Summary
Stiffness plays a central action in plant cell extension. Here, we present a protocol to detect changes in stiffness on the external epidermal cell wall of living plant roots using atomic force microscopy (AFM). We provide generalized instructions for collecting force-distance curves and analysis of stiffness using contact-based mechanical model. With this protocol, and some initial training in AFM, a user is able to perform indentation experiments on 4- and 5-day-old Arabidopsis thaliana and determine stiffness properties.
For complete details on the use and execution of this protocol, please refer to Godon et al.1
Subject areas: Atomic Force Microscopy (AFM), Plant Sciences, Physics
Graphical abstract

Highlights
-
•
Growing Arabidopsis thaliana in flat crystallization plates
-
•
Transferring 4-day-old seedlings into Petri dishes with various Fe and Al metals
-
•
Nanoindentation of seedlings in the transition zone with atomic force microscopy
-
•
Determining elastic properties of stress roots using the trimechanic theory
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Stiffness plays a central action in plant cell extension. Here, we present a protocol to detect changes in stiffness on the external epidermal cell wall of living plant roots using atomic force microscopy (AFM). We provide generalized instructions for collecting force-distance curves and analysis of stiffness using contact-based mechanical model. With this protocol, and some initial training in AFM, a user is able to perform indentation experiments on 4- and 5-day-old Arabidopsis thaliana and determine stiffness properties.
Before you begin
Sterilize seeds and prepare agar before sowing. Then, grow seeds on agar plates for 4 days. This protocol requires a rigorous planning to avoid unavailability of any instrument/facility on the measurement day.
Seed sterilization
Timing: 1.5 h
The surface of Arabidopsis seeds must be sterilized before sowing. The seeds harvested in a natural environment tend to have contamination by bacterial or fungi spores. Thus, sterilization and filtering becomes necessary after harvesting to avoid contamination. All these steps are performed in a sterile hood.
-
1.Pour a small quantity of seeds (an equivalent of around 50 μL or less) in the microfuge tube.
-
a.Label the microfuge tube with the name of the seeds and add the date.
-
b.Cover the label with Parafilm or tape so that the label does not fade out.
-
a.
-
2.
Add 1 mL of washing solution (SDS 0.05%, EtOH 70%) using a sterile pipette tip.
-
3.
Close the cap and gently invert the tube upside down for 2–3 min to expose all the seeds to the liquid solution.
-
4.Remove the liquid from the microfuge tube.
-
a.Place the pipette tip at the bottom of the tube; push twice in the washing solution gently so that seeds distribute along the sidewall.
-
b.Carefully pipette out the liquid with the help of the tip and discard; avoid taking out the seeds!
-
a.
-
5.Wash the seeds with Ethanol.
-
a.Add 1 mL of ethanol (100%).
-
b.Gently shake the microfuge tube for 1 min minimum.
-
c.Pipette out the ethanol the same way as the step 4b; avoid taking out the seeds!
-
a.
-
6.Dry the seeds.
-
a.Keep the microfuge tube open for at least 1 h under the laminar flow hood, or until dry.
-
a.
-
7.Store the seeds.
-
a.Seeds can be kept at 20°C–24°C for short storage, and in 4°C with low hygrometry for long storage.
-
a.
Note: Moisture content affects the germination capacity of seeds. If not dried enough, the moisture content left can damage the outer seed coat, opening it before germination. Sterile seeds can be stored for up to 3 months. When kept for a longer time, the germination rate decreases with time, seeds do not germinate synchronously, or seeds do not germinate at all.
Agar media preparation
Timing: 1 h
Agar medium requires a sterilization step and thus must be prepared in advance.
-
8.
Pour 4 g of agar powder in a 500 mL glass bottle (8 g/L).
-
9.
Add 2.5 g of sucrose to the bottle (5 g/L).
-
10.
Add 490 mL of deionized water and stir to dissolve the powders.
-
11.
Take the bottle to laminar hood and open a new sealed 10 mL pipette.
-
12.
Add 10 mL of the solution nutritive to the bottle in the sterile laminar flow hood.
Note: Keep the nutrient solution in a sterile environment and at 4°C.
-
13.
Label the bottles with the autoclave tape and autoclave (120°C, 30 min).
Note: Do not forget to autoclave microfuge tubes, pipette tips, and wooden toothpicks. After autoclaving, open all the sterilized materials in the clean laminar hood only. It is better to make small batches of agar bottles instead of big bottles (1 L). Repeated heating of the medium leads to a loss of water and an increase in salt concentration.
Medium and culture plate preparation
Timing: 1.5 h
The seedlings need to be grown for four days before nanoindentation experiments. Steps 14–15 explain how to grow seedlings on horizontally standing crystallization plates, a very critical improvement over the usage of vertically positioned Petri dish.
-
14.Heating the agar.
-
a.Normally agar is in solidified form at room temperature. Slightly unscrew/loosen the lid of the bottle without fully opening it.Note: Opening the lid in air will make the solution non-sterile. But remember to loosen the cap slightly; if not, bottle will burst during heating. It is important to wear a protective glove and goggles.
-
b.Place the bottle in the microwave at full power (1450 W).Note: Timing for heating is proportional to the volume of the solution in the bottle. Take out the bottle after 1–2 min, or when it is partially melted, swirl gently for 2–3 min. It helps to even out the heat and dissolve agar completely. The goal is to melt and not boil the agar. If needed, place it back into the microwave for 30–40 s. Excessive heating can lead to water evaporation, which changes the concentration of nutrients.Note: Make sure to wear thermo-protective gloves to handle the hot bottle.
-
a.
-
15.Preparing a culture plate (perform all steps in laminar flow hood, 30 min).
-
a.In a 50 mL polypropylene tube, add 30 mL of melted agar solution and 600 μL of MES buffer solution.
-
b.Add 1 μM of FeCl2 (or 3 μL from a 10 mM stock).
-
c.Gently invert upside down the polypropylene tube to mix well.
-
d.Pour carefully medium solution to the wells up to the brim of the crystallization plate. The number of wells to use depends on the total number of seeds that you need to grow for your daily experiment. Troubleshooting problem 1.
-
e.Allow the medium to cool down at room temperature until solidifies.
-
f.Spread the seeds onto the agar and align them evenly with a sterile wooden stick. Approximately 10–12 seeds in each well.Note: If more seeds are poured, they will have less space to grow and reduced amount of nutrients available.
-
g.Slightly dip the seeds inside agar so that the seeds have better contact with agar. This helps the roots to grow downwards and straight.
-
h.Pour some water in 2 or 3 empty wells of the culture plate to keep high hygrometry for the next 4 days.
-
i.Seal the plate with the micro-pore tape. Porous nature of the tape allows aeration and prevents water evaporation outside the plate.
-
j.Place the plate in the Peltier-cooled incubator for next 4 days with a day/night cycle of 18/6 h and corresponding temperature of 24°C/21°C.
