Summary
Hepatocellular carcinoma (HCC) is a prevalent malignant tumor, with a growing incidence and death rate worldwide. The aims and challenges of treating HCC include targeting the tumor, entering the tumor tissue, inhibiting the spread and growth of tumor cells. M27-39 is a small peptide isolated from the antimicrobial peptide Musca domestica cecropin (MDC), whereas HTPP is a liver-targeting, cell-penetrating peptide obtained from the circumsporozoite protein (CSP) of Plasmodium parasites. In this study, M27-39 was modified by HTPP to form M(27–39)-HTPP, which targeted tumor penetration to treat HCC. Here, we revealed that M(27–39)-HTPP had a good ability to target and penetrate the tumor, effectively limit the proliferation, migration, and invasion, and induce the apoptosis in HCC. Notably, M(27–39)-HTPP demonstrated good biosecurity when administered at therapeutic doses. Accordingly, M(27–39)-HTPP could be used as a new, safe, and efficient therapeutic peptide for HCC.
Subject areas: Oncology, Chemistry, Cancer
Graphical abstract

Highlights
-
•
M(27–39)-HTPP effectively targets hepatocellular carcinoma
-
•
M(27–39)-HTPP potently inhibited the growth of HCC in vivo and in vitro
-
•
M(27–39)-HTPP exerts anti-tumor effects by acting on mitochondria
Oncology; Chemistry; Cancer
Introduction
Hepatocellular carcinoma (HCC) is an important cancer worldwide with over 900,000 new cases in 2020 and an estimated 830,000 deaths, ranking sixth in incidence and third in mortality globally.1 The occurrence of HCC is high in China.2,3 The number of new cases in 2020 accounted for 46% of the global number of new cases and the age of onset tended to be younger. Currently, surgery, chemotherapy, and radiation therapy are well-established therapeutic strategies for HCC.4,5,6 However, most patients are diagnosed in the middle and late stages, and often miss the best treatment opportunity.7,8 HCC has become a major health problem worldwide owing to its late detection, increasing incidence, and high mortality.9,10,11 Therefore, novel, safe, and efficient anti-HCC drugs must be developed and their mechanisms of action must be elucidated.12,13
Biological targeted therapy, which uses targeted molecules as carriers to transport radionuclides, toxins, or chemical drugs as warheads, is gaining attention in cancer research.14,15 The advantages of a targeting strategy include high specificity, high selectivity, reduced medication dosage, improved pharmacological efficacy, and reduced toxicity and side effects, which open up a wide range of possibilities for the treatment of HCC.16,17,18 However, because most targeted drug delivery systems have very large molecular weights, they have low cell permeability and cannot penetrate the tumor to kill tumor cells.19 Cell-penetrating peptides (CPPs) are short, less-toxic peptides that can permeate cellular membranes and effectively deliver bioactive payloads into cells.20,21,22 For the past 20 years, CPPs have been frequently employed as ligands in drug delivery systems to improve drug aggregation deep within tumors as a way to treat cancer.23,24
Heparin sulfate proteoglycan (HSPG) is a complex macromolecule mainly found on the cell surface and in the extracellular matrix of liver cells.25 Glypican3 (GPC3) is a member of the HSPG family. Studies have shown that GPC3 is abundant in HCC tissues, but not in normal liver tissues.26,27 HTPP is a liver-targeting CPPs derived from the circumsporozoite protein (CSP) of Plasmodium parasites.28,29 Previous studies have revealed that HTPP can precisely bind to the HSPG to acquire liver-targeting properties and penetrate the cell membrane.30,31 Therefore, we speculated that it might also be targeted to GPC3, and could penetrate tumors.
Antimicrobial peptides (AMPs) have several remarkable advantages, including small size, tumor penetration, and good biocompatibility.32 Antimicrobial peptides can destroy cell membranes or act on intracellular targets through cell membranes and have a unique mechanism of action, which makes it challenging to produce drug resistance against and do not damage normal cells of higher animals.33 Owing to the unsatisfactory development and clinical outcomes of conventional antibacterial and anti-tumor medications, the aforementioned features of antimicrobial peptides indicate a good future for their application and development in cancer therapy.
In a previous study, the antibacterial peptide Musca domestica cecropin (MDC), which has antibacterial and anti-tumor activities, was screened and obtained from M. domestica larvae.34 When its structure-activity relationship was assessed, the peptide M27-39, with higher anti-HCC activity and smaller molecules, was mined and obtained.35 Therefore, we envisaged the fusion of HTPP and M27-39 to produce a novel peptide that has the targeted penetration effect of HTPP and anti-tumor effect of M27-39.
Whether the newly synthesized peptides retain the respective structures of HTPP and M27-39 and can target and penetrate HCC tissue and exhibit anti-tumor actions remains uncertain. Therefore, the purpose of this study was to develop a cell transmembrane peptide chimera that can target and penetrate HCC in vitro and in vivo to exert anti-tumor effects and analyze the mechanism of this peptide’s anti-HCC action.
Results
Structural analysis and synthesis of M(27–39)-HTPP
The various connection modalities of HTPP and M27-39 were examined using ExPASy tools, and computer-aided targeted antimicrobial peptide chimera creation was applied. By binding HTPP at the C-terminus of M27-39 without adding a linker peptide in the middle, the original structure of HTPP was preserved. HTPP consequently joined the C-terminus of M27-39 (Figure S1A). The molecular weight of M(27–39)-HTPP was comparable to the theoretical molecular weight determined by MALDI-MS, and the purity of M(27–39)-HTPP was 97.46% by RP-HPLC (Figures S1B and S1C). M(27–39)-HTPP was expected to have both the targeted penetration impact of HTPP and the boosting effect of M27-39 on the apoptosis of HepG2 cells based on the structure of M(27–39)-HTPP.
In vitro and in vivo targeted penetration experiments sections
To determine whether M(27–39)-HTPP selectively bound to HepG2 cells, M(27–39)-HTPP and M27-39 were labeled with FITC dye. HepG2 cells were treated with these peptides, and the fluorescence in HepG2 cells was examined. Green (FITC) fluorescence in the M(27–39)-HTPP group was greater than that in the M27-39 group (Figures 1B and 1C). HepG2, L02, MCF7, HCT116, and A549 cells were treated with FITC-M(27–39)-HTPP. The green (FITC) fluorescence in HepG2 cells was greater than that in other cells. These results suggest that M(27–39)-HTPP could target HepG2 cells (Figures 1D and 1E). To confirm that M(27–39)-HTPP had tumor penetration capability, FITC-M(27–39)-HTPP was used to treat HepG2 3D tumor spheroids to assess the penetration capability; M27-39 was used as a control. After a certain time, CLSM z stack scanning revealed that M(27–39)-HTPP in tumor spheres was markedly higher than that of M27-39 (Figure 1F), indicating that M(27–39)-HTPP could accumulate more effectively in HepG2 3D tumor spheres. Owing to the cell-penetrating features of the HTPP peptide, the penetration capability of M(27–39)-HTPP was markedly improved compared with that of M27-39. HepG2 cells were immunofluorescently stained to confirm the specific binding location of M(27–39)-HTPP in HepG2 cells. Confocal laser microscopy revealed a clear yellow color at the coexisting sites of green FITC-M(27–39)-HTPP and red GPC3 (Figure 1G). To rule out nonspecific binding of HTPP/M(27–39)-HTPP to HepG2 cells, siRNA experiments were performed. GPC3 siRNA transfection significantly reduced the expression level of GPC3 in HepG2 cells (Figure 1H), and the fluorescence intensity of siRNA + HTPP/M(27–39)-HTPP group was significantly reduced compared with HTPP/M(27–39)-HTPP group (Figures 1I and 1J). It suggests that M(27–39)-HTPP target HepG2 cells by binding to GPC3 receptor.
Figure 1.
Evaluation of in vitro targeted uptake of M(27–39)-HTPP
(A) Localization of M(27–39)-HTPP in HepG2 cells.
(B) Assessment of M(27–39)-HTPP and M27-39 uptake in HepG2 cells (Microscopes).
(C) Assessment of M(27–39)-HTPP and M27-39 uptake in HepG2 cells (Flow cytometry).
(D) Assessment of M(27–39)-HTPP uptake HepG2, L02, MCF7, HCT116 and A549 cells (Microscopes).
(E) Assessment of M(27–39)-HTPP uptake HepG2, L02, MCF7, HCT116 and A549 cells (Flow cytometry).
(F) Comparison of penetration of M(27–39)-HTPP and M27-39 into HepG2 tumor spheres.
(G) M(27–39)-HTPP colocalizes with GPC3.
(H) GPC3 expression after transfection with GPC3 siRNA.
(I) HepG2 cells incubated with FITC-HTPP by pre-treating with siRNA interference (Flow cytometry).
(J) HepG2 cells incubated with FITC-M(27–39)-HTPP by pre-treating with siRNA interference (Flow cytometry).∗p< 0.05; ∗∗p< 0.01; ∗∗∗p< 0.001. as compared with the negative control group. #p< 0.05; ##p< 0.01; ###p< 0.001. ▲p< 0.05; ▲▲p< 0.01; ▲▲▲p< 0.001. ★p< 0.05; ★★p< 0.01; ★★★p< 0.001. N.S., not significant. Data are presented as the mean ± SD.
