Abstract
The d105 dominant-negative mutant form of the herpes simplex virus 1 (HSV-1) single-stranded DNA-binding protein, ICP8 (d105 ICP8), inhibits wild-type viral replication, and it blocks both viral DNA replication and late gene transcription, although to different degrees (M. Gao and D. M. Knipe, J. Virol. 65:2666–2675, 1991; Y. M. Chen and D. M. Knipe, Virology 221:281–290, 1996). We demonstrate here that this protein is also capable of preventing the formation of intranuclear prereplicative sites and replication compartments during HSV infection. We defined three patterns of ICP8 localization using indirect immunofluorescence staining of HSV-1-infected cells: large replication compartments, small compartments, and no specific intranuclear localization of ICP8. Cells that form large replication compartments replicate viral DNA and express late genes. Cells that form small replication compartments replicate viral DNA but do not express late genes, while cells without viral replication compartments are incapable of both DNA replication and late gene expression. The d105 ICP8 protein blocks formation of prereplicative sites and large replication compartments in 80% of infected cells and formation of large replication compartments in the remaining 20% of infected cells. The phenotype of d105 suggests a correlation between formation of large replication compartments and late gene expression and a role for intranuclear rearrangement of viral DNA and bound proteins in activation of late gene transcription. Thus, these results provide evidence for specialized machinery for late gene expression within replication compartments.
Herpes simplex virus 1 (HSV-1) is a double-stranded DNA virus with a 150-kbp genome. During productive infection, it replicates in the nucleus of the host cell. HSV gene expression occurs in a temporally regulated cascade in which viral genes are transcribed in a specific order, and their expression is tightly controlled by other HSV gene products. The immediate-early (IE) genes are transcribed first after viral DNA is deposited in the nucleus and encode multiple activators of viral gene expression. These proteins stimulate expression of the early (E) genes, which include the viral DNA replication proteins. Seven viral proteins are required for HSV DNA replication: the helicase-primase complex (UL5, UL8, and UL52), the origin-binding protein (UL9), the single-stranded DNA-binding protein (UL29 or ICP8), the DNA polymerase (UL30), and the polymerase processivity factor (UL42). In addition to these seven proteins, unknown host cell factors are likely to be required, as it has not been possible to initiate origin-dependent HSV DNA replication outside an intact cell nucleus. Once viral DNA has been replicated, the late (L) genes are expressed. DNA replication and late gene expression have always been considered to be tightly linked, and it had been difficult to separate the two processes genetically (36).
Viral DNA replication in HSV-infected cells occurs within specific regions of the cell nucleus. When the locations of viral replication proteins and viral DNA are visualized in infected cells by indirect immunofluorescence or with a viral protein fused to the green fluorescent protein (GFP), they are all observed to congregate in compartments that start as small dots early in infection (termed prereplicative sites) and grow to eventually fill the nucleus (8, 33; T. J Taylor, E. E. McNamee, and D. M. Knipe, unpublished data). These globular structures were named replication compartments (33). Viral DNA replication (8) and much of the late gene transcription occur within the boundaries of these structures (21, 32, 34, 39). When viral replication is blocked, the replication proteins are still targeted to and remain in the small punctate prereplicative sites (33). A study of binucleate cells demonstrated that the shape and location of replication compartments within the nucleus are determined by the host cell nuclear architecture (9). It has also been demonstrated that some of the prereplicative sites observed early in infection are localized adjacent to the nuclear ND10 sites (26, 30, 40). ND10 sites are nuclear matrix-associated complexes containing PML, Sp100, and other proteins (1, 11, 22, 41). Their function in uninfected cells is not yet known, but in HSV-infected cells they are disrupted by proteasomal degradation in a pathway requiring the HSV immediate-early gene product ICP0 (12, 13, 28, 29). It is thought that ND10 proteins serve as markers of a specific site of viral DNA deposition upon entry into the cell nucleus, as the genomes of other DNA viruses are also known to be present next to ND10 (19).
ND10 sites serve as the initial site of HSV DNA deposition, and immediate-early and early gene transcription occurs at these sites (30). Once the immediate-early genes are expressed, ICP0 leads to the disruption of the ND10 sites, but the viral DNA and viral replication proteins remain, and prereplicative sites are formed adjacent to the original ND10 site locations (2, 30, 40). Viral DNA replication is initiated here, and replication compartments are formed (2, 30, 33, 40).
ICP8, the single-stranded DNA-binding protein, is an essential part of the DNA replication machinery (5, 7). ICP8 plays several additional roles in the HSV lytic life cycle. Genetic studies have demonstrated that ICP8 is required for the localization of viral replication and cellular proteins to replication compartments (3, 8). ICP8 is also implicated in the regulation of gene expression by exerting a negative effect on transcription from the parental genome (16–18) and a positive effect on late gene expression from progeny genomes (15).
This report continues the description of the d105 mutant of ICP8 and this mutant's ability to differentially affect DNA replication and late gene expression. The d105 ICP8 mutant contains a deletion near its C terminus (residues 1082 to 1169), which leaves the nuclear localization signal intact. Previous reports have demonstrated that d105 ICP8 acts as a dominant-negative repressor of wild-type ICP8 activity (6, 15). When expressed in Vero cells, either by transient transfection or in a stably transfected cell line, d105 ICP8 inhibits the replication of wild-type virus by 50- to 100-fold. d105 ICP8 can bind single-stranded DNA with an affinity similar to that of wild-type ICP8, and transfection of large amounts of the wild-type ICP8 gene can overcome its inhibitory effect, indicating that d105 ICP8 is most likely acting as a competitive inhibitor of wild-type ICP8. d105 ICP8 manifests its repression of ICP8 function with effects on both DNA replication and late gene expression. To study these effects, we isolated a cell line that stably expresses the d105 ICP8 protein (V2.6 cell line) (15). When V2.6 cells are infected with wild-type HSV-1, there is a fivefold reduction in DNA replication and a 50- to 100-fold reduction in late gene expression (15), which is manifested at the transcriptional level (6). Previous experiments have demonstrated that the 50- to 100-fold repression of late gene expression in V2.6 cells is far greater than what would occur in normal infection with DNA replication levels reduced to 20% of the wild-type level (6, 15). Here, we demonstrate that the d105 ICP8 protein fails to localize to replication compartments and prevents wild-type ICP8 and the other replication proteins and transcription factors from localizing to prereplicative sites and replication compartments as well.
