Abstract
VP22, the 301-amino-acid phosphoprotein product of the herpes simplex virus type 1 (HSV-1) UL49 gene, is incorporated into the tegument during virus assembly. We previously showed that highly modified forms of VP22 are restricted to infected cell nuclei (L. E. Pomeranz and J. A. Blaho, J. Virol. 73:6769–6781, 1999). VP22 packaged into infectious virions appears undermodified, and nuclear- and virion-associated forms are easily differentiated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (J. A. Blaho, C. Mitchell, and B. Roizman, J. Biol. Chem. 269:17401–17410, 1994). As VP22 packaging-associated undermodification is unique among HSV-1 tegument proteins, we sought to determine the role of VP22 during viral replication. We now show the following. (i) VP22 modification occurs in the absence of other viral factors in cell lines which stably express its gene. (ii) RF177, a recombinant HSV-1 strain generated for this study, synthesizes only the amino-terminal 212 amino acids of VP22 (Δ212). (iii) Δ212 localizes to the nucleus and incorporates into virions during RF177 infection of Vero cells. Thus, the carboxy-terminal region is not required for nuclear localization of VP22. (iv) RF177 synthesizes the tegument proteins VP13/14, VP16, and VHS (virus host shutoff) and incorporates them into infectious virions as efficiently as wild-type virus. However, (v) the loss of VP22 in RF177 virus particles is compensated for by a redistribution of minor virion components. (vi) Mature RF177 virions are identical to wild-type particles based on electron microscopic analyses. (vii) Single-step growth kinetics of RF177 in Vero cells are essentially identical to those of wild-type virus. (viii) RF177 plaque size is reduced by nearly 40% compared to wild-type virus. Based on these results, we conclude that VP22 is not required for tegument formation, virion assembly/maturation, or productive HSV-1 replication, while the presence of full-length VP22 in the tegument is needed for efficient virus spread in Vero cell monolayers.
The herpesvirus tegument has historically been described as an amorphous protein layer located between the nucleocapsid and the envelope (50). However, many recent publications propose a more tightly controlled organization of this structure (33, 37, 55). Four viral proteins which compose the bulk of the tegument's mass (27, 28, 53) are encoded by genes that lie in a consecutive stretch in the unique long segment of the herpes simplex virus type 1 (HSV-1) genome. UL46, UL47, UL48, and UL49 encode VP11/12 (55), VP13/14 (38, 55), VP16 (9, 44), and VP22 (21), respectively. VP22 is the most abundant tegument protein in virus particles (27). Additionally, the virion host shutoff (VHS) protein, encoded by the UL41 gene (43), is also incorporated into virus particles (29, 30, 49, 51). Of these tegument proteins, only VP16 has been shown to provide an essential function in structural assembly of the virus in tissue culture (3, 49, 54, 55).
The five tegument proteins mentioned above are phosphoproteins (5, 24), which may undergo additional posttranslational modifications at various stages of the infection cycle. VP11/12, VP13/14, VP16, VP22, and VHS have all been shown to incorporate radiolabeled phosphate in infected-cell extracts (5, 20, 51), while VP11/12, VP13/14, VP16, and VP22 can be radiolabeled in isolated virus particles (32). Phosphorylated forms of VP22 can be differentiated based on their migration in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and the fastest-migrating form of VP22 is observed in purified virion preparations (5, 20, 45). VP13/14 and, to a lesser extent, VP22 were reported to be glycosylated in virion particles (39). VP13/14 and VP22 are nucleotidylated (5, 6), and VP22 is mono(ADP-ribosyl)ated (5, 48). While the roles of each of these modifications during infection remain unknown, recent results suggest that virion protein phosphorylation may regulate the assembly and dissociation of the tegument (41).
A number of studies have examined the role of individual tegument proteins in virion assembly. Following infection with a recombinant virus expressing two copies of VP22, VP13/14 levels were decreased in purified virions, presumably to compensate for the increased levels of packaged VP22 (33). Virions devoid of VP11/12 exhibit increased incorporation of VP13/14 (55). In recombinant viruses in which synthesis of VP13/14 or both VP13/14 and VP11/12 is abolished, increased amounts of VP16 are incorporated into virions (55). Taken together, these results suggest that the total amount of tegument protein incorporated into the virion is constant but the relative ratio of individual tegument proteins is flexible.
The present study was designed to examine the function of VP22 during infection, focusing on its potential role in tegument assembly. RF177, a recombinant strain of HSV-1 generated for the purposes of this study, synthesized low levels of a truncated form of VP22 (Δ212). Tegument proteins VP13/14, VP16, and VHS were synthesized and incorporated into RF177 virions at levels comparable to wild-type virus, while full-length VP22 was absent from purified RF177 particles. No obvious structural differences between RF177 and wild-type virus particles were observed in electron microscopic analyses. Single-step growth kinetics of RF177 in Vero cells were nearly identical to those of wild-type virus, although RF177 exhibited a slight reduction in titer at very late times postinfection. Finally, RF177 demonstrated smaller plaques than wild-type virus in Vero monolayers. Based on these results, we conclude that (i) tegument formation, virion assembly and maturation, and productive HSV-1 replication can occur in the absence of full-length VP22 and (ii) full-length VP22 in the viral tegument is required for efficient virus spread in Vero cells.
MATERIALS AND METHODS
Cells and virus.
African green monkey kidney (Vero) and thymidine kinase-negative (tk−) human osteosarcoma (143B) cells were obtained from the American Type Culture Collection and passaged in Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% fetal bovine serum. VP22-expressing V49 and 14349 and control V202 and 14320 cells, generated as described below, were passaged in DMEM supplemented with 5% fetal bovine serum and G418 (1 mg/ml; Gibco-BRL). HSV-1(Δ305) (47), from which 500 bp of the tk locus have been deleted, HSV-1(R2507) (46), and the prototype HSV-1(F) (14) were provided by Bernard Roizman, University of Chicago. R2507 was derived from Δ305 and contains a chimeric α27-tk cassette inserted approximately 200 bp upstream from the initiation of the UL48 open reading frame (46). To obtain virus stocks, subconfluent monolayer Vero or V49 cultures (approximately 3 × 106 cells) were inoculated with virus at a multiplicity of infection (MOI) of 0.01 for 2 h at 37°C in 199V medium (Life Technologies) supplemented with 1% newborn calf serum (NBCS). The inoculum was then removed, fresh DMEM supplemented with 5% NBCS was added, and the cells were incubated at 37°C in 5% CO2. Virus stocks were prepared once the infection reached a cytopathic effect (CPE) of 100% and counted on Vero cells, and aliquots were stored at −80°C. All MOIs were derived from the number of PFU of virus on Vero cells.
Plasmids. (i) Plasmids constructed for the generation of VP22-expressing cell lines.
Plasmid pBabeNeo has been described elsewhere (40). pRB251 (3) contains the coding region of the HSV-1(F) UL49 gene from the BglII site at 106750 to the EcoRV site at 105107 (35) and has been used in previous studies (5). The 1,643-bp BglII-EcoRI fragment of pRB251 was inserted into the BamHI and EcoRI sites of pBabeNeo to generate pJB133 (A. Ireland and J. A. Blaho, unpublished results). In this plasmid, VP22 expression is directed from the Moloney murine leukemia virus long terminal repeat. However, a portion of the viral UL49 promoter still remains in the construct, which might facilitate enhanced expression during HSV-1 infection.
(ii) Plasmids constructed for insertional mutagenesis of UL49.
pJB177 was constructed as a two-step clone for the generation of a recombinant virus possessing a deletion in the carboxy-terminal coding region of UL49. First, the BamF (Fig. 1, line 2) fragment of HSV-1(F) was digested with PstI and BamHI (Fig. 1, line 3), and the resulting 4.4-kb fragment containing the UL49 gene was cloned into pGEM-3Z (Promega) to generate pJB175. pJB175 was digested with NsiI and XhoI, and the 1,344-bp NsiI-XhoI fragment of pEGFP-C1 (Clontech), containing a cytomegalovirus promoter-driven green fluorescence protein (GFP) cassette, was inserted to yield pJB177 (Fig. 1, line 5). This strategy left 45 bp at the 3′ end of the UL49 gene (designated by a solid rectangle in Fig. 1, lines 5 and 6) and inserts a stop codon after the histidine at amino acid 212. Additionally, no effort was made to introduce a new polyadenylation sequence after the stop codon. pJB142 contains the BamQ (Fig. 1, line 2) fragment of HSV-1(F) inserted into the BamHI site of pGEM-3Z (A. Ireland and J. A. Blaho, unpublished results) and was used to repair the 500-bp deletion in the tk locus of RD177 (Fig. 1, line 5).
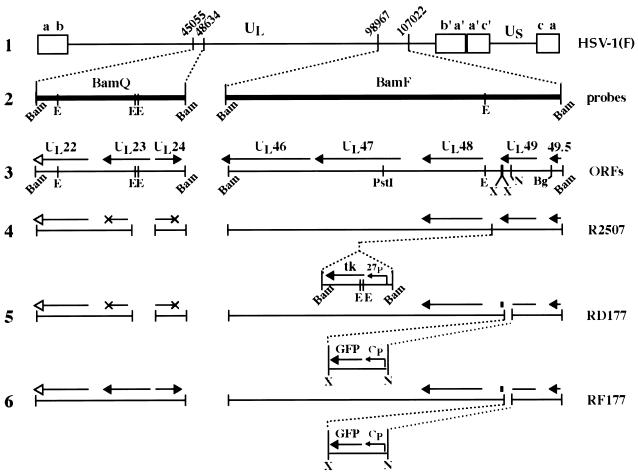
FIG. 1.
Schematic representations of the HSV-1 genome and viruses used in this study. The wild-type HSV-1 genome is shown in line 1. The unique long (UL), unique short (Us), and terminal repeat segments (a, b, c, a′, b′, and c′) are indicated. The coordinates of the BamQ and BamF fragments in the viral genome are indicated, and these regions are expanded in line 2. Line 3 shows the predicted open reading frames in the BamQ and BamF regions of wild-type virus. Solid arrowheads mark translation stop sites and show the direction of transcription; arrow length indicates approximate transcript size. The UL22 transcript designated with an open arrow possesses a translational stop site beyond the boundaries of the BamQ fragment. R2507 (line 4) is a derivative of the parental strain Δ305. Thus, R2507 possesses a deletion in the BamQ fragment which removes 500 bp, including a portion of the UL23 gene encoding the viral tk and a portion of the UL24 gene. These disrupted transcripts are denoted by an X at the locations of their stop sites in the wild-type genome. In R2507, a 1.8-kb tk expression cassette containing the viral tk gene under control of the ICP27 promoter (27p) has been inserted approximately 200 bp upstream of the UL48 start site (46). RD177 (line 5) and RF177 (line 6) were generated for the present study as described in Materials and Methods. Both viruses express the GFP under control of a cytomegalovirus promoter (Cp). The small solid rectangles in lines 5 and 6 indicate that the GFP cassette was inserted 45 bp 5′ of the UL49 stop site. The deletion in the BamQ locus present in R2507 and RD177 was repaired in RF177. Restriction sites: Bam, BamHI; E, EcoRV; X, XhoI; N, NsiI; Bg, BglII.
VP22-expressing and control cell lines.