To assess the hepatoma targeting ability of M(27–39)-HTPP in vivo, the VISQUE in vivo Smart-LF imaging system was used to examine the biodistribution of Cy5.5 labeled M27–39 and M(27–39)-HTPP following intravenous administration. For M(27–39)-HTPP, a bright spot was observed in a part of the liver from 3 h post-injection onward, with the fluorescence signal intensity increasing with time, reaching a maximum at 6 h. A fluorescence signal was still observed at 24 h. For the M27-39 group, the fluorescence signal of the liver was always weak and almost disappeared at 24 h (Figure 2A). All the main organs, including the heart, lung, liver, spleen, and kidney, were collected and imaged in vitro 6 h after injection. From the in vitro images, compared with M27–39, M(27–39)-HTPP had a higher accumulation in the hepatoma tissue, and the fluorescence intensity of the hepatoma tissue was markedly higher than that of other tissues (Figures 2B–2D). After the animals were killed, frozen sections of the liver tissue were stained to assess the penetration effectiveness of M(27–39)-HTPP. Similarly M(27–39)-HTPP showed higher accumulation in tumor tissues (Figure 2E), indicating that M(27–39)-HTPP can penetrate tumors. These results suggested that M(27–39)-HTPP has a good capacity to target HepG2 cells in vitro and in vivo and enter the interior of the tumor.
Figure 2.
In vivo targeted penetration of M(27–39)-HTPP
(A) Imaging of small animals in vivo.
(B–D) Comparison of the fluorescence intensity of each organ in M(27–39)-HTPP group, and comparison of the fluorescence intensity of liver between M(27–39)-HTPP and M27-39 group.
(E and F) Localization of M(27–39)-HTPP and M27-39 in mouse liver. ∗p< 0.05; ∗∗p< 0.01; ∗∗∗p< 0.001. as compared with the negative control group. N.S., not significant. Data are presented as the mean ± SD.
In vitro anti-tumor assays
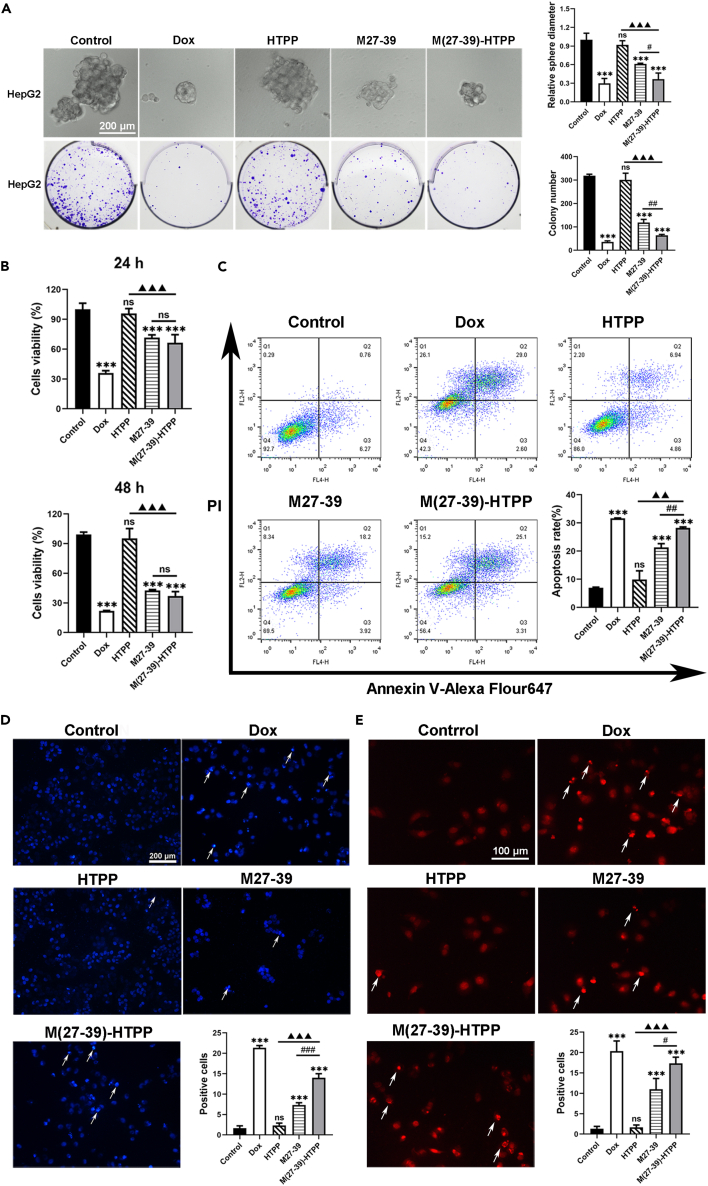
CCK-8, 3D HepG2 tumor spheroid formation, and colony formation assays were performed to explore the effects of M(27–39)-HTPP on tumor growth in vitro. Cell viability was evaluated using CCK-8 assay at different time points after treatment. As shown, M(27–39)-HTPP or M27-39 after treatment caused cell death, with cell viabilities of 66.35% and 71.58%, respectively, 24 h after treatment. After 48 h of treatment, cell viability was 37.01% and 42.50%, respectively, suggesting that M(27–39)-HTPP considerably inhibited the proliferation of HepG2 cells (Figure 3A). In additional, the M(27–39)-HTPP group showed markedly decreased HepG2 tumor spheroid formation and colony formation abilities compared with the control groups (Figure 3B). These findings suggest that M(27–39)-HTPP decreased the proliferation, spheroid formation, and colony formation abilities of HepG2 cells. Several methods were used to determine whether M(27–39)-HTPP decreased HepG2 cell survival by inducing apoptosis. For the apoptosis of HepG2 cells, when treated with M(27–39)-HTPP, the early apoptosis and late apoptosis stages were 3.31% and 25.1%, respectively (Figure 3C). However, when treated with M27-39, the early apoptosis and late apoptosis stages were 3.92% and 18.92%, respectively (Figure 3C). Chromatin was also found to shrink markedly after incubation with M(27–39)-HTPP for 24 h, suggesting cell apoptosis (Figures 3D and 3E). P53, Bax, Caspase-9, Caspase-3, PARP mRNA and protein expression increased of pro-apoptotic factors; however, Bcl-2 mRNA and protein expression decreased (Figures 4A and 4B). The content of protein Cyt-C (mitochondria removal) increased after M(27–39)-HTPP treatment (Figure 4B). Clearly, M(27–39)-HTPP had a better ability to promote the death of HepG2 cells.
Figure 3.
In vitro inhibition of HepG2 cells by M(27–39)-HTPP
(A) Cell viability of the HepG2 cells treated with various drugs.
(B) Inhibition of tumor globule and colony formation in HepG2 by M(27–39)-HTPP.
(C) Apoptosis of the HepG2 cells induced by M(27–39)-HTPP, statistical analysis of apoptosis rate.
(D and E) HepG2 cells were stained for apoptotic cells, using and hoechst33324 and TUNEL assay staining; arrows, representative apoptotic cells with hoechst33324-positive and TUNEL-positive. ∗p< 0.05; ∗∗p< 0.01; ∗∗∗p< 0.001. As compared with the negative control group. #p< 0.05; ##p< 0.01; ###p< 0.001. ▲p< 0.05; ▲▲p< 0.01; ▲▲▲p< 0.001. N.S., not significant. Data are presented as the mean ± SD.
Figure 4.
Effect of M(27–39)-HTPP on the expression of apoptosis-related genes and proteins in HepG2 cells
(A) Relative mRNA expression of P53, Bcl-2, Bax, Caspase-9, Caspase-3 and PARP in HepG2 cells.
(B) Western blot and relative protein expression of P53, Bcl-2, Bax, Cyt-C, Cleaved Caspase-9, Cleaved Caspase-3 and Cleaved PARP in HepG2 cells. ∗p< 0.05; ∗∗p< 0.01; ∗∗∗p< 0.001. As compared with the negative control group. #p< 0.05; ##p< 0.01; ###p< 0.001. ▲p< 0.05; ▲▲p< 0.01; ▲▲▲p< 0.001. N.S., not significant. Data are presented as the mean ± SD.
