Abstract
Malignancies involving the central nervous system present unique challenges for diagnosis and monitoring due to the difficulties and risks of direct biopsies and the low specificity and/or sensitivity of other techniques for assessment. In recent years, liquid biopsy of the cerebrospinal fluid (CSF) has emerged as a convenient alternative that combines minimal invasiveness with the ability to detect disease-defining or therapeutically actionable genetic alterations from circulating tumor DNA (ctDNA). Since CSF can be obtained by lumbar puncture, or an established ventricular access device at multiple time points, ctDNA analysis enables initial molecular characterization and longitudinal monitoring throughout a patient's disease course, promoting optimization of treatment regimens.
This review outlines some of the key aspects of ctDNA from CSF as a highly suitable approach for clinical assessment, the benefits and drawbacks, testing methods, as well as potential future advancements in this field. We anticipate wider adoption of this practice as technologies and pipelines improve and envisage significant improvements for cancer care.
Graphical abstract
Introduction
The acquisition of somatic genetic alterations in cancer forms the basis of genomic testing in oncology, by determining critical aspects of a tumor's biology that may have diagnostic and prognostic relevance, permitting disease monitoring and, most importantly, directing therapeutic decision-making [1], [2], [3]. Traditionally, the clinical approach has been to directly test the DNA from biopsied or resected tumor samples, but this is uniquely challenging for cancers involving the central nervous system (CNS). Brain biopsies carry inherent risks that can preclude the acquisition of ample tissue for initial molecular characterization and subsequent monitoring, resulting in a compelling demand for alternative testing approaches. With the advent of liquid biopsies, molecular testing of cancer patients now includes analysis of biofluids from accessible compartments [4]. Relevant analytes within these fluids include circulating tumor DNA (ctDNA), circulating tumor cells, RNAs, proteins and extracellular vesicles [5]. The obvious advantages of liquid biopsy are the ease of specimen procurement compared to invasive surgery, the ability to capture a more comprehensive molecular profile of the tumor with less sampling bias, and the capacity for dynamic longitudinal monitoring. Biofluids that are being actively studied in this context, and proposed for routine clinical utilization, include plasma and cerebrospinal fluid (CSF) [6], [7], [8], [9], [10], [11], [12].
For CNS tumors, genomic testing of plasma has proven to be highly insensitive [9,[13], [14], [15], [16]]. The blood-brain barrier restricts the transport of molecules between the CNS and the peripheral blood and severely limits the detection of cancer-related biomarkers in plasma. By contrast, CSF has unique attributes that are beneficial for genetic assessment. Through close contact with the brain and spinal cord, the CSF is recipient to a wide range of products shed or secreted by cells. The very low cellularity of CSF limits the dilution of ctDNA by genomic DNA from non-neoplastic cells and leads to higher variant allelic frequencies (VAF) with greater sensitivity to detect ctDNA in primary brain tumors compared to plasma [9,[13], [14], [15]]. In regard to metastatic cancers to the CNS, which are approximately tenfold more common than primary CNS tumors [17], the results appear better than those of primary brain malignancies, partly because of a more aggressive biology than lower grade primary brain tumors, but also due to their direct contact with CSF in cases of leptomeningeal dissemination [7,12]. Furthermore, given the protection afforded by the blood brain barrier, sampling of CSF may provide information that is distinct from non-CNS biofluids or peripheral tissues, particularly in the context of variable penetration of targeted therapies and in tumors that may exhibit specific selective pressures as they survive in their protective environment [18,19]. All of these features make CSF liquid biopsies an ideal substrate for detecting and tracking molecular alterations across a wide variety of CNS malignancies and may enhance clinical decision-making and therapeutic options for patients when used in conjunction with other available clinical data.
Cerebrospinal fluid as a source for ctDNA testing
CSF is chiefly produced by the choroid plexus within the ventricles of the brain (400–600 mL daily in the adult) and is estimated to be exchanged 3–5 times per day [20]. Its flow is pulsatile, coursing through the ventricular system with multidirectional movement to the subarachnoid space. Absorption occurs via the arachnoid granulations and lymphatic channels within the dural meninges and cranial/ spinal nerve sheaths, wherein it is directed to head, cervical and peri‑spinal lymph nodes [21], [22], [23]. CSF mixes with interstitial fluid within the CNS parenchyma and at the interface with blood vessels; CSF may therefore collect ctDNA through direct tumor contact when there is leptomeningeal or vascular involvement and potentially through its relationship with interstitial fluid [24], [25], [26]. Thus, ctDNA that is secreted from living cells, or released from dying tumor cells in the CNS parenchyma or its coverings, may enter into the CSF and be readily sampled by lumbar puncture (LP) of the thecal sac or a ventricular access device (e.g. Ommaya ventricular port) [27]. Although genetic alterations from both leptomeningeal disease and intraparenchymal tumors may be detected in the CSF, success rates are greater when there is leptomeningeal or direct ventricular involvement [7,12,[28], [29], [30], [31]]. Similar to observations in plasma ctDNA studies, analysis of ctDNA from CSF allows a more comprehensive assessment of the genomic landscape of heterogeneous tumors compared to direct tissue sampling of a lesion, which may reflect only focal changes [6,9,[32], [33], [34], [35]].
The unique composition of CSF provides distinct advantages for ctDNA testing over other body fluids. Under normal conditions, CSF is paucicellular, with a leukocyte count of less than 5 cells per mm3. CSF therefore has minimal total cell-free DNA (cfDNA), unlike plasma which receives a large contribution from normal turnover of hematopoetic cells. As a result, in neoplastic conditions where ctDNA is shed by the tumor, ctDNA in CSF will appear highly enriched and undiluted, compared to ctDNA in plasma, where it may constitute only a minor proportion in a sea of total cfDNA [13]. Tumor-derived genetic changes can be detected at much higher variant allelic fractions (VAF) than plasma and thus can be captured using routine NGS assays and pipelines, often without the need for very high read depths or other significant modifications as would be needed for plasma. Furthermore, given that CSF usually has negligible cfDNA, the amount of ctDNA in CSF can reflect the severity of CNS tumor burden, with greater ctDNA levels portending a worse prognosis [36,37].
Given the ease of accessibility, and the rapid replenishment of CSF, CSF sampling enables longitudinal and regular monitoring of CNS tumors. For instance, glioblastomas invariably recur after resection with evolution of the mutational profile, yet only 10–25% of patients are re-sampled at recurrence [37], [38], [39], [40], [41], [42] and remain genomically uncharacterized after initial surgery. While ctDNA analysis from CSF in patients with high grade gliomas is not currently the standard of care, tracking the genomic changes within CSF by measuring ctDNA levels, mutational profile and corresponding VAF of glioma patients after resection may predict disease progression [37,39]. More studies are warranted in this area, however longitudinal molecular surveillance using ctDNA from CSF could circumvent ambiguities in imaging interpretation and the need for risky tissue re-biopsies.
Technically, CSF collection is a relatively safe and easy procedure to perform with few, but notable, potential complications [43]. The most common complications of a lumbar puncture (LP) include short-term post procedure headache, experienced by 10–30% of patients, and variable discomfort or pain. Less commonly, spinal hematoma, particularly in patients who are anticoagulated or are thrombocytopenic, and infection may be encountered. In patients with abnormally high intracranial pressure from mass effect, edema, or obstructive hydrocephalus, a lumbar puncture may lead to cerebral herniation and is therefore a relative procedural contraindication. A careful clinical evaluation is important to prevent or mitigate these complications. The use of image guided techniques, such as ultrasound, fluoroscopy, or computer tomography, may be highly valuable to reduce the number of attempts, decrease chances of bloody taps and reduce the time and discomfort during the procedure, particularly for more challenging cases such as pediatric patients, individuals with prior spine surgeries, those with high body mass index, scoliosis or significant degenerative disease.
