Abstract
Human immunodeficiency virus type 1 (HIV-1) variants that use the coreceptor CCR5 for entry (R5; macrophage tropic) predominate in early infection, while variants that use CXCR4 emerge during disease progression. Some late-stage variants use CXCR4 alone (X4; T-cell tropic), while others use both CXCR4 and CCR5 (R5X4; dualtropic). It has been proposed that dualtropic R5X4 strains are intermediates in the evolution from R5 to X4, and we hypothesized that a dualtropic primary-isolate quasispecies might contain variants that represent the spectrum of coreceptor use in vivo. We generated a panel of 35 functional full-length env clones from the primary-isolate quasispecies of a dualtropic prototype strain, HIV-1 89.6PI. Thirty of the functional env clones (86%) were R5X4, four (11%) were R5, and one (3%) was predominantly X4. V3 to V5 sequences did not reveal clustering by coreceptor usage, and no specific sequence motif or V3 charge pattern corresponded to coreceptor utilization. Complete sequencing of seven functionally divergent Env proteins revealed ≥98.7% homology and conservation of structurally important domains. Chimeras between the R5X4 89.6 prototype and an R5 variant indicated that multiple regions contributed to the use of CXCR4, while chimeras with the X4 variant implicated a single residue in V4 in CCR5 use. These results confirm, at the molecular level, both that dualtropic variants are a predominant component of late-stage syncytium-inducing isolates and that variants restricted to each coreceptor coexist with dualtropic species in vivo. Coreceptor-restricted minority variants may reflect residual R5 species from earlier in disease as well as emerging X4 variants.
Human immunodeficiency virus type 1 (HIV-1) entry requires interaction of the viral envelope glycoprotein with cellular CD4 and a seven-transmembrane G protein-coupled chemokine receptor (reviewed in reference 1). Differential use of distinct chemokine receptors by HIV-1 isolates has largely explained differences in viral tropism and cytopathogenicity. While several chemokine receptors and orphan receptors can mediate entry in vitro, the principal HIV-1 coreceptors are the β-chemokine receptor CCR5 and the α-chemokine receptor CXCR4. Non-syncytium-inducing (NSI) HIV-1 variants that use CCR5 (R5; macrophage [M] tropic) play a crucial role in sexual, blood-borne, and vertical transmission and are the predominant viral population immediately after seroconversion and during asymptomatic infection (26, 37). Syncytium-inducing (SI) variants that use CXCR4 evolve in about 50% of individuals with AIDS, coincident with CD4+ T-cell decline, and the emergence of SI variants is strongly associated with accelerated disease (32). Recent studies with SCID mice and lymphoid tissue ex vivo have suggested that the emergence of CXCR4 use may be responsible for, rather than simply a consequence of, enhanced immune destruction (13, 22).
SI variants may use CXCR4 alone (X4; T-cell-line [T] tropic) or in addition to CCR5 (R5X4; dualtropic). While the dominance of R5 strains early in infection and the emergence of CXCR4-using variants later in disease are widely recognized, several important questions regarding the relationship between R5, R5X4, and X4 variants in vivo remain incompletely resolved. (i) Since new infections are initiated by M-tropic NSI R5 strains, and not by X4 or R5X4 strains even when these variants are present in late-stage transmitters, does this reflect the universal persistence of R5 variants alongside the CXCR4-using SI strains as they emerge late in disease (26, 34)? (ii) Although several recent studies document the ability of late-stage primary-isolate swarms to use both CCR5 and CXCR4 (10, 25), does this reflect mainly the coexistence of both X4 and R5 variants within the swarm or mainly dualtropic R5X4 variants? (iii) Similarly, while many late-stage SI strains are clearly R5X4 (8, 28), single-coreceptor X4 isolates are well described (2, 33) but their place in viral evolution is not clear. It may be that R5X4 strains represent intermediates in the transition from R5 to X4 (11), but it is not known whether X4 variants would eventually emerge from R5X4 populations in vivo given sufficient time. Thus, the spectrum of individual variants that coexist within the late-stage SI quasispecies is an important question that may offer insights into aspects of pathogenesis.
HIV-1 89.6PI is a dualtropic primary isolate from the blood of an individual with AIDS (8). An infectious molecular clone derived from this primary isolate provided the first indication that both macrophage tropism and the T-cell-line-tropic SI phenotype could be a feature of a single virus and not the result of multiple variants within a primary-isolate quasispecies. These characteristics have since been linked to the use of CCR5 and CXCR4, respectively, and this well-characterized infectious clone is widely used as an R5X4 prototype. We hypothesized that the 89.6PI quasispecies might contain R5 variants reflecting earlier stages of infection and X4 variants related to disease progression in vivo. Identifying and analyzing the spectrum of quasispecies within this isolate may thus provide insight into the steps involved in the phenotypic transition from R5 to R5X4 and possibly to X4 that occurs during HIV-1 pathogenesis. To address the relationship between R5, R5X4, and X4 variants within a dualtropic viral swarm, we evaluated the coreceptor usage pattern of a panel of related full-length 2.5-kb envelopes that we cloned from the original 89.6 primary-isolate quasispecies.
Construction of a panel of related env genes from an R5X4 primary isolate.
