SUMMARY
OBJECTIVE:
The objective of this study, carried out at the university hospital of the Federal University of Rio Grande, was to assess whether the treatment of chronic hepatitis C with direct-acting antivirals and the sustained virological response will affect the metabolic influences of the hepatitis C virus and whether these effects will vary according to genotypes and virus load.
METHODS:
This is an intervention pre-post study, carried out from March 2018 to December 2019, evaluating 273 hepatitis C virus patients treated with direct-acting antivirals. Inclusion criteria included being monoinfected with hepatitis C virus and achieving sustained virological response . Exclusion criteria included the presence of decompensated cirrhosis or co-infected with hepatitis B virus or human immunodeficiency virus. Genotypes, genotype 1 subtypes, and hepatitis C virus viral load were analyzed. Glucose metabolism was evaluated by the Homeostasis Model Assessment-insulin resistance indices: Homeostasis Model Assessment-β, TyG, and HbA1c, measured at the beginning of treatment and in sustained virological response. Statistical analysis with a T test by paired comparison of the means of the variables in the pretreatment and in the sustained virological response.
RESULTS:
Homeostasis Model Assessment-insulin resistance analysis: there were no significant differences between pretreatment and sustained virological response. Homeostasis Model Assessment-β analysis: significant increase in genotype 1 patients (p<0.028). TyG index analysis: significant increase in genotype 1b (p<0.017), genotype 3 (p<0.024), and genotype non-1 with low viral load (p<0.039). HbA1c analysis: significant decrease in genotype 3 (p<0.001) and genotype non-1 patients with low viral load (p<0.005).
CONCLUSION:
We detected significant metabolic influences after sustained virological response: impairment in lipid profile and improvements in the glucose metabolism. We found significant differences in genotype dependence, genotype 1 subtypes, and viral load.
KEYWORDS: Genotype, Hepatitis C, Lipid metabolism, Insulin resistance, Viral load
INTRODUCTION
Hepatitis C virus (HCV) can induce insulin resistance (IR) regardless of the severity of liver disease, demonstrating that this virus can induce this metabolic effect even in the preliminary stages of the disease 1 . The influence of HCV on lipid metabolic pathways has also been demonstrated, with evidence of significant changes after sustained virological response (SVR), suggesting a direct viral effect 2 . The viral cycle depends on cholesterol metabolism in host cells, which causes hypolipidemia during chronic infection 3 . The possible influence of HCV genotypes on lipid and glucose metabolism remains not well defined 4 . Overall, there is a great variability in the geographical distribution of HCV genotypes. In Brazil, the most prevalent genotype is 1 (G1) (64%), followed by genotype 3 (33%), and genotypes 2 and 4 (3%) 5 . There is evidence that the eradication of G1 is more beneficial in relation to IR than the eradication of genotypes 2 and 3 3,4 .
In addition to the genotype, some studies associate elevated levels of viral load (VL) with the presence of IR in patients with chronic hepatitis C 3,6 . However, other studies found no association between VL value, IR, and type 2 diabetes mellitus (T2DM) 7,1 .
The most commonly used method to evaluate the influence of SVR on glucose metabolism employs the Homeostasis Model Assessment (HOMA) indexes. The HOMA index estimates IR (HOMA-IR) and cell-β function (HOMA-β) 8 . Another proposed method for measuring IR is the TyG index, a product of fasting triglyceride levels and blood glucose 9 . Glycated hemoglobin (HbA1c) 10 , widely used in daily practice, provides an additional analysis of metabolic effects.
Considering the effectiveness of direct-acting antivirals (DAAs) and the association of HCV with systemic disorders, it can be expected that the eradication of the virus will improve metabolic parameters and reduce the rates of IR and T2DM in patients with chronic infection 11 . However, the effect of SVR on glucose control is still not clear 12–14 . The present study aimed to compare glucose and lipid metabolism in patients with chronic hepatitis C treated with DAAs in pretreatment and SVR, with special attention to genotypes and VL.
METHODS
This was an intervention pre-post study that initially evaluated 481 patients eligible for treatment with DAAs, treated at the Dr. Miguel Riet Corrêa Jr. University Hospital of the Federal University of Rio Grande (FURG) from March 2018 to December 2019. The study was approved by the ethics and research committee in the health area of FURG (CEPAS), under the process number: 23116.00516/2018-56. The CAAE registration number generated on the Brazil platform is 82698018.0.00005324. All patients read and signed free and informed consent forms before participating in the study. The treatment followed the inclusion and duration criteria of the Clinical Protocol and Therapeutic Guidelines for Hepatitis C and Co-infections 2018/2019 15 .
