Abstract
Background
Likelihood of Neisseria gonorrhoeae infection in women exposed to male sex partners with increasing N. gonorrhoeae burdens and enhancement by Chlamydia trachomatis is not defined.
Methods
We identified men with urethritis and their regular female sex partners. Exposure to N. gonorrhoeae burdens in men was compared in N. gonorrhoeae-infected versus -uninfected partners. Association of N. gonorrhoeae infection in women with burdens in male partners was estimated using logistic regression. Association of C. trachomatis coinfection and N. gonorrhoeae burdens in women adjusted for burdens in male partners was estimated by linear regression.
Results
In total, 1816 men were enrolled; 202 had ≥2 partners, 91 who confirmed monogamy and were enrolled; 77% were married. Seventy were partners of N. gonorrhoeae-infected men; 58 (83%) were N. gonorrhoeae infected, 26 (45%) C. trachomatis coinfected. Infected women had partners with 9.3-fold higher N. gonorrhoeae burdens than partners of uninfected women (P = .0041). Association of N. gonorrhoeae infection in women with upper quartiles of N. gonorrhoeae burdens in partners increased (odds ratios ≥ 2.97)compared to the first quartile (P = .032). N. gonorrhoeae burdens in C. trachomatis-coinfected women were 2.82-fold higher than in C. trachomatis-uninfected women (P = .036).
Conclusions
N. gonorrhoeae infections increased in women whose partners were infected with higher N. gonorrhoeae burdens. C. trachomatis coinfection was associated with increased N. gonorrhoeae burdens in women.
Keywords: Chlamydia trachomatis infection in women, Neisseria gonorrhoeae
Exposure to male gonococcal burdens was compared in gonococcal-infected versus -uninfected monogamous female partners. Gonococcal infections increased in women whose partners were infected with higher burdens. Chlamydia coinfection in women was also associated with increased gonococcal burdens in the women.
The impact of Neisseria gonorrhoeae burdens in infected men with urethritis upon the likelihood of female sex partners becoming infected with N. gonorrhoeae is not defined. An increase in the infectious dose of pathogenic bacteria is often associated with a corresponding increase in the likelihood of infection and generally is considered axiomatic [1]. However, individual strains of N. gonorrhoeae vary in their ability to escape innate host defense mechanisms [2]; therefore, N. gonorrhoeae burdens required to infect may also vary [3]. We examined exposure of women to large burdens of N. gonorrhoeae present in gonococcal urethritis in men to circumvent potential differences in infectious potential of clinical isolates to override natural immunity and thereby identify dose as a major variable.
N. gonorrhoeae is second to Chlamydia trachomatis as the most common cause of bacterial sexually transmitted infection (STI); the 2 often coinfect [4–6]. In certain geographic locales, N. gonorrhoeae may be expanding into the C. trachomatis niche because of increasing N. gonorrhoeae antimicrobial resistance [7]. Frequent coexistence of dual infection suggests that a biological variable(s) may increase N. gonorrhoeae burdens present during infection when C. trachomatis is present [8, 9]. Coinfection is reported in 10%–40% of persons with N. gonorrhoeae infection in the United States and United Kingdom [10]. Coinfection may preclude spontaneous clearance of N. gonorrhoeae infection [11]; persons repeatedly infected with N. gonorrhoeae are more likely coinfected [12]. Consequently, coinfection may prolong the burden of N. gonorrhoeae in the clinical and epidemiological reservoir. We examined the effect of C. trachomatis coinfection in women on their N. gonorrhoeae burden to better understand increased N. gonorrhoeae infection in C. trachomatis-infected women. Abnormal vaginal flora, that include diminished lactobacilli and increases in anaerobic bacteria, is a risk factor for the development of N. gonorrhoeae/C. trachomatis infection [13–15]. We examined relative abundance of lactobacilli and bacterial vaginosis (BV)-like flora in women infected with N. gonorrhoeae and or C. trachomatis in the context of exposure to and transmission of N. gonorrhoeae.
METHODS
Participants
Men
Our study was conducted from April 2011 to August 2015, at the Institute of Dermatology, Chinese Academy of Medical Sciences, STD Clinic, Nanjing, China. In total, 1816 Chinese speaking men, ≥18 years old, were evaluated for symptomatic urethritis, defined by urethral discharge and/or dysuria. The Institutional Review Boards of the Institute of Dermatology, (approval No. 2009-62), the University of Massachusetts Chan Medical School (No. 13448) and Boston University School of Public Health (No. H28858) approved the study; all participants provided written informed consent. Men who indicated they had unprotected sex with 2 or more women were asked to identify their regular partner(s) with whom they had 1 or more episodes of unprotected vaginal intercourse in the 30 days before to 30 days after symptomatic onset of urethritis (the spread period). Two groups were identified: (1) men diagnosed with gonorrhea microscopically by Gram's stain of urethral specimens with polymorphonuclear neutrophils (PMNs) plus gram-negative intracellular diplococci (GNIDs); and (2) men with presumptive nongonococcal urethritis (NGU; PMNs without GNIDs).
Women
Chinese speaking women, identified as regular sex partners by men, were eligible if they were ≥18 years of age, stated they were not human immunodeficiency virus (HIV) infected, and indicated that they had been monogamous in the recent 30 days with the man who identified them (exclusion criteria for women are in Supplementary Data). Baseline characteristics included: demographic information; contraception use; a history of STIs and reports of genital symptoms. A vaginal speculum examination was performed and specimens collected for laboratory testing. Cervicovaginal lavage (CVL) was also performed [16]; fluid was collected and stored at −80°C for additional microbial testing. A bimanual pelvic examination was also performed.
Laboratory Testing
Microbiology and Molecular Methods: Men
First-voided urine specimens and urethral swabs were collected. Gram stains were performed and swabs streaked onto modified Thayer–Martin medium (Zhuhai DL Biotech Co. Ltd). Gonococci (N. gonorrhoeae) were identified by colonial morphology, Gram's stain and oxidase testing.
Urethral exudates were inoculated into liquid culture media (Mycoplasma IST2; bioMerieux) to identify Ureaplasma species and Mycoplasma hominis. Urine specimens were examined for C. trachomatis by polymerase chain reaction (PCR; DAAN Gene Co. Ltd.). Mycoplasma genitalium and Trichomonas vaginalis PCRs were performed as previously described [17]. N. gonorrhoeae colony forming unit (CFU) equivalents/mL were quantitated in male urine specimens using a modification of asymmetric quantitative polymerase chain reaction (qPCR) [18] (also described in Supplementary Data). Urine specimens from men identified as having NGU, who had monogamous partners (see below), were confirmed negative for N. gonorrhoeae by PCR (DAAN Gene Co. Ltd.).
