Highlights
-
•
In this study, we aimed to reveal the biological significance of hypoxia in promoting tumor progression and tumor cell state transition. Hypoxia is characterized in most solid tumors ascribed to aberrant vascularization and limited blood supply. Specifically, PDAC tumors are often subjected into basal and classical subtypes, according to tumor cell states, which distinguish the biological behavior tendency of PDAC tumor cells. Currently, the formation process of the intertumoral and intratumoral heterogeneity remains unrevealed. Through multiple bioinformatic analysis, we revealed expression levels of hypoxia signature linked to a basal/squamous-like tumor cell state and a more aggressive phenotype in pancreatic adenocarcinoma.
-
•
Through single-cell RNA-seq analysis, we revealed that upregulation of hypoxia signature expression levels was linked to classical-basal-squamous tumor cell state transition in PDAC tumors. Cell-cell ligand-receptor analysis suggested hypoxic PDAC tumor cells derives a secreting signaling network leading to tumor cell dedifferentiation and extra-cellular matrix formation. Through bulk RNA sequencing data analysis, we found overexpression of hypoxia signatures were related to poor prognosis of PDAC. We further revealed overexpression hypoxia signatures were related to TP53 mutation. P63, as a specific biomarker for squamous-like PDAC, were found specifically overexpressed in hypoxic PDACs. In conclusion, our findings revealed the biological role and clinical significance of hypoxia signatures and LDHA in PDAC, laying a foundation for further research on pancreatic malignancy management.
Keywords: Pancreatic adenocarcinoma, Hypoxia, TP53, Keratinization, TP63, LDHA
Abstract
Pancreatic ductal adenocarcinoma (PDAC) is a highly heterogeneous cancer with limited understanding of its classification and tumor microenvironment. Here, by analyzing single-nucleus RNA sequencing of 43, 817 tumor cells from 15 PDAC tumors and non-tumor, we find that hypoxia signatures were heterogeneous across samples and were potential regulators for tumor progression and more aggressive phenotype. Hypoxia-high PDAC tends to present a basal/squamous-like phenotype and has significantly increased outgoing signaling, which enhances tumor cell stemness and promotes metastasis, angiogenesis, and fibroblast differentiation in PDAC. Hypoxia is related to an extracellular matrix enriched microenvironment, and increased possibility of TP53 mutation in PDAC. TP63 is a specific marker of squamous-like phenotype, and presents elevated transcriptome levels in most hypoxia PDAC tumors. In summary, our research highlights the potential linkage of hypoxia, tumor progression and genome alteration in PDAC, leading to further understand of the formation of inter-tumoral and intra-tumoral heterogenous in PDAC. Our study extends the understanding of the diversity and transition of tumor cells in PDAC, which provides insight into future PDAC management.
Introduction
Pancreatic adenocarcinoma (PDAC) is the most common type of pancreatic cancer (PC), known for its high mortality rate, has increasing incidence and mortality worldwide [1]. Due to the heterogeneity and complex stromal microenvironment, patients with pancreatic cancer respond differentially to treatments including chemotherapy and immunotherapy, leading to limited therapeutic effects and poor overall prognosis [2]. However, the processes involved in the formation of the heterogeneity intra-tumor and inter-tumor remain unclear. A major cell type within the PDAC stroma is the cancer-associated fibroblast (CAF) [3]. Previous studies revealed that enrichment of CAF in the microenvironment leads to poor prognosis, and showed potential linkage with basal-like PDAC subtype [4,5].
Two large studies [6,7] reported gene expression subtypes of PDAC, extending the subtypes previously described by Collisson et al. [8]. The squamous samples of Bailey et al. showed distinct overlap with the basal-like samples defined by Moffitt et al., while the Bailey et al. pancreatic progenitor and Collisson et al. classical group largely overlapped the classical samples defined by Moffitt et al., suggesting that PDAC tumors can be consistently classified into a basal-like/squamous group and a classical/progenitor group. immunogenic and aberrantly differentiated exocrine (ADEX) or exocrine-like subtypes showed distinct overlap with the low-purity samples, suggesting that these subtypes may reflect gene expression from non-neoplastic cells.
Basal and progenitor subtypes were suggested to be more adapted to hypoxic environments, as they are associated with increased expression of genes involved in glycolysis and reduced oxidative phosphorylation [8], such as carbonic anhydrase 9 (CA9) and glucose transporter 1 (GLUT1), making them better suited to surviving in hypoxic environments [9].
Hypoxia is characterized in most solid tumors and is ascribed to aberrant vascularization and limited blood supply [10]. Accumulating evidence indicates that hypoxia is a key promoter for adaption and selection of cancer cells to surrounding conditions, thus inducing changes that are suitable for tumor progression [5]. Specifically, PDAC tumors are often subjected to basal and classical subtypes, according to tumor cell states, which distinguish the biological behavior tendency of PDAC tumor cells. Dominant tumor cell subtypes are shown in individual PDAC samples [11], however, recent studies showed continuous and convertible tumor cell states at tumor cell level [12]. Thus, it is hypothesized that the formation of the heterogeneity in PDAC is environment-derived [13,14], which might be a down-stream event of hypoxia.
There is a clear consensus in studies in which tumor genomes have been sequenced and analyzed that hypoxia is associated with tumor genomic instability [15]. Hypoxia has been found to cause DNA damage in the form of replication stress indirectly, and stimulates the production of reactive oxygen species (ROS) in mitochondria, which in turn stabilizes Hypoxia-Inducible Factor 1 alpha (HIF-1α) [16]. P53 protein is a hypoxia regulator, with elevated expression level in hypoxia-exposed conditions, down-regulates the activity of HIF-1α and induces cell apoptosis [17]. Previous studies showed hypoxia selects for cell with mutant TP53 [18].
In the current study, we reported that hypoxia is related to cell state transition, leading to a developed and aggressive phenotype, which may be the major event in the formation of the complex eco-system in PDAC. Hypoxia signatures-enriched PDAC patients are characterized by poor prognosis, most TP53 mutants. Squamous signature TP63 were found elevated in hypoxia PDAC, indicating a potential adeno-squamous transition in PDAC, induced by hypoxia. Constantly, we found that hypoxic PDAC showed distinct overlap with basal/squamous subtype defined by former studies [6], [7], [8], indicating novel hypoxia signatures like LDHA could classify PDAC subtypes accurately. In summary, our study highlights the important role of hypoxia in oncogenesis and tumor progression, extends the understanding of the diversity and transition of tumor cells in PDAC, which provides insight onto future PDAC management.
Results
Gene module defined PDAC tumor cell subgroups in single nucleus sequencing profiles
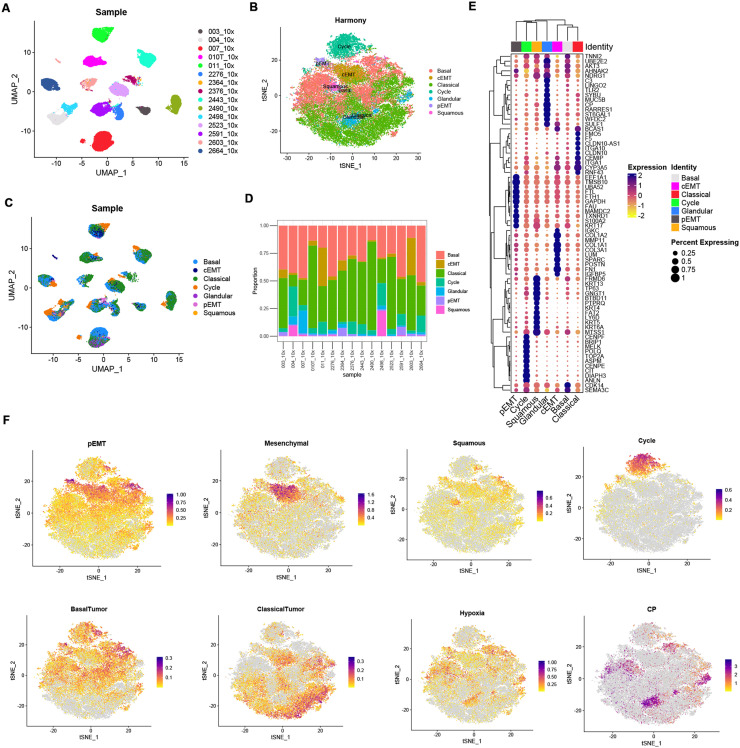
We analyzed a published single-nucleus sequencing dataset to explore the heterogeneity in pancreatic adenocarcinomas at single-cell level [19]. Totally 43, 817 tumor cells from 15 PDAC tumors without neoadjuvant treatments were extracted and re-clustered using Seurat pipeline, Notably, we found that tumor cell features vary from sample sources (Fig. 1A and Supplementary Figure 1). To identify the shared features across patients, we integrated tumor cells from different samples using ‘Harmony’. 7 major tumor cell sub-types with the expression of known marker genes and gene sets were identified including Cycle (TOP2A, ANLN), Basal (LAMC2, VIM), Classical (CLDN10, ITGA10), pEMT (S100A2, KRT17), cEMT (COL3A1, IGFBP5), Squamous (LY6D, KRT13) and Glandular (CP, MUC5B) (Fig. 1B and Supplementary Table 2). Proportion of tumor cell subtype components of each sample were shown in Fig. 1C and Fig. 1D. Consistent with previous studies in other tumors, tumor cells showed distinct intertumoral heterogeneity featured with patient-specific clusters [20,21]. Expression levels of specific tumor cell markers were shown in Fig. 1E. Module scores of individual cells were shown in feature plots in Fig. 1F.