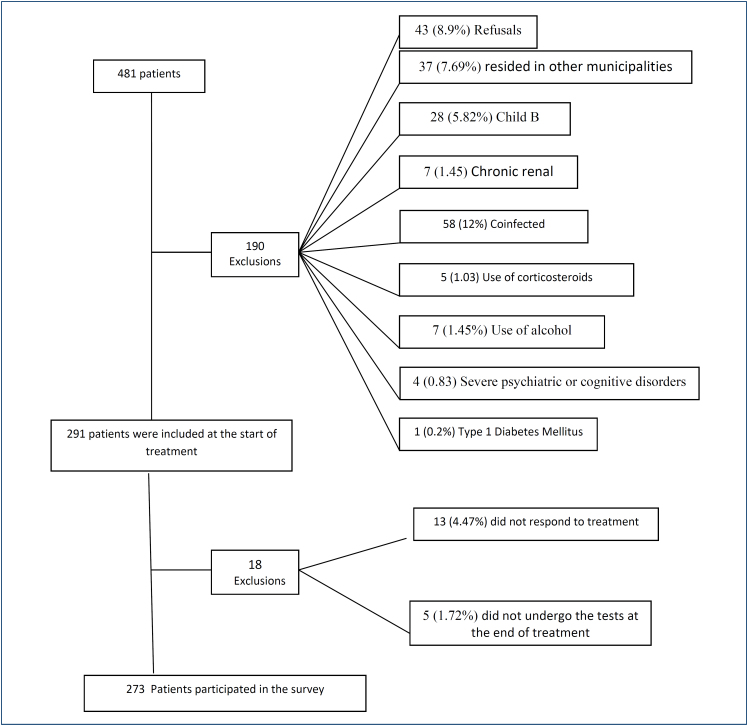
Inclusion criteria were as follows: monoinfected by HCV, achieving SVR, and living in the Rio Grande or São José do Norte municipalities. Exclusion criteria were as follows: the presence of decompensated cirrhosis, patients co-infected with hepatitis B virus or human immunodeficiency virus (HIV), severe psychiatric or cognitive disorders, chronic renal failure, type 1 DM, steroid or anabolic use, and alcohol consumption. According to these criteria, Figure 1 shows the number of patients who were included in this study. The diagnosis of T2DM, pre-diabetes, and normoglycemic followed the criteria adopted in the Guidelines of the Brazilian Society of Diabetes, 2019–2020 10 .
Figure 1. Flowchart of excluded patients.
The genotypes and subtypes of G1 and VL were evaluated by the real-time polymerase chain reaction technique. Undetectable VL, after the 12th week of treatment, was considered as SVR 15 .
For the evaluation of glucose metabolism, the following indices were used: HOMA-IR 16 , HOMA-β 16 , TyG 9 , and HbA1c. The parameters evaluated were measured at the beginning of treatment and in SVR. For comparison analysis, the patients were subdivided into groups according to genotypes and subtypes of genotype 1 and VL to demarcate their respective influences on glycidic and lipid metabolism.
Preliminary data analysis consisted of checking the frequency in search of extreme values, categorization, and creation of derived variables, and then using the T test to compare means. Next, the analysis of the means and their respective standard deviations of the variables in the pretreatment and the SVR were carried out. The confidence interval of the means was also calculated, followed by the T test for paired comparison of the means, adopting a p<0.05 as significant. All these analyses were performed using the statistical package.
This project was approved by the ethics and research committee in the health area of FURG (CEPAS) under the process number: 23116.00516/2018-56.
RESULTS
A total of 273 patients participated in this study. The mean age was 57 years old, 70.7% were white, and 52.7% were male. Regarding liver injury, 78 (28.6%) were cirrhotic, and 92 (33.7%) were classified as F0 or F1. About the glucose profile, 125 (45.8%) patients were prediabetic and 50 (18.3%) were diabetic. As for laboratory tests, we highlight a significant increase in the values of triglycerides, total cholesterol (TC), and low-density lipoproteins (LDL), but not in high-density lipoproteins (HDL) in SVR (Table 1).
Table 1. Anthropometric and laboratory data and comparison parameters.
| n (%) | Pretreatment | SVR | p-value | ||
|---|---|---|---|---|---|
| Age (mean±SD) | 57.03 (±11.11) | ||||
| ≤54 years | 98 (35.9) | ||||
| 55–64 years | 113 (41.4) | ||||
| ≥65 years | 62 (22.7) | ||||
| Color | |||||
| White | 193 (70.7) | ||||
| Nonwhite | 80 (29.3) | ||||
| Genre | |||||
| Male | 144 (52.7) | ||||
| Female | 129 (47.3) | ||||
| Weight (mean±SD) | 73.54 (±14.69) | 0.127 | |||
| WC Abdominal (mean±SD) | 73.21 (±14.72) | 94.04 (±11.99) | 0.206 | ||
| Adequate | 73 (29.3) | 94.38 (±11.87) | |||
| Inadequate | 200 (70.7) | ||||
| BMI (mean±SD) | 25.59 (±4.76) | 27.73 (±4.84) | 0.093 | ||
| ≤24.9 (81) | 81 (29.7) | ||||
| 25–29.9 (116) | 117 (42.9) | ||||
| ≥30 (74) | 75 (27.5) | ||||
| Glycemic profile (mean±SD) | 101.84 (±27.29) | 107.79 (±31.70) | 0.976 | ||
| With normal blood glucose | 98 (35.9) | ||||
| With prediabetes | 125 (45.8) | ||||
| With diabetes | 50 (18.3) | ||||
| Glycemic/genotypes (mean±SD) | |||||
| Genotype 1 (146) | 105.02 (±21.01) | 104.27 (±22.23) | 0.541 | ||
| enotype 2 (38) | 115.47 (±42.31) | 116.00 (±37.23) | 0.889 | ||
| Genotype 3 (87) | 109.28 (±28.10) | 110.25 (±40.57) | 0.775 | ||
| Profile triglycerides (mean±SD) | |||||
| Genotype 1 (146) | 101.87 (±66.30) | 105.30 (±66.47) | 0.416 | ||
| Genotype 2 (38) | 104.95 (±49.45) | 109.51 (±54.57) | 0.563 | ||
| Genotype 3 (87) | 79.85 (±31.56) | 100.94 (±76.15) | 0.031 | ||
| Laboratories (mean±SD) | |||||
| Total cholesterol | 166.37 (±36.91) | 185.52 (±38.41) | <0.001 | ||
| LDL | 97.30 (±33.86) | 111.80 (±36.66) | <0.001 | ||
| HDL | 49.59 (±14.23) | 50.26 (±13.66) | 0.652 | ||
| Triglycerides | 95.44 (±55.67) | 104.47 (±67.50) | 0.006 | ||
BMI: body mass index; WC: waist circumference; HbA1c: glycated hemoglobin; HDL: high-density lipoprotein; LDL: low-density lipoprotein; SD: standard deviation; SVR: sustained viral response.
