Abstract
To date there is no clinically approved adjuvant to drive a protective T-helper cell 17 (Th17) immune response against Mycobacterium tuberculosis (Mtb). Trehalose Dimycolate (TDM) is a glycolipid molecule found in the cell wall of Mtb and similar species. Our team has discovered novel synthetic TDM derivatives that target Mincle receptors and when presented on the surface of amine functionalized silica nanoparticles (A-SNPs) adopt the requisite supramolecular structure for Mincle receptor agonism. Here we describe the preparation and characterization methods for these critical silica nanoparticles (SNPs) co-loaded with Mincle agonists (MAs) and a model antigen. In this work, A-SNPs with a particle diameter of 246 ± 11 nm were prepared and examined for co-adsorption of two synthetic MAs along with ovalbumin (OVA). Due to the insolubility of the studied MAs in aqueous environment, aggregation of the MAs made separation of the adjuvant-loaded SNPs from the free-form MAs via centrifugation very challenging. To facilitate separation, we synthesized modified SNPs with comparable amine surface functionalization to the original A-SNPs, but with a superparamagnetic iron oxide core (M-A-SNPs), to allow for magnetic separation. We also substituted Alexa Fluor 488-labeled ovalbumin (AF 488 OVA) for the un-tagged OVA to improve the sensitivity of our quantitation method. A RP-HPLC method was developed to simultaneously determine the amount of adsorption of both the Mincle adjuvant and the model antigen to the A-SNPs. AF488 OVA demonstrated higher than 90% adsorption, with or without the co-adsorption of MAs. Likewise, MAs exhibited higher than 80% adsorption in the presence or absence of antigen. The developed formulations were tested in vitro using murine RAW cells and human peripheral blood mononuclear cells, exhibiting good cytokine induction in both cell lines. Results from these studies indicate that A-SNPs could be used as a customizable presentation platform to co-deliver antigens along with different MAs of varying structural features and biophysical properties.
Keywords: Silica nanoparticles, Mincle, adjuvant, Antigen, Co-adsorption, Magnetic separation, Co-delivery
Graphical abstract
1. Introduction
Pattern recognition receptor (PRR) binding and activation represent a key first step implicated in the protection against pathogens that cause a significant disease burden including Mycobacterium tuberculosis (Mtb), Staphylococcus aureus, and others [1]. C-type lectin receptors (CLRs) are important PRRs involved in pathogen detection and innate immune activation leading to induction of adaptive immunity [2]. Therefore, targeted stimulation of CLRs holds promise for driving a superior protective immune response against these diseases [3, 4]. It is hypothesized that protection against major intracellular pathogens, such as Mtb, requires the induction of potent antigen-specific CD4+ T cell responses [5, 6]. Ongoing research efforts seek to co-deliver antigens and adjuvants with various delivery systems to enhance both cell-mediated and humoral immunity. After several decades typified by the use of aluminum salts as the main adjuvant approved for boosting responses to the adsorbed antigen, there is now a growing list of adjuvants and multicomponent nanoparticle-based vaccine formulations emerging which represent promising approaches for improving next-generation vaccines [7].
Poor uptake of soluble protein antigens reduces vaccine immunogenicity and efficacy. As a result, nanoparticle delivery platforms have emerged as the leading candidates for improving both efficacy and safety profiles of subunit vaccines [8, 9]. It is important for the safety of a vaccine that the adjuvant(s) used do not trigger adverse side effects or systemic reactogenicity. One strategy for preventing this is to physically link or co-deliver the adjuvant and antigen. This can be accomplished by directly linking or confining both components via association with a particle carrier. This holds the dual benefit of limiting the introduction of soluble antigens and adjuvants and enabling co-delivery of the vaccine components to the same cell to produce a more robust and specific immune response [7, 10].
Previous studies have revealed the criticality of inflammatory cytokines and co-stimulatory molecules like CD40 and CD80/86 help bridge innate and adaptive immunity resulting in higher numbers of antigen-specific CD4+ and CD8+ T cells [11]. A major challenge underlying the use of nanomaterials in vaccines lies in identifying particle designs that can efficiently target and activate antigen-presenting cells (APCs), especially dendritic cells (DCs) [9]. The physical linkage between adjuvants and antigen was postulated to reflect the need for both components to target the same APCs, thus enhancing antigen uptake and presentation [7]. This is particularly important when targeting Mtb, which requires the induction of potent antigen specific CD4+ T cell responses [12]. Targeting of resident lymph node (LN) DCs by free antigen prior to their activation by CLR adjuvant molecules should be avoided to elicit potent Th1/Th17 anti-mycobacterial responses. Early DC uptake of soluble antigen can result in nonactivated antigen-pulsed DCs which do not effectively support the survival and differentiation of CD4 Th1/Th17 effector memory cells [7, 13].
SNPs can be engineered to adopt a wide array of shapes, sizes, and customized surface functionalities with variable porosity and payload capacity, which makes them effective carriers of both antigens and adjuvants. Recent studies have demonstrated that antigen and/or adjuvant-loaded SNPs can drive both humoral and cellular immune responses while maintaining good biocompatibility and a low cost of production [9, 14–18]. Antigen loaded SNPs accumulate in APCs in the draining LN after injection leading to dramatically enhanced induction of antigen-specific B and T cell responses as compared to soluble vaccine formulations [9, 19]. For example, cationic SNPs were able to efficiently co-load and present multiple copies of a negatively charged oligonucleotide adjuvant and Ovalbumin (OVA) antigen through electrostatic interactions. That strategy enhanced immunogenic responses [20]. The FDA has classified silica as “Generally Recognized as Safe” and silica has been used as a food additive and in cosmetics. Amorphous silica particles are degraded into water-soluble orthosilicic acid (Si(OH)4) that can be excreted in the urine. According to several reports, SNPs at concentrations less than or equal to 20 mg/ml exhibit no cytotoxicity or inflammatory responses [21]. Despite prior reports showing favorable properties, the co-delivery of Mincle ligands and antigens on SNPs has not been described previously.
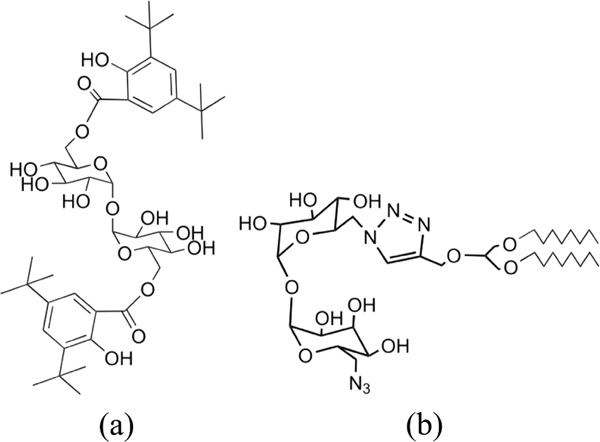
Trehalose dimycolate (TDM) is a glycolipid molecule found in the cell wall of Mycobacterium tuberculosis (Mtb) and similar species [22]. We recently reported that a lipidated trehalose scaffold could potentially be used to generate new libraries of potent TDM derivatives with improved physicochemical properties for application as vaccine adjuvants [3, 23]. In an extension of this work we have synthesized an asymmetric MA using a robust click chemistry-based process allowing for the synthesis and testing of an asymmetric lipidated triazolyl-functionalized trehalose derivative, UM1128 [24]. In this report, UM1128 was compared to UM1024, a potent diaryl trehalose derivative [23, 24], in terms of loading efficiency on A-SNPs and biological activity in vitro either in the presence or absence of a model antigen. The chemical structures of the studied MAs are presented in Figure 1. Furthermore, a custom-designed procedure to produce and characterize SNP-based vaccine delivery platforms is described. First, a mixture of A-SNPs and the amphiphilic MAs were deposited as a thin film in a round-bottomed flask by evaporation of the organic solute. Then, the MAs were adsorbed onto the surface of A-SNPs via bath sonication in aqueous media. This was followed by the presentation of a model antigen, OVA or Alexa Fluor 488-labeled OVA (AF488 OVA), to the A-SNPs to allow for adsorption through the association of the negatively charged antigen to the positively charged A-SNP. MAs were observed as insoluble aggregates in aqueous solutions that pellet with the A-SNPs during centrifugation making the separation of the A-SNPs-adsorbed MAs from the free-form MAs aggregates challenging. This separation issue was resolved by using superparamagnetic iron oxide nanoparticles with a silica shell (M-A-SNPs) of comparable surface functionalization, surface area, and zeta potential to the A-SNPs. The use of M-A-SNPs in place of A-SNPs allowed for separation of loaded particles from free-form MAs using a magnet and thus an accurate determination of adjuvant adsorption to A-SNPs in both the presence and absence of the model antigen. The presence of MAs interferes with commonly used protein quantitation techniques such as the bicinchoninic acid assay (BCA), so a different analytical technique was required to measure the adsorption of OVA to A-SNPs in this vaccine formulation. In addition to the preparation protocol, a RP-HPLC separation method was developed which enabled simultaneous quantitation of AF488 OVA and MAs co-adsorption to M-A-SNPs. The developed RP-HPLC method permitted separation and concurrent quantitation of both analytes from a mixed sample by using a combination of UV and fluorescence detection. Antigen demonstrated high adsorption, with no indication the presence of MAs interfered with its adsorption efficiency. MAs also exhibited high adsorption in the presence or absence of antigen.
