Summary
Although meropenem, colistin, and tigecycline are recognized as the last-line antibiotics for multidrug-resistant Gram-negative bacteria (MDR-GN), the emergence of mobile resistance genes such as blaNDM, mcr, and tet(X) severely compromises their clinical effectiveness. Developing novel antibiotic adjuvants to restore the effectiveness of existing antibiotics provides a feasible approach to address this issue. Herein, we discover that a Food and Drug Administration (FDA)-approved drug daunorubicin (DNR) drastically potentiates the activity of last-resort antibiotics against MDR-GN pathogens and biofilm-producing bacteria. Furthermore, DNR effectively inhibits the evolution and spread of colistin and tigecycline resistance. Mechanistically, DNR and colistin combination exacerbates membrane disruption, induces DNA damage and the massive production of reactive oxygen species (ROS), ultimately leading to bacterial cell death. Importantly, DNR restores the effectiveness of colistin in Galleria mellonella and murine models of infection. Collectively, our findings provide a potential drug combination strategy for treating severe infections elicited by Gram-negative superbugs.
Subject areas: Medical microbiology, Microbiology
Graphical abstract

Highlights
-
•
DNR resensitizes drug-resistant bacteria to the last-line antibiotics
-
•
DNR thwarts the evolution and transmission of antimicrobial resistance
-
•
DNR can directly bind to MCR-1 protein and reduce its expression
-
•
DNR plus colistin exacerbates membrane damage and the over-production of ROS
Medical microbiology; Microbiology
Introduction
Antibiotic resistance has become the biggest threat to human health in the 21st century.1,2 Gram-negative bacteria (GNB) show higher intrinsic resistance to many Gram-positive bacteria-active antibiotics because these drugs cannot penetrate the GNB outer membrane.3,4 Therefore, infectious diseases caused by GNB are more challenging to treat in clinic.5,6 Carbapenems are one of the main treatments for severe drug-resistant GNB infections. However, New Delhi metallo-β-lactamase1 (NDM-1) was discovered in 2009, and it has spread widely in more than 70 countries around the world, seriously weakening the efficacy of carbapenems.7 To combat the increasingly multidrug-resistant (MDR) GNB, colistin, and tigecycline have been applied in clinical treatment successively as the last line of defense. In clinical practice, the cationic cyclic peptide antibiotic colistin can be used for the treatment of various infectious diseases, including peritonitis, sepsis, and post-burn infections.8,9 By contrast, complicated skin and skin structure infections (cSSSI) or complex intra-abdominal infections (cIAI) in adults are treated with tigecycline, the first medication in the glycyltetracycline class approved by FDA in 2005.10 In recent years, plasmid-mediated mcr-111 and tet(X3/X4)12 were discovered in 2015 and 2019, respectively, which can be transferred to Escherichia coli, Klebsiella pneumoniae, and other clinical pathogens, resulting in the widespread of colistin/tigecycline resistance. Heretofore, there are no effective drugs available to deal with complex drug-resistant bacterial infections. Therefore, new strategies are urgently needed to counteract the antimicrobial resistance (AMR) threat.
Antibiotic therapy is currently the dominant treatment for bacterial infections.13 However, the current research and development capabilities of new antibiotics are seriously insufficient, and finding new antibiotics with distinct targets is getting harder, especially for GNB.14 Besides, high costs and long cycles limit the development of new medicines.15 Only a small number of novel antibiotics such as daptomycin have been approved by FDA in recent decades.16 Comparatively, identifying novel antibiotic adjuvants from previously approved compounds (PACs) offers a viable strategy for preserving antibiotic efficacy.17,18,19 Moreover, the low cost and good safety, as well as the favorable pharmacokinetic properties of PACs, make them attractive antibiotic adjuvant candidates for translational application. Historically, classical β-lactamase inhibitors such as clavulanate, sulbactam, and tazobactam, effectively prevent the hydrolysis of antibiotics by inhibiting β-lactamase activity,20 which obtain proven success in the clinic over the past decades. Additionally, some progress has been achieved in the development of new antibiotic adjuvants in recent years. For instance, the natural substance aspergillomarasmine A (AMA), which was once an angiotensin-converting enzyme (ACE) inhibitor, recovers the efficacy of meropenem against NDM-positive Enterobacteriaceae.21 Owing to the substitution of bismuth on zinc ions in the active center of NDM-1, bismuth nitrate shows excellent synergistic antibacterial action with meropenem.22 Additionally, by removing bacterial biofilms, triclosan improves the antibacterial action of tobramycin against Pseudomonas aeruginosa.23 Apparently, combining antibiotics with their potentiators screened from PACs offers a new pipeline to defeat the increasing resistance crisis.
Daunorubicin (DNR) is an FDA-approved antitumor drug for treating acute myeloid or lymphocytic leukemia.24,25 Several previous studies have partially demonstrated the antibacterial effects of DNR against Gram-positive and Gram-negative bacteria,26,27 while its therapeutic potential as an antibiotic adjuvant remains elusive. Here, we discovered that DNR significantly enhances the effectiveness of the last-resort antibiotics against MDR GNB. Their combination effectively reduces the bacterial loads of drug-resistant bacteria, inhibits the formation of biofilm, and removes mature biofilm bacteria. In addition, DNR remarkably suppresses the evolution and spread of colistin/tigecycline resistance. Mechanistic studies showed that DNR assists colistin to destroy the integrity of the cell membrane of drug-resistant bacteria, subsequently causes DNA damage, induces oxidative damage, and eventually leads to bacterial cell death. Importantly, DNR improves the in vivo effectiveness of colistin in animal infection models. The identification of DNR as an antibiotic adjuvant provides a new solution to alleviate the growing crisis of bacterial resistance.
Results
DNR displays excellent synergistic activity with meropenem/colistin/tigecycline
Using checkerboard assays, we first investigated the synergistic action of DNR with eight kinds of antibiotics against MDR E. coli B2 (blaNDM-5 + mcr-1) or tigecycline-resistant E. coli B3-1 (tet(X4)). The results showed that DNR potentiated the activity of meropenem, rifampicin, colistin and tigecycline (fractional inhibitory concentration index [FICI]: 0.375, 0.375, 0.125, and 0.188, respectively), of which the synergistic activity with colistin was the strongest, followed by tigecycline, whereas had only weak synergistic activity or no activity with the other four antibiotics (FICI > 0.5) (Figure 1). To reveal whether the synergistic activity of DNR with meropenem, colistin, or tigecycline was only effective on resistant bacteria, we measured the potentiating activity of DNR in engineered E. coli DH5α (PUC19-blaNDM-5/mcr-1/tet(X4)), empty E. coli DH5α (PUC19) and two reference strains. Interestingly, we found that DNR and the above three antibiotics displayed strong synergistic activity against drug-resistant bacteria, while weak or even no synergistic activity for sensitive bacteria, implying that the inhibition of specific resistance determinants is related to the action of DNR (Figures 2 and S1). Furthermore, we evaluated the synergistic effects of DNR and colistin/tigecycline against four clinical isolates of drug-resistant pathogens, including E. coli G92 (mcr-1), K. pneumoniae D120 (mcr-8), K. pneumoniae YZSH26 (tet(X4)), and Acinetobacter baumannii C222 (tet(X6)). As expected, DNR substantially potentiated colistin/tigecycline activity against these clinical strains (Figure 3). Of particular note is colistin, whose minimum inhibitory concentrations (MICs) against mobile colistin resistance (MCR)-positive bacteria were drastically decreased from 4 to 0.004 μg/mL (1,000-fold) at sub-inhibitory concentrations of DNR. Meanwhile, great potentiation of DNR to colistin/tigecycline was also observed in other tested mcr-1 or tet(X4)-carrying clinical isolates (Figure S2). Consistently, the synergistic effect of DNR and colistin was higher than that of tigecycline, and the antibacterial activity of colistin was enhanced by more than 16-fold under one-quarter MIC of DNR.
Figure 1.
Checkerboard assay of the synergistic effect of DNR with various antibiotics against multidrug-resistant (MDR) E. coli B2 or with tigecycline against tet(X4)-carrying E. coli B3-1
Dark blue areas show greater cell density. The data show the average optical density (OD) of two biological replicates (600 nm). An FIC index of less than 0.5 is used to define synergy.
Figure 2.
Potentiating effect of DNR on three clinically important antibiotics against engineered drug-resistant and sensitive bacteria
Dark blue areas show greater cell density. The data show the average optical density (OD) of two biological replicates (600 nm). An FIC index of less than 0.5 is used to define synergy.
Figure 3.
Synergistic activity between DNR with colistin or tigecycline against clinical resistant strains
Dark blue areas show greater cell density. The data show the average OD of two biological replicates (600 nm). An FIC index of less than 0.5 is used to define synergy.