Vero (V49 and V202) or 143B (14349 and 14320) cells were transfected with pJB133 (V49 and 14349) or pBabeNeo (V202 and 14320) using Dotap (Vero) or Fugene (143B) (Boehringer Mannheim) according to the manufacturer's specifications. At 2 (Vero) or 3 (143B) days posttransfection, medium was changed to DMEM supplemented with 5% fetal bovine serum (FBS) and G418 at 2 mg/ml (Vero) or 0.3 mg/ml (143B). After 16 days (Vero) or 24 days (143B) of selection, surviving cells were cloned into separate wells, expanded, and tested for VP22 expression by immunoblotting and immunofluorescence. VP22-expressing cells were recloned at least twice to ensure monoclonality. For both V49 and 14349 cells, medium was changed to 5% FBS containing G418 at 1 mg/ml 1 month after the initial transfection.
Isolation of viral DNA for transfections and Southern blots.
Viral DNA was prepared from roller bottles of Vero cells as follows. Approximately 2 × 108 Vero cells were infected at 0.01 PFU/cell. At 2 days postinfection (p.i.), cells exhibiting 100% CPE were scraped into the medium, pelleted at 350 × g for 5 min, rinsed once in phosphate-buffered saline (PBS), and resuspended in 2.0 ml of T10E50 (10 mM Tris-HCl, 50 mM EDTA [pH 8.0]) containing 100 μl of 10% Nonidet P-40 (Sigma). Cells were incubated on ice for 15 min and then treated in a Dounce homogenizer with pestle B (loose) five times. Nuclei were pelleted again at low speed for 5 min and discarded. Then 100 μl of 20% SDS–50 μl of 20-mg/ml proteinase K (Sigma)–10 μl of 5-mg/ml RNase A (Sigma) was added to the supernatant (cytoplasmic extract) and incubated for 1 h at 37°C. This mixture was extracted twice with phenol, once with phenol-chloroform (50:50, vol/vol), and once with chloroform only and precipitated at −20°C for at least 1 h from ethanol after adding sodium acetate to 0.3 M. The precipitate was pelleted at 17,000 × g for 15 min and resuspended in 150 μl of T10E0.1 (10 mM Tris-HCl, 0.1 mM EDTA [pH 8.0]). DNA concentrations were determined from UV absorption measured at an optical density at 260 nm.
DNA transfections and recombinational mutagenesis.
Schematic maps of the HSV-1 genome, the UL49 and UL23 loci, and all recombinant viruses used in this study are presented in Fig. 1. R2507 (Fig. 1, line 4) and pJB177 DNA were used in a modified CaCl2 transfection protocol (25) to generate RD177 (Fig. 1, line 5). Approximately 15.0 μg each of viral and pJB177 DNAs were combined with sterile distilled H2O to a total volume of 250 μl. Then 250 μl of 2×HEPES-buffered saline (280 mM NaCl, 10 mM KCl, 1.5 mM Na2HPO4, 12 mM glucose, 50 mM HEPES [pH 7.05]) and 25 μl of 2.5 M CaCl2 were added to DNA and incubated at room temperature for 15 min. This mixture was added to 25-cm2 flasks of 50% confluent V49 cells, split the previous day into 5.0 ml of DMEM supplemented with 5% NBCS. After 3.5 h at 37°C in 5% CO2, DNA-containing medium was aspirated, and cells were washed once in DMEM and shocked with DMEM containing 15% glycerol for 90 s. Glycerol-containing medium was aspirated, and cells were washed once in DMEM and incubated for 6 days in DMEM containing 5% NBCS, at which point GFP was detected in viral plaques by fluorescence microscopy. Pooled virus was prepared from infected V49 cells, grown in 14349 cells, and selected in medium containing bromodeoxyuridine (100 μg/ml) (Sigma). Following bromodeoxyuridine selection, GFP-positive plaques were purified a second time in V49 cells, and the integrity of the resultant virus, RD177 (Fig. 1, line 5), was confirmed by Southern hybridization (data not shown).
To repair the 500-bp deletion in the UL23 locus of RD177, genomic viral DNA was purified and cotransfected into V49 cells with pJB142 as described above. Transfectants were selected in 14349 cells in DMEM containing 5% NBCS supplemented with hypoxanthine-aminopterin-thymidine (Gibco-BRL). The resulting virus, RF177 (Fig. 1, line 6), was plaque purified three times prior to confirmation of the genotype by Southern hybridization using the BamF and BamQ probes (Fig. 1, line 2) as described below. RF177 possesses a 230-bp deletion of the 3′ end of the UL49 gene from the NsiI to the XhoI sites into which the 1,344-bp GFP expression cassette was inserted. This insertion generates a stop codon directly after the histidine at amino acid 212 of VP22, and accordingly, this virus expresses only the amino-terminal 212 amino acids of VP22, termed Δ212. It should be noted that this insertion does not generate a GFP fusion protein.
Analysis of viral DNA by Southern hybridizations.
V49 cells were grown in 25-cm2 flasks, and DNA was isolated as above except that amounts were scaled down fivefold. Approximately 7.5 μg of each viral DNA was used in Southern blots. The DNA hybridization protocol used in the study was a modification of that which was described previously (31). Viral DNAs were digested for 16 h prior to loading a 0.8% Tris–phosphate–EDTA–agarose gel (100 V for 2 h). The separated DNAs were transferred by capillary action to nylon membranes as recommended by the manufacturer (Zetaprobe; Bio-Rad). Prehybridization was done at 65°C in 30% formamide (spectroscopic grade; EM Science)–6× saline sodium citrate–1% SDS–0.5% skim milk (Carnation nonfat dry milk)–0.2 mg of freshly boiled salmon sperm DNA per ml for 30 min prior to the addition of approximately 106 cpm of freshly boiled, nick-translated (NEN Dupont) 32P-labeled probe and hybridization at 65°C for at least 12 h. The stringent washes (twice) of the blot were done at 65°C for 1 h in 20% formamide–4× saline sodium citrate–1% SDS, followed by autoradiography on Kodak X-OMAT film at −80°C.
Immunological reagents.
RGST49 is a rabbit polyclonal antibody directed against a GST-VP22 fusion protein (5). Affinity-purified RGST49 antibody was generated as described previously (45) and used at dilutions of 1:10 for immunofluorescence and 1:50 for immunoblotting. Hybridoma supernatant containing G49 monoclonal antibody specific for VP22 has been described previously (45). Monoclonal antibody specific for α-tubulin was obtained from Sigma and was used at a dilution of 1:500 for immunofluorescence. Anti-VP16 (1–21) monoclonal antibody was purchased from Transduction Laboratories and used at a dilution of 1:500. R220/5 polyclonal antiserum to VP13/14 (39) (a kind gift from David Meredith) was used at a dilution of 1:500 (5). Anti-VHS polyclonal antiserum was a kind gift from Sully Read, University of Missouri-Kansas City, and was used at a dilution of 1:500. Anti-gD antibody 1103 was obtained from Goodwin Cancer Research Laboratories. Fluorescein isothiocyanate-conjugated anti-rabbit immunoglobulin G (IgG; heavy and light chains) IgG and Texas Red-conjugated anti-mouse IgG were purchased from Vector Laboratories (Santa Cruz, Calif.) and used at a dilution of 1:100 in 1% bovine serum albumin (BSA). Alexa568-conjugated highly cross-adsorbed anti-rabbit IgG (Molecular Probes A-11036) was used at a dilution of 1:250 in 1% BSA.
Indirect immunofluorescence and microscopy.
Vero, V49, V202, 14349, and 14320 cells were prepared for indirect immunofluorescence as previously described (45). Briefly, cells rinsed twice in PBS were fixed in 2.5% methanol-free formaldehyde (Polysciences, Inc.) for 20 min at room temperature, rinsed twice again in PBS, and permeabilized in 100% acetone at −20°C for 3 to 5 min. Infected cells were incubated for 16 h in 1% BSA (Sigma) supplemented with 10 μg of pooled human Ig (Sigma) per ml. The primary antibodies used for immunofluorescence studies were diluted as described above and added for 1 h. After extensive rinsing with PBS, the appropriate secondary antibody was added and incubated for 45 min. Finally, the cells were preserved in a 0.1% solution of Mowoil (Sigma) with 2.5% DABCO (Sigma) used as an antibleaching agent under a fresh coverslip and sealed with nail polish. Cells were visualized on an Olympus IX70/IX-FLA inverted fluorescence microscope, and images were acquired using a Sony DKC-5000 digital photo camera linked to a PowerMac G3 and processed through Adobe Photoshop version 4.0.
Infected whole-cell extracts.
Whole-cell extracts were prepared from approximately 106 cells in 140 mM NaCl–3 mM KCl–10 mM Na2HPO4–1.5 mM KH2PO4 (pH 7.5) (PBS) containing protease inhibitors [10 mM l-1-chlor-3-(4-tosylamido)-7-amino-2-heptanon-hydrochloride (TLCK), 10 mM l-1-chlor-3(4-tosylamido)-4-phenyl-2-butanone (TPCK), and 100 mM phenylmethylsulfonyl fluoride]. Extracts of the infected cells were pelleted by low-speed centrifugation and resuspending the pellet in PBS containing 1.0% Triton X plus protease inhibitors. Lysis by sonication was performed using a Bronson sonifier.
Analysis of viral replication kinetics.
Single-step growth curves were performed as follows. Approximately 106 Vero cells grown to 100% confluence in 25-cm2 flasks were incubated for 1 h with 5.0 PFU per cell, rinsed in PBS, and incubated at 37°C. At the appropriate times p.i., stocks were prepared from each flask and titered in duplicate on Vero cells. Each point on the resulting growth curve (see Fig. 6) represents the average titer calculated from three complete independent experiments. The stocks of RF177 and HSV-1(F) used for the growth curves were prepared in Vero cells.
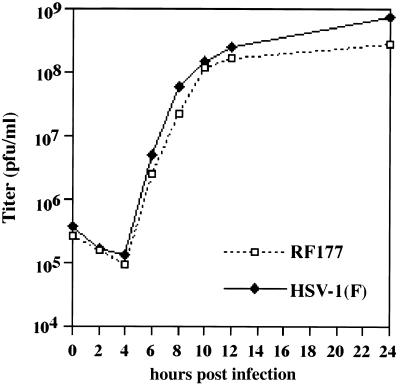
FIG. 6.
HSV-1(F) and RF177 single-step growth curves in Vero cells. Confluent monolayers of Vero cells were infected at an MOI of 5.0 PFU per cell as described in Materials and Methods. At 0, 2, 4, 6, 8, 10, 12, and 24 h p.i., stocks were prepared and counted in duplicate on Vero cells. Each point represents the average of three independent experiments. HSV-1(F) growth is indicated by a solid line and solid diamonds, and RF177 growth curve is shown as a dotted line and open squares.
Giemsa staining and determination of plaque size on Vero cells.
Confluent monolayers of Vero cells in 25-cm2 flasks (approximately 106 cells) were infected with 80 PFU of virus for 48 h in DMEM supplemented with 5% NBCS containing 10 μg of human Ig per ml (Sigma), as described above, until well-formed and isolated plaques arose. Cells were rinsed twice in PBS, fixed in methanol for 5 min at room temperature, and stained for 20 min using KaryoMAX Giemsa stain (Gibco-BRL) diluted 1:10 in distilled H2O. The diameters of 50 random RF177 or HSV-1(F) plaques were measured on an Olympus CK-2 inverted light microscope. In control experiments, plaques were also fixed for indirect immunofluorescence and stained with anti-gD monoclonal antibody.
Preparation of purified [32P]orthophosphate-labeled virions.
