Abstract
Microbial consortia under anaerobic conditions are involved in oxidizing organic matter in the sludge to produce methane gas. However, in developing countries like Kenya, these microbes have not been fully identified to target them for the efficient harnessing of biofuel. This study collected wet sludge from two anaerobic digestion lagoons 1 and 2 that were operational during sampling at Kangemi Sewage Treatment Plant, in Nyeri County, Kenya. DNA was extracted from samples using commercially available ZymoBIOMICS™ DNA Miniprep Kit and sequenced using Shotgun metagenomics. Samples were analyzed using MG-RAST software (Project ID: mgp100988), which allowed for identifying microorganisms directly involved in various stages of methanogenesis pathways. The study found hydrogenotrophic methanogens, such as Methanospirillum (32%), Methanobacterium (27%), Methanobrevibacter (27%), and Methanosarcina (32%), being predominant in the lagoon communities, whereas acetoclastic microorganisms such as the Methanoregula (22%) and the acetate oxidazing bacteria such as Clostridia (68%) were the key microbes for that pathway in the sewage digester sludge. Furthermore, Methanothermobacter (18%), Methanosarcina (21%), Methanosaeta (15%), and Methanospirillum (13%) carried out the methylotrophic pathway. In contrast, Methanosarcina (23%),Methanoregula (14%), methanosaeta (13%), and methnanoprevibacter (13%) seemed to play an important role in the final step of methane release. This study concluded that the sludge produced from the Nyeri-Kangemi WWTP harbors microbes with significant potential for biogas production. The study recommends a pilot study to investigate the efficiency of the identified microbes for biogas production.
Keywords: Sewage sludge, Methanogens, Hydrogenotrophic, Acetoclastic, Methylotrophic
Graphical abstract
1. Introduction
The microbial community in sludge possesses great potential as a bio-energy source. Despite such favourable industrial opportunities, appropriate microbial identification techniques have not been applied to exploit it; therefore, the sludge has been considered a ‘black box′ with possible unexploited biotechnological reactions [1]. Culture and isolation-dependent methods do not present the actual methanogenesis reaction in sludge since most microorganisms within the sludge communities cannot be cultured in vitro [2]. Great advancements in microbial studies have been made in recent years, and nucleic acid-based molecular methods can identify methanogenic microorganisms by DNA sequencing of their ribosomal RNA (r RNA) genes without the need to isolate the microorganisms [3].
Currently, hydrolysis, acidogenesis, acetogenesis, and the methanogenesis are the main steps involved in converting organic matter into methane [4]. The hydrolysis, acidogenesis, acetogenesis, are driven by a wide spectrum of microbiomes except for the last step, methanogenesis which are exclusively driven by a specific group of archaea known as methanogens. These steps form the three main pathways involved in methanogenesis and the final step of the release of methane gas; (i) the hydrogenotrophic pathway, which uses the H2/CO2 substrate where the CO2 binds to methanofuran, and it is broken down to formyl-methanofuran in the presence of H2. This process is catalyzed by the enzyme formyl-methanofuran dehydrogenase [5]. The formal part of formyl-methanofuran is transferred to coenzyme tetrahydromethanopterin forming formyl-tetrahydromethanopterin catalyzed by enzyme formyl transferase [6]. The formyl-tetrahydro-methanofuran is broken down to methyl-tetrahydromethanopterin catalyzed by coenzyme F420. A methyltransferase-catalyzed reaction allows the methyl group of methyl-tetrahydromethanopterin to be transferred to coenzyme M [7]. (ii) The acetoclastic pathway, where acetate is activated to acetyl-CoA by the action of acetate kinase or activity of acetyl-CoA synthetase. The acetyl-CoA molecule is then dismutated using the enzyme acetyl-CoA decarbonylase, where the carbonyl group is oxidized to carbon dioxide while the methyl group is reduced to methane [8]. (iii) Methylotrophic pathway utilizes methanol and methylated amines as substrates. The methyl group is transferred to the corrinoid protein by the methyltransferase. The coronoid protein is then channeled through the methanogenic pathways in the methyl -CoM stage, where they are finally reduced to methane [9]. (iv) The final step of methane production involves methyl-coenzyme M reductase and two coenzymes: N-7 mercapto heptanoyl threonine phosphate (HS-HTP) and coenzyme F430 [10].
Many microorganisms responsible for the methanogenesis process have been reported in the literature. The archaea such as Methanobacteria, Methanocella, Methanococcus, Methanomicrobia, Methanopyra, Methanosarcina and Methanomassiliicoccus has been identified as the main methanogens [11]. Among these, Methanosarcina has been proposed to be capable of carrying out the final step in methanogenesis of the release of methane [12]The advancement in metagenomics sequencing has favourably enabled a better understanding of the sludge microbiome community and microorganisms responsible for converting organic matter to methane. Even though metagenomics potential has been in the past restricted to archaea, metagenomics sequencing studies have shown that this conclusion underestimates the methanogens potential and the microbes involved [13]. With the growing technology, recent studies have revealed that we are beginning to understand the methanogenesis process and the microbes involved in the process [11].
In Kenya, the biotechnological production of energy from wastewater sludge is a potential venture for a relatively efficient, low-cost wastewater-sludge treatment system. The Nyeri Water and Sewerage Company (NYEWASCO)-Kangemi wastewater treatment plant is one of Kenya's modern and best-managed WWTP. The treatment plant has simple processes designed as trickling filters, sedimentation tanks, anaerobic lagoons, and maturation ponds. The sludge treatment process contains; (i) the desludging chamber, a tank that separates sludge and the liquid components through hydraulic pressure and desludges after every three (3) hours. In the wastewater treatment process, the raw sludge from the different desludging chambers is then pumped into the (ii) sludge well, where sludge is allowed to settle before it is pumped to (iii) the sludge lagoons. The lagoons have a volume of 25000 m3 with an Organic Loading Rate (OLR) averaging 2.01 kg/m3.d and a hydraulic retention time (HRT) of 90 days. The sludge lagoons are digestion tanks where anaerobic digestion takes place for three to four months. Here the vegetation and scum are allowed to accumulate over time as part of the biological treatment of sludge and later on are removed. The treated sludge is then allowed through the underground valve to the drying beds by gravity. The dry beds are fitted with concrete slabs with spacing between the water from dewatering to infiltrating to the ground. The sludge is allowed to dry for a month during the wet season and fourteen days during the dry season before being sold to farmers for agricultural land application. The plant produces between 75 and 250 tons of dried-up sludge per month. The sludge is sold at USD 5 per ton, making a profit of around USD 330 to USD 1100 per month from sewage sludge (NYEWASCO, 2007). Its digester system can be upgraded to include biogas production, which is currently lacking. However, it is important to have prior baseline information on the profile of the microbial composition of the sludge from the WWTP. In addition, identifying the microorganisms which metabolize the organic compounds in the wastewater sludge to produce the energy (methane) is vital [13]. This will provide tangible evidence for a cheaper alternative energy source for the Nyeri-Kangemi WWTP and provide information on the biological properties and possible application of biotechnology, including genetic modification of methanogenic organisms for technical applications.
This study identified microorganisms such as protists, bacteria, and archaea in the wastewater sludge from the Nyeri-Kangemi WWTP using the metagenomics method. The shotgun metagenomics techniques was used to characterize the microorganisms. The genes were predicted using the de novo gene prediction pathways [3] and provide microbial diversity and helped to detect their abundances in the sludge samples. The functional methanogenic annotation was performed by classifying predicted metagenomics proteins into protein families using sequence or hidden Markov models (HMM) databases [14].
2. Materials and methods
2.1. Study site
The study was conducted at Nyeri Water and Sanitation Company Limited at Kangemi Sewage Treatment Works. The treatment works are located in Nyeri county, Nyeri township. They are approximately 4 km from the Nyeri town center, at 0°25′S, 36°58″E. The Kangemi Wastewater Treatment Plant has a capacity of over 50,000 m³ per day. It serves approximately 200,000 people [15].
2.2. Sampling
The samples for DNA analyses were collected from the two operational sludge digestion lagoons 1 and 2. Lagoon 1 was still fresh at two weeks old, while lagoon 2 was four months old ready to be drained to the dry beds. Grab samples were collected from six points, each using a 2.5-L container that was acid washed and rinsed with sterile water. The samples were then pooled in one sterile bucket and mixed well using an acid-washed and sterilized shovel. Sub-samples of 5 ml from the composites were collected into 10 ml sterile cryogenic tubes fitted with a cap. This was done for both sludge digestion lagoons 1 and 2. The samples from lagoons were then labelled as 1a,1b,1c, 1d and 1e while lagoon 2 were labelled as 2a,2b,2c, 2d and 2e. After that, 5 ml of the DNA/RNA shield was added to each of the samples to preserve the nucleic acids at ambient temperature and inactivate microorganisms. The samples were stored in a cool box under ice, transported to Kenya Agricultural and Livestock Research Organisation (KALRO), Njoro, and stored at −20 °C before DNA extraction.
