Abstract
Pancreatic cancer exhibits a poor prognosis due to the lack of early diagnostic biomarkers and the resistance to conventional chemotherapy. CD44 has been known as a cancer stem cell marker and plays tumor promotion and drug resistance roles in various cancers. In particular, the splicing variants are overexpressed in many carcinomas and play essential roles in the cancer stemness, invasiveness or metastasis, and resistance to treatments. Therefore, the understanding of each CD44 variant’s (CD44v) function and distribution in carcinomas is essential for the establishment of CD44-targeting tumor therapy. In this study, we immunized mice with CD44v3–10-overexpressed Chinese hamster ovary (CHO)-K1 cells and established various anti-CD44 monoclonal antibodies (mAbs). One of the established clones (C44Mab-3; IgG1, kappa) recognized peptides of the variant-5-encoded region, indicating that C44Mab-3 is a specific mAb for CD44v5. Moreover, C44Mab-3 reacted with CHO/CD44v3–10 cells or pancreatic cancer cell lines (PK-1 and PK-8) by flow cytometry. The apparent KD of C44Mab-3 for CHO/CD44v3–10 and PK-1 was 1.3 × 10−9 M and 2.6 × 10−9 M, respectively. C44Mab-3 could detect the exogenous CD44v3–10 and endogenous CD44v5 in Western blotting and stained the formalin-fixed paraffin-embedded pancreatic cancer cells but not normal pancreatic epithelial cells in immunohistochemistry. These results indicate that C44Mab-3 is useful for detecting CD44v5 in various applications and is expected to be useful for the application of pancreatic cancer diagnosis and therapy.
Keywords: CD44, CD44 variant 5, monoclonal antibody, flow cytometry, immunohistochemistry
1. Introduction
Pancreatic cancer has become the third leading cause of death in men and women combined in the United States in 2023 [1]. The development of pancreatic cancer has been explained by four common oncogenic events, including KRAS, CDKN2A, SMAD4, and TP53 [2,3]. However, pancreatic cancer shows a heterogeneity in drug response and clinical outcomes [4]. Therefore, detailed understanding of pancreatic cancers has been required to improve patient selection for current therapies and to develop novel therapeutic strategies. An integrated genomic analysis of pancreatic ductal adenocarcinomas (PDAC) was performed and defined four subtypes, including squamous, pancreatic progenitor, immunogenic, and aberrantly differentiated endocrine exocrine (ADEX), which correspond to the histopathological characteristics [5]. Additionally, various marker proteins have been investigated for the early diagnostic and drug responses of pancreatic cancers [6]. Studies have suggested that CD44 plays important roles in malignant progression of tumors through its cancer stemness and metastasis-promoting properties [7,8].
CD44 is a type I transmembrane glycoprotein that is expressed as a wide variety of isoforms in various types of cells. [9]. The variety of isoforms is produced by the alternative splicing of CD44 mRNA. The CD44 standard isoform (CD44s) is the smallest isoform of CD44 (85–95 kDa); it is presented on the membrane of most vertebrate cells. CD44s mRNA is assembled by the first five and the last five constant region exons [10]. The CD44 variant isoforms (CD44v) are produced by the alternative splicing of middle variant exons (v1–v10) and the standard exons of CD44s [11]. CD44v is heavily glycosylated, leading to various molecular weights (~250 kDa) owing to N-glycosylation and O-glycosylation [12]. Both CD44s and CD44v (pan-CD44) are known as hyaluronic acid (HA) receptors that mediate cellular homing, migration, adhesion, and proliferation [13].
CD44v is overexpressed in carcinomas and induce metastatic properties [14,15]. A growing body of evidence suggests that CD44v plays critical roles in the promotion of tumor invasion, metastasis, cancer-initiating properties [16], and resistance to chemo- and radiotherapy [7,17]. Reports indicated the important functions of each variant’s exon-encoded region. The v3-encoded region functions as a co-receptor for receptor tyrosine kinases [18]. Since the v3-encoded region possesses heparan sulfate moieties, it can recruit to heparin-binding epidermal growth factor-like growth factor (HB-EGF) and fibroblast growth factors (FGFs). Furthermore, the v6-encoded region forms a ternary complex with HGF and its receptor c-MET, which is essential for its activation [19]. Additionally, oxidative stress resistance is mediated by the v8–10-encoded region through binding with a cystine–glutamate transporter (xCT) subunit [20]. Therefore, establishment and characterization of mAbs that recognize each CD44v is thought to be essential for understanding each variant’s function and development of CD44-targeting tumor diagnosis and therapy. However, the function and distribution of the variant-5-encoded region in tumors has not been fully understood.
Our group established the novel anti-pan-CD44 mAbs, C44Mab-5 (IgG1, kappa) [21] and C44Mab-46 (IgG1, kappa) [22] using the Cell-Based Immunization and Screening (CBIS) method and immunization with the CD44v3–10 ectodomain, respectively. Both C44Mab-5 and C44Mab-46 have epitopes within the standard exon (1 to 5)-encoding sequences [23,24,25]. Furthermore, we showed that both C44Mab-5 and C44Mab-46 are applicable to flow cytometry and immunohistochemistry in oral [21] and esophageal squamous cell carcinomas (SCC) [22]. We have also investigated the antitumor effects of core-fucose-deficient C44Mab-5 in mouse xenograft models of oral SCC [26]. In this study, we developed a novel anti-CD44v5 mAb, C44Mab-3 (IgG1, kappa), by the CBIS method and evaluated its applications, including flow cytometry, Western blotting, and immunohistochemical analyses.
2. Materials and Methods
2.1. Cell Lines
Chinese hamster ovary (CHO)-K1 and mouse multiple myeloma P3X63Ag8U.1 (P3U1) cell lines were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). The human pancreas cancer cell lines PK-1 and PK-8 were obtained from the Cell Resource Center for Biomedical Research Institute of Development, Aging and Cancer at Tohoku University. These cells were cultured in Roswell Park Memorial Institute (RPMI)-1640 medium (Nacalai Tesque, Inc., Kyoto, Japan) supplemented with 100 U/mL penicillin, 100 μg/mL streptomycin, 0.25 μg/mL amphotericin B (Nacalai Tesque, Inc.), and 10% heat-inactivated fetal bovine serum (FBS; Thermo Fisher Scientific, Inc., Waltham, MA, USA). All the cells were grown in a humidified incubator at 37 °C with 5% CO2.
2.2. Plasmid Construction and Establishment of Stable Transfectants
CD44v3–10 open reading frame was obtained from the RIKEN BRC through the National Bio-Resource Project of the MEXT, Japan. CD44s cDNA was amplified using the HotStar HiFidelity Polymerase Kit (Qiagen Inc., Hilden, Germany) and LN229 (a glioblastoma cell line) cDNA as a template. CD44v3–10 and CD44 cDNAs were subcloned into pCAG-Ble-ssPA16 vectors with a signal sequence and N-terminal PA16 tag of 16 amino acids (GLEGGVAMPGAEDDVV) [21,27,28,29,30]; this can be detected by NZ-1, which was originally developed as an anti-human podoplanin mAb [31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46]. The pCAG-Ble/PA16-CD44s and pCAG-Ble/PA16-CD44v3–10 vectors were transfected into CHO-K1 cells using a Neon transfection system (Thermo Fisher Scientific, Inc.), which offers an innovative electroporation method that utilizes a proprietary biologically compatible pipette tip chamber to generate a more uniform electric field for a significant increase in transfection efficiency and cell viability. By the limiting dilution method, CHO/CD44s and CHO/CD44v3–10 clones were finally established.
2.3. Hybridomas
The female BALB/c mice were purchased from CLEA Japan (Tokyo, Japan). All animal experiments were approved by the Animal Care and Use Committee of Tohoku University (Permit number: 2019NiA-001) and performed according to relevant guidelines and regulations to minimize animal suffering and distress in the laboratory. The mice were intraperitoneally immunized with CHO/CD44v3–10 (1 × 108 cells) and Imject Alum (Thermo Fisher Scientific Inc.) as an adjuvant. After the three additional immunizations per week, a booster injection was performed two days before harvesting the spleen cells of immunized mice. The hybridomas were established by the fusion of splenocytes and P3U1 cells using polyethylene glycol 1500 (PEG1500; Roche Diagnostics, Indianapolis, IN, USA). RPMI-1640 supplemented with hypoxanthine, aminopterin, and thymidine (HAT; Thermo Fisher Scientific Inc.) was used for the selection of hybridomas. The supernatants, which are negative for CHO-K1 cells and positive for CHO/CD44v3–10 cells, were selected by flow cytometry using SA3800 Cell Analyzers (Sony Corp. Tokyo, Japan).
2.4. Enzyme-Linked Immunosorbent Assay (ELISA)
Fifty-eight synthesized peptides, covering the CD44v3–10 extracellular domain [23], were synthesized by Sigma-Aldrich Corp. (St. Louis, MO, USA). The peptides (1 µg/mL) were immobilized on Nunc Maxisorp 96-well immunoplates (Thermo Fisher Scientific Inc.). Plate washing was performed with phosphate-buffered saline (PBS) containing 0.05% (v/v) Tween 20 (PBST; Nacalai Tesque, Inc.). After blocking with 1% (w/v) bovine serum albumin (BSA) in PBST, C44Mab-3 (10 µg/mL) was added to each well. Then, the wells were further incubated with peroxidase-conjugated anti-mouse immunoglobulins (1:2000 dilution; Agilent Technologies Inc., Santa Clara, CA, USA). One-Step Ultra TMB (Thermo Fisher Scientific Inc.) was used for enzymatic reactions. An iMark microplate reader (Bio-Rad Laboratories, Inc., Berkeley, CA, USA) was used to mesure the optical density at 655 nm.