Fig. 1.
Single-cell landscape of 15 pancreatic adenocarcinoma samples without pre-resection treatment. (A) PDAC cell states various from patients. (B) Integrating and reclustering of PDAC cells suggested 7 major clusters. (C) UMAP plot showing tumor cell components, grouped by sample. (D) Stacked bar plot showing tumor cell components, grouped by sample. (E) Expression of marker genes of each major cluster, showed by dot plot. (F) Expression of marker gene modules and genes showed by feature plot.
Gene module signatures express differentially in PDAC and adjacent normal tissues
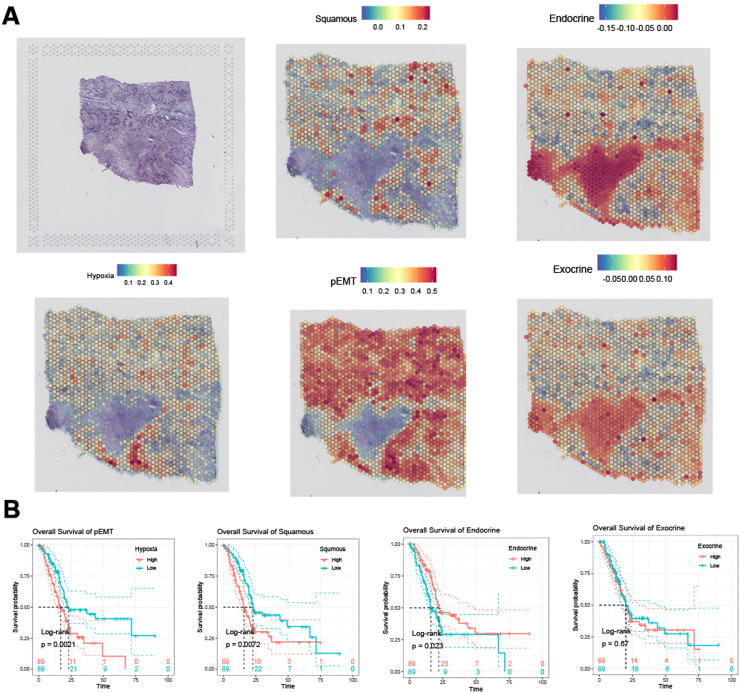
Due to the auto-digestive feature of pancreatic normal tissue, RNA sequencing on normal pancreas tissue is limited [22]. To define the relative expression of gene module signatures in PDAC and adjacent normal tissues, we analyzed a spatial transcriptome profile obtained from a pan-cancer study. ssGSEA were applied to score the gene module expression, which has been proven to be robust for spatial transcriptome data. Hypoxia, squamous, pEMT gene modules were higher expressed in PDAC tumor tissues, while endocrine and exocrine gene modules were higher expressed in adjacent normal tissues (Fig. 2A). We then evaluated the association between gene modules and PDAC survival outcome. TCGA-PAAD patients were divided into two groups according to median ssGSEA score of specific gene modules, respectively. Briefly, pEMT and squamous gene modules were found significantly related to shorter overall survival interval, while endocrine gene module showed significant relationship with longer overall survival interval, and exocrine gene module showed no significant relationship with overall survival, according to log-rank test, threshold p<0.05 (Fig. 2B).
Fig. 2.
(A) Spatial expression pattern of gene modules defining PDAC cell states. (B) Impact of defined gene modules on overall survival of TCGA-PAAD patients.
PDAC samples consist of tumor cells at different status of differentiation
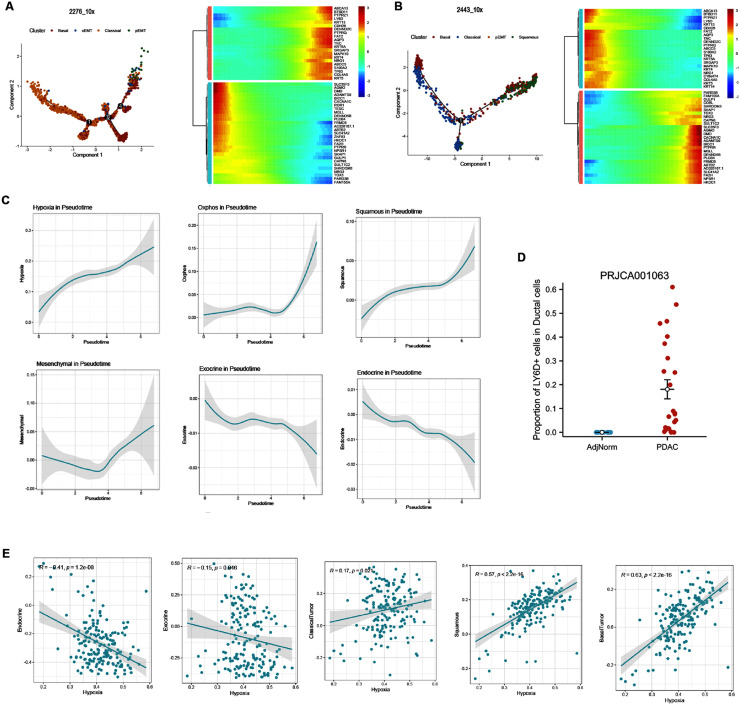
To reveal the differentiation process in PDAC, we explored the gene expression pattern along tumor cell state transition using pseudotime trajectory analysis. As shown in Fig. 1, sample 2276_10x and sample 2443_10x showed distinct basal/squamous-like features. Tumor cells from sample 2276_10x and sample 2443_10x was mainly composed of cell states including classical, basal, pEMT and cEMT, while sample 2443_10x was mainly composed with cell states including classical, basal, pEMT and squamous. We found classical tumor cells mainly located at the origin of this trajectory, while pEMT, cEMT and squamous tumor cells mainly located at the terminal (Fig. 3A and Fig. 3B). Genes involved in the normal pancreatic secretion were decreased gradually, while malignant signatures including S100A2, TP63, KRT6A increased along the trajectory (Fig. 3A and Fig. 3B). Also, we analyzed the dynamic alteration of gene patterns along this trajectory. Notably, gene patterns associated to hypoxia and squamous patterns increased constantly, contrasted to the sequential decrease in endocrine and exocrine gene patterns (Fig. 3C). These results revealed that the process of PDAC cell state transition was dynamic and continuous, with coexistence of several states exhibited by individual cells. For example, features of the squamous epithelium including TP63, LY6D and KRT6A were found in sample 2276_10x, although this sample did not exhibit the characteristics of pancreatic squamous cell carcinoma (PASC). Also, exocrine and endocrine features were found in tumor cells.
Fig. 3.
