Abstract
Rothmund-Thomson syndrome (RTS) is a rare autosomal-recessive disorder with clinical features consisting of rash, poikiloderma, sparse hair, short stature, juvenile cataracts, skeletal abnormalities, and cancer predisposition. Genetic studies involving detection of pathogenic RECQL4 variants provide the diagnostic certitude. Osteosarcoma was found in two-thirds RECQL4-mutated RTS patients, while hematological malignancies were rarely reported. The variant diversity of RECQL4 gene has not been fully identified and mutations associated with hematologic malignancies are not well described. In this study, we presented a pedigree of RTS from a Chinese family, among which the proband was diagnosed with de novo myelodysplastic syndrome (MDS). Comprehensive medical examination and chromosome karyotyping were performed on the proband. Whole exome sequencing (WES) was performed on the proband, his sister and his mother. The familial cosegregation of sequence variants derived from WES was conducted by polymerase chain reaction–based Sanger sequencing. Structures of candidate RECQL4 mutants were done by in silico analysis to assess pathogenicity. Three novel RECQL4 germline variants, including c.T274C, c.G3014A, and c.G801C, were identified by WES and validated by Sanger sequencing. Prediction of conformation indicated that the structural stability of human RECQL4 protein was largely affected with these variants. The co-occurring U2AF1 p.S34F and TP53 p.Y220C mutations might contribute to the development of MDS. Our study expands the mutational spectrum of RECQL4 and provides underlying molecular mechanism for the development of MDS in RTS patients.
Keywords: Myelodysplastic Syndrome, Pedigree, RECQL4, Rothmund-Thomson Syndrome, Whole Exome Sequencing
1. INTRODUCTION
Rothmund-Thomson syndrome (RTS), a rare autosomal-recessive disorder, is spanning clinical features of facial rash (poikiloderma), sparse hair, sparse eyelashes and/or eyebrows, short stature, juvenile cataracts, skeletal and dental abnormalities, as well as cancer predisposition. The diagnosis of RTS is dependent on clinical findings and/or biallelic pathogenic variants in RECQL4 or ANAPC1 by molecular genetic testing.1 Up to now, 2 subtypes of RTS have been recognized. Type I RTS is the less common (35%–40%) subtype with clinical features including poikiloderma, ectodermal dysplasia, and juvenile cataracts. No RECQL4 mutation for this type has been identified. Type II RTS is the more common (60%–65%) with clinical features of poikiloderma, congenital bone defects, and increased risk for cancer.2 Type II RTS is due to compound heterozygous or homozygous mutations of RECQL4. Approximately 60% of RTS patients are caused by RECQL4 mutation and 10% by ANAPC1 mutation.1 The genetic variants of the remaining 30% patients are still unknown.
RECQL4 encodes a DNA helicase, which unwinds double-stranded DNA and RNA-DNA hybrids into single-stranded DNA templates. Through modulating DNA replication, transcription, and damage response, RECQL4 maintains the integrity and stability of genome.3,4 According to the Human Gene Mutation Database (HGMD, professional 2021.04) (http://www.hgmd.cf.ac.uk/ac/index.php), more than 100 RECQL4 mutations have been reported in RTS. However, due to the low-throughput limitation of Sanger sequencing in the past, the variant diversity of RECQL4 gene has not been fully identified.
Cancer is a common complication of RTS. Osteosarcoma is found in two-thirds RECQL4-mutated RTS patients. However, few hematological malignancies which mostly are lymphomas have been reported.5 The underlying molecular mechanisms of hematological malignancies developed in RTS are unknown. Previous studies represented that different mutation site could affect the development and phenotype of disease.6,7 Co-occurring mutation might synergistically promote the pathogenesis of hematological malignancies. In the era of next-generation sequencing, new technological platform such as whole exome sequencing (WES) could help to describe the mutational atlas of RTS with hematological disorder.8,9
Here we report an RTS patient who was diagnosed with de novo myelodysplastic syndrome (MDS). We used WES to identify new disease-causing variants. Novel RECQL4 germline mutations were confirmed by Sanger sequencing of both genomic DNA (gDNA) from peripheral blood and hair follicle. Protein structure was predicted to estimate the harmful effect of missense variant to spatial conformation and function of RECQL4.
2. MATERIALS AND METHODS
2.1. Study design and participants
This study was approved by the Ethics Committee of Ruijin Hospital, Shanghai Jiaotong University School of Medicine (Approval number #2021-173). A Chinese family with 1 member diagnosed as RTS was enrolled. All participants gave written informed consent. Comprehensive medical histories, blood and hair follicle samples were collected from all the accessible family members.