Figure 1.
The structures of the MAs studied (a) UM1024 and (b) UM1128.
The developed formulations were tested in vitro in murine RAW cells and human peripheral blood mononuclear cells (hPBMCs) where they exhibited good cytokine induction. The tested formulations also showed a good ability to engage the Mincle receptor. The SNPs-based vaccine antigen and adjuvant co-delivery formulation described here can be easily customized to co-deliver antigens along with different MAs of varying structural features and biophysical properties with the potential to improve both humoral and cell-mediated immunity in an in vivo system, which is currently under investigation.
2. Materials and methods
2.1. Materials
All chemicals used were of Analytical Reagent grade and all solvents were of HPLC grade. Trifluoroacetic acid (TFA) and ammonium hydroxide were obtained from J.T. Baker, NJ, USA. Water for irrigation (WFI) was obtained from Baxter Healthcare Corp, IL, USA. Methanol, anhydrous ethanol, isopropyl alcohol, Triton X-100, and sodium hydroxide were obtained from Fisher Scientific, NJ, USA. Tetraethyl orthosilicate (TEOS), and (3-Aminopropyl)triethoxysilane (APTES) were obtained from Sigma-Aldrich, MO, USA. FeCl3 anhydrous (98%) was obtained from Alfa Aesar, MA, USA. FeCl2-4H2O and citric acid monohydrate (CA) were obtained from Acros Organics, USA. OVA and AF488 OVA were purchased from Invitrogen by ThermoFisher Scientific, OK, USA.
2.2. Synthesis of MAs
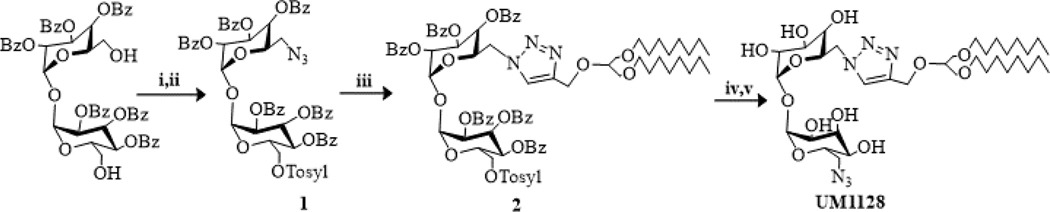
Reactions were monitored by TLC-analysis on Merck Silica gel 60 F254 plates and visualized by UV at 254 nm and dipping in vanillin (vanillin/water/ethanol/sulfuric acid, 0.2 g:5 mL:5 mL:1 mL) or phosphomolybdic acid in ethanol (PMA) and developed with heat. The two compounds were confirmed to be >95% pure by TLC, NMR, and high-performance liquid chromatography-mass spectrometry (HPLC-MS) analyses. 1H and 13C NMR spectra were recorded on a Bruker 500 MHz instrument and were referenced to TMS or a solvent peak. High-resolution HPLC-MS analysis was obtained on an Agilent 6520 Q-TOF mass spectrometer utilizing electrospray ionization source in positive or negative mode. Chromatography was performed on Grace or Biotage automated medium pressure chromatography instruments with preloaded Buchi silica gel cartridges. UM1024 was synthesized according to our previous report [23] and UM1128 was synthesized as described in Scheme 1 [24].
Scheme 1: Reagents and reaction conditions.
i) Tosyl-Cl, TEA, DCM ii) NaN3, DMF, 65 °C. iii) 1-(3-(octyloxy)-2-(prop-2-yn-1-xyloxyl)propoxy)octane), CuSO4, Na ascorbate, dioxane/water, 65 °C. iv) NaN3, DMF, 65 °C v) NaOMe, methanol.
Synthesis of 2-((6-(azidomethyl)-3,4,5-tris(benzoyloxy)tetrahydro-2H-pyran-2-yl)oxy)-6-((tosyloxy)methyl)tetrahydro-2H-pyran-3,4,5-triyl tribenzoate (1). 2,2′,3,3′,4,4′-Hexa-benzoyl-α,α-D-trehalose [25] (6 g, 6.2 mmol) was combined in dichloromethane (0.1M) with p-toluene sulfonyl chloride (3.3 g, 17 mmol) at ambient temperature and triethyl amine (3.3 g, 17 mmol) was added dropwise. Upon reaction completion the solution was poured into ice water (50 mL) and extracted with MTBE (2 × 75 mL). The combined organic extracts were concentrated and the resulting mixture was purified by chromatography on silica gel (linear gradient of 0% to 50% ethyl acetate/heptane) yielding 2,2′,3,3′,4,4′-Hexa-O-Benzoyl-6,6′-bis(p-toluensulfonyl)-α,α-D-trehalose (5.6 g, 4 mmol) as a white solid. This intermediate was combined in DMF (0.3 M) with NaN3 (0.404 g, 0.62 mmol) and heated to 65 oC and monitored by TLC (50% ethyl acetate/heptene). After 6 hours the bis-tosyl intermediate had been consumed and the reaction was cooled to room temperature, poured into ice water (50 mL) and extracted with MTBE (2 × 75 mL). The combined organic extracts were concentrated and the resulting mixture was purified by chromatography on silica gel (linear gradient of 0% to 50% ethyl acetate/heptane) to yield 1 as a white solid (1.575 g, 1.3 mmol, 32%) in addition to the diaza product as a solid (0.972 g, 0.95 mmol, 22%). 1H NMR (400 MHz, CDCl3) δ 8.09 (d, J = 7.9 Hz, 2H), 7.90 (d, J = 7.27 Hz, 2H), 7.83 (d, J = 7.93 Hz, 1H), 7.73 (d, J = 7.93 Hz, 1H), 7.64 (d, J = 7.93 Hz, 1H), 7.56 (t, J = 7.27 Hz, 1H), 7.50–7.27 (m, 7H), 7.22 (d, J = 7.27 Hz, 1H), 6.25–6.09 (m, 1H), 5.63–5.45 (m, 2H), 4.20–3.98 (m, 1H), 3.64–3.47 (m, 1H), 2.95–2.75 (m, 1H), 2.35 (s, 1H); 13C NMR (100MHz, CDCl3) δ 165.5, 165.1, 165.0, 164.9, 164.4, 144.9, 133.6, 133.5, 133.2, 132.2, 129.9, 129.8, 129.7, 129.5, 128.9, 128.7, 128.6, 128.5, 128.4, 128.3, 128.0, 97.7, 71.1, 71.0, 70.1, 70.0, 69.7, 69.0, 68.0, 67.7, 66.3, 49.9, 21.6. Synthesis of 2-((4-(((1,3-bis(octyloxy)propan-2-yl)oxy)methyl)-1H-1,2,3-triazol-1-yl)methyl)-6-((3,4,5-tris(benzoyloxy)-6-((tosyloxy)methyl)tetrahydro-2H-pyran-2-yl)oxy)tetrahydro-2H-pyran-3,4,5-triyl tribenzoate (2). Compound 1 (0.100 g, 0.090 mmol) was dissolved in Dioxane/water (5:1, 0.015M). 1-(3-(octyloxy)-2-(prop-2-yn-1-yloxy)propoxy)octane (0.030 g, 0.10 mmol), Cu(II)SO4 (0.003 g, 0.02 mmol), and sodium ascorbate (0.011 g, 0.05 mmol) were added and the reaction was heated to 50oC and monitored by TLC. Upon completion the reaction was cooled to room temperature, filtered and chromatographed on silica gel (linear gradient of 0% to 50% ethyl acetate/heptane) to yield 2 as a white solid in (0.075 g, 0.05 mmol, 58%). 1H NMR (400 MHz, CDCl3) δ 8.07 (t, J = 8.3 Hz, 4H), 7.86 (d, J = 6.73 Hz, 4H), 7.80 (d, J = 7.52 Hz, 2H), 7.66 (d, J = 7.52 Hz, 2H), 7.60 (d, J = 7.52 Hz, 2H), 7.57–7.24 (m, 18H), 7.18 (d, J = 8.31 Hz, 2H), 6.20–6.05 (m, 2H), 5.57 (d, J =3.60Hz, 1H), 5.49–5.41 (m, 2H), 5.32–5.24 (m,2H), 5.15 (t, J = 9.21 Hz, 1H), 4.82 (s, 2H), 4.33–4.26 (m, 1H), 4.15–4.03 (m, 2H), 4.01–3.93 (m,1H), 3.80–3.73 (m, 1H), 3.62–3.38 (m, 9H), 2.33 (s, 3H), 1.99–1.86 (m, 1H), 1.62–1.50 (m, 4H), 1.37–1.17 (m, 19H), .86 (t,J =6.54Hz,6H); 13C NMR (100MHz, CDCl3) δ 165.4, 165.0, 164.9, 164.6, 164.4, 146.1, 144.8, 71.6, 70.9, 70.8, 70.7, 70.6, 70.1, 70.0, 68.7, 68.1, 67.6, 66.1, 63.7, 49.2, 31.8, 29.6, 29.4, 29.2, 26.1, 22.6, 21.6, 14.1. Synthesis of 2-(azidomethyl)-6-((6-((4-(((1,3-bis(octyloxy)propan-2-yl)oxy)methyl)-1H-1,2,3-triazol-1-yl)methyl)-3,4,5-trihydroxytetrahydro-2H-pyran-2-yl)oxy)tetrahydro-2H-pyran-3,4,5-trio (UM1128). Intermediate 2 (0.075 g, 0.05 mmol) was dissolved in DMF (0.02M) followed by the addition of NaN3 (0.016 g, 0.25 mmol). The reaction was heated to 65oC for 15 hours, cooled to ambient temperature, poured into ice water (10 mL) and extracted with ethyl acetate (2× 20 mL). The combined organic extracted were concentrated and the residue was dissolved in methanol (5 mL). NaOMe (17uL, 0.30 mmol) was added and the reaction was stirred until complete by TLC (30% methanol in chloroform). The reaction was concentrated to 1/3 volume and the product isolated by chromatography on silica gel (linear gradient of 0% to 40% MeOH/DCM) to yield UM1128 (0.032 g, 0.04 mmol, 86%). 1H NMR (400 MHz, CDCl3) δ 7.78 (s, 1H), 5.07 (d, J = 5.31 Hz, 1H), 4.83–4.68 (m, 3H), 4.61–4.48 (m, 1H), 4.22 (t, J = 8.35 Hz, 1H), 4.02–3.69 (m, 11H), 3.58–3.27 (m, 11H), 3.11 (t, J = 9.87 Hz, 1H), 1.61–1.52 (m, 3H), 1.37–1.19 (m, 18H), .87 (t, J = 7.02 Hz, 6H); 13C NMR (100MHz, CDCl3) δ 144.8, 93.5, 86.7, 82.9, 79.1, 72.5, 72.4, 71.5, 71.3, 71.1, 70.8, 70.7, 70.2, 70.0, 63.1, 51.2, 41.8, 31.6, 29.2, 29.1, 29.0, 25.8, 22.4, 13.7.
