Abstract
High prevalence of obesity is attributable in part to consumption of highly palatable, fat-rich foods. However, the mechanism controlling dietary fat intake is largely unknown. In this study we investigated the role of brain-derived neurotrophic factor (BDNF) in the control of dietary fat intake in a mouse model that mimics the common human Val-to-Met (Val66Met) polymorphism that impairs BDNF release via the regulated secretory pathway. BdnfMet/Met mice gained weight much faster than wild-type (WT) mice and developed severe obesity due to marked hyperphagia when they were fed HFD. Hyperphagia in these mice worsened when the fat content in their diet was increased. Conversely, mice lacking leptin exhibited similar hyperphagia on chow and HFD. When 2 diets were provided simultaneously, WT and BdnfMet/Met mice showed a comparable preference for the more palatable diet rich in either fat or sucrose, indicating that increased hyperphagia on fat-rich diets in BdnfMet/Met mice is not due to enhanced hedonic drive. In support of this interpretation, WT and BdnfMet/Met mice increased calorie intake to a similar extent during the first day after chow was switched to HFD; however, WT mice decreased HFD intake faster than BdnfMet/Met mice in subsequent days. Furthermore, we found that refeeding after fasting or nocturnal feeding with HFD activated TrkB more strongly than with chow in the hypothalamus of WT mice, whereas TrkB activation under these 2 conditions was greatly attenuated in BdnfMet/Met mice. These results indicate that satiety factors generated during HFD feeding induce BDNF release to suppress excess dietary fat intake.
Keywords: val66Met polymorphism, dietary fat intake, obesity, regulated BDNF release, TrkB activation
Diet-induced obesity is the most common form of human obesity, which largely results from over consumption of calorie-dense, highly palatable foods such as fat-rich foods (1, 2). The cause of the overconsumption is complex and includes cultural, sociological, psychological, and physiological factors (3–6). The physiological factors could include attenuated homeostatic regulation of energy balance or enhanced hedonic drives in response to ingestion of palatable foods. Hedonic drives are the result of sensory pleasure and activation of the reward pathway during consumption of palatable foods and are likely the neural basis for food preference (6, 7). However, the precise biochemical pathways or genetic factors controlling intake of dietary fats remains largely unknown.
Brain-derived neurotrophic factor (BDNF) is well known for its role in neuronal development and synaptic function (8–11). It exerts its biological actions mainly through its receptor TrkB (tropomyosin receptor kinase B) (10, 12). Substantial evidence has also established a crucial role for BDNF and TrkB in the control of energy balance (13, 14). Mutations that impair BDNF-TrkB signaling cause marked hyperphagia and severe obesity both in mice and humans (15–26). One mouse mutant with reduced expression of TrkB displays increased hyperphagia, following a switch from a chow diet to a high-fat diet (HFD) (16), suggesting that the BDNF-TrkB pathway is involved in the control of dietary fat intake.
Several BDNF polymorphisms, including the common rs6265 polymorphism that results in a valine to methionine substitution at codon 66 (27), have been associated with human obesity in large-scale genome-wide association studies (28–33). The allele frequency of the Val66Met polymorphism is 19.1% in the world population and the highest (44.5%) in Asians according to the NCBI SNP database. The Val-to-Met substitution is in the prodomain of the BDNF precursor and impairs BDNF release through the regulated secretory pathway (27, 34). In this study, using the BdnfMet allele that mimics the rs6265 polymorphism and produces a normal amount of BDNF in the hippocampus (34), we showed that BdnfMet/Met mice displayed increased hyperphagia on fat-rich diets without enhanced hedonic drive and quickly developed severe obesity on HFD. We further found that HFD feeding more strongly activated TrkB than chow feeding in the hypothalamus of wild-type (WT) but not BdnfMet/Met mice. These findings suggest that HFD feeding induces regulated BDNF release to suppress excessive intake of dietary fats.
Materials and Methods
Animals
All animals were from the C57BL/6 background. The BdnfMet knockin mouse strain (34) was kindly provided by Dr Francis Lee. We produced BdnfMet/Met mice and WT (BdnfVal/Val) through 2 breeding programs. In the first breeding program, intercrosses of BdnfMet/Val mice produced WT, BdnfMet/Val, and BdnfMet/Met mice, which were used in the experiments shown in Fig. 1 and Supplementary Fig. S1 (35). In the second breeding program, we set up WT × WT and BdnfMet/Met × BdnfMet/Met crosses using offspring initially from the first breeding program and then also from homozygous crosses. To avoid the genetic background of the 2 breeding lines deviating from each other, we refreshed breeding parents using offspring from BdnfMet/Val × BdnfMet/Val crosses once every 2 years. Mice from the second breeding program were used in all other experiments. C57BL/6J ob/ob mice (strain No. 000632) and age-matched C57BL/6J WT mice were purchased from the Jackson Laboratory. Mice were maintained at 22 °C with a 12/12-hour light/dark cycle and had free access to water and food. Information on different diets used in the study is summarized in Supplementary Table S1 (35). All animal procedures were approved by the Animal Care and Use Committee at UF Scripps Biomedical Research, University of Florida.
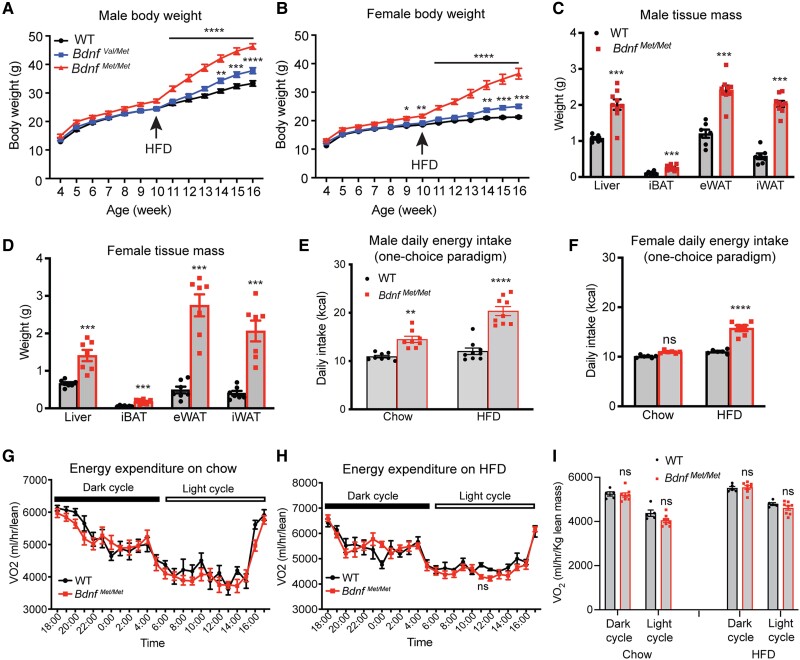
Figure 1.
