Abstract
Background
Rapid diagnostic tests based on detection of histidine-rich proteins (HRPs) are widely used for malaria diagnosis, but parasites carrying pfhrp deletions can evade detection and are increasing in frequency in some countries. Models aim to predict conditions under which pfhrp2 and/or pfhrp3 deletions will increase, but a key parameter—the fitness cost of deletions—is unknown.
Methods
We removed pfhrp2 and/or pfhrp3 from a Malawian parasite clone using gene editing approaches) and measured fitness costs by conducting pairwise competition experiments.
Results
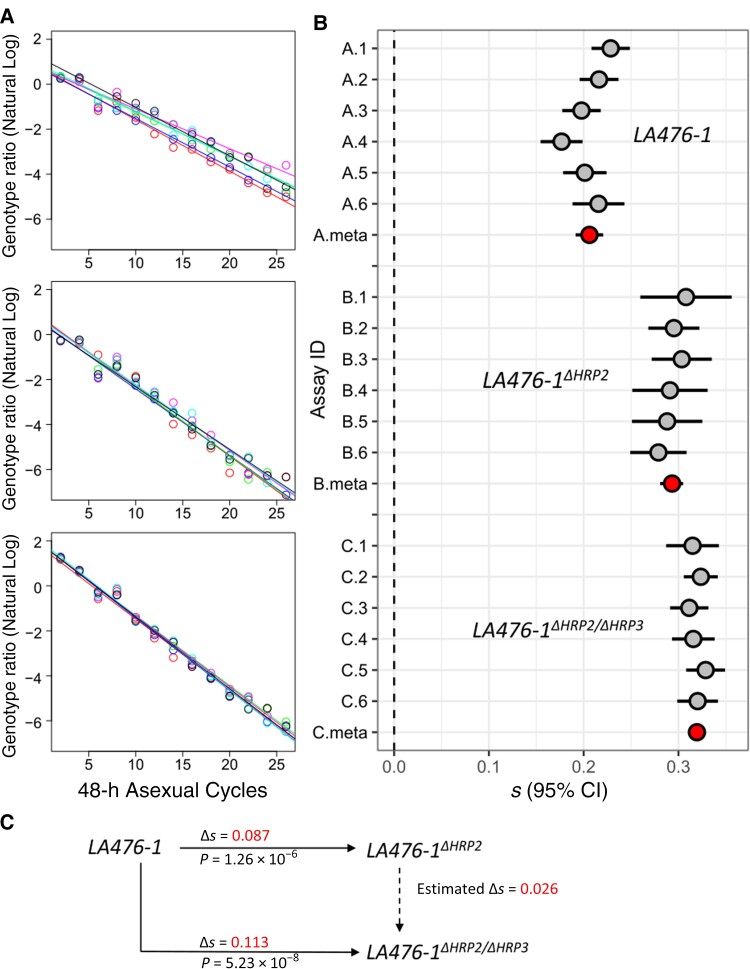
We observed significant fitness costs of 0.087 ± 0.008 (1 standard error) per asexual cycle for pfhrp2 deletion and 0.113 ± 0.008 for the pfhrp2/3 double deletion, relative to the unedited progenitor parasite. Selection against deletions is strong and comparable to that resulting from drug resistance mutations.
Conclusions
Prior modeling suggested that diagnostic selection may drive increased frequency of pfhrp deletions only when fitness costs are mild. Our experiments show that costs of pfhrp deletions are higher than these thresholds, but modeling and empirical results can be reconciled if the duration of infection is short. These results may inform future modeling to understand why pfhrp2/3 deletions are increasing in some locations (Ethiopia and Eritrea) but not in others (Mekong region).
Keywords: Rapid diagnostic tests (RDTs), adaptation, gene copy number, selection, Plasmodium falciparum
The authors used isogenic gene-edited parasites carrying deletions of histidine-rich proteins (HRP) 2 or HRP2 and HRP3 to show that these deletions impose significant fitness costs. This may counteract positive selection for these deletions imposed by HRP-based rapid diagnostic tests.
Rapid diagnostic tests (RDTs) provide a practical way of detecting malaria (Plasmodium falciparum)–infected individuals that can be reliably administered by public health workers with minimal training [1]. These tests use a drop of blood from a finger prick that is applied to a paper strip. The sample flows laterally through a capture line of antibodies against target protein motifs, and the presence of a band indicates an infection. In many endemic areas RDTs are now replacing the reference standard diagnostic approach—microscopy—which requires extensive training and validation to provide consistent results. The most widely used RDTs rely on detection of histidine-rich proteins (HRPs), which are among the most abundant malaria proteins and are produced across the asexual lifecycle [1]. HRP-based RDTs detect products of the pfhrp2 gene (chromosome 8) but also show weaker detection of proteins produced by the pfhrp3 gene (chromosome 13), which shows extensive homology [2]. HRP-based RDTs show high sensitivity, because antibodies target repeat motifs in a highly expressed protein. HRP-based RDTs comprise the majority of the 345 million tests used annually, making this diagnostic tool a critical component of efforts to control and eliminate malaria from endemic areas [1, 3].
Ideally, diagnostic assays should target essential genes. It is clear that pfhrp2 and 3 genes are not essential for parasite survival, because parasites with deletions of pfhrp2 (Dd2, D10) and pfhrp3 (HB3) grow readily in the laboratory [4]. Furthermore, parasites lacking pfhrp2 and/or pfhrp3 (21.6% of parasites lacking both genes) were first observed in Peru [5] and since then have been documented in several countries in South America and sub-Saharan Africa [3, 6–10]. Prevalence of deletions is difficult to assess accurately, because both pfhrp2 and pfhrp3 are situated in highly variable telomeric regions, so polymerase chain reaction–based tests are prone to false-negatives [2, 11]. Because parasites bearing pfhrp2 and pfhrp3 deletions evade detection by HRP-based RDTs, there are concerns that selection for diagnostic evasion could drive increases in frequencies of HRP-deleted parasites. Consistent with this, parasites bearing pfhrp2 and pfhrp3 deletions are now commonly found in the horn of Africa (Eritrea and Ethiopia) [3, 10] and there are strong signals of selection surrounding the pfhrp2 deletion, indicative of recent positive selection for this deletion [3].