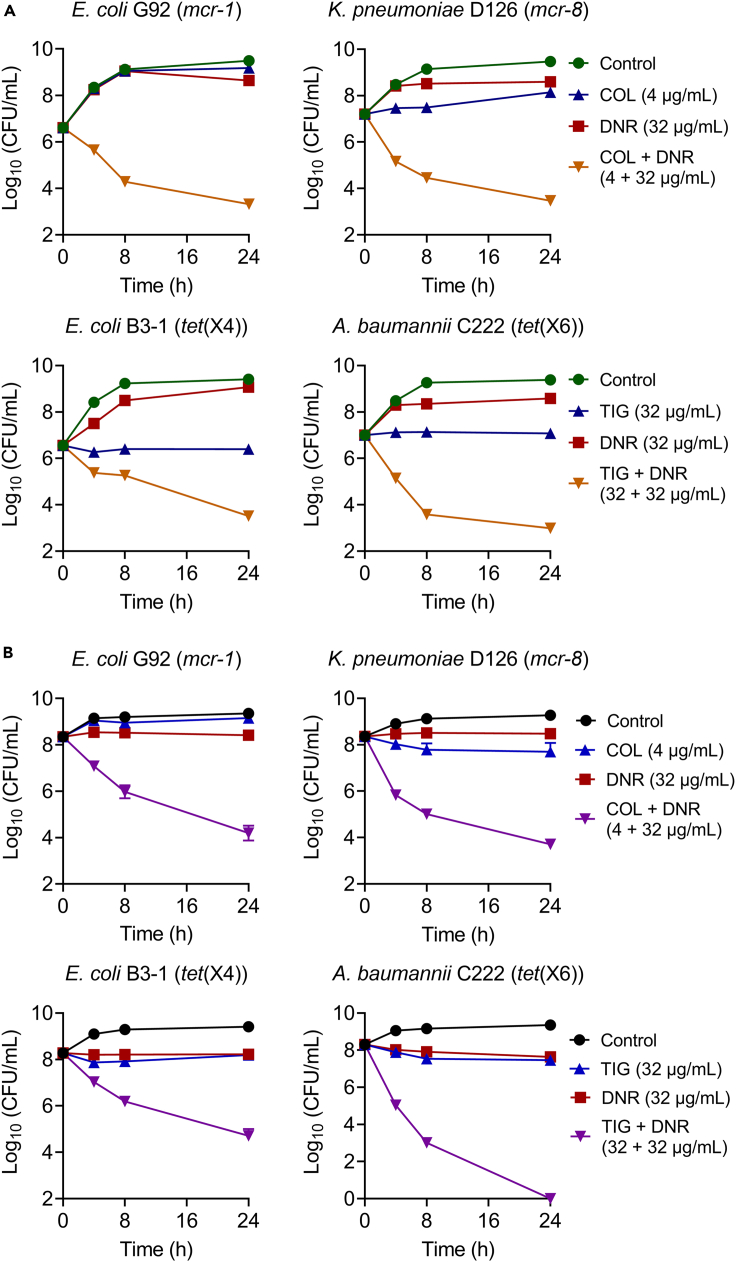
The time-dependent killing curves of individual and combined drugs were performed to investigate their bactericidal effect. We found that the three drugs alone, including DNR, colistin, and tigecycline, displayed only slight bacteriostatic activity, while the combination of DNR with colistin/tigecycline greatly reduced the bacterial loads of mcr/tet(X)-positive strains in the exponential and stationary phase (Figure 4).
Figure 4.
DNR and colistin/tigecycline combinations display strong bactericidal activity against drug-resistant pathogens in exponential phase
(A) and stationary phase (B)Time-kill curves of E. coli G92 (mcr-1), K. pneumoniae D120 (mcr-8), E. coli B3-1 (tet(X4)), and A. baumannii C222 (tet(X6)) in Luria-Bertani broth (LB) in the presence of DNR or colistin or tigecycline alone, or their combination during 24 h. Data were presented as mean ± SD from three biological replicates. CFU, colony-forming unit.
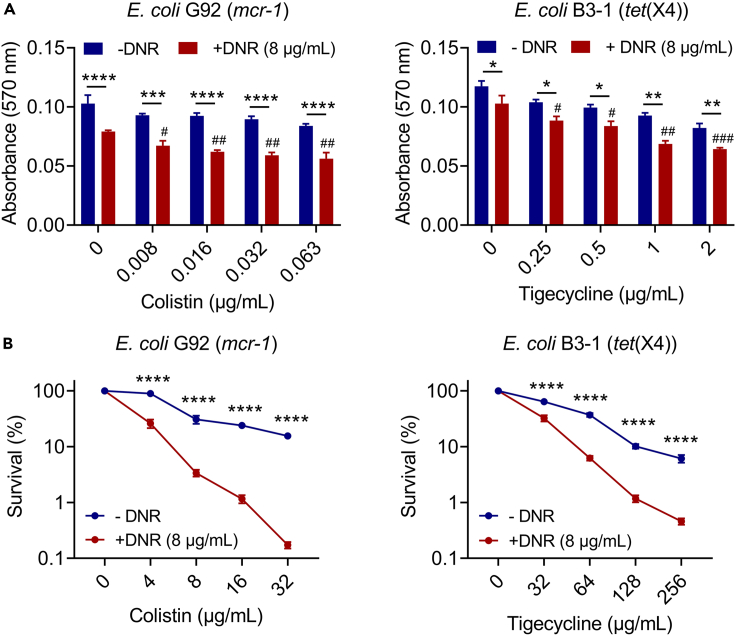
It is suggested that biofilms can greatly resist the bactericidal activity of antibiotics, thus leading to clinical failures.28,29 In light of this, we next investigated the synergistic impact of DNR on the development of biofilm and the destruction of established biofilm using crystal violet staining. As shown in Figure 5A, we found that the presence of DNR greatly enhanced the inhibiting effect of antibiotics on biofilm formation compared to colistin/tigecycline alone. Notably, in the biofilm formation assays, low concentrations of drug combinations have no direct bactericidal activity, indicating that killing the bacteria was not the cause of the suppression of biofilm production at these doses. Moreover, the combined use of DNR and colistin/tigecycline also exhibited a synergistic effect in removing mature biofilms (Figure 5B). Among them, the combination of DNR and the highest concentration of colistin/tigecycline reduced the survival rate of biofilm bacteria to less than 1%. To sum up, these findings show that DNR effectively enhances the antibacterial activity of multiple last-resort antibiotics, particularly for colistin against MDR Gram-negative pathogens and biofilm-producing bacteria.
Figure 5.
Combined use of DNR and colistin/tigecycline effectively inhibits biofilm formation of drug-resistant bacteria and eliminates the mature biofilms
Effects of DNR on the biofilm formation (A) and established biofilm eradication (B) of E. coli G92 (mcr-1) or E. coli B3-1 (tet(X4)) in the presence of increasing concentrations of colistin or tigecycline. Survival of biofilm-encased bacteria in (B) were normalized to blank control. Data were presented as mean ± SD from three biological replicates, and statistical significance was determined by two-way ANOVA (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001) or one-way ANOVA (colistin plus DNR versus DNR (8 μg/mL), #p < 0.05, ##p < 0.01, ###p < 0.001).
Stability and safety evaluation of the combined use of DNR with colistin/tigecycline
Many candidate drugs are very active in vitro but work poorly in vivo, and the influence of various factors under physiological conditions may be an important reason.30 To investigate the therapeutic potential of drug combinations,31 we initially assessed the stabilization of DNR and colistin/tigecycline combinations in the existence of various salt ions and serum, which would affect the in vivo efficacy of medicines. After the addition of 10 mM Na+, K+, EDTA, 10% serum or 10% DMEM into the Mueller-Hinton Broth (MHB), the potentiation of DNR to antibiotics retained and more active in the presence of EDTA and serum; however, divalent ions such as 10 mM Ca2+ and Mg2+ caused the complete loss of the synergistic activity (FICI = 2.0) (Table S4). The alteration of activity in the existence of EDTA and divalent ions is closely related to bacterial membrane permeability,32 suggesting that the synergistic effect of DNR with antibiotics may be associated with the damage of cell membrane.
Since the safety of drugs is of importance for translation application, we next assessed the hemolytic activity and acute toxicity of the drug combinations. As shown in Figure S3, the combined use of DNR (0–64 μg/mL) and colistin/tigecycline displayed a negligible hemolytic effect on mammalian RBCs, with a maximum of only about 2%. In addition, we examined the acute toxicity of DNR and colistin combination in a murine model. Compared with the colistin group, no significant body weight changes in mice after administration with DNR and colistin were observed (Figure S4). Additionally, blood routine and biochemical assays were also carried out. The blood-related parameters were all within the normal range (Tables S5 and S6). These results indicate that the combination of colistin/tigecycline and DNR displays superior antibacterial activity as well as biocompatibility even under physiological conditions.
