Abstract
Primary envelopment of several herpesviruses has been shown to occur by budding of intranuclear capsids through the inner nuclear membrane. By subsequent fusion of the primary envelope with the outer nuclear membrane, capsids are released into the cytoplasm and gain their final envelope by budding into vesicles in the trans-Golgi area. We show here that the product of the UL34 gene of pseudorabies virus, an alphaherpesvirus of swine, is localized in transfected and infected cells in the nuclear membrane. It is also detected in the envelope of virions in the perinuclear space but is undetectable in intracytoplasmic and extracellular enveloped virus particles. Conversely, the tegument protein UL49 is present in mature virus particles and absent from perinuclear virions. In the absence of the UL34 protein, acquisition of the primary envelope is blocked and neither virus particles in the perinuclear space nor intracytoplasmic capsids or virions are observed. However, light particles which label with the anti-UL49 serum are formed in the cytoplasm. We conclude that the UL34 protein is required for primary envelopment, that the primary envelope is biochemically different from the final envelope in that it contains the UL34 protein, and that perinuclear virions lack the tegument protein UL49, which is present in mature virions. Thus, we provide additional evidence for a two-step envelopment process in herpesviruses.
Herpesvirus virions consist of four morphologically differentiable components: the inner nucleoprotein core, which contains the genomic linear double-stranded DNA; the icosahedral capsid, comprising 150 hexamers and 12 pentamers; the tegument, which appears as an amorphous material in the electron microscope; and the envelope, a cell-derived lipid bilayer in which virus-encoded (glyco)proteins are embedded (35). Herpesvirus infection is initiated by the interaction of viral envelope glycoproteins with cellular surface components acting as virus receptors, followed by fusion of the lipid envelope with the cellular plasma membrane (29, 39). Capsids are released into the cytoplasm and transported to the nuclear pores, where they transmit their DNA into the nucleus. Whereas these entry steps during herpesvirus infection are undisputed, herpesvirus egress is still a matter of debate.
It was proposed for herpes simplex virus type 1 (HSV-1) that after intranuclear assembly, capsids bud through the inner nuclear membrane, thereby acquiring an envelope; the complete virion then traverses the secretory pathway, and membrane glycoproteins present in the primary envelope are modified during transit through the different endoplasmic reticulum (ER) and Golgi compartments (19). However, primarily morphological and limited biochemical evidence from different herpesvirus—host cell systems, e.g., pseudorabies virus (PrV), varicella-zoster virus (VZV), and human cytomegalovirus (HCMV), indicated a different virion morphogenesis pathway. For these viruses, it was proposed that the primary envelope of perinuclear virions is lost by fusion with the outer nuclear membrane and that capsids are thereby released into the cytoplasm. This notion could explain the high number of naked intracytoplasmic capsids. Moreover, stages of budding into vesicles in the trans-Golgi region were frequently observed, indicating acquisition of a secondary (and final) envelope by this mechanism (13, 15, 33, 43, 46). Recent biochemical data obtained with mutated viral glycoproteins which were specifically targeted to the ER and Golgi region also favored this model for HSV-1 (6, 44). However, conclusive evidence is still lacking.
We recently demonstrated that the envelope of perinuclear pseudorabies virus virions is morphologically distinct from that of mature virions (15) in that it lacks the characteristic surface projections (spikes) which, by immunogold labeling, have been shown to consist of viral glycoproteins. Moreover, the appearance of tegument material in perinuclear virions is morphologically distinct from that of intracytoplasmic and extracellular virions after final envelopment (15). Thus, perinuclear virions and intracytoplasmic and extracellular virions clearly have different appearances.
The molecular basis for viral egress is only incompletely understood. It has been shown for PrV and HSV-1 that the UL20 and gK proteins play a role in egress (1, 2, 9, 11, 16, 17, 18, 24, 42). Moreover, we recently demonstrated an important role for PrV glycoproteins E, I, and M in this process (3). In the absence of the latter three glycoproteins, intracytoplasmic capsids accumulate in association with amorphous material which labels with an antibody against the tegument protein UL49 (4). In the absence of the PrV UL3.5 protein, intracytoplasmic capsids accumulate and secondary envelopment does not occur (12).
Recently, egress of HSV-1 capsids from the nucleus has been found to require the UL34 protein (36), a putative membrane protein (32). Secondary structure analysis of the deduced amino acid sequence of the HSV-1 UL34 protein shows that it contains a C-terminal hydrophobic region which could serve as a transmembrane anchor. However, a previously suggested signal sequence (32) is probably not present. Surprisingly, the UL34 protein has also been detected in mature HSV-1 virions and has been proposed to interact with cytoplasmic dynein during virus entry. This interaction should target incoming capsids to the nuclear membrane (45). Since UL34 is proposed to be a membrane protein, its presence in mature extracellular virions could be interpreted differently when considering the different egress scenarios. In the single-envelopment model, UL34 should be present in the envelope of perinuclear virions and retained through passage through the secretory pathway. In the double-envelopment model, UL34 has to be present in the trans-Golgi vesicles to finally appear in mature virions. We wanted to analyze the localization and function of the UL34 homolog of PrV, an alphaherpesvirus of swine, which in many ways is similar to HSV-1 (30). To this end, we sequenced the UL34 gene, prepared a monospecific antiserum, established a cell line constitutively expressing UL34, and isolated a UL34 deletion mutant.
MATERIALS AND METHODS
Viruses and cells.
All PrV mutants are based on the wild-type laboratory strain PrV-Ka (21). PrV-9112C2 expresses the glycoprotein B (gB) gene of bovine herpesvirus 1 (BHV-1) instead of the PrV gB gene. In vitro, this virus is phenotypically similar to PrV-Ka (26). gB-negative (gB−) PrV was propagated on gB-expressing cells (34). Viruses were grown on rabbit kidney (RK13) cells in Eagle's minimum essential medium supplemented with 10% fetal calf serum.
Sequencing.
For sequence determination, SalI subfragments of BamHI fragment 1 were cloned into pUC19 and termini were sequenced. The region of interest was found to reside on subfragments 1B and 1C. Fragment Sal-1C was subcloned as 668-bp NruI/SalI and 454-bp NcoI/SalI fragments. Fragment Sal-1B was subcloned as PstI fragments of 891 and ca. 3,600 bp. The 891-bp PstI fragment was further shortened by cleavage with SphI and SmaI. Moreover, a 1,262-bp SalI/HinfI fragment was obtained (see Fig. 1A and B for locations of fragments and cleavage sites). Sequencing was performed using the dideoxy chain termination method (38) with double-stranded plasmid DNA and pUC-specific primers (ThermoSequenase primer cycle sequencing kit with 7-deaza-dGTP; Amersham Pharmacia Biotech, Freiburg, Germany) on an automatic sequencer (LI-COR 4200; MWG Biotech, Ebersberg, Germany). Ambiguous sequences were resequenced manually with a T7 sequencing kit and deaza-G/A T7 sequencing mixtures (Amersham Pharmacia Biotech) to verify correct sequences (22). The boundary between subfragments Sal-1B and Sal-1C was sequenced with specific primers (Sal-1B→Sal-1C: 5′-GATGAGCTTCAGGCGTTGGACCAGG-3′, nucleotides 759 to 735 in GenBank accession no. AJ276165 [see below]; Sal-1C→Sal-1B: 5′-GCCCTATAAAGTCCGCCGCCCGACC-3′, nucleotides 557 to 581 in GenBank accession no. AJ276165). Sequence analysis was performed with the GCG software package (Wisconsin Package version 10.0; Genetics Computer Group, Madison, Wis.).