2.5. Flow Cytometry
CHO-K1, CHO/CD44v3–10, PK-1, and PK-8 were obtained using 0.25% trypsin and 1 mM ethylenediamine tetraacetic acid (EDTA; Nacalai Tesque, Inc.). The cells were incubated with C44Mab-3, C44Mab-46, or blocking buffer (control) (0.1% BSA in PBS) for 30 min at 4 °C. Then, the cells were treated with Alexa Fluor 488-conjugated secondary antibody (Cell Signaling Technology, Inc., Danvers, MA, USA) for 30 min at 4 °C. The data were analyzed using the SA3800 Cell Analyzer and SA3800 software ver. 2.05 (Sony Corp.).
2.6. Determination of Dissociation Constant (KD) via Flow Cytometry
CHO/CD44v3–10 and PK-1 cells were treated with serially diluted C44Mab-3 (0.01–10 µg/mL). Then, the cells were incubated with Alexa Fluor 488-conjugated secondary antibody. Fluorescence data were analyzed using BD FACSLyric and BD FACSuite software version 1.3 (BD Biosciences, Franklin Lakes, NJ, USA). The KD was determined by the fitting binding isotherms to built-in one-site binding models of GraphPad Prism 8 (GraphPad Software, Inc., La Jolla, CA, USA).
2.7. Determination of KD via Surface Plasmon Resonance (SPR)
Measurement of KD between C44Mab-3 and the epitope peptide was performed using SPR. C44Mab-3 was immobilized on the sensor chip CM5 according to the manufacturer’s protocol by Cytiva (Marlborough, MA, USA). C44Mab-3 (10 μg/mL in acetate buffer (pH 4.0; Cytiva)) was immobilized using an amine coupling reaction. The surface of the flow cell 2 of the sensor chip CM5 was treated with 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide and N-hydroxysuccinimide (NHS), followed by the injection of C44Mab-3. The KD between C44Mab-3 and the epitope peptide (CD44p311–330) was determined using Biacore X100 (Cytiva). A single cycle kinetics method was used to measure the binding signals. The data were analyzed by 1:1 binding kinetics to determine the association rate constant (ka) and dissociation rate constant (kd) and KD using Biacore X100 evaluation software (Cytiva).
2.8. Western Blot Analysis
The total cell lysates (10 μg of protein) were separated on 5–20% polyacrylamide gels (FUJIFILM Wako Pure Chemical Corporation, Osaka, Japan). The separated proteins were transferred onto polyvinylidene difluoride (PVDF) membranes (Merck KGaA, Darmstadt, Germany). The blocking was performed with 4% skim milk (Nacalai Tesque, Inc.) in PBST. The membranes were incubated with 10 μg/mL of C44Mab-3, 10 μg/mL of C44Mab-46, 0.5 μg/mL of NZ-1, or 1 μg/mL of an anti-β-actin mAb (clone AC-15; Sigma-Aldrich Corp.) and then incubated with peroxidase-conjugated anti-mouse immunoglobulins (diluted 1:1000; Agilent Technologies, Inc.) for C44Mab-3, C44Mab-46, and anti-β-actin. Anti-rat immunoglobulins (diluted 1:1000; Agilent Technologies, Inc.) conjugated to peroxidase was used for NZ-1. The chemiluminescence signals were obtained with ImmunoStar LD (FUJIFILM Wako Pure Chemical Corporation) and detected using a Sayaca-Imager (DRC Co., Ltd., Tokyo, Japan).
2.9. Immunohistochemical Analysis
One formalin-fixed paraffin-embedded (FFPE) oral SCC tissue was obtained from Tokyo Medical and Dental University [47]. FFPE sections of pancreatic carcinoma tissue arrays (Catalog number: PA241c and PA484) were purchased from US Biomax Inc. (Rockville, MD, USA). Pancreas adenocarcinoma tissue microarray with adjacent normal pancreas tissue (PA241c) contains 6 cases of pancreas adenocarcinoma with matched adjacent normal pancreas tissue, with quadruple cores per case. One oral SCC tissue was autoclaved in citrate buffer (pH 6.0; Nichirei biosciences, Inc., Tokyo, Japan), and pancreatic carcinoma tissue arrays were autoclaved in EnVision FLEX Target Retrieval Solution High pH (Agilent Technologies, Inc.) for 20 min. After blocking with SuperBlock T20 (Thermo Fisher Scientific, Inc.), the sections were incubated with C44Mab-3 (1 μg/mL) and C44Mab-46 (1 μg/mL) for 1 h at room temperature. Then, the sections were incubated with the EnVision+ Kit for mouse (Agilent Technologies Inc.) for 30 min. The color was developed using 3,3′-diaminobenzidine tetrahydrochloride (DAB; Agilent Technologies Inc.). Hematoxylin (FUJIFILM Wako Pure Chemical Corporation) was used for the counterstaining. A Leica DMD108 (Leica Microsystems GmbH, Wetzlar, Germany) was used to examine the sections and obtain images.
3. Results
3.1. Development of an Anti-CD44v5 mAb, C44Mab-3
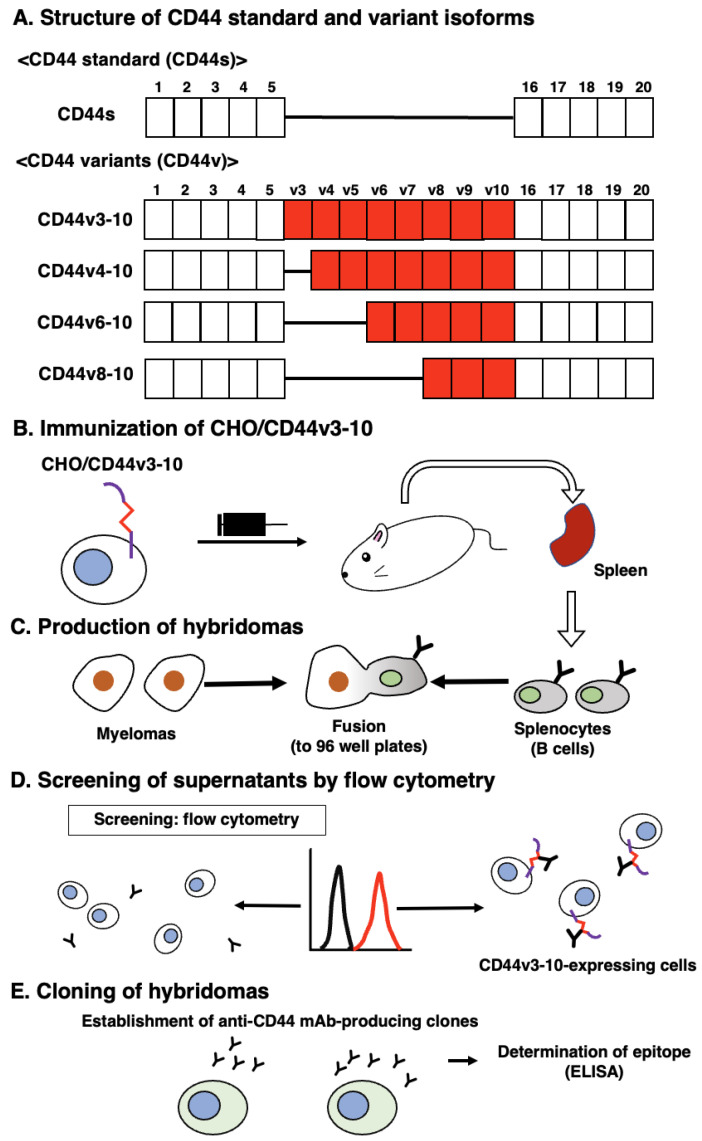
In the CBIS method, we used a stable transfectant (CHO/CD44v3–10 cells) as an immunogen (Figure 1). Mice were immunized with CHO/CD44v3–10 cells, and hybridomas were seeded into 96-well plates. The supernatants, which are negative for CHO-K1 cells and positive for CHO/CD44v3–10 cells, were selected using flow-cytometry-based high throughput screening. By limiting dilution, anti-CD44-mAb-producing clones were finally established. Among them, C44Mab-3 (IgG1, kappa) was shown to recognize both CD44p311–330 (AYEGNWNPEAHPPLIHHEHH) and CD44p321–340 peptides (HPPLIHHEHHEEEETPHSTS), which correspond to the variant-5-encoded sequence (Table 1 and Supplementary Figure S1). In contrast, C44Mab-3 did not recognize other CD44v3–10 extracellular regions. These results indicated that C44Mab-3 specifically recognizes the CD44 variant-5-encoded sequence.
Figure 1.
A schematic illustration of anti-human CD44 mAbs production. (A) Structure of CD44. CD44s mRNA is assembled by the first five (1 to 5) and the last five (16 to 20) exons and translates CD44s. The mRNAs of CD44 variants are produced by the alternative splicing of middle variant exons and translate multiple CD44v such as CD44v3–10, CD44v4–10, CD44v6–10, and CD44v8–10. (B) CHO/CD44v3–10 cells were intraperitoneally injected into BALB/c mice. (C) The splenocytes and P3U1 cells were fused and the hybridomas were produced. (D) The screening was conducted by flow cytometry using parental CHO-K1 and CHO/CD44v3–10 cells. (E) After cloning and additional screening, a clone (C44Mab-3 (IgG1, kappa)) was established. Furthermore, the binding epitope was determined by enzyme-linked immunosorbent assay (ELISA) using peptides that cover the extracellular domain of CD44v3–10.
