ABSTRACT
Methicillin-resistant Staphylococcus aureus (MRSA) infections cause substantive morbidity and mortality in neonates. Using publicly available resources from the National Center of Biotechnology Information (NCBI) and Food and Drug Administration’s (FDA) GalaxyTrakr pipeline, we illustrate the dynamics of MRSA colonization and infection in neonates. Over 217 days of prospective surveillance, analyses revealed concurrent MRSA transmission chains affecting 11 of 17 MRSA-colonized patients (65%), with two clusters that demonstrated intervals of more than a month among the appearance of isolates. All MRSA infected neonates (n = 3) showed previous colonization with the infecting strain. GalaxyTrakr clustering of the NICU strains, in the context of 21,521 international isolates deposited in NCBI’s Pathogen Detection Resource, revealed NICU isolates to be distinct from adult MRSA strains seen locally and internationally. Clustering of the NICU strains within an international context enhanced the resolution of strain clusters and supported the rule-out of suspected, local transmission events within the NICU. Analyses also identified sequence type 1535 isolates, emergent in the Middle East, carrying a unique SCCmec with fusC and aac(6’)-Ie/aph(2’’)-1a that provided a multidrug-resistant phenotype. NICU genomic pathogen surveillance, leveraging public repositories and outbreak detection tools, supports rapid identification of cryptic MRSA clusters, and can inform infection prevention interventions for this vulnerable patient population. Results demonstrate that sporadic infections in the NICU may be indicative of hidden chains of asymptomatic transmission best identified with sequenced-based approaches.
KEYWORDS: antibiotic resistance, genomic epidemiology, MRSA, NICU, nosocomial disease, pathogen detection, Staphylococcus aureus
INTRODUCTION
Staphylococcus aureus is a common cause of health care associated infections, including sepsis, respiratory, skin, and soft tissue infections (1). Neonates, preterm, and low-birthweight infants have increased risks for methicillin-resistant Staphylococcus aureus (MRSA) colonization and infection given their lack of protective colonizing microbiota and immune immaturity (2). MRSA colonization is a well-recognized risk factor for infection in neonatal intensive care unit (NICU) patients. Nosocomial and neonatal transmission of S. aureus has been documented from colonized health care workers, parents, and environmental reservoirs (3). Efforts to reduce infections use culture- or molecular-based detection to identify MRSA-colonized infants to implement strategies for infant decolonization, and to reduce further transmission (4). These efforts rely on sensitive strain-typing techniques to distinguish endemic, outbreak, and community-acquired strains.
MRSA strains frequently harbor broad-spectrum resistance to beta-lactam and other antibiotic classes. Resistant phenotypes are commonly conferred by a mobile staphylococcal cassette chromosome mec (SCCmec) that encodes an alternative peptidoglycan transpeptidase homolog of PBP2, PBP2A, with reduced affinity for beta-lactam antibiotics, particularly cephalosporins (5). Many SCCmecs, as well as non-mec SCCs, encode various other resistance genes, making SCCs important contributors to multidrug resistance.
Whole-genome sequencing (WGS) enables outbreak investigations for MRSA and other pathogens through high-resolution strain tracking (6), a method that provides higher resolution information over targeted molecular strain-typing (7). When combined with epidemiologic data, WGS enables definitive clonal assessments and improved identification of direct transmission events (8). Applied to NICU populations, WGS has improved the detection of health care worker transmission events (9), and supported more effective MRSA screening programs (10). Importantly, WGS can also rule-out nosocomial transmission events, providing reassurance to staff and more efficient utilization of hospital resources in infection prevention (11).
Hospital MRSA investigations using public repositories of internationally sequenced strains increase the resolution of clonal associations, including to support rule out determinations for local strain outbreak clusters (12, 13). Among U.S.-based genomic surveillance programs, the FDA GenomeTrakr Network, and CDC’s PulseNet use validated bacterial WGS methods for outbreak investigation in food safety and public health. While genomic surveillance at the hospital level is less common, the tools provided by these national programs can lessen the costs and resources needed to implement robust genomic surveillance methods within a nosocomial setting (14).
MRSA strains with similar resistance phenotypes in colonized and infected infants raised concerns for asymptomatic and cryptic transmission events. To investigate, genomic analyses of NICU MRSA isolates over a 341-day period identified multiple cryptic transmission chains, including ones associated with active infections. Nationally available tools for outbreak detection leveraged 10,145 internationally deposited MRSA genomes in the NCBI Pathogen Detection resource with the validated GalaxyTrakr platform. Analyses significantly improved the phylogenetic resolution of local strain clusters, improved rule-out determinations, and provided a well-validated pipeline to reduce the costs and resources needed to conduct high-resolution cluster analyses in a hospital setting.
MATERIALS AND METHODS
IRB study protocol and data collection.
The study was conducted under IRB protocol 2011-P-002883 (PI: Bry, Partners Healthcare). The Crimson LIMS was used for sample retrieval. Patient data and microbiological test results were retrieved using the Partners Research Patient Data Registry (15). Data used for analyses were deidentified.
Study design and clinical interventions.