Advantages of CSF cfDNA-based assays over conventional CSF analyses
Historically, CSF analysis has been an integral part of the assessment of patients with suspected CNS malignancy. For solid tumors, cytologic analysis is considered the gold standard for diagnosing malignancy, a process that relies on the morphologic identification of the neoplastic cells in the fluid. In some cases, detection of abnormal CSF protein markers may also be used as an adjunct to cytology, such as α-fetoprotein, β-HCG, PLAP and CEA, in CNS germ cell tumors. For hematologic malignancies, including primary CNS lymphomas or leukemic involvement, both cytology and flow cytometry can be combined to assist in diagnosis. The overall utility of CSF for diagnosing malignancy, however, has suffered from several limitations. A major weakness is that diagnosis has relied on the shedding of malignant cells in CSF, which may be intermittent and in very low quantity. Cytologic interpretation of the morphologic features as strictly malignant may be highly subjective and influenced by differences in fixation and stability of the sample. False negative cytopathology is exceedingly common, with reported sensitivity as low as 45%, even with repeated sampling [44]. In the context of this limited cell yield, molecular and other ancillary studies may also be precluded to aid in the diagnosis and monitoring.
One of the advantages of analyzing cfDNA is that genetic alterations may be detected even when CSF shows benign or negative cytologic findings. In our hands, using a clinical validation cohort of 148 CSF samples submitted for suspicion of CNS malignancy, we compared the performance of ctDNA with the corresponding cell pellet from the same collection. Comprehensive sequencing by MSK-IMPACTTM, a targeted hybrid-capture NGS panel, demonstrated detection of somatic alterations in 71% of all ctDNA samples that were successfully sequenced (72%, n = 106). Among the genetically positive cases with concurrent cytologic assessment, one third had negative/benign morphologic findings (most commonly primary brain tumors). Additionally, sequencing of cfDNA from CSF consistently detected mutations in samples with positive cytology (42/45, 93%) [45]. Overall, ctDNA demonstrated superior sequencing performance compared to genomic DNA from the cell pellet with a 1.6-fold higher mutational rate and three times greater mean VAF in direct comparisons. Other groups have demonstrated similar superior sensitivity of cancer detection using ctDNA testing of CSF over standard CSF cytologic approaches [29,46], however the clinical implications of this, and specifically whether the presence of CSF ctDNA is representative of leptomeningeal dissemination of disease, is uncertain and an area of active research.
Principles and methodologies for CSF ctDNA testing
For diagnostic purposes, ctDNA is highly valuable to characterize tumor-specific mutations, copy number and structural alterations. For primary CNS tumors, profiling ctDNA alone may facilitate the molecular diagnosis and subclassification of diffuse gliomas by the genomic analysis of key genetic alterations in IDH1, IDH2, TP53, ATRX, TERT, H3F3A and HIST1H3B. Similarly, detection of specific mutations and fusions may aid in the diagnosis of metastatic tumors and provide a target for treatment. For instance, the diagnosis of leptomeningeal lung metastasis during the early stages of involvement may be challenging given the limited sensitivity of CSF cytology and specificity of MRI [47]. The finding of specific mutations in EGFR, BRAF, HER2 or MET, and fusions involving ALK, RET, ROS and NTRK1/2/3 may facilitate and supplement the diagnosis, as well as qualify patients for treatment with United States Food and Drug Administration (FDA) approved agents [7,48,49]. Further, the identification of drug resistance mutations arising in patients being treated with targeted agents is of unique value, particularly considering the differential selective pressure of tumors in immune-privileged sites and the evolution of resistance mechanisms that are distinct from those identified in peripheral tissue sites [49,50]. The ability to monitor CSF regularly, may lead to the early identification of resistance mutations and allow the timely implementation of treatments tailored to the evolving disease. While in many cases, the genomic profile may not be tumor-specific, the detection of somatic mutations alone may provide sufficient evidence of involvement by a neoplastic process and enable personalized means of disease monitoring. Mutations may also be detected initially in tissue, subsequently quantified in the CSF, and then used for clinical diagnosis of CSF involvement and for monitoring tumor burden over the disease course [14,37,51].
Similar to plasma, cfDNA from CSF is highly fragmented and the amount shed by tumor may vary considerably, depending on the tumor location, the type of tumor, the extent of involvement and many other factors. Therefore, testing methodologies and assay designs must take into consideration the template fragmentation, particularly when designing amplicon-based assays, and technical finetuning to deliver robust results even in the context of very low nucleic acid yields.
As previously mentioned, in contrast to plasma, the total proportion of ctDNA in CSF is high compared to total cfDNA. This may allow testing even with assays that are not ultrasensitive, at least at the time of initial diagnosis. Two general strategies are commonly considered to study genomic alterations, one of which is the targeted approach in which single or a few key genetic alterations are queried. Targeted sequencing can be extremely sensitive and may be ideal for diagnosis and monitoring of genomic variants that are known. The second strategy is the broad approach, either using large genomic panels, whole exome or whole genome. The advantages of a comprehensive approach lie in the ability to identify novel changes that may occur during treatment, such as resistance mechanisms or evolution and emergence of other alterations. This method also allows for broad screening of a wide array of alterations from any malignancy that may involve the CNS. Disadvantages include typically longer turnaround times, lower overall sensitivity, which may not be suitable for assessment of certain alterations or for longitudinal monitoring and detection of very early recurrences. Depending on the intent of the assay, different technologies are available for use and are summarized in the methods section below.
Pre-analytic variables that influence successful ctDNA detection
How biofluid specimens are collected and handled are fundamental to the success of cfDNA assays. Numerous factors that increase degradation of ctDNA or dilute ctDNA are recognized in plasma, but unfortunately such data is lacking for ctDNA in CSF. We outline a list of pre-analytic issues (i.e. occurring prior to testing) that may affect the success of ctDNA assays in CSF.
Variables before collection: tumor type and location
The type of tumor and the proximity to the CSF space have important implications for the detection of genetic alterations in CSF. High grade primary brain tumors, such as glioblastoma, have better detection rates than low grade primary brain tumors [37]. This is likely due to greater release of ctDNA in tumors with higher proliferation rates, cellular turnover and necrosis but also that higher grade gliomas may have more mutations and copy number alterations that increase the chances of detection, particularly with NGS approaches. Several studies have also demonstrated that the proximity and direct accessibility of the tumor to the CSF space is an important factor in the ability to detect tumor derived DNA and that metastatic tumors with leptomeningeal involvement have higher ctDNA than primary brain tumors, allowing better detection of genomic alterations [7,12].
Collection: collection devices, CSF volume, and time interval from collection to sequencing
To date, there are no universally implemented guidelines for collection, storage or preparation of cfDNA from CSF and hence protocols highly reflect current experience and understanding of maintaining ctDNA stability in plasma. The half-life of ctDNA from CSF is unknown and comprehensive stability studies have not been published.
The half-life of ctDNA in plasma ranges between 16 min and 2.5 h and is likely due to degradation by nucleases within the bloodstream and renal clearance. In addition to this limited stability in circulation, there is also dilution that limits overall assessment. Both genomic and cfDNA released from non-neoplastic cells, such as hematopoietic cells in blood, dilute ctDNA, challenging the detection of mutations which may be present below 1% VAF. In the CSF, variable proportions of lymphocytes will be present and, depending on sample procurement, there may be high contamination of cells from blood, albeit significantly lower than that seen in peripheral blood samples. Regardless of the degree of contamination or number of lymphocytes, similar principles of collection that apply to peripheral blood, also apply to the CSF. Preservatives in a variety of collection tubes are used to stabilize ctDNA and limit the release of genomic DNA from non-neoplastic cells. The time between collection to analysis should also be minimized, in accordance to the stability criteria established for the type of collection tube utilized [52]. Streck and CellSave tubes, for instance, have been shown to stabilize ctDNA and minimize release of genomic DNA more effectively than EDTA tubes over longer periods of time, up to approximately 7 days [53]. If standard K2EDTA tubes are used, yields are comparable in the first 48–72 h. In the absence of stabilization media, another alternative is to separate the ctDNA fraction from the cellular fraction by centrifugation within 6 h [53,54].