Table 1.
Determination of the binding epitope of C44Mab-3 by ELISA.
| Peptide | Coding Exon * | Sequence | C44Mab-3 |
|---|---|---|---|
| CD44p21–40 | 2 | QIDLNITCRFAGVFHVEKNG | − |
| CD44p31–50 | 2 | AGVFHVEKNGRYSISRTEAA | − |
| CD44p41–60 | 2 | RYSISRTEAADLCKAFNSTL | − |
| CD44p51–70 | 2 | DLCKAFNSTLPTMAQMEKAL | − |
| CD44p61–80 | 2/3 | PTMAQMEKALSIGFETCRYG | − |
| CD44p71–90 | 2/3 | SIGFETCRYGFIEGHVVIPR | − |
| CD44p81–100 | 3 | FIEGHVVIPRIHPNSICAAN | − |
| CD44p91–110 | 3 | IHPNSICAANNTGVYILTSN | − |
| CD44p101–120 | 3 | NTGVYILTSNTSQYDTYCFN | − |
| CD44p111–130 | 3/4 | TSQYDTYCFNASAPPEEDCT | − |
| CD44p121–140 | 3/4 | ASAPPEEDCTSVTDLPNAFD | − |
| CD44p131–150 | 4/5 | SVTDLPNAFDGPITITIVNR | − |
| CD44p141–160 | 4/5 | GPITITIVNRDGTRYVQKGE | − |
| CD44p151–170 | 5 | DGTRYVQKGEYRTNPEDIYP | − |
| CD44p161–180 | 5 | YRTNPEDIYPSNPTDDDVSS | − |
| CD44p171–190 | 5 | SNPTDDDVSSGSSSERSSTS | − |
| CD44p181–200 | 5 | GSSSERSSTSGGYIFYTFST | − |
| CD44p191–210 | 5 | GGYIFYTFSTVHPIPDEDSP | − |
| CD44p201–220 | 5 | VHPIPDEDSPWITDSTDRIP | − |
| CD44p211–230 | 5/v3 | WITDSTDRIPATSTSSNTIS | − |
| CD44p221–240 | 5/v3 | ATSTSSNTISAGWEPNEENE | − |
| CD44p231–250 | v3 | AGWEPNEENEDERDRHLSFS | − |
| CD44p241–260 | v3 | DERDRHLSFSGSGIDDDEDF | − |
| CD44p251–270 | v3/v4 | GSGIDDDEDFISSTISTTPR | − |
| CD44p261–280 | v3/v4 | ISSTISTTPRAFDHTKQNQD | − |
| CD44p271–290 | v4 | AFDHTKQNQDWTQWNPSHSN | − |
| CD44p281–300 | v4 | WTQWNPSHSNPEVLLQTTTR | − |
| CD44p291–310 | v4/v5 | PEVLLQTTTRMTDVDRNGTT | − |
| CD44p301–320 | v4/v5 | MTDVDRNGTTAYEGNWNPEA | − |
| CD44p311–330 | v5 | AYEGNWNPEAHPPLIHHEHH | + |
| CD44p321–340 | v5 | HPPLIHHEHHEEEETPHSTS | + |
| CD44p331–350 | v5/v6 | EEEETPHSTSTIQATPSSTT | − |
| CD44p341–360 | v5/v6 | TIQATPSSTTEETATQKEQW | − |
| CD44p351–370 | v6 | EETATQKEQWFGNRWHEGYR | − |
| CD44p361–380 | v6 | FGNRWHEGYRQTPREDSHST | − |
| CD44p371–390 | v6/v7 | QTPREDSHSTTGTAAASAHT | − |
| CD44p381–400 | v6/v7 | TGTAAASAHTSHPMQGRTTP | − |
| CD44p391–410 | v7 | SHPMQGRTTPSPEDSSWTDF | − |
| CD44p401–420 | v7 | SPEDSSWTDFFNPISHPMGR | − |
| CD44p411–430 | v7/v8 | FNPISHPMGRGHQAGRRMDM | − |
| CD44p421–440 | v7/v8 | GHQAGRRMDMDSSHSTTLQP | − |
| CD44p431–450 | v8 | DSSHSTTLQPTANPNTGLVE | − |
| CD44p441–460 | v8 | TANPNTGLVEDLDRTGPLSM | − |
| CD44p451–470 | v8/v9 | DLDRTGPLSMTTQQSNSQSF | − |
| CD44p461–480 | v8/v9 | TTQQSNSQSFSTSHEGLEED | − |
| CD44p471–490 | v9 | STSHEGLEEDKDHPTTSTLT | − |
| CD44p481–500 | v9/v10 | KDHPTTSTLTSSNRNDVTGG | − |
| CD44p491–510 | v9/v10 | SSNRNDVTGGRRDPNHSEGS | − |
| CD44p501–520 | v10 | RRDPNHSEGSTTLLEGYTSH | − |
| CD44p511–530 | v10 | TTLLEGYTSHYPHTKESRTF | − |
| CD44p521–540 | v10 | YPHTKESRTFIPVTSAKTGS | − |
| CD44p531–550 | v10 | IPVTSAKTGSFGVTAVTVGD | − |
| CD44p541–560 | v10 | FGVTAVTVGDSNSNVNRSLS | − |
| CD44p551–570 | v10/16 | SNSNVNRSLSGDQDTFHPSG | − |
| CD44p561–580 | v10/16 | GDQDTFHPSGGSHTTHGSES | − |
| CD44p571–590 | 16/17 | GSHTTHGSESDGHSHGSQEG | − |
| CD44p581–600 | 16/17 | DGHSHGSQEGGANTTSGPIR | − |
| CD44p591–606 | 17 | GANTTSGPIRTPQIPEAAAA | − |
+, OD655 ≥ 0.3; −, OD655 < 0.1. * The CD44 exon-encoded regions are illustrated in Figure 1.
3.2. Flow Cytometric Analysis of C44Mab-3 to CD44-Expressing Cells
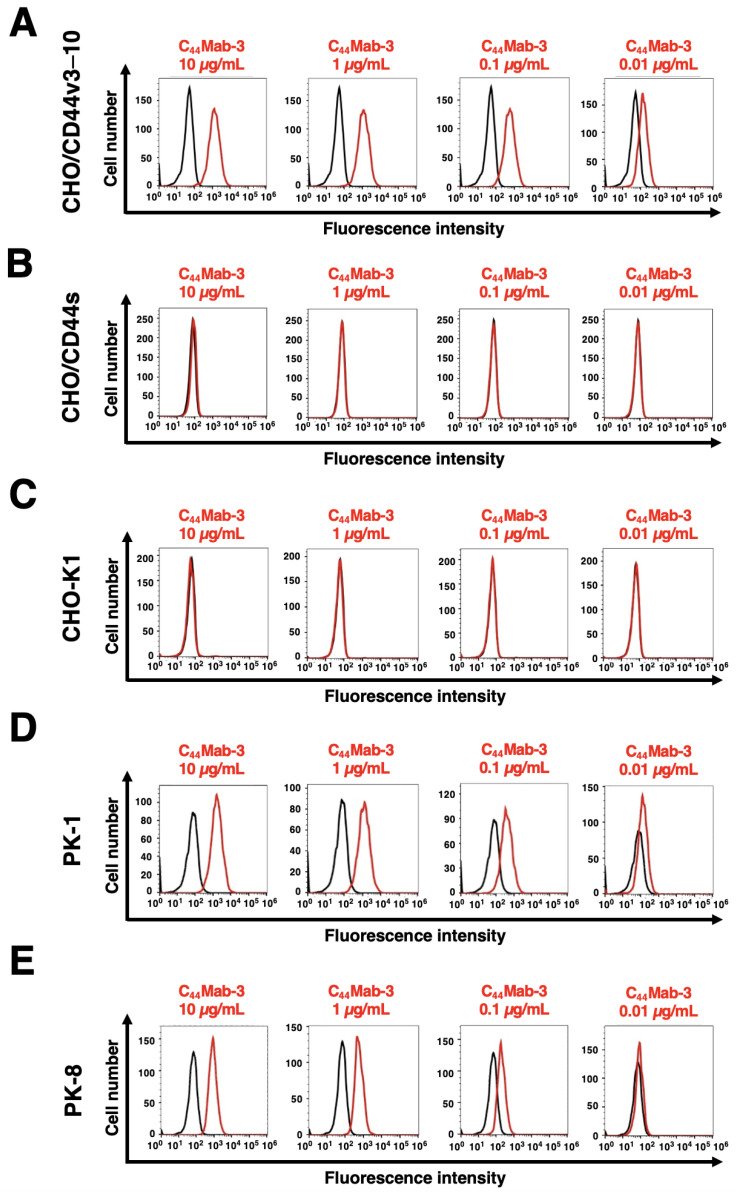
We next investigated the reactivity of C44Mab-3 against CHO/CD44v3–10 and CHO/CD44s cells by flow cytometry. C44Mab-3 recognized CHO/CD44v3–10 cells in a dose-dependent manner (Figure 2A) but do not recognize either CHO/CD44s (Figure 2B) or CHO-K1 (Figure 2C) cells. An anti-pan-CD44 mAb, C44Mab-46 [22], recognized CHO/CD44s cells (Supplementary Figure S2). Furthermore, C44Mab-3 also recognized pancreatic cancer cell lines, such as PK-1 (Figure 2D) and PK-8 (Figure 2E), in a dose-dependent manner.
Figure 2.