We used high-fidelity PCR to make 50 full-length env clones from the dualtropic 89.6PI viral swarm (8). We selected this primary isolate because the infectious molecular clone derived from it is widely employed as an R5X4 prototype in studies of tropism and coreceptor use as well as in both in vitro and in vivo studies of pathogenesis (11, 14, 16). The panel of related env clones was generated from the same cellular DNA pool from which the original 89.6 infectious clone was isolated by lambda phage cloning (8). To ensure that each clone represented a distinct proviral molecule, rather than potentially multiple amplification products of the same template, we used separate aliquots of template DNA in independent PCRs, and only one env clone from each amplification reaction was utilized. Amplification was carried out with primers 5′-AGA AAG AGA AGA AGA CAG TGG CAA TGA-3′ and 5′-TAG CCC TTC CAG TCC CCC CTT TTC TTT TAA-3′ using rTth-XL polymerase (Perkin-Elmer, Foster City, Calif.). Reaction mixtures were heated to 95°C for 1 min, followed by 35 cycles at 94°C for 1 min, 54°C for 5 min, and 72°C for 7 min, and a final 10-min extension at 72°C. Amplification products were treated with Pfu polymerase (Stratagene, La Jolla, Calif.) to generate blunt ends, purified, and ligated into pCR-Blunt (Invitrogen, Carlsbad, Calif.) downstream of the T7 promoter. Clones carrying properly sized and oriented 2.5-kb env inserts were identified by restriction analysis. For clarity, the uncloned primary isolate is referred to as 89.6PI, the env gene from the 9.7-kb infectious molecular clone is referred to as 89.6, and each clone is designated by a number from 1 to 50.
Coreceptor fusion patterns of 89.6-related env clones.
env clones were tested for fusion with CCR5 and CXCR4 in a cell-cell fusion assay that relies on a T7 polymerase-expressing recombinant vaccinia virus and a T7-driven luciferase reporter system, as described previously (11, 29). Within each experiment, controls included effector cells lacking envelope and target cells with CD4 but no chemokine receptor, and 10-fold enhancement of fusion relative to controls was considered positive.
Thirty-five of the 50 clones (70%) encoded Env proteins that were functional for fusion with at least one major coreceptor (Table 1). The others failed to achieve ≥10-fold-greater luciferase expression compared to control cells without Env when mixed with CD4 and at least one coreceptor and also failed to achieve ≥10-fold-greater luciferase expression when mixed with cells expressing CD4 and one coreceptor than when mixed with cells expressing CD4 alone. For most env clones that did not achieve this threshold, there was little luciferase expression or none at all. Those clones were considered nonfunctional and were not investigated further. None of the Env proteins fused with a coreceptor independent of CD4 or used CD4 in the absence of a coreceptor (data not shown).
TABLE 1.
Full-length Env clones generated from the 89.6PI quasispecies
| Coreceptor use patterna | No. of clones | % of all clones | % of functional clones |
|---|---|---|---|
| CCR5 and CXCR4 (R5X4) | 30 | 60 | 86 |
| CCR5 (R5) | 4 | 8 | 11 |
| CXCR4 (X4) | 1 | 2 | 3 |
| Nonfunctional | 15 | 30 |
Coreceptor use was based on cell-cell fusion and, for selected Env clones, was confirmed by pseudotype infections.
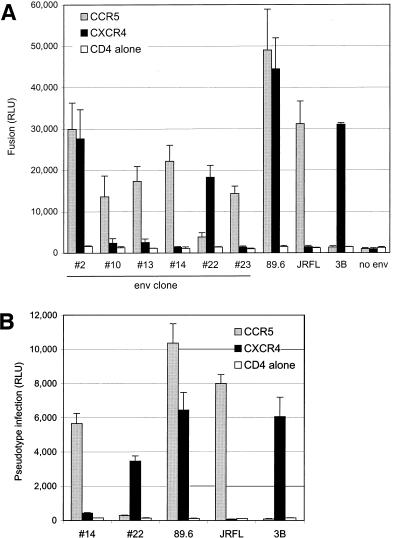
The majority (86%) of functional envelopes from the 89.6PI swarm used both CCR5 and CXCR4 for fusion and so were R5X4, like the prototype 89.6 molecular clone (Table 1). For most, the levels of fusion with the two coreceptors were similar. In addition, we found a minority of variants that were restricted to one individual coreceptor. Four of the functional genes (11%) used CCR5 but not CXCR4 (clones 10, 13, 14, and 23). Only one variant (3%) was selective for CXCR4 (clone 22). Fusion mediated by the functionally divergent Env proteins, along with one representative R5X4 Env (clone 2), is shown in Fig. 1A. These results demonstrate the presence of viral species with distinct coreceptor usage within the dualtropic 89.6PI quasispecies.
FIG. 1.
Fusion and infection mediated by functionally divergent env clones within the 89.6PI quasispecies. (A) Cloned env genes were analyzed for CCR5 and CXCR4-mediated fusion in a cell-cell fusion assay, along with the R5 and X4 prototypes JRFL and 3B, respectively. The env gene designated 89.6 is derived from the full-length infectious clone described previously (8, 11). Only data for the five variants with distinct R5 or X4 patterns and one representative R5X4 env variant are shown in this graph. Each envelope-coreceptor combination was tested a minimum of three times. (B) Pseudotype virions generated with one R5 and one X4 env variant were used to confirm coreceptor selectivity in infection. To generate pseudotype virions, env genes were cotransfected with an env-defective HIV-1 plasmid that carries the luciferase reporter gene in place of nef. U87 cells were transfected with CD4 and the indicated chemokine receptor, infected with equal amounts of each pseudotype virus based on p24 antigen content, and lysed 3 days later for measurement of luciferase expression. Pseudotype infections were tested in three independent experiments.
Env-mediated pseudotype virion infections.