Attempts to model how HRP-based diagnosis of malaria can drive increase in frequency of pfhrp2 and pfhrp3 deletions can be useful (1) to clarify why deletions are at high frequencies in some countries but not others and (2) to prioritize countries where surveillance of deletion frequency is warranted [12, 13]. Current models suggest that low prevalence and a high proportion of infected people seeking treatment are the most important drivers. However, one important parameter that is currently unknown is the fitness cost of deletions: high costs will retard the rate of spread of deletions [12]. Current models suggest that pfhrp2 and pfhrp3 deletions are unlikely to increase in frequency unless comparative fitness of deletion mutants is >90% compared with wild-type parasites. In these simulations, parasites carrying deleted HRP2 were assigned reduced “comparative fitness” compared with wild-type parasites, by reducing their contribution to the infectious pool available to infect mosquitoes each generation.
This project was designed to empirically determine fitness costs of pfhrp2 and pfhrp3 deletions. We generated parasite clones with pfhrp2 and pfhrp2/3 double deletions from a recently isolated Malawian parasite clone using clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (CRISPR/Cas9) methods, and we then measured the fitness of blood-stage parasites using competitive growth assays and used amplicon sequencing assays to determine the outcome of competition. Our results reveal fitness costs of 0.087 per asexual cycle for pfhrp2 deletions and 0.1135 for the pfhrp2/3 double deletion.
METHODS
Parasites
We used a parasite isolate (LA476) collected from a Malawian patient in 2008 as part of a cross-sectional study approved by the College of Medicine Research and Ethics Committee, University of Malawi. In this isolate, pfhrp2 and pfhrp3 are located just outside the telomeric regions (following Otto et al [14]) of chromosomes 8 and 13, and flanking regions show extensive sequence variation and rearrangements [2]. We therefore sequenced the LA476-1 genome to ensure that guide RNAs and flanking sequences could be designed to accurately match this particular parasite isolate.
CRISPR/Cas9 Methods for Generating Deletions
We designed a plasmid for CRISPR/Cas9 deletion of pfhrp2 and pfhrp3 using CRISPR/Cas9 approaches (Figure 1A). The guide RNAs were designed to target positions in the LA476-1 genome (coordinates based on homology with 3D7 genome, PlasmoDB release 46) flanking pfhrp2 and pfhrp3. Plasmids were constructed by Genscript, and homology arms and guide RNA (gRNA) sequences were checked using Sanger sequencing. The homology region consists of 935–1031–base pair (bp) flanking regions surrounding the region targeted for deletion. We designed single-guide RNA (sgRNA) sequences (Table 1) at the 5’, 3’, or mid-regions of the genes and used Cas-OFFinder (version 2.4) software to search for potential off-target sites.
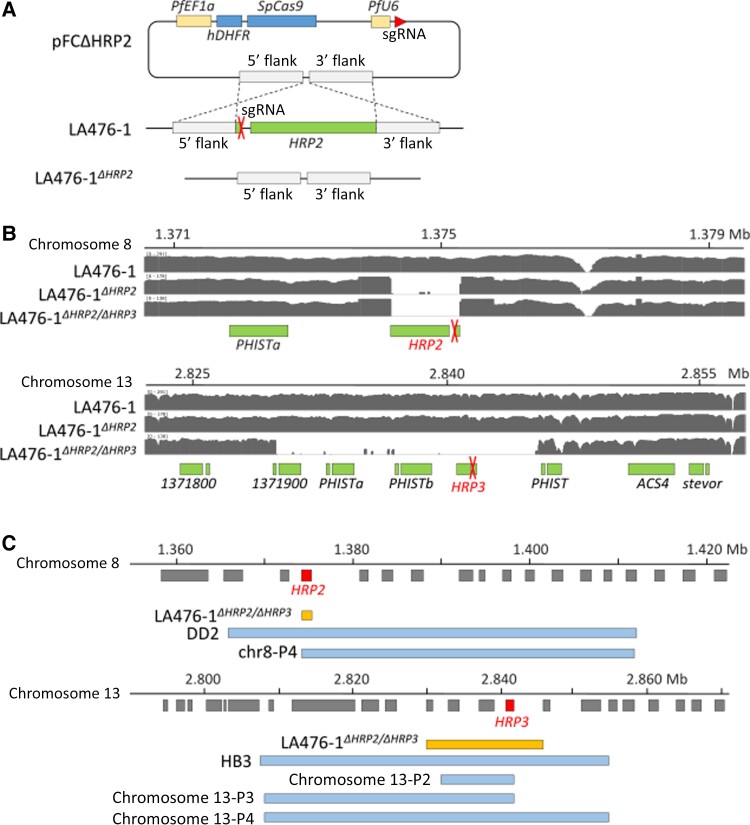
Figure 1.
CRISPR/Cas9–generated pfhrp2 and pfhrp3 deletions. A, Strategy to generate LA476-1ΔHRP2. LA476-1ΔHRP2/ΔHRP3 was generated in the same manner. Plasmids were generated by adding single-guide RNA (sgRNA) and homology arms flanking the upstream and downstream regions of pfhrp2 or pfhrp3 to the pFC plasmid previously reported by Goswami et al [15]. B, Integrative Genomics Viewer (IGV) plots showing pfhrp2 and pfhrp3 deletions for parasites generated. sgRNA target loci were labeled with red X’s. C, Schematic comparisons of deletions in field isolates and CRISPR-generated deletions. DD2 and HB3 are laboratory strains known to have histidine-rich protein (HRP) 2 (DD2) or HRP3 (HB3) deletions. Note: chr8-P4 in the chromosome 8 panel, and chr13-P2, chr13-P3, and chr13-P4 in the chromosome13 panel indicate field samples from Ethiopia grouped by pfhrp2 or pfhrp3 deletion structural profile [3].