The HOMA-IR index did not have significant differences between pretreatment and SVR (Table 2). Using the HOMA-β, we found a significant increase in those with G1 (89.29–103.97; p=0.028), a trend to significance in genotype 3 (87.51–99.61; p=0.058), and in subgroup G1 with low VL (85.15–96.34; p=0.05), there was a significant increase (Table 1).
Table 2. Analysis of the homeostasis model assessment-insulin resistance index, homeostasis model assessment-β cell index, TyG index, and HbA1c in relation to viral characteristics.
| HOMA-IR pretreatment Mean (SD) |
HOMA-IR pretreatment 95%CI |
HOMA-IR SVR Mean (SD) |
HOMA-IR SVR 95%CI |
p-value | HOMA-β pretreatment Mean (SD) |
HOMA-β pretreatment 95%CI |
HOMA-β SVR Mean (SD) |
HOMA-β SVR 95%CI |
p-value | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotypes (G) | Genotype (G) | ||||||||||||
| Genotype 1 (146) | 2.42 (±1.76) | 2.13–2.72 | 2.65 (±1.87) | 2.37–2.99 | 0.079 | Genotype 1 (146) | 89.29 (±62.85) | 79.42–99.37 | 103.97 (±79.97) | 91.27–117.30 | 0.028 | ||
| G1 a (67) | 2.36 (±1.60) | 2.02–2.79 | 2.53 (±1.83) | 2.12–3.00 | 0.457 | G1 a (67) | 85.81 (±53.67) | 73.25–99.82 | 98.27 (±91.03) | 79.46–121.54 | 0.223 | ||
| G1 b (78) | 2.63 (±2.44) | 2.14–3.19 | 3.06 (±3.76) | 2.34–3.94 | 0.098 | G1 b (78) | 91.12 (±62.94) | 77.56–105.43 | 104.16 (±61.73) | 90.66–117.47 | 0.086 | ||
| Genotype 2 (38) | 2.75 (±2.03) | 2.17–3.40 | 2.53 (±1.80) | 2.00–3.10 | 0.323 | Genotype 2 (38) | 79.34 (±54.38) | 63.93–99.16 | 72.78 (±43.84) | 59.75–87.58 | 0.516 | ||
| Genotype 3 (87) | 2.56 (±2.33) | 2.09–3.14 | 2.96 (±3.50) | 2.34–3.80 | 0.117 | Genotype 3 (87) | 87.51 (±56.82) | 76.00–101.42 | 99.61 (±57.16) | 87.76–11.01 | 0.058 | ||
| Viral load (VL) | 2.51 (±1.80) | 2.22–2.84 | 2.68 (±2.05) | 2.36–3.06 | 0.300 | Viral load (VL) | 87.85 (±65.71) | 76.86–99.40 | 98.62 (±79.30) | 86.28–113.14 | 0.147 | ||
| ≥600,000 – High (132) | 2.51 (±1.81) | 2.22–2.85 | 2.69 (±2.06) | 2.34–3.07 | 0.294 | ≥600,000 – High (132) | 87.40 (±65.76) | 77.61–98.48 | 99.23 (±79.29) | 86.33–113.98 | 0.108 | ||
| ≤599,999 – Low (131) | 2.50 (±1.87) | 2.09–2.93 | 2.76 (±1.80) | 2.33-3.16 | 0.239 | ≤599,999 – Low (131) | 92.17±70.27 | 77.17–108.12 | 110.80±94.10 | 90.41–134.05 | 0.089 | ||
| Genotype 1 (G1) | 2.31 (±1.66) | 1.96–2.77 | 2.52 (±1.98) | 2,10–3.07 | 0.148 | Genotype 1 (G1) | 85.15±52.49 | 72.99–99.07 | 96.34±60.84 | 82.02–110.66 | 0.050 | ||
| High VL (76) | 2.54 (±1.74) | 2.10–3.04 | 2.60 (±2.38) | 2.38–3.29 | 0.813 | High VL (76) | 82.02±59.63 | 66.46–98.06 | 81.67±49.57 | 68.98–94.74 | 0.966 | ||
| Low VL (66) | 2.67 (±2.57) | 2.11–3.32 | 3.00 (±3.55) | 2.35–3.85 | 0.181 | Low VL (66) | 87.40±53.29 | 75.30–100.29 | 99.14±57.62 | 86.12–114.00 | 0.080 | ||
| Non-1 Genotype (GN1) | Non-1 Genotype (GN1) | ||||||||||||
| High VL (55) | High VL (55) | ||||||||||||
| Low VL (70) | Low VL (70) | ||||||||||||
|
TyG pretreatment
Mean (SD) |
TyG pretreatment
95%CI |
TyG
RVS Mean (SD) |
TyG
RVS 95%CI |
p-value |
HbA1c pretreatment