DNR suppresses the evolution and spread of mobile colistin/tigecycline resistance
In the long-term struggle with antibiotics, the bacterial resistance would continue to increase after a series of evolutions.33 Therefore, using resistance development and mutation preventive concentration (MPC) assays, we investigated DNR’s potential to prevent the evolution of colistin or tigecycline resistance. After 24 days of continuous passages, we found that DNR substantially slowed the increase of the MICs of colistin/tigecycline against passaged bacteria (Figure 6A). In addition, DNR supplementation dose-dependently reduced the MPC values of the two antibiotics against the corresponding resistant bacteria, implying that DNR can impede the emergence and evolution of colistin/tigecycline resistance (Figure 6B).
Figure 6.
DNR prevents the development of colistin/tigecycline resistance
(A) Resistance development assays of E. coli G92 (mcr-1) and E. coli B3-1 (tet(X4)) after sequential passages with colistin or tigecycline alone or in combination with DNR, respectively.
(B) MPC values of colistin or tigecycline in the presence of increasing concentrations of DNR against E. coli G92 (mcr-1) and E. coli B3-1 (tet(X4)), respectively. Data were obtained from two biological replicates.
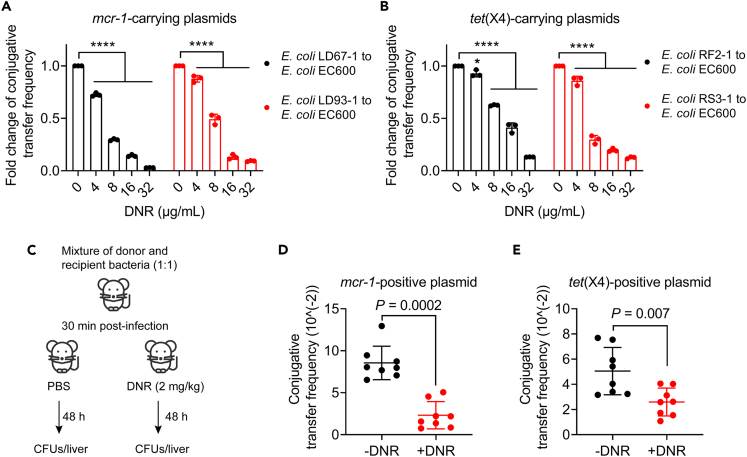
Considering that mcr-1 and tet(X4) genes can spread widely by conjugative plasmids, ultimately conferring resistance to susceptible bacteria,34,35 we next explored the effect of DNR on the conjugation frequency of colistin/tigecycline resistance plasmids. Interestingly, DNR significantly reduced the conjugation frequency of mcr-1/tet(X4)-harboring plasmids from clinical strains to the recipient bacteria E. coli EC600 in a concentration-dependent manner (Figures 7A and 7B). Furthermore, we evaluated the inhibitory effect of DNR on conjugal transfer in a murine model (Figure 7C). Consistently, the conjugation transfer frequency was also significantly reduced in vivo after DNR administration (Figures 7D and 7E). According to these findings, we conclude that DNR may be able to suppress the emergence and spread of mobile colistin/tigecycline resistance.
Figure 7.
DNR blocks the conjugative transfer of mcr-1/tet(X4)-carrying plasmids both in vitro and in murine models
(A and B) Fold change (FC) of conjugative transfer frequency of mcr-1-carrying plasmids (A) or tet(X4)-carrying plasmids (B) from clinical isolates to the recipient bacteria E. coli EC600 in the presence of increasing concentrations of DNR. Data were shown as mean ± SD, and statistical significance was determined by nonparametric one-way ANOVA.
(C) Schematic representation of conjugative transfer infection murine model.
(D and E) Conjugation frequency of mcr-1/tet(X4)-positive plasmids in vivo after treatment with DNR (2 mg/kg). Mann-Whitney U test was used to calculate p values.
Synergistic antibacterial mechanisms of DNR and colistin
Considering that colistin is a membrane-active antibiotic,36 we first evaluated whether DNR could enhance the destructive activity of colistin on the cell membrane. Bacterial membrane integrity was determined using the propidium iodide (PI) probe, which can only cross the damaged membrane. Expectedly, DNR alone significantly increased the fluorescence value excited by PI dye in a concentration-dependent manner, and when it was used in combination with colistin, the fluorescence units were significantly higher than those of colistin or DNR acting alone (Figures 8A and 8B), indicating the enhanced membrane permeability under DNR and colistin combination. Consistently, increased β-galactosidase activity exposed to DNR or their combination was observed, demonstrating that β-galactosidase was released extracellularly due to membrane disruption (Figures 8C and 8D). In connection with the opposite effects of divalent cations and EDTA on the potentiation of DNR to colistin, we reasoned that the increased membrane permeability and membrane damage by DNR in colistin-resistant bacteria is of great importance for their synergistic activity. Given that DNR and colistin combination exhibited superior membrane disruption in mcr-1-positive bacteria, we further investigated the interaction between DNR and MCR-1 protein using in silico docking analysis. Interestingly, docking analysis using MCR-1 as receptor and DNR as ligand showed that DNR had a high affinity with MCR-1, with −11.3 kcal/mol binding energies. Specifically, DNR can bind to MCR-1 protein by Van del Waals (Gly67, Ser89, Met265, and Asn267), hydrogen bond with Thr68, and pi-cation with Tyr72 (Figure 8E). In addition, we constructed point mutant strains at key sites and compared the synergy of DNR and colistin in these strains. The results showed that the synergistic effects of DNR with colistin against E. coli BL21 (pET28a) harboring the MCR-1 mutants (Ser89Ala, Pro266Ala, and Asn267Ala) were significantly reduced (Figure S5), indicating the important roles of these binding sites this interaction.
Figure 8.
DNR exacerbates bacterial membrane damage in together with colistin
(A and B) Bacterial membrane permeability of E. coli G92 (mcr-1) exposed to DNR (A), colistin alone, or colistin plus DNR (8 μg/mL) (B).
(C and D) β-galactosidase activity in E. coli G92 (mcr-1) after exposure to DNR (C), colistin alone, or colistin plus DNR (8 μg/mL) (D). Data were presented as mean ± SD from three biological replicates. two-way ANOVA was used to evaluate statistical significance between colistin plus DNR and colistin (∗∗∗∗p < 0.0001). Statistical significance of colistin plus DNR versus DNR (8 μg/mL) was determined by one-way ANOVA, and shown as ###p < 0.001, ####p < 0.0001.
(E) Interactions between DNR and the MCR-1 combining location residues.
In order to further explore the mechanisms of DNR and colistin synergism, we performed a transcriptomic analysis of E. coli G92 (mcr-1) after treatment with colistin alone or in combination with DNR. As a consequence, 243 up-regulated genes and 306 down-regulated genes were identified (Figure 9A). Gene ontology (GO) terms and kyoto encyclopedia of genes and genomes (KEGG) pathway enrichment analyses were applied to explore the roles of DEGs. SOS response, nitrate reductase complex, and nitrate reductase activity were the most common terms in biological process, cellular component, and molecular function enrichment (Figure 9B). Additionally, those significantly up-regulated genes were mainly involved in phenylalanine metabolism, galactose metabolism, nucleotide excision repair, and purine metabolism (Figure 9C), while the significantly down-regulated genes were enriched in nitrogen metabolism, quorum sensing, and ATP-binding cassette (ABC) transporter pathways (Figure 9D). The following RT-qPCR analysis indicated that the expression profiles of these representative genes in the above pathways were consistent with the transcriptional results (Figure S6A). Furthermore, DNR was revealed to inhibit the expression level of colistin resistance gene mcr-1 in a concentration-dependent manner (Figure S6B), which further explains why DNR and colistin combination has a stronger synergistic activity against drug-resistant bacteria than sensitive bacteria.
Figure 9.
Effect of DNR on mRNA expression in mcr-positive bacteria
(A) Volcano maps of gene expression changes in E. coli G92 (mcr-1) after exposure to colistin plus DNR compared to colistin alone. Differentially expressed genes (DEGs) were identified by gene-expression-level analysis with p values of ≤0.05 and fold change values ≥2 (log2FC ≥ 1 or log2 FC ≤ −1).
(B) Gene ontology (GO) terms of significantly DEGs in E. coli G92 (mcr-1). (C and D) KEEG functional enrichment analysis of up-regulated (C) and down-regulated (D) genes.
Notably, the addition of DNR remarkably enhanced the SOS response of damaged bacteria, and the significantly up-regulated genes, including recN, recA, dinB, dinI, sulA, etc. were all involved in this function, implying that DNR may induce DNA damage. Thus, we next investigated the effect of DNR on bacterial DNA. Using RT-qPCR, we found that the expression of recA gene, which encodes a key protein participating in the homologous DNA repair process, was significantly up-regulated with increasing DNR concentration (Figure 10A). To verify these results, checkerboard assays and killing curves were performed in ΔrecA strain. As a result, stronger synergistic activity between DNR and colistin was observed in ΔrecA strains compared with the parent isolate (Figures 10B–10D), indicating that bacterial DNA damage contributes to their synergistic effect.
Figure 10.
