Abstract
Background
Heterogeneous vancomycin-intermediate Staphylococcus aureus (hVISA) compromise the clinical efficacy of vancomycin. The hVISA isolates spontaneously produce vancomycin-intermediate Staphylococcus aureus (VISA) cells generated by diverse and intriguing mechanisms.
Objective
To characterize the biomolecular profile of clinical hVISA applying genomic, transcriptomic and metabolomic approaches.
Methods
39 hVISA and 305 VSSA and their genomes were included. Core genome-based Bayesian phylogenetic reconstructions were built and alterations in predicted proteins in VISA/hVISA were interrogated. Linear discriminant analysis and a Genome-Wide Association Study were performed. Differentially expressed genes were identified in hVISA-VSSA by RNA-sequencing. The undirected profiles of metabolites were determined by liquid chromatography and hydrophilic interaction in six CC5-MRSA.
Results
Genomic relatedness of MRSA associated to hVISA phenotype was not detected. The change Try38 → His in Atl (autolysin) was identified in 92% of the hVISA. We identified SNPs and k-mers associated to hVISA in 11 coding regions with predicted functions in virulence, transport systems, carbohydrate metabolism and tRNA synthesis. Further, capABCDE, sdrD, esaA, esaD, essA and ssaA genes were overexpressed in hVISA, while lacABCDEFG genes were downregulated. Additionally, valine, threonine, leucine tyrosine, FAD and NADH were more abundant in VSSA, while arginine, glycine and betaine were more abundant in hVISA. Finally, we observed altered metabolic pathways in hVISA, including purine and pyrimidine pathway, CoA biosynthesis, amino acid metabolism and aminoacyl tRNA biosynthesis.
Conclusions
Our results show that the mechanism of hVISA involves major changes in regulatory systems, expression of virulence factors and reduction in glycolysis via TCA cycle. This work contributes to the understanding of the development of this complex resistance mechanism in regional strains.
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) is responsible for a wide range of infections in humans,1 have disseminated worldwide and, are associated with high morbidity and mortality rates.2 Vancomycin is the mainstay of treatment for severe MRSA infections,3 however, low-level glycopeptide resistance in S. aureus has been reported since 1997,4 including the emergence of vancomycin-intermediate S. aureus (VISA) and heterogeneous-VISA (hVISA), compromising the clinical efficacy of vancomycin.5 Isolates with intermediate resistance to vancomycin (VISA) exhibit MIC values of 4–8 mg/L, while hVISA phenotype shows MIC within the susceptible range (≤2 mg/L)6; the hVISA strains contain subpopulations with decreased susceptibility to vancomycin that are not detected by routine susceptibility methods.7,8 Notably, the hVISA strains are associated with vancomycin therapeutic failure and persistent infections.9–11
The frequency of hVISA phenotype appears to be increasing, and is more common among MRSA belonging to ST239 and ST5, two hospital-acquired MRSA lineages prevalent in Asia, South America and Europe.12,13 In Latin America, data related to the prevalence of hVISA is scarce, and is mainly associated with sporadic case reports, particularly in Argentina,14–16 Chile17 and Brazil.18 We previously investigated the hVISA phenotype among a collection of 1189 clinical MRSA isolates recovered from patients in hospitals from nine Latin American countries (2006–2008 and 2011–2014).19–21 We found only 39 MRSA isolates (3.3%) that were classified as hVISA by glycopeptide resistance detection (GRD) and Etest Macro-method (MET), however, only six of these isolates were confirmed as hVISA by the population analysis profile/area under the curve (PAP/AUC). Of note, 95% of the hVISA isolates belonged to the clonal complex (CC) 5. Furthermore, when we searched for changes in proteins associated to hVISA (WalKR, VraTSR, RpoB, RpoC and TcaA), we observed two common amino acid substitutions compromising the 90% of the hVISA isolates; WalK (Leu-14 → Ile) and VraT (Glu-156 → Gly).21
The genetic mechanisms involved in the adaptive response of S. aureus leading to decreased vancomycin susceptibility are complex and not fully understood.22 Vancomycin-intermediate Staphylococcus aureus (VISA) and hVISA strains show common phenotypic features particularly affecting the cell envelope, including cell wall thickening, increased production of peptidoglycan, augmented amounts of D-Ala-D-Ala residues, reduced peptidoglycan cross-linking, reduced autolytic activity and altered expression of PBPs.8,23 Since several pathways and multiple gene changes are involved, genetic-based methods for the detection of their resistance phenotype are not available.22 Nevertheless, the combination of several ‘omics’ technologies (genomics, transcriptomics, proteomics and metabolomics approaches) have proven to be useful in elucidating specific genetic signatures, pathways and metabolic adaptations involved in the progression of complex phenotypes, including VISA. Here, we aimed to characterize the biomolecular profile of clinical isolates of hVISA MRSA recovered from Latin America through the combination of genomics, transcriptomic and metabolomics approaches.
Materials and methods
Bacterial isolates and whole genome sequencing
We included a total of 344 MRSA isolates recovered from Argentina, Brazil, Chile, Colombia, Ecuador, Guatemala, Mexico, Peru and Venezuela as part of two prospective studies of adult patients with S. aureus infection between 2006 and 2008,20 and 2011 and 2014.19 On the basis of the GRD test, 39 isolates were designated as hVISA and 305 as vancomycin-susceptible (VSSA).21 All isolates (Table S1 available as Supplementary data at JAC Online) were sequenced on an Illumina platform, as previously described.21
Phylogenetic analysis
Genome assemblies of 352 genomes; including the 344 MRSA from this study and eight reference genomes (Mu3, Mu50, ATCC29213, FPR3757, N315, TW20, JKD6009, JKD6008) were annotated with RAST,24 and the core genome at 95% was determined using Roary.25 The identified orthogroups were aligned with MUSCLE,26 SNPs were obtained with SNP-sites27 and Bayesian reconstruction analysis was performed using BEAST v.2.6.328 with a GTR + gamma model of heterogeneity, constant population, strict clock and 100 million steps. A maximum credibility clades tree was obtained with 70 million burn in, a 0.5 posterior probability and mean heights.
The phylogenetic relationships of 257 MRSA genomes belonging to CC5, was obtained with a core genome at 99% with Roary25 including references: ATCC29213, N315, Mu3, Mu50 and FPR3757 as outgroup. Phylogenetic matrix determination and Bayesian phylogenetic reconstruction configuration was the same as before. All trees were plotted and annotated in iTol29 and BEAST runs were performed in the CIPRES30 servers.
Identification of polymorphisms and genetic regions potentially associated with the development of hVISA phenotype
We performed pairwise SNP determination of each of the 165 (hVISA and VSSA) genomes belonging to ST5 with N315 genome as reference, using Mummer.31 Then, we built a matrix of the presence and absence of variants across all the evaluated samples, and performed a linear discriminant analysis (LDA) using Python’s scikit-learn32 module to discriminate the genomes with hVISA and VSSA phenotypes. LDA scaling factors were used to identify SNPs. Additionally, we performed a Genome-Wide Association Study (GWAS) of all isolates using PySeer33 over k-mers with sizes between 9 and 100, including phylogenetic distance based on the previous tree and selecting those k-mers with lqrt-P ≤ 1.4 × 10−8 (alpha value of 0.01 after Bonferroni correction). Selected k-mers were annotated against N315 reference genome (VSSA), followed by comparisons against closed genomes of hVISA strains UCL420, UA851 and UE1097.