Mean (SD) |
HbA1c pretreatment
95%CI |
HbA1c
RVS Mean (SD) |
HbA1c
RVS 95%CI |
p-value | ||||
| Genotypes (G) | Genotypes | ||||||||||||
| Genotype 1 (144) | 4.56 (±0.26) | 4.52–4.60 | 4.58 (±0.27) | 4.53–4.62 | 0.510 | Genotype 1 (146) | 5.63 (±0.92) | 5.49–5.80 | 5.54 (±0.78) | 5.42–5.69 | 0.144 | ||
| G1 a (66) | 4.58 (±0.24) | 4.51–4.63 | 4.56 (±0.31) | 4.49–4.64 | 0.651 | G1 a (68) | 5.78 (±1.02) | 5.54–6.04 | 5.69 (±0.98) | 5.46–5.94 | 0.391 | ||
| G1 b (74) | 4.51 (±0.25) | 4.46–4.57 | 4.57 (±0.30) | 4.51–4.64 | 0.017 | G1 b (78) | 5.59 (±0.77) | 5.43–5.79 | 5.47 (±0.66) | 5.33–5.61 | 0.155 | ||
| Genotype 2 (37) | 4.63 (±0.31) | 4,53–4.73 | 4.64 (±0.29) | 4.54–4.74 | 0.771 | Genotype 2 (38) | 5.83 (±0.94) | 5.56–6.15 | 5.91 (±1.05) | 5.59–6.27 | 0.400 | ||
| Genotype 3 (83) | 4.48 (±0.23) | 4,43–4.52 | 4.54 (±0.33) | 4.48–4.62 | 0.024 | Genotype 3 (87) | 5.85 (±1.06) | 5.65–6.11 | 5.54 (±1.08) | 5.34–5.79 | 0.001 | ||
| Viral load (VL) | 4.55 (±0.27) | 4,51–4.60 | 4.56 (±0.31) | 4.51–4.62 | 0.639 | Viral load (VL) | 5.71 (±0.91) | 5.57–5.88 | 5.61 (±1.02) | 5.44–5.80 | 0.117 | ||
| ≥600,000 – High (128) | 4.55 (±0.27) | 4,50–4.60 | 4.56 (±0.31) | 4.50–4.62 | 0.634 | ≥600,000 – High (132) | 5.72 (±0.92) | 5.56–5.88 | 5.61 (±1.03) | 5.45–5.80 | 0.119 | ||
| ≤599,999 – Low (127) | 5.56 (±0.26) | 4.50–4.62 | 4.55 (±0.27) | 4.49–4.61 | 0.813 | ≤599,999 – Low (131) | 5.68 (±0.97) | 5.47–5.92 | 5.53 (±0.88) | 5.35–5.75 | 0.108 | ||
| Genotype 1 (G1) | 4.56 (±0.26) | 4.50–4.63 | 4.59 (±0.26) | 4.52–4.65 | 0.389 | Genotype (G1) | 5.58 (±0.83) | 5.40–5.79 | 5.52 (±0.63) | 5.37–5.67 | 0.462 | ||
| G1+High VL (76) | 4.54 (±0.28) | 4.46–4.62 | 4.57 (±0.37) | 4.47–4.67 | 0.323 | G1+High VL (76) | 5.78 (±0.84) | 5.58–6.01 | 5.73 (±1.20) | 5.44–6.05 | 0.609 | ||
| G1+Low VL (66) | 4.51 (±0.26) | 4.45–4.57 | 4.57 (±0.29) | 4.51–4.64 | 0.039 | G1+Low VL (66) | 5.90 (±1.14) | 5.65–6.19 | 5.59 (±0.99) | 5.38–5.85 | 0.005 | ||
| Non-1 Genotype (GN1) | Non-1 Genotype (GN1) | ||||||||||||
| GN1+High VL (55) | GN1+High VL (55) | ||||||||||||
| GN1+Low VL (68) | GN1+Low VL (70) | ||||||||||||
|
HOMA-IR pretreatment
Mean (SD) |
HOMA-IR pretreatment
95%CI |
HOMA-IR
SVR Mean (SD) |
HOMA-IR
SVR 95%CI |
p-value |
HOMA-β pretreatment
Mean (SD) |
HOMA-β pretreatment
95%CI |
HOMA-β
SVR Mean (SD) |
HOMA-β
SVR 95%CI |
p-value | ||||
| Genotypes (G) | Genotype (G) | ||||||||||||
| Genotype 1 (146) | 2.42 (±1.76) | 2.13–2.72 | 2.65 (±1.87) | 2.37–2.99 | 0.079 | Genotype 1 (146) | 89.29 (±62.85) | 79.42–99.37 | 103.97 (±79.97) | 91.27–117.30 | 0.028 | ||
| G1 a (67) | 2.36 (±1.60) | 2.02–2.79 | 2.53 (±1.83) | 2.12–3.00 | 0.457 | G1 a (67) | 85.81 (±53.67) | 73.25–99.82 | 98.27 (±91.03) | 79.46–121.54 | 0.223 | ||
| G1 b (78) | 2.63 (±2.44) | 2.14–3.19 | 3.06 (±3.76) | 2.34–3.94 | 0.098 | G1 b (78) | 91.12 (±62.94) | 77.56–105.43 | 104.16 (±61.73) | 90.66–117.47 | 0.086 | ||
| Genotype 2 (38) | 2.75 (±2.03) | 2.17–3.40 | 2.53 (±1.80) | 2.00–3.10 | 0.323 | Genotype 2 (38) | 79.34 (±54.38) | 63.93–99.16 | 72.78 (±43.84) | 59.75–87.58 | 0.516 | ||
