Abstract
Incidence of visceral leishmaniasis (VL) in the Indian subcontinent (ISC) has declined by more than 95% since initiation of the elimination program in 2005. As the ISC transitions to the postelimination surveillance phase, an accurate measurement of human-vector contact is needed to assure long-term success. To develop this tool, we identified PagSP02 and PagSP06 from saliva of Phlebotomus argentipes, the vector of Leishmania donovani in the ISC, as immunodominant proteins in humans. We also established the absence of cross-reactivity with Phlebotomus papatasi saliva, the only other human-biting sand fly in the ISC. Importantly, by combining recombinant rPagSP02 and rPagSP06 we achieved greater antibody recognition and specificity than single salivary proteins. The receiver operating characteristics curve for rPagSP02 + rPagSP06 predicts exposure to Ph. argentipes bites with 90% specificity and 87% sensitivity compared to negative control sera (P >.0001). Overall, rPagSP02 + rPagSP06 provides an effective surveillance tool for monitoring vector control efforts after VL elimination.
Keywords: India, biomarker, elimination campaign, Phlebotomus argentipes, sand fly salivary proteins, surveillance, visceral leishmaniasis
Two Phlebotomus argentipes saliva proteins are not recognized by sera of individuals bitten by Ph. papatasi and retain their immunogenicity as recombinant molecules. Combined, they deliver a specific and sensitive biomarker of vector exposure for surveillance after visceral leishmaniasis elimination.
Visceral leishmaniasis (VL) is a lethal protozoan disease [1, 2]. In 2006–2010, more than 40 000 VL cases were reported per year from the Indian subcontinent, accounting for 73% of cases worldwide [2]. In 2005, the governments of India, Bangladesh, and Nepal established a program to eliminate VL as a public health problem, defined as a reported incidence of less than 1 case per 10 000 population [3, 4]. The main pillars of the elimination campaign include improved surveillance, early case detection and rapid treatment, and vector control by indoor residual spraying (IRS) [3, 4]. The combined impact of these efforts helped decrease the reported incidence by more than 95% since its last peak in 2007, with the proportion of global VL cases coming from the Indian subcontinent falling to 18% [5, 6]. Nepal and Bangladesh reached the elimination target in recent years, whereas the incidence in India remains above target in several endemic blocks [5–7].
In the Indian subcontinent, there are only 2 anthropophilic sand fly species, Phlebotomus argentipes and Phlebotomus papatasi, with the latter lacking vectorial competence for Leishmania donovani [8], although both species remain susceptible to insecticides [9–11]. The elimination program was launched based on the assumption that VL transmission in the Indian subcontinent was purely anthroponotic [12], and that the sand fly vector, Ph. argentipes, is predominantly endophilic and endophagic [8]. Nevertheless, the impact of IRS is being questioned based on the lack of a demonstrable impact on sand fly densities in sprayed compared to unsprayed villages, and the high sand fly density in outdoor locations not targeted by IRS [13, 14]. Entomological studies suggest that Ph. argentipes is substantially more exophilic and exophagic than previously supposed, possibly resulting from a shift in their response to long-term insecticide pressure, reinforcing the limitation of IRS in controlling vector populations [13–17]. Collectively, these observations point to major knowledge gaps in our understanding of the biology of Ph. argentipes which need to be addressed to ensure that VL control is maintained in India [14–18]. Moreover, as VL cases continue to decrease, human surveillance alone becomes inadequate, and new tools designed to monitor changes in sand fly-human contact will be indispensable for appropriate evaluation of current interventions, ensuring long-term sustainability of VL elimination efforts.
As sand flies bite humans to obtain a blood meal, they egest a repertoire of sand fly salivary proteins (SSP) into the human host [19]. Some of these SSP are immunogenic and have been recognized as suitable biomarkers of exposure to bites of several Leishmania vectors in endemic areas [20–28]. To date, 2 studies used Ph. argentipes salivary gland homogenate (SGH) to assess the antibody response in individuals living in VL-endemic areas in Bihar, India [25, 26]. To overcome potential cross-reactivity, the group preadsorbed human sera against SGH of Ph. papatasi. This approach is labor intensive and not amenable to quality control and reproducibility, highlighting the limitations of using SGH as a field surveillance tool [19, 25, 26]. Recently, Ph. argentipes PagSP06 was identified as an immunodominant SSP in humans and its recombinant form was tested against sera collected from Bangladesh [28]. Here, we validate the immunodominance of PagSP06 in humans and identify another immunodominant protein, PagSP02. We demonstrate that a composite biomarker of the 2 recombinant proteins exhibits a performance superior to that of individual recombinant proteins or SGH. Additionally, we validate the specificity of the rPagSP02 + rPagSP06 composite biomarker against sera of individuals bitten by Ph. papatasi, delivering a reliable, sensitive, and specific tool to measure the intensity of Ph. argentipes bites in humans. Implementation of this tool in a field setting will inform vector-control strategies in support of the VL elimination campaign.
METHODS
Study Design
Fifty-two serum samples were collected in November 2019 from inhabitants of Rahardiyara, Pahadichack, and Maksoodpur, 3 VL-endemic (EN) villages in Bihar, India. Serum samples from individuals living in the United States, a VL-nonendemic (NE) area where Phlebotomus sand flies are absent, were obtained from the National Institutes of Health (NIH) Blood Bank (n = 14). To select our true negative controls, NE samples were prescreened by western blot against Ph. argentipes SGH and those showing nonspecific background were excluded. To assess potential cross-reactivity against the other human-biting sand fly in the region, Ph. papatasi, we included serum samples (n = 12) from individuals exposed to Ph. papatasi bites in Iraq (IR) where Ph. argentipes is absent. IR samples (n = 5) were further selected based on their strong recognition of multiple proteins against Ph. papatasi SGH western blot and serum availability. IR samples were collected from American soldiers 6 months after deployment to Iraq during 2003–2004 and tested for antibodies against Ph. papatasi SSP [29]. EN and IR serum samples were collected according to human protocols approved by the institutional review boards of the University of California San Francisco (protocol No. 19-29535) and All India Institute of Medical Sciences-Patna, and Uniformed Services University of the Health Sciences, William Beaumont Army Medical Center and Walter Reed National Military Medical Center (protocol No. G183ZU 357741).