MATERIALS AND METHODS
Cells and viruses.
Vero (American Type Culture Collection [ATCC]) and CV-1 (ATCC) monkey kidney cells were grown and maintained as described previously (20). The S2 and V2.6 cell lines expressing wild-type ICP8 and d105 ICP8, respectively, were derived from Vero cells (14, 15). The C8 and C105 cell lines expressing ICP8 and d105 ICP8, respectively, were derived from CV-1 cells, as described below. All transformed cell lines were grown in Dulbecco's modification of Eagle's medium (DMEM; Media Tech, Herndon, Va.)–10% fetal bovine serum containing 500 μg of G418 (GIBCO) per ml. The HSV-1 wild-type strain KOS was propagated and titered as described previously (20). The 8GFP virus containing an ICP8-GFP fusion in the ICP8 locus of HSV-1 wild-type strain KOS1.1 was propagated on S2 cells, and the titers of the virus were determined on both Vero cells and S2 cells (Taylor et al., unpublished). The n212 ICP0 mutant virus containing a nonsense mutation in the ICP0 gene of HSV-1 KOS (4) was kindly provided by Priscilla Schaffer (University of Pennsylvania). The titer of the n212 virus was determined by titration on a complementing cell line (U20S cells). The n212 virus was used a multiplicity of infection (MOI) of 2, which was shown to be sufficient for the formation of replication compartments (data not shown).
Plasmids.
The p8B-S (ICP8 gene in a BamHI-SacI fragment), pSVneo (neomycin gene driven by the simian virus 40 promoter) plasmids were described previously (14). The pSVd105 (d105 ICP8 gene driven by the simian virus 40 promoter) plasmid was described previously (15).
Isolation of stably transfected cell lines.
The C8 CV-1 cell line expressing wild-type ICP8 and the C105 CV-1 cell line expressing d105 ICP8 were constructed by transfecting CV-1 cells with either p8B-S and pSVneo or pSVd105 and pSVneo plasmids, respectively. Cells were incubated in medium containing G418 (500 μg/ml) until colonies of cells formed. Colonies of cells were picked, expanded, and tested for either complementation of an ICP8 mutant virus (C8 cells) or repression of wild-type virus infection (C105 cells).
Infections.
Infections were performed at an MOI of 2 PFU per cell. Virus was diluted in cold phosphate-buffered saline (PBS) containing 0.1% glucose and 1% heat-inactivated newborn calf serum and incubated with cells for 1 h at 37°C. The overlay medium was then changed to medium 199 containing 1% heat-inactivated calf serum. In infections containing n-butyrate to block the host cell cycle in the G1 phase (37), the growth medium was replaced with medium 199 containing 1% calf serum supplemented with 100 μM n-butyric acid and 20 mM HEPES buffer (pH 7.6) 12 to 15 h prior to infection as described previously (40). A 100× n-butyric acid–HEPES solution was made up fresh for each experiment. For bromodeoxyuridine (BrdU) incorporation, a 100× (10 mM) stock was made up in DMEM and frozen in aliquots. BrdU was added to the medium 30 min prior to harvesting cells.
Indirect immunofluorescence and antibodies.
Cells were grown on glass coverslips in 24-well plates. At the times indicated, the coverslips were washed in PBS, and the cells were fixed in 2% formaldehyde in PBS for 5 min and then permeabilized in 100% acetone at −20°C for 2 min. When necessary, cells were treated with 4 N HCl for 10 min to expose the BrdU epitopes. Cells were then incubated with the indicated primary antibodies for 30 min at 37°C, washed three times, and incubated with either fluorescein- or rhodamine-labeled secondary antibodies for 30 min at 37°C. The coverslips were washed three times and mounted on glass slides in glycerol gelatin (Sigma) containing 1.3 mg of p-phenyldiamine (Sigma) per ml to reduce photobleaching. The 3-83 rabbit antiserum against ICP8 was described previously (21) and was used at a 1:300 dilution. The ICP8 monoclonal antibody (MAb) 39S (38) was prepared from ascitic fluid samples from animals inoculated with 39S hybridoma cells originally obtained from the ATCC and was used at a 1:30 dilution. The ICP4 MAb 58S a gift from Neal DeLuca, University of Pittsburgh, was used at a 1:20 dilution. The gC MAb C3 a gift from Joseph Glorioso, University of Pittsburgh, was used at a 1:100 dilution. The PML rabbit antiserum, a gift from Anne Dejean, Institut Pasteur, was used at a 1:200 dilution. The BrdU MAb was purchased from Becton Dickinson and used at a 1:10 dilution. Rhodamine isothiocyanate (RITC)- and fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit and RITC- and FITC-conjugated goat anti-mouse secondary antibodies were all purchased from ICN and used at a 1:100 dilution.
Microscopy.
All microscopic images were obtained with a Zeiss Axioplan 2 microscope, captured with a Hamamatsu ORCA digital camera, colorized and processed with Improvision Openlab software, and printed with Adobe Photoshop.
RESULTS
Construction of the transformed cell lines.
All previous experiments examining d105 ICP8 repression of viral replication had been performed in the Vero-derived S2 and V2.6 cells (6, 15). In this work, we used multiple cell lines (Table 1) expressing either wild-type ICP8 or d105 ICP8. It was necessary to generate the CV-1-derived C8 and C105 cell lines for experiments involving the use of a cell cycle inhibitor (n-butyrate) to block cellular DNA replication so that specifically viral DNA replication could be visualized, as Vero cells are not susceptible to n-butyrate (40; E. McNamee and D. M. Knipe, data not shown). After constructing the C8 and C105 cell lines, we determined that the amount of ICP8 protein expressed by these cells after infection was very similar to the amount expressed in S2 and V2.6 cells (15; McNamee and Knipe, data not shown).
TABLE 1.
Cell lines used in this study
| Cell line | Parental cell | Contransfected viral genea |
|---|---|---|
| S2 | Vero | Wild-type ICP8 |
| C8 | CV-1 | Wild-type ICP8 |
| V2.6 | Vero | d105 ICP8 |
| C105 | CV-1 | d105 ICP8 |
Parental cell line was contransfected with pSVneo and the indicated viral gene.
The d105 ICP8 block in viral replication is not overcome at late times in infection.