| Genotype 3 (87) | 2.56 (±2.33) | 2.09–3.14 | 2.96 (±3.50) | 2.34–3.80 | 0.117 | Genotype 3 (87) | 87.51 (±56.82) | 76.00–101.42 | 99.61 (±57.16) | 87.76–11.01 | 0.058 | ||
| Viral load (VL) | 2.51 (±1.80) | 2.22–2.84 | 2.68 (±2.05) | 2.36–3.06 | 0.300 | Viral load (VL) | 87.85 (±65.71) | 76.86–99.40 | 98.62 (±79.30) | 86.28–113.14 | 0.147 | ||
| ≥600,000 – High (132) | 2.51 (±1.81) | 2.22–2.85 | 2.69 (±2.06) | 2.34–3.07 | 0.294 | ≥600,000 – High (132) | 87.40 (±65.76) | 77.61–98.48 | 99.23 (±79.29) | 86.33–113.98 | 0.108 | ||
| ≤599,999 – Low (131) | 2.50 (±1.87) | 2.09–2.93 | 2.76 (±1.80) | 2.33-3.16 | 0.239 | ≤599,999 – Low (131) | 92.17±70.27 | 77.17–108.12 | 110.80±94.10 | 90.41–134.05 | 0.089 | ||
| Genotype 1 (G1) | 2.31 (±1.66) | 1.96–2.77 | 2.52 (±1.98) | 2,10–3.07 | 0.148 | Genotype 1 (G1) | 85.15±52.49 | 72.99–99.07 | 96.34±60.84 | 82.02–110.66 | 0.050 | ||
| High VL (76) | 2.54 (±1.74) | 2.10–3.04 | 2.60 (±2.38) | 2.38–3.29 | 0.813 | High VL (76) | 82.02±59.63 | 66.46–98.06 | 81.67±49.57 | 68.98–94.74 | 0.966 | ||
| Low VL (66) | 2.67 (±2.57) | 2.11–3.32 | 3.00 (±3.55) | 2.35–3.85 | 0.181 | Low VL (66) | 87.40±53.29 | 75.30–100.29 | 99.14±57.62 | 86.12–114.00 | 0.080 | ||
| Non-1 Genotype (GN1) | Non-1 Genotype (GN1) | ||||||||||||
| High VL (55) | High VL (55) | ||||||||||||
| Low VL (70) | Low VL (70) | ||||||||||||
|
TyG pretreatment
Mean (SD) |
TyG pretreatment
95%CI |
TyG
RVS Mean (SD) |
TyG
RVS 95%CI |
p-value |
HbA1c pretreatment
Mean (SD) |
HbA1c pretreatment
95%CI |
HbA1c
RVS Mean (SD) |
HbA1c
RVS 95%CI |
p-value | ||||
| Genotypes (G) | Genotypes | ||||||||||||
| Genotype 1 (144) | 4.56 (±0.26) | 4.52–4.60 | 4.58 (±0.27) | 4.53–4.62 | 0.510 | Genotype 1 (146) | 5.63 (±0.92) | 5.49–5.80 | 5.54 (±0.78) | 5.42–5.69 | 0.144 | ||
| G1 a (66) | 4.58 (±0.24) | 4.51–4.63 | 4.56 (±0.31) | 4.49–4.64 | 0.651 | G1 a (68) | 5.78 (±1.02) | 5.54–6.04 | 5.69 (±0.98) | 5.46–5.94 | 0.391 | ||
| G1 b (74) | 4.51 (±0.25) | 4.46–4.57 | 4.57 (±0.30) | 4.51–4.64 | 0.017 | G1 b (78) | 5.59 (±0.77) | 5.43–5.79 | 5.47 (±0.66) | 5.33–5.61 | 0.155 | ||
| Genotype 2 (37) | 4.63 (±0.31) | 4,53–4.73 | 4.64 (±0.29) | 4.54–4.74 | 0.771 | Genotype 2 (38) | 5.83 (±0.94) | 5.56–6.15 | 5.91 (±1.05) | 5.59–6.27 | 0.400 | ||
| Genotype 3 (83) | 4.48 (±0.23) | 4,43–4.52 | 4.54 (±0.33) | 4.48–4.62 | 0.024 | Genotype 3 (87) | 5.85 (±1.06) | 5.65–6.11 | 5.54 (±1.08) | 5.34–5.79 | 0.001 | ||
| Viral load (VL) | 4.55 (±0.27) | 4,51–4.60 | 4.56 (±0.31) | 4.51–4.62 | 0.639 | Viral load (VL) | 5.71 (±0.91) | 5.57–5.88 | 5.61 (±1.02) | 5.44–5.80 | 0.117 | ||
| ≥600,000 – High (128) | 4.55 (±0.27) | 4,50–4.60 | 4.56 (±0.31) | 4.50–4.62 | 0.634 | ≥600,000 – High (132) | 5.72 (±0.92) | 5.56–5.88 | 5.61 (±1.03) | 5.45–5.80 | 0.119 | ||
| ≤599,999 – Low (127) | 5.56 (±0.26) | 4.50–4.62 | 4.55 (±0.27) | 4.49–4.61 | 0.813 | ≤599,999 – Low (131) | 5.68 (±0.97) | 5.47–5.92 | 5.53 (±0.88) | 5.35–5.75 | 0.108 | ||