DNR-induced DNA damage is critical for its synergistic activity with colistin
(A) The expression levels of recA gene in E. coli G92 (mcr-1) after exposure to increasing concentrations of DNR. Data were shown as mean ± SD, and nonparametric one-way ANOVA was used to assess the statistical significance (∗∗p < 0.01, ∗∗∗∗p < 0.0001).
(B) Synergistic activity between colistin and DNR against E. coli J53 and its recA knockout bacteria.
(C) The deletion of the recA gene in E. coli enhances the synergistic bactericidal activity between DNR and colistin.
(D) Checkerboard assay between DNR and colistin against E. coli EC600 and its recA knockout bacteria.
It has been demonstrated that DNA damage increases ROS production,37 which is critical for the activity of bactericidal antibiotics. Thus, using the 2′,7′-dichlorodihydrofluorescein diacetate (DCFH-DA) probe, we assessed the intracellular ROS levels in E. coli G92 treated with DNR or DNR plus colistin. As a result, we observed that DNR markedly and concentration-dependently boosted the generation of ROS. Higher ROS levels were also found under combined treatment than colistin or DNR alone (Figures 11A and 11B). To further demonstrate the roles of ROS generation in the synergism, N-acetyl-L-cysteine (NAC) and thiourea, two ROS scavengers, were introduced to the subsequent checkerboard experiments and time-dependent death assays, respectively. Consistently, the ROS scavengers significantly reduced the potentiation of DNR to colistin (Figures 11C and 11D). Collectively, these findings indicate that the combined use of DNR and colistin increases membrane damage and induces DNA damage and the massive production of ROS, thereby leading to bacterial cell death.
Figure 11.
DNR triggers oxidative damage in mcr-positive bacteria
(A) Reactive oxygen species (ROS) generation in E. coli G92 (mcr-1) treated by varying concentrations of DNR. Nonparametric one-way ANOVA was used to establish statistical significance (∗∗∗∗p < 0.0001).
(B) Combination of DNR (8 μg/mL) and colistin results in higher ROS generation than colistin or DNR alone. Statistical significance was determined by two-way ANOVA (∗∗∗∗p < 0.0001) or one-way ANOVA between colistin plus DNR and DNR (8 μg/mL) (###p < 0.001).
(C) The synergistic antibacterial effect of colistin and DNR is decreased in the presence of the ROS scavengers NAC or thiourea (10 mM).
(D) Checkerboard experiments of colistin and DNR against E. coli G92 (mcr-1) in the absence or presence of ROS scavengers NAC or thiourea (10 mM).
DNR rescues colistin activity against drug-resistant bacteria in vivo
The effectiveness of this combination in one insect and two mammalian infection models was evaluated. Firstly, E. coli G92 (mcr-1) and K. pneumoniae D126 (mcr-8) were injected respectively to construct the infection models of Galleria mellonella, and then the larvae were treated individually or in combination. Although colistin alone was not effective in protecting G. mellonella larvae, combined treatments with colistin (1 or 5 mg/kg) and DNR (1 mg/kg) remarkably increased larval survival (Figures 12A and 12B). Additionally, a mouse model of peritonitis-sepsis elicited by E. coli G92 (mcr-1) further revealed the in vivo synergy. A single dose of combined treatment increased the survival rate of mice, increasing to 50% and 75% under the action of colistin plus DNR (2 + 0.5 mg/kg, 2 + 1 mg/kg), respectively. By contrast, colistin or DNR monotherapy was not effective in treating severe infections caused by MCR-1-positive E. coli (Figure 12C). Finally, a neutropenic mouse infection model was used to evaluate this synergism. The loads of mcr-1-positive bacteria in the mouse thigh muscles were considerably reduced by two combined administrations of colistin plus DNR (2 + 0.5 mg/kg, 2 + 1 mg/kg) (p = 0.0022 and p < 0.001, respectively) (Figure 12D). These data fully demonstrate that DNR restores the therapeutic effectiveness of colistin against mcr-positive bacterial infections in vivo.
Figure 12.
Colistin and DNR combination is effective in one insect and two mammalian infection models
(A and B) Survival rates of G. mellonella larvae (n = 8 per group) infected by E. coli G92 (mcr-1) (A) or K. pneumoniae (mcr-8) (B) under the treatment of colistin (1 or 5 mg/kg), DNR (1 mg/kg) alone or in combination. p values were calculated using log rank (Mantel-Cox) test.
(C) Survival rates of infected mice (n = 8 per group) treated by colistin (2 mg/kg), DNR (1 mg/kg), or their combination (2 + 0.5 or 2 + 1 mg/kg). The mice were infected by E. coli G92 (mcr-1). p values between COL and COL + DNR were calculated using log rank (Mantel-Cox) test.
(D) In comparison to colistin alone, the combination treatment of colistin and DNR considerably decreases bacterial burdens in the thighs of mice (n = 6 per group). CFU, colony-forming unit. p values were determined by Mann-Whitney U test.
Discussion
The increasing incidence of MDR-GN bacterial infections in the clinical setting has drastically compromised the efficacy of traditional antibiotic treatments. As a result, individuals with severe Gram-negative infections have limited therapy alternatives, which deepens the need for novel antibacterial strategies. Repurposing PACs as prospective antibiotic adjuvants is a promising pipeline to enhance the antibacterial activity of current antibiotics, decrease their dosages, and improve their biosafety by restoring the susceptibility of drug-resistant bacteria to antibiotics.38,39 In this study, we demonstrated that the FDA-approved antineoplastic drug DNR is a potent and multifunctional adjuvant for three last-line antibiotics, including meropenem, colistin, and tigecycline, in fighting against blaNDM/mcr/tet(X)-harboring pathogens. Notably, the synergistic activity of DNR on colistin is superior to the other two antibiotics, with an increased activity up to 1,000-fold, indicating that DNR is a super colistin potentiator for MCR-positive bacteria. Furthermore, DNR significantly reduced the conjugative transfer of mcr/tet(X)-plasmids both in vitro and in a mouse model.
The mechanisms of action responsible for the potentiation of DNR to three antibiotics appear to be different. With regard to meropenem, we speculated that DNR may be a novel inhibitor of metallo-β-lactamase because such synergism was only detected in NDM-positive bacteria. With regard to tigecycline, we suspect that it may be related to the different effects of DNR on protein because tigecycline exerts its bactericidal activity mainly by destroying protein. Considering the unprecedented potentiating effect of DNR on colistin, we sought to elucidate the underlying molecular mechanisms. Colistin is a membrane-disrupting antibiotic, and its antibacterial mechanism is currently known to be related to membrane damage.40 In particular, colistin can specifically bind to lipopolysaccharides (LPS) located on the outer cell membrane, thereby resulting in membrane disruption. However, the mcr-1 and its variations encoding phosphoethanolamine transferases decrease the negative charge of lipid A and enable bacteria resistant to colistin. Consistently, our study revealed that DNR significantly enhanced cell membrane permeability in MCR-positive bacteria. Heretofore, several compounds have been reported to restore the susceptibility of resistant bacteria to colistin, such as melatonin,41 SLAP-25,42 and PBT2,43 by preventing the modification of lipid A by MCR-1. Our research also discovered that DNR can decrease the expression of resistance gene mcr-1, which in turn can reduce the modification of lipid A that MCR-1 causes. Moreover, docking analysis showed that DNR was a potential inhibitor of MCR protein. In line with these findings, DNR and colistin exhibited a more synergistic effect against mcr-positive bacteria than mcr-negative bacteria. In addition, we also found that DNR promoted DNA damage and massive production of ROS. Consistent with the antitumor mechanism of DNR-induced DNA double-strand breaks, DNR also caused significant DNA damage in prokaryotic cells. It has been demonstrated that DNA damage stimulates the creation of ROS and that within 2 h, the level of ROS in DNA-damaged cells grew dramatically, which can cause a death pathway resembling apoptosis.44 Accordingly, DNA damage can cause the production of ROS through the H2AX-reduced coenzyme II oxidase 1 (Noxl)/Racl channel.45 Of course, DNA damage and oxidative stress caused by ROS can promote each other, and excess ROS can in turn lead to DNA damage in various forms, such as double-strand breaks, double-strand aberrations, and site mutations. Nevertheless, the detailed mode of action between DNR and various antibiotics remains to be studied.
Acute toxicity tests showed that DNR (5 mg/kg) hardly increases the hepatic and renal toxicity of colistin. Moreover, our animal studies indicated that low doses of DNR (0.5 and 1 mg/kg) combined with colistin successfully treated drug-resistant bacterial infections, suggesting the therapeutic potential of this combination in clinical practice. To further improve drug efficacy and reduce the side effects of DNR, targeted drug delivery46 and chemical structure optimization can be performed. For example, the toxicity of antitumor doxorubicin was significantly reduced when encapsulated in liposomesthe.47 In addition, demethoxy DNR (idarubicin, IDA) with a removed methoxy group at the C4 position remarkably improved its efficacy and reduced its toxicity.48
In conclusion, we reveal that DNR resensitizes MDR Gram-negative pathogens to the last-line antibiotics, and the synergism with colistin is the strongest. DNR significantly enhances membrane permeability of MCR-positive bacteria and induces DNA damage and oxidative stress. The synergistic effect of DNR and colistin is further evidenced in multiple animal models of infection. Together, our study provides a new drug combination strategy for the treatment of increasing MDR Gram-negative bacterial infections.