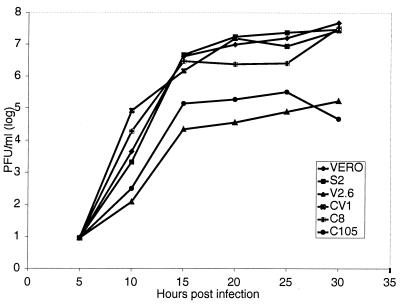
All previous experiments had been performed at 10 hours postinfection (hpi). To ensure that d105 ICP8 was blocking viral replication as opposed to simply delaying infection, we measured the amount of infectious virus present in Vero, S2, V2.6, CV-1, C8, and C105 cells (Table 1) at various times postinfection (Fig. 1). Cells were infected at an MOI of 2, and progeny virus was harvested at 5-h intervals until 30 hpi. The titer of virus produced by each cell line at each time point on Vero cells was then determined. Virus yields in V2.6 cells and C105 cells were 10- to 100-fold lower than those in control cells through 30 hpi. In all experiments, the CV-1-derived cell lines and the Vero-derived cell lines always behaved similarly. This indicated that the effect of d105 ICP8 was not to delay the replication of the virus due to slowed rates of DNA replication and/or late gene expression; instead, it was capable of maintaining a long-term inhibition of virus production.
FIG. 1.
Growth of HSV-1 in Vero, CV-1, S2, C8, V2.6, and C105 cells infected with HSV-1 wild-type strain KOS. All cells were infected at an MOI of 2 and harvested at the indicated times. Viral yield was measured by plaque assay titration of total intracellular plus extracellular virus on Vero cells.
ICP8 does not localize to prereplicative sites and replication compartments in d105 ICP8-expressing cell lines.
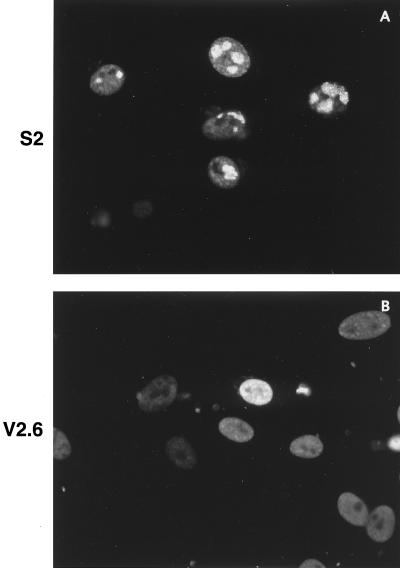
When d105 ICP8 is expressed during HSV-1 infection, it blocks DNA replication and late gene expression of wild-type virus, although to different degrees (15). We hypothesized that the d105 ICP8 mutant may be unable to form the same interactions with the cell that wild-type ICP8 does and thus may not be capable of localizing to the replication compartments containing the seven essential viral replication proteins that form in infected-cell nuclei. To examine the localization of ICP8 in the presence of d105 ICP8, the wild-type ICP8-expressing S2 cells and the d105 ICP8-expressing V2.6 cells were infected with wild-type HSV-1 at an MOI of 2, harvested at 10 hpi, and stained for ICP8 by immunofluorescence. These infection conditions were chosen because at higher MOIs, wild-type ICP8 from the virus is expressed at a level high enough to overcome the competitive inhibition of viral replication by d105 ICP8 (15). At 10 hpi, ICP8 was localized to replication compartments in S2 cells (Fig. 2A), as is normally seen during wild-type viral infections. In the presence of d105 ICP8, ICP8 staining was distributed diffusely throughout the nucleus, with occasional punctate sites in some nuclei (Fig. 2B). Thus, d105 ICP8 exhibited an effect on ICP8 localization, in addition to blocking DNA replication and late gene transcription.
FIG. 2.
Localization of ICP8 in S2 and V2.6 cells. Cells were infected with HSV-1 KOS strain at an MOI of 2 and harvested at 10 hpi. ICP8 was detected with the anti-ICP8 rabbit antiserum 3-83.
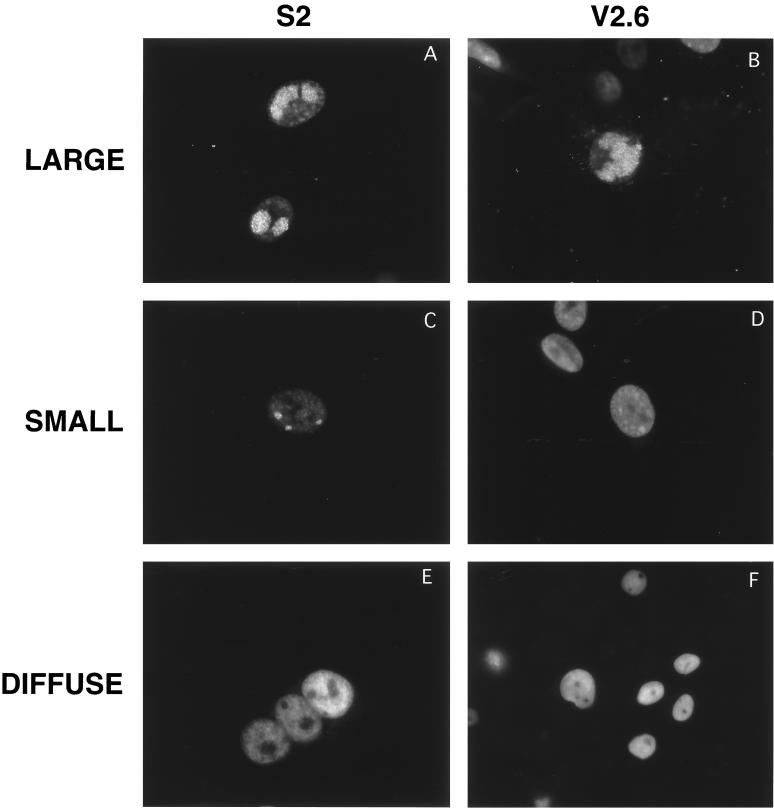
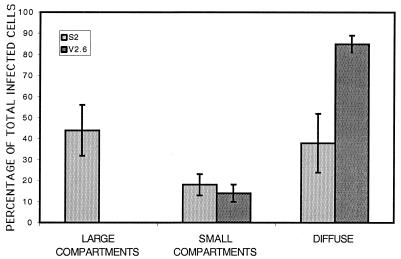
In infected S2 and V2.6 cells, three general patterns of ICP8 staining were observed. These were defined as large replication compartments (Fig. 3A and B), small replication compartments (Fig. 3C and D), and diffuse staining (Fig. 3E and F). When we quantified the type of intranuclear staining in infected cells, we observed that under these conditions nearly one half of the infected S2 cells contained large replication compartments, while fewer cells contained small compartments or diffuse ICP8 (Fig. 4). In contrast, in V2.6 cells, 85% of the infected cells exhibited a diffuse ICP8 distribution (Fig. 4), showing that the lack of replication compartment formation in the d105 ICP8-expressing V2.6 cells was a general observation. The diffuse distribution for ICP8 in V2.6 cells was not simply due to inhibition of viral DNA synthesis because inhibition of viral DNA synthesis by inhibitors or genetic defects causes ICP8 to accumulate in punctate prereplicative sites (33). Thus, d105 ICP8 appeared to block the localization of wild-type ICP8 to both prereplicative sites and replication compartments.