Table 1.
Design of Single-Guide RNA
| Gene | gRNA | Sequence | Outcomea |
|---|---|---|---|
| pfhrp2 | HRP2_gRNA_5’ | TACGTTATCTAACAAAAGTA | Success |
| HRP2_gRNA_3’ | CTATTATTAAATAAATTTAA | Failure | |
| HRP2_ gRNA_mid | AGCTGCATGATGAGCGTGAT | Failure | |
| pfhrp3 | HRP3_gRNA_5’ | TTGTTTAGCAAAAATGCAAA | Failure |
| HRP3_gRNA_3’ | CATGCAGCTGATGCTAATCA | Failure | |
| HRP3_ gRNA_3’-2 | CACGACGATGCCCACCATGA | Failure |
Abbreviation: gRNA, guide RNA.
Success or failure of edits.
We transfected ring-stage parasites with 100 ng of plasmid DNA and selected successful transfectants by treatment with 5-nmol/L WR99210 (Jacobus Pharmaceuticals) for 6 days, and we recovered parasites after approximately 3 weeks. To determine whether recovered parasites contained the deletions expected, we conducted polymerase chain reaction assays using primers as showed in Table 2. We cloned parasites from successful transfection experiments. Recovered parasites were then genome sequenced to define the breakpoints of deletions and to identify any off-target edits elsewhere in the genome.
Table 2.
Primers Used to Validate pfhrp2 or pfhrp3 Deletions
| Gene | Primer Name | Sequence |
|---|---|---|
| pfhrp2 a | Hrp2_Bravo_F | ATGATTCATTATTCTATATTTATAAGGAAGATTAC |
| Hrp2_Bravo_R | CACTTCATGTAT TTATGTATGCAGAAC | |
| pfhrp3 | Hrp3_Check_F | ACGGATTTCATTTTAACCTTCACGA |
| Hrp2_Check_R | GCTCCATCGTGGTGTGCTCCAT |
Primers used for validating pfhrp2 gene deletion were from Jones et al [16].
Measurement of Relative Fitness Costs
We conducted head-to-head competition experiments to determine the impact of pfhrp deletions on parasite fitness in asexual culture. To do this, we competed LA476 and each of our pfhrp deletion mutants against a common competitor (NHP4302; CRISPR edited parasite with 2 synonymous mutations at kelch13 gene locus) [17]. The fitness cost calculated are relative to the common competitor and therefore unitless. All competition experiments were conducted with 6 replicates in 6 well plates. We synchronized parasites to 80% schizont stages by purification through magnetic-activated cell sorting (MACS) purification columns (Miltenyi Biotec); after overnight growth, we plated ring-stage parasites at 50% frequency (0.1% parasitemia) to initiate experiments. We maintained cultures at 0.1–4% parasitemia and collected 80-μL aliquots every 4 days to monitor allele frequency change. We conducted these experiments using complete medium consisting of Roswell Park Memorial Institute (RPMI) 1640 medium supplemented with 2-mmol/L glutamine, 25-mmol/L HEPES, and 50-g/L gentamicin, 0.1-mol/L hypoxanthine, and 0.4% AlbuMAX as a serum source. Parasites were grown at 2% hematocrit and maintained at 37°C with 5% oxygen, 5% carbon dioxide, and 90% nitrogen.
We amplified a 249-bp sequence region of kelch13, spanning the synonymous mutations present in NHP4302, to determine the frequencies of competing parasites within mixtures. We then sequenced amplicons to a high read depth with Illumina MiSeq to quantify allele frequencies, using methods described elsewhere [17].
Statistical Analysis
We plotted the natural log of the parasite ratio (frequency of clone A/frequency of clone B) against time. Outliers were removed using Cook's distance [18] with a cutoff of 4/n (where n is the number of data points). The slope of the best fit linear model provides the selection coefficient (s), a measure of relative fitness of the competing parasites. We used the R package metafor (version 3.0-2) (random-effects model) to compare selection coefficients resulting from pfhrp deletions
Data and Parasite Availability
The gene-edited parasites generated will be made available through BEI/MR4. Raw sequencing data have been submitted to the National Center for Biotechnology Information (NCBI) Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra; under project accession no. PRJNA798076).
RESULTS
Generation of Deletion Mutants
We generated both the LA476Δhrp2 and LA476Δhrp2/hrp3 deletion mutants. However, we were unable to generate the LA476Δhrp3 despite 3 different attempts using different sgRNA (Table 2). The breakpoints of pfhrp2 deletion were the same for both the LA476Δhrp2 and LA476Δhrp2/hrp3 parasites, as expected from the CRISPR/Cas9 design. The deletion spanned 1061 bp (1 374 237–1 375 298) on chromosome 8 and contains only pfhrp2. The pfhrp3 deletion was inadvertently generated using the plasmids designed to make pfhrp2 deletions, reflecting the homology between these 2 genes. The pfhrp3 deletion spans 15 566 bp (2 829 897–2 845 466) on chromosome 13 and contains 3 genes encoding exported proteins of unknown function—PF3D7_1371900, PHISTa (PF3D7_1372000) and PHISTb (PF3D7_1372100)—in addition to pfhrp3 (Figure 1B). For comparison, we show the gene content of naturally occurring deletions at pfhrp2 and pfhrp3 from Ethiopia (Figure 1C) [3].