Limitations of the study
In this study, although we have investigated the effectiveness of DNR and colistin in multiple animal models of infection, further work is still needed to verify the in vivo efficacy and safety of this combination therapy in clinical practice.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals | ||
| Ampicillin | Yuanye Bio-Technology | Cat# S17018 |
| Doxycycline | Yuanye Bio-Technology | Cat# S27317 |
| Ciprofloxacin | Yuanye Bio-Technology | Cat# S17013 |
| Vancomycin | Yuanye Bio-Technology | Cat# Y25829 |
| Rifampicin | Yuanye Bio-Technology | Cat# B25308 |
| Meropenem | Yuanye Bio-Technology | Cat# S31659 |
| Colistin | Yuanye Bio-Technology | Cat# S17057 |
| Tigecycline | Yuanye Bio-Technology | Cat# S24031 |
| Daunorubicin | Yuanye Bio-Technology | Cat# S30893 |
| PBS | Solarbio | Cat# P1020 |
| LB Broth | Solarbio | Cat# L8291 |
| MHB | Solarbio | Cat# M8556 |
| LB agar | Solarbio | Cat# L8290 |
| EDTA | Solarbio | Cat# E1007 |
| Serum | Solarbio | Cat# S9040 |
| DMEM | Solarbio | Cat# 12400024 |
| Glycerol | Solarbio | Cat# G8192 |
| Triton X-100 | Solarbio | Cat# T8200 |
| NaCl | Sigma | Cat# S9888 |
| KCl | Sigma | Cat# P3911 |
| MgCl2 | Sigma | Cat# M8266 |
| CaCl2 | Sigma | Cat# C1016 |
| N-acetylcysteine | Sigma | Cat# N7250 |
| Thiourea | Sigma | Cat# T8656 |
| Methanol | Sigma | Cat# 34860 |
| Crystal violet | Sigma | Cat# C0775 |
| Acetic acid | Sigma | Cat# 1005706 |
| Propidium iodide | Sigma | Cat# P4170 |
| DCFH-DA | Sigma | Cat# D6883 |
| ONPG | Sigma | Cat# N1127 |
| fresh sheep blood cells | Biofeng | Cat# A332-02 |
| DMSO | Solarbio | Cat# D8372 |
| Critical commercial assays | ||
| EASYspinPlus bacterial RNA extraction kit | Vazyme | Cat# 201-02 |
| Experimental models: Organisms | ||
| G. mellonella larvae | Huiyude Biotech Company | N/A |
| BALB/c or ICR mice | Comparative Medicine Center of Yangzhou University | N/A |
| Software and algorithms | ||
| Autodock Vina | Autodock Vina software | https://vina.scripps.edu/ |
| Prism version 9.0 | GraphPad software | https://www.graphpad.com/ |
| Other | ||
| RNA-sequencing data | NCBI | PRJNA818073 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Yuan Liu (liuyuan2018@yzu.edu.cn).
Materials availability
All unique/stable reagents used or generated in this study will be made available on request.
Experimental model and subject details
Bacterial strains and reagents
Table S1 contains a list of the bacterial strains employed in this study. All tested strains were cultivated in a liquid medium, such as MHB, LB Broth, or LB agar plates. The strains that needed to be preserved were placed in 40% glycerol and stored at −80°C.
Mice
Female BALB/c or ICR mice (6–8 weeks old) were purchased from Yangzhou University’s Comparative Medicine Center (Jiangsu, China). The applicable regulations of the Jiangsu Laboratory Animal Welfare and Ethical of Jiangsu Administrative Committee of Laboratory Animals were followed in all mouse-related experiments (permission number, SYXKSU-2007-0005). Jiangsu Association for Science and Technology has granted SCXK-2017-0044 as the license number for the use of experimental animals.
Method details
Antibacterial test
Micro-broth dilution method, with reference to CLSI 2021,49 was used to ascertain the MICs of drugs. In a 96-well plate containing cation-adjusted MH broth (MHB), a twofold dilution series of drug was prepared, and then an equal volume (100 μL/well) of bacterial suspension (1.5 × 106 CFUs/mL) was added. The plates were then kept at 37°C for 18 h. The MICs were defined as the lowest concentration of compound at which no bacterial growth could be observed.
Checkerboard assay
Checkerboard experiment was used to investigate the synergistic activity between DNR and antibiotics according to our previous studies.50,51 Briefly, antibiotics and DNR were diluted into 96-well plates in either horizontal or vertical coordinates, respectively. A microplate reader was used to measure the OD value at 600 nm following 18 h of culture at 37°C (The OD value greater than 0.1 indicates bacterial growth). FICI was calculated as the sum of MICab/MICa and MICba/MICb. MICa and MICb are the corresponding MICs of compounds A and B alone; MICab is the MIC of compound A combined with compound B; MICba is the MIC of compound B combined with compound A. Indicative of synergy is FICI ≤ 0.5.
Checkerboard experiments were carried out to assess the impact of metal ions, EDTA, serum, and DMEM on the stability of the synergistic activity between DNR and colistin/tigecycline. In this process, the MHB containing 10 mM Na+, K+, Ca2+, Mg2+ or EDTA, 10% DMEM, 10% serum was used. To investigate the roles of ROS production, ROS scavengers N-acetylcysteine (NAC, 10 mM) or thiourea (10 mM) were added to MHB. Furthermore, checkerboard assays of colistin and DNR against E. coli J53 and E. coli J53 (ΔrecA) were also performed.
Time-dependent killing curves
Bactericidal curves of drugs against drug-resistant bacteria in the exponential and stationary phases were determined respectively.52 Overnight E. coli G92 (mcr-1) and E. coli B3-1 (tet(X4)) was diluted 1:100 in MHB. Bacterial dilutions were cultured for 2 h (to exponential growth phase) and 4 h (to stationary phase), respectively. Subsequently, bacteria cells were treated with DNR (32 μg/mL) or colistin (4 μg/mL)/tigecycline (32 μg/mL) alone or in combination, and a control group without drugs was set up. At each time point (0, 4, 8 and 24 h), 100 μL of the bacterial culture was pipetted and continuously diluted 10-fold in PBS. The suspension was then plated onto LB agar plates and cultured for an overnight period at 37°C. The number of colonies was counted after 18 h of culture. The time-dependent killing curves of a reference strain E. coli J53 and E. coli J53 (ΔrecA) after exposure to the combination of DNR (32 μg/mL) and colistin (4 μg/mL) were also determined, respectively.
Toxicity analysis
Hemolysis of drugs was measured based on our previous study.53 Briefly, colistin (0 to 16 μg/mL), tigecycline (0 to 128 μg/mL), or the combination of colistin/tigecycline with DNR (0 to 64 μg/mL) were applied to 8% of fresh sheep blood cells for 1 h at 37°C. Triton X-100 (0.2%) was chosen as a positive control and PBS was used as blank control. The formula below was used to calculate the hemolysis rate based on the absorbance of suspension at 576 nm:
Hemolysis (%) = [(OD576 sample-OD576 blank) / (OD576 Triton X-100-OD576 blank)] × 100%.
Acute toxicity of colistin combined with DNR in mice was evaluated over 7 days.54 BALB/c mice were randomly split into two groups (n = 8 per group). A single dose of colistin (10 mg/kg) and a mixture of colistin and DNR (10 + 5 mg/kg) were intraperitoneally administered into mice. Survival of mice was monitored during 7 consecutive days, and the changes in body weight were recorded. On the 7th day, the mice blood was collected for following blood routine and biochemical parameters analysis.
Biofilm inhibition assay
Crystal violet method was used to evaluate DNR's capacity to prevent the formation of biofilm.55
Colistin (0 to 0.063 μg/mL), tigecycline (0 to 2 μg/mL), or the combination of colistin/tigecycline and DNR (8 μg/mL) were added to the E. coli G92 and E. coli B3-1 culture, respectively. After 48 h of incubation, cells were washed three times with 300 μL PBS. Next, 200 μL methanol was added to fix the cells for 15 min. After air-drying, 100 μL 0.1% crystal violet was used to stain for 15 min. Stained biofilm was then rinsed with PBS three times and allowed to air dry naturally. Finally, 100 μL of 33% glacial acetic acid was used to dissolve the stained biofilms. The absorbance of suspension at 570 nm was measured after 30 min of incubation at 37°C.
