Abstract
Background
The CD274 (PD-L1)/PDCD1 (PD-1) immune checkpoint interaction may promote cancer progression, but the expression patterns and prognostic significance of PD-L1 and PD-1 in the colorectal cancer microenvironment are inadequately characterised.
Methods
We used a custom 9-plex immunohistochemistry assay to quantify the expression patterns of PD-L1 and PD-1 in macrophages, T cells, and tumour cells in 910 colorectal cancer patients. We evaluated cancer-specific mortality according to immune cell subset densities using multivariable Cox regression models.
Results
Compared to PD-L1– macrophages, PD-L1+ macrophages were more likely M1-polarised than M2-polarised and located closer to tumour cells. PD-L1+ macrophage density in the invasive margin associated with longer cancer-specific survival [Ptrend = 0.0004, HR for the highest vs. lowest quartile, 0.52; 95% CI: 0.34–0.78]. T cell densities associated with longer cancer-specific survival regardless of PD-1 expression (Ptrend < 0.005 for both PD-1+ and PD-1– subsets). Higher densities of PD-1+ T cell/PD-L1+ macrophage clusters associated with longer cancer-specific survival (Ptrend < 0.005).
Conclusions
PD-L1+ macrophages show distinct polarisation profiles (more M1-like), spatial features (greater co-localisation with tumour cells and PD-1+ T cells), and associations with favourable clinical outcome. Our comprehensive multimarker assessment could enhance the understanding of immune checkpoints in the tumour microenvironment and promote the development of improved immunotherapies.
Subject terms: Cancer microenvironment, Prognostic markers, Tumour immunology, Colorectal cancer
Background
Colorectal cancer is the second leading cause of cancer death worldwide with over 900,000 deaths in 2020 [1]. The assessment of cancer prognosis and treatment is mainly based on the tumour extent and tumour morphology, but the rapidly increasing knowledge on the significance of tumour immune contexture has led to the development of improved immune-related prognostic markers and effective anticancer immunotherapies [2]. In addition to the quantity of immune cells, the activity of immunoregulatory signalling pathways, such as the co-inhibitory pathway of programmed death ligand 1 (PD-L1, CD274) and its receptor programmed death 1 (PD-1, PDCD1), may affect cancer progression. Hereinafter, CD274 and PDCD1 are referred as PD-L1 and PD-1 due to the ubiquity of these “colloquial” protein names. PD-L1 is mainly expressed in macrophages [3], whereas PD-1 is mainly expressed in T cells [4]. The expression of PD-L1 and PD-1 is often upregulated in cancer and their interaction may lead to immunosuppression through T cell exhaustion, thus promoting tumour growth [4]. However, the prognostic significance of immune checkpoint protein expression in many tumour types, including colorectal cancer, has remained controversial [5].
Macrophages are inflammatory cells which have shown to associate with cancer progression [6]. They are commonly categorised into classically activated, M1-polarised, and alternatively activated, M2-polarised macrophages, which differ by their surface antigens, cytokine secretion profiles, and physiological functions. However, instead of two completely distinct subtypes, macrophages are considered to form a continuum of phenotypes that may gradually change their polarisation towards M1-like or M2-like and simultaneously express phenotypic markers for both subtypes. In the tumour microenvironment, M1-like macrophages are thought to have pro-inflammatory effect and be more prevalent in early-stage cancer, whereas the proportion of anti-inflammatory M2-like macrophages increases along cancer progression [6]. Higher M1/M2-like macrophage ratio has been thought to associate with improved cancer-specific survival [7]. The expression patterns of immune checkpoints in macrophages and their clinical value are poorly established in colorectal cancer.
In this study, we used multiplex immunohistochemistry and machine learning-based image analysis to comprehensively characterise PD-L1 and PD-1 immune checkpoint expression in M1-like and M2-like macrophages, T cells, and tumour cells in a large, population-based colorectal cancer cohort of 910 patients. Our primary aim was to (i) evaluate the expression patterns and prognostic significance of PD-L1 and PD-1 in immune cells and tumour cells. As secondary aims, we (ii) clarified the associations between immune checkpoint expression and tumour characteristics, and (iii) investigated the infiltration patterns and prognostic role of M1-like and M2-like polarised macrophages. We hypothesised low expression of PD-L1 and PD-1 immune checkpoints and higher density of M1-like macrophages to associate with favourable colorectal cancer outcome.
Methods
Study population
The study was based on a cohort of 1343 colorectal cancer patients, who underwent a resection for primary colon or rectum carcinoma between January 1, 2000 and December 31, 2015 in Central Finland Central Hospital. The population of the area was on average 270,000 during the study period [8]. The clinical, histopathological, and follow-up data were retrospectively collected from the pathology registry and clinical records of Central Finland Central Hospital. All tumours were screened for DNA mismatch repair (MMR) deficiency and BRAF V600E mutation status with immunohistochemistry [9]. Histological tumour parameters, including tumour differentiation and lymphovascular invasion, were re-evaluated from hematoxylin and eosin-stained whole slides by the study pathologist (J.P.V.). All histological data were analysed blinded to clinical data. We excluded patients who died within 30 days after surgery (N = 40) or received any preoperative oncological treatments (radiotherapy, chemotherapy, or chemoradiotherapy) (N = 243) due to their potential influences on tumour characteristics or immune response [10]. After applying further exclusion criteria of unsuccessful multiplex immunohistochemistry staining or inadequate tumour tissue of both the tumour centre and the invasive margin in tissue microarrays (N = 150), the final cohort comprised samples of 910 colorectal cancer patients.
Multiplex immunohistochemistry
Tissue microarrays were constructed by selecting four 1-mm diameter cores from each tumour [9]. We designed a 9-plex immunohistochemistry panel to identify macrophages with CD68 and CD163, T cells with CD3, and tumour cells with KRT (keratin). PD-L1 and PD-1 immune checkpoint molecules and four macrophage polarisation markers (CD86, CD163, HLA-DR, MRC1) were included in the assay.
The multiplex immunohistochemistry staining was done with Bond-III automated IHC stainer (Leica Biosystems, Buffalo Grove, IL, USA) and Bond Refine Detection kit (DS9800, Leica Biosystems). We used sections of 3.5 µm. Candidate antibodies and suitable dilutions were optimised using conventional immunohistochemistry with 3,3′-Diaminobenzidine (DAB) chromogen in a test tissue microarray consisting of normal colorectal mucosa, colorectal cancer tissue, and tonsil tissue. These antibodies were then combined into a multiplex immunohistochemistry assay. The correspondence of staining patterns of multiplex and conventional immunohistochemistry were visually confirmed in serial sections of a test microarray, and the correspondence was further quantified by manually annotating cells in 10 respective regions (size 200 × 200 µm) of multiplex and conventional immunohistochemistry (Supplementary Fig. S1). The multiplex staining was conducted using a previously validated, cyclic method that uses 3-Amino-9-Ethylcarbazole (AEC) as the chromogen [11]. The workflow for the staining is shown in Supplementary Fig. S2 and the selected monoclonal antibodies along with their dilutions and antigen retrieval conditions are listed in Supplementary Table S1. We used AEC+ high sensitivity substrate (K3469, Dako, Glostrup, Denmark) as the chromogen. After each cycle, the slides were mounted with VectaMount AQ Aqueous Mounting Medium (H-5501, Vector Laboratories, Newark, CA, USA), scanned with 20× objective of NanoZoomer XR (Hamamatsu Photonics, Hamamatsu City, Japan, resolution 0.45 µm/pixel), de-stained with ethanol, and heated to remove the primary and secondary antibodies.
Image analysis
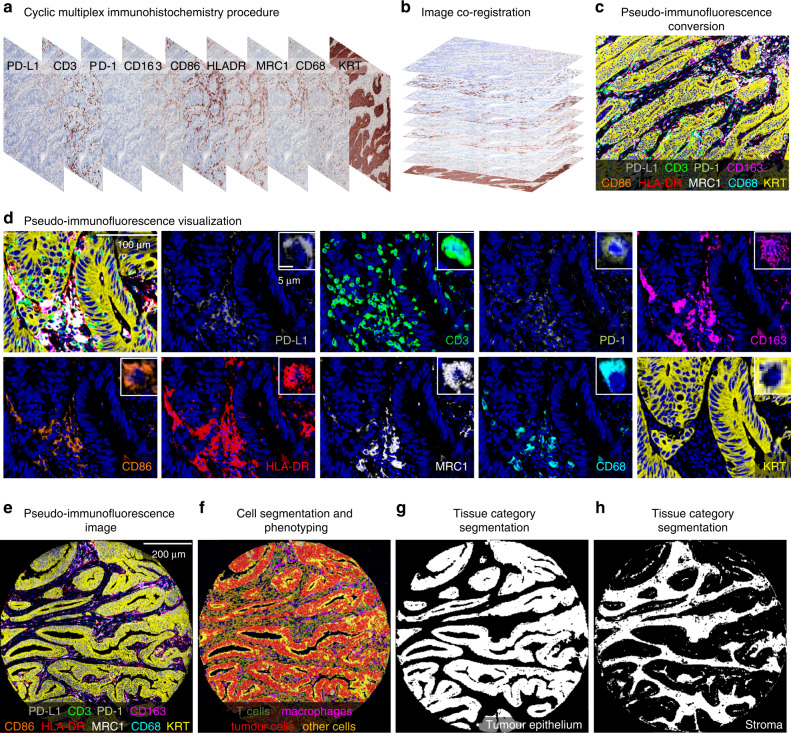
The digitised images of multiplex immunohistochemistry slides were processed with QuPath (version 0.2.3) [12]. Tissue microarray cores were recognised with TMA dearrayer function and separated into single core images. We excluded cores which were folded, included minimal amount of tumour, were necrotic, or comprised less than 50% of the 1-mm diameter core area after all staining cycles. The single core images of all 9 staining cycles were stacked into one 10-channel pseudo-immunofluorescence image (with hematoxylin as the 10th channel) by aligning cell nuclei using the MultiStackReg macro (downloaded from http://bradbusse.net/downloads.html) in ImageJ/Fiji open-source software [13]. This macro enabled the co-registration of the images despite potential minor shifts in tissue during the staining cycles. The conversion of single images into a 10-plex pseudo-immunofluorescence image is illustrated in Fig. 1a–d. The staining intensities were consistent across tissue microarrays (Supplementary Fig. S3), indicating that the assay had performed uniformly.
