Abstract
Microbial tolerance to toxic compounds formed during biomass pretreatment is a significant challenge to produce bio-based products from lignocellulose cost effectively. Rational engineering can be problematic due to insufficient prerequisite knowledge of tolerance mechanisms. Therefore, adaptive laboratory evolution was applied to obtain 20 tolerant lineages of Bacillus subtilis strains able to utilize Distiller's Dried Grains with Solubles-derived (DDGS) hydrolysate. Evolved strains showed both improved growth performance and retained heterologous enzyme production using 100% hydrolysate-based medium, whereas growth of the starting strains was essentially absent. Whole-genome resequencing revealed that evolved isolates acquired mutations in the global regulator codY in 15 of the 19 sequenced isolates. Furthermore, mutations in genes related to oxidative stress (katA, perR) and flagella function appeared in both tolerance and control evolution experiments without toxic compounds. Overall, tolerance adaptive laboratory evolution yielded strains able to utilize DDGS-hydrolysate to produce enzymes and hence proved to be a valuable tool for the valorization of lignocellulose.
Keywords: Adaptive laboratory evolution, Lignocellulose, Enzyme production, Tolerance, Bacillus subtilis
Graphical abstract
Highlights
-
•
Steam explosion and enzymatic hydrolysis of DDGS yielded hydrolysate-based medium.
-
•
Automated adaptive evolution efficiently created tolerant Bacillus subtilis strains.
-
•
Evolved strains showed improved growth and retained enzyme production.
-
•
Mutations associated to the improved phenotype were identified.
1. Introduction
As governments and organizations around the globe have committed to comply with ambitious climate goals, the production of Dried Distillers Grains with Solubles (DDGS) is expected to rise in the coming years (IEA, 2021; RFA, 2020; Shukla et al., 2022). Being a major by-product of the bioethanol industry, the selling of DDGS as animal feed is of vital importance to the economic viability of the bioethanol industry (Chatzifragkou and Charalampopoulos, 2018). However, as the animal feed market is expected to be saturated, there is a growing need for first generation bioethanol plants to convert DDGS into alternative high value products to diversify their outputs and secure the profitability of their assets (Beri et al., 2020; Chatzifragkou and Charalampopoulos, 2018).
In recent years, there has been an increased focus on the use of DDGS as a potential substrate for microbial fermentation to generate value to the bioethanol production process. The rich nutritional composition of DDGS in terms of carbon, nitrogen, and other micronutrients is expected to be an ideal starting point for the bio-manufacturing of a variety of products, including organic acids, biofuels, and hydrolytic enzymes (Iram et al., 2020).
While DDGS contains a considerable amount of carbohydrates, pretreatment of DDGS is required to fractionate and hydrolyze the lignocellulosic fibers to release the fermentable sugars. A multitude of chemical, physical, and biological pretreatment methods and conditions are described in literature (Iram et al., 2020). Often, biomass pretreatment involves the undesirable formation of lignocellulose-derived by-products, which have negative effects on fermentation and lead to a decrease in overall sugar yield. These inhibitory compounds include, amongst others, furan derivatives, organic acids, and phenolic compounds (van der Pol et al., 2016). Detoxification of lignocellulosic hydrolysate is possible, but leads to increased costs and concomitant loss of sugars.
The development of microbial strains with increased tolerance has the potential to minimize the degree of detoxification required. Unfortunately, tolerance is a complex trait, involving the coordinated action of hundreds of genes (Lastiri-Pancardo and Utrilla, 2017). As biomass hydrolysates contain a plethora of different toxic compounds, rational design is challenging due to the lack of prerequisite knowledge. Even if the exact tolerance mechanism can be rationally engineered, it can be challenging to fine-tune the expression of the genes involved to deliver the desired phenotype (Sandberg et al., 2019).
In contrast, tolerance adaptive laboratory evolution (TALE) enables strain optimization without a priori knowledge about the genetic changes necessary to increase tolerance towards hydrolysate-associated inhibitory compounds. By serial passaging of cells in conditions with increasing selective pressure, one can select cells, which have acquired beneficial mutations for the chosen environment. The recent development of using of automated liquid handler systems allow crucial dynamic control of the applied stress during the experiment to maintain a strong selection pressure, while not crashing the cultures (Sandberg et al., 2019).
In this study, TALE was applied as a tool to generate Bacillus subtilis strains with improved tolerance towards the toxic compounds present in DDGS-hydrolysate. B. subtilis (and other closely related species like Bacillus licheniformis) is recognized as one of the work horses in both industrial biotechnology and academia, due to its robustness in industrial fermentations, well-defined endogenous metabolism, distinct genetic background combined with established and emerging genetic manipulation tools, and GRAS-status (generally recognized as safe) (Liu et al., 2013). However, previous work has shown that B. subtilis is severely inhibited by compounds commonly present in biomass hydrolysate (van der Maas et al., 2021). Although ALE has been used to obtain tolerant strains for species like Saccharomyces cerevisiae, Escherichia coli, Clostridium thermocellum and Corynebacterium glutamicum, this method has scarcely been applied to yield B. subtilis strains tolerant towards hydrolysate-associated inhibitory compounds (Almario et al., 2013; Linville et al., 2013; Qin et al., 2016; Wang et al., 2018).
Prior to the TALE experiment, steam explosion pretreatment and enzymatic hydrolysis of the DDGS biomass were performed to produce the DDGS-hydrolysate. Subsequently, the inhibitory effect of compounds present in the pretreated hydrolysate medium was assessed. The TALE experiments were executed with four independent clonal biological replicates for each of the five starting strains. To select the best performing evolved strains a growth screening and a subsequent protein production screening were performed. Lastly, whole-genome-sequencing data of selected evolved isolates were compared and key mutations related to the improved phenotypes were identified.
2. Material and methods
2.1. DDGS pretreatment and enzymatic hydrolysis
DDGS was kindly provided by United Wisconsin Grain Producers (UWGP, Friesland, WI, USA). At UWGP, a process technology called Maximized Stillage Co-products technology MSC™ is applied. In this setup, part of the protein which is left after the bioethanol fermentation step is separated and sold as a high-value protein product (Jessen, 2015). As a result, the DDGS used in this study contains less protein and more fibers compared to conventional DDGS (21% vs 31% crude protein content) (Belyea et al., 2004). Prior to steam explosion, the DDGS biomass was soaked in 0.5% w w−1 sulfuric acid for 20 min. The DDGS slurry (solid/liquid ratio of 20% w w−1) was filtered through cotton fabric using a pressure press (Fischer HP25-M). Wet solids were pretreated in batches of approximately 2.2 kg in a 10 L steam explosion reactor (190 °C, 5 min incubation, 12.5 bar, Knislinge Mekaniska Verkstad AB designed by Process- & Industriteknik AB, Lund University, Sweden) as described by Palmqvist et al. (1996). Batches of pretreated slurry were pooled and subsequently separated by solid-liquid filtration. Wet slurry was stored at −20 °C prior to being subjected to enzymatic hydrolysis.
The pretreated DDGS biomass was diluted to 25% solid content (w V−1) by adding 0.1 M sodium citrate buffer (final pH 5.1). The enzymatic hydrolysis was performed using an enzyme loading of 25 FPU g−1 cellulose (Optimash® F200, DuPont Nutrition and Bioscience). Suspended solutions were incubated in glass bottles (VWR) for 48 h, placed on a horizontal Bottle Roller system housed in an incubator (Thermo ScientificTM, 80 rpm, 50 °C). After the enzymatic hydrolysis, the sugar-rich liquid was filter-sterilized, and used as a carbon and nitrogen source in subsequent experiments. Glucose, xylose, arabinose, acetic acid, 5-HMF, and furfural concentrations were quantified using a HPLC system (RI, Dionex Ultimate 3000, Germany) equipped with an Aminex HPX-87H column (300 × 7.8 mm, Bio-Rad, USA) eluted with 0.005 M H2SO4 at 50 °C for 50 min with a flow rate of 0.6 mL min−1. Total phenolic compounds were determined by a colorimetric method using a Folin-Ciocalteu reagent (Singleton and Rossi, 1965).