2.2. Chromosome karyotyping
Chromosomal structural anomalies in the proband were detected by the G-banding technique according to the routine laboratory procedure.
2.3. DNA extraction and quality control
For each participant, gDNA was extracted from peripheral blood and hair follicle by DNeasy Blood & Tissue Kit (Qiagen #69504) according to the manufacturer’s instruction. Purified DNA was collected and stored at −20°C. DNA degradation and contamination were monitored on 1% agarose gels. DNA concentration was measured by Qubit® DNA Assay Kit in Qubit® 2.0 Flurometer (Invitrogen, Carlsbad, CA, USA).
2.4. Whole exome sequencing
The gDNA samples of the proband (IV.1), his mother (III.6), and his sister (IV.2) were accessed for WES. Following the library preparation and adaptor ligation, the Agilent SureSelect Human All Exon V6 Kit (Agilent Technologies, Santa Clara, CA, USA) was used for exome capture. The Illumina Novaseq platform (Illumina Inc., San Diego, CA, USA) was utilized for gDNA sequencing in Personal Biotechnology Co., Ltd (Shanghai, China) based on the manufacturer’s procedure. The original fluorescence image files were transformed to FASTQ files (raw data) by base calling. After quality control, high-quality reads were subsequently aligned to the human reference genome (UCSC hg19/hg38) to get BAM files. Then, SAM tools and Picard tools were used to select BAM files and perform duplicate marking, local realignment, and base quality recalibration. Duplicate reads were discarded for variants calling. GATK (v4.0) was utilized to detect SNPs and InDels. Control-FREEC was used to detect CNVs.
2.5. Polymerase chain reaction and Sanger sequencing
The nucleotide sequence of REQCL4 gene (NM_004260) was collected from the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov). Primer3 (http://primer3.ut.ee/) was used to design specific primers flanking the expected variation. Primer sequence to verify RECQL4 variants are shown in Table S1, http://links.lww.com/BS/A54. Polymerase chain reaction (PCR) was performed and agarose gel electrophoresis was run to confirm the presence and correct size of the amplified sequence. Afterward, variants identified by WES were confirmed by Sanger sequencing.
2.6. Structural analysis of protein mutant
The structure of wild-type RECQL4 was obtained from the dataset in alphafold (alphafold.ebi.ac.uk/entry/O94761). The predicted structures of RECQL4 mutants were calculated by the Center for High-Performance Computing at Shanghai Jiao Tong University. The first-ranked predicted structure was visualized, aligned, and analyzed through PyMoL software. The overall stability of protein with combined mutants was predicted by FoldX software.
3. RESULTS
3.1. Patient characteristics
A 42-year-old man (the proband) was diagnosed with de novo MDS in May 2021. Complete blood count revealed pancytopenia, including normochromic normocytic anemia (hemoglobin 6.9 g/dL), thrombocytopenia (2000/μL), and leukopenia (2060/μL). Bone marrow (BM) morphology displayed 2% blasts, dyserythropoiesis, dysplastic neutrophils, micromegakaryocytes, and multinucleated megakaryocytes. Iron-stained BM aspirate showed 57% ring sideroblasts. Immunophenotype analysis showed abnormal myeloid maturation with dim expression of CD38 and absent expressional heterogeneity of CD13 and CD33 on myeloid progenitors. Cytogenetic study revealed a complex karyotype: 45~48, XY, del (5) (q13q34), +del (8q24), del(11q23) [8]/46, XY [2] (Figure S1A, http://links.lww.com/BS/A54). WES and Sanger sequencing revealed germline TP53 missense variant “c.C98G: p.P33R” (Figure S1B–E, http://links.lww.com/BS/A54). Molecular mutational profile showed somatic mutations of U2AF1 p.S34F (Variant Allele Frequency, VAF 36.9%) and TP53 p.Y220C (VAF 76.6%) (Figure S1F–G, http://links.lww.com/BS/A54). Based on these findings, the patient was diagnosed as de novo MDS with ring sideroblasts with multilineage dysplasia (MDS-RS-MLD). Other clinical symptoms of the proband included rash, poikiloderma, sparse scalp hair, eyelashes, eyebrows, short stature, and hyperkeratosis (Table 1). Reviewing the patient’s medical history, he was diagnosed with pulmonary artery stenosis at age of 6, psoriasis at 32, diabetes mellitus at 42, and infertile. As no available donor for allogeneic hematopoietic stem cell transplantation, the patient was treated with lenalidomide as maintenance.