The overall volume utilized for isolation of ctDNA is an important factor in the total yield. Various studies have shown successful genomic profiling from ctDNA isolated from 2 to 3 mL of fluid, similar to the volumes generally collected for standard cytologic assessment [10,55]. Specific requirements for testing remain assay-dependent and may be higher if very high sensitivity is sought. When needed, greater volumes could be safely obtained to increase total DNA yield and match the necessary input for various assays and specific technologic and performance requirements.
DNA extraction methods
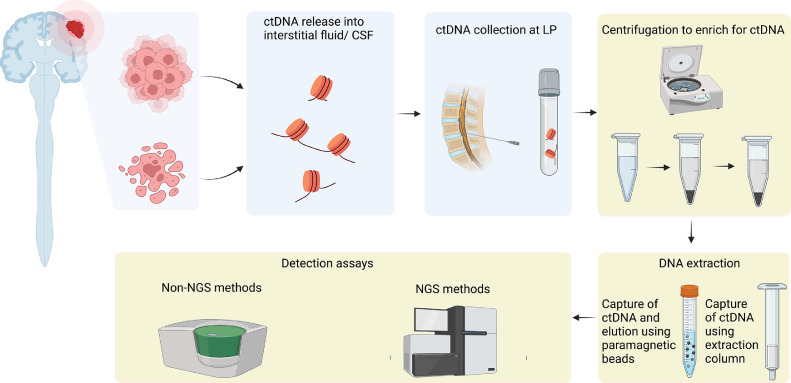
Similar to cfDNA isolation from plasma, the process generally involves initial double centrifugation of the sample to separate cellular and fluid components (Fig. 1): the first spin removes the cellular fraction at a low speed to minimize unwanted release of genomic DNA; the supernatant is subsequently spun at a higher speed to remove any remaining cellular debris [45,56]. Methods to extract cfDNA vary and may apply different principles including binding of DNA molecules to magnetic beads, organic chemicals or silica gel membranes. This may lead to high variability with respect to recovery efficiency, fragment profile and performance. Commercially available kits, such as the MagMax Cell-Free DNA Isolation Kit (Thermo Fisher Scientific, Waltham, MA) rely on cfDNA binding to silica paramagnetic beads [8,45] and have been used effectively. An alternative approach is the use of spin-column methods (minicolumn), such as the QIAamp Circulating Nucleic Acid Kit (Qiagen GmbH, Hilden, Germany), which binds nucleic acids to immobilized silica in the column. Hickmann and colleagues report that this kit provides better results than the Polymer Mediated Enrichment (PME) free circulating DNA Extraction Kit (Analytik Jena, Germany), which captures cfDNA using polymer beads [56]. At this time, the most optimal approach is not well established, highlighting the importance of an adequate validation process for a method that suits the needs of the study in question.
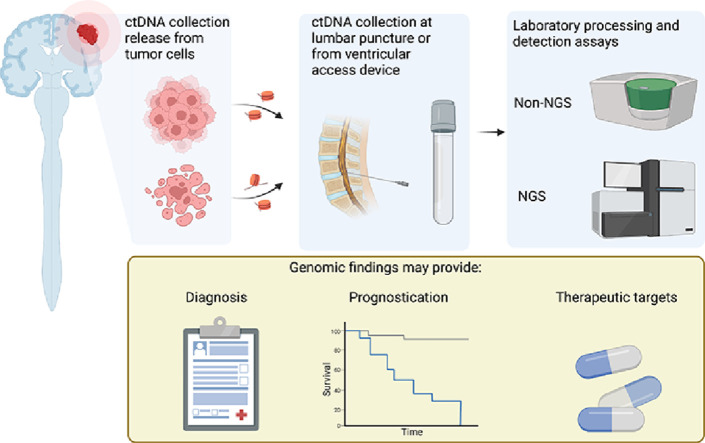
Fig. 1.
Release of ctDNA from tumor cells occurs upon cell death as well as being secreted from tumor cells. ctDNA that enters CSF can then be collected from a ventricular access device or at lumbar puncture. Following DNA extraction, and further processing, specific mutations can be detected using non-NGS approaches or broader mutational profiles using NGS assays. Created with BioRender.com.
Genomic assays
Assays commonly utilized fall into two principal categories: targeted PCR-based non-NGS assays and massive parallel sequencing, which are discussed below and summarized in Fig. 2. Essentially, any method that is used for ctDNA profiling in plasma may also be used for CSF testing. Depending on the technique utilized, analysis may go far beyond single nucleotide substitution detection to encompass detection of alterations in gene copy number, structural rearrangements, analysis of methylation patterns, calculations of tumor mutation burden, analysis of mutational signatures, and assessment of tumor heterogeneity through analysis of allelic fractions. The assays can be optimized to allow high sensitivity, however, the ultimate sensitivity achieved is dependent on the amount of DNA input of the assay. For instance, although many assays for detection of variants in cfDNA quote sensitivities as low as 0.01% and beyond, the achievement of this sensitivity requires the availability of an effective template of more than 1000 genome equivalents for detection (∼10 ng of DNA).
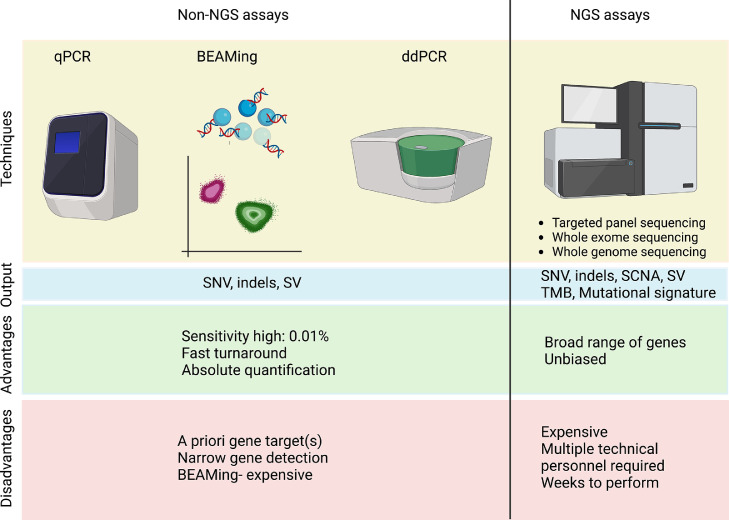
Fig. 2.
Outline of various assays for ctDNA detection in CSF. These can be divided into non-NGS and NGS-based assays. Non-NGS methods are limited by few specific gene targets per test, but benefit from higher sensitivity and faster turnaround than NGS tests. NGS technologies offer the greatest breadth for profiling CSF mutations. SNV- single nucleotide variant, SCNA- somatic copy number alteration, SV- structural variant, TMB- tumor mutational burden. Created with BioRender.com.
Non-NGS approaches
Several PCR-based technologies have been utilized for assessment of ctDNA from CSF including real time PCR, digital PCR, ARMS (amplification refractory mutation system- not discussed here) and BEAMing (beads, emulsion, amplification, and magnetics).
Real-time PCR (qPCR)
Real-time PCR, also known as quantitative PCR, monitors the amplification of a target PCR product per amplification cycle and allows absolute quantification of the product. The limit of detection of qPCR can be as low as 0.01%. In 2014, vu-Han and colleagues utilized custom-designed real time-PCR probes to target SMARCB1 in CSF samples of individuals with atypical teratoid rhabdoid tumors and without germline SMARCB1 mutations [57]. From seven patient samples, the group identified the specific mutations from the tumor samples using multiplex ligand-dependent probe amplification/ primer-walking PCR and then Sanger sequencing. Once these mutations were determined per patient, tumor-specific primers were designed, tested for specificity and the most optimal primers chosen for real-time PCR. Although only two CSF samples had detectable SMARCB1 mutations, which may have been due to variable sample/ DNA quality, DNA concentration and primer design, proof-of-principle for this technique was established.
Other mutations have also been tested with this method. For instance, EGFR mutations have been identified with targeted multiplex qPCR methods on CSF using the Roche Cobas Mutation Test v2 for EGFR mutations [58]. Although qPCR is rapid, it is low throughput and best suited for well characterized alterations as the assay will only detect the specific mutations it has been designed for.