2.3. Preparation of A-SNPs
A-SNPs were prepared according to a modified Stöber method [26, 27] by adding a premixed ethanol solution (51 mL) with ammonium hydroxide aqueous solution (30%) (6 mL) to a TEOS solution (2.6 ml) in 10 mL ethanol under stirring in a 100 mL round bottom flask. The mixture was stirred for 1 hour and further sonicated for 10 min. The flask was then charged with 65 μL TEOS and an equal volume of APTES. The reaction solutions were left to stir overnight at 120 rpm and room temperature. The obtained particles were isolated by centrifugation (3500 rcf, 30 min) and washed several times with ethanol and water for irrigation (WFI) to remove any unreacted APTES. After washing, particles were dried under reduced pressure at 60 °C for 8 hours using a vacuum oven.
2.4. Preparation of M-A-SNPs
M-A-SNPs were prepared in four steps; Step1: superparamagnetic iron oxide nanoparticle (SPION) seeds with diameters of about 8 nm were prepared according to the Massart’s co-precipitation method with some modifications [28]. A mixture of 5.19 g of anhydrous FeCl3 and 3.12 g of FeCl2.4H2O were dispersed in 25 mL WFI and sonicated for 1 hour. This mixture was added, portion wise, into 250 mL of a solution of 1.5 M NaOH and thermostatized at 50 °C in a double-neck 1000 mL round bottom flask. The mixture was left to react for 30 min under N2 atmosphere with mechanical stirring. The sudden change in the pH affects the iron ion solution leading to the formation of black iron oxide nanocrystals. These magnetic nanoparticles were separated by magnetic decantation and then washed with WFI several times. The final product was dried and re-dispersed in chloroform at a concentration of 5 mg/mL. Step 2: modifying the surface of the washed SPIONs from step 1 with citric acid to improve their dispersion in the reaction media used in the following steps according to the work of Yao et al [29]. 0.2 g SPIONs were diluted in 40 mL WFI containing 2 g citric acid monohydrate (CA). The CA modified SPIONs (CA-SPIONs) were obtained after ultra-sonication at room temperature for 3 h. Step 3: forming a silica shell around the CA-SPIONs [29]. A mixture containing 0.1 g CA-SPIONs, 10 mL WFI, 40 ml ethanol, and 1 mL of NH4OH was ultra-sonicated and mechanically stirred at room temperature for 30 min before adding another mixture of TEOS (2.5 mL) and ethanol (10 mL) dropwise into the above reaction system using a syringe pump at a rate of 0.1 mL/min. After reaction at room temperature for 6 hours, the magnetic SNPs (M-SNPs) were separated using a magnet and washed with ethanol and WFI several times. Step 4: Modifying the surface of M-SNPs with APTES. A solution of 53 mg M-SNPs and 6 mL WFI was added to a 25 mL round bottom flask and sonicated for 10 min. The reaction flask was then charged with 5.3 μL APTES. The reaction flask was sonicated and stirred for 3 hours at 60 °C. The M-A-SNPs were collected using a magnet and washed two times with WFI and 5 times with ethanol before drying under reduced pressure in a vacuum oven at 60 °C for 8 hours.
2.5. Characterization of the nanoparticles
2.5.1. Size characterization by transmission electron microscopy (TEM)
Electron microscopy was performed at the EMTrix core facility at the Division of Biological Sciences, University of Montana. The surface morphology and microstructures were analyzed using TEM (Hitachi H-7100 transmission electron microscope). For TEM analysis, 5 uL of 0.1 mg/mL nanoparticle solution in WFI was dropped onto a 400-mesh carbon-supported copper grid and dried. The grid was transferred in a TEM holder and inserted into the microscope. The microscope was operated at 75 kV. 5–10 images were acquired with a magnification of at least 15,000X. The average particle diameter and SD was taken for n=21 particles.
2.5.2. Zeta potential
To measure the zeta potential of nanoparticles, dynamic light scattering (DLS, Zetasizer, Malvern) was used with an He-Ne laser (633 nm) at 90 °C collecting optics at 25 °C. 0.1 mg/mL nanoparticle solutions were prepared in 10 mM NaCl (pH 5.5). 700 μL of each solution were transferred in a folded capillary cuvette and used for sample acquisition. Three measurements were recorded and the average was reported.
2.6. Loading MAs and antigen onto the surface of A-SNPs using a custom-designed film coating technique.
Stock solutions of A-SNPs (20 mg/mL) and the MAs, UM1024 and UM1128 (2 mM) were prepared in ethanol and THF:MeOH (9:1, v/v), respectively. Aliquots from the A-SNPs and Mincle ligand stock solutions were transferred quantitatively into 2 ml round-bottom glass vials and mixed for 10 seconds to prepare final concentrations of 20 mg/ml A-SNPs and 0.2 mM MAs, respectively. The thin film formation for these components was achieved by evaporating the organic solvent under reduced pressure using a rotary evaporator connected to a vacuum system at 50 °C. Following the addition of WFI to rehydrate the films, regular bath sonication was used for 60 min at 55–65 °C. An aliquot from the model antigen prepared at 0.2 mg/mL in WFI was then added to the suspended, adjuvant-coated A-SNPs followed by mixing using a rotator for 2 hours at room temperature to allow adsorption of the anionic antigen to the cationic particles, mainly via electrostatic interactions. The final concentration of the antigen in these formulations was 20 Mg/mL.