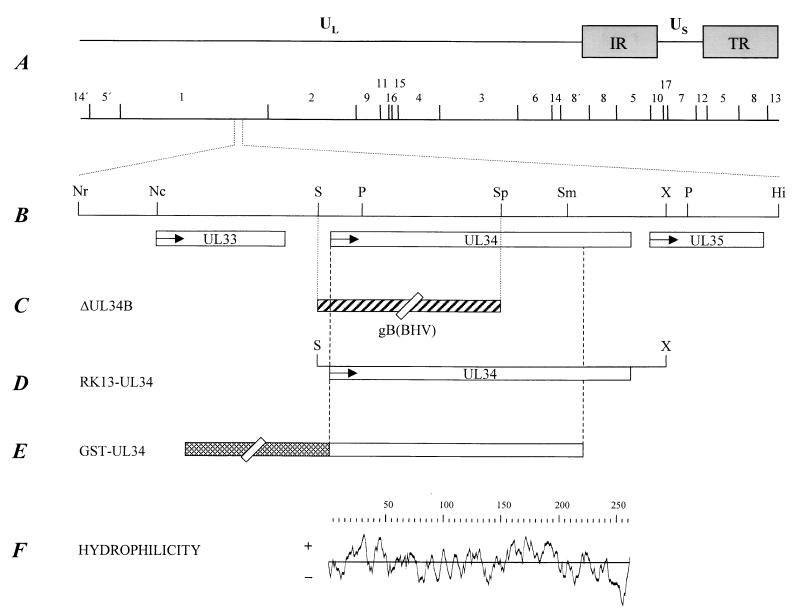
FIG. 1.
Map of the PrV genome. (A) A BamHI restriction fragment is shown below a schematic depiction of the PrV genome. The PrV genome is divided into unique long (UL) and unique short (US) regions by internal and terminal repeats (IR and TR, respectively). (B) Enlarged view of the relevant portion of the genome. The locations of the open reading frames are shown, and transcriptional orientation is indicated by arrows. Relevant cleavage sites are indicated: Nr, NruI; Nc, NcoI; S, SalI; P, PstI; Sp, SphI; Sm, SmaI; X, XhoI; Hi, HinfI. (C) Genomic arrangement in PrV-ΔUL34B, which contains a SalI/SphI deletion in the UL34 gene and a concomitant insertion of the BHV-1 gB gene (the latter not drawn to scale). (D) Genomic fragment used to establish cell line RK13-UL34. (E) Construct used for the expression of UL34 as a GST-UL34 fusion protein. (F) Hydrophilicity plot of the deduced UL34 protein.
Preparation of monospecific antisera.
To obtain an antiserum specific for the UL34 protein, a 681-bp fragment containing codons 1 to 220 of the UL34 gene with an EcoRI site at the 5′ end and an XhoI site at the 3′ end for convenient cloning was created using Platinum Pfx DNA polymerase (Life Technologies, Karlsruhe, Germany). The forward primer was UL34for (5′-CACAGAATTCCATGAGCGGCACCCTGGTCC-3′ [nt 723 to 742], EcoRI site in italics and initiation codon in bold), and the reverse primer was UL34rev (5′-CACACTCGAGACGCGGCCTGGCCCACG-3′ [nt 1384 to 1368], XhoI site in italics). The fragment was cloned into expression vector pGEX-4T-3 (Amersham Biotech Pharmacia) and expressed in Escherichia coli. A ca. 50-kDa glutathione S-transferase (GST) fusion protein was purified by polyacrylamide gel electrophoresis, electroeluted, and used for three immunizations of a rabbit with 100 μg of protein each at 4-week intervals. For preparation of an antiserum specific for the major capsid protein, the product of UL19 gene codons 954 to 1330 (nt 5874 to 7023 in GenBank accession no. L00676) (23) was expressed as a 67-kDa GST fusion protein, electroeluted after polyacrylamide gel electrophoresis, and used for immunization of a rabbit as described above.
Isolation of UL34-expressing cells.
For the construction of a complementing UL34-expressing cell line, a 954-bp SalI/XhoI subfragment of Sal-1B was inserted into pcDNA3 (InVitrogen, Groningen, The Netherlands) to yield pcDNA-UL34. Using SuperFect (Qiagen, Hilden, Germany), RK13 cells were then transfected with pcDNA-UL34, which contains the UL34 gene under the control of the HCMV immediate-early promoter-enhancer. Transfectants were selected with 500 μg of Geneticin (Life Technologies) per ml and tested for UL34 expression by indirect immunofluorescence and Western blotting using the monospecific UL34 antiserum. One cell clone, designated RK13-UL34, was selected for further experiments.
Isolation of a UL34-negative PrV mutant.
For isolation of a UL34-negative PrV mutant, a method devised in our laboratory and designated heterologous cis complementation was used; this method allows easy selection of desired viral mutants (12). It is based on the ability of BHV-1 gB to functionally complement deletion of the PrV gB gene (26), which is lethal to the virus (34). To this end, DNA of gB− PrV was cotransfected with plasmid pΔUL34B (see Fig. 1) into RK13-UL34 cells by calcium phosphate precipitation (14). This plasmid contains a ca. 5.5-kbp BstEII fragment in which the UL34 coding sequence has been partially deleted, including the start codon, and a BHV-1 gB expression unit has been inserted instead (see Fig. 1C). Transfer of the mutated sequences into the viral genome by homologous recombination should result in deletion of the parental UL34 gene and concomitant insertion of the BHV-1 gB gene into the gB− PrV genome. Thus, recombinant viruses should no longer depend on trans-complementation by gB-expressing cells but will rely on UL34-expressing cells in case this protein is essential. After cotransfection, progeny plaques were picked and purified three times, and viral DNA was analyzed by Southern blotting (37). One mutant, designated PrV-ΔUL34B, was randomly chosen for further testing.
Plaque assay and one-step growth analysis.
An assay for plaque formation was performed as described previously (25). For analysis of one-step growth, RK13 and RK13-UL34 cells were infected with PrV-9112C2 or PrV-ΔUL34B at a multiplicity of infection (MOI) of 10. After 1 h at 4°C, the inoculum was removed, prewarmed medium was added, and virus was allowed to penetrate for 1 h at 37°C. The remaining extracellular virus was inactivated by low-pH treatment (9). Immediately thereafter and after 4, 8, 12, 24, and 36 h, cells were scraped into the supernatant. After one freeze (−70°C)-thaw (37°C) cycle, cellular debris was pelleted, and the supernatant was titrated on RK13-UL34 cells. Average values and standard deviations of two independent experiments were calculated.
Virus purification.
For virus purification, cells were infected with wild-type PrV at an MOI of 0.1 and incubated until complete cytopathic effect developed. Remaining intact cells were lysed by freezing (−70°C) and thawing (37°C), cellular debris was removed by low-speed centrifugation, and the virus-containing supernatant was precleared by centrifugation through a 30% sucrose cushion. The pellet was resuspended in TBSal (200 mM NaCl, 2.6 mM KCl, 10 mM Tris-HCl [pH 7.5], 20 mM MgCl2, 1.8 mM CaCl2) and layered onto a discontinuous sucrose gradient (30, 40, and 50% sucrose). Virions accumulated at the boundary between 40 and 50% sucrose and were harvested by aspiration, pelleted, and resuspended in TBSal.
Western blotting and immunofluorescence.
Western blotting of infected-cell lysates or purified virions was performed as described previously (9) using monospecific antisera against the UL34 (dilution, 1:100,000) and UL19 (dilution, 1:1,000,000) proteins, glycoprotein H (gH) (GST-gH; dilution, 1:50,000) (25), and the UL49 protein (dilution 1:100,000) (4) or anti-glycoprotein C (gC) monoclonal antibody (MAb) B16-c8 (dilution, 1:100) (25).