Digital PCR
Digital PCR is a refinement of conventional PCR whereby a sample is partitioned into discrete compartments for amplification of the molecules. After amplification, compartments containing a target sequence are scored as positive or negative based on the presence or absence of a fluorescent tag of the PCR product. Two variations of digital PCR include BEAMing and ddPCR as outlined below.
(1) BEAMing
BEAMing stands for bead, emulsion, amplification, magnetics, which is a sensitive digital PCR approach that incorporates emulsion-based PCR and flow cytometry to detect and quantify DNA mutations. DNA is pre-amplified using target-specific primers and conventional PCR. These templates are then exposed to primers bound to magnetic beads, which are individually separated into microdroplets so that each contains, on average, a single magnetic bead with a template DNA molecule. Every microdroplet undergoes PCR using target-specific primers, resulting in thousands of amplified DNA fragments of a single template DNA molecule per bead. Next, base-pair specific fluorescent primers probes hybridize to the DNA molecules and are subsequently subjected to flow cytometry. The result is a quantification of beads with the mutant and wildtype sequences of interest. This early technology proved successful in the detection of ctDNA but requires a target variant of interest and the costs tend to be higher than ddPCR which can also separate PCR reactions into droplets [59], [60], [61].
(2) Droplet digital PCR
Droplet digital PCR (ddPCR) is a methodology for performing digital PCR that relies on the formation of water-oil emulsion droplets that divide and segregate the sample into distinct partitions. Each sample is fractionated to a maximum of 20,000 droplets where the template is amplified and the product detected based on the specific fluorescent tagging.
Overall, ddPCR is well-adapted to study specific single gene hotspot mutations that may be found in CSF samples, achieving a limit of detection as low as 0.01% per reaction. Rimelen et al. described the advantages in detection of MYD88 L265P mutations in CSF using ddPCR on cfDNA over genomic cell pellet DNA [62]. They showed that in 14 samples tested, for which a diagnosis had been made on histology or MRI and CSF, 3 samples that were negative using genomic DNA were found to harbor the mutation by ddPCR. Hiemcke-Jiwa also replicated the detection of MYD88 L265P in primary CNS lymphomas using ddPCR [63]. For other tumor entities, ddPCR has a high sensitivity to detect targeted mutations based on specific design, such as H3K27M mutations in diffuse midline gliomas (DMG) where brainstem, thalamic, spinal cord lesions may be very difficult to biopsy [39,64]. Assays have also been utilized to detect known resistance mutations such as EGFR T790M [65] in metastatic lung cancer as well as key mutations in breast cancers [36]. Importantly, the VAF of an oncogenic driver mutation from ddPCR can be used to track disease. For instance, it has been shown that the VAF determined by H3K27M ddPCR of CSF could be used to track radiographic and clinical disease progression of patients with DMG [64].
Massive parallel sequencing
Next-generation sequencing technologies, also known as massive parallel sequencing, offer unique advantages over the prior assays described as it changes the focus from the detection of a specific change to the potential characterization of the genomic landscape of a primary tumor or metastasis. NGS technology combines several layers of information to encompass single nucleotide variants (SNV), small insertions/deletions (indels), copy number alterations, structural variants and other markers. While the technical sensitivity for a specific marker may be lower than digital PCR assays, the comprehensiveness of assessment and the ability to detect multiple alterations may ultimately allow similar or higher sensitivity for disease detection.
Targeted NGS gene panels that focus on clinically actionable targets may be used. In our own practice, we use MSK-IMPACT™, a hybrid-capture based assay for detection of genetic alterations in all protein-coding exons and select introns of 505 genes [45]. Although the assay was initially validated for genomic DNA, this assay is now validated for the study of cfDNA from CSF [45]. Taking advantage of the relative purity of ctDNA in CSF samples, we report a high success rate of testing using standard sequencing technology with detection of genomic alterations at high VAF (Fig. 3). Assays that use amplicon-based enrichment, or anchored multiplex PCR prior to NGS may also be used for detection, keeping in mind that the design and validation of the assay must take into account the fragmentation of the template. Both amplicon-based and hybrid-capture NGS methods have been used for detection of mutations, minimal residual disease and tracking tumor evolution over time in CSF samples [8,14,37,49,66]. Despite the relative purity of ctDNA in CSF, high sensitivity assays, similar to those used for ctDNA detection in plasma, would be useful for longitudinal monitoring depending on the clinical context. In instances of glioma and other primary brain tumors, MSK-IMPACT™ testing on CSF detects ctDNA in approximately half of all CSF samples [37,67]. For metastatic tumors with leptomeningeal disease, rates of detection appear to be higher (63%) and as mentioned, this is likely in part because of the proximity of the tumor to the CSF unlike primary brain tumors which are usually parenchymal only [7]. The ability to detect somatic alterations in CSF depends on the amount of tumor-derived DNA that is shed into CSF and this inevitably varies between patients and within patients during the disease course. Furthermore, the detection of ctDNA can be impeded if there is dilution from genomic DNA from non-neoplastic cells within the CSF compartment, such as with inflammatory processes.
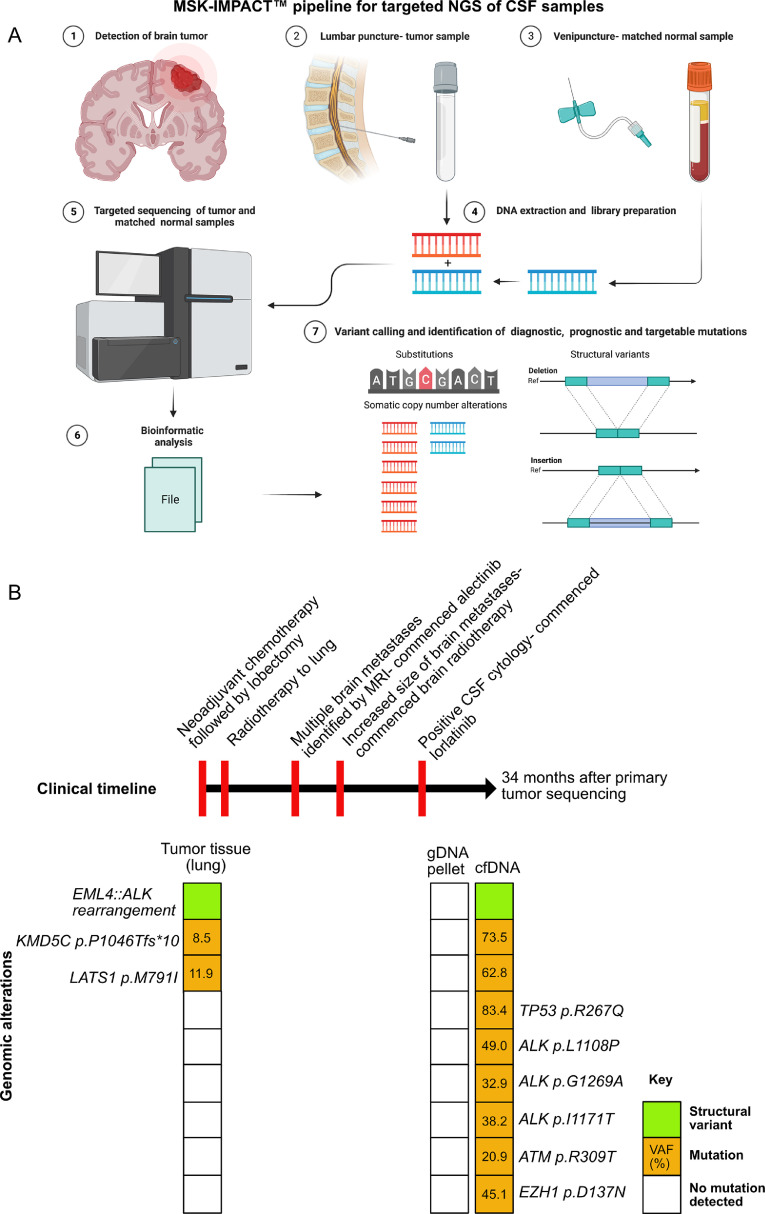
Fig. 3.