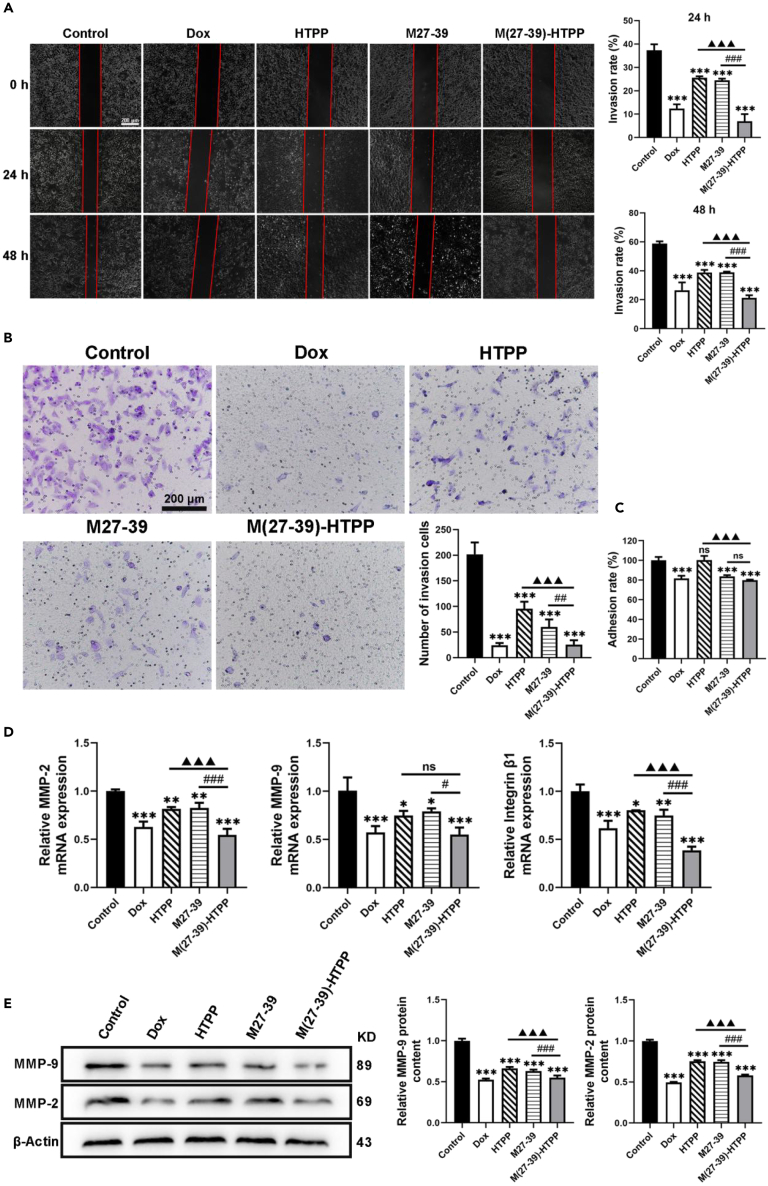
The migration and invasion of HepG2 cells were determined using wound healing and transwell assays, respectively. After 24 and 48 h of incubation, the width of the wound in cells treated with M(27–39)-HTPP and M27-39 was relatively narrow compared with that in the control. Furthermore, compared with the M27-39 group, the M(27–39)-HTPP group had a narrower wound width (Figure 5A). In the transwell assay, after 48 h, treatment with M(27–39)-HTPP and M27-39 in HepG2 cells decreased the invasion number of cells, and the number of HepG2 cells in the M(27–39)-HTPP group was lower (Figure 5B). In the adhesion assay, after 24 h of treatment, M(27–39)-HTPP and M27-39 markedly decreased the adhesion of HepG2 cells (Figure 5C). The mRNA and protein expression levels of in cell migration and adhesion, were detected by RT-PCR and western blot. M(27–39)-HTPP considerably decreased the mRNA expression of MMP-2, MMP-9 and integrin 1, and the protein expression of MMP-2 and MMP-9 (Figures 5D and 5E). M(27–39)-HTPP inhibited the migration, invasion, and adhesion of HepG2 cells. The in vitro anti-tumor results indicated that M(27–39)-HTPP inhibited HepG2 cell growth, promoted apoptosis, and inhibited the migration and invasion of HepG2 cells.
Figure 5.
Effects of M(27–39)-HTPP on migration, invasion and adhesion of HepG2 cells
(A) Wound healing assay measured the migration of the HepG2 cells.
(B) Transwell assay evaluated the invasion of the HepG2 cells.
(C) Adhesion ability of HepG2 cells.
(D) Relative mRNA expression of MMP-2, MMP-9 and Integrin β1 in HepG2 cells.
(E) Western blot and relative protein expression of MMP-2 and MMP-9 in HepG2 cells.∗p< 0.05; ∗∗p< 0.01; ∗∗∗p< 0.001. as compared with the negative control group. #p< 0.05; ##p< 0.01; ###p< 0.001. ▲p< 0.05; ▲▲p< 0.01; ▲▲▲p< 0.001. N.S., not significant. Data are presented as the mean ± SD.
In vivo anti-tumor activity analysis
On the basis of in vitro assays, the in vivo inhibitory effect of M(27–39)-HTPP was investigated in mice with HepG2 hepatoma. As depicted in the image of the livers, large tumors appeared in the control and HTPP groups; however, the livers in the M27-39 group had markedly smaller tumors than those in the control and HTPP groups, and in the M(27–39)-HTPP group tumors even less than the M27-39 group (Figure 6C); this is an excellent illustration of M(27–39)-HTPP, which suppresses cancerous liver tissues. H&E staining of the tumor regions confirmed that the tumor tissue in the M(27–39)-HTPP group was considerably suppressed compared to that in the control group (Figure S2A). Compared with the control or M27-39 groups, immunohistochemical studies revealed increased expression of Caspase-3 and Bax in M(27–39)-HTPP group HCC tumors in situ (Figure S2A), supporting the conclusion that M(27–39)-HTPP slowed tumor development and caused apoptosis. M(27–39)-HTPP also successfully inhibited the proliferation of HCC tumors in situ, according to the proliferating protein Ki67 immunofluorescence results (Figure S2B). RT-PCR and western blotting were performed to investigate the molecular mechanisms involved in M(27–39)-HTPP-induced apoptosis. The results indicated that M(27–39)-HTPP markedly increased the mRNA and protein levels of P53, Bax, Caspase-9, Caspase-3, and PARP and inhibited Bcl-2 expression in the HCC mice (Figures S2C and S2D). Moreover, M(27–39)-HTPP performed better than M27-39. According to the results of an in vivo experiment, M(27–39)-HTPP had a good anti-tumor effect and could effectively inhibit the growth of HCC in mice.
Figure 6.
Anti-tumor effect of M(27–39)-HTPP in vivo
(A and B) Growth status and body weight of nude mice (n = 6).
(C) Hepatoma size in nude mice (n = 6). ∗p< 0.05; ∗∗p< 0.01; ∗∗∗p< 0.001. as compared with the negative control group. #p< 0.05; ##p< 0.01; ###p< 0.001. ▲p< 0.05; ▲▲p< 0.01; ▲▲▲p< 0.001. N.S., not significant. Data are presented as the mean ± SD.
Anti-tumor mechanism assays
To determine whether M(27–39)-HTPP was localized in the mitochondria of HepG2 cells, M(27–39)-HTPP was labeled with FITC dye, mitochondria were labeled with Mito Tracker Deep Red FM, HepG2 cells were treated with M(27–39)-HTPP, and fluorescence in HepG2 cells was examined. An obvious overlap was observed between green and red fluorescence in HepG2 cells after treatment with M(27–39)-HTPP for 2 h (Figure S3A). The ultrastructure of mitochondria following M(27–39)-HTPP treatment was investigated using TEM to confirm whether M(27–39)-HTPP can affect mitochondrial function. The cell membrane of HepG2 cells was incomplete, and many mitochondria in HepG2 cells treated with M(27–39)-HTPP had anamorphic forms (Figure S3B). A swollen mitochondrion may be twice the size of a typical mitochondrion. The cristae shrank, swollen, disintegrated, shattered, and became disordered in each mitochondrion.
Changes in ROS production, mitochondrial membrane potential, NO production, and ATPase activity, all of which are closely associated with apoptosis, were assessed to determine the apoptotic mechanism of M(27–39)-HTPP. To measure alterations in mitochondrial membrane potential, the fluorescent probe, JC-1, which aggregates in the mitochondria to create a polymer (J-aggregates) with red fluorescence signaling and high mitochondrial membrane potential, was used. JC-1 is monomeric with green fluorescence at low mitochondrial membrane potential. The mitochondrial membrane potential of HepG2 cells was markedly reduced by M27-39. M(27–39)-HTPP was more effective than M27-39 at inducing a reduction in the mitochondrial membrane potential (Figures S3C and S3D). The total NO concentration in the control group was markedly lower than that in the M27–39 or M(27–39)-HTPP-treated groups. M(27–39)-HTPP considerably increased NO generation in HepG2 cells (Figure S3F). Inhibition of ATPase activity induces cancer cell death. ATPase activity was assessed in HepG2 cells treated with M(27–39)-HTPP. Compared to the control group, there was a significant decrease in the activity of Na+/K+ ATPase, Ca2+/Mg2+ ATPases, and total ATPases in the M(27–39)-HTPP group (Figure S3G). In addition, M27-39 increased ROS production, and M(27–39)-HTPP exhibited even better effects (Figure S3E). In summary, M(27–39)-HTPP increased ROS production, decreased the mitochondrial membrane potential, increased NO production, and reduced ATPase activity in HepG2 cells, resulting in apoptosis. M27-39 has therapeutic effects; however, it is markedly less effective than M(27–39)-HTPP, which may be because of its low water solubility and poor absorption by HepG2 cells. These experimental findings suggest that M(27–39)-HTPP causes the apoptosis in HepG2 cells by damaging the mitochondrial structure and function.
Biosafety evaluation
CCK8, apoptosis, and ROS in vitro assay results revealed that M(27–39)-HTPP was not obvious toxic to L02, H9C2, and HBZY-1 cells (Figures S4A–S4C). Moreover, there was no increase in alanine transaminase (ALT), glutamic oxaloacetic transaminase (AST), creatinine (CRE), urea nitrogen (BUN), or body weight loss in the M(27–39)-HTPP group mice at the therapeutic dose (Figure S4D). H&E staining of the liver, heart, spleen, lung, and kidney revealed that intravenous injection of M(27–39)-HTPP had no significant influence on the main organs of mice (Figure S4E). According to these findings, M(27–39)-HTPP has low toxicity and is safe in animals and cells.
Discussion
Currently, the treatment of HCC is still associated with great challenges, and the clinical use of anti-HCC drugs leads to remarkable sideeffects.36,37 In addition to conventional methods and agents for HCC treatment, new molecularly targeted therapies are gaining increasing attention.38 Novel molecularly targeted therapies that could specifically target tumor cells with the lowest possible toxicity of toward normal cells and tissues. Therefore, in this study, tumor targeting peptide HTPP was selected to modify tumor therapy peptide M27-39 to synthesize a novel peptide with precise targeting and treatment of HCC. The connection mode is crucial for spatial activity of the fused peptide when two short peptides are fused. Bioinformatics analysis revealed that M(27–39)-HTPP effectively preserved the corresponding structures of M27-39 and HTPP, suggesting that it has the potential for further research.
