Abstract
Human papillomavirus type 16 (HPV16) infects cervical epithelium and is associated with the majority of cervical cancers. The E1∧E4 protein of HPV16 but not those of HPV1 or HPV6 was found to associate with a novel member of the DEAD box protein family of RNA helicases through sequences in its C terminus. This protein, termed E4-DBP (E4-DEAD box protein), has a molecular weight of 66,000 (66K) and can shuttle between the nucleus and the cytoplasm. It binds to RNA in vitro, including the major HPV16 late transcript (E1∧E4.L1), and has an RNA-independent ATPase activity which can be partially inhibited by E1∧E4. E4-DBP was detectable in the cytoplasm of cells expressing HPV16 E1∧E4 (in vivo and in vitro) and could be immunoprecipitated as an E1∧E4 complex from cervical epithelial cell lines. In cell lines lacking cytoplasmic intermediate filaments, loss of the leucine cluster-cytoplasmic anchor region of HPV16 E1∧E4 resulted in both proteins colocalizing exclusively to the nucleoli. Two additional HPV16 E1∧E4-binding proteins, of 80K and 50K, were identified in pull-down experiments but were not recognized by antibodies to E4-DBP or the conserved DEAD box motif. Sequence analysis of E4-DBP revealed homology in its E4-binding region with three Escherichia coli DEAD box proteins involved in the regulation of mRNA stability and degradation (RhlB, SrmB, and DeaD) and with the Rrp3 protein of Saccharomyces cerevisiae, which is involved in ribosome biogenesis. The synthesis of HPV16 coat proteins occurs after E1∧E4 expression and genome amplification and is regulated at the level of mRNA stability and translation. Identification of E4-DBP as an HPV16 E1∧E4-associated protein indicates a possible role for E1∧E4 in virus synthesis.
Human papillomaviruses (HPV) comprise a family of over 70 members which cause epithelial lesions at different histological sites (49). HPV type 1 (HPV1) causes plantar and palmar warts, whereas HPV6 and HPV11 cause genital warts (11). Although genital warts are the most common sexually transmitted disease seen in sexually transmitted disease clinics, the low-risk HPV types (which include HPV1, HPV6, and HPV11) cause predominantly benign tumors and are rarely associated with life-threatening disease (69). High-risk types, such as HPV16, cause flat warts of the cervix and are associated with cervical abnormalities ranging from low-grade squamous intraepithelial lesions to invasive carcinoma (69). HPV DNA is found in over 90% of cervical cancer cases, with HPV16 occurring most often (69). The molecular events which lead to cancer are well understood compared to those which regulate the production of infectious virions. Analysis of papillomavirus late functions has been restricted by the lack of a convenient model system in which to propagate infectious virions in vitro (see references 4 and 14 for reviews).
The virus life cycle is linked to keratinocyte differentiation. Initial infection following a microabrasion or cut leads to cell proliferation and the formation of an expanded population of basal keratinocytes which harbor the viral episome at low copy numbers (14). Events necessary for virus production begin as the infected cell leaves the basal layer. Vegetative viral DNA replication starts in the spinous layer, followed soon after by the synthesis of structural proteins and the assembly of infectious virions (20). Papillomavirus genomes comprise a double-stranded DNA circle of about 8,000 bp which contains eight open reading frames (ORF) in HPV16. Viral gene products are expressed from differentially spliced mRNAs at different times during the migration of the infected cell toward the epithelial surface. With the exception of the virion structural proteins and E4, viral gene products are not readily detected in vivo (15, 21).
E4 proteins are expressed from a spliced mRNA (E1∧E4) (8, 9, 24, 53) and are first detected in naturally occurring lesions in cells in which vegetative viral DNA replication is occurring (6, 20). The proteins persist throughout the late stages of infection and are modified by proteolytic processing and phosphorylation (19, 36, 63). Expression precedes the synthesis of virus structural proteins and the assembly of infectious particles and occurs in cells in which keratinocyte differentiation is inhibited (6, 20). E4 proteins localize in part to cytoplasmic intermediate filaments (IF) in low-grade squamous intraepithelial lesions caused by HPV16 but are also diffusely cytoplasmic and perinuclear (20). As infected cells near the epithelial surface, E4 proteins localize to perinuclear bundles. Although its significance is unclear, expression of the HPV16 E1∧E4 protein (16 E1∧E4) in keratinocytes in monolayer cultures leads to collapse of the cellular IF network (18). 16 E1∧E4 and the collapsed filaments have a perinuclear distribution in these cells (18). Loss of the conserved leucine cluster located toward the N terminus of most E4 proteins leads to nuclear accumulation of truncated E4 gene products in some cells (62).
Although the E1∧E4 proteins of some HPV types can self-associate, no cellular E4-binding proteins have been reported, and the role of E1∧E4 has not been established. The association of E1∧E4 with IF is probably mediated indirectly (22). HPV1 E1∧E4 protein (1 E1∧E4) purified from warts had no demonstrable affinity for keratin monomers or filaments, and neither 1 E1∧E4 nor 16 E1∧E4 could be shown to interact with keratins following cell-free expression or two-hybrid screening (22). To identify cellular proteins which may be involved in IF association and to establish a role for E1∧E4 proteins in the virus life cycle, we searched for cellular targets for E4 and have identified two proteins which can bind 16 E1∧E4 directly. The first of these (E4-IFAP) showed a filamentous staining pattern and may be necessary for E4-IF association. The other, a novel member of the DEAD box protein family (E4-DBP), is described here. E4-DBP was distributed throughout the epithelial cell layers of the normal cervix. The protein had a nucleolar distribution but shuttled between the nucleus and the cytoplasm, localizing exclusively to the cytoplasm at mitosis. E4-DBP showed evidence of cytoplasmic localization in cells expressing full-length E1∧E4 and could be coimmunoprecipitated with E1∧E4 by E4-specific antibodies. The interaction required the C-terminal domain of E4 and did not extend to other members of the DEDH and DEXT box families eukaryotic initiation factor 4A, such as p68 or (eIF4A). Although the function of many DEAD box proteins is uncertain, they are generally involved in regulating translation by affecting splicing, ribosome biogenesis, RNA turnover, or mRNA export. E4-DBP shares homology in its N terminus with DEAD box proteins involved in the control of mRNA stability and ribosome biogenesis. Considering that papillomavirus late gene expression is controlled largely at the level of RNA processing and stability, an in vivo role for the association is suggested.
MATERIALS AND METHODS
Two-hybrid library screening and manipulation of plasmids expressing HPV E1∧E4 proteins.
Two-hybrid screening was carried out as described previously (25) with yeast strain Hfc7 and a HeLa S3 cell cDNA library cloned between the EcoRI and XhoI sites of pGAD GH (insert size, 0.4 to 2.0 kb; Clontech, Palo Alto, Calif.). The 16 E1∧E4 gene was amplified from pMal.16 E1∧E4 (20) using primers CGGGATCCGGAATTCATGGCTGATCCTGCAGCAGCAACG AAG (16E1∧E4forwardA) and GGGGATCCTTATGGGTGTAGTGTTACTATTACAGT (16E1∧E4reverseA) and was cloned between the EcoRI and BamHI sites of pGBT9 or pGAD 424 (Clontech). 16 E1∧E4 mutations in pAP16 (Δ12-16, Δ23-28, Δ27-32, Δ31-36, Δ36-41, Δ41-46, Δ46-51, Δ63-68, Δ73-77, Δ80-83, and Δ84-88) (62) were subcloned into EcoRI/BamHI-digested pGBT9 using primers CGGGATCCGGAATTCATGGCTGATCCTGCAGCAGCA (16E1∧E4 forwardB) and GGAATTCCGGATCCTTATGGGTGTAGTGTTACTATT (16E1∧E4reverseB). 16 E1∧E4 mutant Δ2-6 was prepared from pMal.16E1∧E4 using the alternative forward primer GATCGAATTCATGGCAACGAAGTATCCTCTCC (16E1∧E4forwardΔ2-6) and 16E1∧E4reverseB (see above). Mutants Δ84-88 and Δ86-92 were prepared using the alternative reverse primers GATCGAATTCTTATGGGTGTAGTGTTCGTCCTTTG (16E1∧E4reverse Δ84-88) and GATCGAATTCTTATATTACAGTTAATCCGTCCTTTG (16E1∧E4reverseΔ86-92), respectively, along with 16E1∧E4forwardB (see above). PCR fragments were cloned into pGBT9 following digestion with EcoRI. The shorter forms of 16 E1∧E4 (corresponding to the 16,000-molecular-weight [16K] and 11K proteins of HPV1) were amplified using primer GATCGAATTCAGCACTTGGCCAACCACCCCGCCG (16E1∧E416Kforward) or GATCG AATTCCAGACACCGGAAACCCCTGCCACACC (16E1∧E410Kforward) and the reverse primer 16E1∧E4reverseB (see above). The same primers were used to clone the mutant 16 E1∧E4 genes into pMal-c2X, (New England Biolabs, Beverly, Mass.). The E1∧E4 genes of HPV1 and HPV6 were excised from pGEX.1E1∧E4 and pGEX.6E1∧E4 (17) using EcoRI and cloned into pGBT9. DNA sequencing was used to confirm the integrity of the various constructs and was performed manually or with an ABI automatic sequencing machine.
Identification of E4-DBP cDNAs and cloning of E4-DBP fragments into expression plasmids.
To identify full-length E4-DBP, the EcoRI/XhoI fragment from pGAD.E4-DBP was radioactively labeled with 32P by random priming (Amersham Pharmacia Biotech, Little Chalfont, United Kingdom) and used to screen a HeLa cell cDNA library cloned into pcDNA3.1 (average insert size, 1.22 kb, 107 independent clones; Invitrogen, Groningen, The Netherlands) by colony hybridization (65). The largest of the positive clones (pcDNA.E4-DBP1) was completely sequenced, and the integrity of the E4-DBP ORF was confirmed by carrying out cell-free protein synthesis (TnT system; Promega, Madison, Wis.). The E4-DBP insert was subsequently transferred into the prokaryotic expression vector pET28a (Novagen, Madison, Wis.) after digestion with EcoRI and NotI (pET.E4-DBP). The E4-DBP fragment contained in pGAD.E4-DBP was amplified using primers CCGGAATTCATGGCTCCGACCGAAGCGTCCCAG (DBP/GAD forward) and CCGCTCGAGCGGTTAAATCACTTTGAGGATCTT (DBP/GAD reverse) and was subcloned into pcDNA3.1 after digestion with EcoRI and XhoI. Plasmid pGEX.E4-DBP was prepared by subcloning the EcoRI/XhoI E4-DBP fragment from pGAD.E4-DBP into pGEX4T-3 (Amersham Pharmacia, Uppsala, Sweden), while pMal.E4-DBP was prepared by cloning the same fragment into pMal-c2X. The pGAD 424.keratin 8 and pGAD 424.keratin 18 constructs have been described previously (22). cDNA clones encoding eIF4A (pET3b-4A [56]) and p68 (pt7.7-p68 [41]) were obtained from N. Sonenberg (Department of Biochemistry, McGill University, Montreal, Quebec, Canada) and F. Fuller-Pace (Department of Pathology, University of Dundee, Dundee, United Kingdom) and were used directly in coupled transcription-translation reactions (TnT system) to prepare eIF4A and p68 proteins. All constructs were validated by DNA sequencing as described earlier.