Sand Flies and Mosquitoes
Ph. argentipes (India strain) and Ph. papatasi (Jordan strain) sand flies, were maintained at 26°C, 75% humidity, and 30% sucrose solution. Aedes aegypti (Liverpool strain) and Anopheles albopictus (Nijmegen strain) mosquitoes were maintained at 28°C, 80% humidity, and 10% Karo syrup solution. Mosquitoes and Ph. papatasi sand flies were reared at the Laboratory of Malaria and Vector Research, National Institute of Allergy and Infectious Diseases, NIH. Ph. argentipes sand flies were reared at Walter Reed Army Institute of Research.
Salivary Gland Homogenate
Salivary glands (20–50 pairs) were dissected in 20–50 μL of 1 × phosphate-buffered saline (Lonza) and stored at −80°C until SGH was prepared as described [22].
Production of Recombinant Ph. Argentipes Salivary Proteins
Ph. argentipes salivary gland cDNA library was constructed as described [30]. cDNA encoding Ph. argentipes secreted proteins PagSP02, PagSP06, PagSP07, or PagSP09 were amplified by PCR and cloned into VR2001-TOPO vector [22]. His-tagged plasmids were sequenced and purified from Escherichia coli cells using the NucleoBond PC 2000 plasmid megaprep kit (Takara Bio) then sent for expression in HEK-293F cells. The concentrated supernatant was collected and recombinant proteins were purified by high-performance liquid chromatography (HPLC; Dionex) followed by dialysis [22, 27]. Bands from recombinant proteins were cut from the 12–14% Tris-glycine gel and identified by mass spectrometry (Supplementary Figure 1).
Immunoblotting
Forty micrograms of Ph. argentipes or Ph. papatasi SGH, or 15 μg of recombinant proteins (rPagSP02, rPagSP06, rPagSP07, or rPagSP09) were denatured at 95°C for 5 minutes under nonreducing conditions and separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel-electrophoresis. Proteins were transferred to a 0.45-μm nitrocellulose membrane and blocked overnight at 4°C with 5% nonfat dry milk in Tris-buffered saline with 0.05% Tween 20 (TBST). The next day, serum samples were diluted (1:80) in TBST-M, and each sample was loaded in the mini-protean II multiscreen apparatus (Bio-Rad) and incubated for 3 hours at room temperature, followed by membrane washing. Secondary alkaline phosphatase-conjugated goat anti-human immunoglobulin G heavy and light chains (IgG H+L) (Sigma) was incubated at a 1:8000 dilution for 45 minutes at room temperature. Between steps, membranes were washed 3 times with TBST for 5 minutes each. Alkaline Phosphatase Western Blue Stabilized Substrate (Promega) was added and the reaction was stopped with distilled water water after 1–2 minutes.
Digital Image Analysis
The relative molecular mass was used to identify potential immunogenic/immunodominant targets against Ph. argentipes SGH. Relative density obtained from immunoblot images of Ph. argentipes SGH membranes probed with EN serum samples were analyzed by ImageJ 1.52a software. First, the original images were converted to 8-bit gray scale, and a defined region of interest was assigned to a sample (vertical/individual lane), covering the minimum area to contain that individual lane. Then, the same region of interest was applied to all samples/individual lanes. The area of each peak, corresponding to a single band recognized in that lane, was analyzed using the straight-line tool to separate signal from background. The intensity of each peak, corresponding to the number of pixels in the band, was determined using the wand tool. Values were graphed and a heat-map of immunogenic target proteins was produced.
Enzyme-Linked Immunosorbent Assay
Corning 96-well flat bottom polyvinyl chloride untreated microplates (Costar) were coated with 50 μL of 2 μg/mL of Ph. argentipes SGH or 1 μg/mL of rPagSP02 or rPagSP06, or a combination of 1 μg/mL of each rPagSP02 and rPagSP06. Antigens were diluted in carbonate-bicarbonate buffer, pH 9.6 (Sigma) and plates were incubated overnight at 4°C. Plates were blocked with 200 μL of Ultra-block (Bio-Rad) for 2 hours at room temperature. Fifty microliters of sera were diluted at 1:50 in TBST with 4% bovine serum albumin fraction V fatty acid free (MDMillipore) and incubated for 1 hour at 37°C. Secondary alkaline phosphatase-conjugated goat anti-human IgG (H+L) (Sigma) was incubated at 1:5000 for 1 hour at 37°C. Fifty microliters of p-nitrophenyl phosphate liquid substrate (Sigma) were added to all wells and the optical density (OD) values were recorded at a 405 nm wavelength. After antigen coating and between all incubation steps, plates were washed 6 times with TBST with shaking using the 405 TS Microplate Washer (BioTek). Kinetic curves were used to standardize the enzyme-linked immunosorbent assay (ELISA; Supplementary Figure 2). For our standardized ELISA assay an end point of 1.5 hours was selected.
Statistical Analysis
All graphs and statistical analysis were performed using GraphPad Prism 8.0 software. For ELISA assays, each sample was tested in duplicate in 2–3 independent experiments. Cutoff values were calculated as the mean ± 2 SD of NE or IR controls. Pearson rank correlation test was calculated based on the ELISA OD values between recombinant SSP and Ph. argentipes SGH. Correlation coefficient (r), P values, and the 95% confidence interval (CI) are reported. Receiver operating characteristic (ROC) analysis was performed by plotting the OD values of EN samples for rPagSP02 + rPagSP06 or Ph. argentipes SGH antigens versus NE (n = 10) or IR (n = 5) samples, selected as our true negative controls for the analysis. The ROC cutoff values were selected to the nearest calculated ELISA cutoff values. Kruskal Wallis with Dunn post hoc analysis or Mann-Whitney U post hoc statistical tests were performed as indicated. A P value of ≤ .05 was considered significant.