-
a.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| Arabidopsis thaliana wild-type seeds | Thierry Desnos (CEA/BIAM) | Coler105 |
| Arabidopsis thaliana mutant seeds | Thierry Desnos (CEA/BIAM) | almt151 (∗) |
| Chemicals, peptides, and recombinant proteins | ||
| Agar powder | Sigma-Aldrich | Cat# A7921 (BCBZ7284) |
| Aluminum chloride (AlCl3) | Sigma-Aldrich | Cat# 237051 |
| Ammonium nitrate (NH4NO3) | Sigma-Aldrich | Cat# A3795 |
| Boric acid (H3BO3) | Sigma-Aldrich | Cat# B6768 |
| Calcium chloride dihydrate (CaCl2·2H2O) | Sigma-Aldrich | Cat# C7902 |
| Cobalt(II) chloride hexahydrate (CoCl2·H2O) | Sigma-Aldrich | Cat# C8661 |
| Copper(II) sulfate pentahydrate (CuSO4·5H2O) | Sigma-Aldrich | Cat# C3036 |
| Ethanol absolute | Carlo Erba | Cat# 4145872 |
| Iron chloride (FeCl2) | Sigma-Aldrich | Cat# 44939 |
| Magnesium sulfate heptahydrate (MgSO4·7H20) | Sigma-Aldrich | Cat# M2773 |
| Manganese sulfate monohydrate (MnSO4·H20) | Sigma-Aldrich | Cat# M7899 |
| MES-(2-(N-morpholino)ethanesulfonic acid | Sigma-Aldrich | Cat# M8250 |
| Milli-Q water | Milli-Q | Direct 8 |
| Potassium hydroxide | Sigma-Aldrich | Cat# 221473 |
| Potassium iodide (KI) | Sigma-Aldrich | Cat# 221945 |
| Potassium nitrate (KNO3) | Sigma-Aldrich | Cat# P8291 |
| Potassium phosphate monobasic (KH2PO4) | Sigma-Aldrich | Cat# P5655 |
| Sucrose | Mpbio | Cat# 02 904713-CF |
| Silicon pressure sensitive adhesive (PSA) | NuSil Technology LLC | MED1-1356 |
| Sodium Dodecyl Sulfate (SDS) solution 0.05% | Sigma-Aldrich | Cat# L4390 |
| Sodium molybdate dehydrate (Na2MoO4·2H2O) | Sigma-Aldrich | Cat# M1651 |
| Zinc sulfate heptahydrate (ZnSO4·7H20) | Sigma-Aldrich | Cat# Z1001 |
| Software and algorithms | ||
| AtomicJ, version 2.0 | Hermanowicz et al.2 | https://sourceforge.net/projects/jrobust/ |
| ImageJ, version 1.51j8 | Schneider et al.3 | https://imagej.nih.gov/ij/ |
| NeuronJ_.jar 1.4.3 (with imagescience.jar 3.0.0) | Meijering et al.4 | https://imagescience.org/meijering/software/neuronj/ |
| Prism, v5 or v8 | GraphPad | https://www.graphpad.com/ |
| Other | ||
| AFM cantilevers | Nanoworld AG | PNP-TR-50 |
| AFM Dimension 3100 | Bruker | D3100 |
| AFM Nanowizard IV | JPK | NW4 |
| AFM probe holder | Bruker | DTFML-DD |
| Binocular | Nikon | SMZ800N (C-PS stand) |
| Cover slips (glass 24 × 40 mm) | Agar Scientific | Cat# AGL462440-1 |
| Crystallization plate VDX | Hampton Research | Cat # HR3-140 |
| Glass bottle (500 mL) | Duran, Dutscher | Cat # 818014407 |
| Glass slide StarFrost 3 × 1 in | Knittel Glass | Cat# VS1137#1FKB.01 |
| Horticole Indoor Led, 45 W, 169 LEDs, 276 × 276 × 14 mm, full spectrum | www.cultureindoor.com | Cat# 003-300-370 |
| Laminar flow hood FlowFAST | Faster Air | H 09 SA7010 |
| Microfuge tubes 1.5 mL | Eppendorf | Cat# 0030.120.086 |
| Micro tape Anapore 9.14 m × 1.25 cm | Euromedis | Cat# 135312 |
| Microwave 900W | Severin | mw7825 |
| Peltier-cooled incubator | Memmert | IPP110+ |
| Petri dish 60 × 15 mm | Falcon | Cat# 353004 |
| Pipette motor Ergo one FAST | Starlab | Cat# S7166-0010 |
| Pipette tips 20 μL | TipOne | Cat# S1110-3700 |
| Pipette tips 200 μL | TipOne | Cat# S1111-0700 |
| Pipette tips 1000 μL | TipOne | Cat# S1111-6701 |
| Pipette P20L | Gilson | Cat# FA10003M |
| Pipette P200L | Gilson | Cat# FA10005M |
| Pipette P1000L | Gilson | Cat# FA10006M |
| Polypropylene centrifuge tubes 50 mL | Falcon, Greiner Bio-One | Cat# 227261 |
| Programmable timer | Otio | 93022 |
| Screw thread vials, amber, 12 × 32 mm | Thermo Scientific | Cat# C4013-2 |
| Septa TEF/SIL 8 mm | Thermo Scientific | Cat# C4013-60 |
| Shimadzu 8-425 caps | Thermo Scientific | Cat# C4013-3A |
| Syringe filter 0.22 μm, 33 mm | Millipore Millex-GV | Cat# SLGV033RB |
| Syringe needle 0.8 × 40 mm | Terumo Neolus | Cat# NN-2138R |
| Thermal gloves | VersaTouch | |
| Tweezer Style SM104 4.7″ | Techni-Tool | Cat# 758TW401 |
| Wooden toothpick sticks | N/A | N/A |
(∗) almt151: mutation G881A that results in a defective splicing of intron 3.
Materials and equipment
CRITICAL: When handling the reagents wearing of lab coat, gloves and goggles is advised. Follow the waste disposal rules as per your institute and country’s safety guidelines. Reagents should be stored and prepared according to the manufacturer’s recommendation. Refer to the appropriate material’s safety data sheet.
Preparation of individual stock solutions of each nutrient
-
•
1 M MgSO4·7H2O (246.5 g in 1 L milliQ H20).
-
•
1 M NH4NO3 (80 g in 1 L milliQ H20).
-
•
1 M KNO3 (101.1 g in 1 L milliQ H20).
-
•
1 M CaCl2· 2H2O (147 g in 1 L milliQ H20).
-
•
10 mM KI (166 mg in 100 mL milliQ H20).
-
•
100 mM H3BO3 (618 mg in 100 mL milliQ H20).
-
•
1 M MnSO4· H2O (6.76 g in 40 mL milliQ H20).
-
•
1 M ZnSO4·7H2O (28.7 g in 100 mL milliQ H20).
-
•
25 g/L Na2MoO4·2H2O (1.25g in 50 mL milliQ H20).
-
•
2.5 g/L CuSO4·5H2O (125 mg in 50 mL milliQ H20).
-
•
2.5 g/L CoCl2·6H2O (125 mg in 50 mL milliQ H20).
Filtrate all solutions (0.2 μm) and store at 4°C for months or years unless contaminated.
Composition of the 50× nutrient solution stock (1 L):
| Reagent | Final concentration | Amount |
|---|---|---|
| MgSO4·7H2O | 23.4 mM | 23.4 mL |
| NH4NO3 | 103 mM | 103 mL |
| KNO3 | 94.5 mM | 94.5 mL |
| CaCl2·2H2O | 33.5 mM | 33.5 mL |
| KI | 25 μM | 2.5 mL |
| H3BO3 | 39.3 mM | 39.3 mL |
| MnSO4·H2O | 523 μM | 523 μL |
| ZnSO4·7H2O | 251 μM | 251 μL |
| Na2MoO4·2H2O | 52 μM | 400 μL |
| CuSO4·5H2O | 7.3 μM | 728 μL |
| CoCl2·6H2O | 5.26 μM | 500 μL |
| milliQ H2O | N/A | 701.398 mL |
| Total | N/A | 1 L |
Filtrate (0.2 μm) and store it at 4°C for several months.
Growth solution
| Reagent | Final concentration | Amount |
|---|---|---|
| 50× nutrient solution | N/A | 20 mL |
| Sucrose | 5 g/L | 5 g |
| milliQ H20 | N/A | 960 mL |
| Total | N/A | 980 mL |
Autoclave this solution and keep it in sterile condition, at 20°C–24°C for 6 months. During the protocol, starting at step 3 below, take 980 μL of the growth solution and add 20 μL of MES solution (3.4 mM final concentration); this is the final composition of the “growth solution” which must be used only one day.