Purification of E1∧E4 and E4-DBP proteins for in vitro binding assays and antibody generation.
Maltose-binding protein (MBP), glutathione S-transferase (GST), and His-tagged fusion proteins were expressed from recombinant pGEX (Amersham Pharmacia), pMal (New England Biolabs), or pET (Novagen) vector and purified essentially as described by the manufacturers, except that the bound fusion proteins were washed extensively with 1 M NaCl–50 mM Tris-Cl (pH 8.0) prior to elution from the column (glutathione-Sepharose or amylose resin). E4-DBP was expressed from pET.E4-DBP in Escherichia coli strain BL21(DE3). Cells were grown at 37°C to an optical density at 600 nm of 0.6 in the presence of 100 μg of ampicillin per ml before being induced by the addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Growth was allowed to continue at 30°C for a further 2 h before the cells were pelleted and lysed by sonication in 500 mM NaCl–5 mM imidazole–20 mM Tris-Cl (pH 7.9). His-tagged E4-DBP was purified from the crude lysate using His Bind resin (Novagen) essentially according to the manufacturer's protocols, except that elution was carried out using 500 mM NaCl–1 M imidazole–1 mM β-mercaptoethanol–0.1% Nonidet P-40 (NP-40)–20 mM Tris-Cl (pH 7.9). The purified protein was dialyzed against the same buffer (to avoid precipitation) in the absence of imidazole and snap frozen in aliquots at −70°C.
For ATPase and ATP-binding experiments, E4-DBP was further purified by binding to poly(U)-Sepharose (Sigma, St. Louis, Mo.) on an end-over-end shaker for 1 h at 4°C following dilution of the NaCl concentration to 150 mM [poly(U) binding buffer: 150 mM NaCl, 1 M imidazole, 1 mM β-mercaptoethanol, 0.1% NP-40, 20 mM Tris-Cl (pH 7.9)]. After extensive washing, E4-DBP was eluted using the same buffer containing 500 mM NaCl. In vitro binding assays were carried out with immobilized GST or MBP fusion proteins as described previously (45) and with [35S]methionine-labeled proteins prepared by cell-free expression (E4-DBP, eIF4A, p68, and chloramphenicol acetyltransferase [CAT]; TnT system) or by metabolic labeling of cells in cultures. Proteins binding to GST.16 E1∧E4 or MBP.16 E1∧E4 were eluted by being boiled in sodium dodecyl sulfate (SDS) sample buffer and were visualized by gel electrophoresis and autoradiography.
Monoclonal antibodies to the E1∧E4 protein of HPV16 (TVG402 and TVG405) have been described previously (17, 20). Antibodies to E4-DBP were prepared by immunization of two rabbits with purified GST.E4-DBP expressed from plasmid pGEX.E4-DBP (see above) as previously described (16). Rabbits were immunized at multiple subcutaneous sites using 250 μg of fusion protein in Freund's complete adjuvant. Injections were repeated at 14-day intervals using the same amount of protein in Freund's incomplete adjuvant. Two weeks after the final immunization, the rabbits were bled and the antibody titer was assessed by an enzyme-linked immunosorbent assay with MBP.E4-DBP-coated plates (15). For immunostaining and Western blotting, rabbit antisera was preabsorbed with acetone powder from E. coli strain DH5α expressing GST (from pGEX4T-3) before use (39).
Expression and detection of proteins in tissue culture cells and in formalin-fixed paraffin-embedded tissue.
16 E1∧E4 was expressed in mammalian cells following infection with recombinant vaccinia viruses as described previously (18) or following transfection with pMV11.16 E1∧E4 and pMV11.16ΔLLXLL E1∧E4 using Lipofectamine (Gibco BRL; protocols provided by the manufacturer). Cell lines (SW13 c1.2, COS-7, CV-1, HeLa, and SiHa) were grown in Dulbecco modified Eagle medium containing 10% fetal calf serum (Gibco BRL). The MV11.16 E1∧E4 expression constructs were prepared by amplification of the E1∧E4 gene from plasmid pMal.16 E1∧E4 or pAP16Δ12-16 using primers CGCGAATTCGGATCCCATGGCTGATCCTGCAGCAGCAACG (16E1∧E4forwardC) and CGTCGACGAATTCGTACTATGGGTGTAGTGTTACTATTAC (16E1∧E4reverseC), followed by cloning of the amplified fragment between the BamHI and EcoRI sites of pMV11 downstream of the cytomegalovirus promoter. pMV11 is a modified version of pMV10 (28) created by removal of the lacZ BamHI fragment and insertion of a synthetic polylinker. The sequence inserted into the BamHI site of pMV10 is gGATCCCGGGTACCGAATTCTAgatcc (lowercase letters indicate the pMV10 BamHI site into which the synthetic oligonucleotide was cloned). pMV11 was a gift from Helena Browne, Department of Pathology, University of Cambridge).
Recombinant adenoviruses expressing wild-type or mutant E1∧E4 proteins were prepared by subcloning the HindIII fragment from pMV11.16 E1∧E4 or pMV11.16ΔLLXLL E1∧E4 (which contains the E1∧E4 gene and upstream cytomegalovirus promoter) into the adenovirus shuttle vector pXC15-7 (67). Adenovirus recombinants (rAd.16 E1∧E4) were rescued using the adenovirus type 5 backbone plasmid pJM17 as described previously (51) and were propagated using standard protocols (3).
Detection of the 16 E1∧E4 and E4-DBP proteins in cells in cultures was carried out according to established methods (cells fixed in 5% formaldehyde for 5 min) (17), except that cells were permeabilized with 0.2% Triton X-200 prior to staining. 16 E1∧E4 was detected using antibody TVG405 directly conjugated to Alexa 488 (Pierce-Warriner) or using antibody TVG402 followed by an anti-mouse secondary antibody conjugated to either fluorescein isothiocyanate or Texas red (Amersham Pharmacia Biotech). E4-DBP was detected using anti-rabbit conjugates (Amersham Pharmacia Biotech). The detection of E4-DBP in formalin-fixed paraffin-embedded tissue required epitope retrieval by pressure cooking prior to staining as described by Freeman et al. (29). Immunoprecipitation of E4 complexes from cells infected with rAd.16 E1∧E4 was carried out using TVG402 cross-linked to protein G-Sepharose (39). Western blots obtained with antibodies to E4-DBP or 16 E1∧E4 were visualized using a peroxidase-conjugated second antibody followed by development with the ECL kit reagent (Amersham Pharmacia Biotech).
Analysis of nuclear-cytoplasmic shuttling, ATP binding, and ATPase activity.
CV-1 or COS-7 cells were grown to 70% confluence (as described above) before the addition of actinomycin D (5 μg/ml), cycloheximide (20 μg/ml), or cycloheximide and actinomycin D (same concentrations as those just listed) as described by Pinol-Roma and Dreyfuss (58). Cells were fixed and immunostained with antibodies to E4-DBP, hnRNP A1, or hnRNP C after 1.5, 4.5, or 9 h in the presence of the drug. hnRNP antibodies (58) were a kind gift from G. Dreyfuss (Howard Hughes Medical Institute, Philadelphia, Pa.).
ATP binding was carried out using the ATP analogue 5′-flurosulfonylbenzoyl adenosine (FSBA) according to protocols provided by the manufacturer (Roche Molecular Biochemicals). Briefly, 1 μg of E4-DBP or GST (purified as described above) was incubated in phosphate-buffered saline at 30°C for 20 min in the presence of 0.1 mM FSBA (pH 7.5). After being boiled in SDS sample buffer, FSBA-derivatized proteins were separated by SDS gel electrophoresis prior to Western blotting using an anti-FSBA antibody (Roche). The specificity of binding was confirmed by preincubation of the E4-DBP protein in the presence of 5 mM ATP and 5 mM MgCl2 for 20 min before the addition of FSBA.
E4 competition experiments were carried out by adding 10 μg of purified MBP.16 E1∧E4 fusion protein (approximately a 10-fold molar excess) to the E4-DBP protein prior to incubation with FSBA or by adding 10 μg of purified MBP.16 E1∧E4 in the presence of 5 mM ATP and 5 mM MgCl2. In all cases, preincubation times were 20 min.
ATPase experiments were carried out as described previously with 0.2 μg of E4-DBP or DbpA (31). E. coli 23S rRNA and DbpA were generously provided by F. Fuller-Pace. ATPase reactions were carried out with 50 mM Tris-Cl (pH 7.5)–5 mM MgCl2–1 mM dithiothreitol using 1 μCi of [γ-32P]ATP. For competition experiments, 5 μg of MBP.16 E1∧E4 or 5 μg of bovine serum albumin was added to the reaction mixture prior to the addition of [γ-32P]ATP, and the mixture was preincubated for 30 min at 30°C. E. coli 23S RNA or total HeLa cell RNA (800 ng) was also added to the reaction mixture (where indicated), and the final reaction volume was adjusted to 20 μl.
The binding of E4-DBP to single-stranded or double-stranded DNA or to poly(U)-Sepharose was carried out with 150 mM NaCl–1 mM β-mercaptoethanol–0.1% NP-40–20 mM Tris-Cl (pH 7.9) as described above for the purification of E4-DBP. Bound proteins were eluted using the same buffer containing 50 mM NaCl.
In vitro RNA cross-linking experiments were carried out using 32P-labeled RNA prepared in vitro from plasmids containing the T7 promoter (42). Plasmids containing reconstructed E1∧E4.L1 cDNAs (24) downstream of the T7 promoter of pET7 (Novagen) were kindly provided by S. Stacey (Paterson Institute for Cancer Research, Manchester, United Kingdom). Plasmid pEH165 (lacking the negative regulatory element [NRE]) contains the region of the HPV16 genome between nucleotides 670 and 7171, while pEH1627 contains the region between nucleotides 670 and 7268. Both plasmids lack the region of HPV16 between nucleotides 880 to 3357 and 3631 to 5637, which are absent from the E1∧E4.L1 cDNAs. Plasmid pG63 was used to prepare herpes simplex virus type 1 (HSV-1) IE63 mRNA and has been described previously (42).
Identification of sequence homologies by database searching.
DNA sequences were compared against those in the GenEMBLPlus database (EMBL release 59.0 and GenBank release 112.0) using the FastA DNA sequence analysis program in the Wisconsin Package (version 10.0; Genetics Computer Group, Madison, Wis.). Alignments were carried out using the Megalign program from Lasergene (DNASTAR Inc., Madison, Wis.). Percent similarity and percent divergence were calculated according to the following equations: divergence (i,j) = 100 = 100 × [distance (i,j)/total distance] and similarity (i,j) = 100 × {sum of the matches/[length − gap residues (i) − gap residues (j)]}. Total distance is the sum of all the branch lengths in the phylogenetic tree, while distance (i,j) is the sum of residue distances plus (gap × gap penalty) plus (gap residues × gap length penalty).
Nucleotide sequence accession number.
The EMBL accession number for E4-DBP is AJ276704.