Microbiology and Molecular Methods: Women
Vaginal specimens were examined for yeast forms, T. vaginalis, and clue cells by microscopy. Initial endocervical swab specimens were gram stained to enumerate PMNs/high-powered field and identify GNIDs, and cultured for N. gonorrhoeae. C. trachomatis PCR was performed on a second endocervical swab; a third swab was cultured for Ureaplasma species and M. hominis; and a fourth tested for M. genitalium and T. vaginalis by PCR [17]. A final swab was frozen (−80°C) and, in N. gonorrhoeae-culture–negative women, was tested for N. gonorrhoeae by PCR (DAAN Gene Co. Ltd.). N. gonorrhoeae CFU equivalents were quantitated in CVL specimens [16] by qPCR [18] as indicated above for male urine specimens. To characterize and differentiate vaginal microbiome signatures, CVL specimens from women with: (1) N. gonorrhoeae negative C. trachomatis positive, (2) N. gonorrhoeae positive C. trachomatis negative, or (3) N. gonorrhoeae positive C. trachomatis positive infections, and (4) uninfected with N. gonorrhoeae and C. trachomatis (N. gonorrhoeae negative C. trachomatis negative), were tested for bacterial 16s rRNA V3–V4 hypervariable regions, which were amplified, sequenced, and assigned genera. Total genomic DNA was extracted from CVLs using QuickExtract DNA Extraction Solution (Lucigen). PCR amplification, library construction, sequencing, and analysis were carried out by Novogene Co. In brief, the V3–V4 hypervariable regions of bacterial 16S rRNA genes were amplified using primers 341F (5′-CCTAYGGGRBGCASCAG-3′) and 806R (5′-GGACTACNNGGGTATCTAAT-3′). Barcoded PCR products were sequenced on an Illumina paired-end platform to generate 250-bp paired-end raw reads. Sequences were processed through the Qiime pipeline [19] and operational taxonomic units (OTUs) quantitated against the SILVA 16s database (https://www.arb-silva.de/) [20] clustered at 97%. Alpha diversity scores were calculated for each sample using the Shannon and Simpson indices (https://digitalinsights.qiagen.com/plugins/clc-microbial-genomics-module/) and were reported at the group level. Based solely on the predominant species identified within the samples of each group, community state type (CST) equivalents were also identified. Serologic testing of blood for syphilis (rapid plasma reagin and treponema pallidum particle agglutination) and Herpes simplex (HSV-1 IgM/IgG and HSV-2 IgM/IgG) were also performed.
Data Management and Statistical Analyses
Demographic, clinical, and laboratory data were recorded in a centralized relational structured query language data base with an ASP.NET web front end using standardized data collection and sample tracking forms, which were fax scanned and sent electronically to the Biostatistics and Epidemiology Data Analytics Center at the Boston University School of Public Health (additional methodological details are described in Supplementary Data). Baseline characteristics were compared among the 3 infection groups in women: (1) N. gonorrhoeae positive C. trachomatis positive, (2) N. gonorrhoeae positive C. trachomatis negative, (3) N. gonorrhoeae negative C. trachomatis positive, and the N. gonorrhoeae/C. trachomatis uninfected group (N. gonorrhoeae negative C. trachomatis negative) using ANOVA for testing means and Fisher exact test for testing categorical distributions. Abundance of microbial genera in each of the infection groups was compared by t test with the N. gonorrhoeae/C. trachomatis uninfected group. Geometric mean of N. gonorrhoeae burdens (CFU equivalents) in N. gonorrhoeae-infected men who were partners of N. gonorrhoeae-infected women (including PCR-only N. gonorrhoeae-positive women), was compared by t test with the geometric mean in N. gonorrhoeae-infected men who were partners of N. gonorrhoeae-uninfected women. Total number of coital exposures associated with N. gonorrhoeae infection in women was tested using a nonparametric test [21, 22] for trend. N. gonorrhoeae DNA sequences from dually infected partners were assessed for concordance by N. gonorrhoeae multiantigen sequence typing (NG-MAST; https://pubmlst.org) [23, 24]. The association of N. gonorrhoeae infection status in women with increasing N. gonorrhoeae burdens (qPCR, divided into quartiles) present in male partners and C. trachomatis status in women was estimated using a logistic regression model. Odds ratios (ORs) and 95% confidence intervals (CIs) were estimated for each quartile of male burdens compared to the first quartile. The association of N. gonorrhoeae burdens (log10) present in women with their C. trachomatis status and N. gonorrhoeae burdens (log10) in men was estimated by linear regression. The interaction of C. trachomatis coinfection and N. gonorrhoeae burdens in men was tested using a likelihood ratio test.
RESULTS
Enrollment of Study Participants
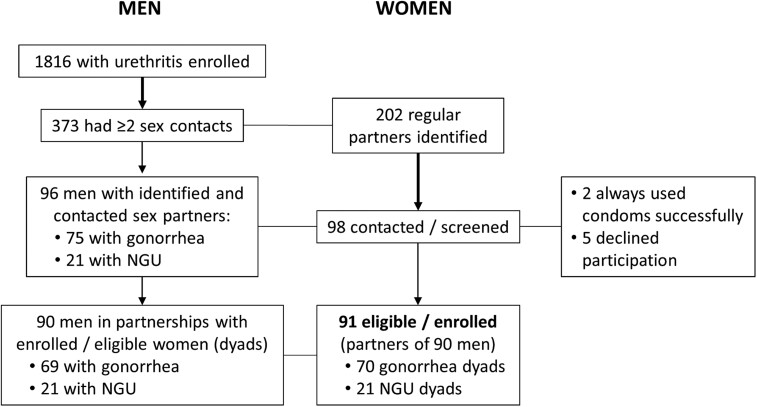
In total, 1816 men were enrolled (Figure 1); 373 had 2 or more sex partners with whom they had unprotected vaginal sex during the spread period; 202 had regular female partners who they identified. Ninety-eight identified matched regular female partners were successfully contacted; all indicated monogamy in the recent 30 days with 96 male partners (2 men each identified 2 regular female partners). Two women were ineligible because their partners always used condoms successfully; 5 declined participation; 91 women were enrolled.
Figure 1.
Men: enrollment (overall) of men with urethritis (having ≥2 sex contacts) and enrolled into this study. Women: identification of regular female sex partners, number contacted and screened, and number eligible and enrolled into this study. Abbreviation: NGU, nongonococcal urethritis.
Microbiology in Men
Microbiologic characterization of urethral specimens from 1816 men is described in Supplementary Data and Supplementary Table 1. In total, 1816 were tested for N. gonorrhoeae, C. trachomatis, M. genitalium, and T. vaginalis; a subset of 1529 were also tested for Ureaplasma species and M. hominis. The prevalence of gonorrhea (and the distribution of other organisms) was maintained in the 96 fully eligible men whose partners had been identified and screened, compared to 373 men who also had 2 or more female partners. Gonorrhea was diagnosed presumptively in 75/96 (78.1%) men by Gram's stain of urethral specimens; all were culture positive for N. gonorrhoeae. Sixty-nine men with gonorrhea matched with 70 partners. N. gonorrhoeae burdens measured by qPCR (Supplementary Data and Supplementary Figure 2) in male urine are shown in Supplementary Table 2. NGU was diagnosed in 21/96 (21.9%) men and confirmed by absence of N. gonorrhoeae by culture and a negative N. gonorrhoeae PCR; 21 men with NGU matched with 21 partners.
Microbiology in Women
Microbiologic characterization of genital specimens from 91 enrolled women is described in Supplementary Data and Supplementary Table 3. In total, 91 were tested for N. gonorrhoeae, C. trachomatis, M. genitalium, and T. vaginalis; a subset of 64 were also tested for Ureaplasma species and M. hominis. Fifty-eight partners of 69 N. gonorrhoeae-infected men (82.9%; 1 infected man had 2 partners) were infected with N. gonorrhoeae; 26 (45%) were coinfected with C. trachomatis. N. gonorrhoeae burdens measured in CVLs by qPCR are shown in Supplementary Table 2. Twelve partners of 21 men with NGU (57.1%) were infected with C. trachomatis; all were negative for N. gonorrhoeae by culture and PCR. A flowchart (Supplementary Figure 2) shows N. gonorrhoeae and C. trachomatis infections in women after contact with infected male sex partners.
