Abstract
The binding of the viral major glycoprotein BLLF1 (gp350/220) to the CD21 cellular receptor is thought to play an essential role during infection of B lymphocytes by the Epstein-Barr virus (EBV). However, since CD21-negative cells have been reported to be infectible with EBV, additional interactions between viral and cellular molecules seem to be probable. Based on a recombinant genomic EBV plasmid, we deleted the gene that encodes the viral glycoprotein BLLF1. We tested the ability of the viral mutant to infect different lymphoid and epithelial cell lines. Primary human B cells, lymphoid cell lines, and nearly all of the epithelial cell lines that are susceptible to wild-type EBV infection could also be successfully infected with the viral mutant in vitro, although the efficiency of infection with BLLF1-negative virus was clearly lower than the one observed with wild-type EBV. Our studies show that the interaction between BLLF1 and CD21 is not absolutely required for the infection of lymphocytes and epithelial cells, indicating that viral molecules other than BLLF1 can mediate the binding of EBV to its target cells. In this context, our results further suggest the hypothesis that additional cellular molecules, apart from CD21, allow virus entry into these cells.
Herpesviruses carry large genomes that code for more than 200 proteins. The complexity of the genomes allows these viruses to adopt different protein expression profiles, e.g., lytic replication versus latent infection (reviewed in reference 25). Herpesviruses encode several glycoproteins that allow binding to different cell types. This phenomenon has been well studied in the case of herpes simplex virus (HSV), for which four different viral receptors have been identified to date (12, 28, 40). These multiple viral receptors may explain the ability of HSV type 1 (HSV-1) to infect cells of various lineages, including epithelial cells and neuronal cells. Other members of the herpesvirus family show a much more restricted cell tropism. The Epstein-Barr virus (EBV), for example, efficiently infects and immortalizes human primary B lymphocytes, but its ability to infect other cells, in particular those of epithelial origin, is quite limited, at least in vitro (25). Infection of B lymphocytes has been shown to be mediated by binding of the N-terminal region of the BLLF1 viral glycoprotein to its receptor, CD21, which also serves as the lymphocyte receptor for the C3d molecule, a member of the complement cascade (11, 29–31). The BLLF1 viral late glycoprotein, also termed gp350/220, is the most abundantly expressed glycoprotein in the viral envelope and the major antigen responsible for stimulating the production of neutralizing antibodies in vivo (46). Not only does the BLLF1 glycoprotein mediate EBV adsorption to CD21, but binding also induces capping of the receptor and endocytosis of the virus into B lymphocytes (43).
The EBV host range is not restricted to B lymphocytes, since the viral genome has been identified in a number of human carcinomas, ranging from nasopharynx carcinoma to gastric adenocarcinomas (25). However, studies of the infection of epithelial cells by EBV have been limited because EBV does not readily infect epithelial cell lines in vitro. More recently, an increasing number of studies have reported on the successful infection of epithelial cells from established cell lines or of primary gastric cells (17, 48). It has been demonstrated that cocultivation of EBV-positive cell lines with various epithelial cell lines, including nasopharynx carcinoma cell lines and keratinocytes, proved to be particularly efficient in infecting target cells with EBV (5, 17).
The mechanism of entry in these cases is not well understood. Whether the CD21 molecule acts as a receptor for EBV on these epithelial cells has been debated. In particular, one group reported a low level of expression of CD21 in the 293 cell line, a cell line of epithelial origin that is easily infectible by EBV, whereas others could not detect any expression of CD21 in these cells (10, 17). According to these reports, it has been proposed that EBV may infect these cells via a CD21-dependent as well as a CD21-independent pathway, suggesting the existence of a second cellular receptor. Such a model indirectly implies that either BLLF1 is able to bind to two different cellular receptors or a viral molecule other than BLLF1 is involved in binding of EBV to its target cells.
To distinguish between these hypotheses, we have constructed an EBV viral mutant lacking the BLLF1 gene and investigated the ability of this mutant to infect various cell lines and primary cells, including known EBV target cells. Our data show that the BLLF1 gene product is dispensable for infection of known EBV target cells and various epithelial cells, suggesting the existence of a different viral ligand that might interact with a cellular receptor besides CD21 on these cells.
MATERIALS AND METHODS
Cells.
The 293 cell line used in this study is a human embryonic epithelial kidney cell line that has been transformed by the introduction of the E1a and E1b genes of the adenovirus type 5 DNA (13). Raji is a human EBV-positive Burkitt's cell line (35). Raji 2.2.5 is an immunoselected mutant of Raji cells that is negative for HLA class II antigen expression due to a deletion in the CIITA gene (1, 20, 42). B lymphocytes from peripheral blood buffy coats were purified on a Ficoll cushion after T-cell rosetting by using sheep erythrocytes as described previously (50). HaCat is an immortalized human keratinocyte cell line that bears mutated alleles of p53 (2, 22), HEp-2 is a human laryngeal carcinoma cell line (ATCC), and HeLa is a human cervix adenocarcinoma cell line (ATCC). Vero is an African green monkey kidney cell line (36), and BHK cells are baby hamster kidney cells (18). All cell lines, with the exception of HeLa cells, were grown in RPMI 1640 medium supplemented with 10% fetal calf serum. HeLa cells were grown in Dulbecco modified Eagle medium–25 mM HEPES supplemented with 10% fetal calf serum (FCS).
Recombinant EBV plasmid.
To generate a BLLF1-negative mutant, the recBCD Escherichia coli strain BJ5183 (16) was transformed with the EBV genome, cloned onto a mini-F factor replicon. This EBV plasmid, p2089, carries the F factor origin of replication, the chloramphenicol resistance gene, the gene for the green fluorescent protein (GFP) under the control of the cytomegalovirus (CMV) promoter, and the hygromycin resistance gene as a selectable marker in eukaryotic cells (8). To introduce a functional deletion of BLLF1, the subclone p58.20 containing the ClaI H fragment from EBV strain B95.8, which encompasses EBV coordinates 88247 to 93260, was used. The XhoI-Bsu36I fragment containing parts of the BLLF1 gene was replaced by the tetracycline resistance gene from the pCP16 plasmid (6) to yield p2192. This cloning step resulted in the deletion of amino acids 79 to 630 of the BLLF1 protein (EBV coordinates 90263 to 91916) and a frameshift of the remaining coding sequences by the insertion of the tetracycline resistance gene. The plasmid p2192 was linearized with AvrII to generate a 6,059-bp fragment consisting of the modified BLLF1 gene flanked by EBV sequences of approximately 1 kb in size. The linearized fragment was transformed into BJ5183 bacteria carrying the whole EBV genome cloned in p2089 to induce homologous recombination via the EBV flanking regions. After double selection with chloramphenicol (final concentration, 15 μg/ml of Luria broth [LB] agar) and tetracycline (final concentration, 10 μg/ml of LB agar), DNA from resistant colonies was prepared and analyzed using various restriction enzymes to confirm correct recombination. Plasmid DNA from positive clones was then electroporated into the recA E. coli strain DH10B (Gibco BRL) for further propagation of the mutant EBV plasmid.
Transfection of cells and stable cell clone selection with hygromycin.
Transfection of cell lines with plasmid DNA was performed using lipid micelles (Lipofectamine; Gibco BRL). The day before transfection, cells were seeded into six-well cluster plates. For transfection, cells were placed in Optimem minimal medium for 2 h and incubated with DNA embedded in lipid micelles for 4 h. For selection of stable cell clones carrying EBV plasmid DNA, cells from one well were transferred to a tissue culture dish, 140 mm in diameter, 1 day posttransfection, and hygromycin was added to the culture medium (100 μg/ml). Three to four weeks later, single outgrowing clones were harvested and expanded. The cell lines were called 293-BLLF1-KO cells.
Southern blot analysis.
DNA extraction from cells, DNA digestion, and hybridization were performed as described previously (8).
Plasmid rescue in E. coli.
Circular EBV plasmid DNA was extracted from 293-BLLF1-KO cells as described previously (14). Extracted DNA was transformed into the E. coli DH10B strain by electroporation (1800 V; 25 μF; 100 Ω). Transformed bacterial clones were selected with medium containing chloramphenicol and tetracycline as described above.
Production of infectious EBV particles.
