP levels indicate the influence of the terminator on mRNA stability and translational efficiency of the upstream transcript. (B) The strength of each phage terminator is expressed as termination activity (%), by calculating the ratio of msfGFP and mCherry fluorescence levels, normalizing for the control construct (no terminator), and converting this value to percentage. (C) Termination activity (%) of all tested terminators in E. coli PIR2, P. aeruginosa PAO1, and P. putida KT2440. Bars and error bars display the mean and standard error of four biological replicates, respectively. Bold labels indicate predicted intrinsic terminators, whereas reference terminators are indicated with green bars. (D) Predicted phage terminator type vs in vivo termination activity. Comparison of the in vivo termination activity (%) between the phage terminators that were predicted to be factor-dependent or intrinsic, factor-independent in different bacterial hosts. The termination activity of the predicted intrinsic terminators is significantly higher than that of the putative factor-dependent terminators in all three hosts (Wilcoxon test, p < 0.0001)
All tested Pseudomonas phage terminators show a broad range of termination activity in E. coli, P. aeruginosa, and P. putida, ranging from 4.96% to 99.8% (Figure 2C). Together, these terminators provide a valuable terminator library of short, 100 bp terminators with weak, medium, and very strong potency. Furthermore, the length of the predicted intrinsic terminators can easily be minimized to their predicted hairpin and U-tract. Interestingly, the predicted intrinsic phage terminators significantly outperform the nonintrinsic terminators in each bacterial species (p < 0.001) (Figure 2D). In addition, all phage terminators show a highly similar ranking in all three hosts, which could indicate a universal functioning in Gram-negative species.
When the phage terminators are compared to the reference terminators, several observations can be made. First, references T7 (WT), T7 (T7), and T1E only show medium to weak termination activity levels compared to the Pseudomonas phage terminators. Interestingly, for terminator T1E only limited read-through of mCherry is observed, but it also has a negative impact on the expression levels of the upstream msfGFP reporter, which considerably reduces its termination activity (Figures S1–S3). Second, reference terminator T0 is known as a strong transcriptional terminator but only ranks as the 10th best terminator out of the 25 tested terminators, despite its established and widespread application. The strongest nine phage terminators (LUZ100 T4, T6, T16, and T19 and LUZ7 T31, T32, T47, T50, and T61) significantly outperform reference terminators T7 (WT), T7 (T7), and T1E in all three hosts (P < 0.05), and most of them even show a significantly increased termination activity compared to the T0 reference terminator (Figure 2). Moreover, it is interesting to note that all nine terminators were predicted as intrinsic terminators with strong hairpin formation (Table 1). We find that, among these terminators, terminators with stronger hairpin structures show enhanced in vivo termination activity in Pseudomonas (r < −0.7, P < 0.05).
Next, we assessed whether the in vivo termination activity of the phage terminators is consistent with the termination efficiencies observed during infection, as captured by ONT-cappable-seq. A positive correlation could be established between the terminator activity of the terminators from phage LUZ7 in E. coli and P. aeruginosa and their average RNA-seq-based TE (Figure S4) (r > 0.6, P < 0.06). This suggests that ONT-cappable-seq data can help to discern terminators that may likely display strong in vivo activities, albeit to a limited extent. It should be noted that, in contrast to the temporal transcriptome data of LUZ7, the LUZ100 ONT-cappable-seq data were obtained after pooling RNA samples from multiple time points during infection.18,19 This pooled approach could potentially introduce biases in the observed transcriptional read-through across infection, which makes a quantitative assessment of the average TE more challenging, perhaps explaining the lack of correlation between the two parameters in LUZ100. In addition, altering the local sequence context of genetic elements is known to have a profound impact on their performance, especially elements that rely on RNA structures.28
LUZ7 T50 Displays Potent Bidirectional Termination
The potential of the top nine phage terminators is further investigated by analyzing their bidirectionality, a desired trait since few bidirectional terminators are currently available.14,29 To this end, all nine phage terminators are cloned in the inverse orientation in the terminator trap. The results show that most phage terminators and all reference terminators display a high level of monodirectionality and possess only weak to mediocre termination activity in the antisense direction (Figure 3A). In contrast to the termination activity of the original phage terminators, which behave largely consistent between hosts, the observed level of antisense termination activity appears more variable. However, one phage terminator, LUZ7 T50, performs exceptionally well in the inverse orientation in all three hosts. This terminator displays antisense termination activity levels of 99.4%, 97.6%, and 96.9% in E. coli, P. aeruginosa, and P. putida, respectively, and can be considered a strong bidirectional terminator.