We examined genome sequences of cloned edited parasites, as described elsewhere [17], for any off-target deletions or mutations relative to the original LA476 parasite. All raw data are in the NCBI database (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA798076). We found only 1 single-nucleotide polymorphism (SNP) (Ser4026Pro) in LA476Δhrp2, located at gene PF3D7_1474200 (unknown function). We investigated sequences from 23 bp upstream to 23 bp downstream from the position of this mutation and compared them with the guide RNA sequence. The statistical significance for the probability that these sequences are targets of the guide sequence were calculated using a formula from Cho et al [19]. Sequences around this SNP showed no similarity to the guide sequence (P > .05), which indicates that this mutation was not an off-target effect of gene editing. Gene PF3D7_1474200 was highly expressed in late-stage gametocytes but not in other blood-stage parasites (data from Malaria Cell Atlas; https://www.sanger.ac.uk/tool/mca/new/). The SNP mutation detected in this gene is unlikely to influence the quantification of parasite fitness in head-to-head experiments.
Results of Competition Experiments
The common competitor outcompeted LA476, LA476Δhrp2, and LA476Δhrp2/hrp3, but the magnitude of the selection coefficient differs in each case, allowing us to determine the relative fitness costs of each of the deletion mutants (Figure 2, Table 3). We observed a mean (standard error [SE]) fitness cost of 0.087 (0.008) for LA476Δhrp2 relative to LA476 (P = 1.26 × 10−6) and 0.113 (0.008) for LA476Δhrp2/hrp3 relative to LA476 (P = 5.23 × 10−8). The double deletion carries a significantly higher fitness cost than the single deletion (P = 5.69 × 10−4), indicating that pfhrp3 deletion also carries a small cost (mean s [SE] = 0.026 [0.006]), if we assume an additive model. We were unable to generate LA476Δhrp3 so cannot determine whether the deletions act in an additive or epistatic manner to determine parasite fitness.
Figure 2.
Measurement of fitness costs. A, Natural log of the parasite ratio against 48-hour life cycle between common competitor (NHP4026) and LA476 (top), LA476Δhrp2 (middle), and LA476Δhrp2/hrp3 (bottom). B, Selection coefficients (s) with 95% confidence intervals (CIs) from 4 sets of competition experiments. Six replicate competition experiments were conducted for each set: gray points show s for each replicate; red points, meta-analysis results for each experimental comparison. Abbreviation: ID, identifier. C, Summary of fitness differences among LA476, LA476Δhrp2, and LA476Δhrp2/hrp3.
Table 3.
Calculation and Summary for Selection Coefficients of All Competition Experimentsa
| Strains in Culture | Assay ID | Mean s (SE)b | R2 | n | P Valuec |
|---|---|---|---|---|---|
| LA476-1 | A.1 | 0.228 (0.0092) | 0.981 | 13 | 5.207 × 10−11 |
| A.2 | 0.216 (0.0091) | 0.983 | 11 | 1.916 × 10−9 | |
| A.3 | 0.198 (0.0092) | 0.974 | 13 | 2.590 × 10−10 | |
| A.4 | 0.177 (0.0101) | 0.962 | 13 | 2.182 × 10−9 | |
| A.5 | 0.201 (0.0103) | 0.972 | 12 | 2.686 × 10−9 | |
| A.6 | 0.216 (0.0121) | 0.969 | 11 | 2.493 × 10−8 | |
| A.meta | 0.206 (0.0074) | NA | 73 | 9.234 × 10−46 | |
| LA476-1ΔHRP2 | B.1 | 0.308 (0.0213) | 0.954 | 11 | 1.566 × 10−7 |
| B.2 | 0.295 (0.0121) | 0.982 | 12 | 3.043 × 10−10 | |
| B.3 | 0.303 (0.0143) | 0.976 | 12 | 1.161 × 10−9 | |
| B.4 | 0.291 (0.0179) | 0.960 | 12 | 1.574 × 10−8 | |
| B.5 | 0.288 (0.0167) | 0.964 | 12 | 8.895 × 10−9 | |
| B.6 | 0.279 (0.0135) | 0.973 | 13 | 3.858 × 10−10 | |
| B.meta | 0.293 (0.0062) | NA | 72 | 5.543 × 10−43 | |
| LA476-1ΔHRP2/ΔHRP3 | C.1 | 0.315 (0.0125) | 0.983 | 12 | 2.232 × 10−10 |
| C.3 | 0.324 (0.0081) | 0.993 | 13 | 3.116 × 10−13 | |
| C.4 | 0.311 (0.0092) | 0.990 | 13 | 1.787 × 10−12 | |
| C.5 | 0.316 (0.0101) | 0.989 | 12 | 2.674 × 10−11 | |
| C.6 | 0.329 (0.0093) | 0.990 | 13 | 1.170 × 10−12 | |
| C.2 | 0.320 (0.0098) | 0.989 | 13 | 2.552 × 10−12 | |
| C.meta | 0.320 (0.0039) | NA | 76 | 8.039 × 10−60 |
Abbreviations: ID, identifier; n, number of time points at which genotype frequencies were measured; NA, not applicable,; R2, correlation coefficient; s, selection coefficient; SE, standard error;
For each combination, we conducted 6 independent competition assays. All competition experiments were conducted versus a common competitor strain (NHP4302), which outcompeted each of the edited parasites examined. Time points with failed sequencing were removed from analysis.
Measured as in Figure 2.
P values represent statistical significance for a test of the null hypothesis that the slope is not different from zero.
DISCUSSION
Our results reveal significant fitness costs of pfhrp2 and pfhrp3 deletions. The pfhrp2 deletion contributes the most to the costs observed, consistent with the higher transcript abundance for pfhrp2 [1], but the pfhrp3 gene also contributes significantly. Overall, the relative fitness costs were a similar order of magnitude to those observed for many drug resistance loci in the laboratory [20].