Summary of the NGS pipeline for testing CSF using MSK-IMPACT™. A: cfDNA from CSF (and gDNA from pellet, if recoverable) are extracted from patients with a suspected CNS tumor. gDNA from blood is used as normal control for matched tumor:normal sequencing. Sequencing data are processed and analyzed for the presence of somatic alterations within the CSF sample including SNV, indels, SCNA and SV. B: Representative example of a clinical case from a 27-year-old woman who was a never smoker with a history of lung adenocarcinoma. The clinical timeline is outlined at the top with a summary of the genomic alterations detected for each sample tested below. VAF for all mutations are expressed as percentages. Testing of the lung tumor demonstrated an EML4::ALK fusion and two mutations by MSK-IMPACT ™. Nearly one year later, she developed brain metastases and received alectinib, an ALK-targeted therapy with additional radiotherapy. Positive CSF cytology indicated progression of disease and therapy switched to lorlatinib, a third generation ALK inhibitor. Following further progression of disease, testing of the CSF was performed via MSK-IMPACT, concurrently sequencing the cfDNA and gDNA from the cell pellet (34 months after the initial primary tumor sequencing). Despite a positive cytology result, testing of the cell pellet did not demonstrate any molecular alterations. By contrast, the cfDNA confirmed the presence of the ALK fusion and all genetic alterations from the initial tumor. Several additional alterations were detected including several ALK kinase domain mutations that are associated with secondary resistance to ALK-inhibitors (ALK p.G1269A, ALK p.I1171T) and indicative of convergent evolution [68,69]. Note the high VAF in cfDNA, which are substantially greater than the lung adenocarcinoma tissue specimen and the absence of alterations detected in the cell pellet. Panel 3A created with Biorender.com.
Aside from targeted NGS panels, whole exome and whole genome sequencing have been applied to CSF, which further expands the breadth of the genome coverage albeit at lower depths. Whole exome sequencing (WES) can highlight the subclonal mutational landscape of medulloblastomas with mutational concordance to the tumor [14]. Particular mention goes to low pass whole genome sequencing, which is a cost-effective means of profiling copy number changes. Studies have shown that this profiling has value in the detection of minimal residual disease for CNS malignancies, such as medulloblastoma, and can correlate with disease burden, enabling clinicians to track therapeutic response [10,36].
By covering large regions of the genome with NGS, additional information can be generated, such as estimation of the tumor mutational burden (TMB) and mutational signatures [37,70]. The United States Food and Drug Administration (FDA) have approved the use of pembrolizumab for patients whose tumor tissue has a TMB that equals or exceeds 10 mutations/ megabase in tissues because of the recognized response to immunotherapy [[71], [72], [73]]. CSF TMB correlates with tissue TMB and holds promise as a metric for informing immunotherapy decisions [35,37].
Future directions
DNA methylation-based testing
DNA methylation profiling of primary CNS tumors is now well-established as an important adjunct to tumor diagnostics [74,75]. Most common approaches use bisulfite conversion on extracted tumor DNA with subsequent array-based analysis, such as the Illumina 850k Infinium array, which can be used to assess the methylation status across thousands of loci in the genome. Based on the profiles of previously characterized tumors, a random forest classifier categorizes the tumor into the highest scoring diagnostic group, which can support or refine tumor classification [76,77]. Other valuable information that is provided includes the MGMT promoter methylation status, which provides better prognostication for IDH-wildtype glioblastoma as well as prediction of therapeutic response to temozolamide, and copy number alterations within the tumor.
Given the strong clinical utility for methylation testing for CNS diagnostics, obtaining methylation profiles from CSF holds promise for diagnostics and prognostication for patients with CNS tumors. Moreover, for cancers that do not harbor informative genomic alterations, methylation-based testing may be useful as a surrogate for disease detection and/ or subclassification of disease. In plasma, methylation testing is being performed on ctDNA with some success. Using nearly 7000 participants divided into training and validation sets of plasma, Liu and colleagues showed that they could detect more than fifty cancer types with increasing sensitivity as cancer stage increased and tissue of origin being accurate in 93% of predicted cases [78]. Others have demonstrated success in classifying intracranial tumors into broad lineages by profiling plasma [79]. However, in CSF, to our knowledge, this has not yet been reported in the literature, and this may be in part because of challenges with low CSF DNA yields as compared to other biofluids.
Fragmentomics
The study of fragment length and profile within a sample is an area of active investigation as a biomarker of cancers in biofluids, particularly in plasma. The fragmentation of cfDNA from tissues is dependent on the chromatin structure, nucleosomal organization, gene expression and the nucleases within the cell of origin. As such, there are regions of the genome that have a predilection for being digested more than others by nucleases, which could provide tissue-of-origin signatures. Fragment properties of particular interest are length, the sequence motif, the structure of the fragment ends and the topology of the fragment (i.e. circular versus linear DNA) [80]. These techniques have been useful in cancer detection as well as determining tissues of origin in cfDNA [81,82]. This could be integrated into the genomic analyses in CSF as another layer of information and assist in tumor detection, classification and monitoring of minimal residual disease because of its high sensitivity.
Conclusion
The data presented in this review emphasizes that there is validation of numerous technologies to detect mutations in CSF ctDNA across a wide variety of cancers and undoubtedly more techniques will emerge that will complement those already mentioned. Challenges that face CSF liquid biopsy partly center on a lack of awareness of this technology, cost/ issues of reimbursement, and the absence of evidence supporting a cost-effective benefit of using these assays for patient care. We anticipate that these knowledge gaps will be filled over time and there will be wider adoption of CSF genomic assays, which will ultimately improve health outcomes for patients with CNS cancers.
CRediT authorship contribution statement
Richard A. Hickman: Writing – original draft. Alexandra M. Miller: Writing – review & editing. Maria E. Arcila: Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Maria E. Arcila has received honoraria or worked on advisory boards for Janssen Global Services, Bristol-Myers Squibb and AstraZeneca, speaker fees from Biocartis and Invivoscribe; has received CME honoraria from OncLive, PeerVoice, Physicians Education Resources, Peerview Institute and Clinical Care Options. All other authors have no other competing interests to declare.
Acknowledgments
This work was supported in part by the Matthew Larson Foundation for Pediatric Brain Tumors through an IronMatt research grant awarded to AMM, the Marie-Josée and Henry R. Kravis Center for Molecular Oncology and the National Cancer Institute Cancer Center Core Grant P30 CA008748. We also thank Cycle for Survival for providing research support (AMM). The views expressed in this article are those of the authors and do not necessarily reflect the views or policies of the Uniformed Services University, the United States Department of Defense, the United States government, or the Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc.