We next determined whether the clones' coreceptor specificity in cell-cell fusion accurately reflected Env-coreceptor-mediated infection, using luciferase reporter virus pseudotypes (9). Two functionally distinct env genes, clones 14 and 22, were subcloned under the control of the cytomegalovirus (CMV) promoter into pCDNA3 (Invitrogen) and cotransfected into 293T cells along with the NL4-3 backbone plasmid (pNL-luc-E−R−) that bears a defective env gene and the luciferase reporter gene in place of nef (9) (kindly provided by N. Landau). Two days later, the supernatant was harvested, clarified by centrifugation, and quantified by p24 antigen content. U87 cells (2 × 105 cells per well in 24-well plates) were transfected with CD4, with or without a coreceptor, and infected the following day with pseudotypes by using 20 ng of p24 antigen, in the presence of Polybrene (5 μg/ml). Luciferase expression was quantified in cell lysates 3 days later. In parallel, pseudotype virions were made with the R5X4 89.6 env, as well as the R5 JRFL and X4 3B prototypes.
As shown in Fig. 1B, pseudotype virions carrying the three 89.6PI-derived variants differed in coreceptor-mediated tropism. Virions carrying env variant 14 infected cells expressing CD4 and CCR5 but not CXCR4, like JRFL. In contrast, virions carrying env variant 22 resembled 3B and infected cells expressing CXCR4 but not CCR5. As expected, virions carrying the prototype 89.6 Env infected cells expressing CD4 with either CCR5 or CXCR4. Thus, Env-mediated infection showed the same selective pattern of coreceptor utilization as did fusion and confirmed the presence of variants restricted to CCR5 or CXCR4 for infection.
Phylogenetic and V3 sequence analysis of related env clones.
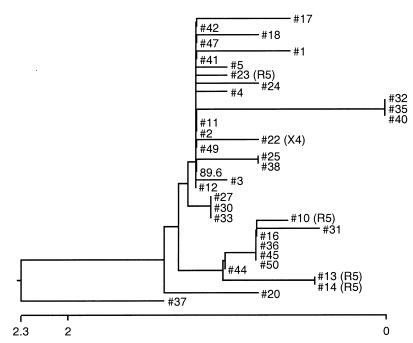
To determine the genetic relatedness among the 36 distinct envelope genes derived from the 89.6PI viral swarm (35 functional clones generated here plus 89.6), we sequenced the V3-to-V5 region of each and compared their predicted amino acid and nucleic acid sequences (Fig. 2). Among all the env variants, the overall nucleic acid homology was ≥97.5%, and 20 distinct sequences were identified. Fourteen sequences were represented by one clone each, two sequences were each represented by two clones, two sequences by three clones each, one sequence by four clones, and one V3-to-V5 sequence was shared by eight of the env variants including the 89.6 prototype. Since each clone was generated independently from separate aliquots of template DNA, they represent independent variants within the quasispecies despite shared V3-to-V5 sequences.
FIG. 2.
Genetic relatedness of all 36 functional env clones from the 89.6PI swarm, determined on the basis of the nucleotide sequences across a ∼600-bp V3-to-V5 region. The dendrogram was generated by using the Clustal method. Clones restricted to CCR5 or CXCR4 are indicated by R5 or X4 to the right of the clone number, while the rest of the env clones, for which no coreceptor usage designation is given, were R5X4.
All env clones with identical V3-to-V5 sequences had the same coreceptor use patterns. However, the R5 env clones as a group did not form a cluster when analyzed by V3-to-V5 amino acid or nucleic acid sequence (Fig. 2), or when the analysis was restricted to V3 alone (data not shown). In fact, only 6 env clones differed from 89.6 within V3. The R5 variants 13 and 14 had a non-charge-altering I327M substitution, two R5X4 variants had a non-charge-altering T305A substitution, and one R5X4 variant had a R300G substitution which lowered the overall charge from +7 in 89.6 to +6 but did not alter coreceptor use. One additional env clone (clone 37) differed from 89.6 at 6 sites in V3 (SIH instead of RLS at residues 308 to 310, and TGD instead of RRN at residues 320 to 322), which resulted in a considerably lower +3 V3 charge but also did not affect the R5X4 phenotype. Thus, the V3 sequence and charge appeared to have little role in regulating differential coreceptor choice among variants in this panel.
Genetic analysis of functionally distinct envelope clones.
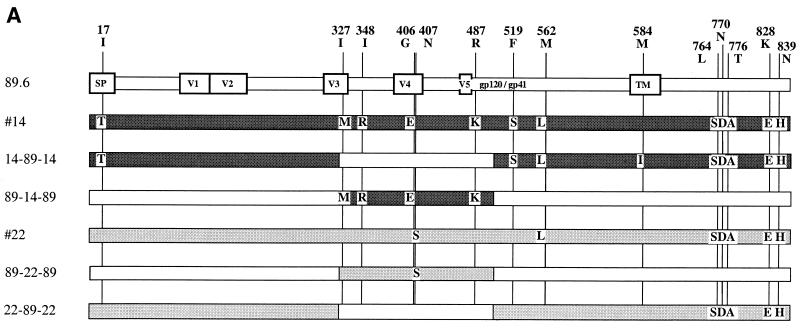
To further address the genetic basis for coreceptor heterogeneity among the related env clones, we obtained full-length 2.5-kb sequences for the functionally divergent subset (X4 clone 22 and R5 clones 10, 13, 14, and 23) and one representative R5X4 variant (clone 2), and compared them with the 89.6 prototype (Fig. 3). During this analysis we identified one error in the published sequence of the 89.6 molecular clone Env (Ala instead of Arg at amino acid 542 in gp41), which has been corrected in GenBank.
FIG. 3.