Other individual stock solutions:
-
•
500 μM phosphate solution KH2PO4 (1 M stock = 13.61 g in 100 mL milliQ H20).
-
•
10 μM FeCl2 solution (10 mM stock = 0.12 g in 100 mL milliQ H20).
-
•
10 μM AlCl3 solution (10 mM stock = 0.13 g in 100 mL milliQ H20). This stock solution should be prepared with caution in a fume hood (use gloves and safety glasses) as the addition of water to the AlCl3 powder releases vapors of hydrochloric acid. Furthermore, this solution must be prepared in a glass beaker as the dissolution of AlCl3 is exothermic. Slowly add the water onto the powder and swirl the beaker until the complete dissolution of the powder (few minutes). Do not autoclave this solution.
-
•
MES Buffer (pH 5.7) (170 mM stock solution = 3.62 g in 100 mL H20); adjust the pH with 10 N KOH.
Filtrate all solutions (0.2 μm) except AlCl3, store all solutions at 4°C.
-
•
Peltier-cooled incubator.
The incubator Memmert IPP+ 110 controls the temperature of a chamber. There is a sidewall opening to accommodate the power cords of the artificial LED box made of a mixture of red, blue, and white color (Figure 1). Both the chamber and LED lights run on a programmable timer with the light being on for 16 h during the day at a controlled temperature of 24°C and 8 h at night without light at 21°C.
Alternatives: Any growth chamber or light box system can be used for growing Arabidopsis thaliana seeds. We have also used natural light, but to maintain homogeneity it is safer to use a reproducible system. Light boxes with LEDs are cheap and easy to find. We have noticed that some LED light boxes do produce a significant heating so by adjusting their height above the growing plates, one can simulate a change in temperature when the light is on (vs off). However, when the light is off, the room temperature should be constant and slightly cooler.
-
•
Pressure sensitive silicon adhesive (NuSil).
Figure 1.
Peltier cooled incubator to grow Arabidopsis thaliana seedlings
Light is provided by an added LED box covering the “full spectrum” of light and made of 196 LEDS: 98 at 630 nm, 42 at 470 nm, 28 at 660 nm, 14 white at 3000 K, and 14 white at 6500 K. A day/night program on the incubator requires the development of a short running sequence of events using a Memmert proprietary software (AtmoCONTROL 2.9.2.0 29/01/2019, provided by Memmert). A caveat is found on the IPP+ 110 when activating the program, it automatically starts at the beginning of the programed cycle (likely the next morning). It implies that the program must be activated 24 h before use.
NuSil is a pressure sensitive adhesive (PSA) that has a property to stretch while polymerizing. It has a composition of 50% of silicone and 50% of ethyl acetate. NuSil has a low viscosity (245 mPa s), which makes it possible to pipet. Ethyl acetate evaporates with time leaving behind only silicone that gets polymerized with time. Troubleshooting problem 2.
CRITICAL: We prefer NuSil over other adhesives because it has a particular characteristic of stretching while drying, which was not reproduced with some other PSAs. For the nanomechanical experiment, it is crucial that the end of the root tip is appropriately fixed on the glass slide since measurements are performed about 500 μm from the root tip. Because of the root capping, the macroscopic rigidity of the root tip was the major source of trouble in the attachment of root tips on glass slides.
Alternatives: The NuSil used in our work is difficult to obtain because of logistical issues with the supplier that hardly delivers small bottles. However, there are alternative PSA available, and we experimented with Silicolease PSA 408/toluene (cat #117149, Elkem, France). Nonetheless, we chose not to pursue it due to its higher viscosity (from 40,000–120,000 mPa·s compared to 245 mPa·s for NuSil) and a compatible tack around 20 kPa (Avantor Sciences).
-
•
Atomic Force Microscopy (AFM) set-up.
Indentation experiments are performed in liquid medium with a Bruker Dimension 3100 AFM. The DTFML-DD probe holder holds triangular Pyrex nitride cantilevers (PNP). The PNP tip has a square pyramidal symmetry, a low sharpness (∼10 nm nominal radius), a half-opening angle of 35°, which was verified by SEM measurements, and a proper spring constant for plant root tissues (nominally 0.08 N/m). The total mass of such a cantilever is about 130 ng, a very negligible mass compared to a single seedling root (∼0.5 mg). AFM runs with a Nanoscope V controller and the Nanoscope 7.3 software using the picoforce module. Indentation of plant root surface was performed with approach-retract curves using a ramp size of 3–4 μm, a scan rate of 0.5 Hz, and 4096 points per curve. Ramp size was adjusted using its z-start position so that an indentation of about 0.5 μm was achieved.
Alternatives: Other AFM instruments can also be used for these experiments on the condition that they have a large enough stage for glass slides. Most modern bio-AFM complies with this specificity.
Step-by-step method details
Culture plate preparation for the transfer of seedlings
Timing: 0.5 h
To stress seedlings with various abiotic metals, seedlings need to be transferred from their original growing conditions to the stress conditions. These Petri dishes are prepared on the day of the experiment.
-
1.Prepare small Petri dishes (60 × 15 mm style). Troubleshooting problem 3.
-
a.Label the bottom of the Petri dish with the stress condition; also include the plating date.
-
b.Warm-up the agar bottle, as described in step 14 (before you begin). Make sure the agar is liquid enough.
-
c.Mix 15 mL of agar and 300 μL of MES buffer in a polypropylene tube.
-
d.If Al stress of 5 μM is needed, then add 7.5 μL of AlCl3 in the polypropylene tube.
-
e.If Fe stress of 10 μM is needed, then add 15 μL of FeCl2 in the polypropylene tube.
-
f.Homogenize the polypropylene tube and deposit the mix in the Petri dish.
-
a.
Note: Always keep the small Petri dishes in the incubator throughout the whole experiment. Seedlings are systematically transferred to a Petri dish for 2 h, even in the absence of metallic stress. Troubleshooting problem 4.
Nanomechanical experiments
Timing: 2 days
Nanoindentation experiments are performed with an AFM using a square-pyramidal tip. Living seedling roots are fixed on glass slides. Steps 2 to 8 describe all the procedures from cantilever attachments to force-distance curve collection. Several steps require the supervision of an AFM expert due to some fragile components of AFM (cantilevers or piezo scanners). Although the procedures are generic, details may vary from one AFM to another.
-
2.Attach cantilever to the AFM scanner (2 min).
-
a.Using the binocular and dedicated tweezer , gently lift the cantilever and place it straight into the groove of the liquid cell holder. Troubleshooting problem 5.
-
b.Attach the liquid cell holder to the piezo scanner.
-
a.
Note: In case of the Dimension 3100, lift the piezo significantly upward (away from the sample). Make sure that the scanner is far from the sample stage so that the tip will not hit the root surface and break.
-
3.Calibrate the AFM system (60 min).
-
a.Perform calibration in air.
-
i.Adjust the laser beam on the cantilever while preserving the highest sum.
-
ii.Perform a contact-based force-distance curve (ramp size of 3 μm, frequency of 0.5 Hz, 4096 points per curve).
-
iii.Determine the deflection sensitivity by measuring the slope of the curve in contact with the surface.
-
iv.Repeat the previous step several times until sensitivity values are constant.
-
v.Perform a thermal tuning to determine the spring constant of the cantilever.
-
vi.Save the computed spring constant and repeat the previous step until the spring constant values are uniform.
-
i.
-
b.Perform calibration in liquid.
-
i.Repeat previous step 3.a but in liquid environment.Note: For the Dimension 3100, only the deflection sensitivity can be performed in liquids.