293-BLLF1-KO cells carrying the BLLF1-negative EBV genome and 293-B95.8/F cells harboring the wild-type EBV plasmid (8) were transfected in six-well cluster plates with an expression plasmid (0.5 μg/well) encoding the BZLF1 gene product that can induce the EBV lytic phase (15). In order to complement the BLLF1-negative mutants, cells were cotransfected with 0.5 μg of the BZLF1 expression plasmid and 1 μg of an expression plasmid derived from pcDNA3.1(+) (Invitrogen) that expresses BLLF1 under the control of a CMV promoter (p2385). At 72 h postinfection, viruses were harvested and filtered through a 1.2-μm-pore-size filter. Virus concentration was achieved by ultracentrifugation at 20,000 × g for 2 h in a fixed-angle rotor. Virus pellets were carefully resuspended in medium, again filtered, and used for infection.
Immunostaining procedures.
Induced cells were washed with phosphate-buffered saline (PBS) and fixed on a glass slide for 15 min with pure acetone. Cells were incubated for 30 min at 37°C with a mouse monoclonal antibody directed either against gp125, a member of the viral capsid antigen complex (Chemicon) (dilution, 1:1,000 in PBS–2% FCS), against BLLF1 (72A1, a supernatant from a hybridoma cell line obtained from the American Type Culture Collection), or gp85 (monoclonal antibody E1D1, kindly provided by L. M. Hutt-Fletcher). After several washes with PBS, cells were incubated with a secondary goat anti-mouse IgG antibody conjugated with the Cy-3 fluorochrome (dilution, 1:100; Dianova). After repeated washings with PBS, cells were embedded in a 10% PBS–glycerol solution, and immunostainings were analyzed by using an Axiovert inverted epifluorescence microscope (Zeiss).
Infection of cells with virus supernatant.
Cell lines were infected with 1 ml of concentrated supernatants containing either BLLF1 mutant virus, BLLF1 mutant virus complemented with the BLLF1 expression plasmid, or wild-type EBV. Target cells were incubated in 24-well cluster plates for 3 days before GFP expression was evaluated by UV microscopy and fluorescence-activated cell sorter (FACS) analysis. The culture medium was supplemented with tetradecanoyl phorbol acetate (TPA) (final concentration, 20 ng/ml) and butyrate (final concentration, 3 mM) at day 2 postinfection to enhance expression of the GFP gene. Primary B lymphocytes (4 × 106) isolated from peripheral blood buffy coats were infected with 1 ml of filtered supernatant and incubated overnight. In combination with WI38 fibroblasts that act as a feeder layer, B cells (1.5 × 105/well) were then plated into 96-well cluster plates and fed once a week with fresh medium. Outgrowing clones were expanded with medium supplemented with 20% FCS.
Inhibition of EBV infection.
The ability of the monoclonal antibody FE8 (34) to inhibit EBV infection was tested using Raji cells, 293 cells, and the 293-TB subclone. Cells (105 of each cell line) were incubated with increasing amounts of mFE8 (0, 0.56, 2.8, and 5.6 μg/ml) for 20 min at 37°C. Afterwards, 0.5 ml of supernatant containing either BLLF1-negative EBV or mutant EBV complemented with BLLF1 was added to the cells and incubated overnight. Supernatant from mock-transfected 293-BLLF1-KO cells was used as a negative control. The next day, supernatant and antibody were removed, and the cells were supplied with fresh medium. After an additional 24 h, GFP-positive cells were evaluated by FACS analysis and UV microscopy.
FACS analysis.
Cells were harvested 3 days after infection, washed twice with PBS–2% FCS solution, and analyzed for GFP fluorescence by flow cytometry using a FACScan (Coulter). Mock-transfected 293-B95.8/F cells (carrying the wild-type EBV plasmid) were used as a negative control. For detection of CD21 surface expression, 3 × 105 cells were harvested, washed with PBS–2% FCS, and incubated for 30 min on ice with different monoclonal antibodies directed against human CD21 (HB5) (ATCC) and FE8 (34); antibodies were used at a concentration of approximately 20 μg/ml). After two washings with PBS-2% FCS, cells were incubated for 30 min with a goat anti-mouse IgG antibody conjugated with the Cy-3 fluorochrome. Cells that were incubated with the second antibody only provided a negative control. After repeated washings with PBS, CD21 expression was evaluated by flow cytometry using a FACScan (Becton Dickinson).
Electron microscopy.
Thin-section preparation of 293-BLLF1-KO cells that had been induced with the BZLF1 expression plasmid was performed as previously described (32). Sections were evaluated at 80 kV using a Zeiss 902 electron microscope.
RT-PCR.
Total RNA was extracted from 106 Raji, HeLa, 293, and 293-TB cells by using the High Pure RNA Isolation Kit (Boehringer Mannheim) according to the manufacturer's protocol. The integrity of isolated RNA was examined by loading 1 μg of RNA onto a 1% agarose-formaldehyde gel. Afterwards, cDNA synthesis was performed with 1 μg of isolated RNA. Oligo(dT)12–18 (100 pmol) was added to each RNA sample, and the mixture was heated to 65°C for 10 min followed by quick chilling on ice. First strand buffer (5×; Gibco BRL), dithiothreitol (final concentration, 10 mM), and dNTP mix (final concentration, 0.5 mM) were added and incubated at 42°C for 2 min. Superscript II reverse transcriptase (200 U) was added, and the mix was incubated for an additional 1 h. The reverse transcription (RT) reaction was stopped by heating to 95°C for 5 min. Subsequently, each PCR was carried out with a 1/10 volume of the RT reaction corresponding to cDNA synthesized from 100 ng of total RNA. The sense primer 5′ GTTGTTCAGGTACCTTCCGC 3′ and the antisense primer 5′ TAGGAAGTGCTGGACACTCG 3′, used to detect CD21-specific transcripts, are described in reference 48 and gave rise to PCR products of 327 bp in size. For each PCR, 0.2 mM dNTPs, 1.5 mM MgCl2, 2 pmol of each primer, 10× buffer, and 2 U of Gold Star DNA polymerase (Eurogentec) were mixed with cDNA in a total volume of 50 μl. The mixture was subjected to 35 cycles of amplification, each cycle consisting of 95°C for 45 s, 55°C for 45 s, and 72°C for 1 min. The PCR products were electrophoresed using a 1% agarose gel.
RESULTS
Construction of a BLLF1-negative EBV mutant strain.
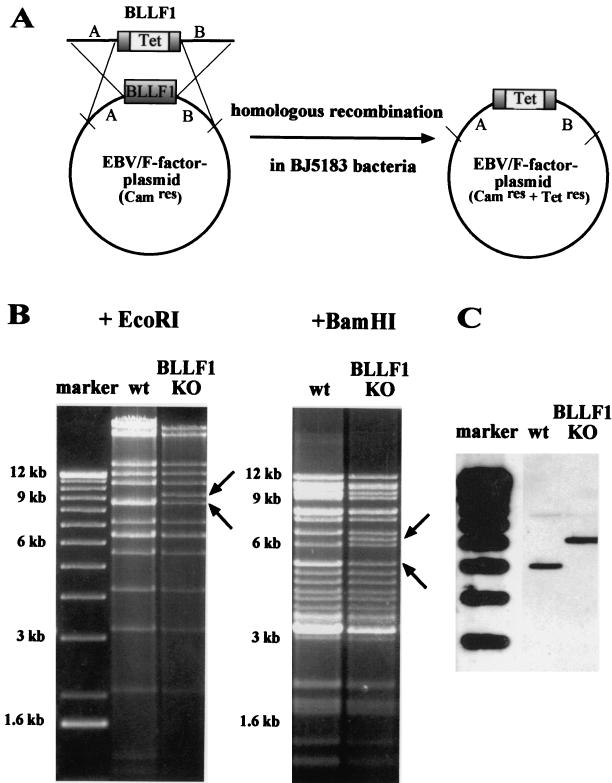
The complete EBV genome has been recently cloned in E. coli with the aid of an F plasmid (8). It also carries the genes for hygromycin resistance in eukaryotic cells and chloramphenicol resistance in E. coli and the gene encoding the enhanced GFP. To construct the BLLF1 knockout mutant, a linear DNA fragment carrying the BLLF1 gene which had been disrupted by insertion of the tetracycline resistance gene was introduced into the wild-type EBV plasmid by homologous recombination in the recA-positive, recBC-negative E. coli strain BJ5183 (Fig. 1A). After selection for chloramphenicol and tetracycline resistance, the EBV plasmid DNA from single colonies was analyzed with several restriction enzymes (data not shown). The BLLF1-negative viral DNA was transformed into the recA E. coli strain DH10B via electroporation. A restriction enzyme analysis of the plasmid DNA from the BLLF1-negative mutant and of wild-type EBV DNA confirmed a correct fragment pattern of the EBV mutant genome (Fig. 1B).
FIG. 1.