Baseline Characteristics in 91 Women
Demographic and Clinical Histories
Baseline characteristics were compared across 3 groups with cervical N. gonorrhoeae and or C. trachomatis infections: (1) N. gonorrhoeae negative C. trachomatis positive; (2) N. gonorrhoeae positive C. trachomatis negative; (3) N. gonorrhoeae positive C. trachomatis positive; and the N. gonorrhoeae/C. trachomatis uninfected group (N. gonorrhoeae negative C. trachomatis negative) (Table 1). Age, race, and college completion were similar across the 4 groups. Overall, 77% (70/91) of women were married (1 cohabited) to their sex partner. Number of days since the last menstrual period did not vary among the groups. Forty-three percent (39/91) used no regular contraception; 3 indicated they had used a condom (with spermicide) as the only means of contraception; 5 others used condoms together with other methods; condom use in 8 women in the 30 days before/after the male partner developed signs/symptoms of urethritis was infrequent (Supplementary Figure 3). Four women reported prior gonorrhea, chlamydial infection, or syphilis; 2 reported genital warts. Fifty-three percent (48/91) reported a prior negative HIV test; the rest had not been tested. Painful intercourse (4 subjects) and abdominal pain (7 subjects) were reported infrequently. Thirty-six percent (33/91) reported prior vaginal infections; there were no differences among the groups. The prevalence of vaginal discharge reported by subjects at the time of the encounter was significantly different among the groups (P = .029); the highest rate was in the group coinfected with N. gonorrhoeae and C. trachomatis (50%) and the lowest rate in the N. gonorrhoeae negative C. trachomatis negative group (10.5%).
Table 1.
Characteristics of Women Infected With Neisseria gonorrhoeae, Chlamydia trachomatis, Both Organisms, or Neither Organism
| Variable | N. gonorrhoeae negative and C. trachomatis negative (n = 19) | N. gonorrhoeae negative and C. trachomatis positive (n = 14) | N. gonorrhoeae positive and C. trachomatis negative (n = 32) | N. gonorrhoeae positive and C. trachomatis positive (n = 26) | P Value |
|---|---|---|---|---|---|
| Baseline | |||||
| Age, y, mean ± SD | 34.58 ± 7.49 | 33.93 ± 7.18 | 31.63 ± 8.22 | 31.46 ± 9.43 | .504 |
| Time since last MP, d, mean ± SD | 16.63 ± 8.47 | 18.79 ± 9.41 | 84.47 ± 270.3 | 26.6 ± 36.25 | .381 |
| Age <40 y | .728 | ||||
| No | 6 (31.6) | 3 (21.4) | 6 (18.8) | 5 (19.2) | |
| Yes | 13 (68.4) | 11 (78.6) | 26 (81.3) | 21 (80.8) | |
| Han race | .205 | ||||
| No | 2 (10.5) | 1 (7.1) | 0 (0.0) | 1 (3.8) | |
| Yes | 17 (89.5) | 13 (92.9) | 32 (100.0) | 25 (96.2) | |
| College or beyond | .226 | ||||
| Less than college | 13 (68.4) | 11 (78.6) | 29 (90.6) | 22 (84.6) | |
| College or beyond | 6 (31.6) | 3 (21.4) | 3 (9.4) | 4 (15.4) | |
| Married or cohabiting | .281 | ||||
| No | 3 (15.8) | 1 (7.1) | 9 (28.1) | 8 (30.8) | |
| Yes | 16 (84.2) | 13 (92.9) | 23 (71.9) | 18 (69.2) | |
| Time since last MP, group | .895 | ||||
| ≤7 d | 3 (15.8) | 3 (21.4) | 3 (9.4) | 5 (20.0) | |
| 8–14 d | 5 (26.3) | 2 (14.3) | 12 (37.5) | 5 (20.0) | |
| 15–21 d | 4 (21.1) | 4 (28.6) | 5 (15.6) | 4 (16.0) | |
| 22–28 d | 6 (31.6) | 4 (28.6) | 8 (25.0) | 7 (28.0) | |
| >28 d | 1 (5.3) | 1 (7.1) | 4 (12.5) | 4 (16.0) | |
| Main contraception | .737 | ||||
| None | 8 (42.1) | 8 (57.1) | 14 (43.8) | 9 (34.6) | |
| Pill | 1 (5.3) | 0 (0.0) | 2 (6.3) | 0 (0.0) | |
| IUD | 6 (31.6) | 5 (35.7) | 7 (21.9) | 10 (38.5) | |
| Tubal ligation | 0 (0.0) | 0 (0.0) | 2 (6.3) | 1 (3.8) | |
| Condom only | 3 (15.8) | 0 (0.0) | 6 (18.8) | 4 (15.4) | |
| Condom with spermicide | 0 (0.0) | 1 (7.1) | 1 (3.1) | 0 (0.0) | |
| Emergency contraception | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (3.8) | |
| Other | 1 (5.3) | 0 (0.0) | 0 (0.0) | 1 (3.8) | |
| Ever tested ± for gonorrhea | .373 | ||||
| Never tested | 2 (10.5) | 2 (14.3) | 8 (25.0) | 9 (34.6) | |
| No | 17 (89.5) | 12 (85.7) | 23 (71.9) | 16 (61.5) | |
| Yes | 0 (0.0) | 0 (0.0) | 1 (3.1) | 1 (3.8) | |
| Ever tested ± for Chlamydia | .677 | ||||
| Never tested | 5 (26.3) | 3 (21.4) | 10 (31.3) | 9 (34.6) | |
| No | 14 (73.7) | 11 (78.6) | 20 (62.5) | 14 (53.8) | |
| Yes | 0 (0.0) | 0 (0.0) | 1 (3.1) | 0 (0.0) | |
| DK | 0 (0.0) | 0 (0.0) | 1 (3.1) | 3 (11.5) | |
| Ever tested ± for syphilis | .718 | ||||
| Never tested | 10 (52.6) | 4 (28.6) | 13 (40.6) | 10 (38.5) | |
| No | 9 (47.4) | 10 (71.4) | 18 (56.3) | 16 (61.5) | |
| Yes | 0 (0.0) | 0 (0.0) | 1 (3.1) | 0 (0.0) | |
| Ever had genital warts | .022 | ||||
| No | 19 (100.0) | 12 (85.7) | 32 (100.0) | 26 (100.0) | |
| Yes | 0 (0.0) | 2 (14.3) | 0 (0.0) | 0 (0.0) | |
| Ever had positive HIV test | .627 | ||||
| Never tested | 10 (52.6) | 5 (35.7) | 17 (53.1) | 10 (38.5) | |
| No | 9 (47.4) | 9 (64.3) | 14 (43.8) | 16 (61.5) | |
| DK | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| Painful intercourse | .259 | ||||
| No | 19 (100.0) | 12 (85.7) | 31 (96.9) | 25 (96.2) | |
| Yes | 0 (0.0) | 2 (14.3) | 1 (3.1) | 1 (3.8) | |
| Abdominal pain | .662 | ||||
| No | 18 (94.7) | 14 (100.0) | 28 (87.5) | 24 (92.3) | |
| Yes | 1 (5.3) | 0 (0.0) | 4 (12.5) | 2 (7.7) | |
