Abstract
Infected-cell protein 0 (ICP0), the product of the herpes simplex virus (HSV) immediate-early (IE) α0 gene, is a promiscuous transactivator of viral early (E) and late (L) gene expression. HSV mutants lacking ICP0 function are severely deficient in viral growth and protein synthesis, particularly at low multiplicities of infection. Early in the infectious process in vitro, ICP0 protein accumulates in distinct domains within the nucleus to form characteristic structures active in the transcription of viral genes. However, following infection of primary trigeminal ganglion cells in vitro with a recombinant HSV mutant that expresses only ICP0, we observed that ICP0 protein accumulated in the characteristic intranuclear distribution only in the nuclei of Schwann cells; neurons in the culture did not accumulate ICP0 despite expression of ICP0 RNA in those cells. The same phenomenon was observed in PC12 cells differentiated to assume a neuronal phenotype. In primary neurons in culture, the amount of ICP0 protein could be increased by pharmacologic inhibition of calcium-activated protease (calpain) activity or by inhibition of protein phosphatase 2B (calcineurin). The failure of ICP0 protein to accumulate in the nucleus of neurons suggests that one mechanism which may impair efficient replication of the virus in neurons, and thus favor the establishment of viral latency in those cells, may be found in the cell-specific processing of that IE gene product.
Herpes simplex virus type 1 (HSV-1) is a double-stranded DNA virus with a broad host range. Viral gene expression in lytic infection proceeds in a tightly regulated temporal cascade of immediate-early (IE), early (E), and late (L) gene products that are expressed sequentially (24, 60). Four of the viral IE gene products (ICP0, ICP4, ICP22, and ICP27) act to regulate subsequent viral gene expression at both transcriptional and posttranscriptional levels (52, 53, 60). The initial expression of IE genes in epithelial cells depends on binding of the viral tegument protein VP16 to TAATGARAT sequences in the IE gene promoter elements (26, 33, 34, 48), a process that requires collaboration with host cell factors (Oct-1 and C1). In natural infection, virions released after primary infection of epithelial cells in skin or mucous membranes are carried by retrograde axonal transport to the sensory ganglion, where the virus may replicate in neurons or supporting cells but ultimately establishes a characteristic latent state exclusively in the sensory neurons of the ganglion (22). HSV replicates efficiently in a wide variety of cell types in a cell cycle-independent fashion, but replication is less efficient in nondividing cells such as neurons (30, 46). Several differences between neurons and other cell types may explain the relatively inefficient replication of HSV in neurons. Because of the limited availability of substrates required for DNA replication in neurons, HSV must provide enzymes for the synthesis of those substrates (e.g., ribonucleotide reductase and thymidine kinase). Mutants that are defective in these enzymes replicate normally in nonneuronal cells but poorly or not at all in neurons (2, 10). Wild-type viral replication also depends on cell-specific regulators of viral gene expression that may have limited expression or restricted intracellular distribution in neurons (35, 36) or the cell-specific expression of inhibitors of IE gene expression (28, 29). It has also been suggested previously that viral DNA synthesis may serve as a critical regulatory branch point between the lytic and latent pathways of infection in neurons (45).
Among the HSV IE genes, ICP0 is a potent promiscuous transactivator of gene expression that has been demonstrated to stimulate expression of HSV IE, E, and L genes, as well as a number of other eukaryotic genes, in transient-transfection assays (16, 41, 49, 51). Although ICP0 expression is not essential for HSV replication in Vero cells, recombinant viruses with both copies of the ICP0 gene deleted show greatly increased particle-to-PFU ratios, reduced expression of viral IE proteins, and impaired viral replication, particularly at low multiplicities of infection (MOIs) (6, 7, 14, 54, 58). Extranuclear sites of ICP0 activity in the regulation of RNA translation have been proposed elsewhere (27), but the principal function of ICP0 as a transactivator of gene expression is a function that requires ICP0 protein in the nucleus. In accord with this function, newly synthesized ICP0 protein translocates early in infection from the cytoplasm to the nucleus, where it accumulate in discrete domains (19, 39) that appear to serve as the site of initial viral genome deposition and early viral replication (38, 40). These nuclear domains (ND10s), which can be identified in uninfected cells by the presence of specific protein components (9, 12), become disrupted and dispersed in an ICP0-dependent fashion as lytic infection proceeds (18, 19). ICP0 colocalizes with ICP4 in the nucleus of infected cells (31), and ICP27 plays a critical role in determining the localization of ICP0 protein (64, 65). But additional HSV IE gene products are not required for ICP0 localization to the nucleus of infected cells, as expression of ICP0 alone from a plasmid results in the nuclear localization of the protein product (64).
We report here that, upon infection of primary neurons in culture, both wild-type HSV and two HSV mutants designed to express only ICP0 in the absence of other IE genes accumulate ICP0 protein within the nucleus only of nonneuronal cells. The block to ICP0 protein accumulation is not at the level of transcription, because ICP0 RNA can be detected both by in situ hybridization and by single-cell reverse transcription-PCR (RT-PCR) of infected neurons, but rather appears to involve protein processing since pharmacologic blockade of either calcium-activated protease (calpain) or protein phosphatase 2B (calcineurin) results in the intranuclear accumulation of ICP0 in neurons. ICP0 expression plays an important role in viral gene transactivation and viral replication. The failure of ICP0 to accumulate in the nucleus of neurons may be one factor that favors the establishment of latency in neurons following viral infection.
MATERIALS AND METHODS
Virus constructs.
Wild-type virus infections were carried out with a laboratory strain, KOS, of HSV-1. The construction of the deletion mutant TOZ.1 has been described previously (32). This viral recombinant has deletions of both copies of ICP4 (11), ICP22 (genomic positions 133372 to 133465), ICP27 (42), and the viral host shutoff gene UL41 (genomic positions 91606 to 92124) and has an ICP0 promoter::lacZ expression cassette in the UL41 locus. The IE gene coding for ICP47 has been left intact in this recombinant, and the ICP4 IE promoter has been substituted for the native thymidine kinase promoter, resulting in a disruption of the HSV UL24 gene. The recombinant was propagated on ICP4-ICP27-producing 7B cells, purified by three rounds of limiting dilution, and verified by Southern blotting (32). A second mutant, QOZHG, was also used. The virus QOZHG (ICP4− ICP27−::HCMV IEp-GFP β-ICP22 β-ICP47 UL41−::ICP0p-lacZ) was created by a genetic cross of the virus TOZ.1 (ICP4− ICP27− ICP22− UL24−::ICP4p-tk UL41−::ICP0p-lacZ) with the virus d106 (ICP4− ICP27−::HCMV IEp-GFP β-ICP22 β-ICP47; a kind gift of N. DeLuca, Pittsburgh, Pa.) (55). Green fluorescent protein (GFP)-positive fluorescent plaques, which stained blue with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside), were purified by three rounds of limiting dilution. The genomic structure of the QOZHG recombinant was confirmed by Southern blot analysis.
Cell culture.