-
ii.Wait for half an hour after adding the growth solution to thermally stabilize the AFM cantilever.
-
iii.Set the sensitivity values obtained in liquid as final values.
-
iv.If the AFM system can perform thermal tuning with frequencies below 1 kHz, then also perform the thermal tune in liquid, otherwise, keep the spring constant obtained during the previous calibration in air.
-
i.
-
a.
-
4.Fixing a plant root on a glass slide (5 min).
-
a.Glass slide set up (2 min for the set-up and 1.5 min for sealing).
-
i.Take two standard glass slides plus two rectangular cover slips and assemble them as shown in Figure 2.
-
ii.Take a micropore tape and place it over the cover slip to bind to the glass slide.Note: Make sure that both cover slips are at the same height in the middle of the slide and that 2/3rd of the width is touching the slide and 1/3rd of it stays outside.
-
iii.Take a third glass slide and place it in the middle of the other two glass slides such that a little rectangular space is left in between (Figure 3).
-
iv.At one end of the glass slide deposit a drop of silicone adhesive (approx. 100–150 μL).
 CRITICAL: Never store NuSil in any clear plastic container like Eppendorf or Falcon tubes. Always store it in dark glass bottle, due to the presence of ethyl acetate solvent.
CRITICAL: Never store NuSil in any clear plastic container like Eppendorf or Falcon tubes. Always store it in dark glass bottle, due to the presence of ethyl acetate solvent. -
v.After depositing NuSil on the glass slide, spread it with the help of another cover slip (Figure 3) dragging upwards to the other end. When we spread a drop of NuSil on the glass slide, it has the thickness of the cover slip and is evenly distributed.
 CRITICAL: A constant thickness of glue is important for reducing the height fluctuation of deposited plant. Do not add more than two drops of NuSil. More amount will lead to longer polymerization time.
CRITICAL: A constant thickness of glue is important for reducing the height fluctuation of deposited plant. Do not add more than two drops of NuSil. More amount will lead to longer polymerization time. -
vi.After spreading NuSil, wait for 20–25 s, so that the adhesive starts polymerizing, yet it is still wet for the sealing purpose.Note: The goal is to place the root onto the adhesive when it is semi-solid so that it stays on the surface and does not dip into the adhesive. It might be tempting to place the plant sooner. Sometimes the plant can dip inside the NuSil and can affect its living state, in particular due the presence of un-evaporated ethyl acetate.
-
vii.Take one seedling from the Petri dish and, using a dedicated tweezer (NuSil is a glue and it is difficult to fully remove from the tweezer. Thus, a dedicated tweezer should be used to NuSil threads sealing); gently lift it from the shoot area preferably under the cotyledon without pinching the seedling.
 CRITICAL: if not lifted properly, this might lead to breakage of the plant into two pieces. In case of longer roots, it might be stuck inside the agar gel and extra caution is required to extract the seedling from the agar. Always lift from the cotyledon part because that area has the center of mass. This way the root can balance itself during the transfer from the Petri dish to the glass slide.
CRITICAL: if not lifted properly, this might lead to breakage of the plant into two pieces. In case of longer roots, it might be stuck inside the agar gel and extra caution is required to extract the seedling from the agar. Always lift from the cotyledon part because that area has the center of mass. This way the root can balance itself during the transfer from the Petri dish to the glass slide.
-
i.
-
b.Depositing a seedling on the glass slide.
- i.
-
ii.Lay down the root straight to enable the indentation in the transition zone. In case the root is bent around the transition zone it is difficult to perform indentation around that area. Troubleshooting problem 7.
-
iii.Fastening the seedling root by sealing it with the semi-solid adhesive present on the glass slide, starting from the top, then the middle and the very end (root tip) to prevent any movement of the root (Figure 4). Troubleshooting problem 8.
 CRITICAL: Seal only 50–100 μm of the root tip area. It preserves the transition zone from any presence of glue.
CRITICAL: Seal only 50–100 μm of the root tip area. It preserves the transition zone from any presence of glue. -
iv.Immediately cover entirely the plant root (at least 100 μL) with growth solution (square #2, Figure 3) to prevent drying during measurement. Make sure to cover the root tip first as it is the most sensitive part of the plant. The system is ready for measurements (Figure 4).
 CRITICAL: Do not use water to cover the root, as it will generate an osmotic imbalance that causes excess water to enter into the cell, which in turn either break or escape from the fastening NuSil bands.
CRITICAL: Do not use water to cover the root, as it will generate an osmotic imbalance that causes excess water to enter into the cell, which in turn either break or escape from the fastening NuSil bands.
-
a.
-
5.Nano-Indentation on the living root tissues (max 25 min).
 CRITICAL: Although the seedling is alive, the horizontal position is not its favorite. Consequently, the experiment should be performed as quickly as possible. A time longer than 30 min gave us unpredictable results as global stress may occur within the root.
CRITICAL: Although the seedling is alive, the horizontal position is not its favorite. Consequently, the experiment should be performed as quickly as possible. A time longer than 30 min gave us unpredictable results as global stress may occur within the root.-
a.Before positioning the plant root under the AFM cantilever, retract the piezo to a safe distance between the cantilever and the plant root.
-
b.Lift the scanner head carefully and insert the glass slide with the plant. Buffer is already on the top of the root, thus carefully lower the scanner head so that the cantilever will not bend upon contact with water (adding a drop of buffer on the cantilever is usually necessary to have a smooth insertion in liquid).
-
c.Orient the glass slide, so that the root orientation is close to vertical and its tip is on the top of the camera (Figure 4).
-
d.The target working area is located approximately 500 μm away from the root tip. This distance can be estimated knowing the length of the PNP cantilever (about 200 μm). Use the stage positioning controls to move the cantilever on the right zone.
 CRITICAL: Difficulty in using PNP cantilever: because the seedling root is pseudo-cylindrical and the PNP tip height rather small (<3.5 μm), it is not advised to probe the root beyond the longitudinal median line of the root. Note that there is a small setback of the tip of PNP cantilever (4 μm), see Figure 5.
CRITICAL: Difficulty in using PNP cantilever: because the seedling root is pseudo-cylindrical and the PNP tip height rather small (<3.5 μm), it is not advised to probe the root beyond the longitudinal median line of the root. Note that there is a small setback of the tip of PNP cantilever (4 μm), see Figure 5. -
e.Focus the camera so that the root surface is clearly visible.
-
f.Wait for a few minutes for the laser to stabilize.
-
g.Center laser spot at (0,0) position. Keep deflection set point to 2.5 V.
-
h.Make a new folder for each node and mention its co-ordinate in the filename.
-
i.Lower the piezo down manually until the root is clearly visible, then start engaging.
-
j.When the surface is detected, then run continuous force-distance (FD) curves to adjust the ramp parameters. With the Dimension 3100, we have to modify the z-ramp start so that our 3 μm-long ramp could be performed correctly. With our open-loop scanner, we did not use a trigger force threshold, and consequently, the ramp must be manually adjusted prior a full record.
-
k.With appropriate FD curves, start collecting and recording curves.
-
l.Because of the limited time (before plant roots display additional stress), we can only probe a limited number of areas. Our best protocol contains 16 physically different areas (called nodes) and each node is composed of a matrix of 2 × 2 FD curves, using the autoramp tool with 50 nm of distance between each point (Figure 6).
-
m.Withdraw the cantilever every time after the recording of the curves and engage again after changing the piezo offset co-ordinates. Troubleshooting problem 9.Alternatives: Modern AFM instruments use force-volume mode that allows the collection of a matrix of nodes with a single FD curve per node. The analysis of data will thus be slightly different as in our protocol (see the quantification and statistical analysis section below).
-
n.After completion of all 16 nodes, for the record, take a screen shot of the plant root under the cantilever.