Fig. 1. Multiplex immunohistochemistry panel and image analysis.
a Digitised multiplex immunohistochemistry image from each staining cycle for one tumour. b, c Image co-registration based on aligning the hematoxylin layers to merge individual images into one 10-channel pseudo-immunofluorescence image. d example images showing the expression patterns of each marker merged and separately. e−h pseudo-immunofluorescence image (e), machine learning-based cell segmentation and phenotyping into T cells, macrophages, tumour cells, and other cells (f), and mask images of the categorisation of tissue into tumour epithelium and stroma (g, h).
The pseudo-immunofluorescence images were analysed with QuPath, utilising previously validated [14], supervised machine learning algorithms. We identified cells with cell detection function and calculated additional smoothed object features to improve subsequent cell phenotyping. Cells were phenotyped into T cells, macrophages, tumour cells, and other cells using the object classifier function built in QuPath, based on the random forests algorithm. For training, example cells were annotated as follows: (1) all CD3 expressing cells were phenotyped as T cells; (2) cells expressing CD68 [15] and/or CD163 [7] were identified as macrophages in line with a prior study [16]; (3) KRT expressing cells were phenotyped as tumour cells; and (4) cells negative for CD3, CD68, CD163 and KRT were classified as other cells. Cell data yielded phenotypes, marker intensities, and coordinates for each cell. To classify tissue categories, QuPath was trained to identify tissue segments of tumour epithelium and stroma using the built-in pixel classifier function. Necrotic regions, empty white space without any tissue, and regions of partial core loss (the hematoxylin staining from the first staining cycle not corresponding with the hematoxylin staining form the last staining cycle) were excluded. The workflow for cell and tissue analyses is described in Supplementary Fig. S4 and the example images from cell segmentation and tissue categorisation in QuPath are represented in Fig. 1e–h.
Immune cell phenotyping and classification
Cell level data were further processed with RStudio (version 1.3.1093) and R statistical programming (version 4.0.3, R Core Team).
Macrophages were classified according to PD-L1 expression and M1/M2 polarisation state. To categorise macrophages according to their polarisation, we calculated a polarisation index for each macrophage consistent with a prior study [16]. First, we converted the intensities of all four macrophage markers into percentiles across all macrophages and calculated a polarisation index by reducing the intensities of M2-like macrophage markers from the intensities of M1-like macrophage markers [formula: (CD86 + HLA-DR) − (CD163 + MRC1); with marker names denoting intensity percentiles]. Using this formula, macrophages with higher polarisation index values were considered more M1-polarised, while those with lower values were considered more M2-polarised. For downstream analyses, the index values were divided into ordinal quartile categories (Q1–Q4) across all macrophages in 910 colorectal cancer cases. As in a prior study [16], macrophages in the lowest group (Q1) were classified as M1-like macrophages and those in the highest group (Q4) were classified as M2-like macrophages. Macrophages in the middle groups (Q2–Q3), covering 50% of the cells, were left unclassified to limit this analysis to the most strongly polarised M1-like and M2-like macrophages. For downstream survival analyses, we calculated the densities of various macrophage subsets in the tumour centre and the invasive margin and categorised the densities into ordinal quartile categories (Q1–Q4). For variables including over 25% zero densities, all zero values were categorised into the lowest group Q1. The remaining values were divided equally into groups Q2–Q4. Ordinal quartile categories were also similarly defined for other immune cell variables.
In addition to macrophages, PD-L1 expression was evaluated in tumour epithelial cells, based on the weighted histoscore method. For each cell, we classified the staining intensity (negative, weak, intermediate, or high), and for each case, we calculated the percentage (0–100%) of tumour cells within each category separately in the tumour centre and the invasive margin. In cases with multiple tissue microarray cores from one region, we calculated the mean values. Finally, PD-L1 histoscore was calculated as follows: PD-L1 histoscore = [(1 × percentage of weakly stained cells)+(2 × percentage of moderately stained cells)+(3 × percentage of strongly stained cells)]. The possible range for values was from 0 (all cells negative) to 300 (all cells strongly positive) [17]. For survival analyses, we calculated the mean histoscore for each tumour and categorised the tumours into negative or positive using a cut-off value of ≥5 for the positivity.
T cells were further subdivided into PD-1 positive or negative based on a fixed cut-off value for their cytoplasmic PD-1 staining intensity (40 intensity units).
To analyse spatial interactions between immune and tumour cells, we used the spatstat (2.2–0) package to calculate nearest neighbour distances (NNDs), which measure the distance from a specific point (e.g., macrophage) to its closest neighbour point of specific category (e.g., tumour cell). For visualisation, scaled intensities of macrophage polarisation markers as a function of NND from tumour cells were plotted with ggplot2 (3.3.3) package using generalised additive model smoothing [formula y ~ s(x)]. For further spatial analyses, we calculated the density of PD-1+/PD-L1+ clusters defined as PD-1+ T cell located within 20-μm distance from the closest PD-L1+ macrophage. The radius was selected in order to identify cells with capability for direct cell-cell interaction consistent with prior reports [9, 18].
Statistical analysis
Statistical analyses were performed in RStudio using packages corrplot (0.90), forestplot (2.0.1), ggpubr (0.4.0), gmodels (2.18.1), spatstat (2.1–0), survival (3.2–7), survminer (0.4.9), and tidyverse (1.3.1).
We used the Wilcoxon rank-sum test for dichotomous variables and the Kruskal–Wallis test for variables with three or more categories to evaluate the associations of continuous immune cell density variables with patient characteristics. The associations of categorical immune cell density variables with patient characteristics were tested with crosstabulation and the Chi-square test to evaluate the statistical significance. We examined the correlations between immune cell densities by calculating Spearman’s correlation coefficients.
As our main analysis, we used univariable and multivariable Cox proportion hazard regression models to measure hazard ratio (HR) point estimates and 95% confidence intervals (CIs) for cancer-specific and overall survival. Cancer-specific survival was considered as the primary survival endpoint, and it was defined as the time from surgery to colorectal cancer death or the end of follow-up. Overall survival was defined as the duration from surgery to death of any cause or the end of follow-up. The total number of deaths was 525 (58%) including 250 (31%) cancer-specific deaths. The median follow-up time for censored cases was 10.1 years (IQR 6.6–13.1). We limited the follow-up to 10 years, considering that most colorectal cancer deaths occur within that period. Schoenfeld residual plots supported the proportionality of hazards during most of the follow-up period up to 10 years. Multivariable models included the following pre-determined indicator variables (with the reference category listed first): sex (male, female), age (<65, 65–75, >75), year of operation (2000–2005, 2006–2010, 2011–2015), tumour location (proximal colon, distal colon, rectum), American Joint Committee on Cancer (AJCC) stage (I–II, III, IV), tumour grade (well/moderately differentiated, poorly differentiated), lymphovascular invasion (negative, positive), MMR status (proficient, deficient), BRAF status (wild-type, mutant). Kaplan–Meier method was used to visualise the estimates of cancer-specific survival, and the statistical significance was tested with the Log-rank test. A P value less than 0.005 was considered statistically significant, in accordance with the recommendation of an expert panel [19].
Results
Image analysis
We analysed 3190 tissue microarray cores from 910 colorectal cancer patients (mean 3.5 per patient, SD 0.69; tumour centre: mean 1.8, SD 0.39; invasive margin: mean 1.7, SD 0.51). The supervised machine learning algorithms yielded data for 21,503,560 cells including 9,136,506 tumour cells, 1,788,538 macrophages, and 1,582,095 T cells. Macrophages were further phenotyped into PD-L1+ and PD-L1− and M1- and M2-like subpopulations. T cells were phenotyped into PD-1+ and PD-1− subpopulations. The median immune cell densities in the tumour centre and the invasive margin were 512 and 808 cells/mm2 for all macrophages, 33 and 64 cells/mm2 for PD-L1+ macrophages and 390 and 591 cells/mm2 for T cells, respectively. Core-to-core correlations for immune cell densities were good or moderate in both the tumour centre and the invasive margin (Supplementary Fig. S5).