Figure 3.
LUZ7 T50 is a strong, bidirectional terminator. (A) The termination activity of the top nine phage terminators and reference terminators is assessed with the terminator trap in E. coli PIR2, P. aeruginosa PAO1, and P. putida KT2440 in the sense (gray) and antisense (green) orientations. Bars and error bars display the mean and standard error of four biological replicates, respectively. (B) ONT-cappable-seq data track of the late infection stage transcriptome of LUZ7 showing bidirectional transcriptional termination activity by terminator LUZ7 T50, located between a convergent gene pair. The LUZ7 terminator T50 sequence contains an A-tract and U-tract on either side of the hairpin (green color), suggesting the intrinsic bidirectional nature of the phage terminator.
Notably, the ONT-cappable-seq transcriptional landscape can also help elucidate the bidirectional nature of terminators. Indeed, transcriptome data of LUZ7 already implied that LUZ7 T50 efficiently halts transcripts that originate from both strands (Figure 3B), consistent with the findings in this experiment. Moreover, in its native genomic context, the phage terminator resides between a pair of convergent genes, and the terminator hairpin is flanked by adenine-rich and uridine-rich stretches, displaying the characteristics of a canonical bidirectional intrinsic terminator. Interestingly, a recent study in E. coli showed that bidirectional transcription termination between convergent gene pairs can be driven by head-on RNAP collisions.30 Given that convergent gene pairs are also ubiquitous in the densely coded genomes of phages, the bidirectional termination activity at these sites in their native genomic context, including LUZ7 T50, T61, and T32, might be orchestrated by similar RNAP conflicts.
Potent Transcriptional Terminators Improve Expression Levels of the Gene of Interest
Transcriptional terminators prevent read-through of the RNA polymerase, but they also stabilize mRNA molecules and thus improve expression levels of the upstream coding sequence.4,5,31 This observation could also be made for the screened phage terminators in E. coli and P. putida, where the nine strongest terminators all show significantly higher msfGFP levels compared to a control construct without terminator (P < 0.05) (Figures S1–S3). Moreover, an inverse relationship between msfGFP and mCherry levels can be observed in E. coli, indicating that stronger terminators with reduced mCherry levels generate higher msfGFP levels (r > 0.3, P < 0.01) (Figure 4), which contradicts the work by He et al.5 By contrast, a positive relationship was observed for P. aeruginosa and P. putida (Figure S5). Due to the design of our terminator trap and the calculation of termination activity, these results reflect the combined effect of transcriptional termination by the terminator and its mRNA protection capacity. A possible strategy to decouple these two properties is the use of RNase sites in the terminator trap as shown by Cambray et al.28 and He et al.5 However, the study of RNase sites in Pseudomonas species is limited compared to E. coli and could potentially prevent the correct comparison of terminator strength in these hosts.32
Figure 4.
Terminators with less mCherry read-through show increased msfGFP expression levels. (A) The msfGFP and mCherry expression levels of all phage terminators and reference terminators were assessed with the terminator trap in E. coli PIR2. Bars and error bars display the mean and standard error of four biological replicates, respectively. (B) A significant negative correlation between msfGFP expression levels and mCherry expression levels is observed for the tested terminators in E. coli PIR2. Dots represent the mean normalized fluorescence levels of four biological replicates for each terminator.