2.2. Plasmid and strain construction
2.2.1. Strain overview
The TALE experiments were performed using three B. subtilis strains, B. subtilis 168 (DSMZ 402), PY79 KO7, and PY79 KO7s (Table 1) (“Bacillus Genetic Stock Center”, Zeigler et al., 2008). Both PY79 derivatives, hold seven deletions (ΔnprE, ΔaprE, Δepr, Δmpr, ΔnprB, Δvpr, Δbpr) to prevent produced proteins from being degraded by native extracellular proteases (Zeigler, 2016). Additionally, the sporulation gene sigF has been deleted in PY79 KO7s. For each of the three background strains, a xylose-consuming variant was constructed to increase the utilization of C5 sugars present in the DDGS hydrolysate. In these variants, three alterations were made. First, the heterologous xylose isomerase gene xylA and the xylulokinase gene xylB from E. coli, and the homologous xylose transport gene araE, were co-overexpressed by using a replicative plasmid (pHT315:pSCG7-SG46-araE:p43-xylA-xylB (Arantes and Lereclus, 1991)). Second, the genomic native promoter and Shine Dalgarno-sequence of the araE gene were exchanged with a promoter and Shine Dalgarno-sequence associated with high expression (Guiziou et al., 2016). Third, the gene encoding for the transcriptional repressor protein of araE, called araR, was deleted. Hence, six strains were made available, of which five were selected for the TALE experiment, being the B. subtilis 168 wild type (BS168), the xylose-consuming variant of BS168 (BS168 xyl), the PY79 KO7 wild type (KO7), the PY79 KO7s wild type (KO7s), and the xylose-consuming variant of KO7s (KO7s xyl).
Table 1.
Overview of constructed strains and plasmids used in this study.
| Strain | Genotype | Source |
|---|---|---|
| E. coli DH5α | φ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 56 hsdR17 (rk− mk+) supE44 thi-1 gyrA relA1 | Lab collection |
| B. subtilis 168 WT | trpC2 | DSM23778 |
| BS168 | B. subtilis 168 WT, pHT315 | This study |
| BS168 xyl | BS168::ΔaraR::PSGC14-SG46-araE, pJD18 | This study |
| BS168 amyQ | BS168, glms::P3P-amyQ::cm | This study |
| BS168 xyl amyQ | BS168 xyl, glms::P3P-amyQ::cm | This study |
| B. subtilis KO7 WT | ΔnprE, ΔaprE, Δepr, Δmpr, ΔnprB, Δvpr, Δbpr | Zeigler (2016) |
| KO7 | B. subtilis KO7 WT, pJD13 | This study |
| KO7 xyl | KO7::ΔaraR:: PSGC14-SG46-araE, pJD18 | This study |
| KO7 amyQ | KO7, glms::P3P-amyQ::cm | This study |
| KO7 xyl amyQ | KO7 xyl, glms::P3P-amyQ::cm | This study |
| B. subtilis KO7s | ΔnprE, ΔaprE, Δepr, Δmpr, ΔnprB, Δvpr, Δbpr, ΔsigF | Zeigler (2016) |
| KO7 | B. subtilis KO7s, pJD13 | This study |
| KO7s xyl | KO7s::ΔaraR:: PSGC14-SG46-araE, pJD18 | This study |
| KO7s amyQ | KO7s, glms::P3P-amyQ::cm | This study |
| KO7s xyl amyQ |
KO7s xyl, glms::P3P-amyQ::cm |
This study |
|
Plasmid |
Description |
Source |
| pJOE8999 | Plasmid containing Pmanp-cas9, pUC ori (E. coli), pE194ts ori (B. subtilis) and kanamycin resistance marker | (J, 2016) |
| pJD2 | pJOE8999 derivative containing araR homology regions | This study |
| pJD5 | pJOE8999- derivative containing ParaE homology regions | This study |
| for indel PSGC14-SG46 | Guiziou et al. (2016) | |
| pHT315 | Replicative plasmid containing pHT315 ori, ColE1 ori, ampicillin resistance, erythromycin resistance marker | Cavin et al. (1998) |
| pJD18 | pHT315 derivative containing PSCG7-SG46-araE and P43-xylA-xylB | This study |
| pProUSER13C1B | ProUSER2.0 plasmid containing P3P, glmS homology regions, and chloramphenicol resistance marker | Falkenberg et al. (2021) |
| pJD21 | pProUSER13C1B derivative for integrating P3P-amyQ downstream of glmS | This study |
During strain construction, bacteria were routinely grown in LB media at 37 °C at 250 rpm or on LB agar plates at 37 °C. When required, antibiotics were supplemented to the media (100 μg mL−1 ampicillin (Amp) for E. coli strains, and 5 μg mL−1 chloramphenicol (Cam), 5 μg mL−1 kanamycin (Kan), 10 μg mL−1 erythromycin (Ery), and 100 μg mL−1 spectinomycin (Spc) for B. subtilis strains).
2.2.2. User cloning and ProUSER vectors
The pJOE8999 and pHT315 plasmids were made by using the USER cloning system (Nour-Eldin et al., 2006). All DNA fragments, pJOE8999 and pHT315 backbones were amplified by PCR using Phusion U Hot Start DNA Polymerase, which is able to amplify DNA fragments containing uracil. The ProUSER2.0 plasmids were digested by AsiSI and Nt.BbvCI restriction enzymes (New England BioLabs, United States) as described in (Falkenberg et al., 2021). The specific digestion of the ProUSER2.0 plasmid by AsiSI and Nt.BbvCI, creates 6 and 8 bp single-stranded DNA overhangs, which enabled USER cloning using the nicked backbones. DNA fragments and plasmid backbones were gel purified (NucleoSpin PCR clean-up gel extraction kit, Macherey-Nagel) and used for USER cloning as previously described (Falkenberg et al., 2021).
2.2.3. Transformation
E. coli transformations were performed using cloned vectors as described in (Inoue et al., 1990) to yield the required plasmid material. B. subtilis strains (BS168, KO7, and KO7s) were either transformed by natural or induced competence. The use of natural competence was carried out as described previously (Vojcic et al., 2012), with the exception that histidine was not added to the SM1, and SM2 media and cells were recovered for at least 2 h prior to exposing them to antibiotics. To induce competence, a mannitol-inducible comKS-cassette was inserted downstream of the glms locus of B. subtilis (unpublished material), and transformation was carried out as previously described (Rahmer et al., 2015).
2.3. Start strain construction
Deletions of the ΔaraR gene and substitutions of the promoter region (PSGC14-SG46-araE) were made using a shuttle vector (pJOE8999) (Altenbuchner, 2016). The vector carries a mannose-inducible cas9 gene system, capable of introducing a double-strand break at its target site. Repair by homologous recombination, using an engineered template containing both upstream and downstream flanking regions of the target site can be used to delete or substitute a desired DNA sequence. As such, both upstream and downstream flanking regions of the araR gene and the promoter region of the native araE gene were amplified using compatible USER primers and cloned into an amplified pJOE8999 backbone by using USER cloning as described above (pJD2, pJD5, Table 1). The replicative plasmid pJD18 was constructed to co-overexpress xylA, xylB, and araE, and subsequently transformed into the xylose-consuming variants (Table 1).
Post evolution, a mannitol-inducible comKS-cassette was inserted downstream of the glmS locus in both the starting strains and selected evolved isolates by transformation of pSIJ1005 (unpublished material). Subsequently, an amylase expression cassette (amyQ, B. amyloliquefaciens) was integrated into the same glmS region by transformation of pJD21, replacing the comKS-cassette. Strong expression of amyQ was expected when integrating the strong constitutive promoter P3P into the glmS region (Table 1) (Falkenberg et al., 2021). The genomic deletions/substitutions and plasmids sequences were verified using a Mix2Seq Sanger sequencing kit (Eurofins Genomics, Luxembourg).
2.4. TALE and ALE experiments
During the TALE experiment, a modified M9-based medium (called M9extra) was supplemented with increasing amounts of DDGS hydrolysate. The M9extra medium contained the following: monosaccharides as carbon source (approximately the same as DDGS-hydrolysate: 30 g L−1 glucose, 10 g L−1 xylose, 5 g L−1 arabinose), M9-salts (12.8 g L−1 Na2HPO4 ·7H2O, 3 g L−1 KH2PO4, 0.5 g L−1 NaCl, 1 g L−1 NH4Cl, 2 mM MgSO4, 0.1 mM CaCl2), supplemented with 50 μM FeCl3, a trace element solution (12.5 μM MnCl2·4H2O, 2.1 μM CoCl2 ·6H2O, 8.5 μM ZnSO4 ·7H2O, 0.6 μM CuCl2 ·2H2O, 0.8 μM H3 Bo3, 1.05 μM NiCl2·6H2O, 1.25 μM NaMoO4 ·2H2O), and 50 mg L−1 L-tryptophan. In addition, media were supplemented with antibiotics (10 μg mL−1 erythromycin). The added hydrolysate was supplemented with the same concentration of the components present in the M9 extra medium (M9-salts, FeCl3, trace element solution, tryptophan, antibiotics) to obtain a constant and targeted selection pressure throughout the TALE.
