SUMMARY
Several ebolaviruses cause outbreaks of severe disease. Vaccines and monoclonal antibody cocktails are available to treat Ebola virus infections, but not Sudan virus or other ebolaviruses. Current cocktails contain antibodies that cross-react with the secreted soluble glycoprotein (sGP) that absorbs virus-neutralizing antibodies. By sorting memory B cells from EBOV infection survivors, we isolated two broadly reactive anti-GP monoclonal antibodies, 1C3 and 1C11, that potently neutralize, protect rodents from disease, and lack sGP cross-reactivity. Both antibodies recognize quaternary epitopes in trimeric ebolavirus GP. 1C11 bridges adjacent promoters via the fusion loop. 1C3 has a tripartite epitope in the center of the trimer apex. One 1C3 antigen-binding fragment anchors simultaneously to the three receptor-binding sites in the GP trimer, and separate 1C3 paratope regions interact differently with identical residues on the three protomers. A cocktail of both antibodies completely protected nonhuman primates from EBOV and SUDV disease, indicating their potential clinical value.
Keywords: antibody therapeutic, Bundibugyo virus, cryo-EM, Ebola virus, ebolavirus, medical countermeasure, monoclonal antibody, nonhuman primate, Sudan virus
eTOC blurb:
Two broadly neutralizing, human survivor antibodies form a therapeutic cocktail that protects non-human primates from otherwise lethal Ebola and Sudan virus disease. The cryo-EM structure illustrates mechanism of neutralization by recognition of quaternary epitopes at complementary sites. One antibody, 1C3, simultaneously blocks all three receptor-binding sites in the GP trimer.
Graphical Abstract

INTRODUCTION
Viruses of the Ebolavirus genus have caused over two dozen human disease outbreaks, and each outbreak has been unpredictable in location, timing, scale, and identity of the etiologic virus. Six distinct ebolaviruses are known, with Ebola virus (EBOV), Sudan virus (SUDV), and Bundibugyo virus (BDBV) having caused more frequent episodes of severe disease (Feldmann et al., 2020). EBOV caused sustained outbreaks in Western Africa in 2013–2016 and in the Democratic Republic of the Congo in 2018–2020, with the virus re-emerging in survivors of both outbreaks in 2021 (CDC, 2021a; Keita et al., 2021; World Health Organization, 2021). SUDV has caused five outbreaks since its discovery in 1976, and BDBV was linked to two outbreaks, one in 2007 and one in 2012 (CDC, 2021b). The GP amino-acid sequences of SUDV and BDBV differ from that of EBOV by 50% and 30%, respectively.
Medical countermeasures are currently only available against EBOV. Efforts to control the 2013–2016 and 2018–2020 outbreaks included extensive vaccination with rVSVΔG-ZEBOV-GP (Ervebo), a vaccine approved by the U.S. Food and Drug Administration (Ollmann Saphire, 2020), and Ad26.ZEBOV/MVA-BN-Filo (Zabdeno/Mvabea), a vaccine approved by the European Medicines Agency (European Commission, 2020). Antibody-based therapeutics analyzed in the Pamoja Tulinde Maisha (PALM) randomized controlled phase II/III clinical trial—performed during the 2018–2020 outbreak—were also EBOV-specific (PREVAIL II Writing Group et al., 2016); (Mulangu et al., 2019). This trial associated treatment with monoclonal antibody (mAb) mAb114 or REGN-EB3 (a combination of three mAbs) with significantly increased survival when compared to the ZMapp control arm, especially in Ebola virus disease (EVD) patients with low viral loads (Mulangu et al., 2019). Both treatments have been approved by the U.S F.D.A.. These treatments target the EBOV glycoprotein (GP), which is the prominent antigen on the virion surface. GP is a trimer of heterodimers, consisting of GP1 and GP2. The GP1 subunit contains a mucin-like-domain (MLD) and the receptor-binding site (RBS); GP2 is a classical class I fusion protein and mediates virion-cell fusion. Distinct ebolaviruses vary significantly in genomic sequence and, as a result, are also antigenically distinct (Kuhn et al., 2020), with natural infection or vaccination eliciting single ebolavirus-specific responses at the polyclonal level. Consequently, individual antibodies that cross-react with multiple ebolaviruses, though rare, are of heightened interest for development of broadly applicable therapeutics and as design primers for the development of vaccines that induce pan-ebolavirus protection.
Multiple studies of anti-ebolavirus mAbs have sought to identify the epitopes that lead to greatest protection and broadest reactivity. Studies of several therapeutic candidates (Pascal et al., 2018; Qiu et al., 2016) and a comprehensive study across the field (Saphire et al., 2018) indicate that maximal efficacy could be achieved via antibody combinations of those that neutralize ebolaviruses through anchoring to the core of GP with those that offer both neutralizing activity and Fc-mediated activity by interacting with the receptor-binding head of GP1. Therapeutic combinations that offer broader reactivity against a variety of ebolaviruses would have broader therapeutic utility and could be deployed prior to identification of the etiologic agent during an Ebola disease outbreak (Bornholdt et al., 2019; Gilchuk et al., 2018).
We previously identified and characterized individual anti-EBOV mAbs that emerged in human survivors of the 2013–2016 EVD outbreak within the first year post-infection (Davis et al., 2019). For this study, we examined later samples from the same patient cohort. We identified two mAbs, 1C3 and 1C11, that asymmetrically recognize the EBOV GP trimer, potently neutralize diverse ebolaviruses, individually protected laboratory mice and domesticated guinea pigs, and together protected crab-eating macaques from SUDV infection and rhesus monkeys from EBOV infection. Using cryogenic electron microscopy (cryo-EM), we explain the structural basis for the broad reactivity of these antibodies.
RESULTS
Initial characterization of cross-reactive mAbs from human EVD survivors
We previously reported the development of specific antibody responses over time in human survivors of the 2013–2016 EVD outbreak (Davis et al., 2019). In that study, we observed persistent B-cell activation and increases in serum antibody avidity for up to 2 years after EVD onset. Somatic hypermutation and neutralizing activity increased steadily over time in the months following infection. We therefore focused our mAb discovery efforts on samples drawn at late time points (≈2 years) after infection. Staining of peripheral blood mononuclear cells (PBMCs) from these samples revealed a rare population of cross-reactive immunoglobulin G (IgG) memory B cells that bound to both EBOV and SUDV GP (Figure S1A). Paired heavy-chain (VH domain) and light-chain (VL domain) sequences were cloned from single B cells from two donors and expressed as mAbs. Purified mAbs from donor EVD5 at 24 months after hospital discharge were named “5.24.xx”, whereas purified mAbs from donor EVD9 at 20 months after hospital discharge were named “9.20.xx.” The majority of these mAbs are bound to EBOV, SUDV, and BDBV GP as determined by enzyme-linked immunosorbent assay (ELISA) (Figure S1B). All mAbs bound to EBOV GP ectodomain lacking the MLD (GPΔmucin; Figure S1C), indicating that their epitopes lie outside this region. MAbs were first screened at biosafety level (BSL) 2 for EBOV neutralization activity using a “biologically contained Renilla luciferase-expressing VP30 gene-deleted EBOV (Davis et al., 2019; Halfmann et al., 2008) (EbolaΔVP30-RenLuc virus; Figure S1E). Plaque reduction neutralization titers (PRNT) for promising mAbs identified in these assays were determined using infectious BDBV, EBOV, and SUDV under BSL-4 conditions (Honnold et al., 2014). Nine antibodies from the panel neutralized EBOV (Figure 1A, Figure S1B). Although all antibodies bound to SUDV and BDBV GP by ELISA (Figure S1B), only five neutralized SUDV, and three of these five also neutralized BDBV. Competition analysis divided the neutralizing antibodies into two groups (Figure 1B). These groups were preliminarily mapped to the GP chalice bowl and GP2 fusion loop, predicated on presence or absence of soluble GP (sGP) binding (Figure S1C), competition analysis with reference mAbs, and binding to previously described EBOV GP variants (Figure S1D).
Figure 1. Initial characterization of neutralizing monoclonal antibodies (mAbs) cloned from Ebola virus (EBOV) and Sudan virus (SUDV) glycoprotein (GP) dual-binding memory B cells.
(A) Neutralization of live EBOV, SUDV, or BDBV by the indicated monoclonal antibodies (mAbs) was assessed by plaque assay. Left: Neutralization of EBOV by varying concentrations of each mAb. Right: Summary table showing the 50 or 80% plaque reduction neutralization test (PRNT50 or PRNT80) for each mAb against the indicated viruses. (B) Grouping of the neutralizing mAbs into competition groups. The numbers in the table represent the binding of fluorescently labeled mAbs (columns) to EBOV GP-expressing cells in the presence of excess unlabeled competitor mAbs (rows). Binding in the presence of each competitor is expressed as a percentage of the fluorescence signal in the absence of competitor. mAbs were classified as soluble glycoprotein (sGP)-binding (+) or non-binding (−) by enzyme-linked immunosorbent assay (ELISA). mAb 9.20.1C3 was unique within the chalice bowl competition group in being unable to bind to sGP (yellow highlight).
Within the fusion loop group, mAb 5.24.1C11 (henceforth referred to as “1C11”) was the strongest neutralizer of EBOV, SUDV, and BDBV of the entire panel and was therefore selected for further study. To complement 1C11, which binds to the base of GP, we chose a second, non-competing antibody specific for the GP head region. Our previous work has shown that antibodies directed at this head region are the most effective at inducing immune effector functions (e.g. antibody-dependent cell cytotoxicity) that contribute to protection from EBOV infection (Saphire et al., 2018). MAb 9.20.1C3 (“1C3”) was the most potent head specific mAb at neutralizing EBOV and SUDV, the two ebolaviruses responsible for the most human cases and outbreaks. Although 1C3 did not neutralize BDBV, it did bind to recombinant BDBV GP in our screening assay (Fig S1B), suggesting it might still be able to contribute to protection against BDBV infection through Fc-dependent effector mechanisms.
To evaluate whether the mAb pair can neutralize the virus synergistically, we next performed in vitro neutralization assays with 1C3 and 1C11 cocktails using recombinant VSV pseudovirus (rVSV). MAb synergy in the combined cocktail was quantified with CompuSyn software to estimate the combination index (CI), a measurement that evaluates the effect of a drug combination vs. drugs individually (Chou, 2010). CI values were calculated for each tested concentration of the cocktail, and CI values < 1 were considered as evidence of synergy (CI=1 – additive effect; CI>1 – antagonism). The dose reduction index (DRI) was also calculated, indicating the fold-reduction in effective dose for individual mAbs in the cocktail. In one of three independent experiments to analyze synergy between 1C3 and 1C11 in the neutralization of rVSV/EBOV GP, at 50% effect level (Fa = 0.5), CI = 0.63 indicating synergism. The IC50 of 1C3 and 1C11 in combination, was lower than the IC50 of each mAb alone, with dose reduction of 2.64-fold for 1C3 and 3.93-fold for 1C11 (Table S1).
Cryogenic electron microscopy structure of mAbs 1C3 and 1C11 bound to EBOV glycoprotein
To investigate the structural basis for the broad reactivity of 1C3 and 1C11, we prepared a complex of EBOV GPΔmucin with 1C3 antigen-binding fragment (Fab) and 1C11 single-chain variable fragment (scFv) and determined the structure of the complex to 3.59-Å resolution using single-particle cryo-EM (Figure 2, and S2; Table S2). We also determined the crystal structure of unbound 1C3 Fab to 2.15-Å resolution to aid in model building (Table S3). Both variable and constant domains of the 1C3 Fab were visible in the crystal structure, but only its variable antigen-binding portions were well-ordered in the cryo-EM reconstruction.
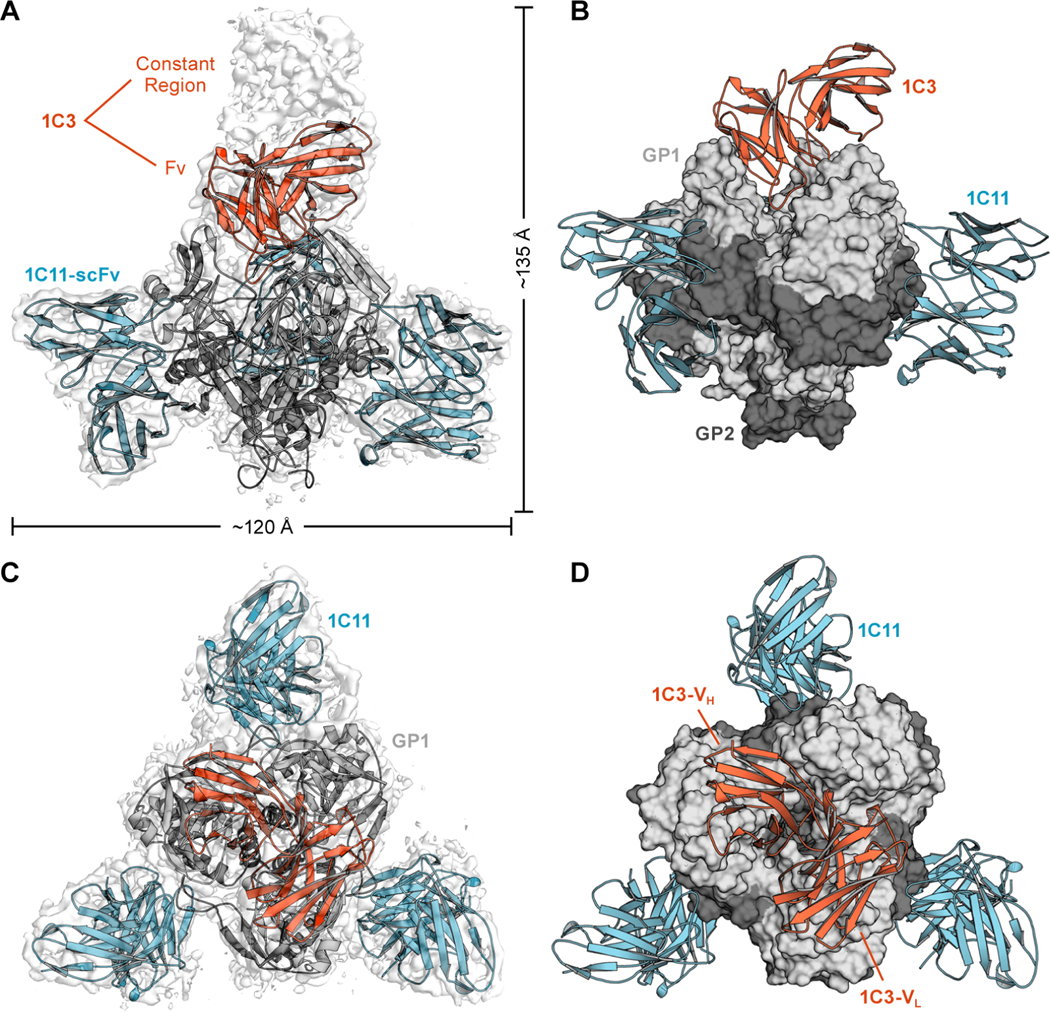
Figure 2. Cryogenic transmission electron microscopy structure of 1C3–1C11-Ebola virus (EBOV) glycoprotein (GP) complex.