RESULTS
Sera From Inhabitants of VL-Endemic Areas in India Recognize Ph. Argentipes But Not Ph. Papatasi Salivary Proteins
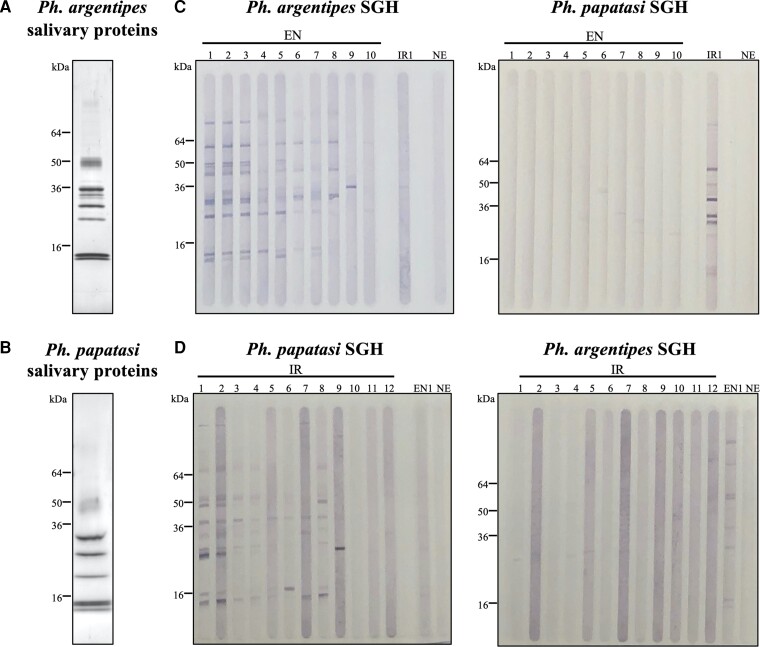
In VL-endemic regions of the Indian subcontinent, Ph. argentipes is prevalent while Ph. papatasi is less abundant [14]. To determine potential cross-reactivity between the SSP of each sand fly species (Figure 1A and 1B) we screened sera collected from Indian individuals living in VL-endemic areas of Bihar (EN, n = 1–10), individuals exposed to Ph. papatasi bites in Iraq where Ph. argentipes is absent (IR, n = 1–12), and 1 of several prescreened nonendemic (NE) control serum from the United States (where Phlebotomus species are absent) against SGH of Ph. argentipes or Ph. papatasi (Figure 1C and 1D and Supplementary Figure 3A). Western blot analysis established that EN sera recognized multiple bands in Ph. argentipes SGH that were not detected by representative IR and NE samples and displayed low or no recognition of Ph. papatasi SSP (Figure 1C). Furthermore, some bands, corresponding to 1 or more Ph. argentipes SSP, were recognized by most EN sera indicative of their immunodominance. Similarly, IR sera strongly recognized Ph. papatasi but not Ph. argentipes SSP (Figure 1D). This establishes that individuals bitten by Ph. argentipes do not recognize SSP of Ph. papatasi and vice versa. Together with the observed immunodominance of distinct proteins in Ph. argentipes saliva, this pointed to the feasibility of finding a marker specific to Ph. argentipes bites.
Figure 1.
Sera from individuals living in VL-endemic areas in the Indian subcontinent recognize native salivary proteins from Phlebotomus argentipes but not from Phlebotomus papatasi. Coomassie blue staining showing the pattern of native Ph. argentipes (A) or Ph. papatasi (B) salivary proteins. Immunoblots run with 40 µg of SGH from Ph. argentipes (C) or from Ph. papatasi (D) sand flies. To assess cross-reactivity between the 2 vector species, western blot membranes were probed with reactive human serum samples from individuals living in a VL-endemic area (EN), Bihar, India, where Ph. argentipes is prevalent, or from individuals exposed to Ph. papatasi bites in Iraq (IR). A sample from an individual living in the United States, a VL-nonendemic area where Phlebotomus sand flies are absent, was used as a negative control (NE). Abbreviations: kDa, kilodalton, relative molecular mass; SGH, salivary gland homogenate; VL, visceral leishmaniasis.
PagSP02 and PagSP06 Are Immunodominant Antigens in Ph. argentipes Saliva
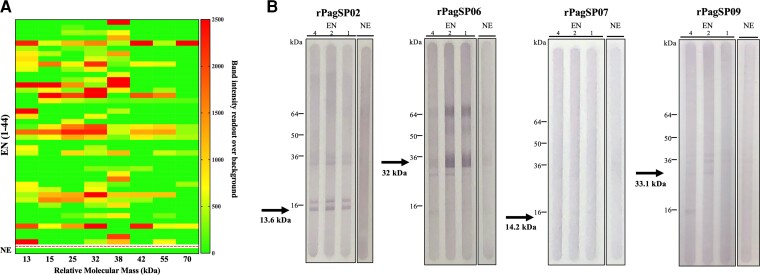
To identify immunodominant proteins from Ph. argentipes saliva we screened a larger sample of EN serum (n = 44) against Ph. argentipes SGH by western blot (Supplementary Figure 3B). The heat map shows relative band intensities corresponding to antibody recognition of different Ph. argentipes SSP for the 44 EN sera (Figure 2A). Overall, 82% of EN sera had antibodies against 1 or several Ph. argentipes SSP while none were recognized by NE control sera (Figure 2A and Supplementary Figure 3A and 3B). Based on band patterns, we identified 5 immunogenic proteins at 13 kDa, 15 kDa, 25 kDa, 32 kDa, and 38 kDa that were recognized by 48%, 34%, 45%, 61%, and 61% of EN sera, respectively (Figure 2A).
Figure 2.
Selection and prioritization of immunodominant Phlebotomus argentipes salivary targets. A, Heat map showing the relative density obtained from immunoblots against Ph. argentipes SGH probed with sera from VL-endemic individuals (EN, n = 44). The frequency and strength of recognition of SGH proteins was used to prioritize immunogenic targets according to their predicted relative molecular mass. Dotted line delineates EN samples from the nonendemic control sample (NE). Scale bar depicts arbitrary units of band intensity readout over background. B, The immunogenicity of Ph. argentipes recombinant proteins, rPagSP02, rPagSP06, rPagSP07, or rPagSP09, was tested by probing the membrane with serum samples (EN, n = 3) from individuals reactive to Ph. argentipes SGH selected from Figure 1B, or a NE control (n = 1). The relative molecular mass of the native Ph. argentipes proteins is shown below each respective arrow. Western blots are representative of 2 independent experiments. Abbreviations: kDa, kilodalton, relative molecular mass; SGH, salivary gland homogenate; VL, visceral leishmaniasis.