PD-L1 expression patterns
PD-L1 expression was detected mainly in macrophages. Of macrophages, 20% were positive and 80% were negative for PD-L1. In macrophage subsets defined by four polarisation markers, PD-L1 expression was enriched in M1-like macrophages. Of PD-L1+ macrophages, 33% were M1-like, 16% were M2-like, and the remaining 51% were less strongly polarised mixed phenotype macrophages. Correspondingly, PD-L1+ macrophage density and M1-like macrophage density showed a moderate positive correlation (R = 0.54), while the correlation between the densities of PD-L1+ macrophages and M2-like macrophages was weak (R = 0.24) (Supplementary Fig. S6). Conversely, PD-L1− macrophage densities showed a stronger correlation with M2-like macrophage densities (R = 0.67) than M1-like macrophage densities (R = 0.29). Median densities for both PD-L1+ and PD-L1− macrophages were higher in tumour stromal (110/mm2 for PD-L1+ and 991/mm2 for PD-L1−) than intraepithelial regions (11.5/mm2 for PD-L1+ and 119/mm2 for PD-L1−).
The clinicopathological characteristics according to PD-L1+ and PD-L1− macrophage densities are shown in Table 1. Higher density of PD-L1+ macrophages associated with proximal tumour location (P = 0.0012), low stage, poor tumour differentiation, absent lymphovascular invasion, MMR deficiency, and BRAF mutation (all P < 0.0001). Higher density of PD-L1− macrophages associated with high stage (P = 0.0049). The associations of macrophage polarisation with clinicopathological characteristics are visualised in Supplementary Fig. S7.
Table 1.
Demographic and clinical characteristics of colorectal cancer cases according to PD-L1+ and PD-L1– macrophage densities.
| Characteristic | Total N | Overall cell density (cells/mm2) Median (25–75th percentiles) |
|||
|---|---|---|---|---|---|
| PD-L1+ macrophages | P | PD-L1− macrophages | P | ||
| All cases | 910 (100%) | 59 (16–170) | 560 (380–780) | ||
| Sex | 0.50 | 0.47 | |||
| Male | 464 (51%) | 60 (17–160) | 570 (390–790) | ||
| Female | 446 (49%) | 59 (15–190) | 560 (370–770) | ||
| Age (years) | 0.18 | 0.95 | |||
| <65 | 247 (27%) | 54 (15–160) | 580 (360–790) | ||
| 65–75 | 331 (36%) | 58 (15–150) | 560 (390–750) | ||
| >75 | 332 (36%) | 74 (18–230) | 550 (370–800) | ||
| Tumour location | 0.0012 | 0.19 | |||
| Proximal colon | 445 (49%) | 74 (20–220) | 580 (420–790) | ||
| Distal colon | 332 (36%) | 51 (12–140) | 530 (360–750) | ||
| Rectum | 133 (15%) | 38 (15–160) | 560 (360–750) | ||
| AJCC stage | <0.0001 | 0.0049 | |||
| I | 151 (17%) | 84 (19–210) | 500 (340–670) | ||
| II | 342 (38%) | 84 (26–210) | 540 (390–790) | ||
| III | 301 (33%) | 47 (11–130) | 590 (420–800) | ||
| IV | 116 (13%) | 26 (6.6–100) | 650 (370–890) | ||
| Tumour grade | <0.0001 | 0.73 | |||
| Low-grade (well to moderately differentiated) | 760 (84%) | 55 (15–160) | 560 (370–780) | ||
| High-grade (poorly differentiated) | 150 (16%) | 104 (24–310) | 570 (410–760) | ||
| Lymphovascular invasion | <0.0001 | 0.20 | |||
| No | 719 (79%) | 70 (21–190) | 550 (380–770) | ||
| Yes | 191 (21%) | 32 (6.5–100) | 600 (380–820) | ||
| MMR status | <0.0001 | 0.20 | |||
| MMR proficient | 772 (85%) | 48 (12–150) | 550 (370–780) | ||
| MMR deficient | 137 (15%) | 190 (60–350) | 590 (460–770) | ||
| BRAF status | <0.0001 | 0.046 | |||
| Wild-type | 763 (84%) | 52 (14–160) | 550 (370–770) | ||
| Mutant | 147 (16%) | 98 (37–290) | 590 (440–790) | ||
AJCC American Joint Committee on Cancer, MMR mismatch repair.
In addition to macrophages, we investigated the PD-L1 expression in tumour cells using the histoscore method. Most of the tumours were negative for PD-L1 or showed very weak expression (histoscore < 5), and only 66 (7%) of the tumours were classified as PD-L1 positive (histoscore ≥ 5). Similar to high PD-L1+ macrophage density, PD-L1 positivity in tumour cells associated with proximal tumour location, poor differentiation, MMR deficiency and BRAF mutation (all P < 0.0001), but there were no associations with stage or lymphovascular invasion (Supplementary Table S2).
PD-1 expression patterns
We evaluated PD-1 expression in T cells. Of all detected T cells, 22% were positive for PD-1. Higher T cell density associated with low stage [P = 0.00025 (tumour centre), P < 0.0001 (invasive margin)], high tumour grade (P < 0.0001, P = 0.0032), MMR deficiency (both P < 0.0001) and BRAF mutation (P < 0.0001, P = 0.062). The findings were mainly similar for PD-1+ and PD-1− T cells (Supplementary Fig. S8). The density of PD-L1+ macrophages positively correlated with the densities of both PD-1+ T cells (R = 0.64) and PD-1− T cells (R = 0.50) (Supplementary Fig. S6).
Survival analyses
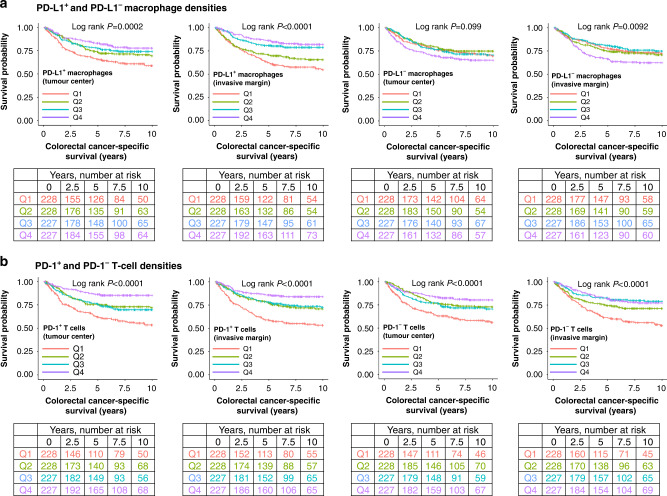
We first examined the prognostic role of total (CD68+/CD163+) macrophage population, which did not reach statistical significance either in the tumour centre or the invasive margin in univariable (Supplementary Fig. S9, Table 2) or multivariable analyses (Table 2) (all P > 0.005). When macrophages were classified according to their PD-L1 expression, higher density of PD-L1+ macrophages associated with improved cancer-specific survival in both the tumour centre and the invasive margin in univariable analyses (Fig. 2a, Table 2). The prognostic value remained significant in the invasive margin in multivariable analysis (Ptrend = 0.0004, HR for Q4 vs. Q1 0.52, 95% CI 0.34–0.78) (Table 2). The full multivariable Cox regression models for PD-L1+ and PD-L1− macrophage densities are shown in Supplementary Table S3. When tumour epithelial and stromal compartments were examined separately, higher PD-L1+ macrophage density associated with longer cancer-specific survival in both tumour intraepithelial and stromal compartments of the tumour centre and the invasive margin in univariable analyses (all Ptrend < 0.005) (Supplementary Fig. S10, Supplementary Table S4). These associations remained significant in the invasive margin in multivariable analyses (Supplementary Table S4). Higher density of PD-L1− macrophages in the invasive margin tended to associate with poor survival but did not reach statistical significance in either univariable or multivariable analysis (Ptrend > 0.005) (Table 2). PD-L1 expression in tumour cells did not significantly associate with survival (Supplementary Table S5). We further evaluated the prognostic significance of PD-L1+ and PD-L1− macrophage densities in relation to MMR status and stage. The survival associations of PD-L1+ or PD-L1− macrophage densities did not significantly differ by MMR status (Supplementary Table S6) or stage (Supplementary Tables S7, S8) in univariable or multivariable analysis (all Pinteraction > 0.005).
Table 2.