Mutant EBV lacking its major glycoprotein BLLF1. (A) Construction of the BLLF1-negative EBV mutant genome by homologous recombination in the recBCD E. coli strain BJ5183. The diagram shows schematically the wild-type EBV genome carrying the BLLF1 gene and a linearized fragment in which most of the BLLF1 sequence was replaced by the gene for tetracycline resistance (Tet). Since this linearized fragment used for targeted allelic exchange also carries EBV sequences that flank the BLLF1 gene, homologous recombination with the wild-type EBV F factor plasmid can take place. After transformation of the BLLF1/Tet fragment into the bacterial strain BJ5183, clones carrying recombinant EBV plasmids were selected for chloramphenicol (Cam) and tetracycline resistance. (B) Restriction fragment analysis of EBV BLLF1-KO DNA in comparison with wild-type B95.8/F factor DNA (wt). BLLF1-negative EBV plasmid DNA was purified from a chloramphenicol- and tetracycline-resistant DH10B E. coli clone and digested with EcoRI or BamHI. The restriction pattern of BLLF1-KO mutant DNA was compared with the one obtained for wild-type EBV DNA. Modified DNA fragments are indicated by arrows. As expected from the predicted BamHI restriction pattern, the introduction of the tetracycline resistance gene into the BLLF1 gene leads to the appearance of an additional 6.2-kb BamHI fragment and the disappearance of one of the two 5-kb fragments that are seen after digestion of wild-type EBV DNA. Similarly, the EcoRI restriction pattern obtained after digestion of the BLLF1 mutant EBV DNA shows a predicted additional fragment of 9.3 kb that correlates with the disappearance of one of the two 8.5-kb fragments obtained when wild-type EBV DNA is analyzed. (C) Southern blot analysis of 293-BLLF1-KO cells harboring the BLLF1-negative EBV mutant strain compared to 293-B95.8/F cells carrying the wild-type EBV plasmid. Total genomic DNA from these cells was digested with the BamHI restriction enzyme. After agarose gel electrophoresis and Southern blotting, the DNA was hybridized with a probe derived from the BLLF1/Tet fragment that was used to introduce the BLLF1 KO mutation. The probe spanning nucleotide coordinates 88335 to 93167 of the B95.8 EBV sequence with a deletion of 90263 to 91916 (see Materials and Methods) identifies a characteristic 6.2-kb signal in DNA from 293-BLLF1-KO cells, compared to a 5-kb band observed in 293 cells carrying wild-type virus (see also panel B). The faint signals at 7.9 kb that are observed with BLLF1-negative virus and with wild-type virus represent remaining BLLF1 sequences that partially hybridize with the probe.
Establishment of a 293 cell clone that stably produces BLLF1-negative EBV virions.
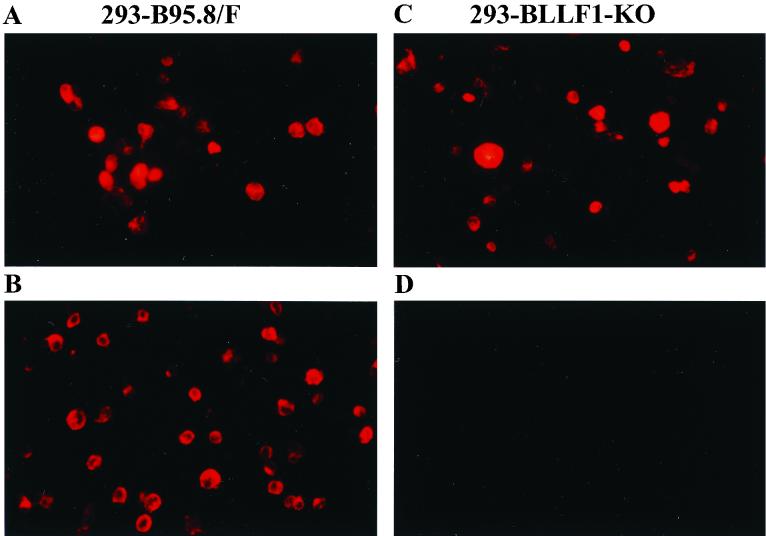
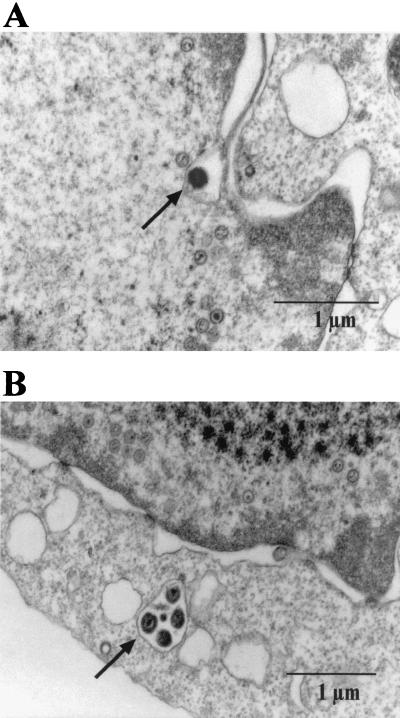
The production of virus stocks containing BLLF1-negative EBV requires the establishment of a permissive cell line that carries the mutant EBV plasmid. To this aim, the BLLF1-negative EBV genome was transfected into 293 cells, and stable hygromycin-resistant cell clones that carried the mutated EBV plasmid episomally were selected. Fig. 1C shows a Southern blot analysis of genomic DNA from one of the hygromycin-resistant clones, using as a probe the BLLF1/Tet fragment that had been introduced into the EBV genome (see also Fig. 1A). This cell clone was termed 293-BLLF1-KO and was used for all the following experiments. The 293-BLLF1-KO cells contain the correct BLLF1 knockout genome, as confirmed by a DNA rescue experiment. Plasmid DNA was extracted from these cells and transformed into the E. coli strain DH10B via electroporation. After selection for chloramphenicol and tetracycline resistance, DNA from outgrowing colonies was again examined by restriction enzyme analysis and proved to yield the same pattern as shown in Fig. 1B (data not shown). The cell clone was then tested for the ability to support the lytic cycle of EBV. To this aim, cells were transiently transfected with an expression plasmid encoding the immediate-early protein BZLF1. Three days after induction, immunostaining of the induced cells with an antibody directed against gp125, a lytic viral protein of the VCA complex, and with an antibody directed against another viral late gene product, the glycoprotein gp85, was performed. Approximately 16% of the 293-BLLF1-KO cells proved to be positive for the expression of the late gene products gp125 and gp85 (Fig. 2 and data not shown). Immunostaining of the induced 293-BLLF1-KO cells with the neutralizing monoclonal antibody 72A1, directed against the N terminus of the BLLF1 glycoprotein, did not yield any signal, as expected for a BLLF1 knockout mutant (Fig. 2). The maturation and egress of the BLLF1-KO viral particles were analyzed by electron microscopy using lytically induced 293-BLLF1-KO cells, as shown in Fig. 3. Viral capsids could be observed in the nucleus and in the cytoplasm after budding through the nucleus membrane.
FIG. 2.
Lytically induced 293-BLLF1-KO cells do not show any expression of the BLLF1 gene product. 293-B95.8/F cells carrying wild-type EBV DNA as a positive control and 293-BLLF1-KO cells carrying BLLF1-negative DNA were induced with BZLF1, harvested 3 days after induction, and fixed in pure acetone. Expression of gp125 and BLLF1 was detected by immunostaining using a monoclonal antibody directed either against gp125 or against the BLLF1 protein. In a second step, staining was visualized by incubating the cells with a goat anti-mouse IgG antibody conjugated with the Cy-3 fluorochrome. 293-B95.8/F cells show positive staining for gp125 (A) and BLLF1 (B), whereas induced 293-BLLF1-KO cells only show expression of gp125 (C) and are negative for the BLLF1 protein (D).
FIG. 3.
Maturation of EBV virions is not dependent on BLLF1 expression. 293-BLLF1-KO cells were induced by transient transfection of BZLF1 and harvested 3 days after induction for preparation of thin sections. Electron microscopy analysis of these cell sections show different stages of EBV maturation. (A) A viral capsid budding from the nucleus membrane in a 293-BLLF1-KO cell is shown. (B) Viral capsids that are transported to the plasma membrane are shown. Viral capsids are indicated by arrows.
The BLLF1-negative EBV mutant is still able to infect and immortalize primary B lymphocytes.
