Summary
During postnatal development, both the maturing microbiome and the host immune system are susceptible to environmental perturbations such as antibiotic use. The impact of timing in which antibiotic exposure occurs was investigated by treating mice from days 5–9 with amoxicillin or azithromycin, two of the most commonly prescribed medications in children. Both early-life antibiotic regimens disrupted Peyer’s patch development and immune cell abundance, with a sustained decrease in germinal center formation and diminished intestinal immunoglobulin A (IgA) production. These effects were less pronounced in adult mice. Through comparative analysis of microbial taxa, Bifidobacterium longum abundance was found to be associated with germinal center frequency. When re-introduced to antibiotic-exposed mice, B. longum partially rescued the immunological deficits. These findings suggest that early-life antibiotic use affects the development of intestinal IgA-producing B cell functions and that probiotic strains could be used to restore normal development after antibiotic exposure.
Subject areas: Microbiology, Developmental biology
Graphical abstract
Highlights
-
•
Early-life antibiotic exposure (ELAE) alters intestinal microbiome composition
-
•
ELAE decreased levels of Bifidobacterium longum
-
•
ELAE negatively impacts intestinal germinal center immune responses
-
•
Restoring B. longum may offset the ELAE negative effects on immunologic development
Microbiology; Developmental biology
Introduction
The natural colonization of infants with commensal microbes is highly associated with crucial gastrointestinal functions including nutrient absorption and development of the mucosal immune system.1,2,3 Although the composition and function of the infant microbiota are subject to multiple factors, including gestational age, mode of delivery, and diet, one of the most important is early-life exposure to antibiotics.4,5,6 Antibiotics can substantially skew infant microbiota composition.7,8 Disturbance of such colonization patterns early in life can have long-lasting host effects including predisposing to the development of immune-mediated diseases, including atopic dermatitis, food allergies, asthma, and type 1 diabetes, in both experimental models and in human children.9,10,11,12,13,14,15
The establishment of stable microbial communities within the gastrointestinal tract during early-life is closely synchronized with host growth and immune development.16,17 The intestine, the site of interaction with both commensal microbiota and ingested antigens and pathogens, represents an important compartment of host immunity. Immunoglobulin (Ig) A is the principal antibody produced at all mucosal sites, including the intestine, and contributes to maintenance of immune homeostasis by binding nutritional and microbial antigens within the gastrointestinal tract.18,19,20
The primary sites for intestinal antigen uptake and presentation in the gut are the Peyer’s patches, which are major components of the gut-associated lymphoid tissue (GALT).21,22 Peyer’s patches are surrounded by a follicle-associated epithelium, which is the interface between the GALT and the luminal microenvironment.22 Early in life, the number of Peyer’s patches is low but increases with immune system maturation.21 Germ-free animals have a reduced number of Peyer’s patches and less than 10% of the number of IgA-producing B cells present in animals with a conventional microbiota.23 Production of all Ig isotypes is reduced in germ-free animals,24 highlighting the importance of the intestinal microbiota in the development of the host GALT and immunity.
Early-life antibiotic exposures during postnatal development have lasting effects on microbiota composition; studies in both humans and mice have found altered microbial composition to be associated with both enhanced disease susceptibility and modified immune status.4,25,26,27,28,29,30,31 We now report a series of five experiments that show that constituents of the early-life microbiota composition are critical to intestinal immune maturation. We found that disruption of the conserved host-microbe cross talk by early-life antibiotic exposure leads to a sustained deficit in GALT-associated tissue morphology and immune function but that there is potential for reversal with specific restoration.
Results
Early-life antibiotic exposure disrupts Peyer’s patch immunological development
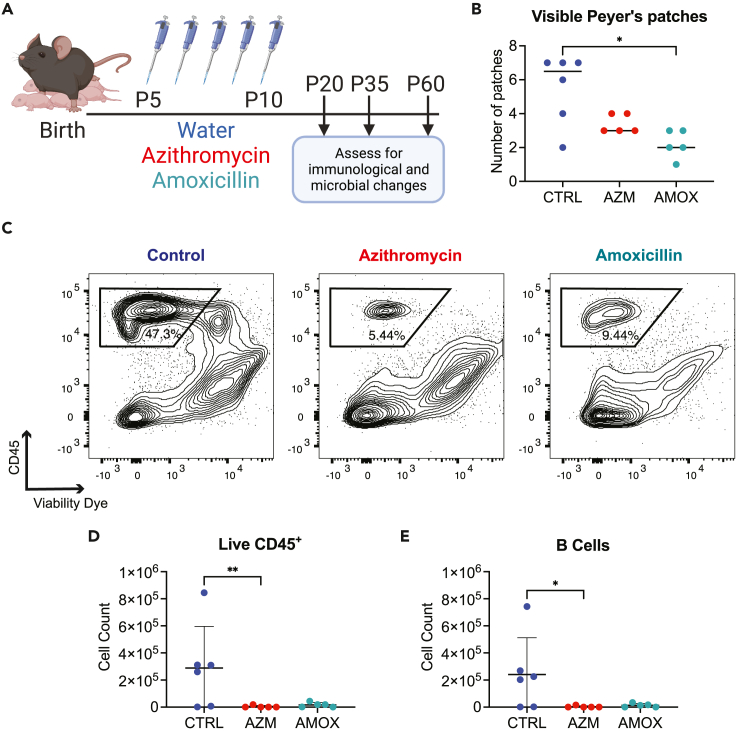
To investigate the impact of early-life antibiotic exposure on gastrointestinal immune development, we first conducted an experiment previously developed for administering pharmacologically relevant doses of azithromycin or amoxicillin to mouse pups via oral administration over a five-day period (Figure 1A).31 Upon our first evaluation of these mice on postnatal day (P) 20 of life, 10 days after antibiotic exposure ceased, we observed fewer intestinal Peyer’s patches were detected in the mice exposed to early-life antibiotics relative to control mice. This difference was statistically significant between control mice and those that received amoxicillin (Figure 1B). Using flow cytometry, we found that by P20, previously antibiotic-exposed mice had dramatically reduced leukocyte (CD45+) populations in the Peyer’s patches (Figure 1C), and this was most evident for B lymphocytes, which normally constitute the majority of the CD45+ population at this mucosal immune site (Figures 1D and 1E). These data indicate that early-life antibiotic exposure interferes with establishment of GALT immune structure and cellularity before weaning age is reached.
Figure 1.
Antibiotic exposure during early life reduces Peyer’s patch hematopoietic cells and lymphocytes at day of life 20
(A) 5-day-old pups were dosed with azithromycin (AZM) (30 mg/kg of body weight), amoxicillin (AMOX) (100 mg/kg of body weight), or water as a control (CTRL). Antibiotics were prepared as a suspension in water and administered orally via a pipette tip once a day for 5 days (volumes were adjusted based on each pup’s body weight to ensure the same dose was given). Mice were sacrificed when they were 20 days old (P20).
(B) Visible Peyer’s patches were counted along the length of the entire small intestine. Single-cell suspensions of Peyer’s patches were prepared for staining with fluorescently tagged antibodies.
(C) Flow cytometry plots showing the P20 Peyer’s patch live CD45+ cell populations after gating on lymphocytes and singlets.
(D) Live CD45+ cell counts from the same representative flow plots shown in panel C.
(E) B cell counts from the same samples; B cells were defined as CD19+B220+ cells. All data shown are from a single representative experiment of two independent experiments; panel B shows the median, and D and E show the mean value ±standard error of mean (SEM); ∗ indicates p < 0.05, ∗∗p < 0.01.