(A). Side view. Cryogenic electron microscopy (cryo-EM) map in transparent grey and ribbon model of the complex. A single 1C3 antigen-binding fragment (Fab; orange) binds into the center of the glycoprotein (GP) trimer, with density visible for the entire Fab (grey), with the variable fragment (Fv) modeled in orange. (B) Molecular surface of EBOV GP in grey, with a single bound 1C3 reaching down into the chalice bowl. (C) Top view. Cryo-EM map in transparent grey. Three 1C11 antibodies, Fv modeled in blue, bind to GP, bridging the fusion loop to an adjacent GP protomer. (D) Molecular surface with the single 1C3 Fab (orange ribbon) extending diagonally across the entire GP trimer.
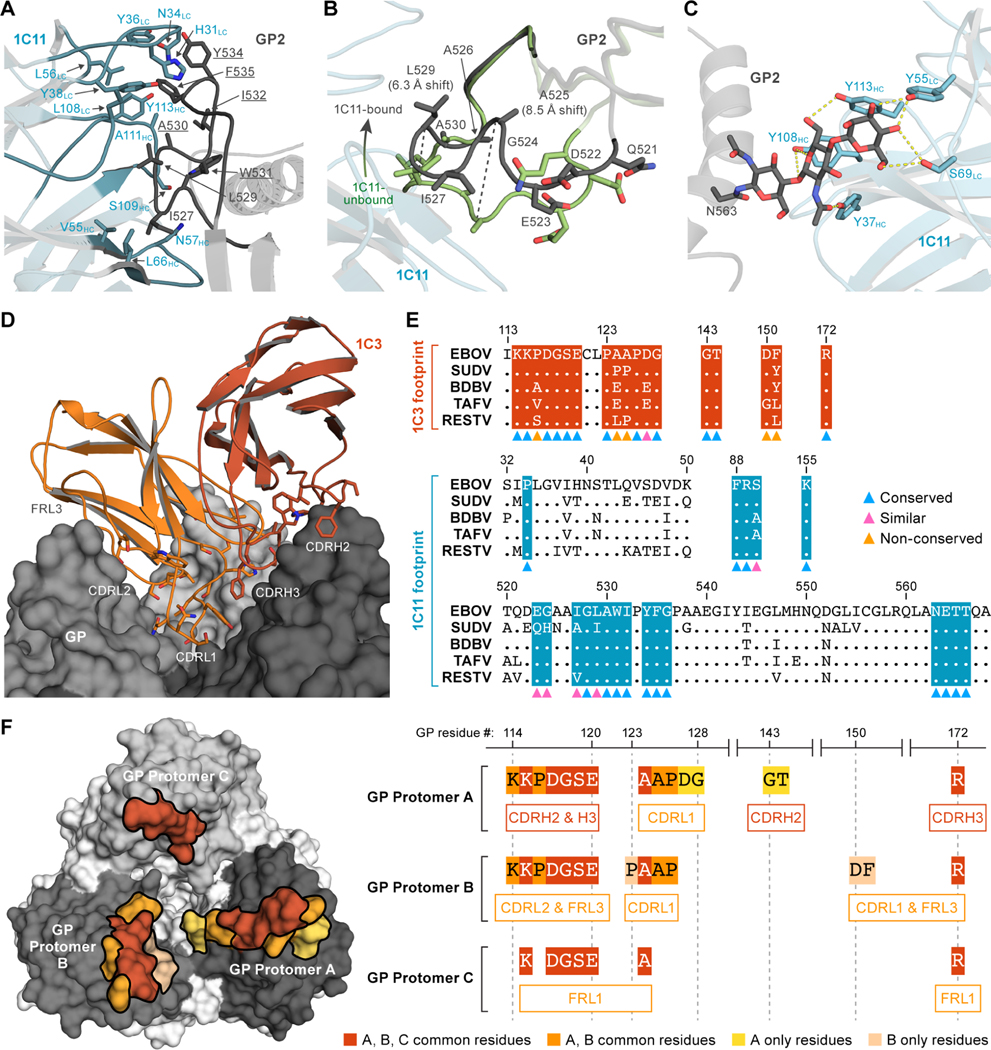
In that structure, three copies of 1C11 are simultaneously bound to the GP trimer with a binding site centered over the highly conserved hydrophobic fusion peptide of the internal fusion loop (Figure 3A). The footprint bridges two adjacent GP monomers in the trimer and includes surface area on both GP1 and GP2 (Figure 2 and S3). This binding mechanism suggests that 1C11 likely neutralizes EBOV by preventing conformational changes required for fusion of virion and host membranes. Each 1C11 scFv buried ≈673 Å 2 of surface area on one GP protomer and ≈408 Å 2 on another. The majority of the contacts are between non-polar side chains of the antibody complementarity determining regions (CDRs) and the hydrophobic fusion peptide of EBOV GP2. The paratope of 1C11 forms a hydrophobic pocket that pulls the fusion peptide slightly away from the surface of the GP relative to its position in unbound GP (6 Å for L529 and 8.5 Å for A525) (Figure 3B). 1C11 also makes several hydrogen bonds with GP, including with the N-linked glycan at position 563 (Figure 3C). The epitope recognized by 1C11 partially overlaps that of other pan-ebolavirus antibodies ADI-15878 (West et al., 2018) and 6D6 (Milligan et al., 2018) but is shifted slightly higher on the GP relative to the other footprints and avoids the poorly conserved regions below the fusion loop (Figure S3). As a result, 1C11 contacts GP1 residues 34, 88–90, and 155 and the GP2 residues 523–524, 527–532, 534–536, and 563–566 (Figure 3A, 3E).
Figure 3. Ebola virus (EBOV) glycoprotein (GP) interactions with 1C11 and 1C3.EBOV GP is shown in cartoon representation.
(A–C) or surface representation (D, F). 1C11 single-chain variable fragment (scFv) is shown in blue, and 1C3 antigen-binding fragment (Fab) is shown in orange. The international immunogenetics information system (IMGT) numbering scheme is used for mAbs 1C11 and 1C3. (A) Closeup of 1C11-fusion loop interaction with key side chains of 1C11 (blue) and GP (grey) illustrated. GP residues that are conserved among five ebolaviruses (EBOV, SUDV, BDBV, RAFV, and RESTV) are underlined. (B) 1C11 draws the fusion loop away from the core of GP upon binding. The conformation of the fusion loop in an unbound GP (PDB:5JQ3) is shown in green, and the 1C11-bound conformation is shown in black. (C) The N-linked glycan at position 563 and the 1C11 complementarity determining region (CDR) side chains that interact with it are illustrated. Side chains within hydrogen bonding range to the glycan are indicated by dashed yellow lines. (D) 1C3 and the head region of EBOV GP are shown in closeup, including the long CDRL1 loop of 1C3 which extends deep into the EBOV GP “chalice”. (E) The footprints of 1C3 and 1C11 are highlighted on the sequence alignment of five major ebolaviruses. Residues were labeled by colored triangles at the bottom of the alignment to show the conservation (blue - conserved, pink - similar, yellow - non-conserved). The footprint of 1C3 contains five non-conserved residues, and the footprint of 1C11 contains only conserved or highly similar residues. (F) On the left, EBOV GP surfaces that directly contact 1C3 are shown in various shades of orange as follows: 1C3-bound residues in common with all three GP protomers are colored in dark orange, residues bound by only two protomers (A and B) are colored in medium orange, and residues unique to a single protomer are colored in light orange and yellow. On the right, the three separate portions of the tripartite 1C3 footprint on the GP protomers A, B, and C are illustrated. For example, residues 114–120 on protomer A are bound by CDRs H2 and H3, while the same residues 114–210 on protomer B are bound by 1C3 CDRL2 and framework region 3. CDRL1 of 1C3 simultaneously contacts residues 124–126 on both protomers A and B.
The other antibody, 1C3, instead binds to the top of GP, near the RBS within the GP1 “head” domain (Figure 3D). Other head-binding antibodies have been described (Cohen-Dvashi et al., 2020; Corti et al., 2016), including the EBOV-specific therapeutic mAb 114. In contrast to the 3:3 stoichiometry of the mAb 114-GP complex (Misasi et al., 2016), 1C3 has a unique binding stoichiometry. Our cryo-EM structure reveals that only one 1C3 Fab anchors to the GP trimer, simultaneously contacting all three GP1 monomers in the center of the GP chalice (Figure 2, Figure 3D). The single copy of 1C3 buries ≈470 Å 2 on the first GP protomer in the trimer, ≈380 Å 2 on the second, and ≈230 Å 2 on the third.
Some of the same residues are bound by 1C3 across the three GP protomers, whereas others differ. Residues 115, 117–120, 124, and 172 of all three GP protomers are bound by 1C3 (Figure 3F). For example, residues 117–120 on protomer A are bound by 1C3 CDRH2 and CDRH3, whereas the same residues on protomer B are bound by 1C3 CDRL2 and framework region (FR) L3 and on protomer C by 1C3 FRL1. Residues K114, P116, A125, and P126 are bound by 1C3 in two of the three GP protomers A and B. GP residues 125–126 are bound by residues 31–33 in CDRL1 in GP protomer A, and by residue N35 in CDRL1 in GP protomer B. In protomer A alone, residues D127 and G128 are bound by CDRL1 and residues G143 and T144 are contacted by CDRH2. In protomer B alone, residue D150 is bound by CDRL1, and residue F151 is bound by FRL3. Interestingly, the buried surface on GP is contributed by 33.3% heavy chain and 66.7% light chain. The majority contribution by the light chain contact is unusual for a human antibody, and likely results from the longer, 11-residue CDRL1 that extends into the bottom of the GP “chalice” to interact with two GP protomers A and B in its path. The 1C3 light chain has 86% identity with the IGKV 4–1 germline. CDR somatic mutations in L1 and L2 include Q27H, N37D, and Y38F. Framework changes include M4L, V13A, N22S, L39F, Y55S, D74V, S77N, S79G, and D86H.
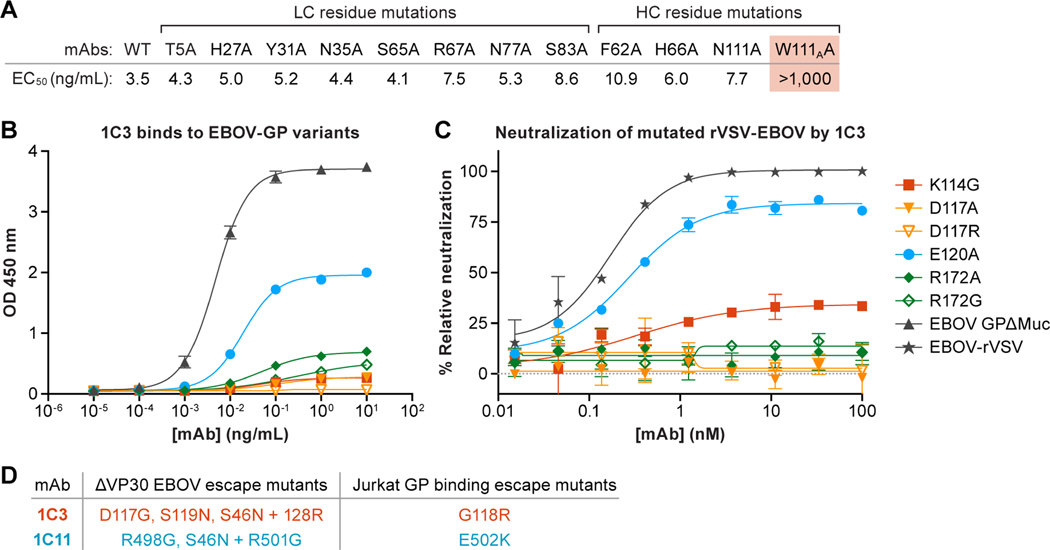
We performed mutagenesis of the antibody CDRs to determine which amino-acid residues were key for 1C3 interaction. Based on contacts observed in the cryo-EM structure, eight amino-acid residue changes in the CDRs and four changes in FRs were introduced and evaluated for binding to GP (Figure 4A). Only one of the single amino-acid residue changes, however, W111AA in CDRH3, substantially affects binding to GP. The side-chain nitrogen of W111A forms a hydrogen bond with the backbone carbonyl of D117 on GP1, though the majority of the W111A side chain is packed into a hydrophobic pocket of the antibody itself. Thus, the W111A change may alter the conformation of CDRH3, as well as removing the hydrogen bond. The observation that the majority of the single amino-acid residue changes do not affect binding suggests the involvement of multiple CDRs in the interaction; total binding energy could be distributed more diffusely across each of the three different CDR–GP1 binding interfaces.
Figure 4. Binding and neutralization analysis of monoclonal antibodies (mAbs) 1C3.The IMGT numbering scheme is used.
(A) Amino-acid residue changes in 1C3. Only one change in 1C3, W111A, compromised tight binding of Ebola virus (EBOV) glycoprotein (GP). All other single amino-acid residue changes could be accommodated, presumably by other key contacts remaining. (B and C) Changes in GP residues. (B) Point changes in GP at 3 of 4 key residues result in reduced binding by 1C3, presumably because these single amino acids form more than one contact point each in the tripartite epitope. (C) Point changes at all four key residues result in reduced neutralization of recombinant vesicular stomatitis virus expressing EBOV GP (rVSV-EBOV) by 1C3. (D) Viral escape mutants from 1C3 and 1C11. Left column: Mutations identified in plaque-purified ΔVP30 Ebola virus isolates grown in the presence of 1C3 or 1C11. Right column: Mutations identified in Jurkat cell lines expressing randomly mutated EBOV GP that were selected for loss of binding to 1C3 or 1C11 by FACS sorting.
We next changed contact sites in the GP1 receptor-binding head. GP1 residues D117, E120, and R172 exist in all three contact sites in the three-protomer, single-Fab footprint (Figure 3F). Residues K114, P116, and P126 are located in the footprints of two protomers (here designated A and B). Residue D127 exists only in the footprint on protomer A, whereas residue P123 exists only in the footprint on protomer B. Identified amino-acid residues of interest were evaluated for both binding and neutralization of recombinant vesicular stomatitis virus (rVSV) expressing EBOV GP containing individual amino-acid residue changes (Figure 4B–C). We observe that mutations of K114, D117, E120, and R172 affect the activity of 1C3. Specifically, 1C3 loses neutralizing activity against rVSV expressing GP D117A, D117R, R172A, and R172G. 1C3 achieves only 30% neutralization of rVSV bearing K114G and 80% neutralization of rVSV bearing E120A (compared to wild-type rVSV). P116G, P123G, P126G, or D127A change does not affect neutralization by 1C3 (Figure S4A). Binding of insect cell-expressed GP variants is affected in a similar manner (Figure 4B). The changes of key residues that are located at the center of 1C3 footprint and are contacted by 1C3 on two or three protomers are more likely to affect the mAb’s neutralization (Figure S4B). When tested against Jurkat cell lines expressing EBOV GP variants, 1C3 loses affinity to GP bearing the previously described amino-acid residue change G118E (Figure S1D). These results indicate that residues presented to 1C3 by two or three GP protomers are critical for 1C3 activity. Simultaneous anchoring to and blockage of all three NPC1 binding sites on GP suggests that 1C3 likely neutralizes EBOV by preventing receptor binding (Figure S5).
As further confirmation of these results, we examined viral escape from 1C3 and 1C11 using a biologically-contained EBOV replication system (ΔVP30 EBOV) (Halfmann et al., 2008). We found that culture of virus in the presence of subneutralizing amounts of 1C3 leads to escape mutants with substitutions at positions 117, 119, or 128 (Figure 4D). Similarly, when we sorted Jurkat cells expressing a library of mutant EBOV GP proteins for mutants that lost binding to 1C3 but maintained high expression, the cell lines we obtained all had point mutations at position 118 (G118R). Importantly, when we selected for 1C11 escape mutations, these localized to a separate area of GP (residues in or near the furin cleavage site at positions 497–501; Figure 4D). As expected, escape mutants for one antibody are fully neutralized by the other, suggesting that a cocktail of 1C3 and 1C11 would provide coverage for viral escape.