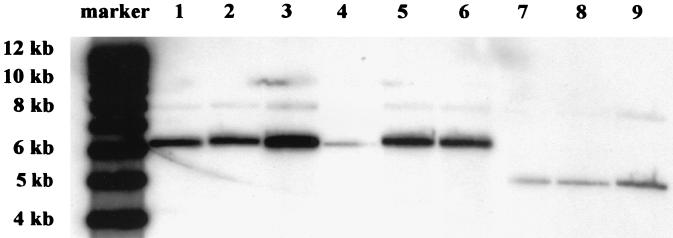
Previous work has shown that preincubation of primary B lymphocytes with BLLF1 peptides encompassing the binding motif for the cellular CD21 molecule, prior to incubation with EBV, abolished the outgrowth of EBV-transformed B cells (29). To address the question of whether primary B lymphocytes can be infected and immortalized with the BLLF1-negative EBV mutant, we performed infection assays with human primary B lymphocytes as target cells. The cells were incubated with supernatants containing either wild-type EBV, BLLF1-negative EBV, or mutant EBV complemented with BLLF1 in trans. The complementation of the BLLF1 mutant phenotype was achieved by transient cotransfection of 293-BLLF1-KO cells with BZLF1 and an expression plasmid encoding BLLF1 under the control of the CMV promoter. The expression of the BLLF1 protein was confirmed by immunostaining as described above (data not shown). As shown in Table 1, we could successfully establish EBV-positive immortalized B lymphocytes using supernatants containing BLLF1-negative EBV, although the frequency of immortalization was reduced compared to immortalization by wild-type EBV. Southern blot analysis of genomic DNA extracted from three independent clones, using cell clones derived from wild-type EBV infection as a positive control, confirmed the BLLF1-negative status of the immortalized B-cell clones (Fig. 4).
TABLE 1.
Transforming frequencies of primary B lymphocytes after infection with EBV supernatants
| Virus supernatant | No. of positive wells/wells plated | Immortalization efficiency (%) |
|---|---|---|
| Wild-type EBV | 27/27 | 100 |
| BLLF1-negative EBV | 4/27 | 15 |
| BLLF1-complemented EBV | 27/27 | 100 |
FIG. 4.
Southern blot analysis of total DNA from immortalized B-cell clones that were established after infection with either BLLF1-KO EBV (lanes 1 to 3), complemented BLLF1-KO EBV (lanes 4 to 6), or wild-type EBV (lanes 7 to 9). Ten micrograms of extracted DNA was digested with BamHI, separated by gel electrophoresis, and blotted as described previously (6). Hybridization was performed with the probe consisting of the BLLF1/Tet fragment that was used for homologous recombination as described in the legend to Fig. 1C. As expected, hybridization with DNA from B-cell clones derived after infection with BLLF1-negative EBV or transiently complemented mutant EBV showed a 6.2-kb signal corresponding to the fragment that carries the mutated BLLF1 gene (lanes 1 to 6). Weaker signals of 7.9 kb represent remaining flanking BLLF1 sequences that are partially recognized by the probe. B-cell lines established with virus stocks derived from the 293-B95.8/F cells (lanes 7 to 9) showed the predicted 5-kb band representing wild-type BLLF1 sequences.
BLLF1-negative EBV infects Raji cells at a lower frequency.
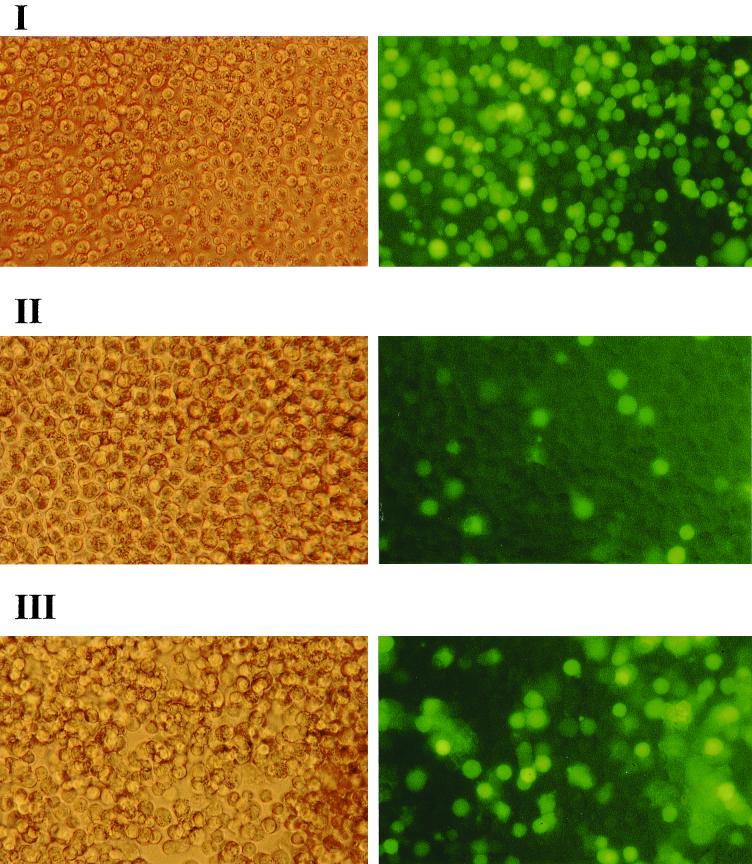
Our experiments with primary B cells suggested that BLLF1 is dispensable for the infection of human B cells. In order to extend this observation, we performed infection experiments with the various supernatants (see above) using Raji cells, a Burkitt lymphoma cell line, as target cells. GFP-positive Raji cells could be observed after incubation with the supernatant from induced 293-BLLF1-KO cells, as shown in Fig. 5. FACS analysis revealed an efficiency of infection that was a factor of 10 to 12 lower with BLLF1-negative virus stocks than the one obtained with wild-type EBV or with mutant viruses that were complemented with BLLF1. The observation that Raji cells can also be successfully infected with BLLF1-negative EBV further demonstrates that the deletion of BLLF1 reduces but does not abrogate virus entry into B cells. Therefore, an alternative pathway, independent of BLLF1, most likely mediates virus entry into these cells.
FIG. 5.
Infection of Raji cells by various EBV stocks, including BLLF1-negative EBV, as detected by GFP expression. Raji cells (105) were incubated with 1 ml of supernatant containing either wild-type EBV (I), mutant EBV lacking BLLF1 (II), or mutant EBV complemented with a BLLF1 expression plasmid (III). Successful infection was detected after 72 h by GFP expression using an Axiovert inverted epifluorescence microscope, as shown in the right panels. The left panels show the corresponding cells via phase-contrast light microscopy (magnification, ×200).
It has been suggested that HLA class II molecules act as a coreceptor for infection of B cells by EBV (23). We wanted to know whether these molecules participate in mediating infection of Raji cells with the BLLF1-negative EBV mutant. To this aim, we used the Raji 2.2.5 cell line that does not show any detectable expression of class II antigens (1, 20). These cells were incubated with supernatants containing either BLLF1-negative EBV virions, BLLF1-complemented mutant EBV, or wild-type EBV as a control. All three supernatants proved to be infectious, as indicated by GFP expression (Table 2). The efficiency of infection, dependent on the supernatants used, decreased by a factor of about 5 to 9 compared to the rates of infection observed with common Raji cells (Table 2), indicating that HLA class II molecules are not absolutely required for successful infection of Raji cells even in the absence of the BLLF1 glycoprotein.
TABLE 2.
Susceptibility of cell lines to infection with wild-type EBV and BLLF1-negative EBVa
| Cell line | % Positive for wild-type EBV | % Positive for BLLF1-negative EBV | % Positive for BLLF1-complemented EBV | Ratio (complemented virus/BLLF1-negative virus) |
|---|---|---|---|---|
| Raji | 77.0 | 6.2 | 60.0 | 9.7 |
| Raji 2.2.5 | 11.2 | 0.7 | 12.9 | 18.4 |
| 293 | 6.2 | 1.8 | 4.5 | 2.5 |
| 293-TB | 18.3 | 5.6 | 15.0 | 2.7 |
| HaCat | 0.0 | 0.0 | 0.0 | |
| HeLa | 0.0 | 0.0 | 0.0 | |
| HEp-2 | 10−3 | 0.0 | 10−3 | |
| Vero | 0.1 | 0.12 | 0.11 | 0.92 |
| BHK | 0.0 | 0.0 | 0.0 |
The percentages of GFP-positive cells, observed after incubation with various viral supernatants, are indicated. The numbers given here represent a mean value obtained from three independent experiments of infection.