For immunofluorescence, transfected and infected RK13 cells were fixed with 3% paraformaldehyde for 20 min and permeabilized with 3% paraformaldehyde–0.3% Triton X-100 for 10 min. Alternatively, cells were fixed with 80% ethanol. Cells were analyzed as described previously (31) using an LSM 510 laser scanning microscope (Zeiss, Oberkochen, Germany).
EM and immunolabeling.
For routine electron microscopy (EM), noninfected RK13 and RK13-UL34 cells and PrV-infected RK13 cells were fixed for 60 min with 2.5% glutaraldehyde buffered in 0.1 M sodium cacodylate (pH 7.2), 300 mosmol; Merck, Darmstadt, Germany). They were then scraped off the plate, pelleted by low-speed centrifugation, and embedded in low-melting-point agarose (Biozym, Oldendorf, Germany). Small pieces were postfixed in 1.0% aqueous OsO4 (Polysciences Europe, Eppelheim, Germany) and stained with uranyl acetate. After stepwise dehydration in ethanol, the cells were cleared in propylene oxide, embedded in Glycid Ether 100 (Serva, Heidelberg, Germany), and polymerized at 59°C for 4 days.
For intracellular labeling of viral proteins, cells were fixed with 0.5% glutaraldehyde in phosphate-buffered saline (PBS) (pH 7.2) for 30 min, embedded in low-melting point agarose, and postfixed in the above fixative for 30 min. Thereafter, samples were blocked with 0.5 M NH4Cl in PBS for 60 min, washed in PBS, stained in 0.5% aqueous uranyl acetate for 15 min, dehydrated in ethanol under progressive lowering of temperature, embedded in the acrylic resin Lowicryl K4M (Lowi, Waldkraiburg, Germany) at −35°C, and polymerized by UV light (λ, 360 nm) (7). Postembedding labeling of ultrathin sections was performed after blocking of surfaces with 1% cold-water fish gelatin–0.02 M glycine–1% bovine serum albumin fraction V (Sigma, Deisenhofen, Germany) in PBS by either overnight incubation at 4°C or 2 h of incubation at room temperature with monospecific anti-UL34 or anti-UL49 serum diluted in PBS-BSA. Diluted gold-tagged goat anti-rabbit antibodies or protein A-gold (GAR10 or PAG10; British BioCell International, Cambridge, United Kingdom) was added for 60 min at room temperature, and excess antibodies were removed by washing. The specificity of the reaction was controlled on uninfected and infected RK13 cells by using a gold conjugate without a primary antibody and by using non-herpesvirus protein-specific antibodies (anti-Newcastle disease virus antibodies). Ultrathin sections of conventionally embedded material and labeled Lowicryl sections, counterstained with uranyl acetate and lead salts, were examined with a model 400T electron microscope (Philips, Eindhoven, The Netherlands).
Nucleotide sequence accession number.
The sequence obtained has been deposited in GenBank under accession no. AJ276165.
RESULTS
Sequence and expression of PrV UL34.
The sequences of the UL33, UL34, and UL35 genes of PrV were established (GenBank accession no. AJ276165) (Fig. 1). The properties are listed in Table 1. All three genes reside in colinear positions with their homologs in the genomes of HSV-1 (28), equine herpesvirus 1 (EHV-1) (40), and VZV (8). Secondary structure prediction for the UL34 protein suggested that it does not contain an N-terminal hydrophobic region which could function as a signal sequence but does have a potential C-terminal membrane anchor (Fig. 1F). Thus, the UL34 protein would represent a type II C-terminally anchored membrane protein similar to the US9 protein of PrV (5, 27).
TABLE 1.
Properties of identified PrV genes
| Gene | ORF starta | ORF stopa | No. of amino acids | Predicted mass (kDa) | % Identity to homologous deduced proteins of:
|
||
|---|---|---|---|---|---|---|---|
| EHV-1 | HSV-1 | VZV | |||||
| UL33 | 218 | 565 | 115 | 12.7 | 46 | 54 | 42 |
| UL34 | 724 | 1,512 | 262 | 28.1 | 51 | 54 | 49 |
| UL35 | 1,567 | 1,878 | 103 | 11.5 | 41 | 31 | 39 |
Start and stop positions of the open reading frame (ORF) are indicated by the nucleotide number in the sequence of GenBank accession no. AJ276165 (first nucleotide of ATG to last nucleotide of stop codon).
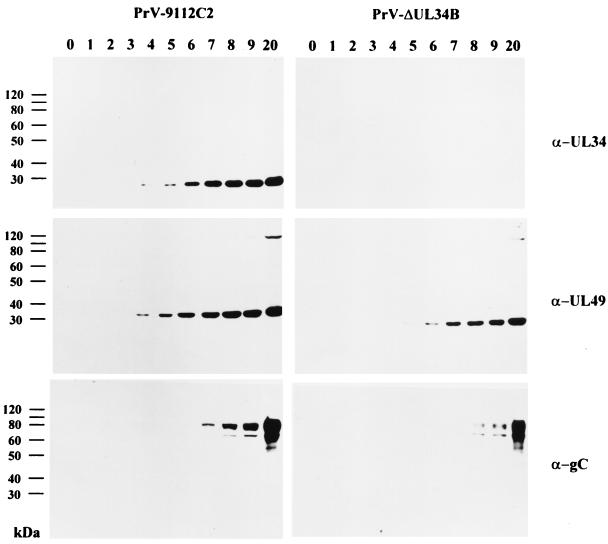
To monitor the expression of the UL34 protein in virus-infected cells, RK13 cells were infected at an MOI of 10 with either PrV-9112C2 or PrV-ΔUL34B and incubated on ice for 1 h. Thereafter, the inoculum was removed, and prewarmed medium was added. Immediately (time zero) and at different times thereafter, cell lysates were prepared and analyzed by Western blotting using the anti-UL34 serum. As shown in Fig. 2, upper left panel, the ca. 28-kDa UL34 protein was first detected 4 h after infection with PrV-9112C2, and the amount increased until 7 h postinfection (p.i.). For comparison, parallel blots were probed with the anti-UL49 serum (Fig. 2, middle left panel) and an anti-gC MAb (Fig. 2, lower left panel). The 33-kDa UL49 protein, which is homologous to HSV-1 VP22, was present in PrV-9112C2-infected-cell lysates as early as 3 h p.i., and the amount increased until 8 h p.i. At late times after infection, a higher-molecular-weight protein which might represent an oligomeric form of the protein was also specifically detected by the antiserum. The late gC protein was not detected before 6 h after PrV-9112C2 infection, and the amounts increased thereafter.
FIG. 2.
Expression kinetics. RK13 cells were infected at an MOI of 10 with PrV-9112C2 (left panels) or PrV-ΔUL34B (right panels). Cell lysates were prepared at the indicated times (hours) and analyzed in Western blots using monospecific antiserum against the UL34 or UL49 protein and a gC-specific MAb.
The PrV UL34 protein is not detected in extracellular virions.