While no other studies have directly examined relative fitness costs of pfhrp2 and pfhrp3 deletion, several other studies are informative, First, Yang et al deleted pfhrp2 to elucidate the transcriptional consequences and function of HRP2 protein and noted that “the growth of the parasite in the host was not affected by the deletion of Pfhrp2 compared with the growth of the wild type.” However, these authors also determined extensive alteration in gene expression in parasites with deleted pfhrp2, suggesting some significant metabolic impact. We note that fitness consequences on the order of s = 0.1 may be difficult to discern without competitive growth assays.
Wellems et al [21] examined the progeny from a genetic cross between HB3 (pfhrp3 deleted) and 3D7 (no deletions), and they found strong selection for progeny carrying intact pfhrp3 in the progeny isolated, consistent with pfhrp3 deletion, or neighboring deleted genes, carrying a fitness cost. Similarly, a cross between Dd2 (pfhrp2 deleted) and HB3 (pfhrp3 deleted) suggested selection against pfhrp3 deletion but not against the pfhrp2 deletion [22].
Zhang et al [23] conducted a mutagenesis study using the piggyBAC transposon system to map parasite genes that are essential and/or result in reduced fitness when disrupted. Both pfhrp2 and pfhrp3 tolerated disruptions by multiple transposons, confirming that they are nonessential (mutagenesis index scores of 1 for both genes [23, supplementary table 5]); furthermore, transposon disruption had little impact on competitive growth in pooled growth assays, suggesting that disabling these genes results in limited relative fitness costs (mutagenesis fitness scores of −1.914 for pfhrp2 and −1.811 for pfhrp3 [23, supplementary table 5]).
The underlying causes of slower growth in parasites bearing deletions will require improved understanding of the functions of HRP2 and HRP3 proteins. Strangely, while these genes are among the most highly expressed in the blood-stage malaria parasites, we currently know rather little about their function [1]. Some results suggest that HRP2 is involved in heme metabolism [24, 25]; the HRP2 protein has a heme-binding site [26], and deletions of pfhrp2 results in significant underexpression or overexpression of several genes involved in hemozoin conversion [27]. This feature of pfhrp2 biology seems most likely to affect fitness in pfhrp2-deleted parasites. Other data suggest that HRP2 is involved in capillary sequestration and cerebral malaria as pfhrp2 alters the binding properties of erythrocytes [1, 28].
How do our empirical measures of the fitness costs of HRP deletions match with predictions from mathematical modelling? The balance of relative fitness costs, which tend to retard spread, and selection for diagnostic evasion are expected to determine spread of pfhrp2/3 deletions. Models suggest that a low prevalence of malaria and a high proportion of infected people seeking treatment are risk factors for spread of pfhrp2 deletions [12]. These models were missing estimates of fitness costs resulting from pfhrp2 and pfhrp3 deletions, but they determined that deletions were unlikely to spread if “comparative fitness” pfhrp2 and pfhrp3 deletions were <90%. The comparative fitness parameter used in modeling work is not directly comparable to the selection coefficients we have calculated to quantify fitness costs in our experimental work, so it is important to compare these parameters. Watson et al [12] incorporated comparative fitness in their models by reducing the contribution of HRP2 deleted parasites to the infectious pool available to infect mosquitoes.
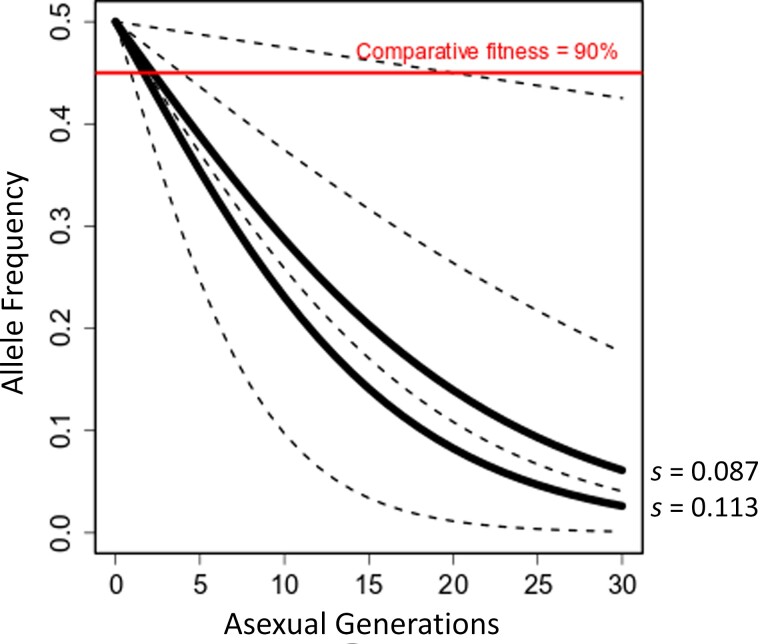
Figure 3 examines the relationship between fitness costs measured per asexual parasite cycle and comparative fitness. The proportion of HRP-deleted parasites drops progressively each asexual cycle. Using the fitness costs determined in our experiments, proportions of pfhrp-deleted parasites drop below the 90% comparative fitness threshold after 1–2 asexual cycles (2–4 days) and show comparable fitness of 50% after 9 or 12 asexual generations for pfhrp2/3 or pfhrp2 deletions, respectively. Given the high fitness costs of pfhrp deletions, the duration of infection before transmission to mosquitoes will be critical for determining the proportion of HRP-deleted parasites available for transmission.
Figure 3.