Flow cytometry using C44Mab-3 against CD44-expressing cells. CHO/CD44v3–10 (A), CHO/CD44s (B), CHO-K1 (C), PK-1 (D), and PK-8 (E) cells were treated with 0.01–10 µg/mL of C44Mab-3, followed by treatment with Alexa Fluor 488-conjugated anti-mouse IgG (Red line). The black line represents the negative control (blocking buffer).
3.3. Determination of the Binding Affinity of C44Mab-3 by Flow Cytometry to CD44-Expressing Cells and SPR with the Epitope Peptide
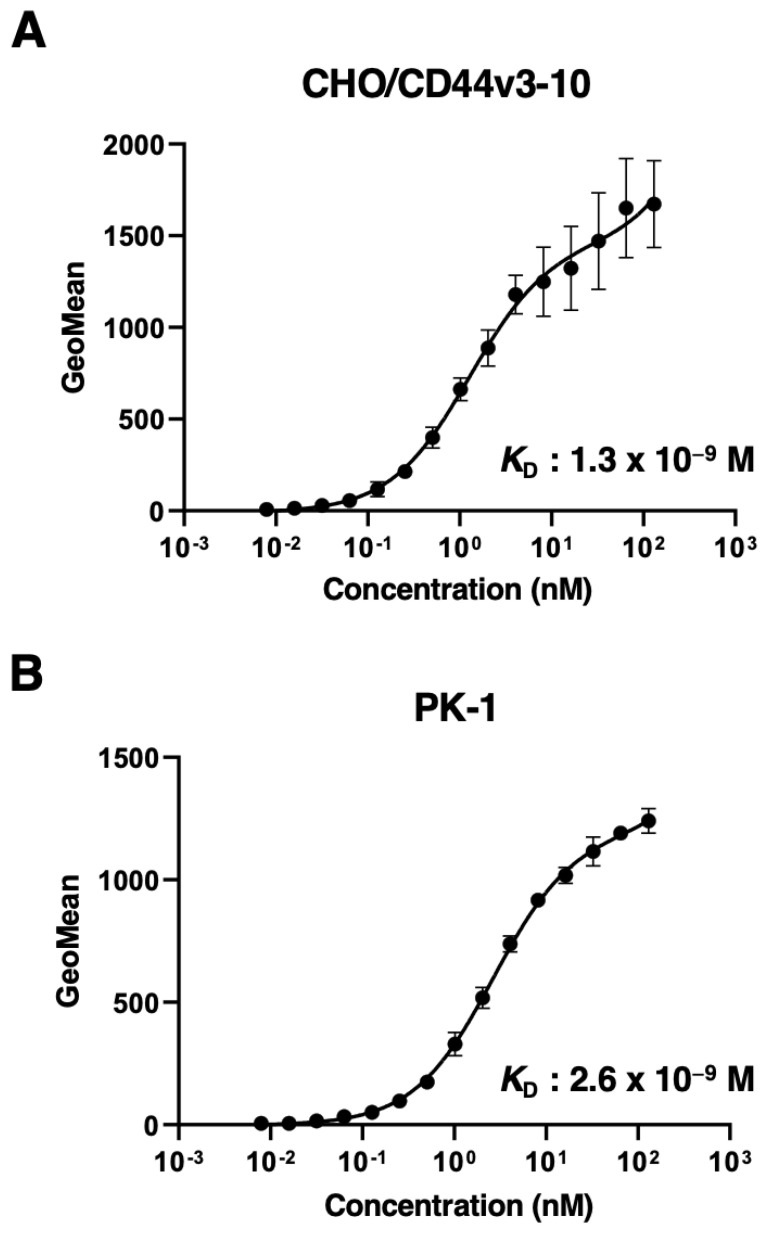
Next, we determined the binding affinity of C44Mab-3 to CHO/CD44v3–10 and PK-1 using flow cytometry. As shown in Figure 3, the KD of CHO/CD44v3–10 and PK-1 was 1.3 × 10−9 M and 2.6 × 10−9 M, respectively, indicating that C44Mab-3 possesses high affinity for CD44v3–10 and endogenous CD44v5-expressing cells.
Figure 3.
The binding affinity of C44Mab-3 to CD44-expressing cells. CHO/CD44v3–10 (A) and PK-1 (B) cells were suspended in 100 µL of serially diluted C44Mab-3 at the indicated concentrations. Then, cells were treated with Alexa Fluor 488-conjugated secondary antibody. Fluorescence data were collected and the apparent dissociation constant (KD) was calculated using GraphPad PRISM 8. Error bars represent means ± SDs.
We also measured the KD of C44Mab-3 with the epitope peptide (CD44p311–330) using Biacore X100. The binding kinetics and measured values are summarized in Supplementary Figure S3. The KD of CD44p311–330 was 5.5 × 10−6 M.
3.4. Western Blot Analysis
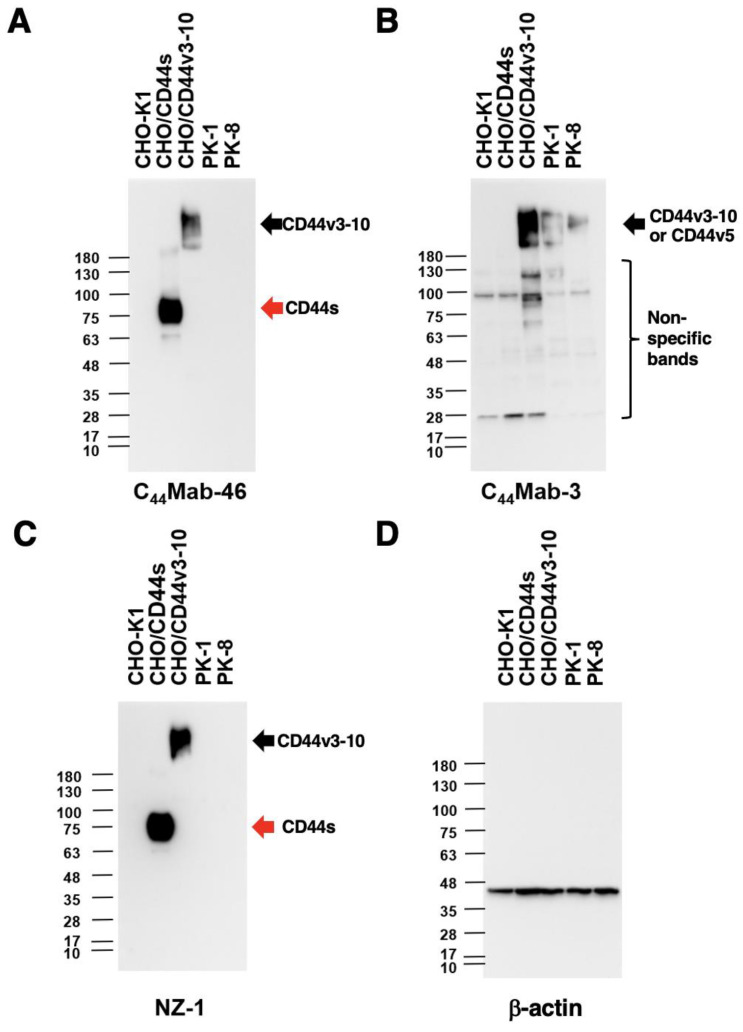
We next performed Western blot analysis to investigate the sensitivity of C44Mab-3. Total cell lysates from CHO-K1, CHO/CD44s, CHO/CD44v3–10, PK-1, and PK-8 were analyzed. As shown in Figure 4A, an anti-pan-CD44 mAb, C44Mab-46, recognized the lysates from both CHO/CD44s (~75 kDa) and CHO/CD44v3–10 (>180 kDa). C44Mab-3 detected CD44v3–10 as bands of more than 180-kDa. Furthermore, C44Mab-3 detected endogenous CD44v5-containing CD44v in PK-1 and PK-8 cells. However, C44Mab-3 did not detect any bands from lysates of CHO-K1 and CHO/CD44s cells (Figure 4B). An anti-PA16 tag mAb (NZ-1) recognized the lysates from both CHO/CD44s (~75 kDa) and CHO/CD44v3–10 (>180 kDa) (Figure 4C). These results indicated that C44Mab-3 specifically detects exogenous CD44v3–10 and endogenous CD44v5-containing CD44v.
Figure 4.
Western blot analysis using C44Mab-3. The cell lysates of CHO-K1, CHO/CD44s, CHO/CD44v3–10, PK-1, and PK-8 (10 µg) were electrophoresed and transferred onto polyvinylidene fluoride (PVDF) membranes. The membranes were incubated with 10 µg/mL of C44Mab-46 (A), 10 µg/mL of C44Mab-3 (B), 0.5 µg/mL of an anti-PA16 tag mAb (NZ-1) (C), and 1 µg/mL of an anti-β-actin mAb (D). Then, the membranes were incubated with anti-mouse immunoglobulins conjugated with peroxidase for C44Mab-46, C44Mab-3, and anti-β-actin. Anti-rat immunoglobulins conjugated with peroxidase were used for NZ-1. The red arrows indicate CD44s (~75 kDa). The black arrows indicate CD44v3–10 or CD44v5 (>180 kDa).