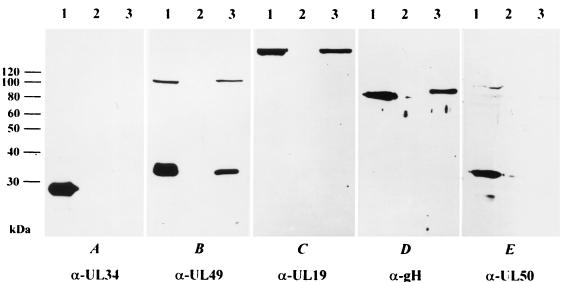
Extracellular PrV particles were purified by sucrose step gradient centrifugation. They were then lysed and analyzed by Western blotting. No reactivity with the anti-UL34 serum was observed in lysates of purified virions (Fig. 3A, lane 3), whereas the UL34 protein was readily demonstrated in lysates of infected cells (Fig. 3A, lane 1). In lysates of mock-infected cells, no signal was observed (Fig. 3A, lane 2). Thus, UL34 does not appear to be a structural component of PrV virions. In contrast, the UL49 protein was present in infected cells and purified virions (Fig. 3B, lanes 1 and 3), as was the UL19 (major capsid) protein (Fig. 3C, lanes 1 and 3) and gH (Fig. 3D, lanes 1 and 3). The virus-encoded dUTPase, a nuclear protein which previously had been shown to be absent from virus particles (20), was detected in infected-cell lysates (Fig. 3E, lane 1) but not in purified virions (Fig. 3E, lane 3).
FIG. 3.
Western blot of infected cells and purified virions. Lysates of RK13 cells infected with wild-type PrV (lanes 1) or mock infected (lanes 2) were analyzed by Western blotting, as were purified wild-type PrV virions (lanes 3). Blots were probed with monospecific antisera against the UL34, UL49, UL19, gH, and UL50 proteins.
The PrV UL34 protein is present in the nuclear membranes of transfected and infected cells.
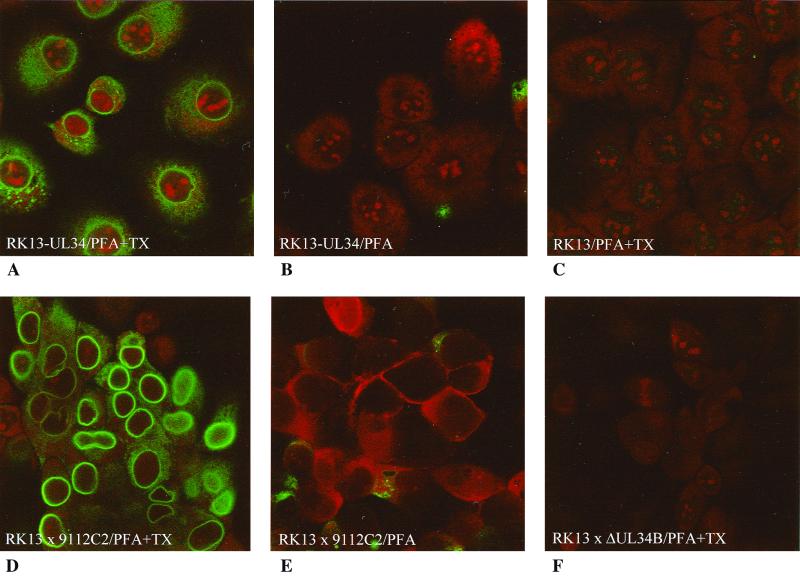
To analyze the intracellular localization of the PrV UL34 protein with and without ongoing virus infection, cell clones stably expressing UL34 were isolated using the monospecific anti-UL34 serum for immunofluorescence screening. A representative cell clone, RK13-UL34, was further analyzed. Besides producing diffuse staining in the cytoplasm with a sometimes speckled appearance, the anti-UL34 serum also produced perinuclear fluorescence in permeabilized RK13-UL34 cells (Fig. 4A); however, it did not react with nonpermeabilized cells (Fig. 4B) or normal RK13 cells (Fig. 4C). After infection with PrV-9112C2, bright perinuclear fluorescence was observed (Fig. 4D). Weaker cytoplasmic fluorescence, probably explained by UL34 being synthesized at cytoplasmic ribosomes, also occurred (see below). Nonpermeabilized RK13 cells infected with PrV-9112C2 showed only weak cytoplasmic staining, which was probably due to cellular damage and subsequent accessibility of the interior of the cell to the antibodies (Fig. 4E). Permeabilized RK13 cells infected with PrV-ΔUL34B did not react with the anti-UL34 serum (Fig. 4F).
FIG. 4.
Immunofluorescence of transfected and infected RK13 cells. RK13-UL34 (A and B) or RK13 (C to F) cells were analyzed either directly (A to C) or after infection with either PrV-9112C2 (D and E) or PrV-ΔUL34B (F). Cells were fixed with 3% paraformaldehyde (PFA) and permeabilized with 3% PFA–0.3% Triton X-100 (PFA+TX), as indicated. Assay results were monitored by laser scanning microscopy.
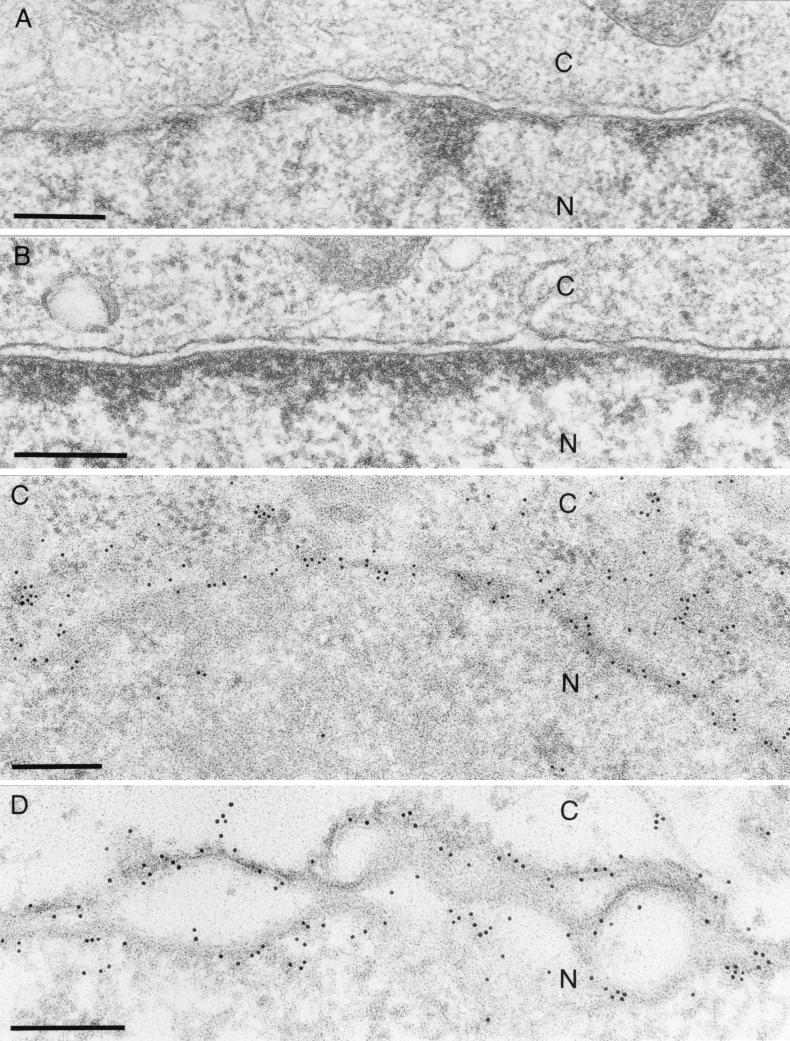
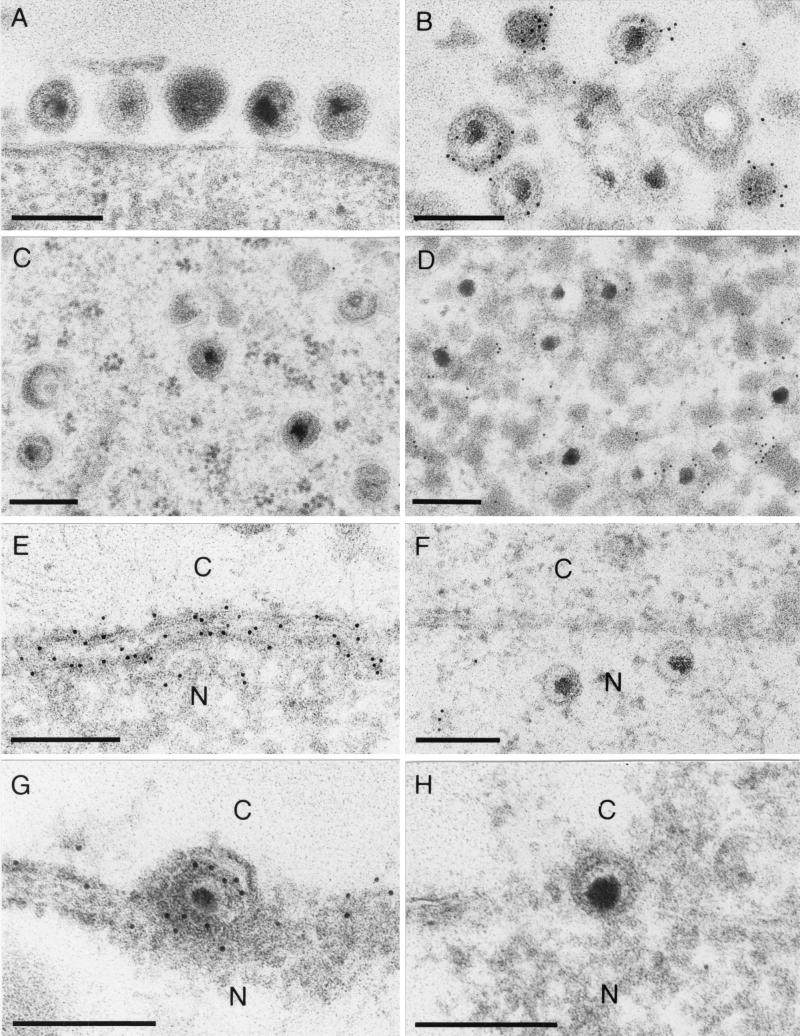
Immunogold labeling of RK13-UL34 cells demonstrated specific labeling of the nuclear membrane, with labeling detected in both lamellae (Fig. 5C and D). Label was also detected dispersed in the cytoplasm and associated with free ribosomes (Fig. 5C). Thus, in constitutively expressing cells, the UL34 protein is present in the nuclear membrane in the absence of other viral gene products. Virus-expressed UL34 also localized to the nuclear membrane (Fig. 6E). Interestingly, virus particles in the perinuclear space showed heavy labeling of UL34 in their membranes, demonstrating that these particles carry the UL34 protein in the envelope (Fig. 6G). However, no labeling of intracytoplasmic (Fig. 6C) or extracellular (Fig. 6A) enveloped particles was observed, indicating that UL34 is not present in their envelopes. This result correlates with the Western blot analysis of purified extracellular virions. Thus, by two different methods, Western blotting and EM with immunogold labeling, we were not able to detect the UL34 protein in mature virions carrying their final envelopes, whereas UL34 was clearly present in the envelopes of perinuclear virions.