Conclusions
ONT-cappable-seq provides a comprehensive approach to identify transcriptional terminators in phage genomes. In this work, we validated 19 phage terminators, identified by ONT-cappable-seq, in an in vivo terminator trap in three Gram-negative hosts. Eleven of these showed an intrinsic hairpin structure, while all intrinsic terminators performed equally well or better than the included reference terminators. Despite the strong capability of ONT-cappable-seq to identify terminators, there is no strong overall correlation between the terminator efficiency as calculated from the transcriptional landscape and the in vivo validation experiment. This discrepancy could be attributed to the different natures of the two experiments, context effects of the surrounding sequences of the terminators, or the interference of unknown phage factors that could play a role in transcriptional termination.28 Furthermore, it is important to note that a standard terminator length of 100 bp was used for the in vivo validation assay, instead of minimizing the terminator sequences to their predicted hairpin and U-trail. In a very compact genome like those of phages, the potential consequence hereof is that we might unintentionally have included other regulatory sequences within this 100 bp that affect the results. Nevertheless, ONT-cappable-seq is proven to be a valuable approach to efficiently mine genomes for functional transcriptional terminators and can, under some experimental designs, give an estimate on the strength of identified terminators in vivo.
Eight potent monodirectional terminators and one strong bidirectional terminator were identified in this work, significantly outperforming current established references terminators in nonmodel hosts. Moreover, these phage terminators do not share conserved sequences and are short in length (100 bp) with the possibility of further minimization. As such, they form a valuable addition to the genetic part toolbox of the popular SynBio host P. putida and important human pathogen P. aeruginosa, and it is likely that they will show similar performance in related Gram-negative species, as they also perform consistently in the more distantly related E. coli. In view of these findings, we argue that phage genomes harbor many more unidentified transcriptional terminators, which are tailored to their host and could rapidly expand the genetic parts toolboxes of numerous nonmodel hosts.
Methods
Bacterial Strains
For this work, E. coli PIR2 (Invitrogen, catalog # C111110), P. putida KT244033 and P. aeruginosa PAO134 were used. For transformation purposes, cells were cultured in standard LB medium with the appropriate antibiotics (as mentioned in the following sections) and incubated at 37 °C (E. coli and P. aeruginosa) or 30 °C (P. putida).
Identification of Phage Terminators Using ONT-cappable-seq
The transcriptional landscapes of phage LUZ7 and phage LUZ100 were previously identified using ONT-cappable-seq.18,19 In short, RNA was extracted at different time points during phage infection and subjected to library preparation. For LUZ7, the different time points were sequenced individually to obtain a temporal transcriptional overview. By contrast, LUZ100 RNA samples were pooled together prior to library preparation to improve cost efficiency and obtain a global transcriptional view. After sequencing, data analysis and transcript boundary detection was performed according to the workflow described by Putzeys et al. (https://github.com/LoGT-KULeuven/ONT-cappable-seq). In this way, phage transcription termination sites (TTSs) and their associated terminator sequences were identified by marking genomic positions with a local accumulation of 3′ read ends. Only the positions with an average read coverage reduction, indicated as termination efficiency (TE), of at least 20% across the candidate TTSs were annotated as terminators. In this work, previously published ONT-cappable-seq data of LUZ7 (GEO accession number GSE196845)18 and LUZ100 (GEO accession number GSE211961) (Table S1) were used to select a subset of 19 terminators that display a wide variety of TEs (Table 1). From this set, 11 phage terminators exhibited features of canonical, intrinsic terminators, as predicted by ARNold.23 The eight remaining terminator sequences were considered to be nonintrinsic terminators. Next, the phage terminator sequences were delineated by extracting the −60 to +40 region surrounding their annotated TTSs, as listed in Table 1.