2.3. DNA extraction
DNA was extracted using the commercially available ZymoBIOMICS™ DNA Miniprep Kit (Zymo Research), designed to extract DNA from a wide array of sample inputs, including sewage sludge, which is immediately ready for metagenomics analyses. The quality of the extracted DNA was analyzed using Nanodrop 2000c Spectrophotometer (Thermo, USA) to determine the concentration and purity of the DNA to ensure the recommended minimum DNA concentration of 10 ng/μL and a purity level of optical density (OD) 260/280 above 1.8. The OD260/280 > 1.8 was attained [16]. It was only grab samples 1c and 1b from lagoon 1 and samples 2a and 2c from lagoon 2 that attained the required DNA purity level. The total extracted DNA was then stored in PCR tubes and labelled as 1c, 1b, 2a and 2c. They were then transported on ice to Inqaba Biotechnical Industries (Pty) Ltd (South Africa) for whole-genome shotgun metagenomics analysis.
2.4. Methanogen phylogenetic analysis
Methanogenic specific phylogenetic characterization was achieved by HMM search of methanogenic related sequences in the MG-RAST analysis platform by focusing on genes encoding to (1) methyl CoM reductase (mcr) responsible for final methane release; (2) formyl methanofuran dehydrogenase (fmd) that releases CO2; (3) dehydrogenase/acetyl-CoA synthase (cdh) that releases acetate; (4) methyltransferase (mta) that releases methanol and (5) the methylamine methyltransferase (mtm, Mtb, mtt) that release fatty acids: all involved in the methanogenesis process. The methanogenic taxonomy was obtained by phylogenetic tree placement of the best-obtained hits [11].
The methanogenic organism's compositional similarity from the different sampling sites was comparatively assessed using the Bray-Curtis measure of beta diversity [17], which compared all pairwise taxonomic abundances between each sample using R vegan, a statistical analysis software.
3. Results
3.1. General functional profiles of the two lagoons
To explore the metabolic potential of the studied community, a detailed analysis of metagenomics sequences was performed against the KO and the SEED Subsystems within the MG-RAST pipeline. An average of 68363 functional hits were detected in all the annotated reads. Genes annotated to metabolism were recorded the highest in the sludge samples (Fig. 1). The functional categories were not significantly different (F (3,48) = 0.768, p = 0.5176) p < 0.05) in the four samples, even though replicates 1b and 1c from lagoon 1 samples recorded a higher functional abundance compared to replicates 2a and 2c from lagoon 2 samples. This indicates that lagoon 1 was functionally more active compared to lagoon 2.
Fig. 1.
Composition of the functional categories across the samples.
Analysis of the commonly used community describers such as; diversity, evenness, and richness indices showed that all the samples were quite similar as far as the indices are concerned. The most functionally diverse and even was sample 2a community (Shannon–Wiener index—1.117, Pielou index 0.5091), while the least one was sample 1b metagenome (Shannon–Wiener index—1.112, Pielou index 0.507) (Table 1). Even though samples in lagoon 1 had a higher abundance of species with functional genes, lagoon 2 recorded a higher functionally diverse and evenly distributed species. Furthermore, similarly to RefSeq Bray–Curtis distances calculation, some samples are similar to each other, e.g., 1c and 1b (95%), 1b and 2c (81%), and samples 1c and 2c (77%). In contrast, samples 2a and 1c (39%) were the most different from most analyzed metagenomes, followed by samples 2a and 1b (42%). Even though samples 2a and 2c were collected from the same lagoon 2, they were relatively dissimilar at 56%. This can be attributed to sampling error when collecting the grab sampling, or samples 2c and 2a were collected from a section that was not well mixed. The other possible explanation is biasness during sub-sampling in lagoon 2.
Table 1.
Common alpha diversity indices of the samples.
| Sample 2a | Sample 2c | Sample 1c | Sample 1b | |
|---|---|---|---|---|
| Individuals | 143270 | 368760 | 591151 | 537534 |
| Dominance_(D) | 0.41720.5828 | 0.4167 | 0.4169 | 0.4182 |
| Simpson_1-D | 1.117 | 0.5833 | 0.5831 | 0.5818 |
| Shannon_H | 0.5091 | 1.116 | 1.116 | 1.112 |
| Evenness_e^H/S | 0.6232 | 0.5087 | 0.5088 | 0.507 |
| Equitability_J | 0.4756 | 0.6227 | 0.6229 | 0.6209 |
| Fisher_alpha | 0.4765 | 0.4399 | 0.4241 | 0.4272 |
3.2. Functional profiles of relevant metagenomics pathways
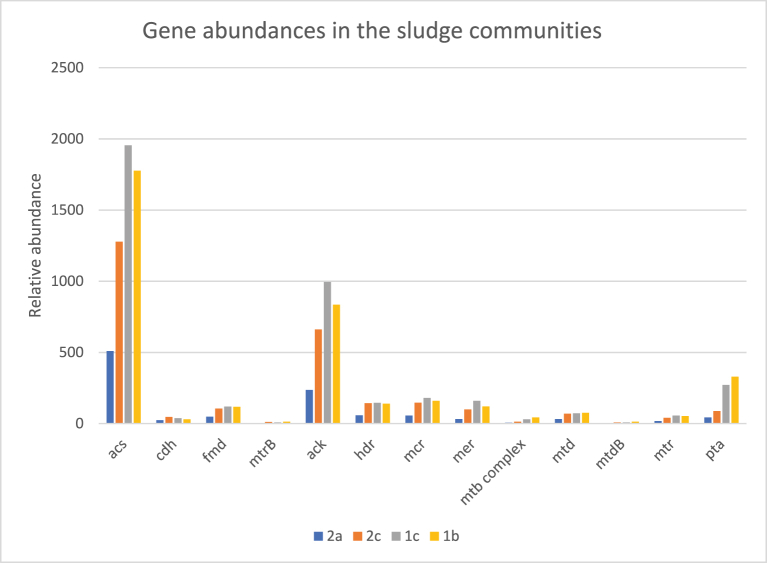
The study focused on functional analysis relevant to the metagenomics pathways. The relative abundance of methanogenesis-related genes was presented from the functional annotations of SEED subsystems analyzed by MG-RAST server. Annotation were run against methanogenesnesis related enzymes namely; formylmethanofuran dehydrogenase (fmd); formylmethanofuran-H4MPT formyltransferase (ftr); methenyl-H4MPT cyclohydrolase (mch); methylene-5,6,7,8-H4MPT dehydrogenase (mtd); H2-forming N5,N10-methylene-H4MPT dehydrogenase (hmd); 5,10-methylene-H4MPT reductase (mer); H4MPT-methyltransferase (mtr); acetate kinase (ack); phosphate acetyltransferase (pta); acetyl-CoA synthetase (acs); CO dehydrogenase/acetyl CoA synthase (cdh); methanol-specific methyltransferase complex (mta); methylamine-specific methyltransferase complex (mtb); the CoB-CoM heterodisulfide reductase (hdr); and the methyl CoM reductase (mcr) that participates in the final release of methane gas.
The hits relating to the methane production genes were not significantly different in the four composite samples (F (3, 64) = 0.6745, p = 0.2517) p < 0.05. Even though the highest number of hits were recorded in sample 1c (4037) and the least hits in 2a (1065), sample 2a community was the most functionally diverse and even with (Shannon–Wiener index—1.68, Pielou index 0.4167) while the least one was sample 1b metagenome (Shannon–Wiener index—1.112, Pielou index 0.507) (Table 2).
Table 2.
Alpha diversity indices for the total methane functional categories.
| Sample 2a | Sample 2c | Sample 1c | Sample 1b | |
|---|---|---|---|---|
| Individuals | 1065 | 2706 | 4037 | 3701 |
| Dominance_D | 0.2888 | 0.2932 | 0.306 | 0.295 |
| Simpson_1-D | 0.7112 | 0.7068 | 0.694 | 0.705 |
| Shannon_H | 1.69 | 1.662 | 1.605 | 1.653 |
| Evenness_e^H/S | 0.4167 | 0.4053 | 0.3831 | 0.4019 |
| Equitability_J | 0.6587 | 0.6479 | 0.6259 | 0.6446 |
| Fisher_alpha | 2.084 | 1.773 | 1.668 | 1.69 |
Bray–Curtis similarity calculation indicated a 93% similarity in the sequence profiles of the selected genes between samples 1b and 1c, probably because they came from the same lagoon. Interestingly, samples 1b and 2b were 84% similar even though they belonged to different lagoons. Samples 1c and 2c were also considerably similar (80%). The lagoon two samples 2a and 2c were different with a similarity value of 56%.
Among the methane pathway selected genes, the acs genes were over-represented followed by the ack and the pta genes (Fig. 2). Microbial consortia annotated to cdh genes were the most diverse and evenly distributed (Shannon–Wiener index—1.368, Pielou index 0.983). The least diverse consortia was annotated from pta genes (Shannon–Wiener index—1.149, Pielou index 0.7891). The microbial profiles consortia of the mcr gene responsible for the final release of methane recorded relatively moderate hits of 540 with a relatively diverse profiles (Shannon–Wiener index—1.317, Pielou index 0.9335) (Table 3).
Fig. 2.
Relative abundance of hits annotated to the different methanogenesis related genes.
Table 3.