FIG. 3.
Examples of different patterns of intranuclear localization of replication proteins in S2 and V2.6 cells infected with HSV-1 KOS strain. Cells were stained with the ICP8 antiserum 3-83. S2 cells expressing wild-type ICP8 (left) and V2.6 cells expressing d105 ICP8 (right) were used. Examples of large compartments (A and B), small compartments (C and D), and diffuse staining (E and F) are depicted.
FIG. 4.
Percentages of infected cells in each class of intranuclear localization of replication proteins. Cells were stained by immunofluorescence for ICP8 with 3-83 rabbit serum, and infected cells were counted and classified as containing large compartments, small compartments, or diffuse staining. The percentage of each in S2 and V2.6 cells are shown. More than 400 cells were counted for each cell line in three separate experiments with at least two coverslips of infected cells per sample per experiment. The error bars show standard deviations of the results of three experiments.
d105 ICP8 prevents wild-type ICP8 and other replication proteins from localizing to replication compartments.
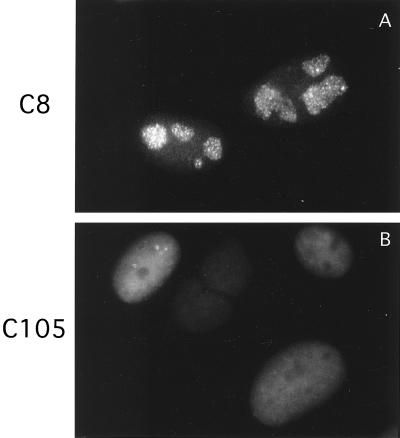
Figure 2 demonstrated that the ICP8 visualized in those experiments was not localizing to replication compartments. However, the anti-ICP8 antibody used was reactive with both wild-type and d105 ICP8. Thus, it was conceivable that the d105 ICP8 was obscuring the wild-type ICP8. To ensure that this was not the case, we infected C8 and C105 cells with an HSV-1 recombinant expressing an ICP8-GFP fusion protein, known to localize to replication compartments as efficiently as wild-type ICP8 (Taylor et al., unpublished). The infected cells were fixed, and GFP was visualized. As observed previously by immunofluorescence, the ICP8-GFP fusion protein localized to replication compartments in S2 cells and was diffuse in V2.6 cell nuclei (Fig. 5). This demonstrated that d105 ICP8 was actually capable of blocking the localization of wild-type ICP8 to replication compartments. Similarly, infected C8 and C105 cells were also stained by immunofluorescence for ICP4, the major viral transactivator (Fig. 6), and UL42, the polymerase accessory factor (data not shown), two proteins known to be localized to replication compartments. Staining for these proteins showed them in replication compartments in both S2 and C8 cells but diffusely distributed in V2.6 and C105 cells.
FIG. 5.
Localization of the ICP8-GFP fusion protein after infection with the 8GFP virus in C8 and C105 cells. The 8GFP-infected cells were harvested and fixed.
FIG. 6.
Localization of ICP4. Localization of IE protein ICP4 in C8 and C105 cells after HSV-1 KOS infection. The cells were stained for ICP4 with the MAb 58S.
DNA replication occurs in large and small compartments.
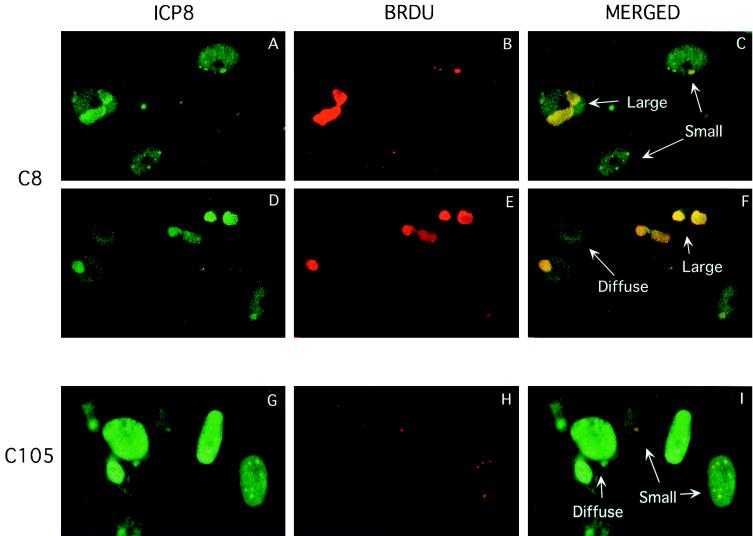
Having shown that formation of large replication compartments was blocked in d105 ICP8-expressing cells, we wished to determine the location of the residual viral DNA synthesis (20% of wild-type levels) observed in these cells. This was accomplished using indirect immunofluorescence to visualize the incorporation of the nucleoside analog BrdU into replicating DNA. To visualize viral DNA synthesis specifically, it was necessary to block the host cell's DNA replication. This can be done with n-butyrate, which stops the cell cycle in the G1 phase but does not have any effect on HSV DNA replication (37). Unfortunately, n-butyrate did not efficiently block the cell cycle in Vero cells (results not shown), so we used CV-1 cells, which have been shown to be susceptible to n-butyrate (40; data not shown). We used the C8 and C105 cell lines expressing wild-type ICP8 and d105 ICP8, respectively, to identify sites of viral DNA replication. In both C8 and C105 cell lines, we observed BrdU labeling only in those cells that contained defined replication compartments, as seen by ICP8 staining (Fig. 7). Both small and large replication compartments supported DNA replication. BrdU labeling was also seen in the few large compartments in C105 cells (data not shown). In the numerous C105 cells that contained diffuse ICP8, BrdU incorporation was not observed (Fig. 7G to I). C8 cells containing diffuse ICP8 also did not contain replicating viral DNA (Fig. 7D to F). Thus, the DNA replication observed in the C105 cells within small replication compartments and the few large replication compartments may account for the 20% level of wild-type DNA replication previously observed by biochemical analyses (15; data not shown).