Biofilm eradication assay
Overnight E. coli G92 and E. coli B3-1 were diluted 1:100 into LB broth and incubated for 4 h at 37°C. A 96-well flat-bottom plate was filled with an equal volume of MHB and 100 μL of bacterial solution. The planktonic bacteria were removed after a 48-h incubation period at 37°C. Colistin (0 to 32 μg/mL), tigecycline (0 to 256 μg/mL), and 8 μg/mL DNR were used to eliminate the mature biofilms, either alone or in combination. After 2 h incubation at 37°C, the remaining cells were dispersed by sonication for 20 min. The mixture was then redissolved, diluted and plated on LB agar plates overnight at 37°C. The colonies were counted after 18 h.
Resistance development study
E. coli G92 and E. coli B3-1 overnight cultures were diluted 1:100 into LB broth containing 0.5 × MIC of colistin/tigecycline or combined with 0.25 × MIC DNR. After 12 h of incubation, bacterial culture was diluted 1:100 and added to a new medicated medium to continue the next generation. Every four passages, the MIC of the cultures was examined. The serial passaging was continued for 24 days.
Mutation preventive concentration assay
Colistin/tigecycline or its combination with DNR were present at various concentrations on LB agar plates. On the corresponding resistant agar plates, 100 μL of E. coli G92 and E. coli B3-1 (1.0 × 1010 CFUs) were plated, and the plates were then incubated at 37°C. Bacterial growth was monitored after 72 h, and the drug's MPC was defined as the lowest concentration that could prevent resistance development (mutant colonies).56
Conjugation assays
Conjugation assays were performed by measuring the conjugation frequency between donor and recipient in the absence or presence of DNR.57 E. coli LD93-1 and E. coli LD67-1 were the mcr-1 donors, whereas E. coli RS3-1 and E. coli RF2-1 were the tet(X4) donors. Bacteria were grown at 37°C until their OD600 value reached 0.5. In a total amount of 2 mL, the donor and recipient were combined in a 1:1 ratio, and then various concentrations of DNR (0 to 32 μg/mL) was added to the mixture. The mixture was serially diluted and plated on LB agar plates with single- or double-drug following an 18 h incubation period at 37°C. According to bacterial CFUs, conjugators and conjugation frequencies were calculated.
Referring to the above results, E. coli LD67-1 was chosen as the mcr-1 donor, and E. coli RF2-1 was chosen as the tet(X4) donor for the following murine models. A 200 μL injection of a mixture of donor and recipient microorganisms was given to female ICR mice (6-8 weeks old) in the right abdominal cavity. After 30 min infection, 200 μL PBS or DNR (2 mg/kg) was intraperitoneally delivered (n = 8 per group), respectively. The mice were slaughtered by cervical dislocation 48 h after infection. To measure CFUs titers, the liver was removed, homogenized, continuously diluted, and plated on LB agar plates with single- or double-drugs.
Membrane permeability and ROS level measurement
E. coli G92 overnight cultures were centrifuged and resuspended with PBS to obtain an OD600 of 0.5. Then, fluorescent dyes were incubated with the bacterial suspension for 15 min at dark. 190 μL of probe-labeled bacterial cells were placed to a 96-well plate after 30 min of incubation, then 10 μL of colistin (0 to 4 μg/mL), DNR (0 to 16 μg/mL), or a combination of colistin and DNR (8 μg/mL) were added. The fluorescence intensity was recorded after 1 h of incubation using a microplate reader (Tecan, Mannedorf, Switzerland).
Cell membrane integrity
To assess the integrity of the cell membrane, bacterial cells were stained with PI (0.5 μM).42 At wavelengths of 535 nm for excitation and 615 nm for emission, the fluorescence intensity was measured.
Total ROS production
2,7-Dichlorodihydrofluorescein diacetate (DCFH-DA, 10 μM) was used to measure ROS levels.58 Cells were rinsed three times with PBS and resuspended in PBS after being treated with ROS probes for 30 min. The fluorescence intensity was measured at the excitation wavelength of 488 nm and the emission wavelength of 525 nm.
β-Galactosidase activity assay
100 μL suspension was taken after treatment with colistin (0 to 4 μg/mL), DNR (0 to 16 μg/mL), or a combination of colistin and DNR (8 μg/mL) for 1 h. After adding 3 mM of O-nitrophenyl β-D-galactopyranoside (ONPG), the absorbance of suspension at 420 nm was measured following a 30-min incubation period at 37°C.
Docking analysis
The crystal structure of the MCR-1 protein (PDB: 5YLF) was used as a template. Using the Autodock Vina tool, molecular docking between MCR-1 and DNR was carried out without the use of water molecules. Discovery Studio 4.5 displays the interactions of DNR with the residues of the binding sites in MCR-1 as a two-dimensional graphic. The MCR-1 mutants (Ser69Ala, Pro266Ala and ASN267Ala) were constructed in E. coli BL21 (pET28a-MCR-1) using optimized primers (Table S7).
Transcriptomic analysis
Overnight E. coli G92 were diluted 1:100 in LB broth and cultured at 37°C for 2 h (until they reached the exponential phase). Then, cells were treated with colistin (4 μg/mL) alone or in combination with DNR (32 μg/mL) for 8 h. Total RNA was extracted using the EASYspinPlus bacterial RNA extraction kit. Subsequently, Hiseq2000 was sequenced with TruseqSBS kit v3HS (200 cycles) with a read length of 2 × 100 (PE100). Filter the original sequencing readings and map the reference genome for E. coli K-12. Using the FPKM (Fragments Per Kilobase of transcript per Million mapped reads) method, differentially expressed genes with a P-value ≤ 0.05 and a FC value ≥ 2 (log2 FC ≥ 1 or log2 FC ≤ −1) were identified. Genes were then subjected to functional enrichment analysis, including GO enrichment and KEGG enrichment.
RT-qPCR analysis
RT-qPCR analysis was performed by the SYBR Green I chimeric fluorescence method using the 7500 Fast Real-Time PCR System under the conditions of pre-denaturation (95°C, 30 s) and 40 cycles (95°C, 10 s; 60°C, 30 s). The change of mRNA expression after combined treatment was determined by 2−ΔΔCt method.
Galleria mellonella infection model
G. mellonella larvae were randomly separated into four groups (n = 8 per group), and subsequently infected with either E. coli G92 or K. pneumoniae suspension (10 μL, 1.0 × 105 CFU per larva, respectively) at the right caudal foot. PBS, colistin (1 or 5 mg/kg), DNR (1 mg/kg), or colistin and DNR (1 + 1 mg/kg or 5 + 1 mg/kg) (10 μL) were administered to the larva's left caudal foot at one hour after infection, respectively. The survival rates of larvae were monitored regularly for 7 days.
Mouse peritonitis-sepsis infection model
E. coli G92 suspension (200 μL, 1.0 × 108 CFU per mouse) was intraperitoneally administered into female BALB/c mice (n = 8 per group). At one-hour post-infection, PBS, colistin (1 mg/kg or 5 mg/kg), DNR (1 mg/kg), or the combination of colistin and DNR (1 + 1 mg/kg or 5 + 1 mg/kg) were intraperitoneally injected, respectively. The survival rates of mice were recorded continuously for 7 days.
Neutropenic mouse thigh infection model
Female BALB/c mice (n = 6 per group) were intraperitoneally injected with cyclophosphamide at doses of 150 and 100 mg/kg on days 4 and 1 before infection, respectively. E. coli G92 (100 μL, 1.0 × 105 CFU per mouse) was then administered intramuscularly to the right thigh of mice. Then, 100 μL of PBS, colistin (2 mg/kg), DNR (1 mg/kg), or their combinations (2 + 0.5 mg/kg or 2 + 1 mg/kg) were administered intraperitoneally at one-hour post-infection. At 48 h post-infection, mice were slaughtered by cervical dislocation and the thigh muscles were removed, diluted and plated on LB agar plates. Following an 18 h incubation period at 37°C, bacterial colonies were counted accordingly.
Quantification and statistical analysis
GraphPad Prism version 9.0 was used to analyze all data, which is displayed as mean ± SD. Unpaired t-test or one-way ANOVA for multiple comparisons were carried out for the in vitro testing. In animal models, statistical significance of survival rate and bacterial load were analyzed by log-rank (Mantel-Cox) test or Mann-Whitney U test, respectively. P < 0.05 was defined as a significant difference.
Acknowledgments
This study was supported by the National Key Research and Development Program of China (2021YFD1801000 and 2018YFA0903400), National Natural Science Foundation of China (32222084, 32172907, and 32002331), Jiangsu Agricultural Science and Technology Innovation Fund (CX(21)2010), a project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD) and 111 Project D18007.
Author contributions
Y.L. and Z.W. designed and supervised the project. J.C. and T.D. performed experiment, analyzed data, and drafted the manuscript. J.S. helped perform experiments. C.C. helped analyze data. Y.L. and J.C. wrote and revised the manuscript. All of the authors read and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: May 4, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.106809.
Contributor Information
Zhiqiang Wang, Email: zqwang@yzu.edu.cn.
Yuan Liu, Email: liuyuan2018@yzu.edu.cn.
Supplemental information
Data and code availability
-
•
This paper does not report original code.