-
o.Move the piezo upward to a safe distance and discard the glass slide.
-
p.Repeat from step 4 for additional seedlings.
-
q.At the end of the day, remove the cantilever holder by lifting the piezo scanner gently.
-
r.On a bench, clean the tip by pouring 100 μL of propanol over. Repeat twice.
-
s.Remove extra propanol with the help of filter paper placed on the edge of the holder.
-
t.Add 100 μL ethanol for further cleaning.
-
u.We can keep the probe inside the holder for future experiments. Always recalibrate before experiment.
-
a.
-
6.Prepare plant roots for checking their growth status (10 min).
-
a.With the remaining roots, not taken for the indentation experiment, transferred them to the Petri dishes containing stressed medium or control medium, try to align them properly and mark the position of the root tip with a pen. Troubleshooting problem 10.
-
b.Use the binoculars to mark the root end for next-day visual assessment.
-
c.Observe phenotype (root extension) after 24 h and take a photograph.
-
a.
-
7.Root length measurement (2 days after the initial experiment).
-
a.Measure the root length 48 h after the transfer to have a clear distinction in the root extension phenotype.Alternatives: Classically, we measure the root length contiguously to AFM measurements. It is also possible to dedicate a specific experiment (excluding AFM) purely for root length measurements.
-
b.Take pictures of the whole roots under stressed and non-stressed conditions. Do not forget to add a ruler in the field of view to calibrate the length measurement with ImageJ.
-
c.Export these pictures to imageJ where they can be opened with the NeuronJ plugin. The NeuronJ plugin allows you to trace the root paths with the computer mouse. The default standard length unit is in pixels.Note: NeuronJ requires an 8-bit gray scale image in a GIF format, so use ImageJ to transform your picture prior to opening it with NeuronJ.
-
d.If NeuronJ is not installed yet, please follow the steps e-f first, otherwise jump to step g.
-
e.Install NeuronJ from https://imagescience.org/meijering/software/neuronj/. Download both jar files: NeuronJ_.jar (version 1.4.3) and imagescience.jar (version 3.0.0).
-
f.Move the two .jar files into the plugins directory of ImageJ software installed on your computer (see the installation manual of ImageJ for your specific computer system).
-
g.Open a gif image of your plant root.
-
h.Add a tracing by selecting the leftmost menu of NeuronJ in Figure 7. First, click at one end of a root and wait a few seconds until the plugin sets up (do not click again).
-
i.After 2–3 s, move the mouse following the root shape and the plugin should automatically track the root feature.Note: Sometimes the plugin loses track of the root, then just click on the last pixel of the root where the tracking was lost. It concatenates a new tracking segment from that click and proceeds like before. You can click as many times as needed to follow perfectly the root.
-
j.When you reach the end of the root, then double-click to exit the tracking.
-
k.Click on the measure-tracing icon to display the results (Figure 7).Note: You can perform the entire tracing of all roots first and, only at the end, select the measurement. Then, you can export all the data using the plugin menu. It provides the global average length of all roots and displays their individual values. It is then possible to save this data as text files.
-
l.Save the results of NeuronJ in text file or spread sheet format. Also, save the labeled path in PNG format, for the record.
-
m.Reopen ImageJ with the original image that contains the ruler and calibrate (set scale) the pixel length of your image. Report the conversion factor to your spreadsheet result to convert NeuronJ pixel size into mm.
-
n.Use any graphical software to plot histograms or boxplots of the root lengths, we used Prism 5.
-
a.
-
8.
Data analysis (15 min per plant).
Figure 2.
Assembling the glass slides to mount plant seedlings for nanoindentation analysis with AFM
Cover slips glued to the green Starfrost glass slides are colored in light gray. A small piece of micropore tape (dark gray) is used to fix the cover slips (for clarity on the left side is shown, but both sides should be fixed in the same way).
Figure 3.
Principle of mounting plant seedlings on glass slides
The goal of this setup is to deposit a uniform layer of NuSil glue on the middle glass slide. The adhesive layer shown in light gray and labeled with the number 1. A long primary root in orange and two green cotyledons schematizes the seedling. The seedling is finally covered with a growth medium shown in light transparent blue color and labeled with the number 2. Cover slips are in light gray whereas micropore tapes are in dark gray.
Figure 4.
Picture showing how the root should be laid down for AFM measurements
The sealing with NuSil should be very thin so that cantilever support is not touching the sealing bands. After sealing carefully, place the middle glass slide of Figure 3 under the piezo scanner. At this point, we need to rotate the glass slide manually so that the root tip is positioned under the cantilever as seen through the camera.
Figure 5.
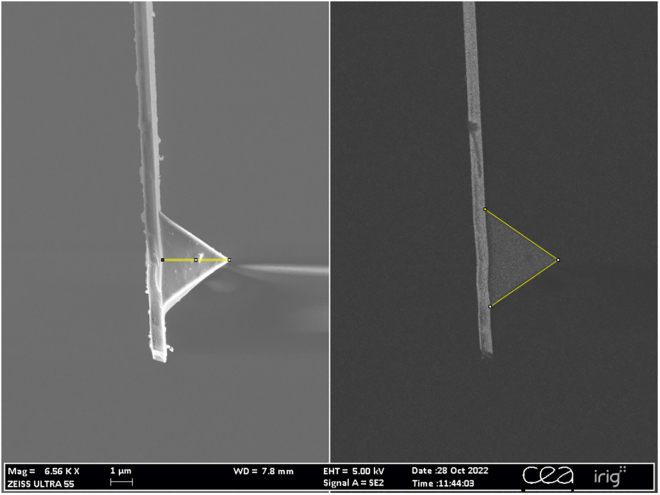
Scanning electron microscopy image of a PNP-TR cantilever used in the AFM experiment
A secondary electron image is on the left and a backscattered electron image is on the right. Tip height and opening angle are highlighted in yellow and were measured using ImageJ: 3.05 μm in height (left) and an opening angle of 69.27°. The manufacturer’s data indicates a tip height of 3.5 μm and a half-opening angle of 35°. Image is magnified 6560× under a tension of 5 kV and a working distance of 7.8 mm.
Figure 6.
Pictorial version of the matrix strategy used to collect force-distance curves
The measurement starts at position 0 and proceed according to blue arrows. Offsets of the piezo scanner at each node of the matrix are indicated below (values in μm). At each node, a submatrix of 2 × 2 FD curves are recorded. Do not forget to update filenames at each node in order to keep track of the matrix (it helps during the data analysis).
Figure 7.

Partial menu of the NeuronJ plugin
Click on the icon to add (1st left) or to remove (2nd left) a tracing. To remove a tracing, it must be finalized, i.e., ended by a double click. To measure tracing data click on the last-right icon.
The reproducibility of the experimental protocol is of prime interest. In particular, measurements of stress and non-stress plant roots must be performed in the same manner because there is a link between the elastic constant and the experimental indentation depth. Therefore, we tried to keep everything as constant as possible to perform our nanoindentation experiments. In our protocol, we used the contact-based mechanical model of Sneddon7 and a novel extension known as the trimechanic theory8 to determine the elastic property of the external cell wall. This model allows us to extract the effective Young’s modulus by fitting the force-indentation curve, after correcting the FD curve.9 The relationship between the force (F) and the indentation (Z) is described by:
where Ê is the effective Young’s modulus () and E the Young’s modulus with η the Poisson’s ratio, and α the half-opening angle of the squared pyramidal tip (35° for PNP tips). Sample FD curves of stressed and non-stressed plant roots are shown in Figure 8. The trimechanic theory is used to fit the indentation curves in multiple depth zones and to extract the effective Young’s modulus as well as the corresponding stiffness.8
Alternatives: AtomicJ, an open source package,2 was also used in our study, and several methods can also be used to obtain the Young’s modulus values e.g., manufacturer software (Bruker, JPK, …).2 A graphical interface for calculating the Young’s modulus is available with the Force V1.1.0 program.10 Details of the computation steps of FD curve analysis are described by Domke & Radmacher.11 It is possible to linearize the force-indentation curve and then fitting with a simple linear regression9; this process can be obtained with basic programing skills in Matlab (or equivalent) or simply with Excel spreadsheets. A recent review on determining elastic properties of biological object has been recently released.12 One of the main differences between all these alternative fitting methods is the strategy to obtain the contact point, which influence greatly the numerical values of E. Nevertheless, when comparing measurements between control and a stress condition, it is reasonable to choose any model, on the condition of always using it the same way for all FD curves.