We took advantage of the availability of the Ph. argentipes transcriptome and proteome [30] to select Ph. argentipes SSP based on relative molecular mass, specifically targeting potential candidates that were highly and more frequently recognized by EN sera (Figure 2A). Based on a predicted relative molecular mass within 13 to 38 kDa and similarity to immunogenic SSP from other species [30], we selected 4 transcripts coding for Ph. argentipes SSP PagSP02, PagSP06, PagSP07, and PagSP09 (Supplementary Figure 1A). Plasmids coding for rPagSP02, rPagSP06, rPagSP07, and rPagSP09 were then expressed in HEK293 mammalian cells and purified by HPLC (Supplementary Figure 1B). The 4 recombinant proteins were tested by western blot using 3 reactive EN human sera (EN samples 4, 2, and 1 in Figure 1C. EN sera recognized rPagSP02 and rPagSP06 recombinant proteins and 1 or multiple rPagSP02 and rSP06 dimers observed in the Coomassie gel (Figure 2B and Supplementary Figure 1B), while no or low antibody response was observed against rPagSP07 or rPagSP09, respectively.
rPagSP02 and rPagSP06 Combined Deliver a Sensitive Biomarker Specific to Ph. argentipes Bites
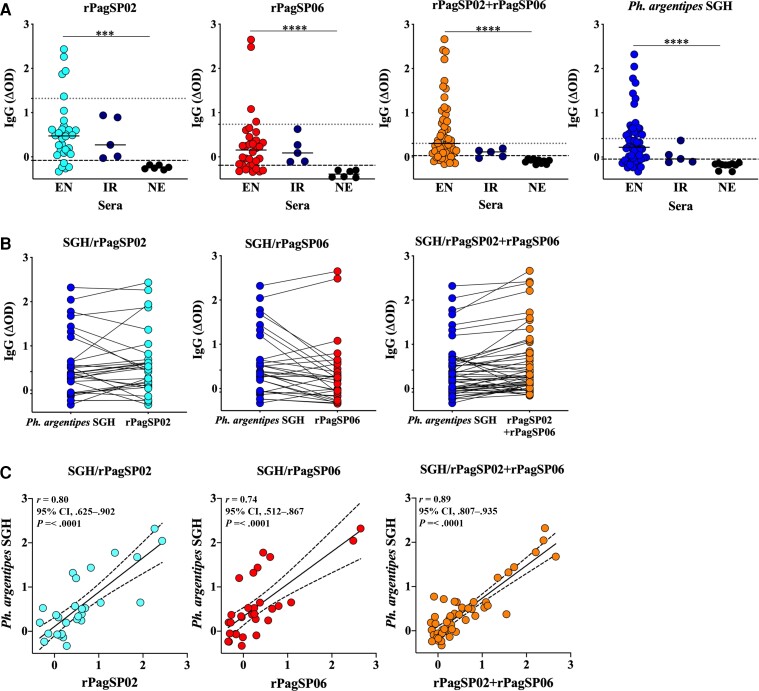
To standardize an ELISA-based biomarker assay for high-throughput screening of exposure to Ph. argentipes bites, we compared the antibody response of up to 52 EN sera against rPagSP02, rPagSP06, or a combination of rPagSP02 + rPagSP06. We observed that 84%, 78%, and 79% of EN individuals displayed an OD above the cutoff value for NE control sera (mean + 2 SD) for rPagSP02 (P ≤ .001), rPagSP06 (P ≤ .0001), or rPagSP02 + rPagSP06 (P ≤ .0001), respectively, which were comparable to that observed against Ph. argentipes SGH (P ≤ .0001) at 71% (Figure 3A). Surprisingly, combining rPagSP02 + rPagSP06 lowered the reactivity of IR sera obtained from individuals bitten by Ph. papatasi, better reproducing the response against Ph. argentipes SGH. The outperformance of rPagSP02 + rPagSP06 compared to either rPagSP02 or rPagSP06 is further reinforced by their one-to-one equivalence in the response to Ph argentipes SGH (Figure 3B). Furthermore, a strong correlation was observed for the antibody response of EN sera to Ph. argentipes SGH and rPagSP02 + rPagSP06 (P < .0001; r = 0.89; 95% CI, .807–.935) that was weaker when using rPagSP02 (P < .0001; r = 0.80; 95% CI, .625–.902) or rPagSP06 (P < .0001; r = 0.74; 95% CI, .512–.867) alone (Figure 3C). Importantly, sera of individuals from Cambodia that had been bitten by Aedes (Supplementary Figure 4A) and Anopheles (Supplementary Figure 4B) mosquitoes, in areas where Ph. argentipes is absent, did not recognize rPagSP02 + rPagSP06 (Supplementary Figure 4C), further validating rPagSP02 + rPagSP06 as a composite biomarker specific to Ph. argentipes bites in humans.
Figure 3.