Identification of amino acid changes in predicted proteins associated with the development of VISA phenotype
We performed a comprehensive systematic literature search in PubMed, including articles published until September 2019 applying the following terms: vancomycin-intermediate Staphylococcus aureus, VISA, heterogeneous vancomycin-Intermediate Staphylococcus aureus, hVISA, Staphylococcus aureus with reduced vancomycin susceptibility, glycopeptide-intermediate Staphylococcus aureus, heterogeneous glycopeptide-intermediate Staphylococcus aureus and hGISA. We identifying a set of 54 proteins previously associated to the hVISA/VISA phenotype including the 11 that we analysed in our previous study (Table S2). The amino acid sequences of these proteins in the 344 genomes were aligned with N315. To determine the difference in the proportion of each amino acid substitution in VSSA and hVISA groups a chi-square test was performed.
Transcriptome analysis by RNA-seq
For RNA-seq analysis, we included the hVISA isolates UCL420 (ST5), UA851 (ST5) and UE1097 (ST1341), which were also assigned as hVISA by PAP/AUC and the VSSA isolates UCL739 (ST5), UA863 (ST5) and UE218 (ST239), which were collected in the same country of the hVISA isolates and were VSSA genetically related (by phylogenomic analyses) to the hVISA isolates (Figure S1). Reference strains Mu3 and N315 were also included. RNA was obtained using the Ribopure RNA purification kit (Ambion, Life Technologies). Three independent biological replicates were performed for each sample. cDNA libraries were made using the TrueSeq RNA Sample Preparation System v.2 (Illumina) and were sequenced on Illumina platform. A count table for 2670 genes was obtained by EDGE-pro v.1.3.134 was used to identify differentially expressed genes (DEG), setting the threshold of Benjamini–Hochberg adjusted P < 0.05.35 DEG were obtained by pairwise comparisons (between hVISA/VISA pairs and also comparing to N315) and identifying the DEG associated to hVISA phenotype.
Metabolic fingerprinting of hVISA and VSSA isolates
Metabolic fingerprinting of ST5 hVISA (UA851 and UCL420) and ST5 VSSA (UA863 and UCL739) isolates and the reference strains Mu3 and N315 was performed. To increase metabolite coverage, we performed liquid chromatography–quadruple time of flight LC–MS (LC–QTOF–MS) and hydrophilic interaction–liquid chromatography (HILLIC–LC–QTOF–MS) as previously described.36,37 The intracellular metabolites were obtained at mid-exponential phase in trypticase soy broth. The supernatants were used for chromatographic analysis (five biological replicates per sample). The compounds were analysed and inspected manually using Agilent MassHunter Profinder B08.0. The data were filtered for presence in all samples and reproducibility by determining the coefficient of variation (CV) in the quality control samples, and molecular features with CV > 20% were removed.
The comparisons between groups were performed using multivariate (MVA) and uni-variate (UVA) analysis using SIMCA 16.0 (Umetrics, Umea, Sweden) and MatLab (R2019b, Mathworks, Inc., Natick, USA), respectively. The P value in UVA was determined using Mann–Whitney U-test with post hoc false discovery rate correction. MVA consisted of a principal component analysis and a supervised OPLS-DA model. Statistically significant metabolites were defined as follows: UVA, those with P ≤ 0.05 and in MVA, those with variance important in projection (VIP) 0 > 1 with Jack-knife confident interval (JK) not including 0.38 Statistically significant metabolites were annotated using Molecular Formula Generator, and CEU Mass Mediator tool,39 which was later used to compare against HMDB (http://hmdb.ca), LipidMaps (http://lipidMAPS.org), Metlin (http://metlin.scripps.edu) and KEGG databases.40 Significant metabolites were further analysed by MS/MS analysis to confirm the metabolite’s identity. Enrichment of metabolic pathways among identified metabolites was performed using MetaboAnalyst v.5.0.41
Adherence assay
Biofilm production was determined for the 39 hVISA strains and 38 selected VSSA isolates using a previously described42 with minor modifications. Briefly, 195 μL of BHI broth and 5 μL of a bacterial suspension (0.5 McFarland standard) were dispensed into sterile plastic 96-well plate. Plates were incubated for 24 hours at 37°C and washed three times with sterile saline (0.89%). Adherent bacteria were fixed with methanol 99% and stained with crystal violet. Elution of the stain was performed with glacial acetic acid (30%). The OD of each well was measured at 570 nm using a TECAM reader.
Results
hVISA strains are phylogenetically associated with their MRSA lineage rather than the resistance phenotype
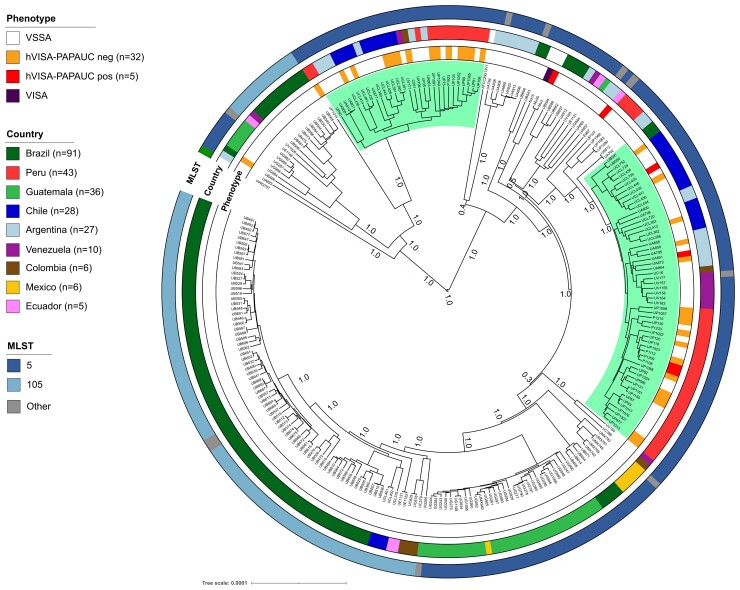
From a phylogenetic tree including the 344 genomes and eight reference genomes (Figure S2), we did not observe specific genetic lineages associated with the hVISA phenotype. However, further exploration of the phylogenetics including only CC5 genomes (257; including 37 out of 39 hVISA) showed that hVISA strains were separated in two different subclades (Figure 1). The first clade included 30 genomes, 13 with hVISA phenotype, while the second clade included 58 genomes, 21 hVISA, including those confirmed by PAP/AUC. Therefore, our findings suggest that there are not evident genomic traits potentially associated with the hVISA phenotype.
Figure 1.