References
- 1.Martincorena I., Campbell P.J. Somatic mutation in cancer and normal cells. Science. 2015;349(6255):1483–1489. doi: 10.1126/science.aab4082. (New York, NY) [DOI] [PubMed] [Google Scholar]
- 2.Kato S., Kim K.H., Lim H.J., Boichard A., Nikanjam M., Weihe E., et al. Real-world data from a molecular tumor board demonstrates improved outcomes with a precision N-of-One strategy. Nat Commun. 2020;11(1):4965. doi: 10.1038/s41467-020-18613-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berger M.F., Mardis E.R. The emerging clinical relevance of genomics in cancer medicine. Nat. Rev. Clin. Oncol. 2018;15(6):353–365. doi: 10.1038/s41571-018-0002-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tivey A., Church M., Rothwell D., Dive C., Cook N. Circulating tumour DNA — Looking beyond the blood. Nat. Rev. Clin. Oncol. 2022;19(9):600–612. doi: 10.1038/s41571-022-00660-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alix-Panabières C., Pantel K. Liquid biopsy: from discovery to clinical application. Cancer Discov. 2021;11(4):858–873. doi: 10.1158/2159-8290.CD-20-1311. [DOI] [PubMed] [Google Scholar]
- 6.Rose Brannon A., Jayakumaran G., Diosdado M., Patel J., Razumova A., Hu Y., et al. Enhanced specificity of clinical high-sensitivity tumor mutation profiling in cell-free DNA via paired normal sequencing using MSK-ACCESS. Nat. Commun. 2021;12(1):3770. doi: 10.1038/s41467-021-24109-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pentsova E.I., Shah R.H., Tang J., Boire A., You D., Briggs S., et al. Evaluating cancer of the central nervous system through next-generation sequencing of cerebrospinal fluid. J. Clin. Oncol. 2016;34(20):2404–2415. doi: 10.1200/JCO.2016.66.6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pan C., Diplas B.H., Chen X., Wu Y., Xiao X., Jiang L., et al. Molecular profiling of tumors of the brainstem by sequencing of CSF-derived circulating tumor DNA. Acta Neuropathol. 2019;137(2):297–306. doi: 10.1007/s00401-018-1936-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pan W., Gu W., Nagpal S., Gephart M.H., Quake S.R. Brain tumor mutations detected in cerebral spinal fluid. Clin. Chem. 2015;61(3):514–522. doi: 10.1373/clinchem.2014.235457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu A.P.Y., Smith K.S., Kumar R., Paul L., Bihannic L., Lin T., et al. Serial assessment of measurable residual disease in medulloblastoma liquid biopsies. Cancer Cell. 2021;39(11) doi: 10.1016/j.ccell.2021.09.012. 1519-30.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seoane J., De Mattos-Arruda L., Le Rhun E., Bardelli A., Weller M. Cerebrospinal fluid cell-free tumour DNA as a liquid biopsy for primary brain tumours and central nervous system metastases. Ann. Oncol. 2019;30(2):211–218. doi: 10.1093/annonc/mdy544. [DOI] [PubMed] [Google Scholar]
- 12.Wang Y., Springer S., Zhang M., McMahon K.W., Kinde I., Dobbyn L., et al. Detection of tumor-derived DNA in cerebrospinal fluid of patients with primary tumors of the brain and spinal cord. Proc. Natl Acad. Sci. 2015;112(31):9704–9709. doi: 10.1073/pnas.1511694112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mattos-Arruda D., Mayor R., Ng C.K., Weigelt B., Martínez-Ricarte F., Torrejon D., et al. Cerebrospinal fluid-derived circulating tumour DNA better represents the genomic alterations of brain tumours than plasma. Nat. Commun. 2015;6(1):1–6. doi: 10.1038/ncomms9839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Escudero L., Llort A., Arias A., Diaz-Navarro A., Martínez-Ricarte F., Rubio-Perez C., et al. Circulating tumour DNA from the cerebrospinal fluid allows the characterisation and monitoring of medulloblastoma. Nat. Commun. 2020;11(1):1–11. doi: 10.1038/s41467-020-19175-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Izquierdo E., Proszek P., Pericoli G., Temelso S., Clarke M., Carvalho D.M., et al. Droplet digital PCR-based detection of circulating tumor DNA from pediatric high grade and diffuse midline glioma patients. Neuro Oncol. Adv. 2021;3(1) doi: 10.1093/noajnl/vdab013. vdab013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bettegowda C., Sausen M., Leary R.J., Kinde I., Wang Y., Agrawal N., et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014;6(224) doi: 10.1126/scitranslmed.3007094. 224ra24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nabors L.B., Ammirati M., Bierman P.J., Brem H., Butowski N., Chamberlain M.C., et al. Central nervous system cancers. J. Natl. Compr. Cancer Netw. 2013;11(9):1114–1151. doi: 10.6004/jnccn.2013.0132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Puhalla S., Elmquist W., Freyer D., Kleinberg L., Adkins C., Lockman P., et al. Unsanctifying the sanctuary: challenges and opportunities with brain metastases. Neuro Oncol. 2015;17(5):639–651. doi: 10.1093/neuonc/nov023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boire A., Brastianos P.K., Garzia L., Valiente M. Brain metastasis. Nat. Rev. Cancer. 2020;20(1):4–11. doi: 10.1038/s41568-019-0220-y. [DOI] [PubMed] [Google Scholar]
- 20.Sakka L., Coll G., Chazal J. Anatomy and physiology of cerebrospinal fluid. Eur. Ann. Otorhinolaryngol. Head Neck Dis. 2011;128(6):309–316. doi: 10.1016/j.anorl.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 21.Zhang E.T., Inman C.B., Weller R.O. Interrelationships of the pia mater and the perivascular (Virchow-Robin) spaces in the human cerebrum. J. Anat. 1990;170:111–123. [PMC free article] [PubMed] [Google Scholar]
- 22.Weller R.O., Kida S., Zhang E.T. Pathways of fluid drainage from the brain–morphological aspects and immunological significance in rat and man. Brain Pathol. 1992;2(4):277–284. doi: 10.1111/j.1750-3639.1992.tb00704.x. [DOI] [PubMed] [Google Scholar]
- 23.Zhang E.T., Richards H.K., Kida S., Weller R.O. Directional and compartmentalised drainage of interstitial fluid and cerebrospinal fluid from the rat brain. Acta Neuropathol. 1992;83(3):233–239. doi: 10.1007/BF00296784. [DOI] [PubMed] [Google Scholar]
- 24.Albargothy N.J., Johnston D.A., MacGregor-Sharp M., Weller R.O., Verma A., Hawkes C.A., et al. Convective influx/glymphatic system: tracers injected into the CSF enter and leave the brain along separate periarterial basement membrane pathways. Acta Neuropathol. 2018;136(1):139–152. doi: 10.1007/s00401-018-1862-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carare R.O., Aldea R., Agarwal N., Bacskai B.J., Bechman I., Boche D., et al. Clearance of interstitial fluid (ISF) and CSF (CLIC) group—Part of Vascular Professional Interest Area (PIA) Cerebrovascular disease and the failure of elimination of Amyloid-β from the brain and retina with age and Alzheimer's disease-Opportunities for Therapy. Alzheimer's Dement. Diagn. Assess. Dis. Monit. 2020;12(1):e12053. doi: 10.1002/dad2.12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szentistvanyi I., Patlak C.S., Ellis R.A., Cserr H.F. Drainage of interstitial fluid from different regions of rat brain. Am. J. Physiol. Ren. Physiol. 1984;246(6) doi: 10.1152/ajprenal.1984.246.6.F835. F835-F44. [DOI] [PubMed] [Google Scholar]
- 27.Wan J., Massie C., Garcia-Corbacho J., Mouliere F., Brenton J.D., Caldas C., et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat. Rev. Cancer. 2017;17(4):223–238. doi: 10.1038/nrc.2017.7. [DOI] [PubMed] [Google Scholar]