Protective efficacy of mAbs 1C3 and 1C11, individually and in combination, in rodents
The broad specificity and neutralization capacity of 1C3 and 1C11, coupled with their asymmetric binding footprints and likely mechanisms of complementary interaction suggested that they could together serve as an effective therapeutic against EBOV and SUDV. We first evaluated the protective efficacy of these antibodies individually in the laboratory mouse model of EVD. Mice were treated with 1C3 or 1C11 24 hours prior to exposure via the intraperitoneal (IP) route with 100 plaque-forming units (PFU) of mouse-adapted EBOV. We chose pre-treatment in the mouse model to confirm that the mAbs had in vivo protective efficacy before moving to more stringent post-infection treatment regimens in larger animal models. A human IgG1 antibody (specific for influenza A virus) was used as a control. In mice, 1C3 and 1C11 provided 90–100% protection from death when administered at a dose of 25 μg or higher, as compared to 10% survival in animals given no treatment or given a control (Figure 5A).
Figure 5. Protective efficacy of human monoclonal antibodies (mAbs) against lethal ebolavirus infection in laboratory mice and domesticated guinea pigs.
(A) Survival of mice treated with 1C3 or 1C11 prior to Ebola virus (EBOV) exposure. mAbs were administered 24 h prior to exposure with 100 plaque-forming units (PFU) of mouse-adapted EBOV. A human immunoglobulin G against influenza A virus (IgG1) mAb was used as a control. Animal survival was assessed twice daily for 21 d. n = 10 mice were studied per treatment condition. (B) Survival of STAT1 KO mice treated with 1C3 or 1C11 after EBOV/BDBV-GP exposure. Groups of STAT1 KO mice at five animals per group were injected with the indicated mAbs by the intraperitoneal route at 24 h after BDBV chimeric virus challenge. Kaplan-Meier survival curve is shown. Each group was compared with phosphate-buffered saline (PBS) control (Mantel-Cox test). (C and D) Groups of guinea pigs at five animals per group were injected with indicated mAbs by the intraperitoneal route at 1 or 3 days after EBOV (C) or Sudan virus (SUDV) (D) exposure. Kaplan-Meier survival curves, body weights, illness score curves, and viremia levels are shown. For analysis of survival, each group was compared to PBS mock control (Mantel-Cox test). In panels representing viremia, each dot corresponds to an individual serum sample. Short horizontal lines indicate the mean value of titers. The dotted horizontal lines show the detection limit. For analysis of viremia data, serum samples collected on different days were pooled together in each experimental group. Samples without detectable virus were arbitrarily assigned the viremia level values corresponding to the detection limit (102 PFU/mL). One-way analysis of variance (ANOVA) with Dunnett correction was used for multiple comparisons between each group and PBS mock control.
Antibodies were next tested for protection against BDBV using the replication-competent fully infectious chimeric virus EBOV/BDBV-GP which have the envelope represented by BDBV GP and the rest of the proteins from EBOV (Ilinykh et al., 2018). STAT-1 knockout mice were exposed to 1,000 PFU of EBOV/BDBV-GP via IP and 24 hours later treated with 0.5 mg 1C3, 0.5 mg 1C11 or the 1C3/1C11 cocktail (0.25 mg each; 0.5 mg total). All 1C3-treated animals died or were found moribund and were euthanized. 1C11 alone conferred 80% protection, and the 1C3/1C11 cocktail provided 100% protection from death (Figure 5B, and Figure S6).
The antibodies were further tested against EBOV in the domesticated guinea pig model of EVD, which is considered a stringent model of EVD. Animals were exposed IP with 10,000 PFU of guinea-pig-adapted (GPA) EBOV (GPA-EBOV) and then treated IP with 1C3, 1C11, or a combination of the two mAbs (1C3 + 1C11; 1:1 ratio) on day 1 or day 3 post-exposure. Neither mAb individually resulted in sufficient protection, but the combination of both antibodies administered on day 3 protected 4 out of 5 animals (80%) against death. Further, the administration of the combination resulted in absence of viremia by day 6, whereas animals given individual mAbs or no treatment had peak viremia on day 6 (Figure 5B). Administration of the cocktail at day 1 was substantially less protective than treatment at day 3, suggesting that having high mAb concentrations during the late stages of infection may be key to protection in this model.
The antibodies were also tested against SUDV in domesticated guinea pigs. Animals were exposed IP with 4,000 PFU GPA-SUDV. 1C3 alone was able to protect 4 out of 5 guinea pigs (80%) from death when delivered IP on Day 3 post-exposure at 10 mg 1C3 per animal. In contrast, only marginal (20%) protection against SUDV was achieved with 1C11. Treatment of animals with 1C3 resulted in lower SUDV titers compared to those treated with 1C11; no significant difference was observed between 1C11 and phosphate-buffered saline (PBS) control groups (Figure 5C). MAb 1C3 was slightly less potent at neutralizing SUDV in vitro than 1C11 (Fig. 1), yet it showed more protective efficacy in vivo. This suggests that factors besides viral neutralization (e.g. recruitment of Fc effector functions) may be important for protection.
Protective efficacy of combinatorial mAb 1C3 and 1C11 treatment in nonhuman primates
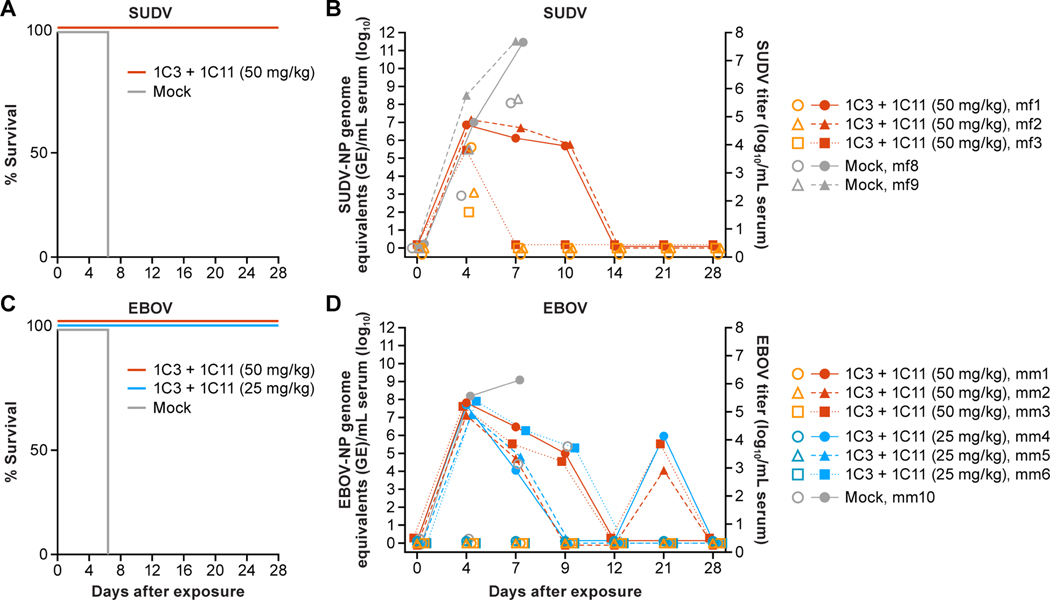
We next evaluated the ability of the combination of 1C3 and 1C11 to protect against disease in nonhuman primate (NHP) models of EVD and Sudan virus disease (SVD). The combination was first tested in the SUDV (Gulu variant) model in crab-eating macaques. On Day 4 or Day 7 post-exposure, a dose of 50 mg/kg (25 mg/kg of each antibody) was administered intravenously. All three treated NHPs survived (Figure 6A) and had minor clinical signs during the expected peak of disease. SUDV titers ranged from 2–3.81 log10 per mL of serum on Day 4 and just prior to administration of the first dose. However, none of these NHPs had detectable virus on Day 7 and prior to treatment with the second dose of mAb combination. In contrast, 8.33 and 7.78 log10 of SUDV were detected terminally in the sera of the untreated control NHPs on Day 7 and Day 8, respectively. Complete clearance of SUDV viremia was achieved in treated NHPs by Day 14 as determined by the absence of any detectable SUDV genome as measured by real-time reverse transcription polymerase chain reaction (RT-qPCR) in sera (Figure 6C).
Figure 6. Efficacy evaluation of 1C3+1C11 combination in lethal nonhuman primate (NHP) models of Sudan virus disease and Ebola virus disease (EVD).
(A) Complete survival of crab-eating macaques (n=3) exposed to Sudan virus (SUDV) following treatment with 50 mg/kg of 1C3+1C11 on Day 4 or Day 7 post-exposure, as compared to mock-treated control nonhuman primates (NHPs; n=2). (C) Complete survival of rhesus monkeys treated with 50 mg/kg or 25 mg/kg of 1C3+1C11 on Day 4 or Day 7 following EBOV exposure as compared to a mock-treated NHP (n=1) (C). (B and D) Infectious titers (log10 per mL serum) and viral genome equivalents (GE) per mL serum are shown over time for each animal following SUDV (C) and EBOV (D) exposure.
The mAb combination was next tested against EBOV in rhesus monkeys at both high (50 mg/kg; 25 mg/kg of each antibody) and low (25 mg/kg; 12.5 mg/kg of each antibody) doses. Protection from severe disease and death was achieved at both doses in treated NHPs as evidenced by mild clinical signs (temporary inappetence, slightly diminished activity), and 100% survival, whereas the mock (PBS)-treated exposed control animal succumbed to acute disease on Day 8 (Figure 6B). Treated NHPs in both high-dose and low-dose combination groups presented with similarly mild clinical signs and subtle shifts in clinical pathology blood parameters, making the two treatment groups (high-dose and low-dose) phenotypically indistinguishable. Although all treated NHPs cleared EBOV viremia by Day 12 as determined by RT-qPCR, a small amount of RNA remained detectable on Day 21 (Figure 6D).
DISCUSSION
The unpredictable timing, location, and causative viruses of Ebola disease outbreaks means that unvaccinated populations could become infected. Additionally, it is possible to become infected with an ebolavirus against which available vaccines do not offer protection. Antibody therapies have been evaluated as medical countermeasures for unvaccinated people, but all of them are EBOV-specific. Further, all antibody treatments thus far clinically evaluated include components that cross-react to the abundantly shed form of the viral GP (sGP), with some of the mAbs having higher affinity for sGP over the viral-surface GP that would be the target for neutralization (Pallesen et al., 2016). As sGP is believed to act in part as an immune decoy, the ability of an mAb to discriminate between sGP and full-length GP has traditionally been considered an asset for a therapeutic candidate.
Finally, mAb114 and REGN-EB3, are offered at a 50 mg/kg and 150 mg/kg dose, respectively (Food and Drug Administration, 2020a, 2020b). A recently published investigational mAb cocktail including antibodies EBOV-442 and EBOV-515 protected against EBOV in NHPs at a 30 mg/kg total dose (20 mg/kg of EBOV-515 and 10 mg/kg of EBOV-442) (Gilchuk et al., 2021). mAbs 1C3 and 1C11, which were protective in NHPs against EBOV at 12.5 mg/kg each. mAb114 consists of a single monoclonal antibody administered once intravenously, but it is not effective against SUDV. Treatment with REGN-EB3 requires three doses of 150 mg/kg consisting of atoltivimab, maftivimab and odesivimab (50 mg/kg of each monoclonal antibody) intravenously. High quantities of therapeutic mAbs are more expensive and may pose logistical challenges with regard to production (higher costs) and supply chain (number of vials required). Furthermore, any long-term adverse effects caused by high concentrations of circulating exogenous mAbs remain unknown, including the possibility of enhanced selection of viral escape mutants and impact on endogenous immune responses. mAbs 1C3 and 1C11 were protective in NHPs against SUDV in two doses at 25 mg/kg each. This was the lowest dose tested in this study. The mAb cocktail containing EBOV-442 and EBOV-515 protected NHPs against SUDV in two doses at a 30 mg/kg total dose (a 2:1 mixture of EBOV-515 and EBOV-442) (Gilchuk et al., 2021). The outcome of this NHP study suggests that protection of NHPs from EBOV- and SUDV-induced disease may be conferred at even lower dosages of the 1C3 and 1C11 combination evaluated here, and may potentially protect against disease using a single administration. Future studies are geared towards answering these questions and include the longitudinal assessment of immune responses of treated NHPs long after recovery.
Previous broad analyses of individual antibodies against ebolaviruses indicated that GP-specific antibodies against the core and head of GP could provide synergistic activities in a therapeutic combination (Gilchuk et al., 2020; Pascal et al., 2018; Saphire et al., 2018). Among antibodies against the head, nearly all cross-react with sGP; identification of neutralizing antibodies against the head that do not cross-react has been a formidable challenge.
In this study, we analyzed a pool of human mAbs from EVD survivors and identified two antibodies, 1C11 and 1C3, that bound the desired core and head epitopes, respectively, were specific for GP, did not cross-react with sGP, offered potent neutralizing activity in vitro against SUDV as well as EBOV, and were efficacious in both laboratory mouse and domesticated guinea pig models of EVD. We next determined that a combination formed by the 1C11–1C3 pair could protect NHPs against EBOV- and SUDV-induced disease, in doses as low as 25 mg/kg (12.5 mg/kg of each mAb).
To determine the mode of recognition by this antibody combination, we determined the cryo-EM structure of the ternary 1C3–1C11–EBOV-GP complex. The cryo-EM structure reveals that 1C11 anchors to the fusion loop, recognizing residues conserved among ebolaviruses and, in a 3:3 stoichiometry, with each copy of 1C11 bound to one of the GP protomers in the trimer, anchoring adjacent protomers together. Surprisingly, the other antibody, 1C3, binds in a unique format: recognition of the GP1 head in a 1:3 stoichiometry, with one single 1C3 Fab simultaneously in contact with all three protomers in the GP trimer. Residues from all three copies of the GP protomer together comprise the antibody footprint, with key contact residues spread across the tripartite antibody footprint.
The requirement of all three GP protomers to be assembled together in their natural oligomeric arrangement strongly indicates that 1C3 was elicited by GP, and not the abundantly shed sGP. The asymmetric binding mode of a single 1C3 to a GP trimer is rare and not previously noted among antibodies against EBOV or other trimeric GPs. As previously described, FVM04, an antibody against EBOV GP, also appears to bind as a single antibody to the GP1 head, as visualized on low-resolution two-dimensional EM imaging (Howell et al., 2016). However, the footprint of FVM04 appears contained on a single protomer, and FVM04 also cross-reacts with sGP. It is the angle of FVM04’s approach to GP that sterically inhibits other copies of FVM04 from binding. In contrast, 1C3 does not bind to sGP. Hence, the single-protomer portion of its tripartite footprint is insufficient for recognition.