Phylogeny of CC5 MRSA isolates. Bayesian phylogenomic tree using the core genome of 257 Clonal complex 5 (CC5) genomes. Highlighted branches represent two groups of hVISA isolates. Annotation rings (from inner to outer) show the vancomycin phenotype, the country and the ST of the isolate. Bootstrap values are showed in each branch. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
Polymorphisms in predicted proteins associated with carbohydrate metabolism, transport systems and virulence in the hVISA phenotype
To assess the genomic features possibly associated with the hVISA phenotype, we sought to identify SNPs perhaps associated with the hVISA phenotype by two alignment free genomic approaches: LDA and GWAS. The first was based on differences in the proportion of SNPs between 127 VSSA and 39 hVISA ST5 isolates (N315 as reference). We identified a total of 19 904 SNPs, from which 986 had a difference in proportion higher than 30% among the groups of strains (VSSA and hVISA) and were located in 286 coding regions and in 93 non-coding regions, which allowed the LDA to separate the phenotypes, except for one VSSA isolate that grouped with the hVISA (Figure S3a, Table S3). The SNPs with >50% difference in proportion in hVISA strains (n = 11) were present in 11 CDS annotated as related to amino acid metabolism (n = 3), carbohydrate metabolism (n = 2) and one of the following functional groups: nitrogen metabolism, virulence, cell wall catabolism, metal transport, a phage and a hypothetical ATP binding protein (Figure S3b and Table S4).
The GWAS analysis included all the genomes as for LDA, however, the results were aligned to the closed genomes of hVISA UCL420, UA851 and UE1097. We identified 4130 statistically significant k-mers of different lengths, which were mapped over 162 genomic regions (115 coding and 47 non-coding) of the N315, UCL420, UE1097 and UA851 genomes (Figure S4). The coding regions where k-mers aligned were annotated as hypothetical proteins (n = 32), mobile elements (n = 29), nucleic acid metabolism (n = 18), virulence elements (n = 16), transport elements (n = 8), amino acid metabolism (n = 4), transcriptional regulators (n = 3), carbohydrate metabolism (n = 3) and phage related (n = 2) (Table S5). Finally, by comparing the results from both approaches, we identified 11 common coding regions. Three of them were related to virulence, three to transport systems, two related to carbohydrate metabolism, two with unknown function and one associated to tRNA synthesis (Table 1).
Table 1.
Annotations of coding regions identified both by GWAS and LDA approaches
| Functional category | Gene name | Annotation |
|---|---|---|
| Virulence | fnbA | Fibronectin binding protein FnbA |
| sdrC | MSCRAMM family adhesin SdrC | |
| sdrE | MSCRAMM family adhesin SdrE | |
| Transport system | pstC | Phosphate ABC transporter permease subunit C |
| AID38744.1 | YjiH: uncharacterized membrane protein, contains nucleoside recognition GATE domain | |
| fhuC | FepC: ABC-type cobalamin/Fe3+-siderophores transport system | |
| Carbohydrate metabolism | hemL1 | Glutamate-1-semialdehyde 2,1-aminomutase |
| Amino acid metabolism | AID38605.1 | Has three different domains: amino acid adenylation domain, a peptide synthase and Lys2B |
| tRNA synthesis | truB | tRNA pseudouridine synthase B (PRK14123) |
| Unknown function | Hypothetical | Conserved hypothetical protein |
| AID39364.1 | Uncharacterized DUF86 domain-containing protein-like Bacillus subtilis YutE |
Amino acid substitutions in predicted regulatory systems proteins potentially associated with hVISA phenotype
In our previous study, we searched the genomes for the most common alterations in predicted proteins associated with the hVISA phenotype (WalKR, VraTSR, GraSR, RpoB, RpoC, TcaA and CapP).21 Nonetheless, many different alterations potentially associated with VISA/hVISA have also been documented in several studies. Therefore, we conducted a systematic literature review where we focused mostly on studies comparing hVISA and VSSA isogenic strains and those including mutagenesis experiments. We selected a total of 43 proteins related to hVISA phenotype and amino acid substitutions in these proteins were searched in the 344 MRSA genomes (Table S6). We detected a total of 2962 amino acid substitutions in 36 proteins that exhibited predicted functions associated with cell wall biogenesis, DNA/RNA processing and membrane biosynthesis. A total of 14 proteins presented amino acid changes that were statistically significant associated with either hVISA or VSSA (P ≤ 0,05) (Table S7). It is worth noting that Try-38 → His in Atl was identified in 92% of the hVISA isolates.
Subsequently, we aimed to determine the frequency of isolates of each phenotype (VSSA, hVISA PAP/AUC negative and PAP/AUC positive) that harbour the amino acid changes in proteins associated to hVISA. Overall, we identified that the most common changes in all hVISA isolates were mainly related to the autolysin (Atl), as well as the two regulatory systems associated to stress cell wall response; WalK and VraT, as described before (Figure S5).
Adhesion, pathogenesis and cell wall genes were differentially expressed in hVISA strains
Next, to evaluate possible differences in gene expression as a mechanism related to the development of hVISA phenotype, we performed an RNA-seq analysis of three hVISA confirmed by PAP/AUC (UCL420, UA851 and UE1097) compared to three VSSA isolates (UCL739, UA863 and UE218) and the reference strains Mu3 and N315 (Table 2).
Table 2.
Characteristics of pair of strains (hVISA and VSSA) selected for transcriptomic and metabolic analysis
| hVISA | VSSA | ||||||
|---|---|---|---|---|---|---|---|
| Strain | Clone-ST | Country | PAP/AUC | Strain | Clone-ST | Country | PAP/AUC |
| UCL420 | CHC-5 | Chile | 1.03 | UCL739 | CHC-5 | Chile | 0.26 |
| UA851 | CHC-5 | Argentina | 1.04 | UA863 | CHC-5 | Argentina | 0.34 |
| UE1097b | BR-1341a | Ecuador | 1.07 | UE218b | BR-239 | Ecuador | 0.39 |
| Mu3 | NYJ-5 | Japan | — | N315 | BR-5 | Japan | 0.35 |
CHC: Chilean/Cordobes; BR: Brazilian; NYJ: New York/Japan.
Single locus variant of ST239.
Strains UE1097 and UE218 were not included in metabolic analysis.
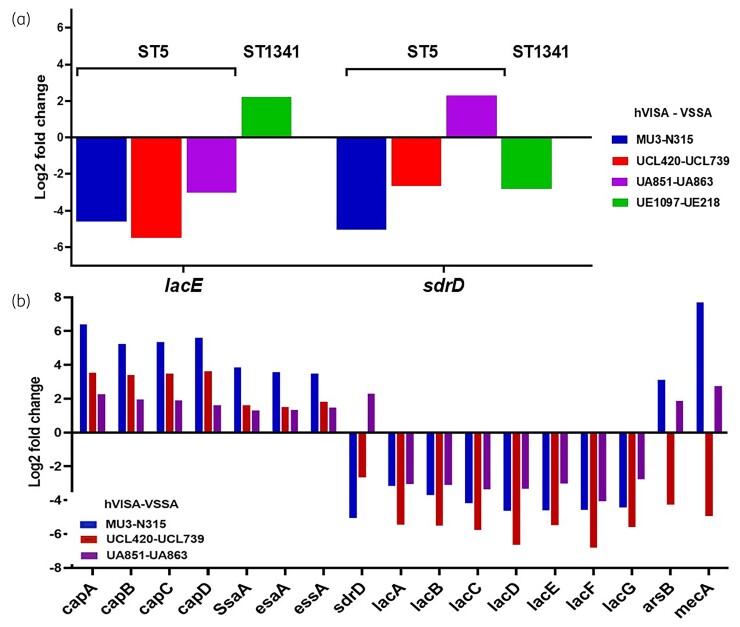
First, we compared the results of each hVISA strain with VSSA isolates belonging to the same sequence type (ST) and phylogenetically related SNPs based in core genome analyses (Figure S1), for a total of four combinations (hVISA-VSSA pairs and reference strain). We identified between 189 and 502 statistically significant DEG, from which between 88 to 300 were downregulated genes and 101 to 202 were upregulated genes (P < 0.05) (Figure 2). The sdrD and lacE genes were differentially expressed in all four pairs (Figure 3a). SdrD, is a cell wall anchored protein, member of the serine-aspartate repeat family (SdrC, SdrD and SdrE), which are implicated to promote biofilm formation.43 Therefore, we tested the capability of biofilm formation of our isolates and we observed that the hVISA isolates were stronger biofilm producers than VSSA isolates (Figure S6), while the lacCDEFG operon is involved in the transport and metabolism of lactose and galactose from D-lactose-6-phosphate.44 In fact, these changes might be influenced by the genetic lineage.