Amino acid alignment of functionally diverse env gene sequences. Full-length sequences were determined for the four R5 env clones (clones 10, 13, 14, and 23), one X4 env clone (clone 22), and one representative R5X4 env clone derived from this pool (clone 2). Sequences were aligned with that of the original R5X4 89.6 clone. R5 clones 13 and 14 had identical Env sequences.
The R5X4 variant analyzed (clone 2) was identical to 89.6 in V3 to V5 but differed in several amino acids outside this region (Fig. 3), while two R5 clones had identical full-length Env sequences (clones 13 and 14). Overall, there was ≥98.7% amino acid homology among fully sequenced Env proteins. All amino acids implicated in direct CD4 binding were conserved among the seven variants (19), and 15 of 18 residues implicated in CCR5 binding were conserved as well (23). The 7 Env proteins analyzed shared 29 of 30 potential N-linked glycosylation sites. The exception was one potential site at N138 that was lacking in the R5X4 variant 2 and the R5 variant 23 but was present in the others, and so was not linked to coreceptor specificity. Unexpectedly, one of the R5 env clones (clone 23) had a single extra cysteine in gp120 (Y434C), confirmed by repeated sequencing. Since this Env supports fusion, the mechanism by which this presumably unpaired cysteine residue allows proper gp120 expression and function is uncertain.
Compared to 89.6, differences in gp120 sequence among the R5 clones were most prominent in the signal peptide and V regions, as expected (Fig. 3). Only one V3 difference was found among the functionally divergent Env proteins (R5 env clones 13 and 14), and that was a single substitution (I330M) that did not affect the strong positive 89.6 V3 charge. Importantly, there was no sequence motif common to the R5 variants compared with the R5X4 env variants that might suggest a common regulator of X4 utilization. The one X4 clone (clone 22) revealed only a single-amino-acid difference in gp120 compared with 89.6, in V4 (N410S), suggesting that this residue may contribute to CCR5 utilization in the background of this CXCR4 use.
Unexpectedly, sequencing of the portion corresponding to gp41 showed that all of the env variants were quite similar to one another but differed from 89.6 in several locations (Fig. 3). All six gp41 variants shared three amino acid differences compared with 89.6, and five of the six shared an additional three distinct residues. This suggests that the 89.6 env gene is the most divergent among the variants cloned from the primary-isolate swarm. Since the gp41 differences did not correlate with R5, R5X4, or X4 patterns, however, they are not likely to be responsible for coreceptor choice among these variants.
Chimeric envelopes and molecular determinants of coreceptor use.
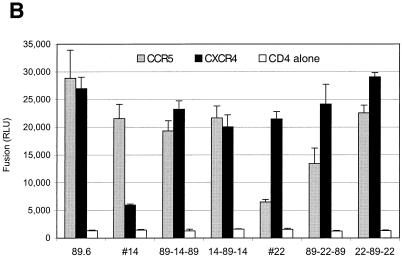
Since sequences did not show any clear linkage to coreceptor use, in order to begin to address the molecular regulation of individual coreceptor usage among related variants of a dualtropic swarm, we generated two pairs of reciprocal recombinants. We selected one R5 clone (clone 14) and one X4 clone (clone 22) and exchanged env domains with the 89.6 R5X4 env by using overlap extension PCR (Fig. 4A). Three overlapping fragments were amplified from each env clone using rTth-XL polymerase. A ∼1,000-bp 5′ fragment was amplified with the same upstream primer used for PCR cloning and downstream primer 5′-CTT ATT ATG TTT CTT CTT GCA TAA-3′; a ∼550-bp V3-to-V5 fragment was amplified using upstream primer 5′-TTA TGC AAG AAG AAA CAT AAT AAG-3′ and downstream primer 5′-GCC CTG GTG GGT GCT ACT CCT ATT-3′; and a ∼1,400-bp fragment encompassing the 3′ portion of gp120 and gp41 was amplified with upstream primer 5′-AAT AGG AGT AGC ACC CAC CAG GGC-3′ and the downstream primer used for PCR cloning of env. Reaction mixtures were heated to 95°C for 1 min, followed by 25 cycles at 94°C for 30 s, 48°C for 30 s, and 72°C for 1 min, with a final extension of 72°C for 10 min. After gel purification, the 5′ and middle fragments were mixed to generate chimeric combinations and reamplified using the 5′ outer primer and 3′ middle fragment primer. The process was repeated using the purified products of this reaction and the 3′ fragment amplified with the 5′ and 3′ outer primers. The final amplification products were treated with Pfu polymerase and ligated downstream of the T7 promoter into pCR-Blunt. Clones carrying properly sized and oriented envelope inserts were identified by restriction analysis and verified by complete sequencing.
FIG. 4.
Coreceptor usage of chimeras generated between related env genes with distinct phenotypes. (A) Construction of recombinant env genes. Chimeras were made between the R5X4 89.6 env molecular clone and the R5 variant 14, and between 89.6 and the X4 variant 22. Overlap extension PCR was used to exchange the indicated regions, which introduced the amino acid changes noted. (B) Coreceptor selectivity of chimeric env variants. The env chimeras were tested for CCR5 and CXCR4 utilization based on cell-cell fusion.
