Summary
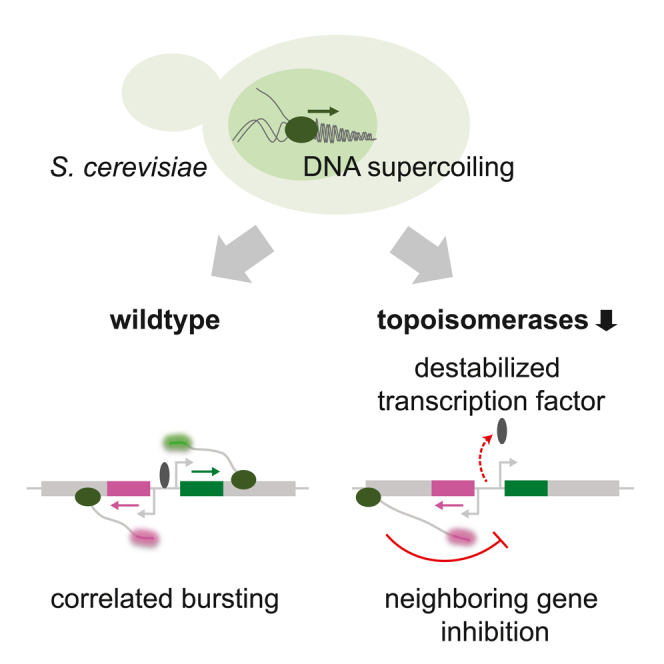
DNA supercoiling has emerged as a major contributor to gene regulation in bacteria, but how DNA supercoiling impacts transcription dynamics in eukaryotes is unclear. Here, using single-molecule dual-color nascent transcription imaging in budding yeast, we show that transcriptional bursting of divergent and tandem GAL genes is coupled. Temporal coupling of neighboring genes requires rapid release of DNA supercoils by topoisomerases. When DNA supercoils accumulate, transcription of one gene inhibits transcription at its adjacent genes. Transcription inhibition of the GAL genes results from destabilized binding of the transcription factor Gal4. Moreover, wild-type yeast minimizes supercoiling-mediated inhibition by maintaining sufficient levels of topoisomerases. Overall, we discover fundamental differences in transcriptional control by DNA supercoiling between bacteria and yeast and show that rapid supercoiling release in eukaryotes ensures proper gene expression of neighboring genes.
Keywords: DNA supercoiling, transcriptional bursting, topoisomerases, single-molecule live-cell imaging, budding yeast
Graphical abstract
Highlights
-
•
Transcription of neighboring GAL genes is coupled, and supercoils impede coupling
-
•
Buildup of supercoils from transcription inhibits transcription at adjacent genes
-
•
Inhibition occurs through destabilized DNA binding of the transcription factor
-
•
Wild-type yeast contains sufficient topoisomerase levels to prevent inhibition
Patel et al. use single-molecule visualization of transcription of neighboring genes at the GAL locus in budding yeast to elucidate how DNA supercoils regulate transcription dynamics in eukaryotes. Accumulation of DNA supercoils from transcription impedes transcription at adjacent genes, but wild-type yeast prevents inhibition by sufficient supercoiling release.
Introduction
During transcription, the movement of RNA polymerase along the DNA generates negative supercoils behind RNA polymerase and positive supercoils in front, as described by the twin-supercoiled-domain model.1,2,3 Transcription-generated supercoils can, in turn, enhance or impede the transcriptional process: negative supercoils facilitate transcription initiation by enabling promoter melting and enhancing the binding of regulatory factors, whereas positive supercoils aid the elongation of RNA polymerases by destabilizing DNA-bound proteins.4,5,6,7,8,9 However, excessive negative or positive supercoils can also repress transcription.10,11,12,13,14 In E. coli, the topoisomerase gyrase is limiting, such that at highly transcribed genes, the dynamic accumulation and release of positive supercoils stochastically switch genes off and on, thereby causing transcriptional bursting.10 Whether eukaryotic topoisomerase levels are limiting and how positive and negative DNA supercoiling control transcriptional bursting in eukaryotes is still unknown.
Transcription-generated supercoils can propagate along the DNA and may activate or deactivate adjacent genes. For multiple bacterial species, negative supercoils generated behind polymerase enhance transcription of upstream divergent genes, whereas positive supercoils in front of polymerase inhibit the transcription of downstream tandem and convergent genes.15,16,17,18,19 Similar mechanisms were proposed in eukaryotes, but direct in vivo evidence is lacking.20,21,22,23,24,25,26,27 In contrast to bacterial DNA, eukaryotic DNA is wrapped in nucleosomes, which may buffer excess positive supercoils to limit their dissipation.28,29 However, chromatin does not absorb negative supercoils.30 Accordingly, negative supercoils propagate up to 1.5 kb around the transcription start site of transcribed genes.23 Whether in vivo negative supercoils enhance the transcription of divergent genes and whether positive supercoils are efficiently buffered by nucleosomes to prevent the inhibition of tandem and convergent genes remain unclear. Additionally, since these previous studies relied on population-based transcription assays, it is unclear whether the transcriptional bursting of adjacent genes is temporally coupled in single cells and how DNA supercoils affect their temporal relationship.
Supercoiling-mapping studies observed that gene bodies are positively supercoiled, and promoters are negatively supercoiled.23,31,32,33 Promoters are maintained in a negatively supercoiled state by restricting topoisomerase TOP1 activity to gene bodies.34 In addition, mammalian genomes contain large negatively supercoiled domains of actively transcribed genes.33 However, since the mapping of supercoils at the single-cell level is technically challenging, little is known about the differences in supercoiling states between cells at a single time point or how supercoiling dynamics affect the transcription dynamics of single eukaryotic cells over time.
In this study, we used the closely positioned and highly expressed divergent (GAL1-GAL10) and tandem (GAL10-GAL7) gene pairs of the GAL gene cluster in S. cerevisiae to investigate how DNA supercoiling affects transcriptional bursting of neighboring eukaryotic genes (Figure 1A). Using single-molecule dual-color imaging, we found that transcriptional bursting of the GAL gene pairs is temporally correlated inside single cells and that yeast topoisomerases are essential for maintaining the correlation. Topoisomerase degradation results in transcription inhibition at neighboring genes that is likely a consequence of the accumulation of both positive and negative supercoils. The resulting transcription inhibition is caused by destabilized binding of the transcription factor (TF) Gal4. Moreover, we find that wild-type (WT) budding yeast has sufficient concentrations of topoisomerases to minimize the inhibition of DNA supercoiling on transcription, implying that DNA supercoils play different regulatory roles on gene transcription in prokaryotes versus eukaryotes.
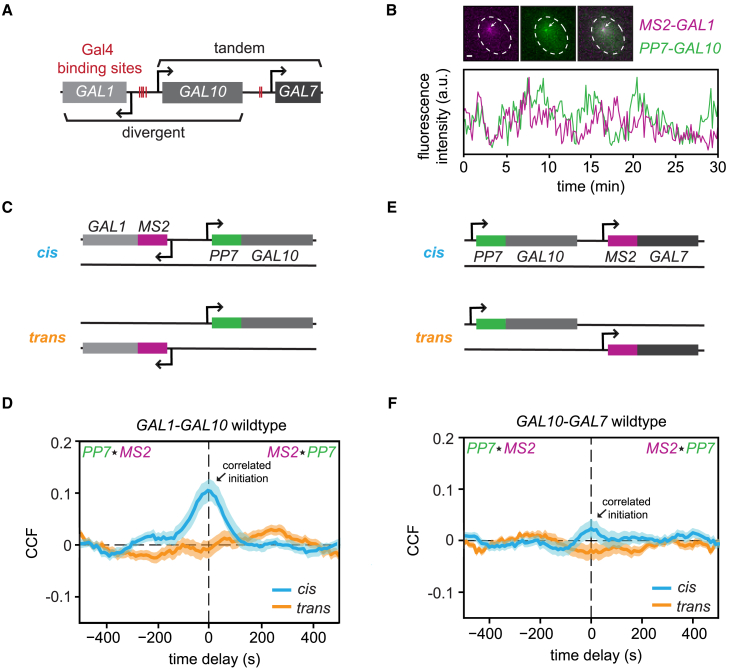
Figure 1.
Transcriptional bursting of the divergent and tandem GAL genes is temporally coupled
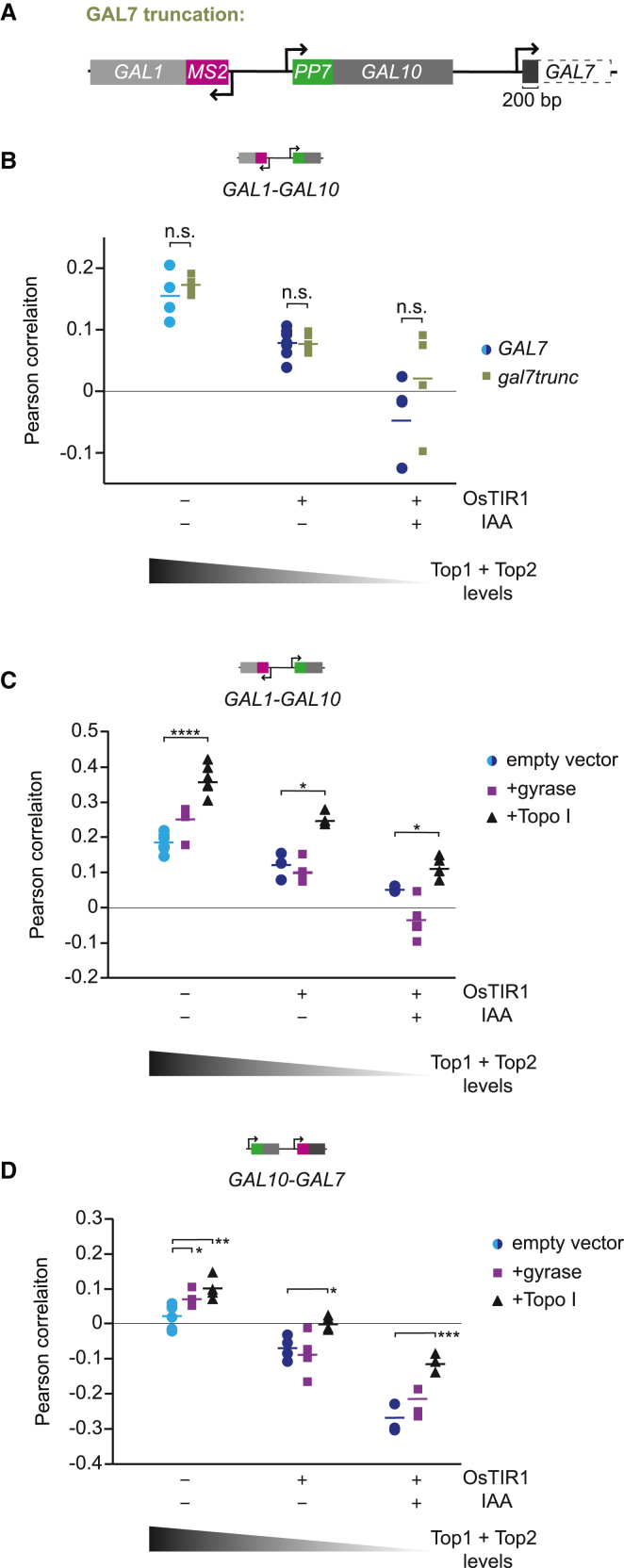
(A) Schematic of GAL gene cluster in yeast. Red lines indicate the binding sites of the transcription factor, Gal4. The gene lengths of GAL1, GAL10, and GAL7 are approximately 1.5, 2.1, and 1.1 kb, respectively, with intergenic distances of 669 and 726 bp. All three genes are highly transcriptionally active in galactose-containing media and produce high levels of DNA supercoils.31 In galactose, the antisense transcripts at this locus are not transcribed.35
(B) Example images of MS2-GAL1 (magenta), PP7-GAL10 (green), and merged (gray) transcription sites (TSs), indicated by arrows (top). Scale bars, 1 μm. Example traces of the quantified fluorescence intensities (arbitrary units) of the MS2-GAL1 and PP7-GAL10 TSs (bottom).
(C) Nascent transcription of GAL1-GAL10 is visualized either on the same allele (cis) or different alleles (trans).
(D) MS2-PP7 cross-correlation of GAL1-GAL10 in the cis (blue, n = 179 cells) and trans (orange, n = 162 cells) configuration. Shaded area indicates SEM. MS2⋆PP7 indicates the CCF of MS2(t) to PP7(t+τ) and PP7⋆MS2 of MS2(t−τ) to PP7(t).
(E) Same as (C) for GAL10-GAL7.
(F) Same as (D) for GAL10-GAL7. Cis: n = 148 cells; trans: n = 125 cells.
See also Figures S1 and S2 and Video S1.
Results
Transcriptional bursting of the divergent and tandem GAL genes is temporally coupled
To understand how neighboring GAL genes are dynamically transcribed inside single cells, we first visualized nascent transcription of the divergent GAL gene pair with single-molecule resolution in live cells by inserting 12xMS2V6 repeats and 14xPP7 repeats at the 5′ of GAL1 and GAL10, respectively (Figures 1B and 1C).36,37 Upon transcription, these repeats form loops that are specifically bound by the fluorescently tagged MS2 and PP7 coat proteins, allowing for nascent RNA visualization at the endogenous loci in living cells. The divergent genes were labeled on the same chromosome (cis configuration) or on two different chromosomes (trans configuration) (Figure 1C) to distinguish between local environment effects and extrinsic noise effects, such as correlations generated by cell-to-cell variations.
To determine whether the transcriptional bursting of the GAL1-GAL10 genes was temporally coupled, we imaged live cells that were induced with galactose, quantified the intensities of the MS2-GAL1 and PP7-GAL10 transcription sites (TSs), and computed the MS2-PP7 cross-correlation function (CCF) (Figures 1B, 1D, and S1A–S1C). The CCF of the MS2 and PP7 time traces was calculated by shifting one trace with respect to another trace by defined time delays, yielding a measure of similarity between MS2-GAL1 and PP7-GAL10 signals at various time delays. The MS2-PP7 CCF of the cis-labeled genes displayed a defined peak at time delay zero, indicating that GAL1 and GAL10 initiate together more than expected by random chance (Figure 1D). The CCF decayed to zero at time delays of approximately −100 and +100 s, which was in accordance with the decay of GAL1 and GAL10 from the auto-correlation functions (ACFs) (Figures S1B and S1C), indicating that after a simultaneous burst, GAL1 and GAL10 transcription is uncorrelated. The CCF of the trans-labeled divergent genes yielded a flat line (Figure 1D), as expected for independently expressed, uncorrelated genes on different chromosomes. To understand the magnitude of the correlation at zero time delay, we quantified the normalized transcriptional overlap, which represents the percentage of co-occurring GAL1-GAL10 transcription events when GAL10 is active (STAR Methods).38 Although we observed a substantial random transcriptional overlap (66% ± 2%) for the trans control, the overlap of the cis configuration was significantly higher (79% ± 1%) (Figure S2C), demonstrating that the transcription of the divergent genes is more correlated in the cis configuration than in the trans and that transcription initiation of divergent genes on the same chromosome is temporally coupled.