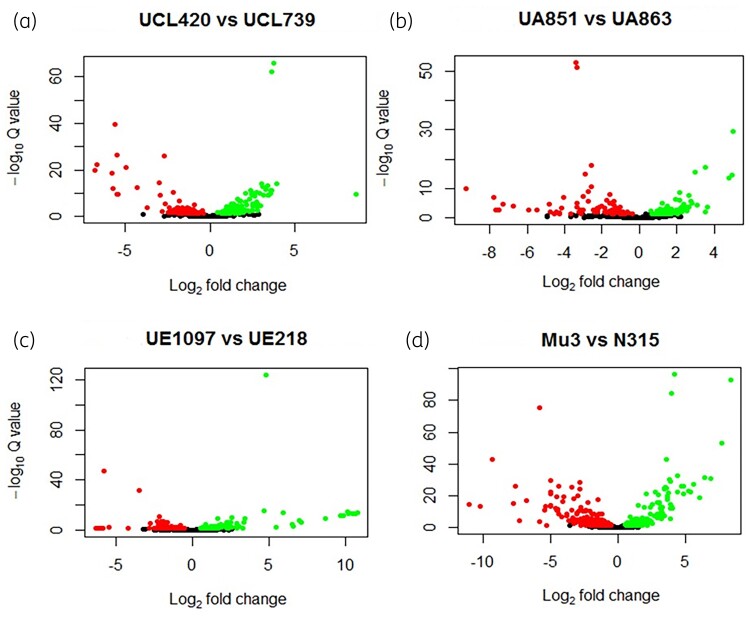
Figure 2.
Volcano plot of DEG between hVISA versus VSSA. The differential expressed genes (P value < 0.05) are represented for each pair of transcriptomes comparison (hVISA versus VSSA). Dots indicate genes downregulated or upregulated in hVISA when compared with VSSA. (a) UCL420 versus UCL739 pair showed a total of 283 DEG, 166 genes were downregulated and 117 upregulated in UCL420 (hVISA). (b) UA851 versus UA863 showed a total of 189 DEG; 88 genes were downregulated and 101 upregulated in UCL851 (hVISA). (c) UE1097 versus UE218 showed a total of 205 DEG; 91 genes were downregulated and 114 upregulated in UE1097 (hVISA). (d) Mu3 versus N315 showed a total of 502 DEG; 300 genes were downregulated and 202 upregulated genes in Mu3 (hVISA). This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
Figure 3.
DEG between hVISA and VSSA transcriptomes. (a) Two common DEG in the four hVISA/VSSA transcriptome pairs are showed. (b) A total of 21 genes in common for all the three comparisons were found; 17 out of the 21 are represented and the other four hypothetical proteins are not shown. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
In a second analysis, we restricted the comparison to the ST5 strains, including three hVISA strains (UCL420, UA851 and Mu3) and three VSSA strains (UCL739, UA863 and N315). We found 21 statistically significant DEG: the capABCD genes (capsular polysaccharides)45; the autolysin (ssaA); adhesin (sdrD), secretion-system (esaA, essA); membrane protein (ssaA); antimonite conferring resistance gene (arsB)46; the mecA, encoding the PBP2a47; lacABCDEF operon (Figure 3b) and four hypothetical proteins.
Further, an additional approach was used to compare the RNA-seq results of each strain against the N315 transcriptome and identify common DEG among hVISA no differentially expressed on VSSA. We identified 10 DEG exclusively in hVISA. Seven were overexpressed in hVISA, among them were: capE, esaD (Ess secretion-system),48phoH (phosphate regulon) and agrD (precursor of the intracellular adhesion polysaccharide). While only the ssl12 (Staphylococcus superantigen-associated protein), the glvC (phosphotransferase) and cidA (murein hydrolase) were under-expressed in hVISA.
In summary, we observed that genes related to cellular adhesion (sdrD), pathogenesis (capABCDE, esaA, essA, esaD) and cell wall recycling (ssaA) were overexpressed in hVISA isolates, while those related to lactose and galactose metabolism (lacABCDEFG) were downregulated.
Amino acid and carbohydrate metabolism is reduced in hVISA strains
We intended to identify metabolic pathways likely involved with the hVISA phenotype. Thus, we determined the metabolomic fingerprinting of two CC5 strains and their related VSSA, as well as the reference strains N315 and Mu3, identifying differential metabolic profiles potentially associated to the hVISA phenotype for these strains (Figure S7). We identified 104 metabolites with statistically significant differences between them (Table S8) by UVA and MVA. The results show that 61 of the 104 metabolites were annotated as fatty acids, followed by amino acids and nucleosides—cofactors with seven and six metabolites, respectively. Among the fatty acids, 35 were annotated as glycerophospholipids (major component of cell membrane lipids), and some signalling precursors and lipoproteins, from which 29 were more abundant among VSSA and 10 in hVISA. Additionally, we found that glycerolipids {such as diacylglycerol [DG(32:0) and DG(18:0)]} and glycerol were more abundant in hVISA than in their VSSA counterparts. In contrast, monoglycosyldiacylglycerol [MGDG(38:9)], which is involved in cell membrane synthesis, was 28% more abundant in VSSA.49
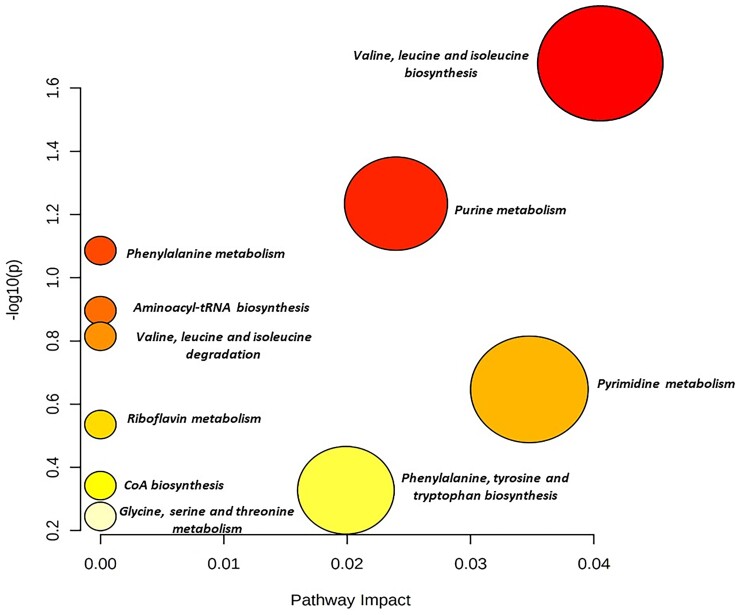
The amino acids valine, threonine, leucine and tyrosine were more abundant in VSSA than hVISA. These amino acids are synthesized from pyruvate and allow the synthesis of other aromatic amino acids such as tryptophan, phenylalanine and indole50 (Table S8). In contrast, arginine, glycine and betaine (serine and glycine derivative) were more abundant in hVISA. Additionally, nucleosides and FAD, NADH cofactors, were more abundant in VSSA. Enrichment analysis of the metabolite set allowed the identification of 10 altered metabolic pathways in hVISA strains, such as the purine and pyrimidine pathway, CoA biosynthesis (glycolysis), metabolism of amino acids such as phenylalanine, valine, leucine, glycine, serine, threonine and isoleucine and aminoacyl tRNA biosynthesis (Figure 4 and Figure S8).