One pair of recombinants exchanged all four gp120 residues differing between 89.6 and the R5 variant 14, except for one amino acid in the signal peptide (Fig. 4A). Introduction of these amino acids from 89.6 into the R5 clone 14 (chimera 14-89-14) conferred on it the ability to use CXCR4 in addition to CCR5 (Fig. 4B). However, replacing these residues in 89.6 with sequences from the R5 clone 14 (89-14-89) did not abrogate the ability of 89.6 to use both CXCR4 and CCR5. When the chimeric env clones were subcloned into CMV-driven vectors and used to make pseudotype virions, identical results were seen for coreceptor-mediated infection (data not shown). These results indicate that these gp120 residues are partially responsible for regulating CXCR4 usage among these related variants, but that other determinants contribute as well. Since the only other differences between 89.6 and clone 14 are in the signal peptide and in gp41, this suggests that coreceptor selectivity among these related species is complex and dependent on cooperation among multiple domains, including regions not generally considered typical tropism determinants.
We then tested the pair of chimeras made between 89.6 and X4 variant 22. Since the only gp120 difference between variant 22 and 89.6 is a single change in V4 (N407S), we used the same overlap extension primer pair described above, which resulted in exchange of this residue only (Fig. 4A). Replacing Ser 407 of the X4 env variant 22 with the 89.6 Asn residue (22-89-22) resulted in an env chimera that fused with both CCR5 and CXCR4 (Fig. 4B), implicating this residue in the regulation of CCR5 use. However, changing the 89.6 sequence to that of clone 22 (89-22-89) reduced, but did not completely block, the use of CCR5. Pseudotype virions generated with the chimeric env variants showed that the same pattern was true for infection (data not shown). Thus, this V4 residue is responsible in part for regulating CCR5 use in these related variants. However, other regions of Env, presumably in gp41, appear also to contribute to or to modulate the coreceptor specificity encoded by gp120.
Biological significance of heterogeneous coreceptor use within the 89.6PI swarm.
In this study we addressed the biological and molecular relationship among R5, R5X4, and X4 variants within a dualtropic primary-isolate quasispecies. This is important because the relationship between R5, R5X4, and X4 HIV-1 variants in vivo is not well understood, yet the evolution from R5 to R5X4, and possibly to X4, is a major determinant of pathogenesis. Our results show that at the molecular level (i) R5 variants persist alongside X4-using strains, implicating a source for R5 viruses in person-to-person transmission by advanced patients, and (ii) CXCR4-restricted X4 variants may ultimately emerge from the R5X4 pool, indicating that R5X4 variants may be evolutionary intermediates rather than the ultimate end result of evolution in vivo. Furthermore, (iii) the heterogeneous biological spectrum was independent of Env sequence and charge patterns traditionally recognized as tropism and coreceptor determinants, indicating that biological analysis, and not the sequence alone, is necessary to understand these relationships in vivo.
Most of the functional env genes in this panel of genetically related clones efficiently used both CCR5 and CXCR4. In addition to the R5X4 majority, however, we also found a few variants that were selective for an individual coreceptor. The functional diversity of low-frequency minority variants within dualtropic quasispecies has not been studied previously, and we chose to analyze this primary isolate because the R5X4 prototype infectious molecular clone and the env gene derived from it are widely used for in vitro, ex vivo, and in vivo studies (6, 11, 14, 16). The role played by R5X4 dualtropic variants in HIV-1 pathogenesis still requires clarification. Several groups have suggested that most SI primary isolates are dualtropic and use both R5 and X4 (10, 25, 28, 34). However, this has been based mainly on dual coreceptor use by uncloned viral swarms, or on a limited number of biologically cloned isolates. The predominance of R5X4 env clones that we found within this late-stage SI isolate demonstrates at the molecular level that it is indeed composed mainly of dualtropic R5X4 variants and is not predominantly a mix of M-tropic R5 and T-tropic X4 variants.
Coexisting with the R5X4 majority, a small group of env clones within the swarm used only CCR5. R5 variants predominate early in infection, and the emergence of CXCR4 utilization occurs as a late event. Thus, the genetically related R5 variants within the 89.6 quasispecies may reflect variants from earlier in the R5-to-R5X4 phenotypic evolution. While it is possible that R5 species would eventually be replaced completely by variants competent for CXCR4 use, our findings suggest that single-coreceptor R5 variants persist alongside R5X4. The persistence of CCR5-restricted minority variants goes along with the observation that this is the virus subtype involved in HIV-1 transmission, even from late-stage individuals who harbor mainly R5X4 or even X4 species.
We also found one env clone that was restricted to CXCR4. In contrast to the widely recognized transition from R5 to R5X4, the relationship between R5X4 and X4 variants in vivo is less certain. The discovery that many late-stage SI strains are R5X4 has led some to suggest that R5X4 species are the selectively advantaged phenotype and the end result of evolution in vivo. On the other hand, X4 primary isolates are well described (2, 33), and it has been hypothesized that R5X4 variants are transitional species in the evolution from R5 to X4 (11). Since the selective forces driving phenotypic evolution in vivo are poorly understood, it is not known whether X4 variants would eventually emerge in most individuals given sufficient time. The presence of X4 species within this swarm, albeit at low frequency, is consistent with such a pattern and could reflect early evidence of X4 emergence. The high degree of genetic similarity among the env variants does not allow for formal evolutionary analysis that could definitively show a temporal relationship between the R5, R5X4, and X4 variants, however. Of note, we recently analyzed the env variants contained within primary-isolate viral swarms obtained from blood of three late-stage AIDS patients, and we found that all of them harbored X4 minority variants, even though only one of the three primary isolates had a SI phenotype (29). Thus, it is possible that viral evolution in vivo at the molecular level may precede the appearance of phenotypic features evident in bulk populations.