3.5. Immunohistochemical Analysis Using C44Mab-3 against Tumor Tissues
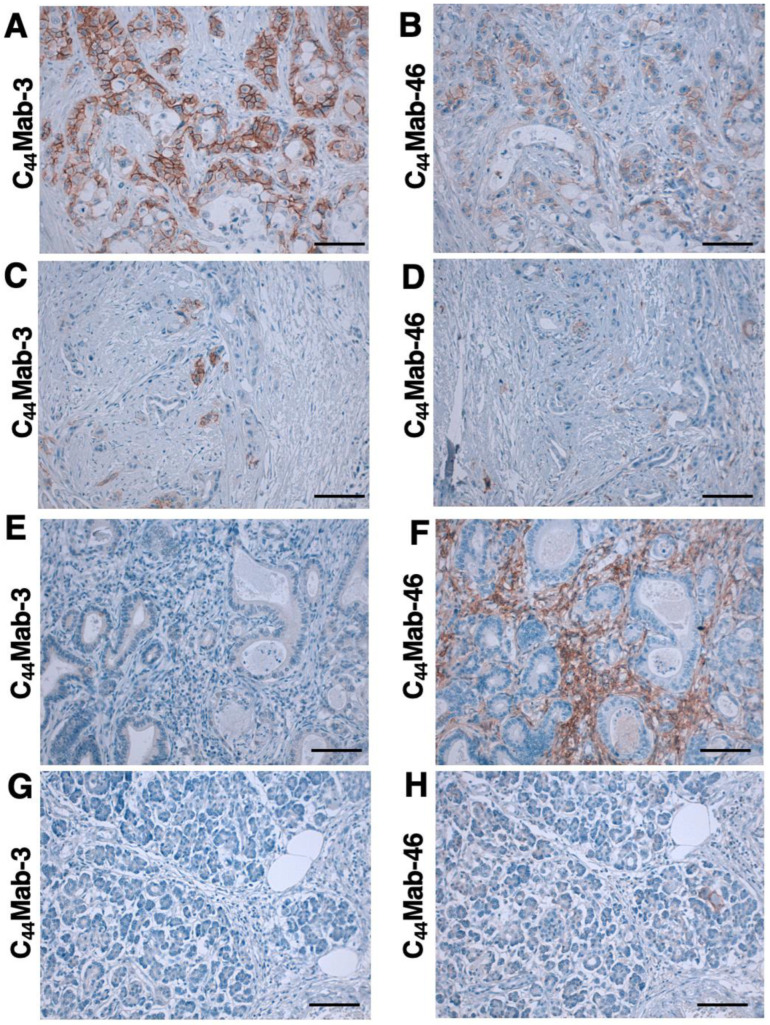
We next examined whether C44Mab-3 could be used for immunohistochemical analyses using FFPE sections. We first examined the reactivity of C44Mab-3 and C44Mab-46 in an oral SCC tissue. As shown in Supplementary Figure S4, C44Mab-3 exhibited a clear membranous staining and could clearly distinguish tumor cells from stromal tissues. In contrast, C44Mab-46 stained both. We then investigated the reactivity of C44Mab-3 and C44Mab-46 in pancreatic carcinoma tissue arrays. Although we performed the antigen retrieval using citrate buffer (pH 6.0) for pancreatic carcinoma tissue arrays in the same way as with an oral SCC tissue, weak staining was observed. Therefore, we next used EnVision FLEX Target Retrieval Solution High pH for the antigen retrieval procedure; C44Mab-3 showed clear membranous staining in pancreatic carcinoma cells with a relatively larger cytoplasm (Figure 5A). C44Mab-46 also stained the same type of pancreatic carcinoma cells (Figure 5B). The staining intensity of C44Mab-3 was much stronger than that of C44Mab-46 (Figure 5A,B). Furthermore, diffusely spread tumor cells in the stroma were stained by both C44Mab-3 and C44Mab-46 (Figure 5C,D). In contrast, both C44Mab-3 and C44Mab-46 did not stain the typical ductal structure of PDAC (Figure 5E,F). In addition, stromal staining using C44Mab-46 was observed in several tissues (Figure 5F). Importantly, normal pancreatic epithelial cells were not stained by C44Mab-3 (Figure 5G). A similar staining pattern was also observed in another tissue array (Supplementary Figure S5). We summarized the data of immunohistochemical analyses in Table 2; C44Mab-3 stained 8 out of 20 cases (40%) (PA484, Figure 5) and 2 out of 6 cases (33%) (PA241c, Supplementary Figure S5) of pancreatic carcinomas. These results indicated that C44Mab-3 could be useful for immunohistochemical analysis of FFPE tumor sections and could recognize a specific type of pancreatic carcinoma.
Figure 5.
Immunohistochemical analysis using C44Mab-3 and C44Mab-46 against pancreatic adenocarcinomas and normal pancreatic tissues. After antigen retrieval, serial sections of pancreatic carcinoma tissue arrays (Catalog number: PA484) were incubated with 1 µg/mL of C44Mab-3 or C44Mab-46, followed by treatment with the Envision+ kit. The color was developed using 3,3′-diaminobenzidine tetrahydrochloride (DAB), and the sections were counterstained with hematoxylin. Scale bar = 100 µm. (A–F) pancreatic adenocarcinomas; (G,H) normal pancreas tissues.
Table 2.
Immunohistochemical analysis using C44Mab-3 against pancreatic carcinoma tissue arrays.
| Tissue Array | Age | Sex | Organ | Pathology Diagnosis | TNM | Grade | Stage | Type | C44Mab-3 |
|---|---|---|---|---|---|---|---|---|---|
| PA241c | 66 | F | Pancreas | Adenocarcinoma | T2N0M0 | 1 | I | malignant | + |
| 66 | F | Pancreas | Adjacent normal pancreas tissue | – | |||||
| 54 | F | Pancreas | Adenocarcinoma | T3N0M0 | 2 | II | malignant | – | |
| 54 | F | Pancreas | Adjacent normal pancreas tissue | – | |||||
| 44 | M | Pancreas | Adenocarcinoma | T3N0M0 | 2 | II | malignant | – | |
| 44 | M | Pancreas | Adjacent normal pancreas tissue | – | |||||
| 59 | M | Pancreas | Adenocarcinoma | T2N0M0 | 3 | I | malignant | – | |
| 59 | M | Pancreas | Adjacent normal pancreas tissue | – | |||||
| 63 | F | Pancreas | Adenocarcinoma | T2N0M0 | 3 | I | malignant | + | |
| 63 | F | Pancreas | Adjacent normal pancreas tissue | – | |||||
| 53 | F | Pancreas | Adenocarcinoma | T3N0M0 | 3 | II | malignant | – | |
| 53 | F | Pancreas | Adjacent normal pancreas tissue | – | |||||
| PA484 | 35 | M | Pancreas | Normal pancreas tissue | - | - | - | normal | – |
| 38 | F | Pancreas | Normal pancreas tissue | - | - | - | normal | – | |
| 38 | M | Pancreas | Normal pancreas tissue | - | - | - | normal | – | |
| 60 | M | Pancreas | Adenocarcinoma | T3N0M0 | 2 | II | malignant | – | |
| 68 | F | Pancreas | Adenocarcinoma | T2N0M0 | 2 | I | malignant | + | |
| 54 | F | Pancreas | Adenocarcinoma | T3N0M0 | 2 | II | malignant | – | |
| 42 | F | Pancreas | Adenocarcinoma | T3N0M0 | 2 | II | malignant | – | |
| 65 | M | Pancreas | Adenocarcinoma | T3N0M0 | 2 | II | malignant | – | |
| 75 | F | Pancreas | Adenocarcinoma | T3N0M1 | 2 | IV | malignant | – | |
| 57 | M | Pancreas | Adenocarcinoma | T3N0M0 | 3 | II | malignant | + | |
| 44 | M | Pancreas | Adenocarcinoma | T3N0M0 | 3 | II | malignant | – | |
| 47 | M | Pancreas | Adenocarcinoma | T3N0M0 | - | II | malignant | – | |
| 41 | M | Pancreas | Adenocarcinoma | T4N1M0 | 2 | III | malignant | – | |
| 64 | F | Pancreas | Adenocarcinoma | T3N0M0 | 2 | II | malignant | – | |
| 58 | F | Pancreas | Adenocarcinoma | T3N0M0 | 3 | II | malignant | – | |
| 47 | F | Pancreas | Adenocarcinoma | T3N1M0 | 3 | III | malignant | + | |
| 78 | M | Pancreas | Adenocarcinoma | T2N0M0 | 3 | I | malignant | + | |
| 49 | M | Pancreas | Adenocarcinoma | T3N0M0 | 2 | II | malignant | + | |
| 53 | F | Pancreas | Adenocarcinoma | T3N0M0 | 3 | II | malignant | + | |
| 60 | M | Pancreas | Adenocarcinoma | T2N0M0 | 3 | I | malignant | + | |
| 57 | F | Pancreas | Adenocarcinoma | T2N0M0 | 3 | I | malignant | – | |
| 61 | M | Pancreas | Mucinous adenocarcinoma | T3N0M1 | 2 | IV | malignant | – | |
| 69 | M | Pancreas | Undifferentiated carcinoma | T2N0M0 | - | I | malignant | + |
+, OD655 ≥ 0.3; −, OD655 < 0.1.
4. Discussion
PDAC is the most common type of pancreatic cancer and has extremely poor prognosis, with a 5-year survival rate of approximately 10% [48]. Advances in therapy have only achieved incremental improvements in overall outcome but can provide notable benefits for undefined subgroups of patients. PDACs are heterogenous neoplasms with various histology [4] and heterogenous molecular landscapes [5]. Therefore, the identification of early diagnostic markers and therapeutic targets in each group has been desired. In this study, we developed C44Mab-3 using the CBIS method (Figure 1) and determined its epitope as variant-5-encoded region of CD44 (Table 1). Then, we showed the usefulness of C44Mab-3 for multiple applications, including flow cytometry (Figure 2 and Figure 3), Western blotting (Figure 4), and immunohistochemistry of PDAC (Figure 5).