Figure 4.
Metabolite set enrichment analysis. Metabolic pathways are represented as circles according to the scores obtained in the enrichment (y-axis) and topology (pathway impact, x-axis) analyses using MetaboAnalyst v.5.0. Darker circles indicate the most significant metabolite changes in each biochemical pathway. The size of the circle corresponds to the pathway impact score and correlates with the metabolites involved. Pathways were annotated by numbering when P values calculated from enrichment analysis were 0.05. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
Discussion
The molecular mechanism leading to the hVISA phenotype is not fully understood and previous descriptions show that multiple genetic factors might be involved.22 Some of the most frequently reported genes associated to VISA are those related to cell envelope regulatory systems (vraTSR, graSR and walKR), membrane-associated protein (tcaA), capsular polysaccharide proteins (capP) and the β-subunit of the RNA polymerase (rpoBC). Additionally, reversible phosphorylation and de-phosphorylation of proteins by Stk1/Stp1 and protease ClpP8,51–53 have been associated with the phenotype. Thus, we aimed to perform a broad omics analyses of hVISA strain recovered from a region where vancomycin use is rampant and little knowledge is known about this phenotype.
The hVISA phenotype has been described mainly in CC5 and ST239 isolates,12 which are also the most common lineages in hVISA from Latin America.21 Beyond the association of the hVISA phenotype to specific MRSA lineages, our findings show that this phenotype does not possess a conserved genetic and transcriptomic fingerprinting. However, applying multiomic approaches, we have been able to characterize the important pathways that might contribute to the S. aureus cellular adaptation.
Of note, 90% of the hVISA isolates belonging to the Chilean/Cordobes clone and ST5 exhibited changes mainly in the global regulators WalK and VraT. Many studies have reported that changes in WalK lead to low susceptibility to vancomycin22 particularly those affecting the WalK histidine kinase domain and, also, in the HAMP domain of the protein.54 On the other hand, VraTSR modulates at least 46 genes involved in cell wall biosynthesis,55,56 so the substitutions in VraT are associated with a decrease in vancomycin susceptibility.57 It has also been reported that alterations in cell wall and membrane biosynthesis are associated with low susceptibility to vancomycin and daptomycin.23,58 Indeed, the highest number of amino acid changes in our hVISA isolates were in predicted proteins involved in cell wall biogenesis. The substitution Tyr-38 → His in Atl, was present in 35 (92%) isolates and several other changes were observed in the autolysin LytM. Thus, it is tempting to speculate that an altered autolytic activity is a characteristic of our hVISA strains.
Our transcriptional analyses indicated that genes associated with cellular adhesion and capsule homeostasis were differentially expressed in our hVISA strains.8 We found that the capABCD gene cluster was differentially expressed in CC5 hVISA strains. Using GWAS and LDA, we identified SNPs and k-mers implicating fnbA and sdrCE (coding for fibronectin binding protein and microbial surface components recognizing adhesive matrix molecules and adhesins, respectively) in the hVISA phenotype.
Our metabolomic analysis suggested important metabolic adaptations of hVISA strains such as increased abundance of betaine (involved in glycine and glycerol synthesis) and decreased synthesis of aromatic amino acids such as tryptophan and phenylalanine, and other amino acids synthesized from pyruvate (valine, leucine and isoleucine). These adaptations are associated with alterations in the glycolysis pathway and in the TCA cycle. A reduced TCA activity in VISA strains has been observed,59 and it has been shown that altering the functionality of the TCA cycle reduces bacterial growth, increases antibiotic tolerance and favours biofilm formation.60 This alteration may be explained because TCA catabolic by-product intermediates such as ammonia (synthesized from arginine in urea cycle) may play a fundamental role in the regulation of pH homeostasis in early stages of formation.37
Our study has several limitations. The relatively small number of isolates for the transcriptomics and metabolomics assays and the inclusion of a reduced number of VSSA strains to identify amino acid changes related to hVISA phenotype, precludes robust generalizations. It is also important to note that metabolomics profiles might hide signals due to the redundancy within the bacterial metabolic pathways, in cases in which one pathway might be affected by others that could compensate for them. Additionally, we did not find a clear correlation between the different omics approaches because of the small number of hVISA strains and lack of clonal diversity of the sample (95% from CC5).
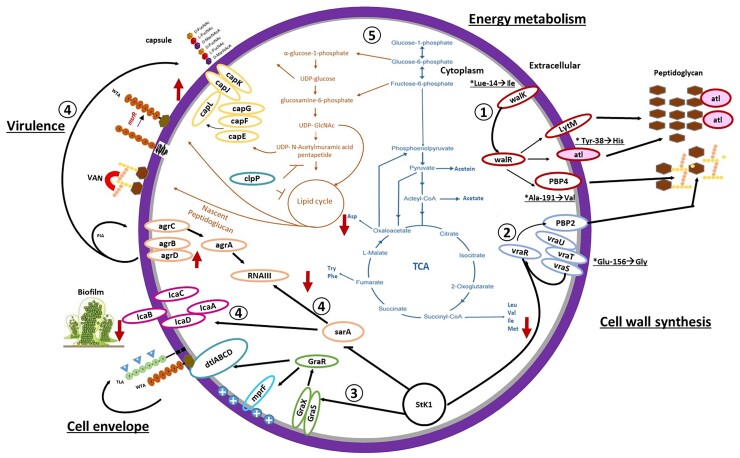
In summary, our description of the hVISA phenotype from 39 Latin American MRSA strains, support the notion of a multifactorial mechanism for the hVISA phenotype that involves: (i) major changes in bacterial cell wall synthesis via the WalKR and VraTSR regulatory systems regulating expression of proteins such as LytM, Atl, PBP4 and PBP2; (ii) increased biofilm formation (icaABCD) and expression of virulence factors [capsule (capABCDEFG), adhesins (sdrD)] by Agr and SarA and (iii) reduction in glycolysis via the TCA cycle (Figure 5).