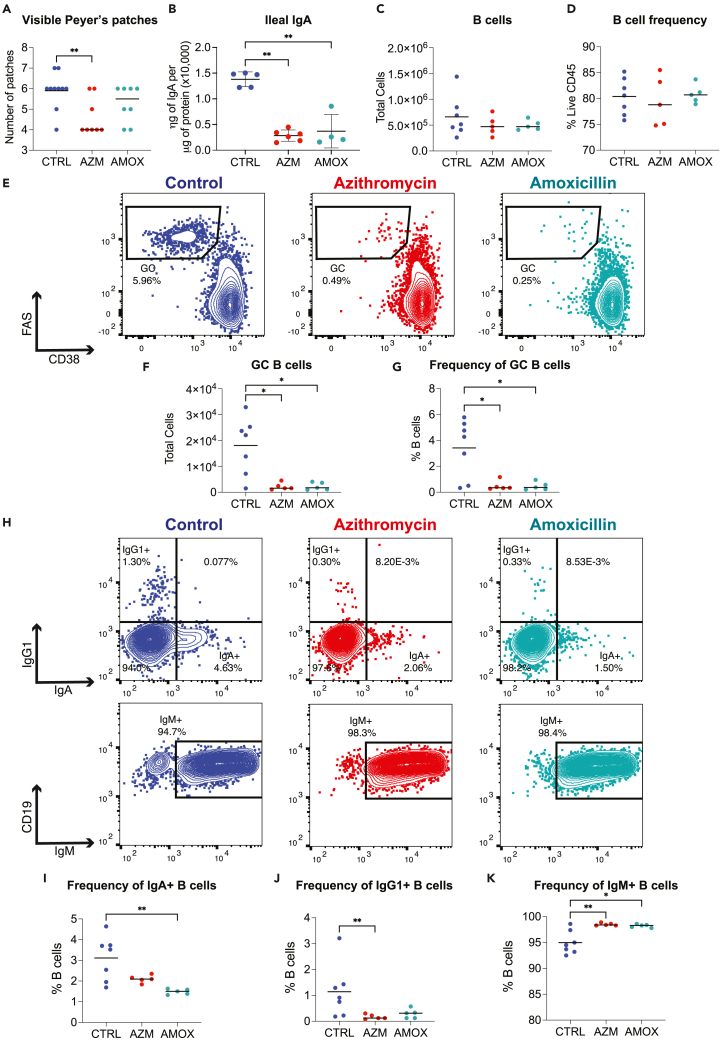
In a second set of experiments, we sought to ascertain the duration of these antibiotic effects by looking at a later point in murine development. To address this question, we repeated the experimental design outlined in Figure 1A and euthanized mice to evaluate their immune status at P35 and P60. Data in Figure 2 summarize experiments of mice euthanized on P35 from one representative experiment of three. At P35, the number of visible Peyer’s patches in the azithromycin-treated mice remained significantly reduced relative to control animals; however, there was greater recovery of visible Peyer’s patches in those that had been similarly exposed to amoxicillin (Figure 2A). Both groups of antibiotic-exposed mice had significant reductions in IgA levels in ileal tissues relative to control animals (Figure 2B), but total Peyer’s patch cell count and B cell frequency were no longer affected by the antibiotic exposure (Figures 2C and 2D), and data not shown). However, germinal center B cells in the antibiotic-treated mice were dramatically reduced in both total count and frequency (Figures 2E–2G) consistent with the reduced IgA levels in the ileum (Figure 2B). Next, to determine the impact of the reduced germinal center formation, we assessed for changes in immunoglobulin class switch recombination (CSR) in the Peyer’s patch B cells of antibiotic-treated mice, using intracellular flow cytometry with staining for IgM, IgG1, and IgA. We found that the antibiotic-treated mice had reduced CSR based on lower IgA and IgG1 expression and significantly increased IgM expression in B cells (Figures 2H–2K). To assess impact on CSR in more distant lymphoid tissue, we examined the mesenteric lymph node B cell populations (Figures S1A–S1D). As there were essentially no differences, we concluded that the CSR changes did not extend beyond the Peyer’s patches.
Figure 2.
Germinal center formation is altered at day of life 35 after early-life antibiotic exposure
At P35, mice were sacrificed, and Peyer’s patches were homogenized into single-cell suspensions and lymphocytes characterized using flow cytometry.
(A) Visible Peyer’s patches along the length of the small intestine were counted.
(B–D) IgA was quantitated in ileal tissue using an IgA-specific ELISA and normalized to total protein levels in the sample, as determined by BCA assay. Abundance and proportions of (C, D) B cells were assessed by flow cytometry.
(E–G) Representative flow cytometry plots of Peyer’s patch germinal center (GC) B cells and (F, G) summary data for GC B cells counts and frequency are plotted. Peyer’s patch single-cell suspensions were fixed and permeabilized after surface staining to permit intercellular staining of IgM, IgG1, and IgA.
(H–K) Representative flow plots for intracellular Ig staining. Summary plots for percent B cells positive for (I) IgA, (J) IgG1, or (K) IgM. All data shown are from a single representative experiment of three independent experiments; plot A shows the median, and all other plots show the mean value ±SEM; ∗ indicates p < 0.05, ∗∗p < 0.01.
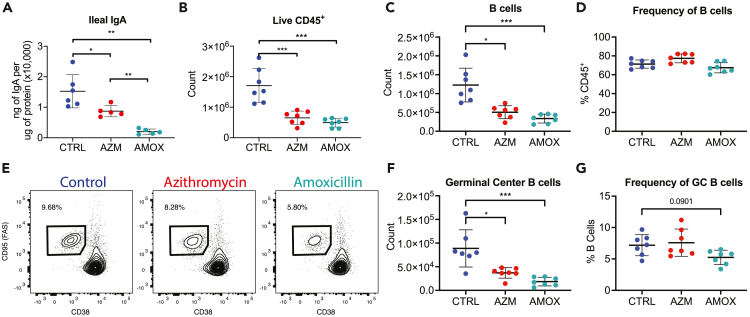
When we assessed mice at P60, 50 days after cessation of the antibiotic treatment, we found that early-life exposure to antibiotics had an enduring impact on GALT composition and function. Both azithromycin and amoxicillin resulted in reduced production of ileal IgA (Figure 3A) as well as decreased abundances of Peyer’s patches, B cells, and germinal center B cells (Figures 3B and 3C). Consistent with the P35 data, there was minimal impact on the frequency of B cells in either group of antibiotic-exposed mice (Figure 3D). There was a marked reduction in germinal center B cell count (Figure 3F), and in the amoxicillin-exposed mice a reduced frequency of germinal center B cells remained; however, this no longer reached statistical significance (Figures 3E–3G). Thus, these experiments showed persistent changes through P60 in Peyer’s patch germinal center formation and immune cell populations following early-life exposure to each of the two antibiotics, with clear functional deficits in IgA production.
Figure 3.
Peyer’s patch germinal center deficits persist for at least 7 weeks after antibiotic exposure
(A–D) Mice were euthanized at P60, IgA was quantitated in ileal tissue (A), and Peyer’s patches were evaluated using flow cytometry. Plots show absolute abundance of (B) live CD45+ cells and (C) B cells, (D) frequency of B cells.
(E–G) Representative flow cytometry plots for Peyer’s patch GC B cell populations and summary plots for GC B cell (F) counts and (G) proportions. All data shown are from a single representative experiment of two independent experiments; plots show the mean value ±SEM; ∗ indicates p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
To further evaluate the impact of early-life antibiotic exposure on GALT immunity, we studied mesenteric lymph nodes at P35 (Figures S2A–S2J) and at P60 (Figures S2K–S2P). At P35, in the antibiotic-exposed mice, trends similar to those described above in the Peyer’s patches were observed, with reduced leukocyte counts (Figure S2B) and reduced total B cell numbers and germinal B cell populations, as well as reduced T lymphocyte counts (Figures S2D, S2F, S2H, and S2J). At P60, consistent with observations in Peyer’s patches (Figures 3E–3G), we found reduced frequency of germinal center B cells in the mesenteric lymph nodes of amoxicillin-exposed mice (Figure S2O). This was the only immune perturbation we measured in the mesentric lymph nodes (MLNs) as the other immune changes detected at P35 had all recovered by P60. These studies indicate that the changes in GALT immunity associated with early-life antibiotic exposure were related to germinal center formation and that the effects were most durable after the amoxicillin exposure.
To assess changes in peripheral B cell immunity, splenic B cells were analyzed at P35; no differences in total leukocyte or lymphocyte populations were observed in this secondary lymphoid organ (Figure S3). However, we found that serum IgA levels were reduced at P35 in both groups of mice exposed to early-life antibiotics (Figure S3B). Evaluating follicular (FO) and marginal zone (MZ) B cells in spleen at P35, we found that the antibiotic exposure did not lead to any significant differences in these B cell compartments (Figures S4A–S4C). These data provide evidence that the observed immune changes after the early-life antibiotic treatment were largely restricted to the GALT and did not reflect a more substantial systemic immune perturbation.
Next, in the third series of experiments, we asked whether the timing of the antibiotic exposure was critical for the observed differences in immunological development. To do so, we studied adult mice (6 weeks of age) that were born in the same facility as the young experimental mice and exposed them to the same antibiotics at the same levels normalized for total body weight, for the same duration (five days), by oral gavage. In contrast to the observed changes in immune composition of animals treated as neonates, we found no significant differences in lymphocyte populations 25 days after the antibiotic exposure ended in adult mice (Figures S5A–S5I). One antibiotic effect observed was that mice that were exposed to azithromycin as adults had significantly (∼25%) reduced serum IgA levels (Figure S5M), which was less than the 50% reduction observed in young mice also 25 days after antibiotic exposure (Figure S2B). Exposure to either antibiotic in adult mice resulted in a decrease in ileal IgA levels, but neither was significantly less than control (Figure S5N). Taken together, these results indicate that the immunological effects initiated by antibiotic exposure are substantially greater in mice exposed to antibiotics at a young age. This supports the hypothesis that a window of immunological development exists in which the host has enhanced susceptibility to antibiotic-induced perturbations.31,32
Early-life antibiotic exposure disrupts gut microbiota
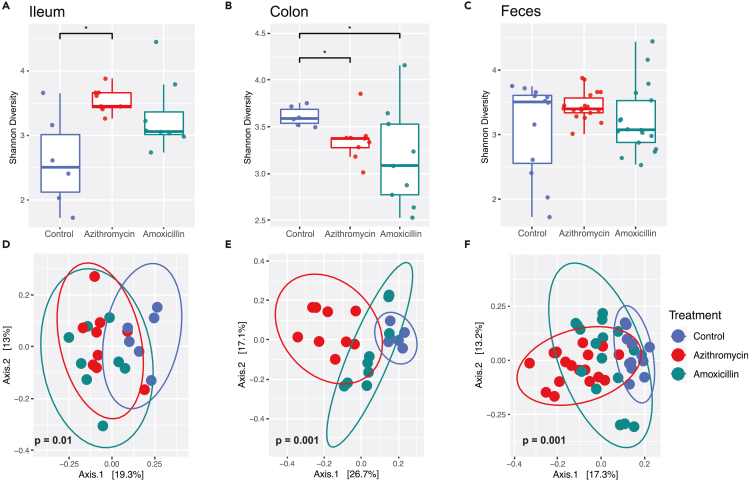
Given the substantial impact of early-life antibiotic exposure on GALT humoral immunity, we next sought to evaluate the changes induced in the intestinal microbiota at P35. Examining microbial populations using 16S rRNA sequencing revealed alpha-diversity (Shannon) scores were increased in the ileum of azithromycin-exposed mice (Figure 4A), reduced in the colon for both groups of antibiotic-exposed mice (Figure 4B), and unchanged in the feces (Figure 4C), relative to control mice. In all three sites, microbial population structures associated with treatment were distinct from control, as determined by unweighted UniFrac analysis (Figures 4D–4F). These results indicate that a brief therapeutic course of antibiotics early in life results in sustained changes in microbial composition across intestinal sites 25 days after the exposure ended.