Trigeminal ganglia (TG) from 17-day rat embryos were dissociated with 0.25% trypsin–1 mM EDTA for 30 min at 37°C with constant shaking and then plated on poly-d-lysine (molecular weight, 70,000 to 150,000; Sigma, St. Louis, Mo.)-coated coverslips at 105 cells/well in a 24-well plate (Falcon, Lincoln Park, N.J.) in 500 μl of defined NeuroBasal medium (GIBCO-BRL, Rockville, Md.) supplemented with 1× B027 (GIBCO), 1× GlutaMAX (GIBCO), 0.5 μg of AlbuMAX II (GIBCO) per ml, 100 U of penicillin per ml, and 100 ng of 7.0s nerve growth factor (NGF) (Sigma) per ml. 5-Fluoro-2′-deoxyuridine (10 μM; Sigma) and uridine (10 μM; Sigma) were added at day 8 to inhibit the growth of dividing cells. The cells were infected at 10 days after plating in 250 μl of medium for 1 h at 37°C with gentle agitation every 20 min. PC12 cells (a gift of J. Chen, University of Pittsburgh) were grown at 37°C in 10% CO2 in Dulbecco's modified Eagle medium (GIBCO) supplemented with 7.5% fetal calf serum (GIBCO) and 7.5% horse serum (GIBCO) on rat tail collagen-coated tissue culture dishes (Falcon) or on coated plastic coverslips (Sarstedt, Newton, N.C.) in 24-well plates. PC12 cells were differentiated to assume a neuronal phenotype by the addition of 100 ng of 2.5s NGF (Sigma) per ml to culture medium containing 1% serum and plated at less than 2 × 104 cells/cm2. Cells were infected with virus 7 days after NGF treatment, in a manner identical to that described for dissociated TG cells.
Immunohistochemistry.
Cells were fixed in Histochoice (Amresco, Solon, Ohio) for 20 min at room temperature (r.t.) and incubated with the primary antibody at 1:1,000 in phosphate-buffered saline (PBS) containing 1% goat serum for 2 h at r.t. or 4°C overnight, followed by a Cy3-conjugated secondary antibody (1:3,000; Sigma) for 1 h at r.t. The anti-ICP0 rabbit polyclonal antibody was the kind gift of R. Courtney (Pennsylvania State University School of Medicine), and the anti-HSV rabbit polyclonal antibody was purchased from Accurate Chemical & Scientific Corp. (Westbury, N.Y.). For detection of β-galactosidase expression, cells were fixed with 4% paraformaldehyde for 10 min at r.t., rinsed for 5 min with PBS twice, and then incubated in X-Gal (Promega, Madison, Wis.)-containing solution for 2 h at 37°C. For detection of GFP, the cells were fixed with 4% paraformaldehyde and observed under epifluorescence. Images were acquired with a Xillix digital camera attached to a personal computer-based image analysis system (MCID, St. Catharines, Ontario, Canada) using the same settings for experimental cultures and mock-infected controls in each experiment.
In situ hybridization.
A plasmid containing the PstI-MluI fragment in the BamHI fragment B of HSV-1 spanning exon 3 of ICP0, obtained from J. Spivack, was linearized with EcoRI and transcribed with T7 RNA polymerase using digoxigenin-UTP (Roche Boehringer Mannheim, Indianapolis, Ind.) to label the transcript, followed by alkaline hydrolysis. The length of the probe was between 200 and 800 bp. Cells were fixed in 4% paraformaldehyde for 10 min, treated with 1% HCl in PBS for 5 min, and then acetylated with acetic anhydride in triethanolamine for 10 min. After equilibration in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 5 min, the cells were dehydrated in graded ethanols and air dried. Hybridization with the digoxigenin-labeled riboprobe (0.5 μg/ml) was carried out overnight at 56°C in 15 μl of hybridization solution (75% formamide, 10% dextran sulfate, 2× SSC, 1× PBS, 1× Denhardt's solution, and 0.02% tRNA). Following hybridization, the coverslips were rinsed three times with 2× SSC for 5 min, digested with RNase A (200 μg/ml in 10 mM Tris-HCl–0.5 M NaCl), rinsed sequentially with 2× SSC and 1× SSC at 56°C, and then washed for 1 h in 1× SSC at 56°C. After blocking with 5% normal goat serum in PBS for 1 h, the digoxigenin-labeled probe was localized with an alkaline phosphatase-conjugated antidigoxigenin antibody (1:5,000; Boehringer Mannheim) and detected with 5-bromo-4-chloro-3-indolylphosphate and nitroblue tetrazolium.
Northern and Western blotting.
For protein detection, cells were lysed in buffer (1× PBS, 1% NP-40, 0.5% sodium deoxycholate, 100 μg of phenylmethylsulfonyl fluoride per ml, and 20 μg of leupeptin per ml), and the protein was separated on a sodium dodecyl sulfate (SDS)–7.5% polyacrylamide gel, transferred to a nitrocellulose membrane, and detected with the anti-ICP0 primary antibody (1:1,000), followed by a peroxidase-conjugated secondary antibody (1:5,000; Sigma) and chemiluminescence detection with ECL (Amersham). Equivalent amounts of total protein were loaded in each lane. Total RNA was extracted from infected cells using the RNeasy minikit (Qiagen, Valencia, Calif.) according to the manufacturer's instructions. Ten micrograms of total RNA per lane was separated in a 1.2% agarose gel containing 0.22 M formaldehyde, transferred to a GeneScreen Plus membrane (NEN, Boston, Mass.), fixed by UV cross-linking, washed in 1% SDS–1× SSC at 77°C for 30 min, prehybridized in hybridization solution [5× SSC, 1% SDS, 10% dextran sulfate, 0.5% dry milk, 25 μg of poly(A) per ml, 25 μg of poly(C) per ml, 200 μg of sheared salmon sperm DNA per ml), and then hybridized overnight at 77°C with the [32P]CTP-labeled ICP0 riboprobe (2 × 106 cpm/ml). The radioactive riboprobe for ICP0 was prepared using the same methods described above for the digoxigenin-labeled probe that was used for the in situ hybridization, except that the alkaline hydrolysis step was omitted. Following hybridization, the membrane was washed for 1 h at 77°C in 5× SSC–0.5% dry milk–1% SDS, twice for 30 min in 2× SSC–0.1% SDS, and twice for 30 min in 0.5× SSC at 77°C. The radioactively labeled probe was detected with a phosphorimager (Molecular Dynamics, Sunnyvale, Calif.).
Single-cell RT-PCR for ICP0.
Individual infected trigeminal neurons identified by the expression of GFP were isolated in a pulled and beveled microcapillary using a Burleigh MIS-5000 micromanipulator (Fisher, New York, N.Y.) and an IM-6-2 microinjector (Narishige, Tokyo, Japan). The single cells were individually placed into thin-walled PCR tubes (Perkin-Elmer, Foster City, Calif.) containing 21 μl of cell lysis buffer (0.5% NP-40 and 10 U of RNase inhibitor). Lysis was accomplished by one round of freezing and thawing followed by incubation for 5 min at 72°C. Seven microliters of the single-cell lysate was saved for RT control, and RT was performed on the remaining 13 μl using the Sensiscript reverse transcriptase kit from Qiagen. Briefly, the RT reaction was performed for 1 h at 37°C by addition of 1× RT buffer containing 0.5 mM (each) dATP, dCTP, dGTP, and dTTP; 10 U of RNase inhibitor; 2.5 μM random hexamer (Perkin-Elmer); and 1 μl of Sensiscript reverse transcriptase. The reaction was terminated by incubation at 95°C for 5 min followed by rapid chilling on ice. PCR was then carried out in a 100-μl reaction volume using a hot-start PCR kit from Qiagen. Of the RT product (cDNA), 10 to 20% was used in a PCR containing 1× PCR buffer; 2 mM MgCl2; 200 μM (each) dATP, dCTP, dGTP, and dTTP; 0.2 μM primers; 2.5 U of HotStarTaq DNA polymerase; and 10 μCi of [32P]dCTP. The PCR was performed in a Perkin-Elmer Thermal Cycler 480 and consisted of a 15-min preincubation at 95°C; 40 cycles of denaturing (94°C, 1 min), annealing (60°C, 1 min), and extension (72°C, 1 min); and final extension for 10 min at 72°C. The PCR product was separated on a 5% acrylamide gel and detected by autoradiography. The appropriate size of the product was determined by comparison to a positive-control PCR product. The primers used for mouse β-actin and ICP0 were chosen because the size of the product distinguishes RNA-derived signal (intron absent, short amplification product) from genomic DNA-derived signal (intron present, larger product). The oligonucleotide primer pairs were as follows: β-actin (amplification fragment, 359 bp), 5′-GGTTCCGATGCCCTGAGGCTC-3′ (sense) and 5′-ACTTGCGGTGCACGATGGAGG-3′ (antisense); ICP0 (amplification fragment, 157 bp), 5′-TTCGGTCTCCGCCTGAGAGT-3′ (sense) and 5′-GACCCTCCAGCCGCATACGA-3′ (antisense), as described in reference 37.