Table 1.
Clinical characteristics of the proband in relation to clinical signs of RTS.
| Clinical sign of RTS | |
|---|---|
| Erythema on the cheeks and face | Present |
| Poikiloderma | Present |
| Sparse scalp hair, eyelashes, and/or eyebrows | Present |
| Small size, usually symmetric for height and weight | Present |
| Gastrointestinal disturbance as a young child: chronic vomiting and diarrhea | Absent |
| Dental abnormalities: hypoplastic teeth, enamel defects, delayed tooth eruption | Absent |
| Nail abnormalities: dysplastic or poorly formed nails | Absent |
| Hyperkeratosis (soles of the feet) | Present |
| Cataracts | Absent |
| Skeletal abnormalities: radial ray defects, ulnar defects, absent or hypoplastic patella, osteopenia, abnormal trabeculation | Absent |
| Cancers | Present |
RTS = Rothmund-Thomson syndrome.
3.2. Identification of novel germline RECQL4 variants
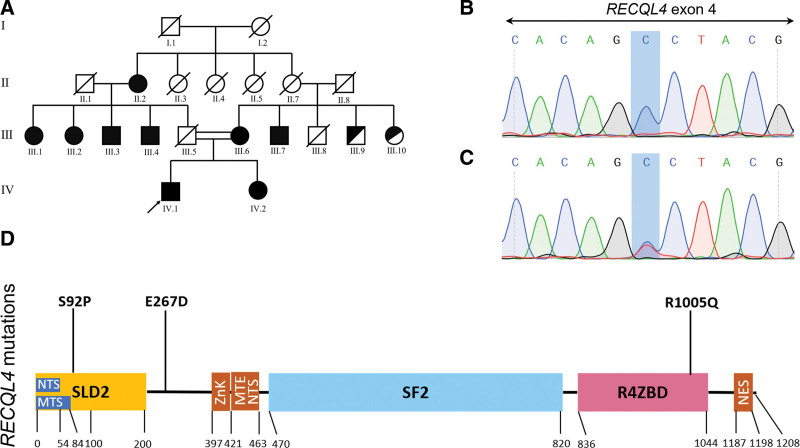
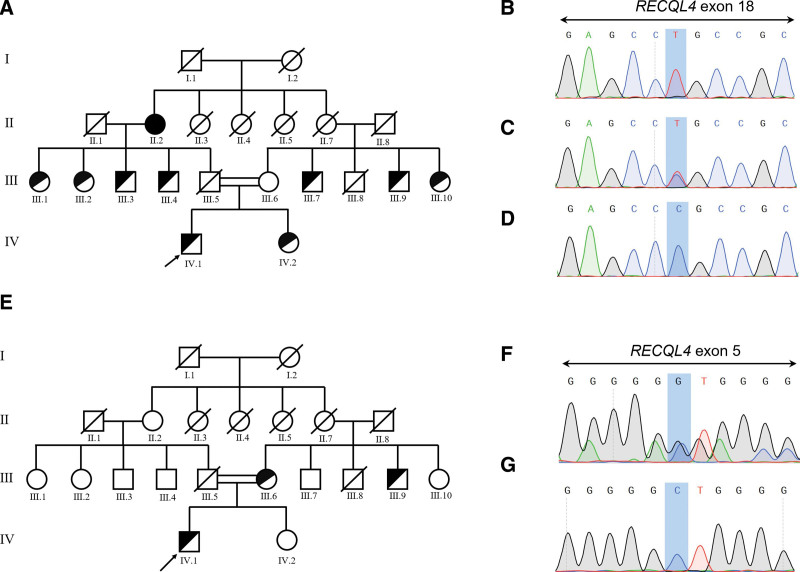
Considering the clinical characteristics, we wondered whether the patient had any underlying germline defects. We used WES to access the possible genetic mutations. A primary variant calling on WES data unveiled 30620 variants, of which 24,663 variants located in coding regions. By removing non-synonymous mutations, 3 novel RECQL4 variants left. “c.T274C: p.S92P” in exon 4 was homozygous missense variant of the proband (IV.1), while his sister and mother also had this homozygous variant (Fig. 1A). “c.G3014A: p.R1005Q” in exon 18 was heterozygous missense variant for the proband, heterozygous for his sister, wild-type for his mother and homozygous for the grandmother (Fig. 2A). His sister, mother, grandmother, as well as homozygous RECQL4-mutated aunts/uncles had clinical signs of erythema on the face, poikiloderma, sparse scalp hair, eyelashes, and/or eyebrows, small size and hyperkeratosis of soles. The heterozygous missense variant “c.G801C: p.E267D” in exon 5 in the proband might also contribute to disease development in a compound heterozygous form. The missense variant “c.G801C: p.E267D” was carried by his mother, while the sister was wild-type (Fig. 2E). The HGMD was checked to support the novelty of these variants. Existence of the c.T274C, c.G3014A, and c.G801C germline variants was confirmed through PCR-Sanger sequencing of both blood and hair follicle DNA (Fig. 1B–C, 2B–D, 2F–G). The location of p.S92P variant in SLD2 domain and p.R1005Q variant in conserved SF2 helicase domain suggested they might impair the DNA helicase function of RECQL4 (Fig. 1D).