An anti-CD44v5 mAb (clone VFF-8) was previously developed and is mainly used for the immunohistochemical analyses of tumors [49]. The epitope of VFF-8 was determined as IHHEHHEEEETPHSTST in the v5-encoded region by ELISA [50]. As shown in Table 1, C44Mab-3 recognized both CD44p311–330 and CD44p321–340 peptides, which commonly possess the HPPLIHHEHH sequence. The epitope of C44Mab-3 partially shares that of VFF-8. Further investigation of the detailed epitope mapping is required. In addition, CD44 is known to be heavily glycosylated [12], and the glycosylation pattern is thought to depend on the host cells. Since the epitope of C44Mab-3 does not contain serine or threonine, the recognition of C44Mab-3 is thought to be independent of the glycosylation.
Immunohistochemistry using VFF-8 and conventional RT-PCR analyses were performed against PDAC [49]. VFF-8 recognized PDAC but not normal pancreas cells. Furthermore, the RT-PCR analysis revealed that the exon v5 appeared in the chain containing at least v4–10 in 80% of PDACs and the cell lines tested. The authors discussed that one of the major differences between normal and PDAC was the linkage of CD44v5 to the CD44v6-containing chain [49]. Our immunohistochemical analysis also support this finding (Figure 5A,C,G). Furthermore, we found that C44Mab-3 could detect atypical types of PDAC, including metaplastic and diffusely invaded tumor cells (Figure 5A,C). In contrast, C44Mab-3 did not stain a typical ductal structure of PDAC (Figure 5E) and normal pancreatic epithelial cells (Figure 5G). In addition to conventional PDAC, the World Health Organization has classified nine histological subtypes of PDAC, which further highlight the morphologic heterogeneity of PDAC [4]. It is worthwhile to investigate whether CD44v5 is expressed in a specific subtype of PDAC in a future study.
Large-scale genomic analyses of PDACs defined four subtypes: (1) squamous; (2) pancreatic progenitor; (3) immunogenic; and (4) ADEX, which correlate with histopathological characteristics [5]. Among them, the squamous subtype is characterized as being enriched for TP53 and KDM6A mutations and having upregulation of the ∆Np63 transcriptional network, hypermethylation of pancreatic endodermal determinant genes, and a poor prognosis [5]. ∆Np63 is known as a marker of basal cells of stratified epithelium and SCC [51]; it is also reported to regulate HA metabolism and signaling [52]. Specifically, ΔNp63 directly regulates the expression of CD44 through p63-binding sites that are located in the promoter region and in the first intron of CD44 gene [52]. Therefore, CD44 transcription could be upregulated in ΔNp63-positive PDAC. However, the mechanism of the variant 5 inclusion during alternative splicing remains to be determined.
Clinical trials of anti-pan-CD44 and variant-specific CD44 mAbs have been conducted [53]. An anti-pan-CD44 mAb, RG7356, exhibited an acceptable safety profile in patients with advanced solid tumors expressing CD44. However, the study was terminated due to no evidence of a clinical and pharmacodynamic dose-response relationship with RG7356 [54]. A clinical trial of a humanized anti-CD44v6 mAb bivatuzumab−mertansine drug conjugate was conducted. However, it failed due to severe skin toxicities [55,56]. The efficient accumulation of mertansine was most likely responsible for the high toxicity [55,56]. Although CD44v5 is not detected in normal pancreatic epithelium by C44Mab-3 (this study) and VFF-8 [49], CD44v5 could be detected in normal lung, skin, gastric, and bladder epithelium by VFF-8 [50]. For the development of the therapeutic use of C44Mab-3, further investigations are required to reduce the toxicity to the above tissues.
We previously converted a mouse IgG1 subclass of mAbs into IgG2a mAb and produced defucosylated mAbs using fucosyltransferase-8-deficient CHO-K1 cells. The defucosylated IgG2a mAbs showed potent antibody-dependent cellular cytotoxicity in vitro and suppressed tumor xenograft growth [26,57,58,59,60,61,62,63]. Therefore, the production of a class-switched and defucosylated version of C44Mab-3 is required to evaluate the antitumor activity in vivo.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/antib12020031/s1, Figure S1, Determination of the binding epitope of C44Mab-3 by ELISA. Figure S2, Recognition of CHO/CD44s and CHO/CD44v3–10 by C44Mab-46 using flow cytometry. Figure S3, Measurement of dissociation constants (KD) between C44Mab-3 and the epitope peptide using SPR. Figure S4, Immunohistochemical analysis using C44Mab-3 and C44Mab-46 against oral squamous cell carcinoma tissue. Figure S5, Immunohistochemical analysis using C44Mab-3 and C44Mab-46 against pancreatic adenocarcinomas and normal pancreatic tissues.
Author Contributions
Y.K. (Yuma Kudo), H.S. and T.T. performed the experiments. M.K.K. and Y.K. (Yukinari Kato) designed the experiments. H.S. and Y.K. (Yuma Kudo) analyzed the data. Y.K. (Yuma Kudo), H.S. and Y.K. (Yukinari Kato) wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The animal study protocol was approved by the Animal Care and Use Committee of Tohoku University (Permit number: 2019NiA-001) for studies involving animals.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in the article and supplementary material.
Conflicts of Interest
The authors have no conflict of interest to declare.
Funding Statement
This research was supported in part by Japan Agency for Medical Research and Development (AMED) under Grant Numbers: JP22ama121008 (to Y.K.), JP22am0401013 (to Y.K.), JP22bm1004001 (to Y.K.), JP22ck0106730 (to Y.K.), and JP21am0101078 (to Y.K.) and by the Japan Society for the Promotion of Science (JSPS) Grants-in-Aid for Scientific Research (KAKENHI) grant nos. 21K20789 (to T.T.), 22K06995 (to H.S.), 21K07168 (to M.K.K.), and 22K07224 (to Y.K.).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Siegel R.L., Miller K.D., Wagle N.S., Jemal A. Cancer statistics, 2023. CA Cancer J. Clin. 2023;73:17–48. doi: 10.3322/caac.21763. [DOI] [PubMed] [Google Scholar]
- 2.Jones S., Zhang X., Parsons D.W., Lin J.C., Leary R.J., Angenendt P., Mankoo P., Carter H., Kamiyama H., Jimeno A., et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Waddell N., Pajic M., Patch A.M., Chang D.K., Kassahn K.S., Bailey P., Johns A.L., Miller D., Nones K., Quek K., et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518:495–501. doi: 10.1038/nature14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taherian M., Wang H., Wang H. Pancreatic Ductal Adenocarcinoma: Molecular Pathology and Predictive Biomarkers. Cells. 2022;11:3068. doi: 10.3390/cells11193068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bailey P., Chang D.K., Nones K., Johns A.L., Patch A.M., Gingras M.C., Miller D.K., Christ A.N., Bruxner T.J., Quinn M.C., et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 6.Espinet E., Klein L., Puré E., Singh S.K. Mechanisms of PDAC subtype heterogeneity and therapy response. Trends Cancer. 2022;8:1060–1071. doi: 10.1016/j.trecan.2022.08.005. [DOI] [PubMed] [Google Scholar]
- 7.Hassn Mesrati M., Syafruddin S.E., Mohtar M.A., Syahir A. CD44: A Multifunctional Mediator of Cancer Progression. Biomolecules. 2021;11:1850. doi: 10.3390/biom11121850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zöller M. CD44: Can a cancer-initiating cell profit from an abundantly expressed molecule? Nat. Rev. Cancer. 2011;11:254–267. doi: 10.1038/nrc3023. [DOI] [PubMed] [Google Scholar]
- 9.Fox S.B., Fawcett J., Jackson D.G., Collins I., Gatter K.C., Harris A.L., Gearing A., Simmons D.L. Normal human tissues, in addition to some tumors, express multiple different CD44 isoforms. Cancer Res. 1994;54:4539–4546. [PubMed] [Google Scholar]
- 10.Yan Y., Zuo X., Wei D. Concise Review: Emerging Role of CD44 in Cancer Stem Cells: A Promising Biomarker and Therapeutic Target. Stem Cells Transl. Med. 2015;4:1033–1043. doi: 10.5966/sctm.2015-0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen C., Zhao S., Karnad A., Freeman J.W. The biology and role of CD44 in cancer progression: Therapeutic implications. J. Hematol. Oncol. 2018;11:64. doi: 10.1186/s13045-018-0605-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mereiter S., Martins Á.M., Gomes C., Balmaña M., Macedo J.A., Polom K., Roviello F., Magalhães A., Reis C.A. O-glycan truncation enhances cancer-related functions of CD44 in gastric cancer. FEBS Lett. 2019;593:1675–1689. doi: 10.1002/1873-3468.13432. [DOI] [PubMed] [Google Scholar]