Figure 4.
Antibiotic exposure alters intestinal microbiota composition
(A–F) Intestinal tract samples were collected from mice at sacrifice at P35, and fecal samples were collected after weaning at P25. DNA was extracted from the intestinal and fecal samples and subjected to 16S rRNA sequencing. Shannon diversity, an alpha-diversity metric, was calculated for samples from (A) ileum, (B) colon, and (C) feces. Unweighted UniFrac, a beta diversity metric is plotted for (D) ileum, (E) colon, and (F) the feces. Statistical significance for Shannon diversity was determined using a non-parametric t test. Histogram shows 95% confidence interval with a line at the mean; ∗ indicates p < 0.05. Comparisons of community structures from control and antibiotic-exposed mice were determined using an Adonis test in R using the Phyloseq and Vegan packages. For each sampling site, the percent sample variation explained by antibiotic treatment was 23.6% (ileum); 37.8% (colon); and 46.4% (fecal). Ellipses show 95% confidence interval for the sample group. p values indicating statistical significance are shown on the corresponding principal component plots.
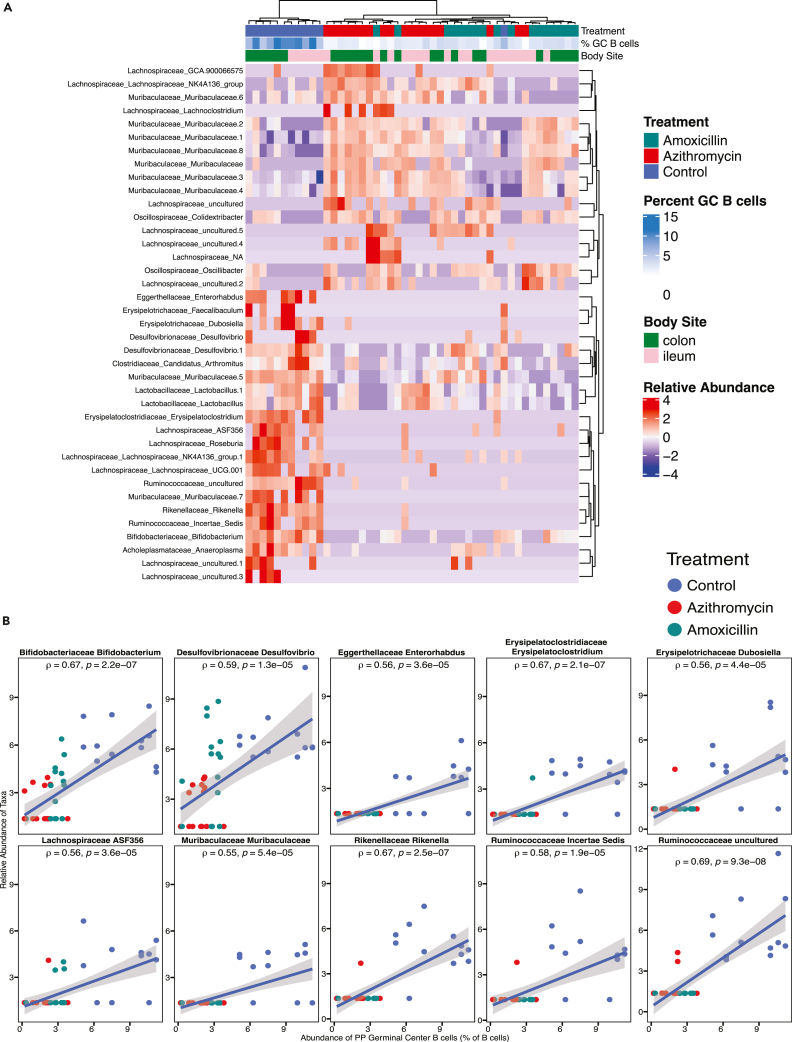
Next, we asked whether any specific intestinal microbiota changes were associated with the disruption of the Peyer’s patch immune cell populations. We used DESeq2 to identify amplicon sequence variants (ASVs) significantly associated with germinal center B cell frequencies by comparing the control mice to the azithromycin- or amoxicillin-exposed mice (p value <0.01). Unsupervised hierarchical clustering of the samples showed differences at P35 related to the experimental group and germinal center B cell frequency (Figure 5A). The segregation shows a set of organisms whose relative abundance is positively associated with germinal center B cell frequency and depleted in antibiotic-exposed mice relative to control mice. Conversely, a set of organisms whose relative abundance is negatively associated with germinal center B cell frequency is enriched in antibiotic-exposed mice (Figure 5A). While comparison between samples from control animals and antibiotic-treated mice reveals the clearest ASV differences, dissimilarities also segregate the amoxicillin- and azithromycin-exposed groups into distinct clusters (Figure 5A). Using a stricter statistical threshold to identify specific taxa (p < 0.0001, rho >0.5), we found 10 unique ASVs strongly correlated with increased germinal center B cell frequency (Figure 5B). These ASVs are all found to be enriched in control mice and positively correlated with germinal center B cell frequency. These data raise the question as to whether or not these strains have immunomodulatory effects that could be exploited to restore germinal center formation after antibiotic exposure.
Figure 5.
ASVs that correlate with Peyer’s patch germinal center B cell frequency
(A) Spearman correlations were performed to identify ileal and colonic ASVs at P35 that significantly correlate with Peyer’s patch germinal center B cell frequencies at p value <0.01. Samples are color-coded by treatment group, a scale is used to display the percent of germinal center B cells, and ASVs are shown in a heatmap with unsupervised hierarchical clustering. The heatmap shows a clear segregation of ASVs that are largely absent in control mice (the top 17 ASVs) and highly abundant in control mice (the bottom 22 ASVs).
(B) Spearman correlation plots are shown for ASVs that significantly correlated with germinal center B cell frequency at p value <0.0001. Rho (ρ) and p value are reported for each ASV, and each dot represents a unique colon or ileal sample color-coded by treatment group. Dataset includes 47 intestinal samples from 24 unique mice.
Restoration of Peyer’s patch development after antibiotic treatment
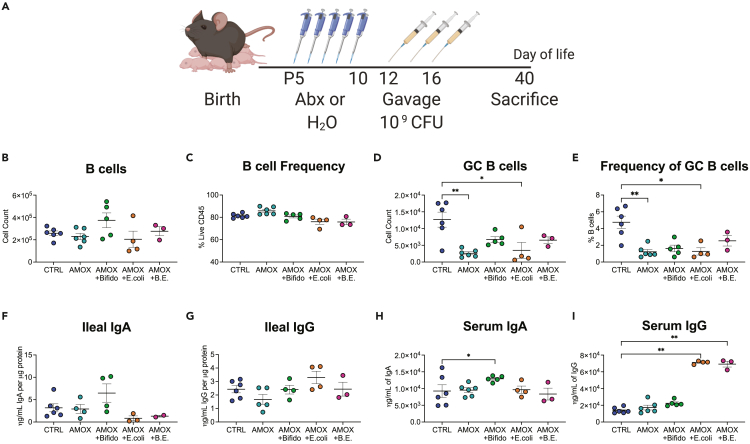
To assess whether we could restore geminal center formation in antibiotic-exposed mice, we studied Bifidobacterium species, organisms whose abundance highly correlated with Peyer’s patch germinal center frequency (Figure 5B), that were substantially depleted by the antibiotics (Figure S6) and that are known to be important early-life commensals in both mice and humans.33,34 In a fourth set of experiments, we used B. longum isolated from human infant stool.4 We chose to test B. longum as Bifidobacterium species are commonly used as probiotic strains in both humans and mice.35,36,37 We introduced 109 colony-forming unit (CFU) of the B. longum strain to the amoxicillin-exposed pups via gavage three times starting on P12 (72 h after the last antibiotic dose) to determine whether restoring depleted bacteria would be sufficient to restore Peyer’s patch immune development to the level of control mice (Figure 6A). For this experiment, we used early-life amoxicillin-treated hosts since amoxicillin exposure had the more consistent and sustained impact on GALT immunity. We used Escherichia coli MG1655 as a control organism since E. coli was not identified (Figure 5) to significantly correlate with germinal center B cell frequency and because it is a proficient colonizer of the murine intestine.38 An unweighted UniFrac analysis, performed on the fecal samples from the experimental and control mice (Figure S7), revealed that the mice gavaged with B. longum, E. coli, or both were still distinct in composition from control mice.
Figure 6.