- 28.Cheok S.K., Narayan A., Arnal-Estape A., Gettinger S., Goldberg S.B., Kluger H.M., et al. Tumor DNA mutations from intraparenchymal brain metastases are detectable in CSF. JCO Precis. Oncol. 2021;5:163–172. doi: 10.1200/PO.20.00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.White M.D., Klein R.H., Shaw B., Kim A., Subramanian M., Mora J.L., et al. Detection of leptomeningeal disease using cell-free DNA from cerebrospinal fluid. JAMA Netw. Open. 2021;4(8) doi: 10.1001/jamanetworkopen.2021.20040. e2120040-e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ying S., Ke H., Ding Y., Liu Y., Tang X., Yang D., et al. Unique genomic profiles obtained from cerebrospinal fluid cell-free DNA of non-small cell lung cancer patients with leptomeningeal metastases. Cancer Biol. Ther. 2019;20(4):562–570. doi: 10.1080/15384047.2018.1538614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ballester L.Y., Glitza Oliva I.C., Douse D.Y., Chen M.M., Lan C., Haydu L.E., et al. Evaluating circulating tumor DNA from the cerebrospinal fluid of patients with melanoma and leptomeningeal disease. J. Neuropathol. Exp. Neurol. 2018;77(7):628–635. doi: 10.1093/jnen/nly046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gerlinger M., Rowan A.J., Horswell S., Larkin J., Endesfelder D., Gronroos E., et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sottoriva A., Spiteri I., Piccirillo S.G., Touloumis A., Collins V.P., Marioni J.C., et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc. Natl Acad. Sci. 2013;110(10):4009–4014. doi: 10.1073/pnas.1219747110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Mattos-Arruda L., Mayor R., Ng C.K.Y., Weigelt B., Martínez-Ricarte F., Torrejon D., et al. Cerebrospinal fluid-derived circulating tumour DNA better represents the genomic alterations of brain tumours than plasma. Nat. Commun. 2015;6:8839. doi: 10.1038/ncomms9839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo W., Jin L., Liang J., Lin G., Zheng J., Zhou D., et al. Detection of mutation profiles and tumor mutation burden of cerebrospinal fluid circulating DNA by a cancer genomic panel sequencing in glioma patients. Clin. Chim. Acta. 2022;534:81–92. doi: 10.1016/j.cca.2022.07.001. [DOI] [PubMed] [Google Scholar]
- 36.Fitzpatrick A., Iravani M., Mills A., Childs L., Alaguthurai T., Clifford A., et al. Assessing CSF ctDNA to improve diagnostic accuracy and therapeutic monitoring in breast cancer leptomeningeal metastasis. Clin. Cancer Res. 2022;28(6):1180–1191. doi: 10.1158/1078-0432.CCR-21-3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller A.M., Shah R.H., Pentsova E.I., Pourmaleki M., Briggs S., Distefano N., et al. Tracking tumour evolution in glioma through liquid biopsies of cerebrospinal fluid. Nature. 2019;565(7741):654–658. doi: 10.1038/s41586-019-0882-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson B.E., Mazor T., Hong C., Barnes M., Aihara K., McLean C.Y., et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science. 2014;343(6167):189–193. doi: 10.1126/science.1239947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cantor E., Wierzbicki K., Tarapore R.S., Ravi K., Thomas C., Cartaxo R., et al. Serial H3K27M cell-free tumor DNA (cf-tDNA) tracking predicts ONC201 treatment response and progression in diffuse midline glioma. Neuro Oncol. 2022;24(8):1366–1374. doi: 10.1093/neuonc/noac030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen Y.R., Sole J., Ugiliweneza B., Johnson E., Burton E., Woo S.Y., et al. National trends for reoperation in older patients with glioblastoma. World Neurosurg. 2018;113 doi: 10.1016/j.wneu.2018.01.211. e179-e89. [DOI] [PubMed] [Google Scholar]
- 41.Tully P.A., Gogos A.J., Love C., Liew D., Drummond K.J., Morokoff A.P. Reoperation for recurrent glioblastoma and its association with survival benefit. Neurosurgery. 2016;79(5):678–689. doi: 10.1227/NEU.0000000000001338. [DOI] [PubMed] [Google Scholar]
- 42.Ramkissoon L.A., Pegram W., Haberberger J., Danziger N., Lesser G., Strowd R., et al. Genomic profiling of circulating tumor DNA from cerebrospinal fluid to guide clinical decision making for patients with primary and metastatic brain tumors. Front. Neurol. 2020;11 doi: 10.3389/fneur.2020.544680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ruff R.L., Dougherty J.H., Jr. Complications of lumbar puncture followed by anticoagulation. Stroke. 1981;12(6):879–881. doi: 10.1161/01.str.12.6.879. [DOI] [PubMed] [Google Scholar]
- 44.Enting R.H. Leptomeningeal neoplasia: epidemiology, clinical presentation, CSF analysis and diagnostic imaging. Cancer Treat. Res. 2005;125:17–30. doi: 10.1007/0-387-24199-x_2. [DOI] [PubMed] [Google Scholar]
- 45.Bale T.A., Yang S.-.R., Solomon J.P., Nafa K., Middha S., Casanova J., et al. Clinical experience of cerebrospinal fluid–based liquid biopsy demonstrates superiority of cell-free DNA over cell pellet genomic DNA for molecular profiling. J. Mol. Diagn. 2021;23(6):742–752. doi: 10.1016/j.jmoldx.2021.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao Y., He J.-.Y., Zou Y.-.L., Guo X.-.S., Cui J.-.Z., Guo L., et al. Evaluating the cerebrospinal fluid ctDNA detection by next-generation sequencing in the diagnosis of meningeal Carcinomatosis. BMC Neurol. 2019;19(1):1–9. doi: 10.1186/s12883-019-1554-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ozcan G., Singh M., Vredenburgh J.J. Leptomeningeal metastasis from non-small cell lung cancer and current landscape of treatments. Clin. Cancer Res. 2022 doi: 10.1158/1078-0432.CCR-22-1585. [DOI] [PubMed] [Google Scholar]
- 48.Yang H., Cai L., Zhang Y., Tan H., Deng Q., Zhao M., et al. Sensitive detection of EGFR mutations in cerebrospinal fluid from lung adenocarcinoma patients with brain metastases. J. Mol. Diagn. 2014;16(5):558–563. doi: 10.1016/j.jmoldx.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 49.Li Y., Jiang B., Yang J., Zhang X., Zhang Z., Ye J., et al. Unique genetic profiles from cerebrospinal fluid cell-free DNA in leptomeningeal metastases of EGFR-mutant non-small-cell lung cancer: a new medium of liquid biopsy. Ann. Oncol. 2018;29(4):945–952. doi: 10.1093/annonc/mdy009. [DOI] [PubMed] [Google Scholar]
- 50.Shih D.J., Nayyar N., Bihun I., Dagogo-Jack I., Gill C.M., Aquilanti E., et al. Genomic characterization of human brain metastases identifies drivers of metastatic lung adenocarcinoma. Nat. Genet. 2020;52(4):371–377. doi: 10.1038/s41588-020-0592-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.De Mattos-Arruda L., Siravegna G. How to use liquid biopsies to treat patients with cancer. ESMO Open. 2021;6(2) doi: 10.1016/j.esmoop.2021.100060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Diehl F., Schmidt K., Choti M.A., Romans K., Goodman S., Li M., et al. Circulating mutant DNA to assess tumor dynamics. Nat. Med. 2008;14(9):985–990. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kang Q., Henry N.L., Paoletti C., Jiang H., Vats P., Chinnaiyan A.M., et al. Comparative analysis of circulating tumor DNA stability In K(3)EDTA, streck, and cellsave blood collection tubes. Clin. Biochem. 2016;49(18):1354–1360. doi: 10.1016/j.clinbiochem.2016.03.012. [DOI] [PubMed] [Google Scholar]
- 54.Merker J.D., Oxnard G.R., Compton C., Diehn M., Hurley P., Lazar A.J., et al. Circulating tumor DNA analysis in patients with cancer: american Society of Clinical Oncology and College of American Pathologists joint review. Arch. Pathol. Lab. Med. 2018;142(10):1242–1253. doi: 10.5858/arpa.2018-0901-SA. [DOI] [PubMed] [Google Scholar]
- 55.Takayasu T., Shah M., Dono A., Yan Y., Borkar R., Putluri N., et al. Cerebrospinal fluid ctDNA and metabolites are informative biomarkers for the evaluation of CNS germ cell tumors. Sci. Rep. 2020;10(1):14326. doi: 10.1038/s41598-020-71161-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hickmann A.-.K., Frick M., Hadaschik D., Battke F., Bittl M., Ganslandt O., et al. Molecular tumor analysis and liquid biopsy: a feasibility investigation analyzing circulating tumor DNA in patients with central nervous system lymphomas. BMC Cancer. 2019;19(1):1–12. doi: 10.1186/s12885-019-5394-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vu-Han T.L., Frühwald M.C., Hasselblatt M., Kerl K., Nagel I., Obser T., et al. Identifying molecular markers for the sensitive detection of residual atypical teratoid rhabdoid tumor cells. Cancer Genet. 2014;207(9):390–397. doi: 10.1016/j.cancergen.2014.05.008. [DOI] [PubMed] [Google Scholar]