A potently neutralizing antibody against HIV gp120, PG9, has also been described (Walker et al., 2009). A single copy of PG9 binds the viral glycoprotein apex and contacts two gp120 protomers (McLellan et al., 2011). Although three possible PG9 epitopes exist on gp120 (between protomers A-B, B-C, and C-A), binding of one copy appears to sterically block the other sites (Julien et al., 2013). PGT145 and CAP256-VRC25.26, like 1C3, target the HIV-1 trimer with one antibody bound per trimer (Gorman et al., 2020; Lee et al., 2017; Liu et al., 2017). PG9, PGT145 and CAP256-VRC25.26 each have unusually long CDR3s (> 28 residues) that are likely required to contact the protein surface through the extensive glycosylation of HIV-1 gp120 apex, although contact with glycan moieties is also involved in their trimer recognition. The EBOV GP apex has fewer glycans relative to HIV-1 gp120 and consequently 1C3 has a CDRH3 that is only 17 amino acids long. Similarly antibody PIA174 also targets the parainfluenza virus III (PIV3) trimer with a stoichiometry of one antibody per spike (Stewart-Jones et al., 2018). PIA174 also has a 17 amino acid CDRH3.
Uniquely for 1C3, glycoprotein recognition is achieved through greater involvement of other CDRs including those from the light chain. The HIV mAbs that have 1:1 Fab:trimer stoichiometry like 1C3 appear to have higher potency (Gorman et al., 2020; Lee et al., 2017; Liu et al., 2017). Such correlation further suggests the advantage of including the asymmetrical head binder 1C3 as part of a therapeutic cocktail. Among the HIV-neutralizing mAbs analyzed thus far CAP256-VRC25.26 and PGT151, aV2-apex and fusion-peptide binder, respectively, have the highest potency (Gorman et al., 2020; Lee et al., 2017; Liu et al., 2017). It would be interesting to see if this pair would be an effective therapeutic cocktail.
In summary, we show that mAbs 1C3 and 1C11 are highly protective against severe disease and death due to EBOV and SUDV infections in NHPs and confer protection through a unique recognition of quaternary epitopes of the ebolavirus GP. Effective EBOV-specific mAb-based therapeutics are now standard of care after the PALM RCT; however, the PALM results sounded a note of caution: in patients with high viral loads, case-fatality rates remained > 60% even with these effective agents (Mulangu et al., 2019). The unique binding footprint, absence of binding to sGP, and potential for dose-reduction make this 1C3/1C11 combination an attractive developmental strategy for consideration in improving outcomes for EVD patients. Furthermore, the ebolaviral breadth provided by the combination may simplify preparedness, therapeutic pre-positioning, and agile public health and clinical response strategies in regions at risk for either EBOV or SUDV outbreaks.
Limitations of the Study
One potential limitation of our study is the relatively small numbers of NHPs per group as part of this pilot. However, we would be amiss not to point out that the evaluation of two dosages of the 1C3 and 1C11 combination against EBOV results in a total n=6 NHPs combined for the efficacy evaluation of the 1C3 and 1C11 combination resulting in complete survival overall. Future studies will include additional NHPs for testing of the dosages described herein, and evaluation of single and lower dosages of the combination
MATERIALS AND METHODS
RESOURCE AVAILABILITY
Lead contact and Materials availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Erica Ollmann Saphire (erica@lji.org).
Data and code availability
Coordinates and structure factors of 1C3 Fab have been deposited in the Protein Data Bank under accession number 7N6P. Cryo-EM map of the 1C3 and 1C11 bound EBOV GP trimer complex has been deposited to the EMDB with accession codes EMD-25471. Fitted coordinates have been deposited to the Protein Data Bank with accession codes 7SWD. Other data are available from the corresponding author upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Samples from human subjects
Four survivors of the 2013–2016 Western African Ebola virus (EBOV) disease (EVD) outbreak who were treated at Emory University Hospital, Atlanta, Georgia, USA, were enrolled in a long-term study of their immune responses following hospital discharge (Davis et al., 2019). All 4 subjects gave informed consent, and all studies described here were approved by Emory University’s human subjects committee (Institutional Review Board protocol #IRB00076700). Patients provided blood samples starting at the time of their discharge from the hospital, and at approximately 1, 3, 6, 12, 18, and 24 months thereafter. Peripheral blood mononuclear cells (PBMC) were isolated from these samples and stored over liquid nitrogen. All work with these blood samples was performed under biosafety level 2 or higher (BSL-2+) conditions. The mAbs described in this study were isolated from two donors, EVD5 and EVD9, from blood samples drawn 24 or 20 months post hospital discharge, respectively, hence mAbs derived from these samples are named 5.24.XXX or 9.20.XXX.
Cell lines
Vero (CCL-81), Vero E6 (CRL-1586) and Jurkat (clone E6–1) cells were obtained from ATCC. Vero cells were cultured in high-glucose Dulbecco’s modified Eagle’s medium containing L-glutamine (DMEM, Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FSC, Omega Scientific, Tarzana, CA) and 1% penicillin-streptomycin solution. Cells were maintained at 37°C in a humidified atmosphere with 5% CO2. Vero E6 cells (green monkey kidney epithelial) were maintained at 5% CO2 at 37°C either in Eagle’s minimal essential medium supplemented with L-glutamine, Penicillin/streptomycin, non-essential amino acids and 10% FCS, or in Dulbecco’s Modified Eagle Medium supplemented with glucose, L-glutamine, sodium pyruvate and 10% heat inactivated, sterile-filtered FCS. Jurkat E6–1 cells (human acute T cell leukemia; male) were maintained in 5%CO2 at 37°C in RPMI-1640 medium supplemented with 100 U/ml Penicillin/Streptomycin and 2 mM L-glutamine (Gibco), and 10% heat inactivated fetal calf serum (Hyclone).Drosophila S2, ExpiCHO-S and Expi293F cell lines were obtained from ThermoFisher Scientific. Drosophila S2 cells were cultured in Schneider’s Drosophlia medium at 27°C in stationary flasks. Stable cell lines were adapted to serum free conditions and maintained with shaking at 27°C. ExpiCHO cells were cultured in ExpiCHO expression medium and maintained at 37°C in a humidified atmosphere with 8% CO2. Expi293F cells (human embryonic kidney epithelial; female) were maintained on orbital shakers at 8% CO2 at 37°C in Expi293 medium (Gibco).
Viruses
In the EBOV mouse challenge study, Bray stock passed once on Vero E6 cells [Mp3, Vp2, Mp9, ppGH, and Vp1] (Bray et al., 1999) was used. Chimeric EBOV/BDBV-GP (GenBank: MH464888) (Ilinykh et al., 2018) was used in mouse protection studies against BDBV in STAT1 KO mice. Guinea pig-adapted EBOV/Mayinga (EBOV-GA) (Cross et al., 2015) and guinea pig-adapted SUDV/Boniface (SUDV-GA) (GenBank: KT878488) (Wong et al., 2015) were used in the domesticated guinea pig model studies of EBOV disease and SUDV disease. Ebola virus/H. sapiens-tc/COD/1995/Kikwit-9510621 (EBOV; NR-50306, Lot 9510621, BEI Resources, USA) was used in NHP challenge studies of EBOV disease. SUDV (strain Gulu) originated from a 35-yearr-old male patient who had died on 16 October 2000 was used in NHP challenge studies of SUDV disease.
Mouse model
Female BALB/c mice, aged 6 to 8 weeks, were purchased from Charles River Laboratory. Five-to seven-week-old STAT1 knockout female mice were acquired from Taconic Biosciences. The animal experiments were performed at Galveston National Laboratory ABSL-4 facility in accordance with NIH guidelines, the Animal Welfare Act, and US federal law and approved by the Institutional Animal Care and Use Committee.
Guinea pig model
Five-to-six week-old female Hartley strain domesticated guinea pigs (Cavia porcellus (Linnaeus, 1758)) were obtained from Charles River Laboratories. Exposure studies were conducted under maximum containment in an animal biosafety level 4 (ABSL-4) facility of the Galveston National Laboratory, University of Texas Medical Branch.
Nonhuman primate (NHP) model
Seven Chinese rhesus monkeys (Macaca mulatta (Zimmermann, 1780)) of both sexes, ages 4–6 years, weighing 4.0–5.5 kg were acquired through WorldWide Primates, Inc. Eight healthy, adult crab-eating (aka cynomolgus) macaques (Macaca fascicularis Raffles, 1821) of Chinese origin (4 female, 4 male) were used in the SUDV challenge study.
METHOD DETAILS
Isolation, production, and initial characterization of human monoclonal antibodies (mAbs)
To identify EBOV/SUDV cross-reactive memory B cells from survivor blood samples, 60 million PBMC were thawed and stained with lymphocyte makers as well as labelled EBOV and Sudan virus (SUDV) glycoprotein (GP) ectodomains. EBOV GP ectodomain was produced as described (Hashiguchi et al., 2015). BDBV GP ectodomain (Cat #0505–015) and SUDV GP ectodomain (Cat #0502–015) were obtained from IBT Bioservices, Rockville, MD, USA. EBOV GP was labelled using the Alexa 488 protein labeling kit (A10235, ThermoFisher, Waltham, MA, USA) and SUDV GP was labelled using the Alexa 647 protein labelling kit (A20173, ThermoFisher). Sorts were performed on a FACS Aria II instrument under BSL2 conditions. EBOV GP+, SUDV GP+ cells were identified within the following gate: live/dead negative, singlets, CD3−, CD20+, CD19+, IgD−. mAbs used for FACS were as previously described (Davis et al., 2019). Single cells were sorted into PCR plates and antibody variable gene segments were amplified by real-time reverse transcription polymerase chain reaction (RT-qPCR) using a template-switching rapid amplification of complementary DNA (cDNA) ends (RACE) approach (Davis et al., manuscript in preparation). Gene segments were cloned into AbVec6W vectors ((Davis et al., 2019); modified from plasmids described by (Tiller et al., 2008)) and expressed in expi293 cells. MAb supernatants were screened initially by enzyme-linked immunosorbent assay (ELISA) on MaxiSorp plates coated with 30 nanograms per well of the following capture reagents: total immunoglobulin G (IgG) capture antibody (expression control), EBOV GP ectodomain, EBOV soluble glycoproteins (sGP), EBOV GPΔmucin (Lee et al., 2008), SUDV GP ectodomain (IBT Bioservices), or Bundibugyo virus (BDBV) GP ectodomain (IBT Bioservices). mAbs that screened positive for GP antigen binding were purified by protein A chromatography and evaluated for neutralizing activity. Two promising mAbs were isolated and named 5.24.1C11 (“1C11”) and 9.20.1C3 (“1C3”).
Ebolavirus glycoprotein variant ELISAs
Variants were produced via site-directed mutagenesis, using a plasmid encoding mucin-deleted EBOV GP, and transfected into Drosophila S2 cells (Thermo Fisher Scientific). Strep-tag purified, mucin-deleted EBOV GP variants were diluted in 0.1 M carbonate–bicarbonate buffer at a concentration of 4 μg/mL. High-binding 96-well ELISA plates (VWR) were coated with this solution and incubated at ambient temperature for 1 h, followed by blocking with 3% bovine serum albumin (BSA) (Sigma Aldrich) diluted in phosphate-buffered saline (PBS) overnight. Serial dilutions of mAbs were applied to the wells and incubated at ambient temperature for 1 h. The antibodies were detected using a goat anti-human IgG Fc-conjugated secondary antibody diluted 1:5,000 (Thermo Fisher Scientific) and 1-Step Ultra TMB substrate (Thermo Fisher Scientific). Color development was monitored and quenched with 1 M sulfuric acid (Thermo Fisher Scientific). Absorbance was measured at 450 nm using a Tecan plate reader (Spark 10M Multimode Plate Reader). EC50 values were calculated using Prism 9.0 after using sigmoidal dose-response nonlinear regression analysis.
Human mAb-screening neutralization assay
Initial identification of neutralizing antibodies was performed in 96-well format using biologically-contained EbolaΔVP30 virus expressing Renilla luciferase, as previously described (Davis et al., 2019; Halfmann et al., 2008). EbolaΔVP30-RenLuc viruses expressing EBOV GP (H.sapiens-tc/GIN/2014/Gueckedou-C07), Sudan GP (H.sapiens-tc/Sudan/1976/Boniface-R4142L), or BDBV GP (H.sapiens-tc/UGA/2007/Butalya-811250) were incubated with 10 μg/mL of mAb for 2 h at 37°C and then inoculated onto Vero cells (ATCC, Manassas VA, USA) expressing EBOV VP30 (Halfmann et al., 2008). Luciferase expression, measured in relative light units (RLU), was measured 3 days later using EnduRen luciferase substrate (Promega), on a Tecan M1000 Pro plate reader. Neutralizing mAbs that reduced EBOV RLU levels >50% were considered neutralizing, and further screened using live virus.
Neutralization assays using live EBOV, SUDV, and BDBV
Neutralization of live viruses by mAbs was assessed by plaque assay under BSL-4 conditions as described (Davis et al., 2019; Honnold et al., 2014). Briefly, viruses were incubated with 2-fold serial dilutions of mAbs and added to Vero E6 cells (ATCC) in 6-well plates. The endpoint titer was determined to be the highest dilution with plaque reduction neutralization test (PRNT) producing a ≥50 or ≥80% reduction (PRNT50, PRNT80) in the number of plaques observed in control wells. EBOV Zaire 1995 (Kikwit strain) was used for neutralization testing. For SUDV, the Boniface strain was used. For BDBV, the Uganda 2007 strain was used.
Neutralization assay using recombinant vesicular stomatitis virus (rVSV)
Neutralization assay was performed with rVSV expressing enhanced green fluorescent protein (eGFP) and EBOV (Makona variant) GP (rVSV-EBOV), as previously described (Wong et al., 2010). Titrated virus was incubated with serially diluted mAbs at 37°C for 1 h before addition to confluent Vero cell (ATCC CCL-81) monolayers in 96-well plates (Thermo Fisher Scientific). Following infection, cells were incubated for 20 h at 37°C in 5% CO2. The cells were then fixed in 4% paraformaldehyde (PFA) (Electron Microscopy Sciences) and nuclei were stained with 2 μg/mL Hoechst 33342 stain (Thermo Fisher Scientific). Plates were imaged using a CellInsight CX5 imager (Thermo Fisher Scientific), and infection was quantified by automated enumeration of total cells and those expressing eGFP. Infection was normalized to the average number of cells infected with rVSV-EBOV without antibodies. Data are presented as the relative infection for each concentration of antibody.
When testing for mAb synergy by the mAb cocktail, the rVSV neutralization assay was performed using the constant ratio combination design based on pre-determined IC50 values of 1C3 and 1C11 (Chou, 2010). Data was analyzed with CompuSyn software to calculate the combination index (CI) (Chou, 2010). CI values were calculated for each tested concentration of the cocktail, and CI values < 1 were considered as evidence of synergy (CI=1 – additive effect; CI>1 – antagonism). Dose reduction index (DRI) was also calculated indicating the fold-reduction in effective doses for individual mAbs in the cocktail.
Competition assays
Competition binding assays were performed on Jurkat E6–1 cells (ATCC) expressing EBOV GP from strain H.sapiens-wt/GIN/2014/Kissidougou-C15 (Jurkat GP cells) (Davis et al., 2019) as described. Briefly, Jurkat GP cells were pre-incubated with 100 μg/mL unlabeled competitor mAb on ice, then test mAbs labeled with the Alexa 488 protein labelling kit (ThermoFisher) were added at 2 μg/mL. After washing, mAb binding to cells was assessed by flow cytometry. In Figure 1B, test mAb binding in the presence of competitor mAb is expressed as a percentage of the fluorescence signal observed in the absence of competitor. In Figure S1D, the percentage blocking of reference mAb binding by the competitor mAb is shown.