Among the 36 functional envelope genes derived from the 89.6PI swarm, there was a high degree of homology in the V3-to-V5 region. However, no common sequence pattern, which might suggest either that R5 viruses were more closely related or that they shared common determinants of coreceptor selectivity, distinguished the R5 from the R5X4 variants. Furthermore, the V3 region, surprisingly, did not appear to regulate the coreceptor specificity of these related species, since the V3 sequences of the R5 env variants did not differ from the highly charged V3 sequence of 89.6. This is in contrast to prototype NSI R5 strains that display a relatively low V3 charge pattern (5, 36) and differs from the central role identified for V3 in many other studies of tropism and coreceptor choice (4, 7, 20, 31, 35). In fact, only one V3 difference was identified among all the functionally distinct Env proteins, and this non-charge-altering substitution was not the principal coreceptor determinant. Interestingly, a previous report analyzed a set of molecular clones derived contemporaneously from an infected individual and also found marked differences in cell tropism despite identical V3 sequences (15). Together, these results demonstrate considerable flexibility in biological characteristics for any given V3 sequences. Thus, while the V3 pattern may evolve over time from a low-charge pattern typical of NSI, M-tropic, R5 strains to a high-charge, SI, T-tropic X4 profile (27), at any given time variants with a range of tropisms and other biological characteristics may still be retained within the population represented by that V3 pattern.
Since no correlation was found between coreceptor choice and V3 sequences, genetic mapping was done and revealed complex determinants of coreceptor choice. While extensive studies have focused on Env determinants of tropism and coreceptor selectivity, this is, to our knowledge, the first study to address coreceptor determinants among naturally occurring, related yet functionally distinct variants within a quasispecies. In analyzing the X4 versus the R5X4 phenotype (i.e., CCR5 use on the background of CXCR4 utilization), an important although not absolute role in CCR5 usage was suggested for a single residue in V4. In analyzing the R5 versus R5X4 phenotype (i.e., CXCR4 use on the background of CCR5 utilization), sequences in gp120 and determinants elsewhere appeared to contribute to CXCR4 use. It is well recognized that multiple Env regions can contribute to tropism and coreceptor use in addition to or independently of V3, particularly V1/V2 and V4/V5 (3, 18). Except for a single residue in the signal peptide, however, the only other differences between 89.6 and the R5 env variant tested were in gp41. This suggests that elements in the transmembrane subunit can, in conjunction with determinants in gp120, modulate coreceptor use. Determinants in gp41 have not previously been reported to specifically influence viral tropism and coreceptor use, and most of the gp41 sequence differences were shared by R5, R5X4, and X4 variants, so it is not yet apparent how they contribute to regulation of coreceptor specificity. Nevertheless, these data support the idea that gp160 is a highly cooperative molecule where specific amino acids may have very different effects depending on the context and the remainder of the protein.
In previous studies we analyzed recombinants between 89.6 and unrelated R5 and X4 prototype strains, and there also we found that multiple Env regions contributed to both cell tropism and coreceptor choice (17, 30). The present data are consistent with those results and confirm that determinants of coreceptor use in 89.6 and its related variants differ from those identified in prototype R5 and X4 strains. The ability of multiple regions to enable dual tropism or coreceptor use suggests that for this strain the dual phenotype is “dominant” and that members of this swarm may be rather plastic in their ability to use the major coreceptors. Genetic analysis has typically considered structural elements that confer specific coreceptor use. Alternatively, it is possible that the Env core is intrinsically able to interact with both coreceptors, and that structural determinants which regulate coreceptor choice interfere with the efficient use of one or the other. Thus, the 89.6 Env may possess a structure that is inherently less susceptible to downregulatory influences of other domains. In addition to CCR5 and CXCR4, 89.6 can use an unusually large range of other seven-transmembrane domain receptors for fusion (6, 11, 12, 24), which may also be consistent with a highly plastic and adaptable coreceptor utilization capacity.
In vivo, aspects of pathogenesis that are linked to differences in coreceptor use include the propensity of CCR5-using strains to participate in person-to-person transmission and macrophage-dependent sequelae such as neurological or pulmonary disease, and the enhanced ability of CXCR4-using strains to deplete CD4+ T cells. A limitation of this study is that it is not known what level of coreceptor use in vitro corresponds with coreceptor use and biologically related properties in vivo, and there may be differences between coreceptor function in vitro and in vivo (14). In addition, coreceptor use in vitro may vary depending on the cell types in which it is tested, expression level, and whether fusion or infection is used as the readout (12, 21, 24). Our studies with pseudotype viruses confirm that the coreceptor specificity observed in fusion also reflects coreceptor use for infection in vitro. It will be useful to compare the pathogenic consequences of these viral variants using recombinant viruses incorporating these env genes in studies of pathogenesis ex vivo or in vivo with animal models. In addition, we anticipate that this panel of genetically related but functionally distinct env variants generated from the 89.6PI dualtropic prototype strain will provide a valuable tool with which to better understand the structural basis for coreceptor choice of naturally occurring viruses that emerge in vivo.
Acknowledgments
We thank D. Williams for expert technical assistance, N. Landau for pNL-luc-E−R−, and B. Hahn, S. Isaacs, D. Kolson, and R. Doms for valuable advice and discussions.
This work was supported by NIH grants HL 58004 and AI 35502.