- 58.Kawahara A., Abe H., Murata K., Ishii H., Azuma K., Takase Y., et al. Screening system for epidermal growth factor receptor mutation detection in cytology cell-free DNA of cerebrospinal fluid based on assured sample quality. Cytopathology. 2019;30(2):144–149. doi: 10.1111/cyt.12660. [DOI] [PubMed] [Google Scholar]
- 59.Vessies D., Greuter M., van Rooijen K., Linders T., Lanfermeijer M., Ramkisoensing K., et al. Performance of four platforms for KRAS mutation detection in plasma cell-free DNA: ddPCR, Idylla, COBAS z480 and BEAMing. Sci. Rep. 2020;10(1):1–9. doi: 10.1038/s41598-020-64822-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dressman D., Yan H., Traverso G., Kinzler K.W. Vogelstein B. Transforming single DNA molecules into fluorescent magnetic particles for detection and enumeration of genetic variations. Proc. Natl. Acad. Sci. U. S. A. 2003;100(15):8817–8822. doi: 10.1073/pnas.1133470100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Diehl F., Li M., Dressman D., He Y., Shen D., Szabo S., et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc. Natl Acad. Sci. 2005;102(45):16368–16373. doi: 10.1073/pnas.0507904102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rimelen V., Ahle G., Pencreach E., Zinniger N., Debliquis A., Zalmaï L., et al. Tumor cell-free DNA detection in CSF for primary CNS lymphoma diagnosis. Acta Neuropathol. Commun. 2019;7(1):1–3. doi: 10.1186/s40478-019-0692-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hiemcke-Jiwa L.S., Minnema M.C., Radersma-van Loon J.H., Jiwa N.M., de Boer M., Leguit R.J., et al. The use of droplet digital PCR in liquid biopsies: a highly sensitive technique for MYD88 p.(L265P) detection in cerebrospinal fluid. Hematol. Oncol. 2018;36(2):429–435. doi: 10.1002/hon.2489. [DOI] [PubMed] [Google Scholar]
- 64.Panditharatna E., Kilburn L.B., Aboian M.S., Kambhampati M., Gordish-Dressman H., Magge S.N., et al. Clinically relevant and minimally invasive tumor surveillance of pediatric diffuse midline gliomas using patient-derived liquid biopsy. Clin. Cancer Res. 2018;24(23):5850–5859. doi: 10.1158/1078-0432.CCR-18-1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Suryavanshi M., Jaipuria J., Panigrahi M.K., Goyal N., Singal R., Mehta A., et al. CSF cell-free DNA EGFR testing using DdPCR holds promise over conventional modalities for diagnosing leptomeningeal involvement in patients with non-small cell lung cancer. Lung Cancer. 2020;148:33–39. doi: 10.1016/j.lungcan.2020.07.034. [DOI] [PubMed] [Google Scholar]
- 66.Aldea M., Hendriks L., Mezquita L., Jovelet C., Planchard D., Auclin E., et al. Circulating tumor DNA analysis for patients with oncogene-addicted NSCLC with isolated central nervous system progression. J. Thorac. Oncol. 2020;15(3):383–391. doi: 10.1016/j.jtho.2019.11.024. [DOI] [PubMed] [Google Scholar]
- 67.Miller A.M., Szalontay L., Bouvier N., Hill K., Ahmad H., Rafailov J., et al. Next-generation sequencing of cerebrospinal fluid for clinical molecular diagnostics in pediatric, adolescent and young adult brain tumor patients. Neuro Oncol. 2022;24(10):1763–1772. doi: 10.1093/neuonc/noac035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Toyokawa G., Hirai F., Inamasu E., Yoshida T., Nosaki K., Takenaka T., et al. Secondary mutations at I1171 in the ALK gene confer resistance to both Crizotinib and Alectinib. J. Thorac. Oncol. 2014;9(12):e86–e87. doi: 10.1097/JTO.0000000000000358. [DOI] [PubMed] [Google Scholar]
- 69.Recondo G., Mezquita L., Facchinetti F., Planchard D., Gazzah A., Bigot L., et al. Diverse resistance mechanisms to the third-generation ALK inhibitor lorlatinib in ALK-Rearranged Lung Cancer. Clin. Cancer Res. 2020;26(1):242–255. doi: 10.1158/1078-0432.CCR-19-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Deng Z., Cui L., Li P., Ren N., Zhong Z., Tang Z., et al. Genomic comparison between cerebrospinal fluid and primary tumor revealed the genetic events associated with brain metastasis in lung adenocarcinoma. Cell Death Dis. 2021;12(10):1–10. doi: 10.1038/s41419-021-04223-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Guibert N., Jones G., Beeler J.F., Plagnol V., Morris C., Mourlanette J., et al. Targeted sequencing of plasma cell-free DNA to predict response to PD1 inhibitors in advanced non-small cell lung cancer. Lung Cancer. 2019;137:1–6. doi: 10.1016/j.lungcan.2019.09.005. [DOI] [PubMed] [Google Scholar]
- 72.Wang Z., Duan J., Cai S., Han M., Dong H., Zhao J., et al. Assessment of blood tumor mutational burden as a potential biomarker for immunotherapy in patients with non–small cell lung cancer with use of a next-generation sequencing cancer gene panel. JAMA Oncol. 2019;5(5):696–702. doi: 10.1001/jamaoncol.2018.7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Marabelle A., Fakih M., Lopez J., Shah M., Shapira-Frommer R., Nakagawa K., et al. Association of tumour mutational burden with outcomes in patients with advanced solid tumours treated with pembrolizumab: prospective biomarker analysis of the multicohort, open-label, phase 2 KEYNOTE-158 study. Lancet Oncol. 2020;21(10):1353–1365. doi: 10.1016/S1470-2045(20)30445-9. [DOI] [PubMed] [Google Scholar]
- 74.Capper D., Jones D.T., Sill M., Hovestadt V., Schrimpf D., Sturm D., et al. DNA methylation-based classification of central nervous system tumours. Nature. 2018;555(7697):469–474. doi: 10.1038/nature26000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pratt D., Sahm F., Aldape K. DNA methylation profiling as a model for discovery and precision diagnostics in neuro-oncology. Neuro Oncol. 2021;23(Supplement_5):S16–S29. doi: 10.1093/neuonc/noab143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Karimi S., Zuccato J.A., Mamatjan Y., Mansouri S., Suppiah S., Nassiri F., et al. The central nervous system tumor methylation classifier changes neuro-oncology practice for challenging brain tumor diagnoses and directly impacts patient care. Clin. Epigenet. 2019;11(1):1–10. doi: 10.1186/s13148-019-0766-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wu Z., Abdullaev Z., Pratt D., Chung H.J., Skarshaug S., Zgonc V., et al. Impact of the methylation classifier and ancillary methods on CNS tumor diagnostics. Neuro Oncol. 2021;24(4):571–581. doi: 10.1093/neuonc/noab227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu M.C., Oxnard G.R., Klein E.A., Swanton C., Seiden M.V. Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann. Oncol. 2020;31(6):745–759. doi: 10.1016/j.annonc.2020.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nassiri F., Chakravarthy A., Feng S., Shen S.Y., Nejad R., Zuccato J.A., et al. Detection and discrimination of intracranial tumors using plasma cell-free DNA methylomes. Nat. Med. 2020;26(7):1044–1047. doi: 10.1038/s41591-020-0932-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lo Y.D., Han D.S., Jiang P., Chiu R.W. Epigenetics, fragmentomics, and topology of cell-free DNA in liquid biopsies. Science. 2021;372(6538):eaaw3616. doi: 10.1126/science.aaw3616. [DOI] [PubMed] [Google Scholar]
- 81.Bao H., Wang Z., Ma X., Guo W., Zhang X., Tang W., et al. An ultra-sensitive assay using cell-free DNA fragmentomics for multi-cancer early detection. Mol. Cancer. 2022;21(1):1–7. doi: 10.1186/s12943-022-01594-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu Y. At the dawn: cell-free DNA fragmentomics and gene regulation. Br. J. Cancer. 2022;126(3):379–390. doi: 10.1038/s41416-021-01635-z. [DOI] [PMC free article] [PubMed] [Google Scholar]