| Ever had vaginal infections | .845 | ||||
| No | 12 (63.2) | 9 (64.3) | 17 (53.1) | 14 (53.8) | |
| Yes | 6 (31.6) | 5 (35.7) | 11 (34.4) | 11 (42.3) | |
| DK | 1 (5.3) | 0 (0.0) | 4 (12.5) | 1 (3.8) | |
| Vaginal discharge patient report | .029 | ||||
| No | 17 (89.5) | 8 (57.1) | 23 (71.9) | 13 (50.0) | |
| Yes | 2 (10.5) | 6 (42.9) | 9 (28.1) | 13 (50.0) | |
| Speculum examinations | |||||
| Vaginal discharge examiner | .516 | ||||
| No | 1 (5.3) | 1 (7.1) | 2 (6.3) | 0 (0.0) | |
| Yes | 18 (94.7) | 13 (92.9) | 30 (93.8) | 26 (100.0) | |
| Inflammation of the cervix | .254 | ||||
| Absent | 12 (63.2) | 5 (35.7) | 14 (43.8) | 8 (30.8) | |
| Minimal | 6 (31.6) | 5 (35.7) | 12 (37.5) | 9 (34.6) | |
| Moderate | 1 (5.3) | 4 (28.6) | 6 (18.8) | 9 (34.6) | |
| Cervical discharge examiner | .505 | ||||
| No | 1 (5.3) | 1 (7.1) | 1 (3.1) | 0 (0.0) | |
| Yes | 18 (94.7) | 13 (92.9) | 31 (96.9) | 26 (100.0) | |
| Cervical discharge [purulent]: amount | .818 | ||||
| Scant | 2 (50.0) | 4 (66.7) | 9 (56.3) | 5 (33.3) | |
| Moderate | 2 (50.0) | 2 (33.3) | 6 (37.5) | 9 (60.0) | |
| Large | 0 (0.0) | 0 (0.0) | 1 (6.3) | 1 (6.7) | |
| Vaginal/cervical microscopy/serologies | |||||
| Vaginal wet prep (WP) | .121 | ||||
| Abnormal | 4 (21.1) | 8 (57.1) | 10 (31.3) | 12 (46.2) | |
| Normal | 15 (78.9) | 6 (42.9) | 22 (68.8) | 14 (53.8) | |
| WP abnormal: trichomonas | .208 | ||||
| Positive | 0 (0.0) | 0 (0.0) | 0 (0.0) | 2 (7.7) | |
| Negative | 19 (100.0) | 14 (100.0) | 32 (100.0) | 24 (92.3) | |
| WP abnormal: yeast | .154 | ||||
| Positive | 0 (0.0) | 1 (7.1) | 0 (0.0) | 0 (0.0) | |
| Negative | 19 (100.0) | 13 (92.9) | 32 (100.0) | 26 (100.0) | |
| WP abnormal clue cell | .222 | ||||
| Positive | 4 (21.1) | 7 (50.0) | 10 (31.3) | 12 (46.2) | |
| Negative | 15 (78.9) | 7 (50.0) | 22 (68.8) | 14 (53.8) | |
| Cervical Gram's stain for PMNs | .019 | ||||
| 0 PMNs | 0 (0.0) | 0 (0.0) | 3 (9.4) | 0 (0.0) | |
| 1–4 PMNs | 11 (57.9) | 6 (42.9) | 11 (34.4) | 4 (15.4) | |
| 5–9 PMNs | 5 (26.3) | 6 (42.9) | 8 (25.0) | 7 (26.9) | |
| ≥10 PMNs | 3 (15.8) | 2 (14.3) | 10 (31.3) | 15 (57.7) | |
| Cervical Gram's stain for GNIDs | .003 | ||||
| Positive | 0 (0.0) | 0 (0.0) | 7 (21.9) | 9 (34.6) | |
| Negative | 19 (100.0) | 14 (100.0) | 25 (78.1) | 17 (65.4) | |
| RPR | .826 | ||||
| Negative | 18 (94.7) | 14 (100.0) | 31 (96.9) | 24 (92.3) | |
| Positive | 1 (5.3) | 0 (0.0) | 1 (3.1) | 2 (7.7) | |
| TPPA | .683 | ||||
| Negative | 18 (100.0) | 14 (100.0) | 30 (93.8) | 24 (92.3) | |
| Positive | 0 (0.0) | 0 (0.0) | 2 (6.3) | 2 (7.7) | |
| HSV-1 IgG | .414 | ||||
| Negative | 1 (5.6) | 0 (0.0) | 3 (9.4) | 0 (0.0) | |
| Positive | 17 (94.4) | 14 (100.0) | 29 (90.6) | 26 (100.0) | |
| HSV-1 IgM | .084 | ||||
| Negative | 18 (100.0) | 14 (100.0) | 32 (100.0) | 23 (88.5) | |
| Positive | 0 (0.0) | 0 (0.0) | 0 (0.0) | 3 (11.5) | |
| HSV-2 IgG | .883 | ||||
| Negative | 14 (77.8) | 10 (71.4) | 21 (65.6) | 18 (69.2) | |
| Positive | 4 (22.2) | 4 (28.6) | 11 (34.4) | 8 (30.8) | |
| HSV-2 IgM | .873 | ||||
| Negative | 18 (100.0) | 14 (100.0) | 30 (93.8) | 25 (96.2) | |
| Positive | 0 (0.0) | 0 (0.0) | 2 (6.3) | 1 (3.8) | |
Data are No. (%) except where indicated.
Abbreviations: DK, don't know; GNID, gram-negative intracellular diplococci; HIV, human immunodeficiency virus; HSV, herpes simplex virus; IgG, immunoglobulin G; IUD, intrauterine device; MP, menstrual period; PMN, polymorphonuclear neutrophil; RPR, rapid plasma reagin; TPPA, Treponema pallidum particle agglutination; WP, wet prep.
Baseline characteristics were compared among the 3 infection groups in women N. gonorrhoeae negative C. trachomatis positive, N. gonorrhoeae positive C. trachomatis negative, N. gonorrhoeae positive C. trachomatis positive, and the uninfected group N. gonorrhoeae negative C. trachomatis negative using ANOVA for testing means and Fisher exact test for testing categorical distributions.
Speculum and Physical Examinations
Ninety-six percent (87/91) of women had vaginal discharge on speculum examination: the quality or amount did not vary across the 4 groups (Supplementary Table 4 and Table 1). Ninety-seven percent (88/91) of women had a cervical discharge: the quality (purulent or mucoid) or amount did not vary across the groups. No genital lesions were identified. Bimanual pelvic examinations were normal (all findings negative) in greater than 91% of subjects; uterine tenderness was elicited in 8 subjects (8.7%).
Vaginal and Cervical Analyses
Results of microscopic vaginal smears (yeast, T. vaginalis, and clue cells) are summarized in Table 1 and Supplementary Table 5. There was no significant difference (P = .353) in the prevalence of clue cells among groups with abnormal wet preps. The numbers of PMNs seen by microscopy on a cervical Gram stain was significantly different (P = .019) among the groups: 25/58 (43%) of N. gonorrhoeae or N. gonorrhoeae/C. trachomatis coinfected women had ≥10 PMNs/high-powered field versus 3/19 (7%) of N. gonorrhoeae-negative C. trachomatis-negative women. Only 28% (16/58) of women infected either with N. gonorrhoeae or coinfected with N. gonorrhoeae and C. trachomatis had GNIDs seen on a Gram stain (Table 1).