The TALE experiments were started with four independent clonal biological replicates for each of the five starting strains (TALE#1-TALE#20) Table S1). In parallel, 20 independent bacterial cultures were passaged during the late exponential growth phase for an average of 330 generations using an automated liquid handler platform (15 mL, 37 °C, 1200 rpm magnetic stirrer) as described previously (Mohamed et al., 2017). OD600nm measurements were performed periodically to determine growth rates and time of passaging of all ALEs (Tecan Sunrise plate reader). Each ALE flask, i.e. batch, was passed into a new flask with fresh medium around an OD600 of 0.9 (equal to 3.86 for a benchtop spectrophotometer with a 1 cm light path) at the late exponential phase. Once a set growth rate was reached, DDGS hydrolysate supplementation to the M9extra media was increased by steps of 5% (v v−1). Periodically, aliquots of samples were frozen in 25% glycerol solution and stored at −80 °C for analysis. At the end of the evolution, strains were able to grow reproducibly at 95–100% hydrolysate supplemented media.
In addition, a control ALE was performed with four independent clonal biological replicates for BS168 xyl and KO7s xyl grown in M9 extra medium (ALE#21 – ALE#28, Table S1), to identify mutations related to media- or cultivation-specific adaptation (LaCroix et al., 2015).
2.5. Characterization of starting strains
Starting strains were streaked on LB agar plates (supplemented with 10 μg mL−1 erythromycin) and grown overnight (16h, 37 °C). Subsequently, one single colony of the start strain was inoculated in 800 μL of LB medium (supplemented with 10 μg mL−1 erythromycin) and grown for 9 h into 96 microtiter deep-well plates. The growth screening experiments were started by inoculating 5 μL of the pre-culture in 295 μL of a modified M9-based medium (called M9extra) supplemented with increasing amounts of DDGS hydrolysate.
2.6. Growth screening experiment
Starting strains and evolved populations were streaked on LB agar plates (supplemented with 10 μg mL−1 erythromycin), and grown overnight (16h, 37 °C). Subsequently, one single colony of the start strain and 21 single colonies of the 20 different evolved populations were inoculated in 800 μL of LB medium (supplemented with 10 μg mL−1 erythromycin) and grown for 9 h in 96 microtiter deep-well plates. Next, 5 μL of the cultures were transferred in 800 μL of 50% (v v−1) M9-extra/hydrolysate and grown overnight (16h, 37 °C) in 96 plates to increase the selection pressure during the pre-culture progressively. The growth screening experiments were started by inoculating 5 μL of the overnight pre-culture in 295 μL of 100% hydrolysate-based medium. Growth performance was assessed qualitatively by incubation in a growth profiler 960 for 24 h (250 rpm, 37 °C, CR1496dg, EnzyScreen BV, Leiden, The Netherlands). The growth profiler system extracts red pixel values from images during the growth experiment, which are used as a proxy for relative growth in this study.
2.7. Amylase production screening experiment
Sixty of the selected clones were transformed, representing all TALE experiments except for one replicate (TALE #4).Transformed clones were inoculated from frozen glycerol stocks on LB agar plates (supplemented with 10 μg mL−1 erythromycin) and grown overnight (16h, 37 °C). Cell pre-cultures were prepared as described in section 2.4. For screening experiments, cultures were started by inoculation of 5 μL of overnight pre-culture in 195 μL of 100% hydrolysate-based medium. Cells were incubated in 96 well microtiter plates, using a microplate spectrophotometer (BioTek ELx808). After 24 h, cultures were centrifuged at 6000 g for 5 min at 4 °C, the pellet was discarded, and the supernatant was used for further analysis.
Additionally, amylase production experiments were performed in Axygen 24-well deep well plate (Corning Life Science, Corning, New York, USA) for both hydrolysate-based medium and a rich synthetic media, called Cal18-2. Cal18-2 media (Rasmussen, Bjoernvad, & Diers, 2000) contained the following: (40 g L−1 yeast extract (LP0021B, Thermofisher Scientific), 1.3 g L−1 MgSO4 ·7H2O, 50 g L−1 maltodextrin (DE 13–17 (Sigma-Aldrich, Saint Louis, MO, USA)), 20 g L−1 NaH2PO4 ·2H2O, 6.7 mL L−1 Na2MoO4 stock solution (2.0 g L−1), 6.7 mL L−1 trace metal solution (consisting of 4.48 g L−1 MnSO4 ·H2O, 3.33 g L−1 FeCl3·6H2O, 0.625 g L−1 CuSO4 ·5H2O, 7.12 g L−1 ZnSO4 ·7H2O), and 100 μL L−1 Pluronic L-61 (Sigma-Aldrich, Saint Louis, MO, USA), adjusted to pH = 6). Strains were streaked on LB agar plates (supplemented with 10 μg mL−1 erythromycin), and grown overnight (16h, 37 °C). Cell pre-cultures were prepared as described above, with the exception that Cal 18-2 pre-cultures were grown in Cal 18-2 medium instead of 50% hydrolysate-based medium. Amylase production experiments started at an initial OD600 of 0.1, and incubated for 24 or 48 h at 20 °C or 37 °C, depending on the experiment. Subsequently, cultures were centrifuged at 6000 g for 5 min at 4 °C, the pellet was discarded, and the supernatant was used for further analysis.
The amylase assay was adapted from Xiao et al. (2006). Culture supernatants were diluted in 100 mM phosphate buffer (pH = 5.9) solution. Subsequently, 10 μL of the diluted supernatant was mixed with 40 μL starch solution (2g L−1 in 100 mM phosphate buffer (pH = 5.9). The samples were incubated at 65 °C for 10 min and stopped by adding 50 μL of 1 M HCl. The reactions were mixed with 50 μL iodine solution consisting of 5 mM I2 and 50 mM KI. The absorbance values were measured at 580 nm.
2.8. Whole-genome Re-sequencing and analysis
Start clones and selected evolved isolates of each TALE and ALE experiment were selected and prepared for whole-genome re-sequencing. Genomic DNA was extracted from overnight cultures (grown in LB medium) using the MasterPure Gram Positive DNA Purification Kit (Lucigen). Quality was assessed by evaluating Abs260nm Abs280nm−1 using a Nanodrop (Thermo Fisher scientific, USA). DNA concentration was measured using a Qubit ds-DNA broad range assay (Thermo Fisher scientific, USA), and paired-end sequencing libraries were generated using the Illumina 300 cycle (150 bp x 2) kit (San Diego, CA, USA). Sequencing was performed on an Illumina NextSeq 500/550 system (Illumina, USA). The average coverage for each sample was over 60. Genome sequencing reads were analyzed using the in-house mutation calling pipeline called “ALE mut pipeline” to generate lists of mutations for each evolved strain (Phaneuf et al., 2019a). The reference strain used for this analysis was B. subtilis 168 with the GenBank accession number NC_000964 and B. subtilis PY79 with the GenBank accession number CP006881.1. Start strain-specific mutations present before evolution were excluded from the analysis.
3. Results and discussion
Industrial-scale manufacturing of bio-based products and energy by microbial cell factories using lignocellulose will play an important role to realize the global sustainability agenda (IEA, 2021; Noorman and Heijnen, 2017; Shukla et al., 2022). However, the use of lignocellulose as a fermentation feedstock is currently hampered by toxic compounds that are released during the biomass pretreatment process. Consequently, the development of microbial strains with increased tolerance towards these toxic compounds is indispensable to realize second-generation bio-manufacturing processes.
3.1. DDGS-based hydrolysate
Steam explosion is one of the most widely studied pretreatment strategies in both lab-scale and different pilot plants, and is considered a cost-effective method near commercialization (Galbe and Wallberg, 2019). As it is an established method, steam explosion pretreatment and a subsequent enzymatic hydrolysis were applied to valorize the DDGS biomass used in this study. The obtained DDGS-based hydrolysate contained sugars, protein, and inhibitory compounds, including furan derivatives, organic acid, and phenolic compounds (Table 2). While the presence of both sugars and protein makes DDGS an appealing starting point for microbial fermentation, the formation of toxic by-products poses a well-known hurdle for B. subtilis that needs to be overcome (Iram et al., 2020; van der Maas et al., 2021).