FIG. 7.
Colocalization of ICP8 and sites of DNA synthesis in C8 (A to F) and C105 (G to I) cells. Examples of the three classes of intranuclear ICP8 localization and visualization of DNA replication are shown. ICP8 is shown in green, and BrdU is shown in red. Rabbit antiserum 3-83 was used to visualize ICP8, and anti-BrdU was used to visualize BrdU. The yellow staining in the merged images demonstrates the sites of overlap in the staining for ICP8 and BrdU.
Late gene expression in C105 cells correlated with the formation of large replication compartments.
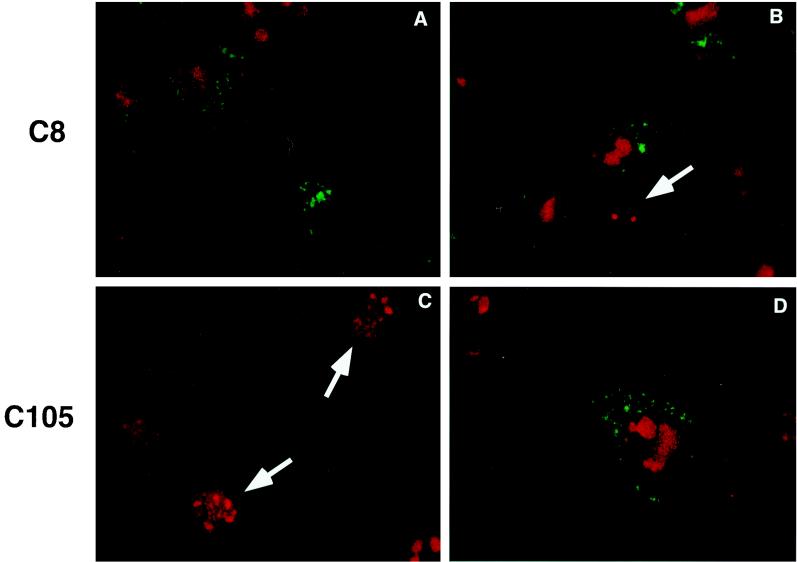
Biochemical analyses had demonstrated that d105 ICP8 reduced viral DNA replication and late gene transcription to 20 and 2% of wild-type levels, respectively (6, 15). Previously, it had been believed that these two viral processes were completely linked, but these data indicated that there is an additional factor beyond DNA replication required for successful viral late gene expression. We used immunofluorescence to examine the relationship between replication compartment formation and late gene expression in both C8 and C105 cells. Infected cells were fixed at 10 hpi and stained with antibodies specific for ICP8 and the late glycoprotein, gC. In both the C8 and C105 cell populations, gC staining was observed only in those cells that formed large replication compartments (Fig. 8). Thus, the viral DNA replication in small compartments was not sufficient to stimulate late gene expression. Thus, the limited number of large replication compartments in d105 ICP8-expressing cells explained the near-total lack of late gene expression.
FIG. 8.
Examples of different classes of intranuclear localization of ICP8 and gC expression in these cells. Cells were infected with HSV-1 KOS and dual labeled with antibodies to ICP8 (3-83) and gC (C3). gC is shown in green, and ICP8 is shown in red. (A and B) Two different fields of C8 cells, demonstrating the expression of gC in cells that form large compartments but not in cells with small compartments (arrow). (C and D) Two fields of C105 cells showing expression of gC in one cell with large compartments and the lack of gC expression in cells with small compartments or diffuse staining.
ICP8 localizes near ND10 sites in a minor population of d105 ICP8-expressing cells.
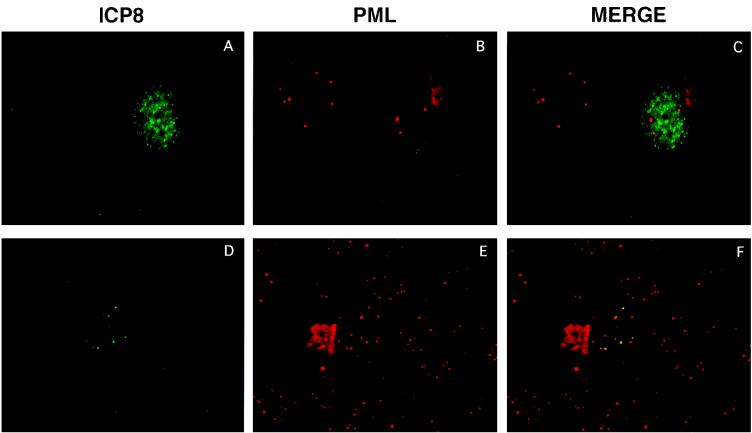
d105 ICP8 blocked prereplicative site and replication compartment formation in 80% of infected cells, and as a result of the lack of formation of large replication compartments, DNA replication and late gene expression were inhibited. To define the site of ICP8 localization in the remaining cells, we determined if ICP8 could localize to ND10 sites in the presence of d105 ICP8. To avoid the ICP0-induced disruption of ND10 epitopes such as PML, we infected cells with an ICP0 mutant virus, n212. After harvesting at 10 hpi, the cells were processed for immunofluorescence and double labeled with antibodies specific for ICP8 and PML. In C105 cells that showed specific sites of ICP8 localization, these sites were located adjacent to the punctate PML staining (Fig. 9A to C), indicating that in these cells, d105 ICP8 did not block localization of wild-type ICP8 to sites near ND10. However, most of the cells showed no specific intranuclear localization of ICP8 (Fig. 9A to C). Therefore, while most d105-expressing cells do not contain ICP8 localized to any specific intranuclear sites, including those near ND10, in the small population of cells that do contain specific sites of ICP8 localization, these sites do correlate with ND10 sites.
FIG. 9.