The NICU at Brigham and Women’s Hospital is a 66-bed facility providing specialized level II care for newborns with noncritical conditions such as hypoglycemia or in need of IV fluid treatment, and level III urgent care for newborns undergoing surgery or as needed for sustained life support. The unit contains distinct areas for levels II and III care. Neonates are transferred between unit areas when medically appropriate. For a 558-day period over 2018 to 2019, MRSA isolates from weekly nasal-perirectal swab screening and from diagnostic cultures were evaluated for relatedness of their phenotypic susceptibility profiles to evaluate suspicious outbreak clusters. Retrospective analyses triggered a prospective genomic surveillance period that started on day 342 to day 558 during which MRSA isolates from all nasal-perirectal swab cultures and diagnostic cultures were sequenced, in addition to the previously collected strains. Medical records for NICU patients during the surveillance period were evaluated for clinical diagnoses, reasons for NICU admission, and additional culture-based analyses, including antimicrobial resistance profiles. In the context of the clinical genomic surveillance program, only patient surveillance and diagnostic strains were evaluated. Nonclinical samples were not collected from patient family members, NICU personnel, nor the environment. Neonates colonized or infected with MRSA were placed in single rooms and managed with Contact Precautions.
Sample collection.
BD BBL CultureSwabs (Becton Dickinson, San Jose, CA) were used for nasal-perirectal (NP) screening for S. aureus. Swabs were cultured to colistin nalidixic acid (CNA) blood agar for 18 to 24 h at 37°C. S. aureus was identified by colony morphology, Gram stain, positive catalase, and coagulase assays (Thermo Fisher Scientific). Diagnostic cultures with S. aureus were evaluated similarly. Collected isolates were preserved at −80°C in 2 mL Cryovials (Corning, Corning, NY). In the context of the clinical genomic surveillance program, and the IRB protocol under which it operates, only patient surveillance and diagnostic strains were evaluated.
Antibiotic resistance testing.
S. aureus isolates were tested clinically for antibiotic resistance by Vitek 2 with Susceptibility Card AST-GP78 (bioMérieux, Durham, NC). Microdilution testing for fusidic acid resistance was done by broth microdilution according to CLSI guidelines (16). MRSA isolates were called by resistance to cefoxitin.
Genome sequencing.
All isolate genomes were sequenced using Illumina NextSeq as described previously (Illumina, San Diengo, CA) (14, 15). MLST 1535 isolates were additionally sequenced using Oxford Nanopore GridIon sequencing (Oxford Nanopore Technologies, Oxford, UK). The libraries were prepared using rapid sequencing kit RBK004 and run on a MIN106D flow cell (R9.4.1) for 48 h according to the manufacturer’s instructions. The genomes were de novo assembled using both the both Nanopore and the NextSeq reads with Unicycler v.0.4.8 (17).
Bioinformatic analyses.
Patient data were analyzed using Python with the packages Matplotlib and SciPy (18, 19). Isolate genome sequencing reads were uploaded to the NCBI Sequence Read Archive for inclusion in the Pathogen Detection program (Table S3), which includes single nucleotide polymorphism (SNP) clustering, virulence gene detection, antimicrobial resistance gene detection, and isolate metadata (12). An additional 21,521 genomes, available at the time of analysis, were included in cluster detection. Phylogenetic results were visualized using the Pathogen Detection Isolates Browser.
A total of 10,387 S. aureus genomes with available assemblies and Illumina sequencing data at NCBI Short Read Archive (SRA) underwent further analysis (Table S4). Whole-genome MLST was performed using the program tblastn and coding sequences from RefSeq genome NC_007795.1 (20, 21). The threshold for inclusion was an e-value ≤0.001 and a length between 0.5× and 1.5× the reference sequence coding length. Gene matches were retained if there was at most one high-quality hit in each genome for all genomes in a downsampled set of all S. aureus isolate genomes available in the NCBI Pathogen Detection program. Gene sequences were only kept if they matched the reference sequence length to allow for efficient SNP calculation across thousands of genomes. Genes were excluded if present in <90% of genomes, resulting in a database of 2,021 genes. Genomes with <90% of these genes represented within the criteria above were excluded, resulting in a final database representing 10,145 genomes from the 10,387 starting genomes. From this set of genomic information, allelic genomic differences normalized for analyzed sequence length were calculated to determine the closest set of 25 genomes to NICU MRSA isolates, keeping all tied for 25th closest. SNP matrices were then calculated using GalaxyTrakr with the CFSAN SNP Pipeline (13, 22). The reference genome for SNP calling was the internal SKESA assembly with the highest N50 score (23). Maximum likelihood phylogenies calculated using FastTree v. 2.1.10 and visualized using the Interactive Tree of Life (24, 25).
Each strain’s MLST was calculated using the reference sequences and definitions from pubMLST.org (26). SCCmec types were determined using the tools and guidelines from SCCmec Finder (27). Virulence gene profiles and antibiotic resistance gene profiles were taken from the analysis at the NCBI Pathogen Detection Program (12).
Annotation of the MLST 1535 SCCmec region used the PATRIC RASTtk-enabled Genome Annotation Service (28). Open reading frames within this sequence were confirmed or identified using blastp searches against the nonredundant protein sequences database at NCBI (21). Cassette chromosome recombinase (ccr) gene identity was investigated using blastp and representative alleles using hits above 80% identity and length (29, 30).
Statistical analyses.
Chi-squared (χ2) and Mann-Whitney U tests to evaluate differences in clinical parameters among MRSA-colonized versus noncolonized patients were calculated using SciPy 1.9.3 (https://scipy.org/). P-values for cases of multi-hypothesis testing were adjusted using the Benjamini-Hochberg procedure to determine the false discovery rate (FDR) (31).
RESULTS
Over a 341-day period, MRSA NICU isolates were evaluated for similarities among their phenotypic antibiotic resistance profiles. Two suspected outbreaks during this period, based on strain phenotypic similarities, triggered prospective genomic surveillance of all MRSA isolates from days 342 to 558, in addition to sequencing of prior available isolates from days 1 to 341 (Fig. 1; Fig. S1). Over the entire 558-day period, 58 MRSA isolates, including 55 screening and three diagnostic, were collected from 22 NICU patients. Of the isolates from diagnostic cultures, patient 12’s isolate originated from eye discharge, while patient 14’s and 16’s isolates originated from epidermal abscesses. During the 217-day prospective surveillance period, 17 neonates of 518 admitted during the prospective genomic surveillance period had MRSA isolates identified, representing 3.3% of NICU patients.