2.7. Determination of co-adsorption of MAs and antigen to M-A-SNPs
2.7.1. Samples preparation
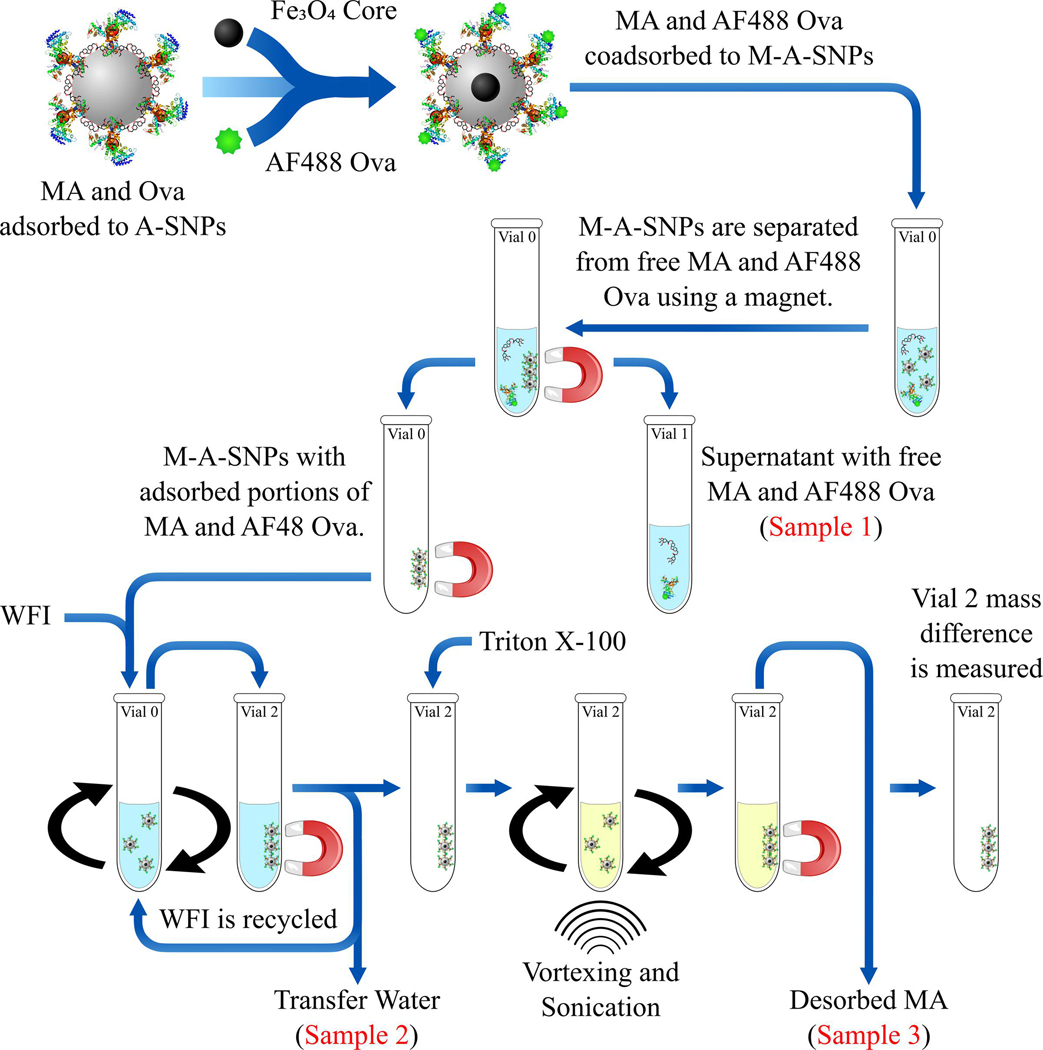
The MAs and AF488 OVA were co-adsorbed to M-A-SNPs following the same procedures described in section 2.6. 400 μL from each sample were transferred to a 2 mL glass vial after vortexing to suspend the M-A-SNPs. To separate the liquid in each sample from the magnetic particles, the vials were placed against a STEMCELL Technologies EasyEights EasySep magnetic separation tube rack for 1 minute. All of the liquid was removed and transferred to separate 2 mL glass vials (Sample 1). 50 μL from Sample 1 was diluted 4 times in 1% Triton X-100 and vortexed for 10 seconds before running the samples on RP-HPLC. An equal volume of WFI (400 μL) was added to the original vials containing only the M-A-SNPs (after removal of the supernatant), followed by resuspension of the particles by vortexing for 10 seconds. Immediately the homogenously suspended particles in WFI were transferred to pre-weighed glass vials. The liquid was separated from the beads using a magnet, transferred back to the original vial, vortexed to resuspend any SNPs remaining in the original vial, and subsequently transferred back to the pre-weighed glass vial along with the suspended SNPs. The previous two steps were repeated by recycling the same liquid until no more particles could be transferred. Then the liquid only was transferred to different glass vials after separating the beads using a magnet (Sample 2). 50 μL from Sample 2 were diluted 4x in 1% Triton X-100 and vortexed for 10 seconds before running the samples on RP-HPLC. 100 μL 1% Triton X-100 were added to the particles left after the previous step in the same vial to extract the MAs. This vial was briefly sonicated and then vortexed for 10 seconds. The liquid extract was separated from the particles using a magnet and transferred into a 2 ml HPLC vial. The extraction step was repeated three more times each with 100 μL 1% Triton X-100 and all the extracts were combined in a single vial and analyzed by HPLC undiluted (Sample 3). The vial left after the previous step containing the M-A-SNPs beads was dried off under reduced pressure for 2 hours, then weighed again to determine the mass of M-A-SNPs transferred.
2.7.2. Simultaneous quantitation of MAs and AF488 OVA using RP-HPLC
Quantitation was performed using a Waters Acquity Arc UHPLC chromatograph system (MA, USA) equipped with a quaternary solvent manager-R, FTN-R sample manager and injector valve with 20 μL loop, a 2475 FLR detector, and 2998 PDA detector. Empower 3 for LC systems [Rev.B.03.01-SR1 (317)] software was used for instrument control, data analysis and acquisition. Separation and quantitation were made on a Waters Symmetry C18 column, 100A, 5 μm, 4.6 mm x 250 mm. A mixture of sol-A: 0.1% TFA in water, sol-B: IPA, and sol-C: 0.1% TFA in methanol in gradient elution mode (0–1 min. sol-A: 50–50, sol-B:20–20; 1–5 min. sol-A: 50–0, sol-B: 20–90; 5–7 min. sol-A: 0–0, sol-B: 90–90; 7–8 min. sol-A: 0–50, sol-B: 90–20; 8–12 min. sol-A: 50–50, sol-B: 20–20) was used, (Figure S1). Flow rate was maintained at 0.6 mL min−1. All determinations were performed at ambient temperature. The system was equilibrated and saturated with the mobile phase for 30 min before the injection of the solutions. Quantification was achieved for the MAs with PDA detection at 216 or 254 nm and for the AF488 OVA with a fluorescence detector at 520 nm after excitation at 495 nm. 15 μL of the solutions were injected in duplicates. All samples were quantitated by peak area based on interpolation from a five-point dilution series in Triton X-100 of the corresponding standard.
2.8. Transgenic HEK cell SEAP Assays.
Human and mouse Mincle expressing HEK cells were obtained from Invivogen (San Deigo, CA). Cells were cultured according to the manufacturer’s instructions in DMEM with 10% FBS, 50 U/ml penicillin, 50 mg/ml streptomycin, 100 mg/ml Normocin, 2 mM L-glutamine, 30 μg/ml blasticidin, 1 ng/ml puromycin and 1x HEK-Blue™ CLR Selection. For assay, indicated formulations were serially diluted into a 96-well tissue culture plate. HEK cells were applied to the plates at a density of 3×105 cells/well and incubated for 18–24 h at 37°C. Cell supernatants were harvested and analyzed via the manufacturer’s instructions using Hek-BlueTM Detection. SEAP activity was assessed by reading the optical density (OD) at 620–655 nm with a microplate reader; data are expressed as the fold change in OD over vehicle treated cells.
2.9. Cell culture and cytokine analysis
Raw264.7 cells were obtained from ATCC (Manassas, VA). Cells were cultured in 50% DMEM with 10%FBS/50% AIMV media (Gibco). Cells were added to serially diluted compounds in AIMV media at 3×105 cells/well. Peripheral blood samples were collected from healthy adult donors. The samples were collected after approval by the University of Montana Institutional Review Board, and signed written informed consent was obtained from each donor. PBMCs were isolated from peripheral blood using Ficoll-Paque. Cells were added at 5×106 cells/well in RPMI with 5% human plasma to the serially diluted formulations. Supernatants were harvested from treated cells 18–24 h post-cell application. Supernatants were analyzed using either a DuoSet ELISA (R&D Systems, Minneapolis, MN) for human IL6 or a Luminex multiplex panel for analytes TNFα, IL-1β, IL-6, IFNγ, IL12p70 and IL-23 (R&D Systems) per the manufacturer’s instructions. Multiplex analysis was performed using a Luminex 200 instrument (Luminex Corporation). ELISAs were read on a SpectraMax® M5 Multi-Mode Microplate Reader at 450 nm and raw OD plotted directly.
3. Results and discussion
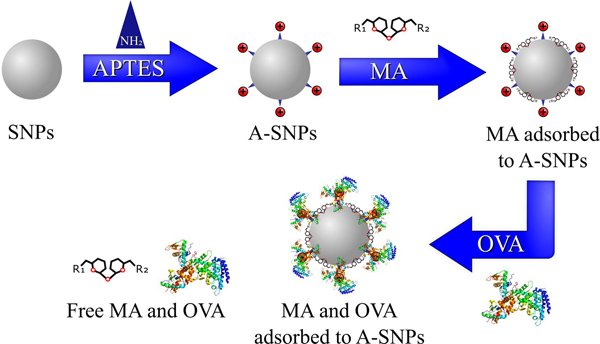
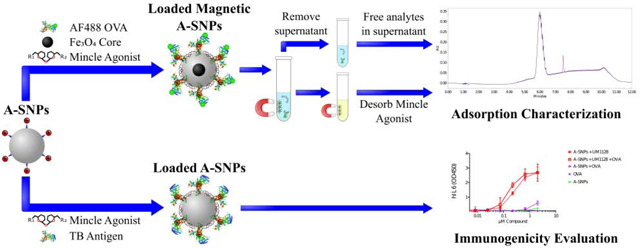
Few nanoparticle delivery systems are capable of delivering MAs in a fashion that can present them to their targeted receptors in an active conformation while at the same time achieve a synchronized targeting and activation of DCs by both the antigen and the Mincle adjuvant [7]. The scope of this work focused mainly on the preparation and characterization of MAs and antigen co-loaded A-SNPs as a novel vaccine candidate. In particular, this report describes the preparation and characterization of MAs, A-SNPs, and M-A-SNPs, then, co-loading of MAs and a model antigen onto the surface of the A-SNPs or M-A-SNPs as illustrated in Figure 2. In addition to the preparation protocol, methods for separation and accurate quantitation of co-adsorption of both components to the magnetic particles are described. Furthermore, we included the synthesis and characterization of UM1128, a novel synthetic TDM derivative.
Figure 2.
Overview of the MAs and antigen co-adsorption process to A-SNPs. MAs were adsorbed to A-SNPs first using a lipid film-like procedure followed by absorbing the antigen later using gentle conditions to maintain it conformational stability.