Cervicovaginal Microbiomes
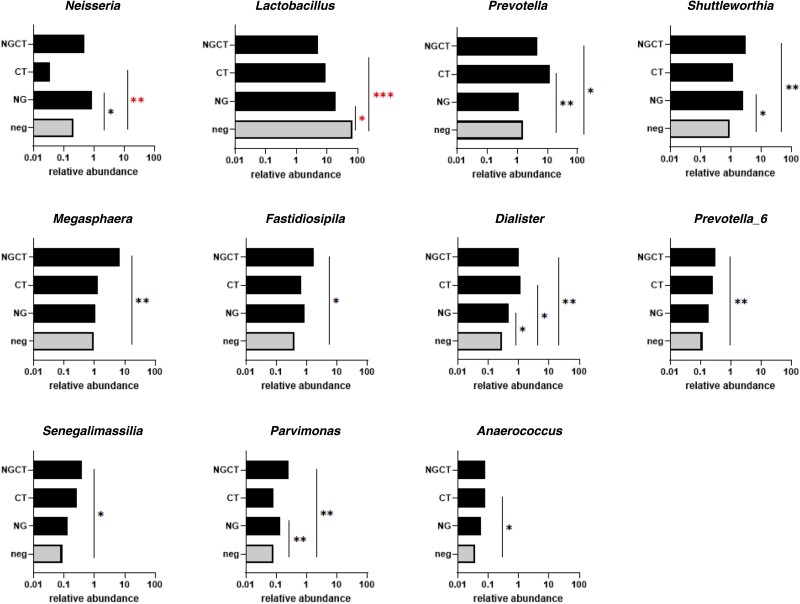
Detailed composition of vaginal microbiomes is presented in Supplementary Table 6. OTUs were normalized using a standard of sequence number corresponding to the sample with the least sequences. Normalized OTU data from the (1) N. gonorrhoeae negative C. trachomatis positive, (2) N. gonorrhoeae positive C. trachomatis negative, and (3) N. gonorrhoeae positive C. trachomatis positive infected groups were each compared with data from group (4), the N. gonorrhoeae negative C. trachomatis negative set of samples in which the abundance of Lactobacillus (using median values: 66% of 1107 total genera and 80% of the top 25 genera) was normal [25]. The top 25 genera (based on median levels of normalized OTU data) comprised greater than 95% mean total abundance in all 4 groups (Supplementary Table 5). Predominance of several major genera in each infection group was increased compared to N. gonorrhoeae-negative C. trachomatis-negative women, mostly in women with N. gonorrhoeae/C. trachomatis coinfection (Figure 2 and Supplementary Table 5). Alpha diversity indicated significant differences between the N. gonorrhoeae-negative C. trachomatis-negative group and the N. gonorrhoeae, C. trachomatis, and N. gonorrhoeae/C. trachomatis groups (Supplementary Table 7). CST 4 equivalent was most prevalent overall (55/91): N. gonorrhoeae (19/32); N. gonorrhoeae/C. trachomatis (22/26); C. trachomatis (8/14); and N. gonorrhoeae-negative C. trachomatis-negative group (6/19). The N. gonorrhoeae-negative C. trachomatis-negative group was predominantly CST 3 equivalent (11/19; Lactobacillus iners). The distribution of CSTs by category are shown in Supplementary Table 8.
Figure 2.
The relative abundance of 11 (of the top 25) bacterial genera measured in Neisseria gonorrhoeae (NG)-infected and/or Chlamydia trachomatis (CT)-infected cervicovaginal lavage specimens that had values significantly different from specimens with neither organism (neg), calculated by t test. Normalized read values of operational taxonomic units (OTUs), quantitated against the SILVA 16s database (https://www.arb-silva.de/) [20] that clustered at 97% OTUs were measured in specimens infected with both N. gonorrhoeae and C. trachomatis (NGCT), C. trachomatis, or N. gonorrhoeae and compared with values in specimens with neither organism (neg). *P < .05, **P < .01, and ***P < .001 denote abundance values that are significantly greater than neither organism (neg). Asterisks denote values significantly greater or less than neg.
Syphilis and HSV Serologies
Four women had positive syphilis serologies (3 rapid plasma reagin and 4 treponema pallidum particle agglutination). HSV antibody results are shown in Table 1; 27/91 (30%) women had measurable HSV-2 IgG antibody; there were no differences among the groups.
Likelihood of Infection in Women Exposed to N. gonorrhoeae-Infected Partners
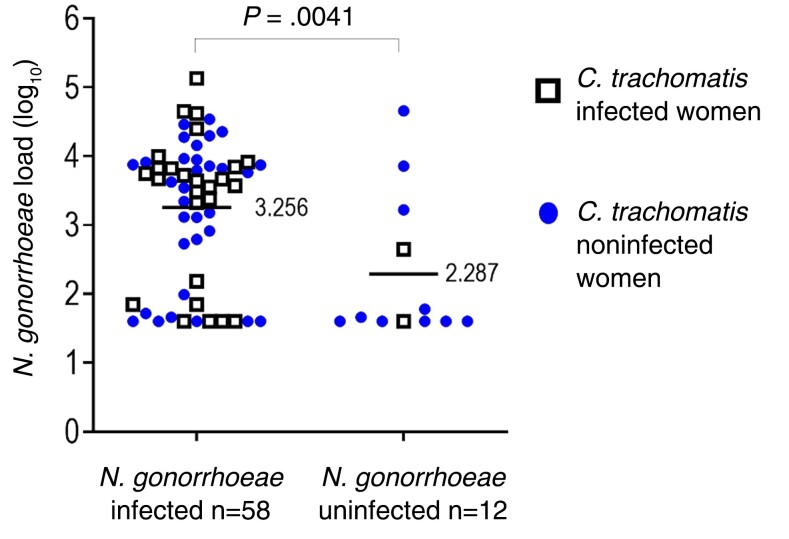
Overall, N. gonorrhoeae-infected women were partners of men with 9.3-fold higher mean N. gonorrhoeae burden (log10 = 3.26) versus mean N. gonorrhoeae burden (log10 = 2.29) in partners of uninfected women (Figure 3) (P = .0041).
Figure 3.
The load (log10 of colony forming unit equivalents) of Neisseria gonorrhoeae in men and N. gonorrhoeae infection status in 70 exposed monogamous female sex partners. Comparison of the means of N. gonorrhoeae burdens in men whose partners were N. gonorrhoeae-infected versus N. gonorrhoeae-uninfected partners was calculated by t test.
Coital Exposures of Women to Gonorrhea-Infected Male Sex Partners
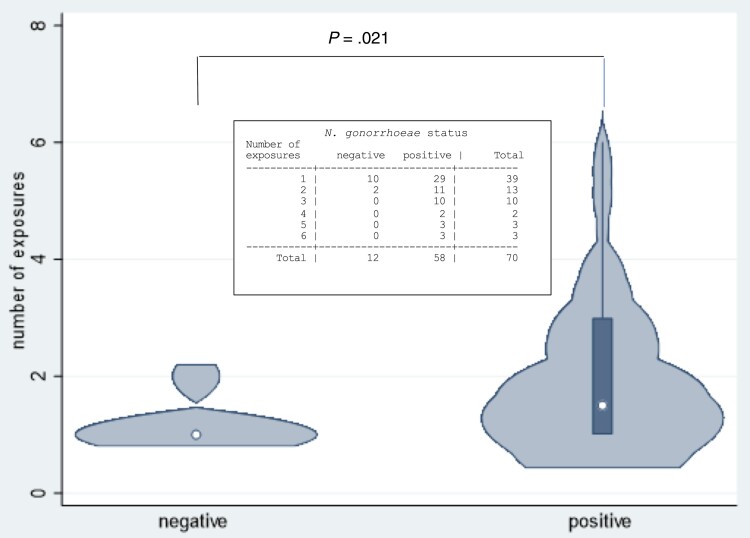
There was a relationship, overall, between increased numbers of coital exposures and likelihood of N. gonorrhoeae infection in women (Figure 4 and Supplementary Figure 3) (P = .021). Exposures where a condom was used were excluded from analysis.
Figure 4.
The number of coital exposures between Neisseria gonorrhoeae-infected men and their female sex partners and the resultant number of infections in women. The total number of exposures associated with N. gonorrhoeae infection in women was tested using a nonparametric test of trend that examined if the rate of N. gonorrhoeae increased with increasing number of exposures.
Genotypes in Gonococcal (N. gonorrhoeae) Partnerships
Forty-six NG-MAST ST types were seen across partnerships where both members were N. gonorrhoeae infected (Supplementary Table 9); 57/58 (98%) N. gonorrhoeae dually infected couples shared an identical NG-MAST ST.