BdnfMet/met mice develop hyperphagic obesity on a high-fat diet (HFD). A, Body weight of male wild-type (WT), BdnfVal/Met, and BdnfMet/Met mice. n = 10-15 mice per genotype; 2-way analysis of variance (ANOVA) (mixed-effects analysis) with Bonferroni multiple comparison test: P less than .0001 for genotype; **P less than .01, ***P less than .001, and ****P less than .0001 when compared with WT mice. B, Body weight of female WT, BdnfVal/Met, and BdnfMet/Met mice. n = 11-15 mice per genotype; 2-way ANOVA (mixed-effects analysis) with Bonferroni multiple comparison test: P less than .0001 for genotype; *P less than .05, **P less than .01, ***P less than .001, and ****P less than .0001 when compared with WT mice. C and D, Weight of individual tissues dissected from male and female mice after 6 weeks of HFD feeding. n = 10-15 mice per group; ***P less than .001 by unpaired t test. E, Daily calorie intake of male mice fed either chow or HFD. Measurements were taken during periods of chow and HFD feeding. n = 8-9 mice per group; 2-way ANOVA with Bonferroni multiple comparison test: P less than .0001 for both genotype and diet and P equal to .0017 for interaction between diet and genotype; **P less than .01 and ****P less than .0001. F, Daily calorie intake of female mice fed either chow or HFD. Measurements were taken during periods of chow and HFD feeding. n = 6 mice per group; 2-way ANOVA with Bonferroni multiple comparison test: P less than .0001 for both genotype and diet and P less than .0001 for interaction between diet and genotype; ns, no significance and ****P less than .0001. G, Oxygen consumption (VO2) of male mice on chow. n = 5-8 mice per group; 2-way repeated measures (RM) ANOVA: P equal to .3397 for genotype. H, VO2 of male mice on HFD. n = 5-8 mice per group; 2-way RM ANOVA: P equal to .5678 for genotype. I, VO2 of male mice fed either chow or HFD during light and dark cycle. n = 6-8 mice per group; 2-way RM ANOVA with Sidak multiple comparison test: ns, no significance. All data are shown as mean ± SEM.
Antibodies
Rabbit polyclonal antibody against phospho-TrkB (Y817) (catalog No. ab81288, RRID: AB_1641129) was from Abcam. Rabbit polyclonal antibodies against total TrkB (catalog No. 4606, RRID: AB_2267470), total signal transducer and activator of transcription 3 (STAT3) (catalog No. 12640, RRID: AB_2629499), and phospho-STAT3 (Tyr705) (catalog No. 9145, RRID: AB_2491009) were purchased from Cell Signaling Technology. Mouse monoclonal anti-β-Actin (catalog No. A5441, RRID: AB_476744) was from Sigma-Aldrich. Secondary goat anti-rabbit horseradish peroxidase (catalog No. A16110, RRID: AB_2534782) and goat anti-mouse horseradish peroxidase (catalog No. 31430, RRID: AB_228307) antibodies were purchased from ThermoFisher Scientific.
Immunoblotting Analysis
Mouse tissues were harvested and homogenized in lysis buffer containing 1% Triton X-100, 50 mM Hepes (pH 7.4), 137 mM NaCl, and proteinase inhibitor cocktail (Roche). In general, approximately 20 μg of total protein was resolved with sodium dodecyl sulfate–polyacrylamide gel electrophoresis and electrotransferred to polyvinylidene fluoride membranes. Primary antibodies were incubated with membranes at 4°C overnight. Secondary antibodies were incubated with membranes at room temperature for 1 hour. An electrochemiluminescence detection system was applied to develop signals.
Physiological Measurements
Measurement of body weight was carried out weekly. For daily food intake, mice were singly housed for at least 1 week to reduce stress response, and food was provided on the bottom of the cage and weighed daily. Only one type of diet was provided in most food intake measurements (one-choice paradigm). When a 2-choice paradigm of food intake was performed, 2 types of diets in 2 separate plastic trays were provided. All food intake measurements were carried out in the home cage for 1 week. Average daily calorie intake was calculated based on average food intake measured over the last 5 days. Body composition was assessed using a Minispec LF-50/mq 7.5 NMR Analyzer (Brucker Optics). Oxygen consumption (VO2) and locomotor activity were monitored with a comprehensive laboratory animal monitoring system (CLAMS, Columbus Instruments).
Quantitative Reverse-Transcription Polymerase Chain Reaction and Brain-derived Neurotrophic Factor Enzyme-linked Immunosorbent Assay
Quantitative reverse-transcription polymerase chain reaction was performed as previously described (36). In brief, mouse tissues were homogenized in TRIzol (Invitrogen), and total RNA was extracted, reverse-transcribed to complementary DNA with random primers, and quantified with real-time polymerase chain reaction. For BDNF measurement, hypothalami were dissected from mice at age 10 weeks. BDNF levels in these samples were measured using a ChemiKine BDNF Sandwich enzyme-linked immunosorbent assay (ELISA) Kit (Sigma-Aldrich No. CYT306, AB_10683049) according to the manufacturer's instructions.
Intraperitoneal Glucose Tolerance Test and Intraperitoneal Insulin Tolerance Test
Intraperitoneal glucose tolerance test (IPGTT) and intraperitoneal insulin tolerance test (IPITT) were performed as described previously (36). For IPGTT, animals were fasted overnight (16 hours) and received an intraperitoneal injection of glucose (1.5 g/kg body weight). For IPITT, animals were fasted for 6 hours and injected intraperitoneally with insulin (1.0 U/kg body weight). Blood was collected from the tail vein at different time points after glucose or insulin injection. A portable glucometer (OneTouch Ultra; LifeScan) was used to measure glucose concentrations.
Mouse Leptin Administration
For Fos induction in response to leptin treatment, the experiment was performed as described previously (18). Two hours after intraperitoneal injection of leptin, mice were perfused with 4% paraformaldehyde. One-fourth of brain sections were used for Fos immunohistochemistry. Fos-positive neurons were counted using Stereo Investigator software (MicroBrightField Inc).
Fasting and Refeeding Experiments
For fasting and refeeding studies, 10-week-old female mice were single-housed for at least 1 week to reduce experimental stress. After acclimation, mice were fasted overnight in cages with Alpha-Dri bedding to eliminate any caloric intake during fasting. After fasting, mice were refed either chow or HFD for 1 hour. Hypothalami were collected immediately after 1 hour of refeeding for subsequent analyses.
Statistical Analyses
Statistical analysis was performed using unpaired t test, one-way analysis of variance (ANOVA), or 2-way ANOVA (GraphPad Prism 9 for MacOS).