FIG. 5.
EM of RK13-UL34 cells. RK13 (A) and RK13-UL34 (B to D) cells were embedded in Glycid Ether 100 and analyzed directly (A and B) or embedded in Lowicryl K4M and analyzed after incubation with the monospecific anti-UL34 serum (C and D). C, cytoplasm; N, nucleus. Bar, 250 nm.
FIG. 6.
EM of RK13 cells infected with wild-type PrV. Cells were embedded in Lowicryl K4M and reacted with monospecific antiserum against the UL34 protein (A, C, E, and G) or the UL49 protein (B, D, F, and H). (A and B) Mature extracellular virions. (C and D) Intracytoplasmic enveloped virions. (E and F) Nuclear membrane. (G and H) perinuclear virions. C, cytoplasm; N, nucleus. Bar, 250 nm.
The PrV UL49 protein is present in intracytoplasmic and extracellular virions but absent from perinuclear virus particles.
The UL49 protein of PrV, a homolog of the HSV-1 tegument protein VP22, has recently been shown to be present in the large intracytoplasmic inclusions formed during infection of noncomplementing cells with a PrV-gE/I/M− triple mutant (4). To assay for the presence of UL49 in perinuclear or mature virions, immunogold labeling of thin sections was performed with the monospecific anti-UL49 serum (Fig. 6B, D, F, and H). The anti-UL49 serum labeled intracytoplasmic (Fig. 6D) and extracellular (Fig. 6B) virions but did not detect any protein in perinuclear virus particles (Fig. 6H). Moreover, no UL49 protein was found in or adjacent to the nuclear membrane (Fig. 6F). Thus, perinuclear virions contain UL34 and lack UL49, and intracytoplasmic and extracellular virions lack UL34 and contain UL49.
The PrV UL34 protein is essential for productive replication.
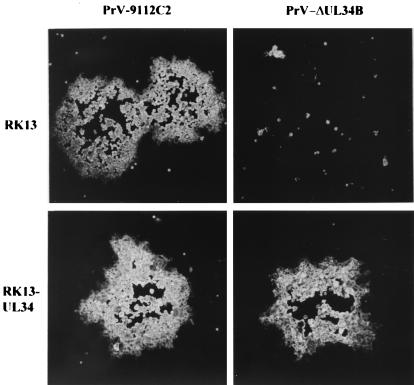
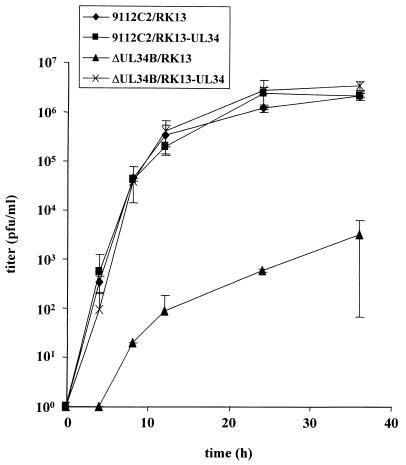
To assay for the function of the UL34 protein during PrV replication, a UL34 deletion mutant was isolated on RK13-UL34 cells by use of a heterologous complementation assay. The genome of the resulting mutant, PrV-ΔUL34B, was analyzed by Southern blot hybridization and found to contain the expected fragment pattern (data not shown). To assay for the absence of UL34 in mutant virus-infected cells, RK13 cells were infected with PrV-ΔUL34B which had been propagated on UL34-expressing cells. At various times p.i., the expression of UL34 was monitored by Western blotting of infected-cell lysates. As shown in Fig. 2, upper right panel, no UL34 protein was detectable at any time point, whereas the UL49 tegument protein (Fig. 2, middle right panel) and gC (Fig. 2, lower right panel) were readily demonstrable but appeared with a delay of ca. 2 h compared to the results for PrV-9112C2-infected cells. That UL34 was indeed required for productive PrV replication could be shown by plaque assays and one-step growth kinetics. In the absence of the UL34 protein, PrV produced only infected single cells or small foci of infection, and no further spread occurred (Fig. 7). Plaque formation was restored on RK13-UL34 cells. In one-step growth (Fig. 8), after infection of RK13-UL34 cells with PrV-ΔUL34B, levels of infectious progeny similar to those obtained with PrV-9112C2-infected RK13 or RK13-UL34 cells were obtained. However, titers of PrV-ΔUL34B grown on RK13 cells were approximately 3 orders of magnitude lower (Fig. 8). Thus, the UL34 protein is necessary for efficient productive PrV replication.