We evaluated the efficiency of M(27–39)-HTPP targeted uptake of ability. In the cellular uptake test, M(27–39)-HTPP uptake by HepG2 cells was markedly higher than that of M27-39, and HepG2 cells showed a higher uptake of M(27–39)-HTPP than L02, MCF7, HCT116, and A549 cells. Tumor spheroids are useful 3D models for assessing tumor biology owing to their form and biological milieu, which are comparable to those of solid tumors.39,40,41 As a result, a 3D HepG2 tumor spheroid model was created to simulate M(27–39)-HTPP in tumors. When the fluorescence in the cell spheres was evaluated, the M(27–39)-HTPP group was found to have more aggregation in the HepG2 tumor spheres than the M27-39 group and penetrated the cell spheres more effectively. In vivo imaging technology is now widely employed in medical research for tumor diagnosis, tumor targeting, and other purposes.42,43 In both hepatoma model fluorescence imaging and hepatoma histological section fluorescence imaging, M(27–39)-HTPP showed substantial accumulation in the hepatoma tissue. Numerous studies have revealed that GPC3 is upregulated in HCC, suggesting its potential significance as a biomarker for the disease.44,45 M(27–39)-HTPP and GPC3 were co-located in laser confocal detection of HepG2 cells, and the binding of M(27–39)-HTPP to HepG2 cells was also reduced after siRNA transfection, indicating that M(27–39)-HTPP interacts with HepG2 cells by specific targeting of GPC3. These findings indicate that M(27–39)-HTPP can precisely target and penetrate the hepatoma tissues.
Tumor cells can continue to split and develop if the signaling mechanisms associated with apoptosis do not function properly. To effectively treat tumors, tumor cell growth must be stopped and apoptosis should be promoted. In CCK-8, 3D HepG2 tumor spheroid formation, and colony formation assays showed that M(27–39)-HTPP had an inhibitory effect on HepG2 cell growth. Further assays, M(27–39)-HTPP markedly increased the apoptosis of HepG2 cells. Together, immune evasion, invasion, and metastasis make it difficult to treat HCC.46 Cell scratch, adhesion, and invasion experiments were performed to evaluate the effects of M(27–39)-HTPP on the metastasis and invasion of HepG2 cells. Based on these results, M(27–39)-HTPP effectively inhibited metastasis and invasion of HepG2 cells. In a hepatoma model of HepG2 cells, M(27–39)-HTPP effectively suppressed tumor growth.
The effects of M(27–39)-HTPP on apoptosis and the expression of important proteins in linked pathways were explored at the molecular level. Mitochondria, as semi-autonomous organelles, not only provide energy to organisms through oxidative phosphorylation but also engage in the stress-induced intrinsic apoptotic pathway.47 Mitochondria have become a focus in cancer treatment because they release cytochrome c to regulate cell death.48 Bax is a mitochondrial outer-membrane effector protein.49 Bcl-2 is an anti-apoptotic protein found on the outer membrane of mitochondria that acts as an apoptosis suppressor.50 Bcl-2 binds to Bax and prevents it from causing the opening of the mitochondrial voltage-dependent anion channel (VDAC), which prevents the release of cytochrome c from the mitochondria.51,52 Increased Bax levels can rupture the outer mitochondrial membrane, causing the release of cytochrome c. Cytochrome c triggers caspase-9 in the cytoplasm, which causes the formation of apoptotic bodies.53,54 Apoptotic bodies promote the activation of caspase-3, which is the main executioner of apoptotic death, further activating PARP, and sending a DNA break signal to induce cell apoptosis.55 Traditionally, P53, a tumor suppressor protein, has been thought to be a crucial factor in tumor suppression because it promotes apoptosis, cellular senescence, and cell cycle progression.56 M(27–39)-HTPP enhanced apoptosis through P53 overexpression. Through the Bax-induced caspase-mediated pro-apoptotic signaling pathway, M(27–39)-HTPP markedly improves the anti-cancer activity. The expression levels of pro-apoptotic proteins from the Bax pathway, including cytochrome c, Cleaved Caspase-9, Cleaved Caspase-3, and Cleaved PARP, were elevated, whereas the expression of anti-apoptotic Bcl-2 decreased. In addition, M(27–39)-HTPP effectively inhibited the expression of Ki67 in tumor tissues. Therefore, M(27–39)-HTPP effectively inhibited HCC growth in mice so as to achieve the purpose of treating HCC.
Mitochondria are essential for cellular energy metabolism, and tumor cells have a higher energy demand. Therefore, the destruction of mitochondrial structure and function triggers the mitochondrial apoptosis pathway, thereby promoting tumor cell apoptosis.57,58 Thus, M(27–39)-HTPP has a superior therapeutic ability against HepG2 cells owing to mitochondrial swelling and deformation, high ROS generation, damage to the mitochondrial membrane, low ATPase activity, NO generation, and activation of the caspase-related cell apoptosis pathway. According to our findings, M(27–39)-HTPP triggers apoptosis by activating the mitochondrial pathway.
Owing to low cell selectivity or other reasons, the existing anti-tumor drugs will also have great toxicity to normal cells while treating tumors, thus causing toxic side effects in other organs or even the whole body. Therefore, preliminary evaluations of the safety and toxicity of M(27–39)-HTPP were performed. M(27–39)-HTPP has low toxicity and is safe as a potential anti-tumor medication.
In conclusion, we report a targeting transmembrane peptide M(27–39)-HTPP of HCC that exhibits good biosecurity. M(27–39)-HTPP has been demonstrated to target hepatoma, penetrate tumors, inhibit the proliferation of HepG2 cells, and promote the apoptosis of HepG2 cells in vitro and in vivo. These findings suggest that the M(27–39)-HTPP peptide may be a safe and effective therapeutic strategy for HCC.
Limitations of the study
In this study, although M(27–39)-HTPP has good targeting ability for HCC, the specific targeting mechanism has not been thoroughly studied. Future studies are needed to investigate the pharmacokinetics of M(27–39)-HTPP in mice, because it is critical for its in vivo safety and dosing interval. In addition, to further study the anti-HCC effect of M(27–39)-HTPP, a variety of HCC cell lines should be selected for future research.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbi Polyclonal Bax | Boster | Cat#BA0315-2 |
| Rabbit monoclonal Bcl-2 | Abcam | Cat#ab182858; RRID:AB_2715467 |
| Rabbit monoclona Cyt-C | Abcam | Cat#ab133504; RRID:AB_2802115 |
| Rabbit monoclonal P53 | Abcam | Cat#ab32389; RRID:AB_776981 |
| Rabbit monoclonal Caspase-9 | Abcam | Cat#ab202068; RRID:AB_2889070 |
| Rabbit monoclonal Caspase-3 | Abcam | Cat#ab184787; RRID:AB_2827742 |
| Rabbit monoclonal PARP | Abcam | Cat#ab191217; RRID:AB_2861274 |
| Rabbit monoclonal GPC3 | Abcam | Cat#ab66596; RRID:AB_1141042 |
| Mouse monoclonal Ki67 | Boster | Cat#M00254-8 |
| Rabbit monoclonal MMP-9 | Abcam | Cat#ab76003; RRID:AB_1310463 |
| Rabbit monoclonal MMP-2 | Abcam | Cat#ab92536; RRID:AB_10561597 |
| Mouse monoclonal β-actin | Beyotime | Cat#AA128; RRID:AB_2861213 |
| Goat Anti-Rabbit | Beyotime | Cat#A0239; RRID:AB_2893355 |
| Goat Anti-Mouse | Beyotime | Cat#A0216; RRID:AB_2860575 |
| Cy3-Goat Anti-Rabbit | Beyotime | Cat#A0516; RRID:AB_2893015 |
| Cy3-Goat Anti-Mouse | Beyotime | Cat#A0521; RRID:AB_2923334 |
| Chemicals, peptides, and recombinant proteins | ||
| M(27–39)-HTPP | Beijing Scilight Biotechnology Ltd | Ac-VAQQAANVAATLKNSRSLGENDDGNNEDNEKLR-NH2 |
| M27-39 | Beijing Scilight Biotechnology Ltd | Ac-VAQQAANVAATLK-NH2 |
| HTPP | Beijing Scilight Biotechnology Ltd | Ac-CNSRSLGENDDGNNEDNEKLR-NH2 |
| GPC3 siRNA | Guangzhou Agee Biotechnology Co., Ltd. | 5′-GGCUCUGAAUCUUGGAAUUTT-3′ |
| Doxorubicin (DOX) | MP Biomedicals LLC | Cat#92-1101 |
| DMF | Aladdin | Cat#68-12-2 |
| DMSO | Aladdin | Cat#909910-43-6 |
| DifcoTM Skim Milk | BD Difco | Cat#232100 |
| Bovine serum albumin (BSA) | Beyotime | Cat#ST2249 |
| Cy5.5-NHS | Apoptosis and Epigenetics Company | Cat#A8103 |
| Critical commercial assays | ||
| One-step TUNEL kit | Beyotime | Cat#C1089 |
| DCFH-DA | Beyotime | Cat#S0033S |
| JC-1 | Beyotime | Cat#C2003S |
| Annexin V Alexa Fluor 647/PI apoptosis kit | 4A Biotech | Cat#FXP023 |
| DAPI | Beyotime | Cat#C1002 |
| Hoechst 33342 | Beyotime | Cat#C1025 |
| Mito-Tracker Red CMXRos | Beyotime | Cat#C1049B |
| NO assay kit | Beyotime | Cat#S0021S |
| Oligonucleotides | ||
| β-actin-F | Tsingke Biotechnology | GGCTGTATTCCCCTCCATCG |
| β-actin-R | Tsingke Biotechnology | CCAGTTGGTAACAATGCCATGT |
| Bcl-2-F | Tsingke Biotechnology | GTCGCTACCGTCGTGACTTC |
| Bcl-2-R | Tsingke Biotechnology | CAGACATGCACCTACCCAGC |
| Bax-F | Tsingke Biotechnology | TGAAGACAGGGGCCTTTTTG |
| Bax-R | Tsingke Biotechnology | AATTCGCCGGAGACACTCG |
| P53-F | Tsingke Biotechnology | AAGACAGGCAGACTTTTCGC |
| P53-R | Tsingke Biotechnology | CACGAACCTCAAAGCTGTCC |
| caspase-3-F | Tsingke Biotechnology | ATGGAGAACAACAAAACCTCAGT |
| caspase-3-R | Tsingke Biotechnology | TTGCTCCCATGTATGGTCTTTAC |
| caspase-9-F | Tsingke Biotechnology | GACGCTCTGCTGAGTCGAG |
| caspase-9-R | Tsingke Biotechnology | GGTCTAGGGGTTTAACAGCCTC |
| PARP-F | Tsingke Biotechnology | GGCAGCCTGATGTTGAGGT |
| PARP-R | Tsingke Biotechnology | GCGTACTCCGCTAAAAAGTCAC |
| MMP 9-F | Tsingke Biotechnology | GAGACTCTACACCCAGGACG |
| MMP 9-R | Tsingke Biotechnology | GAAAGTGAAGGGGAAGACGC |
| MMP 2-F | Tsingke Biotechnology | GTGTTCTTTGCAGGGAATGAAT |
| MMP 2-R | Tsingke Biotechnology | ACGACGGCATCCAGGTTATC |
| Integrin β1 F | Tsingke Biotechnology | CAAGCAGGGCCAAATTGTGG |
| Integrin β1 R | Tsingke Biotechnology | CCTTTGCTACGGTTGGTTACATT |
| Biological samples | ||
| Fetal Bovine Serum | Bioind | Cat#04-007-1A |
| DMEM medium | Gibco | Cat#11995-065 |
| Experimental models: Cell lines | ||
| HepG2 cells | China Center for Type Culture Collection | N/A |
| Experimental models: Organisms/strains | ||
| Male athymic Balb/c mice | Guangdong Medical Animal Experiment Center | N/A |
| Software and algorithms | ||
| ImageJ | Schindelin et al. | https://imagej.nih.gov/ij/ |
| GraphPad Prism8.0.2 | GraphPad | https://www.graphpad.com/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Xuemei Lu (luxuemei@gdpu.edu.cn).