Univariable and multivariable Cox regression models for colorectal cancer-specific and overall survival according to the total, PD-L1+, and PD-L1− macrophage densities in the tumour centre and the invasive margin.
| No. of cases | Colorectal cancer-specific survival | Overall survival | |||||
|---|---|---|---|---|---|---|---|
| No. of events | Univariable HR (95% CI) |
Multivariable HR (95% CI) |
No. of events | Univariable HR (95% CI) |
Multivariable HR (95% CI) |
||
| Tumour centre | |||||||
| Macrophage density | |||||||
| Q1 | 228 | 64 | 1 (referent) | 1 (referent) | 116 | 1 (referent) | 1 (referent) |
| Q2 | 228 | 56 | 0.85 (0.59–1.21) | 0.81 (0.56–1.18) | 109 | 0.93 (0.71–1.21) | 0.92 (0.70–1.20) |
| Q3 | 227 | 63 | 0.97 (0.69–1.38) | 0.84 (0.59–1.20) | 107 | 0.93 (0.72–1.21) | 0.84 (0.64–1.10) |
| Q4 | 227 | 59 | 0.93 (0.65–1.32) | 0.80 (0.56–1.16) | 117 | 1.03 (0.80–1.33) | 0.84 (0.65–1.10) |
| Ptrend | 0.87 | 0.29 | 0.81 | 0.17 | |||
| PD-L1+ macrophage density | |||||||
| Q1 | 228 | 83 | 1 (referent) | 1 (referent) | 134 | 1 (referent) | 1 (referent) |
| Q2 | 228 | 61 | 0.68 (0.49–0.94) | 0.81 (0.57–1.13) | 106 | 0.72 (0.56–0.93) | 0.82 (0.64–1.07) |
| Q3 | 227 | 54 | 0.59 (0.42–0.83) | 0.75 (0.53–1.06) | 102 | 0.68 (0.53–0.88) | 0.73 (0.56–0.95) |
| Q4 | 227 | 44 | 0.47 (0.33–0.69) | 0.63 (0.43–0.93) | 107 | 0.70 (0.54–0.90) | 0.72 (0.55–0.94) |
| Ptrend | <0.0001 | 0.015 | 0.0050 | 0.011 | |||
| PD-L1– macrophage density | |||||||
| Q1 | 228 | 59 | 1 (referent) | 1 (referent) | 110 | 1 (referent) | 1 (referent) |
| Q2 | 228 | 52 | 0.86 (0.59–1.25) | 0.75 (0.51–1.09) | 111 | 1.01 (0.77–1.31) | 0.93 (0.71–1.22) |
| Q3 | 227 | 58 | 0.97 (0.68–1.39) | 0.88 (0.61–1.27) | 108 | 0.99 (0.73–1.29) | 0.93 (0.71–1.21) |
| Q4 | 227 | 73 | 1.31 (0.93–1.85) | 0.85 (0.60–1.21) | 120 | 1.18 (0.91–1.53) | 0.88 (0.67–1.14) |
| Ptrend | 0.085 | 0.62 | 0.25 | 0.35 | |||
| Invasive margin | |||||||
| Macrophage density | |||||||
| Q1 | 228 | 67 | 1 (referent) | 1 (referent) | 113 | 1 (referent) | 1 (referent) |
| Q2 | 228 | 62 | 0.94 (0.66–1.32) | 1.16 (0.81–1.66) | 118 | 1.07 (0.83–1.39) | 1.19 (0.91–1.56) |
| Q3 | 227 | 47 | 0.67 (0.46–0.97) | 0.92 (0.62–1.36) | 100 | 0.84 (0.64–1.09) | 0.98 (0.74–1.28) |
| Q4 | 227 | 66 | 0.99 (0.70–1.39) | 0.93 (0.66–1.32) | 118 | 1.05 (0.81–1.36) | 0.98 (0.76–1.28) |
| Ptrend | 0.54 | 0.46 | 0.83 | 0.57 | |||
| PD-L1+ macrophage density | |||||||
| Q1 | 228 | 92 | 1 (referent) | 1 (referent) | 136 | 1 (referent) | 1 (referent) |
| Q2 | 228 | 71 | 0.75 (0.55–1.02) | 1.07 (0.78–1.48) | 116 | 0.82 (0.64–1.06) | 1.02 (0.79–1.32) |
| Q3 | 227 | 43 | 0.43 (0.30–0.61) | 0.68 (0.47–0.99) | 97 | 0.64 (0.50–0.83) | 0.85 (0.65–1.12) |
| Q4 | 227 | 36 | 0.34 (0.23–0.50) | 0.52 (0.34–0.78) | 100 | 0.62 (0.48–0.80) | 0.69 (0.52–0.91) |
| Ptrend | <0.0001 | 0.0004 | <0.0001 | 0.0053 | |||
| PD-L1– macrophage density | |||||||
| Q1 | 228 | 55 | 1 (referent) | 1 (referent) | 113 | 1 (referent) | 1 (referent) |
| Q2 | 228 | 58 | 1.08 (0.74–1.56) | 1.33 (0.91–1.94) | 108 | 0.98 (0.75–1.28) | 1.08 (0.83–1.42) |
| Q3 | 227 | 50 | 0.88 (0.60–1.29) | 1.02 (0.69–1.50) | 101 | 0.86 (0.66–1.13) | 0.95 (0.72–1.25) |
| Q4 | 227 | 79 | 1.53 (1.08–2.16) | 1.36 (0.96–1.92) | 127 | 1.21 (0.94–1.56) | 1.13 (0.88–1.46) |
| Ptrend | 0.038 | 0.21 | 0.27 | 0.52 | |||
The densities were divided into ordinal quartile categories from low (Q1) to high (Q4).
Multivariable Cox proportional hazards regression models were adjusted for sex (male, female), age (<65, 65–75, >75), year of operation (2000–2005, 2006–2010, 2011–2015), tumour location (proximal colon, distal colon, rectum), stage (I–II, III, IV), tumour grade (well/moderately differentiated, poorly differentiated), lymphovascular invasion (negative, positive), MMR status (proficient, deficient), and BRAF status (wild-type, mutant).
Ptrend values were calculated by using the four categories of immune cell densities as continuous variables in univariable and multivariable Cox proportional hazard regression models.
CI confidence interval, HR hazard ratio.
Fig. 2. Kaplan–Meier survival analysis.
Cancer-specific survival curves for the PD-L1+ and PD-L1− macrophage (a) and PD-1+ and PD-1− T cell (b) densities in the tumour centre and the invasive margin. The densities were divided into ordinal quartiles from low (Q1) to high (Q4). Statistical significance was determined with Log-rank test.
Higher T cell density associated with better cancer-specific survival regardless of PD-1 expression in both univariable (Fig. 2b, Table 3) and multivariable analyses (Table 3). In multivariable analyses, the HR for high PD-1+ T cell density in the tumour centre (Q4 vs. Q1) was 0.46 (95% CI: 0.30–0.71), while the HR for high PD-1− T cell density (Q4 vs. Q1) was 0.48 (95% CI: 0.32–0.70) and the HR for high T cell density (Q4 vs. Q1) was 0.49 (95% CI: 0.33–0.73). Full multivariable Cox regression models with all variables are shown in Supplementary Table S9.
Table 3.
Univariable and multivariable Cox regression models for cancer-specific and overall survival according to the densities of total, PD-1+, and PD-1– T cells in the tumour centre and the invasive margin.
| No. of cases | Colorectal cancer-specific survival | Overall survival | |||||
|---|---|---|---|---|---|---|---|
| No. of events | Univariable HR (95% CI) |
Multivariable HR (95% CI) |
No. of events | Univariable HR (95% CI) |
Multivariable HR (95% CI) |
||
| Tumour centre | |||||||
| T cell density | |||||||
| Q1 | 228 | 88 | 1 (referent) | 1 (referent) | 146 | 1 (referent) | 1 (referent) |
| Q2 | 228 | 58 | 0.55 (0.40–0.77) | 0.72 (0.51–1.01) | 103 | 0.58 (0.45–0.74) | 0.68 (0.53–0.88) |
| Q3 | 227 | 56 | 0.54 (0.39–0.76) | 0.67 (0.47–0.94) | 100 | 0.57 (0.44–0.74) | 0.62 (0.48–0.80) |
| Q4 | 227 | 40 | 0.38 (0.26–0.55) | 0.49 (0.33–0.73) | 100 | 0.56 (0.43–0.72) | 0.59 (0.45–0.77) |
| Ptrend | <0.0001 | 0.0003 | <0.0001 | <0.0001 | |||
| PD-1+ T cell density | |||||||
| Q1 | 228 | 91 | 1 (referent) | 1 (referent) | 147 | 1 (referent) | 1 (referent) |
| Q2 | 228 | 57 | 0.55 (0.39–0.76) | 0.82 (0.58–1.16) | 103 | 0.60 (0.47–0.77) | 0.78 (0.60–1.01) |
| Q3 | 227 | 63 | 0.59 (0.43–0.82) | 0.75 (0.54–1.04) | 109 | 0.63 (0.49–0.81) | 0.68 (0.53–0.87) |
| Q4 | 227 | 31 | 0.28 (0.18–0.42) | 0.46 (0.30–0.71) | 90 | 0.49 (0.38–0.64) | 0.57 (0.43–0.76) |
| Ptrend | <0.0001 | 0.0006 | <0.0001 | <0.0001 | |||
| PD-1– T cell density | |||||||
| Q1 | 228 | 86 | 1 (referent) | 1 (referent) | 145 | 1 (referent) | 1 (referent) |
| Q2 | 228 | 55 | 0.54 (0.38–0.76) | 0.68 (0.48–0.96) | 103 | 0.58 (0.45–0.75) | 0.69 (0.54–0.90) |
| Q3 | 227 | 61 | 0.61 (0.44–0.85) | 0.67 (0.48–0.94) | 106 | 0.63 (0.49–0.81) | 0.61 (0.47–0.79) |
| Q4 | 227 | 40 | 0.39 (0.27–0.57) | 0.48 (0.32–0.70) | 95 | 0.54 (0.42–0.70) | 0.55 (0.42–0.71) |
| Ptrend | <0.0001 | 0.0002 | <0.0001 | <0.0001 | |||
| Invasive margin | |||||||
| T cell density | |||||||
| Q1 | 228 | 94 | 1 (referent) | 1 (referent) | 146 | 1 (referent) | 1 (referent) |
| Q2 | 228 | 67 | 0.66 (0.48–0.90) | 0.83 (0.60–1.14) | 103 | 0.71 (0.56–0.91) | 0.80 (0.62–1.03) |
| Q3 | 227 | 38 | 0.34 (0.23–0.49) | 0.53 (0.36–0.78) | 100 | 0.51 (0.39–0.66) | 0.65 (0.50–0.86) |
| Q4 | 227 | 43 | 0.39 (0.27–0.56) | 0.58 (0.40–0.85) | 100 | 0.58 (0.45–0.75) | 0.64 (0.48–0.83) |
| Ptrend | <0.0001 | 0.0005 | <0.0001 | 0.0003 | |||
| PD-1+ T cell density | |||||||
| Q1 | 228 | 97 | 1 (referent) | 1 (referent) | 141 | 1 (referent) | 1 (referent) |
| Q2 | 228 | 57 | 0.52 (0.38–0.72) | 0.81 (0.58–1.14) | 112 | 0.71 (0.55–0.90) | 0.90 (0.70–1.17) |
| Q3 | 227 | 55 | 0.49 (0.35–0.68) | 0.83 (0.58–1.17) | 101 | 0.61 (0.47–0.79) | 0.82 (0.63–1.08) |
| Q4 | 227 | 33 | 0.29 (0.19–0.43) | 0.52 (0.34–0.79) | 95 | 0.56 (0.43–0.72) | 0.70 (0.53–0.93) |
| Ptrend | <0.0001 | 0.0043 | <0.0001 | 0.011 | |||
| PD-1– T cell density | |||||||
| Q1 | 228 | 94 | 1 (referent) | 1 (referent) | 144 | 1 (referent) | 1 (referent) |
| Q2 | 228 | 60 | 0.60 (0.43–0.82) | 0.80 (0.58–1.12) | 105 | 0.66 (0.51–0.85) | 0.77 (0.59–0.99) |
| Q3 | 227 | 43 | 0.40 (0.28–0.58) | 0.62 (0.43–0.90) | 101 | 0.60 (0.46–0.77) | 0.75 (0.57–0.97) |
| Q4 | 227 | 45 | 0.42 (0.29–0.60) | 0.61 (0.42–0.88) | 99 | 0.58 (0.45–0.76) | 0.65 (0.50–0.85) |
| Ptrend | <0.0001 | 0.0027 | <0.0001 | 0.0021 | |||
The densities were divided into ordinal quartile categories from low (Q1) to high (Q4).