Purified [32P]orthophosphate-labeled virions were isolated as follows. Approximately 2 × 108 Vero cells were infected with RF177 or HSV-1(F) at 5.0 PFU per cell. Infected cells were incubated in HPO4-depleted DMEM (ICN) supplemented with 100 μCi of [32P]orthophosphate between 8 and 14 h p.i., and cells were incubated for an additional 10 h in DMEM plus 5% FBS. At 24 h p.i., extracellular virus was separated from the medium in a Beckman swinging-bucket SW27 rotor and L7 high-speed centrifuge (90 min at 46,000 × g), resuspended in 500 μl of 1 mM PO4 buffer (19 mM NaH2PO4, 81 mM Na2HPO4 [pH 7.4]), and sonicated for 5 s. Cytoplasmic virus (53) was purified as previously described (5). Briefly, cells separated from extracellular medium were rinsed once in PBS and incubated on ice for 10 min in 1.0 ml of PO4 buffer prior to being treated four times using pestle B (loose) in a Dounce homogenizer. Sucrose was added to a final concentration of 0.25 M, and nuclei were removed after a 10-min low-speed spin at 4°C. The cytoplasmic fraction was centrifuged at 100,000 × g for 1 h in a swinging-bucket SW55 rotor to remove large cellular debris. This partially clarified cytoplasmic extract was sonicated for 5 s. Cytoplasmic and extracellular virions were loaded onto separate dextran T10 (Pharmacia) gradients prepared in PO4 buffer (10 to 30%, wt/vol) and centrifuged for 1 h at 72,000 × g in a swinging-bucket SW40Ti rotor. Intact virion particles were removed, and urea was added to a final concentration of 0.5 M, sonicated, and pelleted in a swinging-bucket SW55 rotor at 180,000 × g for 30 min. Both preparation strategies (cytoplasmic and extracellular) yielded approximately 106 PFU of virion stock per ml.
Denaturing gel electrophoresis and immunoblotting.
The protein concentrations of infected cell extracts were determined using a modified Bradford assay (Bio-Rad) according to the manufacturer's specifications. For purified virions, protein concentrations were determined using a Lowry assay (Bio-Rad DC reagent) after solubilizing virions in 10% SDS; it should be noted that this assay is approximately fivefold less sensitive than the Bradford protocol. Equal amounts of infected-cell protein or virions were separated in SDS–15% polyacrylamide gels cross-linked with N,N′-diallyltartardiamide (DATD) (7), electrically transferred to nitrocellulose, and probed as indicated in the figure legends and text. Horseradish peroxidase-conjugated anti-rabbit or anti-mouse IgG (Amersham) secondary antibodies were diluted 1:1,000 in PBS and incubated with the blots for 1 h. Specific viral bands were detected following development with enhanced chemiluminescence reagents (Amersham) and autoradiography at 25°C using X-OMAT film (Eastman Kodak, Rochester, N.Y.). Alkaline phosphatase-conjugated goat anti-rabbit and anti-mouse IgG secondary antibodies were used at 1:500 in PBS and purchased from Southern Biotech (Birmingham, Ala.). Prestained molecular size markers (Gibco-BRL) were included in all acrylamide gels (lanes not shown in figures).
TEM.
For transmission electron microscopy (TEM) to directly observe intracellular and extracellular virus particles during replication, 2 × 108 Vero cells in roller bottles were infected with either RF177 or HSV-1(F) at an MOI of 0.01 PFU per cell for 48 h. Preparation of cells for TEM was done essentially as previously described (56). All chemical reagents were purchased from Electron Microscopic Sciences. Cells were scraped into medium, rinsed twice with PBS, and fixed for 3 h in 3.0% glutaraldehyde in 0.2 M cacodylate buffer (pH 7.3). Cells were rinsed twice again in PBS for 10 min and then postfixed for 1 h in 1% OsO4 in 0.2 M cacodylate buffer (pH 7.3). After incubating for 16 h in PBS, cells were dehydrated by sequential 10-min incubations in increasing concentrations of ethanol up to 100%. Cells were embedded in Epon at 58°C for 20 h. Sections (1 μm) were cut, stained with methylene blue and azure II, and examined by light microscopy prior to ultrathin (approximately 100 to 200 nm) sectioning. Ultrathin sections were stained with uranyl acetate-lead citrate and examined on a JEM 100CX transmission electron microscope. At least 30 cell profiles (54) from each infection were examined for characteristics of viral infection, including chromatin margination, nucleolar segregation, membrane duplication, and the presence of intra- and extracellular virions (50).
Computer analysis and imaging.
All restriction endonuclease mapping and determinations of predicted open reading frames, sizes, and amino acid contents were performed using DNA Strider version 1.2. Immunoblots, autoradiograms, and TEM negatives were digitized at 600 to 2,400 dots per inch using an AGFA Arcus II scanner linked to a Macintosh G3 PowerPC workstation. Raw digital images, saved as tagged image files (TIF) using Adobe Photoshop version 5.0, were organized into figures using Adobe Illustrator version 7.1. Grayscale or color prints of figures were obtained using a Codonics dye sublimation printer.
RESULTS
Our previous studies focused on the localization of VP22 during wild-type HSV-1 infection and on biochemically characterizing VP22 isolated from infected cells (45). The goal of this study is to determine whether a recombinant HSV-1 strain which does not synthesize full-length VP22 is capable of productive replication and assembly of infectious virion particles in cultured Vero cells. Initial attempts to target mutations to UL49 focused on deleting the region between the NcoI site at 106593 and the NcoI site at 105825, which lie 202 bp upstream and 566 bp downstream from the site of UL49 transcription initiation, respectively (35). This mutation was expected to remove enough of the 5′ end of UL49 to result in a nonfunctional gene. Several strategies used to delete this 768-bp NcoI fragment from the genome of HSV-1 were unsuccessful (data not shown). The following study presents the only HSV-1 strain published to date which does not synthesize full-length VP22.
VP22 localizes to the nucleus in stable VP22-expressing cell lines in the absence of other viral proteins.
Based on previous unsuccessful attempts to engineer mutations within the HSV-1 UL49 locus (4; L. E. Pomeranz and J. A. Blaho, unpublished results), we suspected that a complementing cell line may be necessary for the propagation of a recombinant HSV-1 defective for VP22 synthesis. Cell lines which stably express VP22 were produced from Vero (V49) and 143B (14349) cells after transfection with pJB133 as described in Materials and Methods. pJB133 contains HSV-1 sequence from the BglII site 359 bp upstream to the EcoRV site 341 bp downstream from the UL49 coding region (35) cloned into pBabeNeo. Vero and 143B cells were also transfected with pBabeNeo alone to generate V202 and 14320 for use as controls. Approximately 50 to 60% fewer pJB133-transfected cells survived selection than control cells transfected with vector alone, suggesting that VP22 may be toxic to cells. Neomycin-resistant colonies were cloned, and cells were tested for the presence of VP22 by indirect immunofluorescence (Fig. 2) and full-length protein by immunoblotting (data from selection by immunoblot not shown, but see Fig. 4A, lane 3).
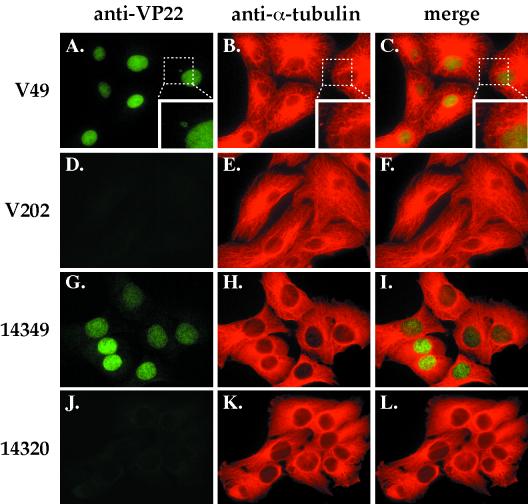
FIG. 2.
Indirect immunofluorescence of stable VP22-expressing (V49 and 14349) and control neomycin-resistant (V202 and 14320) cell lines doubly labeled with antibodies specific for VP22 and α-tubulin. Cells were fixed, permeabilized, and stained with affinity-purified RGST49 and monoclonal antibody to α-tubulin as described in Materials and Methods. (A, D, G, and J) VP22 staining; (B, E, H, and K) α-tubulin staining; (C, F, I, and L) merged images. A larger view of the area designated by the dotted boxes in A, B, and C is shown in the inset at the lower right of each image.
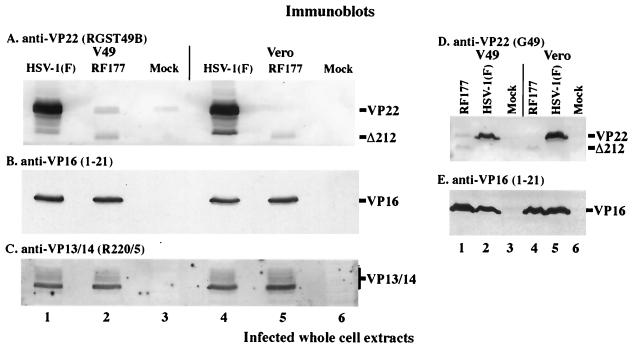
FIG. 4.
Immunoblots of whole-cell extracts prepared from infected V49 and Vero cells. Whole-cell extracts were prepared from HSV-1(F)-, RF177-, or mock-infected V49 and Vero cells. Equal amounts of protein from each extract were separated in 15% DATD–acrylamide, transferred to nitrocellulose, and probed with affinity-purified RGST49 (A), anti-VP16 (B and E), anti-VP13/14 (C), and G49 (D) antibodies as described in Materials and Methods. Secondary antibodies were conjugated to alkaline phosphatase (A and D) or horseradish peroxidase (B, C, and E). The locations of full-length VP22 synthesized during HSV-1(F) infection and from VP22-expressing V49 cells (VP22) and the faster-migrating, truncated form of VP22 (Δ212) expressed during RF177 infection are indicated to the right of panels A and D. VP16 marks the location of VP16 protein in panels B and E, and a vertical bar labeled VP13/14 marks the glycosylated forms of VP13/14 detected by the R220/5 antibody in panel C.
Transient expression of VP22 was reported to stabilize microtubules (16). Thus, it was of interest to determine whether the expression of VP22 in our cell lines would have an effect on the cellular microtubule network. For indirect immunofluorescence, V49, V202, 14349, and 14320 cells grown on coverslips were fixed, permeabilized, and stained with affinity-purified RGST49 antibody specific for VP22 and a monoclonal antibody specific for α-tubulin as described in Materials and Methods. VP22 was detected in the nuclei of V49 and 14349 cells but not V202 and 14320 controls (Fig. 2, compare A and C with D and F; G and I with J and L). No significant differences in microtubule organization were observed in the VP22-expressing cells compared with controls (Fig. 2B, C, E, F, H, I, K, and L). Interestingly, round cytoplasmic bodies exhibiting VP22 immune reactivity could be observed in a small percentage of V49 cells (inset, Fig. 2A). Similar observations have been reported during transient expression of a VP22-GFP fusion protein in COS-1 cells (15).
From these results, we conclude the following. VP22 can be stably maintained in at least two unique cell types; V49 is derived from a monkey kidney cell line, and 14349 is derived from a human osteosarcoma line. Furthermore, VP22 localization in the nuclei of the two cell types examined does not require the presence of other viral proteins. Finally, VP22 synthesis in V49 and 14349 cells does not have an obvious effect on cellular microtubules, since neither colocalization of VP22 with microtubules nor any rearrangement of microtubules was observed in VP22-expressing cells. This may be due to the relatively low levels of VP22 synthesized in the V49 and 14349 cells, compared with the high levels observed during transient transfection (16).
Generation of recombinant virus RF177, which synthesizes Δ212 and not full-length VP22.