As shown in Figure 1, the MAs, UM1024 and UM1128 have distinct structural features as well as different physicochemical properties and critical packing parameters. The packing parameter takes into account the volume of the hydrophobic chain, the equilibrium area per molecule at the aggregate interface, and the length of the hydrophobic chain [30]. UM1024, a brartemicin derivative, has been reported to induce strong vaccine mediated humoral and cell mediated Th17-type response against the TB antigen, M72 making it a strong candidate for use as a TB vaccine adjuvant [23]. UM1128 is an asymmetric lipidated trehalose derivative made via an efficient click chemistry, which also has a free azide group at the 6 position. UM1128 has a lower hydrophobicity, and a smaller packing parameter than UM1024 [24]. These two novel MAs were evaluated for loading efficiency on A-SNPs in the presence or absence of a model antigen as well as biological activity in vitro.
3.1. Preparation of M-A-SNPs and particle characterization
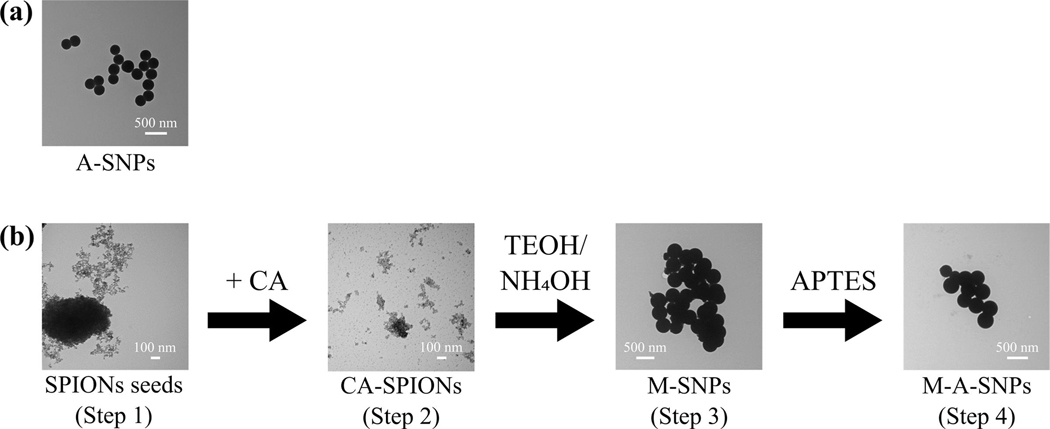
The studied MAs are water insoluble and consequently self-assemble into aggregates when in aqueous environment presenting unique challenges for evaluation of loading efficiency. M-A-SNPs of comparable surface functionalization, surface area, and zeta potential to A-SNPs (Table 1, Figure 3) were prepared to allow for separation of the A-SNPs-loaded MAs from the free-form aggregates and facilitate accurate determination of adjuvant adsorption to A-SNPs in both the presence and absence of the model antigen. M-A-SNPs were prepared in four steps as illustrated in Figure 3b. Step1: preparation of the superparamagnetic iron oxide nanoparticle seeds with diameters of about 8 nm according to the Massart’s co-precipitation method [28]. Step 2: modifying the surface of the SPIONs from step 1 with citric acid to improve their dispersion in the reaction media in the following step according to Yao et al. work [29]. Step 3: forming a silica shell around the CA-SPIONs to obtain M-SNPs [29]. Step 4: Modifying the surface of M-SNPs with APTES to get a cationic surface on the magnetic particles resembling that of A-SNPs. Post-functionalization showed SNP size remained similar, while surface zeta-potential was tunable via functionalization. The successful surface functionalization with APTES was confirmed by measuring the zeta potential for the modified particles and comparing it with the zeta potential of bare SNPs, (Table 1).
Table 1.
Characterization of the developed SNPs-based formulations. Zeta-potential was determined for SNPs formulations diluted 10 X in 10 mM NaCl. Zeta potential is given as the mean ± SEM (n =3).
| SNPs | SNPs Conc. [mg/mL] | MA | MA Conc. (mM) | OVA Conc. [μg/mL] | Particle diameter ± SD (nm) | Total surface area (nm2) | pH | zeta potential ± SD (mV) |
|---|---|---|---|---|---|---|---|---|
| Bare SNPs | 20.0 | --- | --- | --- | --- | --- | 5.5 | − 58 ± 4 |
| Bare M-SNPs | 20.0 | --- | --- | --- | --- | --- | 5.5 | − 48 ± 9 |
| A-SNPs | 20.0 | --- | --- | --- | 246 ± 11 | 2.7 × 1017 | 5.4 | 52 ± 3 |
| M-A-SNPs | 20.0 | --- | --- | — | 362 ± 29 | 1.7 × 1017 | 5.3 | 46 ± 5 |
|
| ||||||||
| A-SNPs | 20.0 | UM1024 | 0.2 | --- | --- | --- | 5.3 | 58 ± 4 |
| A-SNPs | 20.0 | UM1128 | 0.2 | --- | --- | --- | 5.5 | 55 ± 4 |
| A-SNPs | 20.0 | UM1024 | 0.2 | 20 | --- | --- | 5.3 | 32 ± 3 |
| A-SNPs | 20.0 | UM1128 | 0.2 | 20 | --- | --- | 5.3 | 33 ± 4 |
Figure 3.
TEM images for (a) A-SNPs and (b) preparation of M-A-SNPs including Step1: preparation of the superparamagnetic iron oxide nanoparticle seeds. Step 2: modifying the surface of the SPIONs from step 1 with citric acid to improve their dispersion in the reaction media. Step 3: forming a silica shell around the CA-SPIONs to obtain M-SNPs. Step 4: Modifying the surface of M-SNPs with APTES to get a cationic surface on the magnetic particles.
3.2. Simultaneous determination of adsorption of MAs and antigen to M-A-SNPs using HPLC
The studied MAs can be classified as trehalose glycolipids with an amphiphilic nature [31]. Although we suspect that hydrogen bonds play a role in adhesion of the weakly charged MAs to A-SNPs, it is likely that van der waals and hydrophobic interaction forces contribute to the rapid and efficient coating of the A-SNPs surface. In addition, the amphiphilic properties of these compounds aided their coating onto the surface of A-SNPs using a lipid film-like procedure, [32]. In this procedure, A-SNPs suspended in ethanol were mixed with the MA solution in THF: MeOH (9:1, v/v), then thoroughly dried under reduced pressure to form a thin film surface coated on a round-bottom flask or glass vial. It is reported that surface enrichment with amphiphilic lipids may be sensitive to ionic strength [33]. WFI was used as the rehydration diluent to prevent any possible interference from inorganic salts with the Mincle ligand’s packing properties, which might affect their adsorption to A-SNPs. Different sonication conditions and temperatures were investigated and 60 min of sonication at 55–65°C was found to be optimal. After resuspension of the MA-coated A-SNPs, the model antigen was added and adsorbed to the MA-coated A-SNPs by end-over-end mixing at room temperature for 2 hours. Of note, it was determined that trehalose glycolipids tend to aggregate in aqueous environment and adhere strongly to the glass wall of the container during formulation development, which was minimized significantly by adsorbing them onto the high surface area provided by SNPs.
OVA is an anionic monomeric phosphoglyco-protein with a molecular weight of 44.5 kDa and an isoelectric point (IEP) of 4.5 [34]. These physicochemical properties make OVA a good model antigen for the purposes of TB antigen adsorption to A-SNPs. 20 ng/ml of either OVA or AF488 OVA adsorbed to a similar degree (higher than 90%) onto 5 mg/ml A-SNPs, which corresponds to a mass ratio of 1:250 w/w. This was confirmed after spinning down the particles via centrifugation at 600 rcf for 10 min and checking for the free, un-adsorbed antigen using micro BCA assay (data not shown). It should be noted that the presence of MAs interfered with the micro BCA assay and it was not possible to determine the free antigen for samples containing MAs. To solve this problem, AF488 OVA was used as the model antigen so we could reliably quantitate the protein at low concentration ranges (0.3–20 μg/ml) using HPLC coupled with both PDA and fluorescence detectors. This also allowed for simultaneous quantitation of the studied MAs and AF488 OVA in the same HPLC run using a single analytical method.
3.2.1. Samples preparation
As mentioned in the previous section, the separation of the free antigen and MAs was made feasible by using M-A-SNPs instead of A-SNPs (Figure 4). Different solvents were tried in order to dissolve both the MAs and antigen while avoiding the use of organic solvents such as THF, acetone, ethanol and methanol which can precipitate the antigen (water wasn’t a suitable choice to dissolve the water insoluble MAs). Solutions composed of surfactants, Tween 80 or Triton X-100, in WFI were tested and found capable of dissolving both components. Triton X-100 was ultimately chosen as it didn’t interfere with the peak resolution of UM1128, which doesn’t have a strong chromophore, and resulted in a better peak shape for AF488 OVA during HPLC analysis, (Figure 5). As illustrated in Figure 4, a magnet was used to separate the supernatant (Sample 1), which should show any free MAs or antigen, from the M-A-SNPs-adsorbed components. Then, the M-A-SNPs were resuspended in water and transferred to a clean vial to separate them from any of the free-form components that might be sticking to the walls of the original glass vial. This was accomplished using several washes while recycling the water wash in order to avoid diluting the analytes (Sample 2). MAs were then extracted from the separated beads obtained after the previous steps using Triton X-100 several times and the extracts were collected and analyzed undiluted (Sample 3).