RESULTS
Identification of cellular proteins which associate with 16 E1∧E4 by two-hybrid screening.
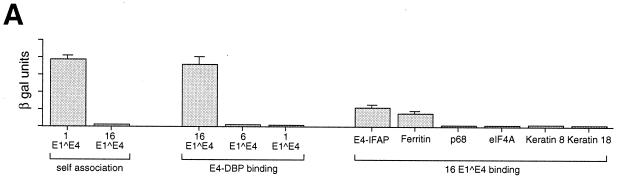
The only biological effect attributed to the 16 E1∧E4 protein is its association with keratin filaments, which occurs following the expression of 16 E1∧E4 in epithelial cells (18, 61, 62). As the 16 E1∧E4 protein does not bind keratins directly (22) (Fig. 1A), a yeast two-hybrid screen was carried out with the initial aim of identifying cellular proteins involved in keratin association. As cytokeratin binding occurs in cervical epithelial cell lines, screening was carried out with a HeLa cell library (in pGAD GH). Seven different pGAD GH clones were identified. The most promising binders activated the β-galactosidase reporter gene to levels similar to those observed when the 1 E1∧E4 proteins were coexpressed together as activation and binding domain fusions (Fig. 1A). Self-association of 1 E1∧E4 (but not 16 E1∧E4) has been well characterized with the two-hybrid approach (2, 22). The strong 16 E1∧E4 binders accounted for over half of the identified clones (17 out of 32 colonies) and, although heterogeneous in size, all contained related inserts (as determined by restriction enzyme digestion). No stimulation of reporter gene activation was apparent in the absence of 16 E1∧E4. DNA sequencing of several of the strong 16 E1∧E4 binders identified in frame with the Gal4 activation domain a DNA insert of approximately 560 bp encoding a novel member of the DEAD box protein family (Fig. 1B). This protein was named E4-DBP (E4-DEAD box protein).
FIG. 1.
(A) Association of 16 E1∧E4 with cellular proteins. 16 E1∧E4, HPV6 E1∧E4, and 1 E1∧E4 were expressed from pGBT9 and examined for their ability to interact with themselves or with cellular proteins expressed from pGAD GH. In each case, the extent of association is indicated by the relative levels of β-galactosidase (β gal) activity, which is shown on the y axis (see Materials and Methods). Individual binding experiments were carried out in triplicate, and error bars are shown above each column. 1 E1∧E4 but not 16 E1∧E4 self-associated. 16 E1∧E4 but not 6 E1∧E4 or 1 E1∧E4 associated with E4-DBP. 16 E1∧E4 also associated with E4-IFAP but not with other DEAD box proteins, such as p68 and eIF4A. No association could be demonstrated with either keratin 8 or keratin 18 monomers. (B) Sequence analysis of the E4-DBP-binding protein. The complete E4-DBP DNA sequence is shown beneath the amino acid sequence of the protein which it encodes. Motifs characteristic of the DEAD box protein family are boxed, and functions associated with these regions are indicated beneath the boxes. The E4-DBP initiation codon and the 2 bases which follow were predicted from comparison with other sequences identified in the GenEMBLPlus database. Homology with these sequences (GenBank accession no. MM1638, MMAA3862, and HS27112) is indicated at the top of the figure (dashes indicate exact matches). The rest of the sequence was derived from DNA sequence analysis of the pcDNA.E4-DBP1 clone isolated here (see Results). The underlining indicates the extent of the E4-DBP region typically present in the pGAD.E4-DBP clones. The sequence of the plasmid–E4-DBP junction is shown for two of the pGAD.E4-DBP clones and for pGEX.E4-DBP. The 5′ end of the insert present in pcDNA.E4-DBP1 is shown by an arrow.
To obtain a full-length E4-DBP clone for functional studies, the pGAD.E4-DBP1 insert (EcoRI/XbaI fragment) was used to probe a HeLa cell cDNA library cloned into pcDNA3.1. Of the 30 clones recovered, the largest contained a 1,799-bp insert (pcDNA.E4-DBP1) whose 5′ end (5′-GGCACCCGAG …) (Fig. 1B) began 20 bp upstream of that in pGAD.E4-DBP1 (Fig. 1B) and encoded a protein with a predicted molecular weight of 50,432. This insert was subcloned into pET28a to allow the expression of E4-DBP as a His-tagged fusion protein. FastA analysis of the GenEMBLPlus DNA sequence database subsequently identified three homologous expressed sequence tags (ESTs), MM1638, MMAA3862, and HS27112 (Fig. 1B), whose 5′ ends extended beyond that of our largest cDNA clone. Two of these contained a Kozak consensus initiation codon in the same reading frame as E4-DBP (47). From this analysis, full-length E4-DBP was predicted to contain two additional N-terminal amino acids that were not expressed in our pET.E4-DBP construct and to encode a protein with a predicted molecular weight of 50,635. E4-DBP cDNA fragments have been reported in a wide range of human tissues, including testis, placenta, liver, retina, and T lymphocytes (EMBL release 59.0 and GenBank release 112.0).
Six other proteins were found to bind E1∧E4 in the two-hybrid screen, but the level of β-galactosidase activation in each case was lower than that with E4-DBP (Fig. 1A). Although the significance of these interactions has not been ascertained, antibodies to one of these proteins (E4-IFAP) showed a filamentous staining pattern in epithelial cells (data not shown). Neither 1 E1∧E4 nor 6 E1∧E4 showed any association with E4-DBP in two-hybrid assays (Fig. 1A). Both proteins (6 E1∧E4 is closely related to 11 E1∧E4) have been reported to self-associate in yeast, however (through C-terminal sequences [2, 7]), and this association may interfere with the detection of cellular partners. Ferritin, which is a common artifact of two-hybrid screening, was represented five times among the weak 16 E1∧E4 binders.
E4-DBP associates specifically with 16 E1∧E4 through sequences in its C terminus.
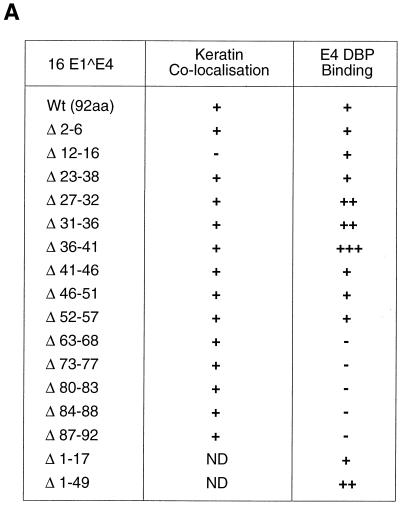
To determine which regions of 16 E1∧E4 are important for E4-DBP association, a series of mutant E1∧E4 genes (62) were subcloned into pGAD GH and screened for interactions. Deletions affecting the last 27 amino acids of 16 E1∧E4 abolished binding (Fig. 2A). Deletions in the central portion of the protein, which included part of the proline-rich region and the PKKHRR motif, enhanced binding (Fig. 2A). Two shorter forms of the HPV16 E4 protein which were analogous to the 16K (deletion of residues 1 to 17) and 11K (deletion of residues 1 to 49) E4 proteins of HPV1 were also found to be able to bind E4-DBP, the latter with greater efficiency than the full-length gene product (Fig. 2A). No significant differences were apparent between the expression levels of the different mutants (as judged by SDS gel electrophoresis and Western blotting). These results indicate that the C terminus of the protein, which in HPV1 and HPV11 is necessary for dimerization, is responsible for the association of E4-DBP with 16 E1∧E4. Considering the great variation in primary amino acid sequences among E4 proteins (23), it is possible that the E1∧E4 proteins of different types have evolved different functions.
FIG. 2.
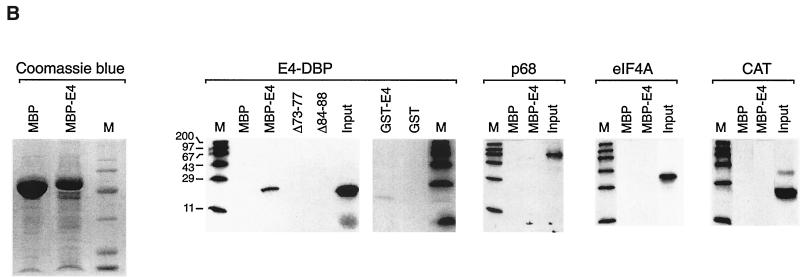
(A) Association with E4-DBP requires the C terminus of 16 E1∧E4. A series of 16 E1∧E4 deletions were tested for their ability to associate with the E4-binding domain of E4-DBP using the yeast two-hybrid system. All five C-terminal mutants failed to interact with 16 E1∧E4. Loss of the N-terminal half of the protein or loss of a sequence within the proline-rich domain enhanced the association. Mutant Δ36-41 (which has lost the putative nuclear localization sequence) bound most strongly. The ability of individual mutants to associate with keratin IF (as previously reported [62]) is also shown. +++, very strong binding; ++, strong binding; +, binding; −, no binding; ND, not determined. (B) 16 E1∧E4 can associate with E4-DBP in vitro. Purified MBP and MBP.16 E1∧E4 (lane MBP-E4) following SDS gel electrophoresis and staining with Coomassie blue are shown at the left of the figure. These proteins were used to assess the extent of interaction with E4-DBP, p68, eIF4A, and CAT. Putative E4-binding proteins were labeled with [35S]methionine in a cell-free expression system and visualized by SDS gel electrophoresis and autoradiography. The input lanes contained 25% of the total protein used in each experiment (either E4-DBP, p68, eIF4A, or CAT, indicated above each autoradiograph). The fraction that specifically bound to immobilized MBP or MBP.16 E1∧E4 is indicated in the adjacent lanes (MBP, MBP-E4, or specific MBP-E4 mutants). Only E4-DBP showed any measurable association with 16 E1∧E4 in these in vitro binding assays (as found by a two-hybrid screen; Fig. 1A). GST.16 E1∧E4 also bound E4-DBP, although the association was much weaker (GST and GST-E4 lanes). MBP E4 mutants Δ73-77 and Δ84-88 were unable to bind E4-DBP. Molecular weight markers (in thousands) are shown in lanes M.
The ability of E1∧E4 to associate with E4-DBP was confirmed using two different approaches. The region of E4-DBP involved in the association with E1∧E4 in yeast was expressed from pET28a using a rabbit reticulocyte lysate and shown to bind MBP.16 E1∧E4 immobilized on amylose resin (Fig. 2B). E4-DBP also bound immobilized GST.16 E1∧E4, although in this case the association was much weaker (Fig. 2B). Neither MBP.16 E1∧E4 nor GST.16 E1∧E4 bound irrelevant proteins, such as CAT, and neither bound the related DEAD box proteins p68 and eIF4A. Similarly, no evidence for p68 or eIF4A binding could be detected in the yeast two-hybrid system (Fig. 1A), suggesting that binding does not occur simply via the conserved helicase motifs (Fig. 2B). MBP.16 E1∧E4 mutants (Δ73-77 and Δ84-88) did not bind E4-DBP (Fig. 2B).