Bifidobacterium longum can partially restore germinal center B cells following antibiotic exposure
(A–E) 5-day-old pups were dosed with amoxicillin (100 mg/kg of body weight) or water as a control by oral ingestion. Aged-matched amoxicillin-exposed mice were mixed at birth and distributed across four litters. On P12, control and some amoxicillin-exposed mice were gavaged with anaerobic transport medium, while other litters of amoxicillin-exposed mice were gavaged with 5–10 × 109 CFU Bifidobacterium longum in anaerobic transport media, 5–10 × 109 CFU Escherichia coli in LB broth, or a mixture of both bacteria gavaged separately. Mice were gavaged two more times on P14 and P16. Mice were sacrificed 24 days after the last gavage, at P40, and (B) absolute counts and (C) frequency of B cells and (D, E) germinal center B cells isolated from the Peyer’s patches of experimental mice.
(F–I) Ileal IgA, (G) Ileal IgG, (H) serum IgA, and (I) serum IgG were quantified using IgA- or IgG-specific ELISAs. Data shown are from a single experiment; a one-way parametric ANOVA was used to test for significance, and all groups were compared to both control and amoxicillin-exposed mouse groups, plots show the mean value ±SEM; ∗ indicates p < 0.05, ∗∗p < 0.01.
Analysis of immune subsets in the GALT showed that B lymphocyte abundances and frequencies in Peyer’s patches were variable (Figures 6B–6E). No treatment group was significantly different from the control group, but the two groups of mice that received E. coli after amoxicillin treatment had lower levels of B cells compared to those that received amoxicillin alone (Figure 6C). As expected from our earlier experiments (Figure 1, Figure 2, Figure 3), mice receiving early-life amoxicillin treatment had significantly fewer germinal center B cells in their Peyer’s patches relative to control mice (Figures 6D and 6E). Introduction of E. coli alone did not change this significant decrease. (Figures 6D and 6E). In comparison, after inoculation with B. longum, the significant decrease in germinal center B cell abundance and frequency associated with antibiotics was partially reversed (Figures 6D and 6E); this was the first indication that gavaging with B. longum might have a beneficial immunological effect in this model. In the mice exposed to amoxicillin, reconstitution with B. longum significantly increased serum IgA compared to control (Figure 6H), with a trend toward increased ileal IgA (Figure 6F). In contrast, amoxicillin-exposed mice gavaged with E. coli had significant increases in serum IgG levels (Figure 6I) and a slight increase in ileal IgG that did not reach statistical significance (Figure 6G), but not in IgA (Figures 6F and 6H). Taken together, these data indicate a partial restorative effect of B. longum on germinal center B cells and IgA production and increased IgG from the E. coli inoculations.
Human intestinal microbiota composition modulates germinal center B cell frequency and abundance
Finally, we sought to examine if intestinal microbial composition is sufficient to determine the local immune landscape. To address this question, in a fifth set of experiments, breeder pairs of germ-free C57BL6 mice were inoculated with fecal samples from seven human donors varying in age. These conventionalized breeders were established in cages based on the donor inoculum they received, and the resultant F1 progeny were evaluated for association between intestinal microbial composition and GALT immune landscape. We also examined the impact of introduction of diverse human microbiota on circulating antibody titers. Select pathogen free (SPF) mice that possess a murine microbiota were used as immunological controls for these experiments. Representation of the abundances of taxa present in the F1 progeny (Figure S8A) indicates the diversity in the human-derived and control SPF microbiota. Although there is heterogeneity within the fecal samples of mice that received the same inoculum (intragroup diversity), contrasting the 8 groups shows that the inter-group diversity (between donor inocula) is much greater. Compared to SPF mice, the recipient mice showed heterogeneity in number of live CD45+ cells within Peyer’s patches (Figure S8B). Frequencies of T and B lymphocytes within Peyer’s patches were mostly similar regardless of donor (Figures S8C and S8D). Germinal center B cell frequency showed the greatest heterogeneity across human donors relative to SPF (Figure S8E). These data underscore the importance of microbial composition in influencing development of germinal centers.
We next asked whether human-associated ASVs were significantly correlated with germinal center B cell frequencies. By unsupervised hierarchical clustering, the samples show strong alignment according to fecal donor source (Figure S9). Consistent with the earlier murine data (Figure 5), the commensal organisms Lachnospiraceae, Desulfovibrionaceae, Erysipelotrichaceae, and Ruminococcaceae were positively associated with germinal center B cell frequency (Figure S9). These results indicate that overlapping organisms in both human and mouse fecal samples had similar associations with Peyer’s patch germinal center B cell frequency.
Discussion
The presented experiments explore the relationship between early-life microbiota and GALT immunological development. We first showed that administration of clinically relevant antibiotic regimens during early-life leads to disruption of the developing microbiome and altered GALT development. Both the antibiotic exposures and the age of the mouse during the exposure influenced the magnitude of the disruption. Our experiments across multiple animal facilities and cohorts of mice indicated that Peyer’s patches were the site of greatest disruption with germinal center B cells consistently affected. As expected,31,39,40 early-life antibiotic exposure altered the composition of the intestinal microbiota relative to control mice (Figure 4), which may be the intermediate mechanism by which two antibiotics with differing antimicrobial spectra induced similar immunological changes. These data are consistent with prior work indicating that early-life antibiotic exposures greatly influence host immunity.13,27,29,31,41 Some of the experimental replicates of these studies were conducted in parallel to the studies reported in Borbet et al. (2022),31 which provides experimental consistency between the two bodies of work. However, the datasets in each of the studies were non-overlapping and unique. Importantly, we found that the effects are specific to early life because analogous antibiotic treatment of adult mice did not result in disruption of the GALT-based immunity. These findings, consistent with prior work,13,17,24,42 indicate a critical early-life window for GALT immunological development, with a finite window of vulnerability to perturbation.
Our results suggest that the composition of the early-life microbiota is an important regulator of germinal center and GALT immunity, a hypothesis supported by the finding that individual human fecal donors differ in their ability to induce germinal center B cells after inoculation of germ-free mice (Figure S8). These fecal-transfer experiments provide a link between intestinal microbiota composition and regulation of Peyer’s patch immune development. GALT function and germinal center formation can be modified by differing fecal microbiota compositions, and these data indicate that human-derived fecal microbes contain taxa that influence germinal center reactions.
Our analysis of animals that were exposed to antibiotics early in life revealed several ASVs significantly correlated with germinal center B cell frequencies (Figures 4 and S9). Furthermore, the observation that gavage with B. longum following antibiotic exposure could influence Peyer’s patch cell populations (Figure 6) provides further evidence that early-life microbial composition is an important determinant of immune development. Together with the taxonomic data (Figure 5), the restorative impact of B. longum on germinal center formation is consistent with the broader goal of restoring normal immune development following early-life antibiotic exposures.
The loss of germinal center B cells we identified is consistent with prior studies in mice showing decreased serum and stool IgA levels following early-life antibiotic exposure.32 Antibiotic exposures have been shown to reduce the levels of short-chain fatty acids (SCFAs) in both humans and mice.27,43,44,45 SCFAs support B cell differentiation of plasma cells in mice, indicating the potential involvement of microbial metabolites in regulating germinal center responses.46 In other studies, antibiotic exposure before weaning lowered antibody titers in mice vaccinated with mycobacterial (Bacille Calmette-Guérin vaccine), pneumococcal (Pneumococcal conjugate vaccine 13), meningococcal (serogroup B or C) antigens or the Infanrix six antigens but had no effects on antibody responses to vaccination in adult mice,41 a finding consistent with antibiotics influencing humoral immune responses in an age-dependent manner. Our results provide further mechanistic understanding of the early-life window of vulnerability related to B cell response maturation.
The benefits of B. longum in early-life immune development are complementary to reports that reduced Bifidobacterium in human infants correlates with increased inflammation in the intestine and systemically, with increased CD4+ T cell differentiation to Th2 and Th17 cell types.36,37,47 When children were given Bifidobacterium for 21 days, fecal levels of Bifidobacterium were slightly increased and total and poliovirus-specific IgA in the feces were elevated,48 consistent with immune adjuvancy. In other experiments, administering Bifidobacterium led to suppression of Th2 and enhanced Treg responses in an ovalbumin-model of food allergy49 and increased levels of IgA in lactating mice.35 When breastfed infants were given Bifidobacterium infantis supplementation (a subspecies of B. longum) during the first 60 days of life, they had more immunoregulatory cells including Tregs, increased interleukin-10 (IL-10) production, and reduced Th2/Th17 induction compared to infants who did not receive the supplementation.37 These changes were linked to the increased abundance of genes involved in human milk oligosaccharide (HMO) metabolism in the microbiota of these infants.37 Bifidobacterium species, both in the gut of infants and in culture, metabolize HMOs to lactate and SCFAs, particularly acetate.37,50,51,52,53 Acetate promotes IgA intestinal responses by facilitating vitamin A synthesis by gut epithelial and dendritic cells,54,55 SCFAs promote Treg induction, which would increase IL-10 and transforming growth factor β (TGF-β) levels, cytokines that promote B cell class CSR to IgA.19,54 Therefore, we suggest that Bifidobacterium metabolites may be responsible for promoting gut IgA responses in infants. This point is further supported by a recent study of Canadian children that found antibiotic use during infancy was associated with increased incidence of asthma. This was not the case if the children were breastfed, as breastfeeding correlated positively with both fucosylated HMO levels in the milk and B. longum abundance in feces.56,57 Our current findings provide further support for the immunomodulatory effects of Bifidobacterium species, particularly pertaining to humoral immune responses in the gut, and specifically their relationship to germinal center formation.