The data of these experiments listed in Table 2 most likely do not represent definitive numbers, since it is not possible to determine the virus titer of the BLLF1-KO mutant by standard infection assays. Although 293-BLLF1-KO cells and BLLF1-complemented 293-BLLF1-KO cells exhibit approximately the same efficiency of induction of the lytic cycle, as observed by immunostaining assays (data not shown), we cannot exclude completely the possibility that BLLF1 plays a role in virus assembly and egress. Nevertheless, our experiments demonstrate that virus entry into B cells takes place even in the absence of the major EBV attachment glycoprotein.
BLLF1 is dispensable for the infection of various epithelial cells.
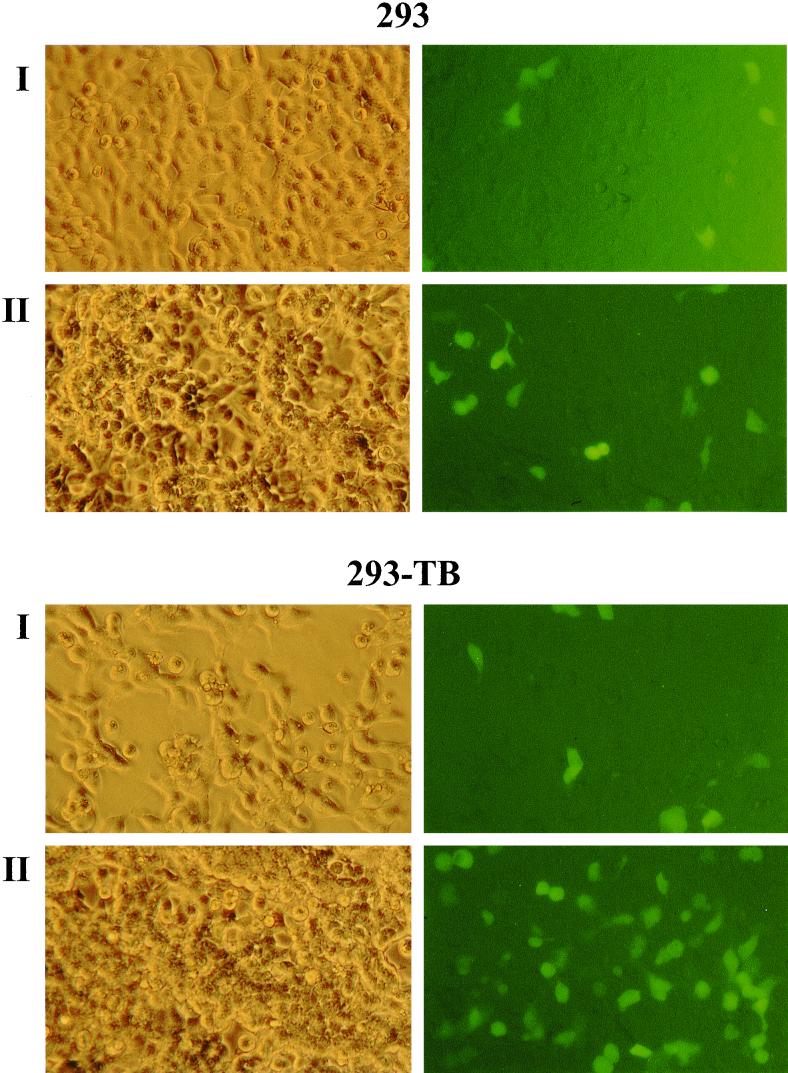
Since it has been reported that epithelial cells can be infected with EBV, we wanted to investigate whether the infection of epithelial cells depends on the expression of BLLF1. Supernatants from induced 293-BLLF1-KO cells were used to infect 293 cells, which have been shown to be infectible by EBV in vitro (10, 17). Our 293 cell line proved to be rather heterogeneous with regard to the efficiency of EBV infection (data not shown). Therefore, four rounds of single-cell selection were performed under limiting dilution conditions, and 30 clones of each round were tested for the ability to become infected with EBV. One of these 293 subclones, termed 293-TB, proved to be about three times more susceptible to EBV infection than the parental 293 population (see Table 2). The 293 cells and the 293-TB cells were infected with the BLLF1-KO mutant virus. Figure 6 shows GFP-positive 293 cells 3 days after incubation of the cells with supernatants containing the BLLF1 mutant virus. The results presented here show that both the parental 293 cells and the 293-TB cells can be infected with the BLLF1-negative EBV mutant. In the presence of BLLF1, the rate of infection increased up to threefold, as listed in Table 2 (see also Fig. 6). Infection with wild-type EBV revealed numbers of GFP-positive cells comparable to those observed with BLLF1-complemented mutant EBV (Table 2).
FIG. 6.
Infection of 293 cells and of 293-TB cells with viral supernatants containing either BLLF1-negative EBV (I) or mutant EBV complemented with a BLLF1 expression plasmid (II). GFP expression was detected 3 days after incubation of the cells with viral supernatants, as shown in the right panels for each cell type.
To investigate the relevance of this observation made with 293 cells, we further analyzed different cell lines for characteristics of their susceptibilities to infection by EBV. As described above, HaCat cells, a human keratinocyte cell line, the human laryngeal carcinoma cell line HEp-2, HeLa cells, and the nonhuman cell lines Vero and BHK were infected with supernatants containing either wild-type EBV, mutant EBV negative for BLLF1, or mutant EBV complemented with BLLF1. Vero cells, which are known to be readily infectible with HSV-1 (36), could be infected with all three virus stocks in a comparable manner (Table 2). Infection of HEp-2 cells with EBV was inefficient and could be observed only with wild-type EBV. In the case of the cell lines HaCat, HeLa, and BHK, no GFP-positive cells could be detected, suggesting that these cells are resistant to wild-type EBV infection as well as to infection with BLLF1-negative EBV.
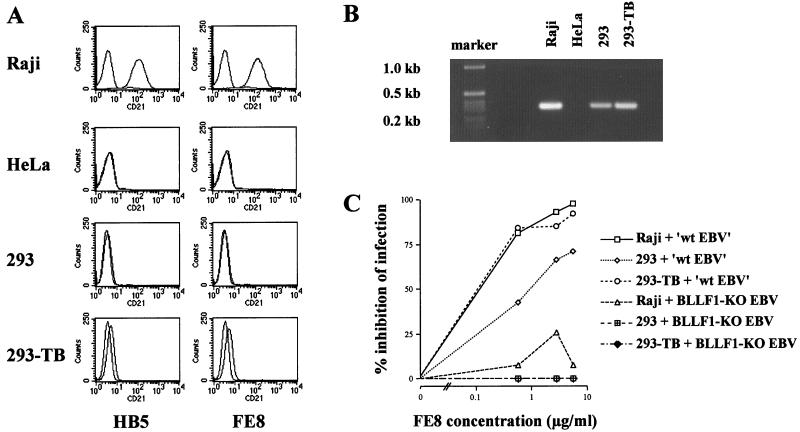
Due to contradictory reports about the expression of CD21 on epithelial cells (10, 17), both the parental 293 cells and the particular 293-TB subclone were analyzed for CD21 surface expression (Fig. 7A). Using different monoclonal antibodies directed against CD21 (see Materials and Methods), we could observe only a minimal shift for 293-TB cells, whereas 293 cells did not show any CD21 surface expression by FACS analysis. In addition, we performed RT-PCR using primers specific for the short consensus repeats (SCR) 1 and 2 of CD21 (48), which had been shown to be necessary for binding of EBV (4). cDNA from Raji cells was used as a positive control, and cDNA from HeLa cells, which had been shown to be resistant to EBV infection, was used as a negative control. As shown in Fig. 7B, positive signals could be observed for Raji cells, 293 cells, and 293-TB cells. HeLa cells proved to be negative for the detection of CD21-specific mRNA transcripts. Since FACS analysis might not be sensitive enough to detect minute amounts of CD21 molecules on the cell surface, we performed an infection inhibition experiment using the monoclonal antibody FE8, which had been demonstrated to block EBV infection of B cells as efficiently as the OKB7 monoclonal antibody (31, 34). Raji cells, 293 cells, and 293-TB cells were preincubated with increasing concentrations of FE8 and subsequently incubated with supernatants containing either BLLF1-negative EBV or mutant EBV complemented with BLLF1. As a control, Raji cells were preincubated in parallel with the HB5 antibody, which also detects CD21 but does not block EBV entry (45). Two days postinfection, cells were harvested and GFP-positive cells were evaluated by FACS analysis and UV microscopy. UV microscopy was preferentially used with virus stocks containing BLLF1-negative EBV due to low infection efficiencies in the course of this experiment. The results of the blocking experiments are illustrated in Fig. 7C. As expected, infection of Raji cells with BLLF1-complemented mutant EBV can be strongly inhibited by the FE8 antibody, whereas blocking of CD21 shows no or little influence on the infection by EBV lacking the BLLF1 glycoprotein. Pretreatment of Raji cells with the control antibody HB5 led to a decrease of about 28% in the rate of infection with complemented mutant virus, whereas no inhibition of BLLF1-KO EBV infection was observed (data not shown). Results of the control experiment using the HB5 antibody suggest that variations in the range observed after BLLF1-KO EBV infection of Raji cells might be considered not significant and probably reflect unspecific hindrances. Taking this observation into account, our results indicated that the infection of Raji cells with BLLF1-negative EBV was independent of the interaction between BLLF1 and CD21. This assumption also seems to apply to 293 and 293-TB cells, since we did not observe a difference in infection efficiency with our BLLF1 mutant strain after application of the FE8 antibody. However, using BLLF1-complemented EBV, infection of the original 293 cells as well as the 293-TB cells can also be inhibited, as observed in the case of Raji cells (Fig. 7C). This indicates that minute amounts of CD21 on the surfaces of the different 293 cells might mediate virus entry via interaction with BLLF1.