Table 2.
Composition of hydrolysate-based medium.
| Compound | Concentration (g L−1) |
|---|---|
| Glucose | 29.1 ± 1.83a |
| Xylose | 9.9 ± 0.67a |
| Arabinose | 5.1 ± 0.33a |
| Protein | 10.5 ± 0.56** |
| 5-HMF | 0.3 ± 0.01** |
| Furfural | 0.9 ± 0.10** |
| Acetic acid | 2.0 ± 0.44** |
| Phenolic compounds | 3.3 ± 0.46** |
Standard deviation of *3 replicates from different hydrolysate batches and **6 technical replicates from the pooled hydrolysate used throughout the study.
3.2. Characterization of starting strains
To confirm the inhibitory effect of DDGS-hydrolysate on the growth performance of B. subtilis, an initial strain characterization was performed using BS168, BS168 xyl, KO7, KO7s and KO7s xyl (Table 1). All starting strains showed decreased growth performance with increasing hydrolysate supplementation (Fig. S1). It is noteworthy that slightly shorter lag phase were observed at 10%–40% hydrolysate supplementation (v v−1) compared to the base medium without hydrolysate for the starting strains, indicating that some nutrients present in the hydrolysate medium might be beneficial to the cells. In this context, the trade-off between the advantage of extra nutrients and disadvantage of harmful toxic compounds present in the hydrolysate vary between the starting strains. Cells have several energy-demanding adaptation mechanisms to tolerate toxic compounds, amongst others, pH homeostasis, maintaining the barrier function of the cell membrane and induction of global cellular stress responses and inhibitor degradation (Ibraheem and Ndimba, 2013). Upon higher supplementation of hydrolysate, it is possible that more energy is diverted from growth to various detoxification mechanisms, leading to a decrease in overall biomass accumulation. A minor increase in red values (pixel values that were used as a proxy for relative cell density) were observed for BS168 grown in 100% hydrolysate after 24 h for unknown reasons. Nonetheless, an overall decrease in growth rate and increase in lag phase showed that none of the initial starting strains could cope with high amounts of hydrolysate supplementation, confirming the inhibitory effect of lignocellulosic hydrolysates and the need to develop strains that are more tolerant.
3.3. Fitness trajectory during TALE and ALE experiments
As tolerance towards inhibitory compounds is challenging to rationally engineer, tolerance adaptive laboratory evolution (TALE) can offer an elegant alternative to obtain tolerant strains without a priori knowledge about the genes responsible for the desired phenotype. Serial passaging of cells in media with increasing amounts of hydrolysate (i.e. selective pressure) allowed for selection of strains that have acquired beneficial mutations. TALE has previously been shown to be an effective tool to obtain strains tolerant towards hydrolysate-associated inhibitory compounds (Almario et al., 2013; Linville et al., 2013; Qin et al., 2016; Wang et al., 2018). In parallel to TALE, it is an informative practice to include adaptive laboratory evolution (ALE) experiments involving a constant cultivation condition to serve as a control. Mutational analysis of both TALE and ALE experiments allows for differentiating between adaptive mutations related to tolerance and those related to basic media components and/or cultivation conditions (Mohamed et al., 2017).
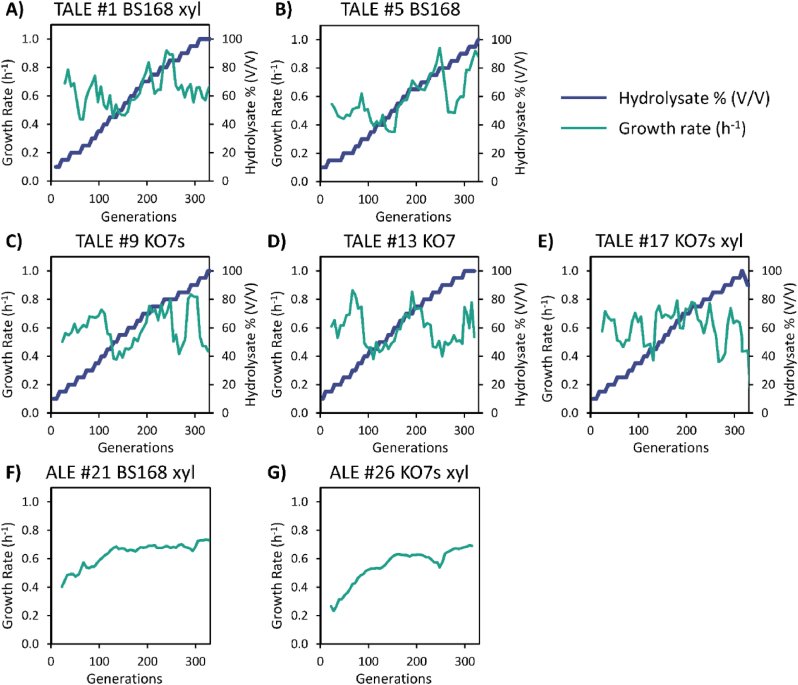
During the TALE experiments, population growth rates fluctuated in response to increasing hydrolysate supplementation (Fig. 1A–E). These fluctuations are observed frequently in TALE experiments and have been previously defined as restorative shifts (Sandberg et al., 2014). Upon exposure to higher amounts of hydrolysate, specific inhibition as well as general stress responses can occur, which can result in reallocating resources from cell growth or inhibiting a growth coupled process directly. Through repeated growth and passage cycles, adaptive mutations arise in the population, which overcome such specific inhibition or reallocate the general stress response more efficiently. These mutations enable the adapted cell to move back towards the pre-inhibited physiological state. Specific tolerance responses have been shown to be more energy efficient than global stress responses (Lastiri-Pancardo and Utrilla, 2017). In contrast, ALE control experiments of the BS168 xyl and KO7s xyl backgrounds displayed a more constant increase in growth rate (Fig. 1F and G). While the initial growth rate was lower for KO7s xyl background compared to the BS168 xyl background (±0.23 h−1 vs ±0.4 h−1), both had similar final growth rates at the end of the experiment (±0.7 h−1). After approximately 330 generations, all TALE experiments successfully yielded strains with increased tolerance to lignocellulosic hydrolysate, being able to grow reproducibly in 95–100% hydrolysate supplemented media.
Fig. 1.
Population growth rates and hydrolysate supplementation concentrations over the course of a set of representative TALE and ALE experiments. Plots for the remaining TALE and ALE replicates are shown in Fig. S2 and Fig. S3.
3.4. Growth screening of evolved and starting strains
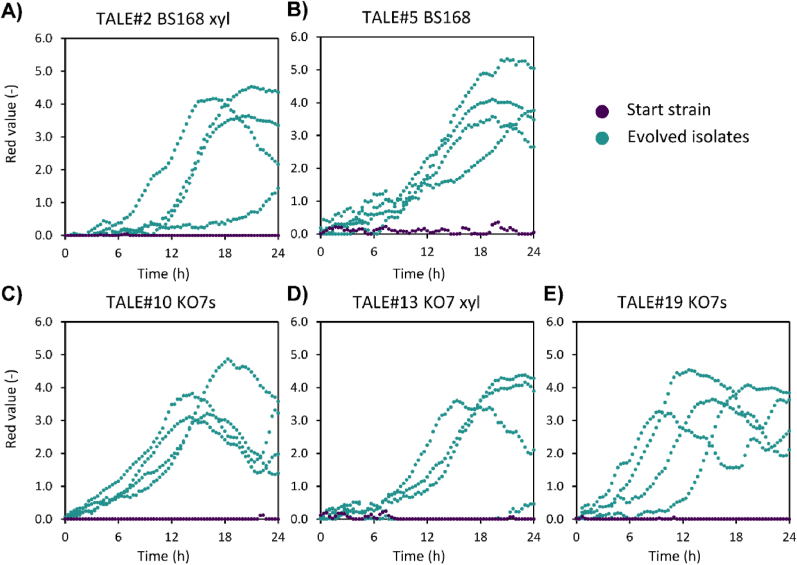
In order to select isolates with increased growth performance, heterogeneous evolved populations were grown on plates and single colonies were picked for an initial screen based on growth in a 100% hydrolysate-based medium. For all 20 independent TALE experiments, growth in 100% hydrolysate could be reproduced for multiple endpoint isolates, while no growth was observed in any of the starting strains (Fig. 2). Growth performance for 21 isolates per TALE experiment was qualitatively assessed and up to four isolates were selected for the amylase production screening.