Figure 1.
RECQL4 c.T274C: p.S92P variant in the RTS pedigree. (A) Pedigree of a consanguineous family with 9 members affected by c.T274C: p.S92P mutation. The proband was indicated by an arrow. Family members marked completely black carried homozygous variant, others only half in black carried heterozygous variant. (B–C) The presence of RECQL4 c.T274C: p.S92P in homozygous state (B) (chromatograph of IV.1 as representative) and heterozygous state (C) (chromatograph of III.9 as representative). (D) Mutational landscape in association with human RECQL4 protein domains, including the SLD2 (yellow) and R4ZBD domains (pink). MTE = mitochondrial exclusion, MTS = mitochondrial targeting signal, NES = nuclear export signal, NTS = nuclear targeting signal, R4ZBD = RECQL4 zinc-binding domain, ZnK = zinc knuckle.
Figure 2.
RECQL4 c.G3014A: p.R1005Q and c.G801C: p.E267D variants in the pedigree with RTS. (A) Pedigree of the family affected by RECQL4 c.G3014A: p.R1005Q variant. The proband indicated by an arrow. Family members marked completely black carried homozygous variant, others only half in black carried heterozygous variant. (B–D) The presence of RECQL4 c.G3014A: p.R1005Q in homozygous state (B) (chromatograph represented by II.2), heterozygous state (C) (chromatograph represented by IV.1), and wild-type (D) (chromatograph represented by III.6). (E) Pedigree of the family affected by RECQL4 c.G801C: p.E267D variant. (F–G) The presence of RECQL4 c.G801C: p.E267D in heterozygous state (F) (chromatograph represented by IV.1) and wild-type (G) (chromatograph represented by IV.2).
3.3. Structural prediction of RECQL4 mutants
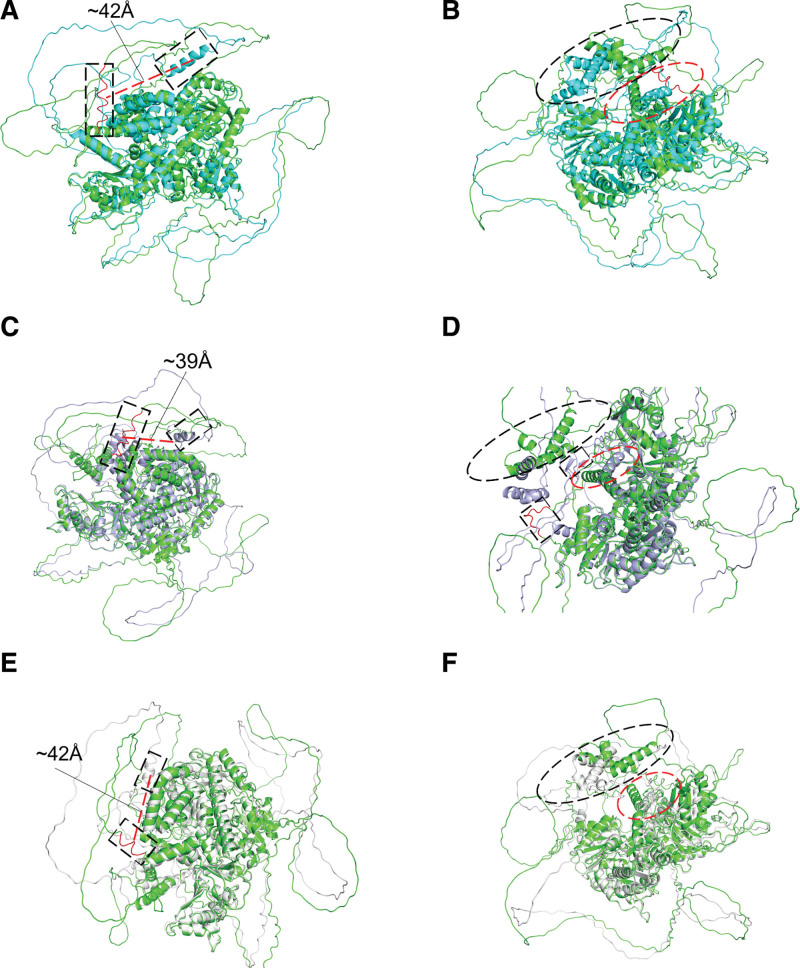
To illustrate whether the novel variants could affect protein function, we predicted the spatial conformation change of RECQL4 mutants. The most significant change in S92P structure is the formation of an α spiral by the residues from P103 to T116, which constructed a flexible loop in wild-type (Fig. 3A). Additionally, the α spiral space of M1-A55 and L373-F390 region could not be fully fitted (Fig. 3B). The most significant change of E267D structure was found in the residues of D104-T116, which formed a flexible loop region in wild-type but converted to an α helix in E267D structure (Fig. 3C). Furthermore, the loop region from R355 to Y363 in wild-type converted to 2 small β sheets in E267D structure. Similar to S92P mutant, the α helix in M1-A55 and L373-F390 region could not be fully superposed (Fig. 3D). The most distinct change in structure of R1005Q is G106-N111 domain. In R1005Q structure, the wild-type flexible loop (G106-N111) was converted to form a short α helix (Fig. 3E). Additionally, M1-A55 and L373-F390 region could also not be fully fitted in the α spiral space (Fig. 3F). Furthermore, each individual had a distinctive mutant