Figure 4.
Overview of the separation process using AF488 OVA and M-A-SNPs. A magnet was used to separate the supernatant (Sample 1), which should show any free MAs or antigen. Then, the M-A-SNPs were resuspended in WFI and transferred to a clean vial to separate them from any of the free-form components. This was accomplished using several washes while recycling the water wash (Sample 2). MAs were then extracted from the separated beads obtained after the previous steps using Triton X-100 several times and the extracts were collected and analyzed undiluted (Sample 3).
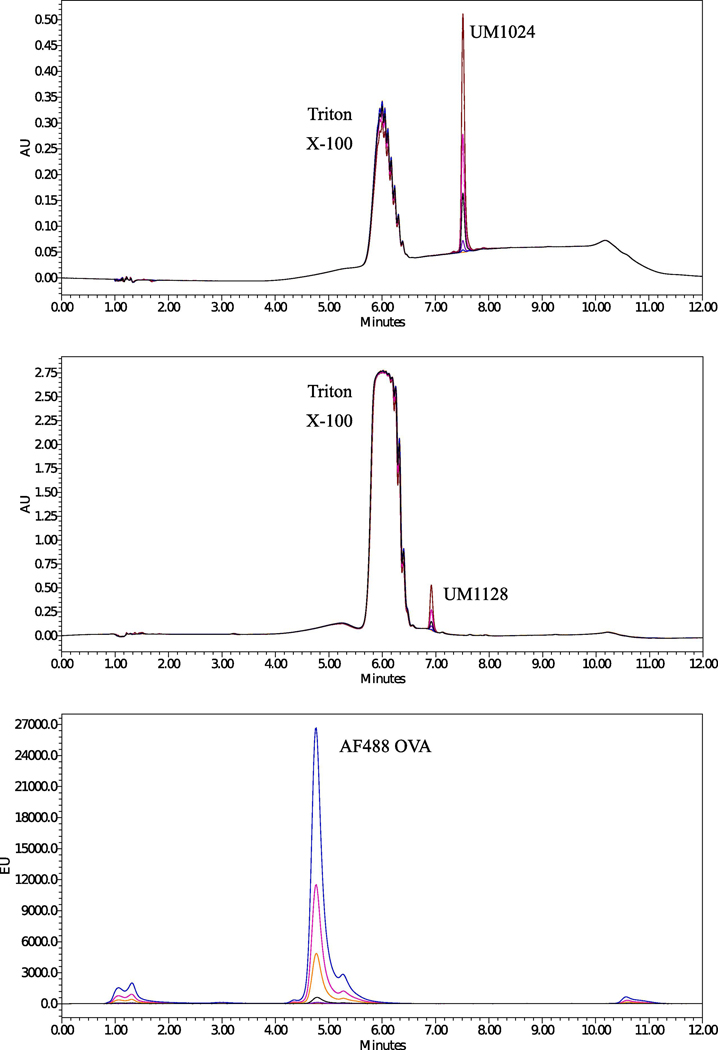
Figure 5.
RP-HPLC chromatograms for (a) UM1024 standards at 254 nm (b) UM1128 standards at 216 nm, and (c) AF488 OVA standards at 520 nm after excitation at 495 nm. The analytes were dissolved in 1% Triton X-100 and analyzed under the described chromatographic conditions.
3.2.2. RP-HPLC analysis
Several trials were carried out to obtain a simple, accurate, and reliable LC method for the simultaneous determination of the studied MAs in combination with the model antigen. Different columns with varying length and internal diameter were evaluated and a satisfactory separation was obtained on a Waters Symmetry C18 column (100 Å, 5 μm, 4.6 mm x 250 mm). Different mobile phase compositions and gradient elution methods were tried to achieve good performance in terms of peak shape and selectivity for the model antigen as well as different MAs with varying structural features, (Figure 5 and Figure S2). The use of buffer was not necessary in this method and the aqueous phase consisted of 0.1% TFA in WFI (sol-A). Methanol, acetonitrile, and IPA were examined as organic modifiers and a combination of IPA (sol-B) and 0. 1% TFA methanol (sol-C) were found to be more suitable for achieving good peak shapes for the MAs with avoiding a high backpressure if only IPA was used. Different gradient elution modes were tested to achieve proper separation of the cited analytes in a reasonable run time, less than 12 min. The described gradient elution mode (0–1 min. sol-A: 50–50, sol-B:20–20; 1–5 min. sol-A: 50–0, sol-B: 20–90; 5–7 min. sol-A: 0–0, sol-B: 90–90; 7–8 min. sol-A: 0–50, sol-B: 90–20; 8–12 min. sol-A: 50–50, sol-B: 20–20) was selected, (Figure S1). Different flow rates were studied and a flow rate of 0.6 mL/min was found to be optimum under the conditions evaluated. A linear relationship was established by plotting the peak areas against the corresponding analyte concentration in the range of 2–200 μM and 10–200 μM for UM1024 and UM1128, respectively, as well as 0.3–20 μg/mL for AF488 OVA, (Figure S3). Statistical analysis of the data gave high value (0.9999) for the correlation coefficient (R2) of the regression equation, as well as a small relative standard deviation and % Error except for UM1128 that is lacking a strong chromophore (Table 2).This data indicates the good linearity of the calibration graphs. LOD was considered as the minimum concentration with a signal to noise ratio of at least three (S/N~3), while LOQ was taken as a minimum concentration with a signal to noise ratio of at least ten (S/N~10). The results of the system suitability tests assure the adequacy of the proposed RP-HPLC method for routine analysis of the studied analytes (Table 2). The system suitability data was summarized in Table S1. The values of the capacity factor (k’) indicate that all peaks are well resolved with respect to the void volume. The values of % RSD of five consecutive injections performed for each analyte show good injection repeatability. The tailing factor (T) reflects good peak symmetry. The theoretical plate numbers (N) demonstrate good column efficiency. The lower retention time of UM1128 (6.8 min) in comparison with UM1024 (7.5 min) as shown by RP-HPLC using a Cl8 column indicates the higher polarity for the asymmetric molecule, UM1128 (Figure 5).
Table 2.
Characteristics and results of the RP-HPLC method.
| Item | UM1024 | UM1128 | AF488 OVA |
|---|---|---|---|
| Detection wavelength (nm) | 254 | 216 | 486 (Ex) and 520 (Em) |
| Linearity range | 2–200 μM | 10–200 μM | 0.3–20 μg/mL |
| LOD (μg mL−1 ) | 1.14 | 1.85 | 0.24 |
| LOQ (μg mL−1 ) | 3.44 | 5.61 | 0.72 |
| Intercept (a) | −4165.0 | −4684.3 | −3581717 |
| Slope (b) | 9474.3 | 3206.2 | 37389187 |
| Correlation coefficient (R2) | 0.9999 | 0.9999 | 0.9999 |
No antigen or MA were detected in sample 1 (supernatant) or sample 2 (wash) for all of the tested formulations, which indicates a high loading efficiency of both components onto A-SNPs. Furthermore, AF488 OVA showed adsorption higher than 90% to A-SNPs, relative to an AF488 OVA only control that was processed similarly, where the presence of MAs didn’t interfere with these adsorption rates, (Table 3). Analyzing sample 3 for the extracted MAs revealed that UM1024 showed 87.9 ± 5.7 % adsorption in the presence of AF488 OVA and 81.8 ± 3.2 % adsorption in the absence of AF488 OVA whereas UM1128 showed 88.0 ± 3.5 % adsorption in the presence of AF488 OVA and 85.5 ± 11.1 % adsorption in the absence of AF488 OVA, (Table 3). The excellent co-adsorption results indicate that A-SNPs can be used as a co-delivery platform for anionic antigens along with different MAs of different structural features and biophysical properties.
Table 3.
% adsorption of 200 μM MAs and 20 μg/ml AF488 OVA to 20 mg/ml M-A-SNPs.