Inhibitor treatment.
TG cultures were pretreated for 4 h with the lysosome inhibitor choloroquine (Sigma), the calpain inhibitor PD 150606 (5 μM; Calbiochem, La Jolla, Calif.), or the proteasome inhibitor lactacystin (10 μM; Calbiochem). PD 150606 and lactacystin were dissolved in 100% dimethyl sulfoxide (Sigma) first and then added into the tissue culture solution at less than 1% of total volume. The cells were then washed twice with Dulbecco's modified Eagle medium and infected with QOZHG virus at an MOI of 5 for 1 h. The virus-containing medium was then replaced with medium containing the inhibitors (or the same concentration of dimethyl sulfoxide alone for the controls) for an overnight incubation. For inhibition of calcineurin activity, cyclosporine (200 ng/ml; Novartis, East Hanover, N.J.) was added into the culture medium 12 h after infection (MOI of 5), and the experiment was terminated 4 h later.
RESULTS
ICP0 accumulates in the nucleus of Schwann cells but not in the nucleus of primary neurons in culture.
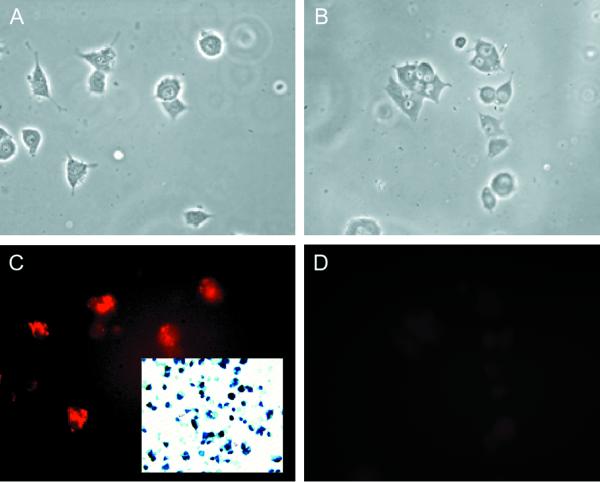
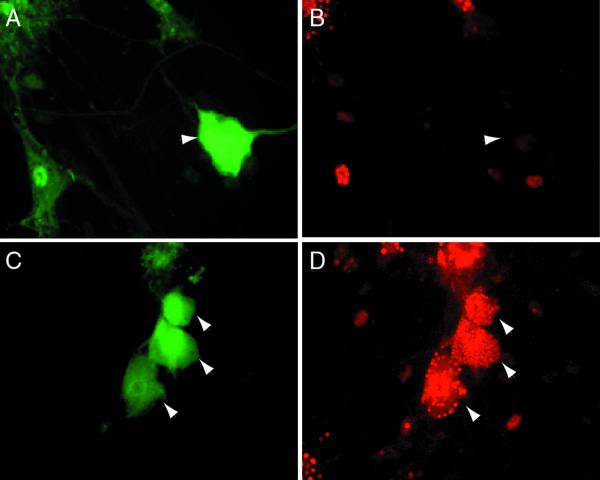
Dissociated TG cultures contain a mixed population of neurons, identified by their characteristic morphology and immunostaining for neuron-specific proteins including the neuronal isoform of microtubule-associated protein 2 (MAP2) and neurofilament proteins and supporting cells that include Schwann cells, satellite cells, and fibroblasts. Infection of these cultures with either wild-type virus or the nonneurotoxic ICP0-expressing recombinant TOZ or QOZHG at an MOI of 5 resulted in the expression of ICP0 protein in the characteristic punctate intranuclear distribution exclusively in nonneuronal cells, not in neurons (Fig. 1). The majority of both neurons and Schwann cells in the culture were infected, as determined by X-Gal staining for lacZ expression (Fig. 2, inset). Neurons are clearly defined in these cultures by the characteristic morphology and the presence of a bright halo on phase microscopy, and the identity of the ICP0-immunoreactive cells as nonneuronal was confirmed by immunocytochemical staining of neurons with an antibody to MAP2 (Fig. 2). The amount of ICP0 immunoreactivity increased over the first 8 h of infection and then remained unchanged from 16 to 24 h postinfection after infection with the replication-incompetent mutant TOZ (data not shown). Similar results were found 5 h after infection of the cultures with wild-type KOS, where the ICP0 accumulated in the nucleus of Schwann cells but not in neurons despite the widespread infection of both neurons and Schwann cells in the culture (Fig. 3). The distribution of ICP0 at later time points after infection with replicating virus could not be determined because of the cytolytic effects of the replicating virus. No ICP0 immunoreactivity was seen with mock-infected cells.
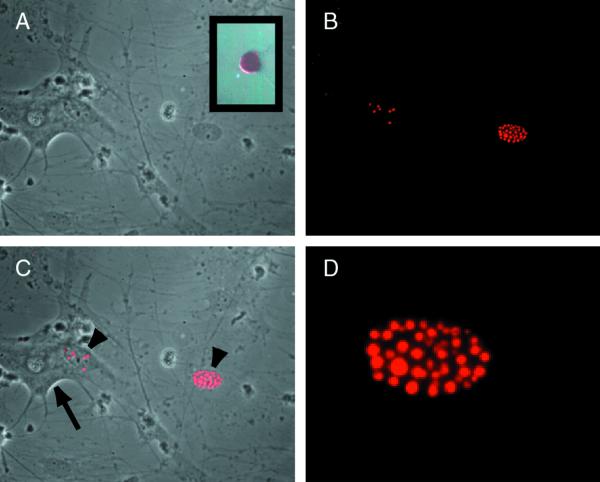
FIG. 1.
ICP0 protein accumulates in the nuclei of nonneuronal cells in dissociated TG cultures. Sixteen hours after infection with TOZ (MOI of 5), ICP0 is found in the nucleus of Schwann cells (arrowheads) but not neurons (arrow, identified by the characteristic appearance) in the cultures (phase contrast [A], fluorescence [B], and merged image [C]). At higher power, the immunostaining in the nucleus of the nonneuronal cells has the characteristic punctate appearance as described previously (D). ICP0 RNA can be detected in infected neurons by in situ hybridization with a digoxigenin-labeled riboprobe (inset, panel A).
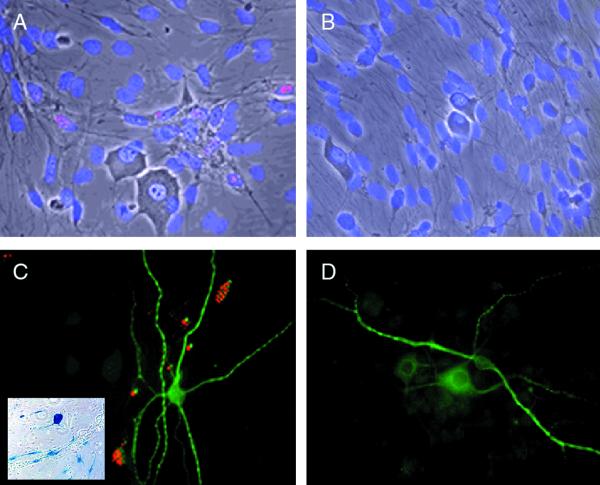
FIG. 2.