In the generation of RF177, GFP production was used as a marker to screen for recombinant virus. The presence of GFP in HSV-1-infected cells does not have any deleterious effects on viral replication (23). The mutation of the UL49 locus was generated as follows (Fig. 1). HSV-1 sequence from the PstI site at 102653 to the BamHI site at 107022 was inserted into pGEM3Z (Promega) to produce pJB175. The UL49 gene in pJB175 was disrupted from the NsiI site at 105760 to the XhoI site at 105530 by insertion of the 1,344-bp NsiI-XhoI fragment of pEGFP-C1 (Clontech) to yield pJB177. Introduction of the GFP expression cassette into the NsiI and XhoI sites in pJB177 resulted in the generation of a stop codon after amino acid 212 in VP22. As described in Materials and Methods, R2507 contains a tk expression cassette 200 bp upstream from the start site of UL48 (Fig. 1, line 4) (46). Recombination between pJB177 and R2507 results in removal of the tk cassette between UL48 and UL49 and the insertion of the GFP expression cassette into the 3′ end of UL49 (Fig. 1, line 5). Thus, the resulting virus, RD177, should express GFP and not tk. This strategy left 45 bp at the 3′ end of the UL49 gene (designated by a solid rectangle in Fig. 1, lines 5 and 6) and inserts a stop codon after the histidine at amino acid 212. pJB177 and R2507 DNA were cotransfected into V49 cells, and RD177 was selected for loss of tk activity in 14349 cells. GFP-positive plaques were purified twice, and the insertion of the GFP cassette into BamF was confirmed by Southern hybridization (data not shown). The tk locus of RD177 was subsequently repaired to wild type by cotransfection of RD177 genomic DNA with pJB142 to generate virus RF177 (Fig. 1, line 6). The integrity of the UL49 and UL23 loci in RF177 was confirmed by Southern hybridization (Fig. 3). Viral DNAs were digested with EcoRV and probed with BamF or BamQ as described in Materials and Methods.
FIG. 3.
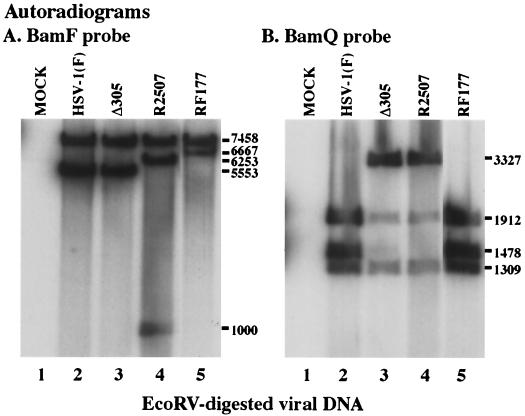
Autoradiographic images of Southern hybridizations using the BamF (A) and BamQ (B) probes. EcoRV-digested DNA from mock- (lane 1), HSV-1(F)- (lane 2), Δ305- (lane 3), R2507- (lane 4), or RF177- (lane 5) infected V49 cells was separated in a 0.8% agarose gel and probed with the BamF fragment, containing the UL49 gene, or the BamQ fragment, containing the UL23 gene, as described in Materials and Methods. The size of EcoRV fragments hybridizing to BamF or BamQ probes appear to the right as molecule weights. DNA from several independent RF177 plaques was analyzed, and lane 5 shows the result for the virus isolate used throughout our studies.
The BamF probe hybridizes with a 7,458-bp band in DNA purified from all viruses (Fig. 3A) and to a 5,553-bp band in HSV-1(F) and Δ305, as expected (Fig. 3A, lane 2 and 3). The insertion of the 1,800-bp tk expression cassette introduces two additional EcoRV sites into R2507 (Fig. 1, line 4). This increases the size of the 5,553-bp fragment to 6,253 bp and generates a new 1,000-bp band (Fig. 3A, lane 4). The BamF probe recognizes a 6,667-bp fragment in RF177 (Fig. 3A, lane 5), consistent with a 230-bp deletion in the UL49 gene and insertion of the 1,344-bp GFP expression cassette in this virus (Fig. 1, line 6).
The BamQ probe hybridizes with 1,912- and 1,309-bp bands in all viral DNAs (Fig. 3B) and to a 3,327-bp band in Δ305 and R2507, as expected (Fig. 3B, lane 3 and 4). The BamQ probe recognizes a 1,478-bp fragment in RF177 (Fig. 3B, lane 5) which is also detected in the wild-type HSV-1(F) DNA (Fig. 3B, lane 2). We did not detect any contamination of RF177 (Fig. 3B, lane 5) with its parental strain RD177 or with strain R2507, used in the initial recombination experiment. From these results, we conclude that RF177 possesses the desired 1,344-bp GFP insertion in the UL49 locus (Fig. 1). In addition, RF177 contains a wild-type tk (UL23) locus.
RF177 expresses low levels of modified Δ212 and does not require V49 cells for late-gene expression.
The next series of experiments examine the biochemistry and subcellular distribution of VP22 synthesized during RF177 infection. To study the accumulation of late viral proteins during RF177 replication, V49 and Vero cells were mock infected or infected with RF177 or HSV-1(F) at an MOI of 1.0 for 24 h. Whole-cell extracts were prepared from infected cells, and equal amounts of protein were separated in 15% DATD–acrylamide gels, transferred to nitrocellulose, and probed with antibodies specific for VP22 (affinity-purified RGST49 or G49), VP16 [VP16 (1-21)], and VP13/14 (R220/5) as described in Materials and Methods. In these studies, VP22 refers to full-length protein synthesized by V49 cells and HSV-1(F), while Δ212 indicates the truncated protein synthesized by RF177. The results are presented in Fig. 4.
Mock-infected V49 cells synthesized VP22 (Fig. 4A, lane 3), which migrated at the same location as full-length VP22 produced during HSV-1(F) infection (Fig. 4A, lane 1). An increase in the amount of cell-expressed VP22 was observed when V49 cells were infected with RF177 (Fig. 4A, lane 2). However, the amount of VP22 synthesized in both mock- and RF177-infected V49 cells did not approach the level of VP22 synthesized during HSV-1(F) infection (Fig. 4A, compare lane 1 with lanes 2 and 3). In addition, at least two electrophoretic forms of full-length VP22 were observed in mock- and RF177-infected V49 cells (Fig. 4A, lane 2 and 3). Multiple forms of VP22 have previously been observed in extracts prepared from infected (5, 20, 45) and transfected (19, 20) cells. Furthermore, it has been demonstrated that the slowest-migrating form of full-length VP22 is sensitive to alkaline phosphatase treatment (20). The slower-migrating forms were dependent on cellular kinases, and in vivo [32P]orthophosphate labeling indicated that the main phosphorylation sites mapped to the amino portion, including serines 71, 72, and 73, with minor phosphorylation detected in the carboxy portion (19, 20). In contrast to mock-infected V49 cells, fast-migrating full-length VP22 was the most abundant form observed during RF177 infection (Fig. 4A, compare lanes 2 and 3). These results do not exclude the possibility that the fast isoform of full-length VP22 seen during RF177 infection has the same mobility as that seen in uninfected V49 cells.
A novel band migrating at approximately 32,000 Da was observed in RF177-infected (Fig. 4A, lane 2) but not mock- or HSV-1(F)-infected V49 cells. The presence of this band is consistent with the fact that insertion of the GFP cassette in RF177 preserves 636 bp in the 5′ coding region UL49 (Materials and Methods). RF177 is expected to synthesize the amino-terminal 212 amino acids of VP22, and this truncated protein is therefore termed Δ212. At least two forms of Δ212 are observed in RF177-infected V49 cells, and the faster-migrating form of Δ212 appears to be more abundant than the slower form (Fig. 4A, lane 2). It should be noted that the predicted size of full-length VP22 is 32,000 Da (35), but the protein consistently migrates closer to 38,000 Da (53). Many HSV-1 proteins, e.g., ICP4 (8), show an inconsistency between their predicted and observed sizes in SDS-PAGE, presumably due to their high proline content or the presence of posttranslational modifications (7). The predicted size of Δ212 is close to 24,000 Da. A consequence of the carboxy-terminal truncation is that the percentage of total protein mass contributed by proline residues is changed from 11% in full-length VP22 to 13% in Δ212 (Materials and Methods). Thus, the higher percentage of proline residues in Δ212 may increase the difference between its predicted and observed masses.
To analyze protein synthesis during RF177 replication in the absence of full-length VP22, infections were performed in Vero cells (Fig. 4A, lane 6). As expected, full-length VP22 was detected following HSV-1(F) infection but not during RF177 (Fig. 4A, lane 5) or mock (Fig. 4A, lane 6) infection of the (non-VP22-expressing) Vero cells. The Δ212 protein was observed during RF177 infection of Vero cells (Fig. 4A, lane 5). However, the accumulation of faster-migrating Δ212 was slightly reduced compared to that observed during RF177 infection of V49 cells (Fig. 4A, compare lane 5 with lane 2). This finding is significant since it is the first indication that RF177 does not require VP22-complementing cells to produce Δ212 protein. However, as with the V49 cells, the amount of Δ212 detected following RF177 infection was substantially less than that of VP22 in the HSV-1(F)-infected cells.
Due to the strikingly low levels of Δ212 observed in RF177-infected V49 and Vero cells, we next compared the accumulation of two representative late proteins during HSV-1(F) and RF177 infection. The membrane in Fig. 4A was reprobed with antibodies specific for the VP16 (Fig. 4B) and VP13/14 (Fig. 4C) tegument proteins. Nearly identical levels of VP16 and VP13/14 were observed in RF177- (lane 2 and 5) and HSV-1(F)- (lane 1 and 4) infected V49 and Vero cells. As expected, no mock-infected cell proteins exhibited reactivity with the anti-VP16 or VP13/14 antibodies (Fig. 4B and C, lanes 3 and 6).
To confirm that Δ212 protein levels were reduced compared to the amount of full-length VP22 expressed during HSV-1(F) infection, a similar immunoblot was probed using G49 anti-VP22 monoclonal antibody (Fig. 4D). G49 is directed against an epitope on the amino-terminal portion of VP22 (L. E. Pomeranz, L. Gillim, and J. A. Blaho, unpublished results). In addition, the majority of epitopes recognized by RGST49 antibodies are located in the VP22 amino terminus (L. E. Pomeranz, L. Gillim, and J. A. Blaho, unpublished results). Strong immune reactivity of G49 was observed with HSV-1(F)-derived full-length VP22 (Fig. 4D, lanes 2 and 5), as expected (45). G49 reactivity with Δ212 (Fig. 4D, lane 1 and 4) was nearly identical to that observed using RGST49 (Fig. 4A, lane 2 and 5). No reactivity of G49 with mock-infected proteins was detected, as expected (Fig. 4D, lane 3 and 6). As above, similar amounts of VP16 were present in RF177- and HSV-1(F)-infected cell extracts (Fig. 4E). This finding supports the hypothesis that the reduced Δ212 signal in Fig. 4A results from less protein and not from reduced reactivity of the truncated protein with RGST49 polyclonal antibody.
From these results, we conclude the following. (i) Modification of full-length VP22 expressed from stably transfected V49 cells occurs in the absence of other viral proteins. An equal ratio exists between the low- and high-mobility forms of VP22 in uninfected V49 cells. However, an increase in a faster-migrating form of full-length VP22 expressed by V49 cells is observed during RF177 infection. (ii) VP22-complementing cells are not required for Δ212 synthesis, since it is produced during RF177 infection of Vero cells. (iii) The ability of RF177 to synthesize at least two late viral proteins is not dependent on the presence of full-length VP22 because similar amounts of VP16 and VP13/14 are detected in RF177- and HSV-1(F)-infected Vero cells. The wild-type levels of these late proteins further indicate that the mutation of the UL49 locus in RF177 does not significantly affect expression from either the UL48 or UL47 locus. Together, these results introduce an HSV-1 system which can be used to study viral replication in the absence of full-length VP22.