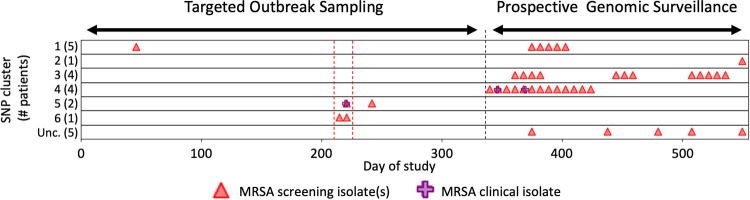
FIG 1.
NICU outbreak SNP cluster occurrence. x axis indicates day during the analysis period. “Targeted Outbreak Sampling” refers to microbiologic-only assessments of NICU strains from days 1 to 341. Triangles and crosses indicate all strains among patients who were also included in later genomic analyses. “Prospective Genomic Surveillance” over days 342 to 558 indicates the period of prospective genomic analyses of new strains. y axis indicates individual NCBI Pathogen Detection SNP clusters; numbers in parentheses indicate number of NICU strains from this study that occurred in the broader NCBI cluster, as follows: SNP clusters (MLST, SCCmec type): 1, PDS000067849.1 (8, IVa[2B]); 2, PDS000069849.1 (72, IVa[2B]); 3, PDS000069850.1 (5, II[2A]); 4, PDS000069851.1 (5, IVa[2B]); 5, PDS000069882.1 (1535, unclassified SCCmec); 6, PDS000069883.1 (8, IVa[2B]); Unc., unclustered isolates with unrelated MLST and SCCmec types. Red dashed lines show a suspected outbreak during targeted outbreak sampling, and a black dashed line marks when prospective surveillance began.
Factors associated with MRSA colonization in neonates included prematurity and low birthweight (Table 1, q = 0.002; Table S1). MRSA-colonized patients also had longer average lengths of stay in the NICU than noncolonized patients, 52.5 ± 34.8 days versus 20.0 ± 24.8 days (P < 0.001). Prematurity (defined as <37 completed weeks of gestation) was also associated with longer NICU lengths of stay regardless of MRSA colonization status, 29.7 ± 29.3 days versus 6.8 ± 7.2 days (Table S2).
TABLE 1.
26 hypotheses from all ICD10 code groups met the prerequisite number of five individuals from the group with MRSA and 20 from the group without MRSA
| ICD10 code group | Definition | Proportion with MRSA (n = 22) |
Proportion without MRSA (n = 501) |
p value MRSA associationa | q value MRSA associationb | p value association with P07 |
|---|---|---|---|---|---|---|
| H35 | Other retinal disorders | 0.50 (11) | 0.04 (22) | <0.001 | <0.001 | <0.001 |
| P27 | Chronic respiratory disease originating in the perinatal period | 0.41 (9) | 0.07 (36) | <0.001 | <0.001 | <0.001 |
| P61 | Other perinatal hematological disorders | 0.73 (16) | 0.24 (120) | <0.001 | <0.001 | <0.001 |
| R63 | Symptoms and signs concerning food and fluid intake | 0.73 (16) | 0.25 (127) | <0.001 | <0.001 | <0.001 |
| P07 | Disorders related to short gestation and low birth wt, not elsewhere classified | 1.0 (22) | 0.61 (304) | <0.001 | 0.002 | NAc |
| Q25 | Congenital malformations of great arteries | 0.36 (8) | 0.11 (54) | 0.001 | 0.003 | 0.967 |
| P74 | Other transitory neonatal electrolyte and metabolic disturbances | 0.41 (9) | 0.13 (66) | 0.001 | 0.003 | 0.012 |
| P81 | Other disturbances of temp regulation of newborn | 0.82 (18) | 0.43 (216) | 0.001 | 0.003 | <0.001 |
| P28 | Other respiratory conditions originating in the perinatal period | 0.95 (21) | 0.65 (325) | 0.006 | 0.018 | <0.001 |
| P59 | Neonatal jaundice from other and unspecified causes | 0.73 (16) | 0.43 (214) | 0.011 | 0.028 | <0.001 |
| P05 | Slow fetal growth and fetal malnutrition | 0.36 (8) | 0.15 (73) | 0.014 | 0.032 | 0.001 |
P values are calculated using χ2 and q values are P values that are false discovery rate adjusted.
Significant results (q value ≤ 0.05) are shown. Values are shown in ascending q value score.
NA, not applicable.
Comparative analyses with 21,521 S. aureus genomes in NCBI’s Pathogen Detection Resource assigned 91.2% of the sequenced NICU MRSA isolates to six international SNP clusters (Fig. 1). Clustered isolates were found in 17 of 22 MRSA-colonized patients. The remaining five patients all had short NICU admissions with only one isolate obtained via routine MRSA screening (Fig. S1). These five isolates showed no relationship to any S. aureus deposited in NCBI, including previously submitted isolate genomes from adult patients at the hospital, and not linked temporally to any transmission chains.