Materials availability
This study did not generate new unique reagents.
Experimental model and subjectdetails
Cell lines and cell culture
The human HCC cell line HepG2 and L02 were provided by the China Center for Type Culture Collection, Wuhan University (Wuhan, China). HBZY-1, H9C2, A549, HCT116, and MCF7 cells were provided by the Shanghai Cell Bank of the Chinese Academy of Sciences (Shanghai, China). All cells were cultured in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin (GIBCO, New York, NY, USA) at 37°C/5% CO2.
Animals
The use of the animals and experimental protocols were approved by the Guidelines for the Care and Use of Experimental Animals, the Guangdong Pharmaceutical University (SYXK (Yue) 2017-0125), and the Guangdong Pharmaceutical University Animal Care and Use Committee, China). 4–6 weeks old Male athymic Balb/c nude mice (4–6 weeks old) weighing approximately 15–18 g of were purchased from the Guangdong Medical Animal Experiment Center and maintained in an SPF animal housing facility.
Method details
Structural analysis and synthesis of M(27–39)-HTPP
ExPASY tools were used to investigate the structures of M27-39 and HTPP, the ligations of the two peptides were examined and the subsequent synthesis was carried out.
In vitro cellular uptake
HepG2 cells (5 × 105 cells/well) were seeded in 6-well plates and cultured overnight. The cells were then treated with FITC-M(27–39)-HTPP (320 μg/mL) and culture medium containing FITC-M27-39 (120 μg/mL) and cultured for another 30 min HepG2, L02, MCF7, HCT116, and A549 cells (5 × 105 cells/well) were seeded in 6-well plates and cultured overnight. The cells were then treated with culture medium containing FITC-M(27–39)-HTPP and cultured for another 30 min. Finally, fluorescence intensity was detected using an inverted microscope (LEICA, Wetzlar, Germany) and flow cytometry (BD FACSCalibur, New Jersey, USA).
3D HepG2 tumor spheroid penetration
HepG2 cells (1,000 cells/well) were seeded in 96-well clear round bottom ultra low attachment microplate (Corning, New York, USA) and grown at 37 °C for 7 days to attain a diameter of approximately 200 μm. During culture, the medium was changed every three days. FITC-M(27–39)-HTPP (320 μg/mL) and M27-39 (120 μg/mL) were added to cell culture medium and incubated for 1 h. After washing with PBS and fixing with 4% paraformaldehyde, 3D tumor spheroids were scanned from the top to the middle at 10-μm intervals using a confocal microscope (Olympus, Tokyo, Japan).
GPC3 siRNA transfection
GPC3 siRNA sequence 5′-GGCUCUGAAUCUUGGAAUUTT-3′ was transfected to HepG2 cells before they were added to FITC-HTPP/M(27–39)-HTPP and incubated for 30 min. The HepG2 cells were treated with negative control (NC, scrambled sequence). The cells were digested and gathered for flow cytometry examination.
Determination of cell viability
Cell Count kit-8 (CCK8) assay was performed to measure cell proliferation. The liver cancer cell line, HepG2, was seeded into 96-well plates at a density of 5 × 104 cells/well. Cells were cultured in high-glucose DMEM for 24hat 37 °C and 5% CO2. Dox (2 μg/mL), HTPP (200 μg/mL), M27-39 (120 μg/mL), and M(27–39)-HTPP (320 μg/mL) were added to cells and incubated for 24 and 48 h. Thereafter, 10 μL of CCK8 (APExBIO, Houston, Texas, USA) was added to each well. OD values were determined using a microplate reader at a wavelength of 490 nm.
3D HepG2 tumor growth assay
HepG2 cells (1000 cells/well) were seeded in a 96-well clear round bottom ultra low attachment microplate (Corning, New York, USA) and grown at 37 °C with 5% CO2. Dox (2 μg/mL), HTPP (200 μg/mL), M27-39 (120 μg/mL), and M(27–39)-HTPP (320 μg/mL) were added to the cells three days later and incubated for 10 days. An inverted microscope (LEICA, Wetzlar, Germany) was used to obtain images from various wells.
Colony formation assay
HepG2 cells were seeded in 6-well culture plates and grown in equal quantities. Dox (2 μg/mL), HTPP (200 μg/mL), M27-39 (120 μg/mL), and M(27–39)-HTPP (320 μg/mL) were added to the cells three days later and incubated for 15 days. The solution was changed every three days during incubation. Cells were fixed in 4% paraformaldehyde and stained with 0.1% crystal violet at 37 °C for 15 min. Images were captured and saved for subsequent analysis.
Apoptosis of HepG2 cells
HepG2 cells were seeded in 6-well culture plates at similar numbers and allowed to develop. The cells were treated with Dox (2 μg/mL), HTPP (200 μg/mL), M27-39 (120 μg/mL), and M(27–39)-HTPP (320 μg/mL). Hoechst 33342 (Beyotime, Shanghai, China) staining was used to assess nuclear changes in HepG2 cells. Images from numerous wells were obtained using an inverted microscope (LEICA, Wetzlar, Germany). To confirm whether M(27–39)-HTPP could induce apoptosis, HepG2 cells were cultured in a medium containing M(27–39)-HTPP for 24 h. The cells were then stained using the Annexin V Alexa Fluor647/PI apoptosis kit (4A Biotech, Beijing, China) and analyzed by flow cytometry (BD FACSCalibur, New Jersey, USA). One-step TUNEL kit (Beyotime, Shanghai, China) was used to detect the apoptosis of HepG2 cells after 24 h of drug treatment.
Wound healing assays
HepG2 cells were seeded in 6-well plates at a density of 1 × 106 cells/well and allowed to reach approximately 95% confluence after 24 h of growth. Cells were scraped with a 10 μL pipette tip to generate a scratch wound and rinsed twice with PBS. The cells were then treated with culture medium containing M(27–39)-HTPP (320 μg/mL), M27-39 (120 μg/mL), HTPP (200 μg/mL), and Dox (2 μg/mL) and the gap distance was measured at the beginning and after 24 or 48 h. Images were captured in different wells using an inverted microscope (LEICA, Wetzlar, Germany). The gap distance was quantitatively evaluated using ImageJ software (Rawak Software Inc., Stuttgart, Germany).
Adhesion assay
HepG2 cells were seeded in 6-well plates. The cells were treated with culture medium containing HTPP (200 μg/mL), M(27–39)-HTPP (320 μg/mL), M27-39 (120 μg/mL), Dox (2 μg/mL) and cultured for another 24 h. The 96-well plates were pre-coated with 80 μL of 1 mg/mL Matrigel (BD, New Jersey, USA) for 4 h. The cells were digested and seeded in 96-well plates at a density of 5 × 104 cells/well for 2 h. Thereafter, 10 μL of CCK8 (0.25 mg/mL) was added to each well. OD values were determined using a microplate reader at a wavelength of 490 nm. Adhesion rate = OD value of administration group/OD value of the blank group × 100%.