Dynamic trajectory of cell state transition in sample 2276_10x (A) and sample 2443_10x (B). (C) Dynamic of cell state gene modules along the trajectory of tumor cell type transition. (D) Single-cell analysis of dataset PRJCA001063 showed LY6D+ squamous-like ductal cells were widely observed in PDAC tissues, without detection in adjacent normal tissues. (E) Hypoxia is related to the transition of PDAC cell states include endocrine, exocrine, classical, basal, pEMT, cEMT, and squamous in TCGA-PAAD cohort.
In another single-cell RNA sequencing profile from 24 PDAC tumor samples and 11 control pancreases without any treatment, we found that 0–60% PDAC ductal cells showed non-zero expression of squamous signature LY6D, while none of normal ductal cells was LY6D positive (Fig. 3D). Thus, we further validated that transcriptome of squamous signatures are widely expressed in PDAC tumor cells.
To further validate the correlation between hypoxia and tumor cell gene expression, we calculated the Pearson correlation between GSVA scores of hypoxia signatures with exocrine, endocrine, classical tumor, basal tumor and squamous tumor signatures in TCGA-PAAD cohort, respectively. Hypoxia genes showed negative correlation with endocrine (r=−0.41, p<0.00001) and exocrine gene signatures (r=−0.15, p<0.05), while showed positive correlation with classical tumor (r = 0.17, p<0.05), basal tumor (r = 0.57, p<0.00001) and squamous tumor (r = 0.63, p<0.00001) signatures, consistent with our findings in single-cell analysis (Fig. 3E).
Enhanced cell-cell secreting interactions were detected among hypoxia-enriched PDAC cells
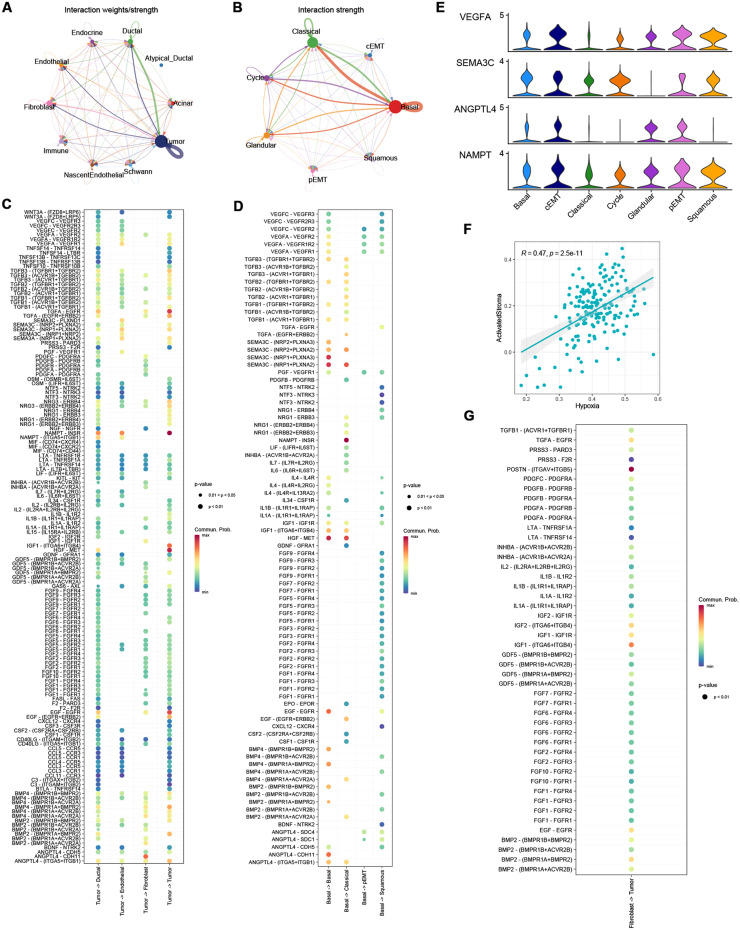
Signaling interactions within PDAC tumor cells were explored using ‘CellChat’ based on well-known ligand-receptor pairs. The strength of signaling patterns indicated that cross-talks among tumor cells were specifically enriched (Fig. 4A), with most outgoing signaling derived by hypoxia-enriched tumor cells, especially basal cells (Fig. 4B). A recent study reported an extracellular vesicle network (EVNet) derived by pancreatic cancer stem cells, which enhances angiogenesis, modulates antitumour immune response, promote cancer-associated fibroblast differentiation, establishes a premetastatic niche and confer metastatic properties to non-metastatic cancer cells [23]. Consistent to former findings, we found a signaling secreting network mainly derived by hypoxia-enriched basal PDAC cells. Outgoing signaling from tumor cells to ductal cells, endothelial cells and fibroblasts was shown in Fig. 4C, basal PDAC cell derived intra-tumor cell signaling were shown in Fig. 4D. Notably, legends VEGFA, SEMA3C, ANGPTL4 and NAMPT were significantly higher expressed in basal, pEMT, cEMT and squamous tumor cells, compared with classical tumor cells (Fig. 4E, MAST test, p<0.00001). Previous study reported that tumor derived signaling could promotes cancer-associated fibroblast differentiation and pericyte-fibroblast transition (PFT), like PDGF signaling [24] and TGFB signaling [25]. Notably, a recent study reported ANGPTL4-CDH11 promotes CAF differentiation and ECM deposition in pancreatic cancer in mice [26]. Moreover, GSVA score of hypoxia signatures and cancer-associated fibroblasts showed a moderate correlation in TCGA-PAAD cohort (Fig. 4F). In another PDAC study, the authors subtyped PDAC into classical/basal and ECM-enriched/immune-enriched subtypes according to specific gene expression. Notably, they reported a distinct correlation between basal tumor subtype and ECM-enriched stroma subtype [4]. However, the mechanisms of the formation of the relationship remains to be revealed, our results indicated hypoxia may serve as a driving factor of a basal-like PDAC phenotype and ECM-enriched microenvironment. Also, we accessed the legend-receptor signaling from fibroblasts to tumor cells, and was shown in Fig. 4G, indicating that fibroblasts may also be involved in promoting the aggressive phenotype of tumor cells in hypoxic PDACs.
Fig. 4.
Legend-receptor interaction analysis suggests enhanced secretion signals from hypoxia-enriched tumor cells induced increasingly malignant behavior of PDAC cells. Strong cell-cell signaling were found between Tumor cells and Tumor cells, fibroblasts and endothelial cells (A-B), specifically derived by hypoxia signature enriched basal PDAC cells. Tumor-cell derived signaling were shown in (C). Basal PDAC cell derived signaling among tumor cell subtypes were shown in (D). (E) VEGFA, SEMA3C, ANGPTL4 and NAMPT were significantly higher expressed in basal, pEMT, cEMT, and squamous cells, compared with classical cells. Otherwise, a moderate correlation between CAF signatures and basal PDAC signatures was detected in TCGA-PAAD (F), interactions between fibroblasts and PDAC tumor cells were shown in (G).