Separate from the results with Bifidobacterium, we had differing results when E. coli alone, or in conjunction with B. longum, was used for restoration. Among other Proteobacteria, Escherichia species are early colonizers of the mammalian intestine when oxygen levels are high in the first weeks of life.47,58,59 Later blooms of the Enterobacteriaceae are an indication of microbial dysbiosis,4,59 which has been positively associated with both metabolic diseases60 and intestinal disease states.61,62 In our studies, we observed increased circulating IgG levels in mice that received E. coli alone or E. coli and B. longum together, with no discernable impact on IgG in animals exposed to B. longum only. Anti-commensal IgG responses,63 and IgG+ plasma cells,64 have been associated with intestinal inflammation. In our model, we have not shown that the antibody responses are commensal specific but that exposure to E. coli or related species in this early-life window appeared to exert an opposing effect to B. longum by initiating IgG responses. This may be indicative of a more inflammatory, or at least, less beneficial outcome.
Collectively, the results from these five experiments indicate a crucial role microbial composition plays in early-life regulation of germinal center induction and IgA levels and provide further evidence that B. longum may play a role in the process. Restoration of intestinal microbes following antibiotic exposure may have benefits for the evolution of B cell responses, and these results suggest that a closer look at other probiotic strains and combinations to restore GALT immune development following early-life antibiotic exposure is necessary. Further studies are needed to determine the precise windows of vulnerability for detrimental impacts of antibiotics and to define opportunities for restoration of immune tone in patients. It is also critical to dissect the precise mechanisms underlying the immune changes following antibiotic regimens, to leverage restoration as a translational intervention in the future.
Limitations of the study
Some limitations for this work include the need to test a greater range of time periods during early life in which the murine host is susceptible to antibiotic perturbation, as well as to test more probiotic strains and replicating these experiments in differing groups of mice. Some limitations for our flow cytometric analysis of Peyer’s patches in these experiments involved pooling 4–6 patches from each individual mouse, so we are unable to dissect compartmentalized contributions that may vary among different sites along the small intestine. The antibiotic exposures used in these experiments, which show parallel disruption of Peyer’s patch immune development, have reproducible impact on the germinal center responses in different animal facilities and with differing batches of parental mice. This point is significant because the parental/founder microbiomes differ across facilities and groups of mice, but the consistent phenotype suggests that multiple taxa may be involved in the microbe-host interactions that influence early-life gut immune education. However, analysis of our results is limited in that some of the immunomodulatory effects associated with antibiotic exposure appear susceptible to differences in the founding microbiota as observed in Figures 6F and 6G where we did not observe the amoxicillin-driven IgA deficits we had seen in other animal facilities. This suggests that particular aspects of microbe and Peyer’s patch immunomodulation remain uncharacterized in these studies. This phenomenon could involve redundant antigens or metabolites present in multiple bacterial strains that likely are involved in intestinal immune development.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-mouse CD45 BV650 (30-F11) | Biolegend | 103151 |
| anti-mouse CD45 PerCPCy5.5 (30-F11) | BD | 550994 |
| anti-mouse/human CD45R/B220 PerCPCy5.5 (RA3-6B2) | Biolegend | 103234 |
| anti-mouse CD45R/B220 APC (RA3-6B2) | eBioscience | Cat # 17-0452-81 |
| anti-mouse CD45R/B220 PE-Cy7 (RA3-6B2) | eBioscience | 25-0452-82 |
| anti-mouse IgG1 APC (A85-1) | BD | 560089 |
| anti-mouse IgA PE (mA-6E1) | eBioscience | 12-4204-82 |
| Fab Anti-Mouse IgM FITC | Jackson Immuno Research Labs | 115-097-020 |
| anti-mouse CD95 (FAS) FITC (SA367H8) | Biolegend | 152606 |
| anti-mouse CD38 PE (90) | Biolegend | 102707 |
| anti-mouse CD3 BV785 (17A2) | Biolegend | 100232 |
| anti-mouse CD3 BV510 (145-2C11) | BD | 563024 |
| anti-mouse CD19 BV510 (6D5) | Biolegend | 115546 |
| anti-mouse CD19 e450 (1D3) | eBioscience | 48-0193-80 |
| anti-mouse CD2⅓5 APC/Cy7 (7E9) | Biolegend | 123417 |
| anti-mouse CD23 PE/Cy7 (B3B4) | eBioscience | 25–0232082 |
| Zombie UV Fixable Viability Dye | Biolegend | 423108 |
| Bacterial Strains | ||
| Bifidobacterium longum | Blaser Lab; Bokulich et al. (2016)4 | HMXZ001 |
| Escherichia coli | Dr. Giulio Quarta | MG1655 |
| Biological Samples | ||
| Mouse fecal pellets | This study | |
| Mouse ileal samples | This study | |
| Mouse cecal samples | This study | |
| Mouse colonic samples | This study | |
| Mouse serum samples | This study | |
| Human fecal samples | Blaser Lab | |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Amoxicillin | Sigma Aldrich | A8523 |
| Azithromycin Oral Suspension | Teva | 00093-2026-23 |
| Anerobic transport medium | Anaerobic Systems | AS-911 |
| Bifidobacterium selective agar | Anaerobic Systems | AS-6423 |
| Bifidus Selective Medium | Sigma Aldrich | Cat # 90273 |
| Critical Commercial Assays | ||
| DNeasy PowerSoil-HTP 96 Well Soil DNA Isolation Kit | Qiagen | Cat # 12955-4 |
| DNeasy PowerLyzer Powersoil kit | Qiagen | Cat # 12855-100 |
| Quant-iT PicoGreen dsDNA assay kit | Life Technologies | Cat #P11496 |
| Qiaquick PCR purification kit | Qiagen | Cat # 28104 |
| IgA Mouse Uncoated ELISA Kit | Invitrogen | Cat # 88-50450-77 |
| IgG (Total) Mouse Uncoated ELISA Kit with Plates | Invitrogen | Cat # 88-50400-22 |
| BCA Assay | Pierce | Cat # 23225 |
| Deposited Data | ||
| 16S rRNA data | This study | Early-life microbiome composition and GALT immune development - ID 14759 |
| Experimental Models: Organisms/Strains | ||
| C57BL/6J Mice | The Jackson Laboratory | Strain #:000664, RRID:IMSR_JAX:000664 |
| C57BL/6JRj Mice | Javier | |
| C57BL/6NTac | Taconic Bioscience | C57BL/6NTac Germ Free |
| Oligonucleotides | ||
| V4 16S rRNA Universal Primers | Walters et al. (2016)65 | F515/R806 |
| Software and Algorithms | ||
| DESeq2 package | Love et al., (2014)66 | version 1.30.1 |
| QIIME2 | Bolyen et al., (2019)67 | QIIME2-2022.2.1 |
| Prism 9 for macOS | GraphPad Software | Version 9.5.1 |
| qiime2R | Bisanz et al., (2018)68 | version 0.99.6 |
| vegan | Oksanen et al., (2022)69 | Version 2.5–7 |
| ComplexHeatmap | Gu et al., (2016)70 | Version 2.6.2 |
| MAAFT | Katoh et al., (2013)71 | Version 7 |
| DADA2 | Callahan et al., (2016)72 | version 0.99.8 |
| FlowJo | BD | Version 10.8.1 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Martin J. Blaser (martin.blaser@cabm.rutgers.edu).
Materials availability
B. longum HMXZ001 will be made available upon request; however, there are restrictions on HMXZ001 due to Material Transfer Agreement (MTA).
Experimental model and study participant details
Mice
C57BL/6J mice were procured from Jackson Laboratories at New York University School of Medicine or Javier (France) for conducting experiments at the University of Zurich. C57BL/6 germ-free mice were acquired from Taconic Biosciences and housed at the germ-free facility at NYU School of Medicine. The isolator underwent monthly bacterial contamination evaluations using quantitative PCR for bacterial 16S rRNA. The mice were provided ad libitum access to food and water and were maintained on a 12-h light/dark cycle and given rodent standard lab chow. For germ-free mice, all food and water were sterilized via autoclaving. All mouse experiments followed federal and institutional regulations and were approved by the New York University Langone Institutional Animal Care and Use Committee (IACUC protocol IA16-00785). Similarly, the University of Zurich’s animal experimentation procedures were reviewed and approved by the Zurich Cantonal Veterinary Office (licenses ZH170/2014 and ZH086/2020).
Isolation and culture of B. longum
The B. longum strain, referred to as HMXZ001, utilized in this study, was obtained from a frozen stool sample from a 6-month-old infant.4 The fecal sample was diluted and streaked on Bifidus Selective Medium and maintained in anaerobic conditions at 37°C for 48 h. The pure cultures were obtained by picking single colonies and re-streaking. The bacteria was identified using microscopic morphology traits and PCR confirmation with a targeted Bifidobacterium genus-specific 16S rRNA primer pair (5′CTCCTGGAAACGGGTGG-3′/5′ GGTGTTCTTCCCGATATCTACA-3′).73 The 16S rRNA PCR product was sequenced and NCBI BLAST was utilized to confirm that this strain belonged to the B. longum species. Subsequently, B. longum was cultured on Bifidobacterium selective agar (Anaerobic Systems) in an anaerobic chamber at 37°C, and agar plates were passaged every 48–72 h.