ICP0 does not accumulate in the nucleus of neurons in culture. The identity of the neurons was confirmed by immunostaining with an antibody to the neuron-specific antigen MAP2 (C and D). TOZ (A and C)- and mock (B and D)-infected cultures, 16 h postinfection, show ICP0 in a punctate pattern in the nucleus of Schwann cells but not of neurons in the infected cultures (A and C). X-Gal staining (inset, panel C) demonstrates that the majority of the cells in the culture, including neurons, were infected and expressed the lacZ transgene. Panels A and B are fused phase and fluorescent images of cultures counterstained with DAPI (4′,6′-diamidino-2-phenylindole).
FIG. 3.
The pattern of accumulation of ICP0 after infection with replicating virus is similar to the pattern seen after infection with TOZ. Five hours after infection with wild-type KOS (MOI of 5), ICP0 is demonstrated by immunohistochemistry in the nucleus of nonneuronal cells (B) but is not seen in neurons (arrows, panels A and B). Wild-type KOS infected virtually all the cells in the culture, demonstrated by robust expression of HSV gC in neurons (D). Panels A and C are the phase micrographs corresponding to the fluorescent images in panels B and D.
ICP0 RNA is expressed in neurons.
ICP0 RNA could be detected by in situ hybridization in a small number of infected neurons (Fig. 1, inset) despite the absence of ICP0 protein staining. However, a much greater proportion of neurons were infected, as determined by X-Gal staining and GFP expression, than expressed ICP0 RNA detectable by in situ hybridization. We therefore proceeded to use single-cell RT-PCR to explore whether ICP0 RNA could be detected in individual infected neurons. For these experiments, the TG cultures were infected with QOZHG, which, like recombinant TOZ, does not express ICP4, ICP22, or ICP27 and has ICP41 (viral host shutoff function) deleted but in addition has the GFP under the control of the human cytomegalovirus IEp allowing us to identify infected neurons in culture. Eighteen hours after infection with QOZHG, infected GFP-expressing neurons identified by their characteristic green fluorescence (approximately 80% of the neurons in the culture appeared to express GFP) were individually removed from the coverslip using a micropipette, and the RNA from the isolated cell was prepared for RT of β-actin or ICP0 using primers specific for the RNA. In this experiment (which was repeated twice), β-actin RNA could be detected in approximately half (6 of 12 and 7 of 16) of the individual cells that were examined, and ICP0 RNA was detected in half (three of six and three of seven) of the β-actin-positive samples (Fig. 4). These data suggest that at least half of the infected neurons in the culture express ICP0 RNA, despite the fact that ICP0 immunostaining was rarely found in neurons.
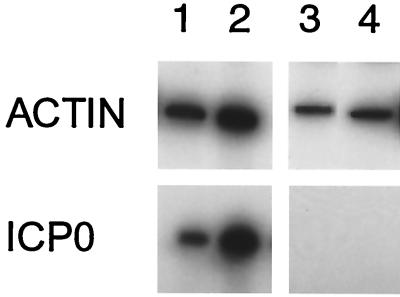
FIG. 4.
ICP0 RNA can be detected in approximately half of individual infected neurons by single-cell RT-PCR of infected neurons. Individual infected neurons identified by green fluorescence were isolated, and RT-PCR for β-actin was used to confirm RNA recovery. Approximately 50% of β-actin RNA-positive cells also showed ICP0 RNA by RT-PCR. Each lane represents the results of PCR amplification of cDNA obtained from a single identified infected cell in which β-actin was detected (upper panels). Results from four representative infected cells, two with detectable ICP0 RNA (lanes 1 and 2) and two without detectable ICP0 RNA (lanes 3 and 4), are shown. All of the uninfected cells (identified by the absence of fluorescence) in which β-actin RNA was detected were negative for ICP0 RNA (data not shown).
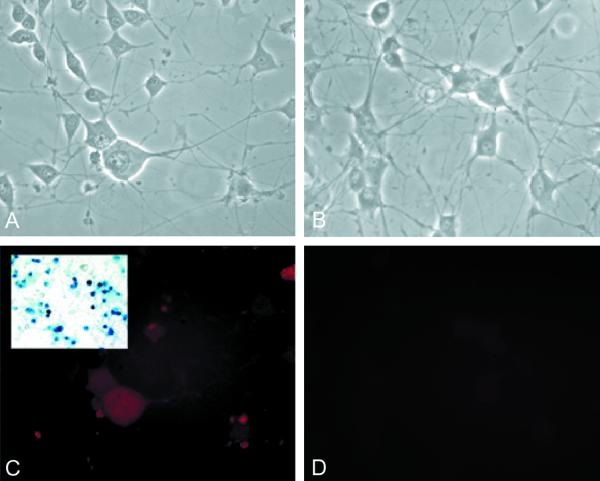
The accumulation of ICP0 protein is reduced in differentiated PC12 cells compared to undifferentiated PC12 cells.
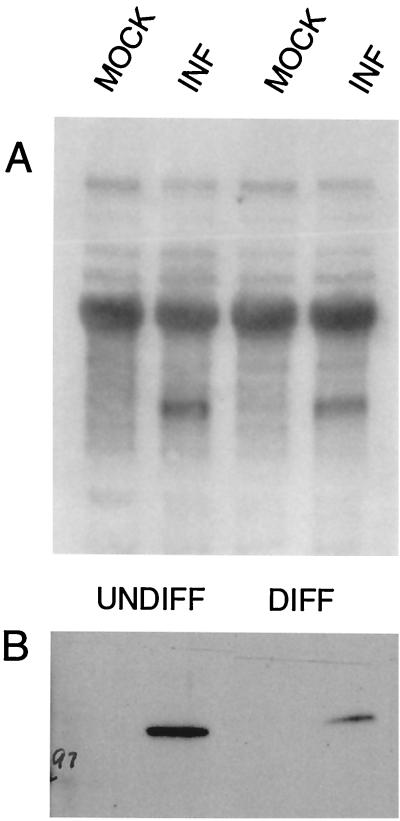
PC12 cells can be stimulated to assume a neuronal phenotype by exposure to NGF. Undifferentiated PC12 cells infected with wt virus or TOZ at an MOI of 3 demonstrated the punctate nuclear distribution similar to that seen with nonneuronal cells in TG cultures (Fig. 5). In contrast, differentiated PC12 cells showed only diffuse staining that was barely distinguishable from that of control uninfected cells stained in a similar fashion (Fig. 6). The difference in protein expression and distribution did not result from a failure of infection or transcription. The vast majority of cells in both differentiated and undifferentiated PC12 cell cultures were infected by TOZ as demonstrated by X-Gal staining for lacZ expression (Fig. 5 and 6, insets). A single 2.7-kb band of ICP0 RNA was isolated from infected undifferentiated and differentiated PC12 cell cultures, and similar amounts were detected in infected differentiated and undifferentiated PC12 cell cultures as determined by Northern blotting (Fig. 7A). By Western blotting, ICP0 protein could be detected as a single 110-kDa band in both differentiated and undifferentiated cultures, but the amount was less in infected differentiated than in undifferentiated cultures (Fig. 7B). These results suggested that the rate of RNA synthesis is similar in differentiated and undifferentiated cells, but that protein turnover is more rapid in differentiated PC12 cells.
FIG. 5.
ICP0 protein accumulates in a punctate nuclear pattern in undifferentiated PC12 cells. Undifferentiated PC12 cells 16 h postinfection with TOZ (A and C) show the punctate nuclear pattern of ICP0 immunohistochemistry characteristic of nonneuronal cells. Virtually all the cells have been infected as demonstrated by X-Gal staining (inset, panel C). Mock-infected cultures are shown in panels B and D. (A and B) Phase microscopy. (C and D) Fluorescence.