Results
Development of Hyperphagic Obesity in BdnfMet/Met Mice on a High-fat Diet
We fed offspring mice from BdnfVal/Met × BdnfVal/Met crosses a chow diet (chow, Teklad Diets 2920x, 16% calories from fat; Supplementary Table S1) (35) after weaning and then an HFD (Research Diets D12492, 60% calories from fat; Supplementary Table S1) (35) at age 10 weeks. BdnfMet/Met mice were slightly heavier than WT (BdnfVal/Val) mice at age 10 weeks (Fig. 1A and 1B); however, regardless of sex, BdnfMet/Met and heterozygous BdnfVal/Met mice developed severe and modest obesity on HFD in merely 6 weeks, respectively (Fig. 1A-1D). In addition, BdnfMet/Met mice developed glucose intolerance and impaired insulin sensitivity on the HFD (Supplementary Fig. S1A-S1D) (35). These observations are consistent with a previous study (37).
To reveal the cause for the development of obesity in BdnfMet/Met mice, we measured food intake and energy expenditure on either chow or HFD. Male, but not female, BdnfMet/Met mice ingested modestly more calories than sex-matched WT mice on chow (Fig. 1E and 1F); however, male and female BdnfMet/Met mice both became extremely hyperphagic on the HFD (Fig. 1E and 1F; eg, 12.03 kcal/day for male WT vs 20.35 kcal/day for male BdnfMet/Met) and there was an interaction between diet and genotype both in the male and female mice (P = .0017 for males and P < .0001 for females). VO2 measurement revealed comparable energy expenditure in WT and BdnfMet/Met mice on either chow or HFD (Fig. 1G-1I). Furthermore, BdnfMet/Met mice had normal physical activity (Supplementary Fig. S1E and S1F) (35) and normal BDNF levels in the hypothalamus (Supplementary Fig. S1G) (35). Therefore, the obese phenotype observed in BdnfMet/Met mice is solely attributable to increased energy consumption.
Increased Hyperphagia of BdnfMet/Met Mice on Fat-rich Diets
As BdnfMet/Met mice displayed much more marked hyperphagia on the HFD than on chow, we asked if there is a relationship between hyperphagia and fat content in diets. We measured daily calorie intake when BdnfMet/Met and WT mice were fed a low-fat diet (LFD, Research Diets D12450J, 10% calories from fat), moderate-fat diet (MFD, Research Diets D17100401, 20% calories from fat), or another HFD (HFD45, Research Diets D08091803, 45% calories from fat) in addition to chow and HFD (see Supplementary Table S1) (35). BdnfMet/Met mice did not show hyperphagia when fed an LFD but became more and more hyperphagic with increasing dietary fat content (Fig. 2A). We observed a positive correlation between the extent of hyperphagia and percentage of calories from fat (Fig. 2B). As a diet with higher fat content generally has higher energy density, BdnfMet/Met mice may become more hyperphagic because of increasing energy density; however, BdnfMet/Met mice were hyperphagic on chow (16% calories from fat) but not on the LFD (Fig. 2A) despite higher energy density in the LFD (3.8 kcal/g) than in chow (3.1 kcal/g; see Supplementary Table S1) (35). These results demonstrate the importance of BDNF in the control of dietary fat intake.
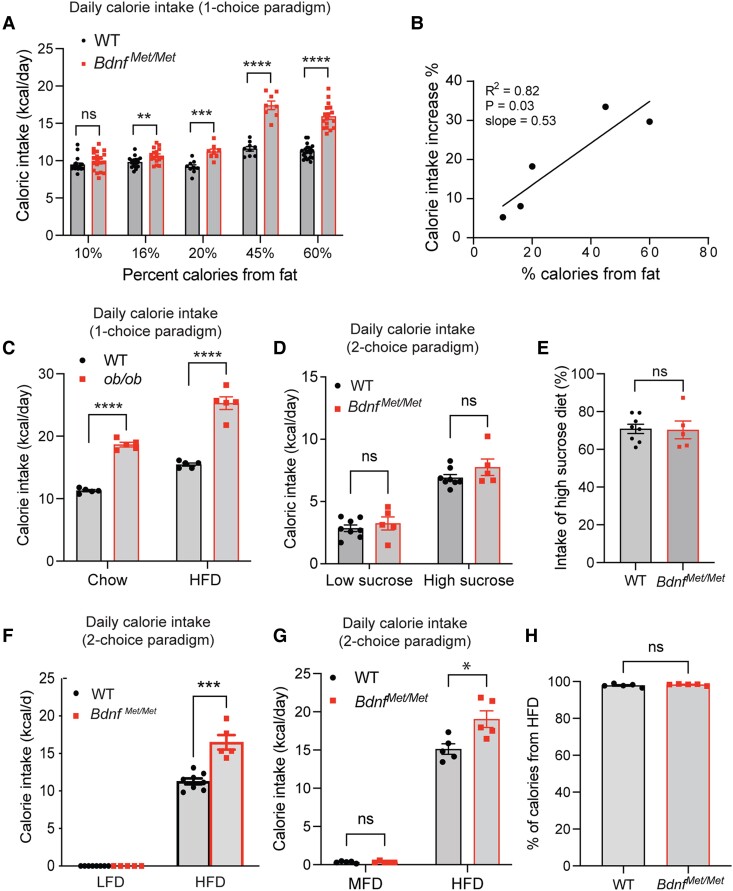
Figure 2.
Brain-derived neurotrophic factor (BDNF) Val66Met polymorphism selectively increases intake of fat-rich diets. All food intake measurements were carried out in young and age-matched wild-type (WT) and BdnfMet/Met mice that were raised on chow and had similar body weights. A, Daily calorie intake of WT and BdnfMet/Met mice on diets with various percentages of calories from fat. Only one type of diet was provided for each test (1-choice paradigm), and the measurements were compiled from several batches of mice. n = 8-22 mice per group; not significant (ns), **P less than .01, ***P less than .001, and ****P less than .0001 by unpaired t test. B, Correlation of dietary fat content and hyperphagia in BdnfMet/Met mice. Percentage of calorie intake increase was calculated by the formula (daily calorie intake by BdnfMet/Met mice – daily calorie intake by WT mice)/daily calorie intake by WT mice%. C, Daily calorie intake of 6-week-old male ob/ob and WT mice fed either chow or high-fat diet (HFD) in the 1-choice paradigm test. n = 5 mice per group; 2-way analysis of variance (ANOVA) with Bonferroni multiple comparison test: P less than .0001 for both genotype and diet and P equal to .0444 for interaction between diet and genotype; ****P less than .0001. D, Daily intake of low-fat, low-sucrose (LFLS) and LFHS when both diets were provided simultaneously (2-choice paradigm). n = 5-8 mice per group; not significant by unpaired t tests. E, Percentage of calorie intake from LFHS. n = 5-8 female mice per group; not significant by unpaired t tests. F, Daily intake of low-fat diet (LFD) and HFD in the 2-choice paradigm test. n = 7-8 female mice per group; ***P less than .001 by unpaired t test. G, Daily intake of moderate-fat diet (MFD) and HFD in the 2-choice paradigm test. n = 5 male mice per group; *P less than .05 and not significant by unpaired t test. H, Percentage of calorie intake from HFD. n = 5 male mice per group; not significant by unpaired t test. All data are shown as mean ± SEM.