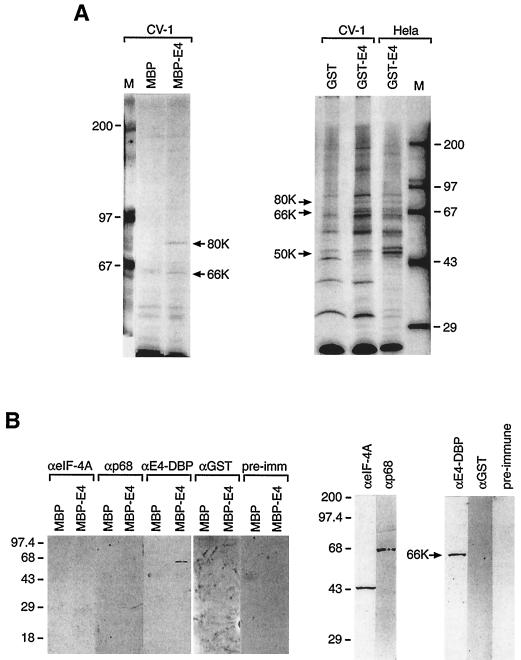
To confirm that 16 E1∧E4 can bind full-length E4-DBP, pull-down experiments were carried out using [35S]methionine-labeled cell extracts. Both GST.16 E1∧E4 and MBP.16 E1∧E4 bound two proteins, of 66K and 80K, in extracts of CV-1 and HeLa cells (Fig. 3A) which were not bound by either GST or MBP alone. GST.16 E1∧E4 also bound a 50K protein (Fig. 3A). Polyclonal antibodies raised to the E4-binding domain of E4-DBP (GST.E4-DBP fusion) reacted specifically with the 66K protein band (Fig. 3B). Neither p68 nor eIF4A associated with MBP.16 E1∧E4 in pull-down experiments (Fig. 3B). The 80K and 50K bands were not detected using antibodies to E4-DBP (Fig. 3B) or to the conserved DEAD box motif (see Materials and Methods).
FIG. 3.
(A) Characterization of 16 E1∧E4-binding proteins in epithelial cell extracts. Cellular proteins from CV-1 or HeLa cells were metabolically labeled with [35S]methionine before being bound to MBP.16 E1∧E4, GST.16 E1∧E4, or MBP or GST control proteins immobilized on agarose. Cellular proteins which associated specifically with 16 E1∧E4 were subsequently identified by SDS gel electrophoresis and autoradiography. Both MBP.16 E1∧E4 and GST.16 E1∧E4 interacted with cellular proteins of 80K and 66K, which were not bound by either MBP or GST alone (arrows). An additional protein of 50K was bound by GST.16 E1∧E4. Lanes M, molecular weight markers. (B) Western blotting identifies the 66K E4-binding protein as E4-DBP. (Left panel) MBP.16 E1∧E4-binding proteins (identified in panel A) were fractionated by SDS gel electrophoresis and Western blotted using antibody to eIF4A, p68, E4-DBP, or GST or using preimmune serum (pre-imm) from the rabbit used to prepare the E4-DBP antiserum. A 66K band was apparent only when antibodies raised to E4-DBP were used in pull-down experiments carried out with MBP.16 E1∧E4. This band was not detectable when pull-down experiments were carried out with MBP alone. (Right panels) Antibody specificity was confirmed by carrying out Western blotting against the total protein extracts used for each pull-down experiment. Antibodies to eIF4A and p68 detected proteins of the predicted sizes (45K and 68K). Antibodies raised against E4-DBP detected a protein of 66K. Preimmune serum and antibodies to GST did not stain any protein bands. Numbers at left of panels indicate molecular weights (in thousands).
E4-DBP is a nucleolar protein which shuttles between the nucleus and the cytoplasm.
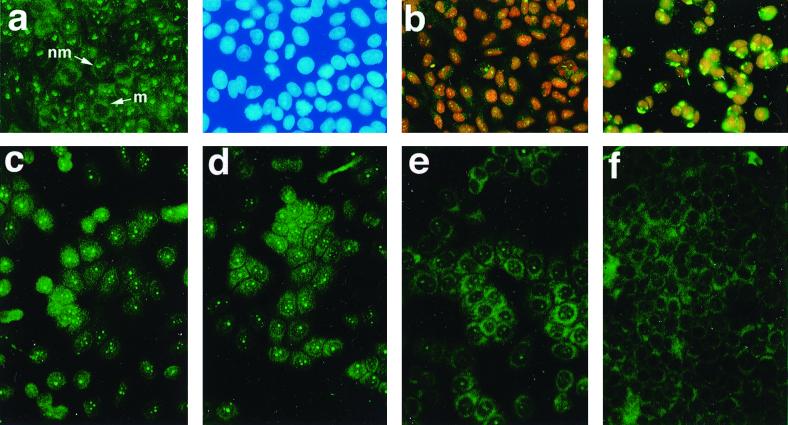
E4-DBP extracted from CV-1 cells migrated as a 66K polypeptide (Fig. 3B) and as a 62K species when expressed from pET.E4-DBP (see Fig. 8). In both cases, the apparent molecular weight was higher than that predicted from the sequence. A 66K E4-DBP protein was also found in HeLa, SiHa, HaCat (human keratinocyte), and COS-7 cells. E4-DBP was predominantly nucleolar (as determined by phase-contrast imaging), although some cytoplasmic staining was also apparent (Fig. 4a). Staining was completely lost when the E4-DBP antibody was incubated in the presence of MBP.E4-DBP but not MBP prior to immunodetection (data not shown).
FIG. 8.
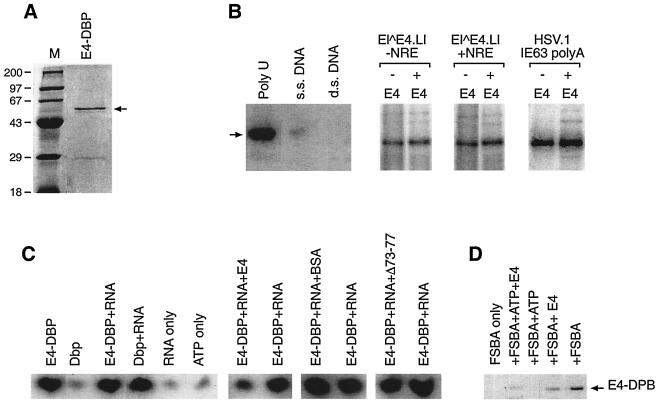
Functional analysis of E4-DBP and consequences of association with 16 E1∧E4. (A) Full-length E4-DBP was expressed from pET28a and purified by nickel ion-affinity chromatography and binding to poly(U)-Sepharose. The purified protein is shown (arrow) alongside molecular weight markers (M; in thousands) following gel electrophoresis and staining with Coomassie blue. (B) Purified E4-DBP (as shown in panel A) was mixed with poly(U)-Sepharose or Sepharose coated with either single-stranded (ss) or double-stranded (ds) DNA. Immobilized E4-DBP was then eluted and detected following gel electrophoresis and Western blotting using anti E4-DBP sera (lanes Poly U, s.s. DNA, and d.s. DNA). The position of the E4-DBP band is indicated by an arrow. E4-DBP bound poly(U)-Sepharose. Weak binding was seen with ss DNA, but an association with ds DNA was not detected. Binding to specific mRNAs is shown in the panels on the right. The primary HPV16 late mRNA (E1∧E4.L1), with (+) or without (−) the NRE, was examined for its ability to bind E4-DBP in an RNA cross-linking assay. E4-DBP could bind both mRNA species as well as a control mRNA derived from HSV-1 (far right). The addition of MBP.16 E1∧E4 had no effect on the ability of E4-DBP to bind RNA. (C) Release of phosphate from [γ-32P]ATP by E4-DBP in the presence or absence of RNA. E4-DBP showed ATPase activity which was not dependent on the addition of RNA. A control DEAD box protein (Dbp) required RNA. No ATPase activity was apparent in RNA-only or ATP-only controls. The addition of MBP.16 E1∧E4 reduced the level of ATPase activity exhibited by E4-DBP. No similar reduction in ATPase activity was apparent when bovine serum albumin or the Δ73-77 mutant protein was added. (D) E4-DBP (arrow) binds the ATP analogue FSBA, and binding is reduced following the addition of either ATP, E4, or both. Complexes between FSBA and E4-DBP were detected by Western blotting using an anti-FSBA antibody (see Materials and Methods) following SDS gel electrophoresis.
FIG. 4.
(a and b) Localization of E4-DBP in uninfected epithelial cells and in cells infected with vacc.16 E1∧E4. (a) E4-DBP (FITC) has a predominantly nucleolar distribution in COS-7 epithelial cells (left panel), although some cytoplasmic fluorescence can also be seen. Nonnucleolar staining is apparent in cells undergoing cell division. m, mitotic cell; nm, nonmitotic cell. Staining with 4′,6′-diamidino-2-phenylindole is also shown (right panel). (b) In CV-1 cells, E4-DBP is predominantly nucleolar (left panel) but is apparent in the cytoplasm following infection with vacc.16 E1∧E4 (right panel). E4-DBP was detected with fluorescein isothiocyanate, and cells were counterstained with propidium iodide. (c to f) E4-DBP shuttles between the nucleus and the cytoplasm. (c) Distribution of E4-DBP in CV-1 cells at time zero. (d) At 1.5 h after treatment with actinomycin D, E4-DBP had a more diffuse nuclear distribution, although nucleolar staining was still apparent. (e and f) After 4.5 h, the protein was predominantly cytoplasmic (e), and by 9 h, E4-DBP was no longer detectable in the nucleoli (f). Images were taken at a magnification of ×40.
16 E1∧E4 is predominantly cytoplasmic in low-grade cervical lesions (20), suggesting that binding to E4-DBP may take place in the cytoplasm. Nucleolar proteins such as nucleolin/C23 and B23/No38 shuttle between the nucleus and the cytoplasm (5), and following the expression of 16 E1∧E4 from a vaccinia virus recombinant (vacc.16 E1∧E4 [18]), the level of E4-DBP in the cytoplasm was found to increase (Fig. 4b). Although this result was a consequence of vaccinia virus infection (data not shown), it suggests that E4-DBP, like nucleolin, may also undergo nuclear-cytoplasmic shuttling.
Vaccinia virus infection results in the rapid shutoff of host cell protein synthesis, an effect which may account for the apparent increase in cytoplasmic E4-DBP levels. To examine this idea, cells were treated with cycloheximide, which inhibits the elongation of polypeptide chains by inhibiting the peptidyltransferase activity of the 60S ribosomal subunits. E4-DBP remained predominantly nuclear under such conditions, suggesting that the cytoplasmic accumulation of E4-DBP is not simply a consequence of the inhibition of protein synthesis. RNA binding proteins, such as hnRNP A1, A2, and E, which promote RNA processing and export, undergo nuclear-cytoplasmic shuttling in association with RNA (58). Following treatment with RNA polymerase II inhibitors, such as actinomycin D, such proteins rapidly accumulate in the cytoplasm. To examine the localization of E4-DBP under such conditions, CV-1 cells were treated with 5 μg of actinomycin D per ml, and the movement of E4-DBP was monitored over time. After 1.5 h, E4-DBP showed a diffuse nuclear and cytoplasmic distribution, although association with nucleoli was still apparent (Fig. 4d; time zero shown in Fig. 4c). At 4.5 h after the inhibition of transcription, E4-DBP was almost exclusively cytoplasmic (Fig. 4e), and after 9 h, nucleoli were no longer visible (Fig. 4f). hnRNP A1 also relocated to the cytoplasm under these conditions, while hnRNP C, which is retained in the nucleus by virtue of a retention sequence, did not (data not shown). Identical results were obtained when actinomycin D and cycloheximide were used together. These studies suggest that E4-DBP shuttles from the nucleus to the cytoplasm and that it probably has a cytoplasmic function in addition to its role in the nucleus.