Multivariable Cox proportional hazards regression models were adjusted for sex (male, female), age (<65, 65–75, >75), year of operation (2000–2005, 2006–2010, 2011–2015), tumour location (proximal colon, distal colon, rectum), stage (I–II, III, IV), tumour grade (well/moderately differentiated, poorly differentiated), lymphovascular invasion (negative, positive), MMR status (proficient, deficient), and BRAF status (wild-type, mutant).
Ptrend values were calculated by using the four categories of immune cell densities as continuous variables in univariable and multivariable Cox proportional hazard regression models.
CI confidence interval, HR hazard ratio.
Considering potential interactions between PD-1 and PD-L1, we next evaluated the prognostic value of PD-1+ T cell density in tumour groups defined by PD-L1+ macrophage densities or tumour cell PD-L1 expression. These analyses indicated that the survival associations of PD-1+ T cells did not statistically significantly differ according to the density of PD-L1+ macrophages or PD-L1 expression in tumour cells in univariable (Supplementary Fig. S11, Supplementary Tables S10, S11) or multivariable analyses (Supplementary Tables S10, S11) (all Pinteraction > 0.005).
In secondary analyses, we investigated the prognostic significance of M1-like and M2-like macrophage densities. Higher M1-like macrophage density associated with longer cancer-specific survival in both the tumour centre and the invasive margin in univariable analyses (Supplementary Fig. S9, Supplementary Table S12). The prognostic value remained significant in the tumour centre in multivariable analyses [Ptrend = 0.0004, HR for high (Q4 vs. Q1) 0.54, 95% CI: 0.38–0.78] (Supplementary Table S12). Higher M2-like macrophage density in the tumour centre associated with longer cancer-specific survival and higher M1:M2-like macrophage density ratio in the tumour centre and the invasive margin associated with worse cancer-specific survival in univariable analyses (Supplementary Fig. S9, Supplementary Table S12) but did not reach statistical significance in multivariable analysis (Ptrend > 0.005). To evaluate the prognostic value of PD-L1 expression in differently polarised macrophage subpopulations, we calculated macrophage densities based on subpopulations defined by both PD-L1 expression and polarisation state (Supplementary Table S13). We found that higher PD-L1+ M1-like macrophage density in both the tumour centre and the invasive margin predicted longer cancer-specific survival in univariable and multivariable models (multivariable Ptrend = 0.0005 and Ptrend = 0.0008, respectively), while the densities of PD-L1− M1-like macrophages, PD-L1+ M2-like macrophages, or PD-L1− M2-like macrophages did not significantly associate with the survival in multivariable models (all Ptrend > 0.005). These results indicate that higher densities of PD-L1+ and M1-like macrophages associate with favourable prognosis, which is further highlighted when PD-L1 expression and polarisation are examined simultaneously.
Spatial analyses
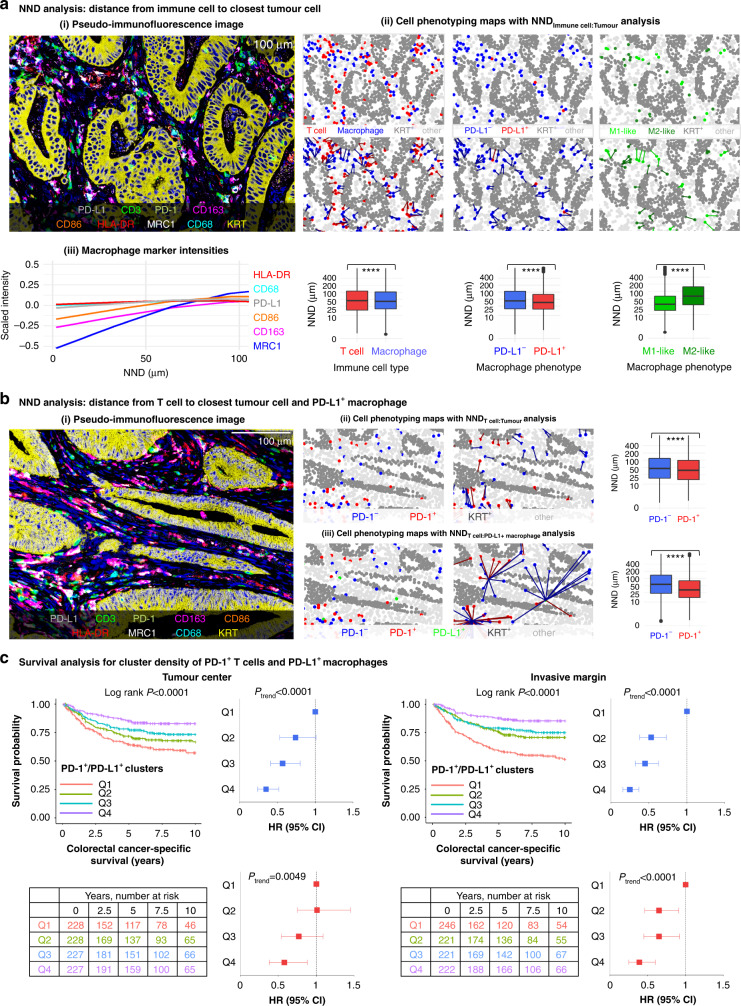
We characterised the spatial arrangement of immune cells by measuring the average distances from immune cells to the nearest tumour cell with NND analysis. Macrophages were located 4.6% closer to tumour cells than T cells on average. Of macrophages, PD-L1+ macrophages were located 24% closer to tumour cells than PD-L1− macrophages (P < 0.0001) and M1-like macrophages were 48% closer to tumour cells than M2-like macrophages (P < 0.0001) (Fig. 3a). To further visualise these findings, we plotted scaled intensities of macrophage polarisation markers and PD-L1 as a function of NND from tumour cells using generalised additive model smoothing (Fig. 3a). This plot showed that M2-like macrophage markers (MRC1 and CD163) had lower scaled intensities at tumour cell proximity than other macrophage markers. The Kaplan–Meier curves showed that the spatial proximity of macrophages (total, PD-L1+, or PD-L1−) or T cells (total, PD-1+, or PD-1−) with tumour cells, as measured with mean NNDs from immune cell to the closest tumour cell, did not significantly associate with cancer-specific survival (P > 0.005) (Supplementary Fig. S12).