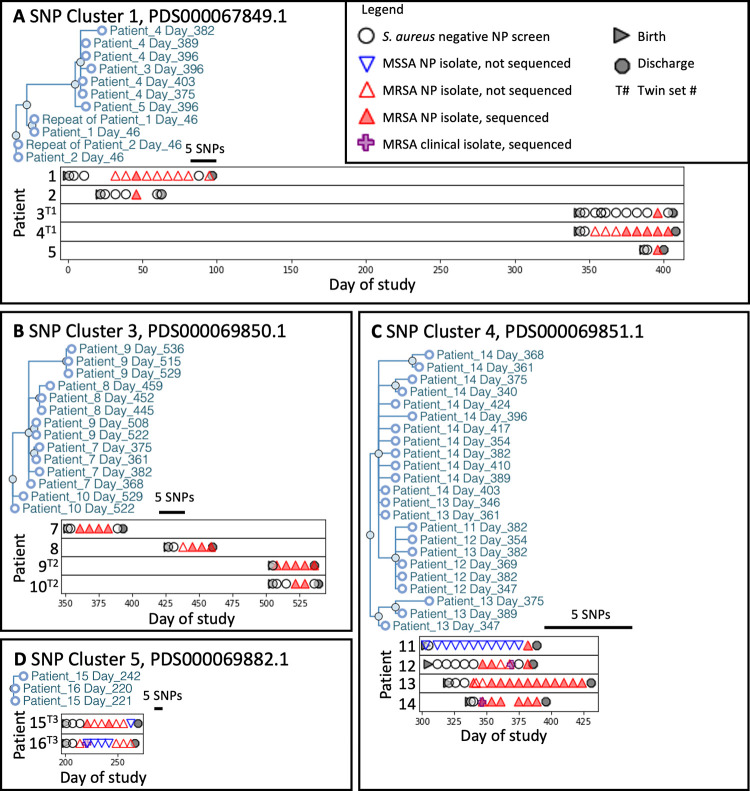
SNP clusters 1, 3, 4, and 5, involved multiple patients while clusters 2 and 6 involved only a single patient (Fig. 2). Among clusters affecting multiple patients, clusters 4 and 5 occurred within single time periods (Fig. 1). In contrast, clusters 1 and 3 occurred across distinct time periods without any patient overlap. The patient isolates in cluster 1 occurred in two intervals separated by 259 days, while strains in cluster 3 occurred across three time periods separated by 49 and 56 days. Among the two other NCBI-associated clusters, cluster 2 included one NICU isolate that clustered with an isolate reported from another institution but was not clonally related (Fig. S2). Cluster 6 included two clonally related isolates from the same patient that were separated by 5 SNPs (Fig. S3). In total, 15 of 22 (68%) MRSA-colonized NICU patients, including all twins and cases of invasive disease, occurred in clusters 1, 3, 4, and 5.
FIG 2.
SNP cluster analyses reveal patient outbreaks and occurrence over time. Panels show NICU MRSA strain phylogenetic trees from the NCBI Pathogen Genomes isolates browser with addition of the anonymized patient ID and day of isolation (A to D). Graphs below each tree show the associated patients and strain occurrences relative to surveillance or diagnostic cultures. Though patient isolates fell within the broader NCBI SNP clusters, no other closely related strains from other institutions fell within these regions of the cluster SNP tree. Bar in each graph indicates a five SNPs distance.
To leverage nationally validated tools for NICU surveillance, we used the GalaxyTrakr pipeline for cluster detection (13). To enhance the genomic context and resolution of strain clusters, analysis incorporated 10,145 S. aureus genomes from the NCBI SRA that had been deposited by other institutions. Whole-genome MLST SNPs of the external genomic data set identified strains closely related to sequenced NICU and adult nosocomial MRSA isolates from Brigham and Women’s Hospital, and that were incorporated in the cluster trees. Cluster calls and tree topologies generated by the GalaxyTrakr SNP Pipeline showed 100% concordance with the NCBI Pathogen Detection tools (Fig. 3; Tables S3 andS4; Fig. S2 to S20) (22). Clusters 1, 3, 4, 5, and 6 demonstrated <10 SNP differences among strains within a cluster and were more distant to their nearest neighboring isolate within the NCBI SRA. Longitudinal isolates from the same patient averaged 3 (±2.2) SNP differences. Of the clusters with isolates from more than two patients, clusters 1 and 3 appeared over multiple time periods and averaged 7.4 (±5.9) and 6.6 (±3.6) SNPs distance. Cluster 4 appeared over a single continuous time period and averaged <2.6 (±1.7) SNPs, a SNP distance less than clusters 1 and 3 (Mann-Whitney P < 0.001). These results indicate close phylogenetic linkage among cluster isolates within their discrete time frames of detection, while clusters with longer time frames of appearance, as seen in clusters 1 and 3, demonstrated higher accumulated SNPs, suggesting potential endemic populations that evolved over time.
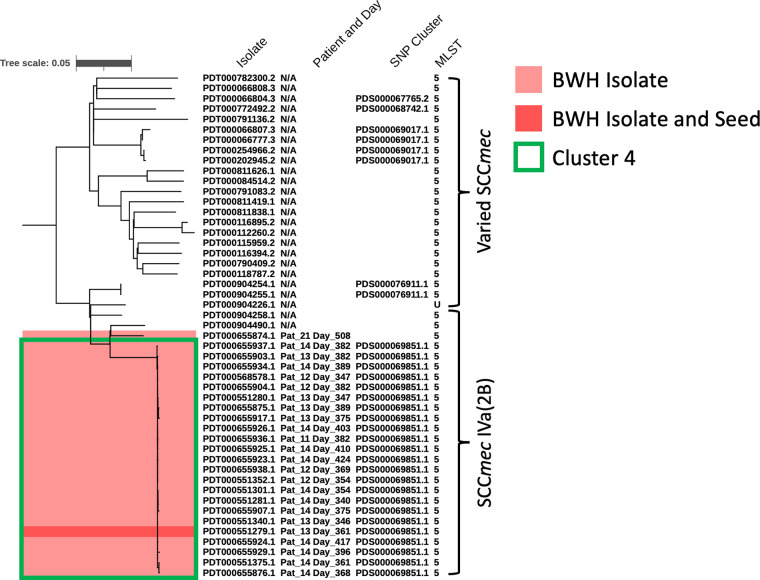
FIG 3.