Maximizing marker coverage and specificity by using a combination of 2 immunogenic Phlebotomus argentipes recombinant proteins, rPagSP02 and rPagSP06. A, Total IgG ELISA against 1 µg/mL of rPagSP02, 1 µg/mL of rPagSP06, a combination of 1 µg/mL of rPagSP02 and rPagSP06 proteins, or 2 µg/mL of Ph. argentipes SGH. Antigens were tested against serum samples (1:50 dilution) from individuals living in VL-endemic areas where Ph. argentipes is prevalent (EN, n = 30–52), in the United States, a nonendemic area for VL where Phlebotomus sand flies are absent (NE, n = 6), or from individuals exposed to Phlebotomus papatasi bites in Iraq (IR, n = 5). Cutoff values were calculated as the mean ± 2 SD of NE controls (black dashed line) or IR controls (gray dotted line). Each sample was tested in duplicate. Mann-Whitney U test. B, Comparison between OD values of Ph. argentipes SGH and each of rPagSP02, rPagSP06, or rPagSP02 + rPagSP06. C, Pearson rank correlation test between Ph. argentipes SGH and each of rPagSP02, rPagSP06, or rPagSP02 + rPagSP06. Correlation coefficient (r), P values, and 95% CI are provided. Representative data of 2–3 independent experiments. OD values were normalized by subtracting the mean value of control wells containing no antigen and a pool of sera with high reactivity against Ph. argentipes SGH (n = 3). Normalized values are represented as ΔOD. ***P ≤ .001, ****P ≤ .0001; a P value of ≤ .05 was considered significant. Abbreviations: CI, confidence interval; ELISA, enzyme-linked immunosorbent assay; IgG, immunoglobulin G; OD, optical density; SGH, salivary gland homogenate; VL, visceral leishmaniasis.
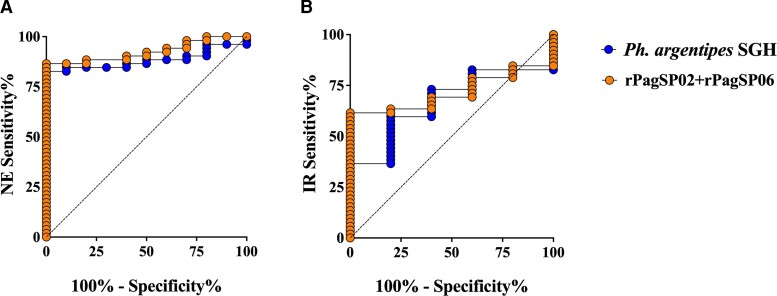
Next, we calculated the sensitivity and specificity of rPagSP02 + rPagSP06 compared to Ph. argentipes SGH. ROC curve analyses using our NE or IR as true negatives indicated that rPagSP02 + rPagSP06 discriminated between EN and NE (Figure 4A) or EN and IR (Figure 4B) sera as effectively as Ph. argentipes SGH. Compared to NE sera, rPagSP02 + rPagSP06 ELISA performed at a predicted sensitivity of 86.54% (95% CI, 74.73%–93.92%) and specificity of 90% (95% CI, 50.58%–99.49%), with an area under the curve (AUC) value of 0.92 (P >.0001) at a calculated cutoff value of > −0.03 (Table 1). Additionally, when ROC curve analysis was performed against IR sera as our true negative controls, rPagSP02 + rPagSP06 had a significantly higher AUC of 0.86 (P = .007, cutoff > 0.34) compared with an AUC of 0.66 for Ph. argentipes SGH (P = .215, cutoff > 0.38; Table 1). Collectively, these data provide strong evidence of a sensitive and species-specific ELISA assay that combines 2 immunodominant Ph. argentipes salivary antigens, rPagSP02 + rPagSP06, as an effective composite biomarker of human exposure to Ph. argentipes bites.
Figure 4.
rPagSP02 + rPagSP06 salivary proteins combined represent a sensitive and specific marker of vector exposure to Phlebotomus argentipes bites. A and B, The probability of positivity of serum samples from individuals living in VL-endemic areas where Ph. argentipes is prevalent (EN) was calculated against either (A) control samples from the United States, a VL-nonendemic area (NE) or (B) control samples from individuals exposed to Phlebotomus papatasi bites in Iraq (IR) by receiver operating characteristics curves for Ph. argentipes SGH and rPagSP02 + rPagSP06 antigens. Representative data of 3 independent experiments. Optical density (OD) values were normalized by subtracting the mean value of control wells containing no antigen and a pool of sera with high reactivity against Ph. argentipes SGH (n = 3). Normalized values are represented as ΔOD. A P value of ≤ .05 was considered significant. Abbreviations: SGH, salivary gland homogenate; VL, visceral leishmaniasis.
Table 1.
Table Summarizing ROC Values Calculated for Each Antigen
| ROC Curves Calculation | Antigen | AUC | Cutoff | Sensitivity, % (95% CI) | Specificity, % (95% CI) | P Value |
| Nonendemic controls | Ph. argentipes SGH | 0.88 | > −0.03 | 73.08 (59.75–83.23) | 100 (72.25–100) | .0001 |
| rPagSP02 + rPagSP06 | 0.92 | > −0.03 | 86.54 (74.73–93.92) | 90 (59.58–99.49) | > .0001 | |
| Iraq controls | Ph. argentipes SGH | 0.66 | > 0.38 | 36.54 (24.80–50.13) | 100 (56.55–100) | .2145 |
| rPagSP02 + rPagSP06 | 0.86 | > 0.34 | 48.08 (35.10–61.31) | 100 (56.55–100) | .0074 |
Abbreviations: AUC, area under the curve; CI, confidence interval; ROC, receiver operating characteristic; SGH, salivary gland homogenate.
DISCUSSION
Although VL incidence in the Indian subcontinent has fallen steeply, elimination as a public health problem does not eradicate leishmanial infection, and vector control will remain crucial to sustain long-term low incidence. This is of particular relevance when elimination targets are reached, and attention and resources inevitably diminish, requiring a rethinking of strategies to maintain surveillance and detect outbreaks [31, 32]. In addition, because of low VL incidence, prohibitively large sample sizes would be required if cases are used as the outcome measure to evaluate the efficacy of interventions highlighting the urgent need for alternative tools to support the next postelimination phase of the program [31–33].
Saliva from arthropod vectors have become prominent as surveillance tools in various field studies [20–23, 25, 26, 28, 34, 35]. A specific and sensitive biomarker of human exposure to Ph. argentipes bites remains a priority for VL-endemic areas in the Indian subcontinent. A direct measure of vector-human contact can be used to address questions regarding human [32, 36, 37] and sand fly [14–17] behavior, potential infection reservoirs [18, 38–40], and the effectiveness of IRS and other vector control modalities [33, 41].