Because the transcriptional overlap of the trans was already substantial, we investigated whether the difference between the cis and trans overlap would increase in conditions with lower galactose concentrations with reduced transcriptional activity, where the random overlap may be lower (Figures S1A–S1C). Despite the reduced transcriptional activity, both the cis and the trans transcriptional overlaps remained the same (Figures S2A–S2C), indicating that the overlap is similar across conditions and confirming that the normalized transcriptional overlap is independent of the transcriptional activity.
The cis and trans correlations were corroborated using single-molecule RNA fluorescence in situ hybridization (smFISH) with probes hybridizing to the MS2 and PP7 loops.39 As a measure for correlated transcription, we computed the Pearson correlation coefficient of the nascent transcript number at the MS2-GAL1 and PP7-GAL10 TSs across thousands of transcriptionally active cells (Figure S2D). Consistent with the live-cell results, GAL1-GAL10 transcription shows a higher Pearson correlation in the cis (R = 0.20 ± 0.02) than in the trans (R = 0.12 ± 0.02) configuration. When positioned on the same chromosome, the divergent GAL1-GAL10 genes thus initiate simultaneously, more than by random chance.
Next, the same approach was used for the tandem GAL10-GAL7 genes (Figures 1E, S1D–S1F, S2E, and S2F). The cis-labeled tandem genes are weakly correlated at time delay zero (Figure 1F) with a transcriptional overlap that is 6.0% ± 0.3% higher than the trans control (Figure S2E). This modest difference could not be confirmed by smFISH (Figure S2F), presumably because it is obscured by extrinsic noise. Overall, live-cell imaging and smFISH indicate that transcription of the divergent and tandem GAL gene pairs in WT yeast is temporally coupled.
Degradation of topoisomerases results in refractory periods
In yeast, transcription-generated supercoiling levels are managed by topoisomerases, Top1 and Top2, which can release both positive and negative supercoils.40 To investigate how DNA supercoiling affects the transcription dynamics and the temporal correlation of the GAL gene pairs, we perturbed DNA supercoiling levels by conditionally degrading endogenous Top1 and Top2 using the auxin-inducible degron system41 (Figure 2A). Strains containing degron-tagged Top1 and Top2, without OsTIR1 expression, showed a similar GAL1-GAL10 correlation as WT, suggesting that tagging does not influence their function (Figure S2D). Homozygous expression of OsTIR1, even without the addition of auxin (−IAA), resulted in basal degradation (44% ± 1%) of the degron-tagged topoisomerases, as measured using the cMyc-tag on Top2 (Figures S2G and S2H). Top1 did not contain a cMyc-tag to monitor its degradation, but since the degron tag was the same, we assumed that its degradation was similar. The addition of auxin (+IAA) resulted in almost complete degradation (89% ± 2%) within 15 min (Figures S2G and S2H). We will refer to the basal and complete degradation conditions as partial (−IAA) and full (+IAA) topoisomerase degradation.
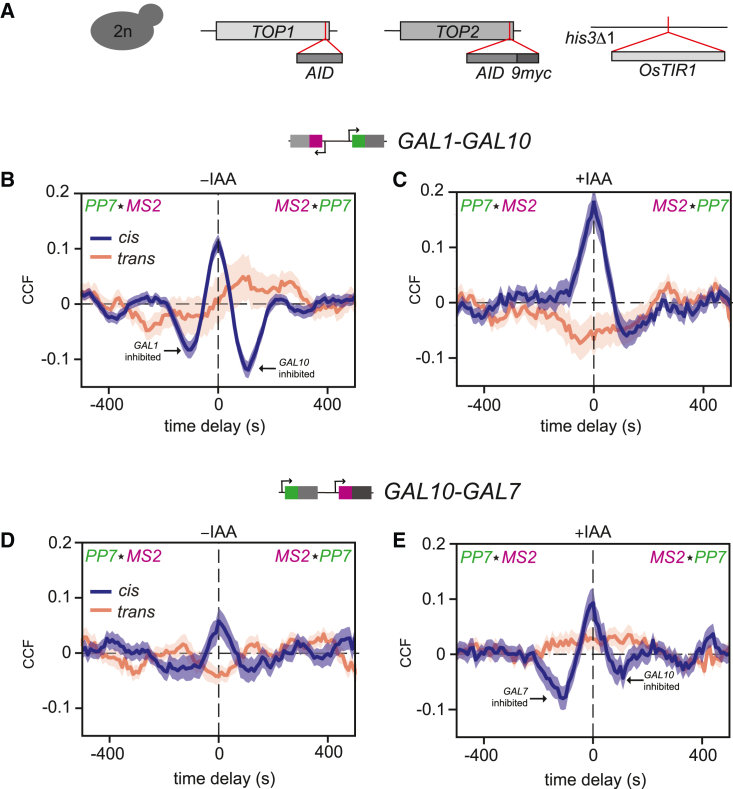
Figure 2.
Degradation of topoisomerases results in refractory periods
(A) Schematic depicting the homozygous tagging of diploid yeast (2n) of endogenous TOP1 and TOP2 with an auxin-inducible degron, and homozygous OsTIR1 insertion at the his3Δ1 locus.
(B and C) MS2-PP7 cross-correlation of the GAL1-GAL10 in cells without (−IAA, left, n = 214 cells [cis], n = 126 cells [trans]) and with auxin (+IAA, right, n = 158 cells [cis], n = 112 cells [trans]) for the cis (dark blue) and trans (orange). Arrows indicate example valleys indicating refractory periods. Shaded area indicates SEM.
(D and E) Same as (B) and (C) for GAL10-GAL7. −IAA: n = 118 cells (cis), 102 cells (trans), +IAA: n = 143 cells (cis), n = 128 cells (trans).
See also Figures S1 and S2.
To understand how DNA supercoiling affects the transcription dynamics of the GAL genes, we performed live-cell imaging of GAL1-GAL10 and GAL10-GAL7 transcription on partial and full topoisomerase degradation (Figures S1A and S1D). Both topoisomerase degradation conditions resulted in a complete loss of GAL gene transcription in a large fraction of the population (Figures S2I and S2J). The active fraction expressing both GAL1 and GAL10 on topoisomerase degradation is lower in the trans than in the cis configuration (Figure S2I), suggesting that transcription activation may be impaired at divergent genes with different gene lengths from MS2/PP7 addition. In subsequent analysis, only transcriptionally active cells were analyzed, but these represented only 30% of the population in some conditions. In these active cells, transcription levels were up to 50% reduced after topoisomerase degradation, as measured from the inverse ACF amplitudes (Figure 3A). Topoisomerase degradation thus causes a large reduction in GAL gene transcription in the population.
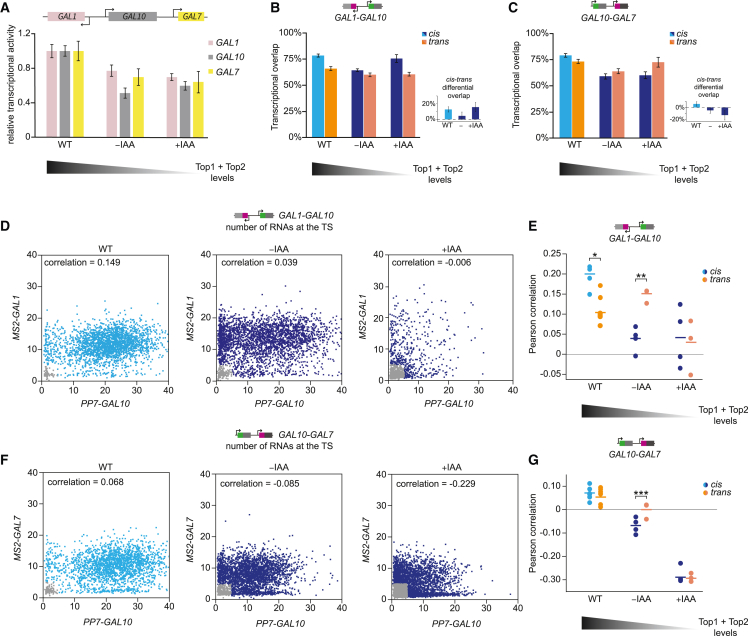
Figure 3.
Degradation of topoisomerases reduces the simultaneous initiation of neighboring genes
(A) Relative transcriptional activity of GAL1, GAL10, and GAL7 in WT and partial (−IAA) and full (+IAA) topoisomerase degradation conditions, calculated by taking the inverse of the ACF amplitudes. WT is normalized to 1. Error bars indicate SEM.
(B and C) Transcriptional overlap of cis and trans conditions for WT, −IAA, and +IAA conditions for (B) GAL1-GAL10 and (C) GAL10-GAL7 computed from the CCF at zero time delay from live-cell experiments. Error bars indicate SEM. The insets show the cis-trans overlap difference for each condition.
(D) Example scatterplots of the number of nascent transcripts at MS2-GAL1 and PP7-GAL10 TSs, determined by smFISH, in WT (left, n = 2,602 cells), −IAA (middle, n = 3,096 cells), and +IAA (right, n = 1,112 cells) for the cis. Each datapoint represents a cell. Scatter plots show one representative replicate out of multiple replicate experiments (n = 4;4;4, from left to right). From these plots, the GAL1-GAL10 active fraction (Figure S2P) and the GAL1-GAL10 Pearson correlation was calculated (E). Gray data points represent transcriptionally inactive cells that were excluded from the Pearson correlation coefficient calculation.
(E) Pearson correlation coefficients of GAL1-GAL10 nascent transcription from smFISH for cis and trans conditions in WT, −IAA, and +IAA conditions. Each circle represents a single smFISH replicate experiment, as shown in (D) (n = 4;6;4;3;4;3, from left to right with each replicate >500 cells). Horizontal lines represent means. Significance was calculated between cis and trans for each condition. Only significant bars are shown. ∗p < 0.05; ∗∗p < 0.01, determined by two-tailed t test.
(F) Same as (D), for MS2-GAL7 and PP7-GAL10 in WT (left, n = 2,865 cells), −IAA (n = 3,815 cells), and +IAA (right, n = 4,544 cells).
(G) Same as (E), for GAL10-GAL7 (n = 7;7;4;3;4;3, from left to right). ∗∗∗p < 0.001.
See also Figures S1–S3.
Next, we explored how topoisomerase degradation affected the GAL gene coupling by calculating the CCFs. For GAL1 and GAL10, partial topoisomerase degradation introduced valleys in the CCF around −100 and +100 s time delays that were not present in the trans (Figure 2B). These valleys indicate that 100 s after a correlated burst, simultaneous transcription of the two genes is observed less often than expected by random chance, suggesting that supercoils that accumulate from transcription inhibit subsequent transcription of its neighbor. Such periods of lower transcription initiation rates immediately after a transcriptional burst have been referred to as refractory periods.42,43,44 In this manuscript, we employ the same nomenclature, regardless of whether the refractory period follows the transcription of the gene itself or neighboring genes. A GAL1 transcriptional burst, either alone or together with GAL10, thus causes a refractory period for GAL10 and vice versa. The CCF also showed recurring peaks every 200 s, indicating periodicity (Figure 2B), likely resulting from the refractory period. Similar but weaker valleys were observed in the ACFs, most prominently for GAL10, indicating GAL10 transcription may also weakly inhibit itself (Figures S1B and S1C). On full topoisomerase degradation, the refractory periods and periodicity for GAL1-GAL10 were partially alleviated (Figure 2C), possibly due to the concurrent reduction in the transcriptional activity of all GAL genes (Figure 3A). As bursts become less frequent (Figure S2M), inhibition from a neighboring gene is expected to interfere less with transcription patterns. In both partial and full depletion conditions, inhibition was stronger for GAL10 than for GAL1 (Figures 2B and 2C).
For GAL10-GAL7, similar refractory periods were observed, with especially strong GAL7 inhibition after full topoisomerase degradation (Figures 2D and 2E). Because of the well-established inhibitory role of positive supercoils, we expect that positive supercoils generated from GAL10 elongation may inhibit subsequent GAL7 initiation.10,19,32 Interestingly, a similar but much weaker valley at −100 s delay is already visible in WT (Figure 1F), suggesting that WT topoisomerase levels are just sufficient to minimize supercoiling-mediated inhibition. Overall, these data indicate that topoisomerase degradation causes refractory periods where transcription of a GAL gene inhibits subsequent transcription at neighboring genes.
Because the inhibition effects appear dependent on topoisomerase concentration, we constructed strains with heterozygous (1xOsTIR1) instead of homozygous (2xOsTIR1) OsTIR1 expression to explore the concentration dependence further. In these strains, both Top1 and Top2 were cMyc-tagged, confirming that both were equally degraded (Figures S2G and S2H). Heterozygous rather than homozygous OsTIR1 expression not only reduced the basal degradation to 23% ± 1% but also showed slightly less degradation on auxin addition (Figure S2H). These strains yielded intermediate phenotypes (Figures S2N and S2O). For GAL1-GAL10, refractory periods were stronger at partial than at full topoisomerase degradation with more GAL10 than GAL1 inhibition (Figures S1B, S1C, and S2N), whereas for GAL7-GAL10, refractory periods were stronger at full degradation with more GAL7 than GAL10 inhibition (Figures S1E, S1F, and S2O). Overall, these results indicate that topoisomerases are important for maintaining high GAL transcription and that transcription-generated DNA supercoiling accumulation limits subsequent transcription of neighboring genes. These inhibitory effects depend on topoisomerase concentration and gene orientation.
Degradation of topoisomerases reduces the simultaneous initiation of neighboring GAL genes
The refractory periods at neighboring genes upon supercoiling accumulation suggested that the divergent and tandem GAL genes initiate together less often than in WT. Although the amplitude of the CCF-peak appeared to increase in topoisomerase-deficient conditions compared with WT (Figures 1D, 1F, and 2B–2F), suggesting increased co-bursting, this amplitude reflects the correlation during both active and inactive periods and is thus confounded by reduced transcriptional activity (Figure 3A). To assess co-bursting during active periods only, we quantified the normalized transcriptional overlap of GAL1-GAL10 and GAL10-GAL7 upon topoisomerase degradation. We observed that the overlap of GAL1-GAL10 cis, compared with trans, was reduced upon partial topoisomerase degradation (Figure 3B). Similar to the effects on the refractory periods, this loss in overlap was alleviated at full degradation conditions (Figure 3B). For GAL10-GAL7, the cis overlap progressively decreased compared with trans with increased degradation (Figure 3C). The reduced overlap indicated that rapid release of DNA supercoils by topoisomerases is essential to maintain coupling of the GAL genes.