International MRSA strain data sets support effective rule out of local NICU suspected transmission events. Phylogenetic tree showing the genomic assembly, patient and day of isolation, NCBI SNP cluster, and MLST of the strains in and most closely related to cluster 4. GalaxyTrakr was used to calculate the phylogenetic relationships based genome sequence read data from NCBI SRA, and a phylogeny calculated with FastTree. NICU patient strains are shown in red, with the seed used identify the closest related strains dark red. A box is drawn around Cluster 4 (PDS000069851.1). The dendrogram is midpoint rooted for clarity.
The incorporation of external MRSA genomic information also facilitated the rule-out of potential transmission events. While patient 21’s MRSA isolate showed an identical antibiogram, MLST, and SCCmec type to other MRSA isolates in NICU cluster 4, GalaxyTrakr analyses showed this strain to be more closely related to an isolate from an external institution and not part of the immediate set of cluster 4-associated NICU isolates, allowing patient 21 to be ruled out within the transmission chain. (Fig. 3; Fig. S6 and S11).
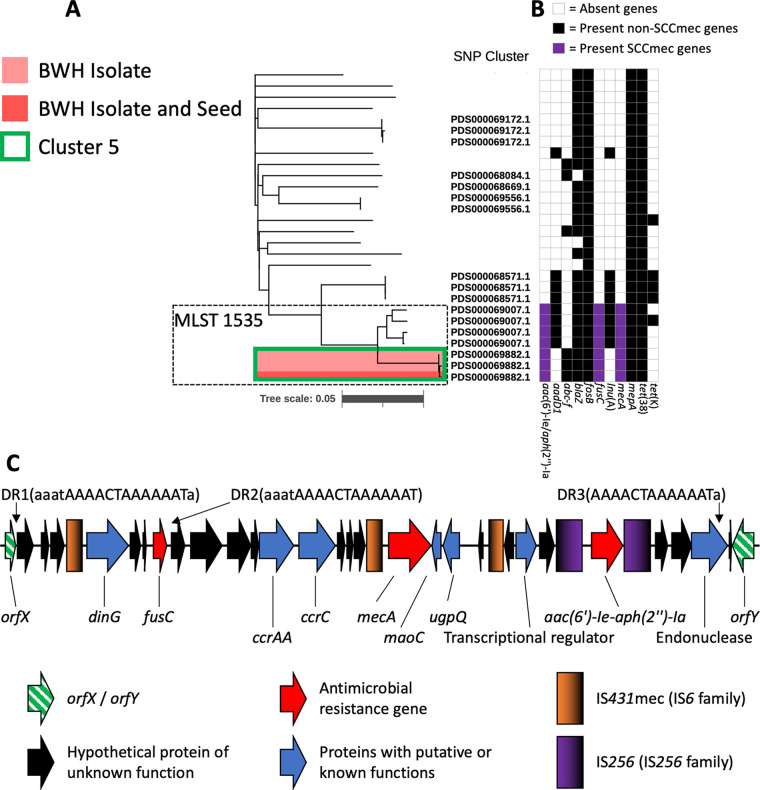
Cluster 5 isolates included nasal-perirectal swab isolates from twin patients 15 and 16, and an epidermal abscess isolate cultured from patient 16. The isolates belonged to MLST 1535 (Fig. 4A), a sequence type most commonly seen in the Middle East (30, 32). The twin’s family had recently immigrated from the Middle East. The MLST 1535 genomes share the presence of three antibiotic resistance genes; aac(6’)-Ie/aph(2’’)-1a, fusC, and mecA (Fig. 4B). High-resolution closed genomes of the cluster 5 isolates revealed an SCCmec region containing these resistance genes flanked by multiple mobilizing insertion sequences and novel repetitive elements that had not been resolvable in the draft-level genomic data (Fig. 4C). The SSCmec contains ccrC and ccrAA with mecA allele C2, fusC in the J3 region, and an IS256 flanked integron in the J1 region that contains aac(6′)-Ie-aph(2″)-Ia (5). Mobilizing IS431mec and IS256 were each found multiple times in this region, as well as a set of three direct repeats sharing a 13 base-long core sequence not found elsewhere in the chromosome. The cluster 5 isolates demonstrated fusidic acid resistance at 8 μg/mL, oxacillin resistance at 2 or 0.5 μg/mL, and gentamicin resistance at ≥16 μg/mL (Table S5).
FIG 4.
Genomic analyses identify NICU MLST 1535 strains with international clusters similar resistance gene profiles and variant SCCmecs. A SNP-based phylogeny of the 25 closest related strains to cluster 5, produced the same as Fig. 3. (A) Strains outside the MLST 1535 box are MLST 15. Antibiotic resistance gene profiles show the stepwise acquisition and loss of resistance genes (B). A high-resolution genomic map of the SCCmec region from NCBI cluster 5 isolates closed by long-read genome sequencing (C). Gradients in the IS element keys show directionality of the similar sequences. A 13 base AAACTAAAAAAT core sequence (DR1 TO DR3) with variable adjacent homologous sequence is directly repeated (DR) three times. This sequence does not appear elsewhere in the genome.