FIG. 6.
ICP0 fails to accumulate in the nucleus of differentiated PC12 cells. PC12 cells differentiated by exposure to NGF to assume a neuronal phenotype show only faint diffuse cytoplasmic immunostaining 16 h after infection with TOZ (C). Mock-infected cultures are shown in panels B and D. Virtually all the cells have been infected as demonstrated by X-Gal staining (inset, panel C). (A and B) Phase microscopy. (C and D) Fluorescence.
FIG. 7.
ICP0 RNA is expressed in both differentiated and undifferentiated PC12 cells, but the amount of protein is reduced in differentiated cells. Undifferentiated (UNDIFF) or differentiated (DIFF) PC12 cells were infected with TOZ at an MOI of 5, and RNA and protein were extracted 16 h after infection. Representative Northern (A) and Western (B) blots of RNA and protein from the infected (INF) and mock-infected PC12 cells are shown. The experiment was independently repeated four times, with similar results.
ICP0 protein accumulation in neurons is increased by inhibition of calcineurin or calpains in culture.
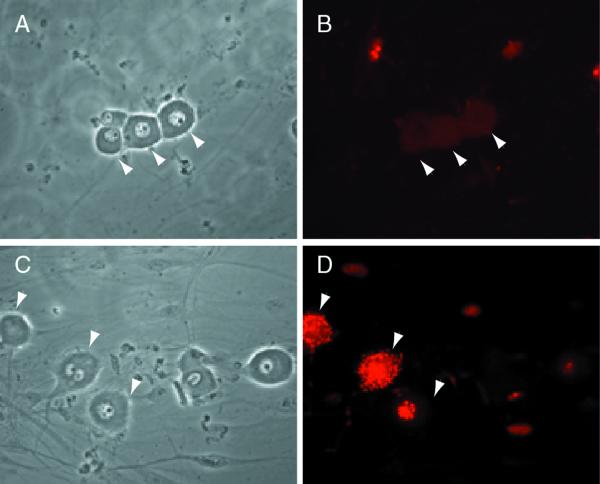
Because the combined data from the PC12 cell and TG culture experiments suggested that ICP0 RNA was expressed in both neuronal and nonneuronal cells but that the protein failed to accumulate in the nucleus in either neurons or in PC12 cells that had been differentiated to a neuronal phenotype, we examined the effect of adding inhibitors of proteolysis to infected cell cultures. There is evidence that proteasome activity is required to stimulate the progression of HSV infection (20), but inhibition of the proteasome pathway with lactacystin did not increase the number of neurons exhibiting intracellular accumulation of ICP0 protein (17 of 1,400 neurons counted on a single well). However, inhibition of calpain activity by PD 150606 resulted in a dramatic increase in the number of neurons that accumulated ICP0 (320 of 800 in a single culture). The amount of ICP0 increased in both the nucleus and the cytoplasm and accumulated in aggregates in both sites, and it appeared in most cases that ICP0 accumulated in the nucleus first and then in the cytoplasm, since many cells had nuclear or nuclear and cytoplasmic ICP0 but almost none demonstrated significant cytoplasmic accumulation in the absence of ICP0 in the nucleus. Similar effects were achieved by the inhibition of protein phosphatase 2B (calcineurin) by the addition of cyclosporine (632 of 1,380 neurons counted). Both inhibition of proteolysis and inhibition of phosphatase resulted in a punctate pattern of ICP0 in the nucleus and accumulation of aggregates of ICP0 in the cytoplasm as well (Fig. 8 and 9).
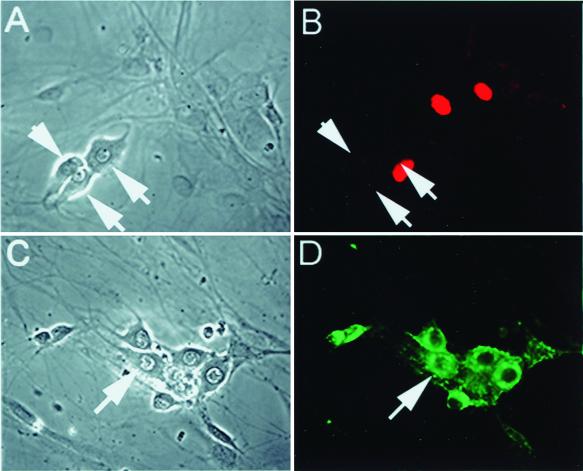
FIG. 8.
Inhibition of calpain activity results in accumulation of ICP0 in neurons. Sixteen hours after infection of primary TG cultures with QOXHG at an MOI of 5, little immunoreactivity is seen in untreated neurons (arrowheads, panels A and B). Neurons (arrowheads) treated with PD 150606 to inhibit calpain activity show marked nuclear and cytoplasmic accumulation of immunoreactive ICP0 protein (C and D). Panels A and C show the phase micrographs corresponding to the fluorescent immunocytochemical images shown in panels B and D.
FIG. 9.
Inhibition of calcineurin activity results in accumulation of ICP0 in neurons. Sixteen hours after infection of primary TG cultures with recombinant QOZHG at an MOI of 5, no ICP0 immunoreactivity is seen in infected neurons (arrowheads). One neuron, identified by its characteristic morphology and identified as infected by the expression of GFP, is shown in panels A and B. A Schwann cell in the same culture is seen in the lower left of those panels. TG cultures treated with cyclosporine to inhibit calcineurin activity show marked accumulation of ICP0 in the nucleus and cytoplasm of infected neurons (arrowheads, panels C and D). Panels A and C are fluorescent micrographs of GFP expression. Panels B and D are cultures stained for ICP0.
DISCUSSION
The early localization of ICP0 in a punctate pattern in the nucleus of infected cells is characteristic of HSV infection. Because of the important role that ICP0 plays as a transcriptional regulator of HSV gene expression, and the evidence that ICP0 mutants that are impaired in their ability to accumulate in the nucleus replicate poorly (14), nuclear localization of the protein is likely to be important in the regulation of that function. The observation that in neurons (and PC12 cells differentiated to a neuronal phenotype) ICP0 protein does not accumulate in the nucleus identifies one important cell-specific aspect of HSV IE gene product behavior that is different in neurons compared to other cells.
Nuclear ICP0 colocalizes with nuclear substructures (ND10s) recognizable by specific immunostaining for Sp100, identified by antiserum obtained from patients with primary biliary cirrhosis, or PML, identified by the presence of antibodies to it in acute promyelocytic leukemia and a fusion partner of the retinoic acid receptor, as well several other proteins (reviewed in reference 38). ND10s are found in a wide variety of cell types but are not ubiquitous. In particular, these structures are absent from neurons and cell lines of neuronal lineage (9). It has been suggested that association of viral genomes and newly transcribed proteins with proteins in ND10 loci may promote the replication of HSV genomes in those cells. HSV-associated ubiquitin-specific protease, which binds strongly to ICP0 and removes ubiquitin adducts from proteins, is an important component of ND10s (21). The ability of ICP0 to bind to HSV-associated ubiquitin-specific protease has been suggested to play a role in the activation of gene expression and stimulation of virus replication (20), since the disruption of ND10s which occurs during HSV infection correlates with ICP0 and proteasome-dependent degradation of several PML isoforms (18). Although in transient-transfection assays ICP0 accumulates in a punctate nuclear distribution that includes but is not exclusive to immunocytochemically defined ND10s (50), viral RNA is found associated with ND10 before protein synthesis (40), which appears to serve as an early site of DNA virus deposition for HSV, adenovirus, and cytomegalovirus (1, 25). Indirect evidence strongly supports the conclusion that these nuclear structures play an important role in viral replication (38). However, the lack of ND10 structures in neurons cannot account for the failure of ICP0 protein to accumulate in punctate structures in neurons, because increasing the amount of ICP0 protein by inhibition of either calpains, lysosomal enzymes, or calcineurin activity in neurons allows ICP0 to accumulate in the characteristic punctate intranuclear pattern in those cells.