Alpha diversity of the methanogenesis related genes.
| Acs | Cdh | Fmd | mtrB | Ack | Hdr | mcr | Mer | mtb complex | Mtd | mtdB | mtr | pta | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Individuals | 5518 | 138 | 391 | 35 | 2727 | 484 | 540 | 411 | 92 | 248 | 30 | 165 | 730 |
| Dominance_D | 0.2911 | 0.2591 | 0.2697 | 0.2756 | 0.2929 | 0.2719 | 0.2791 | 0.2992 | 0.3419 | 0.2668 | 0.308 | 0.2795 | 0.3577 |
| Simpson_1-D | 0.7089 | 0.7409 | 0.7303 | 0.7244 | 0.7071 | 0.7281 | 0.7209 | 0.7008 | 0.6581 | 0.7332 | 0.692 | 0.7205 | 0.6423 |
| Shannon_H | 1.291 | 1.368 | 1.341 | 1.322 | 1.286 | 1.335 | 1.317 | 1.272 | 1.192 | 1.347 | 1.231 | 1.317 | 1.149 |
| Evenness_e^H/S | 0.9095 | 0.982 | 0.9555 | 0.9381 | 0.9047 | 0.9497 | 0.9335 | 0.892 | 0.8232 | 0.9614 | 0.8559 | 0.9335 | 0.7891 |
| Equitability_J | 0.9315 | 0.9869 | 0.9671 | 0.9539 | 0.9277 | 0.9627 | 0.9503 | 0.9176 | 0.8597 | 0.9716 | 0.8878 | 0.9503 | 0.8291 |
| Fisher_alpha | 0.422 | 0.7701 | 0.6203 | 1.164 | 0.4605 | 0.5971 | 0.5859 | 0.6147 | 0.8528 | 0.6773 | 1.24 | 0.739 | 0.5572 |
Enzymes included; formylmethanofuran dehydrogenase (fmd); formylmethanofuran-H4MPT formyltransferase (ftr); methenyl-H4MPT cyclohydrolase (mch); methylene-5,6,7,8-H4MPT dehydrogenase (mtd); H2-forming N5,N10-methylene-H4MPT dehydrogenase(hmd); 5,10-methylene-H4MPT reductase (mer); H4MPT-methyltransferase (mtr); acetate kinase (ack); phosphate acetyltransferase (pta); acetyl-CoA synthetase (acs); CO dehydrogenase/acetyl CoA synthase(cdh); methanol-specific methyltransferase complex (mta); methylamine-specific methyltransferase complex (mtb); the CoB-CoM heterodisulfide reductase (hdr); and the methyl CoM reductase (mcr).
Among the samples, hits annotated to the mcr genes responsible in the final release of methene gas were significantly higher in composite sample 1c with 179 hits and 1b with 159 hits both in lagoon one. Sample 2c recorded 146 hits while the lowest abundance was recorded in sample 2a with only 56 hits (Fig. 2).
3.3. Organisms responsible for the last step of methanogenesis
This analysis revealed profiles annotated to the mcr (methyl CoM reductase) genes that participate in the final release of methane gas. These were found to be substantially different from the composite samples analyzed. The analysis of variance (ANOVA) to compare the means of the different samples revealed that there was a significant difference in the abundance of the different samples (F (3,112) = 2.779, p = 0.0436) at p < 0.05. The Dunn's post hoc test revealed no significance difference in all the samples except between sample 1c and 2a (Table 4).
Table 4.
Dunn's Post Hoc test for mcr profiles.
| Sample 2a | Sample 2c | Sample 1c | Sample 1b | |
|---|---|---|---|---|
| 2a | 0.09728 | 0.02642 | 0.09338 | |
| 2c | 0.09728 | 0.5742 | 0.9843 | |
| 1c | 0.02642 | 0.5742 | 0.5877 | |
| 1b | 0.09338 | 0.9843 | 0.5877 |
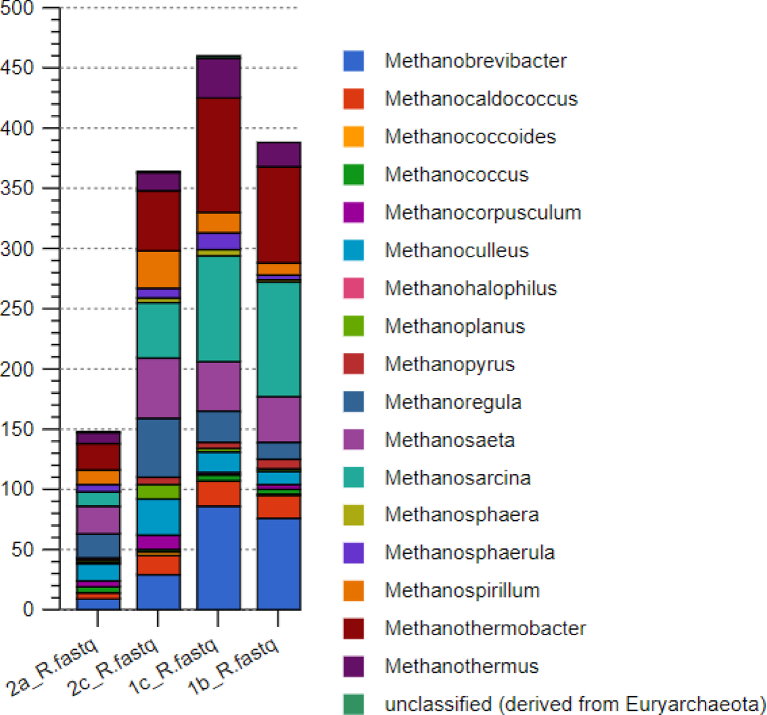
Among the mcr profiles, sample 1c recorded the highest hits, while sample 2a recorded the lowest. Lagoon 1 samples recorded a relatively higher taxa score of 24 each, while lagoon 2 samples recorded a taxa score of 22 for sample 2 c and 21 for sample 2a. There was a generally low dominance among the lagoons ranging between a D value of 0.07753 to 0.08682. Sample 2a was the most diverse and even (Shannon–Wiener index—2.743, Pielou index 0.7304), while sample 1b (Shannon–Wiener index—2.674, Pielou index 0.6042) was the least diverse and least even (Table 5).
Table 5.
Diversity of species annotated to for the mcr genes.
| Sample 2a | Sample 2c | Sample 1c | Sample 1b | |
|---|---|---|---|---|
| Taxa_S | 21 | 22 | 24 | 24 |
| Individuals | 148 | 364 | 460 | 388 |
| Dominance_D | 0.07979 | 0.07862 | 0.07753 | 0.08682 |
| Simpson_1-D | 0.9202 | 0.9214 | 0.9225 | 0.9132 |
| Shannon_H | 2.743 | 2.739 | 2.73 | 2.674 |
| Evenness_e^H/S | 0.7304 | 0.7029 | 0.6474 | 0.6042 |
| Equitability_J | 0.8968 | 0.886 | 0.8632 | 0.8415 |
| Fisher_alpha | 6.684 | 5.149 | 5.381 | 5.657 |
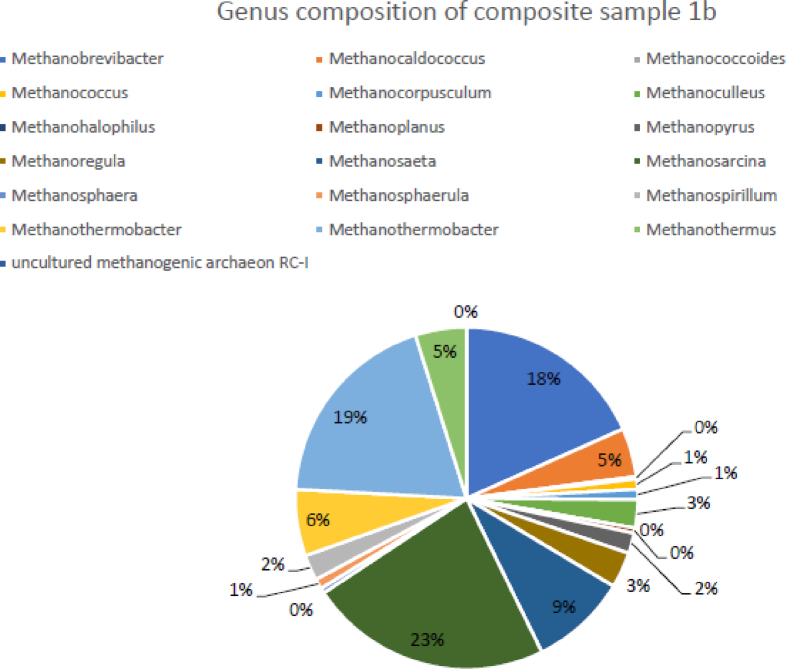
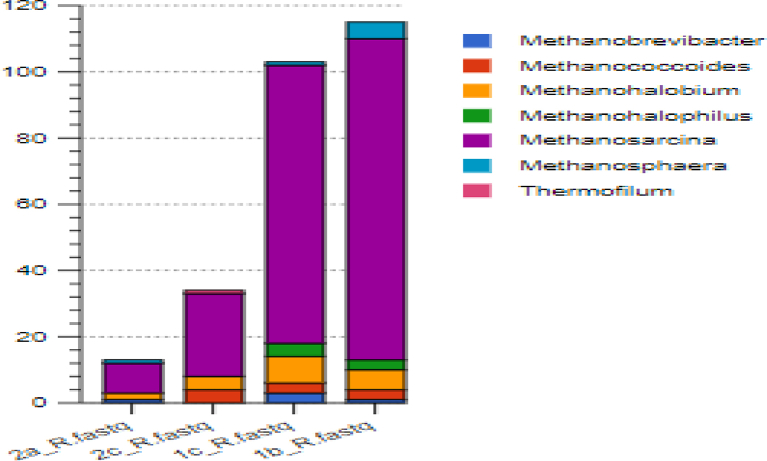
The composite sample 1b was dominated by the Methanosarcina (23%), which is known to produce methane using all the three metabolic pathways for methanogenesis (Gunsalus et al., 2016). The thermophilic hydrogenotrophic Methanothermobacter (19%) and Methanobrevibacter (18%) were the subdominant groups in this sample (Fig. 3). The Metharnosarcina barker (15%) was most of the annotated species, followed by Methernothermobacter thermautotrophicus (14%) and Methanobrevibacter smithii (13%).