Fig. 2.
Growth performance of starting strains and four selected isolates per one representative TALE experiment grown in 100% hydrolysate-based medium: A) BS168 xyl, B) BS168, C) KO7s, D) KO7, and E) KO7s xyl backgrounds. Plots for selected isolates from the remaining replicate TALE experiments are shown in Fig. S4.
3.5. Amylase production screening
Although TALE can be used to obtain a phenotype that is able to grow, it does not imply that tolerant evolved strains also have improved capabilities to make the product of interest (Lennen et al., 2019). Protein secretion in B. subtilis involves numerous components, including secretory translocases, chaperones, protein synthesis elements, and proteases, which all could have been affected during the TALE experiments (Zhang et al., 2020).
To confirm that evolved strains are not only able to grow, but also capable of producing protein using second-generation carbon sources, an amylase expression cassette was inserted into 61 evolved isolates, which were previously selected during the growth screening (Section 3.4). Subsequently, protein production was evaluated based on the amylase activity of culture supernatant of both a hydrolysate-based medium and a defined, rich, non-toxic, synthetic medium (Cal18-2) (Rasmussen et al., 2000).
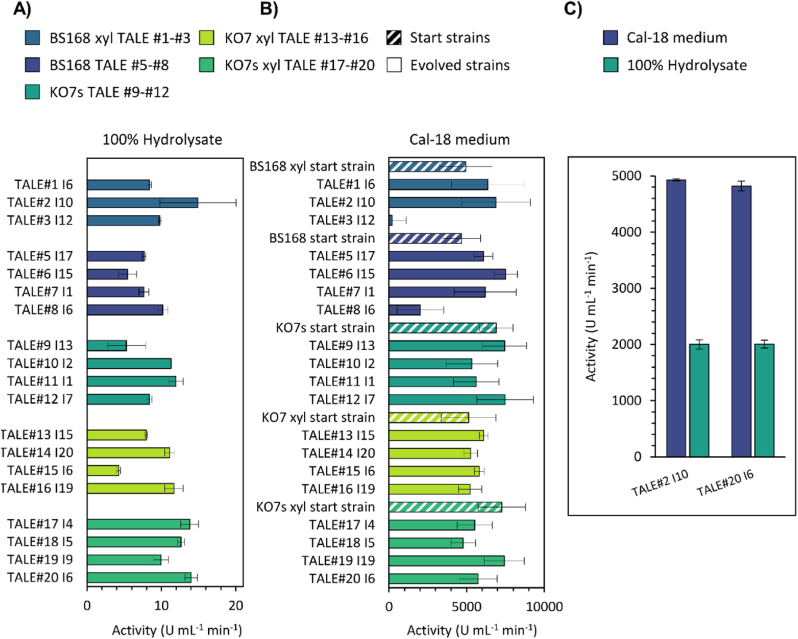
First, evolved isolates were screened for amylase production using a 100% hydrolysate-based medium in 96 well plates (Fig. 3A, Fig. S5). Second, amylase production of both starting strains and selected evolved strains was compared by experiments using Cal18-2 medium in 24 deep well plates (Cal 18-2) (Fig. 3B). Although some evolved strains showed lower amylase activity levels than the starting strains, the production of most of the evolved isolates were either higher or in the same range. Altogether, for most evolved strains, no trade-off between the improved tolerance towards biomass hydrolysate and ability to produce protein was observed (Fig. 3B).
Fig. 3.
Amylase activity of culture supernatant as a measure for enzyme production. A) The amylase production screening was performed in a 96 well plate-format, in which evolved isolates were incubated in 100% hydrolysate-based medium for 24 h. Only the values of the highest producing evolved isolates are shown. B) A verification experiment performed in 24 well plate-format, to determine whether a trade-off exist between tolerance and enzyme production. Starting strains and evolved isolates were incubated in Cal18-2 medium for 48h (B). Comparisons of amylase production for two of the most promising evolved strains in either Cal18-2 medium or the hydrolysate. Cells were incubated in 24 wells for 48h (C). Error bars represent the mean ± s.d of (A) n = two technical duplicates measured during the assay); (B) n = two biological duplicates; (C) n = four biological replicates).
Third, we did a comparative experiment for the most promising evolved strains from each background strain (B. subtilis 168 and B. subtilis PY79 KO7s), and found that production in the hydrolysate was 40% of the production in the industrially relevant Cal18-2 medium, which is a promising result for further developments towards industrial implementation, given the lower sugar and protein content in the hydrolysate used in the experiments.
3.6. Whole-genome Re-sequencing
Subsequent to the screening of the evolved strains, whole-genome re-sequencing is a critical step in the TALE process to decipher the mutational background associated to strains with an improved phenotype in the applied environment. Bioinformatics tools allow comparison of re-sequencing data of both ancestral and multiple independently evolved replicates to determine converged mutations that are likely causal to the improved phenotype (Phaneuf et al., 2019b).
Following this approach, starting strains and selected evolved isolates were send for whole-genome-resequencing (Table S1). During the quality assessment, one isolate was excluded from further analysis due to sequencing issues. For the remaining 19 evolved isolates, the number of acquired mutations ranged from four to thirteen.
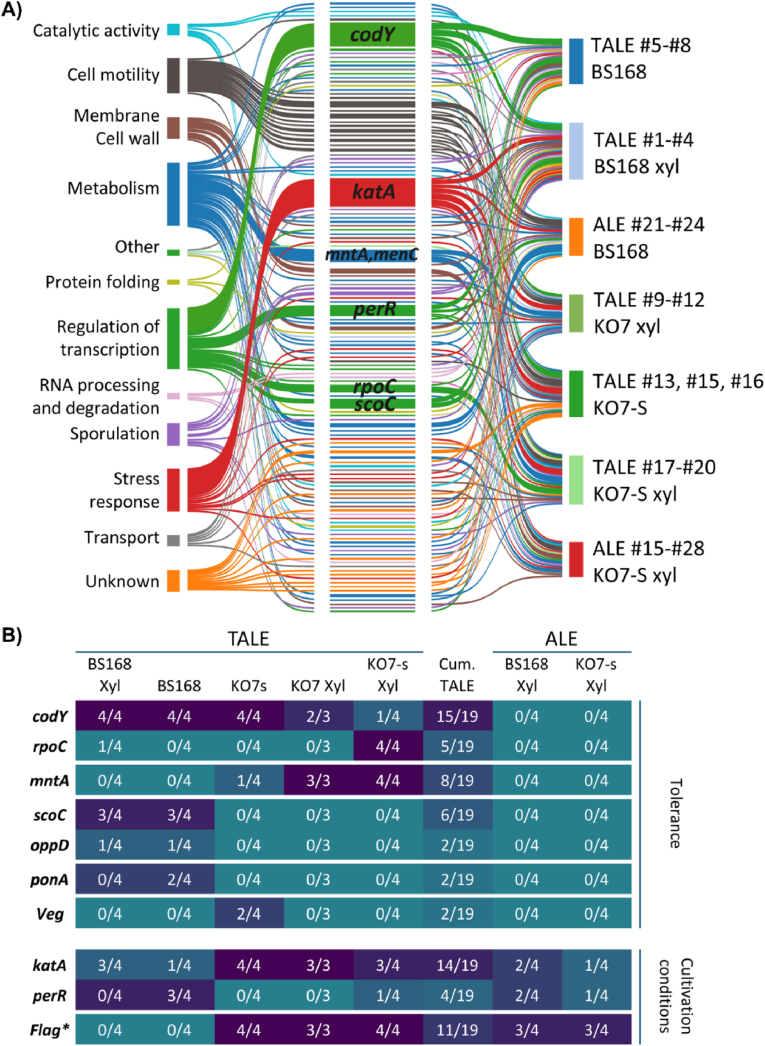
Functional annotations of all mutated genes or genetic regions are summarized based on GO terms in a Sankey diagram (Fig. 4A, (“BSubCyc")). Almost two-third of the mutated genes are involved in stress response mechanisms, metabolism, transcriptional regulation, sporulation, and/or cell motility, highlighting their importance for the improved phenotype. In addition, converged mutations were identified by comparing clonal isolates of 19 TALE experiments and 8 control ALE experiments. Genes or genetic regions that showed a mutation in >2 independent TALE or ALE experiments, in either replicates from the same or different backgrounds, were considered converged mutations and are summarized in Fig. 4B. Shared mutations in genes or genetic regions across the independent replicates strongly suggest that they are adaptive (Sandberg et al., 2019). As such, converged mutations that were acquired exclusively during the TALE experiments are likely to be related to tolerance and included the genes codY, mntA, scoC, rpoC, ponA, veg, and oppD. In contrast, mutations in isolates of both TALE and ALE experiments are likely linked to the cultivation conditions and/or media composition. These included genes associated to protection against oxidative stress (katA, perR), and flagellar structure. Overall, there does not seem to be a clear difference between the converged mutations observed for the BS168/BS168 xyl background strains or KO7x/KO7s xyl background strains. This would suggest that the obtained tolerance is not related to the potential to consume more xylose. A more detailed overview of all observed mutations per isolate is given in Table S3.