We asked if this increased hyperphagia on fat-rich diets is common to monogenic mouse mutants associated with obesity. We measured daily calorie intake of leptin-deficient ob/ob mice when fed either chow or an HFD. Interestingly, ob/ob mice showed a similar degree of hyperphagia on chow and HFD compared with WT mice (Fig. 2C; 65% increase on chow vs 71% increase on HFD), although there is a weak but statistically significant interaction between diet and genotype (P = .0444). Consistent with this finding, leptin signaling did not appear impaired in the hypothalamus of BdnfMet/Met mice as leptin administration activated STAT3 and induced Fos expression comparably in hypothalami of WT and BdnfMet/Met mice (Supplementary Fig. S2) (35). These results suggest that BDNF is more specific than leptin for the control of dietary fat intake.
Normal Hedonic Drive for Palatable Food in BdnfMet/Met Mice
Increased hyperphagia on fat-rich diets could result from increased hedonic drives due to changes in the reward pathway or the sensory system. If so, BdnfMet/Met mice should display increased hyperphagia or increased food preference for a highly palatable high-sucrose diet. We simultaneously provided both a low-fat low-sucrose diet (LFLS, the same as LFD, containing 6.5% sucrose) and a low-fat high-sucrose diet (LFHS, containing 16.4% sucrose; see Supplementary Table S1) (35) to mice in a 2-choice paradigm of food intake assay. Both BdnfMet/Met mice and WT littermates preferred LFHS over LFLS (Fig. 2D); however, BdnfMet/Met mice did not ingest significantly more LFHS than WT mice (Fig. 2E).
To specifically examine food preference for an HFD, we measured calorie intake in WT and BdnfMet/Met mice in the 2-choice paradigm in which LFD and HFD were provided simultaneously. WT and BdnfMet/Met mice both exclusively ate the HFD when LFD and HFD were provided (Fig. 2F). Because preference for the HFD reached 100% in WT mice, the test could not determine if food preference for fat-rich diets is elevated in BdnfMet/Met mice. To address this issue, we compared an MFD and HFD in the 2-choice test. WT and BdnfMet/Met mice ate small and comparable amounts of the MFD (Fig. 2G), and percentages of ingested calories from the HFD in the 2 genotypes were identical at 98% (Fig. 2H), indicating that food preference for fat-rich diets is normal in BdnfMet/Met mice. Collectively, our results indicate that BDNF is required for the control of dietary fat intake rather than hedonic eating.
Attenuated Suppression of High-Fat Diet Intake in BdnfMet/Met Mice
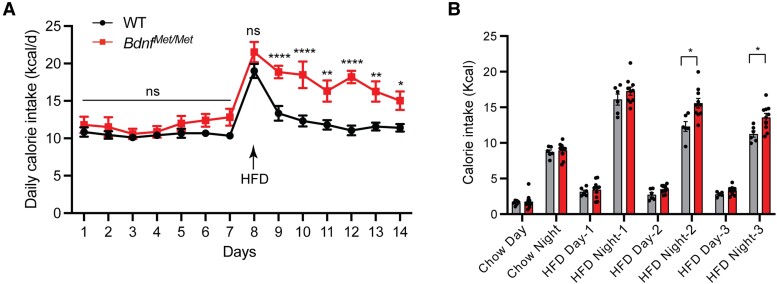
To examine the initial response of BdnfMet/Met mice to an HFD, we singly housed a cohort of naive mice, measured daily calorie intake on chow for a week, replaced chow with an HFD, and measured daily calorie intake on the HFD for another week. During the first day of exposure to HFD, BdnfMet/Met mice and WT mice both greatly elevated their calorie intake due to hedonic drives (Fig. 3A). Interestingly, WT mice quickly reduced calorie intake the following day; however, the reduction in BdnfMet/Met mice was smaller and slower than in WT mice, leading to significantly higher calorie intake in BdnfMet/Met mice than in WT mice (Fig. 3A). The hyperphagia occurred only during the dark cycle when most food is ingested (Fig. 3B). These results suggest there is an appetite-suppressing mechanism after the initial burst of energy intake on HFD exposure in WT mice and that the balance between hedonic drives and this late appetite-suppressing regulation determines the magnitude of hyperphagia on an HFD in subsequent days. The results also indicate that BDNF plays a critical role in this late appetite-suppressing mechanism that regulates dietary fat intake.
Figure 3.
Late suppression of high-fat diet (HFD) intake is attenuated in BdnfMet/met mice. A, Daily calorie intake during transition from chow to HFD. n = 11 female wild-type (WT) mice and 8 female BdnfMet/Met mice; 2-way analysis of variance (ANOVA) (mixed-effects analysis) with Bonferroni multiple comparison test: P equal to .0022 for genotype; ns, no significance, *P less than .05, **P less than .01, and ****P less than .0001. B, Day and night calorie intake during transition from chow to HFD. n = 6 WT mice and 8 BdnfMet/Met mice; 2-way ANOVA with Sidak multiple comparison test: P equal to .0042 for genotype; *P less than .05. All data are shown as mean ± SEM.