Binding to Jurkat GP variant cell lines
Jurkat cell lines expressing EBOV GP K115N, G118E, T144A, T144M, G224D, I527N, I532M, or D632N were previously described (Davis et al., 2019). Binding of the promising mAbs to these cell lines was measured by flow cytometry (for geometric mean channel fluorescence [GMCF]). Binding was normalized to the level of GP expression on each cell line using a reference non-neutralizing antibody specific for the mucin-like domain (MLD), 2.1.6F2 (Davis et al., 2019). Binding of the promising mAbs to each variant cell line was expressed as a percentage of the binding to wild-type EBOV GP by this formula: 100% * [(novel mAb binding to variant cell line)/(2.1.6F2 binding to variant cell line)] / [(novel mAb binding to wild-type Jurkat-GP cells)/(2.1.6F2 binding to wild-type JurkatGP cells)].
Identification of viral escape mutants
To generate escape mutants, GFP-expressing EbolaΔVP30 virus (~200 PFU) was incubated with 2-fold dilutions of mAb starting at 10 μg/mL at 37 °C for 1 hour. Vero VP30 cells in 96-well plates were incubated with the virus-antibody mixture and monitored for 10 days after infection. At this time, virus samples were harvested at the highest concentration of mAb (0.156 μg/ml) in which nearly all the cells were GFP positive (an indication of virus infection). Virus in these samples was passaged on Vero VP30 cells in presence of 0.625 μg/ml of mAb, four times the original antibody concentration. Cells were monitored again for virus infection by the visualization of GFP and harvested once nearly all the cells were GFP positive. Virus from this passage was diluted (10-fold) and incubated with a higher concentration of mAb (2.5 μg/ml) at 37 °C for 1 hour and then added to Vero VP30 cells previously seeded in 12-well tissue culture plates. After incubation for 1 hour, the cells were washed to remove unbound virus, and then overlaid with 1.25% methylcellulose medium (M0512, Sigma). Individual plaques (10–15) were picked 7–8 days after infection and amplified on Vero VP30 cells to generate stock viruses. Up to 15 individual plaque-picked escape mutant viruses were isolated and the GP was sequenced (Halfmann et al., 2008).
Identification of Jurkat GP binding escape mutants
Jurkat cells expressing randomly-mutagenized EBOV GP were generated as described (Davis et al., 2019). Briefly, EBOV GP (strain H.sapiens-wt/GIN/2014/Kissidougou-C15) expressed in a lentiviral vector was randomly mutagenized via error-prone rolling circle replication (Fujii et al., 2006) using Phi29 polymerase (25640010, Cytiva, Marlborough, MA, USA) in the presence of manganese. Jurkat E6–1 cells were infected with this lentiviral stock at low MOI to ensure a single lentiviral integration event per cell. GP-expressing cells were sorted on a FACS Aria II for loss of binding to either Alexa 488 or Alexa 647 labelled 1C3 or 1C11 while retaining binding to Alexa 405 labelled 2.1.6F2 (Alexa 405 NHS Ester, A30000, ThermoFisher). Cells were then single cell cloned using ClonaCell TCS medium (03814, Stemcell Technologies, Vancouver, CA). Individual clones were analyzed by flow cytometry to confirm loss of binding to either 1C3 or 1C11 and retention of binding to 2.1.6F2 and GP was sequenced from RNA isolated from each Jurkat GP clone. Each escape mutation was identified in two separate sorts from at least 3 individual cell lines.
Protein structure determination
A 1C11 single-chain variable fragment (scFv) was ordered from Genscript; this plasmid contained the appropriate heavy-chain (VH domain), (G4S)4 linker, light-chain (VL domain), and C-terminal Strep-tag sequences. The DNA was cloned into a pMT-puro vector, and the protein was expressed in Drosophila S2 cells. Protein was purified from the culture supernatant using a pre-packed StrepTrap HP column (Cytiva Life Sciences) followed by cleavage of the Strep-tag at an enterokinase cleavage site using EKMax (Thermo Fisher Scientific). The tagless protein was further purified using a Superdex 75 Increase 10/300 GL size-exclusion chromatography (SEC) column (Cytiva Life Sciences).
For structure determination, recombinant 1C3 IgG was expressed in Chinese hamster (Cricetulus griseus) ovary (CHO) cells (Thermo Fisher Scientific) by co-transfecting heavy-chain and light-chain-encoding expression vectors. The protein was purified from the culture supernatant using a pre-packed HiTrap Protein A HP column (Cytiva Life Sciences). Purified IgG was digested to antigen-binding fragment (Fab) by incubating with 2% papain (Thermo Fisher Scientific) for 4 h at 37°C, followed by reaction quenching with 50mM iodoacetamide (Sigma). The Fab was purified from the reaction mixture using a Mono Q 5/50 GL column (Cytiva Life Sciences) followed by further purification using a Superdex 75 Increase 10/300 GL SEC column (Cytiva Life Sciences). 1C3 Fab was screened for crystallization using a Douglas Instruments Oryx8, and the protein crystallized in a solution of 100 mM CHES/sodium hydroxide pH 9.5, 40% polyethylene glycol (PEG) 600 at 12 mg/mL. Diffraction data to 2.15-Å resolution were collected at beamline 12–2 at the Stanford Synchrotron Radiation Lightsource, and the structure was solved by molecular replacement using a homology model made with SWISS-MODEL (Waterhouse et al., 2018) as a search model. Two Fab molecules were contained in the asymmetric unit of the P212121 crystals. Molecular replacement, model building, and structure refinement were carried out using the PHENIX suite of programs (Adams et al., 2010) and COOT (Emsley et al., 2010). The final model was validated using the MolProbity server (Williams et al., 2018).
EBOV GPΔmucin was expressed in Drosophila S2 cells using a single plasmid encoding a C-terminally Strep-tagged construct. EBOV GPΔmucin was purified using a pre-packed StrepTrap HP column (Cytiva Life Sciences), followed by cleavage of the Strep-tag at an enterokinase cleavage site, using EKMax (Thermo Fisher Scientific). The tagless protein was further purified using a Superose 6 Increase 10/300 GL SEC column (Cytiva Life Sciences).
A GP-1C11 scFv-1C3 Fab ternary complex was obtained by incubating EBOV GPΔmucin with a 3-fold molar excess of both 1C11 scFv and 1C3 Fab overnight followed by purification using a Superdex 200 Increase 10/300 GL SEC column (Cytiva Life Sciences). Specimens for cryogenic electron microscopy (cryo-EM) imaging were prepared by applying the complex solution to freshly plasma-cleaned Holey carbon C-flat 2/1 400 mesh copper grids (Electron Microscopy Sciences), followed by blotting and plunge-freezing into liquid ethane using a Vitrobot Mark IV (Thermo Fisher Scientific). 1uL of lauryl maltose neopentyl glycol (Anatrace) was mixed with 3uL of the sample just prior to grid application to improve angular distribution.
TEM images were collected automatically using EPU on a Titan Halo 300 kV electron microscope (Thermo Fisher Scientific) at a magnification of 96,000x with a Falcon 3EC camera (Thermo Fisher Scientific) at a dose rate of 0.7 e− / pixel • s for a total dose of ≈40 e− / A2. Beam-induced motion was corrected using MotionCor2 (Zheng et al., 2017), and contrast transfer function (CTF) parameters were estimated using Gctf (Zhang, 2016). Initial reference-free particle-picking, 2-dimensional (2D) classification, and 3D reconstruction were carried out using cisTEM (Grant et al., 2018). The resulting reconstruction was imported into Relion 3 (Zivanov et al., 2018) for template-based particle-picking, 2D classification, 3D classification, and 3D refinement. A forward scatter (FSC) cutoff of 0.143 was used for overall resolution determination. The crystal structures of EBOV GPΔmucin and 1C3 Fab, as well as a homology model of 1C11 scFv made using SWISS-MODEL (Waterhouse et al., 2018) , were docked into the reconstruction using Chimera (Pettersen et al., 2004), and the model was refined using PHENIX real space refine (Adams et al., 2010) and COOT (Emsley et al., 2010) with geometry restraints generated from the crystal structures/homology model. The final model was validated using MolProbity server (Williams et al., 2018).
Mouse protection studies, EBOV, USAMRIID
Groups of 10 laboratory mice were studied for each of the 3 treatment arms. 1C11, 1C3, or control (a human IgG1 antibody [specific for influenza A virus]) was administered via the intraperitoneal (IP) route 24 h prior to exposure. Mice were exposed as described (Davis et al., 2019) via the IP route with 100 plaque-forming units (PFU) of mouse-adapted EBOV. (This is the Bray stock passed once on Vero E6 cells [Mp3, Vp2, Mp9, ppGH, and Vp1] (Bray et al., 1999). Animals were monitored for 21 days post-exposure.
Mouse protection studies, BDBV in STAT1 KO Mice, UTMB
Prior to experimentation, animals were given at least one week to acclimatize to the Galveston National Laboratory ABSL-4 facility. Five- to seven-week-old STAT1 knockout female mice (Taconic Biosciences) were placed in the ABSL-4 facility in the Galveston National Laboratory. Groups of five animals were injected intraperitoneally with 1,000 PFU of the EBOV/BDBV-GP chimeric virus (Ilinykh et al., 2018) in 100 μL of PBS. Twenty-four hours later, animals were injected with mAbs by the intraperitoneal route using 0.5 mg in 100 μL of PBS per animal. Animals treated with 100 μL PBS served as controls. Animal observation procedure was performed as described (Ilinykh et al., 2018). The overall observation period lasted for 28 days.
Testing of antibody efficacy in the domesticated guinea pig model, UTMB
Groups of 5 animals were exposed IP with 10,000 PFU of GPA-EBOV or 4,000 PFU of GPA-SUDV in 0.1 mL of PBS. The single 1C3 or 1C11 antibodies (10 mg) or in combination (5 mg for each mAb) were delivered by the intraperitoneal route 1 or 3 days post-exposure in 1 mL of PBS. The control groups were treated with PBS. Blood collection was performed 3, 6, 9, 12, and 28 days after exposure to analyze viremia titer in the serum samples. Animals were monitored for disease signs, survival, and weight loss as described (Ilinykh et al., 2020). The overall observation period lasted for 28 days.
Analysis of viremia by plaque assay, domesticated guinea pig model
Virus titration was performed on serum samples collected from the guinea pigs in Vero E6 cells by plaque assay as described previously (Ilinykh et al., 2018). The duplicate 10-fold serial dilutions of sera were adsorbed to Vero E6 cell monolayers in 96-well plates for 1 hour, covered with 100 μL 0.45% methylcellulose (Sigma-Aldrich) overlay, and incubated at 37°C for 6 days. The overlay was removed, cell monolayers were fixed with formalin for 24 hours, washed 3 times with PBS, and blocked for 1 hour with 5% dry milk in PBS containing 0.1% TWEEN-20 (PBST). The plaques were immunostained with rabbit anti-GP primary antibody (IBT Bioservices) followed by goat anti-rabbit secondary antibody labeled with horseradish peroxidase (Southern Biotech). Both antibodies were diluted at 1:1,000 in 5% dry milk in PBST. Virus plaques were visualized by staining with the AEC substrate kit (Abcam).
Ethics and approvals
Experimental procedures involving nonhuman primates (NHPs) and infectious EBOV were conducted within the BSL-4 laboratory at the Integrated Research Facility at Fort Detrick (IRF-Frederick), National Institutes for Allergy and Infectious Diseases (NIAID), Division of Clinical Research (DCR), National Institutes of Health (NIH). The IRF-Frederick is accredited (000777) by the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC), approved for Laboratory Animal Welfare (D16–00602) by the Public Health Service (PHS), and registered (51-F-0016) with the United States Department of Agriculture (USDA). The study was approved by the NIAID DCR Animal Care and Use Committee (ACUC) and followed the recommendations provided in The Guide for the Care and Use of Laboratory Animals (National Research Council et al., 2010), the American Veterinary Medical Association (AVMA) guidelines for the euthanasia of animals and followed the Animal Research: Reporting of In Vivo Experiments (ARRIVE) guidelines 2.0: Updated guidelines for reporting animal research (Percie du Sert et al., 2020).
UTMB facilities used in this work are also accredited by the AAALAC and adhere to principles specified in the eighth edition of the Guide for the Care and Use of Laboratory Animals, National Research Council.The animal protocols for testing of mAbs in mice and guinea pigs were approved by the UTMB Institutional Animal Care and Use Committee (IACUC) in compliance with the Animal Welfare Act and other applicable federal statutes and regulations relating to animals and experiments involving animals.
NHP exposure and treatment, EBOV, IRF-Frederick
The protective efficacy of 1C3 and 1C11 (both anti-EBOV GP, Zalgen Labs, Germantown, MD, USA) was evaluated against EBOV in a small proof-of-concept study without a statistically powered analysis of small group differences. Health of animals was examined upon arrival at the IRF-Frederick and twice thereafter when sedated during baseline blood collections prior to virus exposure. All NHPs were deemed suitable for the study despite elevated leukocyte counts in 3 animals without evidence of apparent clinical disease. The NHPs were single-housed and acclimated within the ABSL-4 for 27 days prior to EBOV exposure. Water was offered ad libitum and NHPs were fed High Protein Monkey Diet (No. 5045, LabDiet, St. Louis, MO, USA) supplemented daily with fresh fruits and vegetables. There were no concomitant medications administered to NHPs after study initiation, except for offering an electrolyte solution (Gatorade) diluted equal parts with water.
Three NHPs each were randomly assigned to 2 experimental groups with stratification based on gender, age and body weight, and received combination of 1C3 and 1C11 at 50 mg/kg (25 mg/kg of each mAb) or 25 mg/kg (12.5 mg/kg of each mAb). One NHP served as a mock-treated control and received an equivalent volume of PBS. Study termination, originally scheduled at 28 days post-exposure, was extended to approximately 100 days to monitor the health status of survivors. All staff remained strictly blinded to the experimental grouping and treatment administered until completion of all post-life analyses to reduce bias.
Virus exposure
On Day 0, all NHPs were sedated via intramuscular (IM) injection of 15 mg/kg of Ketamine HCl (KetaThesia, Henry Schein, USA) using a randomized order. The skin above the right deltoid muscle group was clipped prior to needle injection of 0.5 mL containing a target dose of 1,000 PFU of Ebola virus/H. sapiens-tc/COD/1995/Kikwit-9510621 (EBOV; NR-50306, Lot 9510621, BEI Resources, USA) diluted in sterile phosphate buffered saline (PBS; Gibco, USA). The skin around the injection site was circled with a waterproof pen to facilitate cage-side inspection of the area. A dose of 3,550 PFU was determined based on the mean titer calculated from 6 replicate plaque assay titrations of the inoculum using both, a methylcellulose and crystal violet assay and an agarose and neutral red assay (Shurtleff et al., 2012), performed on the day of exposure of NHPs.
Antibody administration
Antibodies 1C3 and 1C11 were stored at −20°C until thawed on the day of administration. A fresh aliquot of the same lot was used for all NHPs of the same group and for both administrations. Antibodies were diluted in sterile PBS (Gibco, USA), aseptically drawn up in 20-mL syringes and capped until use. On Day 4 and Day 7, the NHPs received their respective antibody dose intravenously (IV) via a temporary 22-gauge catheter (Introcan Safety catheter, Braun, USA), aseptically placed in the right (Day 4) or left (Day 7) great saphenous vein and delivered slowly as a bolus using a syringe pump (Medex Medfusion 2010, Medex, Inc., USA) at a rate of 3 mL per min. The dosages for administration on Day 4 or Day 7 were calculated based on individual body weights obtained on Day 0 or Day 4, respectively. The total volumes of antibodies administered ranged from 5.9–18.4 mL. An equivalent, averaged volume of 11.2 mL of sterile PBS (Gibco, USA) was administered to the mock-treated control NHP. The administration time ranged from 3–6 min after which the IV catheter was removed and NHPs returned to their cage for recovery.