REFERENCES
- 1.Berger E A, Murphy P M, Farber J M. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu Rev Immunol. 1999;17:657–700. doi: 10.1146/annurev.immunol.17.1.657. [DOI] [PubMed] [Google Scholar]
- 2.Björndal Å, Deng H K, Jansson M, Fiore J R, Colognesi C, Karlsson A, Albert J, Scarlatti G, Littman D R, Fenyö E M. Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. J Virol. 1997;71:7478–7487. doi: 10.1128/jvi.71.10.7478-7487.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carrillo A, Ratner L. Cooperative effects of the human immunodeficiency virus type 1 envelope variable loops V1 and V3 in mediating infectivity for T cells. J Virol. 1996;70:1310–1316. doi: 10.1128/jvi.70.2.1310-1316.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan S Y, Speck R F, Power C, Gaffen S L, Chesebro B, Goldsmith M A. V3 recombinants indicate a central role for CCR5 as a coreceptor in tissue infection by human immunodeficiency virus type 1. J Virol. 1999;73:2350–2358. doi: 10.1128/jvi.73.3.2350-2358.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chesebro B, Wehrly K, Nishio J, Perryman S. Macrophage-tropic human immunodeficiency virus isolates from different patients exhibit unusual V3 envelope sequence homogeneity in comparison with T-cell-tropic isolates: definition of critical amino acids involved in cell tropism. J Virol. 1992;66:6547–6554. doi: 10.1128/jvi.66.11.6547-6554.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choe H, Farzan M, Sun Y, Sullivan N, Rollins R, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The β-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 7.Cocchi F, DeVico A L, Garzino-Demo A, Cara A, Gallo R C, Lusso P. The V3 domain of the HIV-1 gp120 envelope glycoprotein is critical for chemokine-mediated blockade of infection. Nat Med. 1996;2:1244–1247. doi: 10.1038/nm1196-1244. [DOI] [PubMed] [Google Scholar]
- 8.Collman R, Balliet J W, Gregory S A, Friedman H, Kolson D L, Nathanson N, Srinivasan A. An infectious molecular clone of an unusual macrophage-tropic and highly cytopathic strain of human immunodeficiency virus type 1. J Virol. 1992;66:7517–7521. doi: 10.1128/jvi.66.12.7517-7521.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Connor R I, Chen B K, Choe S, Landau N R. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology. 1995;206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 10.Connor R I, Sheridan K E, Ceradini D, Choe S, Landau N R. Change in coreceptor use correlates with disease progression in HIV-1-infected individuals. J Exp Med. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. A dual-tropic HIV-1 isolate that uses fusin and the β-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 12.Edinger A L, Hoffman T L, Sharron M, Lee B, Yi Y, Choe W, Kolson D L, Mitrovic B, Zhou Y, Faulds D, Collman R G, Hesselgesser J, Horuk R, Doms R W. An orphan seven-transmembrane domain receptor expressed widely in the brain functions as a coreceptor for human immunodeficiency virus type 1 and simian immunodeficiency virus. J Virol. 1998;72:7934–7940. doi: 10.1128/jvi.72.10.7934-7940.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glushakova S, Grivel J C, Fitzgerald W, Sylwester A, Zimmerberg J, Margolis L B. Evidence for the HIV-1 phenotype switch as a causal factor in acquired immunodeficiency. Nat Med. 1998;4:346–349. doi: 10.1038/nm0398-346. [DOI] [PubMed] [Google Scholar]
- 14.Glushakova S, Yi Y J, Grivel J C, Singh A, Schols D, De Clercq E, Collman R G, Margolis L. Preferential coreceptor utilization and cytopathicity by dual-tropic HIV-1 in human lymphoid tissue ex vivo. J Clin Investig. 1999;104:R7–R11. doi: 10.1172/JCI7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Groenink M, Andeweg A C, Fouchier R A M, Broersen S, Van der Jagt R C M, Schuitemaker H, de Goede R E Y, Bosch M L, Huisman H G, Tersmette M. Phenotype-associated env gene variation among eight related human immunodeficiency virus type 1 clones: evidence for in vivo recombination and determinants of cytotropism outside the V3 domain. J Virol. 1992;66:6175–6180. doi: 10.1128/jvi.66.10.6175-6180.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karlsson G B, Halloran M, Li J, Park I W, Gomila R, Reimann K A, Axthelm M K, Iliff S A, Letvin N L, Sodroski J. Characterization of molecularly cloned simian-human immunodeficiency viruses causing rapid CD4+ lymphocyte depletion in rhesus monkeys. J Virol. 1997;71:4218–4225. doi: 10.1128/jvi.71.6.4218-4225.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim F M, Kolson D L, Balliet J W, Srinivasan A, Collman R G. V3-independent determinants of macrophage tropism in a primary human immunodeficiency virus type 1 isolate. J Virol. 1995;69:1755–1761. doi: 10.1128/jvi.69.3.1755-1761.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koito A, Harrowe G, Levy J A, Cheng-Mayer C. Functional role of the V1/V2 region of human immunodeficiency virus type 1 envelope glycoprotein gp120 in infection of primary macrophages and soluble CD4 neutralization. J Virol. 1994;68:2253–2259. doi: 10.1128/jvi.68.4.2253-2259.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwong P D, Wyatt R, Robinson J, Sweet R W, Sodroski J, Hendrickson W A. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Brien W A, Koyanagi Y, Namazie A, Zhao J-Q, Diagne A, Idler K, Zack J A, Chen I S Y. HIV-1 tropism for mononuclear phagocytes can be determined by regions of gp120 outside the CD4-binding domain. Nature. 1990;348:69–73. doi: 10.1038/348069a0. [DOI] [PubMed] [Google Scholar]