Gonococcal (N. gonorrhoeae) Burden in Men and Likelihood (or Risk) of N. gonorrhoeae Infection in Women
The unadjusted estimated association of N. gonorrhoeae infection in women and quartile of N. gonorrhoeae burdens in men (Table 2)was significantly different, overall (P = .032). The highest risk was exposure to the third and fourth quartiles versus the first quartile (OR = 10.82; 95% CI, 1.17–100.44 and OR = 10.18; 95% CI, 1.09–94.83, respectively). When adjusted for C. trachomatis infection in women, the adjusted estimated association (aORs) of women acquiring N. gonorrhoeae infection remained similar with exposure to each successive quartile (Table 2; overall P = .052); the highest risk was again exposure to the third and fourth quartiles versus the first quartile (aOR = 8.66; 95% CI, .90–83.26 and aOR = 10.17; 95% CI, 1.06–97.08, respectively). The unadjusted estimated association of N. gonorrhoeae and C. trachomatis coinfection versus C. trachomatis-negative status in N. gonorrhoeae-infected women showed a positive trend (OR = 4.06; 95% CI, .82–20.20), which persisted when adjusted for N. gonorrhoeae burdens in male sex partners (OR = 3.47; 95% CI, .65–18.59; Table 2).
Table 2.
Logistic Regression Model That Examined Factors Associated With Neisseria gonorrhoeae Infection in Women
| Association With N. gonorrhoeae Infection in Women | Unadjusted Odds Ratios (95% CI) | Adjusteda Odds Ratios (95% CI) |
|---|---|---|
| N. gonorrhoeae burden, quartiles in male sex partner | ||
| 1st quartile (ref) | 1 | 1 |
| 2nd quartile | 2.97 (0.62–14.22) | 2.70b (.54–13.46) |
| 3rd quartile | 10.82 (1.17–100.44) | 8.66b (.90–83.26) |
| 4th quartile | 10.18 (1.09–94.83) P = .032 |
10.17b (1.06–97.08) P = .052 |
| Chlamydia trachomatis status, positive vs negative, in women | 4.06 (.82–20.20) P = .087 |
3.47c (.65–18.59) P = .146 |
Abbreviations: CI, confidence interval; ref, reference.
Model estimates below remained similar after additional adjustment for nonsignificant factors: male C. trachomatis status; male Mycoplasma genitalium status and male age.
Adjusted for C. trachomatis infection in women.
Adjusted for N. gonorrhoeae burdens in male sex partners.
Chlamydia (C. trachomatis) Infection in Women and the Burden of Gonococcal (N. gonorrhoeae) Infection
N. gonorrhoeae burdens in female sex partners of infected men was associated with burdens in their infected male partners (β coefficient [regression slope] = 0.527, P < .001), adjusted for C. trachomatis status in women (Table 3). The estimated association within C. trachomatis-positive women was slightly greater (β = 0.65, P < .001) than within C. trachomatis-negative women (β = 0.45, P < .001) but the difference (interaction) was not significant (P = .306) (Supplementary Figure 4). The number of organisms in N. gonorrhoeae-infected women was also associated with female C. trachomatis status; 2.82-fold higher in C. trachomatis-coinfected versus C. trachomatis-uninfected women (β = 0.451, P = .036), adjusted for N. gonorrhoeae burdens in male partners (Table 3).
Table 3.
Linear Regression Model That Examined Factors Associated With Neisseria gonorrhoeae Burdens in Women
| Association of N. gonorrhoeae Burdens (log10) in Women | Unadjusted β Coefficient (95% CI) | Adjusteda β Coefficient (95% CI)b |
|---|---|---|
| N. gonorrhoeae burdens, log10, in male sex partners | 0.549 (.354–.743) P < .001 |
0.527b (.336–.718) P < .001 |
| Chlamydia trachomatis status, positive vs negative, in women | 0.575 (.076–1.075) P = .25 |
0.451c,d (.031–.871) P = .036 |
Abbreviation: CI, confidence interval.
Model estimates below are similar after additional adjustment for nonsignificant factors: male C. trachomatis status, male Mycoplasma genitalium status, and male age.
Adjusted for C. trachomatis status in women.
Adjusted for burdens of N. gonorrhoeae in male sex partners.
Fold increase: 100.451 = 2.82.
DISCUSSION
Our study showed that the prevalence of N. gonorrhoeae infection in men with urethritis increased by a third in men who indicated they had 2 or more sex partners, and mirrors studies that have shown an increased risk of gonorrhea in men with multiple female sex partners [26–28]. Even so, transmission of N. gonorrhoeae differs markedly, depending on directionality; it was approximately 4-fold lower from women to men (19%–22% [29, 30]), compared to 83% transmission from men to women in our study involving predominantly Chinese married couples. Men with symptomatic N. gonorrhoeae urethritis possess 3.7 × 106 DNA copies per urethral swab by qPCR [31] compared to lower N. gonorrhoeae burdens reported in infected women: 2.0 × 104 copies per mL (vaginal swabs) [32] and 1.45 × 105 CFU/mL (CVLs) [33].
The risk of N. gonorrhoeae infection in women was driven primarily by exposure to increasing burdens in male partners. Nonetheless, we also demonstrated a trend in the likelihood of N. gonorrhoeae infection in women when C. trachomatis infection was present; the odds ratio was decreased by 15% after adjustment for N. gonorrhoeae burdens in male sex partners. Earlier, we reported 73% transmission of N. gonorrhoeae from men to women in the US; coinfection with C. trachomatis (42%) [34] was similar to 45% in our current study. Transmission studies often use concordant infection (both members of the dyad infected) as the measure of transmission (in either direction). In a US study that examined C. trachomatis transmission across dyads, which also included subjects with N. gonorrhoeae infection, the percent of C. trachomatis concordant dyads when women were N. gonorrhoeae infected was significantly higher than in C. trachomatis-concordant dyads with N. gonorrhoeae-uninfected women [35]. In a second US study, 100% of dyads with N. gonorrhoeae-infected women were C. trachomatis concordant [36].
N. gonorrhoeae burdens in infected women range widely [33, 37] but have not been shown to increase during the period of infection and are borderline higher when women are coinfected with C. trachomatis [37]. When adjusted for N. gonorrhoeae burdens in male partners, we found that coinfected women had almost a 3-fold higher N. gonorrhoeae burden compared to women infected with N. gonorrhoeae alone. This suggests that N. gonorrhoeae may flourish when C. trachomatis is present in women and that an increase in the N. gonorrhoeae burden takes place after infection. Killing of gonococci by macrophages and neutrophils in vitro is impaired by type I interferons [38]. We speculate that induction of type I interferons by C. trachomatis [39, 40] may contribute to impaired clearance of N. gonorrhoeae in the setting of coinfection.
Molecular analysis of vaginal microbiome indicated that N. gonorrhoeae/C. trachomatis-negative specimens in our study contained a normal abundance of lactobacilli [25], principally L. iners (CST 3 equivalent), similar to normal microbiomes otherwise reported in women of Asian ethnicity [41]. Diversity indices indicated significant differences between the N. gonorrhoeae, C. trachomatis, and N. gonorrhoeae/C. trachomatis groups compared to the N. gonorrhoeae-negative/C. trachomatis-negative group, which aligns with the traditional diagnosis of BV [42, 43] and a meta-analysis of vaginal microbiota studies [44] that included reports that diagnosed BV exclusively by sequence analysis [45–48]. Several longitudinal studies have established that a high prevalence of BV elevates the risk for incident (subsequent) N. gonorrhoeae and/or C. trachomatis genital infection [13–15].