FIG. 7.
Expression of CD21 in Raji cells and different epithelial cell lines. (A) FACS analysis of Raji cells, HeLa cells, 293 cells, and 293-TB cells for human CD21 surface expression. Cells (3 × 105 for each cell line) were incubated with two anti-CD21 monoclonal antibodies (HB5 and FE8) and subsequently incubated with a Cy3-conjugated goat anti-mouse IgG antibody. As negative controls, each cell sample was incubated with the second antibody only. The results shown are representative of two independent experiments. (B) RT-PCR analysis. cDNA (100 ng) was used to amplify CD21-specific mRNA transcripts in Raji cells, HeLa cells, 293 cells, and 293-TB cells. (C) Infection inhibition experiment using the monoclonal antibody FE8. Cells (105) were incubated with increasing amounts of the antibody for 20 min at 37°C. As indicated, 0.5 ml of supernatant containing either complemented mutant EBV (designated ‘wt EBV’) or BLLF1-negative EBV was added and incubated for 24 h. Afterwards, supernatant and antibody were removed, and fresh medium was added to the cells. After an additional 24 h, cells were evaluated for GFP expression. As controls, Raji cells were incubated with increasing amounts of the HB5 antibody, as described for FE8, prior to infection with BLLF1-KO EBV or complemented BLLF1 mutant virus (data not shown).
Other epithelial cell lines used in this series of experiments were negative for CD21 surface expression by FACS analysis. Vero cells that are susceptible to EBV infection did not show any CD21 expression. Since the rates of infection of Vero cells listed in Table 2 suggest that BLLF1 plays a minor role, if any, in the mode of virus entry into this cell line, the CD21 status of these cells was not further investigated by other means.
DISCUSSION
EBV codes for an envelope glycoprotein, BLLF1, which is known to play an important role in infection of B cells by binding to the CD21 protein. EBV is also able to infect epithelial cells in vitro (39) and in vivo. The first evidence for EBV infecting an epithelial target in vivo was the finding that the viral genome is regularly present in nasopharyngeal carcinomas (25, 51). Many other examples of tumors of epithelial origin, also apart from the nasopharynx, were found to be EBV positive, e.g., salivary gland carcinomas (37) and gastric tumors of the more common adenocarcinoma type (25, 38). However, the route whereby EBV enters epithelial target cells in vivo has not been elucidated to date. It has been reported that some human epithelial cell lines express low levels of CD21 in vitro, but in nasopharyngeal carcinoma cells, CD21 is not detectable (25). Therefore, the question of whether an additional EBV receptor exists on epithelial cells has been addressed by several groups (10, 48, 49). The existence of a second EBV receptor might also point to an additional viral molecule, apart from BLLF1, that interacts with this receptor.
To further investigate the mode of EBV entry into its target cells, we have constructed a recombinant EBV strain lacking a functional BLLF1 gene. This mutant virus strain was not impaired with regard to the expression of late genes, such as those for gp125 and gp85, after induction of the lytic cycle. Maturation of virus proved to be normal as analyzed by electron microscopy, strongly suggesting that formation of viral particles is not dependent on the expression of BLLF1. Complementation experiments with a BLLF1 expression plasmid gave rise to fully infectious particles, demonstrating that the modifications introduced into the viral genome were confined to the BLLF1 gene. The host range of the BLLF1 mutant viruses was evaluated by using different cell lines as target cells. Supernatants containing BLLF1-negative EBV were incubated with human primary B lymphocytes, Raji cells, and various epithelial cell lines.
We show here that cell lines that can become infected with wild-type EBV are also susceptible to infection with BLLF1-negative EBV (Table 2), except for the HEp-2 cell line, in which infection could be observed with wild-type EBV only. Since the rate of infection of HEp-2 cells with wild-type virus was extremely low, no conclusion can be drawn with regard to the susceptibility of this cell line to infection with BLLF1-negative EBV. Although the efficiency of infection was less with BLLF1-negative EBV, primary B lymphocytes and Raji cells could be successfully infected. In addition, the HLA class II-negative Raji 2.2.5 cell line could be infected with the BLLF1 mutant virus strain. However, the rate of infection decreased with both wild-type and BLLF1-negative EBV compared to that for normal Raji cells.
The results of the experiments using primary B lymphocytes and Raji cells are contradictory to previous reports, since binding of BLLF1 to CD21 on B cells was thought to be a prerequisite for virus entry. By preincubating B cells with multimeric BLLF1-derived peptides, Nemerow et al. showed that inhibition of the interaction between BLLF1 and CD21 abolishes the ability of EBV to infect and transform B cells (29). In addition, Tanner et al. observed blocking of EBV adsorption by using either soluble BLLF1 or a derivative of the BLLF1 protein in which the amino terminus was deleted (44). However, preincubation of B cells with multimeric BLLF1 peptides or soluble BLLF1 protein might lead to unspecific steric hindrances that impede access of other viral ligands to the target cell. This hypothesis could provide an explanation for the different conclusions made by these groups and us. Our results clearly show that BLLF1 is dispensable for infection and immortalization of B lymphocytes in vitro as well as for infection of lymphoid cells like Raji cells and the HLA class-II negative Raji 2.2.5 subclone. These data suggest the existence of an additional EBV molecule which is able to partially complement the BLLF1-negative phenotype and mediates binding of EBV to its target cells. From the CD21 blocking experiments with Raji cells, we can further conclude that entry of EBV lacking its major glycoprotein most likely involves an additional receptor besides CD21 on B cells.
Alternative binding mechanisms are known for other herpesviruses, such as the alphaherpesviruses HSV-1 and -2. The gC glycoprotein of HSV-1, e.g., is required for the initial attachment of the virus via cell surface heparan sulfate. A second glycoprotein, gB, also contributes, although to a lesser extent, to the binding of heparan sulfate. Mutant viruses lacking gC were more impaired in binding than mutants depleted of gB (21). Apart from these glycoproteins, HSV-1 encodes another viral envelope glycoprotein, gD. This glycoprotein interacts with additional human receptors, including HVEM or HveA, a member of the tumor necrosis factor receptor family (28), and HveC, the herpesvirus entry protein C, which shows homologies to the poliovirus receptor (12). Recognition of one of these receptors by gD is thought to trigger fusion of the viral envelope with the cell membrane. A related mode of infection is known from the study of the human cytomegalovirus, a betaherpesvirus. In this case, the gC-II glycoprotein complex was found to be the major component that possesses the ability to bind to immobilized heparin (19). In addition, the most abundant envelope glycoprotein, gB, is also able to interact with heparan sulfate (3, 7). Besides this initial interaction, gB associates with a different class of receptors, e.g., annexin II (33) or other yet-undefined nonheparin components (3).
There are several EBV envelope glycoproteins that are known to be important for mediating virus entry. The complex consisting of the glycoproteins gp85 and gp25, e.g., triggers fusion of the virus envelope with the cell membrane (26, 47). A third glycoprotein of this complex, gp42, has been reported to bind to HLA class II molecules (24, 41). These molecules have been proposed to function as a coreceptor required for EBV infection of B lymphocytes (23). However, our results, in agreement with those of Faggioni et al. (9), show that HLA class II molecules at least are not absolutely required for the infection of Raji cells. In addition, since the BLLF1-negative strain is still able to infect the HLA class II-negative Raji clone, it seems most likely that HLA class II molecules also do not play a crucial role in mediating the BLLF1-independent mode of infection. Nevertheless, a role of gp42 in this infectious pathway cannot be excluded so far. There are still a number of EBV glycoproteins, e.g., gN, gM, gp78, and gp150, with yet-unknown functions that might contribute to the pathway of EBV entry in the absence of BLLF1.