We improved the sensitivity and specificity of an assay to detect exposure to bites of Ph. argentipes in humans by using a combination of 2 immunodominant Ph. argentipes SSP, PagSP02 and PagSP06, belonging to the SP15- and SP32-like family of proteins, respectively [30]. Importantly, using sera of individuals bitten by Ph. papatasi in Iraq, where Ph. argentipes is absent [42], we detected weakly cross-reactive antibodies against recombinant rPagSP02 and rPagSP06 when tested individually, suggesting similarities with SP15- and SP32-like proteins from Ph. papatasi, respectively, supported by previous observations against rPagSP06 [28]. However, when rPagSP02 and rPagSP06 were combined, the level of cross-reactive antibodies was considerably diminished. rPagSP02 + rPagSP06 did not recognize antibodies from sera of individuals exposed to mosquitoes, in areas where Ph. argentipes is absent [42, 43], strongly suggesting the validity of this composite biomarker as a specific indicator of exposure to Ph. argentipes bites.
A recent study targeted the human antibody response against 3 Ph. argentipes recombinant proteins, rPagSP04, rPagSP05, and rPagSP06, in individuals living in Bangladesh [28]. The authors observed that rPagSP06 had the strongest correlation (r = 0.76) against Ph. argentipes SGH. Importantly, a weak correlation (r = 0.43) was observed when endemic sera was screened against Ph. argentipes and Ph. papatasi SGH, but the reactivity of rPagSP06 in individuals bitten by Ph. papatasi was not investigated in the study [28]. Overall, these data support our findings of the immunodominance of rPagSP06 and the observed low cross-reactivity between SSP of the 2 vectors.
A handful of field reports established the short persistence of antisaliva antibodies in the absence of sand fly bites [25, 26, 29, 44, 45], with titers diminishing as fast as 30 days for Ph. argentipes [25, 26, 29, 44, 45]. Correspondingly, antibodies against Ph. argentipes SGH increased above baseline 180 days after potential reexposure to Ph. argentipes fly bites [25]. These studies indicate that antisaliva antibodies are transient, and suggest that their levels provide a good measure of the intensity and duration of exposure to sand fly bites. Interestingly, the percent of positivity against Ph. argentipes salivary antigens was lower in individuals living in Bangladesh compared to India, with only 36% and 40% of individuals developing antibodies against rPagSP06 or Ph. argentipes SGH, respectively [28], compared to a positivity of 78% and 79% against rPagSP06 or rPagSP02 + rPagSP06, respectively, in our study. Of note, previous studies in India showed a positivity range of 63%–82% against Ph. argentipes SGH [25, 26], comparable to our data. Differences in the rate of positivity against Ph. argentipes saliva presumably reflect the intensity of sand fly exposure in the months prior to sample collection. In Bihar, India, Ph. argentipes seasonality is bimodal comprising a small peak at the beginning of the season around March-April, and a major peak by June-August, followed by a gradual decline in sand fly abundance until the end of the season in October-December [13, 17, 46]. Furthermore, compared to Ph. argentipes, the relative abundance of Ph. papatasi in most areas of Bihar is less than 5% with the latter exhibiting a sporadic distribution [13, 15, 17]. These properties make appropriately chosen salivary antigens from Ph. argentipes appealing targets as surveillance tools to estimate recent exposure to vector bites.
An important limitation in this study is the relatively low number of endemic sera tested. Plans are underway to test rPagSP02 + rPagSP06 in a large-scale longitudinal study of endemic villages in India over a follow-up period of 2–3 years. This information will help us evaluate the persistence and fluctuation of antisalivary antibodies in sera from humans living in VL-endemic areas in Bihar, India, throughout several seasons of Leishmania transmission. Furthermore, to validate the use of this tool on a wide scale, a comparative analysis of rPagSP02 and rPagSP06 salivary transcripts from wild-caught Ph. argentipes sand flies across the species distribution range warrants further investigation. This will establish the presence of significant polymorphisms and how they may affect the efficacy of the composite marker.
In summary, we developed a sand-fly-based tool to assess the intensity of vector-human contact in VL-endemic areas of India. This composite biomarker can be used to accurately measure the success of current vector control strategies and to improve vector management approaches. Such tools can also evaluate infection risk and rapidly address outbreaks, both critical to overcoming last mile challenges to VL elimination in the Indian subcontinent.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Contributor Information
Eva Iniguez, Vector Molecular Biology Section, Laboratory of Malaria and Vector Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Rockville, Maryland, USA.
Samiran Saha, Department of Biotechnology, Institute of Science, Visva Bharati University, Bolpur, West Bengal, India.
Georgios Petrellis, Vector Molecular Biology Section, Laboratory of Malaria and Vector Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Rockville, Maryland, USA; Laboratory of Microbiology, Parasitology, and Hygiene, University of Antwerp, Antwerp, Belgium.
Claudio Menenses, Vector Molecular Biology Section, Laboratory of Malaria and Vector Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Rockville, Maryland, USA.
Samantha Herbert, Vector Molecular Biology Section, Laboratory of Malaria and Vector Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Rockville, Maryland, USA.
Yvonne Gonzalez-Rangel, Vector Molecular Biology Section, Laboratory of Malaria and Vector Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Rockville, Maryland, USA.
Tobin Rowland, Entomology Branch, Walter Reed Army Institute of Research, Silver Spring, Maryland, USA.
Naomi E Aronson, Infectious Diseases Division, Department of Medicine, Uniformed Services University of the Health Sciences, Bethesda, Maryland, USA.
Clair Rose, Department of Parasitology, Liverpool School of Tropical Medicine, Liverpool, United Kingdom.
Lee Rafuse Haines, Department of Parasitology, Liverpool School of Tropical Medicine, Liverpool, United Kingdom.
Alvaro Acosta-Serrano, Department of Parasitology, Liverpool School of Tropical Medicine, Liverpool, United Kingdom; Department of Vector Biology, Liverpool School of Tropical Medicine, Liverpool, United Kingdom.
Tiago D Serafim, Vector Molecular Biology Section, Laboratory of Malaria and Vector Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Rockville, Maryland, USA.
Fabiano Oliveira, Vector Molecular Biology Section, Laboratory of Malaria and Vector Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Rockville, Maryland, USA.
Sridhar Srikantiah, Bihar Technical Support Program, CARE India Solutions for Sustainable Development, Patna, India.