To confirm this overlap reduction, we performed smFISH experiments after topoisomerase degradation. Similar to the live-cell experiments, topoisomerase degradation strongly reduced the transcriptional activity and transcriptionally active fraction (Figures 3D, 3F, S2P, and S2Q). However, in contrast to the transcriptional overlap measure, we noticed that the Pearson correlation is not a normalized measure but depended on transcriptional activity. In the trans control, with reduced transcriptional activity at partial and full degradation, we also observed a decrease in the GAL1-GAL10 and GAL10-GAL7 correlations (Figures 3E and 3G). Nevertheless, partial degradation significantly decreased the correlation of the cis compared with the trans for both gene pairs (Figures 3E and 3G). The decreased GAL1-GAL10 and GAL10-GAL7 correlations were also corroborated in untagged WT strains using gene-specific smFISH probes (Figures S3A and S3B). The basal Top1 and Top2 degradation and its associated phenotype could partially be rescued by the addition of the antagonist, auxinole, confirming the specificity of the effects (Figures S3C–S3N). These results indicate that the divergent and tandem GAL genes initiate together less frequently when supercoils accumulate at the locus. For the divergent genes, the reduced correlation challenges previous models, which had predicted that in eukaryotes, the accumulation of negative supercoils in divergent promoters enhances the correlation.22,23,24,25,45 The specific reduction of the GAL1-GAL10 overlap upon partial topoisomerase degradation (Figure 3B) instead suggests a more complicated model at this locus, where DNA supercoiling accumulation results in mutual inhibition between GAL1 and GAL10 and reduced simultaneous transcription.
To understand why supercoiling accumulation reduced simultaneous transcription, we focused on the transcriptional activities of the three GAL genes. Complete topoisomerase degradation reduced the transcription of all three genes, whereas partial topoisomerase degradation inhibited GAL10 transcription more than GAL1 and GAL7 (Figure 3A). Analysis of the bursting parameters using binarized MS2/PP7 traces revealed that topoisomerase inhibition results in shorter-duration, lower-intensity, and lower-frequency bursts for all three genes (Figures S2K–S2M). Burst intensity was already maximally affected at partial degradation (Figure S2K), but burst duration and frequency showed topoisomerase dose-dependent effects (Figures S2L and S2M), with the largest effects at GAL10. The uneven GAL10 inhibition was supported by smFISH, in which the percentage of actively transcribing cells upon partial topoisomerase degradation was reduced more for GAL10 than for GAL1 (Figure S2P). The GAL10 ACF also showed a prominent refractory period, which was not as evident for GAL1 and GAL7 (Figures S1B, S1C, S1E, and S1F). The specific inhibition of GAL10 may explain the GAL1-GAL10 correlation loss upon partial topoisomerase degradation. We conclude that topoisomerases ensure correlated transcription between the GAL gene pairs.
Transcription of GAL7 inhibits GAL10 transcription upon partial topoisomerase depletion
We hypothesized that the disproportionate inhibition of GAL10 transcription upon partial topoisomerase degradation (Figure 3A) is caused by interference from the highly expressed downstream gene GAL7. To test this hypothesis, we used two complementary approaches to limit the possible effects of GAL7 transcription (Figures 4A and 4B).
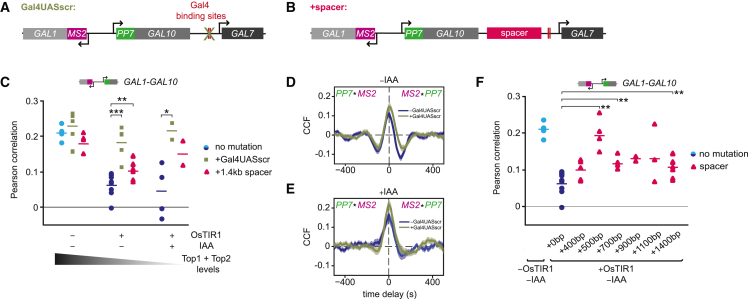
Figure 4.
Transcription of GAL7 inhibits GAL10 transcription in partial topoisomerase degradation conditions
(A and B) Schematic of the GAL1-GAL10 cis-labeled locus with: (A) scrambled Gal4UAS sites (Gal4UASscr) in the GAL7 promoter and (B) insertion of a spacer sequence in the GAL10-GAL7 intergenic region.
(C) Pearson correlation coefficients of GAL1-GAL10 nascent transcription from smFISH of −OsTIR (blue circles), −IAA and + IAA (navy circles) cells with Gal4UASscr (green squares) and insertion of a 1.4 kb spacer (magenta triangles). Horizontal lines represent mean. Each symbol represents a single smFISH replicate experiment (n = 4;6;4;7;6;5;4;2;2, from left to right with each replicate >500 cells). Significance was calculated between without and with Gal4UASscr/spacer for each condition. Only significant bars are shown. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001, determined by two-tailed t test.
(D and E) Overlay of MS2-PP7 cross-correlation of GAL1-GAL10 in topoisomerase-deficient cells (top, −IAA, n = 224 cells; bottom, +IAA, n = 135 cells) with Gal4UASscr (green) and with WT Gal4UAS (navy; same as Figure 2C). Shaded area indicates SEM.
(F) Same as (C) for GAL1-GAL10 with increasing spacer sequence lengths (pink triangles) (n = 4;7;5;4;4;2;3;8).
See also Figure S4.
First, transcription of GAL7 was abolished by scrambling both Gal4 upstream activating sequences (UASs) in the GAL7 promoter (Figures 4A and S4A). Abrogating GAL7 transcription in this mutant was expected to increase GAL10 expression and therefore increase the correlation between GAL1 and GAL10. As predicted, smFISH after partial and full topoisomerase degradation showed an increase in the GAL10 active fraction (Figure S4I) and a rescue in the GAL1-GAL10 correlation to WT (Figure 4C). Live-cell imaging of the GAL1-GAL10 genes upon loss of GAL7 transcription showed dampened valleys at +100 s time delay and loss of periodicity in the GAL10 ACF and GAL1-GAL10 CCF, revealing a weaker GAL10 refractory period, which was most evident in the partial degradation condition (Figures 4D, 4E, and S4B–S4G). GAL7 inhibition also partially rescued the GAL1-GAL10 transcriptional overlap at zero time delay after partial degradation (Figure S4H). In full degradation conditions, where transcription inhibition from neighboring genes was already less evident (Figures 2C and 3B), the GAL1-GAL10 overlap was unaffected by GAL7 inhibition (Figure S4H). These results demonstrate that transcription of GAL7 inhibits GAL10 transcription when topoisomerase levels are reduced. Interestingly, in cells with WT topoisomerase levels (−OsTIR1), elimination of GAL7 transcription did not affect the GAL10 active fraction (Figure S4I), nor the GAL1-GAL10 correlation (Figure 4C), suggesting that WT cells possess sufficient topoisomerase levels to prevent the inhibition of GAL7 on GAL10 transcription.
As a second method to test whether GAL7 transcription inhibits GAL10 transcription, a 1.4 kb spacer sequence was inserted between GAL10 and GAL7 to dissipate transcription-generated supercoils (Figure 4B). In cells with partial topoisomerase degradation, the addition of a spacer increased the GAL10 active fraction (Figure S4J) and partially rescued the GAL1-GAL10 correlation (Figure 4C), corroborating supercoiling-mediated inhibition of GAL10. Similar to the Gal4 UAS perturbation, spacer addition did not affect the GAL1-GAL10 correlation in cells with WT topoisomerase levels (−OsTIR1) (Figure 4C). Moreover, the insertion of spacers with various lengths revealed an optimal intergenic distance of 500 bp that fully rescues the GAL1-GAL10 correlation with partial rescues at other distances (Figure 4F). The reason for this optimal distance is unclear. Overall, these perturbations demonstrate that GAL7 transcription inhibits GAL10 transcription in partial topoisomerase depletion, but not in WT conditions.
Transcription inhibition at the GAL locus is caused by both positive and negative supercoils
We next explored how GAL10 transcription is inhibited by GAL7 transcription. We hypothesized that GAL10 is inhibited either by negative supercoils traveling upstream of the GAL7 promoter or by positive supercoils generated by GAL10 that have limited space to dissipate while GAL7 is being transcribed. To distinguish between these two models, we truncated the GAL7 gene body from 1,100 to 200 bp to specifically reduce the amount of GAL7 transcription-generated negative supercoils without changing the distance for GAL10 supercoils to dissipate (Figure 5A). Productive transcription of the truncation was verified by smFISH (Figures S5A and S5B). In cells with WT topoisomerase levels (−OsTIR1) and with partial topoisomerase degradation, GAL7 truncation did not affect the GAL1-GAL10 correlation nor the GAL1/GAL10 active fraction (Figures 5B and S5C). On full topoisomerase degradation, we observed a small rescue of the fraction of GAL1-GAL10 transcribing cells (Figure S5C), but the correlation was not significantly changed (Figure 5B). These results suggest that upon partial degradation, GAL10 is mainly inhibited by positive supercoils generated by its own transcription. Binding of the transcriptional machinery in the GAL7 promoter may thus create a barrier that limits dissipation of transcription-generated positive DNA supercoils. In full-degradation conditions, negative supercoils from GAL7 transcription may also contribute to the inhibition.
Figure 5.
Transcription inhibition at the GAL locus is caused by both positive and negative supercoils
(A) Schematic of the GAL1-GAL10 cis-labeled locus with a truncation of the GAL7 gene body from 1,100 to 200 bp.
(B) Pearson correlation coefficients of GAL1-GAL10 nascent transcription by smFISH for WT GAL7 (blue) and GAL7 truncation (green) in −OsTIR1, −IAA, and +IAA cells. Each symbol represents a single smFISH replicate experiment (n = 5;5;7;5;6;4, from left to right with each replicate >500 cells). Statistical significance between WT and truncated GAL7 was determined by two-tailed t test.
(C) Same as (B) for GAL1-GAL10 genes with ectopic expression of gyrase (purple squares) and Topo I (black triangles) (n = 6;4;7;3;6;3;3;4;4, from left to right). Significance was calculated between empty vector and gyrase/Topo I for each condition. Only significant bars are shown. ∗p < 0.05; ∗∗∗∗p < 0.0001, determined by two-tailed t test.
(D) Same as (C) for GAL10-GAL7 (n = 6;3;4;5;4;4;3;3;3). ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
See also Figure S5.
To further dissect inhibition by negative and positive supercoiling, we ectopically overexpressed bacterial topoisomerases gyrase or DNA topoisomerase I (Topo I), which selectively relieve positive or negative supercoils, respectively, and were shown to work in yeast.31,46,47 Relieving excess positive supercoils with ectopic gyrase expression only weakly increased the GAL10-GAL7 gene correlation in cells with WT topoisomerase levels (Figure 5D) but did not affect the correlation of GAL1-GAL10 or GAL10-GAL7 in partial or full topoisomerase degradation conditions (Figures 5C and 5D). In contrast, overexpression of bacterial Topo I to relax excess negative supercoils considerably increased the correlation of both gene pairs (Figures 5C and 5D). A small subpopulation exhibited higher GAL gene expression than WT (Figures S5F and S5G), partly explaining the increased GAL1-GAL10 and GAL10-GAL7 correlations. In these overexpression experiments, the timing of Topo I induction coincides with the timing of the observed effects on the correlations, arguing against possible confounding indirect effects (Figures S5D and S5E). Taken together, we conclude that at the GAL locus, transcription is likely inhibited by an accumulation of both negative and positive supercoils.
Supercoiling-mediated inhibition is caused by neither altered chromatin structures nor increased R-loops
Previous studies have shown that supercoiling accumulation can change nucleosome stability.7,9 To gain mechanistic insight into the supercoiling-mediated transcription inhibition by nucleosome position and stability, we performed micrococcal nuclease digestion, followed by sequencing (MNase-seq) in haploid cells (Figures S6A and S6B) using high and low MNase concentrations to map stable and fragile nucleosomes, respectively.48 Fragile nucleosomes are partially unwrapped nucleosomes, bound by the remodeler RSC (remodeling the structure of chromatin), that also occur at the Gal4 UASs in the GAL1-GAL10 promoter.48,49,50
In partial topoisomerase degradation conditions, the position of stable and fragile nucleosomes in the GAL gene promoters was unchanged (Figures S6C and S6D), despite the transcription inhibition in these conditions. Only at full topoisomerase degradation did minor shifts in the nucleosome position appear at the locus, for example at the GAL1 TATA. Similarly, only full topoisomerase degradation resulted in less well-positioned stable nucleosomes genome-wide (Figure S6E). However, since these effects were not observed on partial topoisomerase degradation, we conclude that changed nucleosome positioning or stability is not the main cause of the observed transcription inhibition on supercoiling accumulation.
Next, we reasoned that the accumulation of negative supercoils in gene bodies may lead to the formation of R-loops,11,31,51,52 which may inhibit transcription initiation or cause premature termination. To test the influence of R-loops, we ectopically overexpressed human RNaseH (hsRNH1), which was shown to reduce R-loops in yeast.53 Contrary to our hypothesis, resolving potential excess R-loops did not rescue the GAL1-GAL10 correlation nor the GAL1/GAL10 active fraction by smFISH (Figures S6F and S6G). We therefore conclude that supercoiling-mediated transcription inhibition of the GAL genes is caused by neither altered chromatin structures nor increased R-loops.
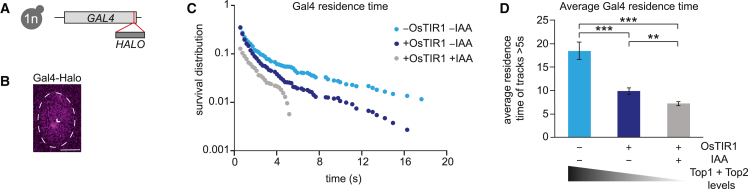
Accumulation of DNA supercoils reduces the Gal4 DNA residence time
Population-based ChIP measurements have suggested that DNA supercoiling accumulation reduces TF binding to DNA.13 To investigate whether supercoiling accumulation affects the binding stability of the TF Gal4, we used single-molecule tracking (SMT) of Gal4-HaloTag to measure the DNA residence time of individual Gal4 molecules in living cells (Figures 6A and 6B). Similar to our previous measurements,35 the semi-log survival probability distribution in WT suggested two populations with different residence times (Figure 6C). We showed that the long-bound molecules are specifically bound TFs that correlate with active transcription.35 Comparison of the survival probability distributions between cells with decreasing topoisomerase levels revealed a progressive reduction in the Gal4 residence times (Figure 6C). Fitting of the survival distributions revealed that WT and partial topoisomerase degradation were best fit by a different function (exponential + power-law) than full degradation (biexponential) and that the fits deviated from the data at long residence times, especially after topoisomerase degradation (Figures S7A–S7C). We therefore refrained from model fitting and simply calculated the average residence time of long-bound molecules (>5 s) (Figure 6D). Partial and full topoisomerase degradation conditions exhibited shorter Gal4 residence times on the DNA. Since Gal4 residence time has been directly linked to the GAL gene burst duration,35 these results suggest that the accumulation of DNA supercoils inhibit transcription by reducing the residence time of Gal4.