FIG. 7.
Plaque formation. Monolayers of RK13 and RK13-UL34 cells were infected under plaque assay conditions with PrV-9112C2 or PrV-ΔUL34B. At 2 days p.i., plaques were fixed in 80% ethanol, stained with an anti-gC MAb, and photographed.
FIG. 8.
One-step growth. RK13 and RK13-UL34 cells were infected with PrV-9112C2 or PrV-ΔUL34B and harvested at different times p.i., and titers were determined on RK13-UL34 cells. Average values from two independent experiments are shown. Vertical lines indicate standard deviations.
The PrV UL34 protein is required for primary envelopment.
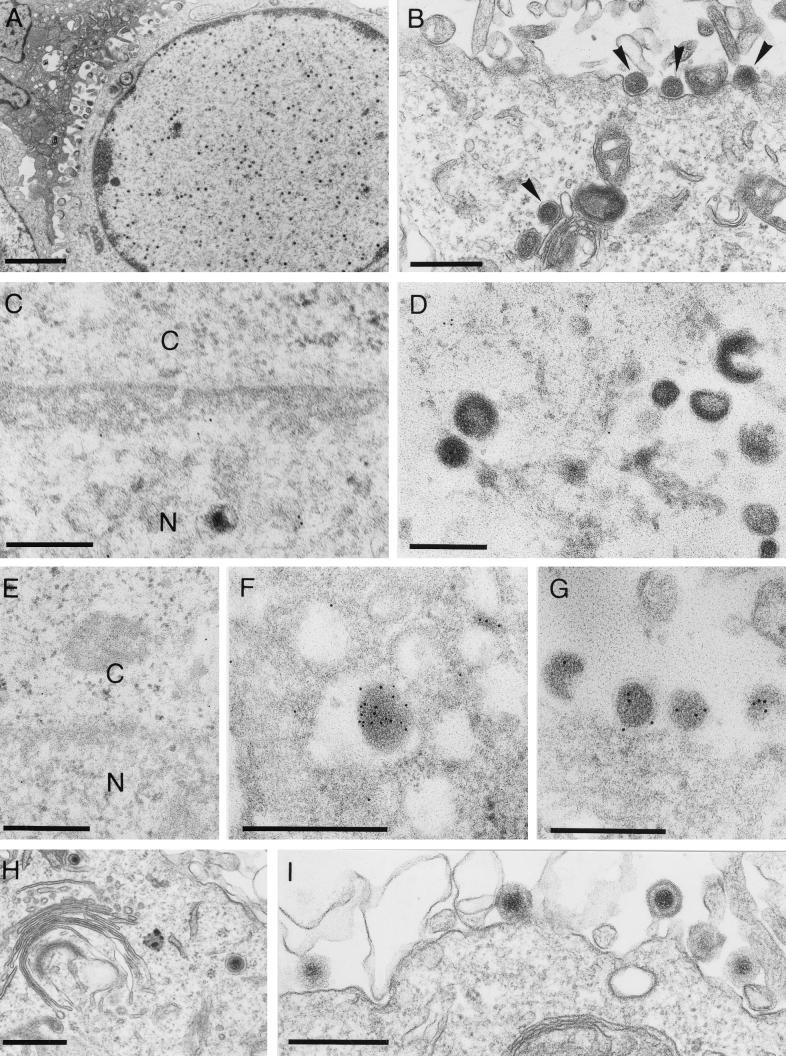
To locate the replication defect in PrV-ΔUL34B, RK13 (Fig. 9A to G) and RK13-UL34 (Fig. 9H and I) cells were infected at an MOI of 0.5 with PrV-ΔUL34B which had been propagated on UL34-expressing cells and were analyzed electron microscopically 14 h p.i. After infection of RK13 cells, neither perinuclear virions nor any intracytoplasmic or extracellular capsids or virions were observed, whereas intranuclear capsids were readily visualized (Fig. 9A). However, light (L) particles were formed in the cytoplasm and released (Fig. 9B). As expected, no specific labeling with the anti-UL34 serum was detected in PrV-ΔUL34B-infected RK13 cells in either the nuclear membrane (Fig. 9C) or intracytoplasmic L particles (Fig. 9D). In contrast, the anti-UL49 serum decorated intracytoplasmic (Fig. 9F) and extracellular (Fig. 9G) L particles, indicating that they contain the UL49 protein. As expected, the anti-UL49 serum did not label the nuclear membrane (Fig. 9E). In RK13-UL34 cells, all stages of virion maturation were observed after infection with PrV-ΔUL34B, including secondary envelopment in the cytoplasm (Fig. 9H) and release of mature virions (Fig. 9I). Also, labeling with the anti-UL34 serum was detected (data not shown). Thus, the absence of UL34 arrests viral morphogenesis before budding of capsids through the inner lamella of the nuclear membrane. However, the UL34 protein is apparently not required for envelopment of the tegument in the cytoplasm resulting in L particles.
FIG. 9.
EM of PrV-ΔUL34B-infected cells. RK13 (A to G) and RK13-UL34 (H and I) cells were infected with PrV-ΔUL34B at an MOI of 0.5 and analyzed 14 h p.i. Infected cells show the presence of nucleocapsids in the nucleus (A) and the production and release of L particles (B, arrowheads). After incubation with the anti-UL34 serum, no labeling of either the nuclear membrane (C) or the L particles (D) was observed. In contrast, the anti-UL49 serum decorated intracytoplasmic (F) and extracellular (G) L particles but not the nuclear membrane (E). In RK13-UL34 cells, all stages of virion maturation were detected, including secondary envelopment (H) and release of mature virions (I). Bars, 2.0 μm in panel A and 500 nm in panels B to I. C, cytoplasm; N, nucleus.
DISCUSSION
The mechanism and molecular basis for the envelopment and egress of herpesvirus particles are still largely unknown. It is generally accepted that after intranuclear capsid assembly and packaging of genomic DNA, capsids bud at the inner nuclear membrane into the perinuclear space, thereby acquiring a first envelope (35). However, controversy exists about the following steps in egress. Whereas for HSV-1 retention of the primary envelope and transport of the complete virus particle through the ER and Golgi region in the secretory pathway were suggested (19), for PrV, VZV, and HCMV it was proposed that the primary envelope is lost by fusion with the outer leaflet of the nuclear membrane or ER and that naked capsids are released into the cytoplasm. They then acquire their second (and final) envelope by budding into vesicles in the trans-Golgi region which contain processed viral glycoproteins (13, 15, 33, 43, 46).
We show here that the PrV UL34 protein localizes to the nuclear membrane when expressed either without other viral genes in stably transfected cells or during virus infection. Moreover, UL34 is present in the envelope of virions found in the perinuclear space. It is, however, undetectable in intracytoplasmic or extracellular virus particles, indicating either selective loss of UL34 during passage through the secretory pathway in the single-envelopment model or loss by fusion of the primary envelope with the outer leaflet of the nuclear membrane; the latter appears to be the more probable scenario. Interestingly, UL34 localizes to both leaflets of the nuclear membrane in transfected and infected cells. By the absence or presence of UL34, first and final envelopes can be distinguished clearly.
Retention of the primary envelope in the single-envelopment model also implies that tegument proteins are incorporated into virions in the nucleus. However, we detected the prominent tegument protein UL49 (homologous to HSV-1 VP22) exclusively in intracytoplasmic and extracellular virions or L particles, which lack capsids. No labeling with the anti-UL49 serum was detected in enveloped virions in the perinuclear space. Thus, UL49 appears to be either enriched in mature particles or added only during secondary envelopment. Taken together, these data clearly favor the deenvelopment-reenvelopment way of herpesvirus egress.