Caryn Bern, Department of Epidemiology and Biostatistics, University of California, San Francisco, San Francisco, California, USA.
Jesus G Valenzuela, Vector Molecular Biology Section, Laboratory of Malaria and Vector Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Rockville, Maryland, USA.
Shaden Kamhawi, Vector Molecular Biology Section, Laboratory of Malaria and Vector Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Rockville, Maryland, USA.
Notes
Acknowledgments . We thank the Protein Expression Laboratory at NCI-Frederick (Frederick, MD) for the expression of recombinant proteins and the Research Technologies Branch at National Institutes of Health, National Institute of Allergy and Infectious Diseases (NIH, NIAID) for analysis and identification of recombinant proteins by mass-spectrometry; and Michael P. Fey, PhD at the Biostatistics Research Branch at NIH, NIAID for his statistical input on this manuscript. Yonas Gebremicale at the Laboratory of Malaria and Vector Research, NIH, NIAID reared and provided the Anopheles albopictus and Aedes aegypti mosquitoes used for salivary gland dissections and immunoblotting. We thank Mara Short for the dissection of Anopheles albopictus salivary glands for the study.
Financial support . This work was supported by the NIH, NIAID Intramural Research Program; the Bill and Melinda Gates Foundation (grant number INV-008856/OPP1196454); and the Global Emerging Infections Surveillance Section, Armed Forces Health Surveillance Branch, Department of Defense (grant number PRoMIS ID P0024_17_HS).
References
- 1. World Health Organization . Global vector control response 2017–2030. https://www.who.int/publications/i/item/9789241512978. Accessed 22 March 2022.
- 2. Alvar J, Vélez ID, Bern C, et al. Leishmaniasis worldwide and global estimates of its incidence. PLoS One 2012; 7:e35671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. World Health Organization . Regional strategic framework for elimination of kala-azar from the South-East Asia region (2005–2015) SEA-VBC 85 2005. New Delhi, India: World Health Organization: Regional Office for South-East Asia. [Google Scholar]
- 4. World Health Organization . Regional strategic framework for elimination of kala-azar from the South-East Asia region (2011–2015) SEA-CD 239 2012. New Delhi, India: World Health Organization Regional Office for South-East Asia.
- 5. World Health Organization . Global health observatory. Leishmaniasis. https://apps.who.int/neglected_diseases/ntddata/leishmaniasis/leishmaniasis.html. Accessed 22 January 2022.
- 6. Ruiz-Postigo JA, Jain S, Mikhailov A, et al. Global leishmaniasis surveillance: 2019–2020, a baseline for the 2030 roadmap. Wkly Epidemiol Rec 2021; 35:401–19. [Google Scholar]
- 7. Gill N, Pandey D, Roy N, Jain DNAS. Kala-azar in India—progress and challenges towards its elimination as a public health problem. Wkly Epidemiol Rec 2021; 2022:267–79. [Google Scholar]
- 8. Chowdhury R, Kumar V, Mondal D, et al. Implication of vector characteristics of Phlebotomus argentipes in the kala-azar elimination programme in the Indian sub-continent. Pathog Glob Health 2016; 110:87–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rocha DA, Costa LMD, Pessoa GDC, Obara MT. Methods for detecting insecticide resistance in sand flies: a systematic review. Acta Trop 2021; 213:105747. [DOI] [PubMed] [Google Scholar]
- 10. National Vector Borne Disease Control Programme . Accelerated plan for kala-azar elimination 2017; 2017:80. https://nvbdcp.gov.in/WriteReadData/l892s/Accelerated-Plan-Kala-azar1-Feb2017.pdf.
- 11. Deb R, Singh RP, Mishra PK, et al. Impact of IRS: four-years of entomological surveillance of the Indian visceral leishmaniases elimination programme. PLoS Negl Trop Dis 2021; 15:e0009101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Singh N, Mishra J, Singh R, Singh S. Animal reservoirs of visceral leishmaniasis in India. J Parasitol 2013; 99:64–7. [DOI] [PubMed] [Google Scholar]
- 13. Poche DM, Garlapati RB, Mukherjee S, et al. Bionomics of Phlebotomus argentipes in villages in Bihar, India with insights into efficacy of IRS-based control measures. PLoS Negl Trop Dis 2018; 12:e0006168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Poche D, Garlapati R, Ingenloff K, Remmers J, Poche R. Bionomics of phlebotomine sand flies from three villages in Bihar, India. J Vector Ecol 2011; 36(Suppl 1):S106–17. [DOI] [PubMed] [Google Scholar]
- 15. Poche DM, Poche RM, Mukherjee S, et al. Phlebotomine sandfly ecology on the Indian subcontinent: does village vegetation play a role in sandfly distribution in Bihar, India? Med Vet Entomol 2017; 31:207–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Poché DM, Torres-Poché Z, Garlapati R, Clarke T, Poché RM. Short-term movement of Phlebotomus argentipes (Diptera: Psychodidae) in a visceral leishmaniasis-endemic village in Bihar, India. J Vector Ecol 2018; 43:285–92. [DOI] [PubMed] [Google Scholar]
- 17. Poche RM, Garlapati R, Elnaiem DE, Perry D, Poche D. The role of palmyra palm trees (Borassus flabellifer) and sand fly distribution in northeastern India. J Vector Ecol 2012; 37:148–53. [DOI] [PubMed] [Google Scholar]
- 18. Singh OP, Hasker E, Boelaert M, Sacks D, Sundar S. Xenodiagnosis to address key questions in visceral leishmaniasis control and elimination. PLoS Negl Trop Dis 2020; 14:e0008363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Andrade BB, Teixeira CR. Biomarkers for exposure to sand flies bites as tools to aid control of leishmaniasis. Front Immunol 2012; 3:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Marzouki S, Kammoun-Rebai W, Bettaieb J, et al. Validation of recombinant salivary protein PpSP32 as a suitable marker of human exposure to Phlebotomus papatasi, the vector of leishmania major in Tunisia. PLoS Negl Trop Dis 2015; 9:e0003991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Carvalho AM, Fukutani KF, Sharma R, et al. Seroconversion to Lutzomyia intermedia LinB-13 as a biomarker for developing cutaneous leishmaniasis. Sci Rep 2017; 7:3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Teixeira C, Gomes R, Collin N, et al. Discovery of markers of exposure specific to bites of Lutzomyia