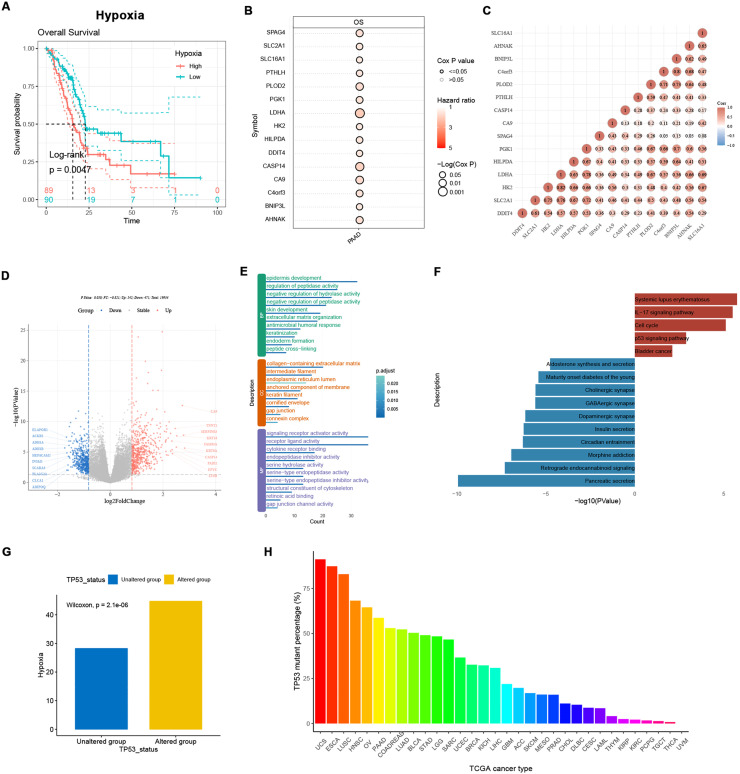
Hypoxia signatures divided TCGA-PAAD patients into two distinct groups
To further detect the significance of hypoxia signatures in the clinical management of PDAC, 178 TCGA-PAAD patients were divided into two groups, according to median GSVA scores of hypoxia signatures (Supplementary Figure 3). Patients with higher hypoxia signature expression showed significantly shorter overall survival, compared with hypoxia-low patients (Fig. 5A, log-rank test, p<0.05). Among the 46 hypoxia genes, 15 genes showed significant correlation with worse overall survival, according to uni-cox tests (Fig. 5B, p<0.05). Notably, LDHA had the strongest prognostic ability and was significantly correlated to other 14 genes, as a well-defined core hypoxia-related gene in previous PDAC studies (Fig. 5C). Differentially expressed genes (DEGs) were detected using ‘limma’. Consistent with our findings in single-cell analysis, novel basal markers like S100A2 and squamous markers like KRT6A and LY6D were significantly overexpressed in hypoxia-high patients, while genes related to normal secretary functions of pancreas like INSM1 and PLA2G2A were significantly down-regulated (Fig. 5D and Supplementary Table 3). GO enrichment showed the up-regulated genes were significantly enriched in extracellular matrix organization and keratinization, suggesting a distinct relationship between hypoxia and ECM formating, squamous-like tumor phenotype, and enhanced tight junction, which may lead to chemotherapy resistance (Fig. 5E). KEGG pathway enrichment analysis showed that p53 signaling pathway was significantly activated in hypoxia-high patients (Fig. 5F). GSVA score of hypoxia signatures in TP53 mutants was significantly higher than TP53 wild-types in TCGA-PAAD cohort (Fig. 5G, Wilcoxon test, p<0.05). P53 was reported to be hypoxia regulator in tumors, inducing cell apoptosis in hypoxic conditions. Thus, we hypothesized that TP53 mutation in cancers is induced by hypoxia via positive selection, rather than a random event. Barplot in Fig. 5H showed the mutation frequency of TP53 across TCGA cancer types. TP53 had higher mutation rate in squamous carcinomas, compared to adenocarcinomas. Among adenocarcinoma, TP53 showed highest mutation rate in PAAD, CRC and LUAD. Pancreatic cancer as a typical Ischemic and hypoxic tumor is more likely to suffer from TP53 mutation.
Fig. 5.
Hypoxia signatures reflects two distinct PDAC subgroups in TCGA-PAAD. TCGA-PAAD patients were divided into two groups according to the median GSVA score of hypoxia signatures. (A) Hypoxia-high patients showed worse overall survival compared with hypoxia-low patients. (B) 15 hypoxia signatures significantly associated with overall survival of TCGA-PAAD by uni-cox test. (C) Correlated expression of 15 hypoxia signatures in TCGA-PAAD cohort. (D) Volcano plot showing differentially expressed genes between hypoxia-high and hypoxia-low group. (E) Enriched GO-terms of genes overexpressed in hypoxia-high group (BP = Biological Process; CC = Cellular Component; MF Molecular Function). (F) Bar plot showing KEGG terms enriched. (G) Hypoxia signatures were higher expressed in TP53 mutants in TCGA-PAAD. (H) Barplot showing TP53 mutation frequency in each TGCA study.
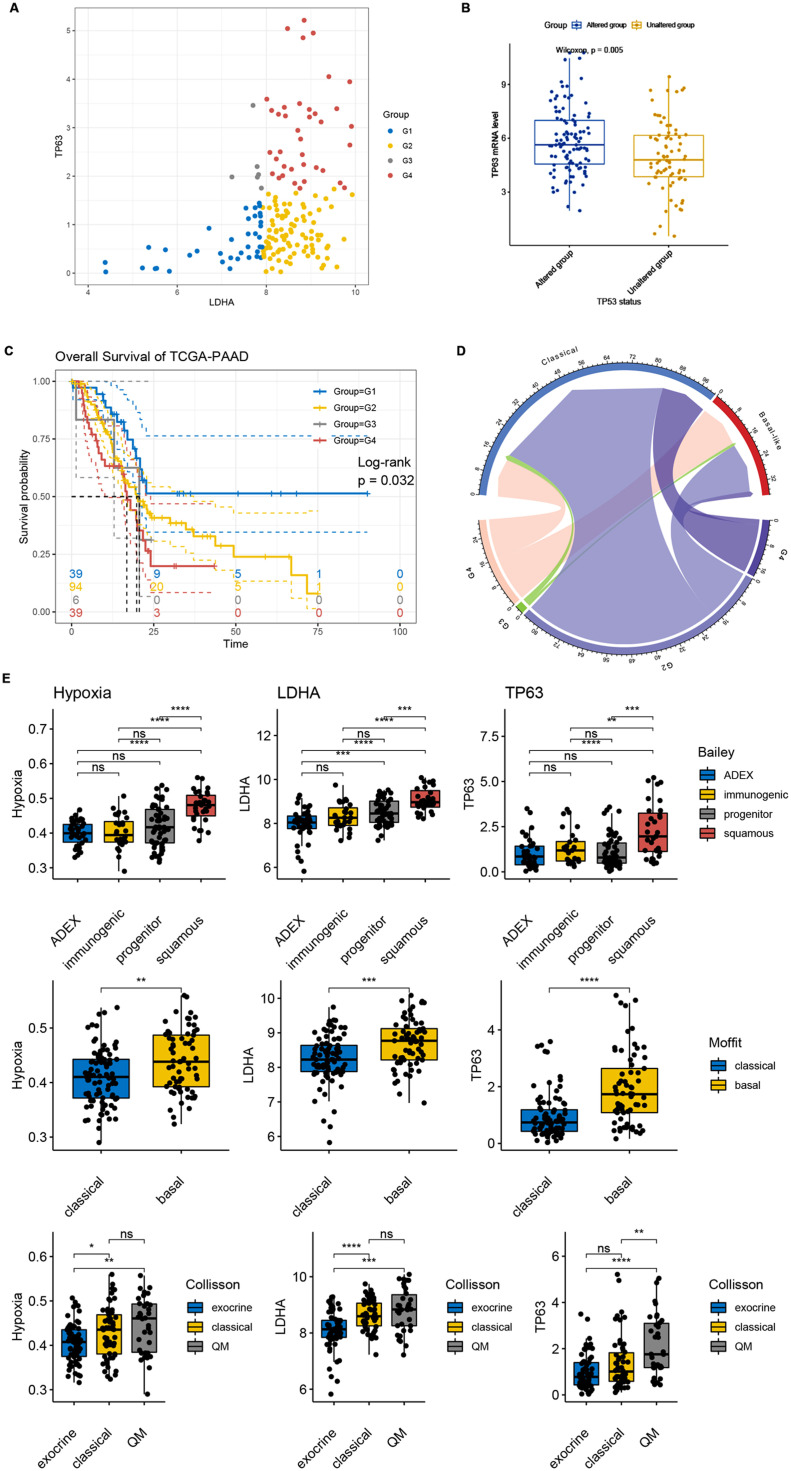
As indicated in former analysis, we found that overexpression of hypoxia signatures and LDHA was related to the transition from classical to basal/squamous-like PDAC tumor cells at single-cell level. Thus, we further evaluated the effect of hypoxia gene LDHA and squamous signature TP63 in distinguishing PAAD characteristics in TCGA-PAAD cohort. TCGA-PAAD patients were divided into 4 groups according to 25th quantile expression level of LDHA and 75th quantile expression of TP63. Interestingly, we observed G3 patients (LDHA low and TP63 high) accounted for a small proportion of the total patients (6/178, 3.37%). Consistent with our findings in snRNA-seq data analysis, these results further validated the necessity of hypoxia in the formation of basal/squamous-like pancreatic adenocarcinoma at bulk RNA-seq level. As shown in Fig. 6B, squamous signature TP63 were significantly overexpressed in TP53 mutants, compared with TP53 wildtypes in TCGA-PAAD cohort, further stated the strong promoting effect of hypoxia in the positive selection of TP53 mutation and leading to a basal/squamous-like subtype. Distinct difference in survival outcomes between G1 and G2, G3, G4 was detected, indicating that limited expression of hypoxia signature LDHA leads to a less hazardous phenotype (Fig. 6C, log-rank test, p<0.05). We then compared outcomes of our grouping-system according to LDHA and TP63 expression with a published grouping-system in 137 TCGA-PAAD patients with annotated Moffit classification [6], and found that G4 patients were more likely to be basal-like (Fig. 6D). Thus, we compared the GSVA score of hypoxia signatures, LDHA and TP63 expression levels of published PDAC subtypes in TCGA-PAAD cohort. Briefly, higher hypoxia score, LDHA and TP63 expression were found in progenitor and squamous subtypes (Bailey et al.) [7](Fig. 6E), basal subtype (Moffit et al.) [6] (Fig. 6F), classical and QM subtypes (Collison et al.) [8] (Fig. 6G) (Wilcoxon tests, *p<0.05, **p<0.001, ***p<0.0001, ****p<0.00001, ns: not significant), indicating that hypoxia signature defines a developed and risky PDAC phenotype. Also, we performed the similar grouping strategy to TCGA-PAAD patients using LDHA and LY6D, for LY6D and TP63 were both indicated as novel squamous-like biomarkers of PDAC in our snRNA-seq analysis. Similar findings were observed and shown in Supplementary Figure 4. These finding suggested that LDHA overexpression was related to classical to basal/squamous-like PDAC tumor cell state transition. Moreover, we further evaluated the relationship between LDHA expression and clinicopathological features of TCGA-PAAD patients. 