Fig. 3.
Genus level profiles of mcr genes annotated profiles in sample 1b
In sample 1c, Methanothermobacter (19%) was dominant; while Methanobrevibacter (17%) and Methanosarcina (18%) were subdominants (see Fig. 4). Metharnosarcina barkeri (13%) and Methernothermobacter thermautotrophicus (13%) formed the abundant species in this composite sample.
Fig. 4.
Genus level profiles of mcr genes annotated profiles in sample 1c.
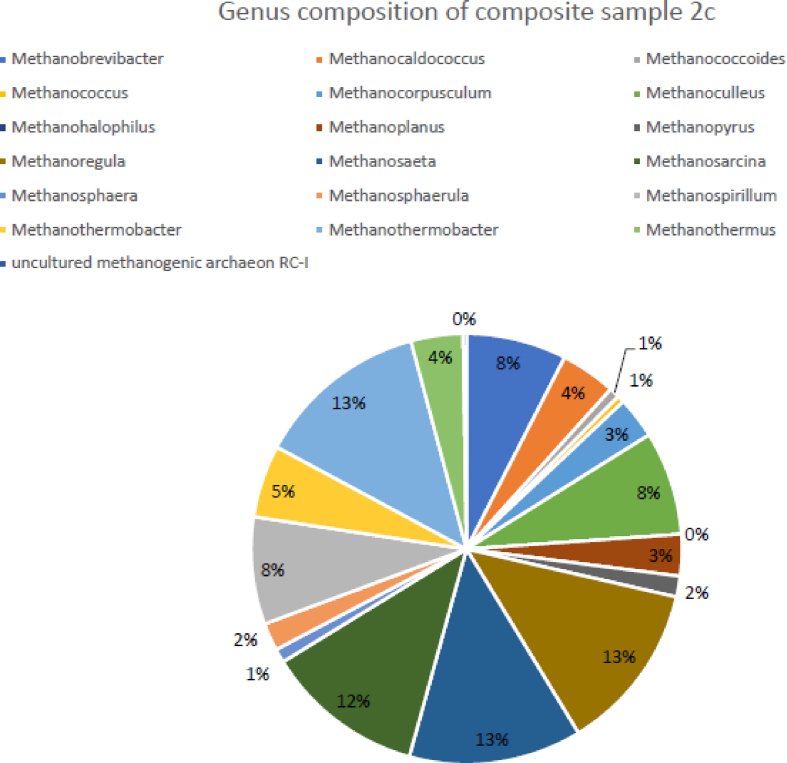
In sample 2c consortium, the dominant groups were the Methanothermobacter (13%), Methanosaeta (13%), Methanoregula (13%), and the Methanosarcina (12%) (Fig. 5). At the species level, mcr gene hits annotated to Methanosaeta thermophile (14%), and Methanoregula boonei (13%) were the most abundant composite sample.
Fig. 5.
Genus level profiles of mcr genes annotated profiles in sample 2c.
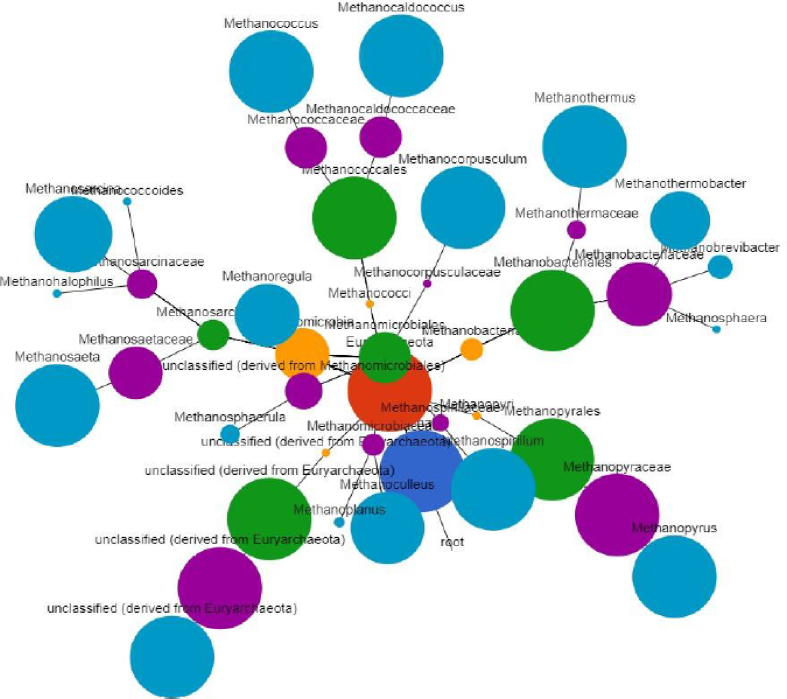
In sample 2a consortium, Methanosaeta (15%), Methanothermobacter (14%), and the Methanosarcina (13%) were the dominant genera. The Methanosaeta thermophile (16%) and Methanoregula boonei (14%) were the abundant species in sample 2a. Fig. 6 indicates a Cytoscape phylogenetic relationship between the mcr-annotated methanogens. Fig. 7 gives the abundance of the mcr-annotated methanogens in the different samples.
Fig. 6.
Cytoscape phylogenetic tree diagram for archaea species annotated to mcr methanogenesis genes responsible for the last release of methane gas.
Fig. 7.
The abundance of the mcr-annotated methanogens in the different samples.
3.4. Organisms responsible for the hydrogenotrophic pathway
An analysis of profiles annotated to formylmethanofuran dehydrogenase subunits A, C, and E (fmdACE) was done since fmd acts as an indicator of the hydrogenotrophic pathway. There were no significant differences in the abundance of hits at p < 0.05 level for the four samples (F (3, 364) = 1.393, p = 0.2447). More hits were recorded in samples 1c (796), followed by 1b (793) drawn from lagoon 1. Lagoon 2 samples recorded lower hits: 2c (671) and 2a (329).
In accordance with taxa, sample 2c was richer with a score of 67, followed by 1c (65), 1b (59), and the lowest was sample 2a with a score of 44. Sample 2c consortium was the most diverse (Shannon–Wiener index—3.203), while sample 2a was the least diverse (Shannon–Wiener index—3.203). Nevertheless, the organisms recorded a lower evenness, with sample 2a having a Pielou index of 0.4431, 2c (0.3672), 1b (0.3578), and the lowest even was sample 1c (0.3047), as shown in Table 6.
Table 6.
Diversity indices of profiles annotated to fmd gene.
| Sample 2a | Sample 2c | Sample 1c | Sample 1b | |
|---|---|---|---|---|
| Taxa_S | 44 | 67 | 65 | 59 |
| Hits | 329 | 671 | 796 | 793 |
| Dominance_D | 0.08683 | 0.07142 | 0.09577 | 0.08378 |
| Simpson_1-D | 0.9132 | 0.9286 | 0.9042 | 0.9162 |
| Shannon_H | 2.97 | 3.203 | 2.986 | 3.05 |
| Evenness_e^H/S | 0.4431 | 0.3672 | 0.3047 | 0.3578 |
| Fisher_alpha | 13.65 | 18.52 | 16.74 | 14.74 |
| Berger-Parker | 0.2036 | 0.1744 | 0.2186 | 0.1866 |
Bray-Curtis similarity index recorded a higher similarity score of 91% between the microbial consortium of samples 1b and 1c probably because they belonged to the same lagoon. Sample 2a was 65% similar to sample 2c even though they came from the same lagoon. The lowest similarity score was between samples 2a and 1b at 49%.