Figure 6.
Supercoiling accumulation reduces the Gal4 residence time on DNA
(A) Schematic of the C-terminal HaloTag at the endogenous GAL4 in haploid yeast (1n).
(B) Representative image of a yeast cell showing a single JFX650-labeled Gal4-HaloTag molecule (arrowhead). Scale bars, 2 μm.
(C) Survival probability distributions for Gal4 residence times in −OsTIR −IAA (323 cells, 706 tracks; 33,520 particles) and +OsTIR −IAA (244 cells, 1,378 tracks; 53,430 particles), and +OsTIR +IAA (107 cells, 187 tracks; 13,465 particles) cells.
(D) Average residence time of Gal4 calculated from trajectories with residence times >5 s for the indicated conditions. Errors indicate SEM, determined by bootstrapping with 300 repeats. ∗∗p < 0.01; ∗∗∗p < 0.001.
See also Figures S6 and S7 and Video S2.
DNA supercoils inhibit neighboring genes of all orientations genome-wide
To understand whether supercoiling-mediated inhibition is also observed genome-wide, we performed cross-linking analysis of cDNAs, followed by sequencing (CRAC-seq) of HTP(His6-TEV-ProteinA)-tagged Rpb1, the largest subunit of RNA Pol II, to obtain high-resolution genome-wide maps of nascent transcripts for WT, partial and full degradation conditions (Figures S7D and S7E). To determine whether genes were inhibited by transcription of their neighbor, we analyzed changes in the RNA Pol II CRAC signal between partial and full topoisomerase degradation and compared both with WT (Figures S7F–S7H, also see legends). Genes were classified by the distance to the neighboring gene (Figure S7I). Genes with a close-by neighbor (100–300 bp) showed a significant downregulation on full topoisomerase degradation, which was less prominent (and less or not significant) at longer distances (>400 bp). This inhibition at longer distances is in line with the observed inhibition at the GAL genes for intergenic distances greater than 700 bp. However, the GAL genes showed a stronger supercoiling-mediated decrease in transcription than observed genome-wide, possibly due to the higher transcriptional activity and therefore likely higher DNA supercoiling production compared with most other yeast genes.54
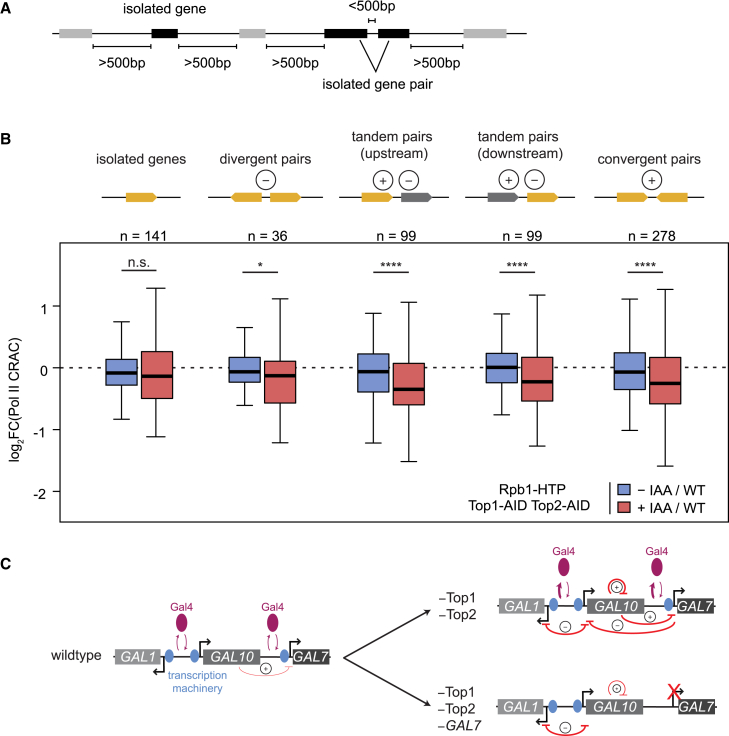
To analyze whether the orientation of the neighboring gene is important for its inhibitory effect, gene pairs were grouped according to their orientations (Figure 7A). We used a threshold of 500 bp for selecting “isolated” groups, since a longer distance threshold yielded an insufficient number of gene pairs (Figure S7J). At this threshold, there was no significant difference for isolated genes without a neighbor between partial and full topoisomerase degradation, although these isolated genes were weakly but significantly downregulated, compared with WT (Figure 7B). We therefore used the partial versus full degradation comparison to analyze the different gene groups. We observed a significant downregulation in transcription for isolated gene pairs of all orientations (Figure 7B). For isolated divergent genes, transcription is likely inhibited by negative supercoils from its neighbor. For tandem pairs, both positive supercoiling and negative supercoiling may contribute to the inhibition, similar to GAL10-GAL7. For isolated convergent gene pairs, the most likely source of inhibition is positive supercoils. Overall, we conclude that the supercoiling-mediated inhibition observed at the GAL locus occurs genome-wide at neighboring genes of all orientations, providing further evidence that both positive and negative supercoiling accumulations inhibit transcription.
Figure 7.
DNA supercoils inhibit neighboring genes genome-wide
(A) Schematic of isolated genes and gene pairs without a neighbor within 500 bp.
(B) Log2 fold change of Rpb1 CRAC signal in −IAA and +IAA conditions, compared with WT conditions for the gene pair orientations indicated at the top. The − and + signs indicate negative and positive DNA supercoils that are expected to inhibit transcription. Gene number and significance are indicated. The box indicates quartiles, the horizontal tick line the median, and the whiskers 1.5 times the interquartile range. Significance of partial versus full topoisomerase degradation is shown: ∗p < 0.05; ∗∗∗∗p < 0.0001, determined by paired t test. Significance of −IAA versus WT, from left to right: p = 0.021; p = 0.51; p = 0.042; p = 0.71; p = 0.031. Significance of +IAA versus WT, from left to right: p = 0.021; p = 0.041; p = 8.8e−6; p = 9.3e−4; p = 2.8e−6.
(C) Schematic of the proposed model. In WT (left), topoisomerase levels are sufficient to minimize supercoiling-mediated inhibition of the GAL genes. We only detect very weak inhibition of GAL7 transcription, likely from positive supercoils from GAL10 transcription. Upon topoisomerase degradation (right, top), accumulation of negative and positive supercoils from transcription inhibit transcription of neighboring genes by destabilizing binding of the transcription factor Gal4. Abrogation of GAL7 transcription (right, bottom) removes the transcription machinery barrier in the GAL7 promoter to allow dissipation of GAL10 supercoils, thereby partially rescuing GAL1-GAL10 co-bursting.
See also Figure S7.
Discussion
In this study, we combined single-molecule transcription imaging at neighboring GAL genes with targeted perturbations to expose how transcription-generated DNA supercoiling shapes transcription dynamics in budding yeast. We find that in WT, topoisomerases are sufficient to minimize supercoiling-mediated inhibition of the highly expressed GAL genes (Figure 7C, left). On topoisomerase degradation, the accumulation of both positive and negative DNA supercoils in the locus causes a temporally restricted expression pattern, where transcription initiation of a gene occurs simultaneously with its neighbor but is also inhibited by its neighbor during subsequent transcription events (Figure 7C, right top). This supercoiling-mediated refractory period results in a loss of correlated transcription of the GAL genes. Moreover, upon partial topoisomerase degradation, GAL10 is strongly inhibited by its own supercoils that cannot dissipate if the downstream GAL7 gene is transcribed (Figure 7C, right bottom), further reducing simultaneous GAL1-GAL10 transcription. Supercoiling accumulation inhibits transcription by destabilizing the binding of the TF Gal4. Overall, our data reveal that rapid supercoiling release is crucial to maintain high transcription levels and coordinate the transcription dynamics of neighboring eukaryotic genes.
In WT, transcriptional bursting of both the GAL gene pairs is coupled (Figures 1D and 1F). The degree of coupling is modest, but similar in magnitude to the co-bursting observed at paralogous genes in Drosophila.55 Similar co-expression of closely positioned divergent genes56,57,58 has fueled the prediction that negative supercoiling induces correlated transcription at neighboring genes similar to bacteria.21,24,27,59 Although we cannot exclude that DNA supercoiling contributes by a small degree to the simultaneous initiation in WT, our data suggest that excess supercoiling at the GAL locus mostly impedes, rather than facilitates, transcription initiation (Figures 2, 3, and S2). Instead, we propose that coupled GAL1-GAL10 initiation mostly originates from Gal4 binding to the shared UASs (Figure 1A). Since fluctuations in Gal4 binding directly cause fluctuations in GAL10 transcription,60 we expect that once Gal4 binds, it simultaneously activates GAL1 and GAL10. In addition, looping or 3D proximity of the GAL1-10 and GAL7 promoters may facilitate correlated Gal4 binding also at the tandem GAL genes.61,62 3D interactions between shared enhancers and co-regulated promoters cause co-bursting in Drosophila, perhaps by allowing coordinated TF binding.55 In line with this model, correlated TF binding has been observed for the serum response factor (SRF), resulting in correlated transcription of its target genes.57 Finally, simultaneous transcription initiation of adjacent genes may be caused by long-distance activation of TFs, TF clustering, or TF activity gradients.56,63,64,65
At the GAL locus, we find that the accumulation of both positive and negative supercoiling impedes transcription. The functional relevance of this inhibition is underscored by an increase in yeast fitness when GAL7 is relocated from the GAL locus to a different chromosome.66 In prokaryotes and eukaryotes, the inhibitory role of positive supercoils is well-established10,19,67 and is consistent with reduced genome-wide transcription of convergent and tandem genes on topoisomerase degradation (Figure 7B). Inhibition from positive supercoils is already weakly evident in WT cells at the tandem GAL10-GAL7 genes (Figure 1F). In addition, our data suggest that transcription inhibition also occurs by negative supercoils (Figures 4D, 5C, 5D, 7B, and S5C) In line with this, in mouse embryonic stem cells, a transient accumulation of negative supercoils during base excision repair was recently found to inhibit transcription and on release, causing increased noise fluctuations.68 The level of negative supercoiling thus requires careful regulation by topoisomerases, since low levels of negative supercoils enhance transcription,4,5,6,22,45,69 but hypernegative supercoiling is inhibitory.11,70,71,72 Transcription inhibition after topoisomerase degradation thus occurs through the accumulation of both positive and negative supercoils.
Positive supercoiling accumulation may inhibit transcription with a similar mechanism as in bacteria, by inhibiting both transcription initiation and elongation.10 How negative supercoils inhibit transcription was so far less clear. Here, we observe that topoisomerase degradation reduces the residence time of Gal4 (Figure 6), as well as the burst duration of the GAL genes (Figure S2L). Since the Gal4 dwell time determines the burst duration,60 it is conceivable that supercoiling-mediated transcription inhibition is caused by destabilized Gal4 binding. We envision that torsional stress in the DNA fiber from either positive or negative supercoiling results in a faster release of DNA-bound factors. Destabilization may also occur for other DNA-interacting proteins, such as factors of the preinitiation complex. Other mechanisms may also contribute to supercoiling-mediated transcription inhibition, such as small nucleosome occupancy changes on full topoisomerase degradation. Negative supercoils can cause the formation of alternative DNA structures such as Z-DNA, quadruplexes, or DNA cruciform,73 which may limit preinitiation complex formation. Moreover, supercoils have been suggested to affect transcription post-initiation, for example, by forming excess R-loops that cause premature termination.11,51 However, we do not find evidence for R-loop-mediated transcription inhibition or slowed elongation. We therefore propose that the DNA supercoiling at the GAL locus predominantly causes inhibition of transcription initiation by destabilized Gal4 binding, rather than inhibition of post-initiation steps.
Unlike in bacteria, our data suggest that in budding yeast, topoisomerases are not present at limiting concentrations for transcription dynamics. First, the refractory period observed upon topoisomerase depletion is not or only weakly present in WT (Figure 1). Second, inhibition of GAL7 transcription or spacer addition in WT has no effect (Figure 4C). This fundamental difference in supercoiling regulation between bacteria and yeast may be the result of differences in the topoisomerase enzymes, as bacterial topoisomerases are specialized for positive or negative supercoils, whereas eukaryotic topoisomerases relieve both.74 This difference in topoisomerase enzymes may also explain why transcription in bacteria is mostly inhibited by positive supercoils, whereas transcription in yeast is inhibited by both. In addition, eukaryotes may buffer positive supercoils by nucleosomes.28,29 Nevertheless, our data indicate that eukaryotic topoisomerases are also not present in large excess since 25% basal degradation of topoisomerases already causes transcriptional effects. Topoisomerase levels may be tightly controlled to ensure that DNA supercoiling accumulation remains at a level that is beneficial for transcription while limiting harmful effects. The weak valleys in the GAL10-GAL7 CCF in WT (Figure 1F) suggest that topoisomerase levels are at the tipping point of this balance.
In more complex eukaryotes, transcription-generated negative supercoils contribute to cohesin extrusion and may therefore facilitate the formation of topologically associating domains (TADs).33,75,76 This mechanism assumes propagation of negative supercoils over much larger genomic distances than 1.5 kb, the distance at which negative supercoils were initially thought to spread.23,77 Although mammalian genes are spaced much further apart than that in yeast, supercoiling-dependent cohesin extrusion may cause supercoiling-effects from adjacent genes at larger distances. In more complex eukaryotes, the accumulation of negative supercoils during nuclear processes, such as base excision repair,68 could influence the transcription of genes throughout the TAD. Overall, our single-cell live-cell approach highlights how efficient release of torsional stress is necessary to prevent transcriptional inhibition of neighboring eukaryotic genes.
Limitations of the study
The lack of methods to measure DNA supercoils in single cells has prevented us from mechanistically linking the single-cell transcription dynamics to local DNA supercoiling changes. The population-based nature of current supercoiling assays limits the interpretation for a mixed population of inactive cells, and dynamically transcribing active cells, which may cancel out any effects of positive and negative supercoils within single cells.
Additionally, bleaching and phototoxicity limited the amount of excitation light and the number of time frames during which transcription could be measured in live cells, which likely introduced a detection limit. Interpretation of noisy and short traces from individual cells was therefore challenging. The current cross-correlation approach reliably detects average enrichments or depletions but may potentially mask rare subpopulations.