178 patients were divided into two groups according to their median LDHA mRNA level, and notably, LDHA expression were significantly correlated to higher pathological T stage (Chi-square test, p = 0.025) and higher histologic grade (Chi-square test, p<0.001). This result indicated that LDHA expression was associated with PDAC tumor cell dedifferentiation and was consistent with our former findings, suggesting the potential usage of hypoxia signature LDHA in PDAC classification and prognosis.
Fig. 6.
LDHA and TP63 improved prediction of PDAC prognosis and suggested consecutiveness and transferability between PDAC subtypes. (A) Scatter plot showing the correlation between LDHA expression and TP63 expression in TCGA-PAAD patients. TCGA-PAAD patients were divided into 4 groups (G1, G2, G3 and G4) according to their quantile 25 LDHA expression and quantile 75 TP63 expression. Patients with low expression of LDHA and high expression of TP63 (G3, n = 6) accounted for a small fraction of the total patients (6/178, 3.37%). (B) TP63 expression was significantly up-regulated in TP53 mutants, compare with TP53 wild-types in TCGA-PAAD cohort, Wilcox test, p<0.00001. (C) G1 patients with low expression of both LDHA and TP63 showed best prognosis while G4 patients with high expression of both LDHA and TP63 showed worst prognosis, Log-rank test, p<0.05. (D) Chord plot showing the linkage between the grouping according to LDHA and TP63 expression and Moffit PDAC-subtype system in TCGA-PAAD cohort. (E-G) Differential GSVA score of hypoxia signatures, expression of LDHA and TP63 in published PDAC-subtype systems.
LDHA expression associates with PDAC differentiation and prognosis
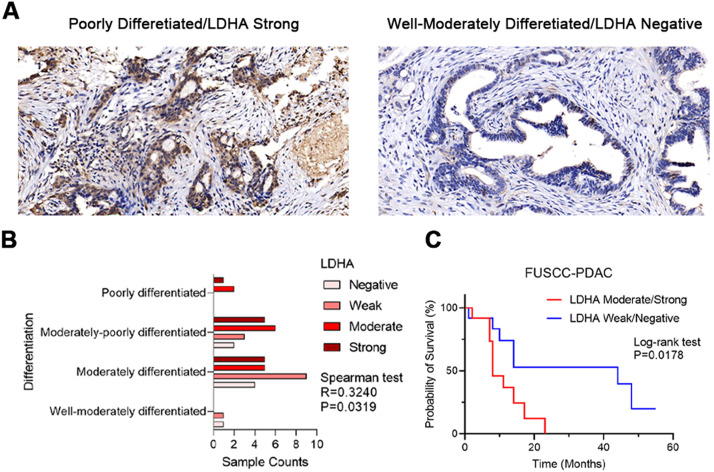
To further validate our findings, we performed immunohistochemical (IHC) staining on 44 PDAC samples underwent resection at Fudan University Shanghai Cancer Center (FUSCC). Spearman correlation test was performed to check the linkage between ranks of LDHA IHC staining intensity and PDAC pathological differentiation status, and log-rank test was performed to check the correlation between LDHA expression and overall survival outcomes. Similarly, higher LDHA IHC staining intensity was found associated with poorer differentiation (Fig. 7B, Spearman test, R = 0.3240, p<0.05) and unfavorable overall survival outcomes (Log-rank test, p<0.05) in FUSCC cohort. Typical immunohistochemistry (IHC) staining images of LDHA-high and LDHA-low PDAC with annotated differentiation status were shown in Fig. 7A. These results further revealed that high expression level hypoxia signature LDHA is associated with poor differentiation of PDAC, leading to unfavorable survival outcomes.
Fig. 7.
IHC staining validated LDHA expression as a prognostic biomarker related with tumor cell dedifferentiation in 44 FUSCC PDAC tumors. (A) Representative IHC staining images of LDHA correlated with PDAC differentiation status. (B) The IHC staining intensity of LDHA was significantly correlated with poor differentiation of PDAC in FUSCC cohort, Spearman correlation test, R = 0.3240, p = 0.0319. (C) The IHC staining intensity of LDHA was significantly correlated with poor prognosis of PDAC in FUSCC cohort, Log-rank test, p = 0.0178. Patients were divided into two groups (group1=LDHA moderate/strong, group2=LDHA weak/negative) according to their LDHA staining intensity.
Discussion
Hypoxia is a constantly evolving participant in tumor growth and progression, orientating tumor fate, rather than a random by-product of the cellular milieu due to uncontrolled tumor growth [27]. In this study, we analyzed snRNA-seq profiles of 15 primary PDACs and identified hypoxia as a representative marker for PDAC progression. We found hypoxia signatures differentially expressed in classical, basal, pEMT, cEMT and squamous PDAC tumor cells, and showed elevated transcriptome levels along the cell state transition. Hypoxic tumor cell prefers to exhibit basal-like behavior. Enhanced outgoing signaling secreted by hypoxic tumor cells promotes metastasis, angiogenesis and cancer-associated fibroblast differentiation. Moreover, there are significant differences in clinicopathological characteristics between hypoxic and non-hypoxic PDACs, according to hypoxia signatures at transcriptome level.
Specifically, TP53 mutations were most found in hypoxic PDAC tumors. Transcription factor p53 is a negative regulator of hypoxia signaling, which is overexpressed in hypoxic conditions, suppresses HIF-1α activity and induces cell apoptosis. Thus, cells with TP53 mutation may be selected in hypoxic conditions. In summary, hypoxia in PDAC leads to a basal-like tumor phenotype with high probability of TP53 mutated and extracellular matrix enriched.