combination. We predicted the overall stability by total energy of protein though FoldX software. The S92P mutant had the most stable structure, then followed by wild-type, S92P-R1005Q, and S92P-E267D-R1005Q. The S92P-E267D mutant combination, which possibly presented in the proband and his mother, had the most unstable structure (Figure S3, http://links.lww.com/BS/A54). Spatial conformations of RECQL4 mutants may change obviously and further disturb the DNA helicase function.
Figure 3.
Visualization of wild-type RECQL4 and mutant RECQL4 protein structure by PyMoL. (A–B) Structure variation from wild-type RECQL4 (green cartoon) to RECQL4 p. S92P (cyan cartoon), RMSD = 1.007 (789 to 789 atoms). Flexible loop (red in dotted box) constructed by the residues from P103 to T116 in wild-type converted to form an α spiral (cyan in dotted box) in S92P structure, with the spatial position offset by about 42Å (red dotted line) (A). The α spiral space of M1-A55 (black dotted ellipse) and L373-F390 region (red dotted ellipse) in S92P mutant (B). (C–D) Structure variation from wild-type RECQL4 (green cartoon) to RECQL4 p. E267D (light blue cartoon), RMSD = 1.017 (810 to 810 atoms). The flexible loop region constructed by D104-T116 in wild-type (red in dotted box) formed an α helix (light blue in dotted box) in E267D structure, with the spatial position offset by about 39 Å (red dotted line) (C). A loop region of the residues from R355 to Y363 (red in dotted box) in wild-type became two small β sheets (light blue in dotted box) in E267D structure. The α helix of M1-A55 (black dotted ellipse) and L373-F390 region (red dotted ellipse) in E267D mutant (D). (E–F) Structure variation from wild-type RECQL4 (green cartoon) to RECQL4 p. R1005Q (white cartoon), RMSD = 1.102 (792 to 792 atoms). The wild-type flexible loop (G106-N111) (red in dotted box) formed a short α helix (white in dotted box) with a spatial position offset about 42Å (red dotted line) in R1005Q structure (E). M1-A55 (black dotted ellipse) and L373-F390 region in the α spiral space (red dotted ellipse) of R1005Q structure (F).
3.4. RECQL4 germline variants in association with hematological malignancies
The novel RECQL4 variants and co-occurring U2AF1 p.S34F and TP53 p.Y220C mutations might promote the onset of de novo MDS in the proband. Furthermore, we summarized that RECQL4 germline variants of specific sites were in association with hematological malignancies, especially for lymphoma in RTS or RAPADILINO syndrome (Table 2). Somatic RECQL4 mutation is rare (Figure S2, http://links.lww.com/BS/A54) in hematological malignancies according to data originated from Cbioportal (http://www.cbioportal.org).16,17 We concluded that the differential mutation sites of RECQL4 and co-occurring mutations might affect the development of hematological malignancy in RTS.