Could the absence of ICP0 in the nucleus of cells with the neuronal phenotype result from alternative splicing of the gene product? The ICP0 gene has three coding sequences; the second exon encodes a zinc binding ring finger domain, while the third exon encodes sequences which are required for nuclear localization (13, 15) and the subsequent transactivation of viral gene expression (15). Failure to splice out the second intron results in a product of 262 residues and a predicted molecular mass of 41 kDa which has a dominant-negative effect on viral gene transcription in transient-transfection assays (61, 62) and which is found after HSV infection of cell types in varying amounts (17). However, it is not likely that alternative splicing in differentiated PC12 cells accounts for the failure of ICP0 to accumulate in the nucleus of neurons or PC12 cells differentiated to a neuronal phenotype, because only a single protein band of 105 kDa was found in infected PC12 cultures, whether differentiated or undifferentiated. Analysis of the first intron demonstrates the existence of four alternative splice variants (8). In addition to the full-length 775-codon mRNA, the variant proteins are predicted to contain 152, 90, and 87 amino acids. None of these smaller splice variants were identified by Western blotting of infected PC12 cultures. It therefore seems unlikely that alternative splicing accounts for the difference in distribution of ICP0 between neuronal and nonneuronal cells. A temperature-sensitive mutant ICP4 protein prevents the nuclear localization of ICP0 (31), but in the current experiments, we have used an ICP4-deletion virus, and nuclear localization in nonneuronal cells infected with this recombinant is unimpaired.
ICP0 protein is extensively modified by posttranslational phosphorylation and nucleotidylylation (3, 4, 43). UL13 contributes to, but is not the sole kinase responsible for, ICP0 phosphorylation (47), and posttranslational processing of ICP0 varies from cell line to cell line. The observation that inhibition of phosphatase activity increases the amount of ICP0 protein in the cell, with accumulation in punctate aggregates both within the nucleus and within the cytoplasm, suggests that phosphorylation may protect newly synthesized ICP0 from rapid degradation, both in neurons and in nonneuronal cells. Phosphorylation is a common pathway used to protect cellular proteins from a variety of degradation pathways, as has been demonstrated previously for structural proteins (23), enzymes involved in signal transduction (5), and transcriptional regulators (44). We have not directly demonstrated that phosphorylation protects ICP0 from degradation; similar accumulation of increased amounts of ICP0 was observed after inhibition of calcium-activated neutral protease activity, which suggests that calpain activity may be one major path of ICP0 degradation, particularly in neurons. Importantly, no protection was afforded by the complete block of the proteasome pathway.
This report is limited to observations of ICP0 distribution in neurons and other cells in culture. As with the situation in tissue culture, we did not observe intranuclear ICP0 immunostaining in neurons in vivo at 1 to 5 days following infection by corneal scarification (data not shown). However, this negative finding is difficult to interpret because the punctate pattern of ICP0 immunostaining that has been reported in many different cell types in culture has not been described (and was not observed by us) even in nonneuronal cells in the TG in vivo. Pharmacologic manipulations that increase ICP0 accumulation in neuronal nuclei in vitro are not possible in vivo. Nonetheless, our observations add to the substantial published literature regarding ICP0 distribution in cells in vitro, illuminating a potentially important difference between neurons and nonneuronal cells in this regard.
The biology of HSV infection is cell type specific, although the molecular mechanisms underlying this specificity have not been fully established. Both primary neurons (59) and neuroblastoma cells (30) in culture are markedly nonpermissive for HSV infection, defined by cytopathic effect and antigen expression. There is substantial evidence that cell-specific transcriptional regulatory activators (26, 34) or inhibitors may influence the expression of HSV IE genes in early infection (28, 29), but the subcellular localization of cellular gene products may also regulate HSV infection. It has recently been reported, for instance, that the C1 host cell factor, an intranuclear protein which collaborates in enhancing viral IE gene expression in a wide variety of cell types, is redistributed from the cytoplasm to the nucleus of neurons under the influence of stimuli that reactivate latent HSV genomes (35). The failure of the ICP0 protein to accumulate in the nucleus of neurons may play a similar role in the primary infection. Herpesviruses characteristically naturally establish a life long latent state in neurons, and a pseudolatent state can be established in nonneuronal cells in vitro by preventing the virus from replicating during initial infection (56, 57, 63). The failure of ICP0 to translocate to the nucleus of neurons suggests a potential mechanism for the inhibition of HSV replication in neurons that may favor the establishment of a latent state in those cells.
ACKNOWLEDGMENTS
This work was supported by grants from the NIH (J.C.G. and D.J.F.), the Department of Veteran's Affairs (M.M. and D.J.F.), and the GenVec Corporation (J.C.G. and D.J.F.).
We acknowledge the excellent technical assistance of XiaoPing Hu in cell culture and immunocytochemistry and of Simon Watkins in imaging.
REFERENCES
- 1.Ahn J H, Hayward G S. The major immediate-early proteins IE1 and IE2 of human cytomegalovirus colocalize with and disrupt PML-associated nuclear bodies at very early times in infected permissive cells. J Virol. 1997;71:4599–4613. doi: 10.1128/jvi.71.6.4599-4613.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aurelian L, Kokuba H, Smith C. Vaccine potential of a herpes simplex virus type 2 mutant deleted in the PK domain of the large subunit of ribonucleotide reductase. Vaccine. 1999;17:1951–1963. doi: 10.1016/s0264-410x(98)00470-8. [DOI] [PubMed] [Google Scholar]
- 3.Blaho J A, Mitchell C, Roizman B. An amino acid sequence shared by the herpes simplex virus 1 alpha regulatory proteins 0, 4, 22, and 27 predicts the nucleotidylylation of the UL21, UL31, UL47, and UL49 gene products. J Biol Chem. 1994;269:17401–17410. [PubMed] [Google Scholar]
- 4.Blaho J A, Mitchell C, Roizman B. Guanylylation and adenylylation of the alpha regulatory proteins of herpes simplex virus require a viral beta or gamma function. J Virol. 1993;67:3891–3900. doi: 10.1128/jvi.67.7.3891-3900.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brondello J M, Pouyssegur J, McKenzie F R. Reduced MAP kinase phosphatase-1 degradation after p42/p44MAPK-dependent phosphorylation. Science. 1999;286:2514–2517. doi: 10.1126/science.286.5449.2514. [DOI] [PubMed] [Google Scholar]