Observations, procedures, and endpoint criteria
All animals were given physical examinations, and blood was collected at the time of virus exposure and at 4, 7, 9, 12, 21, and 28 days after exposure. All NHPs were sedated via IM injection of 15 mg/kg of Ketamine HCl (KetaThesia, Henry Schein, USA), underwent a physical examination including measurement of body weight and temperature, and phlebotomy via venipuncture of the central vein. Serum separator and tubes containing K3 ethylenediamine tetraacetic acid (EDTA) or sodium citrate were utilized for blood collection (Vacuette; Greiner Bio-One, USA). Following virus exposure, animals were observed twice daily and scored for disease progression. Briefly, a score was assigned based on the NHP’s activity and responsiveness (0), slightly subdued (1), withdrawn (2), temporarily recumbent (3), or persistently recumbent (4). A score of 2 or higher triggered an increase in observations to 3 times a day. Euthanasia was required when NHPs scored either 4, or 3 and exhibited a rectal body temperature of equal or less than 34°C.
EBOV detection
Titers of EBOV were determined in sera using an Avicel-based crystal violet stain plaque assay on Vero E6 cell culture monolayers (ATCC CRL-1586)) with a limit of detection (LOD) of 100 PFU. Briefly, 10-fold dilutions of sera were adsorbed to Vero E6 cell monolayers in triplicate and covered with 2.5% Avicel overlay. After 7 days of incubation at 37°C and 5% CO2, a 0.2% crystal violet stain was added, and plaques were counted after 24 hours of incubation. Sera were inactivated in TRIzol LS according to the manufacturer’s instructions (Thermo Fisher Scientific, Waltham, MA, USA), removed from the BSL-4 and nucleic acid isolated using QIAamp Viral RNA Mini Kit (Qiagen, Germantown, MD, USA). Standard curve quantitation of EBOV nucleic acid was conducted using the BEI Resources Critical Reagents Program (CRP) EZ1 RT-PCR kit assay in accordance with manufacturer’s instructions (Trombley et al., 2010) and analyzed on an Applied Biosystems 7500 FastDx Real-Time PCR instrument (Thermo Fisher Scientific, Waltham, MA, USA). Results were transformed into log10 genome equivalents (GEq) per mL of sample with a lower limit of quantitation of 1 × 102 GEq per reaction.
Hematology, serum chemistry, and coagulation for EBOV-exposed NHP samples
A complete blood count (CBC) with reticulocytes and leukocyte differential was performed on a Sysmex XT-2000iV hematology instrument (Sysmex America, NY, USA). Plasma and serum were obtained after separation at room temperature for 10 min followed by centrifugation at 1,800 x g. Serum chemistry was analyzed on a Piccolo Xpress analyzer (Abaxis, NJ, USA) using the Piccolo general chemistry 13 panel, which included analysis of alanine aminotransferase, albumin, alkaline phosphatase, amylase, aspartate aminotransferase, calcium, creatinine, gamma-glutamyl transferase, glucose, total bilirubin, total protein, blood urea nitrogen, and uric acid. Blood collected in sodium citrate tubes was centrifuged for 15 min at 1,500 x g, and the prothrombin time and activated partial thromboplastin time measured within 4 h of collection on a CS-2500 system automated coagulation analyzer (Sysmex America, NY, USA).
NHP exposure and treatment, SUDV, UTMB
Eight healthy, adult crab-eating (aka cynomolgus) macaques (Macaca fascicularis Raffles, 1821) of Chinese origin (4 female, 4 male), ranging in age from ≈4.5–6.5 years and weighing ≈3.1–7.7 kg, were assigned to a treatment group or control group as determined by randomization. The duration of this study was 35 days.
Virus exposure, SUDV
SUDV (strain Gulu) originated from a 35-year-old male patient who had died on 16 October 2000. The study exposure material was from the second Vero E6 cell passage of SUDV. Briefly, the first passage at UTMB consisted of inoculating CDC 808892 (CDC passage 1 of SUDV isolate 200011676) at a multiplicity of infection (MOI) of 0.001 onto Vero E6 cells (ATCC CRL-1586). The cell supernatants were subsequently harvested at Day 7 post-infection and stored at −80°C as ≈1-mL aliquots. No detectable mycoplasma or endotoxin levels were measured (˂0.5 EU/mL). Animals were exposed intramuscularly in the left quadricep with a 1,000 PFU target dose (actual dose 1,363 PFU) of SUDV.
Antibody administration
Treatment was initiated IV at Day 4 and Day 7 after SUDV exposure, with three animals receiving 1C3 (25 mg/kg) and three animals receiving a combination of 1C3 and 1C11 (25 mg/kg each antibody, 50 mg/kg total dose). The 2 remaining animals served as SUDV positive controls and were administered a mock treatment of PBS.
Observations, procedures, and endpoint criteria
All animals were given physical examinations, and blood was collected at the time of virus exposure and at 4, 7, 10, 14, 21, 28, and 35 days after virus exposure. Animals were monitored daily and scored for disease progression with an internal filovirus endpoint scoring sheet approved by the UTMB IACUC. The scoring changes measured from baseline included posture and activity level, attitude and behavior, food intake, respiration, and disease manifestations, such as visible rash, hemorrhage, or ecchymosis. A score of ≥9 indicated that an animal met the criteria for euthanasia.
SUDV detection
On procedure days, 100 μL of blood collected in K2-EDTA tubes was centrifuged and added to 600 μL of AVL viral lysis buffer with 6 μL carrier RNA (Qiagen) for RNA extraction. All blood samples were inactivated in AVL viral lysis buffer prior to removal from the BSL-4 laboratory. Subsequently, RNA was isolated from blood using the QIAamp viral RNA kit (Qiagen), according to the manufacturer’s instructions.
Primers targeting the L gene of SUDV were used for RT-qPCR with the following probe: FAM-5=CAT CCA ATC AAA GAC ATT GCG A 3=-TAMRA; (Life Technologies).Viral RNA was detected using the CFX96 detection system (Bio-Rad Laboratories) in one-step probe RT-qPCR kits (Qiagen) with the following cycle conditions: 50°C for 10 min, 95°C for 10 seconds, and 40 cycles of 95°C for 10 seconds and 59°C for 30 seconds. Threshold cycle (CT) values representing viral genomes were analyzed with CFX Manager software, and the data are shown as genome equivalents (GEq) per milliliter. To create the GEq standard, RNA from SUDV stocks was extracted, and the number of SUDV L genomes was calculated using Avogadro’s number and the molecular weight of the SUDV genome. Limit of detection was 1 × 103 GEq/mL.
Virus titration was performed by plaque assay using Vero E6 cells (ATCC CRL-1586) from all plasma samples as previously described (Pascal et al., 2018). Briefly, increasing 10-fold dilutions of the samples were adsorbed to Vero E6 cell monolayers in duplicate wells (200 μL) and overlaid with 0.8% agarose in 2X Eagle’s minimum essential medium (EMEM) with 10% fetal bovine serum (FBS). After 6 days of incubation at 37°C with 5% CO2, neutral red stain was added and plaques were counted after 48 hours of incubation. The limit of detection for this assay was 25 PFU/mL.
Hematology and serum biochemistry for SUDV-exposed NHP samples
Total white blood cell counts, white blood cell differentials, red blood cell counts, platelet counts, hematocrit values, total hemoglobin concentrations, mean cell volumes, mean corpuscular volumes, and mean corpuscular hemoglobin concentrations were analyzed from blood collected in tubes containing EDTA using a laser-based hematologic analyzer (Beckman Coulter). Serum samples were tested for concentrations of alanine aminotransferase, albumin, alkaline phosphatase, amylase, aspartate aminotransferase, C-reactive protein, calcium, creatinine, gammaglutamyltransferase, glucose, total protein, blood urea nitrogen, and uric acid, and by using a Piccolo point-of-care analyzer and Biochemistry Panel Plus analyzer discs (Abaxis).
QUANTIFICATION AND STATISTICAL ANALYSIS
In ebolavirus glycoprotein variant ELISA, EC50 values were calculated using Prism 9.0 (GraphPad) after using sigmoidal dose-response nonlinear regression analysis. Neutralization half-maximal inhibitory concentration (IC50) titers for neutralization assay using recombinant vesicular stomatitis virus (rVSV) were calculated using non-linear regression [inhibitor] versus normalized response curve fit using Prism 9.0 (GraphPad). In mouse studies, survival curves were estimated using the Kaplan-Meier method. In mouse BDBV challenge studies, each group was compared with phosphate-buffered saline (PBS) control (Mantel-Cox test) and one-way analysis of variance (ANOVA) with Dunnett correction was used for multiple comparisons between each group and PBS mock control. In guinea pig studies, statistical analyses and generation of graphs were performed using Prism 6.05 (GraphPad). Guinea pig survival data were analyzed by log-rank (Mantel-Cox) test. One-Way analysis of variance (ANOVA) was used for statistical analysis of viremia levels. In NHP studies, survival curves were estimated using the Kaplan-Meier method.
Supplementary Material
Figure S1. Isolation and screening of monoclonal antibodies (mAbs) from human survivors after Ebola virus (EBOV) exposure, related to Figure 1.
(A) Staining of immunoglobulin G (IgG) memory B cells from Ebola virus disease patients with fluorescently labeled EBOV glycoprotein (GP) and Sudan virus (SUDV) GP probes. (B) monoclonal antibodies (mAbs) cloned from EBOV GP and SUDV GP dual-binding cells were screened for binding to recombinant EBOV GP, SUDV GP, and Bundibugyo virus (BDBV) GP by enzyme-linked immunosorbent assay (ELISA). Nine mAbs that neutralized luciferase-expressing EBOV reporter virus are marked with asterisks. (C) mAbs were further screened by ELISA for binding to full-length EBOV GP ectodomain, EBOV GPΔmucin, or EBOV soluble GP (sGP). (D) Initial screening neutralization assay. Representative neutralization screening assay using biologically-contained EBOV. Luciferase-expressing EBOV lacking the VP30 gene was incubated with the indicated monoclonal antibodies (mAbs) at 10 μg/mL for 2 h at 37°C, then added to VP30-expressing Vero cell cultures in duplicate wells. Renilla luciferase activity was measured 3 days later. mAbs were considered neutralizing if the luciferase signal was reduced by >90% compared to the average signal in six wells incubated with virus plus negative control antibody. Negative control: VP35-specific mAb. (E) Table showing initial mapping of the binding sites of the nine promising neutralizing mAbs. Left section: The promising mAbs (rows) were used as competitors to inhibit the binding of four reference mAbs (columns) to Jurkat cells expressing EBOV GP. Light-shaded cells indicate competition between a promising mAb and a reference mAb for binding. Right columns: The nine mAbs (rows) were tested for binding to previously described Jurkat cell lines expressing EBOV GP variants with the indicated single amino-acid changes. Expression-normalized binding is shown as a percentage of binding to wild-type EBOV GP. Dark-shaded cells indicate significant loss of binding to a given GP variant.
Figure S2. Cryo-EM local resolution estimation of antibody cocktail in complex with EBOV GP Trimer, related to Figure 2.
The local resolution of the full map is estimated by Relion, showing the side view (A) and the top view (B) of the mAbs and EBOV GP complex. (C) Zooming in on 1C3 variable region and its contacting region on GP. (D) Zooming in on 1C11 scFv and its contacting region on GP.
Figure S3. Broadly neutralizing antibody footprint comparison, related to Figure 3.
The crystal structure of Ebola virus (EBOV) glycoprotein (GP) (PDB 5JQ3) is shown in surface representation with GP1 and GP2 colored light and dark gray, respectively. (A) The footprint of 1C11 is shown in blue, the footprint of 6D6 is shown in magenta, and their shared footprint is shown in purple. (B) The footprint of 1C11 is shown in blue, the footprint of ADI-15878 is shown in yellow, and their shared footprint is shown in green. (C) A comparative sequence alignment to illustrate contact differences among 1C11 and the above four broad neutralizing mAbs. Residues were labeled by colored triangles at the bottom to show the conservation (blue - conserved, pink - similar, yellow - non-conserved).
Figure S4. Neutralization analysis of mAbs 1C3, related to Figure 4.
(A) Point changes at four residues, P116, P123, P126, and D127 did not affect neutralization of rVSV-EBOV by 1C3. (B) GP residues in 1C3 contact sites that were changed in Figure 4B, 4C, and S4A were labeled on the top view surface representation of EBOV GP trimer. The residues that did not affect neutralization were labeled in the left panel with circles, with colors corresponding to Figure S4A. The residues that affected neutralization were labeled in the right panel with stars, with colors corresponding to Figure 4C.
Figure S5. The binding of 1C3 Fab blocks three possible directions of NPC1-C access to EBOV GP, related to Figure 3 and 4.
(A) Mab 1C3 Fab (light chain in yellow; heavy chain in orange) in complex with GP (GP1 in grey, GP2 in white), superimposed with NPC1 (dark blue) in complex with GP (PDB: 5F1B). (B) The footprints of mAb 1C3 and NPC1-C on GP surface viewing from the top. Surface uniquely bound by 1C3 is in orange. Surface uniquely bound by NPC1 is in blue. The ten shared residues are in purple.
Figure S6. Mouse protection studies, BDBV in STAT1 KO Mice, UTMB, related to Figure 5.