Transwell invasion assays
The upper chamber was pre-coated with 80 μL of 1 mg/mL Matrigel (BD, New Jersey, USA) for 12 h. Thereafter, 1 × 105 HepG2 cells were resuspended in 200 μL of DMEM without FBS and seeded in the upper well of a Transwell chamber. DMEM supplemented with 20% FBS was added to the lower chamber. The cells were cultured at 37 °C and 5% CO2 for 48 h. The non-migrated cells were removed and the migrated cells were fixed with 4% paraformaldehyde for 30 min. Migrated cells were stained with 0.5% crystal violet for 15minat 37 °C. The number of migrated cells was determined under an inverted microscope (LEICA, Wetzlar, Germany).
Mitochondrial colocalization
HepG2 cells (5 × 105cells/well) were seeded in a glass-bottomed cell culture dish and co-cultured with culture medium containing FITC-M(27–39)-HTPP (320 μg/mL). After staining with Mito Tracker Deep Red FM (Beyotime, Shanghai, China), the HepG2 cells were fixed with 4% paraformaldehyde. Laser confocal imaging was performed using a confocal laser scanning microscope (LEICA, Wetzlar, Germany).
Transmission electron microscopy (TEM) imaging of the mitochondrial ultrastructure
HepG2 cells were seeded in cell culture flasks at a density of approximately 95%. M(27–39)-HTPP (320 μg/mL) was added to the cells and incubated for 24 h. Thereafter, 2.5% glutaraldehyde was added and the cells were fixed overnight at 4 °C. Changes in mitochondrial microstructure were observed using TEM (Jeol, Tokyo, Japan).
Mitochondrial membrane potential detection
The mitochondrial membrane potential of HepG2 cells was assessed using a mitochondrial membrane potential assay kit with the JC-1 (Beyotime, Shanghai, China). Briefly, 5 × 105 cells were harvested and stained with JC-1 dye for 20minat 37 °C. The cells were then washed twice and analyzed using fluorescence microscopy (LEICA, Wetzlar, Germany) and flow cytometry (BD FACSCalibur, New Jersey, USA). The ratio of red to green fluorescence intensity was calculated.
ROS measurement
HepG2 cells were seeded in 6-well plates (5 × 105 cells/well) and treated with HTPP (200 μg/mL), M27-39 (120 μg/mL), or M(27–39)-HTPP (320 μg/mL) for 24 h. After treatment, the cells were incubated with 20 μL 2,7–Dichlorodi-hydrofluorescin diacetate (DCFH-DA, Beyotime, Shanghai, China) in DMEM for 25minat 37 °C. The cells were immediately analyzed using a flow cytometer (BD FACSCalibur, New Jersey, USA).
Measurement of nitric oxide (NO), ATPase activity measurement
The total NO concentration was determined by measuring the concentrations of nitrate and nitrite, a stable metabolite of NO, using the Griess assay and the total nitric xxide assay kit (Beyotime, Shanghai, China). ATPase levels were estimated using a standard kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, China). The activities of Na+/K+-ATPase, Ca2+/Mg2+-ATPase, and total ATPase were estimated.
Establishment of a mouse hepatoma model
All animal experimental protocols were approved by the Ethics Committee for the Use of Experimental vvvAnimals of the Guangdong Pharmaceutical University. To establish an orthotopic tumor model, mice were injected with 2 × 106 HepG2 cells in the left lobe of the liver. Mice were treated with 5 mg/kg Dox or 35 mg/kg HTPP or 20 mg/kg M27-39 or 55 mg/kg M(27–39)-HTPP via caudal vein once every 48 h for 4 weeks.
In vivo fluorescence imaging
Cy5.5 labeled samples were prepared in the following manner: 5 mg of M(27–39)-HTPP or M27-39 was mixed with 0.6 mg of Cy5.5-NHS ester and 5 mL of DMF in the dark at 25 °C for 6 h Cy5.5 labeled M(27–39)-HTPP or M27-39 was collected via ultrafast centrifugation at 12000 rpm and purified five times with ethanol/water (3:1, v/v) before vacuum drying. HCC mice were subsequently administered a tail vein injection of 0.2 mL of Cy5.5 labeled M(27–39)-HTPP (320 μg/mL) or M27-39 (120 μg/mL). The in vivo fluorescent signals at 1, 3, 6, 12, and 24 h were recorded using a VISQUE In Vivo Smart-LF imaging system. At 6 h, the mice were killed and their major organs (heart, liver, spleen, lung, and kidney) were collected for ex vivo imaging.
In vivo anti-tumor activity and histological analysis
Tumor volume is a therapeutic index and the body weight of mice is a measure of toxicity. The tumor size was determined using the following equation:
The main organ tissues (heart, liver, spleen, lung, and kidney) were collected, washed with saline, immersed in 4% paraformaldehyde, dehydrated, fixed, embedded in paraffin, sectioned, and stained with hematoxylin-eosin (H&E). Finally, the histopathological changes were also observed.
Histology and immunohistochemistry (IHC)
Specimens were fixed overnight in 10% formalin, dehydrated in a graded ethanol series, and embedded in paraffin. Histological analysis was performed using 5 μm paraffin-embedded sections stained with H&E. IHC was performed using the antibodies listed in key resources table. Antigen retrieval was achieved by warming deparaffinized sections in 10 mM citrate buffer (pH 6.0) for 10 min, followed by a 10-min incubation in 10% H2O2 dissolved in methanol to quench the endogenous peroxidase activity. The slides were then incubated with primary antibodies overnight at 4 °C, followed by incubation with secondary HRP-conjugated antibodies for 1hat 37 °C. The slides were counterstained with hematoxylin and images showing protein expression were obtained using a microscope.
Immunofluorescence (IF)
Immunofluorescence analysis was performed using antibodies listed in key resources table. HepG2 cells were seeded on coverslips in 24-well plates for immunofluorescence analysis. After overnight incubation, cells were treated with FITC-M(27–39)-HTPP (320 μg/mL) and fixed with cold 100% methanol for 20 min. At 37 °C, nonspecific sites were blocked with 1% BSA, followed by overnight incubation with the primary antibody. The cells were washed with PBS, incubated for 1hat 37 °C with Cy3-conjugated secondary antibodies, and then washed with PBS. Cell nuclei were stained with DAPI (Meilunbio, Dalian, China) for 5 min before washing with PBS. Finally, fluorescence microscopy (LEICA, Wetzlar, Germany) was used to detect the fluorescence intensity.
Antigen retrieval was achieved by warming deparaffinized sections in 10 mM citrate buffer (pH 6.0) for 10 min, followed by a 10-min incubation in 10% H2O2 dissolved in methanol to quench the endogenous peroxidase activity. At 37 °C, nonspecific sites were blocked with 1% BSA, followed by overnight incubation with the primary antibody. The cells were washed with PBS, incubated for 1hat 37 °C with Cy3-conjugated secondary antibodies, and washed with PBS. Cell nuclei were stained with DAPI for 5 min before washing with PBS. Finally, fluorescence microscopy was used to detect fluorescence intensity.
Quantitative RT-PCR
RNA isolation and RT-PCR were performed according to the manufacturer’s protocol. Total RNA extracted from each tumor was quantified and subjected to reverse transcription as previously described. Fold changes in the mRNA levels of the target genes relative to the endogenous cyclophilin control were calculated. Briefly, the cycle threshold (¼Ct) value of each target gene was subtracted from that of the Ct value of the housekeeping gene (DCt). The target gene DDCt was calculated as the DCt of the target gene minus the DCt of the control. Thefold change in mRNA expression was calculated as 2DDCt. The primer sequences used are listed in key resources table.
Western blot
HepG2 cells and hepatoma tissues were lysed in lysis buffer. After quantifying the lysate protein concentration using a BCA Kit (Beyotime, Shanghai, China), the proteins were loaded for sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to a PVDF membrane (Merck Millipore, Germany). Subsequently, the membranes were blocked with 5% nonfat milk for 1 h and incubated with the primary and secondary antibodies listed in key resources table. The samples were analyzed using BeyoECL Moon (Beyotime, Shanghai, China).
Quantification and statistical analysis
Quantification of wound healing, Western blot, IHC and IF single plane images was performed using ImageJ software (Rawak Software Inc., Stuttgart, Germany). GraphPad Prism (GraphPad Software Inc., San Diego, CA, USA) 8.0.2 software tool was used for statistical analysis. The experimental results are reported as the mean ± SD. One-way analysis of variance (ANOVA) was used to compare the groups. ∗p< 0.05; ∗∗p< 0.01; ∗∗∗p< 0.001. as compared with the negative control group. N.S., not significant.
Acknowledgments
This work was financially supported by National Key R&D Program of China (2018YFC1603900); National Natural Science Foundation of China (32070509 and 31501894); Guangdong Basic and Applied Basic Research Foundation (2021A1515220119); Shenzhen Science and Technology Program (JSGG20220606141800001, JCYJ20220530150412027), ShenzhenFund for Guangdong Provincical High level Clinical Key Specialties (SZGSP006), Sanming Project of Medicine in Shenzhen (SZSM202011008), Shenzhen Key Medical Discipline Construction Fund (SZXK066).
Author contributions
X.L., S.G., and X.J. designed the study; J.W. and R.D. performed the research; B.Z. and J.Y. participated in raising the experimental animals and prepared the tissue sections. J.W., Y.X., S.G., and X.J. analyzed the data. J.W., R.D., and X.L. wrote the manuscript. All the authors reviewed and approved the final version of the manuscript.
Declaration of interests
The patent in question is held by X.L./Guangdong Pharmaceutical University, ZhaoXia Chen, S.G., Lun Zhang, Along Liu, Yanan Tang, Jie Wang, Wenbin Liu, X.J., Jiayong Zhu. (Patent No. ZL201810196235.0). The authors declare no conflict of interest.
Published: April 29, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.106766.