Culture of E. coli
The strain MG1655 was grown on LB agar plates at 37°C overnight (10–12 h) or in liquid broth for 3–8 h and maintained in a logarithmic phase.
Method details
Antibiotic treatment
Between days 5 and 10 after birth, mice were administered either sterile water as a control, or an oral suspension of either 30 mg/kg of azithromycin (Teva) or 100 mg/kg of amoxicillin (Sigma). The desired daily mg/kg dose was achieved by adjusting the volume of antibiotic suspension based on the weight of each mouse. To administer the dose, a 10uL pipette was used, and the pups consumed the suspension by suckling from the pipette tip. It was ensured that the antibiotic suspension was well-resuspended before treating each pup, and a new suspension was prepared every two days. These experiments were conducted in different rooms at the NYU animal facility and the University of Zurich to avoid colony-specific phenotypes. The dams were not treated during these experiments, and whenever feasible, age-matched pups were intentionally mixed between dams before antibiotic treatment on day 5 to avoid litter-driven effects.
Gavage of antibiotic treated mice
B. longum was collected from two selective agar plates after 48 h of growth and transferred into 1 mL of anerobic transport medium (Anaerobe Systems). The bacteria-containing medium was then drawn into a syringe within the anaerobic chamber and transported directly on ice to the animal facility for gavage. For the E. coli strain MG1655, one plate was grown on LB agar plates at 37°C overnight (10–12 h), and the cells were collected into 3 mL of LB broth for gavage. Pure cultures of bacteria were gavaged using autoclaved 24G gavage needles. To determine CFUs, dilutions of inoculum from 106 to 1010 were plated and colonies were counted after 48 h for B. longum and 12–16 h for E. coli.
Fecal transfer to germ-free mice
To prepare the human fecal samples for transfer, 50 mg of fecal matter was added to 1 mL of Beef Broth medium (10.0g Beef Extract, 10.0g Peptone, 5.0g NaCl in 1.0L of water, pH 7.2) supplemented with 20% sterile glycerol. The fecal matter was disrupted by vortex-mixing with a 1 mL sterile syringe without a needle attached. The mixture was then divided into 250uL aliquots and immediately frozen.
When the mice were ready to receive the fecal samples, the samples were thawed on ice and diluted 1:10 in fresh Beef Broth medium without glycerol. The samples were then gavaged into germ-free mice using disposable 18G gavage needles (Fine Science Tools). Fecal samples were collected periodically from the recipient mice and their offspring to assess the engraftment and stability of the microbial community over time.
Flow cytometric analysis of mouse tissues
Spleens, mesenteric lymph nodes, and Peyer’s patches were removed from mice, and then homogenized over a 70-micron nylon mesh in RPMI medium supplemented with 10% FCS, penicillin, and streptomycin. To remove red blood cells from spleens, ACK lysis was performed using a solution containing 150 mM ammonium chloride, 10 mM potassium carbonate, and 0.1 mM EDTA. The resulting cell suspensions were stained with fluorescently tagged antibodies and analyzed using a BD LSRII flow cytometer at the NYU School of Medicine Flow Cytometry Core or a BD Fortessa at the University of Zurich Flow Cytometry Core. Data analysis was performed using FlowJo software (v10.8.1, BD).
Immunoglobulin quantitation
To determine mouse IgA and IgG levels, we used antibody specific ELISAs (Invitrogen). We measured immunoglobulin levels in both serum and ileal tissue homogenate. The immunoglobulin concentrations in the ileal tissue homogenate were normalized to the total protein levels in the sample, which were quantified using the BCA assay.
DNA isolation and 16S rRNA library preparation
The DNA was extracted from gastrointestinal samples using either the DNeasy PowerSoil HTP 96 Kit or the DNeasy PowerLyzer Powersoil kit (Qiagen). The V4 region of the bacterial 16S rRNA gene was amplified in triplicate with barcoded fusion primers (F515/R806).65 After amplification, the triplicates were pooled, and DNA was quantified with Quant-iT PicoGreen from Invitrogen. Up to 96 samples were then combined in equal amounts of 300 ng and purified using the QIAquick PCR purification kit from Qiagen. The DNA was quantified with a Qubit Fluorometer from Life Technologies. The samples were finally pooled at an equal molar concentration of 50 nM and sequenced using an Illumina MiSeq platform at the Genome Technology Center at New York University School of Medicine.
Quantification and statistical analysis
16S rRNA analysis
To process and analyze the data, we used the Quantitative Insights into Microbial Ecology (QIIME2, version 2022.02).67 Sequencing reads were trimmed and denoised using DADA2,72 and aligned using MAFFT.71 We assigned taxonomy using Silva 138 (released December 2019). To determine beta diversity (unweighted UniFrac), we performed analyses using the QIIME2 pipeline and generated plots in R using ggPlot2. For alpha diversity (Observed ASVs) and taxonomic abundance plots, we used the Phyloseq package. We determined statistical significance between community structures using an Adonis test in R with the Phyloseq and Vegan packages. We reported statistically significant p values in the principal component plots. To generate differential abundance plots and heatmaps, we used the DESeq2 and Complex Heatmap packages in R.66,70 We determined differentially abundant ASVs by contrasting control and antibiotic-exposed mice and selecting for ASVs that were significant using a Benjamini Hochberg false discovery rate (FDR) of p < 0.01.
Statistical testing
We performed statistical analysis using Graphpad Prism 9.5.1 software. To test for statistical significance, we used a Kruskal-Wallis test (one-way non-parametric ANOVA). We considered differences statistically significant when p < 0.05. We indicate significance levels with asterisks: ∗ for p < 0.05, ∗∗ for p < 0.01, ∗∗∗ for p < 0.001, and ∗∗∗∗ for p < 0.0001. All plots show the mean value per treatment group ±standard error of the mean (SEM).
Acknowledgments
The authors thank Drs. Matthew Wipperman for input on 16S rRNA analysis, Giulio Quarta for providing Escherichia coli strain MG1655, and Leopoldo Segal, Juan Lafaille, and Shruti Naik for intellectual contributions. Graphical abstract was created using BioRender. TCB was supported by the National Institutes of Health (TL1TR001447, T32ES007324, T32AI007180) and the Bernard Levine Immunology Fellowship. This work was supported by NIH U01AI22285 and the Foundation Leducq TransAtlantic Network and Emch and Sergei Zlinkoff foundations for M.J.B; NIH R01HL125816 for SBK; and the Swiss National Science Foundation (BSCGIO 462 157841/1) for A.M. Flow cytometry technologies were provided by the NYU Langone Cytometry and Cell Sorting Laboratory, and 16SrRNA sequencing was provided by the NYU Genome Technology Center and was supported in part by grant P30CA016087 from the National Institutes of Health/National Cancer Institute.
Author contributions
Conceptualization, TCB, AM, SBK, and MJB; Formal analysis, TCB, SBK, and MJB; Investigation, TCB, MBP, JL, MLH, YSY, XZ, EN, KJ, BJM, VR, and MJH; Resources, XSZ; Writing-original draft, TCB, MBP, SBK, and MJB; Writing-review and editing, TCB, BJM, XSZ, SBK, and MJB; Supervision, AM, SBK, and MJB; Funding acquisition, AM, SBK, and MJB.
Declaration of interests
The authors declare no competing financial or conflicts of interest.
Published: May 4, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.106810.
Contributor Information
Sergei B. Koralov, Email: sergei.koralov@nyulangone.org.
Martin J. Blaser, Email: martin.blaser@cabm.rutgers.edu.
Supplemental information
Data and code availability
16S rRNA data have been deposited at QIITA and are publicly available as of the date of publication: https://qiita.ucsd.edu/study/description/14759. This paper does not report original code.