| Genotype 1 (G1) | 4.56 (±0.26) | 4.50–4.63 | 4.59 (±0.26) | 4.52–4.65 | 0.389 | Genotype (G1) | 5.58 (±0.83) | 5.40–5.79 | 5.52 (±0.63) | 5.37–5.67 | 0.462 | ||
| G1+High VL (76) | 4.54 (±0.28) | 4.46–4.62 | 4.57 (±0.37) | 4.47–4.67 | 0.323 | G1+High VL (76) | 5.78 (±0.84) | 5.58–6.01 | 5.73 (±1.20) | 5.44–6.05 | 0.609 | ||
| G1+Low VL (66) | 4.51 (±0.26) | 4.45–4.57 | 4.57 (±0.29) | 4.51–4.64 | 0.039 | G1+Low VL (66) | 5.90 (±1.14) | 5.65–6.19 | 5.59 (±0.99) | 5.38–5.85 | 0.005 | ||
| Non-1 Genotype (GN1) | Non-1 Genotype (GN1) | ||||||||||||
| GN1+High VL (55) | GN1+High VL (55) | ||||||||||||
| GN1+Low VL (68) | GN1+Low VL (70) | ||||||||||||
HOMA-IR: homeostasis model assessment-insulin resistance; HOMA-β: homeostasis model assessment-β cell; TyG: product of triglycerides and glucose; HbA1c: glycated hemoglobin; SD: standard deviation; SVR: sustained viral response; CI: confidence interval.
Table 2 shows a significant increase in the TyG index in G1b patients (4.51–4.57; p=0.017), genotype 3 (4.48–4.54; p=0.024), and non-1 genotype (N1G) with low VL (4.51–4.57; p=0.039). Regarding HbA1c values in pretreatment and SVR, there was a significant decrease in patients with genotype 3 (5.85–5.54; p=0.001) and N1G with low VL (5.90–5.59; p=0.005).
DISCUSSION
It is estimated that approximately two-thirds of patients with chronic hepatitis C may experience extrahepatic manifestations, which are especially important in metabolic alterations 17 . Some clinical studies have suggested improvements, after antiviral treatment, in glucose metabolism 11,18 . In addition, the effect of HCV modulating the metabolic pathways of intrahepatic cholesterol biosynthesis to promote viral replication can generate significant changes in lipid metabolism in SVR 2 . However, positive and negative metabolic changes were described with treatment with DAAs 11,19 , and a better understanding of this issue is necessary in different populations.
We draw attention to the fact that 64.1% of the patients surveyed have lipid metabolism disorders, while in the general Brazilian population, it is estimated that 7.5–18.5% are prediabetic and 7.7% diabetic 20 , which is in accordance with the hypothesis of the association of HCV with IR and T2DM and in agreement with other authors 5,21 .
In this study, after SVR, there was a significant increase in TC, LDL, and triglycerides, but not in HDL. Studies conducted with patients with genotype 1 mostly observed a significant increase in the rates of TC, LDL, and triglycerides after SVR 22,23 . Similarly, Jain et al. 24 , in a prospective study with 50 individuals, all HCV genotype 3, demonstrated that TC and LDL increased significantly with SVR, but there were no changes in HDL and triglycerides. As antiviral treatment would affect lipid metabolism, it is not well established 2 . We highlight that the worsening observed in lipid metabolism makes an evolutionary control of this profile in patients who obtained SVR necessary.