Δ212 localizes to Vero cell nuclei during RF177 infection.
The next experiment was designed to examine the subcellular distribution of Δ212 during RF177 infection. Vero and V49 cells, grown on coverslips, were mock infected or infected with RF177 or HSV-1(F) at an MOI of 1.0. At 24 h p.i., cells were fixed for indirect immunofluorescence and stained with antibody specific to VP22 (RGST49) as described in Materials and Methods. The results (Fig. 5) were as follows.
FIG. 5.
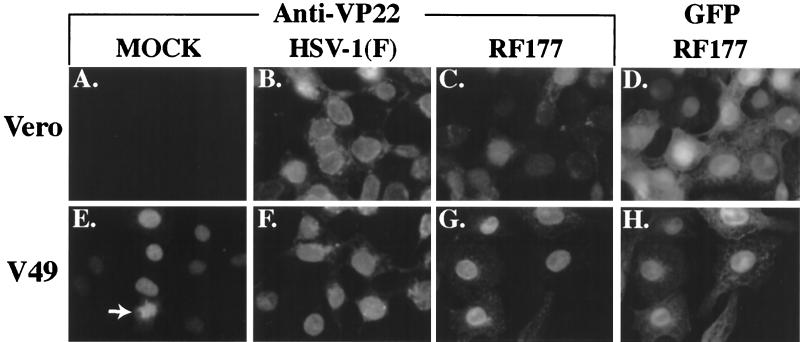
Indirect immunofluorescence and autofluorescence of infected Vero and V49 cells. Vero (A to D) and V49 (E to H) cells were mock infected (A and E) or infected with HSV-1(F) (B and F) or RF177 (C, D, G, and H) at an MOI of 1.0 for 24 h. Cells were then fixed, permeabilized, and stained with affinity-purified RGST49 antibody specific for VP22 (A to C and E to G). GFP autofluorescence of RF177-infected cells is shown in D and H. The arrow in E indicates VP22-associated chromatin in a dividing, mock-infected V49 cell.
VP22 synthesized during HSV-1(F) infection of Vero cells was detected almost exclusively in the nucleus at 24 h p.i. (Fig. 5B), consistent with our original finding that VP22 exhibits a nuclear distribution late in infection (45). Δ212 also exhibited nuclear staining in RF177-infected Vero cells (Fig. 5C). However, the Δ212 staining intensity was low compared with the levels of full-length VP22 seen during HSV-1(F) infection (compare Fig. 5B and C), consistent with our immunoblotting results (Fig. 4A, compare lanes 4 and 5). The immunoblot data presented in Fig. 4 indicate that only Δ212, not VP22, is observed during RF177 infection of Vero cells (Fig. 4A, lane 5). Therefore, the RGST49 immunofluorescence observed in Fig. 5C results exclusively from Δ212. Autofluorescence of GFP produced by RF177 was observed in both the cytoplasm and nuclei of infected Vero cells (Fig. 5D). As expected, no VP22 immune reactivity was detected in mock-infected Vero cells (Fig. 5A).
Mock-infected V49 cells also exhibited VP22 immune reactivity in the nucleus (Fig. 5E). The nuclei in uninfected V49 cells appeared slightly smaller than the nuclei of either HSV-1(F) or RF177-infected cells (compare Fig. 5E with 5F to H). Similar differences in the size of mock- and infected-cell nuclei have been described previously and are the consequence of the effect of viral replication on chromosomal organization (2, 50). In addition, mitotic V49 cells were observed, and these harbored VP22-immune-reactive chromatin (Fig. 5E, arrow), indicating that formaldehyde fixation and acetone permeabilization do not dislodge mitotic cells from coverslips (15). No differences in VP22 staining were observed in HSV-1(F)-infected Vero and V49 cells (compare Fig. 5B and F). GST49-reactive staining in the nuclei of RF177-infected V49 cells was significantly stronger than that observed in RF177-infected Vero cells (compare Fig. 5C and G). This is due to the combination of Δ212 expressed during RF177 infection and full-length VP22 expressed by V49 cells and can be readily confirmed by immunoblotting experiments (Fig. 4A, compare lanes 2 and 5). As with Vero cells, GFP produced during RF177 infection was detected in both the cytoplasm and nuclei of V49 cells.
From these results, we conclude the following. (i) Δ212 localizes to the cell nucleus during infection, inasmuch as both Δ212 and full-length VP22 are present almost exclusively in the nuclei of Vero cells at 24 h p.i. Thus, while the observed intensities of Δ212 and VP22 were different, the subcellular localizations of the two proteins were identical. (ii) The carboxy-terminal region of VP22 is not required for its nuclear localization, since Δ212, which lacks the last 89 amino acids of VP22, is detected in the nuclei of infected Vero cells. Recent data from at least two independent groups suggested that the carboxy half of VP22 was associated with its nuclear retention in uninfected cells (17, 22). We therefore considered the possibility that Δ212 might be absent from the nucleus during RF177 infection, but this was not the case. En suite with our previous data (45), (iii) the detection of VP22 in the nucleus during HSV-1(F) infection does not require that the infection by synchronized.
Kinetics of RF177 growth is nearly identical to that of HSV-1(F) in high-multiplicity infections of Vero cells.
The results in Fig. 4 showed that the late tegument proteins VP16 and VP13/14 produced during RF177 infection accumulate to the same extent in (non-VP22-expressing) Vero cells as those of wild-type virus at 24 h p.i. The goal of this experiment was to document the kinetics of RF177 replication. Vero cells were infected with either RF177 or HSV-1(F) at an MOI of 5.0 PFU per cell, and single-step growth curves were determined as described in Materials and Methods. We wish to note that the stock of RF177 used in these growth experiments was prepared in Vero (not V49) cells. The titer values at each time point presented in Fig. 6 are the averages of three independent experiments.
The growth kinetics of RF177 were nearly indistinguishable from that of HSV-1(F) up to 12 h p.i. However, at the 24-h time point, RF177 titers consistently exhibited an approximately 0.5 log reduction compared to HSV-1(F). The average titer of RF177 at 24 h p.i. was 2.8 × 108 PFU per ml, compared to 7.3 × 108 PFU per ml for HSV-1(F). To determine whether this effect at late times was dependent on the MOI, we performed a low-MOI (0.5 PFU per cell) infection (data not shown). Under these conditions, the difference between RF177 and HSV-1(F) titers at the 24-h time point increased to nearly 1 log.
From the results presented in Fig. 4 and 6, we conclude that RF177 productively replicates in Vero cells, inasmuch as VP16 and VP13/14 levels and RF177 growth kinetics are nearly identical to those of wild-type HSV-1(F). While we observe minor differences between the yields of RF177 and HSV-1(F) at late times in Vero cells, significant amounts of infectious RF177 virus are produced. Thus, these findings are not consistent with a growth defect in RF177. Accordingly, we must conclude that full-length VP22 is not required for productive HSV-1 replication in Vero cells.
RF177 exhibits a reduced plaque size compared with HSV-1(F) following growth in cultured Vero cells.
The results described above suggest that differences in the growth properties of RF177 and HSV-1(F) are only discernable at late infection times or under conditions of low-multiplicity infection. While the differences in RF177 and HSV-1(F) single-step growth kinetics were minor, the lower yields of RF177 obtained with less input virus suggest a decreased replication efficiency during multiple rounds of infection. Consistent with this model, we routinely observed that RF177 plaques appeared smaller than those produced by HSV-1(F). To quantitate these differences, Vero cells were infected at a low multiplicity with either RF177 or HSV-1(F) for 48 h and stained with Giemsa, and the sizes of individual plaques were measured as described in Materials and Methods. Additionally, RF177 and HSV-1(F) plaques were fixed for indirect immunofluorescence and stained with monoclonal antibody specific for gD. Two representative Giemsa-stained and one anti-gD-stained plaque from each virus are shown in Fig. 7.
FIG. 7.
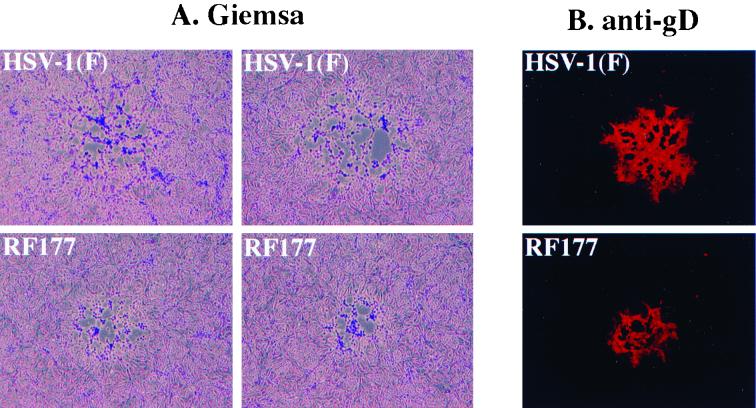
Phase contrast (A) and indirect immunofluorescence images (B) of HSV-1(F) and RF177 plaque morphologies in Vero cells. Confluent monolayers of Vero cells were infected at low multiplicity for 2 days, fixed, and stained with Giemsa (A) or anti-gD antibody (B) as described in Materials and Methods. Two representative Giemsa-stained and one gD-stained plaque of HSV-1(F) and RF177 are shown.
The plaque morphologies of RF177 and HSV-1(F) were essentially identical following either Giemsa (Fig. 7A) or anti-gD (Fig. 7B) staining, since neither virus was highly syncytial and both had only a limited number of multinucleated cells. However, the RF177 plaques were considerably smaller than those of HSV-1(F). Measurements of plaque diameters indicated that the RF177 plaques were actually 62% of the size of the plaques formed by HSV-1(F). From the size reduction of the RF177 plaques, we conclude that one consequence of the loss of the carboxy-terminal region of VP22 is that RF177 appears to have a deficiency in its ability to spread in Vero cells. RF177 plaque size increased when the virus was plated on V49 cells, indicative of VP22 complementation, but the diameters were not as large as those with HSV-1(F) (L. E. Pomeranz and J. A. Blaho, unpublished results). Likely this is due to the small amount of VP22 produced in RF177-infected V49 cells compared to the level in HSV-1(F)-infected cells (Fig. 4). Due to the absence of a syncytial phenotype with RF177, it is unlikely that the spreading defect is at the level of membrane fusion. Our finding of a defect in Vero cell-to-cell spread associated with RF177 is consistent with its tendency to generate lower virus yields than HSV-1(F) following multistep growth experiments.
Virion phosphoproteins and tegument proteins VP13/14, VP16, and VHS but not VP22 or minor virion proteins are efficiently incorporated into purified RF177 virions.
The results above indicated that no full-length VP22 could be detected in whole-cell extracts of RF177-infected Vero cells (Fig. 4), and a consequence of this seems to be reduced cell-to-cell spreading of RF177 (Fig. 7). The goal of the following set of experiments was to determine whether the loss of VP22 in infected cells has any effect on the formation of RF177 virions. Since VP22 is one of the most highly labeled phosphoproteins observed in purified HSV-1 particles (24), we set out to document the phosphorylation patterns of RF177 virion proteins. Vero cells were infected with RF177 or HSV-1(F) in the presence of [32P]orthophosphate, and radiolabeled virions were purified at 24 h p.i. from either cytoplasmic extracts or growth medium as described in Materials and Methods. Virion proteins were separated in 15% DATD–acrylamide, transferred to nitrocellulose, and probed with antibodies specific for VP13/14, VP16, VHS, and VP22 prior to autoradiography. In control experiments, electrophoretically separated proteins derived from extracellular virions were visualized by silver staining. Cellular (cytoplasmic) and extracellular (growth medium) virions were isolated and compared in order to confirm the integrity of our preparations, since both strategies yielded approximately the same amount of infectious virion particles per microliter of virion stock. The results (Fig. 8) were as follows.