-
•
RNA-sequencing data have been deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive database (PRJNA818073) and are publicly available as of the date of publication.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Kuehn B. Antibiotic resistance challenge. JAMA. 2018;320:1851. doi: 10.1001/jama.2018.16587. [DOI] [PubMed] [Google Scholar]
- 2.Antimicrobial Resistance Collaborators. Ikuta K.S., Sharara F., Swetschinski L., Aguilar G.R., Gray A., Han C., Bisignano C., Rao P., Wool E., et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022;399:629–655. doi: 10.1016/S0140-6736(21)02724-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Domalaon R., Idowu T., Zhanel G.G., Schweizer F. Antibiotic hybrids: the next generation of agents and adjuvants against Gram-negative pathogens? Clin. Microbiol. Rev. 2018;31:000777–000817.e117. doi: 10.1128/CMR.00077-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.MacNair C.R., Tsai C.N., Brown E.D. Creative targeting of the Gram-negative outer membrane in antibiotic discovery. Ann. N. Y. Acad. Sci. 2020;1459:69–85. doi: 10.1111/nyas.14280. [DOI] [PubMed] [Google Scholar]
- 5.Ropponen H.K., Richter R., Hirsch A.K.H., Lehr C.M. Mastering the Gram-negative bacterial barrier - chemical approaches to increase bacterial bioavailability of antibiotics. Adv. Drug Deliv. Rev. 2021;172:339–360. doi: 10.1016/j.addr.2021.02.014. [DOI] [PubMed] [Google Scholar]
- 6.Ruppé É., Woerther P.L., Barbier F. Mechanisms of antimicrobial resistance in Gram-negative bacilli. Ann. Intensive Care. 2015;5:61. doi: 10.1186/s13613-015-0061-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumarasamy K.K., Toleman M.A., Walsh T.R., Bagaria J., Butt F., Balakrishnan R., Chaudhary U., Doumith M., Giske C.G., Irfan S., et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect. Dis. 2010;10:597–602. doi: 10.1016/S1473-3099(10)70143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biswas S., Brunel J.M., Dubus J.C., Reynaud-Gaubert M., Rolain J.M. Colistin: an update on the antibiotic of the 21st century. Expert Rev. Anti Infect. Ther. 2012;10:917–934. doi: 10.1586/eri.12.78. [DOI] [PubMed] [Google Scholar]
- 9.Nang S.C., Azad M.A.K., Velkov T., Zhou Q.T., Li J. Rescuing the last-line polymyxins: achievements and challenges. Pharmacol. Rev. 2021;73:679–728. doi: 10.1124/pharmrev.120.000020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stein G.E., Babinchak T. Tigecycline: an update. Diagn. Microbiol. Infect. Dis. 2013;75:331–336. doi: 10.1016/j.diagmicrobio.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 11.Liu Y.Y., Wang Y., Walsh T.R., Yi L.X., Zhang R., Spencer J., Doi Y., Tian G., Dong B., Huang X., et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 2016;16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 12.Sun J., Chen C., Cui C.Y., Zhang Y., Liu X., Cui Z.H., Ma X.Y., Feng Y., Fang L.X., Lian X.L., et al. Plasmid-encoded tet(X) genes that confer high-level tigecycline resistance in Escherichia coli. Nat. Microbiol. 2019;4:1457–1464. doi: 10.1038/s41564-019-0496-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Czaplewski L., Bax R., Clokie M., Dawson M., Fairhead H., Fischetti V.A., Foster S., Gilmore B.F., Hancock R.E.W., Harper D., et al. Alternatives to antibiotics-a pipeline portfolio review. Lancet Infect. Dis. 2016;16:239–251. doi: 10.1016/S1473-3099(15)00466-1. [DOI] [PubMed] [Google Scholar]
- 14.Baym M., Stone L.K., Kishony R. Multidrug evolutionary strategies to reverse antibiotic resistance. Science. 2016;351:aad3292. doi: 10.1126/science.aad3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Payne D.J., Gwynn M.N., Holmes D.J., Pompliano D.L. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discov. 2007;6:29–40. doi: 10.1038/nrd2201. [DOI] [PubMed] [Google Scholar]
- 16.Robbel L., Marahiel M.A. Daptomycin, a bacterial lipopeptide synthesized by a nonribosomal machinery. J. Biol. Chem. 2010;285:27501–27508. doi: 10.1074/jbc.R110.128181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Douafer H., Andrieu V., Phanstiel O., 4th, Brunel J.M. Antibiotic adjuvants: make antibiotics great again. J. Med. Chem. 2019;62:8665–8681. doi: 10.1021/acs.jmedchem.8b01781. [DOI] [PubMed] [Google Scholar]
- 18.Liu Y., Li R., Xiao X., Wang Z. Antibiotic adjuvants: an alternative approach to overcome multi-drug resistant Gram-negative bacteria. Crit. Rev. Microbiol. 2019;45:301–314. doi: 10.1080/1040841X.2019.1599813. [DOI] [PubMed] [Google Scholar]
- 19.Wright G.D. Antibiotic adjuvants: rescuing antibiotics from resistance. Trends Microbiol. 2016;24:862–871. doi: 10.1016/j.tim.2016.06.009. [DOI] [PubMed] [Google Scholar]
- 20.Ding Y., Li Z., Xu C., Qin W., Wu Q., Wang X., Cheng X., Li L., Huang W. Fluorogenic probes/inhibitors of beta-lactamase and their applications in drug-resistant bacteria. Angew. Chem. Int. Ed. 2021;60:24–40. doi: 10.1002/anie.202006635. [DOI] [PubMed] [Google Scholar]
- 21.King A.M., Reid-Yu S.A., Wang W., King D.T., De Pascale G., Strynadka N.C., Walsh T.R., Coombes B.K., Wright G.D. Aspergillomarasmine A overcomes metallo-beta-lactamase antibiotic resistance. Nature. 2014;510:503–506. doi: 10.1038/nature13445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang R., Lai T.P., Gao P., Zhang H., Ho P.L., Woo P.C.Y., Ma G., Kao R.Y.T., Li H., Sun H. Bismuth antimicrobial drugs serve as broad-spectrum metallo-beta-lactamase inhibitors. Nat. Commun. 2018;9:439. doi: 10.1038/s41467-018-02828-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maiden M.M., Hunt A.M.A., Zachos M.P., Gibson J.A., Hurwitz M.E., Mulks M.H., Waters C.M. Triclosan is an aminoglycoside adjuvant for eradication of Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 2018;62:001466-18–e218. doi: 10.1128/AAC.00146-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aubel-Sadron G., Londos-Gagliardi D. Daunorubicin and doxorubicin, anthracycline antibiotics, a physicochemical and biological review. Biochimie. 1984;66:333–352. doi: 10.1016/0300-9084(84)90018-x. [DOI] [PubMed] [Google Scholar]
- 25.Krauss A.C., Gao X., Li L., Manning M.L., Patel P., Fu W., Janoria K.G., Gieser G., Bateman D.A., Przepiorka D., et al. FDA approval summary: (daunorubicin and cytarabine) liposome for injection for the treatment of adults with high-risk acute myeloid leukemia. Clin. Cancer Res. 2019;25:2685–2690. doi: 10.1158/1078-0432.CCR-18-2990. [DOI] [PubMed] [Google Scholar]
- 26.Gumpert J., Dornberger K., Smith T.H. Antimicrobial activities of daunorubicin and adriamycin derivatives on bacterial and protoplast type L-form cells of Bacillus subtilis 170, Escherichia coli B, and Proteus mirabilis VI. Structure--activity relationship. Z. Allg. Mikrobiol. 1982;22:687–692. doi: 10.1002/jobm.3630221002. [DOI] [PubMed] [Google Scholar]
- 27.Hind C.K., Dowson C.G., Sutton J.M., Jackson T., Clifford M., Garner R.C., Czaplewski L. Evaluation of a library of FDA-approved drugs for their ability to potentiate antibiotics against multidrug-resistant Gram-negative pathogens. Antimicrob. Agents Chemother. 2019;63:007699–007719.e819. doi: 10.1128/AAC.00769-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koo H., Allan R.N., Howlin R.P., Stoodley P., Hall-Stoodley L. Targeting microbial biofilms: current and prospective therapeutic strategies. Nat. Rev. Microbiol. 2017;15:740–755. doi: 10.1038/nrmicro.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan J., Bassler B.L. Surviving as a community: antibiotic tolerance and persistence in bacterial biofilms. Cell Host Microbe. 2019;26:15–21. doi: 10.1016/j.chom.2019.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Breij A., Riool M., Cordfunke R.A., Malanovic N., de Boer L., Koning R.I., Ravensbergen E., Franken M., van der Heijde T., Boekema B.K., et al. The antimicrobial peptide SAAP-148 combats drug-resistant bacteria and biofilms. Sci. Transl. Med. 2018;10 doi: 10.1126/scitranslmed.aan4044. [DOI] [PubMed] [Google Scholar]