Finally, the Gal4 residence times measurements represented binding events to all genomic loci, and not specifically to the GAL locus. Nevertheless, since Gal4 only binds to 15 genomic binding sites,78 of which 6 are positioned at the GAL gene cluster, topoisomerase depletion likely reduces the Gal4 residence time at the GAL locus.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| IgG from rabbit serum | Sigma-Aldrich | Cat# I5006; RRID: AB_1163659 |
| c-Myc polyclonal | Thermo Fischer Scientific | Cat# PA5-85185; RRID: AB_2792331 |
| PGK1 monoclonal | Thermo Fischer Scientific | Cat# 459250;RRID: AB_2532235 |
| IRDye® 800CW anti-mouse | Li-cor | Cat# 925-32210; RRID: AB_2687825 |
| IRDye® 800CW anti-rabbit | Li-cor | Cat# 926-32211; RRID: AB_621843 |
| Chemicals, peptides, and recombinant proteins | ||
| Yeast Nitrogen Base w/o AA, Carbohydrate & w/ AS (YNB) (Powder) | US Biological | Cat# Y2025 |
| Drop-out Mix Complete w/o Yeast Nitrogen Base (Powder) | US Biological | Cat# D9515 |
| Bacto™ Agar | Thermo Fischer Scientific | Cat#214030 |
| Bacto™ Peptone | Thermo Fischer Scientific | Cat# 211677 |
| Bacto™ Yeast Extract, technical | Thermo Fischer Scientific | Cat# 288620 |
| D-Glucose | Sigma-Aldrich | Cat# 8270-10KG |
| D-Raffinose | Bio-Connect Life Sciences | Cat# OR06197_2kg |
| D-Galactose | Sigma-Aldrich | Cat# G0750-500G |
| 1× Tris-EDTA buffer pH 8.0 | Invitrogen | Cat# 12090015 |
| D-Sorbitol | Sigma-Aldrich | Cat# S6021 |
| Potassium phosphate monobasic (powder) | Sigma-Aldrich | Cat# P9791 |
| Potassium phosphate dibasic (powder) | Sigma-Aldrich | Cat# P8281 |
| B-Mercaptoethanol | Sigma-Aldrich | Cat# M6250 |
| Lyticase from Arthrobacter luteus (powder) | Sigma-Aldrich | Cat# L2524 |
| Ribonucleoside Vanadyl Complex (RVC; liquid) | NEB | Cat# S1402S |
| Formamide (deionized) | Sigma-Aldrich | Cat# F9037 |
| UltraPure™ SSC, 20X | Thermo Fisher Scientific | Cat# 15557044 |
| Dextran sulfate sodium salt | Sigma-Aldrich | Cat# 67578 |
| ProLong® Gold Antifade Mountant with DAPI | Invitrogen | Cat# P36935 |
| Sodium chloride (NaCl) | Sigma-Aldrich | Cat# S9888 |
| Glycerol | Sigma-Aldrich | Cat# G5516 |
| Phosphate-buffered saline (PBS) | Thermo Fisher Scientific | Cat# 18912014 |
| MyTaq Red Mix 2x | Bioline | Cat#: BIO-25044 |
| DMSO | Sigma | Cat#: D4540 |
| DNase I recombinant, RNase-free | Sigma-Aldrich (Roche) | Cat# 04716728001 |
| cOmplete EDTA-free protease inhibitor cocktail tablets | Sigma-Aldrich (Roche) | Cat# 11873580001 |
| Pefabloc SC-Protease-Inhibitor | Carl Roth | Cat# A154.3 |
| Dynabeads M-280 Tosylactivated | Thermo Fisher Scientific | Cat# 14204 |
| RNace-It Ribonuclease Cocktail | Agilent | Cat# 400720 |
| Recombinant GST-TEV protease | Challal et al.79 | N/A |
| Guanidine hydrochloride | Sigma-Aldrich | Cat# G4505 |
| Ni-NTA Agarose | Qiagen | Cat# 30230 |
| Imidazole | Sigma-Aldrich | Cat# I0125 |
| RNaseOUT Recombinant Ribonuclease Inhibitor | Thermo Fisher Scientific | Cat# 10777019 |
| T4 RNA Ligase 2, truncated KQ | NEB | Cat# M0373L |
| T4 Polynucleotide Kinase | NEB | Cat# M0201L |
| T4 RNA Ligase 1 (ssRNA Ligase) | NEB | Cat# M0204L |
| Proteinase K, recombinant, PCR grade | Sigma-Aldrich (Roche) | Cat# 03115887001 |
| SuperScript IV Reverse Transcriptase | Thermo Fisher Scientific | Cat# 18090050 |
| Exonuclease I | NEB | Cat# M0293S |
| RNase H | NEB | Cat# M0297S |
| LA Taq | Takara | Cat# RR002M |
| Zymolase 100T | US biological | Cat# Z1004.250 |
| Micrococcal nuclease | Sigma-Aldrich | Cat# N5386-200UN |
| Sorbitol | Sigma Aldrich | Cat# 1077581000 |
| Ammonium acetate solution | Sigma Aldrich | Cat# A2706 |
| NP-40 | Sigma Aldrich | Cat# 92016 |
| SDS | Sigma Aldrich | Cat# L3771 |
| Spermidine | Sigma Aldrich | Cat# S0266 |
| Phenol/chloroform (PCI 15:14:1) | Sigma-Aldrich | Cat# P2069-100ML |
| RNaseA/T1 | Thermo Fisher Scientific | Cat# EN0551 |
| Agarose MP | Sigma Aldrich | Cat# 11388991001 |
| 3-indole acetic acid (IAA, auxin) | Sigma Aldrich | Cat# I3750-100G-A |
| NuPAGE™ SDS Running buffer 20x | Thermo Fisher Scientific | Cat# LA0041 |
| NuPAGE™ 3-8%Tris-Acetate protein gels | Thermo Fisher Scientific | Cat#EA0375PK2 |
| Nitrocellulose membrane | Bio-rad | Cat# 1620112 |
| Auxinole | Sigma Aldrich | Cat# SML3231-25MG |
| JFX650 dye | Grimm et al.80 | N/A |
| Critical commercial assays | ||
| LightCycler FastStart DNA Master SYBR Green I | Roche | Cat# 12239364001 |
| LightCycler 480 SYBR Green I Master | Roche | Cat# 04887352001 |
| Qubit dsDNA HS Assay Kit | Thermo Fisher Scientific (Invitrogen) | Cat# Q32851 |
| Vivacon 500 | Sartorius | Cat# VN01H22 |
| QIAquick PCR Purification Kit | Qiagen | Cat# 28104 |
| PCR Isolate II PCR and Gel Kit | Bioline | Cat# BIO-52060 |
| Bioanalyzer High Sensitivity DNA kit | Agilent | Cat# 5067-4626 |
| ISOLATE II Plasmid Mini Kit | Bioline | Cat# BIO-52057 |
| KAPA HTP Library Preparation Kit | KAPA Biosystems | Cat# 07961901001 |
| Deposited data | ||
| MNase-seq data | this study | GEO: GSE196945 |
| Pol II CRAC-seq | this study | GEO: GSE217963 |
| Raw images and Western blots | this study | Mendeley Data: https://doi.org/10.17632/z2w34669gj.1 |
| Experimental models: Organisms/strains | ||
| Please refer to Table S1 | this study | N/A |
| Oligonucleotides | ||
| Please refer to Tables S3, S4, and S5 | this study | N/A |
| Recombinant DNA | ||
| Please refer to Table S2 | this study | N/A |
| Software and algorithms | ||
| ImageJ | Schneider et al.81 | https://imagej.nih.gov/ij/index.html |
| Python | custom code |
https://doi.org/10.5281/zenodo.7820986 https://doi.org/10.5281/zenodo.7820895 https://doi.org/10.5281/zenodo.7820931 https://doi.org/10.5281/zenodo.7821005 |
| Bowtie2 | Langmead and Salzberg82 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| MATLAB (MatTrack v6) | Mazza et al.,83 kind gift from David Ball | https://doi.org/10.5281/zenodo.7821136 |
| DESeq2 | Love et al.84 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| RStudio | RStudio | RRID: SCR_000432 |
| Affinity Designer | Serif | https://affinity.serif.com/en-us/designer/; RRID: SCR_016952 |
| Other | ||
| "Megatron" W5 UV crosslinking unit | UVO3 Ltd | https://www.uvo3.co.uk/ |
| Mixer Mill MM 400 | Retsch | Cat# 20.745.0001 |
| Gelfree 8100 Fractionation Station | Expedeon | Cat# 48100 |
| 18 mm round cover slips coated with poly-L-lysine | Neuvitro | Cat# GG-18-1.5-pll |
| 25mm round cover glasses (#1.5, thickness) | VWR | Cat# 631-0172 |
| 25mm round cover glasses HI D=0.17m | Zeiss | Cat# 000000-1787-996 |
| Wash-N-Dry coverslip rack | Sigma Aldrich | Cat# Z688568-1EA |
| 400 mL tall glass beakers | Novodirect | Cat# 15439093 |
| Microscope slides, SuperFrost® | VWR | Cat# ISO8037/I |
| Attofluor™ Cell Chamber, for microscopy | Thermofisher Scientific | Cat# A7816 |
| Dumont Horlogemakers pincet Gebogen Nr. 7 (forceps, tweezers) | Vos Medisch | Cat# 1121 |
| Cell density meter | VWR | Cat#634-0882 |
| Parafilm® M (4 inches wide) | Merck | P7668 |
| Zeiss Plan-Apochromat 40×/1.40NA Oil | Zeiss | Cat# 420762-9900-000 |
| Zeiss 4-alpha Plan-Apochromat 100×/1.46NA Oil | Zeiss | Cat# 420792-9800-000 |
| Zeiss alpha Plan-Apochromat 100x 1.57NA oil | Zeiss | Cat# 420792-9771-000 |
| SPECTRA X light engine WL:360–680 nm | Lumencor | N/A |
| ORCA Flash 4v3 digital sCMOS camera | Hamamatsu | Cat# C13440-20CU |
| UNO Top stage incubator and objective heater | Okolab | N/A |
| Excitation filters, emission flters and dichroic mirrors | See method details | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Tineke L. Lenstra (t.lenstra@nki.nl).
Materials availability
Plasmids and yeast strains generated in this study are available upon reasonable request from the lead contact with a completed Materials Transfer Agreement.
Experimental model and subject details
Yeast strains, plasmids, and oligos
Haploid yeast cells (Saccharomyces cerevisiae) of BY4741 and BY4742 backgrounds were transformed and mated to obtain the BY4743 diploids listed in Table S1. 12xMS2V6 loops were integrated at 5’ GAL1 with a PCR product containing loxP-kanMX-loxP and at 5’ GAL7 with loxP2272-kanMX-loxP2272, a loxP mutant to prevent recombination with WT loxP sequence. The kanMX was excised with inducible CRE recombinase. Plasmids containing the MS2 and PP7 coat proteins, fused to mScarlet and GFPEnvy, respectively (pTL174 and pTL333), were digested with PacI and integrated at the ura30 locus. Auxin-inducible degron tags at TOP1 and TOP2 were amplified from YTL738 or pTL398 and integrated at the endogenous loci. Plasmid containing OsTIR1 (pTL231) was digested with PacI and integrated at the his31 locus. Gal4UASscr, GAL7 truncation and spacer mutations were made using CRISPR/Cas9.85 The spacer sequence included convergent ADH1t and CUT60t terminator sequences to prevent transcriptional interference. All integrations were checked with PCR and sequencing. Gyrase and Topo I were ectopically expressed from plasmids. smFISH experiments with gene-specific probes upon topoisomerase degradation and CRAC-seq experiments were performed in haploid cells with W303 background. Cells were grown at 30°C in synthetic media. Strains, plasmids and oligos used to construct the strains can be found in Tables S1, S2, and S3, respectively.
Method details
Live-cell imaging of transcription dynamics
Live-cell imaging of transcription dynamics was performed as previously described in Donovan et al.60 and Brouwer et al.86 with minor modifications. Cells were grown at 30°C for at least 14 h in synthetic complete media supplemented with 2% raffinose. The cells were imaged after 30 min galactose induction at 30°C at mid-log (optical density, OD600 0.2–0.4) on a coverslip with an agarose pad consisting of synthetic complete media and 2% galactose. For indole-3-acetic acid (IAA; auxin) treatment, cells were treated with galactose for 30 min and with 500 μM for 15 min before imaging. For auxinole treatment, cells were grown for at least 14 h in 500 μM auxinole and induced with galactose for 30 min before imaging.
Imaging was performed on a setup consisting of an inverted microscope (Zeiss AxioObserver), an alpha Plan-Apochromat 100x 1.46NA oil objective, an sCMOS camera (Hamamatsu ORCA Flash 4v3) with a dual bandpass dichroic (Chroma 59012bs), a 570 nm longpass beamsplitter (Chroma T565lpxr-UF1), and 515/30 and 600/52 emission filters (Semrock FF01-515/30-25 and Semrock FF01-600/52-25), an UNO Top stage incubator and objective heater (OKOlab) at 30°C, LED excitation at 470/24 nm and 550/15 nm (SpectraX, Lumencor) at 0.20% and 0.40% power with an ND2 filter, resulting in a 62 mW/cm2 and 413 mW/cm2 excitation intensity. Wide-field images of GFPEnvy and mScarlet signals were acquired sequentially to prevent spectral crosstalk. Images were recorded at 10s interval for 30 min, with 9 z-stacks (Δz 0.5 μm) and 200 ms exposure using the Micro-Manager software, version 1.4.87 For each condition, at least 3 replicate datasets were acquired with a total at least 100 cells.
Single-molecule FISH
Yeast cultures were grown to mid-log (OD600 0.5) in 25 mL synthetic complete media with 2% raffinose and 2% galactose and smFISH was performed as previously described with minor modifications.39,60 For auxinole treatment, cells were grown in synthetic complete media with 500 μM auxinole and 2% galactose. For the auxin timepoints, 100mL cultures were grown to OD600 0.4 before being divided into 4×25 mL cultures and treated with 500 μM auxin for specified amount of time before fixation. If timepoint is not specified, cells were treated with auxin for 60 min. Cells were harvested at the same time after auxin addition to ensure the same OD.
Cells were fixed with 5% paraformaldehyde (Electron Microscopy Sciences, 15714-S) for 20 min, washed three times with buffer B (1.2 M sorbitol and 100 mM potassium phosphate buffer pH 7.5) and then spheroplasted with 300 units of lyticase (Sigma-Aldrich, L2524-25KU). Cells were then immobilized on poly-L-lysine-coated coverslips (Neuvitro) and permeabilized with 70% ethanol. Coverslips were hybridized for 4 h at 37°C with hybridization buffer containing 10% dextran sulfate, 10% formamide, 2×SSC, and 5 pmol of fluorescent probes. For FISH targeting the PP7 and MS2 repeats, four PP7 probes labeled with Quasar570 and 48 MS2 probes labeled with Quasar670 dyes were used. For FISH targeting GAL1, GAL10 or GAL7, 48 probes labeled with Quasar570 (GAL1 and GAL7) or Quasar670 (GAL10) were used (Table S4). Coverslips were washed 2× for 30 min with 10% formamide, 2×SSC at 37°C, then 1× with 2×SSC, and 1× for 5 min with PBS at room temperature. Coverslips were mounted on microscope slides using ProLong Gold mounting media with DAPI (Thermo Fisher, P36934).