With regard to epithelial cells, we could show that supernatants containing BLLF1-negative EBV infect human 293 cells as well as the 293-TB subclone that had been selected for its higher susceptibility to EBV infection. As was already observed in the case of B lymphocytes, the rate of infection was lower in the absence of BLLF1. After complementation of the BLLF1-negative virus in trans, the rate of infection of 293 cells and the 293 subclone reached the level observed with wild-type EBV. These observations again refer to the existence of an additional EBV envelope protein that is able to partially compensate for the lack of BLLF1 function during the infection of human epithelial cells.
In the past, the expression of CD21 on epithelial cells and its role in EBV infection of these cells have been debated. It has been shown that some epithelial cell lines, such as 293 or primary epithelial cells that line the pharyngeal mucosa, especially in less-differentiated stages, express relatively low levels of CD21 (10, 49). Infection of these epithelial cells was believed to be mediated by binding of BLLF1 to CD21, as is known to be the case for B cells. In our hands, 293 cells were found to be negative for CD21 surface expression by FACS analysis. However, in contrast to the results of Imai et al. (17), we were able to detect CD21 mRNA by performing RT-PCR. This finding might suggest that the cell surface expression of CD21 could be regulated in a posttranscriptional way. The data from the infection inhibition experiment showed that blocking of CD21 led to a decrease in 293 and 293-TB cell infection by BLLF1-complemented mutant EBV. This observation rather supports the hypothesis that small amounts of CD21 molecules might mediate entry of wild-type EBV into this cell line. In the case of the 293-TB subclone, the higher susceptibility to EBV infection, compared to the parental 293 cell population, can be most likely explained by selection for the surface expression of CD21 molecules on these cells. On the other hand, infection of 293 cells and 293-TB cells with BLLF1-negative EBV could not be inhibited by blocking of the CD21 receptor. Therefore, we could demonstrate, as observed for Raji cells, that attachment of BLLF1-KO EBV also occurs via an additional receptor on epithelial cells. A very recent work by Molesworth et al. suggests a possible binding activity for gH in the mode of EBV entry into CD21-negative epithelial cells (27). Regarding our data on infection of 293 cells, this report might offer an attractive model for an interaction of a viral glycoprotein different from BLLF1 with a so-far-unknown receptor on epithelial cells. So far, no conclusions about the identity of such a viral ligand can be drawn from our results.
In summary, we could demonstrate that an Epstein-Barr virus strain that lacks its major attachment protein BLLF1 is able to infect human B lymphocytes and epithelial cells in vitro using an as yet undetermined viral ligand. Moreover, our data show that such an additional viral binding protein most likely interacts with a cellular receptor different from CD21 on B cells and on epithelial cells, such as 293 cells. These findings might prove relevant with regard to the elucidation of the pathway used during EBV infection in vivo.
ACKNOWLEDGMENTS
We thank Katja Dunckelmann and Bärbel Jungnickl for technical and photographic assistance with EM analysis. We also thank Elisabeth Kremmer for supplying monoclonal antibody 72AI and for help in determination of the FE8 antibody concentration. We further thank Wolfgang M. Prodinger for kindly supplying FE8 and HB5 antibodies and Lindsey Hutt-Fletcher for providing the monoclonal antibody E1D1. We thank Anna Meier and Olivier Gires for help with the FACS analysis.
This work was supported by Public Health Service grant CA70723 and grants Ha 1354/3 and SFB 455 from the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Accolla R S. Human B cell variants immunoselected against a single Ia antigen subset have lost expression of several Ia antigen subsets. J Exp Med. 1983;157:1053–1058. doi: 10.1084/jem.157.3.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boukamp P, Petrussevska R T, Breitkreutz D, Hornung J, Markham A, Fusenig N E. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J Cell Biol. 1988;106:761–771. doi: 10.1083/jcb.106.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyle K A, Compton T. Receptor-binding properties of a soluble form of human cytomegalovirus glycoprotein B. J Virol. 1998;72:1826–1833. doi: 10.1128/jvi.72.3.1826-1833.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carel J C, Myones B L, Frazier B, Holers V M. Structural requirements for C3d,g/Epstein-Barr virus receptor (CR2/CD21) ligand binding, internalization, and viral infection. J Biol Chem. 1990;265:12293–12299. [PubMed] [Google Scholar]
- 5.Chang Y, Tung C H, Huang Y T, Lu J, Chen J Y, Tsai C H. Requirement for cell-to-cell contact in Epstein-Barr virus infection of nasopharyngeal carcinoma cells and keratinocytes. J Virol. 1999;73:8857–8866. doi: 10.1128/jvi.73.10.8857-8866.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cherepanov P P, Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- 7.Compton T, Nowlin D M, Cooper N R. Initiation of human cytomegalovirus infection requires initial interaction with cell surface heparan sulfate. Virology. 1993;193:834–841. doi: 10.1006/viro.1993.1192. [DOI] [PubMed] [Google Scholar]
- 8.Delecluse H J, Hilsendegen T, Pich D, Zeidler R, Hammerschmidt W. Propagation and recovery of intact, infectious Epstein-Barr virus from prokaryotic to human cells. Proc Natl Acad Sci USA. 1998;95:8245–8250. doi: 10.1073/pnas.95.14.8245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Faggioni A, Zompetta C, Cirone M, Barile G, Frati L, Accolla R. Superinfection by Epstein-Barr virus of a subset of Raji cells is independent of HLA class-II antigens. Int J Cancer. 1990;45:989. doi: 10.1002/ijc.2910450536. [DOI] [PubMed] [Google Scholar]
- 10.Fingeroth J D, Diamond M E, Sage D R, Hayman J, Yates J L. CD21-dependent infection of an epithelial cell line, 293, by Epstein-Barr virus. J Virol. 1999;73:2115–2125. doi: 10.1128/jvi.73.3.2115-2125.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fingeroth J D, Weis J J, Tedder T F, Strominger J L, Biro P A, Fearon D T. Epstein-Barr virus receptor of human B lymphocytes is the C3d receptor CR2. Proc Natl Acad Sci USA. 1984;81:4510–4514. doi: 10.1073/pnas.81.14.4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geraghty R J, Krummenacher C, Cohen G H, Eisenberg R J, Spear P G. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science. 1998;280:1618–1620. doi: 10.1126/science.280.5369.1618. [DOI] [PubMed] [Google Scholar]
- 13.Graham F L, Smiley J, Russell W C, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 14.Griffin B E, Bjorck E, Bjursell G, Lindahl T. Sequence complexity of circular Epstein-Barr virus DNA in transformed cells. J Virol. 1981;40:11–19. doi: 10.1128/jvi.40.1.11-19.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hammerschmidt W, Sugden B. Identification and characterization of oriLyt, a lytic origin of DNA replication of Epstein-Barr virus. Cell. 1988;55:427–433. doi: 10.1016/0092-8674(88)90028-1. [DOI] [PubMed] [Google Scholar]
- 16.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 17.Imai S, Nishikawa J, Takada K. Cell-to-cell contact as an efficient mode of Epstein-Barr virus infection of diverse human epithelial cells. J Virol. 1998;72:4371–4378. doi: 10.1128/jvi.72.5.4371-4378.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jarrett O, Macpherson I. The basis of the tumorigenicity of BHK 21 cells. Int J Cancer. 1968;3:654–662. doi: 10.1002/ijc.2910030514. [DOI] [PubMed] [Google Scholar]