Groups of STAT1 KO mice at five animals per group were injected with the indicated mAbs by the intraperitoneal route at 24 h after BDBV chimeric virus challenge. Body weight, and illness score curves are shown.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| 9.20.1C3 | This study | |
| 5.24.1C11 | This study | |
| Goat anti-human IgG Fc–conjugated secondary antibody | ThermoFisher Scientific | Catalog # 31413 |
| Goat anti-rabbit secondary antibody labeled with horseradish peroxidase | Southern Biotech | 4030–05 |
| Rabbit anti EBOV GP pAb | IBT Bioservices | Cat#0301–015 |
| Goat anti rabbit HRP | Southern Biotech | Cat#4050–05; RRID: AB_2795955 |
| Bacterial and virus strains | ||
| VSV-deltaG-GFP | Karafast | Cat# EH1020 |
| EbolaΔVP30 virus luciferase | PMID: 31104840 | N/A |
| Guinea pig-adapted EBOV/Mayinga (EBOV-GA) | PMID: 26038397 | N/A |
| Guinea pig-adapted SUDV/Boniface (SUDV-GA) | PMID: 26491156 | GenBank: KT878488 |
| Chimeric EBOV/BDBV-GP | PMID: 30060231 | GenBank: MH464888 |
| Ebola virus/H. sapiens-tc/COD/1995/Kikwit-9510621 (EBOV) | BEI Resources | NR-50306 |
| Chemicals, peptides, and recombinant proteins | ||
| Corning® 96-well Half Area Clear Flat Bottom Polystyrene High Bind Microplate | VWR | 29442–318 |
| 96-Well Clear Bottom Plates, Corning®, Plates with Lids, Tissue Culture Treated | Fisher | 3603 |
| Sulfuric Acid 17N | Fisher Scientific | A300–212 |
| Lonza Insect Xpress Liquid media | VWR | 12001–622 |
| d-Desthiobiotin | Sigma Aldrich | D1411–1G |
| Biolock Biotin Blocking Solution | Iba | 2–0205-250 |
| Bovine Serum Albumin | Sigma Aldrich | A9647–500G |
| Puromycin | InvivoGen | ant-pr-5b |
| Hoechst 33342, Trihydrochloride, Trihydrate, 100 mg | Thermo/invitrogen | H1399 |
| Blotting grade blocker (nonfat dry milk) | Biorad | 1706404 |
| Tween 20 | Fisher Scientific | BP337–500 |
| 0.45% Methylcellulose | Sigma Aldrich | |
| OptiMem Media | Thermofisher Scientific | 31985070 |
| Iodoacetamide | Sigma | I6125–5G |
| Immobilized papain | ThermoFisher Scientific | Cat#20341 |
| EkMax Enterokinase | ThermoFisher Scientific | E18001 |
| 16% Paraformaldehyde (formaldehyde) aqueous solution | Electron Microscopy Sciences | Cat#15710 |
| AEC Substrate Kit | Abcam | Ab64252 |
| Maxisorp flat bottom ELISA plates | ThermoFisher | 442404 |
| Goat anti-human IgG (H+L) capture antibody | Jackson ImmunoResearch | 109–005-088 |
| SUDV GP ectodomain | IBT Bioservices | 0502–015 |
| BDBV GP ectodomain | IBT Bioservices | 0505–015 |
| Recombinant SUDV GPΔTM (aa: 33–637, Boniface 1976) | this study | N/A |
| Recombinant BDBV GP ΔTM (aa: 33–640, Uganda 2007) | this study | N/A |
| Recombinant EBOV GP ΔTM (aa: 33–637, Mayinga 1976) | this study | N/A |
| Recombinant EBOV GP ΔMuc (aa: 33–637, d312–463, Mayinga 1976) | this study | N/A |
| Recombinant SUDV GPΔMuc (aa: 33–637, d314–462, Boniface 1976) | this study | N/A |
| Recombinant BDBV GP ΔMuc (aa: 33–640, d313–460, Uganda 2007) | this study | N/A |
| ExpiCHO Expression Medium | Thermofisher | A2910002 |
| Expi-Fectamine-CHO Transfection kit | Thermo Fisher Scientific | A29129 |
| Dulbecco’s Modified Eagle Medium High Glucose with Glutamax supplement | Thermo Fisher Scientific | 10566016 |
| Lauryl maltose neopentyl glycol | Anatrace | NG310 |
| Phosphate-buffered formalin | ThermoFisher Scientific | Cat#245–684 |
| Tween-20 | Sigma-Aldrich | Cat#P9416–100ML |
| Minimal Essential Medium | ThermoFisher Scientific | Cat#11095–080 |
| Dulbecco’s phosphate-buffered saline | Corning | Cat#21–031-CV |
| Fetal Bovine Serum | HyClone | Cat#SH30910.03HI-ST |
| Penicillin-streptomycin | Invitrogen | Cat#15140122 |
| TRIzol LS Reagent | Thermo Fisher Scientific | 10296010 |
| Critical commercial assays | ||
| ExpiFectamine CHO transfection kit | ThermoFisher | Cat# A29129 |
| 1-Step Ultra TMB Substrate Solution | Thermofisher Scientific | 34029 |
| AEC substrate | Abcam | Cat#ab64252 |
| BEI Resources Critical Reagents Program EZ1 RT-PCR (TaqMan) assay kit | BEI Resources | Trombley, A. R. et al. 2010 |
| QIAamp Viral RNA Mini Kit | QIAGEN | N/A |
| Quantifast Probe RT-PCR kit | QIAGEN | Cat#204456 |
| Deposited Data | ||
| Structure of 9.20.1C3 Fab | This study | PDB: 7N6P |
| Structure of EBOV GP lacking the mucin-like domain with 1C11 scFv and 1C3 Fab bound | This study | PDB: 7SWD |
| Structure of EBOV GP lacking the mucin-like domain with 1C11 scFv and 1C3 Fab bound (cryo-EM) | This study | EMD-25471 |
| Experimental Models: Cell Lines | ||
| Drosophila: Schneider 2 | ThermoFisher Scientific | Cat#R69007; RRID:CVCL_Z232 |
| ExpiCHO-S | ThermoFisher Scientific | A29127; RRID:CVCL_5J31 |
| Vero | ATCC | CCL-81; RRID:CVCL_0059 |
| Jurkat, Clone E6–1 | ATCC | TIB-152; RRID:CVCL_0367 |
| Expi293F | ThermoFisher Scientific | A14527; RRID:CVC L_D615 |
| Vero-E6 | ATCC | CRL-1586; RRID:CVCL_YZ66 |
| Grivet (Chlorocebus aethiops) Vero E6 cells | BEI Resources | N/A |
| Experimental Models: Organisms/strains | ||
| Guinea pig: Hartley | Charles River Laboratories | Cat#051; RRID: NCBITaxon_10141 |
| Mouse: 129S6/SvEv-Stat1tm1Rds (STAT1 KO) | Taconic Biosciences | Cat#TAC:2045; RRID: IMSR_TAC:2045 |
| Chinese rhesus monkeys (Macaca mulatta) | WorldWide Primates, Inc | N/A |
| Recombinant DNA | ||
| Empty vector: phCMV3 | Genlantis | Genlantis Cat# P003300 |
| Plasmid: EBOV-makona-K114A | this study | N/A |
| Plasmid: EBOV-makona-K114G | this study | N/A |
| Plasmid: EBOV-makona-P116A | this study | N/A |
| Plasmid: EBOV-makona-D117A | this study | N/A |
| Plasmid: EBOV-makona-D117R | this study | N/A |
| Plasmid: EBOV-makona-E120A | this study | N/A |
| Plasmid: EBOV-makona-P123G | this study | N/A |
| Plasmid: EBOV-makona-P126G | this study | N/A |
| Plasmid: EBOV-makona-R172A | this study | N/A |
| Plasmid: EBOV-makona-R172G | this study | N/A |
| Empty vector: pMT-puro bip | Addgene | Addgene #17923 |
| Plasmid: EBOV-mayinga-K114A | this study | N/A |
| Plasmid: EBOV-mayinga-K114G | this study | N/A |
| Plasmid: EBOV-mayinga-P116A | this study | N/A |
| Plasmid: EBOV-mayinga-D117A | this study | N/A |
| Plasmid: EBOV-mayinga-D117R | this study | N/A |
| Plasmid: EBOV-mayinga-E120A | this study | N/A |
| Plasmid: EBOV-mayinga-P123G | this study | N/A |
| Plasmid: EBOV-mayinga-P126G | this study | N/A |
| Plasmid: EBOV-mayinga-R172A | this study | N/A |
| Plasmid: EBOV-mayinga-R172G | this study | N/A |
| Plasmid: 1C3 light chain | this study | N/A |
| Plasmid: 1C3 heavy chain | this study | N/A |
| Plasmid: 1C11 light chain | this study | N/A |
| Plasmid: 1C11 heavy chain | this study | N/A |
| 1C11 scFv containing VH, VL | Genscript | N/A |
| Software and algorithms | ||
| Coot | Emsley et al., 2010 | RRID: SCR_014222; https://www2.mrc-Imb.cam.ac.uk/personal/pemsley/coot/ |
| PHENIX | Liebschner et al., 2019 | RRID: SCR_014224; https://www.phenix-online.org |
| Phenix.refine | Adams et al., 2010 | RRID: SCR_016736 |
| PyMOL version 2.3.2 | Schrödinger, LLC | RRID: SCR_000305; https://www.schrodinger.com/pymol |
| UCSF Chimera | Petterson et al., 2004 | RRID: SCR_004097; https://www.cgl.ucsf.edu/chimera/ |
| PDBePISA version 1.52 | EMBL-EBI | http://www.ebi.ac.uk/pdbe/pisa/ |
| cisTem | Grant et al., 2018 | https://cistem.org |
| EPU | Thermo Fisher | https://www.thermofisher.com/ |
| Swiss-model | Waterhouse et al.,2018 | https://swissmodel.expasy.org/ |
| Relion | Zivanov et al., 2018 | https://www3.mrc-lmb.cam.ac.uk/relion/index.php?title=Main_Page |
| Molprobity | Williams et al. 2018 | http://molprobity.biochem.duke.edu/ |
| GraphPad Prism 6.05, 9 | GraphPad Software, Inc. | GraphPad Prism http://www.graphpad.com |
| CFX Maestro version 1.1 | BioRad | CFX Maestro |
| Other | ||
| Spark 10M Multimode Plate Reader | Tecan Life Sciences | Cat# 30086375 |
| Corning® 96-well Half Area Clear Flat Bottom Polystyrene High Bind Microplate | VWR | Cat# 29442–318 |
| 96-Well Clear Bottom Plates, Corning®, Plates with Lids, Tissue Culture Treated | Fisher | Cat# 3603 |
| Microseal 96 well PCR plates | BioRad | Cat# MSP9601 |
| StrepTrap High Performance column | Cytiva Life Sciences | Cat# 28907547 |
| HiTrap Protein A High Performance column | Cytiva Life Sciences | Cat# 17040301 |
| Mono Q 5/50 GL | Cytiva Life Sciences | Cat# 17516601 |
| Superdex 200 Increase 10/300 GL column | Cytiva Life Sciences | Cat# 28990944 |
| Superdex 75 Increase 10/300 GL column | Cytiva Life Sciences | Cat# 29148721 |
| Superdex 6 Increase 10/300 GL column | Cytiva Life Sciences | Cat# 29091596 |
| Cellinsight CX5 High-Content Screening (HCS) Platform | Thermofisher Scientific | Thermo Scientific Cat#CX51110 |
| OctetRed384 | FortéBio | https://www.moleculardevices.com |
| Oryx 8 | Douglas Instruments | www.douglas.co.uk |
| Titan Halo 300kV electron microscope with Falcon 3EC camera | Thermofisher Scientific | https://www.thermofisher.com/ |
| Holey carbon C-flat 2/1 400 mesh copper grids | Electron Microscopy Sciences | https://www.emsdiasum.com/ |
| Vitrobot Mark IV | Thermofisher Scientific | https://www.thermofisher.com/us/en/home/electron-microscopy/products/sample-preparation-equipment-em/vitrobot-system.html |
| CFX96 Touch Real-Time PCR Detection System | BioRad | Cat# 1855195 |
Highlights.
Two survivor antibodies provide broad protection, without secreted sGP cross-reactivity
Cryo-EM structure reveals the quaternary epitopes of the antibodies
One, at chalice center, simultaneously binds all three monomers in the GP trimer
The cocktail of two protects non-human primates from Ebola and Sudan virus disease
ACKNOWLEDGMENTS
We thank Sharon Schendel for editing the manuscript, and Dawid Zyla and Haoyang Li for help with model validation. We gratefully acknowledge our funding from National Institute of Allergy and Infectious Diseases (NIAID) U19 AI142790, Consortium for Immunotherapeutics against Emerging Viral Threats (E.O.S, C.W.D.), DARPA contract W31P4Q-14-1-0010 (C.W.D.), and U19AI109762 (T.W.G). Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (P30GM133894). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NIGMS or NIH.
We thank all the staff of the National Institutes of Health (NIH) NIAID Division of Clinical Research (DCR) Integrated Research Facility at Fort Detrick (IRF-Frederick), Frederick, MD, USA, who supported this study-in particular, Kaleb Sharer, Russel Byrum, Jennifer Jackson, Sarah Klim, Danny Ragland, Marisa St. Claire, and Lisa Hensley. This work was supported in part through Laulima Government Solutions, LLC, prime contract and Battelle Memorial Institute prime contract with the U.S. National Institute of Allergy and Infectious Diseases (NIAID) under Contract No. HHSN272201800013C and No. HHSN272200700016I, respectively (M.R.H., G.W.,). J.H.K. performed this work as an employee of Tunnell Government Services (TGS), a subcontractor of Laulima Government Solutions, LLC under Contract No. HHSN272201800013C. This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261201500003I, Task Order No. HHSN26100043 and Contract No. 75N91019D00024, Task Order No. 75N91019F00130 (I.C.). The views and conclusions contained in this document are those of the authors and should not be interpreted as necessarily representing the official policies, either expressed or implied, of the U.S. Department of Health and Human Services or of the institutions and companies affiliated with the authors. The study protocol was reviewed and approved by the NIH NIAID DCR IRF-Frederick Animal Care and Use Committee in compliance with all applicable federal regulations governing the protection of animals and research. We thank Anya Crane (NIH NIAID DCR IRF-Frederick) for critically editing the manuscript and Jiro Wada (NIH NIAID DCR IRF-Frederick) for helping with figure preparation.
We thank the University of Texas Medical Branch (UTMB) Animal Resource Center for husbandry support of laboratory animals and Daniel Deer and Kevin Melody for assistance with the animal studies. This study was supported in part by the U.S. Department of Health and Human Services, NIH grant UC7AI094660 to the UTMB for BSL-4 operations in support of the Galveston National Laboratory. Opinions, interpretations, conclusions, and recommendations are those of the authors and are not necessarily endorsed by the UTMB.