16 E1∧E4 can associate with the cytoplasmic form of E4-DBP in human epithelial cells.
When expressed in cultured epithelial cells following transfection (from pMV11.16 E1∧E4), 16 E1∧E4 had a diffuse distribution in the cytoplasm and was associated in part with the cytokeratin network (18, 62) (Fig. 5a, c, and d). A similar pattern was found in vivo in cervical lesions caused by HPV16 (20), while in cells lacking intermediate filaments (SW13 cl.2 cells [66]), the protein was diffuse (Fig. 5e, g, and h). In SW13 cl.2 cells and, to a lesser extent, in COS-7 cells, both 16 E1∧E4 and E4-DBP were detected in the cytoplasm (Fig. 5a-d and 5e-h). In light of this result and because E4-DBP shuttles between the nucleus and the cytoplasm, we sought to establish whether 16 E1∧E4 associates with cytoplasmic E4-DBP in cervical epithelial cells.
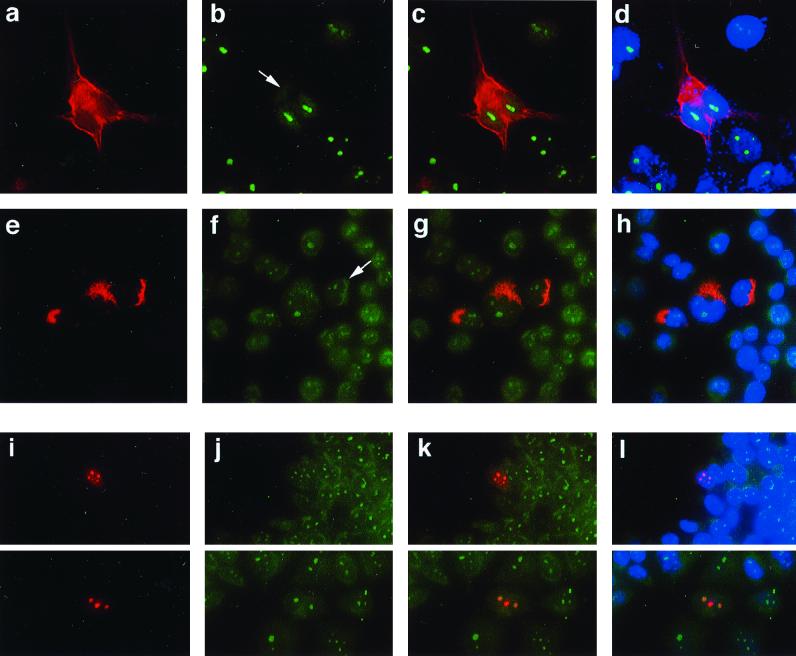
FIG. 5.
(a to d) Localization of E4-DBP in epithelial cells expressing 16 E1∧E4. (a and b) Localization of 16 E1∧E4 (a, red) and E4-DBP (b, green) following expression of 16 E1∧E4 in COS-7 cells by transfection. E4-DBP staining is apparent in the cytoplasm (arrow) in 16 E1∧E4-expressing cells. (c and d) Double (E1∧E4 and E4-DBP) (c) and triple (E1∧E4, E4-DBP, and 4′,6′-diamidino-2-phenylindole) (d) stains. (e to h) Localization of 16 E1∧E4 and E4-DBP in IF-negative cells. (e and f) Localization of 16 E1∧E4 (e, red) and E4-DBP (f, green) following expression of 16 E1∧E4 in SW13 cl.2 cells, which lack IF. Cytoplasmic E4-DBP staining is marked by an arrow in 16 E1∧E4-expressing cells. (g and h) Double (g) and triple (h) stains (as described above). (i to l) Localization of 16 E1∧E4 ΔLLXLL mutant and E4-DBP in IF-negative cells. (i and j) Localization of 16 E1∧E4 ΔLLXLL (i, red) and E4-DBP (j, green) in SW13 cl.2 cells, which lack IF. (k and l) Double (k) and triple (l) stains (as described above). Loss of the LLXLL motif causes relocation of 16 E1∧E4 to the nucleoli in SW13 cl.2 cells.
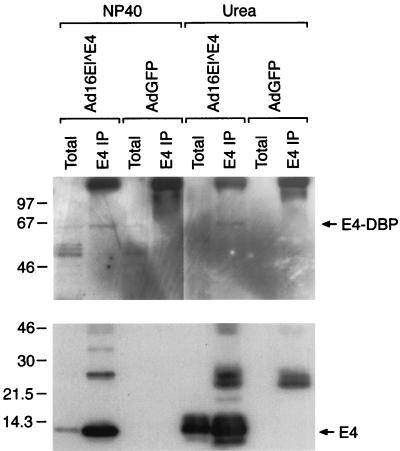
SiHa cells were infected with a recombinant adenovirus expressing 16 E1∧E4 (rAd.16 E1∧E4) or a control adenovirus expressing green fluorescent protein (rAd.GFP), and cell extracts were prepared using buffers containing either NP-40 or urea. SiHa cells were chosen for these experiments because they are infected efficiently with recombinant adenoviruses (100% infection at 30 PFU/cell). E4-DBP was present in both the NP-40 and the urea fractions (as determined by Western blotting) and could be readily immunoprecipitated using antibody to 16 E1∧E4 (rabbit polyclonal antibody to MBP.16 E1∧E4) (Fig. 6). E4-DBP was not precipitated using a rabbit polyclonal antibody to MBP (control) and was not precipitated from cells infected with rAd.GFP (Fig. 6).
FIG. 6.
Detection of complexes between 16 E1∧E4 and E4-DBP in cervical epithelial cells. SiHa cells were infected at 30 PFU/cell with recombinant adenoviruses expressing either 16 E1∧E4 (lanes Ad16E1∧E4) or GFP (lanes AdGFP). At 24 h after infection, cells were lysed with either NP-40 or 9 M urea, and the E4 proteins were immunoprecipitated using TVG402 immobilized on protein A-Sepharose (lanes E4 IP). Total protein extracts were run in adjacent lanes (lanes Total). The gels were Western blotted and probed either with a rabbit polyclonal antibody to E4-DBP (upper blot) or with a rabbit polyclonal antibody to 16 E1∧E4 (lower blot). The expected positions of E4-DBP (66K) and 16 E1∧E4 (10K monomer) are indicated by arrows at the right. E4-DBP could be detected only in E4 immunoprecipitates from cells expressing 16 E1∧E4 (NP-40 extracts) and not when immunoprecipitation was carried out with cells expressing GFP. The 66K E4-DBP protein was difficult to detect in total protein extracts of SiHa cells (lanes Total). Numbers at left indicate molecular weights (in thousands).
The expression of mutant 16 E1∧E4 lacking the leucine-rich N-terminal region (from pMV11.16 ΔLLXLL E1∧E4) resulted in an altered distribution of the protein. The leucine cluster is lost during in vivo infection by HPV1 (13, 19, 20), and it is thought that this modification may also occur in HPV16-infected cervical lesions (20). In SW13 cl.2 cells, which lack cytoplasmic IF (which participate in tethering 16 E1∧E4 in the cytoplasm), the mutant E1∧E4 protein localized primarily to the nucleoli and had a distribution that was indistinguishable from that of E4-DBP (Fig. 5I, J, K, and L). As nucleolar 16 E1∧E4 is not seen in vivo, we did not attempt to precipitate E4–E4-DBP complexes from nucleoli. Our results suggest that the two proteins associate within the cell and that for wild-type 16 E1∧E4, this association probably occurs in the cytoplasm.
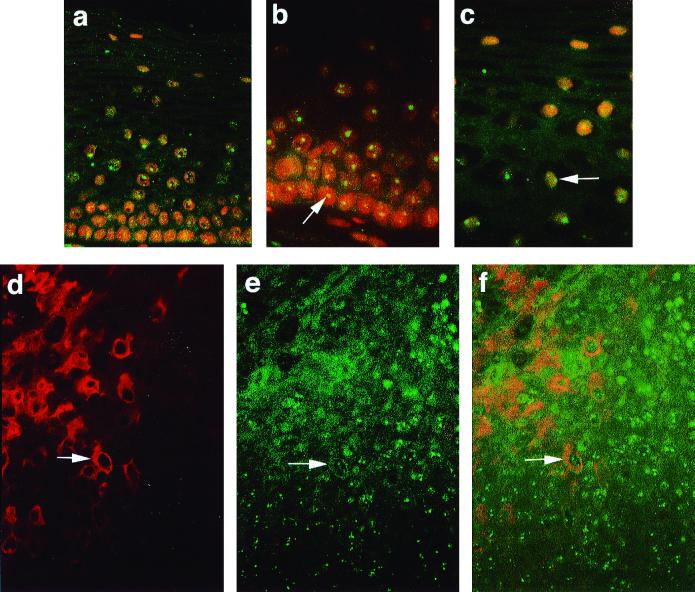
In cervical lesions caused by HPV16, the E1∧E4 protein is predominantly cytoplasmic (as determined by immunostaining; see Fig. 7d) (20). 16 E1∧E4 is first detected in sporadic cells of the spinous layer which support viral genome amplification, and the protein persists into the surface layers of the epithelium (20). In uninfected cervical cells, E4-DBP was detectable in the spinous and granular layers and had a predominantly nucleolar distribution, as seen in cells in cultures (Fig. 7a, b, and c). In HPV-infected tissue, cells expressing 16 E1∧E4 often lacked nucleolar E4-DBP (Fig. 7d, e, and f), even though nuclear degeneration is retarded in these cells (20). Evidence of cytoplasmic E4-DBP was apparent in the upper differentiated layers in some cells, although E4-DBP staining was generally weak (in formalin-fixed paraffin-embedded tissue).
FIG. 7.
(a to c) Distribution of E4-DBP in uninfected cervical cells. (a) E4-DBP (green) is found throughout the living layers of the normal cervix. (b and c) Nucleolar staining is seen in the basal layer (b, arrow) and in terminally differentiating cells (c, arrow). Sections were counterstained with propidium iodide (red). (d to f) Distribution of E1∧E4 and E4-DBP in low-grade lesions caused by HPV16. E4-DBP is present throughout the thickness of the infected epidermis. (d and e) Staining was carried out using antibody to 16 E1∧E4 (d, red) or E4-DBP (e, green). (f) The E4-DBP–E1∧E4 stains are merged. E4-DBP does not undergo dramatic relocation to the cytoplasm in E4-infected cells, although cytoplasmic E4-DBP can be detected (arrows).