Regarding the influence of genotypes on IR, when SVR was calculated, the HOMA-IR index did not detect significant differences between genotypes, but it was observed that the HOMA-β index improved significantly in patients with G1. This finding is in agreement with the research by Huang et al. 25 , in which 72.3% of the patients were G1, and also verified a significant improvement in β-cell function, suggesting that this genotype may have an important action on the β cells of the pancreas. Regarding the TyG index, we saw worsening in patients with G1b and genotype 3, suggesting a more hypolipidemic action of these agents. About HbA1c, there was significant improvement only in patients with genotype 3, suggesting a greater beneficial metabolic effect of SVR in these individuals. This result is in accordance with the research by Jain et al. 24 . However, we agree with the statement that more studies are needed for a better understanding of this phenomenon 3 .
In relation to the different genotypic influences on IR, there are specific associations with genotypes, but the causal relationship remains unclear 4 . Thus, we see that these results are complex and sometimes contradictory, suggesting that the analysis of the influences of genotypes may be obscured by other factors acting on glucose metabolism. However, from a practical point of view, the improvement of HbA1c obtained with SVR in patients with genotype 3 suggests the importance of this treatment on glucose metabolism.
When analyzing the isolated influence of VL, no significant role was detected in relation to glucose metabolism. Other researchers 3,6 indicated that higher VL levels are associated with the presence of IR in patients with chronic hepatitis C. In contrast, in another study 7 with nondiabetic patients, IR was not associated with VL. The present study agrees with these findings, because even in a population of normoglycemic, prediabetic, and diabetic patients, no association between SVR and change in glucose metabolism was found. However, when we analyzed the association of G1 with low VL, we observed a significant increase in HOMA-β, improvement in the TyG index, and HbA1c of these patients. These associations with low VL could suggest a more deleterious and permanent effect on individuals with high VL, who did not improve in any index studied and, in any association, investigated.
Regarding limitations, the high percentage of overweight and obese patients may have made it difficult to find a more generalized beneficial effect of SVR in this sample. There may have been diet-related variations for the analysis of fasting triglyceride levels, which could affect the reliability of the TyG index, making it difficult to interpret the effects of decreased lipolysis suppression with HCV cure. Another limitation is the relatively short follow-up time of these patients.
The observations of this study suggest variable metabolic influences after SVR, indicating differences in the effects of HCV genotypes, genotype 1 subtypes, and VL in specific situations but not in the entire sample, with possible damage to the lipid profile and benefits in the glucose profile of these individuals.
Footnotes
Funding: none.
REFERENCES
- 1.Drazilova S, Janicko M, Skladany L, Kristian P, Oltman M, Szantova M, et al. Glucose metabolism changes in patients with chronic hepatitis C treated with direct acting antivirals. Can J Gastroenterol Hepatol. 2018;2018:6095097–6095097. doi: 10.1155/2018/6095097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meissner EG, Lee YJ, Osinusi A, Sims Z, Qin J, Sturdevant D, et al. Effect of sofosbuvir and ribavirin treatment on peripheral and hepatic lipid metabolism in chronic hepatitis C virus, genotype 1-infected patients. Hepatology. 2015;61(3):790–801. doi: 10.1002/hep.27424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang ML. Metabolic alterations and hepatitis C: from bench to bedside. World J Gastroenterol. 2016;22(4):1461–1476. doi: 10.3748/wjg.v22.i4.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thompson AJ, Patel K, Chuang WL, Lawitz EJ, Rodriguez-Torres M, Rustgi VK, et al. Viral clearance is associated with improved insulin resistance in genotype 1 chronic hepatitis C but not genotype 2/3. Gut. 2012;61(1):128–134. doi: 10.1136/gut.2010.236158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferreira CT, Silveira TR. Hepatites virais: aspectos da epidemiologia e da prevenção. Rev Bras Epidemiol. 2004;7(4):473–487. doi: 10.1590/S1415-790X2004000400010. [DOI] [Google Scholar]
- 6.Hsu CS, Liu CJ, Liu CH, Wang CC, Chen CL, Lai MY, et al. High hepatitis C viral load is associated with insulin resistance in patients with chronic hepatitis C. Liver Int. 2008;28(2):271–277. doi: 10.1111/j.1478-3231.2007.01626.x. [DOI] [PubMed] [Google Scholar]