Previous studies revealed the potential linkage between tumor cell state and the complex extracellular compartments, while the mechanisms were not well understood [6]. Carlo et al. subjected PDAC epithelia into basal/classical subtype and PDAC stroma into ECM-enriched/immune-enriched subtypes with the assistance of laser capture microdissection (LCM) and RNA sequencing [4]. They suggested a strong association between an ECM-rich stroma and basal-like epithelium while immune-rich stroma occurred more often in association with classical epithelia. A hypothesis is that tumor cell phenotype is driven by stroma in PDAC, however, the origin of cancer-associated fibroblasts remains unclear [28]. Emerging evidence indicated that tumor cells could induce ECM formation in tumor microenvironment [29], for example, a previous study showed that in patient-derived xenografts (PDXs) microenvironments dominated by fibrosis and immune infiltration were regulated by tumor-stroma crosstalk [30]. These insights suggested that the ECM-enriched microenvironment in PDAC may be driven by tumor cells, although PDAC is often consisted of mostly stromal compartments [31]. Our results suggested that in hypoxic tumor cell derived secreting signals regulates fibroblasts differentiation and deposition, thus leading to an ECM-enriched tumor microenvironment. The key driver of the formation of PDAC microenvironment needs to be further discussed, which is important for further intervention studies.
Moreover, we found that squamous signatures like TP63 were widely expressed in PDAC tumor cells. TP63 is a specific marker for squamous cancers, which is well-defined in former studies [32,33]. However, in some adenocarcinomas, TP63 overexpression was also found [34]. We found that TP63 overexpression is a common event in hypoxic PDAC tumors, with or without showing the squamous cell carcinoma pathological characteristics. It was reported that adenocarcinoma and squamous cell carcinoma are transformable in specific conditions, especially in lung adenocarcinomas and lung squamous cell carcinomas, which leads to chemotherapy resistance [35]. Our study revealed that squamous-like characteristics were widely found in hypoxic PDAC, similar to findings in non-small cellular lung cancers.
In summary, hypoxia leads to a basal-like tumor phenotype, with high probability of p53 mutation and an ECM-enriched microenvironment. We hypothesized that hypoxia is an early-stage trigger of the complex PDAC ecosystem. However, activation of hypoxia-related pathways acts as a hub regulator of oncogenesis and links with multiple oncogenic pathways involved in PDAC development. Previous studies revealed novel regulators and mechanisms in hypoxic PDAC microenvironment formation, for example, KLF4 was found downregulate LDHA expression and inhibit hypoxia in PDAC, and further effect the global expression patterns of hypoxia-regulated genes, leading to heterogeneity of PDAC tumors. Due to limited understanding of the upstream regulation of hypoxia in tumors, lack of precise therapeutic strategies for hypoxic tumors remains a major problem in PDAC management. It was reported that small molecular drugs such as aspirin and Tamoxifen could block the growth of tumors by inhibiting the expression or activity of HIF-1α, leading to a decline in tumor growth [36]. Our results suggested inhibiting hypoxia participants in early-stage of PDAC may lead to a less aggressive tumor phenotype and improve survival outcomes of PDAC patients.
Pancreatic adenocarcinoma is a typical low-purity tumor. In bulk RNA sequencing analysis, the characterization of tumoral components is affected by non-tumoral components such as tumor-associated fibroblasts, leading to limited understand on tumor development and classification. Single-cell and single-nucleus sequencing has a preference for the capture of different cell types, like tumor cells, fibroblasts and immune cells, leading to bias in the estimation of tumor components. By integrating single-nucleus and bulk sequencing analysis, we separated PDAC tumor cells and described their evolution aspects using single-nucleus RNA sequencing data, and further analyzed the relationship of hypoxia and tumor components using bulk RNA sequencing data, made up for the deficiency of these two sequencing strategies.
In conclusion, despite the fact that the ecosystem of PDAC is complex and remains to be understood, hypoxia provides insights on the formation of this diversity. Hypoxic PDAC tumors are most basal/squamous-like, with p53 mutation and extracellular matrix enriched, leading to unfavorable survival outcomes and lack of therapeutic strategies. Through multiple bioinformatic analysis, we parsed the formation and characteristics of hypoxic-related PDAC ecosystem, thus provide evidence for studies on future PDAC management.
Methods
Single-nucleus RNA-seq data accessing and processing
The processed single-nucleus RNA-seq data of fifteen treatment-naïve patients had pancreas resection and were pathologically were collected from https://singlecell.broadinstitute.org/single_cell/study/SCP1089/human-treatment-naive-pdac-snuc-seq. Cells from 15 PDAC tumors without neoadjuvant treatment were included in this study. Totally 43, 817 PDAC tumor cells were extracted according to the corresponding annotation published [19].
Gene expressions were normalized by the global-scaling normalization method “LogNormalize” in Seurat (v4.1.1), which normalized the gene expressions for each cell by the total expression, followed by multiplying by a scale factor (10,000 by default) and log-transformation. To remove batch effect among the samples, data integration was performed using ‘Harmony’ (0.1.0), with parameter lambda=0.5, dim.use=1:30, theta=2.
To identify marker genes of each cell type, the “FindAllMarkers” function from Seurat was applied to identify differentially expressed genes using ‘MAST’ (v1.20.0) [37]. Only significantly upregulated genes (FDR < 0.05) with 0.3 log fold change and 0.3 minimum expression fraction were retained as marker genes.
Module scoring and developmental trajectory analysis
Scores for gene sets of interest were calculated based on the average relative expression and implemented in the Seurat function ‘AddModuleScore’. Gene sets for tumor subtypes and other PDAC components were obtained from previously published studies [38,39]. Clusters identified by Seurat were annotated according to module scores and specific gene expression. Pseudotime analysis were performed using R package ‘monocle’ (v2.26.0) [40] with default parameters.
Cell–cell communication
To identify and visualize the cell cross-talk among malignant clusters or between tumor cells and other cell types, the R package ‘CellChat’ (v1.1.3) was used according to the developer's vignette (https://github.com/sqjin/CellChat) [41].
Validation cohort
Processed data of PRJCA001063 was downloaded from zenodo [10.5281/zenodo.3969339], which had undergone quality control and annotated [11]. Ductal cells were extracted according to corresponding annotation published. The threshold for LY6D+ ductal cell identification is counts LY6D>1, using the ‘WhichCells’ function of ‘Seurat’.
Spatial transcriptome data accessing and processing
Spatial transcriptome data for an individual PDAC sample was downloaded from the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) with accession number GSE203612. Seurat was applied to process this data. In detail, we used ssGSEA method to score the individual spots of the spatial transcriptome data via ‘GSVA’ (v1.42.0) R package [42].
TCGA-PAAD patients subgrouping
Hypoxic scores of TCGA-PAAD patients were calculated using ‘ssGSEA’ method, according to the expression of the 46 hypoxic genes. TCGA-patients were divided into two distinct group according to their median hypoxia score.
RNA-sequencing expression (level 3) profiles, somatic mutation data and clinical information of TCGA patients were downloaded from cBioPortal (https://www.cbioportal.org) and GDC Data Portal (https://portal.gdc.cancer.gov/). Somatic status of TCGA-PAAD patients were visualized using ‘maftools’ (v2.10.05).
RNA-sequencing expression (level 3) profiles and corresponding clinical information for PACA-AU were downloaded from the ICGC dataset(https://dcc.icgc.org/releases/current/Projects). Log-rank test was used to compare differences in survival between these groups. The timeROC(v 0.4) analysis was used to compare the predictive accuracy of each gene.
Identification of differentially expressed genes and functional analysis
Differentially expressed genes (DEGs) between hypoxia-high patients and hypoxia-low patients were identified using ‘limma’ (v3.50.3). The standard for determining the significance of DEGs was set as P < 0.05 and |log2FC|>1. Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment of the DEGs were performed using R package ‘ClusterProfiler’ (v4.2.2) [43].
Immunohistochemical staining
The 44 clinical tissue samples used in this study for immunohistochemical staining were obtained from patients pathologically diagnosed with PDAC and underwent resection at Fudan University Shanghai Cancer Center (FUSCC). Patient consent and approval from the Institutional Research Ethics Committee were obtained prior to the study. Postsurgical follow up of patients was performed as previously described. Overall survival (OS) was defined as the time interval from surgery to death.