- 13.Slevin M., Krupinski J., Gaffney J., Matou S., West D., Delisser H., Savani R.C., Kumar S. Hyaluronan-mediated angiogenesis in vascular disease: Uncovering RHAMM and CD44 receptor signaling pathways. Matrix Biol. 2007;26:58–68. doi: 10.1016/j.matbio.2006.08.261. [DOI] [PubMed] [Google Scholar]
- 14.Naor D., Wallach-Dayan S.B., Zahalka M.A., Sionov R.V. Involvement of CD44, a molecule with a thousand faces, in cancer dissemination. Semin. Cancer Biol. 2008;18:260–267. doi: 10.1016/j.semcancer.2008.03.015. [DOI] [PubMed] [Google Scholar]
- 15.Günthert U., Hofmann M., Rudy W., Reber S., Zöller M., Haussmann I., Matzku S., Wenzel A., Ponta H., Herrlich P. A new variant of glycoprotein CD44 confers metastatic potential to rat carcinoma cells. Cell. 1991;65:13–24. doi: 10.1016/0092-8674(91)90403-L. [DOI] [PubMed] [Google Scholar]
- 16.Guo Q., Yang C., Gao F. The state of CD44 activation in cancer progression and therapeutic targeting. FEBS J. 2021;289:7970–7986. doi: 10.1111/febs.16179. [DOI] [PubMed] [Google Scholar]
- 17.Morath I., Hartmann T.N., Orian-Rousseau V. CD44: More than a mere stem cell marker. Int. J. Biochem. Cell Biol. 2016;81:166–173. doi: 10.1016/j.biocel.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 18.Bennett K.L., Jackson D.G., Simon J.C., Tanczos E., Peach R., Modrell B., Stamenkovic I., Plowman G., Aruffo A. CD44 isoforms containing exon V3 are responsible for the presentation of heparin-binding growth factor. J. Cell Biol. 1995;128:687–698. doi: 10.1083/jcb.128.4.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Orian-Rousseau V., Chen L., Sleeman J.P., Herrlich P., Ponta H. CD44 is required for two consecutive steps in HGF/c-Met signaling. Genes Dev. 2002;16:3074–3086. doi: 10.1101/gad.242602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishimoto T., Nagano O., Yae T., Tamada M., Motohara T., Oshima H., Oshima M., Ikeda T., Asaba R., Yagi H., et al. CD44 variant regulates redox status in cancer cells by stabilizing the xCT subunit of system xc(-) and thereby promotes tumor growth. Cancer Cell. 2011;19:387–400. doi: 10.1016/j.ccr.2011.01.038. [DOI] [PubMed] [Google Scholar]
- 21.Yamada S., Itai S., Nakamura T., Yanaka M., Kaneko M.K., Kato Y. Detection of high CD44 expression in oral cancers using the novel monoclonal antibody, C(44)Mab-5. Biochem. Biophys. Rep. 2018;14:64–68. doi: 10.1016/j.bbrep.2018.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goto N., Suzuki H., Tanaka T., Asano T., Kaneko M.K., Kato Y. Development of a Novel Anti-CD44 Monoclonal Antibody for Multiple Applications against Esophageal Squamous Cell Carcinomas. Int. J. Mol. Sci. 2022;23:5535. doi: 10.3390/ijms23105535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takei J., Asano T., Suzuki H., Kaneko M.K., Kato Y. Epitope Mapping of the Anti-CD44 Monoclonal Antibody (C44Mab-46) Using Alanine-Scanning Mutagenesis and Surface Plasmon Resonance. Monoclon. Antibodies Immunodiagn. Immunother. 2021;40:219–226. doi: 10.1089/mab.2021.0028. [DOI] [PubMed] [Google Scholar]
- 24.Asano T., Kaneko M.K., Takei J., Tateyama N., Kato Y. Epitope Mapping of the Anti-CD44 Monoclonal Antibody (C44Mab-46) Using the REMAP Method. Monoclon. Antibodies Immunodiagn. Immunother. 2021;40:156–161. doi: 10.1089/mab.2021.0012. [DOI] [PubMed] [Google Scholar]
- 25.Asano T., Kaneko M.K., Kato Y. Development of a Novel Epitope Mapping System: RIEDL Insertion for Epitope Mapping Method. Monoclon. Antibodies Immunodiagn. Immunother. 2021;40:162–167. doi: 10.1089/mab.2021.0023. [DOI] [PubMed] [Google Scholar]
- 26.Takei J., Kaneko M.K., Ohishi T., Hosono H., Nakamura T., Yanaka M., Sano M., Asano T., Sayama Y., Kawada M., et al. A defucosylated antiCD44 monoclonal antibody 5mG2af exerts antitumor effects in mouse xenograft models of oral squamous cell carcinoma. Oncol. Rep. 2020;44:1949–1960. doi: 10.3892/or.2020.7735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kato Y., Yamada S., Furusawa Y., Itai S., Nakamura T., Yanaka M., Sano M., Harada H., Fukui M., Kaneko M.K. PMab-213: A Monoclonal Antibody for Immunohistochemical Analysis Against Pig Podoplanin. Monoclon. Antibodies Immunodiagn. Immunother. 2019;38:18–24. doi: 10.1089/mab.2018.0048. [DOI] [PubMed] [Google Scholar]
- 28.Furusawa Y., Yamada S., Itai S., Sano M., Nakamura T., Yanaka M., Fukui M., Harada H., Mizuno T., Sakai Y., et al. PMab-210: A Monoclonal Antibody Against Pig Podoplanin. Monoclon. Antibodies Immunodiagn. Immunother. 2019;38:30–36. doi: 10.1089/mab.2018.0038. [DOI] [PubMed] [Google Scholar]
- 29.Furusawa Y., Yamada S., Itai S., Nakamura T., Yanaka M., Sano M., Harada H., Fukui M., Kaneko M.K., Kato Y. PMab-219: A monoclonal antibody for the immunohistochemical analysis of horse podoplanin. Biochem. Biophys. Rep. 2019;18:100616. doi: 10.1016/j.bbrep.2019.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Furusawa Y., Yamada S., Itai S., Nakamura T., Takei J., Sano M., Harada H., Fukui M., Kaneko M.K., Kato Y. Establishment of a monoclonal antibody PMab-233 for immunohistochemical analysis against Tasmanian devil podoplanin. Biochem. Biophys. Rep. 2019;18:100631. doi: 10.1016/j.bbrep.2019.100631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kato Y., Kaneko M.K., Kuno A., Uchiyama N., Amano K., Chiba Y., Hasegawa Y., Hirabayashi J., Narimatsu H., Mishima K., et al. Inhibition of tumor cell-induced platelet aggregation using a novel anti-podoplanin antibody reacting with its platelet-aggregation-stimulating domain. Biochem. Biophys. Res. Commun. 2006;349:1301–1307. doi: 10.1016/j.bbrc.2006.08.171. [DOI] [PubMed] [Google Scholar]
- 32.Chalise L., Kato A., Ohno M., Maeda S., Yamamichi A., Kuramitsu S., Shiina S., Takahashi H., Ozone S., Yamaguchi J., et al. Efficacy of cancer-specific anti-podoplanin CAR-T cells and oncolytic herpes virus G47Delta combination therapy against glioblastoma. Mol. Ther. Oncolytics. 2022;26:265–274. doi: 10.1016/j.omto.2022.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ishikawa A., Waseda M., Ishii T., Kaneko M.K., Kato Y., Kaneko S. Improved anti-solid tumor response by humanized anti-podoplanin chimeric antigen receptor transduced human cytotoxic T cells in an animal model. Genes Cells. 2022;27:549–558. doi: 10.1111/gtc.12972. [DOI] [PubMed] [Google Scholar]
- 34.Tamura-Sakaguchi R., Aruga R., Hirose M., Ekimoto T., Miyake T., Hizukuri Y., Oi R., Kaneko M.K., Kato Y., Akiyama Y., et al. Moving toward generalizable NZ-1 labeling for 3D structure determination with optimized epitope-tag insertion. Acta Crystallogr. D Struct. Biol. 2021;77:645–662. doi: 10.1107/S2059798321002527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaneko M.K., Ohishi T., Nakamura T., Inoue H., Takei J., Sano M., Asano T., Sayama Y., Hosono H., Suzuki H., et al. Development of Core-Fucose-Deficient Humanized and Chimeric Anti-Human Podoplanin Antibodies. Monoclon. Antibodies Immunodiagn. Immunother. 2020;39:167–174. doi: 10.1089/mab.2020.0019. [DOI] [PubMed] [Google Scholar]
- 36.Fujii Y., Matsunaga Y., Arimori T., Kitago Y., Ogasawara S., Kaneko M.K., Kato Y., Takagi J. Tailored placement of a turn-forming PA tag into the structured domain of a protein to probe its conformational state. J. Cell Sci. 2016;129:1512–1522. doi: 10.1242/jcs.176685. [DOI] [PubMed] [Google Scholar]
- 37.Abe S., Kaneko M.K., Tsuchihashi Y., Izumi T., Ogasawara S., Okada N., Sato C., Tobiume M., Otsuka K., Miyamoto L., et al. Antitumor effect of novel anti-podoplanin antibody NZ-12 against malignant pleural mesothelioma in an orthotopic xenograft model. Cancer Sci. 2016;107:1198–1205. doi: 10.1111/cas.12985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaneko M.K., Abe S., Ogasawara S., Fujii Y., Yamada S., Murata T., Uchida H., Tahara H., Nishioka Y., Kato Y. Chimeric Anti-Human Podoplanin Antibody NZ-12 of Lambda Light Chain Exerts Higher Antibody-Dependent Cellular Cytotoxicity and Complement-Dependent Cytotoxicity Compared with NZ-8 of Kappa Light Chain. Monoclon. Antibodies Immunodiagn. Immunother. 2017;36:25–29. doi: 10.1089/mab.2016.0047. [DOI] [PubMed] [Google Scholar]