Table 2.
Germline RECQL4 mutations associated with hematological malignancies.
| Patient number | Mutations | Effect | Mutation location | Syndrome | Cancer type | Onset age (y) |
|---|---|---|---|---|---|---|
| 1 | c.806G > A10 | p.Trp269X | N-terminus | RAPADILINO | Lymphoma | 24 |
| 2 | c.1048_1049delAG11 | p.Arg350fsX | N-terminus | RTS | Lymphoma | 34 |
| 3 | c.1390 + 2delT12 | p.Ala420_Ala463del | NTS/MTE/ZnK | RAPADILINO | Lymphoma | 25 |
| 4 | c.1650del713 | p.Ala551fsX | SF2 | RTS | tMDS/tAML | 31 |
| c.2269C>T13 | p.Gln757X | SF2 | ||||
| 5 | c.1704 + 1G > A14 | Missplicing | SF2 | RTS | Lymphoma/ALL | 9 |
| c.1919_1924del TCACAG14 |
p.Leu640_Ala642del insP |
SF2 | ||||
| 6 | c.1913T > C10 | p.Leu638Pro | SF2 | RTS | Lymphoma | 2 |
| c.2419ins510 | p.Arg807fsX | SF2 | ||||
| 7 | c.2492_2493delAT15 | p.His831fsX | C-terminus | BGS | Lymphoma | 7 |
| c.2506_2518del1315 | p.Trp836fsX | R4ZBD |
Mutation type: (del) deletion; (>) nucleotide change from; (X) premature stop codon; (fs) frameshift; (ins) insertion. Mutation location: NTS = nuclear targeting signal, MTE = mitochondrial exclusion, ZnK = zinc knuckle, SF2 = super family2 helicase domains, R4ZBD = RECQL4 zinc-binding domains. Syndrome: RAPADILINO = RAdial hypoplasia/aplasia, PAtellar hypoplasia/aplasia = cleft or highly arched PAlate, DIarrhea and DIslocated joints, LIttle size (>2 SDs below the mean in height) and LImb malformation, and slender NOse and NOrmal intelligence, BGS = Baller-Gerold syndrome, RTS = Rothmund-Thomson syndrome. Cancer type: lymphoma; tMDS = therapy-related myelodysplastic syndrome, tAML = therapy-related acute myeloid leukemia.
4. DISCUSSION
In this study, we reported a Chinese pedigree with 3 pathogenic variants of RECQL4 gene, all of which were novel missense variants. Prediction of conformation indicated that the structural stability of human RECQL4 protein was largely affected by these variants. To the best of our knowledge, this is the first case of de novo MDS reported in RTS patients. Our study expands the genetic spectrum and molecular mechanism of hematological malignancies in patients with RTS.
RECQL4 is a member of DNA helicases which promotes DNA unwinding to affect all aspects of DNA metabolism.4,18 Disturbing their expression and biochemical activity results in genomic instability, disease, and cancer predisposition.1 In the present study, we reported p.S92P missense variant located in the N-terminus SLD2 domain of RECQL4. The SLD2 domain of RECQL4 is important to initiate DNA synthesis by recruiting replication factors to replication origin. A significant number of disease-associated mutations have been reported in SLD2 motif5 and mutations in SLD2 domain to abolish RECQL4 function may be lethal.19,20 Another novel p.R1005Q mutation located in the RECQL4 zinc-binding domain (R4ZBD). Although mutations in RZ4BD domain did not reduce DNA-binding affinity, the Δ944–1032 deletion variant showed a reduction in the velocity of helicase activity to 53% of the wild-type protein.21 Prediction of protein conformation further revealed all the 3 novel variants might affect the stability of RECQL4, although p.E267D was not in the functional motif. Our data underline that these 3 variants might impair the function of RECQL4 and emphasize that prediction of protein structure could help to estimate the harmful effects of novel variants.