| Formulation | % antigen adsorption | % adjuvant adsorption |
|---|---|---|
| M-A-SNPs + UM1024 + AF488 OVA | 95.6 ± 0.1a | 87.9 ± 5.7a |
| M-A-SNPs + UM1024 | 81.8 ± 3.2a | |
| M-A-SNPs + UM1128 + AF488 OVA | 95.8 ± 7.9a | 88.0 ± 3.5a |
| M-A-SNPs + UM1128 | 85.5 ± 11.1a |
Mean and relative standard deviation for 3 determinations
3.3. Immunogenicity of formulations
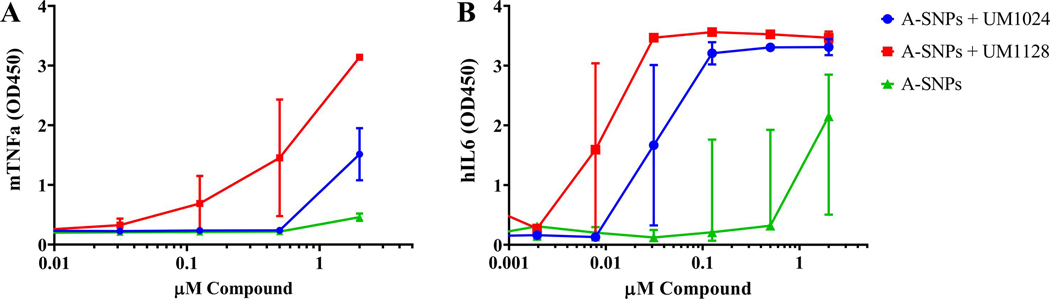
Biologically active formulations for MAs are very limited. The most commonly cited presentation in literature is plate coated for these water insoluble molecules. Plate coated presentation entails dissolving compounds in a suitable solvent such as ethanol or isopropyl alcohol, applying a solution to a plate and allowing the solvent to evaporate leaving the compound on the plate. Although, the plate coating method is typically used for initial compound screening and SAR studies it is not readily transferrable to an in vivo system. Figure S4 shows human IL-6 production from plate coated, DMSO and A-SNPs formulations of UM1024 where A-SNPs showed a very good potency for the loaded water insoluble adjuvant. Thus, we have explored the use of SNPs-based formulations to get around the MAs water insolubility issue and deliver MAs both in vitro and in vivo. MAs induce inflammatory cytokines, such as tumor necrosis factor α (TNFα) and interleukin (IL-6), and promote the development of a Th1/Th17 immune response in association with innate cytokines such as IL-6, IL-1β, IL-12 and IL-23 [3, 23]. To check the ability of the new SNPs-based formulations for driving such responses, cryopreserved PBMCs were thawed, treated with UM1024-adsorbed A-SNPs and incubated at 37°C. The supernatant was harvested 24 hours later and analyzed for TNFα, IL23, IL-1β, INFγ, IL12p70, and IL-6 via a multiplex cytokine array (Luminex). UM1024-loaded A-SNPs were able to drive a strong induction of those cytokines from human PBMCs in comparison to the unloaded A-SNPs (Figure S5), demonstrating that A-SNPs are effective at delivering MA to the targeted receptors/cells. It is worth mentioning that we have not noted any cytotoxicity in vitro with the addition of A-SNPs or MAs loaded A-SNPs at the doses evaluated. All cells appeared healthy following incubation with these formulations (in comparison to untreated PBMCs). Furthermore, we conducted additional studies with human PBMCs assessing viability using Cell Titer Glo® 2.0. where A-SNPs did not impact Cell viability after incubation with freshly isolated hPBMCs for 24 hours, (Figure S6). Of note, we have not observed overt toxicity to date with in vivo studies in mice using up to 2000 μg dose of A-SNPs (data not shown). To compare the immunostimulatory activity of UM1024 and UM1128 adsorbed to A-SNPs in both murine and human cell lines, A-SNPs formulations were assessed for cytokine production from mouse monocytic cell line RAW 264.7. Some very interesting structural features correlating with activity were immediately obvious. UM1128 showed a higher potency than UM1024 in both cell types (Figure 6). In addition, the co-adsorption of OVA along with UM1128 onto A-SNPs didn’t affect the hIL6 induction in hPBMCs, (Figure S7).
Figure 6.
TNFα cytokine production from mouse RAW 264.7 cells (a) and IL6 cytokine production in hPBMCs (b) in response to stimulation with serially diluted 20 μM UM1128 or UM1024 adsorbed to 2 mg/mL A-SNPs. Two replicates (from two independent donors) mean +/− SEM.
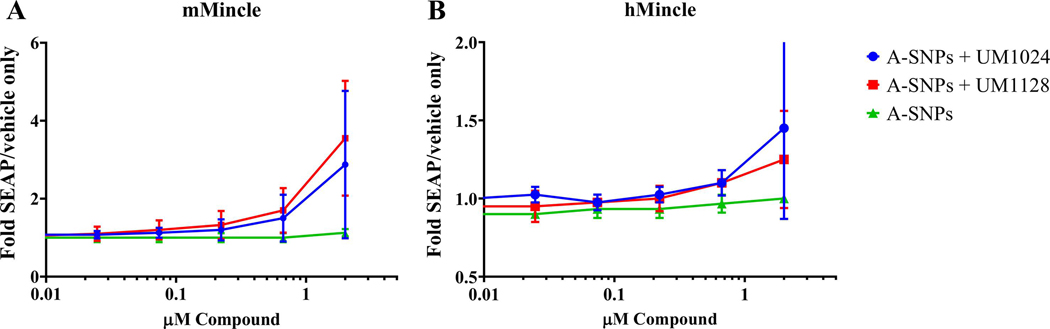
The Mincle-selective activity of UM1024 and UM1128 adsorbed to A-SNPs was evaluated using human embryonic kidney (HEK) cells expressing the human or mouse Mincle receptor along with an NF-KB-driven secreted embryonic alkaline phosphatase (SEAP) reporter. Both UM1024 and UM1128 induced production of SEAP in a dose dependent manner clearly demonstrating these formulations are Mincle receptor specific (Figure 7).
Figure 7.
Activation of Mincle in response to MAs-coated A-SNPs. HEK cells transfected with (a) mouse Mincle, or (b) human Mincle. An NF-KB-driven SEAP reporter were added to the indicated formulations in 96 well plates and incubated with the compounds for 18–24 h followed by assessment of the supernatants for SEAP levels. Data are represented as fold change in OD650 over vehicle treated cells. Three replicates (from three independent donors) mean +/− SEM.
These new formulations are intriguing based on the novelty of this platform, the ability to formulate insoluble PRR agonists such as MAs, the observed efficient antigen and adjuvant coadsorption, and its strong potential for achieving a synchronized delivery of antigens and adjuvants. Testing the release kinetics of MAs-loaded A-SNPs as well as testing the humoral and cell-mediated immune responses in combination with a clinical-stage Mtb antigen, M72 [35], are currently under investigation. The preliminary data that we collected so far clearly demonstrates the efficacy of this delivery system for inducing both cell mediated (Th17) and humoral immunity. We found that MAs and antigen co-adsorbed on A-SNPs at various coating densities elicited significantly increased antigen-specific antibody concentrations over the matched control groups in a dose-dependent manner. Furthermore, Splenocytes from MAs and antigen-loaded A-SNPs-vaccinated mice were found to produce IL-17 in response to TB antigen whole protein restimulation 5 days post-tertiary of vaccination (Unpublished data).
Conclusions
Innovative formulations and delivery systems are urgently needed for the co-delivery of antigens and adjuvant to promote humoral and cell-mediated immunity. Of particular interest are safe and scalable delivery methods for insoluble PRR adjuvants, such as MAs. Here, we describe a simple, but effective procedure for the preparation and thorough characterization of A-SNPs loaded with MAs and a model antigen. In addition, we developed a RP-HPLC method to assess the adsorption of the studied adjuvants and antigens on A-SNPs. These SNP-based formulations have a strong potential to achieve an efficient co-delivery of MAs of different structural features along with antigen for synchronized targeting and activation of antigen presenting cells. Our model antigen demonstrated higher than 90% adsorption to M-A-SNPs, while the presence of MAs didn’t interfere with antigen adsorption efficiency. In addition, the MAs demonstrated more than 80% adsorption to M-A-SNPs in both the presence or absence of the model antigen. The formulations were tested in vitro in murine RAW cells and hPBMCs where they exhibited strong cytokine induction from both cell types. The tested formulations also demonstrated the ability to engage the Mincle receptor. In this fashion, A-SNPs can be used as a versatile platform for tunable delivery of MAs and/or other PRR ligands and antigens. This could be very beneficial when co-delivery of antigen and adjuvant are desired.
Supplementary Material
Highlights.
Nanoparticles can achieve an efficient co-delivery of adjuvants and antigens to immune cells.
First reported use of A-SNPs for co-delivery of synthetic Mincle agonist (adjuvant) and antigen.
A-SNPs are capable of presenting MAs in the proper spatial arrangement and surface density for enhanced immune response.
Novel RP-HPLC method was developed to assess the loading efficiency of Mincle agonist and antigen to A-SNPs.
This SNPs-based vaccine formulation demonstrated good bioactivity in vitro from PBMCs and RAW cells.
ACKNOWLEDGMENTS:
This work was supported by an NIAID Adjuvant Discovery Program Contract (HHSN272201400050C). The authors acknowledge University of Montana Core Services of the Center for Biomolecular Structure and Dynamics (CBSD) and the Department of Chemistry and Biochemistry supported by the National Institutes of General Medical Science (NIH CoBRE grant P20GM103546) for NMR and MS instrumentation. The authors acknowledge the Pharmaceutical Chemistry Department at Cairo University for allowing Walid Abdelwahab to get his postdoctoral training at the University Of Montana Center for Translational Medicine. The authors also acknowledge the EMTrix core facility at the division of biological sciences, University of Montana for TEM characterization.
Abbreviations:
- AF488
Alexa Fluor 488
- APCs
antigen presenting cells
- APTES
(3-Aminopropyl)triethoxysilane
- A-SNPs
amine-functionalized silica nanoparticles
- CA
citric acid monohydrate
- CLR
C-type lectin receptor
- DCs
dendritic cells
- DLS
dynamic light scattering
- DPBS
Dulbecco’s phosphate buffered saline
- ELISA
enzyme-linked immunosorbent assay
- EM
electron microscopy
- FBS
fetal bovine serum
- IL
interleukin
- LN
lymph nodes
- OD
optical density
- hPBMCs
human peripheral blood mononuclear cells
- MA
Mincle agonist
- Mincle
Macrophage Inducible C-type lectin receptor
- Mtb
Mycobacterium tuberculosis
- PDI
polydispersity index
- PBS
phosphate buffer saline
- PRR
pattern recognition receptor
- OVA
ovalbumin
- RP-HPLC
reverse phase-high performance liquid chromatography
- RPMI
Roswell Park Memorial Institute medium
- SNPs
silica nanoparticles
- TEOS
Tetraethyl orthosilicate
- TNFα
tumor necrosis factor α
- TLR
toll-like receptor
- TFA
Trifluoroacetic acid
- WFI
sterile water for irrigation
- UM
University of Montana
- UV/Vis
Ultraviolet/visible light spectra
Footnotes
DISCLOSURES:
The authors have nothing to disclose.