longipalpis, the vector of leishmania infantum chagasi in Latin America. PLoS Negl Trop Dis 2010; 4:e638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sumova P, Sima M, Spitzova T, et al. Human antibody reaction against recombinant salivary proteins of Phlebotomus orientalis in Eastern Africa. PLoS Negl Trop Dis 2018; 12:e0006981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Souza AP, Andrade BB, Aquino D, et al. Using recombinant proteins from Lutzomyia longipalpis saliva to estimate human vector exposure in visceral leishmaniasis endemic areas. PLoS Negl Trop Dis 2010; 4:e649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Clements MF, Gidwani K, Kumar R, et al. Measurement of recent exposure to Phlebotomus argentipes, the vector of Indian visceral leishmaniasis, by using human antibody responses to sand fly saliva. Am J Trop Med Hyg 2010; 82:801–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gidwani K, Picado A, Rijal S, et al. Serological markers of sand fly exposure to evaluate insecticidal nets against visceral leishmaniasis in India and Nepal: a cluster-randomized trial. PLoS Negl Trop Dis 2011; 5:e1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Marzouki S, Abdeladhim M, Abdessalem CB, et al. Salivary antigen SP32 is the immunodominant target of the antibody response to Phlebotomus papatasi bites in humans. PLoS Negl Trop Dis 2012; 6:e1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sumova P, Sanjoba C, Willen L, et al. PpSP32-like protein as a marker of human exposure to Phlebotomus argentipes in Leishmania donovani foci in Bangladesh. Int J Parasitol 2021; 51:1059–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Aronson NE, Oliveira F, Gomes R, et al. Antibody responses to Phlebotomus papatasi saliva in American soldiers with Cutaneous leishmaniasis versus controls. Front Trop Dis 2022; 2:766273. [Google Scholar]
- 30. Anderson JM, Oliveira F, Kamhawi S, et al. Comparative salivary gland transcriptomics of sandfly vectors of visceral leishmaniasis. BMC Genom 2006; 7:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bindroo J, Priyamvada K, Chapman LAC, et al. Optimizing village-level targeting of active case detection to support visceral leishmaniasis elimination in India. Front Cell Infect Microbiol 2021; 11:648847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Priyamvada K, Bindroo J, Sharma MP, et al. Visceral leishmaniasis outbreaks in Bihar: community-level investigations in the context of elimination of kala-azar as a public health problem. Parasit Vectors 2021; 14:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kumar A, Saurabh S, Jamil S, Kumar V. Intensely clustered outbreak of visceral leishmaniasis (kala-azar) in a setting of seasonal migration in a village of Bihar, India. BMC Infect Dis 2020; 20:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kearney EA, Agius PA, Chaumeau V, Cutts JC, Simpson JA, Fowkes FJI. Anopheles salivary antigens as serological biomarkers of vector exposure and malaria transmission: a systematic review with multilevel modelling. Elife 2021; 10:e73080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Maldonado-Ruiz LP, Montenegro-Cadena L, Blattner B, Menghwar S, Zurek L, Londono-Renteria B. Differential tick salivary protein profiles and human immune responses to lone star ticks (Amblyomma americanum) from the wild vs. a laboratory colony. Front Immunol 2019; 10:1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Perry D, Dixon K, Garlapati R, Gendernalik A, Poche D, Poche R. Visceral leishmaniasis prevalence and associated risk factors in the saran district of Bihar. India, from 2009 to July of 2011. Am J Trop Med Hyg 2013; 88:778–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Govil D, Sahoo H, Pedgaonkar SP, Chandra Das K, Lhungdim H. Assessing knowledge, attitudes, and preventive practices related to kala-A: a study of rural Madhepura, Bihar, India. Am J Trop Med Hyg 2018; 98:857–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Molina R, Ghosh D, Carrillo E, et al. Infectivity of post-kala-azar dermal leishmaniasis patients to sand flies: revisiting a proof of concept in the context of the kala-azar elimination program in the Indian subcontinent. Clin Infect Dis 2017; 65:150–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kushwaha AK, Scorza BM, Singh OP, et al. Domestic mammals as reservoirs for Leishmania donovani on the Indian subcontinent: possibility and consequences on elimination. Transbound Emerg Dis 2021; 69:268–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mondal D, Bern C, Ghosh D, et al. Quantifying the infectiousness of post-kala-azar dermal leishmaniasis toward sand flies. Clin Infect Dis 2019; 69:251–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Garlapati R, Iniguez E, Serafim TD, et al. Towards a sustainable vector-control strategy in the post kala-azar elimination era. Front Cell Infect Microbiol 2021; 11:641632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Coleman RE, Burkett DA, Putnam JL, et al. Impact of phlebotomine sand flies on U.S. Military operations at Tallil air base, Iraq: 1. Background, military situation, and development of a “leishmaniasis control program”. J Med Entomol 2006; 43:647–62. [DOI] [PubMed] [Google Scholar]
- 43. Loyer M, Depaquit J, Gay F. A new cavernicolous sand fly from Cambodia: Idiophlebotomus nicolegerae n. sp. (Diptera: Psychodidae). Acta Trop 2016; 155:43–50. [DOI] [PubMed] [Google Scholar]
- 44. Lakhal-Naouar I, Mukbel R, DeFraites RF, et al. The human immune response to saliva of Phlebotomus alexandri, the vector of visceral leishmaniasis in Iraq, and its relationship to sand fly exposure and infection. PLoS Negl Trop Dis 2021; 15:e0009378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Oliveira F, Giorgobiani E, Guimaraes-Costa AB, et al. Immunity to vector saliva is compromised by short sand fly seasons in endemic regions with temperate climates. Sci Rep 2020; 10:7990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Picado A, Das ML, Kumar V, et al. Phlebotomus argentipes seasonal patterns in India and Nepal. J Med Entomol 2010; 47:283–6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.