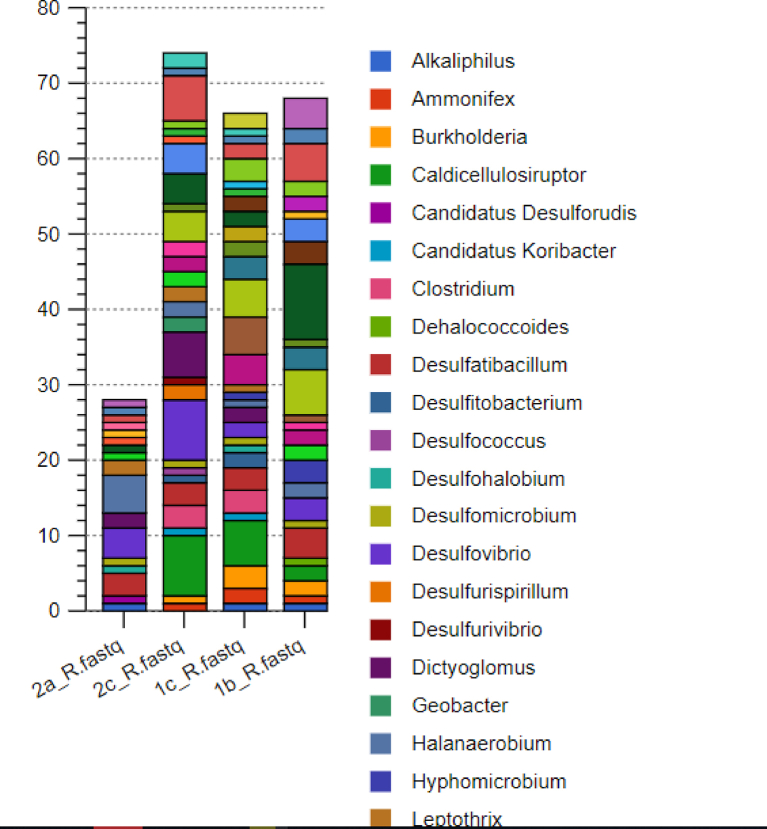
The fmd profiles were mainly from the archaea (90.17%). Among the archaea, Methanosarcina (32%) and Methanothermobacter (27%) dominated sample 1b consortia. A similar scenario was observed in sample 1c with Methanothermobacter (31%) and Methanosarcina (29%). Sample 2c was dominated by Methanobrevibacter (20%), while Methanoregula (13%), Methanosarcina (12%), and Methanobrevibacter (11%) were the subdominant genera. Sample 2a was similar to 2c with Methanospirillum (22%), Methanosarcina (18%), Methanoregula (14%), and Methanothermobacter (11%), as shown in Fig. 8. Fig. 9 summarizes the abundance of hydrogenotrophic archaea community in the four samples.
Fig. 8.
Genera composition of archaea profiles from fmd annotation.
Fig. 9.
Abundance of the different genera of archaea in the hydrogenetrophic.
The Nitrosococcus (22%) dominated sample 1b bacterial consortium among the bacteria. Caldocellulosiruptor (9%), Methylococcus (7%), Methylocella (7%), and Methylobasillus (6%) dominated the sample 1c bacterial community. Caldocellulosiruptor (9%) and Desulfurivibrio (10%) were the dominant bacterial groups in sample 2c consortium. Halanaerobium (17%) was dominant in sample 2a, while Desulfovibrio (14%) and Desulfatibacillum (10%) were the subdominant groups (Fig. 10). Fig. 11 summarizes all the hydrogenotrophic microbes annotated by the fmdACE genes for the bacteria genera.
Fig. 10.
Genera composition of bacterial profiles from fmd annotation.
Fig. 11.
Abundance of the different genera of bacteria in the hydrogenetrophic pathway.
The archaeal species dominated in all the samples with Methanobrevibacter smithii (19%, 22%) Methanosarcina berkeri (16%, 18%) in samples 1b and 1c respectively while Methanospirillum hungatei (17%, 20%) and Methanoregula boonei (12%, 13%) dominated in samples 2c and 2a respectively.
3.5. Organisms responsible for the acetoclastic pathway
In order to identify microorganisms involved in the acetoclastic pathway, an analysis of phylogenetic assignments of the D subunit of Carbon monoxide dehydrogenase/acetyl- CoA synthase (cdhD) was done as it is directly involved in the transmission of a methyl group from acetate during acetoclastic methanogenesis (Thauer et al., 2008).
When considering species dominance, the highest D value was recorded in sample 1c (0.7033), followed by 1b (0.5674), 2c (0.2471), and the lowest was sample 2a (0.1919). The trend in D values was the same as the total number of hits as lagoon 1 recorded higher values than lagoon 2. Nonetheless, sample 2a consortium was the most diverse and even (Shannon–Wiener index—2.142, Pielou index 0.5677), with 2c recording Shannon–Wiener index of 2.064 and Pielou index 0.3581). Samples 1b recorded Shannon–Wiener index of 1.268 and a Pielou index of 0.187. The trend here is that samples in lagoon 2 were the most diverse and even compared to samples in lagoon 1 (Table 7).
Table 7.
Diversity indices for cdh profiles.
| Sample 2a | Sample 2c | Sample 1c | Sample 1b | |
|---|---|---|---|---|
| Taxa_S | 15 | 22 | 19 | 19 |
| Hits | 76 | 162 | 351 | 205 |
| Dominance_D | 0.1919 | 0.2471 | 0.7033 | 0.5674 |
| Simpson_1-D | 0.8081 | 0.7529 | 0.2967 | 0.4326 |
| Shannon_H | 2.142 | 2.064 | 0.8942 | 1.268 |
| Evenness_e^H/S | 0.5677 | 0.3581 | 0.1287 | 0.187 |
When considering taxa in the cdh profiles, sample 2c consortium recorded the highest richness with a score of 22, with the lowest recorded in sample 2a with a score of fifteen 15. Lagoon 1 samples recorded the same score of 19. Nevertheless, the highest number of total hits was recorded in samples from lagoon 1, with 1c recording 351 hits while 1b had 205 hits. Lagoon 2 samples had 162 hits (2c), while the lowest hits were recorded in 2a (76). A test to compare the means of the abundance between the four samples revealed no significant difference (F (3, 104) = 0.4574, p = 0.7123).
Bray-Curtis similarity index recorded a higher similarity score between samples 1b and 1c. The profiles in lagoon 2a were 60% similar to that of 2c. The lowest similarity was recorded in between sample 2a and 1c at 28%. Sample 2c was 42% like sample 1c.
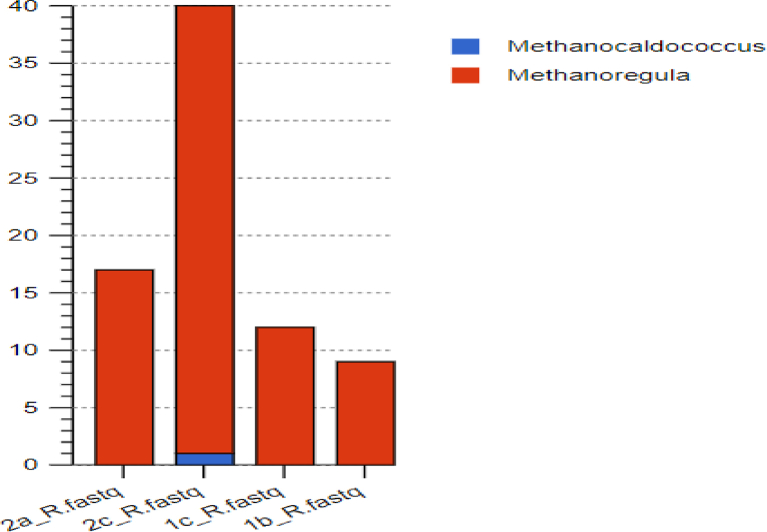
The cdh consortium structure was made up of archaea (8.79%) and bacteria (91.21%). Only two genera of archaea were recorded in the cdh-annotated profiles with one Methanocaldococcus recorded sample 2c only. The Methanoregula dominated in all the samples 2a, 2c, 1c and 1b (Fig. 12). Fig. 13 summarizes the acetoclastic archaea in the samples.
Fig. 12.
Abundance of the different genera of archaea in the acetoclastic pathway.
Fig. 13.
Composition of archaeal cdh-annotated profiles in the composite samples.