Regulated Brain-derived Neurotrophic Factor Release in Response to High-Fat Diet Refeeding
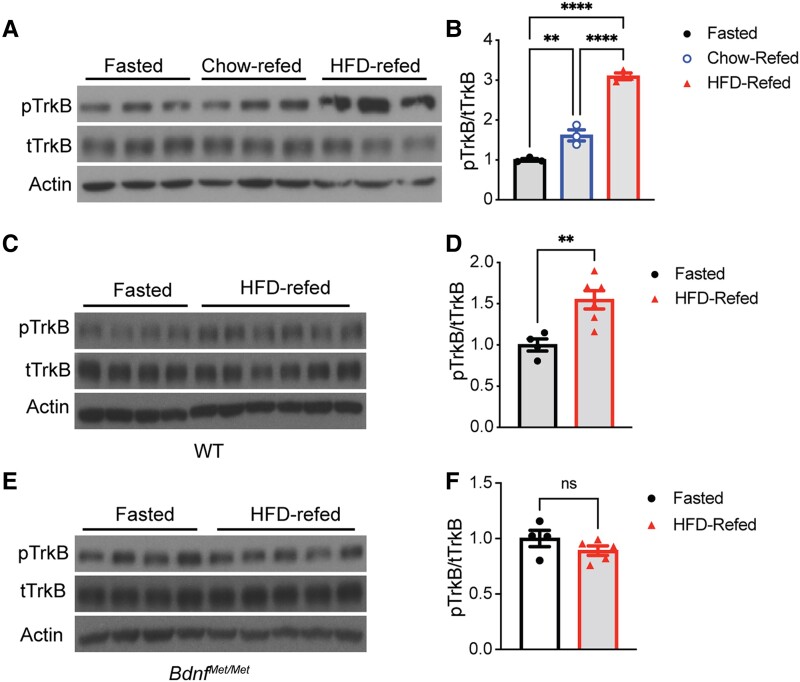
As the Val66Met polymorphism impairs BDNF release through the regulated secretory pathway (27, 34), the observation that BdnfMet/Met mice showed increased hyperphagia on fat-rich diets suggests that regulated BDNF release is essential for the control of dietary food intake. To seek additional evidence for this notion, we sought to determine whether BDNF is released in the hypothalamus in response to an HFD feeding. We first determined if BDNF is quickly released in response to refeeding after overnight food deprivation. Because there are currently no sensitive methods available to directly detect BDNF release in vivo, we monitored TrkB activation as an indicator of BDNF release. Once TrkB is activated by BDNF, several tyrosine (Tyr) residues are phosphorylated, including phospho-Tyr816, which serves as the binding site for phospholipase Cγ1 (10, 14). We fasted WT mice overnight, refed them either chow or an HFD for 1 hour, and measured levels of phosphorylated TrkB (pTrkB) in hypothalamic extracts using immunoblotting analysis with specific antibodies against pTrkB (Tyr816) and total TrkB (Supplementary Fig. S3) (35). Because male and female BdnfMet/Met mice both responded similarly to HFD feeding in weight gain (see Fig. 1A and 1B), this experiment was carried out only in one sex of mice. Chow and HFD refeeding both triggered a significant increase in hypothalamic pTrkB levels (Fig. 4A and 4B). Interestingly, HFD refeeding induced much stronger TrkB activation than chow refeeding (see Fig. 4A and 4B).
Figure 4.
Refeeding stimulates tropomyosin receptor kinase B (TrkB) activation that is dependent on regulated brain-derived neurotrophic factor (BDNF) release. A and B, Immunoblotting analysis and quantification of TrkB activation in hypothalami of 10-week-old female wild-type (WT) mice that were fasted overnight and then refed either chow or HFD for 1 hour. One-way analysis of variance (ANOVA) with Tukey multiple comparison test: **P less than .01, ***P less than .001, and ****P less than .0001. C and D, Immunoblotting analysis and quantification of TrkB activation in hypothalami of 10-week-old female WT mice that were fasted overnight and then refed a high-fat diet (HFD) for 1 hour. **P less than .01 by unpaired t test. E and F, Immunoblotting analysis and quantification of TrkB activation in hypothalami of 10-week-old female BdnfMet/Met mice that were fasted overnight and then refed HFD for 1 hour. ns, no significance by unpaired t test. All data are shown as mean ± SEM.
The Val66Met substitution impairs regulated BDNF secretion in neurons without altering BDNF production and the sequence of mature BDNF (27, 38) (see Supplementary Fig. S1G) (35). Thus, TrkB activation in response to HFD refeeding is expected to be attenuated in BdnfMet/Met mice if refeeding stimulates neurons to release BDNF. Indeed, we found HFD refeeding robustly increased hypothalamic pTrkB levels in WT mice (Fig. 4C and 4D) but not in BdnfMet/Met mice (Fig. 4E and 4F). These results indicate that HFD refeeding quickly and strongly stimulates BDNF release in the hypothalamus.
Regulated Brain-derived Neurotrophic Factor Release in Response to Nocturnal High-fat Diet Feeding
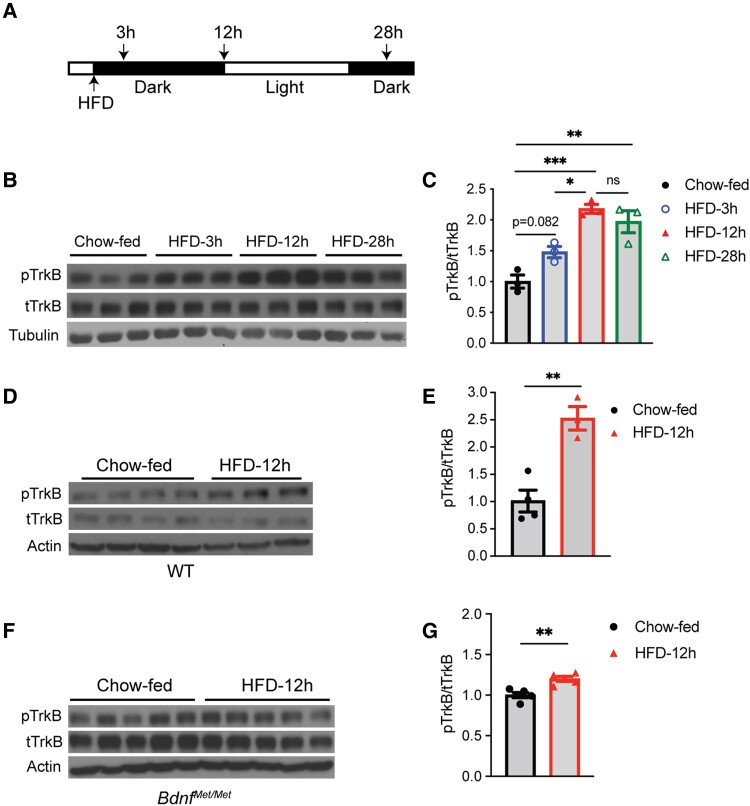
We next tested if daily HFD feeding induces BDNF release and subsequent TrkB activation in the hypothalamus. We maintained WT mice on chow, switched mice to an HFD at the start of a dark cycle for various lengths of time, collected hypothalami, and used chow-fed mice at time 0 as controls (Fig. 5A). Nocturnal HFD feeding for 3 or 12 hours stimulated hypothalamic TrkB activation, as indicated by an increased ratio of pTrkB over total TrkB (Fig. 5B and 5C). Circadian rhythms were not responsible for the change in TrkB activation because nocturnal chow feeding for 12 hours did not significantly alter levels of hypothalamic TrkB activation (Supplementary Fig. S4C and S4D) (35). However, TrkB activation fell back to a level comparable to chow-fed mice after 24 hours of HFD feeding (Supplementary Fig. S4A and S4B) (35), which was after a 12-hour light cycle when mice sleep and do not eat much. These results indicate that signaling through TrkB in the hypothalamus coincides with feeding activities to regulate energy intake during HFD feeding. In support of this argument, we found that hypothalamic TrkB activation was again elevated at the 28-hour time point when mice had been awake and fed an HFD for 4 hours (Fig. 5B and 5C).
Figure 5.