- 31.Zipperer A., Konnerth M.C., Laux C., Berscheid A., Janek D., Weidenmaier C., Burian M., Schilling N.A., Slavetinsky C., Marschal M., et al. Human commensals producing a novel antibiotic impair pathogen colonization. Nature. 2016;535:511–516. doi: 10.1038/nature18634. [DOI] [PubMed] [Google Scholar]
- 32.Liu Y.F., Wang L., Bu C.P., Wang G.Q., Zhang Y.H., Fang S.M., Shi W.Z. Synthesis of luminescent ag nanoclusters with antibacterial activity. J. Nanomater. 2015;2015:1–8. [Google Scholar]
- 33.Wistrand-Yuen E., Knopp M., Hjort K., Koskiniemi S., Berg O.G., Andersson D.I. Evolution of high-level resistance during low-level antibiotic exposure. Nat. Commun. 2018;9:1599. doi: 10.1038/s41467-018-04059-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li R., Lu X., Peng K., Liu Z., Li Y., Liu Y., Xiao X., Wang Z. Deciphering the structural diversity and classification of the mobile tigecycline resistance gene tet(X)-bearing plasmidome among bacteria. mSystems. 2020;5:001344–001420.e220. doi: 10.1128/mSystems.00134-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang R., van Dorp L., Shaw L.P., Bradley P., Wang Q., Wang X., Jin L., Zhang Q., Liu Y., Rieux A., et al. The global distribution and spread of the mobilized colistin resistance gene mcr-1. Nat. Commun. 2018;9:1179. doi: 10.1038/s41467-018-03205-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aslan A.T., Akova M. The role of colistin in the era of new beta-lactam/beta-lactamase inhibitor combinations. Antibiotics. 2022;11:277. doi: 10.3390/antibiotics11020277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Evert B.A., Salmon T.B., Song B., Jingjing L., Siede W., Doetsch P.W. Spontaneous DNA damage in Saccharomyces cerevisiae elicits phenotypic properties similar to cancer cells. J. Biol. Chem. 2004;279:22585–22594. doi: 10.1074/jbc.M400468200. [DOI] [PubMed] [Google Scholar]
- 38.Bernal P., Molina-Santiago C., Daddaoua A., Llamas M.A. Antibiotic adjuvants: identification and clinical use. Microb. Biotechnol. 2013;6:445–449. doi: 10.1111/1751-7915.12044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Y., Tong Z., Shi J., Li R., Upton M., Wang Z. Drug repurposing for next-generation combination therapies against multidrug-resistant bacteria. Theranostics. 2021;11:4910–4928. doi: 10.7150/thno.56205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sabnis A., Hagart K.L., Klöckner A., Becce M., Evans L.E., Furniss R.C.D., Mavridou D.A., Murphy R., Stevens M.M., Davies J.C., et al. Colistin kills bacteria by targeting lipopolysaccharide in the cytoplasmic membrane. Elife. 2021;10 doi: 10.7554/eLife.65836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Y., Jia Y., Yang K., Tong Z., Shi J., Li R., Xiao X., Ren W., Hardeland R., Reiter R.J., Wang Z. Melatonin overcomes MCR-mediated colistin resistance in Gram-negative pathogens. Theranostics. 2020;10:10697–10711. doi: 10.7150/thno.45951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Song M., Liu Y., Huang X., Ding S., Wang Y., Shen J., Zhu K. A broad-spectrum antibiotic adjuvant reverses multidrug-resistant Gram-negative pathogens. Nat. Microbiol. 2020;5:1040–1050. doi: 10.1038/s41564-020-0723-z. [DOI] [PubMed] [Google Scholar]
- 43.De Oliveira D.M.P., Bohlmann L., Conroy T., Jen F.E.C., Everest-Dass A., Hansford K.A., Bolisetti R., El-Deeb I.M., Forde B.M., Phan M.D., et al. Repurposing a neurodegenerative disease drug to treat Gram-negative antibiotic-resistant bacterial sepsis. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.abb3791. [DOI] [PubMed] [Google Scholar]
- 44.Lam S.J., O'Brien-Simpson N.M., Pantarat N., Sulistio A., Wong E.H.H., Chen Y.Y., Lenzo J.C., Holden J.A., Blencowe A., Reynolds E.C., Qiao G.G. Combating multidrug-resistant Gram-negative bacteria with structurally nanoengineered antimicrobial peptide polymers. Nat. Microbiol. 2016;1 doi: 10.1038/nmicrobiol.2016.162. [DOI] [PubMed] [Google Scholar]
- 45.Kang M.A., So E.Y., Simons A.L., Spitz D.R., Ouchi T. DNA damage induces reactive oxygen species generation through the H2AX-Nox1/Rac1 pathway. Cell Death Dis. 2012;3:e249. doi: 10.1038/cddis.2011.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Allen T.M., Cullis P.R. Liposomal drug delivery systems: from concept to clinical applications. Adv. Drug Deliv. Rev. 2013;65:36–48. doi: 10.1016/j.addr.2012.09.037. [DOI] [PubMed] [Google Scholar]
- 47.Batist G., Ramakrishnan G., Rao C.S., Chandrasekharan A., Gutheil J., Guthrie T., Shah P., Khojasteh A., Nair M.K., Hoelzer K., et al. Reduced cardiotoxicity and preserved antitumor efficacy of liposome-encapsulated doxorubicin and cyclophosphamide compared with conventional doxorubicin and cyclophosphamide in a randomized, multicenter trial of metastatic breast cancer. J. Clin. Oncol. 2001;19:1444–1454. doi: 10.1200/JCO.2001.19.5.1444. [DOI] [PubMed] [Google Scholar]
- 48.Fields S.M., Koeller J.M. Idarubicin: a second-generation anthracycline. DICP. 1991;25:505–517. doi: 10.1177/106002809102500511. [DOI] [PubMed] [Google Scholar]
- 49.Humphries R., Bobenchik A.M., Hindler J.A., Schuetz A.N. Overview of changes to the clinical and laboratory standards institute performance standards for antimicrobial susceptibility testing, M100, 31st edition. J. Clin. Microbiol. 2021;59 doi: 10.1128/JCM.00213-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu Y., Jia Y., Yang K., Li R., Xiao X., Wang Z. Anti-HIV agent azidothymidine decreases Tet(X)-mediated bacterial resistance to tigecycline in Escherichia coli. Commun. Biol. 2020;3:162. doi: 10.1038/s42003-020-0877-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tong Z., Xu T., Deng T., Shi J., Wang Z., Liu Y. Benzydamine reverses TMexCD-TOprJ-mediated high-level tigecycline resistance in Gram-negative bacteria. Pharmaceuticals. 2021;14:907. doi: 10.3390/ph14090907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen S., Liu D., Zhang Q., Guo P., Ding S., Shen J., Zhu K., Lin W. A marine antibiotic kills multidrug-resistant bacteria without detectable high-level resistance. ACS Infect. Dis. 2021;7:884–893. doi: 10.1021/acsinfecdis.0c00913. [DOI] [PubMed] [Google Scholar]
- 53.Shi J., Chen C., Wang D., Tong Z., Wang Z., Liu Y. Amphipathic peptide antibiotics with potent activity against multidrug-resistant pathogens. Pharmaceutics. 2021;13:438. doi: 10.3390/pharmaceutics13040438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu Y., Tong Z., Shi J., Jia Y., Deng T., Wang Z. Reversion of antibiotic resistance in multidrug-resistant pathogens using non-antibiotic pharmaceutical benzydamine. Commun. Biol. 2021;4:1328. doi: 10.1038/s42003-021-02854-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mwangi J., Yin Y., Wang G., Yang M., Li Y., Zhang Z., Lai R. The antimicrobial peptide ZY4 combats multidrug-resistant Pseudomonas aeruginosa and Acinetobacter baumannii infection. Proc. Natl. Acad. Sci. USA. 2019;116:26516–26522. doi: 10.1073/pnas.1909585117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Deng T., Jia Y., Tong Z., Shi J., Wang Z., Liu Y. Bismuth drugs reverse Tet(X)-conferred tigecycline resistance in Gram-negative bacteria. Microbiol. Spectr. 2022;10 doi: 10.1128/spectrum.01578-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jia Y., Yang B., Shi J., Fang D., Wang Z., Liu Y. Melatonin prevents conjugative transfer of plasmid-mediated antibiotic resistance genes by disrupting proton motive force. Pharmacol. Res. 2022;175 doi: 10.1016/j.phrs.2021.105978. [DOI] [PubMed] [Google Scholar]
- 58.Chen X., Zhong Z., Xu Z., Chen L., Wang Y. 2',7'-Dichlorodihydrofluorescein as a fluorescent probe for reactive oxygen species measurement: forty years of application and controversy. Free Radic. Res. 2010;44:587–604. doi: 10.3109/10715761003709802. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
This paper does not report original code.
-
•
RNA-sequencing data have been deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive database (PRJNA818073) and are publicly available as of the date of publication.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.