Figure 5.
Schematic representation of the molecular changes of hVISA isolates belonging to ST5. (1) Regulation of the WalKR system on the bacterial wall synthesis proteins LytM, PBP4 and the autolysin Atl that presents Y38H change. (2) hVISA isolates present the E156G change in VraT of the VraTSR system that regulates PBP2 transpeptidase, essential in the synthesis of the bacterial wall. (3) Stk1 regulation of the GraSR system that regulates cell envelope cargo via the MprF and DtlABCD proteins. (4) The SarA regulator controls the expression of the IcaABCD operon (biofilm formation) and the expression of virulence factors, such as the capsule and proteins secreted by Agr. (5) reduction of amino acids involved in the glycolysis cycle and TCA that allows the synthesis of amino acids, lipids, purines and pyrimidines. (*) Predominant genetic changes in hVISA isolates. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
Ethics statement
Ethical approval was obtained from the Ethics Review Committee Universidad El Bosque (UEB 410–2016), Acta No. 012-2016.
Supplementary Material
Acknowledgements
We are indebted to Catalina Espitia and Angie Hernandez for technical assistance on the cellular preparation for metabolomic analysis, and Sebastian Solano and Aura M. Echeverri for technical assistant on sample preparation for RNA-seq analysis.
The following hospitals participated in the collection of isolates: Colombia; Bogotá: Fundación Salud Bosque; Hospital Simón Bolívar; Clínica de Occidente; Clínica del Niño; Clínica Shaio; Hospital El Tunal; Hospital Santa Clara; Hospital Occidente de Kennedy; Hospital Universitario Clínica San Rafael; Clínica San Pedro Claver; Hospital San Ignacio; Fundación Santa Fe de Bogotá; Clínica Infantil Colsubsidio; Clínica Saludcoop Jorge Piñeros Corpas; Instituto Nacional de Cancerología; Cali: Centro Médico Imbanaco; Clínica Saludcoop Occidente Cali; Hospital Universitario del Valle; Medellín: Hospital Pablo Tobón Uribe; Clínica Saludcoop Juan Luis Londoño; Bucaramanga: Clínica La Foscal; Fundación Cardiovascular; Neiva: Hospital Universitario Hernando Moncaleano; Pereira: Hospital Universitario San Jorge; Ecuador: Hospital Vozandes, Hospital Eugenio Espejo, Hospital Baca Ortiz, Hospital Carlos Andrade Marín, Hospital General de las Fuerzas Armadas, Hospital Carlos Andrade Marín; Perú: Hospital Guillermo Almenara; Hospital Alberto Sabogal Laboratorio Clínico Carlos Carrillo; Hospital Nacional Sergio Bernales; Instituto Nacional de Enfermedades Neoplásicas; Hospital Nacional Hipólito Unanue; Venezuela: Clínica La Floresta, Centro Médico de Caracas and Hospital Vargas de Caracas; Argentina; Buenos Aires: Hospital de Clínicas, Hospital Británico, Sanatorio Mater Dei; Brazil; São Paulo: Hospital do Servidor Publico Estadual de São Paulo; Chile; Santiago: Escuela de Medicina, Pontificia Universidad Católica de Chile; Hospital Sótero del Río; Concepción: Hospital Guillermo Grant Benavente; Guatemala, Hospital Roosevelt; Mexico, Hospital Civil de Guadalajara, Centro Universitario Ciencias de la Salud, Universidad de Guadalajara.
Contributor Information
Betsy E Castro, Molecular Genetics and Antimicrobial Resistance Unit, Universidad El Bosque, Bogota, Colombia.
Rafael Rios, Molecular Genetics and Antimicrobial Resistance Unit, Universidad El Bosque, Bogota, Colombia.
Lina P Carvajal, Molecular Genetics and Antimicrobial Resistance Unit, Universidad El Bosque, Bogota, Colombia.
Mónica L Vargas, Molecular Genetics and Antimicrobial Resistance Unit, Universidad El Bosque, Bogota, Colombia.
Mónica P Cala, Metabolomics Core Facility-MetCore, Vice-Presidency for Research, Universidad de los Andes, Bogotá, Colombia.
Lizeth León, Metabolomics Core Facility-MetCore, Vice-Presidency for Research, Universidad de los Andes, Bogotá, Colombia.
Blake Hanson, Center for Infectious Diseases, School of Public Health, University of Texas Health Science Center at Houston, Houston, TX, USA.
An Q Dinh, Center for Infectious Diseases, School of Public Health, University of Texas Health Science Center at Houston, Houston, TX, USA; Division of Infectious Diseases, Houston Methodist Hospital, Houston, TX, USA; Center for Research in Genetics and Genomics—CIGGUR, GENIUROS Research Group, School of Medicine and Health Sciences, Universidad Del Rosario, Bogotá, Colombia.
Oscar Ortega-Recalde, Center for Research in Genetics and Genomics—CIGGUR, GENIUROS Research Group, School of Medicine and Health Sciences, Universidad Del Rosario, Bogotá, Colombia.
Carlos Seas, Instituto de Medicina Tropical Alexander Von Humboldt, Universidad Peruana Cayetano Heredia, Lima, Peru.
Jose M Munita, Millennium Initiative for Collaborative Research on Bacterial Resistance (MICROB-R), Santiago, Chile; Genomics and Resistant Microbes (GeRM) Group. Clínica Alemana de Santiago, Universidad del Desarrollo School of Medicine, Santiago, Chile.
Cesar A Arias, Division of Infectious Diseases, Houston Methodist Hospital, Houston, TX, USA; Center for Infectious Diseases, Houston Methodist Research Institute, Houston, TX, USA.
Sandra Rincon, Molecular Genetics and Antimicrobial Resistance Unit, Universidad El Bosque, Bogota, Colombia.
Jinnethe Reyes, Molecular Genetics and Antimicrobial Resistance Unit, Universidad El Bosque, Bogota, Colombia.
Lorena Diaz, Molecular Genetics and Antimicrobial Resistance Unit, Universidad El Bosque, Bogota, Colombia; Millennium Initiative for Collaborative Research on Bacterial Resistance (MICROB-R), Santiago, Chile; Genomics and Resistant Microbes (GeRM) Group. Clínica Alemana de Santiago, Universidad del Desarrollo School of Medicine, Santiago, Chile.
Funding
This work was supported by Minciencias (COL130874455850) to L.D. and Universidad El Bosque, PCI 9510–2017 to L.P.C. C.A.A. is supported by the National Institute of Allergy and Infectious Diseases (NIAID) of the NIH (award numbers K24AI121296, R01AI134637, R01AI48342 and P01AI152999). B.H. is supported by the National Institute of Allergy and Infectious Diseases (NIAID) of the NIH (award number K01AI148593). J.M.M. is supported by grant FONDECYT regular Project No. 1171805, Ministry of Education, Government of Chile and the Millennium Science Initiative, Ministry of Economy, Development and Tourism, Chile.
Transparency declarations
C. A. A. has received grant support from Merck, MeMed Diagnostics and Entasis Therapeutics. All other authors have none to declare.
Supplementary data
Figures S1 to S8 and Tables S1 to S8 are available as Supplementary data at JAC Online.