Figure 8.
Sample of force-distance curves and their fitting using AtomicJ software
Approach curves are in gray with a plain line for the stress condition (10 μM FeCl2 + 10 μM AlCl3) and with a dashed line for the control condition (no metals). The fitting was capped to 5 nN and is highlighted by a red line. From the stress-condition fit (plain line), the Young’s modulus value is 506 kPa whereas it is 60 kPa for the control condition (dashed lines).
Filtering and collecting fitting data from FD curves are described in the quantification and statistical analysis section. In the end, our protocol provides a single average value (Elasticity, Stiffness, Depth, …) per plant as provided by the trimechanic theory.8 The protocol is applied with the same experimental conditions on three or four plants per day, usually four times, providing a total of about 10–15 plants per condition. Once all the experimental conditions are tested, a boxplot is produced and statistical tests can be applied. A sample of a box-and-whiskers plot drawn with Prism v5 is shown in Figure 9.
Figure 9.
Box-and-whiskers plot of elastic properties of WT seedling roots in absence or presence of aluminum stress
Elasticity is represented by the effective Young’s modulus (Ê) in kPa units. Fe0Al0 indicates the nutrient solution without Fe and without Al. Fe0Al5 indicates the nutrient solution without Fe and with 5 μM of Al. Each box edge represents the first and third quartiles around the median. The whiskers represent the min and max values for each group. The label inserted within a box indicates the total number of measured plants. Exact p-values are obtained according to the Mann-Whitney non-parametric t-test. Results indicate that there are no significant differences in the elasticity of seedling roots grown in absence or in presence of 5 or 10 μM Al.
Expected outcomes
AFM and nanoindentation have the power to quantify variation in the mechanical response of biological tissues such as plant roots in this protocol. Changes in mechanical response can be a consequence of internal stress (oxidative damages) or external abiotic stress (metal toxicity).
The current protocol allows the measuring of about eight plant roots per day. However, it requires a precise organization to avoid waiting for a full 2 h at each specific stress. Note that a 2-h transfer step is always included even in the absence of stress condition (control conditions). The protocol is designed for plant roots but could be easily adapted to other plant organs such as hypocotyls, meristems, or leaves. This protocol could also be adapted for measuring putative change in stiffness connected with the gravitropism effect on plant roots. We have already noticed a correlation between root bending and increased stiffness during our measurements (unpublished data).
The current protocol leads to the measurement of force-distance curves that are translated into force-indentation curves using a contact-based mechanical model. Here we used the model of Sneddon7 as well as a very recent extension called the trimechanic theory.8 The force-distance curves allow us to extract effective elastic constants (effective Young’s modulus). Only the trimechanic theory allows us to obtain the effective elastic constants, stiffness, and associated deformability. In addition, the trimechanic theory analyzes the global stiffness response of the root along the indentation depth.8 This protocol was used to determine the stiffness of plant roots during an abiotic stress caused by the additional presence of Fe and/or Al metal (in preparation). The protocol could be adapted to any kind of metallic stress or complex stresses allying metals, chelator, or pH changes that can be translated into a chemical change, which in turn can be added to the agar composition in a Petri dish.
Indentation on living plants has clear advantages over isolated cells. In particular, it has been demonstrated that fixed root on glass slides remained alive for the duration of an AFM experiment.13 The living status of roots has been tested during this present protocol and neither the fixation system with NuSil nor the horizontally positioned seedlings prevent roots from growing. We have shown that roots continue to grow at least 2 h in such a position. Nevertheless, although root growth is a macroscopic sign of living tissue, it is not a clear indication of the lack of putative stress within the root.
In our protocol, a square pyramidal tip is used to determine the stiffness of plant root surfaces. The protocol could be adapted to any kind of tip shape including paraboloid or spherical tips as well as other types of indentation systems.14 In addition, the same protocol has been used for a preliminary test of rheology on Arabidopsis thaliana roots (unpublished data). Moreover, this methodology is also applicable to decellularized plants. A preliminary decellularization experiment has been tested and suggests that the following protocol can also be applied on decellularized plant roots.
Quantification and statistical analysis
In this protocol, we adopted a hierarchical approach for analyzing experimental data. With the final version of the protocol, 16 data points (nodes) are collected on each single plant. At each data node, redundant data is collected using a 2 × 2 matrix (4 FD curves). We test the homogeneity of data by evaluating the extracted elastic properties from all these four curves. If any value is not within 2 sigma from the mean, then the corresponding FD curve is removed. If more than two FD curves are removed in a node, then the node is removed. If half of the nodes of a plant are removed, then the whole plant data is discarded. A distribution histogram of effective Young’s modulus values obtained at each node of all plants revealed, most of the time, a log-normal distribution. This is not particularly rare in biology.15 Consequently, we decided to compute the geometric mean of all nodes of a plant and provide this value as the final result of this single plant. A quick survey of other averaging methods did not change the overall conclusions of the work.
Finally, but not less important, is the use of an appropriate statistical test (we used non-parametric Mann-Whitney) and a low threshold for the p-value calculation (α = 0.01). The threshold is not yet classical in biology, but not rare16 and in our opinion, it should become a standard due to the inherent variability of biological materials. Equally important is to provide the exact p-value in the manuscripts so that the reader may also perform their own interpretation of the significance of presented data. The critical assumption of t-tests is that data used in the statistical test are obtained from a random sampling, in other words, that these data are independent. This is a very sensitive issue since “true” independent experiments are unreachable under practical laboratory constraints (money, time, instrument availability,…). Thus, it is critical to identify what a reasonable independent experiment is. In our case, we decided that a single experiment was the characterization of a single plant. Consequently, the data used in the statistical tests are average values of elastic properties obtained on several single plants. In practice, we performed experiments on different days with a certain number of plants: usually our results cover at least 4 independent days in which 2 to 4 plants are measured.
Limitations
Our protocol uses classical force-distance curves to measure nanomechanical properties of roots. We used a contact-based mechanical model to obtain the elasticity of plant roots7 and a recently modified version, called the trimechanic theory.8 Alternative analysis could be performed such as shell models,17 hyper-elastic models,18 or visco-elastic models.19 Nevertheless, the protocol intends to focus on the handling of root samples. The same protocol remains valid with other mechanical model. Illustrated details about similar AFM measurements can be found elsewhere.20,21
Our protocol was applied on Arabidopsis thaliana seedlings but could be adopted to any seedling roots having a few cm in length. There are nevertheless various limitations concerning the use of AFM. Most modern instruments use small round Petri dishes as a sample stage. In some AFM systems, it has been difficult to use standard rectangular glass slides. Indeed, to orient the plant root according to the fixed position of the cantilever, it is easier to rotate a glass slide.
A main advantage of our protocol was the use of the Dimension 3100 AFM since it operates on a large motorized platform that can accommodate very large samples such as plant roots. The downside of this instrument is its end of commercial life with no possible upgrade. The AFM used in this protocol has some peculiarity due to the open-loop scanner such as the lack of a performant force trigger set-up for fore-distance curves acquisition. In addition, due to unfocused laser beam, modern small cantilevers such as PF-QNM-LC cannot be used. Nevertheless, modern instruments will not suffer with such problems.