FIG. 8.
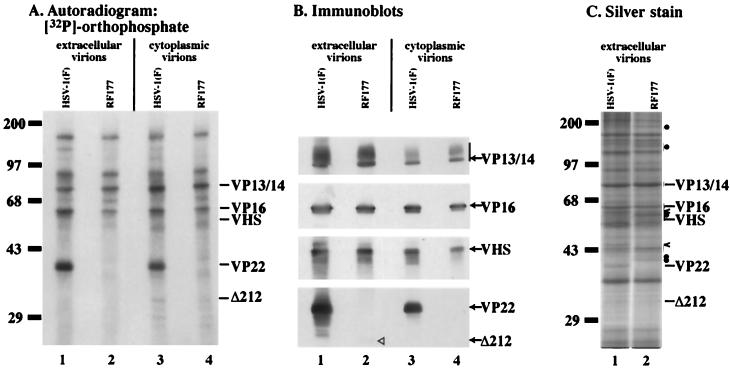
Autoradiographic images (A), immune reactivities (B), and silver staining patterns (C) of [32P]orthophosphate-labeled HSV-1(F) and RF177 virion proteins. Vero cells were infected with HSV-1(F) or RF177 in phosphate-free medium containing 100 μCi of [32P]orthophosphate, and radiolabeled virions were purified from the extracellular growth medium (lane 1 and 2) and the cytoplasm (lane 3 and 4). Virion proteins were separated in a 15% DATD–acrylamide gel, transferred to nitrocellulose, exposed to film for autoradiography, probed with antibodies specific for the indicated tegument proteins, or stained with silver as described in Materials and Methods. The locations of VP13/14, VP16, VHS, full-length VP22 (VP22), and Δ212 are indicated on the right. A vertical bar labeled VP13/14 marks the glycosylated forms of VP13/14. The locations of prestained molecular weight markers are indicated on the left of panels A and C. In panel C, carets mark HSV-1(F)-specific virion bands, while solid circles mark RF177-specific bands.
As expected, HSV-1(F) proteins in virions purified from either the cell cytoplasm (Fig. 8A, lane 3) or growth medium (Fig. 8A, lane 1) exhibited previously defined [32P]orthophosphate labeling profiles (24). A highly labeled band with the same mobility as full-length VP22 was observed in both preparations of HSV-1(F) virions. However, this phosphoprotein was absent from extracellular (Fig. 8A, lane 2) and cytoplasmic (Fig. 8A, lane 4) RF177 virions. Additionally, no radiolabeled RF177-specific bands were observed in the predicted location of Δ212 mobility. No other significant differences in labeled proteins were observed between HSV-1(F) and RF177.
The immune reactivities of VP13/14, VP16, VHS, and VP22 were identical in extracellular (Fig. 8B, lane 1) and cytoplasmic (Fig. 8B, lane 3) HSV-1(F) virion preparations. Extracellular and cytoplasmic RF177 virions exhibited staining patterns similar to those of HSV-1(F) with VP13/14, VP16, and VHS (Fig. 8B, lane 2 and 4). Comparable levels of slow-migrating, glycosylated VP13/14 (39) were observed in both types of HSV-1(F) and RF177 virion preparations. A small but detectable amount of Δ212 was seen in extracellular RF177 (Fig. 8B, lane 2). Δ212 could not be detected in the RF177 cytoplasmic virion preparation (Fig. 8B, lane 4). Minor differences in the total protein amounts (Fig. 8B, lanes 3 and 4 compared to lanes 1 and 2) likely hindered our ability to observe the nonabundant Δ212 protein in the cytoplasmic preparation.
Consistent with the immunostaining results in Fig. 8B, silver staining analyses demonstrated that HSV-1(F) and RF177 produced similar amounts of the major virion proteins, including VP13/14 and VP16 (Fig. 8C). At least three bands in HSV-1(F) virions were not observed in RF177, including a band with the same mobility as VP22. Conversely, at least five bands could be detected in RF177 virions which were not present in HSV-1(F). We have not determined specifically which virion proteins are redistributed between RF177 and HSV-1(F).
From these results, we conclude the following. (i) Infectious virion particles can be produced in the absence of full-length VP22. Loss of full-length VP22 does not significantly affect (ii) RF177 egress, since substantial quantities of infectious virus were purified from the extracellular growth medium or (iii) the phosphorylation profile of other proteins in RF177 particles. (iv) Full-length VP22 is not required for optimal incorporation and processing of the VP13/14, VP16, and VHS tegument phosphoproteins. (v) The truncated form of VP22, Δ212, is incorporated into infectious virus particles which are capable of exiting cells. While we did not observe radiolabeled Δ212, the low levels of the protein in virions make it difficult to assess the significance of this result. However, we cannot rule out the possibility that, even at low levels, the amino-terminal portion of VP22, i.e., Δ212, may have an important function during infection, including involvement in tegument assembly or viral egress. Finally, (vi) loss of full-length VP22 from RF177 appears to be compensated for by alterations in the distribution of several unknown minor virion proteins. It is likely that the relative amounts of existing virion proteins changed between HSV-1(F) and RF177. While RF177 may contain less of at least two proteins observed in HSV-1(F), it has an apparent increase in the amounts of at least five proteins. The striking feature of RF177 is that the distribution of the most abundant virion proteins, including VP13/14 and VP16, does not change.
RF177 virion assembly and structure observed by TEM are identical to those of HSV-1(F).
No significant differences were observed in the profile of tegument VP13/14, VP16, and VHS proteins from purified RF177 virions compared to wild-type HSV-1(F). However, we did detect differences in the distributions of various minor virion proteins between the two viruses. The goal of this experiment was to document the process of RF177 assembly at the electron microscopic level. Vero cells were infected with RF177 or HSV-1(F) (MOI of 0.01) for 48 h and then prepared for and examined by TEM as described in Materials and Methods. Representative fields from TEM analysis showing intracellular RF177 and extracellular RF177 and HSV-1(F) particles are presented in Fig. 9.
FIG. 9.
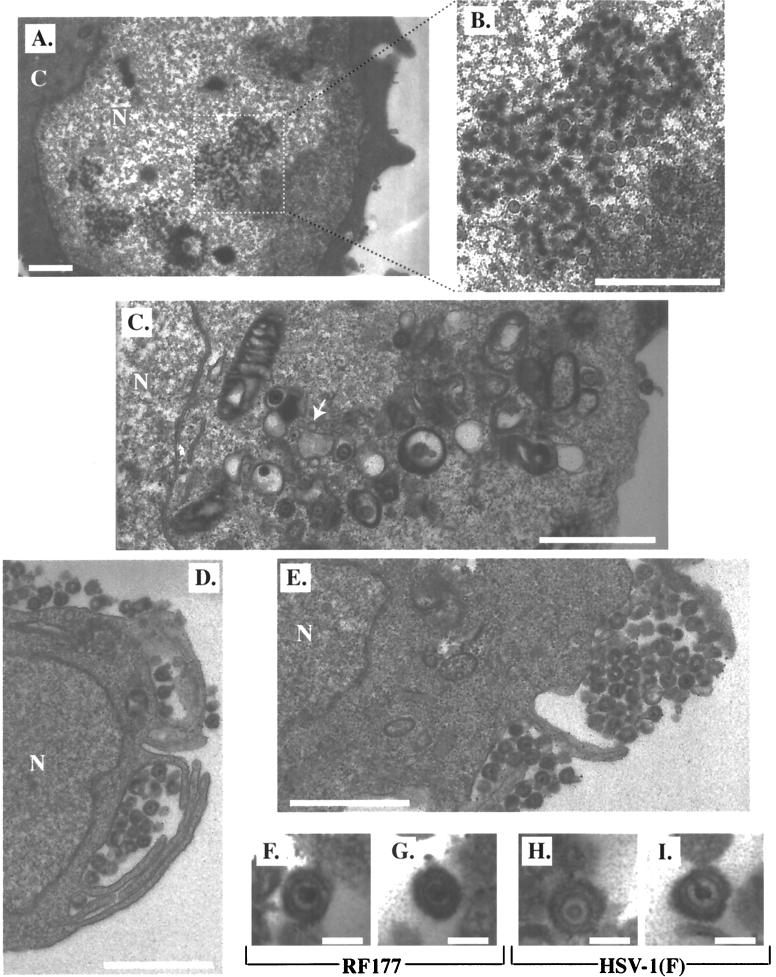
Electron microscopic images of infected Vero cells. Vero cells were infected with RF177 or HSV-1(F) at an MOI of 0.01 PFU per cell, fixed in glutaraldehyde at 48 h p.i., and prepared for electron microscopy as described in Materials and Methods. Representative sections of RF177-infected Vero cells are shown in panels A to E. Panel A is magnified ×10,000 (bar, 1 μm). Panel B is an enlargement of the area enclosed by the dotted square in panel A (bar, 1 μm). Panel C is magnified ×19,000 (bar, 1 μm); the white arrow indicates an empty capsid. Panels D and E are magnified ×19,000 (bar, 1 μm). Two representative RF177 and HSV-1(F) virus particles are shown in panels F and G and panels H and I, respectively. Panels F to I are magnified ×100,000 (bar, 100 nm). C, cytoplasm; N, nucleus.
Dense bodies in the nuclei of RF177-infected Vero cells were observed, and fully formed nucleocapsids accumulated in these regions (Fig. 9A). Upon closer examination, these nucleocapsids proved to contain darkly stained (electron-dense) regions in their centers, consistent with their containing HSV genomic DNA (Fig. 9B). Enveloped viruses were observed inside cytoplasmic vesicles (Fig. 9C). While we also observed naked RF177 capsids in the cytoplasm, their number did not differ from that seen in HSV-1(F)-infected Vero cells (data not shown). The inner regions of a few nucleocapsids in both the RF177 and HSV-1(F) preparations appeared to have sharply delineated borders and small areas devoid of electron-dense material which were unlike empty capsids (Fig. 9C). These particles likely lost their DNA during the thin-sectioning procedure. Enveloped viruses were observed within large vacuoles in the cytoplasm (Fig. 9C and D) or outside the plasma membrane of the cell (Fig. 9D and E). Finally, individual extracellular RF177 (Fig. 9F and G) and HSV-1(F) (Fig. 9H and I) virions were examined at ×100,000 magnification. No significant differences between the two viruses were observed at the ultramicroscopic level. The average diameters of both the RF177 and HSV-1(F) particles were identical (approximately 150 nm).
We conclude that RF177 capsid assembly in the nucleus and viral egress occur in a manner indistinguishable from those in HSV-1(F) by TEM analysis. Taking the results of Fig. 8 and 9 together, it appears that the lack of full-length VP22 is compensated for by other tegument proteins, not including VP13/14, VP16, or VHS, and the resulting infectious RF177 virion does not exhibit any assembly or structural defects compared to HSV-1(F) virus particles.
DISCUSSION
The aim of this study was to gain insight into the function of the VP22 protein during HSV-1 infection. We generated a recombinant virus which synthesizes low levels of truncated VP22 and examined its replication and competence in virion assembly. The significant features of our results are as follows.