- 19.Kari B, Gehrz R. A human cytomegalovirus glycoprotein complex designated gC-II is a major heparin-binding component of the envelope. J Virol. 1992;66:1761–1764. doi: 10.1128/jvi.66.3.1761-1764.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koch W, Candeias S, Guardiola J, Accolla R, Benoist C, Mathis D. An enhancer factor defect in a mutant Burkitt lymphoma cell line. J Exp Med. 1988;167:1781–1790. doi: 10.1084/jem.167.6.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laquerre S, Argnani R, Anderson D B, Zucchini S, Manservigi R, Glorioso J C. Heparan sulfate proteoglycan binding by herpes simplex virus type 1 glycoproteins B and C, which differ in their contributions to virus attachment, penetration, and cell-to-cell spread. J Virol. 1998;72:6119–6130. doi: 10.1128/jvi.72.7.6119-6130.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lehman T A, Modali R, Boukamp P, Stanek J, Bennett W P, Welsh J A, Metcalf R A, Stampfer M R, Fusenig N, Rogan E M, et al. p53 mutations in human immortalized epithelial cell lines. Carcinogenesis. 1993;14:833–839. doi: 10.1093/carcin/14.5.833. [DOI] [PubMed] [Google Scholar]
- 23.Li Q, Spriggs M K, Kovats S, Turk S M, Comeau M R, Nepom B, Hutt-Fletcher L M. Epstein-Barr virus uses HLA class II as a cofactor for infection of B lymphocytes. J Virol. 1997;71:4657–4662. doi: 10.1128/jvi.71.6.4657-4662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Q, Turk S M, Hutt-Fletcher L M. The Epstein-Barr virus (EBV) BZLF2 gene product associates with the gH and gL homologs of EBV and carries an epitope critical to infection of B cells but not of epithelial cells. J Virol. 1995;69:3987–3994. doi: 10.1128/jvi.69.7.3987-3994.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller G. Epstein-Barr virus: biology, pathogenesis, and medical aspects. In: Fields B N, Knipe D M, editors. Virology. 2nd ed. Vol. 2. New York, N.Y: Raven press; 1990. [Google Scholar]
- 26.Miller N, Hutt-Fletcher L M. A monoclonal antibody to glycoprotein gp85 inhibits fusion but not attachment of Epstein-Barr virus. J Virol. 1988;62:2366–2372. doi: 10.1128/jvi.62.7.2366-2372.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Molesworth S J, Lake C M, Borza C M, Turk S M, Hutt-Fletcher L M. Epstein-Barr virus gH is essential for penetration of B cells but also plays a role in attachment of virus to epithelial cells. J Virol. 2000;74:6324–6332. doi: 10.1128/jvi.74.14.6324-6332.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Montgomery R I, Warner M S, Lum B J, Spear P G. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell. 1996;87:427–436. doi: 10.1016/s0092-8674(00)81363-x. [DOI] [PubMed] [Google Scholar]
- 29.Nemerow G R, Houghten R A, Moore M D, Cooper N R. Identification of an epitope in the major envelope protein of Epstein-Barr virus that mediates viral binding to the B lymphocyte EBV receptor (CR2) Cell. 1989;56:369–377. doi: 10.1016/0092-8674(89)90240-7. [DOI] [PubMed] [Google Scholar]
- 30.Nemerow G R, Mold C, Schwend V K, Tollefson V, Cooper N R. Identification of gp350 as the viral glycoprotein mediating attachment of Epstein-Barr virus (EBV) to the EBV/C3d receptor of B cells: sequence homology of gp350 and C3 complement fragment C3d. J Virol. 1987;61:1416–1420. doi: 10.1128/jvi.61.5.1416-1420.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nemerow G R, Wolfert R, McNaughton M E, Cooper N R. Identification and characterization of the Epstein-Barr virus receptor on human B lymphocytes and its relationship to the C3d complement receptor (CR2) J Virol. 1985;55:347–351. doi: 10.1128/jvi.55.2.347-351.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ozel M, Pauli G, Gelderblom H R. The organization of the envelope projections on the surface of HIV. Arch Virol. 1988;100:255–266. doi: 10.1007/BF01487688. [DOI] [PubMed] [Google Scholar]
- 33.Pietropaolo R L, Compton T. Direct interaction between human cytomegalovirus glycoprotein B and cellular annexin II. J Virol. 1997;71:9803–9807. doi: 10.1128/jvi.71.12.9803-9807.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prodinger W M, Schwendinger M G, Schoch J, Kochle M, Larcher C, Dierich M P. Characterization of C3dg binding to a recess formed between short consensus repeats 1 and 2 of complement receptor type 2 (CR2; CD21) J Immunol. 1998;161:4604–4610. [PubMed] [Google Scholar]
- 35.Pulvertaft R J V. Cytology of Burkitt's tumour (African lymphoma) Lancet. 1964;i:238–240. doi: 10.1016/s0140-6736(64)92345-1. [DOI] [PubMed] [Google Scholar]
- 36.Rhim J S, Schell K, Creasy B, Case W. Biological characteristics and viral susceptibility of an African green monkey kidney cell line (Vero) Proc Soc Exp Biol Med. 1969;132:670–678. doi: 10.3181/00379727-132-34285. [DOI] [PubMed] [Google Scholar]
- 37.Saemundsen A K, Albeck H, Hansen J P, Nielsen N H, Anvret M, Henle W, Henle G, Thomsen K A, Kristensen H K, Klein G. Epstein-Barr virus in nasopharyngeal and salivary gland carcinomas of Greenland Eskimoes. Br J Cancer. 1982;46:721–728. doi: 10.1038/bjc.1982.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shibata D, Weiss L M. Epstein-Barr virus-associated gastric adenocarcinoma. Am J Pathol. 1992;140:769–774. [PMC free article] [PubMed] [Google Scholar]
- 39.Sixbey J W, Vesterinen E H, Nedrud J G, Raab-Traub N, Walton L A, Pagano J S. Replication of Epstein-Barr virus in human epithelial cells infected in vitro. Nature (London) 1983;306:480–483. doi: 10.1038/306480a0. [DOI] [PubMed] [Google Scholar]
- 40.Spear P G. Entry of alphaherpesviruses into cells. Semin Virol. 1993;4:167–180. [Google Scholar]
- 41.Spriggs M K, Armitage R J, Comeau M R, Strockbine L, Farrah T, Macduff B, Ulrich D, Alderson M R, Mullberg J, Cohen J I. The extracellular domain of the Epstein-Barr virus BZLF2 protein binds the HLA-DR beta chain and inhibits antigen presentation. J Virol. 1996;70:5557–5563. doi: 10.1128/jvi.70.8.5557-5563.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Steimle V, Otten L A, Zufferey M, Mach B. Complementation cloning of an MHC class II transactivator mutated in hereditary MHC class II deficiency (or bare lymphocyte syndrome) Cell. 1993;75:135–146. [PubMed] [Google Scholar]
- 43.Tanner J, Weis J, Fearon D, Whang Y, Kieff E. Epstein-Barr virus gp350/220 binding to the B lymphocyte C3d receptor mediates adsorption, capping, and endocytosis. Cell. 1987;50:203–213. doi: 10.1016/0092-8674(87)90216-9. [DOI] [PubMed] [Google Scholar]
- 44.Tanner J, Whang Y, Sample J, Sears A, Kieff E. Soluble gp350/220 and deletion mutant glycoproteins block Epstein-Barr virus adsorption to lymphocytes. J Virol. 1988;62:4452–4464. doi: 10.1128/jvi.62.12.4452-4464.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tedder T F, Clement L T, Cooper M D. Expression of C3d receptors during human B cell differentiation: immunofluorescence analysis with the HB-5 monoclonal antibody. J Immunol. 1984;133:678–683. [PubMed] [Google Scholar]
- 46.Thorley-Lawson D A, Poodry C A. Identification and isolation of the main component (gp350-gp220) of Epstein-Barr virus responsible for generating neutralizing antibodies in vivo. J Virol. 1982;43:730–736. doi: 10.1128/jvi.43.2.730-736.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yaswen L R, Stephens E B, Davenport L C, Hutt-Fletcher L M. Epstein-Barr virus glycoprotein gp85 associates with the BKRF2 gene product and is incompletely processed as a recombinant protein. Virology. 1993;195:387–396. doi: 10.1006/viro.1993.1388. [DOI] [PubMed] [Google Scholar]
- 48.Yoshiyama H, Imai S, Shimizu N, Takada K. Epstein-Barr virus infection of human gastric carcinoma cells: implication of the existence of a new virus receptor different from CD21. J Virol. 1997;71:5688–5691. doi: 10.1128/jvi.71.7.5688-5691.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Young L S, Clark D, Sixbey J W, Rickinson A B. Epstein-Barr virus receptors on human pharyngeal epithelia. Lancet. 1986;i:240–242. doi: 10.1016/s0140-6736(86)90776-2. [DOI] [PubMed] [Google Scholar]
- 50.Zeidler R, Meissner P, Eissner G, Lazis S, Hammerschmidt W. Rapid proliferation of B cells from adenoids in response to Epstein-Barr virus infection. Cancer Res. 1996;56:5610–5614. [PubMed] [Google Scholar]
- 51.zur Hausen H, Schulte-Holthausen H, Klein G, Henle W, Henle G, Clifford P, Santesson L. EB-virus DNA in biopsies of Burkitt tumors and anaplastic carcinomas of the nasopharynx. Nature (London) 1970;228:1056–1057. doi: 10.1038/2281056a0. [DOI] [PubMed] [Google Scholar]