DISCUSSION
Genomic-epidemiologic analyses of MRSA isolates from NICU surveillance and diagnostic cultures identified multiple cryptic outbreak clusters and transmission events. All MRSA-colonized neonates had been admitted to the NICU for prematurity (Table 1; Table S1), a factor that with increased lengths of stay elevated risks for MRSA colonization and infection. NICU-specific outbreak strains were found in 65% of MRSA-colonized patients (11 of 17 neonates) during the prospective surveillance period. These outbreaks fell below thresholds of detection using standard phenotypic assessments with clinical microbiologic data, and prospectively discovered using WGS-based techniques.
Three cryptic MRSA outbreaks occurred during the period of prospective genomic surveillance (Fig. 1, 2). Clusters 1, 3, and 4 accounted for isolates in 11 of 17 MRSA-colonized patients (65%), including two patients who developed invasive MRSA infections. Genomic analyses provided high-resolution information to resolve clusters and to also exclude putative transmission events in cases where strains co-occurring in time had identical microbiologic phenotypes. Importantly, clusters 1 and 3 occurred over discontinuous time periods and would not have been identified as related using traditional epidemiologic techniques (Fig. 1). Isolates in clusters 1 and 3 also demonstrated SNP differences of up to 20 within their respective clusters than among the cluster 4 isolates that occurred over the single time period (Table 2; Fig. S4 to S6) (14, 33).
TABLE 2.
CFSAN SNP pipeline average differences within clusters and from isolates to the nearest outliera
| Cluster | No. of isolates in cluster | Avg. Pairwise SNPs, within cluster | Std. | Avg. Pairwise SNPs, nearest neighbor | Std. |
|---|---|---|---|---|---|
| 1 | 9 | 7.4 | 5.9 | 102.4 | 1.3 |
| 3 | 14 | 6.6 | 3.6 | 173.5 | 2.4 |
| 4 | 24 | 2.6 | 1.7 | 163.1 | 1.8 |
| 5 | 3 | 3.0 | 0.8 | 88.7 | 1.2 |
| 6 | 2 | 2 | NA | 193.0 | 1.0 |
| Longitudinal | 45 | 3.0 | 2.2 | NA | NA |
Cluster 2 had only one isolate from this study and was excluded from this analysis. Longitudinal samples include pairwise comparisons of isolates from the same patient.
Of the patients in cluster 4 who developed invasive disease from the colonizing strain, patient 12 had positive MRSA surveillance cultures 3 weeks prior to infection, while patient 14’s infection was diagnosed on the same day as positive surveillance cultures (Fig. 2). Longitudinal isolates from the same patient also showed relatedness, with an average of 3 SNPs difference over time (Table 2). MRSA surveillance cultures thus inform risk for invasive disease with a colonizing strain.
The isolate from patient 21 showed identical microbiologic phenotypes to strains in cluster 4 but a distinct SNP profile and phylogenetic placement, allowing it to be ruled out as part of a potential transmission chain (Fig. 3). The higher resolution analyses enabling this distinction used 10,145 publicly available Illumina-sequencing based genomic data sets from NCBI to resolve subclusters. The external MRSA genomic data were able to clarify this rule-out determination in comparison to the preliminary analysis, which uses a semiarbitrary cutoff of SNPdifferences. In contrast, the lack of intervening related strains within cluster 1 strengthens the hypothesis that cluster 1 represents a strain endemic to the NICU (Fig. 2). Thus, incorporation of large genomic data sets from geographically diverse regions enhances the interpretability of local genomic investigations of nosocomial outbreaks (14).
We also provide new, high-resolution genomic data for MLST 1535 isolates, including the fusidic acid-resistant strains occurring in this study (Table S5). Recent observations of SCC regions encoding mecA with additional resistance genes, including fusC, raises concerns globally for single vectors that can mobilize a multidrug-resistant phenotype among S. aureus. The SCCmec sequence from the closed genomes were highly similar to draft SCCmec sequences reported by Senok, et al. (Fig. 4) (30). The closed genomes harbored IS431mec and IS256 sequences and additional direct repeats within the SCCmec that may facilitate further accrual of resistance genes via transposition or chromosomal recombination events (34). WGS data obtained in the context of hospital outbreak investigations thus have the ability to enhance our epidemiologic and mechanistic understanding of emerging forms of antimicrobial resistance, particularly when deposited into curated public databases that provide real-time access and analysis of genome-level content.
Our studies have focused on application of open source genomic tools used in public health and international surveillance efforts to support hospital-based genomic surveillance programs. SNP findings and phylogenetic trees generated by the validated GalaxyTrakr pipelines (13) showed robust performance placing strains within international outbreak clusters and resolving subcluster relationships to inform local outbreak calls. As centralized and validated computational resources reduce the requirement for locally maintained resources and expertise, analyses interrogating thousands of geographically diverse genomes can be conducted in reasonable time frames to support hospital programs in infection prevention. By reducing the complexity to implement these systems, local efforts can focus on critical questions regarding strain transmission, reservoirs, and risk factors among patients, family members, staff, equipment, and additional environmental sources. Timely reporting of transmission events, with supporting epidemiologic analyses, can then support targeted interventions to reduce and halt further spread.
We demonstrate the application of longitudinal genomic surveillance for MRSA in a NICU setting using nationally validated tools and international repositories to improve outbreak detection, including cryptic transmission events missed by standard epidemiologic and clinical microbiologic assessments. The use of strain genomic data in public repositories, such as the Pathogen Detection resources at NCBI, increase the resolution of hospital clusters as well as the accuracy of rule-out determinations. Analyses provide a global context for uncommon strains that may be seen in an increasingly connected world.
Data availability.