Walid M. Abdelwahab, Writing, Conceptualization, Methodology, Visualization, Validation Alexander Riffey, Investigation, Methodology, Visualization, Writing - Review & Editing Cassie Buhl, Investigation, Methodology, Writing - Review & Editing Craig Johnson, Investigation, Resources Kendal Ryter, Project administration, Conceptualization, Writing - Review & Editing Jay T. Evans, Conceptualization, Writing - Review & Editing, Project administration, Funding acquisition, Supervision David J. Burkhart, Conceptualization, Writing - Review & Editing, Project administration, Funding acquisition, Supervision
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES:
- 1.Mortaz E, et al. , Interaction of Pattern Recognition Receptors with Mycobacterium Tuberculosis. J Clin Immunol, 2015. 35(1): p. 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kingeter LM and Lin X, C-type lectin receptor-induced NF-kB activation in innate immune and inflammatory responses. Cellular & Molecular Immunology, 2012. 9(2): p. 105–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith AJ, et al. , Species-Specific Structural Requirements of Alpha-Branched Trehalose Diester Mincle Agonists. Frontiers in immunology, 2019. 10: p. 338–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bermejo-Jambrina M, et al. , C-Type Lectin Receptors in Antiviral Immunity and Viral Escape. Frontiers in Immunology, 2018. 9(590). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wozniak TM, et al. , Plasmid interleukin-23 (IL-23), but not plasmid IL-27, enhances the protective efficacy of a DNA vaccine against Mycobacterium tuberculosis infection. Infection and immunity, 2006. 74(1): p. 557–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amelio P, et al. , Mixed Th1 and Th2 Mycobacterium tuberculosis-specific CD4 Tcell responses in patients with active pulmonary tuberculosis from Tanzania. PLoS Negl Trop Dis, 2017. 11(7): p. e0005817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kamath AT, et al. , Synchronization of Dendritic Cell Activation and Antigen Exposure Is Required for the Induction of Th1/Th17 Responses. The Journal of Immunology, 2012. 188(10): p. 4828. [DOI] [PubMed] [Google Scholar]
- 8.Sokolova V, et al. , The use of calcium phosphate nanoparticles encapsulating Toll-like receptor ligands and the antigen hemagglutinin to induce dendritic cell maturation and T cell activation. Biomaterials, 2010. 31(21): p. 5627–33. [DOI] [PubMed] [Google Scholar]
- 9.An M, et al. , Silica Nanoparticle as a Lymph Node Targeting Platform for Vaccine Delivery. ACS Appl Mater Interfaces, 2017. 9(28): p. 23466–23475. [DOI] [PubMed] [Google Scholar]
- 10.Fox CB, et al. , Working together: interactions between vaccine antigens and adjuvants. Ther Adv Vaccines, 2013. 1(1): p. 7–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujii S-I, et al. , The linkage of innate to adaptive immunity via maturing dendritic cells in vivo requires CD40 ligation in addition to antigen presentation and CD80/86 costimulation. The Journal of experimental medicine, 2004. 199(12): p. 1607–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diogo GR, et al. , Immunization With Mycobacterium tuberculosis Antigens Encapsulated in Phosphatidylserine Liposomes Improves Protection Afforded by BCG. Frontiers in Immunology, 2019. 10(1349). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamborg M, et al. , Elucidating the mechanisms of protein antigen adsorption to the CAF/NAF liposomal vaccine adjuvant systems: Effect of charge, fluidity and antigen-to-lipid ratio. Biochimica et Biophysica Acta (BBA) - Biomembranes, 2014. 1838(8): p. 2001–2010. [DOI] [PubMed] [Google Scholar]
- 14.Mody KT, et al. , Mesoporous silica nanoparticles as antigen carriers and adjuvants for vaccine delivery. Nanoscale, 2013. 5(12): p. 5167–79. [DOI] [PubMed] [Google Scholar]
- 15.Taki A. and Smooker P, Small Wonders-The Use of Nanoparticles for Delivering Antigen. Vaccines (Basel), 2015. 3(3): p. 638–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu T, et al. , Silica nanorattle with enhanced protein loading: a potential vaccine adjuvant. J Colloid Interface Sci, 2013. 400: p. 168–74. [DOI] [PubMed] [Google Scholar]
- 17.Heidegger S, et al. , Immune response to functionalized mesoporous silica nanoparticles for targeted drug delivery. Nanoscale, 2016. 8(2): p. 938–48. [DOI] [PubMed] [Google Scholar]
- 18.Cha BG, Jeong JH, and Kim J, Extra-Large Pore Mesoporous Silica Nanoparticles Enabling Co-Delivery of High Amounts of Protein Antigen and Toll-like Receptor 9 Agonist for Enhanced Cancer Vaccine Efficacy. ACS Cent Sci, 2018. 4(4): p. 484–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bradbury MS, et al. , Clinically-translated silica nanoparticles as dual-modality cancer-targeted probes for image-guided surgery and interventions. Integrative biology : quantitative biosciences from nano to macro, 2013. 5(1): p. 74–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Toda T. and Yoshino S, Enhancement of ovalbumin-specific Th1, Th2, and Th17 immune responses by amorphous silica nanoparticles. International Journal of Immunopathology and Pharmacology, 2016. 29(3): p. 408–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choi J, et al. , Comparison of cytotoxic and inflammatory responses of photoluminescent silicon nanoparticles with silicon micron-sized particles in RAW 264.7 macrophages. Journal of Applied Toxicology, 2009. 29(1): p. 52–60. [DOI] [PubMed] [Google Scholar]
- 22.Hunter RL, et al. , Multiple roles of cord factor in the pathogenesis of primary, secondary, and cavitary tuberculosis, including a revised description of the pathology of secondary disease. Ann Clin Lab Sci, 2006. 36(4): p. 371–86. [PubMed] [Google Scholar]
- 23.Ryter KT, et al. , Aryl Trehalose Derivatives as Vaccine Adjuvants for Mycobacterium tuberculosis. Journal of Medicinal Chemistry, 2020. 63(1): p. 309–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abdelwahab WAM, D. B, Evans J, Johnson C, Ryter KT, Smith A, Immunogenic trehalose compounds and uses thereof as vaccine adjuvants. 2019: PCT/US2019/020373. [Google Scholar]
- 25.Jiang YL, et al. , Synthesis and evaluation of trehalose-based compounds as novel inhibitors of cancer cell migration and invasion. Chem Biol Drug Des, 2015. 86(5): p. 1017–29. [DOI] [PubMed] [Google Scholar]
- 26.Stöber W, Fink A, and Bohn E, Controlled growth of monodisperse silica spheres in the micron size range. Journal of Colloid and Interface Science, 1968. 26(1): p. 62–69. [Google Scholar]
- 27.Abdelwahab WM, Phillips E, and Patonay G, Preparation of fluorescently labeled silica nanoparticles using an amino acid-catalyzed seeds regrowth technique: Application to latent fingerprints detection and hemocompatibility studies. Journal of Colloid and Interface Science, 2018. 512: p. 801–811. [DOI] [PubMed] [Google Scholar]
- 28.Vaz AM, et al. , Synthesis and characterization of biocatalytic gamma-Fe2O3@SiO2 particles as recoverable bioreactors. Colloids Surf B Biointerfaces, 2014. 114: p. 11–9. [DOI] [PubMed] [Google Scholar]
- 29.Yao Q, et al. , Surface arming magnetic nanoparticles with amine N-halamines as recyclable antibacterial agents: Construction and evaluation. Colloids Surf B Biointerfaces, 2016. 144: p. 319–326. [DOI] [PubMed] [Google Scholar]
- 30.Cullis PR, Hope MJ, and Tilcock CPS, Lipid polymorphism and the roles of lipids in membranes. Chemistry and Physics of Lipids, 1986. 40(2): p. 127–144. [DOI] [PubMed] [Google Scholar]
- 31.Khan AA, Stocker BL, and Timmer MS, Trehalose glycolipids--synthesis and biological activities. Carbohydr Res, 2012. 356: p. 25–36. [DOI] [PubMed] [Google Scholar]
- 32.Meng H, et al. , Use of a lipid-coated mesoporous silica nanoparticle platform for synergistic gemcitabine and paclitaxel delivery to human pancreatic cancer in mice. ACS Nano, 2015. 9(4): p. 3540–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bershteyn A, et al. , Polymer-supported lipid shells, onions, and flowers. Soft matter, 2008. 4(9): p. 1787–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strixner T. and Kulozik U, Handbook of Food Proteins. Handbook of Food Proteins, 2011(222): p. 1–432. [Google Scholar]
- 35.Penn-Nicholson A, et al. , Safety and immunogenicity of candidate vaccine M72/AS01E in adolescents in a TB endemic setting. Vaccine, 2015. 33(32): p. 4025–4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.