(i) Modification of nuclear VP22 occurs in the absence of other viral proteins in cell lines which stably express VP22. This conclusion is based on two independent observations. First, VP22 appears to localize exclusively to the nucleus in two unique VP22-expressing cells types. Second, multiple electrophoretic forms of VP22 are detected upon immediate extraction of uninfected cells. This last result implicates a cellular function(s) in the generation of these multiple forms of VP22. We previously reported that fast-migrating forms of VP22 accumulate in cells late in HSV-1 infection (45). Infection of the VP22-expressing cells with RF177, which does not express full-length VP22, identified changes in VP22 modification associated with viral replication. Since the amount of fast-migrating, cell-derived VP22 increases only after RF177 infection (Fig. 4), this accumulation is regulated by a virus-specific function. Taken together, these results indicate that a combination of cellular and viral factors evoke the complete spectrum of VP22 modified forms during productive infection. In addition, it is conceivable that VP22 and Δ212 may associate during the course of RF177 infection of V49 cells. At this time, we do not know whether the multiple forms of VP22 arise from differential transcription, translation, or posttranslational modification.
One possible explanation is that multiple forms of VP22 result from different levels of phosphorylation. VP22 can be phosphorylated on serine residues in vitro, and it has been proposed that the high-mobility (fast) form of VP22, which is packaged into virion particles (5), is dephosphorylated (19, 20, 41). The incorporation of [32P]orthophosphate into virion-associated full-length VP22 reported in this study (Fig. 8) and by a number of other researchers (5, 24, 32) indicates that the fast-migrating form of VP22 still possesses some type of phosphate-containing modification. VP22 can be nucleotidylated (5, 6) and mono(ADP-ribosyl)ated (5, 48). These modifications present alternatives to simple phosphorylation and may contribute to the observed phosphate incorporation of virion-associated VP22. While modified forms of VP22 have been described using transient-expression techniques (20), this is the first report that multiple forms of VP22 are maintained in cells which stably express VP22.
(ii) RF177 productively infects Vero cells in the absence of full-length VP22. This conclusion is based on the observation that RF177 and HSV-1(F) exhibit identical single-step growth kinetics and produce virion particles of the same size. This finding is consistent with the report that the UL49 homologue in bovine herpesvirus 1 is dispensable for viral replication in cultured cells (34). However, recent findings suggest that the bovine herpesvirus 1 VP22 may be functionally distinct from its HSV-1 homologue (26). It is not known why the levels of Δ212 produced by RF177 are so low. Two possible explanations are that Δ212 is not properly polyadenylated or that the truncated protein is much less stable.
(iii) The truncated segment of VP22, Δ212, localizes to the nucleus during RF177 infection of Vero cells. While the intensity of Δ212 observed by indirect immunofluorescence after RF177 infection was not as strong as that observed for VP22 in HSV-1(F)-infected Vero cells, both proteins clearly localize to the nucleus. This result indicates that the carboxy-terminal region of VP22 is not required for its nuclear localization. The finding is somewhat unexpected, since the carboxy-terminal half (22), or portions thereof (17), of VP22 has been implicated in the nuclear association of VP22 in transient-expression assays. VP22's nuclear localization is regulated during synchronized infection (45) and may be influenced by the cell cycle (15). It is conceivable that viral proteins may participate in VP22's translocation to the nucleus during HSV-1 infection, while other mechanisms may be used to bring VP22 into the nucleus in uninfected cells. Recent computer analyses predict that the amino-terminal portion of VP22 might possess a nuclear localization signal (26), and our findings support this theory. It has been reported that a GFP-VP22 fusion protein, which would be too large to passively diffuse through the nuclear pore complex, produced during transient transfection is almost exclusively cytoplasmic and that there is a cytoplasmic-to-nuclear partitioning when the nuclear membrane breaks down during mitosis (15). When this GFP-VP22 fusion is synthesized in the context of a viral infection, the chimera remains cytoplasmic, with a small number of cells at the periphery of plaques showing nuclear localization (18). We have observed the tight association of full-length VP22 with chromatin in our V49 cells which are actively dividing {undergoing mitosis} (Fig. 5). However, the significance of this mitotic association of VP22 with chromatin during the course of productive HSV-1 replication is not clear, inasmuch as HSV-1 inhibits the cell cycle during infection (13, 52).
(iv) RF177 synthesizes the late VP13/14, VP16, and VHS tegument proteins and incorporates them into virus particles as efficiently as wild-type virus in cultured Vero cells. From previous reports (33, 55), we expected that the distribution of these proteins might change due to the loss of full-length VP22 in RF177 virions. It appears that the loss of VP22 has been compensated for by a redistribution of other, unknown, minor virion proteins. These proteins fall into at least four molecular weight ranges based on their electrophoretic mobilities. The migration of the first group is greater than 170,000 Da. The second group is in the range of 110,000 to 140,000 Da and may include the UL37 gene product (1, 36). The third group migrates between 55,000 and 65,000 Da and may include the UL13 gene product (10). The fourth group migrates between 35,000 and 45,000 Da and may include UL16 (42). Studies designed to determine whether these redistributed virion proteins are actually UL37, UL13, or UL16 are under way. While tegument proteins are obvious candidates for these unknown proteins, we cannot exclude the possibility that the distribution of envelope glycoproteins might change in the absence of VP22. Since RF177 virions are infectious, it is unlikely that capsid proteins have been affected.
The dramatically small amount of Δ212 in RF177 virions indicates that it does not contribute significantly to the overall mass of the virus particle. Nonetheless, we cannot exclude the possibility that this remaining amino-terminal portion of VP22 performs an essential function during RF177 infection. Even the small amount of Δ212 present in RF177 virions may contribute a required structural function to the virion. Alternatively, VP22 may participate enzymatically in a function for which the Δ212 portion is still active. The presence of high- and low-mobility forms of Δ212 provides evidence that the truncation mutant, like full-length VP22, is modified during infection, and this modification may be associated with its functional activities.
(v) The presence of full-length VP22 in the tegument is required for efficient viral spread in infected Vero cell monolayers. The reduced plaque size of RF177 is reminiscent of the previously reported reduction in viral spread for three other HSV-1 deletion viruses. Deletion of either UL13, UL51, or UL14, which are components of virus particles (10–12), leads to a decrease in plaque size (3, 10, 11). Since we observed wild-type distributions of VP13/14, VP16, and VHS, it is unlikely that these proteins are responsible for the phenotype of RF177. Thus, the RF177 spreading defect either is due primarily to the loss of VP22 function or results from the secondary consequence of changes in the RF177 virion protein profile. It is possible that the minor virion proteins whose distributions differ between HSV-1(F) and RF177 could participate in virus spreading, and VP22 may functionally interact with these proteins. In addition, future experiments will determine whether the Δ212 protein itself might have a deleterious effect on viral cell-to-cell spread.
In light of the similar single-step growth kinetics observed for RF177 and HSV-1(F), the inability of RF177 to spread efficiently from cell to cell suggests that the absence of VP22 in the virion leads to a cumulative effect during multiple rounds of RF177 replication. While VP22 may function late in infection, one interpretation of our results is that full-length VP22 may be needed during entry or postentry stages of infection. Currently, RF177 infection is the only model for HSV-1 replication in the absence of full-length VP22. This virus should continue to be a useful reagent in future studies designed to elucidate the biological functions of VP22 during HSV-1 infection.
ACKNOWLEDGMENTS
We thank Ronald Gordan for guidance and advice, Ronald Uson (Mount Sinai School of Medicine) for expert technical assistance during electron microscopic analyses, and Sully Read (University of Missouri-Kansas City) for graciously providing anti-VHS polyclonal antiserum. Bernard Roizman (University of Chicago) provided HSV-1(Δ305), HSV-1(R2507), and a low-passage isolate of the parental HSV-1(F).
These studies were supported in part by grants from the U.S. Public Health Service (AI38873), the American Cancer Society (JFRA 634), and an unrestricted grant from the National Foundation for Infectious Diseases. L.E.P. is a United States Public Health Service Predoctoral Trainee (GM08553). J.A.B. is a Markey Research Fellow and thanks the Lucille P. Markey Charitable Trust for their support.