References
- 1.Sanidad K.Z., Zeng M.Y. Neonatal gut microbiome and immunity. Curr. Opin. Microbiol. 2020;56:30–37. doi: 10.1016/j.mib.2020.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thaiss C.A., Zmora N., Levy M., Elinav E. The microbiome and innate immunity. Nature. 2016;535:65–74. doi: 10.1038/nature18847. [DOI] [PubMed] [Google Scholar]
- 3.Zheng D., Liwinski T., Elinav E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020;30:492–506. doi: 10.1038/s41422-020-0332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bokulich N.A., Chung J., Battaglia T., Henderson N., Jay M., Li H., D Lieber A., Wu F., Perez-Perez G.I., Chen Y., et al. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci. Transl. Med. 2016;8:343ra82. doi: 10.1126/scitranslmed.aad7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gschwendtner S., Kang H., Thiering E., Kublik S., Fösel B., Schulz H., Krauss-Etschmann S., Heinrich J., Schöler A., Schloter M., Standl M. Early life determinants induce sustainable changes in the gut microbiome of six-year-old children. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-49160-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gasparrini A.J., Wang B., Sun X., Kennedy E.A., Hernandez-Leyva A., Ndao I.M., Tarr P.I., Warner B.B., Dantas G. Persistent metagenomic signatures of early-life hospitalization and antibiotic treatment in the infant gut microbiota and resistome. Nat. Microbiol. 2019;4:2285–2297. doi: 10.1038/s41564-019-0550-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fouhy F., Ross R.P., Fitzgerald G.F., Stanton C., Cotter P.D. Composition of the early intestinal microbiota: knowledge, knowledge gaps and the use of high-throughput sequencing to address these gaps. Gut Microb. 2012;3:203–220. doi: 10.4161/gmic.20169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Korpela K., Salonen A., Virta L.J., Kekkonen R.A., Forslund K., Bork P., De Vos W.M. Intestinal microbiome is related to lifetime antibiotic use in Finnish pre-school children. Nat. Commun. 2016;7:10410–10418. doi: 10.1038/ncomms10410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang X.S., Yin Y.S., Wang J., Battaglia T., Krautkramer K., Li W.V., Li J., Brown M., Zhang M., Badri M.H., et al. Maternal cecal microbiota transfer rescues early-life antibiotic-induced enhancement of type 1 diabetes in mice. Cell Host Microbe. 2021;29:1249–1265.e9. doi: 10.1016/j.chom.2021.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aversa Z., Atkinson E.J., Schafer M.J., Theiler R.N., Rocca W.A., Blaser M.J., LeBrasseur N.K. Association of infant antibiotic exposure with childhood health outcomes. Mayo Clin. Proc. 2021;96:66–77. doi: 10.1016/j.mayocp.2020.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Metsälä J., Lundqvist A., Virta L.J., Kaila M., Gissler M., Virtanen S.M. Mother's and offspring's use of antibiotics and infant allergy to cow's milk. Epidemiology. 2013;24:303–309. doi: 10.1097/EDE.0b013e31827f520f. [DOI] [PubMed] [Google Scholar]
- 12.Risnes K.R., Belanger K., Murk W., Bracken M.B. Antibiotic exposure by 6 months and asthma and allergy at 6 years: findings in a cohort of 1,401 US children. Am. J. Epidemiol. 2011;173:310–318. doi: 10.1093/aje/kwq400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Russell S.L., Gold M.J., Willing B.P., Thorson L., McNagny K.M., Finlay B.B. Perinatal antibiotic treatment affects murine microbiota, immune responses and allergic asthma. Gut Microb. 2013;4:158–164. doi: 10.4161/gmic.23567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wernroth M.L., Fall K., Svennblad B., Ludvigsson J.F., Sjölander A., Almqvist C., Fall T. Early childhood antibiotic treatment for otitis media and other respiratory tract infections is associated with risk of type 1 diabetes: a nationwide register-based study with sibling analysis. Diabetes Care. 2020;43:991–999. doi: 10.2337/dc19-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borbet T.C., Zhang X., Müller A., Blaser M.J. The role of the changing human microbiome in the asthma pandemic. J. Allergy Clin. Immunol. 2019;144:1457–1466. doi: 10.1016/j.jaci.2019.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ouwehand A., Isolauri E., Salminen S. The role of the intestinal microflora for the development of the immune system in early childhood. Eur. J. Nutr. 2002;41(Suppl 1):I32–I37. doi: 10.1007/s00394-002-1105-4. [DOI] [PubMed] [Google Scholar]
- 17.Gensollen T., Iyer S.S., Kasper D.L., Blumberg R.S. How colonization by microbiota in early life shapes the immune system. Science. 2016;352:539–544. doi: 10.1126/science.aad9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cerutti A. The regulation of IgA class switching. Nat. Rev. Immunol. 2008;8:421–434. doi: 10.1038/nri2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hand T.W., Reboldi A. Production and function of immunoglobulin A. Annu. Rev. Immunol. 2021;39:695–718. doi: 10.1146/annurev-immunol-102119-074236. [DOI] [PubMed] [Google Scholar]
- 20.Macpherson A.J., Geuking M.B., McCoy K.D. Homeland security: IgA immunity at the frontiers of the body. Trends Immunol. 2012;33:160–167. doi: 10.1016/j.it.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 21.Reboldi A., Cyster J.G. Peyer's patches: organizing B-cell responses at the intestinal Frontier. Immunol. Rev. 2016;271:230–245. doi: 10.1111/imr.12400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jung C., Hugot J.P., Barreau F. Peyer's patches: the immune sensors of the intestine. Int. J. Inflam. 2010;2010 doi: 10.4061/2010/823710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cerutti A., Rescigno M. The biology of intestinal immunoglobulin A responses. Immunity. 2008;28:740–750. doi: 10.1016/j.immuni.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cahenzli J., Köller Y., Wyss M., Geuking M.B., McCoy K.D. Intestinal microbial diversity during early-life colonization shapes long-term IgE levels. Cell Host Microbe. 2013;14:559–570. doi: 10.1016/j.chom.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Borbet T.C., Blaser M.J. Host genotype and early life microbiota alterations have additive effects on disease susceptibility. Mucosal Immunol. 2019;12:586–588. doi: 10.1038/s41385-019-0157-1. [DOI] [PubMed] [Google Scholar]
- 26.Stokholm J., Blaser M.J., Thorsen J., Rasmussen M.A., Waage J., Vinding R.K., Schoos A.M.M., Kunøe A., Fink N.R., Chawes B.L., et al. Maturation of the gut microbiome and risk of asthma in childhood. Nat. Commun. 2018;9:141. doi: 10.1038/s41467-017-02573-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang X.S., Li J., Krautkramer K.A., Badri M., Battaglia T., Borbet T.C., Koh H., Ng S., Sibley R.A., Li Y., et al. Antibiotic-induced acceleration of type 1 diabetes alters maturation of innate intestinal immunity. Elife. 2018;7 doi: 10.7554/eLife.37816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arrieta M.C., Stiemsma L.T., Dimitriu P.A., Thorson L., Russell S., Yurist-Doutsch S., Kuzeljevic B., Gold M.J., Britton H.M., Lefebvre D.L., et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci. Transl. Med. 2015;7:307ra152. doi: 10.1126/scitranslmed.aab2271. [DOI] [PubMed] [Google Scholar]
- 29.Zhang X., Borbet T.C., Fallegger A., Wipperman M.F., Blaser M.J., Muller A. An antibiotic-impacted microbiota compromises the development of colonic regulatory T cells and predisposes to dysregulated immune responses. mBio. 2021;12 doi: 10.1128/mBio.03335-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goethel A., Turpin W., Rouquier S., Zanello G., Robertson S.J., Streutker C.J., Philpott D.J., Croitoru K. Nod2 influences microbial resilience and susceptibility to colitis following antibiotic exposure. Mucosal Immunol. 2019;12:720–732. doi: 10.1038/s41385-018-0128-y. [DOI] [PubMed] [Google Scholar]
- 31.Borbet T.C., Pawline M.B., Zhang X., Wipperman M.F., Reuter S., Maher T., Li J., Iizumi T., Gao Z., Daniele M., et al. Influence of the early-life gut microbiota on the immune responses to an inhaled allergen. Mucosal Immunol. 2022;15:1000–1011. doi: 10.1038/s41385-022-00544-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruiz V.E., Battaglia T., Kurtz Z.D., Bijnens L., Ou A., Engstrand I., Zheng X., Iizumi T., Mullins B.J., Müller C.L., et al. A single early-in-life macrolide course has lasting effects on murine microbial network topology and immunity. Nat. Commun. 2017;8:518. doi: 10.1038/s41467-017-00531-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Russell D.A., Ross R.P., Fitzgerald G.F., Stanton C. Metabolic activities and probiotic potential of bifidobacteria. Int. J. Food Microbiol. 2011;149:88–105. doi: 10.1016/j.ijfoodmicro.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 34.Kujawska M., La Rosa S.L., Roger L.C., Pope P.B., Hoyles L., McCartney A.L., Hall L.J. Succession of Bifidobacterium longum strains in response to a changing early life nutritional environment reveals dietary substrate adaptations. iScience. 2020;23 doi: 10.1016/j.isci.2020.101368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fukushima Y., Kawata Y., Mizumachi K., Kurisaki J., Mitsuoka T. Effect of bifidobacteria feeding on fecal flora and production of immunoglobulins in lactating mouse. Int. J. Food Microbiol. 1999;46:193–197. doi: 10.1016/s0168-1605(98)00183-4. [DOI] [PubMed] [Google Scholar]