Physical properties of E4-DBP and functional consequences of its association with E4.
DEAD box proteins are generally thought to be ATP-dependent RNA (or DNA) helicases, although only a few have actually been shown experimentally to unwind RNA (30). Mutational analysis of the conserved motifs has shown that the GXGKT motif is required for ATP binding, while the DEAD motif is required for ATP hydrolysis (30). The DEAD and SAT motifs are thought to couple ATP hydrolysis to RNA unwinding, while the HRIGR motif is required for association with RNA (30). The region of E4-DBP involved in E4 binding contains the GXGXT and DEAD motifs (Fig. 1B), suggesting that binding would interfere with ATP hydrolysis rather than RNA association. E4-DBP (expressed from pET.E4-DBP) (Fig. 8A) bound to poly(U) and could be purified from bacterial extracts on poly(U)-Sepharose (Fig. 8B). The protein also bound to HPV16 late mRNAs (and control RNAs derived from HSV-1), although this binding was not dependent on the presence of the HPV16 NRE located downstream of the L1 ORF (Fig. 8B). E1∧E4 itself showed no affinity for either DNA or RNA and did not appear to inhibit E4-DBP–RNA binding (Fig. 8B). E4-DBP bound single-stranded DNA poorly but did not associate with double-stranded DNA (Fig. 8B). Following purification [by Ni-nitrilotriacetic acid and poly(U) affinity chromatography], E4-DBP exhibited ATPase activity which was not dependent on the presence of endogenous RNA (Fig. 8C). The addition of MBP.16 E1∧E4 reduced the level of ATPase activity slightly but did not abolish it (Fig. 8C). The E. coli DEAD box protein DbpA was used as a control in these experiments. In this case, helicase activity was totally dependent on the addition of E. coli 23S RNA and was not inhibited by MBP.16 E1∧E4 (Fig. 8C). To determine if the reduction in the ATPase activity of E4-DBP was due to a direct inhibition of ATP binding by the E4 protein, E4-DBP was incubated with the ATP analogue FSBA in the presence or absence of MBP.16 E1∧E4. FSBA was found to specifically bind E4-DBP, and binding could be inhibited by the addition of ATP (Fig. 8D). MBP.16 E1∧E4 (but not MBP) also reduced FSBA binding (Fig. 8D). We conclude that the presence of the full-length E4 protein can interfere with ATP binding and ATPase activity but is not sufficient to completely block these functions in vitro.
E4-DBP has a predicted role in the regulation of mRNA stability and ribosome biogenesis.
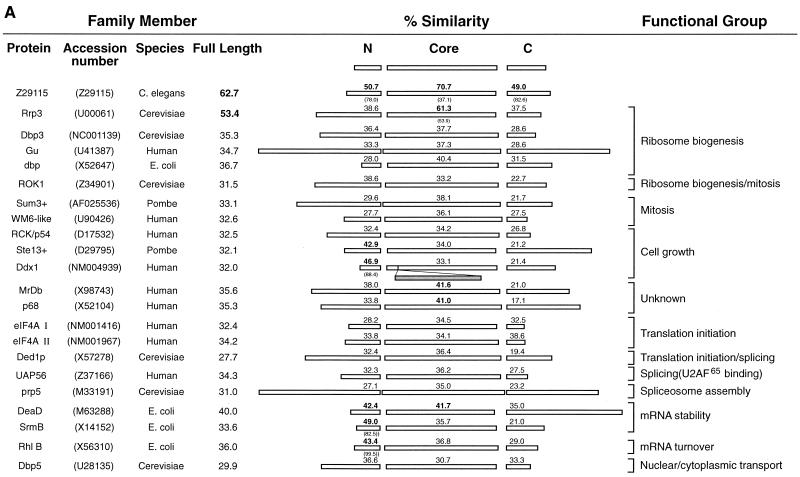
To gain insight into the function of E4-DBP, the full-length (1,799-bp) E4-DBP sequence was compared with 1,890,571 sequences contained in the GenEMBLPlus database. Several EST sequences were identified with 100% (or close to 100%) identity to E4-DBP, but no exact matches were found among proteins with known functions. Six ESTs showed homology to the 5′ end of E4-DBP, and two of these differed from our sequence in that they lacked a C at position 35 (Fig. 1B). A second sequence discrepancy was apparent at nucleotide 932, although this location was outside the region which encoded the E4-binding domain. Both discrepancies occurred in runs of C's and probably represent sequencing errors in the ESTs. DEAD box proteins typically have roles in RNA processing, such as ribosome biogenesis, splicing, translation initiation, RNA transport, and mRNA stability (30), although most have been only poorly characterized. Homology between E4-DBP and other DEAD box proteins was generally strongest within the central core domain (Fig. 9A). The N- and C-terminal extensions are thought to confer specialized functions on the different DEAD box proteins (30), and we were surprised to find that E4-DBP showed greatest similarity in these extensions to three E. coli proteins involved in the regulation of mRNA stability or mRNA degradation (SrmB, DeaD [43], and RhlB [59]) (Fig. 9C). Although the homology between E4-DBP and DeaD extended into the central core domain, this region of E4-DBP was more closely related to the Rrp3 protein of Saccharomyces cerevisiae (55) (Fig. 9C). Both Rrp3 and SrmB have been implicated in ribosome biogenesis. Like SrmB, DeaD, and RhlB, the human Ddx1 protein also shared homology with E4-DBP in its N-terminal extension. Ddx1 and MrDb are frequently upregulated along with myc in human cancers and are thought to confer a proliferative advantage to cells (1, 26, 37). Only one protein showed homology with E4-DBP throughout its length (Caenorhabditis elegans protein; GenBank accession number Z29115) (Fig. 9A). No function has yet been assigned to this protein. Three putative nuclear localization sequences (40) were identified in E4-DBP. The C terminus is rich in arginine, lysine, and glycine residues. In the nucleolar protein C23, a similar region has been reported to be necessary for RNA binding (32). The C-terminal KKRKGR peptide constitutes a putative membrane retention sequence (71).
FIG. 9.
Prediction of the function of E4-DBP by comparison with other members of the DEAD box protein family. (A) The E4-DBP sequence was divided into N-terminal, core (region between the GSGKT and HRVGRTAR motifs), and C-terminal domains before being compared with sequences of other members of the DEAD box protein family (listed at the left along with GenBank/EMBL accession numbers). For each domain, the percent similarity with E4-DBP (see Materials and Methods) is indicated above the box, and the percent divergence (where appropriate) is shown below it. The size of each box indicates the relative size of each domain in the different proteins. Percent similarity obtained from comparison of full-length proteins is shown in the column labeled “Full Length.” Where protein functions are known, they are indicated at the right. Overall similarity was strongest with Z29115 and the yeast Rrp3 protein, which is involved in ribosome biogenesis. (B) When only the N-terminal domain was compared, E. coli proteins (DeaD and RhlB) involved in mRNA turnover showed greater homology than Rrp3. Ddx1 also showed homology in this region. (C) In the core region, Rrp3 showed a higher level of homology to E4-DBP than any of these proteins. E4-DBP was more closely related to DeaD and Rrp3 (in terms of conservation of amino acids and gap positions) than to its closest human neighbor, MrDb. Conserved amino acids are boxed, and motifs characteristic of the DEAD box protein family are shaded.
DISCUSSION
HPV16 late-gene expression is controlled largely at the level of translation (68). The differentiation-dependent promoter (p670) which resides in the E7 ORF is activated during the migration of an infected cell toward the epithelial surface (38), leading to an increase in E1 and E2 and to the production of the major transcript encoding L1. Splice site selection and the presence of mRNA instability elements regulate E4, E2, and E1 mRNA levels, while transcripts encoding L1 (which initiate from the same promoter) are regulated at the level of poly(A) site selection and by cellular proteins which affect splice site activity and mRNA stability (reviewed in reference 14). E1∧E4 transcripts are highly abundant in papillomas, even though they contain NREs, which are found in all HPV mRNAs that terminate at the early poly(A) site (44). These sequences destabilize E6 and E7 transcripts early in infection and ensure that the E6 and E7 proteins are expressed at low levels (44). Transcripts encoding L1 also contain NREs, which are thought to prevent the accumulation of the virus structural proteins in the lower layers of the epidermis (12, 70). The best characterized of these destabilizes late transcripts by promoting rapid loss of the poly(A) tail (46). As transcripts for both E1∧E4 and L1 initiate from p670, the expression of E1∧E4 is predicted to correlate closely with the first appearance of unstable L1 transcripts. The association of 16 E1∧E4 with a putative mRNA binding-protein is potentially important given the extent to which papillomavirus late-gene expression is controlled at the posttranscriptional level.
DEAD box proteins contain a central core domain linked to N- and C-terminal regions with variable sequences (30). E4-DBP contained all eight motifs characteristic of DEAD box helicases (30), including the Walker A and Walker B motifs required for ATP binding and hydrolysis. Similar motifs are found in SF1 and SF3 helicases (35), such as the HPV16 E1 protein, and in AAA-positive proteins, such as MCM2, dynein motor proteins, and certain proteosomal subunits (54). DEAD and DEAH box proteins are SF2 helicases (34, 35), usually with a role in RNA processing. With the exception of eIF4A, which is involved in unwinding mRNAs at their 5′ ends (64), the roles of most DEAD box proteins are poorly defined. Several have been found to affect cell proliferation (1, 10, 26, 27, 37, 60, 72), although in most cases, the direct effect on RNA processing which gives rise to this phenotype is not known. Among proteins with known functions, E4-DBP shares the greatest homology with Rrp3 (55) in its central domain and with the E. coli DEAD box protein SrmB (43) in its N-terminal extension. Like Rrp3, SrmB was originally thought to be involved in ribosome biogenesis but is now also known to play an important role in regulating mRNA stability (43). Two other E. coli DEAD box proteins involved in mRNA turnover also showed homology with E4-DBP in their N termini. RhlB is a component of the E. coli degradosome (59) while DeaD is involved in regulating mRNA stability and ribosome biogenesis (43). The homology did not extend to E. coli DEAD box proteins not involved in RNA turnover (such as Dbp [31]) and was largely confined to the region of E4-DBP necessary for E4 binding. Although we are not certain of the exact role of E4-DBP, its nucleolar distribution and homologies suggest involvement in ribosome biogenesis (57). Its abilities to shuttle in and out of the nucleus and to be sequestered in the cytoplasm at the site of vaccinia virus replication (data not shown) indicate an additional role. Similarity with SrmB and RhlB suggests involvement in the regulation of mRNA turnover.