Footnotes
DECLARATION OF INTERESTS
R.A., C.D. and E.O.S. are inventors on a patent relating to the antibodies described in this work. All other authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornholdt ZA, Herbert AS, Mire CE, He S, Cross RW, Wec AZ, Abelson DM, Geisbert JB, James RM, Rahim MN, et al. (2019). A Two-Antibody Pan-Ebolavirus Cocktail Confers Broad Therapeutic Protection in Ferrets and Nonhuman Primates. Cell Host Microbe 25, 49–58.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray M, Davis K, Geisbert T, Schmaljohn C, and Huggins J (1999). A mouse model for evaluation of prophylaxis and therapy of Ebola hemorrhagic fever. J. Infect. Dis 179 Suppl 1, S248–58. [DOI] [PubMed] [Google Scholar]
- CDC (2021a). History of Ebola Virus Disease (EVD) Outbreaks. [Google Scholar]
- CDC (2021b). Ebola Virus Disease Distribution Map: Cases of Ebola Virus Disease in Africa Since 1976. [Google Scholar]
- Chou T-C (2010). Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 70, 440–446. [DOI] [PubMed] [Google Scholar]
- Cohen-Dvashi H, Zehner M, Ehrhardt S, Katz M, Elad N, Klein F, and Diskin R (2020). Structural Basis for a Convergent Immune Response against Ebola Virus. Cell Host Microbe 27, 418–427.e4. [DOI] [PubMed] [Google Scholar]
- Corti D, Misasi J, Mulangu S, Stanley DA, Kanekiyo M, Wollen S, Ploquin A, Doria-Rose NA, Staupe RP, Bailey M, et al. (2016). Protective monotherapy against lethal Ebola virus infection by a potently neutralizing antibody. Science 351, 1339–1342. [DOI] [PubMed] [Google Scholar]
- Davis CW, Jackson KJL, McElroy AK, Halfmann P, Huang J, Chennareddy C, Piper AE, Leung Y, Albariño CG, Crozier I, et al. (2019). Longitudinal Analysis of the Human B Cell Response to Ebola Virus Infection. Cell 177, 1566–1582.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010). Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Commission (2020). Vaccine against Ebola: Commission grants new market authorisations. [Google Scholar]
- Feldmann H, Sprecher A, and Geisbert TW (2020). Ebola. N. Engl. J. Med 382, 1832–1842. [DOI] [PubMed] [Google Scholar]
- Food and Drug Administration (2020a). FDA Approves First Treatment for Ebola Virus. [Google Scholar]
- Food and Drug Administration (2020b). FDA Approves Treatment for Ebola Virus. [Google Scholar]
- Fujii R, Kitaoka M, and Hayashi K (2006). Error-prone rolling circle amplification: the simplest random mutagenesis protocol. Nat. Protoc 1, 2493–2497. [DOI] [PubMed] [Google Scholar]
- Gilchuk P, Kuzmina N, Ilinykh PA, Huang K, Gunn BM, Bryan A, Davidson E, Doranz BJ, Turner HL, Fusco ML, et al. (2018). Multifunctional pan-ebolavirus antibody recognizes a site of broad vulnerability on the ebolavirus glycoprotein. Immunity 49, 363–374.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchuk P, Murin CD, Milligan JC, Cross RW, Mire CE, Ilinykh PA, Huang K, Kuzmina N, Altman PX, Hui S, et al. (2020). Analysis of a Therapeutic Antibody Cocktail Reveals Determinants for Cooperative and Broad Ebolavirus Neutralization. Immunity 52, 388–403.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchuk P, Murin CD, Cross RW, Ilinykh PA, Huang K, Kuzmina N, Borisevich V, Agans KN, Geisbert JB, Zost SJ, et al. (2021). Pan-ebolavirus protective therapy by two multifunctional human antibodies. Cell 184, 5593–5607.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman J., Chuang G-Y., Lai Y-T., Shen C-H., Boyington JC., Druz A., Geng H., Louder MK., McKee K., Rawi R., et al. (2020). Structure of Super-Potent Antibody CAP256-VRC26.25 in Complex with HIV-1 Envelope Reveals a Combined Mode of Trimer-Apex Recognition. Cell Rep. 31, 107488. [DOI] [PubMed] [Google Scholar]
- Grant T, Rohou A, and Grigorieff N (2018). cisTEM, user-friendly software for single-particle image processing. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann P, Kim JH, Ebihara H, Noda T, Neumann G, Feldmann H, and Kawaoka Y (2008). Generation of biologically contained Ebola viruses. Proc. Natl. Acad. Sci. U. S. A 105, 1129–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashiguchi T, Fusco ML, Bornholdt ZA, Lee JE, Flyak AI, Matsuoka R, Kohda D, Yanagi Y, Hammel M, Crowe JE Jr, et al. (2015). Structural basis for Marburg virus neutralization by a cross-reactive human antibody. Cell 160, 904–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honnold SP, Bakken RR, Fisher D, Lind CM, Cohen JW, Eccleston LT, Spurgers KB, Maheshwari RK, and Glass PJ (2014). Second generation inactivated eastern equine encephalitis virus vaccine candidates protect mice against a lethal aerosol challenge. PLoS One 9, e104708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell KA, Qiu X, Brannan JM, Bryan C, Davidson E, Holtsberg FW, Wec AZ, Shulenin S, Biggins JE, Douglas R, et al. (2016). Antibody Treatment of Ebola and Sudan Virus Infection via a Uniquely Exposed Epitope within the Glycoprotein Receptor-Binding Site. Cell Rep. 15, 1514–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilinykh PA, Graber J, Kuzmina NA, Huang K, Ksiazek TG, Crowe JE Jr, and Bukreyev A (2018). Ebolavirus Chimerization for the Development of a Mouse Model for Screening of Bundibugyo-Specific Antibodies. J. Infect. Dis 218, S418–S422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilinykh PA, Huang K, Santos RI, Gilchuk P, Gunn BM, Karim MM, Liang J, Fouch ME, Davidson E, Parekh DV, et al. (2020). Non-neutralizing Antibodies from a Marburg Infection Survivor Mediate Protection by Fc-Effector Functions and by Enhancing Efficacy of Other Antibodies. Cell Host Microbe 27, 976–991.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien J-P, Lee JH, Cupo A, Murin CD, Derking R, Hoffenberg S, Caulfield MJ, King CR, Marozsan AJ, Klasse PJ, et al. (2013). Asymmetric recognition of the HIV-1 trimer by broadly neutralizing antibody PG9. Proc. Natl. Acad. Sci. U. S. A 110, 4351–4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keita AK, Koundouno FR, Faye M, Düx A, Hinzmann J, Diallo H, Ayouba A, Le Marcis F, Soropogui B, Ifono K, et al. (2021). Resurgence of Ebola virus in 2021 in Guinea suggests a new paradigm for outbreaks. Nature 597, 539–543. [DOI] [PubMed] [Google Scholar]
- Kuhn JH, Amarasinghe GK, and Perry DL (2020). Filoviridae. In Howley PM, Knipe DM, Whelan SPJ, ed. (Wolters Kluwer/Lippincott Williams & Wilkins; ), pp. 449–503. [Google Scholar]
- Lee JE., Fusco ML., Hessell AJ., Oswald WB., Burton DR., and Saphire EO. (2008). Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature 454, 177–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Andrabi R, Su C-Y, Yasmeen A, Julien J-P, Kong L, Wu NC, McBride R, Sok D, Pauthner M, et al. (2017). A Broadly Neutralizing Antibody Targets the Dynamic HIV Envelope Trimer Apex via a Long, Rigidified, and Anionic β-Hairpin Structure. Immunity 46, 690–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Acharya P, Dolan MA, Zhang P, Guzzo C, Lu J, Kwon A, Gururani D, Miao H, Bylund T, et al. (2017). Quaternary contact in the initial interaction of CD4 with the HIV-1 envelope trimer. Nat. Struct. Mol. Biol 24, 370–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan JC, Parekh DV, Fuller KM, Igarashi M, Takada A, and Saphire EO (2018). Structural Characterization of Pan-Ebolavirus Antibody 6D6 Targeting the Fusion Peptide of the Surface Glycoprotein. J. Infect. Dis 219, 415–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misasi J, Gilman MSA, Kanekiyo M, Gui M, Cagigi A, Mulangu S, Corti D, Ledgerwood JE, Lanzavecchia A, Cunningham J, et al. (2016). Structural and molecular basis for Ebola virus neutralization by protective human antibodies. Science 351, 1343–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulangu S, Dodd LE, Davey RT Jr, Tshiani Mbaya O, Proschan M, Mukadi D, Lusakibanza Manzo M, Nzolo D, Tshomba Oloma A, Ibanda A, et al. (2019). A Randomized, Controlled Trial of Ebola Virus Disease Therapeutics. N. Engl. J. Med 381, 2293–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Research Council, Division on Earth and Life Studies, Institute for Laboratory Animal Research, and Committee for the Update of the Guide for the Care and Use of Laboratory Animals (2010). Guide for the Care and Use of Laboratory Animals: Eighth Edition (National Academies Press; ). [Google Scholar]
- Ollmann Saphire E (2020). A Vaccine against Ebola Virus. Cell 181, 6. [DOI] [PubMed] [Google Scholar]
- Pallesen J, Murin CD, de Val N, Cottrell CA, Hastie KM, Turner HL, Fusco ML, Flyak AI, Zeitlin L, Crowe JE, et al. (2016). Structures of Ebola virus GP and sGP in complex with therapeutic antibodies. Nature Microbiology 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascal KE, Dudgeon D, Trefry JC, Anantpadma M, Sakurai Y, Murin CD, Turner HL, Fairhurst J, Torres M, Rafique A, et al. (2018). Development of clinical-stage human monoclonal antibodies that treat advanced Ebola virus disease in nonhuman primates. J. Infect. Dis 218, S612–S626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percie du Sert N, Hurst V, Ahluwalia A, Alam S, Avey MT, Baker M, Browne WJ, Clark A, Cuthill IC, Dirnagl U, et al. (2020). The ARRIVE guidelines 2.0: updated guidelines for reporting animal research. BMJ Open Sci 4, e100115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- PREVAIL II Writing Group, Multi-National PREVAIL II Study Team, Davey RT Jr, Dodd L, Proschan MA, Neaton J, Neuhaus Nordwall J, Koopmeiners JS, Beigel J, Tierney J, et al. (2016). A randomized, controlled trial of ZMapp for Ebola virus infection. N. Engl. J. Med 375, 1448–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Audet J, Lv M, He S, Wong G, Wei H, Luo L, Fernando L, Kroeker A, Bovendo HF, et al. (2016). Two-mAb cocktail protects macaques against the Makona variant of Ebola virus. Sci. Transl. Med [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saphire EO, Schendel SL, Fusco ML, Gangavarapu K, Gunn BM, Wec AZ, Halfmann PJ, Brannan JM, Herbert AS, Qiu X, et al. (2018). Systematic Analysis of Monoclonal Antibodies against Ebola Virus GP Defines Features that Contribute to Protection. Cell 174, 938–952.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shurtleff AC, Biggins JE, Keeney AE, Zumbrun EE, Bloomfield HA, Kuehne A, Audet JL, Alfson KJ, Griffiths A, Olinger GG, et al. (2012). Standardization of the filovirus plaque assay for use in preclinical studies. Viruses 4, 3511–3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart-Jones GBE, Chuang G-Y, Xu K, Zhou T, Acharya P, Tsybovsky Y, Ou L, Zhang B, Fernandez-Rodriguez B, Gilardi V, et al. (2018). Structure-based design of a quadrivalent fusion glycoprotein vaccine for human parainfluenza virus types 1–4. Proc. Natl. Acad. Sci. U. S. A 115, 12265–12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller T, Meffre E, Yurasov S, Tsuiji M, Nussenzweig MC, and Wardemann H (2008). Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J. Immunol. Methods 329, 112–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trombley AR, Wachter L, Garrison J, Buckley-Beason VA, Jahrling J, Hensley LE, Schoepp RJ, Norwood DA, Goba A, Fair JN, et al. (2010). Comprehensive panel of real-time TaqMan polymerase chain reaction assays for detection and absolute quantification of filoviruses, arenaviruses, and New World hantaviruses. Am. J. Trop. Med. Hyg 82, 954–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, et al. (2018). SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West BR, Moyer CL, King LB, Fusco ML, Milligan JC, Hui S, and Saphire EO (2018). Structural Basis of Pan-Ebolavirus Neutralization by a Human Antibody against a Conserved, yet Cryptic Epitope. MBio 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CJ, Headd JJ, Moriarty NW, Prisant MG, Videau LL, Deis LN, Verma V, Keedy DA, Hintze BJ, Chen VB, et al. (2018). MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong AC, Sandesara RG, Mulherkar N, Whelan SP, and Chandran K (2010). A forward genetic strategy reveals destabilizing mutations in the Ebolavirus glycoprotein that alter its protease dependence during cell entry. J. Virol 84, 163–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (2021). Ebola outbreak 2021-North Kivu. [Google Scholar]
- Zhang K. (2016). Gctf: Real-time CTF determination and correction. J. Struct. Biol 193, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache J-P, Verba KA, Cheng Y, and Agard DA (2017). MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJ, Lindahl E, and Scheres SH (2018). New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Isolation and screening of monoclonal antibodies (mAbs) from human survivors after Ebola virus (EBOV) exposure, related to Figure 1.
(A) Staining of immunoglobulin G (IgG) memory B cells from Ebola virus disease patients with fluorescently labeled EBOV glycoprotein (GP) and Sudan virus (SUDV) GP probes. (B) monoclonal antibodies (mAbs) cloned from EBOV GP and SUDV GP dual-binding cells were screened for binding to recombinant EBOV GP, SUDV GP, and Bundibugyo virus (BDBV) GP by enzyme-linked immunosorbent assay (ELISA). Nine mAbs that neutralized luciferase-expressing EBOV reporter virus are marked with asterisks. (C) mAbs were further screened by ELISA for binding to full-length EBOV GP ectodomain, EBOV GPΔmucin, or EBOV soluble GP (sGP). (D) Initial screening neutralization assay. Representative neutralization screening assay using biologically-contained EBOV. Luciferase-expressing EBOV lacking the VP30 gene was incubated with the indicated monoclonal antibodies (mAbs) at 10 μg/mL for 2 h at 37°C, then added to VP30-expressing Vero cell cultures in duplicate wells. Renilla luciferase activity was measured 3 days later. mAbs were considered neutralizing if the luciferase signal was reduced by >90% compared to the average signal in six wells incubated with virus plus negative control antibody. Negative control: VP35-specific mAb. (E) Table showing initial mapping of the binding sites of the nine promising neutralizing mAbs. Left section: The promising mAbs (rows) were used as competitors to inhibit the binding of four reference mAbs (columns) to Jurkat cells expressing EBOV GP. Light-shaded cells indicate competition between a promising mAb and a reference mAb for binding. Right columns: The nine mAbs (rows) were tested for binding to previously described Jurkat cell lines expressing EBOV GP variants with the indicated single amino-acid changes. Expression-normalized binding is shown as a percentage of binding to wild-type EBOV GP. Dark-shaded cells indicate significant loss of binding to a given GP variant.
Figure S2. Cryo-EM local resolution estimation of antibody cocktail in complex with EBOV GP Trimer, related to Figure 2.
The local resolution of the full map is estimated by Relion, showing the side view (A) and the top view (B) of the mAbs and EBOV GP complex. (C) Zooming in on 1C3 variable region and its contacting region on GP. (D) Zooming in on 1C11 scFv and its contacting region on GP.
Figure S3. Broadly neutralizing antibody footprint comparison, related to Figure 3.
The crystal structure of Ebola virus (EBOV) glycoprotein (GP) (PDB 5JQ3) is shown in surface representation with GP1 and GP2 colored light and dark gray, respectively. (A) The footprint of 1C11 is shown in blue, the footprint of 6D6 is shown in magenta, and their shared footprint is shown in purple. (B) The footprint of 1C11 is shown in blue, the footprint of ADI-15878 is shown in yellow, and their shared footprint is shown in green. (C) A comparative sequence alignment to illustrate contact differences among 1C11 and the above four broad neutralizing mAbs. Residues were labeled by colored triangles at the bottom to show the conservation (blue - conserved, pink - similar, yellow - non-conserved).
Figure S4. Neutralization analysis of mAbs 1C3, related to Figure 4.
(A) Point changes at four residues, P116, P123, P126, and D127 did not affect neutralization of rVSV-EBOV by 1C3. (B) GP residues in 1C3 contact sites that were changed in Figure 4B, 4C, and S4A were labeled on the top view surface representation of EBOV GP trimer. The residues that did not affect neutralization were labeled in the left panel with circles, with colors corresponding to Figure S4A. The residues that affected neutralization were labeled in the right panel with stars, with colors corresponding to Figure 4C.
Figure S5. The binding of 1C3 Fab blocks three possible directions of NPC1-C access to EBOV GP, related to Figure 3 and 4.
(A) Mab 1C3 Fab (light chain in yellow; heavy chain in orange) in complex with GP (GP1 in grey, GP2 in white), superimposed with NPC1 (dark blue) in complex with GP (PDB: 5F1B). (B) The footprints of mAb 1C3 and NPC1-C on GP surface viewing from the top. Surface uniquely bound by 1C3 is in orange. Surface uniquely bound by NPC1 is in blue. The ten shared residues are in purple.
Figure S6. Mouse protection studies, BDBV in STAT1 KO Mice, UTMB, related to Figure 5.
Groups of STAT1 KO mice at five animals per group were injected with the indicated mAbs by the intraperitoneal route at 24 h after BDBV chimeric virus challenge. Body weight, and illness score curves are shown.
Data Availability Statement
Coordinates and structure factors of 1C3 Fab have been deposited in the Protein Data Bank under accession number 7N6P. Cryo-EM map of the 1C3 and 1C11 bound EBOV GP trimer complex has been deposited to the EMDB with accession codes EMD-25471. Fitted coordinates have been deposited to the Protein Data Bank with accession codes 7SWD. Other data are available from the corresponding author upon request.