Comparison of fitness costs with comparative fitness used for modeling. Graph shows the consequences of fitness costs on the frequency of competing parasites within a single blood-stage infection. Solid black lines show the allele frequency of parasites with histidine-rich protein (HRP) 2 (upper line) or HRP2/3 deletions (lower line) in competition with wild-type parasites, using the selection coefficients (s) determined in this study; black dotted lines, trajectories assuming fitness costs of 0.01, 0.05, 0.1, and 0.2, respectively (top to bottom); red line, 90% comparative fitness threshold used for modeling [12]. Parasites with HRP2 deletions remain above the 90% comparative fitness threshold for only 1–2 asexual cycles.
Our fitness cost estimates (s = 0.087 for pfhrp2 alone and s = 0.1135 for the double deletion) fall below this 90% comparative fitness threshold from modeling when the durations of blood-stage infection is >2 asexual cycles (4 days) (Figure 3). We have direct measures of patent period (time from mosquito inoculation to detectable blood stream parasitemia) from studies of human volunteers experimentally infected with malaria via infected mosquitoes [29]. These studies determined that patent period ranged from 7 to 12 days. Given that this includes approximately 6 day of parasite development in the liver, the time from emerge of merozoites from the liver to patency is 1–6 days (1–3 asexual cycles). While these estimates are broadly consistent with the window of 1–2 asexual cycles (2–4 days), we recognize that the time to mosquito transmission is likely to be considerably longer than this, particularly in regions of low transmission.
Two other considerations complicate comparison of experimental measures of fitness and modeling assumptions. First, our experiments measured fitness costs of asexual blood-stage parasites, which are just one component of parasite fitness However, levels of competing blood-stage parasites may provide a poor predictor of numbers of gametocytes from competing parasites available to infect mosquitoes. Consistent with this, experiments using rodent malaria parasite clones mixed in different proportions have suggested that minority clones may produce more gametocytes and achieve higher transmission to mosquitoes than majority clones [30]. Second, compensatory changes elsewhere in the genome may mitigate the fitness costs of HRP deletions, as occurs during evolution of drug resistance [31, 32]. As a consequence, fitness costs of newly generated deletions in laboratory maintained parasites may be larger than to those in parasites circulating in nature.
Why are HRP deletions increasing in frequency in some locations but not others? These results are consistent with the idea that deletions will start spreading only when selection for diagnostic evasion outweighs the metabolic costs resulting from deletion, and they may help explain the patchy distributions of pfhrp2 deletions. As transmission is reduced, resulting in higher numbers of symptomatic patients seeking treatment, we expect progressively more locations to reach the tipping point where benefits of pfhrp2/3 deletions outweigh the costs, leading to an increase in deletion frequency and reduced utility of HRP2-based RDTs. Incorporating estimates of fitness costs can aid in generating models to predictive prevalence of pfhrp2/3 deletions and prioritize regions for surveillance.
In some locations, pfhrp2 and pfhrp3 deletions appear to have spread without selection from widespread RDT use. For example, in Amazonia, pfhrp2 and 3 deletions were first documented in 2010, and in examined samples collected in 2003–2008, before widespread deployment of HRP2 RDTs [5]. We suggest two related explanations for this. First, deleterious copy number variants may be more common in parasite populations with very low effective population size (Ne), because selection purging such alleles is weak. Consistent with such a genetic drift–based explanation, epidemic low-Ne South American P. falciparum populations carry more and larger insertions and deletions than endemic African and Asian populations with larger Ne [33]. Second, South American parasite populations typically show highly clonal population structure, with strong linkage disequilibrium between genes on different chromosomes [34, 35]. With such population structure, deleterious mutations may spread by hitchhiking with successful multilocus genotypes. A reduction in population size, increase in genetic drift, and linkage disequilibrium in populations targeted by malaria control efforts will therefore also lower the barriers against spread of deleterious pfhrp2/3 deletions.
The current study had some limitations. We determined fitness during the asexual blood stage, and we cannot rule out that fitness costs may be observed elsewhere during the malaria lifecycle. However, expression of pfhrp2 and pfhrp3 is high only in the blood stages. Neither gene is expressed in liver-stage parasites, and pfhrp2 transcripts were not detected in any mosquito stages, while pfhrp3 shows low expressions only in activated sporozoites [36]. Therefore, pfhrp2 and pfhrp3 seem unlikely to affect parasite fitness outside the blood stages. A related caveat is that in vitro culture differs in many respects from infection within patients [37], so some caution in interpreting these results is needed.
We were unable to edit parasites containing only pfhrp3 deletions, so we cannot measure the cost of pfhrp3 deletions directly. Assuming an additive model, we estimate that cost (ΔHRP3) = cost (ΔHRP2/ΔHRP3) − cost (ΔHRP2). However, if we assume that HRP2 and HRP3 play a similar role in parasite biology, and are partially redundant, then positive epistasis might be a likely outcome—cost (ΔHRP2) + cost(ΔHRP3) > cost (ΔHRP2/ΔHRP3). If ΔHRP3 carries a substantial cost without accompanying ΔHRP2, this might explain the abundance of parasites in the horn of Africa carrying only pfhrp3 deletions.
We used a chloroquine (CQ)–sensitive African parasite for these experiments, although in South American and African countries pfhrp2/3 deletions have arisen in CQ-resistant parasites populations. There is some evidence that CQ-resistance and pfhrp2 may be functionally linked [1], because pfhrp2 has a heme binding domain [26] and is thought to mediate conversion of heme to hemozoin in the vacuole [27]. CQ-resistant parasites have modified heme metabolism pathways [38] and altered food vacuole pH [39], and HRP-2 mediated hemozoin conversion is pH dependent [24]. One possibility is that pfhrp2-mediated heme-to-hemozoin conversion is not used in CQ-resistant parasites, and that deletion of pfhrp2/3 will therefore have lower cost to CQ-resistant parasites. Conducting parallel pfhrp2 and pfhrp3 deletion experiments in a CQ-resistant genetic background would be of considerable interest to test this hypothesis.