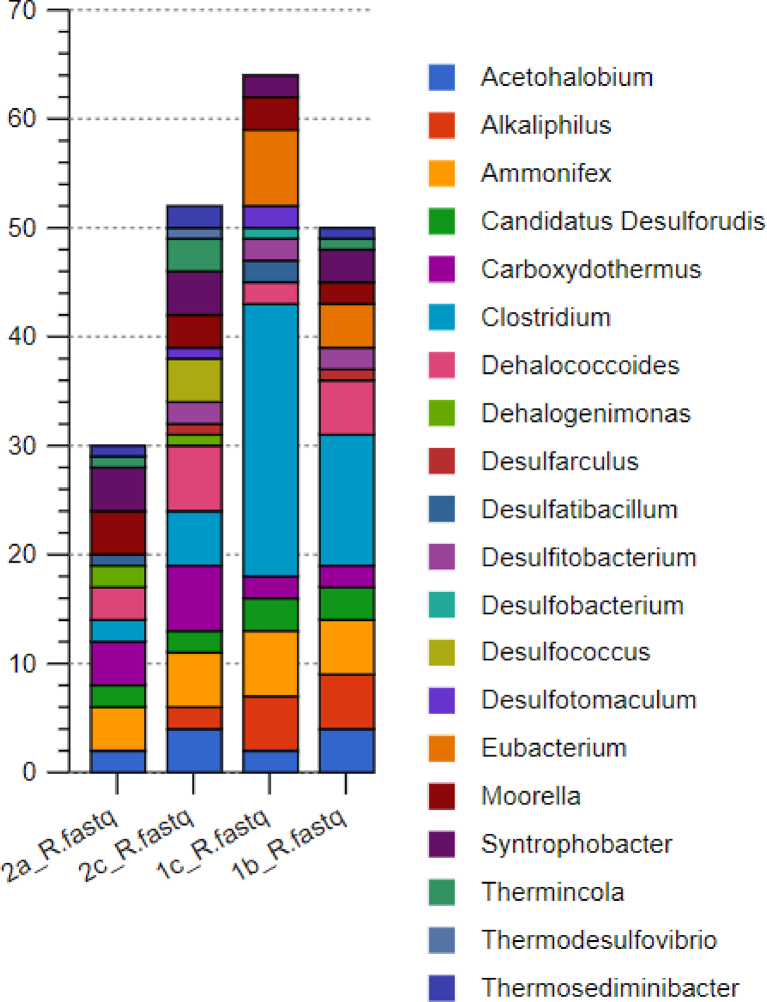
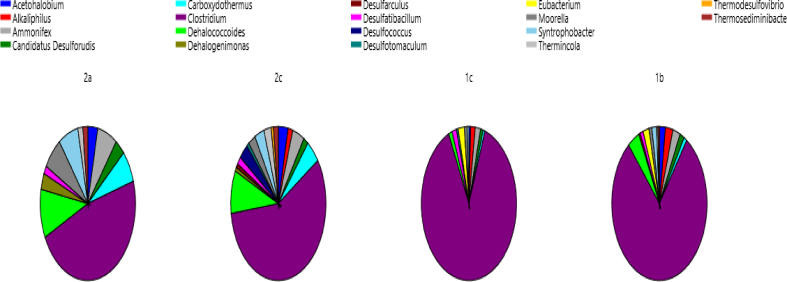
Among the acetoclastic bacterial consortium, Clostridium dominated in the entire sample with 79%, 88%, 57% and 47% in samples 1b, 1c, 2c and 2a respectively (see Fig. 14). Fig. 15 summarizes the acetoclastic bacteria in the sample.
Fig. 14.
Abundance of the different genera of bacteria in the acetoclastic pathway.
Fig. 15.
Composition of bacterial cdh-annotated profiles in the four samples.
Species Clostridum difficile was the most abundant at 79% in sample 1b and 84% in sample 1c. Clostridum difficile also dominated in sample 2c (43%) and 2a (37%) with Methanoregula boonei forming the subdominant group at 24% in sample 2c and 22% in sample 2a.
3.6. Organisms responsible for the methylotrophic pathway
Utilization of methanol or methylamines is the third commonly recognized methanogenic pathway, which contains genes of methanol and mono-, di- and trimethylamine methyltransferases (mta, mtm, Mtb, mtt, respectively). A comparison of the available domain profile sequences of mtaB, mtmB, mtbB, and mttB showed that their abundances were high in sample 1c with 34% of the total hits, followed by sample 1b (32%), 2c (25%) and the least in 2a (9%). The hits in this category were composed of bacteria (3%) and archaea (97%).
Analysis of variance (ANOVA) was carried out to compare the differences in the means of the abundances of profiles in the different sampling points, and there was a significant difference (F (3, 208) = 3.3103), P = 0.02763 between the samples at p < 0.05). A post hoc test was carried out to determine which samples were significantly different from each other and significant differences was noted between sample 2a and 2c, 2a and 1c, 2a and 1b while all the other samples reported no significant differences (Table 8).
Table 8.
Dunn's post hoc test for the methylotrophic pathway’ profiles (ANOVA, 0.006088, p < 0.05).
| 2a | 2c | 1c | 1b | |
|---|---|---|---|---|
| 2a | 0.04315 | 0.00407 | 0.006088 | |
| 2c | 0.04315 | 0.3951 | 0.471 | |
| 1c | 0.00407 | 0.3951 | 0.8968 | |
| 1b | 0.006088 | 0.471 | 0.8968 |
Taxa richness was higher in sample 1c with a score of 43 and lowest in 2a with a score of 31. Generally, dominance was low across the samples with a D value ranging between 0.07018 and 0.07672 with the highest in 2c and lowest in sample 1b. The most diverse consortium was in sample 1b (Shannon–Wiener index—3.022) and 1c (Shannon–Wiener index—3.006). Despite sample 2a being the least diverse (Shannon–Wiener index—2.958), it was the most even consortium with Pielou index of 0.6215 while the least even was sample 1c with the Pielou index of 0.4697 (Table 9).
Table 9.
Diversity indices for methylotrophic pathway’ profiles.
| Sample 2a | Sample 2c | Sample 1c | Sample 1b | |
|---|---|---|---|---|
| Taxa_S | 31 | 38 | 43 | 41 |
| Individuals | 166 | 439 | 601 | 575 |
| Dominance_D | 0.07579 | 0.07672 | 0.07115 | 0.07018 |
| Simpson_1-D | 0.9242 | 0.9233 | 0.9289 | 0.9298 |
| Shannon_H | 2.958 | 2.98 | 3.006 | 3.022 |
| Evenness_e^H/S | 0.6215 | 0.5179 | 0.4697 | 0.5008 |
| Equitability_J | 0.8615 | 0.8191 | 0.7991 | 0.8138 |
| Fisher_alpha | 11.24 | 9.984 | 10.6 | 10.1 |
Among the archaea, the most dominant group was the Methanosarcina (21%) while Methanobrevibecter (19%), Methanothermobacter (18%) and Methanosaeta (15%) were subdominant groups in sample 1b. The same trend is seen in sample 1c with Methanosarcina (29%), while Methanobrevibecter (15%), and Methanothermobacter (15%) forming the sub dominant groups. For the lagoon 2 samples, Methanoregula (18%), Methanosarcina (16%) and Methanospirillum (15%) dominated sample 2c consortium while Methanosarcina (22%), Methanosaeta (15%), Methanoregula (15%), and Methanospirillum (13%) were the dominant group in sample 2c archaeal community (Fig. 16). Fig. 17 summarizes the methylotrophic archaea in the samples.
Fig. 16.
Abundance of the different genera of archaea in the methylotrophic pathways.
Fig. 17.
Composition of the archaea community in the methylotrophic pathway’ profiles.
The Nitrosococcus (33%) and the Methylococcus (33%) dominated the bacterial community in sample 1b. The Methylococcus (31%) and Methylobacterium (31%) dominated sample 1c consortium. In lagoon 2, Thermincola (25%), Methylobacterium (25%), Burkolderia (13%) and Methylovorus (13%) dominated in sample 2c. Methylococcus were the only group found in sample 2a (Fig. 18). Fig. 19 summarizes all the methylotrophic bacteria in the different samples.
Fig. 18.
Abundance of the different genera of bacteria in the methylotrophic pathway.
Fig. 19.
Composition of the bacterial community in the methylotrophic pathway’ profile.
Methanosarcina barkeri (14%, 15%) and Methernothermobacter themeratotrophicus (13%, 13%) were the most abundant in sample 1b and 1c respectively. Methanoregula boonei (15%, 16%) and Methanospirillum hungatei (14%, 14%) were the dominant groups in lagoon 2 (samples 2c and 2b respectively).
4. Discussion
The whole-genome metagenomic analysis is a useful approach for comprehensively describing complex microbial communities [11] Various tools are available for metagenomic analysis to enable different insights into the environmental community function and performance. This study applied a commonly used metagenomic analytical tool (MG-RAST) to describe and compare four composite sample sequences through deep shotgun metagenomics. The MG-RAST pipeline features allowed the functional structure of the representative samples' phylogenetic placement of methanogenesis-related genes as described in [18].
The metagenomic analysis with the MG-RAST pipeline offered an insight into the metagenomic community structure and the abundance of genes involved in methanogenesis. However, it should be noted that this is a general approach and, therefore, difficult to determine interactions between microorganisms involved in each pathway [11]. MG-RAST provides an analysis platform where KO, SEED subsystems, and the RefSeq databases are explored to allow for a more detailed view and identify a specific function with the simultaneous assignment to a taxonomic group.
The abundance of archaea in current study corresponded well with the proportions of the different samples' functional annotations related to methanogenesis. Even though higher hits were recorded in lagoon 1compared to the aged lagoon 2 for all the genes responsible for the different methanogenesis pathways, this is probably attributed to the fact that the sludge in lagoon 2 has stabilized over time, and therefore more microbes can now thrive. Lagoon 2 recorded a higher diversity of the profiles except in the methylotrophic pathway, where lagoon 1 recorded both higher numbers of hits and diversity of organisms (Fig. 16). This can be attributed to more algal blooms on the liquid surface of lagoon one because some of the one-carbon compounds used by methylotrophs, such as methanol and Trimethylamine N oxide (TMAO), are produced by phytoplankton [19].
All the pathways recorded higher diversity with Shannon Wiener indices above 2.5 except for the hydrogenotrophic pathway, which had a lower diversity of organisms but recorded the highest hits compared to the other pathways. Similar results were recorded by Ref. [11]. The metagenome had a relatively high abundance of genes of the hydrogenotrophic pathway despite a low abundance of Archaea in the samples analyzed.