Terminator Trap Design and Vector Construction
To analyze all phage terminators in vivo, fragments were cloned in the terminator trap design of Lammens et al.24 using the SEVAtile shuffling method. First, all genetic parts (PEM7, BCD2, msfgfp, BCD1, and mcherry (Table S3)) and terminators were amplified by tail-PCR (Table S2), after which they were assembled in the pBG vector backbone using a single-pot restriction-ligation protocol with BsaI and T4 DNA ligase (Thermo Fisher Scientific, USA). All assembled constructs were introduced in E. coli PIR2 with heat-shock transformation and selective plating on LBKan50.35 Several transformants of each construct were analyzed with colony PCR and confirmed by Sanger sequencing using vector primers insertDes2_F and SEVA_PS2. Next, correctly assembled vectors were purified and integrated in the Tn7 site of P. aeruginosa PAO1 and P. putida KT2440 using coelectroporation with pTNS2 and selective plating on LBGm30 or LBGm10, respectively, as described elsewhere.36,37
Fluorimetric Assay
To determine the termination activity of each phage terminator in vivo, fluorimetric assays were performed based on msfGFP and mCherry fluorescence intensity. First, overnight cultures of four biological replicates of each sample were prepared in M9 minimal medium containing 1× M9 salts (BD Biosciences, Belgium), 0.2% citrate (Sigma-Aldrich), 2 mM MgSO4 (Sigma-Aldrich, Belgium), 0.1 mM CaCl2 (Sigma-Aldrich), 0.5% casein amino acids (LabM, Neogen Company), and the appropriate antibiotic. The following day, all cultures were diluted 20-fold in fresh medium in a 96-well COSTAR plate (Corning, USA) and incubated at the appropriate temperature for 2–3 h. Next, end point measurements of cell growth and fluorescence intensity were performed in the CLARIOstar Plus microplate reader (BMG Labtech, Germany). Cell growth was determined at OD600, msfGFP fluorescence intensity at 485 nm (ex)/528 nm (em), and mCherry fluorescence intensity at 570 nm (ex)/620 nm (em). All fluorescence measurements were normalized for OD600, corrected for background fluorescence levels of the corresponding wild-type strains, and converted to nM equivalents of 5(6)-carboxyfluorescein (Sigma-Aldrich) as described by Lammens et al.24 The data were analyzed and visualized using JMP 15 Pro (JMP, version 15) (SAS Institute Inc., Cary, NC).
Acknowledgments
This project received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 Research and Innovation Programme (Grant Agreement 819800) and from the Fonds voor Wetenschappelijk Onderzoek Vlaanderen (FWO) as part of the CELL-PHACTORY Project (Grant G096519N). M.B. was funded by a grant from the Special Research Fund (iBOF/21/092).
Glossary
Abbreviations
- TSS
transcription start site
- TTS
transcription termination site
- TE
termination efficiency
- AU
absolute units
- ONT
Oxford Nanopore Technologies
- 5(6)-FAM
5(6)-carboxyfluorescein
- BCD
bicistronic design
- WT
wild-type
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acssynbio.3c00101.
Supplementary figures showing msfGFP expression levels, mCherry expression levels, and termination activity of all tested terminators in E. coli, P. putida, and P. aeruginosa, the correlation between msfGFP and mCherry levels, and the correlation with the original ONT-cappable-seq data; supplementary tables listing the specifications of the used ONT-cappable-seq data set, the primers used in this work, and the DNA sequences of all genetic parts used in this work (PDF)
Author Contributions
† E.-M.L. and L.P. are joint first authors. The work was conceptualized by E.-M.L., L.P., M.B., and R.L. All experimental work was performed by E.-M.L. and L.P.. The manuscript was written by E.-M.L. and L.P. with revisions of M.B. and R.L.
The authors declare no competing financial interest.