- 21.Ohagen A, Li L, Rosenzweig A, Gabuzda D. Cell-dependent mechanisms restrict the HIV type 1 coreceptor activity of US28, a chemokine receptor homolog encoded by human cytomegalovirus. AIDS Res Hum Retrovir. 2000;16:27–35. doi: 10.1089/088922200309575. [DOI] [PubMed] [Google Scholar]
- 22.Picchio G R, Gulizia R J, Wehrly K, Chesebro B, Mosier D E. The cell tropism of human immunodeficiency virus type 1 determines the kinetics of plasma viremia in SCID mice reconstituted with human peripheral blood leukocytes. J Virol. 1998;72:2002–2009. doi: 10.1128/jvi.72.3.2002-2009.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rizzuto C D, Wyatt R, Hernández-Ramos N, Sun Y, Kwong P D, Hendrickson W A, Sodroski J. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science. 1998;280:1949–1953. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- 24.Rucker J, Edinger A L, Sharron M, Samson M, Lee B, Berson J F, Yi Y, Margulies B, Collman R G, Doranz B J, Parmentier M, Doms R W. Utilization of chemokine receptors, orphan receptors, and herpesvirus-encoded receptors by diverse human and simian immunodeficiency viruses. J Virol. 1997;71:8999–9007. doi: 10.1128/jvi.71.12.8999-9007.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scarlatti G, Tresoldi E, Bjorndal A, Fredriksson R, Colognesi C, Deng H K, Malnati M S, Plebani A, Siccardi A G, Littman D R, Fenyo E M, Lusso P. In vivo evolution of HIV-1 co-receptor usage and sensitivity to chemokine-mediated suppression. Nat Med. 1997;3:1259–1265. doi: 10.1038/nm1197-1259. [DOI] [PubMed] [Google Scholar]
- 26.Schuitemaker H, Kootstra N A, de Goede R E Y, de Wolf F, Miedema F, Tersmette M. Monocytotropic human immunodeficiency virus type 1 (HIV-1) variants detectable in all stages of HIV-1 infection lack T-cell line tropism and syncytium-inducing ability in primary T-cell culture. J Virol. 1991;65:356–363. doi: 10.1128/jvi.65.1.356-363.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shankarappa R, Gupta P, Learn G H, Jr, Rodrigo A G, Rinaldo C R, Jr, Gorry M C, Mullins J I, Nara P L, Ehrlich G D. Evolution of human immunodeficiency virus type 1 envelope sequences in infected individuals with differing disease progression profiles. Virology. 1998;241:251–259. doi: 10.1006/viro.1997.8996. [DOI] [PubMed] [Google Scholar]
- 28.Simmons G, Wilkinson D, Reeves J D, Dittmar M T, Beddows S, Weber J, Carnegie G, Desselberger U, Gray P W, Weiss R A, Clapham P R. Primary, syncytium-inducing human immunodeficiency virus type 1 isolates are dual-tropic and most can use either Lestr or CCR5 as coreceptors for virus entry. J Virol. 1996;70:8355–8360. doi: 10.1128/jvi.70.12.8355-8360.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singh A, Besson G, Mobasher A, Collman R G. Patterns of chemokine receptor fusion cofactor utilization by human immunodeficiency virus type 1 variants from the lungs and blood. J Virol. 1999;73:6680–6690. doi: 10.1128/jvi.73.8.6680-6690.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smyth R J, Yi Y, Singh A, Collman R G. Determinants of entry cofactor utilization and tropism in a dualtropic human immunodeficiency virus type 1 primary isolate. J Virol. 1998;72:4478–4484. doi: 10.1128/jvi.72.5.4478-4484.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Speck R F, Wehrly K, Platt E J, Atchison R E, Charo I F, Kabat D, Chesebro B, Goldsmith M A. Selective employment of chemokine receptors as human immunodeficiency virus type 1 coreceptors determined by individual amino acids within the envelope V3 loop. J Virol. 1997;71:7136–7139. doi: 10.1128/jvi.71.9.7136-7139.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tersmette M, Gruters R A, de Wolf F, de Goede R E Y, Lange J M A, Schellekens P T A, Goudsmit J, Huisman H G, Miedema F. Evidence for a role of virulent human immunodeficiency virus (HIV) variants in the pathogenesis of acquired immunodeficiency syndrome: studies on sequential HIV isolates. J Virol. 1989;63:2118–2125. doi: 10.1128/jvi.63.5.2118-2125.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tscherning C, Alaeus A, Fredriksson R, Björndal Å, Deng H K, Littman D R, Fenyö E M, Albert J. Differences in chemokine coreceptor usage between genetic subtypes of HIV-1. Virology. 1998;241:181–188. doi: 10.1006/viro.1997.8980. [DOI] [PubMed] [Google Scholar]
- 34.Valentin A, Albert J, Fenyo E M, Asjo B. Dual tropism for macrophages and lymphocytes is a common feature of primary human immunodeficiency virus type 1 and 2 isolates. J Virol. 1994;68:6684–6689. doi: 10.1128/jvi.68.10.6684-6689.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang W K, Dudek T, Essex M, Lee T H. Hypervariable region 3 residues of HIV type 1 gp120 involved in CCR5 coreceptor utilization: therapeutic and prophylactic implications. Proc Natl Acad Sci USA. 1999;96:4558–4562. doi: 10.1073/pnas.96.8.4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhong P, Peeters M, Janssens W, Fransen K, Heyndrickx L, Vanham G, Willems B, Piot P, Van der Groen G. Correlation between genetic and biological properties of biologically cloned HIV type 1 viruses representing subtypes A, B, and D. AIDS Res Hum Retrovir. 1995;11:239–248. doi: 10.1089/aid.1995.11.239. [DOI] [PubMed] [Google Scholar]
- 37.Zhu T, Mo H, Wang N, Nam D S, Cao Y, Koup R A, Ho D D. Genotypic and phenotypic characterization of HIV-1 in patients with primary infection. Science. 1993;261:1179–1181. doi: 10.1126/science.8356453. [DOI] [PubMed] [Google Scholar]