A more worrisome limitation was the use of NuSil pressure sensitive adhesive. Its availability is unpredictable and we have tried alternatives but with moderate or no success. A key advantage of NuSil is its slow polymerization (min) and low viscosity, which allows us to manipulate with standard pipettes. For alternative, the key property to look for is “pressure sensitive adhesive”. Some commercial alternatives can be found and were tested in our lab but never applied during this protocol.
The extensibility of the protocol to other possible stresses is limited to experimental conditions that must be translated into a chemical change of agar material (the central step of this protocol). For instance, insoluble compounds may not be homogeneously distributed in the agar and therefore the outcome of the phenotype may be unpredictable. In such cases, a different growing methodology should be used such as directly in liquid solutions.
Troubleshooting
Problem 1
Classically, seedlings grow in large Petri dishes that are oriented vertically (step 15). However, to perform nanoindentation experiments, it is critical to have straight root ends; thus preventing artefact from curling effects as seen on Figure 10.
Figure 10.
Example of curled Arabidopsis thaliana seedlings grown almost vertically on agar Petri dish
Because our experiments do not use additional phosphate than the one already present in the agar, it is possible that adding Pi should also reduce the curling effect. The black lines locate the end of the root tip when deposited in the Petri dish. If the root grows, the root tip will extend beyond the black line.
Potential solution
-
•
Flat horizontal crystallization plates (Figure 11) are ideal replacement and suitable for growing plants for 4-5day-old only.
Figure 11.
Example of a crystallization plate used to grow Arabidopsis thaliana seedlings
The plate is on the left and a zoom over a single well on the right. Agar is deposited in some wells and seeds are sown under a laminar hood. No more than 10 seeds should be sown in each well to allow enough space for roots to grow. Some wells are filled with water to provide humidity in the plate. The size of these wells is adapted for 2 cm long seedlings (4-5-day-old for Arabidopsis).
Problem 2
In Materials and Equipment, Pressure sensitive silicon adhesive: NuSil adhesive tends to polymerize with time when stored in any plastic material. If they are stored with a half-empty bottle for longer duration, the air inside the bottle can lead to drying of the adhesive.
Potential solution
-
•
NuSil must be stored in glass aliquots of 2 mL. One of the great advantages of NuSil is its low viscosity that allows relatively easy manipulation. These glass tubes are filled up to the brim with NuSil, then sealed with a rubber-seal cap tightly. To fill the vials, it is necessary to use a sealed syringe instead of a classical pipette tip. This way they are stored in small quantities and last long. Over a three-year period (about 110 experiments), almost ½ of the 1 L bottle of NuSil was used. Use dark-glass bottles because it is the native storage condition of ethyl acetate.
Problem 3
We sometimes observed that one side of the Petri dish displays one phenotype and another side a different phenotype (1). We attributed this phenomenon to a lack of homogeneous distribution of metals on the Petri dishes.
Potential solution
-
•
It is important to mix the added metal solutions well into the liquid agar. This can be slightly tricky, as agar must be liquid enough to mix but not too hot.
Problem 4
Timing of the experiment is critical (1). Because we want to compare physiological and mechanical responses from different experimental conditions, it is critical to keep a strict timing (age of plants and duration of stress). One of the difficulties is that pausing is not possible after starting the experiment (since the sowing). The 2 h transfer period imposes a strict agenda in the progress of the main experiment.
Potential solution
-
•
Make a monthly and weekly calendar with planned experiments.
-
•
Include a detailed daily calendar of the AFM measurement day to avoid any loss of time. Timing included in this protocol is defined after several months of practice; it is thus underestimated for a newcomer.
Problem 5
Improper fitting of the cantilever into the probe holder might lead to errors during AFM calibration steps: sensitivity and force constant of cantilever (Step 2a). Sometimes, the baseline at the beginning of the FD curve is not flat and may hint about poor cantilever positioning in the holder or a poorly fixed root (Figure 12). In case of incorrect sensitivity characterization, the thermal tuning will provide an erroneous cantilever spring constant.
Figure 12.
Example of a FD curve with a bent “baseline” (between 1 and 3 μm)
The approach and retract curves are in red and blue, respectively. The total ramp size is 3 μm while the maximum applied force is about 13 nN. Sometimes such a bending is a sign of a poor set-up in the optical AFM system. Other reasons could also explain the non-flat baseline such as a poorly fixed root on the NuSil or a cantilever is still in contact with the surface at the end of the retract ramp.
Potential solution
-
•
Do not hesitate to remove the probe holder and reposition the cantilever delicately into the groove of the holder. Perform the calibration again. There is no escape from the calibration steps, any improper action will systematically impact all the AFM measurements and their interpretation.
-
•
We suggest the reader to access a complete description of calibration steps here22 and particularly the supplementary information.
Problem 6
Pay attention to the face of the root that is in contact with agar during the transfer time of 2 h (step 4.b.i). Because it is transferred just for 2 h, the side touching the metal-excess agar is most likely to experience the full stress level than the whole root itself.
Potential solution
-
•
It is best to present the side of the root that was in contact with agar to the top access of the glass slide, i.e., facing the tip of the cantilever. We have experienced a reduced heterogeneity in our data after such trick.
Problem 7
The curl effect makes the root rounded and more stretched (Figure 13); it complicates measurements and their interpretation (step 4.b.ii). In some cases, part of the root might be under the cantilever, which will not allow the cantilever to take the measurements properly as it might not touch the desired area. Note that the cantilever support may also touch the NuSil glue before the tip is reaching the surface of the root.
Figure 13.
Image showing a bend root fixed on a glass slide
We found that such bending leads to high elasticity values of the external cell wall structure. The curvature often comes from the growing condition, likely induced by gravitropism. This is likely the most inconvenient aspect of growing seedlings in agar plates.
Potential solution
-
•
In any case, bent roots do experience various mechanical stresses and should not be measured, unless it is a property specifically sought.
Problem 8
Sealing by adhesive is done such that the height shall remain constant throughout the slide (step 4.b.iii). With an excess of adhesive, the tip may touch the adhesive during the positioning under the AFM scanner or the edge of cantilever support may also touch the adhesive that will bias nanoindentation measurements.
Potential solution
-
•
The sealing starts from the top just to make sure of the stiffening of the glue. A minimum of three seals is required so that the root does not move.
-
•
See Figure 14 as an example for fastening the very tip of the root
Figure 14.
Optical magnification of deposited seedling roots on NuSil adhesive over a glass slide
A thin needle (bright color on the left) is used to fasten the position of the root on the adhesive by picking hardening adhesive over the root. Several fastening strips are required along the root but keep a significant room for the investigation zone, which is about 500 μm from the root tip (about five times the root diameter).
Problem 9
Performing a large-scale lateral displacement on the top of a root (step 5.m). Because of the inherent curvature and topography of a plant root, it is not advised to move over long distances (> 1 μm) using piezo scanner offsets.
Potential solution
-
•
We found that withdrawing the cantilever works best on our system. It will avoid any tip damage.
-
•
Re-engage at each measurement node. This practice helps in a better landing of the tip at the correct spot, fewer plant surface scratches, and less biological material deposition on the tip. All these parameters lead to better data acquisition.
Problem 10
The reading of the plant root phenotype (step 6.a) in our experiment is the root extension (measured in mm). Sometimes, plant seeds have different germination “power” and some seedlings may grow slower or faster.
Potential solution
-
•
Always try to transfer roots of similar length so that their growth rate would be similar. This will help in overcoming any errors when measuring root length the next days.