Supplementary Material
References
- Nielsen A. A. K.; Segall-Shapiro T. H.; Voigt C. A. Advances in Genetic Circuit Design: Novel Biochemistries, Deep Part Mining, and Precision Gene Expression. Curr. Opin. Chem. Biol. 2013, 17 (6), 878–892. 10.1016/j.cbpa.2013.10.003. [DOI] [PubMed] [Google Scholar]
- de Felippes F.; McHale M.; Doran R. L.; Roden S.; Eamens A. L.; Finnegan E. J.; Waterhouse P. M. The Key Role of Terminators on the Expression and Post-Transcriptional Gene Silencing of Transgenes. Plant J. 2020, 104 (1), 96–112. 10.1111/tpj.14907. [DOI] [PubMed] [Google Scholar]
- Kelly C. L.; Taylor G. M.; Šatkutė A.; Dekker L.; Heap J. T. Transcriptional Terminators Allow Leak-Free Chromosomal Integration of Genetic Constructs in Cyanobacteria. Microorganisms 2019, 7 (8), 263. 10.3390/microorganisms7080263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanishi M.; Ito Y.; Kintaka R.; Imamura C.; Katahira S.; Ikeuchi A.; Moriya H.; Matsuyama T. A Genome-Wide Activity Assessment of Terminator Regions in Saccharomyces cerevisiae Provides a “Terminatome” Toolbox. ACS Synth. Biol. 2013, 2 (6), 337–347. 10.1021/sb300116y. [DOI] [PubMed] [Google Scholar]
- He Z.; Duan Y.; Zhai W.; Zhang X.; Shi J.; Zhang X.; Xu Z. Evaluating Terminator Strength Based on Differentiating Effects on Transcription and Translation. ChemBioChem 2020, 21 (14), 2067–2072. 10.1002/cbic.202000068. [DOI] [PubMed] [Google Scholar]
- Lammens E.-M.; Nikel P. I.; Lavigne R. Exploring the Synthetic Biology Potential of Bacteriophages for Engineering Non-Model Bacteria. Nat. Commun. 2020, 11 (1), 5294. 10.1038/s41467-020-19124-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley L. A.; Guss A. M. Approaches to Genetic Tool Development for Rapid Domestication of Non-Model Microorganisms. Biotechnol. Biofuels 2021, 14 (1), 1–17. 10.1186/s13068-020-01872-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson A. J.; Wieden H.-J. Rapid Generation of Sequence-Diverse Terminator Libraries and Their Parameterization Using Quantitative Term-Seq. Synth. Biol. 2019, 4 (1), 1–9. 10.1093/synbio/ysz026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viana de Siqueira G. M.; Guazzaroni M.-E. A Termination-Aware Vector Design Improves Heterologous Gene Expression in Pseudomonas putida. bioRxiv 2022, 10.1101/2022.11.22.517491. [DOI] [Google Scholar]
- Amarelle V.; Sanches-Medeiros A.; Silva-Rocha R.; Guazzaroni M. E. Expanding the Toolbox of Broad Host-Range Transcriptional Terminators for Proteobacteria through Metagenomics. ACS Synth. Biol. 2019, 8 (4), 647–654. 10.1021/acssynbio.8b00507. [DOI] [PubMed] [Google Scholar]
- Feike D.; Korolev A. V.; Soumpourou E.; Murakami E.; Reid D.; Breakspear A.; Rogers C.; Radutoiu S.; Stougaard J.; Harwood W. A.; Oldroyd G. E. D.; Miller J. B. Characterizing Standard Genetic Parts and Establishing Common Principles for Engineering Legume and Cereal Roots. Plant Biotechnol. J. 2019, 17 (12), 2234–2245. 10.1111/pbi.13135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian C.; Zhang Y.; Li J.; Wang Y. Benchmarking Intrinsic Promoters and Terminators for Plant Synthetic Biology Research. BioDesign Res. 2022, 2022, 9834989. 10.34133/2022/9834989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen A. A. K.; Der B. S.; Shin J.; Vaidyanathan P.; Paralanov V.; Strychalski E. A.; Ross D.; Densmore D.; Voigt C. A. Genetic Circuit Design Automation. Science 2016, 352 (6281), aac7341. 10.1126/science.aac7341. [DOI] [PubMed] [Google Scholar]