High-fat diet (HFD) feeding induces tropomyosin receptor kinase B (TrkB) activation. A, Diagram of experimental design. HFD feeding started at the beginning of a dark cycle. Hypothalami were dissected before HFD feeding (chow-fed) or 3, 12, or 28 hours after HFD feeding. B and C, Immunoblotting analysis and quantification of TrkB activation in hypothalami of female wild-type (WT) mice fed HFD for various lengths of time. One-way analysis of variance (ANOVA) with Tukey multiple comparison test: ns, no significance, *P less than .05, **P less than .01, and ***P less than .001. D and E, Immunoblotting analysis and quantification of TrkB activation in hypothalami of female WT mice that were fed HFD for 12 hours. **P less than .01 by unpaired t test. F and G, Immunoblotting analysis and quantification of TrkB activation in hypothalami of female BdnfMet/Met mice that were fed HFD for 12 hours. **P less than .01 by unpaired t test. All data are shown as mean ± SEM.
We next questioned whether hypothalamic TrkB activation during nocturnal HFD feeding resulted from regulated BDNF release as had occurred during HFD refeeding following fasting. We found that 12-hour nocturnal HFD feeding strongly stimulated hypothalamic TrkB activation in WT mice (Fig. 5D and 5E; 116% increase), and the stimulation was drastically attenuated in BdnfMet/Met mice (Fig. 5F and G; 15% increase). Furthermore, nocturnal HFD feeding for 12 hours did not alter the expression both of Bdnf and Ntrk2 (encoding TrkB receptor) genes in the hypothalamus (Supplementary Fig. S5) (35). These results indicate that nocturnal HFD feeding stimulates BDNF-expressing neurons to release BDNF and subsequent TrkB activation in the hypothalamus.
Discussion
In this study we found that BdnfMet/Met mice, which mimic the human Val66Met polymorphism (34), displayed increasing hyperphagia on diets with increasing fat content, but not on either an LFD or a palatable high-sucrose diet. Consequently, these mice developed severe hyperphagic obesity on an HFD. Given the high frequency of the BDNF Met polymorphism in the global human population (39–43) and ready availability of fat-rich foods in modern society (44), it would be extremely important to investigate whether people homozygous for the BDNF Met variant have an elevated appetite for fat-rich foods. If they do, our findings suggest that these people are at an especially high risk for developing obesity, which could be curtailed substantially by the strict avoidance of fat-rich foods. Furthermore, SH2B adaptor protein 1 (SH2B1) is a signaling component downstream of TrkB and is involved in activation of mitogen-activated protein kinase (45). Loss-of-function mutations and polymorphisms in the SH2B1 gene have been associated with human obesity (32, 46). It would be interesting to determine whether people with obesity-associated SH2B1 polymorphisms display an elevated appetite for fat-rich foods, although SH2B1 also regulates insulin and leptin signaling pathways.
Little is known about the genetic factors or physiological processes that control dietary fat intake. Many genes that are crucial for the regulation of satiety on chow do not appear to have a role in the control of dietary fat intake. For example, mice deficient in cholecystokinin (CCK) (47), CCK1 receptor (48), leptin (this study), and leptin receptor (3) do not exhibit increased hyperphagia on fat-rich diets compared with chow. However, Mc4r−/− knockout mice do display markedly increased hyperphagia for an extended period after a switch from chow to a fat-rich diet (49, 50). Furthermore, inhibition of melanocortin signaling has been shown to increase consumption of dietary fats (51–54). Surprisingly, food preference for fat-rich diets and high-sucrose diets is reduced in Mc4r−/− mice (3), indicating that hedonic drives for palatable diets are impaired in these mice. Thus, it is likely that the MC4R is required for the brain to respond to specific satiety signals produced during consumption of dietary fats. Alternatively, Mc4r−/− mice may ingest more fat-rich diets to compensate for the impairment in the reward pathway to reach a certain reward value.
Like Mc4r−/− mice, BdnfMet/Met mice also displayed increased consumption of dietary fats; however, our results indicate that deficits in BDNF do not affect hedonic drives for palatable food. First, food preference for an HFD or high-sucrose diet was not altered in BdnfMet/Met mice. Second, calorie intake during the first-day exposure to the HFD was similar in WT and BdnfMet/Met mice. These results indicate that BDNF is essential for animals to suppress excess intake of dietary fats in response to satiety signals specific to fat-rich diets. As BDNF infusion into the brain suppresses hyperphagia on fat-rich diets in mice deficient in MC4R signaling (16), BDNF and MC4R could be at the same pathway to control intake of dietary fats. Why did BdnfMet/Met mice fail to ingest more calories than WT mice during the first-day exposure to the HFD despite impaired activation of hypothalamic TrkB in response to HFD feeding? It is possible that the initial hedonic drive for HFD is too strong to be overcome by the satiety signals.
BDNF is released from neurons through a constitutive secretory pathway and a regulated secretory pathway (55). We found that HFD feeding more strongly induced BDNF release in the hypothalamus than chow feeding and that BdnfMet/Met mice, in which regulated BDNF release is impaired (34), had deficits in the control of dietary fat intake. These findings suggest that BDNF released through the regulated secretory pathway activates TrkB-mediated signaling cascades to suppress dietary fat intake.
When the gene for BDNF or TrkB is deleted in some brain neurons, the resulting mutant mice display hyperphagia on chow (15, 16, 19, 56, 57). In these mutant mice TrkB activation induced by BDNF released through either a constitutive or regulated secretory pathway is disrupted. Based on the findings on regulated BDNF release in this study, these mutant mice are expected to be more hyperphagic on fat-rich diets than on chow, and this is what has been observed (16). It remains to be determined whether constitutive BDNF release plays a role in the control of dietary fat intake.
Our results show that HFD feeding induces stronger TrkB activation in the hypothalamus than chow feeding, which is dependent on regulated BDNF release. Several questions about TrkB activation in response to HFD feeding remain to be answered in future studies: What are satiety signals specific to dietary fat intake? What are the signals responsible for stimulating BDNF release on HFD feeding? What are the identities of BDNF-expressing and TrkB-expressing neurons that regulate dietary fat intake? Addressing these questions will greatly enrich our knowledge of the regulation of dietary fat intake and provide novel targets to develop interventions to counter diet-induced obesity.
Acknowledgments
We thank Dr Francis S. Lee at Weill Medical College of Cornell University for the BdnfMet/Met mouse strain.
Abbreviations
- ANOVA
analysis of variance
- BDNF
brain-derived neurotrophic factor
- HFD
high-fat diet
- IPGTT
intraperitoneal glucose tolerance test
- IPITT
intraperitoneal insulin tolerance test
- LFD
low-fat diet
- LFHS
low-fat, high-sucrose diet
- LFLS
low-fat, low-sucrose diet
- MFD
moderate-fat diet
- pTrkB
phosphorylated tropomyosin receptor kinase B
- SH2B1
SH2B adaptor protein 1
- STAT3
signal transducer and activator of transcription 3
- TrkB
tropomyosin receptor kinase B
- Tyr
tyrosine
- VO2
oxygen consumption
- WT
wild-type
Contributor Information
Xiangyang Xie, Department of Neuroscience, UF Scripps Biomedical Research, University of Florida, Jupiter, Florida 33458, USA.