Fig. 3. Spatial analysis of immune cells using the nearest neighbour distance (NND) function and the cancer-specific survival analysis for PD-1/PD-L1 cluster density.
a pseudo-immunofluorescence image from a tumour site showing all nine markers (i). Cell phenotyping maps with nearest neighbour distance analysis from each immune cell to the closest tumour cell and boxplots visualising the distribution of nearest neighbour distances across all tumour images (N = 3,190). The statistical significance was tested with Wilcoxon rank-sum test. ****P < 0.0001 (ii). The expression levels of various phenotypic markers in macrophages according to the distance to the closest tumour cell. The plots are based on 1,582,095 T cells and 1,788,538 macrophages (iii). b pseudo-immunofluorescence image from a tumour site showing all nine markers (i). Cell phenotyping maps and nearest neighbour distance analyses from each PD-1+ and PD-1– T cell to the closest tumour cell (ii) and to the closest PD-L1+ macrophage (iii). Boxplots visualise the distribution of nearest neighbour distances across all tumour images (N = 3,190). The significance was tested with Wilcoxon rank-sum test. ****P < 0.0001. c Kaplan–Meier survival curves and Cox proportion hazards regression models for cancer-specific survival for PD-1+ T cell/PD-L1+ macrophage cluster densities in 910 patients in the tumour centre and the invasive margin. One cluster is composed of one PD-1+ T cell with at least one PD-L1+ macrophage within a 20 µm radius. The cluster densities were divided into ordinal quartiles from low (Q1) to high (Q4). Statistical significance for Kaplan–Meier survival estimates were determined with Log-rank test. Univariable (blue) and multivariable (red) Cox proportional hazards regression models are represented as forest plots with HRs along with their 95% CIs as whiskers. Multivariable Cox proportional hazards regression models were adjusted for sex (male, female), age (<65, 65–75, >75), year of operation (2000–2005, 2006–2010, 2011–2015), tumour location (proximal colon, distal colon, rectum), stage (I–II, III, IV), tumour grade (well/moderately differentiated, poorly differentiated), lymphovascular invasion (negative, positive), MMR status (proficient, deficient), and BRAF status (wild-type, mutant).
For T cells, we measured NNDs to the closest tumour cell and to the closest PD-L1+ macrophage to evaluate the possibility for PD-1−PD-L1 interactions. We found that PD-1+ T cells were located closer to both tumour cells (difference 13%, P < 0.0001) and PD-L1+ macrophages (difference 35%, P < 0.0001) than PD-1− T cells (Fig. 3b). To evaluate the prognostic value of PD-1+ T cells co-localised with PD-L1+ macrophages, we calculated the density of PD-1+/PD-L1+ clusters. The mean cluster densities in the tumour centre and the invasive margin were 34 clusters/mm2 and 47 clusters/mm2, respectively. Higher density of PD-1+/PD-L1+ clusters associated with longer cancer-specific survival in both the tumour centre and the invasive margin in univariable and multivariable analyses (Fig. 3c). The prognostic value was slightly stronger in the invasive margin, in which the multivariable HR for high cluster density (Q4 vs. Q1) was 0.39 (95% CI 0.25–0.60, Ptrend < 0.0001).
Discussion
In the present study, we used multiplex immunohistochemistry combined with digital image analysis and quantitative density and spatial analysis to comprehensively characterise the expression of PD-L1 (CD274) and PD-1 (PDCD1) immune checkpoints in the colorectal cancer microenvironment. This analysis, conducted in a large, population-based cohort of 910 colorectal cancer patients expands the knowledge on immune checkpoint expression patterns, their prognostic value, and associations with T cell and macrophage infiltration and may help to develop cancer therapies.
The expression of PD-L1 and PD-1 is often elevated in cancer and their interaction may repress the cytotoxic activity of T cells and thus promote tumour immune escape [4]. However, the prognostic value of PD-L1 is incompletely established in colorectal cancer and prior studies have reached contradictory conclusions [20]. We found that PD-L1 expression was more frequently present in macrophages than in tumour cells. Although PD-L1 is commonly thought to have immunosuppressive effect, we found that higher density of PD-L1+ macrophages in the invasive margin associated with longer colorectal cancer-specific survival independent of stage, grade, and MMR status. Our finding is concordant with some prior studies which have reported association between high PD-L1 expression in immune cells and better survival [21–23] and adds to these findings by more accurately defining phenotypes of the immune cells expressing PD-L1. In our study, PD-L1 expression in tumour cells did not associate with prognosis. Prior colorectal cancer studies have reported inconsistent deductions of the prognostic value of PD-L1+ expression in tumour cells [22–27]. Divergent results may be due to the small percentage of PD-L1 positive tumours and variability in antibody selection, quantification methods, or setting cut-off value for the positivity.
The detection and phenotyping of macrophages are challenging because of their plasticity and the lack of specific or standardised immunohistochemical markers for various subpopulations [7]. We used multiplex immunohistochemistry instead of conventional single-plex chromogenic staining, enabling us to phenotype cells with multimarker combinations and to analyse spatial relationships between cells. To separate M1-like and M2-like polarisation states, we utilised two markers for both phenotypes, in line with two previous studies [16, 28], and only included the extremes of the polarisation spectrum in our main analyses. We found that higher density of M1-like macrophages in the tumour centre associated with longer cancer-specific survival, which is in line with some previous studies [29–31], while M2-like macrophages tended to associate with shorter survival, but not significantly in multivariable models.
In analyses combining PD-L1 expression and polarisation status of macrophages, PD-L1+ macrophages were more likely M1-polarised than M2-polarised, which could partly explain their comparable spatial arrangement and prognostic value. The associations of macrophage polarisation phenotypes with PD-L1 expression are not well-characterised, but may be affected by the cytokine environment [32]. In particular, IFNG can induce M1-like macrophage polarisation and PD-L1 expression in macrophages [33, 34]. However, other studies have found that certain cytokines, such as IL6 and IL10, increase PD-L1 expression but drive macrophage polarisation towards an M2-phenotype [33, 35, 36]. Our analyses suggested that the prognostic value of macrophages was influenced by both the polarisation phenotype (M1-like vs. M2-like) and PD-L1 expression, with PD-L1+ M1-like phenotype showing the strongest association with a favourable outcome.
Some recent studies have reported the co-localisation of immune cells with tumour cells to be strongly prognostic [16, 37–39]. Interestingly, macrophage subtypes frequently located in closer proximity to tumour cells (PD-L1+ and M1-like macrophages) also showed the strongest associations with favourable clinical outcome. Shorter average distance of M1-like than M2-like macrophages from the closest tumour cell has been reported also in prior studies of gastric [28], lung [29] and pancreatic [16] cancers. Based on these findings, we hypothesise that the greater co-localisation between macrophages and tumour cells could increase the probability of cell contacts, thus allowing enhanced anti-tumoral macrophage function.
Our study addressed the prognostic significance of T cell subsets defined by PD-1 expression. We found that higher densities of both PD-1+ and PD-1− T cells showed strong associations with favourable survival, and their HR point estimates were very close to that of the overall T cell population. This supports the strong prognostic significance of T cells in general [40], and indicates that it appears to be independent of PD-1 expression by T cells. Our findings were in line with some prior studies in colorectal cancer that have reported associations between higher PD-1+ cell densities and longer survival [22, 24, 25].
To evaluate the possibility of PD-1/PD-L1 interactions occurring in the tumour microenvironment, we assessed the correlations between the densities of immune checkpoint expressing (PD-1+ and PD-L1+) cells, as well as their co-localisation. In line with prior reports [21, 22, 24], we found a moderate correlation between PD-L1+ macrophage and T cell densities. Furthermore, this correlation was higher for PD-1+ T cells than for PD-1− T cells. A prior study [25] reported that mismatch repair deficient colorectal tumours with high PD-1 expression associated with worse recurrence-free survival if PD-L1 expression was high, while high expression of PD-1 associated with prolonged survival if PD-L1 expression was low. In our cohort, such findings were not identified, as higher PD-1+ T cell densities associated with favourable outcome regardless of PD-L1 expression in tumour cells or PD-L1+ macrophage densities. Furthermore, higher density of PD-1+ T cell/PD-L1+ macrophage clusters strongly associated with longer cancer-specific survival.
In contrast to our hypotheses, higher densities of PD-L1+ macrophages and PD-1+ T cells individually, as well as higher density of PD-1+ T cell/PD-L1+ macrophage clusters associated with favourable prognostic impact. It has been well known that certain cancer risk factors (such as obesity and Lynch syndrome genetic mutations) may associate with better clinical outcomes among patients with a given cancer type. Those apparent paradoxical findings can be explained by interpersonal heterogeneity of cancer [41]. Our findings may be explained by the strong induction of these immune checkpoints as a compensatory response to generally elevated anti-tumour inflammatory reaction in immunologically hot tumours [22, 38]. Favourable outcome of high immune checkpoint expression could also be related to IFNG, which is secreted mainly by activated infiltrating T cells and NK cells. High IFNG expression is correlated with both PD-L1 and PD-1 expression and associates with better colorectal cancer prognosis [24]. Furthermore, gut microbiota e.g., Fusobacterium nucleatum, might associate with T cell count, PD-1 and PD-L1 expression, and colorectal cancer prognosis [42].