- Ju X.; Li D.; Liu S. Full-Length RNA Profiling Reveals Pervasive Bidirectional Transcription Terminators in Bacteria. Nat. Microbiol. 2019, 4 (11), 1907–1918. 10.1038/s41564-019-0500-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dar D.; Shamir M.; Mellin J. R.; Koutero M.; Stern-Ginossar N.; Cossart P.; Sorek R. Term-Seq Reveals Abundant Ribo-Regulation of Antibiotics Resistance in Bacteria. Science 2016, 352 (6282), aad9822. 10.1126/science.aad9822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan B.; Boitano M.; Clark T. A.; Ettwiller L. SMRT-Cappable-Seq Reveals Complex Operon Variants in Bacteria. Nat. Commun. 2018, 9 (1), 3676. 10.1038/s41467-018-05997-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grünberger F.; Ferreira-Cerca S.; Grohmann D. Nanopore Sequencing of RNA and CDNA Molecules in Escherichia coli. RNA 2022, 28 (3), 400–417. 10.1261/rna.078937.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putzeys L.; Boon M.; Lammens E.-M.; Kuznedelov K.; Severinov K.; Lavigne R. Development of ONT-Cappable-Seq to Unravel the Transcriptional Landscape of Pseudomonas Phages. Comput. Struct. Biotechnol. J. 2022, 20, 2624–2638. 10.1016/j.csbj.2022.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putzeys L.; Poppeliers J.; Boon M.; Lood C.; Vallino M.; Lavigne R. Transcriptomics-Driven Characterization of LUZ100, a T7-like Pseudomonas Phage with Temperate Features. mSystems 2023, e0118922. 10.1128/msystems.01189-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temme K.; Hill R.; Segall-Shapiro T. H.; Moser F.; Voigt C. A. Modular Control of Multiple Pathways Using Engineered Orthogonal T7 Polymerases. Nucleic Acids Res. 2012, 40 (17), 8773–8781. 10.1093/nar/gks597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts J. W. Mechanisms of Bacterial Transcription Termination. J. Mol. Biol. 2019, 431 (20), 4030–4039. 10.1016/j.jmb.2019.04.003. [DOI] [PubMed] [Google Scholar]
- Lorenz R.; Bernhart S. H.; Höner zu Siederdissen C.; Tafer H.; Flamm C.; Stadler P. F.; Hofacker I. L. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011, 6 (1), 26. 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naville M.; Ghuillot-Gaudeffroy A.; Marchais A.; Gautheret D. ARNold: A Web Tool for the Prediction of Rho-Independent Transcription Terminators. RNA Biol. 2011, 8 (1), 11–13. 10.4161/rna.8.1.13346. [DOI] [PubMed] [Google Scholar]
- Lammens E.-M.; Boon M.; Grimon D.; Briers Y.; Lavigne R. SEVAtile: A Standardised DNA Assembly Method Optimised for Pseudomonas. Microb. Biotechnol. 2022, 15, 370. 10.1111/1751-7915.13922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutalik V. K.; Guimaraes J. C.; Cambray G.; Lam C.; Christoffersen M. J.; Mai Q. A.; Tran A. B.; Paull M.; Keasling J. D.; Arkin A. P.; Endy D. Precise and Reliable Gene Expression via Standard Transcription and Translation Initiation Elements. Nat. Methods 2013, 10 (4), 354–360. 10.1038/nmeth.2404. [DOI] [PubMed] [Google Scholar]
- Silva-Rocha R.; Martínez-García E.; Calles B.; Chavarría M.; Arce-Rodríguez A.; De Las Heras A.; Páez-Espino A. D.; Durante-Rodríguez G.; Kim J.; Nikel P. I.; Platero R.; De Lorenzo V. The Standard European Vector Architecture (SEVA): A Coherent Platform for the Analysis and Deployment of Complex Prokaryotic Phenotypes. Nucleic Acids Res. 2013, 41 (D1), D666–D675. 10.1093/nar/gks1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mairhofer J.; Wittwer A.; Cserjan-Puschmann M.; Striedner G. Preventing T7 RNA Polymerase Read-through Transcription—A Synthetic Termination Signal Capable of Improving Bioprocess Stability. ACS Synth. Biol. 2015, 4 (3), 265–273. 10.1021/sb5000115. [DOI] [PubMed] [Google Scholar]