- 7.Tsochatzis E, Manolakopoulos S, Papatheodoridis GV, Hadziyannis E, Triantos C, Zisimopoulos K, et al. Serum HCV RNA levels and HCV genotype do not affect insulin resistance in nondiabetic patients with chronic hepatitis C: a multicentre study. Aliment Pharmacol Ther. 2009;30(9):947–954. doi: 10.1111/j.1365-2036.2009.04094.x. [DOI] [PubMed] [Google Scholar]
- 8.Song Y, Manson JE, Tinker L, Howard BV, Kuller LH, Nathan L, et al. Insulin sensitivity and insulin secretion determined by homeostasis model assessment and risk of diabetes in a multiethnic cohort of women: the Women’s Health Initiative Observational Study. Diabetes Care. 2007;30(7):1747–1752. doi: 10.2337/dc07-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vasques AC, Novaes FS, Oliveira Mda S, Souza JR, Yamanaka A, Pareja JC, et al. TyG index performs better than HOMA in a Brazilian population: a hyperglycemic clamp validated study. Diabetes Res Clin Pract. 2011;93(3):e98–100. doi: 10.1016/j.diabres.2011.05.030. [DOI] [PubMed] [Google Scholar]
- 10.Sociedade Brasileira de Diabetes Diretrizes da sociedade Brasileira de diabetes 2019-2020. Alamedas. 2019;8(2):178–180. [Google Scholar]
- 11.Hum J, Jou JH, Green PK, Berry K, Lundblad J, Hettinger BD, et al. Improvement in glycemic control of type 2 diabetes after successful treatment of hepatitis C virus. Diabetes Care. 2017;40(9):1173–1180. doi: 10.2337/dc17-0485. [DOI] [PubMed] [Google Scholar]
- 12.Li J, Gordon SC, Rupp LB, Zhang T, Trudeau S, Holmberg SD, et al. Sustained virological response to hepatitis C treatment decreases the incidence of complications associated with type 2 diabetes. Aliment Pharmacol Ther. 2019;49(5):599–608. doi: 10.1111/apt.15102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alsebaey A, Elhelbawy M, Abdel-Razek W, Hashim M, Elshenawy H, Waked I. HCV treatment with direct acting antivirals improves the insulin sensitivity. Expert Rev Anti Infect Ther. 2019;17(9):749–754. doi: 10.1080/14787210.2019.1653184. [DOI] [PubMed] [Google Scholar]
- 14.Lanini S, Bartolini B, Taibi C, Agresta A, Garbuglia AR, Montaldo C, et al. Early improvement of glycaemic control after virus clearance in patients with chronic hepatitis C and severe liver fibrosis: a cohort study. New Microbiol. 2019;42(3):139–144. [PubMed] [Google Scholar]
- 15.Ministério da Saúde. Brasil Protocolo clínico e diretrizes terapêuticas para hepatite C e coinfecções. Secr Vigilância em Saúde Dep DST, Aids e Hepatites Virais. 2018:1–72. [Google Scholar]
- 16.Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28(7):412–419. doi: 10.1007/BF00280883. [DOI] [PubMed] [Google Scholar]
- 17.Cacoub P, Desbois AC, Comarmond C, Saadoun D. Impact of sustained virological response on the extrahepatic manifestations of chronic hepatitis C: a meta-analysis. Gut. 2018;67(11):2025–2034. doi: 10.1136/gutjnl-2018-316234. [DOI] [PubMed] [Google Scholar]
- 18.Ciancio A, Bosio R, Bo S, Pellegrini M, Sacco M, Vogliotti E, et al. Significant improvement of glycemic control in diabetic patients with HCV infection responding to direct-acting antiviral agents. J Med Virol. 2018;90(2):320–327. doi: 10.1002/jmv.24954. [DOI] [PubMed] [Google Scholar]
- 19.Shehab Eldin W, Nada A, Abdulla A, Eldeen SS. The effect of hepatitis c virus eradication with new direct acting antivirals on glucose homeostasis in non-diabetic Egyptian patients. J Diabetes Metab. 2017;8(10):8–11. [Google Scholar]
- 20.Iser BPM, Pinheiro PC, Malta DC, Duncan BB, Schmidt MI. Prediabetes and intermediate hyperglycemia prevalence in adults and associated factors, Health National Survey. Cien Saude Colet. 2021;26(2):531–540. doi: 10.1590/1413-81232021262.34852020. [DOI] [PubMed] [Google Scholar]
- 21.Serfaty L. Metabolic manifestations of hepatitis c virus: diabetes mellitus, dyslipidemia. Clin Liver Dis. 2017;21(3):475–486. doi: 10.1016/j.cld.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 22.Morales AL, Junga Z, Singla MB, Sjogren M, Torres D. Hepatitis C eradication with sofosbuvir leads to significant metabolic changes. World J Hepatol. 2016;8(35):1557–1563. doi: 10.4254/wjh.v8.i35.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doyle MA, Galanakis C, Mulvihill E, Crawley A, Cooper CL. Hepatitis C direct acting antivirals and ribavirin modify lipid but not glucose parameters. Cells. 2019;8(3):252–252. doi: 10.3390/cells8030252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jain A, Kalra BS, Srivastava S, Chawla S. Effect of sofosbuvir and daclatasvir on lipid profile, glycemic control and quality of life index in chronic hepatitis C, genotype 3 patients. Indian J Gastroenterol. 2019;38(1):39–43. doi: 10.1007/s12664-019-00935-w. [DOI] [PubMed] [Google Scholar]
- 25.Huang JF, Huang CF, Yeh ML, Dai CY, Hsieh MH, Yang JF, et al. The outcomes of glucose abnormalities in chronic hepatitis C patients receiving interferon-free direct antiviral agents. Kaohsiung J Med Sci. 2017;33(11):567–571. doi: 10.1016/j.kjms.2017.07.003. [DOI] [PubMed] [Google Scholar]