Dual staining of ICP8 and PML in C105 cells infected with the ICP0 null mutant virus n212. A representative cell that formed small replication compartments dually stained with antibodies against ICP8 (39S) and PML. The yellow staining in the merged image indicates colocalization of the two proteins.
DISCUSSION
We conducted these experiments to explore the nature of the defect in viral replication induced by the dominant-negative mutant d105 ICP8, but the results have also provided information about the role of replication compartments in viral growth. The d105 mutant ICP8 blocks localization of wild-type ICP8 and other viral proteins to replication structures in most infected cells, which explains at least in part, the ability of d105 ICP8 to reduce viral DNA synthesis and late gene expression. Late gene expression correlated with formation of large replication compartments, indicating that progeny viral DNA seems to undergo a change in location or molecular contacts in the large replication compartments that allow increased transcription of late genes.
In the population of d105 ICP8-expressing cells where no specific replication protein localization is observed, approximately 80% of the infected cells, d105 ICP8 appears to prevent the formation of prereplicative sites and replication compartments. The primary effect of d105 in these cells is likely to be on localization rather than inhibition of DNA synthesis because many prior studies have shown that inhibition of DNA synthesis by antiviral drugs or genetic defects in other HSV proteins causes ICP8 to localize to prereplicative sites (3, 8, 25, 27, 33). The d105 ICP8 defect could be due to a defect in localization per se or an interaction with a viral or cellular protein involved in prereplicative site formation.
Approximately 20% of d105 ICP8-expressing cells do form small compartments, indicating that there may be a second site of d105 ICP8 inhibition. This second block occurs after the formation of prereplicative sites and the initiation of DNA synthesis but before late gene expression. These blocks at different stages would have different effects on the status of DNA replication in the cells. When d105 ICP8 blocks prior to the assembly of replication proteins, the replication proteins remain diffusely distributed and there is no DNA replication. This could account for the fivefold reduction in viral DNA replication seen in the presence of d105 ICP8. In the population of cells where the first block can be overcome, a limited amount of DNA is replicated in the small replication compartments that form in these cells. This may account for the 20% level of DNA replication that remains in the presence of d105 ICP8.
The two d105 ICP8-induced blocks in the viral life cycle would have different effects on late gene expression than on DNA replication. In this case, blocking either before prereplicative site formation or after the initiation of DNA synthesis is sufficient to completely prevent late gene expression in those cells. The only cells that can support late gene expression are those in which there is no inhibition of large replication compartment formation. This subpopulation of cells may have lost the ability to express the mutant d105 ICP8. In those cells that maintain DNA replication yet lack late gene expression, there may be insufficient DNA replication to trigger the initiation of late gene expression. Alternatively, there may be a joint signal that acts to stimulate both the formation of large compartments and late gene expression through separate pathways. The level of late gene expression in d105 ICP8-expressing cells is lower than that in cells infected with wild-type virus and treated with phosphonoacetic acid to reduce DNA synthesis to 20% of control levels (6, 15). This supports the latter hypothesis that there is a common signal that controls both large replication compartment formation and late gene expression and this signal is inhibited by d105 ICP8. One possible mechanism may be a rearrangement of the structure of replicating DNA which allows interactions with different proteins. This could involve increased access to DNA by replication proteins and transcription factors, therefore stimulating DNA replication and late gene transcription.
There are a number of ways in which d105 ICP8 may exert its dominant-negative activity. These include the following: (i) binding other viral proteins and preventing their proper intranuclear localization; (ii) binding viral DNA but not viral proteins, therefore displacing wild-type ICP8; (iii) lack of the ability to bind an unknown cellular factor required for proper targeting of viral DNA replication proteins; or (iv) a combination of mechanisms ii and iii. We believe that d105 ICP8 is capable of binding to other viral proteins, because it is recognized by the 39S antibody, which specifically recognizes ICP8 when it is in replication complexes with DNA and the other replication proteins (S. L. Uprichard and D. M. Knipe, unpublished data). In addition, d105 ICP8 can bind DNA in vitro with the same affinity as that of wild-type ICP8 (15). Therefore, d105 ICP8 appears to be capable of forming normal interactions with herpesvirus replication proteins and viral DNA. This leaves the loss of ability to interact with a cellular protein. d105 ICP8 may be able to interact with viral proteins initially but lacks the ability to maintain those interactions during a rearrangement of proteins required for the formation of either prereplicative sites or replication compartments. Each of these possibilities is feasible, and further studies into the relationships between ICP8 and both host cell proteins and viral proteins are under way.
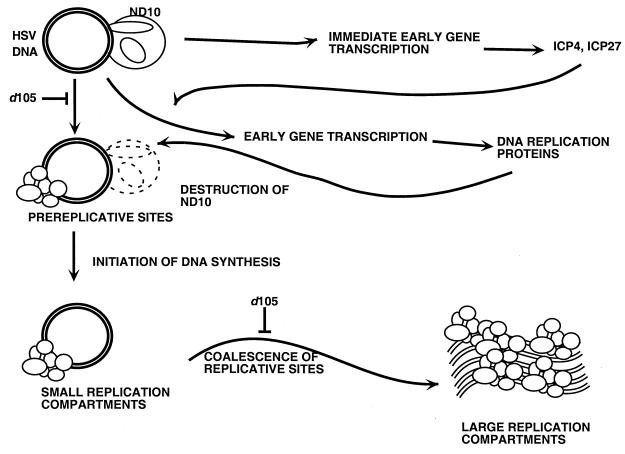
Model for nuclear events in HSV-infected cells.
These results and previous studies have led to a model of nuclear events in HSV-infected cells (Fig. 10), and we can use this to identify steps in the progression of infection where d105 ICP8 may be exerting its inhibitory effect. Initially, the viral DNA is uncoated from the capsid, transported into the nucleus, and localized adjacent to ND10 proteins (30). Immediate-early and early gene transcription occurs here, and ICP0 causes disruption of ND10 structures (12, 13, 28, 29). Once the replication proteins are synthesized, they assemble on the viral DNA to form prereplicative sites and replication is initiated (40). This leads to the formation of small replication compartments. A limited amount of DNA replication occurs here, but this accumulation of primary replication machinery is not sufficient to initiate late gene expression. An unknown stimulus allows the formation of larger replication compartments, either by the growth of smaller compartments or the coalescence of multiple small compartments (Taylor et al., unpublished). Large compartments contain new interactions between viral DNA and cellular and/or viral proteins. ICP8 and specifically the region deleted in the d105 mutation are required for this transformation. The formation of large compartments allows high levels of DNA replication and initiation of late gene transcription. After late genes are expressed and all viral DNA is replicated, the progeny genomes are packaged into capsids and they exit the nucleus.