Fig. 4.
Mutational analysis of evolved isolates. A) Sankey diagram showing the relation between gene function, gene name, and the strain backgrounds. B) Heat map of converged mutated genes across independent TALE and ALE experiments for different backgrounds. Numbers indicate the number of evolved isolates in which mutations in the given genes or genetic regions were observed. In the cumulative TALE column, the total number of evolved isolates in which mutations were observed for all TALE experiments are shown. *Genes associated with the flagellar structure are pooled, these include fliG (TALE#9), fliR (TALE#9, #10, ALE #28), fliH (TALE #12, ALE #21), fliF (TALE #15, ALE #23), fliL (TALE #13), fliK (TALE #16, ALE #27), fliY, (TALE #17), fliZ (TALE #18, ALE #24), fliP (TALE #19), flhA (ALE #26).
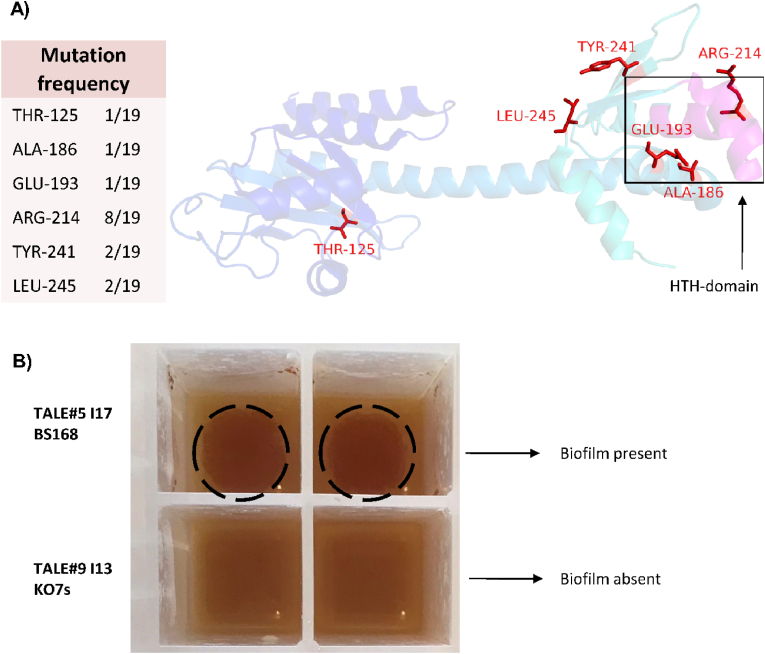
The most frequently mutated gene, specific for only the TALE experiments, was the transcription factor codY. Remarkably, all eight BS168 strains were found to have amino acid substitutions for exactly the same arginine-214 residue (R214X). As R214 is part of the highly conserved DNA-binding domain (202–222), called the HTH-motif, the observed substitutions will likely influence the transcriptional regulation by CodY. Previous studies showed that substitutions of arginine-214 led to a reduced ability of CodY to bind to the target genes. In addition, it has been shown that the context of the HTH-motif of DNA binding domains may influence the recognition and specificity of HTH-mediated protein-DNA interactions (Joseph et al., 2005). Considering the conserved nature of the codY gene, it is possible that the SNPs observed outside of the HTH-domain in the KO7, KO7s, and KO7s xyl backgrounds (T125I, A186T, E193G, 2xY241C, L245P, L245P), could still influence the regulatory function of the protein (Fig. 5A). As the pleiotropic regulator CodY is involved in regulating the expression of several hundred genes (Sonenshein, 2007) (Fig. S6, subtiwiki), it is not straightforward to uncover which gene(s) regulated by CodY that could be responsible for the improved phenotype.
Fig. 5.
A) Structure of the CodY protein (PDB ID:5loe). Mutated amino acids and their side-chains are indicated in red and the HTH DNA-binding domain is colored pink. Structure was visualized using PyMOL (Schrödinger LLC). Numbers in the table indicate the number of evolved isolates carrying mutations in the specific residue. B) Pictures of end isolates TALE#5 I17 and TALE#9 I13 after 24h growth in cal-18 medium. The dark brown patches of biofilm were only present for TALE#5 I17. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
As many independent converged mutations suggest to have a causal relation with the improved tolerance of the evolved isolates, it is interesting to reflect on the replicates which did not share the mutations in the codY gene (TALE#16, TALE#17, TALE#18, TALE#19) and attempt to find mutations which could have a similar functional outcome related to tolerance. The evolved isolates of TALE #17, TALE#18, TALE#19, and TALE#20 all showed the exact similar SNP in the rpoC gene (125,619, C→T) (Fig. 4B, Table S3). The expression of rpoC is regulated by CodY and known to regulate stress response genes, including those of extracytoplasmic function (ECF), σ factors (σM, σW, and σX), and the general stress σ factor (σB) (Lee et al., 2013). Moreover, the end isolate of TALE #16 only showed eight SNPs, of which only five were not shared with other evolved isolates. Intriguingly, this included the HTH-domain global regulator gene cymR. Transcriptome analysis of ΔcymR mutants has shown that many genes related to stress response were altered (Even et al., 2006). Next to codY, TALE-specific converged mutations included mntA, scoC, rpoC, ponA, veg and oppD, which are disucssed in more detail in the supplementary information.
Next, converged mutations that were acquired in both TALE and control ALE experiments were identified. Except for TALE#2, all TALE experiments yielded evolved isolates that were either mutated in the promoter region of the vegetative catalase, katA, or its transcriptional repressor perR. Mutations in the katA promoter were either around the −10/-35 regions, or in the regulatory domain called the Per box. In contrast, mutations in the perR gene led to substitutions across the whole length of the gene. In addition, six out of the eight evolved isolates derived from control ALE experiments also showed mutations in either the promoter region of katA or perR. This suggests that the observed mutations are linked to the cultivation conditions of the experiment and the M9extra-medium rather than tolerance towards biomass hydrolysates. As the mutations are so dominantly present across the different evolution experiments, a higher expression of the vegetative catalase katA may be advantageous by protecting the cell from stress-derived oxidative damage. Apparently, the increased ability to ward off reactive oxygen species is not only beneficial when cultivating cells in biomass hydrolysate, but also in the M9extra medium.
Eleven different genes encoding for parts of the flagella structure were mutated in both TALE and ALE experiments, and hence are likely to play a role in improving growth in the applied cultivation conditions. Mutations included eight deletions, seven introductions of stop codons, and three substitutions. Although the evolved isolates of the BS168/BS168 xyl backgrounds did acquire mutations in genes of the flagella structure during the ALE control experiment, these were absent when TALE conditions were applied. It is worth mentioning that the absence/presence of flagella-associated mutations had a clear impact on the phenotype and ability to form a biofilm (Fig. 5C). It has been extensively described that both the biosynthesis and the operation of flagella represents a considerable cost for the host and that loss of flagella functions can be generally beneficial in well-mixed cultivations (Lara and Gossett). As such, improved growth has been observed in both evolutionary studies and rationally designed cell factories (Mohamed et al., 2020).