We identified 21% of N. gonorrhoeae-infected women who had underlying C. trachomatis infection (their partners were free of C. trachomatis; Supplementary Figure 2), similar to the 22% in our earlier study [34]. We speculate that these women had been exposed earlier to previously infected regular partners whose infections may have been asymptomatic and had cleared spontaneously [49] or had undergone treatment. Prior infection in these women with C. trachomatis may not have been apparent until they became infected with N. gonorrhoeae [34, 50].
Our study has limitations. We used qPCR to quantitate N. gonorrhoeae in male urine; N. gonorrhoeae burdens measured in urines collected from men with symptomatic urethritis can be as much as 3 log10 lower compared to those measured using urethral swabs [31], potentially diminishing accuracy. We showed that N. gonorrhoeae burdens measured in male urine and cervicovaginal swabs were not different, similar to a study that showed that men and women have similar N. gonorrhoeae burdens measured in vaginal swabs, anorectal swabs from men and women, and male first voided urines [32]. An additional limitation was the comparison of multiple factors and characteristics using statistical hypothesis testing. There was the potential of false-positive findings based merely on statistical significance. The estimates of association (odds ratios or regression coefficients) provided a best assessment of the strength of the associations while the CIs provided an assessment of variability of these measures.
In summary, our study showed that women who are exposed to large N. gonorrhoeae burdens such as occur in men with symptomatic urethritis are more likely to become infected; coinfection with Chlamydia increases their burden of N. gonorrhoeae.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Contributor Information
Xiaohong Su, Sexually Transmitted Disease Clinic, Institute of Dermatology, Chinese Academy of Medical Sciences and Peking Union Medical College, Nanjing, China.
Wenjing Le, Sexually Transmitted Disease Clinic, Institute of Dermatology, Chinese Academy of Medical Sciences and Peking Union Medical College, Nanjing, China.
Xiaofeng Zhu, Sexually Transmitted Disease Clinic, Institute of Dermatology, Chinese Academy of Medical Sciences and Peking Union Medical College, Nanjing, China.
Sai Li, Sexually Transmitted Disease Clinic, Institute of Dermatology, Chinese Academy of Medical Sciences and Peking Union Medical College, Nanjing, China.
Baoxi Wang, Sexually Transmitted Disease Clinic, Institute of Dermatology, Chinese Academy of Medical Sciences and Peking Union Medical College, Nanjing, China.
Guillermo Madico, Environmental Health and Safety, Boston University, Boston, MA, USA.
Zhaoyan Yang, Biostatistics and Epidemiology Data Analytics Center, Boston University School of Public Health, Boston, MA, USA.
Christine E Chaisson, Biostatistics and Epidemiology Data Analytics Center, Boston University School of Public Health, Boston, MA, USA.
Robert E McLaughlin, Institute for Life Science Entrepreneurship, Union, NJ, USA.
Sumanth Gandra, Division of Infectious Diseases, Department of Medicine, UMass Chan Medical School, Worcester, MA, USA.
Jungwon Yoon, Division of Infectious Diseases, Department of Medicine, UMass Chan Medical School, Worcester, MA, USA.
Bo Zheng, Division of Infectious Diseases, Department of Medicine, UMass Chan Medical School, Worcester, MA, USA.
Lisa A Lewis, Division of Infectious Diseases, Department of Medicine, UMass Chan Medical School, Worcester, MA, USA.
Sunita Gulati, Division of Infectious Diseases, Department of Medicine, UMass Chan Medical School, Worcester, MA, USA.
George W Reed, Division of Infectious Diseases, Department of Medicine, UMass Chan Medical School, Worcester, MA, USA.
Sanjay Ram, Division of Infectious Diseases, Department of Medicine, UMass Chan Medical School, Worcester, MA, USA.
Peter A Rice, Division of Infectious Diseases, Department of Medicine, UMass Chan Medical School, Worcester, MA, USA.
Notes
Acknowledgments. We thank study participants who were also patients at the Nanjing STD Clinic. We also thank health care providers, administrative and laboratory staff at the Clinic and the Institute of Dermatology, Chinese Academy of Medical Sciences and Peking Union Medical College. In addition, we thank Ellen Klein who assisted with the design of the case report forms used in this study.
Financial support. This work was supported by the National Institutes of Health, National Institute of Allergy and Infectious Diseases (grant number U19 AI084048 to P. A. R.).
References
- 1. Johnson B. OSHA infectious dose white paper. Appl Biosaft 2003; 8:160–5. [Google Scholar]
- 2. Liu Y, Feinen B, Russell MW. New concepts in immunity to Neisseria gonorrhoeae: innate responses and suppression of adaptive immunity favor the pathogen, not the host. Front Microbiol 2011; 2:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hobbs MM, Sparling PF, Cohen MS, Shafer WM, Deal CD, Jerse AE. Experimental gonococcal infection in male volunteers: cumulative experience with Neisseria gonorrhoeae strains FA1090 and MS11mkC. Front Microbiol 2011; 2:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dubbink JH, de Waaij DJ, Bos M, et al. Microbiological characteristics of Chlamydia trachomatis and Neisseria gonorrhoeae infections in South African women. J Clin Microbiol 2016; 54:200–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Papadogeorgakis H, Pittaras TE, Papaparaskevas J, Pitiriga V, Katsambas A, Tsakris A. Chlamydia trachomatis serovar distribution and Neisseria gonorrhoeae coinfection in male patients with urethritis in Greece. J Clin Microbiol 2010; 48:2231–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rowley J, Vander Hoorn S, Korenromp E, et al. Chlamydia, gonorrhoea, trichomoniasis and syphilis: global prevalence and incidence estimates, 2016. Bull World Health Organ 2019; 97:548–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shaw SY, Elliott LJ, Nowicki DL, et al. Comparing the ecological niches of chlamydial and gonococcal infections in Winnipeg, Canada: 2007–2016. Sex Transm Dis 2021; 48:837–43. [DOI] [PubMed] [Google Scholar]
- 8. Vonck RA, Darville T, O'Connell CM, Jerse AE. Chlamydial infection increases gonococcal colonization in a novel murine coinfection model. Infect Immun 2011; 79:1566–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gulati S, Beurskens FJ, de Kreuk BJ, et al. Complement alone drives efficacy of a chimeric antigonococcal monoclonal antibody. PLoS Biol 2019; 17:e3000323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Creighton S, Tenant-Flowers M, Taylor CB, Miller R, Low N. Co-infection with gonorrhoea and chlamydia: how much is there and what does it mean? Int J STD AIDS 2003; 14:109–13. [DOI] [PubMed] [Google Scholar]
- 11. Mensforth S, Ayinde OC, Ross J. Spontaneous clearance of genital and extragenital Neisseria gonorrhoeae: data from GToG. Sex Transm Infect 2020; 96:556–61. [DOI] [PubMed] [Google Scholar]
- 12. Wijers J, Hoebe C, Dukers-Muijrers N, Wolffs P, van Liere G. The characteristics of patients frequently tested and repeatedly infected with Neisseria gonorrhoeae. Int J Environ Res Public Health 2020; 17:1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Abbai NS, Reddy T, Ramjee G. Prevalent bacterial vaginosis infection—a risk factor for incident sexually transmitted infections in women in Durban, South Africa. Int J STD AIDS 2016; 27:1283–8. [DOI] [PubMed] [Google Scholar]
- 14. Bautista CT, Wurapa EK, Sateren WB, Morris SM, Hollingsworth BP, Sanchez JL. Association of bacterial vaginosis with chlamydia and gonorrhea among women in the U.S. Army. Am J Prev Med 2017; 52:632–9. [DOI] [PubMed] [Google Scholar]