FIG. 10.
Model of intranuclear events in cells infected with HSV.
The correlation between formation of large replication compartments and late gene expression provides further evidence that specialized transcriptional machinery is assembled in replication compartments for late gene expression (32). Prior evidence for late transcriptional machinery in HSV-1-infected cells includes the localization of ICP4 (21, 24, 34), host RNA polymerase II (24, 35), ICP27 (10), and ICP22 (24, 35) to replication compartments. This work shows that ICP8 plays a role in promoting the formation of this new late transcriptional machinery in the large compartments.
Potential implication for HSV infection of neuronal cells.
The late transcriptional machinery may also play a role in expression of immediate-early and early genes under certain conditions. We and others have shown that immediate-early and early gene expression in neuronal cells is stimulated by DNA replication (23, 31). The transcriptional sites near the ND10 sites (Fig. 10) may be absent or unavailable in neurons, making transcription of immediate-early and early genes very inefficient. However, if sufficient viral gene products are expressed to allow viral DNA replication and formation of large replication compartments, this may allow transcription of the viral genome in these compartments and greatly increase expression of immediate-early and early mRNAs as well as late mRNAs. Thus, the specialized transcriptional machinery may play a role in late gene transcription in permissive cells and in expression of all HSV genes in nonpermissive neuronal cells. d105 ICP8 provides us with a unique tool to probe the role of ICP8 interactions with both cell and viral proteins in the formation of replication compartments, DNA replication, and late gene expression.
ACKNOWLEDGMENTS
We thank David Spezzano for technical assistance in isolation of the C8 and C105 cell lines and William Lucas for helpful discussions.
This work was supported by NIH grant CA26345 from the National Cancer Institute.
REFERENCES
- 1.Ascoli C A, Maul G G. Identification of a novel nuclear site. J Cell Biol. 1991;112:785–795. doi: 10.1083/jcb.112.5.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burkham J, Coen D M, Weller S K. ND10 protein PML is recruited to herpes simplex virus type prereplicative sites and replication compartments in the presence of viral DNA polymerase. J Virol. 1998;72:10100–10107. doi: 10.1128/jvi.72.12.10100-10107.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bush M, Yager D R, Gao M, Weisshart K, Marcy A I, Coen D M, Knipe D M. Correct intranuclear localization of herpes simplex virus DNA polymerase requires the viral ICP8 DNA-binding protein. J Virol. 1991;65:1082–1089. doi: 10.1128/jvi.65.3.1082-1089.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cai W Z, Schaffer P A. Herpes simplex virus type 1 ICP0 plays a critical role in the de novo synthesis of infectious virus following transfection of viral DNA. J Virol. 1989;63:4579–4589. doi: 10.1128/jvi.63.11.4579-4589.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Challberg M D. A method for identifying the viral genes required for herpesvirus DNA replication. Proc Natl Acad Sci USA. 1986;83:9094–9098. doi: 10.1073/pnas.83.23.9094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Y M, Knipe D M. A dominant mutant form of the herpes simplex virus ICP8 protein decreases viral late gene transcription. Virology. 1996;221:281–290. doi: 10.1006/viro.1996.0377. [DOI] [PubMed] [Google Scholar]
- 7.Conley A J, Knipe D M, Jones P C, Roizman B. Molecular genetics of herpes simplex virus. VII. Characterization of a temperature-sensitive mutant produced by in vitro mutagenesis and defective in DNA synthesis and accumulation of gamma polypeptides. J Virol. 1981;37:191–206. doi: 10.1128/jvi.37.1.191-206.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Bruyn Kops A, Knipe D M. Formation of DNA replication structures in herpes virus-infected cells requires a viral DNA binding protein. Cell. 1988;55:857–868. doi: 10.1016/0092-8674(88)90141-9. [DOI] [PubMed] [Google Scholar]
- 9.de Bruyn Kops A, Knipe D M. Preexisting nuclear architecture defines the intranuclear location of herpesvirus DNA replication structures. J Virol. 1994;68:3512–3526. doi: 10.1128/jvi.68.6.3512-3526.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Bruyn Kops A, Uprichard S L, Chen M, Knipe D M. Comparison of the intranuclear distributions of herpes simplex virus proteins involved in different viral functions. Virology. 1998;252:162–178. doi: 10.1006/viro.1998.9450. [DOI] [PubMed] [Google Scholar]
- 11.Dyck J A, Maul G G, Miller W H J, Chen J D, Kakizuka A, Evans R M. A novel macromolecular structure is a target of the promyelocyte-retinoic acid receptor oncoprotein. Cell. 1994;76:333–343. doi: 10.1016/0092-8674(94)90340-9. [DOI] [PubMed] [Google Scholar]
- 12.Everett R D, Freemont P, Saitoh H, Dasso M, Orr A, Kathoria M, Parkinson J. The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J Virol. 1998;72:6581–6591. doi: 10.1128/jvi.72.8.6581-6591.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Everett R D, Maul G G. HSV-1 IE protein VMW 110 causes redistribution of PML. EMBO J. 1994;13:5062–5069. doi: 10.1002/j.1460-2075.1994.tb06835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao M, Knipe D M. Genetic evidence for multiple nuclear functions of the herpes simplex virus ICP8 DNA-binding protein. J Virol. 1989;63:5258–5267. doi: 10.1128/jvi.63.12.5258-5267.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao M, Knipe D M. Potential role for herpes simplex virus ICP8 DNA replication protein in stimulation of late gene expression. J Virol. 1991;65:2666–2675. doi: 10.1128/jvi.65.5.2666-2675.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Godowski P J, Knipe D M. Identification of a herpes simplex virus function that represses late gene expression from parental viral genomes. J Virol. 1985;55:357–365. doi: 10.1128/jvi.55.2.357-365.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Godowski P J, Knipe D M. Mutations in the major DNA-binding protein gene of herpes simplex virus type 1 result in increased levels of viral gene expression. J Virol. 1983;47:478–486. doi: 10.1128/jvi.47.3.478-486.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Godowski P J, Knipe D M. Transcriptional control of herpesvirus gene expression: gene functions required for positive and negative regulation. Proc Natl Acad Sci USA. 1986;83:256–260. doi: 10.1073/pnas.83.2.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishov A M, Maul G G. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J Cell Biol. 1996;134:815–826. doi: 10.1083/jcb.134.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knipe D M. Cell growth transformation by herpes simplex virus. Prog Med Virol. 1982;28:114–144. [PubMed] [Google Scholar]