- 6.Cai W, Schaffer P A. Herpes simplex virus type 1 ICP0 regulates expression of immediate-early, early, and late genes in productively infected cells. J Virol. 1992;66:2904–2915. doi: 10.1128/jvi.66.5.2904-2915.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai W Z, Schaffer P A. Herpes simplex virus type 1 ICP0 plays a critical role in the de novo synthesis of infectious virus following transfection of viral DNA. J Virol. 1989;63:4579–4589. doi: 10.1128/jvi.63.11.4579-4589.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carter K L, Roizman B. Alternatively spliced mRNAs predicted to yield frame-shift proteins and stable intron 1 RNAs of the herpes simplex virus 1 regulatory gene alpha 0 accumulate in the cytoplasm of infected cells. Proc Natl Acad Sci USA. 1996;93:12535–12540. doi: 10.1073/pnas.93.22.12535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho Y, Lee I, Maul G G, Yu E. A novel nuclear substructure, ND10: distribution in normal and neoplastic human tissues. Int J Mol Med. 1998;1:717–724. doi: 10.3892/ijmm.1.4.717. [DOI] [PubMed] [Google Scholar]
- 10.Coen D M, Kosz-Vnenchak M, Jacobsen J G. Thymidine kinase-negative herpes simplex virus mutants establish latency in mouse trigeminal ganglia but do not reactivate. Proc Natl Acad Sci USA. 1989;86:4736–4740. doi: 10.1073/pnas.86.12.4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeLuca N A, McCarthy A M, Schaffer P A. Isolation and characterization of deletion mutants of herpes simplex virus type 1 in the gene encoding immediate-early regulatory protein ICP4. J Virol. 1985;56:558–570. doi: 10.1128/jvi.56.2.558-570.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dyck J A, Maul G G, Miller W H, Jr, Chen J D, Kakizuka A, Evans R M. A novel macromolecular structure is a target of the promyelocyte-retinoic acid receptor oncoprotein. Cell. 1994;76:333–343. doi: 10.1016/0092-8674(94)90340-9. [DOI] [PubMed] [Google Scholar]
- 13.Everett R D. Analysis of the functional domains of herpes simplex virus type 1 immediate-early polypeptide Vmw110. J Mol Biol. 1988;202:87–96. doi: 10.1016/0022-2836(88)90521-9. [DOI] [PubMed] [Google Scholar]
- 14.Everett R D. Construction and characterization of herpes simplex virus type 1 mutants with defined lesions in immediate early gene 1. J Gen Virol. 1989;70:1185–1202. doi: 10.1099/0022-1317-70-5-1185. [DOI] [PubMed] [Google Scholar]
- 15.Everett R D. A detailed mutational analysis of Vmw110, a trans-acting transcriptional activator encoded by herpes simplex virus type 1. EMBO J. 1987;6:2069–2076. doi: 10.1002/j.1460-2075.1987.tb02472.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Everett R D. Transactivation of transcription by herpes virus products: requirement for two HSV-1 immediate-early polypeptides for maximum activity. EMBO J. 1984;3:3135–3141. doi: 10.1002/j.1460-2075.1984.tb02270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Everett R D, Cross A, Orr A. A truncated form of herpes simplex virus type 1 immediate-early protein Vmw110 is expressed in a cell type dependent manner. Virology. 1993;197:751–756. doi: 10.1006/viro.1993.1651. [DOI] [PubMed] [Google Scholar]
- 18.Everett R D, Freemont P, Saitoh H, Dasso M, Orr A, Kathoria M, Parkinson J. The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J Virol. 1998;72:6581–6591. doi: 10.1128/jvi.72.8.6581-6591.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Everett R D, Maul G G. HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J. 1994;13:5062–5069. doi: 10.1002/j.1460-2075.1994.tb06835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Everett R D, Meredith M, Orr A. The ability of herpes simplex virus type 1 immediate-early protein Vmw110 to bind to a ubiquitin-specific protease contributes to its roles in the activation of gene expression and stimulation of virus replication. J Virol. 1999;73:417–426. doi: 10.1128/jvi.73.1.417-426.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Everett R D, Meredith M, Orr A, Cross A, Kathoria M, Parkinson J. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO J. 1997;16:1519–1530. doi: 10.1093/emboj/16.7.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fink D J, DeLuca N A, Glorioso J C. Gene transfer to neurons using herpes simplex virus-based vectors. Annu Rev Neurosci. 1996;19:265–287. doi: 10.1146/annurev.ne.19.030196.001405. [DOI] [PubMed] [Google Scholar]
- 23.Greenwood J A, Troncoso J C, Costello A C, Johnson G V. Phosphorylation modulates calpain-mediated proteolysis and calmodulin binding of the 200-kDa and 160-kDa neurofilament proteins. J Neurochem. 1993;61:191–199. doi: 10.1111/j.1471-4159.1993.tb03555.x. [DOI] [PubMed] [Google Scholar]
- 24.Honess R W, Roizman B. Regulation of herpesvirus macromolecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J Virol. 1974;14:8–19. doi: 10.1128/jvi.14.1.8-19.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishov A M, Maul G G. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J Cell Biol. 1996;134:815–826. doi: 10.1083/jcb.134.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones K A, Tjian R. Sp1 binds to promoter sequences and activates herpes simplex virus ‘immediate-early’ gene transcription in vitro. Nature. 1985;317:179–182. doi: 10.1038/317179a0. [DOI] [PubMed] [Google Scholar]
- 27.Kawaguchi Y, Bruni R, Roizman B. Interaction of herpes simplex virus 1 α regulatory protein ICP0 with elongation factor 1δ: ICP0 affects translational machinery. J Virol. 1997;71:1019–1024. doi: 10.1128/jvi.71.2.1019-1024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kemp L M, Dent C L, Latchman D S. Octamer motif mediates transcriptional repression of HSV immediate-early genes and octamer-containing cellular promoters in neuronal cells. Neuron. 1990;4:215–222. doi: 10.1016/0896-6273(90)90096-x. [DOI] [PubMed] [Google Scholar]
- 29.Kemp L M, Latchman D S. Regulated transcription of herpes simplex virus immediate-early genes in neuroblastoma cells. Virology. 1989;171:607–610. doi: 10.1016/0042-6822(89)90631-4. [DOI] [PubMed] [Google Scholar]
- 30.Kennedy P G, Clements G B, Brown S M. Differential susceptibility of human neural cell types in culture to infection with herpes simplex virus. Brain. 1983;106:101–119. doi: 10.1093/brain/106.1.101. [DOI] [PubMed] [Google Scholar]
- 31.Knipe D M, Smith J L. A mutant herpesvirus protein leads to a block in nuclear localization of other viral proteins. Mol Cell Biol. 1986;6:2371–2381. doi: 10.1128/mcb.6.7.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krisky D M, Marconi P C, Oligino T J, Rouse R J, Fink D J, Cohen J B, Watkins S C, Glorioso J C. Development of herpes simplex virus replication-defective multigene vectors for combination gene therapy applications. Gene Ther. 1998;5:1517–1530. doi: 10.1038/sj.gt.3300755. [DOI] [PubMed] [Google Scholar]
- 33.Kristie J, Roizman B. Host cell protein binds to the cis-acting site required for virion-mediated induction of herpes simplex virus 1 α genes. Proc Natl Acad Sci USA. 1987;84:71–75. doi: 10.1073/pnas.84.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kristie T M, Roizman B. DNA-binding site of major regulatory protein α4 specifically associated with the promoter-regulatory domains of a genes of herpes simplex virus type I. Proc Natl Acad Sci USA. 1986;83:4700–4704. doi: 10.1073/pnas.83.13.4700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kristie T M, Vogel J L, Sears A E. Nuclear localization of the C1 factor (host cell factor) in sensory neurons correlates with reactivation of herpes simplex virus from latency. Proc Natl Acad Sci USA. 1999;96:1229–1233. doi: 10.1073/pnas.96.4.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lillycrop K A, Liu Y Z, Theil T, Moroy T, Latchman D S. Activation of the herpes simplex virus immediate-early gene promoters by neuronally expressed POU family transcription factors. Biochem J. 1995;307:581–584. doi: 10.1042/bj3070581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lynas C, Laycock K A, Cook S D, Hill T J, Blyth W A, Maitland N J. Detection of herpes simplex virus type 1 gene expression in latently and productively infected mouse ganglia using the polymerase chain reaction. J Gen Virol. 1989;70:2345–2355. doi: 10.1099/0022-1317-70-9-2345. [DOI] [PubMed] [Google Scholar]