Jessica Houtz, Department of Neuroscience, UF Scripps Biomedical Research, University of Florida, Jupiter, Florida 33458, USA.
Guey-Ying Liao, Department of Neuroscience, UF Scripps Biomedical Research, University of Florida, Jupiter, Florida 33458, USA.
Yuting Chen, Department of Neuroscience, UF Scripps Biomedical Research, University of Florida, Jupiter, Florida 33458, USA; Skaggs Graduate School of Chemical and Biological Sciences, The Scripps Research Institute, Jupiter, Florida 33458, USA.
Baoji Xu, Department of Neuroscience, UF Scripps Biomedical Research, University of Florida, Jupiter, Florida 33458, USA; Skaggs Graduate School of Chemical and Biological Sciences, The Scripps Research Institute, Jupiter, Florida 33458, USA.
Financial Support
This work was supported by the National Institutes of Health (grant Nos. R01 DK103335 and R01 DK105954 to B.X.) and a postdoctoral fellowship from the American Heart Association (No. AHA827718 to J.H.).
Author Contributions
X.X. and B.X. designed the research; X.X., J.H., G.Y.L., Y.C., and B.X. performed and analyzed the data; and X.X., J.H., and B.X. wrote the paper.
Data Availability
Original data generated and analyzed during this study are included in this published article or in the data repositories listed in “References.”
References
- 1. Stein CJ, Colditz GA. The epidemic of obesity. J Clin Endocrinol Metab. 2004;89(6):2522–2525. [DOI] [PubMed] [Google Scholar]
- 2. NCD Risk Factor Collaboration (NCD-RisC) . Trends in adult body-mass index in 200 countries from 1975 to 2014: a pooled analysis of 1698 population-based measurement studies with 19.2 million participants. Lancet. 2016;387(10026):1377–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Panaro BL, Cone RD. Melanocortin-4 receptor mutations paradoxically reduce preference for palatable foods. Proc Natl Acad Sci U S A. 2013;110(17):7050–7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang Y, Beydoun MA. The obesity epidemic in the United States—gender, age, socioeconomic, racial/ethnic, and geographic characteristics: a systematic review and meta-regression analysis. Epidemiol Rev. 2007;29(1):6–28. [DOI] [PubMed] [Google Scholar]
- 5. Tomiyama AJ. Stress and obesity. Annu Rev Psychol. 2019;70(1):703–718. [DOI] [PubMed] [Google Scholar]
- 6. Morton GJ, Meek TH, Schwartz MW. Neurobiology of food intake in health and disease. Nat Rev Neurosci. 2014;15(6):367–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lutter M, Nestler EJ. Homeostatic and hedonic signals interact in the regulation of food intake. J Nutr. 2009;139(3):629–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hofer M, Pagliusi SR, Hohn A, Leibrock J, Barde YA. Regional distribution of brain-derived neurotrophic factor mRNA in the adult mouse brain. EMBO J. 1990;9(8):2459–2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yan Q, Rosenfeld RD, Matheson CR, et al. Expression of brain-derived neurotrophic factor protein in the adult rat central nervous system. Neuroscience. 1997;78(2):431–448. [DOI] [PubMed] [Google Scholar]
- 10. Chao MV. Neurotrophins and their receptors: a convergence point for many signalling pathways. Nat Rev Neurosci. 2003;4(4):299–309. [DOI] [PubMed] [Google Scholar]
- 11. Waterhouse EG, Xu B. New insights into the role of brain-derived neurotrophic factor in synaptic plasticity. Mol Cell Neurosci. 2009;42(2):81–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huang EJ, Reichardt LF. Neurotrophins: roles in neuronal development and function. Annu Rev Neurosci. 2001;24(1):677–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rios M. BDNF and the central control of feeding: accidental bystander or essential player? Trends Neurosci. 2013;36(2):83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xu B, Xie X. Neurotrophic factor control of satiety and body weight. Nat Rev Neurosci. 2016;17(5):282–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rios M, Fan G, Fekete C, et al. Conditional deletion of brain-derived neurotrophic factor in the postnatal brain leads to obesity and hyperactivity. Mol Endocrinol. 2001;15(10):1748–1757. [DOI] [PubMed] [Google Scholar]
- 16. Xu B, Goulding EH, Zang K, et al. Brain-derived neurotrophic factor regulates energy balance downstream of melanocortin-4 receptor. Nat Neurosci. 2003;6(7):736–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liao GY, Li Y, Xu B. Ablation of TrkB expression in RGS9-2 cells leads to hyperphagic obesity. Mol Metab. 2013;2(4):491–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liao GY, An JJ, Gharami K, et al. Dendritically targeted Bdnf mRNA is essential for energy balance and response to leptin. Nat Med. 2012;18(4):564–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. An JJ, Liao GY, Kinney CE, Sahibzada N, Xu B. Discrete BDNF neurons in the paraventricular hypothalamus control feeding and energy expenditure. Cell Metab. 2015;22(1):175–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ozek C, Zimmer DJ, De Jonghe BC, Kalb RG, Bence KK. Ablation of intact hypothalamic and/or hindbrain TrkB signaling leads to perturbations in energy balance. Mol Metab. 2015;4(11):867–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yeo GSH, Hung CCC, Rochford J, et al. A de novo mutation affecting human TrkB associated with severe obesity and developmental delay. Nat Neurosci. 2004;7(11):1187–1189. [DOI] [PubMed] [Google Scholar]
- 22. Gray J, Yeo GSH, Cox JJ, et al. Hyperphagia, severe obesity, impaired cognitive function, and hyperactivity associated with functional loss of one copy of the brain-derived neurotrophic factor (BDNF) gene. Diabetes. 2006;55(12):3366–3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Han JC, Liu QR, Jones MP, et al. Brain-derived neurotrophic factor and obesity in the WAGR syndrome. N Engl J Med. 2008;359(9):918–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Montague CT, Farooqi IS, Whitehead JP, et al. Congenital leptin deficiency is associated with severe early-onset obesity in humans. Nature. 1997;387(6636):903–908. [DOI] [PubMed] [Google Scholar]
- 25. Farooqi IS, Yeo GS, Keogh JM, et al. Dominant and recessive inheritance of morbid obesity associated with melanocortin 4 receptor deficiency. J Clin Invest. 2000;106(2):271–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Farooqi IS, O'Rahilly S. Monogenic obesity in humans. Annu Rev Med. 2005;56(1):443–458. [DOI] [PubMed] [Google Scholar]
- 27. Egan MF, Kojima M, Callicott JH, et al. The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell. 2003;112(2):257–269. [DOI] [PubMed] [Google Scholar]