Imaging was performed on two similar microscopes consisting of an inverted microscope (Zeiss AxioObserver), a Plan-Apochromat 40x 1.4NA oil DIC UV objective, a 1.60x optovar, and an sCMOS camera (Hamamatsu ORCA Flash 4v3). For Quasar570, a 562 nm longpass dichroic (Chroma T562lpxr), 595/50 nm emission filter (Chroma ET595/50m) and 550/15 nm LED excitation at full power (Spectra X, Lumencor) were used. For Quasar670, a 660 nm longpass dichroic (Semrock FF660-Di02-25x36 or Chroma T660lpxrxt), 697/60 nm emission filter (Chroma ET697/60m) and 640/30 nm LED excitation at full power (Spectra X, Lumencor) were used. For DAPI, either a 410nm/490nm/570nm/660nm dichroic (Chroma vcgr-spx-p01-PC), a 430/35 nm, 512/45 nm, 593/40 nm, 665 nm longpass emission filter (Chroma vcgr-spx-p01-EM) or a 425 nm longpass dichroic (Chroma T425lpxr) and a 460/50 nm emission filter (Chroma ET460/50m) and LED excitation at 395/25 nm at 25% power (Spectra X, Lumencor) were used. For each sample and each channel, we utilized the Micro-Manager software, version 1.4 to acquire at least 50 fields-of-view, each consisting of a 21 z-stack (Δz 0.3 μm) at 25 ms exposure for DAPI and 250 ms exposure for Quasar570 and Quasar670. For the smFISH experiments with the untagged topoisomerase-deficient haploids, all imaging settings were the same except a 1.25× optovar was used and each field-of-view consisted of 13 z-stack (Δz 0.5 μm).
Western blot
Yeast cultures were grown to mid-log (OD600nm 0.4) in 25 mL synthetic complete media with 2% raffinose and 2% galactose. For auxinole treatment, cells were grown in synthetic complete media with 500 μM auxinole and 2% galactose. The cells were treated with 500 μM auxin for 15, 30, or 60 min. Cells were harvested at the same time to ensure the same OD. Cells were washed with PBS twice and then incubated in 200 mM NaOH for 10 min. The cells were pelleted and resuspended in 2× SDS-PAGE solvent (4% SDS, 20% glycerol, 0.1 M DTT, 0.125 M Tris-HCl pH 7.5 and Roche EDTA-free protease inhibitor cocktail) and boiled at 95°C for 5min. The lysates were centrifuged, the supernatant was collected and snap-frozen in liquid nitrogen and stored at −80°C.
To determine the loading volume, samples were first checked with a dot blot. The same WT control strain was used to ensure similar loading between experiments. For the western blot, samples were run on a 3-8% Tris-acetate gel (Thermo Fisher Scientific, EA0375PK2) at 100V for 2 hours and wet transferred (Bio-Rad, 1703930) on a nitrocellulose membrane at 300 mA for 4 hours. The membrane was washed with PBS for 5 min, blocked with 5% milk, dissolved in PBS, for 1 h at 18-22°C and incubated in 2% milk dissolved in TBS-T containing 1:1000 dilution of anti-cMyc (Thermo Fisher Scientific, #MA1-980) or anti-PGK (Thermo Fisher Scientific, #PA5-28612) primary antibodies, at 4°C for 14 hours. The membrane was washed with PBS for 5 min three times and incubated with 2% milk dissolved in TBS-T containing fluorescent anti-mouse (LI-COR, 926-32210) or anti-rabbit (LI-COR, 926-32211) secondary antibodies for 1 h at 18-22°C in the dark.
MNase-seq
Preparation and analysis of mono-nucleosomal DNA was performed as described previously60,88 with minor modifications and with two biological replicates. Haploid cells were grown in synthetic complete media with 2% raffinose or 2% galactose from OD600 0.3 to OD600 1.0, fixed in 1% paraformaldehyde, washed with 1 M sorbitol, treated with spheroplasting buffer (1M sorbitol, 1 mM β-mercaptoethanol, 10 mg/mL zymolyase 100T (US biological, Z1004.250)) and washed twice with 1 M sorbitol. Spheroplasted cells were treated with 0.01171875 or 0.1875 U micrococcal nuclease (Sigma-Aldrich, N5386-200UN) in digestion buffer (1 M sorbitol, 50 mM NaCl, 10 mM Tris pH 7.4, 5 mM MgCl2, 0.075% NP-40, 1 mM β-mercaptoethanol, 0.5 mM spermidine) at 37°C. After 45 min, reactions were terminated on ice with 25 mM EDTA and 0.5% SDS. Samples were treated with proteinase K for 1 h at 37°C and decrosslinked overnight at 65°C. Digested DNA was extracted with phenol/chloroform (PCI 15:14:1), precipitated with NH4-Ac, and treated with 0.1 mg/mL RNaseA/T1. The extent of digestion was checked on a 3% agarose gel.
Sequencing libraries were prepared using the KAPA HTP Library Preparation Kit (07961901001, KAPA Biosystems) using 1 mg of input DNA, 5 mL of 10 mM adapter, double-sided size selection before and after amplification using 10 cycles. Adapters were created by ligation of the Universal adapter to individual sequencing adapters (Table S5). Libraries were checked on Bioanalyzer High Sensitivity DNA kit (Agilent) and sequencing was performed on a NextSeq550.
Single-molecule tracking of Gal4
Cells were grown at 30°C for at least 14 h in synthetic complete media, supplemented with 2% raffinose and 2% galactose. At mid-log (optical density, OD600 0.2–0.4), cells were treated with 5 pM (H3-HaloTag cells) or 500 pM (Gal4-HaloTag cells) of JFX650 dye80 and incubated at 30°C for 15 minutes. The cells were washed with warm media and immobilized on a coverslip with an agarose pad consisting of synthetic complete media with 2% raffinose and 2% galactose.
The cells were imaged on ELYRA.P1 (Zeiss) equipped with an incubator (Pecon) and Scanning Stage Piezo 130x100 (Zeiss). We used an alpha Plan-Apochromat 100x 1.57NA oil objective (Zeiss) and a filter set (Zeiss LBF 405/488/642). The cells were excited simultaneously with Highly Inclined Laminated Optical (HILO) sheet illumination mode with 488 nm and 640 nm using 1.6 W/cm2 and 2 mW/cm2 excitation intensities, respectively. Images were captured with 30 ms exposure at 200 ms interval for 1,000 time points. The emission was split in two channels (TV1 and TV2) using a duolink splitter (Zeiss) holding a filter set with a BS642 dichroic beamsplitter (Zeiss) and BP495–550 and LP655 emission filters (Semrock) onto two EM-CCD iXon DU 897 cameras (Andor).
CRAC-seq
For Pol II CRAC experiments, 2 L per condition of cells with endogenously HTP-tagged RBP1 and AID-tagged TOP1, TOP2 genes were grown to exponential phase in synthetic media lacking tryptophane at 30°C and harvested at OD600 = 0.6. Depletion of Top1-AID and Top2-AID was induced by treatment with 5 mM auxin for 1 hour before harvesting. Processing of the CRAC-seq data was performed as previously described in Candelli et al.89 and Challal et al.79
Quantification and statistical analysis
Statistical details for individual experiments have been provided in the figure legends.
Analysis of live-cell transcription dynamics
For image analysis of transcription dynamics, the intensity calculation and tracking of the transcription sites was calculated as previously described in Donovan et al.60 using a custom Python script (https://doi.org/10.5281/zenodo.7820895 with dependencies from https://doi.org/10.5281/zenodo.7820931). The images were maximum intensity projected and then corrected for xy-drift in the stage using an affine transformation. Cells were segmented using Otsu thresholding and watershedding. The intensity of the TS was calculated for each color by fitting a 2D Gaussian mask after local background subtraction as described previously.90 To detect the TSs, initial intensity thresholds of 9 and 7 standard deviations (SD) from the mean background was used for PP7 and MS2 signals, respectively. For frames where no TS was detected, a second fit was made in the vicinity of the initial detected spots using lower intensity thresholds of 6 and 4 SD from the mean background for PP7 and MS2, respectively. If no TSs were detected in a frame after the second fit, the intensity was measured at the xy-coordinates of the previous frame. The tracking within each cell was inspected visually, and the endpoint of each trace was manually set at the last frame where a TS was visible. Dividing cells and cells in which TSs were not reliably detected were excluded from the analysis. Only the cells that exhibited both PP7 and MS2 signals were considered for analysis. Cells with only signal in one channel were inspected but exhibited insufficient coat protein levels in the other channel for reliable analysis. For each cell, the background was estimated by fitting a Lorentzian distribution to intensities measured at four points at a fixed distance from the TS in each frame in the same cell. The mean background was subtracted from the intensity trace to obtain background-subtracted intensity traces. Active fraction was computed by accounting for cells that exhibited both PP7 and MS2 signals for at least 600 seconds. Cells where we did not detect transcription sites above background were classified as inactive.
To determine the on and off periods, the fluorescence signal was binarized by setting a threshold that was a specific standard deviation above the MS2 and PP7 background intensities. To determine a binarization threshold that captured the correct bursting kinetics, the sum of squared residuals between the ACFs of the binary signals (range 1.0-5.0, steps of 0.25) and the ACF of the analog fluorescence signal between 10s and 100s was calculated. The minimal residual was found at threshold values of 2.75 and 4.50 standard deviations above background, with residuals of 0.0025 and 0.0010 for MS2 and PP7, respectively. The burst intensity was measured for frames where the binarized signal was on. The burst duration and burst frequency (defined as inverse of time between bursts) were calculated from the binarized data, and bursts with a duration of a single frame were considered as errors from the binarization and were excluded. Reported error bars and significance was calculated from bootstrapping with 1000 repetitions.
ACF, CCF, and transcriptional overlap
For each time trace, autocorrelation (ACF) and crosscorrelation functions (CCF) were computed as
| (Equation 1) |
where <·> denotes the time average, δa(t) = a(t) - ⟨a(t)⟩ and a(t) and b(t) can be combinations of the MS2 and PP7 time traces.60,90 Correlation functions were computed using fast Fourier transforms and upon shifting the two signals, non-overlapping ends were trimmed. The functions were normalized for each trace individually. To correct for non-stationary effects (i.e. photobleaching, cell cycle, etc.), the global mean signal was used to calculate corrections, which were then subtracted. For single-trace correlation functions, each point was given a weight corresponding to the number of overlapping time intervals (τ) from the signals used in its computation. Correlation functions from single time traces were averaged together to reach statistical convergence. Bootstrapping was performed with 10,000 repetitions to obtain standard error of the mean correlation functions (SEM).
We used the ACFs and CCFs to calculate the normalized transcriptional overlap (called fractional overlap by Rodriguez et al.),38 which provides an estimate of the fraction of bursts of one gene, which co-occur with the bursts of another gene. We normalized the cross-correlation functions of the GAL1-GAL10 and GAL10-GAL7 genes by their respective ACFs:
| (Equation 2) |
where Gab(τ) represents the CCFs of GAL1-GAL10 or GAL10-GAL7. Ga(0) represents the ACF amplitude at = 0 of GAL10 in GAL1-GAL10 or GAL10-GAL7 pair. Each trace was normalized before the traces were averaged together.
To estimate the amplitude of the CCF at = 0, we fit the CCF with a Gaussian. The measured ACF amplitude at τ = 0 is overestimated due to shot noise, so to estimate the representative amplitudes Ga(0) and Gb(0), we fit a line through the first 4 to 10 (omitting τ = 0) values of Ga and Gb. The fit with the best coefficient of determination was used to extrapolate the values of Ga(0).
Rodriguez et al. presented the transcriptional overlap calculation for a model with assumptions that the transcriptional events are square pulses of equal duration and height and are uniformly distributed over time. To confirm that this calculation can be applied for bursts with trapezoidal transcription events that are exponentially distributed, we simulated a 4-state model (ON-ON, ON-OFF, OFF-ON, OFF-OFF) for a gene pair where the promoter states are correlated, similar to GAL1-GAL10. We find that at lower transcription rates, the calculated normalized transcriptional overlap deviates from the theoretical values. However, for highly correlated gene pairs, as observed in the real data, the calculated transcriptional overlap from Equation 2 matches the theoretical transcriptional overlap between the two genes.
smFISH analysis
For smFISH image analysis, a custom-written Python script was used to detect, localize, and classify the spots (https://doi.org/10.5281/zenodo.7820986 with dependencies from https://doi.org/10.5281/zenodo.7820931). Cells and nuclei were segmented using Otsu thresholding and watershedding. Spots were localized by fitting a 3D Gaussian mask after local background subtraction.90 Cells in which no spots were detected were excluded from further analysis since a visual inspection indicated that these cells were not properly segmented or were improperly permeabilized. For each cell, the TS was defined as the brightest nuclear spot and the number of RNAs at each TS was determined by normalizing the intensity of each TS with the median fluorescent intensity of the cytoplasmic RNAs detected in all cells. Cells were further subclassified based on their cell cycle stage using the integrated DAPI intensity of each cell calculated from the maximum intensity projection images.91 A distribution of nuclear DAPI intensities was fit with a bimodal Gaussian model. The TS intensity was only analyzed in G1 cells, with nuclear intensities [1 SD, 0.75× SD] around the mean of the first peak.
Cells with fewer than 5 RNAs at the TS were classified as inactive, and cells with 5 or more RNAs at the TS were classified as active cells. Subsequently, the fraction of active cells for each gene and the Pearson correlation coefficients of the active cells were determined for various conditions. For smFISH experiments with GAL10, GAL1, and GAL7 probes, spots were fit using 2D fitting, and the threshold to classify as an active cell was set to 2.5.
Western blot quantification
The fluorescence signal of Western blots was quantified using ImageJ.81 A region of interest (ROI) was outlined around the largest sample and the same ROI was used for all samples on the membrane. The background was calculated by averaging the intensities of eight ROIs on the membrane where there was no signal present. The integrated intensities of the samples were background-subtracted and normalized to a no-OsTIR1 control strain to determine degradation upon OsTIR1 addition and auxin addition.
Quantification of eGFP-Topo I fluorescence
To quantify the Topo I-eGFP fluorescence, cells were segmented using Otsu thresholding and watershedding. The fluorescence of each pixel within the cell was integrated and subtracted by the mean background of the image. The background was defined as the pixels unoccupied by the cell masks. The background-subtracted intensities were normalized by the area of the cell.