Contributor Information
Shuiqing Gui, Email: guishuiqing@163.com.
Xiaobao Jin, Email: jinxf2001@163.com.
Xuemei Lu, Email: luxuemei@gdpu.edu.cn.
Supplemental information
Data and code availability
Data reported in this paper will be shared by the lead contact upon request. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA. Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Li G., Zhao L. Sorafenib-loaded hydroxyethyl starch-TG100-115 micelles for the treatment of liver cancer based on synergistic treatment. Drug Deliv. 2019;26:756–764. doi: 10.1080/10717544.2019.1642418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Y., Chen X., He D., Zhou Y., Qin L. Surface-modified nanoerythrocyte loading DOX for targeted liver cancer chemotherapy. Mol. Pharm. 2018;15:5728–5740. doi: 10.1021/acs.molpharmaceut.8b00881. [DOI] [PubMed] [Google Scholar]
- 4.Xu J., Cheng X., Tan L., Fu C., Ahmed M., Tian J., Dou J., Zhou Q., Ren X., Wu Q., et al. Microwave responsive nanoplatform via P-selectin mediated drug delivery for treatment of hepatocellular carcinoma with distant metastasis. Nano Lett. 2019;19:2914–2927. doi: 10.1021/acs.nanolett.8b05202. [DOI] [PubMed] [Google Scholar]
- 5.Gong X., Zheng Y., He G., Chen K., Zeng X., Chen Z. Multifunctional nanoplatform based on star-shaped copolymer for liver cancer targeting therapy. Drug Deliv. 2019;26:595–603. doi: 10.1080/10717544.2019.1625467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yokoyama-Mashima S., Yogosawa S., Kanegae Y., Hirooka S., Yoshida S., Horiuchi T., Ohashi T., Yanaga K., Saruta M., Oikawa T., Yoshida K. Forced expression of DYRK2 exerts anti-tumor effects via apoptotic induction in liver cancer. Cancer Lett. 2019;451:100–109. doi: 10.1016/j.canlet.2019.02.046. [DOI] [PubMed] [Google Scholar]
- 7.Harada T., Matsumoto S., Hirota S., Kimura H., Fujii S., Kasahara Y., Gon H., Yoshida T., Itoh T., Haraguchi N., et al. Chemically modified antisense oligonucleotide against ARL4C inhibits primary and metastatic liver tumor growth. Mol. Cancer Therapeut. 2019;18:602–612. doi: 10.1158/1535-7163.MCT-18-0824. [DOI] [PubMed] [Google Scholar]
- 8.Guo Z., Zhou Y., Yang J., Shao X. Dendrobium candidum extract inhibits proliferation and induces apoptosis of liver cancer cells by inactivating Wnt/beta-catenin signaling pathway. Biomed. Pharmacother. 2019;110:371–379. doi: 10.1016/j.biopha.2018.11.149. [DOI] [PubMed] [Google Scholar]
- 9.Castelli G., Pelosi E., Testa U. Liver cancer: molecular characterization, clonal evolution and cancer stem cells. Cancers. 2017;9:127. doi: 10.3390/cancers9090127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y., Liang B., Song X., Wang H., Evert M., Zhou Y., Calvisi D.F., Tang L., Chen X. Loss of apc cooperates with activated oncogenes to induce liver tumor formation in mice. Am. J. Pathol. 2021;191:930–946. doi: 10.1016/j.ajpath.2021.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Llovet J.M., Kelley R.K., Villanueva A., Singal A.G., Pikarsky E., Roayaie S., Lencioni R., Koike K., Zucman-Rossi J., Finn R.S. Hepatocellular carcinoma. Nat. Rev. Dis. Prim. 2021;7:6. doi: 10.1038/s41572-020-00240-3. [DOI] [PubMed] [Google Scholar]
- 12.Han H., Wang L., Liu Y., Shi X., Zhang X., Li M., Wang T. Combination of curcuma zedoary and kelp inhibits growth and metastasis of liver cancer in vivo and in vitro via reducing endogenous H2S levels. Food Funct. 2019;10:224–234. doi: 10.1039/c8fo01594e. [DOI] [PubMed] [Google Scholar]
- 13.Shao J., Lu J., Zhu W., Yu H., Jing X., Wang Y.L., Wang X., Wang X.J. Derepression of LOXL4 inhibits liver cancer growth by reactivating compromised p53. Cell Death Differ. 2019;26:2237–2252. doi: 10.1038/s41418-019-0293-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.An H.W., Li L.L., Wang Y., Wang Z., Hou D., Lin Y.X., Qiao S.L., Wang M.D., Yang C., Cong Y., et al. A tumour-selective cascade activatable self-detained system for drug delivery and cancer imaging. Nat. Commun. 2019;10:4861. doi: 10.1038/s41467-019-12848-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu C., Chen X., Huang Y., Chen Y. Co-administration of iRGD with peptide HPRP-A1 to improve anticancer activity and membrane penetrability. Sci. Rep. 2018;8:2274. doi: 10.1038/s41598-018-20715-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ding Z., Wang D., Shi W., Yang X., Duan S., Mo F., Hou X., Liu A., Lu X. In vivo targeting of liver cancer with tissue- and nuclei-specific mesoporous silica nanoparticle-based nanocarriers in mice. Int. J. Nanomed. 2020;15:8383–8400. doi: 10.2147/IJN.S272495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang M., Xie F., Wen X., Chen H., Zhang H., Liu J., Zhang H., Zou H., Yu Y., Chen Y., et al. Therapeutic PEG-ceramide nanomicelles synergize with salinomycin to target both liver cancer cells and cancer stem cells. Nanomedicine. 2017;12:1025–1042. doi: 10.2217/nnm-2016-0408. [DOI] [PubMed] [Google Scholar]
- 18.Zhang P., Wang J., Chen H., Zhao L., Chen B., Chu C., Liu H., Qin Z., Liu J., Tan Y., et al. Tumor microenvironment-responsive ultrasmall nanodrug generators with enhanced tumor delivery and penetration. J. Am. Chem. Soc. 2018;140:14980–14989. doi: 10.1021/jacs.8b09396. [DOI] [PubMed] [Google Scholar]
- 19.Wang M.D., Hou D.Y., Lv G.T., Li R.X., Hu X.J., Wang Z.J., Zhang N.Y., Yi L., Xu W.H., Wang H. Targeted in situ self-assembly augments peptide drug conjugate cell-entry efficiency. Biomaterials. 2021;278:121139. doi: 10.1016/j.biomaterials.2021.121139. [DOI] [PubMed] [Google Scholar]
- 20.Richardson A., Muir L., Mousdell S., Sexton D., Jones S., Howl J., Ross K. Modulation of mitochondrial activity in HaCaT keratinocytes by the cell penetrating peptide Z-Gly-RGD(DPhe)-mitoparan. BMC Res. Notes. 2018;11:82. doi: 10.1186/s13104-018-3192-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu J., Li J., Wang H., Liu C.B. Mitochondrial-targeted penetrating peptide delivery for cancer therapy. Expet Opin. Drug Deliv. 2018;15:951–964. doi: 10.1080/17425247.2018.1517750. [DOI] [PubMed] [Google Scholar]
- 22.Cerrato C.P., Pirisinu M., Vlachos E.N., Langel Ü. Novel cell-penetrating peptide targeting mitochondria. Faseb. J. 2015;29:4589–4599. doi: 10.1096/fj.14-269225. [DOI] [PubMed] [Google Scholar]
- 23.Grau M., Walker P.R., Derouazi M. Mechanistic insights into the efficacy of cell penetrating peptide-based cancer vaccines. Cell. Mol. Life Sci. 2018;75:2887–2896. doi: 10.1007/s00018-018-2785-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalmouni M., Al-Hosani S., Magzoub M. Cancer targeting peptides. Cell. Mol. Life Sci. 2019;76:2171–2183. doi: 10.1007/s00018-019-03061-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao W., Xu Y., Liu J., Ho M. Epitope mapping by a Wnt-blocking antibody: evidence of the Wnt binding domain in heparan sulfate. Sci. Rep. 2016;6:26245. doi: 10.1038/srep26245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ho M., Kim H. Glypican-3: a new target for cancer immunotherapy. Eur. J. Cancer. 2011;47:333–338. doi: 10.1016/j.ejca.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Montalbano M., Georgiadis J., Masterson A.L., McGuire J.T., Prajapati J., Shirafkan A., Rastellini C., Cicalese L. Biology and function of glypican-3 as a candidate for early cancerous transformation of hepatocytes in hepatocellular carcinoma (Review) Oncol. Rep. 2017;37:1291–1300. doi: 10.3892/or.2017.5387. [DOI] [PubMed] [Google Scholar]
- 28.Lu X., Jin X., Huang Y., Wang J., Shen J., Chu F., Mei H., Ma Y., Zhu J. Construction of a novel liver-targeting fusion interferon by incorporation of a Plasmodium region I-plus peptide. BioMed Res. Int. 2014;2014:261631. doi: 10.1155/2014/261631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang J., Lu X., Jin X., Zeng W., Zhu J. Comparative pharmacokinetics, tissue distribution, excretion of recombinant liver-targeting interferon with IFN α2b administered intramuscular in rats. Curr. Pharmaceut. Biotechnol. 2015;16:911–919. [PubMed] [Google Scholar]
- 30.Lu X., Wang J., Jin X., Huang Y., Zeng W., Zhu J. Targeting mechanism of a novel liver-targeting interferon IFN-CSP involves liver heparan sulfate proteoglycan. Curr. Drug Deliv. 2016;13:528–533. doi: 10.2174/1567201812666150827123602. [DOI] [PubMed] [Google Scholar]