Nearly two-thirds RTS patients with biallelic pathogenic RECQL4 variants developed osteosarcoma. However, it was reported that cancer risk for individuals with monoallelic RECQL4 pathogenic variants was not significantly different with estimates obtained from SEER data.22 Although RECQL4 germline mutations also promote hereditary predisposition and familial clustering of hematopoietic neoplasms such as MDS, leukemia, and lymphoma,23 the incidence is relatively rare. We summarized hematological malignancies in RTS patients which might be in association with specific mutational sites of RECQL4 (Table 2). Patient 4 was diagnosed with therapy-related MDS which was secondary to osteosarcoma. Furthermore, in the present pedigree, only the proband developed MDS. RECQL4 “loss-of-function” mutation could result in genomic instability and increase the hereditary susceptibility of MDS. Additional genetic events seemed to be involved in the full penetrance of MDS for patients with RTS. To our best knowledge, no recurrent mutation except for RECQL4 has been reported in patients with MDS and concurrent RTS.13,24–27 Our molecular genetic study showed that somatic U2AF1 p.S34F and TP53 p.Y220C mutations, and germline TP53 p.P33R mutation co-occurred in the proband. Somatic U2AF1 p.S34F mutation and TP53 p.Y220C mutation are recurrently present in de novo MDS.28,29 Thus, these co-occurred mutations might contribute to forming complex chromosomal karyotype and promoting the development of MDS. Further analyses of the pathologic function of differential RECQL4 mutational sites and co-occurring genetic events will facilitate to understand the development of hematopoietic neoplasms in RTS.
In conclusion, we reported 3 novel RECQL4 pathogenic variants in RTS and explored the association of RECQL4 mutations with hematopoietic malignancies. Our results further provide underlying mechanism for the development of hematopoietic malignancies in RTS.
ACKNOWLEDGMENTS
This work was supported by National Natural Science Foundation of China (grant numbers 81770106, 82170206).
AUTHOR CONTRIBUTIONS
Chuanhe Jianga, Hao Zhanga, Chuxian Zhao contributed equally to this study.
C.J., H.Z., and C.Z. designed the experiments, reviewed literatures, analyzed the data, and wrote the paper. L.W. diagnosed the patient. Z.P. and X.H. proposed and designed the study, interpreted the results, wrote the manuscript, and oversaw the project. All authors reviewed and approved the manuscript.
Supplementary Material
Footnotes
Conflict of interest: The authors declare that they have no conflict of interest.
All data generated or analyzed in this study are included in this published article and additional files. The whole exome sequencing raw data generated in this study is available in the NCBI Sequence Read Archive (SRA) database (http://www.ncbi.nlm.nih.gov/bioproject/824973; BioProject ID: PRJNA824973).
This work was supported by National Natural Science Foundation of China (grant numbers 81770106, 82170206).
REFERENCES
- [1].Wang LL, Plon SE. Rothmund-Thomson Syndrome In: GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993. 1999 Oct 6 [updated 2020 Jun 4]. [PubMed] [Google Scholar]
- [2].Wang LL, Gannavarapu A, Kozinetz CA, et al. Association between osteosarcoma and deleterious mutations in the RECQL4 gene in Rothmund–Thomson syndrome. JNCI: J National Cancer Institute. 2003;95(9):669–674. [DOI] [PubMed] [Google Scholar]
- [3].Ajeawung NF, Nguyen TTM, Lu L, et al. Mutations in ANAPC1, encoding a scaffold subunit of the anaphase-promoting complex, cause Rothmund-Thomson syndrome type 1. Am J Hum Genet. 2019;105(3):625–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Croteau DL, Popuri V, Opresko PL, Bohr VA. Human RecQ helicases in DNA repair, recombination, and replication. Annu Rev Biochem. 2014;83:519–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Xu X, Chang CW, Li M, Liu C, Liu Y. Molecular mechanisms of the RECQ4 pathogenic mutations. Front Mol Biosci. 2021;8:791194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Yu C, Li J, Yuan Z, Liu S, Zou L. Two novel mutations affecting the same splice site of PKD1 correlate with different phenotypes in ADPKD. Ren Fail. 2014;36(5):687–693. [DOI] [PubMed] [Google Scholar]