MNase-seq analysis
MNase-seq was analyzed with a custom python script (https://doi.org/10.5281/zenodo.7821005). Paired-end 2×75-bp reads were aligned to the reference genome SacCer3 using Bowtie 2.82 Nucleosome dyads were found by taking the middle of each paired read of insert size between 95 and 225 bp and were smoothed with a 31-bp window.88 The minimal read-length of 95 bp ensured inclusion of subnucleosomal particles (fragile nucleosomes) rather than regulatory factors, which is especially important when using low MNase digestion conditions.
To determine the position of the +1 nucleosome for each gene, the coverage was determined in a 4000 bp window around the annotated TSS. For each gene, the coverage was summed and smoothed using a Gaussian filter with 40 bp window. The peaks were determined using a peak calling function. The +1 nucleosome was defined as the first peak after the minimum of the smoothed coverage. To compute the metagene plot, genes were aligned at the +1 nucleosome based on classifications in the unperturbed condition and the coverages were summed and normalized by the number of genes.
Analysis of Gal4 single-molecule tracking
Single-molecule tracking movies were analyzed using a custom MATLAB software based on MatTrack (version 6, https://doi.org/10.5281/zenodo.7821136).60,83 Dividing cells were excluded from the analysis. To determine the region of interest for the analysis, the nucleus was labeled with PP7 coat protein, fused to GFPEnvy to aid segmentation. The Gal4-HaloTag labeling density is such that 1-2 molecule were labeled in each nucleus, making it unlikely to have multiple labeled molecules in one position. Only molecules that were tracked in more than 4 frames were considered to be bound. To determine whether a molecule was bound or diffusing, a threshold was used based on tracking of histone H3. Tracking of histone H3 showed that 99% of single molecules had a frame-to-frame displacement of < 0.35 μm at 200 ms interval. These maximum displacements were used to determine whether Gal4 particles are chromatin bound or diffusing. The cumulative distribution of dwell times of bound Gal4 molecules (survival probability plot) was corrected for bleaching based on the photobleaching kinetics of the bound histone population.92 Briefly, the histone (H3-HaloTag) SMT data, acquired using the same conditions as Gal4-HaloTag SMT data, is fitted to a family of exponentials and the exponential distribution of the longer component is used to normalize the Gal4-HaloTag survival. The residence time distributions were computed from the photobleaching-corrected survival distributions and the average was calculated for bound tracks that were greater than 5 s in duration.
CRAC-seq analysis
Computational analyses of the CRAC-seq experiments were performed with ad hoc scripts in the R Studio environment. Differential gene expression analyses were carried out using the DESeq2 package.84 The wildtype condition presented in Figure S7 was previously published in Aiello et al.53
Acknowledgments
We thank Bas van Steensel for helpful discussions and guidance, Fred van Leeuwen and Stefano G. Manzo for critical reading of the manuscript, and members of the T.L.L. lab for helpful feedback. We thank Evelina Tutucci for sharing the 12xMS2V6 plasmid (Addgene plasmid 104390), Daniela Delneri for the loxP-mutant plasmid, Benjamin Albert and David Shore for Top1-AID, Top2-AID W303 haploids, and Joaquim Roca for gyrase (pSTS77) and Topo I (YEptopA-PGAL1) plasmids. We thank David Ball for assistance with SMT analysis and the NKI Research High Performance Computing Facility and Genomics Core Facility for assistance. This work was supported by an institutional grant of the Dutch Cancer Society and of the Dutch Ministry of Health, Welfare and Sport; the Dutch Research Council (NWO, 016.Veni.192.071 and gravitation program CancerGenomiCs.nl); Oncode Institute, which is partly financed by the Dutch Cancer Society; and the European Research Council (ERC Starting grant 755695 BURSTREG). U.A. was supported by the French Ministry for Education and Research, by the Foundation ARC pour la Recherche sur le Cancer, and by the EUR G.E.N.E. (reference #ANR-17-EURE-0013), which is part of the Université Paris Cité IdEx #ANR-18-IDEX-0001 funded by the French Government through its “Investments for the Future” program.
Author contributions
Conceptualization, H.P.P. and T.L.L.; data curation, H.P.P. and U.A.; formal analysis, H.P.P., S.C., W.P., I.B., U.A., and T.L.L.; funding acquisition, I.B., T.L.L., and D.L.; investigation, H.P.P.; methodology, H.P.P. and T.L.L.; software, H.P.P., S.C., W.P., I.B., U.A., and T.L.L.; supervision, D.L. and T.L.L.; visualization, H.P.P. and T.L.L.; writing – original draft, H.P.P. and T.L.L.; writing – review & editing, H.P.P., T.L.L., S.C., W.P., I.B., U.A., and D.L.
Declaration of interests
The authors declare no competing interests.
Published: May 18, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.molcel.2023.04.015.
Supplemental information
Data and code availability
-
•
The sequencing (MNase-seq and Pol II CRAC-seq) data from this publication have been deposited and are publicly available as of the date of publication. Accession numbers and DOI are listed in the key resources table. Microscopy data is available upon request.
-
•
All original code for the live-cell, smFISH, SMT and MNase-seq analysis has been deposited to Zenodo. DOI are listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Liu L.F., Wang J.C. Supercoiling of the DNA template during transcription. Proc. Natl. Acad. Sci. USA. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsao Y.P., Wu H.Y., Liu L.F. Transcription-driven supercoiling of DNA: direct biochemical evidence from in vitro studies. Cell. 1989;56:111–118. doi: 10.1016/0092-8674(89)90989-6. [DOI] [PubMed] [Google Scholar]
- 3.Wu H.Y., Shyy S.H., Wang J.C., Liu L.F. Transcription generates positively and negatively supercoiled domains in the template. Cell. 1988;53:433–440. doi: 10.1016/0092-8674(88)90163-8. [DOI] [PubMed] [Google Scholar]
- 4.Kouzine F., Sanford S., Elisha-Feil Z., Levens D. The functional response of upstream DNA to dynamic supercoiling in vivo. Nat. Struct. Mol. Biol. 2008;15:146–154. doi: 10.1038/nsmb.1372. [DOI] [PubMed] [Google Scholar]
- 5.Mizutani M., Ohta T., Watanabe H., Handa H., Hirose S. Negative supercoiling of DNA facilitates an interaction between transcription factor IID and the fibroin gene promoter. Proc. Natl. Acad. Sci. USA. 1991;88:718–722. doi: 10.1073/pnas.88.3.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parvin J.D., Sharp P.A. DNA topology and a minimal set of basal factors for transcription by RNA polymerase II. Cell. 1993;73:533–540. doi: 10.1016/0092-8674(93)90140-l. [DOI] [PubMed] [Google Scholar]
- 7.Sheinin M.Y., Li M., Soltani M., Luger K., Wang M.D. Torque modulates nucleosome stability and facilitates H2A/H2B dimer loss. Nat. Commun. 2013;4 doi: 10.1038/ncomms3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tabuchi H., Handa H., Hirose S. Underwinding of DNA on binding of yeast TFIID to the TATA element. Biochem. Biophys. Res. Commun. 1993;192:1432–1438. doi: 10.1006/bbrc.1993.1576. [DOI] [PubMed] [Google Scholar]
- 9.Teves S.S., Henikoff S. Transcription-generated torsional stress destabilizes nucleosomes. Nat. Struct. Mol. Biol. 2014;21:88–94. doi: 10.1038/nsmb.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chong S., Chen C., Ge H., Xie X.S. Mechanism of transcriptional bursting in bacteria. Cell. 2014;158:314–326. doi: 10.1016/j.cell.2014.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drolet M. Growth inhibition mediated by excess negative supercoiling: the interplay between transcription elongation, R-loop formation and DNA topology. Mol. Microbiol. 2006;59:723–730. doi: 10.1111/j.1365-2958.2005.05006.x. [DOI] [PubMed] [Google Scholar]
- 12.Ma J., Bai L., Wang M.D. Transcription under torsion. Science. 2013;340:1580–1583. doi: 10.1126/science.1235441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pedersen J.M., Fredsoe J., Roedgaard M., Andreasen L., Mundbjerg K., Kruhøffer M., Brinch M., Schierup M.H., Bjergbaek L., Andersen A.H. DNA topoisomerases maintain promoters in a state competent for transcriptional activation in Saccharomyces cerevisiae. PLoS Genet. 2012;8 doi: 10.1371/journal.pgen.1003128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roedgaard M., Fredsoe J., Pedersen J.M., Bjergbaek L., Andersen A.H. DNA topoisomerases are required for preinitiation complex assembly during GAL gene activation. PLoS One. 2015;10 doi: 10.1371/journal.pone.0132739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen C.-C., Chou M.-Y., Huang C.-H., Majumder A., Wu H.-Y. A cis-spreading nucleoprotein filament is responsible for the gene silencing activity found in the promoter relay mechanism. J. Biol. Chem. 2005;280:5101–5112. doi: 10.1074/jbc.M411840200. [DOI] [PubMed] [Google Scholar]
- 16.El Hanafi D., Bossi L. Activation and silencing of leu-500 promoter by transcription-induced DNA supercoiling in the Salmonella chromosome. Mol. Microbiol. 2000;37:583–594. doi: 10.1046/j.1365-2958.2000.02015.x. [DOI] [PubMed] [Google Scholar]
- 17.El Houdaigui B., Forquet R., Hindré T., Schneider D., Nasser W., Reverchon S., Meyer S. Bacterial genome architecture shapes global transcriptional regulation by DNA supercoiling. Nucleic Acids Res. 2019;47:5648–5657. doi: 10.1093/nar/gkz300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rhee K.Y., Opel M., Ito E., Hung Sp., Arfin S.M., Hatfield G.W. Transcriptional coupling between the divergent promoters of a prototypic LysR-type regulatory system, the ilvYC operon of Escherichia coli. Proc. Natl. Acad. Sci. USA. 1999;96:14294–14299. doi: 10.1073/pnas.96.25.14294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sobetzko P. Transcription-coupled DNA supercoiling dictates the chromosomal arrangement of bacterial genes. Nucleic Acids Res. 2016;44:1514–1524. doi: 10.1093/nar/gkw007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ancona M., Bentivoglio A., Brackley C.A., Gonnella G., Marenduzzo D. Transcriptional bursts in a nonequilibrium model for gene regulation by supercoiling. Biophys. J. 2019;117:369–376. doi: 10.1016/j.bpj.2019.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brackley C.A., Johnson J., Bentivoglio A., Corless S., Gilbert N., Gonnella G., Marenduzzo D. Stochastic model of supercoiling-dependent transcription. Phys. Rev. Lett. 2016;117 doi: 10.1103/PhysRevLett.117.018101. [DOI] [PubMed] [Google Scholar]
- 22.Dunaway M., Ostrander E.A. Local domains of supercoiling activate a eukaryotic promoter in vivo. Nature. 1993;361:746–748. doi: 10.1038/361746a0. [DOI] [PubMed] [Google Scholar]
- 23.Kouzine F., Gupta A., Baranello L., Wojtowicz D., Ben-Aissa K., Liu J., Przytycka T.M., Levens D. Transcription dependent dynamic supercoiling is a short-range genomic force. Nat. Struct. Mol. Biol. 2013;20:396–403. doi: 10.1038/nsmb.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyer S., Beslon G. Torsion-mediated interaction between adjacent genes. PLoS Comput. Biol. 2014;10 doi: 10.1371/journal.pcbi.1003785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Naughton C., Corless S., Gilbert N. Divergent RNA transcription: a role in promoter unwinding? Transcription. 2013;4:162–166. doi: 10.4161/trns.25554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang G.-Z., Lercher M.J., Hurst L.D. Transcriptional coupling of neighboring genes and gene expression noise: evidence that gene orientation and noncoding transcripts are modulators of noise. Genome Biol. Evol. 2011;3:320–331. doi: 10.1093/gbe/evr025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnstone C.P., Galloway K.E. Supercoiling-mediated feedback rapidly couples and tunes transcription. Cell Rep. 2022;41 doi: 10.1016/j.celrep.2022.111492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bancaud A., Wagner G., Conde E Silva N., Lavelle C., Wong H., Mozziconacci J., Barbi M., Sivolob A., Le Cam E., Mouawad L., et al. Nucleosome chiral transition under positive torsional stress in single chromatin fibers. Mol. Cell. 2007;27:135–147. doi: 10.1016/j.molcel.2007.05.037. [DOI] [PubMed] [Google Scholar]
- 29.Bancaud A., Conde e Silva N., Barbi M., Wagner G., Allemand J.F., Mozziconacci J., Lavelle C., Croquette V., Victor J.M., Prunell A., et al. Structural plasticity of single chromatin fibers revealed by torsional manipulation. Nat. Struct. Mol. Biol. 2006;13:444–450. doi: 10.1038/nsmb1087. [DOI] [PubMed] [Google Scholar]
- 30.Kaczmarczyk A., Meng H., Ordu O., van Noort J.V., Dekker N.H. Chromatin fibers stabilize nucleosomes under torsional stress. Nat. Commun. 2020;11 doi: 10.1038/s41467-019-13891-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Achar Y.J., Adhil M., Choudhary R., Gilbert N., Foiani M. Negative supercoil at gene boundaries modulates gene topology. Nature. 2020;577:701–705. doi: 10.1038/s41586-020-1934-4. [DOI] [PubMed] [Google Scholar]
- 32.Guo M.S., Kawamura R., Littlehale M.L., Marko J.F., Laub M.T. High-resolution, genome-wide mapping of positive supercoiling in chromosomes. eLife. 2021;10 doi: 10.7554/eLife.67236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naughton C., Avlonitis N., Corless S., Prendergast J.G., Mati I.K., Eijk P.P., Cockroft S.L., Bradley M., Ylstra B., Gilbert N. Transcription forms and remodels supercoiling domains unfolding large-scale chromatin structures. Nat. Struct. Mol. Biol. 2013;20:387–395. doi: 10.1038/nsmb.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baranello L., Wojtowicz D., Cui K., Devaiah B.N., Chung H.J., Chan-Salis K.Y., Guha R., Wilson K., Zhang X., Zhang H., et al. RNA polymerase II regulates topoisomerase 1 activity to favor efficient transcription. Cell. 2016;165:357–371. doi: 10.1016/j.cell.2016.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lenstra T.L., Coulon A., Chow C.C., Larson D.R. Single-molecule imaging reveals a switch between spurious and functional ncRNA transcription. Mol. Cell. 2015;60:597–610. doi: 10.1016/j.molcel.2015.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lenstra T.L., Larson D.R. Single-molecule mRNA detection in live yeast. Curr. Protoc. Mol. Biol. 2016;113:14.24.1–14.24.15. doi: 10.1002/0471142727.mb1424s113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tutucci E., Vera M., Biswas J., Garcia J., Parker R., Singer R.H. An improved MS2 system for accurate reporting of the mRNA life cycle. Nat. Methods. 2018;15:81–89. doi: 10.1038/nmeth.4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rodriguez J., Ren G., Day C.R., Zhao K., Chow C.C., Larson D.R. Intrinsic dynamics of a human gene reveal the basis of expression heterogeneity. Cell. 2019;176:213–226.e18. doi: 10.1016/j.cell.2018.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patel H.P., Brouwer I., Lenstra T.L. Optimized protocol for single-molecule RNA FISH to visualize gene expression in S. cerevisiae. Star Protoc. 2021;2 doi: 10.1016/j.xpro.2021.100647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sperling A.S., Jeong K.S., Kitada T., Grunstein M. Topoisomerase II binds nucleosome-free DNA and acts redundantly with topoisomerase I to enhance recruitment of RNA Pol II in budding yeast. Proc. Natl. Acad. Sci. USA. 2011;108:12693–12698. doi: 10.1073/pnas.1106834108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nishimura K., Fukagawa T., Takisawa H., Kakimoto T., Kanemaki M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat. Methods. 2009;6:917–922. doi: 10.1038/nmeth.1401. [DOI] [PubMed] [Google Scholar]