To our knowledge, this is the first study examining the PD-L1/PD-1 expression along with macrophage polarisation using multimarker analysis. However, the findings of this study should be interpreted with some caution. First, the analyses were conducted using tissue microarrays, which may not fully represent the immune infiltration in the whole tumour area [43]. To increase the validity of our results, we analysed multiple cores from each tumour (average: 3.5 cores) and the cores were selected from different sites representing average immune cell infiltrates. Reasonably good core-to-core correlations indicated that the number of analysed tissue microarray cores was adequate. By using tissue microarrays, we could examine 910 tumours cost-efficiently and select only representative tumour areas to be stained and analysed. Second, PD-L1 staining patterns may differ between antibody clones. We decided to use a well-validated clone (E1L3N) that is frequently applied for clinical use in evaluating PD-L1 status in lung cancer [44]. However, the optimal antibody for colorectal cancer is yet to be determined. Third, macrophage phenotyping requires several markers and there are no single consensus markers for M1/M2 polarisation states. We used two polarisation markers for both M1 and M2 subpopulations and determined the polarisation states in line with prior reports [16, 45]. Fourth, the cell detection algorithm of QuPath (image analysis software that was utilised) is not able to segment cell membrane, and we used cytoplasmic staining intensities for markers that are expressed in either cell cytoplasm or cell membrane. Fifth, most of the patients were non-Hispanic white, and we excluded patients with preoperative treatment, which led to underrepresentation of rectal cancers. Therefore, the applicability of the results for patients of different ethnicities and with preoperative treatments needs to be confirmed by independent studies. Furthermore, the patients were operated along a period of 16 years, during which the cancer treatments have developed. To mitigate possible bias related to this, we included year of operation as a covariate in multivariable survival models. Sixth, the information on extramural venous invasion and perineural invasion were not available, although they are strong prognostic indicators in colorectal cancer. Seventh, although multiplex immunohistochemistry analysis enabled detailed immune cell characterisation, the method is laborious and would need further automation and validation to be applied as a biomarker in clinical setting.
This study has important strengths. Our study included a large, comprehensively analysed [9, 22, 46–49] population-based cohort evaluated in accordance with the latest guidelines. All tumours were also screened for MMR status and BRAF mutation status representing two key molecular features of colorectal cancer. Multiplex immunohistochemistry staining together with machine learning-based image analysis enabled accurate phenotyping of each cell with multimarker combination in a single batch, facilitating the consistency and reproducibility of the analysis. Flow cytometry and RNA sequencing methods are other commonly used myeloid cell phenotyping methods but, unlike multiplex immunohistochemistry, they do not provide the spatial information of the cells.
In conclusion, this study shows that higher density of PD-L1 expressing macrophages and their spatial proximity with PD-1 expressing T cells associate with prolonged survival of colorectal cancer patients. Our results highlight the utility of detailed multimarker analysis in understanding the role of PD-L1 and PD-1 expression in cancer immune escape and developing improved immunotherapies.
Supplementary information
Author contributions
Conceptualisation: HE, SAV, SO, JAN, JB, TK, JPV. Data curation: HE, MA, OH, E-VW, TTS, JB, JPM, JPV. Funding acquisition: HE, JPM, JPV. Investigation: HE, MA, SAV, OH, E-VW, TTS, JB, JPM, TK, JPV. Methodology: HE, SAV, SO, JAN, MCL, JPV. Formal analysis: HE, JPV. Resources: JB, JPM, TK, JPV. Supervision: TK, JPV. Visualisation: HE, SAV, MCL, JPV. Writing – original draft: HE, JPV. Writing – review & editing: All authors.
Funding
This study was funded by Cancer Foundation Finland (59-5619 to J.P. Väyrynen) and Emil Aaltonen Foundation (220022K to H. Elomaa). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Open Access funding provided by University of Oulu including Oulu University Hospital.
Data availability
The datasets generated and/or analysed during this study are not publicly available. The sharing of data will require approval from relevant ethics committees and/or biobanks. Further information including the procedures to obtain and access data of Finnish Biobanks are described at https://finbb.fi/en/fingenious-service.
Competing interests
JAN reports grants from NanoString, Akoya Biosciences and Illumina outside the submitted work. TTS is the CEO and co-owner of Healthfund Finland Oy and reports interview honoraria from Boehringer Ingelheim Finland and Amgen Finland. The other authors declare that they have no conflicts of interest.
Ethics approval and consent to participate
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the hospital administration and the ethics board (Dnro13U/2011, 1/2016 and 8/2020) and the National Supervisory Authority for Welfare and Health (Valvira). The need to obtain informed consent from the study patients was waived (Valvira Dnro 3916/06.01.03.01/2016).
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41416-023-02238-6.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Fridman WH, Zitvogel L, Sautès-Fridman C, Kroemer G. The immune contexture in cancer prognosis and treatment. Nat Rev Clin Oncol. 2017;14:717–34. doi: 10.1038/nrclinonc.2017.101. [DOI] [PubMed] [Google Scholar]
- 3.Llosa NJ, Cruise M, Tam A, Wicks EC, Hechenbleikner EM, Taube JM, et al. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov. 2015;5:43–51. doi: 10.1158/2159-8290.CD-14-0863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pauken KE, Torchia JA, Chaudhri A, Sharpe AH, Freeman GJ. Emerging concepts in PD-1 checkpoint biology. Semin Immunol. 2021;52:101480. doi: 10.1016/j.smim.2021.101480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ni X, Sun X, Wang D, Chen Y, Zhang Y, Li W, et al. The clinicopathological and prognostic value of programmed death-ligand 1 in colorectal cancer: a meta-analysis. Clin Transl Oncol. 2019;21:674–86. doi: 10.1007/s12094-018-1970-9. [DOI] [PubMed] [Google Scholar]
- 6.Boutilier AJ, Elsawa SF. Macrophage polarization states in the tumor microenvironment. Int J Mol Sci. 2021;22:6995. doi: 10.3390/ijms22136995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jayasingam SD, Citartan M, Thang TH, Mat Zin AA, Ang KC, Ch’ng ES. Evaluating the polarization of tumor-associated macrophages into M1 and M2 phenotypes in human cancer tissue: technicalities and challenges in routine clinical practice. Front Oncol. 2019;9:1512. doi: 10.3389/fonc.2019.01512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Väyrynen V, Wirta E-V, Seppälä T, Sihvo E, Mecklin J-P, Vasala K, et al. Incidence and management of patients with colorectal cancer and synchronous and metachronous colorectal metastases: a population-based study. BJS Open. 2020;4:685–92. doi: 10.1002/bjs5.50299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elomaa H, Ahtiainen M, Väyrynen SA, Ogino S, Nowak JA, Friman M, et al. Prognostic significance of spatial and density analysis of T lymphocytes in colorectal cancer. Br J Cancer. 2022;127:514–23. doi: 10.1038/s41416-022-01822-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nagtegaal ID, Marijnen CAM, Kranenbarg EK, Mulder-Stapel A, Hermans J, van de Velde CJH, et al. Short-term preoperative radiotherapy interferes with the determination of pathological parameters in rectal cancer. J Pathol. 2002;197:20–7. doi: 10.1002/path.1098. [DOI] [PubMed] [Google Scholar]
- 11.Banik G, Betts CB, Liudahl SM, Sivagnanam S, Kawashima R, Cotechini T, et al. High-dimensional multiplexed immunohistochemical characterization of immune contexture in human cancers. Methods Enzymol. 2020;635:1–20. doi: 10.1016/bs.mie.2019.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bankhead P, Loughrey MB, Fernández JA, Dombrowski Y, McArt DG, Dunne PD, et al. QuPath: open source software for digital pathology image analysis. Sci Rep. 2017;7:16878. doi: 10.1038/s41598-017-17204-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–82. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Väyrynen JP, Lau MC, Haruki K, Väyrynen SA, Dias Costa A, Borowsky J, et al. Prognostic significance of immune cell populations identified by machine learning in colorectal cancer using routine hematoxylin and eosin-stained sections. Clin Cancer Res. 2020;26:4326–38. doi: 10.1158/1078-0432.CCR-20-0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chistiakov DA, Killingsworth MC, Myasoedova VA, Orekhov AN, Bobryshev YV. CD68/macrosialin: not just a histochemical marker. Lab Invest. 2017;97:4–13. doi: 10.1038/labinvest.2016.116. [DOI] [PubMed] [Google Scholar]
- 16.Väyrynen SA, Zhang J, Yuan C, Väyrynen JP, Dias Costa A, Williams H, et al. Composition, spatial characteristics, and prognostic significance of myeloid cell infiltration in pancreatic cancer. Clin Cancer Res. 2021;27:1069–81. doi: 10.1158/1078-0432.CCR-20-3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jensen K, Krusenstjerna-Hafstrøm R, Lohse J, Petersen KH, Derand H. A novel quantitative immunohistochemistry method for precise protein measurements directly in formalin-fixed, paraffin-embedded specimens: analytical performance measuring HER2. Mod Pathol. 2017;30:180–93. doi: 10.1038/modpathol.2016.176. [DOI] [PubMed] [Google Scholar]