- 31.Liu A., Gui S., Zhang L., Chen Z., Tang Y., Xiao M., Wang J., Liu W., Jin X., Zhu J., Lu X. Production of bioactive liver-targeting interferon Mu-IFN-CSP by soluble prokaryotic expression. Amb. Express. 2017;7:192. doi: 10.1186/s13568-017-0493-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ray T., Kar D., Pal A., Mukherjee S., Das C., Pal A. Molecular targeting of breast and colon cancer cells by PAR1 mediated apoptosis through a novel pro-apoptotic peptide. Apoptosis. 2018;23:679–694. doi: 10.1007/s10495-018-1485-4. [DOI] [PubMed] [Google Scholar]
- 33.Li L., Shi Y., Cheng X., Xia S., Cheserek M.J., Le G. A cell-penetrating peptide analogue, P7, exerts antimicrobial activity against Escherichia coli ATCC25922 via penetrating cell membrane and targeting intracellular DNA. Food Chem. 2015;166:231–239. doi: 10.1016/j.foodchem.2014.05.113. [DOI] [PubMed] [Google Scholar]
- 34.Lu X.M., Jin X.B., Zhu J.Y., Mei H.F., Ma Y., Chu F.J., Wang Y., Li X.B. Expression of the antimicrobial peptide cecropin fused with human lysozyme in Escherichia coli. Appl. Microbiol. Biotechnol. 2010;87:2169–2176. doi: 10.1007/s00253-010-2606-3. [DOI] [PubMed] [Google Scholar]
- 35.DENG Rui Z.J.-l., Xue-mei L.U. Screening and structure-activity relationship analysis of anti-tumor derived peptides based on Musca domestica cecropin. China Biotechnol. 2021;41:14–22. doi: 10.13523/j.cb.2105028. [DOI] [Google Scholar]
- 36.Liu H., Ling C.C., Yeung W.H.O., Pang L., Liu J., Zhou J., Zhang W.Y., Liu X.B., Ng T.P.K., Yang X.X., et al. Monocytic MDSC mobilization promotes tumor recurrence after liver transplantation via CXCL10/TLR4/MMP14 signaling. Cell Death Dis. 2021;12:489. doi: 10.1038/s41419-021-03788-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang X., Gan G., Wang X., Xu T., Xie W. The HGF-MET axis coordinates liver cancer metabolism and autophagy for chemotherapeutic resistance. Autophagy. 2019;15:1258–1279. doi: 10.1080/15548627.2019.1580105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fu J., Wang H. Precision diagnosis and treatment of liver cancer in China. Cancer Lett. 2018;412:283–288. doi: 10.1016/j.canlet.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 39.He X., Chen X., Liu L., Zhang Y., Lu Y., Zhang Y., Chen Q., Ruan C., Guo Q., Li C., et al. Sequentially triggered nanoparticles with tumor penetration and intelligent drug release for pancreatic cancer therapy. Adv. Sci. 2018;5:1701070. doi: 10.1002/advs.201701070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Y., Huo Y., Yao L., Xu Y., Meng F., Li H., Sun K., Zhou G., Kohane D.S., Tao K. Transcytosis of nanomedicine for tumor penetration. Nano Lett. 2019;19:8010–8020. doi: 10.1021/acs.nanolett.9b03211. [DOI] [PubMed] [Google Scholar]
- 41.Ivanov D.P., Parker T.L., Walker D.A., Alexander C., Ashford M.B., Gellert P.R., Garnett M.C. In vitro co-culture model of medulloblastoma and human neural stem cells for drug delivery assessment. J. Biotechnol. 2015;205:3–13. doi: 10.1016/j.jbiotec.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 42.Arami H., Khandhar A.P., Tomitaka A., Yu E., Goodwill P.W., Conolly S.M., Krishnan K.M. In vivo multimodal magnetic particle imaging (MPI) with tailored magneto/optical contrast agents. Biomaterials. 2015;52:251–261. doi: 10.1016/j.biomaterials.2015.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Na J.H., Koo H., Lee S., Min K.H., Park K., Yoo H., Lee S.H., Park J.H., Kwon I.C., Jeong S.Y., Kim K. Real-time and non-invasive optical imaging of tumor-targeting glycol chitosan nanoparticles in various tumor models. Biomaterials. 2011;32:5252–5261. doi: 10.1016/j.biomaterials.2011.03.076. [DOI] [PubMed] [Google Scholar]
- 44.Capurro M., Wanless I.R., Sherman M., Deboer G., Shi W., Miyoshi E., Filmus J. Glypican-3: a novel serum and histochemical marker for hepatocellular carcinoma. Gastroenterology. 2003;125:89–97. doi: 10.1016/s0016-5085(03)00689-9. [DOI] [PubMed] [Google Scholar]
- 45.Zhu Z.W., Friess H., Wang L., Abou-Shady M., Zimmermann A., Lander A.D., Korc M., Kleeff J., Büchler M.W. Enhanced glypican-3 expression differentiates the majority of hepatocellular carcinomas from benign hepatic disorders. Gut. 2001;48:558–564. doi: 10.1136/gut.48.4.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Song Z., Liu T., Chen J., Ge C., Zhao F., Zhu M., Chen T., Cui Y., Tian H., Yao M., et al. HIF-1alpha-induced RIT1 promotes liver cancer growth and metastasis and its deficiency increases sensitivity to sorafenib. Cancer Lett. 2019;460:96–107. doi: 10.1016/j.canlet.2019.06.016. [DOI] [PubMed] [Google Scholar]
- 47.Burke P.J. Mitochondria, bioenergetics and apoptosis in cancer. Trends Cancer. 2017;3:857–870. doi: 10.1016/j.trecan.2017.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhan W., Liao X., Li L., Chen Z., Tian T., Yu L., Chen Z. In vitro mitochondrial-targeted antioxidant peptide induces apoptosis in cancer cells. OncoTargets Ther. 2019;12:7297–7306. doi: 10.2147/OTT.S207640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ekert P.G., Read S.H., Silke J., Marsden V.S., Kaufmann H., Hawkins C.J., Gerl R., Kumar S., Vaux D.L. Apaf-1 and caspase-9 accelerate apoptosis, but do not determine whether factor-deprived or drug-treated cells die. J. Cell Biol. 2004;165:835–842. doi: 10.1083/jcb.200312031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moldoveanu T., Follis A.V., Kriwacki R.W., Green D.R. Many players in BCL-2 family affairs. Trends Biochem. Sci. 2014;39:101–111. doi: 10.1016/j.tibs.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bao T., Ke Y., Wang Y., Wang W., Li Y., Wang Y., Kui X., Zhou Q., Zhou H., Zhang C., et al. Taraxasterol suppresses the growth of human liver cancer by upregulating Hint1 expression. J. Mol. Med. 2018;96:661–672. doi: 10.1007/s00109-018-1652-7. [DOI] [PubMed] [Google Scholar]
- 52.Hockenbery D.M. Targeting mitochondria for cancer therapy. Environ. Mol. Mutagen. 2010;51:476–489. doi: 10.1002/em.20552. [DOI] [PubMed] [Google Scholar]
- 53.Ben Safta T., Ziani L., Favre L., Lamendour L., Gros G., Mami-Chouaib F., Martinvalet D., Chouaib S., Thiery J. Granzyme B-activated p53 interacts with Bcl-2 to promote cytotoxic lymphocyte-mediated apoptosis. J. Immunol. 2015;194:418–428. doi: 10.4049/jimmunol.1401978. [DOI] [PubMed] [Google Scholar]
- 54.Guo X., Yang N., Ji W., Zhang H., Dong X., Zhou Z., Li L., Shen H.M., Yao S.Q., Huang W. Mito-bomb: targeting mitochondria for cancer therapy. Adv. Mater. 2021;33:e2007778. doi: 10.1002/adma.202007778. [DOI] [PubMed] [Google Scholar]
- 55.Vizetto-Duarte C., Custódio L., Gangadhar K.N., Lago J.H.G., Dias C., Matos A.M., Neng N., Nogueira J.M.F., Barreira L., Albericio F., et al. Isololiolide, a carotenoid metabolite isolated from the brown alga Cystoseira tamariscifolia, is cytotoxic and able to induce apoptosis in hepatocarcinoma cells through caspase-3 activation, decreased Bcl-2 levels, increased p53 expression and PARP cleavage. Phytomedicine. 2016;23:550–557. doi: 10.1016/j.phymed.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 56.Zhang J., Jiang Y., Li Y., Li W., Zhou J., Chen J., Shang Z., Gu Q., Wang W., Shen T., Hu W. Micelles modified with a chitosan-derived homing peptide for targeted intracellular delivery of ginsenoside compound K to liver cancer cells. Carbohydr. Polym. 2020;230:115576. doi: 10.1016/j.carbpol.2019.115576. [DOI] [PubMed] [Google Scholar]
- 57.Wang W., Dong X., Liu Y., Ni B., Sai N., You L., Sun M., Yao Y., Qu C., Yin X., Ni J. Itraconazole exerts anti-liver cancer potential through the Wnt, PI3K/AKT/mTOR, and ROS pathways. Biomed. Pharmacother. 2020;131:110661. doi: 10.1016/j.biopha.2020.110661. [DOI] [PubMed] [Google Scholar]
- 58.Zhang L., Li S., Wang R., Chen C., Ma W., Cai H. Cytokine augments the sorafenib-induced apoptosis in Huh7 liver cancer cell by inducing mitochondrial fragmentation and activating MAPK-JNK signalling pathway. Biomed. Pharmacother. 2019;110:213–223. doi: 10.1016/j.biopha.2018.11.037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data reported in this paper will be shared by the lead contact upon request. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.