In short, mutations associated with increased tolerance included global regulators (codY, scoC, rpoC, ponA, veg) and more specific genes (transporters mntA and oppD). One of the known key advantages of TALE studies is the discovery of specific SNPs in global regulators, responsible for the tuning of a multitude of gene expression levels that restore robust growth (Sandberg et al., 2019). Although alterations in global regulator genes render it challenging to provide a molecular mechanistic basis for the improved phenotypes, mutations present in many independent experiments (e.g., codY) are highly likely to be causal and can be used as targets for rational strain design. Identifying the mechanistic impact of the identified mutations on gene expression using additional -omics approaches, such as RNA sequencing, would give valuable insights for future metabolic engineering work. Current work can also be expanded to evolutionary studies focusing on individual inhibitory compounds present in biomass hydrolysate to unravel general and specific chemical tolerance mechanisms systematically. Moreover, additional rational design and evolutionary engineering can generate strains even closer to their biological limit. Nevertheless, as industrial implementation requires high concentration of sugars and concomitantly higher amounts of inhibitors, the realization of a cost-effective bioprocess using lignocellulose demands a multidisciplinary approach looking beyond the cell, fine-tuning both pretreatment, detoxification, and fermentation conditions to keep the concentration of inhibitory compounds within the limits of biology.
4. Conclusions
This study demonstrated that TALE could efficiently be applied to obtain B. subtilis strains with increased tolerance to biomass hydrolysate-associated inhibitory compounds. Production experiments showed that evolved isolates did not only show improved growth performance, but were also able to produce protein. Whole-genome resequencing of independently evolved isolates revealed codY is likely related to the improved tolerance, while katA and loss of flagella functions are expected to be associated to the employed cultivation conditions. As B. subtilis is a well-known industrial chassis organism, the obtained results are relevant for the use of lignocellulose for the manufacturing of bio-based products.
CRediT authorship contribution statement
Jasper L.S.P. Driessen: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – original draft. Josefin Johnsen: Data curation, Investigation, Methodology, Writing – review & editing. Ivan Pogrebnyakov: Investigation. Elsayed T.T. Mohamed: Visualization, Writing – review & editing. Solange I. Mussatto: Conceptualization, Funding acquisition. Adam M. Feist: Conceptualization, Methodology, Resources, Writing – review & editing. Sheila I. Jensen: Conceptualization, Funding acquisition, Investigation, Methodology, Project administration, Supervision, Writing – review & editing. Alex T. Nielsen: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was funded by the Novo Nordisk Foundation within the framework of the Fermentation-based Biomanufacturing Initiative (FBM), grant number: NNF17SA0031362. We further acknowledge funding from the Independent Research Foundation Denmark (grant no. 7017-00321B) and Novo Nordisk Foundation (NNF), Denmark (grant number NNF20CC0035580). We would like to thank International Flavors & Fragrances Inc. for kindly providing the Optimash® F200, the United Wisconsin Grain Producers for kindly providing the DDDS biomass, Francesco Reggianini for his support with some of the experimental work, and Christian Roslander and Mats Galbe for their advice and support during the steam explosion pretreatment (Lund University). Graphical abstract was created with BioRender.com.
Handling Editor: Mattheos Koffas
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mec.2023.e00223.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
Data availability
Data will be made available on request.
References
- Almario M.P., Reyes L.H., Kao K.C. Evolutionary engineering of Saccharomyces cerevisiae for enhanced tolerance to hydrolysates of lignocellulosic biomass. Biotechnol. Bioeng. 2013;110:2616–2623. doi: 10.1002/bit.24938. [DOI] [PubMed] [Google Scholar]
- Altenbuchner J. Editing of the Bacillus subtilis genome by the CRISPR-Cas9 system. Appl. Environ. Microbiol. 2016;82:5421–5427. doi: 10.1128/AEM.01453-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arantes O., Lereclus D. Construction of cloning vectors for Bacillus thuringiensis. Gene. 1991;108:115–119. doi: 10.1016/0378-1119(91)90495-W. [DOI] [PubMed] [Google Scholar]
- Beri D., York W.S., Lynd L.R., Peña M.J., Herring C.D. Development of a thermophilic coculture for corn fiber conversion to ethanol. Nat. Commun. 2020:1–11. doi: 10.1038/s41467-020-15704-z. 2020 111 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BSubCyc | Encyclopedia of Bacillus Subtilis Subtilis Genes and Metabolism [WWW Document], n.d. URL https://bsubcyc.org/(accessed 1.September.2022).
- Cavin J.F., Dartois V., Diviès C. Gene cloning, transcriptional analysis, purification, and characterization of phenolic acid decarboxylase from Bacillus subtilis. Appl. Environ. Microbiol. 1998;64:1466–1471. doi: 10.1128/aem.64.4.1466-1471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatzifragkou A., Charalampopoulos D. Sustainable Recovery and Reutilization of Cereal Processing By-Products. 2018. Distiller's dried grains with solubles (DDGS) and intermediate products as starting materials in biorefinery strategies. [DOI] [Google Scholar]
- Even S., Burguière P., Auger S., Soutourina O., Danchin A., Martin-Verstraete I. Global control of cysteine metabolism by CymR in Bacillus subtilis. J. Bacteriol. 2006;188:2184–2197. doi: 10.1128/JB.188.6.2184-2197.2006/SUPPL_FILE/SUPPLEMENTARY_DATA.ZIP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkenberg K.B., Mol V., Sainz De La A., Larrea M., Pogrebnyakov I., Nørholm M.H.H., Nielsen A.T., Jensen S.I. The ProUSER2.0 toolbox: genetic parts and highly customizable plasmids for synthetic biology in Bacillus subtilis. ACS Synth. Biol. acssynbio.1c00130. 2021 doi: 10.1021/ACSSYNBIO.1C00130. [DOI] [PubMed] [Google Scholar]
- Galbe M., Wallberg O. Pretreatment for biorefineries: a review of common methods for efficient utilisation of lignocellulosic materials. Biotechnol. Biofuels. 2019:1–26. doi: 10.1186/S13068-019-1634-1. 2019 121 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiziou S., Sauveplane V., Chang H.J., Clerté C., Declerck N., Jules M., Bonnet J. A part toolbox to tune genetic expression in Bacillus subtilis. Nucleic Acids Res. 2016;44:7495–7508. doi: 10.1093/nar/gkw624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibraheem O., Ndimba B.K. Molecular adaptation mechanisms employed by ethanologenic bacteria in response to lignocellulose-derived inhibitory compounds. Int. J. Biol. Sci. 2013;9:598–612. doi: 10.7150/ijbs.6091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IEA . vol. 224. Int. Energy Agency; 2021. (Net Zero by 2050: A Roadmap for the Global Energy Sector). [Google Scholar]
- Inoue H., Nojima H., Okayama H. High efficiency transformation of Escherichia coli with plasmids. Gene. 1990;96:23–28. doi: 10.1016/0378-1119(90)90336-. [DOI] [PubMed] [Google Scholar]
- Iram A., Cekmecelioglu D., Demirci A. Distillers' dried grains with solubles (DDGS) and its potential as fermentation feedstock. Appl. Microbiol. Biotechnol. 2020 doi: 10.1007/s00253-020-10682-0. [DOI] [PubMed] [Google Scholar]
- Joseph P., Ratnayake-Lecamwasam M., Sonenshein A.L. A region of Bacillus subtilis CodY protein required for interaction with DNA. J. Bacteriol. 2005;187:4127–4139. doi: 10.1128/JB.187.12.4127-4139.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCroix R.A., Sandberg T.E., O'Brien E.J., Utrilla J., Ebrahim A., Guzman G.I., Szubin R., Palsson B.O., Feist A.M. Use of adaptive laboratory evolution to discover key mutations enabling rapid growth of Escherichia coli K-12 MG1655 on glucose minimal medium. Appl. Environ. Microbiol. 2015;81:17–30. doi: 10.1128/AEM.02246-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara A., Gossett G. 2020. Minimal Cells: Design, Construction, Biotechnological Applications. n.d. [Google Scholar]