References
- 1. Dayan GH, Mohamed N, Scully ILet al. Staphylococcus aureus: the current state of disease, pathophysiology and strategies for prevention. Expert Rev Vaccines 2016; 15: 1373–92. 10.1080/14760584.2016.1179583 [DOI] [PubMed] [Google Scholar]
- 2. Turner NA, Sharma-Kuinkel BK, Maskarinec SAet al. Methicillin-resistant Staphylococcus aureus: an overview of basic and clinical research. Nat Rev Microbiol 2019; 17: 203–18. 10.1038/s41579-018-0147-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lambert M. IDSA guidelines on the treatment of MRSA infections in adults and children. Am Fam Physician 2011; 84: 455–6. [Google Scholar]
- 4. Hiramatsu K, Aritaka N, Hanaki Het al. Dissemination in Japanese hospitals of strains of Staphylococcus aureus heterogeneously resistant to vancomycin. Lancet 1997; 350: 1670–3. 10.1016/S0140-6736(97)07324-8 [DOI] [PubMed] [Google Scholar]
- 5. McGuinness WA, Malachowa N, DeLeo FR. Vancomycin resistance in Staphylococcus aureus. Yale J Biol Med 2017; 90: 269–81. [PMC free article] [PubMed] [Google Scholar]
- 6. CLSI . Performance Standards for Antimicrobial Susceptibility Testing—Thirty-Second Edition: M100. 2022. [Google Scholar]
- 7. Lee MY, Lee WI, Kim MHet al. Etest methods for screening heterogeneous vancomycin-intermediate Staphylococcus aureus (hVISA) strains. Curr Microbiol 2020; 77: 3158–67. 10.1007/s00284-020-02123-y [DOI] [PubMed] [Google Scholar]
- 8. Howden BP, Davies JK, Johnson PDRet al. Reduced vancomycin susceptibility in Staphylococcus aureus, including vancomycin-intermediate and heterogeneous vancomycin-intermediate strains: resistance mechanisms, laboratory detection, and clinical implications. Clin Microbiol Rev 2010; 23: 99–139. 10.1128/CMR.00042-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kim T, Kim ES, Park SYet al. Phenotypic changes of methicillin-resistant Staphylococcus aureus during vancomycin therapy for persistent bacteraemia and related clinical outcome. Eur J Clin Microbiol Infect Dis 2017; 36: 1473–81. 10.1007/s10096-017-2956-1 [DOI] [PubMed] [Google Scholar]
- 10. Yang CC, Sy CL, Huang YCet al. Risk factors of treatment failure and 30-day mortality in patients with bacteremia due to MRSA with reduced vancomycin susceptibility. Sci Rep 2018; 8: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Howden BP, Johnson PDR, Ward PB. Isolates with low-level vancomycin resistance associated with persistent methicillin-resistant Staphylococcus aureus bacteremia. Antimicrob Agents Chemother 2006; 50: 3039–47. 10.1128/AAC.00422-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shariati A, Dadashi M, Al Met al. Global prevalence and distribution of vancomycin resistant, vancomycin intermediate and heterogeneously vancomycin intermediate Staphylococcus aureus clinical isolates: a systematic review and meta-analysis. Sci Rep 2020; 10: 1–16. 10.1038/s41598-019-56847-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang S, Sun X, Chang Wet al. Systematic review and meta-analysis of the epidemiology of vancomycin-intermediate and heterogeneous vancomycin-intermediate Staphylococcus aureus isolates. PLoS ONE 2015; 10: e0136082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Errecalde L, Ceriana P, Gagetti Pet al. [First isolation in Argentina of community-acquired methicillin-resistant Staphylococcus aureus with intermediate susceptibility to vancomycin and nonsusceptibility to daptomycin]. Rev Argent Microbiol 2013; 45: 99–103. [DOI] [PubMed] [Google Scholar]
- 15. Di Gregorio S, Perazzi B, Ordoñez AMet al. Clinical, microbiological, and genetic characteristics of heteroresistant vancomycin-intermediate Staphylococcus aureus Bacteremia in a teaching hospital. Microb Drug Resist 2015; 21: 25–34. 10.1089/mdr.2014.0190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Di Gregorio S, Fernandez S, Cuirolo Aet al. Different vancomycin-intermediate Staphylococcus aureus phenotypes selected from the same ST100-hVISA parental strain. Microb Drug Resist 2017; 23: 44–50. 10.1089/mdr.2016.0160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vega F, Domínguez M, Bello Het al. Isolation of Staphylococcus aureus hetero-resistant to vancomycin (hVISA) in the Regional Hospital of Concepción. Chile. Rev Chil Infectol 2015; 32: 588–90. 10.4067/S0716-10182015000600017 [DOI] [PubMed] [Google Scholar]
- 18. de Oliveira Silveira AC, Da Cunha GR, Caierão Jet al. Molecular epidemiology of heteroresistant vancomycin-intermediate Staphylococcus aureus in Brazil. Brazilian J Infect Dis 2015; 19: 466–72. 10.1016/j.bjid.2015.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Arias CA, Reyes J, Carvajal LPet al. A prospective cohort multicenter study of molecular epidemiology and phylogenomics of Staphylococcus aureus bacteremia in nine Latin American countries. Antimicrob Agents Chemother 2017; 61: e00816-17. 10.1128/AAC.00816-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Reyes J, Rincón S, Díaz Let al. Dissemination of methicillin-resistant Staphylococcus aureus USA300 sequence type 8 lineage in Latin America. Clin Infect Dis 2009; 49: 1861–7. 10.1086/648426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Castro BE, Berrio M, Vargas MLet al. Detection of heterogeneous vancomycin intermediate resistance in MRSA isolates from Latin America. J Antimicrob Chemother 2020; 75: 2424–31. 10.1093/jac/dkaa221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hu Q, Peng H, Rao X. Molecular events for promotion of vancomycin resistance in vancomycin intermediate Staphylococcus aureus. Front Microbiol 2016; 13:1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Howden BP, Peleg AY, Stinear TP. The evolution of vancomycin intermediate Staphylococcus aureus (VISA) and heterogenous-VISA. Infect Genet Evol 2014; 21: 575–82. 10.1016/j.meegid.2013.03.047 [DOI] [PubMed] [Google Scholar]
- 24. Overbeek R, Olson R, Pusch GDet al. The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res 2014; 42: D206–14. 10.1093/nar/gkt1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Page AJ, Cummins CA, Hunt Met al. ROARY: rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015; 31: 3691–3. 10.1093/bioinformatics/btv421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Edgar RC. Muscle: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 2004; 5: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Page AJ, Taylor B, Delaney AJet al. SNP-sites: rapid efficient extraction of SNPs from multi-FASTA alignments. Microb Genomics 2016; 2: e000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bouckaert R, Heled J, Kühnert Det al. Beast 2: a software platform for Bayesian evolutionary analysis. PLoS Comput Biol 2014; 10: 1–6. 10.1371/journal.pcbi.1003537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Letunic I, Bork P. Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 2019; 47: W256–9. 10.1093/nar/gkz239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Miller MA, Pfeiffer W, Schwartz T. The CIPRES science gateway: enabling high-impact science for phylogenetics researchers with limited resources. Proceedings of the 1st Conference of the Extreme Science and Engineering Discovery Environment: Bridging from the eXtreme to the Campus and Beyond2012: 1–8. [Google Scholar]