All sequence data are available under NCBI Bioproject PRJNA278886. Accessions from this study are listed in Table S4.
ACKNOWLEDGMENTS
We thank Mary Delaney for clinical microbiologic support, the Wadsworth Center Advanced Genomic Technologies Center (AGTC) for sequencing, and Beth Flanigan, MD, in the BWH Department of Pediatric Newborn Medicine for helpful review of results.
The work of J.N.W. was supported by the National Center for Biotechnology Information of the National Library of Medicine (NLM), and the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health, and the work of J.W.C. by the BWH Clinical Microbiology Fellowship. Studies were supported by P30 DK034854 (Bry). Sequencing at the AGTC was supported by FDA Cooperative Agreements 5U18FD006229 and 1U18FD006763 (Wolfgang).
Footnotes
Supplemental material is available online only.
Contributor Information
Lynn Bry, Email: LBRY@bwh.harvard.edu.
Carey-Ann D. Burnham, Pattern Bioscience
REFERENCES
- 1.Nelson MU, Gallagher PG. 2012. Methicillin-resistant Staphylococcus aureus in the neonatal intensive care unit. Seminars in Perinatology 36:424–430. doi: 10.1053/j.semperi.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dong Y, Glaser K, Speer CP. 2018. New threats from an old foe: methicillin-resistant Staphylococcus aureus infections in neonates. Neonatology 114:127–134. doi: 10.1159/000488582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson J, Quach C. 2017. Outbreaks in the neonatal ICU: a review of the literature. Curr Opin Infect Dis 30:395–403. doi: 10.1097/QCO.0000000000000383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gerber SI, Jones RC, Scott MV, Price JS, Dworkin MS, Filippell MB, Rearick T, Pur SL, McAuley JB, Lavin MA, Welbel SF, Garcia-Houchins S, Bova JL, Weber SG, Arnow PM, Englund JA, Gavin PJ, Fisher AG, Thomson RB, Vescio T, Chou T, Johnson DC, Fry MB, Molloy AH, Bardowski L, Noskin GA. 2006. Management of outbreaks of methicillin-resistant Staphylococcus aureus infection in the neonatal intensive care unit: a consensus statement. Infection Control and Hospital Epidemiology 27:139–145. doi: 10.1086/501216. [DOI] [PubMed] [Google Scholar]
- 5.International Working Group on the Classifications of Staphylococcal Cassette Chromosome Elements. 2009. Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob Agents Chemother 53:4961–4967. doi: 10.1128/AAC.00579-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Köser CU, Holden MTG, Ellington MJ, Cartwright EJP, Brown NM, Ogilvy-Stuart AL, Hsu LY, Chewapreecha C, Croucher NJ, Harris SR, Sanders M, Enright MC, Dougan G, Bentley SD, Parkhill J, Fraser LJ, Betley JR, Schulz-Trieglaff OB, Smith GP, Peacock SJ. 2012. Rapid whole-genome sequencing for investigation of a neonatal MRSA outbreak. N Engl J Med 366:2267–2275. doi: 10.1056/NEJMoa1109910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lakhundi S, Zhang K. 2018. Methicillin-resistant Staphylococcus aureus: molecular characterization, evolution, and epidemiology. Clin Microbiol Rev 31. doi: 10.1128/CMR.00020-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.SenGupta DJ, Cummings LA, Hoogestraat DR, Butler-Wu SM, Shendure J, Cookson BT, Salipante SJ. 2014. Whole-genome sequencing for high-resolution investigation of methicillin-resistant Staphylococcus aureus epidemiology and genome plasticity. J Clin Microbiol 52:2787–2796. doi: 10.1128/JCM.00759-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harris SR, Cartwright EJP, Török ME, Holden MTG, Brown NM, Ogilvy-Stuart AL, Ellington MJ, Quail MA, Bentley SD, Parkhill J, Peacock SJ. 2013. Whole-genome sequencing for analysis of an outbreak of meticillin-resistant Staphylococcus aureus: a descriptive study. Lancet Infectious Diseases 13:130–136. doi: 10.1016/S1473-3099(12)70268-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nübel U, Nachtnebel M, Falkenhorst G, Benzler J, Hecht J, Kube M, Bröcker F, Moelling K, Bührer C, Gastmeier P, Piening B, Behnke M, Dehnert M, Layer F, Witte W, Eckmanns T. 2013. MRSA transmission on a neonatal intensive care unit: epidemiological and genome-based phylogenetic analyses. PLoS One 8:e54898. doi: 10.1371/journal.pone.0054898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prosperi M, Veras N, Azarian T, Rathore M, Nolan D, Rand K, Cook RL, Johnson J, Morris JG, Salemi M. 2013. Molecular epidemiology of community-associated methicillin-resistant Staphylococcus aureus in the genomic era: a cross-sectional study. Sci Rep 3:1902. doi: 10.1038/srep01902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.National Center for Biotechnology Information. 2016. The NCBI Pathogen Detection Project [Internet]. National Library of Medicine (US), National Center for Biotechnology Information. https://www.ncbi.nlm.nih.gov/pathogens/. [Google Scholar]