- 21.Knipe D M, Senechek D, Rice S A, Smith J L. Stages in the nuclear association of the herpes simplex virus transcriptional activator protein ICP4. J Virol. 1987;61:276–284. doi: 10.1128/jvi.61.2.276-284.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Korioth F, Gieffers C, Maul G G, Frey J. Molecular characterization of NDP52, a novel protein of the nuclear domain 10, which is redistributed upon virus infection and interferon treatment. J Cell Biol. 1995;130:1–13. doi: 10.1083/jcb.130.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kosz-Vnenchak M, Jacobson J, Coen D M, Knipe D M. Evidence for a novel regulatory pathway for herpes simplex virus gene expression in trigeminal ganglion neurons. J Virol. 1993;67:5383–5393. doi: 10.1128/jvi.67.9.5383-5393.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leopardi R, Ward P L, Ogle W O, Roizman B. Association of herpes simplex virus regulatory protein ICP22 with transcriptional complexes containing EAP, ICP4, RNA polymerase II, and viral DNA requires posttranslational modification by the UL13 protein kinase. J Virol. 1997;71:1133–1139. doi: 10.1128/jvi.71.2.1133-1139.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liptak L, Uprichard S L, Knipe D M. Functional order of assembly of herpes simplex virus DNA replication proteins into prereplicative site structures. J Virol. 1996;70:1759–1767. doi: 10.1128/jvi.70.3.1759-1767.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lukonis C J, Burkham J, Weller S K. Herpes simplex virus type 1 prereplicative sites are a heterogeneous population: only a subset are likely to be precursors to replication compartments. J Virol. 1997;71:4771–4781. doi: 10.1128/jvi.71.6.4771-4781.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lukonis C J, Weller S K. Characterization of nuclear structures in cells infected with herpes simplex virus type 1 in the absence of viral DNA replication. J Virol. 1996;70:1751–1758. doi: 10.1128/jvi.70.3.1751-1758.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maul G G, Everett R D. The nuclear location of PML, a cellular member of the C3HC4 zinc-binding domain protein family, is rearranged during herpes simplex virus infection by the C3HC4 viral protein ICPO. J Gen Virol. 1994;75:1223–1233. doi: 10.1099/0022-1317-75-6-1223. [DOI] [PubMed] [Google Scholar]
- 29.Maul G G, Guldner H H, Spivack J G. Modification of discrete nuclear domains induced by herpes simplex virus type 1 immediate early gene 1 product (ICP0) J Gen Virol. 1993;74:2679–2690. doi: 10.1099/0022-1317-74-12-2679. [DOI] [PubMed] [Google Scholar]
- 30.Maul G G, Ishov A M, Everett R D. Nuclear domain 10 as preexisting potential replication start sites of herpes simplex virus type 1. Virology. 1996;217:67–75. doi: 10.1006/viro.1996.0094. [DOI] [PubMed] [Google Scholar]
- 31.Nichol P F, Chang J Y, Johnson E M, Jr, Olivo P D. Herpes simplex virus gene expression in neurons: viral DNA synthesis is a critical regulatory event in the branch point between the lytic and latent pathways. J Virol. 1996;70:5476–5486. doi: 10.1128/jvi.70.8.5476-5486.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Phelan A, Dunlop J, Patel A H, Stow N D, Clements J B. Nuclear sites of herpes simplex virus type 1 DNA replication and transcription colocalize at early times postinfection and are largely distinct from RNA processing factors. J Virol. 1997;71:1124–1132. doi: 10.1128/jvi.71.2.1124-1132.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quinlan M P, Chen L B, Knipe D M. The intranuclear location of a herpes simplex virus DNA-binding protein is determined by the status of viral DNA replication. Cell. 1984;36:857–868. doi: 10.1016/0092-8674(84)90035-7. [DOI] [PubMed] [Google Scholar]
- 34.Randall R E, Dinwoodie N. Intranuclear localization of herpes simplex virus immediate-early and delayed-early proteins: evidence that ICP 4 is associated with progeny virus DNA. J Gen Virol. 1986;67:2163–2177. doi: 10.1099/0022-1317-67-10-2163. [DOI] [PubMed] [Google Scholar]
- 35.Rice S A, Long M C, Lam V, Spencer C A. RNA polymerase II is aberrantly phosphorylated and localized to viral replication compartments following herpes simplex virus infection. J Virol. 1994;68:988–1001. doi: 10.1128/jvi.68.2.988-1001.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roizman B, Sears A. Herpes simplex viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2231–2296. [Google Scholar]
- 37.Shadan F F, Cowsert L M, Villarreal L P. n-Butyrate, a cell cycle blocker, inhibits the replication of polyomaviruses and papillomaviruses but not that of adenoviruses and herpesviruses. J Virol. 1994;68:4785–4796. doi: 10.1128/jvi.68.8.4785-4796.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Showalter S D, Zweig M, Hampar B. Monoclonal antibodies to herpes simplex virus type 1 proteins, including the immediate-early protein ICP 4. Infect Immun. 1981;34:684–692. doi: 10.1128/iai.34.3.684-692.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spencer C A, Dahmus M E, Rice S A. Repression of host RNA polymerase II transcription by herpes simplex virus type 1. J Virol. 1997;71:2031–2040. doi: 10.1128/jvi.71.3.2031-2040.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Uprichard S L, Knipe D M. Assembly of herpes simplex virus replication proteins at two distinct intranuclear sites. Virology. 1997;229:113–125. doi: 10.1006/viro.1996.8430. [DOI] [PubMed] [Google Scholar]
- 41.Weis K, Rambaud S, Lavau C, Jansen J, Carvalho T, Carmo-Fonseca M, Lamond A, Dejean A. Retinoic acid regulates aberrant nuclear localization of PML-RAR alpha in acute promyelocytic leukemia cells. Cell. 1994;76:345–356. doi: 10.1016/0092-8674(94)90341-7. [DOI] [PubMed] [Google Scholar]