- [7].Sieber OM, Segditsas S, Knudsen AL, et al. Disease severity and genetic pathways in attenuated familial adenomatous polyposis vary greatly but depend on the site of the germline mutation. Gut. 2006;55(10):1440–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Mohamadian M, Ghandil P, Naseri M, Bahrami A, Momen AA. A novel homozygous variant in an Iranian pedigree with cerebellar ataxia, mental retardation, and dysequilibrium syndrome type 4. J Clin Lab Anal. 2020;34(11):e23484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Zhang Y, Qin W, Wang H, Lin Z, Tang Z, Xu Z. Novel pathogenic variants in the RECQL4 gene causing Rothmund-Thomson syndrome in three Chinese patients. J Dermatol. 2021;48(10):1511–1517. [DOI] [PubMed] [Google Scholar]
- [10].Siitonen HA, Sotkasiira J, Biervliet M, et al. The mutation spectrum in RECQL4 diseases. Eur J Hum Genet. 2009;17(2):151–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].van Rij MC, Grijsen ML, Appelman-Dijkstra NM, et al. Rothmund-Thomson syndrome and osteoma cutis in a patient previously diagnosed as COPS syndrome. Eur J Pediatr. 2017;176(2):279–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Siitonen HA, Kopra O, Kaariainen H, et al. Molecular defect of RAPADILINO syndrome expands the phenotype spectrum of RECQL diseases. Hum Mol Genet. 2003;12(21):2837–2844. [DOI] [PubMed] [Google Scholar]
- [13].Carlson AM, Lindor NM, Litzow MR. Therapy-related myelodysplasia in a patient with Rothmund-Thomson syndrome. Eur J Haematol. 2011;86(6):536–540. [DOI] [PubMed] [Google Scholar]
- [14].Simon T, Kohlhase J, Wilhelm C, Kochanek M, De Carolis B, Berthold F. Multiple malignant diseases in a patient with Rothmund-Thomson syndrome with RECQL4 mutations: case report and literature review. Am J Med Genet A. 2010;152A(6):1575–1579. [DOI] [PubMed] [Google Scholar]
- [15].Debeljak M, Zver A, Jazbec J. A patient with Baller-Gerold syndrome and midline NK/T lymphoma. Am J Med Genet A. 2009;149A(4):755–759. [DOI] [PubMed] [Google Scholar]
- [16].Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discovery. 2012;2(5):401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):l1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Lu H, Fang EF, Sykora P, et al. Senescence induced by RECQL4 dysfunction contributes to Rothmund-Thomson syndrome features in mice. Cell Death Dis. 2014;5(5):e1226–e1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Shin G, Jeong D, Kim H, Im JS, Lee JK. RecQL4 tethering on the pre-replicative complex induces unscheduled origin activation and replication stress in human cells. J Biol Chem. 2019;294(44):16255–16265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Xu X, Rochette PJ, Feyissa EA, Su TV, Liu Y. MCM10 mediates RECQ4 association with MCM2-7 helicase complex during DNA replication. EMBO J. 2009;28(19):3005–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kaiser S, Sauer F, Kisker C. The structural and functional characterization of human RecQ4 reveals insights into its helicase mechanism. Nat Commun. 2017;8:15907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Martin-Giacalone BA, Rideau TT, Scheurer ME, Lupo PJ, Wang LL. Cancer risk among RECQL4 heterozygotes. Cancer Genet. 2022;262-263:107–110. [DOI] [PubMed] [Google Scholar]
- [23].Mangaonkar AA, Patnaik MM. Hereditary predisposition to hematopoietic neoplasms: when bloodline matters for blood cancers. Mayo Clin Proc. 2020;95(7):1482–1498. [DOI] [PubMed] [Google Scholar]
- [24].Rizzari C, Bacchiocchi D, Rovelli A, et al. Myelodysplastic syndrome in a child with Rothmund-Thomson syndrome: a case report. J Pediatr Hematol Oncol. 1996;18(1):96–97. [DOI] [PubMed] [Google Scholar]
- [25].Ilhan I, Arikan U, Büyükpamukçu M. Myelodysplastic syndromes and RTS. Pediatr Hematol Oncol. 1996;13(2):197–197. [DOI] [PubMed] [Google Scholar]
- [26].Pianigiani E, De Aloe G, Andreassi A, Rubegni P, Fimiani M. Rothmund-Thomson syndrome (Thomson-type) and myelodysplasia. Pediatr Dermatol. 2001;18(5):422–425. [DOI] [PubMed] [Google Scholar]
- [27].Narayan S, Fleming C, Trainer AH, Craig JA. Rothmund-Thomson syndrome with myelodysplasia. Pediatr Dermatol. 2001;18(3):210–212. [DOI] [PubMed] [Google Scholar]
- [28].Jiang Y, Gao SJ, Soubise B, Douet-Guilbert N, Liu ZL, Troadec MB. TP53 in myelodysplastic syndromes. Cancers (Basel). 2021;13(21):5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Graubert TA, Shen D, Ding L, et al. Recurrent mutations in the U2AF1 splicing factor in myelodysplastic syndromes. Nat Genet. 2011;44(1):53–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.