Statistical analysis
Bioinformatics analysis and associated statistical calculations were performed with R 4.0.3. The Kaplan–Meier survival curves were plotted using the R packages ‘survival’ (v3.4.0) and ‘survminer’ (v0.4.9). Coxph test was used to determine the hazard ratio (HR) and 95% confidence interval of hypoxic gene signatures in TCGA-PAAD through the GSCALite website (http://bioinfo.life.hust.edu.cn/web/GSCALite/) [44].
Funding
This study was jointly supported by the National Natural Science Foundation of China (U21A20374, 82072698, 82103551), Shanghai Municipal Science and Technology Major Project (21JC1401500), Scientific Innovation Project of Shanghai Education Committee (2019-01-07-00-07-E00057), Clinical Research Plan of Shanghai Hospital Development Center (SHDC2020CR1006A), and Xuhui District Artificial Intelligence Medical Hospital Cooperation Project (2021–011).
Ethical statement
This study was approved by the Institutional Research Ethics Committee of Fudan University Shanghai Cancer Center (FUSCC), and all the patients signed written informed consent prior to the investigation.
Table 1.
Comparison of clinical and pathological features between the LDHA-high group and LDHA-low group of TCGA-PAAD, chi-square tests.
| Characteristics | Low expression of LDHA | High expression of LDHA | P value |
|---|---|---|---|
| n | 89 | 89 | |
| Pathologic T stage, n (%) | 0.025 | ||
| T1&T2 | 21 (11.9%) | 10 (5.7%) | |
| T3&T4 | 66 (37.5%) | 79 (44.9%) | |
| Pathologic N stage, n (%) | 0.117 | ||
| N0 | 29 (16.8%) | 20 (11.6%) | |
| N1 | 57 (32.9%) | 67 (38.7%) | |
| Histologic grade, n (%) | < 0.001 | ||
| G1 | 24 (13.6%) | 7 (4%) | |
| G2&G3&G4 | 63 (35.8%) | 82 (46.6%) | |
| Age, n (%) | 0.072 | ||
| <= 65 | 41 (23%) | 53 (29.8%) | |
| > 65 | 48 (27%) | 36 (20.2%) | |
| Gender, n (%) | 0.763 | ||
| Female | 41 (23%) | 39 (21.9%) | |
| Male | 48 (27%) | 50 (28.1%) |
Supplementary Figure 1. UMAP plots of acinar cells, ductal cells, atypical ductal cells and tumor cells of the single-nucleus RNA-seq profile.
Supplementary Figure 2. Oncoplot showing the somatic mutation status of hypoxia-high and hypoxia-low TCGA-PAAD patients, respectively.
Supplementary Figure 3. PCA plot showing hypoxia signature expression features in hypoxia-high group and hypoxia-low group.
Supplementary Figure 4. TGCA-PAAD patients were divided into LDHAlow/LY6Dlow, LDHAlow/LY6Dhigh, LDHAhigh/LY6Dlow and LDHAhigh/LY6Dhigh subgroups, according to their 25th percentile of LDHA mRNA levels and 75th percentile of LY6D mRNA levels. (B) Kaplan-mere plot showing the overall survivals of these 4 subgroups.
Supplementary Table 1. Signatures used for gene module scoring.
Supplementary Table 2. Marker genes identified for each tumor cell subtype.
Supplementary Table 3. Differentially expressed genes (DEGs) in hypoxia-high group, compared with hypoxia-low group in TCGA-PAAD cohort.
Supplementary Table 4. KEGG terms of differentially expressed genes (DEGs) in hypoxia-high group, compared with hypoxia-low group in TCGA-PAAD cohort by gene set enrichment analysis (GSEA).
Supplementary Table 5. Clinical features of the 44 FUSCC-PDAC samples included.
CRediT authorship contribution statement
Mingwei Dong: Methodology, Software, Formal analysis, Writing – original draft, Visualization. Rong Tang: Methodology, Software, Validation, Formal analysis, Writing – original draft, Visualization. Wei Wang: Software, Formal analysis. Jin Xu: Software, Formal analysis. Jiang Liu: Software, Formal analysis. Chen Liang: Validation, Formal analysis. Jie Hua: Validation, Formal analysis. Qingcai Meng: Validation, Formal analysis. Xianjun Yu: Writing – review & editing, Funding acquisition, Resources, Supervision. Bo Zhang: Conceptualization, Writing – review & editing, Funding acquisition, Resources, Supervision. Si Shi: Conceptualization, Writing – review & editing, Funding acquisition, Resources, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
We acknowledge Prof. Aviv Regev (Massachusetts Institute of Technology, Harvard University) and Prof. Itai Yanai (New York University) for sharing their meaningful data which supported our work.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2023.101692.
Contributor Information
Bo Zhang, Email: zhangbo@fudanpci.org.
Si Shi, Email: shisi@fudanpci.org.
Appendix. Supplementary materials
References
- 1.Huang J., Lok V., Ngai C.H., Zhang L., Yuan J., Lao X.Q., et al. Worldwide burden of, risk factors for, and trends in pancreatic cancer. Gastroenterology. 2021;160(3):744–754. doi: 10.1053/j.gastro.2020.10.007. [DOI] [PubMed] [Google Scholar]
- 2.Wood L.D., Canto M.I., Jaffee E.M., Simeone D.M. Pancreatic cancer: pathogenesis, screening, diagnosis, and treatment. Gastroenterology. 2022;163(2):386–402. doi: 10.1053/j.gastro.2022.03.056. e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ho W.J., Jaffee E.M., Zheng L. The tumour microenvironment in pancreatic cancer - clinical challenges and opportunities. Nat. Rev. Clin. Oncol. 2020;17(9):527–540. doi: 10.1038/s41571-020-0363-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maurer C., Holmstrom S.R., He J., Laise P., Su T., Ahmed A., et al. Experimental microdissection enables functional harmonisation of pancreatic cancer subtypes. Gut. 2019;68(6):1034–1043. doi: 10.1136/gutjnl-2018-317706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Riera-Domingo C., Audigé A., Granja S., Cheng W.C., Ho P.C., Baltazar F., et al. Immunity, hypoxia, and metabolism-the ménage à trois of cancer: implications for immunotherapy. Physiol. Rev. 2020;100(1):1–102. doi: 10.1152/physrev.00018.2019. [DOI] [PubMed] [Google Scholar]
- 6.Moffitt R.A., Marayati R., Flate E.L., Volmar K.E., Loeza S.G., Hoadley K.A., et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat. Genet. 2015;47(10):1168–1178. doi: 10.1038/ng.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bailey P., Chang D.K., Nones K., Johns A.L., Patch A.M., Gingras M.C., et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531(7592):47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 8.Collisson E.A., Sadanandam A., Olson P., Gibb W.J., Truitt M., Gu S., et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat. Med. 2011;17(4):500–503. doi: 10.1038/nm.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McDonald P.C., Chafe S.C., Brown W.S., Saberi S., Swayampakula M., Venkateswaran G., et al. Regulation of pH by carbonic anhydrase 9 mediates survival of pancreatic cancer cells with activated KRAS in response to hypoxia. Gastroenterology. 2019;157(3):823–837. doi: 10.1053/j.gastro.2019.05.004. [DOI] [PubMed] [Google Scholar]