In the region required for E4 binding, E4-DBP also showed homology with the human DEAD box protein Ddx1 and with the Schizosaccharomyces pombe protein Ste13+. Although neither has been functionally characterized in terms of its effect on RNA, both have roles in the regulation of cell growth (33, 50). Growth stimulation has also been reported for other family members following their overexpression (MrDb [37], Sum3+ [27], WM6 [72], and ROK1 [60]). 16 E1∧E4 inhibits cell growth when expressed in monolayer cultures (unpublished data), while cells expressing 16 E1∧E4 in vivo lack differentiation markers (e.g., filaggrin and certain keratins). Both effects could result from changes in cellular RNA processing, although further work will be needed to establish this idea. Only a modest reduction in ATP binding and ATPase activity was apparent when E4-DBP was incubated with 16 E1∧E4, however, and without a clear function for E4-DBP, the consequences of the interaction remain speculative. The activity of some DEAD box proteins, such as eIF4A, is stimulated by association with its binding proteins. eIF4B increases the RNA-binding activity of eIF4A and confers specificity for a particular RNA sequence (52). Unlike eIF4B, however, E4 has no apparent affinity for RNA, and while E4-DBP bound RNA well, it showed no apparent sequence specificity (binding different mRNA species equally well). Interestingly, two cellular proteins, of 40K and 65K, have recently been shown to associate with the HPV16 NRE (12). The 65K protein, which is present in nuclear extracts, was identified as U2AF65; this protein also binds to instability elements in hepatitis B virus mRNAs (11). U2AF65 has recently been shown to associate with a novel human DEAD box protein (UAP56) and to induce conformational changes in mRNA. Whether E4-DBP can bind U2AF65 or the 40K cytoplasmic protein remains to be tested.
16 E1∧E4 is predominantly cytoplasmic when expressed in vivo. The association of E4 with cytoplasmic E4-DBP does not suggest involvement in ribosome biogenesis or pre-mRNA splicing (which are established functions of other family members). In cells in monolayer cultures, however, loss of the N terminus of 16 E1∧E4 leads to a cytoplasmic and nuclear distribution, and in some cells, clear colocalization of E1∧E4 to nucleoli is apparent (Fig. 5). N-terminal truncation of the LLXLL region is a modification of E4 which occurs in vivo during differentiation, raising the possibility that E4 may also target the nuclear form of E4-DBP. Like many other E4 proteins, 16 E1∧E4 contains a consensus nuclear localization motif in its central region (PKKHRR in 16 E1∧E4), and late in infection, the protein accumulates at the nuclear periphery (20). E4 is modified by phosphorylation and proteolytic processing during the migration of an infected cell toward the epithelial surface (36), and the function of E4 is likely to be modified as a result. Further work will establish the effect of such changes on E4-DBP binding.
The identification of E4-DBP as a 16 E1∧E4-binding protein suggests a novel role for E1∧E4 in the virus life cycle. Sequence comparison implicates E4-DBP in the regulation of mRNA stability and ribosome biogenesis. Papillomavirus late-gene expression is controlled largely at the level of pre-mRNA processing and mRNA stability, while individual proteins, such as E5 and L1, are expressed from bi- or polycistronic messages. The E4 ORF overlaps the viral E2 ORF, suggesting that their functions may be linked. The E2 protein of HPV5 has recently been shown to bind serine- and arginine-rich (SR) proteins, which are involved in pre-mRNA processing (48). The association of E4 with a DEAD box helicase also indicates a role for E4 in the posttranscriptional control of gene expression.
ACKNOWLEDGMENTS
John Doorbar, Robert C. Elston, and Sawsan Napthine contributed equally to this work.
We thank Tony Minson, John Skehel, and Barklie Clements for providing encouragement during the course of this study.
This work was supported by the Medical Research Council (support given to J.D., R.C.E., S.N., D.J., E.M., and H.M.G.), the Cancer Research Campaign (support given to N.C. and S.S.), Roche Products (support given to P.M.), and the University of Glasgow (support given to J.D.). John Doorbar is a Royal Society university research fellow.
REFERENCES
- 1.Akiyama K, Akao Y, Yokoyama M, Nakagawa Y, Noguchi T, Yagi K, Nishi Y. Expression of two dead box genes (DDX1 and DDX6) is independent of that of MYCN in human neuroblastoma cell lines. Biochem Mol Biol Int. 1999;47:563–568. doi: 10.1080/15216549900201603. [DOI] [PubMed] [Google Scholar]
- 2.Ashmole I, Gallimore P H, Roberts S. Identification of conserved hydrophobic C-terminal residues of the human papillomavirus type 1 E1E4 protein necessary for E4 oligomerisation in vivo. Virology. 1998;240:221–231. doi: 10.1006/viro.1997.8909. [DOI] [PubMed] [Google Scholar]
- 3.Becker T C, Noel R J, Coats W S, Gomez-Foix A M, Alam T, Gerard R D, Newgard C B. Use of recombinant adenovirus for metabolic engineering of mammalian cells. Methods Cell Biol. 1994;43A:161–189. doi: 10.1016/s0091-679x(08)60603-2. [DOI] [PubMed] [Google Scholar]
- 4.Bonnez W. Murine models of human papillomavirus-infected human xenografts. Papillomavirus Rep. 1998;9:27–38. [Google Scholar]
- 5.Borer R A, Lehner C F, Eppenberger H M, Nigg E A. Major nucleolar proteins shuttle between nucleus and cytoplasm. Cell. 1989;56:379–390. doi: 10.1016/0092-8674(89)90241-9. [DOI] [PubMed] [Google Scholar]
- 6.Breitburd F, Croissant O, Orth G. Expression of human papillomavirus type-1 E4 gene products in warts. In: Steinberg B M, Brandsma J L, Taichman L B, editors. Cancer cells. Vol. 5. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1987. pp. 115–122. [Google Scholar]
- 7.Bryan J T, Fife K H, Brown D R. The intracellular expression pattern of the human papillomavirus type 11 E1∧E4 protein correlates with its ability to self associate. Virology. 1998;241:49–60. doi: 10.1006/viro.1997.8965. [DOI] [PubMed] [Google Scholar]
- 8.Chow L T, Nasseri M, Wolinsky S M, Broker T R. Human papillomavirus type 6 and 11 mRNAs from genital condylomata acuminata. J Virol. 1987;61:2581–2588. doi: 10.1128/jvi.61.8.2581-2588.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chow L T, Reilly S S, Broker T R, Taichman L B. Identification and mapping of human papillomavirus type 1 RNA transcripts recovered from plantar warts and infected epithelial cell cultures. J Virol. 1987;61:1913–1918. doi: 10.1128/jvi.61.6.1913-1918.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daga R R, Jimenez J. Translational control of the cdc25 cell cycle phosphatase: a molecular mechanism coupling mitosis to cell growth. J Cell Sci. 1999;112:3137–3146. doi: 10.1242/jcs.112.18.3137. [DOI] [PubMed] [Google Scholar]
- 11.de Villiers E M. Human pathogenic papillomavirus types: an update. In: Zur Hausen H, editor. Human pathogenic papillomaviruses. Heidelberg, Germany: Springer-Verlag; 1994. pp. 1–12. [Google Scholar]
- 12.Dietrich-Goetz W, Kennedy I M, Levins B, Stanley M A, Clements J B. A cellular 65-kDa protein recognizes the negative regulatory element of human papillomavirus late mRNA. Proc Natl Acad Sci USA. 1997;94:163–168. doi: 10.1073/pnas.94.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doorbar J. The E4 proteins and their role in the viral life cycle. In: Lacey C, editor. Papillomavirus reviews: current research on papillomaviruses. Leeds, United Kingdom: Leeds Medical Information, Leeds University Press; 1996. pp. 31–38. [Google Scholar]
- 14.Doorbar J. Late stages of the papillomavirus life cycle. Papillomavirus Rep. 1998;9:119–123. [Google Scholar]
- 15.Doorbar J, Campbell D, Grand R J A, Gallimore P H. Identification of the human papillomavirus-1a E4 gene products. EMBO J. 1986;5:355–362. doi: 10.1002/j.1460-2075.1986.tb04219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doorbar J, Coneron I, Gallimore P H. Sequence divergence yet conserved physical characteristics among the E4 proteins of cutaneous human papillomaviruses. Virology. 1989;172:51–62. doi: 10.1016/0042-6822(89)90106-2. [DOI] [PubMed] [Google Scholar]
- 17.Doorbar J, Ely S, Coleman N, Hibma M, Davies D H, Crawford L. Epitope mapped monoclonal antibodies against the HPV16 E1∧E4 protein. Virology. 1992;187:353–359. doi: 10.1016/0042-6822(92)90327-l. [DOI] [PubMed] [Google Scholar]
- 18.Doorbar J, Ely S, Sterling J, McLean C, Crawford L. Specific interaction between HPV16 E1∧E4 and cytokeratins results in collapse of the epithelial cell intermediate filament network. Nature. 1991;352:824–827. doi: 10.1038/352824a0. [DOI] [PubMed] [Google Scholar]
- 19.Doorbar J, Evans H S, Coneron I, Crawford L V, Gallimore P H. Analysis of HPV1 E4 gene expression using epitope-defined antibodies. EMBO J. 1988;7:825–833. doi: 10.1002/j.1460-2075.1988.tb02881.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doorbar J, Foo C, Coleman N, Medcalf E, Hartley O, Prospero T, Napthine S, Sterling J, Winter G, Griffin H. Characterisation of events during the late stages of HPV16 infection in vivo using high affinity synthetic Fabs to E4. Virology. 1997;238:40–52. doi: 10.1006/viro.1997.8768. [DOI] [PubMed] [Google Scholar]
- 21.Doorbar J, Gallimore P H. Identification of proteins encoded by the L1 and L2 open reading frames of human papillomavirus type 1a. J Virol. 1987;61:2793–2799. doi: 10.1128/jvi.61.9.2793-2799.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doorbar J, Medcalf M, Napthine S. Analysis of HPV1 E4 complexes and their association with keratins in vivo. Virology. 1996;218:114–126. doi: 10.1006/viro.1996.0171. [DOI] [PubMed] [Google Scholar]
- 23.Doorbar J, Myers G. The E4 protein. In: Myers G, Delius H, Icenogel J, Bernard H-U, Baker C, Halpern A, Wheeler C, editors. Human papillomaviruses 1996. III. Los Alamos, N. Mex: Los Alamos National Laboratory; 1996. pp. 58–80. [Google Scholar]
- 24.Doorbar J, Parton A, Hartley K, Banks L, Crook T, Stanley M, Crawford L. Detection of novel splicing patterns in a HPV16-containing keratinocyte cell line. Virology. 1990;178:254–262. doi: 10.1016/0042-6822(90)90401-c. [DOI] [PubMed] [Google Scholar]
- 25.Elston R, Napthine S, Doorbar J. The identification of a conserved binding motif within human papillomavirus type 16 E6 binding peptides, E6AP and E6BP. J Gen Virol. 1998;79:371–374. doi: 10.1099/0022-1317-79-2-371. [DOI] [PubMed] [Google Scholar]