- 13.Gangiredla J, Rand H, Benisatto D, Payne J, Strittmatter C, Sanders J, Wolfgang WJ, Libuit K, Herrick JB, Prarat M, Toro M, Farrell T, Strain E. 2021. GalaxyTrakr: a distributed analysis tool for public health whole genome sequence data accessible to non-bioinformaticians. BMC Genomics 22:114. doi: 10.1186/s12864-021-07405-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Worley J, Delaney ML, Cummins CK, DuBois A, Klompas M, Bry L. 2021. Genomic determination of relative risks for clostridioides difficile infection from asymptomatic carriage in intensive care unit patients. Clin Infect Dis 73:e1727–e1736. doi: 10.1093/cid/ciaa894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nudel K, Zhao X, Basu S, Dong X, Hoffmann M, Feldgarden M, Allard M, Klompas M, Bry L. 2018. Genomics of Corynebacterium striatum, an emerging multi-drug resistant pathogen of immunocompromised patients. Clin Micro Infect 24:1016.e7–1016.e13. doi: 10.1016/j.cmi.2017.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.CLSI. 2018. Methods for dilution antimicrobial susceptiblity tests for bacteria that grow aerobically, 11 ed. Clinical and Laboratory Standards Institute, Wayne, PA, USA. [Google Scholar]
- 17.Gonzalez-Escalona N, Sharma SK. 2020. Closing Clostridium botulinum group I genomes using a combination of short- and long-reads. Front Microbiol 11. doi: 10.3389/fmicb.2020.00239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hunter JD. 2007. Matplotlib: a 2D graphics environment. Computing in Science & Engineering 9:90–95. doi: 10.1109/MCSE.2007.55. [DOI] [Google Scholar]
- 19.Virtanen P, Gommers R, Oliphant TE, Haberland M, Reddy T, Cournapeau D, Burovski E, Peterson P, Weckesser W, Bright J, SJvd W, Brett M, Wilson J, Millman KJ, Mayorov N, Nelson ARJ, Jones E, Kern R, Larson E, Carey CJ, Polat İ, Feng Y, Moore EW, VanderPlas J, Laxalde D, Perktold J, Cimrman R, Henriksen I, Quintero EA, Harris CR, Archibald AM, Ribeiro AH, Pedregosa F, Mulbregt P, Contributors S. 2020. SciPy 1.0 – fundamental algorithms for scientific computing in Python. Nat Methods 17:261–272. doi: 10.1038/s41592-019-0686-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gillaspy AF, Worrell V, Orvis J, Roe BA, Dyer DW, Iandolo JJ. 2006. The Staphylococcus aureus NCTC 8325 Genome, p 381–412. In Fischetti VA, Novick RP, Ferretti JJ, Portnoy DA, Julian I. Rood (eds). Gram-positive pathogens, 2nd ed. ASM Press, Washington, D.C. [Google Scholar]
- 21.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davis S, Pettengill J, Luo Y, Payne J, Shpuntoff A, Rand H, Strain E. 2015. CFSAN SNP Pipeline: an automated method for constructing SNP matrices from next-generation sequence data. PeerJ Computer Science 1:e20. doi: 10.7717/peerj-cs.20. [DOI] [Google Scholar]
- 23.Souvorov A, Agarwala R, Lipman DJ. 2018. SKESA: strategic k-mer extension for scrupulous assemblies. Genome Biol 19:153. doi: 10.1186/s13059-018-1540-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Letunic I, Bork P. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44:W242–245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jolley KA, Bray JE, Maiden MCJ. 2018. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res 3. doi: 10.12688/wellcomeopenres.14826.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaya H, Hasman H, Larsen J, Stegger M, Johannesen TB, Allesøe RL, Lemvigh CK, Aarestrup FM, Lund O, Larsen AR. 2018. SCCmecFinder, a web-based tool for typing of staphylococcal cassette chromosome mec in Staphylococcus aureus using whole-genome sequence data. mSphere 3. doi: 10.1128/mSphere.00612-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brettin T, Davis JJ, Disz T, Edwards RA, Gerdes S, Olsen GJ, Olson R, Overbeek R, Parrello B, Pusch GD, Shukla M, Thomason JA, Stevens R, Vonstein V, Wattam AR, Xia F. 2015. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep 5. doi: 10.1038/srep08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Monecke S, Jatzwauk L, Müller E, Nitschke H, Pfohl K, Slickers P, Reissig A, Ruppelt-Lorz A, Ehricht R. 2016. Diversity of SCCmec elements in Staphylococcus aureus as observed in South-Eastern Germany. PLoS One 11:e0162654. doi: 10.1371/journal.pone.0162654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Senok AC, Somily AM, Slickers P, Raji MA, Garaween G, Shibl A, Monecke S, Ehricht R. 2017. Investigating a rare methicillin-resistant Staphylococcus aureus strain: first description of genome sequencing and molecular characterization of CC15-MRSA. Infection and Drug Resistance 10:307–315. doi: 10.2147/IDR.S145394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc: Series B (Methodological) 57:289–300. [Google Scholar]
- 32.Ankrum A, Hall BG. 2017. Population dynamics of Staphylococcus aureus in cystic fibrosis patients to determine transmission events by use of whole-genome sequencing. J Clin Microbiol 55:2143–2152. doi: 10.1128/JCM.00164-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Orsi RH, Borowsky ML, Lauer P, Young SK, Nusbaum C, Galagan JE, Birren BW, Ivy RA, Sun Q, Graves LM, Swaminathan B, Wiedmann M. 2008. Short-term genome evolution of Listeria monocytogenes in a non-controlled environment. BMC Genomics 9:539. doi: 10.1186/1471-2164-9-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Larsen J, Andersen PS, Winstel V, Peschel A. 2017. Staphylococcus aureus CC395 harbours a novel composite staphylococcal cassette chromosome mec element. J Antimicrob Chemother 72:1002–1005. doi: 10.1093/jac/dkw544. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download jcm.00014-23-s0001.pdf, PDF file, 6.4 MB (6.4MB, pdf)
Data Availability Statement
All sequence data are available under NCBI Bioproject PRJNA278886. Accessions from this study are listed in Table S4.