While our deletion on chromosome 8 contained only pfhrp2, our deletion on chromosome 13 contained 3 genes in addition to pfhrp3 deletions. We cannot determine whether the fitness costs observed result from deletions of pfhrp3 genes or are due to deletion of flanking genes. The extensive variation in the telomeric regions of chromosomes 8 and 13 preclude precise deletion of only pfhrp2 or pfhrp3 genes. It is important to note that naturally occurring pfhrp2 and 3 deletions also contain several deleted flanking genes (Figure 1C). However, the estimated cost of pfhrp3 deletion is quite small (0.026) (Figure 2C) suggesting that neither pfhrp3 nor flanking genes have a large impact on parasite fitness.
In conclusion, these experiments provide direct estimates of fitness costs resulting from pfhrp2 and pfhrp2/3 deletions in isogenic parasites. These costs are significant, and they suggest that parasites bearing pfhrp deletions will be rapidly outcompeted in coinfections with wild-type parasites. Our empirical measures of fitness costs are compatible with modeling results only when the duration of blood-stage infection before infection of mosquitoes is very short. The evolution of diagnostic evasion has strong parallels with drug resistance evolution, where fitness costs may result in loss in resistance alleles in the absence of treatment [40]. However, as with drug resistance, we might also expect there to be strong selection for compensatory changes that restore fitness to parasites carrying pfhrp2 and pfhrp3 deletions in nature [31, 32, 41].
Notes
Acknowledgments. We are grateful to patients who participated in the study. Comments from two anonymous reviewers greatly improved the manuscript.
Author contributions. S. N, X. L., and T. A. designed the experiments; S. C. N collected the parasite isolate and performed its initial characterization; S. N. conducted experiments; X .L. performed all the NGS analysis and data curation; S. N, X. L., and T. A. wrote the original manuscript.
Financial support. This work was supported by the National Institutes of Health (grant R37 AI048071 to T. A.); the National Center for Research Resources (Research Facilities Improvement Program grant C06 RR013556 to Texas Biomedical Research Institute); and the Gates Malaria Partnership (grant to S. C. N. supporting collection of parasite isolates).
Contributor Information
Shalini Nair, Disease Intervention and Prevention Program, Texas Biomedical Research Institute, San Antonio, Texas, USA.
Xue Li, Disease Intervention and Prevention Program, Texas Biomedical Research Institute, San Antonio, Texas, USA.
Standwell C Nkhoma, BEI Resources, American Type Culture Collection, Manassas, Virginia, USA.
Tim Anderson, Disease Intervention and Prevention Program, Texas Biomedical Research Institute, San Antonio, Texas, USA.
References
- 1. Poti KE, Sullivan DJ, Dondorp AM, et al. HRP2: transforming malaria diagnosis, but with caveats. Trends Parasitol 2020; 36:112–26. [DOI] [PubMed] [Google Scholar]
- 2. Gendrot M, Fawaz R, Dormoi J, et al. Genetic diversity and deletion of Plasmodium falciparum histidine-rich protein 2 and 3: a threat to diagnosis of P. falciparum malaria. Clin Microbiol Infect 2019; 25:580–5. [DOI] [PubMed] [Google Scholar]
- 3. Feleke SM, Reichert EN, Mohammed H, et al. Plasmodium falciparum is evolving to escape malaria rapid diagnostic tests in Ethiopia. Nat Microbiol 2021; 6:1289–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kemp DJ, Thompson JK, Walliker D, et al. Molecular karyotype of Plasmodium falciparum: conserved linkage groups and expendable histidine-rich protein genes. Proc Natl Acad Sci U S A 1987; 84:7672–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gamboa D, Ho MF, Bendezu J, et al. A large proportion of P. falciparum isolates in the Amazon region of Peru lack pfhrp2 and pfhrp3: Implications for malaria rapid diagnostic tests. PLoS One 2010; 5:e8091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Berhane A, Anderson K, Mihreteab S, et al. Major threat to malaria control programs by Plasmodium falciparum lacking histidine-rich protein 2, Eritrea. Emerg Infect Dis 2018; 24:462–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Grignard L, Nolder D, Sepúlveda N, et al. A novel multiplex qPCR assay for detection of Plasmodium falciparum with histidine-rich protein 2 and 3 (pfhrp2 and pfhrp3) deletions in polyclonal infections. EBioMedicine 2020; 55:102757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Parr JB, Verity R, Doctor SM, et al. Pfhrp2-deleted Plasmodium falciparum parasites in the Democratic Republic of the Congo: a national cross-sectional survey. J Infect Dis 2017; 216:36–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Thomson R, Parr JB, Cheng Q, et al. Prevalence of Plasmodium falciparum lacking histidine-rich proteins 2 and 3: a systematic review. Bull World Health Organ 2020; 98:558–568F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Menegon M, L'Episcopia M, Nurahmed AM, et al. Identification of Plasmodium falciparum isolates lacking histidine-rich protein 2 and 3 in Eritrea. Infect Genet Evol 2017; 55:131–4. [DOI] [PubMed] [Google Scholar]