- 26.Fang C M, Shi C, Xu Y H. Deregulated c-myc expression in quiescent CHO cells induces target gene transcription and subsequent apoptotic phenotype. Cell Res. 1999;9:305–314. doi: 10.1038/sj.cr.7290029. [DOI] [PubMed] [Google Scholar]
- 27.Forbes K C, Humphrey T, Enoch T. Suppressors of cdc25p overexpression identify two pathways that influence the G2/M checkpoint in fission yeast. Genetics. 1998;150:1361–1375. doi: 10.1093/genetics/150.4.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Forrester A, Farrell H, Wilkinson G, Kaye J, Davis-Poynter N, Minson T. Construction and properties of a mutant of herpes simplex virus type 1 with glycoprotein H coding sequences deleted. J Virol. 1992;66:341–348. doi: 10.1128/jvi.66.1.341-348.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Freeman A, Morris L S, Mills A D, Stoeber K, Laskey R A, Williams G H, Coleman N. Minichromosome maintenance proteins as biological markers of dysplasia and malignancy. Clin Cancer Res. 1999;5:2121–2132. [PubMed] [Google Scholar]
- 30.Fuller-Pace F V. RNA helicases: modulators of RNA structure. Trends Cell Biol. 1994;4:271–274. doi: 10.1016/0962-8924(94)90210-0. [DOI] [PubMed] [Google Scholar]
- 31.Fuller-Pace F V, Nicol S M, Reid A D, Lane D P. DbpA: a DEAD box protein specifically activated by 23S rRNA. EMBO J. 1993;12:3619–3626. doi: 10.1002/j.1460-2075.1993.tb06035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ghisolfi L, Kharrat A, Joseph G, Amalric F, Erard M. Concerted activities of the RNA recognition and the glycine-rich C-terminal domains of nucleolin are required for efficient complex formation with pre-ribosomal RNA. Eur J Biochem. 1992;209:541–548. doi: 10.1111/j.1432-1033.1992.tb17318.x. [DOI] [PubMed] [Google Scholar]
- 33.Godbout R, Packer M, Bie W. Overexpression of a DEAD box protein (DDX1) in neuroblastoma and retinoblastoma cell lines. J Biol Chem. 1998;273:21161–21168. doi: 10.1074/jbc.273.33.21161. [DOI] [PubMed] [Google Scholar]
- 34.Gorbalenya A E, Koonin E V. Helicases: amino acid sequence comparisons and structure-function relationships. Curr Opin Struct Biol. 1993;3:419–429. [Google Scholar]
- 35.Gorbalenya A E, Koonin E V, Wolf Y I. A new superfamily of putative NTP-binding domains encoded by genomes of small DNA and RNA viruses. FEBS Lett. 1990;262:145–148. doi: 10.1016/0014-5793(90)80175-i. [DOI] [PubMed] [Google Scholar]
- 36.Grand R J A, Doorbar J, Smith K J, Coneron I, Gallimore P H. Phosphorylation of the human papillomavirus type 1 E4 proteins in vivo and in vitro. Virology. 1989;170:201–213. doi: 10.1016/0042-6822(89)90367-x. [DOI] [PubMed] [Google Scholar]
- 37.Grandori C, Mac J, Siebelt F, Ayer D E, Eisenman R N. Myc-Max heterodimers activate a DEAD box gene and interact with multiple E box-related sites in vivo. EMBO J. 1996;15:4344–4357. [PMC free article] [PubMed] [Google Scholar]
- 38.Grassmann K, Rapp B, Maschek H, Petry K U, Iftner T. Identification of a differentiation-inducible promoter in the E7 open reading frame of human papillomavirus type 16 (HPV-16) in raft cultures of a new cell line containing high copy numbers of episomal HPV-16 DNA. J Virol. 1996;70:2339–2349. doi: 10.1128/jvi.70.4.2339-2349.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 40.Hicks G R, Raikhel N V. Annu. Rev Cell Dev Biol. 1995;11:155–188. doi: 10.1146/annurev.cb.11.110195.001103. [DOI] [PubMed] [Google Scholar]
- 41.Iggo R D, Lane D P. Nuclear protein p68 is an RNA-dependent ATPase. EMBO J. 1989;8:1827–1831. doi: 10.1002/j.1460-2075.1989.tb03577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ingram A, Phelan A, Dunlop J, Clements J. Immediate early protein IE63 of herpes simplex virus type 1 binds RNA directly. J Gen Virol. 1996;77:1847–1851. doi: 10.1099/0022-1317-77-8-1847. [DOI] [PubMed] [Google Scholar]
- 43.Iost I, Dreyfus M. mRNAs can be stabilized by DEAD-box proteins. Nature. 1994;372:193–196. doi: 10.1038/372193a0. [DOI] [PubMed] [Google Scholar]
- 44.Jeon S, Lambert P F. Integration of HPV16 DNA into the human genome leads to increased stability of E6/E7 mRNAs: implications for cervical carcinogenesis. Proc Natl Acad Sci USA. 1995;92:1654–1658. doi: 10.1073/pnas.92.5.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Keen N, Elston R, Crawford L. Interaction of the E6 protein of human papillomavirus with cellular proteins. Oncogene. 1994;9:1493–1499. [PubMed] [Google Scholar]
- 46.Kennedy I A, Haddow J K, Clements J B. A negative regulatory element in the human papillomavirus type 16 genome acts at the level of late mRNA stability. J Virol. 1991;65:2093–2097. doi: 10.1128/jvi.65.4.2093-2097.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kozak M. Recognition of AUG and alternative initiator codons is augmented by G in position +4 but is not generally affected by the nucleotides in positions +5 and +6. EMBO J. 1997;16:2482–2492. doi: 10.1093/emboj/16.9.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lai M C, Teh B H, Tarn W Y. A human papillomavirus E2 transcriptional activator. The interactions with cellular splicing factors and potential function in pre-mRNA processing. J Biol Chem. 1999;274:11832–11841. doi: 10.1074/jbc.274.17.11832. [DOI] [PubMed] [Google Scholar]
- 49.Laimins L A. The biology of human papillomaviruses: from warts to cancer. Infect Agents Dis. 1993;2:74–86. [PubMed] [Google Scholar]
- 50.Maekawa H, Nakagawa T, Uno Y, Kitamura K, Shimoda C. The ste13+ gene encoding a putative RNA helicase is essential for nitrogen starvation-induced G1 arrest and initiation of sexual development in the fission yeast Schizosaccharomyces pombe. Mol Gen Genet. 1994;244:456–464. doi: 10.1007/BF00583896. [DOI] [PubMed] [Google Scholar]
- 51.McGrory W J, Bautista D S, Graham F L. A simple technique for the rescue of early region 1 mutations into infectious human adenovirus type 5. Virology. 1988;163:614–617. doi: 10.1016/0042-6822(88)90302-9. [DOI] [PubMed] [Google Scholar]
- 52.Methot N, Pickett G, Keene J D, Sonenberg N. In vitro RNA selection identifies RNA ligands that specifically bind to eukaryotic translation initiation factor 4B: the role of the RNA recognition motif. RNA. 1996;2:38–50. [PMC free article] [PubMed] [Google Scholar]
- 53.Nasseri M, Hirochika R, Broker T R, Chow L. A human papillomavirus type 11 transcript encoding an E1∧E4 protein. Virology. 1987;159:433–439. doi: 10.1016/0042-6822(87)90482-x. [DOI] [PubMed] [Google Scholar]
- 54.Neuwald A F, Aravind L, Spouge J L, Koonin E V. AAA+: a class of chaperone-like ATPases associated with assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- 55.O'Day C L, Chavanikamannil F, Abelson J. 18S rRNA processing requires the RNA helicase-like protein Rrp3. Nucleic Acids Res. 1996;24:3201–3207. doi: 10.1093/nar/24.16.3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pause A, Sonenberg N. Mutational analysis of a DEAD box RNA helicase: the mammalian translation initiation factor eIF-4A. EMBO J. 1992;11:2643–2654. doi: 10.1002/j.1460-2075.1992.tb05330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pederson T. The plurifunctional nucleolus. Nucleic Acids Res. 1998;26:3871–3876. doi: 10.1093/nar/26.17.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pinol-Roma S, Dreyfuss G. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature. 1992;355:730–732. doi: 10.1038/355730a0. [DOI] [PubMed] [Google Scholar]
- 59.Py B, Higgins C F, Krisch H M, Carpousis A J. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature. 1996;381:169–172. doi: 10.1038/381169a0. [DOI] [PubMed] [Google Scholar]
- 60.Rhee J Y, Lee W B, Kim J. Characterization and intracellular localization of the Rok1 protein involved in yeast cell division. Mol Cell. 1998;8:68–74. [PubMed] [Google Scholar]
- 61.Roberts S, Ashmole I, Johnson G D, Kreider J W, Gallimore P H. Cutaneous and mucosal human papillomavirus E4 proteins form intermediate filament-like structures in epithelial cells. Virology. 1993;197:176–187. doi: 10.1006/viro.1993.1578. [DOI] [PubMed] [Google Scholar]
- 62.Roberts S, Ashmole I, Rookes S, Gallimore P. Mutational analysis of the human papillomavirus type 16 E1∧E4 protein shows that the C terminus is dispensable for keratin cytoskeleton association but is involved in inducing disruption of the keratin filaments. J Virol. 1997;71:3554–3562. doi: 10.1128/jvi.71.5.3554-3562.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rogel-Gaillard C, Breitburd F, Orth G. Human papillomavirus type 1 E4 proteins differing by their N-terminal ends have distinct cellular locations when transiently expressed in vitro. J Virol. 1992;66:816–823. doi: 10.1128/jvi.66.2.816-823.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rogers G W, Richter N J, Merrick W C. Biochemical and kinetic characterization of the RNA helicase activity of eukaryotic initiation factor 4A. J Biol Chem. 1999;274:12236–12244. doi: 10.1074/jbc.274.18.12236. [DOI] [PubMed] [Google Scholar]
- 65.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 66.Sarria A J, Lieber J G, Nordeen S K, Evans R M. The presence or absence of a vimentin-type intermediate filament network affects the shape of the nucleus in human SW-13 cells. J Cell Sci. 1994;107:1593–1607. doi: 10.1242/jcs.107.6.1593. [DOI] [PubMed] [Google Scholar]
- 67.Schaack J, Langer S, Guo X. Efficient selection of recombinant adenovirus by vectors that express beta-galactosidase. J Virol. 1995;69:3920–3923. doi: 10.1128/jvi.69.6.3920-3923.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schwartz S. Cis-acting negative RNA elements on papillomavirus late mRNAs. Semin Virol. 1998;8:291–300. [Google Scholar]
- 69.Shah K V, Howley P M. Papillomaviruses. In: Fields B N, Knipe D M, Howley P M, editors. Virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2077–2109. [Google Scholar]
- 70.Sokolowski M, Zhao C, Tan W, Schwarz S. AU-rich mRNA instability elements on human papillomavirus type 1 late mRNAs and c-fos mRNAs interact with the same cellular factors. Oncogene. 1997;15:2303–2319. doi: 10.1038/sj.onc.1201415. [DOI] [PubMed] [Google Scholar]
- 71.Teasdale R D, Jackson M R. Signal-mediated sorting of membrane proteins between the endoplasmic reticulum and the Golgi apparatus. Annu Rev Cell Dev Biol. 1996;12:27–54. doi: 10.1146/annurev.cellbio.12.1.27. [DOI] [PubMed] [Google Scholar]
- 72.Warbrick E, Glover D. A Drosophila gene encoding a DEAD box RNA helicase can suppress loss of wee1/mik1 function in Schizosaccharomyces pombe. Mol Gen Genet. 1994;245:654–657. doi: 10.1007/BF00282229. [DOI] [PubMed] [Google Scholar]