- 42.Harper C.V., Finkenstädt B., Woodcock D.J., Friedrichsen S., Semprini S., Ashall L., Spiller D.G., Mullins J.J., Rand D.A., Davis J.R.E., et al. Dynamic analysis of stochastic transcription cycles. PLoS Biol. 2011;9 doi: 10.1371/journal.pbio.1000607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Molina N., Suter D.M., Cannavo R., Zoller B., Gotic I., Naef F. Stimulus-induced modulation of transcriptional bursting in a single mammalian gene. Proc. Natl. Acad. Sci. USA. 2013;110:20563–20568. doi: 10.1073/pnas.1312310110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suter D.M., Molina N., Gatfield D., Schneider K., Schibler U., Naef F. Mammalian genes are transcribed with widely different bursting kinetics. Science. 2011;332:472–474. doi: 10.1126/science.1198817. [DOI] [PubMed] [Google Scholar]
- 45.Kouzine F., Liu J., Sanford S., Chung H.J., Levens D. The dynamic response of upstream DNA to transcription-generated torsional stress. Nat. Struct. Mol. Biol. 2004;11:1092–1100. doi: 10.1038/nsmb848. [DOI] [PubMed] [Google Scholar]
- 46.Trigueros S., Roca J. Failure to relax negative supercoiling of DNA is a primary cause of mitotic hyper-recombination in topoisomerase-deficient yeast cells. J. Biol. Chem. 2002;277:37207–37211. doi: 10.1074/jbc.M206663200. [DOI] [PubMed] [Google Scholar]
- 47.Trigueros S., Roca J. A GyrB-GyrA fusion protein expressed in yeast cells is able to remove DNA supercoils but cannot substitute eukaryotic topoisomerase II. Genes Cells. 2002;7:249–257. doi: 10.1046/j.1365-2443.2002.00516.x. [DOI] [PubMed] [Google Scholar]
- 48.Kubik S., Bruzzone M.J., Jacquet P., Falcone J.-L., Rougemont J., Shore D. Nucleosome stability distinguishes two different promoter types at all protein-coding genes in yeast. Mol. Cell. 2015;60:422–434. doi: 10.1016/j.molcel.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 49.Brahma S., Henikoff S. RSC-Associated Subnucleosomes Define MNase-Sensitive Promoters in Yeast. Mol. Cell. 2019;73:238–249.e3. doi: 10.1016/j.molcel.2018.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Floer M., Wang X., Prabhu V., Berrozpe G., Narayan S., Spagna D., Alvarez D., Kendall J., Krasnitz A., Stepansky A., et al. A RSC/nucleosome complex determines chromatin architecture and facilitates activator binding. Cell. 2010;141:407–418. doi: 10.1016/j.cell.2010.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chedin F., Benham C.J. Emerging roles for R-loop structures in the management of topological stress. J. Biol. Chem. 2020;295:4684–4695. doi: 10.1074/jbc.REV119.006364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim S., Beltran B., Irnov I., Jacobs-Wagner C. Long-distance cooperative and antagonistic RNA polymerase dynamics via DNA supercoiling. Cell. 2019;179:106–119.e16. doi: 10.1016/j.cell.2019.08.033. [DOI] [PubMed] [Google Scholar]
- 53.Aiello U., Challal D., Wentzinger G., Lengronne A., Appanah R., Pasero P., Palancade B., Libri D. Sen1 is a key regulator of transcription-driven conflicts. Mol. Cell. 2022;82:2952–2966.e6. doi: 10.1016/j.molcel.2022.06.021. [DOI] [PubMed] [Google Scholar]
- 54.Xiong L., Zeng Y., Tang R.-Q., Alper H.S., Bai F.-W., Zhao X.-Q. Condition-specific promoter activities in Saccharomyces cerevisiae. Microb. Cell Factories. 2018;17 doi: 10.1186/s12934-018-0899-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Levo M., Raimundo J., Bing X.Y., Sisco Z., Batut P.J., Ryabichko S., Gregor T., Levine M.S. Transcriptional coupling of distant regulatory genes in living embryos. Nature. 2022;605:754–760. doi: 10.1038/s41586-022-04680-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cohen B.A., Mitra R.D., Hughes J.D., Church G.M. A computational analysis of whole-genome expression data reveals chromosomal domains of gene expression. Nat. Genet. 2000;26:183–186. doi: 10.1038/79896. [DOI] [PubMed] [Google Scholar]
- 57.Ebisuya M., Yamamoto T., Nakajima M., Nishida E. Ripples from neighbouring transcription. Nat. Cell Biol. 2008;10:1106–1113. doi: 10.1038/ncb1771. [DOI] [PubMed] [Google Scholar]
- 58.Michalak P. Coexpression, coregulation, and cofunctionality of neighboring genes in eukaryotic genomes. Genomics. 2008;91:243–248. doi: 10.1016/j.ygeno.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 59.Sevier S.A., Levine H. Properties of gene expression and chromatin structure with mechanically regulated elongation. Nucleic Acids Res. 2018;46:5924–5934. doi: 10.1093/nar/gky382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Donovan B.T., Huynh A., Ball D.A., Patel H.P., Poirier M.G., Larson D.R., Ferguson M.L., Lenstra T.L. Live-cell imaging reveals the interplay between transcription factors, nucleosomes, and bursting. EMBO J. 2019;38 doi: 10.15252/embj.2018100809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lainé J.P., Singh B.N., Krishnamurthy S., Hampsey M. A physiological role for gene loops in yeast. Genes Dev. 2009;23:2604–2609. doi: 10.1101/gad.1823609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.O’Sullivan J.M., Tan-Wong S.M., Morillon A., Lee B., Coles J., Mellor J., Proudfoot N.J. Gene loops juxtapose promoters and terminators in yeast. Nat. Genet. 2004;36:1014–1018. doi: 10.1038/ng1411. [DOI] [PubMed] [Google Scholar]
- 63.Chong S., Dugast-Darzacq C., Liu Z., Dong P., Dailey G.M., Cattoglio C., Heckert A., Banala S., Lavis L., Darzacq X., et al. Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science. 2018;361 doi: 10.1126/science.aar2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dobi K.C., Winston F. Analysis of transcriptional activation at a distance in Saccharomyces cerevisiae. Mol. Cell. Biol. 2007;27:5575–5586. doi: 10.1128/MCB.00459-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Karr J.P., Ferrie J.J., Tjian R., Darzacq X. The transcription factor activity gradient (TAG) model: contemplating a contact-independent mechanism for enhancer-promoter communication. Genes Dev. 2022;36:7–16. doi: 10.1101/gad.349160.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lang G.I., Botstein D. A test of the coordinated expression hypothesis for the origin and maintenance of the GAL cluster in yeast. PLoS One. 2011;6 doi: 10.1371/journal.pone.0025290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gartenberg M.R., Wang J.C. Positive supercoiling of DNA greatly diminishes mRNA synthesis in yeast. Proc. Natl. Acad. Sci. USA. 1992;89:11461–11465. doi: 10.1073/pnas.89.23.11461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Desai R.V., Chen X., Martin B., Chaturvedi S., Hwang D.W., Li W., Yu C., Ding S., Thomson M., Singer R.H., et al. A DNA repair pathway can regulate transcriptional noise to promote cell fate transitions. Science. 2021;373 doi: 10.1126/science.abc6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tabuchi H., Hirose S. DNA supercoiling facilitates formation of the transcription initiation complex on the fibroin gene promoter. J. Biol. Chem. 1988;263:15282–15287. [PubMed] [Google Scholar]
- 70.Baaklini I., Usongo V., Nolent F., Sanscartier P., Hraiky C., Drlica K., Drolet M. Hypernegative supercoiling inhibits growth by causing RNA degradation. J. Bacteriol. 2008;190:7346–7356. doi: 10.1128/JB.00680-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dages S., Dages K., Zhi X., Leng F. Inhibition of the gyrA promoter by transcription-coupled DNA supercoiling in Escherichia coli. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-33089-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mizutani M., Ura K., Hirose S. DNA superhelicity affects the formation of transcription preinitiation complex on eukaryotic genes differently. Nucleic Acids Res. 1991;19:2907–2911. doi: 10.1093/nar/19.11.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bacolla A., Wells R.D. Non-B DNA conformations, genomic rearrangements, and human disease. J. Biol. Chem. 2004;279:47411–47414. doi: 10.1074/jbc.R400028200. [DOI] [PubMed] [Google Scholar]
- 74.Pommier Y., Sun Y., Huang S.-Y.N., Nitiss J.L. Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nat. Rev. Mol. Cell Biol. 2016;17:703–721. doi: 10.1038/nrm.2016.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Racko D., Benedetti F., Dorier J., Stasiak A. Transcription-induced supercoiling as the driving force of chromatin loop extrusion during formation of TADs in interphase chromosomes. Nucleic Acids Res. 2018;46:1648–1660. doi: 10.1093/nar/gkx1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Neguembor M.V., Martin L., Castells-García Á., Gómez-García P.A., Vicario C., Carnevali D., AlHaj Abed J., Granados A., Sebastian-Perez R., Sottile F., et al. Transcription-mediated supercoiling regulates genome folding and loop formation. Mol. Cell. 2021;81:3065–3081.e12. doi: 10.1016/j.molcel.2021.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Racko D., Benedetti F., Dorier J., Stasiak A. Are TADs supercoiled? Nucleic Acids Res. 2019;47:521–532. doi: 10.1093/nar/gky1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rhee H.S., Pugh B.F. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell. 2011;147:1408–1419. doi: 10.1016/j.cell.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Challal D., Barucco M., Kubik S., Feuerbach F., Candelli T., Geoffroy H., Benaksas C., Shore D., Libri D. General regulatory factors control the fidelity of transcription by restricting non-coding and ectopic initiation. Mol. Cell. 2018;72:955–969.e7. doi: 10.1016/j.molcel.2018.11.037. [DOI] [PubMed] [Google Scholar]
- 80.Grimm J.B., Xie L., Casler J.C., Patel R., Tkachuk A.N., Falco N., Choi H., Lippincott-Schwartz J., Brown T.A., Glick B.S., et al. A general method to improve fluorophores using deuterated auxochromes. JACS Au. 2021;1:690–696. doi: 10.1021/jacsau.1c00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mazza D., Ganguly S., McNally J.G. Monitoring dynamic binding of chromatin proteins in vivo by single-molecule tracking. Methods Mol. Biol. 2013;1042:117–137. doi: 10.1007/978-1-62703-526-2_9. [DOI] [PubMed] [Google Scholar]
- 84.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Laughery M.F., Hunter T., Brown A., Hoopes J., Ostbye T., Shumaker T., Wyrick J.J. New vectors for simple and streamlined CRISPR-Cas9 genome editing in Saccharomyces cerevisiae. Yeast Chichester Engl. 2015;32:711–720. doi: 10.1002/yea.3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Brouwer I., Patel H.P., Meeussen J.V.W., Pomp W., Lenstra T.L. Single-molecule fluorescence imaging in living Saccharomyces cerevisiae Cells. Star Protoc. 2020;1 doi: 10.1016/j.xpro.2020.100142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Edelstein A.D., Tsuchida M.A., Amodaj N., Pinkard H., Vale R.D., Stuurman N. Advanced methods of microscope control using μManager software. J. Biol. Methods. 2014;1 doi: 10.14440/jbm.2014.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.de Jonge W.J., O’Duibhir E., Lijnzaad P., van Leenen D., Groot Koerkamp M.J., Kemmeren P., Holstege F.C. Molecular mechanisms that distinguish TFIID housekeeping from regulatable SAGA promoters. EMBO J. 2017;36:274–290. doi: 10.15252/embj.201695621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Candelli T., Challal D., Briand J.-B., Boulay J., Porrua O., Colin J., Libri D. High-resolution transcription maps reveal the widespread impact of roadblock termination in yeast. EMBO J. 2018;37 doi: 10.15252/embj.201797490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Coulon A., Ferguson M.L., De Turris V., Palangat M., Chow C.C., Larson D.R. Kinetic competition during the transcription cycle results in stochastic RNA processing. eLife. 2014;3:1–22. doi: 10.7554/eLife.03939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fu X., Patel H.P., Coppola S., Xu L., Cao Z., Lenstra T.L., Grima R. Quantifying how post-transcriptional noise and gene copy number variation bias transcriptional parameter inference from mRNA distributions. eLife. 2022;11 doi: 10.7554/eLife.82493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Garcia D.A., Fettweis G., Presman D.M., Paakinaho V., Jarzynski C., Upadhyaya A., Hager G.L. Power-law behavior of transcription factor dynamics at the single-molecule level implies a continuum affinity model. Nucleic Acids Res. 2021;49:6605–6620. doi: 10.1093/nar/gkab072. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A representative video of an induced yeast cell showing MS2 (magenta) and PP7 (green) transcription sites (TSs), which are outlined by the white boxes. MS2 and PP7 TSs were imaged sequentially at 10s intervals for 180 frames with 9 z-slices (Δz = 0.5 μm). Video is a maximum intensity xy-projection with xz- and yz-projections shown as side views
A representative single-plane video of induced yeast cells expressing Gal4-HaloTag molecules which are sparsely labeled with JFX650 dye. The cells were imaged with 200 ms intervals for 900 frames
Data Availability Statement
-
•
The sequencing (MNase-seq and Pol II CRAC-seq) data from this publication have been deposited and are publicly available as of the date of publication. Accession numbers and DOI are listed in the key resources table. Microscopy data is available upon request.
-
•
All original code for the live-cell, smFISH, SMT and MNase-seq analysis has been deposited to Zenodo. DOI are listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.