- 38.Maul G G. Nuclear domain 10, the site of DNA virus transcription and replication. Bioessays. 1998;20:660–667. doi: 10.1002/(SICI)1521-1878(199808)20:8<660::AID-BIES9>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 39.Maul G G, Guldner H H, Spivack J G. Modification of discrete nuclear domains induced by herpes simplex virus type 1 immediate early gene 1 product (ICP0) J Gen Virol. 1993;74:2679–2690. doi: 10.1099/0022-1317-74-12-2679. [DOI] [PubMed] [Google Scholar]
- 40.Maul G G, Ishov A M, Everett R D. Nuclear domain 10 as preexisting potential replication start sites of herpes simplex virus type-1. Virology. 1996;217:67–75. doi: 10.1006/viro.1996.0094. [DOI] [PubMed] [Google Scholar]
- 41.Mavromara-Nazos P, Silver S, Hubenthal-Voss J, McKnight J L, Roizman B. Regulation of herpes simplex virus 1 genes: alpha gene sequence requirements for transient induction of indicator genes regulated by beta or late (gamma 2) promoters. Virology. 1986;149:152–164. doi: 10.1016/0042-6822(86)90117-0. [DOI] [PubMed] [Google Scholar]
- 42.McCarthy A M, McMahan L, Schaffer P A. Herpes simplex virus type 1 ICP27 deletion mutants exhibit altered patterns of transcription and are DNA deficient. J Virol. 1989;63:18–27. doi: 10.1128/jvi.63.1.18-27.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mitchell C, Blaho J A, Roizman B. Casein kinase II specifically nucleotidylylates in vitro the amino acid sequence of the protein encoded by the alpha 22 gene of herpes simplex virus 1. Proc Natl Acad Sci USA. 1994;91:11864–11868. doi: 10.1073/pnas.91.25.11864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Musti A M, Treier M, Bohmann D. Reduced ubiquitin-dependent degradation of c-Jun after phosphorylation by MAP kinases. Science. 1997;275:400–402. doi: 10.1126/science.275.5298.400. [DOI] [PubMed] [Google Scholar]
- 45.Nichol P F, Chang J Y, Johnson E M, Jr, Olivo P D. Herpes simplex virus gene expression in neurons: viral DNA synthesis is a critical regulatory event in the branch point between the lytic and latent pathways. J Virol. 1996;70:5476–5486. doi: 10.1128/jvi.70.8.5476-5486.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nichol P F, Chang J Y, Johnson E M, Jr, Olivo P D. Infection of sympathetic and sensory neurons with herpes simplex virus does not elicit a shut-off of cellular protein synthesis: implications for viral latency and herpes vectors. Neurobiol Dis. 1994;1:83–94. doi: 10.1006/nbdi.1994.0011. [DOI] [PubMed] [Google Scholar]
- 47.Ogle W O, Ng T I, Carter K L, Roizman B. The UL13 protein kinase and the infected cell type are determinants of posttranslational modification of ICP0. Virology. 1997;235:406–413. doi: 10.1006/viro.1997.8710. [DOI] [PubMed] [Google Scholar]
- 48.O'Hare P, Goding C R, Haigh A. Direct combinatorial interaction between a herpes simplex virus regulatory protein and a cellular octamer-binding factor mediates specific induction of virus immediate-early gene expression. EMBO J. 1988;7:4231–4238. doi: 10.1002/j.1460-2075.1988.tb03320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O'Hare P, Hayward G S. Evidence for a direct role for both the 175,000- and 110,000-molecular-weight immediate-early proteins of herpes simplex virus in the transactivation of delayed-early promoters. J Virol. 1985;53:751–760. doi: 10.1128/jvi.53.3.751-760.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O'Rourke D, Elliott G, Papworth M, Everett R, O'Hare P. Examination of determinants for intranuclear localization and transactivation within the RING finger of herpes simplex virus type 1 IE110k protein. J Gen Virol. 1998;79:537–548. doi: 10.1099/0022-1317-79-3-537. [DOI] [PubMed] [Google Scholar]
- 51.Quinlan M P, Knipe D M. Stimulation of expression of a herpes simplex virus DNA-binding protein by two viral functions. Mol Cell Biol. 1985;5:957–963. doi: 10.1128/mcb.5.5.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roizman B, Sears A. Herpes simplex viruses and their replication. In: Fields B N, Knipe D M, Howley P M, Chanock R M, Hirsch M S, Melnick J L, Monath T P, Roizman B, editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2231–2295. [Google Scholar]
- 53.Roizman B, Sears A E. Herpes simplex viruses and their replication. In: Roizman B, Whitley R J, Lopez C, editors. The Human herpesviruses. New York, N.Y: Raven Press; 1993. pp. 11–68. [Google Scholar]
- 54.Sacks W R, Schaffer P A. Deletion mutants in the gene encoding the herpes simplex virus type 1 immediate-early protein ICP0 exhibit impaired growth in cell culture. J Virol. 1987;61:829–839. doi: 10.1128/jvi.61.3.829-839.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Samaniego L A, Neiderhiser L, DeLuca N A. Persistence and expression of the herpes simplex virus genome in the absence of immediate-early proteins. J Virol. 1998;72:3307–3320. doi: 10.1128/jvi.72.4.3307-3320.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scheck A C, Wigdahl B, De Clercq E, Rapp F. Prolonged herpes simplex virus latency in vitro after treatment of infected cells with acyclovir and human leukocyte interferon. Antimicrob Agents Chemother. 1986;29:589–593. doi: 10.1128/aac.29.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shiraki K, Rapp F. Establishment of herpes simplex virus latency in vitro with cycloheximide. J Gen Virol. 1986;67:2497–2500. doi: 10.1099/0022-1317-67-11-2497. [DOI] [PubMed] [Google Scholar]
- 58.Stow N D, Stow E C. Isolation and characterization of a herpes simplex virus type 1 mutant containing a deletion within the gene encoding the immediate early polypeptide Vmw110. J Gen Virol. 1986;67:2571–2585. doi: 10.1099/0022-1317-67-12-2571. [DOI] [PubMed] [Google Scholar]
- 59.Vahlne A, Nystrom B, Sandberg M, Hamberger A, Lycke E. Attachment of herpes simplex virus to neurons and glial cells. J Gen Virol. 1978;40:359–371. doi: 10.1099/0022-1317-40-2-359. [DOI] [PubMed] [Google Scholar]
- 60.Ward P L, Roizman B. Herpes simplex genes: the blueprint of a successful human pathogen. Trends Genet. 1994;10:267–274. doi: 10.1016/0168-9525(90)90009-u. [DOI] [PubMed] [Google Scholar]
- 61.Weber P C, Kenny J J, Wigdahl B. Antiviral properties of a dominant negative mutant of the herpes simplex virus type 1 regulatory protein ICP0. J Gen Virol. 1992;73:2955–2961. doi: 10.1099/0022-1317-73-11-2955. [DOI] [PubMed] [Google Scholar]
- 62.Weber P C, Wigdahl B. Identification of dominant-negative mutants of the herpes simplex virus type 1 immediate-early protein ICP0. J Virol. 1992;66:2261–2267. doi: 10.1128/jvi.66.4.2261-2267.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wrzos H, Rapp F. Establishment of latency in vitro with herpes simplex virus temperature-sensitive mutants at nonpermissive temperature. Virus Res. 1987;8:301–308. doi: 10.1016/0168-1702(87)90002-5. [DOI] [PubMed] [Google Scholar]
- 64.Zhu Z, Cai W, Schaffer P A. Cooperativity among herpes simplex virus type 1 immediate-early regulatory proteins: ICP4 and ICP27 affect the intracellular localization of ICP0. J Virol. 1994;68:3027–3040. doi: 10.1128/jvi.68.5.3027-3040.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhu Z, Schaffer P A. Intracellular localization of the herpes simplex virus type 1 major transcriptional regulatory protein, ICP4, is affected by ICP27. J Virol. 1995;69:49–59. doi: 10.1128/jvi.69.1.49-59.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]