- Lastiri-Pancardo G.M., Utrilla J. Engineering of Microorganisms for the Production of Chemicals and Biofuels from Renewable Resources. Springer International Publishing; 2017. Evolutionary engineering of microorganisms to overcome toxicity during lignocellulose hydrolysates utilization; pp. 181–200. [DOI] [Google Scholar]
- Lee Y.H., Nam K.H., Helmann J.D. A mutation of the RNA polymerase β′ subunit (rpoC) confers cephalosporin resistance in Bacillus subtilis. Antimicrob. Agents Chemother. 2013;57:56–65. doi: 10.1128/AAC.01449-12/ASSET/82833558-DA11-44B6-A858-F8C8C177932F/ASSETS/GRAPHIC/ZAC9991014440008.JPEG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennen R., Jensen K., Mohammed E., Malla S., Börner R., Chekina K., Özdemir E., Bonde I., Koza A., Maury J., Pedersen L., Schöning L., Sonnenschein N., Palsson B., Sommer M., Feist A., Nielsen A., Herrgård M. Adaptive laboratory evolution reveals general and specific chemical tolerance mechanisms and enhances biochemical production. 2019. bioRxiv 634105. [DOI] [PubMed]
- Linville J.L., Rodriguez M., Land M., Syed M.H., Engle N.L., Tschaplinski T.J., Mielenz J.R., Cox C.D. Industrial robustness: understanding the mechanism of tolerance for the populus hydrolysate-tolerant mutant strain of Clostridium thermocellum. PLoS One. 2013;8 doi: 10.1371/journal.pone.0078829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Liu Y., Shin H.D., Chen R.R., Wang N.S., Li J., Du G., Chen J. Developing Bacillus spp. as a cell factory for production of microbial enzymes and industrially important biochemicals in the context of systems and synthetic biology. Appl. Microbiol. Biotechnol. 2013;97:6113–6127. doi: 10.1007/s00253-013-4960-4. [DOI] [PubMed] [Google Scholar]
- Mohamed E.T., Wang S., Lennen R.M., Herrgård M.J., Simmons B.A., Singer S.W., Feist A.M. Generation of a platform strain for ionic liquid tolerance using adaptive laboratory evolution. Microb. Cell Factories. 2017;16:204. doi: 10.1186/s12934-017-0819-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed E.T., Werner A.Z., Salvachúa D., Singer C.A., Szostkiewicz K., Rafael Jiménez-Díaz M., Eng T., Radi M.S., Simmons B.A., Mukhopadhyay A., Herrgård M.J., Singer S.W., Beckham G.T., Feist A.M. Adaptive laboratory evolution of Pseudomonas putida KT2440 improves p-coumaric and ferulic acid catabolism and tolerance. Metab. Eng. Commun. 2020;11 doi: 10.1016/j.mec.2020.e00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noorman H.J., Heijnen J.J. Biochemical engineering's grand adventure. Chem. Eng. Sci. 2017;170:677–693. doi: 10.1016/J.CES.2016.12.065. [DOI] [Google Scholar]
- Nour-Eldin H.H., Hansen B.G., Nørholm M.H.H., Jensen J.K., Halkier B.A. Advancing uracil-excision based cloning towards an ideal technique for cloning PCR fragments. Nucleic Acids Res. 2006;34:e122. doi: 10.1093/NAR/GKL635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmqvist E., Hahn-Hägerdal B., Galbe M., Larsson M., Stenberg K., Szengyel Z., Tengborg C., Zacchi G. Design and operation of a bench-scale process development unit for the production of ethanol from lignocellulosics. Bioresour. Technol. 1996;58:171–179. doi: 10.1016/S0960-8524(96)00096-X. [DOI] [Google Scholar]
- Phaneuf P.V., Gosting D., Palsson B.O., Feist A.M. Aledb 1.0: a database of mutations from adaptive laboratory evolution experimentation. Nucleic Acids Res. 2019;47:D1164–D1171. doi: 10.1093/nar/gky983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phaneuf P.V., Gosting D., Palsson B.O., Feist A.M. Aledb 1.0: a database of mutations from adaptive laboratory evolution experimentation. Nucleic Acids Res. 2019;47:D1164–D1171. doi: 10.1093/nar/gky983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin D., Hu Y., Cheng J., Wang N., Li S., Wang D. An auto-inducible Escherichia coli strain obtained by adaptive laboratory evolution for fatty acid synthesis from ionic liquid-treated bamboo hydrolysate. Bioresour. Technol. 2016;221:375–384. doi: 10.1016/j.biortech.2016.09.024. [DOI] [PubMed] [Google Scholar]
- Rahmer R., Heravi K.M., Altenbuchner J. Construction of a super-competent Bacillus subtilis 168 using the PmtlA-comKS inducible cassette. Front. Microbiol. 2015;6 doi: 10.3389/FMICB.2015.01431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen M.D., Bjoernvad M.E., Diers I. 2000. Pectate Lyase Fusion for Expression and Secretion of Polypeptides. [Google Scholar]
- Rfa . 2020. Annual Industry Outlook 2020. [Google Scholar]
- Sandberg T.E., Pedersen M., Lacroix R.A., Ebrahim A., Bonde M., Herrgard M.J., Palsson B.O., Sommer M., Feist A.M. Evolution of Escherichia coli to 42 °C and subsequent genetic engineering reveals adaptive mechanisms and novel mutations. Mol. Biol. Evol. 2014;31:2647–2662. doi: 10.1093/MOLBEV/MSU209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg T.E., Salazar M.J., Weng L.L., Palsson B.O., Feist A.M. The emergence of adaptive laboratory evolution as an efficient tool for biological discovery and industrial biotechnology. Metab. Eng. 2019;56:1–16. doi: 10.1016/j.ymben.2019.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla P.R., Skea J., Reisinger A., Slade R., Fradera R., Pathak M., Al A., Malek K., Renée Van Diemen B., Hasija A., Lisboa G., Luz S., Malley J., Mccollum D., Some S. Sixth Assess. Rep. Intergov. Panel Clim. Chang. Summ. Policymakers. 2022. Climate change 2022 mitigation of climate change working group III contribution to the sixth assessment report of the intergovernmental panel on climate change summary for policymakers edited by. [Google Scholar]
- Singleton V.L., Rossi J.A. Colorimetry of total phenolics with phosphomolybdic-phosphotungstic acid reagents. Am. J. Enol. Vitic. 1965;16 [Google Scholar]
- Sonenshein A.L. Control of key metabolic intersections in Bacillus subtilis. Nat. Rev. Microbiol. 2007;5:917–927. doi: 10.1038/nrmicro1772. [DOI] [PubMed] [Google Scholar]
- van der Maas L., Driessen J.L.S.P., Mussatto S.I. Effects of inhibitory compounds present in lignocellulosic biomass hydrolysates on the growth of Bacillus subtilis. Energies. 2021;14:8419. doi: 10.3390/EN14248419. [DOI] [Google Scholar]
- van der Pol E.C., Vaessen E., Weusthuis R.A., Eggink G. Identifying inhibitory effects of lignocellulosic by-products on growth of lactic acid producing micro-organisms using a rapid small-scale screening method. Bioresour. Technol. 2016;209:297–304. doi: 10.1016/j.biortech.2016.03.037. [DOI] [PubMed] [Google Scholar]
- Vojcic L., Despotovic D., Martinez R., Maurer K.H., Schwaneberg U. An efficient transformation method for bacillus subtilis DB104. Appl. Microbiol. Biotechnol. 2012;94:487–493. doi: 10.1007/s00253-012-3987-2. [DOI] [PubMed] [Google Scholar]
- Wang X., Khushk I., Xiao Y., Gao Q., Bao J. Tolerance improvement of Corynebacterium glutamicum on lignocellulose derived inhibitors by adaptive evolution. Appl. Microbiol. Biotechnol. 2018;102:377–388. doi: 10.1007/s00253-017-8627-4. [DOI] [PubMed] [Google Scholar]
- Xiao Z., Storms R., Tsang A. A quantitative starch-iodine method for measuring alpha-amylase and glucoamylase activities. Anal. Biochem. 2006;351:146–148. doi: 10.1016/J.AB.2006.01.036. [DOI] [PubMed] [Google Scholar]
- Zeigler D.R. 2016. https://www.researchgate.net/project/Construction-of-exoprotease-free-marker-free-Bacillus-subtilis-hosts (CONSTRUCTION OF EXOPROTEASE-FREE, MARKER-FREE BACILLUS SUBTILIS HOSTS). Daniel R Zeigler | 2 updates | 1 publications | Research Project [WWW Document]. URL. accessed. [Google Scholar]
- Zeigler D.R., Prágai Z., Rodriguez S., Chevreux B., Muffler A., Albert T., Bai R., Wyss M., Perkins J.B. The origins of 168, W23, and other Bacillus subtilis legacy strains. J. Bacteriol. 2008;190:6983–6995. doi: 10.1128/JB.00722-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Su L., Wu J. 2020. (Recent Advances in Recombinant Protein Production by Bacillus Subtilis). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.