- 28. Frayling TM, Timpson NJ, Weedon MN, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316(5826):889–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Loos RJ, Lindgren CM, Li S, et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat Genet. 2008;40(6):768–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Willer CJ, Speliotes EK, Loos RJ, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41(1):25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Thorleifsson G, Walters GB, Gudbjartsson DF, et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet. 2009;41(1):18–24. [DOI] [PubMed] [Google Scholar]
- 32. Speliotes EK, Willer CJ, Berndt SI, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42(11):937–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Okada Y, Kubo M, Ohmiya H, et al. ; GIANT Consortium . Common variants at CDKAL1 and KLF9 are associated with body mass index in east Asian populations. Nat Genet. 2012;44(3):302–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen ZY, Jing D, Bath KG, et al. Genetic variant BDNF (Val66Met) polymorphism alters anxiety-related behavior. Science. 2006;314(5796):140–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Xu B. Supplementary data for “Genetic Val66Met BDNF Variant Increases Hyperphagia on Fat-rich Diets in Mice.” Mendeley Data. Deposited January 5,2023. https://data.mendeley.com/datasets/6m6nf6njb4/1 [DOI] [PMC free article] [PubMed]
- 36. Mansuy-Aubert V, Xie X, Gong Z, et al. Imbalance between neutrophil elastase and its inhibitor α1-antitrypsin in obesity alters insulin sensitivity, inflammation, and energy expenditure. Cell Metab. 2013;17(4):534–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yang J, Park KW, Cho S. Inhibition of the CD36 receptor reduces visceral fat accumulation and improves insulin resistance in obese mice carrying the BDNF-Val66Met variant. J Biol Chem. 2018;293(34):13338–13348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chen ZY, Patel PD, Sant G, et al. Variant brain-derived neurotrophic factor (BDNF) (Met66) alters the intracellular trafficking and activity-dependent secretion of wild-type BDNF in neurosecretory cells and cortical neurons. J Neurosci. 2004;24(18):4401–4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cargill M, Altshuler D, Ireland J, et al. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat Genet. 1999;22(3):231–238. [DOI] [PubMed] [Google Scholar]
- 40. Shimizu E, Hashimoto K, Iyo M. Ethnic difference of the BDNF 196G/A (val66met) polymorphism frequencies: the possibility to explain ethnic mental traits. Am J Med Genet B Neuropsychiatr Genet. 2004;126B(1):122–123. [DOI] [PubMed] [Google Scholar]
- 41. Choi MJ, Kang RH, Lim SW, Oh KS, Lee MS. Brain-derived neurotrophic factor gene polymorphism (Val66Met) and citalopram response in major depressive disorder. Brain Res. 2006;1118(1):176–182. [DOI] [PubMed] [Google Scholar]
- 42. Itoh K, Hashimoto K, Kumakiri C, Shimizu E, Iyo M. Association between brain-derived neurotrophic factor 196 G/A polymorphism and personality traits in healthy subjects. Am J Med Genet B Neuropsychiatr Genet. 2004;124B(1):61–63. [DOI] [PubMed] [Google Scholar]
- 43. Tsai SJ, Cheng CY, Yu YW, Chen TJ, Hong CJ. Association study of a brain-derived neurotrophic-factor genetic polymorphism and major depressive disorders, symptomatology, and antidepressant response. Am J Med Genet B Neuropsychiatr Genet. 2003;123B(1):19–22. [DOI] [PubMed] [Google Scholar]
- 44. Saltiel AR. New therapeutic approaches for the treatment of obesity. Sci Transl Med. 2016;8(323):323rv322. [DOI] [PubMed] [Google Scholar]
- 45. Qian X, Riccio A, Zhang Y, Ginty DD. Identification and characterization of novel substrates of Trk receptors in developing neurons. Neuron. 1998;21(5):1017–1029. [DOI] [PubMed] [Google Scholar]
- 46. Doche ME, Bochukova EG, Su HW, et al. Human SH2B1 mutations are associated with maladaptive behaviors and obesity. J Clin Invest. 2012;122(12):4732–4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lo CM, King A, Samuelson LC, et al. Cholecystokinin knockout mice are resistant to high-fat diet-induced obesity. Gastroenterology. 2010;138(5):1997–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Donovan MJ, Paulino G, Raybould HE. CCK(1) receptor is essential for normal meal patterning in mice fed high fat diet. Physiol Behav. 2007;92(5):969–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Butler AA, Marks DL, Fan W, Kuhn CM, Bartolome M, Cone RD. Melanocortin-4 receptor is required for acute homeostatic responses to increased dietary fat. Nat Neurosci. 2001;4(6):605–611. [DOI] [PubMed] [Google Scholar]
- 50. Srisai D, Gillum MP, Panaro BL, et al. Characterization of the hyperphagic response to dietary fat in the MC4R knockout mouse. Endocrinology. 2011;152(3):890–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Koegler FH, Schaffhauser RO, Mynatt RL, York DA, Bray GA. Macronutrient diet intake of the lethal yellow agouti (Ay/a) mouse. Physiol Behav. 1999;67(5):809–812. [DOI] [PubMed] [Google Scholar]
- 52. Hagan MM, Rushing PA, Benoit SC, Woods SC, Seeley RJ. Opioid receptor involvement in the effect of AgRP- (83-132) on food intake and food selection. Am J Physiol Regul Integr Comp Physiol. 2001;280(3):R814–R821. [DOI] [PubMed] [Google Scholar]
- 53. Samama P, Rumennik L, Grippo JF. The melanocortin receptor MCR4 controls fat consumption. Regul Pept. 2003;113(1-3):85–88. [DOI] [PubMed] [Google Scholar]
- 54. Boghossian S, Park M, York DA. Melanocortin activity in the amygdala controls appetite for dietary fat. Am J Physiol Regul Integr Comp Physiol. 2010;298(2):R385–R393. [DOI] [PubMed] [Google Scholar]
- 55. Cunha C, Brambilla R, Thomas KL. A simple role for BDNF in learning and memory? Front Mol Neurosci. 2010;3:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Liao GY, Kinney CE, An JJ, Xu B. TrkB-expressing neurons in the dorsomedial hypothalamus are necessary and sufficient to suppress homeostatic feeding. Proc Natl Acad Sci U S A. 2019;116(8):3256–3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. An JJ, Kinney CE, Tan JW, Liao GY, Kremer EJ, Xu B. TrkB-expressing paraventricular hypothalamic neurons suppress appetite through multiple neurocircuits. Nat Commun. 2020;11(1):1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Original data generated and analyzed during this study are included in this published article or in the data repositories listed in “References.”