The results revealed Metharnosarcina as the most abundant archaea in the fresh lagoon one, while Methanospirillum was abundant in the aged lagoon 2. Similar trends are recorded in other studies ([5,20]) [20]. reported the Firmicutes and Nitrospira genera as the predominant bacteria while Methanosaeta, Methanosarcina, and the Methanospirillum dominated the archaea. On the other hand [5], recorded a shift in the composition of archaea from Methanosaeta to Mycobacterium.
The majority of the annotated mcr sequences were assigned taxonomically to the genera Methanoregula and Methanospirilum in both lagoons suggesting these genera play a dominant role in the last step of methane production in the sludge. There is no information linking the acidophilic Methanoregula with the mcr genes as encountered in this study. Other studies suggest Methanospirillum [21]; Methanocorpusculum, Methanobacterium [22]; and Methnanosaeta [23]as the major taxonomic groups assigned to the mcr genes sequences. The Methanospirillum identified in this study is well adapted with a large genome suggesting the presence of unrecognized biochemical/physiological properties that likely extend to the other Methanospirillaceae and include the ability to form the unusual sheath-like structure and to successfully interact with syntrophic bacteria [21].
The phylogenetic placement of hydrogenotrophic pathway organisms was annotated to Methanobrevibacter and Methanosarcina genera in lagoon one, while in lagoon 2, the Methanoregula and Methanospirillum were the dominant hydrogenotrophic genera [11]. reported Methanobrevibacter, Methanomassiliicoccales, Methanoregula, and Methanoculleus as the major contributors to methane production in sewage sludge. The study proved that the hdr genes are found in the methanogenic archaea and acid and thiosulphate-reducing bacteria such as Halanaerobium, sulfate-reducing Desulfovibrio, and the alkene degrading Desulfatibacilum. Other studies ([24,25]) have supported this finding where “methanogenic” genes are also present in other archaea and bacteria [25]. reported the sulfate-reducing Archaeoglobus fulgidus using many enzymes and coenzymes in anaerobic lactic acid oxidation to produce CO2, also used by methanogenic archaea in the reduction of CO2 to methane. Desulfobacterium autotrophicum contains gene clusters for the heterodisulfide reductase [24].
The acetoclastic pathway is the most active and important methanogenesis pathway, especially in sludge anaerobic digesters where acetate contributes two-thirds of the total methane production [8]. The Methanosarcina and Methanosaeta have been described as the genera where acetoclastic methanogenesis occurs [26,27]. The phylogenetic placement of the cdr genes was assigned to Clostridium and Methanoregula as the major taxonomic groups in this category in lagoon one and the aged lagoon two, respectively. The abundance of Clostridium in lagoon 1 (one) acetoclastic pathway consortium compared to methanogens is an interesting phenomenon suggesting the possibility that they play a role in the production of acetate [28]. in their work supported the involvement of some bacteria, such as clostridia, when studying metabolic reconstruction of metagenome-assembled genomes (MAGs) from a thermophilic sewage waste biowaste digester covering the basic functions of the biogas microbial community; consistently identified the uncultured Dethiobacteraceae together with Syntrophaceticus, Tepidanaerobacter, and unclassified Clostridia as members of a potential acetate-oxidizing core community in nine full-scale digesters, whereas acetoclastic methanogens were barely detected. This may be annotated to the fact that acetoclastic methanogens and syntrophic acetate-oxidizing bacteria (SAOB) compete for acetate, a major intermediate in the mineralization of organic matter. The results presented in this study may provide new insights into a remarkable anaerobic digestion ecosystem where members of the Bacteria domain possibly realize acetate catabolism [28]. further demonstrated this by metagenomics and enrichment cultivation, revealing a core community of diverse and novel uncultured acetate-oxidizing bacteria and concluding that their genomic repertoire suggests metabolic plasticity besides the potential for syntrophic acetate oxidation [29]. found that contaminants such as antibiotics limit acetoclastic methanogens, and the resistant syntrophic acetate bacterial oxidants take over from the methanogens. A suggestion has been given that there might be a shift where syntrophic acetate oxidation replaces acetoclastic methanogenesis during thermophilic digestion of bio-waste [30] Nevertheless, there is a need for more studies to quantify syntrophic acetate oxidation versus acetoclastic methanogenesis.
The methylotrophic pathway was dominated by Methanosaeta and Methernothermobacter genera in the lagoons 1. Methanoregula and Methanospirillum were the dominant methylotrophic methane producers in the aged lagoons [31]. reported a similar result in anaerobic digesters with Methanomassiliicoccus, Methanosarcina, metanospirillum, and methanosaeta in the list of organisms representing the methylotrophic pathway. Nitrosococcus, Methylococcus, and Methylobacterium were the abundant bacteria in this freshly prepared lagoon 1, while the Thermincola, Mycobacterium, and Methylococcus were abundant bacteria in the methylotrophic annotated consortium. Kaster et al. [9] suggested the existence of the methylotrophic bacteria that use methanogenic enzymes and coenzymes in their energy metabolism. According toGilmore et al. [32], Methanobacterium, Methanosarcina, Methanosphaera, and Methanocorpusculum are suggested to be capable of methylotrophic, acetoclastic, and hydrogenotrophic methanogenesis. In this study, Methanoregula seemed to be dominating all the three methanogenesis pathways. This may be due to their ability that trends toward energy conservation in genome composition [9].
4.1. Limitation of the study
The study is without limitations, which may affect our results. This study only describes microorganisms of only one treatment plant with an aim of providing a baseline data for possible application of biogas production. Biases may have been introduced during the DNA extraction process but this was minimised by the use of an optimal extraction kit specifically for metagenomics compared to other kits. High throughput illumina sequencing has also been proven to produce good repeatability. The sequencing depth of 3.2 GB may not be deep enough to satisfactorily explore rare species within sludge in lagoons 1 and 2, however relatively high number of sequences ranging between 149, 542,217 and 485,547,217 per sample. The other advantage is that metagenomics sequencing used in this work is neither low throughput nor PCR based. Lastly, an average base pairs ranging from 157 to 172 bps used in this study may be short. For most metagenomics pipelines, including MR-RAST, 100 bps is long enough to identify a species but an optimal of 200 bps id recommended for a trade-off between the rate of under prediction and the production of such reads.
5. Conclusion
The approach presented in this study allowed exploration in detail of complex microbial communities coming from methane-producing environments. Microbial communities in methane-producing sludge environments would be expected to contain a high abundance of genes of different steps of hydrogenotrophic, acetoclastic, and methylotrophic pathways, which optimally are encoded by a few microbes. This view was true for the two lagoons under investigation, especially in the freshly created lagoon 1. Nevertheless, we observed different levels of methanogenesis genes and their dispersion amongst various microorganisms. This was especially apparent for the acetoclastic pathway suggesting that the syntrophic acetate oxidation bacteria are reservoirs of metagenomic genes that contribute to the methane cycle. There was no significant difference in the diversity of acetoclastic and the hydrogenotrophic pathways. However, the methylotrophic pathways were significantly different between the lagoons. The organisms responsible for the last step release of methane gas were significantly different among the four samples. There is a need to quantify the methane produced by the different pathways of methanogenesis. An investigation on the effect of physio chemical factors such as heavy metals and antibiotics on microbes responsible for the different stages of methane production at the plant will also be prudent. The study also recommends a pilot experiment on the efficiency of the main methanogens identified in this study to produce biogas, individually and as a consortium.
Funding
The study was partially funded by Rotary Club of Vienna and Egerton University.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
We appreciate Florian of Rotary Club of Vienna; George Gathungu of NYEWASCO-Kangemi plant; Cyrus Kimani (MSC) of KALRO-Njoro; Victoria Wavinya of Inqaba biotechnologies, and Prof. Nzula Kitaka, Anastacia Muia (Ph.D.), Geoffrey Ongo'ndo (Ph.D.), Priscilla Wangari, Erick Owino, Eddison Musikoyo, and Lewis Maina of Egerton University for their academic and technical advice as the case may be.
Contributor Information
Allan.K. Kimisto, Email: akimisto@gmail.com.
Anastasia W. Muia, Email: anastasia.muia@egerton.ac.ke.
Geoffrey O. Ong'ondo, Email: geoffrey.ongondo@egerton.ac.ke.
Kimani.C. Ndung'u, Email: nkimanicyrus@gmail.com.