- 15. Gallo MF, Macaluso M, Warner L, et al. Bacterial vaginosis, gonorrhea, and chlamydial infection among women attending a sexually transmitted disease clinic: a longitudinal analysis of possible causal links. Ann Epidemiol 2012; 22:213–20. [DOI] [PubMed] [Google Scholar]
- 16. McQuillen DP, Gulati S, Ram S, et al. Complement processing and immunoglobulin binding to Neisseria gonorrhoeae determined in vitro simulates in vivo effects. J Infect Dis 1999; 179:124–35. [DOI] [PubMed] [Google Scholar]
- 17. Li Y, Su X, Le W, et al. Mycoplasma genitalium in symptomatic male urethritis: macrolide use is associated with increased resistance. Clin Infect Dis 2020; 70:805–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Madico G, Quinn TC, Boman J, Gaydos CA. Touchdown enzyme time release-PCR for detection and identification of Chlamydia trachomatis, C. pneumoniae, and C. psittaci using the 16S and 16S-23S spacer rRNA genes. J Clin Microbiol 2000; 38:1085–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010; 7:335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 2007; 73:5261–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jonckheere AR. A distribution-free k-sample test against ordered alternatives. Biometrika 1954; 41:133–45. [Google Scholar]
- 22. Terpstra TJ. The asymptotic normality and consistency of Kendall's test against trend, when ties are present in one ranking. Indag Math 1952; 14:327–33. [Google Scholar]
- 23. Martin IM, Ison CA, Aanensen DM, Fenton KA, Spratt BG. Rapid sequence-based identification of gonococcal transmission clusters in a large metropolitan area. J Infect Dis 2004; 189:1497–505. [DOI] [PubMed] [Google Scholar]
- 24. Jolley KA, Bray JE, Maiden MCJ. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res 2018; 3:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Miller EA, Beasley DE, Dunn RR, Archie EA. Lactobacilli dominance and vaginal pH: why is the human vaginal microbiome unique? Front Microbiol 2016; 7:1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dela H, Attram N, Behene E, et al. Risk factors associated with gonorrhea and chlamydia transmission in selected health facilities in Ghana. BMC Infect Dis 2019; 19:425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Barry PM, Kent CK, Klausner JD. Risk factors for gonorrhea among heterosexuals—San Francisco, 2006. Sex Transm Dis 2009; 36:S62–6. [DOI] [PubMed] [Google Scholar]
- 28. Van Duynhoven YT, van de Laar MJ, Schop WA, Mouton JW, van der Meijden WI, Sprenger MJ. Different demographic and sexual correlates for chlamydial infection and gonorrhoea in Rotterdam. Int J Epidemiol 1997; 26:1373–85. [DOI] [PubMed] [Google Scholar]
- 29. Holmes KK, Johnson DW, Trostle HJ. An estimate of the risk of men acquiring gonorrhea by sexual contact with infected females. Am J Epidemiol 1970; 91:170–4. [DOI] [PubMed] [Google Scholar]
- 30. Hooper RR, Reynolds GH, Jones OG, et al. Cohort study of venereal disease. I: the risk of gonorrhea transmission from infected women to men. Am J Epidemiol 1978; 108:136–44. [DOI] [PubMed] [Google Scholar]
- 31. Priest D, Ong JJ, Chow EPF, et al. Neisseria gonorrhoeae DNA bacterial load in men with symptomatic and asymptomatic gonococcal urethritis. Sex Transm Infect 2017; 93:478–81. [DOI] [PubMed] [Google Scholar]
- 32. van der Veer B, Hoebe C, Dukers-Muijrers N, van Alphen LB, Wolffs PFG. Men and women have a similar Neisseria gonorrhoeae bacterial load: a comparison of three anatomical sites. J Clin Microbiol 2020; 58:e01171-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lowe TL, Kraus SJ. Quantitation of Neisseria gonorrhoeae from women with gonorrhea. J Infect Dis 1976; 133:621–6. [DOI] [PubMed] [Google Scholar]
- 34. Lin JS, Donegan SP, Heeren TC, et al. Transmission of Chlamydia trachomatis and Neisseria gonorrhoeae among men with urethritis and their female sex partners. J Infect Dis 1998; 178:1707–12. [DOI] [PubMed] [Google Scholar]
- 35. Quinn TC, Gaydos C, Shepherd M, et al. Epidemiologic and microbiologic correlates of Chlamydia trachomatis infection in sexual partnerships. JAMA 1996; 276:1737–42. [PubMed] [Google Scholar]
- 36. Schillinger JA, Katz BP, Markowitz LE, et al. Genotype-specific concordance of Chlamydia trachomatis genital infection within heterosexual partnerships. Sex Transm Dis 2016; 43:741–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Stupiansky NW, Van Der Pol B, Williams JA, Weaver B, Taylor SE, Fortenberry JD. The natural history of incident gonococcal infection in adolescent women. Sex Transm Dis 2011; 38:750–4. [DOI] [PubMed] [Google Scholar]
- 38. Andrade WA, Agarwal S, Mo S, et al. Type I interferon induction by Neisseria gonorrhoeae: dual requirement of cyclic GMP-AMP synthase and toll-like receptor 4. Cell Rep 2016; 15:2438–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Barker JR, Koestler BJ, Carpenter VK, et al. STING-dependent recognition of cyclic di-AMP mediates type I interferon responses during Chlamydia trachomatis infection. mBio 2013; 4:e00018-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang Y, Yeruva L, Marinov A, et al. The DNA sensor, cyclic GMP-AMP synthase, is essential for induction of IFN-beta during Chlamydia trachomatis infection. J Immunol 2014; 193:2394–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ravel J, Gajer P, Abdo Z, et al. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci U S A 2011; 108:4680–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Coleman JS, Gaydos CA. Molecular diagnosis of bacterial vaginosis: an update. J Clin Microbiol 2018; 56:e00342-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fredricks DN, Fiedler TL, Marrazzo JM. Molecular identification of bacteria associated with bacterial vaginosis. N Engl J Med 2005; 353:1899–911. [DOI] [PubMed] [Google Scholar]
- 44. Tamarelle J, Thiebaut ACM, de Barbeyrac B, Bebear C, Ravel J, Delarocque-Astagneau E. The vaginal microbiota and its association with human papillomavirus, Chlamydia trachomatis, Neisseria gonorrhoeae and Mycoplasma genitalium infections: a systematic review and meta-analysis. Clin Microbiol Infect 2019; 25:35–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Brotman RM, Shardell MD, Gajer P, et al. Interplay between the temporal dynamics of the vaginal microbiota and human papillomavirus detection. J Infect Dis 2014; 210:1723–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mitra A, MacIntyre DA, Lee YS, et al. Cervical intraepithelial neoplasia disease progression is associated with increased vaginal microbiome diversity. Sci Rep 2015; 5:16865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Oh HY, Kim BS, Seo SS, et al. The association of uterine cervical microbiota with an increased risk for cervical intraepithelial neoplasia in Korea. Clin Microbiol Infect 2015; 21:674.e1–9. [DOI] [PubMed] [Google Scholar]
- 48. van der Veer C, Bruisten SM, van der Helm JJ, de Vries HJ, van Houdt R. The cervicovaginal microbiota in women notified for Chlamydia trachomatis infection: a case–control study at the sexually transmitted infection outpatient clinic in Amsterdam, The Netherlands. Clin Infect Dis 2017; 64:24–31. [DOI] [PubMed] [Google Scholar]
- 49. Lewis J, Price MJ, Horner PJ, White PJ. Genital Chlamydia trachomatis infections clear more slowly in men than women, but are less likely to become established. J Infect Dis 2017; 216:237–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Batteiger BE, Fraiz J, Newhall WJ, Katz BP, Jones RB. Association of recurrent chlamydial infection with gonorrhea. J Infect Dis 1989; 159:661–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.