In PrV, the UL34 protein is not detected in extracellular virions by either Western blotting of virion lysates or immunogold labeling with the monospecific UL34 antiserum. Although it could be argued that UL34 may be modified, e.g., by phosphorylation, as has been shown for the HSV-1 UL34 protein (32), and may then no longer be recognized by our serum, we consider this possibility highly unlikely. The antiserum was prepared against a large portion of the protein, and it reacted very well in Western blotting and immuno-EM analyses (this study) and radioimmunoprecipitation assays (data not shown). Thus, the PrV UL34 protein is apparently absent from extracellular infectious virus and, therefore, should not play a role in PrV entry. This notion is in contrast to the situation for HSV-1, in which the UL34 protein has been described as a minor component of virions which, by interacting with cytoplasmic dynein, is involved in the transport of incoming capsids to the nuclear pore (45). Interestingly, the product of the Epstein-Barr virus BFRF1 gene, a positional homolog of UL34, is also detected in extracellular virions (10).
In the absence of the UL34 protein, neither virions in the perinuclear space nor intracytoplasmic capsids or virions or extracellular virus progeny were observed by EM. Thus, the UL34 protein is required for capsids to leave the nucleus. It is, however, not necessary for the formation of L particles, which contain tegument and envelope but lack capsids. Thus, the secondary envelopment in the cytoplasm obviously requires only tegument protein and trans-Golgi vesicles with viral glycoproteins. This notion correlates with previous findings that a block in capsid assembly does preclude the formation of infectious PrV virions but not the formation of PrV L particles (T. C. Mettenleiter et al., unpublished data). Consequently, L particles are labeled with the anti-UL49 serum but are not stained with the anti-UL34 serum.
The UL34 protein is moderately conserved in sequences within the Alphaherpesvirinae, and positional homologs are present in the other two subfamilies of the Herpesviridae (28). Thus, the described function may apply for all herpesviruses, a notion which appears reasonable for such a crucial step in virion morphogenesis. However, we cannot exclude the possibility of distinct functions for the different UL34 homologs in these viruses or the possibility that the UL34 protein constitutes a multifunctional protein.
The UL34 protein of HSV-1 is phosphorylated by a viral kinase encoded by the US3 gene (32). In PrV, a US3 deletion mutant has been observed to accumulate enveloped virus particles in the perinuclear space (41). Therefore, in the absence of UL34, budding at the inner nuclear membrane does not occur, whereas in the absence of US3 (and possibly a lack of phosphorylation of UL34), deenvelopment at the outer nuclear membrane seems to be blocked. Since UL34 has been shown by us to be located in both leaflets, the phosphorylation state of the UL34 protein may well account for these different functions. Which role the UL34 protein, as a constituent of the primary envelope of perinuclear virions, plays in the fusion process is unclear.
ACKNOWLEDGMENTS
Part of this work was supported by the Deutsche Forschungsgemeinschaft (DFG grant Me 854/5-1).
We thank Christel Möller, Petra Meyer, Uta Hartwig, and Nadine Müller for excellent technical assistance; Gabi Weidt for help with establishing the complementing cell line; Klaus Osterrieder for help with the confocal laser scanning microscope; and Egbert Mundt for immunization of rabbits.
REFERENCES
- 1.Avitabile E, Ward P L, Di Lazzaro C, Torrisi M R, Roizman B, Campadelli-Fiume G. The herpes simplex virus UL20 protein compensates for the differential disruption of exocytosis of virions and viral membrane glycoproteins associated with fragmentation of the Golgi apparatus. J Virol. 1994;68:7397–7405. doi: 10.1128/jvi.68.11.7397-7405.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baines J D, Ward P L, Campadelli-Fiume G, Roizman B. The UL20 gene of herpes simplex virus 1 encodes a function necessary for viral egress. J Virol. 1991;65:6414–6424. doi: 10.1128/jvi.65.12.6414-6424.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brack A R, Dijkstra J M, Granzow H, Klupp B G, Mettenleiter T C. Inhibition of virion maturation by simultaneous deletion of glycoproteins E, I, and M of pseudorabies virus. J Virol. 1999;73:5364–5372. doi: 10.1128/jvi.73.7.5364-5372.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brack A R, Klupp B G, Granzow H, Tirabassi R, Enquist L W, Mettenleiter T C. Role of the cytoplasmic tail of pseudorabies virus glycoprotein E in virion formation. J Virol. 2000;74:4004–4016. doi: 10.1128/jvi.74.9.4004-4016.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brideau A, Banfield B W, Enquist L W. The Us9 gene product of pseudorabies virus, an alphaherpesvirus, is a phosphorylated, tail-anchored type II membrane protein. J Virol. 1998;72:4560–4570. doi: 10.1128/jvi.72.6.4560-4570.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Browne H, Bell S, Minson T, Wilson D. An endoplasmic reticulum-retained herpes simplex virus glycoprotein H is absent from secreted virions: evidence for reenvelopment during egress. J Virol. 1996;70:4311–4316. doi: 10.1128/jvi.70.7.4311-4316.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carlemalm E, Garavito R M, Villiger W. Resin development for electron microscopy and an analysis of embedding at low temperature. J Microsc. 1982;126:123–143. doi: 10.1111/j.1365-2818.1982.tb00358.x. [DOI] [PubMed] [Google Scholar]
- 8.Davison A J, Scott J E. The complete DNA sequence of varicella-zoster virus. J Gen Virol. 1986;67:1759–1816. doi: 10.1099/0022-1317-67-9-1759. [DOI] [PubMed] [Google Scholar]
- 9.Dietz P, Klupp B G, Weiland E, Köllner B, Mettenleiter T C. Pseudorabies virus glycoprotein K requires the UL20 gene product for processing. J Virol. 2000;74:5083–5090. doi: 10.1128/jvi.74.11.5083-5090.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farina A, Santarelli R, Gonella R, Bei R, Muraro R, Cardinali G, Uccini S, Ragona G, Frati L, Faggioni A, Angeloni A. The BFRF1 gene of Epstein-Barr virus encodes a novel protein. J Virol. 2000;74:3235–3244. doi: 10.1128/jvi.74.7.3235-3244.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuchs W, Klupp B G, Granzow H, Mettenleiter T C. The UL20 gene product of pseudorabies virus functions in virus egress. J Virol. 1997;71:5639–5646. doi: 10.1128/jvi.71.7.5639-5646.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuchs W, Klupp B G, Granzow H, Rziha H J, Mettenleiter T C. Identification and characterization of the pseudorabies virus UL3.5 protein, which is involved in virus egress. J Virol. 1996;70:3517–3527. doi: 10.1128/jvi.70.6.3517-3527.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gershon A A, Sherman D L, Zhu Z, Gabel C A, Ambron R T, Gershon M D. Intracellular transport of newly synthesized varicella-zoster virus: final envelopment in the trans-Golgi network. J Virol. 1994;68:6372–6390. doi: 10.1128/jvi.68.10.6372-6390.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graham F L, van der Eb A J. A new technique for the assay of infectivity of human adenovirus. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 15.Granzow H, Weiland F, Jöns A, Klupp B G, Karger A, Mettenleiter T C. Ultrastructural analysis of the replication cycle of pseudorabies virus in cell culture: a reassessment. J Virol. 