- 31. Kurtz S, Phillippy A, Delcher ALet al. Versatile and open software for comparing large genomes. Genome Biol 2004; 5: R12. 10.1186/gb-2004-5-2-r12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pedregosa F, Varoquaux G, Gramfort Aet al. Scikit-learn: machine learning in python. J Mach Learn Res 2011; 12: 2825–30. [Google Scholar]
- 33. Lees JA, Galardini M, Bentley SDet al. Pyseer: a comprehensive tool for microbial pangenome-wide association studies. Bioinformatics 2018; 34: 4310–2. 10.1093/bioinformatics/bty539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Magoc T, Wood D, Salzberg SL. EDGE-pro: estimated degree of gene expression in prokaryotic genomes. Evol Bioinform Online 2013; 9: 127–36. 10.4137/EBO.S11250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014; 15: 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Meyer H, Liebeke M, Lalk M. A protocol for the investigation of the intracellular Staphylococcus aureus metabolome. Anal Biochem 2010; 401: 250–9. 10.1016/j.ab.2010.03.003 [DOI] [PubMed] [Google Scholar]
- 37. Stipetic LH, Dalby MJ, Davies RLet al. A novel metabolomic approach used for the comparison of Staphylococcus aureus planktonic cells and biofilm samples. Metabolomics 2016; 12: 1–11. 10.1007/s11306-016-1002-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rubingh CM, Bijlsma S, Derks EPPAet al. Assessing the performance of statistical validation tools for megavariate metabolomics data. Metabolomics 2006; 2: 53–61. 10.1007/s11306-006-0022-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gil-De-La-Fuente A, Godzien J, Saugar Set al. CEU mass mediator 3.0: a metabolite annotation tool. J Proteome Res 2019; 18: 797–802. 10.1021/acs.jproteome.8b00720 [DOI] [PubMed] [Google Scholar]
- 40. Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes progression. Nucleic Acids Res 2000; 28: 3316–32. 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pang Z, Chong J, Zhou Get al. Metaboanalyst 5.0: narrowing the gap between raw spectra and functional insights. Nucleic Acids Res 2021; 49: W388–96. 10.1093/nar/gkab382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stepanović S, Vuković D, Dakić Iet al. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J Microbiol Methods 2000; 40: 175–9. 10.1016/S0167-7012(00)00122-6 [DOI] [PubMed] [Google Scholar]
- 43. Ajayi C, Åberg E, Askarian Fet al. Genetic variability in the sdrD gene in Staphylococcus aureus from healthy nasal carriers. BMC Microbiol 2018; 18: 1–13. 10.1186/s12866-018-1179-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rosey EL, Oskouian B, Stewart GC. Lactose metabolism by Staphylococcus aureus: characterization of lacABCD, the structural genes of the tagatose 6-phosphate pathway. J Bacteriol 1991; 173: 5992–8. 10.1128/jb.173.19.5992-5998.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rausch M, Deisinger JP, Ulm Het al. Coordination of capsule assembly and cell wall biosynthesis in Staphylococcus aureus. Nat Commun 2019; 10:1404. 10.1038/s41467-019-09356-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ji G, Silver S. Regulation and expression of the arsenic resistance operon from Staphylococcus aureus plasmid pI258. J Bacteriol 1992; 174: 3684–94. 10.1128/jb.174.11.3684-3694.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Reichmann NT, Pinho MG. Role of SCCmec type in resistance to the synergistic activity of oxacillin and cefoxitin in MRSA. Sci Rep 2017; 7: 1–9. 10.1038/s41598-017-06329-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Anderson M, Chen YH, Butler EKet al. Esad, a secretion factor for the Ess pathway in Staphylococcus aureus. J Bacteriol 2011; 193: 1583–9. 10.1128/JB.01096-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hewelt-Belka W, Nakonieczna J, Belka Met al. Untargeted lipidomics reveals differences in the lipid pattern among clinical isolates of Staphylococcus aureus resistant and sensitive to antibiotics. J Proteome Res 2016; 15: 914–22. 10.1021/acs.jproteome.5b00915 [DOI] [PubMed] [Google Scholar]
- 50. DeMars Z, Bose JL. Redirection of metabolism in response to fatty acid kinase in Staphylococcus aureus. J Bacteriol 2018; 200: 1–17. 10.1128/JB.00345-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang Y, Li X, Jiang Let al. Novel mutation sites in the development of vancomycin- intermediate resistance in Staphylococcus aureus. Front Microbiol 2017; 7: 2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Matsuo M, Cui L, Kim Jet al. Comprehensive identification of mutations responsible for heterogeneous vancomycin-intermediate Staphylococcus aureus (hVISA)-to-VISA conversion in laboratory-generated VISA strains derived from hVISA clinical strain Mu3. Antimicrob Agents Chemother 2013; 57: 5843–53. 10.1128/AAC.00425-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cheung A, Duclos B. Stp1 and Stk1: the yin and yang of vancomycin sensitivity and virulence in vancomycin-intermediate Staphylococcus aureus strains. J Infect Dis 2012; 205: 1625–7. 10.1093/infdis/jis255 [DOI] [PubMed] [Google Scholar]
- 54. Cui L, Neoh HM, Shoji Met al. Contribution of vraSR and graSR point mutations to vancomycin resistance in vancomycin-intermediate Staphylococcus aureus. Antimicrob Agents Chemother 2009; 53: 1231–4. 10.1128/AAC.01173-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Qureshi NK, Yin S, Boyle-Vavra S. The role of the staphylococcal VraTSR regulatory system on vancomycin resistance and vanA operon expression in vancomycin-resistant Staphylococcus aureus. PLoS ONE 2014; 15: e85873. 10.1371/journal.pone.0085873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pechous R, Ledala N, Wilkinson BJet al. Regulation of the expression of cell wall stress stimulon member gene msrA1 in methicillin-susceptible or -resistant Staphylococcus aureus. Antimicrob Agents Chemother 2004; 48: 3057–63. 10.1128/AAC.48.8.3057-3063.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Boyle-Vavra S, Yin S, Jo DSet al. Vrat/YvqF is required for methicillin resistance and activation of the VraSR regulon in Staphylococcus aureus. Antimicrob Agents Chemother 2013; 57: 83–95. 10.1128/AAC.01651-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mishra NN, Bayer AS, Weidenmaier Cet al. Phenotypic and genotypic characterization of daptomycin-resistant methicillin-resistant Staphylococcus aureus strains: relative roles of mprF and dlt operons. PLoS ONE 2014; 9: e107426. 10.1371/journal.pone.0107426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gardner Stewart G, Marshall Set al. Metabolic mitigation of Staphylococcus aureus vancomycin intermediate-level susceptibility. Antimicrob Agents Chemother 2017; 62: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Nelson JL, Rice KC, Slater SRet al. Vancomycin-intermediate Staphylococcus aureus strains have impaired acetate catabolism: implications for polysaccharide intercellular adhesin synthesis and autolysis. Antimicrob Agents Chemother 2007; 51: 616–22. 10.1128/AAC.01057-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.