- 10.Tao J., Yang G., Zhou W., Qiu J., Chen G., Luo W., et al. Targeting hypoxic tumor microenvironment in pancreatic cancer. J. Hematol. Oncol. 2021;14(1):14. doi: 10.1186/s13045-020-01030-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peng J., Sun B.F., Chen C.Y., Zhou J.Y., Chen Y.S., Chen H., et al. Single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Res. 2019;29(9):725–738. doi: 10.1038/s41422-019-0195-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Juiz N.A., Iovanna J., Dusetti N. Pancreatic cancer heterogeneity can be explained beyond the genome. Front. Oncol. 2019;9:246. doi: 10.3389/fonc.2019.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ren B., Cui M., Yang G., Wang H., Feng M., You L., et al. Tumor microenvironment participates in metastasis of pancreatic cancer. Mol. Cancer. 2018;17(1):108. doi: 10.1186/s12943-018-0858-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tan Z., Xu J., Zhang B., Shi S., Yu X., Liang C. Hypoxia: a barricade to conquer the pancreatic cancer. Cell. Mol. Life Sci.: CMLS. 2020;77(16):3077–3083. doi: 10.1007/s00018-019-03444-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bhandari V., Li C.H., Bristow R.G., Boutros P.C. Divergent mutational processes distinguish hypoxic and normoxic tumours. Nat. Commun. 2020;11(1):737. doi: 10.1038/s41467-019-14052-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang M., Bolderson E., O'Byrne K.J., Richard D.J. Tumor hypoxia drives genomic instability. Front. Cell. Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.626229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rohwer N., Cramer T. Hypoxia-mediated drug resistance: novel insights on the functional interaction of HIFs and cell death pathways. Drug Resist. Updates: Rev. Comment. Antimicrob. Anticancer Chemother. 2011;14(3):191–201. doi: 10.1016/j.drup.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 18.Zhang C., Liu J., Xu D., Zhang T., Hu W., Feng Z. Gain-of-function mutant p53 in cancer progression and therapy. J. Mol. Cell. Biol. 2020;12(9):674–687. doi: 10.1093/jmcb/mjaa040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hwang W.L., Jagadeesh K.A., Guo J.A., Hoffman H.I., Yadollahpour P., Reeves J.W., et al. Single-nucleus and spatial transcriptome profiling of pancreatic cancer identifies multicellular dynamics associated with neoadjuvant treatment. Nat. Genet. 2022;54(8):1178–1191. doi: 10.1038/s41588-022-01134-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karthaus W.R., Hofree M., Choi D., Linton E.L., Turkekul M., Bejnood A., et al. Regenerative potential of prostate luminal cells revealed by single-cell analysis. Science. 2020;368(6490):497–505. doi: 10.1126/science.aay0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song G., Shi Y., Meng L., Ma J., Huang S., Zhang J., et al. Single-cell transcriptomic analysis suggests two molecularly subtypes of intrahepatic cholangiocarcinoma. Nat. Commun. 2022;13(1):1642. doi: 10.1038/s41467-022-29164-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jun E., Oh J., Lee S., Jun H.R., Seo E.H., Jang J.Y., et al. Method optimization for extracting high-quality RNA from the human pancreas tissue. Transl. Oncol. 2018;11(3):800–807. doi: 10.1016/j.tranon.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruivo C.F., Bastos N., Adem B., Batista I., Duraes C., Melo C.A., et al. Extracellular vesicles from pancreatic cancer stem cells lead an intratumor communication network (EVNet) to fuel tumour progression. Gut. 2022;71(10):2043–2068. doi: 10.1136/gutjnl-2021-324994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hosaka K., Yang Y., Seki T., Fischer C., Dubey O., Fredlund E., et al. Pericyte-fibroblast transition promotes tumor growth and metastasis. Proc. Natl. Acad. Sci. U.S.A. 2016;113(38):E5618–E5627. doi: 10.1073/pnas.1608384113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calon A., Tauriello D.V., Batlle E. TGF-beta in CAF-mediated tumor growth and metastasis. Semin. Cancer Biol. 2014;25:15–22. doi: 10.1016/j.semcancer.2013.12.008. [DOI] [PubMed] [Google Scholar]
- 26.Peran I., Dakshanamurthy S., McCoy M.D., Mavropoulos A., Allo B., Sebastian A., et al. Cadherin 11 promotes immunosuppression and extracellular matrix deposition to support growth of pancreatic tumors and resistance to gemcitabine in mice. Gastroenterology. 2021;160(4):1359–1372. doi: 10.1053/j.gastro.2020.11.044. e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patel A., Sant S. Hypoxic tumor microenvironment: opportunities to develop targeted therapies. Biotechnol. Adv. 2016;34(5):803–812. doi: 10.1016/j.biotechadv.2016.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zeltz C., Primac I., Erusappan P., Alam J., Noel A., Gullberg D. Cancer-associated fibroblasts in desmoplastic tumors: emerging role of integrins. Semin. Cancer Biol. 2020;62:166–181. doi: 10.1016/j.semcancer.2019.08.004. [DOI] [PubMed] [Google Scholar]
- 29.Morita T., Kodama Y., Shiokawa M., Kuriyama K., Marui S., Kuwada T., et al. CXCR4 in tumor epithelial cells mediates desmoplastic reaction in pancreatic ductal adenocarcinoma. Cancer Res. 2020;80(19):4058–4070. doi: 10.1158/0008-5472.CAN-19-2745. [DOI] [PubMed] [Google Scholar]
- 30.Nicolle R., Blum Y., Marisa L., Loncle C., Gayet O., Moutardier V., et al. Pancreatic adenocarcinoma therapeutic targets revealed by tumor-stroma cross-talk analyses in patient-derived xenografts. Cell. Rep. 2017;21(9):2458–2470. doi: 10.1016/j.celrep.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang S., Zheng Y., Yang F., Zhu L., Zhu X.Q., Wang Z.F., et al. The molecular biology of pancreatic adenocarcinoma: translational challenges and clinical perspectives. Signal Transd. Target. Therap. 2021;6(1):249. doi: 10.1038/s41392-021-00659-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Somerville T.D.D., Xu Y., Miyabayashi K., Tiriac H., Cleary C.R., Maia-Silva D., et al. TP63-mediated enhancer reprogramming drives the squamous subtype of pancreatic ductal adenocarcinoma. Cell Rep. 2018;25(7):1741–1755. doi: 10.1016/j.celrep.2018.10.051. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song Q., Yang Y., Jiang D., Qin Z., Xu C., Wang H., et al. Proteomic analysis reveals key differences between squamous cell carcinomas and adenocarcinomas across multiple tissues. Nat. Commun. 2022;13(1):4167. doi: 10.1038/s41467-022-31719-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Napoli M., Wu S.J., Gore B.L., Abbas H.A., Lee K., Checker R., et al. ΔNp63 regulates a common landscape of enhancer associated genes in non-small cell lung cancer. Nat. Commun. 2022;13(1):614. doi: 10.1038/s41467-022-28202-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao Y., Zhang W., Han X., Li F., Wang X., Wang R., et al. YAP inhibits squamous transdifferentiation of Lkb1-deficient lung adenocarcinoma through ZEB2-dependent DNp63 repression. Nat. Commun. 2014;5:4629. doi: 10.1038/ncomms5629. [DOI] [PubMed] [Google Scholar]
- 36.Cortes E., Lachowski D., Robinson B., Sarper M., Teppo J.S., Thorpe S.D., et al. Tamoxifen mechanically reprograms the tumor microenvironment via HIF-1A and reduces cancer cell survival. EMBO Rep. 2019;20(1) doi: 10.15252/embr.201846557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Finak G., McDavid A., Yajima M., Deng J., Gersuk V., Shalek A.K., et al. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 2015;16:278. doi: 10.1186/s13059-015-0844-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barkley D., Moncada R., Pour M., Liberman D.A., Dryg I., Werba G., et al. Cancer cell states recur across tumor types and form specific interactions with the tumor microenvironment. Nat. Genet. 2022;54(8):1192–1201. doi: 10.1038/s41588-022-01141-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peng X.L., Moffitt R.A., Torphy R.J., Volmar K.E., Yeh J.J. De novo compartment deconvolution and weight estimation of tumor samples using DECODER. Nat. Commun. 2019;10(1):4729. doi: 10.1038/s41467-019-12517-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qiu X., Mao Q., Tang Y., Wang L., Chawla R., Pliner H.A., et al. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods. 2017;14(10):979–982. doi: 10.1038/nmeth.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jin S., Guerrero-Juarez C.F., Zhang L., Chang I., Ramos R., Kuan C.H., et al. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 2021;12(1):1088. doi: 10.1038/s41467-021-21246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hänzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformat. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics: J. Integrat. Biol. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu C.J., Hu F.F., Xia M.X., Han L., Zhang Q., Guo A.Y. GSCALite: a web server for gene set cancer analysis. Bioinformatics. 2018;34(21):3771–3772. doi: 10.1093/bioinformatics/bty411. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.