REFERENCES
- 1.Albright A G, Jenkins F J. The herpes simplex virus UL37 protein is phosphorylated in infected cells. J Virol. 1993;67:4842–4847. doi: 10.1128/jvi.67.8.4842-4847.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aubert M, Blaho J A. The herpes simplex virus type 1 regulatory protein ICP27 is required for the prevention of apoptosis in infected human cells. J Virol. 1999;73:2803–2813. doi: 10.1128/jvi.73.4.2803-2813.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barker D E, Roizman B. Identification of three genes nonessential for growth in cell culture near the right terminus of the unique sequences of long component of herpes simplex virus 1. Virology. 1990;177:684–691. doi: 10.1016/0042-6822(90)90534-x. [DOI] [PubMed] [Google Scholar]
- 4.Barker D E, Roizman B. The unique sequence of the herpes simplex virus 1 L component contains an additional translated open reading frame designated UL49.5. J Virol. 1992;66:562–526. doi: 10.1128/jvi.66.1.562-566.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blaho J A, Mitchell C, Roizman B. An amino acid sequence shared by the herpes simplex virus 1 alpha regulatory proteins 0, 4, 22, and 27 predicts the nucleotidylylation of the UL21, UL31, UL47, and UL49 gene products. J Biol Chem. 1994;269:17401–17410. [PubMed] [Google Scholar]
- 6.Blaho J A, Mitchell C, Roizman B. Guanylylation and adenylylation of the alpha regulatory proteins of herpes simplex virus require a viral beta or gamma function. J Virol. 1993;67:3891–3900. doi: 10.1128/jvi.67.7.3891-3900.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blaho J A, Roizman B. Analyses of HSV proteins for posttranslational modifications and enzyme functions. In: Brown S M, Maclean A R, editors. Methods in molecular medicine: herpes simplex virus protocols. Vol. 10. Ottawa, Canada: Humana Press Inc.; 1998. pp. 237–256. [DOI] [PubMed] [Google Scholar]
- 8.Blaho J A, Roizman B. ICP4, the major regulatory protein of herpes simplex virus, shares features common to GTP-binding proteins and is adenylated and guanylated. J Virol. 1991;65:3759–3769. doi: 10.1128/jvi.65.7.3759-3769.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campbell M E, Palfreyman J W, Preston C M. Identification of herpes simplex virus DNA sequences which encode a trans-acting polypeptide responsible for stimulation of immediate early transcription. J Mol Biol. 1984;180:1–19. doi: 10.1016/0022-2836(84)90427-3. [DOI] [PubMed] [Google Scholar]
- 10.Coulter L J, Moss H W, Lang J, McGeoch D J. A mutant of herpes simplex virus type 1 in which the UL13 protein kinase gene is disrupted. J Gen Virol. 1993;74:387–395. doi: 10.1099/0022-1317-74-3-387. [DOI] [PubMed] [Google Scholar]
- 11.Cunningham C, Davison A J, MacLean A R, Taus N S, Baines J D. Herpes simplex virus type 1 gene UL14: phenotype of a null mutant and identification of the encoded protein. J Virol. 2000;74:33–41. doi: 10.1128/jvi.74.1.33-41.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daikoku T, Ikenoya K, Yamada H, Goshima F, Nishiyama Y. Identification and characterization of the herpes simplex virus type 1 UL51 gene product. J Gen Virol. 1998;79:3027–3031. doi: 10.1099/0022-1317-79-12-3027. [DOI] [PubMed] [Google Scholar]
- 13.de Bruyn Kops A, Knipe D M. Formation of DNA replication structures in herpes virus-infected cells requires a viral DNA binding protein. Cell. 1988;55:857–868. doi: 10.1016/0092-8674(88)90141-9. [DOI] [PubMed] [Google Scholar]
- 14.Ejercito P M, Kieff E D, Roizman B. Characterization of herpes simplex virus strains differing in their effects on social behaviour of infected cells. J Gen Virol. 1968;2:357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- 15.Elliott G, O'Hare P. Cytoplasm-to-nucleus translocation of a herpesvirus tegument protein during cell division. J Virol. 2000;74:2131–2141. doi: 10.1128/jvi.74.5.2131-2141.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elliott G, O'Hare P. Herpes simplex virus type 1 tegument protein VP22 induces the stabilization and hyperacetylation of microtubules. J Virol. 1998;72:6448–6455. doi: 10.1128/jvi.72.8.6448-6455.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elliott G, O'Hare P. Intercellular trafficking and protein delivery by a herpesvirus structure protein. Cell. 1997;88:223–233. doi: 10.1016/s0092-8674(00)81843-7. [DOI] [PubMed] [Google Scholar]
- 18.Elliott G, O'Hare P. Live-cell analysis of a green fluorescent protein-tagged herpes simplex virus infection. J Virol. 1999;73:4110–4119. doi: 10.1128/jvi.73.5.4110-4119.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elliott G, O'Reilly D, O'Hare P. Identification of phosphorylation sites within the herpes simplex virus tegument protein VP22. J Virol. 1999;73:6203–6206. doi: 10.1128/jvi.73.7.6203-6206.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elliott G, O'Reilly D, O'Hare P. Phosphorylation of the herpes simplex virus type 1 tegument protein VP22. Virology. 1996;226:140–145. doi: 10.1006/viro.1996.0638. [DOI] [PubMed] [Google Scholar]
- 21.Elliott G D, Meredith D M. The herpes simplex virus type 1 tegument protein VP22 is encoded by gene UL49. J Gen Virol. 1992;73:723–726. doi: 10.1099/0022-1317-73-3-723. [DOI] [PubMed] [Google Scholar]
- 22.Fang B, Xu B, Kock P, Roth J A. Intercellular trafficking of VP22-GFP fusion proteins is not observed in cultured mammalian cells. Gene Ther. 1998;5:1420–1424. doi: 10.1038/sj.gt.3300741. [DOI] [PubMed] [Google Scholar]
- 23.Foster T P, Rybachuk G V, Kousoulas K G. Expression of the enhanced green fluorescent protein by herpes simplex virus type 1 (HSV-1) as an in vitro or in vivo marker for virus entry and replication. J Virol Methods. 1998;75:151–160. doi: 10.1016/s0166-0934(98)00107-4. [DOI] [PubMed] [Google Scholar]
- 24.Gibson W, Roizman B. Proteins specified by herpes simplex virus. Staining and radiolabeling properties of B capsid and virion proteins in polyacrylamide gels. J Virol. 1974;13:155–165. doi: 10.1128/jvi.13.1.155-165.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Graham F L, van der Eb A J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 26.Harms J S, Ren X, Oliveira S C, Splitter G A. Distinctions between bovine herpesvirus 1 and herpes simplex virus type 1 VP22 tegument protein subcellular associations. J Virol. 2000;74:3301–3312. doi: 10.1128/jvi.74.7.3301-3312.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heine J W, Honess R W, Cassai E, Roizman B. Proteins specified by herpes simplex virus. XII. The virion polypeptides of type 1 strains. J Virol. 1974;14:640–651. doi: 10.1128/jvi.14.3.640-651.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Honess R W, Roizman B. Proteins specified by herpes simplex virus. XI. Identification and relative molar rates of synthesis of structural and nonstructural herpes virus polypeptides in the infected cell. J Virol. 1973;12:1347–1365. doi: 10.1128/jvi.12.6.1347-1365.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kwong A D, Frenkel N. The herpes simplex virus virion host shutoff function. J Virol. 1989;63:4834–4839. doi: 10.1128/jvi.63.11.4834-4839.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kwong A D, Kruper J A, Frenkel N. Herpes simplex virus virion host shutoff function. J Virol. 1988;62:912–921. doi: 10.1128/jvi.62.3.912-921.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lagunoff M, Roizman B. Expression of a herpes simplex virus 1 open reading frame antisense to the gamma(1)34.5 gene and transcribed by an RNA 3′ coterminal with the unspliced latency-associated transcript. J Virol. 1994;68:6021–6028. doi: 10.1128/jvi.68.9.6021-6028.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lemaster S, Roizman B. Herpes simplex virus phosphoproteins. II. Characterization of the virion protein kinase and of the polypeptides phosphorylated in the virion. J Virol. 1980;35:798–811. doi: 10.1128/jvi.35.3.798-811.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leslie J, Rixon F J, McLauchlan J. Overexpression of the herpes simplex virus type 1 tegument protein VP22 increases its incorporation into virus particles. Virology. 1996;220:60–68. doi: 10.1006/viro.1996.0286. [DOI] [PubMed] [Google Scholar]
- 34.Liang X, Chow B, Li Y, Raggo C, Yoo D, Attah-Poku S, Babiuk L A. Characterization of bovine herpesvirus 1 UL49 homolog gene and product: bovine herpesvirus 1 UL49 homolog is dispensable for virus growth. J Virol. 1995;69:3863–3867. doi: 10.1128/jvi.69.6.3863-3867.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McGeoch D J, Dalrymple M A, Davison A J, Dolan A, Frame M C, McNab D, Perry L J, Scott J E, Taylor P. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J Gen Virol. 1988;69:1531–1574. doi: 10.1099/0022-1317-69-7-1531. [DOI] [PubMed] [Google Scholar]
- 36.McLauchlan J, Liefkens K, Stow N D. The herpes simplex virus type 1 UL37 gene product is a component of virus particles. J Gen Virol. 1994;75:2047–2052. doi: 10.1099/0022-1317-75-8-2047. [DOI] [PubMed] [Google Scholar]
- 37.McLauchlan J, Rixon F J. Characterization of enveloped tegument structures (L particles) produced by alphaherpesviruses: integrity of the tegument does not depend on the presence of capsid or envelope. J Gen Virol. 1992;73:269–276. doi: 10.1099/0022-1317-73-2-269. [DOI] [PubMed] [Google Scholar]
- 38.McLean G, Rixon F, Langeland N, Haarr L, Marsden H. Identification and characterization of the virion protein products of herpes simplex virus type 1 gene UL47. J Gen Virol. 1990;71:2953–2960. doi: 10.1099/0022-1317-71-12-2953. [DOI] [PubMed] [Google Scholar]
- 39.Meredith D M, Lindsay J A, Halliburton I W, Whittaker G R. Post-translational modification of the tegument proteins (VP13 and VP14) of herpes simplex virus type 1 by glycosylation and phosphorylation. J Gen Virol. 1991;72:2771–2775. doi: 10.1099/0022-1317-72-11-2771. [DOI] [PubMed] [Google Scholar]
- 40.Morgenstern J P, Land H. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morrison E E, Wang Y F, Meredith D M. Phosphorylation of structural components promotes dissociation of the herpes simplex virus type 1 tegument. J Virol. 1998;72:7108–7114. doi: 10.1128/jvi.72.9.7108-7114.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nalwanga D, Rempel S, Roizman B, Baines J D. The UL 16 gene product of herpes simplex virus 1 is a virion protein that colocalizes with intranuclear capsid proteins. Virology. 1996;226:236–242. doi: 10.1006/viro.1996.0651. [DOI] [PubMed] [Google Scholar]
- 43.Oroskar A A, Read G S. Control of mRNA stability by the virion host shutoff function of herpes simplex virus. J Virol. 1989;63:1897–1906. doi: 10.1128/jvi.63.5.1897-1906.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pellett P E, McKnight J L, Jenkins F J, Roizman B. Nucleotide sequence and predicted amino acid sequence of a protein encoded in a small herpes simplex virus DNA fragment capable of trans-inducing alpha genes. Proc Natl Acad Sci USA. 1985;82:5870–5874. doi: 10.1073/pnas.82.17.5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pomeranz L E, Blaho J A. Modified VP22 localizes to the cell nucleus during synchronized herpes simplex virus type 1 infection. J Virol. 1999;73:6769–6781. doi: 10.1128/jvi.73.8.6769-6781.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Poon A P, Roizman B. The phenotype in vitro and in infected cells of herpes simplex virus 1 alpha trans-inducing factor (VP16) carrying temperature-sensitive mutations introduced by substitution of cysteines. J Virol. 1995;69:7658–7667. doi: 10.1128/jvi.69.12.7658-7667.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Post L E, Mackem S, Roizman B. Regulation of alpha genes of herpes simplex virus: expression of chimeric genes produced by fusion of thymidine kinase with alpha gene promoters. Cell. 1981;24:555–565. doi: 10.1016/0092-8674(81)90346-9. [DOI] [PubMed] [Google Scholar]
- 48.Preston C M, Notarianni E L. Poly(ADP-ribosyl)ation of a herpes simplex virus immediate early polypeptide. Virology. 1983;131:492–501. doi: 10.1016/0042-6822(83)90515-9. [DOI] [PubMed] [Google Scholar]
- 49.Read G S, Frenkel N. Herpes simplex virus mutants defective in the virion-associated shutoff of host polypeptide synthesis and exhibiting abnormal synthesis of alpha (immediate early) viral polypeptides. J Virol. 1983;46:498–512. doi: 10.1128/jvi.46.2.498-512.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roizman B, Furlong D. The replication of herpesviruses. In: Fraenkel-Conrat H, Wagner R R, editors. Comprehensive virology. Vol. 3. New York, N.Y: Plenum Press; 1974. pp. 229–403. [Google Scholar]
- 51.Smibert C A, Johnson D C, Smiley J R. Identification and characterization of the virion-induced host shutoff product of herpes simplex virus gene UL41. J Gen Virol. 1992;73:467–470. doi: 10.1099/0022-1317-73-2-467. [DOI] [PubMed] [Google Scholar]
- 52.Song B, Liu J J, Yeh K C, Knipe D M. Herpes simplex virus infection blocks events in the G1 phase of the cell cycle. Virology. 2000;267:326–334. doi: 10.1006/viro.1999.0146. [DOI] [PubMed] [Google Scholar]
- 53.Spear P G, Roizman B. Proteins specified by herpes simplex virus. V. Purification and structural proteins of the herpesvirion. J Virol. 1972;9:143–159. doi: 10.1128/jvi.9.1.143-159.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weinheimer S P, Boyd B A, Durham S K, Resnick J L, O'Boyle D R., II Deletion of the VP16 open reading frame of herpes simplex virus type 1. J Virol. 1992;66:258–269. doi: 10.1128/jvi.66.1.258-269.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Y, McKnight J L. Herpes simplex virus type 1 UL46 and UL47 deletion mutants lack VP11 and VP12 or VP13 and VP14, respectively, and exhibit altered viral thymidine kinase expression. J Virol. 1993;67:1482–1492. doi: 10.1128/jvi.67.3.1482-1492.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou S L, Gordon R E, Bradbury M, Stump D, Kiang C L, Berk P D. Ethanol up-regulates fatty acid uptake and plasma membrane expression and export of mitochondrial aspartate aminotransferase in HepG2 cells. Hepatology. 1998;27:1064–1074. doi: 10.1002/hep.510270423. [DOI] [PubMed] [Google Scholar]