- 11. Parr JB, Anderson O, Juliano JJ, et al. Streamlined, PCR-based testing for pfhrp2- and pfhrp3-negative Plasmodium falciparum. Malar J 2018; 17:137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Watson OJ, Slater HC, Verity R, et al. Modelling the drivers of the spread of Plasmodium falciparum hrp2 gene deletions in sub-Saharan Africa. Elife 2017; 6:25008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Watson OJ, Verity R, Ghani AC, et al. Impact of seasonal variations in Plasmodium falciparum malaria transmission on the surveillance of pfhrp2 gene deletions. Elife 2019; 8:e40339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Otto TD, Böhme U, Sanders M, et al. Long read assemblies of geographically dispersed Plasmodium falciparum isolates reveal highly structured subtelomeres. Wellcome Open Res 2018; 3:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Goswami D, Betz W, Locham NK, et al. A replication-competent late liver stage-attenuated human malaria parasite. JCI Insight 2020; 5:e135589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jones S, Subramaniam G, Plucinski MM, et al. One-step PCR: a novel protocol for determination of pfhrp2 deletion status in Plasmodium falciparum. PLoS One 2020; 15:e0236369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nair S, Li X, Arya GA, et al. Fitness costs and the rapid spread of kelch13-C580Y substitutions conferring artemisinin resistance. Antimicrob Agents Chemother 2018; 62:e00605–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cook RD. Detection of influential observation in linear regression. Technometrics 1977; 19:15–8. [Google Scholar]
- 19. Cho SW, Kim S, Kim Y, et al. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res 2014; 24:132–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Stokes BH, Dhingra SK, Rubiano K, et al. Plasmodium falciparum K13 mutations in Africa and Asia impact artemisinin resistance and parasite fitness. Elife 2021; 10:e66277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wellems TE, Walliker D, Smith CL, et al. A histidine-rich protein gene marks a linkage group favored strongly in a genetic cross of Plasmodium falciparum. Cell 1987; 49:633–42. [DOI] [PubMed] [Google Scholar]
- 22. Sepúlveda N, Phelan J, Diez-Benavente E, et al. Global analysis of Plasmodium falciparum histidine-rich protein-2 (pfhrp2) and pfhrp3 gene deletions using whole-genome sequencing data and meta-analysis. Infect Genet Evol 2018; 62:211–9. [DOI] [PubMed] [Google Scholar]
- 23. Zhang M, Wang C, Otto TD, et al. Uncovering the essential genes of the human malaria parasite Plasmodium falciparum by saturation mutagenesis. Science 2018; 360:eaap7847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lynn A, Chandra S, Malhotra P, et al. Heme binding and polymerization by Plasmodium falciparum histidine rich protein II: influence of pH on activity and conformation. FEBS Lett 1999; 459:267–71. [DOI] [PubMed] [Google Scholar]
- 25. Papalexis V, Siomos MA, Campanale N, et al. Histidine-rich protein 2 of the malaria parasite, Plasmodium falciparum, is involved in detoxification of the by-products of haemoglobin degradation. Mol Biochem Parasitol 2001; 115:77–86. [DOI] [PubMed] [Google Scholar]
- 26. Sullivan DJ J, Gluzman IY, Goldberg DE. Plasmodium hemozoin formation mediated by histidine-rich proteins. Science 1996; 271:219–22. [DOI] [PubMed] [Google Scholar]
- 27. Yang Y, Tang T, Feng B, et al. Disruption of Plasmodium falciparum histidine-rich protein 2 may affect haem metabolism in the blood stage. Parasit Vectors 2020; 13:611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Benedetti CE, Kobarg J, Pertinhez TA, et al. Plasmodium falciparum histidine-rich protein II binds to actin, phosphatidylinositol 4,5-bisphosphate and erythrocyte ghosts in a pH-dependent manner and undergoes coil-to-helix transitions in anionic micelles. Mol Biochem Parasitol 2003; 128:157–66. [DOI] [PubMed] [Google Scholar]
- 29. Churcher TS, Sinden RE, Edwards NJ, et al. Probability of transmission of malaria from mosquito to human is regulated by mosquito parasite density in naïve and vaccinated hosts. PLoS Pathog 2017; 13:e1006108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Taylor LH, Walliker D, Read AF. Mixed-genotype infections of malaria parasites: within-host dynamics and transmission success of competing clones. Proc Biol Sci 1997; 264:927–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Durão P, Balbontín R, Gordo I. Evolutionary mechanisms shaping the maintenance of antibiotic resistance. Trends Microbiol 2018; 26:677–91. [DOI] [PubMed] [Google Scholar]
- 32. Nair S, Miller B, Barends M, et al. Adaptive copy number evolution in malaria parasites. PLoS Genet 2008; 4:e1000243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cheeseman IH, Miller B, Tan JC, et al. Population structure shapes copy number variation in Malaria parasites. Mol Biol Evol 2016; 33:603–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Taylor AR, Echeverry DF, Anderson TJC, et al. Identity-by-descent with uncertainty characterises connectivity of Plasmodium falciparum populations on the Colombian-Pacific coast. PLoS Genet 2020; 16:e1009101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Griffing SM, Mixson-Hayden T, Sridaran S, et al. South American Plasmodium falciparum after the malaria eradication era: clonal population expansion and survival of the fittest hybrids. PLoS One 2011; 6:e23486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Real E, Howick VM, Dahalan FA, et al. A single-cell atlas of Plasmodium falciparum transmission through the mosquito. Nat Commun 2021; 12:3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Brown AC, Guler JL. From circulation to cultivation: Plasmodium in vivo versus in vitro. Trends Parasitol 2020; 36:914–26. [DOI] [PubMed] [Google Scholar]
- 38. Shafik SH, Cobbold SA, Barkat K, et al. The natural function of the malaria parasite's chloroquine resistance transporter. Nat Commun 2020; 11:3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wicht KJ, Mok S, Fidock DA. Molecular mechanisms of drug resistance in Plasmodium falciparum malaria. Annu Rev Microbiol 2020; 74:431–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kublin JG, Cortese JF, Njunju EM, et al. Reemergence of chloroquine-sensitive Plasmodium falciparum malaria after cessation of chloroquine use in Malawi. J Infect Dis 2003; 187:1870–5. [DOI] [PubMed] [Google Scholar]
- 41. Schrag SJ, Perrot V. Reducing antibiotic resistance. Nature 1996; 381:120–1. [DOI] [PubMed] [Google Scholar]