References
- 1.Seviour R.J., Nielsen P.H. Microbial Ecology of Activated Sludge. IWA Publishing; 2010. Microbial communities in activated sludge plants; pp. 95–126.https://vbn.aau.dk/en/publications/microbial-communities-in-activated-sludge-plants [Google Scholar]
- 2.Pham V.H., Kim J. Cultivation of unculturable soil bacteria. Trends Biotechnol. 2012;30(9):475–484. doi: 10.1016/j.tibtech.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 3.Giwa A.S., Ali N., Athar M.A., Wang K. Dissecting microbial community structure in sewage treatment plant for pathogens' detection using metagenomic sequencing technology. Arch. Microbiol. 2019;202(4):825–833. doi: 10.1007/s00203-019-01793-y. [DOI] [PubMed] [Google Scholar]
- 4.Bajpai P. Anaerobic Technology in Pulp and Paper Industry; 2017. Basics of Anaerobic Digestion Process; pp. 7–12. [DOI] [Google Scholar]
- 5.Li J.X., Wang L.A., Wang L., Zhan X.Y., Huang C. Exploring the biogas production and microbial community from Co-digestion of sewage sludge with municipal solid waste incineration fresh leachate. Int. J. Environ. Sci. Technol. 2020;18(4):901–912. doi: 10.1007/s13762-020-02884-w. [DOI] [Google Scholar]
- 6.Dubey G., Kollah B., Ahirwar U., Mohanty S.R. Carbon dioxide metabolism and ecological significance of enzyme complex systems in terrestrial ecosystem. Current Life Sciences. 2015;1(2):35–45. [Google Scholar]
- 7.Crable B.R., Plugge C.M., McInerney M.J., Stams A.J. Enzyme Research; 2011. Formate Formation and Formate Conversion in Biological Fuels Production. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.St-Pierre B., Wright A.G. vol. 138. Bioresource Technology; 2013. pp. 277–284. (Metagenomic Analysis of Methanogen Populations in Three Full-Scale Mesophilic Anaerobic Manure Digesters Operated on Dairy Farms in Vermont, USA). [DOI] [PubMed] [Google Scholar]
- 9.Kaster A.K., Goenrich M., Seedorf H., Liesegang H., Wollherr A., Gottschalk G., Thauer R.K. 2011. More than 200 Genes Required for Methane Formation from H2 and CO2 and Energy Conservation Are Present in Methanothermobacter Marburgensis and Methanothermobacter Thermautotrophicus. Archaea, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mashhadi Z. Doctoral dissertation; Virginia Tech: 2010. Identification and Characterization of the Enzymes Involved in Biosynthesis of FAD and Tetrahydromethanopterin in Methanocaldococcus Jannaschii. [Google Scholar]
- 11.Pyzik A., Ciezkowska M., Krawczyk P.S., Sobczak A., Drewniak L., Dziembowski A., Lipinski L. Comparative analysis of deep sequenced methanogenic communities: identification of microorganisms responsible for methane production. Microb. Cell Factories. 2018;17(1) doi: 10.1186/s12934-018-1043-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lyu Z., Shao N., Akinyemi T., Whitman W.B. Methanogenesis. Curr. Biol. 2018;28(13):R727–R732. doi: 10.1016/j.cub.2018.05.021. [DOI] [PubMed] [Google Scholar]
- 13.Desai C., Pathak H., Madamwar D. Advances in molecular and “-omics” technologies to Gauge microbial communities and bioremediation at xenobiotic/anthropogen contaminated sites. Bioresour. Technol. 2010;101(6):1558–1569. doi: 10.1016/j.biortech.2009.10.080. [DOI] [PubMed] [Google Scholar]
- 14.Kim N.K., Lee S.H., Kim Y., Park H.D. Current understanding and perspectives in anaerobic digestion based on genome-resolved metagenomic approaches. Bioresour. Technol. 2022;344 doi: 10.1016/j.biortech.2021.126350. [DOI] [PubMed] [Google Scholar]
- 15.Kariunga S., Kitaka N., Muia W. Transformation towards Sustain-Able and Resilient WASH Services: Proceedings Of the 41st WEDC International Conference. Loughborough University; Nakuru, Kenya: 2018, July. Performance evaluation of Kangemi Sewage Plant in nutrients and organic matter removal, Nyeri Kenya; pp. 9–13. cс WEDC. [Google Scholar]
- 16.Bunu S., Otele D., Alade T., Dodoru R. Determination of serum DNA purity among patients undergoing antiretroviral therapy using nanodrop-1000 spectrophotometer and polymerase chain reaction. Biomed. Biotech. Res. J. (BBRJ) 2020;4(3):214. doi: 10.4103/bbrj.bbrj_68_20. [DOI] [Google Scholar]
- 17.Custodio J.M., Michelini L.J., de Castro M.R.C., Vaz W.F., Neves B.J., Cravo P.V., Napolitano H.B. Structural insights into a novel anticancer sulfonamide chalcone. New J. Chem. 2018;42(5):3426–3434. [Google Scholar]
- 18.Meyer F., Bagchi S., Chaterji S., Gerlach W., Grama A., Harrison T., Paczian T., Trimble W.L., Wilke A. MG-RAST version 4—lessons learned from a decade of low-budget ultra-high-throughput metagenome analysis. Briefings Bioinf. 2017;20(4):1151–1159. doi: 10.1093/bib/bbx105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dinasquet J., Tiirola M., Azam F. Enrichment of bacterioplankton able to utilize one-carbon and methylated compounds in the coastal Pacific Ocean. Front. Mar. Sci. 2018;5:307. https://www.frontiersin.org/articles/10.3389/fmars.2018.00307/full [Google Scholar]
- 20.Díaz E.E., Stams A.J., Amils R., Sanz J.L. Phenotypic properties and microbial diversity of Methanogenic granules from a full-scale upflow anaerobic sludge bed reactor treating brewery wastewater. Appl. Environ. Microbiol. 2006;72(7):4942–4949. doi: 10.1128/aem.02985-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gunsalus R.P., Cook L.E., Crable B., Rohlin L., McDonald E., Mouttaki H., Sieber J.R., Poweleit N., Zhou H., Lapidus A.L., Daligault H.E., Land M., Gilna P., Ivanova N., Kyrpides N., Culley D.E., McInerney M.J. Complete genome sequence of Methanospirillum hungatei type strain JF1. Stand. Gen. Sci. 2016;11(1) doi: 10.1186/s40793-015-0124-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keerthana K.P., Krishnan S.R., Sengali S.R., Srinivasan R., Prabhakaran N., Balaji G., Latha K. Microbiome digital signature of MCR genes–an in silico approach to study the diversity of methanogenic population in laboratory-developed and pilot-scale anaerobic digesters. Acc. Microb. 2019;1(5) doi: 10.1099/acmi.0.000044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ellis J.T., Tramp C., Sims R.C., Miller C.D. Characterization of a Methanogenic community within an algal fed anaerobic digester. ISRN Microb. 2012;2012:1–12. doi: 10.5402/2012/753892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strittmatter A.W., Liesegang H., Rabus R., Decker I., Amann J., Andres S., Henne A., Fricke W.F., Martinez-Arias R., Bartels D., Goesmann A., Krause L., Pühler A., Klenk H., Richter M., Schüler M., Glöckner F.O., Meyerdierks A., Gottschalk G., Amann R. Genome sequence of Desulfobacterium autotrophicumHRM2, a marine sulfate reducer oxidizing organic carbon completely to carbon dioxide. Environ. Microbiol. 2009;11(5):1038–1055. doi: 10.1111/j.1462-2920.2008.01825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liesegang H., Kaster A., Wiezer A., Goenrich M., Wollherr A., Seedorf H., Gottschalk G., Thauer R.K. Complete genome sequence of Methanothermobacter marburgensis , a Methanoarchaeon model organism. J. Bacteriol. 2010;192(21):5850–5851. doi: 10.1128/jb.00844-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fenchel T., King G., Blackburn T. Aquatic sediments. Bacter. Biogeochem. 2012:121–142. doi: 10.1016/b978-0-12-415836-8.00007-4. [DOI] [Google Scholar]
- 27.Vincent S.G., Jennerjahn T., Ramasamy K. Assessment of microbial structure and functions in coastal sediments. Microb. Commun. Coast. Sed. 2021:167–185. doi: 10.1016/b978-0-12-815165-5.00006-6. [DOI] [Google Scholar]
- 28.Dyksma S., Jansen L., Gallert C. Syntrophic acetate oxidation replaces acetoclastic methanogenesis during thermophilic digestion of biowaste. Microbiome. 2020;8(1) doi: 10.1186/s40168-020-00862-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gou M., Wang H., Li J., Sun Z., Nie Y., Nobu M.K., Tang Y. Different inhibitory mechanisms of chlortetracycline and enrofloxacin on mesophilic anaerobic degradation of propionate. Environ. Sci. Pollut. Control Ser. 2019;27(2):1406–1416. doi: 10.1007/s11356-019-06705-7. [DOI] [PubMed] [Google Scholar]
- 30.Campanaro S., Treu L., Rodriguez-Rojas L.M., Kovalovszki A., Ziels R.M., Maus I., Zhu X., Kougias P.G., Basile A., Luo G., Schlüter A., Konstantinidis K.T., Angelidaki I. 2020. New Insights from the Biogas Microbiome by Comprehensive Genome-Resolved Metagenomics of Nearly 1600 Species Originating from Multiple Anaerobic Digesters. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buan N.R. Methanogens: pushing the boundaries of biology. Emerg. Top. Life Sci. 2018;2(4):629–646. doi: 10.1042/etls20180031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gilmore S.P., Henske J.K., Sexton J.A., Solomon K.V., Seppälä S., Yoo J.I., Huyett L.M., Pressman A., Cogan J.Z., Kivenson V., Peng X., Tan Y., Valentine D.L., O'Malley M.A. Genomic analysis of methanogenic archaea reveals a shift towards energy conservation. BMC Genom. 2017;18(1) doi: 10.1186/s12864-017-4036-4. [DOI] [PMC free article] [PubMed] [Google Scholar]