- 39.Ito A., Ohta M., Kato Y., Inada S., Kato T., Nakata S., Yatabe Y., Goto M., Kaneda N., Kurita K., et al. A Real-Time Near-Infrared Fluorescence Imaging Method for the Detection of Oral Cancers in Mice Using an Indocyanine Green-Labeled Podoplanin Antibody. Technol. Cancer Res. Treat. 2018;17:1533033818767936. doi: 10.1177/1533033818767936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tamura R., Oi R., Akashi S., Kaneko M.K., Kato Y., Nogi T. Application of the NZ-1 Fab as a crystallization chaperone for PA tag-inserted target proteins. Protein Sci. 2019;28:823–836. doi: 10.1002/pro.3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shiina S., Ohno M., Ohka F., Kuramitsu S., Yamamichi A., Kato A., Motomura K., Tanahashi K., Yamamoto T., Watanabe R., et al. CAR T Cells Targeting Podoplanin Reduce Orthotopic Glioblastomas in Mouse Brains. Cancer Immunol. Res. 2016;4:259–268. doi: 10.1158/2326-6066.CIR-15-0060. [DOI] [PubMed] [Google Scholar]
- 42.Kuwata T., Yoneda K., Mori M., Kanayama M., Kuroda K., Kaneko M.K., Kato Y., Tanaka F. Detection of Circulating Tumor Cells (CTCs) in Malignant Pleural Mesothelioma (MPM) with the “Universal” CTC-Chip and An Anti-Podoplanin Antibody NZ-1.2. Cells. 2020;9:888. doi: 10.3390/cells9040888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nishinaga Y., Sato K., Yasui H., Taki S., Takahashi K., Shimizu M., Endo R., Koike C., Kuramoto N., Nakamura S., et al. Targeted Phototherapy for Malignant Pleural Mesothelioma: Near-Infrared Photoimmunotherapy Targeting Podoplanin. Cells. 2020;9:1019. doi: 10.3390/cells9041019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fujii Y., Kaneko M., Neyazaki M., Nogi T., Kato Y., Takagi J. PA tag: A versatile protein tagging system using a super high affinity antibody against a dodecapeptide derived from human podoplanin. Protein Expr. Purif. 2014;95:240–247. doi: 10.1016/j.pep.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 45.Kato Y., Kaneko M.K., Kunita A., Ito H., Kameyama A., Ogasawara S., Matsuura N., Hasegawa Y., Suzuki-Inoue K., Inoue O., et al. Molecular analysis of the pathophysiological binding of the platelet aggregation-inducing factor podoplanin to the C-type lectin-like receptor CLEC-2. Cancer Sci. 2008;99:54–61. doi: 10.1111/j.1349-7006.2007.00634.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kato Y., Vaidyanathan G., Kaneko M.K., Mishima K., Srivastava N., Chandramohan V., Pegram C., Keir S.T., Kuan C.T., Bigner D.D., et al. Evaluation of anti-podoplanin rat monoclonal antibody NZ-1 for targeting malignant gliomas. Nucl. Med. Biol. 2010;37:785–794. doi: 10.1016/j.nucmedbio.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Itai S., Ohishi T., Kaneko M.K., Yamada S., Abe S., Nakamura T., Yanaka M., Chang Y.W., Ohba S.I., Nishioka Y., et al. Anti-podocalyxin antibody exerts antitumor effects via antibody-dependent cellular cytotoxicity in mouse xenograft models of oral squamous cell carcinoma. Oncotarget. 2018;9:22480–22497. doi: 10.18632/oncotarget.25132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kamisawa T., Wood L.D., Itoi T., Takaori K. Pancreatic cancer. Lancet. 2016;388:73–85. doi: 10.1016/S0140-6736(16)00141-0. [DOI] [PubMed] [Google Scholar]
- 49.Gansauge F., Gansauge S., Zobywalski A., Scharnweber C., Link K.H., Nussler A.K., Beger H.G. Differential expression of CD44 splice variants in human pancreatic adenocarcinoma and in normal pancreas. Cancer Res. 1995;55:5499–5503. [PubMed] [Google Scholar]
- 50.Heider K.H., Mulder J.W., Ostermann E., Susani S., Patzelt E., Pals S.T., Adolf G.R. Splice variants of the cell surface glycoprotein CD44 associated with metastatic tumour cells are expressed in normal tissues of humans and cynomolgus monkeys. Eur. J. Cancer. 1995;31a:2385–2391. doi: 10.1016/0959-8049(95)00420-3. [DOI] [PubMed] [Google Scholar]
- 51.Rothenberg S.M., Ellisen L.W. The molecular pathogenesis of head and neck squamous cell carcinoma. J. Clin. Investig. 2012;122:1951–1957. doi: 10.1172/JCI59889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Compagnone M., Gatti V., Presutti D., Ruberti G., Fierro C., Markert E.K., Vousden K.H., Zhou H., Mauriello A., Anemone L., et al. ΔNp63-mediated regulation of hyaluronic acid metabolism and signaling supports HNSCC tumorigenesis. Proc. Natl. Acad. Sci. USA. 2017;114:13254–13259. doi: 10.1073/pnas.1711777114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Orian-Rousseau V., Ponta H. Perspectives of CD44 targeting therapies. Arch. Toxicol. 2015;89:3–14. doi: 10.1007/s00204-014-1424-2. [DOI] [PubMed] [Google Scholar]
- 54.Menke-van der Houven van Oordt C.W., Gomez-Roca C., van Herpen C., Coveler A.L., Mahalingam D., Verheul H.M., van der Graaf W.T., Christen R., Rüttinger D., Weigand S., et al. First-in-human phase I clinical trial of RG7356, an anti-CD44 humanized antibody, in patients with advanced, CD44-expressing solid tumors. Oncotarget. 2016;7:80046–80058. doi: 10.18632/oncotarget.11098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Riechelmann H., Sauter A., Golze W., Hanft G., Schroen C., Hoermann K., Erhardt T., Gronau S. Phase I trial with the CD44v6-targeting immunoconjugate bivatuzumab mertansine in head and neck squamous cell carcinoma. Oral Oncol. 2008;44:823–829. doi: 10.1016/j.oraloncology.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 56.Tijink B.M., Buter J., de Bree R., Giaccone G., Lang M.S., Staab A., Leemans C.R., van Dongen G.A. A phase I dose escalation study with anti-CD44v6 bivatuzumab mertansine in patients with incurable squamous cell carcinoma of the head and neck or esophagus. Clin. Cancer Res. 2006;12:6064–6072. doi: 10.1158/1078-0432.CCR-06-0910. [DOI] [PubMed] [Google Scholar]
- 57.Li G., Suzuki H., Ohishi T., Asano T., Tanaka T., Yanaka M., Nakamura T., Yoshikawa T., Kawada M., Kaneko M.K., et al. Antitumor activities of a defucosylated anti-EpCAM monoclonal antibody in colorectal carcinoma xenograft models. Int. J. Mol. Med. 2023;51:18. doi: 10.3892/ijmm.2023.5221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nanamiya R., Takei J., Ohishi T., Asano T., Tanaka T., Sano M., Nakamura T., Yanaka M., Handa S., Tateyama N., et al. Defucosylated Anti-Epidermal Growth Factor Receptor Monoclonal Antibody (134-mG(2a)-f) Exerts Antitumor Activities in Mouse Xenograft Models of Canine Osteosarcoma. Monoclon. Antibodies Immunodiagn. Immunother. 2022;41:1–7. doi: 10.1089/mab.2021.0036. [DOI] [PubMed] [Google Scholar]
- 59.Kawabata H., Suzuki H., Ohishi T., Kawada M., Kaneko M.K., Kato Y. A Defucosylated Mouse Anti-CD10 Monoclonal Antibody (31-mG(2a)-f) Exerts Antitumor Activity in a Mouse Xenograft Model of CD10-Overexpressed Tumors. Monoclon. Antibodies Immunodiagn. Immunother. 2022;41:59–66. doi: 10.1089/mab.2021.0048. [DOI] [PubMed] [Google Scholar]
- 60.Kawabata H., Ohishi T., Suzuki H., Asano T., Kawada M., Suzuki H., Kaneko M.K., Kato Y. A Defucosylated Mouse Anti-CD10 Monoclonal Antibody (31-mG(2a)-f) Exerts Antitumor Activity in a Mouse Xenograft Model of Renal Cell Cancers. Monoclon. Antibodies Immunodiagn. Immunother. 2022;41:320–327. doi: 10.1089/mab.2021.0049. [DOI] [PubMed] [Google Scholar]
- 61.Asano T., Tanaka T., Suzuki H., Li G., Ohishi T., Kawada M., Yoshikawa T., Kaneko M.K., Kato Y. A Defucosylated Anti-EpCAM Monoclonal Antibody (EpMab-37-mG(2a)-f) Exerts Antitumor Activity in Xenograft Model. Antibodies. 2022;11:74. doi: 10.3390/antib11040074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tateyama N., Nanamiya R., Ohishi T., Takei J., Nakamura T., Yanaka M., Hosono H., Saito M., Asano T., Tanaka T., et al. Defucosylated Anti-Epidermal Growth Factor Receptor Monoclonal Antibody 134-mG(2a)-f Exerts Antitumor Activities in Mouse Xenograft Models of Dog Epidermal Growth Factor Receptor-Overexpressed Cells. Monoclon. Antibodies Immunodiagn. Immunother. 2021;40:177–183. doi: 10.1089/mab.2021.0022. [DOI] [PubMed] [Google Scholar]
- 63.Takei J., Ohishi T., Kaneko M.K., Harada H., Kawada M., Kato Y. A defucosylated anti-PD-L1 monoclonal antibody 13-mG(2a)-f exerts antitumor effects in mouse xenograft models of oral squamous cell carcinoma. Biochem. Biophys. Rep. 2020;24:100801. doi: 10.1016/j.bbrep.2020.100801. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available in the article and supplementary material.