- 36.Henrick B.M., Chew S., Casaburi G., Brown H.K., Frese S.A., Zhou Y., Underwood M.A., Smilowitz J.T. Colonization by B. infantis EVC001 modulates enteric inflammation in exclusively breastfed infants. Pediatr. Res. 2019;86:749–757. doi: 10.1038/s41390-019-0533-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Henrick B.M., Rodriguez L., Lakshmikanth T., Pou C., Henckel E., Arzoomand A., Olin A., Wang J., Mikes J., et al. Bifidobacteria-mediated immune system imprinting early in life. Cell. 2021;184:3884–3898.e11. doi: 10.1016/j.cell.2021.05.030. [DOI] [PubMed] [Google Scholar]
- 38.Chang D.E., Smalley D.J., Tucker D.L., Leatham M.P., Norris W.E., Stevenson S.J., Anderson A.B., Grissom J.E., Laux D.C., Cohen P.S., Conway T. Carbon nutrition of Escherichia coli in the mouse intestine. Proc. Natl. Acad. Sci. USA. 2004;101:7427–7432. doi: 10.1073/pnas.0307888101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adami A.J., Bracken S.J., Guernsey L.A., Rafti E., Maas K.R., Graf J., Matson A.P., Thrall R.S., Schramm C.M. Early-life antibiotics attenuate regulatory T cell generation and increase the severity of murine house dust mite-induced asthma. Pediatr. Res. 2018;84:426–434. doi: 10.1038/s41390-018-0031-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Russell S.L., Gold M.J., Reynolds L.A., Willing B.P., Dimitriu P., Thorson L., Redpath S.A., Perona-Wright G., Blanchet M.R., Mohn W.W., et al. Perinatal antibiotic-induced shifts in gut microbiota have differential effects on inflammatory lung diseases. J. Allergy Clin. Immunol. 2015;135:100–109. doi: 10.1016/j.jaci.2014.06.027. [DOI] [PubMed] [Google Scholar]
- 41.Lynn M.A., Tumes D.J., Choo J.M., Sribnaia A., Blake S.J., Leong L.E.X., Young G.P., Marshall H.S., Wesselingh S.L., Rogers G.B., Lynn D.J. Early-life antibiotic-driven dysbiosis leads to dysregulated vaccine immune responses in mice. Cell Host Microbe. 2018;23:653–660.e5. doi: 10.1016/j.chom.2018.04.009. [DOI] [PubMed] [Google Scholar]
- 42.Chung H., Pamp S.J., Hill J.A., Surana N.K., Edelman S.M., Troy E.B., Reading N.C., Villablanca E.J., Wang S., Mora J.R., et al. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 2012;149:1578–1593. doi: 10.1016/j.cell.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Høverstad T., Carlstedt-Duke B., Lingaas E., Norin E., Saxerholt H., Steinbakk M., Midtvedt T. Influence of oral intake of seven different antibiotics on faecal short-chain fatty acid excretion in healthy subjects. Scand. J. Gastroenterol. 1986;21:997–1003. doi: 10.3109/00365528608996411. [DOI] [PubMed] [Google Scholar]
- 44.Li R., Wang H., Shi Q., Wang N., Zhang Z., Xiong C., Liu J., Chen Y., Jiang L., Jiang Q. Effects of oral florfenicol and azithromycin on gut microbiota and adipogenesis in mice. PLoS One. 2017;12 doi: 10.1371/journal.pone.0181690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feng Y., Huang Y., Wang Y., Wang P., Song H., Wang F. Antibiotics induced intestinal tight junction barrier dysfunction is associated with microbiota dysbiosis, activated NLRP3 inflammasome and autophagy. PLoS One. 2019;14 doi: 10.1371/journal.pone.0218384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim M., Qie Y., Park J., Kim C.H. Gut microbial metabolites fuel host antibody responses. Cell Host Microbe. 2016;20:202–214. doi: 10.1016/j.chom.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jordan A., Carding S.R., Hall L.J. The early-life gut microbiome and vaccine efficacy. Lancet. Microbe. 2022;3:e787–e794. doi: 10.1016/S2666-5247. [DOI] [PubMed] [Google Scholar]
- 48.Fukushima Y., Kawata Y., Hara H., Terada A., Mitsuoka T. Effect of a probiotic formula on intestinal immunoglobulin A production in healthy children. Int. J. Food Microbiol. 1998;42:39–44. doi: 10.1016/s0168-1605(98)00056-7. [DOI] [PubMed] [Google Scholar]
- 49.Zhang L.L., Chen X., Zheng P.Y., Luo Y., Lu G.F., Liu Z.Q., Huang H., Yang P.C. Oral Bifidobacterium modulates intestinal immune inflammation in mice with food allergy. J. Gastroenterol. Hepatol. 2010;25:928–934. doi: 10.1111/j.1440-1746.2009.06193.x. [DOI] [PubMed] [Google Scholar]
- 50.Bunesova V., Lacroix C., Schwab C. Fucosyllactose and L-fucose utilization of infant Bifidobacterium longum and Bifidobacterium kashiwanohense. BMC Microbiol. 2016;16:248–260. doi: 10.1186/s12866-016-0867-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kirmiz N., Robinson R.C., Shah I.M., Barile D., Mills D.A. Milk glycans and their interaction with the infant-gut microbiota. Annu. Rev. Food Sci. Technol. 2018;9:429–450. doi: 10.1146/annurev-food-030216-030207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vatanen T., Ang Q.Y., Siegwald L., Sarker S.A., Le Roy C.I., Duboux S., Delannoy-Bruno O., Ngom-Bru C., Boulangé C.L., Stražar M., et al. A distinct clade of Bifidobacterium longum in the gut of Bangladeshi children thrives during weaning. Cell. 2022;185:4280–4297.e12. doi: 10.1016/j.cell.2022.10.011. [DOI] [PubMed] [Google Scholar]
- 53.Frese S.A., Hutton A.A., Contreras L.N., Shaw C.A., Palumbo M.C., Casaburi G., Xu G., Davis J.C.C., Lebrilla C.B., Henrick B.M., et al. Persistence of supplemented Bifidobacterium longum subsp. infantis EVC001 in breastfed infants. mSphere. 2017;2:e00501-17–e00517. doi: 10.1128/mSphere.00501-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goverse G., Molenaar R., Macia L., Tan J., Erkelens M.N., Konijn T., Knippenberg M., Cook E.C.L., Hanekamp D., Veldhoen M., et al. Diet-derived short chain fatty acids stimulate intestinal epithelial cells to induce mucosal tolerogenic dendritic cells. J. Immunol. 2017;198:2172–2181. doi: 10.4049/jimmunol.1600165. [DOI] [PubMed] [Google Scholar]
- 55.Wu W., Sun M., Chen F., Cao A.T., Liu H., Zhao Y., Huang X., Xiao Y., Yao S., Zhao Q., et al. Microbiota metabolite short-chain fatty acid acetate promotes intestinal IgA response to microbiota which is mediated by GPR43. Mucosal Immunol. 2017;10:946–956. doi: 10.1038/mi.2016.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dai D.L.Y., Petersen C., Hoskinson C., Del Bel K.L., Becker A.B., Moraes T.J., Mandhane P.J., Finlay B.B., Simons E., Kozyrskyj A.L., et al. Breastfeeding enrichment of B. longum subsp. infantis mitigates the effect of antibiotics on the microbiota and childhood asthma risk. Med. 2023;4:92–112.e5. doi: 10.1016/j.medj.2022.12.002. [DOI] [PubMed] [Google Scholar]
- 57.Stokholm J., Thorsen J. Can breastfeeding promote an antibiotic-resilient microbiome? Med. 2023;4:67–68. doi: 10.1016/j.medj.2023.01.002. [DOI] [PubMed] [Google Scholar]
- 58.Penders J., Thijs C., Vink C., Stelma F.F., Snijders B., Kummeling I., van den Brandt P.A., Stobberingh E.E. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. 2006;118:511–521. doi: 10.1542/peds.2005-2824. [DOI] [PubMed] [Google Scholar]
- 59.Shin N.R., Whon T.W., Bae J.W. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015;33:496–503. doi: 10.1016/j.tibtech.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 60.Fei N., Zhao L. An opportunistic pathogen isolated from the gut of an obese human causes obesity in germfree mice. ISME J. 2013;7:880–884. doi: 10.1038/ismej.2012.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Morgan X.C., Tickle T.L., Sokol H., Gevers D., Devaney K.L., Ward D.V., Reyes J.A., Shah S.A., LeLeiko N., Snapper S.B., et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mirsepasi-Lauridsen H.C., Vallance B.A., Krogfelt K.A., Petersen A.M. Escherichia coli pathobionts associated with inflammatory bowel disease. Clin. Microbiol. Rev. 2019;32:e00060-18–e00018. doi: 10.1128/CMR.00060-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Castro-Dopico T., Dennison T.W., Ferdinand J.R., Mathews R.J., Fleming A., Clift D., Stewart B.J., Jing C., Strongili K., Labzin L.I., et al. Anti-commensal IgG drives intestinal inflammation and type 17 immunity in ulcerative colitis. Immunity. 2019;50:1099–1114.e10. doi: 10.1016/j.immuni.2019.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Uzzan M., Martin J.C., Mesin L., Livanos A.E., Castro-Dopico T., Huang R., Petralia F., Magri G., Kumar S., Zhao Q., et al. Ulcerative colitis is characterized by a plasmablast-skewed humoral response associated with disease activity. Nat. Med. 2022;28:766–779. doi: 10.1038/s41591-022-01680-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Walters W., Hyde E.R., Berg-Lyons D., Ackermann G., Humphrey G., Parada A., Gilbert J.A., Jansson J.K., Caporaso J.G., Fuhrman J.A., et al. Improved bacterial 16S rRNA gene (V4 and V4-5) and fungal internal transcribed spacer marker gene primers for microbial community surveys. mSystems. 2016;1:e00009-15–e00015. doi: 10.1128/mSystems.00009-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bolyen E., Rideout J.R., Dillon M.R., Bokulich N.A., Abnet C.C., Al-Ghalith G.A., Alexander H., Alm E.J., Arumugam M., Asnicar F., et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bisanz J.E. qiime2R: importing QIIME2 artifacts and associated data into R sessions. Version 0.99. 2018;13 [Google Scholar]
- 69.Oksanen J., Blanchet F.G., Friendly M., Kindt R., Legendre P., McGlinn D., Minchin P., O’Hara R., Simpson G., Solymos P. Vegan: community Ecology package. R package version 2. 2022;2020:5–7. [Google Scholar]
- 70.Gu Z., Eils R., Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 71.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J.A., Holmes S.P. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Matsuki T., Watanabe K., Tanaka R. Genus-and species-specific PCR primers for the detection and identification of bifidobacteria. Curr. Issues Intest. Microbiol. 2003;4:61–69. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
16S rRNA data have been deposited at QIITA and are publicly available as of the date of publication: https://qiita.ucsd.edu/study/description/14759. This paper does not report original code.