1997;71:2072–2082. doi: 10.1128/jvi.71.3.2072-2082.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hutchinson L, Johnson D C. Herpes simplex virus glycoprotein K promotes egress of virus particles. J Virol. 1995;69:5401–5413. doi: 10.1128/jvi.69.9.5401-5413.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hutchinson L, Goldsmith K, Snoddy D, Gosh H, Graham F L, Johnson D C. Identification and characterization of a novel herpes simplex virus glycoprotein, gK, involved in cell fusion. J Virol. 1992;66:5603–5609. doi: 10.1128/jvi.66.9.5603-5609.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jayachandra S, Baghian A, Kousoulas K G. Herpes simplex virus type 1 glycoprotein K is not essential for infectious virus production in actively replicating cells but is required for efficient envelopment and translocation of infectious virions from the cytoplasm to the extracellular space. J Virol. 1997;71:5012–5024. doi: 10.1128/jvi.71.7.5012-5024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson D C, Spear P G. Monensin inhibits the processing of herpes simplex virus glycoproteins, their transport to the cell surface, and the egress of virions from infected cells. J Virol. 1982;43:1102–1112. doi: 10.1128/jvi.43.3.1102-1112.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jöns A, Mettenleiter T C. Identification and characterization of pseudorabies virus dUTPase. J Virol. 1996;70:1242–1245. doi: 10.1128/jvi.70.2.1242-1245.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaplan A S, Vatter A E. A comparison of herpes simplex and pseudorabies viruses. Virology. 1959;7:394–407. doi: 10.1016/0042-6822(59)90068-6. [DOI] [PubMed] [Google Scholar]
- 22.Klupp B G, Mettenleiter T C. Sequence and expression of the glycoprotein gH gene of pseudorabies virus. Virology. 1991;182:732–741. doi: 10.1016/0042-6822(91)90614-h. [DOI] [PubMed] [Google Scholar]
- 23.Klupp B G, Kern H, Mettenleiter T C. The virulence-determining genomic Bam HI fragment 4 of pseudorabies virus contains genes corresponding to the UL15 (partial), UL18, UL19, UL20, and UL21 genes of herpes simplex virus and a putative origin of replication. Virology. 1992;191:900–908. doi: 10.1016/0042-6822(92)90265-q. [DOI] [PubMed] [Google Scholar]
- 24.Klupp B G, Baumeister J, Dietz P, Granzow H, Mettenleiter T C. Pseudorabies virus glycoprotein gK is a virion structural component involved in virus release but not required for entry. J Virol. 1998;72:1949–1958. doi: 10.1128/jvi.72.3.1949-1958.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klupp B G, Mettenleiter T C. Glycoprotein gL-independent infectivity of pseudorabies virus is mediated by a gD-gH fusion protein. J Virol. 1999;73:3014–3022. doi: 10.1128/jvi.73.4.3014-3022.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kopp A, Mettenleiter T C. Stable rescue of a glycoprotein gII deletion mutant of pseudorabies virus by glycoprotein gI of bovine herpesvirus 1. J Virol. 1992;66:2754–2762. doi: 10.1128/jvi.66.5.2754-2762.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kutay U, Hartmann E, Rapoport T. A class of membrane proteins with a C-terminal anchor. Trends Cell Biol. 1993;3:72–75. doi: 10.1016/0962-8924(93)90066-a. [DOI] [PubMed] [Google Scholar]
- 28.McGeoch D J, Dalrymple M A, Davison A J, Dolan A, Frame M C, McNab D, Perry L J, Scott J E, Taylor P. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J Gen Virol. 1988;69:1531–1574. doi: 10.1099/0022-1317-69-7-1531. [DOI] [PubMed] [Google Scholar]
- 29.Mettenleiter T C. Initiation and spread of α-herpesvirus infections. Trends Microbiol. 1994;2:2–4. doi: 10.1016/0966-842x(94)90335-2. [DOI] [PubMed] [Google Scholar]
- 30.Mettenleiter T C. Pseudorabies (Aujeszky's disease) virus: the virus and molecular pathogenesis—state of the art June 1999. Vet Res. 2000;31:99–115. doi: 10.1051/vetres:2000110. [DOI] [PubMed] [Google Scholar]
- 31.Nixdorf R, Klupp B G, Karger A, Mettenleiter T C. Effects of truncation of the carboxy terminus of pseudorabies virus glycoprotein B on infectivity. J Virol. 2000;74:7137–7145. doi: 10.1128/jvi.74.15.7137-7145.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Purves F, Spector D, Roizman B. UL34, the target of the herpes simplex virus US3 protein kinase, is a membrane protein which in its unphosphorylated state associates with novel phosphoproteins. J Virol. 1992;66:4295–4303. doi: 10.1128/jvi.66.7.4295-4303.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Radsak K, Eickmann M, Mockenhaupt T, Bogner E, Kern H, Eis-Hübinger A, Reschke M. Retrieval of human cytomegalovirus glycoprotein B from the infected cell surface for virus envelopment. Arch Virol. 1996;141:557–572. doi: 10.1007/BF01718317. [DOI] [PubMed] [Google Scholar]
- 34.Rauh I, Weiland F, Fehler F, Keil G M, Mettenleiter T C. Pseudorabies virus mutants lacking the essential glycoprotein gII can be complemented by glycoprotein gI of bovine herpesvirus 1. J Virol. 1991;65:621–631. doi: 10.1128/jvi.65.2.621-631.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roizman B. Herpesviridae. In: Fields B N, Knipe D M, Howley P M, editors. Virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2221–2231. [Google Scholar]
- 36.Roller R, Zhou Y, Schnetzer R, Ferguson J, DeSalvo D. Herpes simplex virus type 1 UL34 gene product is required for viral envelopment. J Virol. 2000;74:117–129. doi: 10.1128/jvi.74.1.117-129.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 38.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spear P. Entry of alphaherpesviruses into cells. Semin Virol. 1993;4:167–180. [Google Scholar]
- 40.Telford E, Watson M S, McBride K, Davison A J. The DNA sequence of equine herpesvirus-1. Virology. 1992;189:304–316. doi: 10.1016/0042-6822(92)90706-u. [DOI] [PubMed] [Google Scholar]
- 41.Wagenaar F, Pol J M A, Peeters B, Gielkens A L J, de Wind N, Kimman T G. The US3-encoded protein kinase from pseudorabies virus affects egress of virions from the nucleus. J Gen Virol. 1995;76:1851–1859. doi: 10.1099/0022-1317-76-7-1851. [DOI] [PubMed] [Google Scholar]
- 42.Ward P L, Campadelli-Fiume G, Avitabile E, Roizman B. Localization and putative function of the UL20 membrane protein in cells infected with herpes simplex virus type 1. J Virol. 1994;68:7406–7417. doi: 10.1128/jvi.68.11.7406-7417.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whealy M E, Card J P, Meade R P, Robbins A K, Enquist L W. Effect of brefeldin A on alphaherpesvirus membrane protein glycosylation and virus egress. J Virol. 1991;65:1066–1081. doi: 10.1128/jvi.65.3.1066-1081.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Whiteley A, Bruun B, Minson T, Browne H. Effects of targeting herpes simplex virus type 1 gD to the endoplasmic reticulum and trans-Golgi network. J Virol. 1999;73:9515–9520. doi: 10.1128/jvi.73.11.9515-9520.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ye G-J, Vaughan K T, Vallee R B, Roizman B. The herpes simplex virus 1 UL34 protein interacts with a cytoplasmic dynein intermediate chain and targets nuclear membrane. J Virol. 2000;74:1355–1363. doi: 10.1128/jvi.74.3.1355-1363.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu Z, Gershon M D, Hao Y, Ambron R T, Gabel C A, Gershon A A. Envelopment of varicella-zoster virus: targeting of viral glycoproteins to the trans-Golgi network. J Virol. 1995;69:7951–7959. doi: 10.1128/jvi.69.12.7951-7959.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]