- Cambray G.; Guimaraes J. C.; Mutalik V. K.; Lam C.; Mai Q. A.; Thimmaiah T.; Carothers J. M.; Arkin A. P.; Endy D. Measurement and Modeling of Intrinsic Transcription Terminators. Nucleic Acids Res. 2013, 41 (9), 5139–5148. 10.1093/nar/gkt163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. J.; Liu P.; Nielsen A. A. K.; Brophy J. A. N.; Clancy K.; Peterson T.; Voigt C. A. Characterization of 582 Natural and Synthetic Terminators and Quantification of Their Design Constraints. Nat. Methods 2013, 10 (7), 659–664. 10.1038/nmeth.2515. [DOI] [PubMed] [Google Scholar]
- Wang L.; Watters J. W.; Ju X.; Lu G.; Liu S. Head-on and Co-Directional RNA Polymerase Collisions Orchestrate Bidirectional Transcription Termination. Mol. Cell 2023, 83 (7), 1153–1164. 10.1016/j.molcel.2023.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe D.; Kim K.; Kang M.; Lee S.-G.; Cho S.; Palsson B.; Cho B.-K. Tunable 3′-UTR Valves for Metabolic Flux Control in Escherichia coli. Nucleic Acids Res. 2022, 50 (7), 4171–4186. 10.1093/nar/gkac206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apura P.; Gonçalves L. G.; Viegas S. C.; Arraiano C. M. The World of Ribonucleases from Pseudomonads: A Short Trip through the Main Features and Singularities. Microb. Biotechnol. 2021, 14 (6), 2316–2333. 10.1111/1751-7915.13890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagdasarian M.; Lurz R.; Rückert B.; Franklin F. C. H.; Bagdasarian M. M.; Frey J.; Timmis K. N. Specific-Purpose Plasmid Cloning Vectors II. Broad Host Range, High Copy Number, RSF 1010-Derived Vectors, and a Host-Vector System for Gene Cloning in Pseudomonas. Gene 1981, 16 (1–3), 237–247. 10.1016/0378-1119(81)90080-9. [DOI] [PubMed] [Google Scholar]
- Stover C. K.; Pham X. Q.; Erwin A. L.; Mizoguchi S. D.; Warrener P.; Hickey M. J.; Brinkman F. S. L.; Hufnagle W. O.; Kowalik D. J.; Lagrou M.; Garber R. L.; Goltry L.; Tolentino E.; Westbrock-Wadman S.; Yuan Y.; Brody L. L.; Coulter S. N.; Folger K. R.; Kas A.; Larbig K.; Lim R.; Smith K.; Spencer D.; Wong G. K. S.; Wu Z.; Paulsen I. T.; Reizer J.; Saier M. H.; Hancock R. E. W.; Lory S.; Olson M. V. Complete Genome Sequence of Pseudomonas aeruginosa PAO1, an Opportunistic Pathogen. Nature 2000, 406 (6799), 959–964. 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on Transformation of Escherichia coli with Plasmids. J. Mol. Biol. 1983, 166 (4), 557–580. 10.1016/S0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Choi K. H.; Kumar A.; Schweizer H. P. A 10-min Method for Preparation of Highly Electrocompetent Pseudomonas aeruginosa Cells: Application for DNA Fragment Transfer between Chromosomes and Plasmid Transformation. J. Microbiol. Methods 2006, 64 (3), 391–397. 10.1016/j.mimet.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Choi K. H.; Gaynor J. B.; White K. G.; Lopez C.; Bosio C. M.; Karkhoff-Schweizer R. A. R.; Schweizer H. P. A Tn7-Based Broad-Range Bacterial Cloning and Expression System. Nat. Methods 2005, 2 (6), 443–448. 10.1038/nmeth765. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.