Resource availability
Lead contact
Jean-Luc Pellequer (jean-luc.pellequer@ibs.fr).
Materials availability
This study did not generate new unique reagents.
Data and code availability
This study did not generate/analyze datasets/code.
Acknowledgments
IBS acknowledges integration into the Interdisciplinary Research Institute of Grenoble (IRIG, CEA). This work acknowledges the AFM platform at the IBS and the ANR project BioPhyt - 18-CE20-0023-03 and the support of the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement No 812772, Project Phys2BioMed. We thank Bérangère Moreau (CEA Grenoble DRF/IRIG/DEPHY/MEM/LEMMA) for her help in SEM images. We also thank Isabel Ayala and Lionel Imbert (IBS/NMR) for their support in lab experiments.
Author contributions
H.K. established the current protocol and performed the mechanical measurements. J.-M.T. performed the initial mechanical analysis. A.-E.F. provided critical improvements in the protocol. D.F. performed scanning electron microscopy on PNP and other cantilevers. C.G. established the initial preliminary protocol and performed initial AFM measurements. S.-w.W.C. developed the trimechanic theory used to interpret the mechanical data. T.D. initiated the project in 2014 and provided plant materials. J.-L.P. directed the study and wrote the manuscript with the help of all co-authors.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Thierry Desnos, Email: thierry.desnos@cea.fr.
Jean-Luc Pellequer, Email: jean-luc.pellequer@ibs.fr.
References
- 1.Godon C., Kaur H., Teulon J.-M., Chen S.W.W., Desnos T., Pellequer J.-L. In: Mechanics of Cells and Tissues in Diseases. Lekka M., Navajas D., Radmacher M., Podestà A., editors. Walter de Gruyter GmbH; 2023. Plant root cell wall stiffening induced by a metallic stress; pp. 335–344. [DOI] [Google Scholar]
- 2.Hermanowicz P., Sarna M., Burda K., Gabryś H. AtomicJ: an open source software for analysis of force curves. Rev. Sci. Instrum. 2014;85 doi: 10.1063/1.4881683. [DOI] [PubMed] [Google Scholar]
- 3.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meijering E., Jacob M., Sarria J.C.F., Steiner P., Hirling H., Unser M. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry A. 2004;58:167–176. doi: 10.1002/cyto.a.20022. [DOI] [PubMed] [Google Scholar]
- 5.Balzergue C., Dartevelle T., Godon C., Laugier E., Meisrimler C., Teulon J.-M., Creff A., Bissler M., Brouchoud C., Hagège A., et al. Low phosphate activates STOP1-ALMT1 to rapidly inhibit root cell elongation. Nat. Commun. 2017;8 doi: 10.1038/ncomms15300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaur H., Godon C., Teulon J.-M., Desnos T., Pellequer J.-L. In: Mechanics of Cells and Tissues in Diseases. Lekka M., Navajas D., Radmacher M., Podestà A., editors. Walter de Gruyter GmbH; 2023. Preparation and deposition of plant roots for AFM nanomechanical measurements; pp. 125–138. [DOI] [Google Scholar]
- 7.Sneddon I.N. The relation between load and penetration in the axisymmetric boussinesq problem for a punch of arbitrary profile. Int. J. Eng. Sci. 1965;3:47–57. [Google Scholar]
- 8.Chen S.W.W., Teulon J.M., Kaur H., Godon C., Pellequer J.L. Nano-structural stiffness measure for soft biomaterials of heterogeneous elasticity. Nanoscale Horiz. 2022;8:75–82. doi: 10.1039/D2NH00390B. [DOI] [PubMed] [Google Scholar]
- 9.Carl P., Schillers H. Elasticity measurement of living cells with an atomic force microscope: data acquisition and processing. Pflugers Arch. 2008;457:551–559. doi: 10.1007/s00424-008-0524-3. [DOI] [PubMed] [Google Scholar]
- 10.Lekka M., Laidler P., Gil D., Lekki J., Stachura Z., Hrynkiewicz A.Z. Elasticity of normal and cancerous human bladder cells studied by scanning force microscopy. Eur. Biophys. J. 1999;28:312–316. doi: 10.1007/s002490050213. [DOI] [PubMed] [Google Scholar]
- 11.Domke J., Radmacher M. Measuring the elastic properties of thin polymer films with the atomic force microscope. Langmuir. 1998;14:3320–3325. doi: 10.1021/la9713006. [DOI] [Google Scholar]
- 12.Kontomaris S.V., Stylianou A., Chliveros G., Malamou A. Determining spatial variability of elastic properties for biological samples using AFM. Micromachines. 2023;14 doi: 10.3390/mi14010182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fernandes A.N., Chen X., Scotchford C.A., Walker J., Wells D.M., Roberts C.J., Everitt N.M. Mechanical properties of epidermal cells of whole living roots of Arabidopsis thaliana: an atomic force microscopy study. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2012;85 doi: 10.1103/PhysRevE.85.021916. [DOI] [PubMed] [Google Scholar]
- 14.Routier-Kierzkowska A.L., Weber A., Kochova P., Felekis D., Nelson B.J., Kuhlemeier C., Smith R.S. Cellular force microscopy for in vivo measurements of plant tissue mechanics. Plant Physiol. 2012;158:1514–1522. doi: 10.1104/pp.111.191460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Millet A. A universal model for the Log-normal distribution of elasticity in polymeric gels and its relevance to mechanical signature of biological tissues. Biology. 2021;10:64. doi: 10.3390/biology10010064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kozlova L., Petrova A., Ananchenko B., Gorshkova T. Assessment of primary cell wall nanomechanical properties in internal cells of non-fixed Maize roots. Plants. 2019;8:172. doi: 10.3390/plants8060172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsugawa S., Yamasaki Y., Horiguchi S., Zhang T., Muto T., Nakaso Y., Ito K., Takebayashi R., Okano K., Akita E., et al. Elastic shell theory for plant cell wall stiffness reveals contributions of cell wall elasticity and turgor pressure in AFM measurement. Sci. Rep. 2022;12 doi: 10.1038/s41598-022-16880-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jorba I., Beltrán G., Falcones B., Suki B., Farré R., García-Aznar J.M., Navajas D. Nonlinear elasticity of the lung extracellular microenvironment is regulated by macroscale tissue strain. Acta Biomater. 2019;92:265–276. doi: 10.1016/j.actbio.2019.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peaucelle A., Braybrook S.A., Le Guillou L., Bron E., Kuhlemeier C., Höfte H. Pectin-induced changes in cell wall mechanics underlie organ initiation in Arabidopsis. Curr. Biol. 2011;21:1720–1726. doi: 10.1016/j.cub.2011.08.057. [DOI] [PubMed] [Google Scholar]
- 20.Bovio S., Long Y., Moneger F. Use of atomic force microscopy to measure mechanical properties and turgor pressure of plant cells and plant tissues. J. Vis. Exp. 2019;149 doi: 10.3791/59674. [DOI] [PubMed] [Google Scholar]
- 21.Rauschert I., Benech J.C., Sainz M., Borsani O., Sotelo-Silveira M. Atomic force microscopy to study the physical properties of epidermal cells of live Arabidopsis roots. J. Vis. Exp. 2022;181 doi: 10.3791/63533. [DOI] [PubMed] [Google Scholar]
- 22.Schillers H., Rianna C., Schäpe J., Luque T., Doschke H., Wälte M., Uriarte J.J., Campillo N., Michanetzis G.P.A., Bobrowska J., et al. Standardized Nanomechanical Atomic force microscopy Procedure (SNAP) for measuring soft and biological samples. Sci. Rep. 2017;7:5117. doi: 10.1038/s41598-017-05383-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate/analyze datasets/code.