- 18.Carstens JL, Correa de Sampaio P, Yang D, Barua S, Wang H, Rao A, et al. Spatial computation of intratumoral T cells correlates with survival of patients with pancreatic cancer. Nat Commun. 2017;8:15095. doi: 10.1038/ncomms15095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benjamin DJ, Berger JO, Johannesson M, Nosek BA, Wagenmakers E-J, Berk R, et al. Redefine statistical significance. Nat Hum Behav. 2018;2:6–10. doi: 10.1038/s41562-017-0189-z. [DOI] [PubMed] [Google Scholar]
- 20.Alexander PG, McMillan DC, Park JH. A meta-analysis of CD274 (PD-L1) assessment and prognosis in colorectal cancer and its role in predicting response to anti-PD-1 therapy. Crit Rev Oncol Hematol. 2021;157:103147. doi: 10.1016/j.critrevonc.2020.103147. [DOI] [PubMed] [Google Scholar]
- 21.Lee KS, Kwak Y, Ahn S, Shin E, Oh H-K, Kim D-W, et al. Prognostic implication of CD274 (PD-L1) protein expression in tumor-infiltrating immune cells for microsatellite unstable and stable colorectal cancer. Cancer Immunol Immunother. 2017;66:927–39. doi: 10.1007/s00262-017-1999-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahtiainen M, Wirta E-V, Kuopio T, Seppälä T, Rantala J, Mecklin J-P, et al. Combined prognostic value of CD274 (PD-L1)/PDCDI (PD-1) expression and immune cell infiltration in colorectal cancer as per mismatch repair status. Mod Pathol. 2019;32:866–83. doi: 10.1038/s41379-019-0219-7. [DOI] [PubMed] [Google Scholar]
- 23.Wyss J, Dislich B, Koelzer VH, Galván JA, Dawson H, Hädrich M, et al. Stromal PD-1/PD-L1 expression predicts outcome in colon cancer patients. Clin Colorectal Cancer. 2019;18:e20–38. doi: 10.1016/j.clcc.2018.09.007. [DOI] [PubMed] [Google Scholar]
- 24.Li Y, Liang L, Dai W, Cai G, Xu Y, Li X, et al. Prognostic impact of programed cell death-1 (PD-1) and PD-ligand 1 (PD-L1) expression in cancer cells and tumor infiltrating lymphocytes in colorectal cancer. Mol Cancer. 2016;15:55. doi: 10.1186/s12943-016-0539-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee LH, Cavalcanti MS, Segal NH, Hechtman JF, Weiser MR, Smith JJ, et al. Patterns and prognostic relevance of PD-1 and PD-L1 expression in colorectal carcinoma. Mod Pathol. 2016;29:1433–42. doi: 10.1038/modpathol.2016.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masugi Y, Nishihara R, Yang J, Mima K, da Silva A, Shi Y, et al. Tumour CD274 (PD-L1) expression and T cells in colorectal cancer. Gut. 2017;66:1463–73. doi: 10.1136/gutjnl-2016-311421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Droeser RA, Hirt C, Viehl CT, Frey DM, Nebiker C, Huber X, et al. Clinical impact of programmed cell death ligand 1 expression in colorectal cancer. Eur J Cancer. 2013;49:2233–42. doi: 10.1016/j.ejca.2013.02.015. [DOI] [PubMed] [Google Scholar]
- 28.Huang Y-K, Wang M, Sun Y, Di Costanzo N, Mitchell C, Achuthan A, et al. Macrophage spatial heterogeneity in gastric cancer defined by multiplex immunohistochemistry. Nat Commun. 2019;10:3928. doi: 10.1038/s41467-019-11788-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng X, Weigert A, Reu S, Guenther S, Mansouri S, Bassaly B, et al. Spatial density and distribution of tumor-associated macrophages predict survival in non-small cell lung carcinoma. Cancer Res. 2020;80:4414–25. doi: 10.1158/0008-5472.CAN-20-0069. [DOI] [PubMed] [Google Scholar]
- 30.Edin S, Wikberg ML, Dahlin AM, Rutegård J, Öberg Å, Oldenborg P-A, et al. The distribution of macrophages with a M1 or M2 phenotype in relation to prognosis and the molecular characteristics of colorectal cancer. PLoS One. 2012;7:e47045. doi: 10.1371/journal.pone.0047045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Väyrynen JP, Haruki K, Väyrynen SA, Lau MC, Dias Costa A, Borowsky J, et al. Prognostic significance of myeloid immune cells and their spatial distribution in the colorectal cancer microenvironment. J Immunother Cancer. 2021;9:e002297. doi: 10.1136/jitc-2020-002297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cai H, Zhang Y, Wang J, Gu J. Defects in macrophage reprogramming in cancer therapy: the negative impact of PD-L1/PD-1. Front Immunol. 2021;12:690869. doi: 10.3389/fimmu.2021.690869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Najafi M, Hashemi Goradel N, Farhood B, Salehi E, Nashtaei MS, Khanlarkhani N, et al. Macrophage polarity in cancer: a review. J Cell Biochem. 2019;120:2756–65. doi: 10.1002/jcb.27646. [DOI] [PubMed] [Google Scholar]
- 34.Duffield AS, Ascierto ML, Anders RA, Taube JM, Meeker AK, Chen S, et al. Th17 immune microenvironment in Epstein-Barr virus-negative Hodgkin lymphoma: implications for immunotherapy. Blood Adv. 2017;1:1324–34. doi: 10.1182/bloodadvances.2017007260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang W, Liu Y, Yan Z, Yang H, Sun W, Yao Y, et al. IL-6 promotes PD-L1 expression in monocytes and macrophages by decreasing protein tyrosine phosphatase receptor type O expression in human hepatocellular carcinoma. J Immunother Cancer. 2020;8:1–14. doi: 10.1136/jitc-2019-000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen S, Crabill GA, Pritchard TS, McMiller TL, Wei P, Pardoll DM, et al. Mechanisms regulating PD-L1 expression on tumor and immune cells. J Immunother Cancer. 2019;7:305. doi: 10.1186/s40425-019-0770-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Väyrynen JP, Haruki K, Lau MC, Väyrynen SA, Ugai T, Akimoto N, et al. Spatial organization and prognostic significance of NK and NKT-like cells via multimarker analysis of the colorectal cancer microenvironment. Cancer Immunol Res. 2022;10:215–27. doi: 10.1158/2326-6066.CIR-21-0772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lazarus J, Maj T, Smith JJ, Perusina Lanfranca M, Rao A, D’Angelica MI, et al. Spatial and phenotypic immune profiling of metastatic colon cancer. JCI Insight. 2018;3:e121932. doi: 10.1172/jci.insight.121932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lazarus J, Oneka MD, Barua S, Maj T, Lanfranca MP, Delrosario L, et al. Mathematical modeling of the metastatic colorectal cancer microenvironment defines the importance of cytotoxic lymphocyte infiltration and presence of PD-L1 on antigen presenting cells. Ann Surg Oncol. 2019;26:2821–30. doi: 10.1245/s10434-019-07508-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alexander PG, McMillan DC, Park JH. The local inflammatory response in colorectal cancer - Type, location or density? A systematic review and meta-analysis. Cancer Treat Rev. 2020;83:101949. doi: 10.1016/j.ctrv.2019.101949. [DOI] [PubMed] [Google Scholar]
- 41.Nishihara R, VanderWeele TJ, Shibuya K, Mittleman MA, Wang M, Field AE, et al. Molecular pathological epidemiology gives clues to paradoxical findings. Eur J Epidemiol. 2015;30:1129–35. doi: 10.1007/s10654-015-0088-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao Y, Bi D, Xie R, Li M, Guo J, Liu H, et al. Fusobacterium nucleatum enhances the efficacy of PD-L1 blockade in colorectal cancer. Signal Transduct Target Ther. 2021;6:398. doi: 10.1038/s41392-021-00795-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Giltnane JM, Rimm DL. Technology insight: identification of biomarkers with tissue microarray technology. Nat Clin Pract Oncol. 2004;1:104–11. doi: 10.1038/ncponc0046. [DOI] [PubMed] [Google Scholar]
- 44.Munari E, Zamboni G, Lunardi G, Marconi M, Brunelli M, Martignoni G, et al. PD-L1 expression in non-small cell lung cancer: evaluation of the diagnostic accuracy of a laboratory-developed test using clone E1L3N in comparison with 22C3 and SP263 assays. Hum Pathol. 2019;90:54–9. doi: 10.1016/j.humpath.2019.05.003. [DOI] [PubMed] [Google Scholar]
- 45.Väyrynen JP, Haruki K, Lau MC, Väyrynen SA, Zhong R, Dias Costa A, et al. The prognostic role of macrophage polarization in the colorectal cancer microenvironment. Cancer Immunol Res. 2021;9:8–19. doi: 10.1158/2326-6066.CIR-20-0527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seppälä TT, Böhm JP, Friman M, Lahtinen L, Väyrynen VMJ, Liipo TKE, et al. Combination of microsatellite instability and BRAF mutation status for subtyping colorectal cancer. Br J Cancer. 2015;112:1966–75. doi: 10.1038/bjc.2015.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Porkka N, Lahtinen L, Ahtiainen M, Böhm JP, Kuopio T, Eldfors S, et al. Epidemiological, clinical and molecular characterization of Lynch-like syndrome: A population-based study. Int J Cancer. 2019;145:87–98. doi: 10.1002/ijc.32085. [DOI] [PubMed] [Google Scholar]
- 48.Wirta E-V, Seppälä T, Friman M, Väyrynen J, Ahtiainen M, Kautiainen H, et al. Immunoscore in mismatch repair-proficient and -deficient colon cancer. J Pathol Clin Res. 2017;3:203–13. doi: 10.1002/cjp2.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kellokumpu I, Kairaluoma M, Mecklin J-P, Kellokumpu H, Väyrynen V, Wirta E-V, et al. Impact of age and comorbidity on multimodal management and survival from colorectal cancer: a population-based study. J Clin Med. 2021;10:1751. doi: 10.3390/jcm10081751. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analysed during this study are not publicly available. The sharing of data will require approval from relevant ethics committees and/or biobanks. Further information including the procedures to obtain and access data of Finnish Biobanks are described at https://finbb.fi/en/fingenious-service.